Abstract
Microtubules typically promote nuclear centring during early embryonic divisions in centrosome‐containing vertebrates. In acentrosomal mouse zygotes, microtubules also centre male and female pronuclei prior to the first mitosis, this time in concert with actin. How nuclear centring is brought about in subsequent acentrosomal embryonic divisions has not been studied. Here, using time‐lapse imaging in mouse embryos, we find that although nuclei are delivered to the cell centre upon completion of the first mitotic anaphase, the majority do not remain stationary and instead travel all the way to the cortex in a microtubule‐dependent manner. High cytoplasmic viscosity in 2‐cell embryos is associated with non‐diffusive mechanisms involving actin for subsequent nuclear centring when microtubules again exert a negative influence. Thus, following the first mitotic division, pro‐centring actin‐dependent mechanisms work against microtubule‐dependent de‐centring forces. Disrupting the equilibrium of this tug‐of‐war compromises nuclear centring and symmetry of the subsequent division potentially risking embryonic development. This circuitous centring process exposes an embryonic vulnerability imposed by microtubule‐dependent de‐centring forces.
Keywords: division symmetry, nuclear centring, nucleus positioning, preimplantation development, reproduction
Subject Categories: Cell Adhesion, Polarity & Cytoskeleton; Cell Cycle; Development
A circuitous nuclear centring process imposed by antagonistic microtubule‐ and actin‐based mechanisms risks mitotic divisions in early mouse embryos becoming asymmetric.
Introduction
During preimplantation development, a one‐cell embryo (or zygote) undergoes a series of mitotic divisions, culminating in the formation of a blastocyst. Human reproduction is surprisingly inefficient with many pregnancy failures originating from errors arising during this preimplantation period (Macklon et al, 2002; Hyde & Schust, 2015). Predictably, the more upstream the error, the more devastating the consequences such that errors arising during the first embryonic mitotic division are not compatible with life in either mice or humans (Zhan et al, 2016; Pauerova et al, 2020). Understanding how the embryo regulates the earliest mitotic divisions is therefore critically important.
A striking facet of preimplantation development is the need to switch from meiotic‐ to mitotic‐type divisions. Oocytes are engineered to undergo highly asymmetrica meiotic divisions that preserve the bulk of cytoplasmic contents for supporting embryogenesis. Following entry of the male genome, however, the fertilised oocyte (or zygote) must suddenly reconfigure cell division into a symmetrical mitotic pattern (Clift & Schuh, 2013). Asymmetrical early embryonic divisions have repeatedly been correlated with poor developmental outcomes during assisted reproductive treatments (Hardarson et al, 2001; Racowsky et al, 2011; Desai et al, 2018), yet how division patterning is regulated during early embryonic divisions remains unclear. This is made all the more remarkable when one considers that for at least the first mitotic division in mice, the embryonic genome is largely quiescent and that canonical centrosomal mitotic spindles do not emerge until several divisions later (Courtois et al, 2012; Schneider & Ellenberg, 2019). Hence, the oocyte transitions abruptly from dividing asymmetrically to cleaving symmetrically whilst still under maternal control and with an oocyte‐like acentrosomal machinery. The earliest embryonic mitotic divisions therefore represent a unique and challenging period of regulatory change.
During early embryonic mitotic divisions of non‐mammalian vertebrate organisms, nuclear positioning typically determines spindle positioning, which in turn determines cleavage plane location (Hasley et al, 2017). Positioning the nucleus at the centre of nascent daughter cells prior to nuclear envelope breakdown (NEBD) is therefore critical for ensuring symmetrical division of early embryonic cells (or blastomeres). Previous work has uncovered how oocytes position acentrosomal spindles for achieving asymmetric meiotic divisions (Brunet & Verlhac, 2011) and how pronuclei become centred in preparation for the first mitotic division (Chaigne et al, 2016; Scheffler et al, 2021), but almost nothing is known regarding nuclear positioning beyond this when repetitive mitotic divisions are underway, and centrosomes are lacking in mouse embryos. The actin cytoskeleton is well known to displace the acentrosomal mouse oocyte spindles and with it the cleavage plane to an off‐centre position, thereby producing daughter cells of unequal sizes (Yi & Li, 2012; Holubcova et al, 2013; Almonacid et al, 2014). As with oocytes, the actin cytoskeleton plays a crucial role in centring parental genomes and hence first mitotic spindles in mouse zygotes prior to the embryo's first mitosis (Chaigne et al, 2016; Scheffler et al, 2021). Importantly, microtubules act in concert with actin to centre pronuclei prior to the first mitosis.
In centrosome‐containing embryos such as those from frogs and zebrafish, dynein‐dependent forces acting on astral microtubules centre nuclei immediately following the first mitotic anaphase (Wuhr et al, 2010). It is assumed that following the first mitotic anaphase, nuclei in acentrosomal mouse embryos are also quickly positioned at the cell centres of each blastomere of the now 2‐cell embryo. To gain insight into nuclear centring when embryonic divisions are underway and centrosomes are absent, we here studied nuclear centring following the first mitotic division in mouse embryos. We find that although nuclei arrive at the cell centres of 2‐cell embryos by the end of anaphase, the vast majority then continue travelling towards the cortex before returning to the cell centre ahead of NEBD. Centre‐to‐cortex travel is mediated by microtubules, whereas the reverse journey back to the cell centre is actin‐dependent. Depolymerising microtubules to prevent centre‐to‐cortex displacement has no deleterious effects on the subsequent division and is therefore superfluous. Surprisingly, therefore, and contrary to their better‐known pro‐centring roles, we here find that microtubules are counterproductive to nuclear centring and underpin a vulnerability to symmetry of embryonic divisions and consequently to development.
Results
Biphasic post‐anaphase nuclear movement following first embryonic mitosis
To study the first mitotic division and nuclear centring during the following interphase in real time using confocal imaging, we microinjected one‐cell zygotes with RFP‐tagged histone 2B (H2B‐RFP) cRNA and used the microtubule dye, SiR‐Tubulin, to label chromosomes and spindles, respectively, as before (Wei et al, 2018, 2020; Iljas et al, 2020). We found that ~ 30 min following anaphase onset, chromosomes had become fully separated and cleavage into two daughter cells was well underway with the cleavage plane coinciding with the spindle midzone (Fig 1A). Using time‐lapse imaging of the nuclear pore complex component, Pom121, and immunostaining of nuclear envelope lamins, we find that nuclear formation begins at this time (Appendix Fig S1A and B). After a further 4–5 h, nuclei had become centrally positioned within each daughter cell of the now 2‐cell embryo and remained there for > 10 h (Fig 1A). Unexpectedly, however, most nuclei did not immediately attain a central location within daughter cells following anaphase (Fig 1A). Instead, in 94% of the embryos imaged (47 out of 50), nuclei continued to travel away from one another during telophase/interphase (Fig 1A and B; Movie EV1).
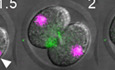
Figure 1. Nuclear movement during and after mouse embryonic first mitotic division.
-
A, BRepresentative time‐lapse images (A) and kymograph (B) of mouse zygotes expressing H2B–RFP (magenta) and labelled with SiR‐Tubulin (green). Shown is an example of marked chromosomal displacement to the cortex.
-
CChanges in chromosome‐to‐cell centre distances (d) over time relative to anaphase onset (n = 50 blastomeres).
-
DA histogram showing the frequencies of maximal d values for individual blastomeres (n = 50 blastomeres).
-
EShown are three representative examples of nuclear tracking from metaphase (dashed white lines) to late 2‐cell interphase. The look‐up table shows the colour scale corresponding to nuclear velocities ranging from 0 to 100% of maximum.
-
FShown are representative time‐lapse images of a zona‐free embryo undergoing the first mitotic division depicting a distinct chromosome‐induced cortical protrusion (see 1 h post‐anaphase onset and corresponding enlarged region).
Data information: In (A, F), time is shown as h post‐metaphase and scale bar = 20 μm. In (C, D), data are shown as mean ± SEM.
