Abstract
Deciphering the relationship between molecules, olfactory receptors (ORs) and corresponding odors remains a challenging task. It requires a comprehensive identification of ORs responding to a given odorant. With the recent advances in artificial intelligence and the growing research in decoding the human olfactory perception from chemical features of odorant molecules, the applications of advanced machine learning have been revived. In this study, Convolutional Neural Network (CNN) and Graphical Convolutional Network (GCN) models have been developed on odorant molecules-odors and odorant molecules-olfactory receptors using a large set of 5955 molecules, 160 odors and 106 olfactory receptors. The performance of such models is promising with a Precision/Recall Area Under Curve of 0.66 for the odorant-odor and 0.91 for the odorant-olfactory receptor GCN models respectively. Furthermore, based on the correspondence of odors and ORs associated for a set of 389 compounds, an odor-olfactory receptor pairwise score was computed for each odor-OR combination allowing to suggest a combinatorial relationship between olfactory receptors and odors. Overall, this analysis demonstrate that artificial intelligence may pave the way in the identification of the smell perception and the full repertoire of receptors for a given odorant molecule.
Subject terms: Chemical biology, Computational biology and bioinformatics
Introduction
Smell is a sense that allows the perception and discrimination of a large number of volatile environmental chemicals in the air by using the nose. It has been observed that smell is involved in the social behavior of many species but also in the location of food, ability to detect dangerous situations like fire, identification of predators, toxic compounds, mate choice and mother-infant recognition1. For humans, olfaction influences our well-being (looking for pleasantness) and play a major role in eating behavior with the perception of food quality and for social communication with the use of fragrance2. The smell impairment has a strong impact on the quality of life and it has been recently highlighted with COVID-19 causing the loss of smell of many individuals3.
The sense of smell is commonly associated with large and diverse families of odorant receptors that detect odor stimuli in the nose and transform them into patterns of neuronal activity that are recognized in the brain4–7.
In humans, it is estimated that millions and perhaps billion of odorant molecules are recognized by around 400 different human olfactory receptors (hORs)8–11. Odorants, commonly present in food, fragrance and cosmetic products, stimulate G-protein-coupled olfactory receptors (ORs) located in the olfactory sensory neurons of the nasal epithelium12,13. It has been reported than the olfactory system uses a combinatorial olfactory receptors code to encode an odor14–16. One odorant can interact with several different ORs and one OR can be activated by a large panel of molecules. Although recent optimizations in functional expression of ORs for the screening of odorant compound libraries have been made, investigating all combinations is still expensive, time consuming and remains therefore a tremendous challenge17.
It is important to notice that the semantic is a source of complexity for the verbal description of odors18,19. Indeed, the description of the odor of a molecule involves several odor attributes, or odor notes, which are “odor objects” i.e., the odors perceived in our environment20–22. Yet, these odors result from the perception of numerous odorant molecules, which increase the difficulty to have reliable odors descriptions.
Despite some experimental studies have identified odorant-OR interactions in some organisms (mainly in mammals and insects)23–26, the link between activation of ORs and odor perception remains limited9,27–31. Considering that the perception depends on chemistry, several studies have attempted to connect odorant physicochemical properties to the olfactory perceptions32–38. Crowd-sourced DREAM Olfaction Prediction Challenge was organized in the aim to predict human olfactory perception for 19 semantic descriptors for odors as well as intensity and pleasantness based on chemical features and machine learning models39. Such analysis can then be used to identify new structural motifs for ligands during large virtual screening campaigns40,41. Recently, artificial intelligence technology using deep neural networks (DNN)42, graph neural networks (GNN)43 or convolutional neural networks (CNN)44,45 have been performed to underlie the relationship between the structure of chemicals and odors. They reported that such machine learning approaches outperformed classical methods applied to chemical-odor relationships.
Based on these observations, we decided to go one step further and to analyze the relationships of chemical-odors and chemical-olfactory receptors based on the chemical structure of odorant using deep learning approaches such as graph neural networks (GNN), and convolutional neural networks (CNN). The relationship between chemicals—olfactory receptors and odor perception is of high interest in the determination of (i) chemical properties—odor relationships, (ii) chemical properties—olfactory receptors relationships and (iii) olfactory receptors—odor relationships. Furthermore, the global chemicals-olfactory receptors-odors relationship has been investigated using a confidence score proposing a combination of receptors that can play a role in the perception of odors.
Materials and methods
Datasets
This study is based on the integration of two different data sets (i) data for chemical-odor relationships and (ii) data for chemical-olfactory receptor relationships.
Chemical-Odor
We extracted chemical-odors from two separate sources: The Good Scents Company (TGSC) Database46 (as of January 2021), and Leffingwell Database47. Both databases contained information linking the compound and its chemical structure to the odor description as several odor notes. From the TGSC database, we got 27,779 chemicals of which 5659 are related to one or several odor notes. From the Leffingwell database, we got 6054 compounds that are related to one or several odors notes. We merged the outcomes from both databases, eliminating duplicated information. Compounds occurring with the same structure (based on Inchi Key encoding48) but with different names (synonyms) were removed. Odor notes from Leffingwell database was matched with TGSC as reference. To limit the complexity of the models and avoid mis-classification due to poor representation of an odor note, odor notes with less than 20 chemicals were not considered in this analysis. After all these steps, we obtained a dataset made up of 5955 compounds and 160 odors. Each compound is related from 1 to at the maximum 10 odor notes using the order proposed by TGSC.
Compound-olfactory receptor
Compounds tested experimentally on olfactory receptors were gathered from different data sources. It included information from OdorDB49, ODORactor50, OlfactionDB51 and from the literature. To the purpose of the study we considered, first, human receptors in the construction of learning models. We collected 74 human olfactory receptors for 365 compounds. In a second step, human receptors that are orthologs to rodent olfactory receptors, and on which bioactivity has been measured, were also included in the learning model development. With the aggregation of this data, we reached a dataset of 445 different compounds tested on 106 different olfactory receptors.
The datasets generated and analysed during the current study are available in the Table S1 in supplementary.
Methods
Global overview of the odorant molecules
To visualize the distribution of the molecules according to their odors and their activity on olfactory receptors, the structure of each molecule was encoded into 1024 ECFP (Extended Connectivity Fingerprint) fingerprints52. Then, the matrix of fingerprint was projected into a 2D map using a reduction technique, UMAP (the Uniform Manifold Approximation and Projection), that was applied recently with smell compounds53. Such projection allows to look over the distribution of the molecules in a 2D space and to map corresponding odors and olfactory receptors associated to each molecule.
Machine learning models
Different machine learning models have been generated in order to assess their performance in the prediction of compound-odor and compound-olfactory receptor relationships. Since one compound can be related to one or more odors, it raised a multi-label classification problem. Consequently, we developed 3 types of models adapted for multiclass: (i) a Random Forest model, based on RDKit descriptors54 and ECFP (Extended Connectivity Fingerprint) fingerprint, (ii) a Convolutional Neural Network (CNN) based on ECFP and (iii) a Graph-based Neural Network (GNN). Random Forest were built using scikit-learn python package55, GNN with DeepChem56 and CNN with Tensorflow57. The evaluation metric used was the Area Under ROC Curve (AUROC) and the Precision/Recall Area Under Curve (PRC-AUC). For CNN and GNN, an internal validation of the models was carried out using a fivefold Cross Validation for each of them.
Random forest (RF) models
For the RF models, in a first strategy, the molecule’s structure was encoded in 154 2D descriptors (using RDKit) and the odor notes labels binarized in 0 or 1. Then, a RF model was built using 500 mtrees and 15 ntry. We optimized the hyperparameters in order to minimize the Out Of Bag Score (OOB Score) and maximize the AUROC score (Table 1).
Table 1.
Parameters considered in the compound-odor Random Forest models.
| n_estimators | criterion | max_depth | min_samples_split | min_samples_leaf | max_features |
|---|---|---|---|---|---|
| 500 | ‘gini’ | 75 | 2 | 1 | 15 |
In a second strategy, the chemical structure was encoded into ECFP fingerprints. This type of fingerprint was chosen because it is a method of vectorial representation of molecules quite similar to the one used for the Graph-based Model (described below). Thus, a 1024-bit ECFP fingerprint was generated for each molecule and then a RF model was performed using the same parameters.
