Abstract
The non-classical human leukocyte antigen (HLA)-G exerts immune-suppressive properties modulating both NK and T cell responses. While it is physiologically expressed at the maternal–fetal interface and in immune-privileged organs, HLA-G expression is found in tumors and in virus-infected cells. So far, there exists little information about the role of HLA-G and its interplay with immune cells in biopsies, surgical specimen or autopsy tissues of lung, kidney and/or heart muscle from SARS-CoV-2-infected patients compared to control tissues. Heterogeneous, but higher HLA-G protein expression levels were detected in lung alveolar epithelial cells of SARS-CoV-2-infected patients compared to lung epithelial cells from influenza-infected patients, but not in other organs or lung epithelia from non-viral-infected patients, which was not accompanied by high levels of SARS-CoV-2 nucleocapsid antigen and spike protein, but inversely correlated to the HLA-G-specific miRNA expression. High HLA-G expression levels not only in SARS-CoV-2-, but also in influenza-infected lung tissues were associated with a high frequency of tissue-infiltrating immune cells, but low numbers of CD8+ cells and an altered expression of hyperactivation and exhaustion markers in the lung epithelia combined with changes in the spatial distribution of macrophages and T cells. Thus, our data provide evidence for an involvement of HLA-G and HLA-G-specific miRNAs in immune escape and as suitable therapeutic targets for the treatment of SARS-CoV-2 infections.
Keywords: HLA-G, SARS-CoV-2, microRNA, Immune cell infiltration, Immune response
Introduction
The human leukocyte antigen (HLA)-G is a non-classical HLA class I molecule, which was first characterized by its expression at the feto-maternal interface. In contrast to classical HLA class I antigens, HLA-G is less polymorphic and exists in seven different isoforms, from which four are membrane bound (HLA-G1 to -G4) and three are soluble forms (sHLA-G5 to -G7) [1]. In addition, some novel HLA-G isoforms have been recently identified, but their functions have not yet been determined [2, 3]. In addition to alternative splicing, sHLA-G1 can be generated by proteolytic cleavage mainly mediated by the activity of matrix metalloproteinases (MMPs) [4]. Under physiologic conditions, HLA-G has a highly restricted tissue distribution and is mainly expressed on immune-privileged organs, i.e., thymus, cornea, testis, erythroblasts, mesenchymal stem cells, and cytotrophoblasts. In these tissues, the most common HLA-G isoforms are HLA-G1, sHLA-G1 and HLA-G5 [5]. HLA-G exerts immune-suppressive properties on CD8+ cytotoxic T lymphocytes (CTL), natural killer (NK) cells, B cells and dendritic cells by interacting with the appropriate immune cell inhibitor receptors, in particular the Ig-like transcript (ILT)-2, ILT-4, the killer cell immunoglobulin-like receptor (KIR)-2DL4, NKG2A/CD94 and CD160 [5–9]. Next to the interactions with its receptors, the HLA-G-associated tolerance induction could be also mediated by intracellular transfer mechanisms, such as trogocytosis, extracellular vesicles (EV), or tunneling nanotubes [10–12].
The expression of HLA-G is tightly regulated at distinct levels [13]. These include a transcriptional up-regulation of HLA-G by various cytokines, such as interferon (IFN)-ƴ and tumor necrosis factor (TNF)-α, and growth factors, like, e.g., the transforming growth factor (TGF)-β, granulocyte–macrophage colony stimulating factor (GM-CSF), and granulocyte colony stimulating factor (G-CSF) [14–16] as well as microenvironmental factors including indoleamine 2,3-dioxygenase (IDO), hypoxia, metabolites and stress factors, such as heat shock and chemicals, [13, 17]. In addition, HLA-G is altered by epigenetic processes mediated by methylation and histone acetylation [18] or by post-transcriptional control [19]. The latter is due to RNA-binding proteins (RBPs) and/or microRNAs (miRNAs) directed against the 3ʹ untranslated region (UTR) or the coding sequence (CDS) of HLA-G, thereby inhibiting the HLA-G expression [20–24]. In addition, the long non-coding RNA HOX transcript anti-sense RNA (HOTAIR) has been shown to downregulate HLA-G expression [25].
During the last two decades, highly variable levels of HLA-G neo-expression leading not only to escape from immune surveillance, but also to tolerogenic responses of transplants have been described in different neoplasms, autoimmune, and inflammatory diseases as well as upon pathogen infection by parasites, bacteria, and viruses [26–35]. In addition, the pathophysiologic neo-expression of HLA-G surface antigens and of sHLA-G isoforms in tumors or upon viral infections was associated with disease progression, poor clinical outcome, and adverse therapy response of patients [36–46]. Recently, evidence accumulated that HLA-G is an emerging susceptibility and/or protection relevant factor for unresolved virus infection and viral resistance [47]. Thus, targeting of HLA-G, its receptors, or HLA-G-relevant molecules might offer a novel therapeutic strategy for malignant tumors and also viral infections.
The severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is a novel RNA beta coronavirus, which causes the coronavirus disease-2019 (COVID-19) [48]. In March 2020, the World Health Organization (WHO) declared COVID-19 as a pandemic public health disease. This disease is associated with infection of the upper respiratory tract with the risk of sustaining a pneumonia and/or an acute respiratory distress syndrome (ARDS) accompanied by a high patients’ morbidity and mortality rate depending on the virus strain [49]. SARS-CoV-2 enters the host cells through its spike protein by binding to the angiotensin-converting enzyme 2 (ACE2) receptor, which is abundantly expressed on alveolar type II epithelial cells of the respiratory tract [50]. SARS-CoV-2 infection often causes an immune deregulation including sustained cytokine production and hyperinflammation, which in turn is associated with disease severity and induces damages to host tissues [51]. Immune profiling of peripheral blood or bronchioalveolar fluid as well as damaged lung tissues has revealed major changes in the immune system of COVID-19 patients [52, 53]. A unique immunological profile was found in the peripheral blood of COVID-19 patients with an increased number of NK cells, but low T cell numbers and overexpression of T cell immunoglobulin and mucin domain (TIM)-3, programmed cell death ligand 1 (PD-L1) and CD69 in both immune effector cells suggesting a hyperactivated and exhausted immune response upon COVID-1 infection [54]. However, an increased understanding of the immunogenicity in combination with immune responses is urgently needed and will provide further information about the pathophysiologic role of SARS-CoV-2 and the clinical manifestation of severe disease [55, 56]. Regarding a putative link between HLA-G and COVID-19 infection, it has been suggested that in an early inflammatory stage, the host might produce the anti-inflammatory cytokine IL-10, which could later enhance HLA-G expression to avoid injuries [57]. These data were further extended by a positive correlation next to IL-10 with IL-6 and IL-8 in the acute phase of the SARS-CoV-2 infection [58]. Thus, HLA-G expression and/or secretion might reflect a negative feedback response to inflammatory processes during viral infections [29]. Despite HLA-G has been suggested to have immune regulatory functions in SARS-CoV-2-infected patients [59–61], a thorough analysis of HLA-G in COVID-19 patients is still lacking. To elucidate the role of HLA-G during COVID-19, membraneous HLA-G expression in organs of SARS-CoV-2-infected patients and respective controls, its spatial context, and interplay with immune cells were determined and correlated with clinical parameters.
Materials and methods
Patients’ characteristics
Formalin-fixed, paraffin-embedded (FFPE) lung tissue samples from 65 SARS-CoV-2-affected patients were collected in Germany between 2020 and 2021 at the Institutes of Pathology of the University Hospital of the Martin-Luther University Halle-Wittenberg, the University Hospital of the Friedrich-Alexander University Erlangen-Nuremberg and of the Hannover Medical School, Hannover. Tissue samples from lungs of patients who died from influenza (n = 12) or from heart attack (n = 10) were collected in the period from 2009 to 2020 and served as controls. In a subset of SARS-CoV-2-infected patients, FFPE tissues from further organs were available (see Table 1). The FFPE tissues derived from autopsies, or in the case of six patients that survived COVID-19, were obtained by interventional tissue resection during the disease. Clinical and laboratory data were collected from the medical records. SARS-CoV-2 and influenza infections were proven by real-time polymerase chain reaction (RT-PCR) during the life-time of the patients. All samples analyzed are summarized in Table 1.
Table 1.
Clinico-pathological characteristics and sample specifications
| Category | SARS-CoV-2 | Influenza | Control |
|---|---|---|---|
| Number of patients | 65 | 10 | 12 |
| Clinical data | |||
| Age min–max (mean) | 36–96 (71) | 34–84 (64) | 54–83 (69) |
| Gender | |||
| Male | 43 | 6 | 7 |
| Female | 22 | 4 | 5 |
| Survival time | |||
| < 7 days | 17 | ||
| > 7 days | 42 | ||
| Survived | 6 | ||
| Tissues analyzed for HLA-G expression | |||
| Lung | 65 | 10 | 12 |
| Brain | 10 | – | – |
| Heart | 12 | – | – |
| Kidney | 21 | – | – |
| Pancreas | 12 | – | – |
| Spleen | 6 | – | – |
| Liver | 6 | – | – |
| Samples analyzed for miRNA expression | 20 | 5 | 5 |
| Samples analyzed by MSI | 26 | 8 | 5 |
| Histological pattern of acute interstitial pneumonia | |||
| Exudative pattern | 12 | – | – |
| Organizing pattern | 52 | – | – |
| Fibrosis | 1 | – | – |
Cohorts were stratified based on COVID-19 death data from the Robert Koch Institute as follows: 1st wave comprises calendar weeks 10–31 in 2020. Calendar weeks 32 in 2020 to 11 in 2021 were selected for the 2nd wave [8]. The 3rd wave was defined as beginning of calendar weeks 12–32 in 2021, and the 4th wave as calendar weeks 33–53 in 2021.