To quantify this nuclear movement, the distance of nuclei from the centre of each daughter cell (d) was plotted over time (Fig 1C). By 0.5 h after anaphase onset, chromosomes travelling away from the cleavage plane arrived close to the centre of each newly formed daughter cell (d = 3.8 ± 2.1 μm). Unexpectedly, nuclei did not retain this central location and continued a trajectory towards the cell cortex with d becoming maximal (12.7 ± 3.2 μm; Fig 1C) by 1.5 h post‐anaphase onset with nuclei travelling all the way out to the cortex in many cases (Fig 1A). Nuclei then changed direction and travelled from the cortex back towards the cell centre reflected in a progressive reduction of d to a minimum of 3.1 ± 1.7 μm by 5 h post‐anaphase onset, a phase we refer to as re‐centring (Fig 1A–C). A frequency distribution of the maximal d value for individual blastomeres (n = 50) showed that only a small minority of embryos (6%) had a maximal d value < 10 μm (Fig 1D). Thus, for the first 5 h following first mitotic anaphase, nuclei in over 90% of embryos undergo substantial centre‐to‐cortex movement followed by movement in the opposite direction before establishing a position at the daughter cell's centre.
Next, we tracked nuclear velocity and found that maximal velocities occurred during anaphase and centre‐to‐cortex movement after which nuclei travelled at low velocity from the cortex back to the cell centre. In some cases, nuclei appeared to follow a loop‐shaped journey although it is unclear whether embryo rotation, which often occurred simultaneously, may have contributed (Fig 1E; Movie EV1 and Appendix Fig S1C). In keeping with high‐velocity travel to the cortex, in zona‐free oocytes—which enabled us to better image the cortex—there was marked outpocketing of the cortex at the point of nuclear impact (Fig 1F). Upon returning to the cell centre, nuclei did not undergo any further net displacement in any particular direction but underwent small oscillations, which lasted throughout interphase. Thus, following the vast majority of first mitotic divisions, nuclear travel involves an initial fast centre‐to‐cortex phase followed by slow re‐centring. Importantly, therefore, nuclei are not delivered to the cell centre at the end of anaphase via the shortest possible route.
Microtubules and F‐actin mediate distinct phases of nuclear movement
We next investigated which of the major drivers of nuclear positioning, microtubules and/or microfilaments (F‐actin; Gundersen & Worman, 2013) might be responsible for the nuclear movement we identified.
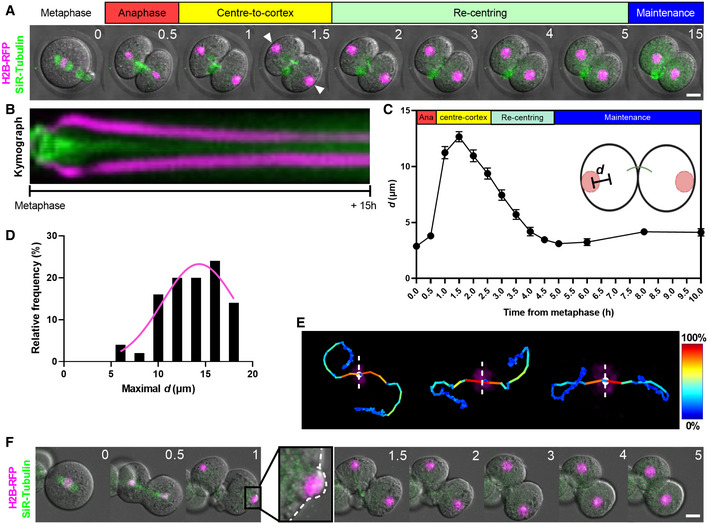
To test the role of microtubules for the initial fast phase of movement from centre‐to‐cortex, we used the microtubule depolymeriser, nocodazole. To avoid disrupting anaphase, nocodazole was added during time‐lapse imaging immediately after chromosomes had segregated and reached daughter cell centres but prior to further displacement to the cortex (Fig 2A; Movie EV2). Unlike dimethyl sulphoxide (DMSO)‐treated controls in which a spindle midzone persisted for 1–2 h (Fig 2B), we found that nocodazole resulted in complete midzone dissolution within 30 min of treatment (Fig 2C) supporting robust microtubule depolymerisation. Notably, nuclei became immobilised at cell centres following nocodazole treatment resulting in stable d values (Fig 2C and D; Movie EV2). Thus, the fast centre‐to‐cortex phase of nuclear movement is dependent on microtubules. We attempted to test the role of actin by adding the actin depolymerising drug, cytochalasin D, immediately following anaphase completion. However, due to actin's indispensable role in mediating cleavage furrow ingression, cytochalasin D resulted in reopening of the cleavage furrow (Appendix Fig S2A and Movie EV2), thereby prohibiting further analyses.
Figure 2. Phase‐specific roles of microtubules and F‐actin during nuclear movement.
-
ASchematic of drug‐treatment regimens for different experiments. Drugs were added either at anaphase completion (B, C) or when nuclei had reached the cortex (E–G).
-
B, CRepresentative time‐lapse images of embryos in which DMSO (B) or nocodazole (C) was added after anaphase completion.
-
DChanges in nucleus‐to‐cell centre distances (d) over time relative to time of drug addition following DMSO (n = 22 blastomeres) or nocodazole treatment (n = 26 blastomeres) at anaphase completion.
-
E–GRepresentative time‐lapse images of embryos in which DMSO (E), nocodazole (F) or cytochalasin D (G) were added when nuclei had travelled to the cortex.
-
HChanges in nucleus‐to‐cell centre distances (d) over time relative to time of drug addition DMSO (n = 20 blastomeres), nocodazole (n = 12 blastomeres) or cytochalasin D treatment (n = 14 blastomeres) after nuclei reached the cortex.
Data information: In (B, C, E–G), time is shown as h post‐drug addition and scale bar = 20 μm. In (D, H), data are shown as mean ± SEM. Statistical significance was assessed by an ordinary two‐way ANOVA with Šidák's multiple comparisons test (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, unremarked datapoints = not significant).
Next, we investigated the slow re‐centring phase of nuclear movement. To do this, we added drugs during time‐lapse imaging after the initial fast phase was complete and nuclei had arrived at the cortex (Fig 2A). We found that nuclei readily underwent re‐centring following microtubule disruption by nocodazole (Fig 2F and H; Movie EV2), similar to DMSO‐treated embryos (Fig 2E and H). Indeed, nuclear re‐centring was advanced in nocodazole‐treated embryos, occurring 1.5–2 h faster than in DMSO‐treated controls (Fig 2H). In complete contrast, following actin disruption using cytochalasin D, nuclei remained stationary at the cell cortex (Fig 2G and H; Movie EV2). We used another actin disruptor, latrunculin B, and again found that nuclear re‐centring was abolished identical to effects seen with cytochalasin D (Appendix Fig S2B and C). Thus, the slow nuclear re‐centring phase requires actin but not microtubules. Interestingly, in contrast to the importance of microtubules during the fast first phase, microtubules are dispensable and even appear to hinder the slow second phase of nuclear movement (Fig 2H).
Nuclear movement plays no physiological role in embryo development
Given that interactions between the cytoskeleton and nuclear envelope modulate gene expression in mouse oocytes (Almonacid et al, 2019), we wondered whether one or more aspects of the marked cytoskeleton‐mediated nuclear movement we identified was physiologically important for subsequent embryo development. We began by investigating the importance of each phase of nuclear movement for the integrity of the subsequent (second) embryonic division.
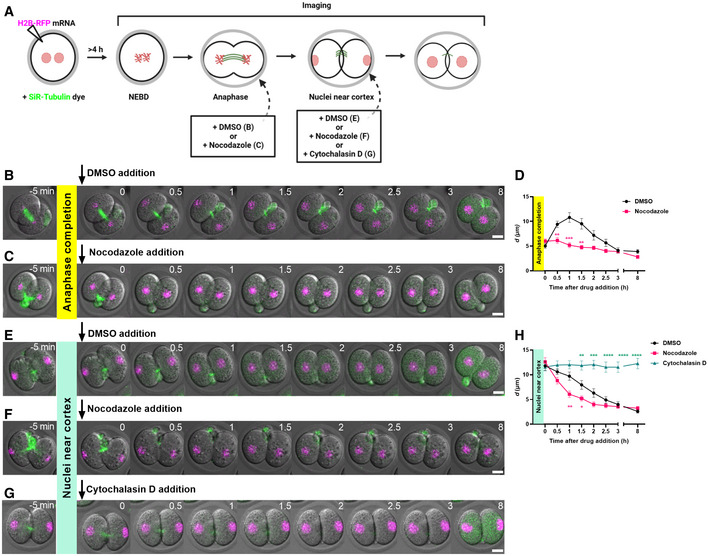
To test whether the first phase of nuclear movement to the cortex might be important, we imaged embryos whose nuclei had been prevented from centre‐to‐cortex displacement by nocodazole. Following nocodazole washout, blastomeres efficiently assembled spindles at the cell centre and completed an unperturbed second mitotic division (n = 15; Fig 3A). Notably, these embryos entered the second mitotic division without first displacing the nucleus to the cortex indicating that such displacement is not necessary.
