Abstract
The properties of two candidate Salmonella typhi-based live oral typhoid vaccine strains, BRD691 (S. typhi Ty2 harboring mutations in aroA and aroC) and BRD1116 (S. typhi Ty2 harboring mutations in aroA, aroC, and htrA), were compared in a number of in vitro and in vivo assays. BRD1116 exhibited an increased susceptibility to oxidative stress compared with BRD691, but both strains were equally resistant to heat shock. Both strains showed a similar ability to invade Caco-2 and HT-29 epithelial cells and U937 macrophage-like cells, but BRD1116 was less efficient at surviving in epithelial cells than BRD691. BRD1116 and BRD691 were equally susceptible to intracellular killing within U937 cells. Similar findings were demonstrated in vivo, with BRD1116 being less able to survive and translocate to secondary sites of infection when inoculated into the lumen of human intestinal xenografts in SCID mice. However, translocation of BRD1116 to spleens and livers in SCID mice occurred as efficiently as that of BRD691 when inoculated intraperitonally. The ability of BRD1116 to increase the secretion of interleukin-8 following infection of HT-29 epithelial cells was comparable to that of BRD691. Therefore, loss of the HtrA protease in S. typhi does not seem to alter its ability to invade epithelial cells or macrophages or to induce proinflammatory cytokines such as IL-8 but significantly reduces intracellular survival in human intestinal epithelial cells in vitro and in vivo.
Salmonella strains harboring genetically defined, attenuating mutations are attracting interest for use as live, single-dose oral vaccines against typhoid and as delivery vehicles for heterologous antigens (4–6, 12–16, 32–35). Following oral ingestion, Salmonella bacteria invade the gut wall (8, 20), promoting neutrophil infiltration of the mucosa (21, 25) and subsequent macrophage recruitment. The ability of Salmonella to invade the epithelium (24) and to survive in macrophages (1) is essential for virulence and may represent a major factor in determining host restriction. Macrophages are believed to play a role in immunity to Salmonella, but they may also facilitate the dissemination of viable bacteria to deeper tissues, such as the liver and spleen (3). Salmonella can also induce apoptosis in macrophages, a factor which may further contribute to virulence (7, 27).
Mutations in a number of different Salmonella genes have been evaluated for the ability to attenuate virulence, including phoP and phoQ, which encode a two-component regulator (13), and crp and cya, which encode the cAMP receptor system (33). Salmonella strains harboring mutations in aro genes, whose products are enzymes involved in the shikimate biosynthesis pathway, are effective oral vaccines (14). Salmonella aro mutants are auxotrophic for certain aromatic compounds such as the three aromatic amino acids tryptophan, tyrosine, and phenylalanine, as well as para-aminobenzoic acid and 2,3-dihydroxybenzoate. Some of these compounds are not available at sufficient levels in mammalian tissues to sustain growth of Salmonella aro mutants, leading to attenuation.
Several S. typhi-based candidate typhoid vaccine strains harboring aro mutations have been evaluated in volunteers (16, 35). Two of these, CVD906 and CVD908, are derivatives of S. typhi strains ISP1820 and Ty2, respectively, which harbor deletion mutations in aroC and aroD (17, 34). Although CVD906 and CVD908 are highly immunogenic in volunteers, viable S. typhi bacteria are detected in the circulation when high doses of vaccine (108 CFU) are given orally. Since Salmonella bacteria are disseminated in the blood inside macrophages, mutations that interfere with macrophage survival may impair this spread.
htrA is a stress response gene in Salmonella that encodes a periplasmic protease that degrades aberrant proteins (19). S. typhimurium htrA strains are attenuated (6) and have an increased sensitivity to hydrogen peroxide. These data have implicated a role for htrA in the survival of S. typhimurium in murine macrophages (1). The CVD908 htrA mutant strain has also recently been evaluated in volunteers (33, 35). Interestingly, the CVD908 htrA mutant strain was highly immunogenic, but, unlike with strain CVD908, bacteria were not detected in the peripheral blood of volunteers. Little is known about how htrA influences S. typhi virulence in vivo, although increased susceptibility to macrophage-mediated killing may be involved.
Pathogenicity, attenuation, and immunogenicity of S. typhi-based vaccines are not easy to monitor outside the human host. It would be useful to have available simple in vivo and in vitro assays that correlate with the behavior of vaccine candidate strains in humans. To address this issue, and to correlate the lack of systemic spread in volunteers with a model infection system, we introduced an htrA mutation into S. typhi BRD691, which harbors deletions in aroA and aroC. We subsequently examined the influence of this mutation using in vitro cell invasion and intracellular survival assays as well as infections in an in vivo model of human small intestine in SCID mice (10, 17, 31).
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in the course of this study are described in Table 1. Bacteria were routinely cultured on L agar or in L broth supplemented with aromatic compounds as previously described (5). Ampicillin was used at a final concentration of 50 μg/ml. All chemicals and antibiotics were obtained from Sigma (Poole, United Kingdom), unless otherwise stated.
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Relevant property(ies) | Reference or source |
|---|---|---|
| S. typhi | ||
| BRD681 | S. typhi Ty2 ΔaroA | 4 |
| BRD691 | S. typhi Ty2 ΔaroAaroC | 4 |
| BRD1116 | BRD691 ΔhtrA | This work |
| E. coli | ||
| HB101 | F′ pro leu thi hsdSB20 recA endA rpsL20 (Strr) | 31 |
| DH5α | endA hsdR17 supE44 thi-1 recA1ϕ80dlacZ ΔM15 | Gibco-BRL |
| SM10λpir | λpir lysogen | 26 |
| Plasmids | ||
| pUC18 | Cloning vehicle | Boehringer Mannheim |
| pTYH1 | pUC18 plus 3.2-kb HindIII fragment of S. typhi htrA | This work |
| pΔTYAH4 | pTYH1 cut with EcoRV and religated | This work |
| pTYHTR | pTYH1 containing 659-bp deletion in htrA | This work |
| pGP704 | Suicide replicon; Apr | 26 |
| pTYHTG1 | pGP704 containing htrA gene with 659-bp deletion | This work |
DNA manipulation techniques.
