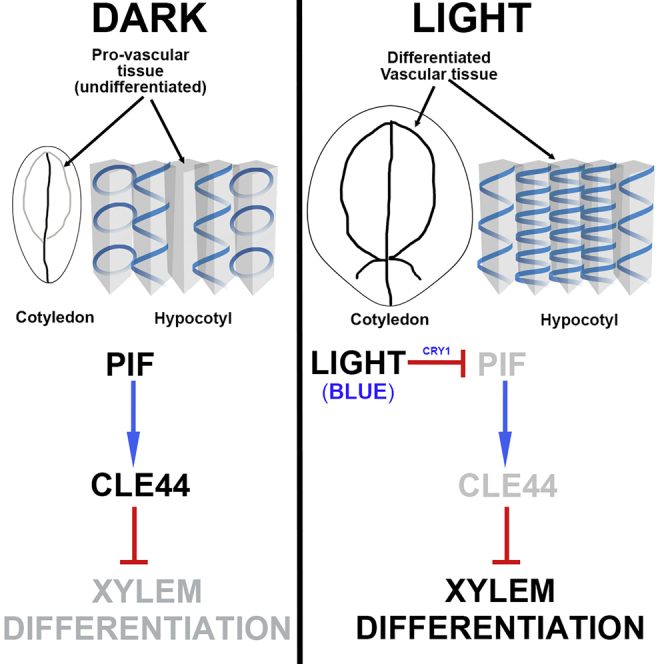
Summary
The balance between cell proliferation and differentiation in the cambium defines the formation of plant vascular tissues. As cambium cells proliferate, subsets of daughter cells differentiate into xylem or phloem. TDIF-PXY/TDR signaling is central to this process. TDIF, encoded by CLE41 and CLE44, activates PXY/TDR receptors to maintain proliferative cambium. Light and water are necessary for photosynthesis; thus, vascular differentiation must occur upon light perception to facilitate the transport of water and minerals to the photosynthetic tissues. However, the molecular mechanism controlling vascular differentiation in response to light remains elusive. In this study we show that the accumulation of PIF transcription factors in the dark promotes TDIF signaling and inhibits vascular cell differentiation. On the contrary, PIF inactivation by light leads to a decay in TDIF activity, which induces vascular cell differentiation. Our study connects light to vascular differentiation and highlights the importance of this crosstalk to fine-tune water transport.
Keywords: plant development, Arabidopsis, vascular development, cell differentiation, xylem, light signaling, signaling transduction, transcriptional regulation, PIF, photomorphogenesis
Graphical abstract
Highlights
-
•
Active CLE peptide TDIF inhibits xylem differentiation in etiolated seedlings
-
•
The expression of the TDIF precursor CLE44 is rapidly inhibited by light
-
•
PIF transcription factors are necessary for TDIF expression in the dark
-
•
Blue light signaling prevents TDIF expression, which promotes xylem differentiation
As water is essential for photosynthesis, plant vascular differentiation must be orchestrated with environmental fluctuations. Ghosh et al. report that xylem cell differentiation is regulated by light in a mechanism that involves the fine-tune regulation of the TDIF precursor CLE44 by PIF transcription factors.
Introduction
Water transport is required for photosynthesis; thus, plant vascular development must proceed such that water demands can be met during formation of new organs. Xylem and phloem, the vascular tissues, are defined in the cambium via precise regulation of the balance between cell proliferation and differentiation. TDIF-PXY/TDR signaling is central to this process (Etchells et al., 2016). Peptide ligand TDIF, encoded by CLE41 and CLE44 (Ito et al., 2006), activates PXY/TDR receptors to maintain proliferation. Cambium cells differentiate to xylem or phloem in the absence of active TDIF-PXY/TDR complexes. Vascular development is stimulated by light. After gemination, light induces photoautotrophic growth and photosynthesis. Consequently, the differentiation of vascular cells must occur to favor the transport of water and minerals from the soil to the green tissues and newly developing organs. Plants perceive light through the action of photoreceptors, which modulate the activity of the transcription factors (TFs) that orchestrate development. Despite the association between light signaling and vascular development, molecular mechanisms linking the two are unknown. Our work shows that vascular differentiation is inhibited in the dark by a mechanism that depends on PIF TFs. Dark-mediated accumulation of PIFs is necessary for CLE44 induction and thus maintenance of undifferentiated vasculature. In illuminated environments, PIF inactivation by photoreceptors causes a decrease in CLE44 expression. This CLE44 decline, in turn, leads to reduced PXY/TDR signaling, which induces the xylem differentiation required to fulfill the water demands associated with photoautotrophic development.
Results and discussion
Light induces xylem differentiation in seedlings
When a seed germinates in darkness (skotomorphogenesis), it adopts etiolated growth, characterized by rapid elongation of the hypocotyl, maintenance of the apical hook to protect the shoot meristem, and inhibition of cotyledon greening and growth (Figure 1A). Here, growth depends on the energy stored in the endosperm nutrient reserves (Kornberg and Beevers, 1957; Penfield et al., 2004). Light perception guides the transition to photoautotrophic development, which inhibits hypocotyl elongation and promotes cotyledon greening and expansion for energy production via photosynthesis. This process is known as deetiolation.
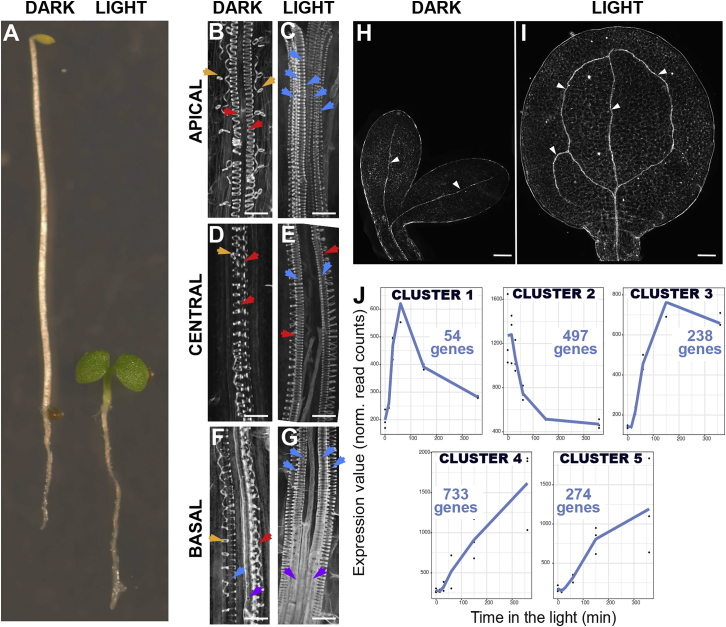
Figure 1.
Light induces vasculature differentiation
(A) Five-day-old seedlings grown in the dark (left) and light (right).
(B–G) SCW deposition arrangements observed in hypocotyls of 5-day-old seedlings grown in the dark (B, D, and F) or light (C, E, and G). (B and C) Apical, (D and E) central, and (F and G) basal regions. Arrows highlight SCW arrangements; annular (orange), helical (red), reticulate (blue), pitted (purple). Scale bars: 10 μm.
(H and I) Differences in cotyledon vein differentiation (white arrowheads) between 5-day-old dark- (H) and light-grown (I) cotyledons. Scale bars: 100 μm.
(J) Genes clustered by transcriptional behavior during seedling deetiolation represented as average value. Data are representative of three independent experiments per time point. See also Data S1.
Water is essential for photosynthesis, and so it must be transported from the root to the newly developing organs via vascular tissues. We hypothesized that during deetiolation, vasculature development must be stimulated by a light-dependent molecular mechanism. We tested this by defining the impact of light on changes to vascular tissues of 5-day-old seedlings grown in both light and dark conditions (Figures 1A–1G).
