Abstract
Purpose
The aim of this study was to clarify the distribution and drug resistance of pathogens causing urinary tract infection (UTI) and to provide a scientific reference for the rational application of antibiotics.
Patients and Methods
The results of bacterial identification and drug sensitivity analysis of midstream urine samples in our hospital from January 2018 to December 2020 were retrospectively analyzed. The data were analyzed using WHONET 5.6 and SPSS 26.0 (IBM) software.
Results
In all, 1786 pathogens were isolated from 13,141 midstream urine culture samples. Of these, 1093 (61.2%) were gram-negative bacteria, mainly Escherichia coli [29.1%] and Klebsiella pneumoniae [14.3%]; 543 (30.4%) were gram-positive bacteria, mainly Enterococcus faecium [16.7%] and Enterococcus faecalis [8.4%]; and 150 (8.4%) were fungal isolates, with the most common being Candida albicans (3.7%). The resistance rates of E. coli to piperacillin/tazobactam (3.4% vs 10.0%, p<0.05), ampicillin/sulbactam (43.0% vs 53.8%, p<0.05), and ciprofloxacin (58.0% vs 72.9%, p<0.05) increased significantly. K. pneumoniae was highly sensitive to ertapenem (100%). Two Enterococcus spp were highly sensitive to tigecycline (100%), and a small number of norvancomycin-resistant strains were found. The drug resistance rate of E. faecium to quinupristin was 6.7%. The drug resistance rates of E. faecalis to furantoin and ampicillin were 4.0% and 4.7%, respectively.
Conclusion
The pathogens that cause UTIs in patients are diverse, with the most common being E. coli. The isolated pathogens exhibited different resistance patterns. Antibiotics should be rationally selected based on the resistance patterns of the pathogens.
Keywords: urinary tract infections, midstream urine culture, pathogenic bacteria distribution, antimicrobial agent, antibiotic resistance
Introduction
Urinary tract infections (UTIs) are common infections that are primarily caused by bacteria and typically require antibiotic treatment.1 However, delays in laboratory diagnosis of the causative pathogens often lead clinicians to initiate empiric antibiotic therapy. In the past 80 years, empirical use, overuse, and misuse of antimicrobial agents have been linked to outbreaks of antimicrobial resistance.2 In recent years, superbugs and multidrug-resistant bacteria have rapidly emerged worldwide and are spreading in every country at a faster rate than expected, which poses a significant threat to human health.3 If not tackled promptly and effectively, the problem of antimicrobial resistance can potentially cause millions of preventable deaths and hundreds of billions of pounds in economic costs each year.4 Considerable research and analysis have been carried out worldwide regarding antibiotic resistance in bacteria. In The Lancet, Cassini et al5 emphasized that the adverse impact of drug-resistant bacterial infections remains one of the greatest challenges to global public health. The analysis of the distribution and drug resistance of UTI pathogens is beneficial to the rational use of antibiotics. However, the distribution and drug resistance are regional and temporal, and the experience of different countries and regions is only for reference.6 Recent studies have indicated that the prevalence of antibiotic resistance in gram-negative uropathogens varies widely across the world. For example, Klebsiella pneumoniae had extensive antibiotic resistance mechanisms, and until 2005, few carbapenem-resistant Klebsiella strains were reported in Europe, but between 2005 and 2015, it appeared in multiple countries at a rate of 40%–60%.7 Even within the same country, the resistance rates of uropathogenic Escherichia coli (UPEC) to imipenem can vary dramatically. One study reported no imipenem-resistant strains of UPEC, while another showed that UPEC were only 50% susceptible to imipenem.8,9 Unfortunately, as the predominant strains of UTIs, UPEC can exert pathogenicity and antibiotic resistance in different individuals and geographic regions by relying on flexible and diverse genomic signatures.10,11 In addition, multidrug-resistant bacteria can transfer genetic elements between bacterial cells, or even the concerted activities of mobile genetic elements that can move within or between DNA molecules, facilitating the acquisition and spread of resistance genes.12,13 The predicted prevalence of multidrug-resistant E. coli resistance by 2030 is 77%.14 Coincidentally, the global change in gram-positive bacteria is equally worrying.
They are emerging as important causative agents of UTIs in elderly patients, pregnant women, and catheterized patients.6,15 To make matters worse, these positive cocci can cause serious nosocomial infections that are difficult to control.16,17 Among reports of nosocomial infections associated with vancomycin-resistant Enterococcus, most of the sequenced isolates belonged to the clonal lineage ST80/CT1013 in Germany, while the Dutch report was ST117, and the Chinese report was ST78, ST192, and ST570.17–19
The distribution and drug resistance of bacteria vary greatly in different countries and regions. Active surveillance can alert people to the spread of resistant strains.20 Therefore, it is of great significance to actively monitor the distribution characteristics and drug resistance of local UTI pathogens, promote the rational use of antibiotics, and reduce the emergence of drug-resistant strains.
In this study, we analyzed the data of pathogen distribution and drug sensitivity of midstream urine culture results in our hospital from 2018–2020. The aim of this study is to provide a reference for the prevention and treatment of UTIs and the rational use of antibiotics.
Materials and Methods
Sample Collection
This retrospective study was conducted in a comprehensive regional medical center in the eastern Chongming District of Shanghai from January 2018 to December 2020.
Only the culture results of the first time were considered when the same strain was cultured in the same patient for two consecutive times. This study was reviewed and approved by the ethics committee of the Chongming Branch of Shanghai Tenth People’s Hospital, Tongji University School of Medicine.