Unless otherwise stated, DNA manipulations were carried out as described by Sambrook et al. (30). Restriction enzymes, plasmid vectors, and buffers were purchased from Boehringer Mannheim or New England Biolabs (Hitchin, United Kingdom). T4 DNA ligase was purchased from Gibco-BRL (Paisley, United Kingdom). Chromosomal DNA of S. typhi was isolated by the method of Hull et al. (18), except that the crude DNA extract was incubated overnight at 50°C in the presence of proteinase K and sodium lauryl sarcosinate. Plasmid DNA was purified by using the WizardPrep system (Promega, Southampton, United Kingdom). DNA fragments were purified from agarose gels by the method of Tautz and Renz (36). Electroporation of S. typhi was carried out as described before (5).
Cloning of the S. typhi htrA gene.
The cloning and sequencing of the S. typhimurium htrA gene has been described previously (19). To isolate the S. typhi htrA gene, BRD691 DNA was purified, cleaved with HindIII and probed with a DNA fragment encoding the S. typhimurium htrA gene. This demonstrated that the S. typhi htrA gene was located on a 3.2-kb fragment sized similarly to the S. typhimurium htrA gene (data not shown). The S. typhi htrA gene was cloned by cleaving with HindIII, ligating the 3.2-kb fragment into the HindIII site of pUC18, and transforming into Escherichia coli HB101. Five hundred ampicillin-resistant colonies were isolated and screened for the presence of the S. typhi htrA gene by using the S. typhimurium htrA gene as the probe. Sixteen positive colonies were isolated, all of which contained the 3.2-kb HindIII fragment of S. typhi DNA (data not shown). One of these plasmids, designated pTYHI, was used for further studies.
Introduction of a defined deletion into the htrA gene of S. typhi.
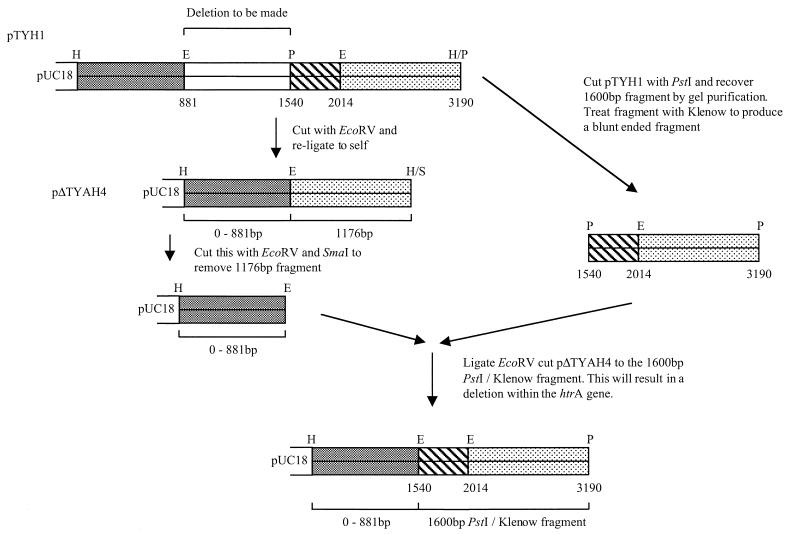
Restriction analysis and DNA sequencing of the S. typhi htrA gene revealed the presence of an EcoRV and PstI site within the coding sequence which could be used to generate a deletion (Fig. 1). pTYHI was first cleaved with EcoRV and then was religated and transformed into E. coli HB101. The resulting plasmid, pΔTYAH4, was digested with EcoRV and SmaI. The 3.57-kb fragment was recovered and ligated with the 1.6-kb PstI fragment purified from pTYHI, introducing a 659-bp deletion (pTYHTR). The suicide replicon pGP704 was used to introduce the htrA deletion into the chromosome of S. typhi (24). Plasmid pTYHTR was digested with HindIII and XmnI, and the resulting 2.53-kb fragment of S. typhi DNA was ligated into pGP704 and used to transform E. coli SM10λpir. A plasmid of the expected size was identified by restriction mapping (data not shown) and designated pTYHTG1.
FIG. 1.
Cloning strategy for the construction of a defined deletion in the S. typhi htrA gene. The final 2.53-kb fragment is cut with HindIII and cloned into pGP704 to produce pTYHTG1. Abbreviations: H, HindIII; E, EcoRV; P, PstI; S, SmaI.
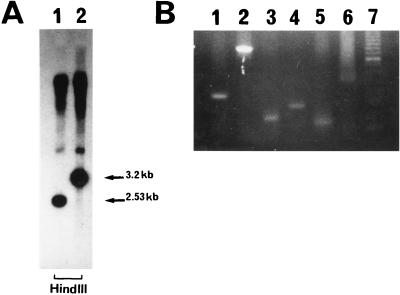
The construction and characterization of the S. typhi Ty2 aroA aroC strain BRD691 has been described previously (5). The plasmid pTYHTG1 was electroporated into BRD691, followed by screening for ampicillin-resistant colonies (i.e., those in which the suicide replicon had potentially become integrated into the chromosome at the htrA gene). Several of the ampicillin-resistant colonies were picked and grown overnight at 37°C in L broth supplemented with aromatic compounds but without ampicillin. The cultures were diluted to yield ∼200 colonies per plate and were spread onto L agar. Several hundred colonies were replica plated to isolate ampicillin-sensitive colonies (i.e., those in which a double recombination event had taken place, resulting in the loss of the suicide replicon). A total of 112 such colonies were screened by PCR with oligonucleotides corresponding to the regions upstream and downstream of the deletion in the htrA gene. One colony, designated BRD1116, produced a DNA fragment of the appropriate size. The genotype of this strain was confirmed by Southern hybridization and by PCR analysis for the aroA, aroC, and htrA genes (Fig. 2).
FIG. 2.