A similar RF protocol was also applied with olfactory receptors using the same parameters.
Convolutional neural network (CNN) model
A convolutional neural network (CNN) model was developed based on ECFP fingerprints encoding. At the difference of RF, CNN is a method based on neuron convolutions. In our CNN model, the architecture of the network is organized as follows: the concatenation of the message is done by 2 layers of dimensions [32, 32] with a rectifier linear unit activation function ‘RELU’, a batch normalization that standardize input data in order to reduce the number of epochs for training network, and finally a maxpooling parameter that reduce spatial size by some operations, preventing overfitting and reducing computational cost. The fully connected neural net consists in layers of a size 128 dots (Dense layer). The readout is done using a softmax function with 160 tasks for odors (and 106 tasks for olfactory receptors) and a Categorical Cross Entropy loss function. The model has been trained on 300 epochs and a 5 folds cross validation was performed (60 epochs for each fold). More information about the CNN implementation can be obtained here58.
Graph-based neural network (GNN) model
We decided to develop a graph-based Neural Network (GNN) model because it is close to the architecture of the model based on molecular graphs. By considering chemical bonds as edges and atoms as nodes, molecules can be represented as graphs. This type of representation can then be used to develop graph-based model. In our study we considered the implementation of a Graph Convolutional Network (GCN)59. GCN consists of message passing layers, followed by a reduce-sum operation to obtain at the end, a fully connected layer. In a first step, each molecule is featured into a set of fixed-length vectors where each vector is calculated for each atom. Once the molecule has been featured, a series of operations consisting of concatenating the message takes place. This is the convolutional part of the model. Then, each molecular graph is reduced to a vector that will yield a fully connected neural network for final prediction. The architecture of the network is as follows: the concatenation of the message is done by 2 layers of dimensions [64, 64] with rectifier linear unit activation function ‘RELU’, a batch normalization that standardize input data in order to reduce the number of epochs for training network, a dropout that omit some units to prevent from overfitting and finally a maxpooling that reduce spatial size, prevent overfitting and reduces computational cost. The fully connected neural net consists of a layer of a size 128 (Dense layer) with RELU activation and batch normalization. The readout is done using a softmax function with 160 tasks for odors (and 106 tasks for olfactory receptors) and a Softmax Cross Entropy loss function.
The model has been trained on 300 epochs and a 5 folds cross validation was performed (60 epochs for each fold).
In addition to this model, a second GCN was created by grouping the odors by categories in order to predict the corresponding categories rather than each odor note individually. Thus, the parameters used for this model are the same as the GCN presented above. The odor notes have been grouped according to the correspondences shown in the Table 2.
Table 2.
Grouping of odor notes in categories having a similar perceptual space.
| Natural | "green", "herbal", "earthy", "natural", "leafy", "oakmoss", "weedy", "grassy", "hay", "ozone", "malty" |
| Woody | "cedar", "woody", "camphoreous", "pine", "fir needle", "sandalwood", "terpenic" |
| Fruity | "fruity", "pineapple", "apple", "currant", "peach", "berry", "grapefruit", "grape", "apricot", "banana", "cherry", "melon", "pear", "plum", "raspberry", "ripe", "strawberry", "coconut", "citrus", "lemon", "orange", "bergamot", "mandarin", "nutty", "hazelnut" |
| Spicy | "peperry", "spicy", "mustard" |
| Fresh | "fresh", "minty", "cooling", "mentholic" |
| Dairy | "milky", "dairy", "cheesy", "creamy", "buttery" |
| Floral | "floral", "rose'7'orangeflower", "chamomile", "lavender", "geranium", "hawthorn", "jasmin", "jasmine", "hyancinth", "lilac", "lily", "lily of the valley", "magnolia", "muguet", "narcissus", "orchid", "savory", "violet", "vetiver", "ylang", 'mimosa', "patchouli", "hyacinth", "tea", "cinnamyl", "orris", "tropical" |
| Alcohol | "alcoholic", "cognac", "brandy", "winey" |
| Spice | "cum in", "cinnamon", "rum my", "clove", "licorice", "tonka", "anisic" |
| Fat | "oily", "fatty", "waxy" |
| Sweet | "sweet", "caramellic", "honey", "vanilla", "almond", "cocoa", "chocolate", "popcorn" |
| Vegetable | "peppery", "radish", "cucumber", "cabbage", "tomato", "celery", "potato", "vegetable", "corn", "mushroom" |
| Marine | "marine", "fishy" |
| Meaty | "meaty", "beefy", "roasted", "smoky" |
| Animal | "animal", "musk", "tallow", "amber" |
| Clean | "clean", "bready", "soapy" |
| Sharp | "pungent", "sharp", "alliaceous", "garlic", "onion", "leathery", "bitter", "sour" |
| Cooked | "cooked", "toasted" |
| Odorless | « odorless » |
| Unpleasant | "musty", "sulfurous", "ammoniacal", "fermented" |
| Balsamic | « balsamic » |
| Large | "ethereal", "aromatic", "phenolic", "coumarinic", "aldehydic", "ketonic", "lactonic" |
| Other | burnt", "coffee", "cortex", "dry", "dusty", " medicinal", "metallic", "gassy ", "juicy", "solvent", "sweaty", "tobacco", "warm", "plastic", " powdery" |
Odor-receptor model
From the two datasets, 383 compounds targeting olfactory receptors and also related to odor notes were identified. It means that for each molecule, odor notes and olfactory receptors correspondence can be highlighted. Given the imbalance in the two data sets and the imbalance in the binary classes (much more negative than positive outcomes), an odor-olfactory receptor pairwise (OORP) score was computed between the odor and receptor information based of the common active compounds using the equation below:
With COiORy being the number of compounds common between an odor (Oi) and an olfactory receptor (ORy), CtotOi being the total number of compounds associated to the odor notes (Oi), and CtotORy the total number of compounds associated to the olfactory receptor (ORy).
The odor notes-olfactory receptor pairwise score is between 0 and 1. The closer to 1 is the score, the more significant is the relation between an olfactory receptor and an odor note.
Results
Global analysis of the data collected
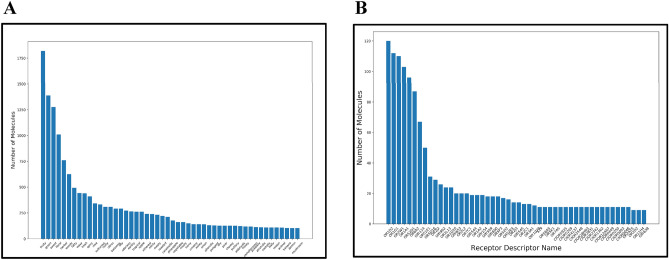
The data collected on chemicals, olfactory receptors and odor notes are very heterogeneous, with many molecules for some odor notes/receptors and very few for others. Fruity is the odor associated with the highest number of molecules (> 1750) (Fig. 1A). More than 1000 molecules are sweet, green and floral. At the difference, less than 200 molecules are associated with mushroom, jasmin or banana. We have to notice that a molecule is usually associated with several odor notes. On average, a molecule has 3, 4 odors which is in agreement with previous studies37,38. Some odor notes could be closely related to each other and an odor note could be a more specific term to a general category of odor note. Like for example banana, melon, pear or apple are specific odor notes but also belong to a more general fruity odor.
Figure 1.
(A) Occurrence of compounds related to odors. (B) Occurrence of compounds related to olfactory receptors.
Similarly, looking on the relation between compounds and olfactory receptors, it is observed that OR1D2, OR1G1, OR2W1, OR1A1, OR52D1, OR6A2 and olfr124 (ortholog to OR2B4 in human) are receptors with more than 50 molecules interacting to them (Fig. 1B). On average a molecule interacts with 3,46 olfactory receptors.