Ethical approval
Autopsies were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. All autopsies were conducted after consent for autopsy was obtained from the deceased or next of kin, or autopsies were requested by the health authorities or by the prosecutor’s office. Each participating center had local ethical approval. Clinical parameters were recovered from the final biopsy/autopsy report or the laboratory information system. Only cases with a positive SARS-CoV-2 test (usually antigen tests from either nasopharyngeal or PCR from either nasopharyngeal swab or tissue) either preclinical, clinical, or post-mortem, were implemented in the analyses. Regarding the cause of death, data were taken from the final autopsy report.
The use of collected FFPE tissue samples and serum/plasma samples from patients who survived COVID-19 was approved by the Ethical Committees of the Medical Faculty of the Martin Luther University Halle-Wittenberg, Halle (2017-81), of the MHH Hannover (9022-BO.K-22c), and of the Medical Faculty of the University Hospital in Erlangen.
Standard morphological evaluation
Hematoxylin and eosin (HE)-stained tissue slides were used to evaluate the specific pattern of damage following SARS-CoV-2 infection with respect to the following histological patterns of acute interstitial pneumonia: acute exudative, organized, and fibrotic pattern [62, 63]. Furthermore, tissue-infiltrating lymphocytes in the alveolar walls were quantified in accordance to the evaluation of stromal tissue-infiltrating lymphocytes (TILs) using the guidelines of the International Working Group for tumor-infiltrating lymphocytes [64].
Immunohistomorphological evaluation
Conventional immunohistochemistry (IHC) was performed on 3 µm thick, consecutive sections of FFPE samples with the Bond Polymer refine detection Kit (Leica, DS9800) according to the manufacturer’s instructions on a fully automated IHC stainer (Leica Bond). For IHC staining, the anti-HLA-G monoclonal antibody (mAb) (Abcam, UK, clone 4H84) was used as recently described [22]. In addition, mAbs directed against CD3 (Labvision, Germany, clone SP7), CD20 (DAKO, California, USA, clone L26), CD56 (Cell marque, Massachusetts, USA, clone MRQ-42) and the SARS-CoV nucleocapsid protein (Rockland Inc., Pennsylvania, USA, clone 200-401-A50) and the spike protein (Abcam, UK, clone ab272504) were employed according to the supplier’s instructions. Sections were examined and imaged with a Zeiss Axiophot microscope (Zeiss, Jena, Germany). Two pathologists (MB and CW), independently and blinded to the clinical data, scored all samples. HLA-G expression was analyzed using a Histoscore as previously described [65]. The relative amount of positively stained cells (%) was multiplied by their intensity from 0 (negative), 1 (weak), 2 (moderate) to 3 (intense) leading to the expression intensity (or H-score) that was further classified as absent (0), low (1–100), intermediate (101–200), or strong (201–300) overall expression.
Multispectral imaging
Multispectral imaging (MSI) was performed using the basic protocol described in Wickenhauser et al. [66]. Two multiplex panels with different mAbs and opal-dye combinations were used. The first panel included anti-PD-L1 clone E1L3N (Cell Signaling E1L3N, 1:150) in combination with Opal690, anti-Foxp3 clone 236A/E/ (Abcam, 1:00) with Opal540, anti-CD3 clone SP7 (ThermoFisher SP7, 1:100) with Opal570, anti-CD163 clone MRQ-26 (Cell Marque, 1:50) with Opal620 and anti-panCK Ab AE1/AE3 (Dako, 1:150) with Opal520. The second panel comprised anti-TIM-3 (Abcam, ab241332, 1:1000) with Opal 520, anti-PD-1 (Biocare Medical, NAT105, 1:50) with Opal 540, anti-CD8 (DAKO, C8/144b, 1:50) with Opal 570, anti-TIGIT (Biozol, USC-PAN056HU01-1, 1:50) with Opal 620, anti-CD69 (Abcam, ab233396, 1:50) with Opal 650 and anti-HLA-G (Abcam, clone 4H84, 1:100) with Opal 690. After counterstaining with DAPI (Akoya Biosciences, Marlborough, MA), the sections were mounted and scanned with the Vectra Polaris System (Akoya Biosciences, Marlborough, MA) and a mean of 18 regions of interest (ROIs) per slide were taken with a 20 × zoom. The inForm software (Version 2.4.10, Akoya Biosciences) was employed to perform cell segmentation and phenotyping. PhenoptrReports scripts were used within R to evaluate the frequency and density of the different cell types as well as their interspatial relationships.
RNA extraction, cDNA synthesis, and qPCR analyses
Two 5 µm thick FFPE tissue slides of lung tissues were subjected to the extraction of total RNA using the MasterPure™ Complete DNA & RNA Purification Kit according to the manufacturer’s protocol (Lucigen, Middleton, WI, USA).
For quantification of the selected HLA-G regulatory miRNAs, template-specific cDNA syntheses were performed with the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, USA) and miR specific stem-loop primers [67] as published by Jasinski-Bergner et al. [22]. Subsequently, the qPCR analyses were performed by calculation of relative copy numbers. The HLA-G non-relevant and highly abundantly expressed miR-3960 served as a house keeping gene. For the qPCR reactions GoTaq® qPCR Master Mix (Promega, Madison, WI, USA) was employed. The sequences of the stem-loop primers as well as of the respective qPCR primers and their amplification settings are listed in Table 2.
Table 2.
Summary of the primer sequences and conditions
| Primer | Sequence (5ʹ⟶3ʹ) | Application | Condition (°C) |
|---|---|---|---|
| miR-148A SL Rkt | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACAAAG | cDNA synthesis | 42 |
| miR-148A qPCR | GCCCTCAGTGCACTACAGA | qPCR | 60 |
| miR-148B SL Rkt | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACAAAG | cDNA synthesis | 42 |
| miR-148B qPCR | GCCCTCAGTGCATCACAGGA | qPCR | 60 |
| miR-152 SL Rkt | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCCAAGT | cDNA synthesis | 42 |
| miR-152 qPCR | GCCCTCAGTGCATGACAGA | qPCR | 60 |
| miR-3960 SL Rkt | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCCCCCG | cDNA synthesis | 42 |
| miR-3960 qPCR | GCCCGGCGGCGGCGGAGGC | qPCR | 60 |
| miR-548q SL Rkt | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCCGCCA | cDNA synthesis | 42 |
| miR-548qfw qPCR | GCCCGCTGGTGCAAAAGTAA | qPCR | 60 |
| miR-628-5p SL Rkt | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCCTCTA | cDNA synthesis | 42 |
| miR-628-5p qPCR | GCCCATGCTGACATATTTAC | qPCR | 60 |
| miR-744-5p SL Rkt | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTGCTGT | cDNA synthesis | 42 |
| miR-744-5p qPCR | GCCCTGCGGGGCTAGGGCTA | qPCR | 60 |
| miR qPCR rev | GTGCAGGGTCCGAGGT | qPCR | 60 |
Statistics
Statistical analyses were performed employing IBM SPSS statistic packages (version 25) or GraphPad Prism9. Kolmogorov–Smirnov test revealed non-parametric data (p < 0.05). The Mann–Whitney U test was used to compare clinical data, frequencies of immune cell subpopulations, and immunohistochemical expression pattern. For correlation analysis Pearson’s correlation was performed. p values < 0.05 were considered statistically significant. All graphs were created using GraphPad Prism 9.
Results
Determination of HLA-G expression in lung tissues from SARS-CoV-2- and influenza-infected patients and histomorphologic normal control lung tissues
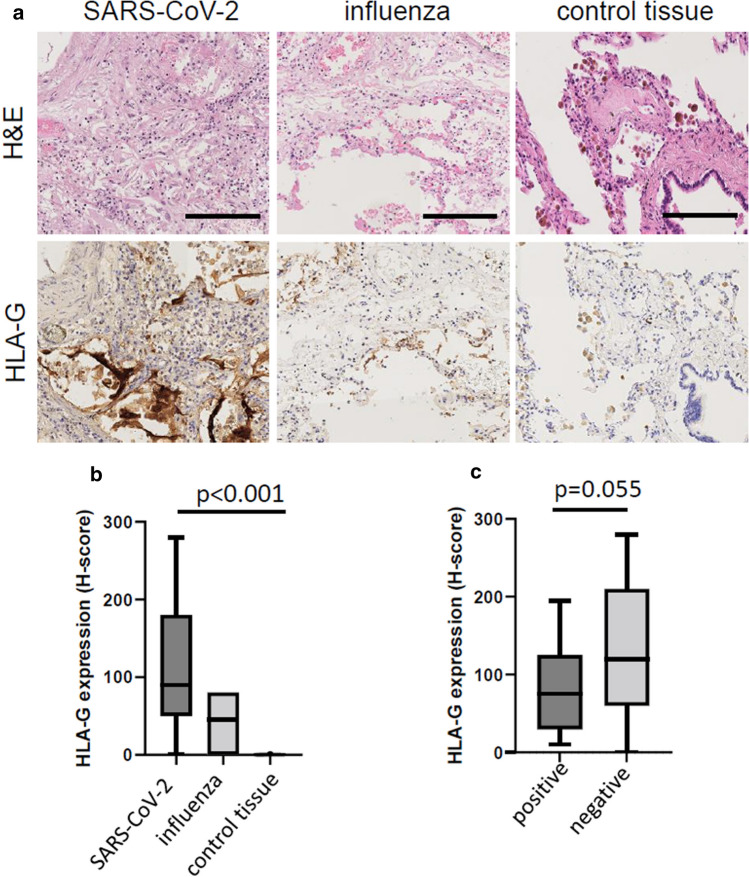
To determine whether SARS-CoV-2 infection induces HLA-G expression, lung tissues from SARS-CoV-2- (n = 65) or influenza- (n = 10) infected patients as well as from respective controls (n = 12) were stained by IHC with the HLA-G-specific mAb 4H83 detecting all major HLA-G isoforms. Representative images of the tissue stainings are shown in Fig. 1a demonstrating a highly heterogeneous HLA-G expression pattern in the lung tissues analyzed with a predominant positivity in pneumocytes and weaker expression in immune cells and some cases of bronchial respiratory epithelia. A comparable staining pattern, but statistically significant higher HLA-G expression levels were found in SARS-CoV-2- compared to lung tissues from influenza-infected patients (Fig. 1b). In contrast, pneumocytes of the lung control samples lacked HLA-G expression, while a few HLA-G-positive immune cells were detected (Fig. 1b). To evaluate whether this HLA-G expression was organ specific or also relevant in other organs receptive to SARS-CoV-2-induced damage, HLA-G expression was determined in representative samples of kidney (n = 21) and heart tissues (n = 12) obtained from SARS-CoV-2-infected patients. All these tissue samples lacked HLA-G expression (data not shown) suggesting a tissue-specific HLA-G neo-expression upon course of COVID-19.
