Abstract
Survivin is a component of the chromosomal passenger complex, which includes Aurora B, INCENP, and Borealin, and is required for chromosome segregation and cytokinesis. We performed a genome-wide screen of deubiquitinating enzymes for survivin. For the first time, we report that USP19 has a dual role in the modulation of mitosis and tumorigenesis by regulating survivin expression. Our results found that USP19 stabilizes and interacts with survivin in HCT116 cells. USP19 deubiquitinates survivin protein and extends its half-life. We also found that USP19 functions as a mitotic regulator by controlling the downstream signaling of survivin protein. Targeted genome knockout verified that USP19 depletion leads to several mitotic defects, including cytokinesis failure. In addition, USP19 depletion results in significant enrichment of apoptosis and reduces the growth of tumors in the mouse xenograft. We envision that simultaneous targeting of USP19 and survivin in oncologic drug development would increase therapeutic value and minimize redundancy.
Keywords: deubiquitinating enzyme, USP19, mitosis, chromosome, survivin, apoptosis, protein turnover, tumor progression
Graphical abstract
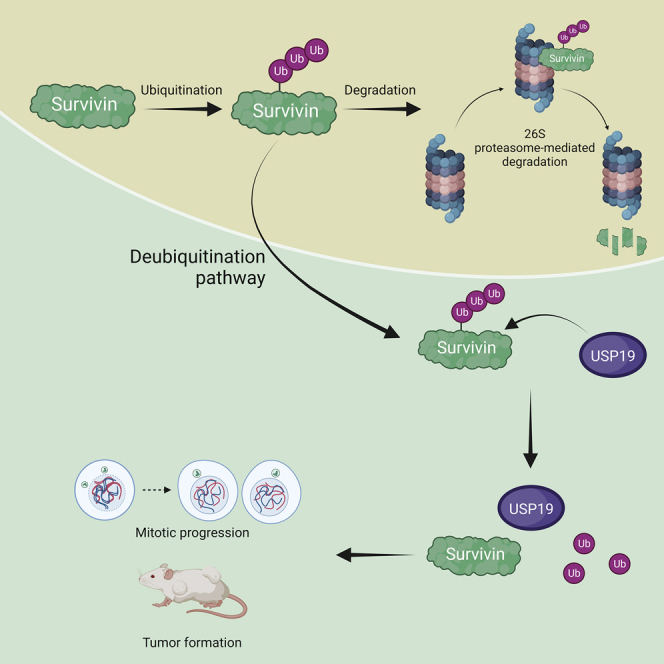
In this study, Chandrasekaran et al. found that USP19 regulated mitotic progression by stabilizing survivin protein. Loss of USP19 in HCT116 cells displayed several mitotic defects and cytokinesis failure. USP19 also regulated oncogenic functions of survivin. The in vivo mouse tumor model demonstrated that USP19 was involved in survivin-mediated tumor progression.
Introduction
Apoptosis is a physiological cell death process that is pivotal for the maintenance of healthy cells and tissues.1,2,3 Dysregulation between the balance of apoptosis and proliferation results in cancer.3 The inhibitor of apoptosis protein (IAP) family is an important family of proteins involved in the blocking of apoptotic cell death.4 Cancer cells use numerous evasion approaches to avoid being detected or eradicated by immune cells. Resistance to apoptosis is one of the imperative evasion strategies through which cancer cells escape detection and enhance proliferation. Hence, proteins associated with the regulation of apoptosis hold significant importance and can be potential targets for tumor therapy.5
The IAP family is characterized by a domain of approximately 70 amino acids known as the baculoviral IAP repeat (BIR).6 The mammalian genome encodes eight IAP family members BIRC1–8. Among these, BIRC5 (the gene that encodes survivin protein) contains a BIR domain that suppresses apoptosis and is organized structurally as a stable dimer.6,7,8,9 Survivin is overexpressed in several cancers10,11,12,13,14 and is the fourth most highly expressed protein in human cancer tissues compared to normal control tissues.15 Survivin dysregulates apoptosis by blocking caspase activation and also alters sensitivity to antitumor drugs, resulting in disease survival or recurrence.5 Owing to its indisputable role in cancer, blocking the survivin oncogene or factors that stabilize the survivin protein by various immunotherapeutic or molecular approaches are promising therapeutic strategies in cancer.5
Survivin is also a key factor expressed in the G2/M phase of the cell cycle. The chromosomal passenger complex (CPC) is a hetero-tetrameric complex that is composed of four subunits: the three regulatory and targeting components survivin, inner centromere protein (INCENP), and Borealin, and the enzymatic component Aurora B.16 CPC targets different locations at differing times during mitotic progression and regulates key mitotic events such as the correction of chromosome-microtubule attachment errors, regulation of the spindle assembly checkpoint, and cytokinesis.17 Survivin associates with microtubules during the early stage of mitosis. Any disruption of survivin-microtubule interactions leads to loss of survivin-dependent anti-apoptotic property that results in cell death during mitosis.18 In cancer cells, a high level of survivin expression overcomes the apoptotic checkpoint and promotes the aberrant progression of transformed cells through mitosis.14
Post-translational modifications such as ubiquitination and deubiquitination are essential processes that regulate protein turnover to maintain cell homeostasis and cellular events.14 Deubiquitinating enzymes (DUBs) are crucial in anticancer therapeutics as they can reverse the process of ubiquitination by removing ubiquitin molecules from cancer-related proteins targeted for degradation.19,20,21 Ubiquitin-specific proteases (USPs) form the largest subfamily of DUBs, comprising approximately 50 members.20 Growing bodies of evidence prove that DUBs are key factors regulating cell division and tumorigenesis.22,23,24,25 A study has reported that USP19 regulates the activity of c-IAP1 and c-IAP2, which are crucial for the correct function of the cell cycle.26 In addition, the ubiquitin-proteasome pathway has been found to regulate survivin degradation in a cell-cycle-dependent manner, and the BIR domain of survivin is essential for maintaining its stability at the G2/M phase.27 Moreover, survivin has been found to undergo deubiquitination by USP9X, and it regulates chromosomal alignment and segregation during the process of mitosis.28 To our knowledge, a screen for DUBs that regulate the survivin protein and its importance in mitosis and tumorigenesis has not been reported. Thus, we initiated this study to screen systematically for potential DUBs that regulate survivin protein in cancer cells and its functions using our recently developed CRISPR-Cas9-mediated DUB knockout (KO) library.29
In the present study, a loss-of-function genome-wide screening of DUBs through the CRISPR-Cas9 system identified USP19 as a novel protein stabilizer of survivin. We demonstrated that USP19 is a potential protein interactor of survivin and regulates survivin protein turnover by preventing its degradation through the ubiquitin-proteasomal pathway. Loss of USP19 leads to several mitotic defects in cancer cells and also suppresses tumor growth. Altogether, our results have elucidated the dual role of USP19 in regulating cancer cell mitosis and tumorigenesis.
Results
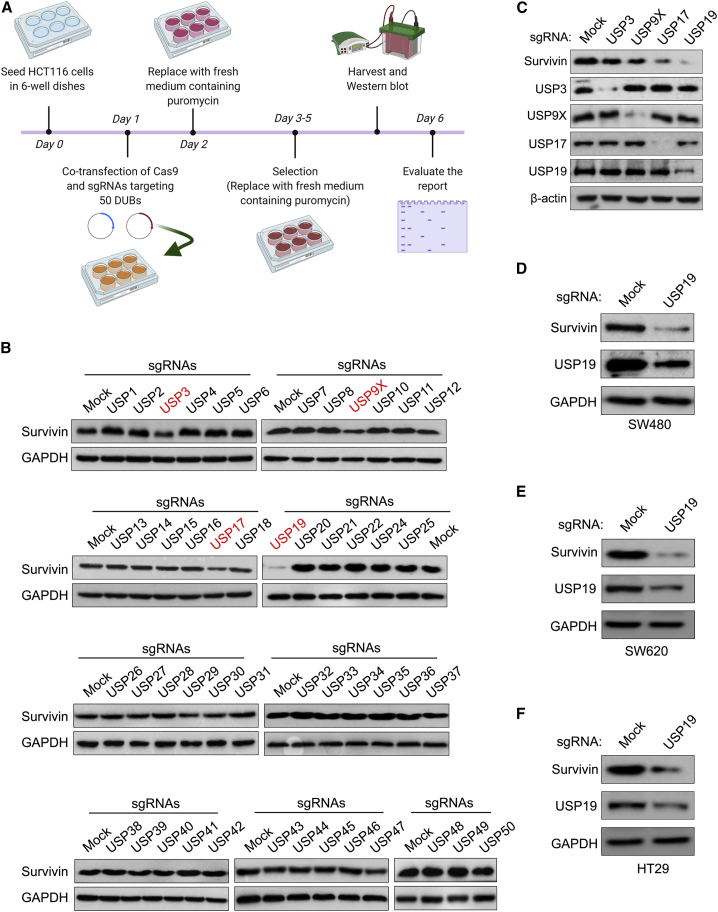
Genome-scale screening of the ubiquitin-specific protease subfamily for survivin protein using the DUB KO library
Recently, we have reported the generation of a CRISPR-Cas9-mediated DUB KO library to screen putative DUBs for the protein of interest.29,30 The single-guide RNAs (sgRNAs) targeting entire sets of genes encoding USP subfamilies along with Cas9 were co-transfected into HCT116 cells. A variation in endogenous survivin protein levels caused by the loss of function of a particular DUB was analyzed by western blotting (Figure 1A). We found that the KO of USP3, USP9X, USP17 and USP19 reduced the endogenous level of survivin protein when compared with mock (Figure 1B). Among these putative DUBs, USP9X has been previously reported to regulate survivin stability by its deubiquitinating activity.28 Interestingly, we noticed that sgRNA targeting USP19 showed a higher reduction in survivin protein levels compared to USP9X (Figure 1B). Moreover, the cross-confirmation of all of the putative DUBs on survivin protein showed USP19 as the strongest candidate compared to other putative DUBs (Figure 1C). To further corroborate the effect of USP19 on survivin protein levels, we transfected sgRNA targeting USP19 in SW480, SW620, and HT29 cells. The USP19 knockdown showed a reduction in survivin protein in all of the cell lines tested (Figures 1D–1F). Therefore, our results suggested that USP19 may be a potential DUB regulating survivin protein.
Figure 1.
CRISPR-Cas9-based genome-scale screening of USP family proteins for survivin
(A) Schematic representation of a DUB knockout (KO) library screening system. Day 0: HCT116 cells were seeded at a density of 3.5 × 105 cells/well on 6-well plates and maintained in RPMI. Day 1: DUB KO library consisting of sgRNAs individually targeting an entire set of USP family members along with Cas9 were co-transfected using Lipofectamine 2000 in HCT116 cells. Day 2: The RPMI medium was replaced with complete medium containing puromycin (1.5 μg/mL). Days 3–5: HCT116 transfected cells were allowed to grow under puromycin selection. Day 6: The transfected HCT116 cells were harvested and western blot was performed using survivin antibody. (B) western blot analysis showing endogenous survivin expression levels. (C) western blot analysis of putative DUB candidates in HCT116 cells. (D–F) Validation of the effect of USP19 on survivin protein expression in (D) SW480, (E) SW620, and (F) HT29 cell lines.
