Abstract
To expand the genomic information of Hypericaceae, particularly on Cratoxylum, we characterized seven novel complete plastid genomes (plastomes) of five Cratoxylum and two of its allied taxa, including C. arborescens, C. formosum subsp. formosum, C. formosum subsp. pruniflorum, C. maingayi, C. sumatranum, Hypericum hookerianum, and Triadenum breviflorum. For Cratoxylum, the plastomes ranged from 156,962 to 157,792 bp in length. Genomic structure and gene contents were observed in the five plastomes, and were comprised of 128–129 genes, which includes 83–84 protein-coding (CDS), 37 tRNA, and eight rRNA genes. The plastomes of H. hookerianum and T. breviflorum were 138,260 bp and 167,693 bp, respectively. A total of 110 and 127 genes included 72 and 82 CDS, 34 and 37 tRNA, as well as four and eight rRNA genes. The reconstruction of the phylogenetic trees using maximum likelihood (ML) and Bayesian inference (BI) trees based on the concatenated CDS and internal transcribed spacer (ITS) sequences that were analyzed separately have revealed the same topology structure at genus level; Cratoxylum is monophyletic. However, C. formosum subsp. pruniflorum was not clustered together with its origin, raising doubt that it should be treated as a distinct species, C. pruniflorum based on molecular evidence that was supported by morphological descriptions.
Subject terms: Biological techniques, Cell biology, Genetics, Molecular biology, Plant sciences
Introduction
The family Hypericaceae Jussieu comprises nine genera and over 500 species worldwide. In general, members of Hypericaceae are further categorized into three different tribes viz. Cratoxyleae Bentham & J.D. Hooker, Hypericeae Choisy, and Vismieae Choicy1. As the smallest tribe in the family, two genera are recognized in Cratoxyleae, viz. Cratoxylum Blume and the monotypic genus, Eliea Cambess2. At present, there are seven accepted species of Cratoxylum Blume (Hypericaceae, Malpighiales), and three of them are recognized with at least two intraspecific identities3. Members of Cratoxylum are native to the tropical Asia region, widespread from India through South China to Malesia and are commonly soughed for their wood as a source of timber and charcoal production4. The great adaptability in harsh environments and fast-growing performance has warrant some of these species as potential replanting species that are useful for peatland rehabilitation strategies5,6.
Despite the potential as useful rehabilitation agents in peat swamp forests, genetic studies on Cratoxylum are limited. Genetic data of Cratoxylum are only restricted to short gene sequences derived from the plastid, mitochondrial and nuclear regions of either C. arborescens (Vahl) Blume or C. cochinchinense (Lour.) Blume as representative species of its genus in the reconstruction of the phylogenetic tree of Malpighiales7–9. The lack of the phylogenetic studies among species of Cratoxylum has hindered our understanding of this genus at its genetic level.
The plastid genome (plastome) is a valuable resource for molecular taxonomy research. Angiosperm plastomes are circular haploid genomes with a large single copy (LSC) region, two inverted repeats (IR), and a small single copy (SSC) region that are typically small in between 110–240 kbp in length10. Recently, researchers have shown great interest in obtaining the complete plastome sequences through next-generation sequencing technique. This is because when compared to short gene sequences, genome-scale datasets that are used in phylogenetics contain larger number of single nucleotide polymorphism, which could contribute to the reconstruction of a well-supported phylogenetic tree11. Owing to the advancement in sequencing technique and the availability of useful bioinformatic programs to aid in the assembly and annotation of the plastomes, to date, many complete plastome sequences have been made available publicly to decipher ambiguous phylogenetic relationships in complicated genera12,13. On the other hand, the nuclear DNA internal transcribed spacer (ITS) region has served to be useful in revealing the biparental inheritance of plants at a nuclear genome level14. Among all nuclear genes, amplification of the ITS sequence is known to be easier, and the genetic information provided is also useful to delimit individuals at an intraspecific level15.
Although records on published complete plastomes are increasing substantially over the years, genome data for Hypericaceae is still lacking. To date, published records on the complete plastome sequences of Cratoxylum are only limited to C. cochinchinense, in which at least three genome sequences of different accessions are available in the NCBI GenBank database (as of September 2021). In view of the need to expand the genomic information of Hypericaceae, we further sequenced and characterized the plastomes of five taxa of Cratoxylum, including C. arborescens, C. formosum subsp. formosum (Jack) Benth. & Hook.f. ex Dyer, C. formosum subsp. pruniflorum (Kurz) Gogelein, C. maingayi Dyer, and C. sumatranum (Jack) Blume, as well as two closely-related species, Hypericum hookerianum Wight & Arn. and Triadenum breviflorum Wall. ex Dyer. In order to reveal the phylogenetic relationship among species of Cratoxylum, we further performed phylogenetic analysis using the plastid protein-coding sequence (CDS) dataset and the nuclear DNA internal transcribed spacer (ITS) sequence region. The findings of this work will serve as important reference for the phylogenetic and evolutionary studies of Hypericaceae and Malpighiales.
