Abstract
The development of CRISPR-Cas9 mediated gene editing technology is revolutionizing molecular biology, biotechnology, and medicine. However, as with other nucleic acid technologies, CRISPR would greatly benefit from chemical modifications that optimize delivery, activity, and specificity of gene editing. Amide modifications at certain positions of short interfering RNAs have been previously shown to improve their RNAi activity and specificity, which motivated the current study on replacement of selected internucleoside phosphates of CRISPR RNAs with amide linkages. Herein we show that amide modifications did not interfere with CRISPR-Cas9 activity when placed in the protospacer adjacent motif (PAM) distal region of CRISPR RNAs. In contrast, modification of the seed region led to loss of DNA cleavage activity at most but not all positions. These results are encouraging for future studies on amides as backbone modifications in CRISPR RNAs.
Graphical Abstract
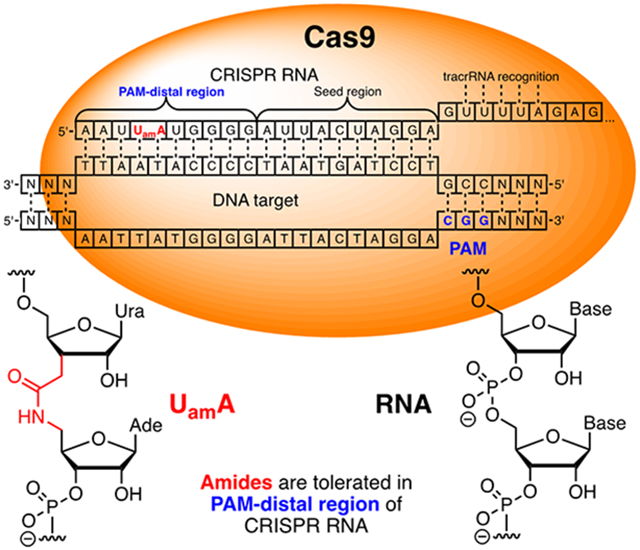
Gene editing using CRISPR (clustered regularly interspaced short palindromic repeats) technology has become a broadly used tool in molecular and synthetic biology and holds exciting promise as a novel therapeutic strategy for treating genetic diseases.1-2 However, as with other nucleic acid driven gene regulation technologies, on-target specificity of CRISPR needs optimization, especially for potential therapeutic applications.3-4 Previous studies have shown that chemical modifications at specific positions in the CRISPR RNA (crRNA) 20-nucleotide guide sequence can significantly reduce off-target DNA cleavage with minimal loss of on-target activity.5-8 For example, replacement of the protospacer adjacent motif (PAM) distal region RNA with DNA nucleotides,5 incorporation of 2′-F,6 locked and bridged7 sugar modifications, and modification of phosphate linkages as 2′-O-methyl-3′-phosphonoacetates8 were well tolerated in the DNA recognition region of crRNAs and, in some cases, reduced undesired cleavage at off-target sites. Other studies have shown that crRNAs can be extensively modified providing that the chemical changes do not interfere with key Cas9-RNA interactions.9-11 In general, modifications were better tolerated in the PAM-distal than in the seed region.5-6, 10 These reports encouraged us to explore if crRNAs could tolerate amide internucleoside linkages (Figure 1) as backbone modifications.
Figure 1.
Structure and sequences of amide-modified crRNAs. The seed region of spacer RNA is underlined, and the repeat region for tracrRNA recognition is shown in outlined font. The position of amide-linked dinucleotide is indicated by bold blue and green fonts.
Our research group previously reported that amides were excellent mimics of internucleoside phosphates in RNA.12 Replacement of individual phosphates with amides was well tolerated in short interfering RNAs13-14 (siRNAs) and even improved the RNAi activity and specificity when placed at certain positions in siRNA guide and passenger strands.14-15 An siRNA guide strand having four consecutive phosphates replaced with amides retained high silencing activity when paired with an optimized amide-modified passenger strand.16 In this Letter, we report the first study of amides as internucleoside linkages in crRNAs. We found that crRNAs modified with amides in the PAM-distal region and at some positions in the seed region retained good Cas9 activity.
