Abstract
Pseudomonas aeruginosa R-type pyocin particles have been described as bacteriocins that resemble bacteriophage tail-like structures. Because of their unusual structure, we reexamined whether they contained nucleic acids. Our data indicated that pyocin particles isolated from P. aeruginosa C (pyocin C) contain DNA. Probes generated from this DNA by the random-primer extension method hybridized to distinct bands in restriction endonuclease-digested P. aeruginosa C genomic DNA. These probes also hybridized to genomic DNA from 6 of 18 P. aeruginosa strains that produced R-type pyocins. Asymmetric PCR, complementary oligonucleotide hybridization, and electron microscopy indicated that pyocin C particles contained closed circular single-stranded DNA, approximately 4.0 kb in length. Examination of total intracellular DNA from mitomycin C-induced cultures revealed the presence of two extrachromosomal DNA molecules, a double-stranded molecule and a single-stranded molecule, which hybridized to pyocin DNA. Sequence analysis of 7,480 nucleotides of P. aeruginosa C chromosomal DNA containing the pyocin DNA indicated the presence of pyocin open reading frames with similarities to open reading frames from filamentous phages and cryptic phage elements. We did not observe any similarities to known phage structural proteins or previously characterized pseudomonal prt genes expressing R-type pyocin structural proteins. These studies demonstrate that pyocin particles from P. aeruginosa C are defective phages that contain a novel closed circular single-stranded DNA and that this DNA was derived from the chromosome of P. aeruginosa C.
Pseudomonas aeruginosa strains produce three distinct families of bacteriocins, designated S, F, and R pyocins (19, 21). They differ by their morphology and mode of killing. Their bactericidal activities are strain specific and have been used as a typing tool for P. aeruginosa strains, along with other typing schemes such as serotyping and phage typing.
The S-type pyocins are like colicins in their structure and mode of action; they have an effector and an immunity component, with the effector component possessing DNase and lipase activity. Four subtypes of S-type pyocin have been identified: S1, S2, S3, and AP41 (10, 39). The genes for S1 and S2 pyocins map near the flaY gene (38), and the genes for AP41 map between the lys-9015 and argF genes (37). The S-type pyocins are proteinase sensitive and cannot be sedimented or observed by electron microscopy, reflecting their small size. The F-type pyocins are curved rods with distal filaments. They vary in their host ranges but are structurally, morphologically, and antigenically similar (23, 24).
R-type pyocins resemble bacteriophage tails of T-even phages, being composed of a contractile sheath, a core, and tail fibers. Five subtypes of R-type pyocins have been identified (R1 to R5), and they differ in host range but are immunologically similar (19). The receptors for R-type pyocins are the lipopolysaccharides or lipooligosaccharides found in the outer membrane of gram-negative bacteria (11). The apparent mode of killing is by pore formation in the membrane and disruption of the membrane potential (44). The genes for R-type pyocin production have been mapped to a 13-kbp fragment located between the trpCD and trpE genes at approximately 35 min of the P. aeruginosa chromosome. This region encodes 15 proteins, PrtA to PrtO, including a positive regulator protein, PrtN. A 16th protein, PrtP, is located between the strA and rifA genes. There is also a negative regulatory protein, PrtR, that is a target for the RecA protein (26), and the PrtN and PrtR proteins control the expression of R-type pyocins. R-type pyocin particles are immunochemically and genetically similar to the tails of temperate P. aeruginosa bacteriophages (19, 21, 40, 41). It has been suggested that the R-type pyocins and Pseudomonas bacteriophages such as PS-17 are the descendants of a common ancestral bacteriophage in which the genes for the head proteins and replicative functions have been lost or were never incorporated for pyocin (40).
Our interest in pyocins relates to their interaction with Neisseria gonorrhoeae and N. meningitidis (4, 28). Gonococcal clones that survive pyocin lysis frequently have modifications of their lipooligosaccharides (9, 28). Physicochemical studies have shown that variants with sequential deletions in lipooligosaccharide sugars can be selected (9, 18). There is similarity between these observations and the antigenic conversions of lipopolysaccharide brought about by temperate phages interacting with Salmonella to modify certain biosynthetic mechanisms previously under the control of the bacterial genome (32). We have become interested in whether similar mechanisms were operative in pyocin interaction with the gonococcus. Past studies based on absorption spectral analysis had concluded that pyocin particles did not contain nucleic acids (20). Other investigators have suggested that R-type pyocin particles contain genetic material (6). This has not been studied by molecular biologic techniques. Therefore, we conducted the present investigation to reexamine whether pyocin particles contain genetic material and to characterize the material. Our results indicate that the pyocin particles contain single-stranded DNA (ssDNA) and that this DNA is complementary to a region in the genome of P. aeruginosa C.
MATERIALS AND METHODS
Bacterial strains, plasmids, and oligonucleotides.
All the bacterial strains, plasmids and oligonucleotides used in this study are listed in Table 1. The P. aeruginosa R-type pyocin typing strains of Farmer and Herman (13) were obtained as a gift from Jerold Sadoff, Walter Reed Army Institute for Research. P. aeruginosa C was the source of pyocin (pyocin C) and chromosomal DNA. Cultures for pyocin isolation were grown in tryptic soy broth (Difco Laboratories, Detroit, Mich.) (30). N. gonorrhoeae was grown at 35°C in a 5% CO2 atmosphere on GC medium base (Difco Laboratories) supplemented with 1% (vol/vol) IsoVitaleX (BBL Microbiology Systems, Cockeysville, Md.). Escherichia coli NM522 and DH5α were used for recombinant constructions and were grown in Luria-Bertani medium. All strains were stored at −70°C in Mueller-Hinton broth (Difco Laboratories) containing 10% glycerol. B. W. Holloway of Monash University, Victoria, Australia, kindly provided us with the cosmid library (33).
TABLE 1.
