Abstract
Advances in high-throughput sequencing techniques and bioinformatic analysis have refuted the “junk” RNA hypothesis that was claimed against non-coding RNAs (ncRNAs). Circular RNAs (circRNAs); a class of single-stranded covalently closed loop RNA molecules have recently emerged as stable epigenetic regulators. Although the exact regulatory role of circRNAs is still to be clarified, it has been proven that circRNAs could exert their functions by interacting with other ncRNAs or proteins in their own physiologically authentic environment, regulating multiple cellular signaling pathways and other classes of ncRNAs. CircRNAs have also been reported to exhibit a tissue-specific expression and have been associated with the malignant transformation process of several hematological and solid malignancies. Along this line of reasoning, this review aims to highlight the importance of circRNAs in Breast Cancer (BC), which is ranked as the most prevalent malignancy among females. Notwithstanding the substantial efforts to develop a suitable anticancer therapeutic regimen against the heterogenous BC, inter- and intra-tumoral heterogeneity have resulted in an arduous challenge for drug development research, which in turn necessitates the investigation of other markers to be therapeutically targeted. Herein, the potential of circRNAs as possible diagnostic and prognostic biomarkers have been highlighted together with their possible application as novel therapeutic targets.
Keywords: CircrRNAs, Breast cancer, Diagnosis, Prognosis, Precision medicine, Heterogeneous tumor, Biomarkers
1. Introduction
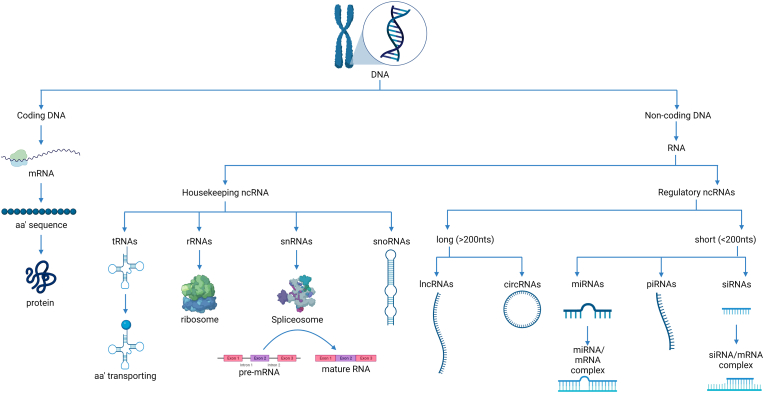
For a long period of time, it was thought that most DNA was coding material that gets translated into protein [1]. However, more than 98% of the human genome was found to be non-coding for proteins while the rest 1-2% encodes ≈21,000 distinct proteins [1]. Lately, the non-coding part of the genome, known as non-coding RNAs (ncRNAs) is classified by function into two categories; housekeeping ncRNAs and regulatory RNAs as presented (Fig. 1) [2]. Housekeeping ncRNAs include transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), small nuclear RNAs (snRNAs), and small nucleolar RNAs (snoRNAs), whereas the regulatory ncRNAs are sub-grouped by length into long ncRNAs (lncRNAs) (>200 nt), and short ncRNAs (<200 nt).
Fig. 1.
Classification of human genome according to coding potential.
This figure represents a classification of human genome (DNA) where DNA includes coding sequences that are transcribed and translated into proteins and non-coding sequences that are only transcribed into RNA. Those RNA molecules are known as non-coding RNAs (ncRNAs) as they lose their coding capacity and to be translated into proteins. ncRNAs are further classified according to its function into housekeeping ncRNAs and regulatory RNAs.
A new subclass of the lncRNAs is the circular RNAs (circRNAs) which have become a matter of interest, due to their rising therapeutic role, high stability profile, high diagnostic and prognostic values, and finally their major contribution to cellular proliferation and the malignant transformation process [3,4].
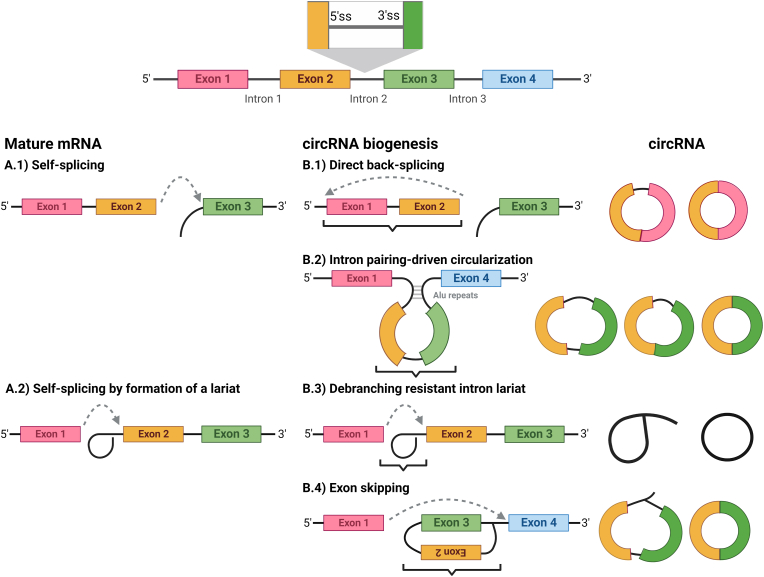
Since most of the eukaryotic genes are interrupted by introns, these introns are removed before protein translation [5]. The transcribed pre-mRNAs undergo certain splicing by the spliceosomes, which are cellular machinery that are able to remove introns to join exons together forming mature RNAs (Fig. 2A) [6]. Hence, splicing can intentionally result in various RNA isoforms since it is highly regulated [7], leading to RNA molecules of different functions, locations and of divergent regulatory roles such as circRNAs [8].
Fig. 2.
CircRNAs Biogenesis.
This is a representative figure showing mature mRNA splicing and corresponding canonical biogenesis of circRNAs which might include B.1) direct back splicing. B.2) Intron pairing driven circularization, B.3) debranching resistant intron lariat and B.4) lariat-driven circularization (exon skipping).
Recently, circRNAs have been casted as novel promising regulators in the field of oncology [9]. It has been reported that circRNAs could behave differently in diverse malignant contexts, in which they could act either as a tumor promotor (oncogenic), or a tumor suppressor circRNA [3,10].
It is notable that unraveling the role of circRNAs in cancer is still a virgin field that needs further investigation [4,11]. Until now, several research groups are focusing on exploring the potential roles of different circRNAs in various solid and humoral malignancies. Yet, the field can still accommodate several research groups to explore the promising role of circRNAs as diagnostic, prognostic biomarkers and/or therapeutic targets or tools in the field of oncology. In the current review, the authors will try to shed the light onto the potential role of circRNAs in breast cancer (BC), which is considered one of the most heterogenous and dominating cancers among females.
2. What are CircRNAs?
CircRNAs are generated by special types of splicing and they are mainly found in the cytoplasm [12]. The term “circRNA” was introduced by Sanger et al. in 1976, when an infectious single-stranded covalently closed RNA molecules in plant viroid was characterized [13]. Yet, in the early 2010s, circRNAs were identified in human cells to be of significant abundance [14,15].
Similar to other lncRNAs, circRNAs can serve as miRNA sponges, protein scaffolds, protein decoys and can even have a translational function [16]. However, unlike lncRNAs, most circRNAs get spliced from coding pre-mRNAs [17]. Additionally, due to them lacking 3′ and 5’ free ends and existing as continuous loops, they are highly resistant to degradation by RNAses [18].
2.1. Biogenesis of circRNA
In-vivo circRNAs are usually originated from protein-coding sequences [19]. The exact mechanisms by which they are produced are shown in Fig. 2 and include I) direct back-splicing [20], II) intron-pairing-driven circularization [21], III) debranching resistant intron lariat [22], and IV) lariat-driven circularization (exon skipping) [23,24]. Typically, pre-mRNAs are spliced by different mechanisms to produce the mature mRNAs, which consist of exons only and are later translated into amino acid sequences [25].
One of the mechanisms by which pre-mRNAs are being regularly spliced is the self-splicing mechanism (Fig. 2A1) [26]. At the end of this process, the free OH′ group at the 5′ splice site (ss) is ligated to the following downstream 3'ss forming a 3′-5′ covalent phosphodiester bond between two consecutive exons [26]. However, when the free OH′ at 5'ss is being peculiarly joined to an upstream 3'ss (in the back), this leads to formation of circRNA that includes exonic parts in its sequence (Fig. 2B1) [20].
Occasionally, if the pre-mRNAs contain two non-adjacent introns containing complementary Arthrobacter luteus (ALU) repeats [27], these repeats hybridize/(pair) together bringing flanking introns to become in close proximity, forming a conformational structure that facilitates back splicing of the pre-mRNA (Fig. 2B2) [28,29].
Moreover, mature mRNAs could be produced by different self-splicing mechanisms by which the 5'ss forms a loop-like structure called “lariat” within the same intron to be excised (Fig. 2A2) [30]. The excised intron is usually de-branched by a certain enzyme that was identified in 1985 [31]. However, if the lariat fail to de-branch, an intronic circRNAs may be formed (Fig. 2B3) [22]. However, in Lariat-driven circRNA biogenesis model, which is also called “exon-skipping”, the lariat is extended to form a loop with another downstream-intron, skipping some exons inside the loop structure (Fig. 2B4) [23]. Then, the resulting circRNA could be processed by canonical splicing or not [19].
CircRNAs could obtain sequence of exons only (exonic circRNAs), introns only (intronic circRNAs), or both (exon-intron circRNAs) [32]. The formation of covalently exonic circRNAs occurs when a 3′ end of an exon (5'ss) is joined to a 5′ end of the same exon (single-exon circRNA), or an upstream exon (multiple-exon circRNA), forming a closed RNA loop. However, single-exon circRNA showed longer length than average exon length [33]. Nevertheless, multiple-exon circRNAs which lack intronic sequence is more frequent form of circRNAs [34].
2.2. CircRNAs nomenclature
Unfortunately, until now there is no consensus agreement that has been established concerning the nomenclature of circRNAs. There are various nomenclature systems used leading to high level of confusion between similar studies [35]. Despite the great efforts done by different research groups for creating central databases compiling all circRNA data, yet the information is not complete and inconsistent creating isolated islands each of them is using their own language and their own nomenclature system to annotate the circRNAs [36].
Currently, there are 4 available resources that could be considered as databases for circRNAs: circBase [37], circBank [38], CIRCpedia [39], and circAtlas [40]. CircBase is the original database and their proposed nomenclature system was based on the species and some numerical codes added by the database. Later, circBank and circAtlas started to provide a more user-friendly annotation for circRNAs where circRNAs names start to include the gene symbol of the transcriptional unit responsible for its generation based on the genomic coordinate references from UCSC resources [41]. However, CIRCpedia started to use a specific denomination for each circRNA, including the original species and an internal number without the reference to the source gene making it totally different from the nomenclature originally proposed by circBase [37].
The problem is not only that the same circRNA might have several names that are nearly similar but differ in suffix-numerical codes such as CircMTO1 (hsa_circ_0007874, hg19: chr6:74175931–74,176,329) which was found to have 11 different names reported until now. Yet, the real disaster that it was reported that some circRNAs that are nearly identical in names and with the same index sometimes correspond to different circRNAs.
Accordingly, such lack of standardization in such virgin field prevents the required validation of experimental results and the detailed understanding about circRNAs' functional roles and thus hinders the deep knowledge required to be understood about circRNAs at such stage.
2.3. CircRNAs interactions with other molecules
2.3.1. CircRNAs-ncRNAs interactions: an endogenous sponge/decoy to microRNAs (miRNAs)
A subclass of the short ncRNAs is microRNAs (miRNAs). miRNAs have an average length of 22 nt and are mostly intragenic; made from introns and few exons of protein coding genes [42].
In fact, miRNAs have been found to transcriptionally regulate the expression of almost 60% of the whole human genome. They are found intracellularly and extracellularly where they can serve as biomarkers for diseases [43,44], malignancies [45], and as a guide for cell-cell interactions [46]. Aberrant expression levels of miRNAs can lead to several malignancies [47]. On the other hand, circRNAs have multiple binding sites for miRNAs [48]. By binding to the miRNAs, circRNAs “sponge” them and prevent them from performing their previously mentioned functions, thus hindering their regulatory functions [32]. Such “sponging” effect could express remarkable impacts on cellular proliferation and cancer progression [32,49]. Using bioinformatics, those miRNA-binding sites can be predicted using web tools such as Circular RNA Interactome (CircInteractome: https://circinteractome.nia.nih.gov/) [50,51]. For instance, circRNF20 harbor miR-487a, acting as miRNA sponge in BC [49]. In non-oncological context, it has also found that circRNA CCDC66 (circCCDC66) acts as a sponge for miR342-3p in vascular smooth muscle cells and ranked as a novel circRNA playing a central role in abdominal aortic aneurysm pathogenesis [52].
2.4. CircRNAs-protein interactions
2.4.1. Protein sponges/decoys
By the few protein-binging sites that circRNAs harbor, they could also bind to proteins and sequester them, acting like an antagonist to hinder their physiological function [53]. One of the most common protein classes that bind to circRNA is RNA-binding proteins (RBP). For instance, circ-TNPO3 inhibits gastric cancer (GC) metastasis by decoying insulin-like growth factor 2 binding protein 3 (IGF2BP3) protein [54]. When circ-TNPO3 sequesters IGF2BP3, the expression of MYC and its target SNAIL was suppressed, decreasing the proliferation and metastasis capacity of GC cells [54]. Nonetheless, it was also reported that in colorectal cancer cell lines, circ-SIRT1 bind to the eukaryotic translation initiation factor 4A3 (EIF4A3), hence blocking its inhibitory effect on epithelial mesenchymal transition and promoting the proliferation and invasion of colorectal cancer cell lines [55].
CircRNAs can also serve as protein decoys by binding to cellular proteins and altering their regular physiological function [56,57]. In NSCLC, circ_0000079 (CiR79) sequesters Fragile X-Related 1 (FXR1) protein hindering its complexation with PRKCI, consequently, inhibiting the induction of this complex for cell proliferation and invasion as shown in Table 1 [58]. In addition, ciR79-FXR1 interaction resulted in a decrease in the SNAIL protein levels which is an essential gene for cancer cell growth [58].
Table 1.
