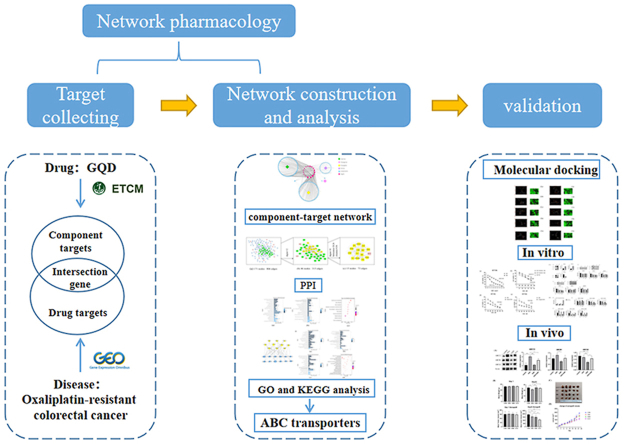
Abstract
Background
Resistance to chemotherapeutic drugs, such as oxaliplatin (OXA), can lead to unsatisfactory chemotherapy results during the treatment of advanced colorectal cancer (CRC). Therefore, we investigated the potential targets and mechanisms of Gegen Qinlian Decoction (GQD) against OXA-resistant CRC through network pharmacology and performed molecular docking and experimental verification.
Methods
We collected potential compounds, targets, and related disease genes from public databases. The pharmacology model of the compound-target-pathway network and protein-protein interaction (PPI) network were established. The potential active components and mechanisms underlying GQD reversing OXA-resistant CRC were systematically predicted, and the key targets were verified by performing molecular docking and in vitro and in vivo experiments.
Results
A total of 160 active ingredients, 407 potential targets, and 406 CRC drug resistance genes were collected. 16 intersecting genes, which included ABCG2 and belonged to 139 active compounds including baicalin and wogonin. They were enriched in 12 signaling pathways, including ABC transport and metabolism. Along with network topology analysis, ABCB1 and ABCC2 were identified as key targets and proved that various active components of GQD combined well with them. GQD alone and synergized with OXA could inhibit the protein and mRNA of ABC transporters in vivo and in vitro, decrease the IC50 of OXA-resistant CRC to OXA with a good synergistic index at different treatment times and concentrations, improve the sensitivity of OXA-resistant CRC to OXA, inhibit drug efflux, decrease tumor volume, and increase the weight of nude mice at the late stage of treatment.
Conclusions
GQD can target ATP-binding proteins, inhibit ABC transporters, reverse OXA resistance, increase the sensitivity of OXA-resistant CRC cells to OXA, decrease tumor volume, alleviate toxic side effects, improve prognosis, and have good synergistic therapeutic effects. These results provide an effective research tool to elucidate ethnomedicine for modernizing refractory diseases.
Keywords: Colorectal cancer, Gegen qinlian decoction, Chemotherapy resistance, Network pharmacology, ABC transporter
Graphical abstract
Highlights
-
•
Active components and potential key targets demonstrated a strong binding activity.
-
•
GQD can inhibit ABC transporter both in vivo and in vitro.
-
•
GQD reverses oxaliplatin resistance in colorectal cancer and synergizes with oxaliplatin.
Colorectal cancer; Gegen qinlian decoction; Chemotherapy resistance; Network pharmacology; ABC transporter.
1. Introduction
Colorectal cancer (CRC) is a major threat to human health. The international cancer research organization GLOBOCAN 2020 reported that CRC ranks second and third in the incidence and mortality of global cancer, respectively (Sung et al., 2021). Almost 50% of patients with CRC have progressive cancer at the time of initial diagnosis (Dekker et al., 2019).
Because of the use of oxaliplatin (OXA) for CRC treatment, overall CRC treatment has improved remarkably. However, acquired drug resistance is still a major reason for treatment failure (Martinez-Balibrea et al., 2015; Meads et al., 2009). The main mechanisms underlying drug resistance include drug inactivation, drug target alteration, drug efflux, DNA damage repair, cell death inhibition, and epithelial–mesenchymal transition (EMT) (Holohan et al., 2013). Guidelines recommend FOLFOX and FOLFIRI regimens as the first-line therapies for intermediate to advanced CRC, with low objective remission rates, chemoresistance, and toxic side effects (Giacchetti et al., 2000; Provenzale et al., 2020; Tauriello and Batlle, 2016). Patients with advanced CRC treated with FOLFOX chemotherapy have a median overall survival (OS) of 19.3 months and a median progression-free survival (PFS) of 9.0 months. Patients with advanced CRC who failed first-line FOLFOX treatment with FOLFIRI had a median OS of 7.8–10.5 months and a median PFS of only 4.3 months (Guo et al., 2016; Neugut et al., 2019).
Therefore, OXA resistance in advanced CRC should be studied and more efficient treatment to improve patient prognosis and quality of life should be found.
Gegen Qinlian Decoction (GQD), an extract from Treatise on Exogenous Febrile Disease, contains Pueraria lobata (Willd.) Ohwi (PLO), Scutellaria baicalensis Georgi (SBG), Coptis chinensis Franch (CCF), and Glycyrrhiza uralensis Fisch (GUF). The plant names were checked on http://www.theplantlist.org. It can clear internal heat and stop diarrhea. It is used for the treatment of damp-heat intestinal diseases. Many in vivo and in vitro studies have shown that GQD can regulate CRC development. The traditional formula Zuojinwan, with Coptis chinensis Franch. as the main ingredient, inhibits the PI3K/Akt/NF-κB signaling pathway to reverse multidrug resistance in OXA-resistant CRC, indicating significant anticancer potency (Huang et al., 2020; Sui et al., 2014). Baicalin inhibits CRC cell growth and promotes apoptosis by inhibiting EMT, stem cells, and the cell cycle (Yang et al., 2020). Dysbiosis of the intestinal flora is an important challenge in the therapeutic progression of CRC, and in MicroSatellite stability/CRC (MSS/CRC), Scutellaria baicalin can increase PD-1 blockade by regulating intestinal flora and tumor microenvironment (Lv et al., 2019; W. Zhang et al., 2021). Baicalein, as a substrate of P-glycoprotein, can inhibit multidrug resistance protein (MDR) activity, and target the TLR4/HIF-1α/VEGF signaling pathway to inhibit CRC (M. Chen et al., 2021; Palko-Labuz et al., 2017). Berberine can be anti-CRC by increasing mucosal permeability, blocking the cell cycle, and inhibiting telomerase activity (Samad et al., 2021). GQD containing the above active ingredients can exert powerful anticancer efficacy in a multi-target and multi-pathway manner with the potential to reverse drug resistance and increase OXA sensitivity. Therefore, we aimed to investigate the targets and molecular mechanisms of action of GQD in alleviating OXA resistance and improving efficacy in CRC using a network pharmacological approach and validated them with molecular docking in vitro and in vivo experiments.
2. Methods
2.1. Database construction
2.1.1. Acquisition of active ingredients and the action targets of Chinese medicine
The database ETCM (Encyclopedia of Traditional Chinese Medicine) (http://www.tcmip.cn/ETCM/) was applied to search for the active ingredients and the action targets of GQD. The herbal ingredients recorded in this database were collected in accordance with the Pharmacopoeia of the People's Republic of China and other literature, and the pharmacokinetic parameters were calculated to predict the potential action targets of the herbal ingredients (Xu et al., 2019).
2.1.2. CRC drug resistance-related genes
"Colorectal Cancer", "Colon cancer", "rectum-colon carcinoma", "chemoresistance", "chemotherapy resistance", "drug resistance", "Oxaliplatin-resistance", and other keywords were searched across the GENE EXPRESSION OMNIBUS (GEO) database, and the dataset GSE42387 was downloaded and analyzed. This dataset collects gene expression profiles of multiple parental and drug-resistant colon cancer cell lines. After the normalization of the raw expression matrix, differentially expressed genes (DEGs) were obtained by paired t-test using the Limma package for parental and drug-resistant cells. The IDs were converted into gene symbols according to the supplemental file. Then, after deleting the duplicate genes and blank probes, up- and down-regulated genes were set with a threshold of P < 0.05 and |log2FC|>1, and the ggscatter package was used to plot differential gene volcanoes and the heatmap package in order to plot the differential gene heat maps.
2.1.3. CRC-resistance genes associated with the active ingredients of GQD
The resistance-associated genes were intersected with the Traditional Chinese medicine (TCM) targets collected by ETCM, and it was considered the main action targets of GQD in alleviating CRC resistance; this part of the genes were both the action targets of GQD and the mechanism targets that induced GQD resistance.
2.2. Bioinformatics analysis
2.2.1. Construction of the GQD Chinese medicine network
Based on the data collected in the abovementioned steps, a visual network of Chinese herbal medicine—active ingredient—the target was constructed using Cytoscape 3.6.0 (https://cytoscape.org/).
2.2.2. Protein–protein interaction network of intersecting genes
The 16 intersecting genes were applied to construct the protein–protein interaction network (PPI) with the BiosGenet plug-in function of Cytoscape, and the network was topologically analyzed using the Analysis Function in Tools in order to extract the sub-network 1 with a threshold of 2 times the median degree, after which the sub-network 1 was analyzed with the CytoNCA plug-in with Degree, Betweenness, and Closeness median as the threshold value and the sub-network 2 was extracted to form the core network and the important targets in this core network.
2.2.3. GO functional enrichment and pathway analysis of GQD-related CRC drug-resistance genes
Gene Ontology (GO) functional enrichment and pathway analysis were performed on 16 intersecting genes with the STRING 11.0 (https://string-db.org/). GO results were classified into 3 categories: biological process (BP), cellular component (CC), and molecular function (MF); both GO and KEGG retained only those results with false discovery rate (FDR) < 0.05. We also performed GO and KEGG analysis of 407 potential TCM targets with the STRING.