Fig. 1.
Evaluation of HLA-G expression in lung tissues from SARS-CoV-2- and influenza-infected patients and controls. HLA-G expression in SARS-CoV-2- or influenza-infected lungs as well as in lungs from control patients was determined by IHC as described in “Material and methods”. a Representative micrographs of HLA-G expression in lung epithelia from SARS-CoV-2- or influenza-infected patients as well as controls are shown. Magnification 1:40; bars indicate a scale of 50 µm. H&E hematoxylin and eosin staining; b, c HLA-G expression is shown using the H-score for the different patients (SARS-CoV-2, n = 65; influenza, n = 10; control patients, n = 12) or for the SARS-CoV-2-infected individuals divided regarding the presence or absence of detectable nucleocapsid antigen by IHC (positive, n = 13; negative, n = 52)
Correlation of HLA-G expression with SARS-CoV-2 nucleocapsid antigen and spike protein expression
Next to HLA-G, the SARS-CoV-2 nucleocapsid and spike protein antigen expression was determined in the lung tissue samples analyzed using IHC. In the group of SARS-CoV-2-infected patients, the nucleocapsid expression was detectable in 23/65 (35.4%), the spike protein in 31/65 (47.7%) of cases. All samples expressing the nucleocapsid were also positive for the spike protein. The expression of the respective markers was detectable in pneumocytes, respiratory epithelia, and lung infiltrating immune cells, but neither in other tissue samples from SARS-CoV-2-infected patients nor in lung epithelium samples of influenza-infected or control patients. All SARS-CoV-2 nucleocapsid antigen-positive lung samples expressed HLA-G, while the 42 nucleocapsid antigen-negative tissues from patients with proven SARS-CoV-2 infection presented a trend towards higher HLA-G expression levels, although statistically non-significant (Fig. 1c).
Inverse correlation of HLA-G expression with HLA-G-specific miRNAs
Building on the miRNA-mediated posttranscriptional regulation of HLA-G expression [20–23, 68], the expression of HLA-G-specific miRNAs known to inhibit HLA-G expression by either binding to the 3’ UTR or CDS was analyzed by qPCR in lung tissues across the samples and correlated with the HLA-G staining intensity (H-score) obtained by IHC (Fig. 1b). The HLA-G protein levels of FFPE specimen were categorized into a HLA-G negative/weak (n = 8; HLA-G negative with H-score = 0; HLA-G weak H-score < 50) and a HLA-G medium/high (n = 17; HLA-G medium H-score > 50 and < 100; HLA-G high H-score > 100) group. A statistically significant, inverse expression of HLA-G-regulating miRNAs and HLA-G protein was detected for miR-744-5p (p = 0.0063) and for miR-152 (p = 0.036), which bind to the CDS and the 3ʹ UTR of the HLA-G mRNA, respectively, whereas the expression of miR-548q (p = 0.0982) and miR-148B (p = 0.2302) did not reach statistical significance (Fig. 2). The miRNAs miR-628 and miR-148A completely lacked an inverse expression to the HLA-G protein (Fig. 2). It is noteworthy that miR-744-5p and miR-152 classified as key regulators of HLA-G in previous studies [23] displayed the highest abundancies, strengthening their importance in the post-transcriptional regulation of HLA-G in tissues of SARS-CoV-2-diseased individuals.
Fig. 2.
Correlation of HLA-G miRNA expression with HLA-G intensity in lung sections of SARS-CoV-2 patients. Expression of the relative copy numbers of HLA-G-regulating miRNAs was determined by qPCR on the lung sections of SARS-CoV-2 patients. The results are shown as Box-Whiskers Plots upon subdivision of the patients into HLA-G negative/weak (n = 8) and HLA-G medium/high (n = 17) based on the H-score obtained by IHC staining (HLA-G negative = 0; HLA-G weak < 50; HLA-G medium > 50 and < 100; HLA-G high > 100). The statistical significance (p value) was determined by calculation of the two sided student’s t test
Effect of SARS-CoV-2-infection on immune cell infiltration in lung tissues and correlation with HLA-G expression levels
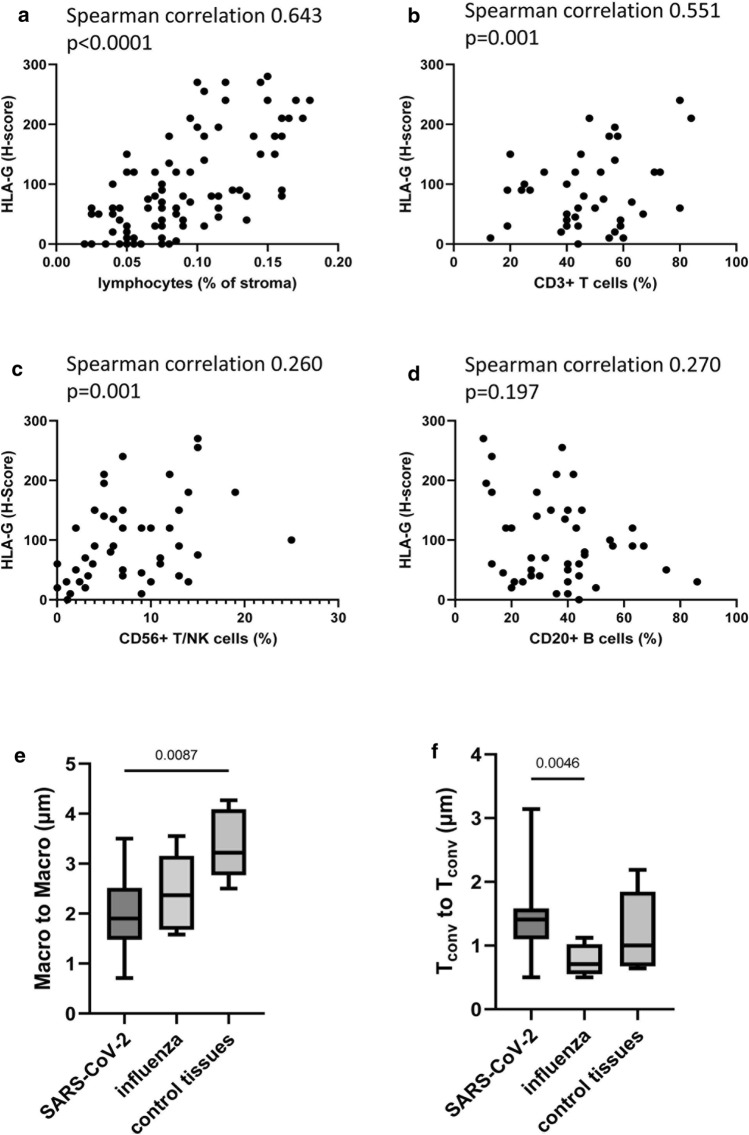
To investigate whether there exists a link between HLA-G expression levels and the cellular environment as well as its spatial organization, the immune cell infiltration of tissues across all samples was assessed by quantification of lymphocyte infiltration using HE staining, conventional IHC and MSI (Fig. 3). An increased immune cell infiltration in the lungs of SARS-CoV-2- and to a lesser extent of influenza-infected patients compared to healthy controls was observed (data not shown). Lymphocyte quantification in tissues from SARS-CoV-2-infected patients demonstrated a statistically significant positive correlation of lung-specific HLA-G expression levels and the immune cell density (Fig. 3a). Furthermore, in these patients, pulmonary HLA-G expression was positively correlated with the frequency of CD3+ T cells and CD56+ T/NK cells analyzed by conventional IHC (Fig. 3b, c). In contrast, there was no correlation between HLA-G expression and the frequency of CD20+ B cells (Fig. 3d). Interestingly, lung tissues with detectable SARS-CoV-2 nucleocapsid antigen showed a slightly lower lymphocyte infiltration (data not shown).
Fig. 3.
Altered composition of the immune cell infiltrate in lung tissues of SARS-CoV-2-infected patients. Lung tissues from the SARS-CoV-2 patients (n = 65) were evaluated for lymphocyte infiltration by HE staining (a) or by conventional IHC using mAbs directed against CD3 for total T cells (b), CD56 for NK or NKT cells (c) and CD20 for B cells (d). The resulting cell frequencies are shown in correlation to the H-score of HLA-G. In addition, the Spearman correlation coefficients and p values are given. Lung tissues from SARS-COV-2-diseased patients were stained by MSI and evaluated as described in "Material and Methods". The minimum distance (in μm) between macrophages and macrophages (e) as well as between conventional T cells (Tconv, i.e. CD3+ Foxp3neg) to Tconv cells (f) is shown as Box-Whiskers plot. The lines represent the median values of the groups
The analysis of the frequency and composition of the immune cell repertoire was extended by a more detailed examination of the spatial distribution between the different immune cells (Fig. 3e, f). The spotwise evaluation of MSI demonstrated no significant differences in the number of infiltrating immune cells, such as CD163+ macrophages or total CD3+ T cells across the samples (data not shown). However, evaluation of the relative distance among the different immune cells in the lung epithelia highlighted that in the SARS-CoV-2 patient group the macrophages were in closer proximity to each other, while the conventional T cells determined as CD3+ Foxp3neg cells were more distant from each other than in the tissues of controls or influenza-infected patients, respectively (Fig. 3e, f).
