Abstract
Lung cancer has a high morbidity and mortality rate, and affected patients have a poor prognosis and low survival. The therapeutic approaches for lung cancer treatment, including surgery, radiotherapy, and chemotherapy, are not completely effective, due to late diagnosis. Although the identification of genetic drivers has contributed to the improvement of lung cancer clinical management, the discovery of new diagnostic and prognostic tools remains a critical issue. Liquid biopsy (LB) represents a minimally invasive approach and practical alternative source to investigate tumor-derived alterations and to facilitate the selection of targeted therapies. LB allows for the testing of different analytes such as circulating tumor cells, extracellular vesicles (EVs), tumor-educated platelets, and cell-free nucleic acids including DNAs, RNAs, and noncoding RNAs (ncRNAs). Several regulatory factors control the key cellular oncogenic pathways involved in cancers. ncRNAs have a wide range of regulatory effects in lung cancers. This review focuses on emerging regulatory ncRNAs, freely circulating in body fluids or shuttled by EVs, such as circular-RNAs, small nucleolar-RNAs, small nuclear-RNAs, and piwi-RNAs, as new biomarkers for early detection, prognosis, and monitoring of therapeutic strategy of lung cancer treatment.
Keywords: extracellular vesicles, liquid biopsy, lung cancer, noncoding RNA, circular-RNA, piwi RNA
Introduction
Lung cancer (LC) is one of the primary causes of mortality worldwide, with about 2,200,000 new cases and around 1,800,000 deaths estimated in the world in 2020 by the Global Cancer Observatory (https://gco.iarc.fr). LC originates from basal epithelial cells of the lung and is classified into two main categories: non-small-cell lung cancer (NSCLC) and small cell lung cancer (SCLC).1 NSCLC is the most prominent form of LC; about 85% of all LC are NSCLC, this subtype represents one of the most frequent causes of cancer-related death.2,3 The prognoses and therapeutic strategies for NSCLC depend on the disease stage at diagnosis. While the surgery remains the standard of care for NSCLC,4 it is often unsuccessful in patients with metastasis. Unfortunately, NSCLC patients are mostly diagnosed at advanced stages since they are frequently asymptomatic at early stages;5,6 thus, finding new biomarkers for early diagnosis is an urgent need.
The understanding of NSCLC pathogenesis has improved through the identification of activating mutations and amplifications of oncogenes, including Kirsten rat sarcoma virus (KRAS),7,8 epidermal growth factor receptor (EGFR),9 and inactivating mutations in tumor suppressive genes, such as p53.10 Nowadays, it is estimated that up to 69% of advanced NSCLC patients carry druggable genetic alterations in different genes, such as EGFR, KRAS, anaplastic lymphoma kinase (ALK), human epidermal growth factor receptor 2 (HER2), c-Ros oncogene 1 (ROS1), or V-raf murine sarcoma oncogene homolog B1 (BRAF).11 The list of exploitable targets is rapidly growing and includes eight different genes with approved targeted therapies, including EGFR, ALK, ROS1, MET, BRAF, KRAS, NTRK, and RET.12 Despite initial responses to targeted therapies, the clinical benefit of these agents is typically limited and virtually all patients progress through the acquisition of multiple on-target and/or off-target resistance mechanisms.13 Therefore, it is important to identify new therapeutic targets for NSCLC.14 In the era of precision medicine, NSCLC clinical management can benefit from a new tool, liquid biopsy (LB).15
LB is a minimally invasive procedure, alternative, or complementary to tissue biopsy, and utilizes body fluids, such as blood, plasma, serum, saliva, or urine to collect elements mirroring the patient-specific disease state.12 LB allows the analysis of circulating biomarkers that reflect systemic tumor burden and represent intra-tumoral heterogeneity. LB includes the analysis of circulating tumor cells (CTC), extracellular vesicles (EVs), tumor-educated platelets (TEPs), and cell-free nucleic acids (cf-NAs) such as circulating tumor DNA (ctDNA) and circulating tumor RNA (ctRNA). The combination of circulating factors transported in the bloodstream has been referred to as ‘tumor circulome’. In the near future, the diagnosis based on cf-NAs could represent a new route in personalized medicine.16,17 Currently, CTCs and ctDNA are the only components of LB approved in clinical practice by the Food and Drug Administration, as biomarkers for diagnosis, prognosis, and monitoring treatment response.
LB is an important source of cf-NAs and provides information about cancer cell mutation profiles, while cell-free noncoding RNAs (ncRNAs) are promising biomarker candidates in the diagnosis and prognosis of cancer.14 The clinical application of LB technology in LC, includes early diagnosis, personalized treatment, prognosis prediction, longitudinal monitoring for cancer progression, and therapeutic response. LB is also considered a promising method for the identification of patients with a high risk of disease progression after curative surgery.18
The analysis of CTCs in patients with early NSCLC reflects their value in early diagnosis. CTC amount increases significantly in most patients with tumor progression.19 CTCs could be used as an additional method to detect ALK gene rearrangement when appropriate tissue biopsy samples could not be obtained.20 Moreover, CTCs are used to supplement the tissue-based EGFR mutation detection in LC and to guide the precision treatment of EGFR.21 LB has also entered in clinical practice as ctDNA-based tumor genotyping for the management of advanced-stage LC.22 It was reported that the plasma ctDNA levels, in NSCLC patients, are higher than in subjects with chronic respiratory inflammation or healthy individuals. Collecting ctDNA from blood is a non-invasive method, with high clinical potential, that can be repeated over time, contributing to the identification of small residual diseases or recurrence rates.23
In advanced NSCLC, the use of ctDNA, which had previously been limited to EGFR mutation detection in patients with inadequate tissue for tumor genotyping and/or for the identification of EGFR, T790M-resistant mutation, has now been extended to other genetic aberrations. Thanks to the use of plasma next-generation sequencing (NGS), this analysis, termed the ‘plasma-first’ approach, is moved to a treatment-naïve setting.3
Recently, it was also developed a new integrated genomic strategy for early-stage LC detection using a ctDNA-based machine-learning platform, named LC likelihood in plasma. This study introduces improvements to Cancer Personalized Profiling by deep Sequencing ctDNA analysis that facilitates screening applications.24 Nowadays, the European Society for Medical Oncology and the International Society for Study of LC have recommended the use of a multigene NGS approach in the molecular evaluation of advanced-stage NSCLC patients.3 Although LB is a growing field in cancer management, some challenges to the sensitivity of CTCs and ctDNA in clinical practice remain. The percentage of ctDNA is often as low as 1% and the number of CTCs is limited. EVs, ctRNA, and TEPs are considered novel tumor circulome elements with great potential at any stage of cancer for adequate clinical management.25 EVs could overcome some limits of LB; they have the advantages to be abundant in blood samples of cancer patients, stable in biofluids, and protect cf-NAs from degradation. Although EVs isolation and quantification are challenging and need standardization, the analysis of the different biological components of LB could be used to explore complementary aspects to illustrate the molecular profiles of LC comprehensively.3
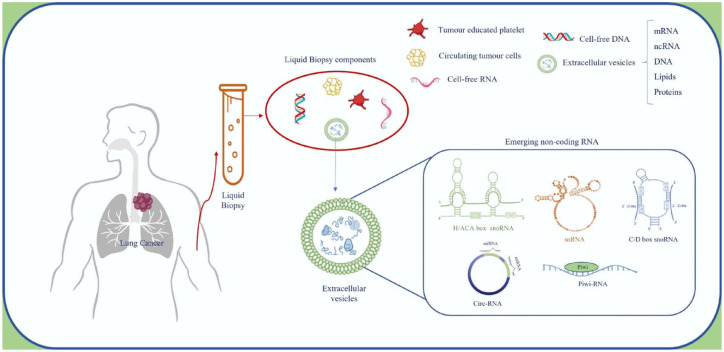
This review focuses on circulating-free or EVs containing emerging regulatory ncRNAs, such as circular-RNAs (CircRNAs), small nucleolar-RNAs (snoRNAs), small nuclear-RNAs (snRNAs), and piwi-interacting RNAs (piRNAs), in LC. These ncRNAs are essential in maintaining the spatial–temporal architecture of transcriptional and translational programs under malignant conditions and have gained the progressive attention of the scientific community. In addition, we discuss the potential roles of EVs and ncRNAs as biomarkers in LC LB (Figure 1).
Figure 1.
LB as a tool for LC management. Schematic representation of LB and EV components, focusing on emerging ncRNAs described in EVs.
EVs, extracellular vesicles; LB, liquid biopsy; ncRNAs, noncoding RNAs.
Extracellular vesicles
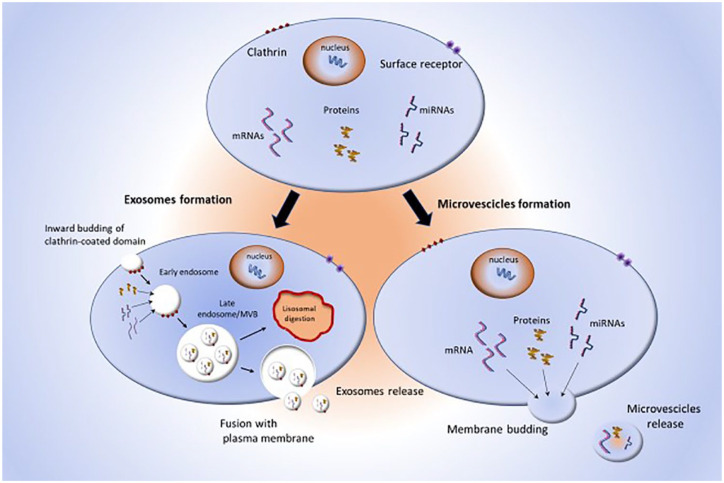
EVs are nanoscale membrane particles, released by all eukaryotic and prokaryotic cytotypes, in physiological and pathological conditions.26–28 EVs carry proteins, metabolites, lipids, and nucleic acids, including DNA fragments and RNAs that can induce phenotypic reprogramming of target cells.29–31 Based on their biogenesis and size, EVs are classified into two main classes: exosomes and microvesicles (MVs; Figure 2).32
Figure 2.