2.3. Molecular docking validation
The 3D structures of the active compounds of GQD were downloaded from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) and the energy minimization calculations were performed with the Chem3D software. The crystal structures of the target proteins were obtained from the PDB Protein Data Bank (https://www.rcsb.org/) and high-resolution, pre-existing ligand molecule, and human-derived crystal structures were selected. The Surflex–Dock docking mode of the Sybyl-x2.1.1 software was used to extract the crystal ligands in order to expose the active pockets, and, after dehydration, hydrogenation, and the removal of terminal residues, docking was verified for the target and active components, and the docking results were measured by Total Score. The higher the Total Score value, the more stable the protein bound to the molecule, and the higher the Total Score, the more stable the binding of the protein to the molecule.
2.4. In vitro and in vivo experiments
2.4.1. Drugs and reagents
OXA was purchased from MCE (HY-17371), and the GQD components were purchased from the Famous Medicine Library of Zhejiang Chinese Medical University (PLO: 201101, Anhui, China; SBG: 201101, Sichuan, China; CCF: 201001, Shanxi, China; GUF: 210101, Xinjiang, China); HCT116 and HCT116/L were purchased from CTCC (Zhejiang, China) and cultured in DMEM (Cienry, CR-12800-S) supplemented with 10% inactivated fetal bovine serum (GIBCO, 10091–141) in an incubator filled with 5% CO2 and 95% atmosphere at 37 °C; trypsin (GIBCO, 25200–056); and CCK8 (GLPBIO, GK10001). Rabbit anti-ABCB1 (#13978), rabbit anti-ABCC2 (#12559), and rabbit anti-ABCG2 (#42078) were purchased from CST (Danvers, MA, USA), and rabbit anti-NA-K-ATPase was purchased from Abbkine (ABP55363).
2.4.2. Sample preparation
PLO (75 g) was soaked in 800 mL of cold water for 30 min and then boil for 20 min, followed by a decoction of 45g SBG, 45 g CCF, and 30 g GUF for 10 min. The dregs were then added to 600 mL of water, decocted for 30 min, filtered again, and then combined with the previous filtrate for concentration. The concentrated filtrate was placed in a vacuum freeze dryer for lyophilization. Based on the calculation, 195 g of the raw drug was yielded. The concentrated filtrate was placed in a vacuum freeze-dryer for lyophilization, and it was calculated that 195 g of the raw drug could yield 29.56 g of the lyophilized powder, indicating a yield of 15.16%. The filtrate was dissolved in DMEM each time and then filtered through the 0.22-μm-filter membrane (Millipore, SLGP033RB) to remove the insoluble impurities.
2.4.3. Analysis of GQD by UPLC-Q-TOF-MS and the identification of its main constituents
Mass spectral data were collected by the Waters SYNAPT G2-Si UPLC-Quadrupole Tandem Time of Flight Mass Spectrometer in positive and negative 2 ion modes. A CORTECS T3 column (2.1 × 100 mm, 1.6-μm particle size) was used, and the column temperature was 35 °C. The following chromatographic conditions were used: mobile phase A was acetonitrile, mobile phase B was 0.1% formic acid water; the elution gradients were as follows: 0–2 min, 5% A; 2–32 min, 5–100% A; 32–33 min, 100% A; 33.5 min, 5% A; 33.5–35 min, 5% A. Flow rate: 0.3 mL/min; injection volume: 2 μL. The mass spectrometry conditions. The ionization mode was electrospray ionization (ESI). The capillary voltage was 3000 V in the positive ionization mode, the desolventizing temperature was 500 °C, and the desolventizing gas flow rate was 1000 L/h. The negative ionization mode was 2500 V, the desolventizing temperature was 400 °C, and the desolventizing gas flow rate was 800 L/h. The cone hole voltage was 40 V, the ion source temperature was 120 °C, the offset source voltage was 80 V, the cone hole gas was 50 L/h, and the materialization gas pressure was 6.0 Bar. Collision energy: 0 V for low energy, 20–50 V for high gradient energy. Collision gas: high purity argon. Three batches of extractions were measured. The data were subsequently processed with the Unifi/Qi software.
2.4.4. Cell viability
Cells (1 × 105/mL) were spread uniformly in a 96-well flat-bottom culture plate and cultured for 24 h, followed by the addition of each gradient of drug action for 12 h and 24 h; the old solution was then aspirated and washed with PBS, and the absorbance was measured at 450 nm with an enzyme marker after adding 10% CCK8 and waiting for the OD value to reach approximately 1.0.
2.4.5. Western blotting
The protein was extracted by Invent Biotechnologies, SM-005, and dissolved in denatured protein lysate (Invent Biotechnologies, WA-009). After high-temperature denaturation, the protein concentration was determined by BCA (Beyotime, P0012). An equal amount of protein from each experimental group was separated on 4–20% SurePAGE (Genscript, M00656) and transferred onto a 0.45-μm PVDF membrane (Millipore, IPVH00010). Then, 5% non-fat dry milk was sealed and then treated with the first antibody overnight at 4 °C. After cleaning the unbound first antibody, the membrane was treated with a second antibody (Epizyme, China) at room temperature for 1 h. Image Lab (Bio-Rad, California, USA) was used for imaging and quantitative analysis. The Stripping Buffer (APPLIYENP, 1650) was used to wash the antibody and re-incubate with the first antibody overnight, followed by imaging and analyses.
2.4.6. RNA extraction and qRT-PCR
Total RNA was extracted by using the RNA Extraction Kit (RT001, Yishan Biotech), and NanoDrop was used to evaluate the concentration and purity of the extract. The total RNA was transformed into cDNA by using a reverse transcription kit (RR047A, Takara), and qRT-PCR was performed as per the TB Green method (RR820A, Takara) under the following condition: 95 °C for 30 s; 95 °C for 5 s, 60 °C for 30 s, 40 cycles. The expression of ABCB1, ABCC2, and ABCG2 was quantified into butler genes β-actin and calculated by the 2-ΔΔCt method.
2.4.7. Rhodamine accumulation
According to different drug-intervention strategies, the cells were incubated with 5 μg/mL Rho-123 (Beyotime, C2007) at 37 °C for 1 h. The cells were then observed under the fluorescence inverted microscope (Zeiss, Axio Observer. A1) and photographed under the same parameter conditions.
2.4.8. Determination of CI
The synergistic effects were determined by calculating the Combination Index (CI) with the CompuSyn software. CI < 1, = 1, or >1 indicates synergistic, additive, or antagonistic effects, respectively (P. Chen et al., 2021; Fransson et al., 2016).
2.4.9. Mice xenograft experiment
The laboratory animal management and ethics committee of the Zhejiang Chinese Medical University approved all animal experiments (No. IACUC-20210816-01). Six-week-old SPF-grade BABL/c nude mice were purchased from Slac Laboratory Animal Co (Shanghai, China). After 1 week of acclimatization feeding, HCT116/L cells, in the logarithmic growth phase, were inoculated in 0.2 mL of sterile saline and adjusted to 1.5 ∗ 107 mice/mL for injection in the axilla of the right forelimb. The mice were then randomly assigned to the control, OXA, GQD, and GQD + OXA groups for chemotherapy treatment when the tumor volume reached 50 mm3. In the control group, 10 mL/kg of physiological saline was administered via gavage every day for 3 weeks. In the OXA group, 5 mg/kg of oxaliplatin was injected intraperitoneally twice a week for 3 weeks. In the GQD group, 5000 mg/kg of GQD was administered via gavage (equivalent to the human dose) every day for 3 weeks; in the OXA + GQD group, 5 mg/kg of oxaliplatin was injected intraperitoneally, and 5000 mg/kg of GQD was administered via gavage every day for 3 weeks. The dosage of GQD is calculated by the equivalent dose ratio between human and animal in terms of body surface area described in Experimental Methodology of Pharmacology (Wei et al., 2010). The equivalent dose ratio is 0.0026, the clinical dose of human is X mg/kg, and the standard weight of adult and mouse is 70 kg and 20 g respectively. The mouse dose conversion formula is X mg/kg × 70 kg × 0.0026/20 g = 9.1X mg/kg. The clinical dosages are PLO 15 g, SBG 9 g, CCF 9 g, and GUF 6 g. The clinical dose of GQD is 39000 mg/70 kg in total. After conversion, the dose of mouse is 5,070 mg/kg. For the convenience of drug configuration, we selected 5000 mg/kg as the final mouse dose. The tumor diameters were routinely measured every 2 days with a caliper, and the differences in the transplantation tumor volumes and tumor growth inhibition rates were compared among the groups. Tumor volumes were calculated with the formula: Length × Width × Width/2. At the end of the experiment, the mice from each group were executed and the tumor tissues were retained for testing.
2.5. Statistical analyses
Statistical analyses were performed with GraphPad Prism 8.3.0. Data were presented as the means ± standard deviation (SD). Differences between the two samples were analyzed by Student’s t-test or unpaired two-sided t-test. One-way analysis of variance (ANOVA) was performed to analyze the data from multiple groups. Each experiment was repeated thrice.∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
3. Results
3.1. GQD contains 160 active ingredients and 407 potential targets
After deleting the active ingredients without potential targets, SBG had 57 active ingredients and 267 targets. CCF had 11 active ingredients and 60 targets. PLO had 29 active ingredients and 223 targets. GUF had 68 active ingredients, 241 targets, 160 active ingredients after deduplication, and 407 potential targets. All active ingredients and target information are shown in Supplementary material 1.
