SUMMARY
Cellular signaling by fibroblast growth factor receptors (FGFRs) is a highly regulated process mediated by specific interactions between distinct subsets of fibroblast growth factor (FGF) ligands and two FGFR isoforms generated by alternative splicing: an epithelial b- and mesenchymal c-isoforms. Here, we investigate the properties of a mini-protein, mb7, developed by an in silico design strategy to bind to the ligand-binding region of FGFR2. We describe structural, biophysical, and cellular analyses demonstrating that mb7 binds with high affinity to the c-isoforms of FGFR, resulting in inhibition of cellular signaling induced by a subset of FGFs that preferentially activate c-isoforms of FGFR. Notably, as mb7 blocks interaction between FGFR with Klotho proteins, it functions as an antagonist of the metabolic hormones FGF19 and FGF21, providing mechanistic insights and strategies for the development of therapeutics for diseases driven by aberrantly activated FGFRs.
Graphical Abstract

In brief
Park et al. show that a de-novo-designed mini-protein, mb7, can specifically recognize c-isoforms of FGFRs. By masking the regions of FGFR that are critical for the FGFR activation, mb7 can potently inhibit cellular signaling by a subset of FGFs that preferentially activate FGFR c-isoform signaling.
INTRODUCTION
Fibroblast growth factors (FGFs) constitute a family of 22 proteins that play critical roles in mediating a variety of biological processes, such as growth, development, and metabolism (Ornitz and Itoh, 2015, 2022). They mediate cellular responses by binding to and stimulating cellular signaling through four members of FGF receptors (FGFRs) that belong to the receptor tyrosine kinase (RTK) superfamily (Lemmon and Schlessinger, 2010). The majority of the family members designated canonical FGFs act in a paracrine or autocrine manner. Canonical FGFs act together with heparin or heparan sulfate proteoglycans (HSPG) to bind to FGFRs and stimulate the dimerization and activation (Schlessinger et al., 2000). On the other hand, the three members of the endocrine FGF subfamily, FGF19, FGF21, and FGF23, that interact weakly with heparin or HSPG bind with high affinity and specificity to Klotho family of receptors that function as the “zip code”-like co-receptors required for FGFR activation and cell signaling (Chen et al., 2018; Goetz et al., 2007; Kurosu et al., 2007; Kuzina et al., 2019; Lee et al., 2018). Ligand-induced dimerization of FGFR extracellular domain (ECD) brings two intracellular tyrosine kinase regions together to initiate auto-transphosphorylation and tyrosine kinase activation (Lemmon and Schlessinger, 2010; Schlessinger, 2000). This leads to the recruitment of signaling molecules by direct complex formation of signaling molecules with activated FGFR or through indirect interactions mediated by closely associated docking proteins such as Frs2 and Shc, specialized in recruiting unique complements of signaling proteins (Schuller et al., 2008; Xu et al., 1998).
The ECDs of FGFRs contain three immunoglobulin (Ig)-like domains: D1, D2, and D3 (Kiselyov et al., 2006; Plotnikov et al., 1999). Structural studies have previously demonstrated that heparin-mediated dimerization occurs through a complex network of interactions in D2 regions, whereas the ligand-receptor interactions occur in the D2–D3 region (Schlessinger et al., 2000). In particular, two distinct isoforms, an epithelial “b” and a mesenchymal “c,” generated by alternative splicing of FGFR1-3 transcripts recognize specifically different FGF subfamilies (Eswarakumar et al., 2005; Ornitz and Itoh, 2001; Ornitz et al., 1996; Zhang et al., 2006). FGF ligands can be broadly categorized based on their isoform-specific FGFR interactions except for FGF1 and FGF2, which are classified as universal FGFs. FGF4, FGF5, FGF6, FGF8, FGF17, FGF18, FGF9, FGF16, FGF20, FGF19, FGF21, and FGF23 exhibit specificities toward the c-isoform, whereas FGF7, FGF3, FGF10, and FGF22 exclusively bind to and signal through the b-isoform (Zhang et al., 2006). These isoform specificities of FGF family members are attributed to the variations in amino acid sequences in their receptor-interacting regions. Since the binding affinities of FGF toward FGFRs are relatively low compared with other ligand-RTK affinities, a few changes in the amino acid sequences of FGFs can significantly affect their specificity toward FGFR isoforms. Most notably, the crystal structure of FGF8b bound to the ligand-binding region of FGFR2c revealed the potential role of the N-terminal region of FGF8b on its specific recognition of FGFR c-isoform (Olsen et al., 2006). Furthermore, the crystal structure of FGF23 in complex with the extracellular region of α-Klotho (KLAECD) and the ligand-binding region of FGFR1c (FGFR1cD2D3) demonstrated a critical role of KLAECD-FGFR1cD2D3 interactions in the FGFR c-isoform specificities of FGF23 signaling (Chen et al., 2018).
Here, we describe the properties of a de-novo-designed mini-protein that potently and selectively interacts with the c-isoforms of FGFRs. The crystal structure of the mini-protein in complex with the third Ig-like domain of FGFR4 (FGFR4D3) reveals the molecular interactions critical for the FGFR c-isoform-specific recognition of mini-protein, which in turn affect its inhibitory activities on the cellular FGFR signaling by multiple FGFs exhibiting c-isoform specificities.
RESULTS
A de-novo-designed mini-protein selectively binds to the c-isoform of FGFRs with high affinity
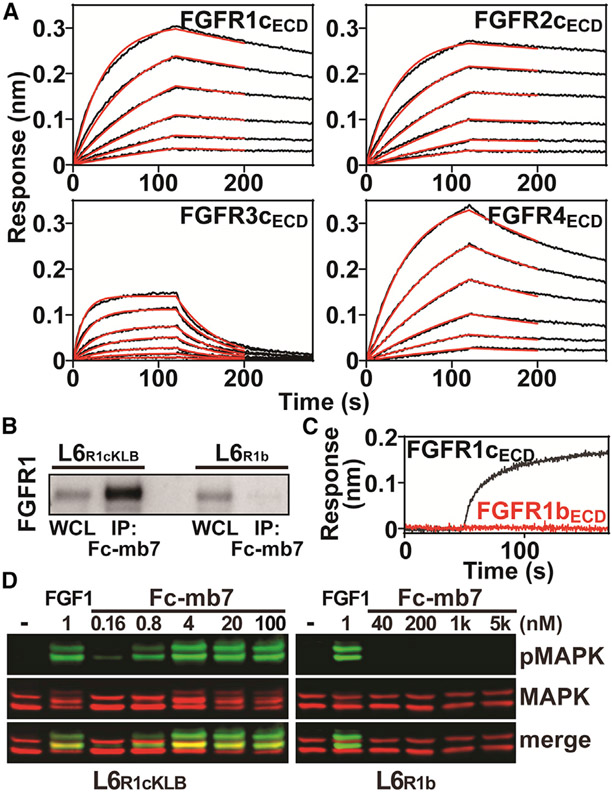
Cao et al. recently described an in silico design strategy enabling the development of a series of mini-proteins targeting specific regions in a variety of cell-surface receptors (Cao et al., 2022). One such molecule, mb7, was designed to bind to the extracellular domain (ECD) of FGFR2c (FGFR2cECD), and its properties were experimentally determined with biophysical and structural studies. Based on the amino acid sequence alignment of all FGFRs, we hypothesized that mb7 could also bind to other FGFRs. We first created a construct of mb7 of which the N terminus is attached to an Fc region of human immunoglobulin G (IgG) (Fc-mb7; Figure S1A), and characterized its binding kinetics for the ECD of each FGFR c-isoform using bio-layer interferometry (BLI) measurements (Figure 1A). Indeed, Fc-mb7 potently binds to FGFR1cECD, FGFR2cECD, and FGFR4ECD, and, to a lesser degree, to FGFR3cECD (Table S1). The differential binding affinity of mb7 to each of the FGFR family members can be accounted for by differences between the amino acid sequences of FGFR ECDs within the mb7-binding region. We have further confirmed these results by using mb7 with His-tag (His-mb7) to determine the binding kinetic parameters for the interactions between His-mb7 and the ligand-binding regions of either FGFR1c (FGFR1cD2D3) or FGFR4 (FGFR4D2D3) using BLI measurements (Figure S1B and Table S1).
Figure 1. mb7 can potently bind to c-isoforms, but not b-isoforms, of FGFRs.
(A) Representative sensorgrams from BLI measurements for the interactions between mb7 and FGFR c-isoforms. Anti-human Fc capture biosensors immobilized with Fc-mb7 were dipped into solutions containing a series of concentrations of FGFR1cECD, FGFR2cECD, FGFR3cECD, or FGFR4ECD. Sensorgrams (black lines) were fitted with a 1:1 binding model (red lines) to calculate binding kinetic parameters.
(B) Equal amounts of whole-cell lysates (WCLs) of L6R1cKLB or L6R1b with matching levels of FGFR expressions were incubated with Fc-mb7 immobilized on protein A beads and the bound proteins were immunoblotted using an anti-FGFR1 antibody.
(C) BLI sensorgrams showing the interactions between Fc-mb7 and either FGFR1cECD (black, 400 nM) or FGFR1bECD (red, 400 nM).
(D) Cellular activities induced by Fc-mb7 in L6R1cKLB and L6R1b cells. L6R1cKLB and L6R1b cells were treated with indicated concentrations of Fc-mb7 and the cell lysates were analyzed with western blot using anti-MAPK and -pMAPK antibodies. See also Table S1 and Figure S1.
Since mb7 was originally designed to bind to the D3 of FGFR2c, we next asked whether mb7 can also bind specifically to c-isoforms of FGFRs. First, we analyzed L6 cells stably co-expressing human FGFR1c and human KLB (L6R1cKLB) or FGFR1b (L6R1b) that were matched for expression levels of FGFRs (Figure 1B, lanes 1 and 3) and compared the amount of each of the FGFRs captured from an equal amount of cell lysates by Fc-mb7 (Figure 1B, lanes 2 and 4). The results shown in Figure 1B indicate that, while Fc-mb7 was able to robustly bind FGFR1c from L6R1cKLB, it was not able to capture FGFR1b from L6R1b in any appreciable amount. In order to confirm this, we immobilized Fc-mb7 on anti-Fc antibody sensors to monitor BLI responses from the ECD of FGFR1b (FGFR1bECD). The BLI sensorgrams shown in Figure 1C indicated that Fc-mb7 was unable to bind to FGFR1bECD at 400 nM, whereas it produced maximum BLI responses upon binding to 400 nM FGFR1cECD, similar to the results shown in Figure 1A. Furthermore, we compared the stimulatory activity of L6R1cKLB and L6R1b upon cell treatments with Fc-mb7. We anticipated that Fc-mb7 would bring two molecules of FGFR1c in proximity, resulting in FGFR dimerization and activation due to the dimeric nature of the Fc-mb7 fusion protein (Figure S1C) that lead to enhanced mitogen-activated protein kinase (MAPK) and FRS2 phosphorylation. Fc-mb7 indeed strongly induced phosphorylation of MAPK (pMAPK) in L6R1cKLB cells even at 4 nM Figure 1D, left panel) whereas monomeric His-mb7 was unable to induce pMAPK in L6R1cKLB cells (Figure S1D). By contrast, Fc-mb7 was unable to activate cellular signaling in L6R1b cells even at very high concentrations (Figure 1D, right panel), validating that mb7 can specifically bind to the c-isoform of FGFRs on the cell surface. It is noteworthy that FGF1 was able to induce robust cellular signaling in either L6R1cKLB or L6R1b cells Figure 1D), confirming the functional integrities of FGFRs in both of these cells. It is also noteworthy that the amino acid sequence of FGFR4ECD is similar to the c-isoforms of FGFRs 1–3, which explains mb7’s ability to bind to FGFR4 as well (Figures 1A and S1B).