Results and discussion
Plastome features
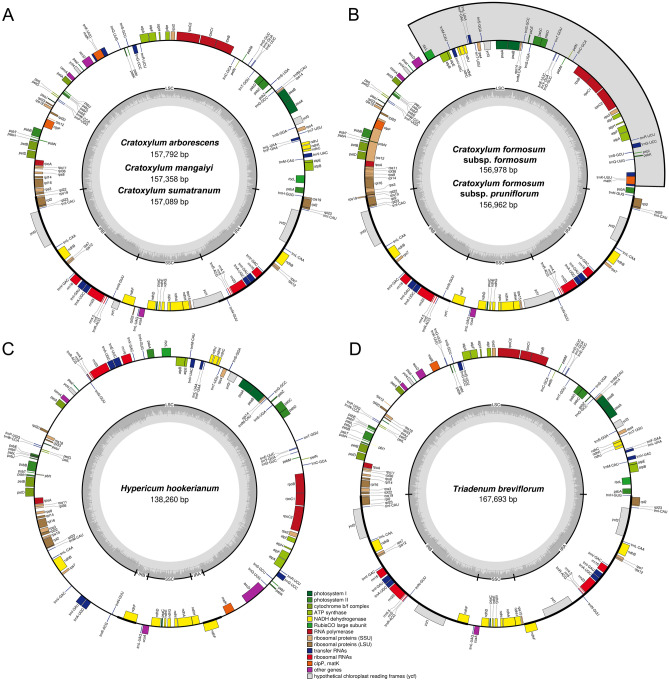
All seven plastomes obtained from this study exhibited a typical quadripartite structure, which comprised of a large single-copy (LSC) and a small single-copy (SSC) region that are separated by a pair of inverted repeats (IR) (Fig. 1). Plastome sizes were between 156,962 bp (C. formosum subsp. pruniflorum and 157,792 bp (C. arborescens) among the five taxa of Cratoxylum, while H. hookerianum and T. breviflorum were 138,260 bp and 167,693 bp in length, respectively (Table 1). A total of 128–129 genes were predicted in the plastome of the five taxa of Cratoxylum, which comprised of 83–84 CDS, 37 tRNA, and eight rRNA genes. Cratoxylum formosum subsp. formosum and C. formosum subsp. pruniflorum were short of one CDS compared to the other three species of Cratoxylum, which was the rpl32 gene that should be located at the SSC region (Table 2). There were 15 genes, including nine CDS and six tRNA genes, that contained one intron, while two genes, clpP and ycf3, contained two introns.
Figure 1.
Genome structure and gene map of the seven taxa of Hypericaceae used in this study. Cratoxylum arborescens, C. maingayi, C. sumatranum (A); Cratoxylum formosum subsp. formosum, C. formosum subsp. pruniflorum (B); Hypericum hookerianum (C); Triadenum breviflorum (D). The inside circle genes are transcribed clockwise, and the outside circle genes are transcribed counter-clockwise. The color codes describe different functional groups of the genes. The thick lines indicate the boundary of the inverted repeats (IRA and IRB), demarcated between the large single-copy (LSC) and small single-copy (SSC) regions. The dark gray area in the inner circle represents genomic GC content, whereas light gray indicates AT content.
Table 1.
General characteristics of complete plastid genomes of the seven taxa of Hypericaceae obtained in this study.
| Collector and collection number | Source of origin | Plastome features | GenBank accession number | ||||||
|---|---|---|---|---|---|---|---|---|---|
| CDS | tRNA | rRNA | Total length (bp) | GC content (%) | Plastome | ITS | |||
| Cratoxulum arborescens | Syazwan; SAS678 | Selangor, Malaysia | 84 | 37 | 8 | 157,792 | 36.1 | MZ703418 | MZ674200 |
| Cratoxylum formosum subsp. formosum | A. Chaveerach; 1089 | Udonthani, Thailand | 83 | 37 | 8 | 156,978 | 36.3 | MZ703419 | MZ674201 |
| Cratoxylum formosum subsp. pruniflorum | A. Chaveerach; 1090 | Udonthani, Thailand | 83 | 37 | 8 | 156,962 | 36.3 | MZ703416 | MZ674202 |
| Cratoxylum maingayi | Syazwan; SAS679 | Selangor, Malaysia | 84 | 37 | 8 | 157,358 | 36.2 | MZ703417 | MZ674203 |
| Cratoxylum sumatranum | A. Chaveerach; 1091 | Udonthani, Thailand | 84 | 37 | 8 | 157,089 | 36.3 | MZ703415 | MZ674204 |
| Hypericum hookerianum | A. Chaveerach; 1092 | Chiang Mai, Thailand | 72 | 34 | 4 | 138,260 | 38.1 | MZ714015 | MZ703053 |
| Triadenum breviflorum | Zhang et al.; TanCM704 | Jiangxi, China | 82 | 37 | 8 | 167,693 | 37.4 | MZ714016 | OM980718 |
Table 2.
Genes predicted in complete plastid genome of the five taxa of Cratoxylum used in this study.
| Category | Group of function | Genes |
|---|---|---|
| Self-replication related genes | Large subunit of ribosome proteins | rpl2(× 2)*, rpl14, rpl16*, rpl20, rpl22, rpl23(× 2), rpl32a, rpl33, rpl36 |
| Small subunit of ribosomal proteins | rps2, rps3, rps4, rps7(× 2)*, rps8, rps11, rps12, rps14, rps15, rps18, rps19(2) | |
| DNA-dependent RNA polymerase | rpoA, rpoB, rpoC1*, rpoC2 | |
| rRNA genes | rrn4.5(× 2), rrn5(× 2), rrn16(× 2), rrn23(× 2) | |
| tRNA gene | trnA-UGC(× 2)*, trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnfM-CAU, trnG-GCC, trnG-UCC*, trnH-GUG, trnI-CAU(× 2), trnI-GAU(× 2)*, trnK-UUU*, trnL-CAA(× 2), trnL-UAA*, trnL-UAG, trnM-CAU, trnN-GUU(× 2), trnP-UGG, trnQ-UUG, trnR-ACG(× 2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC(2), trnV-UAC*, trnW-CCA, trnY-GUA | |
| Photosynthesis related genes | Photosystem I | psaA, psaB, psaC, psaI, psaJ |
| Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| NADH oxidoreductase | ndhA*, ndhB(× 2)*, ndhC, ndhD, ndhE, ndhF(× 2), ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Cytochrome b6/f complex | petA, petB*, petD*, petG, petL, petN | |
| Cytochrome c synthesis | ccsA | |
| ATP synthase | atpA, atpB, atpE, atpF*, atpH, atpI | |
| Rubisco | rbcL | |
| Other genes | Maturase | matK |
| Protease | clpP** | |
| Envelope membrane protein | cemA | |
| Subunit acetyl-CoA-carboxylase | accD | |
| Unknown function genes | Conserved hypothetical chloroplast reading frames | ycf1, ycf2(× 2), ycf3**, ycf4 |
Genes that contain duplicates are indicated in parenthesis. *Indicates gene containing single intron; **Indicates gene containing two introns; aIndicates gene not found in Cratoxylum formosum subsp. formosum and C. formosum subsp. pruniflorum.