3.2. A total of 406 differential genes exist in OXA-resistant CRC
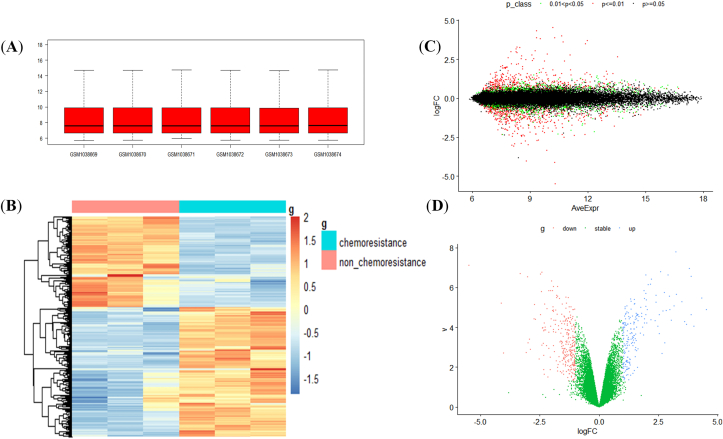
Under the threshold of P value < 0.05 and | log2FC | > 1, there were 406 differential genes, including 244 upregulated genes and 162 downregulated genes (Figure 1A-D). The complete differential genes are shown in Supplementary material 2, and the 80 genes with the largest changes are shown in Table 1.
Figure 1.
Heat map and volcano plot of CRC drug resistance-associated genes (A) Box plots indicate that the dataset was standardized and there were no evident outliers with mean values 7–8 (B) A heat map indicates the clustering of differential genes to measure the similarity of expression between the samples or genes; the horizontal coordinate represents the sample and the vertical coordinate represents the gene; the blue to red color indicates the increasing abundance of gene expression (C) A volcano plot with the horizontal coordinate is the average gene expression of each sample, and the vertical coordinate is the logFC, with the P value set as the standard, P ≤ 0.01 is depicted in red, P ≥ 0.05 is depicted in black, and 0.01 < P < 0.05 is depicted in green (D) The horizontal coordinate is represented as v = -log 10(P.Value) and the vertical coordinate is logFC, red color indicates genes with downregulated expression when compared with the control, the green color indicates genes with a stable expression, and the blue color indicates genes with upregulation.
Table 1.
A total of 80 differential genes of CRC drug resistance showing the greatest change.
| Up-regulated gene | Log2FC | Down-regulated gene | Log2FC |
|---|---|---|---|
| FABP1 | 4.5236033 | INHBA | -5.48198194 |
| AKR1C1 | 3.99521782 | PCP4 | -4.13157658 |
| LOC100289255 | 3.88686321 | BEX2 | -4.03588872 |
| NTS | 3.88473047 | PRKCDBP | -3.33450382 |
| HBE1 | 3.8022812 | LCN2 | -3.06985669 |
| AKR1B10 | 3.68456207 | TCEAL8 | -3.00763389 |
| AKR1B1 | 3.61274775 | ALDH1L2 | -2.99558465 |
| DHRS2 | 3.48788807 | ARHGAP23 | -2.92658064 |
| AGMO | 3.26385778 | KRT80 | -2.90222167 |
| HBG1 | 3.24053395 | CASK | -2.74702818 |
| POTEG | 3.16445429 | C11orf20 | -2.72725738 |
| HHLA2 | 2.97812609 | AMIGO2 | -2.68809036 |
| POTEE | 2.96544267 | FAM129A | -2.52302237 |
| CADPS | 2.90743908 | LOC440335 | -2.46268539 |
| ABCC2 | 2.83665525 | GSTT2 | -2.46044744 |
| ITGB7 | 2.76552061 | IGF2 | -2.41964372 |
| TUBAL3 | 2.76135917 | FSTL1 | -2.40257896 |
| SYTL5 | 2.69869098 | SYBU | -2.40171634 |
| FST | 2.65677931 | ALPP | -2.39295692 |
| POTEB | 2.60306754 | IGFBP1 | -2.36194225 |
| FCGBP | 2.53796827 | KRT34 | -2.3579977 |
| ANKRD30BP2 | 2.47757188 | CXCR4 | -2.34096979 |
| RBP1 | 2.39326631 | NNMT | -2.33379109 |
| CD68 | 2.37192884 | ALPPL2 | -2.31948602 |
| DQX1 | 2.21633075 | NUPR1 | -2.30637846 |
| OTC | 2.1938992 | C4orf7 | -2.30347821 |
| C21orf81 | 2.17515307 | LHFP | -2.28769114 |
| MAGEA2B | 2.0779236 | TFF2 | -2.26722801 |
| PEG10 | 2.02486691 | ARRDC4 | -2.21892961 |
| SOHLH2 | 2.01318061 | TFF1 | -2.1855721 |
| IFI16 | 1.99404873 | TXNIP | -2.1608127 |
| NPSR1 | 1.97655798 | ECHDC2 | -2.11812894 |
| POTED | 1.97180474 | LGALS2 | -2.11206689 |
| ANXA10 | 1.96964627 | IFIT1 | -2.1087158 |
| COL4A5 | 1.92039417 | PI3 | -2.05980214 |
| ABCB1 | 1.90993759 | CCDC85A | -2.05148507 |
| ANKRD20A9P | 1.89309324 | HSPB3 | -2.04360956 |
| AKR1C3 | 1.80381286 | GNG11 | -2.03176537 |
| LRIG1 | 1.79942349 | VIM | -2.0178172 |
| MYO1A | 1.73159146 | GUCY1A3 | -2.00706877 |
3.3. Various active components of GQD target the OXA-resistant CRC
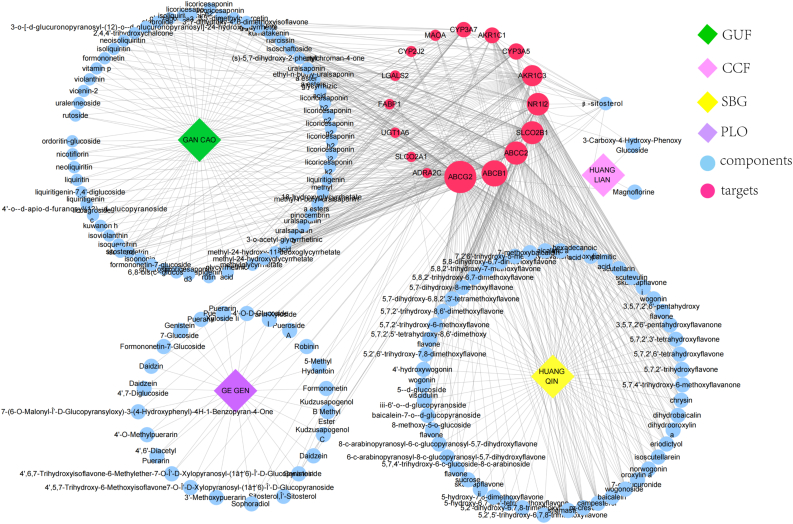
A total of 16 intersecting genes were obtained after intersecting 407 drug targets with 406 drug resistance genes, the upregulated genes were as follows: ABCB1, ABCC2, ABCG2, AKR1C1, AKR1C3, FABP1, NR1I2, SLCO2B1, UGT1A6; the downregulated genes were as follows: ADRA2C, CYP2J2, CYP3A5, CYP3A7, LGALS2, MAOA, and SLCO2A1. The above genes mainly belong to ABC transporter protein, CYP450 family, and solute carrier organic anion transporter family. The CYP450 family is mainly involved in the physiological process of drug catabolism. The solute carrier organic anion transporter family is involved in organic anion transport. The annotation of each gene is shown in Supplementary material 3. As shown in Figure 2, the 16 intersecting genes and their corresponding active compounds were plotted in Cytoscape. The larger the icon, the more active ingredients the gene belongs to, and ABCG2 belongs to the most active ingredients, followed by ABCB1, SLCO2B1, and ABCC2, which indicated that GQD can act on these targets to sensitize CRC chemotherapy.
Figure 2.
The active compounds and the herbal medicines are represented as concentric circles, and the compounds of multiple genera are arranged individually. The red color indicates the intersecting gene, and the greater the size of the icon, more the number of active ingredients belonging to the gene. Blue color indicates the active compound, yellow color indicates Scutellaria baicalensis Georgi, green color indicates Glycyrrhiza uralensis Fisch., pink color indicates Coptis chinensis Franch., and purple color indicates Pueraria lobata (Willd.) Ohwi.
3.4. GO and KEGG pathways show that GQD is closely related to CRC and OXA resistance
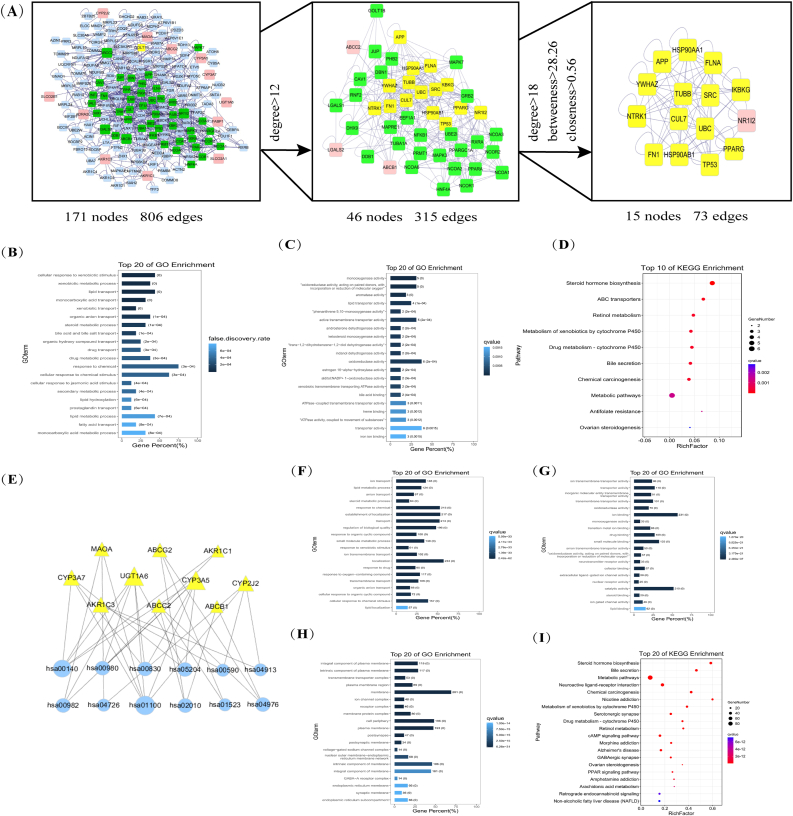
A total of 16 intersecting genes were enriched on 64 BP terms, 30 MF terms, and one CC (GO:0045177) with an FDR of 0.0153, and the complete terms are shown in Supplementary material 4. Figure 3B-D showed only the first 20 BP and MF bars of the 16 intersecting genes enriched. We found that 12 signaling pathways such as steroid hormone biosynthesis and ABC transporters were enriched and were mainly involved in molecular functions and biological processes such as substance transport energy across the membrane. As shown in Figure 3E, steroid hormone biosynthesis and metabolic pathways were the most enriched genes. CYP3A5, CYP2J2, and other metabolism-related genes were the most involved pathways.
