Abstract
O-linked N-acetylglucosamine (O-GlcNAc) is a widespread reversible modification on nucleocytoplasmic proteins that plays an important role in many biochemical processes and is highly relevant to numerous human diseases. The O-GlcNAc modification has diverse functional impacts on individual proteins and glycosites, and methods for editing this modification on substrates are essential to decipher these functions. Herein, we review recent progress in developing methods for O-GlcNAc regulation, with a focus on methods for editing O-GlcNAc with protein- and site-selectivity in cells. The applications, advantages, and limitations of currently available strategies for writing and erasing O-GlcNAc and future directions are also discussed. These emerging approaches to manipulate O-GlcNAc on a target protein in cells will greatly accelerate the development of functional studies and enable therapeutic interventions in the O-GlcNAc field.
Introduction
O-GlcNAc is a monosaccharide post-translational modification (PTM) installed on thousands of nucleocytoplasmic and mitochondrial proteins at multiple serine or threonine residues in eukaryotic cells [1]. O-GlcNAc was first discovered by the Hart group in 1984 [2], and subsequently mapped to more than 5000 proteins across organisms at over 7000 putative glycosites [3]. O-GlcNAc reports on the nutritional state of the cell as the hexosamine biosynthetic pathway (HBP) [4] integrates signals from several nutrient sources, including amino acid, carbohydrate, fatty acid, nucleotide, and energy metabolism, for the generation of UDP-GlcNAc, which is the active donor substrate for protein O-GlcNAcylation (Figure 1). Beyond serving as a nutrient sensor [5], protein O-GlcNAcylation has been reported to play pivotal roles in nearly all major cellular processes, including the cell cycle [6,7], genome maintenance [8,9], epigenetic regulation [10–12], protein turnover [13–15], signaling pathways [16,17], cytoskeletal functions [18,19], and apoptosis [20,21]. Physiologically, dysregulation of O-GlcNAc has been correlated with many diseases, including diabetes [22], cancer [23], cardiovascular disease [24], and neurodegenerative diseases [25].
Figure 1. The O-GlcNAcylation process.
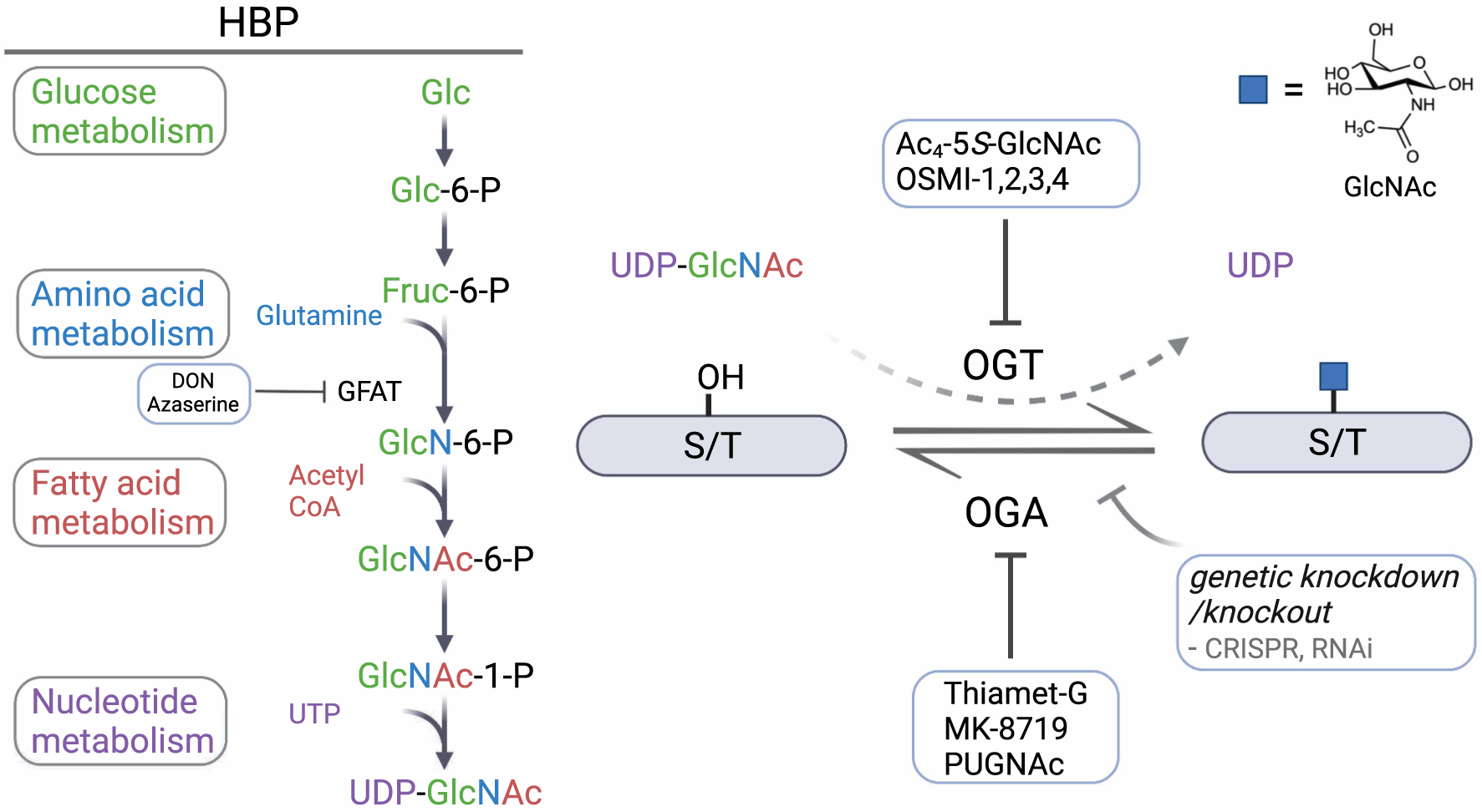
The hexosamine biosynthetic pathway (HBP) starts from taking up glucose (Glc) and generates the high-energy end product UDP-GlcNAc by integrating molecules from multiple metabolic pathways. OGT transfers GlcNAc to Ser/Thr residues on substrate proteins, which can be removed by OGA. Inhibitors targeting GFAT, OGT and OGA, as well as genetic knockdown/knockout approaches are applied to perturbing the O-GlcNAc cycling process globally.
The O-GlcNAc modification itself is primarily governed by a pair of enzymes in mammalian cells, O-GlcNAc transferase (OGT) [26] for installation and a β-N-acetylglucosaminidase, O-GlcNAcase (OGA) [27], for removal. These enzymes are also emerging as crucial therapeutic targets as functional studies for O-GlcNAc progress to potential therapeutic applications [28,29]. The regulation of O-GlcNAc by OGT and OGA is in stark contrast with phosphorylation, a separate PTM that is regulated by hundreds of kinases and phosphatases. The promiscuous activities of OGT and OGA regulate O-GlcNAc levels during the lifespan of their substrates, which can be dynamically written or erased multiple times in response to metabolism and other environmental changes [30]. In addition, these enzymes are present across many tissue types and are essential for embryonic stem cell viability [31] and development in mammals [32].
The study of O-GlcNAc has been facilitated by methods to detect and manipulate the modification. Reliable detection methods, such as metabolic [33] and chemoenzymatic labeling strategies [34], and glycoproteomics [35], have enabled visualization and quantification of the O-GlcNAc modification. In conjunction, developing techniques capable of direct alteration of the O-GlcNAc modification is critical for the purpose of functional analyses and therapeutic interventions in both physiological and pathological processes. Writing and erasing O-GlcNAc from a target protein or even a desired modification site hold promises to monitor biological consequences in both a gain-of-function and loss-of-function manner. To this end, researchers have endeavored to design multiple strategies for tuning O-GlcNAc in the cell on a global scale, which have mostly focused on manipulation of UDP-GlcNAc [36,37] or the pair of cycling enzymes, OGT [38,39] and OGA [40,41]. However, there remain many challenges to achieving protein- and even glycosite-selectivity without perturbing global O-GlcNAc levels in cells, owing to the complexity and dynamics of the O-GlcNAc modification.
