Graphical abstract
Keywords: Druggable protein, Convolutional neural network, Recurrent neural network, Bidirectional long short-term memory, Deep neural network, Model visualization
Highlights
-
•
We developed the first deep learning-based druggable protein classifier for fast and accurate identification of potential druggable proteins.
-
•
Experimental results on a standard dataset demonstrate that the prediction performance of deep learning model is comparable to those of existing methods.
-
•
We visualized the representations of druggable proteins learned by deep learning models, which helps us understand how they work.
-
•
Our analysis reconfirms that the attention mechanism is especially useful for explaining deep learning models.
Abstract
Introduction
The top priority in drug development is to identify novel and effective drug targets. In vitro assays are frequently used for this purpose; however, traditional experimental approaches are insufficient for large-scale exploration of novel drug targets, as they are expensive, time-consuming and laborious. Therefore, computational methods have emerged in recent decades as an alternative to aid experimental drug discovery studies by developing sophisticated predictive models to estimate unknown drugs/compounds and their targets. The recent success of deep learning (DL) techniques in machine learning and artificial intelligence has further attracted a great deal of attention in the biomedicine field, including computational drug discovery.
Objectives
This study focuses on the practical applications of deep learning algorithms for predicting druggable proteins and proposes a powerful predictor for fast and accurate identification of potential drug targets.
Methods
Using a gold-standard dataset, we explored several typical protein features and different deep learning algorithms and evaluated their performance in a comprehensive way. We provide an overview of the entire experimental process, including protein features and descriptors, neural network architectures, libraries and toolkits for deep learning modelling, performance evaluation metrics, model interpretation and visualization.
Results
Experimental results show that the hybrid model (architecture: CNN-RNN (BiLSTM) + DNN; feature: dictionary encoding + DC_TC_CTD) performed better than the other models on the benchmark dataset. This hybrid model was able to achieve 90.0% accuracy and 0.800 MCC on the test dataset and 84.8% and 0.703 on a nonredundant independent test dataset, which is comparable to those of existing methods.
Conclusion
We developed the first deep learning-based classifier for fast and accurate identification of potential druggable proteins. We hope that this study will be helpful for future researchers who would like to use deep learning techniques to develop relevant predictive models.
Introduction
Most marketed drugs are small organic molecules that elicit a therapeutic response by binding to a series of biological macromolecules, such as peptides, proteins, and nucleic acids. Within these macromolecules, proteins are currently the first druggable targets for the design and development of new drugs [1] because over 95% of known drug targets are proteins and coordinate approximately 93% of known drug-target interactions [2]. These proteins are usually referred to as “druggable proteins” and contain structures favouring interactions with drugs/compounds, which is where the original definition of protein druggability was born [3]. In view of a purely structural point, druggability is related to the possibility that a drug/compound binds a particular protein target with high affinity where the distance between them is less than 1 µM [4], [5]. Previous studies have indicated that interactions with drugs/compounds can modulate the function of many classes of druggable proteins, including G protein-coupled receptors, ion channels, nuclear hormone receptors, transporters and enzymes [6], [7]. A number of nonenzymes, such as scaffolding, regulatory and structural proteins, and the proteins related to specific protein–protein interactions, have also become binding targets for drugs in recent years [8]. This suggests that there is still a huge demand for the discovery and characterization of drug targets. In addition, these pharmaceutically useful protein targets are heavily involved in underlying mechanisms related to cardiovascular disease, hypertension, immune system functions, and other diseases [9]. Therefore, rapid and accurate identification of potential druggable proteins is a vital first step in drug discovery and development.
Although scientists and researchers have made great efforts to achieve this goal in recent decades, developing new drugs remains a challenging task due to the complexity, cost, time consumption and low success rate of traditional experimental methods [10]. In general, the development of a single new drug often takes over 12 years and costs on average 2.6 billion USD [11], while only 4% of drug development plans are eventually authorized to produce licenced drugs [12], [13]. Moreover, modern drug development faces a constant increase in costs, while the number of newly approved drugs per year is declining [14]. One of the main reasons for this phenomenon is the lack or incorrect choice of drug targets [15]. Thus, the search for appropriate druggable proteins from the human proteome is a key point in the study of drug targets.
In recent years, various computational strategies for predicting potential druggable proteins have emerged, which commonly use the sequence, structural, and functional features of proteins as input [5], [16], [17], [18], [19] but also system-level properties such as network topological features [20], [21], [22], [23]. Various machine learning (ML) algorithms have been employed to develop in silico models, including support vector machine (SVM) [24], [25], [26], [27], neural network (NN) [28], [29], naive Bayes (NB) [30], [31], logistic regression (LR) [32], hidden Markov model (HMM) [33], random forest (RF) [34], and ensemble methods [35], [36], [37]. In 2016, through in-depth data mining, Jamali et al. proposed an enlarged benchmark dataset for algorithm development, starting a new round of studies to predict potential druggable proteins [38]. They used 443 sequence-derived protein attributes to represent both druggable and nondruggable proteins and six conventional machine learning algorithms to build the predictors, of which NN yielded the highest accuracy of 89.78%. Subsequently, Sun et al. performed a comprehensive comparison of different combinations of protein features and machine learning algorithms using the same dataset in 2018 [39]. Interestingly, they found that the LQL-v using NN obtained the highest training accuracy of 91.10%, but the overlapped 3-gram word2vec using LR afforded the best test accuracy of 89.55%, which was selected as the final predictor. In 2019, Lin et al. chose pseudo-amino acid composition, dipeptide composition and reduced sequence of proteins as input, and each protein was first transformed into a 591-dimensional vector [40]. Then, they exploited the genetic algorithm (GA) to optimize the feature set, and the Bagging algorithm to perform ensemble learning with multiple SVM predictors. Using the 5-fold cross-validation, the Bagging-SVM ensemble predictor provided the best overall accuracy of 93.78%.
As a subdiscipline of ML, deep learning (DL) was developed from artificial neural networks (ANNs) and has grown rapidly in the past two decades [41]. Different from traditional ML algorithms, DL can automatically extract features and learn patterns from various types of data, which makes it particularly suitable for handling large and complex data. The past decades have witnessed an unprecedented number of research studies considering the application of deep learning in the fields of speech recognition, computer vision, and natural language processing [42]. Furthermore, DL algorithms have become typical approaches for drug activity prediction, drug-target interaction identification, medical image analysis, and lead molecule discovery in the field of drug development and biomedicine [43], [44] and for sequence analysis, structure prediction, biomolecular property and function prediction, and systems biology in the field of bioinformatics [45], [46], [47]. Convolutional neural networks (CNNs), recurrent neural networks (RNNs) and deep neural networks (DNNs) are the three most commonly used deep learning architectures. Although deep learning algorithms have been widely applied to drug discovery [48], [49], genomic sequence analysis [50], and protein sequence and structure classification [51], [52] in recent years, to date, there are no methods or tools available for predicting potential druggable proteins using deep learning.
In this study, our objective is to investigate novel deep learning techniques with potential applications for facilitating the discovery of innovative targets. We describe the main strategies of deep learning methods, including protein descriptors, neural network architectures, libraries and toolkits for deep learning modelling and gold-standard datasets for system training and benchmarking. Subsequently, we explored the usefulness of DL-based approaches for predicting druggable proteins from amino acid sequences. Experimental results using the benchmark dataset show that the hybrid neural network architecture achieves the best overall prediction performance. The hybrid model that integrates convolutional-recurrent neural networks (CNN-RNNs) and deep neural networks (DNNs) can predict druggable proteins with, on average, 90.0% accuracy, 89.6% F value, 89.0% recall, 90.5% precision, and 0.800 MCC when tested on the test set 10 times. Across the nonredundant blind test, the best model also achieves ACC, F value, recall, PRE, and MCC values of 84.8%, 85.3%, 91.1%, 80.3% and 0.703, respectively, highlighting the robustness of the model. Finally, we explain the predictions of the deep learning models using the self-attention mechanism and projection-based visualization approach. This investigation is the first study to identify potential druggable proteins using deep learning methods, and we hope to provide a new research strategy for future studies of druggable proteins.
Materials and methods
Datasets
With reference to other works, the public standard dataset established by Jamali et al. [38] was used in this work to construct the training and test datasets, where the positive samples (proteins approved as drug targets by FDA) were extracted from the DrugBank database [53], while the negative samples (proteins that cannot be considered drug targets) were collected from Swiss-Prot according to the methods proposed by Li et al. [25] and Bakheet et al. [26]. All sequences were filtered for the removal of identical and similar sequences. In addition, research and experimental proteins known as drug targets of human origin from the DrugBank and Therapeutic Target Database (TTD) [54] and members of their related families were also eliminated from the negative samples. After the above data processing steps, 1,224 druggable proteins and 1,319 nondruggable proteins were retained as the positive and negative samples, respectively. The deep learning models were fitted to a training dataset containing 80% randomly selected proteins, and their predictive performance was compared and evaluated on a test dataset containing the remaining 20%.