Figure 3.
(A) Protein-protein interaction network. The pink color indicates the 16 intersecting genes, the blue color indicates the other genes in the PPI network where the 16 intersecting genes are located, the green color indicates the gene in the sub-network 1 extracted after Degree >18, and the yellow color indicates the gene in the sub-network 2. The top 20 BP (B) and MF (C) of the 16 intersecting genes enriched. The top 20 BP(F), MF(G), and CC(H) of GQD. The horizontal axis of the bars represents the percentage of intersecting genes in that term; the darker the color, the smaller the false discovery rate (FDR). The values on the bars indicate the number of genes on that pathway and the FDR value. Bubble plot for the top 10 signaling pathways of the 16 intersecting genes (D) and GQD (I). The larger the bubble, the more the number of genes enriched on that pathway; the darker the color, the smaller the FDR(E) A network diagram of the intersecting genes and their pathways. The larger the icon, the more enriched the genes and their pathways.
The 407 targets of GQD were enriched in 1544 BP terms, 313 MF terms, and 133 CCs, mainly clustered in 191 signaling pathways, such as platinum resistance and NF-κB signaling pathway (Figures 3F-3I), which indicated that GQD is involved in many cancer-related pathways and exerts anti-tumor efficacy in a multi-target and pathway manner (see Supplementary material 5 for complete signaling pathways).
The smaller the FDR, the more genes enriched are in pathways and terms, the closer the relationship with GQD in alleviating CRC resistance, and the more these pathways, terms, and genes garner our attention.
3.5. Topology analysis of intersection genes reveals important targets of GQD acting on OXA-resistant CRC
Protein–protein interaction (PPI) network was constructed with the BisoGenet plug-in (Figure 3A), the topology analysis of PPI was performed with the analysis function of tools, and Degree >12 was used as the threshold to extract sub-network 1, and then with CytoNCA plug-in, Degree >18, Betweenness >28.26, Closeness >0.56 as thresholds to extract sub-network 2. A total of 4 intersecting genes (NR1I2, ABCB1, ABCC2, and LGALS2) were in the extracted subnetwork 1, which we considered the key genes. One intersecting gene (NR1I2) in subnetwork 2 was probably the most important one as a key gene. These nodes can play an important role in the PPI network against CRC resistance. NR1I2 leads to drug resistance by inducing ABC transporters (Dong et al., 2017; Rachmale et al., 2022). The transporters are the OXA-resistant CRC-related gene that GQD targets the most. Therefore, we hypothesize that ABC transporters can be an important way for GQD to reverse OXA resistance and increase its sensitivity.
3.6. GQD lyophilized powder contains various chemical composition
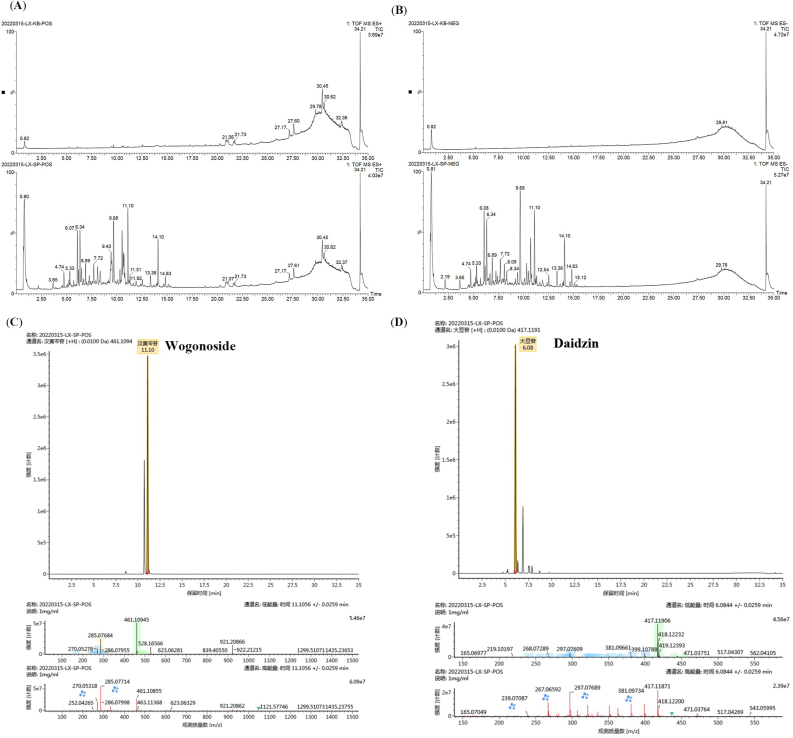
The chemical composition of GQD aqueous extract was analyzed by UPLC-Q-TOF/MS, and the results of the top 10 in descending order of response based on chromatographic information (peak retention time, and response time) and mass spectrometric information (exact relative molecular mass, and adducts) in positive and negative ion modes are shown in Supplementary material 6 (specific mass spectrum information of the compound is shown in Figure 4C-D). The total ion chromatography under different ion modes (Figure 4A-B) showed that there were some differences in the chromatographic peak type and peak number of GQD aqueous extract. A total of 485 components can be detected in the positive ion mode and 390 compounds in the negative ion mode.
Figure 4.
(A) Base peak chromatograms of GQD in the positive ion mode (B) Base peak chromatograms of GQD in the negative ion mode (C) Chromatogram, mass chromatogram, and mass spectrometry of Wogonoside in the positive ion mode (D) Chromatogram and mass spectrometry of Daidzin in the positive ion mode.
3.7. Molecular docking shows that GQD has good binding performance with ABC transporters
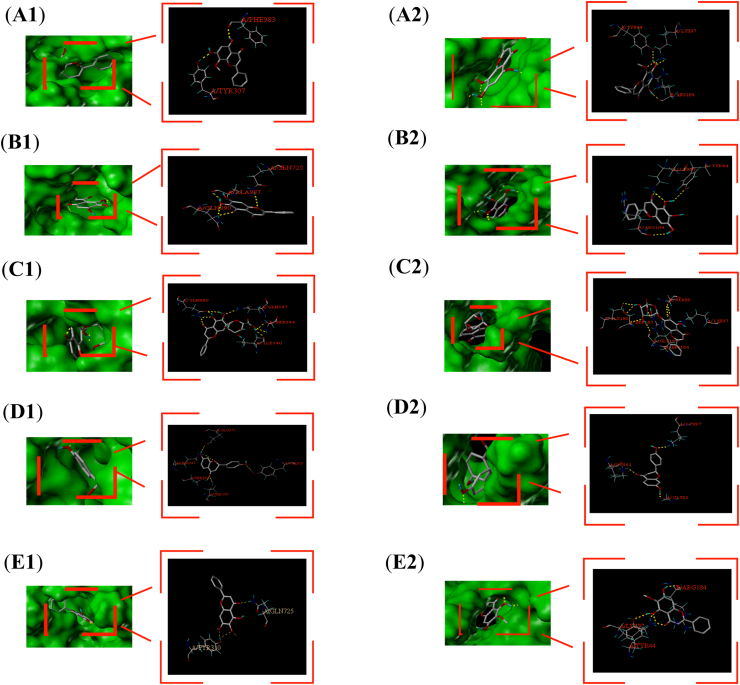
Based on the results of network pharmacology and mass spectrometry, we performed the docking of ABC transporter proteins. Because the human crystal structure of ABCC2 was not included in PDB, and the sequence similarity with homologous proteins was low after homology modeling with SWISS-MODEL, ABCB1 (PDB ID: 7A69) and ABCG2 (PDB ID: 6HBU) were selected for docking. A total of 62 common active components targeting the three ABC proteins were first screened, and then the top 5 active components (wogonin, oroxylin a, baicalein, baicalin, liquiritigenin) and the top active components with mass spectral response numbers were selected for docking according to the number of occurrences of these active components in the whole GQD. Total Score>7 indicates very high binding activity and 3 < Total Score<5 indicates moderate binding activity. The results showed that most of the active ingredients were well bound to the proteins, and the specific docking sites are shown in Figure 5A-E, and the specific binding power is shown in Table 2 (1: incorrect penetration of ligand, the smaller its absolute value, the better; 2: ligand region, the smaller its absolute value, the better).
Figure 5.
Specific binding sites of proteins and active components. 1: ABCB1; 2: ABCG2; A–E: wogonin, baicalein, baicalin, liquiritigenin, oroxylin a.
Table 2.
