Abstract
COVID-19 is a worldwide pandemic caused by SARS-coronavirus-2 (SARS-CoV-2). Less than a year after the emergence of the Covid-19 pandemic, many vaccines have arrived on the market with innovative technologies in the field of vaccinology. Based on the use of messenger RNA (mRNA) encoding the Spike SARS-Cov-2 protein or on the use of recombinant adenovirus vectors enabling the gene encoding the Spike protein to be introduced into our cells, these strategies make it possible to envisage the vaccination in a new light with tools that are more scalable than the vaccine strategies used so far. Faced with the appearance of new variants, which will gradually take precedence over the strain at the origin of the pandemic, these new strategies will allow a much faster update of vaccines to fight against these new variants, some of which may escape neutralization by vaccine antibodies. However, only a vaccination policy based on rapid and massive vaccination of the population but requiring a supply of sufficient doses could make it possible to combat the emergence of these variants. Indeed, the greater the number of infected individuals, the faster the virus multiplies, with an increased risk of the emergence of variants in these RNA viruses. This review will discuss SARS-CoV-2 pathophysiology and evolution approaches in altered transmission platforms and emphasize the different mutations and how they influence the virus characteristics. Also, this article summarizes the common vaccines and the implication of the mutations and genetic variety of SARS-CoV-2 on the COVID-19 biomedical arbitrations.
Abbreviations: ACE2, angiotensin-converting enzyme 2; BIBP, Beijing Institute of biological products; CD, cluster of differentiation; CDC, Centers for Disease Control and Prevention; COVID-19, coronavirus disease 2019; CRP, C-reactive protein; CTD, C-terminal domains; E, envelope; ELISA, enzyme-linked immunosorbent assay; ERGIC, endoplasmic reticulum–Golgi intermediate compartment; ESR, erythrocyte sedimentation rate; FP, fusion-peptide; GCSF, granulocyte colony-stimulating factor; GISAID, Global Initiative on Sharing Avian Influenza Data; GMTs, geometric mean titers; HCoVs, human coronaviruses; ICU, intensive care unit; Ig, immunoglobulin; IL, interleukin; LDH, lactate dehydrogenase; LNPs, lipid nanoparticles; M, membrane; MERS-CoV, Middle East respiratory syndrome coronavirus; mRNA, messenger RNA; N, nucleocapsid; NABs, neutralization antibodies; Nsp, non-structural proteins; NTD, N-terminal domain; nts, nucleotides; ORFs, open reading frames; PANGOLIN, phylogenetic assignment of named global outbreak lineages; PCR, polymerase chain reaction; pp, polyproteins; r, recombinant; RBD, receptor-binding domain; RdRP, RNA dependent RNA polymerase; RDT, rapid diagnostic test; RNA, ribonucleic acid; ROCM, rhino-orbital-cerebral mucormycosis; S, spike; SARS-CoV, severe acute respiratory syndrome coronavirus; sgRNA, subgenomic RNA; SNP, single nucleotide polymorphism; TAG-VE, Technical Advisory Organization on SARS-CoV-2 Virus Evolution; TMPRSS2, transmembrane protease serine-2; TNF, tumor necrosis factor; VOC, variant of concern; VOHC, variants of high consequences; VOI, variant of interest; WHO, World Health Organization; αCoV, Alpha coronavirus; βCoV, Beta coronavirus; γCoV, Gamma coronavirus; δCoV, Delta coronavirus
Keywords: SARS-CoV-2, Mutation, Variants, Pandemic disease, Vaccines
Graphical Abstract
1. Introduction
Coronaviruses are a highly considered group of viruses containing ribonucleic acid (RNA) genetic material that circulates in the animal kingdom. Homo sapiens can be naturally infected with seven types of coronaviruses, although they originated from animals [1]. The triad of extremely infective coronaviruses, including Middle East respiratory syndrome coronavirus (MERS-CoV), severe acute respiratory syndrome coronavirus (SARS-CoV), and SARS-CoV-2, can cause massive respiratory illnesses in humans. In contrast, the four taxa of low-pathogenic human coronavirus, including HKU1, OC43, NL63, and 229E, commonly just cause self-limiting respiratory tract illnesses [2], [3]. Concerning cladogram and genetic relationships of different coronaviruses, MERS-CoV, SARS-CoV, HKU1, OC43, and SARS-CoV-2 are beta coronaviruses, whereas NL63 and 229E are alphacoronaviruses [4].
The origin of the new coronavirus that causes the current corona pandemic outbreaks in the city of Wuhan, China. A few days before the end of 2019, physicians in Wuhan discovered limited cases of atypical pneumonia attributed to the new coronavirus. It may have jumped from an animal source to a human in early November of the same year [5]. It was later discovered that the disease's causative agent is an RNA virus that originates from the same family as the coronavirus that caused SARS at the beginning of the century and the MERS pandemic at the beginning of the previous decade [6].
The recent outbreak of SARS-CoV-2 revealed a hidden reservoir for wild animals of life-threatening viruses and the potential threat of the spread of zoonoses [7]. One of the most populated wet markets that marketed alive animals in Wuhan, China was related to the eruption of SARS-CoV-2 [8]. Bats are the primary repositories of these types of viruses, and relying on genetic analysis, once more bats were accused as native hosts of SARS-CoV2 as it shared 96% of their genes with two SARS-like CoVs, viz. bat-SLCoVZX45 and bat-SL-CoVZX2, from bats [9] although we cannot be sure which animal is the intermediate host [10]. In the beginning, snakes charged as an adapter host of SARS-CoV-2 transmission [11]. Later, pangolins were proposed as potential transitional hosts for SARS-CoV-2 as the genetic symmetry with CoVs in pangolins is high, and the difference is less than 1% [12]. The epidemiology of COVID-19 can be divided into three stages [13]. In the first stage, the analysis conducted by the epidemiologists showed that direct contact was the influencing factor in the transmission of infection from one infected person to another [14], [15]. Later, infection rose to 205 cases outside of Wuhan, in the city of Beijing and Guangdong, and then the disease began to spread, even though Wuhan had been placed under lockdown. On February 10, 2020, 50–80% of cases were clustered around Beijing, Shanghai, Jiangsu, and Shandong in the third stage [16].
SARS-CoV-2 is a single-stranded RNA virus with variable open reading frames (ORFs). Coronavirus genome size ranges from 26 to 32 kb. It consists of a structural gene unit encoding 9680 amino acids (6 – 11 ORFs that encode S, E, M, and N proteins and two large ORF genes (ORF1a and ORF1b) that encode 16 non-structural proteins (NSP), including RNA-dependent RNA polymerase (RdRp)). The remaining ORFs encode structural and accessory proteins of SARS-CoV-2 [17], [18].
Through the WHO, in addition to continuing to monitor the emergence of new variants of SARS-CoV-2 (SARS-CoV-2), there are key priorities for the global response to the variants of concern. These priorities, on top of which is the development of new and modified vaccines; therefore, many vaccines have been approved for widespread use recently [19]. Despite the vaccine effectiveness, these vaccines must be developed to accommodate the new variants, although no effective immune escape has been observed [20].
It was emphasized that the different races or genetic backgrounds of people, and some methods of closure and quarantine, affect rates of death and injury [21]. Furthermore, viral evolution is considered the main contributory factor of the SARS-CoV-2 transmissibility and the severity of infection [22].
Vaccines saved huge numbers of lives in such a short time. Billions have received one or more doses of the vaccine. Continuously, new SARS‐CoV‐2 strains have been emerging. Although vaccination reduces symptomatic cases, hospitalization, and mortality, the rapid mutation of the virus and the emergence of new strains limit the effectiveness of these vaccines [23], [24].
Even though the community has a high vaccination rate, outbreaks can still happen. The number of doses, kind of vaccination, and SARS-CoV-2 variant all impact effectiveness [25]. To attain herd immunity, vaccination rates among the population must be at least 90% [26]. The world always needs to increase the coverage of vaccines in a better way to avoid the spread of more infectious variants and the vaccine's lower effectiveness against them [27].
In this context, this review integrates many aspects of the novel coronavirus with insights into the mutations of SARS-CoV-2 and the impact of these mutations on virus properties. This article provides further information on viral mutations and a summary of the evolution of viruses during the COVID-19 pandemic. Also, this review gives an overview of the various classification and nomenclature systems for COVID-19 variants, standard vaccinations, and the impact of SARS-CoV-2 genetic mutations on the COVID-19 biomedical arbitrations.
2. Structure genomics of SARS-CoV-2
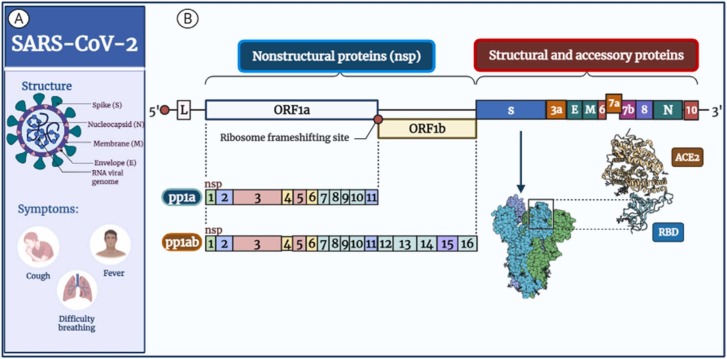
SARS-CoV-2 are icosahedral symmetry-wrapped particles with a diameter of 80–220 nm. Virions have distinct spike projections about 9–12 nm from the virus's envelope, giving it the crown shape under the electron microscope [28]. SARS-CoV-2 has a non-segmented, single (+RNA) strand, the genome of about 29,903 nucleotides (nts) [3], [29], SARS-CoV-2 surface M (membrane) proteins, S (spike), and E (envelope) are implanted in lipid bilayer membrane enveloped by the helical nucleocapsid (N) coating viral RNA [30] ( Fig. 1 A).
Fig. 1.
Diagrams represent SARS-CoV-2 structure and genomic organization. A: Diagrammatic representation of the structural characteristics and symptoms of SARS-CoV-2 as well as its key structural proteins. The spike (S) proteins cover the virion, while the membrane (M) and envelope (E) proteins are located among the S proteins in the viral envelope. Genomic RNA is surrounded by nucleocapsid (N) proteins inside phospholipid bilayers. B: The genomic organization of SARS-CoV-2. The SARS-CoV-2 genome (29,903 nucleotides) contains the 5′ UTR, ORF1a/b encoding 16 nsps for replication, four genes encoding structural proteins (S, E, M, and N proteins), six accessory genes (ORF3a, ORF6, ORF7a, ORF7b, ORF8, and ORF10) encoding accessory proteins and the 3′ UTR.
The SARS-CoV-2 genome has two untranslated regions at the 5′ and 3′ ends and 11 ORFs that encode 27 proteins. ( Table 1 and Fig. 1 B). SARS-CoV-2 shows a genome organization similar to beta coronaviruses. The major ORF (ORF1/ab) comprises around 70% of the genome from the 5′ end and encodes each replicase complex polyproteins pp1a (4405 aa, amino acid) or pp1ab (7096 aa), according to ribosomal frameshift [31] (Fig. 1 B).
Table 1.
SARS CoV-2 gene and encoded proteins [29].
| Gene | Gene_id | Nucleotides | Protein | Protein_id | Amino Acids | |
|---|---|---|---|---|---|---|
| 5′UTR | – | 1.265 = 265 | – | – | – | |
| orf1ab | 43740578 | 266.21555 = 21290 | ORF1ab polyprotein | YP_009724389.1 | 7096 | |
| Non-structural proteins | nsp-1 | 266.805 = 540 | Leader protein | YP_009725297.1 | 1.180 = 180 | |
| nsp-2 | 806.2719 = 1914 | nsp-2 | YP_009725298.1 | 181.818 = 638 | ||
| nsp-3 | 2720.8554 = 5835 | Papain like protease | YP_009725299.1 | 819.2763 = 1945 | ||
| nsp-4 | 8555.10054 = 1500 | nsp-4 | YP_009725300.1 | 2764.3263 = 500 | ||
| nsp-5 | 10055.10972 = 918 | 3 C-like proteinase | YP_009725301.1 | 3264.3569 = 306 | ||
| nsp-6 | 10973.11842 = 870 | nsp-6 | YP_009725302.1 | 3570.3859 = 290 | ||
| nsp-7 | 11843.12091 = 249 | nsp-7 | YP_009725303.1 | 3860.3942 = 83 | ||
| nsp-8 | 12092.12685 = 594 | nsp-8 | YP_009725304.1 | 3943.4140 = 198 | ||
| nsp-9 | 12686.13024 = 339 | nsp-9 | YP_009725305.1 | 4141.4253 = 113 | ||
| nsp-10 | 13025.13441 = 417 | nsp-10 | YP_009725306.1 | 4254.4392 = 139 | ||
| nsp-11 | 13442.13480 = 39 | nsp-11 | ||||
| nsp-12 | 13442.16236 = 2795 | RNA-dependent RNA polymerase | YP_009725307.1 | 4393.5324 = 932 | ||
| nsp-13 | 16237.18039 = 1803 | Helicase | YP_009725308.1 | 5325.5925 = 601 | ||
| nsp-14 | 18040.19620 = 1619 | 3′-to-5′ exonuclease | YP_009725309.1 | 5926.6452 = 527 | ||
| nsp-15 | 19621.20658 = 1038 | EndoRNAse | YP_009725310.1 | 6453.6798 = 346 | ||
| nsp-16 | 20659.21552 = 894 | 2′-O-ribose methyltransferase | YP_009725311.1 | 6799.7096 = 298 | ||
| S | 43740568 | 21563.25384 = 3822 | Surface glycoprotein | YP_009724390.1 | 1273 | |
| ORF3a | 43740569 | 25393.26220 = 828 | ORF3a | YP_009724391.1 | 275 | |
| E | 43740570 | 26245.26472 = 228 | Envelope protein | YP_009724392.1 | 75 | |
| M | 43740571 | 26523.27191 = 669 | Membrane glycoprotein | YP_009724393.1 | 222 | |
| ORF6 | 43740572 | 27202.27387 = 186 | ORF6 | YP_009724394.1 | 61 | |
| ORF7a | 43740573 | 27394.27759 = 366 | ORF7a | YP_009724395.1 | 121 | |
| ORF7b | 43740574 | 27756.27887 = 132 | ORF7b | YP_009725318.1 | 43 | |
| ORF8 | 43740577 | 27894.28259 = 366 | ORF8 | YP_009724396.1 | 121 | |
| N | 43740575 | 28274.29533 = 1260 | Nucleocapsid phosphoprotein | YP_009724397.2 | 419 | |
| ORF10 | 43740576 | 29558.29674 = 117 | ORF10 | YP_009725255.1 | 38 | |
| 3′UTR | – | 29675.29903 = 229 | – | – | – | |
https://www.ncbi.nlm.nih.gov/nuccore/1798174254 [Accessed 19 Jan 2022]
ORF1a encodes polyprotein (pp) 1a containing nsp1–11, while ORF1a and ORF1b together produce pp1ab containing nsp1–16 through a (−1) ribosomal frameshift overreading the stop codon of ORF1a. [32] the nsp-1 mediates RNA processing and replication [32]. The nsp-2 modulates the survival signaling pathway of the host cell, while nsp-3 is believed to separate the translated protein [33], [34]. The nsp-4 and nsp6 contain transmembrane domains and modify endoplasmic reticulum (ER) membranes; nsp-5 participates in the process of polyprotein during replication [35], [36], [37]. The nsp-7 with nsp-8 significantly increased the combination of nsp12 and template-primer RNA, nsp-9 functions as an ssRNA-binding protein, and nsp-10 is critical for the cap methylation of viral mRNAs [35], [36], [37]. The nsp-11 is a protein with an uncertain function [38]. The nsp-12 is an RdRp, a critical component of coronavirus replication/transcription; at the same time, nsp-13 binds with ATP, and the zinc-binding domain participates in the replication and transcription process [39]. The nsp-14 is a 3–5′ exoribonuclease used for proofreading, while nsp-15 has Mn2+-dependent endoribonuclease activity [39].
Virion-forming structural proteins E, N, M, and S; are encoded by the virus's remaining one-third genome from the 3′ end. Coalesced within these genes, ORFs that code for a minimum of 6 accessory proteins are termed ORFs 3a, ORFs 6, ORFs 7a, ORFs 7b, ORFs 8, and ORFs 10 [2], [40]. The N protein coats the viral genome and packs the RNA in the virions as a ribonucleoprotein complex [41]; the E proteins play critical roles in virus invasion and reproduction, including host cell membrane permeability, the virus-host cell interaction, and virus assembly [42], [43]; the M protein remains essential intended for coronavirus assembly, and morphogenesis [44]; The S protein built-in over the surface of the virus, intermediates the virion's attachment to surface receptors of the host cell and manages the fusion of host cell membranes with viral, enabling invasion of a virus into a host cell [45]. The roles of all accessory proteins are unknown, but they may be involved in viral adaptation and host immunity [46].
3. SARS-CoV-2 origin
Since the outbreaks of SARS in 2002 and MERS in 2012, the likelihood of the transmission of coronaviruses to humans from animals has been established [47], [48]. In the early stages of Wuhan's pneumonia epidemic, researchers obtained complete genome sequences from five SARS-CoV-2 infected patients. SARS-CoV shares 79.5% sequence similarity with these genetic sequences. Noticeably, SARS-CoV-2 appears divergent from SARS-CoV. It is a new beta coronavirus infects people [5], [18]. With a sequence similarity of 96.2%, the results revealed that SARS-CoV-2 is thoroughly linked to the BatCov RaTG13 strain. These outcomes suggest that SARS-CoV-2 is of bat origin and may have developed naturally from bat coronavirus; RaTG13 [18], [49].
Although RaTG13 has the maximum genetic match to SARS-CoV-2. However, due to recombination, three further bat viruses (RpYN06, RmYN02, and PrC31) are more highly correlated across much of the genome (especially ORF1ab) and hence share other common ancestors with SARS-CoV-2 [50], [51], [52]. Additionally, SARS-CoV-2 is very closely associated with bat viruses, with 100% identical amino acids in the nsp-7 and E proteins to BAT-SL-CoVZC45, implying that bats oblige as a reservoir host for SARS-CoV-2 or related viruses [29]. Additionally, some researchers have suggested that pangolins (Manis javanica), an endangered tiny mammal, may also have ancestral beta-CoVs related to SARS-CoV-2 [53], [54]. SARS-CoV-2 shares about 85–92% nucleotide sequence identity with these new pangolin CoV genomes. Pangolin coronavirus and SARS-CoV-2 share the amino acid identity of 90.4%, 96.7%, 98.2%, and 100%, respectively, within S, N, M, and E genes [55]. Transmission of SARS-CoV-2 from humans to dogs, tigers, cats, lions, and minks has also been reported [56], [57].
