Significance
Tracking the animal reservoir of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and its variants is important for understanding the current COVID-19 pandemic and preventing future pandemics. Speculations about the source of the omicron variant are abundant, yet experimental evidence has been scarce. Here, we provide the structural information on how omicron recognizes its mouse receptor. Our study demonstrates that the omicron mutations in the receptor-binding region are structurally adapted to mouse angiotensin-converting enzyme 2 (ACE2), informing an understanding of the origin of the omicron variant and the evolution of SARS-CoV-2. It may facilitate epidemiological surveillance of SARS-CoV-2 in animals to prevent future coronavirus pandemics.
Keywords: COVID-19, omicron variant, mouse angiotensin-converting enzyme 2, receptor-binding domain (RBD), X-ray crystallography
Abstract
The sudden emergence and rapid spread of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) omicron variant has raised questions about its animal reservoir. Here, we investigated receptor recognition of the omicron’s receptor-binding domain (RBD), focusing on four of its mutations (Q493R, Q498R, N501Y, and Y505H) surrounding two mutational hotspots. These mutations have variable effects on the RBD’s affinity for human angiotensin-converting enzyme 2 (ACE2), but they all enhance the RBD’s affinity for mouse ACE2. We further determined the crystal structure of omicron RBD complexed with mouse ACE2. The structure showed that all four mutations are viral adaptations to mouse ACE2: three of them (Q493R, Q498R, and Y505H) are uniquely adapted to mouse ACE2, whereas the other one (N501Y) is adapted to both human ACE2 and mouse ACE2. These data reveal that the omicron RBD was well adapted to mouse ACE2 before omicron started to infect humans, providing insight into the potential evolutionary origin of the omicron variant.
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) omicron variant emerged abruptly and spread rapidly around the globe (1–4). Tracking the animal reservoir of SARS-CoV-2 and its variants is important for understanding the current COVID-19 pandemic and preventing future pandemics. Speculations about the source of the omicron variant are abundant, yet experimental evidence has been scarce (5). The interactions between the receptor-binding domain (RBD) of coronavirus spike proteins and their host receptor are among the best systems for understanding coronavirus evolution (6, 7). Both SARS-CoV-2 and closely related SARS-CoV-1 recognize human angiotensin-converting enzyme 2 (ACE2) as their receptor (8–10). Previous research on the receptor recognition of SARS-CoV-1 has provided insight into the animal origin of SARS-CoV-1 (11–15). The RBD of the original SARS-CoV-2 strain (i.e., prototypic RBD) differs from the RBD of a bat coronavirus by only a few residues, supporting a bat origin of the prototypic RBD (16). The omicron RBD (strain BA.2) differs from the prototypic RBD by 16 residues, seven of which are located in the receptor-binding motif (RBM) that directly contacts ACE2 (3). To recover the evolutionary traces left by these RBM mutations, this study compared the structural adaptations of the omicron RBD to ACE2 from human and mouse, two possible sources of omicron (5).
Three virus-binding hotspots have been identified at the interfaces between SARS-CoV-2 RBD and human ACE2 (hACE2) and between SARS-CoV-1 RBD and hACE2 (14, 17, 18). These hotspots center on Lys31 in hACE2 (i.e., hotspot-31), Lys353 in hACE2 (i.e., hotspot-353), and a receptor-binding ridge in the viral RBD (i.e., hotspot-ridge) (Fig. 1A). These virus-binding hotspots are also mutational hotspots for SARS-CoV-1: all of the RBM mutations occurred around the hotspots and impacted the structural stability of the hotspots (13, 14). Establishment of the “hotspots” concept was instrumental in determining the molecular mechanisms by which SARS-CoV-1 was transmitted from palm civets to humans (11–15). The RBM mutations in the SARS-CoV-2 omicron variant are also around the hotspots (Fig. 1A). Curiously, only a few of these omicron mutations enhance the RBD’s affinity for hACE2, while some other mutations reduce it (Fig. 1B) (17). Structural details of the interface between the omicron RBM (strain BA.1) and hACE2 elucidated the role of each of these mutations in binding hACE2 (17). The omicron mutations that reduce the RBD’s affinity for hACE2 are structurally incompatible with hACE2, raising questions about what other species may have mediated the evolution of omicron.
Fig. 1.
Binding interactions between SARS-CoV-2 RBD (from prototypic strain or omicron strain) and ACE2 (from human or mouse). (A) Structure of the interface between prototypic RBM and hACE2 (PDB ID: 6VW1). RBM is in magenta. hACE2 is in green. RBD residues that have undergone mutations from the prototypic strain to the omicron variant (strain BA.2) are shown as sticks. Three mutational hotspots are highlighted: hotspot-353 centers on Lys353 in hACE2, hotspot-31 centers on Lys31 in hACE2, and hotspot-ridge centers on the receptor-binding ridge in hACE2. (B and C) SPR assay for the binding of RBD (from prototypic strain or omicron strain) to ACE2 (from human or mouse). ACE2-Fc was coated to a protein A chip in a fixed direction, and individual RBDs flowed through. Data in B are from one of our recent studies (17), except that the omicron variant (strain BA.2) in this study replaced strain BA.1 in the previous study. Data in C are from the current study. The data in B and C are presented as mean ± SEM (n = 3 or n = 4) on a log scale. A Student’s two-tailed t test was performed to analyze the statistical difference between the RBD on the Left in either panel and each of the other RBDs in the same panel; the results are labeled on top of each bar. The statistical difference between the R493Q mutation and the N477S/R493Q double mutations was also analyzed in C; the result was labeled between the two bars. The horizontal dashed lines represent the measurements for the prototypic RBD in B or the omicron RBD in C and are used for comparison with other measurements in the respective panel. ***P < 0.001; **P < 0.01; *P < 0.05. n.s., statistically not significant, N.D., not detected.