Different functional mode of actions of CircRNAs.
| Function | CircRNA | Biological functions | Interacting miRNA/protein | Disease/Cell type | References |
|---|---|---|---|---|---|
| miRNA sponge | CDR1 (CiRS-7) | Induce the expression of Ubiquitin protein ligase A (UBE2A) | miR-7 | Alzheimer's disease | [69] |
| Cir-ITCH | Regulates the expression of ITCH | miR-7 | Colorectal cancer | [70] | |
| miR-17 | |||||
| miR-214 | |||||
| Circ-LARP4 | Regulates the expression of LATS1 | miR-424 | Gastric cancer | [71] | |
| Circ-TCF25 | Regulates the expression of CDK6 | miR-103a-3p | Bladder cancer | [72] | |
| miR-107 | |||||
| Circ-CER | Regulates the expression of MMP13 | miR-136 | Osteoarthritis | [73] | |
| Circ-PVT1 | Regulates the expression of E2F2 | miR-125 | Gastric cancer | [74] | |
| Circ-001564 | Unknown | miR-29c-3p | Osteosarcoma | [75] | |
| Circ-ZNF609 | Regulates the expression of ATK3 | miR-150-5p | Hirschsprung disease | [76] | |
| Circ-VMA21 | Regulates the expression of XIAP | miR-200c | Intervertebral disc degeneration | [77] | |
| circMTO1 | Regulates the expression of P21 | miR-9 | Hepatocellular carcinoma | [78] | |
| circ_0005986 | Regulates the expression of Notch 1 | miR-129b | Hepatocellular carcinoma | [79] | |
| circHIPK3 | Regulates the expression of IGF-1 and Aquaporin 3 | miR-558 | Bladder cancer Hepatocellular carcinoma |
[80,81] | |
| miR-124 | |||||
| miR-379 | |||||
| circACTA2 | Regulates the expression of α-SMA | miR-548f-5p | Vascular smooth muscle cells | [82] | |
| Protein Sponge/Decoy | Circ-TNPO3 | Suppresses metastasis by regulating MYC and SNAIL | IGF2BP3 | Gastric cancer | [54] |
| Circ_0000079 | Inhibited cell proliferation and invasion | FXR1 | NSCLC | [58] | |
| Protein Scaffold | Circ-NDUFB2 | Inhibits growth and metastasis | TRIM25 | NSCLC | [62] |
| IGF2BPs | |||||
| Circ-PDE4B | Inhibit cartilage degradation | RIC8A and MID1 | Chondrocyte cell | [63] | |
| Circ-DCUN1D4 | Suppresses metastasis | HuR | Lung adenocarcinoma | [64] | |
| Protein Recruitment | Circ-MRPS35 | Tumor suppressor | KAT7 | Gastric cancer | [59] |
| Circ-SIRT1 | Promotes cell Proliferation | eIF4A3 | Colorectal Cancer | [55] | |
| Translatable circRNAs | Circ-EIF3J | Enhance RNA Polymerase II activity | U1 | HEK293 and HeLa cells | [68] |
| Circ-PAIP2 | Enhance RNA Polymerase II activity | U1 | HEK293 and HeLa cells | [68] |
2.4.2. Protein recruitment
CircRNAs could also guide proteins to certain cellular locations in a processes called protein recruitment [57]. It was recently reported that circMRPS35 could potentially repress GC through in-vitro and ex-vivo analysis, where it was found to act as a modular scaffold to recruit histone acetyltransferase KAT7 to the promoters of FOXO1 and FOXO3a genes. This will elicit acetylation of H4K5 in their promoters, and thus govern histone modification [59]. From a non-oncological perspective, it was found that CircCwc27could directly bind to purine-rich element-binding protein A (Pur-α), increase the retention of cytoplasmic Pur-α, and suppress Pur-α recruitment to the promoters of a cluster of Alzheimer related genes, such as dopamine receptor D1, amyloid precursor protein, regulatory inhibitor subunit 1B, protein phosphatase 1, neurotrophic tyrosine kinase receptor type 1, and LIM homeobox 8 [60].
2.5. Protein scaffolds
CircRNAs have different binding sites for proteins [57]. Thus, they can help in protein scaffolding and protein-complex stabilization by facilitating the contact between different proteins [57]. Besides, scaffold formation could initiate nuclear or cytoplamic translocation of the protein through the nuclear pores [61]. For example, circNDUFB2 was found to facilitate the degradation of IGF2BPs by forming a ternary complex between TRIM25 and IGF2BPs, to enhance their interaction [62]. Consequently, IGF2BPs is ubiquitinated and degraded decreasing tumor progression [62]. Furthermore, in human cartilage samples, circPDE4B was found to act as a scaffold for RIC8A and MID1 promoting their association, and resulting in proteasomal degradation of RIC8A and preventing cartilage degradation [63]. Moreover, circDCUN1D4 in lung adenocarcinoma formed a complex with the RNA-binding protein, HuR protein, promoting its cytoplasmic translocation, and suppressing the metastatic potential of cancerous cells [64].
2.6. Translatable circRNAs
Although circRNAs are classified as lncRNAs, they can code for some short peptides [65,66]. The coding circRNAs contain an internal ribosomal entry site (IRES), which allows the translation of some coding sequences within the circRNA [61]. Furthermore, start codon sequence of mRNA was found to be included in many circRNAs molecules [67]. However, the translation of circRNAs usually occurs in a Cap-independent manner since circRNAs do not include 5'cap sequences [65]. Therefore, circRNA translation is more resistant to translation-regulatory proteins, i.e., 4E-BP protein [65].
2.7. Miscellaneous functions
It was also discovered that some exon-intron (EI) circRNAs can regulate their own gene transcription [56,68]. For instance, circEIF3J and circPAIP2, as summarized in Table 1 are nuclear circRNAs that can interact with U1; small nuclear ribonucleoproteins (snRNPs) and, hence, enhance RNA polymerase II activity. This activity in turn increases their parental gene transcription [68]. Nevertheless, circRNAs could exert hybrid function such as what circDCUN1D4 does in lung adenocarcinoma [64]. Beside the aforementioned function in translocating HuR protein, circDCUN1D4 also promotes the interaction between HuR protein and thioredoxin-interacting protein (TXNIP) mRNA by acting as a scaffold for them forming a RNA-protein ternary complex showing a mix of different functions of circRNAs typical functions [64]. However, the functional characterization of circRNAs is still in immense need for further investigation to achieve better understanding [50].
3. Why breast cancer?
3.1. Dominancy of breast cancer among females
BC has recently surpassed lung cancer as the most commonly diagnosed cancer worldwide [83]. In 2020, almost 2.3 million new BC cases were diagnosed, referring to the fact that approximately 1 in every 8 cancer cases diagnosed in 2020 was BC [83]. At the same time, about 658,000 BC-related deaths were reported in 2020 [83]. BC is a highly complex disease. It might be referred to as several diseases that occur in one organ [84,85]. Although it affects only one anatomic site, it is a heterogeneous disease with great phenotypical variability [86].
Several laboratory techniques have been utilized such as immunohistochemistry [87] and gene expression profiling, in order to classify the various subtypes of BC [88]. To date, accumulating evidence supports the fact that this disparity in biological subtypes is concomitant with distinctions in treatment response and disease-specific outcomes [89]. Several factors such as tumor morphology and grade, tumor size, lymph node metastases, and expression of estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor-2 (HER-2/neu) are currently known as important parameters essential for treatment tailoring. However, there is a great necessity to further enhance the understanding of prognostic and diagnostic biomarkers that will streamline the process of precision medicine [90].
Resistance of cancer cells to conventional therapeutic approaches such as chemotherapeutic drugs has necessitated the direction toward the personalized and targeted therapies [91]. Even the responsiveness for targeted therapies and immunotherapy such as immune checkpoint blockades (ICBs) is only effective in a subset of cancer patients [92]. To increase the survival rate in BC patients, novel therapeutic targets should be discovered and investigated to help in uncovering the crosstalk among oncogenic signaling pathways [92,93].
In the second part of this review, the authors will focus on the complexity of cellular and molecular heterogeneity among BC tumors, focusing on the novel role of circRNAs as a new layer of complexity evading BC heterogeneity. Also, this review will crystalize the potential role of circRNAs in BC and its utilization capacity to act as stable non-invasive biomarkers and/or therapeutic tool in BC precision medicine.
4. Heterogeneous nature of BC tumors
4.1. Inter-tumor and intra-tumor heterogeneity in BC
BC molecular heterogeneity is on both levels intra-tumoral and inter-tumoral. Inter-tumor heterogeneity is noted as the presence of various types of BC with different molecular phenotypes among different individuals, whilst intra-tumor heterogeneity is due to the existence of heterogeneous cell populations within an individual tumor mass [94].
Inter-tumor heterogeneity leads to the existence of different BC subtypes which are categorized by their molecular profiles, morphology, and expression of specific biomarkers (ER, PR and HER2) [95]. This results in BC variability with unique biological behaviors and diverse drug resistance and clinical outcomes [96]. On the contrary, intra-tumoral heterogeneity is found within the tumors of the same type [94]. It is the tumor's ability to acclimatize and adjust to new tumor microenvironment (TME) settings. This new setting may be caused by chemotherapy, radiotherapy, severe ischemia, or a decline in nutrients [97]. Consequently, the tumor specimen taken during a biopsy could not be a representative of the entire real tumor composition, because the tumor may comprise phenotypically different cancer cell populations with different properties and resistance to drugs [98]. In fact, genetic mutations and/or epigenetic modifications are the source of intra-tumor heterogeneity, which explains why the same cell types have dissimilar phenotypic variants [97]. Moreover, this gives a reason why TME components (e.g., different populations of cancer fibroblast or stromal heterogeneity), immune system infiltration, or even the dysregulation of the extracellular matrix, may result in variability in tumor composition [98]. Fascinatingly, the histological and genetic profiles of tumors in patients remain unaltered over their lifetime despite their heterogeneity, whether they metastasized, or remained localized, and even at the end-stage disease [99].
One of the ways to decode the BC heterogeneity may be the identification of different cell phenotypes, cell density, or their localization in the tumor. Heterogeneous cells usually have different molecular pictures within individual tumors [98]. However, heterogeneity could also be predicted by the BC grade [95]. Since the grade of BC is considered as an important prognostic factor, it is incorporated in tools such as Nottingham Prognostic Index (NPI) as well as PrognosTILs [100].
4.2. Heterogeneity on the receptors level
BC cells usually have molecular markers such as ER or PR hormone receptors (HR) expression, and ERBB2 gene amplification, formerly known as HER2 receptor [101]. Based on the most recent European Society of Medical Oncology (ESMO) guidelines, BC can be subdivided into four molecular subtypes: 1) luminal A, in which HR are expressed while the HER2 is absent (HR+/HER2−); 2) luminal B, in which ER is positive while HER2 may or may not be expressed (ER+/HER2−/+); 3) HER2 enriched, in which (HR−/HER2+); and 4) basal-like subtype, which is also known as triple-negative BC (TNBC), tumor cells do not possess any of the 3 standard molecular markers (HR−/HER2−) [84,102,103].
The markers of heterogeneity are defined as the difference in the expression of these receptors [95]. For instance, HER2+ tumors are aggressive tumors yet are usually treated with anti-HER2 therapy while ER-positive tumors are mostly well-differentiated, less aggressive, and linked to better post-surgery results [104]. On the other hand, TNBC is a highly heterogeneous cancer in regard to its molecular and phenotypical features [105]. TNBC is the most aggressive and most likely to recur out of those four groups [106]. Based on such sub-classification, the patient's treatment protocol is decided [107], taking into consideration the aforementioned histological and anatomic cancer stage [108]. Collectively, some of BC subtypes have better prognosis and response to treatment, while other subtypes are notoriously aggressive, with a poor prognosis and reaction to treatment that again reflects the heterogeneity of BC tumors [109].
4.3. Heterogeneity on the genetic level
Scientists have exploited genetic heterogeneity to be used as a risk factor used to identify the risk of BC occurrence and even helps in predicting its possible subtype. Mutations in BRCA genes are among the most clearly established risk factors for BC [110,111]. About 12% of BC patients under 40 years of age were found to have BRCA gene mutations [112]. BRCA1 and BRCA2 are considered as BC susceptibility genes [110]. They are tumor-suppressor genes involved in DNA repair and mutations in these genes confer a 45–65% risk of BC by the age of 70 years [111]. TNBC tumors are identified by the high occurrence of germ line BRCA1/2 mutations, almost twice its frequency in BC patients [112].
4.4. Heterogeneity on the epigenetic level
Although genetic changes have been recognized long time ago as a key player in BC development, heterogeneity in epigenetic profile in BC tissues compared to healthy tissues has directed studies to focus on this level of cell regulation as to be a potential initiator for tumor progression [113]. Epigenetic changes such as chromatin remodeling and alteration in expression levels of ncRNAs, all could affect gene expression. Due to availability of methodological approaches, DNA hypermethylation and changes in ncRNAs profile have been studied more intensively in literature, compared to studies on histone modifications in cancer patients [114]. Yet, fluctuation in ncRNAs expression levels has not only correlated to BC hallmarks, but also has been linked to chemotherapeutic resistance [115]. Studies on miRNAs and lately lncRNAs have remarkably increased in the last decades, showing aberrant expression that could be exploited as potential diagnostic and prognostic markers for all cancer types generally and for BC in particular [116]. On the other hand, circRNAs, which are relatively more recent and less investigated compared to miRNAs have shown superior characteristics attributing them a higher potential for clinical application [117]. Stability and conservation of circRNAs which can be secreted from cells and tissue into the serum could be used as a biomarkers to help in disease detection and for treatment response follow-up [14].
5. CircRNAs: Revolution Era in the prognosis and diagnosis of BC
The prognostic and diagnostic biomarkers for different malignancies have been investigated for centuries, however, the survival rate in patients have not become promising enough [118]. As mentioned in the previous section, cancer heterogeneity represents a great challenge in treatment strategies [119,120]. Albeit that, the treatment protocol should be further studied to reach better clinical outcomes [118]. CircRNAs-based biomarkers present an emerging and promising pivotal role in tumor detection, treatment, likelihood of recurrence, and metastasis property prediction [50,121]. Therefore, previous research articles have tackled circRNAs role in tumorigenesis generally and in BC particularly (Table 2) [3,[122], [123], [124], [125], [126], [127], [128], [129]].
Table 2.