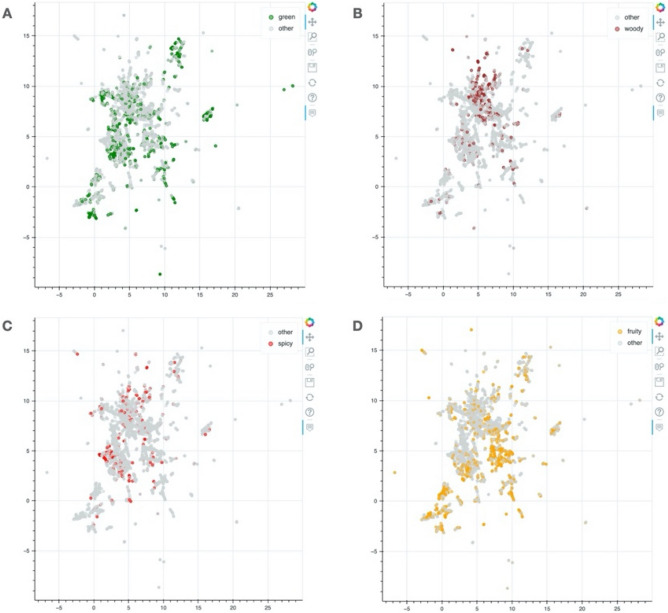
Using a UMAP visualization technique, the relation between chemical structure, odor notes and olfactory receptors can be depicted in an interactive 2D map. It is a way to represent the distribution of molecules in a 2D space. For example, comparing compounds having fruity, spicy, woody and green odor notes (Fig. 2), some compounds are more grouped in some area of the map and others compounds are more spread all over the chemical space. It means that there are some specific structural features for some compounds associated to a specific odor note compared to others odor notes for which it is more general.
Figure 2.
UMAP representation of compounds distribution in a 2D map projection. (A) Compounds with green odor note, (B) Compounds with woody odor note, (C) Compounds with spicy odor note, (D) Compounds with fruity odor note. The UMAP representations were developed with the python package bokeh (v.2.4.3): https://docs.bokeh.org/e60.
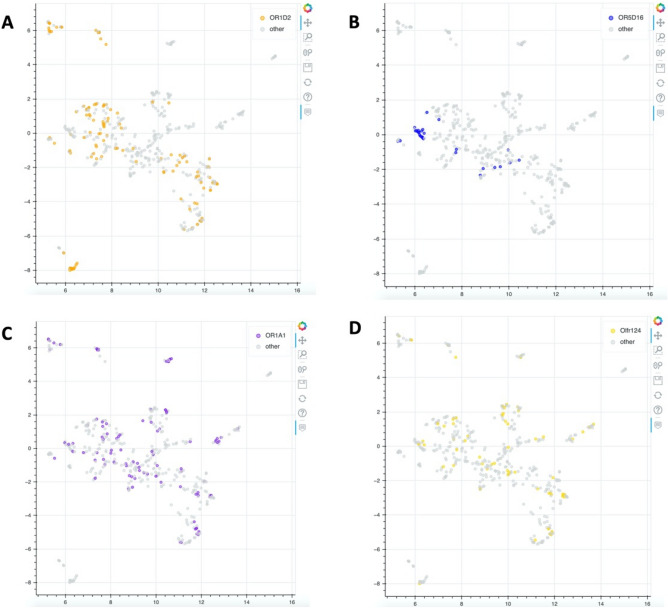
A similar observation can be concluded for some olfactory receptors, notably the OR1D2, OR5D16 for which some bioactive compounds on theses receptors are grouped in some area of the map while others ORs (OR1A1, OR2B4) are more spread over the chemical space (Fig. 3).
Figure 3.
UMAP representation of compounds distribution in a 2D map projection. (A) Compounds active on OR1D2, (B) Compounds active on OR5D16, (C) Compounds active on OR1A1, (D) Compounds active on OR2D4. The UMAP representations were developed with the python package bokeh (v.2.4.3): https://docs.bokeh.org/e60.
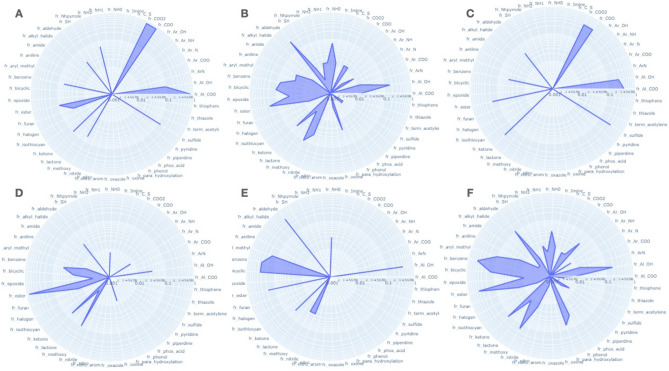
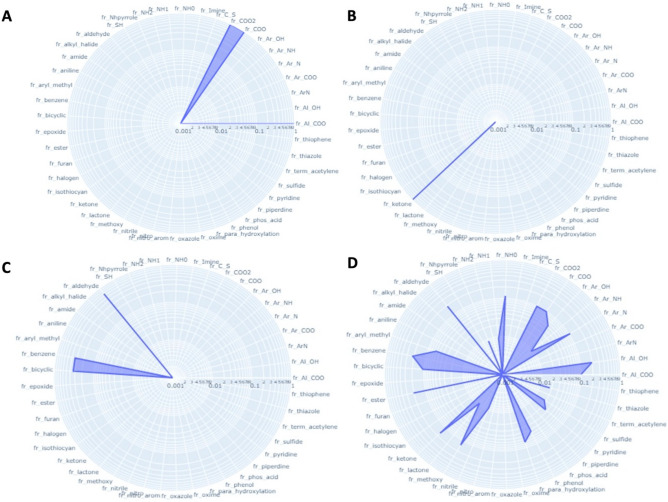
To look over the frequency of chemical groups related to odors and receptors, radar plots have been developed with 62 molecular substructure and group of atoms. Based on these plots, an ensemble of structural features that occur more frequently with some odors but also with some olfactory receptors can be observed (Figs. S1 & S2 in supplementary). For example, a majority of compounds associated to the odor note ‘acidic’ possess a COO group. However, compounds associated to ‘citrus’ odor note are represented by a sparser ensemble of group of atoms (OH, aldehyde, ester, methoxy, NH…). Interestingly, the ‘cheese’ odor note is also highly associated to the presence of a COO group in a compound. Globally, specific odor notes that are associated to a fruit (apple, apricot, banana), a vegetable (celery, cucumber) or a flower (rose, muguet, narcissus) are related to few groups of atoms while general class of odors i.e., fruity, floral, sweet, phenolic encompass larger groups of compounds with a higher diversity in physicochemical properties (Fig. 4).
Figure 4.
Radar plot based on the frequency of physicochemical properties observed on the set compounds associated to an odor note. (A) Acidic, (B) Citrus, (C) Cheese, (D) Apple, (E) Muguet, (F) Floral. The radarplots were developed with the python package plotly (v.5.3.1): https://plotly.com/61.
With olfactory receptors, some specific structural features are also more frequently observed with some ORs while other Ors are less specific and can be impacted by different groups of molecules. For example, a majority of molecules associated to OR52E1 possess a carboxylic group, OR4D6 ligands have a ketone, OR1D3 ligands have a benzene, a bicyclic and an aldehyde group. Similar to odors, it is observed that Ors with a large set of compounds (i.e., OR1G1, OR2W1, OR1A1, OR52D1, OR6A2) are also associated to compounds with diverse groups of atoms (Fig. 5). So, it could be assumed that some Ors are more selective to some ligands with specific features than others Ors that are more general62.
Figure 5.
Radar plot based on the frequency of physicochemical properties observed on the set compounds associated to an olfactory receptor. (A) OR52E1, (B) OR4D6, (C) OR1D3, (D) OR1G1. The radarplots were developed with the python package plotly (v.5.3.1): https://plotly.com/61.
