Abstract
CRISPR has become a cutting-edge research method and holds great potential to revolutionize biotechnology and medicine. However, like other nucleic acid technologies, CRISPR will greatly benefit from chemical innovation to improve activity and specificity for critical in vivo applications. Chemists have started optimizing various components of the CRISPR system; the present Perspective focuses on chemical modifications of CRISPR RNAs (crRNAs). As with other nucleic acid-based technologies, early efforts focused on well-established sugar and backbone modifications (2′-deoxy, 2′-F, 2′-OMe, and phosphorothioates). Some more significant alterations of crRNAs have been done using bicyclic (locked) riboses and phosphate backbone replacements (phosphonoacetates and amides); however, the range of chemical innovation applied to crRNAs remains limited to modifications that have been successful in RNA interference and antisense technologies. The encouraging results given by these tried-and-true modifications suggest that, going forward, chemists should take a bolder approach – research must aim to investigate what chemistry will have most impact on maturing CRISPR as therapeutic and other in vivo technologies. With an eye to the future, this Perspective argues that the complexity of CRISPR presents rich unprecedented opportunities for chemists to synergize advances in synthetic methodology and structural biochemistry to rationally optimize crRNA-protein interactions.
Graphical Abstract
1. INTRODUCTION
CRISPR (clustered, regularly interspaced, short palindromic repeats) RNA and Cas (CRISPR-associated) proteins form a unique RNA-guided bacterial immune defense system developed to cleave DNA of invading viruses and plasmids.1-3 Because CRISPR RNA (crRNA) can be easily programmed to cleave almost any DNA (or RNA in case of Cas13) target, CRISPR-Cas has been adopted for gene editing and has become an important and robust tool with broad applications in molecular biology and genomics.1, 4-6 CRISPR-Cas also has an intriguing potential to become a novel therapeutic approach.5, 7-9 Mechanistic and structural aspects of RNA-Cas complexes have been extensively covered in recent literature2-3, 10-12 and will not be discussed in detail here. The present Perspective focuses on chemical approaches to improve CRISPR-Cas activity and specificity through rational modification of crRNAs for practical applications as research tools and therapeutic modalities.
CRISPR–Cas systems are broadly divided in two major classes and six types.3, 13-15 Class 1 uses multiprotein-crRNA effector complexes and is further divided in types I, III, and IV depending on the specific Cas proteins involved. Class 2 uses a single Cas protein as an endonuclease effector and is further divided in types II, V, and VI.14 Type II, characterized by the signature endonuclease Cas9 is the most extensively studied CRISPR system.16 Accordingly, the majority of the chemical modifications, as will be discussed below, have been studied in Cas9 crRNAs. Recently, crRNAs of Cas12a (type V system) and Cas13d (type VI-D system) have also been modified and will be briefly reviewed.
Cleavage of double-stranded DNA (dsDNA) initiates cellular repair mechanisms that can be repurposed for rational gene editing.1, 4, 17 The predominant pathway of dsDNA break repair is non-homologous end joining (NHEJ). NHEJ is an error-prone repair mechanism that results in small insertions or deletions of DNA at the repair site, the so called indels.17 While NHEJ is usually not the desired pathway for precise gene editing, creations of indels enables a convenient method to quantify CRISPR activity using endonucleases (Surveyor or T7EI) that selectively cleave the mismatched indels created by NHEJ of cleaved DNA targets. Precise gene editing is usually achieved through homology-directed repair (HDR) by co-delivering a dsDNA donor fragment with sequence homologous to target DNA but containing the desired edit.17
This Perspective focuses on recent studies that used chemical modifications to optimize crRNAs. As CRISPR is a relatively young technology, the efforts to modify crRNAs started only in 201418 and are still in early stages. So far chemists have tested mostly the sugar and backbone modifications that have been well-established to work in antisense and RNA interference (for related recent reviews see refs.19-21). This Perspective concludes that given the significant advances in synthetic methodology and structural biochemistry, we are well positioned to pursue more innovative and bolder approaches that would rationally modify crRNAs to optimize CRISPR activity, specificity, and other technological aspects for in vivo applications in medicine and biotechnology.
2. MODIFICATIONS OF Cas9 CRISPR RNAs
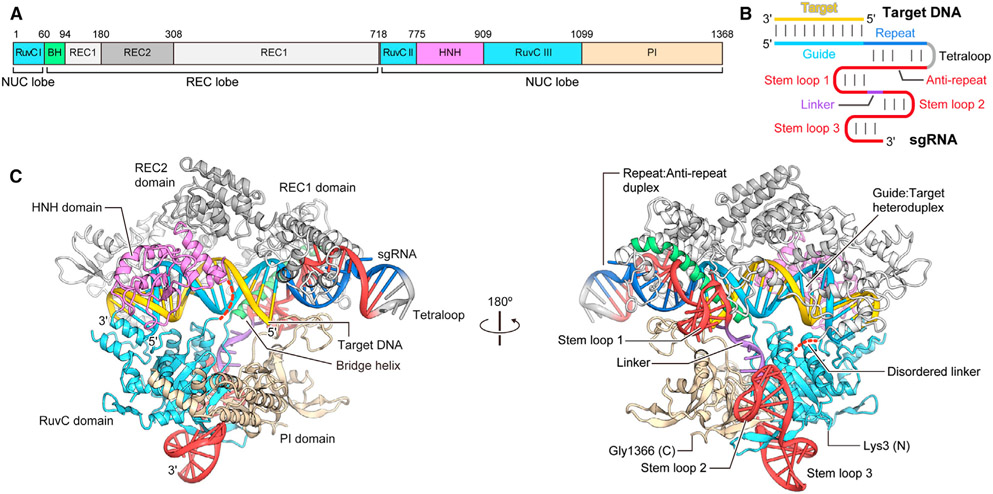
Among various Cas9 proteins, Streptococcus pyogenes Cas9 (SpCas9) is the most studied endonuclease featuring a bilobed architecture built of target recognition (REC lobe, Figure 1) and nuclease (NUC lobe) parts.22 The guide RNA/DNA complex (blue and yellow in Figure 1 B and C) is accommodated at the interface of REC and NUC lobes.
Figure 1.
Structure of the Streptococcus pyogenes Cas9/single guide RNA (sgRNA)/target DNA ternary complex published by Zhang, Nureki and co-workers.22 A Domain organization of SpCas9, BH denotes the bridge helix; B Schematic representation of the sgRNA/DNA complex; C Ribbon representation of the Cas9/sgRNA/DNA complex. Disordered linkers are shown as red dotted lines. Reproduced with permission from ref . 22 Copyright (2014) Elsevier.
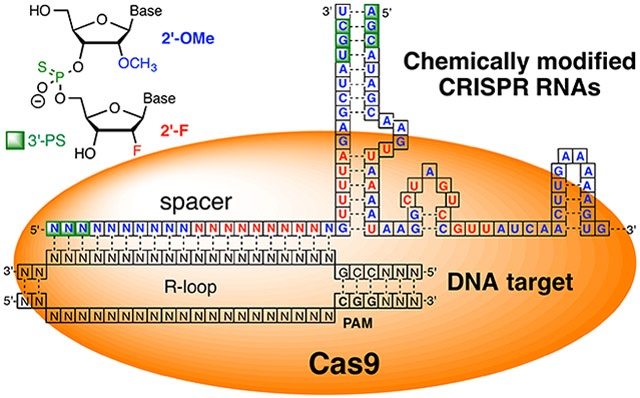
Native Cas9 uses two RNAs, a 42-nucleotide long crRNA and an 80-nucleotide long trans-activating crRNA (tracrRNA), to guide sequence specific recognition and cleavage of dsDNA.10, 23-24 crRNA and tracrRNA form a partially complementary complex (Figure 2), where crRNA carries the DNA recognition sequence called spacer, acquired from past invading viruses and plasmids,1-3 and tracrRNA is responsible for association with Cas9 protein. In genome editing and other technologies it is common to link the two RNAs together through a GAAA tetraloop in a 102-nucleotide long single guide RNA (sgRNA, see Figure 2).23
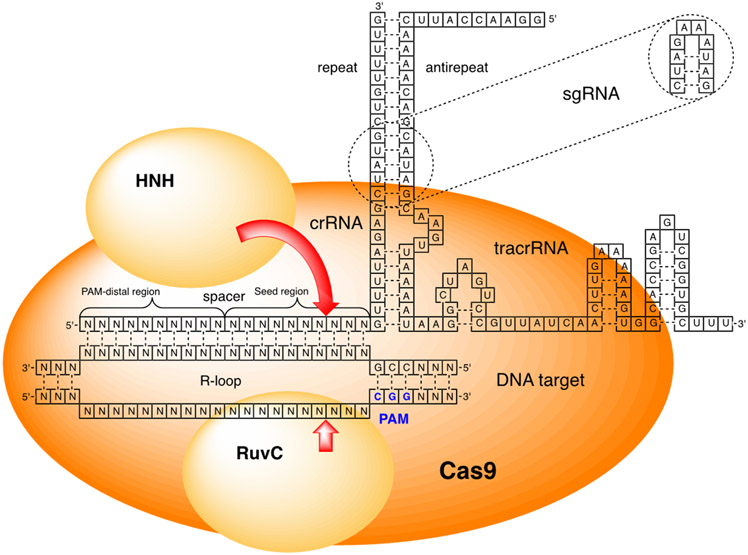
Figure 2.