Schematic representation of EV biogenesis. Donor cells release exosomes and MVs with different mechanisms.
EV, extracellular vesicle; MVs, microvesicles.
Exosomes are lipid bilayer nanovesicles of endosomal origin, with a diameter of 30–150 nm instead, MVs have a diameter of 200–1000 nm and are shed directly from discrete microdomains of the plasma membrane, enriched in phosphatidylserine, cholesterol, and lipid rafts.33
EVs can be collected from various biofluids, such as serum, plasma, saliva, breast milk, nasal secretions, urine, semen, and pathological effusions.6,29,34,35 Several mechanisms of EV release have been described, that involve different machineries such as ESCRT complex, tetraspanins, sphingomyelinases, redistribution of phospholipids, and depolymerization of cytoskeletal actin.26
EVs can be internalized by target cells through receptor-ligand binding, direct fusion with plasma membranes, phagocytosis, micropinocytosis, actin polymerization, and filopodia extension. It has also been reported that EVs can be internalized by target cells as intact vesicles surfing on filopodia.27,36–38 EVs encapsulate their cargo in a phospholipidic bilayer, providing high stability, a long half-life, and resistance to degradation. Their stability is attributed to the lipid components of their membranes, which are enriched in cholesterol, phosphatidylserine, glycosphingolipids, sphingomyelin, and annexin, as compared to the cellular plasma membrane.39
EVs can travel long distances and deliver their cargo to specific cell types through specific ligand–receptor interactions.39
Another intriguing feature of EVs is their potential to cross tissue barriers. Several studies showed their ability to cross the blood–brain barrier. EVs’ stability, addressability, and barrier penetration make them encouraging therapeutic delivery devices.40,41
EV cargos, transported in directed ways to target cells, can act in autocrine, paracrine, and systemic manners. EVs regulate biological processes and crosstalk between cells, playing an important role in cancer.34,42–44 Several reports demonstrate the key role of EVs in cancer progression,45 premetastatic niche formation,46 metastasis,47,48 and drug resistance.49,50 The potential for EVs as biomarkers in LC early detection and progression has been reported as well. EVs have been identified as good biomarkers for monitoring response to LC therapies.26
Since anticancer therapies alter the amount of EVs in biofluids, the dynamic analysis of circulating EVs and their contents can provide real-time information on therapeutic responses.51 EVs promise to be a next-generation diagnostic and therapeutic tool in LC, to monitor immune-checkpoint inhibitors (ICIs) therapy with programmed cell death protein 1 (PD-1) or programmed cell death-ligand 1 (PD-L1) antibodies, a standard treatment for advanced NSCLC. It was reported that specific microRNAs (miRNAs) in plasma EVs are differentially expressed between responders and non-responders’ patients to ICIs and have potential as predictive biomarkers for anti–PD-1/PD-L1 treatment response.52 Recently, de Miguel-Perez et al.53 evaluated EVs containing PD-L1, as a biomarker for the prediction of durable treatment response and survival in patients with NSCLC undergoing treatment with ICIs. An increase in EV-PD-L1 was observed in non-responders’ patients in comparison to responders. These findings indicate that EV-PD-L1 dynamics could be used to stratify patients with advanced NSCLC, who would experience durable benefits from ICIs.53
As aforementioned, EVs are natural intercellular shuttles for NAs such as DNA, and RNAs including ncRNAs.54,55 The DNA packaging into EVs was significantly higher in cancer-derived EVs compared to EVs from noncancer cells. In LC, the combination of RNA/DNA contained in exosomes (exoRNA/DNA) and ctDNA for T790M detection has higher sensitivity and specificity compared to ctDNA alone.56
Recently, Park and colleagues demonstrated that also EVs derived from bronchial washing (BW-EVs) could be used for accurate and frequent genotyping of EGFR mutation in patients with NSCLC. BW-EVs are useful for the early detection of actionable mutations, for the selection of personalized therapy, and for monitoring disease progression. The well-known T790M mutation seems to be detected from BW-EV-DNA with a superior detection rate to plasma-derived ctDNA or tissue biopsy.57 Although the ‘gold standard’ source for LB testing on EGFR is represented by ctDNA, recovered from plasma, recent studies indicate that EVs are a good source of RNA to identify EGFR mutations. EV-RNA and ctDNA can function as independent biological sources in LB of NSCLC patients providing a complementary informative set of tumor dynamics. While ctDNA is predominantly released as a consequence of the apoptotic and necrotic process, EVs are released from viable cells to transmit information to other cells providing a complex cargo available for molecular analysis.58 Moreover, it was demonstrated that EVs carry mutated EGFR as mRNA and protein, mirroring the disease status in metastatic NSCLC. Sensitizing (exon 19 deletion, L858R) and resistance (T790M) mutations were quantified in EV-RNA. The comparison of mutation detection between EV-RNA and ctDNA using digital droplet PCR (ddPCR) indicates that EVs have a better detection rate for exon 19 deletions and L858R point mutation.59 These reports suggest that EV-RNA provides a new tool appropriate for use in clinical practice to investigate the dynamics of common driver EGFR mutations in NSCLC patients receiving TKIs.
Noncoding RNAs
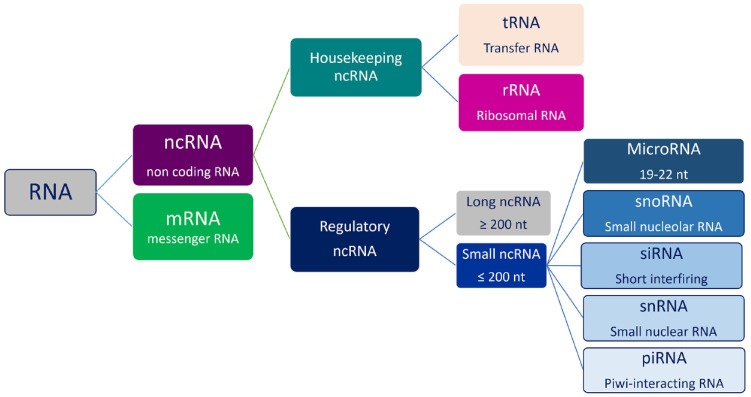
ncRNAs are molecules of RNA with no protein translation potential, involved in physiological and pathological processes.60 Data from genome-wide transcriptional analysis in humans have shown that the amount of protein-coding transcripts account for approximately 2% of the entire genome, while ncRNAs represent about 98% of all genomic output.29,61 Interestingly, it has been reported that the proportion of noncoding regions in the genome increases according to the complexity of organisms, suggesting an important role for these sequences in the physiology and development of organisms.62,63 Studies on these non-protein-coding RNAs have received a lot of interest in many fields, especially in cancer, leading to new hypotheses about cancer biology.64 ncRNAs can be classified as housekeeping and regulatory. Housekeeping ncRNAs are abundantly and ubiquitously expressed in cells, and primarily regulate generic cellular functions. While regulatory ncRNAs are usually considered key regulatory RNA molecules, function as modulators of gene expression at epigenetic, transcriptional, and post-transcriptional levels.65–67
Based on their molecular size, all regulatory ncRNAs are subclassified into small ncRNAs, with transcripts shorter than 200 nucleotides, including miRNAs, snRNAs, snoRNAs, and piRNAs and large ncRNAs, with transcripts longer than 200 nucleotides, that include CircRNAs.68
Despite comprising less than 1% of total cellular ncRNA content, regulatory ncRNAs play crucial roles in transcription, post-transcriptional mechanisms, and translation. Regulatory ncRNA-mediated gene silencing constitutes one important type of epigenetic alteration and has been implicated in several cases of human carcinogenesis. Regulatory ncRNAs (Figure 3) modulate gene expression through various mechanisms.69,70 ncRNAs can influence lung tumorigenesis, and they play an important role in premetastatic niche formation and metastasis of NSCLC.71 Since ncRNAs circulate in biofluid freely or encapsulated in EVs, they are intriguing as potential biomarkers and therapeutic targets in LC LB scenario.
Figure 3.
RNA classification. RNAs are divided into two main classes: mRNAs and ncRNAs. Each category has different components.
ncRNAs, noncoding RNAs.