Urine Culture and Drug Sensitivity Test
Urine was usually sampled in the morning before the antibacterial treatment. After the vulva was cleaned, the clean midstream urine was collected into sterile tubes and sent to the microbiology laboratory within 2 h for quantitative culture.
The urine samples were inoculated onto 5% sheep blood agar, MacConkey, and fungal chromogenic plates (all purchased from Shanghai Komagal Co. Ltd.) using sterile calibrated loops (10 µL). The inoculations were incubated aerobically at 35 °C for 24–48 h, and the colony growths were observed.
Gram-negative species were considered positive for culture if a single bacterial species reached a concentration of ≥ 105 CFU/mL; when the growth of gram-positive bacteria in a urine culture was ≥ 104 CFU/mL, it was defined as positive.
The isolated strains were subjected to colony morphology, Gram staining, and identification using the VITEK 2 Compact automatic microbial identification system (bioMérieux; Craponne, France). Finally, susceptibility tests were performed using the drug-sensitive reaction card (bioMérieux; Craponne, France) supplemented with the Kirby–Bauer disk diffusion method. The interpretation was based on the Clinical and Laboratory Standards Institute (CLSI) 2018 standard.
Escherichia coli ATCC25922, Staphylococcus aureus ATCC29213, Enterococcus faecalis ATCC29212, Pseudomonas aeruginosa ATCC27853, Enterobacter hormaechei ATCC700323, and Klebsiella pneumoniae ATCC700603 were used as quality control strains, provided by Shanghai Clinical Laboratory Center. Quality control strains should be tested with clinical specimens in accordance with standard methods. When changing the drug or culture medium, the corresponding quality control strain must be used for testing. All quality control strains should be monitored continuously for 30 days before routine monitoring (quality control once a week).
Statistical Analysis
The data are shown as the mean ± standard deviation (SD). The susceptibility test results were processed using WHONET 5.6 software (WHO, Geneva, Switzerland). The culture results were analyzed using SPSS 22 software (Chicago, IL, USA), and categorical variables were analyzed using Pearson’s chi-square test and Fisher’s exact test. P < 0.05 indicated significant differences. GraphPad Prism 8 software (GraphPad Software, LaJolla, CA, USA) was used for drawing.
Results
Distribution of the Main Pathogenic Bacteria
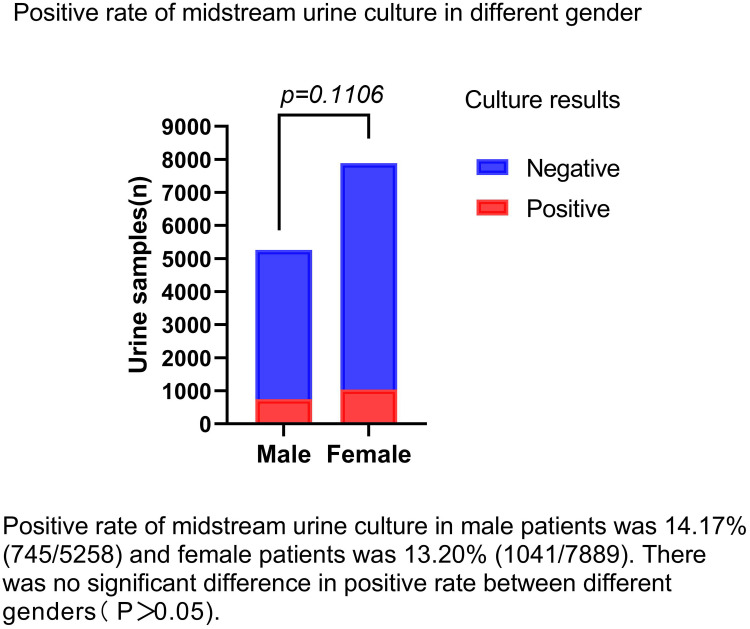
A total of 13,141 urine samples were detected in this study, comprising 1768 samples from the outpatient department and 11,373 samples from the inpatient department. Of these patients, 5274 were male and 7867 were female. Their ages ranged from 22–102 years, with an average age of 73.82 ± 16.14 years. A total of 1786 strains of pathogenic bacteria were isolated from all the midstream urine samples, with a positive rate of 13.6%. The culture-positive rate of male patients was 14.17% (745/5258) and that for female patients was 13.20% (1041/7889). There was no significant difference in positive rate between different genders (P > 0.05), as shown in Figure 1.
Figure 1.
Positive rate of midstream urine culture in different gender.
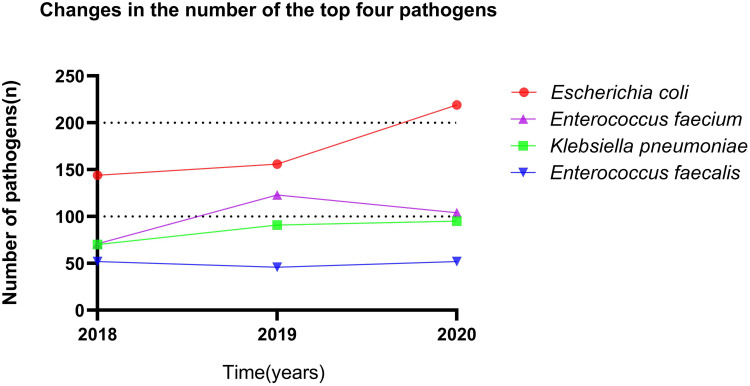
Of all the positive results, 1093 (61.2%) were gram-negative bacteria, 543 (30.4%) were gram-positive bacteria, and 150 (8.4%) were fungi. The top 10 pathogenic bacteria were E. coli (29.1%), E. faecium (16.7%), K. pneumoniae (14.3%), E. faecalis (8.4%), Proteus mirabilis (5.3%), Pseudomonas aeruginosa (4.4%), Candida albicans (3.7%), Candida glabrata (2.8%), Acinetobacter baumannii (2.4%), and Staphylococcus haemolyticus (2.2%). The distribution of pathogenic bacteria in the midstream urine culture is shown in Table 1. We show the number of changes in the top four pathogens from 2018–2020, as shown in Figure 2.