Cartoon representation of SpCas9 protein, crRNA, and tracrRNA complex bound to dsDNA target. Spacer designates the crRNA’s sequence that base pairs and recognizes the DNA target; PAM - protospacer adjacent motif is a short sequence recognized by Cas proteins.
The DNA binding and cleaving by Cas9-RNA complex starts with recognition of a short DNA sequence, 5′-NGG-3′, called protospacer adjacent motif (PAM, where N can be any nucleotide). Cas9 uses two arginine residues to sequence specifically recognize guanidines of 5′-NGG-3′ that enables CRISPR to distinguish foreign DNA targets having PAM from self-DNA (the acquired spacer stored at CRISPR locus) where the is PAM absent. Binding of PAM by Cas9 triggers conformational relaxing and unwinding of dsDNA, first exposing the PAM-proximal nucleotides to recognition by the seed region of spacer (~10 nucleotides) followed by base pairing between the entire 20-nucleotide long 5′-end of crRNA and the target DNA, which leads to displacement of the non-target strand as an R-loop (Figure 2).23-24 Formation of the R-loop is a two-step process proceeding through an intermediate state having only partial unwinding of the PAM-proximal 8-12 base pairs (seed region).25 Mismatches in the seed region destabilize the intermediate state and strongly inhibit R-loop formation. Mismatches in the PAM-distal region are more tolerated and may allow R-loop formation under certain conditions. For example, the fully unwound R-loop can be formed in presence of PAM-distal and even some PAM-proximal mismatches in negatively supercoiled DNA that may be present under physiological conditions during transcription and replication.25 The catalytically inactive intermediate state serves as a checkpoint for mismatched base pairing in spacer-target DNA. Following the R-loop formation, the HNH nuclease domain of Cas9 undergoes conformational change to the final catalytically competent docked state (D state).26-27 Only after docking in the D state does HNH cleave the target strand, while the RuvC nuclease domain cleaves the displaced nontarget strand three base pairs upstream of the PAM (Figure 2).26-27 The conformational change of HNH domain allosterically controls the RuvC domain nuclease activity; both DNA strands are cleaved at the same time.26 Molecular dynamics symulations proposed that the allosteric control starts with PAM binding activating the HNH and RuvC domains, and is followed by allosteric control of DNA cleavage at the target as well at off-target sites.28 The mechanistic complexity of DNA recognition and mismatch checking provides rich opportunities for chemists to modulate Cas9 specificity using chemical modifications of crRNAs.
One of the first proposed and most direct ways of improving the specificity of crRNA was reducing the number of base pairs in the spacer-target DNA complex. Joung and co-workers18 reported that truncating the 3′-end of the sgRNA spacer from 20 to 17 nucleotides retained on-target cleavage activity for most DNA targets, while significantly reducing off-target cleavage. This study used human U2OS.EGFP cells constitutively expressing an EGFP reporter gene and measured indel frequencies using a T7 Endonuclease I (T7EI) mismatch cleavage assay.29 Using high-throughput next generation sequencing (NGS), the authors estimated as much as 5,000 times reduction of the off-target activity in some cases.18 The authors proposed that the observed improvement in specificity might have been due to reduced binding energy between sgRNA and its DNA target. However, later studies showed that truncation of crRNA increased Cas9 specificity by preventing the conformational transition of HNH from the intermediate to docked D state (conformational checkpoint) rather than simply reducing the affinity of crRNA for mismatched targets.27 These studies illustrate the potential of using a mechanistic understanding of how the Cas-crRNA complex recognizes and selects cognate DNA targets to optimize cleavage specificity using rationally designed chemical modifications.
2.1. Synthesis of chemically modified crRNAs
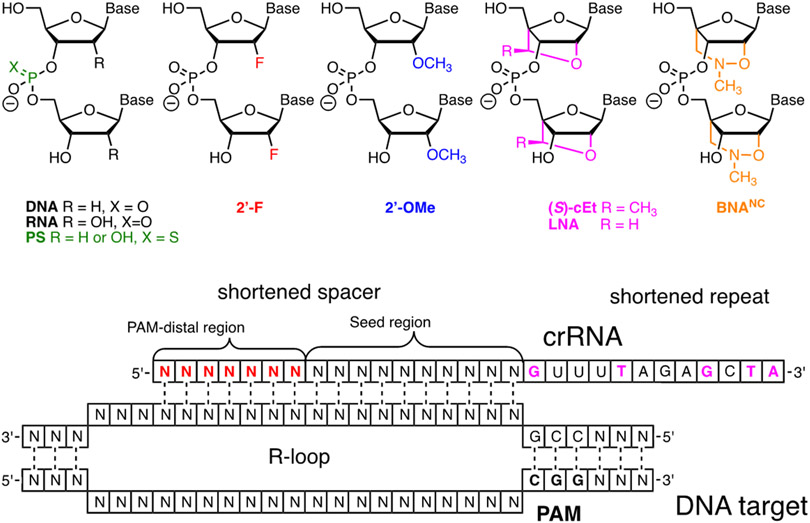
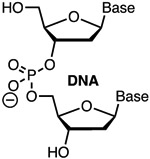
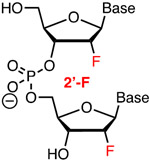
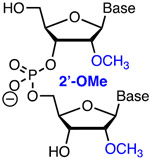
Modification of crRNA requires solid phase chemical synthesis, which, given that the size of these molecules varies from 40- to >100-nucleotides in length, is not a trivial task. The development of phosphoroamidite chemistry, improved 2′-OH protecting groups, and automated solid phase synthesis protocols has enabled commercial synthesis of long RNA molecules.30-31 Hence, a researcher may be able to procure crRNA molecules having common chemical modifications from commercial vendors, though at a relatively high cost. For example, at the time of writing, Dharmacon (Horizon Discovery Biosciences Ltd.) offered HPLC-purified unmodified crRNAs as well as custom made RNAs having 2′-deoxy (DNA), 2′-F, 2′-OMe, phosphorothioate (Figure 3), and a variety of base-modified nucleotides. Incorporation of other modifications discussed below requires the research laboratories to have their own in-house oligonucleotide synthesis capability. Some phosphoroamidite building blocks may be available form commercial vendors (e.g., Glen Research offers LNA phosphoroamidites in addition to 2′-F and 2′-OMe monomers) while more complex and novel modifications will require researcher to make their own modified phosphoroamidites. The latter opens rich opportunities for collaboration between laboratories exploring CRISPR biology and those having expertise in chemical modifications of nucleic acids.
Figure 3.
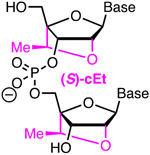
Top: chemical modifications of backbone (phosphorothioate) and ribose tested in crRNAs. Bottom: optimized design of ribose-modified truncated crRNA developed by Bennett, Cleveland, and co-workers.42
2.2. Ribose modifications of Cas9 crRNAs: DNA
In the context of nucleic acid chemistry, replacement of ribonucleotides with DNA counterparts is one of the simplest and most effective crRNA modifications. DNA is cheaper, easier to synthesize, and more stable in biological media than RNA, which are all obvious advantages of the crRNA/DNA chimeras. A disadvantage of DNA is the lack of 2′-OH, which may be problematic at positions where 2′-OH of crRNA or tracrRNA interact with Cas protein. Gagnon and co-workers reported that crRNA having up to 18 5′-terminal ribonucleotides replaced with deoxyribonucleotides retained DNA cleavage activity in an in vitro assay using purified SpCas9.32 However, a study by Xue, Langer, Anderson and co-workers found that DNA modification of guides was less tolerated in assays using HEK293T cells infected with lentiviruses to constitutively express GFP and SpCas9.33 The latter study found that replacement of eight 5′-terminal ribonucleotides of crRNA or sgRNAs with deoxyribonucleotides did not reduce on-target genome editing activity, while significantly lowering off-target activity of Cas9.33 crRNA and sgRNAs with 12 5′-terminal deoxyribonucleotides retained useful activity, but 14 5′-terminal deoxyribonucleotides caused complete loss of activity. In this study, off-target activity of crRNA targeting the VEGF-A gene was reduced from 10-15% off-target indels for all-ribo crRNA to 0-4% off-target indels for crRNA having 10 5′-terminal deoxyribonucleotides.33 GUIDE-seq analysis34 of three other 10-DNA modified crRNA targeting mouse Pcsk9, human EMX1, and human 293 site 4 showed no detectable off-target activity in mouse Hepa1-6 liver cells or human HEK293 cells.33 crRNAs also tolerated 16 3′-terminal deoxyribonucleotides, which led to an optimized crRNA design having eight 5′-terminal and 16 3′-terminal deoxyribonucleotides.33
The discrepancy between results of in vitro and cellular assays is currently not well understood and, most likely, is a notable consequence of the complexity of CRISPR-Cas biology. Other studies discussed below have generally found that modified crRNAs were significantly more active in assays using purified recombinant Cas9 than when endogenous DNA cleavage was assayed in cells.35-36 Furthermore, the results of cellular assays depend on assay format, which is an inherent challenge when trying to compare the effect of chemical modifications observed in various studies. In general, cellular assays fall in one of the two broad groups: assays using cells constitutively expressing a fluorescent reporter gene (typically, EGFP) or assays using endonuclease (Surveyor or T7EI) mismatch cleavage of endogenously edited DNA targets. These assays may also differ by cell line and transfection protocols used, wherein Cas9 may be constitutively expressed in cells or co-delivered with crRNAs. Several studies have also used advanced sequencing techniques to measure on- and off-target activity of modified crRNAs. To help the reader through these complexities, this Perspective briefly mentions the specific assays together with the principal results of each study. The pros and cons of each modification, and the key results and assay formats of each study are also summarized in Table 1. Future studies would greatly benefit from chemists adopting more uniform and well characterized cellular assays that would improve reproducibility of results. For example, HEK293 cells constitutively expressing Cas9 are commercially available and would increase the robustness of assays by eliminating variability in transfection of Cas9 protein or mRNA.