Southern hybridization and PCR analysis demonstrating the presence of deletions in the htrA, aroA, and aroC genes. (A) HindIII-cleaved chromosomal DNA from BRD1116 (lane 1) and BRD691 (lane 2) reacted with the enhanced chemiluminescence-labelled probe. (B) Screening of S. typhi colonies for deletions in aroA, aroC, and htrA by amplifying the deletion region of chromosomal DNA directly from whole cells by PCR. Lanes: 1, BRD1116 carrying 330-bp htrA deletion fragment; 2, BRD691 carrying functional 989-bp htrA fragment; 3, BRD1116 carrying 175-bp deletion aroC fragment; 4, BRD681 carrying 272-bp aroC fragment; 5, BRD1116 carrying 120-bp aroA deletion fragment; 6, CVD908 carrying functional 504-bp aroA fragment; 7, molecular weight standard.
DNA amplification by PCR.
PCR with S. typhi colonies was carried out with Taq DNA polymerase and the GeneAmp kit (Perkin-Elmer Cetus, Norwalk, Conn.) as described previously (5). The oligonucleotides used in the PCR were derived from the 5′ end and a sequence complementary to the 3′ end of the htrA gene. The oligonucleotides BO5710 (5′ GAAATTCATGGCGCTGGGCTCCGGCGTAAT 3′) and BO5711 (5′ TGAGGCCAATTTGCGCGGCGGGTGAGTTC 3′) mapped at bp 721 to 750 and bp 1710 to 1739 of the S. typhi sequence, respectively. The two sets of primers used to amplify regions within the aroA and aroC genes have been described previously (5).
Southern hybridization.
Chromosomal DNA from S. typhi strains was cleaved with HindIII, followed by electrophoretic separation on a 1% agarose slab gel and was finally transferred onto nitrocellulose filters. The filters were hybridized with the PCR-generated DNA fragments encoding the intact htrA gene, which had been labelled using the ECL Random Prime kit (Amersham, Little Chalfont, United Kingdom).
Heat stress response.
Bacteria were grown at 30°C overnight with shaking and were subsequently diluted 1:500, grown to an optical density at 650 nm of 0.1, and transferred to 45°C. An aliquot of 100 μl of the culture was removed, serially diluted in sterile phosphate-buffered saline (PBS), and plated onto L agar in order to calculate viability after 30, 60, 90, 120, and 150 min.
Oxidative stress response.
Bacteria were grown at 30°C overnight with shaking and were subsequently diluted 1:500 into L broth containing 10 mM hydrogen peroxide. Viable counts were taken at 30, 60, 90, 120, and 150 min following the initial subculture.
Cell culture medium.
Dulbecco’s modified essential medium (DMEM) containing l-glutamine and nonessential amino acids supplemented with 10% heat-inactivated fetal calf serum was used to maintain the human adenocarcinoma cell lines Caco-2 and HT-29 and the murine macrophage cell line RAW264.7. RPMI-1640 medium supplemented with 10% fetal calf serum was used to maintain the human monocytic cell line U937.
Invasion and intracellular survival assays.
The invasion and intracellular survival assays were carried out essentially as described by Lee and Falkow (23). Caco-2 and HT-29 cells were dispensed into 24-well tissue culture trays (model 25820; Corning) at 5 × 104 cells per well and were incubated until confluency at 37°C in 5% CO2. RAW264.7 macrophage cells were seeded at 105 cells per well 24 h before infection. Bacterial strains were aerobically grown overnight at 37°C in aro broth. An aliquot of this overnight culture was diluted 1:1,000 in aro broth and aerobically grown at 37°C until late exponential phase (optical density at 650 nm, 0.6 to 0.8). Cells were inoculated with 107 CFU of the bacterial strains suspended in DMEM (Caco-2 and HT-29) or RPMI-1640 (RAW264.7) (multiplicity of infection, 50:1) and incubated for 2, 24, 48, and 72 h. Cells were subsequently washed three times with PBS, and growth of extracellular bacteria was inhibited by replacing the media with DMEM or RPMI-1640 containing 50 μg of gentamicin per ml. Cells were further incubated for 1 h, washed three times with PBS, and lysed by the addition of 1 ml of 0.1% Triton X-100 in PBS for 30 min at 37°C. Lysates were diluted in PBS and plated onto L agar overnight at 37°C for enumeration of viable bacteria. For infection periods longer than 2 h, the medium was replaced after 2 h with DMEM containing 10 μg of gentamicin per ml, until lysis.
Infection of U937 cells was carried out in a similar manner, with the following modifications. The cells were seeded in 25-cm2 flasks (model 430639; Costar) at a concentration of 105 cells/ml and were incubated with phorbol myristate acetate (10 ng/ml) for 48 h. The medium was replaced, and viable cells were enumerated using trypan blue staining. Bacterial strains were resuspended in RPMI-1640 and added to the cells at a multiplicity of infection of 50:1. After a 2-h infection, cells were pelleted and washed three times with PBS and the medium was replaced with RPMI-1640 containing gentamicin (50 μg/ml). After a further 1-h incubation, numbers of viable cells were calculated and the medium was removed. The cells were washed three times with PBS and lysed with PBS–Triton X-100 as described above. Intracellular survival of Salmonella and the effect on growth of U937 were determined over 72-h.
Cell viability assay.
In order to determine the effects of infection with the vaccine strains on the viability of epithelial cells, the monotetrazolium (MTT) assay was used (2).
Cells were seeded in 96-well tissue culture plates at a concentration of 5 × 104 cells per well, in 100 μl of medium. Cells were infected for 1 h at a bacterium/cell ratio of 50:1. The cells were washed three times with sterile PBS, and the medium was replaced with fresh DMEM without phenol red, containing 50 μg of gentamicin per ml. Cells were incubated for 48 h at 37°C. MTT stock solution (10 μl; 5 mg/ml in DMEM without phenol red) was added to the cells and incubated at 37°C for 3 h. Insoluble formazan salt was dissolved by the addition of 100 μl of 20% (wt/vol) sodium dodecyl sulfate in 10 mM HCl overnight at 37°C. Absorbance of the converted dye was measured at a wavelength of 570 nm.
Quantification of IL-8 production by infected epithelial monolayers.
