Significance
Regulation of transcription and pre–messenger RNA (mRNA) splicing is essential for transcript diversity and light-regulated gene expression in plants. Although several transcription factors have been described, only a few splicing factors are known to regulate light signaling pathways. Here, we describe the functional characterization of splicing factor SWAP1, which interacts with two previously characterized splicing factors, SFPS and RRC1, forming a ternary complex. SWAP1 also interacts with photoactivated phytochrome B (phyB). All three single and double mutant combinations display similar hypocotyl lengths under red light. SWAP1 modulates alternative splicing of a large number of genes, and a subset of these genes is coordinately regulated by SFPS, RRC1, and SWAP1. These results highlight the importance of phyB-interacting splicing factors in light-regulated plant development.
Keywords: Arabidopsis, photomorphogenesis, pre-mRNA splicing, splicing factor, phytochrome B
Abstract
Light signals perceived by a group of photoreceptors have profound effects on the physiology, growth, and development of plants. The red/far-red light–absorbing phytochromes (phys) modulate these aspects by intricately regulating gene expression at multiple levels. Here, we report the identification and functional characterization of an RNA-binding splicing factor, SWAP1 (SUPPRESSOR-OF-WHITE-APRICOT/SURP RNA-BINDING DOMAIN-CONTAINING PROTEIN1). Loss-of-function swap1-1 mutant is hyposensitive to red light and exhibits a day length–independent early flowering phenotype. SWAP1 physically interacts with two other splicing factors, (SFPS) SPLICING FACTOR FOR PHYTOCHROME SIGNALING and (RRC1) REDUCED RED LIGHT RESPONSES IN CRY1CRY2 BACKGROUND 1 in a light-independent manner and forms a ternary complex. In addition, SWAP1 physically interacts with photoactivated phyB and colocalizes with nuclear phyB photobodies. Phenotypic analyses show that the swap1sfps, swap1rrc1, and sfpsrrc1 double mutants display hypocotyl lengths similar to that of the respective single mutants under red light, suggesting that they function in the same genetic pathway. The swap1sfps double and swap1sfpsrrc1 triple mutants display pleiotropic phenotypes, including sterility at the adult stage. Deep RNA sequencing (RNA-seq) analyses show that SWAP1 regulates the gene expression and pre–messenger RNA (mRNA) alternative splicing of a large number of genes, including those involved in plant responses to light signaling. A comparative analysis of alternative splicing among single, double, and triple mutants showed that all three splicing factors coordinately regulate the alternative splicing of a subset of genes. Our study uncovered the function of a splicing factor that modulates light-regulated alternative splicing by interacting with photoactivated phyB and other splicing factors.
Light functions not only as an energy source for photosynthesis but also as an environmental signal that modulates growth and development throughout the plant life cycle. To perceive and respond to surrounding light, plants contain an array of photoreceptors with distinct and/or overlapping wavelength perception. These photoreceptors collectively sense the light intensity, color, and duration to optimize growth and development of plants (1, 2). One such photoreceptor, ubiquitous across the plant and bacterial kingdom is “phytochrome” (phy), which perceives and responds to the red/far-red wavelength of the light spectrum (3, 4). In Arabidopsis, phys are encoded by a multigene family consisting of five different genes (PHYA to PHYE) (5). In dark-grown plants, phys are predominantly localized to the cytosol in the inactive red light absorbing (Pr) form, and upon red light illumination, they are photoconverted to the biologically active far-red light absorbing (Pfr) form and then translocated into the nucleus to form discrete nuclear bodies called photobodies (PBs) (3, 4). These PBs are membraneless subnuclear dynamic structures, where phyB undergoes liquid-liquid phase separation and interacts with diverse target proteins to initiate appropriate signaling cascades (6–9). One group of proteins, which were recently identified as interacting with photoactivated phyB and colocalized to PBs, are splicing factors that regulate light-induced pre–messenger RNA (mRNA) alternative splicing (AS) (10).
Pre-mRNA AS is an essential process in eukaryotes that increases the complexity of gene expression by generating multiple forms of mature mRNAs from a single multiexon/intron pre-mRNA (4, 11–13). Both internal and external factors modulate pre-mRNA AS to optimize mRNA diversity (14–19). A recent comprehensive analysis of AS revealed that about 79% of multiexonic protein-coding pre-mRNAs undergo AS in response to different stimuli, suggesting a high prevalence of AS phenomenon in Arabidopsis (20). AS is a tightly regulated and multistep biochemical process accomplished by a highly conserved, dynamic, and flexible macromolecular ribonucleoprotein complex called the spliceosome (13). The functional spliceosome complex consists of a core and a group of trans-acting factors (also known as auxiliary splicing regulatory proteins). The core is made up of ∼200 proteins plus a set of five small nuclear ribonucleoproteins (snRNPs; U1, U2, U4, U5, and U6), whereas trans-acting factors such as SR (Serine/Arginine-rich) proteins and hnRNPs (heterogeneous nuclear ribonucleoproteins) variably interact with the core to guide the spliceosome complex to modulate appropriate AS event(s) (12, 21, 22). Pre-mRNAs destined for AS contain a set of cis-acting elements, which include exonic and intronic splicing enhancers (ESEs and ISEs) as well as exonic and intronic splicing silencers (ESSs and ISSs) (13, 23). Interactions between cis-acting elements and trans-acting factors eventually define the AS event(s) to generate mature mRNA. Positively acting trans-acting factors such as SR proteins typically bind to ESEs/ISEs, promoting splicing, while negatively acting trans-acting factors such as hnRNPs typically bind to ESSs/ISSs, suppressing pre-mRNA splicing. Thus, a cascade of protein–protein and protein–RNA interactions in response to a range of stimuli modulates optimal pre-mRNA splicing (12, 13, 21, 23, 24).
The critical role of phys in modulation of the global gene expression pattern is well documented, and in the past few years, it has been shown that photoactivated phys also regulate pre-mRNA AS (10, 11, 24–29). Among the most crucial AS targets of photoactivated phys are pre-mRNAs encoding several SR proteins and U1/U2 snRNPs, which in turn further control the AS of many target pre-mRNAs (19, 25). Recently, we showed that at least two bona fide splicing factors, SFPS (SPLICING FACTOR FOR PHYTOCHROME SIGNALING) and RRC1 (REDUCED RED-LIGHT RESPONSES IN CRY1CRY2 BACKGROUND1), interact with photoactivated phyB and modulate pre-mRNA splicing of a large number of genes to optimize photomorphogenesis in Arabidopsis (30, 31). In this study, we describe the identification and characterization of an SFPS interacting protein partner, SWAP1 (SUPPRESSOR-OF-WHITE-APRICOT/SURP RNA-BINDING DOMAIN-CONTAINING PROTEIN1; At4g31200), a putative RNA- binding protein, that forms a ternary complex with SFPS and RRC1 to modulate plant responses to red light by regulating the AS of pre-mRNAs of a subset of light-regulated genes.
Results
swap1-1, Like sfps-2 and rrc1-3, Is Hyposensitive to Red Light.
Recently, we showed that SFPS and RRC1 are involved in light-regulated pre-mRNA AS and differential gene expression to modulate photomorphogenesis in Arabidopsis (30, 31). SFPS was first identified in a forward genetic screen of mutants defective in red light signaling, and subsequently, RRC1 was identified as one of the interacting partners of SFPS in an IP-MS (immunoprecipitation followed by mass spectrometry) analysis. The same IP-MS study also identified SWAP1 as another potential interacting partner of SFPS (SI Appendix, Fig. S1 A and B). SWAP1 is an ∼73-kda protein, and an in silico data analysis categorized SWAP1 as an RNA-binding protein containing SWAP and RPR (regulation of nuclear pre-mRNA) domains in its N- and C- termini, respectively (SI Appendix, Fig. S1C).
