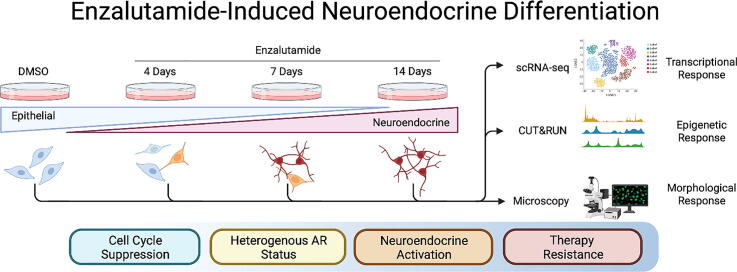
Graphical abstract
Abbreviations: ASI, androgen signaling inhibition; CRPC, castration resistant prostate cancer; CYCL, cycling like; ENZ, enzalutamide; GEMM, genetically engineered mouse model; HNPC, hormone naïve prostate cancer; NE, neuroendocrine; NED, neuroendocrine differentiation; NEL, neuroendocrine like; NEPC, neuroendocrine prostate cancer; TF, transcription factor
Keywords: Neuroendocrine Differentiation (NED), Neuroendocrine Prostate Cancer (NEPC), Androgen Receptor (AR), Single Cell RNA-Seq, Epigenetics, Transdifferentiation
Highlights
-
•
Targeting AR induces NED, cell cycle arrest, and AR signaling repression.
-
•
Histone PTMs correlate with gene expression following AR targeting.
-
•
Time course scRNA-seq reveals dynamic patterns in neuroendocrine activation.
-
•
Enzalutamide treatment induces heterogenous androgen receptor status in cells.
-
•
High-resolution model system recapitulates therapy resistance in prostate cancer.
Abstract
Metastatic and locally advanced prostate cancer is treated by pharmacological targeting of androgen synthesis and androgen response via androgen signaling inhibitors (ASI), most of which target the androgen receptor (AR). However, ASI therapy invariably fails after 1–2 years. Emerging clinical evidence indicates that in response to ASI therapy, the AR-positive prostatic adenocarcinoma can transdifferentiate into AR-negative neuroendocrine prostate cancer (NEPC) in 17–25 % treated patients, likely through a process called neuroendocrine differentiation (NED). Despite high clinical incidence, the epigenetic pathways underlying NED and ASI therapy-induced NED remain unclear. By utilizing a combinatorial single cell and bulk mRNA sequencing workflow, we demonstrate in a time-resolved manner that following AR inhibition with enzalutamide, prostate cancer cells exhibit immediate loss of canonical AR signaling activity and simultaneous morphological change from epithelial to NE-like (NEL) morphology, followed by activation of specific neuroendocrine (NE)-associated transcriptional programs. Additionally, we observed that activation of NE-associated pathways occurs prior to complete repression of epithelial or canonical AR pathways, a phenomenon also observed clinically via heterogenous AR status in clinical samples. Our model indicates that, mechanistically, ASI therapy induces NED with initial morphological change followed by deactivation of canonical AR target genes and subsequent de-repression of NE-associated target genes, while retaining AR expression and transcriptional shift towards non-canonical AR activity. Coupled with scRNA-seq and CUT&RUN analysis, our model system can provide a platform for screening of potential therapeutic agents that may prevent ASI-induced NED or reverse the NED process.
1. Introduction
Prostate cancer is the most frequently diagnosed cancer and second-leading cause of cancer related death for men in the United States [1]. Patients typically present in clinic with localized disease, for which fractionated ionizing radiation (FIR) and/or surgical resection with curative intent are indicated [2]. However, 10–15 % of low/intermediate risk patients and 50 % of high risk patients develop resistance to FIR therapy [3], [4]. For these patients, and patients with metastatic prostate cancer on initial presentation, pharmacological targeting of the androgen receptor (AR) signaling axis with androgen signaling inhibitor (ASI) compounds is the current standard of care [2]. The vast majority of prostate adenocarcinomas arise from the luminal cells of the prostate [5], [6], and as part of the male reproductive system, the prostate and most cell populations comprising the organ are inherently responsive to androgen signaling via AR activity [7]. AR is a nuclear hormone receptor that binds to testosterone and dihydrotestosterone via a ligand binding domain (LBD), undergoes conformational change to expose a nuclear localization signal (NLS) and a DNA binding domain, translocates from the cytoplasm to the nucleus, and binds to chromatin at androgen response element motifs to activate or repress AR-target genes [8], [9]. Current ASI therapy relies on inhibition of testosterone synthesis (e.g., abiraterone acetate inhibits CYP17a, which catalyzes biosynthesis of DHEA, a sex steroid precursor to both testosterone and estrogen [10]), or in the cellular response to testosterone via the AR (e.g., AR antagonist enzalutamide binds the LBD of AR, preventing conformational change and exposure of NLS and DNA binding domain to prevent both nuclear translocation and chromatin binding, respectively [11]). Initial therapeutic response is strong, with up to 50 % reduction in PSA level and tumor burden; however, the response is not durable beyond 22–24 months and biochemical relapse is invariable after 1–2 years [12], resulting in castration resistant prostate cancer (CRPC).
Counterintuitively, the development and subsequent approval of increasingly potent ASI compounds, as well as increasingly earlier deployment of ASI as adjuvant and neoadjuvant therapy strategy, result in increased incidence of ASI therapy-induced neuroendocrine (NE) prostate cancer (NEPC) in up to 25 % of CRPC patients [13]. While the mechanism remains unclear, both clinical and preclinical evidence support that under selective pressure of potent ASI therapy, the epithelial AR-positive adenocarcinoma cells transdifferentiate into small, AR-negative, NE-like (NEL) cells in a process called neuroendocrine differentiation (NED) [14]. NEPC cells are histologically characterized as AR-negative and NE-marker positive with small cell histology similar to small cell lung cancer [15]. The majority of clinical and genetically engineered mouse model (GEMM) data support clonal expansion of luminal cells with concomitant RB1, TP53, and PTEN genomic and/or functional loss [16]. Further, recent combinatorial transcriptomic, epigenomic, and genomic studies indicate that NEPC cells show heterogenous AR status in addition to significant transcriptional reprogramming without significant change in mutational burden, suggesting that NED is a largely epigenetic process and may possess a degree of reversibility [17], [18], [19].
NED has been extensively characterized in vitro and in vivo observationally, but the underlying mechanisms and sequence of events are not yet clear. Both AR-dependent and AR-independent mechanisms have been reported [20], [21], [22], [23]; however, increased incidence of ASI therapy-induced NEPC merit exploration of the role of AR in NED/NEPC. As an epigenetic regulator of differentiation, loss of AR via ASI therapy results in de-repression of stemness and neuronal master regulators BRN2 and SOX2 [22], [24], [25] as well as REST [26], a co-repressor of AR involved in silencing neuronal-associated genes, potentially highlighting an AR-dependent axis. Conversely, AR-independent mechanisms of ASI therapy-induced NED have been reported via ONECUT2 upregulation of HIF1/hypoxia signaling axis [21] or by sustaining growth and proliferation via FGF/FGFR instead of AR [20], suggesting ASI therapy simply selects for cells that develop AR-independent growth/survival mechanisms. Recent evidence has shown heterogenous AR status of NEPC and NEL cells, suggesting both classes of mechanisms likely contribute to therapy induced NED [19] and support designation of five or more subclasses of NEPC. Regardless of route of NED induction, NEL cells share common traits. Neuron-associated transcription factors POU2F3/BRN2, ASCL1, SOX2, FOXA2, epigenetic regulator EZH2, and secretion products NSE, SYP, and CHGA are frequently upregulated [13], while AR-target genes and epithelial markers AR, KLK3/PSA, FOXA1, REST, NKX3.1, KRT8 are frequently repressed [5].
Several in vitro and in vivo model systems have existed for the study of NED and therapy-induced NED, but none were designed with the specific intent of pharmacological screening for prevention of NED or resensitization of NEL cells in the context of ASI exposure of epithelial adenocarcinoma cells. The Owen Witte lab first reported that normal epithelial cells can be reprogrammed to undergo NED with clinically-relevant signatures via functional knockdown of TP53 and RB1 and simultaneous overexpression of AKT1, MYC, and BCL2 [27]. Due to the high alteration rates of TP53 (68.1 %), RB1 (76.6 %), simultaneous TP53/RB1 (59.6 %) in clinical NEPC specimens and PTEN (40.7 %) in mCRPC specimens [28], [29], the majority of commonly used model systems focus on development of NEPC or NEL tumors, but do not necessarily focus on the epigenetic plasticity associated with early cellular response to ASI targeting [30], [31], [32].
Here, we attempt to directly examine the NED process of hormone sensitive LNCaP cells prior to and during the course of enzalutamide-mediated ASI therapy-induced NED by examining dynamic profiles of genome-wide transcriptomics combining advanced technologies, e.g., bulk mRNA-seq, epigenetic cleavage under target and restriction under nuclease (CUT&RUN) sequencing, single cell RNA sequencing (scRNA-seq), and morphological evaluation. Our goal is to develop a model system with the intent to screen compounds for the prevention of NED or resensitization of NEL cells to ASI therapy.
2. Materials and methods
2.1. Cell culture
LNCaP cell line was purchased from ATCC. Routine mycoplasma screening was performed as described previously by Owens et. al. using the LookOut PCR Mycoplasma Detection Kit (Sigma) [33]. Cells were stored as frozen stock in vapor phase of LN2 and thawed prior to use. Cell lines were cultured 3 passages after thawing prior to experimentation and maintained for no longer than 30 total passages. LNCaP cells were cultured in RPMI 1640 (Corning), and All media were supplemented with 10 % FBS (Atlanta Biologicals), 1 mM sodium pyruvate (Corning), penicillin (100 units/mL) and streptomycin (100 μg/mL) combination (Gibco), and 2 mM/L l-glutamine (Corning).