4. Human CoVs (HCoVs) Taxonomy
SARS-CoVs are one of the most common viruses in the family Coronaviridae of the Nidovirales order, Cornidovirineae suborder. Coronaviridae is a family of viruses that is divided into Letovirinae and Orthocoronavirinae. The Letovirinae includes one genus Alphaletovirus, in contrast, the subfamily Orthocoronaviridae is further categorized according to genomic sequence and phylogenetic investigation into four species; α (Alpha)-CoV, β (Beta)-CoV, γ (Gamma)-CoV, and the δ (Delta)-CoV, which contain 19, 14, 5, and 7 species, respectively [58]. The ability of viruses in this order can transcribe numerous 3′-nested subgenomic RNAs (sgRNA), giving the order the name Nido-"nest." [59].
The α- and β CoVs are most usually found in mammals, whereas γ- and δ CoVs are principally found in birds. SARS-CoV-2 is a different lineage in the subgenus Sarbecovirus, according to Chan et al. [60]. Among the four other coronaviruses strains linked to mild symptoms of the common cold in humans previously recognized, HCoV-229E, and HCoV-NL63 which belong to α CoVs, and HCoV-HKU1 and HCoV-OC43 are classified as different subgroup of β CoVs called Embecovirus [61].
Furthermore, other coronaviruses include transmissible gastroenteritis virus [62], infectious bronchitis virus in chickens [63], beluga whale CoV-SW1 [64], bat coronaviruses HKU10 and CDPHE15 [58], and swine acute diarrhea syndrome-CoV [65], have generated pandemic infections in wild and domestic mammals, resulting in high fatality rates and a significant financial burden.
5. SARS-CoV-2 pathogenesis and transmission mechanisms
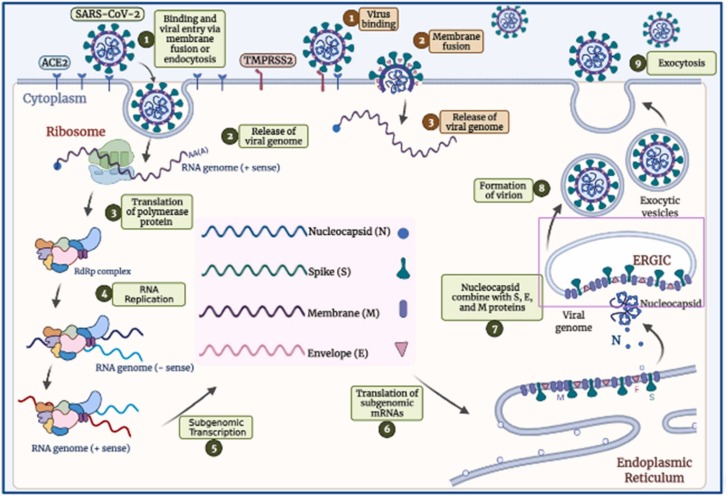
Endocytosis occurs when SARS-CoV-2 infects a host cell either directly by fusing the viral E to the cell membrane or indirectly by fusing the membrane within the endosome [66] ( Fig. 2 ). A RBD (receptor-binding domain) of S protein, found at the C-terminal region of the S1 subunit, binds to receptors on the human cell surface, allowing the virus to enter the cell [2], [67]. ACE2 (Angiotensin-converting enzyme 2) is a key receptor of SARS-CoV-2 that is broadly expressed in the lung, vascular endothelium, intestine, testis, heart, liver, and kidney cells [68], [69], [70].
Fig. 2.
SARS-CoV-2 entrance and replication pathways.
Depending on how SARS-CoV-2 enters the body, different proteases cleave the S2′ site. The two primary proteases involved in S protein activation are cathepsin L and TMPRSS2 (transmembrane protease serine-2) [71]. Clathrin-mediated endocytosis transfers the virus–ACE2 complex to endolysosomes, where cathepsins undertake S2′ cleavage, which requires an acidic environment if the target cell does not produce enough TMPRSS2 [66]. The FP (fusion-peptide) is exposed after cleavage of the S2′ site in both entrance pathways, and detachment of S1 from S2 induces significant conformational changes in the S2 subunit, primarily in heptad repeat-1, driving FP forward to the targeted membrane and commencing membrane union [72], [73]. A viral-cellular fusion hole releases all viral genetic material into the cytoplasm. The infectious RNA is uncoated by proteasomes and translated into replicative enzymes from ORF 1a/b by host ribosomes in the cytoplasm. A virus-encoded protease splits pp1a and pp1b polyproteins into replicase complex nsps, comprising RdRp. These enzymes create full-length (−RNA) from the virus's genome, which is used to transcribe numerous sgRNAs. sgRNAs translate accessory and structural proteins into the ERGIC for virion assembly [45]. Nucleocapsids are reproduced in the cytoplasm and self-assemble into fresh virions within the ERGIC membrane. Finally, infected cells produce new virions through a process known as exocytosis, which can infect other cells [45].
6. Possible variables affecting symptom variability and severity of SARS-CoV-2
6.1. Environmental, clinical, and social factors
Coronaviruses have been known since the SARS-CoV and MERS-CoV epidemics were zoonotic [74]. The clinical indications for moderate patients are dry cough, fever, fatigue, headache, and chest pain. However, severe patients might progress to severe infections (such as pneumonia), ARDS, organ failure, and death [2], [75]. Patients with moderate indications, particularly the elderly or individuals with comorbidity, can exacerbate and give rise to great mortality [75], [76]. Zhang et al. [77] suggest that age and levels of ESR (erythrocyte sedimentation rate), albumin, D-dimer, and IL6 are exhaustively associated with the severity of COVID-19 patients. ESR can be used as a helpful sign for differentiating severe disease courses in the first stage to raise the survival of patients with severe COVID-19. Different studies have shown lymphocytopenia predicts the severity of SARS-CoV-2 and can be utilized as a prognostic marker in COVID-19 patients with severe disease [78], [79], [80], [81]. Other researches have found that lymphocytopenia and increased inflammatory cytokines levels in patients with COVID-19 [2], [75] as well as neutrophil/ cluster of differentiation 8 + (CD8 +) T cell proportion and high IL6 levels [82], [83] as indicators for quick detection of severe COVID-19 patients.
Nonetheless, it was unknown whether the lymphocytopenia seen in COVID-19 individuals is brought on by lymphocyte destruction or tissue infiltration. Huang et al. [76] reported that COVID-19 patients in an ICU (intensive care unit) have apparent higher levels of cytokines containing IL-7, IL-2, IL-10, TNF, and G-CSF (granulocyte- colony-stimulating factor), compared with non-ICU patients. Hyperimmune response in the form of cytokine storm causes death in severe COVID-19 patients [84].
D-dimer, aspartate aminotransferase, albumin, fibrinogen, creatinine, neutrophils, procalcitonin, and platelets were among the laboratory values that might be used to expect the severity of SARS-COV-2 [85], [86]. The lymphocyte count, ferritin, and hemoglobin levels were the most substantial predictive signs of severe COVID-19 [87]. In a particular study, abnormal elevation of liver enzyme levels was shown in many COVID-19 patients [88].
LDH (Lactate dehydrogenase) is an indicator of liver function, and the death risk in severe COVID-19 patients was 3-fold higher with liver dysfunction than in those without liver dysfunction [89]. SARS-COV-2 infection can cause local destruction in the liver and bile duct; the ACE2 receptor is abundantly expressed. LDH is likewise a marker of the severity of idiopathic pulmonary fibrosis, and an inaccessible increase in the levels of LDH with an average level of other liver enzymes is predictive of pneumonia triggered by COVID-19. This level is raised due to tissue damage [90], [91].
Higher CRP (C-reactive protein) levels indicate an acute inflammatory response, and the high levels of lymphocytes are a sign of infection and hyper-immune function [92]. Ferritin level was abnormally high in severe COVID-19 patients who needed mechanical ventilation [93]. Most patients with severe COVID-19 disease require more oxygen to keep their oxygen capacity overhead at 90%. Consequently, hypoxia is closely allied to a decrease in hemoglobin levels. Patients will develop hypoxemia when a significant reduction in hemoglobin levels is found, impacting their prognosis [80], [87]. Research by Samra et al. [94] demonstrated that susceptibility and severity for COVID-19 are higher in patients who have blood group A, while they are lower in persons with blood group O. As a result, group A may require improved personal protection to minimize the infection risk, as well as prompt treatment and proper management if the infection is detected.
6.2. Human genetic effects on COVID-19 susceptibility
In parallel to environmental, clinical, and social factors, different host genetic patterns can affect the susceptibility and response to COVID-19 infection in diverse populations [93], [95]. These connections were studied using genetic studies, whole-exome sequencing, and genome-wide association studies by several consortia such as COVID Host Genetics Initiative (HGI), COVID human genetic effort, Genetics of Mortality In Critical Care (GenOMICC), and commercial genomics service providers such as 23andMe [6], [96], [97], [98].
6.2.1. Angiotensin-converting enzyme 2 (ACE2)
Angiotensin-converting enzyme 2 (ACE2) is a coronavirus receptor, a B0 AT1 amino acid transporter [99]. Single-nucleotide polymorphisms (SNPs) in the gene ACE2 (Xp22.2) have been shown to impact its binding affinity and the expression of SARS-CoV-2, altering susceptibility to SARS-CoV-2 infection [100], [101]. ACE2 variants such as p.Asp427Tyr, p.Pro389His, and p.Met383Thr were found to have slightly low interaction with the S protein [102].
Mohlendick et al. found in 2021 that GG genotypes of the ACE2 splice region variant rs2285666 (Intron 3/4, G>A) are more vulnerable to SARS-CoV-2 infection, with an increased risk of developing severe disease or death from COVID-19 [103]. These findings can also explain the high risk of COVID-19 illness and hospitalization in patients with diabetes, hypertension, cerebral stroke, and coronary heart disease, as rs2285666 SNP is associated with these chronic diseases [104], [105].
Although ACE2 acts as a SARS-CoV-2 receptor and helps in virus entry into the cell, ACE2/Ang (1−7) system acts as an antioxidant and anti-inflammatory agent that protects the lung against ARDS [106]. In diabetic patients, ACE2 expression is reduced due to glycosylation, which can explain the increased COVID-19 severity in these patients [107].
6.2.2. Transmembrane serine protease TMPRSS2
In addition to initiating viral entry, TMPRSS2 cleavage of SARS-CoV-2 S-protein is in charge of immune evasion and increasing infection [108]. In whole-exome sequencing, Ravikanth et al. found that a detrimental mutation in TMPRSS2 (rs12329760, G > A, p. V160M) lowered spike cleavage function and viral entry, resulting in less severe illness [109]. Hou et al. also mentioned another unique but prevalent polymorphism (rs12329760, p.Val160Met) associated with a differential genetic vulnerability to COVID-19 [102]. COVID-19 infection may be shown in higher percentages in males due to the genetic variation in androgen-responsiveness of TMPRSS2 as (rs8134378) and, therefore, the higher expression of SARS-COV-2 [110].
6.2.3. Human Leukocyte Antigen
Susceptibility to and severity of COVID-19 can be affected by Human Leukocyte Antigen-B (HLA-B). HLA-B* 46:01 allele-carrying individuals are more susceptible to COVID-19 as they have the lowest number of predicted binding peptides for SARS-CoV-2. Conversely, the HLA-B* 15:03 allele shows less susceptibility due to high presentation to conserved SARS-CoV-2 peptides [111].
6.2.4. Interferons (IFN)
Life-threatening COVID-19 is associated with loss of function variants of type I interferon (IFN) immunity. This may suggest using type I IFN as an early-stage treatment of COVID-19 [6], [112].
7. SARS-CoV-2 mutations and evolution
Mutations in the SARS-CoV-2 virus, as an RNA virus, arise spontaneously during replication due to the lack of RNA polymerase proofreading activity and thus make far more mistakes [113]. Hundreds of cumulative mutations have existed since the virus's emergence [114]. As mutations occur, new mutants are found. Most SARS-CoV-2 genome mutations have no noticeable effect on the virus's propagation and pathogenicity, thus the disease's course [115]. The main concern about these emerging mutations is a potentially dangerous modification that could increase infection severity or a failure of the currently developing vaccines' effects [116].
The most common mutations are in the S protein gene encoding, linked to viral entry into cells [117]. Among these mutations, D614G (aspartate to glycine in protein position 614) is the most prominent [118]. SARS-CoV-2 isolates from the world have also been shown to have several mutations in the RdRp coding region of the Orf1ab polyprotein gene. The 14408 C>T transition is the most common [22].
There is about 96% sequence similarity between SARS-CoV-2 and SARS-CoV in some encoded proteins, such as coronavirus major proteinase 3-chymotrypsin like protease, RNA-dependent RNA polymerase (RdRp), and papain-like protease. As a result, it was assumed that SARS-CoV-2 would behave similarly to SARS-CoV in terms of human infection and pathogenicity [119]. S1 and S2 are two S protein regions, with S1 required for host cell receptor binding and S2 necessary for membrane fusion. The S1 region contains three C-terminal domains (CTD) and one N-terminal domain (NTD) [120]. The RBD protein in the CTD1 binds to the ACE2, allowing SARS-CoV to adhere to human host cells [121].
SARS-CoV-2 is more pathogenic than SARS-CoV due to its higher affinity for ACE2 than SARS-CoV [122]. Mutagenesis is a molecular biological process that modifies an organism's genetic coding and hence acts as a major source of viral evolution [123]. Mutations in the S protein RBD boosted viral infectivity, indicating that SARS-CoV-2 evolution is regulated by infectivity-based natural selection [115].
With a genomic size of 29.99 kb, SARS-CoV-2 contains more than 28,000 different mutations. Random genetic drifts during replication, ribosome fidelity-related errors in translation, and various errors caused by hostile cellular circumstances triggered by host immune reactions all result in mutations at the molecular level [115]. Furthermore, at the organism level, host gene editing is a dominant mechanism for changes in SARS-CoV-2 [124].
SARS-CoV-2 employs Nsp-12 RdRps to proofread, unlike flu viruses, genetically. SARS-CoV-2 may mutate slower than flu viruses under the same settings. However, SARS-CoV-2 development is governed by transmission. Viral transmission requires infectivity and pathogenicity. However, rapid, widespread, and global SARS-CoV-2 transmission drives significant alterations [125].
The mutation rate of SARS-CoV-2 was assessed by Yang et al. in over 3100 genomic sequences. With 58.4% missense mutations (1224), they found 2096 simple nucleotide variations and seven short deletions. They also discovered a high substitution frequency at locus 23.403, resulting in a change in the 614 amino acids of the spike glycoprotein, which could alter its function [126]. Van Dorp et al. discovered that the S protein is responsible for nearly 80% of SARS-CoV-2 mutations. Because of its adaptability, the Orf1ab gene expresses a significant number of mutations in its regions, resulting in alterations in this protein and the structural and nonstructural proteins Nsp-6, Nsp-11, and Nsp-13, indicating that it is a heavy evolutionary agent [127].
8. Variants of SARS-CoV-2
Variant or "genetic variant" refers to a sort of subclass of an organism that has undergone genetic modification from the main strain but is not sufficiently distinct to be referred to as a distinct strain. A variety exhibiting distinctive physical qualities is referred to as a strain. Simply put, a strain is a virus variety that differs from its parent virus in structure and behavior [128]. The variants of fastest-spreading characteristics were noticed in South Africa, the United Kingdom (UK), and Brazil. Researchers were doubtful that variants could potentially distress numerous genetic changes that change disease severity and escape from immunity defense machinery. In addition, variants may make vaccinated persons may be re-infected with new-fangled variants [129].
8.1. Nomenclatures for the SARS-CoV-2 variants
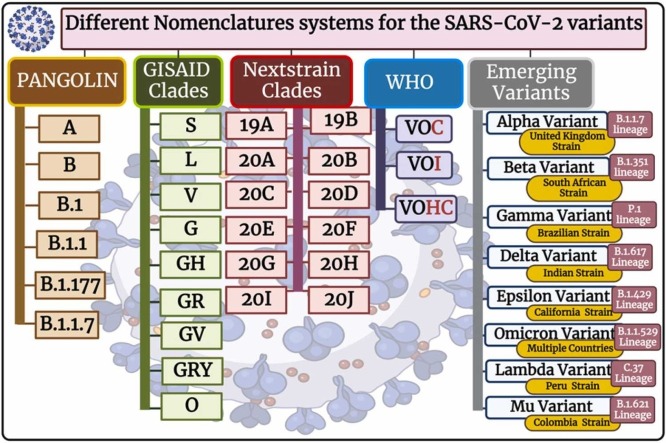
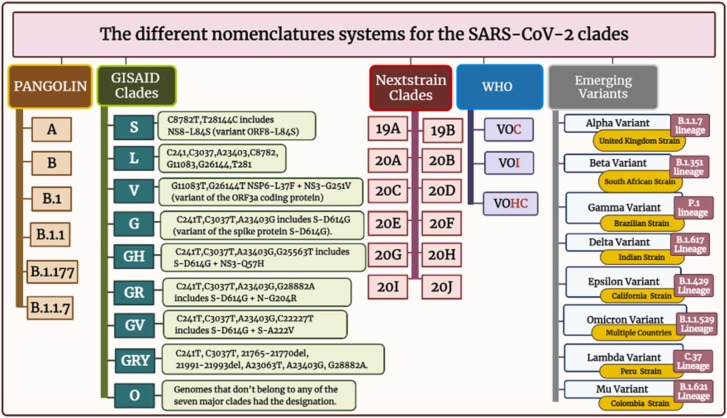
A clade in virology represents groupings of related viruses’ pieces of genetic blueprints, and taxonomy can be used to follow changes in those viruses. The clades explore information about virus infection symptoms severity, progression, treatment responses, and the rate of spread [130]. Different nomenclature systems are inured to nominate different SARS-CoV-2 variations; the most common are PANGO, GISAID, and Nextstrain, in addition to WHO and emerging variants systems [131], [132], [133] ( Fig. 3 ).
Fig. 3.
The different nomenclature systems for the SARS-CoV-2 clades.
8.1.1. PANGOLIN nomenclature
Rambaut et al. devised and built-in April 2020, the terminology in flux proposal for SARS-CoV-2 lineages, likewise identified as PANGOLIN (the phylogenetic assignment of named global outbreak lineages), to document and track SARS-CoV-2 transmission lineages globally [132], [133]. The PANGOLIN nomenclature contains the main lineages of A and B. in addition to B.1 and B.1.1, besides B.1.177 and B.1.1.7 which are furtherly divided (Fig. 3). The PANGOLIN nomenclature highlights viral lineages that are currently circulating as well as those that transfer to new hosts [133]. The A lineage has two nucleotides at 8782 and 28144 in ORF1ab and ORF8, respectively, with the RaTG13 and RmYN02 defined bat viruses. In contrast, lineage B has different nucleotides at these two places. They then went on to define things even more. Subsequently, the SARS-CoV-2 ancestries are given a number value and incline from the A or B lineage [29], [133], [134].