Figure 3. No correlation between nuclear displacement and embryo development.
-
A, BRepresentative time‐lapse images of embryos that have been washed out of either nocodazole after nuclei were prevented from travelling to the cortex after anaphase (A, n = 15 embryos) or cytochalasin D after nuclei had been trapped at the cortex by disrupting actin (B, n = 22 embryos).
-
CGraph comparing the distribution of maximal d values in embryos that developed to the blastocyst stage or not. Each data point represents the mean maximal d value of the two blastomeres from the same embryo (n = 45 embryos).
-
DGraph of time taken to develop to the blastocyst stage in relation to maximal d values (n = 27 embryos).
-
EGraph comparing the distribution of times taken for nuclei to re‐centre in embryos that developed to the blastocyst stage or not (n = 45 embryos).
-
FGraph of time taken to develop to the blastocyst stage in relation to time taken for nuclei to re‐centre (n = 27 embryos).
Data information: In (A, B), time is shown as h post‐drug washout and scale bar = 20 μm. In (C–F), data are shown as mean ± SEM. In (C, E), statistical significance was assessed by Welch's t‐test, ns: not significant. In (D, F), the strength of correlation (r) and statistical significance were assessed using the Pearson correlation test, ns: not significant.
Next, to determine whether the timing of the second phase of movement leading to re‐centring was critical, we imaged embryos that had been washed out of cytochalasin D. We first asked whether there was a restricted time window during which nuclear re‐centring could occur. After nuclei had been trapped at the cortex by cytochalasin D treatment for ~ 12 h, we washed embryos into inhibitor‐free medium and undertook time‐lapse imaging. We found that following inhibitor washout, nuclei in 91% (21 out of 23) of embryos resumed centring and attained a central location within 4–5 h, similar to untreated controls (Appendix Fig S3A and B; Movie EV3). We further found that in 67% (14 out of 21) of embryos in which nuclear re‐centring was successful, blastomeres progressed to the second mitotic division and cleaved symmetrically (Fig 3B; Movie EV3). Thus, embryos can complete nuclear centring during the 2‐cell interphase even after a protracted delay. Moreover, neither nuclear travel to the cortex nor establishment of nuclear centring within a specified period is necessary for the second mitotic division.
To further investigate a potential physiological role for nuclear displacement, we asked whether different modes of nuclear centring in untreated embryos might impact in vitro development to the blastocyst stage. To minimise the detrimental effects of long‐term imaging for 4 days required for tracking blastocyst development, we used SiR‐DNA instead of H2B‐RFP to label DNA as this allowed us to employ less toxic far‐red wavelengths. With this strategy, we obtained high rates of blastocyst formation (80%) despite continuous imaging for 4 days (Movie EV3), comparable to unimaged embryos cultured under standard conditions in our laboratory (Iljas et al, 2020). We first analysed embryos, which formed blastocysts to determine whether the likelihood of blastulation might be correlated with the extent to which nuclei were displaced towards the cortex following the first mitotic anaphase. We found that the range of maximal displacement was very similar for embryos that formed blastocysts as for those that did not (Fig 3C). Moreover, there was no correlation between maximal displacement values and the time taken to form a blastocyst (Fig 3D). Similarly, the speed with which nuclei re‐centred did not appear to impact blastulation as the range of nuclear centring times was indistinguishable between embryos that did or did not form blastocysts (Fig 3E) and there was no correlation between centring time and the time taken for blastulation (Fig 3F).
Collectively, these data do not identify any physiological implications for nuclear displacement to the cortex prior to nuclear centring. This therefore raised the suspicion that by increasing the difficulty of centring, such nuclear movement may instead present a vulnerability that could compromise subsequent divisions.
Pre‐NEBD nuclear positioning determines the degree of division symmetry
To investigate the importance of nuclear centring, we next asked whether failure of nuclear centring prior to nuclear envelope breakdown (NEBD) would impact the next mitotic division or whether events occurring post‐NEBD could compensate. We found that embryos possess robust centring mechanisms as 21 of 23 blastomeres whose nuclei had been trapped at the cortex by cytochalasin D effectively centred their nuclei following drug washout (Fig 3B; Appendix Fig S3A and B). Notably, however, these are not fail‐safe mechanisms as nuclei remained off‐centre in two blastomeres (Appendix Fig S3C).
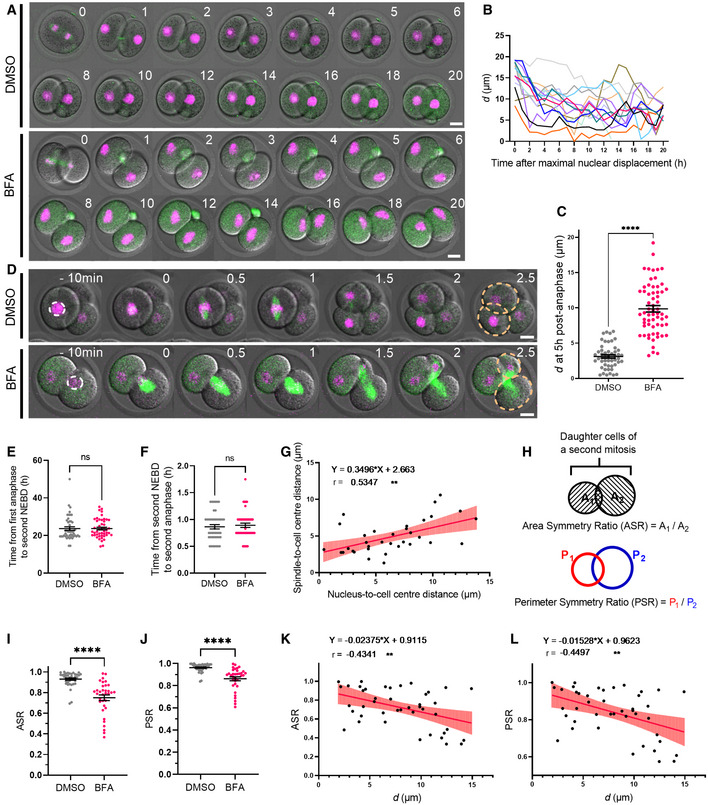
To further investigate the effect of pre‐NEBD nuclear positioning on division symmetry and cell cycle timings, we sought other inhibitors which would disrupt nuclear positioning whilst still allowing cytokinesis to proceed. Brefeldin A (BFA) is an inhibitor of protein trafficking and endomembrane structures previously shown to affect nuclear positioning in oocytes (Wang et al, 2008; Mogessie & Schuh, 2017). We found that BFA‐treated zygotes completed the first mitotic division and exhibited nuclei that travelled to the cortex following anaphase as seen in DMSO‐treated controls (Fig 4A). Although nuclei following BFA treatment then often moved from the cortex to the centre of newly formed blastomeres, they did not maintain a stable position there and instead moved towards the cortex again resulting in a convoluted nuclear path (Fig 4A and B; Movie EV4). Consequently, unlike DMSO‐treated control embryos that typically achieve nuclear centring within 5 h after anaphase (Fig 4A), BFA‐treated embryos do not, resulting in higher d values at this timepoint (Fig 4C). Highly significantly, due to this positioning instability, 70% of BFA‐treated blastomeres entered the second mitotic division with off‐centre nuclei that were > 5 μm away from the cell centre (Fig 4D).
Figure 4. Division symmetry is defined by pre‐NEBD nuclear positioning.