Bacterial strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotides | Genotype, markers, sequence | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | F-, thi-1 f80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rk− mk+) supE44 gyrA96 relA | Bethesda Research Laboratories—Life Technologies |
| E. coli NM522 | F′, lacIq Δ(lacZ)M15 proA+ B+ supE thi Δ(lac-proAB) Δ(hsdMS-mcrB)5 (rk− mk+) mcrBC | New England Biolabs |
| N. gonorrhoeae 1291 | Our collection | |
| P. aeruginosa C | 14 | |
| P. aeruginosa PAO-1 | 35 | |
| Plasmids | ||
| pGEM3zf+ | Ampicillin resistant | Promega Corp. |
| pPSC-1 | Ampicillin resistant | This study |
| pPSC-2 | Ampicillin resistant | This study |
| pPSC-3 | Ampicillin resistant | This study |
| pPSC-4 | Ampicillin resistant | This study |
| Oligonucleotides | ||
| T7 primer | Promega Corp. | |
| SP6 primer | Promega Corp. | |
| PSC-3 OLIGO 1 | 5′-ATG GCT CTT GAT AGT TCG GG-3′ | |
| PSC-3 OLIGO 2 | 5′-CCC GAA CTA TCA AGA GCC AT-3′ | |
| A12 SP6-EXT | 5′-AGT TTC AGC GCG TTG AGT-3′ | |
| A12 SP6-OLIGO 1 | 5′-CGG GAT GAT CTT TAT CGG AA-3′ |
DNA manipulation.
P. aeruginosa chromosomal DNA was prepared by the method of Marmur (25). All plasmid constructs were made in pGEM3zf+ (Promega Corp., Madison, Wis.). T4 DNA ligase and restriction endonucleases were purchased from Bethesda Research Laboratories (Gaithersburg, Md.) and Promega Corp. and used as specified by the suppliers.
Cloning and screening.
The genomic library of P. aeruginosa C was made in pGEM3zf+ as previously described (1). Colonies were screened by colony hybridization with the probes generated by random priming of the DNA extracted from pyocin C.
Southern hybridization.
DNA was digested to completion with appropriate restriction enzymes, fractionated in 0.8% agarose gels, and transferred to either nitrocellulose or nylon membranes (35). Southern blots were hybridized with probes generated by the random-priming method. All blots were washed at 68°C for 1 h with a buffer containing 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.1% sodium dodecyl sulfate (SDS) and then twice at 68°C for 1 h each with a buffer containing 0.5× SSC and 0.1% SDS. Autoradiography was performed with Kodak XAR-5 film (Eastman Kodak Co., Rochester, N.Y.) and a Cronex intensifying screen (DuPont Co., Wilmington, Del.).
DNA sequencing and analysis.
DNA was sequenced by the dideoxynucleotide chain termination method (36) with [35S]ATP by using the Sequenase II kit (Amersham) or with the Applied Biosystem automated sequencer by using fluorescent terminator dye tags at the DNA Sequencing Facility, University of Iowa. Sequences of both strands were obtained. Ambiguities were resolved by resequencing both strands of the region from an independent clone. The sequence was analyzed with various programs of the Wisconsin GCG package (14). Similarity searches against DNA and protein sequence databases were performed with the FASTA (31), BLAST, or BLASTX (2, 3, 15) algorithms.
PCR and asymmetric PCR.
PCR was performed as previously described (34), except that annealing was performed at 62°C. Asymmetric PCR was performed with a 50-fold molar excess of either the T7 or SP6 promoter primers as previously described (27).
Pyocin isolation.
Pyocins were isolated from cultures of P. aeruginosa as previously described (29).
Cesium chloride density gradient centrifugation.
Pyocin C particles were sedimented in a cesium chloride (CsCl) gradient (42). The CsCl was dissolved in 10 mM Tris-HCl (pH 7.5) containing 10 mM MgCl2 and 10 mM MgSO4. The density of the starting solution was 1.3872 g/ml. The gradient was centrifuged in a VTi65 vertical rotor for 18 h at 50,000 rpm at 25°C and was fractionated in 100-μl fractions. Refractometry was performed on the odd-numbered aliquots with a refractometer (Bausch & Lomb, Inc., Rochester, N.Y.). The absorbance at 280 nm and 260 nm was measured in a DU650 spectrophotometer (Beckman Instruments) for the same fractions. Dot hybridization studies were performed with 5-μl aliquots from the odd fractions. Pyocin particles recovered from the gradient were tested for their ability to lyse N. gonorrhoeae 1291, as previously described (9), after 5-μl aliquots were diluted from the odd fractions in 90 μl of sterile 10 mM Tris-HCl (pH 7.5) containing 10 mM MgCl2 and 10 mM MgSO4.
Isolation of DNA from the R-type pyocin of P. aeruginosa C.
Purified pyocin C particles isolated from 6 liters of overnight broth cultures were treated with RNase (50 μg/ml) and DNase (100 μg/ml) to degrade any contaminating nucleic acids. The particles were heated at 100°C for 10 min, cooled on ice, and precipitated with polyethylene glycol 8000 (1:9 of a 40% PEG solution). The particles were then subjected to proteinase K (final concentration, 100 μg/ml) digestion for 1 h at 65°C in 10 mM EDTA–0.5% SDS. This extract was then subjected to phenol-chloroform extraction. The DNA was precipitated with 100% ethanol (1:2, vol/vol) at −20°C overnight. The precipitate was washed with 70% ethanol three times, dried, and stored at −20°C.
Time course study of pyocin DNA induction.
A 25-ml culture of P. aeruginosa C was induced with mitomycin C by a previously published method (29). After the induction, 1.5-ml aliquots were collected at the desired times and the total intracellular DNA was extracted from the bacterial pellet. The genomic DNAs were treated with RNase (100 μg/ml) for 60 minutes at 37°C, Southern blotted to a nylon membrane (35), and probed with a gel-purified 1.7-kb insert from pPSC-3 labeled by random-primer extension. Selected samples were treated with 50% formamide for 15 min at 65°C before being loaded.
Generation of biotinylated probes for in situ hybridizations.
The double-stranded probes were generated with the Bio-Prime labeling kit from Gibco-BRL Life Technologies as recommended by the manufacturer. The strand-specific probes were generated by the following method. The template for the probe (1,743-bp insert DNA from pPSC-3) was isolated by two rounds of gel purification after complete HincII digestion of pPSC-3. The concentration of the 1.7-kbp template DNA was quantified with a Beckman DU-650 spectrophotometer, and 1 μg of template DNA was mixed with a twofold molar excess of primer. The primers used were A12 SP6-EXT and A12 SP6-OLIGO 1 (Table 1). The template-primer mixture was denatured by a 10-min incubation in a 100°C water bath and quickly chilled in an ice-water bath. The mixture was incubated for 10 min at 65°C and allowed to cool to room temperature slowly. Afterwards, 2 μl of 10× Klenow buffer containing dATP, dGTP, TTP, biotinylated dCTP, and 1 μl of Klenow enzyme were added, and the mixture was incubated for 1 h at 37°C. The unincorporated biotinylated dCTP was removed through ethanol precipitation, and the efficiency of labeling was determined.