Although the gene content in plastome of Cratoxylum was consistent across the five taxa examined in this study, there was an inversed gene block arrangement detected in the LSC region, between rbcL and trnK-UUU genes (Fig. 1). The inversed gene block was approximately 55,000 bp in length, containing 28 CDS and 19 tRNA genes. By comparing to other plastomes of closely related families, we identified that the gene arrangement for the gene block in C. cochinchinense, C. formosum subsp. formosum, and C. formosum subsp. pruniflorum was similar to those of Bonnetiaceae, Calophyllaceae, Chrysobalanaceae, and Clusiaceae, i.e. Bonnetia paniculata (GenBank accession no. MK995182), Caraipa heterocarpa (GenBank accession no. MW853787), Garcinia mangostana (GenBank accession no. KX822787), and Licania micrantha (GenBank accession no. KX180080); while the gene arrangement for the gene block in C. arborescens, C. maingayi, and C. sumatrana was identical to those of Podostemaceae, i.e. Marathrum capillaceum (GenBank accession no. MN165813) and Tristicha trifaria (GenBank accession no. MK995179). This finding was congruent with a previous work, in which gene block inversion between rbcL and accD was observed in two clusioid families, including Hypericaceae and Podostemaceae, as well as Papilionoideae16. For H. hookerianum and T. breviflorum, a total of 110 and 127 genes were predicted, including 72 and 82 CDS, 34 and 37 tRNA, as well as four and eight rRNA genes, respectively. The GC content of the plastome for the five taxa of Cratoxylum ranged between 36.1 and 36.3%, while GC content for the plastomes of H. hookerianum and T. breviflorum was 38.1% and 37.4%, respectively.
Short and large sequence repeats
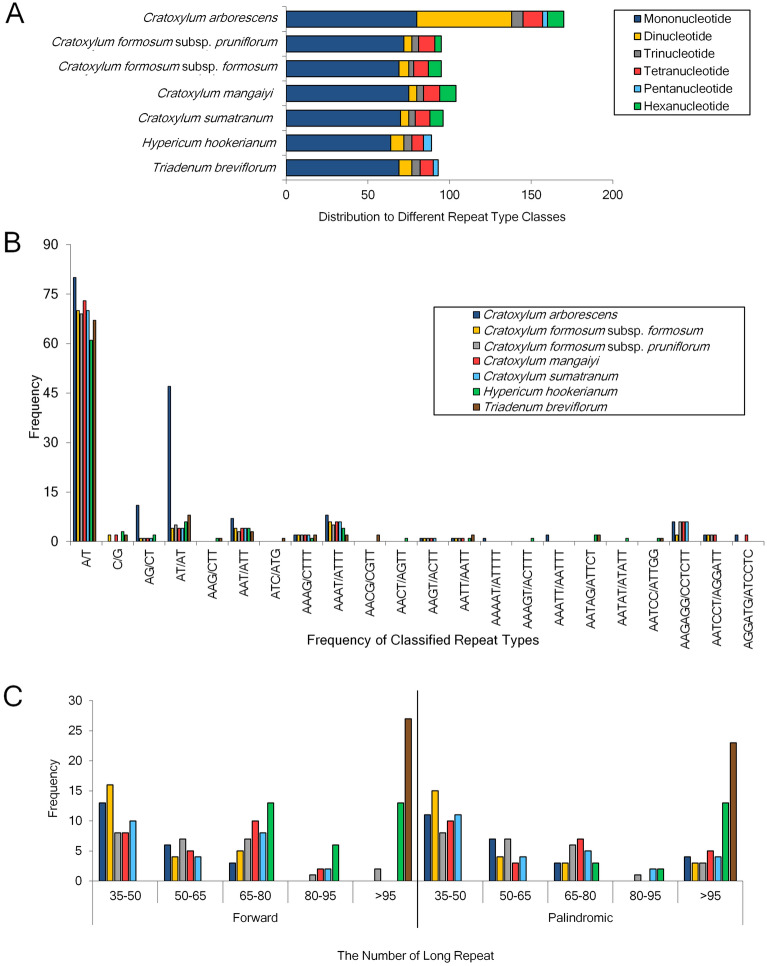
Simple sequence repeats (SSRs) or microsatellites were short tandem repeats of 1–6 nucleotides and motifs at a specific locus are present in all genomes, particularly eukaryotes. Besides being developed as genome markers for the use in marker assisted selection, kinship, breeding, etc., SSRs contribute to the performance of important regulatory functions with the variations in their lengths at the coding regions17,18. In this study, the total SSRs detected in the plastomes of C. arborescens, C. cochinchinense, C. formosum subsp. formosum, C. formosum subsp. pruniflorum, C. maingayi, and C. sumatranum were 170, 103, 95, 95, 104, and 96, respectively (Fig. 2). The mononucleotide repeats were most abundant among all repeat types, ranging between 69 (C. formosum subsp. formosum) and 80 (C. arborescens); the frequency of mononucleotide repeat type A/T was greater than the repeat type C/G. It was worth noting that pentanucleotides were only found present in C. arborescens, including two AAATT/AATTT and one AAAAT/ATTTT repeat type. Large repeats were only recorded in forms of forward as well as palindromic repeats in the six plastomes assessed. All plastomes were identified with 25 each for both repeats, except for C. sumatranum that has 24 forward repeats and 26 palindromic repeats.
Figure 2.
Repeat sequences in the plastid genomes of the seven taxa of Hypericaceae used in this study. Number of different simple sequence repeat types (A); the number of classified SSR repeat units (B); distribution and frequency of long repeat including forward and palindromic repeats (C).
