Significance
In the field of drug delivery, supramolecular materials made from self-assembled prodrugs can greatly improve drug loading efficiencies and sustain release kinetics, especially for poorly soluble, hydrophobic drugs. Here, we report the development of such materials using a small-molecule, proregenerative drug and a low-molecular–weight, hydrophilic polymer. Through experimental characterization and computational modeling, we show that subtle changes in prodrug molecular architecture have dramatic effects on self-assembly morphology, drug release profiles, and cellular internalization. In this structure–property analysis, we develop a library of carriers for our versatile drug of interest while simultaneously establishing fundamental principles in prodrug design and amphiphile self-assembly behavior that can be easily applied to future drug–polymer conjugates.
Keywords: supramolecular, self-assembly, drug delivery
Abstract
Supramolecular self-assemblies of hydrophilic macromolecules functionalized with hydrophobic, structure-directing components have long been used for drug delivery. In these systems, loading of poorly soluble compounds is typically achieved through physical encapsulation during or after formation of the supramolecular assembly, resulting in low encapsulation efficiencies and limited control over release kinetics, which are predominately governed by diffusion and carrier degradation. To overcome these limitations, amphiphilic prodrugs that leverage a hydrophobic drug as both the therapeutic and structure-directing component can be used to create supramolecular materials with higher loading and controlled-release kinetics using biodegradable or enzymatically cleavable linkers. Here, we report the design, synthesis, and characterization of a library of supramolecular polymer prodrugs based on poly(ethylene glycol) (PEG) and the proregenerative drug 1,4-dihydrophenonthrolin-4-one-3-carboxylic acid (DPCA). Structure–property relationships were elucidated through experimental characterization of prodrug behavior in both the wet and dry states using scattering techniques and electron microscopy and corroborated by coarse-grained modeling. Molecular architecture and the hydrophobic-to-hydrophilic ratio of PEG–DPCA conjugates strongly influenced their physical state in water, ranging from fully soluble to supramolecular spherical assemblies and nanofibers. Molecular design and supramolecular structure, in turn, were shown to dramatically alter hydrolytic and enzymatic release and cellular transport of DPCA. In addition to potentially expanding therapeutic options for DPCA through control of supramolecular assemblies, the design principles elaborated here may inform the development of other supramolecular prodrugs based on hydrophobic small-molecule compounds.
Hydrophobic small-molecule drugs continue to dominate the pharmaceutical industry, accounting for an estimated 60% of all compounds currently in research and development (1). In their bioactive form, however, these hydrophobic compounds often exhibit poor bioavailability due to their low aqueous solubility (1, 2). To overcome this challenge, macromolecular prodrugs have been developed by covalently coupling the bioactive compound to a synthetic polymer, peptide, or polysaccharide in an effort to optimize pharmacokinetic profiles (3). Through careful molecular design, prodrugs based on hydrophobic drugs and hydrophilic polymers can be created to self-assemble into supramolecular aggregates due to their amphiphilic nature (3, 4). As drug delivery vehicles, these supramolecular structures offer several advantages over conventional soluble prodrugs, including decreased renal clearance and extended drug release kinetics (5, 6). While many of these attributes may also be achieved through physical encapsulation of drug within nanocarriers, such as polymeric micelles (6), single-chain nanoparticles (7), and surfactant micelles (8), significantly higher drug loading of poorly soluble compounds is often achieved with prodrug aggregates, and the risk of carrier toxicity is eliminated. Recently, we reported on a self-assembling prodrug system using the proregenerative drug 1,4-dihydrophenonthrolin-4-one-3-carboxylic acid (DPCA). Our polymer prodrug design consisted of hydrophobic DPCA conjugated to a hydrophilic, linear poly(ethylene glycol) (PEG) chain via a hydrolyzable ester bond, thereby using DPCA as both the therapeutic payload and structural element. In aqueous solutions above a critical concentration, the PEG–DPCA conjugates assembled into well-defined nanofibers consisting of a drug-rich core and polymeric corona, with tenfold higher loading than previous hydrogel systems (9, 10). Such nonspherical assemblies of prodrug structures have been rarely reported, and clear structure–property principles that would guide their design have yet to be established (3, 4, 11–13).
The full benefits of prodrug delivery systems at large can only be realized through improved understanding of fundamental structure–property behaviors, such as the influence of molecular design on supramolecular aggregate morphology, drug release kinetics, and bioactivity. Insight into such structure–property relationships will in turn directly inform prodrug route of administration, dosing, and ultimate clinical utility. Our drug of interest, DPCA, targets ubiquitously expressed cellular oxygen sensing pathways to induce regeneration of damaged tissues (9, 14). Because the classical wound healing cascade is conserved across organ systems, we hypothesize that treatment with DPCA is likely applicable to a broad array of injuries and degenerative diseases. Therefore, there is considerable motivation to overcome challenges in DPCA delivery and optimize prodrug behavior for multitissue targeting. To fully exploit the therapeutic utility of DPCA and begin to predict the bioactivity and degradation behavior of a model macromolecular prodrug, this study evaluates the fundamental structure–property relationship between molecular architecture, self-assembly, and drug carrier performance. We present a detailed investigation into the assembly and stability of three PEG–DPCA conjugates of varying hydrophobic (DPCA) to hydrophilic (PEG) stoichiometries. Using a variety of wet- and dry-state imaging and scattering techniques as well as coarse-grained modeling, we studied PEG–DPCA prodrugs containing one, two, or three drug molecules per polymer chain (abbreviated P7D1, P7D2, and P7D3). We observed a transition from soluble spherical prodrug aggregates to worm-like micelles with increasing drug content. Furthermore, we explored the relationship between prodrug structure and mechanism of drug release, including partial hydrolysis of multidrug conjugates and susceptibility to enzymatic cleavage. We then found that prodrug molecular architecture influenced cellular internalization, possible drug binding, and in vitro bioactivity. The results of this study highlight the importance of molecular architecture in the performance of PEGylated drugs, suggesting avenues for precise control over drug dosing without sacrificing the stability of assembled, supramolecular objects. Our findings establish clear design principles for the development of future prodrug systems based on DPCA and related hydrophobic drugs.
Results and Discussion
Prodrug Design, Synthesis, and Purification.
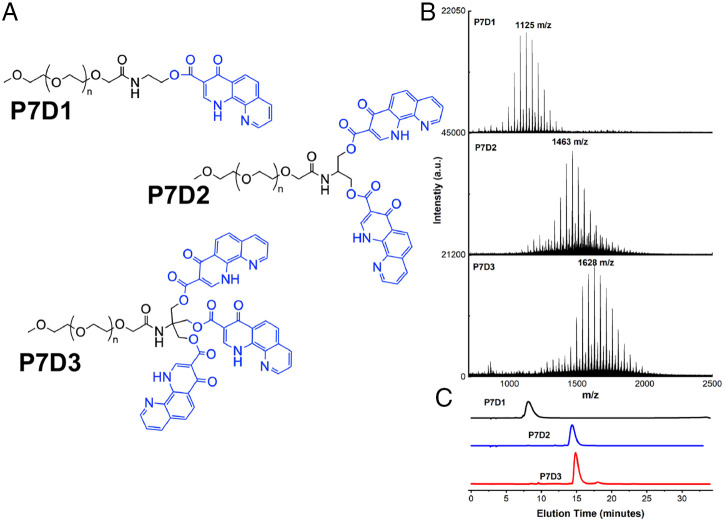
Fig. 1 outlines the molecular design and characterization of the three prodrugs analyzed in this study. Each prodrug consists of a linear monofunctional PEG chain (number average molecular weight (Mn) 750 g/mol) functionalized with a mono-, di-, or trihydroxy end group via an amide linker, allowing for conjugation of one, two, or three molecules of DPCA. The rationale for our prodrug design stems from the hypothesis that intermolecular hydrogen bonding, van der Waals interactions, and π–π stacking interactions among DPCA motifs directs the formation of supramolecular structures with a drug-rich core and a hydrophilic PEG corona that affords colloidal stability and prevents protein adsorption. With respect to drug delivery, the ester bonds of PEG–DPCA allow drug release via hydrolysis when exposed to aqueous environments in vivo.