To understand the biological function of SWAP1, we obtained a homozygous T-DNA (transferred DNA) insertional mutant (swap1-1; SALK_027493) from ABRC (Arabidopsis Biological Resource Center) that displayed almost no expression of SWAP1 (SI Appendix, Fig. S2 A and B) after the T-DNA insertion site. When we examined seedling phenotypes under dark, continuous red, far-red, and blue light conditions, swap1-1 displayed WT (wild-type) sensitivity under dark, far-red, and blue light conditions, while it exhibited long hypocotyl phenotypes under red light (Fig. 1 A–C and SI Appendix, Fig. S2 C–F). In addition, SWAP1pro:SWAP1-GFP/swap1-1 transgenic lines grown under similar red light conditions complemented the swap1-1 mutant phenotype (SI Appendix, Fig. S3 A–D). Although SWAP1 regulates red light responses, the abundance of SWAP1 mRNA and protein was not altered in response to prolonged red light illumination (SI Appendix, Fig. S3 E and F).
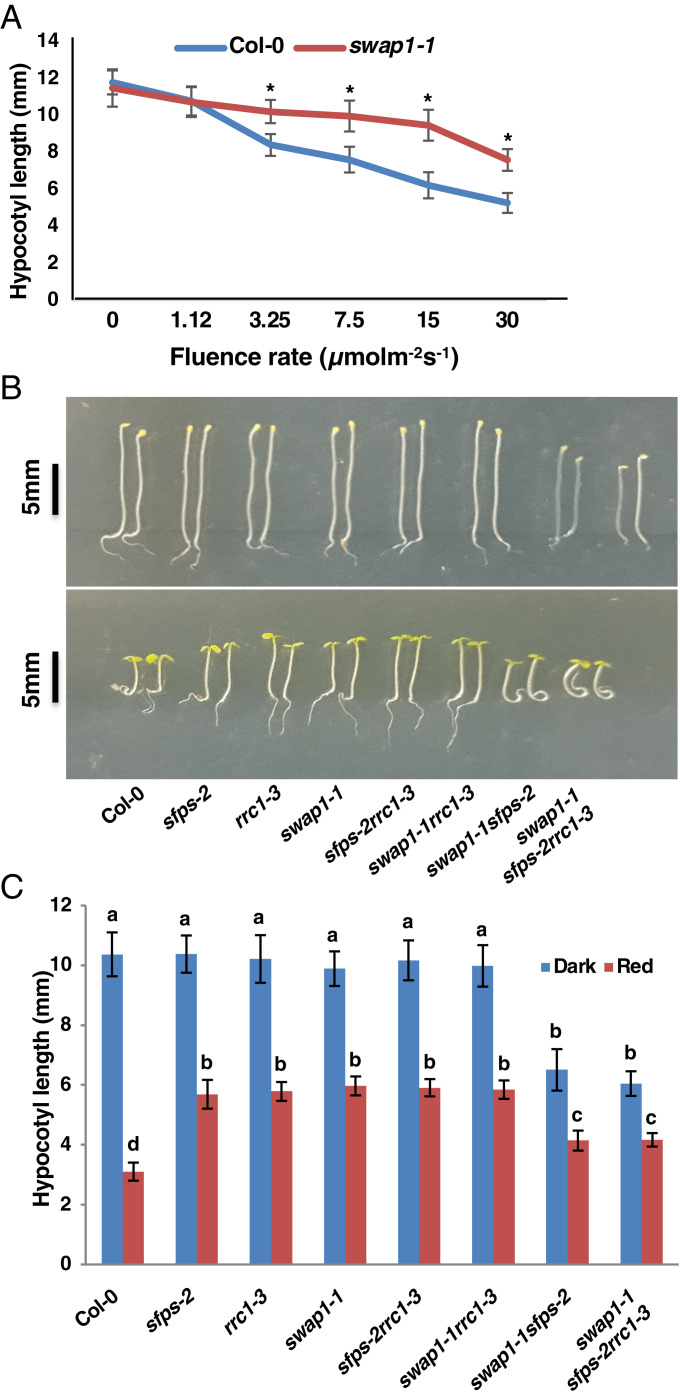
Fig. 1.
swap1-1 is hyposensitive to red light and acts in the same genetic pathway as sfps and rrc1. (A) Fluence rate response curve shows the hypocotyl length of 4-d-old seedlings grown under either continuous dark or continuous red light of different fluence rate. Error bars indicate SEM (n > 30), and asterisks indicate significant difference compared to Col-0 (P < 0.05) based on Student’s t test. (B and C) Digital images of representative seedlings (B) and bar graph showing hypocotyl lengths (C) of different genotypes grown under either continuous dark or continuous red light (7 µmolm−2s−1) for 4 d. Bars indicate mean length in millimeters, and error bars indicate SD. Statistical significance among different genotypes was determined using single-factor ANOVA followed by Tukey’s post hoc analysis and is indicated by different letters.
To test whether SWAP1 participates in the modulation of vegetative to reproductive growth in Arabidopsis, the flowering time of WT and swap1-1 mutant was quantified and compared by counting the number of rosette leaves and days-to-flower under both short-day(8-h light: 16-h dark) and long-day (16-h light: 8-h dark) conditions. The swap1-1 mutant plants flowered earlier than WT plants under both short-day and long-day conditions. On the other hand, SWAP1pro:SWAP1-GFP/swap1-1 transgenic plants flowered at the same time as WT plants (SI Appendix, Figs. S4 and S5). Taken together, these data suggest that SWAP1 is one of the essential regulatory components that modulate red light signaling and day light–independent flowering in Arabidopsis.
swap1-1, sfps-2, and rrc1-3 Function in the Same Genetic Pathway to Regulate Photomorphogenesis.
To investigate the genetic relationships among the three splicing factor mutants, we created three double (swap1sfps, swap1rrc1, and sfpsrrc1) and triple (swap1sfpsrrc1) mutant combinations and examined their seedling phenotypes under red light. swap1rrc1 and sfps-2rrc1 displayed hypocotyl lengths similar to those of the respective single mutants (Fig. 1 B and C), suggesting that they function in the same genetic pathway. However, the swap1-1sfps-2 double and swap1-1sfps-2rrc1-3 triple mutants displayed pleiotropic phenotypes, including short hypocotyls under both dark and red light (Fig. 1 B and C) and delayed onset of true leaves (SI Appendix, Fig. S6). At the juvenile stage, all three single, double, and triple mutants displayed progressively shorter primary root length compared to WT (SI Appendix, Fig. S7). Moreover, swap1-1 single, swap1-1rrc1-3 and swap1-1sfps-2 double, and swap1-1sfps-2rrc1-3 triple mutants displayed shorter inflorescence and silique lengths and sterility at the adult stage compared to WT (SI Appendix, Fig. S8), suggesting that these splicing factors function in other developmental pathways in addition to regulating seedling photomorphogenesis.
SWAP1 Interacts with SFPS and RRC1 and Forms a Ternary Complex.