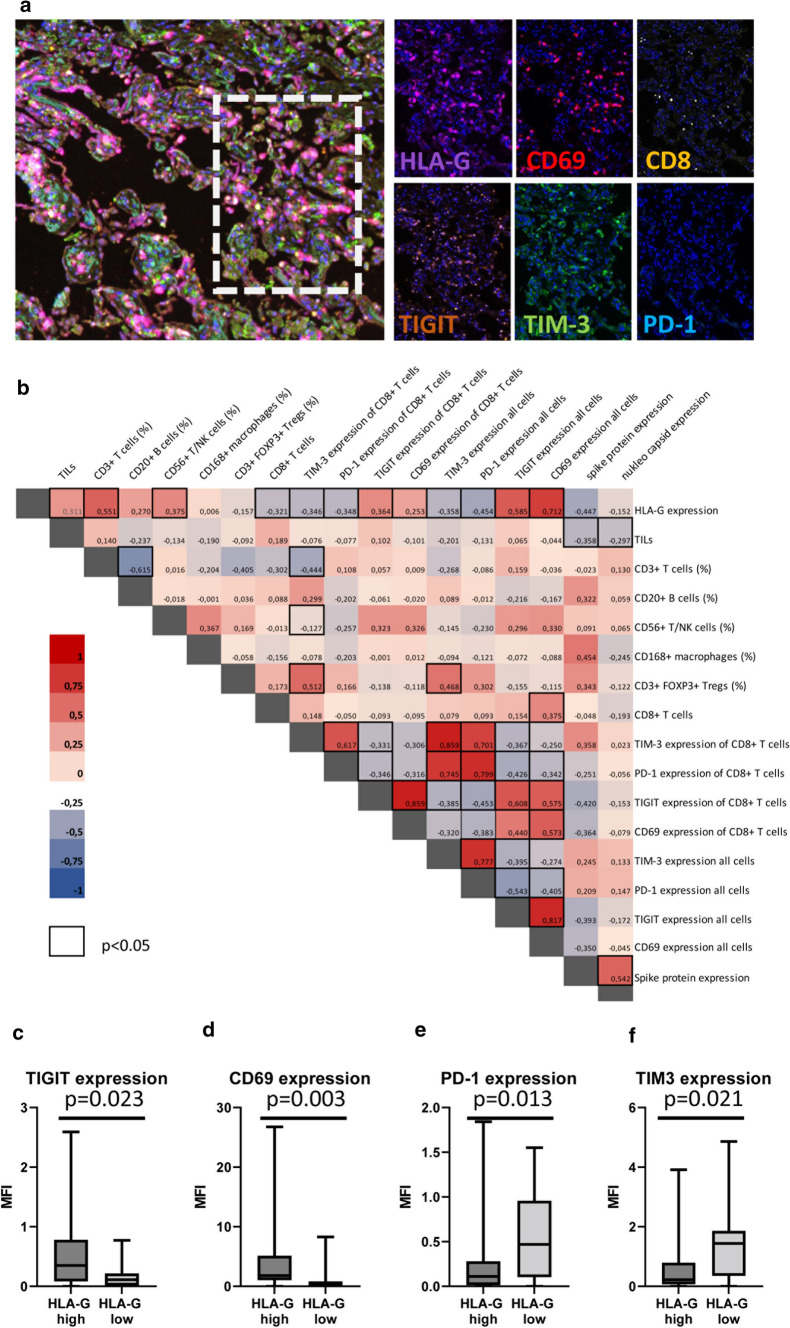
Since viral infections have been shown to influence T cell function by inducing hyperactivation and anergy [69], the frequency of CD8+ T cells as well as the expression of the T cell immunoreceptor with Ig and ITIM domains (TIGIT), TIM-3, programmed cell death protein 1 (PD-1), and CD69 was evaluated as surrogate markers by MSI. Data were correlated with the SARS-CoV-2-induced HLA-G expression as representatively shown in Fig. 4a. Higher HLA-G expression levels correlated with lower CD8+ T cell frequencies (Fig. 4b). These CD8+ T cells showed a significantly increased expression of CD69 and TIGIT (Fig. 4c, d), while the expression of TIM-3 and PD-1 (Fig. 4e, f) was significantly lower in high HLA-G-expressing cases. However, even though the activation marker CD69 was significantly higher expressed in these T cells, CD69+ T cells were more distant to high HLA-G-expressing lung epithelial cells with a minimal distance of 1.58 µm versus 3.08 µm, and an average distance of 4.73 µm versus 118.23 µm.
Fig. 4.
Correlation of the frequency and function of CD8+ T cells to HLA-G expression. MSI staining was performed as described in Materials and Methods and staining patterns of exhaustion/hyperactivation markers were correlated to HLA-G. a Representative multiplex staining of lung tissues from a SARS-CoV-2 patient with a multiplex panel consisting of mAbs directed against HLA-G (violet), CD69 (red), CD8 (yellow), TIGIT (orange), TIM-3 (green), and PD-1 (turquoise). b Pearson correlation map with association to functional markers: Pearson correlation coefficients are represented by different colors defined in the scale bar on the right side of the correlation map. Significant associations are highlighted by a black frame. Correlation of TIGIT (c), CD69 (d), PD-1 (e) and TIM-3 (f) expression to HLA-Ghigh and HLA-Glow expression levels
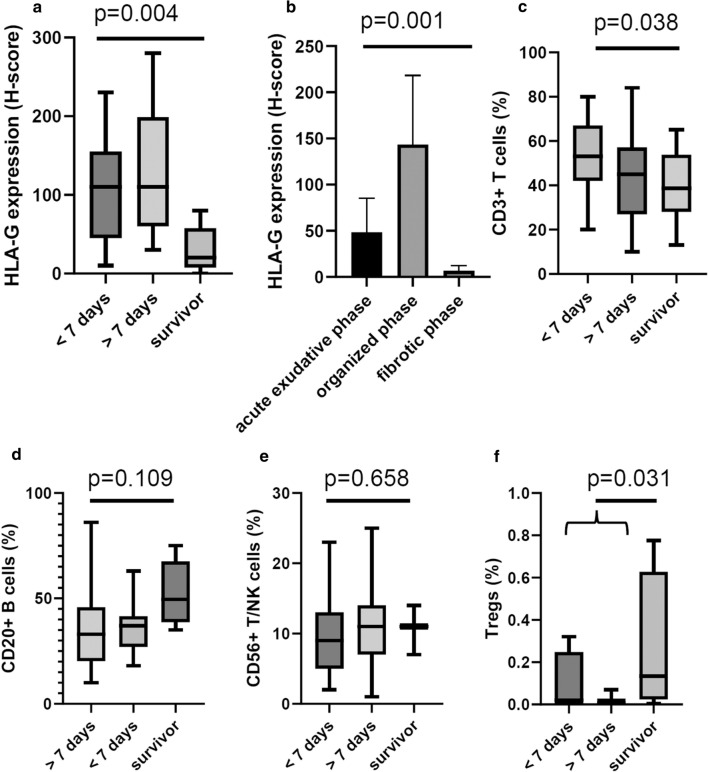
Clinical relevance of HLA-G expression, frequency, and composition of the pulmonary immune cell infiltrate in SARS-CoV-2-infected patients
To determine whether HLA-G expression levels might have a prognostic impact in SARS-CoV-2-infected lungs, the level of HLA-G expression was correlated to the severity of disease. Therefore, the patients with SARS-CoV-2 infection were divided into those who survived and those with early and late disease depending whether death occurred before or after 7 days from the start of respiratory symptoms. In SARS-CoV-2-infected patients HLA-G and nucleocapsid expression levels were associated with the patients’ outcome as shown in Fig. 5. In detail, lung resection tissues of the six SARS-CoV-2 survivors exhibited significantly lower HLA-G levels than lung tissues from deceased patients (Fig. 5a). Among the deceased individuals the HLA-G expression was lower in patients, who survived longer than 7 days compared to patients with a survival of less than 7 days after the first symptoms (Fig. 5a). Interestingly, most patients with detectable SARS-CoV-2 nucleocapsid antigen expression in lung epithelia died early (< 7 days after first symptoms, data not shown). It is noteworthy that lung samples with an early acute interstitial pneumonia pattern (exudative phase) showed lower HLA-G expression levels compared to samples with later inflammatory stages (organized and fibrotic phases) (Fig. 5b).
Fig. 5.
Clinical relevance of HLA-G expression and immune infiltration. HLA-G expression was correlated with the survival of SARS-CoV-2-infected patients (a) as well as to their histological pattern of acute interstitial pneumonia (b), whereas the frequencies of the different immune cell subpopulations were correlated only with survival (c–f), evaluated as the time from the first symptoms to death (< 7 days and > 7 days) or survival
In addition, the correlation of the frequency and localization of lymphocytes to the survival of SARS-CoV-2-infected patients demonstrated a significantly lower frequency of CD3+ T cells in the stroma of lung tissues of survivors (Fig. 5c). In addition, despite not reaching statistical significance, survivors exhibited higher levels of CD20+ and CD56+ cells (Fig. 5d, e). Moreover, a significantly enhanced presence of regulatory T cells (Tregs) determined as CD3+ Foxp3+ cells could be highlighted in the SARS-CoV-2 survivors versus deceased patients (Fig. 5f) although their frequency was low.
Discussion
Recently, HLA-G has been shown to be upregulated by various viruses, such as HCV, HCMV, HPV and HIV [32, 70, 71], which was extended in this report to SARS-CoV-2 infection. We show here for the first time that patients infected with the SARS-CoV-2 and pulmonary disease had a high frequency of membranous HLA-G expression in the lung tissues, predominantly the alveocytes, but not in other tissues suggesting an organ-specific HLA-G neo-expression. Despite sharing a viral pathogenic origin and a similar damage pattern within the lung, the level of HLA-G expression in lung epithelia of COVID-19 patients was higher than that of influenza-diseased patients and might reflect an altered immune response associated with an increased fibrosis score in the lungs of COVID-19 patients [72, 73]. In contrast, lung epithelia of non-virus-infected controls lacked HLA-G expression.