In this review, we highlight recent approaches for writing and erasing O-GlcNAc on a target protein in cells. We first introduce several global in vivo approaches for regulating O-GlcNAc levels. Next, we summarize recent advances in the development of novel tools for O-GlcNAc manipulation with improved molecular precision. Finally, we discuss remaining challenges and potential solutions to editing O-GlcNAc in cells and opportunities for new tools needed in the future.
Global approaches for writing and erasing O-GlcNAc in cells
To measure the phenotypic consequences of O-GlcNAcylation on substrates in cells, a primary approach is to perturb the O-GlcNAcylation process (Figure 1). Methods that target UDP-GlcNAc levels, OGT, and OGA have been widely used to control the writing and erasing steps of O-GlcNAc in cells. Since UDP-GlcNAc is the final product of the HBP, which integrates with almost every metabolic pathway, manipulation of the nutrients, such as glucose or glutamine, or enzymes in the HBP, such as glutamine:fructose-6-phosphate-amidotransferase (GFAT), can influence the final concentration of UDP-GlcNAc and thereby alter the O-GlcNAcylation levels on some target proteins [42]. However, as metabolites and HBP enzymes are also involved in many other physiological processes, these methods to manipulate UDP-GlcNAc levels may interfere with other pathways, making it hard to assign functional contributions to O-GlcNAcylation of a specific protein.
Chemical inhibition of OGT and OGA are also effective ways to control O-GlcNAc in cells (Figure 1). Chemical inhibitors of OGT have been identified by the design of a series of UDP-GlcNAc analogs, such as Ac4-5S-GlcNAc [43], as well as by high-throughput screening (HTS). Walker and colleagues discovered OSMI-1 via HTS [44] and developed its derivative OSMI-4 with low nanomolar inhibitory potency and on-target cellular activity through structure-based evolution [39]. Development of OGA inhibitors is also of great interest, in particular due to their potential therapeutic application to neurodegenerative diseases [40,45]. Thiamet-G is one of the most widely used OGA inhibitors in many in vitro and in vivo studies [40]. Inhibition of OGA by thiamet-G elevates O-GlcNAcylation in cells and in vivo, which reduces phosphorylation of tau in both rat cortex and hippocampus, providing a potential new therapeutic application in the context of Alzheimer’s disease (AD) and the associated tauopathies.
Other inhibitors targeting the HBP, OGT and OGA, as well as genetic knockdown and knockout approaches, such as CRISPR and RNAi, have also been applied to the regulation of O-GlcNAc in cells [46]. Although these approaches have been extensively used in studying the functional roles of O-GlcNAc, they cause perturbations to O-GlcNAc levels in the proteome globally, which makes it hard to distinguish the unique functional contribution of O-GlcNAc on various target proteins or even glycosites. Cells are additionally very sensitive to the disruption of endogenous O-GlcNAcylation homeostasis and incubation with either OGT or OGA inhibitors leads to rapid changes on both protein and RNA levels of OGT [39] and OGA [47]. These systemic cellular methods for manipulating O-GlcNAc levels are effective for measuring phenotypic effects of O-GlcNAc, but must be coupled to one of the more targeted approaches described below to gain deeper mechanistic understandings for the contributions of O-GlcNAc on specific protein substrates.
Approaches for writing and erasing O-GlcNAc on target proteins in cells
To interrogate the functional roles of O-GlcNAc on a desired target protein, producing highly glycosylated or deglycosylated target proteins is necessary. In general, incubation of a purified target protein with OGT in vitro readily affords high O-GlcNAc stoichiometry. The target protein can also be directly co-expressed with OGT in E. coli or insect cells [48]. However, these methods inevitably omit factors from a dynamic, complex cellular environment. Therefore, writing and erasing O-GlcNAc on a target protein in living cells is crucial for functional interrogation in the O-GlcNAc field. Recent advances in proximity-based approaches that use various affinity molecules to write and erase other PTMs on target proteins in living cells [49–54], including ‘proteolysis targeting chimeras (PROTACs)’ for targeted protein degradation [55], have inspired the design of proximity-induced enzymatic reactions for engineering O-GlcNAc. Several research groups, including ours, have made efforts to develop new chemical biology tools for tuning O-GlcNAcylation on a protein of interest (POI) in living cells.
For the purpose of targeted protein O-GlcNAcylation, we recently implemented a nanobody-directed approach to write and erase O-GlcNAc on target proteins in living cells (Figure 2A). This strategy is predicated on engineering the nanobody and the O-GlcNAc cycling enzymes, OGT and OGA, to enhance their recruitment to the desired target protein in living cells [56,57]. Beyond their high affinity and specificity similar to canonical IgG antibodies, the nanobody is small in size (<15 kD), highly soluble and stable, and amenable to intracellular expression and protein engineering [58]. These advantages of nanobodies are well suited to targeting endogenous proteins inside living cells. Leveraging these advantages, we achieved selective O-GlcNAcylation of target proteins in cells by fusion of OGT to a nanobody that recognizes a tag or endogenous protein target. To further improve the protein selectivity and reduce perturbations to non-targeted substrates, full-length OGT with 13.5 helix-turn-helix tetratricopeptide repeats (TPRs) that recognize endogenous substrates was truncated to a shorter form with 4.5 TPRs [OGT(4)]. OGT(4) displayed weaker transferase activity on endogenous substrates, which thereby enhanced the target protein selectivity after fusion of a nanobody by reduction in the off-target effects on global O-GlcNAcylation level. Using a nanobody against the EPEA epitope derived from α-synuclein [59], the nanobody-OGT(4) fusion selectively increased O-GlcNAc on EPEA-tagged target proteins, as well as endogenous α-synuclein in HEK293T cells. Increasing availability of nanobodies that target endogenous proteins has the potential to dramatically broaden the spectrum of endogenous substrates targeted by this system in the future [60,61].
Figure 2. Approaches for writing and erasing O-GlcNAc from target proteins in cells.