Results on ligand-odor notes model
Once, the global analysis of these data was realized, machine learning models were developed to predict in one hand the odor notes and in the other hand the olfactory receptors, associated to a molecule. About the ligand-odor note models, 3 types of models were built i.e., Random Forest, Convolutional Neural Network (CNN) and Graph Convolutional Network (GCN). Based on the AUROC and the PRC-AUC estimation, the GCN showed the best performance of prediction, with an AUROC = 0.96 and a PRC-AUC = 0.49 (Table 3). Random Forest models have inferior performance with both Morgan Fingerprints and RDKit descriptors. CNN model based on Morgan fingerprints was the worst with an AUC = 0.53 and a PRC-AUC = 0.04. So, models based on neural network and graph-type information seems to have better performance. To evaluate the robustness of the models, A fivefold cross validation was performed. Although the AUROC is still high, the PRC-AUC went down to 0.24 respectively. The unbalanced data set might explain this reduction of PRC-AUC performance.
Table 3.
Performance of the 4 models applied on the compound-odor note dataset. The results in bold are the performance on the full dataset. The value in brackets depicts the results of the fivefold cross validation model.
| Compound—Odor note | ||
|---|---|---|
| AUROC | PRC-AUC | |
| GCN | 0.964 [0.82] | 0.497 [0.24] |
| RF-Morgan Fp | 0.631 | 0.222 |
| RF-RDKit Features | 0.644 | 0.241 |
| CNN-Morgan Fp | 0.532 | 0.042 |
In more details, the performance for each odor note, odor notes with high PRC-AUC such as ‘malty’ (0.99), ‘odorless’ (0.89), ‘maple’ (0.85), ‘sandalwood’ (0.84), ‘alcoholic’ (0.83), ‘musk’ (0.83), ‘ambergris’ (0.81) and odors with low performance i.e., ‘tea’ (0.13), ripe (0.18), ‘chocolate’ (0.21), ‘metallic’ (0.21), ‘aromatic’ (0.22) can be identified (supplementary Table S2).
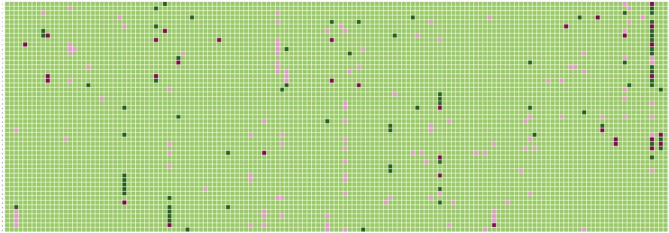
The prediction of odor notes associated for each molecule by the GCN model can also be depicted in a heatmap (supplementary Fig. S3). A representation for a subset of compounds is depicted in Fig. 6.
Figure 6.
Heatmap representation of the performance with the GCN model for a subset of the compound-odor note associations. The odor notes are represented on the X-axis and the compounds on the Y axis. A compound associated to an odor note and correctly classified by the GCN model is colored with a dark green cell. A compound not associated to an odor note and correctly predicted has a light green color. A compound linked to an odor note and wrongly predicted by the model is represented with a pink color and a compound not linked to an odor note and predicted to be associated to this odor note is shown with a red color. The heatmap was developed with the python package seaborn (v.0.11.2): https://seaborn.pydata.org/63,64.
Based on this heatmap, we can observe that many compounds are predicted to ‘sweet’ and the ‘fruity’ odors with a mixed of good and bad prediction. Floral, fresh and herbal odors notes are also general classes of odors with many mis-classified compounds (pink color). For some compounds, the classification is excellent with no misclassification. This is the case for example, for 3-phenyl propyl alcohol which is correctly predicted to the odor note balsamic and sweet; butyl acetate which is related to banana, ethereal, fruity and solvent; (E)-isoeugenyl acetate which is correctly predicted to spicy and clove and (Z)-7-decenal which is predicted to citrus, aldehydic and cucumber among others. However, many compounds have a combination of good and bad predictions. At the opposite, some compounds are wrongly predicted and do not capture the odor note on which it has been associated with. This is the case for “benzyl acetone” which is not predicted by the model to be associated to balsamic and floral but for which the model is predicted the odor of almond and sweet. The model is not able to annotate the animal odor note for skatole compound, neither the fruity, fatty, cheesy, herbal coconut odor note for the 2-nonanone compound.
As some odors might be relatively close in perception (for example citrus vs lemon, cheese vs cheesy), a second GCN model was developed by grouping the 160 odors in 23 categories. The results in Table 4 depicts a good AUROC performance (0.92). Interestingly, the PRC-AUC performance is higher with a score of 0.67 (0.40 in cross validation). Therefore, the GCN model seems more robust and suitable with a reduced number of odors.
Table 4.
Performance of the GCN model applied on the compound-odor dataset grouped on 23 categories. The results in bold are the performance on the full dataset. The value in brackets depicts the results of the fivefold cross validation model.
| Compound—Odor GROUP | ||
|---|---|---|
| AUROC | PRC-AUC | |
| GCN | 0.92 [0.80] | 0.67 [0.40] |
Results on ligands-receptors model
Similarly, to the previous models developed on compound-odor relationships, RF, CNN and GCN models were developed on ligand-receptor information. At the difference of the compound-odors relationships models, the ligand-receptor dataset is smaller containing 365 odorants with known bioactivity on 74 human olfactory receptors. Developing a GCN model on this dataset we obtained a AUROC = 0.98 (0.67 in cross validation) and a PRC-AUC = 0.71 (0.22 in cross validation). The large drop observed for the PRC-AUC in cross validation indicate that the model is not too stable and might be due to a limited size of the data set. Therefore, we decided to enrich our dataset with the integration of chemicals having a bioactivity on rodent olfactory receptors orthologs to human receptors, assuming that they share a similar mechanism of action. With this step, predictive models were developed based on 445 compounds with known bioactivity on 106 olfactory receptors. The performances of the models are presented in Table 5. Again, the GCN model have higher AUROC (0.99) and PRC-AUC (0.91) than the other machine learning models. The GCN model conserved a good AUROC score in cross validation (0.71) and with a better PRC-AUC score (0.4). These results suggest that the model’s performance is dependent on the data inclusion. The scattering of the compound—olfactory receptors information might be a cause of the fall of the PRC-AUC when using a subset of the compound-OR data set.
Table 5.
Performance of the 4 models applied on the compound-olfactory receptors dataset. The results in bold are the performance on the full dataset. The value in brackets depicts the results of the fivefold cross validation model.
| Compound—Receptor (human + orthologs) | ||
|---|---|---|
| AUROC | PRC-AUC | |
| GCN | 0.99 [0.71] | 0.91 [0.40] |
| RF-Morgan Fp | 0.612 | 0.212 |
| RF-RDKit Features | 0.586 | 0.167 |
| CNN-ECFP4 | 0.536 | - |
Looking on the GCN model performance for each OR (Table S3 in supplementary), we observe that many ORs have the maximum AUROC and PRC-AUC score (OR5A2, OR4D6) while others ORs obtained low PRC-AUC (OR56A1, OR52M1, OR56A4). The fact that some ORs have few compounds associated may facilitate the good performance for these odors.
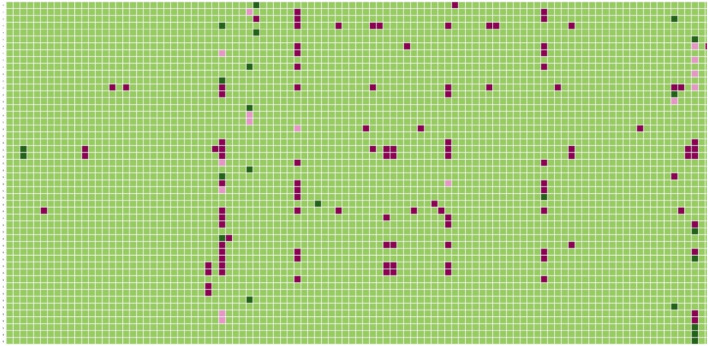
On the heatmap (Fig. S4 in supplementary), we can observe that some ligands are correctly predicted i.e., coffee difuran predicted active on OR1A1, butyrophenone on OR6A2, 4 phenyl-1 butanol on OR1G1, (E)-cinnamyl nitrile on OR1D2 and 4-tert-butyl cyclohexanone active on the human ortholog OR5D16 (olfr73 in mouse). A large set of compounds are wrongly predicted on OR5A1, OR52D1, OR56A2. In fact, these receptors are annotated to molecules with diverse physicochemical features, generating some difficulty to the models to discriminate between true positives and false positives. An example of the heatmap representation is depicted in Fig. 7.