HT-29 cells were seeded in six-well tissue culture plates at a concentration of 1 × 105 to 2 × 105 cells/ml, grown until confluency, and infected as detailed above. At 3, 6, 24, 48, and 72 h after the infection, the supernatant was removed, frozen in liquid nitrogen, and stored at −70°C. Interleukin-8 (IL-8) enzyme-linked immunosorbent assays of the supernatants were subsequently performed as previously described (9), with optimal concentrations of polyclonal goat anti-human IL-8 antibodies (R & D Systems, Minneapolis, Minn.) as capturing antibodies and polyclonal rabbit anti-human IL-8 antibodies (Endogen, Boston, Mass.) as detecting antibodies. Alkaline phosphatase-labelled monoclonal mouse anti-rabbit immunoglobulin G (Sigma) was used as a second-step antibody. Bound alkaline phosphatase was visualized with the substrate p-nitrophenylphosphate (Sigma). The sensitivity of the IL-8 enzyme-linked immunosorbent assay was 50 pg/ml.
Human intestinal xenograft infections.
We recently described a xenogeneic model of human small intestine as a system to study Salmonella infection of nontransformed epithelial cells in vivo (10, 18, 31). Briefly, pieces of human fetal intestine (mean gestational age = 12 weeks) were implanted subcutaneously in CB-17 SCID mice and were allowed to develop for 12 to 16 weeks. During this time the xenografts develop a mucosa which shows morphological and functional similarities to that of pediatric intestine (31). All procedures were performed with the full approval of the Cambridge Local Ethics Committee and in accordance with Home Office guidelines.
Bacteria were grown to late exponential phase and were resuspended in sterile PBS to a concentration of 5 × 108 CFU/ml. SCID mice were subcutaneously inoculated with 0.1 ml of the bacterial strains, using a Microlance 3 needle (25 gauge; Becton Dickinson, Drogheda, Ireland) inserted through the xenograft wall into the lumen. At 6 and 24 h following infection, mice were killed and the xenografts, spleens, and livers were removed. The organs were homogenized in sterile PBS containing 0.1% Triton X-100, serially diluted, and plated onto L agar in order to enumerate bacteria. Spleen and liver counts were pooled to determine translocation. Half of each xenograft was organ cultured in DMEM containing 50 μg of gentamicin per ml for 1 h at 37°C prior to homogenization. Mice without intestinal xenografts that had been inoculated intraperitonally with either 106, 107, or 108 bacteria were killed at 6, 24, 48, and 72 h, and pooled spleen and liver bacterial counts were made.
Statistical analysis.
Data are expressed as means ± standard errors of the means (SEM). Statistical significance of differences between groups was assessed by Mann-Whitney U tests on untransformed data.
RESULTS
Heat and oxidative stress response of BRD691 and BRD1116.
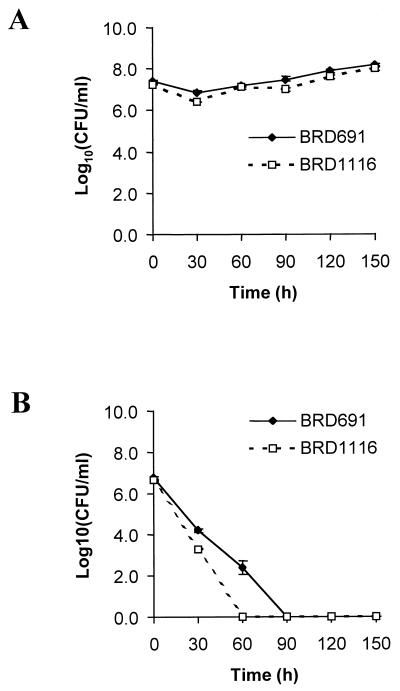
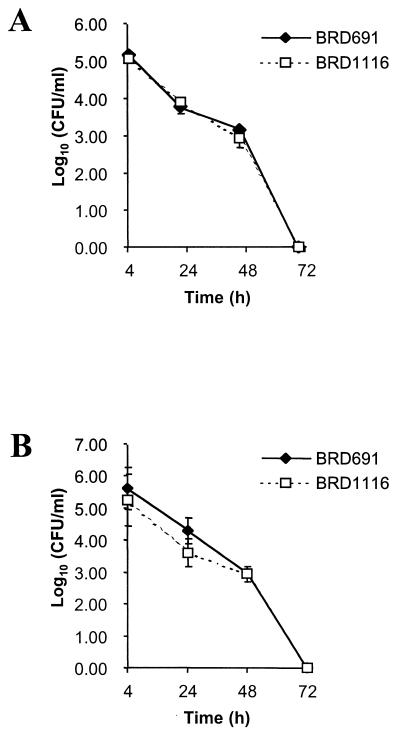
The E. coli HtrA periplasmic protease has been shown to be essential for bacterial growth at elevated temperatures, as it is required to degrade thermally aggregated proteins (22). However, in S. typhimurium the htrA gene is essential for survival in medium containing significant levels of hydrogen peroxide but not for survival at elevated temperatures (19). The ability of BRD1116 to survive increased temperatures and exposure to hydrogen peroxide was therefore examined. BRD1116 exhibited properties similar to those of S. typhimurium htrA for both heat and oxidative shock (Fig. 3) in that there was no significant difference in the ability to withstand increased temperatures but a markedly increased susceptibility to oxidative stress compared with that of BRD691 was observed.
FIG. 3.
The effect of 10 mM H2O2 (A) and a growth temperature of 42°C (B) on the viability of the S. typhi htrA mutant BRD1116 and the parent strain BRD691. Data are means ± SEM (error bars) (n = 6).
Decreased epithelial intracellular survival of BRD1116 compared with BRD691.