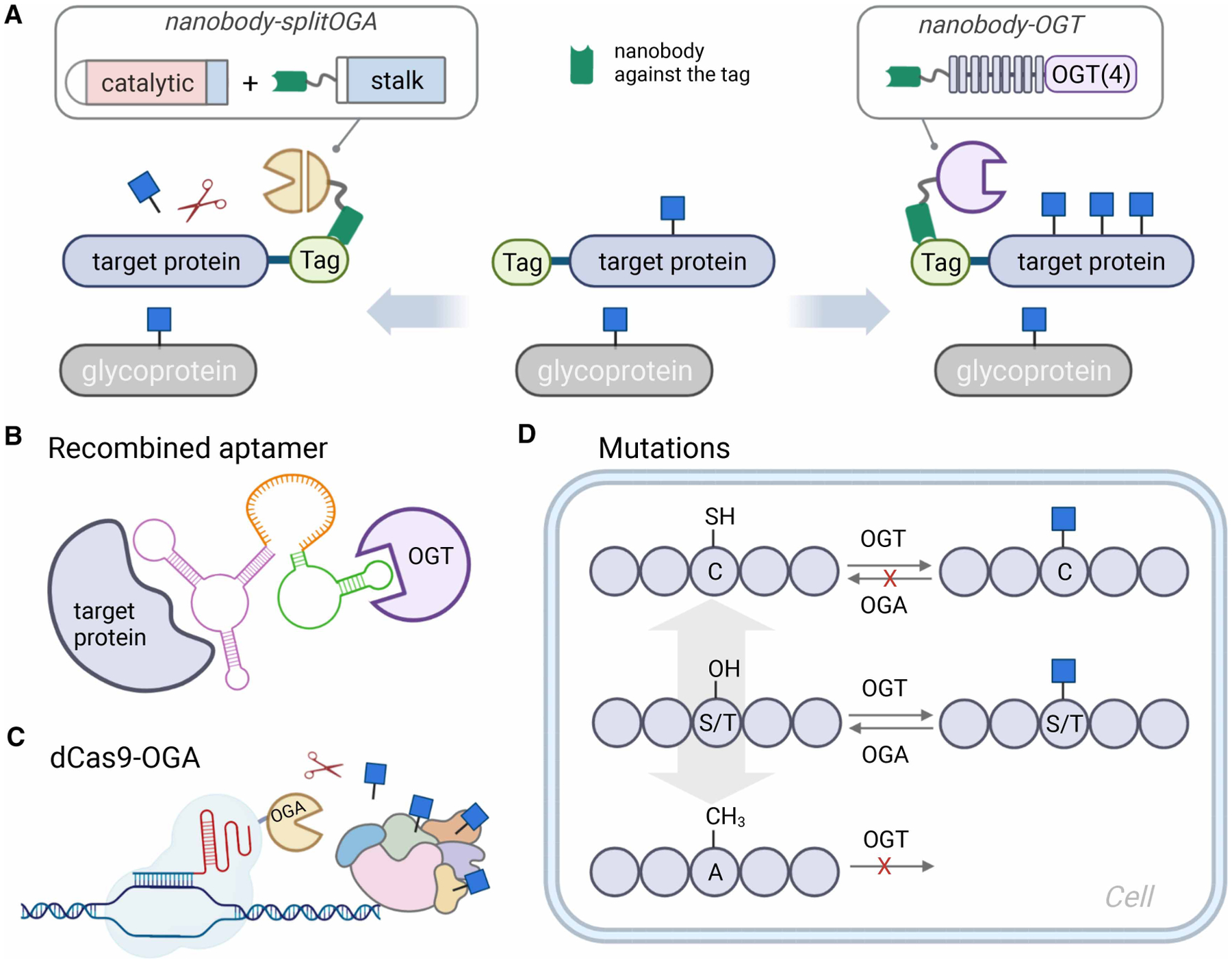
(A) Nanobody-fused engineered OGT [56] and split OGA [57] allow target protein glycosylation and deglycosylation in cells, respectively. (B) A recombined aptamer is designed to recruit endogenous OGT in close proximity to the target protein for increased O-GlcNAcylation [71]. (C) dCas9-OGA fusion allows the O-GlcNAc removal from proteins surrounding a target gene region [72]. (D) Site-directed mutagenesis enables O-GlcNAc engineering at a specific site [90].
Recently, we also reported a nanobody-fused split OGA as a complementary tool for targeted protein deglycosylation in living cells [57]. Since O-GlcNAc modifications can be essential mediators of protein stability, function, and protein–protein interactions, selective removal of O-GlcNAc from a target protein would help evaluate contributions of O-GlcNAc in a loss-of-function manner. To reduce the potential global impacts caused by OGA overexpression, we took advantage of the fact that OGA can be cleaved into two fragments by caspase-3 during apoptosis [62] and designed differentially truncated N- and C-fragments accordingly. After systematic cellular optimization, a nanobody-fused split OGA was generated for targeted O-GlcNAc removal on several O-GlcNAcylated proteins with minimal effects to the surrounding O-GlcNAc proteome. This nanobody-directed O-GlcNAc eraser enabled selective deglycosylation of two transcription factors, c-Jun and c-Fos, to reveal functional roles of O-GlcNAc on protein stability and transcription ability, respectively.
The nanobody-fused OGT and split OGA serve as a complementary toolset for writing and erasing O-GlcNAc on target proteins in cells, thus allowing for the direct dissections of O-GlcNAc functions on a desired protein. Due to the modular design of the nanobody-fused OGT and split OGA, this system can be further integrated with recent developments on nanobody engineering to gain more spatial and temporal precision [63–66] and thereby facilitate the interrogation of O-GlcNAc turnover within different cellular contexts. We also observed that a nanobody with a moderate affinity (Kd = 310 nM) [67] showed better glycosylation or deglycosylation performance than a nanobody with a higher affinity (Kd = 0.59 nM) [68] possibly due to appropriate reversibility [69]. Although both OGT and OGA were engineered to reduce their inherent enzymatic activity and thereby lower impact on the global O-GlcNAcome, the application of this pair of tools still requires overexpression of the nanobody–enzyme fusion in cells.
Direct recruitment of endogenous OGT or OGA to the target protein by heterobifunctional molecules is a promising solution to address this caveat. Using systematic evolution of ligands by exponential enrichment (SELEX) [70], Zhu and Hart obtained an RNA aptamer that binds to OGT [71]. A recombined RNA aptamer composed of this OGT-specific aptamer and an aptamer binding to a desired protein would induce proximity between endogenous OGT and the target protein, leading to elevated O-GlcNAcylation on the target protein (Figure 2B). Using this approach, the authors interrogated the interplay between O-GlcNAcylation and other PTMs, such as phosphorylation and ubiquitination, on the transcription factor β-catenin within cells and discovered a positive correlation between O-GlcNAcylation and its transcriptional activity. A series of RNA aptamers targeting OGT and OGA with different affinities have been selected, which may enlarge the toolkit for writing or erasing O-GlcNAc on individual proteins in the future. However, this design requires development of an aptamer for the desired target protein followed by careful adjustment of the recombined aptamer’s length and orientation to optimize the enzymes’ efficacy for each target protein. Also, while aptamers with higher affinities to OGT or OGA may enhance recruitment of these enzymes to the target protein, reversibility of the aptamer binding event is a key challenge to enable enzymatic turnover and not cause global perturbations to the broader O-GlcNAcome.
Alternative approaches using other protein binders could be explored in the future [61]. Several small molecule chimeras for editing PTMs on a target protein have been recently reported [54], including chimeric small molecules for proximity-directed phosphorylation [51] or dephosphorylation [53]. However, small molecule ligands with good specificity, affinity, and cell-penetrating ability for OGT or OGA beyond inhibitors remain to be discovered.
To specifically modulate O-GlcNAc on proteins surrounding retrotransposon promoters, Boulard et al. [72] recently developed a novel targeted deglycosylation tool with genomic sequence specificity through fusion of inactive dCas9 and OGA (Figure 2C). Using this method, they achieved local chromatin deglycosylation and subsequently reactivated the expression of the targeted retrotransposon family, which reinforced the connection between O-GlcNAcylation of chromatin factors and methylation-dependent silencing on retrotransposons. Similarly, fusions of dCas9 to OGT or OGA have been applied to studies of O-GlcNAc and its effects on gene transcription, though the design awaits full characterizations [73]. Although sgRNA helps locate dCas9-fused OGT or OGA to a desired genomic region, O-GlcNAc alteration may still happen on multiple proteins accessible to the fusion enzymes, which necessitates additional engineering for elevated protein selectivity.
