Abstract
Frequent turnover of dengue virus (DENV) clades is one of the major forces driving DENV persistence and prevalence. In this study, we assess the fitness advantage of nine stable substitutions within the envelope (E) protein of DENV serotypes. Two tandem neighboring substitutions, threonine to lysine at the 226th (T226K) and glycine to glutamic acid at the 228th (G228E) residues in the DENV2 Asian I genotype, enhance virus infectivity in either mosquitoes or mammalian hosts, thereby promoting clades turnover and dengue epidemics. Mechanistic studies indicate that the substitution‐mediated polarity changes in these two residues increase the binding affinity of E for host C‐type lectins. Accordingly, we predict that a G228E substitution could potentially result in a forthcoming epidemic of the DENV2 Cosmopolitan genotype. Investigations into the substitutions associated with DENV fitness in hosts may offer mechanistic insights into dengue prevalence, thus providing a warning of potential epidemics in the future.
Keywords: C‐type lectin, dengue virus, host, mosquito, mutation
Subject Categories: Evolution & Ecology; Microbiology, Virology & Host Pathogen Interaction
Investigations into adaptive mutations associated with DENV fitness in mosquitoes and mammalian hosts offer insight into an evolutionary force resulting in DENV2 epidemics, possibly providing early warning of potential epidemics in the future.

Introduction
Dengue fever is the most widely spread arthropod‐borne viral disease that afflicts tropical and subtropical countries worldwide (Kyle & Harris, 2008; Messina et al, 2014). Both Aedes aegypti and Aedes albopictus mosquitoes act as native vectors to transmit dengue viruses (DENVs) (Cummings et al, 2004). There are four antigenetically distinct DENV serotypes (denoted DENV1 to DENV4), in which considerable genetic variations are phylogenetically categorized as several genotypes (Vasilakis & Weaver, 2008). DENVs have rapidly disseminated within more than one hundred countries in the past 50 years (Brady et al, 2012). It is estimated that 390 million people may be infected by DENVs annually with 96 million manifesting apparent clinical symptoms (Bhatt et al, 2013; Brady & Hay, 2020). The explosive dengue prevalence has been associated with the global expansion of the anthropophilic vectors A. aegypti and A. albopictus, the growth of human populations, uncontrolled urbanization, and the expansion of international commerce and travel (Kyle & Harris, 2008; Brady & Hay, 2020). Nonetheless, a frequent turnover of DENV clades (Rico‐Hesse et al, 1997; Wittke et al, 2002; Messer et al, 2003; Thu et al, 2004), in which a clade disappears after circulating in a region for several years and is replaced by a new clade that sometimes arises and persists for a period with an epidemic, acts as a major force to promote the continuous prevalence of dengue (Messer et al, 2003; Kyle & Harris, 2008; OhAinle et al, 2011; Manokaran et al, 2015). Therefore, dengue epidemics exhibit complex wave‐like dynamics, in which oscillations in DENV prevalence are a characteristic feature (Cummings et al, 2004; Ty Hang et al, 2010). Accumulating evidence indicates that the remarkable genetic diversity of DENVs is a determinant in the process of clade turnover and dengue epidemics (Rico‐Hesse et al, 1997; Leitmeyer et al, 1999; Messer et al, 2003; Kyle & Harris, 2008; Vasilakis & Weaver, 2008; Manokaran et al, 2015).
DENV maintains a lifecycle between human or nonhuman primates and mosquitoes; therefore, its transmission capacity is largely determined by its adaptability to these hosts (Yu et al, 2021). This adaptation often relies on the generation of DENV genetic diversity through the acquisition of mutations in the viral genomes (Cologna & Rico‐Hesse, 2003; OhAinle et al, 2011; Chan et al, 2019). A high rate of intrinsic mutation of DENV results in genetic diversity, thereby promoting disease circulation and dynamics (Vasilakis & Weaver, 2008). Nonetheless, the role of molecular adaptation in the continuous turnover process of DENV clades remains to be understood. In this study, we assessed the adaptation advantage of stable substitutions within the envelope (E) protein of four DENV serotypes and offered insight that molecular adaptations impelled an emerging prevalence of the DENV2 Asian I genotype in Southeast Asia (Ty Hang et al, 2010). Furthermore, our study forecasted that a potential clade turnover mediated by the DENV2 Cosmopolitan genotype might lead to a potential forthcoming dengue epidemic.
Results and Discussion
The impact of stable substitutions in the DENV E protein on viral infectivity
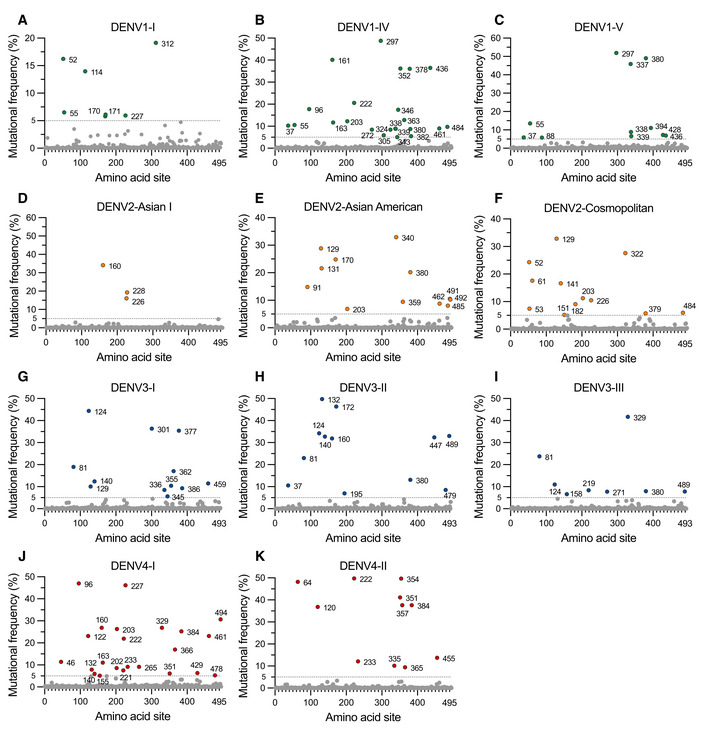
Given that beneficial mutations could be naturally selected in both hosts and mosquitoes, investigations into DENV variants and their dynamics with time might help us understand the evolution and infection trajectory of DENV. The envelope (E) protein is the major determinant of antigenicity and has multiple functions in DENV replication (Mukhopadhyay et al, 2005). We therefore collected 18,715 E protein sequences belonging to different genotypes of 4 DENV serotypes (Appendix Table S1). Subsequently, multiple sequence alignments were performed in each genotype category. Mutational frequency, which was defined as the percentage of the nonconserved residues in each site of the alignment, was calculated in the DENV sequences. The sites with a mutational frequency > 5% were defined as the positions of effective variants. Nonetheless, the assessment of mutational frequency was not performed for several DENV genotypes because of the limitation of sequence availability (Appendix Table S1). Of note, 41, 27, 32, and 34 sites met the criteria for effective variants in the 4 DENV serotypes, respectively (Fig EV1A–K). We next assessed the associations between the occurrence of mutations and the time of tracking. The five‐year occurrence frequency of substitutions was defined as the percentage of sequences carrying a given effective variant within the total sequences of a genotype in five successive years. Thus, we chose the effective variants, in which the occurrence frequency progressively increased from less than 20% (< 20%) before 1995 to more than 80% (> 80%) in 2016–2020, as the stable substitutions representing a dominant amino acid fixed or almost fixed in the DENV contemporary isolates (Fig 1A–D). There were 10 stable substitutions identified in 4 DENV serotypes, including 171T in DENV1‐I, 131Q, 170T, 340T and 380V in DENV2‐Asian American, 226K and 228E in DENV2‐Asian I, 301S and 377I in DENV3‐I, and 265A in DENV4‐I (Figs 1A–D and EV2A–J), implying the potential adaptation advantage of these substitutions in naturally circulating DENVs.
Figure EV1. Mutational frequency of the DENV1‐4 E proteins.
-
A–CThere were 7, 23, and 11 sites of effective variants in DENV1‐I (A), DENV1‐IV (B), and DENV1‐V (C), respectively.
-
D–FThere were 3, 12, and 12 sites of effective variants in DENV2‐Asian I (D), DENV2‐Asian American (E), and DENV2‐Cosmopolitan (F), respectively.
-
G–IThere were 12, 12, and 8 sites of effective variants in DENV3‐I (G), DENV3‐II (H), and DENV3‐III (I), respectively.
-
J, KThere were 23 and 11 sites of effective variants in DENV4‐I (J) and DENV4‐II (K).
Source data are available online for this figure.
Figure 1. Assessing the adaptation advantage of stable substitutions within the DENV E protein.
-
A–DHeat maps of the occurrence frequency of effective variants within each genotype in DENV1 (A), DENV2 (B), DENV3 (C), and DENV4 (D). Residues with the occurrence frequency < 20% before 1995 were shown in the heat maps. Red square frames represent 10 stable substitutions, including 171T in DENV1‐I (A), 131Q, 170T, 340T, and 380V in DENV2‐Asian American (B), 226K and 228E in DENV2‐Asian I (B), 301S and 377I in DENV3‐I (C), and 265A in DENV4‐I (D). Gray boxes represent the data that were not available.