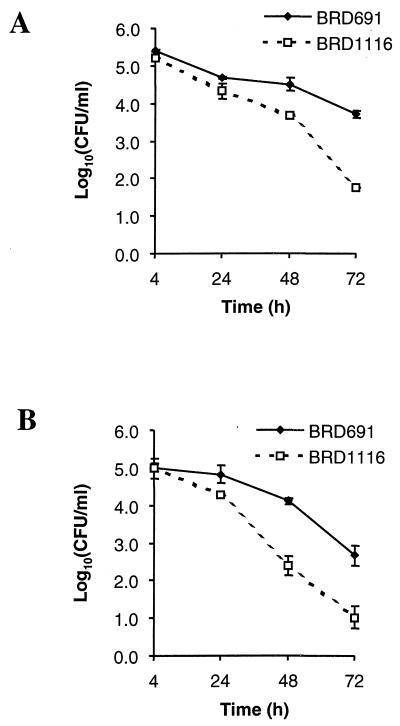
The ability of BRD1116 and BRD691 to invade and survive in the human adenocarcinoma cell lines Caco-2 and HT-29 was examined. These cell lines, which form monolayers and differentiate to different extents when confluent, forming a well-developed brush border membrane in the case of Caco-2, have been used extensively as models for human intestinal epithelium (29). There was no significant difference between BRD691 or BRD1116 invasion of Caco-2 or HT-29 monolayers (Fig. 4), indicating that htrA is not directly involved in the invasion process. After 48 h there was a 10-fold decrease in the numbers of viable BRD1116 recovered in comparison to BRD691. By 72 h ∼100-fold less viable BRD1116 was recovered. These results suggest that the S. typhi HtrA protease is required for prolonged survival within intestinal epithelial cells.
FIG. 4.
Invasion and intracellular survival of BRD691 and BRD1116 in the human intestinal epithelial cell lines Caco-2 (A) and HT-29 (B) over 72 h. Data are means ± SEM (error bars) (n = 6).
The effect of infection with the two S. typhi strains on the viability of Caco-2 and HT-29 intestinal epithelial cells was also examined, with MTT used as a marker of cell viability. There was no significant effect on cell viability following infection with either strain in either cell line after 48 h (Table 2), suggesting that these strains do not cause significant differences in epithelial cell death upon invasion.
TABLE 2.
Viability of Caco-2 and HT-29 cells in response to infection with S. typhi or noninvasive E. coli
| Cell line | % Viable cells (mean ± SEM) after infection witha:
|
||
|---|---|---|---|
| BRD691 | BRD1116 | E. coli HB101 | |
| Caco-2 | 93.42 ± 1.6 | 96.45 ± 7.74 | 98.64 ± 3.76 |
| HT-29 | 91.92 ± 2.20 | 93.43 ± 1.34 | 96.24 ± 3.41 |
Cell viability was assessed 48 h following initial infection, by using the tetrazolium dye MTT. Values are expressed as mean percentages compared to uninfected controls ± SEM (n = 6).
Intracellular survival of BRD1116 and BRD691 within the human monocytic cell line U937 and the macrophage cell line RAW264.7.
The ability of BRD1116 and BRD691 to invade and survive within activated U937 cells (a human monocytic cell line) and the murine macrophage cell line RAW264.7 was then examined. Activated U937 cells have been used extensively as a human macrophage model (28). There was no significant difference between the two strains in their ability to invade and survive within this cell line (Fig. 5A), as both strains were eliminated from the culture within 72 h. There was no significant difference in growth or viability of U937 cells infected with either strain, compared to uninfected controls (data not shown).
FIG. 5.
Invasion and intracellular survival of BRD691 and BRD1116 in the human monocytic cell line U937 (A) and the murine macrophage cell line RAW264.7 (B) over 72 h. Data are means ± SEM (error bars) (n = 6).
There was a three- to fourfold decrease in the numbers of viable BRD1116 recovered from RAW264.7 cells at 4 and 24 h after infection, suggesting a decreased ability of BRD1116 to survive within this cell line. After 48 h however, there was no difference in the numbers of viable bacteria recovered for either strain.
BRD1116 is attenuated in an in vivo model system of human intestine.
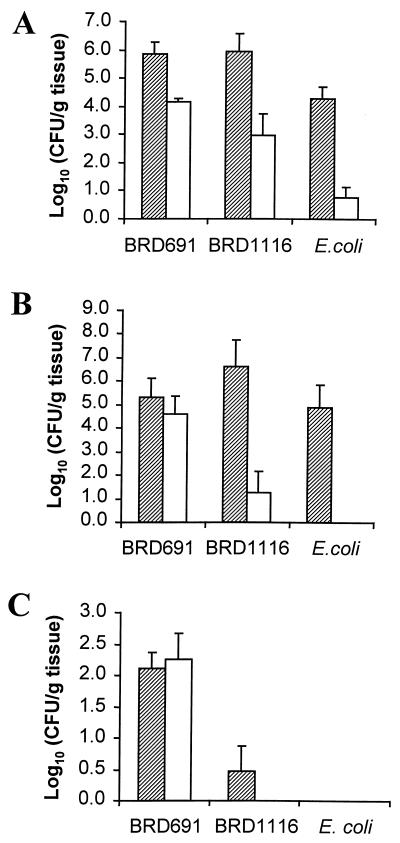
A xenogeneic model of human intestine was used to examine S. typhi invasion, intracellular survival and translocation to secondary sites of infection. Using the gentamicin resistance assay to quantify intracellular bacteria, it was possible to compare the ability of the two vaccine strains to invade the intestinal mucosa. In additional experiments E. coli DH5α was inoculated in vivo into the xenograft lumen to test the ability of the system to exclude a noninvasive, nonpathogenic organism. At 6 h following infection there was no significant difference between BRD1116 and BRD691 either in the levels of total bacteria present within the tissue or in the levels of intracellular bacteria (Fig. 6A). Very few viable E. coli were recovered, indicating that the xenograft mucosa forms a tight monolayer, impermeable to noninvasive organisms. Tissue examined 24 h after infection (Fig. 6B) showed a clear decrease in the numbers of viable intracellular BRD1116 compared to BRD691. Translocation of BRD1116 to secondary sites of infection, e.g., spleen and liver, was significantly reduced compared to that of BRD691 after 6 h and was completely abrogated within 24 h (Fig. 6C).
FIG. 6.
Invasion and intracellular survival of BRD691 and BRD1116 in an in vivo xenograft model of human intestine after 6 h (A) and 24 h (B) of infection. Shaded bars represent total counts in the xenografts, and open bars represent counts obtained from xenografts incubated with gentamicin prior to homogenization. (C) Translocation of bacteria to secondary sites of infection (spleen plus liver counts) over 6 h (shaded bars) and 24 h (open bar). Data are means ± SEM (error bars) (n = 4).