USP19 positively regulates survivin protein levels
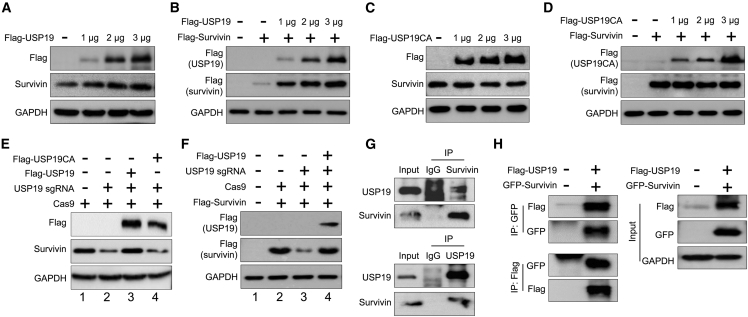
To determine whether USP19 could stabilize the protein levels of survivin, we transfected HCT116 cells with increasing concentrations of FLAG-USP19. We found that USP19 stabilized the endogenous survivin level in a dose-dependent manner (Figure 2A), and a similar result was observed at the exogenous level (Figure 2B). In contrast, the dose-dependent increase of catalytic mutant USP19 (FLAG-USP19C506A), showed no stabilizing effect on survivin protein at both endogenous (Figure 2C) and exogenous (Figure 2D) levels. These results indicate that USP19 may act as a protein stabilizer of survivin through deubiquitinating activity.
Figure 2.
USP19 regulates survivin protein stability
(A) The effect of USP19 on endogenous survivin protein expression was determined in HCT116 cells. (B) Exogenous protein levels of survivin in HCT116 cells were analyzed upon transfection with increasing concentrations of FLAG-USP19, along with a constant amount of FLAG-survivin. (C) The effect of the catalytic mutant form of USP19 (USP19C506A) on endogenous survivin protein was analyzed upon transfection with increasing concentrations of USP19C506A. (D) The effect of USP19C506A on exogenous survivin protein was analyzed upon transfection with increasing concentrations of USP19C506A, along with a constant amount of FLAG-survivin in HCT116 cells. Western blot analysis was performed with the indicated antibodies. (E and F) HCT116 cells were transfected with indicated plasmids to check the reconstitution effect of USP19 in USP19-depleted cells on survivin by western blot analysis at an (E) endogenous or (F) exogenous level. (G and H) Interactions between (G) endogenous and (H) exogenous USP19 and survivin were analyzed in HCT116 cells. Cell lysates were immunoprecipitated with USP19- or survivin-specific antibodies and immunoblotted with indicated antibodies.
Next, we observed that the reduction of survivin expression upon knockdown of USP19 was rescued by the reconstitution with FLAG-USP19 in USP19-depleted cells both at endogenous (Figure 2E, lane 3) and exogenous levels (Figure 2F, lane 4). However, the reconstitution of FLAG-USP19CA in USP19-depleted cells failed to rescue the survivin level endogenously (Figure 2E, lane 4) Altogether, these data suggest that USP19 regulates survivin protein stability.
USP19 interacts and co-localizes with survivin
To delineate the molecular mechanisms by which USP19 regulates the function of survivin protein, we analyzed the interaction between USP19 and survivin under physiological conditions. Endogenous co-immunoprecipitation analysis using specific antibodies against USP19 or survivin demonstrated that USP19 interacts with survivin and vice versa in HCT116 cells (Figure 2G). Consistent with this result, ectopically expressed FLAG-USP19 was co-precipitated with GFP-survivin in HCT116 cells and vice versa (Figure 2H), indicating that USP19 interacts with the survivin protein at both endogenous and exogenous levels.
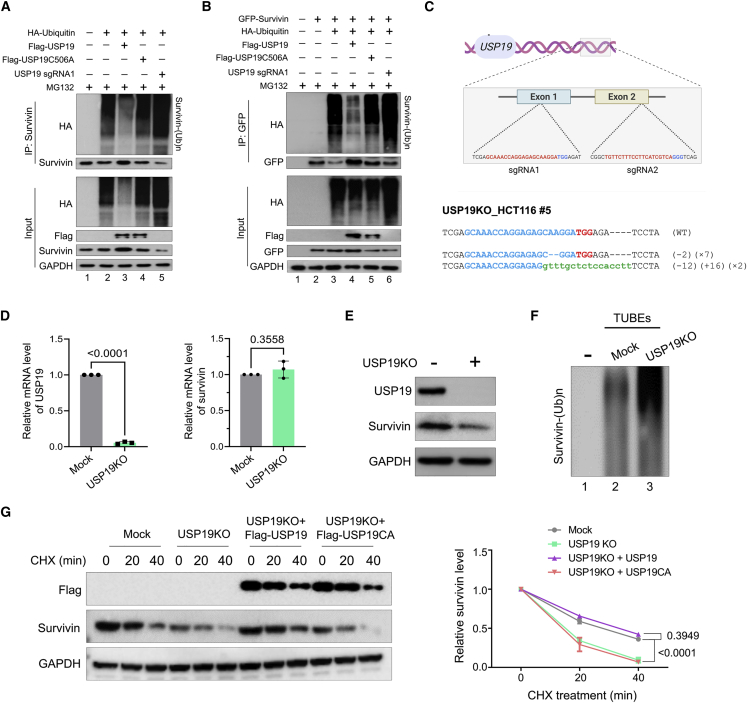
USP19 deubiquitinates and extends the half-life of the survivin protein
Previous studies have demonstrated that survivin undergoes ubiquitin-mediated proteasomal degradation.27,31 To check the effect of USP19 on survivin ubiquitination, we performed an endogenous deubiquitination assay to analyze the polyubiquitination status of survivin by transfecting wild-type (WT) or mutant USP19 or sgRNA targeting USP19 in HCT116 cells. We observed that USP19 WT significantly reduces the polyubiquitination of endogenous survivin compared to mock (Figure 3A, lane 3 versus lane 2) but not USP19CA (Figure 3A, lane 4 versus lane 2). In contrast, USP19 knockdown showed a dramatic increase in the polyubiquitination smear of survivin compared to mock (Figure 3A, lane 2 versus lane 2). Likewise, an exogenous deubiquitination assay also showed a greater reduction in the polyubiquitin smear on survivin in the presence of USP19 but not with USP19CA (Figure 3B).
Figure 3.
USP19 deubiquitinates survivin and extends the half-life of survivin protein
(A and B) The deubiquitinating activity of (A) endogenous and (B) exogenous USP19 on survivin protein was performed in HCT116 cells. HCT116 cells were transfected with indicated constructs and treated with 5 μM MG132 for 6 h before harvesting the samples. Next, survivin-specific antibodies were used to immunoprecipitate endogenous survivin protein or GFP antibody was used to immunoprecipitate exogenous survivin, and the polyubiquitination status of endogenous or exogenous survivin in the presence or absence of FLAG-USP19 or FLAG-USP19C506A was analyzed by western blotting. (C) Schematic representation shows the sgRNA design strategy to target the USP19 gene to generate a stable USP19 KO cell line in HCT116 cells (top panel). The bottom panel shows the Sanger sequence results of the USP19KO clone#5 cell line. (D) The qPCR analysis was performed to validate the USP19KO efficiency and USP19KO effect on survivin mRNA level in HCT116 cells. Data are presented as the means and standard deviations of 3 independent experiments. A 2-tailed t test was used, and the p values are as indicated. (E) Western blot analysis was performed to check the effect of USP19 and survivin protein levels in mock and USP19KO cells. (F) TUBE assay was performed to assess the loss of USP19 on ubiquitination status of survivin in mock and USP19KO HCT116 cells. (G) Mock, USP19KO, and USP19KO reconstituted with either FLAG-USP19 WT or FLAG-USP19C506A transfected HCT116 cells were used to analyze the half-life of survivin. CHX (150 μg/mL) was treated for the indicated time intervals and harvested for western blotting with the indicated antibodies. Data are presented as the means and standard deviations of 3 independent experiments. Two-way ANOVA followed by Tukey’s post hoc test was used with the indicated p values.
To further validate the loss of function of USP19-mediated post-translational regulation of survivin, we generated single-cell-derived USP19 KO clones using the CRISPR-Cas9 system in the HCT116 cell line. To achieve this, we designed two sets of sgRNA1 and sgRNA2 targeting exon 1 and exon 2 of the USP19 gene, respectively (Figure 3C, top panel). The HCT116 cells were co-transfected with sgRNAs targeting USP19 along with Cas9 and the cleavage efficiency was analyzed using a T7E1 assay. As shown in Figure S1A, sgRNA1 showed significant cleavage efficiency exhibiting a higher indel percentage than sgRNA2.
To obtain single cell-derived KO clones, transfected cells were selected with puromycin and diluted into 96-well plates for single-cell clonal selection. Single cell-derived clones were then screened for USP19 gene disruption using a T7E1 assay. The T7E1+ single-cell-derived USP19 KO clones showing cleavages (Figures S1B and S1C) were subjected to western blotting to check for USP19 expression and its effect on survivin protein. As a control batch, mock clones were also subjected to single-cell dilution using scrambled sgRNAs. Western blot analysis revealed that clones #5, #10, and #11 showed complete disruption of USP19 expression (Figure S1D). Moreover, USP19 KO#5 decreased survivin expression significantly compared to mock and other clones. Sanger sequencing results demonstrated that the USP19KO clone#5 (hereafter, USP19KO) displayed out-of-frame mutations (Figure 3C, bottom panel).
We further validated the KO efficiency of USP19 and its effect on survivin by qPCR and western blot. Both mRNA and protein levels of USP19 were completely disrupted in the USP19KO group (Figures 3D and 3E). In addition, the USP19KO clone displayed a significant reduction in survivin protein (Figure 3E) but not at the mRNA level (Figure 3D, right panel), indicating that USP19 does not regulate survivin at the transcriptional level. To further support our result, we used the tandem ubiquitin binding entities (TUBEs) assay, which has a high-affinity probe for ubiquitinated proteins.32 Our data confirmed a high ubiquitin smear on survivin in USP19KO cells compared with mock cells (Figure 3F), indicating that the loss of USP19 enhances the ubiquitination of survivin. These results showed that USP19 regulates survivin expression at the post-translational level.
Based on the above observations, we hypothesized that USP19 may influence survivin protein turnover. To this end, we treated protein synthesis inhibitor cycloheximide (CHX) in the presence or absence of USP19 in HCT116 cells. We observed a dramatic reduction in the half-life of survivin in USP19KO cells compared to mock (Figure 3G). The reduced half-life of the survivin protein was rescued when USP19KO cells were reconstituted with FLAG-USP19, while USP19CA had no effect (Figure 3G). Collectively, these data indicated that USP19 regulated survivin protein turnover by deubiquitinating activity.