Expansion and contraction of the IR regions
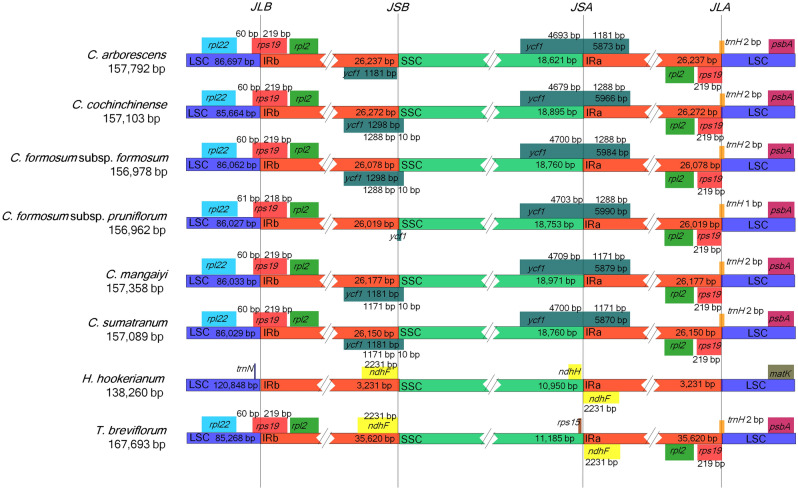
The genes adjacent to the IR junctions in the plastome of the six taxa of Cratoxylum examined displayed identical gene content (Fig. 3). The genes adjacent to the junction LSC/IRb (JLB) were rpl22 and rps19, with rps19 located across JLB. However, for the junction LSC/IRa (JLA), rps19 was intact in the IRa region, while trnH of the LSC region was recorded crossing over JLA. The ycf1 gene was placed across the junction SSC/IRa (JSA) for all six taxa of Cratoxylum examined, but for the junction SSC/IRb (JSB), only the ycf1 gene of four taxa of Cratoxylum, including C. cochinchinense, C. formosum subsp. formosum, C. maingayi, and C. sumatranum, were detected placing across JSB. The ycf1 of C. arborescens was still intact in the IRb region, while ycf1 gene of C. formosum subsp. pruniflorum was identified to be short in length and presumed to be a pseudogene, was located in the SSC region. When analyzed together with the six taxa of Cratoxylum, the gene content adjacent to the IR junctions in H. hookerianum and T. breviflorum exhibited some variations when compared to Cratoxylum. At JLA and JLB, the gene contents of T. breviflorum was similar to those of Cratoxylum, in which rps19 and trnH were placed across JLB and JLA, respectively. However, for H. hookerianum, the trnN gene was placed in the LSC region, next to JLB, while rbcL of the LSC region was the closest gene next to JLA. Both H. hookerianum and T. breviflorum had the ndhF genes located in the IR regions, adjacent to JSA and JSB, while at the SSC region, ndhA and rps15 were placed next to JSA in H. hookerianum and T. breviflorum, respectively.
Figure 3.
Inverted repeat (IR) border analysis based on the complete plastid genomes of eight taxa of Hypericaceae, including C. arborescens, C. cochinchinense, C. formosum subsp. formosum, C. formosum subsp. pruniflorum, C. maingayi, C. sumatranum, Hypericum hookerianum, and Triadenum breviflorum.
Comparative genomic analysis
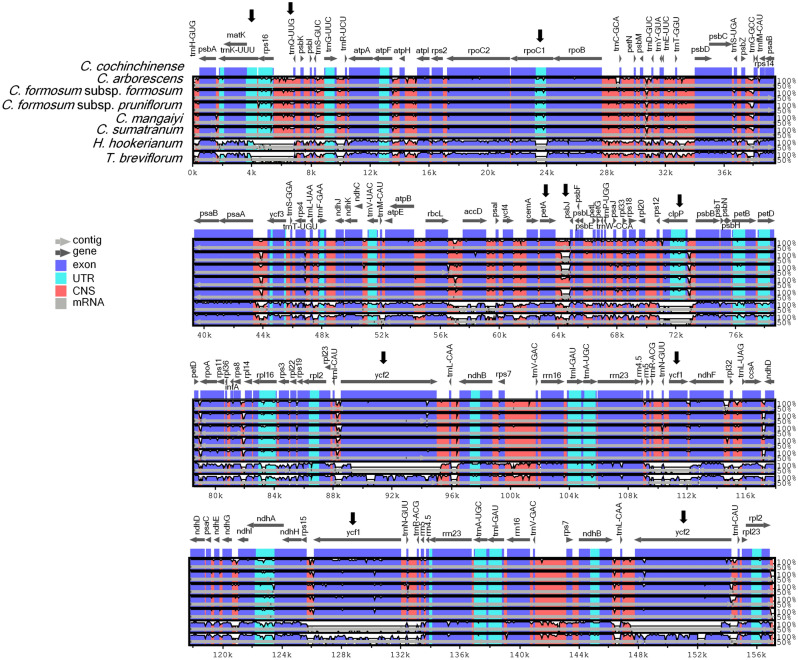
Genome comparison analysis of the complete plastome sequences revealed high conservatism across all taxa of Cratoxylum, with C. cochinchinense as the reference genome (Fig. 4). A small gap that resembles a variation in form of deletion, was observed at the intergenic spacer region psbJ-petA of C. formosum subp. formosum, C. formosum subsp. pruniflorum, and C. sumatranum. When compared to H. hookerianum and T. breviflorum, at least six large gaps could be observed across the plastome, indicating great variations in nucleotide sequence at genus level. These gaps were located at the trnQ-UUG-trnK-UUU, clpP, trnR-ACG-ndhF, trnB-ACG-rps15, and two ycf2 regions. The multiple sequence alignment of the six taxa of Cratoxylum was 169,031 bp in length, containing 1447 singletons and 22,090 parsimony informative sites. There were at least 1678 indel events identified, including a total of 23,364 indel sites.
Figure 4.
Genome comparative analysis of seven taxa of Hypericaceae used in this study, with the complete plastid genome sequence of Cratoxylum cochinchinense (MN399961) as the reference genome. Analysis was conducted using mVISTA under Shuffle-LAGAN mode. Figure legend describes the direction and types of gene regions using color codes. Probability threshold was set at 50%. Black arrows indicate regions that display distinct divergence in the plastid genome.