Crystal structures of mb7 in complex with FGFR4D3 reveal mb7’s unique ability to interact with the hydrophobic groove region in c-isoform of FGFRs
We have previously reported a crystal structure of mb7 in complex with the D3 of FGFR4 (FGFR4D3) confirming the applicability of the in silico design method (PDB: 7N1J) (Cao et al., 2022). We now have obtained an additional crystal structure of mb7 in complex with FGFR4D3 in a new crystallization condition (Table S2). Each of these conditions produced crystals with unique space groups (P63 and P21; Figures S2A-S2C) and the asymmetric unit containing two molecules of FGFR4D3:mb7 complex. Overall structures of FGFR4D3:mb7 complex crystallized in these two conditions are similar to each other, with Cα RMSD of 0.924 Å (Figure S2A). However, the different intermolecular interactions within the crystal packing interfaces of these two structures (Figure S2B) led to the different dimeric arrangements within each of the asymmetric units (Figure S2C). This suggests that the 2:2 arrangements of FGFR4D3:mb7 complex in either of these conditions result from the crystallization process.
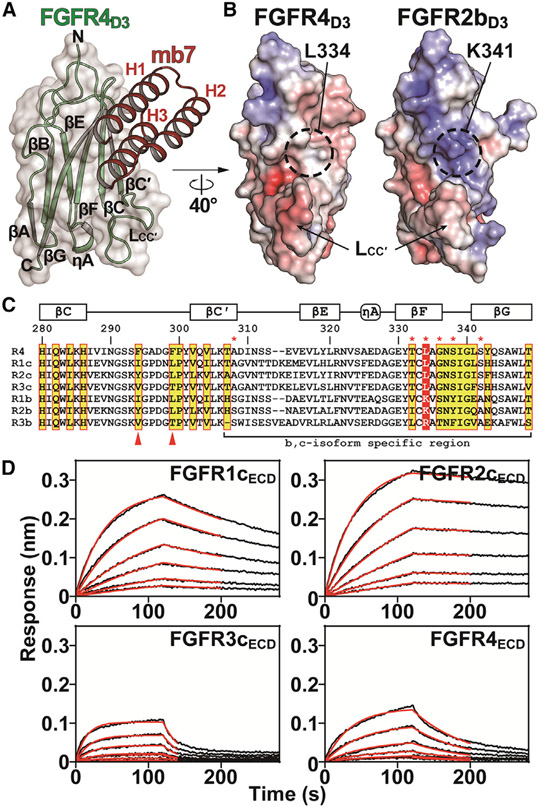
The crystal structure reveals that mb7, having three helix bundle (H1-3), makes extensive interactions with the hydrophobic groove of FGFR4D3 composed of βG-βF-βC-βC′ (βGFCC′) and with the residues in the βC-βC′ loop (LCC′) (Figures 2A and S2D), with an interface area of 828.8 Å2 calculated from the PISA server (Krissinel and Henrick, 2007) and the shape complementarity of 0.67 (Lawrence and Colman, 1993), which are comparable with the values observed for typical antibody-antigen interactions. Most of the residues making contact with mb7 are conserved among all FGFRs according to the amino acid alignment of D3 region in all FGFRs. Strikingly, however, a comparison of electrostatic surface potentials in D3 of all FGFRs reveals significant differences in the surface charge distributions between b- and c-isoforms in βGFCC′ areas (Figures 2B and S3A). Most notably, a positively charged residue strictly conserved among the b-isoforms (e.g., K341 in FGFR2b) positions itself right in the middle of the βGFCC′ surface (Figures 2B and 2C), contributing significantly to the positive surface potential in the βGFCC′ area for the b-isoform of FGFRs. The corresponding residue (e.g., L343 in FGFR2c; Figure 2C) is also strictly conserved as leucine among c-isoforms, resulting in more hydrophobic surface (Figures 2B and 2C), which, in turn, allows for the specific interactions with mb7. In addition, phenyl ring of F347 in FGFR3c corresponding to less bulky hydrophobic residues in other c-isoforms (e.g., L341 in FGFR4, Figure 2C) would make steric clashes with H1 of mb7, and this may provide a reason for the reduced binding affinity between mb7 and FGFR3cECD (Figure 1A; Table S1).
Figure 2. Crystal structure of mb7 in complex with FGFR4D3 reveals how mb7 specifically interacts with c-isoforms of FGFRs.
(A) Crystal structure of FGFR4D3 in complex with mb7. FGFR4D3 and mb7 are shown in green and red cartoons, respectively, with their secondary structural elements labeled. Three α helices of mb7 are labeled as H1, H2, and H3.
(B) Comparison of surface electrostatic potentials for FGFR4D3 and FGFR2bD3. Coordinates of mb7 (from FGFR4D3:mb7 complex) and FGF1 (from FGFR2bD2D3:FGF1 complex; PDB: 3OJM) were omitted for clarity. Locations of L334 in FGFR4 and K341 in FGFR2b are highlighted with dashed circles and LCC′ regions are indicated with arrows.
(C) Amino acid sequence alignment of human FGFRS at the mb7-binding region. The residues corresponding to the FGFR4 residues contacting mb7 are highlighted with yellow boxes with red outlines. Residue numbers in FGFR4, along with the secondary structure elements, are indicated above the alignment. The residues specific to b- or c-isoform of FGFRs are marked with red asterisks above the sequences. Residues corresponding to L334 in FGFR4 are highlighted with a red box. Arrowheads indicate the residues in LCC′ regions that are unique in each subfamily member.
(D) Representative sensorgrams from BLI measurements for the interactions between Fc-mb7TH and FGFR1cECD, FGFR2cECD, FGFR3cECD, or FGFR4ECD. Sensorgrams (black lines) were fitted with a 1:1 binding model (red lines) to calculate binding kinetic parameters. See also Tables S1 and S2, and Figures S2 and S3.
What was not predicted from the computational model of FGFR2cD3:mb7 complex was the extensive interactions between mb7 and LCC′ region in FGFR4D3 (Figures S3B and S3C). This was because mb7 was designed using the crystal structure of FGFR2c with missing coordinates for LCC′ in D3 (PDB:1EV2) (Plotnikov et al., 2000). LCC′ area is disordered in most of the available crystal structures of FGF-bound FGFRs, which suggests that LCC′ region in D3 of FGFR is intrinsically flexible even with a bound FGF. However, clear electron densities for LCC′ were observed in the diffraction data for both of our structures of FGFR4D3:mb7 complex (Figure S3B), suggesting that LCC′ region in D3 became ordered upon mb7 binding. Since there are amino acid sequence differences between LCC′ regions of FGFR subfamily members (Figure 2C), we hypothesized that we would be able to generate a variant of mb7 that recognizes specific subfamily of FGFRs, assuming mb7-LCC′ interactions contribute significantly to the overall binding kinetics. By a thorough inspection of the crystal structure (Figure S3C), we designed a variant of mb7 that contains two amino acid substitutions, K48T/K49H (mb7TH), that would disturb the interactions with LCC′ area in FGFR4D3 but not with LCC′ area in other c-isoforms. When we characterized the binding kinetics between Fc-mb7TH and ECDs of all FGFR c-isoforms, this variant indeed exhibited significant differences in the binding kinetics for FGFR4ECD compared with the wild-type mb7, while the binding kinetics for all other c-isoforms were less affected (Figure 2D; Table S1). Importantly, these results highlight the substantial contribution of mb7-LCC′ interactions to the overall binding between mb7 to FGFR4D3 and suggest a potential strategy for the development of subfamily-biased mb7 variants (Dang et al., 2019).
Cellular signaling induced by FGF1 or FGF2 is not significantly affected by mb7 binding
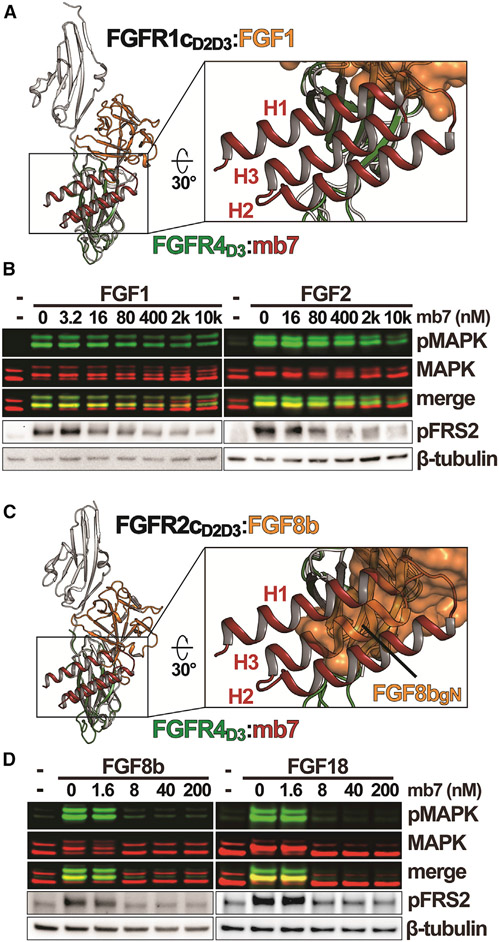
Comparison of our FGFR4D3:mb7 structure with available crystal structures of FGF-bound FGFRs reveals that mb7 binds to a region in FGFR4D3 that partially overlaps with the FGF-binding site (Figure 3A). Since mb7 exhibits high affinities to the D3 of FGFR c-isoforms (Figures 1 and 2; Table S1), we hypothesize that mb7 will sterically prevent FGF1 and FGF2 from binding to FGFR c-isoforms, although the region in mb7 that may potentially compete against FGF1 (or FGF2) for FGFR binding is relatively small.
Figure 3. Inhibitory activities of mb7 on FGFR signaling critically depend on the binding mode unique to each member of paracrine FGFs.
(A) Comparison of FGFR4D3:mb7 and FGFR1c:FGF1 (PDB: 3OJV) structures. The structure of FGFR4D3:mb7 is overlaid to the structure of FGFR1c:FGF1 in reference to the D3 regions. A close-up view shown on the right illustrates the partial overlap between mb7 (red cartoon) and FGF1 (orange surface). FGFR1c and FGFR4 are colored in gray and green, respectively.