An independent test set with a balanced sample size was used to further evaluate the performance of the developed deep learning model. The positive samples and negative samples were obtained from the latest version of the DrugBank database (Version 5.1.8) [55] and a study by Kim et al. [56], respectively. We first removed the sequences present in the standard dataset and then carried out a 20% homology reduction of the remaining sequences using CD-HIT [57]. Any sequence with an amino acid sequence length smaller than 50 AA or greater than 10,000 AA was also omitted. Finally, 224 druggable proteins and 237 nondruggable proteins were retained in the independent test set.
Feature extraction
Word representation of protein sequences
As the two most common encoding methods for deep learning techniques, dictionary and one-hot encoding schemes have been widely used to obtain primary protein sequence information [58], [59]. For the former scheme, each amino acid in a protein sequence is encoded as an ordinal number, and the 20 natural amino acids are sequentially encoded as numbers from 1 to 20 (e.g., ‘A’ is encoded as 1). In this way, each protein sequence is encoded as an L-dimensional numerical vector, where L is the length of the protein. For the latter scheme, the residues in the sequence are encoded as a vector in which only one element is a ‘1′ and the other elements are ‘0′. The ‘1′ in the vector is a representation of the amino acid, and different amino acids use different positions in this vector, so the length of the vectors is 20 as default. In addition, the k-mer operation can be used for encoding methods. For example, each sequence is first divided into different windows with the same length k using the sliding window with a certain stride (usually set as 1), and then the generated windows can be encoded by the above two encoding methods. Since the k-mer operation counts the short peptides of length ‘k’ instead of only residues, it has a long and sparse output vector.
Descriptor representation of protein sequences
When using DL to predict druggable proteins, each protein is represented by a multidimensional numerical vector composed of descriptors that can truly reflect their intrinsic correlation with the target to be predicted. These descriptors define the sequence composition, physicochemical properties and conservation profiles of proteins. Descriptors based on sequence composition reflect the occurrence frequencies of different amino acid types and their combinations in a protein sequence. Descriptors based on physicochemical properties describe protein sequences in terms of the composition, transition and distribution of hydrophobicity, van der Waals volume, polarity, polarizability, charge, secondary structure and solvent accessibility. Descriptors based on conservation profiles reflect the evolutionary information of protein sequences.
Amino acid composition (AAC), dipeptide composition (DC) and tripeptide composition (TC)
Composition is both the simplest and most important descriptor of protein sequences. Different types of compositions, including amino acid, dipeptide and tripeptide compositions, have been proven to be important for the development of classification models in previous studies. For a total of 20 natural amino acid types, the AAC calculates the frequency of each type of amino acid, and a protein sequence is thus converted into a 20-dimensional vector {d1, d2, …, d20}. Dipeptide composition (DC) is calculated using the percentages of the 400 dipeptide combinations {d1, d2, …, d400}. Tripeptide composition (TC) reflects the statistical frequency of any combination of three amino acids, and a protein sequence is converted into an 8000-dimensional vector {d1, d2, …, d8000}.
Composition-transition-distribution (CTD)
As a global sequence descriptor, composition-transition-distribution (CTD) is often used to characterize protein and peptide sequences, encompassing three feature descriptors: composition (C), transition (T) and distribution (D) [60]. The CTD feature represents the amino acid distribution patterns of a specific structural or physicochemical property in a protein sequence. For each physicochemical property, the 20 natural amino acids are classified into three groups. The first composition descriptor set reflects the global percentage of a particular amino acid attribute group in a protein sequence, the second transition descriptor set reflects the percent frequency of transitions between two different groups along the whole protein sequence, and the third distribution descriptor set reflects the distribution patterns of a particular amino acid attribute group in this protein sequence. In CTD, composition, transition and distribution descriptors are encoded as 21-, 21- and 105-dimensional feature vectors, respectively.
Position-specific scoring matrix (PSSM)
In previous studies, the position-specific scoring matrix (PSSM) has been demonstrated to be an effective and powerful feature for encoding the evolutionary information of protein sequences [61], [62]. For a protein sequence, the PSSM is generated by using PSI-BLAST to search against the Swiss-Prot database, with three iterations and an E-value cut-off of 0.001 for multiple sequence alignment. In this process, a matrix consisting of L rows and 20 columns is created, where L is the length of a protein sequence and 20 columns represent the occurrence or substitution of each type of the 20 natural amino acids. Then, a formula is used to make all PSSM matrices size-uniform, which has been described in our earlier study [61]. Ultimately, each protein sequence is transformed into a 20-dimensional vector.
Deep learning methods
There are several different types of neural network architectures related to deep learning. Currently, the most typical and commonly used neural networks include convolutional neural networks (CNNs), recurrent neural networks (RNNs), and deep neural networks (DNNs). In addition, some variants of the above network types, such as the hybrid neural network, consisting of the CNN, RNN and DNN blocks, are frequently employed. The diversity of neural network types and the complexity of neural network architectures have made the deep learning method extremely successful in myriad different fields [42], [63]. An overview of the basic modules of different deep learning architectures is described in detail below.
Convolutional neural networks (CNNs)
CNNs are the most important deep-learning-based approaches for computer vision [64]. The architecture of a typical CNN consists of three types of layers: convolutional, pooling and fully connected layers. The convolutional layer is designed to use the convolution operation for extracting and gathering the information contained in the input data. The commonly used CNN layer is the 2D convolutional layer, which can be described as follows:
| (1) |
where ‘’ is the kernel size, ‘’ is the input with a 2D shape, ‘’ is the bias and ‘’ is the kernel for convolution. In the application of biological sequences, the kernel is simplified to 1D since the sequences do not contain 2D relations, so the representation can be described as:
| (2) |
The 1D kernel can gather the local information from the sequence data but will ignore the information outside the kernel. Pooling is also an important operation in the CNN architecture. The basic principle of the pooling process is to calculate the representative value from fixed-size windows. Thus, the pooling layer can be used to reduce the computational requirements through the network and minimize overlapping by reducing the spatial size of the activation map. In addition to the basic network layers described above, the rectified linear unit (ReLU) activation function and dropout regularization methods are often used in CNNs with the aim of introducing nonlinearities and avoiding overfitting during model training.
Recurrent neural networks (RNNs)
Recurrent neural networks (RNNs) are popular in sequence labelling tasks because they can use previous information in a sequence to process the current input. Long short-term memory (LSTM) neural networks and gated recurrent unit (GRU) neural networks are two special subclasses of RNNs. They are designed to capture sequential information and historical states are used to implement ‘memory’ and ‘forgetting’ mechanisms. For example, in the LSTM, two hidden states, ‘’ and ‘’, are used to update the information along the time ‘’:
| (3) |
| (4) |
| (5) |
where , the generated vectors ‘’ are the temporary vectors, ‘’ is the concatenate operation of two vectors, and ‘’ is the Hadamard product. The ‘memory’ and ‘forgetting’ mechanisms are performed when updating ‘’ in every iteration. The bidirectional LSTM (Bi-LSTM) is an extension of LSTM in which the input sequence is processed forwards and backwards, and then both outputs are concatenated. Bi-LSTM consists of two LSTM layers sharing the same inputs in two directions, but the output ‘’ is determined by the two hidden states ‘’ and ‘’ at the same position in both directions. If no shift is used, the relation is:
| (6) |
where ‘’ is the weight, ‘’ is the activation function, ‘’ is the length of the sequence and ‘’ is the bias.
Deep neural networks (DNNs)
The DNN architecture is the basic framework for deep learning. All deep neural networks were essentially developed based on the traditional machine learning algorithm - artificial neural networks (ANNs), which have been successfully applied in many research and application areas [65], [66], [67], [68]. The main differences between DNNs and ANNs are the number of hidden layers, layer-to-layer connections and feature learning ability when processing different data [69]. In general, the deep neural network is a standard multilayer neural network with more than one hidden layer between the input and output layers, which is commonly called the multilayer perceptron (MLP) [70]. By stacking multiple hidden layers, DNNs can learn high-level abstractions in the input data with a deep architecture composed of multiple nonlinear transformations. Theoretically, DNNs can handle any type of data, but they cannot learn the complex correlations among the features as CNNs and RNNs do. Therefore, for the prediction of druggable proteins, discrete protein descriptors, such as AAC, DC, TC, CTD, and PSSM, are commonly used as inputs to DNNs.
Convolutional-recurrent neural networks (CNN-RNNs)
As the most popular hybrid deep learning methods, CNN-RNNs, which are a kind of quasi-recurrent neural network (QRNN) [71], are proposed to integrate the advantages of CNNs and RNNs and have been successfully applied to various biological applications [72], [73], [74]. The hybrid CNN-RNN leverages the feature extraction capability of CNN and connects the obtained feature representations to an RNN to capture sequential dependencies between the input data.