In this study, we provide biochemical and structural evidence demonstrating that the omicron mutations are better adapted to mouse ACE2 (mACE2) than to hACE2, suggesting that mice mediated the onset of the omicron variant. Our study helps clarify the animal reservoir of the omicron variant and contributes to the understanding of SARS-CoV-2 evolution. The findings may facilitate epidemiological surveillance of SARS-CoV-2 in animals to prevent future coronavirus pandemics.
Results
Two major omicron strains, BA.2 and BA.1, were discovered at about the same time in fall 2021, but BA.2 soon became more dominant in many parts of the world (19). The two strains differ in their RBM by only one residue: BA.2 contains Gly496, whereas BA.1 contains Ser496. In this study, we focused on the omicron BA.2. Among the seven RBM mutations in the omicron RBD, three mutations are located around hotspot-ridge: T478K and E484A slightly enhance the RBD’s affinity for hACE2, but they do not have direct interactions with hACE2 (17), whereas S477N significantly enhances the RBD’s interaction with hACE2 (Fig. 1B) (20). The other four RBM mutations are located around hotspot-353 or hotspot-31: N501Y (located around hotspot-353) significantly enhances the RBD’s affinity for hACE2, Q493R (located around hotspot-31) and Y505H (located around hotspot-353) reduce hACE2 binding, and Q498R (located around hotspot-353) has no significant net impact on hACE2 binding (Fig. 1B). Overall, the omicron RBD binds to hACE2 more tightly than does the prototypic RBD (Fig. 1B), despite some of the omicron mutations contributing unfavorably to hACE2 binding.
In this study, we investigated binding interactions between the omicron RBD and mACE2, focusing on the four mutations (Q493R, Q498R, N501Y, and Y505H) around hotspot-31 or hotspot-353. Our result from the surface plasmon resonance (SPR) assay showed that whereas the prototypic RBD does not bind mACE2, the omicron RBD binds to mACE2 with good affinity (Fig. 1C). To examine the impact of omicron mutations on the RBD’s binding to mACE2, single mutations (corresponding to the RBM mutations from prototype to omicron) could be introduced to the prototypic RBD. However, undetectable binding between the prototypic RBD and mACE2 can negatively impact the reliability of the SPR assay using mutant prototypic RBDs. Hence, we introduced reverse mutations (corresponding to residue changes from omicron to prototype) to the omicron RBD and measured the mutant omicron RBDs’ affinity for mACE2 (Fig. 1C). These reverse mutations (R493Q, R498Q, Y501N, and H505Y) all significantly reduced mACE2 binding, meaning that all of the corresponding mutations in the omicron RBM (Q493R, Q498R, N501Y, and Y505H) enhance mACE2 binding. Taken together, all four omicron mutations located around hotspot-31 or hotspot-353 contribute favorably to mACE2 binding.
To confirm the above SPR data, we performed an omicron pseudovirus entry assay. To this end, we packaged omicron pseudoviruses (i.e., retroviruses pseudotyped with the omicron spike protein) and used them to infect mACE2-expressing cells. We also prepared four mutant omicron pseudoviruses by introducing each of the reverse mutations (R493Q, R498Q, Y501N, or H505Y) to omicron pseudoviruses. All four reverse mutations significantly reduced the efficiency of omicron pseudoviruses in entering mACE2-expressing cells (SI Appendix, Fig. S1). Hence, the result from the pseudovirus entry assay is consistent with the SPR data, indicating that all four omicron mutations located around hotspot-31 or hotspot-353 (Q493R, Q498R, N501Y, and Y505H) contribute favorably to mACE2 binding.
To provide a structural understanding of the role of the above omicron mutations in mACE2 binding, we determined the structure of the interface between the omicron RBD and mACE2. We developed a convenient, reliable, and accurate structural platform for studying the interactions between the RBD from different SARS-CoV-2 (or SARS-CoV-1) strains and ACE2 from different host species (11, 14, 17, 18). Taking advantage of high-quality crystals of the SARS-CoV-1 RBD/hACE2 complex, this platform keeps intact the crystal contact regions involving the core structures from the SARS-CoV-1 RBD and hACE2 and varies the noncrystal contact regions involving the RBM of SARS-CoV-2 RBD and three virus-binding motifs (VBMs) of hACE2. More specifically, a chimeric RBD was constructed to contain the core structure from SARS-CoV-1 RBD and the RBM from the SARS-CoV-2 omicron strain (i.e., chimeric omicron RBD), whereas a chimeric ACE2 was constructed to contain the core structure from hACE2 and three VBMs from mACE2 (i.e., chimeric mACE2) (Fig. 2 A and B). The complex of the chimeric omicron RBD and chimeric mACE2 was crystallized under the same condition as the complex of SARS-CoV-1 RBD and hACE2 but reveals the detailed interactions between the omicron RBM and mACE2 VBMs. The structure of this complex was determined at 2.84 Å (Fig. 2A and SI Appendix, Fig. S2A and Table S1). To provide a comparison, we also determined the structure of the chimeric omicron RBD complexed with hACE2 at 2.38 Å (Fig. 2C and SI Appendix, Fig. S2B and Table S1). Note that omicron RBD from BA.2 strain was used in both structures in the current study, instead of the omicron RBD from BA.1 strain in a recent study (17). To understand the structural adaptions of omicron RBD to mACE2, we compared these two structures along with the structure of the chimeric prototypic RBD/hACE2 that we determined previously (18), focusing on hotspot-31 and hotspot-353.