-
E–HGrowth kinetics and plaque morphology of DENV2‐Asian I 16681 strain (16681, U87411) (E), DENV2‐Asian American DENV2/US/BID‐V1164/1986 strain (BID, EU482568) (F), DENV3‐I PF89/27643 strain (PF89, AY744677) (G), and DENV4‐I H241 strain (H241, AY947539) (H). Vero cells were infected with each mutant at a multiplicity of infection (MOI) of 0.01, and the cell supernatant was collected at Days 1, 3, 5, 7, and 9 for the plaque assay.
Figure EV2. Annual occurrence frequency of 10 stable substitutions in DENV1‐4 E proteins.
-
A–JThe annual occurrence frequency (AOF) of 10 stable substitutions was assessed, including 171T in DENV1‐I (A), 226K and 228E in DENV2‐Asian I (B and C), 131Q, 170T, 340T, and 380V in DENV2‐Asian American (D–G), 301S and 377I in DENV3‐I (H and I), and 265A in DENV4‐I (J).
Source data are available online for this figure.
We therefore assessed the roles of these stable substitutions in DENV pathogenicity and infectivity in native hosts. Several early DENV strains that were identified before 1995, including the DENV1‐I Hawaii strain (Hawaii, EU848545), DENV2‐Asian I 16681 strain (16681, U87411), DENV2‐Asian American BID‐V1164 strain (BID, EU482568), DENV3‐I PF89/27643 strain (PF89, AY744677), and DENV4‐I H241 strain (H241, AY947539), were selected as reference sequences to construct infectious clones. Nonetheless, the infectious clone of DENV1‐I Hawaii strain was not successfully constructed in E. coli. Subsequently, DENV mutants with a single stable substitution were generated based on their parental DENV strains, named 16681‐T226K, 16681‐G228E, BID‐L131Q, BID‐I170T, BID‐M340T, BID‐I380V, PF89‐L301S, PF89‐V377I, and H241‐T265A. These DENV mutants replicated well, albeit with variable kinetics, and formed plaques of similar sizes on Vero cells (Fig 1E–H). Both the infectivity and prevalence of these DENV mutants were assessed in A. aegypti mosquitoes. The DENV loads were significantly enhanced by the single substitutions of threonine (T) with lysine (K) at the 226th residue of the DENV2‐Asian I 16681 strain (16681‐T226K), isoleucine (I) with threonine (T) at the 170th residue of the DENV2‐Asian American BID strain (BID‐I170T), and leucine (L) with serine (S) at the 301st residue of the DENV3‐I PF89 strain (PF89‐L301S) compared with their parental DENVs (Fig 2A–D). Thus, the result indicates the adaptation advantage of these substitutions in DENV infection of mosquitoes.
Figure 2. The impact of stable substitutions in the DENV E protein on viral infectivity.
-
A–DOne‐week‐old mosquitoes were thoracically infected with DENV2‐Asian I (A), DENV2‐Asian American (B), DENV3‐I (C), and DENV4‐I (D) mutants, respectively. A. aegypti mosquitoes were infected intrathoracically with 10 p.f.u. of DENV. The DENV viral loads were assessed at 3 days postinfection via qRT–PCR and were normalized against A. aegypti actin.
-
E–HFour‐week‐old AG6 mice were infected with DENV (2,500 p.f.u.) by footpad inoculation. Blood samples were collected for DENV viremia detection from Day 1 to Day 7. The viral loads from AG6 mice infected with DENV2‐Asian I (E), DENV2‐Asian American (F), DENV3‐I (G), and DENV4‐I (H) were measured by qRT–PCR and were normalized against mouse GAPDH.
-
I, JInfection of DENV2‐Asian I mutants on hu‐moDC (I) and hu‐moMØ (J). The cells were infected with 0.01 MOI of the indicated viruses. The infected cells were detected at 48 h postinfection by qRT–PCR, and DENV viral loads were normalized against human GAPDH.
To address the role of these mutations in DENV pathogenicity in mammalian hosts, we infected the type I and II interferon receptors double‐deficient (ifnar −/− + ifngr −/−) C57BL/6 mouse (AG6 mouse) with either DENV mutants or their parental viruses (Fig 2E–H). The AG6 mouse is an animal model that is commonly used to study DENV infections (Orozco et al, 2012; Zhu et al, 2021; Zhang et al, 2022b). Intriguingly, 16681‐T226K presented slightly lower viremia than the 16681 strain; nonetheless, a glycine (G) to glutamic acid (E) mutation at the 228th residue (16681‐G228E), which is a neighboring substitution of the 226th site, resulted in significantly higher viremia (Fig 2E). We next assessed the impact of these two substitutions on DENV infectivity in primary human immune cells. Consistent with the results from AG6 mice, the 16681‐T226K mutant exhibited slightly lower infectivity than its parental strain (Fig 2I and J); nonetheless, 16681‐G228E significantly enhanced the infectivity of 16681 strain in both human primary monocyte‐derived dendritic cells (hu‐moDC) (Fig 2I) and macrophages (hu‐moMØ) (Fig 2J). Notably, the viremia of the infected mice was not enhanced by the other DENV mutants. Overall, both T226K and G228E substitutions regulate the infectivity and pathogenicity of DENV2‐Asian I in mammalian hosts or mosquitoes.
Compensatory substitutions increase the fitness of DENV2 in transmission cycle
The phylogenetic analysis indicated that the T226K mutation appeared in approximately 1995; nonetheless, the corresponding DENV2 clade with this mutation did not effectively disseminate in nature (Fig 3A). Of note, the G228E substitution sequentially occurred from 1999 based on the T226K mutation. The DENV2 mutants with double substitutions (T226K/G228E) spread rapidly, thus being dominant in isolates of the DENV2 Asian I genotype from 2000 (Fig 3A). To address the fitness of the neighboring mutations in native hosts, we generated a DENV mutant with T226K and G228E substitutions in an infectious clone of the DENV2 16681 strain, named 16681‐T226K/G228E. This DENV mutant grew well on Vero cells and formed plaques of similar size to the parental strain (Appendix Fig S1A and B). Since the cluster with single G228E substitution did not exist in native isolates of the DENV2 Asian I genotype (Fig 3A), we infected AG6 mice with the mutants existing in nature (16681‐T226K and 16681‐T226K/G228E) and the parental 16681 strain. DENV2 with T226K and G228E double substitutions developed higher viremia than DENV2 with a single T226K mutant and parental strain (Fig 3B). Consistent with the viremia results, DENV2‐T226K/G228E strain resulted in higher viral loads in animal tissues (Fig 3C–E) and higher mortality in AG6 mice (Fig 3F). We next assessed both the infectivity and prevalence of these DENV mutants in A. aegypti mosquitoes through blood feeding (Fig 3G). The DENV loads were higher in mosquitoes infected by the DENV2 mutants with a T226K substitution compared with that of the parental 16681 strain (Fig 3H), suggesting that the compensatory substitutions in the E protein rendered more evolutionary fitness advantage to the DENV2 Asian I genotype in native hosts.
Figure 3. The T226K/G228E compensatory substitutions promote the infectivity and prevalence of mammalian hosts and vectors.
-
AThe five‐year occurrence frequency of residues in 226th and 228th sites from < 1995 to 2016–2020.
-
B–FFour‐week‐old AG6 mice were infected with 2,500 p.f.u. of viruses by footpad inoculation. The viremia of infected AG6 mice from Day 1 to Day 7 was measured using qRT–PCR. Two days postinfection, infected mice were euthanized, and the viral loads in the bone marrow (C), spleen (D), and footpad (E) were measured by qRT–PCR. The survival rates of mice (F) were recorded daily.
-
G, HViruses (500 μl, 1 × 106 p.f.u. ml−1) were mixed with fresh human blood (500 μl) for membrane feeding of A. aegypti (G). Mosquito infectivity was determined by qRT–PCR at 8 days after the blood meal. The number at the top of each column represents the infected number/total number (H).
-
I–LMosquitoes were thoracically infected with 10 p.f.u. of viruses. Ten days postinfection, five infected mosquitoes were allowed to bite one 4‐week‐old AG6 mouse. From Day 1 to Day 8, infected mice were subjected to daily biting by naive A. aegypti, and blood samples were collected for DENV2 viremia detection by qRT–PCR (J). Mosquitoes were collected at 8 days after the blood meal for qRT–PCR to compare the infection rate of DENV2 (K, L). The number at the top of each column represents the infected number/total number (L).
Data information: In (B–E), n = 5 biological replicates. In (F), n = 10 biological replicates. In (H, L), one dot represents one mosquito, and the horizontal lines represent the median of the results. In (J), n = 4 biological replicates. In (B–E, J), the data are presented as means ± SEM. The data were analyzed statistically using the Mann–Whitney test (B–E, J). In (F), the data were analyzed statistically using the log‐rank (Mantel–Cox) test. In (H, K, L), the data were analyzed statistically using the Fisher's exact test. The P value represents a comparison between the mutants and the 16681 WT strain. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, n.s., not significant (P > 0.05).
Source data are available online for this figure.
We next investigated whether the compensatory substitutions might influence the DENV2 prevalence via a “mosquito‐AG6 mouse–mosquito” transmission cycle (Fig 3I). The mosquito‐bitten animals presented high viral loads for all the three DENV2 strains (Fig 3J), suggesting that these three DENV2 strains can be efficiently transmitted from infected mosquitoes to mammalian hosts. Over a complete mosquito–host–mosquito transmission cycle, the mosquito infection rate of 16681‐T226K was similar to that of the 16681 parental strain; nonetheless, 16681‐T226K/G228E showed a significantly higher prevalence of mosquito infection than the other two strains (Fig 3K and L), indicating that T226K/G228E compensatory substitutions in the E protein greatly increased the infectivity of DENV2, thereby promoting DENV dissemination through a “host–mosquito” transmission cycle in nature.