(B) Western blot showing inhibitory effects of mb7 on FGF1 and FGF2-induced signaling. L6R1cKLB cells were treated with indicated concentrations of mb7 for 30 min, followed by stimulations with either 0.6 nM FGF1 or FGF2. Levels of MAPK, pMAPK, pFRS2, and β-tubulin were analyzed with western blot.
(C) Structural comparison of FGFR4D3:mb7 and FGFR2c:FGF8b (PDB: 2FDB) complexes. A close-up view on the right shows that H3 of mb7 (red) significantly occludes the binding site of gN of FGF8b (orange surface) on FGFR c-isoform. FGFR2c and FGFR4 are colored in gray and green, respectively.
(D) Inhibitory activities of mb7 on cellular signaling induced by FGF8b and FGF18. L6R1cKLB cells pre-treated with various concentrations of mb7 were stimulated with FGF8b or FGF18. Cell lysates were immunoblotted using anti-MAPK, -pMAPK, -pFRS2, and -β-tubulin. See also Figures S4 and S6.
To test the ability of mb7 to inhibit FGF1 binding to FGFR c-isoforms, we performed a series of pull-down experiments. Solutions containing mb7 and FGF1 with or without heparin decasaccharide (dp10) were incubated with hexa-histidine-tagged FGFR1cD2D3 bound to Ni-NTA resin, and the proteins bound to FGFR1cD2D3 were visualized with Coomassie-stained SDS-PAGE. As anticipated from the structural analyses (Figure 3A), mb7 inhibited FGF1 binding to FGFR1cD2D3 (Figure S4A). However, when 400 μM dp10 was added to the same solution, FGF1 was able to bind to FGFR1cD2D3 in the presence of mb7 (Figure S4A). This indicates that, in the presence of heparin, mb7 is not able to efficiently prevent FGF1 binding to FGFR1c, due to the heparin-mediated enhancement of the FGF1-FGFR1c interaction (Ornitz and Leder, 1992; Spivak-Kroizman et al., 1994). These results also suggest that the inhibition of mb7 of cellular signaling induced by FGF1 may not be efficient, since the activation of cellular signaling by all paracrine FGFs critically depends on the presence of heparin or HSPG on the cell surface.
We next tested the capacity of mb7 to inhibit FGFR signaling induced by FGF1 or FGF2. L6R1cKLB cells were treated with increasing concentrations of mb7 followed by stimulation with FGF1 or FGF2, and the cellular signaling by FGFR was monitored by detecting the phosphorylation levels of downstream molecules in the FGFR signaling pathway (Figure 3B) using western blot analyses. As previously demonstrated, L6R1cKLB cells exhibited a low basal level of phosphorylation and robust cellular responses upon stimulation by FGF1 or FGF2 (Figure S4B) and, therefore, can be used as a simple platform to study cellular signaling by a variety of FGFs, including FGF19 and FGF21, of which cellular responses are dependent on the presence of β-Klotho (Kuzina et al., 2019; Leeet al., 2018). The results shown in Figure 3B indicate that mb7 does not significantly alter the cellular signaling stimulated by either FGF1 or FGF2; partial inhibition of FGFR signaling, as monitored by levels of pFRS2 (on Y436) and pMAPK, was observed in the presence of high concentrations (>1 μM) of mb7. These results are somewhat unexpected, given the high-affinity interactions between mb7 and FGFR1c (Figure 1) and the potent ability of Fc-mb7 in inducing cellular activities in L6R1cKLB cells (Figure 1D), suggesting mb7 can bind to FGFR1c on the cell surface with a high affinity. These are, however, in line with the pull-down assay results shown in Figure S4A, suggesting that the heparin-mediated enhancement of FGF1-FGFR1c interaction can overcome the partial blockade of FGF1-binding site on FGFRD3 by mb7, resulting in the partial inhibition only at the high concentration of mb7. Since we did not add exogenous heparin oligosaccharides in all our cellular experiments, we surmise that the endogenous heparan sulfate or HSPG on the cell surface of L6 cells—which is required for the cellular signaling by paracrine FGFs—may critically affect the inhibitory activities of mb7 against FGF1 and FGF2 signaling.
Cellular signaling induced by FGF8b subfamily members are potently inhibited by mb7
FGF8 family members, FGF8, FGF17, and FGF18, are known to exhibit high FGFR isoform specificities through their own alternative splicing. Previous studies demonstrated how FGF8b can achieve such high specificities toward FGFR c-isoforms; the crystal structure revealed that FGF8b, through its unique N-terminal helix (gN), makes substantial contacts with the hydrophobic groove in D3 of FGFR2c (Olsen et al., 2006). This study also revealed that mutating a single residue within the gN of FGF8b, F32, significantly affects its binding affinity toward FGFR c-isoforms and its biological functions, emphasizing the role of interactions between gN of FGF8b and FGFR c-isoforms.
Comparison of our FGFR4D3:mb7 structure and FGFR2cD2D3: FGF8b structure (PDB: 2FDB) revealed a significant overlap between the mb7-interacting region in FGFR4D3 and FGF8b-interacting region in FGFR2cD3 (Figure 3C), because gN of FGF8b makes extensive interactions with βGFCC′ surface on FGFR2cD3. Notably, the phenyl rings of F32 (within gN) and F93 (in β4–β5 loop of FGF core region) of FGF8b, which are strictly conserved among FGF8b family members, positioned themselves on the βGFCC′ surface inthe exact same place as the sidechain atoms of Y52 and F56 of mb7, respectively (Figure S4C). Therefore, we hypothesized that mb7, via its extensive interaction with βGFCC′ surface on D3 of FGFR c-isoforms, would be able to potently block FGF8b binding to FGFR c-isoforms and inhibit FGFR c-isoform-specific signaling by FGF8b family members.
To test our hypothesis derived from the structural analyses, we investigated the effects of mb7 on the cellular signaling by FGF8b monitored with the levels of pMAPK and pFRS2. Remarkably, as shown in Figure 3D, mb7 was able to potently inhibit FGF8b-stimulated cellular signaling in L6R1cKLB cells, as the levels of both pMAPK and pFRS2 are significantly decreased upon treatments of mb7 at around 8 nM. A similar level of inhibitory potency was also observed for FGF18-induced signaling in L6R1cKLB cells (Figure 3D), which could be attributed to the similarity between the amino acid sequences of FGF8b and FGF18 within the regions that recognize βGFCC′ surfaces on D3 of FGFR c-isoforms. It is noteworthy that both the FGF8b and FGF18 used in these experiments were able to induce robust pMAPK and pFRS2 responses in L6R1cKLB cells (Figure S4B), and that the concentrations of FGF8b and FGF18 used in the inhibition assay were high enough to exert full response in L6R1cKLB cells. It is conceivable that the reason mb7 can achieve such high level of inhibitory potency for FGF8b or FGF18 is because the cellular signaling by FGF8b family members, unlike the signaling by FGF1 or FGF2, depends on their extensive interactions with βGFCC′ surface that mb7 potently binds to. Since, like other paracrine FGFs, FGF8b- and FGF18-induced cellular signaling are significantly dependent on the presence of heparin or heparan sulfate (Chuang et al., 2010; Loo and Salmivirta, 2002), mb7’s ability to block FGF8b-binding site on FGFR1cD3 seems to be potent enough to overcome the heparin-mediated enhancement of the FGF8b-FGFR1c interaction. Moreover, inspection of FGFR2cD2D3:FGF8b structure additionally revealed that, unlike FGF1 and FGF2, FGF8b does not interact with the βC′–βE loop of D3 but instead makes significant interactions with the residues in βC′ strand, rendering D3 of FGFR2c into the structure of canonical IgG (Olsen et al., 2006). Therefore, once mb7 makes extensive interactions with βGFCC′ surface, it would “lock” the conformation of FGFRc D3, thus preventing FGF8b from properly engaging in the hydrophobic groove of FGFRc D3 and inducing FGFR signaling (Figure S4C).
Intrigued by these results, we additionally tested the inhibitory effects of mb7 on FGF5-stimulated cellular activities in L6R1cKLB cells (Figures S4D and S4E). FGF4 family members, including FGF4, FGF5, and FGF6, potently induce cellular activities exclusively related to FGFR c-isoform signaling, although how they achieve such high FGFR isoform specificities remains unknown due to the lack of mechanistic studies on the interactions between FGFRs and FGF4 family members. Interestingly, FGF5-stimulated FGFR signaling in L6R1cKLB cells was potently inhibited by mb7 (Figure S4E), suggesting that FGFR signaling activated by FGF5 may critically depend on FGF5’s interactions with βGFCC′ surface of FGFR1cD3 and that FGF5 family members may recognize FGFR c-isoforms in a similar manner to the way FGF8b family members do.
Mb7 potently blocks Klotho-binding site on FGFR c-isoforms
Members of the endocrine FGF family, FGF19, FGF21, and FGF23, signal through multiple FGFR c-isoforms in the presence of Klotho proteins, which act as high-affinity zip-code-like receptors for each of endocrine FGFs; α-Klotho (KLA) for FGF23 and β-Klotho (KLB) for FGF19 and FGF21 (Chen et al., 2018; Kuzina et al., 2019; Lee et al., 2018). Because the affinities between endocrine FGFs and FGFR c-isoforms are weak, c-isoform specificities of FGFR signaling by endocrine FGFs are attributed to Klotho-FGFR interactions. Notably, the crystal structure of KLAECD in complex with FGF23 and FGFR1cD2D3 revealed how the receptor binding arm (RBA) region in KLA (KLARBA; residues N530–I578) interacts with βGFCC′ surface on FGFR1cD3. The deletion of RBA region from either KLA or KLB abrogated their functions as co-receptors for FGF23- or FGF21-signaling (Chen et al., 2018). We observed that the RBA region in KLB (KLBRBA; residues E531–I582) was disordered in all of our previously determined crystal structures of the extracellular domain of KLB (KLBECD) (Kuzina et al., 2019; Lee et al., 2018). In addition, KLBRBA was susceptible to proteolysis upon long-term storage at 4°C (Figure S5A), although whether this proteolysis occurs in the intact KLB molecules on the cell surface is unknown at the moment. Moreover, when KLBRBA was proteolytically cleaved, it was no longer able to form a stable complex with FGFR1cD2D3, and the presence of FGFR1cD2D3 rendered KLBRBA resistant to such proteolysis (Figure S5A). Based on these observations combined with the results from previous reports, we surmised that KLBRBA interacts with FGFR1cD3 or FGFR4D3 in a similar manner to the way KLARBA does.