Table 1.
Distribution and Composition of 1786 Strains of Pathogenic Bacteria
| Pathogen | Strain (n) | Percentage (100%) |
|---|---|---|
| Gram negative | 1093 | 61.2 |
| Escherichia coli | 519 | 29.1 |
| Klebsiella pneumoniae | 256 | 14.3 |
| Proteus mirabilis | 95 | 5.3 |
| Pseudomonas aeruginosa | 79 | 4.4 |
| Acinetobacter baumannii | 42 | 2.4 |
| Enterobacter cloacae | 32 | 1.8 |
| Morganella morganii | 22 | 1.2 |
| Others* | 48 | 2.7 |
| Gram positive | 543 | 30.4 |
| Enterococcus faecium | 298 | 16.7 |
| Enterococcus faecalis | 150 | 8.4 |
| Staphylococcus haemolyticus | 39 | 2.2 |
| Streptococcus agalactiae | 22 | 1.2 |
| Others** | 34 | 1.9 |
| Fungus | 150 | 8.4 |
| Candida albicans | 66 | 3.7 |
| Candida glabrata | 50 | 2.8 |
| Candida tropicalis | 23 | 1.3 |
| Others*** | 11 | 0.6 |
| Total | 1786 | 100 |
Notes: The data are expressed as numbers and percentages. Other*: Serratia sp. (n = 18), Citrobacter sp. (n = 12), Serratiamarcescens (n = 8), Aeromonas hydrophila (n = 6), Stenotrophomonas maltophilia (n = 3), Pseudomonas putida (n = 1). Other**: Staphylococcusepidermidis (n = 17), Staphylococcus capitis (n = 11), coagulase negative Staphylococcus (n = 6). Other***: Candidakrusei (n = 6), Candidaparapsilosis (n = 5).
Figure 2.
Changes in the number of the top four pathogens.
Resistance of E. coli to Common Antibiotics
Antibiotics with good sensitivity to E. coli were amikacin, imipenem, furantoin, cefotetan, piperacillin/tazobactam, and ertapenem. The drug resistance rates of imipenem and ertapenem showed an increasing trend, but the difference was not significant. Compared with 2019, the drug resistance rates of piperacillin/tazobactam and ampicillin/sulbactam increased significantly in 2020, and the differences between the two groups were significant (P < 0.05). The drug resistance rate of E. coli to ciprofloxacin and levofloxacin reached > 60%. The drug resistance rate of ciprofloxacin increased significantly in 2020 compared with that in 2018 and 2019, and the difference was significant (P < 0.05). (Table 2).
Table 2.
Resistance Rate of Escherichia coli to Antibiotics
| Antibacterial Drugs | 2018 n (R)% | 2019 n (R)% | 2020 n (R)% | 2018–2020 n (R)% |
|---|---|---|---|---|
| n = 144 | n = 156 | n = 219 | n = 519 | |
| Piperacillin/tazobactam | 5 (3.4) | 11 (7.0) | 22 (10.0)a | 38 (7.3) |
| Ceftazidime | 48 (33.9) | 45 (28.9) | 67 (30.5) | 160 (30.8) |
| Cefepime | 32 (22.1) | 30 (19.3) | 52 (23.6) | 114 (22.0) |
| Imipenem | 3 (1.9) | 3 (1.8) | 12 (5.4) | 18 (3.5) |
| Bactrim | 61 (42.3) | 59 (38.0) | 91 (41.5) | 211 (40.7) |
| Ciprofloxacin | 89 (61.8) | 90 (58.0) | 160 (72.9)a,b | 339 (65.3) |
| Gentamicin | 52 (36.1) | 57 (36.7) | 77 (35.1) | 186 (35.8) |
| Tobramycin | 25 (17.3) | 22 (14.1) | 39 (17.8) | 86 (16.6) |
| Ampicillin/Sulbactam | 62 (43.0) | 70 (45.1) | 118 (53.8)a | 250 (48.2) |
| Levofloxacin | 86 (59.7) | 88 (56.7) | 144 (65.7) | 318 (61.3) |
| Nitrofurantoin | 8 (5.3) | 6 (3.7) | 11 (4.8) | 25 (4.8) |
| Ampicillin | 110 (76.4) | 112 (72.2) | 172 (78.5) | 394 (75.9) |
| Amikacin | 7 (4.8) | 4 (2.5) | 5 (2.2) | 16 (3.1) |
| Ceftriaxone | 87 (60.5) | 83 (53.6) | 116 (53.1) | 286 (55.1) |
| Cefotetan | 5 (3.4) | 7 (4.5) | 15 (6.8) | 27 (5.2) |
| Ertapenem | 7 (4.8) | 12 (7.7) | 20 (9.1) | 39 (7.5) |
Notes: n, number of strains; R, drug resistance rate; aCompared with the drug resistance rate in 2018, P < 0.05; bCompared with the drug resistance rate in 2019, P < 0.05.