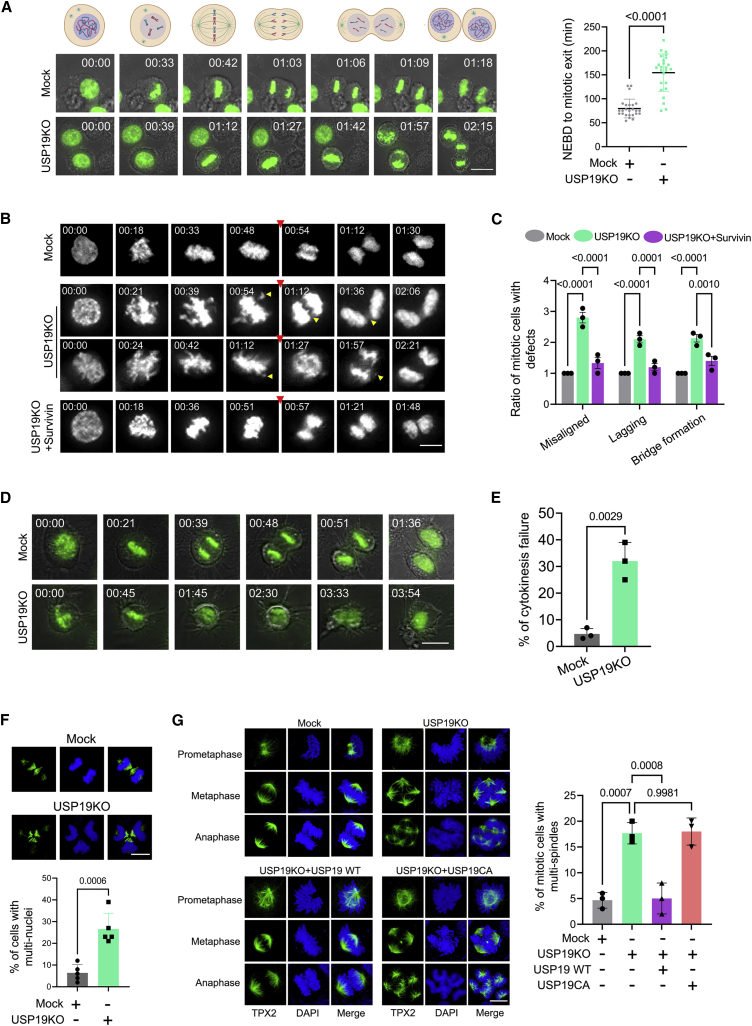
USP19 functions in mitosis
Over the past year, studies have shown that the role of survivin in cancer pathogenesis is not limited to apoptosis inhibition, but is also involved in the regulation of mitosis.33,34 It is known that Taxol binds to microtubules and induces mitotic arrest.35 We treated both mock and USP19KO groups with Taxol and found that Taxol-induced mitotic arrest was observed in the mock group, whereas USP19KO mostly displayed multi-lobed nuclei and severe nuclear abnormalities (Figure S2). These morphologies demonstrated that the loss of USP19 interferes with the spindle checkpoint function in Taxol-treated cells. To validate the effect of USP19 in mitosis, we optimized live-cell imaging of mock or USP19KO cells transfected with GFP-H2B. The nuclear envelope breakdown (NEBD) to the mitotic exit in living cells using time-lapse microscopy revealed that the mitotic timing was delayed in the USP19KO group compared to mock (Figure 4A; Videos S1 and S2).
Figure 4.
USP19 is required for successful mitotic progression
(A–E) Mock or USP19KO HCT116 cells were transfected with GFP-H2B, synchronized using thymidine and released into fresh medium. Cells were then monitored using a time-lapse microscope for 12 h and images were taken at 3-min intervals. The results were from 3 independent experiments. (A) Screenshots taken from the time-lapse microscopy of mitotic cell populations (n = 25). NEBD to the mitotic exit was estimated in the mentioned groups and graphically represented. Representative images were formed by merging GFP-H2B fluorescence and phase-contrast images. Scale bar, 25 μm. Data are presented as the means and standard deviations. A 2-tailed t test was used, and the p value is as indicated. (B) Screenshots taken from time-lapse microscopy at the indicated times from NEBD (occurred at 00:00). The anaphase onset is denoted by the red arrowhead. Yellow arrowheads show misaligned or lagging chromosomes, chromatin bridges, or improperly separated chromosomes. Scale bar, 10 μm. (C) Graphical representation of the quantification of mitotic defects from time-lapse videos (n = 10). Data are presented as the means and standard deviations of 3 independent experiments. Two-way ANOVA followed by Bonferroni post hoc test was used, and p values are as indicated. (D) Time-lapse microscopic images of cells undergoing mitosis and cytokinesis. Representative images were formed by merging GFP-H2B fluorescence and phase-contrast images. Scale bar, 25 μm. (E) Quantification of cytokinesis failure from time-lapse videos (n = 35). Data are presented as the means and standard deviations of 3 independent experiments. A 2-tailed t test was used, and the p value is as indicated. (F) Mock or USP19KO HCT116 cells were stained with TPX2 antibody (n = 5). Immunofluorescence analysis was performed to check bi- or multi-nuclei conditions and graphically represented. Scale bar, 10 μm. Data are presented as the means and standard deviations of 5 independent experiments. A 2-tailed t test was used, and the p value is as indicated. (G) Mock, USP19KO, and USP19KO cells transfected with USP19 WT or USP19CA and immunofluorescence staining was performed using TPX2-specific antibody (n = 25). HCT116 cells showing multi-spindles were counted after immunofluorescence staining and graphically represented. Data are presented as the means and standard deviations of 3 independent experiments. One-way ANOVA followed by Tukey’s post hoc test was used with the indicated p values. Scale bar, 10 μm.
Mitosis in USP19KO HCT116 cells
GFP-H2B was transfected in USP19KO HCT116 cells and then treated with thymidine. GFP was used to visualize chromosome movement. This video displays the presence of misaligned chromosomes and mitotic delay during mitosis. These images were taken every 3 min and they are played back at 5 frames per second. Scale bar, 10 μm.
In the live-cell imaging analysis, the USP19KO group displayed a high frequency of mitotic defects such as chromosome misalignments, lagging chromosomes, and chromatin bridges. However, the overexpression of survivin in USP19KO group displayed reduced mitotic defects compared to USP19KO group (Figures 4B and 4C; Video S3, Video S4, Video S5, Video S6). We also found that USP19KO groups failed cytokinesis at significantly higher rates than mock groups (Figures 4D and 4E; Videos S7 and S8). These phenotypical defects caused by USP19 depletion were again re-confirmed by analyzing fixed cells. Multi-nuclear phenotypes, an evidence of cytokinesis failure, were frequently observed in USP19KO groups at the late telophase (Figure 4F). In addition, spindle multi-polarity occurred frequently in USP19KO mitotic cells (Figure 4G). The defects observed in the USP19KO group were significantly rescued by the overexpression of USP19 WT, but not with catalytically mutant USP19 (Figure 4G), which suggests that USP19 DUB activity is critical during mitotic progression.
Mitosis in mock HCT116 cells
GFP-H2B was transfected in mock HCT116 cells and then treated with thymidine. This video displays the normal mitosis. These images were taken every 3 min and they are played back at 5 frames per second. Scale bar, 10 μm.
Mitosis in USP19KO HCT116 cells
GFP-H2B was transfected in USP19KO HCT116 cells and then treated with thymidine. This video displays the chromosome misalignments, chromosome bridges, and lagging chromosomes. These images were taken every 3 min and they are played back at 5 frames per second. Scale bar, 10 μm.
Mitosis in USP19KO HCT116 cells
GFP-H2B was transfected in USP19KO HCT116 cells and then treated with thymidine. This video displays the chromosome misalignments, chromosome bridges, and lagging chromosomes. These images were taken every 3 min and they are played back at 5 frames per second. Scale bar, 10 μm.
Mitosis in survivin reconstituted in USP19KO HCT116 cells
GFP-H2B was transfected in USP19KO+survivin HCT116 cells and then treated with thymidine. This video displays the normal mitosis. These images were taken every 3 min and they are played back at 5 frames per second. Scale bar, 10 μm.
Mitosis and cytokinesis in mock HCT116 cells
GFP-H2B was transfected in mock HCT116 cells and then treated with thymidine. This video displays normal cytokinesis. These images were taken every 3 min and they are played back at 5 frames per second. Scale bar, 10 μm.
Mitosis and cytokinesis in USP19KO HCT116 cells
GFP-H2B was transfected in USP19KO HCT116 cells and then treated with thymidine. This video displays cytokinesis failure. These images were taken every 3 min and they are played back at 5 frames per second. Scale bar, 10 μm.
USP19 affects survivin functions in mitosis
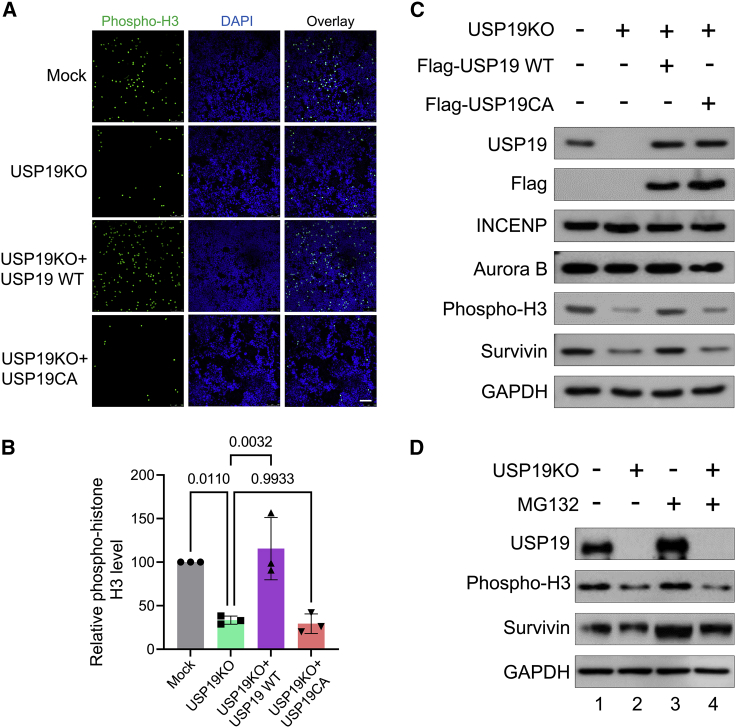
A previous study reported that survivin-depleted cells display a reduction in the levels of phospho-histone H3, which is a substrate of survivin.36 Thus, we questioned whether USP19 could regulate the functions of survivin, and used the phospho-histone H3 as an indicator of survivin downstream signaling. By immunofluorescence staining, we noticed a reduction of histone H3 phosphorylation in USP19KO compared to mock (Figures 5A and 5B). Reconstitution of USP19 WT in the USP19KO group reversed the effect but USP19CA failed to do so (Figures 5A and 5B). Western blot analysis also showed that the depletion of USP19 lowered both phospho-histone H3 and survivin protein levels in nocodazole-treated cells. These levels were restored by the expression of USP19 WT, but not by USP19CA (Figure 5C), suggesting that USP19-mediated deubiquitination modulates survivin stability, subsequently altering its function. We also noticed that USP19 regulates mitotic functions by stabilizing survivin, but not other CPC members INCENP and Aurora B, suggesting the specificity of the USP19 effect on survivin function.