Fig. 1.
(A) Molecular structures of PEG-based prodrugs P7D1, P7D2, and P7D3. In each case, DPCA (blue) is conjugated to a 750 g/mol linear PEG (black) via a hydrolysable ester linkage. (B and C) Successful drug coupling is verified by increasing molecular weight, as seen by MALDI-TOF-MS (B) and elution time in analytical HPLC (C). After purification, free DPCA and PEG as well as partially coupled prodrugs are removed; a.u. arbitrary units.
Detailed synthetic procedures for PEG–DPCA prodrug synthesis can be found in the SI Appendix, Supplementary Information Text. Briefly, (2,2,6,6-Tetramethylpiperidin-1-yl)oxyl (TEMPO)-mediated oxidation of methoxy PEG was followed by 3-[Bis(dimethylamino)methyliumyl]-3H-benzotriazol-1-oxide hexafluorophosphate (HBTU)-mediated coupling of 2-aminoethanol, 2-amino-1,3-propanediol, or 2-amino-2-(hydroxymethyl)-1,3-propanediol to yield mono-, di-, or trihydroxy functional PEG (PEG-[OH]1, PEG-[OH]2, of PEG-[OH]3). DPCA was then conjugated to the terminal hydroxyls of the PEG chains via 1,1′-carbonyldiimidazole–activated esterification to generate the prodrugs P7D1, P7D2, and P7D3 (1H-NMR in SI Appendix, Fig. S4). Before characterization, crude reaction products containing partially coupled prodrugs or free, uncoupled DPCA were purified via semipreparative high-performance liquid chromatography (HPLC) fractionation or silica gel column chromatography. Final products were characterized via analytical HPLC and mass spectrometry (MS), showing a final prodrug purity of >95 mol% P7DX (X represents 1, 2, or 3) content.
Self-Assembly of PEG–DPCA Prodrugs.
Self-assembly of PEG–DPCA prodrugs was achieved by direct dissolution of lyophilized powders into pure water or phosphate-buffered saline (PBS) at 50 °C for 30 min. A sol-gel transition from a gel to liquid-state, was previously reported to occur at 50 °C for P7D3, allowing for uniform mixing to occur, and at this temperature, visual dissolution of all prodrugs is achieved (10). The solution was then cooled to room temperature (25 °C) for 1 h with slight agitation before characterization. Rheological characterization of assembled samples showed that at high concentrations, P7D3 suspensions displayed a high-viscosity, shear-thinning behavior characteristic of entangled nanofiber solutions, as previously reported (10, 15). However, this behavior was not observed in P7D1 and P7D2 samples, which instead exhibited shear responses comparable to water, even at 100 mg/mL (SI Appendix, Fig. S7).
To determine if PEG–DPCA prodrugs formed stable, supramolecular aggregates beyond a critical micelle concentration (CMC)/critical fiber concentration, a Nile Red probe assay was used (16, 17). While the fluorescence of Nile Red is quenched in water, it emits strongly in hydrophobic environments when excited at 490 nm. Thus, partition of the dye into the hydrophobic, DPCA-rich core during self-assembly of prodrug structures is likely to be captured by a nonlinear increase in Nile Red fluorescence with prodrug concentration. This effect was observed for all PEG–DPCA conjugates, suggesting aggregation above a critical concentration in the range of 1 mM. This value is in good agreement with PEG-phenanthroline compounds reported by Nagy et al. (18, 19). Interestingly, the CMC values for PEG–DPCA assemblies do not seem to be directly related to DPCA content (Table 1 and SI Appendix, Fig. S8). This is surprising, as the hydrophobic nature of DPCA is presumed to be the main driving force for self-assembly. We speculate that this result does not arise due to limitations in our experimental technique, as we were able to capture accurate CMC values for the common surfactant Triton-X at lower concentrations (SI Appendix, Fig. S9). We also wondered if the effect could be due to quenching of Nile Red by DPCA; however, no changes in Nile Red fluorescence in the presence of soluble DPCA were observed (SI Appendix, Fig. S9). We also repeated the experiment using pyrene as an alternative probe. Changes in pyrene peak emission at 372 nm and 384 nm were monitored as a function of prodrug concentration at 334-nm excitation. When transitioning from a polar to nonpolar environment, the ratio between peak emission intesities (I372/I384) is expected to decrease (20). We observed a dramatic decrease in I372/I384 peak ratios for all prodrugs at concentrations close to their estimated CMC, suggesting diffusion of pyrene into hydrophobic, DPCA-rich cores (SI Appendix, Fig. S10). While it is possible that closely associating DPCA-rich cores may exclude our fluorescent probes, we have verified through superresolution stochastic imaging (structured illumination microscopy [SIM]) that dye encapsulation is possible within assembled PEG–DPCAs (Fig. 2D). Finally, in the case of P7D3, which was predicted to have the lowest CMC of the prodrugs studied, we observed a loss of shear-thinning properties indicative of nanofiber entanglement below the reported CMC (SI Appendix, Fig. S11). Therefore, we attribute the observed trend showing the lowest CMC value for P7D2, followed by P7D1 and P7D3, to prodrug geometry, thus highlighting an important structure–property relationship. Possibly, the stereochemical limitations for rotation within the spacer molecules for one, two, and three DPCA units can act as a limiting factor for self-assembly. Thus, assemblies of different shapes may occur at the same concentration. It is possible that for both P7D2 and P7D3 prodrugs, the presence of intramolecular interactions between DPCA molecules on the PEG chain increase prodrug solubility and CMC, despite their large, hydrophobic volume fraction, compared to P7D1. This phenomenon would also explain the higher CMC for P7D3 than for P7D2. Indirect evidence of such intramolecular DPCA–DPCA interactions on PEG chains may be gleaned from differences in ultraviolet–visible spectroscopy (UV-Vis) absorption spectra between PEG–DPCAs and free drug. At concentrations below estimated CMC (0.1 mg/mL for all samples), the absorption spectrum for P7D1 closely resembled the shape of saturated aqueous solutions of DPCA (SI Appendix, Fig. S12). Following conjugation of two or three molecules of DPCA to PEG, a red shift in DPCA peak absorption at 316 nm, 333 nm, and 348 nm was found to occur with increasing conjugation rate. Such shifts may be indicative of π–π stacking between intramolecular DPCA because such interactions have been shown to shift the absorbance spectra of other compounds, such as benzene and various chromophores, to longer wavelengths (21, 22). Notably, changes in the concentration of free DPCA in saturated aqueous solutions did not result in peak shifts.
Table 1.
CMC values for polymer prodrugs with one, two, or three DPCA molecules per PEG
| Prodrug structure | Average CMC (P7DX mg/mL) | Average CMC (P7DX mM) |
|---|---|---|
| P7D1 | 2 ± 0.5 | 1.85 |
| P7D2 | 1 ± 0.2 | 0.73 |
| P7D3 | 3.2 ± 0.7 | 2.1 |
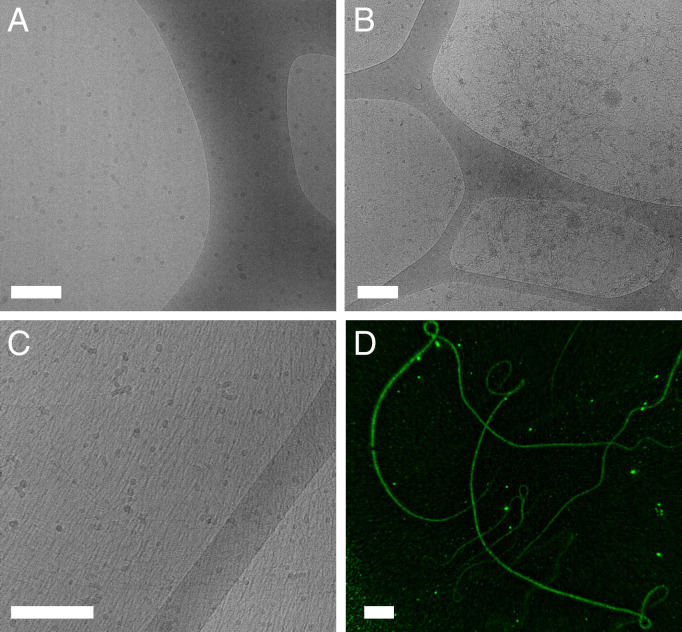
Fig. 2.