To investigate how mb7 might affect the activities of endocrine FGFs, we first compared the crystal structure of KLAECD:FGFR1cD2D3: FGF23 complex (PDB: 5W21) with our FGFR4D3:mb7 structure, as illustrated in Figure 4A. Strikingly, substantial surface area on FGFR1cD3 recognized by KLARBA is also interacting with H3 of mb7, assuming mb7 binds to FGFR1c in the same manner as it binds to FGFR4. This implies that, since KLArBa makes extensive contacts with βGFCC′ surface and LCC′ on FGFR1cD3, most of the area on FGFR1c that KLARBA binds to would be occluded by mb7. Particularly, the hydroxyphenyl ring of Y52 of mb7 occupies exactly the same region in FGFR4D3 as the indole ring of W549 in KLARBA (Figure 4A), making contacts with the hydrophobic groove on FGFR1cD3 composed of residues highly conserved among the c-isoforms, in the same manner as the aforementioned phenyl ring of F32 of FGF8b lies on the surface of FGFR2cD3 (Figure S4C). Moreover, sequence alignment reveals that W549 in KLARBA corresponds to W553 in KLBRBA, implying these residues in Klotho receptors might be critical for their specific binding to FGFR c-isoforms. Based on these structural analyses, we hypothesize that the binding of mb7 on FGFR1cD3 or FGFR4D3 would impose a significant amount of steric hindrance for KLBRBA (or KLARBA), preventing KLBECD (or KLAECD) binding to FGFR1cECD and FGFR4ECD.
Figure 4. Cellular signaling by endocrine FGFs can be achieved by blocking Klotho-FGFR interactions.
(A) Structural comparison between FGFR4D3:mb7 complex and FGF23:FGFR1cD2D3:KLA complex (PDB: 5W21). The structures are overlaid in reference to the D3 of FGFRs. KLARBA (blue) interacts with βGFCC′ surface of FGFR1cD3 (gray) and mb7 (red) interacts with βGFCC′ surface of FGFR4D3 (green). Close-up views highlight the steric clashes between mb7 and KLARBA (upper right), as well as the side chains of Y52 in mb7 and W549 in KLARBA occupying the same hydrophobic groove in FGFR D3 (lower right).
(B) MST-based competition assay with mb7 against FC-KLBECD for FGFRIcECD (green) or FGFR4ECD (red) binding. A series of concentrations of mb7 were added to the fluorescently labeled FGFR1cECD or FGFR4ECD, which were mixed with 1 μM FC-KLBECD. Normalized fluorescence values (Fnorm) plotted against mb7 concentration, shown as individual data points, were fitted with the Hill equation to obtain IC50 values of 37.1 ± 2.39 nM and 84.2 ± 1.28 nM against Fc-KLBECD:FGFR1cECD and Fc-KLBECD:FGFR4ECD complexes, respectively (IC50 values are indicated as average ± variation at 68% confidence). Shaded areas indicate the regions that were used to calculate Fnorm (Fnorm = Fhot/Fcold, blue for Fcold and red for Fhot). The measurements were done in triplicates.
(C and D) Inhibitory activities of mb7 on FGF19-induced cellular activities in L6R1cKLB, L6R4KLB (C), and HEP3B (D) cells as monitored by the levels of phosphorylation of MAPK and FRS2. Cells pre-treated with various concentrations of mb7 were stimulated with FGF19, and the lysates were immunoblotted using anti-MAPK, -pMAPK, -pFRS2, and -β-tubulin. See also Figures S5 and S6.
In order to test this hypothesis, we performed a series of competition assays using MicroScale Thermophoresis (MST), similar to the assay used in our previous study (Lee et al., 2018). Fluorescently labeled FGFR1cECD or FGFR4ECD was mixed with a series of concentrations of mb7 together with a fixed amount (1 μM) of Fc-KLBECD, KLBECD fused to Fc region of human IgG1 that was previously used in our structural studies (Kuzina et al., 2019; Lee et al., 2018). The thermophoretic movements of molecules during the MST experiments were then monitored using the fluorescence signals on either FGFR1ECD or FGFR4ECD. As shown in Figure 4B, we observed significantly large thermophoretic changes coming from the displacement of Fc-KLBECD from FGFR1cECD (green) or FGFR4ECD (red) upon addition of an increasing amount of mb7, allowing us to confidently calculate half maximal inhibitory concentration (IC50) values, 37.1 nM and 84.2 nM for Fc-KLB/FGFR1cECD and Fc-KLB/FGFR4ECD, respectively. In addition, we performed a series of BLI-based competition assay where FGFR1cECD was immobilized on the biosensors and increasing concentrations of mb7 were added, then a fixed concentration of Fc-KLBECD was applied. The resulting BLI sensorgrams shown in Figure S5B indicate that the BLI responses originating from Fc-KLBECD bound to FGFR1cECD decrease as the fraction of mb7-bound FGFR1cECD increases, confirming the results from MST competition assay (Figure 4B) that mb7 binding to FGFR1cECD effectively blocks KLB-FGFR interactions. These results clearly demonstrate that mb7 potently blocks Klotho-binding sites on FGFR c-isoforms and confirm that KLBRBA indeed binds to FGFR1cD3 or FGFR4D3 in a manner similar to the way KLARBA does. We propose, therefore, that mb7, with its high-affinity interactions with FGFR1cD3 and FGFR4D3, will also be able to block KLA-FGFR interactions, which would negatively affect FGF23 signaling.
Cellular signaling by endocrine FGFs can be modulated by mb7
We next investigated the effects of mb7 binding on cellular signaling stimulated by endocrine FGFs. Although the exact mechanism of how endocrine FGFs stimulate dimerization of Klotho:FGFR complex is currently unknown, it was proposed that the heparin—which exhibits negligible binding affinity to endocrine FGFs—might still play a role in the activation of FGFR signaling by endocrine FGFs (Chen et al., 2018; Wu et al., 2007). Structural analyses shown in Figure 4A suggest that mb7 binding to D3 of FGFR c-isoforms would not only block KLA or KLB binding but also prevent endocrine FGFs from being properly engaged in the FGFR-binding site due to steric hindrance. However, since endocrine FGFs do not induce FGFR signaling without Klotho proteins, we surmised that blocking Klotho-binding sites on FGFR1cD3 or FGFR4D3 by mb7 would be sufficient to abolish endocrine FGF-stimulated cellular activities, similarly to the effects of RBA deletion from Klotho receptors. In order to test the effects of mb7 on the cellular signaling by endocrine FGFs, we used FGF19 as an example to monitor its cellular activities in L6 R1cKLB cells as well as L6 cells stably co-expressing FGFR4 and KLB (L6R4KLB). As we previously reported, FGF19 induced robust cellular responses in either L6R1cKLB or L6R4KLB, as monitored by levels of pMAPK and pFRS2 (Figure S5C). We then monitored how FGF19-induced signaling in L6R1cKLB and L6R4KLB were affected upon treatments with mb7. Remarkably, as shown in Figure 4C, mb7 was able to potently inhibit FGF19-stimulated signaling in either L6R1cKLB or L6R4KLB, starting at around 8 nM. This experiment demonstrates that the cellular signaling by FGF19, which is primarily mediated via interactions with KLB (Kuzina et al., 2019), can be potently inhibited by blocking the molecular interactions between KLB and FGFR c-isoforms.
We next asked how mb7 would affect the FGF19-FGFR4 signaling pathway in hepatocellular carcinoma (HCC) cell lines. Aberrant FGF19-FGFR4 signaling through FGF19 overexpression that drives an autocrine loop has previously been implicated in various cancer models (Desnoyers et al., 2008; French et al., 2012) and is now well-established as an oncogenic driver in a subset of HCC patients (Kim et al., 2019). Interestingly, only FGF19/FGFR4/KLB-positive HCC cell lines (e.g., HEP3B and Huh-7) were sensitive to FGFR4-specific tyrosine kinase inhibitors (TKIs) developed independently by multiple groups (Hagel et al., 2015; Joshi et al., 2017; Weiss et al., 2019), implying that FGF19-induced signaling in HCC cells and the proliferation of HCC cells driven by aberrant FGF19 signaling could be inhibited by manipulating the interactions between FGF19, KLB, and FGFR4. We hypothesized that mb7, by blocking the FGFR4-KLB interaction, would inhibit cellular signaling induced by FGF19 in HCC cells expressing FGFR4 and KLB. In order to test our hypothesis, we pre-treated HEP3B cells with a series of concentrations of mb7 followed by stimulation with FGF19 and monitored the levels of pMAPK and pFRS2. Consistent with the previous reports (Desnoyers et al., 2008; Hagel et al., 2015), HEP3B cells that have high levels of FGF19, FGFR4, and KLB showed phosphorylation of FRS2 in unstimulated cells, and addition of FGF19 further enhanced the pFRS2 level, which is also reflected in the pMAPK level (Figure 4D). Similar to the inhibitory effects on FGF19-induced signaling in L6R1cKLB and L6R4KLB (Figure 4C), mb7 potently blocked the cellular response induced by exogenous FGF19 in HEP3B cells, as levels of pFRS2 and pMAPK were decreased to the baseline upon treatment with 40 nM of mb7. Moreover, treatments with high concentrations (~1 mM) of mb7 further decreased pFRS2 levels, suggesting the presence of active FGFR signaling in HEP3B cells other than FGF19-FGFR4-KLB signaling that can be blocked by mb7 at high concentrations. Taken together, these data support our hypothesis that mb7 can potently suppress FGF19-induced FGFR4 signaling in HEP3B cells, which is critically dependent on the interaction between KLB and FGFR4.
DISCUSSION
Targeting specific regions in cell-surface receptors with antibodies is a challenging task when the target area is too small to be properly engaged by bulky proteins. In addition, in silico design of antibodies that target specific surfaces on proteins is difficult because of the intrinsic complexity of the antibody-antigen recognition process. In this report, we showed that mb7, a de-novo-designed mini-protein, recognizes specifically c-isoforms of FGFRs and potently binds to the small hydrophobic groove area of D3, βGFCC′. Moreover, these experiments provide insights on how FGF8b and FGF19 family members achieve high specificities for FGFR c-isoforms and how to modulate their cellular activities.
The primary mechanisms for the inhibitory activities by mb7 are illustrated in Figure 5. The interactions between FGFR c-isoforms and FGF1 family members can be partially masked by mb7; however, mb7 is unable to efficiently inhibit cellular signaling by FGF1 family members due to the heparin-mediated augmentation of FGF1-FGFR1c interactions (Figure 5A). On the other hand, FGF8b family members use their unique gN helix regions to extensively engage in the βGFCC′ surface of FGFR c-isoforms (Olsen et al., 2006), and FGF19 family members, through their C-terminal tail regions, make high-affinity interactions with Klotho receptors, which use their RBA regions to recognize the βGFCC′ surface of FGFR c-isoforms (Chen et al., 2018; Kuzina et al., 2019; Lee et al., 2018). The cellular activities by either FGF8b or FGF19 family members proved to be dependent on the molecular interactions involving the βGFCC′ surface of FGFR D3, since masking that surface of FGFR c-isoforms by mb7 completely abolished the cellular signaling stimulated by either FGF8b, FGF18, or FGF19. Therefore, through its high-affinity interactions with D3 of FGFR c-isoforms, mb7 can potently inhibit c-isoform-specific FGFR signaling either by directly competing against the ligands (Figure 5B, for FGF8b family members) or by competing against the Klotho receptors that are necessary for ligand-binding and activation (Figure 5C, for FGF19 family members).
