Abstract
Vertebrate and invertebrate models of neurodegenerative diseases, such as Alzheimer’s disease, Parkinson’s disease, and amyotrophic lateral sclerosis, have been paramount to our understanding of the pathophysiology of these conditions; however, the brain epigenetic landscape is less well established in these disease models. DNA methylation, histone modifications, and microRNAs are among commonly studied mechanisms of epigenetic regulation. Genome-wide studies and candidate studies of specific methylation marks, histone marks, and microRNAs have demonstrated the dysregulation of these mechanisms in models of neurodegenerative diseases; however, the studies to date are scarce and inconclusive and the implications of many of these changes are still not fully understood. In this review, we summarize epigenetic changes reported to date in the brain of vertebrate and invertebrate models used to study neurodegenerative diseases, specifically diseases affecting the aging population. We also discuss caveats of epigenetic research so far and the use of disease models to understand neurodegenerative diseases, with the aim of improving the use of model organisms in this context in future studies.
Keywords: Neurodegenerative diseases, neurodegeneration, model organisms, neuroepigenetics, DNA methylation, histone modifications, miRNA, Alzheimer’s disease, Parkinson’s disease, amyotrophic lateral sclerosis
Introduction
Neurodegenerative diseases comprise a group of chronic neurological disorders characterized by progressive functional and structural neuronal deterioration, ultimately resulting in death.1 Some of the most common neurodegenerative diseases causing increasing morbidity and mortality in the aging population are Alzheimer’s disease (AD),2 Parkinson’s disease (PD)3 and amyotrophic lateral sclerosis (ALS).4 Model organisms displaying aspects of these diseases have enabled the understanding of some of the molecular mechanisms underpinning their pathologic etiologies as well as the course of neurodegenerative processes.5 Increasing research has shown changes in genomic regulatory markers and machinery in human post-mortem brains from people with these conditions, which may play a role in disease pathogenesis; however, genomic changes are less well established in model organisms that offer the advantage of investigating early stages of disease as opposed to post-mortem end stages, as well as manipulating therapeutic targets.6-8 This review aims to explore genomic regulatory changes reported to date in the brain in model organisms used to study neurodegenerative diseases, and address caveats of their use in neuroepigenetic research, in order to promote better and a more targeted use of disease models.
Pathophysiology, Etiology, and Genetics of Neurodegenerative Diseases
AD, PD, and ALS are each characterized by specific pathological brain features that can only be definitively confirmed post-mortem. These hallmark features include amyloid-β (Aβ) plaques and tau neurofibrillary tangles in AD, α-synuclein aggregates in PD, and inclusions of TAR DNA-binding protein-43 (TDP-43) in ALS. The progression of these individual pathologies is disease-specific, and each can be found in various different regions of the brain; however, regions primarily affected by these pathological features, and hence commonly studied in disease models, include the hippocampus and the cerebral cortex in AD, the substantia nigra in PD, and the motor cortex, brainstem, and spinal cord in ALS.1
The causes of these neurodegenerative conditions are both familial and sporadic in nature. Familial forms are often early-onset (<65 years) due to genetic predisposition. For example, autosomal dominant mutations in the amyloid precursor protein (APP), presenilin 1 (PSEN1) or presenilin 2 (PSEN2) genes result into familial AD; mutations in the genes encoding α-synuclein (SNCA) or leucine rich repeat kinase 2 (LRRK2) cause autosomal-dominant PD; mutations in the superoxide dismutase 1 (SOD1) gene result into familial ALS.1,9 Notably, these genetic mutations can be introduced in animals to model disease phenotypes, which will be discussed later in this review.
The aforementioned autosomal dominant inheritance accounts for a minor proportion of disease incidence for AD, PD and ALS, however; the vast majority of cases are late-onset (>65 years) and sporadic.10 Nevertheless, the existence of a genetic component contributing to sporadic forms is widely accepted given the scientific evidence of recent years, especially propelled by genome-wide association studies that have reported how genetic variation may increase (or decrease) disease susceptibility.1,10 Additionally, studies investigating monozygotic twins (ie, individuals that share the same genetic information) have shown discordance in the onset and progression of these conditions,11-15 suggesting that the genetic contribution to disease etiology and progression goes beyond the genetic sequence, likely including epigenetic mechanisms involved in genomic regulation and expression. DNA methylation, histone modifications and non-coding RNAs are among the most commonly recognized epigenetic mechanisms (Figure 1) associated with enhancing or silencing gene expression, and have been shown to play crucial roles in neurogenesis and early brain development.16 Importantly, changes to these regulatory mechanisms and their associated molecular machinery are observed in post-mortem brain tissue from people exhibiting neurodegenerative diseases, as reviewed by others.6-8
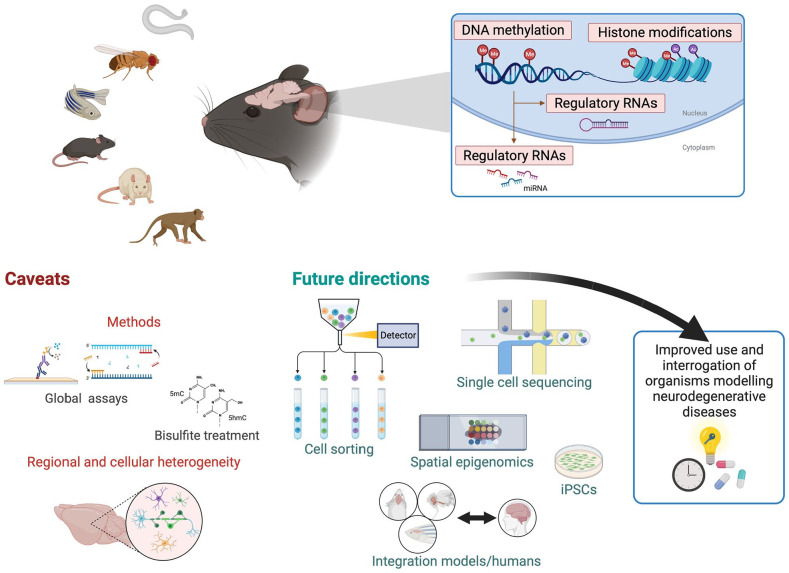
Figure 1.
Vertebrate and invertebrate models useful for epigenetics research of neurodegerative diseases, epigenetic marks investigated to date, common caveats, and future directions to improve the use and benefits of these powerful models. Model organisms used to study neuroepigenetics of neurodegenerative diseases (top left) range from simpler organisms such as nematodes (Caenorhabditis elegans), fruit fly (Drosophila melanogaster) and zebra fish (Danio rerio), rodents (mice and rats), and non-human primates. DNA methylation, histone modifications and regulatory RNAs (predominantly miRNAs) comprise epigenetic processes that have been studied in these models (top right). Caveats of studies to date (bottom left) include methods commonly employed, many assessing global levels of epigenetic modifications (global assays), and—in the case of DNA methylation—the vast majority relying in bisulfite conversion, which does not allow to differentiate between 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC). Of great importance in epigenetics research, regional and cellular heterogeneity have predominantly not been taken into account and/or explored. Future research (bottom right) must mitigate limitations of studies thus far by discerning tissue- and cell-specific signatures, take advantage of immerging single-cell and spatial technologies, integrate findings from model organisms with human studies, and consider additional strategies such as the parallel use of iPSCs. Taken together with the development of better disease models currently underway, future research should aim to improve the use of model organisms for the understanding of epigenetic processes in neurogenerative conditions, and of how and when we should modify and manipulate aspects of human disease, particularly important for drug discovery and testing.
Created with BioRender.com.
Research investigating the role of epigenetic processes in the context of neurodegenerative diseases has exponentially increased in recent years, aiming to understand its implications, and particularly pushed forward with the advancement of technologies facilitating the study of epigenetic changes.
Vertebrate and Invertebrate Models in Neuroscience Research
Vertebrate and invertebrate organisms are distinguished by the presence and absence of a vertebral column, respectively. They are characterized by unique genetic makeups, which give rise to their individual embryonic and developmental processes. Mutual molecular, biological and genomic features between these species exist, however, that make them valuable tools for neuroscience research, including for the study of human neurodegenerative diseases.17 There are currently no animal species that can precisely model the complexity of the human nervous system nor its associated disease states; nevertheless, animal models recapitulating aspects of neurodegenerative disease have been developed, allowing to investigate molecular, biological, and pathological mechanisms, with potential translation to humans.
Several limitations and ethical issues make carrying out brain research in humans very challenging. Human brain tissue used to study neurodegenerative diseases is usually collected post-mortem, often presenting significant brain damage as a consequence of late stages of disease, making it difficult to get a perspective that goes beyond the final stages of disease and understand causal factors and changes that occurred over disease course. The use of animal models hence helps overcoming these challenges by providing more flexibility and facilitating the study of early and progressive changes.
Rodents (eg, mice and rats) and non-human primates are the most popularly used vertebrates to model neurodegenerative diseases. Mice in particular can be easily genetically modified to express disease-specific genetic mutations, thereby phenocopying familial forms and exhibiting disease-associated pathologies. Genetic manipulation can also be performed on rats but to a lesser extent due to reduced feasibility. In addition, rodents can also be chemically induced to recapitulate disease, for example through injections of artificial pathological proteins or extracts from post-mortem human brain tissue. This type of approach results in cognitive impairment in rodent models, consistent with functional impairments seen in the neurodegenerative human brain. More recently, spontaneous models have also been described, where pathology is driven primarily by aging, constituting a promising strategy to recapitulate sporadic forms of disease.18 For instance, the senescence-accelerated mouse prone 8 (SAMP8) model is used to model sporadic AD and is driven primarily by accelerated aging. The model exhibits Aβ and tau pathology as well as cognitive impairments, characteristic of AD.19 Table 1 lists the mouse models of AD reported in this review. Non-human primates, such as monkeys, have many similarities with humans due to their biological proximity, behavioral complexity, and the development of natural neurodegenerative pathology, such as the accumulation of Aβ plaques. This makes them an advantageous model; however, their long lifespan and ethical concerns restrict their use for the study of neurodegenerative diseases.17
Table 1.
Mouse models of Alzheimer’s disease reported in this review.
| Model | Description |
|---|---|
| 3xTg-AD | APP Swedish, MAPT P301L, PSEN1 M146V mutations20 |
| 5xFAD | APP Swedish, Florida, and London mutations, and PSEN1 M146L and L286V mutations21 |
| APPNL-F | APP Swedish and Iberian mutations22 |
| APPNL-G-F | APP Swedish, Arctic, and Iberian mutations22 |
| APPSWE, IND | APP Swedish and Indiana mutations (under Thy1 promoter)23 |
| APP/PS1 | APP Sweden and PSEN1 ΔE9 mutations24 |
| APP/PS1-21 | APP Swedish and PSEN1 L166P mutations25 |
| APP23 | APP Swedish double mutations (K651M and N652L)26 |
| CK-p25 | Overexpression of p25 (a regulator of cyclin-dependent kinase)27 |
| J20 | APP Swedish and Indiana mutations (under PDGF-β promoter)28 |
| OA42i | Oligomeric amyloid β1–42 plus ibotenic acid (oA42i) induced mouse29 |
| P301S | MAPT P301S mutation30 |
| PSAPP | APP Swedish and PSEN1 M146L mutations31 |
| PSEN dKO | Deletions of PSEN1 and PSEN232 |
| SAMP8 | Senescence accelerated mouse developed from AKR/J natural mouse line33 |
| SAMP10 | Senescence accelerated mouse developed from AKR/J natural mouse line34 |
| Tau-22 | MAPT G272V and P301S mutations35 |
| Tg2576 | APP Swedish mutation36 |
Lower vertebrates, such as Danio rerio (zebrafish), have seen an increase in popularity as valuable models in neuroscience in recent years. Some advantages for their use include being small, and thus requiring relatively-simple and small research facilities, the fact that they grow at a fast rate, and how easily they can be genetically manipulated; however, despite exhibiting a DNA methylation system similar to mammals, the absence of gene orthologs for many human genes limits their use.18,37 Invertebrates such as Drosophila melanogaster (fruit fly) and Caenorhabditis elegans (nematode worm) also share many similarities with humans at the molecular level. These organisms have a fully sequenced genome and can also be easily genetically modified; however, their physiology is very different to that of humans, and their DNA methylation profiles are distinct to those in humans, therefore limiting their use as disease models for the study of epigenetics in neurodegenerative disorders.17
DNA Methylation
DNA methylation, the most commonly studied epigenetic mark, regulates gene expression, generally by promoting gene silencing. It involves the covalent transfer of a methyl group from S-adenosyl methionine onto the fifth carbon atom of cytosine nucleotides on DNA. This modified state is known as 5-methylcytosine (5mC) and can be found concentrated at cytosine-guanine-rich regions (or CpG islands) in the DNA. Further modifications can also occur: 5mC can be converted 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC).16 5hmC, in particular, was originally thought to be a transient state, later shown to have an important role on its own, particularly in the brain.38 Of note, as mentioned, DNA modifications present differently in distinct organisms used for the study of neurodegenerative disorders. For example, non-CpG DNA methylation is restricted to vertebrates.39 In C. elegans in particular, DNA methylation was initially considered to be absent, but was later shown to be restricted to adenine N6-methylation (6mA).40
Various studies have investigated changes in global 5mC and 5hmC levels in mouse models of AD; however, the findings reported to date are not consistent between studies (Table 2).23,41-48 Whilst study design caveats may be one reason for this discordance, such as the assessment of a small number of samples, the type of model used in each study may also be a contributing factor. Another important factor that contributes to the complexity of these findings includes differences across distinct brain regions and/or stages of a disease. The brain regions reported to exhibit changes in global 5mC and 5hmC include the hippocampus and the cerebral cortex, which are primarily affected in AD, suggesting vulnerability of pathology-affected brain regions to changes in these methylation marks. In contrast, findings from the cerebellum showed no changes in 5mC and 5hmC in multiple genetic AD mouse models.23,41,48 This parallels the fact that the cerebellum is often spared of pathological changes in the human AD brain,49 and is consistent with DNA methylation studies in humans.50
Table 2.
