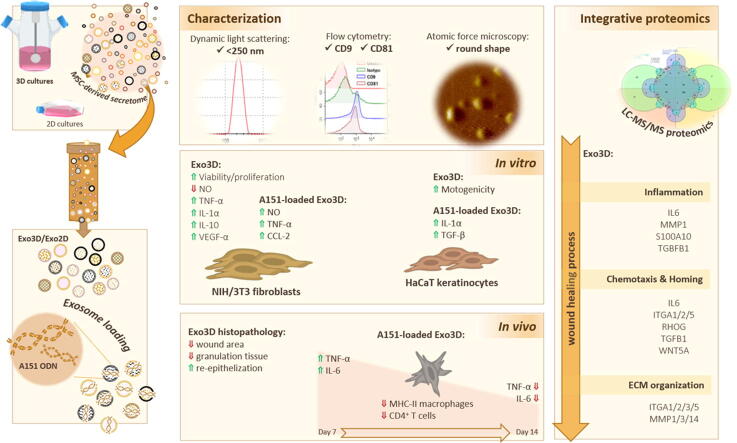
Graphical abstract
Keywords: Exosomes, Immunosuppressive oligodeoxynucleotide loading, Immunomodulation, Proteomics, 3D-cultured mesenchymal stem/stromal cells, Wound healing
Highlights
-
•
The MSC-derived secretome from 3D cultures enhances fibroblast and keratinocyte mitogenic and motogenic capacity in vitro, respectively.
-
•
The cargo of the 3D MSC-derived exosomes (Exo3D) reveals wound healing-related proteins and promotes wound resolution in a wound healing in vivo model.
-
•
Loading MSC-derived exosomes with A151 ODN further reduces the systemic levels of IL-6 and TNF-α pro-inflammatory cytokines at the late stage of wound healing in vivo, crucial for a full regenerated tissue.
-
•
A151-loaded Exo3D have a great potential as a noncellular off-the-shelf therapy for non-healing wound treatment.
Abstract
Introduction
Non-healing wounds remain a major burden due to the lack of effective treatments. Mesenchymal stem cell-derived exosomes (MSC-Exo) have emerged as therapeutic options given their pro-regenerative and immunomodulatory features. Still, little is known on the exact mechanisms mediated by MSC-Exo. Importantly, modulation of their efficacy through 3D-physiologic cultures together with loading strategies continues underexplored.
Objectives
To uncover the MSC-Exo-mediated mechanism via proteomic analyses, and to use 3D-culture and loading technologies to expand MSC-Exo efficacy for cutaneous wound healing.
Methods
MSC-Exo were produced in either 3D or 2D cultures (Exo3D/Exo2D) and loaded with an exogenous immunosuppressive oligodeoxynucleotide (A151 ODN). Both, loaded and naïve exosomes were characterised regarding size, morphology and the presence of specific protein markers; while IPA analyses enabled to correlate their protein content with the effects observed in vitro and in vivo. The Exo3D/Exo2D regenerative potential was evaluated in vitro by assessing keratinocyte and fibroblast mitogenicity, motogenicity, and cytokine secretion as well as using an in vivo wound splinting model. Accordingly, the modulation of inflammatory and immune responses by A151-loaded Exo3D/Exo2D was also assessed.
Results
Exo3D stimulated mitogenically and motogenically keratinocytes and fibroblasts in vitro, with upregulation of IL-1α and VEGF-α or increased secretion of TGF-β, TNF-α and IL-10. In vivo, Exo3D reduced the granulation tissue area and promoted complete re-epithelization of the wound. These observations were sustained by the proteomic profiling of the Exo3D cargo that identified wound healing-related proteins, such as TGF-β, ITGA1-3/5, IL-6, CDC151, S100A10 and Wnt5α. Moreover, when loaded with A151 ODN, Exo3D differentially mediated wound healing-related trophic factors reducing the systemic levels of IL-6 and TNF-α at the late stage of wound healing in vivo.
Conclusion
Our results support the potential of A151-loaded Exo3D for the treatment of chronic wounds by promoting skin regeneration, while modulating the systemic levels of the pro-inflammatory cytokines.
1. Introduction
The success of the wound healing process greatly depends on the correct sequence and timing of three interrelated phases – inflammation, new tissue formation and remodelling – which ends in the formation of a scar tissue [1]. Deviation from the strict regulation process, due to acute injuries or to external factors such as aging, burns, diabetes, vascular and autoimmune diseases [2], [3], leads to faulty healing and chronic/non-healing wounds may arise. Common features shared by chronic wounds include prolonged/excessive inflammation, persistent infections, and the inability of dermal and/or epidermal cells to respond to reparative stimuli. These pathophysiologic phenomena result in the failure of wound healing culminating in a reduced quality of life, amputations, or even early death [4]. Current available therapeutic methods to accelerate wound healing consist in debridement [5], wound dressings [6], skin grafting [7] or the administration of growth factors [8]. Still, its efficacy is limited to the improvement of patient care [9]. As such, regenerative medicine resorting to stem cells and/or their by-products has emerged as an effective therapeutic strategy to overcome incomplete wound resolution.
Mesenchymal stem/stromal cells (MSCs) are multipotent stem cells derived from a variety of tissues that have progressively become valid candidates for developing cell-based therapies for tissue regeneration due to their recognized anti-inflammatory, immunomodulatory, immunosuppressive, and angiogenic properties [10], [11], [12], [13]. This has been extensively demonstrated in several disease models namely rheumatoid arthritis [10], colitis [14], acute liver injury/failure [15], among others [16], [17], [18], where MSCs enabled tissue physiology re-establishment. Such features have been linked to MSCs paracrine activity and extracellular vesicles (EVs) secretion. Indeed, MSCs have emerged as promising candidates for cutaneous wounds treatment, not due to their ability to differentiate into the wounding cells, such as fibroblasts, but mostly through the induction of cellular crosstalks necessary to interrelate the different healing phases [19], [20]. Such induction is mediated by the MSC secretion of wound healing-promoting growth factors, cytokines and chemokines, that reduce wound inflammation and thus, promoting tissue repair [12], [21]. They include PDGF, EGF, FGF, IGF, TGF, IFN, TNF-α, VEGF, SDF-1, IL, HGF, CCL-2 (or MCP-1), LIF, and CCL-20, with immunosuppressive and regenerative activities [21], [22], [23]. In particular, high levels of IL-6, IL-8 and CXCL-1 were found in the secretome of MSCs derived from bone marrow and linked to an enriched cell infiltration, re-epithelialization and angiogenesis by promoting epithelial and endothelial cell migration [22].
Other soluble trophic factors important for the different phases of wound healing have also been specifically identified in the secretome of a specific population of umbilical cord tissue-derived MSCs (UC-MSCs), namely G-CSF, KGF, EGF and FGF-2 [24]. Interestingly, UC-MSC spheroids showed a distinct and more pro-regenerative secretome (CM3D) profile, when compared to that of standard monolayer cultures (CM2D). Trophic factors such as VEGF-α, HGF, MMP-2/9, TGF-β, FGF-2, G-CSF and IL-6 known to regulate tissue homeostasis, granulation tissue formation, and the remodelling stages of wound healing, were found prominently in CM3D. Moreover, MSC-derived CM3D significantly improved fibroblast and keratinocyte migration along with elastin production and capillary maturation in vitro [10], [12]. Likewise, the administration of CM3D to excisional wounds in vivo resulted in a fully regenerated tissue with the re-establishment of a vascular system and faster appearance of mature skin appendages, such as glands and hair follicles, which ultimately sustains the use of 3D-culture strategies to modulate and improve the therapeutic effect of MSCs.
Besides free soluble trophic factors, the MSC secretome is also composed of EVs, such as exosomes, that confer specific intercellular communication, through the transfer of critical biological molecules and genetic information, while protecting their cargo (e.g., growth factors, chemokines, cytokines, nucleic acids, miRNAs, etc…) from degradation [25], [26], [27], [28], [29], [30]. These characteristics confers targeted delivery and, consequently, therapeutic advantages for the EVs. In wound healing, MSC-derived exosomes would be expected to have similar therapeutic effects to the cells that secret them, namely on the modulation of inflammation and immune responses, but their impact on these crucial wound healing processes appears to be less evident [19], [22]. Nevertheless, MSC-derived exosomes have shown to be mainly associated to re-epithelialization [31], [32] and angiogenesis [32], [33]. The cargo of MSC-derived exosomes may contain HECT, CXCL-1/2/8, IFITM-2, HERC-5, and DEFA-1 that are chemoattractants of immune cells, which may induce a better protection against potential infections in damaged tissues [34]. Moreover, exosome-contained cytokines, such as PDGF, G-CSF, VEGF, CCL-2, as well as IL-6/8 were shown to induce AKT pathway in keratinocytes and the presence of Wnt led to β-catenin activation that overall contributes to improved wound healing [31].
In turn, several synthetic immunosuppressive oligodeoxynucleotides (ODN) have been developed to alleviate excessive inflammation and immune activation either by inhibiting immune responses broadly or in a target-specific manner. The broad-acting A151 ODN for example, containing 4 repeats of mammalian telomeric TTAGGG, was first reported in 2003 and initially its suppressive activity thought to be restricted to blocking the interaction between CpG ODN and TLR9 [35]. However, A151 ODN is responsible for metabolically reprogramming immune cells towards immunosuppression via mechanisms, such as binding to STAT or AIM2 proteins or downregulation of PI3K/AKT/mTOR signalling [36], [37], [38]. Within the context of pathophysiological conditions involving prolonged or excessive inflammation, such as non-healing wounds, this ODN can present itself as an added value. As such, in this work, we aimed to clarify the impact of 3D culturing strategies in the modulation of the exosome content, as well as their effect upon loading with A151 ODN, on cutaneous wound healing using in vitro and in vivo methodologies. In addition, an integrative proteomic analysis was adopted to unveil their mediated mechanism. We hypothesised that these approaches boost the exosome action and improve their therapeutic value, therefore avoiding the safety concerns associated with the use of the cells themselves.
2. Materials and methods
2.1. Ethics statement
The Ethics Committee of the Hospital Dr. José de Almeida (Cascais, Portugal) has approved this study where umbilical cord donations were subjected to written informed consents according to European Directive 2004/23/EC under the scope of a research protocol between ECBio - Research & Development in Biotechnology, S.A. and HPP Saúde - Parcerias Cascais, S.A.. All animal experiments were performed in accordance with the EU Directive (2010/63/EU), Portuguese law (DL 113/2013) and all relevant legislations, and approved by Direção Geral de Alimentação e Veterinária (DGAV), Portugal and by the Bilkent Animal Ethics Committee, Turkey.
2.2. UC-MSC isolation and culture
UC-MSCs were isolated from umbilical cord tissue as previously described by [39] and cultured in α-MEM supplemented with 10% FBS under three-dimensional (3D) and two-dimensional (2D) conditions according to [10], [12].
2.3. Conditioned media (CM) preparation
Conditioned media (CM) or secretome from UC-MSCs were produced from cells with similar number of passages. UC-MSC CM from 3D spinner flask cultures (CM3D) was obtained by inoculating a single cell suspension at 1 × 106 cells/mL with 80 rpm stirring rate, and subjected to successive medium adaptations. At day 5, FBS was completely removed from the cell culture. The volume of fresh α-MEM was adjusted to obtain a conditioning volume per cell equivalent to that in the 2D system [10], [12]. To produce UC-MSC CM in 2D monolayer cultures (CM2D), cells were seeded at 1 × 104 cells/cm2 in α-MEM supplemented with 5% FBS and cultured until 90% of confluency was reached. At this point, medium was replaced by α-MEM without FBS, to a final volume of 25 mL/175 cm2. Both CM were collected after 48 h under sterile conditions.