Figure 5. Schematic diagrams describing two distinct mechanisms of FGFR signaling inhibition by mb7.
(A) Partial blockade of FGF1-binding site on FGFRD3 by mb7 results in an ineffective inhibition due to the heparin- or HSPG-mediated enhancements of interactions between FGF1 family members and FGFR1c.
(B) FGF8b family members preferentially bind to FGFR c-isoforms via their unique gN helix region. High-affinity interactions between mb7 and FGFR c-isoforms at the D3 regions where gN helix of FGF8b family members binds to prevent FGF8b family members from activating FGFR signaling.
(C) Klotho proteins use their RBA region to exclusively interact with FGFR c-isoforms. Complete blockade of Klotho-binding site on D3 region of FGFR c-isoforms by mb7 effectively prevents FGF19 family members from activating Klotho-dependent FGFR signaling.
FGF19 is an ileum-derived enterokine that regulates hepatic bile acid synthesis (Inagaki et al., 2005; Kir et al., 2011). Aberrant expression of FGF19 in the liver, however, can lead to HCC where its constitutive signaling promotes uncontrolled proliferation in an autocrine-paracrine manner through FGFR4 (Desnoyers et al., 2008; French et al., 2012; Lin and Desnoyers, 2012; Nicholes et al., 2002; Sawey et al., 2011). Lenvatinib, a TKI targeting multiple kinases including FGFRs that was recently approved by the US Food and Drug Administration (FDA) for patients with unresectable HCC, showed selective and potent antiproliferative activities against FGF19/FGFR4/KLB-expressing HCC cells (Matsuki et al., 2018). However, on-target resistance to TKIs, mainly via gatekeeper mutations, is inevitable. A recent report from a phase 1 study of the FGFR4-specific TKI, fisogatinib, in HCC patients showed that extended treatment with fisogatinib resulted in the development of resistance via mutations in the FGFR4 kinase domain (Hatlen et al., 2019). Therefore, targeting Klotho-FGFR interactions using an agent such as mb7 may provide a viable—yet largely unexplored—orthogonal therapeutic strategy that can be used to overcome resistance in FGF19-driven HCC.
Last, as demonstrated in Figure 2, the structural information presented in this manuscript could be utilized to engineer variants of mb7 that have the specificities for the subfamily of FGFR c-isoforms, similar to the approach previously used (Dang et al., 2019). This will be a key process when the interactions regarding a subfamily of FGFR, e.g., KLB-FGFR4, needs to be blocked while minimally disrupting the interactions regarding other FGFR c-isoforms (e.g., KLB-FGFR1c).
In summary, we present the properties of a de-novo-designed mini-protein specifically targeting FGFR c-isoforms. The degree to which mb7 inhibits the FGF-induced cellular activities is critically determined by the mechanism of c-isoform-specific FGFR signaling unique to each family of FGFs. Therefore, targeting the βGFCC′ surface of D3 of FGFR c-isoforms presents an opportunity to modulate cellular activities by a subset of FGFs.
Limitations of the study
Our structural and biochemical studies demonstrated that mb7, a de-novo-designed molecule, can specifically interact with c-isoforms of FGFR with high binding affinity and potently modulate cellular signaling specific to FGFR c-isoforms. However, since mb7 was originally designed in silico, we cannot exclude the possibility that mb7 may interact with the extracellular region of cell-surface receptors other than FGFRs. While mb7 was previously shown to exhibit little cross-reactivity against a number of proteins having various shapes and surface charges (Cao et al., 2022), it has not yet been rigorously tested against cell-surface proteins containing Ig-like domains with hydrophobic surfaces. Future experiments involving more comprehensive analyses on the cross-reactivity in the cellular context will provide information on its applicability. Moreover, our current results from the biophysical measurements using the purified soluble extracellular domains may not necessarily reflect the values against the intact receptors embedded on the cell membrane. Extracellular matrix components such as HSPG or other cell-surface receptors that were reported to interact with FGFRs (Ornitz and Itoh, 2022), for example, might influence how mb7 interacts with FGFR c-isoforms and affect mb7’s activities on different cells.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Sangwon Lee (s.lee@yale.edu).
Materials availability
All plasmids and cell lines generated in this study are available from the authors upon request.
Data and code availability
This paper analyzes existing, publicly available structures in the Protein DataBank. Their accession codes are listed in the key resources table.
Coordinates and structure factors for the FGFR4D3:mb7 complex in P21 space group are deposited in the Protein Data Bank with the accession code 7TYD.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-FGFR1 | in-house | N/A |
| Mouse monoclonal anti-MAPK | Cell Signaling Technology | Cat# 9107; RRID: AB_10695739 |
| Rabbit monoclonal anti-β-tubulin | Cell Signaling Technology | Cat# 2128; RRID: AB_823664 |
| Rabbit polyclonal anti-phospho-MAPK (pThr202, pTyr204 of p44 MAPK) | Cell Signaling Technology | Cat# 9101; RRID: AB_331646 |
| Rabbit polyclonal anti-phospho-FRS2 (Y436) | R&D systems | Cat# AF5126; RRID: AB_2106234 |
| Alexa Fluor 680 Donkey anti-Mouse IgG antibody | Invitrogen | Cat# A10038; RRID: AB_2534014 |
| IRDye800 CW Donkey anti-Rabbit IgG antibody | LI-COR | Cat# 926-32213; RRID: AB_621848 |
| Bacterial and virus strains | ||
| BL21-Gold(DE3) | Agilent Technologies | Cat# 230132 |
| Chemicals, peptides, and recombinant proteins | ||
| FGF2 | Gibco | Cat# PHG0367 |
| FGF5 | R&D systems | Cat# 237-F5-050 |
| FGF8b | R&D systems | Cat# 423-F8-025 |
| FGF18 | Peprotech | Cat# 100-28 |
| Heparin octa-saccharide (Dp10) | Iduron | Cat# HO10 |
| Heparin sepharose resin (Heparin Sepharose 6 Fast Flow) | GE healthcare | Cat# 17-0998-01 |
| Ni-NTA agarose bead | Qiagen | Cat# 30210 |
| Protein A-HRP | Invitrogen | Cat# 101023 |
| Protein A resin (Rec-Protein A-Sepharose 4B) | Invitrogen | Cat# 101142 |
| Western ECL substrate (Clarity Western ECL Substrate) | Bio-Rad | Cat# 170-5060 |
| Critical commercial assays | ||
| Monolith NT.115 instrument | NanoTemper Technologies | N/A |
| Monolith NT.115 MST Premium Coated Capillaries | NanoTemper Technologies | Cat# MO-K025 |
| Monolith Protein Labeling Kit RED-Tris-NTA dye | NanoTemper Technologies | Cat# MO-L008 |
| Octet RED96 system | FortéBio | N/A |
| Anti-penta-His biosensor (HIS1K Biosensor) | FortéBio | Cat# 18-5120 |
| Anti-human IgG Fc biosensor (AHC Biosensor) | FortéBio | Cat# 18-5060 |
| Deposited data | ||
| Crystal structure of FGFR4D3:mb7 in P21 space group | This paper | PDB: 7TYD |
| Crystal structure of FGFR4D3:mb7 in P63 space group | (Cao et al., 2022) | PDB: 7N1J |
| Crystal structure of FGF1:FGFR1c | (Beenken et al., 2012) | PDB: 3OJV |
| Crystal structure of FGF1:FGFR2b | (Beenken et al., 2012) | PDB: 3OJM |
| Crystal structure of FGF1:FGFR3c | (Olsen et al., 2004) | PDB: 1RY7 |
| Crystal structure of FGF2:FGFR2c | (Ibrahimi et al., 2001) | PDB: 1IIL |
| Crystal structure of FGF23:FGFR1:KLA | (Chen et al., 2018) | PDB: 5W21 |
| Crystal structure of FGFR3b:Fab | (Qing et al., 2009) | PDB: 3GRW |
| Crystal structure of FGF8b:FGFR2c | (Olsen et al., 2006) | PDB: 2FDB |
| Experimental models: Cell lines | ||
| Rat: L6 cells | ATCC | Cat# CRL-1458; RRID:CVCL_0385 |
| Rat: L6R1b cells | This paper | N/A |
| Rat: L6R1cKLB cells | (Lee et al., 2018) | N/A |
| Rat: L6R4KLB cells | (Kuzina et al., 2019) | N/A |
| Human: Hep3B cells (male) | ATCC | Cat# HB-8064 |
| Recombinant DNA | ||
| pCEP4 | Invitrogen | Cat# V04450 |
| pCEP4-FGF19 | This paper | N/A |
| pCEP4-FGFR1bECD (M1–E376) | This paper | N/A |
| pCEP4-FGFR1cECD (M1–E376) | This paper | N/A |
| pCEP4-FGFR4ECD (M1–D369) | This paper | N/A |
| pCEP4-Fc-mb7 | This paper | N/A |
| pCEP4-Fc-mb7TH (K47T, K48H) | This paper | N/A |
| pCEP4-Fc-KLBECD (M30–T983) | (Lee et al., 2018) | N/A |
| pET-28a(+) | Novagen | Cat# 69864 |
| pET-28a(+)-FGF1 (F16–D155) | This paper | N/A |
| pET-28a(+)-FGFR1cD2D3 (T141–R365) | This paper | N/A |
| pET-28a(+)-FGFR4D2D3 (S141–T357) | This paper | N/A |
| pET-28a(+)-FGFR4D3-HIs (S245–D355) | This paper | N/A |
| pET-29b(+) | Novagen | Cat# 69872 |
| pET-29b(+)-His-mb7 | (Cao et al., 2022) | N/A |
| Software and algorithms | ||
| FortéBio Data Analysis HT software 10.0 | FortéBio | N/A |
| Graphpad Prism 8.0 | Graphpad | https://www.graphpad.com |
| Image Studio Lite | LI-COR Biosciences | https://www.licor.com/bio/image-studio-lite/ |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| MO.Affinity Analysis software | NanoTemper Technologies | https://nanotempertech.com/ |
| Phenix | (Adams et al., 2010) | https://phenix-online.org/ |
| MolProbity | (Chen et al., 2010) | http://molprobity.biochem.duke.edu/ |
| PISA server | (Krissinel and Henrick, 2007) | https://www.ebi.ac.uk/pdbe/pisa/ |
| PyMol | Schrödinger | http://www.pymol.org |
| XDS | (Kabsch, 2010) | https://xds.mr.mpg.de/ |
| Other | ||
| Expi293 expression system kit | Gibco | Cat# A14635 |
| QuikChange II site-directed mutagenesis kit | Agilent | Cat# 200523 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
BL21-Gold(DE3) cells (Agilent Technologies) transformed with recombinant pET-28a(+) or pET-29b(+) plasmids were grown in LB media supplemented with 100 μg/mL kanamycin at 37°C. Expi293F cells (Thermo Fisher Scientific) were maintained in a humidified incubator with 8% CO2 at 37°C with Expi293 expression media (Thermo Fisher Scientific). L6 cells stably co-expressing FGFR1c with KLB (L6R1cKLB) or FGFR4 with KLB (L6R4KLB) were maintained in a humidified incubator with 5% CO2 at 37°C with Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS, Thermo Fisher Scientific), 100 U/mL Penicillin-Streptomycin (Thermo Fisher Scientific), 0.1 mg/mL hygromycin B (Thermo Fisher Scientific), and 1 μg/mL puromycin (Thermo Fisher Scientific). L6 cells stably expressing FGFR1b (L6R1b) were maintained in DMEM supplemented with 10% FBS, 100 U/mL Penicillin-Streptomycin, and 1 μg/mL puromycin. HEP3B cells (ATCC) were maintained in a humidified incubator with 5% CO2 at 37°C with Eagle’s Minimum Essential Medium (EMEM, ATCC) supplemented with 10% FBS and 100 U/mL Penicillin-Streptomycin.