There exist a number of publications on the immune cell repertoire and/or its spatial distribution in COVID-19 damaged lungs ranging from macroscopic to single-cell level [74], but a correlation between HLA-G expression, the immune cell composition, and the clinical course has not yet been described. In agreement with published data using targeted spatial transcriptomics, changes in the cell type composition and interactions between macrophages and T cells using IHC and MSI were found in the diseased lungs. This study showed for the first time that the increased immune cell infiltration was associated with high HLA-G-expression levels, which might be due to cytokines secreted by T cells and macrophages, like IL-10, TGF-β and/or IFN-γ, present in high numbers of the lung tissue microenvironment [75, 76]. Furthermore, higher HLA-G expression levels were associated with decreased frequencies of CD69+ CD8+ T cells. CD8+ T cells expressing the activation marker CD69 were more distant located to HLA-Ghigh cells compared to cases with low or no HLA-G expression. In this context, it is noteworthy that HLA-G inhibits T cell activity by binding to the ILT2 and ILT4 receptors, and therefore activated CD8+ T cells in the proximity of HLA-G expressing cells are not functional [77, 78]. The expression of HLA-G and its receptors (ILT-2, ILT-4, KIR-2DL-4) could be also found in peripheral immune cells, like T and B cells as well as monocytes upon SARS-CoV-2 infection [59]. CD8+ T cells have been shown to overexpress CD69 and TIM-3 in the peripheral blood of COVID-19-infected patients compared to healthy controls, which is a characteristic for a hyperactivated/exhausted T cell phenotype. This observation has also clinical relevance, since the hyperactivated T cell status was accompanied by a reduced survival of COVID-19-infected patients [54]. Based on the analysis of PBMC in COVID-19-infected patients, CD8+ T cells exhibit an exhausted phenotype characterized by the surface expression of TIGIT, TIM-3 and/or PD-1 [79, 80]. In our study, TIGIT expression was significantly increased in HLA-G-positive cases, while PD-1 and TIM-3 exhibited a significantly higher surface expression on CD8+ T cells, which reflect an exhausted CD8+ T cell phenotype and mainly confirmed the published results of PBMC analysis of COVID-19-infected individuals. Dysfunction or T cell exhaustion result in deficient T cell responses in COVID-19-infected individuals. This might be driven by a type I IFN-induced transcriptional network regulating the expression of co-inhibitory molecules [81].
In addition, a negative correlation between the expression of HLA-G, the nucleocapsid, and the spike protein was detected, which point to an altered expression of these molecules during the disease and indicate an up-regulation of viral proteins in the early phase of COVID-19. The negative correlation between HLA-G expression and COVID-19 course underlines that HLA-G might be a potential therapeutic target in this disease. This hypothesis is further supported by an association between the HLA-G variant rs9380142 and the susceptibility to SARS-CoV-2 infection [82].
The molecular mechanisms regulating HLA-G expression are highly complex and comprise single nucleotide polymorphisms (SNPs) in the CDS and 3ʹ UTR of the HLA-G gene, epigenetic, transcriptional as well as posttranscriptional regulation. In multiple tumor entities, the expression of HLA-G-regulating miRNAs has been shown to be inversely correlated with HLA-G [21, 23]. In this study, we observed that lung biopsies from COVID-19-diseased patients displaying increased HLA-G protein expression exhibit lower miR-744-5p and miR-152 expression levels suggesting a deregulated HLA-G-specific miRNA expression due to COVID-19 infection. However, further evaluations are required to identify the processes responsible for directly or indirectly leading to a down-regulation of these miRNAs as well as to understand why the other known HLA-G-regulating miRNAs are not inversely expressed.
Other mechanisms resulting in HLA-G neo-expression are related to the composition of the tissue microenvironment, in particular of immune-suppressive inflammatory cytokines, such as IL-10, TGF-β, and IFN-γ, which are able to upregulate and/or enhance HLA-G expression [83]. Therefore, analysis of cytokine secretion by the different immune cell subpopulations in the SARS-CoV-2-infected lungs might help to evaluate their role in HLA-G neo-expression and in the outcome of SARS-CoV-2-infected patients.
Conclusions
We here propose that HLA-G is a major player in the altered immunogenicity of SARS-CoV-2-infected lung epithelia. Thus, HLA-G might serve as prognostic marker and might also pave the way to develop HLA-G as therapeutic target for the treatment of COVID-19 infection by restoring the exhausted immune responses induced by HLA-G.
Acknowledgements
We would like to thank Maria Heise and Nicole Ott for excellent secretarial help.
Abbreviations
- Ab
Antibody
- ACE2
Angiotensin-converting enzyme 2
- ARDS
Acute respiratory distress syndrome
- CDS
Coding sequence
- CoV-2
Coronavirus-2
- COVID-19
Coronavirus disease-2019
- CTL
Cytotoxic T lymphocytes
- DFS
Disease-free survival
- ELISA
Enzyme-linked immunosorbent assay
- FFPE
Formalin-fixed, paraffin-embedded
- G-CSF
Granulocyte colony stimulating factor
- GM-CSF
Granulocyte–macrophage colony stimulating factor
- HLA
Human leukocyte antigen
- HLA-G
Human leukocyte antigen-G
- HE
Hematoxylin and eosin
- HOTAIR
HOX transcript anti-sense RNA
- IDO
Indoleamine 2,3-dioxygenase
- IFN
Interferon
- IHC
Immunohistochemistry
- IL
Interleukin
- mAb
Monoclonal antibody
- miRNA
microRNA
- MMP
Matrix metalloproteinase
- MSI
Multispectral imaging
- NK
Natural killer
- OS
Overall survival
- PD-1
Programmed cell death protein 1
- PD-L1
Programmed death ligand 1
- RBP
RNA-binding protein
- ROI
Region of interest
- RT-PCR
Real-time polymerase chain reaction
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus-2
- sHLA-G1
Soluble HLA-G1
- TGF
Transforming growth factor
- TIGIT
T cell immunoreceptor with Ig and ITIM domains
- TIL
Tissue-infiltrating lymphocyte
- TIM-3
T cell immunoglobulin and mucin domain 3
- TNF
Tumor necrosis factor
- UTR
Untranslated region
- WHO
World Health Organization
Author contributions
BS designed and guided the project and concept and is responsible for the study. Material preparation, data collection including clinical parameters and analysis were performed by SJB, CM, AM, KB, BY, MB, PE, AH, DJ, CW and MB. The first draft of the manuscript was written by BS, MB, CM and SJB prepared the figure. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was supported by the National University Medicine (NUM) network (Grant no. 01KX2121, BS and CW), the Deutsche Forschungsgemeinschaft (DFG JA 3192/1-1 and 496182670, both BS), DAAD GLACIER (BS), Land Sachsen-Anhalt (BS) and the Jackstädt Foundation (SJB).
Data availability
The datasets generated during and/or analyzed during the current study are not publicly available, since no repository exists, but are available from the corresponding authors.
Declarations
Conflict of interest
The authors have declared that they have no relevant financial or non-financial interests to disclose.
Ethics approval
The study was performed with the principles of the declaration of Helsinki. Approval was granted by the Ethics Committee of the Medical Faculty of the Martin Luther University of Halle-Wittenberg, Halle, Germany.
Footnotes
These authors contributed to a comparable extent to the manuscript.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rouas-Freiss N, Khalil-Daher I, Riteau B, Menier C, Paul P, Dausset J, Carosella ED. The immunotolerance role of HLA-G. Semin Cancer Biol. 1999;9(1):3–12. doi: 10.1006/scbi.1998.0103. [DOI] [PubMed] [Google Scholar]
- 2.Tronik-Le Roux D, Renard J, Verine J, Renault V, Tubacher E, LeMaoult J, Rouas-Freiss N, Deleuze JF, Desgrandschamps F, Carosella ED. Novel landscape of HLA-G isoforms expressed in clear cell renal cell carcinoma patients. Mol Oncol. 2017;11(11):1561–1578. doi: 10.1002/1878-0261.12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin A, Zhang X, Zhang RL, Zhang JG, Zhou WJ, Yan WH. Clinical significance of potential unidentified HLA-G isoforms without alpha1 domain but containing intron 4 in colorectal cancer patients. Front Oncol. 2018;8:361. doi: 10.3389/fonc.2018.00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rizzo R, Trentini A, Bortolotti D, Manfrinato MC, Rotola A, Castellazzi M, Melchiorri L, Di Luca D, Dallocchio F, Fainardi E, Bellini T. Matrix metalloproteinase-2 (MMP-2) generates soluble HLA-G1 by cell surface proteolytic shedding. Mol Cell Biochem. 2013;381(1–2):243–255. doi: 10.1007/s11010-013-1708-5. [DOI] [PubMed] [Google Scholar]