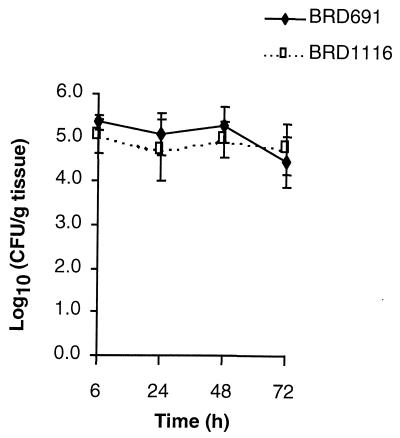
BRD691 and BRD1116 were assessed for the ability to invade and persist within SCID mice that did not carry intestinal xenografts. Mice of the same age as those used for xenograft experiments were infected intraperitonally. Spleen and liver counts (Fig. 7) showed no significant difference in levels of the two bacterial strains over a 72-h period, indicating that the htrA gene is not required for survival within the reticuloendothelial system in this model.
FIG. 7.
Persistence of BRD691 and BRD1116 in spleens and livers of SCID mice infected intraperitonally with 107 bacteria over 72 h. Data are means ± SEM (error bars) (n = 4).
Ability of S. typhi vaccine strains to elicit an increase in the proinflammatory chemokine IL-8 in infected epithelial cells.
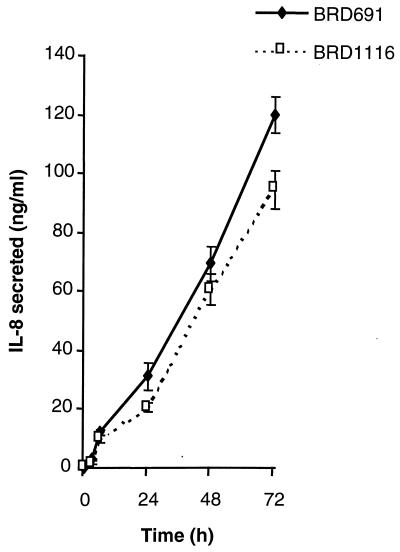
Invasive bacterial strains provoke a cascade of proinflammatory host events following invasion of epithelial cells (9, 21, 25). An important proinflammatory signal molecule during this event is the chemokine IL-8, which is a chemoattractant for neutrophils. The ability of BRD1116 and BRD691 to upregulate the production of IL-8 following infection of HT-29 intestinal epithelial monolayers was compared over a 72-h period (Fig. 8). BRD1116 elicited a slightly smaller IL-8 response than the parent strain at all time points. However, these differences were not found to be significant (P > 0.05).
FIG. 8.
Secretion of IL-8 by HT-29 cells following infection with BRD691 or BRD1116. Data are means ± SEM (error bars) (n = 2).
DISCUSSION
We have examined the properties of two candidate S. typhi live oral typhoid vaccine strains using in vitro and in vivo assays. These assays assessed the response of the strains to heat and oxidative stress, their ability to invade and survive within tissue culture cells, their ability to induce epithelial proinflammatory cytokine production in vitro, and their ability to invade epithelial cells and to disseminate in a human intestinal xenograft model. In common with S. typhimurium htrA mutants, BRD1116 has an increased susceptibility to oxidative stress in comparison to BRD691, the S. typhi aroA aroC parent strain. The ability of BRD1116 to survive in the human macrophage cell line U937 or the murine macrophage cell line over 72 h was not reduced, although there was a reduction in viable BRD1116 bacteria recovered from RAW264.7 cells within the first 24 h of infection, compared to BRD691. BRD1116 was defective in its ability to survive in the human intestinal epithelial cell lines Caco-2 and HT-29 over a 72-h period. These data suggest that the HtrA periplasmic protease is required for survival during the initial invasion of the intestinal mucosa but not the subsequent invasion of recruited macrophages. We recognize, however, that the U937 macrophage cell line manifests impaired nitrite production (28) and in this respect may not be an accurate model of macrophage killing in vivo. The intracellular survival of the two strains may therefore be more accurately observed within the murine macrophage cell line RAW264.7, although killing of S. typhi strains within this cell line is likely to be mainly due to host-restricted mechanisms. A previous report has demonstrated, though, that S. typhimurium htrA mutants have been shown to have an increased susceptibility to killing in isolated murine peritoneal macrophages (1).
Infections of Caco-2 and HT-29 cell lines demonstrated that over 72 h, these cells were capable of inhibiting the growth of internalized S. typhi vaccine strains. Internalized Salmonella bacteria have been widely shown to replicate within the epithelium (24). A recent ultrastructural report showed a simultaneous degradation and replication of intracellular bacteria within Caco-2 cells (37). Mutants of Salmonella that contain deletions in the aro genes are impaired in their ability to replicate, which shifts this equilibrium so that there is a reduction in the number of intracellular bacteria over time. Although the bactericidal mechanisms of epithelial cells are largely unknown, the data shown here suggest that htrA is required by the bacteria to respond to this host response.
In order to compare the survival and replication properties of BRD691 and BRD1116 in nontransformed human intestinal epithelium, a model using human intestine implanted in SCID mice was utilized. The decrease in intracellular survival and translocation exhibited by BRD1116 following intralumenal infection in this system provides additional evidence for epithelium-mediated killing of bacteria in vivo. This is particularly apparent compared to intraperitonal infection, in which no significant difference in the ability of the htrA mutant and the parent strain to infect systemic organs like the spleen and liver was noted. Whereas the intestinal xenografts contain a completely human epithelium and neighboring human intraepithelial lymphocytes, the lamina propria contains ∼70% human cells and 30% murine cells in a chimeric mixture. Moreover, S. typhi is host restricted to humans and does not cause a typhoid-like disease in SCID mice. Thus, although the initial interactions of the invasive bacteria and the xenograft epithelium are reflective of the in vivo cross-talk that may take place following administration of the vaccine strain to humans, the subsequent invasion of murine leukocytes and systemic translocation to other organs may be less instructive. The inability of BRD1116 to spread to the spleen and liver in infected animals may also, for example, be due to preferential killing by CD68+ human macrophages present in the xenograft mucosa (unpublished findings) or may be due to increased host-restricted killing by murine macrophages and neutrophils. The results obtained from this SCID model do, however, provide in vivo evidence for the intestinal mucosa as a site of attenuation of BRD1116.