METHOD DETAILS
Cloning, expression, and purifications
The gene of mb7 with the N-terminal His-tag and a tobacco etch virus (TEV) protease cleavage site was cloned into pET-29b(+) plasmids (Novagen), which was transformed into BL21-Gold (DE3) cells (Agilent Technologies). The cells were grown in LB at 37°C until OD600 reached 0.5, induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG), and grown at 20°C overnight. The cells were collected by centrifugation, then lysed by sonication. His-mb7 was purified to homogeneity using nickel affinity chromatography (Ni-NTA agarose, Qiagen) followed by a size exclusion chromatography (HiLoad 26/200 Superdex 200 pg, Cytiva). For the preparation of mb7 without N-terminal His-tag, purified His-mb7 was incubated with TEV protease at 4°C overnight and subjected to nickel affinity chromatography, followed by a size exclusion chromatography.
The genes of FGF1 (F16-D155), FGFR1cD2D3 (T141–R365), FGFR4D2D3 (S141–T357), and FGFR4D3 (S245–D355, containing C-terminal His-tag) were cloned into pET-28a(+) plasmid (Novagen), which were transformed into BL21-Gold (DE3) cells. The cells were grown in LB at 37°C until OD600 reached 0.5, induced with 1 mM IPTG, and grown at 20°C overnight. The cells were collected by centrifugation and lysed by sonication. FGF1 was purified to homogeneity with affinity chromatography using Heparin Sepharose 6 Fast Flow resins (Cytiva) followed by a size exclusion chromatography. And FGFR1cD2D3, FGFR4D2D3, and FGFR4D3 were refolded from inclusion bodies using the protocol previously published with minor changes (Plotnikov et al., 1999, 2000; Schlessinger et al., 2000) and purified to homogeneity with affinity chromatography using Heparin Sepharose 6 Fast Flow resin (for FGFR1D2D3 and FGFR4D2D3) or Ni-NTA agarose beads (for FGFR4D3), followed by a size exclusion chromatography.
The genes of mb7 with N-terminal mouse heavy chain signal sequence, Fc region of human IgG1, and (GGGGS)× 3 linker (Fc-mb7), FGF19 (L25–K216) with N-terminal mouse heavy chain signal sequence and octa-histidine-tag (His-FGF19), and extracellular domains of FGFR1c (FGFR1cECD, M1–P366), FGFR2c (FGFR2cECD, M1–P373), FGFR3c (FGFR3cECD, M1–G370), FGFR4 (FGFR4ECD, M1–T357), and FGFR1b (FGFR1bECD, M1–P366), each containing the C-terminal octa-histidine-tag were cloned into pCEP4 plasmids (Thermo Fisher Scientific). For Fc-mb7TH, K47T/K48H mutations in mb7 were introduced by QuikChange using Fc-mb7 as a template. The plasmids were transfected into Expi293F cells (Thermo Fisher Scientific) according to the manufacturer’s instructions. The Enhancers 1 and 2 (Thermo Fisher Scientific) were added to the media 18 h after transfection then incubation temperature was changed to 30°C. The media containing secreted proteins were harvested 5 days after transfection. After the centrifugation at 3,000 ×g for 10 min at 4°C, the media containing Fc-mb7 or Fc-mb7TH was incubated with recombinant Protein A Sepharose 4B (Thermo Fisher Scientific), and the protein was eluted from the resin with the buffer containing 100 mM glycine, pH 3.3. The pH of the eluate was immediately neutralized with 100 mM Tris-HCl, pH 8.0, and the buffer was exchanged with phosphate buffered saline (PBS). His-FGF19 and all FGFR extracellular domains were purified with affinity chromatography using Nickel Sepharose Excel (Cytiva) beads followed by a size exclusion chromatography.
Extracellular domain of β-Klotho fused to Fc region of human IgG1 (KLBECD-Fc) was expressed and purified using the protocol described previously (Kuzina et al., 2019; Lee et al., 2018). In brief, HEK293-EBNA cells stably expressing KLBECD-Fc were cultured in DMEM supplemented with 5% FBS for 7 days at 30°C. The KLBECD-Fc in the media was bound to Protein A Sepharose 4B (overnight at 4°C) and eluted with the buffer containing 100 mM glycine (pH 3.3). The eluted fractions were immediately supplemented with 100 mM Tris (pH 7.4), dialyzed against PBS with 5% glycerol, and stored at −80°C until further study.
X-ray crystallography
Purified FGFR4D3 was mixed with a 1.2-fold molar excess of mb7 and subjected to size exclusion chromatography to isolate the FGFR4D3:mb7 complex. Fractions containing the complex were pooled, concentrated to 12 mg/mL, and screened for crystallization using commercially available screening kits using Mosquito Crystal liquid handler (SPT Labtech). Crystals of the FGFR4D3:mb7 complex crystallized in P21 and P63 space groups were obtained with the JCSG + screening solution (Molecular Dimensions) containing 0.1 M Bis-Tris (pH 5.5) and 25% polyethylene glycol (PEG) 3,350 supplemented with 3% 1,5-diaminopentane dihydrochloride (from Additive screen, Hampton Research) and the ProPlex screening solution (Molecular Dimensions) containing 0.2 M sodium chloride, 0.1M MES (pH 6.0), 20% PEG 2,000 monomethyl ether, respectively.
The crystals from both conditions were cryoprotected using the mother liquor supplement with 25% glycerol before being flash-frozen in liquid nitrogen. X-ray diffraction data were collected at the NE-CAT 24ID-E beamline of Advanced Photon Source (Argonne National Laboratory) and processed with XDS (Kabsch, 2010) and HKL2000 (Otwinowski and Minor, 1997). The structures were obtained by molecular replacement with Phaser (McCoy et al., 2007) using the coordinates corresponding to the domain 3 region of FGFR1c (PDB: 1CVS) (Plotnikov et al., 1999) and the coordinates of mb7 as the search model, followed by iterative refinement using PHENIX (Adams et al., 2010; Afonine et al., 2012) and COOT (Emsley et al., 2010). The final structure was validated with MolProbity (Chen et al., 2010). Data collection and refinement statistics are provided in Table S2.
Cell cultures and Western blot analyses
For activation assays, L6R1cKLB cells were serum-starved in DMEM overnight and stimulated with FGF1, FGF2 (Gibco), FGF5 (R&D system), FGF8b (R&D system), FGF18 (Peprotech), FGF19, or Fc-mb7 at 37°C for 5 min. For inhibition assays, L6R1cKLB or L6R4KLB cells were serum-starved in DMEM overnight and treated with a series of concentrations of mb7 at 37°C for 30 min, followed by stimulation with various FGF ligands at the following concentrations: 0.6 nM for FGF1 and FGF2; 3 nM for FGF8b and FGF18; 5 nM for FGF5; and 10 nM for FGF19. Hep3B cells were treated with a series of concentrations of mb7 for 2 h and stimulated with 10 nM of FGF19 for 5 min at 37°C.
The cells were collected, lysed with a lysis buffer containing 50 mM HEPES, 150 mM NaCl, 1 mM EDTA, 1mM EGTA, 25 mM NaF, 10% Glycerol, 1% Triton X-100, 1 mM MgCl2,1 mM Na3VO4, pH 7.5, and cOmplete protease inhibitor cocktail (Roche), followed by a centrifugation to remove un-lysed cells. The supernatant containing the cell lysates was subjected to Western blot analyses with anti-MAPK (Cell Signaling Technology), anti-pMAPK (Cell Signaling Technology), anti-pFRS2 (pTyr436, R&D systems), and anti-β-tubulin (Cell Signaling Technology). Anti-Mouse IgG (Alexa Fluor 680 Donkey anti-Mouse IgG antibody, Invitrogen) and anti-Rabbit IgG (IRDye800 CW Donkey anti-Rabbit IgG antibody, Li-COR) were used as secondary antibodies for anti-MAPK and anti-pMAPK, respectively, which were imaged with Odyssey DLx imaging system (LI-COR). Protein A-HRP (Invitrogen) was used to detect anti-pFRS2 and anti-β-tubulin with chemiluminescence substrate (Bio-rad) which were imaged with iBright FL1000 imaging system (Invitrogen).
Pull-down assays
Fc-mb7 bound to Protein A Sepharose 4B was incubated with the cell lysates of L6R1cKLB or L6R1b at 4°C overnight. The protein A Sepharose 4B was then centrifuged down and washed extensively with lysis buffer. The bound proteins were subjected to Western blot analysis with anti-FGFR1.
For the competition experiment between mb7 and FGF1 to FGFR1cD2D3, Ni-NTA agarose beads were incubated with His-FGFR1cD2D3 and mb7 in the molar ratio of 1:2. The beads were washed with PBS then incubated with excess amounts of FGF1 with or without 400 μM heparin decasaccharide (dp10, iduron). After the beads were washed, the bound proteins were eluted with PBS supplemented with 300 mM imidazole. The inputs and the bound/unbound fractions were analyzed with SDS-PAGE followed by Coomassie staining.
Bio-layer interferometry (BLI) and Microscale thermophoresis (MST) measurements
All the BLI sensorgrams were obtained with the Octet RED96 system (Sartorius). To measure the binding affinities between mb7 and FGFRs, Fc-mb7 (or Fc-mb7TH) was immobilized onto anti-human IgG Fc (AHC) biosensors (Sartorius) which were subsequently dipped into solutions containing a series of concentrations, ranging from 1.56 nM to 100 nM of FGFR1cECD, FGFR2cECD, FGFR3cECD, or FGFR4ECD in 250 mM NaCl, 20 mM HEPES, 1% BSA, and 0.05% Tween 20. In a separate series of experiments, His-mb7 was immobilized onto anti-penta-His (HIS1K) biosensors (Sartorius) which were subsequently dipped into solutions containing a series of concentrations, ranging from 3.91 nM to 250 nM of either FGFR1cD2D3 or FGFR4D2D3 in 300 mM NaCl, 50 mM HEPES, 1% BSA, and 0.05% Tween 20. The kinetic rate constants (kon and koff) and the dissociation constant, KD, were determined by fitting the reference-subtracted sensorgrams globally with the 1:1 Langmuir binding model using FortéBio Data Analysis 10.0 software provided by the manufacturer. For the competition assay between mb7 and KLBECD for FGFR1cECD binding, FGFR1cECD was immobilized onto HIS1K biosensors which were dipped into the solution containing a series of concentrations of mb7. Then the sensors were dipped into the solution containing 200 nM of KLBECD.
