Abstract
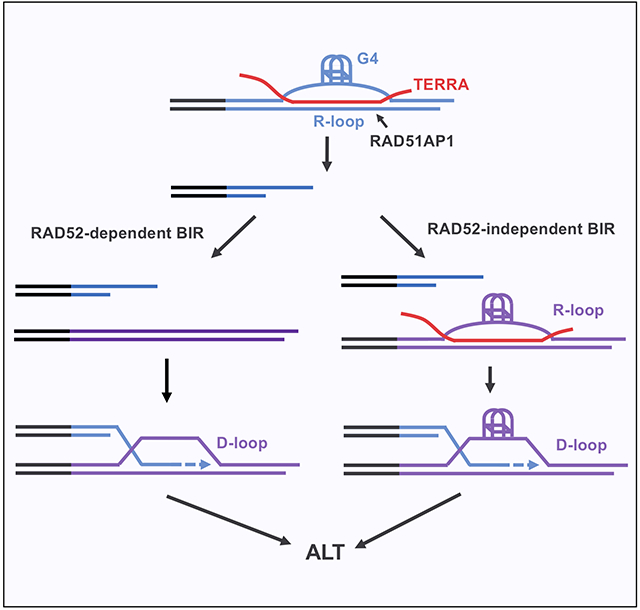
Alternative lengthening of telomeres (ALT), a telomerase-independent process maintaining telomeres, is mediated by break-induced replication (BIR). RAD52 promotes ALT by facilitating D-loop formation, but ALT also occurs through a RAD52-independent BIR pathway. Here, we show that the telomere non-coding RNA TERRA forms dynamic telomeric R-loops and contributes to ALT activity in RAD52 knockout cells. TERRA forms R-loops in vitro and at telomeres in a RAD51AP1-dependent manner. The formation of R-loops by TERRA increases G-quadruplexes (G4s) at telomeres. G4 stabilization enhances ALT even when TERRA is depleted, suggesting that G4s act downstream of R-loops to promote BIR. In vitro, the telomeric R-loops assembled by TERRA and RAD51AP1 generate G4s, which persist after R-loop resolution and allow formation of telomeric D-loops without RAD52. Thus, the dynamic telomeric R-loops formed by TERRA and RAD51AP1 enable the RAD52-independent ALT pathway, and G4s orchestrate an R-to-D loop switch at telomeres to stimulate BIR.
Graphical Abstract
eTOC Blurb
Alternative lengthening of telomere (ALT) occurs through RAD52-dependent and -independent break induced replication (BIR). Yadav et al. show that RAD51AP1 and the telomeric non-coding RNA TERRA promote RAD52-independent ALT by forming dynamic R-loops and increasing G4s at telomeres, enabling an R-to-D loop switch to initiate BIR independently of RAD52.
Introduction
The ability of cancer cells to extend and maintain telomeres is a hallmark of cancer, and it is critical for cancer cells to bypass telomere crisis and obtain replicative immortality. The alternative lengthening of telomeres (ALT) pathway, a telomerase-independent but recombination-dependent mechanism to extend telomeres, is active in a subset of human cancers (Dilley and Greenberg, 2015; Hoang and O’Sullivan, 2020; Sobinoff and Pickett, 2020; Zhang and Zou, 2020). The activation of ALT is prevalent in several cancer types, including sarcomas, glioblastoma, and neuroendocrine pancreatic cancer. Because ALT-positive (ALT+) cancer cells are dependent on ALT to continuously proliferate, inhibition of ALT is a potential therapeutic strategy for treatment of ALT+ tumors (Shay et al., 2012). Hyper activation of ALT also kills ALT+ cancer cells by increasing telomeric replication stress and instability (Lu et al., 2019; Pan et al., 2019; Silva et al., 2019; Zhang et al., 2021). Recent studies have suggested that ALT is a pathway related to break induced replication (BIR), a process that recovers collapsed DNA replication forks (Dilley et al., 2016; Min et al., 2017; Roumelioti et al., 2016; Sobinoff et al., 2017; Zhang et al., 2019). Similar to BIR, ALT occurs through conservative DNA synthesis (Roumelioti et al., 2016). In addition, both BIR and ALT require DNA polymerase δ accessary factors POLD3/4 (Dilley et al., 2016; Roumelioti et al., 2016; Zhang et al., 2019). In yeast, ALT is initiated by the formation of telomeric D-loops through invasion of single-stranded DNA (ssDNA) ends into double-stranded DNA (dsDNA) (Kockler et al., 2021). The human protein RAD52, which is involved in both BIR and ALT (Bhowmick et al., 2016; Min et al., 2017; Sotiriou et al., 2016; Verma et al., 2019; Zhang et al., 2019), promotes the formation of D-loops in telomeric DNA (Zhang et al., 2019). In ALT+ cancer cells, depletion of RAD52 leads to reduced DNA synthesis at telomeres and telomere shortening (Min et al., 2017, 2019; Ozer et al., 2018; Verma et al., 2019; Zhang et al., 2019). These results suggest that RAD52 promotes ALT by generating telomeric D-loops. However, knockout of RAD52 in ALT+ cells does not eliminate ALT activity (Verma et al., 2019; Zhang et al., 2019). Notably, the remaining ALT activity in RAD52 knockout cells is still dependent on POLD3/4, suggesting the existence of a RAD52-independent BIR pathway. These findings raised a question as to how telomeric D-loops are formed in the absence of RAD52.
Telomere repeat containing RNA (TERRA) is a non-coding RNA transcribed from subtelomeric and telomeric regions (Azzalin et al., 2007). TERRA contains the G-rich telomeric repeats and associates with telomeres in cells. In yeast, upregulation of TERRA promotes telomere extension independently of telomerase and suppresses senescence (Graf et al., 2017; Yu et al., 2014). In human ALT+ cancer cells, TERRA is commonly up regulated (Flynn et al., 2015; Lovejoy et al., 2012). Furthermore, TERRA forms R-loops at telomeres, which contribute to the maintenance of ALT telomeres (Arora et al., 2014; Pan et al., 2019; Silva et al., 2021). The replication stress or DNA damage at telomeres is an important trigger for ALT activation. Because R-loops are known to interfere with DNA replication, it was proposed that TERRA-mediated telomeric R-loops promote ALT by inducing replication stress. Indeed, inhibition of TERRA expression at multiple telomeres leads to reduced replication stress at telomeres, decreased ALT activity, and impaired telomere maintenance (Silva et al., 2021). In addition to their ability to induce replication stress, R-loops are also found to play positive roles in DNA repair at DNA double-stranded breaks (DSBs) (Keskin et al., 2014; Marnef and Legube, 2021; Michelini et al., 2017; Ouyang et al., 2021; Teng et al., 2018) and trigger a BIR-like repair process (Chappidi et al., 2020; Tan et al., 2020a). Whether TERRA directly participates in BIR at ALT telomere remains unknown. Nonetheless, the role for TERRA in ALT provides a unique opportunity to investigate how RNA impacts BIR.
The formation of R-loops at DNA breaks and telomeres is regulated by several factors. RAD52 has the ability to generate DNA:RNA hybrids in vitro, and it is implicated in the RNA-templated repair of DSBs (Keskin et al., 2014; Mazina et al., 2017). RAD52 is also important for the R-loop-mediated repair of DSBs induced by reactive oxygen species (ROS) (Tan et al., 2020a). Furthermore, RAD51AP1 promotes R-loop formation in vitro (Ouyang et al., 2021). In cells, RAD51AP1 is required for the formation of R-loops at DSBs, as well as the stimulation of homologous recombination (HR) by transcription or RNA transcripts. Notably, RAD51AP1 enables the invasion of RNA transcripts into double-stranded donor DNA, which facilitates the formation of D-loops by RAD51. DR-loops, an HR intermediate in donor DNA that contains both DNA:DNA and DNA:RNA hybrids, may enhance HR activity in transcribed regions of the genome. Finally, RAD51 was shown to promote TERRA-mediated R-loop formation in vitro and in cells without ALT activity (ALT−) (Feretzaki et al., 2020). Although several proteins have been implicated in DNA:RNA hybrid or R-loop formation, which protein is the primary factor generating TERRA-mediated telomeric R-loops in ALT+ cells is still unknown. Furthermore, whether some of these proteins participate in the BIR at ALT telomeres in an R-loop-dependent manner remains unclear. In addition to R-loops, G-quadruplexes (G4s) also accumulate at ALT telomeres (Amato et al., 2020; Yang et al., 2021). Similar to R-loops, G4s interfere with replication forks (Kumar et al., 2021). Whether TERRA regulates telomeric G4s and whether telomeric G4s play a role in BIR remains elusive.
In this study, we investigated whether TERRA is involved in the RAD52-independent ALT pathway. We found that TERRA forms telomeric R-loops and contributes to ALT activity in RAD52 knockout (KO) cells. The formation of telomeric R-loops by TERRA is dependent on RAD51AP1. Depletion of RAD51AP1 reduced ALT activity in RAD52 KO cells, suggesting that the RAD52-independent ALT pathway is dependent on telomeric R-loops. The formation of telomeric R-loops increases G4s, whereas G4s can stimulate ALT independently of R-loops, suggesting that G4s act downstream of R-loops. In cells, telomeric R-loops are reduced by inhibition of RNA polymerase II (RNAPII), showing their dynamic nature. In vitro, G4s in telomeric DNA persist after R-loops are resolved, and these G4s directly promote D-loop formation in the absence of RAD52. Thus, the dynamic telomeric R-loops formed by TERRA and RAD51AP1 increase G4s at ALT telomeres, opening dsDNA and allowing D-loop formation independently of RAD52. These results provide an explanation of how ALT occurs through the RAD52-independent pathway. Furthermore, they demonstrate an R-to-D loop switch at telomeres orchestrated by G4s, revealing an unexpected mechanism through which RNA stimulates BIR.
RESULTS
TERRA contributes to the RAD52-independent ALT pathway
We and others previously showed that ALT is a bifurcated BIR pathway that operates through both RAD52-dependent and -independent mechanisms (Fig. 1A)(Verma et al., 2019; Zhang et al., 2019). To understand how ALT occurs in the absence of RAD52, we sought to identify factors contributing to the ALT activity in RAD52 KO cells, which have been characterized in our previous study (Zhang et al., 2019). TERRA promotes ALT by forming telomeric R-loops (Arora et al., 2014; Silva et al., 2021). To understand whether TERRA contributes to the ALT activity in RAD52 KO cells, we first tested the localization of TERRA in RAD52 wild-type (WT) and KO U2OS (ALT+) cells. TERRA is localized to ALT-associated PML bodies (APBs) in ALT+ cells (Arora et al., 2014). We used RNA-ISH to detect TERRA, TRF2 antibody to follow telomeres, and PML antibody to visualize PML bodies. In both RAD52 WT and KO cells, TERRA and TRF2 were well colocalized, and some of the TERRA-TRF2 foci were colocalized with PML (Fig. S1A), showing that TERRA localizes to APBs independently of RAD52. Notably, the numbers of APBs were slightly reduced in RAD52 KO cells compared to RAD52 WT cells (Fig. S1B), which is consistent with our previous finding that RAD52-mediated BIR contributes to the efficient formation of APBs (Zhang et al., 2021). Nonetheless, the association of TERRA with APBs remained the same in RAD52 WT and KO cells (Fig. S1B), suggesting that TERRA does not need RAD52 to localize to APBs once they are formed.
Figure 1. TERRA contributes to the RAD52-independent ALT pathway.
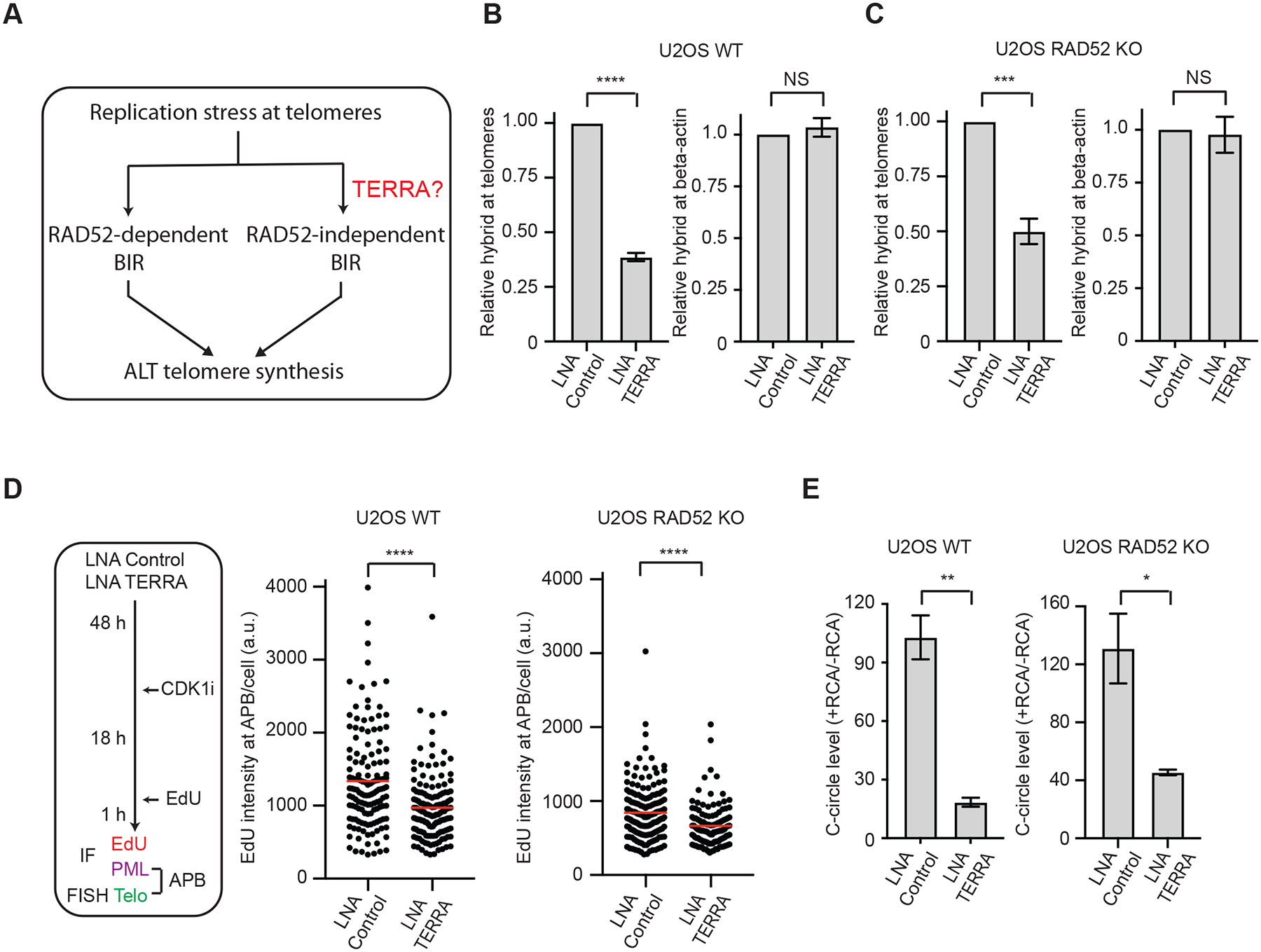
(A) A schematic introducing the bifurcated ALT pathway and the question on the role of TERRA in RAD52-independent ALT. (B-C) U2OS WT (B) and RAD52 KO (C) cells transfected with Control or TERRA LNA were analyzed by DRIP coupled with telomere-specific or β-actinspecific qPCR. Error bars: SEM, n=3 (experimental triplicates); ****: P value <0.0001; ***: P value 0.0001; NS: not significant. (D) The experimental scheme to visualize telomere DNA synthesis at APBs in G2 cells (left). U2OS WT and RAD52 KO cells were transfected with Control or TERRA LNA and synchronized in G2. The EdU signals at APBs were quantified in individual cells (n>120) and normalized by nuclear areas. Red lines: mean values. ****: P value <0.0001. (E) U2OS WT and RAD52 KO cells transfected with Control or TERRA LNA were analyzed by the C-circle assay. The RCA+/RCA-ratios of samples reflect the relative levels of C-circle amplification. Error bars: SEM, n=3 (experimental triplicates); **: P value 0.002, *: P value 0.02.