A recent study on closely related S. typhi htrA live vaccine strains administered to volunteers reported mild diarrhea in a small number of subjects (34). A possible mechanism postulated for these findings was a change in the interaction of the bacteria with the mucosa, resulting in a different cytokine release pattern. One of the important cytokines whose production is increased following invasion by pathogenic bacteria is the neutrophil chemoattractant IL-8 (9, 21, 25). In this study, no significant difference was observed in the ability of BRD1116 and BRD691 to induce epithelial IL-8, which demonstrates that the htrA deletion does not significantly alter the proinflammatory potential of this strain.
BRD1116 and other S. typhi htrA mutant strains are currently undergoing phase 1 clinical trials and may ultimately form the basis of multivalent vaccines expressing heterologous antigens from a number of pathogenic microorganisms. Thus, the ability to ascertain the in vivo role of mutations in htrA and similar genes will be increasingly important. The assays described herein, particularly the intestinal xenograft assay, should prove of particular value in this regard.
ACKNOWLEDGMENTS
This work was primarily supported by the Medeva Vaccine Research Unit. G.D. was supported by a grant from the Wellcome Trust. D.C.L. is a collaborative student funded by the Medical Research Council. L.E. is a recipient of a Career Development Award of the Crohn’s and Colitis Foundation of America.
REFERENCES
- 1.Baumler A J, Kusters J G, Stojiljkovic I, Heffron F. Salmonella typhimurium loci involved in survival within macrophages. Infect Immun. 1994;62:1623–1630. doi: 10.1128/iai.62.5.1623-1630.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carmichael J, Dgraff W G, Gazdar A F, Minna J D, Mitchell J B. Evaluation of a tetrazolium-based semiautomated colorimetric assay—assessment of chemosensitivity testing. Cancer Res. 1987;47:936–942. [PubMed] [Google Scholar]
- 3.Carter P B, Collins F M. The route of enteric infection in normal mice. J Exp Med. 1974;139:1189–1203. doi: 10.1084/jem.139.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chatfield S N, Charles I G, Makoff A J, Oxer M D, Dougan G, Pickard D, Slater D, Fairweather N F. Use of the nirB promoter to direct the stable expression of heterologous antigens in Salmonella oral vaccine strains: development of a single-dose oral tetanus vaccine. Bio/Technology. 1992;10:888–892. doi: 10.1038/nbt0892-888. [DOI] [PubMed] [Google Scholar]
- 5.Chatfield S N, Fairweather N F, Charles I G, Pickard D, Levine M M, Hone D, Posada M, Strugnell R A, Dougan G. Construction of a genetically defined Salmonella typhi Ty2 aroA, aroC mutant for the engineering of a candidate oral typhoid-tetanus vaccine. Vaccine. 1992;10:53–60. doi: 10.1016/0264-410x(92)90420-o. [DOI] [PubMed] [Google Scholar]
- 6.Chatfield S N, Strahan K, Pickard D, Charles I G, Hormaeche C E, Dougan G. Evaluation of Salmonella typhimurium strains harboring defined mutations in htrA and aroA in the murine salmonellosis model. Microb Pathog. 1992;12:145–151. doi: 10.1016/0882-4010(92)90117-7. [DOI] [PubMed] [Google Scholar]
- 7.Chen L M, Kaniga K, Galán J E. Salmonella spp. are cytotoxic for cultured macrophages. Mol Microbiol. 1996;21:1101–1115. doi: 10.1046/j.1365-2958.1996.471410.x. [DOI] [PubMed] [Google Scholar]
- 8.Clark M A, Jepson M A, Simmons N L, Hirst B H. Preferential interaction of Salmonella typhimurium with mouse Peyer’s patch M cells. Res Microbiol. 1994;145:543–552. doi: 10.1016/0923-2508(94)90031-0. [DOI] [PubMed] [Google Scholar]
- 9.Eckmann L, Kagnoff M F, Fierer J. Epithelial cells secrete the chemokine interleukin-8 in response to bacterial entry. Infect Immun. 1993;61:4569–4574. doi: 10.1128/iai.61.11.4569-4574.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eckmann L, Stenson W F, Savidge T C, Lowe D C, Barrett K E, Fierer J, Smith J R, Kagnoff M F. Role of intestinal epithelial cells in the host secretory response to invasive bacteria. J Clin Investig. 1997;100:296–309. doi: 10.1172/JCI119535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fouts T R, Lewis G K, Hone D M. Construction and characterization of a Salmonella typhi-based human immunodeficiency virus type 1 vector vaccine. Vaccine. 1995;13:561–569. doi: 10.1016/0264-410x(94)00016-g. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez C, Hone D, Noriega F R, Tacket C O, Davis J R, Losonsky G, Nataro J P, Hoffman S, Malik A, Nardin E, Sztein M B, Heppner D G, Fouts T R, Isibasi A, Levine M M. Salmonella typhi vaccine strain CVD908 expressing the circumsporozoite protein of Plasmodium falciparum: strain construction and safety and immunogenicity in humans. J Infect Dis. 1994;169:927–931. doi: 10.1093/infdis/169.4.927. [DOI] [PubMed] [Google Scholar]
- 13.Hohmann E L, Oletta C A, Killeen K P, Miller S I. PhoP/phoQ-deleted Salmonella typhi (Ty800) is a safe and immunogenic single-dose typhoid fever vaccine in volunteers. J Infect Dis. 1996;173:1408–1414. doi: 10.1093/infdis/173.6.1408. [DOI] [PubMed] [Google Scholar]
- 14.Hoiseth S K, Stocker B A D. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature (London) 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 15.Hone D M, Harris A M, Chatfield S, Dougan G, Levine M M. Construction of genetically defined double aro mutants of Salmonella typhi. Vaccine. 1991;9:810–816. doi: 10.1016/0264-410x(91)90218-u. [DOI] [PubMed] [Google Scholar]