- 5.Liu L, Wang L, Zhao L, He C, Wang G. The role of HLA-G in tumor escape: manipulating the phenotype and function of immune cells. Front Oncol. 2020;10:597468. doi: 10.3389/fonc.2020.597468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ho GT, Celik AA, Huyton T, Hiemisch W, Blasczyk R, Simper GS, Bade-Doeding C. NKG2A/CD94 is a new immune receptor for HLA-G and distinguishes amino acid differences in the HLA-G heavy chain. Int J Mol Sci. 2020 doi: 10.3390/ijms21124362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carosella ED, Rouas-Freiss N, Tronik-Le Roux D, Moreau P, LeMaoult J. HLA-G: an immune checkpoint molecule. Adv Immunol. 2015;127:33–144. doi: 10.1016/bs.ai.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Amodio G, de Albuquerque RS, Gregori S. New insights into HLA-G mediated tolerance. Tissue Antigens. 2014;84(3):255–263. doi: 10.1111/tan.12427. [DOI] [PubMed] [Google Scholar]
- 9.Xu X, Zhou Y, Wei H. Roles of HLA-G in the maternal-fetal immune microenvironment. Front Immunol. 2020;11:592010. doi: 10.3389/fimmu.2020.592010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin A, Yan WH. Intercellular transfer of HLA-G: its potential in cancer immunology. Clin Transl Immunol. 2019;8(9):e1077. doi: 10.1002/cti2.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown R, Kabani K, Favaloro J, Yang S, Ho PJ, Gibson J, Fromm P, Suen H, Woodland N, Nassif N, Hart D, Joshua D. CD86+ or HLA-G+ can be transferred via trogocytosis from myeloma cells to T cells and are associated with poor prognosis. Blood. 2012;120(10):2055–2063. doi: 10.1182/blood-2012-03-416792. [DOI] [PubMed] [Google Scholar]
- 12.Rebmann V, Konig L, Nardi Fda S, Wagner B, Manvailer LF, Horn PA. The potential of HLA-G-bearing extracellular vesicles as a future element in HLA-G immune biology. Front Immunol. 2016;7:173. doi: 10.3389/fimmu.2016.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castelli EC, Veiga-Castelli LC, Yaghi L, Moreau P, Donadi EA. Transcriptional and posttranscriptional regulations of the HLA-G gene. J Immunol Res. 2014;2014:734068. doi: 10.1155/2014/734068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jorgensen N, Sayed A, Jeppesen HB, Persson G, Weisdorf I, Funck T, Hviid TVF. Characterization of HLA-G regulation and HLA expression in breast cancer and malignant melanoma cell lines upon IFN-gamma stimulation and inhibition of DNA methylation. Int J Mol Sci. 2020 doi: 10.3390/ijms21124307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bukur J, Rebmann V, Grosse-Wilde H, Luboldt H, Ruebben H, Drexler I, Sutter G, Huber C, Seliger B. Functional role of human leukocyte antigen-G up-regulation in renal cell carcinoma. Cancer Res. 2003;63(14):4107–4111. [PubMed] [Google Scholar]
- 16.Moreau P, Flajollet S, Carosella ED. Non-classical transcriptional regulation of HLA-G: an update. J Cell Mol Med. 2009;13(9B):2973–2989. doi: 10.1111/j.1582-4934.2009.00800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garziera M, Scarabel L, Toffoli G. Hypoxic modulation of HLA-G expression through the metabolic sensor HIF-1 in human cancer cells. J Immunol Res. 2017;2017:4587520. doi: 10.1155/2017/4587520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moreau P, Mouillot G, Rousseau P, Marcou C, Dausset J, Carosella ED. HLA-G gene repression is reversed by demethylation. Proc Natl Acad Sci USA. 2003;100(3):1191–1196. doi: 10.1073/pnas.0337539100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verloes A, Spits C, Vercammen M, Geens M, LeMaoult J, Sermon K, Coucke W, Van de Velde H. The role of methylation, DNA polymorphisms and microRNAs on HLA-G expression in human embryonic stem cells. Stem Cell Res. 2017;19:118–127. doi: 10.1016/j.scr.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 20.Manaster I, Goldman-Wohl D, Greenfield C, Nachmani D, Tsukerman P, Hamani Y, Yagel S, Mandelboim O. MiRNA-mediated control of HLA-G expression and function. PLoS ONE. 2012;7(3):e33395. doi: 10.1371/journal.pone.0033395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jasinski-Bergner S, Reches A, Stoehr C, Massa C, Gonschorek E, Huettelmaier S, Braun J, Wach S, Wullich B, Spath V, Wang E, Marincola FM, Mandelboim O, Hartmann A, Seliger B. Identification of novel microRNAs regulating HLA-G expression and investigating their clinical relevance in renal cell carcinoma. Oncotarget. 2016;7(18):26866–26878. doi: 10.18632/oncotarget.8567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jasinski-Bergner S, Stoehr C, Bukur J, Massa C, Braun J, Huttelmaier S, Spath V, Wartenberg R, Legal W, Taubert H, Wach S, Wullich B, Hartmann A, Seliger B. Clinical relevance of miR-mediated HLA-G regulation and the associated immune cell infiltration in renal cell carcinoma. Oncoimmunology. 2015;4(6):e1008805. doi: 10.1080/2162402X.2015.1008805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedrich M, Vaxevanis CK, Biehl K, Mueller A, Seliger B. Targeting the coding sequence: opposing roles in regulating classical and non-classical MHC class I molecules by miR-16 and miR-744. J Immunother Cancer. 2020 doi: 10.1136/jitc-2019-000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reches A, Nachmani D, Berhani O, Duev-Cohen A, Shreibman D, Ophir Y, Seliger B, Mandelboim O. HNRNPR regulates the expression of classical and nonclassical MHC class I proteins. J Immunol. 2016;196(12):4967–4976. doi: 10.4049/jimmunol.1501550. [DOI] [PubMed] [Google Scholar]
- 25.Song B, Guan Z, Liu F, Sun D, Wang K, Qu H. Long non-coding RNA HOTAIR promotes HLA-G expression via inhibiting miR-152 in gastric cancer cells. Biochem Biophys Res Commun. 2015;464(3):807–813. doi: 10.1016/j.bbrc.2015.07.040. [DOI] [PubMed] [Google Scholar]
- 26.Shih Ie M. Application of human leukocyte antigen-G expression in the diagnosis of human cancer. Hum Immunol. 2007;68(4):272–276. doi: 10.1016/j.humimm.2007.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Torun Edis C, Yagci Caglayik D, Uyar Y, Korukluoglu G, Ertek M. Sandfly fever outbreak in a province at Central Anatolia, Turkey. Mikrobiyol Bul. 2010;44(3):431–439. [PubMed] [Google Scholar]
- 28.Catamo E, Zupin L, Crovella S, Celsi F, Segat L. Non-classical MHC-I human leukocyte antigen (HLA-G) in hepatotropic viral infections and in hepatocellular carcinoma. Hum Immunol. 2014;75(12):1225–1231. doi: 10.1016/j.humimm.2014.09.019. [DOI] [PubMed] [Google Scholar]
- 29.Amiot L, Vu N, Samson M. Immunomodulatory properties of HLA-G in infectious diseases. J Immunol Res. 2014;2014:298569. doi: 10.1155/2014/298569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang YH, Zozulya AL, Weidenfeller C, Metz I, Buck D, Toyka KV, Bruck W, Wiendl H. Specific central nervous system recruitment of HLA-G(+) regulatory T cells in multiple sclerosis. Ann Neurol. 2009;66(2):171–183. doi: 10.1002/ana.21705. [DOI] [PubMed] [Google Scholar]
- 31.White SR, Loisel DA, Stern R, Laxman B, Floreth T, Marroquin BA. Human leukocyte antigen-G expression in differentiated human airway epithelial cells: lack of modulation by Th2-associated cytokines. Respir Res. 2013;14:4. doi: 10.1186/1465-9921-14-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu HH, Yan WH, Lin A. The role of HLA-G in human papillomavirus infections and cervical carcinogenesis. Front Immunol. 2020;11:1349. doi: 10.3389/fimmu.2020.01349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong DD, Yang H, Li K, Xu G, Song LH, Fan XL, Jiang XL, Yie SM. Human leukocyte antigen-G (HLA-G) expression in cervical lesions: association with cancer progression, HPV 16/18 infection, and host immune response. Reprod Sci. 2010;17(8):718–723. doi: 10.1177/1933719110369183. [DOI] [PubMed] [Google Scholar]
- 34.Souza DM, Genre J, Silva TG, Soares CP, Rocha KB, Oliveira CN, Jatoba CA, Andrade JM, Moreau P, Medeiros Ada C, Donadi EA, Crispim JC. Upregulation of soluble HLA-G5 and HLA-G6 isoforms in the milder histopathological stages of Helicobacter pylori infection: a role for subverting immune responses? Scand J Immunol. 2016;83(1):38–43. doi: 10.1111/sji.12385. [DOI] [PubMed] [Google Scholar]
- 35.Sabbagh A, Sonon P, Sadissou I, Mendes-Junior CT, Garcia A, Donadi EA, Courtin D. The role of HLA-G in parasitic diseases. HLA. 2018;91(4):255–270. doi: 10.1111/tan.13196. [DOI] [PubMed] [Google Scholar]