As SWAP1 was found to coimmunoprecipitate with SFPS, we investigated whether SWAP1 is a bona fide interacting partner of SFPS and RRC1 through a series of in vitro and in vivo protein–protein interaction assays. Yeast two-hybrid and in vitro pull-down assays using full-length SWAP1, SFPS, and RRC1 indicated that SWAP1 physically interacts with both SFPS and RRC1 (SI Appendix, Fig. S9). An in vivo coimmunoprecipitation (Co-IP) assay performed using the dark- and red light–treated samples indicated that SWAP1 interacts with both SFPS and RRC1 in a light-independent manner (Fig. 2 A and B). Furthermore, Co-IP assays also revealed that the presence or absence of SFPS or RRC1 did not alter the strength of interaction between SWAP1 and RRC1 or SWAP1 and SFPS, respectively (Fig. 2 A and B). Since each of the three proteins physically interacts with each other, these three proteins may form a functional ternary complex to modulate pre-mRNA splicing. To test the formation of the ternary complex, an in vitro pull-down assay was performed using MBP-RRC1 as a bait protein and glutathione S-transferase (GST)–SFPS and an increasing concentration of GST-SWAP1 as prey proteins. The results showed that the association between MBP-RRC1 and GST-SFPS was promoted by increasing amounts of GST-SWAP1, suggesting that these three splicing factors are forming a ternary complex (Fig. 2C).
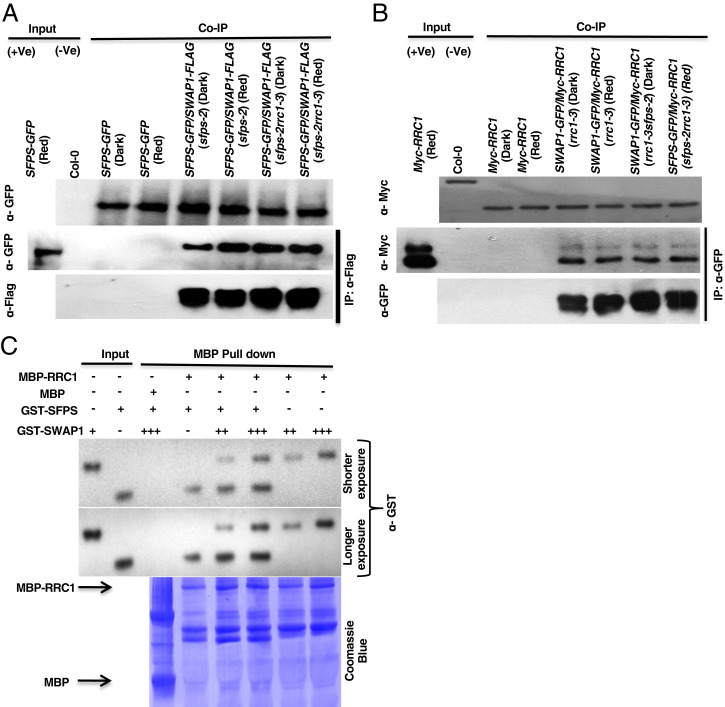
Fig. 2.
SWAP1 interacts with SFPS and RRC1 and forms a ternary complex. (A and B) SWAP1 interacts with SFPS (A) and RRC1 (B) in vivo. Four-day-old seedlings were either kept in the dark or exposed to constant red light for 6 h (7 µmolm−2s−1), and total protein was extracted in native extraction buffer. Then, 1 µg of appropriate antibody bound to Dynabeads (protein A) was added to the protein mixture and incubated for 3 h at 4 °C. After incubation, the beads were pelleted and washed multiple times and proteins were separated on 8 to 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS/PAGE). The presence or absence of specific proteins was detected following immunoblotting with appropriate antibodies. (C) SWAP1, SFPS, and RRC1 proteins formed a ternary complex in vitro. Bacterially expressed and amylose resin-bound MBP and MBP-RRC1 were used as bait proteins to pull down GST-SFPS and GST-SWAP1 prey proteins. For GST-SFPS and GST-SWAP1: +, ∼1 μg; ++, ∼2 μg; and +++, ∼3 μg of protein. The pull-down reaction was incubated for 3 h at 4 °C on a rotating shaker. After the incubation, beads were washed at least five times. Proteins were separated on 10% SDS/PAGE gel, transferred to PVDF membrane, and first immunoblotted with α-GST, followed by Coomassie blue staining of the membrane.
Subcellular localization of SWAP1-GFP revealed that SWAP1 localizes exclusively to the nucleus and forms discrete nuclear speckles of varying sizes (SI Appendix, Fig. S10A). As reported earlier, both SFPS and RRC1 also form discrete nuclear speckles and colocalize with each other (31). Since SWAP1 interacts with both proteins, we examined whether SWAP1 nuclear speckles also colocalize with those of SFPS and RRC1. We prepared SFPS-GFP/SWAP1-mCherry and SWAP1-GFP/RRC1-mCherry double transgenics, and fluorescent confocal imaging was performed using 8-d-old light-grown seedlings. The results show that the SWAP1 nuclear speckles almost always colocalized with nuclear speckles of SFPS and RRC1 (SI Appendix, Fig. S10 B and C). Taken together, these biochemical and imaging data suggest that SWAP1, SFPS, and RRC1 proteins not only colocalize with each other but also physically interact to form a ternary protein complex.
Previously, SFPS and RRC1 were shown to colocalize with 3′ SS (Splice Site) targeting U2-snRNP–associated components such as U2A′, U2AF35A, and U2AF65B in the nucleus (30, 31). As SWAP1 interacts and colocalizes with both SFPS and RRC1, we examined whether SWAP1 colocalizes with U2-snRNP–associated components. Double transgenic lines expressing SWAP1-GFP/U2A′-mCherry, SWAP1-GFP/U2AF35A-mCherry, and SWAP1-GFP/U2AF65B-mCherry were prepared, and root cells from different developmental regions were observed under a confocal fluorescent microscope. Interestingly, the nuclear speckles of SWAP1-GFP invariably colocalized with the nuclear speckles of U2A′-mCherry, U2AF35A-mCherry, and U2AF65B-mCherry (SI Appendix, Fig. S11), suggesting that SWAP1 might regulate 3′ SS selection.
SWAP1 Regulates the Expression of Genes Involved in Light Signaling.
Splicing factors are reported to regulate gene expression in response to a specific stimulus (10, 12, 24). As SWAP1 is predicted to be a splicing factor, we examined whether it modulates gene expression by performing deep RNA sequencing (RNA-seq) of WT and swap1-1 mutant seedlings grown under dark and dark-grown seedlings exposed to 3 h of red light. RNA-seq data were first analyzed using the DESeq2 package, and then enriched Gene Ontology (GO) terms were determined using the GeneCodis4 online tool. A comparison of differentially expressed genes (DEGs) between dark- and red light–treated WT samples identified a total of 6,226 DEGs passing the threshold of fold change (FC) of ≥1.5 and false discovery rate (FDR) of ≤0.05. In contrast, red light–irradiated swap1-1 displayed 7,595 DEGs compared to the dark-treated swap1-1 mutant samples (SI Appendix, Fig. S12A and Dataset S1, I and VII). An in-depth comparison showed a total of 3,932 DEGs between WT and swap1-1 under dark conditions, while under red light, 5,137 genes were found to be differentially expressed between WT and swap1-1 samples (SI Appendix, Fig. S12A and Dataset S1, III and V). A heatmap of the top 1,000 genes shows the gene expression patterns among these samples (SI Appendix, Fig. S12B). GO-term analyses of these comparisons showed significant enrichment of many GO-terms, including alternative mRNA splicing, spliceosome-mediated splicing, RNA metabolism, photomorphogenesis, and red and far-red signal transductions (Dataset S1, II, IV, VI, and VIII). Enrichment of these biologically important GO-terms, as well as differential expression of several genes involved in light signaling, unequivocally highlights the importance of SWAP1 in red light signaling.
SWAP1 Regulates pre-mRNA Splicing in Arabidopsis.