Phylogenetic inference
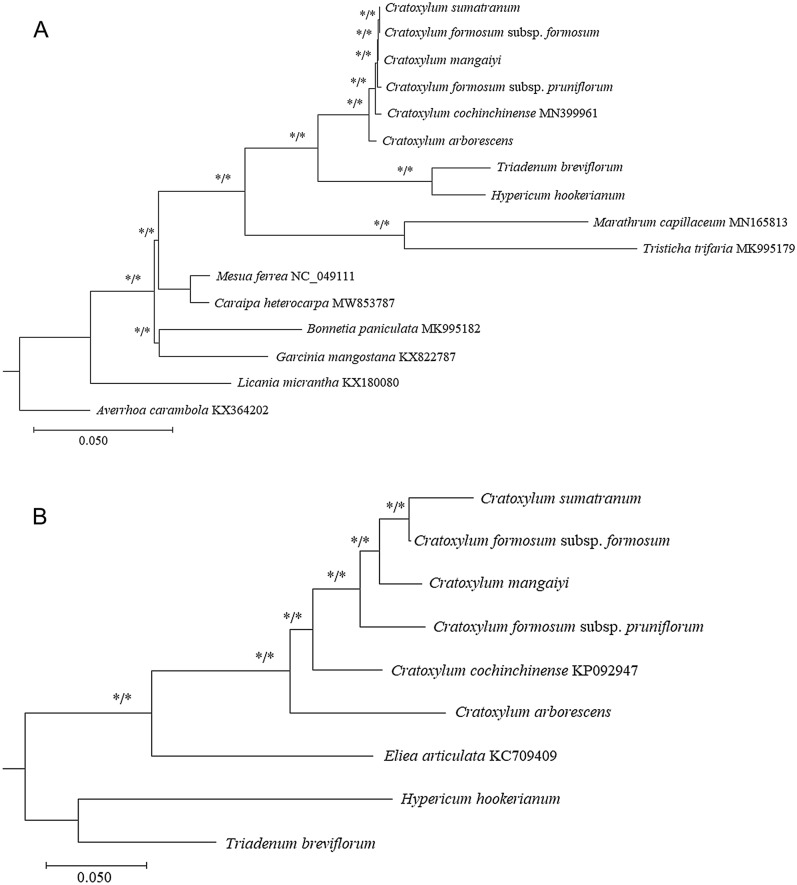
The total length of the multiple sequence alignment of the concatenated CDS dataset was 89,82, while it was 807 bp before trimming and 723 bp after trimming for the ITS dataset. As both the ML and BI tree exhibited identical topology structure in both datasets, only the ML tree was presented in this study, with the BI posterior probability included at each of the branch nodes. In the CDS-tree, all branch nodes were well-supported (BS: ≥ 75; PP: ≥ 0.95); the phylogenetic relationship among all taxa included in the study was well-resolved (Fig. 5A). For Cratoxylum, all six taxa revealed a monophyletic relationship. Cratoxylum arborescens was recorded to diverge first from the other taxa, followed by C. cochinchinense, C. formosum subsp. pruniflorum, and C. maingayi. Cratoxylum formosum subsp. formosum was placed at the tip of the branch with C. sumatranum. For the ITS dataset, a similar tree topology was observed when compared to the tree reconstructed using the CDS-dataset (Fig. 5B); a monophyletic relationship was also observed in Cratoxylum, in which the molecular placement of all six taxa of Cratoxylum in the ITS-tree was identical to those presented in the CDS-tree. The phylogenetic relationship among all taxa of Cratoxylum used in this study was also well-resolved when using the ITS dataset (BS: ≥ 75; PP: ≥ 0.95).
Figure 5.
Phylogenetic trees of Cratoxylum and its allied taxa based on the concatenated protein-coding sequences derived from the plastid genome (A), as well as the nuclear DNA internal transcribed spacer (ITS) sequences (B). The phylogenetic tree was constructed using both maximum likelihood (ML) and Bayesian inference (BI). Bootstrap support (BS) and posterior probabilities (PP) that are considered reliable (BS: ≥ 75; PP: ≥ 0.95) are indicated with an asterisk (*).
Conflicts on taxonomic identity of C. formosum subsp. Pruniflorum
It was noteworthy that C. formosum subsp. pruniflorum was not clustered together with its original, C. formosum subsp. formosum in the phylogenetic trees reconstructed using both the nuclear and plastid regions. Despite the ITS sequences, which are biparental inherited, could indicate possible hybridization in C. formosum subsp. pruniflorum; however, the well-resolved phylogenetic tree based on the maternal inherited plastid genes indicated that C. formosum subsp. formosum and C. formosum subsp. pruniflorum should be treated as two natural groups. Based on the literatures, C. formosum subsp. pruniflorum was first regarded as a distinct species, Hypericum prunifolium Wall19. However, the species has undergone several taxonomic revisions, before it was recognized as a subspecies to C. formosum in 19674. Based on the description, the author emphasized that the key to differentiate between C. formosum subsp. pruniflorum and its original is based on the occurrence of an indumentum; both taxa exhibited high morphological similarities. Although it should not be a key morphological characteristic to differentiate the two taxa, C. formosum subsp. pruniflorum comes with pubescent sepals, while C. formosum subsp. formosum is glabrous at all parts, and they were geographically defined and hardly overlapping. Other morphological characteristics that were proposed to delimit C. formosum subsp. pruniflorum from C. formosum subsp. formosum were—the former has rusty, tomentose young twigs, pedicels and calyx, while the latter is glabrous; the former has short and truncated hypogynous scale, which is 0.7–0.8 mm long, while the latter has a linguiform hypogynous scale that is 2 mm long; the former has capsule that is ovoid-shaped and comes with 54–58 seeds, while the latter has ellipsoid-shaped capsule that is 36–46 seeded20.