DNA methylation and associated machinery in mouse models of Alzheimer’s disease, Parkinson’s disease, and amyotrophic lateral sclerosis.
| Regulation | Brain region | Mouse model | Change | Reference |
|---|---|---|---|---|
| Alzheimer’s disease | ||||
| 5mC | Hippocampus | APPSwe, Ind | ↓ | Do Carmo et al23 |
| 5xFAD | – | Zhang et al41, Griñán-Ferré et al42, Griñán-Ferré et al43, Griñán-Ferré et al44 | ||
| SAMP8 | ↑ | Cosín-Tomás et al45 | ||
| Cerebral cortex | APPSwe, Ind | ↓ | Do Carmo et al23 | |
| APP/PS1 | ↑ | Huang et al46 | ||
| 3xTg-AD | – | Cadena-del-Castillo et al47 | ||
| Prefrontal cortex | 3xTg-AD | – | Zhang et al41 | |
| Cerebellum | APPSwe, Ind | – | Do Carmo et al23 | |
| 3xTg-AD | – | Zhang et al41 | ||
| 5hmC | Hippocampus | APP/PS1 | ↓ | Shu et al48 |
| 3xTg-AD | ↓ | Zhang et al41 | ||
| 5xFAD | – | Griñán-Ferré et al42, Griñán-Ferré et al43, Griñán-Ferré et al44 | ||
| SAMP8 | ↑ | Cosín-Tomás et al45 | ||
| Cerebral cortex | 3xTg-AD | ↑ | Cadena-del-Castillo et al47 | |
| APP/PS1 | ↑ | Huang et al46 | ||
| APP/PS1 | – | Shu et al48 | ||
| Prefrontal cortex | 3xTg-AD | ↓ | Zhang et al41 | |
| Cerebellum | 3xTg-AD | – | Zhang et al41 | |
| APP/PS1 | – | Shu et al48 | ||
| DNMT1 | Hippocampus | 5xFAD | ↓ | Griñán-Ferré et al42 |
| 5xFAD | – | Griñán-Ferré et al44 | ||
| SAMP8 | ↓ | Cosín-Tomás et al45 | ||
| DNMT3a | Hippocampus | 5xFAD | ↑ | Griñán-Ferré et al42 |
| 5xFAD | – | Griñán-Ferré et al44 | ||
| SAMP8 | ↓ | Cosín-Tomás et al45 | ||
| DNMT3b | Hippocampus | 5xFAD | ↑ | Griñán-Ferré et al42, Griñán-Ferré et al44 |
| SAMP8 | – | Cosín-Tomás et al45 | ||
| TET1 | Hippocampus | 5xFAD | ↓ | Griñán-Ferré et al42, Cosín-Tomás et al45 |
| 5xFAD | ↑ | Griñán-Ferré et al44 | ||
| 3xTg-AD | ↓ | Zhang et al41 | ||
| Prefrontal cortex | 3xTg-AD | ↓ | Zhang et al41 | |
| TET2 | Hippocampus | 3xTg-AD | ↓ | Zhang et al41 |
| SAMP8 | ↓ | Cosín-Tomás et al45 | ||
| 5xFAD | – | Griñán-Ferré et al42 | ||
| 5xFAD | ↑ | Griñán-Ferré et al44 | ||
| Prefrontal cortex | 3xTg-AD | ↓ | Zhang et al41 | |
| TET3 | Hippocampus | 3xTg-AD | ↓ | Zhang et al41 |
| Prefrontal cortex | 3xTg-AD | ↓ | Zhang et al41 | |
| MeCP2 | Hippocampus | APP/PS1 | ↑ | Lu et al62 |
| Striatum | APP/PS1 | ↓ | Li et al63 | |
| Parkinson’s disease | ||||
| 5mC | Anterior brain cortex | α-synuclein | ↓ | Desplats et al51 |
| DNMT1 | Anterior brain cortex | α-synuclein | ↓ | Desplats et al51 |
| Substantia nigra | MPTP-induced | ↓ | Zhang et al64 | |
| TET2 | Substantia nigra | MPTP-induced | ↑ | Wu et al65 |
| Amyotrophic lateral sclerosis | ||||
| 5mC | Global brain | SOD1G93A | – | Figueroa-Romero et al52 |
| 5hmC | Global brain | SOD1G93A | ↑ | Figueroa-Romero et al52 |
Abbreviations: 5mC, 5-methylcytosine; 5hmC, 5-hydroxymethylcytosine; DNMT, DNA methyltransferase; MeCP, methyl-CpG-binding protein; TET, ten-eleven translocation enzyme family.
The up arrow (↑) represents upregulation or increased levels. The down arrow (↓) represents downregulation or decreased levels. The dash (-) represents no significant changes identified.
Only a few studies to date have investigated DNA methylation in models of PD and ALS (Table 2). In a mouse model of PD that overexpresses human α-synuclein, Desplats et al51 reported reduced global 5mC levels in the anterior portion of the brain compared to control mice. In a mouse model of ALS bearing the SOD1 G93A mutation, Figueroa-Romero et al52 identified increased 5hmC global levels compared to control mice. Human post-mortem studies investigating brain tissue from individuals with PD53-55 and ALS15,56-58 have revealed aberrant methylation in pathology-affected brain regions. Future studies using mouse models should investigate these changes in pathology-affected brain regions, such as the substantia nigra in PD.
DNA 6mA (methylation of the sixth carbon atom of adenine nucleotides) and RNA N6-methyladenosine (m6A) have also been identified important epigenetic modifications in recent years,59,60 although their functions are still to be clearly defined. Interestingly, a recent study reported reduced DNA 6mA levels in peripheral blood in people with AD when compared to controls, demonstrating its potential as a biomarker.61 Further studies exploring DNA 6mA and RNA m6A are needed, especially studies aiming to understand their role in the brain and how they change in and with disease.
Contrarily to cancer, DNA methylation fluctuations in the brain are much more subtle, thus methods assessing global levels of DNA methylation may not be the most appropriate in this context, which may explain the disparity of reports to date discussed in this section. Quantitative interrogation of selected sites (eg, methylation arrays) and at single-base resolution (eg, next generation sequencing)—which will be considered in the next section—can provide much more valuable insights. Especially when coupled with methodologies involving cell-sorting or single-cell sequencing, these powerful approaches can provide more insightful information, disclose heterogeneity, and likely clarify discordances observed in previous studies.
Genome-wide DNA methylation profiling
In recent years, methylation arrays and sequencing techniques have made feasible DNA methylation analyses on a more extensive—genome-wide—level in both humans and animal models. Of interest, some studies performing genome-wide analyses of mouse models of AD have been conducted using diverse techniques (Table 3).41,48,66-70 Whole-genome bisulfite sequencing (WGBS) is considered the gold standard approach for studying genome-wide methylation at the single-base resolution as it profiles methylation patterns in the entire methylome at both CpG islands and non-CpG regions.71 Zhang et al68 used this method to investigate the genome-wide DNA methylation profile in the cerebral cortex of SAMP8 mice and reported higher methylation levels in introns compared to exons. Three differentially-methylated regions (DMRs) annotated to Dlgap1, Eif2ak2, and Tmem51 showed increased methylation levels in SAMP8 mice in this study, likely associated with AD pathogenesis.68 Zhang et al and colleagues further investigated differential expression of these genes, and reported that Eif2ak2 expression was downregulated due to increased methylation, while Dlgap1 and Tmem51 expression was upregulated.68 These findings emphasize the complexity and diversity of the functional roles of DNA methylation; although DNA methylation is often associated with transcriptional repression, this is not always the case. Not to mention that there is currently an emerging interest in understanding the role of DNA methylation in the regulation of alternative splicing.72
Table 3.
Genome-wide DNA methylation analyses studies of mouse models of Alzheimer’s disease.
| Study | Model | Brain tissue | Modification | Method | Main findings |
|---|---|---|---|---|---|
| Sanchez-Mut et al66 | APP/PS1 (12 mo) 3xTg-AD (18 mo) |
Twelve brain regions, including CA1, CA3 and DG hippocampal subregions, frontal cortex, and hypothalamus | 5mC | MeDIP sequencing, Illumina VeraCode GoldenGate DNA Methylation Mouse Array | Hypermethylation of Sorbs3, Spnb4 and Tbxa2r in the prefrontal cortex of APP/PS1 and 3xTg-AD mice consistent with the hypermethylation of SORBS3, SPTBN4 and TBXA2R in post-mortem human AD brain tissue |
| Cong et al67 | APP/PS1 (11 mo) | Cerebral cortex | 5mC | MeDIP sequencing, Roche NimbleGen, Mouse DNA Methylation 3 ×720 K CpG Island Plus RefSeq Promoter Array | 2346 hypermethylated regions of which 2221 mapped to promoter regions and 485 genes were associated with AD |
| Zhang et al68 | SAMP8 (7 mo) | Cerebral cortex | 5mC | WGBS | 63 DMRs of which 41 regions were hypermethylated and 22 regions were hypomethylated in SAMP8 mice |
| Tang et al69 | PSEN dKO (12 mo) | Hippocampus | 5mC | RRBS | 1094 hypermethylated regions and 1676 hypomethylated regions in PSEN dKO mice |
| Kundu et al70 | APPNL-G-F and APPNL-F (12 mo) | Hippocampus | 5mC | RRBS | 57 DMRs shared between both AD mouse models |
| Shu et al48 | APP/PS1 (12- and 67-wk) | Hippocampus | 5hmC | hMeDIP sequencing | Decrease in total peaks in aged APP/PS1 mice |
| Zhang et al41 | 3xTg-AD (E16.5) | Cortical neurons (fetal brain) | 5hmC | hMeDIP sequencing | Decrease in total peaks in AD group, with most 5hmC marks located in intergenic regions |
Abbreviations: 5hmC, 5-hydroxymethylcytosine; 5mC, 5-methylcytosine; AD, Alzheimer’s disease; CA1, cornu ammonis 1; CA3, cornu ammonis 3; DG, dentate gyrus; dKO, double knock-out; DMR, differentially methylated region; MeDIP, methylated DNA-immunoprecipitation; WGBS, whole genome bisulfite sequencing ; RRBS, reduced representation bisulfite sequencing; hMeDIP, hydroxymethylated DNA-immunoprecipitation.
An alternative method to WGBS, that can also be used to investigate DNA methylation at a genome-wide level, is reduced representation bisulfite sequencing (RRBS). Essentially, a reduced, representative sample of the whole genome at single-base resolution is sequenced, mostly enriched by CpG regions.71 Two studies employed this technique on AD mouse models and identified differentially-methylated regions in the hippocampus enriched for processes relevant to AD and brain homeostatic mechanisms, such as adhesion signaling, cytoskeleton, and synaptic functions.69,70
Research profiling genomic 5hmC have also been conducted in AD mouse models, although still in its infancy.41,48 Two studies used hydroxymethylated DNA-immunoprecipitation sequencing (hMeDIP-seq) to identify changes in 5hmC. Both studies showed a reduction in overall 5hmC in the respective tissues investigated, and identified specific enrichment of differentially-hydroxymethylated regions annotated to genes related to synaptic and neuronal homeostasis, as well as AD pathogenesis.41,48 This suggests that alongside 5mC changes, 5hmC-mediated regulation may also play a critical role in AD pathogenesis and neurodegeneration.
Writers, erasers, and readers
An additional viewpoint of studying DNA methylation mechanisms is looking at the (dys)regulation of the machinery responsible for generating and maintaining it. DNA methyltransferases (DNMTs), known as methylation “writers,” add methyl groups to the DNA, giving rise to 5mC. Among the major DNMTs, DNMT1 is considered the maintenance methylase, and DNMT3a and DNMT3b are considered de novo methylases.16 DNMT3a and DNMT3b are of particular interest in the context of the topic of this review as they introduce methylation marks to DNA over time; however, studies exploring these enzymes in AD mouse models reported conflicting findings (Table 2).42,44,45 For example, studies investigating DNMT1 expression reported either a decrease in hippocampal expression in AD mice,42,45 or no significant changes44 (Table 2). DNMT1 levels were also shown to be decreased in 2 unique mouse models of PD.51,64 It is unclear whether DNMT1 downregulation could be a mutual characteristic across these neurodegenerative diseases and additional studies should investigate this further. DNMT1 is imperative in reproducing DNA methylation after DNA replication and thus helps maintain chromosome stability. A potential consequence for the reported reduction in DNMT1 is a resulting reduction of DNA methylation, that is, hypomethylation in these pathology-affected brain regions.16
The family of ten-eleven translocation (TET) enzymes, known as “erasers,” are methylcytosine dioxygenases that convert 5mC to 5hmC. Research has shown conflicting levels of expression of these enzymes across several mouse models of AD in pathology-affected regions (Table 2).41,42,44,45 In a mouse model of PD (MPTP-induced, that is, generated by inducing the neurotoxin 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine or MPTP, which is highly selective for the substantia nigra and leads to dopaminergic neuron damage), a study demonstrated increased Tet2 gene expression in the substantia nigra compared to control mice.65 As for other epigenetic mechanisms discussed earlier, the relevance of these changes in neurodegenerative conditions is still unclear.
Proteins of the methyl-CpG-binding domain (MBD) family, known as “readers,” recognize and bind specific regions of the genome (usually methylated CpG islands) and recruit chromatin-remodeling proteins to induce changes in DNA transcription; however, these are less well researched.16 Some of the MBD-recognized genomic sites include promoter regions found upstream of genes where transcription is initiated. Of the very few studies investigating this, Bie et al73 and Lu et al62 reported increased hippocampal methyl-CpG-binding protein 2 (MeCP2) levels in rodent models of AD. By using a chromatin immunoprecipitation (ChIP) assay, the teams showed increased cytosine methylation and binding of MeCP2 at the promoter region of the gene encoding neuroligin-1 (Nlgn1) in the hippocampus of Aβ1-40-induced rats73 and APP/PS1 mice62 compared to their respective controls. Neuroligin-1 plays a critical role at synapses, and its dysfunction has been closely associated with AD pathogenesis. Targeting and increasing the expression of Nlgn1 to promote its neuroprotective effects at synapses offers a therapeutic opportunity in slowing down neurodegenerative processes.62,73 A different study demonstrated reduced MeCP2 levels in the striatum of 3-month-old APP/PS1 mice compared to controls, with specific S421 phosphorylation of MeCP2 shown to be increased in the cytoplasm in neurons. The authors suggested that this finding was due to de novo phosphorylation of MeCP2 after AD pathological injury, possibly acting to relieve transcriptional repression.63
The implications of changes in expression of “reader” proteins in the context of neurodegenerative diseases require further investigation.
Histone Modifications
Epigenetic modifications in histones (the proteins that package the DNA into nucleosomes) regulate gene expression by influencing the structure of chromatin and by controlling the binding of regulatory proteins and other effector molecules.16 The core histones that make up the nucleosomes are histones 2A (H2A), 2B (H2B), 3 (H3), and 4 (H4). These can be epigenetically modified through histone modifications to alter chromatin structure and enable chromatin remodeling. Examples of common histone modifications include lysine and arginine methylation, lysine acetylation, and serine and threonine phosphorylation amongst an array of covalent changes.74
Histone acetylation is generally associated with active gene expression at transcription start sites, such as acetylation of lysine at the ninth position of H3 (H3K9ac) and lysine at the 27th position of H3 (H3K27ac).75 Meanwhile, methylated histone marks are often associated with inactive genes. For instance, tri-methylation of lysine at the ninth position of H3 (H3K9me3) and the 27th position of H3 (H3K27me3) are usually distributed across inactive regions and at transcription start sites of inactive genes, respectively. However, some methylated histone marks can be found at active gene loci (eg, H3K4me and H3K4me3). Histone phosphorylation is not as well characterized; nonetheless, H3S10ph and H3T11ph marks seem to have a role in developmental processes.75
Overall, the studies summarized in Table 4 and described in detail in the following sections demonstrate how changes in histone modifications can have detrimental and widespread effects by interfering with the transcription of crucial neuroprotective genes in neurodegenerative conditions.
Table 4.
Changes to histone marks in animal models of Alzheimer’s disease.
| Regulation | Study | Model system (age) | Findings |
|---|---|---|---|
| Histone acetylation | Govindarajan et al78 | Mouse, APP/PS1-21 (15 mo) | ↓ H3K9ac in CA1, CA3, DG, Pir, and M1/2 ↓ H3K14ac in CA3, DG, Pir, Cg and M1/2 ↓ H4K5ac in CA1, CA3, DG, Pir, Cg, and M1/2 ↓ H4K12ac in CA1, CA3, DG and M1/2 ↓ H4K16ac in CA1, CA3, DG |
| Zhang et al79 | Aβ42-induced Drosophila of early age (third instar larvae) and late age (28-dadults) | ↓ H4K16ac in the brain of early and late age models | |
| Lithner et al80 | Mouse, Tg2576 (4 mo) | ↑ H3K14ac in CA3, CTX, PFC | |
| Chatterjee et al81 | Mouse, Tau-22 (12 mo) | ↓ H2BK5K10K15K20ac in dHP | |
| Gräff et al82 | Mouse, CK-p25 (3-6 mo) | ↓ H2BK5ac, H3K14ac, H4K5ac and H4K12ac at multiple genes associated with learning and memory, and synaptic plasticity in HP | |
| Gjoneska et al83 | Mouse, CK-p25 (3 mo) |
H3K27ac peaks in HP (see text for details) |
|
| Klein et al84 | Mouse, P301S Tau (6 and 11 mo) Mouse, CK-p25 (3 mo) |
H3K9ac peaks in HP (see text for details) | |
| Histone methylation | Lithner et al80 | Mouse, Tg2576 (4 mo) | ↑ H3K9me2 in DG, CTX |
| Zheng et al85 | Mouse, 5xFAD (5-6 month) | ↑ H3K9me2 in PFC | |
| Cao et al86 | Mouse, P301S Tau (5-6 mo) | ↑ H3K4me3 in PFC | |
| Gjoneska et al83 | Mouse, CK-p25 (3 mo) |
H3K4me3, H3K4me1, H3K27me3, H3K36me3, H4K20me1, H3K9me3 peaks in HP | |
| Histone phosphorylation | Anderson et al87 | Mouse, 5xFAD (3 mo) | ↓ H3S57ph, H3T58ph and both combined in whole mouse brain homogenates |
Abbreviations: CA1, cornu ammonis 1; CA3, cornu ammonis 3; Cg, cingulate cortex; CTX, cerebral cortex; DG, dentate gyrus; dHP, dorsal hippocampus; HP, hippocampus; M1/2, motor cortex 1 and 2; Pir, piriform cortex; PFC, prefrontal cortex.