Model design for druggable protein prediction
In this work, the model was constructed according to the type of input datasets. The sequential data were encoded by either a built-in encoding mechanism using the dictionary representation or directly a one-hot representation. The encoded array was used in CNN/CNN-RNN layers to yield the processed layer information, which contains the local information from the convolution layers and global information from RNN layers. The discrete feature groups, such as AAC, CTD and PSSM, were processed by using DNN layers. The processed sequential features were reshaped to a 1-D array per sample by using a flattening operation, and all of the feature groups had the same dimensions. We first use the discrete feature groups for prediction separately by using a DNN layer as the projection layer for prediction. Afterwards, all of the combinations of sequential and discrete features were employed for prediction by using a concatenation operation in the feature dimension. Hence, we considered CNN, RNN and CNN-RNN as the core architecture of the hybrid model, and different descriptor combinations of DNN were used as extra information concatenated to the last fully connected layer of core architecture. Together with the combination of the five features (AAC, DC, TC, CTD and PSSM), a total of 31 (i.e., 25-1) DNN models were generated for concatenating the core architecture. Since the concatenation of the submodels will increase the parameters of the hybrid model, the learning rate and epochs should be changed according to the scale of the model. After several tests, the number of epochs is determined by the number of models concatenated before the last fully connected layer:
| (7) |
where ‘’ is the number of models to be connected. For example, using a CNN as the core structure, the ACC and CTD of DNNs will concatenate three outputs before the last fully connected layer; thus, ‘’ is 3, and the number of epochs is 192. The learning rate is determined in a similar way, being set to 5e-5 if only one model is used, 2e-5 if two submodels are used, and 5e-6 if three or more submodels are used. First, the performances of three deep learning models (CNN, RNN and CNN-RNN) with two encoding methods (dictionary and one-hot encoding) were compared using the same training dataset to find the optimal core architecture. Then, the best core architecture was chosen to concatenate the DNN models with different combinations of discrete feature groups. To reduce the effect of random factors in sampling and model fitting, each training procedure was repeated 10 times. Finally, we performed 420 model training procedures, of which 110 were used for a single model and 310 for the hybrid models. The results are shown in detail in the following sections.
Visualizing and understanding deep learning methods
Self-attention mechanism
As a common operation, the attention mechanism is widely used in various nature language processing (NLP) transformer models. The attention mechanism can calculate the similarity between two vectors and modify the weights according to the similarity. Additionally, self-attention focuses on the relations of the inputs from a certain layer for reweighting its outputs, and thus, self-attention is usually used as a neural network layer followed by an RNN layer. Depending on different modelling purposes and data characteristics, there are various algorithms to implement the similarity calculation. For example, the following equation shows the additive self-attention mechanism:
| (8) |
where ‘’, ‘’ and ‘’ are weights that could be updated during the epochs and ‘’ and ‘’ are two bias vectors. Self-attention will calculate all similarity between all variables (i.e., features in this work) and generate the attention weights for further calculation. The attention weights can be used for model interpretation and other downstream analyses.
Visualizing the hidden activity of deep learning models
Using interconnected layers, deep learning models can extract information from data and learn (high-dimensional) higher-level representations in the data that are useful for a given task. However, deep learning models are typically used as “black boxes” because it is difficult to know the specific process by which they make predictions. To explain how the deep learning models make a prediction, we need to understand the evolution of the data in each hidden layer of the model. The nonlinear dimensionality reduction technique uniform manifold approximation and projection (UMAP) has shown potential to provide insightful visual feedback about deep learning models [75]. UMAP projections can visualize the relationships not only between learned representations of input data, but also between artificial neurons.
Metrics for performance evaluation of deep learning methods
In a binary classification task, such as the classification models in this work with druggable and nondruggable two labels, the predictive performance of different models is evaluated by the following metrics:
| (9) |
| (10) |
| (11) |
| (12) |
| (13) |
where TP, FN, TN, and FP represent true positives, false negatives, true negatives, and false positives, respectively. Receiver operating characteristic (ROC) and precision-recall (PR) curves show the performance of classification models, and the area under the curve is often used as a metric to evaluate the performance of binary classification problems. The learning curve refers to a plot of the prediction performance versus the training set size. Usually, both the training and test/validation performance are plotted together to diagnose the bias-variance tradeoff and to assess the complexity of the classification models. For each training epoch, the model accuracy and loss of both the training and validation sets are recorded in the accuracy-loss curve.
Deep learning tool for biological sequence classification
AutoBioSeqpy, a Keras-based [76] deep learning tool, is specifically designed for biological sequence classification [77]. This tool is packaged into powerful and easy-to-use software. The users only need to prepare the input dataset as well as the model template. After that, with a single-line command, autoBioSeqpy can automatically encode the sequence, split the dataset, train and evaluate the model, and generate figures from the results. In addition, autoBioSeqpy can design deep learning models in different types and architectures for DNA, RNA and protein sequences. Currently, the autoBioSeqpy tool has been upgraded to version 2.0, which supports more complex model architectures and incorporates model interpretation and visualization features such as the attention mechanism and UMAP. autoBioSeqpy 2.0 is available at https://github.com/jingry/autoBioSeqpy/tree/2.0/.
Results
The performance comparison of different deep learning models
We first evaluated the predictive performance of 11 different deep learning models, including two CNN models, two RNN models, two CNN-RNN models and five DNN models. For a fair comparison, we tuned the hyperparameters and optimized the architectures of each model to obtain the best possible performance with the training dataset. Table 1 shows the best performance achieved by each of the 11 different deep learning models.
Table 1.
Performance comparison of different deep learning models on the training dataset.
| Encoding | Architecture | ACC (%) | F-value (%) | Recall (%) | PRE (%) | MCC |
|---|---|---|---|---|---|---|
| Dictionary | CNN | 86.8 | 86.4 | 86.8 | 86.8 | 0.741 |
| Onehot | CNN | 87.2 | 86.7 | 85.8 | 87.9 | 0.746 |
| Dictionary | RNN (BiLSTM) | 71.8 | 68.3 | 65.5 | 76.7 | 0.463 |
| Onehot | RNN (BiLSTM) | 69.0 | 63.6 | 61.3 | 75.2 | 0.415 |
| Dictionary | CNN-RNN (BiLSTM) | 88.9 | 88.9 | 85.6 | 92.0 | 0.780 |
| Onehot | CNN-RNN (BiLSTM) | 88.3 | 88.1 | 87.4 | 89.3 | 0.768 |
| AAC | DNN | 88.5 | 87.8 | 85.5 | 90.9 | 0.776 |
| DC | DNN | 87.0 | 87.6 | 89.6 | 85.6 | 0.740 |
| TC | DNN | 83.8 | 85.0 | 91.3 | 79.0 | 0.681 |
| CTD | DNN | 68.1 | 71.2 | 89.5 | 61.3 | 0.387 |
| PSSM | DNN | 84.5 | 84.8 | 91.7 | 77.9 | 0.703 |
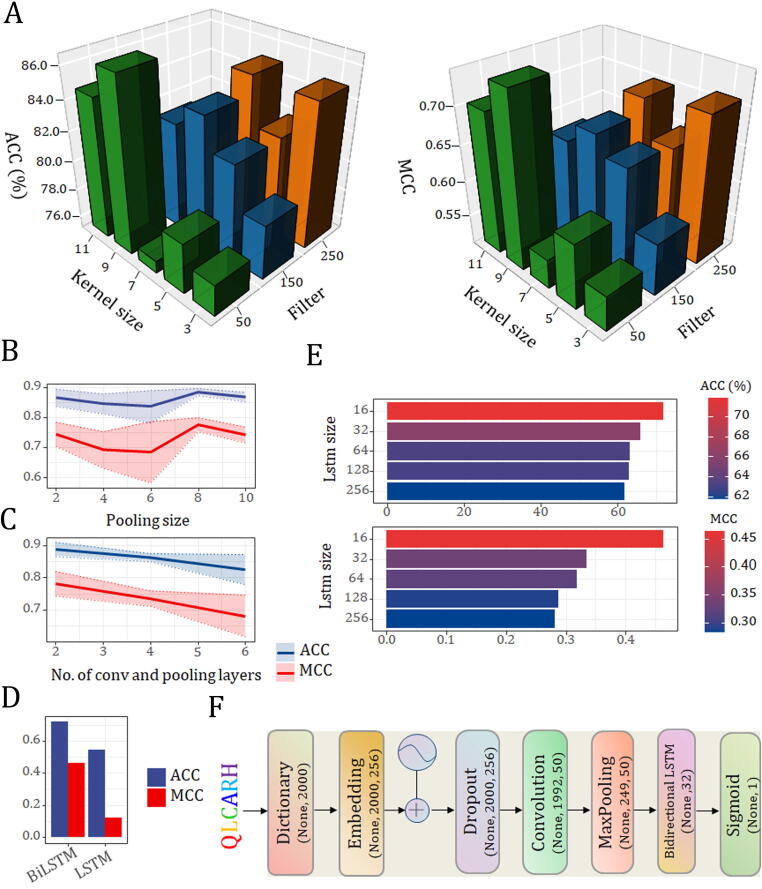
We trained two types of RNN models (LSTM and BiLSTM) on different numbers of hidden cells (Table S1 and S2). Overall, the performance of the RNN (BiLSTM) model was significantly better than that of the RNN (LSTM) model, which again shows that the BiLSTM learns more feature information than the LSTM. When the number of hidden cells was set to 16, the RNN (BiLSTM) model achieved the best performance with MCC values of 0.463 and 0.415 for both encoding methods, respectively. For the convolutional-based models (CNN and CNN-RNN), we mainly optimized their four hyperparameters, including kernel size (3, 5, 7, 9 and 11), number of filters (50, 150 and 250), pooling size (2, 4, 6, 8 and 10) and number of convolutional and pooling layers (2, 3, 4, 5 and 6). Detailed optimization results of the four models are shown in Fig. 1 and Figure S1-S3. The CNN-RNN (BiLSTM) model with the dictionary encoding method obtained the best performance among all core structure candidates, with the highest scores of ACC (88.9%), F value (88.9%), recall (85.6%), PRE (92.0%), and MCC (0.780) (Table 1). This model is constructed from an input layer, an embedding layer, a dropout layer, a convolutional layer, a max pooling layer, a bidirectional LSTM layer, and an output layer (Fig. 1C and 1F). First, the embedding layer transforms the dictionary-encoded input sequence into a 256-dimensional vector representation. A dropout of 0.2 is applied to the embedding layer to prevent overfitting. The convolutional layer has 50 filters, where the kernel size is set to 9 (Fig. 1A). After the convolutional layer, the pooling layer performs max pooling, which outputs the maximum value over a nonoverlapping sliding window of 8. (Fig. 1B). Subsequently, the flattened pooling output is passed to a bidirectional LSTM layer of 16 hidden neurons (Fig. 1D and 1E), which finally connects to a sigmoid activation function that outputs the predicted probability score. Clearly, the dictionary encoding method overperforms the one-hot encoding method on the representation of potential druggable proteins, so the CNN, RNN, and CNN-RNN models with the former achieved better performance than the corresponding models with the latter.