8.1.2. The GISAID nomenclature
The GISAID (Global Initiative on Sharing Avian Influenza Data) [151] has hundreds of full and high-coverage genomes available [135]. The types of SARS-CoV-2 clades, according to GISAID, include S, L, V, G, GH, GR, GV, GRY, and O [136] (Fig. 3 ). At the start of the pandemic, the S and L clades were present. S remained dominant at first, while L was divided into G and V. G was further divided into GR, GH, and GV. GR split into GRY [135]. The letters are the consequence of the mutations that resulted in their branching. Based on polymorphisms at nucleotide locations 8782 and 28144, S stands for SNP at 28144, which results in a serine codon, while L stands for leucine codon [137], [138]. The GR Clade was the most common SARS-CoV-2 clade globally, followed by the GV clade and the GH clade. In republics with extra COVID-19 cases, the clades of D614G are victorious [139]. The more frequently detected clades in COVID-19 cases that were severe or died are the GR clade and GH clade. Disobediently, the G and GV clades were shown to have a much higher incidence among asymptomatic or mildly ill patients. Males have an advanced incidence of severe/dead cases than females, and female patients have a higher frequency of the GR clade. Moreover, the disease severity/the disease rate was more prevalent in the aged than in the adults/children. The GV clade is more frequent in children than adults [137], [138].
8.1.3. Nextstrain nomenclature
The Nextstrain nomenclature currently recognises 12 major clades include: 19 A clade, 19B clade, 20 A clade, 20B clade, 20 C clade, 20D clade, 20E clade, 20 F clade, 20 G clade, 20 H/501Y.V2 clade, 20I/501Y.V1 clade, and 20 J/501Y.V3 clade [140], [141]. The clades are labeled with a letter and the year they're supposed to have arisen. The first strain to be used as a reference within these clades is 19B. However, due to the global travel ban, no more clades were created based on the criteria. As a result, Nextstrain added regional frequency (>30%) and known variations of concern to their main clade designation criteria [142].
8.1.4. WHO nomenclature
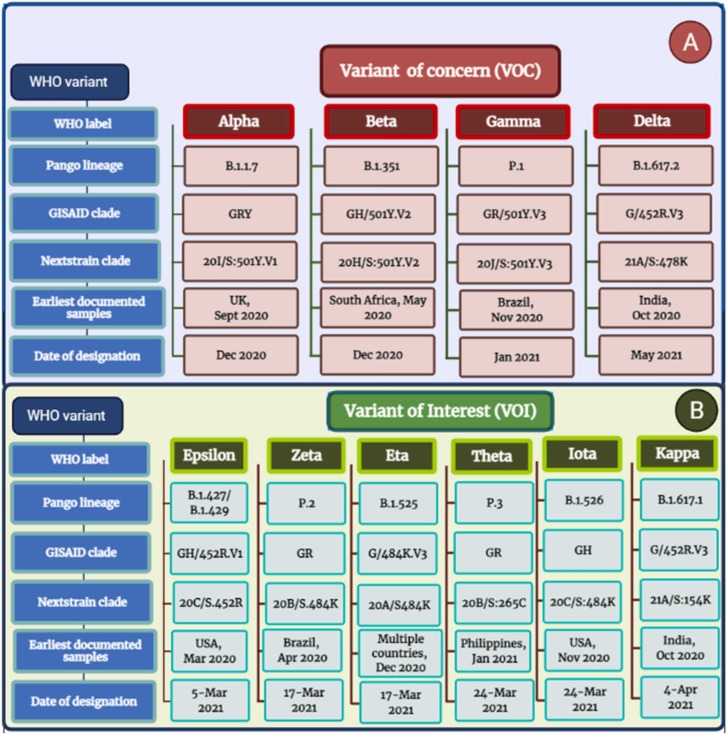
Since the start of the pandemic of COVID-19, the World Health Organization (WHO), more specifically the reference laboratory network, has been tracking this virus's spread and progression. The WHO working group on virus progress was formed in June of the year 2020, with a focus on new-fangled lineage, progress, and implication of different healthcare measures. In addition, the WHO risk-monitoring framework was created globally to track COVID-19 genetic changes, testing and evaluation, and their impact. This collaboration collects, evaluates, and makes decisions for the world according to the situation to help improve health care workers and professional capacities [143], [144]. It's a bit complex; the GISAID, Nextstrain, and Pangolin nomenclatures for SARS-CoV-2 sequences from worldwide. WHO has announced concise, simple labels for the variant SARS-CoV-2 of concern (VOC) and the variant SARS-CoV-2 of interest (VOI), utilizing Greek alphabet letters such as Alpha, then Beta, and also Gamma, and so on ( Fig. 4 A and Fig. 4 B).
Fig. 4.
SARS-CoV-2 Variants of Concern and Variants of Interest.
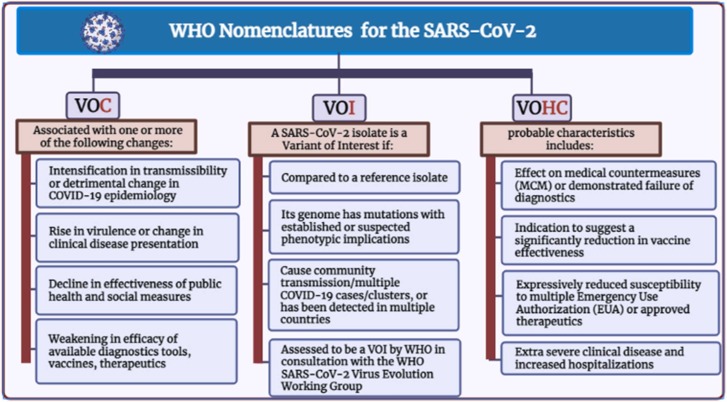
Besides, the variants of high consequences (VOHC) were considered ( Fig. 5). These labels have their merits and cannot replace the current scientific nomenclatures.
Fig. 5.
WHO nomenclature and different variants characteristics.
The current scientific terms can be difficult to say and remember and are prone to misreporting. Accordingly, people frequently refer to variations by their detection locations, which is both stigmatizing and discriminating. To overcome this and to facilitate service connections, WHO invites public governments, media outlets, and others to adopt these new names [143].
Inconsistent with the CDC (Centers for Disease Control and Prevention), the VOC has single or extra alterations that make the virus more easily infect people or spread from person to person, make the virus less responsive to treatments, or affect how well vaccines work against the virus [145]. The associated changes with VOC are included in Fig. (5). The VOC may necessitate sole or extra public health responses, such as reporting to WHO, warning to the CDC, particular geographic initiatives to prevent spread, heightened testing, or studies to discover vaccinations and therapies' performance in combating variation are only some of the options. Depending on the characteristics of the variant, other considerations may include the development of innovative diagnostics or the adjustment of immunizations or medicines [146]. In contrast, the variant of Interest VOI is a genetic mutation linked to fluctuations in binding to a receptor, diminished neutralization by antibodies developed against prior infection or vaccination, lower medical therapy efficacy, probable diagnostic effects, or projected upsurge in infection rates or clinical severity [143].
Fig. (5) includes the features of the SARS-CoV-2 isolate to be a VOI. The VOI may necessitate individual or even more efficient government healthcare measures, for instance, increased sequence surveillance, epidemiological inquiries, and better-quality test center description, to determine the spreading virus, the sternness of the infection, therapeutic efficacy, and vaccine potential protection [143], [146].
Moreover, the VOHC is defined by the CDC as a variant with evidence of diminished effects of deterrence and therapeutic countermeasures (MCMs) compared to prior variants [145]. Probable characteristics of VOHC are included in Fig. (5). Currently, no variants of the SARS-CoV-2 reached the degree of high consequences. However, the VOHC needs to report to the WHO and CDC to stop the spread, improve detection and treatment quality, and enhance vaccine protection [145].
8.1.5. Major Emerging Variants
New-fangled virus strains are surfacing in different areas of the world, with some variants exhibiting even more transmission and infection patterns. Countries including South Africa, the UK, India, and Brazil have increasing zones of concern [147], [148] (Fig. 3).
8.1.5.1. United Kingdom Strain (Alpha Variant)
The B.1.1.7, or alpha lineage, principally arose in the UK throughout September 2020 [149]. This VOC has an S protein alteration at position 501 receptor-binding domain (RBD) to tyrosine (Y) instead of asparagine (N). This mutation is N501Y [130], [148], [150]. Also, alpha Variant mutations, including 69/70 deletion: caused the S protein changes in conformation at P681H due to being befallen impulsively numerous times. The B.1.1.7 lineage has been reported in various nations Since it appeared on December 20, 2020. There is an association between an increase in B.1.1.7 lineage, augmented transmissibility, and death rate than other variants [130], [148], [150]. Though, no signs of illness severity worsening in kids and teens [151].
8.1.5.2. South African Strain (Beta Variant)
The B.1.351 or beta lineage (a.k.a. 20 H/501Y.V2) has numerous alterations such as A701V, D215G, D614G, LAL 242–244 del, K417N, E484K, R246I, D80A, L18F, and N501Y in the S protein, while the residual changes like K1655N, P71L, and T205I N in ORF1a, envelope (E), and viral proteins respectively [152]. Dissimilar to the B.1.1.7 ancestry noticed in the UK, this variant lacks the 69/70 deletion. This modified virus was detected in South Africa in October 2020 and in the USA [153]. Also, B.1.1.7 was recognized in Zambia in 2020 in late December and considered the country a predominant variant. This variant does not alter the severity of the disease, but some mutations affect response to the different strategies of immunizations and neutralization by some polyclonal and monoclonal antibodies derived from Pfizer vaccine recipients (6.5-fold) or Moderna vaccine recipients (8.6-fold) [148], [152], [154].
8.1.5.3. Brazilian Strain (Gamma Variant)
The P.1 or gamma lineage (a.k.a. 20 J/501Y.V3) is a subdivision of the B.1.1.28 variants that were reported first in four tourists from Brazil in Japan. The gamma variant contains the N501Y, E484K, and K417T mutations within RBD, which affect transmission pattern and antigen profile, leading to inappropriate antibody generation via prior infection or vaccinations [152]. In a study on a cluster of cases in Manaus, the P.1 lineage had been found in 42% of the sequenced specimens from late December, in which 75% of the population in this region was infected in October of the year 2020 with SARS-CoV2 [155], [156]. However, the number of cases was increased since mid-December, which raises worries of a probable upsurge in transmissibility and individuals’ re-infection with SARS-CoV-2 [148], [152].
8.1.5.4. Indian Strain (Delta (δ) Variant)
The (δ) B.1.617 Lineage was noticed in October in India in the year 2020, which was named B.1.617, which is also labeled as Delta (δ) variant [157]. Till January 2021, a few cases were reported. On the other hand, by April, the cases are reported in about twenty countries, reaching 50 countries by early May. It has spike mutations D111D (synonymous substitution), G142D P681R, E484Q, and L452R, the last two of which may help it to evade antibodies, among 15 characterizing mutations efficiently. This Delta (δ) variant contains L452R mutation and E484Q mutation; thus, it is named “double mutant [158]. The Delta (δ) variant in the next wave of pandemics starts in 2021 in India [148].
There are increasing rhino-orbital-cerebral mucormycosis (ROCM) cases in COVID-19-infected people, especially in India. Initially, 101 cases of Mucormycosis were reported in COVID-19 patients, of which 82 cases belong to India. By to end of May 2021, the rare fungal disease had killed more than 300 people and infected more than 12,000 in India [159]. Mucormycosis can affect the skin, the sinuses, the CNS, the GIT, the nose, the orbit, the pulmonary, the kidney, the joints, and the heart, but ROCM is the commonest. About 80% of cases have diabetes, while in 76.3% of cases, corticosteroid treatment was given. The gold standard for mucormycosis diagnosis is the clinical characteristics include necrotic, black turbinates easily misguided for dried, crusted blood; hence it’s given the black fungus name.
Additionally, it is difficult to establish a fundamental association between the black fungus and COVID-19 regarding corticosteroids. Nonetheless, several triggers, including high glucose, free iron, low pH, and ketones in the presence of low phagocytic activity of WBC, may precipitate the black fungus in people with COVID-19 concerning corticosteroids which enhances the growth of Mucormycetes. In India especially, reduced hygienic procedures and usage of animal dung in Indian traditional medicine measures provide a source of direct contact with the fungus and increased risk for Mucormycetes [148], [159].
8.1.5.5. California Strain (Epsilon Variant)
The B.1.429 Lineage / CAL 0.20 C Lineage first arose to the consideration of researchers in late 2020 as it surged in California [160]. Lineage B.1.429 has five different alterations, including: in the ORF1ab-gene (D1183Y and I4205V) and S-gene of the S protein (W152C, S13I, L452R). Alteration of concern in practice is L452R alteration. The lineage B.1.429 may be transmitted at higher rates but not explored and need additional research. By WHO, it is labeled as the Epsilon (ε) variant and principally noticed by California researchers in July 2020 in Los Angeles County in 1/1230 collected virus samples [160]. After the increasing numbers of ε-Variants in California. In most US states, the variation has been observed at variable frequencies. Also, small numbers have been noticed in additional countries, and its frequency rapidly declined in February of 2021 [148], [160].
8.1.5.6. Omicron variant
The Technical Advisory Organization on SARS-CoV-2 Virus Evolution (TAG-VE) monitors and analyses SARS-evolution CoV-2's changes to determine their effects. On November 26, 2021, the TAG-VE analyzed the B.1.1.529 COVID-19 (Omicron) variation from South Africa to WHO on November 24 [161].
The infectious picture has three distinct peaks in reported cases in South Africa, the most recent of which was driven by the Delta strain, corresponding with the appearance of the new-fangled B.1.1.529 (Omicron) strain which confirmed infection was in the sample on November 9, of the year2021. The new-fangled B.1.1.529, also known as Omicron, has been classified as a VOC by the WHO, according to the information given, indicating a harmful shift in COVID-19 infections [161]. There are various alterations in this variant. Unlike other VOC, the Omicron has a higher risk of reinfection, decreased response to treatment, and escaping immunization by vaccines [162].
Professor Tulio de Oliveira said Omicron has fifty genetic abnormalities (over 30 genetic alterations in the S protein SARS-CoV-2 and ten in RBD). Global immunization programs are worried about Omicron's RBD (10 changes) having more alterations than delta (2 alterations) [163]. In practice, the infection rate by COVID-19 increased with the discovery of the Omicron new variant infection throughout South Africa's provinces.
The Omicron variant was conveyed in the USA on December 1, 2021. On December 8, as a minimum one, the Omicron novel variant case had been detected in 22 states, with several indicating community transmission. With initial follow-up, no death and single case hospitalization were reported among the 43 instances [161]. Other countries where Omicron has been revealed include Botswana, Belgium, and Hong Kong. Furthermore, numerous republics, for example, England, may introduce a Trace program and Examination to track those who have recently visited a single of the countries where the Omicron variation has been found [163].
This Omicron is detected by the existing polymerase chain reaction (PCR) diagnostics of SARS-CoV-2. The S gene target failure was explored to be used as a marker for Omicron variant undecided sequencing validation. The Omicron variant was found earlier using this method than in prior outbreaks, signifying that it might have a development benefit [161].
8.1.5.7. Peru (Lambda variant)
The Lambda VOI, or the C.37 PANGO lineage, was principally recognized in Lima, Peru, in August 2020. In April 2021, virtually all sequenced genomic separates observed in Peru were recognized. In contrast to the wild-type virus, micro-neutralization experiments after BNT162b2 mRNA and BioNTech-Pfizer immunization showed a substantial 1.6-fold decline in neutralizing tires, indicating higher vulnerability of immunized persons to disease [164].
8.1.5.8. Colombia (Mu variant)
Lineage Mu (B.1.621) is the furthermost newly discovered VOI. The Mu variation was found in 39 republics as of August 30, 2021. Colombia is the center of Mu spread, with the variation being isolated for the first time on January 11, 2021. From March to July 2021, there was a considerable upsurge in Covid-19 cases in Colombia [165]. Throughout the primary phase of the surge, the Gamma version was dominating, but the Mu variant In May, exceeded all other variations, and it has been driving the outbreak in Colombia since then. The mainstream Mu variations have the YY144–145TSN and T95I alterations in the NTD; the N501Y, R346K, E484K transformations in the RBD; and the P681H, D614G, and D950N changes in further segments of the spike [165], [166], [167].
The Mu variant demonstrates a marked confrontation in the case of antibodies provoked via ordinary SARS-CoV-2 and the other type, the BNT162b2 mRNA vaccine. Since Mu innovation is the most important threat to recently evolving variants of the SARS-CoV-2, the Mu variant necessitates more characterization and monitoring [165], [166], [167], [168].
9. Development of COVID-19 vaccines
Once COVID-19 has reached an epidemic level in Wuhan, China. Chinese scientists universally identified the causative virus and shared its genome sequence [169]. This creates a universal competition to generate quickly suitable and effective vaccines utilizing traditional methods and modern next-generation vaccine platforms [170], [171].
The vaccines designed for SARS-CoV-2 are aimed to trigger the body’s immune response to generate antibodies towards S protein or RBD subunits and activate enabled T-lymphocytes [172], [173]. Many viral spikes help bind to the human cells, hence cellular entry and hijacking [174]. Thus, vaccine-generated neutralizing antibodies prevent viral spikes from binding and entering human cells, ending infection [175]. Remarkably, all the current approved COVID-19 vaccines by (US Food and Drug Administration) are highly active in generating these neutralizing antibodies [23], [176].
9.1. COVID-19 vaccine types
During the writing time (January 5, 2022), there were 24 vaccines recorded by National regulatory authorities (NRAs), but only ten of them were fully approved by WHO for use [177] ( Table 2). Six main vaccine types are categorized into two major vaccine platforms, classic and next-generation platform vaccines [178].