(A–D) Cryo-EM micrographs of PEG–DPCA prodrugs prepared at 25 g/L (A–C) and high-resolution SIM imaging of P7D3 at 3 mg/mL (D). For P7D1 (A) and P7D2 (B) samples, spherical assemblies dominate, with the emergence of some worm-like micelles in P7D2. For P7D3 (C), single fibers and bundles are observed, measuring microns in length (D). All self-assembled structures presumably consist of a DPCA-rich core and PEG corona. (Scale bar, 200 nm in A–C or 4 µm in D.)
After indirectly verifying self-assembly using the Nile Red probe assay, the morphology of PEG–DPCA supramolecular structures was visualized through both dry- and wet-state imaging of aqueous suspensions above CMC. In Fig. 2 A and B, micrographs obtained from cryo-electron microscopy (cryo-EM) showed predominately spherical, micelle-like structures for P7D1 and P7D2 with average diameters of 10 nm (median = 8 nm) and 11 nm (median = 9 nm), respectively (SI Appendix, Fig. S13). Deviations between P7D1 and P7D2 dimensions were deemed to be insignificant due to the limitations in image processing. A small population of worm-like micelles with average width of 5 nm was also observed in P7D2. When we examined the rheological behavior of P7D2 solutions, however, we did not observe increases in viscosity or shear-rate dependences indicative of fiber entanglement (SI Appendix, Fig. S7. Therefore, we believe that spherical assemblies are the dominate phase. For P7D1, dynamic light scattering (DLS) at 3g/L suggested the presence of assemblies with number average diameters of 2.2 nm and Z average of 40 nm (SI Appendix, Fig. S14). These values differed significantly from free PEG solutions, which measured <1 nm in diameter (SI Appendix, Fig. S14), and with DPCA unit volume of ∼0.2 nm3 and 0.5 nm3, as determined by estimated bond length for P7D1 and P7D2, respectively, the observed nanoaggregate geometry and size are in good agreement with theoretical predictions (10). When the number of DPCA molecules per PEG chain is increased to three, the sparse worm-like micelle phase visualized in P7D2 begins to strongly dominate (Fig. 2C). Highly aligned fiber scattering was noted in the majority of P7D3 samples due to the formation of near–two-dimensional films during sample preparation, and from these domains, an estimated fiber diameter of 8 nm was determined through image analysis (SI Appendix, Fig. S13). SIM of P7D3 fibers loaded with the lipophilic dye 3,3-diheptyloxacarbocyanine iodide (DiOC7) highlighted the flexibility and length of such fibers not captured by cryo-EM. Dry-state imaging of films cast from aqueous suspensions of all prodrugs obtained via conventional transmission electron microscopy (TEM) also showed similar assembly morphologies (SI Appendix, Fig. S15). To further corroborate findings from dry- and wet-state imaging, small angle x-ray scattering (SAXS) was also performed on aqueous solutions at 25 g/L. The one-dimensional plot of the samples shown in SI Appendix, Fig. S16, indicates a strong structural dependence on the objects depending on the number of DPCA units, all deviating from a PEG reference solution. Despite the difficulty in fitting the curves to a specific geometry, explained also by the distribution of structures seen in EM images and noted in Fig. 2, the cylindrical geometry expected in P7D3 is supported by the rapid upturn at 0.04 Å−1 with a slope ∼−1. The flatter form factor of P7D2 supports dispersed spheres, although the feature size appears to be smaller than that of P7D3. No apparent form factor is demonstrated in P7D1, possibly due to minimal scattering intensity at high q. The constant slope could indicate agglomeration of the expected spherical micelles in the concentrated solution. Overall, these conclusions on wet-state assembly strongly corroborate cryo-EM imaging. For all prodrugs, dispersity in aggregate morphology and size observed in both EM and scattering data likely arise from the polydispersity of the PEG (Fig. 1B) rather than differences in self-assembly behavior. The array of supramolecular objects identified here may have differential clinical utility due to shape-dependent biodistribution patterns and modes of possible delivery (23, 24).
Molecular Dynamics Simulations.
We argue that, in accordance with traditional amphiphile assembly pathways (25), PEG–DPCA self-assembly is driven by two key forces: molecular geometry and the contrasting solvent affinities of hydrophilic PEG and hydrophobic DPCA. To interrogate these driving forces, we conducted coarse-grained molecular dynamics simulations to capture the salient intermolecular interactions and structural features of P7D1, P7D2, and P7D3. Unlike commonly examined amphiphiles, the PEG–DPCA polymer–drug conjugates feature a flexible hydrophilic component (PEG) and a bulky hydrophobic component (one, two, or three DPCAs) (26–30). Using an implicit solvent model, we account for these contrasting solvent affinities with different interaction potentials. Each DPCA molecule is represented as a single bead that is attracted to other DPCA beads, while the PEG chain is modeled as linearly connected volume-excluding beads. A rigid body constraint (31) was used to enforce the relative arrangement of intramolecular DPCA beads to match the lowest energy geometry identified through the Molecular Mechanics 2 (MM2) force field, a commonly applied force field for determining conformations of hydrocarbon chains and small organic molecules (32). The resulting molecular topologies are shown in Fig. 3A and are further described in the SI Appendix, Supplementary Information Text. Simulations were conducted at volume fractions 0.002 < < 0.008 and DPCA attraction strengths of < εA < (Materials and Methods). This volume fraction range was selected to explore concentrations at which micelles and fibers have been observed experimentally, while the explored εA range was chosen based on the range of DPCA–DPCA attraction strength found in DSC measurements of the enthalpy of fusion of pure DPCA. Equilibrated simulations (SI Appendix, Supplementary Information Text) result in a variety of self-assembled structures featuring a DPCA core surrounded by a PEG corona (Fig. 3 D–F, Inset).
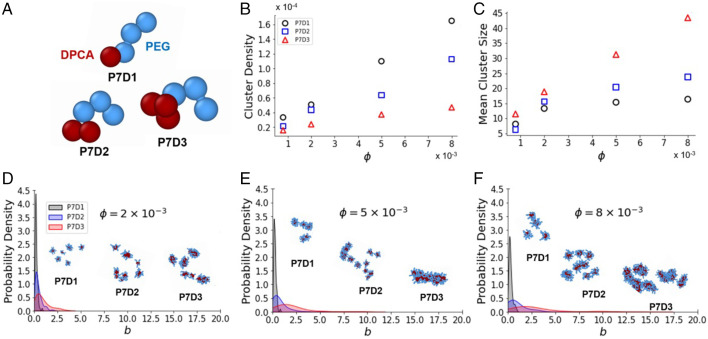
Fig. 3.
(A) Increasing the number of DPCA molecules leads to more fiber-like aggregates for coarse-grained representations of P7D1, P7D2, and P7D3 featuring a flexible polymer chain (blue) appended to one, two, or three DPCA molecules (red). (B and C) Dependence of cluster density (in units of ; Materials and Methods) on volume fraction is noted (B) as well as dependence of mean cluster size (number of molecules per cluster) on volume fraction (C). (D–F) Asphericity distributions ( in units of ) at volume fractions of 0.002 (D), 0.005 (E), and 0.008 (F) highlight differences in assembly morphologies. Simulation snapshots depict spherical micelles formed by P7D1, elongated micelles formed by P7D2, and fibers formed by P7D3. All data are for an attraction strength of εA = 12.5kBT.