The vasculature of a 5-day-old Arabidopsis hypocotyl demonstrates a diarchic organization with two poles of xylem cells and, perpendicular to these, two poles of phloem cells in both light- and dark-grown conditions. A group of procambium or provascular cells occupy the space between xylem and phloem within the vascular cylinder (Figure S1A). Each xylem pole contains one protoxylem cell (outer) and one metaxylem (inner) cell. Protoxylem cells differentiate earlier in development and are characterized by the presence of annular or helical secondary cell wall (SCW) patterning, while metaxylem cells differentiate later as cells with reticulate and pitted SCWs (Turner et al., 2007). Dark-grown seedling hypocotyls exhibited few mature reticulate and pitted metaxylem. If present at all, they were found only adjacent to the hypocotyl-root junction (Figure 1F). By contrast, in light-grown hypocotyls, reticulate metaxylem cells were present along the entire length of the hypocotyl apical-basal axis (Figures 1E and 1G). Thus, in dark-grown hypocotyls, metaxylem was discontinuous, whereas it was continuous in light-grown seedlings. This was most apparent in the middle region of the hypocotyl (Figure 1D). Moreover, the helices of SCW thickenings were looser in both proto and metaxylem in dark-grown seedlings than in those grown in the light (Figures 1B–1G). Importantly, the central procambium or provascular cells between xylem poles only differentiated to metaxylem in light-grown seedlings (Figure 1C). Xylem differentiation in cotyledons was also induced by light. Five-day-old seedlings exhibited cotyledons with a primary vein that connected with the hypocotyl vasculature and extended the length of the cotyledon proximodistal axis. Two secondary veins were also observed at this stage of development that branched from the primary, forming two continuous loops (Figure 1I). In dark-grown seedlings, continuous fully differentiated xylem files were observed only in the primary vein (Figure 1H). By contrast, light-grown seedlings also demonstrated fully differentiated xylem in the secondary loops (Figure 1I). Thus, light promotes xylem differentiation in the hypocotyl and cotyledon.
CLE44 expression is rapidly inhibited by light
To determine how vascular cell differentiation is induced by light, we performed a time course transcriptional analysis of 5-day-old seedlings grown in dark and following a 15-, 30-, 60-, 150-, or 360-min exposure to white light (WL) (Figure S1B). Transcriptional changes were assessed via a two-regression-step approach (Conesa et al., 2006). To improve the identification of biologically meaningful expression trends, we used regression modeling to cluster genes with similar expression profiles (Conesa et al., 2006).
Five clusters were identified, one containing genes rapidly repressed by light, and four others showing different light-induction dynamics (Figure 1J; Data S1). Gene Ontology analysis suggested that clusters 2 and 4 exhibited clear functional specialization, while clusters 1 and 5 shared more ontologies with the others (Figure S1C; Data S2). Strikingly, we found that signaling components of the TDIF-PXY/TDR vascular-differentiation machinery were synchronously downregulated by light. Specifically, the receptor PXL1 and its ligand CLE44 were found among 497 early downregulated genes in cluster 2 (Figures 1J and 2A).
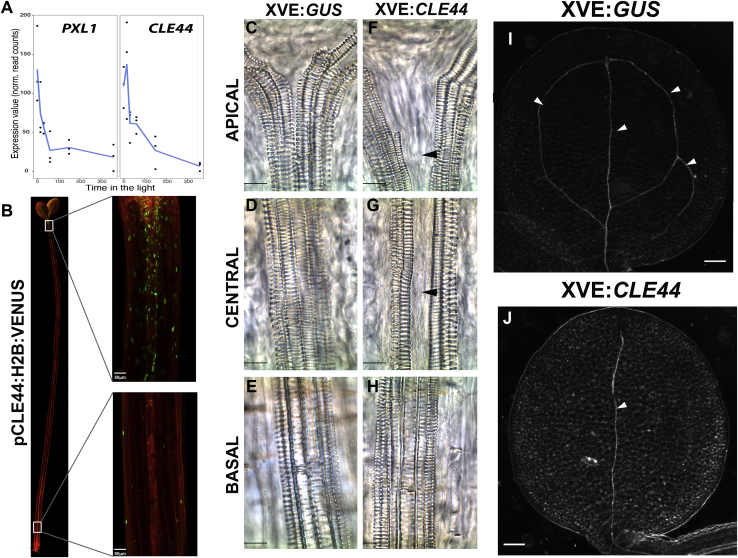
Figure 2.
Light repression of CLE44 induces vascular differentiation
(A) Transcriptional behavior of PXL1 and CLE44 during deetiolation. Data are representative of three independent experiments per time point. See also Data S1.
(B) Confocal analysis of the pCLE44:H2B:VENUS transcriptional reporter in dark-grown seedlings. Image insets show the difference in CLE44 expression between the apical and basal regions of the hypocotyl. Scale bars: 50 μm.
(C–H) Differences in xylem cell differentiation along the length of the hypocotyl between GUS- (C, D, and E) and CLE44- (F, G, and H) overexpressing seedlings grown in the light for 5 days. Scale bars: 10 μm.
(I and J) Differences in cotyledon vein differentiation (white arrowheads) between 5-day-old GUS (I) and CLE44 (J) overexpressing seedlings grown in the light for 5 days. Scale bars: 100μm.
TDIF ligand is derived from CLE41 and CLE44 and perceived by PXY/TDR (Hirakawa et al., 2008), PXL1, and PXL2 receptors (Zhang et al., 2016). As TDIF triggers signaling, we focused our research on understanding its function in response to light. Unlike CLE41, CLE44 expression was light responsive, as confirmed by qRT-PCR (Figure S1D). To examine CLE44 spatial distribution along the hypocotyl, a 2,406-bp CLE44 promoter region was fused to a bright nuclear-localized H2B:VENUS yellow fluorescent protein. Etiolated seedlings demonstrated an apical-basal distribution gradient of CLE44 expression that peaked in the apical part of the organ (Figure 2B). As expected, CLE44 showed an expression maximum in cells surrounding the vascular tissue.
CLE44 overexpression inhibits light-induced xylem differentiation
During secondary growth, one function of TDIF signaling is to repress xylem differentiation (Hirakawa et al., 2008; Ito et al., 2006). If the repression of CLE44 by light is a requirement for light-induced xylem differentiation, maintaining high CLE44 expression in the light may prevent xylem formation. To test this, an estradiol-inducible CLE44 line was created (XVE:CLE44) (Zuo et al., 2000) (Figure S1E). Xylem differentiation was analyzed in XVE:CLE44 and XVE:GUS (control) seedlings grown under constant light in the presence of 17β-estradiol. In controls, the hypocotyl demonstrated clear metaxylem differentiation in the central cells located between xylem poles, with a characteristic high degree of reticulated SCW deposition (Figures 2C–2E). Likewise, the cotyledons showed clear differentiation in the primary vein and distal secondary vein loops (Figure 2I), which were indistinguishable from those of untreated wild-type (WT) seedlings (Figure 1I). Thus, 17β-estradiol treatment alone did not affect vasculature differentiation. By contrast, seedlings overexpressing CLE44 maintained the central provascular cells in an undifferentiated state along most of the hypocotyl apical-basal axis (Figures 2F–2H). The basal region alone demonstrated differentiation of the central cells (Figure 2H). Furthermore, xylem cells failed to differentiate in the cotyledon secondary veins that form the distal loops (Figure 2J). Repression of CLE44 transcription by light is thus required for the induction of xylem differentiation occurring during photomorphogenesis.