Table 2.
| Commercial name | Manufacturer | Country | Vaccine type | Efficacy against |
WHO effective date | |
|---|---|---|---|---|---|---|
| COVID-19 infection |
Severe illness |
|||||
| Covaxin | Bharat Biotech International Limited | India | Inactivated SARS-CoV-2 (Vero cell) | 77.8% | 93.4% | 3/11/2021 |
| Sinopharm | Beijing Institute of Biological Products Co., Ltd. | China | Inactivated SARS-CoV-2 (Vero cell) | 78.1% | 100% | 7/5/2021 |
| Coronavac (Sinovac) | Sinovac Life Sciences Co., Ltd. | China | Inactivated SARS-CoV-2 (Vero cell) | 50.7% (Brazil) 83.5% (Turkey) |
100% | 1/6/2021 |
| Nuvaxovid | Novavax CZ | Czech Republic | SARS-CoV-2 rS | 89.7% (UK) 90.4% (USA, Mexico) |
100% | 20/12/2021 |
| Covovax | Serum Institute of India Pvt. Ltd. | India | SARS-CoV-2 rS protein nanoparticle |
NR | NR | 17/12/2021 |
| Spikevax (Previously Moderna) |
Moderna Biotech | Spain | mRNA Vaccine (Nucleoside modified) |
93.2% | 98.2% | 30/4/2021 |
| Comirnaty | Pfizer/BioNTech GmbH |
Germany | mRNA Vaccine (Nucleoside modified) |
91.3% | 96.7% | 31/12/2020 |
| Janssen (Ad26. COV2-S) | Janssen – Cilag International NV (Janssen) | Belgium | Recombinant Ad26 vector encoding SARS-CoV-2 S | 52.4% | 74.6% | 12/3/2021 |
| Vaxzevria | AstraZeneca/SK Bioscience Co. Ltd | Korea | Recombinant ChAdOx1 vector encoding SARS-CoV-2 S | 74% | 100% | 15/2/2021 |
| Covishield | Serum Institute of India Pvt. Ltd. | India | Recombinant ChAdOx1 vector encoding SARS-CoV-2 S | NR | NR | 15/2/2021 |
S: spike, r: recombinant; Ad: adenovirus; NR: not reported.
9.1.1. Classic platform vaccines that include
9.1.1.1. An inactivated vaccine
The generation of inactivated COVID-19 vaccine in Vero cells involves the proliferation of a live virus using a cell culturing technique and downstream processing for recovery and concentration, followed by inactivation with chemical substances to terminate cellular infectivity and maintain immunogenicity [179].
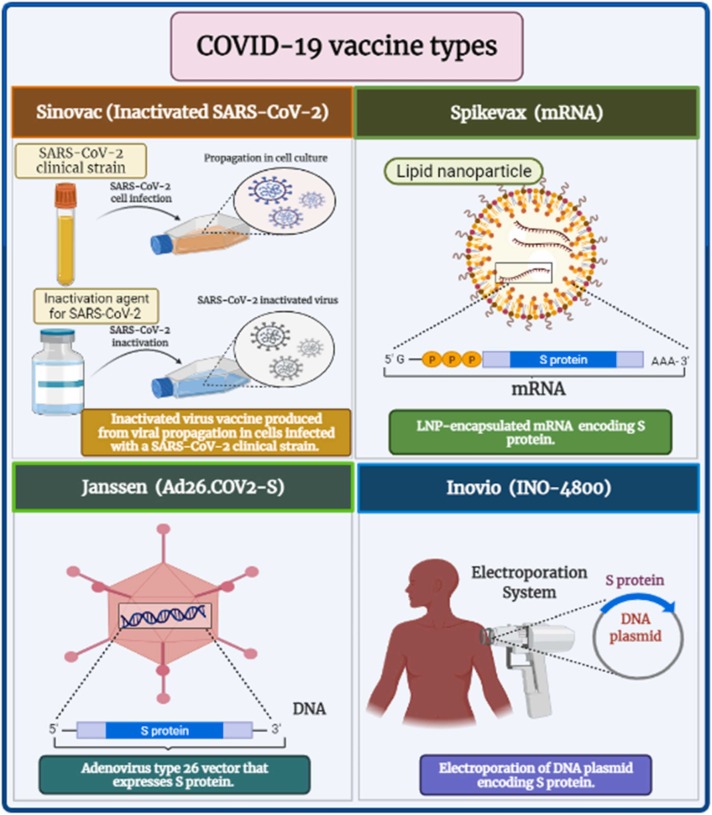
This type activates various immunological reactions in response to the existence of an entire virus with its different surface antigens. Besides, it has low stability and a short action period. Therefore, it evolves an immune response less than a live attenuated virus [181], [182]. Some examples of the currently available inactivated COVID-19 vaccines are Sinopharm, Sinovac ( Fig. 6), and Covaxin. In the production of these vaccines, the β-propiolactone organic compound was used to deactivate the SARS-CoV-2 virus via binding to their genes. Some adjuvants were added to elicit a longer immune response duration [183], [184].
Fig. 6.
COVID-19 vaccine types.
9.1.1.2. Live attenuated vaccine
Live attenuated vaccines are ancient vaccines created by reducing the virulence of a live pathogenic virus [185]. They elicit a long-lasting and high immune response comprising TLRs activation without the addition of adjuvant due to their contents of multivalent antigens that can confer a strong defensive action towards newly developed viral variants [186], [187]. On the other hand, they are unsuitable for immunocompromised patients and difficult to handle during production [188].
Currently, no live-attenuated COVID-19 vaccines are approved. In December 2021, the Codagenix and Serum Institute of India announced launching the first trial of the Covi-Vac live attenuated intranasal vaccine in humans [189].
9.1.1.3. Viral protein subunits-based vaccines
The viral protein vaccine production entails selecting, isolating, and purifying a viral protein that can trigger the immune response and fail to induce any disease[190]. In this COVID-19 vaccine type, the proteinaceous S or RBD subunits are often utilized as immunogenic proteins[191], [192]. Also, adjuvants are often added to trigger a more extended immune response and activate the high production of neutralizing antibodies. Viral subunit vaccines are safer than entire viral vaccines [193]. Both Covovax and Nuvaxovid vaccines contain recombinant (r) S protein nanoparticles of SARS-CoV- 2 that elicit potent humoral and cellular immune actions [194].
In Covovax and Nuvaxovid vaccine production, a genetically engineered baculovirus comprising a gene of SARS-CoV-2 rS is created [195], [196]. After that, the produced S protein is isolated and assembled into lipid nanoparticles (LNPs). Matrix-M was added as an adjuvant to promote immune action [197], [198].
9.1.2. The next-generation platform vaccines include
9.1.2.1. Viral vector established vaccine
A viable replicated, or unreplicated genetically engineered viral vector is constructed to transfer genes of SARS-CoV-2 antigens into the intended host cells. Once the targeted cells are infected and eventually directed to forming a large quantity of SARS-CoV-2 antigens, cellular and humoral immunity are highly triggered [194], [199].
Many viruses have been designed as vectors comprising adenovirus and vaccinia virus. Viral vector-established vaccines are developed by deleting pathogenic genes and sometimes replicating genes [200]. Moreover, viral vectors' transferred genes are not incorporated into the host cellular genome. Therefore, they are harmless. Besides, they elicit an immune response higher than viral protein-based vaccines [201], [202], [203].
Janssen COVID-19 Vaccines (Ad26. COV2-S) comprise a human recombinant replicating-incompetent form of adenoviral vector serotype 26, including the S gene sequence of SARS-CoV-2 [204] (Fig. 6). On the other hand, Vaxzevria and Covishield vaccines contain a Chimpanzee recombinant non-replicating form of adenoviral vector (ChAdOx1) encoding SARS-CoV-2 S [205], [206]. The concern about the high level of neutralizing antibodies in response to familiar human adenoviruses in the Janssen COVID-19 vaccine due to the possibility of a previous immunity against them is limited using a Chimpanzee adenoviral vector in Vaxzevria and Covishield vaccines [205].
9.1.2.2. mRNA constructed vaccine
In this vaccine, viral messenger RNA (mRNA) is constructed to generate proteinaceous S or RBD subunits of SARS-CoV-2 and assimilated inside LNPs to promote its delivery into host cells and prevent degradation [207]. After vaccination, the LNPs enclosed mRNA enters host cells, and then the lipid layer is gradually degraded, and mRNA is released and becomes available for cellular translational machinery to produce its corresponding viral protein, which can elicit both cellular and humoral immunity after trafficking to the plasma membrane [208], [209].
Recently, mRNA-constructed vaccines have been considered a favorable substitute for traditional vaccines due to their high efficiency, safety, low investment, and fast production [210]. Although the previous advantages, their production and utilization are limited due to low stability, ultra-freezing formulation, and sometimes a failure of mRNA delivery into a target human cell [211], [212]. Comirnaty and Moderna vaccines contain LNPs enclosing mRNA-1273 which is constructed through the transcription from its corresponding sequence of DNA template and translated to the proteinaceous S subunit of SARS-CoV-2 [213], [214] (Fig. 6).
9.1.2.3. DNA constructed vaccine
This vaccine technology involves the utilization of bacterial DNA plasmids, which harbor gene sequences for encoding SARS-CoV-2 S or RBD subunits and mammalian-type expression promoters [215], [216], [217]. These plasmids can penetrate the nuclear membrane of B-lymphocytes and eventually encode a protein of interest [218]. Plasmid DNA has been successfully delivered using electroporation [218].
DNA-constructed vaccines are characterized by high stability, ease of shipping, and storage for a long time [219], [220]. On the other hand, the possibility of incorporating transfected genes into the genome of human host cells is one of the significant drawbacks that can lead to a mutation [221].
In August 2021, ZyCoV-D (Indian pharma Zydus Cadila) was India's first DNA vaccine to acquire emergency authorization for COVID-19 [222]. In contrast, INO-4800 (Inovio Pharmaceuticals, China), GX-19 N (Genexine, Korea), and AG0302- COVID-19 (AnGes, Japan) are undertaken the trials of Phase (2/3) [216] (Fig. 6).
10. The impact of the SARS-CoV-2 mutations and genetic diversity on the COVID-19 biomedical arbitrations
SARS-CoV-2 gets mutations when it spreads to new areas. These mutations help SARS-CoV-2 acclimate better within the hosts and new geographical locations. Scientists must closely monitor and track SARS-CoV-2 dynamics, mutations, and genetic diversity to produce more active vaccines. Despite various research, the effects of the host's genetic variables and the genetic variations of SARS-CoV-2 remain unknown [223].
Most SARS-CoV-2 infections are asymptomatic and self-limiting. However, 2% of COVID-19 patients experience serious adverse effects. COVID-19 severity and mortality are largely affected by older age, respiratory, metabolic, immunological, and cardiovascular comorbidities. According to reports, the environment, humans, and animals spread SARS-CoV-2 [224].
The death rates of the COVID-19 virus vary between the countries, generally known to be meaningfully higher in the North area of America and similarly in European countries. Even though numerous opportunities to clarify the alterations in the death rates are confirmed, the difference between virus genomic types and age distribution is still unclear. According to recent evidence, the death rates of SARS-CoV-2 can change significantly depending on the environment. For example, a study showed that the death rate is three times higher outside China, 15.2%, than inside China, 5.6% [225]. Changes in viral infection rates could be due to a combination of reasons, such as isolation, quarantine, the difference in the genetic population, herd immunity, and diverse national approaches permitted for the restrictions of the movement of people [22].
The viral adaptation is underwritten by the RNA virus's mutation frequency, which maintains a balance between genome diversity and genetic information integrity [226].
Plasma from COVID-19-infected convalescent patients is the simplest technique to treat SARS-CoV-2 pandemic infections [227]. In a nutshell, convalescent people's polyclonal NABs (neutralization antibodies) were collected and successfully used to treat newly infected patients [18]. NABs from a donor with a different S protein phenotype may not work for people with a mutant S protein. The NAB of SARS-CoV-2, on the other hand, is likely to be useful for an infected person. The antibodies may enhance pathogenic immunological progressions in COVID-19 patients with various viral genome content [228].
The validation and assessment of SARS-CoV-2 infection employing lateral flow immunochromatography RDT and ELISA are crucial due to COVID-19's influence on the global economy and the necessity to increase COVID-19 laboratory tests. Although the WHO has not validated all antigen and antibody tests, serological studies could help analyze an ongoing SARS-CoV-2 outbreak, track its recurrence rate, and maintain the COVID-19 diagnosis when real-time PCR results are negative [229].
The RDTs for IgG and IgM antibodies will also play an increasingly crucial role in recognizing individuals without symptoms and evaluating the immunity of healthcare workers if the outburst continues. Cross-reactive antibodies are routinely screened using an S protein ELISA, without a doubt. Antibodies to SARS-CoV-2 with distinct epitopes produced by either N or S mutant proteins may lower the positive predictive value of anti-SARS-CoV-2 antibodies [230], [231].
Mutation-generating processes comprise unprompted damages of the nucleic acid, which may be owing to the host enzymes, particular genetic elements, chemical, and physical mutagens. Likewise, recombination events are involved in creating novel variations [22].
With certain exceptions, such as the Nidovirales order, which distinguishes out for having the largest genomic RNA, the RNA polymerase in several viruses cannot proofread.
To allow virus transcription and replication, Nidoviruses have a complex RNA synthesis machinery controlled by non-structural proteins produced as cleavage products of the ORF1a and ORF1b viral polyproteins [29]. The most critical component of the transcription and replication machinery is the SARS-CoV-2 RdRp (nsp-12). The SARS-CoV-2 has a high homology for the nsp-12 compared to the SARS-CoV, indicating that its mode of action and function may be well preserved [29].
By correcting the RdRp's nucleotide incorporation errors, the ExoN improves the RNA synthesis's dependability. When compared to SARS-CoV wild type, genetic inactivation of the coronavirus ExoN resulted in a twenty-one-fold reduction in replication fidelity. Kirchdoerfer also discovered that in the SARS-CoV virus, nsp-7 and nsp-8 were involved in forming a complex with RdRp [232], [233], [234]. Until now, the serious RdRp residues of SARS-CoV intricate in the nsp-7, nsp-8, and ExoN interaction have still to be recognized[235]. The RdRps are regarded as one of the most crucial drug targets of antiviral medications against various viruses. Some RdRp inhibitors, such as Galidesivir, have been regarded as potential targets for SARS-CoV-2 [236], Favipiravir [237], Ribavirin [238], and Remdesivir [239].
Furthermore, drugs, including Simeprevir, Filibuvir, Tegobuvir, and Cepharanthine, have been suggested as potential RdRp inhibitors [240]. Several antiviral medications target SARS-CoV-2's RdRp. As a result, identifying and studying SARS-CoV-2 RdRp mutations is essential to identifying drug-resistant SARS-CoV-2 strains [22].
11. Impression of the foremost variants on the efficiency of vaccine
COVID-19 vaccines that have been approved or are currently being developed are expected to give some protection against emerging virus variations because they elicit a broad immune response involving a variety of cells and antibodies. Therefore, the mutations or the changes in the virus should not make the vaccines unsuccessful. Unpredictably, suppose any of these vaccines are less effective against one or more variants. In that case, it will be conceivable to change the composition of vaccines to defend against these different variants. The world must stop COVID-19 from spreading to prevent mutations that could weaken vaccines [241].
Furthermore, the programs and manufacturers that use the vaccines may also need to adapt to the COVID-19 virus's evolution: for example, vaccines may need to incorporate more than one strain during development, booster shots may be required, and other vaccine alterations may be needed. Likewise, the trials must be maintained and designed to evaluate any changes in efficacy and be of appropriate diversity and scale to empower durable interpretation of the results [241].
11.1. Neutralization of the variations by the convalescent sera
The genuine virus' ability to promote herd immunity and antibody treatment in convalescent or vaccinated individuals under discerning stress of the polyclonal immune response is an important concern for vaccine application [240]. Neutralization tests used SARS-CoV-2 convalescent serum to test novel variations for immune evasion. Surprisingly, the wild isolates B.1.531 and B.1.1.7 used to test convalescent sera by Gavin et al. had reduction ratios of 13.3 and 2.9, respectively [242], [243].
In addition, a study investigated 44 plasma samples from people who have actually been infected with SARS-CoV-2 using various pseudoviruses, including K417N, D614G, N501Y, and E484K, and discovered that nearly 48% of the samples had no noticeable neutralization activity, with only three samples preserving the titers of the inhibitory dilutions giving about 50% of neutralization [244].
11.2. The vaccination stimulated the neutralization of antibodies to the variations
11.2.1. Comirnaty vaccine
The neutralizing ability of sera from vaccinated individuals against different variations was evaluated using infectious cDNA clones and pseudoviruses. Muik et al. recognized the B.1.1.7 pseudoviruses and Wuhan reference strain in serum samples from forty patients who were administered with the Pfizer (BNT162b2) vaccination in the previously published German (Phase 1/ 2 investigation). The neutralization titers against the B.1.1.7 decreased slightly, but the immune sera completely preserved them, suggesting that B.1.1.7 will not affect the Comirnaty vaccine-mediated protection [245].
Pseudoviruses with spike mutations in B.1.1.7 (del69/70, A570D, P681H, and D1118H) reduce sensitivity to vaccination serum, according to Collier et al. According to the data, the sera of ten of the 23 participants who had received a single dose of the Comirnaty vaccination exhibited a decrease in neutralization against the B.1.1.7 type. There was no difference in inability when the sera were tested with pseudoviruses having the A570D and 69/70 deletion mutations [246].
Sera collected after one dosage of the two-dose regimen only exemplifies partial immunity. Compared to the B.1.1.7 variant, the nullification power of the Comirnaty vaccination immune sera against the B.1.351 variation deteriorated significantly. According to Gavin et al., neutralization titers against B.1.351 variants decreased 7.6-fold, while the fold weakening against B.1.1.7 was 3.3 [242], [243].
The responsibility of the N501Y mutant has been the subject of more research. According to Rathnasinghe et al., the sera from six people who had received two doses of the Pfizer vaccination had to neutralize antibody titers equivalent to the greatest neutralization titers observed in convalescent sera against the mouse-adapted viruses with the N501Y mutation [247]. Another study used an isogenic pair of viruses containing the N501 or Y501 S protein to assess serum neutralization in twenty people who had obtained both doses of the Comirnaty vaccine [154]. Both studies demonstrated that N501Y mutation did not affect the possibility of post-vaccination neutralization [248].
11.2.2. Spikevax vaccine
The ability of the SARS-Co-2 Spikevax mRNA vaccine (mRNA-1273) to neutralize variations was comparable to the Comirnaty vaccine. The sera from 12 people who received the Spikevax vaccine were tested for neutralization versus B.1.351, WT, and B.1.1.7 pseudoviruses. The results showed no significant differences between the Comirnaty vaccine and the Spikevax vaccine against the B.1.351 and B.1.1.7 variants [249]. Shen et al. used a pseudovirus experiment to show that the Spikevax vaccine conferred protection against the B.1.1.7 variant in 40 people. According to the study, vaccination sera neutralized the B.1.1.7 variation, whereas the susceptibility was decreased substantially when compared to the D614G version [250].
11.2.3. Vaxzevria vaccine
The ability of the Vaxzevria vaccination to cross-neutralize these variations has piqued curiosity.