Our results suggest a potential epistatic relationship between the substitutions at the 226th and 228th sites. DENVs are mosquito‐borne RNA viruses prone to mutation. A viral population bottleneck in mosquitoes could enable DENVs vulnerable to genetic drift and fitness loss (Weaver et al, 2021). Our results indicate that the T226K substitution rendered DENV2 more transmissible by mosquitoes, but less infective in mammals. A potential explanation for this kind of compensatory substitutions is that the T226K mutation could emerge first due to a DENV2 population bottleneck in mosquitoes at the cost of host infectivity in mammalian hosts. However, the sequentially compensatory G228E substitution reversed the T226K‐mediated reduction of DENV2 infectivity in mammalian hosts, thereby enabling the DENV2 with the T226K and G228E double substitutions to rapidly disseminate in nature. Based on previous epidemiological surveillance, the DENV2 Asian I genotype was first introduced into southern Vietnam in the late 1990s. The DENV2 Asian American genotype, previously dominant in Southeast Asia, was completely replaced by DENV2 Asian I genotype from 2003 to 2007, when there was a much higher dengue incidence that was mostly associated with DENV2 (Ty Hang et al, 2010). In this study, we identified 2 compensatory substitutions emerging throughout the E protein of the DENV2 Asian I genotype isolated in Southeast Asia in approximately 2000, which remain stable in the contemporary isolates. Altogether, these compensatory substitutions in the E protein led to the typical replacement of DENV2 genotypes, which was attributed to an evolutionary fitness adaptation in their native hosts.
Polarity changes in the substitutions determine DENV2 infectivity in both mammalian hosts and mosquitoes
We next aimed to address the mechanism by which the compensatory substitutions in the E protein promote DENV2 infection. Several mammalian C‐type lectins are employed as receptors or attachment factors to facilitate DENV invasion (Tassaneetrithep et al, 2003; Chen et al, 2008). Dendritic cell‐specific intercellular adhesion molecule‐3‐grabbing nonintegrin (DC‐SIGN) is a C‐type lectin that binds to DENVs via high‐mannose glycans on the E protein, and it is an essential attachment factor for DENV infection of dendritic cells, which are the major target cells of DENV in humans (Navarro‐Sanchez et al, 2003; Tassaneetrithep et al, 2003). A previous in silico study revealed that both the 226th and 228th residues are on the interface of DENV E protein and the carbohydrate‐recognition domain (CRD) of DC‐SIGN (Shah et al, 2013). The location of these residues was confirmed by the protein–protein docking protocol in the Rosetta software package and PISA (protein interfaces, surfaces, and assemblies service) (Fig 4A), suggesting that substitutions in both residue sites may regulate DENV infection by tuning the interaction between the DENV E protein and human DC‐SIGN. In the 293T cells ectopically expressing human DC‐SIGN (Appendix Fig S2A), DENV2 mutants with a G228E substitution (16681‐G228E and 16681‐T226K/G228E) showed much higher infectivity; nonetheless, the 16681‐T226K mutation impaired infection compared with that of the parental strain (Fig 4B). The phenotype was also reproduced in hu‐moDCs with abundant DC‐SIGN on the cell surface (Figdor et al, 2002; Fig 4C). Since DC‐SIGN interacts with a structural interface formed by a homodimer of DENV E proteins (Pokidysheva et al, 2006), we therefore exploited purified DENV2 virions to assess the role of these substitutions in DENV E and DC‐SIGN interactions. The T226K substitution reduced, but the G228E substitution oppositely increased, the binding affinity between DC‐SIGN and purified DENV2 virions by a surface plasmon resonance (SPR) assay (Fig 4D). Nonetheless, the DENV2 virions with T226K/G228E compensatory substitutions showed a higher binding affinity for human DC‐SIGN than did the DENV2 parental virions (Fig 4D).
Figure 4. The T226K/G228E compensatory substitutions promote DENV2 infectivity by enhancing the affinity between DENV and C‐type lectins (CTLs).
- The structure and interface of the DENV E and DC‐SIGN CRD (PDB ID 1SL4) complex. The residues, whose distances to 226T or 228G were less than 10 Å, were selected for analysis. In the interface, the interactions of 309R/312R and 226T/228G were shown in the complex structure. The positively‐charged residues in CRD were labeled by red color.
- The 293T cells were transfected with a pcDNA3.1 recombinant plasmid with a human DC‐SIGN gene. The cells transfected by an empty pcDNA3.1 plasmid served as negative controls. Thirty‐six hours after transfection, the cells were infected with DENV2 (0.01 MOI). The DENV2 viral loads were assessed at 48 h postinfection via qRT–PCR and were normalized against human GAPDH.
- hu‐moDCs were infected with 0.01 MOI of DENV2. The infected cells were detected at 48 h postinfection by qRT–PCR.
- The binding affinity between DC‐SIGN and purified DENV2 virions measured by SPR.
- The binding affinity between DC‐SIGN (R309A/R312A) and purified DENV2 virions measured by SPR.
- The structure and interface of the DENV2 E and mosGCTL‐AAEL011408 CRD complex. The residues, whose distances to 226T were less than 10 Å, were selected for analysis. In the interface, the interaction between 66E and 226T was shown in the structure. The negatively‐charged residue in CRD was labeled by blue color.
- The binding affinity between mosGCTL‐AAEL011408 and purified DENV2 virions measured by SPR.
Data information: In (B), n = 6 biological replicates. In (C), n = 5 biological replicates. In (B, C), the data are presented as means ± SEM and were analyzed statistically using an unpaired t‐test. The P value represents a comparison between the mutants and the 16681 WT strain. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, n.s., not significant (P > 0.05).
Source data are available online for this figure.
The substitution of threonine (T) with lysine (K) introduces a positive charge at the 226th residue; nonetheless, the mutation from glycine (G) to glutamic acid (E) oppositely results in a negative charge at the 228th residue. Given that the amino acids within the interfacing region of human DC‐SIGN include multiple residues with a positive charge (Fig 4A), we concluded that the polarity changes of the 2 residues play an essential role in tuning the interactions between DENV E protein and human DC‐SIGN. To address this question, we generated a human DC‐SIGN mutant, in which two arginine (R, positive charge) in the DC‐SIGN‐DENV2 E interfacing region were replaced by alanine (A, neutral polarity; R309A and R312A; Fig 4A). Notably, removal of polarity charges in the interfacing region of human DC‐SIGN offset the effect of either T226K or G228E mutations on DENV2 E‐DC‐SIGN binding (Fig 4E), indicating the essential role of residue polarity in the interaction between human DC‐SIGN and DENV2 E protein.
Mosquito C‐type lectins (mosGCTLs) also play crucial roles in flavivirus infection by facilitating viral entry (Cheng et al, 2010). Multiple A. aegypti mosGCTLs have been identified as key susceptibility factors to facilitate DENV infection by directly interacting with DENV virions, indicating that C‐type lectins act as ligands to promote the DENV infection of mosquitoes (Liu et al, 2014). Nonetheless, the sequences of some mosGCTLs (the original VectorBase number: AAEL000535, AAEL010992, AAEL011607, and AAEL017265) were not available in the current Genome Version (GCA_002204515.1), which might be attributed to the database update in the VectorBase. We next ectopically expressed and then purified 5 available mosquito C‐type lectins in Drosophila S2 cells (Appendix Fig S2B). Of note, a C‐type lectin encoded by AAEL011408, but not the other C‐type lectins, strongly interacted with purified virions of DENV2 16681 strain by an SPR assay (Fig EV3A–E). We next modeled the structure of CRD of A. aegypti mosGCTL‐AAEL011408 by trRosetta (transform‐restrained Rosetta) (Yang et al, 2020; Du et al, 2021; Su et al, 2021). The interfacing residues between DENV2 E protein and CRD of mosGCTL‐AAEL011408 have been predicted by Rosetta and PISA. Intriguingly, the 226th residue in DENV2 E protein, rather than the 228th residue, was located on the binding interface (Fig 4F), suggesting that the 228th substitution is dispensable for the interactions between the DENV2 E protein and the C‐type lectin. Consistent with the aforementioned result, the G228E substitution did not influence the infectivity of the DENV2 16681 strain in A. aegypti mosquitoes (Fig 2A). We next assessed the polarity of interfacing residues in both DENV2 E and mosGCTL‐AAEL011408. A residue (66E) of this mosGCTL presents a negative charge on the binding interface (Fig 4F). The 226th residue in DENV2 E was predicted to neighbor the negatively‐charged residue (Fig 4F). We therefore speculated that the substitution of threonine (T, neutral polarity) by lysine (K, positive charge) at the 226th residue may enhance DENV2 infectivity in mosquitoes by strengthening the binding affinity between the DENV2 E protein and mosquito C‐type lectins. The T226K, but not the G228E substitution, strengthened the binding affinity between mosGCTL‐AAEL011408 and purified DENV2 virions by an SPR assay (Fig 4G). The DENV2 16681 virions with T226K/G228E compensatory substitutions presented a higher binding affinity for mosGCTL‐AAEL011408 than did the DENV2 parental virions, indicating that the mosGCTL‐DENV E interaction plays an important role in DENV infection of mosquitoes. Overall, our results fortified the essential role of C‐type lectins in DENV pathogenesis and infection in its native hosts, indicating that mutation‐mediated changes in binding affinity for these C‐type lectins may largely influence DENV pathogenesis in humans and transmission by mosquitoes.