Fig. 1.
Optimization of hyperparameters and architectures for CNN-RNN (BiLSTM) model with dictionary encoding method. (A) Performance comparison of different filter and kernel size combinations; (B) Performance comparison of different pooling sizes; (C) Performance comparison of different numbers of convolutional and pooling layers; (D) Performance comparison of different RNN architecture types; (E) Performance comparison of different numbers of hidden cells in the bidirectional LSTM layer; (F) Schematic illustration of the optimized model architecture.
All DNN models except for the TC descriptor are constructed from three fully connected layers with 128, 64, and 32 hidden units each. We compared the performance of the same DNN architecture with different protein descriptors (Table 1). Among the five descriptors, AAC had the best overall prediction performance and yielded the highest ACC (88.5%), F value (87.8%), recall (85.5%), PRE (90.9%), and MCC (0.776) scores. We further analysed and visualized the AAC difference between druggable and nondruggable proteins using Composition Profile software [78]. Significant AAC differences were observed between these two classes of proteins, where the druggable proteins appeared to be enriched in isoleucine, lysine and tyrosine (Figure S4).
Twenty models were trained using the five discrete protein descriptors based on four conventional machine learning algorithms: support vector machine (SVM), random forest (RF), naïve Bayes (NB), and K-nearest neighbour (KNN). The Scikit-learn package was used to implement all the models [79]. Fivefold cross-validation was performed to select models among their respective hyperparameters. For SVM, we selected a radial basis function (RBF) kernel, testing different values for regularization parameter C (2-1, 20, …, 25) and kernel width parameter γ (2-5, 2-4, …, 21). The main hyperparameters for RF include the number of decision trees (50, 100, …, 500) and the number of attributes considered at each split (5, 10, …, 40). For KNN, we used the Euclidean distance as a distance function and tested a different number of neighbours (3, 5, …, 21). We compared the prediction results of the DNN models with the SVM, RF, NB and KNN models. Based on the results in Table S3, the DNN constantly outperformed the other algorithms on all protein descriptors except for CTD. RF was the best model for the CTD descriptor, while for other descriptors, it ranked second after the DNN.
Hybrid models further improve predictive performance
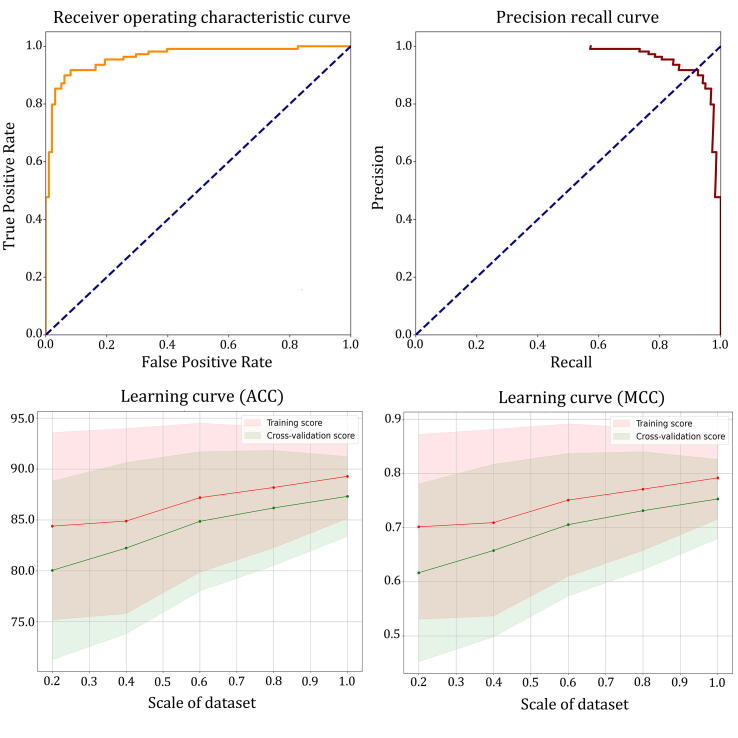
After carefully analysing the prediction results of all single deep learning models, we developed more complex hybrid models that integrate the advantages of different deep learning algorithms. All possible combinations of the five feature groups (AAC, DC, TC, CTD and PSSM) were generated using the concatenated DNN models (details in the section ‘Model design for druggable protein prediction’) and the best core structure (the CNN-RNN (BiLSTM) model with dictionary encoding). Therefore, we designed a total of 31 hybrid models. For each hybrid model, we repeated the training–testing procedure 10 times, and the average of its output was used as the final output for comparison. The performance of hybrid models utilizing various combinations of feature groups is summarized in Table 2. The results indicate that although different combinations of feature groups result in different performances, the hybrid model generally exhibits better performance than a single model. The hybrid model with the ‘DC_TC_CTD’ feature group outperformed other combinations in terms of average ACC (90.0%), F value (89.6%), recall (89.0%), PRE (90.5%), and MCC (0.800). In addition, we employed receiver operator characteristic (ROC) and precision-recall (PR) curves to assess the performance of the best hybrid model (Fig. 2). We calculated the area under the ROC and PR curves, and the values are shown in parentheses under both curves (0.963 for ROC and 0.970 for PR). Then, the learning curves that correspond to the ACC and MCC metrics were plotted to assess the effect of the dataset scale on the predictive performance of the hybrid model. Five different scales (20, 40, 60, 80, and 100%) of subsets were generated by using an external resampling mechanism. Each resampling was repeated 10 times, and each subset was divided into 5 training-test groups for cross-validation so that a total of 250 hybrid models were used for the generation of learning curves. As shown in Fig. 2, the hybrid model becomes increasingly stable as the scale of the dataset increases, but still has relatively good predictive performance on the small sample dataset.
Table 2.