To identify the genetic distance between C. formosum subsp. pruniflorum and its original based on the complete plastome and the ITS sequences, we conducted pairwise distance analysis on the complete plastome and ITS sequences and analyzed them separately. We found that the intraspecific pairwise distance was 0.00351, which was greater than the interspecific pairwise distance between C. formosum subsp. pruniflorum and C. maingayi (0.00159) at plastome level, while intraspecific pairwise distance was 0.0502, which was longer than the interspecific pairwise distance between C. formosum subp. pruniflorum and C. maingayi (0.0358) as well as C. sumatranum (0.0316) at the ITS level. There was no report on natural hybridization in Cratoxylum; despite that the pairwise distance was not a suitable parameter to tell closely related species apart, it was generally accepted that intraspecific pairwise distance of a species should be less than that of the interspecific pairwise distance under a regular basis21,22. On the other hand, the chromosome count in C. cochinchinense is n = 1123, while C. formosum subsp. formosum is known to be n = 724. In general, the chromosome count in diploid plant species was often conserved intraspecifically under natural circumstances25. Cratoxylum cochinchinense was proposed to be conspecific to C. formosum at one time due to their identical morphological features, but the proposal was later denied; morphological variations between the two species were distinct in terms of their tree size, color of the bark, leaf structure, leaf shape, and staminal bundle of the flower4. Thus, we commented that cytology studies on C. formosum subsp. pruniflorum could provide useful insights to the genetic identity of the subspecies when compared to its original. Nevertheless, the finding between conventional taxonomic classification and molecular phylogenetic analysis in Cratoxylum was partly incongruent; the taxonomic identity of C. formosum subsp. pruniflorum to be accepted as a reduced taxon under C. formosum warrants further taxonomic revision on this genus. Based on the molecular evidence in this study, we believed that C. formosum subsp. pruniflorum should be considered as a natural group, and the species name Cratoxylum pruniflorum Kurz should be reinstated.
Conclusion
This study has contributed to the expansion of genome data in Hypericaceae, with the characterization of novel plastomes of five taxa of Cratoxylum, as well as one each from Hypericum and Triadenum. The findings obtained from the well-resolved phylogenetic trees reconstructed using both the CDS and ITS datasets have provided insight to the molecular placement and evolution of Cratoxylum, in which the taxonomic identity of C. formosum subsp. pruniflorum to be recognized as a subspecies under C. formosum was questionable. Nonetheless, the molecular data obtained in this study will be a valuable resource for gaining a better understanding of Hypericaceae taxonomy and phylogeny.
Materials and methods
Plant materials
Fresh leaves of five taxa of Cratoxylum species, including C. arborescens, C. formosum subsp. formosum, C. formosum subsp. pruniflorum, C. maingayi, and C. sumatranum, as well as Hypericum hookerianum and Triadenum breviflorum (Supplementary Fig. 1) were collected from natural populations and ex-situ sites (Table 1). All the experiments were performed in accordance with relevant guidelines and regulations. The identities of each specimen were confirmed by the corresponding authors prior to specimen collection. Leaf specimens were kept in ziplock bags filled with silica gels and transported to respective local laboratories for total genomic DNA extraction.
DNA extraction, genome sequencing and assembly
Total genomic DNA was conducted using DNeasy Plant Mini Kit (QIAGEN, Germany), based on the manufacturer’s protocol. The purity and quantity of the DNA extract were estimated using Qubit™ 4 Fluorometer (Thermo Fisher Scientific, USA). Next-generation sequencing was conducted on an Illumina NovaSeq platform (Illumina, USA) to obtain 350-bp paired-end reads. The NGS QC Toolkit v2.3 was used to trim off the adapter sequences26 and the plastome was assembled using NOVOPlasty v2.7.227 with the rbcL gene of C. cochinchinense (GenBank accession no. MN399961) as the seed sequence. The assembled plastome was annotated and the inverted region junctions were identified using GeSeq v2.0328. The annotated plastome was manually checked for errors. The circular plastome map was visualized using OGDRAW v1.3.129. All the plastome sequences obtained through this study were deposited into the NCBI GenBank database, under the accession number MZ703415—MZ703419, and MZ714015—MZ714016.
Repeat analysis
In order to provide a better understanding between the plastomes of all species of Cratoxylum available online, the complete plastome sequence of C. cochinchinense (GenBank accession no. MN399961) was downloaded from the NCBI GenBank database. Subsequent genome comparative analyses were conducted with the inclusion of the genome data of this species. Using MISA-web, the SSRs of each plastome were identified30. The minimum number of repeat parameters were set for 10, 4, 4, 3, 3, and 3 for mono-, di-, tri-, tetra-, penta-, and hexanucleotide motifs, respectively. The large repeats, which includes the forward, palindromic, reverse, and complement repeats, were identified using REPuter31, in which the minimum repeat size was set at 30 bp and a Hamming distance of 3.
Genome comparative and sequence divergence analyses
To detect the expansion and contraction of the IR region in the plastomes, the boundaries and junctions of the IR regions were visualized using IRscope program32 and further edited using Adobe Photoshop CS6 (Adobe, USA). Genome comparative analysis was carried out using mVISTA33 and genome alignment was performed under Shuffle-LAGAN mode. The plastome sequence of C. cochinchinense (GenBank accession no. MN399961) was selected as the reference genome. The number of polymorphic, parsimony informative, and indel sites present in the multiple genome alignment carried out using MAFFT v734 were also calculated using DnaSP v5.10.0135.