Figure EV3. The binding affinity between 16681 virions and mosquito C‐type lectins.
-
A–EThe binding affinity between purified 16681 virions and mosquito C‐type lectins measured by an SPR assay.
Source data are available online for this figure.
A G228E substitution may result in a forthcoming dengue epidemic of the DENV2 cosmopolitan genotype
The compensatory substitutions in the E protein rendered much higher transmissibility to the DENV2 Asian I genotype, thereby resulting in the turnover of DENV2 lineages by the Asian I genotype to cause a higher dengue incidence in Southeast Asia (Ty Hang et al, 2010). We compared the residues in these 2 sites in the E proteins of the other DENV serotypes. Intriguingly, glutamic acid was stably located at the 229th (229E) residue in DENV1, the 225th (225E) residue in DENV3, and the 228th (228E) residue in DENV4 (Fig EV4A). However, threonine was present at the 226th (226T) residue (224th residue in DENV3) of all isolates of DENV1, DENV3, and DENV4 (Fig EV4A), suggesting that the substitutions of threonine (T) by lysine (K) at the 226th site might offer additional adaptation advantages to the other 3 DENV serotypes in mosquitoes.
Figure EV4. T226K and G228E substitutions in DENV1‐4 E proteins.
- Sequence alignment of DENV E proteins. The consensus sequences of DENV1‐4 genotypes were obtained from Jalview and were aligned by MUSCLE.
- The 5‐year occurrence frequency of 226K in DENV2 Asian I, Asian American, and Cosmopolitan genotypes.
Source data are available online for this figure.
We next assessed the residue presence of these 2 sites in other DENV2 genotypes. Both threonine at the 226th (226T) residue and glycine at the 228th (228G) residue were the most frequently appearing residues in the other DENV2 genotypes (Fig EV4A); nonetheless, the single T226K substitution sporadically appeared in the DENV2 Cosmopolitan genotype according to the surveillance of viral sequences in the epidemic areas (Fig EV4B). Given that the G228E substitution compensatorily occurred with the T226K mutation in the DENV2 Asian I genotype, we speculated that the G228E substitution at the 228th residue potentially occurred in the DENV2 Cosmopolitan genotype under the evolutionary pressure and fitness adaptation and thus might result in potential dengue epidemics caused by lineage replacement. To address this possibility, we modeled the annual occurrence frequency (AOF) of the T226K and G228E substitutions in the DENV2 Cosmopolitan genotype through a Gaussian process regression approach (GPR‐1), which is flexible and precise for modeling time‐series data with limited sample sizes (Kong et al, 2018; Cheng et al, 2019; Fig 5A). In addition to the GPR‐1 model, we employed two parallel approaches, the numerical kinetics and support vector machine regression (SVR) models, as independent verification of the predictions (Cheong et al, 2022; Thornburg et al, 2022; Fig EV5A and B). The predicted AOF of these 2 substitutions fit well with the realistic data of the DENV2 Asian I genotype (Figs 5B and EV5C, and Appendix Table S2), indicating a reliable performance of these prediction models. Subsequently, we predicted the AOF of T226K and G228E in the DENV2 Cosmopolitan genotype in 2019–2040 using these mathematical models. Of note, the G228E substitution might occur within the DENV2 Cosmopolitan genotype from approximately 2025. The AOF of T226K/G228E compensatory substitutions in DENV2 Cosmopolitan genotype might significantly increase from 2025 to 2030 (Figs 5C and EV5D). We next predicted whether the potential prevalence of the DENV2 Cosmopolitan genotype might lead to future dengue epidemics. The prevalence of a DENV2 genotype was defined as the proportion of isolated sequences of a genotype in the total DENV2 sequences. A Gaussian process regression (GPR‐2) was applied to model the prevalence of a DENV2 genotype (Fig 5A). The prevalence predicted by the GPR‐2 model adequately represented the historical occurrence of the DENV2 Asian I genotype (Figs 5D and EV5E). We therefore predicted the annual prevalence of the DENV2 Cosmopolitan genotype from 2025 to 2040 (Fig 5E). One peak of the dengue prevalence caused by the DENV2 Cosmopolitan genotype was forecasted by GPR‐2 in 2035 (Fig 5E). We therefore raised the possibility that the potential epistatic adaptations might impel the emerging prevalence of the DENV2 Cosmopolitan genotype in the future.
Figure 5. Prediction of the annual occurrence frequency of the compensatory substitutions and the prevalence of the DENV2 Cosmopolitan genotype.
- The Gaussian process regression (GPR‐1) for modeling the annual occurrence frequency (AOF) of the T226K/G228E substitutions and the Gaussian process regression (GPR‐2) for determining the correlations between the AOF of T226K/G228E substitutions and the prevalence of a DENV2 genotype. The t denotes the number of years from 1995 (DENV2 Asian I genotype) or from 2019 (DENV2 Cosmopolitan genotype).
- The AOF prediction of the T226K substitution (left panel) and the G228E substitution (right panel) in the DENV2 Asian I genotype.
- The AOF prediction of the T226K substitution (left panel) and the G228E substitution (right panel) in the DENV2 Cosmopolitan genotype.
- The performance of the GPR‐2 in modeling the prevalence of the DENV2 Asian I genotype.
- The prevalence of the DENV2 Cosmopolitan genotype predicted by the GPR‐2 model (orange dashed line + squares). The orange shadow denotes the predicted prevalence range of the DENV2 Cosmopolitan genotype, with the AOF of T226K/G228E substitutions predicted by the numerical kinetics, GPR‐1, and support vector machine (SVR) models serving as the GPR‐2 input data.
Source data are available online for this figure.
Figure EV5. Modeling of the annual occurrence frequency of the T226K/G228E substitutions in DENV2 genotypes and modeling results of the Gaussian process regression (GPR‐2).
- The numerical kinetics modeling process of the annual occurrence frequency of the T226K/G228E substitutions in DENV2 genotypes. The dark gray and orange circles denote the AOF of the T226K and G228E substitutions, respectively. The gray and orange circles denote the AOF of T226 and G228 (100% − (AOF of T226K) and 100% − (AOF of G228E)), respectively. The upper and lower dashed boxes denote the AOF of T226K and G228E substitutions and the AOF of 226T and 228G in Year (t) and Year (t + 1), starting from 1995.
- The support vector machine regression (SVR) modeling process of the annual occurrence frequency of T226K/G228E substitutions in DENV2 genotypes. The gray and orange rectangle boxes denote the input data and output data of the SVR models.
- The AOF prediction of the T226K substitution (left panel) and G228E substitution (right panel) in the DENV2 Asian I genotype.
- The AOF prediction of the T226K substitution (left panel) and G228E substitution (right panel) in the DENV2 Cosmopolitan genotype.
- The regression results for GPR‐2.
- The case number of the DENV2 Asian I genotype from 1995 to 2019.
Source data are available online for this figure.
Dengue exhibits complex oscillations of prevalence, with multiple viral clades circulating in its epidemic areas. The DENV lineages frequently experienced a process of arising and extinction, which was usually associated with a round of expansion of dengue epidemics (Kyle & Harris, 2008). For example, DENV3 strains circulating in Thailand prior to 1992 disappeared from that location and have been replaced by 2 other local lineages (Wittke et al, 2002). Two DENV1 clades were introduced into Myanmar that led to the extinction of an original local DENV1 clade between 1998 and 2000 (Thu et al, 2004, 2005). This replacement preceded the largest outbreak of dengue recorded in Myanmar, in which > 95% of viral isolates recovered from patients were new emerging DENV1 clades (Thu et al, 2004). In addition, the introduction of a genetically distinct Asian American DENV2 strain into the Americas resulted in the replacement, and possibly extinction, of the local American DENV2 lineage (Rico‐Hesse et al, 1997). The replacement events within DENV2 lineages in the Americas were associated with increases in disease incidence and severity (Rico‐Hesse et al, 1997). In this study, we identified two stable mutations that functioned compensatorily with a major contribution to the fitness of the DENV2 Asian I genotype in both hosts and mosquitoes. This fitness adaptation may impel the Asian I lineage viruses to entirely displace the previously dominant Asian American lineage viruses and thus may be an explanation for a large outbreak of DENV2 in Southeast Asia from 2002 to 2006 (Huy et al, 2010; Ty Hang et al, 2010; Limkittikul et al, 2014). Given a much higher fitness of DENV2 clades with these substitutions, the Asian I lineage has been rapidly disseminating from Southeast Asia to China and Australia (Huang et al, 2012; Warrilow et al, 2012; Guo et al, 2013), thus offering insights into an evolutionary force resulting in dramatic expansion of dengue epidemics in nature. Overall, investigation into the potential substitutions that regulate viral fitness in hosts may offer insights into dengue persistent epidemics and thus is critical for early warning of potential epidemics in the future.
Materials and Methods
Acquisition of human blood
Human blood used for mosquito feeding and peripheral blood mononuclear cell (PBMC) isolation were provided and collected at Tsinghua University Hospital by healthy donors who had provided signed informed consent. The donation of human blood samples was approved by the ethics committee at Tsinghua University. Different experiments might use blood samples from different donors.
Mice
C57BL/6 mice deficient in type I and II interferon receptors (AG6 mice) used as the mammalian model of DENV infection were donated by the Institute Pasteur of Shanghai at the Chinese Academy of Sciences and were maintained at Tsinghua University. Four‐week‐old male mice were used for the animal experiments. The mice were bred and maintained in specific pathogen‐free (SPF) laboratory animal rooms at Tsinghua University. The laboratory animal facility has been accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International. All animal operations were approved by the Institutional Animal Care and Use Committee of Tsinghua University and performed in accordance with their recommended practices.