Performance comparison of the hybrid models with different combinations of protein descriptors.
| Core structure (Encoding) | DNN (Feature) | ACC (%) | F-value (%) | Recall (%) | PRE (%) | MCC |
|---|---|---|---|---|---|---|
| AAC | 88.8 | 88.2 | 87.1 | 89.5 | 0.776 | |
| TC | 87.6 | 87.5 | 88.7 | 86.5 | 0.754 | |
| PSSM | 87.5 | 86.8 | 85.9 | 88.0 | 0.751 | |
| DC | 88.2 | 87.9 | 88.8 | 87.3 | 0.766 | |
| CTD | 89.5 | 89.0 | 89.0 | 89.2 | 0.791 | |
| TC_CTD | 88.6 | 88.3 | 89.0 | 87.8 | 0.772 | |
| DC_TC | 88.6 | 88.4 | 89.5 | 87.4 | 0.772 | |
| DC_PSSM | 88.4 | 87.8 | 87.6 | 88.3 | 0.769 | |
| DC_CTD | 88.0 | 87.4 | 87.3 | 87.8 | 0.761 | |
| CTD_PSSM | 87.0 | 86.8 | 87.5 | 86.2 | 0.740 | |
| AAC_TC | 86.6 | 86.1 | 86.2 | 86.0 | 0.732 | |
| TC_PSSM | 87.6 | 87.5 | 89.6 | 85.6 | 0.753 | |
| AAC_CTD | 87.5 | 87.4 | 88.1 | 87.1 | 0.752 | |
| AAC_DC | 88.4 | 88.1 | 88.6 | 87.7 | 0.767 | |
| AAC_PSSM | 88.0 | 88.1 | 88.6 | 87.6 | 0.760 | |
| CNN-RNN | AAC_TC_PSSM | 88.8 | 88.2 | 89.1 | 87.5 | 0.777 |
| (Dictionary) | AAC_TC_CTD | 88.6 | 87.9 | 87.7 | 88.4 | 0.771 |
| TC_CTD_PSSM | 88.4 | 88.1 | 88.2 | 88.2 | 0.768 | |
| AAC_CTD_PSSM | 87.7 | 87.2 | 87.0 | 87.7 | 0.754 | |
| AAC_DC_TC | 89.2 | 89.0 | 90.3 | 87.9 | 0.785 | |
| AAC_DC_PSSM | 88.1 | 87.4 | 86.6 | 88.4 | 0.761 | |
| DC_CTD_PSSM | 88.0 | 87.9 | 89.4 | 86.7 | 0.760 | |
| AAC_DC_CTD | 89.1 | 89.1 | 90.4 | 87.8 | 0.783 | |
| DC_TC_CTD | 90.0 | 89.6 | 89.0 | 90.5 | 0.800 | |
| DC_TC_PSSM | 89.1 | 89.0 | 88.7 | 89.4 | 0.782 | |
| AAC_DC_TC_PSSM | 89.0 | 89.1 | 90.2 | 88.2 | 0.780 | |
| AAC_DC_TC_CTD | 87.9 | 87.6 | 89.6 | 85.8 | 0.760 | |
| AAC_TC_CTD_PSSM | 89.0 | 88.5 | 90.6 | 86.7 | 0.780 | |
| DC_TC_CTD_PSSM | 89.4 | 89.4 | 91.3 | 87.6 | 0.789 | |
| AAC_DC_CTD_PSSM | 88.9 | 88.7 | 90.4 | 87.3 | 0.780 | |
| AAC_DC_TC_CTD_PSSM | 88.5 | 88.4 | 89.1 | 87.8 | 0.771 |
Fig. 2.
ROC, PR, and learning curves for the best hybrid deep learning model on the training dataset.
Performance evaluation with the independent test set
We further evaluated the generalization capabilities of our hybrid model on the previously described independent test set, which was nonredundant at the sequence level. The new dataset consisting of 224 positive samples and 237 negative samples was selected as a blind test. Our final model (architecture: CNN-RNN (BiLSTM) + DNN; feature: dictionary encoding + DC_TC_CTD) demonstrated balanced predictive performance, achieving an overall accuracy of 89.8%, F value of 88.9%, recall of 84.8%, precision of 93.6% and MCC of 0.799. These experimental results verify the efficiency and stability of our proposed method.
Visualizing and understanding deep learning models
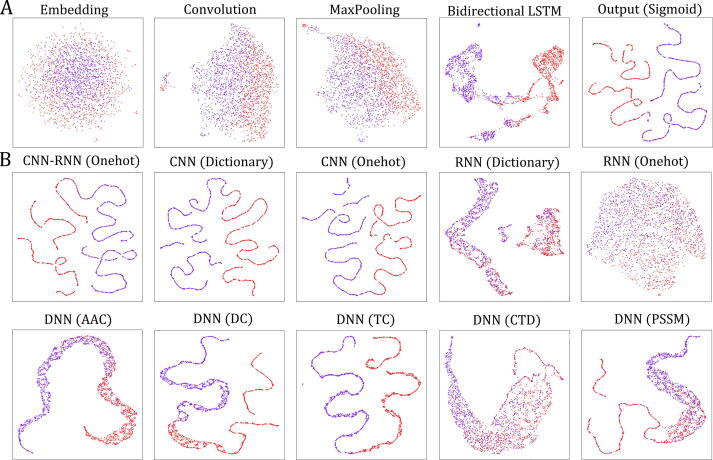
To better understand how the deep learning models work, we utilized the layerUMAP tool [75] to visualize the representations of druggable proteins learned by these models. Here, all samples in the training dataset were grouped into two classes: red points for druggable proteins and purple points for nondruggable proteins. To explain the working mechanisms of the models or to shed light on their internal representations, we first visualized two classes of proteins based on the representations learned by the main hidden layers of the best core structure (CNN-RNN (BiLSTM) model with dictionary encoding). As illustrated in Fig. 3A, the druggable proteins and nondruggable proteins are almost indistinguishable in the first embedding layer but can be very clearly distinguished in the last two hidden layers (bidirectional LSTM layer and output layer). It is clear that these representations become increasingly discriminative along the layer hierarchy during model training. Furthermore, the projections of the output layer representations of the other ten deep learning models are shown in Fig. 3B. As expected, the partitioning effects of the two classes of points in these figures are highly consistent with the predictive performance of the corresponding models.
Fig. 3.
LayerUMAP shows the working mechanism of deep learning models. (A) UMAP projection of inter-layer evolution of the CNN-RNN(BiLSTM) model with dictionary encoding method; (B) UMAP projection of the output layers after training of the ten deep learning models.
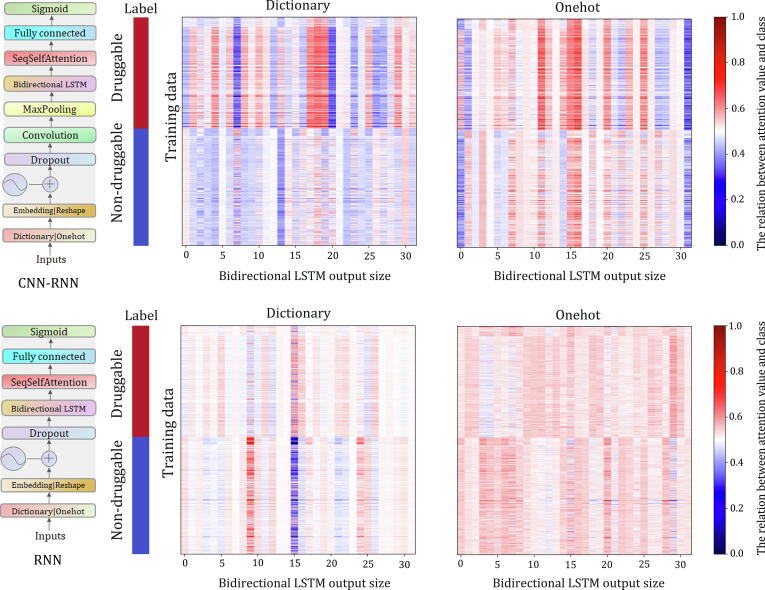
In Table 1, we observe that the CNN-RNN (BiLSTM) model performs significantly better than the RNN (BiLSTM) model for both encoding methods, even though they have the same bidirectional LSTM layer. We can therefore conclude that the convolution-based model can effectively extract predictive features from protein sequences through filter scanning. To further confirm our conclusions, we added a self-attention layer after the bidirectional LSTM layer of the RNN (BiLSTM) and CNN-RNN (BiLSTM) models. We plotted the attention values and self-attention relations to visualize the importance of different hidden neurons in the bidirectional LSTM layers for the classification (Fig. 4). The self-attention heatmaps can reflect the relationship of each protein to the druggable and nondruggable classes. We can see that the difference in attention values between the CNN-RNN (BiLSTM) model for druggable and nondruggable proteins is significantly larger than that of the RNN (BiLSTM) model, thus explaining the difference in their prediction performance.
Fig. 4.
Heat maps show the distributions of the attention values of two bidirectional LSTM layers in the CNN-RNN (BiLSTM) and RNN (BiLSTM) models.