-
ARepresentative time‐lapse image of DNA and SiR‐tubulin in DMSO‐ and BFA‐treated embryos during the first mitotic division and subsequent interphase. Time is shown as h relative to the first mitotic anaphase.
-
BChanges in nucleus‐to‐cell centre distances (d) over time in BFA‐treated embryos, relative to maximal nuclear displacement to the cortex (n = 14 blastomeres).
-
CDot plot of the nucleus‐to‐cell centre distances (d) at 5‐h post‐anaphase in DMSO‐ (n = 50 blastomeres) and BFA‐treated embryos (n = 62 blastomeres).
-
DRepresentative time‐lapse images of the second mitotic division in DMSO‐ and BFA‐treated embryos. Dashed white lines highlight the position of nuclei shortly before NEBD. Dashed yellow lines outline daughter cells. Time is shown as h relative to the second mitotic NEBD.
-
EDot plot of the duration between first mitotic division anaphase onset and second mitotic division NEBD for DMSO‐ (n = 47 blastomeres) and BFA‐treated (n = 47 blastomeres) embryos.
-
FPlot of the duration between NEBD and anaphase of the second mitotic division for DMSO‐ (n = 44 blastomeres) and BFA‐treated (n = 43 blastomeres) embryos.
-
GPlot of the spindle‐to‐cell centre distances against pre‐NEBD nucleus‐to‐cell centre distance (n = 34 blastomeres). Line indicates mathematically regressed fit.
-
HSchematics depicting the calculation of area symmetry ratio (ASR) and perimeter symmetry ratio (PSR). Measurements were undertaken at the middle plane of the confocal Z stack.
-
I, JDot plots of ASR (I) and PSR (J) of DMSO‐ (n = 38) and BFA‐treated (n = 34) daughter cells following the second mitotic division.
-
K, LPlots show the correlation between ASR (K) or PSR (L) and pre‐NEBD nucleus‐to‐cell centre distances (d) (n = 43). Lines indicate mathematically regressed fits.
Data information: In (A, D), scale bar = 20 μm. In (C, E–G, I–L), data are shown as mean ± SEM. In (C), statistical significance was assessed by Welch's t‐test. In (E, F, I, J), statistical significance was assessed by Mann–Whitney test, ****P < 0.0001, ns: not significant. In (G, K, L), the strength of correlation (r) and statistical significance were assessed using the Pearson correlation test. Two‐tailed P‐values are reported (**P < 0.01).
To determine whether off‐centre nuclei delayed onset of the second division, we measured the time interval between anaphase onset of the first division and NEBD of the second division in control and BFA‐treated embryos. We found that NEBD occurred at very similar times relative to anaphase in DMSO‐ and BFA‐treated blastomeres (Fig 4D and E; Movie EV4). The interval between NEBD and anaphase onset for the second mitotic division was also similar regardless of nuclear location, altogether showing that nuclear positioning did not affect cell cycle progression (Fig 4F). The normal timing of cell cycle progression in treated embryos also showed that apart from impacting nuclear position, BFA did not overtly disrupt progression through early cleavage divisions.
Although the second mitotic cell cycle in blastomeres with and without centred nuclei progressed in step with one another, we found that spindles assembled at a position that correlated with the position of nuclei before NEBD (Fig 4D and G); in other words, blastomeres with off‐centre nuclei assembled off‐centre spindles. Significantly, such spindles did not attain a central location by the time of anaphase (Fig 4D). Since the midzone typically determines the plane of cleavage, we next asked whether asymmetrically located spindles resulted in asymmetric divisions, which would become apparent as unequally sized daughter cells. To quantify relative sizes of daughter cells, we first compared the ratios of daughter cell areas, which we refer to as the area symmetry ratio (ASR; Fig 4H); the closer ASR is to 1, the greater the degree of division symmetry. In DMSO‐treated controls with normally centred nuclei prior to NEBD, 36 of 38 embryos had an ASR > 0.8 with an overall mean of 0.93 ± 0.01 (Fig 4I). In stark contrast, only nine of 34 BFA‐treated embryos had an ASR > 0.8 with a mean of 0.75 ± 0.03, significantly lower than controls (Fig 4I). As a second measure of division symmetry, we calculated the ratios of blastomere perimeters, which we refer to as perimeter symmetry ratio (PSR; Fig 4H). As with ASR, PSR was significantly higher and closer to 1 for DMSO‐treated controls (0.96 ± 0.04) than for BFA‐treated (0.86 ± 0.02) embryos (Fig 4J). Notably, ASR and PSR were both inversely correlated with pre‐NEBD nuclear distance from the cell centre (Fig 4K and L).
Collectively, these data show that division symmetry is prescribed by pre‐NEBD nuclear location and cannot be rescued by post‐NEBD spindle positioning. This emphasises the importance of nuclear centring mechanisms and highlights the vulnerability posed by the marked nuclear‐to‐cortex movement we identify here.
2‐cell embryos exhibit high cytoplasmic viscosity and do not employ diffusion‐mediated nuclear centring mechanisms
In prophase I‐arrested oocytes, formin‐mediated actin nucleation generates a pressure gradient that centres nuclei and other large objects like oil droplets via active diffusion (Almonacid et al, 2015; Colin et al, 2020). We therefore asked whether actin‐dependent nuclear centring in 2‐cell embryos occurred by similar diffusion‐mediated pathways. Firstly, we microinjected oil droplets into zygotes and plotted the distance, d, over time for droplets and nuclei following first mitotic anaphase (Fig 5A and B; Movie EV5). As before, we found that nuclei travelled from the cortex to the cell centre covering a total distance of over 10 μm during which d underwent a progressive decline, ultimately becoming < 5 μm by the time nuclei reached the cell centre. In stark contrast, injected oil droplets within the same embryos exhibited only very minor movement towards the centre (< 5 μm) resulting in d values that never declined below 10 μm (Fig 5A and B).
Figure 5. Non‐diffusive nuclear centring and high cytoplasmic viscosity in 2‐cell embryos.
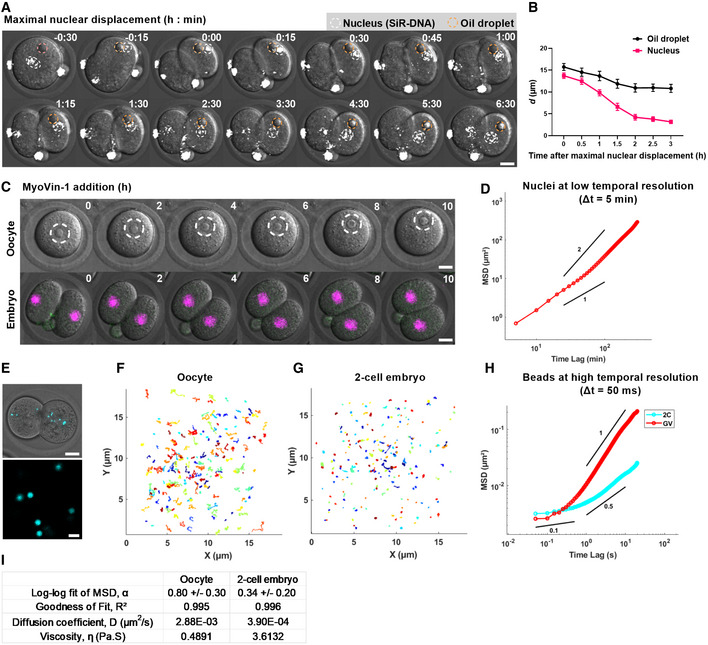
-
ARepresentative time‐lapse images of 2‐cell embryos cultured with SiR‐DNA to label nuclei (white circles) and microinjected with oil droplets (yellow circles). Time is shown as h:min relative to maximal nuclear displacement.
-
BPlot of the distances between oil droplets and nuclei to the cell centre over time (n = 14 oil‐nucleus pairs).
-
CRepresentative time‐lapse images of prophase I‐arrested oocytes (upper panel, dashed circles outline nuclei) (n = 16 oocytes) and 2‐cell embryos microinjected with H2B‐RFP to label DNA (n = 14 embryos) following treatment with MyoVin‐1. Time is shown as h post‐drug addition.
-
DMSD analysis of nuclear displacement during re‐centring (n = 26 blastomeres). Power‐law slopes of 1 and 2 are drawn for comparison.