Cells were seeded at 450,000 cells per 15 cm culture dish and allowed to attach to the dish overnight. The following day, enzalutamide was added at 5 μM concentration diluted from 100 mM stock in DMSO. Dilutions were prepared such that DMSO content in culture plates, including DMSO control, was always 0.1 %. Enzalutamide treatment was performed for 4, 7, or 14 days at 5 μM treatment. Media was refreshed and enzalutamide was re-applied to cell culture dishes every 72 h. DMSO cells were seeded at the same density and treated at same schedule as enzalutamide treatment groups but harvested at 70–80 % confluency.
2.2. Morphological analysis
Percent of NED induction was calculated morphologically from brightfield images acquired at the given day of observation (either 4, 7, or 14 days for enzalutamide groups, or 80 % confluency for DMSO group). Images were acquired on a Nikon TE2000-U microscope with 10x objective. Images were analyzed with ImageJ [34]. NEL cells were defined as cell with neurite extensions longer than twice the length of the cell body as described previously [35].
2.3. RNA isolation and RT-qPCR analysis
LNCaP cells were seeded to either 6 cm or 10 cm dishes at 800,000 or 2,200,000 cells/dish respectively. Cells were allowed to attach for 24 h and then subsequently treated with either enzalutamide (5 μM) or DMSO for specified time period. Cells were then harvested with Trizol reagent (Ambion) and RNA integrity was verified via agarose gel electrophoresis. Promega High Capacity cDNA Reverse Transcription Kit (Promega) was utilized following manufacturer instructions and as described previously [36], [37], [38]. RT-qPCR was performed with FastStart Universal SYBR Green Master Mix (Thermo Fisher Scientific) and detected on a QuantStudio 6 Flex with QuantStudio Real-Time PCR control software (Thermo Fisher Scientific). QuantStudio Design and Analysis software (Thermo Fisher Scientific) was used for data analysis. Technical triplicates were run for all samples, samples without detectable amplification were deemed undetected. Primer sets were validated via melt curve and agarose gel analysis of RT-qPCR product. AR primers were used as described previously [36] and IVL primers were used as described previously [39]. All primer sequences utilized are described in Supplementary Table S1.
2.4. Western blotting
Cells were lysed in RIPA buffer for 1 h on ice. Cell lysates were centrifuged at 12,000 xg for 20 min at 4 °C, and the supernatant was eluted with 5xSDS sampling buffer (100 mM Tris-HCl pH 6.8, 4 % SDS and 20 % glycerol with bromophenol blue) and boiled for 5 min. The proteins were separated through 10 % SDS-PAGE gels and were transferred to a nitrocellulose membrane by means of trans-Blot SD wet transfer cell (Bio-Rad). The membrane was blocked in 5 % skim milk-PBS for 2 hrs, washed, and incubated with primary antibodies in PBS containing 0.1 % Tween 20 (PBST) overnight at 4 °C. Primary antibody was then removed by washing the membrane three times in PBST. The membrane was then incubated with secondary antibody for 1 h at RT. After three washes with PBST, membranes were visualized with Odyssey® XF Imaging System. The intensity of bands was determined with Image Studio™ Lite (LI-COR Biosciences). Antibodies against AR (1:2000, Cell Signaling), ARV7 (1:1000, Abcam), CgA (1:5000, Abcam), NSE (1:2000, Genetex), ASCL1 (1:1000, LS-Bio), NCAM1 (1:1000, Genetex), β-actin (1:5000, Sigma-Aldrich), and LI-COR IRDye 800CW Goat anti-Rabbit IgG (H + L), 0.5 mg (1:20000, LI-COR Biosciences) and LI-COR IRDye 800CW Goat anti-Mouse IgG (H + L), 0.5 mg (1:20000, LI-COR Biosciences) were used for western blotting probing.
2.5. mRNA library preparation for bulk mRNA-seq
Three biological replicates at each time points, undifferentiation and 14 days after the treatment, were collected. Total RNA was first evaluated for its quantity, and quality, using Agilent Bioanalyzer 2100. For RNA quality, a RIN number of 10 was achieved. One hundred nanograms of total RNA was used. cDNA library preparation included mRNA purification/enrichment, RNA fragmentation, cDNA synthesis, ligation of index adaptors, and amplification, following the KAPA mRNA Hyper Prep Kit Technical Data Sheet, KR1352 – v4.17 (Roche Corporate). Each resulting indexed library was quantified, and its quality accessed by Qubit and Agilent Bioanalyzer, and multiple libraries pooled in equal molarity. The pooled libraries were then denatured, and neutralized, before loading to NovaSeq 6000 sequencer at 300 pM final concentration for 100 bp paired-end sequencing (Illumina, Inc.). Approximately 30–40 M reads per library were generated. A Phred quality score (Q score) was used to measure the quality of sequencing. >90 % of the sequencing reads reached Q30 (99.9 % base call accuracy).
2.6. Data analysis of mRNA-seq
The sequencing reads were mapped to the human genome hg38 using a RNA-seq aligner STAR (v2.7.2a) [40] with the parameter: “--outSAMmapqUnique 60”. Uniquely mapped reads were assigned to genes based on GENCODE 31 using featureCounts (v2.0.1) [41] with the following parameters: “-s 2 –p –Q 10 -O”. The genes were filtered out if they had<10 read counts in more than half of samples. Gene expression was normalized by the method of TMM (trimmed mean of M values) and subjected to differential expression analysis using edgeR (v3.20.8) [42], [43]. The differentially expressed genes (DEGs) were defined by the cutoffs of both false discovery rate (FDR) and fold change (FC) in the way of FDR < 0.05 and |log2FC| > 1.
2.7. 10X Genomics single cell RNA sample preparation
On the day of harvest and after image acquisition at the microscope, the 15 cm cell culture dishes were removed from the 37 °C incubator, medium was removed via aspiration, and cells were immediately washed once with 10 mL of 37 °C phosphate buffered saline (PBS), pH 7.4. PBS was removed via aspiration and 3 mL 0.25 % trypsin (Corning) was added to the dishes. Dishes were rotated for complete coverage of the trypsin and incubated on the benchtop for 30 s. Roughly 2 mL of the trypsin was removed via aspiration, and dishes were transferred to a 37 °C incubator for exactly 4 min. 10 mL of 4 °C RPMI supplemented with 10 % FBS was used for resuspension of the trypsinized cell suspension. Viability was checked on Countess II FL automated cell counter to ensure > 90 % viability. Cells were then centrifuged 4 min at 400x g at 4 °C to pellet the cells. Supernatant was aspirated and discarded, then cells were washed with 1 mL 4 °C complete RPMI three times. Final resuspension of cells in 400 μL 4 °C complete RPMI was performed, and cell homogeneity and viability were checked to ensure singlet cells at > 90 % viability would be delivered to Indiana University School of Medicine Chemical Genomics Facility for 10X library preparation.
2.8. 10X Genomics single cell RNA library preparation and sequencing
The single cell gene expression analysis was conducted using a 10X Chromium single cell system (10X Genomics, Inc) and a NovaSeq 6000 sequencer (Illumina, Inc). To maximize quality of cell suspension, cell harvest was optimized to retain > 95 % viability at time of harvest following three washes in RPMI/10 %FBS. Following storage on ice for delivery of cells to 10X sequencing facility, the single cell suspension was subsequently washed a fourth time with RPMI/10 % FBS, recentrifuged, and cell debris, dead cells and cell aggregates were removed by discarding supernatant and resuspending cells. Each clean single cell suspension was then be counted with hemocytometer under microscope for cell number and cell viability. The single cell suspension viability was over > 90 % and minimal cell debris and aggregation were ensured prior to loading. A target of 10,000 cells was used for loading; in practice, following final analysis, between 6,890 and 11,620 individual cells were identified per sample after all processing steps were completed. The cells were applied to a single cell master mix with lysis buffer and reverse transcription reagents, following the Chromium NextGEM Single Cell 3′ Reagent Kits User Guide, CG000204 Rev D (10X Genomics, Inc). Along with the single cell gel beads and partitioning oil, the single cell master mixture containing the single cell suspension was dispensed onto a Single Cell Chip G in separate wells, and the chip loaded to the Chromium Controller for GEM generation and barcoding, followed by cDNA synthesis and library preparation. At each step, the quality of cDNA and library was be examined by Bioanalyzer and Qubit. The resulting library was sequenced in a custom program for 28b plus 91b paired-end sequencing on Illumina NovaSeq 6000. Depending on targeted cell recovery, roughly 33,000 – 82,000 reads per cell were generated and 91 % of the sequencing reads reached Q30 (99.9 % base call accuracy). A Phred quality score (Q score) was used to measure the quality of sequencing.
2.9. Data analysis of scRNA-seq
Cellranger 6.0.1 (https://support.10xgenomics.com/) was utilized to process the raw sequencing data generated. The sequencing reads achieved from FASTQ files were aligned to the human reference genome hg38 with STAR, then traced back to individual cells given the single cell barcodes. Gene expression of individual genes were quantified based on the number of UMIs (unique molecular indices) detected in each cell. The filtered gene-cell barcode matrices generated by cellranger were used for further analysis. Cells with too many detected genes, e.g., unique gene counts over 8000, were filtered out. The gene expression was normalized by total counts of the cell and multiplied by a scaling factor 10,000, followed by log2 transformation. The first 26 principal components (PCs) from the principal component analysis (PCA) were used to cluster cells by a shared nearest neighbor (SNN) modularity optimization-based clustering algorithm [44]. Uniform manifold approximation and projection (UMAP) visualization of the resulting clusters and selected genes were performed with Seurat package [45]. Loom files were generated by velocyto [46]. scVelo [47] was used to generate RNA velocity. Monocle 2 [48] was used to generate trajectory.