CLE44 expression in the dark is regulated by PIFs
Light spectrum influences plant responses (Fankhauser and Chory, 1997); therefore, we considered that CLE44 transcriptional regulation might depend on the perception of specific light wavelengths. Thus, we studied the effect of monochromatic lights on CLE44 expression by exposing dark-grown seedlings to 6 h of blue, red, and far-red lights (BLs, RLs, and FRLs, respectively). Only BL induced reduction of CLE44 expression to levels comparable with WL (Figures 3A and S1D). To better understand the role of light quality in the control of xylem differentiation, we measured the number of differentiated xylem cells present in the apical part of the hypocotyl and their degree of SCW deposition (annular, helical, or reticulate) in seedlings grown under each wavelength. We focused our analysis on the apical part for consistency and because it is the region of highest CLE44 expression (Figure 2B). Monochromatic light treatments produced a reduction in the number of cells undergoing xylem differentiation in all cases compared with WL. However, seedlings grown in BL demonstrated more xylem cells and a higher degree of SCW deposition than the ones grown under RL and FRL (Figures 3B and S2A).
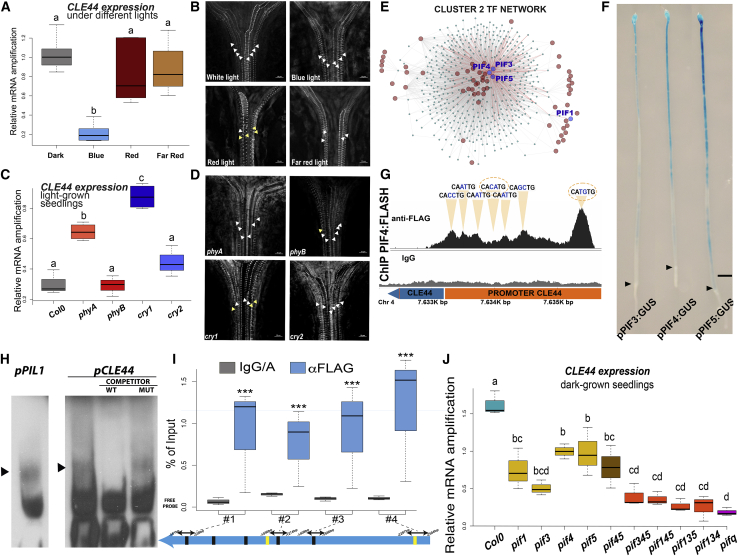
Figure 3.
CLE44 expression is regulated by PIFs
(A) CLE44 expression levels after 6 h of BL, RL, and FRL exposure. Data are representative of three independent experiments and three technical replicates per pair of primers. Values represent mean of expression ± SD. Letters indicate ANOVA + Tukey’s honest significant difference (HSD) pairwise comparison test (p < 0.05).
(B) Orthogonal projections of confocal z stacks representative of hypocotyl xylem cell differentiation of seedling grown under WL (top left), BL (top right), RL (bottom left), and FRL (bottom right). Arrows highlight SCW arrangements; helical (yellow), reticulate (white). Scale bars: 10 μm.
(C) CLE44 expression levels in photoreceptor mutants grown under continuous WL. Data are representative of three independent experiments and three technical replicates per pair of primers. Values represent mean of expression ± SD. Letters indicate ANOVA + Tukey’s HSD pairwise comparison test (p < 0.05).
(D) Orthogonal projections of confocal z stacks representative of hypocotyl xylem cell differentiation in phyA (top left), phyB (top right), cry1 (bottom left), and cry2 (bottom right) mutants grown under continuous WL. Arrows highlight SCW arrangements; helical (yellow), reticulate (white). Scale bars: 10 μm.
(E) Hierarchical representation of the TF network for the genes in cluster 2. Labeled nodes represent the significant TFs identified by the TF2Network software. PIF TFs are indicated.
(F) Expression analysis of PIF3, PIF4, and PIF5 transcriptional GUS reporter lines.
(G) PIF4-FLAG binding regions over CLE44 promoter identified via ChIP-seq. Immunoglobulin G (IgG) indicates the negative control. Orange triangles highlight the location of EBOX elements, dashed ellipses indicate PBE-BOX elements.
(H) Electrophoretic mobility shift assay (EMSA) showing interaction between PIF4 and the G-box elements of pPIL1 (used as control), and the first PBE-BOX found on the CLE44 promoter. WT = unlabelled WT probe, MUT = unlabelled mutated probe.
(I) Direct binding of PIF4 to the promoter of CLE44 by ChIP-qPCR assays. Values obtained from three independent biological replicates and three technical replicates per pair of primers, were normalized to the input and compared against the IgG/A sample. Values represented as percentage of input ± SD. Comparisons between IgG/A and α-FLAG samples were made using Student’s t test (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ns, not significant).
(J) CLE44 expression levels in PIF mutants. Blue, WT; yellow, single; brown, double; orange, triple; purple, quadruple mutants grown in the dark. Data are representative of three independent experiments and three technical replicates per pair of primers. Values represent mean of expression ± SD. Letters indicate ANOVA + Tukey’s HSD pairwise comparison test (p < 0.05).
We reasoned that CLE44 repression by light might be a direct consequence of photoreceptor activity. Thus, we studied CLE44 expression levels in loss of function lines for CRY1 and CRY2 (blue), PHYB (red), and PHYA (far-red) photoreceptors grown in WL. phyA and cry1 mutants demonstrated significantly elevated CLE44 expression compared with WT, phyB, and cry2 (Figure 3C). We also investigated the influence of cry1, cry2, phyA, and phyB mutations in hypocotyl xylem cell differentiation. All photoreceptor mutants had fewer differentiated xylem cells compared with WT. cry1 mutants also displayed significantly lower degrees of SCW deposition (Figures 3D and S2B). Together, these data indicate that the light regulation of CLE44 expression is principally controlled by BL via CRY1 signaling and correlates with xylem differentiation.