The up arrow (↑) represents upregulation or an increase in level. The down arrow (↓) represents downregulation or a decrease in level. CA1, CA3 and DG are regions of the hippocampus.
Histone acetylation is generally associated with transcriptional activation since it reduces the affinity of the histone tail for adjacent nucleosomes and relaxes chromatin structure, allowing transcription machinery to bind to the DNA.75 Immunoassay techniques have been recurrently used to assess global histone acetylation levels in mouse models of AD.45,76,77 One study using this type of approach identified reduced H3 acetylation in 8-month-old SAMP8 mice that was partly restored by exercize.76 Another study focused on age-associated changes in the 3xTg-AD mouse and reported higher H3 and H4 acetylation at 4-, 8-, 11-, and 12-months of age, becoming increasingly discordant with age.77 Global changes in H3ac and H4ac levels should be carefully interpreted, however, as it is not evident how the acetylation marks are distributed in the genome and, thus, how it is being altered.
Various studies have identified changes in histone acetylation marks in the brain, either in whole brain in one study using flies or in defined brain regions in mice (Table 4).78-82 For example, decreased H4K16ac was reported in both mice and flies modeling AD, specifically in hippocampal subregions of APP/PS1-21 mice78 and in the brain of young and old aged Aβ42-induced Drosophila.79 Whilst studies investigating regional or global levels of specific histone marks help understand epigenetic changes, it is critical to identify where these marks are enriched in the genome.
Gräff et al82 characterized specific histone marks at selected brain regions using a conventional approach. The team performed reverse transcriptase-polymerase chain reaction (RT-PCR) to identify H2BK5ac-, H3K14ac-, H4K12ac-, and H4K5ac-immunoprecipitated chromatin at the promoters of selected neuroplasticity and housekeeping genes in the hippocampus of the CK-p25 mouse model of AD. They found reduced H2BK5ac, H3K14ac, H4K12ac and H4K5ac histone marks at numerous candidate genes involved in learning and memory (eg, Bdnf, Cdk5) and synaptic plasticity (eg, GluR1, NR2A, NR2B).82
A study by Gjoneska et al83 profiled 7 histone marks (H3K4me3, H3K4me1, H3K27ac, H3K27me3, H3K36me3, H4K20me1, and H3K9me3) in the hippocampus of CK-p25 mice using ChIP-sequencing (ChIP-seq). The team found 3667 increased and 5056 decreased H3K4me3 peaks at active promoter regions, and 2456 increased and 2154 decreased H3K27ac peaks at active enhancer regions in CK-p25 mice. The increased-levels in enhancer and promoter regions were mainly associated with immune and stimulus-response functions, whereas the decreased-levels were principally associated with synapses and learning-associated functions.83 Similarly, Marzi et al88 performed a histone acetylome-wide association study using ChIP-seq to examine H3K27ac in entorhinal cortex tissues from people with AD, demonstrating extensive variation in H3K27ac across the genome. Some of the hyperacetylated peaks (increased H3K27ac marks) identified were associated with genes related to Aβ and tau pathology and response to hypoxia, whereas some of the hypoacetylated peaks (decreased H3K27ac marks) were associated with genes related to neuronal transmission and synapses.88 Notably, these are similar to the associated pathways discussed by Gjoneska et al,83 where authors further reported mouse-human conservation of chromatin state profiles identified in enhancers in p25-inducible transgenic mice. Specifically, regions orthologous to increased-level enhancers in mouse exhibited immune cell enhancer activity in humans, and orthologs of decreased-level enhancers in mouse corresponded to fetal brain enhancer activity in humans, suggestive of alterations in regulatory regions involved in neuronal plasticity.
More recently, Klein et al84 sought to compare changes in H3K9ac marks between aged human cortices and 2 AD mouse models: MAPT P301S and CK-p25 mice. The study showed spatial patterns in H3K9ac marks that were similar in both AD mouse models and consistent with findings from aged human cortices, reinforcing the utility of these model systems to further explore and modify chromatin regulation changes observed in AD.84
In the substantia nigra of a new mouse model of PD, the MitoPark mouse, Huang et al89 also found disturbed levels of H3K27ac. The MitoPark mouse model reported in this study was generated by conditionally knocking out mitochondrial transcription factor A in dopaminergic neurons, and expressing multiple features of PD. Huang et al89 suggested that hyperacetylation of H3K27 may have arisen due to the mitochondrial dysfunction, that is, generation of the MitoPark mice per se, with the resulting epigenetic changes likely contributing to PD pathogenesis. Further studies are needed for clarification.
Histone methylation is often, but not always, associated with gene and chromatin silencing (eg, H3K9 and H3K27 di- and tri-methylation).75 Studies investigating AD mouse models have shown increased H3K9me2 in the cerebral cortex and DG subregion of the hippocampus of Tg2576 mice80 and the prefrontal cortex of 5xFAD mice85 (Table 4). Post-mortem prefrontal cortex sample from people with AD were also reported to present with higher H3K9me2 compared to healthy controls.85 In contrast, studies in AD mouse models so far showed no significant difference in H3K27me3 levels in the prefrontal cortex, 85,86 M1/M2 motor cortex and S1/S2 somatosensory cortex compared to controls.90
H3K4me and H3K4me3 are frequently found at promoters of active genes and enhance transcription and gene expression.75 A study by Cao et al86 reported increased H3K4me3 but not H3K4me in the prefrontal cortex of the P301S Tau mouse model of AD, mirroring their findings from post-mortem prefrontal cortex tissues from AD patients. In contrast, Dyer et al90 did not find any significant changes in H3K4me3 levels in the M1/M2 motor cortex and S1/S2 somatosensory cortex of 3-, 6-, and 12-month-old APP/PS1 mice.
Recent research has provided evidence for sex-specific differences in specific histone marks (such as H3K4me3, H3K27ac and H3K27me3) in the cortex of the PSAPP mouse model of AD.91 This distribution lies in transcription control regions of genes involved in neuronal functions, which have been associated with cognitive decline in AD patients. Future research should explore whether these changes are also seen in histone writer and eraser enzymes, particularly due to their therapeutical potential.
H3K4me3 has also been implicated in PD models. A study by Nicholas et al92 found decreased H3K4me3 in the striatum of MPTP-induced mice and macaque monkeys. A subsequent study demonstrated the restoration of this histone mark in vitro following treatment with a histone demethylase inhibitor that can cross the blood brain barrier.93 In 6-OHDA-induced PD rats, the same histone demethylase inhibitor rescued dopaminergic neuron loss and motor defects—characteristic features of PD—demonstrating its potential as a therapeutic agent for the treatment of PD.93
Phosphorylation of histones adds a significant negative charge to the nucleosome complex, thereby opening up the chromatin structure. This modification is not as commonly researched as the histone modifications previously discussed. One study, by Anderson et al87 reported decreased H3S57 and H3T58 phosphorylation, either separately or in combination, in the 5xFAD mouse brain (Table 4). Investigating the implications of these histone marks on a genomic and transcriptomic level may be more valuable in identifying the downstream effects, but no studies of such nature have been reported to date.
Histone writers and erasers
A diverse range of enzymes is responsible for the chemical modifications that occur to histones.74 The methyl, acetyl and phosphate groups can be added to histones by “writer” enzymes, such as histone methyltransferases (HMT), acetyltransferases and kinases. These enzymes can be further categorized; for instance, HMTs have high selectivity for lysine and arginine residues, hence they are grouped into lysine methyltransferases (KMTs) and arginine methyltransferases. The chemical groups can also be removed from histones by “eraser” enzymes, such as histone demethylases, deacetylases (HDAC), and phosphatases. These enzymes can also be further categorized; for instance, the HDAC family contains 18 HDAC proteins that can be divided into 2 subcategories and 4 classes in total: classical (classes I, II, and IV) and sirtuins (class III).74
Several studies have identified increased HDAC2 levels in the hippocampus45,82,94 and prefrontal cortex82,94 of AD mouse models and an Aβ42-induced AD Drosophila model79 compared to their respective controls (Table 5). Specifically, Gräff et al82 showed increased HDAC2 binding localized to specific learning and memory, as well as synaptic plasticity genes in the hippocampus of CK-p25 mice compared to controls. Moreover, studies investigating epigenetic changes in the Nlgn1 promoter discussed earlier in this review reported increased HDAC2 binding at this region in the hippocampus of Aβ1-40-induced rats73 and APP/PS1 mice.62 Indeed, NLGN1 is a protein associated with synaptic function and transmission.95 A mouse model of PD bearing the LRRK2 R1441G mutation also was also shown to exhibit increased HDAC2 alongside increased HDAC1 and HDAC3 levels.96 Meanwhile, other HDACs did not show consistent findings between AD model organisms (Table 5).29,45,76,82 Taken together, this suggests that targeting HDAC2 may hold considerable therapeutic potential amongst the other HDACs. Indeed, this modulation of HDAC2 has been recently explored with promising results, with Nakatsuka et al97 reporting amelioration of deficits in long-term potentiation and memory impairment in the hippocampus of APP/PS1 mice following HDAC2 inhibition.
Table 5.
Changes in histone-modifying enzymes in animal models of Alzheimer’s disease and Parkinson’s disease.
| Regulation | Reference | Model system (age) | Findings |
|---|---|---|---|
| Alzheimer’s | |||
| Histone deacetylation | Cosin-Tomas et al45 | Mouse, SAMP8 (2 and 9 mo) | ↑ HDAC1 in HP of both early and late age models ↑ HDAC2 in HP of 2-mo-old SAMP8 mouse only ↓ SIRT1 in HP of both early and late age models ↓ SIRT6 in HP of 9-mo-old SAMP8 mouse only |
| Zhang et al79 | Aβ42-induced Drosophila of early age (third instar larvae) and late age (28-day adults) | ↑ HDAC2 in the brain of both early and late age models | |
| Gräff et al82 | Mouse, CK-p25 (3-6 mo), 5xFAD (6 mo) |
↑ HDAC2 in CA1 and PFC of CK-p25 mouse ↑ HDAC2 in HP and PFC of 5xFAD mouse |
|
| Liu et al94 | Mouse, 3xTg-AD (12 mo) | ↑ HDAC2 in HP | |
| Cosín-Tomás et al76 | Mouse, SAMP8 (8 mo) | ↓ HDAC5 and HDAC6 in HP ↓ SIRT1 in HP |
|
| ArunSundar et al29 | Mouse, oA42i-induced (age not stated) | ↑ HDAC6 in HP | |
| Song et al98 | Mouse, 5xFAD (6 mo) | ↓ SIRT1 in HP | |
| Zhang et al100 | Mouse, APP/PS1 (6 mo) | ↓ SIRT1 in HP | |
| Rodriguez-Ortiz et al99 | Mouse, 3xTg-AD (12 mo) | ↓ SIRT1 in vHP | |
| Histone methylation | Cao et al86 | Mouse, P301S Tau (5-6 mo) | ↑ KMT2A and SETD1B in PFC |
| Zheng et al85 | Mouse, 5xFAD (5-6 mo) | ↑ EHMT1 and EHMT2 in PFC | |
| Parkinson’s | |||
| Histone deacetylation | Kim et al96 | Mouse, LRRK2 (11 mo) | ↑ HDAC1, HDAC2 and HDAC3 |
| Tao et al102 | Mouse, Rotenone-induced (8 wk old) | ↓ SIRT1 in SN | |
Abbreviations: CA1, cornu ammonis 1; EHMT, euchromatic histone-lysine methyltransferase; HDAC, histone deacetylase; HP, hippocampus; KMT, lysine methyltransferase; PFC, prefrontal cortex; SETD; family of SET-domain methyltransferases; SIRT, sirtuin; SN, substantia nigra; vHP ventral hippocampus.
The up arrow (↑) represents upregulation or an increase in level. The down arrow (↓) represents downregulation or a decrease in level.
Multiple studies demonstrated a decrease in Sirtuin-1 levels in the hippocampus of a range of AD mouse models (Table 5).45,76,98-100 Specifically, the decrease reported in 3xTg-AD mice was restricted to the ventral hippocampus and not observed in the dorsal hippocampus.99 The dorsal hippocampus (posterior in primates) is primarily involved in cognitive functions such as learning and memory, whilst the ventral hippocampus (anterior in primates) is associated with emotions and motivation processes; both regions of the brain and their respective functions are affected in AD.101 Furthermore, this decrease was only observed in the 12-month-old 3xTg-AD mice but not in the younger mice, suggesting age-associated effects or influence by stage and progression of the disease.99
In a mouse model of PD induced by rotenone (a pesticide and complex I inhibitor that reproduces features of PD, such as dopaminergic degeneration and α-synuclein inclusions), Tao et al102 showed reduced Sirt1 levels in the substantia nigra, a region particularly affected in the PD brain due to the loss of dopaminergic neurons.
A few studies have also explored specific groups of the KMT subclass of HMTs in AD mouse models. Cao et al86 reported elevated levels of Kmt2a and SET-domain containing 1B histone-lysine methyltransferase (Setd1b) in the prefrontal cortex of P301S Tau mice, with no significant change in Kmt2b, Kmt2c, Kmt2d, and Setd1a. However, in post-mortem prefrontal cortex from individuals with AD, the authors reported a different profile: increased levels of KMT2C, KMT2D, SETD1A, and SETD1B, but unaltered levels of KMT2A and KMT2B compared to control individuals.86 In another study, Zheng et al85 reported increased euchromatic histone-lysine methyltransferase 1 (Ehmt1) and 2 (Ehmt2) levels in the prefrontal cortex of 5xFAD mice. Notably, the upregulation of EHMT1 levels was also observed in human AD prefrontal cortical tissues in this study, although no significant change in EHMT2 levels was found.85 These studies need to be reproduced in other AD mouse models and in humans in order to define the holistic changes in these enzymes and the implications of their dysregulation in AD and other neurodegenerative diseases.
MicroRNAs
Regulatory non-coding RNAs (ncRNAs) comprise transcripts that are not translated into proteins and are key players in gene regulation instead.103 Their main role is the post-transcriptional regulation of gene expression, by affecting the stability and degradation of messenger RNA (mRNA), preventing its translation into proteins. ncRNAs can be categorized into short (<200 nucleotides) and long ncRNAs (>200 nucleotides). This review focuses on one specific subset of small ncRNAs—microRNAs (miRNAs)—which are approximately 21 to 22 nucleotides in length. Changes in brain-enriched miRNAs have been identified in many neurodegenerative diseases, including in studies using animal models, that will be discussed in the next sections.103
Genome-wide microRNA profiling
Microarrays and sequencing techniques have been invaluable for the detection of dysregulated miRNAs in animal models of AD and PD.