Depiction of the specific binding force.
| Protein | Name | Total Score | Crash 1 | Polar 2 |
|---|---|---|---|---|
| ABCG2 | Wogonoside | 9.4548 | -1.6689 | 5.0184 |
| baicalin | 9.1353 | -1.4162 | 4.8086 | |
| Daidzin | 8.1551 | -1.4461 | 3.5646 | |
| 3′-Methoxydaidzin | 7.6355 | -0.9306 | 2.4728 | |
| wogonin | 7.2843 | -1.001 | 3.1565 | |
| oroxylin a | 6.5954 | -0.4413 | 2.9702 | |
| baicalein | 6.3297 | -0.5193 | 2.8369 | |
| liquiritigenin | 6.0909 | -0.8059 | 2.1703 | |
| Puerarin apioside | 5.5249 | -2.8491 | 1.1868 | |
| ABCB1 | baicalin | 6.4172 | -1.986 | 3.873 |
| Puerarin apioside | 6.4406 | -1.5372 | 1.415 | |
| Wogonoside | 6.3314 | -2.0915 | 1.9932 | |
| liquiritigenin | 6.2019 | -0.7235 | 3.7559 | |
| Glycyrrhizinic Acid | 6.1699 | -3.5307 | 1.4341 | |
| wogonin | 4.5987 | -0.7245 | 1.1245 | |
| oroxylin a | 4.4363 | -0.4042 | 2.1211 | |
| baicalein | 4.0608 | -0.6245 | 1.161 |
3.8. GQD inhibits CRC cell proliferation and reverses drug resistance along with OXA
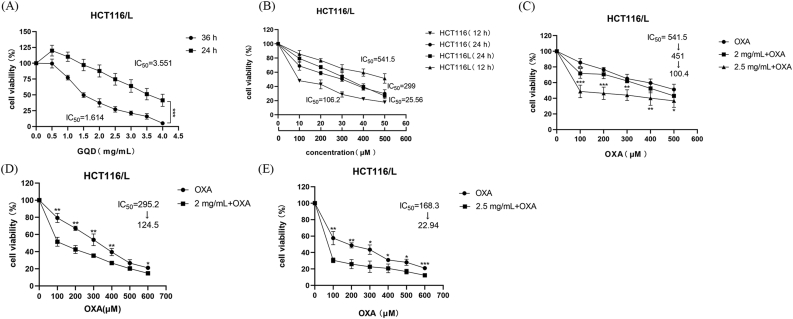
The CCK8 assay showed that GQD and OXA inhibited CRC cell proliferation with increasing time and concentration in HCT116/L. As shown in Figure 6, the half maximal inhibitory concentration (IC50) of GQD was 3.551 mg/mL and 1.614 mg/mL at 24 h and 36 h (Figure 6A), respectively, and the IC50 of OXA was 541.5 μM and 299 μM at 12 h and 24 h (Figure 6B), respectively. After co-incubation with OXA at different concentrations for 12 h and 24 h, GQD significantly enhanced the chemotherapeutic effect of OXA, restored the sensitivity to OXA (Figure 6C-E), and synergistically reversed the resistance index (RI) of OXA (RI = IC50 (HCT116/L)/IC50 (HCT116)). The CI was used to assess whether the combination of GQD and OXA had synergistic effects. As expected, CI values for the combination of different concentrations of OXA and GQD (2 or 2.5 mg/mL) showed significant synergistic effects (CI < 1) (Table 3), with GQD at 2.5 mg/mL showing the strongest synergistic effect with OXA at 100 μM.
Figure 6.
HCT116/L was inhibited with an increase in time, concentration, and combination of OXA and GQD, while GQD could reverse the resistance to OXA. The cell survival rate was detected by CCK8 (A, B): HCT116/L was treated with GQD and OXA for 24 h, 36 h, and 12 h, 24 h, respectively (C): HCT116/L was treated with GQD and OXA for 12 h (D, E): The survival rate of HCT116/L when GQD were co-treated with OXA for 24 h. Data are presented as the mean ± SD. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. 2 mg/mL, 2.5 mg/mL: GQD concentration. 100 μM: OXA concentration.
Table 3.
Combination Index (CI) using the CompuSyn software based on the results illustrated in Figure 6.
| Time(h) | GQD (mg/mL) | Reversal fold of IC50 after GQD | OXA (μM) | Combination Index (CI) |
|---|---|---|---|---|
| 24 | 2 | 2.37108 | 100 | 0.94907 |
| 2.5 | 7.33653 | 100 | 0.77475 | |
| 200 | 0.88926 | |||
| 300 | 0.97931 | |||
| 600 | 0.98810 | |||
| 12 | 2.5 | 5.39342 | 100 | 0.74278 |
| 200 | 0.86189 | |||
| 300 | 0.92831 |
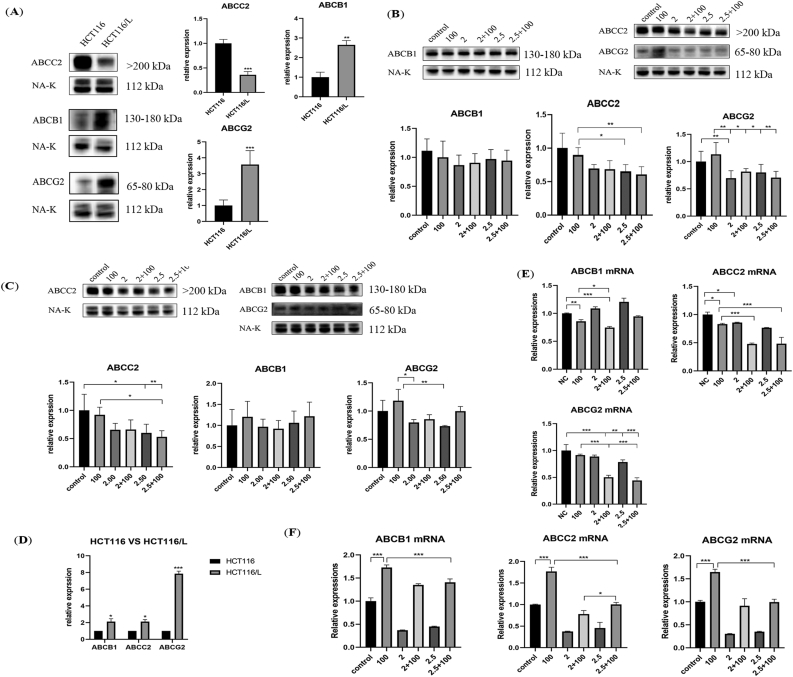
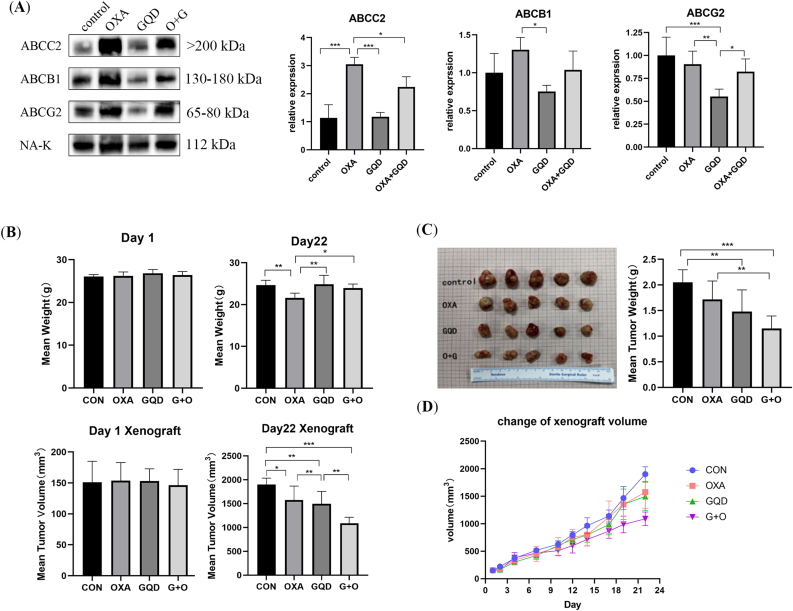
3.9. GQD inhibits the levels of ABC transporter mRNAs and proteins
The WB results showed (Figure 7A) higher levels of ABCB1 and ABCG2 in HCT116/L than in HCT116, whereas the ABCC2 level was decreased. ABCG2 level significantly decreased after incubation with GQD alone and in combination with OXA compared with its increased level after incubation with OXA alone for 12 h (Figure 7B). ABCC2 level also decreased significantly and was negatively correlated with GQD concentration, whereas ABCB1 level decreased but was not statistically significant. After incubation for 24 h (Figure 7C), no significant difference in ABCB1 level was observed; however, compared with OXA alone, it decreased at a GQD concentration of 2 mg/mL. Furthermore, ABCB1 levels increased with the increasing GQD concentration, whereas ABCC2 levels significantly decreased with the increasing TCM concentration. ABCG2 level was significantly decreased when incubated with GQD alone compared with the level when incubated with OXA alone. Though its level was also decreased when incubated with the combination (OXA + GQD), the difference was not significant.
Figure 7.
GQD alone and in synergy with OXA inhibited the protein and mRNA expression of the ABC transporter (A, B) Protein and mRNA expression of ABCB1, ABCC2, and ABCG2 in HCT116 and HCT116/L (C–F) The protein expression and mRNA changes of ABCB1, ABCC2, and ABCG2 when GQD and OXA acted either alone or together for 12 and 24 h. Data are presented as the mean ± SD. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. 2 mg/mL, 2.5 mg/mL: GQD concentration. 100 μM: OXA concentration.
The qRT-PCR results showed different inhibitory effects of GQD at different concentrations and duration of action. ABCB1, ABCC2, and ABCG2 mRNA levels were increased in HCT116/L compared with those in HCT116 (Figure 7D). After 12 h of drug intervention (Figure 7E), ABCB1 mRNA level was significantly decreased when incubated with OXA alone and increased when incubated with TCM alone. ABCB1 mRNA level significantly decreased when incubated with the combination of GQD and OXA (2 mg/mL) compared with the level when incubated with OXA alone. ABCC2 and ABCG2 mRNA levels showed an overall decreasing trend, and compared with the OXA. ABCB1, ABCC2, and ABCG2 mRNA levels were significantly increased after 24 h of OXA intervention (Figure 7F), whereas these levels decreased after GQD intervention alone and reversed the increase induced by OXA when used in combination. In contrast to the protein changes, the ABCC2 mRNA level increased differentially with the increasing TCM concentration.