According to Gavin et al., serum neutralization titers in patients who received the Vaxzevria vaccine at 14 and 28 days following the second dose revealed a 2.5-fold and 2.1-fold decrease against B.1.1.7 strain, correspondingly [242]. They used Vaxzevria vaccine-elicited serum obtained 14 or 28 days after the second dose of vaccine to evaluate the neutralization of a B.1.351 variation at the time [243]. From these data, the Vaxzevria vaccine offers more protection against the B.1.1.7 variant than the B.1.351 variant.
11.2.4. Sinopharm vaccine
The immunological serum of the Sinopharm vaccinations developed by Chinese companies was examined by Gao et al. The analysis uncovered many differences between mRNA vaccines and pseudovirus vaccinations. According to the findings of 24 samples, the neutralizing ability of two immunizations against the B.1.351 variant was lowered by 1.6-fold. GMTs of the immunological serum produced by the BIBP vaccination reduced from 110.9 to 70.9, while the number of people who received the Zhifei vaccine declined from 106.1 to 66.6 [251]. As a result, they stated that the variant of B.1.351 did not influence the protective effects of the two vaccinations.
Phase 3 clinical trials in South Africa of three vaccines from Vaxzevria, Janssen, and Novartis showed that distinct variants affected the vaccines' defensive efficacy. Phase 3 clinical studies of Novavax Inc.'s NVX-CoV2373 in the U.k. revealed an 89.3%-point estimate of vaccination efficacy. Based on PCR data from 56 of the cases treated, efficacy against the U.K. variation was determined to be 85.6% and against the original SARS-CoV-2 strain to be 95.6%. The primary efficacy target of the South African Phase 2b clinical trials was attained with an efficacy of 49.4%. Initial sequencing information was available for 27 of the 44 SARS-CoV-2 cases, with the B.1.351 variant accounting for 92.6% of the total [252]. As a result, there are new worries about the risk of secondary infections and the vaccine's effectiveness in affected individuals.
The vaccination was 66% effective overall in preventing moderate to severe SARS-CoV 2 infections in all participants from various geographical areas and those infected with a novel viral strain during the Phase 3 ENSEMBLE clinical trial for Janssen's SARS-CoV-2 vaccine candidate. In Latin America, 66% of people were protected against SARS-CoV-2 infection, compared to 72% in the U.K. and 57% in South Africa. All SARS-CoV-2 cases in South Africa were almost caused by the B.1.351 strain of the SARS-CoV-2 [253].
According to clinical trials conducted by the Vaxzevria vaccine in the United Kingdom, the vaccine provided protection not only against the original pandemic virus but also against the novel B.1.1.7 strain. Despite the vaccine's great efficacy against non B.1.351 coronavirus variants, the results of the South African study demonstrated that the Vaxzevria vaccine's neutralization of immunological sera against the B.1.351 variant was drastically reduced compared to the original strain. The two-dose Vaxzevria vaccination provides the least protection against SARS-CoV-2 infection produced by the B.1.351 strain of coronavirus [249].
12. Prevention from the future novel versions of COVID-19
Future COVID-19 virus spread can be stopped if the virus replication process is slowed or stopped [241]. Furthermore, the current measures to diminish the transmission, including wearing a mask, hand washing, good ventilation, physical distancing, and avoiding closed settings or crowded places, continue to labor against the innovative variants by reducing the quantity of the transmission of the virus and thus also decreasing the opportunities for the virus mutation [241].
Furthermore, scaling up the rolling out and manufacturing of vaccines as quickly as possible will also be an important way to protect all people before they are infected with the virus or exposed to the risk of novel variants. The high-risk groups should be vaccinated universally to increase protection against recent virus variants worldwide and minimize the risk of virus transmission. Additionally, maintaining equitable access to the COVID-19 vaccines is more dangerous than ever to discourse the evolving pandemic. When more people get vaccinated, we assume the virus circulation will decrease, leading to fewer mutations [241].
Once the public transmission of the SARS-CoV-2 exists, people are revitalized to make the social distance by avoiding the crowds and keeping two meters from others when in public. Particularly, people should sidestep close contact with the ill person. They are also reinvigorated to wear face masks [254].
12.1. The general procedures are endorsed to reduce the spreading of the virus infection
Recommendations include washing hands thoroughly, especially after contacting public surfaces, and using hand sanitizers with at least 60% alcohol [255] as SARS-CoV-2 was deactivated in fifteen seconds when exposed to 80% ethyl alcohol. Also covering the sneeze or cough, avoiding touching the face (in particular eyes, mouth, and nose), confirming that the indoor spaces have enough ventilation [256]. Wearing a facial mask is advised by the WHO as part of an overall approach to reducing SARS-CoV-2 infection in outdoor and indoor situations where interpersonal distance is problematic and extensive propagation, as well as in interior situations with inadequate ventilation [257].
12.2. Effectiveness and booster of vaccines against different COVID-19 strains
A booster dose is a dose of vaccine given just after the primary vaccination series, such as one dose of Ad26. COV2. S or two doses of BNT162b2, ChAdOx1, or mRNA-1273. According to recent research, new variants like Delta and Omicron showed potent immune evasion to neutralizing antibodies produced by primary vaccination or prior infections [258], and that protective effects diminish over time [259].
More breakthrough infections occur in fully immunized individuals due to this deteriorating protection. For instance, in the follow-up tests after six months, the effectiveness in avoiding disease dropped from an initial 90–60%. As a result, the topic of discussion has shifted to the longevity of vaccine-induced protection and the requirement for recurrent booster immunizations [260]. Booster vaccinations provide better defense against the COVID-19 virus. In comparison to two doses of the BNT162b2 vaccine, a third dosage demonstrated 95.3% relative efficacy against COVID-19, according to a phase III trial [261].
In the latter quarter of 2021, when Delta started to take precedence over other variants, many nations began to offer COVID-19 boosters. Although Omicron offered a different problem due to the sharp rise in breakthrough infections by the end of 2021, boosters nevertheless effectively protected against serious illness and hospitalization brought on by Delta. Fortunately, boosters were observed to raise serum anti-spike antibody concentrations and the neutralization titers against Omicron in recipients boosted with BNT162b2, mRNA-1273, or generic mRNA vaccinations [262], [263], [264].
Furthermore, it has been demonstrated that both CoronaVac and mRNA vaccine [265] boosters increase memory B cells specific for anti-receptor binding domains and hasten the production of antibodies that are directed against various variations, including Omicron, in those who have received the vaccines. In boosted people, breakthrough infections of Delta and Omicron are still possible, although the virus loads appear to be lower and the symptoms are slighter [266], [267].
For vulnerable and high-risk groups, booster immunizations are particularly crucial. For instance, studies have shown that to achieve adequate protection against Delta and Omicron, cancer patients undergoing treatment, solid organ transplant recipients, and the elderly [268] require more BNT162b2 or mRNA-1273 boosters [269], [270], [271].
Israel began providing a second booster in January 2022 to elderly and immunocompromised people [272]. In March 2022, second mRNA boosts were also made available in the US to the elderly and those with underlying medical issues. According to a retrospective study conducted in Israel, adults over 60 years old who received a second dosage of BNT162b2 had a lower risk of COVID-19-related hospitalizations and fatalities [273].
The effectiveness against infection with Delta and Omicron was estimated to be 88–93% and 76%, respectively, for patients who got a homologous BNT162b2 booster, whereas the effectiveness against severe disease brought on by Delta and Omicron was evaluated to be 97% and 89%, respectively [274], [275].
Additionally, the efficacy against infection with Delta and Omicron was 94% and 71%, respectively, in ChAdOx1-primed BNT162b2-boosted people [274]. Israeli observational studies also showed that BNT162b2 booster was linked to a decreased risk of SARS-CoV-2 infection in young people, old people, and healthcare workers [276], [277]. Finally, booster dosages were discovered to be very helpful in reducing the risk of infection and serious illnesses brought on by Delta and Omicron variations [278].
13. Conclusion
Globally, COVID-19 swiftly propagates, and it is caused by the zoonotic transmission of SARS-CoV-2 from an animal to becoming a human. This virus affected countries' economic, medical, and public health infrastructures all across the world and resulted in the deaths of a large number of people all over the world due to mutations and new variants development. People from older age groups with underlying health problems and low immunity are more likely to become infected and develop severe symptoms.
Mutations in the S protein and proteins comprising the RDRP were the most common. This mutation leads to different variant expansions. Accordingly, a different nomenclature system, including PANGOLIN, classifies variants into two major classes, A and B. While the next strain divided variants by the year and litter, e.g., 19 A, 19B, 20 A, 20B, and so on. Also, the GISAID classification letter such as V, S, L, G, GR, GH, GRY, GV, and O are consequences of the mutation, and the SNP results in the codon for amino acids e.g., S serine, L leucine.
In contrast, the WHO represents a simple classification and dominant variants as VOC, VOI, and VOHC. Finally, the emerging variants classify variants according to the site where they were first discovered e.g., Alpha variant (UK), Beta variant (South African), Gamma (Brazil), and omicron (South African). Subsequently, different vaccines were developed to prepare the immune system to fight the SARS-CoV-2 virus. They were categorized into two platforms: classic, which includes inactivated, live attenuated, and viral protein subunits-based vaccines, and next-generation platform, which includes viral vector established, mRNA constructed, and DNA constructed vaccines.
The mutations and new variants of SARS-CoV-2 cause increased transmissibility, morbidity, and mortality. They can also escape detection by escape diagnostic tests, show lower susceptibility to therapy with antivirals and antibodies, and finally, reinfect previously recovered and vaccinated people. Lastly, the future COVID-19 variants and widespread prevention strategies necessitate individual efforts as well as worldwide collaborations between scientists, authorities, and the general public to perform global genomic surveillance of SARS-CoV-2 in new cases since it is crucial to rapidly identify any mutation that could result in changes in the phenotypes of the virus. Also, vaccination and global health applications of the general protection measures are endorsed to decrease the spread of infection with COVID-19.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not for profit sectors.
CRediT authorship contribution statement
Ahmed I. Abulsoud: Conceptualization, Methodology, Supervision, Visualization, Writing – original draft. Hussein M. El-Husseiny: Conceptualization, Resources, Supervision, Writing – review & editing. Ahmed A. El-Husseiny: Data curation, Methodology, Writing – original draft. Hesham A. El-Mahdy: Conceptualization, Visualization, Supervision, Writing – original draft, Writing – review & editing. Ahmed Ismail: Conceptualization, Formal analysis, Supervision, Writing – original draft. Samy Y. Elkhawaga: Project administration, Writing – original draft. Emad Gamil Khidr: Investigation, Visualization, Writing – original draft. Doaa Fathi: Conceptualization, Visualization, Writing – original draft. Eman A. Mady: Conceptualization, Visualization, Writing – original draft. Agnieszka Najda: Supervision, Resources, Writing – review & editing. Mohammad Algahtani: Supervision, Resources, Writing – review & editing. Abdulrahman Theyab: Supervision, Resources, Writing – review & editing. Khalaf F Alsharif: Supervision, Resources, Writing – review & editing. Ashraf Albrakati: Project administration, Writing – review & editing. Roula Bayram: Project administration, Writing – review & editing. Mohamed M. Abdel-Daim: Project administration, Writing – review & editing. Ahmed S. Doghish: Conceptualization, Resources, Supervision, Writing – original draft, Writing – review & editing.
Conflict of interest statement
The authors declare they have no conflict of interest.
Acknowledgments
The authors would like to thank Prof. Mahmoud M. Tawfick (Department of Microbiology and Immunology, Faculty of Pharmacy, Heliopolis University) for the professional scientific editing of this manuscript.
Data Availability
No data was used for the research described in the article. This is a Literature Review article without any data or code.
References
- 1.Ye Z.W., Yuan S., Yuen K.S., Fung S.Y., Chan C.P., Jin D.Y. Zoonotic origins of human coronaviruses. Int J. Biol. Sci. 2020;16:1686–1697. doi: 10.7150/ijbs.45472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou X., Wang Y., Gong C., Liu B., Wei G. Production, structural design, functional control, and broad applications of carbon nanofiber-based nanomaterials: a comprehensive review. Chem. Eng. J. 2020 [Google Scholar]
- 3.Doghish A.S., Elkhatib W.F., Hassan E.A., Elkhateeb A.F., Mahmoud E.E., Ahmed M.I., et al. Clinical characteristics of Egyptian male patients with COVID‐19 acute respiratory distress syndrome. Plos One. 2021;16 doi: 10.1371/journal.pone.0249346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forni D., Cagliani R., Clerici M., Sironi M. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017;25:35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lai C.C., Shih T.P., Ko W.C., Tang H.J., Hsueh P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int J. Antimicrob. Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020:370. doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malik Y.S., Sircar S., Bhat S., Sharun K., Dhama K., Dadar M., et al. Emerging novel coronavirus (2019-nCoV)-current scenario, evolutionary perspective based on genome analysis and recent developments. Vet. Q. 2020;40:68–76. doi: 10.1080/01652176.2020.1727993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giri R., Bhardwaj T., Shegane M., Gehi B.R., Kumar P., Gadhave K., et al. When darkness becomes a ray of light in the dark times: understanding the COVID-19 via the comparative analysis of the dark proteomes of SARS-CoV-2, human SARS and bat SARS-like coronaviruses. BioRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 9.Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lorusso A., Calistri P., Petrini A., Savini G., Decaro N. Novel coronavirus (SARS-CoV-2) epidemic: a veterinary perspective. Vet. Ital. 2020;56:5–10. doi: 10.12834/VetIt.2173.11599.1. [DOI] [PubMed] [Google Scholar]
- 11.Ji W., Wang W., Zhao X., Zai J., Li X. Cross-species transmission of the newly identified coronavirus 2019-nCoV. Med Virol. 2020;92:433–440. doi: 10.1002/jmv.25682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yi Y., Lagniton P.N.P., Ye S., Li E., Xu R.H. COVID-19: what has been learned and to be learned about the novel coronavirus disease. Int J. Biol. Sci. 2020;16:1753–1766. doi: 10.7150/ijbs.45134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meskini M., Rezghi Rami M., Maroofi P., Ghosh S., Siadat S.D., Sheikhpour M. An overview on the epidemiology and immunology of COVID-19. J. Infect. Public Health. 2021;14:1284–1298. doi: 10.1016/j.jiph.2021.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lakshmi Priyadarsini S., Suresh M. Factors influencing the epidemiological characteristics of pandemic COVID 19: A TISM approach. Int. J. Healthc. Manag. 2020;13:89–98. [Google Scholar]
- 15.Halaji M., Farahani A., Ranjbar R., Heiat M., Dehkordi F.S. Emerging coronaviruses: first SARS, second MERS and third SARS-CoV-2: epidemiological updates of COVID-19. Infez. Med. 2020;28:6–17. [PubMed] [Google Scholar]
- 16.Epidemiology Working Group for Ncip Epidemic Response CCfDC Prevention. [The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China] Zhonghua Liu Xing Bing. Xue Za Zhi. 2020;41:145–151. doi: 10.3760/cma.j.issn.0254-6450.2020.02.003. [DOI] [PubMed] [Google Scholar]
- 17.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., et al. Addendum: a pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;588 doi: 10.1038/s41586-020-2951-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krause P.R., Fleming T.R., Longini I.M., Peto R., Briand S., Heymann D.L., et al. SARS-CoV-2 Variants and Vaccines. N. Engl. J. Med. 2021;385:179–186. doi: 10.1056/NEJMsr2105280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rossler A., Riepler L., Bante D., von Laer D., Kimpel J. SARS-CoV-2 omicron variant neutralization in serum from vaccinated and convalescent persons. N. Engl. J. Med. 2022 doi: 10.1056/NEJMc2119236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asselta R., Paraboschi E.M., Mantovani A., Duga S. ACE2 and TMPRSS2 variants and expression as candidates to sex and country differences in COVID-19 severity in Italy. Aging (Albany NY) 2020;12:10087–10098. doi: 10.18632/aging.103415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pachetti M., Marini B., Benedetti F., Giudici F., Mauro E., Storici P., et al. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. Transl. Med. 2020;18:179. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramasamy M.N., Minassian A.M., Ewer K.J., Flaxman A.L., Folegatti P.M., Owens D.R., et al. Safety and immunogenicity of ChAdOx1 nCoV-19 vaccine administered in a prime-boost regimen in young and old adults (COV002): a single-blind, randomised, controlled, phase 2/3 trial. Lancet. 2021;396:1979–1993. doi: 10.1016/S0140-6736(20)32466-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosano G., Jankowska E.A., Ray R., Metra M., Abdelhamid M., Adamopoulos S., et al. COVID-19 vaccination in patients with heart failure: a position paper of the Heart Failure Association of the European Society of Cardiology. Eur. J. Heart Fail. 2021;23:1806–1818. doi: 10.1002/ejhf.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirtipal N., Bharadwaj S., Kang S.G. From SARS to SARS-CoV-2, insights on structure, pathogenicity and immunity aspects of pandemic human coronaviruses. Infect. Genet Evol. 2020;85 doi: 10.1016/j.meegid.2020.104502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chung H., He S., Nasreen S., Sundaram M.E., Buchan S.A., Wilson S.E., et al. Effectiveness of BNT162b2 and mRNA-1273 covid-19 vaccines against symptomatic SARS-CoV-2 infection and severe covid-19 outcomes in Ontario, Canada: test negative design study. BMJ. 2021;374:n1943. doi: 10.1136/bmj.n1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phillips N. The coronavirus is here to stay-here’s what that means. Nature. 2021;590:382–384. doi: 10.1038/d41586-021-00396-2. [DOI] [PubMed] [Google Scholar]
- 28.Hu N., Wang H., Qian Q., Jiang Y., Xie J., Zhang D., et al. P-glycoprotein Assoc. Diabetes Mellit. Surviv. Patients Pancreat. Cancer.: 8-year Follow- 2020:53. doi: 10.1590/1414-431X202010168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar S., Nyodu R., Maurya V.K., Saxena S.K. Morphology, genome organization, replication, and pathogenesis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Corona Dis. 2019 (COVID-19): Epidemiol., Pathog., Diagn., Ther. 2020:23. [Google Scholar]
- 31.Helmy Y.A., Fawzy M., Elaswad A., Sobieh A., Kenney S.P., Shehata A.A. The COVID-19 pandemic: a comprehensive review of taxonomy, genetics, epidemiology, diagnosis, treatment, and control. J. Clin. Med. 2020;9:1225. doi: 10.3390/jcm9041225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schubert K., Karousis E.D., Jomaa A., Scaiola A., Echeverria B., Gurzeler L.-A., et al. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat. Struct. Mol. Biol. 2020;27:959–966. doi: 10.1038/s41594-020-0511-8. [DOI] [PubMed] [Google Scholar]
- 33.Yoshimoto F.K. The proteins of severe acute respiratory syndrome coronavirus-2 (SARS CoV-2 or n-COV19), the cause of COVID-19. Protein J. 2020;39:198–216. doi: 10.1007/s10930-020-09901-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Angeletti S., Benvenuto D., Bianchi M., Giovanetti M., Pascarella S., Ciccozzi M. COVID‐2019: the role of the nsp2 and nsp3 in its pathogenesis. J. Med. Virol. 2020;92:584–588. doi: 10.1002/jmv.25719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Littler D.R., Gully B.S., Colson R.N., Rossjohn J. Crystal structure of the SARS-CoV-2 non-structural protein 9. Nsp9. Isc. 2020;23 doi: 10.1016/j.isci.2020.101258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slanina H., Madhugiri R., Bylapudi G., Schultheiß K., Karl N., Gulyaeva A., et al. Coronavirus replication–transcription complex: Vital and selective NMPylation of a conserved site in nsp9 by the NiRAN-RdRp subunit. Proc. Natl. Acad. Sci. USA. 2021:118. doi: 10.1073/pnas.2022310118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krichel B., Falke S., Hilgenfeld R., Redecke L., Uetrecht C. Processing of the SARS-CoV pp1a/ab nsp7–10 region. Biochem. J. 2020;477:1009–1019. doi: 10.1042/BCJ20200029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gadhave K., Kumar P., Kumar A., Bhardwaj T., Garg N., Giri R. Conformational Dynamics of NSP11 Peptide of SARS-CoV-2 Under Membrane Mimetics and Different Solvent Conditions. bioRxiv. 2021:2020.10. 07.330068. [DOI] [PMC free article] [PubMed]
- 39.Brant A.C., Tian W., Majerciak V., Yang W., Zheng Z.-M. SARS-CoV-2: from its discovery to genome structure, transcription, and replication. Cell Biosci. 2021;11:1–17. doi: 10.1186/s13578-021-00643-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ceraolo C., Giorgi F.M. Genomic variance of the 2019–nCoV coronavirus. J. Med. Virol. 2020;92:522–528. doi: 10.1002/jmv.25700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hartenian E., Nandakumar D., Lari A., Ly M., Tucker J.M., Glaunsinger B.A. The molecular virology of coronaviruses. J. Biol. Chem. 2020;295:12910–12934. doi: 10.1074/jbc.REV120.013930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomar P.P.S., Arkin I.T. SARS-CoV-2 E protein is a potential ion channel that can be inhibited by gliclazide and memantine. Biochem. Biophys. Res. Commun. 2020;530:10–14. doi: 10.1016/j.bbrc.2020.05.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mandala V.S., McKay M.J., Shcherbakov A.A., Dregni A.J., Kolocouris A., Hong M. Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers. Nat. Struct. Mol. Biol. 2020;27:1202–1208. doi: 10.1038/s41594-020-00536-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arya R., Kumari S., Pandey B., Mistry H., Bihani S.C., Das A., et al. Structural insights into SARS-CoV-2 proteins. J. Mol. Biol. 2021;433 doi: 10.1016/j.jmb.2020.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boopathi S., Poma A.B., Kolandaivel P. Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. J. Biomol. Struct. Dyn. 2021;39:3409–3418. doi: 10.1080/07391102.2020.1758788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hassan S.S., Choudhury P.P., Uversky V.N., Dayhoff G.W., Aljabali A.A., Uhal B., et al. Variability of Accessory Proteins Rules the SARS-CoV-2 Pathogenicity. bioRxiv. 2020.