Figure 5.
USP19 mimics the role of survivin in mitosis
(A) Mock, USP19KO, and USP19KO cells transfected with USP19 WT or USP19CA. Immunofluorescence staining was performed using phospho-histone H3 antibody. Scale bar, 100 μm. (B) The cells expressing GFP (phospho-histone H3+) were counted and presented graphically. Data are presented as the means and standard deviations of 3 independent experiments. One-way ANOVA followed by Tukey’s post hoc test was used with the indicated p values. (C) Mock, USP19KO, and USP19KO cells transfected with USP19 WT or USP19CA were synchronized by treating with 100 ng/mL nocodazole for 18 h. Western blot analysis was performed with the indicated antibodies. (D) Mock, USP19KO, and USP19KO cells transfected with USP19 WT or USP19CA were synchronized in prometaphase by treating with 100 ng/mL nocodazole for 18 h and then treated with MG132 for 4 h before harvesting. Western blot analysis was performed using indicated antibodies.
Next, we checked the USP19 function on mitotic progression in nocodazole-arrested cells. As shown in Figure 5D, the loss of USP19 reduced the survivin protein level, whereas the MG132-treated USP19KO cells accumulated survivin protein (Figure 5D, lanes 2 and 4). The expression of phospho-histone H3, a mitosis specific marker was also reduced in USP19KO cells (Figure 5D, lanes 2). Interestingly, the reduced expression of phospho-histone H3 could not be regained even after MG132 treatment (Figure 5D, lane 4), suggesting that the loss of USP19 significantly hampered the mitotic progression of the cells. Altogether, we concluded that USP19-mediated deubiquitination of survivin could affect its downstream signaling, which is required for successful mitotic progression.
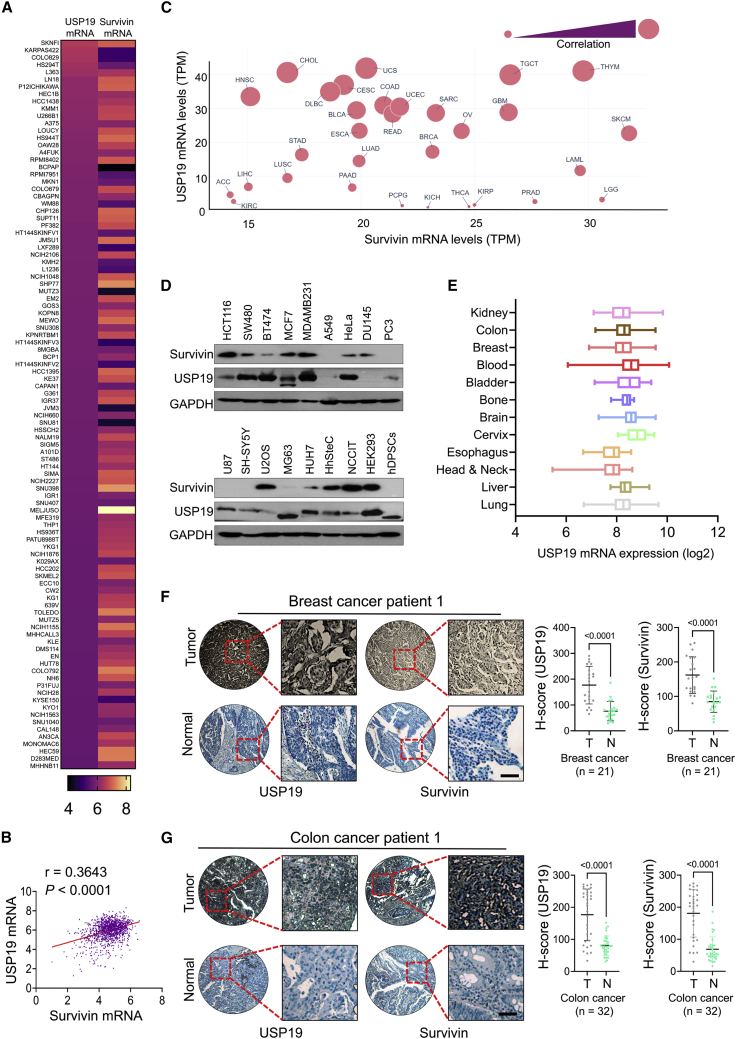
Correlation between USP19 and survivin in cancers
We used the DepMap portal37 to analyze USP19 and survivin mRNA levels from an array of cancers. We observed that high scores for USP19 mRNA expression in any cancer cell line were directly proportional to the mRNA expression of survivin (Figure 6A; Table S1). A scatterplot of the expression patterns of USP19 and survivin produced an r-value of 0.3643 in the overall cancer cell lines, which suggests a positive correlation between USP19 and survivin (Figure 6B; Table S1). The correlation between USP19 and survivin expression profiles using the GEPIA 2 database38 showed that USP19 and survivin were expressed in several cancers and were highly correlated in most of the cancer types (Figure 6C; Table S2). To further validate in silico data, the protein expression of USP19 and survivin were assessed in several cancer cell lines by western blot. We found that both USP19 and survivin are expressed in most of the cancer cell lines (Figure 6D). Next, we used the GENT2 database to analyze the USP19 expression status in cancer tissues.39 Consistent with our previous observations, USP19 was highly expressed in several cancers (Figure 6E; Table S3). However, we could not perform the statistical analysis due to the non-availability of optimal sample size for normal groups from the GENT2 database (Table S4). Altogether, these results strongly suggest that USP19 might be associated with cancer progression.
Figure 6.
USP19 and survivin expression profiles are positively correlated
(A) Heatmap showing the top 100 mRNA expression levels of USP19 and survivin derived from the DepMap portal. Representative samples are arranged from high to low mRNA levels of USP19, and corresponding survivin values are adjusted. (B) Scatterplot between USP19 and survivin mRNA levels from several cancer cell lines. Pearson correlation was performed to estimate the relationship between USP19 and survivin. (C) The mRNA expression profiles of USP19 and survivin were obtained from the GEPIA 2 database and graphically represented using Plotly to assess the correlation between USP19 and survivin. (D) Endogenous protein expression patterns of USP19 and survivin in different cell lines were determined by western blot analysis. In MCF7, MG63, and hDPSCs, the expression of USP19 showed lower molecular weight than the expected size. (E) The mRNA expression data of USP19 was obtained from the GENT2 database and represented graphically. Due to the small sample size for the expression of USP19 in the normal groups obtained from the GENT2 database, the statistical significance between normal and cancer groups could not be calculated. (F and G) Representative immunohistochemical staining images of endogenous USP19 and survivin in human breast cancer (n = 21) and colon cancer (n = 32). All immunohistochemistry (IHC) images were quantified by an H score. A 2-tailed t test was used, and p values are as indicated. Scale bar, 25 μm.
To gain further insights into the expression of USP19 in cancer, we performed an immunohistochemistry analysis in a tissue microarray containing breast and colon cancer tissues and their corresponding normal tissues. We found that both USP19 and survivin were highly upregulated in breast (Figures 6F and S3) and colon cancer tissues when compared to their corresponding normal tissues (Figures 6G and S4). Altogether, USP19 and survivin are upregulated in several cancers, suggesting that the USP19-survivin axis could serve as a novel regulatory target in several cancers.
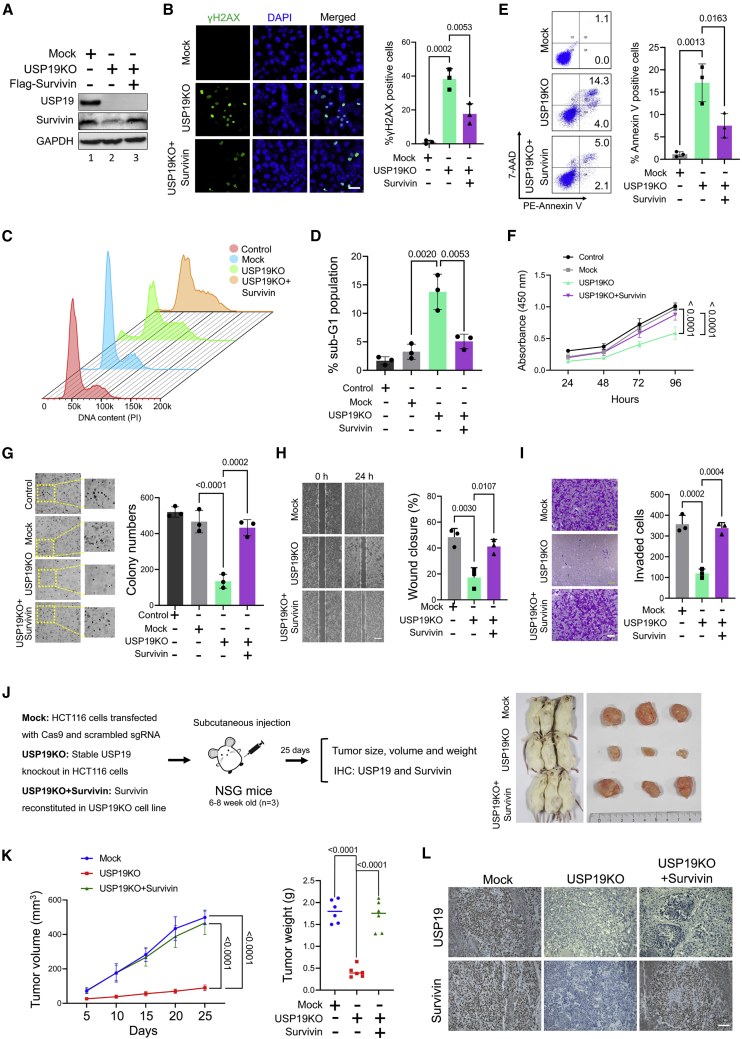
Loss of USP19 induces the DNA damage response and apoptosis
Several reports have concluded that the increased expression of survivin was correlated with the inhibition of apoptosis and chemoresistance in several tumor types.40,41,42,43,44 Interestingly, we also observed that both USP19 and survivin are expressed in several cancer types. Therefore, we investigated the role of USP19-mediated stabilization of survivin on cancer progression in HCT116 cells. To this end, we validated the survivin protein levels in mock and USP19KO cells. Compared to the mock control, USP19KO cells showed reduced levels of survivin protein (Figure 7A, lane 2), while the reconstitution of survivin in the USP19KO group rescued the expression of survivin (Figure 7A, lane 3). Thus, the same cells from Figure 7A were used for the subsequent functional experiments. Immunofluorescence was performed to determine the extent of DNA damage by estimating the canonical DNA damage marker γH2AX. USP19KO cells showed more γH2AX foci formation compared to the mock cells, which were abrogated when survivin was reconstituted (Figure 7B). Next, we investigated whether USP19 depletion induces apoptosis in cancer cells. We examined sub-G1 populations and annexin-V staining in HCT116 cells. USP19 depletion resulted in a significant accumulation of cells within the sub-G1 phase of the cell cycle when compared to the mock cells (Figures 7C and 7D). Moreover, annexin-V/7-AAD staining indicated enhanced cell apoptosis in the USP19KO group compared to the mock (Figure 7E). However, survivin reconstitution in the USP19KO group reverted from the apoptotic phenotype (Figures 7C–7E).