In situ hybridization.
The pyocin particles were concentrated by centrifugation in a Sorvall Super-Centrifuge at 27,500 rpm for 1.5 h. The particles were then treated with DNase I and deposited by whole-mount onto Formvar-coated, carbon-coated, neutrally glow discharged, 200-mesh nickel electron microscopy grids. After a brief fixation with 4% paraformaldehyde in phosphate-buffered saline, the particles on the grids were treated with 0.1 mg of pronase (Sigma) per ml for 1 min and with 0.05 N sodium hydroxide for 4 min. Hybridization was carried out overnight at 30°C in a humid chamber with strand-specific biotinylated probes in a standard hybridization solution (34). The grids were rinsed three times with 2× SSC, and then a streptavidin-gold conjugate (10-nm gold particle size; Streptavidin G10 AuroProbe EM, Amersham Life Sciences, Inc.) was diluted in 1% bovine serum albumin–2× SSC and used overnight at room temperature to localize the hybrids. The grids were rinsed three times in 1% bovine serum albumin–2× SSC and negatively stained with 2% phosphotungstic acid before being visualized with a Hitachi H-7000 transmission electron microscope at an accelerating voltage of 100 kV.
Electron microscopy.
The pyocin particles were visualized with a Hitachi H-7000 microscope. Electron microscopic analysis of the pyocin DNA was performed on polylysine-coated Formvar grids by previously described techniques (7, 45).
Digital image reproduction.
All autoradiographs were digitally scanned with a Apple ColorOne Scanner 600/27 scanner. The brightness and contrast were adjusted in Adobe PhotoShop (version 4.0) and printed on continuous-tone photographic paper from black-and-white negatives.
Nucleotide sequence accession number.
The sequence described in this paper is listed as GenBank accession no. L06240.
RESULTS
Isolation of R-type pyocin particles.
The pyocin particles induced from P. aeruginosa C were isolated from the culture supernatant by the method of Morse et al. (29). The enriched pyocin particles were further purified by centrifugation through a cesium chloride density gradient. The gradient was collected in 0.5-ml fractions, and alternate fractions were analyzed for bactericidal activity against N. gonorrhoeae 1291, and the absorbances at 260 and 280 nm were measured. The maxima for all three parameters plotted to the same fraction (data not shown). The density of the fraction containing the maximum bactericidal activity was measured with a refractometer and determined to be 1.31 g · ml−1. The association of pyocin DNA with the fractions containing bactericidal activity was later confirmed by slot blot hybridization with insert DNA from pPSC3 as the probe (data not shown).
Characterization of pyocin DNA.
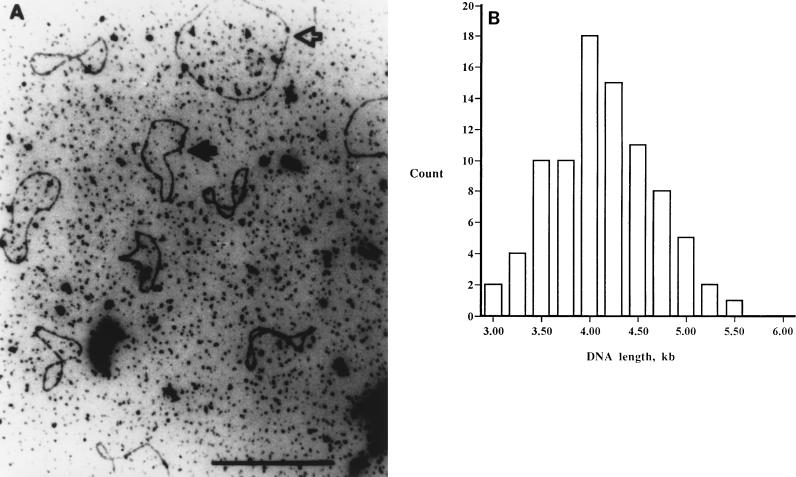
In our initial studies, we sought to determine whether pyocin particles contained nucleic acid. Agarose gel electrophoresis of the ethanol-insoluble material showed a broad band between 2.0 and 2.5 kbp which stained faintly with ethidium bromide (data not shown). We examined the isolated DNA by electron microscopy and observed closed circular molecules (Fig. 1). We determined the average length of the closed circles to be approximately 3.8 kb (n = 86). A probe generated from this nucleic acid by random primer extension with the Klenow enzyme was hybridized to P. aeruginosa C genomic DNA. The Southern analysis demonstrated hybridization to distinct bands in restriction endonuclease-digested genomic DNA (Fig. 2A).
FIG. 1.
(A) Electron micrograph demonstrating that the pyocin DNA is a single-stranded closed circle. The open arrow designates M13, and the solid arrow identifies the pyocin ssDNA. (B) Distribution of the sizes of 86 pyocin ssDNA circles. M13 ssDNA was included in the sample, and measurement of 30 M13 circles served as the standard for estimating the size of the pyocin ssDNA.
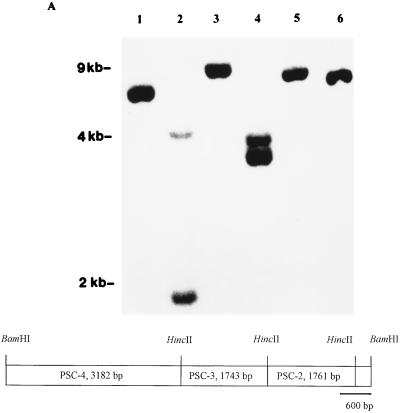
FIG. 2.
(A) Composite figure demonstrating Southern hybridization with a probe generated by random priming of DNA isolated from pyocin particles to EcoRV- (lane 1), HincII- (lane 2), PstI- (lane 3), SalI- (lane 4), XhoI- (lane 5) and BamHI (lane 6)-restricted genomic DNA from P. aeruginosa C. The numbers on the left refer to the relative migration of standards. (B) Restriction map of the 7.5-kb BamHI chromosomal DNA fragment that hybridized to probes generated from the DNA isolated from density gradient-purified pyocin particles. The subcloned HincII fragments are shown.