Mosquitoes
Aedes aegypti (Rockefeller strain) were reared in an incubator (Precision 3758CN, Thermo Scientific) at 28°C and 80% humidity according to published standard rearing procedures. Female mosquitoes aged 8–12 days were subjected for further investigation.
Viruses
Wild‐type strains and mutant viruses of DENV were produced from infectious clones and passaged in Vero cells in virus production‐serum free medium (VP‐SFM; 11681‐020, Gibco). Viruses were titrated using a plaque formation assay (Cheng et al, 2010).
Cells
Vero cells and 293T cells were cultured at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) (11965‐092, Gibco) supplemented with 10% heat‐inactivated fetal bovine serum (16000‐044, Gibco) and 1% antibiotic‐antimycotic (15240‐062, Invitrogen). The Drosophila melanogaster S2 cell line was cultured in Schneider's Drosophila medium (21720‐024, Gibco) supplemented with 10% heat‐inactivated fetal bovine serum and 1% antibiotic–antimycotic. All cell lines were authenticated by the American Type Culture Collection (ATCC) and did not have mycoplasma contamination.
Plaque formation assay
The titers and kinetic features of DENV mutants were determined by a plaque formation assay. Culture supernatants from infected cells were collected at the indicated time points and filtered through a 0.22‐μm filter (SLGP033RB, Millipore) to separate the supernatant. The presence of infectious viral particles in the supernatant was determined using a plaque assay (Cheng et al, 2010). The detection limit in the plaque assay was 10 p.f.u. ml−1 in the cell supernatant.
Sequence data and alignments
A total of 18,715 protein sequences of the DENV1‐4 E (Appendix Table S1) were downloaded from the NIAID Virus Pathogen Database and Analysis Resource (ViPR) (Pickett et al, 2012), which can be accessed at https://www.viprbrc.org/. The sequences were labeled according to the GenBank accession number followed by the year of isolation and isolation country and were categorized by virus genotype referring to ViPR. Sequences within each genotype were subsequently aligned with MUSCLE in the Molecular Evolutionary Genetics Analysis (MEGA) X program. The mutational frequency and occurrence frequency were analyzed in Jalview (http://www.jalview.org/).
Construction of DENV infectious clones
Full‐length genome sequences of the DENV2‐Asian I 16681 strain (16681, U87411), DENV2‐Asian American BID‐V1164 strain (BID, EU482568), DENV3‐I PF89/27643 strain (PF89, AY744677), and DENV4‐I H241 strain (H241, AY947539) were downloaded from the NCBI database. Each DENV genome was divided into four cDNA fragments and separately synthesized by the Tsingke company (Beijing, China). Viral cDNA fragments were assembled into pBR322 (DENV2 16681), pACYC177 (DENV2 BID‐V1164), or pCC1 (DENV4 H241) using the NEBuilder HiFi DNA Assembly Cloning Kit (E5520S, NEB) to achieve the full‐length cDNA infectious clones pBR322‐DENV2‐16681, pACYC‐DENV2‐BID, and pCC1‐DENV4‐H241. Due to a failure to obtain a stable full‐length viral cDNA clone, DENV3 PF89/27643 genomic cDNA was split into two subclones pCC1‐DENV3‐PF89‐F1 and pCC1‐DENV3‐PF89‐F2. Several silent mutations were introduced into DENV3 subclones to generate a unique BsiWI restriction site that was used to assemble full‐length cDNA. A standard mutagenesis method was used to engineer mutations of DENV E protein into the infectious clones as described previously (Zhang et al, 2019). The stability and efficacy of a viral cDNA construction in E. coli are largely dependent on the specific viral genome sequences. As mentioned above, the genome of DENV1‐I Hawaii strain (Hawaii, EU848545) was divided into four cDNA fragments for separate synthesis. However, the infectious clone was not successfully constructed in E. coli.
Recombinant virus recovery
For preparing DENV2 and DENV4 full‐length DNA fragments, the infectious cDNA clone plasmids pBR322‐DENV2‐16681, pACYC‐DENV2‐BID, and pCC1‐DENV4‐H241 were linearized with the appropriate restriction endonucleases. Specifically, pACYC‐DENV2‐BID, and pCC1‐DENV4‐H241 were digested with PacI, and the pBR322‐DENV2‐16681 plasmid was digested with XbaI. For preparing the DENV3‐PF89 full‐length DNA fragment, the viral genome assembly was performed by following a previously described protocol (Zhang et al, 2021). Briefly, pCC1‐DENV3‐PF89‐F1 and pCC1‐DENV3‐PF89‐F2 plasmids were digested with NotI/BsiWI and BsiWI/PacI, respectively. DNA fragments containing the viral genome were separated and purified using the agarose gel extraction method, after which two recovered DNA fragments were ligated with T4 DNA ligase at 16°C overnight to produce the full‐length DNA fragment of DENV3‐PF89. For generating viral RNA transcripts, the full‐length viral DNA fragments prepared above were purified through phenol–chloroform extraction and ethanol precipitation and in vitro transcribed into viral RNAs using the mMESSAGE mMACHINE T7 transcription kit (AM1344, Ambion) according to the manufacturer's instructions. For producing recombinant virus, 3 × 106 Vero cells were seeded into a T‐25 flask and cultured in complete DMEM one day prior to transfection. Subsequently, 5 μg of viral RNA was transfected into cells using the TransIT®‐mRNA Transfection kit (MIR2250, Mirus). Transfected cells were incubated at 37°C with CO2 until the appearance of the cytopathic effect.
Purification of DENV virions
Detailed procedures for purification of DENV virions have been described previously (Liu et al, 2016). In brief, the supernatant of DENV was collected and filtered through a 0.22‐μm filter (SLGP033RB, Millipore) from infected Vero cells at approximately 6 days postinfection. The supernatant was carefully transferred to a clean centrifuge tube and centrifuged at 30,000 g at 4°C for 3 h to pellet the virions. The pellet was washed twice carefully, and VP‐SFM was used to suspend the precipitate. The insoluble components were removed by an additional centrifugation step at 12,000 g at 4°C for 2 min. The virions were aliquoted and stored at −80°C.
Membrane blood feeding
Heparin‐coated tubes (367884, BD Vacutainer) were used to collect fresh human blood from healthy donors, and the operation was performed at the Hospital of Tsinghua University. The blood was centrifuged at 1,000 g for 10 min to separate the serum and blood cells. The serum was incubated at 56°C for 60 min. The blood cells were washed three times using phosphate‐buffered saline (PBS) to remove heparin. The blood cells were resuspended with heat‐inactivated serum, and then the treated blood was mixed with viruses in a feeder of a membrane‐feeding system (Hemotek Limited) at different dilutions based on the experimental design. The female mosquitoes were allowed to feed on the feeder for 30 min in the dark. Engorged female mosquitoes were transferred into new containers and maintained for an additional 8 days. The mosquitoes were subsequently sacrificed for further analysis.
Intrathoracic inoculation of DENV in mosquitoes
Details of the mosquito microinjection procedure have been described previously (Cheng et al, 2010). The mosquitoes were anesthetized on ice for approximately 5 min, and female mosquitoes were selected for microinjection. DENV (10 p.f.u./300 nl) was intrathoracically microinjected into each mosquito. Three days postinfection, mosquitoes were sacrificed, and the viral loads were determined by qRT–PCR.
Mosquito‐AG6 mouse–mosquito transmission cycle
Female A. aegypti mosquitoes were thoracically infected with 10 p.f.u. of DENV. The mosquitoes were separated into netting‐covered cups and starved for 24 h before feeding. Ten days postinfection, each five infected mosquitoes were allowed to bite one 4‐week‐old AG6 mouse. From Day 1 to Day 8, infected mice were subjected to daily biting by naive A. aegypti, and blood samples were collected for DENV viraemia detection by qRT–PCR. The infected mice were anesthetized and placed on top of the cups, and the female mosquitoes were allowed to feed on the mice for 30 min in the dark. Then, the mosquitoes were anesthetized at 4°C, and the engorged mosquitoes were picked out, transferred to new containers and maintained under standard conditions for an additional 8 days. The mosquitoes were subsequently sacrificed for further analysis.
Hu‐moDC and hu‐moMØ generation
Human peripheral blood mononuclear cells (PBMCs) were separated using the Ficoll‐Paque density gradient assay (17144002, GE Healthcare) from donor's fresh blood. Then, PBMCs were suspended in RPMI 1640 medium (22400089, Gibco), distributed into 48‐well plates and incubated for 2 h at 37°C in 5% CO2. For hu‐moDC generation, monocytes were cultured in RPMI 1640 medium supplemented with 10% heat‐inactivated fetal bovine serum, 100 ng/ml granulocyte–macrophage colony‐stimulating factor (GM‐CSF) (34‐8339‐82, eBioscience) and 100 ng/ml interleukin‐4 (IL‐4) (34‐8049‐82, eBioscience) for 7 days. For hu‐moMØ generation, monocytes were cultured in RPMI 1640 medium supplemented with 10% heat‐inactivated fetal bovine serum and 100 ng/ml GM‐CSF for 7 days.