Fig. 2.
Overall structures of chimeric omicron RBD complexed with chimeric mACE2 or hACE2. (A) Overall structure of chimeric omicron RBD complexed with chimeric mACE2. The chimeric omicron RBD contains the core structure (in cyan) from SARS-CoV-1 RBD and RBM (in magenta) from the omicron RBD (strain BA.2). The chimeric mACE2 contains the core structure (in green) from hACE2 and three VBMs (in orange) from mACE2. (B) Sequence alignment between hACE2 and mACE2 in three VBMs (shaded). The centers for hotspot-31 and hotspot-353 (i.e., residues 31 and 353, respectively) are both labeled in red. Residues that directly contact SARS-CoV-2 RBD are labeled in blue. Asterisks indicate positions that have a single, fully conserved residue. Colons indicate positions that have strongly conserved residues. Periods indicate positions that have weakly conserved residues. (C) Overall structure of chimeric omicron RBD complexed with hACE2. hACE2 is in green.
Hotspot-31 stabilizes the center of the RBM/VBMs interface. At the interface between the prototypic RBM and human VBMs (Fig. 3A), both Lys31vbm and Glu35vbm form a hydrogen bond with Gln493rbm. These hydrogen bonds anchor the positively charged Lys31vbm and keep it away from a hydrophobic wall (consisting of several hydrophobic residues, including Phe456rbm and Tyr489rbm). Thus, Gln493rbm plays a critical role in stabilizing the RBM/VBMs interface. At the interface between the omicron RBM and human VBMs (Fig. 3B), the Q493R mutation introduces a positively charged Arg493rbm, disrupting the previous hydrogen bonds with Lys31vbm and Glu35vbm. Instead, due to repulsions with Arg493rbm and the hydrophobic wall, Lys31vbm is forced to point to a new direction with a twisted side chain and forms a hydrogen bond with Gln76vbm with a poor geometry. Hence, the Q493R mutation destabilizes the RBM/VBMs interface, reducing omicron RBD’s affinity for hACE2. In mACE2, residue 31 is an asparagine, replacing Lys31 in hACE2. Thus, at the interface between the omicron RBM and mouse VBMs (Fig. 3C), Arg493rbm forms two bifurcated hydrogen bonds with Asn31vbm, stabilizing the RBM/VBMs interface and enhancing omicron RBD’s affinity for mACE2. Overall, the omicron mutation Q493R around hotspot-31 is structurally adapted to Asn31 in mACE2, but it is structurally incompatible with Lys31 in hACE2.
Fig. 3.
Structural details at mutational hotspot-31. (A) Interface between the prototypic RBM and human VBMs. PDB ID: 6VW1. (B) Interface between the omicron RBM and human VBMs. (C) Interface between the omicron RBM and mouse VBMs. Dotted lines indicate hydrogen bonds. Side chain of 490rbm is not shown for the sake of clarity. Double-ended red arrow indicates repulsion.