- 47.Guan Y., Zheng B., He Y., Liu X., Zhuang Z., Cheung C., et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 48.Hemida M., Perera R., Wang P., Alhammadi M., Siu L., Li M., et al. Middle East Respiratory Syndrome (MERS) coronavirus seroprevalence in domestic livestock in Saudi Arabia. 2010 2013. Eur. 2013;18:20659. doi: 10.2807/1560-7917.es2013.18.50.20659. [DOI] [PubMed] [Google Scholar]
- 49.Zhang C., Zheng W., Huang X., Bell E.W., Zhou X., Zhang Y. Protein structure and sequence reanalysis of 2019-nCoV genome refutes snakes as its intermediate host and the unique similarity between its spike protein insertions and HIV-1. J. Proteome Res. 2020;19:1351–1360. doi: 10.1021/acs.jproteome.0c00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Erxleben C., Adams L.C., Albrecht J., Petersen A., Vahldiek J.L., Thieß H.-M., et al. Improving CT accuracy in the diagnosis of COVID-19 in a hospital setting. Clin. Imaging. 2021;76:1–5. doi: 10.1016/j.clinimag.2021.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lytras S., Hughes J., Martin D., de Klerk A., Lourens R., Pond S., et al. Exploring the natural origins of SARS-CoV-2 in the light of recombination. COVID-19 Research. 2021. [DOI] [PMC free article] [PubMed]
- 52.Zhou H., Ji J., Chen X., Bi Y., Li J., Wang Q., et al. Identification of novel bat coronaviruses sheds light on the evolutionary origins of SARS-CoV-2 and related viruses. Cell. 2021 doi: 10.1016/j.cell.2021.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang N., Yu J., Liu P., Chang J., Ali D., Tian X. Gold nanoparticles synthesized from Curcuma wenyujin inhibits HER-2/neu transcription in breast cancer cells (MDA-MB-231/HER2) Arab. J. Chem. 2020;13:7264–7273. [Google Scholar]
- 54.Shereen M.A., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020;24:91. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiao K., Zhai J., Feng Y., Zhou N., Zhang X., Zou J.-J., et al. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature. 2020;583:286–289. doi: 10.1038/s41586-020-2313-x. [DOI] [PubMed] [Google Scholar]
- 56.Halfmann P.J., Hatta M., Chiba S., Maemura T., Fan S., Takeda M., et al. Transmission of SARS-CoV-2 in domestic cats. N. Engl. J. Med. 2020;383:592–594. doi: 10.1056/NEJMc2013400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sit T.H., Brackman C.J., Ip S.M., Tam K.W., Law P.Y., To E.M., et al. Infection of dogs with SARS-CoV-2. Nature. 2020;586:776–778. doi: 10.1038/s41586-020-2334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.of the International CSG The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Holmes K.V. Coronaviruses (Coronaviridae) Encycl. Virol. 1999:291. [Google Scholar]
- 60.Chan J.F.-W., Kok K.-H., Zhu Z., Chu H., To K.K.-W., Yuan S., et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou Z., Qiu Y., Ge X. The taxonomy, host range and pathogenicity of coronaviruses and other viruses in the Nidovirales order. Anim. Dis. 2021;1:1–28. doi: 10.1186/s44149-021-00005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Doyle L., Hutchings L. A transmissible gastroenteritis in pigs. J. Am. Vet. Med. Assoc. 1946;108:257–259. [PubMed] [Google Scholar]
- 63.Zhao Y., Zhang H., Zhao J., Zhong Q. Jin J-h, Zhang G-z. Evolution of infectious bronchitis virus in China over the past two decades. The. J. Gen. Virol. 2016;97:1566. doi: 10.1099/jgv.0.000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vlasova A.N., Halpin R., Wang S., Ghedin E., Spiro D.J., Saif L.J. Molecular characterization of a new species in the genus Alphacoronavirus associated with mink epizootic catarrhal gastroenteritis. The. J. Gen. Virol. 2011;92:1369. doi: 10.1099/vir.0.025353-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou P., Fan H., Lan T., Yang X.-L., Shi W.-F., Zhang W., et al. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556:255–258. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jackson C.B., Farzan M., Chen B., Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022;23:3–20. doi: 10.1038/s41580-021-00418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 68.Cheng V.C., Lau S.K., Woo P.C., Yuen K.Y. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin. Microbiol. Rev. 2007;20:660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Elshafei A., Khidr E.G., El-Husseiny A.A., Gomaa M.H. RAAS, ACE2 and COVID-19; a mechanistic review. Saudi J. Biol. Sci. 2021;28:6465–6470. doi: 10.1016/j.sjbs.2021.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bosch B.J., Bartelink W., Rottier P.J. Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide. J. Virol. 2008;82:8887–8890. doi: 10.1128/JVI.00415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu S., Xiao G., Chen Y., He Y., Niu J., Escalante C.R., et al. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet. 2004;363:938–947. doi: 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ismail A., Doghish A.S., Elkhatib W.F., Magdy A.M., Mahmoud E.E., Ahmed M.I., et al. Clinical and chest computed tomography features of patients suffering from mild and severe COVID-19 at Fayoum University Hospital in Egypt. Plos One. 2022;17 doi: 10.1371/journal.pone.0271271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. Jama. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang H., Wang X., Fu Z., Luo M., Zhang Z., Zhang K., et al. Potential factors for prediction of disease severity of COVID-19 patients. MedRxiv. 2020.
- 78.Yang X., Yu Y., Xu J., Shu H., Liu H., Wu Y., et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tan L., Wang Q., Zhang D., Ding J., Huang Q., Tang Y.-Q., et al. Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal Transduct. Target. Ther. 2020;5:1–3. doi: 10.1038/s41392-020-0148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Berlin D.A., Gulick R.M., Martinez F.J. Severe covid-19. N. Engl. J. Med. 2020;383:2451–2460. doi: 10.1056/NEJMcp2009575. [DOI] [PubMed] [Google Scholar]
- 81.Hammad R., Elshafei A., Khidr E.G., El-Husseiny A.A., Gomaa M.H., Kotb H.G., et al. Copeptin: a neuroendocrine biomarker of COVID-19 severity. Biomark. Med. 2022;16:589–597. doi: 10.2217/bmm-2021-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu J., Li S., Liu J., Liang B., Wang X., Wang H., et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine. 2020;55 doi: 10.1016/j.ebiom.2020.102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu T., Zhang J., Yang Y., Zhang L., Ma H., Li Z., et al. The potential role of IL-6 in monitoring coronavirus disease 2019. Available at SSRN 3548761. 2020.
- 84.Liu F., Li L., Xu M., Wu J., Luo D., Zhu Y., et al. Prognostic value of interleukin-6, C-reactive protein, and procalcitonin in patients with COVID-19. J. Clin. Virol. 2020;127 doi: 10.1016/j.jcv.2020.104370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang N., Xie T., Ning W., He R., Zhu B., Mao Y. The severity of COVID-19 and its determinants: a systematic review and meta-analysis in China. Sustainability. 2021;13:5305. [Google Scholar]
- 86.Liu W., Tao Z.-W., Wang L., Yuan M.-L., Liu K., Zhou L., et al. Analysis of factors associated with disease outcomes in hospitalized patients with 2019 novel coronavirus disease. Chin. Med. J. 2020;133:1032. doi: 10.1097/CM9.0000000000000775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Qu J., Sumali B., Lee H., Terai H., Ishii M., Fukunaga K., et al. Finding of the factors affecting the severity of COVID-19 based on mathematical models. Sci. Rep. 2021;11:1–7. doi: 10.1038/s41598-021-03632-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Goyal P., Choi J.J., Pinheiro L.C., Schenck E.J., Chen R., Jabri A., et al. Clinical characteristics of Covid-19 in New York city. N. Engl. J. Med. 2020;382:2372–2374. doi: 10.1056/NEJMc2010419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Singh S., Khan A. Clinical characteristics and outcomes of coronavirus disease 2019 among patients with preexisting liver disease in the United States: a multicenter research network study. Gastroenterology. 2020;159:768–771. doi: 10.1053/j.gastro.2020.04.064. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kishaba T., Tamaki H., Shimaoka Y., Fukuyama H., Yamashiro S. Staging of acute exacerbation in patients with idiopathic pulmonary fibrosis. Lung. 2014;192:141–149. doi: 10.1007/s00408-013-9530-0. [DOI] [PubMed] [Google Scholar]
- 91.Yan L., Zhang H.-T., Goncalves J., Xiao Y., Wang M., Guo Y., et al. An interpretable mortality prediction model for COVID-19 patients. Nat. Mach. Intell. 2020;2:283–288. [Google Scholar]
- 92.Alkorashy A.I., Doghish A.S., Abulsoud A.I., Ewees M.G., Abdelghany T.M., Elshafey M.M., et al. Effect of scopoletin on phagocytic activity of U937-derived human macrophages: Insights from transcriptomic analysis. Genomics. 2020;112:3518–3524. doi: 10.1016/j.ygeno.2020.03.022. [DOI] [PubMed] [Google Scholar]
- 93.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Samra S., Habeb M., Nafae R. ABO groups can play a role in susceptibility and severity of COVID-19. Egypt. J. Bronchol. 2021;15:1–5. [Google Scholar]
- 95.Velavan T.P., Pallerla S.R., Ruter J., Augustin Y., Kremsner P.G., Krishna S., et al. Host genetic factors determining COVID-19 susceptibility and severity. EBioMedicine. 2021;72 doi: 10.1016/j.ebiom.2021.103629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Initiative C.-H.G. Mapping the human genetic architecture of COVID-19. Nature. 2021;600:472–477. doi: 10.1038/s41586-021-03767-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pairo-Castineira E., Clohisey S., Klaric L., Bretherick A.D., Rawlik K., Pasko D., et al. Genetic mechanisms of critical illness in COVID-19. Nature. 2021;591:92–98. doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- 98.Shelton J.F., Shastri A.J., Ye C., Weldon C.H., Filshtein-Sonmez T., Coker D., et al. Trans-ancestry analysis reveals genetic and nongenetic associations with COVID-19 susceptibility and severity. Nat. Genet. 2021;53:801–808. doi: 10.1038/s41588-021-00854-7. [DOI] [PubMed] [Google Scholar]
- 99.Gheblawi M., Wang K., Viveiros A., Nguyen Q., Zhong J.C., Turner A.J., et al. Angiotensin-converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system: celebrating the 20th anniversary of the discovery of ACE2. Circ. Res. 2020;126:1456–1474. doi: 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Darbani B. The expression and polymorphism of entry machinery for COVID-19 in human: juxtaposing population groups, gender, and different tissues. Int J. Environ. Res Public Health. 2020:17. doi: 10.3390/ijerph17103433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Devaux C.A., Rolain J.M., Raoult D. ACE2 receptor polymorphism: susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. Microbiol Immunol. Infect. 2020;53:425–435. doi: 10.1016/j.jmii.2020.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hou Y., Zhao J., Martin W., Kallianpur A., Chung M.K., Jehi L., et al. New insights into genetic susceptibility of COVID-19: an ACE2 and TMPRSS2 polymorphism analysis. BMC Med. 2020;18:216. doi: 10.1186/s12916-020-01673-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mohlendick B., Schonfelder K., Breuckmann K., Elsner C., Babel N., Balfanz P., et al. ACE2 polymorphism and susceptibility for SARS-CoV-2 infection and severity of COVID-19. Pharm. Genom. 2021;31:165–171. doi: 10.1097/FPC.0000000000000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pinheiro D.S., Santos R.S., Jardim P., Silva E.G., Reis A.A.S., Pedrino G.R., et al. The combination of ACE I/D and ACE2 G8790A polymorphisms revels susceptibility to hypertension: A genetic association study in Brazilian patients. PLoS One. 2019;14 doi: 10.1371/journal.pone.0221248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yang M., Zhao J., Xing L., Shi L. The association between angiotensin-converting enzyme 2 polymorphisms and essential hypertension risk: A meta-analysis involving 14,122 patients. Renin Angiotensin Aldosterone Syst. 2015;16:1240–1244. doi: 10.1177/1470320314549221. [DOI] [PubMed] [Google Scholar]
- 106.Zou Z., Yan Y., Shu Y., Gao R., Sun Y., Li X., et al. Angiotensin-converting enzyme 2 protects from lethal avian influenza A H5N1 infections. Nat. Commun. 2014;5:3594. doi: 10.1038/ncomms4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wu C., Chen X., Cai Y., Xia J., Zhou X., Xu S., et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. 2020;180:934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., et al. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ravikanth V., Sasikala M., Naveen V., Latha S.S., Parsa K.V.L., Vijayasarathy K., et al. A variant in TMPRSS2 is associated with decreased disease severity in COVID-19. Meta Gene. 2021;29 doi: 10.1016/j.mgene.2021.100930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Alshahawey M., Raslan M., Sabri N. Sex-mediated effects of ACE2 and TMPRSS2 on the incidence and severity of COVID-19; the need for genetic implementation. Curr. Res Transl. Med. 2020;68:149–150. doi: 10.1016/j.retram.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nguyen A., David J.K., Maden S.K., Wood M.A., Weeder B.R., Nellore A., et al. Human leukocyte antigen susceptibility map for severe acute respiratory syndrome coronavirus 2. Virol. 2020:94. doi: 10.1128/JVI.00510-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Povysil G., Butler-Laporte G., Shang N., Weng C., Khan A., Alaamery M., et al. Failure to replicate the association of rare loss-of-function variants in type I IFN immunity genes with severe COVID-19. medRxiv. 2020.
- 113.Fleischmann Jr W.R. Viral genetics. Medical Microbiology 4th edition. 1996.
- 114.Saberinia A., Alinezhad A., Jafari F., Soltany S., Sigari R.A. Oncogenic miRNAs and target therapies in colorectal cancer. Clin. Chim. Acta. 2020;508:77–91. doi: 10.1016/j.cca.2020.05.012. [DOI] [PubMed] [Google Scholar]
- 115.Chen J., Wang R., Wang M., Wei G.-W. Mutations strengthened SARS-CoV-2 infectivity. J. Mol. Biol. 2020;432:5212–5226. doi: 10.1016/j.jmb.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Collier D.A., De Marco A., Ferreira I.A., Meng B., Datir R., Walls A.C., et al. SARS-CoV-2 B. 1.1. 7 escape from mRNA vaccine-elicited neutralizing antibodies. MedRxiv. 2021.
- 117.Kim D., Lee J.-Y., Yang J.-S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921. doi: 10.1016/j.cell.2020.04.011. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Guzzi P.H., Mercatelli D., Ceraolo C., Giorgi F.M. Master regulator analysis of the SARS-CoV-2/human interactome. J. Clin. Med. 2020;9:982. doi: 10.3390/jcm9040982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Morse J.S., Lalonde T., Xu S., Liu W.R. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019–nCoV. Chembiochem. 2020;21:730. doi: 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gui M., Song W., Zhou H., Xu J., Chen S., Xiang Y., et al. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017;27:119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Song W., Gui M., Wang X., Xiang Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.He J., Tao H., Yan Y., Huang S.-Y., Xiao Y. Molecular mechanism of evolution and human infection with SARS-CoV-2. Viruses. 2020;12:428. doi: 10.3390/v12040428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chen J., Wang R., Wei G.-W. Review of the mechanisms of SARS-CoV-2 evolution and transmission. ArXiv. 2021.