To define the characteristics of the pyocin DNA more precisely, we cloned into pGEM3zf+ a 7.5-kbp BamHI fragment of P. aeruginosa C chromosomal DNA that had hybridized with the probe generated from the pyocin DNA. This plasmid was called pPSC-1. This fragment was further restricted with HincII and subcloned into three plasmids, pPSC-2, pPSC-3, and pPSC-4 (Fig. 2B). Hybridization studies indicated that homology to purified pyocin DNA spanned the insert in pPSC-3 completely and overlapped the adjacent portions of the inserts from pPSC-2 and pPSC-4 (data not shown). Dot blot hybridization analysis of genomic DNA from other pyocin-producing P. aeruginosa strains was performed with the insert from pPSC-3 as the probe. This study revealed that genomic DNA from P. aeruginosa A, J, M, N, P, and R of Farmer and Herman (13) had regions homologous to the insert in pPSC-3. The probe did not hybridize to 12 other P. aeruginosa strains that produced different types of pyocins such as F or S. Additionally, this probe did not hybridize to P. aeruginosa PAO-1 genomic DNA, to DNA isolated from selected cosmids containing SpeI fragments used for mapping the genome of strain PAO-1 (33), or to genomic DNA from two P. aeruginosa indicator strains which do not produce R-type pyocin (data not shown).
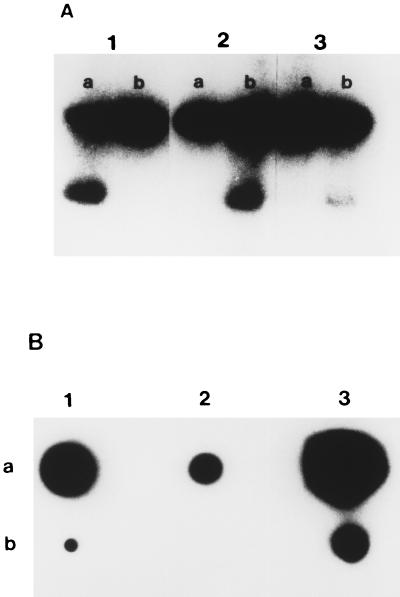
Analysis of the nucleic acid isolated from pyocin particles indicated that it could not be restricted with BamHI, RsaI, PstI, ClaI, PvuI, AvaI, ClaI, Sau3AI, SalI, EcoRI, SphI, HindIII, or HincII. It was degraded by DNase and S1 nuclease but not by mung bean nuclease or RNase. The probe generated from pyocin DNA by random primer extension was hybridized to products from asymmetric PCR of insert in pPSC-3 with T7 and SP6 primers (Fig. 3A). The probe hybridized to the single-strand product from asymmetric PCR with SP6 primer present in excess. Additional experiments with a set of complementary oligonucleotide probes (PSC-3 OLIGO 1 and PSC-3 OLIGO 2 [Table 1]) derived from the sequence of pPSC-3 indicated that pyocin DNA hybridized to only one of the two complementary probes (Fig. 3B).
FIG. 3.
(A) Southern hybridization studies of the single- and double-stranded products produced by asymmetric PCR of the insert from pPSC-3. Row a contains the products of asymmetric PCR with the T7 primer in 50-fold molar excess; row b contains the products of asymmetric PCR with the SP6 primer in 50-fold molar excess. The probe used in panel 1 was the T7 primer, the probe used in panel 2 was the SP6 primer, and the probe used in panel 3 was generated from pyocin ssDNA by random primer extension. (B) Composite showing the results of dot hybridization of a set of complementary probes which were derived from the sequence of a region of pPSC-3 (lane 1, PSC-3 OLIGO 1, lane 2, PSC-3 OLIGO 2) and a probe generated to pyocin DNA by random primer extension (lane 3) of the insert from pPSC-3 (row a) and pyocin DNA (row b). This figure demonstrates that only one of the two complementary primers hybridized to the pyocin DNA, providing further proof that this DNA is single stranded.
Time course of pyocin DNA induction.
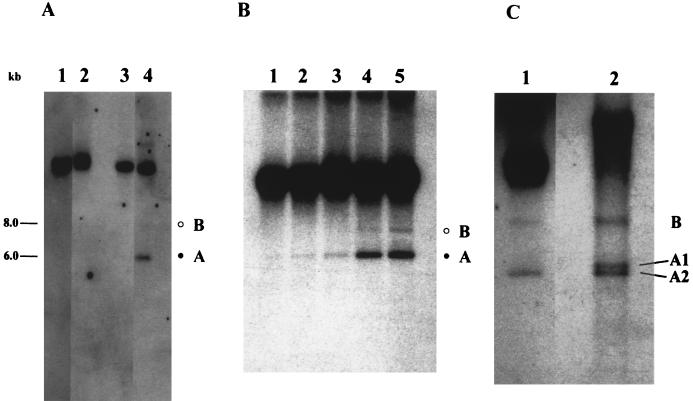
We extracted the total DNA from P. aeruginosa C at various times after induction with mitomycin C and analyzed the DNA by Southern hybridization with the 1.7-kbp fragment of insert DNA from pPSC-3 as the probe. We detected two extrachromosomal DNA elements following the addition of mitomycin C, which we designated A and B (Fig. 4A). These two forms were not observed in uninduced cultures. Both bands were RNase resistant (Fig. 4B), and they did not appear at the same time during pyocin induction; instead, A preceded B. To determine the nature of the extrachromosomal bands, an aliquot of DNA from the 180-min sample was treated with formamide and analyzed by Southern hybridization. The formamide treatment separated the DNA in form A to two distinct bands (A1 and A2), whereas form B remained as a single band (Fig. 4C). We observed that the band corresponding to the chromosomal DNA migrated slower after formamide treatment.
FIG. 4.