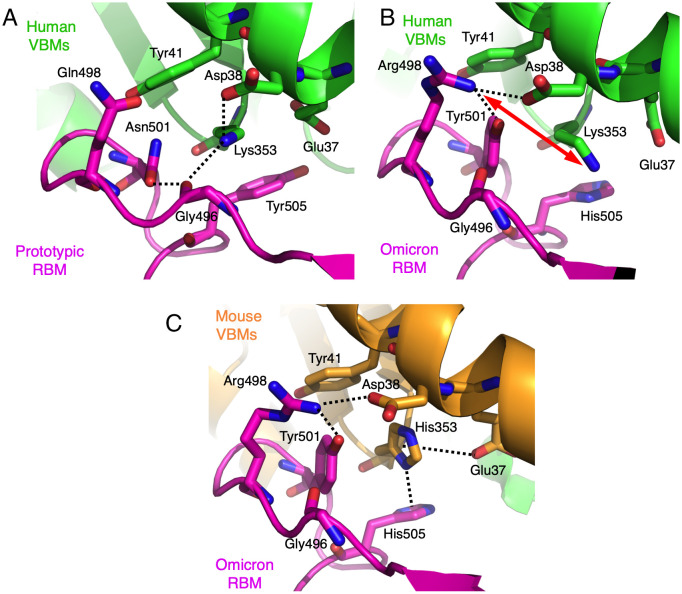
Hotspot-353 stabilizes the RBM/VBMs interface on the side (opposite to the hotspot-ridge side). At the interface between the prototypic RBM and human VBMs (Fig. 4A), the side chain of Lys353vbm is buried in a hydrophobic tunnel; the four tunnel walls consist of the side chains of Tyr41vbm, Glu37vbm, Asn501rbm, and Tyr505rbm. At the end of the tunnel, Lys353vbm forms a salt bridge and a hydrogen bond with Asp38vbm and the main chain of Gly496rbm, respectively. As we previously showed, this tunnel structure is critical for stabilizing the RBM/VBMs interface, and any disturbance of the tunnel structure significantly reduces SARS-CoV-1 RBD’s affinity for hACE2 (21). In the omicron RBM, three mutations occurred around hotspot-353: N501Y, Q498R, and Y505H. Hence, at the interface between the omicron RBM and human VBMs (Fig. 4B), two of the tunnel walls have undergone changes: the newly introduced Tyr501rbm forms a stronger hydrophobic stacking interaction with Lys353vbm than does the previous Asn501rbm, stabilizing the RBM/VBMs interface and enhancing the omicron RBD’s affinity for hACE2. Tyr501rbm also forms an aromatic stacking interaction with Tyr41vbm. Moreover, the newly introduced His505rbm forms a weaker hydrophobic stacking interaction with Lys353vbm than does the previous Tyr505rbm, destabilizing the RBM/VBMs interface and reducing the omicron RBD’s affinity for hACE2. Importantly, the newly introduced Arg498rbm forms a salt bridge with Asp38vbm, forcing Lys353vbm to point to another direction with a twisted side chain. Arg498rbm also forms a hydrogen bond with Tyr501rbm. Hence, the Q498R mutation disrupts the previously stable tunnel structure while forming two new favorable interactions; these favorable changes and unfavorable changes at the RBM/VBMs interface cancel out, and the Q498R mutation has no significant net impact on the omicron RBD’s affinity for hACE2. Therefore, among the three omicron mutations, N501Y is structurally adapted to Lys353 in hACE2, Y505H is structurally incompatible with Lys353 in hACE2, and Q498R is structurally incompatible with Lys353 in hACE2 but is structurally adapted to Asp38 in hACE2.
Fig. 4.
Structural details at mutational hotspot-353. (A) Interface between the prototypic RBM and human VBMs. PDB ID: 6VW1. (B) Interface between the omicron RBM and human VBMs. (C) Interface between the omicron RBM and mouse VBMs. Dotted lines indicate salt bridges, hydrogen bonds, hydrogen-π interaction, or anion-π interaction. Double-ended red arrow indicates repulsion.
Compared with the two interfaces involving human VBMs noted above, hotspot-353 at the interface between the omicron RBM and mouse VBMs has undergone significant changes due to His353 in mACE2 replacing Lys353 in hACE2 (Fig. 4C). His353vbm does not fit into the tunnel structure as well as does Lys353vbm, explaining why the prototypic RBD does not bind mACE2 and why the prototypic SARS-CoV-2 infects mice poorly (22). However, the newly introduced His505rbm forms a hydrogen–π interaction with His353vbm (23). Asp37vbm also forms an anion–π interaction with His353vbm (24). The newly introduced Tyr501rbm forms a π–π stacking with His353vbm (23). All of these interactions stabilize His353vbm. The newly introduced Arg498rbm forms a hydrogen bond and a salt bridge with Tyr501rbm and Asp38vbm, respectively. All of these interactions strengthen the newly structured hotspot-353 at the RBM/VBMs interface and enhance the omicron RBD’s affinity for mACE2, explaining why the omicron RBD binds mACE2 with good affinity (Fig. 1C) and why omicron infects mice efficiently (25). Overall, all three omicron mutations (Q498R, N501Y, and Y505H) around hotspot-353 are structurally adapted to His353 in mACE2.
In addition to the above four mutations located around hotspot-31 or hotspot-353, we also examined the three mutations (S477N, T478K, and E484A) located around hotspot-ridge. To this end, we introduced individual reverse mutations, N477S, K478T, or A484E, into the omicron RBD. N477S and K478T had no significant impact on the omicron RBD’s affinity for mACE2; A484E slightly reduced the omicron RBD’s affinity for mACE2, suggesting that E484A slightly increases the prototypic RBD’s affinity for mACE2 (Fig. 1C). It has been shown that the T478K and E484A mutations have no direct interactions with ACE2, but they may cause small conformational changes in the hotspot-ridge loops, slightly and indirectly impacting RBD/ACE2 binding (17, 20). Moreover, it has been shown that the S477N mutation enhances the prototypic RBD’s affinity for hACE2 (17, 20). Here, our structural data confirmed that Ser477 in the prototypic RBD forms no interaction with hACE2 (SI Appendix, Fig. S3A), whereas Asn477 in the omicron RBD forms hydrogen bonds with the N-terminal Ser19 of hACE2 (SI Appendix, Fig. S3B). The N477S mutation did not significantly affect the omicron RBD’s affinity for mACE2 (Fig. 1C), which was confirmed by the observation that the N477S/R493Q double mutations and the R493Q single mutation reduced the omicron RBD’s affinity with mACE2 to similar extents (Fig. 1C). The lack of interaction between Asn477 in the omicron RBD and mACE2 finds an explanation in our structural data: the N-terminal Ser19 of mACE2 is disordered at the structural interface between the omicron RBM and mACE2 VBMs (SI Appendix, Fig. S3C). The residue differences between hACE2 and mACE2 in residues 20 and 21 may have led to the structural difference between hACE2 and mACE2 in the N-terminal Ser19 (Fig. 2B). Overall, among the omicron mutations around hotspot-ridge, T478K and E484A largely are structurally neutral with hACE2 or mACE2; S477N is structurally adapted to the N terminus of hACE2, but is structurally neutral to the N terminus of mACE2.