2.10. CUT&RUN sample preparation
LNCaP cells were seeded into 15 cm cell culture dishes and treated with enzalutamide or DMSO for 14 days as described above. On the day of harvest, cells were removed from 37 °C incubator and placed on benchtop. Formaldehyde was added to the cell culture media to final concentration of 0.1 % formaldehyde. Medium was mixed and plates were incubated at ambient temperature prior to quenching by addition of glycine to final concentration of 125 mM for 5 min. Media was removed and cells were washed with 10 mL 4 °C PBS. PBS was removed and 3 mL trypsin was added. Cells were detached from the plate and homogenized as described above. Following washes, cells were resuspended in 1 mL complete RPMI at 4 °C supplemented with 10 % DMSO. Cells were then frozen slowly to −80 °C for storage.
2.11. CUT&RUN assay
CUT&RUN was performed with light-fixed LNCaP cells (0.1 % formaldehyde for one minute followed by 125 mM glycine quench) and an automated protocol (autoCUT&RUN) derived from those previously described [49], [50], [51]. In brief, for each CUT&RUN reaction 500 K nuclei [5 million cells/mL prepared in 20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM Spermidine, 1x Roche cOmplete Protease Inhibitor Cocktail] were dispensed to individual wells of a 96-well plate, immobilized onto Concanavalin-A beads (Con-A; EpiCypher #21–1401), and incubated overnight at 4 °C with 0.5 µg of antibody (IgG, H3K4me3, H3K27me3, and H3K27ac antibodies listed in Supplementary Table S2). All antibodies validated to histone post-translation modification (PTM)-defined SNAP-ChIP nucleosome standards as previously [52]. pAG-MNase (EpiCypher #15–1016) was added / activated (2 h @ 4 °C) and CUT&RUN enriched DNA (crosslink reversal not necessary for lightly fixed samples) purified using Serapure beads after mixing at 2:1 (Bead:DNA) ratio. Recovered DNA was quantified using PicoGreen and reactions were normalized to 5 ng DNA (or entirety of the reaction if < 5 ng DNA was recovered) before preparing sequencing libraries (CUTANA CUT&RUN Library Prep kit; EpiCypher #14–1001). All autoCUT&RUN steps were optimized / performed on Tecan Freedom EVO robotics platforms with gentle rocking for incubation steps and magnetic capture for media exchange / washing steps. Sequencing performed on a NextSeq2000, according to manufacturer’s instructions.
2.12. CUT&RUN analysis
Bowtie2 [53] was used to align CUT&RUN sequencing reads on the human genome (hg38). After duplicated reads removed by Picard (https://broadinstitute.github.io/picard/), peak calling of mapped reads was performed using SICER2 [54] with comparison to the background reads from the IgG. The peaks were defined by the cutoff of Bonferroni-adjusted p-value < 0.01. Differential peak calling on histone mark signals between two conditions, before and after the treatment, was performed using the command of sicer_df in the package of SICER2. Genome regions with differential signals were identified by FDR-adj p-value < 0.05 and plotted in the heatmap using deepTools [55]. Signals of histone marks were visualized using bigwig and bedgraph files from CUT&RUN data with SparK [56].
2.13. Functional enrichment analysis
An online tool DAVID [57] was used for functional enrichment analysis for gene ontology (GO) and pathways on selected genes, e.g., either up- or down-regulated DEGs identified by mRNA-seq. Enrichment analysis for cell annotations (Azimuth Cell Types 2021 [58]) and TFs was performed using EnrichR [59], [60] given the DEGs or marker genes detected for individual cell cluster based on scRNA-seq data.
2.14. Statistical analysis
Statistical analysis was performed using GraphPad PRISM for qPCR and western blot data except where otherwise stated. Fold changes for RT-qPCR results were calculated on non-exponentiated ΔΔCt values. Student’s t test was performed between groups. Analysis of single cell data was performed with Cell Ranger, and bulk mRNA-seq was performed as described previously [61].
2.15. Data availability
All sequencing data have been deposited to the GEO database with a super-series number GSE215945, including bulk RNA-seq (GSE215653), single cell RNA-seq (GSE215943), and CUT&RUN (GSE216118).
3. Results
3.1. Enzalutamide treatment of prostate cancer cells induces neuroendocrine differentiation
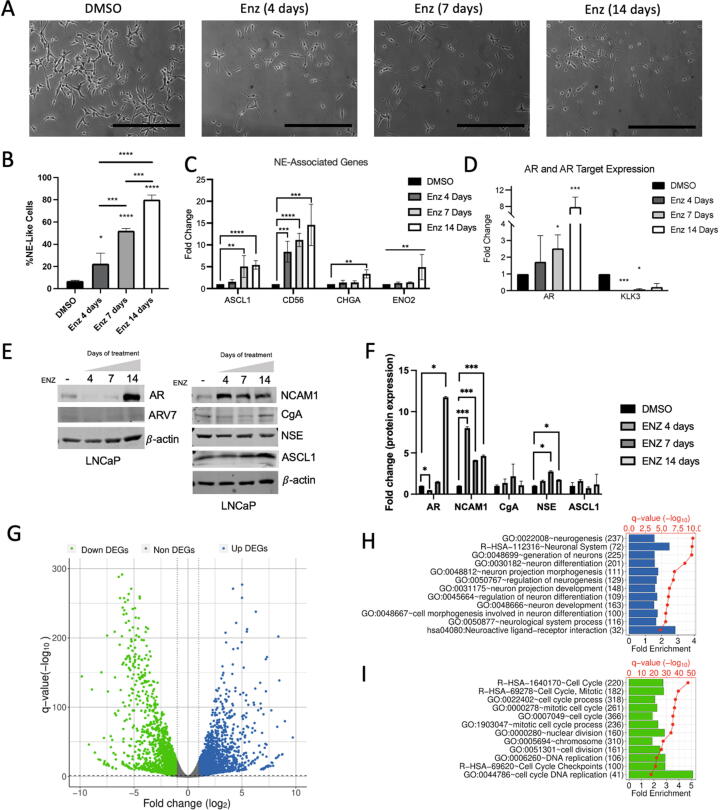
Treatment of hormone-naïve prostate cancer (HNPC) LNCaP cells with enzalutamide resulted in significant morphological change in a dose dependent manner. Typical LNCaP morphology is elliptical cell body with 2–4 polar appendages per cell (Fig. 1A, DMSO). Following enzalutamide treatment, morphological transition to small, round cell bodies with low cytoplasm:nucleus ratio and long neurite-like outgrowths are developed over course of treatment (Fig. 1A). Quantification of morphology indicates a basal level of NEL LNCaP cells at 6.9 %, which increases linearly to 22.5 % (4 days of enzalutamide exposure), to 52.2 % (7 days), and near complete NED induction at 80.1 % (14 days) (Fig. 1B). Quantitative reverse transcription polymerase chain reaction (RT-qPCR) data shows transcriptional upregulation of multiple NE markers genes, namely ASCL1, CD56 (encoding NCAM1 protein), CHGA, and ENO2 (Fig. 1C). Both ENO2 and CHGA was significantly upregulated by day 14 treatment. Chronologically, CD56/NCAM1 showed most rapid upregulation at the transcriptional level, followed by ASCL1, ENO2, and then CHGA (Fig. 1C). Reflecting successful targeting of AR, AR target gene KLK3 / PSA was almost completely silenced immediately following AR targeting with enzalutamide (Fig. 1D). AR, however, was elevated in a dose dependent manner following enzalutamide treatment, with nearly 8-fold increased by day 14 (Fig. 1D). Similarly, a significant 12-fold increase of AR at the protein level occurred at day 14 but was not nearly as pronounced at days 4 and 7 when analyzed via western blot (Fig. 1E, F). Overall, protein-level analysis confirms sequence of events observed with RT-qPCR, in which NCAM1 is upregulated immediately following enzalutamide exposure on day 4, and subsequently AR itself by day 14 (Fig. 1E, F). NCAM1/CD56 exhibited the most significant upregulation of NE-associated markers at both mRNA and protein levels, coinciding with morphological change observed by microscopy by 4 days. As NCAM1/CD56 regulates the interaction between cell membrane and extra cellular matrix in developing neurons and neurites, we were pleasantly surprised to detect the strong correlation between cellular morphology, transcript, and protein level occurring simultaneously after enzalutamide exposure.
Fig. 1.
Enzalutamide treatment of prostate cancer cells induces neuroendocrine differentiation. (A) Brightfield images (scale bar = 100 μM) depicting LNCaP cells with no treatment (DMSO), and enzalutamide treatment over 4, 7, and 14 days. (B) Quantified rate of neuroendocrine differentiation (NED) from A expressed as precent Neuroendocrine (%NE)-like cells. (C-D) RT-qPCR analysis of NE-associated gene expression (C) and AR/AR-target gene expression (D) expressed as fold change. (E) Representative images of western blot for protein-level quantitation of NE-markers and AR / ARv7 in LNCaP cells treated with DMSO or enzalutamide exposure for 4, 7, or 14 days. (F) Quantified densitometry from D. (G) Volcano plot from bulk mRNA-seq data comparing LNCaP cells treated after 14 days enzalutamide exposure vs DMSO control, with upregulated genes in blue and downregulated genes in green. (H, I) Gene ontology (GO) significantly enriched in upregulated (H, blue) and downregulated (I, green) genes upon 14-day enzalutamide exposure. Fold enrichments were shown as bars and statistical significances (-log10(q-value)) were shown as red dots. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Because enzalutamide treatment induced NED in over 80 % of cells at 14 days, we first utilized bulk-level mRNA-seq at this timepoint for an unbiased analysis of the transcriptional reprogramming events in therapy-induced NED. We identified 4,939 differentially expressed genes (DEGs) (Fig. 1G, Supplementary Table S3) following 14-day treatment, and predictably observed significant enrichment of gene ontology (GO) annotations for neuronal-associated development, differentiation, and morphology in upregulated DEGs (Fig. 1H), with corresponding enrichment of cell cycle, DNA replication, and mitotic GO terms in the downregulated DEGs (Fig. 1I). In line with clinical observations of NEPC, gene set enrichment analysis (GSEA) highlighted gene sets associated with neuronal development/differentiation, ErbB pathway, cell:cell junction/adhesion, and signaling pathways for enzalutamide treatment (Fig. S1A), while highlighting cell cycle advancement, and chromatin maintenance for DMSO treatment (Fig. S1B), largely in agreement with observations from GO enrichment analysis described in Fig. 1H/I.