Southern analysis of pyocin DNA synthesis in P. aeruginosa C following mitomycin C addition. (A) Induction of extrachromosomal forms by mitomycin C. Two cultures of P. aeruginosa C were grown in parallel, and mitomycin C was added to one of them. Samples were taken at various times, and total intracellular DNA was analyzed by Southern blot hybridization. Lanes corresponding to 0 min (lanes 1 and 3) and 180 min (lanes 2 and 4) from uninduced (lanes 1 and 2) and induced (lanes 3 and 4) cultures are shown. The two extrachromosomal forms are denoted by the open circle (B) and the solid circle (A). The B band is visible in overexposed autoradiographs. (B) A logarithmic culture of P. aeruginosa C was induced with mitomycin C as described in Materials and Methods. Samples were removed at the designated times postinduction, and total intracellular DNA was extracted. The RNase-treated samples were electrophoresed in a 0.8% agarose gel and analyzed by Southern blot hybridization. The two extrachromosomal forms are denoted by the open circle (B) and the solid circle (A). Lanes: 1, 60 min; 2, 90 min; 3, 120 min; 4, 150 min; 5, 180 min. (C) Formamide treatment of DNA from a mitomycin C-induced culture at 180 min. The undigested DNA was treated with formamide as described in Materials and Methods. The B band was unchanged, whereas the A band separated into two distinct bands, A1 and A2. A gel-purified insert from pPSC-3 was treated in an identical manner, and separation of the double-stranded fragment into single strands was observed. Lanes 1, untreated DNA; 2, 50% formamide-treated DNA.
In situ hybridization experiment.
To investigate the relationship between the pyocin DNA and the pyocin particle, we examined pyocin particle preparations by in situ hybridizations with the complementary probe to pyocin DNA. We examined 15 consecutive fields from in situ hybridization grids probed with same-strand and complementary-strand oligonucleotide probes (A12 SP6-EXT and A12 SP6-OLIGO 1 [Table 1], respectively). In the grid probed with the same-strand probe, we observed gold beads in only 2 (13%) of 15 fields examined. In the fields where beads were present, they were not associated with the pyocin particles (data not shown). In contrast, with the complementary-strand probe, we observed 13 (83%) of 15 fields with gold beads, and these gold beads were associated specifically with pyocin particles (Fig. 5). Similar observations were made when a grid was incubated with probes generated through random hexanucleotide priming. We did not observe binding of the complementary probe in samples that were not treated with pronase and NaOH. Exhaustive electron microscopic analysis failed to reveal the presence of any contaminating filamentous bacteriophages.
FIG. 5.
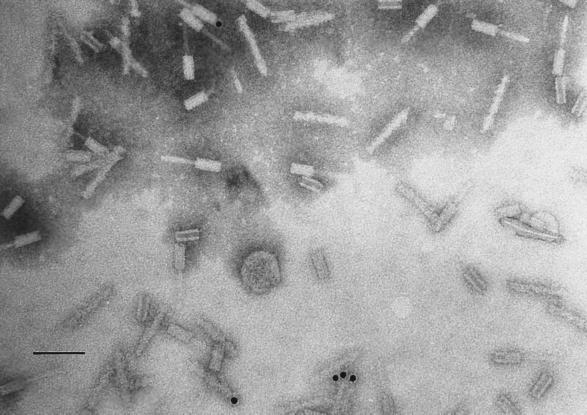
In situ hybridization of whole-mount pyocin particles with a complementary probe to pyocin DNA. The in situ hybridizations were performed as outlined in Materials and Methods. The gold beads were visible in association with the particles only when probed with the antisense probe. The gold beads were found in the core and on the sheath. In some instances, they appeared at one end of the pyocin particle. This indicates tight, specific interaction between the DNA and the particle. Bar, 100 nm.
Sequence analysis.
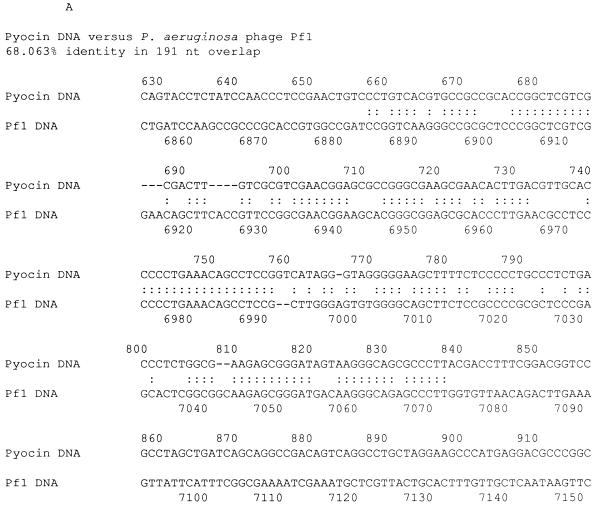
We sequenced the entire pseudomonal DNA in pPSC-1 and determined the length of the insert to be 7,480 nucleotides. We also sequenced a segment of the pyocin DNA isolated from gradient-purified R-type pyocin particles and determined that the sequence was identical to the sequence from pPSC-3 insert DNA (data not shown). We have searched both the DNA and protein sequence databases with the FASTA and BLASTX algorithms. The closest similarity from the FASTA search was a 68% identity over a 191-nucleotide overlap with the genome of Pseudomonas bacteriophage Pf1 (expected P value, 1.4 × 10−19) (17) (Fig. 6A). The overlap region spanned bases 658 to 848 of pyocin DNA and bases 6884 to 7072 of the Pf1 genome. This region of the Pf1 genome is part of an unidentified open reading frame, ORF110, located at the end of the phage genome. The best similarity from the BLASTX search was a 46% identity, and 58% similarity over the first 240 amino acids to a hypothetical protein of 384 amino acids from Vibrio cholerae O1 fs1 phage (expected P value of 1 × 10−51) (12) (Fig. 6B). We did not find any open reading frames with similarity to known phage structural proteins. A putative open reading frame from pyocin DNA (ORF109) showed 28% identity and 39% similarity to ORF7 from the skin element of Bacillus subtilis 168 (43) and 28% identity and 39% similarity to an immunity repressor protein from a B. subtilis temperate phage, phi-105 (8) (Fig. 6B).
FIG. 6.
(A) DNA alignment between pyocin DNA and the genome of P. aeruginosa phage Pf1. The region of the Pf1 genome shown is part of an unknown open reading frame, ORF110, located at the end of the phage genome (17). The alignment was generated by the FASTA algorithm (31). Sequence identities are denoted by colons, and gaps are denoted by dashes. (B) Amino acid sequence alignments. Alignments were performed between pyocin DNA open reading frames and hypothetical open reading frames of phage origin with the GAP algorithm (31). Identical amino acids are indicated by vertical lines, and similar amino acids are indicated by colons and dots.