Table 1.
Summary of chemical modifications of Cas9 crRNAs
| Modification | Prosa | Consb | Reference | Key result | Cells | Assays |
|---|---|---|---|---|---|---|
|
Comav Inexpn Simple | Modstb | Gagnon32 | 18 5′-terminal DNA tolerated in crRNA | In vitro | In vitro |
| Xue, Langer, Anderson33 | 10 5′-terminal DNA in crRNAs abolish off-target activity in Hepa1-6 and HEK293 cells. Optimized crRNA has 8 5′- and 16 3′-DNA | HEK293T cells constitutively expressing GFP and Cas9 | GFP fluorescence, GUIDE-seq | |||
|
Comav Affinity | Modstb | Bennett, Cleveland42 | Seven 5′-terminal 2′-F, part of optimized crRNA that reduced off-target activity | HEK293T, tracrRNA and Cas9 plasmid | Surveyor |
| Anderson58 | Structure-guided 2′-F modification of spacer is tolerated in highly modified sgRNA | In vivo (mouse liver) | TIDE | |||
| Khvorova, Watts, Sontheimer59 | Structure-guided modification of >80% as 2′-OMe or 2′-F in crRNA/tracrRNA increases activity | HEK293T cells constitutively expressing mCherry | mCherry fluorescence, TIDE | |||
|
Comav Inexpn Affinity Metstab | Steric | Bruhn, Porteus41 | Three 3′- and 5′-terminal 2′-OMe slightly increased the activity of sgRNA | K562, CD34+ and T cells, Cas9 plasmid | T7EI, TIDE |
| Anderson58 | Structure-guided 2′-OMe modification is tolerated in invariable region (but not in spacer) of highly modified sgRNA | In vivo (mouse liver) | TIDE | |||
| Khvorova, Watts, Sontheimer59 | Structure-guided modification of >80% as 2′-OMe or 2′-F in crRNA/tracrRNA increases activity | HEK293T cells constitutively expressing mCherry | mCherry fluorescence, TIDE | |||
|
Affinity Metstab | Synth Costly Steric | Bennett, Cleveland42 | Five (S)-cEt in shortened (12-nt) repeat region part of optimized crRNA that reduced off-target activity | HEK293T, tracrRNA and Cas9 plasmid | Surveyor |
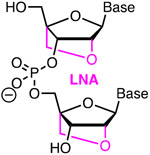
|
Amidite Affinity Metstab | Toxic Costly Steric | Hubbard43 | Up to six LNA are tolerated but less effective than BNANC in reducing off-target activity | U2OS and HeLa cells constitutively expressing Cas9 | T7EI, NGS |
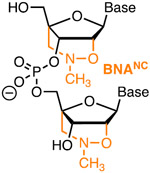
|
Affinity | Synth Costly Steric | Hubbard43 | Up to six BNANC slightly reduce Cas9 on-target activity while broadly reducing off-target activity | U2OS and HeLa cells constitutively expressing Cas9 | T7EI, NGS |
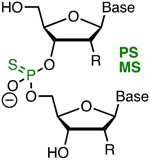
|
Comav Metstab Inexpn Simple Phrmkin | Lowaff Toxic Diast | Bennett, Cleveland42 | Full PS (R = OH) modification of crRNA increased activity ~four-fold | HEK293T, tracrRNA and Cas9 plasmid | Surveyor |
| Bruhn, Porteus41 | Three 3′- and 5′-terminal MS (R = OMe) increased the activity of sgRNA | K562, CD34+ and T cells, Cas9 plasmid | T7EI, TIDE | |||
| Anderson58 | Structure-guided PS modification is tolerated at the 3′- and 5′-ends of highly modified sgRNA | In vivo (mouse liver) | TIDE | |||
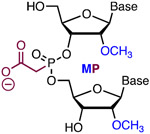
|
Simple Metstab | Costly | Bruhn50 | MP of phosphates 4, 5 and 7; 9-12; and 14 and 16 improved specificity >10-fold | K562, iPS, and CD34+ cells | NGS |
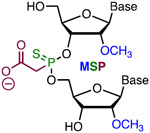
|
Simple Metstab | Costly | Bruhn, Porteus41 | Three 3′- and 5′-terminal MSP strongly increased the activity of sgRNA | K562, CD34+ and T cells, Cas9 plasmid | T7EI, TIDE |
|
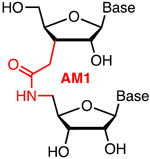
Metstab | Synth Costly | Rozners51 | Individual AM1 were well tolerated in the PAM-distal region of crRNAs | HEK293 cells constitutively expressing Cas9 | T7EI |
Abbreviations for Pros: Affinity – enhanced affinity for complementary nucleic acids, Amidite – amidite is commercially available, Comav – commercially available as an RNA modification, Inexpn – inexpensive to manufacture, Metstab – enhanced metabolic stability, Phrmkin – improved pharmacokinetics, Simple – simple synthesis.
Abbreviations for Cons: Costly – high cost, Diast – diastereomeric mixtures lead to heterogeneous compounds, Lowaff – reduced affinity for complementary nucleic acids, Modstb – modest metabolic stability, Steric – steric bulk may interfere with RNA-protein interactions, Synth – multistep synthesis of monomers required, Toxic – potential toxicity concerns.
2.3. Ribose modifications of Cas9 crRNAs: 2′-F, 2′-OMe and bicyclic riboses
Ribose modifications have been extensively used in antisense oligonucleotides and short interfering RNAs (siRNAs).37-38 The ribose modifications discussed below: 2′-F, 2′-OMe, cEt, LNA, and BNANC share a common advantage of increased affinity for complementary nucleic acids because they drive the ribose towards an RNA-like C3′-endo conformation, which results in a more stable A-type duplexes.37 Among these modifications, 2′-F are the most stabilizing because the strongly electronegative fluorine also enhances Watson-Crick H-bonding and base stacking of the modified duplexes.39 Extensive 2′-F modification is well tolerated in siRNAs where it improves their drug-like properties.40 2′-F, 2′-OMe, and cEt are used to increase metabolic stability of antisense oligonucleotides, while the use of LNA is limited by hepatotoxicity.37 2′-OMe is also used in siRNAs to improve metabolic stability and suppress off-target activity.
In one of the first studies on chemically altered sgRNAs, modification of three 3′- and 5′-terminal nucleotides as 2′-OMe (Figure 3) slightly increased the activity of sgRNA, most likely due to metabolic stabilization of the modified sgRNA, but the benefit was relatively small compared to other modifications (vide supra).41 Another pioneering study by Bennett, Cleveland, and co-workers found that 2′-F modifications were well-tolerated in the PAM-distal region of crRNA’s spacer as measured by Surveyor assay in HEK293T cells (transfected with a plasmid encoding tracrRNA and Cas9 protein).42 These authors developed a truncated 29-nucleotide long crRNA (Figure 3) optimized with seven 5′-terminal 2′-F modifications (the spacer was shortened to 17 nucleotides) combined with five S-constrained ethyl (cEt) substitutions in the shortened (12 nucleotides) repeat region that recognizes tracrRNA.42 The latter enhanced stability of the crRNA-tracrRNA complex, which was critical for the activity of the truncated crRNAs. When targeting VEGF-A, the authors observed a decrease of cleavage at two known off-target sites, the MAX gene and chromosome location 5q14.3, from ~5% for full size unmodified sgRNA to <1% for the optimized 29-nucleotide crRNA; the off-target activity at a third site, chromosome location 22q13.1 was less significant.42
Damha, Gagnon and co-workers reported that fully 2′-F modified crRNAs had similar activity as unmodified crRNAs in an in vitro assay using purified SpCas9.35 However, the fully 2′-F modified crRNA showed little activity in HEK293 cells engineered to stably express GFP and SpCas9.35 Hubbard and co-workers showed that up to six bridged nucleic acid modifications (Figure 3, BNANC) were well tolerated in crRNAs.43 When incorporated as clusters of two or three consecutive modifications BNANC slightly reduced Cas9 on-target activity while broadly reducing off-target DNA cleavage by several orders of magnitude in vitro and in cells. This study used T7EI and NGS assays in U2OS and HeLa cells constitutively expressing Cas9. Interestingly, three consecutive BNANC modifications in the seed region provided the strongest suppression of off-target activity while having relatively modest negative impact on on-target activity. For example, modification of GAA at positions 10-12 (counting from the 2′-end) of WAS crRNA reduced off-target cleavage 10- to 50-fold, while modification of GAA at positions 15-17 of EMX1 crRNA completely eliminated off-target cleavage.43 In both cases the on-target activity was reduced only less that 2-fold. The related locked nucleic acid (LNA) modifications also improved Cas9 specificity, although less than the BNANC modifications. Single-molecule FRET experiments suggested that BNANC modification improved Cas9 specificity by disfavoring the conformational transition of the intermediate to docked state for off-target sequences.43
2.4. Phosphate backbone modifications of Cas9 crRNAs
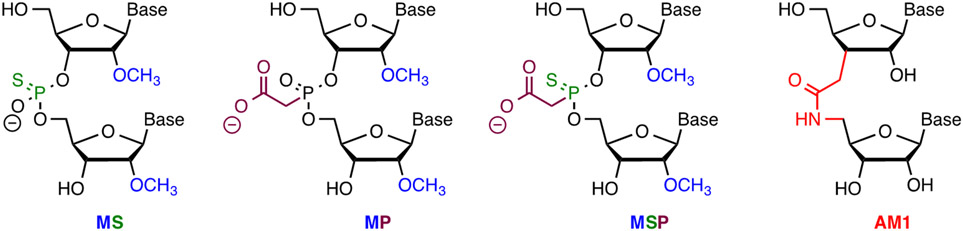
Replacement of non-bridging phosphate oxygen with sulfur creates a phosphorothioate internucleotide linkage (PS, Figure 3),44 one of the most widely used modifications in antisense oligonucleotides and siRNAs.37 PS modifications are easy to synthesize, improve enzymatic stability, but reduce the affinity of modified oligonucleotides for their complementary targets. A minor complication with PS linkages is their chirality that leads to heterogenous diastereomeric mixtures of PS modified oligonucleotides. A major advantage of PS is that they increase oligonucleotide binding to plasma proteins, which significantly improves their pharmacokinetics by slowing down renal excretion. Limited PS modifications are well tolerated in therapeutic DNA and RNA and improve their metabolic stability, delivery, and bioavailability; however, extensive PS modification may lead to toxicity.45-47 PS alone or in combination with other modifications has been used in extensively modified crRNAs, as discussed below.