CircRNAs evading the field of oncology.
| Functional Activity | Cancer Type | circRNA | References |
|---|---|---|---|
| Tumor-Enhancer | Breast cancer | Circ-0044234 | [133] |
| Circ-NOTCH3 | [134] | ||
| Circ-TP63 | [135] | ||
| Circ-0000284 (circHIPK3) | [136,137] | ||
| Circ-0007766 (circ-ERBB2) | [138,139] | ||
| Circ-0019853 (circ-PDCD11) | [140] | ||
| Circ-0084100 (circIKBKB) | [141] | ||
| Circ-0008673 | [142] | ||
| Circ-NOLC1 | [143] | ||
| Circ-0055478 (circPTCD3) | [144] | ||
| Circ-0000073 (circOMA1) | [145] | ||
| Circ-0001944 (circBCBM1) | [146] | ||
| Circ-0103552 | [147] | ||
| Circ-0084927 | [148] | ||
| Circ-RHOT1 | [149] | ||
| Circ-0092338 (circNINL) | [150] | ||
| Circ-CNOT2 | [151] | ||
| Circ-0048764 | [152] | ||
| Circ-0000515 (circRPPH1) | [153,154] | ||
| Circ-0000515 | [155] | ||
| Circ-DNMT1 | [156] | ||
| Circ-PGAP3 | [157] | ||
| Circ-BACH2 | [158] | ||
| Circ-ABCB10 | [159,160] | ||
| Circ-0011946 | [161] | ||
| Circ-gfra1 | [162] | ||
| Circ-0001982 | [163,164] | ||
| Circ-0005230 | [118] | ||
| Circ-IRAK3 | [139,165] | ||
| Circ-0008039 (circFOXO3) | [166] | ||
| Glioma | Circ-TTBK2 | [167] | |
| Circ-NT5E | [168] | ||
| Hepatocellular carcinoma | Circ-ZFR | [169] | |
| Circ-FBLIM1 | [170] | ||
| Circ-HIPK3 | [81] | ||
| Cervical Cancer | Circ-000284 | [171] | |
| Circ-0023404 | [172] | ||
| Lung cancer | Circ-102231 | [173] | |
| Circ-PRKCI | [174] | ||
| Circ- MAN2B2 | [175] | ||
| Circ-0013958 | [176] | ||
| NSCLC | Circ-ZFR | [177] | |
| Circ-0007385 | [[178], [179], [180]] | ||
| Circ-100146 | [181] | ||
| Circ-FOXO3 | [182] | ||
| Cirs-7 (CDR1as) | [183,184] | ||
| Colorectal cancer (CRC) | Circ-CCDC66 | [185] | |
| Circ-HIPK3 | [186] | ||
| Circ-0000069 | [187] | ||
| Circ-001569 | [188] | ||
| Oesophageal squamous cell carcinoma | Cirs-7 (CDR1as) | [189] | |
| Circ-HIPK3 | [190] | ||
| Gastric Cancer | Circ-0047905 | [191] | |
| Oral Squamous Cell Carcinoma (OSCC) | Circ-DOCK1 | [192] | |
| Clear cell renal carcinoma (ccRCC) | Circ-ATP2B1 | [190] | |
| Papillary Thyroid Cancer | Circ-ZFR | [193] | |
| Melanoma | Circ-0084043 (CircADAM9) | [194] | |
| Prostate Cancer (PC-a) | Circ-MYLK | [195] | |
| Gall Bladder Cancer | Circ-HIPK3 | [196] | |
| Head and neck squamous cell carcinoma | Circ-PVT1 | [197] | |
| Tumor-suppressor | Breast cancer | Circ-0006220 | [198] |
| Circ-0120472 (circCCDC85A) | [199] | ||
| Circ-NR3C2 | [200] | ||
| Circ-0000442 | [201] | ||
| Circ-tada2a-E6 | [202] | ||
| Circ-000911 | [203] | ||
| Circ-0072309 | [204] | ||
| Circ-ASS1 | [205] | ||
| Glioblastoma | Circ-FBXW7 | [206] | |
| Circ-SHPRH | [207,208] | ||
| Circ-0001445 (Circ-SMARCA5) | [209] | ||
| Circ-0001946 | [210] | ||
| Hepatocellular carcinoma | Circ-C3P1 | [211] | |
| Circ-MTO1 | [78] | ||
| Circ-0001445 (Circ-SMARCA5) | [212] | ||
| Circ-ZKSCAN1 | [213] | ||
| Circ-ADAMTS13 | [214] | ||
| Circ-ADAMTS14 | [215] | ||
| Circ-0079299 | [216] | ||
| Circ-SMAD2 | [217] | ||
| Gastric Cancer | Circ-TNPO3 | [54] | |
| Circ-YAP1 | [218] | ||
| Circ-0000993 | [219] | ||
| Circ-PSMC3 | [220] | ||
| Circ-FAT1(e2) | [221] | ||
| Circ-0000096 | [222] | ||
| Circ-larp4 | [71] | ||
| Circ-ZFR | [223] | ||
| Lung cancer | Circ-dcun1d4 | [64] | |
| Circ-100395 | [224] | ||
| Circ-0006916 | [225] | ||
| Circ-NOL10 | [226] | ||
| NSCLC | Circ-0000079 (Cir79) | [58] | |
| Circ_0008305 (CircPTK2) | [227] | ||
| Circ-0001649 | [228] | ||
| Circ- ITCH | [229] | ||
| Circ-0043256 (Circacaca) | [230] | ||
| Colorectal cancer (CRC) | Circ-ITGA7 | [231] | |
| Circ-0014717 | [232] | ||
| Circ-ITCH | [70] | ||
| Bladder Cancer | Circ-FNDC3B | [233] | |
| Circ-LPAR1 | [234] | ||
| Cirs-7 (CDR1as) | [235] | ||
| Circ-HIPK3 | [236] | ||
| Circ-ITCH | [237] | ||
| Papillary thyroid cancer | Circ-ITCH | [238] | |
| Ovarian cancer | Circ-ITCH | [239] | |
| Osteosarcoma | Circ-0002052 (Circpappa) | [240] | |
| Oral squamous cell carcinoma | Circ-0008309 | [241] | |
| Cholangiocarcinoma | Circ-0001649 | [242] |
CircRNAs-based diagnostic and prognostic biomarkers are recently used in BC detection, grading, and follow-up since they show BC-tissue specificity and could discriminate cancerous cells from adjacent healthy tissue. Besides, circRNAs may help in differentiation between BC subtypes, giving a fast hint about the treatment protocol to be followed with the patient, and thus, participating in better outcomes. Indeed, prognostic circRNAs themselves could be targeted for therapy, since silencing their expression or exploiting them as therapeutic targets would contribute in enhancing tumor prognosis.
Nonetheless, based on the current understanding of the circRNAs nature, it makes them perfect match for a qualified diagnostic and/or prognostic biomarker. CircRNAs have 4 characteristics that ranks it as a potential biomarker for several oncological diseases including BC. CircRNAs are 1) Stable: this is due to the covalently closed loop structure, therefore making it highly resistant to exonuclease RNase R and more stable and have longer half-live in plasma than any other linear lncRNA [130,131]. 2) Universal: circRNAs have been reported to be the most universal molecule present within the human cell and usually the circRNA is more abundant that its linear form [132]. 3) Specific: circRNAs are reported to be expressed in a tissue-specific manner and in a developmental stage-specific manner which makes them perfect candidates for promising oncological biomarkers [131]. 4) Conservative: circRNAs are evolutionally conserved in different species such as murine models and human beings [131]. In the following section of the review, the authors will discuss the newly discovered prognostic and diagnostics circRNAs used in BC.
5.1. CircRNAs: promising diagnostic biomarkers in BC
The circRNA hsa_circ_0000284 (circHIPK3) was found to be overexpressed in BC tissues compared to its normal counterparts ranking is as potential diagnostic circRNA for BC patients. However, this has further investigations to be validated. Functionally, CircHIPK3 was found to act as a sponge for miR-326. High level circHIPK3 has been associated with poor prognosis. CircHIPK3 was reported to induce cellular proliferation, migration, and invasion ability of the BC cells. In addition, circHIPK3 was found to inhibit the apoptosis of the BC cells. However, this effect is reversed when circHIPK3 was knocked down, highlighting its potential as promising therapeutic target as well [136].
Circ_0008673 was also found to be overexpressed in BC cells. On the molecular level, Circ_0008673 was found to induce CFL2 expression level by sponging miR-153-3p. Due to the high levels of circ_0008673 in BC tissues, elevated levels of CFL2, which is an actin-binding protein, was observed in the BC tissues which leads to increase cell proliferation, migration and invasion capacities of BC cells and decreasing their apoptosis [142]. Collectively, this highlights circHIPK3 and Circ_0008673 as oncogenic mediators and potential diagnostic circRNAs in BC patients.
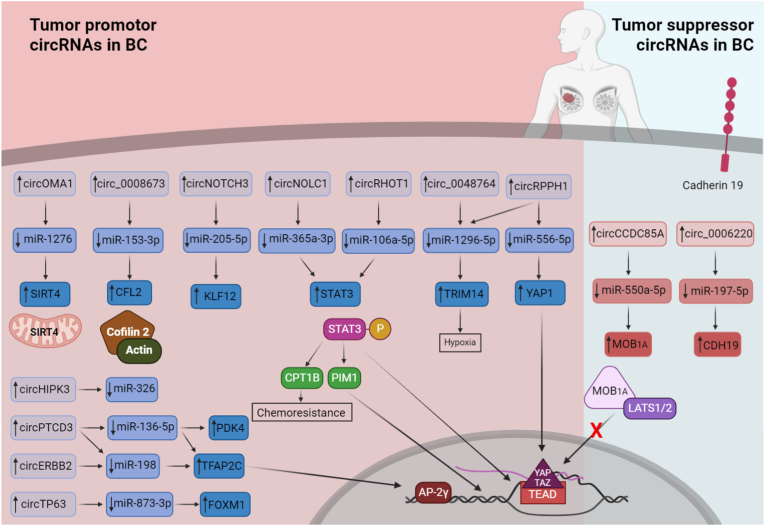
circNOLC1 was also reported to participate in promotion of BC via regulating miR-365a-3p/STAT3 axis as shown in Fig. 3. BC tissues and cell lines have expressed higher level of circNOLC1 compared to normal tissues, eliciting high expression level of STAT3. However, propofol treatment has repressed the expression of both circNOLC1 and STAT3, decreasing BC cells viability [143]. Yet, additional studies have shown that the activation of STAT3 signaling pathway activates CPT1B, which is a cellular molecule that contributes to chemoresistance as represented in Fig. 3 [243]. However, circRHOT1-activated STAT3 is intermediated by sponging miR-106a-5p (Fig. 3) [149]. In a study by Zhang et al, the increase of STAT3 levels via circRHOT1/miR-106a-5p/STAT3 axis has resulted in ferroptosis attenuation in BC cells. However, knockdown of circRHOT1 has retained cell apoptosis ability, increased the levels of ROS and iron in BC cells [149]. Intriguingly, a proto-oncogene PIM1, which is responsible for cell invasion and epithelial-mesenchymal transition (EMT) promotion through upregulating the expression of EMT-related genes in BC, is found to be regulated by the phosphorylated STAT3 as shown in Fig. 3 [243].
Fig. 3.
Different tumor suppressor and tumor promotor circRNAs in BC.
This figure represents an array of tumor suppressor circRNAs and tumor promotor circRNAs with their respective target and their determining role in breast cancer.
hsa_circRNA_0000073 (circOMA1) has been found to be upregulated in BC tissues (Fig. 3). CircOMA1 fosters BC progression by sponging miR-1276, leading to elevation of the mitochondrial matrix protein Sirtuin 4 (SIRT4). SIRT4 has a controversial role in cancer development. It showed anti-tumor activity in lung and colorectal cancer cells, yet, it could act as oncogene in esophageal cancer and BC tissues [145].
Correspondingly, hsa_circ_0000515 (circRPPH1) have been reported previously to participate in TNBC development and progression by regulating miR-556-5p/YAP1 axis [153]. Yet, similar to circ_0048764 [152], circRPPH1 was recently reported to regulate miR-1296-5p/TRIM14 axis in TNBC tissue promoting hypoxia-associated TNBC progression [154].
For hsa_circ_0007766 (circ-ERBB2), it was found to participate in BC development by regulating miR-136-5p/PDK4, miR-136-5p/TFAP2C, or miR-198/TFAP2C axes [138,139]. Similar to circ-ERBB2, hsa_circ_0055478 (circPTCD3) was also found to act as miR-198 sponge. Elevated levels of circPTCD3 in BC tissues and cell line reverse the antiproliferative activity of miR-198, boosting BC development [144].
Intriguingly, circRNAs could be used as a discriminator to differentiate BC subtypes [[133], [134], [135]]. Although hsa_circ_0044234 is upregulated in BC tissues, it remarkably decreased in the TNBC subtype. Therefore, it could be used to discriminate between TNBC and other BC subtypes (being lower in TNBC) with 83.64% specificity and 72.5% sensitivity. Moreover, the downregulation of hsa_circ_0044234 in TNBC cells was found to lead to overexpression of miR-135b-5p by 4.89 units for each unit-decrease in the levels of hsa_circ_0044234. The increase of miR-135b-5p could also lead to decrease of GATA3 expression, which is needed for homologous recombination DNA repair pathway. Thus, TNBC cells usually suffer from the disturbance of this pathway [133]. A study by Guan et al has assured that basal-like BC (BLBC) development and progression involves circ_NOTCH3. Compared to normal breast tissues, BLBC showed higher expression level of circ_NOTCH3 which lead to sponging miR-205-5p and over production of KLF12 protein [134]. Meanwhile, ER+ BC showed higher expression levels of circTP63. circTP63 induces cancer progression via upregulating FOXM1 production by sponging miR-873-3p, which is an inhibitor of FOXM1 mRNA translation [135].
Throughout the past year, few tumor-suppressor circRNAs were discovered. For instance, hsa_circ_0006220 has the ability to suppress TNBC proliferation, migration, and invasion ability by sponging miR-197-5p, which is a natural silencer for CDH19 gene expression. Consequently, hsa_circ_0006220 promotes an increase in cadherin-19 protein cellular levels, retaining its role in maintaining intercellular connections (Fig. 3). However, hsa_circ_0006220 was found to be remarkably downregulated in TNBC tissues compared to other BC subtypes [198].