2.4. Exosome isolation and characterisation
2.4.1. Isolation
Exosomes were isolated from both CM3D (Exo3D) and CM2D (Exo2D) using Exo-spin® kit (Cell Guidance Systems, Cambridge, UK). The kit is based in size exclusion chromatography (SEC) and has been calibrated to purify particles in the range of 30 – 400 nm. The protocol was performed according to the manufacturer’s instructions, with few exceptions as described below. Briefly, for compliance with mass spectrometry (MS) analysis requirements, the precipitation step preceding the purification through the SEC columns was omitted. Instead, after column equilibration with PBS, CM concentrated to a final volume of 1 mL was directly applied to the column. After washing, 5 mL of PBS were added and allowed to drain under gravity to collect the exosome-containing eluate. The exosome-depleted eluate was also collected by adding 7 mL of PBS to the column afterwards. Total protein content was measured using a Bradford method (Bio-Rad Laboratories, Lisbon, Portugal) according with manufacturer’s instructions for microtiter plates. Samples were aliquoted and stored at −80 °C until further use.
2.4.2. A151 ODN loading into exosomes
A151 ODN loading into exosomes was performed with a controlled dehydration-rehydration method adapted from [40]. Briefly, exosomes (Exo3D and Exo2D) and A151 ODN were respectively mixed at 1 μg:1 μg (exosomes:A151 ODN) ratio resulting in A151-loaded Exo3D and A151-loaded Exo2D samples, respectively, and snap-frozen in liquid nitrogen. The mixture was lyophilized overnight with VirTis BenchTop K freeze dryer (SP Scientific, Warminster, PA, USA). For the controlled reconstitution, nuclease-free ddH2O at the 1:10 volume of the original lyophilized solution was added and vigorously vortexed.
2.4.3. Dynamic light scattering (DLS)
The exosome size distribution was analysed by photon correlation spectroscopy (PCS) using a Zetasizer Nano S (Malvern Instruments, Malvern, UK) being measurements made at 25.0 ± 0.1 °C in triplicates for each sample.
2.4.4. Atomic force microscopy (AFM)
Exosomes diluted in PBS were adsorbed onto mica sheets and dried at RT. Images were obtained in non-contact dynamic mode with ambient AFM (NanoMagnetics Instruments, Oxford, UK). The scanning area sizes were 20 × 20 µm. Obtained images were further analysed using NMI ImageAnalyzer (NanoMagnetics Instruments).
2.4.5. Analysis of exosome surface markers by flow cytometry (FC)
Exosomes were captured using carboxyl latex beads (Invitrogen®, Thermo Fisher Scientific) coated with purified anti-human CD63 antibody (Biolegend, San Diego, CA, USA; 1 μL:1 μg ratio, respectively) overnight on a rotator at a low speed. Then, exosome-bead conjugates were incubated with fluorochrome antibodies for human CD9 and CD81 (Biolegend), and their appropriate isotype controls at a concentration of 1 μg/mL for 1 h at RT in the dark. Afterwards, samples were washed and resuspended in 100 μL of PBS to be analysed with NovoCyte flow cytometer (ACEA Biosciences, San Diego, CA, USA).
2.4.6. Proteomic analyses
2.4.6.1. LC-MS/MS analysis
30 μg of total protein of each sample (in triplicate) was prepared for LC-MS/MS analysis with the S-Trap® Micro Spin Column (ProtiFi, Farmingdale, NY, USA) digestion protocol according to the manufacturer’s instructions with slight modifications. 2 μg of purified peptides of each sample was then injected on an Ultimate 3000 RSLC nanoLC (Thermo Scientific®, Bremen, Germany) in-line connected to an LTQ-Orbitrap Elite (Thermo Fisher®). Detailed sample processing methodology and the mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [41] partner repository with the dataset identifier PXD025633.
2.4.6.2. MS data analysis
Data analysis was performed with MaxQuant (version 1.6.8.0) using the Andromeda search engine with default search settings including a false discovery rate set at 1% on the PSM, peptide and protein level. Spectra were searched against the human protein sequences in the Uniprot database (database release version of June 2018), containing 20,960 sequences (www.uniprot.org). The mass tolerance for precursor and fragment ions was set to 4.5 and 20 ppm, respectively, and enzyme specificity was set as C-terminal to arginine and lysine, with a maximum of two missed cleavages. Variable modifications were set to oxidation of methionine residues and acetylation of protein N-termini, while carbamidomethylation of cysteine residues was set as a fixed modification.
2.4.6.3. Ingenuity pathway analysis (IPA)
Gene symbols of the significant proteins identified were uploaded to the IPA server (IPA v2.4 QIAGEN®, Germantown, MD, USA; www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis) for in-depth knowledge analysis using the “Core Analysis” function (Fisher’s Exact Test (FET) p-value: 1e-03). The upstream regulators were predicted by IPA using the default settings. The network explorer of IPA was used to identify interactions among the genes supplied and using these to generate gene–gene interaction networks (GGINs) with shortest literature-supported paths (edges) between genes (nodes). Venn diagrams were generated using the “venn()” function in the R package gplots v3.0.1.1.
2.4.6.4. Immunoblotting analysis
50 µg of total protein from each condition were resolved by SDS-PAGE in 12% polyacrylamide gels prepared as described by [42]. Gels were blotted onto PVDF membranes, which were incubated with primary antibody diluted in 5% BSA [anti-TGF-β and anti-vinculin (VCL) diluted 1:100 and 1:1000, respectively (Abcam, Cambridge, UK), and anti-integrin-α4 (ITGA4) and anti-Wnt-5α diluted 1:100 (Santa Cruz Biotechnology, Heidelberg, Germany)] overnight at 4 °C, washed and incubated with horseradish peroxidase-conjugated anti-mouse (Cytiva, Lisbon, Portugal) or anti-rabbit (Jackson ImmunoResearch, Cambridgeshire, UK) antibodies for 2 h at RT. Immunoreactive bands were detected by enhanced chemiluminescence ECL (Millipore) according to the manufacturer’s instructions and images were recorded using a ChemiDoc XRS System (Bio-Rad Laboratories). Protein loading control was performed with Ponceau S staining.
2.5. In vitro and in vivo evaluation of the effect of exosomes on wound healing
2.5.1. Cell viability assay
Human immortalized keratinocyte cells (HaCaT) and mouse fibroblast cells (NIH/3T3) were seeded at 3.0 × 104 and 1.0 × 104 cells/cm2, respectively, in DMEM (Sigma-Aldrich®) with 4.0 g/L D-(+)-Glucose supplemented with 10% of FBS, at 37 °C, in 5% CO2 humidified atmosphere. Exosome fractions were added to the cells at 1, 5, 25 and 50 µg/mL. Cells were also incubated with complete cell culture medium, DMEM:PBS (3:1) and DMEM with 20% dimethyl sulfoxide (DMSO) as positive, solvent and negative control, respectively. After 48 h, both HaCaT and NIH/3T3 cells were incubated with MTS (Promega, Madison, WI, USA) according to manufacturer’s instructions, for 2 h at 37 °C. Optic density was measured at 570 nm on SPECTROstar Omega (BMG LabTech®, Ortenber, Germany) microplate spectrophotometer. Results were shown as percentage of solvent control, which was considered to be 100%. (n = 3–6).
2.5.2. In vitro scratch assay
HaCaT and NIH/3T3 cells were seeded at 6.0 × 104 and 3.0 × 104 cells/cm2 respectively, in complete cell culture medium as abovementioned. Once at 90% confluence, scratches were performed according to [12]. Cells were incubated in DMEM supplemented either with Exo3D, Exo2D, Exo3D-dep and Exo2D-dep at a concentration of 1 µg/mL corresponding to the highest concentration with negligible cytotoxic effect in both NIH/3T3 and HaCaT cell types. DMEM, DMEM:PBS (3:1) and DMEM with 10% FBS were also tested as negative, solvent and positive controls, respectively. Digital photographs of the scratch area were taken at 0, 8, 20 and 24 h post-scratch, at an amplification of 40 × on Olympus CK30 microscope (Olympus, Tokyo, Japan) and analysed with Motic Images Version 2.0 software (Motic®, Barcelona, Spain; Fig. S1). Cellular migration was quantified by calculating scratch closure, given as the total area occupied by the cells after incubation with the samples in relation to the initial scratch area at 0 h. Three independent experiments were performed for all samples, except for Exo3D-dep in HaCaT where two independent experiments were performed.
2.5.3. In vivo wound healing model
An excisional wound splinting assay was performed according to [12]. Briefly, male 5- to 6-months old Wistar rats (Charles River Laboratories, Barcelona, Spain) were anaesthetized using intraperitoneal (i.p.) injection of ketamine (75 mg/kg; Imalgene®, Merial, Lyon, France) and medetomidine (0.5 mg/kg; Medetor®, Virbac, Burgdorf, Germany). Full-thickness 8-mm wounds were performed and a donut-shaped splint was stitched to prevent skin contraction. Each animal carried four wounds to which 50 μg of total protein content of each sample were applied every 24 h, in a total of 3 applications via subcutaneous injection between the wound margin and the silicone splint of the respective wound, as follows: 1) Exo3D; 2) Exo2D; 3) Exo3D-dep; 4) Exo2D-dep; 5) PBS (solvent vehicle control); and 6) sham (natural wound resolution). Wound area was monitored by taking digital photographs and measured using ImageJ (NIH, Bethesda, MD, USA). The wound area tissue was excised for haematoxylin and eosin (H&E) staining (Fig. S2), which were blindly examined by a pathologist who scored for dermis and epidermis healing phases according to the parameters summarized in Table 1. Four to seven wounds were analysed for each group.
Table 1.
Criteria for histologic scoring of wound healing.
| Score | Dermis stage | Epidermal stage |
|---|---|---|
| 1 | Inflammation | Epidermis not closed |
| 2 | Inflammation and tissue formation | Hyperplasia and hyperkeratosis |
| 3 | Tissue formation | No epidermal changes with no or little hair re-growth |
| 4 | Tissue remodelling | Wound resolved (with hair re-growth) |
| 5 | Wound resolved (with hair re-growth) | --- |
2.6. Evaluation of the effect of loading exosomes with A151 ODN in terms of immunomodulation and wound healing
2.6.1. In vitro cytokine production assay in HaCaT and NIH/3T3 cells
HaCaT and NIH/3T3 cell lines were seeded at 5.0 × 105 cells/well in 12-well plates as abovementioned, and treated with 20 μg/mL of Exo3D, Exo2D, A151-loaded Exo3D and A151-loaded Exo2D. A151 ODN and PBS were used as controls. After 24 h, cytokine ELISA for TGF-β, TNF-α and IL-10 production, RT-qPCR for TGF-β, IL-1α and CCL-2 genes, and Griess Assay for nitric oxide (NO) were performed as described below. A total of two independent experiments were performed.
2.6.2. Determination of cytokine levels by ELISA
PolySorp Nunc-Immuno 96-well plates (Thermo Fisher Scientific) were coated with PBS-diluted antibodies as 50 μL/well overnight at 4 °C. After 2 h at RT of blocking with 5% BSA and 0.05% Tween 20 in PBS followed by 3 washing steps with 0.05% Tween 20 in PBS and 3 washing steps with ddH2O washing steps, cell culture supernatants or blood sera and serially 1:2 diluted recombinant protein standards (50 μL/well) were incubated overnight at 4 °C. The washing steps as abovementioned were performed every incubation period of biotinylated antibodies (50 μL/well) for 2 h and 50 μL/well SA-ALP for 1 h, both at RT. PNPP substrate was then added as 50 μL/well and colour development was followed by frequently taking measurements at 405 nm with the ThermoMax microplate reader (Molecular Devices, San Jose, CA, USA) until the recombinant standards reached saturation.
2.6.3. Determination of gene expression by RT-qPCR
Total RNA isolation and cDNA synthesis were performed with TRIzol Reagent (Thermo Fisher Scientific) and RevertAid RT Reverse Transcription kit (Thermo Fisher Scientific), respectively, according to the instructions of the manufacturers. RT-qPCR reactions were made with LightCycler 480 SYBR Green I Master Mix (Roche Life Science) using the corresponding primers (Table S1) and the reactions performed on LightCycler 480 (Roche Life Science).