Taken together, these results strongly demonstrated that enzalutamide treatment of hormone naïve LNCaP cells induce morphological, transcriptional, and translational changes which recapitulate transdifferentiation of epithelial luminal cells into neuroendocrine-like cells expressing clinically-relevant NE marker genes. Further, unbiased bulk mRNA-seq also confirmed upregulation of multiple neuronal differentiation and neurogenesis related pathways and downregulation of actively mitotic and cycling cells. To understand more about such reprogramming process, and as NED is characterized as epigenetic rather than somatic or structural rearrangements in chromatin, we next examined the correlation between gene expression changes and alterations of canonical histone modifications H3K27ac, H3K4me3, and H3K27me3.
3.2. Epigenetic status of chromatin indicates NE-activation, AR-signaling loss, and inhibition of cell cycle advancement
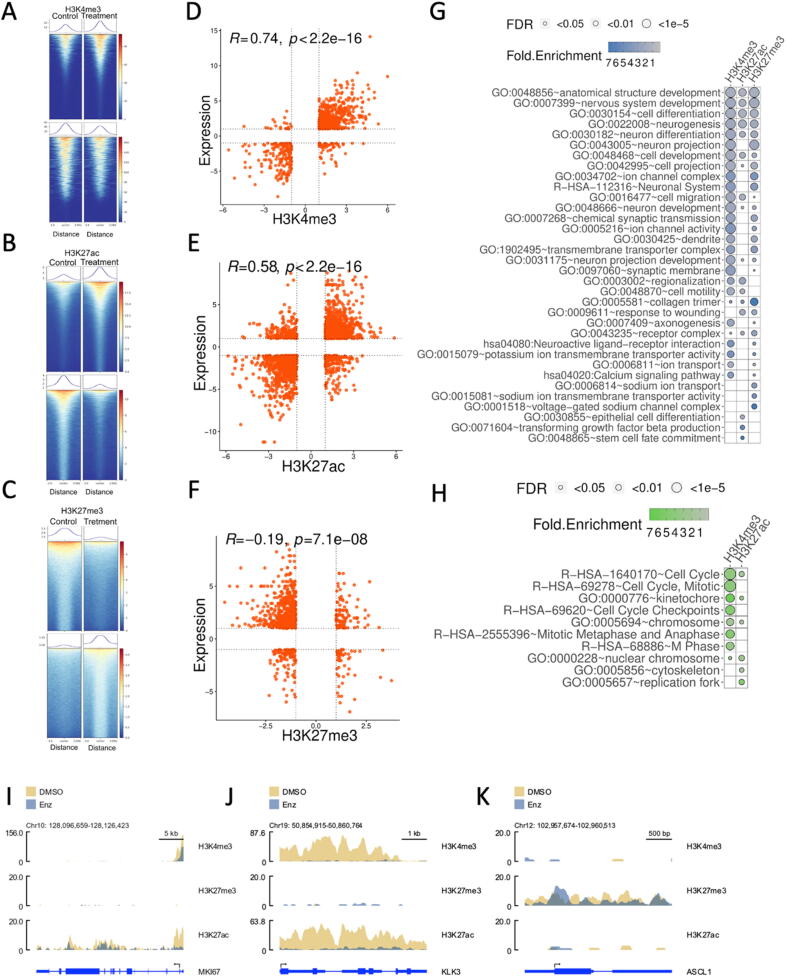
As recent reports have highlighted significant changes in NEPC and neuroendocrine-like (NEL) cells at the chromatin structural level [18], [62], we also utilized cleavage under target and release using nuclease (CUT&RUN) epigenomic profiling technique as traditional ChIP-sequencing input requirement was not amenable to our model system in which cells stop cycling immediately on treatment, and CUT&RUN input requirements are very low [51]. We utilized CUT&RUN to monitor H3K4me3 (indication of active transcription at gene promoter and exons), H3K27ac (canonical activation and euchromatin status), and H3K27me3 (canonical repression). The epigenetic status of H3K4me3, H3K27ac, and H3K27me3 can help to substantiate the differential expression observed via bulk mRNA-seq and RT-qPCR analysis. Additionally, it may suggest distinct mechanisms underlying differential expression patterns.
In general, from 4,547 to 33,529 genome regions underwent remarkable changes with different histone signals (Fig. 2A-C), either up-regulated or down-regulated, while corresponding DEGs simultaneously showed significant changes of gene expression accordingly (Fig. 2D-F). The alternations of H3K4me3 signals exhibited the highest positive correlation coefficient (R = 0.74) with gene expression changes of corresponding genes (Fig. 2D, Supplementary Table S4), followed by H3K27ac (R = 0.58, Fig. 2E, Supplementary Table S5). It is not surprising to see that H3K27me3 had a negative correlation coefficient (R = -0.19) with gene expression (Fig. 2F, Supplementary Table S6). Even though altered H3K4me3, H3K27ac, and H3K27me3 may be associated with different individual genes comprising the total list of DEGs, the functional enrichment analysis on activated DEGs with increased H3K4me3/H3K27ac (or decreased H3K27me3 due to role as a repressive mark) further revealed that all histone marks observed tended to regulate the same neuronal-related functions by targeting same or different genes, e.g., neurogenesis, neuron differentiation, nervous system development, and cell differentiation or development (Fig. 2G). Enhanced H3K4me3 and H3K27ac also shared functions such as cell migration, cell motility, and regionalization, while neuron projection or development, cell projection or specially neuron projection, and dendrite were activated by increased H3K4me3 and reduced H3K27me3 (Fig. 2G). DEGs designated for collagen trimer, response to wounding, or belonging to the receptor complex can be tuned by both growing H3K27ac and abated H3K27me3 (Fig. 2G). Of course, these histone modifications may target unique genes with specific biological functions, e.g., Calcium signaling pathway and axonogenesis by H3K4me3, epithelial cell differentiation by H3K27ac, or sodium ion transport by H3K27me3 (Fig. 2G). Oppositely, signals of H3K4me3 and/or H3k27ac tended to decrease around the repressed genes implicated in cell cycle, rather than AR-target genes and NE-associated genes, in the LNCaP cells after 14 days enzalutamide exposure (Fig. 2H). For example, cell cycle genes, e.g., proliferative marker MKi67 (Fig. 2I) as well as CDK1 and CyclinA2 (CCNA2) (Fig. S2A, S2B), were observed with the similar patterns of epigenetic status. Each of them showed loss of H3K27ac within 1–2 kb of promoter and transcription start site (TSS), with concomitant loss of H3K4me3 at overlapping locations.
Fig. 2.
Epigenetic status of chromatin confirms the strong associations between H3K4me3/H3K27ac/H3K27me3 and gene expression. (A-C) Genome-wide alterations of H3K4me3, H3K27ac, and H3K27me3 before and after enzalutamide treatment. The heatmap shows significant alternations of histone modifications aligned to the peak center, where colors from blue to red represent the histone signals (normalized RPKM × 10 within the bin size of 50 bp) from low to high. The above and below heatmap for each histone modification indicates the increased and decreased signals, respectively, upon the treatment. The curve above each heatmap is the average of histone signals accordingly. (D-E) Positive correlation between gene expression and H3K4me3 (D) or H3K27ac (E). (F) Negative correlation between H3K27me3 and gene expression. (G) Shared or unique GO terms and pathways over-represented in up-regulated DEGs with increased H3K4me3/H3K27ac and/or decreased H3K27me3. (H) GO terms enriched in down-regulated DEGs with decreased H3K4me3 or H3K27ac. (I-K) Differential epigenetic marks around one cell cycle marker Ki67 (I), AR target gene KLK3 (J), and NE master transcription factor ASCL1 (K). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
General consistency was found between bulk mRNA-seq, RT-qPCR and western blot analysis, and epigenetic status for NE-associated genes, AR signaling, and proliferating epithelial associated genes. For example, the dramatic decreases of the expression of AR target genes, e.g., KLK3/PSA (Fig. 2J), KLK2, and FKBP5 (Fig. S2C, S2D), were observed in agreement with significantly universal loss of H3K27ac from the promoter to the gene body. KLK2/3 also showed notable decreases of H3K4me3 which was not observed for FKBP5. KLK3 exhibited loss of H3K4me3 spanning the full gene, while KLK2 was only trimethylated at H3K4 upstream and including the transcription start site. Active gene repression via H3K27me3 was not observed for any AR target genes identified following enzalutamide treatment, suggesting the decreased expression was due to loss of H3K27 acetylation alone.
The expression levels of ASCL1 and SYP, which both are NE markers, were elevated following 14-day treatment. The CUT&RUN analysis suggests that ASCL1 upregulation was mediated predominantly through de-repression by loss of H3K27me3, except for small region close to the transcription start site (Fig. 2K). However, SYP transcription seemed activated by deposition of H3K27ac upon the treatment, indicating that SYP upregulation was due to activation rather than de-repression as observed with ASCL1 (Fig. S2E).