In recent years, a variety of regulatory ncRNAs have been identified, as key modulators of gene expression in different cellular pathways and systems.68,69 Recent findings changed the description of ncRNAs from ‘junk’ transcriptional products to functional regulatory molecules that mediate key processes such as gene expression, chromatin remodeling, transcription, post-transcriptional modifications, and signal transduction.70 The tRNA-derived fragments (tRFs)-based or RNA-related fragments (rRFs)-based therapies might become useful to offer new therapeutic options.72
In the classical view, housekeeping ncRNAs, involved in the maintenance of normal cell functionalities, include transfer RNAs (tRNAs) and ribosome RNAs (rRNAs), which are important in protein translation. This viewpoint was challenged by recent findings on the dysregulated expression of tRNAs in several cancers, including LC.54 The tRNAs are cleaved by angiogenin in tRNA-derived stress-induced RNAs (tiRNAs), which can be further processed by Dicer into tRFs. In LC tissue, tRNAs-Leu and tRNAs-Val are overexpressed, respectively, in 37% and 26% of samples.73 The tRF-Leu-CAG is upregulated in LC and involved in cell proliferation and cell cycle progression, interacting with AURKA protein.74 The tRF-Leu-CAG could be a new diagnostic marker and potential therapeutic target in NSCLC. Specific blockade of some tRFs, such as tRF-Leu-CAG, may have considerable clinical application in suppressing LC cell proliferation and cell cycle progression.73 Recently, it was reported that EVs shuttle tRFs with effects on various cellular processes; the dysregulation of EV-tRFs has been associated with cancer progression and they are potential novel biomarkers for cancer diagnosis.75
The rRNAs, like tRNAs, can be cleaved into small ribosomal rRFs. The function of rRFs is not limited to gene silencing, their structure also plays a key role in rRNA stability.72 The rRNAs are abundant in EVs released by different cancer cell lines; a high-throughput study on human breast cancer cell lines showed that over 80% of RNAs contained in EVs are rRNAs.76 The rRNAs were also detected in EVs collected by different body fluids such as the serum, plasma, urine, and saliva.77
Circular-RNAs
CircRNAs have been identified as a new class of ncRNA with high regulatory potential.78 CircRNAs contained in blood, either free or encapsulated within EVs, have several advantages over canonical linear RNAs as cancer biomarkers.79
CircRNAs, which are found in a large amount within tissues, cells, and body fluids, are aberrantly expressed in cancer tissues and regulate tumor progression. Since they are expressed in stage-specific manners, several studies have shown their potential as helpful diagnostic and prognostic biomarkers for cancers.80
CircRNAs are generated for back-splicing processing from linear pre-messenger RNAs when the 3′ and 5′ ends are ligated to form a continuous loop and covalently closed.79 CircRNAs were first discovered in 1990 when observing that exons of a tumor suppressor gene after their splicing were joined in a different order than their genomic sequence.81 CircRNAs are highly resistant to RNAse activity because of the lack of 5′ and 3′ ends,82 being more stable and having longer half-lives than canonical linear isoforms. In 2015, CircRNAs were described in EVs for the first time, when high-throughput technologies such as genome-wide RNA-seq analyses showed that CircRNAs were enriched in EVs in comparison with parental cells. CircRNAs contained in EVs may be controlled by alteration of associated miRNA levels in parental cells, transferring the biological activity to target cells.83,84
The principal functions of CircRNAs are miRNA inhibition, interaction with RNA-binding proteins, and regulation of parental genes. Particularly, CircRNAs act as miRNA sponges protecting target genes from repression by miRNAs. Notably, one CircRNA can sponge different miRNAs, establishing an intricate and precise regulatory network.85,86
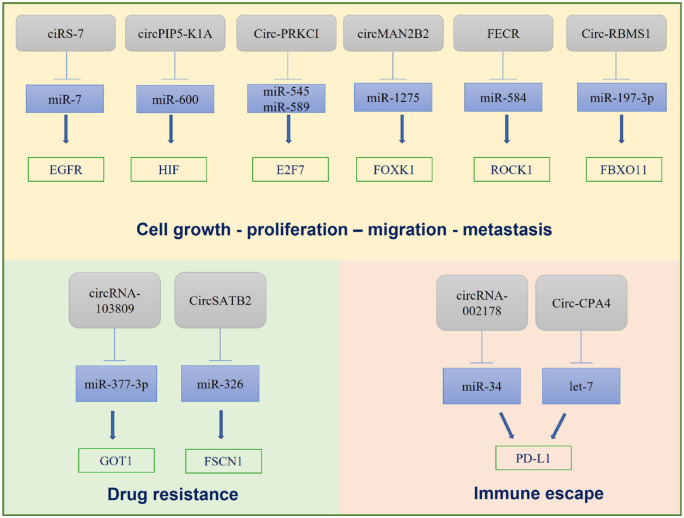
CircRNAs are emerging candidates as biomarkers for diagnosis, prognosis, and therapeutic response of NSCLC in LB.86 CircRNAs are involved in various aspects of LC progression such as cell growth, proliferation, migration, metastasis, drug resistance, and immune escape as summarized in Figure 4. Recently, it was reported that ciRS-7 is associated with NSCLC development. CiRS-7, also called CDR1as, is one of the earliest discovered CircRNAs originating from the back-splicing of CDR1 gene.87 This CircRNA has about 70 miRNA-binding sites and acts as a sponge for miR-7. CiRS-7 is overexpressed in tissues and cell lines of NSCLC and may promote cancer cell proliferation via ciRS-7/miR-7/EGFR/CCNE1/PIK3CD signaling. High levels of ciRS-7 were found to be associated with TNM stage and lymph node metastasis of NSCLC.88
Figure 4.
CircRNAs are involved in different steps of LC progression such as cell growth, migration, metastasis, drug resistance, and immune escape.
CircRNAs, circular RNAs.
Another CircRNA overexpressed in NSCLC tissues compared to adjacent tissues is has_circ_0014130 (circPIP5-K1A), which acts as a sponge for miR-600 and inhibits its activity, upregulating hypoxia-inducible factor 1α, which is involved in tumor proliferation and metastasis.89
The proto-oncogenic CircRNA (circ-PRKCI) originates from the 3q26.2 amplicon, one of the most frequent genomic aberrations in cancer. The increased expression of Circ-PRKCI in lung tissues, caused by amplification of 3q26.2 locus, induces tumorigenesis and progression of LC. Circ-PRKCI functions as a sponge for miR-545 and miR-589 and annuls their suppression of pro-tumorigenic transcription factor E2F7.90
In addition, circMAN2B2 is a product of MAN2B2 mRNA splicing and promotes FOXK1 expression by sponging miR-1275 in LC cells, inducing LC progression.91 FLI1 exonic CircRNA (FECR) is considered an oncogenic driver that helps tumor metastasis by modulating the miR584-ROCK1 pathway. Notably, FECR1 is contained in exosomes collected in the serum of LC patients. Exosomal FECR1 has been associated with poor tumor survival and clinical response to chemotherapy and is useful as a cancer prognostic factor and as a potential biomarker to track LC progression.91
Recently a novel CircRNA-103809/miR-377-3p/GOT1 axis that contributes to cisplatin resistance in NSCLC in vitro and in vivo, has been identified. Targeting this pathway enhances cisplatin sensitivity in NSCLC, and provides new chances for improving the clinical management of NSCLC.92
Moreover, it has been observed that has-CircRNA-002178 can enhance PD-L1 expression in cancer cells inducing T-cell exhaustion via miR-34 inhibition. Tumor cells grow and metastasize by escaping the immune system through different mechanisms, such as PD-L1 expression. PD-L1 is a transmembrane protein, found abundantly expressed in cancer cells, and involved in tumor immune escape by interacting with PD-1.93 CircRNA-002178 has been also detected in exosomes isolated from plasma of LC patients and may be useful as a biomarker for LC early diagnosis. CircRNA-002178 is delivered into CD8+T cells by exosomes to induce PD-1 expression. This CircRNA may promote PD-L1/PD-1 expression in LC.94
Furthermore, Circ-CPA4 is highly expressed in NSCLC relative to normal human bronchial epithelial cells. Circ-CPA4 regulates cell growth, migration, stemness, and drug resistance in NSCLC cells and is involved in CD8+ T-cell inactivation, via the let-7/PD-L1 axis. Circ-CPA4 also positively regulates PD-L1 contained in exosomes.95
Recently, it was demonstrated that CircRNA-102481 is overexpressed in exosomes collected from serum in EGFR-TKIs-resistant patients. The silencing of this CircRNA inhibits EGFR-TKIs-resistant NSCLC cell proliferation and induces apoptosis. CircRNA-102481 overexpression could promote EGFR-TKIs-sensitive NSCLC cell proliferation and inhibit cell apoptosis, which suggests that CircRNA-102481 may contribute to EGFR-TKIs resistance in NSCLC.96 Another CircRNA highly expressed in NSCLC cells is circSATB2, which positively regulates fascin homolog 1, actin-bundling protein 1 (FSCN1) expression via miR-326. CircSATB2 is shuttled by exosomes promoting migration, proliferation, and invasion of NSCLC cells. It also induces aberrant proliferation of normal human bronchial epithelial cells. CircSATB2 is highly expressed in serum exosomes in LC patients.71
The specificity and stability of CircRNAs and the capacity of EVs to interact with target cells while remaining stable in the bloodstream, make CircRNAs shuttled by EVs very attractive as biomarkers for LC early detection.97
Emerging evidence demonstrates that CircRNAs are aberrantly expressed in progressive degenerative lung diseases such as chronic obstructive pulmonary disease (COPD), in which cigarette smoke is considered a crucial risk factor. Recently, it was reported that circ-RBMS1 derived from the RBMS1 gene was higher in COPD patients, and cigarette smoke increased circ-RBMS1 expression in a dose-dependent manner. Circ-RBMS1 directly targeted miR-197-3p and this miRNA targeted FBXO11. In vitro, the knockdown of circ-RBMS1 attenuated cigarette smoke extract induced inflammation and oxidative stress in epithelial cells, via miR-197-3p/FBXO11 axis, suggesting a new insight into the pathogenesis of cigarette smoke-induced COPD.98 Nowadays, the overlapping prevalence of COPD with LC is clear; chronic inflammation and reactive oxygen species can be considered the molecular links between COPD and LC.99 Future findings could demonstrate a key role for circRNAs in this progression.