All MST measurements were performed using the Monolith NT.115 instrument with RedPico/BlueNano double channel (NanoTemper Technologies) with Monolith NT.115 MST Premium Coated Capillaries. Purified FGFR1cECD and FGFR4ECD were fluorescently labeled using the Monolith Protein Labeling Kit RED-Tris-NTA dye (NanoTemper Technologies) according to the instructions provided by the manufacturer. For the competition assays, the thermophoresis of fluorescently labeled (fL)-FGFR1cECD or fl-FGFR4ECD was measured for samples where the concentrations of fl-FGFR ECDs and Fc-KLBECD were kept constant as 10 nM and 1 μM, respectively, with the concentrations of mb7 varying from 0.15 nM to 5,000 nM. The thermophoretic movements of fl-FGFR ECDs in each sample were monitored, with LED power at 5% and MST power at 40%, and the normalized fluorescence intensities (Fnorm), defined as Fhot/Fcold (where Fcold and Fhot refer to the fluorescence intensities averaged over 1 s period before IR laser is on and 4 s after IR laser is on, respectively), for each sample were plotted against the concentrations of mb7. EC50 values were calculated with the Hill model using the MO.Affinity Analysis software (NanoTemper Technologies, v2.3).
QUANTIFICATION AND STATISTICAL ANALYSIS
All immunoblots and measurements presented in this work were repeated at least 3 times with similar results. Western blots were quantified using Image Studio™ Lite software (LI-COR Biosciences) and normalized to controls as indicated in figure legends. All BLI measurements were performed with at least 3 independent series of samples and resulting standard deviations were reported. MST experiments were performed with 3 independent series of samples.
Supplementary Material
Highlights.
A de-novo-designed mini-protein, mb7, selectively binds to the FGFR c-isoform
Mb7 can potently inhibit FGFR signaling induced by a subset of FGFs
Bivalent form of mb7 can activate FGFR c-isoform
ACKNOWLEDGMENTS
This research was funded in part by the National Cancer Institute from the National Institutes of Health (R01 CA258867). This work is based on research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the NIGMS/NIH (P30 GM124165). The Eiger 16M detector on the 24-ID-E beamline is funded by an NIH-ORIP HEI grant (S10OD021527). This research used resources of the Advanced Photon Source (DE-AC02-06CH11357). We thank the Yale Macromolecular X-ray Crystallography Core Laboratory for their assistance with the crystallization and imaging services. Some figures included illustrations created with BioRender.com.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111545.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, and Adams PD (2012). Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr 68, 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beenken A, Eliseenkova AV, Ibrahimi OA, Olsen SK, and Mohammadi M (2012). Plasticity in interactions of fibroblast growth factor 1 (FGF1) N terminus with FGF receptors underlies promiscuity of FGF1. J. Biol. Chem 287, 3067–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Coventry B, Goreshnik I, Huang B, Sheffler W, Park JS, Jude KM, Marković I, Kadam RU, Verschueren KHG, et al. (2022). Design of protein-binding proteins from the target structure alone. Nature 605, 551–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Liu Y, Goetz R, Fu L, Jayaraman S, Hu MC, Moe OW, Liang G, Li X, and Mohammadi M (2018). alpha-Klotho is a non-enzymatic molecular scaffold for FGF23 hormone signalling. Nature 553, 461–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang CY, Lord MS, Melrose J, Rees MD, Knox SM, Freeman C, Iozzo RV, and Whitelock JM (2010). Heparan sulfate-dependent signaling of fibroblast growth factor 18 by chondrocyte-derived perlecan. Biochemistry 49, 5524–5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang LT, Miao Y, Ha A, Yuki K, Park K, Janda CY, Jude KM, Mohan K, Ha N, Vallon M, et al. (2019). Receptor subtype discrimination using extensive shape complementary designed interfaces. Nat. Struct. Mol. Biol 26, 407–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnoyers LR, Pai R, Ferrando RE, Hötzel K, Le T, Ross J, Carano R, D’Souza A, Qing J, Mohtashemi I, et al. (2008). Targeting FGF19 inhibits tumor growth in colon cancer xenograft and FGF19 transgenic hepatocellular carcinoma models. Oncogene 27, 85–97. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswarakumar VP, Lax I, and Schlessinger J (2005). Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 16, 139–149. [DOI] [PubMed] [Google Scholar]
- French DM, Lin BC, Wang M, Adams C, Shek T, Hötzel K, Bolon B, Ferrando R, Blackmore C, Schroeder K, et al. (2012). Targeting FGFR4 inhibits hepatocellular carcinoma in preclinical mouse models. PLoS One 7, e36713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz R, Beenken A, Ibrahimi OA, Kalinina J, Olsen SK, Eliseenkova AV, Xu C, Neubert TA, Zhang F, Linhardt RJ, et al. (2007). Molecular insights into the klotho-dependent, endocrine mode of action of fibroblast growth factor 19 subfamily members. Mol. Cell Biol 27, 3417–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagel M, Miduturu C, Sheets M, Rubin N, Weng W, Stransky N, Bifulco N, Kim JL, Hodous B, Brooijmans N, et al. (2015). First selective small molecule inhibitor of FGFR4 for the treatment of hepatocellular carcinomas with an activated FGFR4 signaling pathway. Cancer Discov. 5, 424–437. [DOI] [PubMed] [Google Scholar]
- Hatlen MA, Schmidt-Kittler O, Sherwin CA, Rozsahegyi E, Rubin N, Sheets MP, Kim JL, Miduturu C, Bifulco N, Brooijmans N, et al. (2019). Acquired on-target clinical resistance validates FGFR4 as a driver of hepatocellular carcinoma. Cancer Discov. 9, 1686–1695. [DOI] [PubMed] [Google Scholar]
- Ibrahimi OA, Eliseenkova AV, Plotnikov AN, Yu K, Ornitz DM, and Mohammadi M (2001). Structural basis for fibroblast growth factor receptor 2 activation in Apert syndrome. Proc. Natl. Acad. Sci. USA 98, 7182–7187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki T, Choi M, Moschetta A, Peng L, Cummins CL, McDonald JG, Luo G, Jones SA, Goodwin B, Richardson JA, et al. (2005). Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2, 217–225. [DOI] [PubMed] [Google Scholar]
- Joshi JJ, Coffey H, Corcoran E,Tsai J, Huang CL, Ichikawa K, Prajapati S, Hao MH, Bailey S, Wu J, et al. (2017). H3B-6527 is a potent and selective inhibitor of FGFR4 in FGF19-driven hepatocellular carcinoma. Cancer Res. 77, 6999–7013. [DOI] [PubMed] [Google Scholar]
- Kabsch W (2010). Xds. Acta Crystallogr. D Biol. Crystallogr 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim RD, Sarker D, Meyer T, Yau T, Macarulla T, Park JW, Choo SP, Hollebecque A, Sung MW, Lim HY, et al. (2019). First-in-Human phase I study of fisogatinib (BLU-554) validates aberrant FGF19 signaling as a driver event in hepatocellular carcinoma. Cancer Discov. 9, 1696–1707. [DOI] [PubMed] [Google Scholar]
- Kir S, Kliewer SA, and Mangelsdorf DJ (2011). Roles of FGF19 in liver metabolism. Cold Spring Harb. Symp. Quant. Biol 76, 139–144. [DOI] [PubMed] [Google Scholar]
- Kiselyov VV, Bock E, Berezin V, and Poulsen FM (2006). NMR structure of the first Ig module of mouse FGFR1. Protein Sci. 15, 1512–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, and Henrick K (2007). Inference of macromolecular assemblies from crystalline state. J. Mol. Biol 372, 774–797. [DOI] [PubMed] [Google Scholar]
- Kurosu H, Choi M, Ogawa Y, Dickson AS, Goetz R, Eliseenkova AV, Mohammadi M, Rosenblatt KP, Kliewer SA, and Kuro-O M (2007). Tissue-specific expression of betaKlotho and fibroblast growth factor (FGF) receptor isoforms determines metabolic activity of FGF19 and FGF21. J. Biol. Chem 282, 26687–26695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzina ES, Ung PMU, Mohanty J, Tome F, Choi J, Pardon E, Steyaert J, Lax I, Schlessinger A, Schlessinger J, and Lee S (2019). Structures of ligand-occupied beta-Klotho complexes reveal a molecular mechanism underlying endocrine FGF specificity and activity. Proc. Natl. Acad. Sci 116, 7819–7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MC, and Colman PM (1993). Shape complementarity at protein/protein interfaces. J. Mol. Biol 234, 946–950. [DOI] [PubMed] [Google Scholar]
- Lee S, Choi J, Mohanty J, Sousa LP, Tome F, Pardon E, Steyaert J, Lemmon MA, Lax I, and Schlessinger J (2018). Structures of beta-klotho reveal a ’zip code’-like mechanism for endocrine FGF signalling. Nature 553, 501–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, and Schlessinger J (2010). Cell signaling by receptor tyrosine kinases. Cell 141, 1117–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin BC, and Desnoyers LR (2012). FGF19 and cancer. Adv. Exp. Med. Biol 728, 183–194. [DOI] [PubMed] [Google Scholar]
- Loo BM, and Salmivirta M (2002). Heparin/Heparan sulfate domains in binding and signaling of fibroblast growth factor 8b. J. Biol. Chem 277, 32616–32623. [DOI] [PubMed] [Google Scholar]