To identify the extent by which SWAP1 modulates pre-mRNA AS in Arabidopsis, in-depth analyses of RNA-seq data were performed using ASpli (Version 2.4.0) (32). To determine the overall differential alternative splicing (DAS) in the mutant, the percentage of inclusion of introns (PIR [Percent Intron Retention]) and exons (PSI [Percent Spliced-In]) was calculated in four systematic combinations: WT_Dark (Col-0_D) versus WT_Red light (Col-0_R), swap1-1_Dark (swap1_D) versus swap1-1_Red light (swap1_R), Col-0_D versus swap1_D, and Col-0_R versus swap1_R. To capture the DAS events with high confidence, only those events that passed through the stringency test of FDR < 0.05, FC ≥ 1.5, and Delta PSI or Delta PIR of ≥0.2 were counted. Col-0_D versus swap1_D analyses identified a total of 7,770 DAS events corresponding to 3,957 gene loci, while Col-0_R versus swap1_R comparison yielded a total of 12,488 DAS events corresponding to 6,123 gene loci (Fig. 3 A and B and Dataset S2, III and V). While all the subcategories of AS events were found in swap1-1 samples, the most predominant was the IR (intron retention) event under both dark- and red light–treated conditions (Fig. 3B and Dataset S2, III and V). Interestingly, analysis of overlapping DAS events between dark- and red light–treated samples identified a total of 3,860 events, corresponding to 50% of dark and 31% of red light events (Fig. 3A), implying a clear shift in SWAP1 targets upon red light illumination.
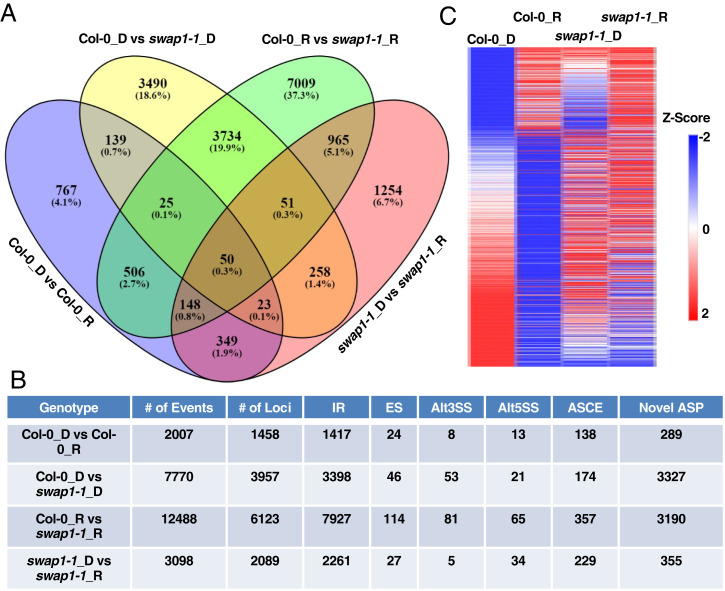
Fig. 3.
SWAP1 modulates pre-mRNA splicing in Arabidopsis. (A) Venn diagram indicating both number and overlapping DAS events in wild-type and swap1-1 mutant seedlings under D- (dark) and R- (red light) treated conditions. (B) Table showing the total number of DAS events and the corresponding gene loci numbers detected in analysis. The total number of DAS events is further subdivided into different categories depending on the type of event. IR, intron retention; ES, exon skipping, Alt3SS, alternative 3'-splice site; Alt5SS, alternative 5'-splice site; ASCE, alternative splicing affecting a consensus exon; Novel ASP, novel Alternative Splicing Pattern. (C) Heatmap of top 1,000 DAS events plotted based on their Z-scores. Z-scores are calculated on the basis of their corresponding individual PSI/PIR values under dark- and red light–treated conditions of wild-type and swap1-1 mutant samples.
Analysis of the scatterplots depicting the splicing efficiency changes among samples indicated that a large number of splicing events showed varying degrees of alterations in swap1-1 compared to WT (SI Appendix, Fig. S13). Likewise, a heatmap comparison of the overall DAS events also revealed that the splicing patterns of the majority of the events in the swap1-1 mutant were largely opposite to that of WT, under both dark- and red light–treated conditions (Fig. 3C). The gene list has photoreceptor genes including phyE and PHOT1 (Phototropin 1), as well as a number of genes that function downstream of these photoreceptors, including PIFs (Phytochrome Interacting Factors), COP1 (Constitutive Photomorphogenic1), B-Box (BBXs), and PAPP2C (Phytochrome Associated Protein Phosphatase type 2C). Furthermore, GO-term analysis showed that a large number of GO-terms were enriched under both dark and light conditions (Dataset S2, IV and VI). GO-terms such as photomorphogenesis, red/far-red light signaling, regulation of flowering, alternative mRNA splicing, and RNA processing were found to be significantly enriched in the dark- or red light–treated samples. We also analyzed the number of genes that were affected only at the pre-mRNA splicing or gene expression or both by plotting the Venn diagram of DEGs and DAS. This analysis revealed that a large number of genes were found to be unique to either pre-mRNA splicing or DEGs, and only a small number of genes were regulated at both the pre-mRNA splicing and gene expression levels (SI Appendix, Fig. S14).
To independently verify the RNA-seq data, we randomly selected five DAS events for RT-qPCR (reverse transcription-quantitative polymerase chain reaction) confirmation. For RT-qPCR, seedlings were grown and treated, and RNA was isolated under the same conditions as RNA-seq samples from three independent biological repeats. As shown in Fig. 4, data generated from RT-qPCR is largely similar to that of RNA-seq analysis, implying that the bulk of RNA-seq data could independently be reproducible and verifiable.
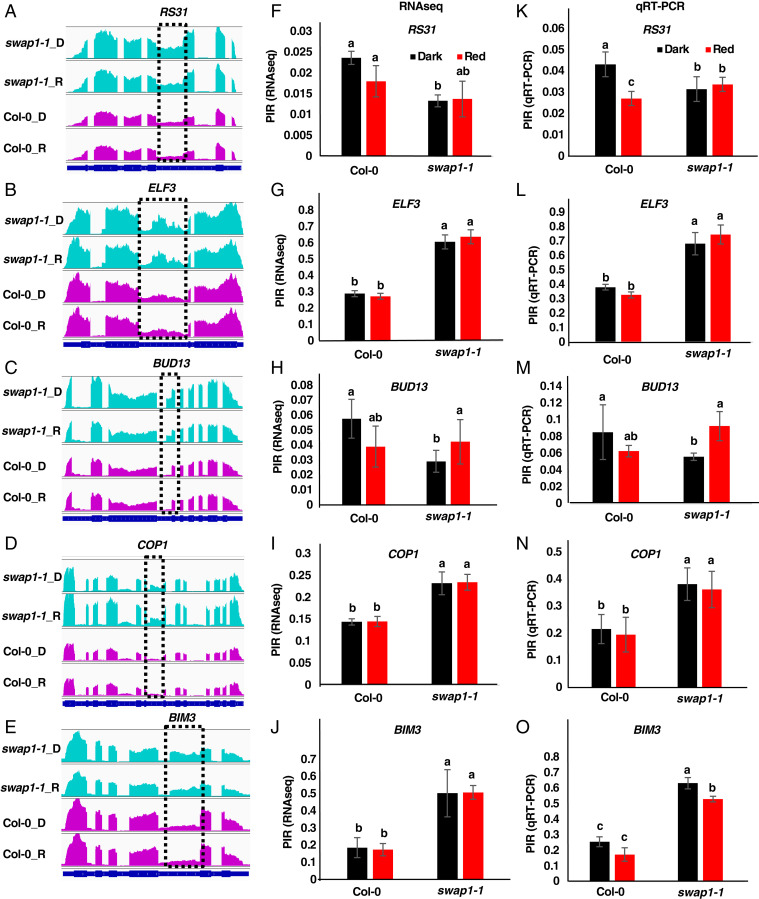
Fig. 4.