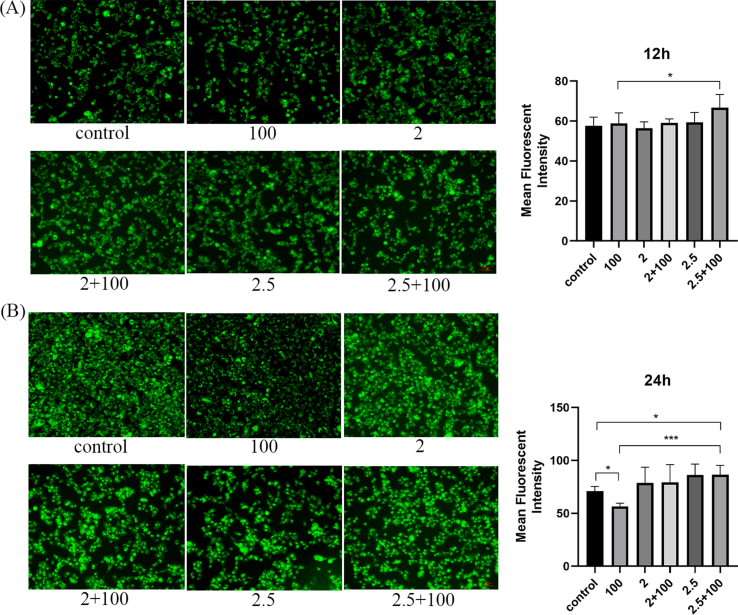
3.10. GQD inhibits drug efflux
The rhodamine accumulation experiment showed that after 24 h of intervention, GQD could significantly increase the intracellular accumulation of rhodamine and inhibit drug efflux compared with OXA intervention (Figure S1B). With the prolongation of drug action time and the increase in intervention concentration, the inhibitory effect of GQD increased remarkably (Figure S1A-B).
3.11. GQD effectively resists OXA-resistant CRC and relieves toxic side effects in vivo
At the end of 3 weeks of treatment, the OXA group showed significant weight loss, whereas the GQD and combination treatment groups showed less weight loss (Figure 8B). At the end of treatment, the tumor volume was significantly reduced in the combination treatment and GQD groups compared with that in the OXA group, indicating that OXA was ineffective in OXA-resistant CRC compared with the combination (Figure 8C). However, OXA combined with GQD could alleviate the toxic side effects caused by chemotherapy, reduce tumor volume, and increase sensitivity to OXA. Moreover, according to the tumor volume change curve (Figure 8D), the therapeutic advantages of GQD + OXA gradually increased with the increasing treatment time. The WB of tumor tissues showed that OXA significantly increased ABCC2 level, whereas GQD alone did not increase its level, and the combination reversed the increasing ABCC2 level. Compared with OXA, GQD alone significantly reduced ABCB1 and ABCG2 levels (Figure 8A).
Figure 8.
GQD inhibits the expression of ABC transporter in vivo and synergizes with OXA to increase the sensitivity to OXA, alleviate the toxic and side effects of OXA, and improve prognosis (A) Xenograft tumors were derived from nude mice and the protein expression levels of ABC transporters were examined and quantified by Western blotting (B) Bodyweight and tumor body changes of different treatment groups on days 1 and 22 after successful tumor establishment (C) A total of 20 xenograft tumors (5 mice/group) from each experimental group (D) Mean tumor size change for each experimental group. Data are presented as the mean ± SD. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
4. Discussion
CRC incidence is high worldwide, including in China (Li et al., 2021). Single chemotherapy for advanced CRC has limitations such as a low effective objective remission rate and several adverse reactions (Guo et al., 2020). Chinese herbal medicine with additive treatment has unique clinical efficacy and advantages in treating malignant tumors such as personalized treatment, overall conditioning, and less toxic side effects (Sun et al., 2021). In this study, we investigated the effect of GQD on improving the effects of CRC chemotherapy and alleviating resistance to OXA using the network pharmacology approach, performed docking validation, and in vivo and in vitro validation of key targets and active compounds. We found that GQD could reduce transporter protein levels and reverse CRC resistance to OXA to understand the therapeutic effect of TCM on CRC, guide clinical practice, and optimize the treatment plan.
GQD is widely used in moist-heat intestinal disorders, and recently several studies have also shown that various active components of GQD, such as baicalin, curcumin, and baicalin, have various anticancer activities and have been used in clinical combination chemotherapy to prevent tumors (Howells et al., 2019; Selvam et al., 2019; Singh et al., 2021; Weng and Goel, 2022). However, a few studies on its effectiveness in treating OXA-resistant CRC are available. The KEGG results showed that GQD acted on multiple cancer-related pathways including the NF-κB signaling, platinum resistance, cancer, PI3K-Akt, ABC transport, IL-17 signaling pathway, and inflammatory bowel disease pathways. The PPI network showed that GQD might have acted via 16 genes including NR1I2, ABCB1, ABCC2, and ABCG2, which act as adjuvants in CRC chemotherapy. Most of which are associated with tumor progression and chemoresistance.
ABCB1, ABCC2, and ABCG2 are ATP-binding transporter proteins whose higher levels confer resistance toward OXA to cancer cells (Po et al., 2020; Y. Wang et al., 2021; Z. Wang et al., 2017; Z. Wang et al., 2020). The activation of aldo-keto reductases in digestive tract cancers contributes to the acquisition of platinum chemoresistance by cancer cells, and the inhibition of AKR1C activity significantly reverses platinum chemoresistance (Chen et al., 2013; Matsunaga et al., 2013). LGALS2 promoted cancer cell adhesion to the vascular endothelium and metastasis to inhibit colon tumor growth ((Barrow et al., 2011; Li et al., 2021). NR1I2 encodes the nuclear receptor Pregnane X, which promotes drug excretion and clearance from the body and mediates the drug-induced upregulation of CYP3A5, and its high expression contributes to the acquisition of resistance in gastrointestinal tumor cells, whereas in vitro and in vitro findings suggest that baicalin significantly inhibits CYP3A5 expression ((Barraclough et al., 2012; Ding et al., 2020; Noll et al., 2016). FABP1 is one of the potential markers of CRC (Ludvigsen et al., 2021; G. L. Zhang, Pan, Huang and Wang, 2019). This shows that GQD is closely associated with anti-CRC chemoresistance.
These 16 intersecting genes are the main targets of glycyrrhetinic acid, licoricesaponin, baicalin and baicalein. Baicalin can treat CRC via non-dependent autophagy leading to cell death, and trigger apoptosis via the TLR4/NF-κB signaling pathway to inhibit migration and enhance antitumor immunity in CRC (M. Chen et al., 2021; Choi et al., 2017). Glycyrrhizic acid affects the Wnt signaling pathway to promote the entry of gerberosin into cells for antitumor activity (Y. Li et al., 2021). Baicalein can prevent CRC by regulating p53 nuclear translocation and promoting apoptosis in colon cancer cells (Feng et al., 2018).
Based on the differential gene KEGG enrichment pathway analysis and molecular docking results, we suggest that the pathway ABC transporters is the most important pathway in CRC resistance inhibition and sensitivity restoration by GQD. The docking results showed that ABCB1 has high docking activity with ABCG2, wogonin, and liquiritigenin, which proved our prediction. Especially, ABCG2 exhibits strong binding activity toward baicalin and baicalein. Outward-bound transporter proteins of the ATP-binding cassette family are the main factors in the acquisition of chemoresistance by tumor cells, and their increased levels cause drug efflux leading to MDR (Amawi et al., 2019; Holohan et al., 2013; Theile and Wizgall, 2021). P-glycoprotein (P-gp) inhibitors, RNA interference, nano drugs, and combination drugs are new strategies against MDR; however, current clinical trials of related transporter protein inhibitors have been unsuccessful and have shown toxic side effects of these drugs (Mohammad et al., 2020; Robey et al., 2018; Tian et al., 2012). Baicalein is a substrate of P-gp and competitively inhibits P-gp to enhance anticancer activity, and flavonoids and alkaloids also modulate P-gp, the main active component of GQD (Cao et al., 2020; Palko-Labuz et al., 2017).
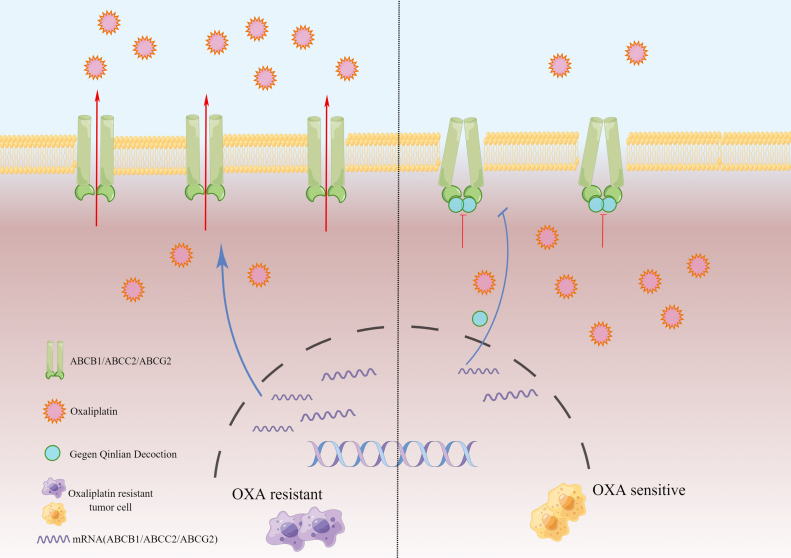
The molecular docking results showed that the ABC transporter protein docked well with GQD activity. The in vivo experiments also showed that GQD reversed CRC resistance to OXA and exerted a therapeutic effect along with OXA, decreasing mRNA and protein levels of ABCB1, ABCC2, and ABCG2, supporting the hypothesis, and it also alleviated the toxic effects of OXA. The WB and qRT-PCR results revealed that GQD alone and in combination with OXA inhibited transporter protein mRNA and protein levels and effectively enhanced the killing effect of OXA on cancer cells. Rhodamine accumulation indicated that GQD significantly increased the intracellular accumulation of rhodamine and inhibited drug efflux, and its effect is correlated to concentration and time. GQD has great potential in reversing resistance to OXA and increasing sensitivity to it. However, according to the present results, we found that the changes of mRNAs and protein after GQD intervention were unsynchronized. After 24 h of GQD intervention, the protein level gradually decreased with the increasing GQD concentration; however, the mRNA level increased. We speculate that GQD plays a certain role in translation and post-translation modifications. Surprisingly, our repeated results showed that the level of ABCC2 mRNA in OXA-tolerant CRC cells increased, whereas the protein level decreased significantly. We suspect that some mechanisms in the process of translation and post-translation modification have prevented this trend. We also suspect it relates to the construction of OXA-resistant cells. However, GQD could still act on the protein or gene target. The changes cancer cells underwent during OXA tolerance need to be explored in the future. Though studies on ABC transporters are increasing, the limitations and side effects of several generations of inhibitors make their clinical application difficult (Mohammad et al., 2020; Robey et al., 2018; Engle & Kumar, 2022). Due to the inhibition of ABC transporters, reversal of resistance to OXA, and safe side effects of GQD, developing new transporter inhibitors is possible.