For a DPCA attraction strength of εA = (the approximate experimental value at room temperature as determined by differential scanning calorimety (DSC); SI Appendix, Fig. S6 and Supplementary Information Text), increasing the DPCA content of the amphiphile resulted in fewer clusters forming per volume (Fig. 3B) but larger clusters (Fig. 3C) at all but the lowest density examined. Moreover, the concentration dependence of the cluster density and mean cluster size was qualitatively distinct for the P7D3 compared with the P7D1 and P7D2 systems. The size of P7D3 clusters increased linearly with concentration, while the cluster density exhibited a sublinear increase. By contrast, P7D1 and P7D2 both exhibited a sublinear increase in the mean cluster size but a nearly linear increase in cluster density (Fig. 3C). The disparate scaling of cluster density and cluster size for P7D3 compared with P7D1 and P7D2 points to a morphological difference in the self-assembled aggregates resulting from topological differences in the amphiphiles.
To further explore these morphological differences, we analyzed the shapes of the self-assembled aggregates by computing the asphericity parameter for each cluster (Materials and Methods). For perfectly spherical clusters, the asphericity parameter is identically zero. Larger values of indicate less spherical morphologies, including fiber-like aggregates (33). The probability density of reveals P7D1 clusters to be largely spherical, with asphericities rarely exceeding a value of = 1.0, even at densities an order of magnitude higher than the experimentally observed CMC (this corresponds to = 0.008 in our simulations; SI Appendix, Table S1). P7D2 clusters were slightly less spherical, while the asphericity distribution of P7D3 exhibited a striking departure from those of P7D1 and P7D2 (SI Appendix, Fig. S17). At the same density ( = 0.008), P7D3 self-assembled into highly aspherical structures, as evidenced by the probability density exhibiting a long tail with asphericities > 10.0 not atypical.
Our simulations confirm the presence of fiber-like structures for P7D3 at densities and attraction strengths where P7D1 and P7D2 are largely limited to spherical aggregates. This finding corroborates the experimental evidence that suggests that P7D3 forms a dominate fraction of nanofibers at the conditions examined. The increased tendency for fiber formation with increasing DPCA content cannot be explained with a simple rescaling of the molecular energies (SI Appendix, Fig. S18) and must therefore be a consequence of the hydrophobic topology. While classical amphiphile self-assembly can be well predicted by calculating the critical packing parameter (CPP), in this manuscript, we report on a “reversed amphiphile-like” structure consisting of a flexible, hydrophilic tail (PEG) and bulky hydrophobic head group (DPCA). We attempted to calculate CPPs for PEG–DPCAs using the dimensions of the molecules represented in coarse-grained modeling while considering DPCA molecules as spheres, yielding overall spherical (P7D1), ellipsoidal (P7D2), or triagonal (P7D3) volumes with an ellipsoidal or spherical cross-section/length. We determined that our values only agreed with CPP assembly predictions on aggregate morphology when PEG chains were assumed to have large surface area (>4 nm2), which may result from high degrees of coiling. Even at this low molecular weight, however, PEG is assumed to exist as a loose coil in water (34). Therefore, we believe that our system may deviate from theories of self-assembly commonly applied to surfactants and block copolymers. To probe this point further, future work will examine the precise role of the geometry of the hydrophobic group in the phase behavior of these systems.
In Vitro Drug Release.
Our motivation for understanding structure–property relationships of PEG–DPCA is to exploit new supramolecular structures to control the delivery of DPCA for therapeutic purposes. DPCA is an inhibitor of 4-prolylhydroxylase (PHD), which regulates the transcription factor hypoxia inducible factor-1α (HIF-1α), which is a key component of the cellular response to hypoxia. Hydroxylation of HIF-1α by PHD triggers its degradation; therefore, the presence of DPCA or other PHD inhibitors offers a route to HIF-1α stabilization. Recently, we showed that in vivo delivery of DPCA led to high levels of HIF-1α and regeneration of soft and hard tissues (9, 10, 35). To correlate the structural findings above to drug release, we investigated the time dependence of DPCA release from P7D1, P7D2, and P7D3. As seen in Fig. 4, the kinetics of release are directly related to prodrug molecular design and supramolecular morphology. Although release of DPCA is presumed to occur through hydrolysis of the PEG–DPCA ester linkage for all prodrugs investigated, two separate regimens were considered, below CMC and above CMC.
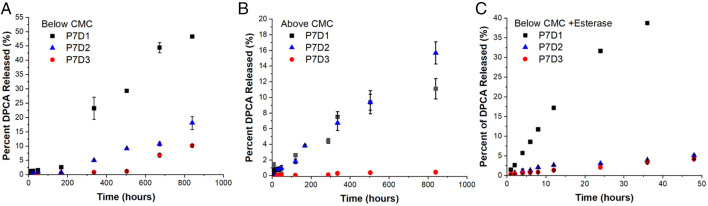
Fig. 4.
(A and B) Drug release from PEG–DPCA prodrugs by hydrolysis in buffered saline was measured below (A) and above (B) CMC. (C) Release of DPCA from P7D1, P7D2, and P7D3 in the presence of esterase enzyme was compared, revealing an acceleration of DPCA release in the presence of esterase and an overall relationship between prodrug hydrophobicity and release kinetics.
Prodrug hydrolysis below CMC.
For soluble (unassembled) polymer prodrugs, the effects of increasing the number of DPCA units at the PEG chain end were observed by monitoring hydrolysis of prodrug solutions below CMC (Fig. 4A). Compared to P7D3, which shows little observable hydrolysis over 2 wk (<2%), hydrolysis of P7D2 and P7D1 appears to result in sustained release kinetics. At the conclusion of 2 wk, hydrolysis of more than 20% of conjugates was observed for P7D1 and ∼5% for P7D2, but 5 wk was required for P7D3 to reach above 5%. Significant differences in prodrug stability can likely be attributed to attractive intramolecular interactions between DPCA molecules, creating an increasingly hydrophobic environment as the number of DPCA molecules per PEG is increased from one to three. A similar relationship between drug conjugation rate and release was noted by Chen et al. in the creation of peptide-based prodrugs containing multiple molecules of doxorubicin (36). The kinetics of hydrolysis below CMC appear to be somewhat inversely related to DPCA content. HPLC and matrix-assisted laser desorption/ionization-time of flight MS (MALDI-TOF-MS) analysis of P7D3 incubated in water revealed the emergence of partially hydrolyzed PEG–DPCA as well as free PEG (SI Appendix, Fig. S19), suggesting that DPCA release may occur in a stepwise fashion through P7D2 and P7D1 intermediates before complete drug release is achieved. Thus, a degradation mechanism forming analogous structures to P7D1/P7D2 from P7D3 likely occurs, further highlighting the importance of these fundamental structure-property analyses to be conducted for multidrug prodrugs.
Prodrug hydrolysis above CMC.
Above CMC (in the self-assembled state), DPCA release was significantly slower for P7D1, but similar kinetics were detected for P7D2 and P7D3 compared to below CMC, where ∼2% of drug was released over the first 4 d and less than 7% after 2 wk (Fig. 4B). Such long-term kinetics have also been observed for filamentous nanostructures formed from peptide–paclitaxel prodrugs (37). The reported slower rate, for especially P7D1, is likely due to decreased exposure of the DPCA ester linkage to water within the hydrophobic core of assembled structures. Compared to hydrolysis data, the release of DPCA from assembled prodrugs appears to be less dependent on drug content. Thus, by modulating the molecular design and self-assembly of prodrugs, drug dosing and release can be controlled.