Following previous studies by others, we chose 36-nucleotide long crRNAs17 targeting the human vascular endothelial growth factor A (VEGF-A)6 and hypoxanthine phosphoribosyl-transferase 1 (HPRT1)17 genes (Figure 1) to explore the effect of replacing selected internucleoside phosphates with amide linkages on activity of Cas9 complexes. Amide-modified crRNAs (Figure 1) were synthesized following our previously reported approach to modified siRNAs13-15 using amide-linked phosphoramidite dimers UG, UA, and AG. The key challenge for crRNAs that tend to be rich in guanosines was the preparation of an amino-G unit of UG and AG dimers (Scheme 1). The synthesis started with installation of 5′-azide functionality in the known protected guanosine 118 followed by cleavage of the isopropylidene group and 2′-O-protection with TOM. Hydrogenation of azide 4 gave an amine that was directly coupled with uridine carboxylic acid 7, prepared as previously reported.13 Alternatively, installation of the diphenylcarbamoyl (DPC) protection and reduction of the azide gave a guanosine amine 6 that was coupled with adenosine carboxylic acid 10, prepared as previously reported.14 Phosphorylation of dimers 8 and 11 with 2-cyanoethyl N,N,N′,N′-tetraisopropylphosphorodiamidite and 4,5-dicyanoimidazole (DCI)19 gave the UG and AG phosphoramidite dimers 9 and 12, respectively. The corresponding UU and UA phosphoramidite monomers were synthesized as previously reported.13
Scheme 1.
Synthesis of amide-linked phosphoramidite dimers for preparation of amide-modified crRNAs.
Amide-modified crRNAs were synthesized and purified as described in Supporting Information, and their effect on Cas9 activity was studied using the T7 Endonuclease 1 (T7E1) mismatch cleavage (EMC) assay.20 Briefly, [HEK-293]Cas9 cells (ATCC, CRL-1573Cas9 cell line) that constitutively express Cas9 protein were reverse transfected with crRNA:tracrRNA duplexes (Figure 1) using Lipofectamine RNAiMax for 48 h (for details, see Supporting Information).17 The genomic DNA was extracted, the target DNA regions were PCR amplified, and the DNA cleavage activity was measured using the EMC assay (Figures S4-S5, Tables S4-S5).17
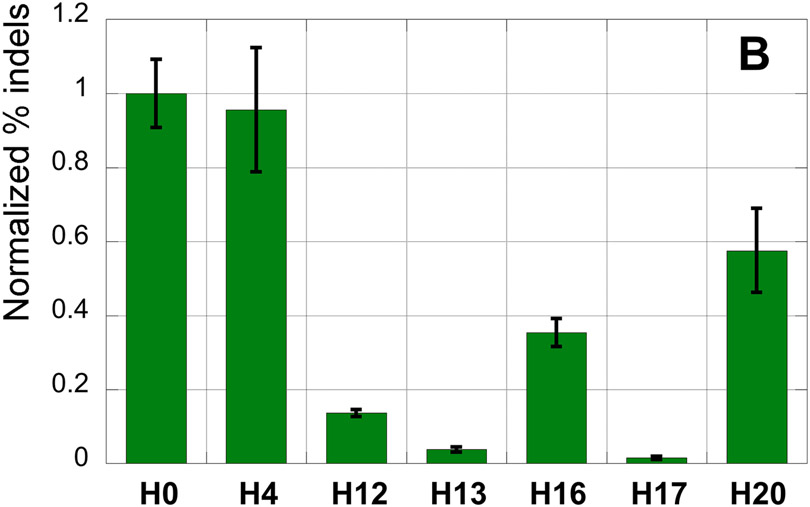
The results, reported as % indels normalized to activity of unmodified crRNAs V0 and H0 (Figure 2), showed that amide modifications in the PAM-distal region in V3 and H4 retained high CRISPR activity. Moving the amide linkage closer to the seed region in V7 and V11 slightly decreased the activity. These results are very encouraging for future studies on amide-modified crRNAs because Ryan et. al. reported that 2′-O-methyl-3′-phosphonoacetate backbone modifications at positions 4, 5, 7, and 11 significantly increased CRISPR on-target specificity.8 Amide modifications of the seed region notably decreased crRNA-guided DNA cleavage activity, which was expected given that previous studies have reported higher sensitivity of the seed region to sugar and backbone modifications.5-6, 10 Interestingly, H16 showed significant CRISPR activity despite having the modification in the center of seed region. This may be promising for future studies as modification of phosphate 16 has increased CRISPR specificity in previous studies.8 Finally, H20 retained more than half of DNA cleavage activity.
Figure 2.
EMC assay results normalized to activity of unmodified crRNAs, V0 and H0: (A) crRNAs targeting VEGF-A gene, and (B) crRNAs targeting HPRT1 gene.