Various studies have performed microarray analyses in the brain of a range of mouse models of AD (Table 6), including Tg2576,104,105 APP23,106 APP/PS1,107-110 5xFAD,111,112 3xTg-AD,113 SAMP8,45,114-116 senescence-accelerated mouse prone 10 (SAMP10),116 and PSEN dKO117 mice. Some of these studies focused on specific miRNAs, whilst others performed detailed investigations of the cellular and molecular pathways affected and targeted by these miRNAs. Higaki et al105 demonstrated upregulation of members of the miR-200 family (miR-141, -200a, -200b, -200c, -429) and miR-183 family (miR-96, -182 and -183) in the cortex of 10-month-old Tg2576 mice. Intriguingly, the upregulation of members of the miR-200 family in Tg2576 mice appeared to be limited to the phase of increasing Aβ plaque deposition. miR-200 family members have been shown to regulate neuronal proliferation, homeostasis and apoptosis, and hence their dysregulation may interfere with these vital regulatory processes.105 Liu et al107 reported downregulated miR-200a, -200b, -182, and -183, in contrast to some of the results reported by Higaki et al105 It is worth noting that the tissue samples were much different between the 2 studies; Liu et al107 investigated the hippocampus, whereas Higaki et al105 looked at cerebral cortex. Another study profiled miRNAs in APP/PS1 mice (whole brain) at 1-, 3-, 6-, and 9-months of age, to determine miRNA expression patterns over development and age.108 The changes in expression of some miRNAs overlapped between age groups, suggesting that these miRNAs may act across different stages of the disease. Some additional studies investigating Aβ-induced rat models of AD118,119 and an Aβ-induced Drosophila model,120 shared similar findings, including changes to neuronal health and vital signaling and regulatory mechanisms such as PI3K/Akt and Jak-STAT cellular pathways.
Table 6.
MicroRNA microarray studies in animal models of Alzheimer’s disease and Parkinson’s disease.
| Study | Model system | Age (months) | Brain tissue | Findings | Focus/Implications of study |
|---|---|---|---|---|---|
| Alzheimer’s disease | |||||
| Wang et al104 | Mouse, Tg2576 | 12 | HP | Several dysregulated miRNAs (eg, miR-124, -125a, -125b) | Upregulated miR-124 associated with synaptic dysfunction and memory loss |
| Higaki et al105 | Mouse, Tg2576 | 10 | CTX | ↑ 7 miRNAs > 2-fold ↓ 1 miRNA > 2-fold |
Upregulated miR-200 and miR-183 family members |
| Schonrock et al106 | Mouse, APP23 | 2, 7, 13 | HP | Several dysregulated miRNAs (eg, miR-409-3p, -148b, -30c, -9, -21) | miRNA deregulation in hippocampal cultures paralleled results from APP23 mouse |
| Liu et al107 | Mouse, APP/PS1 | 9 | HP | ↑ 15 miRNAs > 2-fold ↓ 13 miRNAs > 2-fold |
Downregulated miR-135a and −200b involved in disease pathogenesis and as potential biomarkers of AD |
| Wang et al108 | Mouse, APP/PS1 | 1, 3, 6, 9 | Whole brain | Several dysregulated miRNAs at different age points (eg, miR-342-3p elevated at 1-, 6-, and 9-mo of age) | Eleven aberrantly regulated miRNAs that are conserved in humans and are predicted to be associated with disease pathology, MAPK and TGF-β signaling pathways |
| Li et al109 | Mouse, APP/PS1 | 5 | HP | ↑ 5 miRNAs ↓ 15 miRNAs |
Upregulated miR-574 associated with synaptic plasticity |
| Wang et al110 | Mouse, APP/PS1 | 3, 6 | CTX | ↑ 20 miRNAs ↓ 17 miRNAs |
Upregulated miR-34a associated with apoptosis that may contribute to AD pathogenesis |
| Noh et al111 | Mouse, 5xFAD | 4, 8 | HP | Several dysregulated miRNAs (eg, miR-139, -340, -3470a) | miR-139, -340 and −3470a associated with bio-energy metabolism related genes and metabolic dysfunction |
| Zhang et al112 | Mouse, 5xFAD | 4 | HP | ↑ 21 miRNAs ↓ 6 miRNAs |
Downregulated miR-188 associated with disease neuropathology, neuroinflammation, and synaptic and cognitive impairments |
| Barak et al113 | Mouse, 3xTg-AD | 4, 16 | HP | Several dysregulated miRNAs in both age groups (eg, miR-15a, -34a, -298, -101a, -294) | Predicted pathways upregulated include renal cell carcinoma,
colorectal cancer, chronic myeloid leukemia, and
glioma Predicted pathways downregulated include MAPK signaling, axon guidance and regulation of actin cytoskeleton |
| Cosín-Tomás et al45 | Mouse, SAMP8 | 2, 9 | HP | Several dysregulated miRNAs of which 6 miRNAs overlapped at both ages | Predicted pathways affected include oxidative stress, inflammation, pathological development, cell cycle dysregulation, neurogenesis |
| Zhou et al114 | Mouse, SAMP8 | 3 | HP | ↑ 7 miRNAs > 1.5-fold
↓ 8 miRNAs > 1.5-fold |
Downregulated miR-181c associated with regulation of axon guidance and MAPK signaling among other functional processes |
| Zhang et al115 | Mouse, SAMP8 | 8 | HP | Several dysregulated miRNAs (eg, miR-214-3p, 464-5p, -194-5p, −129a) | Downregulated miR-214-3p associated with autophagy and apoptosis |
| Zhang et al116 | Mouse, SAMP8 | 3 | HP | ↑ 148 miRNAs ↓ 171 miRNAs |
Co-upregulated miRNAs associated with MAPK, insulin and neurotrophin signaling pathways, and regulation of actin cytoskeleton |
| Mouse, SAMP10 | 3 | HP | ↑ 139 miRNAs ↓ 163 miRNAs |
||
| Ham et al117 | Mouse, PSEN dKO | 7, 12, 18 | CTX, HP | Several dysregulated miRNAs in both brain regions | Age-dependent miRNAs in PSEN dKO mice and associated pathways indicate pathological aging |
| Wang et al118 | Rat, Aβ1-42-induced | Age not stated | HP | ↑ 93 miRNAs > 2-fold ↓ 90 miRNAs > 2-fold |
Regulatory interactions between miRNAs and circular RNAs may play important roles in AD pathogenesis |
| Li et al119 | Rat, Aβ25-35-induced | 3, 6 | HP | Several dysregulated miRNAs, for example, miR-30b, -129-5p | Upregulated miR-30b associated with neuronal injury, neuronal loss and neuroinflammation |
| Kong et al120 | Drosophila, Aβ42-induced | Adult flies, 16 days post-treatment | Brain | ↑ 8 miRNAs ↓ 9 miRNAs |
Predicted pathways affected include MAPK signaling, dorso-ventral axis formation, Jak-STAT signaling pathway |
| Parkinson’s disease | |||||
| Asikainen et al121 | C. elegans, α-synuclein | Fourth larval stage | ↑ 5 miRNAs ↓ 7 miRNAs |
Aberrantly regulated miRNAs in all three C. elegans models suggest they may be involved in neuropathophysiological mechanisms in disease | |
| C. elegans, cat-1 mutation | Fourth larval stage | ↑ 2 miRNAs ↓ 3 miRNAs |
|||
| C. elegans, pdr-1 mutation | Fourth larval stage | ↑ 2 miRNAs ↓ 1 miRNA |
|||
| Shen et al122 | C. elegans, HASNA53T OX | Fourth larval stage | ↑ 18 miRNAs ↓ 13 miRNAs |
3 miRNAs (cel-miR-1018, cel-miR-230-3p, and cel-miR-797-5p) were predicted to target orthologs of human K07H8.2 and SLC41A1 | |
Abbreviations: AD, Alzheimer’s disease; CTX, cerebral cortex; HP, hippocampus; Jak-STAT, Janus kinase-signal transducer and activator of transcription; MAPK, mitogen-activated protein kinase; miR or miRNA, microRNA; PD, Parkinson’s disease; TGF- β, transforming group factor β.
The up arrow (↑) represents upregulation or an increase in level. The down arrow (↓) represents downregulation or a decrease in level.
In PD, a study by Asikainen et al and colleagues investigated dysregulation of miRNAs in three C. elegans models bearing the human α-synuclein A53T mutation, or mutations within the vesicular catecholamine transporter (cat-1) or parkin (pdr-1) ortholog, reporting differential expression of several miRNAs in these models, including miR-64 and miR-65 families (Table 6).121 Another study investigated miRNAs in C. elegans overexpressing human mutant α-synuclein and reported the dysregulation of 3 miRNAs when comparing mutants (HASNA53T OX) to controls expressing wildtype human α-synuclein (HASNWT OX).122
miRNA-sequencing (miRNA-seq) is a powerful method for miRNA profiling, which is replacing the use of microarrays, particularly since it provides an unbiased investigation of all miRNAs.123 Three studies to date have conducted miRNA-seq to interrogate miRNAs in the APP/PS1 mouse model of AD (Table 7).124-126 Specifically, Luo et al126 used 2 sibling pairs (transgenic mice and wildtype littermate controls) to identify AD-associated miRNA dysregulation. High-throughput deep miRNA-seq has also been employed on a mouse model of PD bearing the human α-synuclein A53T mutation; the team reported dysregulation of specific miRNAs in the substantia nigra, including miR-144-5p, miR-200a-3p and miR-542-3p.127
Table 7.
MicroRNA sequencing studies in animal models of Alzheimer’s disease and Parkinson’s disease.
| Study | Model system | Age (months) | Brain tissue | Findings | Focus/Implications of study |
|---|---|---|---|---|---|
| Alzheimer’s disease | |||||
| Ma et al124 | Mouse, APP/PS1 | 6, 9 | CTX | ↑ 12 miRNAs at 6 mo, 27 miRNAs at 9 mo ↓ 24 miRNAs at 6 mo, 29 miRNAs at 9 mo |
Interactions between dysregulated miRNAs, circular RNAs and messenger RNAs may play important roles in AD pathogenesis |
| Li et al125 | Mouse, APP/PS1 | 1, 3, 6, 9 | CTX | Several dysregulated miRNAs across all age groups, for example, miR-80 reduced at 6- and 9-mo of age | Interactions between dysregulated miRNAs and messenger RNAs may play important roles in the progression of AD |
| Luo et al126 | Mouse, APP/PS1 | 9 | CTX | Several dysregulated miRNAs, for example, miR-99b-5p, 138-5p, 100-5p | Predicted pathways affected include axon guidance, PI3K/Akt signaling pathway and MAPK signaling pathway |
| Parkinson’s disease | |||||
| Mo et al127 | Mouse, α-synuclein | 12 | Midbrain | ↑ 32 miRNAs > 2-fold ↓ 25 miRNAs > 2-fold |
Candidate miRNAs from miRNA sequencing results were screened in cerebrospinal fluid from PD patients to identify potential biomarkers for PD diagnosis |
Abbreviations: AD, Alzheimer’s disease; CTX, cerebral cortex; MAPK, mitogen-activated protein kinase; miR or miRNA, microRNA; PD, Parkinson’s disease; PI3K/Akt, phosphatidylinositol 3-kinase/protein kinase B; SN, substantia nigra.
The up arrow (↑) represents upregulation or an increase in level. The down arrow (↓) represents downregulation or a decrease in level.
It is worth considering that miRNAs are specific to particular mechanisms within species, therefore it will be crucial to identify and confirm the human counterparts of miRNAs investigated in mice and other organisms. Furthermore, recognizing their associated pathways and targets can make the effective use of these findings in translational research achievable.
Candidate microRNA studies
The vast number of studies found during the literature search for this review consisted of candidate miRNA studies. These studies explored specific miRNAs to identify and confirm their dysregulation in the brain in the context of disease and determine their role in the disease process, using primarily RT-PCR. RT-PCR-based approaches detect miRNAs with high sensitivity and specificity, and are often used to validate microarray expression data or to identify changes identified in post-mortem brain tissues.123 Supplemental Tables S3–S5 summarize studies published to date investigating the dysregulation of miRNAs in AD, PD and ALS, respectively, with the reported miRNAs categorized according to their associated biological processes and pathways. Of note, the pathways and mechanisms described to be affected by these miRNAs are well-known features and drivers of neurodegeneration. When interpreting these findings, it is also important to acknowledge that miRNAs have diverse roles and that their effects on downstream pathways are frequently interconnected.
Many miRNAs have been investigated across several studies with concordant and discordant findings between different diseases. For example, miR-34a,128-131 miR-146a,132-136 and miR-155135-137 have been reported to be upregulated in AD rodent models. MiR-146a has also been shown to be disrupted in the cortex of ALS mice, but downregulated instead.138 In the study, Gomes et al and colleagues suggest that this downregulation of miR-146a in ALS mice may be an early event preceding the upregulation of other inflammatory molecules and pathways at the symptomatic stage of disease.138 The upregulation of miR-146a in AD models, on the other hand, may be involved in neuroprotective mechanisms possibly trying to prevent detrimental neuroinflammation in latter stages of disease. Other mutual miRNAs across neurodegenerative diseases include miR-124 (downregulated in AD139 and PD140 models), miR-34a (upregulated in AD128-131 and PD141 models) and miR-19a (upregulated in AD142 and ALS143 models), suggesting their involvement in common neurodegenerative processes. Interestingly, research on the dysregulation of some of these miRNAs in neuroglial cells has also emerged, including one study which showed miR-146a overexpression switched microglia to its neuroprotective phenotype in vitro and in APP/PS1 mice,144 adding to the body of research confirming the substantial role of glial cells in neurodegenerative diseases.
Caveats and future perspectives
Epigenetics in neurodegenerative diseases, particularly in vertebrate and invertebrate models, is a growing research field, with only limited work having been done to date, mostly focused in mouse models, which we described throughout this review. Additional model organisms, such as Drosophila and C. elegans, and their research potential, have also been discussed. Taken together, the research summarized in this review corroborates the utility of model organisms to study epigenetic regulation in the context of brain disease. There are, however, caveats associated with using these models to study the aforementioned complex diseases, and limitations of using model organisms should be addressed in studies utilizing them. Inconclusive reports to date using these models pose a major challenge in understanding the real importance of epigenetic mechanisms for neurodegenerative processes. The reasons why inconclusive findings have been reported—between different models and when comparing models to human post-mortem brains—are, at least in part, possibly related to whether a given model organism is the most suitable for the biological problem in question. Additional models that better recreate human disease are needed, which may offer the opportunity to overcome these challenges. Recent efforts have started to be put in place to improve existing mouse models in order to, for example, improve the modeling of as many aspects of AD as possible. For instance, Neuner et al and colleagues developed an AD-BXD mouse model by crossing 5xFAD mice with mice from the BXD genetic reference panel.145 Likewise, Yang et al146 crossed the APP/PS1 mouse model of AD with wild-derived strains of mice. The resultant strains improve genetic heterogeneity in the mouse models, consequently better illustrating the extensive genetic and epigenetic variation seen in humans. Furthermore, the MODEL-AD consortium is developing and rigorously characterizing the next generation of AD mouse models, aiming to develop models that closely reflect the sporadic late-onset human form of AD.147 These newer models offer a valuable opportunity to further investigate changes in genomic regulation in AD, and similar efforts for other neurodegenerative disorders should be pursuit.
Importantly, model organisms should be perceived as models, and the fact that they cannot recapitulate all aspects of brain disease should be acknowledged, considered, and embraced in studies making use of them. Model organisms do present many advantages (eg, short life span, tissue accessibility, testing for drug and molecular targets, powerful for functional validations and for understanding biological mechanisms and pathways), and their use should be focused on these strengths. Functional experiments in particular constitute some of the most powerful uses for these models. Indeed, functional studies successfully characterizing epigenetic changes identified in humans have started to emerge. As a follow up of DNA methylation and H3K4me3 changes identified in the gene ANK1 in human AD brains,50,148 its Drosophila ortholog has been functionally characterized, including to determine interactions with tau and Aβ, as well as how it is involved in neurodegeneration and memory processes.149
One limitation specific to epigenetic research using model organisms is that DNA methylation exhibits organism-specificity for certain species, with non-CpG DNA methylation being restricted to vertebrates, which is something important to consider when using invertebrate models.37 Overall, low levels of overall DNA methylation are found in organism such as C. elegans, and D. melanogaster. Despite Danio rerio (also known as zebrafish) being a well-established vertebrate model organism used in neuroscience, which exhibits DNA methylation,150 we did not come across any work to date using this powerful model to study DNA methylation in the context of neurodegenerative processes; future studies should explore genomic regulation processes in zebrafish models of neurodegenerative diseases.18,37 Similarly, non-human primates have many similarities to humans, including the presence of DNA methylation and the development of natural neurodegenerative pathologies17; very few studies have investigated genomic regulation in non-human primates, however, possibly as a result of the challenges associated with investigating non-human primates, such as their long lifespan, as well as ethical constrains in many countries. In fact, only one study to date investigated epigenetic changes in these models.151 Recently, Sato et al152 explored the generation of non-human primate models of AD to overcome some of their lifespan limitations, by adding mutations identified in familial cases of AD. The team focused primarily on the common marmosets (Callithrix jacchus), which exhibit much genetic, physiological, and anatomical proximity to humans. Most importantly, marmosets develop natural senile Aβ plaques and phosphorylated tau pathology in the brain. By inserting genetic mutations, as in other animal models discussed in this review, Sato et al152 reported that this fastens disease onset, facilitating the use of marmosets to study AD. These and additional, newly developed, models of AD, PD, and ALS are described in Table 8. Future studies should explore neuroepigenetic mechanisms in these models.