GQD can improve chemotherapy efficacy by inhibiting transporter proteins, reducing OXA efflux, and reducing toxic side effects of currently existing chemical drugs with its advantages of being natural and less toxic and improving the quality of life of patients with advanced CRC and chemotherapy efficacy.
5. Conclusion
We are the first to discuss and validate the mechanism by which GQD targets and restores sensitivity to OXA against OXA-resistant CRC by combining network pharmacology prediction, molecular docking, and in vitro and in vivo experiments. We show that GDQ reverses CRC resistance to OXA by reducing the levels and efflux of ABC transporters and exhibits potent anti-CRC effects along with OXA, providing a promising approach to elucidate the scientific basis and specific mechanisms of TCM for disease treatment.
Declarations
Author contribution statement
Xiang Lin: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Li Xu: Performed the experiments; Analyzed and interpreted the data.
Huicheng Tan: Analyzed and interpreted the data.
Xinyi Zhang: Contributed reagents, materials, analysis tools or data; Analyzed and interpreted the data.
Li Yao and Huan Shao: Contributed reagents, materials, analysis tools or data.
Xuan Huang: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data.
Funding statement
Xuan Huang was supported by Administration of Traditional Chinese Medicine of Zhejiang Province [2021ZZ013].
Data availability statement
Data will be made available on request.
Declaration of interest’s statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors acknowledge gratitude to all the staff who participated in the study and the authorization of Figdraw.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
Fig.S1.
Rhodamine accumulation experiment of HCT116/L for 12 h (A) and 24 h (B) under the intervention of GQD. Scale bar: 50 µm. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. 2 mg/mL, 2.5 mg/mL: GQD concentration. 100 µM: OXA concentration.
Figure S2.
The mechanism of anti-cancer effect of Gegen Qinlian Decoction (drawn by Figdraw).
References
- Amawi H., Sim H.M., Tiwari A.K., Ambudkar S.V., Shukla S. ABC transporter-mediated multidrug-resistant cancer. Adv. Exp. Med. Biol. 2019;1141:549–580. doi: 10.1007/978-981-13-7647-4_12. [DOI] [PubMed] [Google Scholar]
- Barraclough K.A., Isbel N.M., Lee K.J., Bergmann T.K., Johnson D.W., McWhinney B.C., Staatz C.E. NR1I2 polymorphisms are related to tacrolimus dose-adjusted exposure and BK viremia in adult kidney transplantation. Transplantation. 2012;94(10):1025–1032. doi: 10.1097/TP.0b013e31826c3985. [DOI] [PubMed] [Google Scholar]
- Barrow H., Guo X., Wandall H.H., Pedersen J.W., Fu B., Zhao Q., Yu L.G. Serum galectin-2, -4, and -8 are greatly increased in colon and breast cancer patients and promote cancer cell adhesion to blood vascular endothelium. Clin. Cancer Res. 2011;17(22):7035–7046. doi: 10.1158/1078-0432.CCR-11-1462. [DOI] [PubMed] [Google Scholar]
- Cao Y., Shi Y., Cai Y., Hong Z., Chai Y. The effects of traditional Chinese medicine on P-Glycoprotein-Mediated multidrug resistance and approaches for studying the herb-P-glycoprotein interactions. Drug Metab. Dispos. 2020;48(10):972–979. doi: 10.1124/dmd.120.000050. [DOI] [PubMed] [Google Scholar]
- Chen C.C., Chu C.B., Liu K.J., Huang C.Y., Chang J.Y., Pan W.Y.…Chen L.T. Gene expression profiling for analysis acquired oxaliplatin resistant factors in human gastric carcinoma TSGH-S3 cells: the role of IL-6 signaling and Nrf2/AKR1C axis identification. Biochem. Pharmacol. 2013;86(7):872–887. doi: 10.1016/j.bcp.2013.07.025. [DOI] [PubMed] [Google Scholar]
- Chen M., Zhong K., Tan J., Meng M., Liu C.M., Chen B.…Kwan H.Y. Baicalein is a novel TLR4-targeting therapeutics agent that inhibits TLR4/HIF-1alpha/VEGF signaling pathway in colorectal cancer. Clin. Transl. Med. 2021;11(11):e564. doi: 10.1002/ctm2.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Liu X.Q., Lin X., Gao L.Y., Zhang S., Huang X. Targeting YTHDF1 effectively re-sensitizes cisplatin-resistant colon cancer cells by modulating GLS-mediated glutamine metabolism. Mol. Ther. Oncolytics. 2021;20:228–239. doi: 10.1016/j.omto.2021.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi B.Y., Joo J.C., Lee Y.K., Jang I.S., Park S.J., Park Y.J. Anti-cancer effect of Scutellaria baicalensis in combination with cisplatin in human ovarian cancer cell. BMC Compl. Alternative Med. 2017;17(1):277. doi: 10.1186/s12906-017-1776-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Wang Z., Xie G.F., Li C., Zuo W.W., Meng G., Li J.J. Pregnane X receptor is associated with unfavorable survival and induces chemotherapeutic resistance by transcriptional activating multidrug resistance-related protein 3 in colorectal cancer. Mol. Cancer. 2017;16(1):71. doi: 10.1186/s12943-017-0641-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker E., Tanis P.J., Vleugels J.L.A., Kasi P.M., Wallace M.B. Colorectal cancer. Lancet. 2019;394(10207):1467–1480. doi: 10.1016/S0140-6736(19)32319-0. [DOI] [PubMed] [Google Scholar]
- Ding W., Cao C., Gao Y., Zhou X., Lai Y. Inhibition of CYP3A4 and CYP3A5 expression by scutellarin is not mediated via the regulation of hsa-miR-27a, 27b, 148a, 298 and 451a levels. Xenobiotica. 2020;50(11):1267–1274. doi: 10.1080/00498254.2018.1555344. [DOI] [PubMed] [Google Scholar]
- Engle K., Kumar G. Cancer multidrug-resistance reversal by ABCB1 inhibition: a recent update. Eur. J. Med. Chem. 2022;239 doi: 10.1016/j.ejmech.2022.114542. [DOI] [PubMed] [Google Scholar]
- Feng Q., Wang H., Pang J., Ji L., Han J., Wang Y., Lu L. Prevention of wogonin on colorectal cancer tumorigenesis by regulating p53 nuclear translocation. Front. Pharmacol. 2018;9:1356. doi: 10.3389/fphar.2018.01356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransson A., Glaessgen D., Alfredsson J., Wiman K.G., Bajalica-Lagercrantz S., Mohell N. Strong synergy with APR-246 and DNA-damaging drugs in primary cancer cells from patients with TP53 mutant High-Grade Serous ovarian cancer. J. Ovarian Res. 2016;9(1):27. doi: 10.1186/s13048-016-0239-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacchetti S., Perpoint B., Zidani R., Le Bail N., Faggiuolo R., Focan C.…Levi F. Phase III multicenter randomized trial of oxaliplatin added to chronomodulated fluorouracil-leucovorin as first-line treatment of metastatic colorectal cancer. J. Clin. Oncol. 2000;18(1):136–147. doi: 10.1200/JCO.2000.18.1.136. [DOI] [PubMed] [Google Scholar]
- Guo J., Yu Z., Das M., Huang L. Nano codelivery of oxaliplatin and folinic acid achieves synergistic chemo-immunotherapy with 5-fluorouracil for colorectal cancer and liver metastasis. ACS Nano. 2020;14(4):5075–5089. doi: 10.1021/acsnano.0c01676. [DOI] [PubMed] [Google Scholar]
- Guo Y., Xiong B.H., Zhang T., Cheng Y., Ma L. XELOX vs. FOLFOX in metastatic colorectal cancer: an updated meta-analysis. Cancer Invest. 2016;34(2):94–104. doi: 10.3109/07357907.2015.1104689. [DOI] [PubMed] [Google Scholar]
- Holohan C., Van Schaeybroeck S., Longley D.B., Johnston P.G. Cancer drug resistance: an evolving paradigm. Nat. Rev. Cancer. 2013;13(10):714–726. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- Howells L.M., Iwuji C.O.O., Irving G.R.B., Barber S., Walter H., Sidat Z.…Brown K. Curcumin combined with FOLFOX chemotherapy is safe and tolerable in patients with metastatic colorectal cancer in a randomized phase IIa trial. J. Nutr. 2019;149(7):1133–1139. doi: 10.1093/jn/nxz029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Zhang Z., Li W., Kong F., Yi P., Huang J.…Zhang S. Network pharmacology-based prediction and verification of the active ingredients and potential targets of zuojinwan for treating colorectal cancer. Drug Des. Dev. Ther. 2020;14:2725–2740. doi: 10.2147/DDDT.S250991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Zhao L., Lau Y.S., Zhang C., Han R. Genome-wide CRISPR screen identifies LGALS2 as an oxidative stress-responsive gene with an inhibitory function on colon tumor growth. Oncogene. 2021;40(1):177–188. doi: 10.1038/s41388-020-01523-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Lu B., Luo C., Cai J., Lu M., Zhang Y., Dai M. Incidence, mortality, survival, risk factor and screening of colorectal cancer: a comparison among China, Europe, and northern America. Cancer Lett. 2021;522:255–268. doi: 10.1016/j.canlet.2021.09.034. [DOI] [PubMed] [Google Scholar]