2.6.4. Measurement of NO level by Griess assay
Culture supernatant samples were incubated with Griess reagent (Fluka, St. Gallen, Switzerland) in 1:1 ratio at a final volume of 100 μL for 15 min at RT. Absorbance at 540 nm was measured using Synergy HT microplate reader.
2.6.5. In vivo quantification of pro-inflammatory cytokines and immune cells
Female BALB/C mice, 8 to 10-weeks old were housed in the Bilkent University Animal Facility in the Department of Molecular Biology and Genetics under standard laboratory conditions with free access to commercial standard rodent chow and acidified water ad libitum. After anaesthesia i.p. with ketamine (75 mg/kg) and xylazine (0.5 mg/kg), a full-thickness splinting model was followed as abovementioned using a sterile 6-mm biopsy punch. Animals were randomly divided into groups according to the treatment: 1) Exo3D; 2) Exo2D; 3) A151-loaded Exo3D; 4) A151-loaded Exo2D; 5) PBS (solvent vehicle control), and 6) A151. Each treatment consisted of 30 µg of total protein via subcutaneous injection that was repeated after 48 h. Animals were sacrificed at days 7 (n = 2–3) and 14 (n = 5–7) for systemic cytokine measurements and isolation of spleen cells. Cardiac puncture blood was used for cytokine ELISA. For the analyses of relevant immune cell populations, spleens from BALB/c mice were surgically removed. Single cell suspensions were prepared and counted with the NovoCyte flow cytometer. Gating was performed to assure only live cells and to discriminate doublets (Fig. S3). Finally, splenocytes were stained with relevant antibodies to analyse T-lymphocyte, macrophage and myeloid derived suppressor cell (MDSC) populations and their activating and inhibiting surface markers (Fig. S3).
2.7. Statistical analysis
Statistical analyses were performed in GraphPad Prism v7.0 software (La Jolla, CA, USA). To estimate the significance of the data differences, one-way ANOVA with Tukey’s multiple comparisons post-hoc was used, except for the data of in vitro viability and scratch assays, and in vivo histological scores, where two-way ANOVA with Tukey’s multiple comparisons post-hoc test was performed. Results are presented as means ± standard error of the mean (SEM), except where indicated and p-values are presented for statistically significant results (* p < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001).
3. Results
3.1. Loading of immunosuppressive A151 ODN into exosomes preserves physicochemical characteristics of UC-MSC-derived exosomes regardless of the cell culture system adopted
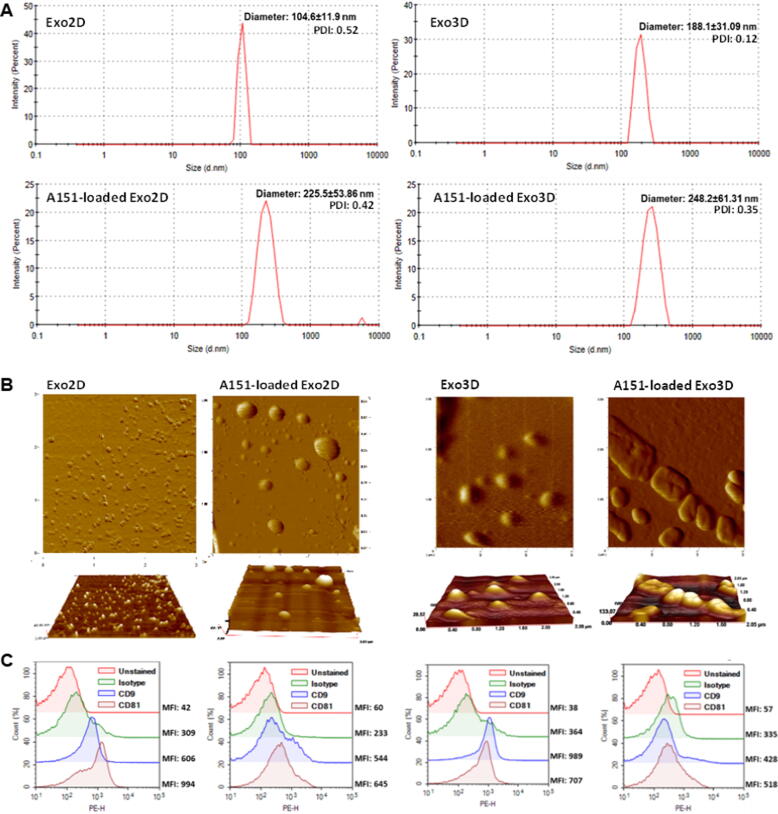
The physicochemical characteristics of exosomes such as size, morphology, and the presence of specific protein markers, e.g., CD9 and CD81, were assessed by comparing exosomes originated from both 2D (Exo2D) and 3D (Exo3D) culture systems before and after A151 ODN loading. Size distribution was performed by DLS in aqueous state and by AFM in dry state.
DLS results revealed that Exo2D were smaller than Exo3D with Z-average diameter of 104.6 ± 11.9 nm and 188.1 ± 31.09 nm, respectively (Fig. 1A). Polydispersity index (PDI) was higher on Exo2D than Exo3D (0.52 vs. 0.12) suggesting a high polydisperse sample [43]. Nevertheless, the presence of only one peak per sample indicated homogenous populations of both exosome samples.
Fig. 1.
Loading of A151 ODN preserves physicochemical characteristics of MSC-derived exosomes. (A) Size distribution of exosomes produced under 3D/2D conditions either naïve (Exo3D/2D) or loaded with A151 ODN (A151-loaded Exo3D/2D) determined by dynamic light scattering (DLS); (B) Representative AFM images of Exo3D/2D and A151-loaded Exo3D/2D dried on mica surface and obtained in non-contact dynamic mode; (C) Flow cytometry analyses of Exo3D/2D and A151-loaded Exo3D/2D captured with anti-CD63 coated latex beads and stained with PE-labelled anti-CD9, anti-CD81 and their appropriate isotype controls. Abbreviations: Exo3D - exosomes derived from UC-MSC cultured in 3D conditions; Exo2D - exosomes derived from UC-MSC cultured in 2D conditions; A151-loaded Exo3D - exosomes produced under 3D culture conditions loaded with A151 ODN; A151-loaded Exo2D - exosomes produced under 2D culture conditions loaded with A151 ODN.
As expected, loading A151 ODN into exosomes led to an increase of the average size, however with different impact on Exo2D and Exo3D (A151-loaded Exo2D and Exo3D, respectively; Fig. 1A). While A151-loaded Exo2D increased twice their average diameter to 225.5 ± 53.86 nm, A151-loaded Exo3D with average size of 248.2 ± 61.31 nm only increased around 30%. Additionally, loading A151 ODN within either Exo2D or Exo3D considerably increased size variation as seen by the standard deviation values (Fig. 1A). However, in contrast to 3D conditions, the PDI value did not differ critically for A151-loaded Exo2D when compared to naïve Exo2D (0.52 vs. 0.42; Fig. 1A). AFM image analysis corroborated the size distribution data revealing that A151 ODN loading into exosomes increased the particle size both in diameter and height, resulting in more heterogenous populations (Fig. 1B). AFM imaging further suggested an increased size (up to 400 nm) and loss of well-rounded shape of the exosomes, when compared to the values obtained in aqueous state with DLS (Fig. 1B). However, it should be noted that in AFM imaging exosomes are attached to a surface and therefore their morphology and size could appear more distorted when compared to that seen aqueous state.
The tetraspanins CD9, CD63 and CD81, besides their roles on membrane organization, are commonly used as exosome surface markers [44]. Thus, to demonstrate their presence on Exo2D and Exo3D and to assess if A151 ODN loading procedure impaired the membrane integrity and bound proteins, exosome surface protein characterisation was performed by FC. Exosomes were captured with anti-CD63-coated carboxyl latex beads and stained with fluorochrome-labelled anti-CD9 and anti-CD81 antibodies. Both CD9 and CD81 were found to be present on both loaded and non-loaded samples (Fig. 1C), with mean fluorescence intensity (MFI) values belonging to the fluorochrome-labelled tetraspanin-specific antibodies. Overall, both UC-MSC-derived Exo2D and Exo3D displayed key exosome-specific surface markers which was not critically affected by A151 ODN loading procedure.
3.2. Exosome cargo reveals wound healing-related proteins
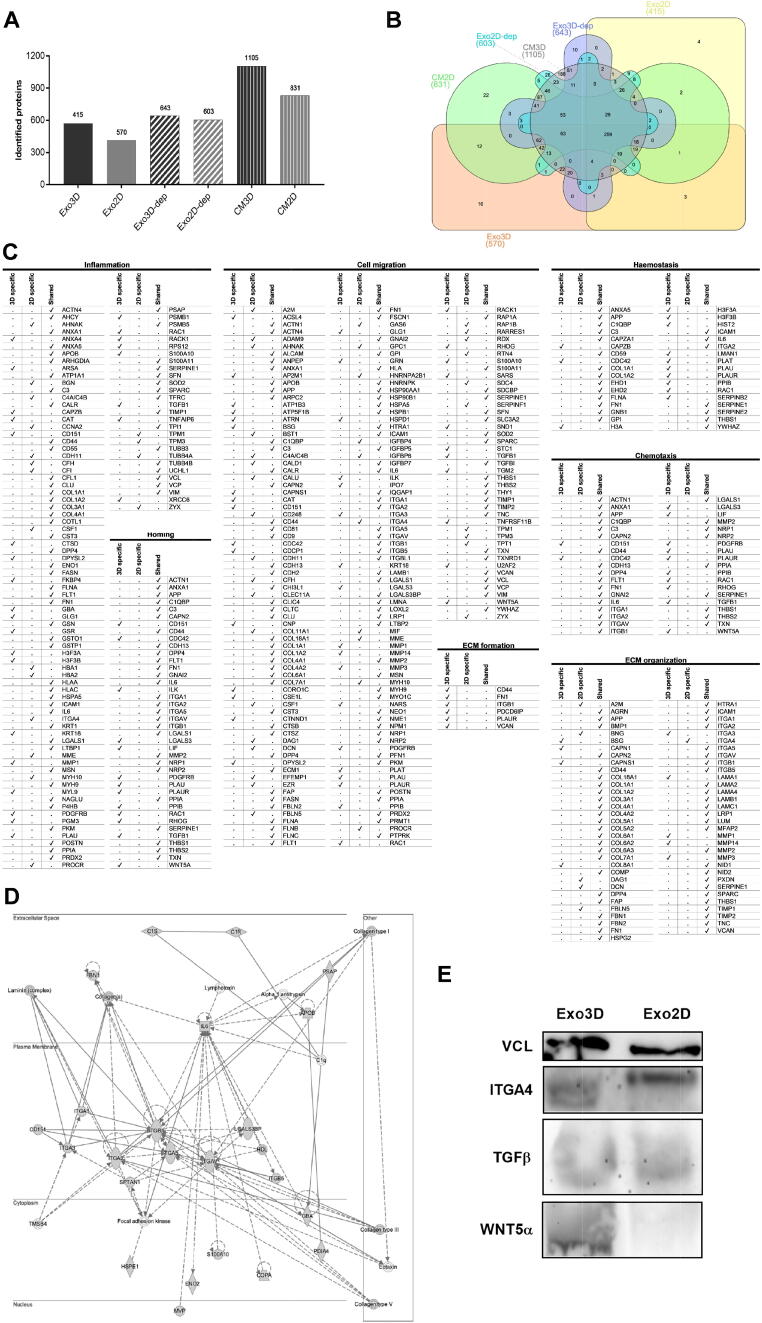
To explore exosomes as therapeutic mediators, the effect of the different culture conditions on the cargo of the exosomes was characterised by MS-based proteomic analyses. The protein content of the UC-MSC secretome (CM2D and CM3D) and of the CM2D and CM3D depleted of exosomes (Exo3D-dep and Exo2D-dep) were analysed as well, as controls.