We have examined the 7,480 bp of pyocin DNA for direct repeats of at least 10 nucleotides with the program REPEAT in the GCG package (14). The program identified 52 direct repeats ranging from 10 to 14 nucleotides. Among these, 12 repeats flanked a length of DNA ranging from 4,101 to 7,366 bp. Of these 12 repeats, 4 flanked approximately 4 kb of chromosomal DNA. A 14-mer direct repeat (CTCCAGGCCCTGGA, 3127 to 3140 and 7241 to 7254) flanked 4,101 bp of chromosomal pyocin DNA (data not shown). The G+C content of the entire 7,480 bp is 55.97%, well below that of the P. aeruginosa genome at 67%.
DISCUSSION
Our studies have demonstrated that R-type pyocin particles induced from P. aeruginosa C contain closed circular ssDNA of approximately 4 kb. The source of this DNA was determined to be a 7.5-kb BamHI fragment of P. aeruginosa genomic DNA. We detected two species of extrachromosomal DNA that hybridized to pyocin DNA sequences. One species appeared to be double stranded, and the other appeared to be single stranded. The entire 7.5-kb fragment was sequenced, and database searches indicated similarities between hypothetical open reading frames from pyocin DNA and two filamentous phages.
The size distribution of the pyocin DNA molecule, as determined by measurements from the electron micrographs, was broad (Fig. 1). Similar measurements of M13 DNA used as a size standard had a narrow size distribution. The broad size distribution was probably due to incomplete denaturation of the DNA prior to microscopy. The isolated pyocin DNA migrated as a broad band between 2 and 2.5 kb in agarose gel electrophoresis, suggesting a high degree of secondary-structure formation. The extensive folding of the pyocin DNA is also suggested by the in situ hybridization experiments where NaOH treatment was required for binding of the oligonucleotide probe.
The Southern blot hybridization experiment (Fig. 2) indicated that a single copy of pyocin DNA was present in the chromosome of P. aeruginosa C. Additionally, direct sequencing of pyocin DNA generated a banding pattern typical of a single template population (data not shown). We interpreted these results to suggest that the pyocin DNA isolated from gradient-purified pyocin particles probably contained a single family of DNA molecules and that the association between the pyocin DNA and the particle was due to a specific interaction. Multiple families of pyocin DNA molecules would have generated multiple bands in Southern blot hybridizations and multiple sequence profiles from direct sequencing of pyocin DNA due to heterogeneous template DNA population. We did not observe results that indicated a heterogenous pyocin DNA population. Further supporting the homogeneity of the pyocin DNA population were the results from dot blot hybridization analysis of P. aeruginosa strains with isolated pyocin DNA as the probe template. These experiments suggested that the hybridization to the pyocin DNA was restricted to R-type pyocin-producing strains of P. aeruginosa (data not shown).
ssDNA phages replicate their DNA through a double-stranded replicative intermediate, typically via a rolling-circle mechanism (4). Southern blot hybridization of intracellular DNA indicated the presence of two extrachromosomal DNA species (Fig. 4A and B). The effect of formamide on these two bands suggested that band A could have been double stranded and band B could have been single stranded (Fig. 4C). The appearance of band A preceded that of band B, suggesting that the latter band may have been derived from the former, similar to ssDNA phage replication. These observations suggest that following mitomycin C induction, the pyocin DNA may have been replicated through a double-stranded replicative intermediate, perhaps by a rolling-circle mechanism. There is a family of interrelated plasmids in gram-positive bacteria that replicate through a single-stranded intermediate by a rolling-circle mechanism (16). These plasmids were also detected in the gram-negative bacteria Helicobacter pylori and Shigella sonnei (22, 47). Our pyocin DNA is unlikely to be a pseudomonal counterpart to these unique plasmids, because our data suggest that the double-stranded form could be the intermediate and the single-stranded form could be the product.
The apparent length of the DNA in band B was estimated to be 8 kb, relative to the migration profile of the 1-kb marker DNA (Gibco-BRL). We have performed the induction experiment four times. On three occasions, we observed the migration pattern presented here, and in the fourth experiment band B migrated at approximately 4 kb while band A remained unchanged. It could be that the 8-kb DNA molecule represents a dimer of the 4-kb pyocin DNA molecule observed in electron micrographs and in the one induction experiment. We do not have a logical explanation for the observations, except that the results could be related to different physiological conditions that favored the production of dimers instead of monomers. Different growth conditions affect the degree of plasmid supercoiling and the extent of plasmid multimer formation.
The in situ hybridization experiments (Fig. 5) suggested a physical association between pyocin DNA and the R-type pyocin particle. The concept that R-type pyocin contains DNA was suggested 25 years ago by Bradley and Dewar (6). They observed both filled and empty R-type pyocin particles in their preparations. The majority of the gold beads we observed were associated with the pyocin particle attached to either the core or the sheath, probably depending on the state of pyocin DNA denaturation. The small number of pyocin particle-associated gold beads probably reflects the high degree of secondary structure that can exist in pyocin DNA. Agarose gel electrophoresis of isolated pyocin DNA suggests that the pyocin DNA is probably highly condensed, either wrapped around a protein core or folded onto itself or both. Alternatively, the low frequency of pyocin DNA association with the particles may reflect the inefficiency of the packaging system. Given that R-type pyocin particles are considered defective phages, the packaging system it has may not be efficient. In situ hybridizations with sense probe (negative control) suggest that our observations with the antisense probe were specific to pyocin DNA.
Exhaustive searches of the databases did indicate a high degree of similarity between pyocin DNA open reading frames and those from both gram-negative and gram-positive phages or cryptic phage elements (Fig. 6B). Filamentous phage fs1 is a recently identified phage from V. cholerae O1. Although it is possible that the DNA isolated from R-type pyocin preparations came from a previously unidentified copurifying filamentous phage, we believe that this is unlikely for the following reasons. First, we did not observe any filamentous phages in our electron micrographs of R-type pyocin particles either before or after CsCl density gradient centrifugation. Second, exhaustive database searches failed to detect any similarities between the 7.5-kb pyocin region of pseudomonal DNA and known structural proteins of pseudomonal filamentous phages (such as Pf1) or other phages. Lastly, Wiseman et al. estimate an equilibrium buoyant density of 1.280 g · ml−1 at 20°C for the filamentous phages Pf1 (46). The pyocin particles sedimented at a higher density (1.31 g · ml−1) and would have been separated from these phages on our gradients. Based on these observations, we believe that the ssDNA we have identified is derived from pyocin C particles and not from a filamentous bacteriophage.