3.3. Time-resolved, single cell analysis reveals differences in morphology, canonical AR-suppression, and activation of NEL transcription signatures
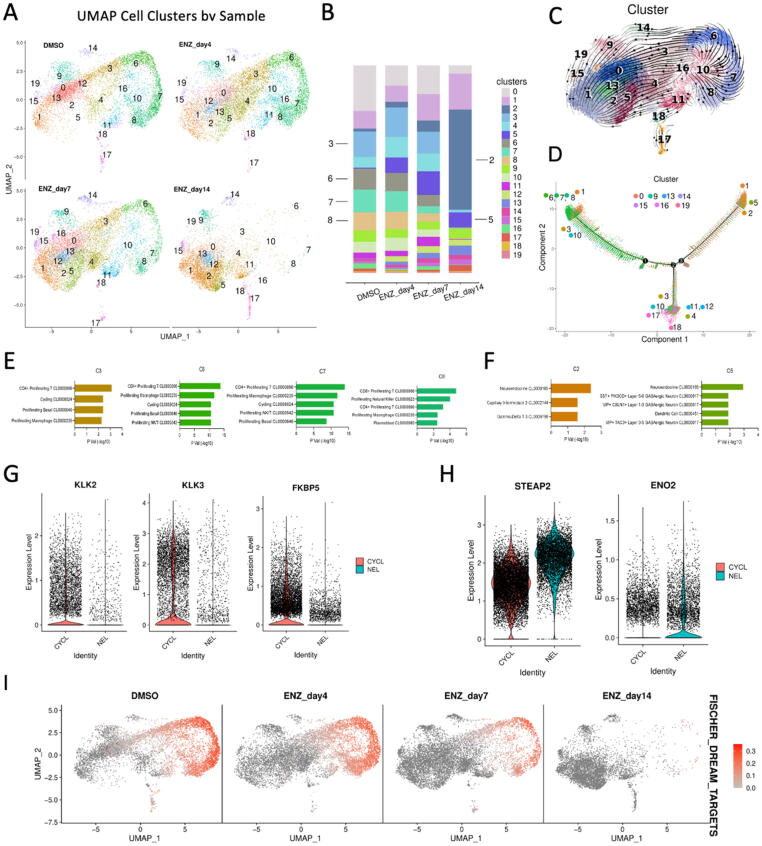
Enzalutamide treatment induced remarkable changes in cellular morphology, epigenetic status of histone modification, subsequent changes in transcriptional programs, and resultant protein expression and neuropeptide secretion in LNCaP cells. Most significantly, each of these phenomena were largely in agreement and were highly reproducible between experiments. Because no somatic or structural changes would be expected in such a short treatment window, clinical data suggest very little structural genomic changes between CRPC/NEPC, and epigenetic status correlated well with altered transcription in our model system, we hypothesized that ASI-induced NED is a largely epigenetic process. To investigate this further, we decided to perform high resolution single cell RNA-sequencing (scRNA-seq) by utilizing the 10X genomics platform and Cell Ranger software [63]. Following the same time course described above, we isolated single cells from DMSO control, 4-day, 7-day, and 14-day enzalutamide treatment of LNCaP cells and subjected them to scRNA-seq. To use a comparative approach to analyze the model system, we also utilized morphological observation as depicted (Fig. 1A) as well as bulk mRNA-seq screening (Fig. 1F).
Distinct cell subtypes were noticed given the scRNA-seq data from DMSO control to after 14-day treatment (Fig. 3A). The populations of cell subtypes were distinguishing across different time points (Fig. 3B). Some clusters were highly specific to the enzalutamide samples from 4 to 14 days, e.g., clusters C2 and C5 (C2/5), while several clusters decreased with the time points, e.g., clusters C3, C6, C7, and C8 (C3/6/7/8), in addition to others which were potentially transient-status cells more common to 4-day and 7-day samples sets, such as C4 comprised mostly by cells of enzalutamide day 4 and day 7 treatment (Fig. 3A-B). Among them, the population of cluster C2 cells was almost exclusively specific to 14-day enzalutamide treatment, while clusters C3/6/7/8 were almost completely exclusive of cells from 14-day enzalutamide treatment. The RNA velocity analysis (Fig. 3C) and trajectory analysis (Fig. 3D) indicated a trajectory split starting from C3/6/7/8 and branching to either C2/5 or to remaining transient clusters. These approaches suggested two potential pathways for the shift, one pathway mediated through C3/6/7/8 to finally NEL C2/5. The other reached to clusters C4/10/11/12/17/18 etc. (Fig. 3C).
Fig. 3.
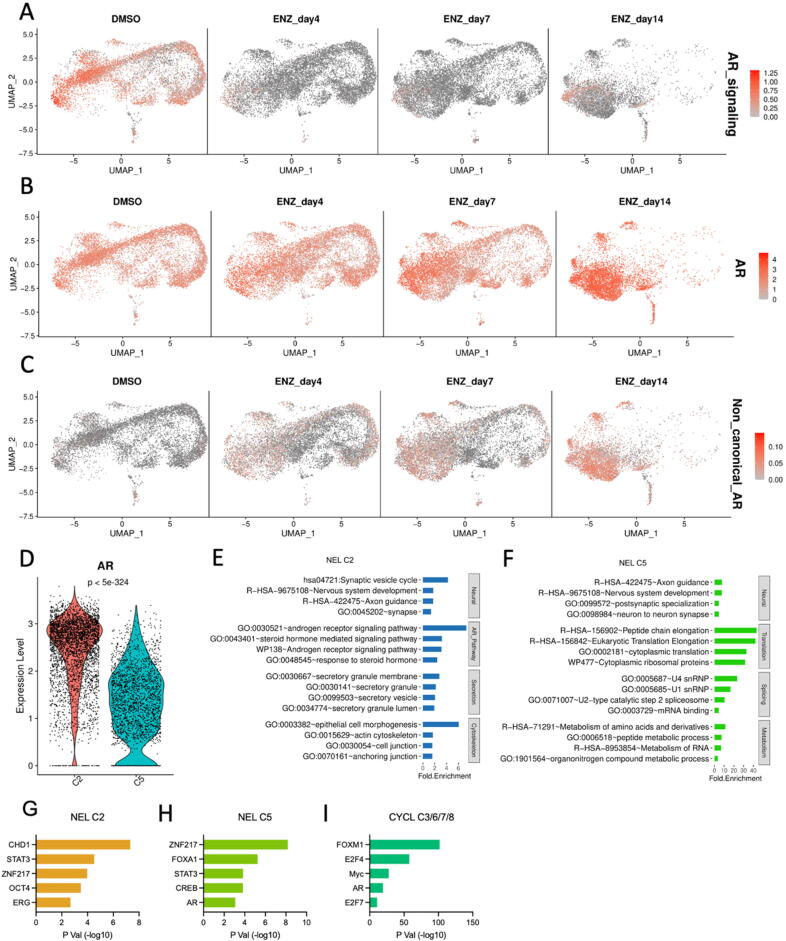
Time-resolved, single cell analysis reveals differences in morphology, canonical AR-suppression, and activation of NEL transcription signatures. (A) UMAP representation of total cell population comprised of DMSO, 4 day, 7 day, and 14 day enzalutamide-treated LNCaP cells stratified by treatment. (B) Contribution of each treatment condition (DMSO, enzalutamide (4, 7, 14 days)) to total cell population for each cluster. (C) RNA Velocity representation of UMAP-stratified cell population. (D) Trajectory analysis exploring two-path split of major groups of cells after treatment. (E-F) Azimuth cell type enrichment for CYCL cell clusters (E) and NEL cell clusters (F). (G-H) Violin plots for AR target genes (G) and NE-associated genes (H) stratified by CYCL and NEL cell clusters. (I) Feature plot of DREAM complex target gene expression in individual cells under each treatment.
To understand the cell subtypes of these distinguished clusters, we performed enrichment analysis on the signature genes identified by gene expression profile from each cluster profile utilizing the Azimuth Cell Types 2021 annotation set [58] (Fig. 3E-F, Fig. S3A-C). Morphological analysis suggested that cells stop cycling almost immediately on enzalutamide treatment, consistent with successful targeting of AR in AR-driven cells, so it was not surprising to observe that clusters C3/6/7/8, which were lost in a dose/time-dependent manner and almost entirely absent in 14-day treated cells, resembled actively cycling and mitotic cells based on Azimuth annotation (Fig. 3E). We named them as cycling or cycling-like cells (CYCL). Conversely, C2/5 as the largest population of cells in the 14-day treatment group resembled neuroendocrine and dendritic cells based on the same annotation database (Fig. 3F), suggesting that they are in fact NEL cells. Confirmed by our scRNA-seq, genes in the downstream of AR signaling pathway, e.g., KLK2/3, or genes mediated by AR, e.g., FKBP5, were repressed in NEL cells (clusters C2/5) compared to CYCL cells (Fig. 3G). Conversely, expression levels of some NE-associated genes were much higher in NEL cells than in CYCL cells, e.g., ENO2 and STEAP2 (Fig. 3H). STEAP2 is known as a gene supporting prostate cancer progression as well[64].
The CYCL cells in C3/6/7/8 also had high enrichment scores of S phase and G2M phase, while NEL cells in C2 showed no enrichment for these cycle stages (Fig. S4A-B). Utilizing the Reactome G0_And_Early_G1 dataset also showed enrichment in C3/6/7/8 exclusively, with no enrichment in the 14-day enzalutamide treated cells (Fig. S4C). We further checked the expression levels of dimerization partner, RB-like, E2F and multi-vulval class B (DREAM) targets [65], as the DREAM complex is a master epigenetic regulator of cell cycle advancement and is frequently dysregulated in cancers [66]. These genes were significantly highly expressed in CYCL cells (C3/6/7/8) in DMSO samples and earlier stages (7–10 days) of the enzalutamide treatment that is completely lost after 14 days (Fig. 3I). Taken together, these results indicate that following enzalutamide treatment, LNCaP cells exhibit loss of cell cycle advancement with a strong gain of NE-associated characteristics both transcriptionally, translationally, and morphologically.