The LC-MS/MS proteomic analysis retrieved 2040 distinct proteins. This analysis also revealed that the content of samples derived from UC-MSC 3D cultures (CM3D, Exo3D and Exo3D-dep) presented more protein diversity, when compared to that of 2D culture samples (CM2D, Exo2D and Exo2D-dep; Fig. 2A). It further allowed the identification of unique CM2D and CM3D sets of proteins that were commonly expressed as shown in the Venn diagram in Fig. 2B. More specifically, a set of 16 and 4 proteins were exclusively identified in Exo3D and in Exo2D, respectively.
Fig. 2.
Proteomic profiling of MSC-derived secretome and exosomes reveals the presence of proteins related to key biological processes for wound healing. (A) Total protein numbers identified in CM3D, CM2D, Exo3D, Exo2D, Exo3D-dep and Exo2D-dep; (B) Venn diagram shows the unique and shared protein numbers among CM3D, CM2D, Exo3D, Exo2D, Exo3D-dep and Exo2D-dep; (C) IPA Disease&Function analysis for pairwise comparison of Exo3D and Exo2D regarding crucial wound healing events; (D) IPA gene–gene interaction network showing the protein–protein interactions of TGF-β with the Exo3D content regarding the functions of cell morphology, cell-to-cell signalling and interaction, and tissue development; (E) Confirmation of selected proteomic data by western blot analysis of VCL, TGFβ, ITGA4 and WNT5A. Abbreviations: CM3D - whole secretome collected from UC-MSC 3D cultures; CM2D - whole secretome collected from UC-MSC 2D cultures; Exo3D - exosomes derived from UC-MSC cultured in 3D conditions; Exo2D - exosomes derived from UC-MSC cultured in 2D conditions; Exo3D-dep – CM3D depleted of exosomes; Exo2D-dep – CM2D depleted of exosomes.
In addition, IPA disease and function analysis for pairwise comparisons was performed considering all identified proteins in both, Exo3D and Exo2D samples since they are likely the underlying causes of potential differential therapeutic effects. Overall, the identified proteins are involved in different biological processes such as metabolic and cellular processes and mostly found in cell parts. However, a deeper analysis was focused on key biological processes involved in all phases of the wound healing, namely inflammation, haemostasis, cell migration, chemotaxis, homing and extracellular matrix (ECM) formation and organization (Fig. 2C). Enrichment analyses highlighted that a higher prevalence of proteins associated to many of these processes were contained within Exo3D, while Exo2D presented lower wound healing phase representation, except for inflammatory response, migration of cells and ECM organization. Regarding the common proteins associated to inflammatory response, cell migration, haemostasis, chemotaxis and homing, normalised abundance analysis evidenced the presence of APP, C3, FN1, IL6, VCL and SERPINE1 in all samples. Concerning the haemostasis process, the proteins CAPZB, CDC42, H3A, H3F3A/B, HIST2, LMAN1, PLAT, PLAU, PLAUR, PPIB, RAC1 and SERPINB2 were only observed in Exo3D. Likewise, CD151, CDC42, ILK, LGALS3, LIF, PDGFRB, PLAU, PLAUR, PPIB, RAC1, RHOG, TGFβ and WNT5A were associated to chemotaxis, homing and cell migration processes; whereas CD151, PDGFRB, PLAU, RAC1 and TGFβ were related to inflammation and only found in Exo3D as well. Moreover, ECM formation-related proteins were only found in Exo3D and include CD44, FN1, ITGB1, PDCD6IP, PLAUR and VCAN. Contrarily, both Exo3D and Exo2D shared proteins associated to the process of ECM organization, namely BMP1, MMP2 and VCAN. While BSG, CAPN1, CAPNS1, COL8A1, ITGA3, LAMA1, and MMP1/3/14 were identified exclusively on Exo3D, unique proteins from Exo2D were A2M, BNG, DAG1, DCN, FBLN5 and ITGA4 (Fig. 2C).
Among those proteins, TGFβ and WNT5A, exclusive for 3D samples, ITGA4, exclusive for 2D samples, and VCL for both samples, were chosen for validation of the proteomic data by western-blot analysis (Fig. 2E). In addition, given the specific presence of TGFβ on Exo3D and their direct role in inflammation, cell migration, chemotactic and homing mechanisms; and in order to add new insights on TGFβ interactions with other targets, an IPA interaction network was performed showing the protein–protein interactions of TGFβ with the Exo3D content regarding the functions of cell morphology, cell-to-cell signalling and interaction, and tissue development (Fig. 2D). Herein, TGFβ showed to be involved on ECM-related processes such as cell adhesion and collagen synthesis, binding and deposition as seen by the interaction with laminin, integrins (ITGA1, ITGA2, ITGA3 and ITGA5) and collagen (types I, III and V). Moreover, this network confirmed the role of TGFβ on i) inflammation by interacting with IL6, a known cytokine involved in inflammatory and immune responses, on ii) haemostasis due to the interaction with S100A10, which is related with the dissolution of fibrin clot, and on iii) cell migration through the communication with CD151 which was found to be specifically related chemotaxis and homing (Fig. 2C-D).
3.3. Samples produced under 3D culture conditions enhance fibroblast mitogenic ability and keratinocyte motogenic capacity in vitro
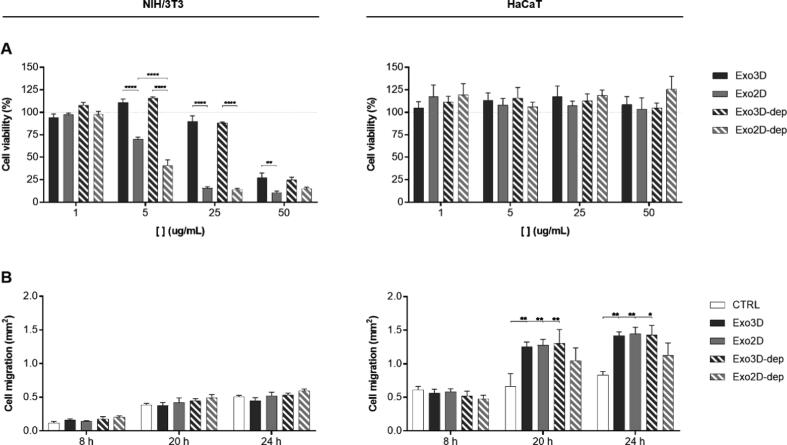
Following the proteomic findings, the effect of UC-MSC-derived exosomes on the viability and migration of keratinocytes (HaCaT) and fibroblasts (NIH/3T3) in vitro was studied by cell viability and scratch assays, respectively (Fig. 3). Besides testing Exo3D and Exo2D samples, UC-MSC secretome depleted of exosomes (Exo3D-dep and Exo2D-dep) were used as controls.
Fig. 3.
Secretome fractions primed by 3D culturing improve fibroblasts mitogenic ability and keratinocytes motogenic capacity in vitro. (A) NIH/3T3 (left) and HaCaT (right) viability evaluated by MTS reduction assay after 48 h of incubation with Exo3D/2D and Exo3D/2D-dep. n = 3–6. (B) Graphic representation of the average area occupied by the NIH/3T3 (left) and HaCaT (right) cells after 8, 20 and 40 h in contact with Exo3D/2D and Exo3D/2D-dep as evaluated by in vitro scratch assay. Culture medium was used as negative control. Data is shown as mean ± SEM. n = 2–3. * p < 0.05, ** p < 0.01, **** p < 0.0001. Abbreviations: Exo3D - exosomes derived from UC-MSC cultured in 3D conditions; Exo2D - exosomes derived from UC-MSC cultured in 2D conditions; Exo3D-dep – CM3D depleted of exosomes; Exo2D-dep – CM2D depleted of exosomes.
Fig. 3A shows that keratinocyte viability was maintained in all conditions (1–50 μg/mL). Conversely, fibroblast viability was significantly promoted by Exo3D and Exo3D-dep (5 and 25 μg/mL) when compared to 2D counterparts. In fact, increasing concentrations of Exo2D and Exo2D-dep led to reduced cell viability to ∼ 10–15%. On the other hand, Figs. 3B and S1 show that both Exo3D and Exo2D promoted the motogenic activity of keratinocytes when compared to control at the latest time-points (20 and 24 h), whereas no effect was observed in fibroblasts. Similar results were obtained with the Exo3D-dep fraction, highlighting the importance of 3D culturing for improving tissue regeneration-related processes.
3.4. Exo3D promotes wound resolution in a wound healing in vivo model
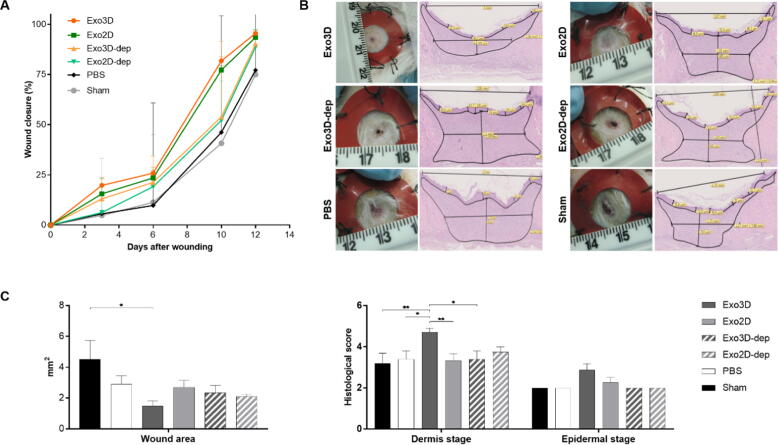
Given the evidence that, besides the distinct cargo profiles, Exo3D and Exo2D differentially mediated the fibroblast and keratinocyte responses in vitro, the effect of cell culture conditions on the therapeutic potency of MSC-derived exosomes using an in vivo wound healing splinting rat model were further evaluated. Macroscopic observations showed that both Exo2D- and Exo3D-treated wounds exhibited accelerated wound closure when compared to both PBS and sham control-treated wounds (Fig. 4A). At day 10, exosome-treated wounds revealed a significant improvement in terms of wound closure when compared with wounds treated with the exosome-depleted fractions of the 2D and 3D secretome (Exo2D-dep and Exo3D-dep), being the percentage of wound closure ∼ 80%, ∼50% and ∼ 40% in Exo-treated wounds, Exo-dep-treated wounds and controls, respectively. Importantly, a critical histological analysis confirmed different wound healing stages between Exo-treated wounds, especially in terms of wound area and dermal regeneration (Figs. 4B-C and S2). Exo3D-treated wounds showed a reduced wound and granulation tissue areas, complete re-epithelization translating for higher dermis and epidermal stage scores, not observed with any other treatment (Fig. 4B-C). In contrast, the high extension of granulation tissue in wounds treated with Exo2D, Exo2D-dep and Exo3D-dep indicates a delayed wound resolution (Figs. 4B-C and S2).
Fig. 4.
Exosomes isolated from UC-MSC 3D cultures (Exo3D) enhance wound healing in vivo. (A) Graph represents the closure of the wounds treated with Exo3D/2D, Exo3D/2D-dep, PBS (solvent vehicle control) and sham (natural wound resolution) control throughout time via macroscopic analyses. Data is shown as mean ± SEM. n = 4–7. (B) Representative images (left) of macroscopic view and (right) histological slices stained by H&E of Exo3D/2D-, Exo3D/2D-dep-, PBS and sham-treated wounds at day 14. (C) Graphs represent the histological wound area and dermis and epidermis stages scores, evaluated in Exo3D/2D-, Exo3D/2D-dep-, PBS- and sham-treated wounds at day 14 via microphotographs histological analyses. Data is shown as mean ± SEM. * p < 0.05, ** p < 0.01. Abbreviations: Exo3D - exosomes derived from UC-MSC cultured in 3D conditions; Exo2D - exosomes derived from UC-MSC cultured in 2D conditions; Exo3D-dep – CM3D depleted of exosomes; Exo2D-dep – CM2D depleted of exosomes.