Crystal structures of Cas9 in complex with single guide RNA and target DNA show that Cas9 makes hydrogen bonding contacts to phosphates 1, 3–5, and 11–19.21-22 These contacts are more extensive in the seed region than in the PAM-distal region, which is consistent with overall higher sensitivity of seed to amide modifications. However, there was no direct correlation as V3 and H4, having putative hydrogen bonding contacts to Cas9 at the amide linkages, were more active than V7 or H20 that were not expected to form hydrogen bonds with Cas9. This is similar to our previous observations that RNAi activity of amide-modified siRNAs did not directly correlate with the hydrogen bonding interactions Ago2 makes to siRNA phosphates.14
To obtain preliminary insights about the effect of amide modifications on CRISPR specificity, we explored cleavage activity of three known off-targets of VEGF-A crRNA: MAX gene at chromosome position 14q23, chromosome position 5q14.3, and chromosome 22q13.1.6 In our experimental system, only 5q14.3 showed significant off-target cleavage (Figure S6). Cleavage of 22q13.1 was detected, but at a level too low for reproducible quantification (<1%), and we could not detect any off-target cleavage of the MAX gene. Quantification of 5q14.3 cleavage (Table S7) showed that amide modifications at V3, V7, and V11, positions that maintained high on-target activity (Figure 2), had a small effect on the off-target activity. After normalization for changes in on-target activity, V3 appeared to slightly decrease the off-target cleavage, while V7 and V11 appeared to slightly increase the off-target activity (Figure S7). While these preliminary results did not reveal a strong positive effect from amide modifications, it is conceivable that modification of other positions may improve Cas9 specificity. Testing amide modifications of other PAM-distal internucleoside linkages will require synthesis of other dimers and expanding the scope of crRNA sequences.
It is also possible that despite reduced activity of seed-modified crRNAs, modifications of certain positions may be beneficial if significant increase in specificity could be achieved. For example, Charpentier and co-workers showed that mutations of R63, R66 and R70 to alanine increased the specificity of Cas9.23 These arginines contact phosphates 12–15 of the seed region of crRNA.22 Replacing phosphates with modified internucleoside linkages, such as amides, is an alternative way to modulate Cas9-crRNA interactions and cleavage specificity. In the present study, V13 and V15 decreased on-target activity without suppressing unwanted off-target cleavage (Table S6). Our current experimental set up did not allow us to test modifications of phosphates 12 and 14 that along with 16 (that retained relatively high on-target activity, Figure 2) remain attractive targets to explore in future.
The present study suggests that, similar to Ago2, Cas9 can use RNA guides that have selected phosphates replaced with amide linkages, though the pattern of tolerance is different for Cas9 than Ago2. While in our previous study Ago2 activity was slightly reduced in both seed and supplementary region,14 in the current study Cas9 shows higher sensitivity to seed modifications but good tolerance to modification of the PAM-distal region. Our study is the first demonstration that a non-ionic non-phosphorous backbone modification is tolerated at some positions in crRNAs. These results are promising for future studies on amide-modified crRNAs because backbone modifications in the PAM-distal region have improved CRISPR specificity in previous studies.8
Supplementary Material
ACKNOWLEDGMENT
This work was supported by National Institutes of Health (R35 GM130207 to E.R.).
Footnotes
Supporting Information. General experimental procedures, synthesis, purification, and MS characterization of crRNAs; experimental details and results of EMC assays; NMR spectra of new compounds.