Oncogenic miR-550a-5p has expressed high intracellular levels in BC tissues leading to increase in their proliferation, migration, and invasion ability by hindering MOB1A activity [199]. Induction of hsa_circ_0120472 (circCCDC85A) expression in BC cells and tissues, which was found to be initially down regulated, has reversed the oncogenic properties of miR-550a-5p (Fig. 3) [199]. MOB1A has shown intriguing role in limiting tumor progression as a part of Hippo signaling pathway. Upon phosphorylation by MST1/2 kinases, MOB1A/1B interact with LATS1/2 kinases forming a complex that leads to subsequent auto phosphorylation of LATS [244]. YAP and its paralog TAZ are key transcriptional regulators that are phosphorylated by LATS kinases leading to their sequestration or degradation, hindering their translocation to the nucleus and prohibiting their contribution to gene expression when bind to TEAD transcription factors (Fig. 3) [244].
5.2. CircRNAs: promising prognostic biomarkers in BC
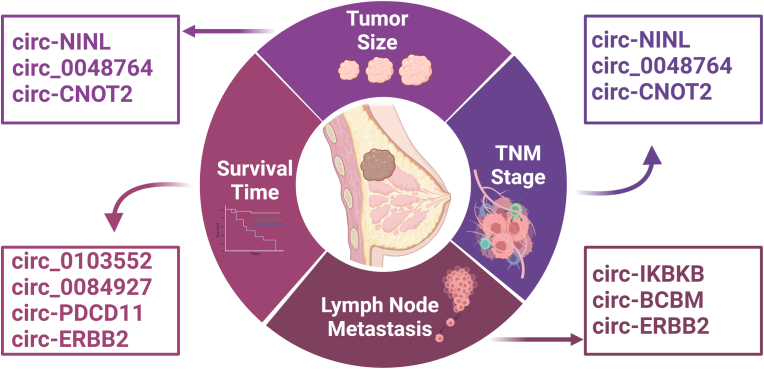
Prognostic biomarkers are crucial when it comes to clinical practice. Since circRNAs are stable, conserved, and easily processed molecules that show high tissue specificity, plentiful studies have investigated the potential role of circRNAs as prognostic biomarkers to help in following the clinicopathological states of BC patients [245]. During the past year, number of circRNAs was correlated to patient survival time (Fig. 4). One of BC tissue-specific circRNAs is circ_0103552, it was found to promote BC progression through sponging miR-515-5p, which leads to upregulation of the angiogenic factor CYR61 promoting BC tumorigenesis [147]. BC patients survival time was found to be shorter for patients who displayed high circ_0103552 expression profile [147]. Similarly, circ_0084927 was correlated with BC prognosis and its downregulation was linked to longer survival period. However, on the molecular level, circ_0084927 sponges miR-142-3p, leading to upregulation of ERC1 protein [148]. Likewise, when the oncogene hsa_circ_0019853 (circ-PDCD11) is upregulated in TNBC, it is to be correlated to unfavorable survival and prognosis since circ-PDCD11 sponges miR-432-5p leading to promotion of LDHA expression. Accordingly, the rate of glucose uptake, lactate generation, ATP synthesis and microenvironment acidification was increased, enhancing aerobic glycolysis [140]. [246]. Nevertheless, expression level of circ-ERBB2 has been negatively correlated with poor TNBC survival by contributing in TNBC cells proliferation and invasion. When circ-ERBB2 cytoplasmic levels increases, it sponges miR-136-5p, leading to upregulation of PDK4 and enhancing of Warburg effect [138].
Fig. 4.
Different promising prognostic circRNAs in BC.
This figure represents an array of circRNAs which are correlated to survival time, tumor size, TNM stage, and lymph node metastasis in BC representing promising BC prognostic biomarkers.
As such, other circRNAs were correlated with larger tumor size, advanced TNM stage, and lymph node metastasis (Fig. 4). Hsa_circ_0092338 (circNINL), circCNOT2 and circ_0048764 were highly correlated with poorer TNM stage and lager tumor size [[150], [151], [152]]. CircNINL has shown elevated expression levels in BC cells compared to normal tissue [150]. It was found to elicit BC tumorigenesis by promoting the transcription of oncogenes which are regulated by β-catenin signaling pathway. This is done by sponging of miR-921 on circNINL leading to upregulation of ADAM9, allowing its interaction with E-cadherin initiating the β-catenin signaling pathway [150]. Likewise, circCNOT2 was found to be upregulated in BC tissues leading to their poor prognosis by promoting the expression of TWIST1, a helix-loop-helix transcription factor, via sponging miR-409-3p [151]. However, circCNOT2 was also linked to low patient's survival rates [151]. Moreover, circ_0048764 partakes in BC tumorigenesis by sponging miR-1296-5p, similar to circRPPH1, leading to elevation of TRIM14 which is known as a cancer-progression promoting factor whose depletion induces apoptosis and results in better cancer prognosis [152].
On the other hand, hsa_circ_0084100 (circIKBKB), hsa_circ_0001944 (circBCBM), and hsa_circ_0007766 (circ-ERBB2) did not only show the potential to predict BC metastatic ability, but also BC intrinsic metastatic property and destination [139,141,146]. A study done by Xu et al., they discovered that circIKBKB overexpression is strongly correlated with BC bone metastasis by NF-κB-mediated mechanism that results in osteoclastogenesis induction. Generally, the synthesis of IκBα, which is a natural NF-κB inhibitor is mediated by activated NF-κB. IκBα bind to N-terminal of NF-κB and hinder its binding to chromatin and export it back to the cytoplasm in a form negative feedback loop. When circIKBKB present in the cytoplasm, it bound to active IKKβ and p65 facilitating their interaction. Thus, instead of formation of p65/IκBα complex and maintaining the negative feedback loop, circIKBKB results in IKKβ-mediated IκBα phosphorylation and promotion of NF-κB pathway which in turn upregulation of many bone-remodeling factors, and thus, induction of osteoclastogenesis and BC bone metastasis [141]. Meanwhile, circBCBM1 expression levels were correlated to BC-brain metastasis in tissues, cell lines and plasma samples. CircBCBM1 sponges miR-125a leading to subsequent increase in BRD4, a member of epigenetic regulator protein family. The elevated BRD4 levels induce MMP9 production via SHH signaling pathway. Secretion of MMP9 typically increased BBB permeability. Therefore, upregulation of BRD4-MMP9 axis facilitated BC-brain metastasis [146]. As such, expression of circ-ERBB2 was linked to lymph node metastasis in ERBB2+/HER2+ BC patients by working as miR-136-5p or miR-198 sponge. Consequently, TFAP2C, also called AP-2γ, is upregulated. TFAP2C is known to be a key mediator in cell cycle and cell apoptosis regulation. When the expression levels of circ-ERBB2 increased, cell proliferation, invasion, and migration was promoted, whereas and cell apoptosis was delayed [139]. In addition, circ-ERBB2 levels were correlated to HER2 receptor status, whether it was present or not [139].
6. Conclusion
This review sheds the light onto a fast-growing field of research. It is quite evident that the field of circRNA has witnessed an enormous growing number of publications in few years. CircRNAs have been casted as pivotal players in the tumorigenic process. CircRNAs have been proven to act as miRNA sponges and have a intertwined interaction with several oncogenic proteins thus affecting several cancer hallmarks such as proliferation, migration, invasion, angiogenesis and apoptosis. CircRNAs have been reported to act as promising therapeutic tools/targets in BC. However, circRNAs have not been well studied as prominent players at the cancer-immune synapse in the TME of BC or any other solid malignancy. This underscores the importance of directing future studies on studying the onco-immunogenic role of circRNAs in BC, and its possible involvement in the crosstalk between cancer and effector immune cells at the TME as well. In BC, circRNAs have been discussed as potential diagnostic and prognostic biomarkers. However, this area still needs further experimental validation before its clinical translation.
7. Future Recommendations
Notwithstanding the need for further understanding for the physiological and pathological roles of circRNAs, it is evident that they are involved in almost all biological processes. The interpretation of the mechanistic points of all circRNAs associated with BC, either functioning as oncogenes, or as tumor suppressors will be a significant addition to further unlocking minds in the realm of BC. Indeed, this could be achieved by deeply comprehending how heterogeneous malicious TNBC operates and the prospect of targeting circRNAs. All the aforementioned details imply the alarming intricacy and convolution of the BC, and how such a multi-layered disease will need intensive research and ingenuity. Finally, the authors highly appraise the great potential of circRNAs and that they would provide a promising therapeutic tool for this predicament, and that with the unraveling and interpreting the deep sea of circRNAs, which may serve as prognostic, diagnostic, and even therapeutic tools, or molecules to be targeted for cell cycle control. Hence, this could be achieved by advanced analysis and investigation in the field.
CRediT authorship contribution statement
Alyaa Dawoud: Data curation, Formal analysis, Literature review, data analysis, original draft preparation, Tables and Figures preparation. Zeina Ihab Zakaria: Data curation, Formal analysis, Literature review, data analysis, original draft preparation. Hannah Hisham Rashwan: Data curation, Formal analysis, Literature review, data analysis, original draft preparation. Maria Braoudaki: Writing, revising the manuscript critically for important intellectual content and editing the final version of the manuscript. Rana A. Youness: Conceptualization, and design of work, writing, revising the manuscript critically for important intellectual content and editing the final version of the manuscript. All authors read, revised and approved the final submitted manuscript.
Abbreviation List
- ALU
Arthrobacter luteus
- BL1
basal-like
- BL2
basal-like 2
- MES
mesenchymal
- MSL
mesenchymal stem-like
- IM
immunomodulatory
- LAR
luminal androgen receptor
- BLIA
basal-like immune-activated
- BLIS
basal-like immunosuppressed
- TME
tumor microenvironment
- ICB
immune checkpoint blockades
- DNA
Deoxyribonucleic acid
- RNA
Ribonucleic acid
- rRNA
Ribosomal RNAs
- tRNAs
Transfer RNAs
- snRNAs
Small nuclear RNAs
- snoRNAs
Small nucleolar RNAs
- pre-mRNA
Precursor messenger RNA
- RNA pol II
RNA polymerase II
- DGCR8
DiGeorge Syndrome Critical Region 8
- HER2
Human Epidermal growth factor Receptor 2
- ERBB2
Erb-b2 receptor tyrosine kinase 2 (ERBB2)
- NSCLC
Non-small cell lung cancer
- EMT
epithelial-mesenchymal transition
- TFAP2C
Target transcription factor activator protein 2C
References
- 1.Richard Boland C. Non-coding RNA: it's not junk. Dig. Dis. Sci. Mar. 2017;62(5):1107–1109. doi: 10.1007/S10620-017-4506-1. 2017 625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Romano G., Veneziano D., Acunzo M., Croce C.M. Small non-coding RNA and cancer. Carcinogenesis. May 2017;38(5):485–491. doi: 10.1093/CARCIN/BGX026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bach D.H., Lee S.K., Sood A.K. Circular RNAs in cancer. Mol. Ther. Nucleic Acids. Jun. 2019;16:118–129. doi: 10.1016/J.OMTN.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han B., Chao J., Yao H. Circular RNA and its mechanisms in disease: from the bench to the clinic. Pharmacol. Ther. Jul. 2018;187:31–44. doi: 10.1016/J.PHARMTHERA.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 5.Lim C.S., Wardell S.J.T., Kleffmann T., Brown C.M. The exon–intron gene structure upstream of the initiation codon predicts translation efficiency. Nucleic Acids Res. May 2018;46(9):4575–4591. doi: 10.1093/NAR/GKY282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karijolich J., Yu Y.-T. vol. 7. 2010. pp. 192–204. (Spliceosomal snRNA Modifications and Their Function). 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Witten J.T., Ule J. Understanding splicing regulation through RNA splicing maps. Trends Genet. Mar. 2011;27(3):89–97. doi: 10.1016/J.TIG.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Z., Burge C.B. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. May 2008;14(5):802–813. doi: 10.1261/RNA.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li F., Yang Q., He A.T., Yang B.B. Circular RNAs in cancer: limitations in functional studies and diagnostic potential. Semin. Cancer Biol. Oct. 2020 doi: 10.1016/J.SEMCANCER.2020.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Li Z., Ruan Y., Zhang H., Shen Y., Li T., Xiao B. Tumor-suppressive circular RNAs: mechanisms underlying their suppression of tumor occurrence and use as therapeutic targets. Cancer Sci. 2019;110(12):3630–3638, Dec. doi: 10.1111/CAS.14211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xin C., Huang F., Wang J., Li J., Chen Q. Roles of circRNAs in cancer chemoresistance (Review) Oncol. Rep. Oct. 2021;46(4):1–11. doi: 10.3892/OR.2021.8176/HTML. [DOI] [PubMed] [Google Scholar]