Photoreceptor signaling relies on the modulation of TF activity that controls the expression of light-responsive genes (Buti et al., 2020). We sought to identify TFs whose activity could explain CLE44 light-transcriptional behavior. We considered that the synchronous behavior of the genes in each transcriptional cluster (Figure 1J) could be explained by the existence of common transcriptional regulators. Ab initio TF binding predictions were undertaken using the TF2Network tool, which predicts potential regulators for a set of co-expressed genes (Kulkarni et al., 2018). The algorithm identified putative regulators for genes in all clusters (Data S3). Cluster 2, which included CLE44, was predicted to be regulated by 65 TFs, with PIF3, PIF5, and PIF4 among the most significant ones, targeting 44, 62, and 153 genes, respectively (Figure 3E; Data S3). Light signaling converges in PIF TFs, which, in turn, inhibit photomorphogenic growth (Lucas and Prat, 2014). In the dark, PIFs accumulate to promote the expression of genes involved in skotomorphogenic growth. The perception of light leads to their inactivation (Park et al., 2004). Downregulation of PIFs upon light perception is key to proper transition to photomorphogenic development (Lucas et al., 2008). Hence, we hypothesized that PIF activity was necessary for the expression of CLE44 in the dark. If correct, CLE44 expression would occur in regions where PIFs are also expressed. We tested this by investigating the expression pattern of transcriptional reporters for PIF3, PIF4, and PIF5, which have contrasting functions in skotomorphogenesis (Lucas and Prat, 2014). All three demonstrated an apical-basal expression gradient with higher expression toward the cotyledons (Figure 3F). PIF3, PIF4, and PIF5 expression thus overlapped with that of CLE44 (Figure 2B). Moreover, we searched for the presence of PIF-binding elements in the CLE44 promoter and identified seven E-BOX elements (−2,500 bp), of which two were PIF BINDING E-BOX elements (PBE-CATGTG) (Martínez et al., 2018; Zhang et al., 2013), implicating PIFs in regulating CLE44 expression in the dark (Figure 3G). We confirmed that PIF4 binds to the first PBE-BOX in the CLE44 promoter in vitro (Figure 3H). Next, we analyzed previously published chromatin immunoprecipitation sequencing (ChIP-seq) data for PIF4:FLAG-overexpressing plants (Pedmale et al., 2015), which showed high-confidence PIF4 binding enrichment over the CLE44 promoter (Figure 3G). The intensity of the binding correlated with the location of the E-BOX elements in the CLE44 promoter. To further validate the PIF4 binding to CLE44 promoter, we performed ChIP-qPCR on light-grown seedlings overexpressing PIF4:FLAG. Four independent regions of the CLE44 promoter consistently displayed enrichment in the antibody sample versus the control, confirming the ChIP-seq results from Pedmale et al. (2015) (Figure 3I). Finally, to determine the role of PIF on CLE44 transcription, we analyzed CLE44 transcript levels in dark-grown seedlings for single (pif1, pif3, pif4, and pif5), double (pif45), triple (pif345, pif145, pif135, and pif134), and quadruple PIF mutants (pifq; pif3, pif4, pif5, and pif7), as it has been shown that the PIF family shows functional redundancy (Leivar et al., 2008; Lorrain et al., 2009; Nozue et al., 2007; Shin et al., 2009). CLE44 transcription was significantly reduced in all single and higher-order PIF mutants (Figure 3J). Thus, our data indicate that CLE44 dark-dependent expression requires the combinatorial activity of different PIFs. Although, CLE41 expression was not affected in pifq mutants grown in the dark compared with WT (Figure S2C), the fact that CLE44 levels are ∼10× higher than CLE41 in etiolated seedlings indicates that CLE44 is the main TDIF precursor and is regulated in a PIF-dependent manner.
CLE44 induction in light-grown seedlings prevents xylem differentiation in WT and pifq mutants
As PIF genes are necessary for CLE44 expression in the dark, we reasoned whether their genetic manipulation would affect xylem differentiation. Indeed, pifq mutants did have greater xylem differentiation levels than WT when grown in the dark, as fewer cells with annular deposition patterns were observed while more with helical and reticular wall arrangements were present (Figures 4A, 4C, and 4P). Regarding cotyledon venation, more than 80% of dark-grown WT seedlings demonstrated undifferentiated secondary veins. By contrast, only 5% remained undifferentiated in pifq cotyledons (Figures 4K, 4N, and 4Q). The phenotype of pifq mutants indicates that the repression of xylem differentiation in darkness is mediated by PIFs. In the light, all WT and pifq xylem files showed reticulate SCW patterns (Figures 4F, 4H, and S2D) and fully differentiated secondary cotyledon veins (Figure S2F).
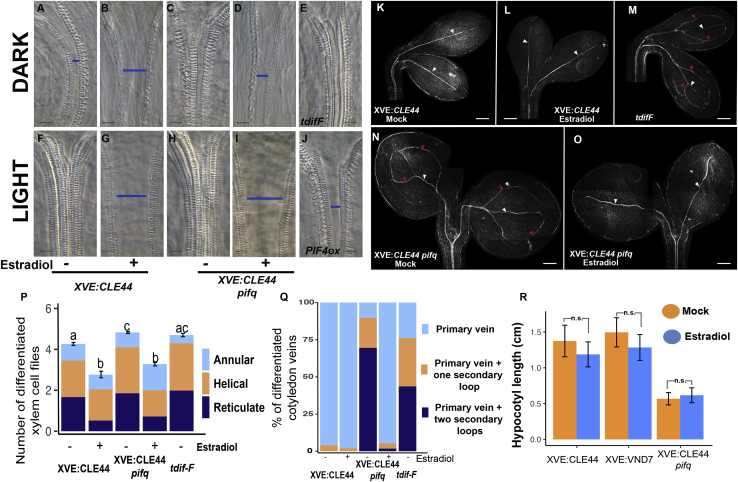
Figure 4.
CLE44 overexpression prevents xylem differentiation in WT and pifq mutants
(A–J) Differences in xylem cell differentiation in 5-day-old dark- (A–E) and light-grown (F–J) hypocotyls where CLE44 and PIF expression is perturbed. Blue bars indicate undifferentiated provascular cells. Scale bars: 10 μm.
(A and B) XVE:CLE44 grown in the dark. (A) Mock and (B) estradiol treated.
(C and D) XVE:CLE44 in pifq background grown in the dark. (C) Mock and (D) estradiol treated.
(E) tdifF mutant grown in the dark.
(F and G) XVE:CLE44 grown in the light. (A) Mock and (B) estradiol treated.
(H and I) XVE:CLE44 in pifq background in the light.
(J) 35S:PIF4:FLAG grown in the light.
(K–O) Differences in cotyledon vein differentiation (white arrowheads) of dark-grown seedlings where CLE44 and PIF expression is perturbed. White arrowheads, differentiated primary vein; red, differentiated secondary vein. Scale bars: 100 μm.
(K and L) XVE:CLE44. (K) Mock and (L) 17β-estradiol treated.
(M) tdifF mutant.
(N and O) XVE:CLE44 in pifq background. (N) Mock and (O) 17β-estradiol treated.
(P) Differences in hypocotyl xylem differentiation between XVE:CLE44; XVE:CLE44 in pifq and tdifF mutants grown in the dark and in the presence of mock and 17β-estradiol. Values (n > 25) represent mean of differentiated cells ± SE. Letters represent ANOVA + Tukey’s HSD statistical test for total number of differentiated cells. Data S5 contains details of the sample size, mean, SE values, and ANOVA + Tukey’s HSD comparisons.
(Q) Differences (as percentage) in cotyledon vein differentiation between XVE:CLE44 and XVE:CLE44 in pifq and tdifF mutants grown in the dark and in the presence of mock and 17β-estradiol (n > 50). Data S5 contains details of the sample size and percentages.
(R) Differences in hypocotyl length between XVE:CLE44 and XVE:CLE44 in pifq and XVE:VND7 dark-grown seedlings in the presence of mock and 17β-estradiol. Values (n > 20) represent mean of hypocotyl length ± SD. Data S5 contains details of the sample size, mean, and SD values. Comparisons between mock and 17β-estradiol treated samples were made using Student’s t test (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ns, not significant).
To determine if the inhibition of xylem differentiation in the dark depends on the PIF-dependent expression of CLE44, we introgressed the XVE:CLE44 transgene into the pifq mutant via crossing. The effect of CLE44 induction in WT and pifq backgrounds grown in either dark or light was determined. CLE44 induction in dark-grown WT seedlings led to fewer xylem files with reduced SCW complexity, as determined by more cells with annular depositions (Figures 4B and 4P). This suggests that the high CLE44 expression observed in dark-grown WT seedlings does not fully saturate the PXY/TDR receptor. Dark-grown pifq seedlings complemented with CLE44 via estradiol induction also showed reduced numbers of xylem-cell files with simpler cell walls (Figures 4D and 4P). In the cotyledons, CLE44 induction inhibited the differentiation of the secondary veins observed in the pifq control sample (Figures 4N and 4O). These results are consistent with CLE44 acting downstream of PIFs, as CLE44 induction suppressed dark-grown pifq vascular phenotypes.