3.5. A151-loaded Exo differentially mediate the production of wound healing-related trophic factors from fibroblasts and keratinocytes
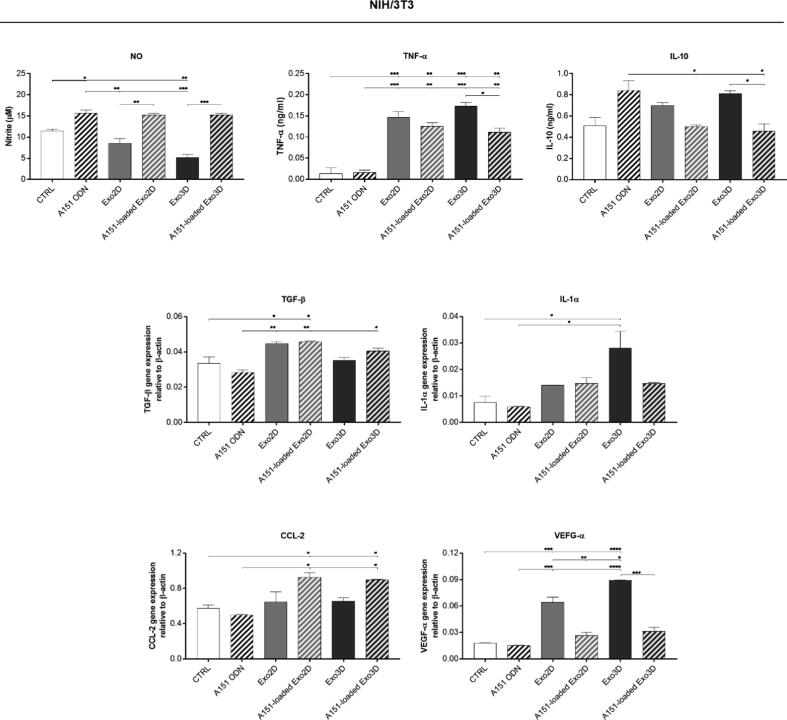
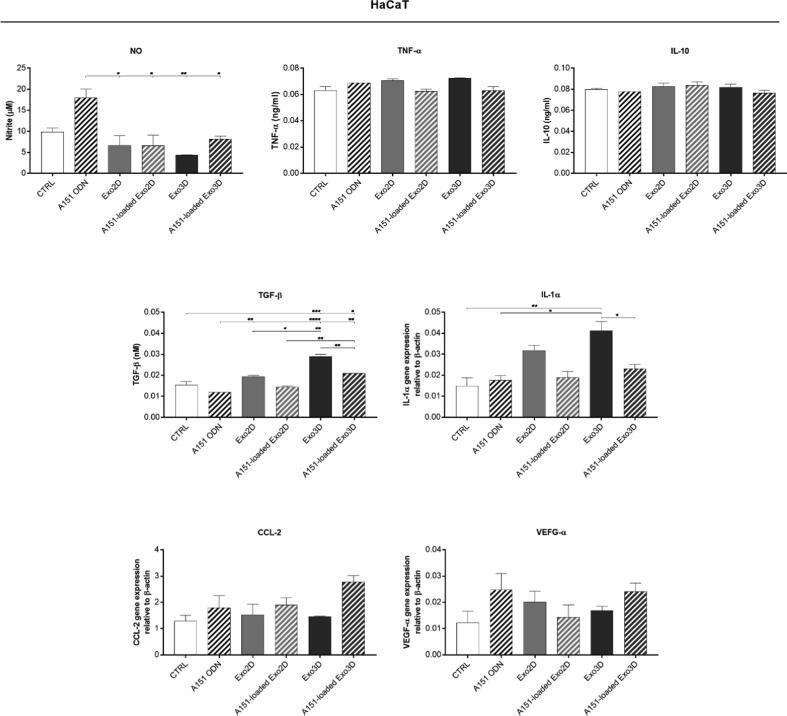
Herein, the production of wound healing-related trophic factors by specific skin cell types was monitored in NIH/3T3 and HaCaT cells, representing fibroblasts and keratinocytes, respectively. Cell incubation with A151 ODN, Exo3D/2D, or A151-loaded Exo3D/2D for 24 h, allowed the determination of the levels of NO, TNF-α, IL-10, TGF-β, IL-1α, CCL-2 and VEGF-α (Figs. 5 and 6).
Fig. 5.
A151 ODN loaded exosomes mediate the production of wound healing-related trophic factors by fibroblasts (NIH/3T3 cell line). Nitric oxide (NO), TNF-α and IL-10 secretions were determined by Griess assay and ELISA, respectively. TGF-β, IL-1α, CCL-2 and VEGF-α expressions, relative to β-actin, were quantified by RT-qPCR. Cells maintained in cell culture medium were used as control (CTRL). Data is shown as mean ± SEM. n = 2. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. Abbreviations: Exo3D - exosomes derived from UC-MSC cultured in 3D conditions; Exo2D - exosomes derived from UC-MSC cultured in 2D conditions; A151 ODN - immunosuppressor A151 oligodeoxynucleotide; A151-loaded Exo3D - exosomes produced under 3D culture conditions loaded with A151 ODN; A151-loaded Exo2D - exosomes produced under 2D culture conditions loaded with A151 ODN.
Fig. 6.
A151 ODN loaded exosomes mediate the release of wound healing-related trophic factors by keratinocytes (HaCaT cell line). Nitric oxide (NO), TNF-α and IL-10 secretions were determined by Griess assay and ELISA, respectively. TGF-β, IL-1α, CCL-2 and VEGF-α expressions, relative to β-actin, were quantified by RT-qPCR. Cells maintained in cell culture medium were used as control (CTRL). Data is shown as mean ± SEM. n = 2. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.Abbreviations: Exo3D - exosomes derived from UC-MSC cultured in 3D conditions; Exo2D - exosomes derived from UC-MSC cultured in 2D conditions; A151 ODN - immunosuppressor A151 oligodeoxynucleotide; A151-loaded Exo3D - exosomes produced under 3D culture conditions loaded with A151 ODN; A151-loaded Exo2D - exosomes produced under 2D culture conditions loaded with A151 ODN.
NO production was decreased in NIH/3T3 upon exposure to Exo3D when compared to control. In contrast, A151 ODN either alone or loaded into exosome samples significantly induced NO production (Fig. 5). Similarly, only A151 ODN alone could induce NO secretion in HaCaT cells (Fig. 6).
Moreover, both loaded and non-loaded Exo3D/2D increased up to ∼8-fold the secretion of the inflammatory cytokine TNF-α in NIH/3T3 cells (Fig. 5); while only Exo3D was responsible for a mild increase of TNF-α in HaCaT cells (Fig. 5). The pro-inflammatory cytokine IL-1α mRNA level was also elevated by Exo3D in NIH/3T3 and HaCaT cells (∼3-fold; Figs. 5 and 6). However, the loaded form of Exo3D showed a suppressive activity of TNF-α in NIH/3T3 cells and of IL-1α in HaCaT cells when compared to naïve Exo3D (Figs. 5 and 6).
Conversely, secretion of the anti-inflammatory cytokine IL-10 was promoted by both Exo3D and A151 ODN alone (∼40%) in fibroblasts (Fig. 5). In the case of keratinocytes, none of the treatments altered IL-10 secretion (Fig. 6).
The production of TGF-β was promoted by all 2D samples (loaded and non-loaded) in fibroblasts, whereas in keratinocytes the same effect was observed as a result of the exposure to both Exo3D samples (loaded and non-loaded; Figs. 5 and 6).
Interestingly, neither A151 ODN alone or naïve Exo2D/3D affected CCL-2 production in fibroblasts nor keratinocytes. However, a significant increase was observed by A151 ODN-loaded forms of both exosomes (Fig. 5) in NIH/3T3. Finally, augmented VEGF-α expression levels were observed in NIH/3T3 cells incubated with Exo3D/2D samples, being Exo3D responsible for the most pronounced effect (Fig. 6).
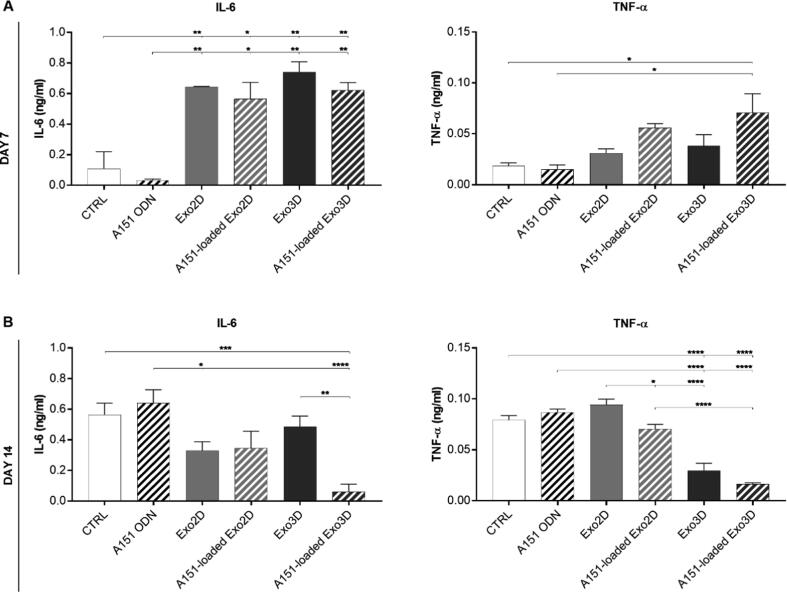
3.6. A151-loaded Exo3D reduced systemic levels of IL-6 and TNF-α cytokines at the late stage of wound healing
Given the excessive inflammatory pathophysiologic condition of chronic wounds preventing their healing, the immunomodulatory effects of loading A151 ODN into exosomes were evaluated in vivo through a wound healing model.
Day 7 blood analyses of mice administered with A151 ODN alone or loaded into exosome samples, as well as, naïve Exo2D and Exo3D resulted in higher IL-6 level in circulation when compared to control-treated group (Fig. 7A). Accordingly, TNF-α levels were also significantly elevated by A151-loaded Exo3D (Fig. 7A). Notably, at day 14, i.e., at a late stage of wound resolution, animals treated with A151-loaded Exo3D showed significantly lower levels of IL-6 and TNF-α (Fig. 7B).
Fig. 7.
A151 ODN loaded exosomes produced under 3D conditions (A151-loaded Exo3D) modulate pro-inflammatory cytokine levels in vivo. Serum levels of IL-6 and TNF-α of mice treated with Exo3D, Exo2D, A151-loaded Exo3D, A151-loaded Exo2D, A151 ODN or PBS (CTRL), as controls, were determined by ELISA at days 7 (A) and 14 (B). Data is shown as mean ± SEM. n = 2–7. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. Abbreviations: Exo3D - exosomes derived from UC-MSC cultured in 3D conditions; Exo2D - exosomes derived from UC-MSC cultured in 2D conditions; A151 ODN - immunosuppressor A151 oligodeoxynucleotide; A151-loaded Exo3D - exosomes produced under 3D culture conditions loaded with A151 ODN; A151-loaded Exo2D - exosomes produced under 2D culture conditions loaded with A151 ODN.