Polymerase chain reaction and Sanger sequencing
To obtain the ITS (ITS1-5.8S-ITS2) sequences of the five taxa of Cratoxylum, as well as H. hookerianum, and T. breviflorum used in this study, polymerase chain reaction (PCR) was carried out using a pair of universal primers, ITS5 5′-GGAAGTAAAAGTCGTAACAAGG-3′ and ITS4 5′-TCCTCCGCTTATTGATATGC-3′36. PCR amplification was conducted on a final reaction volume of 25 µL volume reaction containing 12.5 µL of the 2 × GoTaq® Green Master Mix (Promega, USA), 10 µM of each forward and reverse primers, and 15 ng of DNA template. PCR amplification was programmed with thermal settings of an initial denaturation at 95 °C for 2 min, followed by 30 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s, extension at 72 °C for 1 min, and a final extension at 72 °C for 5 min. The amplicons were verified via gel electrophoresis and viewed under the UV machine prior to be sent for direct Sanger sequencing at both ends using an ABI 3730 DNA Analyzer (Applied Biosystems, USA). The resulting sequences were aligned and manually edited using MEGA737 to obtain the clean sequences that will be subjected to phylogenetic analysis. The ITS sequence obtained from this study were deposited into the NCBI GenBank database under the accession numbers MZ674200—MZ674204, MZ703053, and OM980718.
Phylogenetic reconstruction
The reconstruction of the CDS-based phylogenetic tree was conducted based on the concatenated CDS sequences of 14 taxa, in which eight are from Hypericaceae, while seven closely-related species, Bonnetia paniculata (Clusiaceae; GenBank accession no. MK995182), Caraipa heterocarpa (Calophyllaceae; GenBank accession no. MW853787), Garcinia mangostana (Clusiaceae; GenBank accession no. KX822787), Licania micrantha (Chrysobalanaceae; GenBank accession no. KX180080), Marathrum capillaceum (Podostemaceae; GenBank accession no. MN165813), Mesua ferrea (Calophyllaceae; GenBank accession no. NC_049111), as well as Tristicha trifaria (Podostemaceae; GenBank accession no. MK995179) that belong to Malpighiales were analyzed together. Averrhoa carambola (Oxalidaceae; GenBank accession no. KX364202) of Oxalidales was included as outgroup. Plastome sequences were aligned using MAFFT v734 and phylogenetic analysis was conducted using both maximum likelihood (ML) and Bayesian inference (BI) method. For ML analysis, a generalized-time-reversible (GTR) model with gamma (+ G) (= GTR + G) was set and an ML tree was reconstructed using RAxML v8.2.11 under 1000 bootstrap replicates38. BI analysis was conducted using the MrBayes v3.2.7a39 pipeline available in the CIPRES Science Gateway40. A mixed substitution type and a 4 by 4 nucleotide substitution model were selected for the likelihood model, and a 2,000,000-generation Markov Chain Monte Carlo analysis and four Markov chains were implemented. Data sampling was conducted at every 100 generations, while the first 25% of trees was discarded as burn-in. The final tree results for both analyses were visualized using FigTree v.1.4.441.
The ITS-based phylogenetic trees was reconstructed based on the ITS sequences of seven taxa from Cratoxyleae including the five taxa of Cratoxylum used in this study, C. cochinchinense (GenBank accession no. KP092947), and Eliea articulata (GenBank accession no. KC709409). Hypericum hookerianum and T. breviflorum were included as outgroups. Multiple sequence alignment was conducted using MUSCLE embedded in MEGA736 and the alignment was trimmed using trimAL v1.242 by selecting the gappyout option to reduce the systematic errors produced by poor alignment. ML analysis was conducted using MEGA737, in which the “Find Best DNA/Protein Model (ML)” function embedded in MEGA7 calculated that the Tamura 3-parameter (T92) model with invariant sites included (+ I) (= T92 + I) was the optimal DNA substitution model. All sites were included in the analysis and calculation was conducted with 1000 bootstrap replicates. For BI analysis, calculations were performed using MrBayes v3.2.7a39 following the same parameters and settings as mentioned above.
Supplementary Information
Acknowledgements
This work was supported by the Research and Graduate Studies of Khon Kaen University. The authors thank the Germplasm Bank of Wild Species in Southwest China for providing the DNA material of Triadenum breviflorum for this study.
Author contributions
Conceptualization, S.Y.L., A.C. and R.S.; methodology, R.S., U.A.; formal analysis, R.S., S.K., S.Y.L.; resources, P.S. and S.A.S.; data curation, P.S., S.Y.L.; writing-original draft preparation, R.S., S.K.; writing, review and editing, S.A.S., S.Y.L., A.C.; supervision, T.T., A.C.; funding acquisition, R.S., A.C. All authors have read and agreed to the published version of the manuscript.
Data availability
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, accession number (MZ674200-MZ674204, MZ703053, MZ703415–MZ703419, MZ714015–MZ714016, and OM980718). The raw NGS data that support the findings of this study are available from the corresponding author, A.C., upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-23639-2.
References
- 1.Stevens, P. F. Angiosperm Phylogeny Website. Version 14. http://www.mobot.org/MOBOT/research/APweb (2017).
- 2.Stevens PF. Hypericaceae. In: Klaus K, editor. Flowering Plants·Eudicots. Springer; 2007. pp. 194–201. [Google Scholar]
- 3.POWO. Plants of the World Online. Facilitated by the Royal Botanic Gardens, Kew. https://powo.science.kew.org (2022).