2′-O-Methyl 3′-phosphonoacetate (2′-O-Me 3′-PACE or MP, Figure 4) and 2′-O-methyl 3′-thiophosphonoacetate (2′-O-Me 3′-thioPACE or MPS) are backbone modifications that improve metabolic stability while maintaining high stability of modified RNA duplexes.48-49 Because the negative charge of 2′-O-Me PACE and 2′-O-Me thioPACE linkages resides on the carboxylate residue, these modifications may engage in interactions with Cas proteins that are not accessible for unmodified phosphates. Modification of the three 3′- and 5′-terminal nucleotides of sgRNAs in both ribose and phosphate backbone to 2′-O-Me 3′-PS (MS, Figure 4) or 2′-O-Me 3′-thioPACE (MSP) significantly increased indel formation and editing by homologous recombination of three target genes (IL2RG, HBB, and CCR5) as measured by T7EI assays, tracking of indels by decomposition (TIDE) analysis, and deep sequencing of PCR amplicons in human K562 cells.41 The increased editing efficiency was consistent with enhanced intracellular stability of the end-modified sgRNAs in human cells. Along similar lines, Bennett, Cleveland, and co-workers reported that full PS-modified crRNA was about four times more active than unmodified crRNA, and the activity was further enhanced by addition of five 2′-O-Me modifications (resulting in MS modifications) at each terminus of the full phosphorothioate-modified crRNA.42
Figure 4.
Chemical modifications of phosphate backbone tested in crRNAs.
Bruhn and co-workers systematically replaced the phosphates within the spacer region of sgRNAs one by one with 2′-O-Me 3′-PACE (MP) linkages.50 The results identified three main regions where the MP modification improved Cas9 specificity by an order of magnitude or greater without significant loss of on-target activity. Specifically, modification of phosphates 4, 5 and 7; 9-12; and 14 and 16 provided remarkable improvements in in vitro assays.50 Interestingly, phosphate 15 stood out as non-tolerant to MP modification. Using deep sequencing of PCR amplicons the authors showed that MP modification of positions 5, 7, and 11 gave ~10-fold or greater reduction of off-target activity for IL2RG, VEGFA and HBB sgRNAs in cellular assays (K562, iPS, and CD34+ cells).50 The authors proposed that the MP modifications improved Cas9 specificity by destabilizing sgRNA base pairing with DNA target, which may lead to rejection of off-target sequences having mismatched base pairs during the R-loop formation. However, this proposal was based on discussion of related literature data and was not specifically tested with experiments in this study.
Our research group replaced selected individual phosphates in crRNAs targeting VEGF-A and HPRT1 genes with amide linkages (AM1, Figure 4).51 AM1 linkages, first reported as DNA modifications in 1993,52-53 are surprisingly good mimics of phosphates in RNA54 that are well tolerated in siRNAs55 where they suppress off-target activity when incorporated at the first nucleotide of the passenger strand.56 AM1 having H-bond donor (N-H) and acceptor (C=O) functionality may engage in interactions with Argonaute and Cas proteins that are not accessible for phosphates, which act only as H-bond acceptors.55 T7EI assays in HEK293 cells constitutively expressing Cas9 showed that amide modifications were well tolerated in the PAM-distal regions of these crRNAs, but decreased the DNA cleavage activity when placed in the seed region.51 These preliminary results are encouraging and should stimulate future studies on amides and other novel crRNA modifications as potential tools to modulate CRISPR activity and specificity.
2.5. Combining ribose and phosphate backbone modifications
Several studies have attempted extensive modification of crRNAs by combining various sugar and backbone chemistries.57-59 Smith and co-workers reported that minimal 2′-O-methyl phosphorothioate modification of terminal nucleotides (two 5′-MS in crRNA with two 5′- or 3′-MS in tracrRNA) gave a modest increase of DNA cleavage activity (measured using the T7EI assay) when crRNA and tracrRNA were lipid co-transfected with Cas9 mRNA or Cas9 protein in U2OS and HeLa cells.57 These authors reported a decrease in cell viability with more extensive modification patterns.
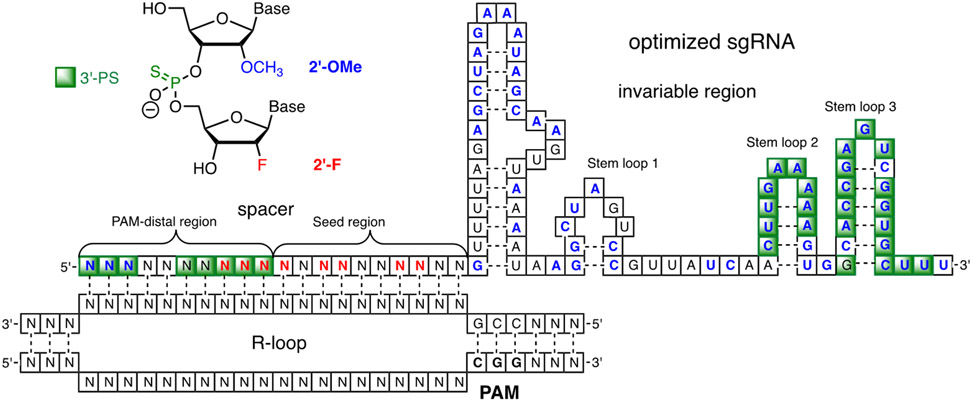
Anderson and co-workers used a structure-guided approach to explore the limits of sgRNA modification with the goal of stabilizing the guide RNA for the in vivo application of CRISPR technology.58 This study used HEK293 cells engineered to stably express GFP and SpCas9 to evaluate the activity of modified sgRNAs. Initially, the authors evaluated modification of the invariable (repeat-antirepeat) and spacer regions separately. Complete modification of the invariable region of sgRNA as 2′-OMe or as phosphorothioate RNA, or modification of all pyrimidines to 2′-F nucleotides, abolished sgRNA activity. The authors concluded that the invariable part of the sgRNA cannot be fully modified, and that even partial modification can significantly lower the activity of sgRNAs. In contrast, structure-guided modification of nucleotides that do not have their 2′-OH groups interacting with Cas9 in crystal structures22, 60 gave a highly active sgRNA having 60 out of 81 invariable region nucleotides modified with 2′-OMe. A similar approach to introducing phosphorothioate linkages in place of phosphates that do not interact with Cas9 gave further improvement of activity of the sgRNA modified with 2′-OMe and PS in the invariable region (Figure 5).58
Figure 5.