Because TERRA promotes ALT by forming telomeric R-loops (Arora et al., 2014; Silva et al., 2021), we next asked whether TERRA retains the ability to form telomeric R-loops in RAD52 KO cells. To address this question, we sought to knock down TERRA with locked nucleic acid (LNA) antisense oligos (Chu et al., 2017) and assess the effects on telomeric R-loops. The TERRA foci detected by RNA in situ hybridization (RNA-ISH), which were susceptible to RNaseA treatment, were reduced by the TERRA LNA (Fig. S1C). Northern dot blot of TERRA confirmed that TERRA levels were reduced (Fig. S1D). The knockdown of TERRA was also confirmed by RT-qPCR using subtelomere-specific primers (Fig. S1E). TERRA knockdown did not alter the cell cycle (Fig. S1F). Next, we measured the levels of telomeric R-loops by DNA:RNA immunoprecipitation (DRIP) coupled with telomere-specific qPCR. The specificity of DRIP was confirmed by RNaseH treatment (Fig. S1G). TERRA knockdown significantly reduced the levels of telomeric R-loops in both RAD52 WT and KO cells (Fig. 1B–C). In contrast, the R-loops at the β-actin gene were not affected by TERRA knockdown (Fig. 1B–C). Thus, TERRA retains the ability to specifically form telomeric R-loops in RAD52 KO cells. As we reported previously (Zhang et al., 2019), the level of C-circles, a marker of ALT activity (Henson et al., 2009), was increased in passaged RAD52 KO cells (Fig. S1H), which probably reflects a compensatory upregulation of the RAD52-independent ALT pathway. To analyze R-loops at endogenous telomeres, we performed proximity ligation assay (PLA) with the S9.6 antibody recognizing DNA:RNA hybrids and TRF2 antibody detecting telomeres. The combination of TRF2 and S9.6 antibodies, but not either antibody alone, generated significant levels of PLA foci (Fig. S1I), confirming the presence of R-loops at telomeres. Using the TRF2-S9.6 PLA, we found that telomeric R-loops were increased in RAD52 KO cells (Fig. S1J). These results raise the possibility that TERRA-mediated telomeric R-loops promote ALT activity in RAD52 KO cells.
To determine whether TERRA contributes to the ALT activity in RAD52 KO cells, we analyzed the ALT telomere synthesis in APBs (ATSA) as we previously described (Zhang et al., 2019). We first knocked down TERRA, then synchronized cells in G2 with the CDK1 inhibitor RO-3306, and finally detected APBs with TRF2 and PML antibodies and DNA synthesis with EdU (Fig. 1D). In both RAD52 WT and KO cells, TERRA knockdown significantly reduced APB-associated DNA synthesis (Fig. 1D). Knockdown of TERRA did not reduce APBs in RAD52 KO cells (Fig. S1K), suggesting that TERRA promotes RAD52-independent ALT in APBs. Furthermore, TERRA knockdown reduced the levels of C-circles in both RAD52 WT and KO cells (Fig. 1E), confirming that the ALT activity in RAD52 KO cells requires TERRA.
To ascertain that the effects of TERRA knockdown are indeed attributed to the reduction in telomeric R-loops, we inducibly overexpressed RNaseH1 in U2OS cells. As reported previously (Arora et al., 2014), overexpression of RNaseH1 reduced C-circle levels (Fig. S1L), confirming that telomeric R-loops contribute to ALT activity. Because RNaseH1 did not completely remove telomeric R-loops, knockdown of TERRA further reduced C-circle levels (Fig. S1L). Importantly, in TERRA knockdown cells, overexpression of RNaseH1 did not affect C-circle levels (Fig. S1L), indicating that the relevant target of RNaseH1 is not present at a sufficient level. Thus, knockdown of TERRA removes the target of RNaseH1 in ALT, confirming that TERRA knockdown affects ALT by reducing telomeric R-loops.
RAD51AP1 promotes formation of TERRA-mediated telomeric R-loops in vitro
A previous study suggested that RAD51AP1 promotes ALT by recruiting RAD52 to APBs (Barroso-Gonzalez et al., 2019). However, whether RAD51AP1 has a RAD52-independent function in ALT is not known. We recently showed that RAD51AP1 directly promotes R-loop formation in vitro and enhances HR by generating R-loops at DSBs in cells (Ouyang et al., 2021). To test whether RAD51AP1 promotes formation of TERRA-mediated telomeric R-loops, we performed in vitro biochemical experiments using purified RAD51AP1 protein and TERRAmimicking RNA oligos (referred to as TERRA). RAD51AP1 bound to TERRA efficiently in electrophoresis mobility shift assay (EMSA) (Fig. 2A and S2A). In the presence of dsDNA plasmids containing telomeric repeats, RAD51AP1 enabled the formation of R-loops by TERRA (Fig. 2B, S2B). The R-loops formed by TERRA and RAD51AP1 were confirmed by RNaseH digestion (Fig. 2B). Furthermore, RAD51AP1 promoted telomeric R-loop formation in a concentration-dependent manner (Fig. S2B). Compared to wild-type RAD51AP1 (RAD51AP1WT), RAD51AP1DBM, a mutant defective for DNA and RNA binding (Dunlop et al., 2012; Ouyang et al., 2021), displayed significantly reduced abilities to bind TERRA and form telomeric R-loops (Fig. S2C–D). These results show that RAD51AP1 is able to promote TERRA-mediated telomeric R-loop formation independently of RAD52.
Figure 2. RAD51AP1 promotes formation of telomeric R-loops and ALT activity.
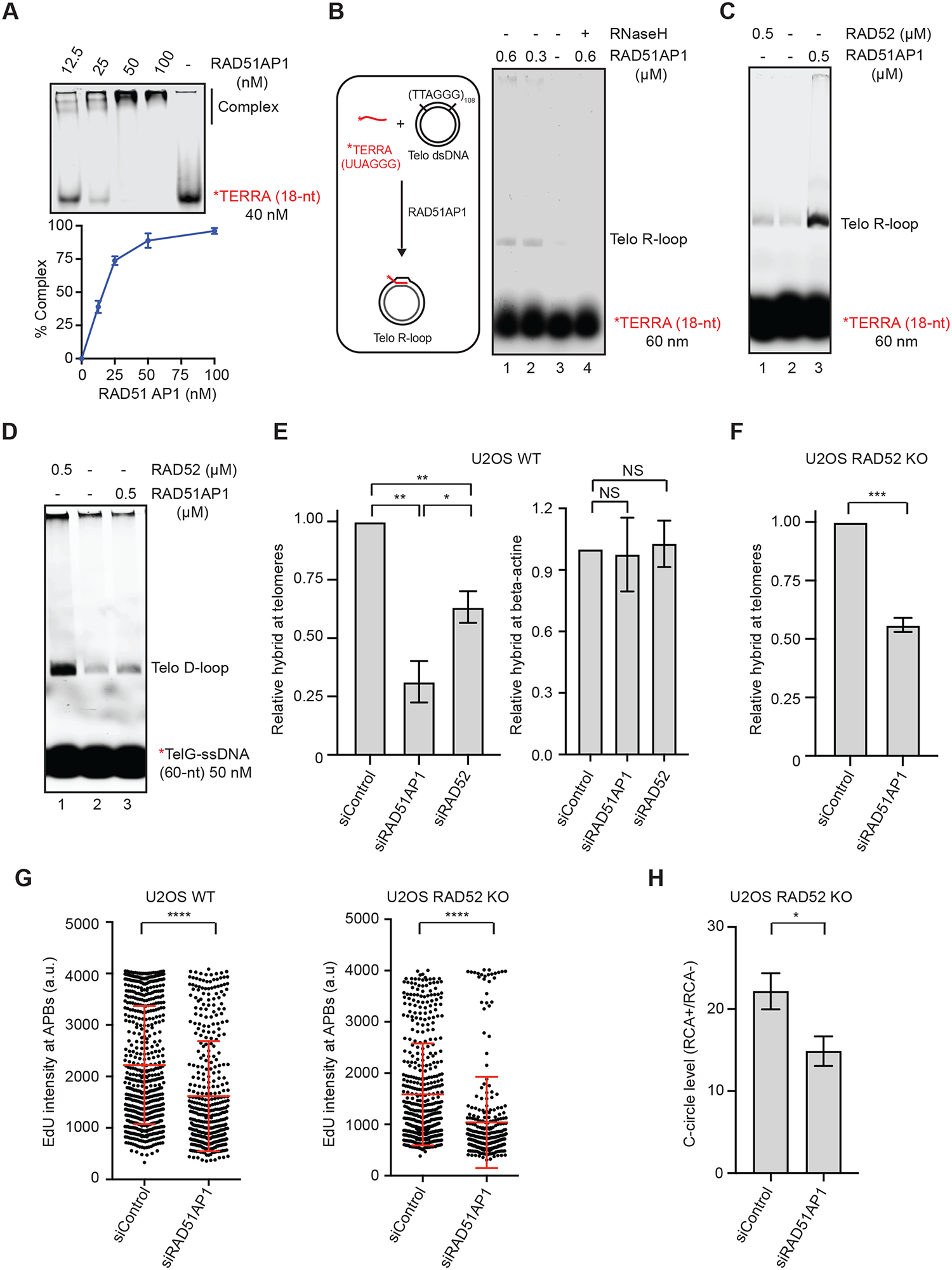
(A) Binding of RAD51AP1 to TERRA. Increasing concentrations of RAD51AP1 were incubated with TERRA-IR800 (18-nt). Reaction products were separated on a native acrylamide gel. The binding of RAD51AP1 to TERRA was quantified by measuring the reduction of free TERRA in the gel. Mean values of two experiments (n=2) are shown. (B) A schematic of the telomeric R-loop formation assay (left); RAD51AP1 was incubated with TERRA-IR800 and dsDNA containing telomeric sequences for 20 min. In lane 4, the sample was treated with RNaseH (0.5 unit) for 5 min after R-loop formation. R-loops were separated from free TERRA on an agarose gel. (C) RAD51AP1 and RAD52 were compared in the R-loop formation assay shown in (B). (D) RAD51AP1 and RAD52 were compared in the telomeric D-loop formation assay. TelG-ssDNAIR800 was incubated with RAD51AP1 or RAD52 and dsDNA containing telomeric sequences for 20 min. Reaction products were separated on an agarose gel. (E) U2OS WT cells transfected with siControl, siRAD51AP1, or siRAD52 were analyzed by DRIP coupled with telomere-specific or β-actin-specific qPCR. Error bars: SEM, n=3 (experimental triplicates); P value 0.001 (siControl vs siRAD51AP1); 0.005 (siControl vs siRAD52); 0.04 (siRAD51AP1 vs siRAD52); NS: not significant. (F) RAD52 KO cells transfected with siControl or siRAD51AP1 were analyzed by DRIP coupled with telomere-specific qPCR. Error bars: SEM, n=3 (experimental triplicates); ***: P value 0.0001. (G) U2OS WT and RAD52 KO cells were transfected with siControl or siRAD51AP1, synchronized in G2, and analyzed for EdU intensity in APBs. The EdU signals at APBs were quantified in individual cells (n>300) and normalized by nuclear areas. Error bars: SD, n=3 (experimental triplicates); ****: P value <0.0001. (H) U2OS RAD52 KO cells transfected with siControl or siRAD51AP1 were analyzed by the C-circle assay. The RCA+/RCA− ratios of samples reflect the relative levels of C-circle amplification. Error bars: SEM, n=3 (experimental triplicates); *: P value 0.01.