- Li Y., Gong Y., Zhang X., Wang J., Cheng Y., Liu F., Dong L. Exploring the synergistic mechanism of Gegen Qinlian Decoction on the Wnt signaling pathway using an integrated strategy of network pharmacology and RNA-seq. J. Ethnopharmacol. 2021;278 doi: 10.1016/j.jep.2021.114283. [DOI] [PubMed] [Google Scholar]
- Ludvigsen M., Thorlacius-Ussing L., Vorum H., Stender M.T., Thorlacius-Ussing O., Honore B. Proteomic characterization of colorectal cancer tissue from patients identifies novel putative protein biomarkers. Curr. Issues Mol. Biol. 2021;43(2):1043–1056. doi: 10.3390/cimb43020074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv J., Jia Y., Li J., Kuai W., Li Y., Guo F.…Li Z. Gegen Qinlian decoction enhances the effect of PD-1 blockade in colorectal cancer with microsatellite stability by remodelling the gut microbiota and the tumour microenvironment. Cell Death Dis. 2019;10(6):415. doi: 10.1038/s41419-019-1638-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Balibrea E., Martinez-Cardus A., Gines A., Ruiz de Porras V., Moutinho C., Layos L., Abad A. Tumor-related molecular mechanisms of oxaliplatin resistance. Mol. Cancer Therapeut. 2015;14(8):1767–1776. doi: 10.1158/1535-7163.MCT-14-0636. [DOI] [PubMed] [Google Scholar]
- Matsunaga T., Hojo A., Yamane Y., Endo S., El-Kabbani O., Hara A. Pathophysiological roles of aldo-keto reductases (AKR1C1 and AKR1C3) in development of cisplatin resistance in human colon cancers. Chem. Biol. Interact. 2013;202(1-3):234–242. doi: 10.1016/j.cbi.2012.09.024. [DOI] [PubMed] [Google Scholar]
- Meads M.B., Gatenby R.A., Dalton W.S. Environment-mediated drug resistance: a major contributor to minimal residual disease. Nat. Rev. Cancer. 2009;9(9):665–674. doi: 10.1038/nrc2714. [DOI] [PubMed] [Google Scholar]
- Mohammad I.S., He W., Yin L. Insight on multidrug resistance and nanomedicine approaches to overcome MDR. Crit. Rev. Ther. Drug Carrier Syst. 2020;37(5):473–509. doi: 10.1615/CritRevTherDrugCarrierSyst.2020025052. [DOI] [PubMed] [Google Scholar]
- Neugut A.I., Lin A., Raab G.T., Hillyer G.C., Keller D., O'Neil D.S.…Hershman D.L. FOLFOX and FOLFIRI use in stage IV colon cancer: analysis of SEER-medicare data. Clin. Colorectal Cancer. 2019;18(2):133–140. doi: 10.1016/j.clcc.2019.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll E.M., Eisen C., Stenzinger A., Espinet E., Muckenhuber A., Klein C., Sprick M.R. CYP3A5 mediates basal and acquired therapy resistance in different subtypes of pancreatic ductal adenocarcinoma. Nat. Med. 2016;22(3):278–287. doi: 10.1038/nm.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palko-Labuz A., Sroda-Pomianek K., Uryga A., Kostrzewa-Suslow E., Michalak K. Anticancer activity of baicalein and luteolin studied in colorectal adenocarcinoma LoVo cells and in drug-resistant LoVo/Dx cells. Biomed. Pharmacother. 2017;88:232–241. doi: 10.1016/j.biopha.2017.01.053. [DOI] [PubMed] [Google Scholar]
- Po A., Citarella A., Catanzaro G., Besharat Z.M., Trocchianesi S., Gianno F., Ferretti E. Hedgehog-GLI signalling promotes chemoresistance through the regulation of ABC transporters in colorectal cancer cells. Sci. Rep. 2020;10(1) doi: 10.1038/s41598-020-70871-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provenzale D., Ness R.M., Llor X., Weiss J.M., Abbadessa B., Cooper G.…Ogba N. NCCN guidelines insights: colorectal cancer screening, version 2.2020. J. Natl. Compr. Cancer Netw. 2020;18(10):1312–1320. doi: 10.6004/jnccn.2020.0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachmale M., Rajput N., Jadav T., Sahu A.K., Tekade R.K., Sengupta P. Implication of metabolomics and transporter modulation based strategies to minimize multidrug resistance and enhance site-specific bioavailability: a needful consideration toward modern anticancer drug discovery. Drug Metab. Rev. 2022;54(2):101–119. doi: 10.1080/03602532.2022.2048007. [DOI] [PubMed] [Google Scholar]
- Robey R.W., Pluchino K.M., Hall M.D., Fojo A.T., Bates S.E., Gottesman M.M. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat. Rev. Cancer. 2018;18(7):452–464. doi: 10.1038/s41568-018-0005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samad M.A., Saiman M.Z., Abdul Majid N., Karsani S.A., Yaacob J.S. Berberine inhibits telomerase activity and induces cell cycle arrest and telomere erosion in colorectal cancer cell line, HCT 116. Molecules. 2021;26(2) doi: 10.3390/molecules26020376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvam C., Prabu S.L., Jordan B.C., Purushothaman Y., Umamaheswari A., Hosseini Zare M.S., Thilagavathi R. Molecular mechanisms of curcumin and its analogs in colon cancer prevention and treatment. Life Sci. 2019;239 doi: 10.1016/j.lfs.2019.117032. [DOI] [PubMed] [Google Scholar]
- Singh S., Meena A., Luqman S. Baicalin mediated regulation of key signaling pathways in cancer. Pharmacol. Res. 2021;164 doi: 10.1016/j.phrs.2020.105387. [DOI] [PubMed] [Google Scholar]
- Sui H., Pan S.F., Feng Y., Jin B.H., Liu X., Zhou L.H.…Li Q. Zuo Jin Wan reverses P-gp-mediated drug-resistance by inhibiting activation of the PI3K/Akt/NF-kappaB pathway. BMC Compl. Alternative Med. 2014;14:279. doi: 10.1186/1472-6882-14-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., He M., Zhang M., Zeng S., Chen L., Zhao H.…Xu H. Traditional Chinese medicine and colorectal cancer: implications for drug discovery. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.685002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- Tauriello D.V.F., Batlle E. Targeting the microenvironment in advanced colorectal cancer. Trends Cancer. 2016;2(9):495–504. doi: 10.1016/j.trecan.2016.08.001. [DOI] [PubMed] [Google Scholar]
- Theile D., Wizgall P. Acquired ABC-transporter overexpression in cancer cells: transcriptional induction or Darwinian selection? Naunyn-Schmiedeberg’s Arch. Pharmacol. 2021;394(8):1621–1632. doi: 10.1007/s00210-021-02112-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C., Ambrosone C.B., Darcy K.M., Krivak T.C., Armstrong D.K., Bookman M.A.…DeLoia J.A. Common variants in ABCB1, ABCC2 and ABCG2 genes and clinical outcomes among women with advanced stage ovarian cancer treated with platinum and taxane-based chemotherapy: a Gynecologic Oncology Group study. Gynecol. Oncol. 2012;124(3):575–581. doi: 10.1016/j.ygyno.2011.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang Y., Qin Z., Cai S., Yu L., Hu H., Zeng S. The role of non-coding RNAs in ABC transporters regulation and their clinical implications of multidrug resistance in cancer. Expet. Opin. Drug Metabol. Toxicol. 2021;17(3):291–306. doi: 10.1080/17425255.2021.1887139. [DOI] [PubMed] [Google Scholar]
- Wang Z., Sun X., Feng Y., Liu X., Zhou L., Sui H.…Li Q. Dihydromyricetin reverses MRP2-mediated MDR and enhances anticancer activity induced by oxaliplatin in colorectal cancer cells. Anti Cancer Drugs. 2017;28(3):281–288. doi: 10.1097/CAD.0000000000000459. [DOI] [PubMed] [Google Scholar]
- Wang Z., Zhan Y., Xu J., Wang Y., Sun M., Chen J.…Xu K. Beta-sitosterol reverses multidrug resistance via BCRP suppression by inhibiting the p53-MDM2 interaction in colorectal cancer. J. Agric. Food Chem. 2020;68(12):3850–3858. doi: 10.1021/acs.jafc.0c00107. [DOI] [PubMed] [Google Scholar]
- Wei W., Wu X., Li Y. 2010. Experimental Methodology of Pharmacology. [Google Scholar]
- Weng W., Goel A. Curcumin and colorectal cancer: an update and current perspective on this natural medicine. Semin. Cancer Biol. 2022;80:73–86. doi: 10.1016/j.semcancer.2020.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H.Y., Zhang Y.Q., Liu Z.M., Chen T., Lv C.Y., Tang S.H.…Huang L.Q. ETCM: an encyclopaedia of traditional Chinese medicine. Nucleic. Acids Res. 2019;47(D1):D976–D982. doi: 10.1093/nar/gky987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Bai H., Sa Y., Zhu P., Liu P. Inhibiting EMT, stemness and cell cycle involved in baicalin-induced growth inhibition and apoptosis in colorectal cancer cells. J. Cancer. 2020;11(8):2303–2317. doi: 10.7150/jca.37242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G.L., Pan L.L., Huang T., Wang J.H. The transcriptome difference between colorectal tumor and normal tissues revealed by single-cell sequencing. J. Cancer. 2019;10(23):5883–5890. doi: 10.7150/jca.32267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., An Y., Qin X., Wu X., Wang X., Hou H., Cao H. Gut microbiota-derived metabolites in colorectal cancer: the bad and the challenges. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.739648. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.