Figure 7.
Heatmap representation of the performance prediction with the GCN model for a subset of the compound-olfactory receptors (ORs) associations. The ORs are represented on the X-axis and the compounds on the Y axis. A compound associated to an OR and correctly classified by the GCN model is colored with a dark green cell. A compound not associated to an OR and correctly predicted has a light green color. A compound linked to an OR and wrongly predicted by the model is represented in a pink color and a compound not linked to an OR and predicted to be associated to this odor is shown with a red color. The heatmap was developed with the python package seaborn (v.0.11.2): https://seaborn.pydata.org/63,64.
Results on receptors-odor notes relationship.
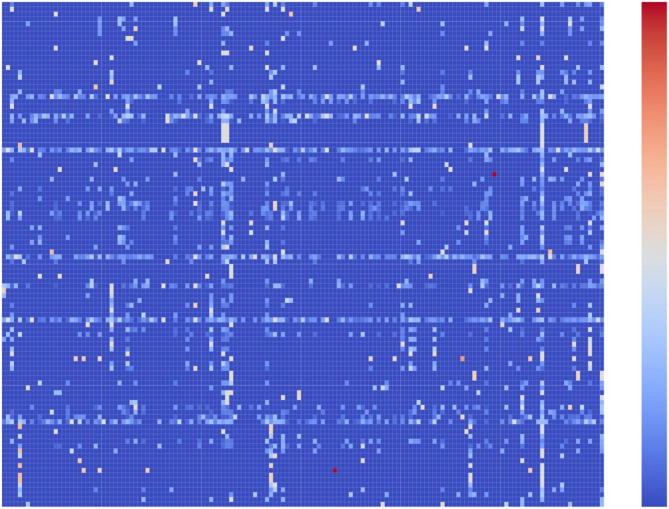
As 357 compounds targeting human olfactory receptors and related to odor notes were identified in our data sets, an odor-olfactory receptor pairwise score between each odor and each receptor i.e., the possible relation between odor notes and receptors, was computed (supplementary Table S4) and represented within a heatmap (Fig. 8). Globally, based on 151 odor notes and 104 ORs, such heatmap allows to suggest relation between olfactory receptors and odor notes due to the number of shared compounds. Some ORs seem more related to some odor notes than others. For example, the corn odor note is uniquely associate to OR1G1. The patchouli odor note is associated to OR5D16 and the cumin odor note is associated to OR1D2. The savory odor note is more associated with the OR1A1 receptor (OORP = 0.51) while waxy and woody odor notes are strongly associated with the OR2AT4 receptor (OORP = 0.51 and OORP = 0.52 respectively). Interestingly such matrix gives a score for each OR on each odor note. It means that a set of ORs can be suggested to a set of odor notes. For example, OR1G1 and OR1D2 are associate to more than 70 odor notes reflecting no high specificity of these ORs to odors. At the difference, OR10A6 is linked to balsamic, floral and hyacinth. OR1E3 is linked to almond, hawthorn, pungent and sweet and OR8D1 is strongly associated to burnt, carmellic, coffee, maple, sugar and sweet. From the literature, some of these potential associations have been confirmed. Triller et al. 2008 mentioned that OR1D2 is highly related to muguet65. Veithen et al. show that OR1D2 might be also related to floral, fruity, citrus66. In our study, in addition to these odor notes, high relation with lactonic, rose and peach are also observed. A patent suggested that the olfactory receptors R52L1, OR52E8, OR52B2, OR5112, OR52E1, OR52A5, OR56A5 are involved in the perception of human sweat67. In addition, it is claimed that chemicals with a carboxylic acids group could be the relation between these ORs and the sweat odor. In our analysis, the olfactory receptors OR117P and OR52B2 contribute in majority with the sweaty odor note.
Figure 8.
Heatmap representation of the odor note-olfactory receptor pairwise score for a set of 383 compounds targeting olfactory receptors and related to odor notes. The ORs are represented on the X-axis and the odor notes on the Y-axis. A red dot represents a high odor note-olfactory receptor pairwise score and a dark blue dot, no odor note-olfactory receptor relationship. The heatmap was developed with the python package seaborn (v.0.11.2): https://seaborn.pydata.org/63–65.
Comparison of models’ performance
To assess the performance of these models, we compared the results of our chemical-odor models to the DREAM Olfaction Prediction Challenge39, and our chemical-odor and OR-odor models to the recent ones reported by Kowalewski et al40.
About the chemical-odor model, we used the same 69 test chemicals from the DREAM Olfaction Prediction Challenge39 to evaluate our model performance. For the odor prediction, we obtained an average balanced accuracy (BA) of 0.71 using as positive the compounds up to the top 10% perception for an odor39 (supplementary Table S5). Compared to the recent AUC of 0.78 obtained by Kowalewski et al. our model has a little lower performance. Looking at the 19 perceptions from DREAM, our models have a relatively good BA (> 0.7) for ‘bakery’, ‘fish’, ‘garlic’, ‘acid’, ‘sweaty’, ‘amonia/urine’ ‘wood’ and ‘grass’. For the other perceptions, the BA is weaker. It can be explained by the fact that matching the 160 odors used in our study to the 19 perceptual odors considered in Keller et al. publication39 might increase the number of false positive rate. For example, the odors “cold”, “decayed” and “warm” are not specifically annotated in our odors collection and grouping some of the odors in our dataset might bring some noise in this comparison exercise.
About the ligand-OR model, Kowalewski et al. used the same external set of 69 chemicals to predict associated olfactory receptors to them. Having only the chemical-ORs prediction from their study (and not the experimental value) we could only compared their prediction to our model’s result for 23 olfactory receptors (supplementary Table S6). Interestingly, half of their prediction was retrieved in our models. In general, there models predicted around 3 times more chemical-OR relationship compared to our model (354 versus 120 chemical-OR predictions) for this set of olfactory receptors.
Finally, about the OR-odor, in the Kowalewski et al. publication, 34 human ORs-perception were predicted. Interestingly, compared to our results, we can observe similar OR-odor note relationships like for example OR52D1 with ‘animal’, ‘sweaty’, ‘rose’ and ‘violet’, OR2B11 with ‘coffee’ and OR2W1 with ‘spicy’, ‘clove’, ‘caramel’ and ‘cheesy’ among others. At the difference for others ORs, we obtained different relationships. For example, our study suggests that OR1A2 contribute in priority with the odors ‘aldehydic’, fatty’, ‘grassy’, ‘hay’, ‘ozone’ whereas in their studied, important relationships between OR1A2 and ‘warm’ and ‘sweet’ were reported. We suggest also that OR1D2, OR1G1, OR52D1 and OR6A2 could contribute to the odor note ‘fishy’ whereas there heatmap showed a higher contribution of OR2T34 and OR51E1.
Overall, the fact that different data sets of ligand-odor notes and ligand-olfactory receptors are used in both studies has probably an impact on the results. Further experiments should help in the precision of these predictive models.
Discussion–conclusion
Using, a large data set of 5955 compounds, 160 odors and 106 olfactory receptors, machine learning models based on artificial intelligence i.e., Random Forest, CNN and GCN approaches were developed. Such models can then be used to predict the odor note(s) and olfactory receptor(s) associated for a new compound using the chemical structure of it. In addition, based the correspondence of odor notes and ORs associated for a set of 389 compounds, a score was computed for each odor note-OR combination allowing to decipher the combinatorial relationship between olfactory receptors and odor notes.