While this study focused on the RBM mutations in the omicron BA.2 strain, spike mutations outside the RBM may also affect receptor binding. In the prototypic strain, Lys417 is the only RBD residue that is located outside the RBM region but directly interacts with hACE2 (26). It has been shown that the K417N mutation in other SARS-CoV-2 strains loses direct contact with hACE2, reducing RBD’s affinity for hACE2 (27). So far no synergistic effect has been reported between the K417N mutation and any of the RBM mutations. In both the omicron BA.1 and BA.2 strains, residue 417 is an asparagine, which is expected to have no direct contact with hACE2 or mACE2. Moreover, spike residues outside the RBD may indirectly affect receptor binding. Previously, we showed that the RBD takes one of two conformations in the entire spike protein—standing up for receptor binding and lying down for immune evasion (28). Recently, we determined the cryo-electron microscopy structure of the spike ectodomain of the omicron BA.1 strain (29). Our structure showed that several spike mutations outside the RBD stabilize the standing-up conformation of the RBD (29), which may facilitate the binding of the spike protein to hACE2 or mACE2. Future studies are needed to elucidate the detailed roles of these non-RBM mutations in the spike protein’s binding to hACE2 or mACE2.
Discussion
Identifying the animal reservoir of SARS-CoV-2 is critical for understanding the evolutionary history of the virus and for preventing future pandemics. Recently, a bat coronavirus was discovered to have remarkably similar RBD to the prototypic SARS-CoV-2 RBD (16), suggesting a bat origin of the prototypic RBD. As SARS-CoV-2 spreads in humans, its RBD has accumulated mutations, many of which facilitate the virus’s immune evasion (30). The SARS-CoV-2 omicron variant has recently become the dominant strain (1–4). The omicron RBM contains significantly more mutations than all of the previous SARS-CoV-2 variants, raising the question of whether nonhuman animals may have mediated its evolution (5). Previously, we identified virus-binding hotspots at coronavirus/receptor interfaces, the same sites that are also often targeted by viral mutations (11, 12, 18, 21). In this study using the hotspot analysis, we investigated how the RBM mutations in omicron are structurally adapted to mACE2.
A common theme surrounding hACE2 as the receptor for coronavirus RBDs is how to place its Lys31 and Lys353 at the RBD/hACE2 interface. On the one hand, both lysines are surrounded by numerous hydrophobic residues; if not neutralized, their positive charges are incompatible with the neighboring hydrophobic residues, destabilizing the RBD/hACE2 interface. On the other hand, if properly neutralized, salt bridges (or strong hydrogen bonds) formed by these lysines can contribute a significant amount of binding energy to the RBD/hACE2 interactions (due to the low dielectric constant in hydrophobic environments). Thus, it is not surprising that for both SARS-CoV-1 and SARS-CoV-2, the majority of the receptor adaptation mutations have occurred surrounding hotspot-31 or hotspot-353, aiming to improve viral accommodations for the two lysines. However, two mutations in the omicron RBM, Q493R and Q498R, introduce two positively charged arginines that disrupt the structures of the two hotspots and force Lys31 and Lys353 to point to a different direction with a twisted side chain. Moreover, the Y505H mutation in the omicron RBM also reduces the support for hotspot-353. Our biochemical data confirm that both Q493R and Y505H significantly reduce the RBD’s affinity for hACE2, whereas Q498R had no significant net impact on hACE2 binding (because it also introduces favorable interactions). Hence, these three mutations cannot be SARS-CoV-2’s adaption to hACE2. In contrast, in mACE2, Lys31 and Lys353 become Asn31 and His353, respectively. These two residue changes, particularly the K353H change, are unfavorable for the binding of coronavirus RBDs, explaining why neither SARS-CoV-1 nor prototypic SARS-CoV-2 uses mACE2 as their receptor (22, 31). However, all three mutations Q493R, Q498R, and Y505H, along with N501Y, stabilize the restructured hotspot-31 and hotspot-353 in mACE2 and increase the omicron RBD’s affinity for mACE2. Overall, both our structural and biochemical data have identified three omicron mutations (Q493R, Q498R, and Y505H) that are uniquely adapted to mACE2 (Table 1).
Table 1.