- 12.Ashwal-Fluss R., et al. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. Oct. 2014;56(1):55–66. doi: 10.1016/J.MOLCEL.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 13.HL S., G K., D R., HJ G., AK K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. U.S.A. 1976;73(11):3852–3856. doi: 10.1073/PNAS.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patop I.L., Wüst S., Kadener S. Past, present, and future of circRNAs. EMBO J. Aug. 2019;38(16) doi: 10.15252/EMBJ.2018100836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salzman J., Gawad C., Wang P.L., Lacayo N., Brown P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. Feb. 2012;7(2) doi: 10.1371/JOURNAL.PONE.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou W.-Y., Cai Z.-R., Liu J., Wang D.-S., Ju H.-Q., Xu R.-H. Circular RNA: metabolism, functions and interactions with proteins. Mol. Cancer. Dec. 2020;19(1):1–19. doi: 10.1186/S12943-020-01286-3. 2020 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Z.-J., Shen J. vol. 14. May 2016. pp. 514–521. (Circular RNA Participates in the Carcinogenesis and the Malignant Behavior of Cancer). 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greene J., et al. Circular RNAs: biogenesis, function and role in human diseases. Front. Mol. Biosci. Jun. 2017;(JUN):38. doi: 10.3389/FMOLB.2017.00038. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D'Ambra E., Capauto D., Morlando M. Exploring the regulatory role of circular RNAs in neurodegenerative disorders. Int. J. Mol. Sci. Nov. 2019;20(21):5477. doi: 10.3390/IJMS20215477. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilusz J.E., Jeremy Wilusz C.E. A 360° view of circular RNAs: from biogenesis to functions. Wiley Interdiscip. Rev. RNA. Jul. 2018;9(4) doi: 10.1002/WRNA.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeck W.R., et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. Feb. 2013;19(2):141–157. doi: 10.1261/RNA.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qu S., et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. Sep. 2015;365(2):141–148. doi: 10.1016/J.CANLET.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 23.Liu L., Wang J., Khanabdali R., Kalionis B., Tai X., Xia S. vol. 14. Dec. 2017. pp. 1715–1721. (Circular RNAs: Isolation, Characterization and Their Potential Role in Diseases). 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelly S., Greenman C., Cook P.R., Papantonis A. Exon skipping is correlated with exon circularization. J. Mol. Biol. 2015;427(15):2414–2417, Jul. doi: 10.1016/J.JMB.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 25.Lee Y., Rio D.C. vol. 84. Jun. 2015. pp. 291–323. (Mechanisms and Regulation of Alternative Pre-mRNA Splicing). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rio D.C. Splicing of pre-mRNA: mechanism, regulation and role in development. Curr. Opin. Genet. Dev. Jan. 1993;3(4):574–584. doi: 10.1016/0959-437X(93)90093-5. [DOI] [PubMed] [Google Scholar]
- 27.Jeck W.R., et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. Feb. 2013;19(2):141–157. doi: 10.1261/RNA.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo J., Tong J., Zheng J. Circular RNAs: a promising biomarker for endometrial cancer. Cancer Manag. Res. 2021;13:1651. doi: 10.2147/CMAR.S290975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilusz J.E. Repetitive elements regulate circular RNA biogenesis. Mobile Genet. Elem. Jan. 2015;5(3):39. doi: 10.1080/2159256X.2015.1045682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carriero S., Damha M.J. Inhibition of pre-mRNA splicing by synthetic branched nucleic acids. Nucleic Acids Res. 2003;31(21):6157–6167, Nov. doi: 10.1093/NAR/GKG824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruskin B., Green M.R. An RNA processing activity that debranches RNA lariats. Science. 1985;229(4709):135–140. doi: 10.1126/SCIENCE.2990042. 80. [DOI] [PubMed] [Google Scholar]
- 32.Ragan C., Goodall G.J., Shirokikh N.E., Preiss T. Insights into the biogenesis and potential functions of exonic circular RNA. Sci. Rep. Feb. 2019;9(1):1–18. doi: 10.1038/s41598-018-37037-0. 2019 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X.O., Bin Wang H., Zhang Y., Lu X., Chen L.L., Yang L. Complementary sequence-mediated exon circularization. Cell. Sep. 2014;159(1):134–147. doi: 10.1016/J.CELL.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 34.Yu C.Y., Kuo H.C. The emerging roles and functions of circular RNAs and their generation. J. Biomed. Sci. Apr. 2019;26(1):1–12. doi: 10.1186/S12929-019-0523-Z. 2019 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vromman M., Vandesompele J., Volders P.J. Closing the circle: current state and perspectives of circular RNA databases. Briefings Bioinf. Jan. 2021;22(1):288–297. doi: 10.1093/BIB/BBZ175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Costa M.C., Enguita F.J. Towards a universal nomenclature standardization for circular RNAs. Non-coding RNA Investig. Jun. 2020;4 doi: 10.21037/NCRI.2020.03.01. 0. 2–2. [DOI] [Google Scholar]
- 37.Glažar P., Papavasileiou P., Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20(11):1666–1670, Nov. doi: 10.1261/RNA.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu M., Wang Q., Shen J., Yang B.B., Ding X. Circbank: a comprehensive database for circRNA with standard nomenclature. RNA Biol. Jul. 2019;16(7):899–905. doi: 10.1080/15476286.2019.1600395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dong R., Ma X.K., Li G.W., Yang L. CIRCpedia v2: an updated database for comprehensive circular RNA annotation and expression comparison. Dev. Reprod. Biol. Aug. 2018;16(4):226–233. doi: 10.1016/J.GPB.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ji P., et al. Expanded expression landscape and prioritization of circular RNAs in mammals. Cell Rep. Mar. 2019;26(12):3444–3460. doi: 10.1016/J.CELREP.2019.02.078. e5. [DOI] [PubMed] [Google Scholar]
- 41.Haeussler M., et al. The UCSC Genome Browser database: 2019 update. Nucleic Acids Res. Jan. 2019;47(D1):D853–D858. doi: 10.1093/NAR/GKY1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Brien J., Hayder H., Zayed Y., Peng C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. Aug. 2018;9(AUG):402. doi: 10.3389/FENDO.2018.00402/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Backes C., Meese E., Keller A. Specific miRNA disease biomarkers in blood, serum and plasma: challenges and prospects. Mol. Diagn. Ther. Jul. 2016;20(6):509–518. doi: 10.1007/S40291-016-0221-4. 2016 206. [DOI] [PubMed] [Google Scholar]
- 44.Zhou S.S., et al. miRNAS in cardiovascular diseases: potential biomarkers, therapeutic targets and challenges. Acta Pharmacol. Sin. 2018;39(7):1073–1084, Jun. doi: 10.1038/aps.2018.30. 2018 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bertoli G., Cava C., Castiglioni I. MicroRNAs: new biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics. 2015;5(10):1122. doi: 10.7150/THNO.11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turchinovich A., Samatov T.R., Tonevitsky A.G., Burwinkel B. Circulating miRNAs: cell-cell communication function? Front. Genet. 2013;4(JUN):119. doi: 10.3389/FGENE.2013.00119/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.MacFarlane L.-A., Murphy P.R. MicroRNA: biogenesis, function and role in cancer. Curr. Genomics. Dec. 2010;11(7):537. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chung C.H., Walter M.H., Yang L., Chen S.C.G., Winston V., Thomas M.A. Predicting genome terminus sequences of Bacillus cereus-group bacteriophage using next generation sequencing data. BMC Genom. 2017 doi: 10.1186/s12864-017-3744-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verduci L., Strano S., Yarden Y., Blandino G. The circRNA–microRNA code: emerging implications for cancer diagnosis and treatment. Mol. Oncol. Apr. 2019;13(4):669. doi: 10.1002/1878-0261.12468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Santer L., Bär C., Thum T. Circular RNAs: a novel class of functional RNA molecules with a therapeutic perspective. Mol. Ther. Aug. 2019;27(8):1350. doi: 10.1016/J.YMTHE.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dudekula D.B., Panda A.C., Grammatikakis I., De S., Abdelmohsen K., Gorospe M. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. Jan. 2016;13(1):34–42. doi: 10.1080/15476286.2015.1128065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang R., Wang Z., Meng G., Hua L. Circular RNA CCDC66 facilitates abdominal aortic aneurysm through the overexpression of CCDC66. Cell Biochem. Funct. Oct. 2020;38(7):830–838. doi: 10.1002/CBF.3494. [DOI] [PubMed] [Google Scholar]
- 53.Wang X., Fang L. Advances in circular RNAs and their roles in breast Cancer. J. Exp. Clin. Cancer Res. Aug. 2018;37(1):1–12. doi: 10.1186/S13046-018-0870-8/TABLES/2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu T., et al. Circular RNA circ-TNPO3 suppresses metastasis of GC by acting as a protein decoy for IGF2BP3 to regulate the expression of MYC and SNAIL. Mol. Ther. Nucleic Acids. Dec. 2021;26:649–664. doi: 10.1016/J.OMTN.2021.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang X., et al. Circ-SIRT1 promotes colorectal cancer proliferation and EMT by recruiting and binding to eIF4A3. Anal. Cell Pathol. 2021;2021 doi: 10.1155/2021/5739769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Momen-Heravi F., Bala S. Circular RNAs in head and neck cancer diagnosis and potential molecular targeting. Otorhinolaryngol. Neck Surg. 2018;3(3) doi: 10.15761/OHNS.1000177. [DOI] [Google Scholar]
- 57.Huang A., Zheng H., Wu Z., Chen M., Huang Y. Circular RNA-protein interactions: functions, mechanisms, and identification. Theranostics. 2020;10(8):3503. doi: 10.7150/THNO.42174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen C., Zhang M., Zhang Y. Circ_0000079 decoys the RNA-binding protein FXR1 to interrupt formation of the FXR1/PRCKI complex and decline their mediated cell invasion and drug resistance in NSCLC. Cell Transplant. Sep. 2020;29 doi: 10.1177/0963689720961070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jie M., et al. CircMRPS35 suppresses gastric cancer progression via recruiting KAT7 to govern histone modification. Mol. Cancer. Mar. 2020;19(1):1–16. doi: 10.1186/S12943-020-01160-2/FIGURES/8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Song C., et al. Circular RNA Cwc27 contributes to Alzheimer's disease pathogenesis by repressing Pur-α activity. Cell Death Differ. Sep. 2021;2021:1–14. doi: 10.1038/s41418-021-00865-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feng Z., et al. Functions and potential applications of circular RNAs in cancer stem cells. Front. Oncol. 2019;9(JUN):500. doi: 10.3389/FONC.2019.00500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li B., et al. circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity. Nat. Commun. Dec. 2021;12(1) doi: 10.1038/S41467-020-20527-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shen S., et al. circPDE4B prevents articular cartilage degeneration and promotes repair by acting as a scaffold for RIC8A and MID1. Ann. Rheum. Dis. Sep. 2021;80(9):1209–1219. doi: 10.1136/ANNRHEUMDIS-2021-219969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liang Y., et al. circDCUN1D4 suppresses tumor metastasis and glycolysis in lung adenocarcinoma by stabilizing TXNIP expression. Mol. Ther. Nucleic Acids. Mar. 2020;23:355–368. doi: 10.1016/J.OMTN.2020.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pamudurti N.R., et al. Translation of CircRNAs. Mol. Cell. Apr. 2017;66(1):9. doi: 10.1016/J.MOLCEL.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu P., et al. Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol. Cancer. Feb. 2020;19(1):1–14. doi: 10.1186/S12943-020-1147-3. 2020 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu C.-Y., Kuo H.-C. The emerging roles and functions of circular RNAs and their generation. J. Biomed. Sci. Apr. 2019;26(1):1–12. doi: 10.1186/S12929-019-0523-Z. 2019 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li Z., et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. Feb. 2015;22(3):256–264. doi: 10.1038/nsmb.2959. 2015 223. [DOI] [PubMed] [Google Scholar]
- 69.Lukiw W.J. Circular RNA (circRNA) in Alzheimer's disease (AD) Front. Genet. 2013;4(DEC) doi: 10.3389/FGENE.2013.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang G., Zhu H., Shi Y., Wu W., Cai H., Chen X. cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/β-catenin pathway. PLoS One. Jun. 2015;10(6) doi: 10.1371/JOURNAL.PONE.0131225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang J., et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol. Cancer. Sep. 2017;16(1) doi: 10.1186/S12943-017-0719-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhong Z., Lv M., Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci. Rep. 2016;6(Aug) doi: 10.1038/SREP30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu Q., et al. Circular RNA related to the chondrocyte ECM regulates MMP13 expression by functioning as a MiR-136 ‘sponge’ in human cartilage degradation. Sci. Rep. Mar. 2016;6 doi: 10.1038/SREP22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen J., et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. Mar. 2017;388:208–219. doi: 10.1016/J.CANLET.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 75.Wang C., Ren M., Zhao X., Wang A., Wang J. Emerging roles of circular RNAs in osteosarcoma. Med. Sci. Mon. Int. Med. J. Exp. Clin. Res. Oct. 2018;24:7043–7050. doi: 10.12659/MSM.912092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Peng L., et al. Circular RNA ZNF609 functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-150-5p in Hirschsprung's disease. Oncotarget. 2017;8(1):808–818. doi: 10.18632/ONCOTARGET.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cheng X., et al. Circular RNA VMA21 protects against intervertebral disc degeneration through targeting miR-200c and X linked inhibitor-of-apoptosis protein. Ann. Rheum. Dis. May 2018;77(5):770–779. doi: 10.1136/ANNRHEUMDIS-2017-212056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Han D., et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. Oct. 2017;66(4):1151–1164. doi: 10.1002/HEP.29270. [DOI] [PubMed] [Google Scholar]