- 36.Lin A, Yan WH. Human leukocyte antigen-G (HLA-G) expression in cancers: roles in immune evasion, metastasis and target for therapy. Mol Med. 2015;21(1):782–791. doi: 10.2119/molmed.2015.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hiraoka N, Ino Y, Hori S, Yamazaki-Itoh R, Naito C, Shimasaki M, Esaki M, Nara S, Kishi Y, Shimada K, Nakamura N, Torigoe T, Heike Y. Expression of classical human leukocyte antigen class I antigens, HLA-E and HLA-G, is adversely prognostic in pancreatic cancer patients. Cancer Sci. 2020;111(8):3057–3070. doi: 10.1111/cas.14514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu HH, Gan J, Xu DP, Li L, Yan WH. Comprehensive transcriptomic analysis reveals the role of the immune checkpoint HLA-G molecule in cancers. Front Immunol. 2021;12:614773. doi: 10.3389/fimmu.2021.614773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wuerfel FM, Huebner H, Haberle L, Gass P, Hein A, Jud SM, Hack CC, Wunderle M, Schulz-Wendtland R, Erber R, Hartmann A, Ekici AB, Beckmann MW, Fasching PA, Ruebner M. HLA-G and HLA-F protein isoform expression in breast cancer patients receiving neoadjuvant treatment. Sci Rep. 2020;10(1):15750. doi: 10.1038/s41598-020-72837-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lazaro-Sanchez AD, Salces-Ortiz P, Velasquez LI, Orozco-Beltran D, Diaz-Fernandez N, Juarez-Marroqui A. HLA-G as a new tumor biomarker: detection of soluble isoforms of HLA-G in the serum and saliva of patients with colorectal cancer. Clin Transl Oncol. 2020;22(7):1166–1171. doi: 10.1007/s12094-019-02244-2. [DOI] [PubMed] [Google Scholar]
- 41.Rizzo R, Gabrielli L, Bortolotti D, Gentili V, Piccirilli G, Chiereghin A, Pavia C, Bolzani S, Guerra B, Simonazzi G, Cervi F, Capretti MG, Fainardi E, Luca DD, Landini MP, Lazzarotto T. Study of soluble HLA-G in congenital human cytomegalovirus infection. J Immunol Res. 2016;2016:3890306. doi: 10.1155/2016/3890306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park Y, Park Y, Lim HS, Kim YS, Hong DJ, Kim HS. Soluble human leukocyte antigen-G expression in hepatitis B virus infection and hepatocellular carcinoma. Tissue Antigens. 2012;79(2):97–103. doi: 10.1111/j.1399-0039.2011.01814.x. [DOI] [PubMed] [Google Scholar]
- 43.Bertol BC, de Araujo JNG, Sadissou IA, Sonon P, Dias FC, Bortolin RH, de Figueiredo-Feitosa NL, de Freitas LCC, de Miranda Henrique SR, de Tarrapp CC, Oliveira Ramos AD, de Luchessi J, Freitas LMZ, Maciel VN, Silbiger and E. A. Donadi, Plasma levels of soluble HLA-G and cytokines in papillary thyroid carcinoma before and after thyroidectomy. Int J Clin Pract. 2020;74(10):e13585. doi: 10.1111/ijcp.13585. [DOI] [PubMed] [Google Scholar]
- 44.Ben Yahia H, Babay W, Bortolotti D, Boujelbene N, Laaribi AB, Zidi N, Kehila M, Chelbi H, Boudabous A, Mrad K, Mezlini A, Di Luca D, Ouzari HI, Rizzo R, Zidi I. Increased plasmatic soluble HLA-G levels in endometrial cancer. Mol Immunol. 2018;99:82–86. doi: 10.1016/j.molimm.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 45.Scarabel L, Garziera M, Fortuna S, Asaro F, Toffoli G, Geremia S. Soluble HLA-G expression levels and HLA-G/irinotecan association in metastatic colorectal cancer treated with irinotecan-based strategy. Sci Rep. 2020;10(1):8773. doi: 10.1038/s41598-020-65424-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Konig L, Kasimir-Bauer S, Hoffmann O, Bittner AK, Wagner B, Manvailer LF, Schramm S, Bankfalvi A, Giebel B, Kimmig R, Horn PA, Rebmann V. The prognostic impact of soluble and vesicular HLA-G and its relationship to circulating tumor cells in neoadjuvant treated breast cancer patients. Hum Immunol. 2016;77(9):791–799. doi: 10.1016/j.humimm.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 47.Xu HH, Shi WW, Lin A, Yan WH. HLA-G 3′ untranslated region polymorphisms influence the susceptibility for human papillomavirus infection. Tissue Antigens. 2014;84(2):216–222. doi: 10.1111/tan.12359. [DOI] [PubMed] [Google Scholar]
- 48.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y, Zhao Y, Li Y, Wang X, Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, Cao Y, Yousif AS, Bals J, Hauser BM, Feldman J, Muus C, Wadsworth MH, Kazer SW, Hughes TK, Doran B, Gatter GJ, Vukovic M, Taliaferro F, Mead BE, Guo Z, Wang JP, Gras D, Plaisant M, Ansari M, Angelidis I, Adler H, Sucre JMS, Taylor CJ, Lin B, Waghray A, Mitsialis V, Dwyer DF, Buchheit KM, Boyce JA, Barrett NA, Laidlaw TM, Carroll SL, Colonna L, Tkachev V, Peterson CW, Yu A, Zheng HB, Gideon HP, Winchell CG, Lin PL, Bingle CD, Snapper SB, Kropski JA, Theis FJ, Schiller HB, Zaragosi LE, Barbry P, Leslie A, Kiem HP, Flynn JL, Fortune SM, Berger B, Finberg RW, Kean LS, Garber M, Schmidt AG, Lingwood D, Shalek AK, Ordovas-Montanes J, H. C. A. L. B. N. E. a. lung-network@humancellatlas.org and H. C. A. L. B. Network SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181(5):1016–1035. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lucas C, Wong P, Klein J, Castro TBR, Silva J, Sundaram M, Ellingson MK, Mao T, Oh JE, Israelow B, Takahashi T, Tokuyama M, Lu P, Venkataraman A, Park A, Mohanty S, Wang H, Wyllie AL, Vogels CBF, Earnest R, Lapidus S, Ott IM, Moore AJ, Muenker MC, Fournier JB, Campbell M, Odio CD, Casanovas-Massana A, Yale IT, Herbst R, Shaw AC, Medzhitov R, Schulz WL, Grubaugh ND, Dela Cruz C, Farhadian S, Ko AI, Omer SB, Iwasaki A. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584(7821):463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park J, Foox J, Hether T, Danko D, Warren S, Kim Y, Reeves J, Butler DJ, Mozsary C, Rosiene J, Shaiber A, Afshinnekoo E, MacKay M, Bram Y, Chandar V, Geiger H, Craney A, Velu P, Melnick AM, Hajirasouliha I, Beheshti A, Taylor D, Saravia-Butler A, Singh U, Wurtele ES, Schisler J, Fennessey S, Corvelo A, Zody MC, Germer S, Salvatore S, Levy S, Wu S, Tatonetti N, Shapira S, Salvatore M, Loda M, Westblade LF, Cushing M, Rennert H, Kriegel AJ, Elemento O, Imielinski M, Borczuk AC, Meydan C, Schwartz RE, Mason CE. Systemic tissue and cellular disruption from SARS-CoV-2 infection revealed in COVID-19 autopsies and spatial omics tissue maps. bioRxiv. 2021 doi: 10.1101/2021.03.08.434433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rebillard RM, Charabati M, Grasmuck C, Filali-Mouhim A, Tastet O, Brassard N, Daigneault A, Bourbonniere L, Anand SP, Balthazard R, Beaudoin-Bussieres G, Gasser R, Benlarbi M, Moratalla AC, Solorio YC, Boutin M, Farzam-Kia N, Descoteaux-Dinelle J, Fournier AP, Gowing E, Laumaea A, Jamann H, Lahav B, Goyette G, Lemaitre F, Mamane VH, Prevost J, Richard J, Thai K, Cailhier JF, Chomont N, Finzi A, Chasse M, Durand M, Arbour N, Kaufmann DE, Prat A, Larochelle C. Identification of SARS-CoV-2-specific immune alterations in acutely ill patients. J Clin Invest. 2021 doi: 10.1172/JCI145853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Varchetta S, Mele D, Oliviero B, Mantovani S, Ludovisi S, Cerino A, Bruno R, Castelli A, Mosconi M, Vecchia M, Roda S, Sachs M, Klersy C, Mondelli MU. Unique immunological profile in patients with COVID-19. Cell Mol Immunol. 2021;18(3):604–612. doi: 10.1038/s41423-020-00557-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ameratunga R, Woon ST, Steele R, Lehnert K, Leung E, Brooks AES. Severe COVID-19 is a T cell immune dysregulatory disorder triggered by SARS-CoV-2. Expert Rev Clin Immunol. 2022;18(6):557–565. doi: 10.1080/1744666X.2022.2074403. [DOI] [PubMed] [Google Scholar]
- 56.Lotfinejad P, Asadzadeh Z, Najjary S, Somi MH, Hajiasgharzadeh K, Mokhtarzadeh A, Derakhshani A, Roshani E, Baradaran B. COVID-19 infection: concise review based on the immunological perspective. Immunol Invest. 2022;51(2):246–265. doi: 10.1080/08820139.2020.1825480. [DOI] [PubMed] [Google Scholar]
- 57.Moreau P, Adrian-Cabestre F, Menier C, Guiard V, Gourand L, Dausset J, Carosella ED, Paul P. IL-10 selectively induces HLA-G expression in human trophoblasts and monocytes. Int Immunol. 1999;11(5):803–811. doi: 10.1093/intimm/11.5.803. [DOI] [PubMed] [Google Scholar]
- 58.Cordeiro JFC, Fernandes TM, Toro DM, da Silva-Neto PV, Pimentel VE, Perez MM, de Carvalho JCS, Fraga-Silva TFC, Oliveira CNS, Argolo JGM, Degiovani AM, Ostini FM, Puginna EF, da Silva JS, Santos I, Bonato VLD, Cardoso CRB, Dias-Baruffi M, Faccioli LH, Donadi EA, Sorgi CA, Fernandes APM, G. On Behalf Of The Immunocovid Study The severity of COVID-19 affects the plasma soluble levels of the immune checkpoint HLA-G molecule. Int J Mol Sci. 2022 doi: 10.3390/ijms23179736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang S, Gan J, Chen BG, Zheng D, Zhang JG, Lin RH, Zhou YP, Yang WY, Lin A, Yan WH. Dynamics of peripheral immune cells and their HLA-G and receptor expressions in a patient suffering from critical COVID-19 pneumonia to convalescence. Clin Transl Immunol. 2020;9(5):e1128. doi: 10.1002/cti2.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fraga-Silva TFC, Maruyama SR, Sorgi CA, Russo EMS, Fernandes APM, de Barros Cardoso CR, Faccioli LH, Dias-Baruffi M, Bonato VLD. COVID-19: integrating the complexity of systemic and pulmonary immunopathology to identify biomarkers for different outcomes. Front Immunol. 2020;11:599736. doi: 10.3389/fimmu.2020.599736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Al-Bayatee NT, Ad'hiah AH. Soluble HLA-G is upregulated in serum of patients with severe COVID-19. Hum Immunol. 2021;82(10):726–732. doi: 10.1016/j.humimm.2021.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ashcroft T, Simpson JM, Timbrell V. Simple method of estimating severity of pulmonary fibrosis on a numerical scale. J Clin Pathol. 1988;41(4):467–470. doi: 10.1136/jcp.41.4.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gagiannis D, Umathum VG, Bloch W, Rother C, Stahl M, Witte HM, Djudjaj S, Boor P, Steinestel K. Antemortem vs postmortem histopathologic and ultrastructural findings in paired transbronchial biopsy specimens and lung autopsy samples from three patients with confirmed SARS-CoV-2. Am J Clin Pathol. 2022;157(1):54–63. doi: 10.1093/ajcp/aqab087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Salgado R, Denkert C, Demaria S, Sirtaine N, Klauschen F, Pruneri G, Wienert S, Van den Eynden G, Baehner FL, Penault-Llorca F, Perez EA, Thompson EA, Symmans WF, Richardson AL, Brock J, Criscitiello C, Bailey H, Ignatiadis M, Floris G, Sparano J, Kos Z, Nielsen T, Rimm DL, Allison KH, Reis-Filho JS, Loibl S, Sotiriou C, Viale G, Badve S, Adams S, Willard-Gallo K, Loi S, T. W. G. International The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann Oncol. 2015;26(2):259–271. doi: 10.1093/annonc/mdu450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hirsch FR, Varella-Garcia M, Bunn PA, Jr, Di Maria MV, Veve R, Bremmes RM, Baron AE, Zeng C, Franklin WA. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol. 2003;21(20):3798–3807. doi: 10.1200/JCO.2003.11.069. [DOI] [PubMed] [Google Scholar]