In addition to RAD51AP1, RAD52 was also shown to promote DNA:RNA hybridization in vitro (Keskin et al., 2014). Using purified RAD52 and RAD51AP1 proteins, we found that RAD51AP1 was significantly more active than RAD52 in forming TERRA-mediated telomeric R-loops (Fig. 2C). Interestingly, RAD52 efficiently promoted telomeric D-loop formation as we previously reported (Zhang et al., 2019), but RAD51AP1 largely lacked this activity (Fig. 2D). These results suggest that RAD52 and RAD51AP1 have distinct activities at telomeres. While RAD52 primarily promotes D-loop formation, RAD51AP1 is more potent for forming R-loops. A recent study suggested that RAD51 promotes the formation of TERRA-mediated telomeric R-loops in ALT− cells (Feretzaki et al., 2020). To compare the activities of RAD51, RAD52 and RAD51AP1 in telomeric R-loop formation, we used biotinylated TERRA to form and capture R-loops in vitro and quantified them with qPCR (Fig. S2E). We found that RAD51AP1 was significantly more active than RAD52 and RAD51 in forming telomeric R-loops (Fig. S2F). These results suggest that RAD51AP1 is likely the primary factor driving the formation of telomeric R-loops.
RAD51AP1 promotes telomeric R-loops and ALT activity in RAD52 KO cells
To test whether RAD51AP1 regulates telomeric R-loops in ALT+ cells, we knocked down RAD51AP1 in U2OS cells with siRNA (Fig. S2G). As detected by telomere-specific DRIP, knockdown of RAD51AP1 significantly reduced the levels of telomeric R-loops (Fig. 2E). Depletion of RAD52 also reduced telomeric R-loops without altering RAD51AP1 levels, but the effect was less pronounced (Fig. 2E, S2H). The levels of R-loops at the β-actin gene were not affected by knockdown of RAD51AP1 or RAD52 (Fig. 2E), showing that they are not required for the formation of baseline R-loops outside of telomeres. While knockdown of RAD51AP1 reduced R-loops at telomeres, it did not significantly decrease TERRA-positive APBs and only modestly reduced TERRA levels (Fig. S2I), suggesting that the decrease of telomeric R-loops is largely attributed to reduced DNA:RNA hybridization. Importantly, knockdown of RAD51AP1 in RAD52 KO cells also resulted in a significant reduction in telomeric R-loops (Fig. 2F), showing that RAD51AP1 promotes the formation of telomeric R-loops even in the absence of RAD52.
To determine whether RAD51AP1 is involved in the RAD52-independent ALT pathway, we knocked down RAD51AP1 in RAD52 WT and KO U2OS cells. Knockdown of RAD51AP1 significantly reduced the DNA synthesis in APBs in both RAD52 WT and KO cells (Fig. 2G), showing that RAD51AP1 indeed contributes to the RAD52-independent ALT activity. Furthermore, knockdown of RAD51AP1 in RAD52 KO cells reduced C-circle levels (Fig. 2H), confirming that RAD51AP1 promotes ALT activity in the absence of RAD52. Consistent with the results in U2OS cells, knockdown of RAD51AP1 in two other ALT+ cell lines, SAOS2 and SKLU1, also reduced telomeric R-loops and ALT activity (Fig. S2J–K), supporting the notion that RAD51AP1 generally promotes ALT through telomeric R-loops. Together, these results suggest that RAD52 and RAD51AP1 have independent functions in ALT. While RAD52 directly promotes the formation of telomeric D-loops, RAD51AP1 can drive ALT independently of RAD52 through R-loop formation.
RAD51AP1 promotes telomere maintenance and proliferation in RAD52 KO cells
Since RAD51AP1 contributes to the ALT activity in RAD52 KO cells, we next tested whether RAD51AP1 is important for telomere maintenance in the absence of RAD52. Using U2OS RAD52 KO cells, we knocked out RAD51AP1 with CRISPR-Cas9 and generated RAD52, RAD51AP1 double KO lines (referred to as RAD52, 51AP1 DKO; Fig. S3A). To assess the maintenance of telomeres, we analyzed telomeres in metaphase spreads using telomere FISH. As we reported previously (Zhang et al., 2019), telomere FISH signals were diminished in RAD52 KO cells compared to WT cells (Fig. 3A). Loss of RAD52 also increased the telomeres displaying signalfree ends (SFEs) (Fig. 3B, S3B), which are indicative of telomere instability. Compared to RAD52 KO cells, RAD52, 51AP1 DKO cells displayed an even more severe loss of telomere FISH signals and even higher levels of SFEs (Fig. 3A–B, S3B), suggesting that RAD51AP1 contributes to telomere maintenance in the absence of RAD52. Using telomere-specific primers and qPCR to quantify telomeric DNA content, we confirmed that knockout of RAD51AP1 in RAD52 KO cells resulted in a loss of telomeric DNA (Fig. 3C). Knockout of RAD51AP1 in RAD52 KO cells also reduced cell growth compared to the parental RAD52 KO cells (Fig. 3D), indicating that RAD51AP1 is important for the proliferation of RAD52 KO cells. Furthermore, we detected more β-gal-positive cells in the RAD52, 51AP1 DKO cell population than in the RAD52 KO cell population (Fig. 3E, S3C), suggesting that loss of RAD51AP1 in RAD52 KO cells increases cellular senescence. Together, these results provide evidence that RAD51AP1 promotes telomere maintenance and prevents replicative senescence in the absence of RAD52.
Figure 3. RAD51AP1 promotes telomere maintenance and proliferation in RAD52 KO cells.
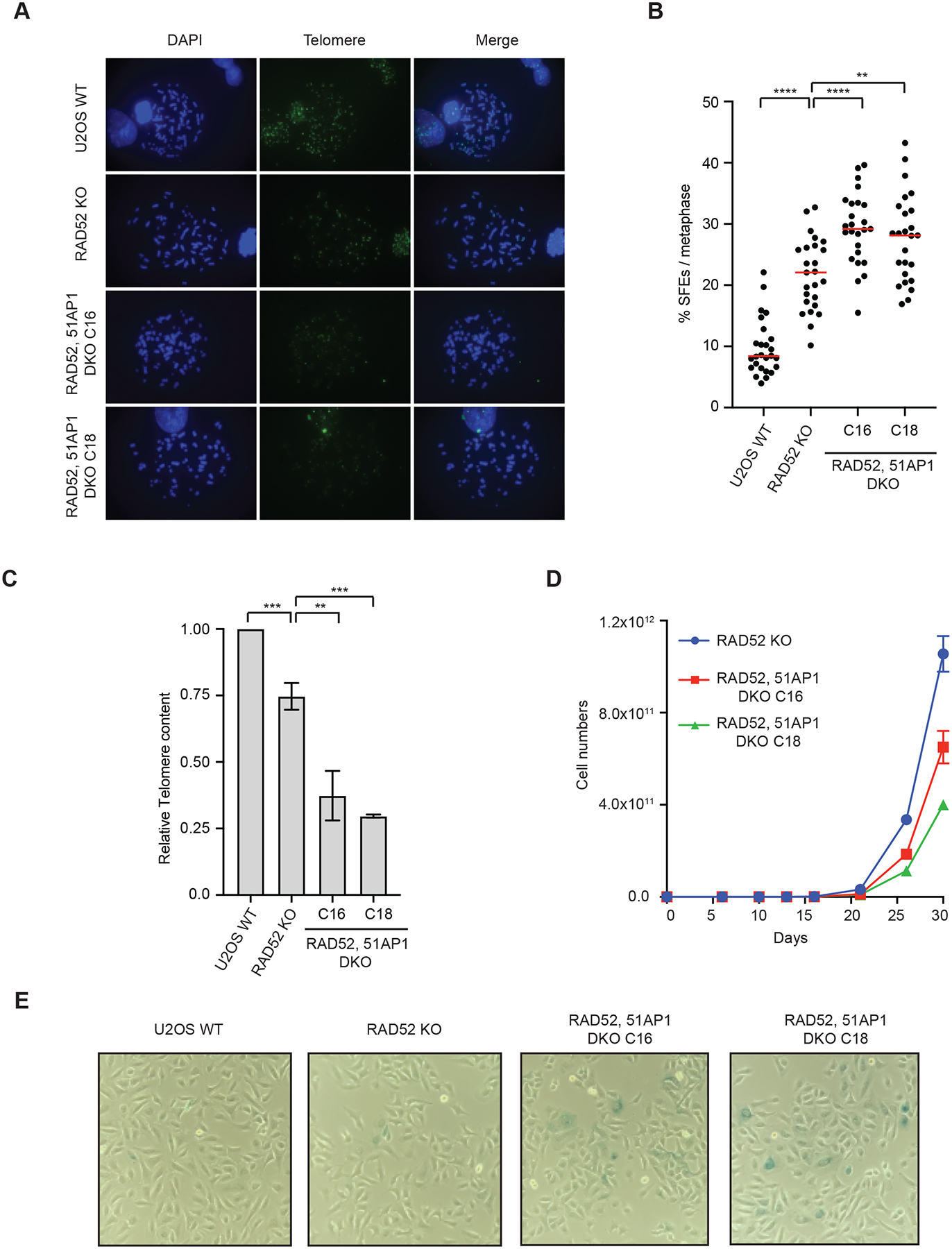
(A) U2OS WT, RAD52 KO, and RAD52, 51AP1 DKO (clones C16 and C18) cells were synchronized in metaphase and telomeric FISH was performed using TelC-488 probe. (B) Signal free ends (SFEs) were quantified from (A). >25 metaphases were quantified from each cell line. Red lines: mean values. ****: P value <0.0001, **: P value 0.003. (C) Telomeres DNA content was quantified in U2OS WT, RAD52 KO, and RAD52, 51AP1 DKO (clones C16 and C18) cells after 1 month of passages. Error bars: SD, n=3 (experimental triplicates); ***: P value 0.001; **: P value 0.003 (D) Cell growth analysis of U2OS WT, RAD52 KO, and RAD52, 51AP1 DKO (clones C16 and C18) cells. (E) Representative images of β -galactosidase staining of U2OS WT, RAD52 KO, and RAD52, 51AP1 DKO (clones C16 and C18) cell populations. All cell lines were passaged in parallel for ~1 month.
Telomeric R-loops increases G4s
Telomeric R-loops promote ALT by inducing replication stress and collapse of replication forks (Arora et al., 2014; Silva et al., 2021). In RAD52 KO cells, while telomeric R-loops can still induce replication stress and fork collapse, telomeric D-loops have to form through a RAD52-independent mechanism. The contributions of TERRA and RAD51AP1 to the RAD52-independent ALT activity raise the possibility that telomeric R-loops may promote D-loop formation in addition to inducing replication stress. One possible mechanism by which telomeric R-loops promote D-loop formation is to open telomeric dsDNA. However, when TERRA forms telomeric R-loops, it hybridizes with the C-rich DNA strand, thereby preventing G-strand DNA end invasion and D-loop formation (Fig. S4A, upper panel). Notably, the G-rich DNA strand of telomere is displaced as ssDNA in telomeric R-loops, which may increase the formation of G4s. The presence of G4s at ALT telomeres may keep dsDNA open and facilitate G-strand DNA end invasion and D-loop formation (Fig. S4A, lower panel). To test whether telomeric R-loops promote G4 formation, we used TERRA, RAD51AP1, and dsDNA plasmids to generate R-loops in vitro, and then measured G4 levels with the anti-G4 BG4 antibody (Fig. 4A). Indeed, a significant increase of G4s was detected when telomeric R-loops were formed in a TERRA-and RAD51AP1-dependent manner in vitro.
Figure 4. Telomeric R-loops increases G4s.