Figure 7.
USP19KO inhibits tumor progression in vitro and in vivo
Mock, USP19KO, and USP19KO cells reconstituted with survivin plasmid in HCT116 cells were used to perform the following experiments. (A) Western blot analysis was performed using USP19- and survivin-specific antibodies. (B) Immunofluorescence analysis was used to measure γH2AX foci formation. Green, γH2AX; blue, nucleus stained by DAPI. Scale bar, 25 μm. The right panel depicts the percentage of γH2AX+ cells. Data are presented as the means and standard deviations of 3 independent experiments. One-way ANOVA followed by Tukey’s post hoc test was used with the indicated p values. (C and D) Flow cytometry was performed with PI staining to measure the DNA content and is represented graphically. Data are presented as the means and standard deviations of 3 independent experiments. One-way ANOVA followed by Tukey’s post hoc test was used with the indicated p values. (E) Flow cytometry was performed to analyze annexin-V and 7-AAD+ cells and graphically represented. Data are presented as the means and standard deviations of 3 independent experiments. One-way ANOVA followed by Tukey’s post hoc test was used with the indicated p values. (F) Cell viability assay was performed using CCK-8 reagent and represented as the absorbance at 450 nm. Data are presented as the means and standard deviations of 3 independent experiments. Two-way ANOVA followed by Tukey’s post hoc test was used with the indicated p values. (G) Colony formation was measured after 14 days. The colony numbers were quantified and are presented graphically (right panel). Data are presented as the means and standard deviations of 3 independent experiments. One-way ANOVA followed by Tukey’s post hoc test was used with the indicated p values. (H) An in vitro scratch assay was performed to assess the migration potential of the groups mentioned. Scale bar, 100 μm. Data are presented as the means and standard deviations of 3 independent experiments. One-way ANOVA followed by Tukey’s post hoc test was used with the indicated p values. (I) Transwell cell invasion assay was performed with the groups mentioned. Scale bar, 200 μm. Invaded cells were quantified using ImageJ software and represented graphically. Data are presented as the means and standard deviations of 3 independent experiments. One-way ANOVA followed by Tukey’s post hoc test was used with the indicated p values. (J) Xenografts were generated by subcutaneously injecting the mentioned cell groups into the right flank of NSG mice (n = 3). Tumor volumes were recorded and stored for IHC experiments. The right panel shows the tumors excised from the mice after the experiment. (K) Tumor volume and weight were calculated and are presented graphically. Data are presented as the means and standard deviations of 6 biological replicates (n = 6). One-way or 2-way ANOVA followed by Tukey’s post hoc test was used with the indicated p values. (L) Xenograft tumors were sectioned and paraffin embedded. Immunohistochemical analysis was performed with the antibodies indicated. Scale bar, 50 μm.
USP19 regulates the oncogenic potential of survivin in vitro and in vivo
To validate the effects of USP19-mediated survivin stability on cancer progression, we performed various assays related to carcinogenesis activity. The cell viability assay showed that the depletion of USP19 decreased cell viability, which was reverted when cells were reconstituted with survivin (Figure 7F). The colony formation assay showed a significant decrease in colony numbers in the USP19KO group when compared to the mock (Figure 7G). Similarly, migration and invasion were also inhibited significantly in the USP19KO group when compared to the mock group (Figures 7H and 7I). However, the colony formation, migration, and invasion of cells were significantly increased when USP19KO groups were reconstituted with survivin (Figures 7G–7I).
Next, we performed in vivo studies to corroborate the effect of USP19 in oncogenic transformation. To this end, NSG mice were subcutaneously injected in the right flanks, with mock, USP19KO, and USP19KO reconstituted with survivin HCT116 cells. The tumor growth was recorded every 5 days for 25 days (Figures 7J and S5). Mice injected with USP19KO cells displayed a significant reduction in tumor volume and weight compared to mice transplanted with mock (Figure 7K). The reconstitution of survivin in USP19KO cells led to a significant increase in tumor volume and weight (Figures 7J and 7K). Furthermore, we performed immunohistochemical analysis to check the status of USP19 and survivin proteins in mouse xenograft tumors. Reduced expression of both USP19 and survivin proteins was observed in the USP19KO tumor xenograft groups compared to mock groups (Figure 7L).
Discussion
Mitosis is a dynamic cellular process that requires a continuous assembly as well as a disassembly of protein complexes. Optimal chromosome segregation includes the attachment of sister kinetochores to microtubules from opposite spindle poles to form bi-oriented chromosomes on the metaphase spindle through the CPC.45 Many DUBs such as USP4, USP7, USP9, USP29, USP35, USP44, USP48, and CYLD have direct or indirect roles during the cell-cycle progression and checkpoint maintenance.19,23,25,46,47,48,49,50 Survivin is an essential component of CPC that ensures CPC centromeric localization, optimal segregation of chromosomes, regulation of microtubule formation, and cytokinesis.51 In addition, survivin exhibits CPC-independent functions during interphase through the regulation of microtubule dynamics.52 Ubiquitination controls dynamic protein-to-protein binding and chromosomal segregation independently of protein degradation. It is known that USP9X, a deubiquitinating enzyme, is required for survivin dissociation from centromeres.28 Interestingly, survivin undergoes ubiquitination through K48 or K63 linkages during mitosis.53 Thus, further studies are necessary to investigate different ubiquitination sites of survivin and its regulation by DUBs during mitotic progression.
In this study, we determined that USP19 can reverse the ubiquitination of the survivin protein and regulate its protein turnover (Figures 1, 2, 3, and 4). Several studies have reported the involvement of survivin in cellular survival, proliferation, cell cycle, and other cellular stress responses.54 Survivin overexpression in cancers is typically linked with a poor prognosis and chemoresistance.55,56 Survivin nuclear expression is known as a prognostic marker in a variety of cancers.57,58 From our study, we demonstrated that USP19-mediated regulation of survivin is critical for proper cytokinesis (Figures 4 and 5) and control over cancer progression (Figures 6 and 7). Thus, the role of USP19 as a survivin protein stabilizer and in cancer cell division provides a novel insight of using USP19 as a potential target in anti-cancer therapy.
Collectively, we report USP19 as a DUB to reverse the proteasomal degradation of the survivin protein. Our data denote that USP19 is a regulator that favors cancer progression by stabilizing the survivin protein. Because of the redundancy of multifaceted interactions and signaling networks, blocking survivin alone may not result in robust antitumor activity. Thus, we suggest that a combinatorial approach of concomitantly targeting survivin and its stabilizer USP19, as well as other pro-apoptotic factors, would overcome such caveats. Such simultaneous targeting would not only offer novel perspectives in rational cancer therapy but also provide broad clinical applications to treat cancer patients regardless of their genetic makeup.
Materials and methods
Cell culture
HCT116, SW480, BT474, MDAMB231, PC3, MG63, and HUH7 cell lines were purchased from the Korean Cell Line Bank (Seoul, South Korea). MCF7, A549, HeLa, DU145, U87MG, SH-SY5Y, U2OS, NCCIT, and HEK293T were purchased from ATCC (Manassas, VA, USA). hDPSCs were purchased from Lonza (Basel, Switzerland). HHSteCs were kindly provided by Yun Soo Bae (Ewha University, Seoul, Korea). HCT116, BT474, MDAMB231, and NCCIT cell lines were maintained in RPMI (Gibco BRL, Rockville, MD, USA) supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin and streptomycin (Gibco) at 37°C in a humidified atmosphere with 5% CO2. SW480, MCF7, A549, HeLa, DU145, PC3, U87MG, SH-SY5Y, U2OS, MG63, HUH7, HHSteCs, and HEK293T cell lines were cultured in DMEM medium (Gibco) supplemented with 10% FBS and 1% penicillin and streptomycin at 37°C in a humidified atmosphere with 5% CO2. hDPSCs were cultured in DPSCBM (Lonza) and 1% penicillin and streptomycin (Gibco) at 37°C in a humidified atmosphere with 5% CO2. The cells were passaged regularly depending on cell confluence.
Transfection
HEK283T cells were transfected with Lipofectamine 2000 (Thermo Fisher, Waltham, MA, USA) following the manufacturer’s recommendations. HCT116 cells were transfected with indicated plasmids using Lipofectamine 3000 (Thermo Fisher) following the manufacturer’s recommendations.
Plasmids and sgRNAs
A mammalian expression vector encoding FLAG-survivin was kindly provided by Prof. Taeg Kyu Kwon (Keimyung University, South Korea). The pEGFP-N1 GFP-survivin plasmid was kindly provided by Prof. Dong-Hoon Jin (Asan Medical Center, South Korea). The pEGFP-N1 GFP-H2B (Addgene [Watertown, MA, USA], no. 11680), pRK-FLAG-USP19 (Addgene no. 78597), pRK5-FLAG-USP19C506A (Addgene no. 36307), and pcDNA-HA-ubiquitin (Addgene no. 18712) were purchased from Addgene. For screening DUB candidates, a plasmid encoding Cas9-2a-mRFP-2a-PAC (puromycin N-acetyl-transferase puromycin resistance gene), and a pRG2 plasmid encoding single guide RNA was purchased from Toolgen (Seoul, South Korea). The sgRNA target sequences were designed using a public tool (www.broadinstitute.org) and cloned into the vectors as described previously.29 Briefly, oligonucleotides containing each target sequence were synthesized (Bioneer, Seoul, South Korea), and T4 polynucleotide kinase was used to add terminal phosphates to the annealed oligonucleotides (Bio-Rad, Hercules, CA, USA). The oligonucleotide sequences used to clone sgRNA-targeting DUBs are listed in Table S5. The vector was digested with the BsaI restriction enzyme and ligated with the annealed oligonucleotides.