Modification pattern of sgRNA optimized by Anderson and co-workers.58
Complete modification of the spacer region or modification of the ten seed nucleotides abolished (in case of 2′-OMe) or significantly decreased (in case of 2′-F) crRNA activity, while modification of the ten PAM-distal region nucleotides was better tolerated with only some decrease in activity. As with the invariable region, structure-guided modification of ten nucleotides that do not interact with Cas9 slightly increased the activity of crRNA compared to the unmodified counterpart. A combination of structure-guided 2′-F modifications and substitution of spacer phosphates that do not interact with Cas9 with phosphorothioate linkages provided a further increase in crRNA activity; however, similar combination of 2′-OMe and phosphorothioate modifications was not tolerated.58
Combining the best modification patterns of invariable and spacer regions in the optimized sgRNA (Figure 5) showed robust DNA cleavage of several targets in mouse liver (>40% indels) which was significantly higher than activity of sgRNA having only three terminal modifications (~20% indels) or unmodified sgRNA (<5% indels). The authors hypothesized that the observed activity enhancement was due to increased metabolic stability of the modified RNA guides.58
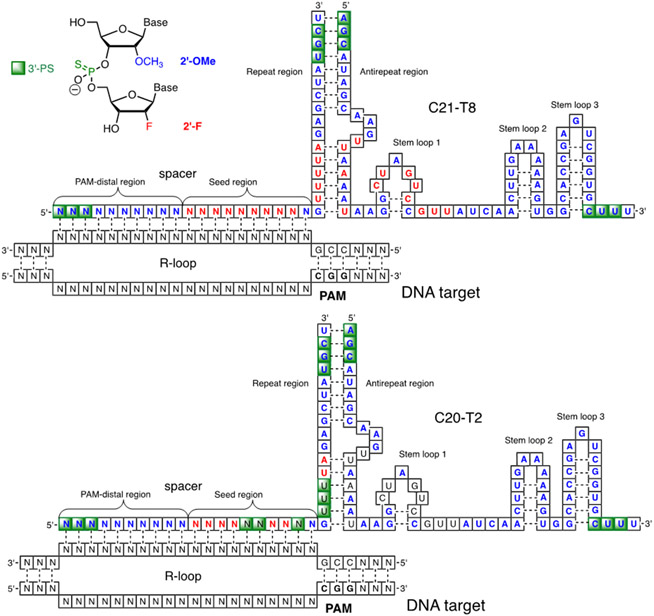
A team led by Khvorova, Watts, and Sontheimer explored the possibility to modify all 2′-OH groups in crRNA and tracrRNA.59 This study used HEK293 cells engineered to stably express GFP followed by an out-of-frame mCherry as fluorescent reporters. DNA cleavage followed by NHEJ repair places mCherry in-frame, resulting in a fluorescence signal. The results were confirmed by TIDE analysis. The fully modified crRNA/tracrRNA complex had all ribonucleotides replaced with 2′-OMe or 2′-F nucleotides (C21-T8, Figure 6) and also had three phosphorothioate linkages at each end of the crRNA and tracrRNA strands.59 2′-F was used at positions that were sensitive to 2′-OMe modifications such as the seed region, repeat-antirepeat helix, and stem loop 1. Remarkably, this fully modified crRNA/tracrRNA complex retained significant, albeit reduced activity in cell assays. The optimal modification pattern (C20-T2, Figure 6) had >80% 2′-OMe or 2′-F nucleotides but retained ribonucleotides with or without phosphorothioate modification at most of the sensitive positions.59 The optimized crRNA/tracrRNA complex (C20-T2) was as potent as or slightly better than the unmodified counterpart in cellular assays.
Figure 6.
Modification patterns of crRNAs and tracrRNAs optimized by Khvorova, Watts, Sontheimer, and co-workers.59
The complexity of crRNAs, diverse chemistries of modifications and their placement patterns, and various assays used in studies discussed above make direct comparison of results difficult. In general, modifications in the PAM-distal region are tolerated better than modifications in the seed region, similar to how Cas9 tolerates mismatches in crRNA/DNA complex.61 This is likely due to the destabilization of intermediate state of DNA target recognition that inhibits R-loop formation similar to the effect of mismatched base pairs discussed above.25 However, notable exceptions have been reported, for example, two or three consecutive BNANC modifications in the seed region had relatively modest negative impact on on-target activity.43 Most of the studies on Cas9 crRNAs used well-established modifications that performed well in antisense and short interfering RNAs and focused on improving metabolic stability. In this context, the success of structure-guided modification illustrates the opportunities for synergy between synthetic and structural chemistry. Going forward, chemists are well positioned to take a bolder approach when considering future innovations for optimizing crRNAs.
3. MODIFICATIONS OF Cas12a AND Cas13 crRNAs
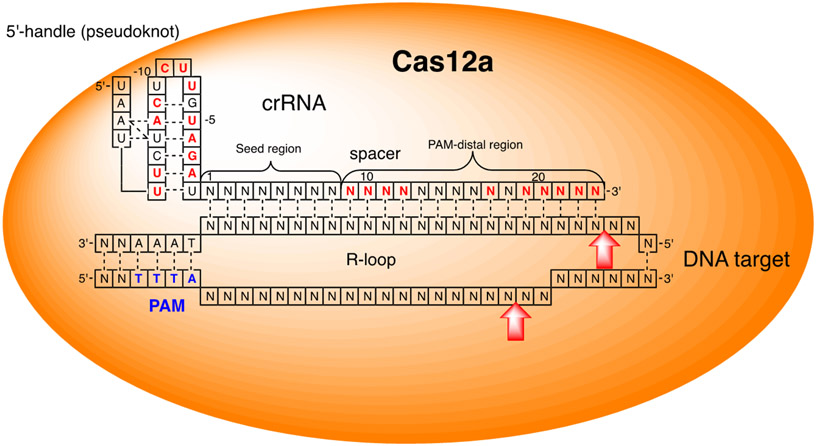
Cas12a (previously known as Cpf1) is another class 2 CRISPR endonuclease.14-15 In contrast to Cas9, Cas12a recognizes a short T-rich PAM (e.g., 5′-TTTA-3′ in Acidaminococcus sp. BV3L6 shown in Figure 7) and uses a single crRNA to introduce staggered DNA double-strand breaks having 4-5-nucleotide long 5′-overhangs.62 Cas12a crRNA (Figure 7) features a short 5′-handle pseudoknot followed by a 23-nucleotide spacer and is notably shorter and structurally simpler than Cas9 sgRNA or crRNA/tracrRNA. Cas12a is more sensitive to mismatches in the seed region of the crRNA-target DNA complex.63-64
Figure 7.
Cartoon representation of Cas12a protein and crRNA complex bound to dsDNA target.
In contrast to Cas9, Cas12a was more sensitive to shortening of the crRNA. Rahdar and co-workers showed that only one nucleotide, the terminal U, could be removed from the 5′-handle pseudoknot of the crRNA (Figure 7) without losing DNA cleavage activity.65 Up to five nucleotides could be removed from the 3′-end, shortening the DNA recognition spacer of Cas12a crRNA to 18 nucleotides.62, 65 This study used the Surveyor nuclease assay in HEK293T cells transfected with modified crRNAs and a DNA plasmid encoding Cas12a. Interestingly, extension of 3′-truncated crRNA with one 2′-OMe cytosine at the 5′-end and addition of a 2′-OMe at the 3′-uridine, together with two phosphorothioate linkages, increased the DNA cleavage activity ~1.7-fold.65 Kim and co-workers extended the 3′-end of crRNA with nine nucleotides and found that 2′-OMe modification (but not 2′-F) of this overhang increased cleavage activity in an in vitro assay and in mouse zygotes.66 As with Cas9, the increased activity of terminally-modified Cas12a crRNAs was most likely due to metabolic stabilization of RNA.
3.1. Ribose modifications of Cas12a crRNAs
As with Cas9 crRNAs, replacement of Cas12a ribonucleotides with their deoxy counterparts is one of the simplest and most cost/benefit effective modifications. In their study discussed above, Rahdar and co-workers also found that modification of the spacer region with 2′-deoxyribonucleotides at the last five 3′-positions and at internal positions 1, 8, and 9 doubled the activity of Cas12a crRNA.65 A study by Kim et. al.36 reported that crRNAs with eight 3′-terminal deoxyribonucleotides retained full DNA cleavage activity and showed improved Cas12a specificity in in vitro assays. Interestingly, while these results were confirmed for cleavage of plasmid DNA in a cellular assay, Cas12a was less tolerant to modification with 2′-deoxyribonucleotides when targeting endogenous DNA. Modification of even four 3′-terminal positions to 2′-deoxyribonucleotides significantly decreased (to <50%) editing at the endogenous locus DNMT1 in HEK293FT cells, as analyzed by targeted amplicon sequencing.36 A single 2′-deoxyribonucleotide modification was tolerated in cellular assays in the PAM-distal region of crRNA (but not in the seed region), and at position eight, even increased the Cas12a activity to ~150%. Modification of the 5′-handle pseudoknot strongly decreased the activity in both in vitro and cellular assays.36 A more recent study also showed that DNA modifications were not tolerated in the 5′-handle pseudoknot of crRNA in an assay that used HEK293T cells stably expressing EGFP.67
Similar to Cas9, Dong and co-workers found that the seed region of Cas12a crRNA was sensitive to modifications while the PAM-distal region tolerated 2′-F nucleotides better than 2′-OMe.68 HEK293T cells were transfected with the modified crRNAs and a DNA plasmid encoding Cas12a. T7EI assays showed that modification of 10 3′-nucleotides to 2′-F did not decrease cleavage activity, while only five 2′-OMe modifications were tolerated (10 2′-OMe 3′-terminal modifications resulted in complete loss of activity). Introduction of two or more 2′-F at the 5′-end of the seed notably reduced cleavage activity. Modifications of the 5′-handle pseudoknot with five or more 2′-F or 2′-OMe 5′-terminal nucleotides or full modification of the crRNA with phosphorothioate linkages resulted in complete loss of activity. However, a crRNA optimized using a structure-guided approach69 and containing 11 and 10 2′-F nucleotides in the 5′-handle pseudoknot and the PAM-distal region, respectively (Figure 7, red nucleotides), retained full activity of the unmodified crRNA.68 Interestingly, using the same 2′-OMe modification pattern resulted in complete loss of activity.