-
ETop panel: an example of fluorescent bead distribution in a 2‐cell embryo. Lower panel: an example of region imaged for bead tracking at high temporal resolution.
-
F, GTrajectories of bead movement over a 20‐s duration in oocytes (F, n = 217 beads from 23 oocytes) and 2‐cell embryos (n = 277 beads from 26 embryos). Each coloured line represents a single bead track.
-
HMSD plot of beads in oocytes (n = 217 beads from 23 oocytes) and 2‐cell embryos (n = 277 beads from 26 embryos). Power‐law slopes of 1, 0.5 and 0.1 are drawn for comparison.
-
IResults obtained from MSD analysis. α represents the slope of the log–log fit of the MSD curve using the first 25% (100 data points). D represents the diffusion coefficient estimated from the linear fit of the MSD curve.
Data information: Scale bar = 20 μm in A, C, E, except 2 μm in the lower panel of E. In (B), data are shown as mean ± SEM. In (D, H), the ensemble‐averaged MSDs are shown.
Source data are available online for this figure.
Next, we tested the involvement of myosin Vb, which is the motor required for diffusive motion in oocytes (Almonacid et al, 2015). Consistent with a lack of diffusive motion during the first embryonic division, we found that a Myosin V‐specific inhibitor, MyoVin‐1 (Islam et al, 2010), consistently disrupted nuclear positioning in prophase I‐arrested oocytes but not in 2‐cell embryos (Fig 5C; Movie EV5). Furthermore, the mean squared displacement (MSD) profile of re‐centring nuclei in MyoVin‐1‐treated embryos was significantly greater than 1 (P < 0.001) and approaches 2 on a log–log scale, characteristic of active rather than diffusive transport (Fig 5D).
Since our results suggest 2‐cell embryos do not exhibit diffusion‐based centring and nuclei do not drift from their position at the cortex for several hours after cytochalasin D treatment (Fig 2G), we wondered whether cytoplasmic resistance to nuclear motion brought about by high cytoplasmic viscosity might be a feature of 2‐cell embryos (Almonacid et al, 2015; Chaigne et al, 2016). To gain insight into this, we studied the rheological properties of the cytoplasm of 2‐cell embryos using live‐cell particle‐tracking microrheology (Wirtz, 2009). For this, we microinjected passive beads and recorded their individual movements using high‐speed video tracking (Fig 5E; Movie EV5). At the ultra‐high temporal resolution of 50 ms, bead movement is considered to be driven by thermal motion and the extent of bead displacement is inversely proportional to the viscosity of the surrounding cytoplasm (Fakhri et al, 2014; Wessel et al, 2015). We tracked bead trajectories in oocytes and 2‐cell embryos and found that beads explored more space during the 20 s recording time in oocytes (Fig 5F; Movie EV5) compared with embryos (Fig 5G). Consistent with this, the mean MSD curve obtained from > 200 bead trajectories showed a more rapid growth with lag time in oocytes than in embryos (Fig 5H). For observation times between 50 ms and 5 s, beads in both groups underwent unbiased random motion with MSD slopes less than 1 on the log–log scale (Fig 5H). On longer time scales from 5 to 20 s, the mean MSD approached 1 in oocytes whereas although an increase was observed in 2‐cell embryos, mean MSD nevertheless remained significantly lower at ~ 0.5, characteristic of constrained motion.
Next, we used the mean diffusion coefficient of the beads estimated from ensemble‐averaged MSDs to calculate cytoplasmic viscosity using the Stokes‐Einstein equation and found that 2‐cell embryos have a cytoplasmic viscosity of 3.6 Pa.S (Fig 5I). This is broadly consistent with the viscosity reported for C. elegans and Drosophila embryos (0.67–1 Pa.S), mouse and human fibroblasts (1–4.5 Pa.S) derived using similar intracellular measurements (Kole et al, 2004; Daniels et al, 2006, 2010; Wessel et al, 2015; Khatri et al, 2022). Notably, we found that the cytoplasmic viscosity of mouse oocytes was 0.5 Pa.S, over 7 times lower than in 2‐cell embryos.
Altogether, these data show that 2‐cell embryos do not centre nuclei using non‐specific diffusion mechanisms employed by oocytes. It may be that the markedly higher cytoplasmic viscosity of embryos is not conducive to diffusive mechanisms.
Depletion of cortical actin is associated with impaired cortex‐to‐centre return
It seemed unlikely that a force acting to centre the nucleus would already be present during the initial centre‐to‐cortex movement since this would require such forces to be countered by microtubule‐dependent ones. We therefore asked whether actin‐dependent centring forces were only activated when nuclei gained proximity to the cortex as seen with pronuclei in zygotes (Scheffler et al, 2021). To monitor changes in F‐actin in real time, we microinjected zygotes with UtrCH‐mCherry and simultaneously labelled DNA with H2B‐mClover. We found that F‐actin signals increased markedly at the region of cortex overlying approaching nuclei and remained elevated whilst nuclei started returning to the centre before dissipating as nuclei moved further away from the cortex (Fig 6A; Movie EV6). Notably, adding cytochalasin D when nuclei had reached the cortex completely abolished localised cortical F‐actin enrichment within 10 min (Fig 6B; Movie EV6). We fixed phalloidin‐treated embryos to study cortical and cytoplasmic actin networks and found that both cytochalasin D and latrunculin B markedly depleted cortical actin whilst also reducing cytoplasmic actin compared with DMSO‐treated controls (Appendix Fig S4E and F). Significantly, we earlier found that these drugs caused nuclei to stagnate at the cortex (see Fig 2G; Appendix Fig S2B) altogether supporting the notion that cortical actin polymerisation promotes nuclear centring.
Figure 6. Actin‐dependent nuclear positioning in 2‐cell embryos.
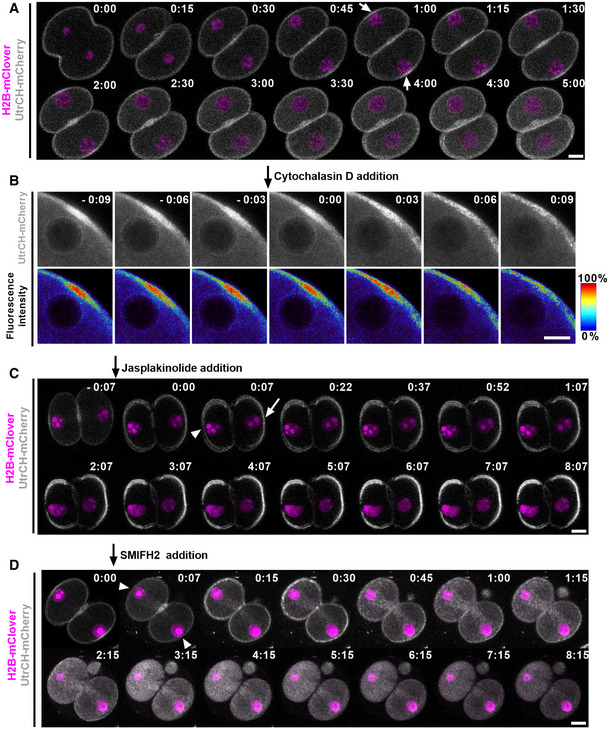
-
ARepresentative time‐lapse images of F‐actin (UtrCH‐mCherry) and nuclei (H2B‐mClover). Time is shown as h:min relative to anaphase onset. Arrows indicate high UtrCH‐mCherry intensity (n > 30 embryos).
-
BRepresentative time‐lapse imaging of F‐actin (UtrCH‐mCherry) after cytochalasin D addition (n = 10 blastomeres). Time is shown as h:min relative to drug addition. The look‐up table shows the colour scale corresponding to fluorescence intensity ranging from 0 to 100% of maximum.
-
C, DRepresentative time‐lapse imaging of F‐actin (UtrCH‐mCherry) after Jasplakinolide (C, n = 11 blastomeres) and SMIFH2 (D, n = 9 blastomeres) addition. Time is shown as h:min relative to drug addition. Arrows indicate increased UtrCH‐mCherry intensity relative to the last frame, and arrowheads indicate decreased UtrCH‐mCherry intensity.