Prodrug enzymatic cleavage below CMC.
Our previous in vivo study using hydrogels based on P7D3 showed that up-regulation of HIF-1α occurred as soon as 1 d after administration (10), suggesting that the rate of drug release in vivo may be much more rapid than in vitro observations conducted in buffered saline. To better understand this discrepancy, we next sought to determine if PEG–DPCA prodrugs are susceptible to enzymatic hydrolysis by carboxyl esterase enzymes, commonly found in mammalian liver cells and blood plasma. As seen in Fig. 4C, the presence of porcine liver esterase (EC 3.1.1.1) significantly increased DPCA release in P7D1, P7D2, and P7D3 solutions below CMC, with the following release kinetics: P7D1 > P7D2 > P7D3. We note that at 24 h, P7D1 shows significant drug hydrolysis (>30%), while P7D2 and P7D3 only reach 5% hydrolysis in the presence of esterase. We speculate that this effect could be attributed to prodrug structure, with intramolecular associations of DPCA in P7D2 and P7D3 altering substrate recognition and/or accessibility to the esterase active site.
Prodrug Bioactivity.
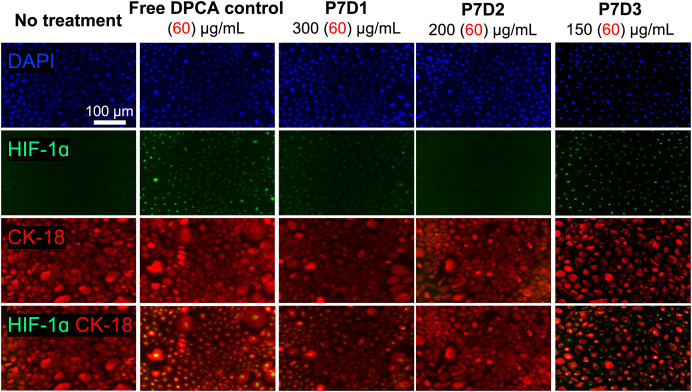
Finally, the ability of prodrugs (P7D1, P7D2, and P7D3) to stabilize HIF-1α was evaluated in primary human amnion endothelial cells (AECs). AECs make up a major component of the fetal membrane and are a potential target for novel regenerative therapies (38). AECs were exposed to medium containing free DPCA, P7D1, P7D2, or P7D3 at a uniform DPCA concentration of 60 µg/mL (below CMC for all prodrugs). We observed that after 4 h, AECs treated with conditioned medium containing P7D3 displayed robust expression of HIF-1α, limited expression following P7D1, and no obvious stabilization in the case of P7D2 compared with free DPCA (Fig. 5). Based on prodrug hydrolysis data (Fig. 4A), we do not expect to observe DPCA release in extracellular medium over this timespan. Therefore, we speculated that the bioactivity of P7D3 may be dependent on cellular internalization because drug release was found to be accelerated when prodrugs were exposed to intracellular esterase (Fig. 4C). Differences in prodrug amphiphilicity and molecular architecture may also preclude passive uptake of P7D1 and P7D2, leading to their inability to stabilize HIF-1α in the present study. To investigate this possibility, we used the inherent fluorescence of DPCA to determine prodrug uptake to assess cellular internalization. Surprisingly, we observed an increase in fluorescence intensity associated with DPCA in live AECs treated with P7D3, but not P7D1 or P7D2 (SI Appendix, Fig. S20). In P7D3-treated cells, DPCA signal was also highly localized to the nucleus, where PHD isoforms PHD1 and PHD3 are known to reside (39). In contrast to the ubiquitously expressed PHD2 isoform, PHD1 and PHD3 are known to exhibit tissue-specific expression patterns, but the presence of PHD3 has been reported in amniotic cells (40). Nuclear localization of factor inhibiting HIF, an alternative regulator of HIF-1α and possible target for DPCA, has also been observed (41, 42). MALDI-TOF-MS of cellular lysates also suggested the presence of intact P7D3 after 24-h incubation with prodrugs. After 48 h of incubation, this signal as well as those from free PEG became more pronounced, suggesting intracellular prodrug cleavage. Lysates from cells incubated with P7D1 showed the weak presence of free PEG, but not P7D1, and no values matching the molecular weight of PEG or prodrugs were found in cells treated with P7D2 (SI Appendix, Fig. S21), further corroborating findings on disparate internalization.
Fig. 5.
Transient stabilization of HIF-1α in AECs treated with growth medium containing free DPCA or PEG–DPCA prodrugs for 4 h. HIF-1α staining (green) is shown overlaid with DAPI nuclear staining (blue) and intermediate filament protein CK-18 (red) to visualize cell boundaries.
Thus, our data suggest a relationship between molecular architecture and biological activity in the PEG–DPCA system. It is well known that cellular uptake is highly dependent on physiochemical properties such as molecular weight, lipophilicity, and protein-binding capacity. Therefore, it is reasonable to assume that alterations in prodrug molecular architecture could lead to disparities in cellular internalization. DPCA is claimed to act as a competitive inhibitor of the 2-oxoglutarate cofactor that is needed for hydroxylation of HIF-1α by PHD enzymes (43–45). However, DPCA is also capable of ligating to the active iron site within PHD enzymes via the phenanthroline and/or carboxylic acid moieties (19, 45). Computational molecular docking of DPCA against HIF hydroxylases has predicted that bidentate iron ligation via the carboxylic acid moiety is preferred, and analogs of DPCA lacking one nitrogen or one pyridine ring exhibit reduced activity (41). Speculating that iron binding by DPCA could be influenced by supramolecular aggregation, we measured the UV-Vis spectra of mixtures of P7D1, P7D2, and P7D3 with iron (II) (FeCl2 (aqueous) at a 1:3 molar ratio of Fe to DPCA). Spectral changes were suggestive of iron binding (SI Appendix, Fig. S22) for all prodrugs and free DPCA at 510 nm and 560 nm, corresponding to phenanthroline- (46, 47) or a ferrozine-type binding (48), respectively. Notably, dramatic differences were noted for the iron-binding spectra of P7D1 and P7D3. Thus, it is possible that prodrug molecular architecture directly affects iron binding, and this may underlie the observed differences in bioactivity. Ultimately, the mechanism of PHD inhibition is known to be complex (49). Therefore, further studies will be needed to investigate the mechanism of action of PEG–DPCAs.
Conclusion
The utility of macromolecular prodrugs created by covalently bonding bioactive compounds to polymers or peptides is well known. Rationally designing these prodrugs to achieve desired biodistribution profiles, release kinetics, and activity, however, remains a significant challenge. In this study, we elucidated the structure–property relationship of PEG-based prodrugs created to deliver the proregenerative drug DPCA. Our general design involved covalently bonding DPCA, a poorly water-soluble PHD inhibitor, to the terminal end of a linear PEG via hydrolysable ester bonds. By controlling drug conjugation per PEG chain, we were able to precisely direct self-assembly behavior to form supramolecular nanostructures visualized in the wet and dry state, confirming a significant dependency on prodrug molecular design and drug carrier performance. Differences in the ability of PEG–DPCA prodrugs to stabilize HIF-1α in human cells suggest a complex relationship between molecular architecture and PHD inhibition. Fundamentally, this study highlights the importance of understanding the relationship between molecular design and self-assembly when designing supramolecular polymer prodrugs. Because HIF-1α is expressed in all cell types, we speculate that the translational potential of these prodrugs will be of interest for many applications in regenerative medicine.
Materials and Methods
Details on synthesis and additional experimental protocols can be found in the SI Appendix, Supplementary Information Text.
Preparation of Self-Assembled PEG–DPCA Structures.