Induction of CLE44 in both WT and pifq seedlings growing in the light also inhibited xylem differentiation in the central vascular cells in the apical hypocotyl (Figures 4G–4I, S2D, and S2E) and in secondary cotyledon veins (Figure S2F). Furthermore, PIF4 overexpression was sufficient to inhibit regular xylem differentiation in the apical hypocotyl (Figure 4J), supporting the concept of CLE44 being a PIF transcriptional target. Dark-grown tdif-F seedlings, which lack the activity of CLE41, -42, -43, and -44 (Smit et al., 2020), show a higher degree of hypocotyl and cotyledon xylem differentiation than WT, highlighting the role of TDIF in this process (Figures 4E, 4M, and S2E).
In the 1960’s, the botanist Katherine Esau described, in her book Anatomy of Seed Plants, how the extent of xylem differentiation depends on the context of organ growth (Esau, 1960). Specifically, xylem displays annular or low degrees of helical thickening in elongating organs, while mature organs show a higher degree of thickening that hinders cell elongation. The differences between dark- and light-grown seedlings described here provide one mechanism by which xylem differentiation is regulated. Given that the induction of CLE44 expression inhibits xylem differentiation, we sought prima facie evidence that xylem differentiation itself is sufficient to impede the elongation process of a whole organ. Overexpression of VASCULAR RELATED NAC-DOMAIN PROTEIN 7 (VND7) induces transdifferentiation of various non-vascular cells into xylem vessels with SCW thickenings (Yamaguchi et al., 2010). To determine whether increased SCW deposition inhibited organ elongation, we induced VND7 expression in dark-grown seedlings. Many hypocotyl cells underwent transdifferentiation to xylem vessels with reticulate SCW deposition (Figure S2G). Surprisingly, significant differences in hypocotyl elongation were not observed. Moreover, the induction of CLE44 also did not induce hypocotyl elongation in WT and pifq plants (Figure 4R). These results support recent findings indicating that the epidermis, and not the vascular tissue, coordinates hypocotyl elongation (Kim et al., 2020; Robinson and Kuhlemeier, 2018).
Collectively, our data propose a model whereby the accumulation of PIFs in the dark inhibits vascular differentiation and SCW deposition by directly activating the expression of CLE44. CLE44 activates PXY/TDR family receptor kinases promoting maintenance of cambium identity and preventing precocious xylem differentiation. The presence of light rapidly promotes PIF inactivation, which, in turn, lowers CLE44 transcription. Under such circumstances, xylem differentiation is induced, thus supporting water transport. Our data suggest that plants rely on CLE44 modulation to prevent precocious xylem differentiation under sub-optimal photosynthetic conditions, and thus avoid the cell-wall deposition of carbon-rich polysaccharides.
Limitations of the study
The active form of CLE peptides is the result of proteolytic processing of precursor proteins (Ito et al., 2006). We show that CLE44 transcription is induced in the dark in a PIF-dependent manner. However, we do not evaluate the effect of light in the proteolytic processing of TDIF precursors. While our analysis manipulating CLE44 expression suggests that CLE44 proteolytic processing does not represent a limiting step, it would be interesting to further investigate if this level of control is also subject to environmental regulation.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-FLAG-M2 | Sigma | F3165; AB_259529 |
| Protein A/G Magnetic Beads | Pierce | 88802 |
| Bacterial and virus strains | ||
| Escherichia coli: ecloni | Lucigen | 60107 |
| Agrobacteria: GV3101pSoup | N/A | N/A |
| Biological samples | ||
| Arabidopsis thaliana – Columbia-0 | N/A | N/A |
| A. thaliana: phyA-211 | Eirini Kaiserli Reed et al., 1993 | phyA-211 |
| A. thaliana: phyB-9 | Dr. Eirini Kaiserli Reed et al., 1993 | phyB-9 |
| A. thaliana: cry1-hb4-b104 | Dr. Eirini Kaiserli Ahmad and Cashmore, 1993 | cry1-hb4-b104 |
| A. thaliana: cry2-1 | Dr. Eirini Kaiserli Guo et al., 1999 | cry2-1 |
| A. thaliana: tdif-F | Dr. Peter Etchells Smit et al., 2020 | tdif-F |
| A. thaliana: pifq | Dr. Jaime F. Martinez García Leivar et al., 2008 | pifq |
| A. thaliana: pif1-2 | Dr. Jaime F. Martinez García Leivar et al., 2008 | pif1-2 |
| A. thaliana: pif3-3 | Dr. Jaime F. Martinez García Leivar et al., 2008 | pif3-3 |
| A. thaliana: pif4-1 | This lab Leivar et al., 2008 | pif3-3 |
| A. thaliana: pif5-3 | Dr. Jaime F. Martinez García Leivar et al., 2008 | pif5-3 |
| A. thaliana: pif1-2pif3-3pif4-1 | Dr. Jaime F. Martinez García Leivar et al., 2008 | pif1-2pif3-3pif4-1 |
| A. thaliana: pif1-2pif3-3pif5-3 | Dr. Jaime F. Martinez García Leivar et al., 2008 | pif1-2 pif3-3 pif5-3 |
| A. thaliana: pif1-2 pif4 pif5-3 | Dr. Jaime F. Martinez García Leivar et al., 2008 | pif1-2 pif4 pif5-3 |
| A. thaliana: pif3-3 pif4 pif5-3 | Dr. Jaime F. Martinez García Leivar et al., 2008 | pif3-3 pif4 pif5-3 |
| A. thaliana: pif4 pif5-3 | Dr. Jaime F. Martinez García Leivar et al., 2008 | pif4 pif5-3 |
| A. thaliana: pif1-1, pif3-3, pif4-2, pif5-3 | Dr. Jaime F. Martinez García Leivar et al., 2008 | pif1-1, pif3-3, pif4-2, pif5-3 |
| A. thaliana: 35S:PIF4:FLASH | Dr. Ullas Pedmalle Pedmale et al., 2015 | 35S:PIF4:FLASH |
| A. thaliana: pPIF3:GUS | NASC | N69166 |
| A. thaliana: pPIF4:GUS | NASC | N69169 |
| A. thaliana: pPIF5:GUS | NASC | N69172A |
| A. thaliana: pCLE44:H2B:VENUS | This paper | pCLE44:H2B:VENUS |
| A. thaliana: XVE:CLE44 | This paper | XVE:CLE44 |
| A. thaliana XVE:CLE44 in pifq | This paper | XVE:CLE44 in pifq |
| Chemicals, peptides, and recombinant proteins | ||
| MS salts | Duchefa | M0222 |
| MES | Duchefa | M1503 |
| 17β-estradiol | Sigma | E8875 |
| X-Gluc | Apollo Scientific | BIMB1021 |
| Critical commercial assays | ||
| TNT Quick | Promega | L1170 |
| LightShift™ Chemiluminescent EMSA | Thermo Scientific | 20148 |
| Streptavidin Magnetic beads | NEB | S1420S |
| RevertAid RT | Thermo Scientific | 10161310 |
| HiFi Polymerase | PCR Bio | PB10.41 |
| Spurr Low Viscosity Embedding kit | Sigma | EM0300 |
| Deposited data | ||
| Time course RNAseq data of seedlings during deetiolation | This paper | GSE178268 |
| ChIPseq data for 35S:PIF4:FLASH | Pedmale et al., 2015 | GSE68193 |
| Oligonucleotides | ||
| See Data S4 for a complete list of all oligonucleotides used in this study | IDT | N/A |
| Recombinant DNA | ||
| pMDC7 | Curtis and Grossniklaus, 2003 | pMDC7 |
| pMCY2 | Emami et al., 2013 | pMCY2 |
| p2R3a-3xVenusYFP-OcsT | Siligato et al., 2016 | p2R3a-3xVenusYFP-OcsT |
| Software and algorithms | ||
| ImageJ 1.44o | National Institute of Health | https://imagej.nih.gov/ij/ |
| R | R Core Team (2021) | https://www.R-project.org/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Miguel de Lucas (Miguel.de-lucas@durham.ac.uk).