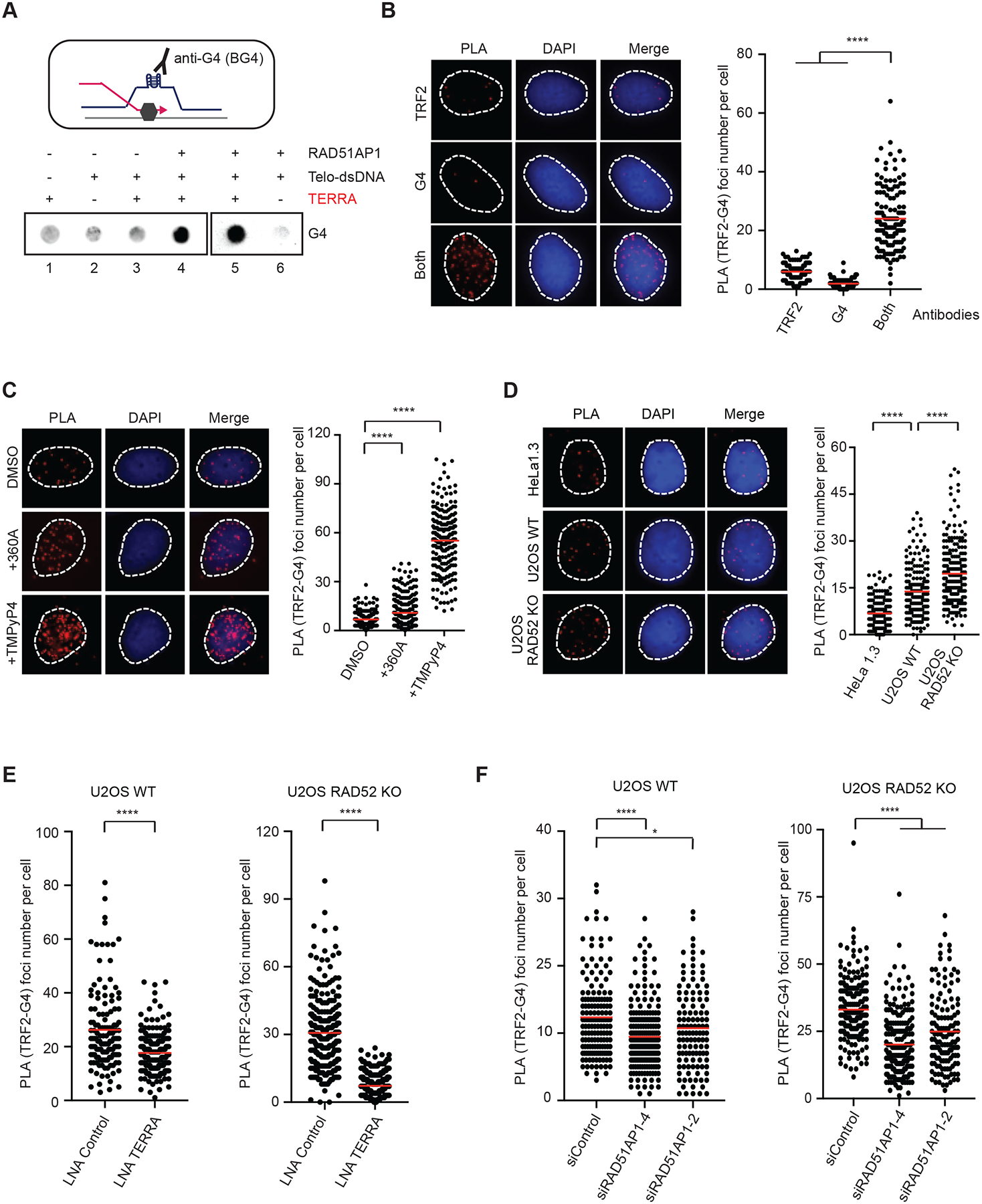
(A) A schematic of G4 formation at an R-loop (upper panel). Telomeric R-loops were generated by incubating TERRA (60-nt, 60 nM), RAD51AP1 (0.5 μM) and dsDNA containing telomeric sequences for 20 min. Reaction products were analyzed for G4s by dot blot using the BG4 antibody. (B) Detection of telomeric G4s by PLA. U2OS cells were analyzed by PLA using anti-G4 (1H6) and anti-TRF2 antibodies, either alone or in combination. Representative images of PLA foci are shown (left) and quantified in individual cells (right) (n>140). Red lines: mean values. ****: P value <0.0001. (C) U2OS cells were treated with 360A (10 μM) or TMPYP4 (4 μM) for 6 hr, and telomeric G4s were analyzed as in (B) (n>220). Red lines: mean values. ****: P value <0.0001. (D) HeLa1.3, U2OS WT and U2OS RAD52 KO cells were analyzed as in (B) (n>190). Red lines: mean values. ****: P value <0.0001. (E) U2OS WT and RAD52 KO cells transfected with Control or TERRA LNA were analyzed as in (B) (n>240). Red lines: mean values. ****: P value <0.0001. (F) U2OS WT and RAD52 KO cells transfected with siControl or two different siRAD51AP1 were analyzed as in (B) (n>130). Red lines: mean values. ****: P value <0.0001, *: P value 0.03. To specifically detect G4s in DNA, all PLA experiments were performed with cells treated with RNaseA (10 μg/ml) at 37 °C for 1 hr.
To detect the G4s at endogenous telomeres, we used TRF2 and G4 antibodies to perform PLA in U2OS cells. The combination of TRF2 and G4 antibodies, but not either antibody alone, generated significant levels of PLA foci (Fig. 4B), confirming the presence of G4s at ALT telomeres. Treatment of cells with G4 stabilizers increased the PLA foci of TRF2 and G4s (Fig. 4C, S4B). Furthermore, ALT+ U2OS cells displayed higher levels of PLA foci than ALT− HeLa1.3 cells, and RAD52 KO further increased PLA foci in U2OS cells (Fig. 4D), suggesting that ALT+ cells have more G4s at telomeres than ALT− cells and the RAD52-independent ALT activity is associated with high levels of G4s. To test the effects of telomeric R-loops on G4s, we knocked down TERRA and RAD51AP1 in RAD52 WT and KO cells. Knockdown of TERRA reduced the TRF2-G4 PLA foci in both RAD52 WT and KO cells (Fig. 4E). Similarly, depletion of RAD51AP1 decreased PLA foci in both RAD52 WT and KO cells (Fig. 4F). In contrast, stabilizing telomeric R-loops in RAD52 KO cells by depleting RNaseH1 or Senataxin (SETX) increased telomeric G4s (Fig. S4C–D) (Arora et al., 2014; Vohhodina et al., 2021). Importantly, overexpression of RNaseH1 reduced TRF2-G4 PLA foci in U2OS cells treated with control LNA, but not in cells treated with the TERRA LNA (Fig. S4E), suggesting that the removal of TERRA-mediated R-loops by RNaseH1 decreases telomeric G4s. Similar to that in U2OS cells, knockdown of RAD51AP1 in SAOS2 and SKLU1 cells also reduced telomeric G4s (Fig. S4F). These results suggest that the formation of telomeric R-loops by TERRA and RAD51AP1 indeed promotes G4 formation at ALT telomeres even in the absence of RAD52.
Dynamic telomeric R-loops and G4s promote ALT in cells
To test whether G4s affect ALT activity in the presence and absence of RAD52, we used pyridostatin (PDS) to stabilize G4s in RAD52 WT and KO cells and analyzed the effects on telomere synthesis (Fig. 5A). Stabilization of G4s increased DNA synthesis in APBs in both RAD52 WT and KO cells (Fig. 5A). Furthermore, G4 stabilization elevated C-circle levels in RAD52 WT and KO cells (Fig. 5B). These results suggest that G4s promote ALT activity even in the absence of RAD52. Because the formation of telomeric R-loops by TERRA and RAD51AP1 increases G4s, we asked whether G4s function downstream of R-loops to stimulate ALT. We knocked down TERRA to reduce telomeric R-loops, and then stabilized spontaneous G4s with PDS and tested the effects on telomere synthesis (Fig. 5C). As expected, stabilizing G4s increased DNA synthesis in APBs, whereas knockdown of TERRA reduced telomere synthesis (Fig. 5C, lanes 1–3). Importantly, G4 stabilization in TERRA knockdown cells significantly rescued telomere synthesis (Fig. 5C, lanes 3–4), suggesting that G4s can promote ALT even when R-loop levels are low. Similarly, stabilization of G4s by PDS also rescued the ALT activity in RAD52 KO cells after RAD51AP1 knockdown (Fig. 5D). These results support the notion that telomeric G4s can act independently and downstream of R-loops to promote ALT.
Figure 5. G4s act downstream of transient R-loops to promote ALT.
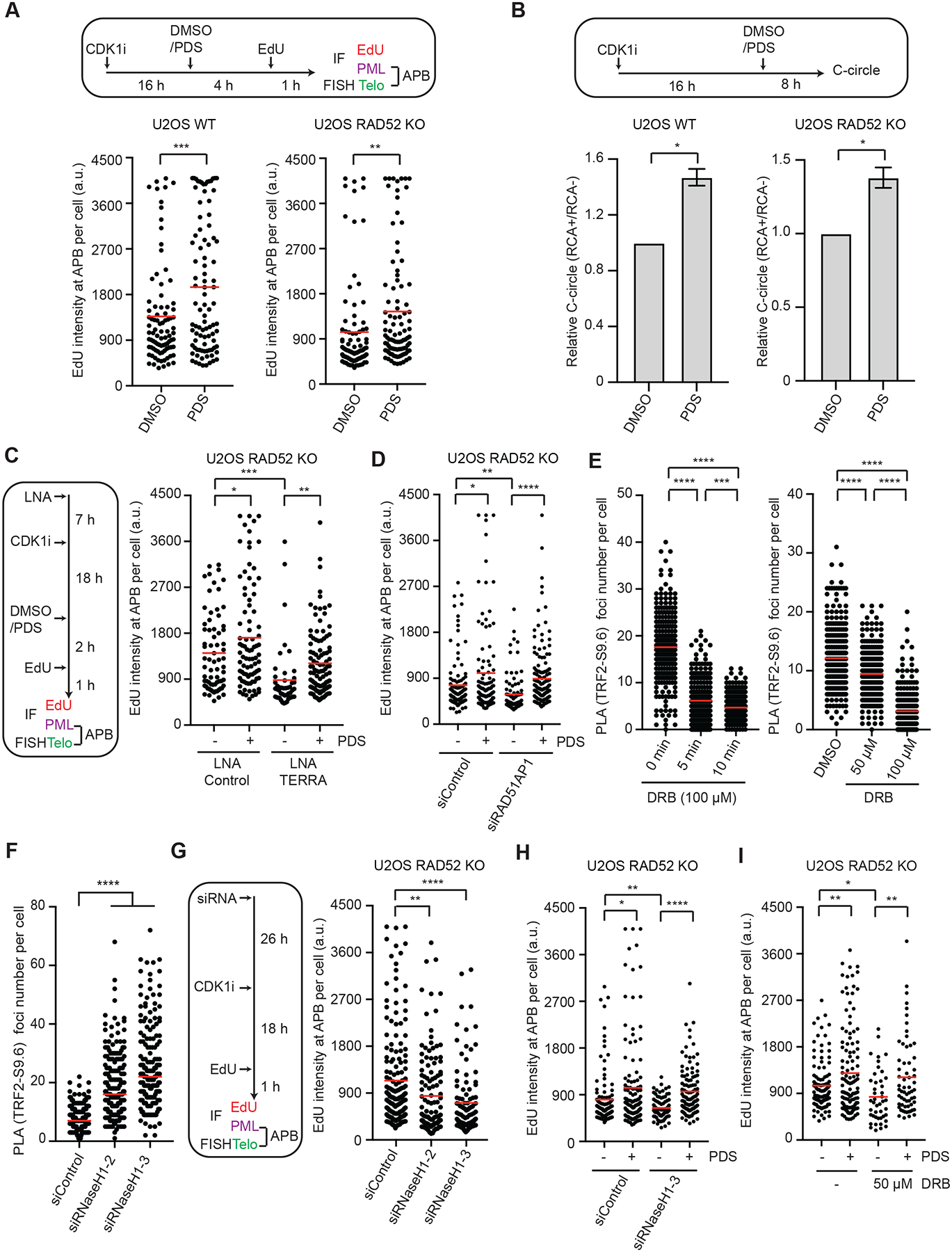
(A) U2OS WT and RAD52 KO cells were synchronized in G2, treated with DMSO or 2.5 μM PDS, and analyzed for EdU intensity at APBs. The EdU signals at APBs were quantified in individual cells (n>200) and normalized by nuclear areas. Red lines: mean intensities. ***: P value 0.0001, **: P value 0.008. (B) U2OS WT and RAD52 KO cells were treated with 5 μM PDS and analyzed by the C-circle assay. The RCA+/RCA-ratios of samples reflect the relative levels of C-circle amplification. Error bars: SEM, n=3 (experimental triplicates); P values 0.01 (left panel) and 0.03 (right panel). (C) U2OS RAD52 KO cells transfected with Control or TERRA LNA were synchronized in G2 and then treated with DMSO or 2.5 μM PDS as indicated. The EdU signals at APBs were quantified in individual cells (n>230) and normalized by nuclear areas. Red lines: mean intensities. *: P value 0.02, **: P value 0.004, ***: P value 0.0001. (D) U2OS RAD52 KO cells transfected with siControl or siRAD51AP1 were synchronized in G2 and then treated with DMSO or 3 μM PDS for 3 hrs. The EdU signals at APBs were quantified in individual cells (n>100) and normalized by nuclear areas. Red lines: mean intensities. *: P value 0.015, **: P value 0.005, ****: P value <0.0001. (E) U2OS RAD52 KO cells were treated with 100 μM DRB for 0, 5, and 10 min (left), or with 0, 50, and 100 μM DRB for 30 min (right). Telomeric DNA:RNA hybrids were quantified by PLA using S9.6 and TRF2 antibodies. PLA foci were quantified in individual cells (n>220). Red lines: means. ***: P value 0.0001, ****: P value <0.0001. (F) U2OS RAD52 KO cells treated with Control or RNaseH1 siRNA were analyzed for telomeric DNA:RNA hybrids by PLA using S9.6 and TRF2 antibodies. PLA foci were quantified in individual cells (n>170). Red lines: means. ****: P value <0.0001. (G) U2OS RAD52 KO cells treated with Control or RNaseH1 siRNA were synchronized in G2 and analyzed for EdU intensity at APBs. The EdU signals at APBs were quantified in individual cells (n≥150) and normalized by nuclear areas. Red lines: mean intensities. **: P value 0.003, ****: P value <0.0001. (H) U2OS RAD52 KO cells transfected with siControl or siRNaseH1 were synchronized in G2 and then treated with DMSO or 3 μM PDS for 3 hrs. The EdU signals at APBs were quantified in individual cells (n>130) and normalized by nuclear areas. Red lines: mean intensities. *: P value 0.019, **: P value 0.007, ****: P value <0.0001. (I) U2OS RAD52 KO cells were synchronized in G2 and then treated with DMSO, 3 μM PDS, and 50 μM DRB for 3 hr. The EdU signals at APBs were quantified in individual cells (n>125) and normalized by nuclear areas. Red lines: mean intensities. *: P value 0.011, **: P value 0.003.
Telomeric G4s may promote ALT by helping D-loop formation. For G-strand DNA end invasion to occur at telomeric R-loops, the DNA:RNA hybrids in R-loops have to be removed (Fig. S4A). The levels of many dynamic R-loops are rapidly reduced after RNAPII inhibition (Crossley et al., 2020). To analyze the dynamics of telomeric R-loops, we performed PLA with TRF2 and S9.6 antibodies in the presence and absence of 5,6-Dichlorobenzimidazole 1-β-Dribofuranoside (DRB), an RNAPII inhibitor. DRB reduced the levels of telomeric R-loops in a time-and concentration-dependent manner (Fig 5E). These results suggest that telomeric R-loops are a dynamic structure that is constantly assembled and disassembled in cells.