Comparison with other methods
As we described in the introduction section, three studies have been carried out to develop predictors with different machine learning algorithms and features using the dataset from Jamili et al. [38], [39], [40]. Here, we also used this standard dataset to compare the performance of our proposed deep learning model with these existing methods. It is worth noting that DrugMiner and Lin et al.’s method were constructed based on the whole dataset, while Sun et al. extracted 10% of the whole dataset to construct the independent test set, and we randomly selected 20% as the independent test set. All results from the four methods are listed in Table 3, the first three of which are directly from the studies of Sun et al. [39] and Lin et al. [40]. As shown in Table 3, the GA-Bagging-SVM with 143 features achieved the best overall predictive performance and provided the highest scores of ACC (93.8%), F value (93.6%), and MCC (0.878). Our proposed deep learning model did not obtain the best overall prediction performance, but its performance was comparable to that of state-of-the-art methods and provided the highest recall rate (94.5%). Since our deep learning models were developed in autoBioSeqpy software [77], many operations, including data reading, parameter initialization, sequence encoding, model loading, training, and evaluation, can be executed automatically with one-line commands, which can save considerable time for users. In addition, compared with the other three methods, our model is more flexible and interpretable, which supports the use of deep learning techniques to develop predictors for the identification of potential druggable proteins. More importantly, this is the first study to use deep learning algorithms, which also provides a new strategy for druggable protein prediction.
Table 3.
Performance comparison of our proposed deep learning model with the existing methods.
| Method | Algorithms | Encoding method | ACC (%) | F-value (%) | Recall (%) | PRE (%) | MCC |
|---|---|---|---|---|---|---|---|
| DrugMiner | NN | 443 features | 89.78 | 90.14 | 90.14 | – | 0.7952 |
| DrugMiner | NN | 130 features | 92.10 | 92.41 | 92.80 | – | 0.8417 |
| Sun et al.’ method | NN | Jamali-v | 88.12 | – | – | – | – |
| Sun et al.’ method | NN | Overlapped 3-gram word2vec | 90.23 | – | – | – | – |
| Sun et al.’ method | NN | LQL-v | 91.93 | – | – | – | – |
| Lin et al.’ method | PseAAC-DPC-RS | 591 features | 90.98 | 90.97 | 87.88 | – | 0.8212 |
| Lin et al.’ method | PseAAC-DPC-RS-GA | 143 features | 93.42 | 93.07 | 92.21 | – | 0.8711 |
| Lin et al.’ method | GA-Bagging-SVM | 143 features | 93.78 | 93.58 | 92.86 | – | 0.8781 |
| Our work | Hybrid DL model | Dictionary + DC_TC_CTD | 92.40 | 91.90 | 94.50 | 89.50 | 0.8490 |
Discussion
The discovery of drug targets is a matter of utmost importance for the development of new drugs [80], and the accurate and effective identification of these targets is a key step in the drug development process. At present, increasing attention has been given to druggable proteins, which play a key role in various diseases and have proven to be one of the most important sources of drug targets [81]. Traditional experimental methods are laborious, time-consuming, expensive, and cannot be used in a high-throughput manner. Therefore, computational methods have become particularly popular in recent years for identifying potential druggable proteins for the design and development of new drugs. In this work, we sought to develop a novel method for predicting druggable proteins directly from primary sequences through the use of deep learning algorithms. We carried out extensive experiments for comparison and presented an in-depth analysis. The experimental results show that the hybrid neural network model achieves better classification performance than the other models tested. The superior performance of the hybrid model shows that (i) neural networks (such as CNNs and CNN-RNNs) can extract high-order information hidden in the sequences; (ii) the use of additional information (e.g. handcrafted protein features) is helpful for improving performance; and (iii) the proposed framework can integrate protein features and deep neural network models well.
As shown in this study, a good deep learning model is related to many factors, including the architecture of the model, hyperparameters, feature engineering, etc. A suitable architecture ensures proper progression of data processing. The hyperparameters can maintain a certain training progress with reasonable loss values and gradients in each epoch. Feature engineering can provide and refine correct information from the current data, even if a part of it has been integrated into the deep learning layers. In addition, the quality of the dataset (e.g., size and noise) will be an important factor. Deep learning methods have demonstrated flexibility in model design and good interpretability in practical applications in several machine learning areas, such as computer vision (CV) and artificial intelligence (AI), but a major reason for this success is the large size of the training data. For example, the number of figures used to train a CV model can exceed 1 million for just one epoch, and using the strategy of manifold learning for some AI problems can generate a nearly infinite number of samples for learning. However, in druggable protein prediction, it is currently difficult to obtain large-scale data for training, so the architecture and parameters of neural networks are simplified and reduced to avoid overfitting, rather than embedding state-of-the-art structures such as various transformer structures to mine the intrinsic information hidden by the sequences. One possible solution is to use transfer learning to use more corrected data but still need a preprovided dataset for parameter initialization. We believe that with the development of experimental techniques in the postgenomic era, the big data generated by the high-throughput experiments on biological systems will be enough for deep learning modelling in this field.
In summary, we focused on recent deep learning applications in druggable protein prediction with methods, tools, and gold-standard datasets that are used to train and test machine learning models. We described several types of protein descriptors that are frequently used in feature engineering, especially for deep learning applications. We introduced some deep learning toolkits and libraries, that can design and train different neural network architectures, generate figures from the results and carry out downstream analysis. Based on the experimental results, we generated an amino acid character dictionary and protein features extracted from sequence information and subsequently implemented these features into a hybrid model that integrated convolutional recurrent neural networks (CNN-RNNs) and deep neural networks (DNNs). The hybrid model achieved the best overall prediction performance and afforded the highest scores of ACC (90.0%), F value (89.6%), recall (89.0%), PRE (90.5%), and MCC (0.800). The innovations of this study are as follows: (i) to the best of our knowledge, we developed the first deep learning-based druggable protein classifier for fast and accurate identification of potential druggable proteins; (ii) different deep learning models and features are employed to develop the classifier; (iii) to better interpret the models and explain their predictions, two novel visualization techniques are introduced to give insight into the function of intermediate feature layers and the operation of the classifier; (iv) our comprehensive benchmark assessment suggests valuable directions for future algorithm development.
Data availability
All datasets and code for this study is freely available online at https://github.com/jingry/autoBioSeqpy/tree/2.0/examples/Druggableproteins.
This article does not contain any studies with human or animal subjects.
Compliance with Ethics Requirements
This article does not contain any studies with human or animal subjects.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work has been supported by the National Natural Science Foundation of China (No. 21803045), Natural Science Foundation of Department of Education of Guizhou Province ([2021]021), and Joint project of Luzhou Municipal People’s Government and Southwest Medical University (2020LZXNYDJ39).
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2022.01.009.
Contributor Information
Runyu Jing, Email: jingryedu@gmail.com.