Lyophilized powders of PEG–DPCAs obtained from semipreparative fractionation were added directly to ultrapure water or 10 mM PBS (pH 7). Solutions were then heated to 50 °C in a water bath for 30 min, with periodic vortexing. Fully soluble, homogeneous solutions were then removed from heat and cooled to 25 °C in a bench-top shaker table for 1 h. Before dilution, all PEG–DPCA solutions were reheated to 50 °C and cooled.
CMC Measurements.
Nile Red.
Nile Red stock solution (0.5 μL of 2.5 mM) prepared in ethanol was added to a microcentrifuge tube, and tubes were shielded from light and left open to allow ethanol to evaporate overnight. Then, 500 μL of aqueous PEG–DPCA suspensions at various concentrations or control solutions was added to each tube. To promote interactions between dye and self-assembled structures, mixtures were vortexed rigorously and placed on a bench-top shaker overnight protected from light. One hundred and fifty microliters of each sample was then transferred to a black 96-well plate, and emission spectra were collected from 520 to 700 nm at 490-nm excitation using a plate reader (Biosystems Synergy H1 microplate reader) and compared to background samples containing no dye. Spectra were normalized to no-dye controls, and peak fluorescence intensity at 670 nm was extracted and graphed versus log of prodrug concentration. The CMC was calculated as the intersection of two lines of best fit for the lower and upper regions of data. SD was determined from at least two experimental replicates.
Pyrene.
For all prodrugs, 0.625 μL of a 4 mM pyrene stock prepared in methanol was added to 1 mL of aqueous solution of prodrug at the specified concentration. Samples were then mixed by pipetting and transferred to quartz cuvettes with 5-mm path length. Emission spectra from 350 nm to 450 nm was recorded at 334 nm excitation on a Photon Technology International PTI QuantaMaster fluorescence/luminescence spectrometer. Excitation and emission slit widths were set to 1 nm. All data were collected from triplicate runs. The average ratio between emission peak intensity at 372 nm and 384 nm was then determined and plotted as a function of prodrug concentration.
Cryo-TEM.
Cryo-TEM imaging was performed on a FEI TALOS F200C microscope with 200 kV accelerating voltage. The images were recorded via an FEI Ceta 16M camera. Lacey carbon grids (300 mesh, Ted Pella, Inc.) were plasma treated by a plasma cleaner (PDC-32G, Harrica Plasma) before sample grid preparation. Vitrified grids were prepared using the FEI Vitrobot Mark IV, an automated plunge-freezing device. A droplet of 3 μL of sample solution at 25 g/L or 7 g/L was deposited onto the plasma-treated lacey grid. The grid was then blotted with blotting paper at 100% humidity and rapidly plunged into liquid ethane. Grids were stored in liquid nitrogen until transferred to the electron microscope for imaging. ImageJ was used to determine particle size and fiber dimensions. For the former structures, all images were converted to black and white, and particles were analyzed for diameter. To determine fiber width, profile plots showing gray scale values were obtained from original images. Fiber diameters were then taken as the distance between local minima.
SIM.
SIM was performed on an Elyra PS.1. Before semipreparative HPLC purification, P7D3 samples were prepared at 3 mg/mL in water containing 1.5 mol% DiO67 dye. To prevent fiber diffusion during imaging and preserve the wet-state structure, a covalent PEG-based hydrogel was used to mount fibers between glass coverslips and slides (50). For embedding, hydrogel precursor Tris(2-aminoethyl)amine–modified, multiarm PEG was added to the P7D3–DiOC7 solutions at 10 wt%. Ten microliters of the solution was then diluted 1:2 with a 100 mg/mL solution of N-hydroxysuccinimide–functionalized multiarm PEG on a glass slide and rapidly mixed with pipetting. After 3 min, a glass coverslip was mounted on top of the slide. Samples were left undisturbed for an additional 10 min to allow for complete gelation before imaging.
Coarse-Grained Modeling.
We represented each DPCA molecule as a single bead. Intermolecular DPCA–DPCA interactions were modeled with a Lennard–Jones (LJ) 6–12 potential that was cut off at (where is the LJ diameter) to capture the attraction between DPCA molecules. The strength of DPCA–DPCA attractions was set by the LJ well depth, εA, a key control parameter in our study. The 750-Da PEG was represented as three linearly connected LJ beads (with the same as the DPCA beads) using the Kremer–Grest model (51). This choice resulted in an average end-to-end distance of our coarse-grained PEG of . The ratio of to the DPCA size (taken to be , the interparticle distance below which all particle pairs repel each other) was then , which is nearly identical to the experimentally estimated value of by Chen et al. (10). The PEG–DPCA and PEG–PEG LJ interactions were cut off at to model purely repulsive interactions (i.e., the Weeks–Chandler–Anderson potential) (52). The LJ well depth of these repulsive interactions was set to the thermal energy .
All simulations were conducted using the graphics processing unit (GPU)-enabled open-source HOOMD-blue simulation package (53). Each simulation consisted of 1,000 molecules and was equilibrated using an annealing procedure described in the SI Appendix, Supplementary Information Text. The volume fraction was defined as the ratio of the solute volume (the product of the number of beads and the volume of each bead ) to the total system volume , that is, . For the spherical bead volume, we took the bead diameter to be .
Simulations were analyzed using the open-source simulation and visualization software OVITO (54). Beads within a cutoff distance of were grouped into the same cluster. The cluster density reported in Fig. 3B was calculated by dividing the number of clusters by the system volume. The mean cluster size reported in Fig. 3C was determined by averaging the number of molecules in a cluster. We chose molecules rather than beads to facilitate comparisons between the assembly of molecules with different DPCA contents.
The asphericity of each cluster was then computed by diagonalizing its gyration tensor :
| [1] |
following the convention (note that the scalar radius of gyration is defined as ). The asphericity parameter is the difference between the largest component and the average of the two smaller components:
| [2] |
All asphericities reported are in units of the LJ bead diameter .
Hydrolysis and Enzymatic Cleavage of PEG–DPCAs.
Solutions of PEG–DPCAs were prepared in PBS following the standard protocol outlined above. At t = 0, a 10 mg/mL stock solution of esterase from porcine liver (Sigma Aldrich, 9016-18-6) was added to +Enzyme samples to achieve a final prodrug concentration of 0.3 mM and enzyme concentration of 1 mg/mL (28 U/mg). −Enzyme samples were diluted to the final prodrug concentration using PBS. Samples were then vortexed and placed in a temperature-controlled bench-top shaker set to 37 °C. At predetermined timepoints, the concentrations of free DPCA and PEG–DPCA were determined using an Agilent 1260 Infinity HPLC in analytical mode with UV detection at 261 nm and gradient mobile phase from 70/30 (vol/vol%) water/acetonitrile (ACN) with 0.1% trifluoroacetic acid (TFA) to 100% ACN over 30 min.
DPCA Release from Self-Assembled PEG–DPCAs.
Solutions of PEG–DPCAs (6.4 mM) were prepared in PBS following the standard protocol outlined above. At t = 0, 0.5 mL of cooled suspensions was loaded into dialysis cassettes with a 2,000 Da molecular weight cutoff. Cassettes were then submerged in a 60-mL PBS bath, sealed, and placed in a temperature-controlled bench-top shaker set to 37 °C. At predetermined timepoints, a 1-mL aliquot was removed from the bath and replaced with fresh PBS. The concentration of DPCA from bath samples was determined using an Agilent 1260 Infinity HPLC in analytical mode with UV detection at 261 nm and gradient mobile phase from 70/30 (vol/vol%) water/ACN with 0.1% TFA to 100% ACN over 30 min.
Cell Culture.