- Matsuki M, Hoshi T, Yamamoto Y, Ikemori-Kawada M, Minoshima Y, Funahashi Y, and Matsui J (2018). Lenvatinib inhibits angiogenesis and tumor fibroblast growth factor signaling pathways in human hepatocellular carcinoma models. Cancer Med. 7, 2641–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ (2007). Phaser crystallographic software. J. Appl. Crystallogr 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholes K, Guillet S, Tomlinson E, Hillan K, Wright B, Frantz GD, Pham TA, Dillard-Telm L, Tsai SP, Stephan JP, et al. (2002). A mouse model of hepatocellular carcinoma: ectopic expression of fibroblast growth factor 19 in skeletal muscle of transgenic mice. Am. J. Pathol 160, 2295–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen SK, Ibrahimi OA, Raucci A, Zhang F, Eliseenkova AV, Yayon A, Basilico C, Linhardt RJ, Schlessinger J, and Mohammadi M (2004). Insights into the molecular basis for fibroblast growth factor receptor autoinhibition and ligand-binding promiscuity. Proc. Natl. Acad. Sci 101, 935–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen SK, Li JYH, Bromleigh C, Eliseenkova AV, Ibrahimi OA, Lao Z, Zhang F, Linhardt RJ, Joyner AL, and Mohammadi M (2006). Structural basis by which alternative splicing modulates the organizer activity of FGF8 in the brain. Genes Dev. 20, 185–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz DM, and Itoh N (2001). Fibroblast growth factors. Genome Biol. 2. REVIEWS3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz DM, and Itoh N (2015). The fibroblast growth factor signaling pathway. Wiley Interdiscip Rev. Dev. Biol 4, 215–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz DM, and Itoh N (2022). New developments in the biology of fibroblast growth factors. WIREs Mech. Dis 14, e1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz DM, and Leder P (1992). Ligand specificity and heparin dependence of fibroblast growth factor receptors 1 and 3. J. Biol. Chem 267, 16305–16311. [PubMed] [Google Scholar]
- Ornitz DM, Xu J, Colvin JS, McEwen DG, MacArthur CA, Coulier F, Gao G, and Goldfarb M (1996). Receptor specificity of the fibroblast growth factor family. J. Biol. Chem 271, 15292–15297. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, and Minor W (1997). Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Plotnikov AN, Hubbard SR, Schlessinger J, and Mohammadi M (2000). Crystal structures of two FGF-FGFR complexes reveal the determinants of ligand-receptor specificity. Cell 101, 413–424. [DOI] [PubMed] [Google Scholar]
- Plotnikov AN, Schlessinger J, Hubbard SR, and Mohammadi M (1999). Structural basis for FGF receptor dimerization and activation. Cell 98, 641–650. [DOI] [PubMed] [Google Scholar]
- Qing J, Du X, Chen Y, Chan P, Li H, Wu P, Marsters S, Stawicki S, Tien J, Totpal K, et al. (2009). Antibody-based targeting of FGFR3 in bladder carcinoma and t(4;14)-positive multiple myeloma in mice. J. Clin. Invest 119, 1216–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawey ET, Chanrion M, Cai C, Wu G, Zhang J, Zender L, Zhao A, Busuttil RW, Yee H, Stein L, et al. (2011). Identification of a therapeutic strategy targeting amplified FGF19 in liver cancer by Oncogenomic screening. Cancer Cell 19, 347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger J (2000). Cell signaling by receptor tyrosine kinases. Cell 103, 211–225. [DOI] [PubMed] [Google Scholar]
- Schlessinger J, Plotnikov AN, Ibrahimi OA, Eliseenkova AV, Yeh BK, Yayon A, Linhardt RJ, and Mohammadi M (2000). Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol. Cell 6, 743–750. [DOI] [PubMed] [Google Scholar]
- Schüller AC, Ahmed Z, Levitt JA, Suen KM, Suhling K, and Ladbury JE (2008). Indirect recruitment of the signalling adaptor Shc to the fibroblast growth factor receptor 2 (FGFR2). Biochem. J 416, 189–199. [DOI] [PubMed] [Google Scholar]
- Spivak-Kroizman T, Lemmon MA, Dikic I, Ladbury JE, Pinchasi D, Huang J, Jaye M, Crumley G, Schlessinger J, and Lax I (1994). Heparin-induced oligomerization of FGF molecules is responsible for FGF receptor dimerization, activation, and cell proliferation. Cell 79, 1015–1024. [DOI] [PubMed] [Google Scholar]
- Weiss A, Adler F, Buhles A, Stamm C, Fairhurst RA, Kiffe M, Sterker D, Centeleghe M, Wartmann M, Kinyamu-Akunda J, et al. (2019). FGF401, A first-in-class highly selective and potent FGFR4 inhibitor for the treatment of FGF19-driven hepatocellular cancer. Mol. Cancer Ther 18, 2194–2206. [DOI] [PubMed] [Google Scholar]
- Wu X, Ge H, Gupte J, Weiszmann J, Shimamoto G, Stevens J, Hawkins N, Lemon B, Shen W, Xu J, et al. (2007). Co-receptor requirements for fibroblast growth factor-19 signaling. J. Biol. Chem 282, 29069–29072. [DOI] [PubMed] [Google Scholar]
- Xu H, Lee KW, and Goldfarb M (1998). Novel recognition motif on fibroblast growth factor receptor mediates direct association and activation of SNT adapter proteins. J. Biol. Chem 273, 17987–17990. [DOI] [PubMed] [Google Scholar]
- Zhang X, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, and Ornitz DM (2006). Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J. Biol. Chem 281, 15694–15700. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This paper analyzes existing, publicly available structures in the Protein DataBank. Their accession codes are listed in the key resources table.
Coordinates and structure factors for the FGFR4D3:mb7 complex in P21 space group are deposited in the Protein Data Bank with the accession code 7TYD.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-FGFR1 | in-house | N/A |
| Mouse monoclonal anti-MAPK | Cell Signaling Technology | Cat# 9107; RRID: AB_10695739 |
| Rabbit monoclonal anti-β-tubulin | Cell Signaling Technology | Cat# 2128; RRID: AB_823664 |
| Rabbit polyclonal anti-phospho-MAPK (pThr202, pTyr204 of p44 MAPK) | Cell Signaling Technology | Cat# 9101; RRID: AB_331646 |
| Rabbit polyclonal anti-phospho-FRS2 (Y436) | R&D systems | Cat# AF5126; RRID: AB_2106234 |
| Alexa Fluor 680 Donkey anti-Mouse IgG antibody | Invitrogen | Cat# A10038; RRID: AB_2534014 |
| IRDye800 CW Donkey anti-Rabbit IgG antibody | LI-COR | Cat# 926-32213; RRID: AB_621848 |
| Bacterial and virus strains | ||
| BL21-Gold(DE3) | Agilent Technologies | Cat# 230132 |
| Chemicals, peptides, and recombinant proteins | ||
| FGF2 | Gibco | Cat# PHG0367 |
| FGF5 | R&D systems | Cat# 237-F5-050 |
| FGF8b | R&D systems | Cat# 423-F8-025 |
| FGF18 | Peprotech | Cat# 100-28 |
| Heparin octa-saccharide (Dp10) | Iduron | Cat# HO10 |
| Heparin sepharose resin (Heparin Sepharose 6 Fast Flow) | GE healthcare | Cat# 17-0998-01 |
| Ni-NTA agarose bead | Qiagen | Cat# 30210 |
| Protein A-HRP | Invitrogen | Cat# 101023 |
| Protein A resin (Rec-Protein A-Sepharose 4B) | Invitrogen | Cat# 101142 |
| Western ECL substrate (Clarity Western ECL Substrate) | Bio-Rad | Cat# 170-5060 |
| Critical commercial assays | ||
| Monolith NT.115 instrument | NanoTemper Technologies | N/A |
| Monolith NT.115 MST Premium Coated Capillaries | NanoTemper Technologies | Cat# MO-K025 |
| Monolith Protein Labeling Kit RED-Tris-NTA dye | NanoTemper Technologies | Cat# MO-L008 |
| Octet RED96 system | FortéBio | N/A |
| Anti-penta-His biosensor (HIS1K Biosensor) | FortéBio | Cat# 18-5120 |
| Anti-human IgG Fc biosensor (AHC Biosensor) | FortéBio | Cat# 18-5060 |
| Deposited data | ||
| Crystal structure of FGFR4D3:mb7 in P21 space group | This paper | PDB: 7TYD |
| Crystal structure of FGFR4D3:mb7 in P63 space group | (Cao et al., 2022) | PDB: 7N1J |
| Crystal structure of FGF1:FGFR1c | (Beenken et al., 2012) | PDB: 3OJV |
| Crystal structure of FGF1:FGFR2b | (Beenken et al., 2012) | PDB: 3OJM |
| Crystal structure of FGF1:FGFR3c | (Olsen et al., 2004) | PDB: 1RY7 |
| Crystal structure of FGF2:FGFR2c | (Ibrahimi et al., 2001) | PDB: 1IIL |
| Crystal structure of FGF23:FGFR1:KLA | (Chen et al., 2018) | PDB: 5W21 |
| Crystal structure of FGFR3b:Fab | (Qing et al., 2009) | PDB: 3GRW |
| Crystal structure of FGF8b:FGFR2c | (Olsen et al., 2006) | PDB: 2FDB |
| Experimental models: Cell lines | ||
| Rat: L6 cells | ATCC | Cat# CRL-1458; RRID:CVCL_0385 |
| Rat: L6R1b cells | This paper | N/A |
| Rat: L6R1cKLB cells | (Lee et al., 2018) | N/A |
| Rat: L6R4KLB cells | (Kuzina et al., 2019) | N/A |
| Human: Hep3B cells (male) | ATCC | Cat# HB-8064 |
| Recombinant DNA | ||
| pCEP4 | Invitrogen | Cat# V04450 |
| pCEP4-FGF19 | This paper | N/A |
| pCEP4-FGFR1bECD (M1–E376) | This paper | N/A |
| pCEP4-FGFR1cECD (M1–E376) | This paper | N/A |
| pCEP4-FGFR4ECD (M1–D369) | This paper | N/A |
| pCEP4-Fc-mb7 | This paper | N/A |
| pCEP4-Fc-mb7TH (K47T, K48H) | This paper | N/A |
| pCEP4-Fc-KLBECD (M30–T983) | (Lee et al., 2018) | N/A |
| pET-28a(+) | Novagen | Cat# 69864 |
| pET-28a(+)-FGF1 (F16–D155) | This paper | N/A |
| pET-28a(+)-FGFR1cD2D3 (T141–R365) | This paper | N/A |
| pET-28a(+)-FGFR4D2D3 (S141–T357) | This paper | N/A |
| pET-28a(+)-FGFR4D3-HIs (S245–D355) | This paper | N/A |
| pET-29b(+) | Novagen | Cat# 69872 |
| pET-29b(+)-His-mb7 | (Cao et al., 2022) | N/A |
| Software and algorithms | ||
| FortéBio Data Analysis HT software 10.0 | FortéBio | N/A |
| Graphpad Prism 8.0 | Graphpad | https://www.graphpad.com |
| Image Studio Lite | LI-COR Biosciences | https://www.licor.com/bio/image-studio-lite/ |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| MO.Affinity Analysis software | NanoTemper Technologies | https://nanotempertech.com/ |
| Phenix | (Adams et al., 2010) | https://phenix-online.org/ |
| MolProbity | (Chen et al., 2010) | http://molprobity.biochem.duke.edu/ |
| PISA server | (Krissinel and Henrick, 2007) | https://www.ebi.ac.uk/pdbe/pisa/ |
| PyMol | Schrödinger | http://www.pymol.org |
| XDS | (Kabsch, 2010) | https://xds.mr.mpg.de/ |
| Other | ||
| Expi293 expression system kit | Gibco | Cat# A14635 |
| QuikChange II site-directed mutagenesis kit | Agilent | Cat# 200523 |