Materials availability
Plasmids generated in this study have been deposited to Addgene, and Arabidopsis lines generated in this study have been deposited to NASC.
Experimental model and subject details
Arabidopsis thaliana
All Arabidopsis plants used for this study were in Columbia-0 background. Photoreceptor mutants were kindly provided by Dr. Eirini Kaiserly (University of Glasgow, UK) and generated as follows: phyA-211 (Reed et al., 1993), phyB-9 (Reed et al., 1993), cry1-hb4-b104 (Ahmad and Cashmore, 1993) and cry2-1 (Guo et al., 1999). Combinations of PIF mutants were kindly provided by Dr. Jaime F. Martinez García (IBMCP-CSIC, Valencia, Spain) (Leivar et al., 2008). Transgenic plants expressing pCLE44:H2B:3xVENUS and XVE:CLE44 were generated via Agrobacterium transformation (Clough and Bent, 1998). XVE:CLE44 in the pifq background was generated via crossing. pPIF3:GUS (N69166), pPIF4:GUS (N69169) and pPIF5:GUS (N69172) expressing lines were obtained from NASC and PIF4:FLASH over-expressing lines were kindly provided by Ullas Pedmale (CSHL-New York, USA) (Pedmale et al., 2015).
Escherichia coli
E.cloni competent bacteria (Lucigen) were used for routine molecular biology. All bacteria were grown in LB medium (Melford). The medium was also supplemented with 25μg.mL−1 kanamycin (Melford), 100 μg.mL−1 carbenicillin (Melford), 50μg.mL−1spectinomycin (Melford) and/or 20μg.mL−1streptomycin (Melford) as required to maintain the different plasmids.
Method details
Arabidopsis growth conditions
For seedling analysis, seeds were surfaced sterilized and plated on Murashige and Skoog (1% MS) medium without sucrose. Seeds were stratified in dark for 3d at 4°C, and then transferred and kept vertical into a Sanyo growth chamber with a PAR light intensity of 40μmol.m−2.s−1 illuminated by a daylight-white fiuorescence lamp (FL40SS ENW/37; Panasonic) in a 24h-light cycle and 21°C of temperature. For light treatments, an ELEXIA lamp (Heliospectra) with 430nm, 660nm and 730nm LED diodes was used to provide BL, RL and FRL treatments at intensities mimicking the ones detected by the fluorescence lamps at those wavelengths (430nm = 5μmol.m−2.s−1, 660nm = 5μmol.m−2.s−1 and 730nm = 1μm.m−2.s−1). For dark-grown seedlings, plates were covered with 3 layers of aluminum foil upon 6h of germination induction with WL. For all the experiments where the induction of the gene was performed via XVE system, sterile seeds were germinated on 1% MS media supplemented with 10μM of 17β-estradiol (SIGMA) for 5 days. Selection of transgenic seedlings was performed in 1% MS medium supplemented with 30μg.mL−1 hygromycin or via mCherry seed fluorescence (Emami et al., 2013).
Vector construction
All primers used in this study are listed in Data S4. For the construction of pCLE44:H2B:VENUS we first amplified the 3xVenus sequence from p2R3a-3xVenusYFP-OcsT (Siligato et al., 2016) using the primers 3xVENUS_F and 3xVENUS_R and introduced via hot fusion reaction (Fu et al., 2014) into HindIII linear pMCY2 (Emami et al., 2013) destination plasmid to create pMCY2_3xVenus. HISTONE 2B nuclear protein was amplified from Arabidopsis cDNA using the primers H2B_F and H2B_R and inserted in the HindIII site via Hot Fusion reaction. Lastly, CLE44 promoter was amplified from genomic Col-0 DNA using the primers pCLE44_H2B_3xVenus_F and R and inserted into the SapI site via Hot Fusion to create the final pCLE44:H2B:3xVenus_pMCY2 plasmid. For the 17β-estradiol inducible CLE44 transgene, we amplified CLE44 coding sequence using the primers CLE44_pMDC7_F and R. The amplified PCR was inserted via hot fusion reaction into the pMCD7 plasmid (Curtis and Grossniklaus, 2003) digested with AscI and PacI, creating the plasmid CLE44_pMDC7.
RNA-seq sample collection and analysis
Arabidopsis seedlings (Col-0) were grown at 21°C, in constant darkness, for 5 days following germination induction in 1% MS medium without sucrose. Tissue from dark-grown seedlings was collected in a dark room under green light. Remaining seedlings were then exposed to 40μmol.m−2.s−1 WL for 15, 30, 60, 150, or 360min. Seedlings were collected at these time points and immediately frozen in liquid nitrogen. Three biological replicates were taken at each time point. mRNA extraction and cDNA synthesis were done as described by (Kumar et al., 2012). The resulting cDNA samples were used to create a library compatible with Illumina sequencing (Kumar et al., 2012). Single-read sequencing was conducted on an Illumina HiSeq 2500 at 50bp SR. Reads were quality filtered as described (Toal et al., 2018) and mapped to the AtRTD2-QUASI transcriptome (Zhang et al., 2017) using Kallisto (Bray et al., 2016; Zhang et al., 2017). Kallisto was run using the default k-mer size of 31 bp, and with the following additional parameters: -b 30 --single -l 187 -s 81. Output files were processed using tximport (Soneson et al., 2016) (type = “kallisto”, countsFromAbundance = “lengthScaledTPM)”. The gene-level count values generated by tximport were then used to identify differentially expressed genes. maSigPro (Conesa et al., 2006) was used to identify genes which changed dynamically over the entire time course, taking all time points into account. Gene-level count values were normalized in DESeq prior to their use in maSigPro. We observed a tendency of the maSigPro package to declare genes expressed at extremely low levels as changing significantly over time. To address this, genes for which 2 or more of the biological replicates at any given time point had “0” values were removed from our analysis. The following parameters were used for the maSigPro analysis: design matrix = 5 degrees of freedom; p.vector (Q = 0.01, MT.adjust = “BH”, counts = TRUE, theta = 2.814765), T.fit (step.method = “backward”, alfa = 0.01); sigs(rsq = 0.5, vars = “all”).Genes which exhibited similar expression profiles over the time course were clustered together using the “see.genes()” function within maSigPro (cluster.method = “hclust”, cluster.data = 1, k = 5). maSigPro automatically determined the optimal number of clusters to group genes into; for our data, this was 9 clusters. However, many of the clusters had extremely similar expression profiles; therefore, we grouped similar clusters together to create 5 final clusters.
qRT-PCR analysis
cDNA was generated as described above. qRT-PCR reactions were run in a RotorgeneQ thermocycler and differential expression was calculated by obtaining the mean CT amplification values from three independent experiments (biological replicates) and from the average of three technical replicates per biological replicate. In each case, amplification was calculated relative to a PP2A control (AT1G69960). All possible pairwise comparisons were tested using ANOVA test followed by TUKEY HSD test (p < 0.05). Primer sequences used for the detection of each transcript are listed in Data S4.
Electrophoresis mobility shift assay (EMSA)
EMSAs were performed according to (Martínez et al., 2018) with minor modifications. DNA fragments for pPIL1 (used as control) and the first PBE-BOX element found on pCLE44 were analyzed by using 5′ biotinylated oligonucleotides (Data S4). PIF4 protein was synthesized using transcription and translation system (TNT-Promega) according to manufacturer’s protocol and using a PCR fragment containing PIF4_CDS under the control of the T7 promoter. Detection of the biotinylated DNA was performed using LightShift™ Chemiluminescent EMSA (Thermo Scientific).