Furthermore, it was reported that circ-FASRA in plasma has diagnostic value for NSCLC and can be used as a biomarker for the detection of non-invasive NSCLC. With the development of second-generation sequencing technology, screening CircRNA differentially expressed in body fluid samples of LC patients and healthy controls can be used to find a new biomarker for diagnosis and prognosis and as a potential therapeutic target.100
Small nuclear RNA
Several diseases such as cancer and chronic lung disease have been associated with snRNAs alterations, acting on the cellular transcriptome. Each snRNA, such as U1, and U2 (the nomenclature arises from high uridine content), is complexed with small nuclear ribonucleoproteins to exert their functions.70 The snRNAs act in spliceosome complexes, recognizing 5′ and 3′intron/exon boundaries during splicing of introns from pre-messenger RNA transcripts.101
snRNAs (<200 nucleotides) include a small group of non-polyadenylated, noncoding transcripts that act in the nucleoplasm. snRNAs are divided into two classes, Sm and Lsm class RNAs, based on common sequence features and protein cofactors. Sm-class RNAs are characterized by a 5′-trimethylguanosine cap, a 3′ stem-loop, and a Sm-site that consists of a binding site for a group of seven Sm proteins, forming a hetero-heptameric ring structure. Lsm-class RNAs consist of a mono-methyl-phosphate cap and a 3′ stem-loop, ending in a stretch of uridines that form the binding site for a hetero-heptameric ring of Lsm proteins. The principal components of Sm-class RNAs are U1, U2, U4, U4atac, U5, U7, U11, and U12, whereas the more studied Sm-class RNAs are U6 and U6atac.102
The high stability of snRNAs in biological samples makes them novel diagnostic biomarkers, supporting prognostic or predictive indicators, and tools to monitor treatment in cancer patients.103
Fragments of U2 snRNA (called RNU2-1f) may be collected in sera of patients with pancreatic, colorectal, and ovarian cancers and cerebrospinal fluid of patients with primary central nervous system lymphoma.104–106 RNU2-1f has also been found in the serum of patients with metastatic melanoma.107 High levels of this snRNA have been associated with tumor progression and after surgical removal of pancreatic and colorectal tumors, RNU2-1f levels have been shown to decrease.101 Furthermore, RNU2 is highly expressed in lung tissue and efficiently exported into circulation, elevated levels of RNU2-1f have been detected in the serum of LC patients. RNU2-1f may be a potential biomarker for LC patients.108 LC snRNAs can regulate alternative splicing to drive genetic and neoplastic disease and alter mRNA profile in TEPs. Platelets are ‘educated’ by their tumor environment, containing a dynamic variety of RNA subsets, including snRNAs. RNA profiles of TEPs allow for the distinction between cancer patients and healthy controls. For instance, RNA profiles of TEPs can be utilized to predict oncogenic status, such as MET or HER2 positivity, and reveal gene mutations in KRAS or EGFR. In addition, U1, U2, and U5 are significantly downregulated in TEP of LC patients compared with healthy controls. TEP U1, U2, and U5 levels can be decreased in LC patients, and their downregulation has been correlated with LC progression. Moreover, TEP U1, U2, and U5 levels may be directly correlated with paired exosomes and TEP from treated patients but not from untreated patients. U1, and U5 but not U2 in platelets can be elevated by exosomes released from apoptotic cells.109
Small nucleolar RNA
snoRNAs are a group of intron-encoded ncRNAs mainly accumulated in nucleoli that consist of 60–300 nucleotides. snoRNAs are grouped into two families called box C/D snoRNAs (SNORDs) and box H/ACA snoRNAs (SNORAs).110
snoRNAs are involved in post-transcriptional modification and maturation of ribosomal RNAs, snRNAs, and other RNAs.111 snoRNAs are involved in several physiological and pathological processes. In addition, snoRNAs have oncogenic or tumor-suppressive functions in different cancers activating invasion, metastasis, angiogenesis, and sustained proliferative signaling or increasing growth suppressors and cell death. Some reports suggest that snoRNAs are associated with p53 regulation. P53 is a well-known tumor suppressor that responds to cellular stresses to regulate the expression of target genes involved in cell cycle arrest, apoptosis, and DNA repair.112 Recently, it was reported that snoRNAs are associated with p53 pathway. In particular, SnoRNA42 is overexpressed in NSCLC and has an oncogenic role by affecting p53 expression.113
Moreover, SNORD78 is upregulated in LC and tumor-initiating cells of LC, suggesting that it might play a role in lung tumorigenesis. SNORD78 is also upregulated in cancer stem cells in NSCLC and is essential for the self-renewal of cancer stem cells in NSCLC.114
It was reported that SNORD46 acts as an oncogene in LC. In vitro silencing of SNORD46 leads to decreased cell viability, invasion, and migration inhibition.115 An in silico analysis of the expression and clinical relevance of SNORDs in human cancer indicates that SNORD46 is negatively correlated with forkhead box O3 (FOXO3), a transcription factor that triggers apoptosis.116
snoRNAs can predict LC progression from the initial stages. The overexpression of SNORD28, SNORA21, SNORA47, SNORD66, SNORA78, and SNORA68 leads to worse overall survival in LC patients and these snoRNAs are differentially expressed between lung tumors of stage I and normal tissue.116 In NSCLC patients, SNORD33, SNORA42, SNORD66, and SNORD78 are overexpressed. Moreover, snoRNAs can be considered LC biomarkers in combination with other miRNAs.117 A panel of biomarkers for LC, comprising miR-21, miR-32, and miR-210 and SNORD66, SNORD78, collected from sputum, was developed to be used as a potential tool for a non-invasive LC diagnosis. It was reported that the combined use of miRNAs and snoRNAs demonstrated higher sensitivity and specificity compared with a single type of ncRNA biomarkers, offering a new approach for LC early detection.118
Interestingly, it was reported that five snoRNAs: SNORA14B, SNORA18, SNORA25, SNORA74A, and SNORD22 were encapsulated in exosomes isolated from the serum of pancreatic cancer patients and conditioned medium of pancreatic cell lines. SNORA74A and SNORA25 are highly expressed in the early stages of pancreatic cancer in comparison with healthy controls.119 These data indicate that exosomal snoRNAs are useful in the diagnosis of cancer; further studies are needed to confirm their role in LC.
Piwi-interacting RNAs
P-element-induced wimpy testis piRNAs are a new investigated class of small ncRNAs.120 piRNAs have been discovered in male gonadal cells, with the central role of protecting germinal cells from transposable elements (TEs), especially of viral origin, and are germline specific.121 piRNAs are small ncRNAs of 26–31 nt; they have a 2′-O-methylation at the 3′ end as a distinctive and exclusive feature of all piRNAs. The precursors of piRNAs are single-stranded transcripts without secondary hairpin structures.122,123 These precursors are generated from precise genomic locations with repetitive elements with a Dicer-independent process. The nascent piRNAs require further post-transcriptional modifications to become mature piRNAs. The biogenesis of piRNAs includes two steps: a primary and secondary amplification cycle referred to as a ‘ping-pong cycle’ in which piRNAs are bound to piwi proteins.124 piRNA biogenesis is an adaptive process that silences active transposons with sequences complementary to piRNA cluster transcripts. piRNAs control and silence TEs to protect the genome since uncontrolled TE expression may lead to a loss of genome integrity.125 Recently, it was reported that piRNAs are widely expressed in somatic cells and human cancer cells. About 30,000 piRNAs were found in the human genome and recent studies have suggested that piRNAs play a role in human cancer pathogenesis. Cancer, stem, and germ cells share key biological characteristics such as the ability for self-renewal and rapid proliferation. Although piRNAs were first described as important regulators in maintaining germline stem cells, it is conceivable that rapidly dividing cancer cells might adopt and utilize self-renewal machinery like germ cells. Recently, a growing number of studies have revealed the role of piRNAs in cancers, introducing a new biological concept in which mechanisms of piRNA-mediated gene regulation specific to germline cells also have oncogenic and tumor suppressive roles.126
piRNAs are expressed in a tissue-specific manner in several human tissues, regulating important signaling pathways at the transcriptional or post-transcriptional level. In malignant cells, piRNAs participate in the epigenetic regulation of DNA123 and are essential for maintaining cancer stemness.127 The aberrant expression of piRNAs and PIWI proteins in cancers might be used as new biomarkers and therapeutic targets for tumor diagnosis and treatment. It was suggested that piRNA-54265 can be used as a biomarker for the early detection and clinical monitoring of colorectal cancer. piRNA-36712 has been described as a novel tumor suppressor and might be a breast cancer prognostic predictor. In addition, piR-823 has been found in cancer cells and plasma, has a role in regulating tumor cell growth, and its expression fluctuates in many cancers, including gastric cancer and multiple myeloma.128 In colorectal cancer, piR-823 downregulation increases cell apoptosis by inducing transcription factor HSF1, an apoptosis activator, and inhibits cell proliferation.129 piRNA-54265 is highly expressed in colorectal cancer tissue and serum; this piRNA activates STAT3 signaling, thereby inducing cancer progression.130 In multiple myeloma, piRNA-823 silencing induces the expression of apoptosis-related genes, modulating DNA methylation.131 Another piRNA, piR-651, is downregulated in patients with Hodgkin lymphoma relative to healthy controls. Furthermore, low levels of piR-651 correlate with poor prognosis in Hodgkin lymphoma patients.132 piRNAs are not easily degradable and can move across the plasma membrane. This feature suggests that piRNAs can easily be detected in body fluids, and recent studies have indicated that piRNAs, contained in EVs released by cancer cells, remain stable in body fluids. Peng and colleagues have demonstrated that piRNAs are contained in EVs from the urine of patients with prostate cancer. The expression of piR349843, piR382289, piR158533, and piR002468 in urinary EVs is increased in prostate cancer patients relative to healthy controls.133
Emerging findings suggest that piRNAs may be considered potential biomarkers for LC diagnosis. It has been shown that piR-hsa-211106 can inhibit the progression of lung adenocarcinomas enhancing chemotherapy sensitivity, suggesting that piR-hsa-211106 is a potential therapeutic target for LC.134 piRNA expression profiles of lung adenocarcinoma tissues and adjacent normal tissues have identified ten piRNAs overexpressed in tumor tissues. Among these, piR-hsa-26925 and piR-hsa-5444 are upregulated in EVs collected from the serum of patients with lung adenocarcinoma.135 These findings indicate that LB can allow for the detection of both free piRNAs and piRNAs contained within EVs. Moreover, piRNAs may become therapeutic and diagnostic tools for various cancer types, including LC.
Conclusion and perspectives
LB is a promising diagnostic tool with several advantages over conventional invasive methods. LB is useful to identify prognostic, diagnostic, and predictive biomarkers in LC not only for its minimal invasiveness, which allows repeating the biopsy within the scope of tumor surveillance and yields genetic information about cancer, considering the tumor heterogeneity and the presence of subclones. LB represents a source of biomarkers including EVs, ctDNA, CTC, and cf-NAs such as ncRNAs. EVs appear to be ideal for LB thanks to the stability of their membrane and their ability to travel in body fluids, at long distances.