Data information: In (A–D), data shown are representative examples of biological replicates as specified. Images are the maximum intensity projection of 3 z‐slices. Scale bars = 20 μm except in (B), scale bar = 10 μm.
To further investigate whether cortical actin polymerisation promotes nuclear re‐centring, we treated embryos with jasplakinolide, which has the opposite effect from cytochalasin D and latrunculin B of polymerising and stabilising actin filaments. We found that in eight out of 11 blastomeres, jasplakinolide completely prevented nuclear centring, whereas in the other three blastomeres, the nuclei rapidly launched towards the cell centre (Fig 6C; Movie EV6). Interestingly, both live imaging of F‐actin and phalloidin staining of fixed embryos showed that following jasplakinolide treatment, the majority of the cortical surface exhibited marked actin enrichment, but there were smaller regions that showed significantly reduced intensity compared with DMSO‐treated controls (Appendix Fig S4E and F). Importantly, cortically located nuclei in live and fixed embryos consistently occupied regions where cortical actin intensity was reduced (Appendix Fig S4A and E).
Since actin assembly relies on nucleators including formin and/or Arp2/3 (Bovellan et al, 2014), we next investigated the effects of their inhibitors, SMIFH2 and CK666, respectively (Rizvi et al, 2009; Hetrick et al, 2013). Adding SMIFH2 when nuclei had been displaced to the cortex led to the immediate disappearance of cortical actin enrichment without a significant decrease in cytoplasmic actin (Appendix Fig S4E and F). SMIFH2‐induced depletion of cortical actin was associated with complete loss of re‐centring as d values remained stable for > 8 h (Fig 6D; Appendix Fig S4D). In contrast, following DMSO treatment which had no effect on cortical actin (Appendix Fig S4E), d values reached a nadir by 3 h coincident with nuclei reaching the cell centre (Appendix Fig S4D and E, and Movie EV6). Unlike SMIFH2, however, Arp2/3 inhibition with CK666 at a dose capable of disrupting spindle localisation in metaphase II‐arrested oocytes (Yi et al, 2011) had no effect on nuclear centring in embryos with d values exhibiting a decline indistinguishable from controls (Appendix Fig S4B and D, and Movie EV6). Collectively, the foregoing show that nuclear movement towards the centre occurs when cortical actin increases and, conversely, that nuclear stagnation at the cortex is strongly associated with reduced actin, altogether supporting the importance of cortical actin for nuclear re‐centring. In contrast, we did not uncover any association between nuclear movement and cytoplasmic actin levels as nuclear stagnation occurred regardless of whether cytoplasmic actin remained normal or was reduced (e.g. compare cytochalasin D with SMIFH2, Appendix Fig S4E and F).
Another key regulator of cortical properties acting in concert with actin is myosin II activity (Cartagena‐Rivera et al, 2016; Chugh et al, 2017). To test the role of myosin II, we inhibited myosin II ATPase activity using blebbistatin when nuclei had been displaced to the cortex. We found this acutely prevented nuclear re‐centring for 3–4 h by which time, nuclei had become centred in controls (Appendix Fig S4C and D, and Movie EV6). Following this delay, we did observe that nuclear movement resumed, which may be due to the photoinactivation of blebbistatin (Kolega, 2004; Sakamoto et al, 2005). As expected, blebbistatin did not alter either cortical or cytoplasmic actin (Appendix Fig S4E and F), suggesting that myosin II is required for efficient centring in concert with cortical actin polymerisation.
Taken together, these data show that nuclear cortex‐to‐centre movement is strongly correlated with cortical actin enrichment and support that cortical actin polymerisation and myosin II are required for displacing nuclei away from the cortex, although we do not exclude a role for cytoplasmic actin.
Microtubule‐based forces antagonise nuclear centring
We were struck by the increased efficiency of nuclear re‐centring following microtubule depolymerisation (Fig 2H), which suggested that microtubules were exerting forces on nuclei directed away from the centre and towards the cortex. We calculated the MSD slope during microtubule‐dependent nuclear movement to the cortex and found it follows a power law of 1.82 ± 0.13 and is significantly greater than 1 (P < 0.001; Fig 7A). Since a slope of 2 on a log–log scale indicates active transport, our result supports that microtubule‐induced forces were indeed acting on the nucleus.
Figure 7. Microtubule‐based forces antagonise nuclear centring.
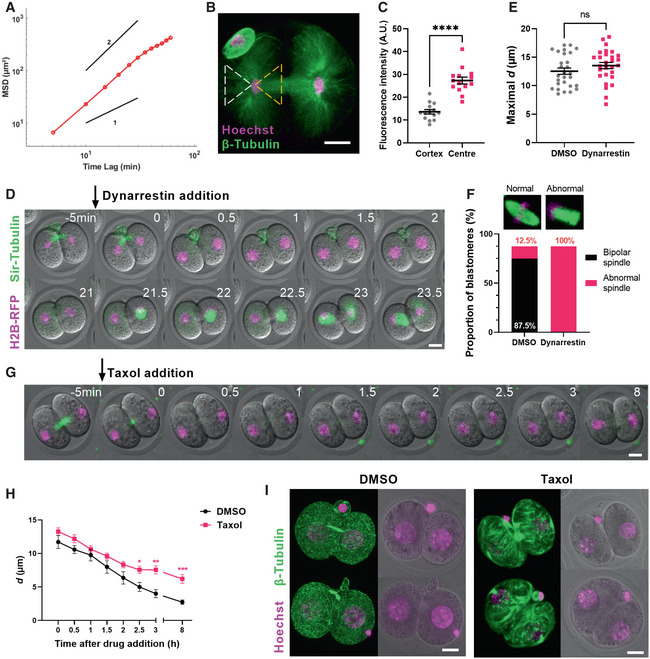
- MSD analysis during centre‐to‐cortex nuclear displacement (n = 18 blastomeres). Power‐law slopes of 1 and 2 are drawn for comparison.
- Shown is a representative 2‐cell embryo immunostained for β‐tubulin (green) and DNA (magenta) at anaphase completion (n = 8 embryos).
- Dot plot of mean β‐tubulin fluorescence intensity within cytoplasmic regions that either lie ahead of the nucleus (cortex‐facing aspect, dashed white triangle in B) or behind the nucleus (cleavage plane‐facing aspect, dashed yellow triangle in B) (n = 14 embryos).
- Representative time‐lapse images of embryos expressing H2B‐RFP and labelled with SiR‐tubulin following the addition of dynarrestin at anaphase completion (n = 14 embryos).
- Dot plot of maximal d values between DMSO‐ and dynarrestin‐treated embryos (n = 28 blastomeres per group).
- The proportions of normal bipolar and abnormal spindles in DMSO‐ and dynarrestin‐treated blastomeres undergoing the second mitosis.
- Representative time‐lapse images of embryos expressing H2B‐RFP and labelled with SiR‐tubulin following the addition of taxol at maximal nuclear displacement.
- Changes in nucleus‐to‐cell centre distances (d) over time relative to the time of drug addition in DMSO‐ (n = 19) and taxol‐ (n = 19) treated 2‐cell blastomeres (mean ± SEM).
- Representative images of embryos immunostained for β‐tubulin and Hoechst following the completion of live imaging (shown in E) in DMSO‐ and taxol‐treated embryos (n = 9 embryos per group). Time is shown as h post‐drug addition. Scale bar = 20 μm.
Data information: In (D, F), time is shown as h post‐drug addition. In (B, D, G, I), scale bar = 20 μm. In (C, E, H), data are shown as mean ± SEM. In (C, E), statistical significance was assessed by Welch's t‐test, ****P < 0.0001, ns = not significant. In (H), statistical significance was assessed by an ordinary two‐way ANOVA with Šidák's multiple comparisons test (*P < 0.05, **P < 0.01, ***P < 0.001, unremarked datapoints = not significant).
We next asked whether these forces acted to pull nuclei towards the cortex or push them away from the cell centre. We immunostained oocytes for β‐tubulin and quantified microtubule fluorescence intensity at the end of first mitotic anaphase (Fig 7B). We quantified fluorescence in the region between the nucleus and cortex and found that it was roughly half as intense as that between nuclei and the forming cleavage plane (Fig 7C). Thus, as nuclei start moving towards the cortex, microtubule density is greatest in the region behind the direction of travel suggesting that microtubules could be pushing nuclei towards the cortex.