ORF109 of pyocin DNA demonstrated high degree of similarity to the transcriptional repressor protein from B. subtilis phage phi-105 (8) and a related cryptic phage element (43). The immunity repressor protein of phi-105 belongs to a family of DNA-binding transcription regulator proteins from B. subtilis temperate phages and is thought to be involved in immunity and regulation of prophage induction (48). Members of this family have an N-terminal helix-turn-helix motif (48). Pyocin ORF109 also has a helix-turn-helix motif at its N terminus, suggesting that ORF109 polypeptide may be a DNA-binding transcription regulator.
We have demonstrated that R-type pyocin particles induced from P. aeruginosa C contained closed circular, ssDNA molecules of chromosomal origin. Sequence analysis of the chromosomal region containing the 4-kb pyocin DNA revealed a number of open reading frames with significant similarity to open reading frames from filamentous phages. In particular, ORF109 had significant similarity to transcriptional repressors from gram-positive phages. It would be interesting to delete either this ORF or prtR and determine the effects on pyocin induction. The association of an ssDNA molecule with the R-type pyocin particle raises interesting questions about its potential role in the antigenic conversion of N. gonorrhoeae lipooligosaccharide molecules.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant AI 18384 from the National Institute of Allergy and Infectious Diseases.
E. Six and M. Feiss provided us with helpful discussions and comments.
REFERENCES
- 1.Abu Kwaik Y, McLaughlin R E, Apicella M A, Spinola S M. Analysis of Haemophilus influenzae type b lipooligosaccharide-synthesis genes that assemble or expose a 2-keto-3-deoxyoctulosonic acid epitope. Mol Microbiol. 1991;5:2475–2480. doi: 10.1111/j.1365-2958.1991.tb02092.x. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S F, Madden T L, Schaumlffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker T A, Wickner S H. Genetics and enzymology of DNA replication in Escherichia coli. Annu Rev Genet. 1992;26:447–477. doi: 10.1146/annurev.ge.26.120192.002311. [DOI] [PubMed] [Google Scholar]
- 5.Blackwell C C, Law J A. Typing of non-serogroupable Neisseria meningitidis by means of sensitivity to R-type pyocins of Pseudomonas aeruginosa. J Infect. 1981;3:370–378. doi: 10.1016/s0163-4453(81)91996-4. [DOI] [PubMed] [Google Scholar]
- 6.Bradley D E, Dewar C A. The structure of phage-like objects associated with non-induced bacteriocinogenic bacteria. J Gen Microbiol. 1966;45:399–408. [Google Scholar]
- 7.Coggins L W, editor. Preparation of nucleic acids for electron microscopy. Washington, D.C: IRL Press; 1987. [Google Scholar]
- 8.Dhaese P, Seurinck J, De Smet B, Van Montagu M. Nucleotide sequence and mutational analysis of an immunity repressor gene from Bacillus subtilis temperate phage F105. Nucleic Acids Res. 1985;13:5441–5455. doi: 10.1093/nar/13.15.5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dudas K C, Apicella M A. Selection and immunochemical analysis of lipooligosaccharide mutants of Neisseria gonorrhoeae. Infect Immun. 1988;56:499–504. doi: 10.1128/iai.56.2.499-504.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duport C, Baysse C, Michel-Briand Y. Molecular characterization of pyocin S3, a novel S-type pyocin from Pseudomonas aeruginosa. J Biol Chem. 1995;270:8920–8927. doi: 10.1074/jbc.270.15.8920. [DOI] [PubMed] [Google Scholar]
- 11.Dyke J, Berk R S. Growth inhibition and pyocin receptor properties of endotoxin from Pseudomonas aeruginosa. Proc Soc Exp Biol Med. 1974;145:1405–1408. doi: 10.3181/00379727-145-38023. [DOI] [PubMed] [Google Scholar]
- 12.Ehara M, Shimodori S, Kojima F, Ichinose Y, Hirayama T, Albert M J, Supawat K, Honma Y, Iwanaga M, Amako K. Characterization of filamentous phages of Vibrio cholerae O139 and O1. FEMS Microbiol Lett. 1997;154:293–301. doi: 10.1111/j.1574-6968.1997.tb12659.x. [DOI] [PubMed] [Google Scholar]
- 13.Farmer J J D, Herman L G. Epidemiological fingerprinting of Pseudomonas aeruginosa by the production of and sensitivity of pyocin and bacteriophage. Appl Microbiol. 1969;18:760–765. doi: 10.1128/am.18.5.760-765.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Genetics Computer Group. Program manual for the Wisconsin package. 8th ed. Madison, Wis: Genetics Computer Group; 1994. [Google Scholar]
- 15.Gish W, States D J. Identification of protein coding regions by database similarity search. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 16.Gruss A, Dusko Ehrlich S. The family of highly interrelated single-stranded deoxyribonucleic acid plasmids. Microbiol Rev. 1989;53:231–241. doi: 10.1128/mr.53.2.231-241.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hill D F, Short N J, Perham R N, Petersen G B. DNA sequence of the filamentous bacteriophage Pf1. J Mol Biol. 1991;218:349–364. doi: 10.1016/0022-2836(91)90717-k. [DOI] [PubMed] [Google Scholar]
- 18.John C M, Griffiss J M, Apicella M A, Mandrell R E, Gibson B W. The structural basis for pyocin resistance in Neisseria gonorrhoeae lipooligosaccharides. J Biol Chem. 1991;266:19303–19311. [PubMed] [Google Scholar]
- 19.Kageyama M, editor. Bacteriocins and bacteriophages in Pseudomonas aeruginosa. Tokyo, Japan: University of Tokyo Press; 1975. [Google Scholar]
- 20.Kageyama M. Studies of pyocin. I. Physical and chemical properties. J Biochem. 1964;55:49–53. doi: 10.1093/oxfordjournals.jbchem.a127839. [DOI] [PubMed] [Google Scholar]