Approaches for writing and erasing specific O-GlcNAc sites on target proteins in cells
O-GlcNAcylation may occur at multiple sites on an individual protein, which may display discrete stoichiometry and dynamics that lead to different functional roles. For example, transcription factor CREB contains multiple glycosites, but only Ser40 is induced by neuronal depolarization [74]. Likewise, two O-GlcNAc sites were identified on NF-κB p65, yet only O-GlcNAcylation on Thr352 is responsible for hyperglycemia-induced NF-κB activation [75]. Due to the promiscuity of OGT and OGA on both substrates and modification sites, heterogenous protein mixtures with varied modification levels will be produced unless there is only one glycosite existing, making it hard to differentiate the contribution of O-GlcNAc on a specific glycosite. Therefore, identification of glycosites and applications of approaches to site-specifically regulate O-GlcNAcylation are of great significance for functional analyses. O-GlcNAc maps to glycosites and their respective proteins have been recently cataloged by two databases [3,76]. Building upon the glycosite maps afforded by mass spectrometry, the most common approach to determine function is via a loss-of-function mutation of the modified Ser/Thr to Ala to permanently prohibit O-GlcNAcylation (Figure 2D) [15,75]. This method is readily conducted on either overexpressed or endogenous substrates, providing an unmodified protein control for functional comparison. Nonetheless, the Ser/Thr to Ala mutation cannot always recapitulate the phenotype resulting from the naturally unmodified target proteins [15]. And for proteins wherein phosphate and O-GlcNAc occupy the same sites, this type of mutation also disrupts potential phosphorylation as well [100].
Compared with removing O-GlcNAc from a known site, adding O-GlcNAc to a specific site is much more challenging, necessitating the development of creative strategies to write O-GlcNAc in the field. Several in vitro methods have been explored to introduce O-GlcNAc or its analogs to a target protein with stoichiometric site-specificity. Expressed protein ligation [77] has been used for synthesis of small proteins (<15 kD) with site-specific O-GlcNAcylation, such as tau [78] or α-synuclein [79–81]. Through this method, Pratt and co-workers discovered that O-GlcNAc inhibits monomeric α-synuclein aggregation with site-specific differences [80]. Separately, Davis and co-workers have developed chemical methods at cysteine to generate proteins with site-specific S-GlcNAc in vitro, which is a non-hydrolyzable O-GlcNAc analog [82,83]. This method chemically transforms a cysteine residue at a specific site to dehydroalanine, which further reacts with a nucleophilic GlcNAc derivative to install the S-GlcNAc modification. Although these methods provide a homogenous O-GlcNAcylated protein product, they are limited to a small range of chemically compatible substrates in vitro.
The genetic code expansion (GCE) technique is also widely employed to site-specifically incorporate diverse unnatural amino acids (UAAs) into target proteins both in vitro and in vivo using an orthogonal tRNA and its synthetase [84]. Therefore, an alternative indirect strategy for O-GlcNAc installation is by the introduction of a reactive warhead, such as an alkyne [85] or ketone [86] via GCE first, followed by the conjugation with a GlcNAc moiety via bioorthogonal chemistries. Separately, Wang and co-workers site-specifically generated reactive dehydroalanine or dehydrobutyrine in live cells by harnessing the sulfur–fluoride exchange reaction (SuFEx) between a latent bioreactive UAA (FSY) and a nearby serine or threonine, which can subsequently react with a thiol-saccharide to prepare glycosylated proteins [87]. Although the second conjugation step with the sugar moiety could be conducted inside cells in principle, current demonstrations of these strategies are performed in vitro using purified proteins. Moreover, to date only O-GlcNAc mimics with artificial chemical linkages can be generated, leading to an incomplete recapitulation of the O-GlcNAcylated target protein.
To co-translationally produce the identical O-GlcNAc modified serine or threonine in cells, scientists have attempted to incorporate an O-GlcNAcylated amino acid directly by GCE. The first glycosylated UAA, tri-acetyl-β-GlcNAc-serine was reported to be genetically encoded into myoglobin in E. coli by an orthogonal Methanococcus jannaschii tyrosyl tRNA synthetase [88], which was retracted later due to non-replicability. Although GCE has been widely applied to install diverse PTMs on specific proteins in cells [89], neither O-GlcNAcylated amino acids nor their derivatives have been successfully incorporated into target proteins in cells directly by GCE to date.
Separately, the van Aalten group recently reported a straightforward genetic recoding approach to install a non-hydrolyzable S-GlcNAc site-specifically on a target protein in live cells (Figure 2D) [90]. Cysteine S-linked GlcNAcylation was identified as a stable PTM in mammals, which can be installed by OGT but is unable to be hydrolyzed by OGA [91,92]. By harnessing the ability of OGT to modify cysteine, simply mutating Ser/Thr to Cys on the known glycosite results in a stable S-GlcNAc modification both in vitro and in vivo. Using CRISPR–Cas9 technology, the authors genetically encoded an endogenous OGA with a S405C mutation, resulting in a much higher GlcNAcylation stoichiometry (70%) due to the non-hydrolysable S-GlcNAcylation. The hyper-S-GlcNAcylated OGAS405C showed a substantially reduced half-life, indicating the functional effects of GlcNAcylation on the stability of OGA. This surprisingly simple approach requires neither the overexpression of O-GlcNAcylation processing enzymes or the treatment of chemical inhibitors, which greatly minimized the global effects on the broader O-GlcNAcylated proteome. Like the Ser/Thr to Ala mutation, cysteine mutation can be easily conducted by either common molecular biology techniques for recombinant proteins or CRISPR–Cas9 gene-editing method for endogenous proteins in the context of complicated in vivo settings. This site-specific approach to directly introduce the GlcNAc modification on a target protein in mammalian cells is readily amenable to other target proteins whose glycosites have been mapped.
Despite the advantages of site-directed mutagenesis for evaluating the contribution of specific glycosites on a target protein, some caveats remain. These approaches require the prior identification of glycosite, which is not always straightforward and requires further biochemical validation [93–95]. Furthermore, Ser/Thr to Cys mutation for stable GlcNAc installation may not be applicable to proteins with many sites of O-GlcNAcylation or those glycosites whose O-GlcNAc stoichiometry cannot be elevated by OGA inhibition. Separately, substitution of the side chains of amino acids to a reactive thiol group of Cys can alter the protein’s chemical properties and thereby influence the protein folding and stability unexpectedly [96,97]. Nonetheless, the generated S-GlcNAc modification can be an adequate structure mimic of the O-GlcNAc modification for functional studies as several labs have shown that S-GlcNAc has a similar conformation to O-GlcNAc when it binds to an O-GlcNAc binding protein, and can be detected by a pan-specific O-GlcNAc antibody [87] and O-GlcNAc ‘reader’ proteins [90], through computational and biochemical analyses [90,98].
Conclusion
Mounting evidence underscores the wide existence and profound biological significance of O-GlcNAcylation on thousands of proteins in the cell. Development of more powerful tools for O-GlcNAc regulation are highly sought in the field to accelerate the molecular dissection of roles for the O-GlcNAc modification. In addition to approaches to globally manipulate O-GlcNAcylation in cells, such as chemical inhibitors and genetic overexpression/knockdown of OGT or OGA, methods reviewed here with protein- or site-specificity have recently expanded the toolkit for O-GlcNAc manipulations and therefore facilitate the dissection of O-GlcNAc functions within various environments.
For engineering protein-selective O-GlcNAcylation [56,57,71], proximity-induced recruitment of OGT and OGA have yielded successful approaches to writing and erasing O-GlcNAc from a target protein. Substrates can be targeted by selective nanobodies or aptamers. To date, nanobody-fused OGT/OGA, recombined RNA aptamers for OGT recruitment, and dCas9-fused OGA allow the installation or removal of O-GlcNAc with increased protein or region specificity without the need of the prior identification of glycosites. However, given the formation of transient enzyme–substrate complex induced by these methods, experiments should be carefully controlled to rule out potential side effects to protein diffusion, stability, and interactions [99]. Incorporation of glycosylated amino acids via GCE is attractive for achieving site-specific glycosylation yet is only applicable in vitro to date [100,101]. Site-directed mutagenesis of the glycosite to Cys or Ala is the only available approach to control O-GlcNAcylation with site-selective precision in cells [68,90]. However, it builds upon prior knowledge of the glycosite localization and is only applicable to a subpopulation of substrates. These methods for target protein and glycosite manipulation in cells are complementary and therefore may be utilized for different target proteins and biological contexts.