- 66.Wickenhauser C, Bethmann D, Feng Z, Jensen SM, Ballesteros-Merino C, Massa C, Steven A, Bauer M, Kaatzsch P, Pazaitis N, Toma G, Bifulco CB, Fox BA, Seliger B. Multispectral fluorescence imaging allows for distinctive topographic assessment and subclassification of tumor-infiltrating and surrounding immune cells. Methods Mol Biol. 2019;1913:13–31. doi: 10.1007/978-1-4939-8979-9_2. [DOI] [PubMed] [Google Scholar]
- 67.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33(20):e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reches A, Berhani O, Mandelboim O. A unique regulation region in the 3' UTR of HLA-G with a promising potential. Int J Mol Sci. 2020 doi: 10.3390/ijms21030900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McLane LM, Abdel-Hakeem MS, Wherry EJ. CD8 T cell exhaustion during chronic viral infection and cancer. Annu Rev Immunol. 2019;37:457–495. doi: 10.1146/annurev-immunol-041015-055318. [DOI] [PubMed] [Google Scholar]
- 70.Rashidi S, Farhadi L, Ghasemi F, Sheikhesmaeili F, Mohammadi A. The potential role of HLA-G in the pathogenesis of HBV infection: Immunosuppressive or immunoprotective? Infect Genet Evol. 2020;85:104580. doi: 10.1016/j.meegid.2020.104580. [DOI] [PubMed] [Google Scholar]
- 71.Schust DJ, Tortorella D, Ploegh HL. HLA-G and HLA-C at the feto-maternal interface: lessons learned from pathogenic viruses. Semin Cancer Biol. 1999;9(1):37–46. doi: 10.1006/scbi.1998.0106. [DOI] [PubMed] [Google Scholar]
- 72.Choreno-Parra JA, Jimenez-Alvarez LA, Cruz-Lagunas A, Rodriguez-Reyna TS, Ramirez-Martinez G, Sandoval-Vega M, Hernandez-Garcia DL, Choreno-Parra EM, Balderas-Martinez YI, Martinez-Sanchez ME, Marquez-Garcia E, Sciutto E, Moreno-Rodriguez J, Barreto-Rodriguez JO, Vazquez-Rojas H, Centeno-Saenz GI, Alvarado-Pena N, Salinas-Lara C, Sanchez-Garibay C, Galeana-Cadena D, Hernandez G, Mendoza-Milla C, Dominguez A, Granados J, Mena-Hernandez L, Perez-Buenfil LA, Dominguez-Cheritt G, Cabello-Gutierrez C, Luna-Rivero C, Salas-Hernandez J, Santillan-Doherty P, Regalado J, Hernandez-Martinez A, Orozco L, Avila-Moreno F, Garcia-Latorre EA, Hernandez-Cardenas CM, Khader SA, Zlotnik A, Zuniga J. Clinical and immunological factors that distinguish COVID-19 from pandemic influenza A(H1N1) Front Immunol. 2021;12:593595. doi: 10.3389/fimmu.2021.593595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schuurman AR, Reijnders TDY, Saris A, Ramirez Moral I, Schinkel M, de Brabander J, van Linge C, Vermeulen L, Scicluna BP, Wiersinga WJ, Vieira Braga FA, van der Poll T. Integrated single-cell analysis unveils diverging immune features of COVID-19, influenza, and other community-acquired pneumonia. Elife. 2021 doi: 10.7554/eLife.69661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schulte-Schrepping J, Reusch N, Paclik D, Bassler K, Schlickeiser S, Zhang B, Kramer B, Krammer T, Brumhard S, Bonaguro L, De Domenico E, Wendisch D, Grasshoff M, Kapellos TS, Beckstette M, Pecht T, Saglam A, Dietrich O, Mei HE, Schulz AR, Conrad C, Kunkel D, Vafadarnejad E, Xu CJ, Horne A, Herbert M, Drews A, Thibeault C, Pfeiffer M, Hippenstiel S, Hocke A, Muller-Redetzky H, Heim KM, Machleidt F, Uhrig A, Bosquillon de Jarcy L, Jurgens L, Stegemann M, Glosenkamp CR, Volk HD, Goffinet C, Landthaler M, Wyler E, Georg P, Schneider M, Dang-Heine C, Neuwinger N, Kappert K, Tauber R, Corman V, Raabe J, Kaiser KM, Vinh MT, Rieke G, Meisel C, Ulas T, Becker M, Geffers R, Witzenrath M, Drosten C, Suttorp N, von Kalle C, Kurth F, Handler K, Schultze JL, Aschenbrenner AC, Li Y, Nattermann J, Sawitzki B, Saliba AE, Sander LE, Deutsche C-OI. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182(6):1419–1440. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guan Z, Song B, Liu F, Sun D, Wang K, Qu H. TGF-beta induces HLA-G expression through inhibiting miR-152 in gastric cancer cells. J Biomed Sci. 2015;22:107. doi: 10.1186/s12929-015-0177-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wastowski IJ, Simoes RT, Yaghi L, Donadi EA, Pancoto JT, Poras I, Lechapt-Zalcman E, Bernaudin M, Valable S, Carlotti CG, Jr, Flajollet S, Jensen SS, Ferrone S, Carosella ED, Kristensen BW, Moreau P. Human leukocyte antigen-G is frequently expressed in glioblastoma and may be induced in vitro by combined 5-aza-2'-deoxycytidine and interferon-gamma treatments: results from a multicentric study. Am J Pathol. 2013;182(2):540–552. doi: 10.1016/j.ajpath.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shiroishi M, Kuroki K, Rasubala L, Tsumoto K, Kumagai I, Kurimoto E, Kato K, Kohda D, Maenaka K. Structural basis for recognition of the nonclassical MHC molecule HLA-G by the leukocyte Ig-like receptor B2 (LILRB2/LIR2/ILT4/CD85d) Proc Natl Acad Sci USA. 2006;103(44):16412–16417. doi: 10.1073/pnas.0605228103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Desgrandchamps F, LeMaoult J, Goujon A, Riviere A, Rivero-Juarez A, Djouadou M, de Gouvello A, Dumont C, Wu CL, Culine S, Verine J, Rouas-Freiss N, Hennequin C, Masson-Lecomte A, Carosella ED. Prediction of non-muscle-invasive bladder cancer recurrence by measurement of checkpoint HLAG's receptor ILT2 on peripheral CD8(+) T cells. Oncotarget. 2018;9(69):33160–33169. doi: 10.18632/oncotarget.26036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rha MS, Jeong HW, Ko JH, Choi SJ, Seo IH, Lee JS, Sa M, Kim AR, Joo EJ, Ahn JY, Kim JH, Song KH, Kim ES, Oh DH, Ahn MY, Choi HK, Jeon JH, Choi JP, Kim HB, Kim YK, Park SH, Choi WS, Choi JY, Peck KR, Shin EC. PD-1-expressing SARS-CoV-2-Specific CD8(+) T cells are not exhausted, but functional in patients with COVID-19. Immunity. 2021;54(1):44–52. doi: 10.1016/j.immuni.2020.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Aghbash PS, Eslami N, Shamekh A, Entezari-Maleki T, Baghi HB. SARS-CoV-2 infection: the role of PD-1/PD-L1 and CTLA-4 axis. Life Sci. 2021;270:119124. doi: 10.1016/j.lfs.2021.119124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sumida TS, Dulberg S, Schupp JC, Lincoln MR, Stillwell HA, Axisa PP, Comi M, Unterman A, Kaminski N, Madi A, Kuchroo VK, Hafler DA. Type I interferon transcriptional network regulates expression of coinhibitory receptors in human T cells. Nat Immunol. 2022;23(4):632–642. doi: 10.1038/s41590-022-01152-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pairo-Castineira E, Clohisey S, Klaric L, Bretherick AD, Rawlik K, Pasko D, Walker S, Parkinson N, Fourman MH, Russell CD, Furniss J, Richmond A, Gountouna E, Wrobel N, Harrison D, Wang B, Wu Y, Meynert A, Griffiths F, Oosthuyzen W, Kousathanas A, Moutsianas L, Yang Z, Zhai R, Zheng C, Grimes G, Beale R, Millar J, Shih B, Keating S, Zechner M, Haley C, Porteous DJ, Hayward C, Yang J, Knight J, Summers C, Shankar-Hari M, Klenerman P, Turtle L, Ho A, Moore SC, Hinds C, Horby P, Nichol A, Maslove D, Ling L, McAuley D, Montgomery H, Walsh T, Pereira AC, Renieri A, Gen OI, Gen CI, Shen X, Ponting CP, Fawkes A, Tenesa A, Caulfield M, Scott R, Rowan K, Murphy L, Openshaw PJM, Semple MG, Law A, Vitart V, Wilson JF, Baillie JK, I. C. Investigators, C.-H. G. Initiative, I. andMe, B. Investigators Genetic mechanisms of critical illness in COVID-19. Nature. 2021;591(7848):92–98. doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- 83.Westin AT, Gardinassi LG, Soares EG, Da Silva JS, Donadi EA, Da Silva Souza C. HLA-G, cytokines, and cytokine receptors in the non-aggressive basal cell carcinoma microenvironment. Arch Dermatol Res. 2022;314(3):247–256. doi: 10.1007/s00403-021-02218-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are not publicly available, since no repository exists, but are available from the corresponding authors.