RT-qPCR confirmation of RNA-seq data. (A–E) IGV (Integrative Genomics Viewer) visualization of read depth graphs depicting the DAS events in (A) RS31, (B) ELF3, (C) BUD13, (D) COP1, and (E) BIM3 as detected from RNA-seq analysis of Col-0 versus swap1-1 samples. (F–J) Intron retention patterns of corresponding genes as obtained from RNA-seq analysis of Col-0 versus swap1-1 samples. (K–O) Quantification of retained introns of the corresponding genes as determined independently by RT-qPCR analysis. Statistical significance between genotypes was calculated using single-factor ANOVA followed by Tukey’s post hoc analysis and is indicated by different letters. Error bars indicate SD.
To investigate the functional relevance of SWAP1 in red light–modulated pre-mRNA splicing, we first determined the DAS events in WT (Col-0_D vs. Col-0_R) and then compared them against the list of DAS events in swap1-1 (swap1_D vs. swap1_R). Col-0_D versus Col-0_R analyses identified a total of 2,007 DAS events corresponding to 1,458 gene loci, while in swap1-1 mutant, a total of 3,098 splicing events corresponding to 2,089 gene loci were found to be altered in response to red light (Fig. 3B and Dataset S2, I, VII). Identification of overlapping DAS events interestingly revealed that only 28% and 18% of the overall WT and swap1-1 AS events overlapped, respectively (Fig. 3A), meaning a very large percentage (∼82%) of DAS events observed in swap1-1 were unique. The gene list includes several genes involved in photomorphogenesis and hypocotyl length elongation (HY2, PIL6, and PAPP2) and flowering time regulation (FCA, VIP4, VEL2, and AGLs), as well as several splicing factor genes including RS40, PRP31, SR31, and SCL30A. Moreover, a GO-term analysis of unique DAS of swap1-1 identified several GO-terms related to photomorphogenesis, red/far-red light signaling, pre-mRNA splicing, and flowering (Dataset S2, II and VIII). These data clearly indicate that SWAP1 plays a major role at the molecular level to optimize the global AS of a large number of direct and indirect target genes in response to red light stimulation.
In our previous studies, we showed that ELF3 (EARLY FLOWERING 3) pre-mRNA is one of the direct targets of both SFPS and RRC1, as both proteins were found to be associated with ELF3 pre-mRNA in vivo and to regulate its AS pattern (30, 31). In our RNA-seq analysis, we identified that ELF3 is also one of the targets of SWAP1-modulated AS (SI Appendix, Fig. S15A and Dataset S2, III, V). We therefore presumed that like SFPS and RRC1, SWAP1 might also associate with ELF3 pre-mRNA. This was tested by performing an RNA-immunoprecipitation assay followed by an RT-qPCR assay by employing growth conditions, light treatments, and methods as described previously (30, 31). It was observed that SWAP1 was indeed associated with various regions of ELF3 pre-mRNA (SI Appendix, Fig. S15B). This clearly implies that ELF3 is one of the direct targets of SWAP1.
SWAP1, SFPS, and RRC1 Control Pre-mRNA Splicing Individually and Coordinately in Binary and Ternary Combinations.
To identify genome-wide AS events regulated by all three splicing factors, we performed AS analyses of RNA-seq data of the three double (swap1sfps, swap1rrc1, and sfpsrrc1) and triple (swap1sfpsrrc1) mutant combinations (Dataset S2, IX–XXIV). In addition, we performed a similar analysis with previously described datasets for sfps-2 and rrc1-3 (30, 31), and all DAS events identified among 36 samples are shown in SI Appendix, Table S1. To investigate the correlation of splicing efficiency changes of DAS events among the single and higher order mutant combinations, we determined the Delta_PSI/PIR of every individual DAS event. As shown in the heatmap (SI Appendix, Fig. S16A), the splicing efficiency of a majority of DAS events was largely similar among the single, double, and triple mutant combinations but opposite to that of WT, under both dark- and red light–treated conditions.
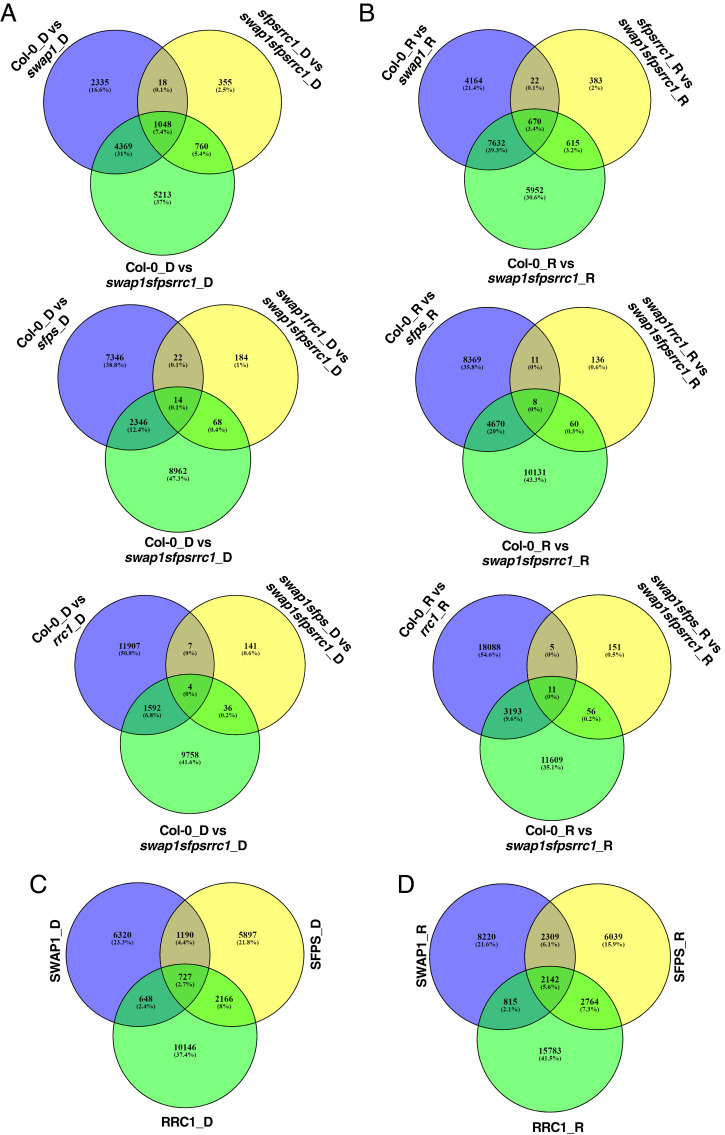
To define the contribution of individual splicing factors in controlling AS in response to light, the DAS events identified from the swap1sfpsrrc1 triple versus WT comparison provide all the possible AS events regulated by all three splicing factors under both dark and light conditions. In addition, we identified the loss-of-function (e.g., swap1 vs. WT for SWAP1) and gain-of-function (e.g., sfpsrrc1 vs. WT for SWAP1) events for each splicing factor. A three-way Venn diagram comparison identifies the total number of events (e.g., 2,335+18+355+4,369+1,048+760 = 8,885 for SWAP1 in dark) regulated by each splicing factor under dark and red light conditions. These analyses revealed that SWAP1, SFPS, and RRC1 regulated 8,885, 9,980, and 13,687 events, respectively, in dark, and 13,486, 13,254 and 21,504 events, respectively, under red light (Fig. 5 A and B).
Fig. 5.