Resistance of K. pneumoniae to Common Antibiotics
K. pneumoniae is most sensitive to ertapenem, followed by amikacin. However, the 3-year resistance rate of imipenem and cefotetan was stable at approximately 45%. Ceftazidime, cefepime, piperacillin, tazobactam, and ampicillin sulbactam all maintained drug resistance rates above 55% (Table 3).
Table 3.
Resistance Rate of Klebsiella pneumoniae to Antibiotics
| Antibacterial Drug | 2018 n (R)% | 2019 n (R)% | 2020 n (R)% | 2018–2020 n (R)% |
|---|---|---|---|---|
| n = 70 | n = 91 | n = 95 | n = 256 | |
| Piperacillin/tazobactam | 42 (60.0) | 51 (56.0) | 48 (50.5) | 141 (55.1) |
| Ceftazidime | 44 (62.9) | 61 (67) | 51 (53.1) | 156 (60.9) |
| Cefepime | 44 (62.9) | 52 (57.1) | 48 (50.0) | 144 (56.3) |
| Imipenem | 34 (48.5) | 43 (47.3) | 40 (42.1) | 117 (45.7) |
| Bactrim | 39 (55.7) | 48 (52.7) | 48 (50.0) | 135 (52.7) |
| Ciprofloxacin | 47 (67.1) | 64 (70.3) | 60 (62.5) | 171 (66.8) |
| Gentamicin | 42 (60.0) | 57 (62.6) | 59 (61.5) | 158 (61.7) |
| Tobramycin | 33 (47.1) | 51 (56.0) | 50 (52.1) | 134 (52.3) |
| Ampicillin/Sulbactam | 49 (70) | 66 (72.5) | 63 (65.6) | 178 (69.5) |
| Levofloxacin | 44 (62.9) | 63 (69.2) | 57 (59.4) | 164 (64.1) |
| Nitrofurantoin | 40 (57.1) | 53 (58.2) | 55 (57.3) | 148 (57.8) |
| Ampicillin | 52 (74.3) | – | – | – |
| Amikacin | 25 (35.7) | 35 (38.5) | 33 (34.4) | 93 (36.3) |
| Ceftriaxone | 50 (71.4) | 67 (73.6) | 61 (63.5) | 178 (69.5) |
| Cefotetan | 31 (44.3) | 42 (46.2) | 45 (46.9) | 118 (46.1) |
| Ertapenem | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
Abbreviations: n, number of strains; R, drug resistance rate; “-”, Not detected.
Sensitivity of Two Gram-Positive Enterococcus spp to Common Antibiotics
There were no resistant strains to tigecycline, high concentrations of streptomycin, and gentamicin in the two gram-positive Enterococcus spp, but a few strains were resistant to vancomycin. E. faecium is extremely resistant to levofloxacin and ciprofloxacin, and it showed good sensitivity to quinupristin, with a drug resistance rate of 6.7% in 3 years, but the drug resistance rate in 2020 was significantly higher than that in 2019, and the difference was significant (P < 0.05). Compared with 2018, the drug resistance rate of furantoin in 2019 and 2020 was on the rise, and the difference was significant (P < 0.05).
The antibiotic resistance rate of E. faecalis remained stable, and its resistance rate to erythromycin, levofloxacin, and ciprofloxacin was more than 66%. The resistance of E. faecalis to norvancomycin, furantoin, and ampicillin was 1.3%, 4.0%, and 4.7%, respectively (Tables 4 and 5).
Table 4.
Drug Resistance Rate of Enterococcus faecium to Antibiotics
| Antibacterial Drug | 2018 n (R)% | 2019 n (R)% | 2020 n (R)% | 2018–2020 n (R)% |
|---|---|---|---|---|
| n = 71 | n = 123 | n = 104 | n = 298 | |
| Ciprofloxacin | 64 (90.1) | 113 (91.7) | 93 (89.4) | 270 (90.6) |
| Quinupristin | 4 (5.6) | 4 (3.3) | 12 (11.5)b | 20 (6.7) |
| Ciprofloxacin | 67 (94.4) | 115 (93.5) | 94 (90.4) | 276 (92.6) |
| Norvancomycin | 3 (4.2) | 3 (2.4) | 7 (6.7) | 13 (4.4) |
| High concentration of streptomycin | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| High concentration of streptomycin | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Levofloxacin | 62 (87.3) | 112 (91.1) | 93 (89.4) | 267 (89.6) |
| Nitrofurantoin | 19 (26.7) | 63 (51.2)a | 54 (51.9)a | 136 (45.6) |
| Ampicillin | 64 (90.1) | 112 (91.1) | 92 (88.5) | 268 (89.9) |
| Tigecycline | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
Notes: n, number of strains; R, drug resistance rate; aCompared with the drug resistance rate in 2018, P < 0.05; bCompared with the drug resistance rate in 2019, P < 0.05.
Table 5.