Structural adaptions of omicron RBD residues to mACE2 or hACE2
| Omicron RBD residues | Omicron residues are adapted to which ACE2 residues? |
|---|---|
| Arg493 | Adapted to Asn31 in mACE2 (Incompatible with Lys31 in hACE2) |
| His505 | Adapted to His353 in mACE2 (Incompatible with Lys353 in hACE2) |
| Arg498 | Adapted to His353 in mACE2 (Incompatible with Lys353 in hACE2); Adapted to Asp38 in both hACE2 and mACE2 |
| Tyr501 | Adapted to Tyr41 in both hACE2 and mACE2; Adapted to Lys353 in hACE2 and His353 in mACE2 |
| Asn477 | Adapted to N-terminal residues in hACE2 (Neutral with N-terminal residues in mACE2) |
| Lys478 | Largely neutral with hACE2 and mACE2 |
| Ala484 | Largely neutral with hACE2 and mACE2 |
Our detailed structural analysis of the RBM mutations provides insight into the host receptor adaptations by the omicron variant. Among the seven RBM mutations, four (Q493R, Q498R, Y505H, and T501Y) are structurally adapted to, three (S477N, T478K, and E484A) are neutral with, and none is incompatible with mACE2; in contrast, two of the RBM mutations (T501Y and S477N) are structurally adapted to, three (T478K, E484A, and Q498R) are neutral with, and two (Q493R and Y505H) are incompatible with hACE2 (Table 1). Overall, the omicron RBD is adapted to mACE2 significantly better than to hACE2. Importantly, our study has identified three RBM mutations (Q493R, Q498R, and Y505H) that are specifically adapted to mACE2, suggesting that these mutations are the evolutionary traces left by the omicron variant.
Our study has implications for the evolution of the omicron variant. Because Asn31 and His353 in ACE2 are the main targets of omicron’s adaptations, we searched all of the currently available ACE2 sequences and found that mice are the only species containing Asn31 and His353 in their ACE2 (SI Appendix, Table S2), suggesting that omicron evolved in mice. Our structural findings are supported by numerous studies that discovered the same mouse-specific mutations Q493R (or a similar Q493K) and Q498R (32–38). It is worth noting that although the prototypic SARS-CoV-2 does not infect mice efficiently, some of the earlier SARS-CoV-2 variants from humans and other animal species had evolved the N501Y mutation that can facilitate SARS-CoV-2’s usage of mACE2 as its receptor (20, 39, 40). Indeed, as the current study has demonstrated, the N501Y mutation is structurally adapted to ACE2 from more than one host species. Hence, a SARS-CoV-2 variant containing the N501Y mutation might have been transmitted from human or another animal species to mice. Afterward, as the SARS-CoV-2 variant spread in mice, mouse-specific RBM mutations (e.g., Q493R, Q498R, and Y505N) evolved, contributing to the emergence of the omicron variant. In addition to infecting humans, the omicron variant may transmit to some other host species whose ACE2 contains VBM residues that are compatible with the binding of the omicron RBD. For example, the ACE2 molecules of some species of rats contain either Asn31 or His353 (SI Appendix, Table S2). More epidemiological surveillance of SARS-CoV-2 in mice and other rodents can help further clarify the evolution and transmission of the omicron variant.
Materials and Methods
Plasmids.
The genes encoding the spike protein of SARS-CoV-2 prototypic strain (GenBank: QHD43416.1), hACE2 (GenBank accession No.: NM_021804), and mACE2 (GenBank accession No. NM_021804) were synthesized (GenScript Biotech). The gene encoding the spike protein of SARS-CoV-2 omicron strain (BA.2; GISAID: EPI_ISL_6795834) was constructed by site-directed mutagenesis of the gene encoding the spike protein of SARS-CoV-2 prototypic strain. The genes encoding the RBD of SARS-CoV-2 prototypic strain (residues 319 to 535), the RBD of SARS-CoV-2 omicron strain (residues 319 to 535), hACE2 (residues 1 to 615), and mACE2 (residues 1 to 615) were constructed from the above full-length genes. The gene encoding the chimeric omicron RBD was constructed by site-directed mutagenesis of the gene encoding the chimeric prototypic RBD (17, 18). The gene encoding the chimeric mACE2 was constructed by site-directed mutagenesis of the gene encoding hACE2 to include VBM sequences from mACE2 (National Center for Biotechnology Information Reference Sequence: NP_001123985.1) (Fig. 2B). The prototypic RBD-His, omicron RBD-His, and omicron RBD-His mutants were subcloned into pLenti-transfer vector (Addgene) with an N-terminal tissue plasminogen activator signal peptide and a C-terminal His tag. hACE2-Fc and mACE2-Fc were subcloned into the same vector except that a C-terminal human immunoglobulin G4 Fc region replaced the His tag. Chimeric omicron RBD-His, hACE2-His, and chimeric mACE2-His were subcloned into pFastBac I vector (Life Technologies) with an N-terminal honey bee melittin signal peptide and a C-terminal His tag.
Protein Expression and Purification.
The prototypic RBD-His, omicron RBD-His, omicron RBD-His mutants, hACE2-Fc, and mACE2-Fc were prepared from 293F mammalian cells (41). Briefly, lentiviral particles were packaged for construction of stable cell lines expressing one of the above proteins. Stable cell lines expressing one of the above proteins were selected in the presence of Puromycin (Gibco). Each of the proteins was collected from cell culture medium, purified on Ni-NTA column for His-tagged proteins or on Protein A column for Fc-tagged proteins, and purified further on Superdex200 gel filtration column (Cytiva).