Identification of DAS events regulated by SFPS, RRC1, and SWAP1. (A and B) Venn diagrams showing SWAP1- (Top), SFPS- (Middle), and RRC1- (Bottom) regulated DAS events under either dark-treated (A) or red-light–treated conditions (B). (C and D) SWAP1-, SFPS-, and RRC1-specific as well as overlapping DAS events regulated by one or two or three splicing factors observed in the dark (C) or red light (D).
Moreover, another three-way comparison of the DAS events regulated by each splicing factor identified the extent to which these splicing factors controlled AS events either individually (e.g., 6,320 in dark or 8,220 under red light for SWAP1) or in a binary or ternary complex under dark and red light conditions (Fig. 5 C and D). These analyses show that all three splicing factors regulated more events under red light compared to dark. Furthermore, these three splicing factors regulated approximately three times more events coordinately under red light compared to dark. Among the events regulated by all three splicing factors in dark and red light conditions, only 11% of events were common and ∼72% were unique to light condition (SI Appendix, Fig. S16B), implying that the ternary complex has a preferential role under light conditions. Overall, these data strongly suggest a crucial role of these splicing factors in reprogramming transcript diversity in response to red light to promote photomorphogenesis.
SWAP1 Interacts with Red/Far-Red–Light Photoreceptor phyB.
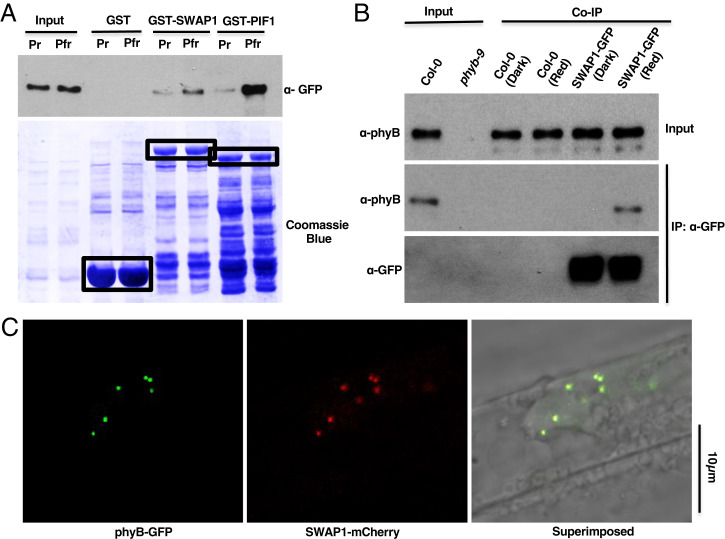
As SWAP1 physically interacts with phyB-interacting SFPS and RRC1, we examined whether SWAP1 also interacts with phyB. Yeast two-hybrid analysis of full-length SWAP1 and phyB showed that these two proteins interact in yeast (SI Appendix, Fig. S17). In addition, an in vitro pull-down assay using bacterially expressed GST-SWAP1 as a bait and full-length phyB-GFP expressed in yeast cells as a prey showed that GST-SWAP1 interacted preferentially with the phyB-Pfr form (Fig. 6A). GST-PIF1 was used as a control, which also interacted strongly with the Pfr form of phyB. To verify the interaction between SWAP1 and phyB, an in vivo Co-IP assay was performed using SWAP1pro:SWAP1-GFP/swap1-1 transgenic seedlings. Immunoprecipitation of SWAP1-GFP followed by probing for native phyB indicated that SWAP1 did interact with the Pfr form of phyB under the in vivo condition, albeit in a light-dependent manner (Fig. 6B). As SWAP1 and phyB physically interacted with each other and formed discrete nuclear speckles and PBs, respectively, we examined whether the nuclear bodies of these two proteins colocalize. When 5-d-old (4 d in the dark and 1 d in white light) SWAP1-mCherry/phyB-GFP double transgenic seedlings were imaged using confocal fluorescent microscopy, almost all the nuclear speckles of SWAP1-mCherry colocalized with phyB-GFP PBs (Fig. 6C). Taken together, these results clearly show that SWAP1 and phyB colocalize within the nucleus and also physically interact under in vivo conditions.
Fig. 6.
SWAP1 interacts with phyB. (A) Bacterially expressed GST-SWAP1 interacts with phyB-GFP expressed in yeast. Bacterially expressed and glutathione beads–bound GST, GST-SWAP1, and GST-PIF1 were mixed with crude extracts of yeast cell–expressed phyB-GFP. One set of tubes was illuminated with far-red light (Pr), and another set was exposed to red light (Pfr) for 10 min. Tubes were then incubated for 2 h at 4 °C. After the incubation, beads were pelleted and washed multiple times, and proteins were separated on 8% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS/PAGE). The presence or absence of specific proteins was detected following immunoblotting with appropriate antibodies. GST, GST-SWAP1 and GST-PIF1 are inicated by black boxes. (B) SWAP1 interacts with phyB under in vivo conditions. Four-day-old seedlings were either kept in the dark or exposed to constant red light for 6 h (7 µmolm−2s−1), and total protein was extracted in native extraction buffer. Then, 1 µg of appropriate antibody bound to Dynabeads (protein A) was added to the protein mixture and incubated for 3 h at 4 °C. After incubation, the beads were pelleted and washed multiple times and proteins were separated on 8% SDS-PAGE. The presence or absence of specific proteins was detected following immunoblotting with appropriate antibodies. (C) Nuclear speckles of SWAP1-mCherry colocalized with phyB-GFP PBs. Four-day-old dark-grown seedlings of 35Spro:SWAP1-mCherry/35Spro:phyB-GFP double transgenics were exposed to 24 h of white light, and then primary root cells were imaged using confocal microscopy.
Discussion
Being sessile, plants are routinely exposed to an ever-changing environment. In response to such changes in their surrounding environment, plants deploy diverse strategies, including changes in growth and developmental pattern(s) to better suit the surrounding conditions. Growth and developmental plasticity in plants (and in other eukaryotes) are often modulated by reprogramming of gene expression globally, which includes pre-mRNA AS and translational regulation (4, 11, 33). The role of AS in transcriptome diversity and gene expression in response to an array of both internal and external stimuli is becoming increasingly evident (4, 11, 12, 24). Light is one such environmental stimulus that shapes plants’ responses by modulating the transcriptome diversity through AS (10, 24, 26). While in the past the core emphasis of light-mediated signaling was at the level of transcriptional regulation of gene expression, several recent deep RNA-seq studies have described the significance of light-mediated changes in AS patterns, transcriptome complexity, and their corresponding effect on plant development (4, 11). Previously, our own study identified the phy interacting splicing factor SFPS positively regulating the phy signaling pathway by intricately modulating the pre-mRNA AS (30). Subsequently, a Co-IP mass spectrometric study of SFPS-GFP identified other proteins, including bona fide splicing factors such as RRC1 (31) and SWAP1 (present study).
In this study, we present several lines of molecular, biochemical, and genetic evidence to demonstrate that SWAP1 is indeed a phyB-interacting bona fide splicing factor, controlling red light–mediated pre-mRNA splicing to regulate photomorphogenesis in collaboration with SFPS and RRC1. Similar to SFPS and RRC1, SWAP1 protein contains an RNA-binding SWAP domain and is categorized as a “splicing regulatory” protein (SI Appendix, Fig. S1C). swap1 mutant displays red light–specific hyposensitivity and flowers early, resembling the sfps and rrc1 mutant phenotypes (Fig. 1 and SI Appendix, Figs. S2–S5) (30, 31). Through a series of in vitro and in vivo protein–protein interaction assays as well as protein colocalization studies, we established that SWAP1 not only colocalizes with SFPS and RRC1 but also interacts with them in a light-independent manner, forming a ternary complex (Fig. 2 and SI Appendix, Figs. S9 and 10). In addition, SWAP1 interaction with SFPS is not dependent on RRC1. Similarly, SWAP1 interaction with RRC1 also does not require SFPS (Fig. 2). Hence, a single combination of two proteins and a ternary complex might target a distinct set of pre-mRNAs, in addition to a subset of overlapping targets. This could be verified by identifying the possible unique as well as overlapping direct pre-mRNAs targets of these splicing factors.