Next, we asked whether the removal of DNA:RNA hybrids is required for the ALT activity in RAD52 KO cells. RNaseH1 is involved in the suppression of telomeric R-loops in ALT+ cells (Arora et al., 2014). We knocked down endogenous RNaseH1 in RAD52 KO cells with two distinct siRNAs and analyzed the effects on telomere DNA synthesis. As reported, knockdown of RNaseH1 increased telomeric DNA:RNA hybrids (Fig. 5F, S5) (Arora et al., 2014). Conversely, knockdown of RNaseH1 reduced the telomere synthesis in APBs (Fig 5G), which suggests that the removal of DNA:RNA hybrids is a necessary step in the RAD52-independent ALT pathway. Notably, while knockdown of RNaseH1 reduced the ALT activity in RAD52 KO cells, this reduction was suppressed by PDS (Fig. 5H), indicating that an increase of G4s could bypass the effects of inefficient DNA:RNA hybrid removal. In addition, DRB also reduced the RAD52-independent ALT, and this reduction was also suppressed by PDS (Fig. 5I), confirming that the reduction in R-loops can be bypassed by an increase in G4s. These results lend further support to the notion that telomeric G4s act downstream of R-loops to promote ALT. Given that both the formation and removal of telomeric R-loops contribute to the ALT activity in RAD52 KO cells, our results suggest that the transient formation of telomeric R-loops, rather than their persistent presence, promotes RAD52-independent ALT.
Dynamic telomeric R-loops and G4s promote D-loop formation in vitro
To biochemically test whether telomeric G4s can persist after R-loop resolution, we used TERRA, RAD51AP1 and dsDNA plasmids to generate R-loops in vitro, and then removed DNA:RNA hybrids with RNaseH (Fig. 6A). As expected, in the absence of RNaseH, DNA:RNA hybrids and G4s were detected in telomeric R-loops by S9.6 and BG4 antibodies, respectively (Fig. 6A). After RNaseH treatment, S9.6 signals disappeared but BG4 signals persisted (Fig. 6A). These results show that the telomeric G4s generated by TERRA and RAD51AP1 can indeed persist after DNA:RNA hybrids are removed.
Figure 6. Dynamic telomeric R-loops promote D-loop formation in the absence of RAD52.
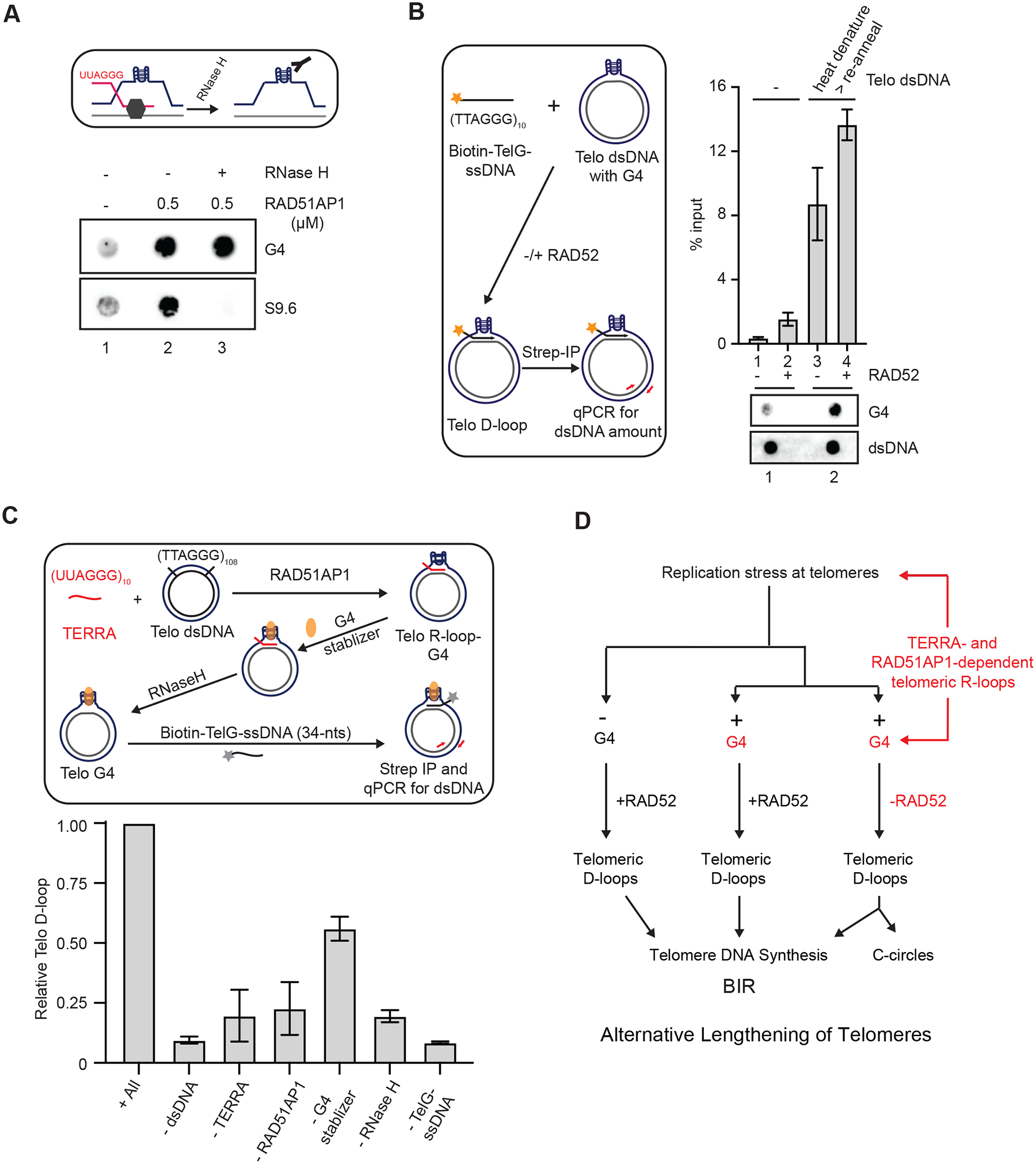
(A) A schematic for testing the presence of G4s in R-loops after removal of DNA:RNA hybrids (upper panel). Telomeric R-loops were generated by incubating TERRA (60-nt, 60 nM), RAD51AP1 (0.5 μM) and dsDNA containing telomeric sequences for 20 min. In lane 3, the sample was treated with RNaseH (0.5 unit) for 5 min after R-loop formation. Reaction products were analyzed for DNA:RNA hybrids and G4s by dot blot using S9.6 and BG4 antibodies, respectively. (B) The experimental scheme to test whether telomeric G4s promote D-loop formation (left). dsDNA containing telomeric sequences was heat denatured and reannealed slowly to increase G4s. The presence of high or low levels of G4s in dsDNA (1 μg) was confirmed by dot blot (bottom panel). The dsDNA harboring high or low levels of G4s was incubated with biotin-TelG-ssDNA (30 nM) in the presence or absence of RAD52 (0.3 μM) for 20 min. Telomeric D-loops were captured with streptavidin beads (10 uL) and quantified by qPCR using primers specific to the dsDNA. Error bars: SEM, n=2. (C) The experimental scheme to reconstitute the TERRA-and RAD51AP1-mediated formation of telomeric D-loops (upper panel). Telomeric R-loops were generated by incubating TERRA (60-nt, 60 nM), RAD51AP1 (0.3 μM) and dsDNA containing telomeric sequences. PDS (1 mM) was added to stabilize G4s, and the DNA:RNA hybrids in R-loops were removed by RNaseH (0.5 unit, 5 min). Subsequently, biotin-TelG-ssDNA was added, and the resulting D-loops were captured with streptavidin beads (10 μL) and quantified by qPCR. Error bars: SEM: n=3. The effects of omitting individual components of the reaction were tested. (D) A model in which the dynamic telomeric R-loops assembled by TERRA and RAD51AP1 promote G4 accumulation and D-loop formation, enabling the RAD52-independent ALT pathway.
To directly test whether G4s can open telomeric dsDNA and enable D-loop formation independently of RAD52, we developed a condition to increase G4s in the dsDNA plasmid containing telomeric repeats. The dsDNA plasmid was heat denatured and slowly reannealed in the presence of 40% PEG-200 and 100 mM KCl. Using the BG4 antibody, we confirmed that this condition significantly increased G4 levels in the dsDNA plasmid (Fig. 6B). We next used a ssDNA oligo containing TTAGGG repeats (Tel-G-ssDNA) and dsDNA plasmids harboring different levels of G4s to carry out D-loop formation assay. In this assay, Tel-G-ssDNA is biotinylated, so that the resulting D-loop-containing plasmids can be captured with streptavidin-coated beads and quantified by qPCR (Fig. 6B). We first validated the D-loop formation assay using purified RAD52 and dsDNA plasmids harboring baseline G4s. As expected, only low levels of D-loops were detected in the absence of RAD52, and RAD52 significantly stimulated D-loop formation (Fig. 6B, lanes 1–2, S6). Importantly, when dsDNA plasmids with high levels of G4 were used, robust D-loop formation was detected even in the absence of RAD52 (Fig. 6B, lane 3). These results demonstrate that telomeric G4s can directly promote D-loop formation independently of RAD52. Nonetheless, RAD52 further increased D-loop formation in the dsDNA plasmid harboring high levels of G4s (Fig. 6B, lane 4), suggesting that the opening of telomeric dsDNA by G4s also facilitates RAD52-mediated D-loop formation.
Finally, we tested whether TERRA and RAD51AP1 can drive the formation of telomeric D-loops in vitro. We first used TERRA, RAD51AP1 and dsDNA plasmids to generate R-loops in vitro, and then stabilized G4s and removed DNA:RNA hybrids with RNaseH (Fig. 6C). The resulting G4-containing dsDNA plasmids without DNA:RNA hybrids were incubated with biotinylated Tel-G-ssDNA, and the D-loop-containing dsDNA plasmids generated by the reaction were captured with streptavidin-coated beads and quantified using qPCR. Using this in vitro assay, we detected TERRA-and RAD51AP1-dependent formation of D-loops (Fig. 6C). As expected, the formation of D-loops is dependent on both Tel-G-ssDNA and dsDNA. Omitting RNaseH reduced D-loop formation, which is consistent with the reduced ALT activity in RAD52 KO cells after RNaseH1 knockdown (Fig. 5G, 6C). Leaving out G4 stabilizer also modestly decreased D-loop levels, supporting the idea that G4s enhance D-loop formation. Most importantly, this TERRA-and RAD51AP1-dependent reaction to form telomeric D-loops occurred in the absence of RAD52, providing direct evidence that TERRA and RAD51AP1 can promote formation of telomeric D-loops independently of RAD52 in vitro.
Discussion
A number of studies suggest that ALT is a BIR-related pathway (Dilley et al., 2016; Min et al., 2017; Roumelioti et al., 2016; Sobinoff et al., 2017; Zhang et al., 2019). One of the critical events to initiate BIR at telomeres is the formation of DSBs. At ALT telomeres, one-ended DSBs may be generated by replication forks that collapse at R-loops and/or G4s (Amato et al., 2020; Arora et al., 2014; Silva et al., 2021; Yang et al., 2021). The nucleolytic processing of R-loops and/or G4s may also generate DSBs at ALT telomeres (Cristini et al., 2019; Lin et al., 2013; Sollier et al., 2014; Yasuhara et al., 2018). Indeed, the telomeric DSBs generated by a fusion of TRF1 and the Fok-I nuclease trigger robust DNA synthesis at ALT telomeres (Dilley et al., 2016). A second important event in BIR is the invasion of ssDNA ends into dsDNA, which gives rise to D-loops (Wu and Malkova, 2021). At ALT telomeres, RAD52 is a key factor promoting D-loop formation. However, ALT activity is not eliminated in RAD52 KO cells, suggesting the existence of a RAD52-independent D-loop forming activity. Our previous study suggested that RAD51 is not critical for the RAD52-independent ALT pathway (Zhang et al., 2019). In this study, we find that TERRA and RAD51AP1 play important roles in this pathway. Several important details of this ALT pathway have emerged from our results. First, RAD51AP1 is a critical factor that promotes the formation of telomeric R-loops by TERRA. Second, in addition to inducing replication stress, TERRA-mediated R-loops may participate in D-loop formation at ALT telomeres. Finally, the TERRA and RAD51AP1-driven pathway can operate independently of RAD52, explaining how telomeric D-loops are formed in RAD52 KO cells. These findings not only shed light on how TERRA, which is upregulated in ALT+ cancer cells, promotes ALT activity, but also provide a revised framework of the ALT pathway (Fig. 6D). This revised framework will help future investigations to further delineate the process of ALT and target ALT in cancer therapy.
TERRA promotes ALT by inducing replication stress (Arora et al., 2014; Silva et al., 2021), and its role in D-loop formation adds a mechanism by which TERRA contributes to ALT activation in cancer cells. It is interesting to note that a number of DNA repair proteins involved in ALT, such as RAD52 and RAD51AP1, have affinities to DNA:RNA hybrids (Keskin et al., 2014; Ouyang et al., 2021). The formation of telomeric R-loops in APBs may facilitate the recruitment of DNA repair proteins. Yeast studies suggested that TERRA is upregulated at short telomeres, which promotes the DNA damage response and telomere extension (Graf et al., 2017). We previously showed that the RAD52-independent ALT activity is progressively increased in RAD52 KO cells as telomeres are shortened (Zhang et al., 2019), raising the possibility that this ALT pathway is stimulated by the upregulation of TERRA at short telomeres.