ChIP-qPCR analysis
35S:PIF4:FLASH seedlings were grown under constant WL conditions for 5 days. Chromatin immunoprecipitation (ChIP) was performed as previously described (Lee et al., 2007) with the following modifications. The crude nuclear pellet of three independent replicates was resuspended in nuclear lysis buffer and sonicated in a Covaris M220 (Woburn, MA, USA) focused-ultrasonicator for 6 min at 6°C with a 5% duty factor. The soluble chromatin solution was incubated with 1 μg of anti-FLAG (Sigma) and IgG/A magnetic beads over-night. Chromatin-antibody complexes were captured with protein A/G magnetic beads (Thermo Fisher Scientific, Waltham, MA, USA). De-crosslinking reaction was performed with Chelex slurry (Bio-Rad, Watford, UK) (Nelson et al., 2006). For the identification of the PIF4 regulated regions, primer pairs were designed to amplify four different regions along the CLE44 promoter. We then performed a comparative analysis between IgG/A and PIF4:FLAG bound chromatin using RotorgeneQ thermocycler. Differential enrichment was estimated a percentage of input.
GUS staining
Plant tissue was fixed in 90% acetone for 30min and washed twice with water before GUS staining. Seedlings were submerged in the GUS staining solution (50mM phosphate buffer, 0.2% Triton TX-100, 1.5mM potassium ferrocyanide, 1.5mM potassium ferricyanide, and 2mM X-Gluc (5-bromo4-chloro-3-indolyl b- D -glucuronide cyclohexylammonium salt dissolved in DMSO; Gold Biotechnology G1281C1) vacuum infiltrated for 5min, and incubated at 37°C in the dark for 2h. Seedlings were washed with 70% ethanol over-night and re-hydrated with a series of diluted ethanol (50, 25, 10% and water). Seedlings were then mounted with Hoyer’s solution on microscope slides. The activity of the GUS reporter gene was observed under a Zeiss Axioscope 2 fiuorescence microscope.
Xylem differentiation analysis
Seedlings were incubated with clearing solution (1% SDS, 200mM NaOH) for 30min and rinsed twice with distilled water. Then transferred to chloral hydrate solution (40g chloral hydrate, 10mL glycerol, 20mL H2O) over-night and then mounted in Hoyer’s solution and observed under a Zeiss Axioscope 2 fiuorescence microscope. The analysis of cotyledon vasculature was performed using a dark field filter.
Hypocotyl cross sections
Five-day-old light-grown hypocotyls incubated overnight at 4°C in fixation buffer (2.5% glutaraldehyde + 2% paraformaldehyde in 0.2M phosphate buffer, pH 7). Dehydration was performed by incubating the sample for 2h in serial dilutions of ethanol (20, 40, 60, 80, 90, and 95%). The sample was plastic embedded by performing the following steps: 12h incubation in 7:1 ethanol: Spurr’s resin (SIGMA), 12h incubation in 3:1 ethanol:Spurr’s resin, 12h incubation in 100% Spurr’s resin, and 12h incubation in Spurr’s resin. The resin was polymerized at 70°C for 12h. Blocks were trimmed, and 4μm cross sections were produced with a Finesse ME+ (Thermo scientific) microtome. Toluidine blue staining (0.1% of Toluidine blue in 0.1M phosphate buffer, pH 6.8) was performed before microscopy analysis.
Confocal analysis
Samples were imaged using a Zeiss 800 with Airscan (Department of Biosciences, Durham University).
Accession numbers
Next-generation DNA sequencing raw and processed data have been deposited into the Gene Expression Omnibus (GEO) with accession number GEO: GSE178268.
Quantification and statistical analysis
Data for quantification of the differences in xylem differentiation (hypocotyl and cotyledon) are represented as mean ± standard error (SE). Number of hypocotyls or cotyledons (n), mean, percentage and SE for each xylem cell differentiation analysis are summarized on Data S5. RNAseq and qRT-PCR analysis were performed in three biological replicates (100-300 seedlings/replicate). qRT-PCR data quantification is presented as mean of expression ± standard deviation (SD). Statistical analyses, indicated in figure legends, were performed using R software. Comparisons between two groups were made using Student’s t test (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ns = not significant). Comparisons between multiple groups were made using ANOVA + Tukey HSD test (p < 0.05). ANOVA + Tukey HSD tables are summarized on Data S5.
Acknowledgments
This work was supported by the start-up package and seedcorn funds from Durham University and by Gatsby Foundation research grants. S.G.’s research is funded via BBSRC Strategic LOLA (BB/V003534/1), and J.F.N.’s research is funded via DTP-BBSRC PhD studentship (BB/M011186/1). The authors thank Prof. Miguel Angel Blázquez (IBMCP-CSIC) for discussions and valuable suggestions that clearly improved the quality of the work and Dr. Eugenio G. Minguet for his advice setting the monochromatic light treatments.
Author contributions
S.G. conducted experiments and data analysis and provided comments. G.M.C.C. conducted data analysis and provided comments. J.F.N. conducted experiments and provided comments. J.P.E. provided comments and resources. M.d.L. conceived the project, conducted data analysis and experiments, and wrote the article. All authors read and approved the article.
Declaration of interests
The authors declare no competing interests.
Published: July 19, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111075.
Supplemental information
Data and code availability
-
•
RNAseq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This paper analyses existing, publicly available data. These accession numbers for the datasets are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to re-analyze the data reported in this paper is available from the lead contact upon request.
References
- Ahmad M., Cashmore A.R. HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature. 1993;366:162–166. doi: 10.1038/366162a0. [DOI] [PubMed] [Google Scholar]
- Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- Buti S., Hayes S., Pierik R. The bHLH network underlying plant shade-avoidance. Physiol. Plant. 2020;169:312–324. doi: 10.1111/ppl.13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. Floral dip: a simplified method forAgrobacterium-mediated transformation ofArabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Conesa A., Nueda M.J., Ferrer A., Talón M. maSigPro: a method to identify significantly differential expression profiles in time-course microarray experiments. Bioinformatics. 2006;22:1096–1102. doi: 10.1093/bioinformatics/btl056. [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emami S., Yee M., Dinneny J.R. A robust family of Golden Gate Agrobacterium vectors for plant synthetic biology. Front. Plant Sci. 2013;4:339. doi: 10.3389/fpls.2013.00339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau K. Anatomy of Seed Plants. Wiley; New York: 1960. Xylem: general structure and cell types. [Google Scholar]
- Etchells J.P., Smit M.E., Gaudinier A., Williams C.J., Brady S.M. A brief history of the TDIF-PXY signalling module: balancing meristem identity and differentiation during vascular development. New Phytol. 2016;209:474–484. doi: 10.1111/nph.13642. [DOI] [PubMed] [Google Scholar]
- Fankhauser C., Chory J. Light control of plant development. Annu. Rev. Cell Dev. Biol. 1997;13:203–229. doi: 10.1146/annurev.cellbio.13.1.203. [DOI] [PubMed] [Google Scholar]
- Fu C., Donovan W.P., Shikapwashya-Hasser O., Ye X., Cole R.H. Hot fusion: an efficient method to clone multiple DNA fragments as well as inverted repeats without ligase. PLoS One. 2014;9:e115318. doi: 10.1371/journal.pone.0115318.t003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Duong H., Ma N., Lin C. The Arabidopsis blue light receptor cryptochrome 2 is a nuclear protein regulated by a blue light-dependent post-transcriptional mechanism. Plant J. 1999;19:279–287. doi: 10.1046/j.1365-313x.1999.00525.x. [DOI] [PubMed] [Google Scholar]