Mosquito C‐type lectins (mosGCTLs) purification in the Drosophila expression system
The plasmids of mosquito C‐type lectin genes in the pMT/Bip‐V5/His A vector for expression in Drosophila S2 cells were maintained in our laboratory. Detailed procedures for protein purification in the Drosophila expression system have been described previously (Liu et al, 2016). Briefly, stable S2 cells expressing C‐type lectins were amplified in 175 cm2 flasks containing Express Five serum‐free medium (10486‐025, Gibco) for protein expression. The cells were induced with 500 μM copper sulfate (CuSO4) for 4 days for protein expression. Subsequently, the supernatant was filtered and the proteins were purified using a TALON Purification Kit (635515, Clontech). The concentration of proteins was measured using protein assay dye (500‐0006, Bio–Rad), and the purity of proteins was verified by SDS‐polyacrylamide gel electrophoresis (PAGE).
Expression and purification of recombinant proteins
The full‐length human DC‐SIGN gene was amplified from the complementary DNA of hu‐moDC, cloned into a pcDNA3.1‐flag expression vector and subsequently transfected using Lipo3000 DNA transfection reagent (L3000015, Invitrogen) for ectopic expression in 293T cells. The genes encoding the extracellular domain of human DC‐SIGN protein and its mutant were cloned into a pET‐28a (+) expression vector (69864, Millipore). The cloning primers are presented in Appendix Table S3. Recombinant DC‐SIGN proteins were expressed in the Escherichia coli BL21 Rosetta strain in an insoluble form in inclusion bodies. The proteins were dissolved in 8 M urea (100 mM Tris–HCl, 50 mM glycine, 8 M urea). Then the purified DC‐SIGN proteins were dialyzed overnight in renaturation buffer (1 mM glutathione, 0.25 mM glutathione disulfide, 50 mM Tris–HCl, 50 mM NaCl, 1% (v/v) glycerol, pH = 8.0). The buffer of DC‐SIGN protein solution was exchanged with PBS and concentrated via a 10‐kD ultrafiltration tube (UFC9010, Millipore). The concentration of refolded DC‐SIGN proteins was determined with protein assay dye (500‐0006, Bio–Rad). The quality of the proteins was analyzed by SDS–PAGE.
Western blotting analysis and antibodies
Cells were washed with PBS and harvested in lysis buffer supplemented with a protease inhibitor cocktail (1862209, Thermo Scientific). The lysates were centrifuged at 12,000 g at 4°C for 10 min to remove debris. Then, the supernatant was denatured in protein loading buffer at 100°C for 10 min. The samples were separated by 10% SDS–PAGE and electrophoretically transferred to polyvinylidene fluoride membranes, which were then blocked and incubated with primary antibody. The following antibodies were used in the experiments: anti‐Flag (8146, 1:2,000 dilution) antibody and anti‐V5 (13202, 1:2,000 dilution) antibody were obtained from Cell Signaling Technology (CST).
Protein–protein docking
The high‐resolution X‐ray 3D crystal structure of the DC‐SIGN CRD (PDB ID 1SL4) was retrieved from the PDB database. The amino acid sequence of CRD of mosGCTL‐AAEL011408 was predicted through SMART (a Simple Modular Architecture Research Tool; Letunic & Bork, 2018; Letunic et al, 2021), and the structure of CRD was predicted by the trRosetta (transform‐restrained Rosetta; Yang et al, 2020; Du et al, 2021; Su et al, 2021). The homology model of the DENV2 16681 E protein was developed using the SWISS‐MODEL Web Server (Bordoli et al, 2009). The DENV2 16681 soluble E protein (aa 1–393) was generated with the mature DENV2 structure (PDB ID 6ZQU) as a template. The protein–protein docking protocol of the Rosetta software package was performed to predict and assess the interactions between DENV2 E and C‐type lectins. Local docking (docking perturbation parameter: 3 Å translation and 8° rotation) was performed to generate 500 candidate structures. The structure models with the lowest total_score were selected for analysis of interface residues. PISA (protein interfaces, surfaces and assemblies service) at the European Bioinformatics Institute (Krissinel & Henrick, 2007) was used to calculate the interface residues of the docked models. All the structures were generated using PyMOL.
Surface plasma resonance (SPR) analysis
The Biacore 8K system was used to perform SPR experiments at 25°C with a flow rate of 30 μl/min in buffer A. The virions were diluted with Hank's balanced salt solution (HBST). For the SPR measurement at pH 4.0, the purified DENV2 virions were coupled to a CM5 sensor chip (BR100530, GE Healthcare) into different channels with more than 1,000 response units through HBST. Then, the proteins were diluted with PBST (101 mM Na2HPO4, 18 mM KH2PO4, 27 mM KCl, 1.37 M NaCl, and 0.05% Tween‐20, pH 7.4), and the serially diluted proteins flowed over the chip surface in PBST buffer. The proteins were regenerated with 2 M MgCl2. Equilibrium dissociation constants (KD) of each pair of interactions were calculated using Biacore Insight Evaluation software (GE Healthcare).
RNA quantification by qRT–PCR
Total RNA was extracted from homogenized mosquitoes, cell lysate and supernatant samples or homogenized mouse tissue samples using the Multisource RNA miniprep kit (AP‐MN‐MS‐RNA‐250, Axygen) and was reverse‐transcribed into cDNA using an iScript cDNA synthesis kit (170‐8890, Bio–Rad). TaqMan qPCR or SYBR Green qPCR amplification kits were used to perform qRT–PCR on the Bio–Rad CFX‐96 Touch Real‐Time Detection System. Gene quantities were normalized against A. aegypti actin (AAEL011197) or glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) for human or mouse samples. The primer sequences are shown in Appendix Table S3.
Modeling of the annual occurrence frequency (AOF) of T226K/G228E substitutions
We modeled the time‐series data of the AOF of T226K/G228E substitutions in the DENV2 Asian I genotype from 1995 to 2019 through the numerical kinetics model, Gaussian process regression (GPR‐1), and support vector machine regression (SVR) approaches.
Numerical kinetics model for the AOF of T226K/G228E substitutions
The numerical kinetics modeling method has been used to model various kinetics systems (Onischenko et al, 2020; Thornburg et al, 2022). We deduced the numerical kinetics models for the AOF of T226K/G228E substitutions (Equations 1 and 2). The deducing procedure is shown in Fig EV5A and Appendix Supplementary Text S1.
| (1) |
| (2) |
where t is the number of years from 1995, ka226 (year−1−kt226) is the rate constant of T226K substitution, kb226 (year−1) is the rate constant of the substitution of lysine with another amino acid at the 226th residue, ka228 (year−1−kt228) is the rate constant of G228E substitution, kb228 (year−1) is the rate constant of the substitution of glutamic acid with another amino acid at the 228th residue, and kt226 and kt228 are the exaptational constants of t. Equations 1 and 2 were solved by using the Runge–Kutta 4th‐order method in Excel (the step was set to 0.1 year). The time‐series data of the AOF of T226K/G228E substitutions in the DENV2 Asian I genotype (Appendix Table S2) were fitted with Equations 1 and 2 by using the nonlinear least square method in Solver in Excel.
Gaussian process regression for the AOF of T226K/G228E substitutions (GPR‐1)
The Gaussian Process Regression (GPR) method is effective in modeling both small sample and complex nonlinear regression systems (Cheng et al, 2019; Mena et al, 2021). We employed a GPR‐1 model to forecast the AOF of T226K/G228E substitutions in DENV2 Asian I (Fig 5A). In modeling the AOF of the T226K substitution, the input and output data were defined as x = {t + 1, the AOF of T226K (t)}T and y = {the AOF of T226K (t + 1)}T. The input and output data of GPR‐1 for the G228E substitution were defined as x = {t + 1, the AOF of T226K (t) and G228E (t)}T and y = {the AOF of G228E (t + 1)}T (Fig 5A and Appendix Table S2). y of a function f(x) can be described by yi = f(xi) + ε, i = 1, 2, 3…25, where ε is the Gaussian independent identically distributed error and f(x) is distributed as a Gaussian process with mean function m(x) and covariance function K(x, x′), which is created by the kernel function. Thus the Gaussian process is expressed as f(x) ~ Gauss process m(x), K(x, x′).
Support vector machine regression (SVR)
The SVR is a nonparametric machine learning tool that is widely used in small sample regression systems (Zhou & Jetter, 2006; Cheong et al, 2022; Zhang et al, 2022a). The input data x and output data y of SVR were the same as those of GPR‐1 (Fig EV5B and Appendix Table S2). The SVR approximates x and f(x) with a function that is formulated in the high‐dimensional feature space: , where f(x) is the response function; ω and b are the weight vector and the bias parameter, respectively; φ(x) is the mapping function that maps x to feature space; α and α* are the Lagrange multiplier; and k(x, xk) is the kernel function. GPR‐1 and SVR were developed using the Regression Learner application in MATLAB R2020b (MathWorks, USA). The Bayesian Optimization (BO) method was used to search for the best hyperparameters (Appendix Table S4) of the models. Using the BO method, we tried different hyperparameter values to minimize the model mean squared error (MSE) between f(x) and y . The optimal hyperparameter values of the GPR‐1 were: exponential kernel function, sigma = 0.004, kernel scale = 0.39 (AOF‐T226K substitution) and exponential kernel function, sigma = 1.17, kernel scale = 0.85 (AOF‐G228E substitution). The optimal hyperparameter values of the SVR model were summarized in Fig EV5B. The AOF of T226K/G228E substitutions predicted by the numerical kinetics, GPR‐1, and SVR models adequately represented the realistic data of the DENV2 Asian I genotype, with Pearson r > 0.97 (P < 0.001) (Appendix Table S5). Subsequently, we forecasted the AOF of T226K and G228E substitutions in the DENV2 Cosmopolitan genotype in 2019–2040 (t = 0–21) by these developed models. For all models, the initial value of the AOF of the T226K substitution was 19.2%. The model parameters of the numerical kinetics model were identical to those of DENV2‐Asian I.