As each of these strategies provides complementary methods to manipulate O-GlcNAc, the combination of multiple approaches will provide a comprehensive understanding of the biological significance of O-GlcNAcylation on a given target protein. Development of new methods for writing and erasing O-GlcNAc on target proteins in cells will serve to further enhance options to manipulate O-GlcNAc in biological settings. Future directions of growth include efforts to extend current in vitro methods with site or protein specificity to the intracellular settings. Along with advances in bioorthogonal chemistries, modified cysteine conversion chemistry holds the promise to introduce GlcNAc site-specifically inside cells [83,102]. Another attractive direction is the introduction of the O-GlcNAc-modified amino acids to a specific site on the target protein via GCE in cells. Separately, identification of non-inhibitive OGT/OGA ligands may be employed in the context of heterobifunctional small molecules by coupling to substrate ligands to afford protein-selective control of O-GlcNAcylation. In the long term, new insights into how OGT and OGA recognize their substrates [103] may spur the generation of novel genetically encoded tools with substrate preferences through direct engineering or evolution of the single pair of enzymes. Future developments will provide additional creative solutions to write and erase O-GlcNAc on a target protein in cells and foretell a bright future for research on O-GlcNAcylation. Since many diseases are featured by abnormal O-GlcNAcylation, like hyper-O-GlcNAcylation in cancers and diabetes, and hypo-O-GlcNAcylation in AD, these approaches may expand opportunities to target O-GlcNAc in the clinic in the future [104].
Perspectives.
O-GlcNAcylation is a ubiquitous reversible sugar modification on Ser/Thr residues, which is controlled by a single pair of enzymes, OGT and OGA. O-GlcNAcylation on different substrates or modification sites shows diverse functions, which are associated with multiple physiological processes and diseases.
Deciphering O-GlcNAc functions on a specific protein or modification site is a long-standing demand in this field. Many cellular approaches with protein or site selectivity are emerging, including proximity-induced reactions and genetic mutagenesis.
The development of new tools with protein and site-specificity in live cells will expand the scope of O-GlcNAc engineering. Heterobifunctional small molecules and glycosylated amino acids via GCE represent two potential advances in the future.
Acknowledgements
This work was supported by the National Institutes of Health (U01CA242098-01, C.M.W.), the Sloan Foundation (C.M.W.), Camille-Dreyfus Teacher Scholar (C.M.W.), and Harvard University. Figures are created with BioRender. com.
Abbreviations
- AD
Alzheimer’s disease
- GCE
genetic code expansion
- GFAT
glutamine: fructose-6-phosphate-amidotransferase
- HBP
Hexosamine biosynthetic pathway
- HTS
high-throughput screening
- OGA
O-GlcNAcase
- O-GlcNAc
O-linked N-acetylglucosamine
- OGT
O-GlcNAc transferase
- POI
protein of interest
- PROTAC
proteolysis targeting chimera
- PTM
post-translational modification
- RNAi
RNA interference
- SELEX
systematic evolution of ligands by exponential enrichment
- SuFEx
sulfur–fluoride exchange reaction
- TPR
helix-turn-helix tetratricopeptide repeats
- UAA
unnatural amino acid.
Footnotes
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Yang X and Qian K (2017) Protein O-GlcNAcylation: emerging mechanisms and functions. Nat. Rev. Mol. Cell Biol 18, 452–465 10.1038/nrm.2017.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torres CR and Hart GW (1984) Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. evidence for O-linked GlcNAc. J. Biol. Chem 259, 3308–3317 10.1016/s0021-9258(17)43295-9 [DOI] [PubMed] [Google Scholar]
- 3.Wulff-Fuentes E, Berendt RR, Massman L, Danner L, Malard F, Vora J et al. (2021) The human O-GlcNAcome database and meta-analysis. Sci. Data 8, 25 10.1038/s41597-021-00810-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marshall S, Bacote V and Traxinger RR (1991) Discovery of a metabolic pathway mediating glucose-induced desensitization of the glucose transport system. Role of hexosamine biosynthesis in the induction of insulin resistance. J. Biol. Chem 266, 4706–4712 [PubMed] [Google Scholar]
- 5.Hart GW (2019) Nutrient regulation of signaling and transcription. J. Biol. Chem 294, 2211–2231 10.1074/jbc.AW119.003226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu C and Li J (2018) O-GlcNAc: a sweetheart of the cell cycle and DNA damage response. Front. Endocrinol. (Lausanne) 9, 415 10.3389/fendo.2018.00415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slawson C, Lakshmanan T, Knapp S and Hart GW (2008) A mitotic GlcNAcylation/phosphorylation signaling complex alters the posttranslational state of the cytoskeletal protein vimentin. Mol. Biol. Cell 19, 4130–4140 10.1091/mbc.E07-11-1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miura Y, Sakurai Y and Endo T (2012) O-GlcNAc modification affects the ATM-mediated DNA damage response. Biochim. Biophys. Acta 1820, 1678–1685 10.1016/j.bbagen.2012.06.013 [DOI] [PubMed] [Google Scholar]
- 9.Ma X, Liu H, Li J, Wang Y, Ding YH, Shen H et al. (2017) Poleta O-GlcNAcylation governs genome integrity during translesion DNA synthesis. Nat. Commun 8, 1941 10.1038/s41467-017-02164-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu D, Cai Y and Jin J (2017) Potential coordination role between O-GlcNAcylation and epigenetics. Protein Cell 8, 713–723 10.1007/s13238-017-0416-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bauer C, Gobel K, Nagaraj N, Colantuoni C, Wang M, Muller U et al. (2015) Phosphorylation of TET proteins is regulated via O-GlcNAcylation by the O-linked N-acetylglucosamine transferase (OGT). J. Biol. Chem 290, 4801–4812 10.1074/jbc.M114.605881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakabe K, Wang Z and Hart GW (2010) β-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc. Natl Acad. Sci. U.S.A 107, 19915–19920 10.1073/pnas.1009023107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruan HB, Nie Y and Yang X (2013) Regulation of protein degradation by O-GlcNAcylation: crosstalk with ubiquitination. Mol. Cell. Proteomics 12, 3489–3497 10.1074/mcp.R113.029751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Y, Liu TW, Cecioni S, Eskandari R, Zandberg WF and Vocadlo DJ (2015) O-GlcNAc occurs cotranslationally to stabilize nascent polypeptide chains. Nat. Chem. Biol 11, 319–325 10.1038/nchembio.1774 [DOI] [PubMed] [Google Scholar]