- 124.Wang R., Hozumi Y., Zheng Y.-H., Yin C., Wei G.-W. Host immune response driving SARS-CoV-2 evolution. Viruses. 2020;12:1095. doi: 10.3390/v12101095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gribble J., Stevens L.J., Agostini M.L., Anderson-Daniels J., Chappell J.D., Lu X., et al. The coronavirus proofreading exoribonuclease mediates extensive viral recombination. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yang X., Lv L., Ye K. Genome-wide mutation landscape of SARS-CoV-2. Preprint (Version 1) available at Research Square. 2020.
- 127.van Dorp L., Acman M., Richard D., Shaw L.P., Ford C.E., Ormond L., et al. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect., Genet. Evol. 2020;83 doi: 10.1016/j.meegid.2020.104351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kuhn J.H., Bao Y., Bavari S., Becker S., Bradfute S., Brister J.R., et al. Virus nomenclature below the species level: a standardized nomenclature for natural variants of viruses assigned to the family Filoviridae. Arch. Virol. 2013;158:301–311. doi: 10.1007/s00705-012-1454-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cosar B., Karagulleoglu Z.Y., Unal S., Ince A.T., Uncuoglu D.B., Tuncer G., et al. SARS-CoV-2 Mutations and their Viral Variants. Cytokine and growth factor reviews. 2021. [DOI] [PMC free article] [PubMed]
- 130.Davies N.G., Jarvis C.I., Edmunds W.J., Jewell N.P., Diaz-Ordaz K., Keogh R.H., et al. Increased hazard of death in community-tested cases of SARS-CoV-2 Variant of Concern 202012/01. MedRxiv. 2021.
- 131.Liu B., Liu K., Zhang H., Zhang L., Bian Y., Huang L. CoV-Seq, a New Tool for SARS-CoV-2 genome analysis and visualization: development and usability study. J. Med. Internet Res. 2020;22 doi: 10.2196/22299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rambaut A., Holmes E.C., O’Toole Á., Hill V., McCrone J.T., Ruis C., et al. Addendum: a dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2021;6:415. doi: 10.1038/s41564-021-00872-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rambaut A., Holmes E.C., O'Toole Á., Hill V., McCrone J.T., Ruis C., et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.GISAID C. lineage nomenclature aids in genomic epidemiology studies of active hCoV-19 viruses. 2020.
- 136.Mercatelli D., Giorgi F.M. Geographic and genomic distribution of SARS-CoV-2 mutations. Front Microbiol. 2020;11:1800. doi: 10.3389/fmicb.2020.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hamed S.M., Elkhatib W.F., Khairalla A.S., Noreddin A.M. Global dynamics of SARS-CoV-2 clades and their relation to COVID-19 epidemiology. Sci. Rep. 2021;11:8435. doi: 10.1038/s41598-021-87713-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Shu Y., McCauley J. GISAID: Global initiative on sharing all influenza data–from vision to reality. Eurosurveillance. 2017;22:30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sengupta A., Hassan S.S., Choudhury P.P., Clade G.R. and clade GH isolates of SARS-CoV-2 in Asia show highest amount of SNPs. Infect., Genet. Evol. 2021;89 doi: 10.1016/j.meegid.2021.104724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bedford T., Hodcroft E., Neher R. Updated Nextstrain SARS CoV-2 clade naming strategy. 〈https://nextstrainorg/blog/2021–01-06-updated-SARS-CoV-2-clade-naming〉 2021.
- 142.Yoo H.-M., Kim I.-H., Kim S. Nucleic Acid Testing of SARS-CoV-2. Int J. Mol. Sci. 2021;22:6150. doi: 10.3390/ijms22116150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Konings F., Perkins M.D., Kuhn J.H., Pallen M.J., Alm E.J., Archer B.N., et al. SARS-CoV-2 variants of interest and concern naming scheme conducive for global discourse. Nat. Microbiol. 2021;6:821–823. doi: 10.1038/s41564-021-00932-w. [DOI] [PubMed] [Google Scholar]
- 144.COVID-19 W.H.O. weekly epidemiological update, 25. Spec. Ed. 2021:ed2021. [Google Scholar]
- 145.Lauring A.S., Malani P.N. Variants of SARS-CoV-2. JAMA. 2021;326:880–. [DOI] [PubMed]
- 146.Control CfD, Prevention. SARS-CoV-2 variant classifications and definitions. 2021. p. 2020.
- 147.Sanyaolu A., Okorie C., Marinkovic A., Haider N., Abbasi A.F., Jaferi U., et al. The emerging SARS-CoV-2 variants of concern. Ther. Adv. Infect. Dis. 2021;8 doi: 10.1177/20499361211024372. 20499361211024372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Aleem A., Akbar Samad A.B., Slenker A.K. Emerging Variants of SARS-CoV-2 And Novel Therapeutics Against Coronavirus (COVID-19). StatPearls. Treasure Island (FL): StatPearls Publishing Copyright © 2021, StatPearls Publishing LLC.; 2021. [PubMed]
- 149.Kim Y., Kim E.-J., Lee S.-W., Kwon D. Review of the early reports of the epidemiological characteristics of the B. 1.1. 7 variant of SARS-CoV-2 and its spread worldwide. Osong Public Health Res Perspect. 2021. [DOI] [PMC free article] [PubMed]
- 150.team Ee Updated rapid risk assessment from ECDC on the risk related to the spread of new SARS-CoV-2 variants of concern in the EU/EEA - first update. Eur. Surveill. 2021:26. doi: 10.2807/1560-7917.ES.2021.26.3.2101211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Brookman S., Cook J., Zucherman M., Broughton S., Harman K., Gupta A. Effect of the new SARS-CoV-2 variant B.1.1.7 on children and young people. Lancet Child Adolesc. Health. 2021;5:e9–e10. doi: 10.1016/S2352-4642(21)00030-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gómez C.E., Perdiguero B., Esteban M. Emerging SARS-CoV-2 variants and impact in global vaccination programs against SARS-CoV-2/COVID-19. Vaccin. (Basel) 2021:9. doi: 10.3390/vaccines9030243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. MedRxiv. 2020.
- 154.Wang P., Casner R.G., Nair M.S., Wang M., Yu J., Cerutti G., et al. Increased resistance of SARS-CoV-2 variant P. 1 to antibody neutralization. Cell host Microbe. 2021;29:747–751. doi: 10.1016/j.chom.2021.04.007. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sabino E.C., Buss L.F., Carvalho M.P., Prete C.A., Crispim M.A., Fraiji N.A., et al. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet. 2021;397:452–455. doi: 10.1016/S0140-6736(21)00183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Faria N.R., Claro I.M., Candido D., Franco L.M., Andrade P.S., Coletti T.M., et al. Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings. Virological. 2021;372:815–821. [Google Scholar]
- 157.Yadav P.D., Sapkal G.N., Abraham P., Ella R., Deshpande G., Patil D.Y., et al. Neutralization of Variant Under Investigation B.1.617.1 With Sera of BBV152 Vaccinees. Clinical Infectious Diseases. 2021. [DOI] [PubMed]
- 158.Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 159.Singh A.K., Singh R., Joshi S.R., Misra A. Mucormycosis in COVID-19: A systematic review of cases reported worldwide and in India. Diabetes Metab. Syndr. 2021;15 doi: 10.1016/j.dsx.2021.05.019. (-) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhang W., Davis B.D., Chen S.S., Martinez J.M.S., Plummer J.T., Vail E. Emergence of a novel SARS-CoV-2 variant in Southern California. Jama. 2021;325:1324–1326. doi: 10.1001/jama.2021.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Team C.C.-R. SARS-CoV-2 B.1.1.529 (Omicron) Variant - United States, December 1-8, 2021. MMWR Morb. Mortal. Wkly Rep. 2021;70:1731–1734. doi: 10.15585/mmwr.mm7050e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.COVID C, Team. SARS-CoV-2 B. 1.1. 529 (Omicron) Variant—United States, December 1–8, 2021. 2021. p. 1731. [DOI] [PMC free article] [PubMed]
- 163.Choudhary O.P., Dhawan M. Priyanka. Omicron variant (B.1.1.529) of SARS-CoV-2: Threat assessment and plan of action. Int J. Surg. 2021;97 doi: 10.1016/j.ijsu.2021.106187. (-) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zuckerman N., Nemet I., Kliker L., Atari N., Lustig Y., Bucris E., et al. The SARS-CoV-2 Lambda variant and its neutralisation efficiency following vaccination with Comirnaty, Israel, April to June 2021. Eur. Surveill. 2021:26. doi: 10.2807/1560-7917.ES.2021.26.45.2100974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Uriu K., Kimura I., Shirakawa K., Takaori-Kondo A., Nakada T.-a, Kaneda A., et al. Neutralization of the SARS-CoV-2 Mu variant by convalescent and vaccine serum. N. Engl. J. Med. 2021 doi: 10.1056/NEJMc2114706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Collier D.A., De Marco A., Ferreira I.A., Meng B., Datir R.P., Walls A.C., et al. Sensitivity of SARS-CoV-2 B. 1.1. 7 to mRNA vaccine-elicited antibodies. Nature. 2021;593:136–141. doi: 10.1038/s41586-021-03412-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hacisuleyman E., Hale C., Saito Y., Blachere N.E., Bergh M., Conlon E.G., et al. Vaccine breakthrough infections with SARS-CoV-2 variants. N. Engl. J. Med. 2021;384:2212–2218. doi: 10.1056/NEJMoa2105000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hu B., Guo H., Zhou P., Shi Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kangabam R., Sahoo S., Ghosh A., Roy R., Silla Y., Misra N., et al. Next-generation computational tools and resources for coronavirus research: from detection to vaccine discovery. Comput. Biol. Med. 2021;128 doi: 10.1016/j.compbiomed.2020.104158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yang J., Wang W., Chen Z., Lu S., Yang F., Bi Z., et al. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature. 2020;586:572–577. doi: 10.1038/s41586-020-2599-8. [DOI] [PubMed] [Google Scholar]
- 173.Chaudhry S.N., Hazafa A., Mumtaz M., Kalsoom U., Abbas S., Kainaat A., et al. New insight on possible vaccine development against SARS-CoV-2. Life Sci. 2020 doi: 10.1016/j.lfs.2020.118421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Vankadari N., Wilce J.A. Emerging COVID-19 coronavirus: glycan shield and structure prediction of spike glycoprotein and its interaction with human CD26. Emerg. Microbes Infect. 2020;9:601–604. doi: 10.1080/22221751.2020.1739565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zuo Z., Wu T., Pan L., Zuo C., Hu Y., Luo X., et al. Modalities and mechanisms of treatment for coronavirus disease 2019. Front. Pharmacol. 2021:2257. doi: 10.3389/fphar.2020.583914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Banerji A., Wickner P.G., Saff R., Stone C.A., Jr, Robinson L.B., Long A.A., et al. mRNA vaccines to prevent COVID-19 disease and reported allergic reactions: current evidence and suggested approach. Allergy Clin. Immunol. Pr. 2021;9:1423–1437. doi: 10.1016/j.jaip.2020.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.WHO. 10 Vaccines Approved for Use by WHO. (Accessed January 5, 2022). 2022.
- 178.Samaranayake L.P., Seneviratne C.J., Fakhruddin K.S. Coronavirus disease 2019 (COVID-19) vaccines: A concise review. Oral Dis. 2021. [DOI] [PMC free article] [PubMed]
- 179.Verdecia M., Kokai-Kun J.F., Kibbey M., Acharya S., Venema J., Atouf F. COVID-19 vaccine platforms: Delivering on a promise? Hum. Vaccin Immunother. 2021;17:2873–2893. doi: 10.1080/21645515.2021.1911204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Chi W.-Y., Li Y.-D., Huang H.-C., Chan T.E.H., Chow S.-Y., Su J.-H., et al. COVID-19 vaccine update: vaccine effectiveness, SARS-CoV-2 variants, boosters, adverse effects, and immune correlates of protection. 2022;29:1–27. doi: 10.1186/s12929-022-00853-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Wang J., Peng Y., Xu H., Cui Z., Williams R.O. The COVID-19 vaccine race: challenges and opportunities in vaccine formulation. AAPS PharmSciTech. 2020;21:1–12. doi: 10.1208/s12249-020-01744-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Sanders B., Koldijk M., Schuitemaker H. Vaccine analysis: strategies, principles, and control. Springer,; 2015. Inactivated Viral Vaccines; pp. 45–80. [Google Scholar]
- 183.Ghasemiyeh P., Mohammadi-Samani S., Firouzabadi N., Dehshahri A., Vazin A. A focused review on technologies, mechanisms, safety, and efficacy of available COVID-19 vaccines. Int. Immunopharmacol. 2021;100 doi: 10.1016/j.intimp.2021.108162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Nims R., Plavsic M. Circovirus inactivation: a literature review. bioprocessing. Journal. 2012:11. [Google Scholar]
- 185.Wang W.-H., Urbina A.N., Lin C.-Y., Yang Z.-S., Assavalapsakul W., Thitithanyanont A., et al. Targets and strategies for vaccine development against dengue viruses. Biomed. Pharmacother. 2021;144 doi: 10.1016/j.biopha.2021.112304. [DOI] [PubMed] [Google Scholar]
- 186.Kumar S., Sunagar R., Gosselin E. Bacterial protein toll-like-receptor agonists: a novel perspective on vaccine adjuvants. Front. Immunol. 2019;10:1144. doi: 10.3389/fimmu.2019.01144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Van Dis E., Sogi K.M., Rae C.S., Sivick K.E., Surh N.H., Leong M.L., et al. STING-activating adjuvants elicit a Th17 immune response and protect against Mycobacterium tuberculosis infection. Cell Rep. 2018;23:1435–1447. doi: 10.1016/j.celrep.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Alsulaiman J.W., Khasawneh A.I., Kheirallah K.A. Could “trained immunity” be induced by live attenuated vaccines protect against COVID-19? Review of available evidence. J. Infect. Dev. Ctries. 2020;14:957–962. doi: 10.3855/jidc.12805. [DOI] [PubMed] [Google Scholar]
- 189.Shah S.S., Patel C.M., Patel D.H., Vadgama P.H., Patel M., Trivedi R. A review on modern use of intranasal vaccination in the treatment of SARS-COV-2. J. Drug Deliv. Ther. 2021;11:263–270. [Google Scholar]
- 190.Syomin B., Ilyin Y. Virus-like particles as an instrument of vaccine production. Mol. Biol. 2019;53:323–334. doi: 10.1134/S0026893319030154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ura T., Yamashita A., Mizuki N., Okuda K., Shimada M. New vaccine production platforms used in developing SARS-CoV-2 vaccine candidates. Vaccin. (Basel) 2021;39:197. doi: 10.1016/j.vaccine.2020.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Meseda C.A., Stauft C.B., Selvaraj P., Lien C.Z., Pedro C., Nuñez I.A., et al. MVA vector expression of SARS-CoV-2 spike protein and protection of adult Syrian hamsters against SARS-CoV-2 challenge. npj Vaccin. 2021;6:1–10. doi: 10.1038/s41541-021-00410-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Khalaj‐Hedayati A., Chua C.L.L., Smooker P., Lee K.W. Nanoparticles in influenza subunit vaccine development: immunogenicity enhancement. Influenza Other Respir. Virus. 2020;14:92–101. doi: 10.1111/irv.12697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Lin H.-T., Chen C.-C., Chiao D.-J., Chang T.-Y., Chen X.-A., Young J.-J., et al. Nanoparticular CpG-adjuvanted SARS-CoV-2 S1 protein elicits broadly neutralizing and Th1-biased immunoreactivity in mice. Int. J. Biol. Macromol. 2021;193:1885–1897. doi: 10.1016/j.ijbiomac.2021.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Mouliou D.S., Pantazopoulos I., Gourgoulianis K.I. Social response to the vaccine against COVID-19: the underrated power of influence. journal of personalized medicine. 2022;12:15. doi: 10.3390/jpm12010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.WHO . World Health Organization,; 2021. Interim recommendations for use of the Novavax NVX-CoV2373 vaccine against COVID-19: interim guidance, 20 December 2021. [Google Scholar]
- 197.Batty C.J., Heise M.T., Bachelder E.M., Ainslie K.M. Vaccine formulations in clinical development for the prevention of severe acute respiratory syndrome coronavirus 2 infection. Adv. Drug Deliv. Rev. 2021;169:168–189. doi: 10.1016/j.addr.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Tsakiri M., Naziris N., Demetzos C. Innovative vaccine platforms against infectious diseases: Under the scope of the COVID-19 pandemic. Int. J. Pharm. 2021;610 doi: 10.1016/j.ijpharm.2021.121212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Dong Y., Dai T., Wei Y., Zhang L., Zheng M., Zhou F. A systematic review of SARS-CoV-2 vaccine candidates. Signal Transduct. Target Ther. 2020;5:1–14. doi: 10.1038/s41392-020-00352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Belete T.M. A review on Promising vaccine development progress for COVID-19 disease. Vacunas. 2020;21:121–128. doi: 10.1016/j.vacun.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Robert-Guroff M. Replicating and non-replicating viral vectors for vaccine development. Curr. Opin. Biotechnol. 2007;18:546–556. doi: 10.1016/j.copbio.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Sauter S.L., Rahman A., Muralidhar G. Non-replicating viral vector-based AIDS vaccines: interplay between viral vectors and the immune system. Curr. HIV Res. 2005;3:157–181. doi: 10.2174/1570162053506900. [DOI] [PubMed] [Google Scholar]
- 203.Callaway E. Coronavirus vaccines leap through safety trials - but which will work is anybody's guess. Nature. 2020;583:669–670. doi: 10.1038/d41586-020-02174-y. [DOI] [PubMed] [Google Scholar]
- 204.Boyarsky B.J., Chiang T.P.-Y., Ou M.T., Werbel W.A., Massie A.B., Segev D.L., et al. Antibody response to the Janssen COVID-19 vaccine in solid organ transplant recipients. Transplantation. 2021;105:e82–e83. doi: 10.1097/TP.0000000000003850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Rijkers G.T., Weterings N., Obregon-Henao A., Lepolder M., Dutt T.S., van Overveld F.J., et al. Antigen Presentation of mRNA-Based and Virus-Vectored SARS-CoV-2 Vaccines. Vaccin. (Basel) 2021;9:848. doi: 10.3390/vaccines9080848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Chaudhary J.K., Yadav R., Chaudhary P.K., Maurya A., Kant N., Rugaie O.A., et al. Insights into COVID-19 vaccine development based on immunogenic structural proteins of SARS-CoV-2, Host Immune Responses, and Herd Immunity. Cells. 2021;10:2949. doi: 10.3390/cells10112949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Chauhan N., Soni S., Gupta A., Aslam M., Jain U. Interpretative immune targets and contemporary position for vaccine development against SARS‐CoV‐2: A systematic review. J. Med. Virol. 2021;93:1967–1982. doi: 10.1002/jmv.26709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Seneff S., Nigh G. Worse than the disease? reviewing some possible unintended consequences of the mRNA Vaccines Against COVID-19. Int. J. Vaccin. Theory, Pract., Res. 2021;2:38–79. [Google Scholar]
- 209.Wadhwa A., Aljabbari A., Lokras A., Foged C., Thakur A. Opportunities and challenges in the delivery of mRNA-based vaccines. Pharmaceutics. 2020;12:102. doi: 10.3390/pharmaceutics12020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Guevara M.L., Persano S., Persano F. Lipid-based vectors for therapeutic mRNA-based anti-cancer vaccines. Curr. Pharm. Des. 2019;25:1443–1454. doi: 10.2174/1381612825666190619150221. [DOI] [PubMed] [Google Scholar]
- 211.Khurana A., Allawadhi P., Khurana I., Allwadhi S., Weiskirchen R., Banothu A.K., et al. Role of nanotechnology behind the success of mRNA vaccines for COVID-19. Nano Today. 2021;38 doi: 10.1016/j.nantod.2021.101142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Uddin M.N., Roni M.A. Challenges of storage and stability of mRNA-based COVID-19 vaccines. Vaccin. (Basel) 2021;9:1033. doi: 10.3390/vaccines9091033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Fortner A., Schumacher D. First COVID-19 vaccines receiving the US FDA and EMA emergency use authorization. Discov. (Craiova) 2021;9 doi: 10.15190/d.2021.1. e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Elkhalifa D., Rayan M., Negmeldin A.T., Elhissi A., Khalil A. Chemically modified mRNA beyond COVID-19: potential preventive and therapeutic applications for targeting chronic diseases. Biomed. Pharmacother. 2022;145 doi: 10.1016/j.biopha.2021.112385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Zhu D., Meng Y., Qimuge A., Bilige B., Baiyin T., Temuqile T., et al. Oral delivery of SARS-CoV-2 DNA vaccines using attenuated Salmonella typhimurium as a carrier in rat. BioRxiv. 2020:2020.07.23.217174. [DOI] [PMC free article] [PubMed]
- 216.Chavda V.P., Hossain M.K., Beladiya J., Apostolopoulos V. Nucleic Acid Vaccines for COVID-19: A Paradigm Shift in the Vaccine Development Arena. Biologics. 2021;1:337–356. [Google Scholar]
- 217.Powers A.M. Vaccine and therapeutic options to control chikungunya virus. Clin. Microbiol. Rev. 2018;31:e00104–e00116. doi: 10.1128/CMR.00104-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Gulce-Iz S., Saglam-Metiner P. In: Immune Response Activation and Immunomodulation. Tyagi R.K., Bisen P.S., editors. IntechOpen; 2019. Current state of the art in DNA vaccine delivery and molecular adjuvants: Bcl-xL anti-apoptotic protein as a molecular adjuvant; pp. 1–16. [Google Scholar]
- 219.Yin Y., Su W., Zhang J., Huang W., Li X., Ma H., et al. Separable Microneedle Patch to Protect and Deliver DNA Nanovaccines Against COVID-19. ACS Nano. 2021;15:14347–14359. doi: 10.1021/acsnano.1c03252. [DOI] [PubMed] [Google Scholar]
- 220.Peletta A., Prompetchara E., Tharakhet K., Kaewpang P., Buranapraditkun S., Techawiwattanaboon T., et al. DNA vaccine administered by cationic lipoplexes or by in vivo electroporation induces comparable antibody responses against SARS-CoV-2 in Mice. Vaccin. (Basel) 2021:9. doi: 10.3390/vaccines9080874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Keshavarz M., Mirzaei H., Salemi M., Momeni F., Mousavi M.J., Sadeghalvad M., et al. Influenza vaccine: Where are we and where do we go? Rev. Med. Virol. 2019;29 doi: 10.1002/rmv.2014. [DOI] [PubMed] [Google Scholar]
- 222.Chavda V.P., Pandya R., Apostolopoulos V. DNA vaccines for SARS-CoV-2: toward third-generation vaccination era. Expert Rev. Vaccin. 2021;20:1549–1560. doi: 10.1080/14760584.2021.1987223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Jiang S., Du L., Shi Z. An emerging coronavirus causing pneumonia outbreak in Wuhan, China: calling for developing therapeutic and prophylactic strategies. Emerg. Microbes Infect. 2020;9:275–277. doi: 10.1080/22221751.2020.1723441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Prevention ECfD, Control. Situation update worldwide, as of 21 April 2020. Eur Cent Dis Prev Control. 2020.