Antibiotic Resistance Rate of Enterococcus faecalis to Antibiotics
| Antibacterial Drug | 2018 n (R)% | 2019 n (R)% | 2020 n (R)% | 2018–2020 n (R)% |
|---|---|---|---|---|
| n = 52 | n = 46 | n = 52 | n = 150 | |
| Ciprofloxacin | 38 (73.1) | 28 (60.9) | 34 (66.7) | 100 (66.7) |
| Quinupristin | 28 (53.9) | – | – | – |
| Erythromycin | 43 (82.7) | 39 (84.8) | 37 (71.2) | 119 (79.3) |
| Norvancomycin | 1 (1.9) | 1 (2.2) | 0 (0) | 2 (1.3) |
| High concentration of streptomycin | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| High concentration of Gentamicin | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Levofloxacin | 38 (73.1) | 28 (60.9) | 34 (65.4) | 100 (66.7) |
| Nitrofurantoin | 2 (3.8) | 1 (2.2) | 3 (5.8) | 6 (4.0) |
| Ampicillin | 4 (7.7) | 1 (2.2) | 2 (3.8) | 7 (4.7) |
| Tigecycline | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
Abbreviations: n, number of strains; R, drug resistance rate; “-”, Not detected.
Discussion
In recent years, UTI-related pathogens and their susceptibility patterns have undergone significant changes worldwide.21 Although the types of antibacterial drugs are constantly updated, drug resistance to pathogens is also concomitantly increasing, and China has a high level of antibiotic resistance due to the overuse of antibiotics.22 The incidence, pathogen spectrum, and drug resistance rates of both community-acquired and hospital-acquired infections vary with geographical environment.23 Empiric treatment of UTIs requires a good knowledge of the local pathogen epidemiological data. Therefore, it is very important to understand the distribution of bacterial infections and the characteristics of drug resistance in this region to ensure the rational use of antibiotics and to reduce the generation of resistant strains. This study analyzed the urine culture results of 13,141 midstream urine samples collected from 2018–2020 in our hospital. Within 3 years of this study, the distribution of pathogenic bacteria showed some notable characteristics of drug resistance.
The main pathogens associated with UTI in this study were 1093 gram-negative bacteria (61.2%), 543 gram-positive bacteria (30.4%), and 150 fungi (8.4%). We found that the most commonly detected pathogens remained consistent, and their ranks remained relatively constant. The detection rate of E. coli was still the first and showed an increasing trend. E. coli was the most common pathogen of UTI, accounting for 29.1%. In this study, the drug resistance rate of E. coli to cefepime and ceftazidime reached 19.3%–33.9%, and the drug resistance rate to ampicillin and quinolones reached > 55%, which limited the efficacy of antibiotics widely used in our clinical work, and may also be the reason for the easy failure of empirical treatment. One-third of community-acquired UTIs caused by E. coli in the human population is caused by ultra-broad-spectrum β-lactamase (ESBLs) strains, which are highly resistant to the antibiotics widely used in the community, limiting the effectiveness of empirical treatment of these infections.24 A β-lactamase inhibitor compound is considered a good control for ESBLs-producing strains. However, the resistance rates of E. coli to piperacillin/tazobactam and ampicillin/sulbactam were 7.3% and 53.8%, respectively, and the resistance rates of both showed an increasing trend in these 3 years. This upward trend may be related to the increasing number of β-lactamase species in recent years; many class α β-lactamase species are less sensitive to sulbactam and tazobactam, and the development of novel β-lactamase inhibitors is expected to reverse the resistance to these drugs.25 In addition, the intensity of use of β-lactamase inhibitor compound preparations is related to the occurrence of drug resistance; therefore, the optimal management of prescription is a recommended method.26
The resistance rate of E. coli to common fluoroquinolones (ciprofloxacin and levofloxacin) was > 60%, and the resistance rate of ciprofloxacin has been significantly increasing year by year. It has been reported that the sensitivity rate of UTI-related E. coli in hospitalized patients to these two drugs is only approximately 30%,27 which was similar to our results. In community-acquired UTIs, the expression of the BLA CTX-M-14 gene of E. coli leads to an increase in drug resistance, and it is necessary to limit the empirical use of cephalosporins and fluoroquinolones for UTI treatments in China.28
Furthermore, because of their disabling and potentially long-lasting side effects, the European Commission has restricted fluoroquinolones to uncomplicated cystitis, and recommended their use only when it is considered inappropriate to treat these infections with other commonly used antimicrobials.29
However, in other studies,30,31 levofloxacin showed good efficacy against E. coli by optimizing dosing regimens and strengthening renal toxicity monitoring while minimizing the toxicity risk. This action may be due to the high bioavailability of levofloxacin, which is maintained at a high concentration in target organs to achieve ideal clinical effects. Therefore, further research is needed to determine whether the high drug resistance rate in the laboratory can be used as the sole indicator of empirical drug use. In addition, carbapenems, cefotetan, and amikacin showed good antibacterial activity against E. coli, while furantoin, as a cheap oral preparation, also had a low drug resistance rate in this study, which is similar to the results of a study in Poland.32 However, a study in India reported that E. coli had a resistance rate of 81.82% to furantoin, which indicates significant differences in antibiotic resistance patterns among different countries and regions.33 In this study, K. pneumoniae is highly sensitive to ertapenem, but the 3-year drug resistance rate of imipenem and cefotetan is approximately 45%, and the drug resistance rate of ceftazidime, cefepime, piperacillin, tazobactam, and ampicillin sulbactam is > 55%. The multiple resistance of K. pneumoniae leads to few antibiotic choices, which poses a great threat to life. K. pneumoniae can produce carbapenemase and gain resistance to carbapenems, and the strain with this characteristic is resistant to penicillin and cephalosporin. Studies34,35 have reported that such drug-resistant bacteria tend to spread. In the case of suspected multidrug-resistant infections, especially UTIs caused by carbapenemase-resistant K. pneumoniae, drug sensitivity tests and molecular biological detection should be conducted as soon as possible to guide the use of antibiotics.34
It is also a common pathogen of nosocomial infection, so the prevention and control measures of nosocomial infections and the management measures of clinical application of antibiotics should be strengthened. In a children’s hospital in China, five patients were infected with OXA-232 carbapenemase-producing K. pneumoniae during hospitalization.35 Strengthening hand hygiene compliance and taking contact precautions are important measures to stop nosocomial outbreaks. In an Italian study,36 the resistance rate of K. pneumoniae to carbapenems and colistin was 76.92% and 73.08%, respectively. In almost the same period, a study from Hebei Province, China, showed that CRKP accounted for only 13.4%–14.6% of all K. pneumoniae isolates collected from 43 tertiary hospitals and was highly susceptible to colistin.37 However, these two studies emphasized that appropriate prevention strategies and rapid and accurate identification of drug-resistant bacteria are essential to control the spread of multidrug-resistant bacterial infections. Immunosuppression, longer stay of hospitalization, ICU admission, antibiotic exposure, surgery, mechanical ventilation, and invasive procedures, such as catheterization or tracheotomy, may account for differences in CRPK infection and resistance rates.38,39 The specimens in our study were mainly collected from general patients in the hospital, and imipenem is the only carbapenem in use in our hospital, which may be the reason for this difference. In a word, infections caused by K. pneumoniae require special attention, and susceptibility may vary by institution and over time.