Antibodies and reagents
Mouse monoclonal antibodies against FLAG (Anti-DDDDK-tag, M185-3L, 1:1,000) (MBL Life Science, Tokyo, Japan), phospho-histone H2AX (Ser139) (Merck (Readington Township, NJ, USA], 05-636), GFP (sc-996, 1:1,000), INCENP (sc-376514, 1:500), ubiquitin (sc-8017, 1:1,000), USP9X (sc-365353, 1:1,000), hemagglutinin (HA) (sc-7392, 1:1,000), glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (sc-32233, 1:1,000), β-actin (sc-47778, 1:1,000), and normal mouse IgG (sc-2025, 1:1,000) were purchased from Santa Cruz Biotechnology (Dallas, TX, USA). Rabbit polyclonal antibodies against survivin (Cell Signaling Technology [Danvers, MA, USA], 2803S), USP19 (Proteintech [Rosemont, IL, USA], 25768-1-AP), USP3 (Genetex [Irvine, CA, USA], GTX128238), USP17/DUB3 (Novus Biologicals [Littleton, CO, USA], NBP1-79745), Aurora B (Bethyl Laboratories [Montgomery, TX, USA], A300-431A), phospho-histone H3 (Cell Signaling Technology, 3377), TPX2 (Novus Biologicals, NB500-179), and 488/594-conjugated secondary antibodies (A21207 and A21203, 1:200) (Invitrogen, Waltham, MA, USA) were used. Protein A/G Plus Agarose beads (sc-2003, Santa Cruz Biotechnology), proteasomal inhibitor MG132 (S2619, Selleckchem, Houston, TX, USA), protein translation inhibitor cycloheximide (239765, Merck), puromycin (12122530, Invitrogen), and DAPI (H-1200, Vector Laboratories, Burlingame, CA, USA) were purchased from the manufacturers indicated.
DUBs screening using DUB KO sgRNA library
We screened the USPs for the regulation of survivin protein level using our previously generated DUB KO sgRNA library set.29 On day 0, HCT116 cells were seeded on 35-mm culture dishes at the density of 3 × 105 cells and incubated overnight. On day 1, HCT116 cells were co-transfected with Cas9 and sgRNA targeting 50 individual DUBs. The next day, puromycin at the concentration of 1.5 μg/mL was added and selected for 3 days. Finally, cells were harvested and the western blot analysis was performed using survivin-specific antibody.
T7E1 assay
Genomic DNA was isolated using DNeasy Blood & Tissue kits (Qiagen, Hilden, Germany), following the manufacturer’s protocol. The region of DNA consisting of the nuclease target site was amplified by performing PCR using hemi-nested or nested primers. PCR amplicons were denatured by heating and annealed to generate heteroduplex DNA, which was treated with 5 U of T7 endonuclease 1 enzyme (New England Biolabs, Ipswitch, MA, USA) for 20 min at 37°C. The cleavage was observed using 1.5% agarose gel electrophoresis. Mutation frequencies in terms of indel percentage were based on band intensity using ImageJ software (NIH) and the following equation: mutation frequency (%) = 100 × (1 − [1 − fraction cleaved]1/2), where the fraction cleaved was the total relative density of the cleavage bands divided by the sum of the relative density of the cleavage and uncut bands. The oligonucleotide sequences used to obtain the PCR amplicon for the T7E1 assay are as follows: USP19 sgRNA1 first PCR: forward 5′-TCCAGTGGTGACTAAGCAGC-3′ and reverse 5′-CCCCAACAGCACCATCTTCTG-3′, USP19 s PCR: 5ʹ-ATTTGGCCACAAAGAGTTGC-3′ and reverse 5′-CCCCAACAGCACCATCTTCTG-3′ and USP19 sgRNA2 first PCR: forward 5′-AAGGACCCATGATAACCTCAGT-3′ and reverse 5′-CTACTGTGCTCAGGGCAGATG-3′, and USP19 sgRNA2 second PCR: forward 5′-GTAGAGACGGGGTTTCACCA-3′ and reverse 5′-CTACTGTGCTCAGGGCAGATG-3′.
Real-time PCR
Total RNA was isolated using Trizol reagent (Favorgen, Kaohsiung, Taiwan). RNA pellets were suspended in 30 μL of nuclease-free water and the RNA concentration was estimated. Total RNA (500 ng) was reverse transcribed into cDNA using the SuperScript III First-Strand Synthesis System (Life Technologies, Carlsbad, CA, USA) with an oligo dT primer. qPCR was performed in triplicate using Fast SYBR Green master mix (Life Technologies) and a StepOnePlus Real-Time PCR System (Life Technologies) with the loading controls: GAPDH-targeting primers (forward primer: 5′-CATGTTCGTCATGGGTGTGAACCA-3′, reverse primer: 5′-AGTGATGGCATGGACTGTGGTCAT-3′), USP19-targeting primers (forward primer: 5′-TAAATCCAAGGCACGATCTGAGG-3′ and reverse primer: 5′-GCTTTGGGGTTACATGCTCCA-3′), and survivin-targeting primers (forward primer: 5′-CAGTGTTTCTTCTGCTTCAAGG-3′ and reverse primer: 5′-CTTATTGTTGGTTTCCTTTGCAT-3′) were used. The relative expressions of USP19 and survivin were normalized with GAPDH and shown using GraphPad Prism 9 (GraphPad Software, San Diego, CA, USA).
Immunoprecipitation assays
The plasmids mentioned were co-transfected into the HCT116 cells. After 48 h, cells were collected and lysed by protein lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 10 mM KCl, 1.5 mM MgCl2, 0.5% NP-40, 10% glycerol, and 1 mM EDTA), and the protein concentration was measured using a Bradford assay (Thermo Fisher). Approximately 3 mg of lysates were immunoprecipitated with antibodies overnight and then incubated with 25 μL of protein A/G Sepharose beads at 4°C for 2 h. Beads were washed with lysis buffer, eluted, and boiled in 2× SDS sample buffer. Immunoprecipitated proteins were detected by western blot analysis with the indicated antibodies.
Deubiquitination assay
The DUB activity of USP19 on survivin protein stability was determined in HCT116 cells. The cells were treated 48 h post-transfection with proteasome inhibitor MG132 for 6 h and harvested. This was followed by immunoprecipitation and western blotting with the indicated antibodies.
Generation of KO cell lines using CRISPR-Cas9
HCT116 cells were co-transfected with sgRNA1 targeting USP19 and Cas9 constructs. Puromycin selection (1.5 μg/mL) was performed for 2 days after transfection. The next day, the cells were seeded into 96-well plates at a density of 0.25 cells per well. After 15 days, rounded single-cell colonies were selected. The selected colonies were trypsinized and re-plated into 24-well cell culture plates. Three days after subculturing, genomic DNA was isolated from single-cell clones and used for the T7E1 assay. T7E1+ clones were further cultured and stored in a liquid nitrogen tank for later use. USP19 and survivin protein levels in the control and USP19 KO groups were determined by western blotting using specific antibodies.
TUBEs assay
The ubiquitination status of survivin protein was analyzed by a TUBEs assay (LifeSensors, Malvern, PA, USA). Briefly, mock and USP19KO HCT116 cells were harvested for protein extraction and incubated with the required amount of TUBEs antibody at 4°C for 3 h. The samples were then subjected to immunoprecipitation analysis followed by immunoblotting with survivin antibody to determine the polyubiquitination status of survivin protein.
Immunofluorescence
HCT116 cells were grown on glass coverslips and incubated at 37°C in a humidified atmosphere with 5% CO2. The cells were then fixed for 15 min at room temperature in 4% paraformaldehyde. After two PBS washes, cells were permeabilized for 5 min in PBS containing 0.1% Triton, thoroughly washed in PBS, and blocked with 3% bovine serum albumin (BSA). Cells were then treated overnight at 4°C with the appropriate primary antibodies diluted in BSA. On the following day, the cells were washed and incubated with appropriate Alexa Fluor 488-conjugated secondary antibodies for 1 h. The cells were finally incubated with DAPI and mounted using VectaShield (Vector Laboratories). Cells were visualized and images were taken using a Leica fluorescence microscope (Leica DM 5000B; Leica CTR 5000; Wetzlar, Germany). All of the data were obtained from three independent experiments, and the identity of all of the samples was removed before the analysis.
Apoptosis assays
To estimate cell apoptosis, the PE annexin-V/7-AAD staining kit (BD Biosciences, Haryana, India) was used according to the manufacturer’s protocol. Briefly, HCT116 cells (mock control, USP19KO, and USP19KO reconstituted with survivin) were seeded in 6-well plates and incubated at 37°C in a humidified atmosphere with 5% CO2. The cells were harvested and washed twice with PBS containing 10% FBS. The required cells were counted and 5 μL annexin-V and 7-AAD were added to the cells, which were then kept in the dark for 15 min. The stained cells were resuspended in the binding buffer and flow cytometry was performed within 1 h. For propidium iodide (PI) staining (BD Biosciences), the cell lines mentioned previously were harvested, washed twice with ice-cold PBS containing 10% FBS, and fixed with 70% ethanol until use. Next, 2 mg/mL RNaseA was added to the cells at 4°C for 15 min followed by 10 μL PI at a concentration of 50 mg/mL at room temperature for 10 min. Finally, DNA content was measured using flow cytometry. Data were analyzed using FACS Diva software (version 8, BD Biosciences).
Time-lapse microscopy
Time-lapse live imaging was performed using the 40× objective lens on a microscope (DeltaVision Core; GE Healthcare, Chicago, IL, USA) equipped with a charge-coupled device camera in a CO2 chamber at 37°C. Mock, USP19KO HCT116 cells, and/or survivin reconstituted with USP19KO group were transfected with GFP-H2B and seeded onto a 4-well chamber (Lab-Tek II Chambered Cover glass, Thermo Fisher). Time-lapse images were taken at 3-min intervals and maximally projected. All of the data were obtained from three independent experiments, and the identity of all of the samples was removed before the analysis.
Cell viability assay
For the cell viability assay, untransfected cells (control), mock, USP19KO, and USP19KO reconstituted with survivin HCT116 cells were seeded at a density of 1 × 104 cells per well in 96-well plates. On the following day, CCK-8 assay reagent (Dojindo Molecular Technologies, Rockville, MD, USA) was added to each well following the manufacturer’s protocol. Absorbance was measured at 450 nm using a spectrophotometer and values were recorded graphically.
Soft agar assay
Untransfected cells (control), mock, USP19KO, and USP19KO reconstituted with survivin HCT116 cells were subjected to a soft agar assay. First, 1% agarose gel and 1× complete DMEM medium were mixed at a 1:1 ratio and plated onto 35-mm plates. The plates were incubated overnight and cells were resuspended in 0.75% agarose with DMEM (1:1 ratio) and seeded at a density of 1 × 104 cells per well. Cells were cultured for 14 days at 37°C in a humidified atmosphere containing 5% CO2. The anchorage-independent colony numbers were stained using crystal violet dye (0.01% diluted in 20% methanol) and counted under light microscopy (IX71, Olympus, Tokyo, Japan) at 4× magnification.
Wound healing assay
Migration activity was analyzed by the wound healing assay. Mock USP19KO and USP19KO reconstituted with survivin HCT116 cells were cultured to near 90% confluence. Scratches were made in the monolayers with a sterile pipette tip in a definite array. The wounded cell layer was washed with PBS and incubated in complete medium. Wound closure was compared at the time intervals indicated using a light microscope. ImageJ software was used for quantification. Migration rates were measured and the wounded area was calculated.