Several ncRNAs have been detected in biofluids. The origin of these molecules might be passively released by dead cells or active secretion via EVs or vesicle-free RNA-binding protein-dependent pathways. Several studies focus on ncRNAs contained in EVs for their high stability, derived from the packaging in a membranous structure that protects them from the degrading effects of RNases. Recent findings suggest that ncRNAs are differentially expressed between disease patients and healthy individuals, supporting their use as potential biomarkers for diagnostic tests. ncRNAs provide useful information regarding tumor burden, treatment responsiveness, and malignant progression.136 The development and diffusion of high-throughput technologies such as NGS and ddPCR have boosted LB as a potential diagnostic tool in cancer.
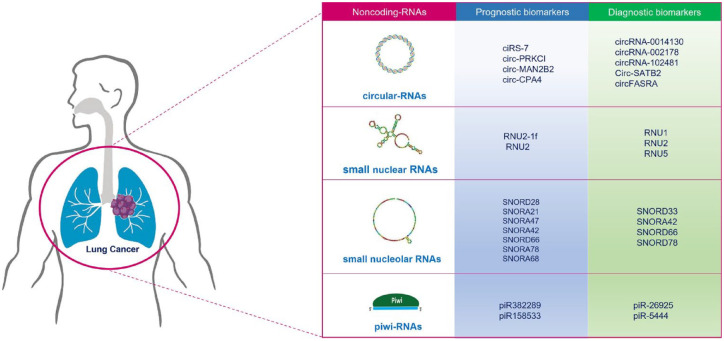
Emerging ncRNA can be useful for their diagnostic and prognostic potential (Figure 5). The structural stability of CircRNAs may have interesting applications for designing drugs that can be delivered free or encapsulated in EVs.137 In addition, EVs containing CircRNAs are internalized by recipient cells, where they affect post-transcriptional regulation of gene expression and can be exploited for their therapeutic potential.138 Some CircRNAs are found to be significantly associated with LC. Many clinical results have demonstrated that CircRNAs can be considered potential biomarkers. Nevertheless, the sample quantities of these studies are limited, and the precise regulatory mechanism of CircRNAs in LC should be better defined.139
Figure 5.
ncRNAs as diagnosis and prognostic biomarkers in LC.
ncRNAs, noncoding RNAs.
The aberrant expression of piRNAs has been associated with cancer progression and metastasis in different cancer types, suggesting their role in cancers as oncogenes or tumor suppressors. Although they can be detected in the blood of cancer patients, few reports have been published about piRNAs contained in EVs.140
Although snoRNAs localize primarily to the nucleus and have previously been considered housekeeping genes, accumulating evidence indicates that snoRNAs have oncogenic roles. Since snoRNAs are abundant and easily detectable in cancers and blood and are functionally relevant in oncogenesis, they may become an important target for cancer therapy.141 Moreover, snoRNAs contained in EVs can be considered novel biomarkers for LC diagnosis.142 snoRNAs might become pivotal elements to improve the knowledge of LC and relevant multimodal tools to ameliorate cancer patient management from their diagnosis to their treatment.143
While the evaluation of actionable mutations in LB has a confirmed clinical value, the use of epigenetic alterations, with few exceptions, has not reached clinical practice yet. Among the different fields where epigenetic factors may play a role as biomarkers, screening and diagnosis are the areas closer to the clinic.144
Since the functions of many ncRNAs are not completely known and EVs shuttle a myriad of biomolecules, EV language remains difficult to decode. Understanding the complex networks of interactions coordinated by ncRNAs would offer a unique chance to design better therapeutic options. Further studies are needed to confirm the potential role of ncRNAs freely circulating in body fluids or contained in EVs as biomarkers in the field of precision oncology.
Currently, few clinical trials (precisely 13) on EVs in LC management have been proposed (www.clinicaltrials.gov), but the number of clinical studies on EV-ncRNAs (as reported in Table 1) in this field growing exponentially.
Table 1.
Clinical studies on ncRNAs as diagnosis and prognostic biomarkers in LC.
| ncRNAs | Up/down regulated in LC | Source | Cases and samples | Diagnostic/prognostic value | Reference |
|---|---|---|---|---|---|
| CircRNAs | |||||
| ciRS-7 | Up | Tissues | 128 pairs of NSCLC and para-cancerous tissues | Prognostic value inferior OS in subjects with highly expressed ciRS-7 (p < 0.05) | de Fraipont et al.,88 Su et al.145 |
| CircRNA-0014130 | Up | Tissues | 46 NSCLC tissues and 46 non-tumor tissue | Diagnostic value sensitivity 87.0% specificity 84.8% | Li et al.,90 Zhang et al.146 |
| Circ-PRKCI | Up | Tissues | 60 pairs of tumor NSCLC tissue and adjacent non-tumor tissue | Prognostic value inferior OS in subjects with highly expressed circ-PRKCI (p < 0.05) | Chi et al.,89 Meng et al.147 |
| Circ-MAN2B2 | Up | Tissues | 5 pairs of NSCLC tissue and adjacent non-tumor tissue | Potentially prognostic | Qiu et al.91 |
| CircRNA-002178 | Up | Tissues exosomes | 105 pairs of LUAD tissues and adjacent noncancerous tissues | Diagnostic value LAC early diagnosis AUC = 0.9956 | Zhu et al.,93 Wang et al.94 |
| Circ-CPA4 | Up | Tissues | 50 pairs of NSCLC tissues and adjacent noncancerous tissues | Potentially prognostic | Hong et al.95 |
| CircRNA-102481 | Up | Exosomes | 58 NSCLC patients | Potentially diagnostic | Yang et al.96 |
| Circ-SATB2 | Up | Tissues exosomes | 59 paired NSCLC and matched normal adjacent tissue | Diagnostic value Early diagnosis AUC = 0.616 (tissue) AUC = 0.660 (exosome) | Zhang et al.71 |
| Circ-FASRA | Up | Tissues plasma | 35 paired NSCLC and matched normal adjacent tissue | Potentially diagnostic and prognostic | Chen et al.100 |
| snRNA | |||||
| U2 | Up | Serum | 62 LC and 96 controls | Diagnostic value sensitivity 79% specificity 79.2 | Mazières et al.108 |
| U1, U2, U5 | Down | TEP | 405 LC and 361 controls | Diagnostic value sensitivity 85.9% specificity 70.1% | Dong et al.109 |
| snoRNA | |||||
| SnoRNA42 | Up | Tissues | 64 NSCLC patients | Prognostic value inferior OS in subjects with highly expressed SNORA42 (p<0.01) | Mei et al.148 |
| SNORA78 | Up | Tissues | 56 primary NSCLC tissues and paired adjacent noncancerous tissues | Prognostic value Inferior OS in subjects with highly expressed SNORD78 (p = 0.0113) | Zheng et al.114 |
| SNORD28 | Up | Tissues | 77 primary NSCLC tissues | Prognostic value Inferior OS in subjects with highly expressed SNORDs/SNORAs (p < 0.001) | Gong et al.,116 Gao et al.,117 Wang et al.139 |
| SNORA21 | |||||
| SNORA47 | |||||
| SNORD66 | |||||
| SNORA78 | |||||
| SNORA68 | |||||
| SNORD33 | Up | Sputum | 46 NSCLC tissues and 55 controls | Diagnostic value AUC = 0.90 | Su et al.118 |
| SNORA42 | |||||
| SNORD66 | |||||
| SNORD78 | |||||
| piRNAs | |||||
| piR-26925 | Up | Exosomes | 70 LUAD patients and 57 healthy controls | Diagnostic value AUC of 0.833 | Li et al.135 |
| piR-5444 | |||||
AUC, are under the curve; CircRNAs, circular RNAs; ncRNAs, noncoding RNAs; NSCLC, non-small-cell lung cancer; piRNAs, piwi-interacting RNAs; snoRNA, small nucleolar RNA; snRNA, small nuclear RNA; TEP, tumor-educated platelet.
The combined analysis of the different components of LB, including the ncRNAs contained in EVs, may potentially help to comprehend the dynamics of molecular alterations and support the clinical decisions in LC management. Overall, while this field may seem to be in infancy, the evidence reported in this review indicates the potential of ncRNAs as rising stars biomarkers and therapeutic targets for precision oncological treatments in LC patients.
Acknowledgments
None.
Footnotes
ORCID iDs: Diego de Miguel-Perez  https://orcid.org/0000-0002-2822-4466
https://orcid.org/0000-0002-2822-4466
Simona Taverna  https://orcid.org/0000-0002-2192-8938
https://orcid.org/0000-0002-2192-8938
Contributor Information
Giuseppe Cammarata, Institute of Translational Pharmacology (IFT), National Research Council (CNR) of Italy, Palermo, Italy.
Diego de Miguel-Perez, Center for Thoracic Oncology, Tisch Cancer Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
Alessandro Russo, Medical Oncology Unit, A.O. Papardo & Department of Human Pathology, University of Messina, Messina, Italy.
Ariel Peleg, Center for Thoracic Oncology, Tisch Cancer Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
Vincenza Dolo, Department of Life, Health and Environmental Sciences, University of L’Aquila, L’Aquila, Italy.
Christian Rolfo, Center for Thoracic Oncology, Tisch Cancer Institute, Icahn School of Medicine at Mount Sinai, One Gustave Levy Place, Box 1079, New York, NY 10029-6574, USA.
Simona Taverna, Institute of Translational Pharmacology (IFT), National Research Council (CNR) of Italy, Via Ugo La Malfa, 153, Palermo 90146, Italy.
Declarations
Ethics approval and consent to participate: Not Applicable.
Consent for publication: Not Applicable.
Author contribution(s): Giuseppe Cammarata: Conceptualization; Data curation; Writing – original draft; Writing – review & editing.
Diego de Miguel-Perez: Writing – original draft; Writing – review & editing.
Alessandro Russo: Data curation; Writing – review & editing.
Ariel Peleg: Writing – review & editing.
Vincenza Dolo: Supervision; Writing – review & editing.
Christian Rolfo: Supervision; Writing – review & editing.
Simona Taverna: Conceptualization; Data curation; Writing – original draft; Writing – review & editing.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
C. R. declares conflict of interest, he is a speaker for Roche, Guardant health, MSD and Astra Zeneca, advisory board for ARCHER, Novartis, Nivata, BMS, Boston Pharmaceutical, EMD Serono, Pfizer. C.R. has research collaboration with LC Research Foundation-Pfizer Grant. All other authors have no conflicts of interest to declare.