- 15.Li X, Zhu Q, Shi X, Cheng Y, Li X, Xu H et al. (2019) O-GlcNAcylation of core components of the translation initiation machinery regulates protein synthesis. Proc. Natl Acad. Sci. U.S.A 116, 7857–7866 10.1073/pnas.1813026116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ong Q, Han W and Yang X (2018) O-GlcNAc as an integrator of signaling pathways. Front. Endocrinol. (Lausanne) 9, 599 10.3389/fendo.2018.00599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nie H, Ju H, Fan J, Shi X, Cheng Y, Cang X et al. (2020) O-GlcNAcylation of PGK1 coordinates glycolysis and TCA cycle to promote tumor growth. Nat. Commun 11, 36 10.1038/s41467-019-13601-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lambert M, Claeyssen C, Bastide B and Cieniewski-Bernard C (2020) O-GlcNAcylation as a regulator of the functional and structural properties of the sarcomere in skeletal muscle: an update review. Acta Physiol. (Oxf.) 228, e13301 10.1111/apha.13301 [DOI] [PubMed] [Google Scholar]
- 19.Tarbet HJ, Dolat L, Smith TJ, Condon BM, O’Brien ET III, Valdivia RH et al. (2018) Site-specific glycosylation regulates the form and function of the intermediate filament cytoskeleton. eLife 7, e31807 10.7554/eLife.31807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi J, Gu JH, Dai CL, Gu J, Jin X, Sun J et al. (2015) O-GlcNAcylation regulates ischemia-induced neuronal apoptosis through AKT signaling. Sci. Rep 5, 14500 10.1038/srep14500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma Z, Vocadlo DJ and Vosseller K (2013) Hyper-O-GlcNAcylation is anti-apoptotic and maintains constitutive NF-κB activity in pancreatic cancer cells. J. Biol. Chem 288, 15121–15130 10.1074/jbc.M113.470047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma J and Hart GW (2013) Protein O-GlcNAcylation in diabetes and diabetic complications. Expert Rev. Proteomics 10, 365–380 10.1586/14789450.2013.820536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parker MP, Peterson KR and Slawson C (2021) O-GlcNAcylation and O-GlcNAc cycling regulate gene transcription: emerging roles in cancer. Cancers (Basel) 13, 1666 10.3390/cancers13071666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright JN, Collins HE, Wende AR and Chatham JC (2017) O-GlcNAcylation and cardiovascular disease. Biochem. Soc. Trans 45, 545–553 10.1042/BST20160164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryan P, Xu M, Davey AK, Danon JJ, Mellick GD, Kassiou M et al. (2019) O-GlcNAc modification protects against protein misfolding and aggregation in neurodegenerative disease. ACS Chem. Neurosci 10, 2209–2221 10.1021/acschemneuro.9b00143 [DOI] [PubMed] [Google Scholar]
- 26.Haltiwanger RS, Blomberg MA and Hart GW (1992) Glycosylation of nuclear and cytoplasmic proteins. Purification and characterization of a uridine diphospho-N-acetylglucosamine:polypeptide β-N-acetylglucosaminyltransferase. J. Biol. Chem 267, 9005–9013 [PubMed] [Google Scholar]
- 27.Gao Y, Wells L, Comer FI, Parker GJ and Hart GW (2001) Dynamic O-glycosylation of nuclear and cytosolic proteins: cloning and characterization of a neutral, cytosolic β-N-acetylglucosaminidase from human brain. J. Biol. Chem 276, 9838–9845 10.1074/jbc.M010420200 [DOI] [PubMed] [Google Scholar]
- 28.Ju Kim E (2020) O-GlcNAc transferase: structural characteristics, catalytic mechanism and small-molecule inhibitors. Chembiochem 21, 3026–3035 10.1002/cbic.202000194 [DOI] [PubMed] [Google Scholar]
- 29.Macauley MS and Vocadlo DJ (2010) Increasing O-GlcNAc levels: an overview of small-molecule inhibitors of O-GlcNAcase. Biochim. Biophys. Acta 1800, 107–121 10.1016/j.bbagen.2009.07.028 [DOI] [PubMed] [Google Scholar]
- 30.Zachara NE, O’Donnell N, Cheung WD, Mercer JJ, Marth JD and Hart GW (2004) Dynamic O-GlcNAc modification of nucleocytoplasmic proteins in response to stress. A survival response of mammalian cells. J. Biol. Chem 279, 30133–30142 10.1074/jbc.M403773200 [DOI] [PubMed] [Google Scholar]
- 31.Shafi R, Iyer SP, Ellies LG, O’Donnell N, Marek KW, Chui D et al. (2000) The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc. Natl Acad. Sci. U.S.A 97, 5735–5739 10.1073/pnas.100471497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang YR, Song M, Lee H, Jeon Y, Choi EJ, Jang HJ et al. (2012) O-GlcNAcase is essential for embryonic development and maintenance of genomic stability. Aging Cell 11, 439–448 10.1111/j.1474-9726.2012.00801.x [DOI] [PubMed] [Google Scholar]
- 33.Wang H and Mooney DJ (2020) Metabolic glycan labelling for cancer-targeted therapy. Nat. Chem 12, 1102–1114 10.1038/s41557-020-00587-w [DOI] [PubMed] [Google Scholar]
- 34.Lopez Aguilar A, Briard JG, Yang L, Ovryn B, Macauley MS and Wu P (2017) Tools for studying glycans: recent advances in chemoenzymatic glycan labeling. ACS Chem. Biol 12, 611–621 10.1021/acschembio.6b01089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu Q and Yi W (2021) Chemistry-assisted proteomic profiling of O-GlcNAcylation. Front. Chem 9, 702260 10.3389/fchem.2021.702260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wellen KE, Lu C, Mancuso A, Lemons JM, Ryczko M, Dennis JW et al. (2010) The hexosamine biosynthetic pathway couples growth factor-induced glutamine uptake to glucose metabolism. Genes Dev. 24, 2784–2799 10.1101/gad.1985910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vasconcelos-Dos-Santos A, Loponte HF, Mantuano NR, Oliveira IA, de Paula IF, Teixeira LK et al. (2017) Hyperglycemia exacerbates colon cancer malignancy through hexosamine biosynthetic pathway. Oncogenesis 6, e306 10.1038/oncsis.2017.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang C, Xie F, Li L, Zhang C, Zhang Y, Ying W et al. (2019) Hepatocyte nuclear factor 1 alpha (HNF1A) regulates transcription of O-GlcNAc transferase in a negative feedback mechanism. FEBS Lett. 593, 1050–1060 10.1002/1873-3468.13381 [DOI] [PubMed] [Google Scholar]
- 39.Martin SES, Tan ZW, Itkonen HM, Duveau DY, Paulo JA, Janetzko J et al. (2018) Structure-based evolution of low nanomolar O-GlcNAc transferase inhibitors. J. Am. Chem. Soc 140, 13542–5 10.1021/jacs.8b07328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuzwa SA, Macauley MS, Heinonen JE, Shan X, Dennis RJ, He Y et al. (2008) A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of tau in vivo. Nat. Chem. Biol 4, 483–490 10.1038/nchembio.96 [DOI] [PubMed] [Google Scholar]
- 41.Gorelik A and Ferenbach AT (2020) CRISPR-Cas9-mediated depletion of O-GlcNAc hydrolase and transferase for functional dissection of O-GlcNAcylation in human cells. bioRxiv 2020.08.19.258079 10.1101/2020.08.19.258079 [DOI] [Google Scholar]
- 42.Chiaradonna F, Ricciardiello F and Palorini R (2018) The nutrient-sensing hexosamine biosynthetic pathway as the hub of cancer metabolic rewiring. Cells 7, 53 10.3390/cells7060053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gloster TM, Zandberg WF, Heinonen JE, Shen DL, Deng L and Vocadlo DJ (2011) Hijacking a biosynthetic pathway yields a glycosyltransferase inhibitor within cells. Nat. Chem. Biol 7, 174–181 10.1038/nchembio.520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ortiz-Meoz RF, Jiang J, Lazarus MB, Orman M, Janetzko J, Fan C et al. (2015) A small molecule that inhibits OGT activity in cells. ACS Chem. Biol 10, 1392–1397 10.1021/acschembio.5b00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Selnick HG, Hess JF, Tang C, Liu K, Schachter JB, Ballard JE et al. (2019) Discovery of MK-8719, a potent O-GlcNAcase inhibitor as a potential treatment for tauopathies. J. Med. Chem 62, 10062–10097 10.1021/acs.jmedchem.9b01090 [DOI] [PubMed] [Google Scholar]