Table 8.
More recent animal models of Alzheimer’s disease and Parkinson’s disease.
| Study | Animal | Model | Description |
|---|---|---|---|
| Alzheimer’s disease | |||
| Neuner et al145 | Mouse | AD-BXD | 5xFAD mouse model of AD crossed with mice from the BXD genetic reference panel |
| Yang et al146 | Mouse | CAST.APP/PS1, WSB.APP/PS1, PWK.APP/PS1 | APP/PS1 mouse model of AD crossed with multiple wild-derived strains of mice (CAST, WSB, or PWK) |
| Baglietto-Vargas et al153 | Mouse | hAβ-KI | Knock-in of wildtype human Aβ under control of mouse App locus |
| Sato et al152 | Common marmosets | PSEN1-ΔE9 | Deletion of exon 9 of PSEN1 gene |
| Dong et al154 | Zebrafish | 7 dpf psen1Q96_K97del/ + larvae | Deletion of 6 nucleotides in the zebrafish psen1 gene |
| Barthelson et al155 | Zebrafish |
psen2T141 _ L142delinsMISLISV
psen2N140fs |
In-frame mutation or frameshift mutation, respectively, introduced at zebrafish psen2 gene |
| Benbow et al156 | C. elegans | Aβ1-42; tau-Tg | Pan-neuronally expresses both the toxic Aβ1-42 peptide and the wildtype 4R1N isoform of human tau |
| Parkinson’s disease | |||
| Dave et al157 | Rat | Pink1 KO, DJ-1 KO, Parkin KO |
Knockout of Pink1, DJ-1, or Parkin genes |
| Pütz et al158 | Drosophila | Mbt-null | Knockout of PAK4 homolog Mushroom bodies tiny |
Abbreviations: AD, Alzheimer’s disease; dpf, days post-fertilization; KI, knock-in; KO, knock-out; Tg, transgenic.
Additional strategies that can mitigate some of the limitations associated to the use of animal models include the use of human induced pluripotent stem cells (iPSC)-derived brain cells (eg, iPSC-derived neurons), including in co-culture systems containing neurons and other major brain cell types (eg, astrocytes and microglia).159 A major advantage of using iPSCs is the fact that it offers the possibility of generating brain cells from any donor, including from sporadic cases of neurodegenerative diseases and even prodromal or asymptomatic individuals. These attractive technologies come with their own limitations, however; for example, as a consequence of their cellular reprograming, involving epigenetic remodeling, iPSC-derived brain cells are transcriptionally and epigenetically similar to immature brain cells.160 This makes them great for studying neuronal development but poses challenges when studying aging-associated diseases. Studies using iPSCs have already contributed greatly to our understanding of certain aspects of neurodegenerative diseases, and, in parallel to studies using animal models, can be very powerful in understanding and validating findings in mice. Indeed, 2 studies from the same laboratory explored DNA methylation changes in the hexanucleotide repeat expansion in the C9orf72 gene known to cause ALS, in human iPSCs and mice. Specifically, Esanov et al and colleagues reported that hypermethylation in the C9orf72 promoter, seen in some ALS patients, was recapitulated in motor neuronal differentiation in 1 iPSC line and in the cortex of only a subset of C9BAC mice similarly to what is observed in the human C9-ALS population.161,162 According to the study using iPSCs, 5mC levels are reduced with reprograming but re-acquired with differentiation into motor neurons.161 In addition to methylation changes in the C9orf72 promoter, the second study provides evidence that hypermethylation of the hexanucleotide repeat itself in the cortex of C9BAC mice increases with age, which aligns with the developmental progression seen in patients.162 Important limitations of these 2 publications by Esanov et al should be considered when interpreting their findings, however; examples include the investigation of a very low number of subjects (human and mouse), and, importantly, the observation of C9orf72 promoter hypermethylation with differentiation in motor neurons from a single iPSC line, as well as the use of a single mouse model. Further studies are needed to validate these findings, particularly to disentangle the complex relationship between 5mC and 5hmC in the context of C9orf72 repeat expansion.
Considering the research covered in this review, a limited number of studies explored enrichment of epigenetic marks to specific functional regions systematically. The study by Gjoneska et al and colleagues, profiling histone modifications in p25-inducible transgenic mice, explored functional enrichment of regulatory elements for changes in promoter and enhancer regions in detail.83 By performing enrichment analysis of gene ontology (GO) categories from differential gene expression analysis in the same mice, Gjoneska et al83 reported that increased-level enhancers and promoters were enriched for immune and stimulus-response functions, and decreased-level enhancers and promoters were enriched for synapse and learning-associated functions in CK-p25 mice. Enrichment of regulatory motifs uncovered distinct regulatory motifs for promoters and enhancers: increased-level peaks exhibited enrichment for NFκB, E2F, PPARG, IRF and PU.1 for both (suggesting targeting of immune regulation), decreased-levels in enhancers enriched for DNA-binding RFX motifs, and decreased-levels in promoters enriched for zinc-finger ZIC motifs. In addition to correlating histone marks with gene expression, the authors went even further by evaluating the effect of increased-levels for enhancer regions in gene expression in cell models and found that 8 out of the 9 increased-level human orthologs tested were indeed able to drive in vitro expression. Also at a functional validation level, Sanchez-Mut et al66 used a custom DNA methylation array specifically containing promoters of genes related to sensory perception, cognition, neuroplasticity, brain physiology and mental disorders, to interrogate relevant regulatory elements in 12 brain regions of APP/PS1 and 3xTg-AD mice. Importantly, hypermethylation of promoter regions for Sorbs3, Spnb4 and Tbxa2r in the prefrontal cortex of APP/PS1 and 3xTg-AD mice was also observed in human frontal cortex (corresponding human orthologs) from individuals at late stages of AD.
One major methodological caveat of DNA methylation studies to date is the use of sodium bisulfite treatment as part of the laboratory techniques to profile DNA methylation, which hinders the ability to differentiate between 5mC and 5hmC.163 This is important because 5hmC is an individual methylation mark alongside 5mC,164 which regulates many relevant brain processes, as mentioned. Oxidative bisulfite conversion can be employed to overcome this issue, where 5hmC is converted to 5fC, and then uracil, thus allowing accurate detection of 5mC alone; 5hmC can be estimated from the quantification of the difference between bisulfite and oxidative bisulfite conversions.165 It should be noted that any approach involving bisulfite conversion damages nucleic acids, resulting in short DNA fragments, however. The uprising of third-generation sequencing (also known as long-read sequencing) technologies, such as nanopore sequencing, provide encouraging alternatives that can overcome limitations of conventional bisulfite sequencing. Despite the excitement associated with the opportunities that forefront sequencing technologies can offer, including next-generation sequencing in addition to long-read sequencing, these methods are realistically not accessible to all research laboratories, mostly because of their high costs, the need for specialized equipment and staff, and the scarcity of standardized bioinformatic pipelines for epigenomic analyses. Methylation arrays are reliable and still widely used alternatives for the interrogation of selected methylation sites across the genome, offering reduced costs, i.e., proving more accessible and feasible for many research groups. Vertebrate arrays, and in particular standardized mouse methylation arrays, also offer insightful venues for studying DNA methylation, and should also be considered for near future studies.
Of great importance, studies in model organisms of neurodegenerative diseases performed so far have not explored epigenetic changes in specific brain subregions and their layers, and in different cell populations. Firstly, different brain regions and their subregions are involved in controlling a diverse range of activities and behaviors and are affected differently in disease. Most importantly, different tissues and cell types have distinct epigenetic profiles, which regulate their unique functions. A range of cells, from neurons to glial cells (such as microglia, astrocytes, and oligodendrocytes), and their respective subtypes, make up the cells in the brain, each with important individual roles. The use of cell deconvolution computational algorithms enables the opportunity to disentangle, at least in part, cell-specific patterns and proportions in studies using bulk samples, improving interpretability and reducing confounding effects of cellular heterogeneity. These computational techniques rely on reference panels and their quality, accuracy, and similarity to the testing samples, however, often revealing not to be the most appropriate approach to employ. The purification of different cell populations using cell sorting techniques, such as fluorescence-activated cell sorting,166 magnetic affinity cell sorting167 and laser capture microdissection, can overcome these limitations.168 Additionally, emerging single-cell technologies offer a powerful and reliable space for refining epigenomic regulation at the level of individual cells, allowing the identification of key individual cell changes and how different cell and their associated changes relate to each other, as well as the identification of relevant cell populations. Combined with the ongoing rise of spatial epigenomics, rendering the possibility to survey distributions in different regions, layers, and cells of the brain, the fast-growing field of epigenomics (and multiomics) technologies promises many breakthroughs and much knowledge expansion in the next few years. Moving forward, it is important that future epigenetic studies discriminate cell-specific profiles, and thus take advantage of state-of-the art methodologies, such as the above-mentioned technologies, to collect more refined and insightful data for the better understanding of neurogenerative diseases and their complexity.
Epigenetic research has gained a lot of attention in recent years, with the implication of epigenetic processes in the brain, and particularly in neurodegenerative diseases, having been an important contributor in better understanding these conditions. Of note, epigenetic mechanisms are dynamic and reversible, changing throughout life, development, aging and disease, and being influenced by the environment. Importantly, this also implicates that they possess high potential as disease biomarkers and drug targets.16
As technologies continue to advance and models continue to improve, more studies taking advantage of cutting-edge laboratory and analytical approaches to survey model organisms and their overlaps with humans are necessary, to grasp a better understanding of how and when we should modify and manipulate aspects of human neurodegenerative diseases that are recapitulated in these models.
Conclusions
Research investigating neurodegenerative diseases, including AD, PD, and ALS, has shown that genetics alone cannot explain disease etiology, and that additional genomic processes, such as epigenetic mechanisms, may also have an important contribution. Understanding the role of these mechanisms in said diseases is still a nascent research field, and more is still to be understood about the consequences of any changes in the expression and activity of epigenetic regulation.
Human studies exploring the epigenetic landscape of brain diseases rely on post-mortem tissue, which usually corresponds to end stages of the disease and often displays significant degeneration, hence the use of model organisms offers the advantage to in understand epigenetic changes at different stages of the neurodegenerative process, including at earlier stages, as well as changes that mirror disease progression.
Throughout this review we discussed a range of vertebrate and invertebrate models that have been used to date to model neurodegenerative diseases to investigate epigenetic changes, which has mostly focused on studying AD. In parallel with advancements of technologies and laboratory methods for epigenomic assessments, increasing efforts in developing innovative and viable disease models are currently underway; it will be paramount to maximize the use of these models to ascertain the genomic dysregulation in neurodegenerative diseases, in order to identify and test effective drug targets and molecules for treating them (Figure 1). Future research expanding on the studies described here, especially studies taking advantage of emerging single-cell and spatial epigenomic technologies, will certainly transform our understanding of neurodegenerative diseases and deliver important insights that can be applied for their alleviation.
Supplemental Material
Supplemental material, sj-docx-1-gae-10.1177_25168657221135848 for The Neuroepigenetic Landscape of Vertebrate and Invertebrate Models of Neurodegenerative Diseases by Thanga Harini Sundaramoorthy and Isabel Castanho in Epigenetics Insights
Acknowledgments
We thank Jonathan Mill for his expertise, mentorship and support throughout all aspects of this project.
Footnotes
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This project was supported in part by funding from the University of Exeter to cover THS’s medical science research project. The remaining funding, including I.C.’s postdoctoral fellowship position at the University of Exeter when this work was developed, was supported by a research grant from Alzheimer’s Research UK (ARUK-PG2018B-016).
Ethical statements and informed consent information (if applicable): Not applicable
ORCID iD: Isabel Castanho  https://orcid.org/0000-0003-1413-626X
https://orcid.org/0000-0003-1413-626X
Supplemental material: Supplemental material for this article is available online.