- 79.Fu L., et al. Hsa_circ_0005986 inhibits carcinogenesis by acting as a miR-129-5p sponge and is used as a novel biomarker for hepatocellular carcinoma. Oncotarget. 2017;8(27):43878–43888. doi: 10.18632/ONCOTARGET.16709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tian F., Wang Y., Xiao Z., Zhu X. [Circular RNA CircHIPK3 promotes NCI-H1299 and NCI-H2170 cell proliferation through miR-379 and its target IGF1] Zhongguo Fei Ai Za Zhi. 2017;20(7):459–467. doi: 10.3779/J.ISSN.1009-3419.2017.07.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen G., Shi Y., Liu M., Sun J. circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis. Feb. 2018;9(2) doi: 10.1038/S41419-017-0204-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sun Y., et al. A novel regulatory mechanism of smooth muscle α-actin expression by NRG-1/circACTA2/miR-548f-5p Axis. Circ. Res. Sep. 2017;121(6):628–635. doi: 10.1161/CIRCRESAHA.117.311441. [DOI] [PubMed] [Google Scholar]
- 83.Sung H., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. May 2021;71(3):209–249. doi: 10.3322/CAAC.21660. [DOI] [PubMed] [Google Scholar]
- 84.Abdel-Latif M., Youness R.A. Why natural killer cells in triple negative breast cancer? World J. Clin. Oncol. Jul. 2020;11(7):464–476. doi: 10.5306/WJCO.V11.I7.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Youness R.A., Gad M.Z. Long non-coding RNAs: functional regulatory players in breast cancer. Non-coding RNA Res. Mar. 2019;4(1):36–44. doi: 10.1016/J.NCRNA.2019.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fragomeni S.M., Sciallis A., Jeruss J.S. Molecular subtypes and local-regional control of breast cancer. Surg. Oncol. Clin. Jan. 2018;27(1):95–120. doi: 10.1016/J.SOC.2017.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nielsen T.O., et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin. Cancer Res. 2004;10(16):5367–5374, Aug. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 88.Perou C.M., et al. Molecular portraits of human breast tumours. Nature. Aug. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 89.Voduc K.D., Cheang M.C.U., Tyldesley S., Gelmon K., Nielsen T.O., Kennecke H. Breast cancer subtypes and the risk of local and regional relapse. J. Clin. Oncol. 2010;28(10):1684–1691, Apr. doi: 10.1200/JCO.2009.24.9284. [DOI] [PubMed] [Google Scholar]
- 90.Sachdev J.C., Sandoval A.C., Jahanzeb M. Update on precision medicine in breast cancer. Cancer Treat Res. 2019;178:45–80. doi: 10.1007/978-3-030-16391-4_2. [DOI] [PubMed] [Google Scholar]
- 91.Tsuruo T., et al. Molecular targeting therapy of cancer: drug resistance, apoptosis and survival signal. Cancer Sci. Jan. 2003;94(1):15–21. doi: 10.1111/J.1349-7006.2003.TB01345.X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Spranger S., Gajewski T.F. Impact of oncogenic pathways on evasion of antitumour immune responses. Nat. Rev. Cancer. Jan. 2018;18(3):139–147. doi: 10.1038/nrc.2017.117. 2018 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hu X., et al. Genetic alterations and oncogenic pathways associated with breast cancer subtypes. Mol. Cancer Res. Apr. 2009;7(4):511–522. doi: 10.1158/1541-7786.MCR-08-0107. [DOI] [PubMed] [Google Scholar]
- 94.Aleskandarany M.A., Vandenberghe M.E., Marchiò C., Ellis I.O., Sapino A., Rakha E.A. Tumour heterogeneity of breast cancer: from morphology to personalised medicine. Pathobiology. May 2018;85(1–2):23–34. doi: 10.1159/000477851. [DOI] [PubMed] [Google Scholar]
- 95.Turashvili G., Brogi E. Tumor heterogeneity in breast cancer. Front. Med. 2017;4(DEC):227. doi: 10.3389/FMED.2017.00227/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fisher R., Pusztai L., Swanton C. Cancer heterogeneity: implications for targeted therapeutics. Br. J. Cancer. Feb. 2013;108(3):479. doi: 10.1038/BJC.2012.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hinohara K., Polyak K. Intratumoral heterogeneity: more than just mutations. Trends Cell Biol. Jul. 2019;29(7):569. doi: 10.1016/J.TCB.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Januškevičienė I., Petrikaitė V. Heterogeneity of breast cancer: the importance of interaction between different tumor cell populations. Life Sci. Dec. 2019;239:117009. doi: 10.1016/J.LFS.2019.117009. [DOI] [PubMed] [Google Scholar]
- 99.Visvader J.E. Cells of origin in cancer. Nat. Jan. 2011;469(7330):314–322. doi: 10.1038/nature09781. 2011 4697330. [DOI] [PubMed] [Google Scholar]
- 100.Sejben A., Nyári T., Zombori T., Cserni G. Comparison of Nottingham prognostic index, PREDICT and PrognosTILs in triple negative breast cancer –a retrospective cohort study. Pathol. Oncol. Res. Oct. 2020;26(4):2443–2450. doi: 10.1007/S12253-020-00846-8/FIGURES/3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yu D., Hung M.C. Overexpression of ErbB2 in cancer and ErbB2-targeting strategies. Oncogene. Dec. 2000;19(53):6115–6121. doi: 10.1038/sj.onc.1203972. 2000 1953. [DOI] [PubMed] [Google Scholar]
- 102.Waks A.G., Winer E.P. Breast cancer treatment. JAMA. Jan. 2019;321(3) doi: 10.1001/JAMA.2018.20751. 316–316. [DOI] [PubMed] [Google Scholar]
- 103.Cardoso F., et al. Early breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. Aug. 2019;30(8):1194–1220. doi: 10.1093/ANNONC/MDZ173. [DOI] [PubMed] [Google Scholar]
- 104.Wang J., Xu B. Targeted therapeutic options and future perspectives for HER2-positive breast cancer. Signal Transduct. Targeted Ther. Sep. 2019;4(1):1–22. doi: 10.1038/s41392-019-0069-2. 2019 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kvokačková B., Remšík J., Jolly M.K., Souček K. Phenotypic heterogeneity of triple-negative breast cancer mediated by epithelial–mesenchymal plasticity. Cancers. May 2021;13(9) doi: 10.3390/CANCERS13092188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xing L., et al. The circRNA circIFI30 promotes progression of triple-negative breast cancer and correlates with prognosis. Aging (Albany NY) Jun. 2020;12(11):10983. doi: 10.18632/AGING.103311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kwa M., Makris A., Esteva F.J. Clinical utility of gene-expression signatures in early stage breast cancer. Nat. Rev. Clin. Oncol. May 2017;14(10):595–610. doi: 10.1038/nrclinonc.2017.74. 2017 1410. [DOI] [PubMed] [Google Scholar]
- 108.Waks A.G., Winer E.P. Breast cancer treatment: a review. JAMA. Jan. 2019;321(3):288–300. doi: 10.1001/JAMA.2018.19323. [DOI] [PubMed] [Google Scholar]
- 109.Yin L., Duan J.J., Bian X.W., Yu S.C. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. Jun. 2020;22(1):1–13. doi: 10.1186/S13058-020-01296-5/TABLES/3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Daly A.A., Rolph R., Cutress R.I., Copson E.R. A review of modifiable risk factors in young women for the prevention of breast cancer. Breast Cancer. 2021;13:241. doi: 10.2147/BCTT.S268401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Giordano S.H. vol. 378. Jun. 2018. pp. 2311–2320. (Breast Cancer in Men). 24. [DOI] [PubMed] [Google Scholar]
- 112.Copson E.R., et al. Germline BRCA mutation and outcome in young-onset breast cancer (POSH): a prospective cohort study. Lancet Oncol. Feb. 2018;19(2):169–180. doi: 10.1016/S1470-2045(17)30891-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Byler S., et al. Genetic and epigenetic aspects of breast cancer progression and therapy. Anticancer Res. 2014;34(3):1071–1077. [PubMed] [Google Scholar]
- 114.Buocikova V., et al. Epigenetics in breast cancer therapy—new strategies and future nanomedicine perspectives. Cancers. Dec. 2020;12(12):3622. doi: 10.3390/CANCERS12123622. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Guo M., Peng Y., Gao A., Du C., Herman J.G. Epigenetic heterogeneity in cancer. Biomark. Res. Oct. 2019;7(1):1–19. doi: 10.1186/S40364-019-0174-Y. 2019 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yerukala Sathipati S., Ho S.Y. Identifying a miRNA signature for predicting the stage of breast cancer. Sci. Rep. Oct. 2018;8(1):1–11. doi: 10.1038/s41598-018-34604-3. 2018 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liang Z.Z., Guo C., Zou M.M., Meng P., Zhang T.T. circRNA-miRNA-mRNA regulatory network in human lung cancer: an update. Cancer Cell Int. May 2020;20(1):1–16. doi: 10.1186/S12935-020-01245-4. 2020 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xu Y., et al. Increased expression of circular RNA circ_0005230 indicates dismal prognosis in breast cancer and regulates cell proliferation and invasion via miR-618/CBX8 signal pathway. Cell. Physiol. Biochem. Dec. 2018;51(4):1710–1722. doi: 10.1159/000495675. [DOI] [PubMed] [Google Scholar]
- 119.Telli M.L., Gradishar W.J., Ward J.H. NCCN guidelines updates: breast cancer. J. Natl. Compr. Cancer Netw. May 2019;17(5):552–555. doi: 10.6004/JNCCN.2019.5006. [DOI] [PubMed] [Google Scholar]
- 120.Baliu-Piqué M., Pandiella A., Ocana A. Breast cancer heterogeneity and response to novel therapeutics. Cancers. Nov. 2020;12(11):3271. doi: 10.3390/CANCERS12113271. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tucker D., Zheng W., Zhang D.-H., Dong X. Circular RNA and its potential as prostate cancer biomarkers. World J. Clin. Oncol. Aug. 2020;11(8):563. doi: 10.5306/WJCO.V11.I8.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Huang J., et al. The dual role of circular RNAs as miRNA sponges in breast cancer and colon cancer. Biomedica. Oct. 2021;9(11):1590. doi: 10.3390/BIOMEDICINES9111590. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xu J., et al. The regulation network and clinical significance of circular RNAs in breast cancer. Front. Oncol. Jul. 2021;11:2660. doi: 10.3389/FONC.2021.691317/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lyu L., et al. Regulatory mechanisms, functions, and clinical significance of CircRNAs in triple-negative breast cancer. J. Hematol. Oncol. Dec. 2021;14(1):1–18. doi: 10.1186/S13045-021-01052-Y/TABLES/2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zeng Y., et al. The biogenesis, function and clinical significance of circular RNAs in breast cancer. Cancer Biol. Med. 2021;18 doi: 10.20892/J.ISSN.2095-3941.2020.0485. 0–0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ghafouri-Fard S., Hussen B.M., Taheri M., Ayatollahi S.A. Emerging role of circular RNAs in breast cancer. Pathol. Res. Pract. Jul. 2021;223 doi: 10.1016/J.PRP.2021.153496. [DOI] [PubMed] [Google Scholar]
- 127.Gong L., Zhou X., Sun J. Circular RNAs interaction with MiRNAs: emerging roles in breast cancer. Int. J. Med. Sci. 2021;18(14):3182. doi: 10.7150/IJMS.62219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Li Z., Chen Z., Hu G.H., Jiang Y. Roles of circular RNA in breast cancer: present and future. Am. J. Transl. Res. 2019;11(7):3945. [Online]. Available:/pmc/articles/PMC6684920/ [PMC free article] [PubMed] [Google Scholar]
- 129.Yu C.-Y., Kuo H.-C. The emerging roles and functions of circular RNAs and their generation. J. Biomed. Sci. Apr. 2019;26(1):1–12. doi: 10.1186/S12929-019-0523-Z. 2019 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Enuka Y., Lauriola M., Feldman M.E., Sas-Chen A., Ulitsky I., Yarden Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. Feb. 2016;44(3):1370–1383. doi: 10.1093/NAR/GKV1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhang Z., Yang T., Xiao J. Circular RNAs: promising biomarkers for human diseases. EBioMedicine. Aug. 2018;34:267–274. doi: 10.1016/J.EBIOM.2018.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mo D., et al. A universal approach to investigate circRNA protein coding function. Sci. Rep. Dec. 2019;9(1) doi: 10.1038/S41598-019-48224-Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Darbeheshti F., et al. Circular RNA hsa_circ_0044234 as distinct molecular signature of triple negative breast cancer: a potential regulator of GATA3. Cancer Cell Int. Dec. 2021;21(1):1–12. doi: 10.1186/S12935-021-02015-6/FIGURES/6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Guan B., Li Q., Zhang H.Z., Yang H.S. circ_NOTCH3 functions as a protooncogene competing with miR-205-5p, modulating KLF12 expression and promoting the development and progression of basal-like breast carcinoma. Front. Oncol. Jan. 2021;10:3086. doi: 10.3389/FONC.2020.602694/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Deng Y., Xia J., Xu Y.E. Circular RNA circTP63 enhances estrogen receptor-positive breast cancer progression and malignant behaviors through the miR-873-3p/FOXM1 axis. Anti Cancer Drugs. 2021;32(1):44–52. doi: 10.1097/CAD.0000000000001010. [DOI] [PubMed] [Google Scholar]
- 136.Luo N., Liu S., Li X., Hu Y., Zhang K. vol. 20. 2021. pp. 1320–1333. (Circular RNA circHIPK3 Promotes Breast Cancer Progression via Sponging MiR-326). 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Qi L., Sun B., Yang B., Lu S. circHIPK3 (hsa_circ_0000284) promotes proliferation, migration and invasion of breast cancer cells via miR-326. OncoTargets Ther. 2021;14:3671–3685. doi: 10.2147/OTT.S299190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Huang Y., Zheng S., Lin Y., Ke L. Circular RNA circ-ERBB2 elevates the Warburg effect and facilitates triple-negative breast cancer growth by the MicroRNA 136-5p/pyruvate dehydrogenase kinase 4 Axis. Mol. Cell Biol. Sep. 2021;41(10) doi: 10.1128/MCB.00609-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhong J., et al. Circular RNA circ-ERBB2 promotes HER2-positive breast cancer progression and metastasis via sponging miR-136-5p and miR-198. J. Transl. Med. Nov. 2021;19(1):1–14. doi: 10.1186/S12967-021-03114-8. 2021 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Xing Z., et al. CircRNA circ-PDCD11 promotes triple-negative breast cancer progression via enhancing aerobic glycolysis. Cell Death Dis. Aug. 2021;7(1):1–8. doi: 10.1038/s41420-021-00604-y. 2021 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Xu Y., et al. Circular RNA circIKBKB promotes breast cancer bone metastasis through sustaining NF-κB/bone remodeling factors signaling. Mol. Cancer. Dec. 2021;20(1):1–18. doi: 10.1186/S12943-021-01394-8/FIGURES/7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhang L., et al. Circ_0008673 regulates breast cancer malignancy by miR-153-3p/CFL2 axis. Arch. Gynecol. Obstet. Jul. 2021:1–10. doi: 10.1007/S00404-021-06149-W/FIGURES/3. [DOI] [PubMed] [Google Scholar]