Although UTIs are mainly caused by gram-negative bacteria, gram-positive bacteria have also become important pathogens. Advanced age, diabetes, pregnancy, and indwelling catheter are all risk factors for infection.40 Gram-positive bacteria can maintain a high affinity for urinary tract epithelial cells by producing a variety of virulence factors, leading to pathogenicity.6 In catheter-related infections, 74% of cases lead to acute and persistent infections through biofilm formation, which is also an important cause of antibiotic resistance.40 E. faecium and E. faecalis accounted for 30.4% of the pathogens in this study, while Enterococcus spp accounted for only 3.5% of the pathogens in a simple UTI pathogen analysis conducted in southern China.41 Because most of the specimens were collected from hospitalized patients, older patients and catheter-related infections may be responsible for the high proportion of gram-positive bacteria in this study.
Both Enterococcus species have low sensitivity to levofloxacin and ciprofloxacin and are characterized by widespread resistance to the most commonly used antimicrobials. Because enterococci are characterized by a natural resistance to many antibiotics and rapidly increasing acquired resistance, it is much more difficult to treat enterococci UTIs than those caused by other bacteria.6 It has been reported that outbreaks of hospital-acquired vancomycin-resistant enterococcal infections, which cause severe nosocomial infections, are increasing worldwide.42 A few vancomycin-resistant strains of E. faecium and E. faecalis were cultured in this study, and it is necessary to strengthen the surveillance, and prevent and control nosocomial infection.
The retrospective nature is a limitation of the current study. In addition, this study was only based on the data reported in the laboratory and failed to conduct genome analysis on the multidrug-resistant strains. Furthermore, we analyzed susceptibility patterns for only the top four urological pathogens. The remaining microbes were not sufficient for analysis in this study. We also did not conduct drug sensitivity tests on the cultured fungal strains.
Conclusion
Our results will help clinicians better understand the microbiological characteristics of UTI pathogens and increased antibiotic resistance rates in this region. In the midstream urine cultures used in this study, E. coli is still the most common pathogen of UTI, followed by E. faecium and K. pneumoniae. Moreover, we noted that the resistance of some pathogens to biotin was increasing. We recommend that bacterial culture and drug sensitivity tests should be conducted before antibiotic use to guide the rational use of antibiotics.
Acknowledgment
We are grateful to all the authors for their contributions to this study.
Funding Statement
This study was supported by the Shanghai Chongming District Innovation Talent Project (Project Number 2020CMCX05) and the Science and Technology Commission of Chongming District, Shanghai Municipality (CKY-2020-24).
Abbreviations
UTIs, Urinary tract infections; K. pneumoniae, Klebsiella pneumoniae; UPEC, Uropathogenic Escherichia coli; CRKP, Carbapenem-resistant Klebsiella pneumoniae.