References
- 1. Gan L, Cookson MR, Petrucelli L, La Spada AR. Converging pathways in neurodegeneration, from genetics to mechanisms. Nat Neurosci. 2018;21:1300-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alzheimer’s Association. 2020 Alzheimer’s disease facts and figures. Alzheimers Dement. 2020. https://alz-journals.onlinelibrary.wiley.com/doi/full/10.1002/alz.12068 [Google Scholar]
- 3. National Institute for Health and Care Excellence. Parkinson’s Disease: How Common is It? Updated 2018. Accessed December 27, 2021. https://cks.nice.org.uk/topics/parkinsons-disease/background-information/prevalence/
- 4. Longinetti E, Fang F. Epidemiology of amyotrophic lateral sclerosis: an update of recent literature. Curr Opin Neurol. 2019;32:771-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dawson TM, Golde TE, Lagier-Tourenne C. Animal models of neurodegenerative diseases. Nat Neurosci. 2018;21:1370-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Berson A, Nativio R, Berger SL, Bonini NM. Epigenetic regulation in neurodegenerative diseases. Trends Neurosci. 2018;41:587-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Coppedè F. Genetics and epigenetics of Parkinson’s disease. ScientificWorldJournal. 2012;2012:489830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Belzil VV, Katzman RB, Petrucelli L. ALS and FTD: an epigenetic perspective. Acta Neuropathol. 2016;132:487-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ferrari R, Kapogiannis D, Huey ED, Momeni P. FTD and ALS: a tale of two diseases. Curr Alzheimer Res. 2011;8:273-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lunnon K, Mill J. Epigenetic studies in Alzheimer’s disease: current findings, caveats, and considerations for future studies. Am J Med Genet B Neuropsychiatr Genet. 2013;162B:789-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gatz M, Reynolds CA, Fratiglioni L, et al. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry. 2006;63:168-174. [DOI] [PubMed] [Google Scholar]
- 12. Nee LE, Lippa CF. Alzheimer’s disease in 22 twin pairs—13-year follow-up: hormonal, infectious and traumatic factors. Dement Geriatr Cogn Disord. 1999;10:148-151. [DOI] [PubMed] [Google Scholar]
- 13. Cook RH, Schneck SA, Clark DB. Twins with Alzheimer’s disease. Arch Neurol. 1981;38:300-301. [DOI] [PubMed] [Google Scholar]
- 14. Goldman SM, Marek K, Ottman R, et al. Concordance for Parkinson’s disease in twins: A 20-year update. Ann Neurol. 2019;85:600-605. [DOI] [PubMed] [Google Scholar]
- 15. Tarr IS, McCann EP, Benyamin B, et al. Monozygotic twins and triplets discordant for amyotrophic lateral sclerosis display differential methylation and gene expression. Sci Rep. 2019;9:8254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yao B, Christian KM, He C, Jin P, Ming GL, Song H. Epigenetic mechanisms in neurogenesis. Nat Rev Neurosci. 2016;17:537-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Drummond E, Wisniewski T. Alzheimer’s disease: experimental models and reality. Acta Neuropathol. 2017;133:155-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mullane K, Williams M. Preclinical models of Alzheimer’s Disease: Relevance and translational validity. Curr Protoc Pharmacol. 2019;84:e57. [DOI] [PubMed] [Google Scholar]
- 19. Griñán-Ferré C, Corpas R, Puigoriol-Illamola D, Palomera-ávalos V, Sanfeliu C, Pallàs M. Understanding epigenetics in the neurodegeneration of Alzheimer’s Disease: SAMP8 Mouse Model. J Alzheimers Dis. 2018;62:943-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Oddo S, Caccamo A, Shepherd JD, et al. Triple-transgenic model of Alzheimer’s disease with plaques and tangles: intracellular Abeta and synaptic dysfunction. Neuron. 2003;39:409-421. [DOI] [PubMed] [Google Scholar]
- 21. Oakley H, Cole SL, Logan S, et al. Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J Neurosci. 2006;26:10129-10140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Saito T, Matsuba Y, Mihira N, et al. Single App knock-in mouse models of Alzheimer’s disease. Nat Neurosci. 2014;17:661-663. [DOI] [PubMed] [Google Scholar]
- 23. Do Carmo S, Hanzel CE, Jacobs ML, et al. Rescue of early bace-1 and global DNA demethylation by S-adenosylmethionine reduces amyloid pathology and improves cognition in an Alzheimer’s model. Sci Rep. 2016;6:34051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jankowsky JL, Fadale DJ, Anderson J, et al. Mutant presenilins specifically elevate the levels of the 42 residue beta-amyloid peptide in vivo: evidence for augmentation of a 42-specific gamma secretase. Hum Mol Genet. 2004;13:159-170. [DOI] [PubMed] [Google Scholar]
- 25. Radde R, Bolmont T, Kaeser SA, et al. Abeta42-driven cerebral amyloidosis in transgenic mice reveals early and robust pathology. EMBO Rep. 2006;7:940-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sturchler-Pierrat C, Abramowski D, Duke M, et al. Two amyloid precursor protein transgenic mouse models with Alzheimer disease-like pathology. Proc Natl Acad Sci U S A. 1997;94:13287-13292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cruz JC, Tseng HC, Goldman JA, Shih H, Tsai LH. Aberrant cdk5 activation by p25 triggers pathological events leading to neurodegeneration and neurofibrillary tangles. Neuron. 2003;40:471-483. [DOI] [PubMed] [Google Scholar]
- 28. Mucke L, Masliah E, Yu GQ, et al. High-level neuronal expression of abeta 1-42 in wild-type human amyloid protein precursor transgenic mice: synaptotoxicity without plaque formation. J Neurosci. 2000;20:4050-4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. ArunSundar M, Shanmugarajan TS, Ravichandiran V. 3,4-dihydroxyphenylethanol assuages cognitive impulsivity in Alzheimer’s disease by attuning HPA-Axis via differential crosstalk of α7 nAChR with MicroRNA-124 and hdac6. ACS Chem Neurosci. 2018;9:2904-2916. [DOI] [PubMed] [Google Scholar]
- 30. Yoshiyama Y, Higuchi M, Zhang B, et al. Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron. 2007;53:337-351. [DOI] [PubMed] [Google Scholar]
- 31. Holcomb L, Gordon MN, McGowan E, et al. Accelerated Alzheimer-type phenotype in transgenic mice carrying both mutant amyloid precursor protein and presenilin 1 transgenes. Nat Med. 1998;4:97-100. 1998. [DOI] [PubMed] [Google Scholar]
- 32. Mirnics K, Norstrom EM, Garbett K, et al. Molecular signatures of neurodegeneration in the cortex of PS1/PS2 double knockout mice. Mol Neurodegener. 2008;3:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yagi H, Katoh S, Akiguchi I, Takeda T. Age-related deterioration of ability of acquisition in memory and learning in senescence accelerated mouse: SAM-P/8 as an animal model of disturbances in recent memory. Brain Res. 1988;474:86-93. 1988. [DOI] [PubMed] [Google Scholar]
- 34. Shimada A, Ohta A, Akiguchi I, Takeda T. Age-related deterioration in conditional avoidance task in the SAM-P/10 mouse, an animal model of spontaneous brain atrophy. Brain Res. 1993;608:266-272. 1993. [DOI] [PubMed] [Google Scholar]
- 35. Schindowski K, Bretteville A, Leroy K, et al. Alzheimer’s disease-like tau neuropathology leads to memory deficits and loss of functional synapses in a novel mutated tau transgenic mouse without any motor deficits. Am J Pathol. 2006;169:599-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hsiao K, Chapman P, Nilsen S, et al. Correlative memory deficits, Abeta elevation, and amyloid plaques in transgenic mice. Science. 1996;274:99-102. [DOI] [PubMed] [Google Scholar]
- 37. de Mendoza A, Lister R, Bogdanovic O. Evolution of DNA methylome diversity in Eukaryotes. J Mol Biol. 2019;432:1687-1705. doi: 10.1016/j.jmb.2019.11.003 [DOI] [PubMed] [Google Scholar]
- 38. Sun W, Zang L, Shu Q, Li X. From development to diseases: the role of 5hmC in brain. Genomics. 2014;104:347-351. [DOI] [PubMed] [Google Scholar]
- 39. de Mendoza A, Poppe D, Buckberry S, et al. The emergence of the brain non-CpG methylation system in vertebrates. Nat Ecol Evol. 2021;5:369-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Greer E, Blanco M, Gu L, et al. DNA Methylation on N6-Adenine in C. elegans. Cell. 2015;161:868-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang Y, Zhang Z, Li L, et al. Selective loss of 5hmC promotes neurodegeneration in the mouse model of Alzheimer’s disease. FASEB J. 2020;34:16364-16382. [DOI] [PubMed] [Google Scholar]
- 42. Griñán-Ferré C, Sarroca S, Ivanova A, et al. Epigenetic mechanisms underlying cognitive impairment and Alzheimer disease hallmarks in 5XFAD mice. Aging. 2016;8:664-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Griñán-Ferré C, Marsal-García L, Bellver-Sanchis A, et al. Pharmacological inhibition of G9a/GLP restores cognition and reduces oxidative stress, neuroinflammation and β-amyloid plaques in an early-onset Alzheimer’s disease mouse model. Aging. 2019;11:11591-11608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Griñán-Ferré C, Izquierdo V, Otero E, et al. Environmental enrichment improves cognitive deficits, AD hallmarks and epigenetic alterations presented in 5xFAD Mouse Model. Front Cell Neurosci. 2018;12:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cosín-Tomás M, Álvarez-López MJ, Companys-Alemany J, et al. Temporal integrative analysis of mRNA and microRNAs expression profiles and epigenetic alterations in female SAMP8, a model of Age-Related cognitive decline. Front Genet. 2018;9:596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Huang Q, Xu S, Mo M, et al. Quantification of DNA methylation and hydroxymethylation in Alzheimer’s disease mouse model using LC-MS/MS. J Mass Spectrom. 2018;53:590-594. [DOI] [PubMed] [Google Scholar]
- 47. Cadena-del-Castillo C, Valdes-Quezada C, Carmona-Aldana F, Arias C, Bermúdez-Rattoni F, Recillas-Targa F. Age-dependent increment of hydroxymethylation in the brain cortex in the triple-transgenic mouse model of Alzheimer’s disease. J Alzheimers Dis. 2014;41:845-854. [DOI] [PubMed] [Google Scholar]
- 48. Shu L, Sun W, Li L, et al. Genome-wide alteration of 5-hydroxymenthylcytosine in a mouse model of Alzheimer’s disease. BMC Genomics. 2016;17:381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Serrano-Pozo A, Frosch MP, Masliah E, Hyman BT. Neuropathological alterations in Alzheimer disease. Cold Spring Harb Perspect Med. 2011;1:a006189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lunnon K, Smith R, Hannon E, et al. Methylomic profiling implicates cortical deregulation of ANK1 in Alzheimer’s disease. Nat Neurosci. 2014;17:1164-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Desplats P, Spencer B, Coffee E, et al. Alpha-synuclein sequesters dnmt1 from the nucleus: a novel mechanism for epigenetic alterations in Lewy body diseases. J Biol Chem. 2011;286:9031-9037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Figueroa-Romero C, Guo K, Murdock BJ, et al. Temporal evolution of the microbiome, immune system and epigenome with disease progression in ALS mice. Dis Model Mech. 2019;13:13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kaut O, Schmitt I, Wüllner U. Genome-scale methylation analysis of Parkinson’s disease patients’ brains reveals DNA hypomethylation and increased mRNA expression of cytochrome P450 2E1. Neurogenetics. 2012;13:87-91. [DOI] [PubMed] [Google Scholar]
- 54. Young JI, Sivasankaran SK, Wang L, et al. Erratum: genome-wide brain DNA methylation analysis suggests epigenetic reprogramming in Parkinson disease. Neurol Genet. 2019;5:e355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Navarro-Sánchez L, Águeda-Gómez B, Aparicio S, Pérez-Tur J. Epigenetic study in Parkinson’s Disease: A pilot analysis of DNA methylation in candidate genes in brain. Cells. 2018;7:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Morahan JM, Yu B, Trent RJ, Pamphlett R. A genome-wide analysis of brain DNA methylation identifies new candidate genes for sporadic amyotrophic lateral sclerosis. Amyotroph Lateral Scler. 2009;10:418-429. [DOI] [PubMed] [Google Scholar]
- 57. Taskesen E, Mishra A, van der Sluis S, et al. Author correction: susceptible genes and disease mechanisms identified in frontotemporal dementia and frontotemporal dementia with amyotrophic lateral sclerosis by DNA-methylation and GWAS. Sci Rep. 2018;8:7789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kim BW, Jeong YE, Wong M, Martin LJ. DNA damage accumulates and responses are engaged in human ALS brain and spinal motor neurons and DNA repair is activatable in iPSC-derived motor neurons with SOD1 mutations. Acta Neuropathol Commun. 2020;8:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ji P, Wang X, Xie N, Li Y. N6-methyladenosine in RNA and DNA: an epitranscriptomic and epigenetic player implicated in determination of Stem Cell Fate. Stem Cells Int. 2018;2018:3256524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li Z, Zhao P, Xia Q. Epigenetic methylations on N6-adenine and N6-adenosine with the same input but different output. Int J Mol Sci. 2019;20:2931. doi: 10.3390/ijms20122931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lv S, Zhou X, Li YM, et al. N6-methyladenine-modified DNA was decreased in Alzheimer’s disease patients. World J Clin Cases. 2022;10:448-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lu X, Yang B, Yu H, et al. Epigenetic mechanisms underlying the effects of triptolide and tripchlorolide on the expression of neuroligin-1 in the hippocampus of APP/PS1 transgenic mice. Pharm Biol. 2019;57:453-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Li P, Quan W, Wang Z, Chen Y, Zhang H, Zhou Y. AD7c-NTP impairs adult striatal neurogenesis by affecting the biological function of MeCP2 in APP/PSl transgenic mouse model of Alzheimer’s Disease. Front Aging Neurosci. 2020;12:616614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhang HQ, Wang JY, Li ZF, et al. DNA methyltransferase 1 is dysregulated in Parkinson’s disease via mediation of miR-17. Mol Neurobiol. 2021;58:2620-2633. [DOI] [PubMed] [Google Scholar]
- 65. Wu TT, Liu T, Li X, et al. TET2-mediated Cdkn2A DNA hydroxymethylation in midbrain dopaminergic neuron injury of Parkinson’s disease. Hum Mol Genet. 2020;29:1239-1252. [DOI] [PubMed] [Google Scholar]
- 66. Sanchez-Mut JV, Aso E, Panayotis N, et al. DNA methylation map of mouse and human brain identifies target genes in Alzheimer’s disease. Brain. 2013;136:3018-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cong L, Jia J, Qin W, Ren Y, Sun Y. Genome-wide analysis of DNA methylation in an APP/PS1 mouse model of Alzheimer’s disease. Acta Neurol Belg. 2014;114:195-206. [DOI] [PubMed] [Google Scholar]
- 68. Zhang S, Qin C, Cao G, Guo L, Feng C, Zhang W. Genome-wide analysis of DNA methylation profiles in a senescence-accelerated mouse prone 8 brain using whole-genome bisulfite sequencing. Bioinformatics. 2017;33:1591-1595. [DOI] [PubMed] [Google Scholar]
- 69. Tang X, Ruan S, Tang Q, et al. DNA methylation profiles in the hippocampus of an Alzheimer’s disease mouse model at mid-stage neurodegeneration. Int J Clin Exp Med. 2018;11:11876-11888. [Google Scholar]
- 70. Kundu P, Torres ERS, Stagaman K, et al. Integrated analysis of behavioral, epigenetic, and gut microbiome analyses in App(nl-G-F), App(NL-F), and wild type mice. Sci Rep. 2021;11:4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wreczycka K, Gosdschan A, Yusuf D, Grüning B, Assenov Y, Akalin A. Strategies for analyzing bisulfite sequencing data. J Biotechnol. 2017;261:105-115. [DOI] [PubMed] [Google Scholar]
- 72. Lev Maor G, Yearim A, Ast G. The alternative role of DNA methylation in splicing regulation. Trends Genet. 2015;31:274-280. [DOI] [PubMed] [Google Scholar]
- 73. Bie B, Wu J, Yang H, Xu JJ, Brown DL, Naguib M. Epigenetic suppression of neuroligin 1 underlies amyloid-induced memory deficiency. Nat Neurosci. 2014;17:223-231. [DOI] [PubMed] [Google Scholar]
- 74. Seto E, Yoshida M. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol. 2014;6:a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kimura H. Histone modifications for human epigenome analysis. J Hum Genet. 2013;58:439-445. [DOI] [PubMed] [Google Scholar]