- 143.Liu Y.-P., Heng J.-Y., Zhao X.-Y., Li E.-Y. The inhibition of circular RNA circNOLC1 by propofol/STAT3 attenuates breast cancer stem cells function via miR-365a-3p/STAT3 signaling. J. Transl. Med. Nov. 2021;19(1):1–16. doi: 10.1186/S12967-021-03133-5. 2021 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhang Z., Hu H., Li Q., Yi F., Liu Y. A novel circular RNA circPTCD3 promotes breast cancer progression through sponging miR-198. Cancer Manag. Res. Nov. 2021;13:8435. doi: 10.2147/CMAR.S256091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Xu L., Xu K., Xiang L., Yan J. Circular RNA OMA1 regulates the progression of breast cancer via modulation of the miR-1276/SIRT4 axis. Mol. Med. Rep. Oct. 2021;24(4):1–11. doi: 10.3892/MMR.2021.12367/HTML. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Fu B., et al. Circular RNA circBCBM1 promotes breast cancer brain metastasis by modulating miR-125a/BRD4 axis. Int. J. Biol. Sci. 2021;17(12):3104. doi: 10.7150/IJBS.58916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Huang Q., He Y., Zhang X., Guo L. Circular RNA hsa_circ_0103552 promotes proliferation, migration, and invasion of breast cancer cells through upregulating cysteine-rich angiogenic inducer 61 (CYR61) expression via sponging MicroRNA-515-5p. Tohoku J. Exp. Med. 2021;255(2):171–181. doi: 10.1620/TJEM.255.171. [DOI] [PubMed] [Google Scholar]
- 148.Gong G., She J., Fu D., Zhen D., Zhang B. Circular RNA circ_0084927 regulates proliferation, apoptosis, and invasion of breast cancer cells via miR-142-3p/ERC1 pathway. Am. J. Transl. Res. 2021;13(5):4120. [Online]. Available:/pmc/articles/PMC8205726/ [PMC free article] [PubMed] [Google Scholar]
- 149.Zhang H., Ge Z., Wang Z., Gao Y., Wang Y., Qu X. Circular RNA RHOT1 promotes progression and inhibits ferroptosis via mir-106a-5p/STAT3 axis in breast cancer. Aging (Albany NY) 2021;13(6):8115. doi: 10.18632/AGING.202608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Xu C., et al. Circular RNA circNINL promotes breast cancer progression through activating β-catenin signaling via miR-921/ADAM9 axis. J. Biochem. Sep. 2021;169(6):693–700. doi: 10.1093/JB/MVAB005. [DOI] [PubMed] [Google Scholar]
- 151.Su Q., Shen H., Gu B., Zhu N. vol. 12. Jan. 2021. pp. 9058–9069. (Circular RNA CNOT2 Knockdown Regulates Twist Family BHLH Transcription Factor via Targeting microRNA 409-3p to Prevent Breast Cancer Invasion, Migration and Epithelial–Mesenchymal Transition). 1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 152.Xie F., Xiong Y., Yan J., Wang L., Yan W. Circular RNA circ_0048764 promotes the development of breast cancer by regulating microRNA-1296-5p/tripartite motif containing 14 axis. Nov. 2021. [DOI] [PMC free article] [PubMed]
- 153.Zhou Y., Liu X., Lan J., Wan Y., Zhu X. Circular RNA circRPPH1 promotes triple-negative breast cancer progression via the miR-556-5p/YAP1 axis. Am. J. Transl. Res. 2020;12(10):6220. [Online]. Available:/pmc/articles/PMC7653573/ [PMC free article] [PubMed] [Google Scholar]
- 154.Jinsihan D., Li D., Zhang M., Feng J., Zhao Q. Hypoxia-associated circular RNA RPPH1 modulates triple-negative breast cancer cell growth via the miR-1296-5p/TRIM14 axis. Biocell. Mar. 2021;45(3):671–684. doi: 10.32604/BIOCELL.2021.012519. [DOI] [Google Scholar]
- 155.Cai F., et al. Hsa_circ_0000515 is a novel circular RNA implicated in the development of breast cancer through its regulation of the microRNA-296-5p/CXCL10 axis. FEBS J. Feb. 2021;288(3):861–883. doi: 10.1111/FEBS.15373. [DOI] [PubMed] [Google Scholar]
- 156.Du W.W., et al. A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene. Jul. 2018;37(44):5829–5842. doi: 10.1038/s41388-018-0369-y. 2018 3744. [DOI] [PubMed] [Google Scholar]
- 157.He D., Yang X., Kuang W., Huang G., Liu X., Zhang Y. The novel circular RNA circ-PGAP3 promotes the proliferation and invasion of triple negative breast cancer by regulating the miR-330-3p/myc Axis. OncoTargets Ther. 2020;13 doi: 10.2147/OTT.S274574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wang X., Xue B., Zhang Y., Guo G., Duan X., Dou D. Up-regulated circBACH2 contributes to cell proliferation, invasion, and migration of triple-negative breast cancer. Cell Death Dis. Apr. 2021;12(5):1–14. doi: 10.1038/s41419-021-03684-x. 2021 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Liang H.F., Zhang X.Z., Liu B.G., Jia G.T., Li W.L. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am. J. Cancer Res. 2017;7(7):1566. [Online]. Available:/pmc/articles/PMC5523036/ [PMC free article] [PubMed] [Google Scholar]
- 160.Yang W., Gong P., Yang Y., Yang C., Yang B., Ren L. Circ-ABCB10 contributes to paclitaxel resistance in breast cancer through let-7a-5p/DUSP7 Axis. Cancer Manag. Res. 2020;12:2327. doi: 10.2147/CMAR.S238513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zhou J., Zhang W.W., Peng F., Sun J.Y., He Z.Y., Wu S.G. Downregulation of hsa_circ_0011946 suppresses the migration and invasion of the breast cancer cell line MCF-7 by targeting RFC3. Cancer Manag. Res. Mar. 2018;10:535. doi: 10.2147/CMAR.S155923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.He R., et al. CircGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J. Exp. Clin. Cancer Res. Oct. 2017;36(1):1–12. doi: 10.1186/S13046-017-0614-1/FIGURES/5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Qiu Z., Wang L., Liu H. Hsa_circ_0001982 promotes the progression of breast cancer through miR-1287-5p/MUC19 axis under hypoxia. World J. Surg. Oncol. Jun. 2021;19(1):1–15. doi: 10.1186/S12957-021-02273-8. 2021 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Tang Y.Y., et al. vol. 36. Nov. 2017. pp. 901–908.https://home.liebertpub.com/dna (Circular RNA Hsa_circ_0001982 Promotes Breast Cancer Cell Carcinogenesis through Decreasing miR-143). 11. [DOI] [PubMed] [Google Scholar]
- 165.Wu J., et al. CircIRAK3 sponges miR-3607 to facilitate breast cancer metastasis. Cancer Lett. Aug. 2018;430:179–192. doi: 10.1016/J.CANLET.2018.05.033. [DOI] [PubMed] [Google Scholar]
- 166.Liu Y., Lu C., Zhou Y., Zhang Z., Sun L. Circular RNA hsa_circ_0008039 promotes breast cancer cell proliferation and migration by regulating miR-432-5p/E2F3 axis. Biochem. Biophys. Res. Commun. Jul. 2018;502(3):358–363. doi: 10.1016/J.BBRC.2018.05.166. [DOI] [PubMed] [Google Scholar]
- 167.Zhang H.Y., Zhang B.W., Zhang Z.B., Deng Q.J. Circular RNA TTBK2 regulates cell proliferation, invasion and ferroptosis via miR-761/ITGB8 axis in glioma. Eur. Rev. Med. Pharmacol. Sci. 2020;24(5):2585–2600. doi: 10.26355/EURREV_202003_20528. [DOI] [PubMed] [Google Scholar]
- 168.Wang R., et al. CircNT5E acts as a sponge of miR-422a to promote glioblastoma tumorigenesis. Cancer Res. Sep. 2018;78(17):4812–4825. doi: 10.1158/0008-5472.CAN-18-0532/653245/AM/CIRCNT5E-ACTS-AS-A-SPONGE-OF-MICRORNA-422A-TO. [DOI] [PubMed] [Google Scholar]
- 169.Tan A., Li Q., Chen L. CircZFR promotes hepatocellular carcinoma progression through regulating miR-3619–5p/CTNNB1 axis and activating Wnt/β-catenin pathway. Arch. Biochem. Biophys. Jan. 2019;661:196–202. doi: 10.1016/J.ABB.2018.11.020. [DOI] [PubMed] [Google Scholar]
- 170.Lai Z., Wei T., Li Q., Wang X., Zhang Y., Zhang S. Exosomal circFBLIM1 promotes hepatocellular carcinoma progression and glycolysis by regulating the miR-338/LRP6 Axis. Sep. 2020. https://home.liebertpub.com/cbr [DOI] [PubMed]
- 171.Ma H.B., Yao Y.N., Yu J.J., Chen X.X., Li H.F. Extensive profiling of circular RNAs and the potential regulatory role of circRNA-000284 in cell proliferation and invasion of cervical cancer via sponging miR-506. Am. J. Transl. Res. 2018;10(2):592. [Online]. Available:/pmc/articles/PMC5835825/ [PMC free article] [PubMed] [Google Scholar]
- 172.Guo J., Chen M., Ai G., Mao W., Li H., Zhou J. Hsa_circ_0023404 enhances cervical cancer metastasis and chemoresistance through VEGFA and autophagy signaling by sponging miR-5047. Biomed. Pharmacother. Jul. 2019;115 doi: 10.1016/J.BIOPHA.2019.108957. [DOI] [PubMed] [Google Scholar]
- 173.Zong L., et al. Increased expression of circRNA_102231 in lung cancer and its clinical significance. Biomed. Pharmacother. Jun. 2018;102:639–644. doi: 10.1016/J.BIOPHA.2018.03.084. [DOI] [PubMed] [Google Scholar]
- 174.Qiu M., et al. The circular RNA circPRKCI promotes tumor growth in lung adenocarcinoma. Cancer Res. Jun. 2018;78(11):2839–2851. doi: 10.1158/0008-5472.CAN-17-2808/653114/AM/THE-CIRCULAR-RNA-CIRCPRKCI-PROMOTES-TUMOR-GROWTH. [DOI] [PubMed] [Google Scholar]
- 175.Ma X., et al. Circular RNA circMAN2B2 facilitates lung cancer cell proliferation and invasion via miR-1275/FOXK1 axis. Biochem. Biophys. Res. Commun. Apr. 2018;498(4):1009–1015. doi: 10.1016/J.BBRC.2018.03.105. [DOI] [PubMed] [Google Scholar]
- 176.Zhu X., et al. hsa_circ_0013958: a circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J. Jul. 2017;284(14):2170–2182. doi: 10.1111/FEBS.14132. [DOI] [PubMed] [Google Scholar]
- 177.Zhang H., Wang X., Hu B., Zhang F., Wei H., Li L. Circular RNA ZFR accelerates non-small cell lung cancer progression by acting as a miR-101-3p sponge to enhance CUL4B expression. Artif. Cell Nanomed. Biotechnol. 2019;47(1):3410–3416. doi: 10.1080/21691401.2019.1652623/SUPPL_FILE/IANB_A_1652623_SM4992.DOCX. [DOI] [PubMed] [Google Scholar]
- 178.Ye Y., Zhao L., Li Q., Xi C., Li Y., Li Z. circ_0007385 served as competing endogenous RNA for miR-519d-3p to suppress malignant behaviors and cisplatin resistance of non-small cell lung cancer cells. Thorac. Cancer. Aug. 2020;11(8):2196–2208. doi: 10.1111/1759-7714.13527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ning Z., et al. Exosomal circ_0007385 enhances non-small cell lung cancer cell proliferation and stemness via regulating miR-1253/FAM83A axis. Anti Cancer Drugs. Jan. 2022;33(1):61–74. doi: 10.1097/CAD.0000000000001103. [DOI] [PubMed] [Google Scholar]
- 180.Ding D., Yang F., Chen Z., Ying J. Circ_0007385 regulates cell proliferation, apoptosis and stemness via targeting miR-493-3p/RAB22A axis in non-small cell lung cancer. Thorac. Cancer. Feb. 2022;13(4):571–581. doi: 10.1111/1759-7714.14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Chen L., et al. Circular RNA 100146 functions as an oncogene through direct binding to miR-361-3p and miR-615-5p in non-small cell lung cancer. Mol. Cancer. Jan. 2019;18(1):1–8. doi: 10.1186/S12943-019-0943-0. 2019 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Yang M., Zheng E., Ni J., Xu X., Jiang X., Zhao G. Circular RNA circFOXO3 facilitate non-small cell lung cancer progression through upregulating HMGB3 via sponging miR-545-3p/miR-506-3p. Tissue Cell. Apr. 2022;75 doi: 10.1016/J.TICE.2021.101702. [DOI] [PubMed] [Google Scholar]
- 183.Chen J., Yang J., Fei X., Wang X., Wang K. CircRNA ciRS-7: a novel oncogene in multiple cancers. Int. J. Biol. Sci. 2021;17(1):379. doi: 10.7150/IJBS.54292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Li Y., Zhang J., Pan S., Zhou J., Diao X., Liu S. CircRNA CDR1as knockdown inhibits progression of non-small-cell lung cancer by regulating miR-219a-5p/SOX5 axis. Thorac. Cancer. Mar. 2020;11(3):537–548. doi: 10.1111/1759-7714.13274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Mo Y., et al. Circular RNA CCDC66 improves murine double minute 4 (MDM4) expression through targeting miR-370 in colorectal cancer. Comput. Math. Methods Med. 2022;2022 doi: 10.1155/2022/7723995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Yan Y., Su M., Qin B. CircHIPK3 promotes colorectal cancer cells proliferation and metastasis via modulating of miR-1207-5p/FMNL2 signal. Biochem. Biophys. Res. Commun. Apr. 2020;524(4):839–846. doi: 10.1016/J.BBRC.2020.01.055. [DOI] [PubMed] [Google Scholar]
- 187.Guo J.N., et al. Comprehensive profile of differentially expressed circular RNAs reveals that hsa_circ_0000069 is upregulated and promotes cell proliferation, migration, and invasion in colorectal cancer. OncoTargets Ther. Dec. 2016;9:7451–7458. doi: 10.2147/OTT.S123220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Xie H., et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. May 2016;7(18):26680–26691. doi: 10.18632/ONCOTARGET.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Huang H., Wei L., Qin T., Yang N., Li Z., Xu Z. Circular RNA ciRS-7 triggers the migration and invasion of esophageal squamous cell carcinoma via miR-7/KLF4 and NF-κB signals. Cancer Biol. Ther. Jan. 2019;20(1):73–80. doi: 10.1080/15384047.2018.1507254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Han Z., et al. Erβ-mediated alteration of circATP2B1 and miR-204-3p signaling promotes invasion of clear cell renal cell carcinoma. Cancer Res. May 2018;78(10):2550–2563. doi: 10.1158/0008-5472.CAN-17-1575. [DOI] [PubMed] [Google Scholar]
- 191.Lai Z., et al. Analysis of co-expression networks for circular RNAs and mRNAs reveals that circular RNAs hsa_circ_0047905, hsa_circ_0138960 and has-circRNA7690-15 are candidate oncogenes in gastric cancer. Cell Cycle. Dec. 2017;16(23):2301–2311. doi: 10.1080/15384101.2017.1380135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wang L., et al. CircDOCK1 suppresses cell apoptosis via inhibition of miR-196a-5p by targeting BIRC3 in OSCC. Oncol. Rep. Mar. 2018;39(3):951–966. doi: 10.3892/OR.2017.6174/DOWNLOAD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Wei H., Pan L., Tao D., Li R. Circular RNA circZFR contributes to papillary thyroid cancer cell proliferation and invasion by sponging miR-1261 and facilitating C8orf4 expression. Biochem. Biophys. Res. Commun. Sep. 2018;503(1):56–61. doi: 10.1016/J.BBRC.2018.05.174. [DOI] [PubMed] [Google Scholar]