Although the results are promising, there are still some limitations and the models will need to be optimized in the aim to increase their performance.
First, the perception of an odor is highly dependent of an individual and odors annotation to a compound are suggestive, depending of ethnicity, alimentary behavior, age68–72. Indeed, the definition of some odor notes might be fuzzy (cheese vs cheesy). Recently, 540 individuals were asked to rate the intensity and pleasantness of 9 musk compounds and their ORs were sequenced in the aim to identify genetic variations that could explain the genetic susceptibility to odor perception73. Furthermore, it is well admitted that an odor results from the perception of a mixture of molecules, which give more complexity in such classification74. Grouping some odors rationally, in more general categories, can improve the performance and the robustness of the GCN models.
Secondly, about the ORs, the number of compounds with known activity on ORs is still low. Mori estimated that more than 400 000 different compounds are odorous to the human nose75. Still, we collected only a couple of hundred of molecules with bioactivity on ORs. Increasing the number of functional ORs experiments for large set of compounds would definitively improve the quality of the models. We have noticed that some ORs are highly investigated and other less9,76. For example, OR1A177, OR1D278, OR1G179, OR2W180, OR2M381 have been reported to be active by more than 100 compounds. At the opposite, there are 72 ORs for which only one compound has been tested active. Developing a GCN model with ORs having enough compounds tested (for example > 5) could improve the model performance on ORs. Another possibility would be to increase the chemical-OR bioactivities by studying the transcriptional profile modulation of ORs in vivo i.e., in olfactory sensory neurons (OSN) in vertebrates. Recent studies have been reported on this direction and identified the full repertoire of receptors activated by a given odorant82,83. Although encouraging, the number of compounds with transcriptional profile is still limited.
In third, the stereochemistry of a molecule is may be not optimal in our data set. It has been reported that stereoisomers of a chemical can be related to different odors84,85. For example, the R-carvone is related to minty odor while its enantiomer, the S-carvone, has a caraway odor77. Although enantiomeric compounds have similar chemical functions, it has been reported that as few as 5% of enantiomer couples have a similar smell86,87. It is possible that the racemic form of some of the compounds, used in this study, has been considered and it might cause a mis classification to some odors.
About machine learning approaches, CNN and GCN are the latest and powerful machine learning approaches. GCN seems to outperform CNN and RF in our study. Many odorant-odor notes models have been described recently. Sharma et al. have reported a model based on 5185 chemical and 542 smell using a Deep Neural Network (DNN) algorithm with promising results42. The performance is a little lower with a AUROC = 0.76. However, one advantage of DNN is, it automatically identifies optimal features overcoming the problem of feature selection. On a more restricted data set (476 chemicals and 21 odor notes), Keller et al. obtained an AUROC of 0.83 based on a Random Forest method39 and Sanchez-Lengeling et al. described a GNN model with an AUROC = 0.89 using 5030 chemicals and 138 smells43. Models based on olfactory receptors are more limited. Kowalewski et al. developed a SVM model using 150 odorants and 34 human olfactory receptors with an AUC = 0.8840. Recently, a conglomerate of artificial intelligence driven prediction engines for olfactory decoding was reported, including odorant-OR interactions predictions based on structure-based approaches88. The models showed good performance with an AUC = 0.87 for ORs and an AUC = 0.94 for smell based on DNN methods.
Overall, these results illustrate the potential of artificial intelligence to decipher the relationship of odorant molecules with olfactory receptors and smell perception. Associating to several previous studies carried out by other research groups18,39–41, our study provides an increase in the knowledge of the links between odor notes, molecular structures of odorants and target olfactory receptors of mammals. Especially, thanks to largest data as well in number of odorants than in number of olfactory receptors, we show that our model is able to correctly connect numerous pairs odorant-OR, and now to predict other new pairs.
However, models based on artificial intelligence can show some limits with odors and receptors that are not well represented by chemicals. As recently pointed by Gerkin89, it is necessary to use a large volume of odorant molecules with the corresponding odorant description as several as odor notes (or odor attributes). Moreover, the molecular properties of the odorants must be described by a large number of molecular descriptors able to report all their structural characteristics.
Expanding the knowledge of our sense of smell by combining different sources of data from chemical biology (proteome-transcriptome) and human perception with advanced computational approaches will move forward the identification of the complete olfactory repertoire associated to the human smell perception.
Supplementary Information
Author contributions
O.T. et K.A. planned the study. R.A. performed the analysis. R.A. and O.T. wrote the main manuscript text. All authors reviewed the manuscript.
Funding
The authors received funding for this work by Agence Nationale de la Recherche, ANR-18-CE21-0006, project MULTIMIX (https://anr.fr/en).
Data availability
The datasets compiled in this study are available for the scientific community in supplementary Table S1. We hope that it will be a good resource for further investigations.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-23176-y.
References
- 1.Zarzo M. The sense of smell: Molecular basis of odorant recognition. Biol. Rev. 2007;82:455–479. doi: 10.1111/j.1469-185X.2007.00019.x. [DOI] [PubMed] [Google Scholar]
- 2.Croy I, Nordin S, Hummel T. Oflactory disorders and quality of life—an updated review. Chem. Senses. 2014;39:185–194. doi: 10.1093/chemse/bjt072. [DOI] [PubMed] [Google Scholar]
- 3.Glezer I, Bruni-Cardoso A, Schechtman D, Malnic B. Viral infection and smell loss: The case of COVID-19. J. Neurochem. 2021;157:930–943. doi: 10.1111/jnc.15197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Menashe I, Lancet D. Variations in the human olfactory receptor pathway. Cell Mol. Life Sc. 2006;63:1485–1493. doi: 10.1007/s00018-006-6111-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Padmanabhan K, et al. Centrifugal inputs to the main Olfactory bulb revealed through whole brain circuit-mapping. Front. Neuroanat. 2019;12:115. doi: 10.3389/fnana.2018.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sato T, et al. Architecture of odor information processing in the olfactory system. Anat. Sci. Int. 2008;83:195–206. doi: 10.1111/j.1447-073X.2007.00215.x. [DOI] [PubMed] [Google Scholar]
- 7.Murthy VN. Olfactory maps in the brain. Ann. Rev. Neurosci. 2011;34:233–258. doi: 10.1146/annurev-neuro-061010-113738. [DOI] [PubMed] [Google Scholar]
- 8.Breer H. Olfactory receptors: Molecular basis for recognition and discrimination of odors. Anal. Bioanal. Chem. 2003;377:427–433. doi: 10.1007/s00216-003-2113-9. [DOI] [PubMed] [Google Scholar]
- 9.Saito H, Chi Q, Zhuang H, Matsunami H, Mainland JD. Odor coding by a mammalian receptor repertoire. Sci Signal. 2009;2:1–14. doi: 10.1126/scisignal.2000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tromelin A. Odour perception: A review of an intricate signalling pathway: Olfactory system and odour perception. Flavour Fragr J. 2016;31:107–119. [Google Scholar]
- 11.Bushdid C, Magnasco MO, Vosshall LB, Keller A. Human can discriminate more than 1 trillion olfactory stimuli. Science. 2014;44:1370–1372. doi: 10.1126/science.1249168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buck L, Axel R. A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 13.DeMaria S, Ngai J. The cell biology of smell. J. Cell Biol. 2010;191:443–452. doi: 10.1083/jcb.201008163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polak EH. Mutiple profile-multiple receptor site model for vertebrate olfaction. J. Theor. Biol. 1973;40:469–484. doi: 10.1016/0022-5193(73)90005-2. [DOI] [PubMed] [Google Scholar]
- 15.Malnic B, Hirono J, Sato T, Buck LB. Combinatorial receptor codes for odors. Cell. 1999;96:713–723. doi: 10.1016/s0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- 16.Furudono Y, Sone Y, Takizawa K, Hirono J, Sato T. Relationship between peripheral receptor code and perceived odor quality. Chem. Senses. 2009;34:151–158. doi: 10.1093/chemse/bjn071. [DOI] [PubMed] [Google Scholar]
- 17.Zhuang HY, Matsunami H. (2007) Synergism of accessory factors in functional expression of mammalian odorant receptors. J. Biol. Chem. 2009;282:15284–15293. doi: 10.1074/jbc.M700386200. [DOI] [PubMed] [Google Scholar]
- 18.Gutierrez ED, Dhurandhar A, Keller A, Meyer P, Cecchi GA. Predicting natural language descriptions of mono-molecular odorants. Nat. Commun. 2018;9:4979. doi: 10.1038/s41467-018-07439-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thieme, A., Korn, D., Alves, V., Muratov, E., Tropsha, A. Novel classification of mono-molecular odorants using standardized semantic profiles. (2022).