Jiesi Luo, Email: ljs@swmu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Kandoi G., Acencio M.L., Lemke N. Prediction of druggable proteins using machine learning and systems biology: a mini-review. Front Physiol. 2015;6:366. doi: 10.3389/fphys.2015.00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Santos R., Ursu O., Gaulton A., Bento A.P., Donadi R.S., Bologa C.G., et al. A comprehensive map of molecular drug targets. Nat Rev Drug Discov. 2017;16(1):19–34. doi: 10.1038/nrd.2016.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keller T.H., Pichota A., Yin Z. A practical view of 'druggability'. Curr Opin Chem Biol. 2006;10(4):357–361. doi: 10.1016/j.cbpa.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 4.Radusky L, Defelipe LA, Lanzarotti E, Luque J, Barril X, Marti MA, et al. TuberQ: a Mycobacterium tuberculosis protein druggability database. Database (Oxford) 2014; 2014:bau035. https://doi.org/10.1093/database/bau035. [DOI] [PMC free article] [PubMed]
- 5.Ghadermarzi S., Li X., Li M., Kurgan L. Sequence-Derived Markers of Drug Targets and Potentially Druggable Human Proteins. Front Genet. 2019;10:1075. doi: 10.3389/fgene.2019.01075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dorsam R.T., Gutkind J.S. G-protein-coupled receptors and cancer. Nat Rev Cancer. 2007;7(2):79–94. doi: 10.1038/nrc2069. [DOI] [PubMed] [Google Scholar]
- 7.Usha T., Shanmugarajan D., Goyal A.K., Kumar C.S., Middha S.K. Recent Updates on Computer-aided Drug Discovery: Time for a Paradigm Shift. Curr Top Med Chem. 2018;17(30):3296–3307. doi: 10.2174/1568026618666180101163651. [DOI] [PubMed] [Google Scholar]
- 8.Makley L.N., Gestwicki J.E. Expanding the number of 'druggable' targets: non-enzymes and protein-protein interactions. Chem Biol Drug Des. 2013;81(1):22–32. doi: 10.1111/cbdd.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamanishi Y., Kotera M., Kanehisa M., Goto S. Drug-target interaction prediction from chemical, genomic and pharmacological data in an integrated framework. Bioinformatics. 2010;26(12):i246–i254. doi: 10.1093/bioinformatics/btq176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui W., Aouidate A., Wang S., Yu Q., Li Y., Yuan S. Discovering Anti-Cancer Drugs via Computational Methods. Front Pharmacol. 2020;11:733. doi: 10.3389/fphar.2020.00733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan H.C.S., Shan H., Dahoun T., Vogel H., Yuan S. Advancing Drug Discovery via Artificial Intelligence. Trends Pharmacol Sci. 2019;40(8):592–604. doi: 10.1016/j.tips.2019.06.004. [DOI] [PubMed] [Google Scholar]
- 12.Munos B. Lessons from 60 years of pharmaceutical innovation. Nat Rev Drug Discov. 2009;8(12):959–968. doi: 10.1038/nrd2961. [DOI] [PubMed] [Google Scholar]
- 13.Paul S.M., Mytelka D.S., Dunwiddie C.T., Persinger C.C., Munos B.H., Lindborg S.R., et al. How to improve R&D productivity: the pharmaceutical industry’s grand challenge. Nat Rev Drug Discov. 2010;9:203–214. doi: 10.1038/nrd3078. [DOI] [PubMed] [Google Scholar]
- 14.Csermely P., Korcsmáros T., Kiss H.J.M., London G., Nussinov R. Structure and dynamics of molecular networks: a novel paradigm of drug discovery: a comprehensive review. Pharmacol Ther. 2013;138(3):333–408. doi: 10.1016/j.pharmthera.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Butcher S.P. Target discovery and validation in the post-genomic era. Neurochem Res. 2003;28(2):367–371. doi: 10.1023/a:1022349805831. [DOI] [PubMed] [Google Scholar]
- 16.Fauman E.B., Rai B.K., Huang E.S. Structure-based druggability assessment–identifying suitable targets for small molecule therapeutics. Curr Opin Chem Biol. 2011;15(4):463–468. doi: 10.1016/j.cbpa.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y.-T., Li Y.i., Huang Z.-f., Xu Z.-J., Yang Z., Chen Z.-x., et al. Multi-algorithm and multi-model based drug target prediction and web server. Acta Pharmacol Sin. 2014;35(3):419–431. doi: 10.1038/aps.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu T., Altman R.B. Identifying druggable targets by protein microenvironments matching: application to transcription factors. CPT Pharmacometrics Syst Pharmacol. 2014;3(1):93. doi: 10.1038/psp.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michel M., Homan E.J., Wiita E., Pedersen K., Almlöf I., Gustavsson A.-L., et al. In silico Druggability Assessment of the NUDIX Hydrolase Protein Family as a Workflow for Target Prioritization. Front Chem. 2020;8 doi: 10.3389/fchem.2020.00443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emig D, Ivliev A, Pustovalova O, Lancashire L, Bureeva S, Nikolsky Y, et al. Drug target prediction and repositioning using an integrated network-based approach. PLoS One 2013; 8:e60618. https://doi.org/10.1371/journal.pone.0060618. [DOI] [PMC free article] [PubMed]
- 21.Li Z.-C., Zhong W.-Q., Liu Z.-Q., Huang M.-H., Xie Y., Dai Z., et al. Large-scale identification of potential drug targets based on the topological features of human protein-protein interaction network. Anal Chim Acta. 2015;871:18–27. doi: 10.1016/j.aca.2015.02.032. [DOI] [PubMed] [Google Scholar]
- 22.Mousavian Z., Masoudi-Nejad A. Drug-target interaction prediction via chemogenomic space: learning-based methods. Expert Opin Drug Metab Toxicol. 2014;10(9):1273–1287. doi: 10.1517/17425255.2014.950222. [DOI] [PubMed] [Google Scholar]
- 23.Lempiäinen H., Brænne I., Michoel T., Tragante V., Vilne B., Webb T.R., et al. Network analysis of coronary artery disease risk genes elucidates disease mechanisms and druggable targets. Sci Rep. 2018;8(1) doi: 10.1038/s41598-018-20721-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han L.Y., Zheng C.J., Xie B., Jia J., Ma X.H., Zhu F., et al. Support vector machines approach for predicting druggable proteins: recent progress in its exploration and investigation of its usefulness. Drug Discov Today. 2007;12(7-8):304–313. doi: 10.1016/j.drudis.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 25.Li Q., Lai L. Prediction of potential drug targets based on simple sequence properties. BMC Bioinf. 2007;8:353. doi: 10.1186/1471-2105-8-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bakheet T.M., Doig A.J. Properties and identification of human protein drug targets. Bioinformatics. 2009;25:451–457. doi: 10.1093/bioinformatics/btp002. [DOI] [PubMed] [Google Scholar]
- 27.Huang C., Zhang R., Chen Z., Jiang Y., Shang Z., Sun P., et al. Predict potential drug targets from the ion channel proteins based on SVM. J Theor Biol. 2010;262(4):750–756. doi: 10.1016/j.jtbi.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 28.Zhang G.L., Khan A.M., Srinivasan K.N., August J.T., Brusic VLADIMIR. Neural models for predicting viral vaccine targets. J Bioinform Comput Biol. 2005;03(05):1207–1225. doi: 10.1142/s0219720005001466. [DOI] [PubMed] [Google Scholar]
- 29.Niwa T. Prediction of biological targets using probabilistic neural networks and atom-type descriptors. J Med Chem. 2004;47(10):2645–2650. doi: 10.1021/jm0302795. [DOI] [PubMed] [Google Scholar]
- 30.Nidhi, Glick M., Davies J.W., Jenkins J.L. Prediction of biological targets for compounds using multiple-category Bayesian models trained on chemogenomics databases. J Chem Inf Model. 2006;46(3):1124–1133. doi: 10.1021/ci060003g. [DOI] [PubMed] [Google Scholar]
- 31.Gonen M. Predicting drug-target interactions from chemical and genomic kernels using Bayesian matrix factorization. Bioinformatics. 2012;28(18):2304–2310. doi: 10.1093/bioinformatics/bts360. [DOI] [PubMed] [Google Scholar]
- 32.Yang J., He S., Zhang Z., Bo X. NegStacking: drug-target interaction prediction based on ensemble learning and logistic regression. IEEE/ACM Trans Comput Biol Bioinform. 2021;18(6):2624–2634. doi: 10.1109/TCBB.2020.2968025. [DOI] [PubMed] [Google Scholar]
- 33.González-Díaz H., Cruz-Monteagudo M., Molina R., Tenorio E., Uriarte E. Predicting multiple drugs side effects with a general drug-target interaction thermodynamic Markov model. Bioorg Med Chem. 2005;13(4):1119–1129. doi: 10.1016/j.bmc.2004.11.030. [DOI] [PubMed] [Google Scholar]
- 34.Shi H., Liu S., Chen J., Li X., Ma Q., Yu B. Predicting drug-target interactions using Lasso with random forest based on evolutionary information and chemical structure. Genomics. 2019;111(6):1839–1852. doi: 10.1016/j.ygeno.2018.12.007. [DOI] [PubMed] [Google Scholar]
- 35.Ezzat A., Wu M., Li X.L., Kwoh C.K. Drug-target interaction prediction using ensemble learning and dimensionality reduction. Methods. 2007;129:81–88. doi: 10.1016/j.ymeth.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 36.Zhang W., Liu F., Luo L., Zhang J. Predicting drug side effects by multi-label learning and ensemble learning. BMC Bioinf. 2015;16:365. doi: 10.1186/s12859-015-0774-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang W., Zou H., Luo L., Liu Q., Wu W., Xiao W. Predicting potential side effects of drugs by recommender methods and ensemble learning. Neurocomputing. 2016;173:979–987. doi: 10.1016/j.neucom.2015.08.054. [DOI] [Google Scholar]
- 38.Jamali A.A., Ferdousi R., Razzaghi S., Li J., Safdari R., Ebrahimie E. DrugMiner: comparative analysis of machine learning algorithms for prediction of potential druggable proteins. Drug Discov Today. 2016;21(5):718–724. doi: 10.1016/j.drudis.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 39.Sun T., Lai L., Pei J. Analysis of protein features and machine learning algorithms for prediction of druggable proteins. Quant Biol. 2018;6(4):334–343. doi: 10.1007/s40484-018-0157-2. [DOI] [Google Scholar]