Chimeric omicron RBD-His, hACE2-His, and chimeric mACE2-His were prepared from sf9 insect cells using the Bac-to-Bac system (Life Technologies) (18). Briefly, the His-tagged proteins were harvested from cell culture medium, purified on Ni-NTA column, and purified further on Superdex200 gel filtration column (Cytiva).
SPR Assay.
Binding interactions between RBDs and ACE2 molecules were measured by SPR using a Biacore S200 system (Cytiva) (17). Briefly, ACE2-Fc was immobilized to a protein A chip (Cytiva). Serial dilutions of purified recombinant RBD-His flowed through (protein concentrations ranging from 20 to 3,200 nM) in a running buffer composed of 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.4, 150 mM NaCl, 3 mM ethylenediaminetetraacetic acid, and 0.05% tween 20. Biacore Evaluation Software (Cytiva) was used for calculating the binding kinetics.
Pseudovirus Entry Assay.
Pseudovirus entry assay was performed as described previously (41). Briefly, HEK293T cells were cotransfected with a plasmid encoding the omicron spike protein or one of its mutants, a helper plasmid (encoding HIV-1 gag and pol proteins), and a reporter plasmid (encoding a luciferase reporter gene). Pseudoviruses were collected 72 h after transfection and then were used to infect HEK293T cells stably expressing mACE2. The pseudoviruses and target cells were incubated together at 37 °C for 2 h. Subsequently, the medium was changed to fresh medium, followed by incubation for another 48 h. Cells were then washed with phosphate-buffered saline buffer and lysed. Aliquots of cell lysates were transferred to plates, followed by the addition of luciferase substrate. Relative light units were measured using an EnSpire plate reader (PerkinElmer). The amounts of pseudovirus particles were quantified through Western blot analysis of p24 levels and used to calibrate pseudovirus entry efficiencies.
Crystallization and Structure Determination.
The complexes of the chimeric omicron RBD and hACE2 and of the chimeric omicron RBD and chimeric mACE2 were each purified on gel filtration chromatography. Crystals of each of the complexes were grown at room temperature over wells containing 100 mM Tris (pH 8.5), 20 to 26% PEG 6000, and 100 mM NaCl. X-ray diffraction data were collected at beamline 12–1 of Stanford Synchrotron Radiation Lightsource (SSRL). HKL2000 was used for data processing (42). Both of the structures were determined by molecular replacement using the structure of prototypic chimeric RBD complexed with hACE2 as the search template (Protein Data Bank [PDB] accession code 6VW1). PHENIX and CCP4 were used for molecular replacement and model refinement (43, 44). COOT was used for model building (45). PYMOL (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.) was used for making structural figures. Structure data and refinement statistics are shown in SI Appendix, Table S1.
Supplementary Material
Acknowledgments
This study was supported by NIH grant Nos. R01AI089728 and R01AI110700 (to F.L.) and R35GM118047 (to H.A.). The Biacore S200 system was supported by NIH Office of Research Infrastructure Programs grant No. 1S10OD021539. We thank Linfen Huang for technical support and Clyde Smith at beamline 12-1 of SSRL (contract number DE-AC02-76SF00515) for assistance in data collection.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2206509119/-/DCSupplemental.
Data, Materials, and Software Availability
Crystal structure data have been deposited in the Protein Data Bank (www.rcsb.org): PDB accession No. 7UFK (46) is for chimeric omicron RBD (strain BA.2) complexed with hACE2 and PDB accession No. 7UFL (47) is for chimeric omicron RBD (strain BA.2) complexed with chimeric mACE2. All study data are included in the article and/or SI Appendix.
References
- 1.Karim S. S. A., Karim Q. A., Omicron SARS-CoV-2 variant: A new chapter in the COVID-19 pandemic. Lancet 398, 2126–2128 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maslo C., et al. , Characteristics and outcomes of hospitalized patients in South Africa during the COVID-19 Omicron wave compared with previous waves. JAMA 327, 583–584 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saxena S. K., et al. Characterization of the novel SARS-CoV-2 Omicron (B.1.1.529) variant of concern and its global perspective. J. Med. Virol. 94, 1738–1744 (2022). [DOI] [PubMed] [Google Scholar]
- 4.Gu H., et al. , Probable transmission of SARS-CoV-2 Omicron variant in quarantine hotel, Hong Kong, China, November 2021. Emerg. Infect. Dis. 28, 460–462 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mallapaty S., Where did Omicron come from? Three key theories. Nature 602, 26–28 (2022). [DOI] [PubMed] [Google Scholar]
- 6.Li F., Receptor recognition mechanisms of coronaviruses: A decade of structural studies. J. Virol. 89, 1954–1964 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li F., Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 3, 237–261 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou P., et al. , A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wan Y., Shang J., Graham R., Baric R. S., Li F., Receptor recognition by the novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 94, e00127-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li W., et al. , Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426, 450–454 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li F., Structural analysis of major species barriers between humans and palm civets for severe acute respiratory syndrome coronavirus infections. J. Virol. 82, 6984–6991 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li F., Receptor recognition and cross-species infections of SARS coronavirus. Antiviral Res. 100, 246–254 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li F., Li W., Farzan M., Harrison S. C., Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 309, 1864–1868 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Wu K., Peng G., Wilken M., Geraghty R. J., Li F., Mechanisms of host receptor adaptation by severe acute respiratory syndrome coronavirus. J. Biol. Chem. 287, 8904–8911 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li W., et al. , Animal origins of the severe acute respiratory syndrome coronavirus: Insight from ACE2-S-protein interactions. J. Virol. 80, 4211–4219 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Temmam S., et al. , Bat coronaviruses related to SARS-CoV-2 and infectious for human cells. Nature 604, 330–336 (2022). Correction in: Nature 607, E19 (2022). [DOI] [PubMed] [Google Scholar]