Availability of data and materials: Not Applicable.
References
- 1. Thakur SK, Singh DP, Choudhary J. Lung cancer identification: a review on detection and classification. Cancer Metastasis Rev 2020; 39: 989–998. [DOI] [PubMed] [Google Scholar]
- 2. Rijavec E, Coco S, Genova C, et al. Liquid biopsy in non-small cell lung cancer: highlights and challenges. Cancers (Basel) 2019; 12: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rolfo C, Mack P, Scagliotti GV, et al. Liquid biopsy for advanced NSCLC: a consensus statement from the international association for the study of lung cancer. J Thorac Oncol Off Publ Int Assoc Study Lung Cancer 2021; 16: 1647–1662. [DOI] [PubMed] [Google Scholar]
- 4. Raman V, Yang C-FJ, Deng JZ, et al. Surgical treatment for early stage non-small cell lung cancer. J Thorac Dis 2018; 10: S898–S904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gridelli C, Rossi A, Carbone DP, et al. Non-small-cell lung cancer. Nat Rev Dis Prim 2015; 1: 15009. [DOI] [PubMed] [Google Scholar]
- 6. Taverna S, Giallombardo M, Gil-Bazo I, et al. Exosomes isolation and characterization in serum is feasible in non-small cell lung cancer patients: critical analysis of evidence and potential role in clinical practice. Oncotarget 2016; 7: 28748–28760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kerk SA, Papagiannakopoulos T, Shah YM, et al. Metabolic networks in mutant KRAS-driven tumours: tissue specificities and the microenvironment. Nat Rev Cancer 2021; 21: 510–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Román M, López I, Guruceaga E, et al. Inhibitor of differentiation-1 sustains mutant KRAS-driven progression, maintenance, and metastasis of lung adenocarcinoma via regulation of a FOSL1 network. Cancer Res 2019; 79: 625–638. [DOI] [PubMed] [Google Scholar]
- 9. Masuda K, Horinouchi H, Tanaka M, et al. With an EGFR mutation and high PD-L1 expression. J Cancer Res Clin Oncol 2021; 147: 245–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhou Y, Guo D, Zhang Y. Association of MicroRNA-21 with P53 at mutant sites R175H and R248Q, clinicopathological features, and prognosis of NSCLC. Mol Ther oncolytics 2020; 19: 208–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wheeler DA, Wang L. From human genome to cancer genome: the first decade. Genome Res 2013; 23: 1054–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lindeman NI, Cagle PT, Aisner DL, et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: guideline from the college of American pathologists, the international association for the study of lung cancer, and the association for molecular pathology. J Thorac Oncol Off Publ Int Assoc Study Lung Cancer 2018; 13: 323–358. [DOI] [PubMed] [Google Scholar]
- 13. Russo A, Cardona AF, Caglevic C, et al. Overcoming TKI resistance in fusion-driven NSCLC: new generation inhibitors and rationale for combination strategies. Transl Lung Cancer Res 2020; 9: 2581–2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Szilágyi M, Pös O, Márton É, et al. Circulating cell-free nucleic acids: main characteristics and clinical application. Int J Mol Sci 2020; 21: 6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pisapia P, Costa JL, Pepe F, et al. Next generation sequencing for liquid biopsy based testing in non-small cell lung cancer in 2021. Crit Rev Oncol Hematol 2021; 161: 103311. [DOI] [PubMed] [Google Scholar]
- 16. Connors D, Allen J, Alvarez JD, et al. International liquid biopsy standardization alliance white paper. Crit Rev Oncol Hematol 2020; 156: 103112. [DOI] [PubMed] [Google Scholar]
- 17. Rolfo C, Castiglia M, Hong D, et al. Liquid biopsies in lung cancer: the new ambrosia of researchers. Biochim Biophys Acta 2014; 1846: 539–546. [DOI] [PubMed] [Google Scholar]
- 18. Li W, Liu J-B, Hou L-K, et al. Liquid biopsy in lung cancer: significance in diagnostics, prediction, and treatment monitoring. Mol Cancer 2022; 21: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lin D, Shen L, Luo M, et al. Circulating tumor cells: biology and clinical significance. Signal Transduct Target Ther 2021; 6: 404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lyu M, Zhou J, Ning K, et al. The diagnostic value of circulating tumor cells and CtDNA for gene mutations in lung cancer. Onco Targets Ther 2019; 12: 2539–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tong B, Xu Y, Zhao J, et al. Prognostic role of circulating tumor cells in patients with EGFR-mutated or ALK-rearranged non-small cell lung cancer. Thorac Cancer 2018; 9; 640–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rolfo C, Russo A. Liquid biopsy for early stage lung cancer moves ever closer. Nat Rev Clin Oncol 2020; 17: 523–524. [DOI] [PubMed] [Google Scholar]
- 23. Herath S, Sadeghi Rad H, Radfar P, et al. The role of circulating biomarkers in lung cancer. Front Oncol 2021; 11: 801269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chabon JJ, Hamilton EG, Kurtz DM, et al. Integrating genomic features for non-invasive early lung cancer detection. Nature 2020; 580: 245–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu J, Hu S, Zhang L, et al. Tumor circulome in the liquid biopsies for cancer diagnosis and prognosis. Theranostics 2020; 10: 4544–4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Reclusa P, Taverna S, Pucci M, et al. Exosomes as diagnostic and predictive biomarkers in lung cancer. J Thorac Dis 2017; 9: S1373–S1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cocozza F, Grisard E, Martin-Jaular L, et al. SnapShot: extracellular vesicles. Cell 2020; 182: 262.e1–262.e1. [DOI] [PubMed] [Google Scholar]
- 28. Ragni E, Banfi F, Barilani M, et al. Extracellular vesicle-shuttled MRNA in mesenchymal stem cell communication. Stem Cells 2017; 35: 1093–1105. [DOI] [PubMed] [Google Scholar]
- 29. Xu R, Rai A, Chen M, et al. Extracellular vesicles in cancer - implications for future improvements in cancer care. Nat Rev Clin Oncol 2018; 15: 617–638. [DOI] [PubMed] [Google Scholar]
- 30. Galvano A, Taverna S, Badalamenti G, et al. Detection of RAS mutations in circulating tumor DNA: a new weapon in an old war against colorectal cancer. A systematic review of literature and meta-analysis. Ther Adv Med Oncol 2019; 11: 1758835919874653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rigogliuso S, Donati C, Cassarà D, et al. An active form of sphingosine kinase-1 is released in the extracellular medium as component of membrane vesicles shed by two human tumor cell lines. J Oncol 2010; 2010: 509329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol 2018; 19: 213–228. [DOI] [PubMed] [Google Scholar]
- 33. Ratajczak MZ, Ratajczak J. Extracellular microvesicles/exosomes: discovery, disbelief, acceptance, and the future? Leukemia 2020; 34: 3126–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pucci M, Reclusa Asiain P, Durendez Saez E, et al. Extracellular vesicles as mirna nano-shuttles: dual role in tumor progression. Target Oncol 2018; 13: 175–187. [DOI] [PubMed] [Google Scholar]
- 35. Taverna S, Giusti I, D’Ascenzo S, et al. Breast cancer derived extracellular vesicles in bone metastasis induction and their clinical implications as biomarkers. Int J Mol Sci 2020; 21: 3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Heusermann W, Hean J, Trojer D, et al. Exosomes surf on filopodia to enter cells at endocytic hot spots, traffic within endosomes, and are targeted to the ER. J Cell Biol 2016; 213: 173–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Reclusa P, Verstraelen P, Taverna S, et al. Improving extracellular vesicles visualization: from static to motion. Sci Rep 2020; 10: 6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mineo M, Garfield SH, Taverna S, et al. Exosomes released by K562 chronic myeloid leukemia cells promote angiogenesis in a Src-dependent fashion. Angiogenesis 2012; 15: 33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Maas SLN, Breakefield XO, Weaver AM. Extracellular vesicles: unique intercellular delivery vehicles. Trends Cell Biol 2017; 27: 172–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu P, Zhang B, Ocansey DKW, et al. Extracellular vesicles: a bright star of nanomedicine. Biomaterials 2021; 269: 120467. [DOI] [PubMed] [Google Scholar]
- 41. Passiglia F, Caglevic C, Giovannetti E, et al. Primary and metastatic brain cancer genomics and emerging biomarkers for immunomodulatory cancer treatment. Semin Cancer Biol 2018; 52: 259–268. [DOI] [PubMed] [Google Scholar]
- 42. Monteleone F, Taverna S, Alessandro R, et al. SWATH-MS based quantitative proteomics analysis reveals that curcumin alters the metabolic enzyme profile of CML cells by affecting the activity of MiR-22/IPO7/HIF-1alpha axis. J Exp Clin Cancer Res 2018; 37: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fontana S, Saieva L, Taverna S, et al. Contribution of proteomics to understanding the role of tumor-derived exosomes in cancer progression: state of the art and new perspectives. Proteomics 2013; 13: 1581–1594. [DOI] [PubMed] [Google Scholar]
- 44. Pucci M, Taverna S, Reclusa P, et al. Exosomes in semen: opportunities as a new tool in prostate cancer diagnosis. Transl Cancer Res 2017; 6: S1331–S1338. [Google Scholar]
- 45. de Miguel Pérez D, Rodriguez Martínez A, Ortigosa Palomo A, et al. Extracellular vesicle-MiRNAs as liquid biopsy biomarkers for disease identification and prognosis in metastatic colorectal cancer patients. Sci Rep 2020; 10: 3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wortzel I, Dror S, Kenific CM, et al. Exosome-mediated metastasis: communication from a distance. Dev Cell 2019; 49: 347–360. [DOI] [PubMed] [Google Scholar]