- 40.Lin J., Chen H., Li S., Liu Y., Li X., Yu B. Accurate prediction of potential druggable proteins based on genetic algorithm and Bagging-SVM ensemble classifier. Artif Intell Med. 2019;98:35–47. doi: 10.1016/j.artmed.2019.07.005. [DOI] [PubMed] [Google Scholar]
- 41.Wen B., Zeng W.F., Liao Y., Shi Z., Savage S.R., Jiang W., et al. Deep Learning in Proteomics. Deep Learn Proteomics Proteomics. 2020;20(21-22):1900335. doi: 10.1002/pmic.201900335. https://doi.org/10.1002/pmic.v20.21-2210.1002/pmic.201900335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 43.Cao C., Liu F., Tan H., Song D., Shu W., Li W., et al. Deep Learning and Its Applications in Biomedicine. Genom Proteomics Bioinformat. 2018;16(1):17–32. doi: 10.1016/j.gpb.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patel L., Shukla T., Huang X., Ussery D.W., Wang S. Machine Learning Methods in Drug Discovery. Molecules. 2020;25:5277. doi: 10.3390/molecules25225277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muzio G., O'Bray L., Borgwardt K. Biological network analysis with deep learning. Brief Bioinform. 2021;22:1515–1530. doi: 10.1093/bib/bbaa257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Naseer S., Hussain W., Khan Y.D., Rasool N. Optimization of serine phosphorylation prediction in proteins by comparing human engineered features and deep representations. Anal Biochem. 2021;615:114069. doi: 10.1016/j.ab.2020.114069. [DOI] [PubMed] [Google Scholar]
- 47.Naseer S., Ali R.F., Khan Y.D., Dominic P.D.D. iGluK-Deep: computational identification of lysine glutarylation sites using deep neural networks with general pseudo amino acid compositions. J Biomol Struct Dyn. 2021:1–14. doi: 10.1080/07391102.2021.1962738. [DOI] [PubMed] [Google Scholar]
- 48.Zhao T., Hu Y., Valsdottir L.R., Zang T., Peng J. Identifying drug-target interactions based on graph convolutional network and deep neural network. Brief Bioinform. 2021;22:2141–2150. doi: 10.1093/bib/bbaa044. [DOI] [PubMed] [Google Scholar]
- 49.Wen M., Zhang Z., Niu S., Sha H., Yang R., Yun Y., et al. Deep-Learning-Based Drug-Target Interaction Prediction. J Proteome Res. 2017;16(4):1401–1409. doi: 10.1021/acs.jproteome.6b00618. [DOI] [PubMed] [Google Scholar]
- 50.Deep learning for genomics. Nat Genet. 2019;51:1. https://doi.org/10.1038/s41588-018-0328-0. [DOI] [PubMed]
- 51.Szalkai B., Grolmusz V. Near Perfect Protein Multi-Label Classification with Deep Neural Networks. Methods. 2018;132:50–56. doi: 10.1016/j.ymeth.2017.06.034. [DOI] [PubMed] [Google Scholar]
- 52.Almagro Armenteros J.J., Sønderby C.K., Sønderby S.K., Nielsen H., Winther O., Hancock J. DeepLoc: prediction of protein subcellular localization using deep learning. Bioinformatics. 2017;33(21):3387–3395. doi: 10.1093/bioinformatics/btx431. [DOI] [PubMed] [Google Scholar]
- 53.Law V., Knox C., Djoumbou Y., Jewison T., Guo A.C., Liu Y., et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res. 2014;42(D1):D1091–D1097. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu F., Han B., Kumar P., Liu X., Ma X., Wei X., et al. Update of TTD: Therapeutic Target Database. Nucleic Acids Res. 2010;38(suppl_1):D787–D791. doi: 10.1093/nar/gkp1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim B., Jo J., Han J., Park C., Lee H. In silico re-identification of properties of drug target proteins. BMC Bioinf. 2017;18:248. doi: 10.1186/s12859-017-1639-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fu L., Niu B., Zhu Z., Wu S., Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Veltri D., Kamath U., Shehu A., Hancock J. Deep learning improves antimicrobial peptide recognition. Bioinformatics. 2018;34(16):2740–2747. doi: 10.1093/bioinformatics/bty179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou J., Troyanskaya O.G. Predicting effects of noncoding variants with deep learning-based sequence model. Nat Methods. 2015;12(10):931–934. doi: 10.1038/nmeth.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rao H.B., Zhu F., Yang G.B., Li Z.R., Chen Y.Z. Update of PROFEAT: a web server for computing structural and physicochemical features of proteins and peptides from amino acid sequence. Nucleic Acids Res. 2011;39:W385–W390. doi: 10.1093/nar/gkr284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luo J., Yu L., Guo Y., Li M. Functional classification of secreted proteins by position specific scoring matrix and auto covariance. Chemom Intell Lab Syst. 2012;110(1):163–167. doi: 10.1016/j.chemolab.2011.11.008. [DOI] [Google Scholar]
- 62.Kabir M., Arif M., Ahmad S., Ali Z., Swati Z.N.K., Yu D.J. Intelligent computational method for discrimination of anticancer peptides by incorporating sequential and evolutionary profiles information. Chemom Intell Lab Syst. 2018;182:158–165. doi: 10.1016/j.chemolab.2018.09.007. [DOI] [Google Scholar]
- 63.Wainberg M., Merico D., Delong A., Frey B.J. Deep Learning in Biomedicine. Nat Biotechnol. 2018;36(9):829–838. doi: 10.1038/nbt.4233. [DOI] [PubMed] [Google Scholar]
- 64.Tulbure A.-A., Tulbure A.-A., Dulf E.-H. A review on modern defect detection models using DCNNs - Deep convolutional neural networks. J Adv Res. 2022;35:33–48. doi: 10.1016/j.jare.2021.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rostamian H., Lotfollahi M.N. Statistical modeling of aspirin solubility in organic solvents by Response Surface Methodology and Artificial Neural Networks. Phys A. 2020;540:123253. doi: 10.1016/j.physa:2019.123253. [DOI] [Google Scholar]
- 66.Rostamian H., Lotfollahi M.N. A novel statistical approach for prediction of thermal conductivity of CO2 by Response Surface Methodology. Phys A. 2019;527:121175. doi: 10.1016/j.physa.2019.121175. [DOI] [Google Scholar]
- 67.Esfe M.H., Tatar A., Ahangar M.R.H., Rostamian H. A comparison of performance of several artificial intelligence methods for predicting the dynamic viscosity of TiO2/SAE 50 nano-lubricant. Physica E Low Dimens Syst Nanostruct. 2018;96:85–93. doi: 10.1016/j.physe.2017.08.019. [DOI] [Google Scholar]
- 68.Esfe M.H., Rostamian H., Esfandeh S., Afrand M. Modeling and prediction of rheological behavior of Al2O3-MWCNT/5W50 hybrid nano-lubricant by artificial neural network using experimental data. Phys A. 2018;510:625–634. doi: 10.1016/j.physa.2018.06.041. [DOI] [Google Scholar]
- 69.Miotto R., Wang F., Wang S., Jiang X., Dudley J.T. Deep learning for healthcare: review, opportunities and challenges. Brief Bioinform. 2018;19:1236–1246. doi: 10.1093/bib/bbx044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Juez-Gil M., Erdakov I.N., Bustillo A., Pimenov D.Y. A regression-tree multilayer-perceptron hybrid strategy for the prediction of ore crushing-plate lifetimes. J Adv Res. 2019;18:173–184. doi: 10.1016/j.jare.2019.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bradbury J, Merity S, Xiong C, Socher R. Quasi-recurrent neural networks. arXiv preprint arXiv:1611.01576, 2016. https://arxiv.org/abs/1611.01576.
- 72.Quang D., Xie X. DanQ: a hybrid convolutional and recurrent deep neural network for quantifying the function of DNA sequences. Nucleic Acids Res. 2016;44(11) doi: 10.1093/nar/gkw226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pan X., Rijnbeek P., Yan J., Shen H.B. Prediction of RNA-protein sequence and structure binding preferences using deep convolutional and recurrent neural networks. BMC Genomics. 2018;19:511. doi: 10.1186/s12864-018-4889-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu L., Liu F., Li Y., Luo J., Jing R. DeepT3_4: A Hybrid Deep Neural Network Model for the Distinction Between Bacterial Type III and IV Secreted Effectors. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.605782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McInnes L, Healy J. UMAP: uniform manifold approximation and projection for dimension reduction. 2018. Preprint at https://arxiv.org/abs/1802.03426.
- 76.Chollet F. Keras, GitHub. 2015. https://github.com/fchollet/keras.
- 77.Jing R, Li Y, Xue L, Liu F, Li M, Luo J. autoBioSeqpy: A Deep Learning Tool for the Classification of Biological Sequences. J. Chem. Inf. Model. 2020; 60:3755–64. https://doi.org/10.1021/acs.jcim.0c00409. [DOI] [PubMed]
- 78.Vacic V., Uversky V.N., Dunker A.K., Lonardi S. Composition Profiler: A tool for discovery and visualization of amino acid composition differences. BMC Bioinf. 2007;8:211. doi: 10.1186/1471-2105-8-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., et al. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011;12:2825–2830. https://jmlr.org/papers/v12/pedregosa11a.html [Google Scholar]
- 80.Lindsay M.A. Finding new drug targets in the 21st century. Drug Discov Today. 2005;10(23-24):1683–1687. doi: 10.1016/S1359-6446(05)03670-6. [DOI] [PubMed] [Google Scholar]
- 81.Hopkins A.L., Groom C.R. The druggable genome. Nat Rev Drug Discov. 2002;1(9):727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets and code for this study is freely available online at https://github.com/jingry/autoBioSeqpy/tree/2.0/examples/Druggableproteins.
This article does not contain any studies with human or animal subjects.