- 46.Ma J, Wu C and Hart GW (2021) Analytical and biochemical perspectives of protein O-GlcNAcylation. Chem. Rev 121, 1513–1581 10.1021/acs.chemrev.0c00884 [DOI] [PubMed] [Google Scholar]
- 47.Zhang Z, Tan EP, VandenHull NJ, Peterson KR and Slawson C (2014) O-GlcNAcase expression is sensitive to changes in O-GlcNAc homeostasis. Front. Endocrinol. (Lausanne) 5, 206 10.3389/fendo.2014.00206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao H, Shi M, Wang R, Wang C, Shao C, Gu Y et al. (2018) A widely compatible expression system for the production of highly O-GlcNAcylated recombinant protein in Escherichia coli. Glycobiology 28, 949–957 10.1093/glycob/cwy077 [DOI] [PubMed] [Google Scholar]
- 49.Caussinus E, Kanca O and Affolter M (2011) Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nat. Struct. Mol. Biol 19, 117–121 10.1038/nsmb.2180 [DOI] [PubMed] [Google Scholar]
- 50.Lepeta K, Roubinet C, Kanca O, Ochoa-Espinosa A, Bieli D, Cabernard C et al. (2021) In vivo regulation of fluorescent fusion proteins by engineered kinases. bioRxiv 2021.03.26.433940 10.1101/2021.03.26.433940 [DOI] [Google Scholar]
- 51.Siriwardena SU, Munkanatta Godage DNP, Shoba VM, Lai S, Shi M, Wu P et al. (2020) Phosphorylation-inducing chimeric small molecules. J. Am. Chem. Soc 142, 14052–7 10.1021/jacs.0c05537 [DOI] [PubMed] [Google Scholar]
- 52.Kanner SA, Shuja Z, Choudhury P, Jain A and Colecraft HM (2020) Targeted deubiquitination rescues distinct trafficking-deficient ion channelopathies. Nat. Methods 17, 1245–1253 10.1038/s41592-020-00992-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamazoe S, Tom J, Fu Y, Wu W, Zeng L, Sun C et al. (2020) Heterobifunctional molecules induce dephosphorylation of kinases-a proof of concept study. J. Med. Chem 63, 2807–2813 10.1021/acs.jmedchem.9b01167 [DOI] [PubMed] [Google Scholar]
- 54.Modell AE, Lai S, Nguyen TM and Choudhary A (2021) Bifunctional modalities for repurposing protein function. Cell Chem. Biol 28, 1081–1089 10.1016/j.chembiol.2021.06.005 [DOI] [PubMed] [Google Scholar]
- 55.Burslem GM and Crews CM (2020) Proteolysis-targeting chimeras as therapeutics and tools for biological discovery. Cell 181, 102–114 10.1016/j.cell.2019.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramirez DH, Aonbangkhen C, Wu HY, Naftaly JA, Tang S, O’Meara TR et al. (2020) Engineering a proximity-directed O-GlcNAc transferase for selective protein O-GlcNAcylation in cells. ACS Chem. Biol 15, 1059–1066 10.1021/acschembio.0c00074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ge Y, Ramirez DH, Yang B, D’Souza AK, Aonbangkhen C, Wong S et al. (2021) Target protein deglycosylation in living cells by a nanobody-fused split O-GlcNAcase. Nat. Chem. Biol 17, 593–600 10.1038/s41589-021-00757-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheloha RW, Harmand TJ, Wijne C, Schwartz TU and Ploegh HL (2020) Exploring cellular biochemistry with nanobodies. J. Biol. Chem 295, 15307–15327 10.1074/jbc.REV120.012960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.De Genst EJ, Guilliams T, Wellens J, O’Day EM, Waudby CA, Meehan S et al. (2010) Structure and properties of a complex of α-synuclein and a single-domain camelid antibody. J. Mol. Biol 402, 326–343 10.1016/j.jmb.2010.07.001 [DOI] [PubMed] [Google Scholar]
- 60.Wilton EE, Opyr MP, Kailasam S, Kothe RF and Wieden HJ (2018) sdAb-DB: the single domain antibody database. ACS Synth. Biol 7, 2480–2484 10.1021/acssynbio.8b00407 [DOI] [PubMed] [Google Scholar]
- 61.Harmansa S and Affolter M (2018) Protein binders and their applications in developmental biology. Development 145, dev148874 10.1242/dev.148874 [DOI] [PubMed] [Google Scholar]
- 62.Butkinaree C, Cheung WD, Park S, Park K, Barber M and Hart GW (2008) Characterization of β-N-acetylglucosaminidase cleavage by caspase-3 during apoptosis. J. Biol. Chem 283, 23557–23566 10.1074/jbc.M804116200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Farrants H, Tarnawski M, Muller TG, Otsuka S, Hiblot J, Koch B et al. (2020) Chemogenetic control of nanobodies. Nat. Methods 17, 279–282 10.1038/s41592-020-0746-7 [DOI] [PubMed] [Google Scholar]
- 64.Yu D, Lee H, Hong J, Jung H, Jo Y, Oh BH et al. (2019) Optogenetic activation of intracellular antibodies for direct modulation of endogenous proteins. Nat. Methods 16, 1095–1100 10.1038/s41592-019-0592-7 [DOI] [PubMed] [Google Scholar]
- 65.Jedlitzke B, Yilmaz Z, Dorner W and Mootz HD (2020) Photobodies: light-activatable single-domain antibody fragments. Angew. Chem. Int. Ed. Engl 59, 1506–1510 10.1002/anie.201912286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tang JCY, Drokhlyansky E, Etemad B, Rudolph S, Guo B, Wang S et al. (2016) Detection and manipulation of live antigen-expressing cells using conditionally stable nanobodies. eLife 5, e15312 10.7554/eLife.15312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fridy PC, Li Y, Keegan S, Thompson MK, Nudelman I, Scheid JF et al. (2014) A robust pipeline for rapid production of versatile nanobody repertoires. Nat. Methods 11, 1253–1260 10.1038/nmeth.3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kirchhofer A, Helma J, Schmidthals K, Frauer C, Cui S, Karcher A et al. (2010) Modulation of protein properties in living cells using nanobodies. Nat. Struct. Mol. Biol 17, 133–138 10.1038/nsmb.1727 [DOI] [PubMed] [Google Scholar]
- 69.Ramirez DH, Ge Y and Woo CM (2021) O-GlcNAc engineering on a target protein in cells with nanobody-OGT and nanobody-splitOGA. Curr. Protoc 1, e117 10.1002/cpz1.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tuerk C and Gold L (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510 10.1126/science.2200121 [DOI] [PubMed] [Google Scholar]
- 71.Zhu Y (2019) Regulating O-GlcNAcylation on Specific Proteins Using RNA Aptamers, Johns Hopkins University [Google Scholar]
- 72.Boulard M, Rucli S, Edwards JR and Bestor TH (2020) Methylation-directed glycosylation of chromatin factors represses retrotransposon promoters. Proc. Natl Acad. Sci. U.S.A 117, 14292–8 10.1073/pnas.1912074117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Parker MP, Fedosyuk H, Novikova LV, Slawson C and Peterson KR (2020) Regulation of GATA-1-controlled genes by O-GlcNAcylation in erythroid cells. Blood 136, 46–47 10.1182/blood-2020-141429 [DOI] [Google Scholar]