Rahdar and co-workers found that the 3′-truncated crRNA, where five 3′-terminal nucleotides were 2′-F-modified, increased the DNA cleavage activity ~1.5-fold.65 Further addition of 2′-F, 2′-OMe, or PS modifications to the PAM-distal region resulted in decrease of activity, with a notable exception of cEt that was well-tolerated at position 9. Multiple 2′-F nucleotide modifications in the 5′-handle pseudoknot decreased the DNA cleavage activity; however, a single cEt modification was well-tolerated at several positions, most notably at A(−12), C(−11), U(−7), or U(−5).65
More recently, Gagnon, Damha and co-workers reported similar results that crRNAs optimized using structure-guided approach and containing up to 13 2′-F nucleotides in the 5′-handle pseudoknot had activity similar to the unmodified crRNA.67 This study also showed that individual 2′-NH2 modifications of the positions that tolerated the 2′-F nucleotides in the 5′-handle pseudoknot decreased the Cas12a activity. Collectively, these studies emphasize the importance of using structural information as a guide for chemical modification of complex and highly folded crRNAs.
3.2. Phosphate backbone modifications of Cas12a crRNAs
In contrast to Cas9, full phosphorothioate (PS) modification of Cas12a resulted in ~70% loss of functionality.65 The seed region was most sensitive to backbone PS modifications with the 5′-handle and PAM-distal regions being more tolerant. Structure-guided PS modification of positions that do not interact with Cas12a gave a crRNA containing two terminal PS at the 5′-handle and six alternating PS in the PAM-distal region, which retained 80% of activity of the unmodified crRNA.65
3.3. Modifications of Cas13 crRNAs
In contrast to the Cas9 and Cas12a complexes discussed above, Cas13 proteins cleave RNA.14-15 Most recently, Sanjana and co-workers reported that 3′-terminal modifications of Cas13d crRNA increased mRNA cleavage activity in RfxCas13d-expressing HEK293FT cells as monitored by flow cytometry quantification of the corresponding protein expression.70 This study showed that 3′-extension of crRNAs with an inverted thymidine, three 2′-OMe uridines, or introduction of two 3′-PS linkages increased mRNA knockdown, while more extensive 5′- and internal modification decreased or even completely abrogated Cas13d activity. Interestingly, combining 2′-OMe and PS did not lead to further improvement compared to individual modifications. The authors suggested that the improvement of activity was most likely due to metabolic stabilization of crRNA by the 3′-modifications, while the loss activity was attributed to disrupting the Cas13-crRNA interactions by 5′- and internal modifications.70
4. SUMMARY AND OUTLOOK
As expected for a relatively young technology, chemists have only recently started contributing to optimization of CRISPR RNA components. The first study to truncate Cas9 crRNA was published in 201418 and was followed by two studies in 201541-42 reporting the effect of the first ribose and backbone modifications on Cas9 activity and specificity. While the overall number of detailed studies reviewed above is still below 20, several common trends and notable discrepancies have emerged among the various studies. The latter are most likely rooted in the complexity of CRISPR-Cas9 compared to other more mature nucleic acid technologies, such as antisense and RNA interference.
In general, there are significant differences among various studies, which is most likely caused by differences in assays used (cell lines, delivery methods, etc.) and reflects the complexity of CRISPR biology. For example, in human cells one needs to deliver not only the guide RNAs, but also the Cas proteins; thus, the results may be strongly influenced by delivery method and may vary depending on whether a Cas9 plasmid, Cas9 mRNA, or a cell line constitutively expressing Cas9 protein was used. It appears that results of in vitro assays do not correlate with cellular assays; typically, chemical modifications are much better tolerated in in vitro assays, which does not bear out in cellular assays. While the specific factors that cause the discrepancy of results between in vitro and in cell assays are poorly understood, a recent study of Cas12a activity showed that it may be related to accessibility and/or conformation (e.g., supercoiling) of the target DNA.36 In this study, DNA of transfected plasmid was cleaved similarly as in vitro, whereas cleavage of the same endogenous target (DNMT1) was strongly inhibited by the chemical modifications.36 Nevertheless, most of the studies agree that the PAM-distal region of Cas9 and Cas12a tolerates chemical modifications much better than the seed region. The latter can be modified using a structure-guided approach that avoids altering functional groups that interact with Cas proteins (e.g., 2′-OH or phosphate).
The repeat-antirepeat region of crRNA/tracrRNA shows variable sensitivity to modifications – some parts are highly sensitive while others can be modified relatively easily. Cas12a crRNA has a smaller and tightly folded repeat, the 5′-handle pseudoknot, which appears to be especially sensitive to chemical modifications, but the 3′-terminal and some internal positions can be modified without loss or even with some increase of DNA cleavage activity.
When modifying crRNAs, nucleic acid chemists have mostly used the well-established tried-and-true sugar and backbone modifications, such as 2′-F, 2′-OMe, and phosphorothioates, alone or in combination with each other. Overall, 2′-F modifications are better tolerated than 2′-OMe, though the specific pattern varies between studies. As in antisense and RNA interference approaches in the past, these modifications have improved metabolic stability of crRNA and, in some cases, specificity of DNA cleavage; however, they have not yielded groundbreaking improvements over simpler approaches such as shortening crRNAs or replacing ribonuclotides with deoxy counterparts. It appears that the future innovation lies in bolder approaches that will use more complex chemistries to rationally optimize crRNA-protein interactions. Going forward, chemists need to move beyond the tried-and-true 2′-F, 2′-OMe, and phosphorothioate modifications and focus on innovation that will capitalize on rich structural information to optimize crRNA-protein interactions. There are several backbone modifications that showed promising biophysical properties in early studies,37-38 for example formacetal, thioformacetal, and methylene(methylimino) internucleotide linkages, but have not been evaluated in biological assays and may provide new ways of improving siRNAs and crRNAs.
Several studies reviewed below have used structure-based approaches to identify crRNA nucleotides that should not be modified to maintain Cas-crRNA interactions critical for DNA cleavage activity. However, few, if any, have attempted to integrate structural and mechanistic information to rationally design chemical modification for engineering RNA-protein interactions to improve activity or specificity. Considering the advances in structural biology and synthetic organic chemistry, we have the means to advance from simple sugar modifications developed in the last century to rational engineering of RNA chemistry and further develop CRISPR as impactful research tools and novel therapeutic approaches. The complexity of CRISPR presents rich opportunities for chemical innovation that synergizes advances in synthetic methodology and structural biochemistry to rationally optimize CRISPR technology.
Finally, the impact of chemical modifications is likely to expand beyond crRNAs of Cas nucleases to novel CRISPR tools and technologies. In this respect, the first chemically modified RNA guides of base editors71 and prime editors72 have recently been reported.73-74 Adenine or cytosine base editors are catalytically inactive Cas proteins fused to adenine or cytosine deaminases that convert A·T to G·C or C·G to T·A base pairs, respectively. Optimized base editors can correct single point mutations.71 Prime editors are Cas9 nickases (mutated to cleave only one DNA strand) fused to reverse transcriptases.72 Prime editors use prime editing guide RNA (pegRNA) that has both spacer for recognition of the DNA target and a 3′-extension that encodes a template for reverse transcriptase to synthesize the edited DNA product.72 Because base and prime editors do not create dsDNA breaks they cause less off-target editing than Cas9 mediated HDR.
Xue and co-workers reported that modification of the first and last three nucleotides of sgRNA with 2′-OMe and PS linkages improved the activity of adenine base editor in HEK293T cells, presumably by metabolic stabilization of the sgRNA.73 pegRNAs having 2′-OMe and PS modifications between the first and last three nucleotides have also been reported.74 However, chemical synthesis of modified pegRNAs that may range from ~130 to >200 nucleotides in length will be a challenging task even for experts in RNA synthesis.
Taken together, the studies above have demonstrated the potential of chemical modifications to optimize CRISPR efficiency and specificity. However, these studies have only scratched the surface of what we can do in terms of both modification chemistry and targets that we can modify. For example, CRISPR specificity and versatility (PAM sequences that can be targeted) have been optimized by engineering Cas9. It is conceivable that protein and RNA chemical modifications could synergize in controlling and enhancing CRISPR applications in new and unprecedented ways.
Funding Sources
The work on RNA chemical modifications in E.R. lab was supported by National Institutes of Health (R35 GM130207 to E.R.).