AECs were isolated from the placental fetal membranes of patients undergoing elective caesarean delivery at University of California San Francisco (UCSF) Moore Women’s Hospital. The fetal membranes were deidentified and considered as discarded human specimens exempt from institutional review board approval. After multiple washes with sterile ice-cold Hank’s balanced salt solution (Gibco) supplemented with 1% penicillin/streptavidin (Gibco) and 0.1% amphotericin B (Gibco), the amnion layer of the fetal membranes was mechanically separated from the chorion layer and cut into 1 × 1 cm pieces. The amnion pieces were then transferred into 50-mL conical tubes containing 20 mL of prewarmed 0.25% trypsin/ethylenediaminetetraacetic acid (EDTA) (Gibco) and incubated at 37 °C for 10 min with gentle shaking. The obtained cells at this step were discarded to exclude blood clots and cellular debris. The remaining amniotic membrane pieces were transferred into new tubes containing trypsin/EDTA and incubated at 37 °C for 30 min with gentle shaking for the second digestion. After neutralizing the enzymatic digestion with complete culture media (Dulbecco's Modified Eagle Medium (DMEM)/F12 [Gibco] supplemented with 10% fetal bovine serum [Gibco], 1% penicillin/streptavidin, 0.1% amphotericin B, and 10 ng/mL epidermal growth factor [PeproTech]), the solution was filtered through a 100-μm cell strainer and centrifuged at 200 × g for 5 min at 4 °C. The obtained cell pellets were resuspended with complete medium and cultured in a tissue culture plate.
In Vitro Bioactivity of PEG–DPCA.
The day before PEG–DPCA treatment, AECs were seeded in a 96-well plate at a density of 2 × 105 cells/mL. The following day, solutions of PEG–DPCA were prepared by dissolving each PEG–DPCA powder in prewarmed complete medium at predetermined concentrations, sterile filtering using a 0.22-µm syringe filter, and adding to cells at 100 µL per well. Four hours after treatment, the bioactivity of the PEG–DPCAs was evaluated by immunostaining of HIF-1α. For immunostaining, AECs were fixed with 4% paraformaldehyde for 10 min and permeabilized with 0.3% Triton X-100 for 20 min at room temperature. To prevent nonspecific binding of antibodies, 5% goat blocking serum was added to cells for 1 h. Cells were then incubated with primary anti-HIF-1α (1:300 dilution, ab179483, Abcam) and anti-cytokeratin-18 (CK-18; 1:200 dilution, MA119039, Thermo Scientific), as a specific marker of AECs, at 4 °C overnight. After multiple washes with PBS, cells were treated with secondary antibodies (Alexa Fluor 488 [1:200, A11008, Invitrogen] and Alex Fluor 594 [1:200, A11005, Invitrogen]) for 45 min at room temperature. Cell nuclei were counterstained with 4′,6‐diamidino‐2‐phenylindole (DAPI) in antifade reagent (ProLong Diamond Antifade mountant with DAPI, Life Technologies). The stained cells were observed using a Keyence BZX-800 microscope. For prodrug internalization studies, cells were seeded in glass-bottom 24-well plates at 2 × 105 cells/mL and treated with soluble prodrugs as described. Prodrug concentration was normalized to 0.2 mg/mL DPCA to allow for accurate detection. After 4 h, cells were washed three times with warm PBS and imaged at 405 nm excitation. To isolate cell lysates for MALDI-TOF-MS analysis, cells were initially seeded at 1 × 105 cells/mL in six-well plates and grown to confluency. After incubation with prodrugs, cells were washed five times with warm PBS and trypsinized. Cell pellets were isolated via centrifugation and washed three times with warm PBS. Pellets were then dispersed in 200 μL of lysis buffer (1 M NaCl in 50 mM Tris-HCl buffer, pH 7.5) and sonicated for 30 min. Cellular debris was then removed by centrifugation at 8,000 × g for 5 min. Lysates were then run on MALDI-TOF-MS following standard sample preparation (SI Appendix, Supplementary Information Text).
Supplementary Material
Acknowledgments
We thank Dr. Tomoko Ogiyama and Erika Salzman for contributions to DPCA and imidazole-DPCA synthesis and Dr. Jaffar Hasnain and Gautam Bordia for helpful discussions. SAXS experiments were performed at Beamline 7.3.3 of the Advanced Light Source at Lawrence Berkeley National Laboratory. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the US Department of Energy under Contract Number DE-AC02-05CH11231. We also thank Dr. Andrew Jimenez for SAXS data processing and Dr. Chenhui Zhu for data collection. This work was supported by the NSF Graduate Research Fellowship under Grants DGE 1752814 and DGE 2146752, NIH Grants RO1DE021104 and R01EB022031, and US Department of Defense Grant W81XWH1910468. We acknowledge NIH Grant S10OD024998 for funding of the AV-600 MHz NMR instrument and the NIH S10 program under Award Number 1S10OD018136-01 for Elyra PS.1 imaging. We thank members of the Healy, Chang, and Streets laboratories at University of California, Berkeley, for instrument access and assistance with rheology, fluorimetry, and cell imaging. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. W.X. and D.P. thank the primary support of NSF through the University of Delaware Materials Research Science and Engineering Center, grant DMR-2011824.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2208593119/-/DCSupplemental.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Fahr A., Liu X., Drug delivery strategies for poorly water-soluble drugs. Expert Opin. Drug Deliv. 4, 403–416 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Dahan A., Miller J. M., The solubility–permeability interplay and its implications in formulation design and development for poorly soluble drugs. AAPS J. 14, 244–251 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rautio J., Meanwell N. A., Di L., Hageman M. J., The expanding role of prodrugs in contemporary drug design and development. Nat. Rev. Drug Discov. 17, 559–587 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Su H., et al. , The role of critical micellization concentration in efficacy and toxicity of supramolecular polymers. Proc. Natl. Acad. Sci. U.S.A. 117, 4518–4526 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Croy S. R., Kwon G. S., Polymeric micelles for drug delivery. Curr. Pharm. Des. 12, 4669–4684 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Ahmad Z., Shah A., Siddiq M., Kraatz H.-B., Polymeric micelles as drug delivery vehicles. RSC Advances 4, 17028–17038 (2014). [Google Scholar]
- 7.Aiertza M. K., Odriozola I., Cabañero G., Grande H.-J., Loinaz I., Single-chain polymer nanoparticles. Cell. Mol. Life Sci. 69, 337–346 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawrence M. J., Surfactant systems: Their use in drug delivery. Chem. Soc. Rev. 23, 417–424 (1994). [Google Scholar]
- 9.Zhang Y., et al. , Drug-induced regeneration in adult mice. Sci. Transl. Med. 7, 290ra92 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng J., Amin D., Latona J., Heber-Katz E., Messersmith P. B., Supramolecular polymer hydrogels for drug-induced tissue regeneration. ACS Nano 13, 5493–5501 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin R., Cheetham A. G., Zhang P., Lin Y. A., Cui H., Supramolecular filaments containing a fixed 41% paclitaxel loading. Chem. Commun. (Camb.) 49, 4968–4970 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vemula P. K., et al. , Prodrugs as self-assembled hydrogels: A new paradigm for biomaterials. Curr. Opin. Biotechnol. 24, 1174–1182 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Sufeng Z., et al. , An inflammation-targeting hydrogel for local drug delivery in inflammatory bowel disease. Sci. Transl. Med. 7, 300ra128 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeFrates K. G., Franco D., Heber-Katz E., Messersmith P. B., Unlocking mammalian regeneration through hypoxia inducible factor one alpha signaling. Biomaterials 269, 120646 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Afifi H., Karlsson G., Heenan R. K., Dreiss C. A., Solubilization of oils or addition of monoglycerides drives the formation of wormlike micelles with an elliptical cross-section in cholesterol-based surfactants: A study by rheology, SANS, and cryo-TEM. Langmuir 27, 7480–7492 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Goodwin A. P., Mynar J. L., Ma Y., Fleming G. R., Fréchet J. M. J., Synthetic micelle sensitive to IR light via a two-photon process. J. Am. Chem. Soc. 127, 9952–9953 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Poelma S. O., et al. , Controlled drug release to cancer cells from modular one-photon visible light-responsive micellar system. Chem. Commun. (Camb.) 52, 10525–10528 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagy M., Zsuga M., Rácz D., Kéki S., Synthesis of phenanthroline-terminated polymers and their Fe(II)-complexes. J. Polym. Sci. A Polym. Chem. 48, 2709–2715 (2010). [Google Scholar]