Modeling of the prevalence of a DENV2 genotype
A Gaussian process regression approach (GPR‐2) was used to model the correlations between the AOF of T226K/G228E substitutions and the prevalence of a DENV2 genotype via the available data of the DENV2 Asian I genotype from 1998 to 2019 (Fig 5A). The input and output data of GPR‐2 were defined as x = {t, the AOF of T226K/G228E at time t}T and y = {the prevalence of DENV2 Asian I genotype}T (Appendix Table S6). The output data y = {the prevalence of DENV2 Asian I genotype}T of the GPR‐2 model were preprocessed before training: the prevalence of the DENV2 Asian I genotype in 1995 was subtracted from that in 1998–2019. Subsequently, the prevalence of the DENV2 Asian I genotype in 1995 was added to the predicted values from 1998 to 2019 as the GPR‐2 model output data. We detected and deleted the outlier output data using the isoutlier function in MATLAB R2020b before training. GPR‐2 was trained using the Regression Learner application in MATLAB R2020b. The optimal hyperparameters were searched by the BO method: rational quadratic kernel function, sigma = 0.002, kernel scale = 2.28. We used leave‐one‐out cross‐validation to assess the performance of the GPR‐2 model. Then we predicted the annual prevalence of the DENV2 Cosmopolitan genotype in 2025–2040 by the trained GPR‐2. The input data were x = {t (the number of years from 2019), the AOF of T226K/G228E (predicted by the numerical kinetics, GPR‐1, and SVR models)}T. We obtained the changes in the prevalence of the DENV2 Cosmopolitan genotype based on 2019 by the trained GPR‐2 model. Subsequently, we added the prevalence of the DENV2 Cosmopolitan genotype in 2019 to the predicted value. The output data were the annual prevalence of the DENV2 Cosmopolitan genotype from 2025 to 2040.
Quantification and statistical analysis
The mice used were bred and maintained in specific pathogen‐free (SPF) laboratory animal rooms at Tsinghua University under the same conditions and randomly divided into different groups during the experiment. Mosquitoes of the same age were used, and those that died before the measurement were excluded from the analysis. The investigators were not blinded to the allocation during the experiments or to the outcome assessment. No statistical methods were used to predetermine the sample size. Descriptive statistics are provided in the figure legends. The Kruskal–Wallis analysis of variance was conducted to detect any significant variation among replicates. If no significant variation was detected, the results were pooled for further comparison. Given the nature of the experiments and the types of samples, the differences between continuous variables were assessed using a nonparametric Mann–Whitney test or unpaired t tests. Differences in mosquito infection rates were analyzed using Fisher's exact test. The survival rates of the infected mice were statistically analyzed using the log‐rank (Mantel–Cox) test. All analyses were performed using GraphPad Prism statistical software or IBM SPSS statistical software.
Biosafety statement
All experiments involving dengue virus were conducted in Biosafety Level 2 laboratory (BSL‐2) approved by both Beijing Municipal Health Commission (approval number: 2021‐006) and Institutional Animal Care and Use Committee of Tsinghua University (approval number: 20‐CG1).
Author contributions
Lu Chen: Conceptualization; resources; data curation; software; formal analysis; validation; investigation; visualization; methodology; project administration; writing – review and editing. Xianwen Zhang: Conceptualization; data curation; supervision; validation; methodology; writing – review and editing. Xuan Guo: Conceptualization; data curation; software; formal analysis; validation; visualization; methodology; writing – review and editing. Wenyu Peng: Conceptualization; data curation; software; formal analysis; validation; visualization; writing – review and editing. Yibin Zhu: Supervision; funding acquisition; validation; investigation; visualization. Zhaoyang Wang: Investigation; methodology. Xi Yu: Supervision; methodology. Huicheng Shi: Validation; visualization; methodology; writing – review and editing. Yuhan Li: Software; methodology. Liming Zhang: Software; methodology. Lei Wang: Methodology. Penghua Wang: Supervision; validation; writing – review and editing. Gong Cheng: Conceptualization; resources; supervision; funding acquisition; writing – original draft; project administration; writing – review and editing.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Supporting information
Source Data for Expanded View and Appendix
Appendix
Expanded View Figures PDF
PDF+
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Acknowledgements
We thank the core facilities of the Center for Life Sciences and Center of Biomedical Analysis for technical assistance (Tsinghua University). We acknowledge the Protein Preparation and Characterization Core Facility of Tsinghua University Branch of China National Center for Protein Sciences Beijing and Zi Yang for providing the facility support. This work was funded by grants from the National Key Research and Development Plan of China (2021YFC2300200, 2020YFC1200104, and 2018YFA0507202), The National Natural Science Foundation of China (32188101, 31825001, 81730063, 81961160737, and 82102389), Tsinghua University Spring Breeze Fund (2020Z99CFG017), Shenzhen San‐Ming Project for prevention and research on vector‐borne diseases (SZSM201611064), the Yunnan Cheng gong expert workstation (202005AF150034), Innovation Team Project of Yunnan Science and Technology Department (202105AE160020), Tsinghua‐Foshan Innovation Special Fund (2022THFS6124), and Young Elite Scientists Sponsorship Program (2021QNRC001).
EMBO reports (2022) 23: e55671
Data availability
This study includes no data deposited in external repositories.
References
- Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O et al (2013) The global distribution and burden of dengue. Nature 496: 504–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T (2009) Protein structure homology modeling using SWISS‐MODEL workspace. Nat Protoc 4: 1–13 [DOI] [PubMed] [Google Scholar]
- Brady OJ, Hay SI (2020) The global expansion of dengue: How Aedes aegypti mosquitoes enabled the first pandemic arbovirus. Annu Rev Entomol 65: 191–208 [DOI] [PubMed] [Google Scholar]
- Brady OJ, Gething PW, Bhatt S, Messina JP, Brownstein JS, Hoen AG, Moyes CL, Farlow AW, Scott TW, Hay SI (2012) Refining the global spatial limits of dengue virus transmission by evidence‐based consensus. PLoS Negl Trop Dis 6: e1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KWK, Watanabe S, Jin JY, Pompon J, Teng D, Alonso S, Vijaykrishna D, Halstead SB, Marzinek JK, Bond PJ et al (2019) A T164S mutation in the dengue virus NS1 protein is associated with greater disease severity in mice. Sci Transl Med 11: eaat7726 [DOI] [PubMed] [Google Scholar]
- Chen S‐T, Lin Y‐L, Huang M‐T, Wu M‐F, Cheng S‐C, Lei H‐Y, Lee C‐K, Chiou T‐W, Wong C‐H, Hsieh S‐L (2008) CLEC5A is critical for dengue‐virus‐induced lethal disease. Nature 453: 672–676 [DOI] [PubMed] [Google Scholar]
- Cheng G, Cox J, Wang P, Krishnan MN, Dai J, Qian F, Anderson JF, Fikrig E (2010) A C‐type lectin collaborates with a CD45 phosphatase homolog to facilitate West Nile virus infection of mosquitoes. Cell 142: 714–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Ramchandran S, Vatanen T, Lietzén N, Lahesmaa R, Vehtari A, Lähdesmäki H (2019) An additive Gaussian process regression model for interpretable non‐parametric analysis of longitudinal data. Nat Commun 10: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong J‐H, Wang SC, Park S, Porembka MR, Christie AL, Kim H, Kim HS, Zhu H, Hyung WJ, Noh SH et al (2022) Development and validation of a prognostic and predictive 32‐gene signature for gastric cancer. Nat Commun 13: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cologna R, Rico‐Hesse R (2003) American genotype structures decrease dengue virus output from human monocytes and dendritic cells. J Virol 77: 3929–3938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings DA, Irizarry RA, Huang NE, Endy TP, Nisalak A, Ungchusak K, Burke DS (2004) Travelling waves in the occurrence of dengue haemorrhagic fever in Thailand. Nature 427: 344–347 [DOI] [PubMed] [Google Scholar]
- Du Z, Su H, Wang W, Ye L, Wei H, Peng Z, Anishchenko I, Baker D, Yang J (2021) The trRosetta server for fast and accurate protein structure prediction. Nat Protoc 16: 5634–5651 [DOI] [PubMed] [Google Scholar]
- Figdor CG, van Kooyk Y, Adema GJ (2002) C‐type lectin receptors on dendritic cells and Langerhans cells. Nat Rev Immunol 2: 77–84 [DOI] [PubMed] [Google Scholar]
- Guo X, Zhao Q, Wu C, Zuo S, Zhang X, Jia N, Liu J, Zhou H, Zhang J (2013) First isolation of dengue virus from Lao PDR in a Chinese traveler. Virol J 10: 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J‐H, Su C‐L, Yang C‐F, Liao T‐L, Hsu T‐C, Chang S‐F, Lin C‐C, Shu P‐Y (2012) Molecular characterization and phylogenetic analysis of dengue viruses imported into Taiwan during 2008–2010. Am J Trop Med Hyg 87: 349–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huy R, Buchy P, Conan A, Ngan C, Ong S, Ali R, Duong V, Yit S, Ung S, Te V et al (2010) National dengue surveillance in Cambodia 1980‐2008: Epidemiological and virological trends and the impact of vector control. Bull World Health Organ 88: 650–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong D, Chen Y, Li N (2018) Gaussian process regression for tool wear prediction. Mech Syst Signal Process 104: 556–574 [Google Scholar]
- Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797 [DOI] [PubMed] [Google Scholar]
- Kyle JL, Harris E (2008) Global spread and persistence of dengue. Annu Rev Microbiol 62: 71–92 [DOI] [PubMed] [Google Scholar]
- Leitmeyer KC, Vaughn DW, Watts DM, Salas R, Villalobos I, Chacon D, Ramos C, Rico‐Hesse R (1999) Dengue virus structural differences that correlate with pathogenesis. J Virol 73: 4738–4747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P (2018) 20 years of the SMART protein domain annotation resource. Nucleic Acids Res 46: D493–D496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Khedkar S, Bork P (2021) SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res 49: D458–D460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limkittikul K, Brett J, L'Azou M (2014) Epidemiological trends of dengue disease in Thailand (2000–2011): a systematic literature review. PLoS Negl Trop Dis 8: e3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhang F, Liu J, Xiao X, Zhang S, Qin C, Xiang Y, Wang P, Cheng G (2014) Transmission‐blocking antibodies against mosquito C‐type lectins for dengue prevention. PLoS Pathog 10: e1003931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Liu Y, Nie K, Du S, Qiu J, Pang X, Wang P, Cheng G (2016) Flavivirus NS1 protein in infected host sera enhances viral acquisition by mosquitoes. Nat Microbiol 1: 16087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manokaran G, Finol E, Wang C, Gunaratne J, Bahl J, Ong EZ, Tan HC, Sessions OM, Ward AM, Gubler DJ et al (2015) Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science 350: 217–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mena GE, Martinez PP, Mahmud AS, Marquet PA, Buckee CO, Santillana M (2021) Socioeconomic status determines COVID‐19 incidence and related mortality in Santiago, Chile. Science 372: eabg5298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer WB, Gubler DJ, Harris E, Sivananthan K, De Silva AM (2003) Emergence and global spread of a dengue serotype 3, subtype III virus. Emerg Infect Dis 9: 800–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messina JP, Brady OJ, Scott TW, Zou C, Pigott DM, Duda KA, Bhatt S, Katzelnick L, Howes RE, Battle KE et al (2014) Global spread of dengue virus types: Mapping the 70 year history. Trends Microbiol 22: 138–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Kuhn RJ, Rossmann MG (2005) A structural perspective of the flavivirus life cycle. Nat Rev Microbiol 3: 13–22 [DOI] [PubMed] [Google Scholar]
- Navarro‐Sanchez E, Altmeyer R, Amara A, Schwartz O, Fieschi F, Virelizier JL, Arenzana‐Seisdedos F, Desprès P (2003) Dendritic‐cell‐specific ICAM3‐grabbing non‐integrin is essential for the productive infection of human dendritic cells by mosquito‐cell‐derived dengue viruses. EMBO Rep 4: 723–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- OhAinle M, Balmaseda A, Macalalad AR, Tellez Y, Zody MC, Saborío S, Nuñez A, Lennon NJ, Birren BW, Gordon A et al (2011) Dynamics of dengue disease severity determined by the interplay between viral genetics and serotype‐specific immunity. Sci Transl Med 3: 114ra128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onischenko E, Noor E, Fischer JS, Gillet L, Wojtynek M, Vallotton P, Weis K (2020) Maturation kinetics of a multiprotein complex revealed by metabolic labeling. Cell 183: 1785–1800.e26 [DOI] [PubMed] [Google Scholar]
- Orozco S, Schmid MA, Parameswaran P, Lachica R, Henn MR, Beatty R, Harris E (2012) Characterization of a model of lethal dengue virus 2 infection in C57BL/6 mice deficient in the alpha/beta interferon receptor. J Gen Virol 93: 2152–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett BE, Sadat EL, Zhang Y, Noronha JM, Squires RB, Hunt V, Liu M, Kumar S, Zaremba S, Gu Z et al (2012) ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res 40: D593–D598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokidysheva E, Zhang Y, Battisti AJ, Bator‐Kelly CM, Chipman PR, Xiao C, Gregorio GG, Hendrickson WA, Kuhn RJ, Rossmann MG (2006) Cryo‐EM reconstruction of dengue virus in complex with the carbohydrate recognition domain of DC‐SIGN. Cell 124: 485–493 [DOI] [PubMed] [Google Scholar]
- Rico‐Hesse R, Harrison LM, Salas RA, Tovar D, Nisalak A, Ramos C, Boshell J, De Mesa MTR, Nogueira RM, Da Rosa AT (1997) Origins of dengue type 2 viruses associated with increased pathogenicity in the Americas. Virology 230: 244–251 [DOI] [PubMed] [Google Scholar]
- Shah M, Wadood A, Rahman Z, Husnain T (2013) Interaction and inhibition of dengue envelope glycoprotein with mammalian receptor DC‐sign, an in‐silico approach. PLoS One 8: e59211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H, Wang W, Du Z, Peng Z, Gao SH, Cheng MM, Yang J (2021) Improved protein structure prediction using a new multi‐scale network and homologous templates. Adv Sci 8: 2102592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassaneetrithep B, Burgess TH, Granelli‐Piperno A, Trumpfheller C, Finke J, Sun W, Eller MA, Pattanapanyasat K, Sarasombath S, Birx DL et al (2003) DC‐SIGN (CD209) mediates dengue virus infection of human dendritic cells. J Exp Med 197: 823–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornburg ZR, Bianchi DM, Brier TA, Gilbert BR, Earnest TM, Melo MC, Safronova N, Sáenz JP, Cook AT, Wise KS et al (2022) Fundamental behaviors emerge from simulations of a living minimal cell. Cell 185: 345–360.e28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thu HM, Lowry K, Myint TT, Shwe TN, Han AM, Khin KK, Thant KZ, Thein S, Aaskov J (2004) Myanmar dengue outbreak associated with displacement of serotypes 2, 3, and 4 by dengue 1. Emerg Infect Dis 10: 593–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thu HM, Lowry K, Jiang L, Hlaing T, Holmes EC, Aaskov J (2005) Lineage extinction and replacement in dengue type 1 virus populations are due to stochastic events rather than to natural selection. Virology 336: 163–172 [DOI] [PubMed] [Google Scholar]
- Ty Hang VT, Holmes EC, Veasna D, Quy NT, Tinh Hien T, Quail M, Churcher C, Parkhill J, Cardosa J, Farrar J et al (2010) Emergence of the Asian 1 genotype of dengue virus serotype 2 in Viet Nam: in vivo fitness advantage and lineage replacement in South‐East Asia. PLoS Negl Trop Dis 4: e757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilakis N, Weaver SC (2008) The history and evolution of human dengue emergence. Adv Virus Res 72: 1–76 [DOI] [PubMed] [Google Scholar]
- Warrilow D, Northill JA, Pyke AT (2012) Sources of dengue viruses imported into Queensland, Australia, 2002–2010. Emerg Infect Dis 18: 1850–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC, Forrester NL, Liu J, Vasilakis N (2021) Population bottlenecks and founder effects: Implications for mosquito‐borne arboviral emergence. Nat Rev Microbiol 19: 184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittke V, Robb T, Thu H, Nisalak A, Nimmannitya S, Kalayanrooj S, Vaughn D, Endy T, Holmes E, Aaskov J (2002) Extinction and rapid emergence of strains of dengue 3 virus during an interepidemic period. Virology 301: 148–156 [DOI] [PubMed] [Google Scholar]
- Yang J, Anishchenko I, Park H, Peng Z, Ovchinnikov S, Baker D (2020) Improved protein structure prediction using predicted interresidue orientations. Proc Natl Acad Sci U S A 117: 1496–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Shan C, Zhu Y, Ma E, Wang J, Wang P, Shi P‐Y, Cheng G (2021) A mutation‐mediated evolutionary adaptation of Zika virus in mosquito and mammalian host. Proc Natl Acad Sci U S A 118: e2113015118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Xie X, Xia H, Zou J, Huang L, Popov VL, Chen X, Shi P‐Y (2019) Zika virus NS2A‐mediated virion assembly. MBio 10: e02375‐19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Liu Y, Liu J, Bailey AL, Plante KS, Plante JA, Zou J, Xia H, Bopp NE, Aguilar PV et al (2021) A trans‐complementation system for SARS‐CoV‐2 recapitulates authentic viral replication without virulence. Cell 184: 2229–2238.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhao L, Jiang J, Zheng J, Yang L, Li Y, Zhou J, Liu T, Xu J, Lou W (2022a) Multiplexed nanomaterial‐assisted laser desorption/ionization for pan‐cancer diagnosis and classification. Nat Commun 13: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhu Y, Liu Z, Peng Y, Peng W, Tong L, Wang J, Liu Q, Wang P, Cheng G (2022b) A volatile from the skin microbiota of flavivirus‐infected hosts promotes mosquito attractiveness. Cell 185: e2516 [DOI] [PubMed] [Google Scholar]
- Zhou D‐X, Jetter K (2006) Approximation with polynomial kernels and SVM classifiers. Adv Comput Math 25: 323–344 [Google Scholar]
- Zhu Y, Zhang C, Zhang L, Yang Y, Yu X, Wang J, Liu Q, Wang P, Cheng G (2021) A human‐blood‐derived microRNA facilitates flavivirus infection in fed mosquitoes. Cell Rep 37: 110091 [DOI] [PubMed] [Google Scholar]