ABBREVIATIONS
- Cas
CRISPR-associated protein
- CRISPR
clustered, regularly interspaced, short palindromic repeats
- crRNA
CRISPR RNA
- EGFP
enhanced green fluorescent protein
- GFP
green fluorescent protein
- HDR
homology-directed repair
- LNA
locked nucleic acid
- NGS
next generation sequencing
- NHEJ
non-homologous end joining
- PACE
3′-phosphonoacetate
- PAM
protospacer adjacent motif
- tracrRNA
trans-activating crRNA
- T7EI
T7 Endonuclease I
- thioPACE
3′-thiophosphonoacetate
- TIDE
tracking of indels by decomposition
REFERENCES
- (1).Wright AV; Nunez JK; Doudna JA, Biology and applications of CRISPR systems: Harnessing nature's toolbox for genome engineering. Cell 2016, 164, 29–44. [DOI] [PubMed] [Google Scholar]
- (2).Jiang F; Doudna JA, CRISPR-Cas9 Structures and Mechanisms. Annu. Rev. Biophys 2017, 46, 505–529. [DOI] [PubMed] [Google Scholar]
- (3).Hille F; Richter H; Wong SP; Bratovič M; Ressel S; Charpentier E, The Biology of CRISPR-Cas: Backward and Forward. Cell 2018, 172, 1239–1259. [DOI] [PubMed] [Google Scholar]
- (4).Liu G; Lin Q; Jin S; Gao C, The CRISPR-Cas toolbox and gene editing technologies. Mol. Cell 2022, 82, 333–347. [DOI] [PubMed] [Google Scholar]
- (5).Katti A; Diaz BJ; Caragine CM; Sanjana NE; Dow LE, CRISPR in cancer biology and therapy. Nat. Rev. Cancer 2022, 22, 259–279. [DOI] [PubMed] [Google Scholar]
- (6).Zhang F, Development of CRISPR-Cas systems for genome editing and beyond. Q. Rev. Biophys 2019, 52, e6. [Google Scholar]
- (7).Doudna JA, The promise and challenge of therapeutic genome editing. Nature 2020, 578, 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Li B; Niu Y; Ji W; Dong Y, Strategies for the CRISPR-Based Therapeutics. Trends Pharmacol. Sci 2020, 41, 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Hendriks D; Clevers H; Artegiani B, CRISPR-Cas Tools and Their Application in Genetic Engineering of Human Stem Cells and Organoids. Cell Stem Cell 2020, 27, 705–731. [DOI] [PubMed] [Google Scholar]
- (10).Wang H; La Russa M; Qi LS, CRISPR/Cas9 in Genome Editing and Beyond. Annu. Rev. Biochem 2016, 85, 227–264. [DOI] [PubMed] [Google Scholar]
- (11).Raper AT; Stephenson AA; Suo Z, Sharpening the Scissors: Mechanistic Details of CRISPR/Cas9 Improve Functional Understanding and Inspire Future Research. J. Am. Chem. Soc 2018, 140, 11142–11152. [DOI] [PubMed] [Google Scholar]
- (12).Wang JY; Pausch P; Doudna JA, Structural biology of CRISPR–Cas immunity and genome editing enzymes. Nat. Rev. Microbiol 2022, DOI: 10.1038/s41579-022-00739-4. [DOI] [PubMed] [Google Scholar]
- (13).Makarova KS; Wolf YI; Alkhnbashi OS; Costa F; Shah SA; Saunders SJ; Barrangou R; Brouns SJJ; Charpentier E; Haft DH; Horvath P; Moineau S; Mojica FJM; Terns RM; Terns MP; White MF; Yakunin AF; Garrett RA; van der Oost J; Backofen R; Koonin EV, An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol 2015, 13, 722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Shmakov S; Smargon A; Scott D; Cox D; Pyzocha N; Yan W; Abudayyeh OO; Gootenberg JS; Makarova KS; Wolf YI; Severinov K; Zhang F; Koonin EV, Diversity and evolution of class 2 CRISPR–Cas systems. Nat. Rev. Microbiol 2017, 15, 169–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Koonin EV; Makarova KS; Zhang F, Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol 2017, 37, 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Wang H; La Russa M; Qi LS, CRISPR/Cas9 in Genome Editing and Beyond. Annu. Rev. Biochem 2016, 85, 227–264. [DOI] [PubMed] [Google Scholar]
- (17).Xue C; Greene EC, DNA Repair Pathway Choices in CRISPR-Cas9-Mediated Genome Editing. Trends Genet. 2021, 37, 639–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Fu Y; Sander JD; Reyon D; Cascio VM; Joung JK, Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol 2014, 32, 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Cai W; Wang M, Engineering nucleic acid chemistry for precise and controllable CRISPR/Cas9 genome editing. Science Bulletin 2019, 64, 1841–1849. [DOI] [PubMed] [Google Scholar]
- (20).Wu J; Yin H, Engineering guide RNA to reduce the off-target effects of CRISPR. J. Genet. Genom 2019, 46, 523–529. [DOI] [PubMed] [Google Scholar]
- (21).Filippova J; Matveeva A; Zhuravlev E; Stepanov G, Guide RNA modification as a way to improve CRISPR/Cas9-based genome-editing systems. Biochimie 2019, 167, 49–60. [DOI] [PubMed] [Google Scholar]
- (22).Nishimasu H; Ran FA; Hsu PD; Konermann S; Shehata SI; Dohmae N; Ishitani R; Zhang F; Nureki O, Crystal Structure of Cas9 in Complex with Guide RNA and Target DNA. Cell 2014, 156, 935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Jinek M; Chylinski K; Fonfara I; Hauer M; Doudna JA; Charpentier E, A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Gasiunas G; Barrangou R; Horvath P; Siksnys V, Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U.S.A 2012, 109, E2579–E2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Ivanov IE; Wright AV; Cofsky JC; Palacio Aris KD; Doudna JA; Bryant Z, Cas9 interrogates DNA in discrete steps modulated by mismatches and supercoiling. Proc. Natl. Acad. Sci. U. S. A 2020, 117 , 5853–5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Sternberg SH; La France B; Kaplan M; Doudna JA, Conformational control of DNA target cleavage by CRISPR-Cas9. Nature 2015, 527, 110–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Dagdas YS; Chen JS; Sternberg SH; Doudna JA; Yildiz A, A conformational checkpoint between DNA binding and cleavage by CRISPR-Cas9. Science Advances 2017, 3, eaao0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Nierzwicki Ł; Arantes PR; Saha A; Palermo G, Establishing the allosteric mechanism in CRISPR-Cas9. WIREs Comput. Mol. Sci 2021, 11, e1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Cho SW; Kim S; Kim JM; Kim J-S, Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol 2013, 31, 230–232. [DOI] [PubMed] [Google Scholar]
- (30).Kelley ML; Strezoska Z; He K; Vermeulen A; Smith A. v. B., Versatility of chemically synthesized guide RNAs for CRISPR-Cas9 genome editing. J. Biotechnol 2016, 233, 74–83. [DOI] [PubMed] [Google Scholar]
- (31).Glazier DA; Liao J; Roberts BL; Li X; Yang K; Stevens CM; Tang W, Chemical Synthesis and Biological Application of Modified Oligonucleotides. Bioconjugate Chem. 2020, 31, 1213–1233. [DOI] [PubMed] [Google Scholar]
- (32).Kartje ZJ; Barkau CL; Rohilla KJ; Ageely EA; Gagnon KT, Chimeric guides probe and enhance Cas9 biochemical activity. Biochemistry 2018, 57, 3027–3031. [DOI] [PubMed] [Google Scholar]
- (33).Yin H; Song C-Q; Suresh S; Kwan S-Y; Wu Q; Walsh S; Ding J; Bogorad RL; Zhu LJ; Wolfe SA; Koteliansky V; Xue W; Langer R; Anderson DG, Partial DNA-guided Cas9 enables genome editing with reduced off-target activity. Nat. Chem. Biol 2018, 14, 311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Tsai SQ; Zheng Z; Nguyen NT; Liebers M; Topkar VV; Thapar V; Wyvekens N; Khayter C; Iafrate AJ; Le LP; Aryee MJ; Joung JK, GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol 2015, 33, 187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).O’Reilly D; Kartje ZJ; Ageely EA; Malek-Adamian E; Habibian M; Schofield A; Barkau CL; Rohilla KJ; DeRossett LB; Weigle AT; Damha MJ; Gagnon KT, Extensive CRISPR RNA modification reveals chemical compatibility and structure-activity relationships for Cas9 biochemical activity. Nucleic Acids Res. 2019, 47, 546–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Kim H; Lee W.-j.; Oh Y; Kang S-H; Hur JK; Lee H; Song W; Lim K-S; Park Y-H; Song B-S; Jin YB; Jun B-H; Jung C; Lee D-S; Kim S-U; Lee SH, Enhancement of target specificity of CRISPR–Cas12a by using a chimeric DNA–RNA guide. Nucleic Acids Res. 2020, 48, 8601–8616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Wan WB; Seth PP, The Medicinal Chemistry of Therapeutic Oligonucleotides. J. Med. Chem 2016, 59, 9645–9667. [DOI] [PubMed] [Google Scholar]