- 4.Gogelein AJF. A revision of the genus Cratoxylum Bl (Guttiferae) Blumea. 1967;15:453–475. [Google Scholar]
- 5.Zhuang X. Rehabilitation and development of forest on degraded hills of Hong Kong. For. Ecol. Manag. 1997;99:197–201. doi: 10.1016/S0378-1127(97)00205-3. [DOI] [Google Scholar]
- 6.Suwito D, Suratman, Poedjirahajoe E. The potency of Cratoxylum arborescens Blume (Geronggang) and Combrecarpus rotundatus Dans (Tumih) as natural regeneration in degraded tropical peat swamp forest. JISDeP. 2021;2:272–289. doi: 10.46456/jisdep.v2i3.185. [DOI] [Google Scholar]
- 7.Tokuoka T, Tobe H. Phylogenetic analyses of Malpighiales using plastid and nuclear DNA sequences, with particular reference to the embryology of Euphorbiaceae sens. str. J. Plant Res. 2006;119:599–616. doi: 10.1007/s10265-006-0025-4. [DOI] [PubMed] [Google Scholar]
- 8.Wurdack KJ, Davis CC. Malpighiales phylogenetics: Gaining ground on one of the most recalcitrant clades in the angiosperm tree of life. Am. J. Bot. 2009;96:1551–1570. doi: 10.3732/ajb.0800207. [DOI] [PubMed] [Google Scholar]
- 9.Ruhfel BR, et al. Phylogeny of the clusioid clade (Malpighiales): Evidence from the plastid and mitochondrial genomes. Am. J. Bot. 2011;98:306–325. doi: 10.3732/ajb.1000354. [DOI] [PubMed] [Google Scholar]
- 10.Wicke S, Schneeweiss GM, Depamphilis CW, Müller KF, Quandt D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011;76:273–297. doi: 10.1007/s11103-011-9762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc. Natl. Acad. Sci. U.S.A. 2010;107:4623–4628. doi: 10.1073/pnas.0907801107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ji Y, et al. Plastome phylogenomics, biogeography, and clade diversification of Paris (Melanthiaceae) BMC Plant Biol. 2019;19:543. doi: 10.1186/s12870-019-2147-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mehmood F, Rahim A, Heidari P, Ahmed I, Poczai P. Comparative plastome analysis of Blumea, with implications for genome evolution and phylogeny of Asteroideae. Ecol. Evol. 2021;11:7810–7826. doi: 10.1002/ece3.7614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neves SS, Forrest LL. Plant DNA sequencing for phylogenetic analyses: From plants to sequences. Methods Mol. Biol. 2011;718:183–235. doi: 10.1007/978-1-61779-276-2_10. [DOI] [PubMed] [Google Scholar]
- 15.Álvarez IJFW, Wendel JF. Ribosomal ITS sequences and plant phylogenetic inference. Mol. Phylogenet. Evol. 2003;29:417–434. doi: 10.1016/S1055-7903(03)00208-2. [DOI] [PubMed] [Google Scholar]
- 16.Jin DM, Jin JJ, Yi TS. Plastome structural conservation and evolution in the clusioid clade of Malpighiales. Sci. Rep. 2020;10:9091. doi: 10.1038/s41598-020-66024-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ganie SH, Das PUSD, Sharma MP. Authentication of medicinal plants by DNA markers. Plant Gene. 2015;4:83–89. doi: 10.1016/j.plgene.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Avvaru AK, Saxena S, Sowpati DT, Mishra RK. MSDB: A comprehensive database of simple sequence repeats. Genome Biol. Evol. 2017;9:1797–1802. doi: 10.1093/gbe/evx132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wallich, N. n. 7276 Hypericum prunifolium. Numer. List [Wallich]. 245 (1832).
- 20.Biswas SN. Notes on Cratoxylum formosum ssp. pruniflorum (Kurz) Gog. (Hypericaceae Sensu Stricto) Nelumbo. 1973;15:167–169. [Google Scholar]
- 21.Meier R, Shiyang K, Vaidya G, Ng PK. DNA barcoding and taxonomy in Diptera: A tale of high intraspecific variability and low identification success. Syst. Biol. 2006;55:715–728. doi: 10.1080/10635150600969864. [DOI] [PubMed] [Google Scholar]
- 22.Zhou YF, et al. Gene flow and species delimitation: A case study of two pine species with overlapping distributions in southeast China. Evol. Int. J. Org. Evol. 2010;64:2342–2352. doi: 10.1111/j.1558-5646.2010.00988.x. [DOI] [PubMed] [Google Scholar]
- 23.Michel, C. Cratoxylum (PROSEA). Plant Use English.https://uses.plantnet-project.org/e/index.php?title=Cratoxylum_(PROSEA)&oldid=327154 (2017).
- 24.Robson NKB. Hypericaceae in Flora Malesiana—Series I. Spermatophyta. Alpha Edition; 1974. pp. 1–29. [Google Scholar]
- 25.Cai ZQ, Zhang T, Jian HY. Chromosome number variation in a promising oilseed woody crop, Plukenetia volubilis L. (Euphorbiaceae) Caryologia. 2013;66:54–58. doi: 10.1080/00087114.2013.780442. [DOI] [Google Scholar]
- 26.Patel RK, Jain M. NGS QC toolkit: A toolkit for quality control of next generation sequencing data. PLoS One. 2012;7:e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dierckxsens N, Mardulyn P, Smits G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017;45:e18. doi: 10.1093/nar/gkw955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tillich M, et al. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017;45:W6–W11. doi: 10.1093/nar/gkx391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greiner S, Lehwark P, Bock R. Organellar Genome DRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019;47:W59–W64. doi: 10.1093/nar/gkz238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beier S, Thiel T, Münch T, Scholz U, Mascher M. MISA-web: A web server for microsatellite prediction. Bioinformatics. 2017;33:2583–2585. doi: 10.1093/bioinformatics/btx198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurtz S, et al. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amiryousefi A, Hyvӧnen J, Poczai P. IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 2018;34:3030–3031. doi: 10.1093/bioinformatics/bty220. [DOI] [PubMed] [Google Scholar]
- 33.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katoh K, Rozewicki J, Yamada KD. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 36.White TJ, Bruns TD, Lee SB, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols: A Guide to Methods and Applications. Academic Press; 1990. pp. 315–322. [Google Scholar]
- 37.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ronquist F, Teslenko M, Mark PVD, Huelsenbeck J. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller, M. A., Pfeiffer, W. & Schwartz, T. The CIPRES science gateway: A community resource for phylogenetic analyses. in Proceedings of the 2011 TeraGrid Conference: Extreme Digital Discovery 41. 10.1145/2016741.2016785 (2011)
- 41.Rambaut, A. FigTree. http://tree.bio.ed.ac.uk (2018).
- 42.Capella-Gutiérrez S, Silla-Martinez JM, Gabaldon T. TrimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, accession number (MZ674200-MZ674204, MZ703053, MZ703415–MZ703419, MZ714015–MZ714016, and OM980718). The raw NGS data that support the findings of this study are available from the corresponding author, A.C., upon reasonable request.