- 74.Rexach JE, Clark PM, Mason DE, Neve RL, Peters EC and Hsieh-Wilson LC (2012) Dynamic O-GlcNAc modification regulates CREB-mediated gene expression and memory formation. Nat. Chem. Biol 8, 253–261 10.1038/nchembio.770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang WH, Park SY, Nam HW, Kim DH, Kang JG, Kang ES et al. (2008) NFκb activation is associated with its O-GlcNAcylation state under hyperglycemic conditions. Proc. Natl Acad. Sci. U.S.A 105, 17345–17350 10.1073/pnas.0806198105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ma J, Li Y, Hou C and Wu C (2021) O-GlcNAcAtlas: a database of experimentally identified O-GlcNAc sites and proteins. Glycobiology 31, 719–723 10.1093/glycob/cwab003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thompson RE and Muir TW (2020) Chemoenzymatic semisynthesis of proteins. Chem. Rev 120, 3051–3126 10.1021/acs.chemrev.9b00450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schwagerus S, Reimann O, Despres C, Smet-Nocca C and Hackenberger CP (2016) Semi-synthesis of a tag-free O-GlcNAcylated tau protein by sequential chemoselective ligation. J. Pept. Sci 22, 327–333 10.1002/psc.2870 [DOI] [PubMed] [Google Scholar]
- 79.Marotta NP, Lin YH, Lewis YE, Ambroso MR, Zaro BW, Roth MT et al. (2015) O-GlcNAc modification blocks the aggregation and toxicity of the protein (-synuclein associated with Parkinson’s disease. Nat. Chem 7, 913–920 10.1038/nchem.2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Levine PM, Galesic A, Balana AT, Mahul-Mellier AL, Navarro MX, De Leon CA et al. (2019) α-Synuclein O-GlcNAcylation alters aggregation and toxicity, revealing certain residues as potential inhibitors of Parkinson’s disease. Proc. Natl. Acad. Sci. U. S. A 116, 1511–1519 10.1073/pnas.1808845116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lewis YE, Galesic A, Levine PM, De Leon CA, Lamiri N, Brennan CK et al. (2017) O-GlcNAcylation of (-synuclein at serine 87 reduces aggregation without affecting membrane binding. ACS Chem. Biol 12, 1020–1027 10.1021/acschembio.7b00113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lercher L, Raj R, Patel NA, Price J, Mohammed S, Robinson CV et al. (2015) Generation of a synthetic GlcNAcylated nucleosome reveals regulation of stability by H2A-Thr101 GlcNAcylation. Nat. Commun 6, 7978 10.1038/ncomms8978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dadova J, Galan SR and Davis BG (2018) Synthesis of modified proteins via functionalization of dehydroalanine. Curr. Opin. Chem. Biol 46, 71–81 10.1016/j.cbpa.2018.05.022 [DOI] [PubMed] [Google Scholar]
- 84.de la Torre D and Chin JW (2021) Reprogramming the genetic code. Nat. Rev. Genet 22, 169–184 10.1038/s41576-020-00307-7 [DOI] [PubMed] [Google Scholar]
- 85.Kaya E, Gutsmiedl K, Vrabel M, Muller M, Thumbs P and Carell T (2009) Synthesis of threefold glycosylated proteins using click chemistry and genetically encoded unnatural amino acids. Chembiochem 10, 2858–2861 10.1002/cbic.200900625 [DOI] [PubMed] [Google Scholar]
- 86.Liu H, Wang L, Brock A, Wong CH and Schultz PG (2003) A method for the generation of glycoprotein mimetics. J. Am. Chem. Soc 125, 1702–1703 10.1021/ja029433n [DOI] [PubMed] [Google Scholar]
- 87.Yang B, Wang N, Schnier PD, Zheng F, Zhu H, Polizzi NF et al. (2019) Genetically introducing biochemically reactive amino acids dehydroalanine and dehydrobutyrine in proteins. J. Am. Chem. Soc 141, 7698–7703 10.1021/jacs.9b02611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang Z, Gildersleeve J, Yang YY, Xu R, Loo JA, Uryu S et al. (2004) A new strategy for the synthesis of glycoproteins. Science 303, 371–373 10.1126/science.1089509 [DOI] [PubMed] [Google Scholar]
- 89.Chen H, Venkat S, McGuire P, Gan Q and Fan C (2018) Recent development of genetic code expansion for posttranslational modification studies. Molecules 23, 1662 10.3390/molecules23071662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gorelik A, Bartual SG, Borodkin VS, Varghese J, Ferenbach AT and van Aalten DMF (2019) Genetic recoding to dissect the roles of site-specific protein O-GlcNAcylation. Nat. Struct. Mol. Biol 26, 1071–1077 10.1038/s41594-019-0325-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Maynard JC, Burlingame AL and Medzihradszky KF (2016) Cysteine S-linked N-acetylglucosamine (S-GlcNAcylation), a new post-translational modification in mammals. Mol. Cell. Proteomics 15, 3405–3411 10.1074/mcp.M116.061549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rao FV, Dorfmueller HC, Villa F, Allwood M, Eggleston IM and van Aalten DM (2006) Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis. EMBO J. 25, 1569–1578 10.1038/sj.emboj.7601026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Palaniappan KK and Bertozzi CR (2016) Chemical glycoproteomics. Chem. Rev 116, 14277–14306 10.1021/acs.chemrev.6b00023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ma J and Hart GW (2014) O-GlcNAc profiling: from proteins to proteomes. Clin. Proteomics 11, 8 10.1186/1559-0275-11-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Woo CM, Iavarone AT, Spiciarich DR, Palaniappan KK and Bertozzi CR (2015) Isotope-targeted glycoproteomics (IsoTaG): a mass-independent platform for intact N- and O-glycopeptide discovery and analysis. Nat. Methods 12, 561–567 10.1038/nmeth.3366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chung HS, Wang SB, Venkatraman V, Murray CI and Van Eyk JE (2013) Cysteine oxidative posttranslational modifications: emerging regulation in the cardiovascular system. Circ. Res 112, 382–392 10.1161/CIRCRESAHA.112.268680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hoyt EA, Cal PMSD, Oliveira BL and Bernardes GJL (2019) Contemporary approaches to site-selective protein modification. Nat. Rev. Chem 3, 147–171 10.1038/s41570-019-0079-1 [DOI] [Google Scholar]
- 98.De Leon CA, Levine PM, Craven TW and Pratt MR (2017) The sulfur-linked analogue of O-GlcNAc (S-GlcNAc) is an enzymatically stable and reasonable structural surrogate for O-GlcNAc at the peptide and protein levels. Biochemistry 56, 3507–3517 10.1021/acs.biochem.7b00268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kuey C, Larocque G, Clarke NI and Royle SJ (2019) Unintended perturbation of protein function using GFP nanobodies in human cells. J. Cell Sci 132, jcs.234955 10.1242/jcs.234955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fahmi NE, Dedkova L, Wang B, Golovine S and Hecht SM (2007) Site-specific incorporation of glycosylated serine and tyrosine derivatives into proteins. J. Am. Chem. Soc 129, 3586–3597 10.1021/ja067466n [DOI] [PubMed] [Google Scholar]
- 101.Matsubara T, Iijima K, Watanabe T, Hohsaka T and Sato T (2013) Incorporation of glycosylated amino acid into protein by an in vitro translation system. Bioorg. Med. Chem. Lett 23, 5634–5636 10.1016/j.bmcl.2013.08.035 [DOI] [PubMed] [Google Scholar]
- 102.Wright TH, Bower BJ, Chalker JM, Bernardes GJ, Wiewiora R, Ng WL et al. (2016) Posttranslational mutagenesis: a chemical strategy for exploring protein side-chain diversity. Science 354, aag1465 10.1126/science.aag1465 [DOI] [PubMed] [Google Scholar]
- 103.Stephen HM, Adams TM and Wells L (2021) Regulating the regulators: mechanisms of substrate selection of the O-GlcNAc cycling enzymes OGT and OGA. Glycobiology 31, 724–733 10.1093/glycob/cwab005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhu Y and Hart GW (2021) Targeting O-GlcNAcylation to develop novel therapeutics. Mol. Aspects Med 79, 100885 10.1016/j.mam.2020.100885 [DOI] [PubMed] [Google Scholar]