- 47. Taverna S, Pucci M, Giallombardo M, et al. Amphiregulin contained in NSCLC-exosomes induces osteoclast differentiation through the activation of EGFR pathway. Sci Rep 2017; 7: 3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science 2020, 367: eaau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Giallombardo M, Taverna S, Alessandro R, et al. Exosome-mediated drug resistance in cancer: the near future is here. Ther Adv Med Oncol 2016; 8: 320–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Taghvimi S, Vakili O, Soltani Fard E, et al. Exosomal microRNAs and long noncoding RNAs: novel mediators of drug resistance in lung cancer. J Cell Physiol 2022; 237: 2095–2106. [DOI] [PubMed] [Google Scholar]
- 51. Zhou E, Li Y, Wu F, et al. Circulating extracellular vesicles are effective biomarkers for predicting response to cancer therapy. EBioMedicine 2021; 67: 103365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shukuya T, Ghai V, Amann JM, et al. Circulating microRNAs and extracellular vesicle-containing microRNAs as response biomarkers of anti-programmed cell death protein 1 or programmed death-ligand 1 therapy in NSCLC. J Thorac Oncol Off Publ Int Assoc Study Lung Cancer 2020; 15: 1773–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. de Miguel-Perez D, Russo A, Arrieta O, et al. Extracellular vesicle PD-L1 dynamics predict durable response to immune-checkpoint inhibitors and survival in patients with non-small cell lung cancer. J Exp Clin Cancer Res 2022; 41: 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Qiu Y, Li P, Zhang Z, et al. Insights into exosomal non-coding RNAs sorting mechanism and clinical application. Front Oncol 2021; 11: 664904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cammarata G, Barraco N, Giusti I, et al. Extracellular vesicles-CeRNAs as ovarian cancer biomarkers: looking into CircRNA-MiRNA-MRNA code. Cancers (Basel) 2022; 14: 3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Castellanos-Rizaldos E, Grimm DG, Tadigotla V, et al. Exosome-based detection of EGFR T790M in plasma from non-small cell lung cancer patients. Clin Cancer Res 2018; 24: 2944–2950. [DOI] [PubMed] [Google Scholar]
- 57. Park J, Lee C, Eom JS, et al. Detection of EGFR mutations using bronchial washing-derived extracellular vesicles in patients with non-small-cell lung carcinoma. Cancers (Basel) 2020; 12: 2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pasini L, Notarangelo M, Vagheggini A, et al. Unveiling mutational dynamics in non-small cell lung cancer patients by quantitative EGFR profiling in vesicular RNA. Mol Oncol 2021; 15: 2423–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Purcell E, Owen S, Prantzalos E, et al. Epidermal growth factor receptor mutations carried in extracellular vesicle-derived cargo mirror disease status in metastatic non-small cell lung cancer. Front cell Dev Biol 2021; 9: 724389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li J, Liu C. Coding or noncoding, the converging concepts of RNAs. Front Genet 2019; 10: 496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mattick JS. Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep 2001; 2: 986–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Morceau F, Chateauvieux S, Gaigneaux A, et al. Long and short non-coding RNAs as regulators of hematopoietic differentiation. Int J Mol Sci 2013; 14: 14744–14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet 2006; 15: R17–R29. [DOI] [PubMed] [Google Scholar]
- 64. Sansone P, Savini C, Kurelac I, et al. Packaging and transfer of mitochondrial DNA via exosomes regulate escape from dormancy in hormonal therapy-resistant breast cancer. Proc Natl Acad Sci U. S. A. 2017; 114: E9066–E9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014; 157: 77–94. [DOI] [PubMed] [Google Scholar]
- 66. Peschansky VJ, Wahlestedt C. Non-coding RNAS as direct and indirect modulators of epigenetic regulation. Epigenetics 2014; 9: 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res 2007; 17: 556–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hombach S, Kretz M. Non-coding RNAs: classification, biology and functioning. Adv Exp Med Biol 2016; 937: 3–17. [DOI] [PubMed] [Google Scholar]
- 69. Slack FJ, Chinnaiyan AM. The role of non-coding RNAs in oncology. Cell 2019; 179: 1033–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer 2018; 18: 5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhang N, Nan A, Chen L, et al. Circular RNA circSATB2 promotes progression of non-small cell lung cancer cells. Mol Cancer 2020; 19: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rosace D, López J, Blanco S. Emerging roles of novel small non-coding regulatory RNAs in immunity and cancer. RNA Biol 2020; 17: 1196–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Huang S-Q, Sun B, Xiong Z-P, et al. The dysregulation of TRNAs and TRNA derivatives in cancer. J Exp Clin Cancer Res 2018; 37: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Shao Y, Sun Q, Liu X, et al. TRF-leu-CAG promotes cell proliferation and cell cycle in non-small cell lung cancer. Chem Biol Drug Des 2017; 90: 730–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Weng Q, Wang Y, Xie Y, et al. Extracellular vesicles-associated TRNA-derived fragments (TRFs): biogenesis, biological functions, and their role as potential biomarkers in human diseases. J Mol Med (Berl) 2022; 100: 679–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Jenjaroenpun P, Kremenska Y, Nair VM, et al. Characterization of RNA in exosomes secreted by human breast cancer cell lines using next-generation sequencing. PeerJ 2013; 1: e201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Abramowicz A, Story MD. The long and short of it: the emerging roles of non-coding RNA in small extracellular vesicles. Cancers (Basel) 2020; 12: 1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Taby R, Issa J-PJ. Cancer epigenetics. CA Cancer J Clin 2010; 60: 376–392. [DOI] [PubMed] [Google Scholar]
- 79. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013; 495: 333–338. [DOI] [PubMed] [Google Scholar]
- 80. Zhang Z, Yang T, Xiao J. Circular RNAs: promising biomarkers for human diseases. EBioMedicine 2018; 34: 267–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Dragomir M, Calin GA. Circular RNAs in cancer - lessons learned from microRNAs. Front Oncol 2018; 8: 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Nigro JM, Cho KR, Fearon ER, et al. Scrambled exons. Cell 1991; 64: 607–613. [DOI] [PubMed] [Google Scholar]
- 83. Li J, Yang J, Zhou P, et al. Circular RNAs in cancer: novel insights into origins, properties, functions and implications. Am J Cancer Res 2015; 5: 472–480. [PMC free article] [PubMed] [Google Scholar]
- 84. Wang S, Zhang K, Tan S, et al. Circular RNAs in body fluids as cancer biomarkers: the new frontier of liquid biopsies. Mol Cancer 2021; 20: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Chen L-L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol 2020; 21: 475–490. [DOI] [PubMed] [Google Scholar]
- 86. Fanale D, Taverna S, Russo A, et al. Circular RNA in exosomes. Adv Exp Med Biol 2018; 1087: 109–117. [DOI] [PubMed] [Google Scholar]
- 87. Zhang X, Yang D, Wei Y. Overexpressed CDR1as functions as an oncogene to promote the tumor progression via MiR-7 in non-small-cell lung cancer. Onco Targets Ther 2018; 11: 3979–3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. de Fraipont F, Gazzeri S, Cho WC, et al. Circular RNAs and RNA splice variants as biomarkers for prognosis and therapeutic response in the liquid biopsies of lung cancer patients. Front Genet 2019; 10: 390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Chi Y, Luo Q, Song Y, et al. Circular RNA CircPIP5K1A promotes non-small cell lung cancer proliferation and metastasis through MiR-600/HIF-1α regulation. J Cell Biochem 2019; 120: 19019–19030. [DOI] [PubMed] [Google Scholar]
- 90. Li C, Zhang L, Meng G, et al. Circular RNAs: pivotal molecular regulators and novel diagnostic and prognostic biomarkers in non-small cell lung cancer. J Cancer Res Clin Oncol 2019; 145: 2875–2889. [DOI] [PubMed] [Google Scholar]
- 91. Qiu M, Xia W, Chen R, et al. The circular RNA CircPRKCI promotes tumor growth in lung adenocarcinoma. Cancer Res 2018; 78: 2839–2851. [DOI] [PubMed] [Google Scholar]
- 92. Li L, Li W, Chen N, et al. FLI1 exonic circular RNAs as a novel oncogenic driver to promote tumor metastasis in small cell lung cancer. Clin Cancer Res an Off J Am Assoc Cancer Res 2019; 25: 1302–1317. [DOI] [PubMed] [Google Scholar]
- 93. Zhu X, Han J, Lan H, et al. A novel circular RNA Hsa_circRNA_103809/MiR-377-3p/GOT1 pathway regulates cisplatin-resistance in non-small cell lung cancer (NSCLC). BMC Cancer 2020; 20: 1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Wang J, Zhao X, Wang Y, et al. CircRNA-002178 act as a CeRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis 2020; 11: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Hong W, Xue M, Jiang J, et al. Circular RNA circ-CPA4/ Let-7 MiRNA/PD-L1 axis regulates cell growth, stemness, drug resistance and immune evasion in non-small cell lung cancer (NSCLC). J Exp Clin Cancer Res 2020; 39: 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Yang B, Teng F, Chang L, et al. Tumor-derived exosomal CircRNA_102481 contributes to EGFR-TKIs resistance via the MiR-30a-5p/ROR1 axis in non-small cell lung cancer. Aging (Albany. NY) 2021; 13: 13264–13286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Huang L, Rong Y, Tang X, et al. Circular RNAs are promising biomarkers in liquid biopsy for the diagnosis of non-small cell lung cancer. Front Mol Biosci 2021; 8: 625722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Qiao D, Hu C, Li Q, et al. Circ-RBMS1 knockdown alleviates CSE-induced apoptosis, inflammation and oxidative stress via up-regulating FBXO11 through MiR-197-3p in 16HBE cells. Int J Chron Obstruct Pulmon Dis 2021; 16: 2105–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Osei ET, Florez-Sampedro L, Timens W, et al. Unravelling the complexity of COPD by MicroRNAs: it’s a small world after all. Eur Respir J 2015; 46: 807–818. [DOI] [PubMed] [Google Scholar]