- 76. Cosín-Tomás M, Alvarez-López MJ, Sanchez-Roige S, et al. Epigenetic alterations in hippocampus of SAMP8 senescent mice and modulation by voluntary physical exercise. Front Aging Neurosci. 2014;6:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Walker MP, LaFerla FM, Oddo SS, Brewer GJ. Reversible epigenetic histone modifications and bdnf expression in neurons with aging and from a mouse model of Alzheimer’s disease. Age (Dordr). 2013;35:519-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Govindarajan N, Agis-Balboa RC, Walter J, Sananbenesi F, Fischer A. Sodium butyrate improves memory function in an Alzheimer’s disease mouse model when administered at an advanced stage of disease progression. J Alzheimers Dis. 2011;26:187-197. [DOI] [PubMed] [Google Scholar]
- 79. Zhang H, Karisetty BC, Bhatnagar A, et al. Tip60 protects against amyloid-β-induced transcriptomic alterations via different modes of action in early versus late stages of neurodegeneration. Mol Cell Neurosci. 2020;109:103570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lithner CU, Lacor PN, Zhao WQ, et al. Disruption of neocortical histone H3 homeostasis by soluble aβ: implications for Alzheimer’s disease. Neurobiol Aging. 2013;34:2081-2090. [DOI] [PubMed] [Google Scholar]
- 81. Chatterjee S, Cassel R, Schneider-Anthony A, et al. Reinstating plasticity and memory in a tauopathy mouse model with an acetyltransferase activator. EMBO Mol Med. 2018;10:e8587. doi: 10.15252/emmm.201708587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Gräff J, Rei D, Guan JS, et al. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature. 2012;483:222-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Gjoneska E, Pfenning AR, Mathys H, et al. Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer’s disease. Nature. 2015;518:365-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Klein HU, McCabe C, Gjoneska E, et al. Epigenome-wide study uncovers large-scale changes in histone acetylation driven by tau pathology in aging and Alzheimer’s human brains. Nat Neurosci. 2019;22:37-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Zheng Y, Liu A, Wang ZJ, et al. Inhibition of EHMT1/2 rescues synaptic and cognitive functions for Alzheimer’s disease. Brain. 2019;142:787-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Cao Q, Wang W, Williams JB, Yang F, Wang ZJ, Yan Z. Targeting histone K4 trimethylation for treatment of cognitive and synaptic deficits in mouse models of Alzheimer’s disease. Sci Adv. 2020;6:eabc8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Anderson KW, Mast N, Pikuleva IA, Turko IV. Histone H3 Ser57 and Thr58 phosphorylation in the brain of 5XFAD mice. FEBS Open Bio. 2015;5:550-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Marzi SJ, Leung SK, Ribarska T, et al. A histone acetylome-wide association study of Alzheimer’s disease identifies disease-associated H3K27ac differences in the entorhinal cortex. Nat Neurosci. 2018;21:1618-1627. [DOI] [PubMed] [Google Scholar]
- 89. Huang M, Lou D, Charli A, et al. Mitochondrial dysfunction-induced H3K27 hyperacetylation perturbs enhancers in Parkinson’s disease. JCI Insight. 2021;6:e138088. doi: 10.1172/jci.insight.138088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Dyer M, Phipps AJ, Mitew S, Taberlay PC, Woodhouse A. Age, but not amyloidosis, induced changes in global levels of histone modifications in susceptible and disease-resistant neurons in Alzheimer’s disease model mice. Front Aging Neurosci. 2019;11:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Casciaro F, Persico G, Rusin M, et al. The Histone H3 k4me3, k27me3, and k27ac genome-wide distributions are differently influenced by Sex in Brain Cortexes and Gastrocnemius of the Alzheimer’s Disease PSAPP Mouse Model. Epigenomes. 2021;5:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Nicholas AP, Lubin FD, Hallett PJ, et al. Striatal histone modifications in models of levodopa-induced dyskinesia. J Neurochem. 2008;106:486-494. [DOI] [PubMed] [Google Scholar]
- 93. Mu MD, Qian ZM, Yang SX, Rong KL, Yung WH, Ke Y. Therapeutic effect of a histone demethylase inhibitor in Parkinson’s disease. Cell Death Dis. 2020;11:927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Liu D, Tang H, Li XY, et al. Targeting the HDAC2/HNF-4A/miR-101b/AMPK pathway rescues Tauopathy and dendritic abnormalities in Alzheimer’s disease. Mol Ther. 2017;25:752-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Südhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455:903-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Kim T, Song S, Park Y, Kang S, Seo H. HDAC inhibition by valproic acid induces neuroprotection and improvement of PD-like behaviors in LRRK2 R1441G transgenic mice. Exp Neurobiol. 2019;28:504-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Nakatsuka D, Izumi T, Tsukamoto T, et al. Histone deacetylase 2 knockdown ameliorates morphological abnormalities of dendritic branches and spines to improve synaptic plasticity in an APP/PS1 transgenic mouse model. Front Mol Neurosci. 2021;14:782375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Song Y, Hu M, Zhang J, Teng ZQ, Chen C. A novel mechanism of synaptic and cognitive impairments mediated via microRNA-30b in Alzheimer’s disease. EBioMedicine. 2019;39:409-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Rodriguez-Ortiz CJ, Baglietto-Vargas D, Martinez-Coria H, LaFerla FM, Kitazawa M. Upregulation of miR-181 decreases c-Fos and SIRT-1 in the hippocampus of 3xTg-AD mice. J Alzheimers Dis. 2014;42:1229-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Zhang QS, Liu W, Lu GX. miR-200a-3p promotes b-Amyloid-induced neuronal apoptosis through down-regulation of SIRT1 in Alzheimer’s disease. J Biosci. 2017;42:397-404. [DOI] [PubMed] [Google Scholar]
- 101. Coupé P, Manjón JV, Lanuza E, Catheline G. Lifespan changes of the human brain in Alzheimer’s Disease. Sci Rep. 2019;9:3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Tao H, Liu Y, Hou Y. miRNA-384-5p regulates the progression of Parkinson’s disease by targeting SIRT1 in mice and SH-SY5Y cell. Int J Mol Med. 2019;45:441-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Maciotta S, Meregalli M, Torrente Y. The involvement of microRNAs in neurodegenerative diseases. Front Cell Neurosci. 2013;7:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Wang X, Liu D, Huang HZ, et al. A novel MicroRNA-124/PTPN1 signal pathway mediates synaptic and memory deficits in Alzheimer’s disease. Biol Psychiatry. 2018;83:395-405. [DOI] [PubMed] [Google Scholar]
- 105. Higaki S, Muramatsu M, Matsuda A, et al. Defensive effect of microRNA-200b/c against amyloid-beta peptide-induced toxicity in Alzheimer’s disease models. PLoS One. 2018;13:e0196929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Schonrock N, Ke YD, Humphreys D, et al. Neuronal microRNA deregulation in response to Alzheimer’s disease amyloid-beta. PLoS One. 2010;5:e11070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Liu CG, Wang JL, Li L, Xue LX, Zhang YQ, Wang PC. MicroRNA-135a and -200b, potential biomarkers for Alzheimer׳s disease, regulate β secretase and amyloid precursor protein. Brain Res. 2014;1583:55-64. [DOI] [PubMed] [Google Scholar]
- 108. Wang LL, Min L, Guo QD, et al. Profiling microRNA from brain by microarray in a transgenic mouse model of Alzheimer’s Disease. Biomed Res Int. 2017;2017:8030369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Li F, Wei G, Bai Y, et al. MicroRNA-574 is involved in cognitive impairment in 5-month-old APP/PS1 mice through regulation of neuritin. Brain Res. 2015;1627:177-188. [DOI] [PubMed] [Google Scholar]
- 110. Wang X, Liu P, Zhu H, et al. miR-34a, a microRNA up-regulated in a double transgenic mouse model of Alzheimer’s disease, inhibits bcl2 translation. Brain Res Bull. 2009;80:268-273. [DOI] [PubMed] [Google Scholar]
- 111. Noh H, Park C, Park S, Lee YS, Cho SY, Seo H. Prediction of miRNA-mRNA associations in Alzheimer’s disease mice using network topology. BMC Genomics. 2014;15:644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Zhang J, Hu M, Teng Z, Tang YP, Chen C. Synaptic and cognitive improvements by inhibition of 2-AG metabolism are through upregulation of microRNA-188-3p in a mouse model of Alzheimer’s disease. J Neurosci. 2014;34:14919-14933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Barak B, Shvarts-Serebro I, Modai S, et al. Opposing actions of environmental enrichment and Alzheimer’s disease on the expression of hippocampal microRNAs in mouse models. Transl Psychiatry. 2013;3:e304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Zhou H, Zhang R, Lu K, et al. Deregulation of miRNA-181c potentially contributes to the pathogenesis of AD by targeting collapsin response mediator protein 2 in mice. J Neurol Sci. 2016;367:3-10. [DOI] [PubMed] [Google Scholar]
- 115. Zhang Y, Li Q, Liu C, et al. MiR-214-3p attenuates cognition defects via the inhibition of autophagy in SAMP8 mouse model of sporadic Alzheimer’s disease. Neurotoxicol. 2016;56:139-149. [DOI] [PubMed] [Google Scholar]
- 116. Zhang R, Zhang Q, Niu J, et al. Screening of microRNAs associated with Alzheimer’s disease using oxidative stress cell model and different strains of senescence accelerated mice. J Neurol Sci. 2014;338:57-64. [DOI] [PubMed] [Google Scholar]
- 117. Ham S, Kim TK, Lee S, Tang YP, Im HI. MicroRNA profiling in aging brain of PSEN1/PSEN2 double knockout mice. Mol Neurobiol. 2018;55:5232-5242. [DOI] [PubMed] [Google Scholar]
- 118. Wang Z, Xu P, Chen B, et al. Identifying circRNA-associated-ceRNA networks in the hippocampus of aβ1-42-induced Alzheimer’s disease-like rats using microarray analysis. Aging. 2018;10:775-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Li L, Xu Y, Zhao M, Gao Z. Neuro-protective roles of long non-coding RNA MALAT1 in Alzheimer’s disease with the involvement of the microRNA-30b/CNR1 network and the following PI3K/AKT activation. Exp Mol Pathol. 2020;117:104545. [DOI] [PubMed] [Google Scholar]
- 120. Kong Y, Wu J, Yuan L. MicroRNA expression analysis of adult-onset Drosophila Alzheimer’s disease model. Curr Alzheimer Res. 2014;11:882-891. [PubMed] [Google Scholar]
- 121. Asikainen S, Rudgalvyte M, Heikkinen L, et al. Global microRNA expression profiling of Caenorhabditis elegans Parkinson’s disease models. J Mol Neurosci. 2010;41:210-218. [DOI] [PubMed] [Google Scholar]
- 122. Shen L, Wang C, Chen L, Wong G. Dysregulation of MicroRNAs and PIWI-Interacting RNAs in a Caenorhabditis elegans Parkinson’s disease model overexpressing human α-Synuclein and influence of tdp-1. Front Neurosci. 2021;15:600462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Watanabe Y, Maekawa M. Chapter 2 - methods and strategies to determine epigenetic variation in human disease. In: Tollefsbol TO, ed. Epigenetics in Human Disease. 1st ed. Academic Press; 2012;7-27. [Google Scholar]
- 124. Ma N, Pan J, Ye X, Yu B, Zhang W, Wan J. Whole-transcriptome analysis of APP/PS1 Mouse Brain and identification of circRNA-miRNA-mRNA networks to investigate AD pathogenesis. Mol Ther Nucleic Acids. 2019;18:1049-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Li ZR, Liu R, Zeng L, Jiang HL, Ashraf G. MicroRNA and mRNA profiling of cerebral cortex in a transgenic mouse model of Alzheimer’s disease by RNA sequencing. Neural Regen Res. 2021;16:2099-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Luo H, Wu Q, Ye X, et al. Genome-wide analysis of miRNA signature in the APPswe/PS1ΔE9 mouse model of alzheimer’s disease. PLoS One. 2014;9:e101725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Mo M, Xiao Y, Huang S, et al. MicroRNA expressing profiles in A53T mutant alpha-synuclein transgenic mice and Parkinsonian. Oncotarget. 2017;8:15-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Sarkar S, Jun S, Rellick S, Quintana DD, Cavendish JZ, Simpkins JW. Expression of microRNA-34a in Alzheimer’s disease brain targets genes linked to synaptic plasticity, energy metabolism, and resting state network activity. Brain Res. 2016;1646:139-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Zhang YL, Xing RZ, Luo XB, et al. Anxiety-like behavior and dysregulation of miR-34a in triple transgenic mice of Alzheimer’s disease. Eur Rev Med Pharmacol Sci. 2016;20:2853-2862. [PubMed] [Google Scholar]
- 130. Xu Y, Chen P, Wang X, Yao J, Zhuang S. miR-34a deficiency in APP/PS1 mice promotes cognitive function by increasing synaptic plasticity via AMPA and NMDA receptors. Neurosci Lett. 2018;670:94-104. [DOI] [PubMed] [Google Scholar]
- 131. Jian C, Lu M, Zhang Z, et al. miR-34a knockout attenuates cognitive deficits in APP/PS1 mice through inhibition of the amyloidogenic processing of APP. Life Sci. 2017;182:104-111. [DOI] [PubMed] [Google Scholar]
- 132. Wang G, Huang Y, Wang LL, et al. MicroRNA-146a suppresses ROCK1 allowing hyperphosphorylation of tau in Alzheimer’s disease. Sci Rep. 2016;6:26697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Nakano M, Kubota K, Kobayashi E, et al. Bone marrow-derived mesenchymal stem cells improve cognitive impairment in an Alzheimer’s disease model by increasing the expression of microRNA-146a in hippocampus. Sci Rep. 2020;10:10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Mai H, Fan W, Wang Y, et al. Intranasal administration of miR-146a Agomir rescued the pathological process and cognitive impairment in an AD mouse model. Mol Ther Nucleic Acids. 2019;18:681-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Arena A, Iyer AM, Milenkovic I, et al. Developmental expression and dysregulation of miR-146a and miR-155 in Down’s Syndrome and Mouse Models of Down’s syndrome and Alzheimer’s Disease. Curr Alzheimer Res. 2017;14:1305-1317. [DOI] [PubMed] [Google Scholar]
- 136. De D, Mukherjee I, Guha S, et al. Rheb-mTOR activation rescues aβ-induced cognitive impairment and memory function by restoring miR-146 activity in glial cells. Mol Ther Nucleic Acids. 2021;24:868-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Guedes JR, Custódia CM, Silva RJ, de Almeida LP, Pedroso de Lima MC, Cardoso AL. Early miR-155 upregulation contributes to neuroinflammation in Alzheimer’s disease triple transgenic mouse model. Hum Mol Genet. 2014;23:6286-6301. [DOI] [PubMed] [Google Scholar]
- 138. Gomes C, Cunha C, Nascimento F, Ribeiro JA, Vaz AR, Brites D. Cortical neurotoxic astrocytes with early ALS pathology and miR-146a deficit replicate gliosis markers of symptomatic SOD1G93A mouse model. Mol Neurobiol. 2019;56:2137-2158. [DOI] [PubMed] [Google Scholar]
- 139. Li AD, Tong L, Xu N, et al. miR-124 regulates cerebromicrovascular function in APP/PS1 transgenic mice via c1ql3. Brain Res Bull. 2019;153:214-222. [DOI] [PubMed] [Google Scholar]
- 140. Kanagaraj N, Beiping H, Dheen ST, Tay SS. Downregulation of miR-124 in MPTP-treated mouse model of Parkinson’s disease and MPP iodide-treated MN9D cells modulates the expression of the calpain/cdk5 pathway proteins. Neuroscience. 2014;272:167-179. [DOI] [PubMed] [Google Scholar]
- 141. Horst CH, Schlemmer F, de Aguiar Montenegro N, et al. Signature of aberrantly expressed microRNAs in the striatum of rotenone-induced Parkinsonian Rats. Neurochem Res. 2018;43:2132-2140. [DOI] [PubMed] [Google Scholar]