- 21.Kageyama M, Shimomiya T, Aihara Y, Kobayashi M. Characterization of a bacteriophage related to R-type pyocins. J Virol. 1979;32:951–957. doi: 10.1128/jvi.32.3.951-957.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kleanthous H, Clayton C L, Tabaqchali S. Characterization of a plasmid from Helicobacter pylori encoding a replication protein common to plasmids in gram-positive bacteria. Mol Microbiol. 1991;5:2377–2389. doi: 10.1111/j.1365-2958.1991.tb02084.x. [DOI] [PubMed] [Google Scholar]
- 23.Kuroda K, Kageyama M. Biochemical properties of a new flexuous bacteriocin, pyocin F1, produced by Pseudomonas aeruginosa. J Biochem. 1979;85:7–19. doi: 10.1093/oxfordjournals.jbchem.a132332. [DOI] [PubMed] [Google Scholar]
- 24.Kuroda K, Kageyama M, Maeda T, Fujime S. Physicochemical properties of pyocin F1. J Biochem. 1979;85:21–28. doi: 10.1093/oxfordjournals.jbchem.a132313. [DOI] [PubMed] [Google Scholar]
- 25.Marmur J. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 26.Matsui H, Sano Y, Ishihara H, Shinomiya T. Regulation of pyocin genes in Pseudomonas aeruginosa by positive (prtN) and negative (prtR) regulatory genes. J Bacteriol. 1993;175:1257–1263. doi: 10.1128/jb.175.5.1257-1263.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCabe P C. Production of single stranded DNA by asymmetric PCR. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols. New York, N.Y: Academic Press, Inc.; 1989. pp. 13–21. [Google Scholar]
- 28.Morse S A, Apicella M A. Isolation of a lipopolysaccharide mutant of Neisseria gonorrhoeae using an R-type bacteriocin from Pseudomonas aeruginosa: an analysis of the antigenic and biologic differences. J Infect Dis. 1982;145:206–218. doi: 10.1093/infdis/145.2.206. [DOI] [PubMed] [Google Scholar]
- 29.Morse S A, Vaughan P, Johnson D, Iglewski B H. Inhibition of Neisseria gonorrhoeae by a bacteriocin from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1976;10:354–362. doi: 10.1128/aac.10.2.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pau W S, Terry C S. The effect of medium composition upon the production of pyocin. J Appl Bacteriol. 1976;41:369–377. doi: 10.1111/j.1365-2672.1976.tb00648.x. [DOI] [PubMed] [Google Scholar]
- 31.Pearson W R, Lipman D J. Improved tools for biological sequence analysis. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rapin A M C, Kalckar H M. The relation of bacteriophage attachment to lipopolysaccharide structure. In: Weinbaum G, Kadis S, Ajl S J, editors. Microbial toxins. IV. New York, N.Y: Academic Press, Inc.; 1971. pp. 267–307. [Google Scholar]
- 33.Ratnaningsih E, Dharmsthiti V, Krishnapillai V, Morgan A, Sinclair M, Holloway B. A combined physical and genetic map of Pseudomonas aeruginosa PAO. J Gen Microbiol. 1990;136:2351–2357. doi: 10.1099/00221287-136-12-2351. [DOI] [PubMed] [Google Scholar]
- 34.Saiki R K. Amplification of genomic DNA. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols. New York, N.Y: Academic Press, Inc.; 1989. pp. 13–21. [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 36.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sano Y, Kageyama M. Genetic determinant of pyocin AP41 as an insert in the Pseudomonas aeruginosa chromosome. J Bacteriol. 1984;158:562–570. doi: 10.1128/jb.158.2.562-570.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sano Y, Matsui H, Kobayashi M, Kageyama M. Molecular structures and functions of pyocins S1 ans S2 in Pseudomonas aeruginosa. J Bacteriol. 1993;175:2907–2916. doi: 10.1128/jb.175.10.2907-2916.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sano Y, Matsui H, Kobayashi M, Kageyama M. Pyocins S1 and S2, bacteriocins of Pseudomonas aeruginosa. In: Silver S, Chakrabarty A M, Iglewski B, Kaplan S, editors. Pseudomonas: biotransformations, pathogenesis, and evolving biotechnology. Washington, D.C: ASM Press; 1990. pp. 352–358. [Google Scholar]
- 40.Shinomiya T, Ina S. Genetic comparison of bacteriophage PS17 and Pseudomonas aeruginosa R-type pyocin. J Bacteriol. 1989;171:2287–2292. doi: 10.1128/jb.171.5.2287-2292.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shinomiya T, Shiga S, Kikuchi A, Kageyama M. Genetic determinant of pyocin R2 in Pseudomonas aeruginosa PAO. II. Physical characterization of pyocin R2 genes using R-prime plasmids constructed from R68.45. Mol Gen Genet. 1983;189:382–389. doi: 10.1007/BF00325899. [DOI] [PubMed] [Google Scholar]
- 42.Szybalski W. Use of cesium sulfate for equilibrium density gradient centrifugation. Methods Enzymol. 1968;12:330–360. [Google Scholar]
- 43.Takemaru K, Mizuno M, Sato T, Takeuchi M, Kobayashi Y. Complete nucleotide sequence of a skin element excised by DNA rearrangement during sporulation in Bacillus subtilis. Microbiology. 1995;141:323–327. doi: 10.1099/13500872-141-2-323. [DOI] [PubMed] [Google Scholar]
- 44.Uratani Y, Hoshino T. Pyocin R1 inhibits active transport in Pseudomonas aeruginosa and depolarizes membrane potential. J Bacteriol. 1984;157:632–636. doi: 10.1128/jb.157.2.632-636.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams R C. Use of polylysine for absorption of nucleic acids and enzymes to electron microscope specimen films. Proc Natl Acad Sci USA. 1977;74:2311–2315. doi: 10.1073/pnas.74.6.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wiseman R L, Berkowitz S A, Day L A. Different arrangements of protein subunits and single-stranded circular DNA in the filamentous bacterial viruses fd and Pf1. J Mol Biol. 1976;102:549–561. doi: 10.1016/0022-2836(76)90333-8. [DOI] [PubMed] [Google Scholar]
- 47.Yasukawa H, Hase T, Sakai A, Masamune Y. Rolling-circle replication of the plasmid pKYM isolated from a gram-negative bacterium. Proc Natl Acad Sci USA. 1991;88:10282–10286. doi: 10.1073/pnas.88.22.10282. . (Erratum, 89:4220, 1992.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zahler S A. Temperate bacteriophages of Bacillus subtilis. In: Calendar R, editor. The bacteriophages. Vol. 1. New York, N.Y: Plenum Press; 1988. pp. 559–592. [Google Scholar]