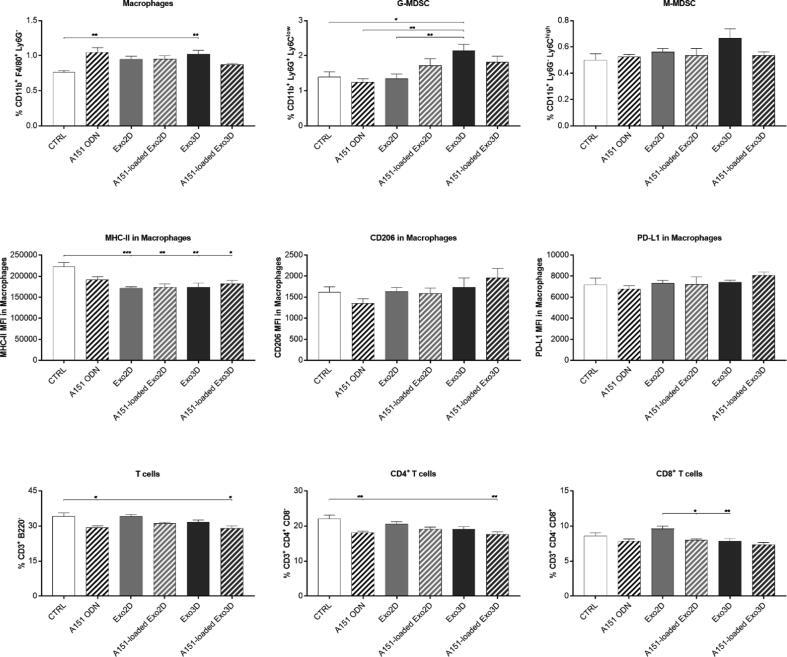
Furthermore, at day 14, all the treatments caused a slight elevation in macrophage numbers, but only statistically significant for A151 ODN alone and Exo3D when compared to the control (Fig. 8). While no alterations were observed in M-MDSC population, Exo3D significantly elevated G-MDSC level (Fig. 8). Interestingly, despite not reducing macrophage population, both loaded and non-loaded Exo3D/2D significantly decreased MHC-II surface expression in macrophages (Figs. 8 and S4). However, no change in the anti-inflammatory macrophage (M2 type) markers CD206 and PD-L1 was observed (Fig. 8). This suggests that especially Exo3D, but also Exo2D, exerted their systemic immunomodulatory effects through G-MDSC induction and reduction of antigen presentation capacity of macrophages rather than promoting M2 type macrophage polarization. Additionally, A151-loaded Exo3D significantly reduced T-cell populations (Figs. 8 and S4), however differential effects were observed in terms of subpopulations. While promoting a noticeable decrease in the CD4+ T-cell population (Figs. 8 and S4), no effect was observed on CD8+ T-cells (Figs. 8 and S4), therefore, showing the specific suppression of CD4+ T-cell type by A151-loaded Exo3D. Overall, all samples were able to cause significant systemic immunosuppression at late stages of wound healing as reflected by the reduction of inflammatory capacity of macrophages and higher G-MDSC numbers. Importantly, only A151-loaded Exo3D were also able to reduce pro-inflammatory cytokine levels in circulation and T-lymphocyte numbers.
Fig. 8.
A151 ODN loaded exosomes produced under 3D conditions (A151-loaded Exo3D) reduced inflammatory capacity of macrophages and T-lymphocyte cell numbers at late stages of wound healing. Differences in spleen macrophage and T-cell populations at day 14 of mice treated with Exo3D, Exo2D, A151-loaded Exo3D, A151-loaded Exo2D, A151 ODN or PBS (CTRL) analysed by flow cytometry. Data is shown as mean ± SEM. n = 5–7. * p < 0.05, ** p < 0.01, *** p < 0.001. Abbreviations: Exo3D - exosomes derived from UC-MSC cultured in 3D conditions; Exo2D - exosomes derived from UC-MSC cultured in 2D conditions; A151 ODN – immunosuppressor A151 oligodeoxynucleotide; A151-loaded Exo3D - exosomes produced under 3D culture conditions loaded with A151 ODN; A151-loaded Exo2D - exosomes produced under 2D culture conditions loaded with A151 ODN.
4. Discussion
The dogma that MSCs promote tissue regeneration only if they are present at the injury site appears to have been demystified. Over the past few years, increasing evidence has uncovered a fundamental role of the MSC secretome as an active substance capable of modulating cellular responses and signalling pathways, thus promoting tissue repair [45], [46]. Important data further disclosed that the MSC secretome itself may present different contents as a consequence of cell origin, cell culture conditions or cellular microenvironment [47]. Indeed, although most of the studies performed so far have been conducted using secretome from MSCs cultured in traditional static monolayer cultures (2D), priming MSCs by key factors or by 3D culturing has also proven to significantly enhance its therapeutic efficacy, namely in the context of cutaneous wound healing [48]. Understanding the factors contained in the MSC secretome responsible for the observed therapeutic effects has led to the study of the different components of the secretome, namely the exosomes. Several publications suggest that exosomes may be the active players in the regeneration process, more than the fraction containing the soluble trophic factors [49], [50]. Exosomes have been reported as being intrinsically involved in several important biological processes such as inflammation, immune response, cell survival, migration and proliferation, angiogenesis, and matrix remodelling [22]. For the treatment of cutaneous wounds, the few studies reporting the use of MSC-derived exosomes describe their contribution through the improvement of the migration of fibroblasts and keratinocytes that impacts on the re-epithelization and ECM deposition processes [19], [31], [33], [51], [52], [53]. However, usually non-healing/chronic wounds suffer from exacerbated inflammatory and immune responses that prevent their healing. In this context, loading molecules into exosomes presents itself as a strategy for modulating these processes that are often jeopardized in the context of disease. As such, we hypothesised that 3D culture conditions and loading with an exogenous immunosuppressive ligand (A151 ODN) could potentiate the functional effects of exosomes secreted by UC-MSCs, and ultimately support their application for the treatment of cutaneous wounds.
In a first approach, exosomes derived from UC-MSC 3D and 2D cultures were characterised concerning size distribution and surface proteins, both before and after A151 ODN loading. Although the morphology of UC-MSC-derived exosomes was influenced by the cell culture system and loading methodology, the presence of CD63, CD9 and CD81 exosome surface markers was not compromised, enabling its use for in vitro and in vivo studies.
Moreover, as changes in cell environment admittedly influence the cell responses including exosome secretion [54], we first evaluated the impact of the different culture conditions on the cargo of the UC-MSC exosomes and its effect in wound healing. It has been previously reported that MSCs cultured as spheroids showed induced expression of genes related to the adhesion to endothelial cells, the inhibition of tumour properties and the modulation of inflammatory reactions, namely CXCR-4, IL-24 or TSG-6 and PGE2, respectively [55], [56], [57], [58]. Key proteins for tissue regeneration, such as VEGF-α, TGF-β1, FGF-2, KGF, EGF and HGF, have also been identified in the secretome of UC-MSCs cultured under 3D conditions [12], [24]. Herein, 3D culture conditions led to a higher protein number and wider protein diversity contained within the exosomes than the 2D environment as shown by proteomic analysis. Moreover, despite most of the proteins is linked to essential and basic cellular metabolic and structure, a deeper analysis focusing on biological processes characteristic of the wound healing phases unveiled distinct proteomic profiles between Exo3D and that of Exo2D. Exo3D contained unique proteins associated to chemotaxis and homing (e.g., CD151, LIF, RHOG, TGFβ and WNT5A), and ECM formation (e.g., ITGB1, PLAUR and VCAN); whereas no proteins related to these biological functions were uniquely identified in Exo2D. Both Exo3D and Exo2D shared proteins associated with inflammation, cell migration and ECM organization, such as IL6, or VCL, however CD151, S100A10, TGFβ, MMP1, MMP14, MMP3 were specifically found in Exo3D while CSF1 and ITGA4 in Exo2D. Importantly, similar observations have been described by several authors that further linked exosomes' MoA with AKT, ERK, Notch, STAT-3, and Wnt/β-catenin signalling pathways [22]. IL-6 has shown to be present in exosomes and to display different roles during wound healing [59], [60], [61], [62]. Shabbir et al. also reported that MSC exosomes were able to activate signalling pathways Akt, ERK, and STAT3 that mediated these events by the secretion of HGF, IGF-1, NGF and SDF-1 [33]. On the other hand, Fang et al described that UC-MSC exosomes negatively regulated TGF-β/SMAD-2 pathway in fibroblasts, which culminated in faster wound closure with reduced scar tissue formation [52].
The effect of the different proteomic profiles between Exo3D and Exo2D was further demonstrated in vitro and in vivo. Exo3D showed significantly higher mitogenic activity over fibroblasts and when compared to Exo2D; whereas both exosome samples resulted in enhanced motogenicity of keratinocytes. Furthermore, although either Exo2D or Exo3D stimulated the cytokine production of TGF-β and IL-1α in keratinocytes, and of TNF-α and VEGF-α in fibroblasts, only Exo3D showed a marked effect on fibroblasts through the secretion of IL-1α and IL-10. Along with TNF-α, IL-1α secreted mainly by keratinocytes but also by fibroblasts have been described to be elevated during inflammation, and to have a supportive role on re-epithelization [63], [64]. On the other hand, TGF-β is responsible for the induction of numerous processes during wound healing including granulation tissue formation, keratinocyte proliferation and migration, collagen production and deposition [65]. Also, VEGF, which is a recognized mediator of angiogenesis, is responsible for promoting immune cell recruitment to the wound [63]. Finally, the anti-inflammatory cytokine IL-10 has been described to promote scarless healing [66]. As such, the secretion of IL-10 by fibroblasts exposed to Exo3D could foresee the better resolution of the wounds observed in the wound healing in vivo model. In fact, Exo3D-treated wounds showed a reduced wound area, improved re-epithelization and maturation of the granulation tissue when compared to the other treatments.
Several other factors counteracting the exacerbated inflammatory and immune responses typical of chronic wounds are also implicated in the wound healing process. Nitric oxide (NO) is recognized by its role in the wound healing early stages, but the inhibition of NO synthase enzymes is associated to pathologic conditions. Lower NO levels have been found in chronic wounds caused by diabetes or malnutrition [67]. Similarly, the monocyte/macrophage chemoattractant CCL-2 (or MCP-1), which promotes macrophage infiltration to the wound, is also lacking in chronic wounds [68]. On the other hand, persistent high levels of TNF-α are associated with delayed wound healing [69], [70]. Huang et al. [70] reported that treating wounds of diabetic rats with TNF-α antagonist improved the wound healing process by promoting keratinocyte migration and reducing M1 type macrophages infiltration. Herein, the culture conditions did not exert a significant effect on the secretion of any of these factors by NIH/3T3 or HaCaT cells. However, loading of A151 ODN into exosomes increased the secretion of NO, TNF-α and CCL-2 by fibroblasts evidencing that the therapeutic effect of exosomes could be potentiated by A151 ODN. In fact, in vivo, the immunomodulatory treatments appeared to exacerbate the inflammatory phase of wound healing, evident by the elevated levels of IL-6 and TNF-α, at an early stage, i.e., day 7. This response has been reported as essential so that healing can occur [59]. However, at day 14, the animals treated with A151-loaded Exo3D showed significant lower levels of serum IL-6 and TNF-α. Besides regulating leukocyte infiltration and angiogenesis [60], low levels of IL-6 at wound resolution state are related to collagen remodelling thus promoting a scarless tissue upon resolution [61]. At the cellular level, loading with an immunosuppressive ligand further resulted in the systemic suppression of immune cells at the late stage. Both A151 ODN and A151-loaded exosomes decreased MHC-II expression, hence the inflammatory capacities of macrophages. Exo3D also elevated the granulocytic MDSC population while A151 ODN and A151-loaded Exo3D decreased CD4+ T-cell numbers. In fact, MDSCs have shown to enhance cutaneous wound healing by inflammatory modulation, through their immunosuppressive function, but also by promoting cell proliferation upon fibrocyte differentiation [71]. Overall, these results imply that subcutaneous injection of A151-loaded Exo3D can cause systemic immunosuppression at the late stages of wound healing which may help prevent the development of non-closure chronic wounds.