- 17.Geng Q., et al. , Structural basis for human receptor recognition by SARS-CoV-2 Omicron VARIANT BA.1. J. Virol. 96, e0024922 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shang J., et al. , Structural basis of receptor recognition by SARS-CoV-2. Nature 581, 221–224 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawaoka Y., et al. , Characterization and antiviral susceptibility of SARS-CoV-2 Omicron/BA.2. Res. Sq., 10.21203/rs.3.rs-1375091/v1 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zahradník J., et al. , SARS-CoV-2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution. Nat. Microbiol. 6, 1188–1198 (2021). [DOI] [PubMed] [Google Scholar]
- 21.Wu K., et al. , A virus-binding hot spot on human angiotensin-converting enzyme 2 is critical for binding of two different coronaviruses. J. Virol. 85, 5331–5337 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dinnon K. H. III, et al. , A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature 586, 560–566 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao S. M., Du Q. S., Meng J. Z., Pang Z. W., Huang R. B., The multiple roles of histidine in protein interactions. Chem. Cent. J. 7, 44 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang D. X., Wang M. X., Exploring anion-π interactions and their applications in supramolecular chemistry. Acc. Chem. Res. 53, 1364–1380 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Shuai H., et al. , Emerging SARS-CoV-2 variants expand species tropism to murines. EBioMedicine 73, 103643 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lan J., et al. , Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581, 215–220 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Laffeber C., de Koning K., Kanaar R., Lebbink J. H. G., Experimental evidence for enhanced receptor binding by rapidly spreading SARS-CoV-2 variants. J. Mol. Biol. 433, 167058 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shang J., et al. , Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U.S.A. 117, 11727–11734 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye G., Liu B., Li F., Cryo-EM structure of a SARS-CoV-2 Omicron spike protein ectodomain. Nat. Commun. 13, 1214 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harvey W. T., et al. ; COVID-19 Genomics UK (COG-UK) Consortium, SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 19, 409–424 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li W., et al. , Efficient replication of severe acute respiratory syndrome coronavirus in mouse cells is limited by murine angiotensin-converting enzyme 2. J. Virol. 78, 11429–11433 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong L. R., et al. , Eicosanoid signalling blockade protects middle-aged mice from severe COVID-19. Nature 605, 146–151 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gu H., et al. , Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science 369, 1603–1607 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leist S. R., et al. , A mouse-adapted SARS-CoV-2 induces acute lung injury and mortality in standard laboratory mice. Cell 183, 1070–1085.e12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gawish R., et al. , ACE2 is the critical in vivo receptor for SARS-CoV-2 in a novel COVID-19 mouse model with TNF- and IFNγ-driven immunopathology. eLife 11, e74623 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang K., et al. , Q493K and Q498H substitutions in Spike promote adaptation of SARS-CoV-2 in mice. EBioMedicine 67, 103381 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun S., et al. , Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2. Nat. Commun. 12, 5654 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zakhartchouk A. N., et al. , Immunogenicity of a receptor-binding domain of SARS coronavirus spike protein in mice: Implications for a subunit vaccine. Vaccine 25, 136–143 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang L., et al. , Ten emerging SARS-CoV-2 spike variants exhibit variable infectivity, animal tropism, and antibody neutralization. Commun. Biol. 4, 1196 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Virtanen J., et al. , Experimental infection of mink with SARS-COV-2 Omicron variant and subsequent clinical disease. Emerg. Infect. Dis. 28, 1286–1288 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geng Q., et al. , Novel virus-like nanoparticle vaccine effectively protects animal model from SARS-CoV-2 infection. PLoS Pathog. 17, e1009897 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Otwinowski Z., Minor W., Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 43.Liebschner D., et al. , Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winn M. D., et al. , Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Zhang W., et al. , Crystal structure of chimeric omicron RBD (strain BA.2) complexed with human ACE2. Protein Data Bank. https://www.rcsb.org/structure/7ufk. Deposited 22 March 2022.
- 47.Zhang W., et al. , Crystal structure of chimeric omicron RBD (strain BA.2) complexed with chimeric mouse ACE2. Protein Data Bank. https://www.rcsb.org/structure/7ufl. Deposited 22 March 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Crystal structure data have been deposited in the Protein Data Bank (www.rcsb.org): PDB accession No. 7UFK (46) is for chimeric omicron RBD (strain BA.2) complexed with hACE2 and PDB accession No. 7UFL (47) is for chimeric omicron RBD (strain BA.2) complexed with chimeric mACE2. All study data are included in the article and/or SI Appendix.