Ethics Approval
The study was approved by the Ethics Committee of Chongming Branch of Shanghai Tenth People’s Hospital, Shanghai2 and the Tongji University School of Medicine, Shanghai, China (No. 2020111205). The ethics committee waived the need for written informed consent provided by participants due to the retrospective nature of this study. Patients’ anonymous information was provided from the microbiology hospital laboratory, which isolated the strains. The study completely followed the principles outlined in the Declaration of Helsinki.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Wagenlehner FME, Bjerklund Johansen TE, Cai T., et al. Epidemiology, definition and treatment of complicated urinary tract infections. Nat Rev Urol. 2020;17(10):586–600. doi: 10.1038/s41585-020-0362-4 [DOI] [PubMed] [Google Scholar]
- 2.Christaki E, Marcou M, Tofarides A. Antimicrobial resistance in bacteria: mechanisms, evolution, and persistence. J Mol Evol. 2020;88(1):26–40. doi: 10.1007/s00239-019-09914-3 [DOI] [PubMed] [Google Scholar]
- 3.Chokshi A, Sifri Z, Cennimo D, et al. Global contributors to antibiotic resistance. J Glob Infect Dis. 2019;11(1):36–42. doi: 10.4103/jgid.jgid_110_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson M, Schulze K, Cassini A, et al. A governance framework for development and assessment of national action plans on antimicrobial resistance. Lancet Infect Dis. 2019;19(11):e371–e384. doi: 10.1016/S1473-3099(19)30415-3 [DOI] [PubMed] [Google Scholar]
- 5.Cassini A, Högberg LD, Plachouras D, et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect Dis. 2019;19(1):56–66. doi: 10.1016/S1473-3099(18)30605-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gajdács M, Ábrók M, Lázár A, et al. Increasing relevance of Gram-positive cocci in urinary tract infections: a 10-year analysis of their prevalence and resistance trends. Sci Rep. 2020;10(1):17658. doi: 10.1038/s41598-020-74834-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. 2017;41(3):252–275. doi: 10.1093/femsre/fux013 [DOI] [PubMed] [Google Scholar]
- 8.Raeispour M, Ranjbar R. Antibiotic resistance, virulence factors and genotyping of Uropathogenic Escherichia coli strains. Antimicrob Resist Infect Control. 2018;7(1):118. doi: 10.1186/s13756-018-0411-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khonsari MS, Behzadi P, Foroohi F. The prevalence of type 3 fimbriae in Uropathogenic Escherichia coli isolated from clinical urine samples. Meta Gene. 2021;28:100881. doi: 10.1016/j.mgene.2021.100881 [DOI] [Google Scholar]
- 10.Hozzari A, Behzadi P, Kerishchi Khiabani P, et al. Clinical cases, drug resistance, and virulence genes profiling in Uropathogenic Escherichia coli. J Appl Genet. 2020;61(2):265–273. doi: 10.1007/s13353-020-00542-y [DOI] [PubMed] [Google Scholar]
- 11.Behzadi P. Classical chaperone-usher (CU) adhesive fimbriome: uropathogenic Escherichia coli (UPEC) and urinary tract infections (UTIs). Folia Microbiol (Praha). 2020;65(1):45–65. doi: 10.1007/s12223-019-00719-x [DOI] [PubMed] [Google Scholar]
- 12.Partridge SR, Kwong SM, Firth N, et al. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev. 2018;31(4):e00088–17. doi: 10.1128/CMR.00088-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rozwadowski M, Gawel D. Molecular Factors and Mechanisms Driving Multidrug Resistance in Uropathogenic Escherichia coli —An Update. Genes. 2022;13(8):1397. doi: 10.3390/genes13081397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alvarez-Uria G, Gandra S, Mandal S, et al. Global forecast of antimicrobial resistance in invasive isolates of Escherichia coli and Klebsiella pneumoniae. Int J Infect Dis. 2018;68:50–53. doi: 10.1016/j.ijid.2018.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang L, Huang C, Yan Y, et al. Urinary tract infection etiological profiles and antibiotic resistance patterns varied among different age categories: a retrospective study from a tertiary general hospital during a 12-year period. Front Microbiol. 2022;12:813145. doi: 10.3389/fmicb.2021.813145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ulrich N, Vonberg R-P, Gastmeier P. Outbreaks caused by vancomycin-resistant Enterococcus faecium in hematology and oncology departments: a systematic review. Heliyon. 2017;3(12):e00473. doi: 10.1016/j.heliyon.2017.e00473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bender JK, Hermes J, Zabel LT, et al. Controlling an unprecedented outbreak with vancomycin-resistant Enterococcus faecium in Germany, October 2015 to November 2019. Microorganisms. 2022;10(8):1603. doi: 10.3390/microorganisms10081603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gast KB, van Oudheusden AJG, Murk JL, et al. Successful containment of two vancomycin-resistant Enterococcus faecium (VRE) outbreaks in a Dutch teaching hospital using environmental sampling and whole-genome sequencing. J Hosp Infect. 2021;111:132–139. doi: 10.1016/j.jhin.2021.02.007 [DOI] [PubMed] [Google Scholar]
- 19.Zhou W, Zhou H, Sun Y, et al. Characterization of clinical enterococci isolates, focusing on the vancomycin-resistant enterococci in a tertiary hospital in China: based on the data from 2013 to 2018. BMC Infect Dis. 2020;20(1):356. doi: 10.1186/s12879-020-05078-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ripabelli G, Lucia. SM, Scutellà M, Felice V, Manuela T. Escherichia coli Carbapenem-Resistant KPC- and TEM-Producing ST131 Isolated from a Hospitalized Patient with Urinary Tract Infection: first Isolation in Molise Region, Central Italy, July 2018. Microb Drug Resist. 2020;26(1):38–45. doi: 10.1089/mdr.2019.0085 [DOI] [PubMed] [Google Scholar]