5. Conclusion
This study identifies exosomes as being the active substance in MSCs and highlights the importance of the culture conditions for the production of improved cell-based therapeutic products by revealing the enriched protein cargo of Exo3D. It also establishes that the exosomes play a critical role in cutaneous wound healing in vitro and in vivo and further suggest the exosomes’ MoA by identifying the abundant proteins within their cargo. It shows that TGF-β, ITGA1-3/5, IL-6, CDC151, S100A10 and Wnt5α are specifically present in Exo3D supporting the observed Exo3D effects in improving cell migration, viability, and proliferation in vitro, and wound resolution promotion in vivo, the key properties of cutaneous wound healing. Importantly, the present study demonstrates for the first time that loading an immunosuppressive ODN on the harvested exosomes might aid in wound healing by mediating the production of wound healing-related trophic factors from fibroblasts and keratinocytes and via systemic immunosuppression at the late stages of wound healing that unblocks the non-healing process of chronic wounds. As such, exosomes derived from UC-MSCs 3D cultures loaded with exogenous A151 ODN may represent itself as a noncellular off-the-shelf therapeutic modality for non-healing wound treatment.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD025633 (http://www.ebi.ac.uk/pride/archive/projects/PXD025633).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This study was partially supported by FCT (PTDC/MED-TOX/29183/2017, PTDC/MED-QUI/31721/2017 and PTDC/SAU-SER/30197/2017), by strategic funding for iMed.ULisboa (UIDB/04138/2020 and UIDP/04138/2020), for UnIC (UID/00051/2020) and for iBiMED (UID/BIM/04501/2020), and by European Union, Quadro de Referência Estratégico Nacional (QREN), Fundo Europeu de Desenvolvimento Regional (FEDER) and Programa Operacional Fatores de Competitividade (COMPETE) (POCI-01-0145-FEDER-007628 and LISBOA-01-0145- FEDER-030197). VIB Proteomics Core and COST Actions (EU Framework Programme Horizon 2020) CA16113 and CA16119 are also acknowledged. The authors thank Dr. Tânia Carvalho for her valuable assistance on histological analysis. SPC also acknowledges EFIS-IL short-term fellowship.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2022.01.013.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Martin P. Wound Healing-Aiming for Perfect Skin Regeneration. Science. 1997;276(80):75–81. doi: 10.1126/science.276.5309.75. [DOI] [PubMed] [Google Scholar]
- 2.Lindholm C., Searle R. Wound management for the 21st century: combining effectiveness and efficiency. Int Wound J. 2016;13:5–15. doi: 10.1111/iwj.12623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duscher D., Barrera J., Wong V.W., Maan Z.N., Whittam A.J., Januszyk M., et al. Stem Cells in Wound Healing: The Future of Regenerative Medicine? A Mini-Review. Gerontology. 2016;62(2):216–225. doi: 10.1159/000381877. [DOI] [PubMed] [Google Scholar]
- 4.Frykberg R.G., Banks J. Challenges in the Treatment of Chronic Wounds. Adv Wound Care. 2015;4(9):560–582. doi: 10.1089/wound.2015.0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han G., Ceilley R. Chronic Wound Healing: A Review of Current Management and Treatments. Adv Ther. 2017;34(3):599–610. doi: 10.1007/s12325-017-0478-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones V., Grey J.E., Harding K.G. Wound dressings. BMJ. 2006;332(7544):777–780. doi: 10.1136/bmj.332.7544.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun B.K., Siprashvili Z., Khavari P.A. Advances in skin grafting and treatment of cutaneous wounds. Science (80-) 2014;346(6212):941–945. doi: 10.1126/science.1253836. [DOI] [PubMed] [Google Scholar]
- 8.Smiell J.M., Wieman T.J., Steed D.L., Perry B.H., Sampson A.R., Schwab B.H. Efficacy and safety of becaplermin (recombinant human platelet-derived growth factor-BB) in patients with nonhealing, lower extremity diabetic ulcers: a combined analysis of four randomized studies. Wound Repair Regen. 1999;7(5):335–346. doi: 10.1046/j.1524-475X.1999.00335.x. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y., Dulchavsky D.S., Gao X., Kwon D., Chopp M., Dulchavsky S., et al. Wound Repair by Bone Marrow Stromal Cells through Growth Factor Production. J Surg Res. 2006;136(2):336–341. doi: 10.1016/j.jss.2006.07.037. [DOI] [PubMed] [Google Scholar]
- 10.Miranda J.P., Camões S.P., Gaspar M.M., Rodrigues J.S., Carvalheiro M., Bárcia R.N., et al. The Secretome Derived From 3D-Cultured Umbilical Cord Tissue MSCs Counteracts Manifestations Typifying Rheumatoid Arthritis. Front Immunol. 2019;10 doi: 10.3389/fimmu.2019.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bárcia R.N., Santos J.M., Filipe M., Teixeira M., Martins J.P., Almeida J., et al. What Makes Umbilical Cord Tissue-Derived Mesenchymal Stromal Cells Superior Immunomodulators When Compared to Bone Marrow Derived Mesenchymal Stromal Cells? Stem Cells Int. 2015;2015:1–14. doi: 10.1155/2015/583984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santos J.M., Camões S.P., Filipe E., Cipriano M., Barcia R.N., Filipe M., et al. Three-dimensional spheroid cell culture of umbilical cord tissue-derived mesenchymal stromal cells leads to enhanced paracrine induction of wound healing. Stem Cell Res Ther. 2015;6(1) doi: 10.1186/s13287-015-0082-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Becker A.D., Riet I.V. Homing and migration of mesenchymal stromal cells: How to improve the efficacy of cell therapy? World J Stem Cells. 2016;8(3):73. doi: 10.4252/wjsc.v8.i3.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Legaki E., Roubelakis M.G., Theodoropoulos G.E., Lazaris A., Kollia A., Karamanolis G., et al. Therapeutic Potential of Secreted Molecules Derived from Human Amniotic Fluid Mesenchymal Stem/Stroma Cells in a Mice Model of Colitis. Stem Cell Rev Reports. 2016;12(5):604–612. doi: 10.1007/s12015-016-9677-1. [DOI] [PubMed] [Google Scholar]
- 15.Zagoura D.S., Roubelakis M.G., Bitsika V., Trohatou O., Pappa K.I., Kapelouzou A., et al. Therapeutic potential of a distinct population of human amniotic fluid mesenchymal stem cells and their secreted molecules in mice with acute hepatic failure. Gut. 2012;61(6):894–906. doi: 10.1136/gutjnl-2011-300908. [DOI] [PubMed] [Google Scholar]
- 16.Teixeira FG, Carvalho MM, Panchalingam KM, Rodrigues AJ, Mendes-Pinheiro B, Anjo S, et al. Impact of the Secretome of Human Mesenchymal Stem Cells on Brain Structure and Animal Behavior in a Rat Model of Parkinson’s Disease. Stem Cells Transl Med 2017;6:634–46. 10.5966/sctm.2016-0071. [DOI] [PMC free article] [PubMed]
- 17.Hassan Famian M, Montazer Saheb S, Montaseri A. Conditioned Medium of Wharton’s Jelly Derived Stem Cells Can Enhance the Cartilage Specific Genes Expression by Chondrocytes in Monolayer and Mass Culture Systems. Adv Pharm Bull 2017;7:123–30. 10.15171/apb.2017.016. [DOI] [PMC free article] [PubMed]
- 18.Sevivas N., Teixeira F.G., Portugal R., Araújo L., Carriço L.F., Ferreira N., et al. Mesenchymal Stem Cell Secretome: A Potential Tool for the Prevention of Muscle Degenerative Changes Associated With Chronic Rotator Cuff Tears. Am J Sports Med. 2017;45(1):179–188. doi: 10.1177/0363546516657827. [DOI] [PubMed] [Google Scholar]
- 19.Camões SP, Santos JM, Carvalho F, Miranda JP. Mesenchymal Stem Cells for Cutaneous Wound Healing. In: Rodrigues G, Roelen BAJ, editors. Concepts Appl. Stem Cell Biol., 2020, p. 247–67. 10.1007/978-3-030-43939-2_13. [DOI]
- 20.Jahoda C.AB., Reynolds A.J. Hair follicle dermal sheath cells: unsung participants in wound healing. Lancet. 2001;358(9291):1445–1448. doi: 10.1016/S0140-6736(01)06532-1. [DOI] [PubMed] [Google Scholar]
- 21.Hu M.S., Borrelli M.R., Lorenz H.P., Longaker M.T., Wan D.C. Mesenchymal Stromal Cells and Cutaneous Wound Healing: A Comprehensive Review of the Background, Role, and Therapeutic Potential. Stem Cells Int. 2018;2018:1–13. doi: 10.1155/2018/6901983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Casado-Díaz A., Quesada-Gómez J.M., Dorado G. Extracellular Vesicles Derived From Mesenchymal Stem Cells (MSC) in Regenerative Medicine: Applications in Skin Wound Healing. Front Bioeng Biotechnol. 2020;8:1–19. doi: 10.3389/fbioe.2020.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoang D.H., Nguyen T.D., Nguyen H.-P., Nguyen X.-H., Do P.T.X., Dang V.D., et al. Differential Wound Healing Capacity of Mesenchymal Stem Cell-Derived Exosomes Originated From Bone Marrow, Adipose Tissue and Umbilical Cord Under Serum- and Xeno-Free Condition. Front Mol Biosci. 2020;7 doi: 10.3389/fmolb.2020.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miranda J.P., Filipe E., Fernandes A.S., Almeida J.M., Martins J.P., De La Fuente A., et al. The Human Umbilical Cord Tissue-Derived MSC Population UCX ® Promotes Early Motogenic Effects on Keratinocytes and Fibroblasts and G-CSF-Mediated Mobilization of BM-MSCs when Transplanted In Vivo. Cell Transplant. 2015;24(5):865–877. doi: 10.3727/096368913X676231. [DOI] [PubMed] [Google Scholar]
- 25.Wu P., Zhang B., Shi H., Qian H., Xu W. MSC-exosome: A novel cell-free therapy for cutaneous regeneration. Cytotherapy. 2018;20(3):291–301. doi: 10.1016/j.jcyt.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 26.Shi Y., Shi Hui, Nomi Adnan, Lei-lei Zhang, Zhang Bin, Qian Hui. Mesenchymal stem cell–derived extracellular vesicles: a new impetus of promoting angiogenesis in tissue regeneration. Cytotherapy. 2019;21(5):497–508. doi: 10.1016/j.jcyt.2018.11.012. [DOI] [PubMed] [Google Scholar]
- 27.Lai R.C., Arslan F., Lee M.M., Sze N.S.K., Choo A., Chen T.S., et al. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res. 2010;4(3):214–222. doi: 10.1016/j.scr.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Z., Yang J., Yan W., Li Y., Shen Z., Asahara T. Pretreatment of Cardiac Stem Cells With Exosomes Derived From Mesenchymal Stem Cells Enhances Myocardial Repair. J Am Heart Assoc. 2016;5 doi: 10.1161/JAHA.115.002856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cosenza S., Toupet K., Maumus M., Luz-Crawford P., Blanc-Brude O., Jorgensen C., et al. Mesenchymal stem cells-derived exosomes are more immunosuppressive than microparticles in inflammatory arthritis. Theranostics. 2018;8(5):1399–1410. doi: 10.7150/thno.21072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li T., Yan Y., Wang B., Qian H., Zhang X.u., Shen L.i., et al. Exosomes Derived from Human Umbilical Cord Mesenchymal Stem Cells Alleviate Liver Fibrosis. Stem Cells Dev. 2013;22(6):845–854. doi: 10.1089/scd.2012.0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang B, Wang M, Gong A, Zhang X, Wu X, Zhu Y, et al. HucMSC-Exosome Mediated-Wnt4 Signaling Is Required for Cutaneous Wound Healing. Stem Cells 2015;33:2158–68. 10.1002/stem.1771. [DOI] [PubMed]