Transwell invasion assay
Cell invasion was assessed using 0.8 μm Transwell chambers coated with Matrigel (Corning, NY, USA) according to the manufacturer’s protocol. Briefly, 3 × 104 cells were suspended in 500 μL serum-free DMEM medium and placed into 24-well chambers. Next, 750 μL complete medium was added and the cells were incubated at 37°C with 5% CO2. After 24 h, the cells from the upper surface of the insert were scraped off, and the cells on the lower surface were fixed using ice-cold methanol. The cells were then stained with crystal violet, and the average number of cells were counted using light microscopy.
Xenograft tumor experiment
Male NOD scid gamma (NSG) mice (6 weeks old) were used for the animal experiment. The animal study was approved by the IACUC. All of the mice were housed under an artificial 12 h light/dark cycle with access to food and water ad libitum. Animals were randomized into three groups (six mice per group) and mock, USP19KO, and USP19KO reconstituted with survivin HCT116 cells (4.0 × 106) were subcutaneously injected into the right flank of each mouse. The volume of the xenografts was measured every 5 days for 25 days and estimated using the formula V = D × d2/2, where D is the long axis, and d is the short axis of the tumor. At the end of the study, all of the mice were sacrificed by CO2 asphyxiation, and tumor grafts were harvested. The weight of the harvested tumors was measured, and the tumor tissues from the mouse xenografts were subjected to immunohistochemical analysis.
Immunohistochemistry
For the tissue microarray experiment, we purchased colon cancer (n = 32) and breast cancer (n = 21) tissues from ISU Abxis (Gyeonggi-do, South Korea). Formalin-fixed and paraffin-embedded tissue samples were processed and incubated with USP19 (Proteintech) and survivin (Cell Signaling Technology) antibodies following the manufacturers’ recommendations. These tissue samples were counterstained with hematoxylin, dehydrated, and mounted. Xenograft tumor tissue samples were fixed in 4% paraformaldehyde overnight at 4°C. Tissues were then dehydrated in 70% ethanol and processed in paraffin. The samples were stained with USP19 and survivin antibodies. Imaging of tumor sections on slides was done on a Leica DM5000 B.
Gene expression profiling interactive analysis
The DepMap portal, GEPIA 2, and GENT2 containing RNA expression data, which include several tumor and normal samples from the TCGA and GTEx databases, were used in this study.37,38,39 We used single- or multi-gene analyses and correlation analysis of different cancer types to analyze the expression profiles of USP19 and survivin.
Statistical analysis
Results were documented as means and standard deviations from at least three independent experiments (unless otherwise specified in the respective figure legends). Comparisons between two sets were analyzed using the Student’s t test. Experiments involving three or more groups were examined by one-way or two-way analyses of variance followed by Tukey’s post hoc test. A p value <0.05 was considered statistically significant. All of the statistical analyses were performed in GraphPad Prism 9 software.
Acknowledgments
We are grateful to all members of the S.R. and K.-S.K. labs, especially Jencia Carminha Colaco, for technical support. Figures 1A and 3C (top panel) and the graphical abstract were created with Biorender.com. This research was supported by the National Research Foundation of Korea (NRF) (2021M3A9H3015389), the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Korean government (MSIT) (2022M3A9E4016936 and 2022M3A9E4082648), and the Korean Fund for Regenerative Medicine (KFRM) grant funded by the Korea government (the Ministry of Science and ICT, the Ministry of Health and Welfare) (22A0304L1-01).
Author contributions
A.P.C., A.T., and S.R. designed the study and analyzed and interpreted the data. A.P.C., and A.T. wrote the manuscript, and S.R. assisted them. A.P.C., and A.T. conducted all of the experiments, and N.P., N.S., J.K.K., K.K., C.-H.P., and S.-H.H. assisted them. K.-S.K. and S.R. procured financial support. All of the authors reviewed the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2022.07.019.
Contributor Information
Kye-Seong Kim, Email: ks66kim@hanyang.ac.kr.
Suresh Ramakrishna, Email: suri28@hanyang.ac.kr.
Supplemental information
References
- 1.Zhang C., Liu Y. Targeting cancer with sesterterpenoids: the new potential antitumor drugs. J. Nat. Med. 2015;69:255–266. doi: 10.1007/s11418-015-0911-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun L., Peng Q., Qu L., Gong L., Si J. Anticancer agent icaritin induces apoptosis through caspase-dependent pathways in human hepatocellular carcinoma cells. Mol. Med. Rep. 2015;11:3094–3100. doi: 10.3892/mmr.2014.3007. [DOI] [PubMed] [Google Scholar]
- 3.Su Z., Yang Z., Xu Y., Chen Y., Yu Q. Apoptosis, autophagy, necroptosis, and cancer metastasis. Mol. Cancer. 2015;14:48. doi: 10.1186/s12943-015-0321-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brady S.W., Zhang J., Tsai M.-H., Yu D. PI3K-independent mTOR activation promotes lapatinib resistance and IAP expression that can be effectively reversed by mTOR and Hsp90 inhibition. Cancer Biol. Ther. 2015;16:402–411. doi: 10.1080/15384047.2014.1002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pereira S.S., Monteiro M.P., Antonini S.R., Pignatelli D. Apoptosis regulation in adrenocortical carcinoma. Endocr. Connect. 2019;8:R91–R104. doi: 10.1530/EC-19-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cossu F., Milani M., Mastrangelo E., Lecis D. Targeting the BIR domains of inhibitor of apoptosis (IAP) proteins in cancer treatment. Comput. Struct. Biotechnol. J. 2019;17:142–150. doi: 10.1016/j.csbj.2019.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X., Duan N., Zhang C., Zhang W. Survivin and tumorigenesis: molecular mechanisms and therapeutic strategies. J. Cancer. 2016;7:314–323. doi: 10.7150/jca.13332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altieri D.C. Survivin – the inconvenient IAP. Semin. Cell Dev. Biol. 2015;39:91–96. doi: 10.1016/j.semcdb.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bagrezaei F., Hassanshahi G., Mahmoodi M., Khanamani Falahati-Pour S., Mirzaei M.R. Expression of inhibitor of apoptosis gene family members in bladder cancer tissues and the 5637 tumor cell line. Asian Pac. J. Cancer Prev. 2018;19:529–532. doi: 10.22034/APJCP.2018.19.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaiswal P.K., Goel A., Mittal R.D. Survivin: a molecular biomarker in cancer. Indian J. Med. Res. 2015;141:389–397. doi: 10.4103/0971-5916.159250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grossman D., McNiff J.M., Li F., Altieri D.C. Expression and targeting of the apoptosis inhibitor, survivin, in human melanoma. J. Invest. Dermatol. 1999;113:1076–1081. doi: 10.1046/j.1523-1747.1999.00776.x. [DOI] [PubMed] [Google Scholar]
- 12.Cai Y., Ma W., Huang X., Cao L., Li H., Jiang Y., Lu N., Yin Y. Effect of survivin on tumor growth of colorectal cancer in vivo. Int. J. Clin. Exp. Pathol. 2015;8:13267–13272. [PMC free article] [PubMed] [Google Scholar]
- 13.He X., Yang K., Wang H., Chen X., Wu H., Yao L., Ma S. Expression and clinical significance of survivin in ovarian cancer: a meta-analysis. PLoS One. 2018;13 doi: 10.1371/journal.pone.0194463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jha K., Shukla M., Pandey M. Survivin expression and targeting in breast cancer. Surg. Oncol. 2012;21:125–131. doi: 10.1016/j.suronc.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 15.Velculescu V.E., Madden S.L., Zhang L., Lash A.E., Yu J., Rago C., Lal A., Wang C.J., Beaudry G.A., Ciriello K.M., et al. Analysis of human transcriptomes. Nat. Genet. 1999;23:387–388. doi: 10.1038/70487. [DOI] [PubMed] [Google Scholar]
- 16.Santarelli A., Mascitti M., Lo Russo L., Sartini D., Troiano G., Emanuelli M., Lo Muzio L. Survivin-based treatment strategies for squamous cell carcinoma. Int. J. Mol. Sci. 2018;19:971. doi: 10.3390/ijms19040971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carmena M., Wheelock M., Funabiki H., Earnshaw W.C. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol. 2012;13:789–803. doi: 10.1038/nrm3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie S., Ogden A., Aneja R., Zhou J. Microtubule-binding proteins as promising biomarkers of paclitaxel sensitivity in cancer chemotherapy. Med. Res. Rev. 2016;36:300–312. doi: 10.1002/med.21378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chandrasekaran A.P., Suresh B., Sarodaya N., Ko N.-R., Oh S.-J., Kim K.-S., Ramakrishna S. Ubiquitin specific protease 29 functions as an oncogene promoting tumorigenesis in colorectal carcinoma. Cancers. 2021;13:2706. doi: 10.3390/cancers13112706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haq S., Das S., Kim D.-H., Chandrasekaran A.P., Hong S.-H., Kim K.-S., Ramakrishna S. The stability and oncogenic function of LIN28A are regulated by USP28. Biochim. Biophys. Acta Mol. Basis Dis. 2019;1865:599–610. doi: 10.1016/j.bbadis.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 21.Poondla N., Chandrasekaran A.P., Kim K.-S., Ramakrishna S. Deubiquitinating enzymes as cancer biomarkers: new therapeutic opportunities? BMB Rep. 2019;52:181–189. doi: 10.5483/BMBRep.2019.52.3.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Du T., Li H., Fan Y., Yuan L., Guo X., Zhu Q., Yao Y., Li X., Liu C., Yu X., et al. The deubiquitylase OTUD3 stabilizes GRP78 and promotes lung tumorigenesis. Nat. Commun. 2019;10:2914. doi: 10.1038/s41467-019-10824-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park J., Kwon M.-S., Kim E.E., Lee H., Song E.J. USP35 regulates mitotic progression by modulating the stability of Aurora B. Nat. Commun. 2018;9:688. doi: 10.1038/s41467-018-03107-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dietachmayr M., Rathakrishnan A., Karpiuk O., von Zweydorf F., Engleitner T., Fernández-Sáiz V., Schenk P., Ueffing M., Rad R., Eilers M., et al. Antagonistic activities of CDC14B and CDK1 on USP9X regulate WT1-dependent mitotic transcription and survival. Nat. Commun. 2020;11:1268. doi: 10.1038/s41467-020-15059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Antao A.M., Kaushal K., Das S., Singh V., Suresh B., Kim K.-S., Ramakrishna S. USP48 governs cell cycle progression by regulating the protein level of Aurora B. Int. J. Mol. Sci. 2021;22:8508. doi: 10.3390/ijms22168508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mei Y., Hahn A.A., Hu S., Yang X. The USP19 deubiquitinase regulates the stability of c-IAP1 and c-IAP2. J. Biol. Chem. 2011;286:35380–35387. doi: 10.1074/jbc.M111.282020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao J., Tenev T., Martins L.M., Downward J., Lemoine N.R. The ubiquitin-proteasome pathway regulates survivin degradation in a cell cycle-dependent manner. J. Cell Sci. 2000;113 Pt 23:4363–4371. doi: 10.1242/jcs.113.23.4363. [DOI] [PubMed] [Google Scholar]