- 19.Kuki A., Nagy M., Nagy L., Zsuga M., Kéki S., Ligand size distribution of phenanthroline-functionalized polyethylene glycol-iron(II) complexes determined by electrospray ionization mass spectrometry and computer simulation. J. Am. Soc. Mass Spectrom. 21, 1561–1564 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Li H., Hu D., Liang F., Huang X., Zhu Q., Influence factors on the critical micelle concentration determination using pyrene as a probe and a simple method of preparing samples. R. Soc. Open Sci. 7, 192092 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q., et al. , High performance shape memory polyimides based on π–π interactions. J. Mater. Chem. A Mater. Energy Sustain. 3, 352–359 (2015). [Google Scholar]
- 22.Koren A. B., Curtis M. D., Francis A. H., Kampf J. W., Intermolecular interactions in π-stacked conjugated molecules. Synthesis, structure, and spectral characterization of alkyl bithiazole oligomers. J. Am. Chem. Soc. 125, 5040–5050 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Geng Y., et al. , Shape effects of filaments versus spherical particles in flow and drug delivery. Nat. Nanotechnol. 2, 249–255 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Champion J. A., Mitragotri S., Role of target geometry in phagocytosis. Proc. Natl. Acad. Sci. U.S.A. 103, 4930–4934 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Israelachvili J. N., Intermolecular and Surface Forces (Academic Press, Elsevier, ed. 3, 2011). [Google Scholar]
- 26.Maibaum L., Dinner A. R., Chandler D. W., Micelle formation and the hydrophobic effect. J. Phys. Chem. B 108, 6778–6781 (2004). [Google Scholar]
- 27.Woo H.-J., Carraro C., Chandler D., Assembly of extended interfaces and micelles: Charge frustrated models of amphiphilic mixtures. Faraday Discuss. 104, 183–191 (1996). [Google Scholar]
- 28.Glotzer S. C., Solomon M. J., Anisotropy of building blocks and their assembly into complex structures. Nat. Mater. 6, 557–562 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Israelachvili J. N., Mitchell D. J., Ninham B. W., Theory of self-assembly of hydrocarbon amphiphiles into micelles and bilayers. J. Chem. Soc. Faraday Trans. II 2, 1525–1568 (1976). [Google Scholar]
- 30.Santos A. P., Panagiotopoulos A. Z., Determination of the critical micelle concentration in simulations of surfactant systems. J. Chem. Phys. 144, 044709 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Nguyen T. D., Phillips C. L., Anderson J. A., Glotzer S. C., Rigid body constraints realized in massively-parallel molecular dynamics on graphics processing units. Comput. Phys. Commun. 182, 2307–2313 (2011). [Google Scholar]
- 32.Allinger N. L., Conformational analysis. 130. MM2. A hydrocarbon force field utilizing V1 and V2 torsional terms. J. Am. Chem. Soc. 99, 8127–8134 (2002). [Google Scholar]
- 33.Arkın H., Janke W., Gyration tensor based analysis of the shapes of polymer chains in an attractive spherical cage. J. Chem. Phys. 138, 54904 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Alessi M. L., Norman A. I., Knowlton S. E., Ho D. L., Greer S. C., Helical and coil conformations of poly(ethylene glycol) in isobutyric acid and water. Macromolecules 38, 9333–9340 (2005). [Google Scholar]
- 35.Nagai K., et al. , An injectable hydrogel-formulated inhibitor of prolyl-4-hydroxylase promotes T regulatory cell recruitment and enhances alveolar bone regeneration during resolution of experimental periodontitis. FASEB J. 34, 13726–13740 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Z., et al. , Controlled release of free doxorubicin from peptide–drug conjugates by drug loading. J. Control. Release 191, 123–130 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Chakroun R. W., et al. , Fine-tuning the linear release rate of paclitaxel-bearing supramolecular filament hydrogels through molecular engineering. ACS Nano 13, 7780–7790 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Beck V., Lewi P., Gucciardo L., Devlieger R., Preterm prelabor rupture of membranes and fetal survival after minimally invasive fetal surgery: A systematic review of the literature. Fetal Diagn. Ther. 31, 1–9 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Strowitzki M. J., Cummins E. P., Taylor C. T., Protein hydroxylation by hypoxia-inducible factor (HIF) hydroxylases: Unique or ubiquitous? Cells 8, 384 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soilleux E. J., et al. , Use of novel monoclonal antibodies to determine the expression and distribution of the hypoxia regulatory factors PHD-1, PHD-2, PHD-3 and FIH in normal and neoplastic human tissues. Histopathology 47, 602–610 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Banerji B., et al. , The inhibition of factor inhibiting hypoxia-inducible factor (FIH) by β-oxocarboxylic acids. Chem. Commun. (Camb.) 43, 5438–5440 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Wang Y., Zhong S., Schofield C. J., Ratcliffe P. J., Lu X., Nuclear entry and export of FIH are mediated by HIF1α and exportin1, respectively. J. Cell Sci. 131, jcs219782 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hirsilä M., Koivunen P., Günzler V., Kivirikko K. I., Myllyharju J., Characterization of the human prolyl 4-hydroxylases that modify the hypoxia-inducible factor. J. Biol. Chem. 278, 30772–30780 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Semenza G. L., HIF-1 and mechanisms of hypoxia sensing. Curr. Opin. Cell Biol. 13, 167–171 (2001). [DOI] [PubMed] [Google Scholar]
- 45.Franklin T. J., Morris W. P., Edwards P. N., Large M. S., Stephenson R., Inhibition of prolyl 4-hydroxylase in vitro and in vivo by members of a novel series of phenanthrolinones. Biochem. J. 353, 333–338 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Agustina E., et al. , Simple and precise quantification of iron catalyst content in carbon nanotubes using UV/visible spectroscopy. ChemistryOpen 4, 613–619 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee T. S., Kolthoff I. M., Leussing D. L., Reaction of ferrous and ferric iron with 1,10-phenanthroline. I. Dissociation constants of ferrous and ferric phenanthroline. J. Am. Chem. Soc. 70, 2348–2352 (1948). [DOI] [PubMed] [Google Scholar]
- 48.Shaked Y., Kustka A. B., Morel F. M. M., Erel Y., Simultaneous determination of iron reduction and uptake by phytoplankton. Limnol. Oceanogr. Methods 2, 137–145 (2004). [Google Scholar]
- 49.Fong G.-H., Takeda K., Role and regulation of prolyl hydroxylase domain proteins. Cell Death Differ. 15, 635–641 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Kwak G., et al. , Sustained exosome-guided macrophage polarization using hydrolytically degradable PEG hydrogels for cutaneous wound healing: Identification of key proteins and miRNAs, and sustained release formulation. Small 18, e2200060 (2022). [DOI] [PubMed] [Google Scholar]
- 51.Kremer K., Grest G. S., Dynamics of entangled linear polymer melts: A molecular‐dynamics simulation. J. Chem. Phys. 92, 5057 (1998). [Google Scholar]
- 52.Weeks J. D., Chandler D., Andersen H. C., Role of repulsive forces in determining the equilibrium structure of simple liquids. J. Chem. Phys. 54, 5237 (2003). [Google Scholar]
- 53.Anderson J. A., Glaser J., Glotzer S. C., HOOMD-blue: A Python package for high-performance molecular dynamics and hard particle Monte Carlo simulations. Comput. Mater. Sci. 173, 109363 (2020). [Google Scholar]
- 54.Stukowski A., Visualization and analysis of atomistic simulation data with OVITO—the open visualization tool. Model. Simul. Mater. Sci. Eng. 18, 015012 (2009). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.