- 21.Bilal H, Khan MN, Rehman T, et al. Antibiotic resistance in Pakistan: a systematic review of past decade. BMC Infect Dis. 2021;21(1):244. doi: 10.1186/s12879-021-05906-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian L, Zhang Z, Sun Z. Antimicrobial resistance trends in bloodstream infections at a large teaching hospital in China: a 20-year surveillance study (1998-2017). Antimicrob Resist Infect Control. 2019;8(1):86. doi: 10.1186/s13756-019-0545-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tandogdu Z, Wagenlehner FM. Global epidemiology of urinary tract infections. Curr Opin Infect Dis. 2016;29(1):73–79. doi: 10.1097/QCO.0000000000000228 [DOI] [PubMed] [Google Scholar]
- 24.Larramendy S, Deglaire V, Dusollier P, et al. Risk Factors of Extended-Spectrum Beta-Lactamases-Producing Escherichia coli Community Acquired Urinary Tract Infections: a Systematic Review. Infect Drug Resist. 2020;13:3945–3955. doi: 10.2147/IDR.S269033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hofer U. Novel metallo-β-lactamase inhibitors. Nat Rev Microbiol. 2022;20(3):125. [DOI] [PubMed] [Google Scholar]
- 26.Cusini A, Herren D, Bütikofer L, et al. Intra-hospital differences in antibiotic use correlate with antimicrobial resistance rate in Escherichia coli and Klebsiella pneumoniae: a retrospective observational study. Antimicrob Resist Infect Control. 2018;7(1):89. doi: 10.1186/s13756-018-0387-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Q, Zhang H, Wang Y, et al. Antimicrobial susceptibilities of aerobic and facultative gram-negative bacilli isolated from Chinese patients with urinary tract infections between 2010 and 2014. BMC Infect Dis. 2017;17(1):192. doi: 10.1186/s12879-017-2296-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jia P, Zhu Y, Li X, et al. High prevalence of extended-spectrum beta-lactamases in Escherichia coli strains collected from strictly defined community-acquired urinary tract infections in adults in China: a multicenter prospective clinical microbiological and molecular study. Front Microbiol. 2021;12:663033. doi: 10.3389/fmicb.2021.663033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.European Medicines Agency. Disabling and potentially permanent side effects lead to suspension or restrictions of quinolone and fluoroquinolone antibiotics. Quinolone and fluoroquinolone Article-31 referral; 2019. Available from: https://www.ema.europa.eu/en/documents/referral/quinolone-fluoroquinolone-article-31-referral-disabling-potentially-permanent-side-effects-lead_en.pdf. Accessed October 26, 2022.
- 30.Canouï E, Kerneis S, Morand P, et al. Oral levofloxacin: population pharmacokinetics model and pharmacodynamics study in bone and joint infections. J Antimicrob Chemother. 2022;77(5):1344–1352. doi: 10.1093/jac/dkac031 [DOI] [PubMed] [Google Scholar]
- 31.Cojutti PG, Ramos-Martin V, Schiavon I, et al. Population pharmacokinetics and pharmacodynamics of levofloxacin in acutely hospitalized older patients with various degrees of renal function. Antimicrob Agents Chemother. 2017;61(3). doi: 10.1128/AAC.02134-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kot B, Wicha J, Grużewska A, et al. Virulence factors, biofilm-forming ability, and antimicrobial resistance of urinary Escherichia coli strains isolated from hospitalized patients. Turk J Med Sci. 2016;46(6):1908–1914. doi: 10.3906/sag-1508-105 [DOI] [PubMed] [Google Scholar]
- 33.Prakash D, Saxena RS. Distribution and antimicrobial susceptibility pattern of bacterial pathogens causing urinary tract infection in urban community of Meerut city, India. ISRN Microbiol. 2013;2013:749629. doi: 10.1155/2013/749629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chapelle C, Gaborit B, Dumont R, et al. Treatment of UTIs Due to Klebsiella pneumoniae Carbapenemase-Producers: how to Use New Antibiotic Drugs? A Narrative Review. Antibiotics. 2021;10(11). doi: 10.3390/antibiotics10111332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yin D, Dong D, Li K, et al. Clonal dissemination of OXA-232 carbapenemase-producing Klebsiella pneumoniae in neonates. Antimicrob Agents Chemother. 2017;61(8). doi: 10.1128/AAC.00385-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Di Tella D, Tamburro M, Guerrizio G, et al. Molecular Epidemiological Insights into Colistin-Resistant and Carbapenemases-Producing Clinical Klebsiella pneumoniae Isolates. Infect Drug Resist. 2019;12:3783–3795. doi: 10.2147/IDR.S226416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang N, Zhan M, Liu J, et al. Prevalence of carbapenem-resistant Klebsiella pneumoniae infection in a Northern Province in China: clinical characteristics, drug resistance, and geographic distribution. Infect Drug Resist. 2022;15:569–579. doi: 10.2147/IDR.S347343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lou T, Du X, Zhang P, et al. Risk factors for infection and mortality caused by carbapenem-resistant Klebsiella pneumoniae: a large multicentre case-control and cohort study. J Infect. 2022;84(5):637–647. doi: 10.1016/j.jinf.2022.03.010 [DOI] [PubMed] [Google Scholar]
- 39.Li J, Li Y, Song N, et al. Risk factors for carbapenem-resistant Klebsiella pneumoniae infection: a meta-analysis. J Glob Antimicrob Resist. 2020;21:306–313. doi: 10.1016/j.jgar.2019.09.006 [DOI] [PubMed] [Google Scholar]
- 40.Shrestha LB, Baral R, Khanal B. Comparative study of antimicrobial resistance and biofilm formation among Gram-positive uropathogens isolated from community-acquired urinary tract infections and catheter-associated urinary tract infections. Infect Drug Resist. 2019;12:957–963. doi: 10.2147/IDR.S200988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong CKM, Kung K, Au-Doung PLW, et al. Antibiotic resistance rates and physician antibiotic prescription patterns of uncomplicated urinary tract infections in southern Chinese primary care. PLoS One. 2017;12(5):e0177266. doi: 10.1371/journal.pone.0177266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao W, Howden BP, Stinear TP. Evolution of virulence in Enterococcus faecium, a hospital-adapted opportunistic pathogen. Curr Opin Microbiol. 2018;41:76–82. doi: 10.1016/j.mib.2017.11.030 [DOI] [PubMed] [Google Scholar]