- 28.Vong Q.P., Cao K., Li H.Y., Iglesias P.A., Zheng Y. Chromosome alignment and segregation regulated by ubiquitination of survivin. Science. 2005;310:1499–1504. doi: 10.1126/science.1120160. [DOI] [PubMed] [Google Scholar]
- 29.Das S., Chandrasekaran A.P., Suresh B., Haq S., Kang J.-H., Lee S.-J., Kim J., Kim J., Lee S., Kim H.H., et al. Genome-scale screening of deubiquitinase subfamily identifies USP3 as a stabilizer of Cdc25A regulating cell cycle in cancer. Cell Death Differ. 2020;27:3004–3020. doi: 10.1038/s41418-020-0557-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chandrasekaran A.P., Kaushal K., Park C.H., Kim K.S., Ramakrishna S. USP32 confers cancer cell resistance to YM155 via promoting ER-associated degradation of solute carrier protein SLC35F2. Theranostics. 2021;11:9752–9771. doi: 10.7150/thno.63806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y.B., Gao X., Deeb D., Brigolin C., Zhang Y., Shaw J., Pindolia K., Gautam S.C. Ubiquitin-proteasomal degradation of antiapoptotic survivin facilitates induction of apoptosis in prostate cancer cells by pristimerin. Int. J. Oncol. 2014;45:1735–1741. doi: 10.3892/ijo.2014.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hjerpe R., Aillet F., Lopitz-Otsoa F., Lang V., England P., Rodriguez M.S. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 2009;10:1250–1258. doi: 10.1038/embor.2009.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mita A.C., Mita M.M., Nawrocki S.T., Giles F.J. Survivin: key regulator of mitosis and apoptosis and novel target for cancer therapeutics. Clin. Cancer Res. 2008;14:5000–5005. doi: 10.1158/1078-0432.CCR-08-0746. [DOI] [PubMed] [Google Scholar]
- 34.Altieri D.C. Survivin, versatile modulation of cell division and apoptosis in cancer. Oncogene. 2003;22:8581–8589. doi: 10.1038/sj.onc.1207113. [DOI] [PubMed] [Google Scholar]
- 35.Hornick J.E., Bader J.R., Tribble E.K., Trimble K., Breunig J.S., Halpin E.S., Vaughan K.T., Hinchcliffe E.H. Live-cell analysis of mitotic spindle formation in taxol-treated cells. Cell Motil. Cytoskeleton. 2008;65:595–613. doi: 10.1002/cm.20283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olsen A.L., Davies J.M., Medley L., Breen D., Talbot D.C., McHugh P.J. Quantitative analysis of survivin protein expression and its therapeutic depletion by an antisense oligonucleotide in human lung tumors. Mol. Ther. Nucleic Acids. 2012;1:e30. doi: 10.1038/mtna.2012.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehár J., Kryukov G.V., Sonkin D., et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang Z., Kang B., Li C., Chen T., Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park S.-J., Yoon B.-H., Kim S.-K., Kim S.-Y. GENT2: an updated gene expression database for normal and tumor tissues. BMC Med. Genomics. 2019;12:101. doi: 10.1186/s12920-019-0514-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Small S., Keerthivasan G., Huang Z., Gurbuxani S., Crispino J.D. Overexpression of survivin initiates hematologic malignancies in vivo. Leukemia. 2010;24:1920–1926. doi: 10.1038/leu.2010.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng K.Y., Wang Z.L., Gu Q.Y., Hao M. Survivin overexpression is associated with aggressive clinicopathological features in cervical carcinoma: a meta-analysis. PLoS One. 2016;11 doi: 10.1371/journal.pone.0165117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Véquaud E., Desplanques G., Jézéquel P., Juin P., Barillé-Nion S. Survivin contributes to DNA repair by homologous recombination in breast cancer cells. Breast Cancer Res. Treat. 2016;155:53–63. doi: 10.1007/s10549-015-3657-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garg H., Suri P., Gupta J.C., Talwar G.P., Dubey S. Survivin: a unique target for tumor therapy. Cancer Cell Int. 2016;16:49. doi: 10.1186/s12935-016-0326-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Altieri D.C. Survivin, cancer networks and pathway-directed drug discovery. Nat. Rev. Cancer. 2008;8:61–70. doi: 10.1038/nrc2293. [DOI] [PubMed] [Google Scholar]
- 45.Nye A.C., Rajendran R.R., Stenoien D.L., Mancini M.A., Katzenellenbogen B.S., Belmont A.S. Alteration of large-scale chromatin structure by estrogen receptor. Mol. Cell. Biol. 2002;22:3437–3449. doi: 10.1128/MCB.22.10.3437-3449.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gilberto S., Peter M. Dynamic ubiquitin signaling in cell cycle regulation. J. Cell Biol. 2017;216:2259–2271. doi: 10.1083/jcb.201703170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wickström S.A., Masoumi K.C., Khochbin S., Fässler R., Massoumi R. CYLD negatively regulates cell-cycle progression by inactivating HDAC6 and increasing the levels of acetylated tubulin. EMBO J. 2010;29:131–144. doi: 10.1038/emboj.2009.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reddy S.K., Rape M., Margansky W.A., Kirschner M.W. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature. 2007;446:921–925. doi: 10.1038/nature05734. [DOI] [PubMed] [Google Scholar]
- 49.Liu H., Zhang H., Wang X., Tian Q., Hu Z., Peng C., Jiang P., Wang T., Guo W., Chen Y., et al. The deubiquitylating enzyme USP4 cooperates with CtIP in DNA double-strand break end resection. Cell Rep. 2015;13:93–107. doi: 10.1016/j.celrep.2015.08.056. [DOI] [PubMed] [Google Scholar]
- 50.Chandrasekaran A.P., Woo S.H., Sarodaya N., Rhie B.H., Tyagi A., Das S., Suresh B., Ko N.R., Oh S.J., Kim K.-S., Ramakrishna S. Ubiquitin-specific protease 29 regulates Cdc25A-mediated tumorigenesis. Int. J. Mol. Sci. 2021;22:5766. doi: 10.3390/ijms22115766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nogueira-Ferreira R., Vitorino R., Ferreira-Pinto M.J., Ferreira R., Henriques-Coelho T. Exploring the role of post-translational modifications on protein–protein interactions with survivin. Arch. Biochem. Biophys. 2013;538:64–70. doi: 10.1016/j.abb.2013.07.027. [DOI] [PubMed] [Google Scholar]
- 52.Giodini A., Kallio M.J., Wall N.R., Gorbsky G.J., Tognin S., Marchisio P.C., Symons M., Altieri D.C. Regulation of microtubule stability and mitotic progression by survivin. Cancer Res. 2002;62:2462–2467. [PubMed] [Google Scholar]
- 53.Kong Y., Wang Z., Huang M., Zhou Z., Li Y., Miao H., Wan X., Huang J., Mao X., Chen C. CUL7 promotes cancer cell survival through promoting Caspase-8 ubiquitination. Int. J. Cancer. 2019;145:1371–1381. doi: 10.1002/ijc.32239. [DOI] [PubMed] [Google Scholar]
- 54.Altieri D.C. Targeted therapy by disabling crossroad signaling networks: the survivin paradigm. Mol. Cancer Ther. 2006;5:478–482. doi: 10.1158/1535-7163.MCT-05-0436. [DOI] [PubMed] [Google Scholar]
- 55.Nigam J., Chandra A., Kazmi H.R., Parmar D., Singh D., Gupta V., M N. Expression of survivin mRNA in gallbladder cancer: a diagnostic and prognostic marker? Tumor Biol. 2014;35:9241–9246. doi: 10.1007/s13277-014-2200-x. [DOI] [PubMed] [Google Scholar]
- 56.Kim Y.-H., Kim S.-M., Kim Y.-K., Hong S.-P., Kim M.-J., Myoung H. Evaluation of survivin as a prognostic marker in oral squamous cell carcinoma. J. Oral Pathol. Med. 2010;39:368–375. doi: 10.1111/j.1600-0714.2009.00844.x. [DOI] [PubMed] [Google Scholar]
- 57.Simonetti O., Lucarini G., Rubini C., Lazzarini R., DI Primio R., Offidani A. Clinical and prognostic significance of survivin, AKT and VEGF in primary mucosal oral melanoma. Anticancer Res. 2015;35:2113–2120. [PubMed] [Google Scholar]
- 58.Grabowski P., Griss S., Arnold C.N., Hörsch D., Göke R., Arnold R., Heine B., Stein H., Zeitz M., Scherübl H. Nuclear survivin is a powerful novel prognostic marker in gastroenteropancreatic neuroendocrine tumor disease. Neuroendocrinology. 2005;81:1–9. doi: 10.1159/000084892. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mitosis in USP19KO HCT116 cells
GFP-H2B was transfected in USP19KO HCT116 cells and then treated with thymidine. GFP was used to visualize chromosome movement. This video displays the presence of misaligned chromosomes and mitotic delay during mitosis. These images were taken every 3 min and they are played back at 5 frames per second. Scale bar, 10 μm.
Mitosis in mock HCT116 cells
GFP-H2B was transfected in mock HCT116 cells and then treated with thymidine. This video displays the normal mitosis. These images were taken every 3 min and they are played back at 5 frames per second. Scale bar, 10 μm.
Mitosis in USP19KO HCT116 cells
GFP-H2B was transfected in USP19KO HCT116 cells and then treated with thymidine. This video displays the chromosome misalignments, chromosome bridges, and lagging chromosomes. These images were taken every 3 min and they are played back at 5 frames per second. Scale bar, 10 μm.
Mitosis in USP19KO HCT116 cells
GFP-H2B was transfected in USP19KO HCT116 cells and then treated with thymidine. This video displays the chromosome misalignments, chromosome bridges, and lagging chromosomes. These images were taken every 3 min and they are played back at 5 frames per second. Scale bar, 10 μm.
Mitosis in survivin reconstituted in USP19KO HCT116 cells
GFP-H2B was transfected in USP19KO+survivin HCT116 cells and then treated with thymidine. This video displays the normal mitosis. These images were taken every 3 min and they are played back at 5 frames per second. Scale bar, 10 μm.
Mitosis and cytokinesis in mock HCT116 cells
GFP-H2B was transfected in mock HCT116 cells and then treated with thymidine. This video displays normal cytokinesis. These images were taken every 3 min and they are played back at 5 frames per second. Scale bar, 10 μm.
Mitosis and cytokinesis in USP19KO HCT116 cells
GFP-H2B was transfected in USP19KO HCT116 cells and then treated with thymidine. This video displays cytokinesis failure. These images were taken every 3 min and they are played back at 5 frames per second. Scale bar, 10 μm.