- 20.Kaeppler K. Crossmodal associations between olfaction and vision: Color and shape visualizations of odors. Chemosens. Percept. 2018;11:95–111. [Google Scholar]
- 21.Barwich AS. A critique of olfactory objects. Front. Psychol. 2019;10:1337. doi: 10.3389/fpsyg.2019.01337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas-Danguin T, Sinding C, Romagny S, El Mountassir F, Atanasova B, Le Berre E, Le Bon AM, Coureaud G. The perception of odor objects in everyday life: A review on the processing of odor mixtures. Front. Psychol. 2014;5:504. doi: 10.3389/fpsyg.2014.00504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of drosophila odorant receptors in vivo. PLoS Biol. 2006;4:240–257. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yarmolinsky DA, Zuker CS, Ryba NJP. Common sense about taste: From mammals to insects. Cell. 2009;139:234–244. doi: 10.1016/j.cell.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sinakevitch I, Bjorklund GR, Newbern JM, Gerkin RC, Smith BH. Comparative study of chemical neuroanatomy of the olfactory neuropil in mouse, honey bee, and human. Biol. Cybern. 2018;112:127–140. doi: 10.1007/s00422-017-0728-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis RL. Olfactory learning. Neuron. 2004;44:31–48. doi: 10.1016/j.neuron.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 27.Benbernou N, et al. Functional analysis of a subset of canine olfactory receptor genes. J. Hered. 2007;98:500–505. doi: 10.1093/jhered/esm054. [DOI] [PubMed] [Google Scholar]
- 28.Araneda RC, Peterlin Z, Zhang X, Chesler A, Firestein S. A pharmacological profile of the aldehyde receptor repertoire in rat olfactory epithelium. J. Physiol. 2004;555:743–756. doi: 10.1113/jphysiol.2003.058040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacquier V, Pick H, Vogel H. Characterization of an extended receptive ligand repertoire of the human olfactory receptor OR17-40 comprising structurally related compounds. J. Neurochem. 2006;97:537–544. doi: 10.1111/j.1471-4159.2006.03771.x. [DOI] [PubMed] [Google Scholar]
- 30.Krautwurst D, Yau KW, Reed RR. Identification of ligands for olfactory receptors by functional expression of a receptor library. Cell. 1998;95:917–926. doi: 10.1016/s0092-8674(00)81716-x. [DOI] [PubMed] [Google Scholar]
- 31.Wetzel CH, et al. Functional expression and characterization of a drosophila odorant receptor in a heterologous cell system. Proc. Natl. Acad. Sci. USA. 2001;98:9377–9380. doi: 10.1073/pnas.151103998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pashkovski SL, et al. Structure and flexibility in cortical representations of odour space. Nature. 2020;583:253–258. doi: 10.1038/s41586-020-2451-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keller A, Vosshall LB. Olfactory perception on chemically diverse molecules. BMC Neurosci. 2016;17:55. doi: 10.1186/s12868-016-0287-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kraft P, Bajgrowicz JA, Denis C, Frater G. Odds and trends: Recent developments in the chemistry of odorants. Angew. Chem. 2000;39:2981–3010. doi: 10.1002/1521-3773(20000901)39:17<2980::aid-anie2980>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 35.Khan RM, et al. Predicting odor pleasantness from odorant structure: Pleasantness as a reflection of the physical world. J. Neurosci. 2007;27:10015–10023. doi: 10.1523/JNEUROSCI.1158-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castro JB, Ramanathan A, Chennubhotla CS. Categorical dimensions of human odor descriptor space revealed by non-negative matrix factorization. PLoS ONE. 2013;8:1. doi: 10.1371/journal.pone.0073289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez-Mayorga K, et al. Characterization of a comprehensive flavor database. J. Chemometr. 2011;25:550–560. [Google Scholar]
- 38.Tromelin A, Chabanet C, Audouze K, Koensgen F, Guichard E. Multivariate statistical analysis of a large odorants database aimed at revealing similarities and links between odorants and odors. Flav. Frag. J. 2018;33:106–126. [Google Scholar]
- 39.Keller A, et al. Predicting human olfactory perception from chemical features of odor molecules. Sciences. 2017;355:820–826. doi: 10.1126/science.aal2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kowalewski J, Huynh B, Ray A. A system-wide understanding of the Human olfactory percept chemical space. Chem. Senses. 2021;46:1. doi: 10.1093/chemse/bjab007. [DOI] [PubMed] [Google Scholar]
- 41.Kowalewski J, Ray A. Predicting human olfactory perception from activities of odorant receptors. iScience. 2020;23:101361. doi: 10.1016/j.isci.2020.101361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharma A, Kumar R, Ranjta S, Varadwaj PK. SMILES to Smell: decoding the structure-odor relationship of chemical compounds using the deep neural network approach. J. Chem. Inf. Model. 2021;61:676–688. doi: 10.1021/acs.jcim.0c01288. [DOI] [PubMed] [Google Scholar]
- 43.Sanchez-Lengeling B, Wei JN, Lee BK, Gherkin RC, Asputu-Guzik A, Wiltschko AB. Machine learning for scent: learning generalizable perceptual representations of small molecules. Arxiv. 2019;1910:10685. [Google Scholar]
- 44.Tran, N., Kepple, D., Sergey, A. S., & Koulakov, A. A. DeepNose: Using artificial neural networks to represent the space of odorants. In Proceedings of 36th International Conference on Machine Learning, Long Beach, California, PMLR97 (2019).
- 45.Jing Y, Bian Y, Hu Z, Wang L, Xie XQ. Deep learning for drug desing: An Artificial Intelligence paradigm for drug discovery in the big data era. AAPS. 2018;20:58. doi: 10.1208/s12248-018-0210-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.The Good Scents Company, Available online: http://www.thegoodscentscompany.com/.
- 47.Leffingwell & Associates. Flavor-Base. 9th Edition. Available online: http://www.leffingwell.com/ flavbase.htm.
- 48.Goodman JM, Pletnev I, Thiessen P, Bolton E, Heller SR. InChI version 1.06: now more than 99.99% reliable. J. Cheminf. 2021;13:40. doi: 10.1186/s13321-021-00517-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skoufos E, Marenco L, Nadkarni PM, Miller PL, Shepherd GM. Olfactory receptor database: A sensory chemoreceptor resource. Nucl. Acis Res. 2000;28:341–343. doi: 10.1093/nar/28.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu X, Su X, Wang F, Huang Z, Wang Q, Li Z, et al. ODORactor: A web server for deciphering olfactory coding. Bioinformatics. 2011;27:2302–2303. doi: 10.1093/bioinformatics/btr385. [DOI] [PubMed] [Google Scholar]
- 51.Modena D, Trentini M, Corsini M, Bombaci A, Giorgetti A. OlfactionDB: A database of olfactory receptors and their ligands. Adv. Life Sci. 2011;1:1–5. [Google Scholar]
- 52.Rogers D, Hahn M. Extended-connectivity fingerprints. J. Chem. Inf. Model. 2010;50:742–754. doi: 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
- 53.Rugard M, Jaylet T, Taboureau O, Tromelin A, Audouze K. Smell compounds classification using UMAP to increase knowledge of odors and molecular structures linkages. PLoS ONE. 2021;16:e0252486. doi: 10.1371/journal.pone.0252486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Landrum, G. 2010. RDKit: Open-source cheminformatics. https://www.rdkit.org (2010).