Fig. 4.
ASI-induced NED suppresses canonical AR signaling, cell cycle advancement, but not AR expression. (A-C) Heatmap of total cell population for DMSO and enzalutamide treated LNCaP cells showing enrichment for AR signaling score (B), direct transcription of AR gene itself (B), and non-canonical AR signaling (C). (D) Violin plot depicting AR expression at mRNA level for NEL C2 and C5. (E-F) DAVID enrichment of terms associated with more highly expressed gene signatures in NEL C2 (E) or NEL C5 (F). (G-I) TFs enriched in NEL C2 (G), NEL C5 (H), and CYCL C3/6/7/8 (I), separately.
3.4. ASI-induced NED induces shift from canonical to noncanonical AR signaling pathways and development of NE-like cells with heterogenous AR status
ASI therapy targets either the biosynthesis of testosterone or the AR-mediated cellular response to testosterone. Therefore, we utilized the scRNA-seq data to check AR status and AR signaling pathway simultaneously. Immediately following enzalutamide treatment, AR signaling sharply decreased for 4-day, 7-day, and 14-day treatment groups (Fig. 4A). This was driven largely by AR-target genes KLK2, KLK3, and FKBP5 suppression immediately following enzalutamide treatment. However, while enzalutamide treatment largely suppressed androgen signaling and transcription of canonical AR-target genes as expected, we were surprised to observe that transcription of AR was upregulated roughly 4-fold based on bulk mRNA-seq quantitation (Fig. 4B).
Heterogenous AR status and activation of noncanonical AR signaling pathway activation in enzalutamide resistant cell lines has recently been reported [19], [67]. To assess whether our model system was able to recapitulate the canonical / noncanonical AR signaling switch described by He et. al., we calculated a noncanonical AR signaling score and indeed observed dose/time-dependent activation in enzalutamide-treated cells (Fig. 4C). Surprisingly, despite using a single cell line in our experiment, we were able to identify-two NEL clusters, C2 and C5, in our dataset that demonstrated variable AR gene expression, with NEL C2 exhibiting higher AR expression than NEL C5 (Fig. 4D). As AR upregulation followed long term exposure of LNCaP cells to enzalutamide (Fig. 4B), we next sought to determine if the differential AR status in NEL C2 and C5 resulted in any functional consequence to the cells, or if instead transcriptionally inactive AR was simply higher in C2 and C5. Towards this end, we first identified DEGs highly expressed in either C2 or C5 then performed DAVID enrichment analysis on these DEGs. Distinct cellular functions were recognized to be enriched in C2 and C5, respectively (Fig. 4E, 4F). Both NEL clusters indicated the enrichment for nervous system development, axon guidance, and other neuronal/neuroendocrine related functions, but the genes from C2 and C5 implicated in these pathways and functions are totally different. Moreover, NEL C2 showed NE-associated secretory processes as well as cytoskeletal/morphological related processes (Fig. 4E). Conversely, NEL C5 exhibited enrichment in the process of splicing, translation, and metabolism (Fig. 4F). Together, these results suggest that two NEL clusters distinguished by heterogenous AR status exhibit clearly common and distinct biological functional processes.
To evaluate epigenetic regulatory mechanism underlying, we performed enrichment analysis of transcription factor (TF) motifs against the signature genes comprising C2 (NEL/AR + ), C5 (NEL/AR-), and CYCL cells (C3/6/7/8), respectively. ZNF217 and STAT3, both associated strongly with NEPC, were enriched in C2 and C5 signature genes (Fig. 4G, 4H). As comparison, AR, as well as E2F, FOXM1, and Myc motifs were over-represented in the signature genes of CYCL cells (Fig. 4I). Observation of different TFs between C2 and C5 further strengthened the heterogeneity of AR status in NEL cells. For example, AR and CREB motifs were significantly enriched in signature genes of C5 (Fig. 4H), while the more stem-associated Yamanaka factor OCT4 was observed in NEL C2 (Fig. 4G). While differential motif enrichment does not rule out C5 simply as a transitory state, it does recapitulate multiple clinical reports that have demonstrated AR signaling retention within NEL cells and suggests NEL phenotype does not fundamentally require abrogation or AR or AR signaling. Interestingly, while AR motif was enriched only for NEL C5 (Fig. 4H), NEL C2 indicated higher AR expression (Fig. 4D), potentially suggesting cells of NEL C2 are attempting to regain AR signaling through compensatory AR upregulation.
Enzalutamide treatment of LNCaP cells represents a homogenous in vitro model system, but surprisingly can recapitulate heterogenous response to pharmacological targeting of AR by showing differential AR expression, compensatory AR reactivation at both the transcript and protein level, subpopulation-specific retention of AR target gene expression and AR signaling score, and two distinct NEL cell populations with opposing AR status. The high resolution afforded by single-cell sequencing platform collectively demonstrates that even within a single cell line and only 14 days of enzalutamide exposure, subpopulations of cells modulate AR status, AR signaling, and NEL transdifferentiation, resembling the same heterogeneity observed in both clinical samples and preclinical models.
4. Discussion
4.1. NED model systems and time-resolved analysis of ASI-induced NED recapitulates clinically relevant transcription signatures and provides sequence specificity
Clinical presentation of NEPC tissues can be mixed morphology or can be predominantly NE, also called focal NEPC. Multiple studies indicate underlying RB1, TP53, and PTEN genomic or functional loss are present in clinical NEPC samples without necessarily being present at the same frequency in CRPC progenitor cells [13]. ASI selective pressure is hypothesized to select for cells with genomic perturbation of these genes prior to NED and progression to NEPC. However, it is nearly impossible to generate mechanistic data on clinically available samples alone, and model systems are necessary. Normal epithelial tissue has been reprogrammed to undergo NED following functional abrogation of TP53 and RB1, and PTEN (via AKT1 overexpression), but the experiment was performed with underlying overexpression of BCL2 and MYC [27]. Cells with AKT1, MYC, and BCL2 overexpression retained prostatic lineage marker expression until functional loss of RB1 and TP53 was introduced; activation of NE-associated transcriptional programs required RB1 and TP53 [27]. The TRAMP mouse model, with prostate-specific, SV40 T-mediated functional loss RB1 and P53, spontaneously generates prostatic adenocarcinoma and can be induced to develop NEPC following castration [68], [69]. NEL tumors have also been reported in mice with loss of both PTEN and RB1, with more aggressive features when TP53 is also lost [70]. Clinically, NEPC tumors harbor significantly higher rates of RB1 and TP53 loss [29], [71], yet there is still no current answer if TP53 and RB1 loss are required for NED induction or, instead, they simply enable the more rapid proliferation of NEL cells compared to NEL cells without these alterations [72]. Supporting this, TRAMP mice show increased incidence of NEL tumor development from 5 % − 53 % following castration [73]. In LNCaP cells, functional knockout of both TP53 and RB1 showed no development of NEL features in vitro or in vivo; further, AR and SOX2 expression changes were not detected, indicating TP53 and RB1 loss may be necessary, but not sufficient, for NED [74]. LNCaP cells are TP53 and RB1-competent, but do harbor one deleted allele of PTEN [75]. Our model system demonstrates in single cell resolution that following pharmacological targeting of AR, activation of NE-associated programs can occur independently of RB1/TP53 loss or repression of epithelial markers. Further, we demonstrate a significant upregulation of AR gene transcription, likely as compensatory mechanism for pharmacological AR-inhibition, resulting in a significant increase in AR at the protein level by day 14. We also detect that even in TP53/RB1-competent background, transcription of common NE marker genes is detectable after 4 days of enzalutamide exposure. We also demonstrated that NE marker NCAM1 (gene, CD56), a neuronal cell adhesion marker, is rapidly upregulated by day 4 at both the mRNA and protein levels, consistent with the rapid development of NEL morphology also observed by day 4. These results suggest a sequence of events in which initial morphological change occurs rapidly following pharmacological AR targeting, followed by functional upregulation of master regulator transcription factors, followed by compensatory AR upregulation. While the model system utilized for this research only replicates the NED process in vitro, these results may support the model of NEPC development in which NEL cells, or cells undergoing NED process, retain receptivity to androgen-mediated growth and survival signals, and NEL cells that regain proliferative capacity in an AR-dependent manner are then selected for under potent AR targeting.
Neuronal splicing factor SRRM4 is a marker of NEPC, and overexpression of SRRM4 in LNCaP cells has been reported to be required for development NEL cellular morphology and transcriptional programs in vitro and in vivo [76]; however, we did not detect change in SRRM4 expression, and while SRRM4 clearly mediates progression of established NEPC tumor cells [23], our data demonstrate that SRRM4 is not required for initial stages of enzalutamide-induced lineage plasticity. Further, a switch to NEL morphology clearly preceded full activation of NE-associated transcriptional change, although this merits further exploration of the functional consequence. By observing the initial steps of therapy-induced NED alone, our reported model system can rule out the confounding variables that would otherwise be ubiquitous in observations based solely on clinical samples.