- 194.Chen Z., et al. Knockdown of circ_0084043 suppresses the development of human melanoma cells through miR-429/tribbles homolog 2 axis and Wnt/β-catenin pathway. Life Sci. Feb. 2020;243 doi: 10.1016/J.LFS.2020.117323. [DOI] [PubMed] [Google Scholar]
- 195.Dai Y., et al. Circular RNA myosin light chain kinase (MYLK) promotes prostate cancer progression through modulating mir-29a expression. Med. Sci. Mon. Int. Med. J. Exp. Clin. Res. May 2018;24:3462. doi: 10.12659/MSM.908009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Kai D., Yannian L., Yitian C., Dinghao G., Xin Z., Wu J. Circular RNA HIPK3 promotes gallbladder cancer cell growth by sponging microRNA-124. Biochem. Biophys. Res. Commun. Sep. 2018;503(2):863–869. doi: 10.1016/J.BBRC.2018.06.088. [DOI] [PubMed] [Google Scholar]
- 197.Verduci L., et al. The oncogenic role of circPVT1 in head and neck squamous cell carcinoma is mediated through the mutant p53/YAP/TEAD transcription-competent complex. Genome Biol. Dec. 2017;18(1) doi: 10.1186/S13059-017-1368-Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Shi Y., Han T., Liu C. CircRNA hsa_circ_0006220 acts as a tumor suppressor gene by regulating miR-197-5p/CDH19 in triple-negative breast cancer. Ann. Transl. Med. 2021;9(15) doi: 10.21037/ATM-21-2934. 1236-1236, Aug. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Meng L., et al. Circular RNA circCCDC85A inhibits breast cancer progression via acting as a miR-550a-5p sponge to enhance MOB1A expression. Breast Cancer Res. Dec. 2022;24(1):1–13. doi: 10.1186/S13058-021-01497-6/FIGURES/7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Fan Y., et al. CircNR3C2 promotes HRD1-mediated tumor-suppressive effect via sponging miR-513a-3p in triple-negative breast cancer. Mol. Cancer. Dec. 2021;20(1):1–22. doi: 10.1186/S12943-021-01321-X/FIGURES/8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Zhang X.Y., Mao L. Circular RNA Circ_0000442 acts as a sponge of MiR-148b-3p to suppress breast cancer via PTEN/PI3K/Akt signaling pathway. Gene. Jan. 2021;766:145113. doi: 10.1016/J.GENE.2020.145113. [DOI] [PubMed] [Google Scholar]
- 202.Xu J.-Z., et al. circTADA2As suppress breast cancer progression and metastasis via targeting miR-203a-3p/SOCS3 axis. Cell Death Dis. Feb. 2019;10(3):1–16. doi: 10.1038/s41419-019-1382-y. 2019 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Wang H., Xiao Y., Wu L., Ma D. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-000911/miR-449a pathway in breast carcinogenesis. Int. J. Oncol. Mar. 2018;52(3):743–754. doi: 10.3892/IJO.2018.4265/HTML. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Yan L., Zheng M., Wang H. Circular RNA hsa_circ_0072309 inhibits proliferation and invasion of breast cancer cells via targeting miR-492. Cancer Manag. Res. 2019;11:1033–1041. doi: 10.2147/CMAR.S186857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Hou J.C., et al. Circular RNA circASS1 is downregulated in breast cancer cells MDA-MB-231 and suppressed invasion and migration. Epigenomics. Feb. 2019;11(2):199–213. doi: 10.2217/EPI-2017-0167. [DOI] [PubMed] [Google Scholar]
- 206.Yang Y., et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J. Natl. Cancer Inst. 2018;110(3) doi: 10.1093/JNCI/DJX166. Mar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Zhang M., et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. Mar. 2018;37(13):1805–1814. doi: 10.1038/S41388-017-0019-9. [DOI] [PubMed] [Google Scholar]
- 208.Begum S., Yiu A., Stebbing J., Castellano L. Novel tumour suppressive protein encoded by circular RNA, circ-SHPRH, in glioblastomas. Oncogene. Jul. 2018;37(30):4055–4057. doi: 10.1038/S41388-018-0230-3. [DOI] [PubMed] [Google Scholar]
- 209.Barbagallo D., et al. CircSMARCA5 inhibits migration of glioblastoma multiforme cells by regulating a molecular Axis involving splicing factors SRSF1/SRSF3/PTB. Int. J. Mol. Sci. Feb. 2018;19(2) doi: 10.3390/IJMS19020480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Li X., Diao H. Circular RNA circ_0001946 acts as a competing endogenous RNA to inhibit glioblastoma progression by modulating miR-671-5p and CDR1. J. Cell. Physiol. Aug. 2019;234(8):13807–13819,. doi: 10.1002/JCP.28061. [DOI] [PubMed] [Google Scholar]
- 211.Zhong L., et al. Circular RNA circC3P1 suppresses hepatocellular carcinoma growth and metastasis through miR-4641/PCK1 pathway. Biochem. Biophys. Res. Commun. May 2018;499(4):1044–1049. doi: 10.1016/J.BBRC.2018.03.221. [DOI] [PubMed] [Google Scholar]
- 212.Li Z., et al. Using circular RNA SMARCA5 as a potential novel biomarker for hepatocellular carcinoma. Clin. Chim. Acta. May 2019;492:37–44. doi: 10.1016/J.CCA.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 213.Li J., Bao S., Wang L., Wang R. CircZKSCAN1 suppresses hepatocellular carcinoma tumorigenesis by regulating miR-873-5p/downregulation of deleted in liver cancer 1. Dig. Dis. Sci. Dec. 2021;66(12):4374–4383. doi: 10.1007/S10620-020-06789-Z. [DOI] [PubMed] [Google Scholar]
- 214.Qiu L., et al. Circular RNA profiling identifies circADAMTS13 as a miR-484 sponge which suppresses cell proliferation in hepatocellular carcinoma. Mol. Oncol. Feb. 2019;13(2):441–455. doi: 10.1002/1878-0261.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Song C., et al. The competing endogenous circular RNA ADAMTS14 suppressed hepatocellular carcinoma progression through regulating microRNA-572/regulator of calcineurin 1. J. Cell. Physiol. Mar. 2019;234(3):2460–2470. doi: 10.1002/JCP.26764. [DOI] [PubMed] [Google Scholar]
- 216.Zheng H., et al. A circular RNA hsa_circ_0079929 inhibits tumor growth in hepatocellular carcinoma. Cancer Manag. Res. 2019;11:443–454. doi: 10.2147/CMAR.S189338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Zhang X., Luo P., Jing W., Zhou H., Liang C., Tu J. circSMAD2 inhibits the epithelial-mesenchymal transition by targeting miR-629 in hepatocellular carcinoma. OncoTargets Ther. May 2018;11:2853–2863. doi: 10.2147/OTT.S158008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Liu H., et al. Circular RNA YAP1 inhibits the proliferation and invasion of gastric cancer cells by regulating the miR-367-5p/p27 Kip1 axis. Mol. Cancer. Oct. 2018;17(1) doi: 10.1186/S12943-018-0902-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Zhong S., et al. Circular RNA hsa_circ_0000993 inhibits metastasis of gastric cancer cells. Epigenomics. Oct. 2018;10(10):1301–1313. doi: 10.2217/EPI-2017-0173. [DOI] [PubMed] [Google Scholar]
- 220.Rong D., et al. CircPSMC3 suppresses the proliferation and metastasis of gastric cancer by acting as a competitive endogenous RNA through sponging miR-296-5p. Mol. Cancer. Feb. 2019;18(1) doi: 10.1186/S12943-019-0958-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 221.Fang J., et al. A novel circular RNA, circFAT1(e2), inhibits gastric cancer progression by targeting miR-548g in the cytoplasm and interacting with YBX1 in the nucleus. Cancer Lett. Feb. 2019;442:222–232. doi: 10.1016/J.CANLET.2018.10.040. [DOI] [PubMed] [Google Scholar]
- 222.Li P., et al. Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br. J. Cancer. Feb. 2017;116(5):626–633. doi: 10.1038/BJC.2016.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Liu T., et al. Circular RNA-ZFR inhibited cell proliferation and promoted apoptosis in gastric cancer by sponging miR-130a/miR-107 and modulating PTEN. Cancer Res. Treat. 2018;50(4):1396–1417, Oct. doi: 10.4143/CRT.2017.537. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 224.Chen D., Ma W., Ke Z., Xie F. CircRNA hsa_circ_100395 regulates miR-1228/TCF21 pathway to inhibit lung cancer progression. Cell Cycle. , Aug. 2018;17(16):2080–2090. doi: 10.1080/15384101.2018.1515553/SUPPL_FILE/KCCY_A_1515553_SM9475.DOCX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Dai X., et al. RNA-binding protein trinucleotide repeat-containing 6A regulates the formation of circular RNA circ0006916, with important functions in lung cancer cells. Carcinogenesis. Jul. 2018;39(8):981–992. doi: 10.1093/CARCIN/BGY061. [DOI] [PubMed] [Google Scholar]
- 226.Nan A., et al. Circular RNA circNOL10 inhibits lung cancer development by promoting SCLM1-mediated transcriptional regulation of the humanin polypeptide family. Adv. Sci. (Weinheim, Baden-Wurttemberg, Ger. Jan. 2018;6(2) doi: 10.1002/ADVS.201800654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Wang L., et al. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-β-induced epithelial-mesenchymal transition and metastasis by controlling TIF1γ in non-small cell lung cancer. Mol. Cancer. Sep. 2018;17(1) doi: 10.1186/S12943-018-0889-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Liu T., Song Z., Gai Y. Circular RNA circ_0001649 acts as a prognostic biomarker and inhibits NSCLC progression via sponging miR-331-3p and miR-338-5p. Biochem. Biophys. Res. Commun. Sep. 2018;503(3):1503–1509. doi: 10.1016/J.BBRC.2018.07.070. [DOI] [PubMed] [Google Scholar]
- 229.Li Z., Guo X., Gao S. Circ-ITCH correlates with less advanced tumor features as well as prolonged survival, and it inhibits cells proliferation but promotes apoptosis in non-small cell lung cancer. Transl. Cancer Res. Sep. 2019;8(5):1672–1679. doi: 10.21037/TCR.2019.08.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Tian F., Yu C.T., Ye W.D., Wang Q. Cinnamaldehyde induces cell apoptosis mediated by a novel circular RNA hsa_circ_0043256 in non-small cell lung cancer. Biochem. Biophys. Res. Commun. Nov. 2017;493(3):1260–1266. doi: 10.1016/J.BBRC.2017.09.136. [DOI] [PubMed] [Google Scholar]
- 231.Yang G., et al. Circ-ITGA7 sponges miR-3187-3p to upregulate ASXL1, suppressing colorectal cancer proliferation. Cancer Manag. Res. 2019;11:6499–6509. doi: 10.2147/CMAR.S203137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Wang F., Wang J., Cao X., Xu L., Chen L. Hsa_circ_0014717 is downregulated in colorectal cancer and inhibits tumor growth by promoting p16 expression. Biomed. Pharmacother. Feb. 2018;98:775–782. doi: 10.1016/J.BIOPHA.2018.01.015. [DOI] [PubMed] [Google Scholar]
- 233.Liu H., et al. Invasion-related circular RNA circFNDC3B inhibits bladder cancer progression through the miR-1178-3p/G3BP2/SRC/FAK axis. Mol. Cancer. Nov. 2018;17(1) doi: 10.1186/S12943-018-0908-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Lin G., et al. circLPAR1 is a novel biomarker of prognosis for muscle-invasive bladder cancer with invasion and metastasis by miR-762. Oncol. Lett. Mar. 2019;17(3):3537–3547. doi: 10.3892/OL.2019.9970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Yuan W., et al. Circular RNA Cdr1as sensitizes bladder cancer to cisplatin by upregulating APAF1 expression through miR-1270 inhibition. Mol. Oncol. Jul. 2019;13(7):1559–1576. doi: 10.1002/1878-0261.12523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Li Y., et al. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. Sep. 2017;18(9):1646–1659. doi: 10.15252/EMBR.201643581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Yang C., et al. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol. Cancer. Jan. 2018;17(1) doi: 10.1186/S12943-018-0771-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Wang M., Chen B., Ru Z., Cong L. CircRNA circ-ITCH suppresses papillary thyroid cancer progression through miR-22-3p/CBL/β-catenin pathway. Biochem. Biophys. Res. Commun. Sep. 2018;504(1):283–288. doi: 10.1016/J.BBRC.2018.08.175. [DOI] [PubMed] [Google Scholar]
- 239.Hu J., et al. The circular RNA circ-ITCH suppresses ovarian carcinoma progression through targeting miR-145/RASA1 signaling. Biochem. Biophys. Res. Commun. Oct. 2018;505(1):222–228. doi: 10.1016/J.BBRC.2018.09.060. [DOI] [PubMed] [Google Scholar]
- 240.rong Zhang P., Ren J., shan Wan J., Sun R., Li Y. Circular RNA hsa_circ_0002052 promotes osteosarcoma via modulating miR-382/STX6 axis. Hum. Cell. Jul. 2020;33(3):810–818. doi: 10.1007/S13577-020-00335-9. [DOI] [PubMed] [Google Scholar]
- 241.Abi A., Ghaedi K., Khosravi A., Hayat S.M.G. Circular RNAs and glioma: small molecule with big actions. Curr. Mol. Med. Jun. 2020;21(1):25–44. doi: 10.2174/1566524020666200610171139. [DOI] [PubMed] [Google Scholar]
- 242.Xu Y., et al. Downregulated circular RNA hsa_circ_0001649 regulates proliferation, migration and invasion in cholangiocarcinoma cells. Biochem. Biophys. Res. Commun. Feb. 2018;496(2):455–461. doi: 10.1016/J.BBRC.2018.01.077. [DOI] [PubMed] [Google Scholar]
- 243.Ma J.H., Qin L., Li X. Role of STAT3 signaling pathway in breast cancer. Cell Commun. Signal. Feb. 2020;18(1) doi: 10.1186/S12964-020-0527-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Boopathy G.T.K., Hong W. Role of Hippo pathway-YAP/TAZ signaling in angiogenesis. Front. Cell Dev. Biol. 2019;7(APR):49. doi: 10.3389/FCELL.2019.00049/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.He X., et al. Circular RNAs: their role in the pathogenesis and orchestration of breast cancer. Front. Cell Dev. Biol. Mar. 2021:431. doi: 10.3389/FCELL.2021.647736. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Li J., et al. Circular HER2 RNA positive triple negative breast cancer is sensitive to Pertuzumab. Mol. Cancer. Sep. 2020;19(1):1–18. doi: 10.1186/S12943-020-01259-6/FIGURES/6. [DOI] [PMC free article] [PubMed] [Google Scholar]