Contrary to the above findings, it is well documented in other models that nuclear movement during early embryonic divisions involves microtubule‐based pulling mediated by the minus‐end directed microtubule motor, dynein (Hasley et al, 2017). To test whether pulling forces might be involved, we treated embryos with the recently characterised small‐molecule dynein inhibitor, dynarrestin (Hoing et al, 2018), after first mitotic anaphase was complete but prior to the start of centre‐to‐cortex movement. Notably, we found that nuclear movement towards the cortex remained completely intact in dynarrestin‐treated embryos (Fig 7D and E; Movie EV7). These embryos underwent NEBD without positioning defects, although the second mitosis could not be completed due to abnormal spindle structure (Fig 7F; Movie EV7), as expected with dynein's critical role in spindle bipolarity (van Heesbeen et al, 2014). Therefore, dynein is required for spindle organisation but not its positioning, which is determined by pre‐NEBD nuclear positioning.
The foregoing support that microtubule‐dependent forces directed towards the cortex are responsible for displacing nuclei to the cortex. We reasoned that similar microtubule‐dependent forces persisted during re‐centring, thereby explaining the faster re‐centring we observed following nocodazole‐induced depolymerisation (Fig 2H). To test this further, we treated embryos during time‐lapse imaging with either DMSO or taxol after nuclei had been displaced to the cortex in 2‐cell embryos (Fig 7G, Movie EV7). Although the initial phase of re‐centring was comparable in both groups, we found that taxol severely impeded final re‐centring such that nuclear distances from the centre remained significantly higher than in DMSO‐treated embryos (Fig 7H). To visualise the effect of taxol on the microtubule network, we immunostained 2‐cell embryos that had been treated with taxol and found that the latter induced the formation of dense microtubule bundles that traversed blastomeres contrasting with the diffuse short‐branching pattern seen with DMSO treatment (Fig 7I, Movie EV7). We reasoned that the same microtubule pattern was not seen during live imaging of taxol‐treated embryos likely because SiR‐tubulin dye and paclitaxel target the same β‐tubulin binding site (Lukinavicius et al, 2018).
Collectively, the foregoing show that microtubules deter nuclearre‐centring by exerting forces directed away from the cell centre.
Discussion
Here, we study how nuclei become centred during interphase between the first and second mitotic divisions in mouse embryos. Although nuclear positioning has been studied during embryonic mitotic divisions in other model systems such as flies, fish and frogs, to our knowledge, this is the first in‐depth analysis of nuclear positioning in a mammalian model. Prior analyses focussed on centring of pronuclei in one‐cell mouse zygotes, which occurs after fertilisation but before the onset of mitotic divisions (Courtois et al, 2012; Yamagata & FitzHarris, 2013; Chaigne et al, 2016; Reichmann et al, 2018; Scheffler et al, 2021). By introducing small‐molecule inhibitors whilst live embryos were being imaged, we were able to target specific phases of embryonic division without disrupting the preceding phase. Thus, to study the role of microtubules during nuclear travel to the cortex, we introduced nocodazole after anaphase was complete since earlier introduction would prevent anaphase from even occurring. Moreover, to investigate the role of actin during re‐centring, we introduced actin‐disrupting agents after nuclei had reached the cortex as earlier treatments prevented cytokinesis.
Early mitotic divisions in mouse embryos occur without canonical centriole‐containing centrosomes and are symmetrical, representing a dramatic shift from the preceding highly asymmetrical meiotic divisions in oocytes. In contrast to somatic cells in which centrosome‐nucleated astral microtubules immediately establish stable nuclear positions at the centre of daughter cells after cytokinesis, we find that nuclei in much larger acentrosomal mouse blastomeres do not immediately become stably positioned at the cell centre. Instead, we discover that although nuclei form close to the cell centre at the end of anaphase, they then undergo marked displacement, very often travelling all the way to the cortex, before moving back to the cell centre in preparation for the ensuing (second) mitosis.
Microtubules have predominantly been found to play pro‐centring roles in embryos (Hasley et al, 2017). In frogs and fish, forces mediated by the minus‐end directed microtubule motor, dynein, position nuclei at the cell centre so that equal‐sized blastomeres are generated during the ensuing division (Wuhr et al, 2010). In mouse zygotes, microtubules also promote pronuclear centring in a dynein‐dependent manner (Scheffler et al, 2021). In stark contrast, here we find that microtubules in 2‐cell mouse embryos exert de‐centring effects with nocodazole‐induced depolymerisation completely preventing nuclear displacement to the cortex. Microtubules continue to exert an off‐centring effect during nuclear return to the cell centre as re‐centring occurs faster following microtubule depolymerisation. Conversely, stabilising microtubules with taxol during this return leg, permanently impedes nuclear re‐centring. Thus, microtubules in 2‐cell mouse embryos actively de‐centre nuclei. Notably, whilst microtubules do not affect nuclear positioning in oocytes, antagonistic interactions between the microtubule‐actin networks and a detrimental effect of taxol on nuclear positioning have been reported in oocytes (Almonacid et al, 2015, 2019). Exactly how microtubules move nuclei to the cortex remains to be elucidated, but our data suggest that unlike other systems, dynein‐dependent pulling is not a major player as no effects were seen following dynein inhibition and microtubules concentrate on the lagging aspect of nuclei rather than on the cortex‐facing aspect.
Microtubules collaborate with actin filaments to position pronuclei in mouse zygotes as well as in embryos of other organisms (Deshpande & Telley, 2021). In mouse zygotes, male pronuclear centring is initiated by actin and completed by microtubules (Scheffler et al, 2021). In stark contrast, our data uncover that 2‐cell embryos are solely reliant on actin for nuclear re‐centring with microtubules exerting negative effects. Unlike oocytes which use a non‐specific, diffusion‐based nuclear centring mechanism based on actin vesicles, we found that 2‐cell embryos employ a nucleus‐targeted and actin‐dependent mechanism involving the motor myosin II rather than myosin Vb. Intracellular microrheology measurements suggest that mouse 2‐cell embryos exhibit higher cytoplasmic viscosity than mouse oocytes and other cell types (Kole et al, 2004; Daniels et al, 2006; Wessel et al, 2015; Khatri et al, 2022). Since myosin Vb has been proposed to fluidise the oocyte cytoplasm (Ahmed et al, 2018), it is possible that its inactivity in 2‐cell embryos contributes to a more viscous cytoplasm. Further details regarding high cytoplasmic viscosity in 2‐cell embryos and its physiological significance remain to be explored.
Multiple actomyosin assemblies co‐exist, including bulk cytoplasmic networks and the actomyosin cortex, and the specific contributions of each one to organelle positioning are difficult to dissect (Sakamoto et al, 2020). Our data suggest that launching the nuclei away from the cell periphery in 2‐cell embryos likely initially requires cortical actin. However, it is possible that contraction of the actomyosin cortex may also set the cytoplasm in motion (Illukkumbura et al, 2020). Cytoplasmic mechanisms such as retrograde actin flow in association with nuclear envelope proteins and hydrodynamic centring forces could be involved to facilitate fine centring and maintenance of the nuclei at cell centres (Ierushalmi et al, 2020; Luxton et al, 2010; Munro et al, 2004). The absence of chemicals capable of selectively targeting individual actin networks limited our ability to investigate the contribution of cytoplasmic actin and other factors.
Errors in embryonic divisions impact all subsequent divisions making the earliest divisions the most influential for development. We propose that by displacing nuclei away from their intended destination, the nuclear movement we identify after the embryo's earliest mitotic division constitutes a major Achilles heel for embryo development. In keeping with this, we find no physiological role for off‐centre nuclear displacement as preventing such displacement using nocodazole has no deleterious impact on embryo development to the advanced blastocyst stage, albeit we cannot rule out the possibility that more subtle defects could manifest post‐blastulation. Importantly, there are no post‐NEBD mechanisms for sensing and re‐positioning the spindle as seen in other cell types (Yeh et al, 1995; O'Connell & Wang, 2000). Consequently, as with other models, the position of the nucleus defines where the spindle assembles and cleavage occurs in keeping with which we find that downstream divisions become asymmetric when nuclei remain off‐centre. Interestingly, human embryos exhibiting asymmetrical early cleavage divisions are only half as likely as symmetrically cleaving embryos to develop to the blastocyst stage (Desai et al, 2018). Notably, nuclear displacement and re‐centring can be seen during time‐lapse imaging of human IVF embryos (Vera‐Rodriguez et al, 2015; Desai et al, 2018). Collectively, therefore, marked nuclear displacement following the very first embryonic division exposes a marked vulnerability during embryo development. It is possible that conditions that compromise embryonic integrity, such as ageing or sub‐optimal embryo culture conditions in the IVF laboratory, could expose this vulnerability and compromise pregnancy success.