In this study, we observed an interesting ‘division of labor’ between RAD52 and RAD51AP1 at ALT telomeres. While RAD52 has a robust activity in telomeric D-loop formation, its activity in generating telomeric R-loops is significantly weaker than RAD51AP1. In contrast, RAD51AP1 efficiently promotes telomeric R-loop formation, but only shows a neglectable activity in forming telomeric D-loops. These results suggest that RAD52 and RAD51AP1 play distinct roles at ALT telomeres. It is plausible that RAD52 directly generates D-loops during BIR, whereas RAD51AP1 enables TERRA to form R-loops and indirectly promotes D-loop formation. The formation of telomeric R-loops and G4s may also enhance the RAD52-mediated D-loop formation, allowing RAD52 and RAD51AP1 to function in concert (Fig. 6D). It is interesting to note that RAD51AP1 interacts with RAD52 and helps recruit RAD52 to APBs (Barroso-Gonzalez et al., 2019). Thus, while RAD52 and RAD51AP1 have distinct activities in the ALT pathway, they also function cooperatively. Consistent with a recent report (Feretzaki et al., 2020), we find that RAD51 can promote telomeric R-loop formation in vitro. However, the R-loop forming activity of RAD51 is significantly lower than that of RAD51AP1. Previous studies by others and us suggested that RAD51 is not critical for the ALT pathway in human cells (Dilley et al., 2016; Min et al., 2017; Zhang et al., 2019). It is possible that RAD51 generates telomeric R-loops in ALT− cells, but this role is taken over by RAD51AP1 at ALT telomeres. Consistent with this possibility, RAD51AP1 but not RAD51 was identified as an ALT-specific, telomere-associated protein by a proteomic study (Garcia-Exposito et al., 2016). Notably, RAD51AP1 is not needed for the formation of baseline R-loops at the β-actin gene, indicating that RAD51AP1 may be specifically involved in stress-induced R-loop formation. We speculate that the high replication stress at ALT telomeres may enable RAD51AP1 to function as the primary R-loop-forming factor in this specific context.
Previous studies by others and us suggest that DNA:RNA hybrids directly participate in DSB repair as repair intermediates (McDevitt et al., 2018; Michelini et al., 2017; Storici et al., 2007; Tan et al., 2020a). We recently showed that the RNA transcripts at a transcriptionally active locus form DNA:RNA hybrids in donor DNA upon DSB formation, promoting the invasion of ssDNA ends through DR-loops (Ouyang et al., 2021). However, the DR-loop model cannot explain the function of telomeric R-loops in ALT because TERRA prevents the invasion of G-rich ssDNA ends, which is necessary for ALT (Fig. S4A). Instead, we find that the formation of telomeric R-loops increases G4s. TERRA and RAD51AP1 directly promote G4 formation in vitro, and both are required for maintaining G4s at ALT telomeres, suggesting that R-loops act upstream of G4s in the ALT pathway. Telomeric R-loops are reduced by inhibition of RNAPII but increased by depletion of RNaseH1, showing that telomeric R-loops are dynamic. Importantly, loss of RNaseH1 reduces telomere synthesis in RAD52 KO cells, indicating that the removal of DNA:RNA hybrids at telomeres is also required of RAD52-independent ALT activity. Together, these results put forth a model in which the dynamic R-loops at ALT telomeres generate relatively persistent G4s to open dsDNA, leading to an R-to-D loop switch. It should be noted that R-loop accumulation in E. coli promotes constitutive stable DNA replication (cSDR), a process resembling BIR (Hong et al., 1995). The R-to-D loop switch may be evolutionarily conserved even in prokaryotes.
The model of R-to-D loop switch raises several questions. How are the dynamics of telomeric R-loops regulated? How are G4s regulated at telomeres? Can the R-to-D loop switch occur outside of telomeres? Several ALT regulators, such as RNaseH1, FANCM, and BLM, are implicated in R-loop removal and may affect R-loop dynamics (Arora et al., 2014; Pan et al., 2019; Silva et al., 2019; Tan et al., 2020b). The G4-forming and -binding protein RIF1 preferentially associates with ALT telomeres (Moriyama et al., 2017; Silverman et al., 2004). Notably, while the G4s in telomeric DNA facilitate D-loop formation during BIR, they need to be resolved during lagging strand synthesis. Several helicases that suppress G4s, such as PIF1, FANCJ, BLM, WRN, and RTEL1 (Drosopoulos et al., 2015; Kotsantis et al., 2020; Ribeyre et al., 2009; Wu et al., 2008), are also involved in ALT or telomere recombination (Li et al., 2021; Saini et al., 2013; Wilson et al., 2013; Zhang et al., 2021). The temporally regulated interplays between R-loops and D-loops proposed in the R-to-D loop switch model may help explain the DNA repair functions of RNAs in specific chromosomal and cellular contexts. In addition to telomeres, the R-to-D loop switch may occur in transcribed sequences prone to from secondary structures, promoting BIR or HR. Together, the models of R-to-D loop switch and DR-loops provide possible mechanisms by which RNAs promote D-loop formation during homology-dependent DNA repair, shedding light on the functions of RNAs in maintaining genomic integrity.
In an accompanying paper by Kaminski et al. (Kaminski, 2022), the authors also show that RAD51AP1 binds TERRA, promotes formation of telomeric R-loops, and facilitates D-loop formation in telomeric DNA. Notably, they observed not only an activity of RAD51AP1 in forming TERRA-mediated R-loops, but also a stimulatory effect of RAD51P1 and RNA on RAD51-mediated D-loop formation in non-telomeric DNA, consistent with our previous finding (Ouyang et al., 2021). This paper also shows that the resolution of R-loops by SETX is required for ALT activity. Furthermore, through a proteomic approach, this study reveals a function of RAD51AP1 and its associated proteins PDS5a, ZMYND8, and BRG1 in suppressing TERRA accumulation and transcription-replication conflicts at telomeres. Together, this study and ours highlight the importance of RAD51AP1 and dynamic telomeric R-loops in telomere stability, D-loop formation, and ALT.
STAR * METHODS
RESOURCE AND AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Lee Zou (zou.lee@mgh.harvard.edu; Massachusetts General Hospital Cancer Center, Harvard Medical School, Boston, MA, USA).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Raw western blot and Immunofluorescence data have been deposited at Mendeley and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PML | Abcam | Cat# ab96051; RRID: AB_10679887 |
| TRF2 | Novus Biology | Cat# NB110-57130; RRID: AB_844199 |
| G-quadruplex (BG4) | Millipore Sigma | Cat# MABE917; RRID: AB_2750936 |
| G-quadruplex (1H6) | Millipore Sigma | Cat# MABE1126 |
| S9.6 | Millipore Sigma | Cat# MABE1095; RRID: AB_2861387 |
| RAD52 | Abcam | Cat# ab124971; RRID: AB_10971685 |
| Biotin | Abcam | Cat# ab53494; RRID: AB_867860 |
| GAPDH | Millipore | Cat# ABS16; RRID: AB_10806772 |
| Ku80 | NeoMarker | Cat# MS-285 P; RRID: AB_62299 |
| RAD51AP1 | Proteintech | Cat#11255-1-AP; RRID: AB_2300786 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| T4 Polynucleotide Kinase | NEB | M0201S |
| Proteinase K | Thermo Fischer | 25530049 |
| RNase H | NEB | M0297 |
| RNase A | Millipore Sigma | R6513 |
| CDK1i (RO-3306) | Selleckchem | S7747 |
| BSA | NEB | B9000S |
| Click-iT EdU Alexa Fluor 488 Imaging Kit | Thermo Fischer | C10337 |
| Formamide | Sigma | 47671-1L-F |
| 20x SSC | Invitrogen | AM9763 |
| Azide-PEG3-Biotin | Sigma | 762024-10MG |
| phi29 DNA Polymerase | NEB | M0269L |
| dNTP Set (100 mM) | Thermo Fischer | 10297018 |
| PureLink Genomic DNA Mini Kit | Thermo Fischer | K182002 |
| ULTRAhyb hybridization buffer | Invitrogen | AM8669 |
| LNA Control and LNA TERRA | Qiagen | Custom made |
| DRB | Millipore Sigma | D1916 |
| Deposited data | ||
| Raw western blot and immunofluorescence data | This study | https://data.mendeley.com/datasets/vhz6phhb6c/draft?a=1e79524c-52e6-4716-a154-4a7d3885ecc7 |
| Oligonucleotides | ||
| Oligos used in this study | This study | Table S1 |
| Software and Algorithms | ||
| Fiji | ImageJ | https://imagej.net/software/fiji/ |
| NIS element viewer | Nikon | https://www.nikoninstruments.com/Products/Software/NIS-Elements-Advanced-Research/NIS-Elements-Viewer |
| Image Lab | BioRad | http://www.bio-rad.com/en-us/product/image-lab-software |
| Image studio | Li-COR | https://www.licor.com/bio/image-studio/ |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell culture
U2OS, HeLa1.3, and U2OS-derived RAD52 KO and RAD52, 51AP1 DKO cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS), 2 mM Glutamine and 1% penicillin/streptomycin. SAOS2 and SKLU1 cell lines were cultured in McCoy’s 5A and DMEM/F-12, respectively, with 10 % FBS, 2 mM Glutamine, and 1% penicillin/streptomycin.
METHOD DETAILS
Generation of KO cell lines
RAD51AP1 was knocked out in U2OS RAD52 KO cells using CRISPR-Cas9 and sgRNAs. Two sgRNAs targeting the exons 3 and 5 of RAD51AP1 splicing variant 1 were expressed from the vector pU6-(BbsI)_CBh-Cas9-T2A-mCherry (from the laboratory of R. Kuehn via Addgene). Cells co-transfected with plasmids expressing the gRNAs and a plasmid expressing the puromycin resistant gene were selected with 1 μg/ml puromycin for 5 days. Single cells were sorted by FACS into 96-well plates and grown for two weeks. Deletion of RAD51AP1 was confirmed by western blot using RAD51AP1 antibody. RAD52 KO cells were previously described (Zhang et al., 2019). U2OS RAD52, 51AP1 DKO clones C16 and C18 were used in indicated experiments. The DNA sequences encoding the gRNAs targeting RAD51AP1 are shown in Table S1.
RNA interference
siRNA transfections were done by reverse transfection with Lipofectamine RNAiMax (Invitrogen). All siRNAs were transfected at 5 nM or as stated in figure legends. LNA transfections were done similarly to siRNA transfections (50 or 100 nM). RAD51AP1–4 (used in all experiments unless specified). The sequences of the siRNAs and LNAs used in this study are shown in Table S1. The same protocol was applied to all cell lines tested.
Cell cycle analysis
Cells were transfected with LNAs for 30 hr and incubated with 10 mM EdU for 15 min. EdU was labeled with fluorescent dye picolyl azide by click-it reaction. DNA was stained with propidium iodide. Cell cycle distribution of cells was determined by FACS.
ALT telomere synthesis in APBs (ATSA) assay
Assay was performed as previously described (Zhang et al., 2019). Briefly, synchronized G2 cells were incubated with 20 mM EdU for 1 hr. Cells on coverslips were treated with pre-extraction buffer (0.1% Triton X-100, 20 mM HEPES-KOH pH 7.9, 50 mM NaCl, 3 mM MgCl2, 300 mM sucrose), fixed with PFA and cold methanol (−20°C), then permeabilized in PBS with 0.5%Triton X-100 and blocked with block solution (1xPBS containing 0.05% Tween-20 and 3% BSA). Cells were incubated with primary antibody overnight, washed with PBST (1xPBS containing 0.05% Tween-20), and incubated with secondary antibodies conjugated to fluorophores in the same solution. For telomere FISH, cells were fixed with PFA, blocked with PBG (0.2% [w/v] cold-water fish gelatin, 0.5% [w/v] BSA in PBS), dehydrated in 70%, 85%, and 100% ethanol, and allowed to air dry completely. Cells were incubated with hybridization solution (70% formamide, 2x SSC, 2 mg/ml BSA, 10% dextran sulfate) with 100 nM PNA probe TelC-FITC (F1009, PNA Bio), denatured for 10 min at 85°C, then incubated at room temperature overnight. Cells were washed with wash solution 1 (70% formamide, 2x SSC) and wash solution 2 (2x SSC, 0.1% tween-20) containing DAPI. Images were captured with a Nikon 90i microscope. The same protocol was applied to all cell lines tested.
C-circle assay
C-circle assay was performed as previously described (Zhang et al., 2019). Briefly, genomic DNA was extracted with PureLink Genomic DNA Mini Kit (K182002). Genomic DNA (16 ng) was diluted in 1 X φ29 buffer (NEB) supplemented with 0.2 mg/ml BSA, 0.1% Tween, 4 mM dithiothreitol (DTT), and 1 mM dNTPs without dCTP. Each genomic DNA sample was tested in the presence and absence of 7.5 U φ29 DNA polymerase (NEB). Samples were incubated at 30°C for 8 hr followed by 20 min at 65°C. The levels of telomeric DNA were quantified by qPCR. The primers used in this assay are shown in Table S1.
TERRA RNA-ISH
TERRA RNA-ISH was performed as in (Chu et al., 2017). Briefly, cells were incubated in CSK buffer (100 mM NaCl, 300 mM Sucrose, 3 mM MgCl2, 10 mM PIPES pH 7, 0.5% Triton X-100, 10 mM Vanadyl Ribonucleoside Complex), fixed with PFA, dehydrated by 70%, 85%, 100% of ethanol, and then air dried. Cells were hybridized in buffer (50% formamide, 2x SSC, 2mg/ml BSA, 10% dextran sulfate, 10 mM Vanadyl Ribonucleoside Complex) with 20 nM PNA probe TelC-FITC (F1009, PNA Bio) at 42°C overnight. Cells were washed three times with wash buffer 1 (2x SSC + 50% formamide) and then three times with 2x SSC buffer containing DAPI. Images were captured with a Nikon 90i microscope.
Dot blot assay
For TERRA detection by dot blot, total RNA isolated from cells was dotted on nylon membrane with a Bio-Dot SF microfiltration apparatus (BioRad), air dried, and crosslinked with UV (120 mJ/cm2). Membrane was preincubated with ULTRAhyb buffer (Invitrogen AM8669) and incubated with pre-denatured IR800-TelC or GAPDH probe at 42°C overnight. Membrane was washed three times with 2X SSC containing 0.1 % SDS and then three times with 2X SSC. Membrane was imaged using an Odyssey scanner (LI-COR Biosciences). For G4 detection using antibody, UV crosslinked DNA on membrane was blocked by 5% milk in PBS buffer and then incubated with primary antibody (BG4) at 4°C overnight. Membrane was washed three times with PBST buffer and then incubated with anti-flag antibody in 5% milk for 1 hr. Membrane was washed three times and incubated with secondary antibody, washed and imaged using ECL and ChemiDoc (BioRad).