- 32.Zhang J., Guan J., Niu X., Hu G., Guo S., Li Q., et al. Exosomes released from human induced pluripotent stem cells-derived MSCs facilitate cutaneous wound healing by promoting collagen synthesis and angiogenesis. J Transl Med. 2015;13(1) doi: 10.1186/s12967-015-0417-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shabbir A., Cox A., Rodriguez-Menocal L., Salgado M., Badiavas E.V. Mesenchymal Stem Cell Exosomes Induce Proliferation and Migration of Normal and Chronic Wound Fibroblasts, and Enhance Angiogenesis In Vitro. Stem Cells Dev. 2015;24(14):1635–1647. doi: 10.1089/scd.2014.0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mardpour S., Hamidieh A.A., Taleahmad S., Sharifzad F., Taghikhani A., Baharvand H. Interaction between mesenchymal stromal cell-derived extracellular vesicles and immune cells by distinct protein content. J Cell Physiol. 2019;234(6):8249–8258. doi: 10.1002/jcp.27669. [DOI] [PubMed] [Google Scholar]
- 35.Gursel I., Gursel M., Yamada H., Ishii K.J., Takeshita F., Klinman D.M. Repetitive Elements in Mammalian Telomeres Suppress Bacterial DNA-Induced Immune Activation. J Immunol. 2003;171(3):1393–1400. doi: 10.4049/jimmunol.171.3.1393. [DOI] [PubMed] [Google Scholar]
- 36.Shirota H., Gursel M., Klinman D.M. Suppressive Oligodeoxynucleotides Inhibit Th1 Differentiation by Blocking IFN-γ- and IL-12-Mediated Signaling. J Immunol. 2004;173(8):5002–5007. doi: 10.4049/jimmunol.173.8.5002. [DOI] [PubMed] [Google Scholar]
- 37.Kaminski J.J., Schattgen S.A., Tzeng T.-C., Bode C., Klinman D.M., Fitzgerald K.A. Synthetic Oligodeoxynucleotides Containing Suppressive TTAGGG Motifs Inhibit AIM2 Inflammasome Activation. J Immunol. 2013;191(7):3876–3883. doi: 10.4049/jimmunol.1300530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yazar V, Kilic G, Bulut O, Canavar Yildirim T, Yagci FC, Aykut G, et al. A suppressive oligodeoxynucleotide expressing TTAGGG motifs modulates cellular energetics through the mTOR signaling pathway. Int Immunol 2020;32:39–48. 10.1093/intimm/dxz059. [DOI] [PMC free article] [PubMed]
- 39.Martins J.P., Santos J.M., Almeida J.M., Filipe M.A., de Almeida M.V.T., Almeida S.C.P., et al. Towards an advanced therapy medicinal product based on mesenchymal stromal cells isolated from the umbilical cord tissue: quality and safety data. Stem Cell Res Ther. 2014;5(1) doi: 10.1186/scrt398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gursel M, Gursel I. Development of CpG ODN Based Vaccine Adjuvant Formulations. In: Thomas S, editor. Vaccine Des. Methods Protoc. Vol. 2 Vaccines Vet. Dis., vol. 1404, New York, NY: Springer New York; 2016, p. 289–98. 10.1007/978-1-4939-3389-1_20. [DOI] [PubMed]
- 41.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 2019;47:D442–50. 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed]
- 42.Miranda J., Bakheit M.A., Liu Z., Yin H., Mu Y., Guo S., et al. Development of a recombinant indirect ELISA for the diagnosis of Theileria sp. (China) infection in small ruminants. Parasitol Res. 2006;98(6):561–567. doi: 10.1007/s00436-005-0105-8. [DOI] [PubMed] [Google Scholar]
- 43.Danaei M., Dehghankhold M., Ataei S., Hasanzadeh Davarani F., Javanmard R., Dokhani A., et al. Impact of Particle Size and Polydispersity Index on the Clinical Applications of Lipidic Nanocarrier Systems. Pharmaceutics. 2018;10(2):57. doi: 10.3390/pharmaceutics10020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andreu Z., Yáñez-Mó M. Tetraspanins in extracellular vesicle formation and function. Front Immunol. 2014;5:442. doi: 10.3389/fimmu.2014.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gunawardena T.N.A., Rahman M.T., Abdullah B.J.J., Abu Kasim N.H. Conditioned media derived from mesenchymal stem cell cultures: The next generation for regenerative medicine. J Tissue Eng Regen Med. 2019;13(4):569–586. doi: 10.1002/term.2806. [DOI] [PubMed] [Google Scholar]
- 46.Konala V.B.R., Mamidi M.K., Bhonde R., Das A.K., Pochampally R., Pal R. The current landscape of the mesenchymal stromal cell secretome: A new paradigm for cell-free regeneration. Cytotherapy. 2016;18(1):13–24. doi: 10.1016/j.jcyt.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ferreira J.R., Teixeira G.Q., Santos S.G., Barbosa M.A., Almeida-Porada G., Gonçalves R.M. Mesenchymal stromal cell secretome: Influencing therapeutic potential by cellular pre-conditioning. Front Immunol. 2018;9:1–17. doi: 10.3389/fimmu.2018.02837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu G.-Y., Liu Y., Lu Y., Qin Y.-R., Di G.-H., Lei Y.-H., et al. Short-term memory of danger signals or environmental stimuli in mesenchymal stem cells: implications for therapeutic potential. Cell Mol Immunol. 2016;13(3):369–378. doi: 10.1038/cmi.2015.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lai R.C., Chen T.S., Lim S.K. Mesenchymal stem cell exosome: a novel stem cell-based therapy for cardiovascular disease. Regen Med. 2011;6(4):481–492. doi: 10.2217/rme.11.35. [DOI] [PubMed] [Google Scholar]
- 50.EL Andaloussi S., Mäger I., Breakefield X.O., Wood M.J.A. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12(5):347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 51.Hu L., Wang J., Zhou X., Xiong Z., Zhao J., Yu R., et al. Exosomes derived from human adipose mensenchymal stem cells accelerates cutaneous wound healing via optimizing the characteristics of fibroblasts. Sci Rep. 2016;6(1) doi: 10.1038/srep32993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fang S, Xu C, Zhang Y, Xue C, Yang C, Bi H, et al. Umbilical Cord-Derived Mesenchymal Stem Cell-Derived Exosomal MicroRNAs Suppress Myofibroblast Differentiation by Inhibiting the Transforming Growth Factor-β/SMAD2 Pathway During Wound Healing. Stem Cells Transl Med 2016;5:1425–39. 10.5966/sctm.2015-0367. [DOI] [PMC free article] [PubMed]
- 53.Wang X., Jiao Y., Pan Y., Zhang L., Gong H., Qi Y., et al. Fetal Dermal Mesenchymal Stem Cell-Derived Exosomes Accelerate Cutaneous Wound Healing by Activating Notch Signaling. Stem Cells Int. 2019;2019:1–11. doi: 10.1155/2019/2402916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vizoso F., Eiro N., Cid S., Schneider J., Perez-Fernandez R. Mesenchymal Stem Cell Secretome: Toward Cell-Free Therapeutic Strategies in Regenerative Medicine. Int J Mol Sci. 2017;18:1852. doi: 10.3390/ijms18091852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frith J.E., Thomson B., Genever P.G. Dynamic Three-Dimensional Culture Methods Enhance Mesenchymal Stem Cell Properties and Increase Therapeutic Potential. Tissue Eng Part C Methods. 2010;16(4):735–749. doi: 10.1089/ten.tec.2009.0432. [DOI] [PubMed] [Google Scholar]
- 56.Bartosh T.J., Ylostalo J.H., Mohammadipoor A., Bazhanov N., Coble K., Claypool K., et al. Aggregation of human mesenchymal stromal cells (MSCs) into 3D spheroids enhances their antiinflammatory properties. Proc Natl Acad Sci. 2010;107(31):13724–13729. doi: 10.1073/pnas.1008117107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Potapova I.A., Brink P.R., Cohen I.S., Doronin S.V. Culturing of Human Mesenchymal Stem Cells as Three-dimensional Aggregates Induces Functional Expression of CXCR4 That Regulates Adhesion to Endothelial Cells. J Biol Chem. 2008;283(19):13100–13107. doi: 10.1074/jbc.M800184200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.YlÖstalo J.H., Bartosh T.J., Coble K., Prockop D.J. Human Mesenchymal Stem/Stromal Cells Cultured as Spheroids are Self-activated to Produce Prostaglandin E2 that Directs Stimulated Macrophages into an Anti-inflammatory Phenotype. Stem Cells. 2012;30(10):2283–2296. doi: 10.1002/stem.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gallucci R.M., Sugawara T., Yucesoy B., Berryann K., Simeonova P.P., Matheson J.M., et al. Interleukin-6 Treatment Augments Cutaneous Wound Healing in Immunosuppressed Mice. J Interf Cytokine Res. 2001;21(8):603–609. doi: 10.1089/10799900152547867. [DOI] [PubMed] [Google Scholar]
- 60.Lin Z.-Q., Kondo T., Ishida Y., Takayasu T., Mukaida N. Essential involvement of IL-6 in the skin wound-healing process as evidenced by delayed wound healing in IL-6-deficient mice. J Leukoc Biol. 2003;73(6):713–721. doi: 10.1189/jlb.0802397. [DOI] [PubMed] [Google Scholar]
- 61.Liechty K.W., Adzick N.S., Crombleholme T.M. Diminished interleukin 6 (IL-6) production during scarless human fetal wound repair. Cytokine. 2000;12(6):671–676. doi: 10.1006/cyto.1999.0598. [DOI] [PubMed] [Google Scholar]
- 62.Yew T.-L., Hung Y.-T., Li H.-Y., Chen H.-W., Chen L.-L., Tsai K.-S., et al. Enhancement of Wound Healing by Human Multipotent Stromal Cell Conditioned Medium: The Paracrine Factors and p38 MAPK Activation. Cell Transplant. 2011;20(5):693–706. doi: 10.3727/096368910X550198. [DOI] [PubMed] [Google Scholar]
- 63.Behm B., Babilas P., Landthaler M., Schreml S. Cytokines, chemokines and growth factors in wound healing. J Eur Acad Dermatol Venereol. 2012;26:812–820. doi: 10.1111/j.1468-3083.2011.04415.x. [DOI] [PubMed] [Google Scholar]
- 64.Eming S.A., Martin P., Tomic-Canic M. Wound repair and regeneration: Mechanisms, signaling, and translation. Sci Transl Med. 2014;6(265) doi: 10.1126/scitranslmed.3009337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barrientos S., Stojadinovic O., Golinko M.S., Brem H., Tomic-Canic M. Growth factors and cytokines in wound healing. Wound Repair Regen. 2008;16:585–601. doi: 10.1111/j.1524-475X.2008.00410.x. [DOI] [PubMed] [Google Scholar]
- 66.King A., Balaji S., Le L.D., Crombleholme T.M., Keswani S.G. Regenerative Wound Healing: The Role of Interleukin-10. Adv Wound Care. 2014;3(4):315–323. doi: 10.1089/wound.2013.0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Luo J.-D., Chen A.F. Nitric oxide: a newly discovered function on wound healing. Acta Pharmacol Sin. 2005;26(3):259–264. doi: 10.1111/j.1745-7254.2005.00058.x. [DOI] [PubMed] [Google Scholar]
- 68.Wood S., Jayaraman V., Huelsmann E.J., Bonish B., Burgad D., Sivaramakrishnan G., et al. Pro-inflammatory chemokine CCL2 (MCP-1) promotes healing in diabetic wounds by restoring the macrophage response. PLoS ONE. 2014;9(3):e91574. doi: 10.1371/journal.pone.0091574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ashcroft G.S., Jeong M.-J., Ashworth J.J., Hardman M., Jin W., Moutsopoulos N., et al. Tumor necrosis factor-alpha (TNF-α) is a therapeutic target for impaired cutaneous wound healing. Wound Repair Regen. 2012;20(1):38–49. doi: 10.1111/j.1524-475X.2011.00748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang S.-M., Wu C.-S., Chiu M.-H., Wu C.-H., Chang Y.-T., Chen G.-S., et al. High glucose environment induces M1 macrophage polarization that impairs keratinocyte migration via TNF-α: An important mechanism to delay the diabetic wound healing. J Dermatol Sci. 2019;96(3):159–167. doi: 10.1016/j.jdermsci.2019.11.004. [DOI] [PubMed] [Google Scholar]
- 71.Ou L., Shi Y., Dong W., Liu C., Schmidt T.J., Nagarkatti P., et al. Kruppel-like factor KLF4 facilitates cutaneous wound healing by promoting fibrocyte generation from myeloid-derived suppressor cells. J Invest Dermatol. 2015;135(5):1425–1434. doi: 10.1038/jid.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD025633 (http://www.ebi.ac.uk/pride/archive/projects/PXD025633).