- 225.Baud D., Qi X., Nielsen-Saines K., Musso D., Pomar L., Favre G. Real estimates of mortality following COVID-19 infection. Lancet Infect. Dis. 2020;20:773. doi: 10.1016/S1473-3099(20)30195-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Domingo E. Quasispecies Theory in Virology. Virol. 2002;76:463–465. doi: 10.1128/JVI.76.1.463-465.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Kruse R.L. Therapeutic strategies in an outbreak scenario to treat the novel coronavirus originating in Wuhan. China F1000Res. 2020;9:72. doi: 10.12688/f1000research.22211.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Jiang S., Hillyer C., Du L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol. 2020;41:355–359. doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.WHO . World Health Organization; 2020. Laboratory Testing for Coronavirus Disease ( COVID-19) in Suspected Human Cases: Interim Guidance, 19 March 2020. [Google Scholar]
- 230.Xiao S.Y., Wu Y., Liu H. Evolving status of the 2019 novel coronavirus infection: proposal of conventional serologic assays for disease diagnosis and infection monitoring. J. Med. Virol. 2020;92:464. doi: 10.1002/jmv.25702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Gorse G.J., Donovan M.M., Patel G.B. Antibodies to coronaviruses are higher in older compared with younger adults and binding antibodies are more sensitive than neutralizing antibodies in identifying coronavirus‐associated illnesses. J. Med. Virol. 2020;92:512–517. doi: 10.1002/jmv.25715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Ogando N.S., Ferron F., Decroly E., Canard B., Posthuma C.C., Snijder E.J. The curious case of the nidovirus exoribonuclease: its role in RNA synthesis and replication fidelity. Front. Microbiol. 2019;10:1813. doi: 10.3389/fmicb.2019.01813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Eckerle L.D., Becker M.M., Halpin R.A., Li K., Venter E., Lu X., et al. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10:1–9. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., et al. Structure of RNA-dependent RNA polymerase from 2019-nCoV, a major antiviral drug target. BioRxiv. 2020.
- 236.Lim S.-Y., Osuna C., Lakritz J., Chen E., Yoon G., Taylor R., et al. Open Forum Infectious Diseases. Oxford University Press,; 2017. Galidesivir, a direct-acting antiviral drug, abrogates viremia in rhesus macaques challenged with Zika virus. [Google Scholar]
- 237.Furuta Y., Gowen B.B., Takahashi K., Shiraki K., Smee D.F., Barnard D.L. Favipiravir (T-705), a novel viral RNA polymerase inhibitor. Antivir. Res. 2013;100:446–454. doi: 10.1016/j.antiviral.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Morgenstern B., Michaelis M., Baer P.C., Doerr H.W., Cinatl J., Jr Ribavirin and interferon-β synergistically inhibit SARS-associated coronavirus replication in animal and human cell lines. Biochem. Biophys. Res. Commun. 2005;326:905–908. doi: 10.1016/j.bbrc.2004.11.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X., et al. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. MBio. 2018;9:e00221–18. doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Andreano E., Piccini G., Licastro D., Casalino L., Johnson N., Paciello I., et al. SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. BioRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 241.WHO. The effects of virus variants on COVID-19 vaccines. 2021. 2021.
- 242.Supasa P., Zhou D., Dejnirattisai W., Liu C., Mentzer A.J., Ginn H.M., et al. Reduced neutralization of SARS-CoV-2 B. 1.1. 7 variant by convalescent and vaccine sera. Cell. 2021;184:2201–2211. doi: 10.1016/j.cell.2021.02.033. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Zhou D., Dejnirattisai W., Supasa P., Liu C., Mentzer A.J., Ginn H.M., et al. Evidence of escape of SARS-CoV-2 variant B. 1.351 from natural and vaccine-induced sera. Cell. 2021;184:2348–2361. doi: 10.1016/j.cell.2021.02.037. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., et al. SARS-CoV-2 501Y. V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021;27:622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- 245.Rathnasinghe R., Jangra S., Cupic A., Martínez-Romero C., Mulder L.C., Kehrer T., et al. The N501Y mutation in SARS-CoV-2 spike leads to morbidity in obese and aged mice and is neutralized by convalescent and post-vaccination human sera. MedRxiv. 2021 [Google Scholar]
- 246.Muik A., Lui B.G., Wallisch A.-K., Bacher M., Muehl J., Reinholz J., et al. Neutralization of SARS-CoV-2 Omicron pseudovirus by BNT162b2 vaccine-elicited human sera. medRxiv. 2021.
- 247.Collier D., Meng B., Ferreira I., Datir R., Temperton N.J., Elmer A., et al. Impact of SARS-CoV-2 B. 1.1. 7 Spike variant on neutralisation potency of sera from individuals vaccinated with Pfizer vaccine BNT162b2. MedRxiv. 2021 [Google Scholar]
- 248.Xie X., Zou J., Fontes-Garfias C.R., Xia H., Swanson K.A., Cutler M., et al. Neutralization of N501Y mutant SARS-CoV-2 by BNT162b2 vaccine-elicited sera. BioRxiv. 2021 doi: 10.1038/s41591-021-01270-4. [DOI] [PubMed] [Google Scholar]
- 249.Bian L., Gao F., Zhang J., He Q., Mao Q., Xu M., et al. Effects of SARS-CoV-2 variants on vaccine efficacy and response strategies. Expert Rev. Vaccin. 2021;20:365–373. doi: 10.1080/14760584.2021.1903879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Shen X., Tang H., McDanal C., Wagh K., Fischer W., Theiler J., et al. SARS-CoV-2 variant B. 1.1. 7 is susceptible to neutralizing antibodies elicited by ancestral spike vaccines. Cell host Microbe. 2021;29:529–539. doi: 10.1016/j.chom.2021.03.002. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Huang B., Dai L., Wang H., Hu Z., Yang X., Tan W., et al. Neutralization of SARS-CoV-2 VOC 501Y. V2 by human antisera elicited by both inactivated BBIBP-CorV and recombinant dimeric RBD ZF2001 vaccines. BioRxiv. 2021.
- 252.Novavax. Novavax COVID-19 vaccine demonstrates 89.3% efficacy in UK phase 3 trial. 2021.
- 253.Johnson J. Johnson announces single-shot Janssen COVID-19 vaccine candidate met primary endpoints in interim analysis of its phase 3 ENSEMBLE trial. Jan 29, 2021. 2021.
- 254.Honein M.A., Christie A., Rose D.A., Brooks J.T., Meaney-Delman D., Cohn A., et al. Summary of guidance for public health strategies to address high levels of community transmission of SARS-CoV-2 and related deaths, December 2020. MMWR Morb. Mortal. Wkly Rep. 2020;69:1860–1867. doi: 10.15585/mmwr.mm6949e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Hirose R., Ikegaya H., Naito Y., Watanabe N., Yoshida T., Bandou R., et al. Survival of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and influenza virus on human skin: importance of hand hygiene in coronavirus disease 2019 (COVID-19) Clin. Infect. Dis. 2021;73:e4329–e4335. doi: 10.1093/cid/ciaa1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Lindsley W.G., Derk R.C., Coyle J.P., Martin S.B., Jr., Mead K.R., Blachere F.M., et al. Efficacy of portable air cleaners and masking for reducing indoor exposure to simulated exhaled SARS-CoV-2 Aerosols - United States, 2021. MMWR Morb. Mortal. Wkly Rep. 2021;70:972–976. doi: 10.15585/mmwr.mm7027e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Rahimi Pordanjani S., Hasanpour A., Askarpour H., Bastam D., Rafiee M., Khazaei Z., et al. Aspects of epidemiology, pathology, virology, immunology, transmission, prevention, prognosis, diagnosis, and treatment of COVID-19 pandemic: a narrative review. Int J. Prev. Med. 2021;12:38. doi: 10.4103/ijpvm.IJPVM_469_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Davies M.A., Kassanjee R., Rousseau P., Morden E., Johnson L., Solomon W., et al. Outcomes of laboratory‐confirmed SARS‐CoV‐2 infection in the Omicron‐driven fourth wave compared with previous waves in the Western Cape Province, South Africa. 2022. [DOI] [PMC free article] [PubMed]
- 259.Andrews N., Tessier E., Stowe J., Gower C., Kirsebom F., Simmons R., et al. Duration of protection against mild and severe disease by Covid-19. vaccines. 2022;386:340–350. doi: 10.1056/NEJMoa2115481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Thomas S.J., Moreira E.D., Jr, Kitchin N., Absalon J., Gurtman A., Lockhart S., et al. Saf. Effic. BNT162b2 mRNA Covid-19 Vaccin. 6 Mon. 2021;385:1761–1773. doi: 10.1056/NEJMoa2110345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Moreira E.D., Jr, Kitchin N., Xu X., Dychter S.S., Lockhart S., Gurtman A., et al. Safety and efficacy of a third dose of BNT162b2 COVID-19. vaccine. 2022;386:1910–1921. doi: 10.1056/NEJMoa2200674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Muik A., Lui B.G., Wallisch A.-K., Bacher M., Mühl J., Reinholz J., et al. Neutralization SARS-CoV-2 Omicron BNT162b2 mRNA Vaccin. Hum. sera. 2022;375:678–680. doi: 10.1126/science.abn7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Accorsi E.K., Britton A., Fleming-Dutra K.E., Smith Z.R., Shang N., Derado G., et al. Association between 3 doses of mRNA COVID-19 vaccine and symptomatic infection caused by the SARS-CoV-2 Omicron and Delta variants. 2022;327:639–651. doi: 10.1001/jama.2022.0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Lustig Y., Gonen T., Meltzer L., Gilboa M., Indenbaum V., Cohen C., et al. Superior immunogenicity and effectiveness of the third compared to the second BNT162b2 vaccine dose. 2022:1–7. [DOI] [PubMed]
- 265.Muecksch F., Wang Z., Cho A., Gaebler C., Ben Tanfous T., DaSilva J., et al. Increased memory B cell potency and breadth after a SARS-CoV-2 mRNA boost. 2022:1–7. [DOI] [PMC free article] [PubMed]
- 266.Kuhlmann C., Mayer C.K., Claassen M., Maponga T., Burgers W.A., Keeton R., et al. Breakthrough infections with SARS-CoV-2 omicron despite mRNA vaccine booster dose. 2022;399:625–626. doi: 10.1016/S0140-6736(22)00090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Levine-Tiefenbrun M., Yelin I., Alapi H., Katz R., Herzel E., Kuint J., et al. Viral loads of Delta-variant SARS-CoV-2 breakthrough infections after vaccination and booster with BNT162b2. 2021;27:2108–2110. doi: 10.1038/s41591-021-01575-4. [DOI] [PubMed] [Google Scholar]
- 268.Collier D.A., Ferreira I.A., Kotagiri P., Datir R.P., Lim E.Y., Touizer E., et al. Age-related immune response heterogeneity to SARS-CoV-2 vaccine BNT162b2. 2021;596:417–422. doi: 10.1038/s41586-021-03739-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Shapiro L.C., Thakkar A., Campbell S.T., Forest S.K., Pradhan K., Gonzalez-Lugo J.D., et al. Efficacy of booster doses in augmenting waning immune responses to COVID-19 vaccine in patients with cancer. 2022;40:3–5. doi: 10.1016/j.ccell.2021.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Naranbhai V., Denis K.J.S., Lam E.C., Ofoman O., Garcia-Beltran W.F., Mairena C.B., et al. Neutralization breadth SARS-CoV-2 Viral Var. Prim. Ser. Boost. SARS-CoV-2 Vaccin. Patients Cancer. 2022;40:103–108. doi: 10.1016/j.ccell.2021.12.002. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Zeng C., Evans J.P., Chakravarthy K., Qu P., Reisinger S., Song N.-J., et al. COVID-19 mRNA booster vaccines elicit strong protection against SARS-CoV-2 Omicron variant in patients with cancer. 2022;40:117–119. doi: 10.1016/j.ccell.2021.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Bar-On Y.M., Goldberg Y., Mandel M., Bodenheimer O., Amir O., Freedman L., et al. Protection by a fourth dose of BNT162b2 against omicron in Israel. 2022;386:1712–1720. doi: 10.1056/NEJMoa2201570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Arbel R., Sergienko R., Friger M., Peretz A., Beckenstein T., Yaron S., et al. Eff. a Second BNT162b2 Boost. Vaccin. Hosp. death COVID-19 adults aged 60 years. 2022:1–5. doi: 10.1038/s41591-022-01832-0. [DOI] [PubMed] [Google Scholar]
- 274.Andrews N., Stowe J., Kirsebom F., Toffa S., Rickeard T., Gallagher E., et al. Covid-19 Vaccin. Eff. Omicron (B. 1. 1. 529) Var. 2022;386:1532–1546. doi: 10.1056/NEJMoa2119451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Tartof S.Y., Slezak J.M., Puzniak L., Hong V., Frankland T.B., Ackerson B.K., et al. Effectiveness of a third dose of BNT162b2 mRNA COVID-19 vaccine in a large US health system: a retrospective cohort study. 2022;9 doi: 10.1016/j.lana.2022.100198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Arbel R., Hammerman A., Sergienko R., Friger M., Peretz A., Netzer D., et al. BNT162b2 Vaccin. Boost. Mortal. Covid. 2021;19(385):2413–2420. doi: 10.1056/NEJMoa2115624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Muhsen K., Maimon N., Mizrahi A., Varticovschi B., Bodenheimer O., Gelbshtein U., et al. Effects of BNT162b2 Covid-19 vaccine booster in long-term care facilities in Israel. 2022;386:399–401. doi: 10.1056/NEJMc2117385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Chi W.-Y., Li Y.-D., Huang H.-C., Chan T.E.H., Chow S.-Y., Su J.-H., et al. COVID-19 vaccine update: vaccine effectiveness, SARS-CoV-2 variants, boosters, adverse effects, and immune correlates of protection. J. Biomed. Sci. 2022;29:82. doi: 10.1186/s12929-022-00853-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article. This is a Literature Review article without any data or code.