- 16.Hone D M, Tacket C O, Harris A M, Kay B, Losonsky G, Levine M M. Evaluation in volunteers of a candidate live oral attenuated Salmonella typhi vector vaccine. J Clin Investig. 1992;90:412–420. doi: 10.1172/JCI115876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang G T-J, Eckmann L, Savidge T C, Kagnoff M F. Infection of human intestinal epithelial cells with invasive bacteria upregulates apical intercellular adhesion molecule-1 (ICAM-1) expression and neutrophil adhesion. J Clin Investig. 1996;98:572–583. doi: 10.1172/JCI118825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hull R A, Gill R E, Hsu P, Minshew B H, Falkow S. Construction and expression of recombinant plasmids encoding type 1 and mannose resistant pili from a urinary tract infection Escherichia coli isolate. Infect Immun. 1981;33:933–938. doi: 10.1128/iai.33.3.933-938.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson K, Charles I G, Dougan G, Pickard D, O’Gaora P, Costa G, Ali T, Miller I, Hormaeche C. The role of a stress-response protein in Salmonella typhimurium virulence. Mol Microbiol. 1991;5:401–407. doi: 10.1111/j.1365-2958.1991.tb02122.x. [DOI] [PubMed] [Google Scholar]
- 20.Jones B D, Ghori N, Falkow S. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer’s patches. J Exp Med. 1994;180:15–23. doi: 10.1084/jem.180.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jung H C, Eckmann L, Yang S-K, Panja A J, Fierer J, Morzycka-Wroblewska E, Kagnoff M F. A distinct array of proinflammatory cytokines is expressed in human colon epithelial cells in response to bacterial invasion. J Clin Investig. 1995;95:55–65. doi: 10.1172/JCI117676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laskowska E, Kuczyflska-Wicnik D, Skrko-Glonek J, Taylor A. Degradation by proteases Lon, Clp and HtrA, of Escherichia coli proteins aggregated in vivo by heat shock; HtrA protease action in vivo and in vitro. Mol Microbiol. 1996;22:555–571. doi: 10.1046/j.1365-2958.1996.1231493.x. [DOI] [PubMed] [Google Scholar]
- 23.Lee C A, Falkow S. The ability of Salmonella to enter mammalian cells is affected by bacterial growth state. Proc Natl Acad Sci USA. 1990;87:4304–4308. doi: 10.1073/pnas.87.11.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leung K Y, Finlay B B. Intracellular replication is essential for the virulence of Salmonella typhimurium. Proc Natl Acad Sci USA. 1991;88:11470–11474. doi: 10.1073/pnas.88.24.11470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCormick B A, Colgan S P, Delp-Archer C, Miller S I, Madara J L. Salmonella typhimurium attachment to human intestinal epithelial monolayers: transcellular signalling to subepithelial neutrophils. J Cell Biol. 1993;123:895–907. doi: 10.1083/jcb.123.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monack D M, Raupach B, Hromockyi A E, Falkow S. Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc Natl Acad Sci USA. 1996;95:9833–9838. doi: 10.1073/pnas.93.18.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ralph P. Macrophage cell lines. In: Weir D M, editor. Handbook of experimental immunology. 4th ed. 2. Cellular immunology. Oxford, United Kingdom: Blackwell Scientific Publications; 1986. pp. 45.1–45.15. [Google Scholar]
- 29.Rousset M. The human colon carcinoma cell lines HT-29 and Caco-2. Two in vitro models for the study of intestinal differentiation. Biochimie. 1986;68:1035–1040. doi: 10.1016/s0300-9084(86)80177-8. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 31.Savidge T C, Morey A L, Ferguson D J P, Fleming K A, Shmakov A N, Phillips A D. Human intestinal development in a severe-combined immunodeficient xenograft model. Differentiation. 1995;58:361–371. doi: 10.1046/j.1432-0436.1995.5850361.x. [DOI] [PubMed] [Google Scholar]
- 32.Strugnell R, Dougan G, Chatfield S N, Charles I G, Fairweather N F, Tite J, Li J L, Beesley J, Roberts M. Characterization of a Salmonella typhimurium aro vaccine strain expressing the P.69 antigen of Bordetella pertussis. Infect Immun. 1992;60:3994–4002. doi: 10.1128/iai.60.10.3994-4002.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tacket C O, Hone D M, Curtiss III R, Kelly S M, Losonsky G, Guers L, Harris A M, Edelman R, Levine M M. Comparison of the safety and immunogenicity of Salmonella typhi strains in adult volunteers. Infect Immun. 1992;60:536–541. doi: 10.1128/iai.60.2.536-541.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tacket C O, Hone D M, Losonsky G A, Guers L, Edelman R, Levine M M. Clinical acceptability and immunogenicity of CVD 908 Salmonella typhi vaccine strain. Vaccine. 1992;10:443–446. doi: 10.1016/0264-410x(92)90392-w. [DOI] [PubMed] [Google Scholar]
- 35.Tacket C O, Sztein M B, Losonsky G A, Wasserman S S, Nataro J P, Edelman R, Pickard D, Dougan G, Chatfield S N, Levine M M. Safety of live oral Salmonella typhi vaccine strains with deletions in htrA and aroC aroD and immune response in humans. Infect Immun. 1997;65:452–456. doi: 10.1128/iai.65.2.452-456.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tautz D, Renz M. An optimised freeze-squeeze method for the recovery of DNA fragments from agarose gels. Anal Biochem. 1992;132:14–19. doi: 10.1016/0003-2697(83)90419-0. [DOI] [PubMed] [Google Scholar]
- 37.Wells C L, van de Westerlo E M A, Jechorek R P, Erlandsen S L. Intracellular survival of enteric bacteria in cultured human enterocytes. Shock. 1996;6:27–34. doi: 10.1097/00024382-199607000-00007. [DOI] [PubMed] [Google Scholar]
- 38.Yang S, Eckmann L, Panja A, Kagnoff M F. Differential and regulated expression of C-X-C, C-C, and C-chemokines by human colon epithelial cells. Gastroenterology. 1997;113:1214–1223. doi: 10.1053/gast.1997.v113.pm9322516. [DOI] [PubMed] [Google Scholar]