- 142. Zheng K, Hu F, Zhou Y, et al. miR-135a-5p mediates memory and synaptic impairments via the rock2/Adducin1 signaling pathway in a mouse model of Alzheimer’s disease. Nat Commun. 2021;12:1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Marcuzzo S, Bonanno S, Kapetis D, et al. Up-regulation of neural and cell cycle-related microRNAs in brain of amyotrophic lateral sclerosis mice at late disease stage. Mol Brain. 2015;8:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Liang C, Zou T, Zhang M, et al. MicroRNA-146a switches microglial phenotypes to resist the pathological processes and cognitive degradation of Alzheimer’s disease. Theranostics. 2021;11:4103-4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Neuner SM, Heuer SE, Huentelman MJ, O’Connell KMS, Kaczorowski CC. Harnessing genetic complexity to enhance translatability of Alzheimer’s disease mouse models: A path toward Precision Medicine. Neuron. 2019;101:399-411.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Yang HS, Onos KD, Choi K, et al. Natural genetic variation determines microglia heterogeneity in wild-derived mouse models of Alzheimer’s disease. Cell Rep. 2021;34:108739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Oblak AL, Forner S, Territo PR, et al. Model organism development and evaluation for late-onset Alzheimer’s disease: MODEL-AD. Alzheimers Dement (N Y). 2020;6:e12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Smith AR, Smith RG, Macdonald R, et al. The histone modification H3K4me3 is altered at the ANK1 locus in Alzheimer’s disease brain. Future Sci OA. 2021;7:FSO665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Higham JP, Malik BR, Buhl E, et al. Alzheimer’s disease associated genes ankyrin and Tau cause shortened lifespan and memory loss in Drosophila. Front Cell Neurosci. 2019;13:260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Goll MG, Halpern ME. Chapter 5 - DNA methylation in Zebrafish. In: Cheng X, Blumenthal RM. eds. Progress in Molecular Biology and Translational Science. Academic Press; 2011;193-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Lardenoije R, van den Hove DLA, Havermans M, et al. Age-related epigenetic changes in hippocampal subregions of four animal models of Alzheimer’s disease. Mol Cell Neurosci. 2018;86:1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Sato K, Sasaguri H, Kumita W, et al. A non-human primate model of familial Alzheimer’s disease. bioRxiv. bioRxiv. 2020. doi: 10.1101/2020.08.24.264259 [DOI] [Google Scholar]
- 153. Baglietto-Vargas D, Forner S, Cai L, et al. Generation of a humanized aβ expressing mouse demonstrating aspects of Alzheimer’s disease-like pathology. Nat Commun. 2021;12:2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Dong Y, Newman M, Pederson SM, Barthelson K, Hin N, Lardelli M. Transcriptome analyses of 7-day-old zebrafish larvae possessing a familial Alzheimer’s disease-like mutation in psen1 indicate effects on oxidative phosphorylation, ECM and MCM functions, and iron homeostasis. BMC Genomics. 2021;22:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Barthelson K, Pederson SM, Newman M, Jiang H, Lardelli M. In-Frame and frameshift mutations in zebrafish presenilin 2 affect different cellular functions in young adult brains. J Alzheimers Dis Rep. 2021;5:395-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Benbow SJ, Strovas TJ, Darvas M, Saxton A, Kraemer BC. Synergistic toxicity between tau and amyloid drives neuronal dysfunction and neurodegeneration in transgenic C. elegans. Hum Mol Genet. 2020;29:495-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Dave KD, De Silva S, Sheth NP, et al. Phenotypic characterization of recessive gene knockout rat models of Parkinson’s disease. Neurobiol Dis. 2014;70:190-203. [DOI] [PubMed] [Google Scholar]
- 158. Pütz SM, Kram J, Rauh E, et al. Loss of p21-activated kinase Mbt/PAK4 causes Parkinson-like phenotypes in Drosophila. Dis Model Mech. 2021;14. doi: 10.1242/dmm.047811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Penney J, Ralvenius WT, Tsai LH. Modeling Alzheimer’s disease with iPSC-derived brain cells. Mol Psychiatry. 2020;25:148-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Imm J, Pishva E, Ali M, et al. Characterization of DNA methylomic signatures in induced pluripotent stem cells during neuronal differentiation. Front Cell Dev Biol. 2021;9:647981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Esanov R, Belle KC, van Blitterswijk M, et al. C9orf72 promoter hypermethylation is reduced while hydroxymethylation is acquired during reprogramming of ALS patient cells. Exp Neurol. 2016;277:171-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Esanov R, Cabrera GT, Andrade NS, et al. A C9ORF72 BAC mouse model recapitulates key epigenetic perturbations of ALS/FTD. Mol Neurodegener. 2017;12:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Nestor C, Ruzov A, Meehan R, Dunican D. Enzymatic approaches and bisulfite sequencing cannot distinguish between 5-methylcytosine and 5-hydroxymethylcytosine in DNA. Biotechniques. 2010;48:317-319. [DOI] [PubMed] [Google Scholar]
- 164. Hahn MA, Szabó PE, Pfeifer GP. 5-hydroxymethylcytosine: a stable or transient DNA modification? Genomics. 2014;104:314-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Fukuzawa S, Takahashi S, Tachibana K, Tajima S, Suetake I. Simple and accurate single base resolution analysis of 5-hydroxymethylcytosine by catalytic oxidative bisulfite sequencing using micelle incarcerated oxidants. Bioorg Med Chem. 2016;24:4254-4262. [DOI] [PubMed] [Google Scholar]
- 166. Martin D, Xu J, Porretta C, Nichols CD. Neurocytometry: Flow cytometric sorting of specific neuronal populations from human and Rodent Brain. ACS Chem Neurosci. 2017;8:356-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Holt LM, Stoyanof ST, Olsen ML. Magnetic cell sorting for in vivo and in vitro astrocyte, neuron, and microglia analysis. Curr Protoc Neurosci. 2019;88:e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Blalock EM, Buechel HM, Popovic J, Geddes JW, Landfield PW. Microarray analyses of laser-captured hippocampus reveal distinct gray and white matter signatures associated with incipient Alzheimer’s disease. J Chem Neuroanat. 2011;42:118-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Liu H, Chu W, Gong L, Gao X, Wang W. MicroRNA-26b is upregulated in a double transgenic mouse model of Alzheimer’s disease and promotes the expression of amyloid-β by targeting insulin-like growth factor 1. Mol Med Rep. 2016;13:2809-2814. [DOI] [PubMed] [Google Scholar]
- 170. Liu Y, Zhang Y, Liu P, et al. MicroRNA-128 knockout inhibits the development of Alzheimer’s disease by targeting PPARγ in mouse models. Eur J Pharmacol. 2019;843:134-144. [DOI] [PubMed] [Google Scholar]
- 171. Tang Y, Bao JS, Su JH, Huang W. MicroRNA-139 modulates Alzheimer’s-associated pathogenesis in SAMP8 mice by targeting cannabinoid receptor type 2. Genet Mol Res. 2017;16. doi: 10.4238/gmr16019166 [DOI] [PubMed] [Google Scholar]
- 172. Liang C, Mu Y, Tian H, et al. MicroRNA-140 silencing represses the incidence of Alzheimer’s disease. Neurosci Lett. 2021;758:135674. [DOI] [PubMed] [Google Scholar]
- 173. Zhang L, Fang Y, Zhao X, et al. MiR-204 silencing reduces mitochondrial autophagy and ROS production in a murine AD model via the TRPML1-activated STAT3 pathway. Mol Ther Nucleic Acids. 2021;24:822-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Wang CN, Wang YJ, Wang H, et al. The anti-dementia effects of donepezil involve miR-206-3p in the hippocampus and Cortex. Biol Pharm Bull. 2017;40:465-472. [DOI] [PubMed] [Google Scholar]
- 175. Xiao G, Chen Q, Zhang X. MicroRNA-455–5p/CPEB1 pathway mediates aβ-related learning and memory deficits in a mouse model of Alzheimer’s disease. Brain Res Bull. 2021;177:282-294. [DOI] [PubMed] [Google Scholar]
- 176. Li SH, Gao P, Wang LT, et al. Osthole stimulated neural stem cells differentiation into neurons in an Alzheimer’s disease cell model via upregulation of MicroRNA-9 and rescued the functional impairment of hippocampal neurons in APP/PS1 transgenic mice. Front Neurosci. 2017;11:340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Zhu HC, Wang LM, Wang M, et al. MicroRNA-195 downregulates Alzheimer’s disease amyloid-β production by targeting bace1. Brain Res Bull. 2012;88:596-601. [DOI] [PubMed] [Google Scholar]
- 178. Sha S, Shen X, Cao Y, Qu L. Mesenchymal stem cells-derived extracellular vesicles ameliorate Alzheimer’s disease in rat models via the microRNA-29c-3p/BACE1 axis and the Wnt/β-catenin pathway. Aging. 2021;13:15285-15306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Wang Y, Shi M, Hong Z, Kang J, Pan H, Yan C. MiR-130a-3p has protective effects in Alzheimer’s disease via targeting DAPK1. Am J Alzheimers Dis Other Demen. 2021;36:15333175211020572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Jiang Y, Xu B, Chen J, et al. Micro-RNA-137 inhibits Tau hyperphosphorylation in Alzheimer’s disease and targets the CACNA1C gene in transgenic mice and human neuroblastoma SH-SY5Y Cells. Med Sci Monit. 2018;24:5635-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Liang C, Zhu H, Xu Y, et al. MicroRNA-153 negatively regulates the expression of amyloid precursor protein and amyloid precursor-like protein 2. Brain Res. 2012;1455:103-113. [DOI] [PubMed] [Google Scholar]
- 182. Wu Q, Yuan X, Bai J, et al. MicroRNA-181a protects against pericyte apoptosis via directly targeting FOXO1: implication for ameliorated cognitive deficits in APP/PS1 mice. Aging. 2019;11:6120-6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Lee K, Kim H, An K, et al. Replenishment of microRNA-188-5p restores the synaptic and cognitive deficits in 5XFAD mouse model of Alzheimer’s Disease. Sci Rep. 2016;6:34433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Yang K, Feng S, Ren J, Zhou W. Upregulation of microRNA-196a improves cognitive impairment and alleviates neuronal damage in hippocampus tissues of Alzheimer’s disease through downregulating LRIG3 expression. J Cell Biochem. 2019;120:17811-17821. [DOI] [PubMed] [Google Scholar]
- 185. Wang X, Xu Y, Zhu H, Ma C, Dai X, Qin C. Downregulated microRNA-222 is correlated with increased p27Kip(1) expression in a double transgenic mouse model of Alzheimer’s disease. Mol Med Rep. 2015;12:7687-7692. [DOI] [PubMed] [Google Scholar]
- 186. Qian Q, Zhang J, He FP, et al. Down-regulated expression of microRNA-338-5p contributes to neuropathology in Alzheimer’s disease. FASEB J. 2019;33:4404-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Liu CG, Wang JL, Li L, Wang PC. MicroRNA-384 regulates both amyloid precursor protein and β-secretase expression and is a potential biomarker for Alzheimer’s disease. Int J Mol Med. 2014;34:160-166. [DOI] [PubMed] [Google Scholar]
- 188. He C, Su C, Zhang W, Wan Q. MiR-485-5p alleviates Alzheimer’s disease progression by targeting PACS1. Transl Neurosci. 2021;12:335-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Shi R, Zhang S, Cheng G, Yang X, Zhao N, Chen C. Ginsenoside rg1 and acori graminei Rhizoma attenuates neuron cell apoptosis by promoting the expression of miR-873-5p in Alzheimer’s disease. Neurochem Res. 2018;43:1529-1538. [DOI] [PubMed] [Google Scholar]
- 190. Zong Y, Yu P, Cheng H, et al. MiR-29c regulates NAV3 protein expression in a transgenic mouse model of Alzheimer׳s disease. Brain Res. 2015;1624:95-102. [DOI] [PubMed] [Google Scholar]
- 191. Yang G, Song Y, Zhou X, et al. MicroRNA-29c targets β-site amyloid precursor protein-cleaving enzyme 1 and has a neuroprotective role in vitro and in vivo. Mol Med Rep. 2015;12:3081-3088. [DOI] [PubMed] [Google Scholar]
- 192. Wang H, Liu J, Zong Y, et al. MiR-106b aberrantly expressed in a double transgenic mouse model for Alzheimer’s disease targets TGF-β type II receptor. Brain Res. 2010;1357:166-174. [DOI] [PubMed] [Google Scholar]
- 193. Huang W, Li Z, Zhao L, Zhao W. Simvastatin ameliorate memory deficits and inflammation in clinical and mouse model of Alzheimer’s disease via modulating the expression of miR-106b. Biomed Pharmacother. 2017;92:46-57. [DOI] [PubMed] [Google Scholar]
- 194. Dong J, Liu Y, Zhan Z, Wang X. MicroRNA-132 is associated with the cognition improvement following voluntary exercise in SAMP8 mice. Brain Res Bull. 2018;140:80-87. [DOI] [PubMed] [Google Scholar]
- 195. El Fatimy R, Li S, Chen Z, et al. MicroRNA-132 provides neuroprotection for tauopathies via multiple signaling pathways. Acta Neuropathol. 2018;136:537-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Deng Y, Zhang J, Sun X, et al. MiR-132 improves the cognitive function of rats with Alzheimer’s disease by inhibiting the MAPK1 signal pathway. Exp Ther Med. 2020;20:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Cong L, Cong Y, Feng N, Liang W, Wu Y. Up-regulated microRNA-132 reduces the cognition-damaging effect of sevoflurane on Alzheimer’s disease rats by inhibiting FOXA1. Genomics. 2021;113:3644-3652. [DOI] [PubMed] [Google Scholar]
- 198. Su C, Yang X, Lou J. Geniposide reduces α-synuclein by blocking microRNA-21/lysosome-associated membrane protein 2A interaction in Parkinson disease models. Brain Res. 2016;1644:98-106. [DOI] [PubMed] [Google Scholar]
- 199. Zhou J, Zhao Y, Li Z, et al. MiR-103a-3p regulates mitophagy in Parkinson’s disease through Parkin/Ambra1 signaling. Pharmacol Res. 2020;160:105197. [DOI] [PubMed] [Google Scholar]
- 200. Chiu CC, Yeh TH, Chen RS, et al. Upregulated expression of MicroRNA-204-5p leads to the death of dopaminergic cells by targeting DYRK1A-Mediated apoptotic signaling cascade. Front Cell Neurosci. 2019;13:399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Cai M, Chai S, Xiong T, et al. Aberrant expression of circulating MicroRNA leads to the dysregulation of alpha-synuclein and other pathogenic genes in Parkinson’s disease. Front Cell Dev Biol. 2021;9:695007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Su Y, Deng MF, Xiong W, et al. MicroRNA-26a/Death-Associated protein kinase 1 signaling induces synucleinopathy and dopaminergic neuron degeneration in Parkinson’s disease. Biol Psychiatry. 2019;85:769-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Liang H, Ding B, Liang J, Shi X, Jiang X, Gao Y. MicroRNA-10a inhibits A30P α-synuclein aggregation and toxicity by targeting proapoptotic protein BCL2L11. Int J Clin Exp Pathol. 2018;11:624-633. [PMC free article] [PubMed] [Google Scholar]
- 204. Li D, Yang H, Ma J, Luo S, Chen S, Gu Q. MicroRNA-30e regulates neuroinflammation in MPTP model of Parkinson’s disease by targeting nlrp3. Hum Cell. 2018;31:106-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Li Y, Fang J, Zhou Z, et al. Downregulation of lncRNA BACE1-AS improves dopamine-dependent oxidative stress in rats with Parkinson’s disease by upregulating microRNA-34b-5p and downregulating BACE1. Cell Cycle. 2020;19:1158-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Fan Y, Zhao X, Lu K, Cheng G. LncRNA BDNF-AS promotes autophagy and apoptosis in MPTP-induced Parkinson’s disease via ablating microRNA-125b-5p. Brain Res Bull. 2020;157:119-127. [DOI] [PubMed] [Google Scholar]
- 207. Zhang G, Chen L, Liu J, et al. HIF-1α/microRNA-128-3p axis protects hippocampal neurons from apoptosis via the axin1-mediated Wnt/β-catenin signaling pathway in Parkinson’s disease models. Aging. 2020;12:4067-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Gong X, Huang M, Chen L. Mechanism of miR-132-3p promoting neuroinflammation and dopaminergic neurodegeneration in Parkinson’s Disease. ENEURO. 2022;9. doi: 10.1523/ENEURO.0393-21.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Wang ZH, Zhang JL, Duan YL, Zhang QS, Li GF, Zheng DL. MicroRNA-214 participates in the neuroprotective effect of resveratrol via inhibiting α-synuclein expression in MPTP-induced Parkinson’s disease mouse. Biomed Pharmacother. 2015;74:252-256. [DOI] [PubMed] [Google Scholar]
- 210. Ma X, Zhang H, Yin H, et al. Up-regulated microRNA-218-5p ameliorates the damage of dopaminergic neurons in rats with Parkinson’s disease via suppression of LASP1. Brain Res Bull. 2021;166:92-101. [DOI] [PubMed] [Google Scholar]
- 211. Zhang Y, Xu W, Nan S, Chang M, Fan J. MicroRNA-326 inhibits apoptosis and promotes proliferation of dopaminergic neurons in Parkinson’s disease through suppression of KLK7-Mediated MAPK signaling pathway. J Mol Neurosci. 2019;69:197-214. [DOI] [PubMed] [Google Scholar]
- 212. Hu YB, Zhang YF, Wang H, et al. MiR-425 deficiency promotes necroptosis and dopaminergic neurodegeneration in Parkinson’s disease. Cell Death Dis. 2019;10:589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Wu Q, Xi DZ, Wang YH. MicroRNA-599 regulates the development of Parkinson’s disease through mediating LRRK2 expression. Eur Rev Med Pharmacol Sci. 2019;23:724-731. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-gae-10.1177_25168657221135848 for The Neuroepigenetic Landscape of Vertebrate and Invertebrate Models of Neurodegenerative Diseases by Thanga Harini Sundaramoorthy and Isabel Castanho in Epigenetics Insights