- 55.Pedregosa F, et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 56.Wu Z, Ramsundar B, Feinberg E, Gomes J, Geniesse CP, et al. MoleculeNet: a benchmark for molecular machine learning. Chem. Sci. 2017;9:513–530. doi: 10.1039/c7sc02664a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abadi, M., Agarwal, A., Barham, P., Brevdo, E., & Chen, Z., et al. TensorFlow: Large-scale machine learning on heterogeneous systems. (2015). http://download.tensorflow.org/paper/whitepaper2015.pdf.
- 58.Ilyas N, Shahzad A, Kim K. Convolutional neural network-based image crowd counting: Review, categorization, analysis and performance evaluation. Sensors. 2019;20:43. doi: 10.3390/s20010043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kearnes S, McCloskey K, Berndl M, Pande V, Riley P. Molecular graph convolutions: Moving beyond fingerprints. J. Comput Aided Mol. Des. 2016;30:595–608. doi: 10.1007/s10822-016-9938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bokeh Development Team. Bokeh: Python library for interactive visualization. (2014). http://www.bokeh.pydata.org.
- 61.Plotly Technologies Inc. Collaborative data science publisher: Plotly technologies Inc. place of publication: Montréal, Qc (2015) URL, 2015. https://plot.ly.
- 62.Massberg D, Hatt H. Human olfactory receptors: Novel cellular functions outside of the nose. Physiol. Rev. 2018;98:1739–1763. doi: 10.1152/physrev.00013.2017. [DOI] [PubMed] [Google Scholar]
- 63.Waskom ML. Seaborn: Statistical data visualization. JOSS. 2021;6(60):3021. [Google Scholar]
- 64.Hunter JD. Matplotlib: A 2D graphics environment. Comput Sci. Eng. 2007;9(3):90–95. [Google Scholar]
- 65.Triller A, Boulden EA, Churchill A, Hatt H, Englund J, Spehr M, Sell CS. Odorant-receptor interactions and odor percept: A chemical perspective. Chem Biodivers. 2008;5:862–886. doi: 10.1002/cbdv.200890101. [DOI] [PubMed] [Google Scholar]
- 66.Veithen A, Wilin F, Philippeau M, Chatelain P. OR1D2 is a broadly tuned human olfactory receptor. Chem. Senses. 2015;40:262–263. [Google Scholar]
- 67.Chatelain, P., Veithen, A. Olfactory receptors involved in the perception of sweat carboxylic acids and the use thereof. Patent EP3004157B1. 2013.
- 68.Young JM, Trask BJ. The sense of smell: Genomics of vertebrate odorant receptors. Hum. Mol. Gen. 2002;11:1153–1160. doi: 10.1093/hmg/11.10.1153. [DOI] [PubMed] [Google Scholar]
- 69.Knape K, Beyer A, Stary A, Buchbauer G, Wolschann P. Genomics of selected human odorant receptors. Monatshefte Fur Chemie. 2008;139:1537–1544. [Google Scholar]
- 70.Ferdenzi C, Roberts SC, Schirmer A, Delplanque S, Cekic S, Porcherot C, Cayeux I, Sander D, Grandjean D. Variability of affective responses to odors: Culture, gender, and olfactory knowledge. Chem. Senses. 2013;38:175–186. doi: 10.1093/chemse/bjs083. [DOI] [PubMed] [Google Scholar]
- 71.Wackermannova M, Pinc L, Jebavy L. Olfactory sensitivity in mammalian species. Physiol. Res. 2016;65:369–390. doi: 10.33549/physiolres.932955. [DOI] [PubMed] [Google Scholar]
- 72.Trimmer C, et al. Genetic variation across the human olfactory receptor repertoire alters odor perception. Proc. Natl. Acad Sci. USA. 2019;116:9475–9480. doi: 10.1073/pnas.1804106115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mainland J. Identifying key olfactory receptors in odor perception using machine learning. Chem. Senses. 2020;45:141–141. [Google Scholar]
- 74.Thomas-Danguin T, et al. The perception of odor objects in everyday life: a review on the processing of odor mixtures. Front. Psychol. 2014;5:504. doi: 10.3389/fpsyg.2014.00504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mori K. Grouping of odorant receptors: Odour maps in the mammalian olfactory bulb. Biochem Soc Trans. 2003;31:134–136. doi: 10.1042/bst0310134. [DOI] [PubMed] [Google Scholar]
- 76.Trimmer C, Mainland JD. Simplifying the Odor Landscape. Chem. Senses. 2017;42:177–179. doi: 10.1093/chemse/bjx002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Geithe C, Protze J, Kreuchwig F, Krause G, Krautwurst D. Structural determinants of conserved enantiomer-selective carvone binding pocket in the human odorant receptor OR1A1. Cell Mol. Life Sci. 2017;74:4209–4229. doi: 10.1007/s00018-017-2576-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Triller A, Boulden EA, Churchill A, Hatt H, Englund J, Spehr M, Sell CS. Odorant-receptor interactions and odor percept: A chemical perspective. Chem. Biodivers. 2008;5(6):862–886. doi: 10.1002/cbdv.200890101. [DOI] [PubMed] [Google Scholar]
- 79.Sanz G, Schlegel C, Pernollet JC, Briand L. Comparison of odorant specificity of two human olfactory receptors from different phylogenetic classes and evidence for antagonism. Chem. Senses. 2005;30:69–80. doi: 10.1093/chemse/bji002. [DOI] [PubMed] [Google Scholar]
- 80.Oh SJ. Computational evaluation of interactions between olfactory receptor OR2W1 and its ligands. Genomics Inform. 2021;19:e9. doi: 10.5808/gi.21026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Noe F, Polster J, Geithe C, Kotthoff M, Schieberle P, Krautwurst D. OR2M3: A highly specific and narrowly tuned human odorant receptor for the sensitive detection of onion key food odorant 3-mercapto-2-methylpentan-1-ol. Chem. Senses. 2017;42:195–210. doi: 10.1093/chemse/bjw118. [DOI] [PubMed] [Google Scholar]
- 82.Von der Weid B, et al. Large-scale transcriptional profiling of chemosensory neurons identifies receptor-ligand pairs in vivo. Nat. Neurosci. 2015;18:1455–1463. doi: 10.1038/nn.4100. [DOI] [PubMed] [Google Scholar]
- 83.Jiang Y, Gong NN, Hu XS, Ni MJ, Pasi R, Matsunani H. Molecular profiling of activated olfactory neurons identifies odorant receptors for odors in vivo. Nat. Neurosci. 2015;18:1446–1454. doi: 10.1038/nn.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Laska M. Olfactory discrimination ability of human subjects for enantiomers with an isopropenyl group at the chiral center. Chem. Senses. 2004;29:143–152. doi: 10.1093/chemse/bjh019. [DOI] [PubMed] [Google Scholar]
- 85.Laska M, Teubner P. Olfactory discrimination ability for homologous series of aliphatic alcohols and aldehydes. Chem. Senses. 1999;24:263–270. doi: 10.1093/chemse/24.3.263. [DOI] [PubMed] [Google Scholar]
- 86.Brookes JC, Horsfield AP, Stoneham AM. Odour character differences for enantiomers correlate with molecular flexibility. J. R. Soc. Interface. 2009;6:75–86. doi: 10.1098/rsif.2008.0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Genva M, Kemene TK, Deleu M, Lins L, Fauconnier ML. Is it possible to predict the odor of a molecule on the basis of its structure? Int. J. Mol. Sci. 2019;20:3018. doi: 10.3390/ijms20123018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gupta R, et al. OdoriFy: A conglomerate of Artificial Intelligence-driven prediction engines for olfactory decoding. J. Biol. Chem. 2021;297:100956. doi: 10.1016/j.jbc.2021.100956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gerkin RC. Parsing sage and rosemary in time: The machine learning race to crack olfactory perception. Chem. Senses. 2021;46:1. doi: 10.1093/chemse/bjab020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets compiled in this study are available for the scientific community in supplementary Table S1. We hope that it will be a good resource for further investigations.