4.2. Specific subpopulations of ASI-treated cells indicate dependency of ASI-induced NED on AR pathway and divergence of NEL cells into distinct clusters
Recent reports of patient-derived NEPC samples have highlighted heterologous AR status and noncanonical signaling [19], suggesting that AR potentially serves a functional role in cells even under enzalutamide treatment or potentially drive enzalutamide resistance [67]. The results of our study suggest that both of these features can be recapitulated in vitro with a simple 2D cell culture-based platform when coupled with high resolution analytical methods. We observed two distinctly NEL clusters following enzalutamide treatment. C2, which retained higher level of AR expression and showed enrichment of AR pathway related terms, also showed NE-associated secretory processes as well as cytoskeletal/morphological related processes, which would be in general agreement with histological staining for neuropeptide secretion and small cell morphology. Conversely, NEL C5 exhibited enrichment of splicing, translation, and metabolism terms, which may indicate that NEL C5 has either undergone more significant deviation away from parental line or that NEL C5 represents a more plastic/transitory state with variable cellular processes serving role as quiescent cancer stem cell or therapy-resistant cell via plasticity in key cellular functions. The discrepancy between C2 and C5 may represent the frequently clinically observed divergence of canonical AR-/NE + NEPC tumor cells from more recently described AR+/NE + cells. This lineage split of NEL cells results in a hypothesis that genetic perturbations on different genes or pathways may lead to the ability to regain proliferative capacity and result in further divergence of the C2 and C5 cell populations into more distinct subcategories of NEL cells. This avenue merits further explanation and may be experimentally determined by introduction of clinically relevant PTEN, TP53, or RB1 loss via inducible knockdown with or without extension of time course.
4.3. Utility of in vitro ASI-induced NED model system as aid for resistance-focused drug screening
By observing the early stages of ASI therapy-induced NED in a single cell and time-resolved manner, and subsequently characterizing the morphological, transcriptional, and epigenetic sequences involved, we sought to develop a model system that can be applied to pharmacological development beyond target engagement and growth suppression but can be placed in the clinically relevant context of disease progression. Loss of AR protein and transcriptional activity are hallmarks of NEPC, but here we have successfully demonstrated that they are not necessary for initial activation of NE-associated transcriptional signatures and change in morphology. Our model system has already provided evidence that canonical NEPC characteristics – genomic or functional TP53 and RB1 loss, SRMM4-mediated alternative expression patterns, and upregulated EZH2 transcript and activity – may contribute more to the maintenance of NEL phenotype and rapid growth/proliferation than to the immediate development of plasticity and enzalutamide-response.
Our goal with this model system is to provide a platform for the screening of novel pharmacologically active compounds that may inhibit components of the cellular plasticity process in the context of ASI therapy-induced NED in prostate cancer cells. Significant value exists in distinction of compounds that target existing NEPC metastases versus compounds that would inhibit NED development, and availibility of such compounds could present significant ramifications for patient stratification.
5. Conclusion
Our model system provides a high-resolution and time-resolved approach to screening for drugs to either inhibit the ASI therapy-induced NED process or to resensitize developing NE-like cells to ASI therapy and delay or prevent progression of NEPC. We recognize that this model does provide value by focusing on a significantly clinically relevant unmet need of deciphering the underlying mechanisms of ASI therapy-induced NED even though it doesn’t fully capture clinical NEPC development. Logical extensions of this system are: 1) Performance on representative panel of HNPC and CRPC cell lines, 2) Performance on GEMM tissue under ASI therapy or castration, and 3) Inclusion of ATAC-seq or high-throughput proteomics to confirm transcriptional changes with resultant functional heterochromatic repression or altered protein content following transcriptional dysregulation.
Funding
This work was supported by the U.S. Army Medical Research Acquisition Activity, Prostate Cancer Research Program (W81XWH-12–1-0346, W81XWH-13–1-0398, and W81XWH-16–1-0394) and NIH/NCI R01CA212403, Indiana University Simon Comprehensive Cancer Center (IUSCCC, NIH/NCI P30CA082709) Near-Miss Initiative, and Purdue University Center for Cancer Research (PUCCR, NIH/NCI P30CA082709) Small Grants. All bioinformatics analysis was conducted by the Collaborative Core for Cancer Bioinformatics (C3B) shared by IUSCCC and PUCCR with additional support from the Walther Cancer Foundations. Andrew Asberry was supported by drug discovery trainee fellowship under NIH T32 Fellowship Grant NIH T32GM125620.
CRediT authorship contribution statement
Andrew Michael Asberry: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Resources, Data curation, Writing – original draft, Writing – review & editing. Sheng Liu: Methodology, Software, Validation, Formal analysis, Investigation, Resources, Data curation, Visualization, Writing – review & editing. Hye Seung Nam: Methodology, Validation, Investigation, Writing – review & editing. Xuehong Deng: Methodology, Investigation, Writing – review & editing. Jun Wan: Conceptualization, Methodology, Software, Validation, Formal analysis, Resources, Data curation, Writing – review & editing, Supervision, Project administration, Funding acquisition. Chang-Deng Hu: Conceptualization, Methodology, Validation, Formal analysis, Resources, Data curation, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Authors thank all members from Labs of Dr. Chang-Deng Hu and Dr. Jun Wan for constructive suggestions. We also thank Dr. Xiaoling Xuei, Dr. Fang Fang, and Patrick McGuire in the Center of Medical Genomics at Indiana University School of Medicine for technical support and 10X library preparation. Graphical Abstract created with BioRender.com.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.10.031.
Contributor Information
Jun Wan, Email: junwan@iu.edu.
Chang-Deng Hu, Email: hu1@purdue.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 2.Mohler J.L., et al. Prostate Cancer, Version 2.2019, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2019;17(5):479–505. doi: 10.6004/jnccn.2019.0023. [DOI] [PubMed] [Google Scholar]
- 3.Khuntia D., Reddy C.A., Mahadevan A., Klein E.A., Kupelian P.A. Recurrence-free survival rates after external-beam radiotherapy for patients with clinical T1–T3 prostate carcinoma in the prostate-specific antigen era: What should we expect? Cancer. 2004;100(6):1283–1292. doi: 10.1002/cncr.20093. [DOI] [PubMed] [Google Scholar]
- 4.Viani G.A., Stefano E.J., Afonso S.L. Higher-Than-Conventional Radiation Doses in Localized Prostate Cancer Treatment: A Meta-analysis of Randomized, Controlled Trials. Int J Radiat Oncol. 2009;74(5):1405–1418. doi: 10.1016/j.ijrobp.2008.10.091. [DOI] [PubMed] [Google Scholar]
- 5.Dong B., et al. Single-cell analysis supports a luminal-neuroendocrine transdifferentiation in human prostate cancer. Commun Biol. 2020;3(1):778. doi: 10.1038/s42003-020-01476-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karthaus W.R., et al. Regenerative potential of prostate luminal cells revealed by single-cell analysis. Science. 2020;368(6490):497–505. doi: 10.1126/science.aay0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cunha G.R., et al. Development of the human prostate. Differentiation. 2018;103:24–45. doi: 10.1016/j.diff.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan M.E., Li J., Xu H.E., Melcher K., Yong E. Androgen receptor: structure, role in prostate cancer and drug discovery. Acta Pharmacol Sin. 2015;36(1):3–23. doi: 10.1038/aps.2014.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu X., et al. Structural Insights of Transcriptionally Active, Full-Length Androgen Receptor Coactivator Complexes. Mol Cell. 2020 doi: 10.1016/j.molcel.2020.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rowlands M.G., et al. Esters of 3-Pyridylacetic Acid That Combine Potent Inhibition of 17.alpha.-Hydroxylase/C17,20-Lyase (Cytochrome P45017.alpha.) with Resistance to Esterase Hydrolysis. J Med Chem. 1995;38(21):4191–4197. doi: 10.1021/jm00021a008. [DOI] [PubMed] [Google Scholar]
- 11.Tran C., et al. Development of a Second-Generation Antiandrogen for Treatment of Advanced Prostate Cancer. Science. 2009;324(5928):787–790. doi: 10.1126/science.1168175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beer T.M., et al. Enzalutamide in Metastatic Prostate Cancer before Chemotherapy. N Engl J Med. 2014;371(5):424–433. doi: 10.1056/NEJMoa1405095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aggarwal R., et al. Clinical and Genomic Characterization of Treatment-Emergent Small-Cell Neuroendocrine Prostate Cancer: A Multi-institutional Prospective Study. J Clin Oncol. 2018;36(24):2492–2503. doi: 10.1200/JCO.2017.77.6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu C.-D., Choo R., Huang J. Neuroendocrine Differentiation in Prostate Cancer: A Mechanism of Radioresistance and Treatment Failure. Front Oncol. 2015;5 doi: 10.3389/fonc.2015.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Butler W., Huang J. Neuroendocrine cells of the prostate: Histology, biological functions, and molecular mechanisms. Precis Clin Med. 2021;4(1):25–34. doi: 10.1093/pcmedi/pbab003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berman-Booty L.D., Knudsen K.E. Models of neuroendocrine prostate cancer. Endocr Relat Cancer. 2015;22(1):R33–R49. doi: 10.1530/ERC-14-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conteduca V., et al. Circulating tumor cell heterogeneity in neuroendocrine prostate cancer by single cell copy number analysis. Npj Precis Oncol. 2021;5(1):76. doi: 10.1038/s41698-021-00211-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berchuck J.E., et al. Detecting neuroendocrine prostate cancer through tissue-informed cell-free DNA methylation analysis. Clin Cancer Res. 2021 doi: 10.1158/1078-0432.CCR-21-3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davies A., et al. An androgen receptor switch underlies lineage infidelity in treatment-resistant prostate cancer. Nat Cell Biol. 2021;23(9):1023–1034. doi: 10.1038/s41556-021-00743-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bluemn E.G., et al. Androgen Receptor Pathway-Independent Prostate Cancer Is Sustained through FGF Signaling. Cancer Cell. 2017;32(4):474–489.e6. doi: 10.1016/j.ccell.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi W.W., Boland J.L., Lin J. ONECUT2 as a key mediator of androgen receptor-independent cell growth and neuroendocrine differentiation in castration-resistant prostate cancer. Cancer Drug Resist. 2022 doi: 10.20517/cdr.2021.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bishop J.L., et al. The Master Neural Transcription Factor BRN2 Is an Androgen Receptor-Suppressed Driver of Neuroendocrine Differentiation in Prostate Cancer. Cancer Discov. 2017;7(1):54–71. doi: 10.1158/2159-8290.CD-15-1263. [DOI] [PubMed] [Google Scholar]