- Hirakawa Y., Shinohara H., Kondo Y., Inoue A., Nakanomyo I., Ogawa M., Sawa S., Ohashi-Ito K., Matsubayashi Y., Fukuda H. Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc. Natl. Acad. Sci. USA. 2008;105:15208–15213. doi: 10.1073/pnas.0808444105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Nakanomyo I., Motose H., Iwamoto K., Sawa S., Dohmae N., Fukuda H. Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science. 2006;313:842–845. doi: 10.1126/science.1128436. [DOI] [PubMed] [Google Scholar]
- Kim S., Hwang G., Kim S., Thi T.N., Kim H., Jeong J., Kim J., Kim J., Choi G., Oh E. The epidermis coordinates thermoresponsive growth through the phyB-PIF4-auxin pathway. Nat. Commun. 2020;11:1053. doi: 10.1038/s41467-020-14905-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg H.L., Beevers H. A mechanism of conversion of fat to carbohydrate in Castor beans. Nature. 1957;180:35–36. doi: 10.1038/180035a0. [DOI] [PubMed] [Google Scholar]
- Kulkarni S.R., Vaneechoutte D., Van de Velde J., Vandepoele K. TF2Network: predicting transcription factor regulators and gene regulatory networks in Arabidopsis using publicly available binding site information. Nucleic Acids Res. 2018;46:e31. doi: 10.1093/nar/gkx1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., Ichihashi Y., Kimura S., Chitwood D.H., Headland L.R., Peng J., Maloof J.N., Sinha N.R. A high-throughput method for Illumina RNA-Seq library preparation. Front. Plant Sci. 2012;3:202. doi: 10.3389/fpls.2012.00202/abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., He K., Stolc V., Lee H., Figueroa P., Gao Y., Tongprasit W., Zhao H., Lee I., Deng X.W. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. The Plant Cell. 2007;19:731–749. doi: 10.1105/tpc.106.047688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E., Oka Y., Liu T., Carle C., Castillon A., Huq E., Quail P.H. Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 2008;18:1815–1823. doi: 10.1016/j.cub.2008.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S., Trevisan M., Pradervand S., Fankhauser C. Phytochrome interacting factors 4 and 5 redundantly limit seedling de-etiolation in continuous far-red light. Plant J. 2009;60:449–461. doi: 10.1111/j.1365-313x.2009.03971.x. [DOI] [PubMed] [Google Scholar]
- Lucas M. de, Prat S. PIFs get BRright: PHYTOCHROME INTERACTING FACTORs as integrators of light and hormonal signals. New Phytol. 2014;202:1126–1141. doi: 10.1111/nph.12725. [DOI] [PubMed] [Google Scholar]
- Lucas M. de, Davière J.M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blázquez M.A., Titarenko E., Prat S. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- Martínez C., Espinosa-Ruíz A., de Lucas M., Bernardo-García S., García S.B., Franco-Zorrilla J.M., Prat S. PIF4-induced BR synthesis is critical to diurnal and thermomorphogenic growth. EMBO J. 2018;37:e99552. doi: 10.15252/embj.201899552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J.D., Denisenko O., Sova P., Bomsztyk K. Fast chromatin immunoprecipitation assay. Nucleic acids research. 2006;34 doi: 10.1093/nar/gnj004. e2–e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue K., Covington M.F., Duek P.D., Lorrain S., Fankhauser C., Harmer S.L., Maloof J.N. Rhythmic growth explained by coincidence between internal and external cues. Nature. 2007;448:358–361. doi: 10.1038/nature05946. [DOI] [PubMed] [Google Scholar]
- Park E., Kim J., Lee Y., Shin J., Oh E., Chung W.-I., Liu J.R., Choi G. Degradation of phytochrome interacting factor 3 in phytochrome-mediated light signaling. Plant Cell Physiol. 2004;45:968–975. doi: 10.1093/pcp/pch125. [DOI] [PubMed] [Google Scholar]
- Pedmale U.V., Huang S.-S.C., Zander M., Cole B.J., Hetzel J., Ljung K., Reis P.A.B., Sridevi P., Nito K., Nery J.R., et al. Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell. 2015;164:233–245. doi: 10.1016/j.cell.2015.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S., Rylott E.L., Gilday A.D., Graham S., Larson T.R., Graham I.A. Reserve mobilization in the Arabidopsis endosperm fuels hypocotyl elongation in the dark, is independent of Abscisic Acid, and requires PHOSPHOENOLPYRUVATE CARBOXYKINASE1. Plant Cell. 2004;16:2705–2718. doi: 10.1105/tpc.104.024711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J.W., Nagpal P., Poole D.S., Furuya M., Chory J. Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell. 1993;5:147–157. doi: 10.1105/tpc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson S., Kuhlemeier C. Global compression reorients cortical microtubules in Arabidopsis hypocotyl epidermis and promotes growth. Curr. Biol. 2018;28:1794–1802.e2. doi: 10.1016/j.cub.2018.04.028. [DOI] [PubMed] [Google Scholar]
- Shin J., Kim K., Kang H., Zulfugarov I.S., Bae G., Lee C.-H., Lee D., Choi G. Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA. 2009;106:7660–7665. doi: 10.1073/pnas.0812219106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siligato R., Wang X., Yadav S.R., Lehesranta S., Ma G., Ursache R., Sevilem I., Zhang J., Gorte M., Prasad K., et al. MultiSite gateway-compatible cell type-specific gene-inducible system for plants. Plant Physiol. 2016;170:627–641. doi: 10.1104/pp.15.01246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit M.E., McGregor S.R., Sun H., Gough C., Bågman A.M., Soyars C.L., Kroon J.T., Gaudinier A., Williams C.J., Yang X., et al. A PXY-mediated transcriptional network integrates signaling mechanisms to control vascular development in Arabidopsis. Plant Cell. 2020;32:319–335. doi: 10.1105/tpc.19.00562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson C., Love M.I., Robinson M.D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 2016;4:1521. doi: 10.12688/f1000research.7563.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toal T.W., Ron M., Gibson D., Kajala K., Splitt B., Johnson L.S., Miller N.D., Slovak R., Gaudinier A., Patel R., et al. Regulation of root Angle and gravitropism. G3. 2018;8:3841–3855. doi: 10.1534/g3.118.200540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner S., Gallois P., Brown D. Tracheary element differentiation. Annu. Rev. Plant Biol. 2007;58:407–433. doi: 10.1146/annurev.arplant.57.032905.105236. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M., Goué N., Igarashi H., Plant M.O. VASCULAR-RELATED NAC-DOMAIN6 and VASCULAR-RELATED NAC-DOMAIN7 effectively induce transdifferentiation into xylem vessel elements under control of an induction system. Plant Physiol. 2010;153:906–914. doi: 10.1104/pp.110.154013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Mayba O., Pfeiffer A., Shi H., Tepperman J.M., Speed T.P., Quail P.H. A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 2013;9:e1003244. doi: 10.1371/journal.pgen.1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Lin X., Han Z., Qu L.-J., Chai J. Crystal structure of PXY-TDIF complex reveals a conserved recognition mechanism among CLE peptide-receptor pairs. Cell Res. 2016;26:543–555. doi: 10.1038/cr.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Calixto C.P.G., Marquez Y., Venhuizen P., Tzioutziou N.A., Guo W., Spensley M., Entizne J.C., Lewandowska D., ten Have S., et al. A high quality Arabidopsis transcriptome for accurate transcript-level analysis of alternative splicing. Nucleic Acids Res. 2017;45:5061–5073. doi: 10.1093/nar/gkx267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J., Niu Q.W., Chua N.H. Technical advance: an estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 2000;24:265–273. doi: 10.1046/j.1365-313x.2000.00868.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
RNAseq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This paper analyses existing, publicly available data. These accession numbers for the datasets are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to re-analyze the data reported in this paper is available from the lead contact upon request.