Cell senescence assay
β-galactose staining was performed using the Millipore Sigma CS0030 kit. Briefly, cells were washed, fixed, stained overnight at 37°C, and imaged using Eco-scope.
Telomere FISH
Cells were incubated for 5 h in 0.1 μg/mL KaryoMAX colcemid (Gibco 15212–012), collected and incubated in 75 mM KCl for 30 minutes, then fixed three times in methanol:acetic acid (3:1). Following fixation, cells were dropped onto microscope slides and metaphase spreads were allowed to dry overnight then rehydrated in 1x PBS, followed by fixation in 4% paraformaldehyde for 5 min. Slides were then washed in 1x PBS, dehydrated in an ethanol series (70%, 95%, 100%) pre-chilled to −20°C and air dried. FISH was performed with TelC-Alexa488 (PNA Bio F1004) in hybridization buffer [70% formamide, 10 mM Tris pH 7.2, 0.5% including blocking buffer pH 7.5 (100 mM maleic acid, 150 mM NaCl, 10% Roche blocking reagent #11096176001] preheated to 80°C. Slides were denatured with the probe at 80°C, then allowed to incubate overnight at RT in a humid chamber. Next, slides were washed twice in hybridization wash A (10 mM Tris-HCl pH 7.2, 0.1% BSA, 70% formamide) and three times in hybridization wash B (0.1 M Tris-HCl pH 7.2, 0.15 M NaCl, 0.08% Tween-20). The second hybridization wash B contained DAPI (Life Technologies D1306) at a 1:1000 concentration. Slides were then dehydrated in an ethanol series (70%, 95%, 100%) pre-chilled to −20°C, air dried and mounted using Prolong gold anti-fade mountant (Invitrogen P36930). Images were captured using NIS element software with a Nikon i90 microscope. Images were scored manually. Statistically analysis was performed using GraphPad Prism 9.
TERRA RT-qPCR
Total RNA was extracted using Direct-zol RNA kit (Zymo-R2070). On column DNase I digestion was performed according to the kit’s instruction. 1ug total RNA was digested with 1uL RNasefree DNase I (NEB) at 37C for 30 min. The RNA was recovered using Monarch® RNA Cleanup Kit (NEB). 250ng eluted RNA was treated with dsDNase (Thermo Fisher) according to the manufacturer’s instruction before reverse transcription was performed using SuperScript™ III Reverse Transcriptase (Thermo Fisher) with GAPDH and TERRA specific primers. The qPCR was then carried out using PowerUp™ SYBR™ Green Master Mix (Thermo Fisher) with TERRA_10q and GAPDH specific primer pairs. The fold change was calculated by ΔΔCt method using GAPDH qPCR as normalization reference.
Telomeric DNA:RNA immunoprecipitation (DRIP)
Telomeric DRIP was performed as in (Yasuhara et al., 2018) with some modifications. Genomic DNA was isolated using PureLink genomic DNA kit (Invitrogen). Genomic DNA (10 ug) was fragmented by sonication in IP buffer (50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 0.5 mM EDTA, 0.5% NP40) and incubated in the S9.6 antibody (10 μg) overnight. Protein G dynabeads were added to samples and incubated for 2 hr. Beads were washed three times with IP buffer and eluted in buffer containing 100 mM NaHCO3 and 1% SDS. Eluted fractions were cleaned using PCR elution column and analyzed by qPCR using telomeric primers shown in Table S1.
Proximity ligation assay (PLA)
PLA assay was performed as in (Matos et al., 2020). Briefly, cells were treated with CSK extraction buffer (0.2% Triton X-100, 20 mM HEPES-KOH pH 7.9, 100 mM NaCl, 3 mM MgCl2, 300 mM sucrose, 1 mM EGTA), fixed with PFA and methanol then permeabilized with 1x PBS containing 0.5% Triton-x100. Cells were treated with RNaseA (10 ug/ml) for 1 hr at 37 °C then washed and blocked with 3% BSA in PBST buffer for 1 hr. Cells were incubated with the primary antibodies diluted at 1:500 at 4°C overnight. After three washes with 1x PBST, cells were incubated with anti-mouse minus and anti-rabbit plus PLA probes (PLA kit from Sigma) at 37°C for 1 hr. Cells were washed with PLA buffer A and incubated with ligation buffer containing ligase (PLA kit) for 30 min at 37°C, then washed and incubated with amplification buffer with polymerase (PLA kit) at 37°C for 1 hr. Cells were washed twice with PLA buffer B (PLA kit) and then three times with PBST buffer containing DAPI. Images were captured with a Nikon 90i microscope. The same protocol was applied to all cell lines tested.
Protein purification
RAD52 was purified as in (Zhang et al., 2019). RAD51AP1WT and RAD51AP1DBM were purified as in (Ouyang et al., 2021). RAD51 recombinant protein was purchased from Abcam (ab63808).
Electrophoresis mobility shift assay (EMSA)
Single-stranded telomeric DNA and RNA oligos labeled with IRDye-800 at the 5’ end were incubated with proteins in buffer C (25 mM Tris-HCl (pH7.5), 1 mM EDTA, 1 mM DTT, 50 μg/ml BSA and 50 mM KCl) at 37°C for 15 min. Reaction products were loaded onto 6% PAGE-TBE gels and resolved at 4°C (Nguyen et al., 2017). Gels were imaged using an Odyssey scanner (LI-COR Biosciences).
Telomeric D-loop assay
A Telomeric ssDNA oligo labeled at the 5’ end was incubated with proteins in buffer (35 mM Tris-HCl, 1 mm DTT, 5 mM MgCl2, 50 mg/ml BSA, 50 mM KCl) in absence or presence of 2 mM ATP or AMP-PNP, and/or 2 mM CaCl2). The pTELO plasmid, which contains 660 bp of telomeric sequences, was added to the reactions at 10 nM, and samples were incubated at 30°C for 15 min. After the incubation, 1 mg/ml proteinase K, 0.5% SDS and 0.5 mM EDTA was added, and reactions continued for 5 min. Reaction products were resolved in 1% agarose gel with TAE buffer. Gels were analyzed using an Odyssey scanner (LI-COR Biosciences). For the D-loop assay coupled with qPCR, biotin-TelG-ssDNA and the pTELO plasmid were incubated with proteins to generate D-loops. The resulting D-loops were captured using Streptavidin C1 conjugatedmagnetic beads. D-loops were quantified by qPCR using primers specific to the pTELO plasmid. For the D-loop assay using G4-containing dsDNA plasmids, pTELO was heat denatured and then cooled down slowly in presence of 100 mM KCl and 40% PEG-200. The G4s in dsDNA was quantified by dot blot. The oligos used in these assays are shown in Table S1.
Telomeric R-loop assay
R-loop formation assay was done similarly to the D-loop assay described above except that a telomeric ssRNA oligo (TERRA) labeled at the 5’ end was used. R-loops were also generated with a biotin-TERRA oligo, the pTELO plasmid and proteins. The resulting R-loops were captured with streptavidin-C1 magnetic beads and quantified by qPCR using primers specific to pTELO.
Combined telomeric R-loop and D-loop assay
Telomeric R-loops were generated by incubating unlabeled TERRA (60-nt, 20 nM), pTELO plasmid (10 nM), and RAD51AP1 (0.5 μM) at 30°C for 20 min. G4 stabilizer (360A; 10 μM) was then added, and reactions continued for another 5 min. Samples were treated with RNaseH and RNaseA (0.5 units each) for 5 min, and Biotin-TelG ssDNA (36 nts, 10 nM) was added. The resulting D-loops were captured with streptavidin-C1 magnetic beads and quantified by qPCR using primers specific to pTELO.
QUANTIFICATION AND STATISTICAL ANALYSIS
The colocalization of PML, EdU and telomere FISH signals and PLA foci were quantified MATLAB R2020a software together with the Image Processing Toolbox (Simoneau et al., 2021). For EMSA, D-loop, and R-loop assays, gels were analyzed using an Odyssey scanner (LI-COR Biosciences) and Fiji software. Unpaired Student’s t test is used for statistical analysis as shown in figure legends.
Supplementary Material
Highlights.
TERRA and RAD51AP1 contribute to the ALT activity in RAD52 KO cells.
RAD51AP1 promotes telomeric R-loop formation in vitro and in ALT+ cells.
Telomeric R-loops increase G4s at telomeres and are dynamic in cells.
Telomeric G4s persist without R-loops and enable D-loop formation without RAD52.
Acknowledgments
We thank Dr. R. O’Sullivan for communicating results before publication. We thank members of the Dyson, Lan, and Zou labs for discussions, and Dr. M.-M. Genois for help with cell cycle analysis and Dr. R. Y.-C. Hsu for providing lentivirus expressing RNaseH1-GFP. L.Z. is the James & Patricia Poitras Endowed Chair in Cancer Research. This work is supported by grants from the NIH (CA263934 and CA218856) to L.Z.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interests
All authors declare no competing interests. L.Z. is a member of the advisory board of Molecular Cell.
Inclusion and Diversity
We support inclusive, diverse, and equitable conduct of research.
References
- Amato R, Valenzuela M, Berardinelli F, Salvati E, Maresca C, Leone S, Antoccia A, and Sgura A (2020). G-quadruplex Stabilization Fuels the ALT Pathway in ALT-positive Osteosarcoma Cells. Genes (Basel) 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R, Lee Y, Wischnewski H, Brun CM, Schwarz T, and Azzalin CM (2014). RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat Commun 5, 5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, and Lingner J (2007). Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318, 798–801. [DOI] [PubMed] [Google Scholar]
- Barroso-Gonzalez J, Garcia-Exposito L, Hoang SM, Lynskey ML, Roncaioli JL, Ghosh A, Wallace CT, de Vitis M, Modesti M, Bernstein KA, et al. (2019). RAD51AP1 Is an Essential Mediator of Alternative Lengthening of Telomeres. Mol Cell 76, 11–26 e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmick R, Minocherhomji S, and Hickson ID (2016). RAD52 Facilitates Mitotic DNA Synthesis Following Replication Stress. Mol Cell 64, 1117–1126. [DOI] [PubMed] [Google Scholar]
- Chappidi N, Nascakova Z, Boleslavska B, Zellweger R, Isik E, Andrs M, Menon S, Dobrovolna J, Balbo Pogliano C, Matos J, et al. (2020). Fork Cleavage-Religation Cycle and Active Transcription Mediate Replication Restart after Fork Stalling at Co-transcriptional R-Loops. Mol Cell 77, 528–541 e528. [DOI] [PubMed] [Google Scholar]
- Chu HP, Cifuentes-Rojas C, Kesner B, Aeby E, Lee HG, Wei C, Oh HJ, Boukhali M, Haas W, and Lee JT (2017). TERRA RNA Antagonizes ATRX and Protects Telomeres. Cell 170, 86–101 e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristini A, Ricci G, Britton S, Salimbeni S, Huang SN, Marinello J, Calsou P, Pommier Y, Favre G, Capranico G, et al. (2019). Dual Processing of R-Loops and Topoisomerase I Induces Transcription-Dependent DNA Double-Strand Breaks. Cell Rep 28, 3167–3181 e3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley MP, Bocek MJ, Hamperl S, Swigut T, and Cimprich KA (2020). qDRIP: a method to quantitatively assess RNA-DNA hybrid formation genome-wide. Nucleic Acids Res 48, e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilley RL, and Greenberg RA (2015). ALTernative Telomere Maintenance and Cancer. Trends Cancer 1, 145–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilley RL, Verma P, Cho NW, Winters HD, Wondisford AR, and Greenberg RA (2016). Break-induced telomere synthesis underlies alternative telomere maintenance. Nature 539, 54–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosopoulos WC, Kosiyatrakul ST, and Schildkraut CL (2015). BLM helicase facilitates telomere replication during leading strand synthesis of telomeres. J Cell Biol 210, 191–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlop MH, Dray E, Zhao W, San Filippo J, Tsai MS, Leung SG, Schild D, Wiese C, and Sung P (2012). Mechanistic insights into RAD51-associated protein 1 (RAD51AP1) action in homologous DNA repair. J Biol Chem 287, 12343–12347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feretzaki M, Pospisilova M, Valador Fernandes R, Lunardi T, Krejci L, and Lingner J (2020). RAD51-dependent recruitment of TERRA lncRNA to telomeres through R-loops. Nature 587, 303–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RL, Cox KE, Jeitany M, Wakimoto H, Bryll AR, Ganem NJ, Bersani F, Pineda JR, Suva ML, Benes CH, et al. (2015). Alternative lengthening of telomeres renders cancer cells hypersensitive to ATR inhibitors. Science 347, 273–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Exposito L, Bournique E, Bergoglio V, Bose A, Barroso-Gonzalez J, Zhang S, Roncaioli JL, Lee M, Wallace CT, Watkins SC, et al. (2016). Proteomic Profiling Reveals a Specific Role for Translesion DNA Polymerase eta in the Alternative Lengthening of Telomeres. Cell Rep 17, 1858–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf M, Bonetti D, Lockhart A, Serhal K, Kellner V, Maicher A, Jolivet P, Teixeira MT, and Luke B (2017). Telomere Length Determines TERRA and R-Loop Regulation through the Cell Cycle. Cell 170, 72–85 e14. [DOI] [PubMed] [Google Scholar]
- Henson JD, Cao Y, Huschtscha LI, Chang AC, Au AY, Pickett HA, and Reddel RR (2009). DNA C-circles are specific and quantifiable markers of alternative-lengthening-oftelomeres activity. Nat Biotechnol 27, 1181–1185. [DOI] [PubMed] [Google Scholar]