- (38).Freier SM; Altmann KH, The ups and downs of nucleic acid duplex stability: structure-stability studies on chemically-modified DNA:RNA duplexes. Nucleic Acids Res. 1997, 25, 4429–4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Patra A; Paolillo M; Charisse K; Manoharan M; Rozners E; Egli M, 2'-Fluoro RNA Shows Increased Watson-Crick H-Bonding Strength and Stacking Relative to RNA: Evidence from NMR and Thermodynamic Data. Angew. Chem., Int. Ed 2012, 51, 11863–11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Manoharan M; Akinc A; Pandey RK; Qin J; Hadwiger P; John M; Mills K; Charisse K; Maier MA; Nechev L; Greene EM; Pallan PS; Rozners E; Rajeev KG; Egli M, Unique Gene-Silencing and Structural Properties of 2'-Fluoro-Modified siRNAs. Angew. Chem., Int. Ed 2011, 50, 2284–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Hendel A; Bak RO; Clark JT; Kennedy AB; Ryan DE; Roy S; Steinfeld I; Lunstad BD; Kaiser RJ; Wilkens AB; Bacchetta R; Tsalenko A; Dellinger D; Bruhn L; Porteus MH, Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol 2015, 33, 985–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Rahdar M; McMahon MA; Prakash TP; Swayze EE; Bennett CF; Cleveland DW, Synthetic CRISPR RNA-Cas9-guided genome editing in human cells. Proc. Natl. Acad. Sci. U. S. A 2015, 112, E7110–E7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Cromwell Christopher R; Krysler Amanda R; Hubbard Basil P; Sung K; Park J; Kim Seong K; Jovel J, Incorporation of bridged nucleic acids into CRISPR RNAs improves Cas9 endonuclease specificity. Nat Commun 2018, 9, 1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Eckstein F, Developments in RNA chemistry, a personal view. Biochimie 2002, 84, 841–848. [DOI] [PubMed] [Google Scholar]
- (45).Shen X; Corey DR, Chemistry, mechanism and clinical status of antisense oligonucleotides and duplex RNAs. Nucleic Acids Res. 2018, 46, 1584–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Setten RL; Rossi JJ; Han S. p., The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov 2019, 18, 421–446. [DOI] [PubMed] [Google Scholar]
- (47).Lieberman J, Tapping the RNA world for therapeutics. Nat. Struct. Mol. Biol 2018, 25, 357–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Threlfall RN; Torres AG; Krivenko A; Gait MJ; Caruthers MH, Synthesis and biological activity of phosphonoacetate- and thiophosphonoacetate-modified 2'-O-methyl oligoribonucleotides. Org. Biomol. Chem 2012, 10, 746–754. [DOI] [PubMed] [Google Scholar]
- (49).Dellinger DJ; Sheehan DM; Christensen NK; Lindberg JG; Caruthers MH, Solid-Phase Chemical Synthesis of Phosphonoacetate and Thiophosphonoacetate Oligodeoxynucleotides. J. Am. Chem. Soc 2003, 125, 940–950. [DOI] [PubMed] [Google Scholar]
- (50).Ryan DE; Taussig D; Steinfeld I; Phadnis SM; Lunstad BD; Singh M; Vuong X; Okochi KD; McCaffrey R; Olesiak M; Roy S; Yung CW; Curry B; Sampson JR; Bruhn L; Dellinger DJ, Improving CRISPR-Cas specificity with chemical modifications in single-guide RNAs. Nucleic Acids Res. 2018, 46, 792–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Kotikam V; Gajula PK; Coyle L; Rozners E, Amide Internucleoside Linkages Are Well Tolerated in Protospacer Adjacent Motif-Distal Region of CRISPR RNAs. ACS Chem. Biol 2022, 17, 509–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Idziak I; Just G; Damha MJ; Giannaris PA, Synthesis and hybridization properties of amide-linked thymidine dimers incorporated into oligodeoxynucleotides. Tetrahedron Lett. 1993, 34, 5417–5420. [Google Scholar]
- (53).De Mesmaeker A; Waldner A; Lebreton J; Hoffmann P; Fritsch V; Wolf RM; Freier SM, Amides as a new type of backbone modifications in oligonucleotides. Angew. Chem., Int. Ed. Engl 1994, 33, 226–229. [Google Scholar]
- (54).Selvam C; Thomas S; Abbott J; Kennedy SD; Rozners E, Amides as Excellent Mimics of Phosphate Linkages in RNA. Angew. Chem., Int. Ed 2011, 50, 2068–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Mutisya D; Hardcastle T; Cheruiyot SK; Pallan PS; Kennedy SD; Egli M; Kelley ML; Smith Anja van B.; Rozners E, Amide linkages mimic phosphates in RNA interactions with proteins and are well tolerated in the guide strand of short interfering RNAs. Nucleic Acids Res. 2017, 45, 8142–8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Hardcastle T; Novosjolova I; Kotikam V; Cheruiyot SK; Mutisya D; Kennedy SD; Egli M; Kelley ML; van Brabant Smith A; Rozners E, A Single Amide Linkage in the Passenger Strand Suppresses Its Activity and Enhances Guide Strand Targeting of siRNAs. ACS Chem. Biol 2018, 13, 533–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Basila M; Kelley ML; Smith A. v. B., Minimal 2'-O-methyl phosphorothioate linkage modification pattern of synthetic guide RNAs for increased stability and efficient CRISPR-Cas9 gene editing avoiding cellular toxicity. PLoS One 2017, 12, e0188593/1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Yin H; Song C-Q; Suresh S; Wu Q; Walsh S; Rhym LH; Mintzer E; Bolukbasi MF; Zhu LJ; Kauffman K; Mou H; Oberholzer A; Ding J; Kwan S-Y; Bogorad RL; Zatsepin T; Koteliansky V; Wolfe SA; Xue W; Langer R; Anderson DG, Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat. Biotechnol 2017, 35, 1179–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Mir A; Alterman JF; Hassler MR; Debacker AJ; Hudgens E; Echeverria D; Brodsky MH; Khvorova A; Watts JK; Sontheimer EJ, Heavily and fully modified RNAs guide efficient SpyCas9-mediated genome editing. Nat. Commun 2018, 9, 2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Jiang F; Zhou K; Ma L; Gressel S; Doudna JA, A Cas9-guide RNA complex preorganized for target DNA recognition. Science 2015, 348, 1477–1481. [DOI] [PubMed] [Google Scholar]
- (61).Klein M; Eslami-Mossallam B; Arroyo DG; Depken M, Hybridization Kinetics Explains CRISPR-Cas Off-Targeting Rules. Cell Reports 2018, 22, 1413–1423. [DOI] [PubMed] [Google Scholar]
- (62).Zetsche B; Gootenberg JS; Abudayyeh OO; Slaymaker IM; Makarova KS; Essletzbichler P; Volz SE; Joung J; van der Oost J; Regev A; Koonin EV; Zhang F, Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell 2015, 163, 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Kim D; Kim J; Hur JK; Been KW; Yoon S.-h.; Kim J-S, Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol 2016, 34, 863–868. [DOI] [PubMed] [Google Scholar]
- (64).Kleinstiver BP; Tsai SQ; Prew MS; Nguyen NT; Welch MM; Lopez JM; McCaw ZR; Aryee MJ; Joung JK, Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat. Biotechnol 2016, 34, 869–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).McMahon MA; Prakash TP; Cleveland DW; Bennett CF; Rahdar M, Chemically Modified Cpf1-CRISPR RNAs Mediate Efficient Genome Editing in Mammalian Cells. Mol. Ther 2018, 26, 1228–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Ha D-I; Lee JM; Lee N-E; Kim D; Ko J-H; Kim Y-S, Highly efficient and safe genome editing by CRISPR-Cas12a using CRISPR RNA with a ribosyl-2′-O-methylated uridinylate-rich 3′-overhang in mouse zygotes. Exp. Mol. Med 2020, 52, 1823–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Ageely EA; Chilamkurthy R; Jana S; Abdullahu L; O’Reilly D; Jensik PJ; Damha MJ; Gagnon KT, Gene editing with CRISPR-Cas12a guides possessing ribose-modified pseudoknot handles. Nat. Commun 2021, 12, 6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Li B; Zhao W; Luo X; Zhang X; Zeng C; Dong Y; Li C; Dong Y, Engineering CRISPR-Cpf1 crRNAs and mRNAs to maximize genome editing efficiency. Nat. Biomed. Eng 2017, 1, 0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Yamano T; Nishimasu H; Zetsche B; Hirano H; Slaymaker Ian M.; Li Y; Fedorova I; Nakane T; Makarova Kira S.; Koonin Eugene V.; Ishitani R; Zhang F; Nureki O, Crystal Structure of Cpf1 in Complex with Guide RNA and Target DNA. Cell 2016, 165, 949–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Méndez-Mancilla A; Wessels H-H; Legut M; Kadina A; Mabuchi M; Walker J; Robb GB; Holden K; Sanjana NE, Chemically modified guide RNAs enhance CRISPR-Cas13 knockdown in human cells. Cell Chem. Biol 2022, 29, 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Rees HA; Liu DR, Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet 2018, 19, 770–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Anzalone AV; Randolph PB; Davis JR; Sousa AA; Koblan LW; Levy JM; Chen PJ; Wilson C; Newby GA; Raguram A; Liu DR, Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Jiang T; Henderson JM; Coote K; Cheng Y; Valley HC; Zhang X-O; Wang Q; Rhym LH; Cao Y; Newby GA; Bihler H; Mense M; Weng Z; Anderson DG; McCaffrey AP; Liu DR; Xue W, Chemical modifications of adenine base editor mRNA and guide RNA expand its application scope. Nat. Commun 2020, 11, 1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Nelson JW; Randolph PB; Shen SP; Everette KA; Chen PJ; Anzalone AV; An M; Newby GA; Chen JC; Hsu A; Liu DR, Engineered pegRNAs improve prime editing efficiency. Nat. Biotechnol 2022, 40, 402–410. [DOI] [PMC free article] [PubMed] [Google Scholar]