REFERENCES
- (1).Jinek M; Chylinski K; Fonfara I; Hauer M; Doudna JA; Charpentier E, A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Doudna JA, The promise and challenge of therapeutic genome editing. Nature 2020, 578, 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Vakulskas CA; Behlke MA, Evaluation and Reduction of CRISPR Off-Target Cleavage Events. Nucleic Acid Ther. 2019, 29, 167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Wu J; Yin H, Engineering guide RNA to reduce the off-target effects of CRISPR. J. Genet. Genom 2019, 46, 523–529. [DOI] [PubMed] [Google Scholar]
- (5).Yin H; Song C-Q; Suresh S; Kwan S-Y; Wu Q; Walsh S; Ding J; Bogorad RL; Zhu LJ; Wolfe SA; et al. , Partial DNA-guided Cas9 enables genome editing with reduced off-target activity. Nat. Chem. Biol 2018, 14, 311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Rahdar M; McMahon MA; Prakash TP; Swayze EE; Bennett CF; Cleveland DW, Synthetic CRISPR RNA-Cas9-guided genome editing in human cells. Proc. Natl. Acad. Sci. U. S. A 2015, 112, E7110–E7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Cromwell CR; Krysler AR; Hubbard BP; Sung K; Park J; Kim SK; Jovel J, Incorporation of bridged nucleic acids into CRISPR RNAs improves Cas9 endonuclease specificity. Nat. Commun 2018, 9, 1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Ryan DE; Taussig D; Steinfeld I; Phadnis SM; Lunstad BD; Singh M; Vuong X; Okochi KD; McCaffrey R; Olesiak M; et al. , Improving CRISPR-Cas specificity with chemical modifications in single-guide RNAs. Nucleic Acids Res. 2018, 46, 792–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Mir A; Alterman JF; Hassler MR; Debacker AJ; Hudgens E; Echeverria D; Brodsky MH; Khvorova A; Watts JK; Sontheimer EJ, Heavily and fully modified RNAs guide efficient SpyCas9-mediated genome editing. Nat. Commun 2018, 9, 2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Yin H; Song C-Q; Suresh S; Wu Q; Walsh S; Rhym LH; Mintzer E; Bolukbasi MF; Zhu LJ; Kauffman K; et al. , Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat. Biotechnol 2017, 35, 1179–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).O’Reilly D; Kartje ZJ; Ageely EA; Malek-Adamian E; Habibian M; Schofield A; Barkau CL; Rohilla KJ; DeRossett LB; Weigle AT; et al. , Extensive CRISPR RNA modification reveals chemical compatibility and structure-activity relationships for Cas9 biochemical activity. Nucleic Acids Res. 2019, 47, 546–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Kotikam V; Rozners E, Amide-Modified RNA: Using Protein Backbone to Modulate Function of Short Interfering RNAs. Acc. Chem. Res 2020, 53, 1782–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Mutisya D; Selvam C; Lunstad BD; Pallan PS; Haas A; Leake D; Egli M; Rozners E, Amides are excellent mimics of phosphate internucleoside linkages and are well tolerated in short interfering RNAs. Nucleic Acids Res. 2014, 42, 6542–6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Mutisya D; Hardcastle T; Cheruiyot SK; Pallan PS; Kennedy SD; Egli M; Kelley ML; Smith Anja van B.; Rozners E, Amide linkages mimic phosphates in RNA interactions with proteins and are well tolerated in the guide strand of short interfering RNAs. Nucleic Acids Res. 2017, 45 (14), 8142–8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Hardcastle T; Novosjolova I; Kotikam V; Cheruiyot SK; Mutisya D; Kennedy SD; Egli M; Kelley ML; van Brabant Smith A; Rozners E, A Single Amide Linkage in the Passenger Strand Suppresses Its Activity and Enhances Guide Strand Targeting of siRNAs. ACS Chem. Biol 2018, 13, 533–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Kotikam V; Viel JA; Rozners E, Synthesis and biological activity of short interfering RNAs having several consecutive amide internucleoside linkages. Chem. Eur. J 2020, 26, 685–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Jacobi AM; Rettig GR; Turk R; Collingwood MA; Zeiner SA; Quadros RM; Harms DW; Bonthuis PJ; Gregg C; Ohtsuka M; et al. , Simplified CRISPR tools for efficient genome editing and streamlined protocols for their delivery into mammalian cells and mouse zygotes. Methods 2017, 121–122, 16–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Faurel-Paul E; Yoshida K; Sépulcre M; Dhimane H; Merrer YL, Synthesis of Three Regioisomeric (7-Methoxychromenonyl) methyl Guanosine 5′-Phosphates. Synth. Commun 2009, 39, 459–474. [Google Scholar]
- (19).Smicius R; Engels JW, Preparation of Zwitterionic Ribonucleoside Phosphoramidites for Solid-Phase siRNA Synthesis. J. Org. Chem 2008, 73, 4994–5002. [DOI] [PubMed] [Google Scholar]
- (20).Cho SW; Kim S; Kim JM; Kim J-S, Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol 2013, 31, 230–232. [DOI] [PubMed] [Google Scholar]
- (21).Jiang F; Zhou K; Ma L; Gressel S; Doudna JA, A Cas9-guide RNA complex preorganized for target DNA recognition. Science 2015, 348, 1477–1481. [DOI] [PubMed] [Google Scholar]
- (22).Nishimasu H; Ran FA; Hsu PD; Konermann S; Shehata SI; Dohmae N; Ishitani R; Zhang F; Nureki O, Crystal Structure of Cas9 in Complex with Guide RNA and Target DNA. Cell 2014, 156, 935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Bratovič M; Fonfara I; Chylinski K; Gálvez EJC; Sullivan TJ; Boerno S; Timmermann B; Boettcher M; Charpentier E, Bridge helix arginines play a critical role in Cas9 sensitivity to mismatches. Nat. Chem. Biol 2020, 16, 587–595. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.