- 100. Chen T, Liu Y, Li C, et al. Tumor-derived exosomal CircFARSA mediates M2 macrophage polarization via the PTEN/PI3K/AKT pathway to promote non-small cell lung cancer metastasis. Cancer Treat Res Commun 2021; 28: 100412. [DOI] [PubMed] [Google Scholar]
- 101. Köhler J, Schuler M, Gauler TC, et al. Circulating U2 small nuclear RNA fragments as a diagnostic and prognostic biomarker in lung cancer patients. J Cancer Res Clin Oncol 2016; 142: 795–805. [DOI] [PubMed] [Google Scholar]
- 102. Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol 2007; 8: 209–220. [DOI] [PubMed] [Google Scholar]
- 103. Valadkhan S, Gunawardane LS. Role of small nuclear RNAs in eukaryotic gene expression. Essays Biochem 2013; 54: 79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Baraniskin A, Zaslavska E, Nöpel-Dünnebacke S, et al. Circulating U2 small nuclear RNA fragments as a novel diagnostic biomarker for primary central nervous system lymphoma. Neuro Oncol 2016; 18: 361–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Kuhlmann JD, Baraniskin A, Hahn SA, et al. Circulating U2 small nuclear RNA fragments as a novel diagnostic tool for patients with epithelial ovarian cancer. Clin Chem 2014; 60: 206–213. [DOI] [PubMed] [Google Scholar]
- 106. Baraniskin A, Nöpel-Dünnebacke S, Ahrens M, et al. Circulating U2 small nuclear RNA fragments as a novel diagnostic biomarker for pancreatic and colorectal adenocarcinoma. Int J Cancer 2013; 132: E48–E57. [DOI] [PubMed] [Google Scholar]
- 107. Kuhlmann JD, Wimberger P, Wilsch K, et al. Increased level of circulating U2 small nuclear RNA fragments indicates metastasis in melanoma patients. Clin Chem Lab Med 2015; 53: 605–611. [DOI] [PubMed] [Google Scholar]
- 108. Mazières J, Catherinne C, Delfour O, et al. Alternative processing of the U2 small nuclear RNA produces a 19-22nt fragment with relevance for the detection of non-small cell lung cancer in human serum. PLoS One 2013; 8: e60134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Dong X, Ding S, Yu M, et al. Small nuclear RNAs (U1, U2, U5) in tumor-educated platelets are downregulated and act as promising biomarkers in lung cancer. Front Oncol 2020; 10: 1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Cui C, Liu Y, Gerloff D, et al. NOP10 predicts lung cancer prognosis and its associated small nucleolar RNAs drive proliferation and migration. Oncogene 2021; 40: 909–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Liang J, Wen J, Huang Z, et al. Small nucleolar RNAs: insight into their function in cancer. Front Oncol 2019; 9: 587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Abel Y, Rederstorff M. SnoRNAs and the emerging class of SdRNAs: multifaceted players in oncogenesis. Biochimie 2019; 164: 17–21. [DOI] [PubMed] [Google Scholar]
- 113. Wang G, Li J, Yao Y, et al. Small nucleolar RNA 42 promotes the growth of hepatocellular carcinoma through the P53 signaling pathway. Cell death Discov 2021; 7: 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Zheng D, Zhang J, Ni J, et al. Small nucleolar RNA 78 promotes the tumorigenesis in non-small cell lung cancer. J Exp Clin Cancer Res 2015; 34: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Braicu C, Zimta A-A, Harangus A, et al. The function of non-coding RNAs in lung cancer tumorigenesis. Cancers (Basel) 2019; 11: 605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Gong J, Li Y, Liu C-J, et al. A pan-cancer analysis of the expression and clinical relevance of small nucleolar RNAs in human cancer. Cell Rep 2017; 21: 1968–1981. [DOI] [PubMed] [Google Scholar]
- 117. Gao L, Ma J, Mannoor K, et al. Genome-wide small nucleolar RNA expression analysis of lung cancer by next-generation deep sequencing. Int J Cancer 2015; 136: E623–E629. [DOI] [PubMed] [Google Scholar]
- 118. Su Y, Guarnera MA, Fang H, et al. Small non-coding RNA biomarkers in sputum for lung cancer diagnosis. Mol Cancer 2016; 15: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Kitagawa T, Taniuchi K, Tsuboi M, et al. Circulating pancreatic cancer exosomal RNAs for detection of pancreatic cancer. Mol Oncol 2019; 13: 212–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Ozata DM, Gainetdinov I, Zoch A, et al. PIWI-interacting RNAs: small RNAs with big functions. Nat Rev Genet 2019; 20: 89–108. [DOI] [PubMed] [Google Scholar]
- 121. Tóth KF, Pezic D, Stuwe E, et al. The PiRNA pathway guards the germline genome against transposable elements. Adv Exp Med Biol 2016; 886: 51–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 2009; 10: 126–139. [DOI] [PubMed] [Google Scholar]
- 123. Siddiqi S, Matushansky I. Piwis and piwi-interacting RNAs in the epigenetics of cancer. J Cell Biochem 2012; 113: 373–380. [DOI] [PubMed] [Google Scholar]
- 124. Zimta A-A, Sigurjonsson OE, Gulei D, et al. The malignant role of exosomes as nanocarriers of rare RNA species. Int J Mol Sci 2020; 21: 5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Jing Z, Xi Y, Yin J, et al. Biological roles of PiRNAs in colorectal cancer. Gene 2021; 769: 145063. [DOI] [PubMed] [Google Scholar]
- 126. Weng W, Li H, Goel A. Piwi-interacting RNAs (PiRNAs) and cancer: emerging biological concepts and potential clinical implications. Biochim Biophys Acta Rev Cancer 2019; 1871: 160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Cheng Y, Wang Q, Jiang W, et al. Emerging roles of PiRNAs in cancer: challenges and prospects. Aging (Albany. NY). 2019; 11: 9932–9946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Sabbah NA, Abdalla WM, Mawla WA, et al. PiRNA-823 is a unique potential diagnostic non-invasive biomarker in colorectal cancer patients. Genes (Basel) 2021; 12: 598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Feng J, Yang M, Wei Q, et al. Novel evidence for oncogenic PiRNA-823 as a promising prognostic biomarker and a potential therapeutic target in colorectal cancer. J Cell Mol Med 2020; 24: 9028–9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Mai D, Zheng Y, Guo H, et al. Serum PiRNA-54265 is a new biomarker for early detection and clinical surveillance of human colorectal cancer. Theranostics 2020; 10: 8468–8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Yan H, Wu Q-L, Sun C-Y, et al. PiRNA-823 contributes to tumorigenesis by regulating de novo DNA methylation and angiogenesis in multiple myeloma. Leukemia 2015; 29: 196–206. [DOI] [PubMed] [Google Scholar]
- 132. Cordeiro A, Navarro A, Gaya A, et al. PiwiRNA-651 as marker of treatment response and survival in classical hodgkin lymphoma. Oncotarget 2016; 7: 46002–46013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Peng Q, Chiu PK-F, Wong CY-P, et al. Identification of PiRNA targets in urinary extracellular vesicles for the diagnosis of prostate cancer. Diagnostics (Basel, Switzerland) 2021; 11: 1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Liu Y, Dong Y, He X, et al. PiR-Hsa-211106 inhibits the progression of lung adenocarcinoma through pyruvate carboxylase and enhances chemotherapy sensitivity. Front Oncol 2021; 11: 651915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Li J, Wang N, Zhang F, et al. PIWI-interacting RNAs are aberrantly expressed and may serve as novel biomarkers for diagnosis of lung adenocarcinoma. Thorac Cancer 2021; 12: 2468–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Haranguş A, Berindan-Neagoe I, Todea DA, et al. Noncoding RNAs and liquid biopsy in lung cancer: a literature review. Diagnostics (Basel, Switzerland) 2019; 9: 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Ma S, Kong S, Wang F, et al. CircRNAs: biogenesis, functions, and role in drug-resistant tumours. Mol Cancer 2020; 19: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Xu Y, Kong S, Qin S, et al. Exosomal CircRNAs: sorting mechanisms, roles and clinical applications in tumors. Front Cell Dev Biol 2020; 8: 581558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Wang C, Tan S, Li J, et al. CircRNAs in lung cancer - biogenesis, function and clinical implication. Cancer Lett 2020; 492: 106–115. [DOI] [PubMed] [Google Scholar]
- 140. Xiao L, Wang J, Ju S, et al. Disorders and roles of TsRNA, SnoRNA, SnRNA and PiRNA in cancer. J Med Genet 2022. [DOI] [PubMed] [Google Scholar]
- 141. Toden S, Zumwalt TJ, Goel A. Non-coding RNAs and potential therapeutic targeting in cancer. Biochim Biophys Acta Rev Cancer 2021; 1875: 188491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Li C, Qin F, Hu F, et al. Characterization and selective incorporation of small non-coding RNAs in non-small cell lung cancer extracellular vesicles. Cell Biosci 2018; 8: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Mourksi N-E-H, Morin C, Fenouil T, et al. SnoRNAs offer novel insight and promising perspectives for lung cancer understanding and management. Cells 2020; 9: 541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Palanca-Ballester C, Rodriguez-Casanova A, Torres S, et al. Cancer epigenetic biomarkers in liquid biopsy for high incidence malignancies. Cancers (Basel) 2021; 13: 3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Su C, Han Y, Zhang H, et al. CiRS-7 targeting MiR-7 modulates the progression of non-small cell lung cancer in a manner dependent on NF-KB signalling. J Cell Mol Med 2018; 22: 3097–3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Zhang S, Zeng X, Ding T, et al. Microarray profile of circular RNAs identifies Hsa_circ_0014130 as a new circular RNA biomarker in non-small cell lung cancer. Sci Rep 2018; 8: 2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Meng Y, Sun W-L, Wang M, et al. Circular RNA Circ-prkci promotes lung cancer progression by binding to MicroRNA-1324 to regulate MECP2 expression. Eur Rev Med Pharmacol Sci 2020; 24: 10557–10565. [DOI] [PubMed] [Google Scholar]
- 148. Mei Y-P, Liao J-P, Shen J, et al. Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis. Oncogene 2012; 31: 2794–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]