One of the characteristics of the splicing factors is their association with the components of one of the U-snRNPs, which enables them to be in proximity to the intron-exon junction of the target (10, 24). SWAP1, like SFPS and RRC1, colocalized with the components that recognize 3′SS and recruit U2-snRNP to the pre-mRNA branchpoint (SI Appendix, Fig. S11) (30, 31). Deep RNA-seq analysis revealed that SWAP1 modulated pre-mRNA AS as well as gene expression, under both dark- and red light–treated conditions (Fig. 3 and SI Appendix, Fig. S12). However, when gene loci related to DAS and DEGs were compared, only a small fraction of them were found to be overlapping (SI Appendix, Fig. S14). A possible explanation could be that SWAP1 might first regulate the AS of general splicing and transcription factors, which then modulate the gene expression of downstream genes. Therefore, at least some of the physiological alterations observed in swap1 could be due to the combined effect of direct and indirect molecular events.
SWAP1 was found to control pre-mRNA AS of genes not only under red light–treated conditions but also in the dark, albeit ∼40% fewer genes than under red light (Fig. 3). However, only a small number of genes were found to overlap between dark- and red light–treated conditions. Several light-signaling, hypocotyl length, and flowering time modulator genes were found to be selectively spliced differently in response to red light treatment in swap1-1 (Fig. 3 and Dataset S2, III and V). These data clearly imply that SWAP1 largely targets unique gene loci under two different conditions to modulate optimal plant responses to surrounding ambient light signals. Furthermore, GO analysis of differentially spliced genes revealed that the enrichment of GO-terms was related to splicing, spliceosomes, RNA processing, red and far-red light signaling, photomorphogenesis, circadian rhythm, and regulation of flowering (Dataset S2, IV and VI). As SWAP1, SFPS, and RRC1 interact and form a ternary complex, we identified a subset of coregulated splicing events to evaluate the possible functional role of this complex. These data also revealed that the subsets of splicing events/genes are coregulated by binary interaction between the splicing factors and/or by the ternary complex in dark and red light conditions (Fig. 5). Therefore, it is evident that all three proteins target unique pre-mRNAs/genes in addition to a small subset of common targets to cooperatively modulate plant growth and development.
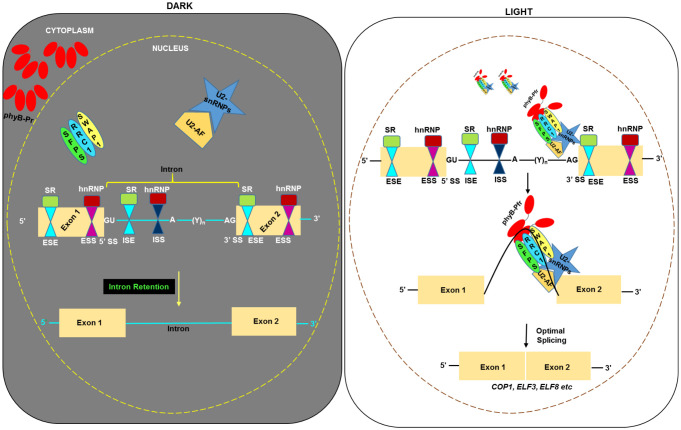
Several phy-interacting splicing factors/regulators have been identified in both Arabidopsis (SFPS, RRC1, SWAP1, NOT9B, and SWELLMAP2) (26, 28, 30, 31) and Physcomitrella patens (PphnRNP-H1, PphnRNP-F1) (34, 35). Since SFPS, RRC1, and SWAP1 also colocalize within phyB PBs (Fig. 6) (30, 31) and phyB PBs have been shown to be liquid-liquid phase separated droplets (8, 9), it is possible that these splicing factors are recruited by phyB in these phase-separated environments in a concentrated manner to modulate pre-mRNA AS and gene regulation (Fig. 7). This is consistent with recent data showing that the blue light photoreceptor cryptochrome 2 (CRY2) interacts and colocalizes with N6-methyladenosine RNA methyltransferase enzymes in CRY2 PBs (36), implying that PBs might be sites for mRNA metabolism and pre-mRNA splicing (7). In response to red light perception, phys are known to modulate the protein abundance of numerous downstream interactor proteins (4); however, this does not appear to be the case with any of the three splicing factors (SI Appendix, Fig. S3F) (30, 31). Therefore, it is possible that the interaction of these splicing factors with phys might result in some biochemical changes within splicing factors, leading to altered target-binding capacity and/or activity. Large-scale proteomic studies have identified multiple phosphorylation sites within SFPS and RRC1 (37, 38). Therefore, phys might induce phosphorylation and/or other posttranslational modification of these splicing factors within the PBs, leading to altered target identification and/or protein activity. Alternatively, phys might directly bind to these splicing factors and sequester their activities as has been shown for PIFs (39–41). These possibilities need to be tested in the future to uncover biochemical mechanisms by which phys/splicing factor complexes regulate pre-mRNA splicing and light-regulated developmental processes.
Fig. 7.
Model of red light–modulated pre-mRNA splicing through the SFPS-RRC1-SWAP1 ternary complex. Left: In the dark, biologically inactive phyB-Pr remained in the cytosol and therefore did not interact with nuclear localized splicing factors or spliceosome complex. Nevertheless, splicing factors remained biologically active and targeted hundreds of pre-mRNAs to modulate their splicing in the dark. Right: In response to red light irradiation, photoconverted and biologically active phyB-Pfr moieties migrated into the nucleus and interacted with splicing factors. This possibly led to biochemical changes within the splicing factors, resulting in targeting of a different set of pre-mRNAs to promote optimal photobiological responses in seedlings/plants. SR, serine/arginine-rich proteins; Yn, poly-pyrimidine tract; hnRNP, heterogeneous nuclear ribonucleoprotein.
Materials and Methods
Plant Materials and Growth Conditions.
All seeds used in this study were in Col-0 background. Seeds were first surface sterilized, plated on MS (without sucrose) medium, and stratified at 4 °C for 4 d. Plates were exposed to 3 h of continuous white light and then transferred to growth chambers with appropriate light/dark conditions. For hypocotyl length measurement at the seedling stage, digital images of 4-d-old seedlings were obtained, and length was measured by using the ImageJ tool.
Adult plants were grown by transplanting ∼10-d-old seedlings to the pots containing Promix soil (Premier Tech Horticulture) and transferred to growth chambers with different light regimens at 22 °C. To study flowering time, plants were grown under either long-day (16-h light/8-h dark) or short-day (8-h light/16-h dark) conditions at 22 °C. Postbolting rosette leaf number as well as number of days to flowering were counted when inflorescence reached ∼1 cm. Detailed information is provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank members of the E.H. laboratory for critical reading of the manuscript. This work was supported by grants from the NSF (MCB-2014408 to E.H. and MCB-2014542 to A.S.N.R.). The authors acknowledge the Texas Advanced Computing Center at The University of Texas at Austin for providing high-performance computing, visualization, and database resources that have contributed to the research results reported in this paper.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2214565119/-/DCSupplemental.