- 23.Zhang X., et al. SRRM4 Expression and the Loss of REST Activity May Promote the Emergence of the Neuroendocrine Phenotype in Castration-Resistant Prostate Cancer. Clin Cancer Res. 2015;21(20):4698–4708. doi: 10.1158/1078-0432.CCR-15-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kregel S., et al. Sox2 Is an Androgen Receptor-Repressed Gene That Promotes Castration-Resistant Prostate Cancer. PLoS ONE. 2013;8(1):e53701. doi: 10.1371/journal.pone.0053701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mu P., et al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53 - and RB1 -deficient prostate cancer. Science. 2017;355(6320):84–88. doi: 10.1126/science.aah4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Svensson C., et al. REST mediates androgen receptor actions on gene repression and predicts early recurrence of prostate cancer. Nucleic Acids Res. 2014;42(2):999–1015. doi: 10.1093/nar/gkt921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park JW et al., “Reprogramming normal human epithelial tissues to a common, lethal neuroendocrine cancer lineage,” p. 6, 2018. [DOI] [PMC free article] [PubMed]
- 28.Conteduca V., et al. Clinical features of neuroendocrine prostate cancer. Eur J Cancer. 2019;121:7–18. doi: 10.1016/j.ejca.2019.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson D., et al. Integrative Clinical Genomics of Advanced Prostate Cancer. Cell. 2015;161(5):1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bishop J.L., et al. PD-L1 is highly expressed in Enzalutamide resistant prostate cancer. Oncotarget. 2015;6(1):234–242. doi: 10.18632/oncotarget.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang S., et al. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell. 2003;4(3):209–221. doi: 10.1016/S1535-6108(03)00215-0. [DOI] [PubMed] [Google Scholar]
- 32.Zhou Z., et al. Synergy of p53 and Rb Deficiency in a Conditional Mouse Model for Metastatic Prostate Cancer. Cancer Res. 2006;66(16):7889–7898. doi: 10.1158/0008-5472.CAN-06-0486. [DOI] [PubMed] [Google Scholar]
- 33.Owens J.L., et al. PRMT5 Cooperates with pICln to Function as a Master Epigenetic Activator of DNA Double-Strand Break Repair Genes. iScience. 2020;23(1) doi: 10.1016/j.isci.2019.100750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Owens JL et al., “Targeting protein arginine methyltransferase 5 (PRMT5) suppresses radiation-induced neuroendocrine differentiation and sensitizes prostate cancer cells to radiation,” Mol. Cancer Ther., p. molcanther.MCT-21-0103-A.2021, Jan. 2022, doi: 10.1158/1535-7163.MCT-21-0103. [DOI] [PMC free article] [PubMed]
- 36.Deng X., et al. Protein arginine methyltransferase 5 functions as an epigenetic activator of the androgen receptor to promote prostate cancer cell growth. Oncogene. 2017;36(9):1223–1231. doi: 10.1038/onc.2016.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsu C.-C., Hu C.-D. Transcriptional activity of c-Jun is critical for the suppression of AR function. Mol Cell Endocrinol. 2013;372(1–2):12–22. doi: 10.1016/j.mce.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang H.-T., Zhang D., Zha Z.-G., Hu C.-D. Transcriptional activation of PRMT5 by NF-Y is required for cell growth and negatively regulated by the PKC/c-Fos signaling in prostate cancer cells. Biochim Biophys Acta. 2014;1839(11):1330–1340. doi: 10.1016/j.bbagrm.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saha K., Adhikary G., Eckert R.L. MEP50/PRMT5 Reduces Gene Expression by Histone Arginine Methylation and this Is Reversed by PKCδ/p38δ Signaling. J Invest Dermatol. 2016;136(1):214–224. doi: 10.1038/JID.2015.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dobin A., et al. STAR: ultrafast universal RNA-seq aligner. Bioinforma Oxf Engl. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinforma Oxf Engl. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 42.McCarthy D.J., Chen Y., Smyth G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40(10):4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinforma Oxf Engl. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waltman L., van Eck N.J. A smart local moving algorithm for large-scale modularity-based community detection. Eur Phys J B. 2013;86(11):471. doi: 10.1140/epjb/e2013-40829-0. [DOI] [Google Scholar]
- 45.Stuart T., et al. Comprehensive integration of single cell data. Genomics. 2018 doi: 10.1101/460147. preprint. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.La Manno G., et al. RNA velocity of single cells. Nature. 2018;560(7719):494–498. doi: 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bergen V., Lange M., Peidli S., Wolf F.A., Theis F.J. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat Biotechnol. 2020;38(12):1408–1414. doi: 10.1038/s41587-020-0591-3. [DOI] [PubMed] [Google Scholar]
- 48.Trapnell C., et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32(4):381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marunde M.R., et al. Nucleosome conformation dictates the histone code. Genomics. 2022 doi: 10.1101/2022.02.21.481373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yusufova N., et al. Histone H1 loss drives lymphoma by disrupting 3D chromatin architecture. Nature. 2021;589(7841):299–305. doi: 10.1038/s41586-020-3017-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skene P.J., Henikoff J.G., Henikoff S. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat Protoc. 2018;13(5):1006–1019. doi: 10.1038/nprot.2018.015. [DOI] [PubMed] [Google Scholar]
- 52.Shah R.N., et al. Examining the Roles of H3K4 Methylation States with Systematically Characterized Antibodies. Mol Cell. 2018;72(1):162–177.e7. doi: 10.1016/j.molcel.2018.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zang C., Schones D.E., Zeng C., Cui K., Zhao K., Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinforma Oxf Engl. 2009;25(15):1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramírez F., Dündar F., Diehl S., Grüning B.A., Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucl Acids Res. 2014;42(Web Server issue):W187–191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kurtenbach S., William Harbour J. SparK: A Publication-quality NGS Visualization Tool. Bioinformatics. 2019 doi: 10.1101/845529. preprint. [DOI] [Google Scholar]
- 57.Dennis G., et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):P3. [PubMed] [Google Scholar]
- 58.Hao Y., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184(13):3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen E.Y., et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kuleshov M.V., et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucl Acids Res. 2016;44(W1):W90–97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beketova E., et al. Protein Arginine Methyltransferase 5 Promotes pICln-Dependent Androgen Receptor Transcription in Castration-Resistant Prostate Cancer. Cancer Res. 2020;80(22):4904–4917. doi: 10.1158/0008-5472.CAN-20-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taavitsainen S., et al. Single-cell ATAC and RNA sequencing reveal pre-existing and persistent cells associated with prostate cancer relapse. Nat Commun. 2021;12(1):5307. doi: 10.1038/s41467-021-25624-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zheng G.X.Y., et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8(1):14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen W.-J., et al. Regulatory Roles of Six-Transmembrane Epithelial Antigen of the Prostate Family Members in the Occurrence and Development of Malignant Tumors. Front Cell Dev Biol. 2021;9 doi: 10.3389/fcell.2021.752426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fischer M., Grossmann P., Padi M., DeCaprio J.A. Integration of TP53, DREAM, MMB-FOXM1 and RB-E2F target gene analyses identifies cell cycle gene regulatory networks. Nucleic Acids Res. 2016;44(13):6070–6086. doi: 10.1093/nar/gkw523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sadasivam S., DeCaprio J.A. The DREAM complex: master coordinator of cell cycle-dependent gene expression. Nat Rev Cancer. 2013;13(8):585–595. doi: 10.1038/nrc3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.He Y., et al. A noncanonical AR addiction drives enzalutamide resistance in prostate cancer. Nat Commun. 2021;12(1):1521. doi: 10.1038/s41467-021-21860-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Greenberg N.M., et al. Prostate cancer in a transgenic mouse. Proc Natl Acad Sci. 1995;92(8):3439–3443. doi: 10.1073/pnas.92.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kaplan-Lefko P.J., et al. Pathobiology of autochthonous prostate cancer in a pre-clinical transgenic mouse model. Prostate. 2003;55(3):219–237. doi: 10.1002/pros.10215. [DOI] [PubMed] [Google Scholar]
- 70.Ku S.Y., et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science. 2017;355(6320):78–83. doi: 10.1126/science.aah4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Beltran H., et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med. 2016;22(3):298–305. doi: 10.1038/nm.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen R., Dong X., Gleave M. Molecular model for neuroendocrine prostate cancer progression. BJU Int. 2018;122(4):560–570. doi: 10.1111/bju.14207. [DOI] [PubMed] [Google Scholar]
- 73.Sulsenti R., et al. Repurposing of the Antiepileptic Drug Levetiracetam to Restrain Neuroendocrine Prostate Cancer and Inhibit Mast Cell Support to Adenocarcinoma. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.622001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nyquist M.D., et al. Combined TP53 and RB1 Loss Promotes Prostate Cancer Resistance to a Spectrum of Therapeutics and Confers Vulnerability to Replication Stress. Cell Rep. 2020;31(8) doi: 10.1016/j.celrep.2020.107669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vlietstra R.J., van Alewijk D.C., Hermans K.G., van Steenbrugge G.J., Trapman J. Frequent inactivation of PTEN in prostate cancer cell lines and xenografts. Cancer Res. 1998;58(13):2720–2723. [PubMed] [Google Scholar]
- 76.Li Y., et al. Establishment of a neuroendocrine prostate cancer model driven by the RNA splicing factor SRRM4. Oncotarget. 2017;8(40) doi: 10.18632/oncotarget.19916. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data have been deposited to the GEO database with a super-series number GSE215945, including bulk RNA-seq (GSE215653), single cell RNA-seq (GSE215943), and CUT&RUN (GSE216118).