- Hoang SM, and O’Sullivan RJ (2020). Alternative Lengthening of Telomeres: Building Bridges To Connect Chromosome Ends. Trends Cancer 6, 247–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong X, Cadwell GW, and Kogoma T (1995). Escherichia coli RecG and RecA proteins in R-loop formation. EMBO J 14, 2385–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski N W. AR; Kwon Y; Lynskey ML; Bhargava R; Barrosso-Gonzalez J; Garcia-Exposito L; He B; Xu M; Melhaharevu D; Watkins SC; Modesti M; Nesvizhskii AI; Zhang H; Sung P; O’Sullivan RJ (2022). RAD51AP1 rgulates ALT-HDR through chromatindirected hoeostasis of TERRA. Molecular Cell In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskin H, Shen Y, Huang F, Patel M, Yang T, Ashley K, Mazin AV, and Storici F (2014). Transcript-RNA-templated DNA recombination and repair. Nature 515, 436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kockler ZW, Comeron JM, and Malkova A (2021). A unified alternative telomerelengthening pathway in yeast survivor cells. Mol Cell 81, 1816–1829 e1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotsantis P, Segura-Bayona S, Margalef P, Marzec P, Ruis P, Hewitt G, Bellelli R, Patel H, Goldstone R, Poetsch AR, et al. (2020). RTEL1 Regulates G4/R-Loops to Avert Replication-Transcription Collisions. Cell Rep 33, 108546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar C, Batra S, Griffith JD, and Remus D (2021). The interplay of RNA:DNA hybrid structure and G-quadruplexes determines the outcome of R-loop-replisome collisions. Elife 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Wang H, Jehi S, Li J, Liu S, Wang Z, Truong L, Chiba T, Wang Z, and Wu X (2021). PIF1 helicase promotes break-induced replication in mammalian cells. EMBO J 40, e104509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Sampathi S, Dai H, Liu C, Zhou M, Hu J, Huang Q, Campbell J, Shin-Ya K, Zheng L, et al. (2013). Mammalian DNA2 helicase/nuclease cleaves G-quadruplex DNA and is required for telomere integrity. EMBO J 32, 1425–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy CA, Li W, Reisenweber S, Thongthip S, Bruno J, de Lange T, De S, Petrini JH, Sung PA, Jasin M, et al. (2012). Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet 8, e1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, O’Rourke JJ, Sobinoff AP, Allen JAM, Nelson CB, Tomlinson CG, Lee M, Reddel RR, Deans AJ, and Pickett HA (2019). The FANCM-BLM-TOP3A-RMI complex suppresses alternative lengthening of telomeres (ALT). Nat Commun 10, 2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marnef A, and Legube G (2021). R-loops as Janus-faced modulators of DNA repair. Nat Cell Biol 23, 305–313. [DOI] [PubMed] [Google Scholar]
- Matos DA, Zhang JM, Ouyang J, Nguyen HD, Genois MM, and Zou L (2020). ATR Protects the Genome against R Loops through a MUS81-Triggered Feedback Loop. Mol Cell 77, 514–527 e514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazina OM, Keskin H, Hanamshet K, Storici F, and Mazin AV (2017). Rad52 Inverse Strand Exchange Drives RNA-Templated DNA Double-Strand Break Repair. Mol Cell 67, 19–29 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDevitt S, Rusanov T, Kent T, Chandramouly G, and Pomerantz RT (2018). How RNA transcripts coordinate DNA recombination and repair. Nat Commun 9, 1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelini F, Pitchiaya S, Vitelli V, Sharma S, Gioia U, Pessina F, Cabrini M, Wang Y, Capozzo I, Iannelli F, et al. (2017). Damage-induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks. Nat Cell Biol 19, 1400–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Wright WE, and Shay JW (2017). Alternative Lengthening of Telomeres Mediated by Mitotic DNA Synthesis Engages Break-Induced Replication Processes. Mol Cell Biol 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Wright WE, and Shay JW (2019). Clustered telomeres in phase-separated nuclear condensates engage mitotic DNA synthesis through BLM and RAD52. Genes Dev 33, 814–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama K, Lai MS, and Masai H (2017). Interaction of Rif1 Protein with G-Quadruplex in Control of Chromosome Transactions. Adv Exp Med Biol 1042, 287–310. [DOI] [PubMed] [Google Scholar]
- Nguyen HD, Yadav T, Giri S, Saez B, Graubert TA, and Zou L (2017). Functions of Replication Protein A as a Sensor of R Loops and a Regulator of RNaseH1. Mol Cell 65, 832–847 e834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang J, Yadav T, Zhang JM, Yang H, Rheinbay E, Guo H, Haber DA, Lan L, and Zou L (2021). RNA transcripts stimulate homologous recombination by forming DR-loops. Nature 594, 283–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozer O, Bhowmick R, Liu Y, and Hickson ID (2018). Human cancer cells utilize mitotic DNA synthesis to resist replication stress at telomeres regardless of their telomere maintenance mechanism. Oncotarget 9, 15836–15846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Chen Y, Biju B, Ahmed N, Kong J, Goldenberg M, Huang J, Mohan N, Klosek S, Parsa K, et al. (2019). FANCM suppresses DNA replication stress at ALT telomeres by disrupting TERRA R-loops. Sci Rep 9, 19110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeyre C, Lopes J, Boule JB, Piazza A, Guedin A, Zakian VA, Mergny JL, and Nicolas A (2009). The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet 5, e1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roumelioti FM, Sotiriou SK, Katsini V, Chiourea M, Halazonetis TD, and Gagos S (2016). Alternative lengthening of human telomeres is a conservative DNA replication process with features of break-induced replication. EMBO Rep 17, 1731–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini N, Ramakrishnan S, Elango R, Ayyar S, Zhang Y, Deem A, Ira G, Haber JE, Lobachev KS, and Malkova A (2013). Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature 502, 389–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay JW, Reddel RR, and Wright WE (2012). Cancer. Cancer and telomeres--an ALTernative to telomerase. Science 336, 1388–1390. [DOI] [PubMed] [Google Scholar]
- Silva B, Arora R, Bione S, and Azzalin CM (2021). TERRA transcription destabilizes telomere integrity to initiate break-induced replication in human ALT cells. Nat Commun 12, 3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva B, Pentz R, Figueira AM, Arora R, Lee YW, Hodson C, Wischnewski H, Deans AJ, and Azzalin CM (2019). FANCM limits ALT activity by restricting telomeric replication stress induced by deregulated BLM and R-loops. Nat Commun 10, 2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman J, Takai H, Buonomo SB, Eisenhaber F, and de Lange T (2004). Human Rif1, ortholog of a yeast telomeric protein, is regulated by ATM and 53BP1 and functions in the S-phase checkpoint. Genes Dev 18, 2108–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoneau A, Xiong R, and Zou L (2021). The trans cell cycle effects of PARP inhibitors underlie their selectivity toward BRCA1/2-deficient cells. Genes Dev 35, 1271–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobinoff AP, Allen JA, Neumann AA, Yang SF, Walsh ME, Henson JD, Reddel RR, and Pickett HA (2017). BLM and SLX4 play opposing roles in recombination-dependent replication at human telomeres. EMBO J 36, 2907–2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobinoff AP, and Pickett HA (2020). Mechanisms that drive telomere maintenance and recombination in human cancers. Curr Opin Genet Dev 60, 25–30. [DOI] [PubMed] [Google Scholar]
- Sollier J, Stork CT, Garcia-Rubio ML, Paulsen RD, Aguilera A, and Cimprich KA (2014). Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol Cell 56, 777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiriou SK, Kamileri I, Lugli N, Evangelou K, Da-Re C, Huber F, Padayachy L, Tardy S, Nicati NL, Barriot S, et al. (2016). Mammalian RAD52 Functions in Break-Induced Replication Repair of Collapsed DNA Replication Forks. Mol Cell 64, 1127–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storici F, Bebenek K, Kunkel TA, Gordenin DA, and Resnick MA (2007). RNA-templated DNA repair. Nature 447, 338–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Duan M, Yadav T, Phoon L, Wang X, Zhang JM, Zou L, and Lan L (2020a). An R-loop-initiated CSB-RAD52-POLD3 pathway suppresses ROS-induced telomeric DNA breaks. Nucleic Acids Res 48, 1285–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Wang X, Phoon L, Yang H, and Lan L (2020b). Resolution of ROS-induced G-quadruplexes and R-loops at transcriptionally active sites is dependent on BLM helicase. FEBS Lett 594, 1359–1367. [DOI] [PubMed] [Google Scholar]
- Teng Y, Yadav T, Duan M, Tan J, Xiang Y, Gao B, Xu J, Liang Z, Liu Y, Nakajima S, et al. (2018). ROS-induced R loops trigger a transcription-coupled but BRCA1/2-independent homologous recombination pathway through CSB. Nat Commun 9, 4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma P, Dilley RL, Zhang T, Gyparaki MT, Li Y, and Greenberg RA (2019). RAD52 and SLX4 act nonepistatically to ensure telomere stability during alternative telomere lengthening. Genes Dev 33, 221–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vohhodina J, Goehring LJ, Liu B, Kong Q, Botchkarev VV Jr., Huynh M, Liu Z, Abderazzaq FO, Clark AP, Ficarro SB, et al. (2021). BRCA1 binds TERRA RNA and suppresses R-Loop-based telomeric DNA damage. Nat Commun 12, 3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MA, Kwon Y, Xu Y, Chung WH, Chi P, Niu H, Mayle R, Chen X, Malkova A, Sung P, et al. (2013). Pif1 helicase and Poldelta promote recombination-coupled DNA synthesis via bubble migration. Nature 502, 393–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, and Malkova A (2021). Break-induced replication mechanisms in yeast and mammals. Curr Opin Genet Dev 71, 163–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Shin-ya K, and Brosh RM Jr. (2008). FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol Cell Biol 28, 4116–4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SY, Chang EYC, Lim J, Kwan HH, Monchaud D, Yip S, Stirling PC, and Wong JMY (2021). G-quadruplexes mark alternative lengthening of telomeres. NAR Cancer 3, zcab031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuhara T, Kato R, Hagiwara Y, Shiotani B, Yamauchi M, Nakada S, Shibata A, and Miyagawa K (2018). Human Rad52 Promotes XPG-Mediated R-loop Processing to Initiate Transcription-Associated Homologous Recombination Repair. Cell 175, 558–570 e511. [DOI] [PubMed] [Google Scholar]
- Yu TY, Kao YW, and Lin JJ (2014). Telomeric transcripts stimulate telomere recombination to suppress senescence in cells lacking telomerase. Proc Natl Acad Sci U S A 111, 3377–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JM, Genois MM, Ouyang J, Lan L, and Zou L (2021). Alternative lengthening of telomeres is a self-perpetuating process in ALT-associated PML bodies. Mol Cell 81, 1027–1042 e1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JM, Yadav T, Ouyang J, Lan L, and Zou L (2019). Alternative Lengthening of Telomeres through Two Distinct Break-Induced Replication Pathways. Cell Rep 26, 955–968 e953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JM, and Zou L (2020). Alternative lengthening of telomeres: from molecular mechanisms to therapeutic outlooks. Cell Biosci 10, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw western blot and Immunofluorescence data have been deposited at Mendeley and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PML | Abcam | Cat# ab96051; RRID: AB_10679887 |
| TRF2 | Novus Biology | Cat# NB110-57130; RRID: AB_844199 |
| G-quadruplex (BG4) | Millipore Sigma | Cat# MABE917; RRID: AB_2750936 |
| G-quadruplex (1H6) | Millipore Sigma | Cat# MABE1126 |
| S9.6 | Millipore Sigma | Cat# MABE1095; RRID: AB_2861387 |
| RAD52 | Abcam | Cat# ab124971; RRID: AB_10971685 |
| Biotin | Abcam | Cat# ab53494; RRID: AB_867860 |
| GAPDH | Millipore | Cat# ABS16; RRID: AB_10806772 |
| Ku80 | NeoMarker | Cat# MS-285 P; RRID: AB_62299 |
| RAD51AP1 | Proteintech | Cat#11255-1-AP; RRID: AB_2300786 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| T4 Polynucleotide Kinase | NEB | M0201S |
| Proteinase K | Thermo Fischer | 25530049 |
| RNase H | NEB | M0297 |
| RNase A | Millipore Sigma | R6513 |
| CDK1i (RO-3306) | Selleckchem | S7747 |
| BSA | NEB | B9000S |
| Click-iT EdU Alexa Fluor 488 Imaging Kit | Thermo Fischer | C10337 |
| Formamide | Sigma | 47671-1L-F |
| 20x SSC | Invitrogen | AM9763 |
| Azide-PEG3-Biotin | Sigma | 762024-10MG |
| phi29 DNA Polymerase | NEB | M0269L |
| dNTP Set (100 mM) | Thermo Fischer | 10297018 |
| PureLink Genomic DNA Mini Kit | Thermo Fischer | K182002 |
| ULTRAhyb hybridization buffer | Invitrogen | AM8669 |
| LNA Control and LNA TERRA | Qiagen | Custom made |
| DRB | Millipore Sigma | D1916 |
| Deposited data | ||
| Raw western blot and immunofluorescence data | This study | https://data.mendeley.com/datasets/vhz6phhb6c/draft?a=1e79524c-52e6-4716-a154-4a7d3885ecc7 |
| Oligonucleotides | ||
| Oligos used in this study | This study | Table S1 |
| Software and Algorithms | ||
| Fiji | ImageJ | https://imagej.net/software/fiji/ |
| NIS element viewer | Nikon | https://www.nikoninstruments.com/Products/Software/NIS-Elements-Advanced-Research/NIS-Elements-Viewer |
| Image Lab | BioRad | http://www.bio-rad.com/en-us/product/image-lab-software |
| Image studio | Li-COR | https://www.licor.com/bio/image-studio/ |