Data, Materials, and Software Availability
RNA-seq data were deposited into the Gene Expression Omnibus (GEO) database (accession No. GSE214299) (42). Source data are provided with this paper. Arabidopsis mutants and transgenic lines, as well as plasmids and antibodies generated during the current study, are available from the corresponding author upon reasonable request.
References
- 1.Kathare P. K., Huq E., “Signals | Light signaling in plants” in Encyclopedia of Biological Chemistry III, Jez J., Ed. (Elsevier, Oxford, ed. 3, 2021), pp. 78–89. [Google Scholar]
- 2.Paik I., Huq E., Plant photoreceptors: Multi-functional sensory proteins and their signaling networks. Semin. Cell Dev. Biol. 92, 114–121 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Legris M., Ince Y. Ç., Fankhauser C., Molecular mechanisms underlying phytochrome-controlled morphogenesis in plants. Nat. Commun. 10, 5219 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng M.-C., Kathare P. K., Paik I., Huq E., Phytochrome signaling networks. Annu. Rev. Plant Biol. 72, 217–244 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clack T., Mathews S., Sharrock R. A., The phytochrome apoprotein family in Arabidopsis is encoded by five genes: The sequences and expression of PHYD and PHYE. Plant Mol. Biol. 25, 413–427 (1994). [DOI] [PubMed] [Google Scholar]
- 6.Van Buskirk E. K., Decker P. V., Chen M., Photobodies in light signaling. Plant Physiol. 158, 52–60 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quail P. H., Photobodies reveal their secret. Nat. Plants 7, 1326–1327 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Chen D., et al. , Integration of light and temperature sensing by liquid-liquid phase separation of phytochrome B. Mol. Cell 82, 3015–3029.e6 (2022). [DOI] [PubMed] [Google Scholar]
- 9.Lee S., Huq E., Phase separation at the heart of “heat” sensing. Mol. Cell 82, 2916–2918 (2022). [DOI] [PubMed] [Google Scholar]
- 10.Kathare P. K., Huq E., Light-regulated pre-mRNA splicing in plants. Curr. Opin. Plant Biol. 63, 102037 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu S.-H., Gene expression regulation in photomorphogenesis from the perspective of the central dogma. Annu. Rev. Plant Biol. 65, 311–333 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Chaudhary S., et al. , Alternative splicing and protein diversity: Plants versus animals. Front Plant Sci 10, 708 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee Y., Rio D. C., Mechanisms and regulation of alternative pre-mRNA splicing. Annu. Rev. Biochem. 84, 291–323 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlaen R. G., et al. , The spliceosome assembly factor GEMIN2 attenuates the effects of temperature on alternative splicing and circadian rhythms. Proc. Natl. Acad. Sci. U.S.A. 112, 9382–9387 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanchez S. E., et al. , A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature 468, 112–116 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Jones M. A., et al. , Mutation of Arabidopsis Spliceosomal Timekeeper Locus1 causes circadian clock defects. Plant Cell 24, 4066–4082 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X., et al. , SKIP is a component of the spliceosome linking alternative splicing and the circadian clock in Arabidopsis. Plant Cell 24, 3278–3295 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petrillo E., et al. , A chloroplast retrograde signal regulates nuclear alternative splicing. Science 344, 427–430 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartmann L., et al. , Alternative splicing substantially diversifies the transcriptome during early photomorphogenesis and correlates with the energy availability in Arabidopsis. Plant Cell 28, 2715–2734 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang R., et al. , A high resolution single molecule sequencing-based Arabidopsis transcriptome using novel methods of Iso-seq analysis. Genome Biol. 23, 149 (2022) . [DOI] [PMC free article] [PubMed]
- 21.Wilkinson M. E., Charenton C., Nagai K., RNA splicing by the spliceosome. Annu. Rev. Biochem. 89, 359–388 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Barta A., Kalyna M., Reddy A. S. N., Implementing a rational and consistent nomenclature for serine/arginine-rich protein splicing factors (SR proteins) in plants. Plant Cell 22, 2926–2929 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu X.-D., Ares M. Jr., Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 15, 689–701 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng Y.-L., Tu S.-L., Alternative splicing and cross-talk with light signaling. Plant Cell Physiol. 59, 1104–1110 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Shikata H., et al. , Phytochrome controls alternative splicing to mediate light responses in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111, 18781–18786 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan T., Heng Y., Wang W., Li J., Deng X. W., SWELLMAP 2, a phyB-interacting splicing factor, negatively regulates seedling photomorphogenesis in Arabidopsis. Front. Plant Sci. 13, 836519 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Godoy Herz M. A., et al. , Light regulates plant alternative splicing through the control of transcriptional elongation. Mol. Cell 73, 1066–1074.e3 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Schwenk P., et al. , Uncovering a novel function of the CCR4-NOT complex in phytochrome A-mediated light signalling in plants. eLife 10, e63697 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shikata H., et al. , The RS domain of Arabidopsis splicing factor RRC1 is required for phytochrome B signal transduction. Plant J. 70, 727–738 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Xin R., et al. , SPF45-related splicing factor for phytochrome signaling promotes photomorphogenesis by regulating pre-mRNA splicing in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 114, E7018–E7027 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xin R., Kathare P. K., Huq E., Coordinated regulation of pre-mRNA splicing by the SFPS-RRC1 complex to promote photomorphogenesis. Plant Cell 31, 2052–2069 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mancini E., Rabinovich A., Iserte J., Yanovsky M., Chernomoretz A., Corrigendum to: ASpli: Integrative analysis of splicing landscapes through RNA-seq assays. Bioinformatics 37, 2609–2616 (2021). [DOI] [PubMed] [Google Scholar]
- 33.Paik I., Yang S., Choi G., Phytochrome regulates translation of mRNA in the cytosol. Proc. Natl. Acad. Sci. U.S.A. 109, 1335–1340 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shih C.-J., et al. , Heterogeneous nuclear ribonucleoprotein H1 coordinates with phytochrome and the U1 snRNP complex to regulate alternative splicing in Physcomitrella patens. Plant Cell 31, 2510–2524 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin B.-Y., Shih C.-J., Hsieh H.-Y., Chen H.-C., Tu S.-L., Phytochrome coordinates with a hnRNP to regulate alternative splicing via an exonic splicing silencer. Plant Physiol. 182, 243–254 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X., et al. , A photoregulatory mechanism of the circadian clock in Arabidopsis. Nat. Plants 7, 1397–1408 (2021). [DOI] [PubMed] [Google Scholar]
- 37.Sugiyama N., et al. , Large-scale phosphorylation mapping reveals the extent of tyrosine phosphorylation in Arabidopsis. Mol. Syst. Biol. 4, 193 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakagami H., et al. , Large-scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol. 153, 1161–1174 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoo C. Y., et al. , Direct photoresponsive inhibition of a p53-like transcription activation domain in PIF3 by Arabidopsis phytochrome B. Nat. Commun. 12, 5614 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park E., Kim Y., Choi G., Phytochrome B., Phytochrome B requires PIF degradation and sequestration to induce light responses across a wide range of light conditions. Plant Cell 30, 1277–1292 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park E., et al. , Phytochrome B inhibits binding of phytochrome-interacting factors to their target promoters. Plant J. 72, 537–546 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.P. K. Kathare et al., RNA-seq data for “SWAP1-SFPS-RRC1 splicing factor complex modulates pre-mRNA splicing to promote photomorphogenesis in Arabidopsis.” GEO. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE214299. Accessed 19 October 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data were deposited into the Gene Expression Omnibus (GEO) database (accession No. GSE214299) (42). Source data are provided with this paper. Arabidopsis mutants and transgenic lines, as well as plasmids and antibodies generated during the current study, are available from the corresponding author upon reasonable request.