Materials & Methods
Mice
Mice having a Balb/cA × C57BL/6J F1 background (CB6F1) were obtained by crossing Balb/cA female and C57BL/6J males. All animals were housed in a specific pathogen‐free environment in filter‐top cages and fed a standard diet.
Embryo, oocyte collection and culture
A 4‐week‐old B6CBF1 female mice were used for experiments and 3–6‐month‐old males were used for mating. All mice were housed in a pathogen‐free environment in filter‐top cages and fed a standard diet at the University of Queensland Centre for Clinical Research (UQCCR) animal facility. All animal work complied with the ethical regulations as approved by the Animal Ethics Committee at the University of Queensland.
To induce follicular development, female mice were given an intraperitoneal injection of 7.5 international units (IU) pregnant mare serum gonadotrophin (PMSG, Pacific vet) dissolved in saline, as previously described (Wei et al, 2018, 2020; Zhou et al, 2019; Iljas et al, 2020; Subramanian et al, 2020; Greaney et al, 2021). After 48 h, 5 IU of human chorionic gonadotropin (hCG, ProSpec, HOR‐250) was administered in the same manner to induce ovulation. Upon injection, female mice were caged overnight with fertile wild‐type males. 20 h following hCG injection, female mice were sacrificed and fertilised zygotes released in pre‐warmed M2 medium (Sigma‐Aldrich, M7167) containing hyaluronidase (Sigma‐Aldrich, 385931) to remove cumulus cells.
To collect oocytes, female mice were given a single PMSG injection and sacrificed 48 h later. Oocytes were collected from punctured ovaries and denuded in handling medium supplemented with 100 μM 3‐isobutyl‐1‐methylxanthine (IBMX) (Greaney et al, 2021). Oocytes were then transferred into IBMX‐containing M16 medium (Fig 4C) or washed into IBMX‐free M16 medium to resume meiosis (Appendix Fig S4B).
RNA preparation and microinjection
Microinjection of zygotes was performed similar to previously described for prophase‐arrested oocytes (Wei et al, 2018, 2020; Zhou et al, 2019; Iljas et al, 2020; Subramanian et al, 2020; Greaney et al, 2021). Briefly, cRNA was produced using the mMESSAGE mMACHINE High Yield Capped RNA Transcription Kit (Ambion) using linearised DNA template, as previously verified. Constructs used in this paper were H2B‐RFP (200 ng/μl), H2B‐mClover (200 ng/μl), UtrCH‐mCherry (700 ng/μl) and Pom121‐mScarlet (800 ng/μl).
Zygotes were stabilised onto a holding pipette (inner diameter 15 μm, outer diameter 75 μm, 35° bend; The Pipette Company) through suction. Microinjection needles were pulled from capillary tubes (Harvard Apparatus, GC150F‐15) using a vertical pipette puller (P30 vertical micropipette puller, Sutter Instruments). The microinjection needle was advanced across the zona pellucida and oolemma into the zygote cytoplasm, aided by a brief electrical pulse delivered by an intracellular electrometer (IE59425IA, Warner Instruments). The injected volume was approximately 5% of the zygote volume, delivered using a Pneumatic PicoPump (PV‐820, World Precision Instruments). Oil droplets were delivered in the same manner using organic olive oil. The rate of zygote death following microinjection was consistently below 10%.
Following microinjection, zygotes were transferred into pre‐equilibrated potassium simplex optimisation medium (KSOM) (MR‐107‐D; Merck) under embryo‐tested light mineral oil (Sigma‐Aldrich) at 37°C in 5% CO2 atmosphere for > 4 h to allow protein expression.
Time‐lapse microscopy
KSOM medium was prepared in glass‐bottom dishes (MatTek) with or without a pre‐added inhibitor and overlaid with mineral oil. For experiments involving drug addition during live imaging, a large KSOM droplet of 200 μl was prepared and 2 μl drug was added on the confocal stage without moving the dish. To visualise microtubules, SiR‐Tubulin dye (Cytoskeleton, Inc.) was pre‐added to the medium at a final concentration of 300 nM, a concentration used in similar embryo live imaging studies (Paim & FitzHarris, 2019). To visualise chromosomes and nuclei, SiR‐DNA dye (Cytoskeleton, Inc.) was used at a final concentration of 250 nM, similar to other mouse embryo studies (Levasseur et al, 2019; Scheffler et al, 2021). We favour the SiR dyes for long‐term imaging experiments as they are excited in the far‐red channel, which in combination with low laser power and ultra‐sensitive detection system minimise phototoxicity.
Fluorescence and brightfield images were simultaneously acquired using a Leica TCS SP8 confocal microscope equipped with a 20×/0.75 NA or 40×/1.0 NA Apochromat water‐immersion objective. An automated pump cap (water‐immersion micro dispenser, Leica; automated pump mp‐x controller, Bartels Mikrotechnik) was fitted on the objective to ensure water immersion for extended imaging periods. The dish was enclosed in a stage‐mounted incubation chamber to maintain conditions of 37°C and 5% CO2 in air. Temperature fluctuation was minimised by a stage‐mounted polycarbonate incubator enclosing the entire microscope (Life Imaging Services). The Leica TCS SP8's Super Z Galvo stage allowed ultra‐rapid movement in the z‐plane, and adaptive focus control function was used to auto‐focus throughout extended imaging. Images were acquired every 5–30 min, covering a 50 μm‐Z stack (every 5 μm). Specific imaging interval and duration parameters are specified in the figures.
Drug treatments
Stock solutions of all small‐molecule inhibitors were made in DMSO at 5–50 mM. To depolymerise microtubules, nocodazole (Sigma‐Aldrich, SML1665) was added to the media during imaging at a final concentration of 10 μM. To inhibit actin polymerisation, Cytochalasin D (Sigma‐Aldrich, C8273) and Latrunculin B (Abcam, ab144291) were added to the media at a final concentration of 5 μM. To stabilise microtubule polymerisation, Paclitaxel (Sigma‐Aldrich, T7402) was used at 20 μM. To stabilise actin polymerisation, Jasplakinolide (Cayman Chemicals, 11705) was used at a final concentration of 500 nM. To inhibit secretion and endomembrane trafficking, BFA (Sigma‐Aldrich, B5936) was used at a concentration of 10 μM. To inhibit myosin V, MyoVin‐1 (Calbiochem, 475984) was used at a concentration of 30 μM. To inhibit myosin II, Blebbistatin (Sigma‐Aldrich, B0560) was used at 100 μM. To inhibit the FH2 domain activity, SMIFH2 (Sigma‐Aldrich, S4826) was used at 25 μM. To inhibit the Arp 2/3 complex, CK666 (Enzo Life Sciences, ALX‐270‐506‐M002) was used at 100 μM. To inhibit dynein, Dynarrestin (Sigma‐Aldrich, SML2332) was used at 25 μM. As a control, DMSO, which was used to dissolve all inhibitors, was added to KSOM medium at comparable concentrations to the drugs.
All inhibitors were added during time‐lapse imaging at specific times as indicated in corresponding figure legends to investigate their effects after mitotic exit with the exception of BFA and MyoVin‐1, which were added at the late zygote stage when imaging commenced. For washout experiments described in Fig 2, embryos were washed into fresh KSOM medium before the onset of second cleavage (~ 12 h after drug addition) by transferring them through serial drops of medium.
Immunofluorescence microscopy and fluorescence intensity quantification
Embryos were fixed and stained similar to described previously for oocytes (Wei et al, 2018, 2020; Zhou et al, 2019; Iljas et al, 2020; Subramanian et al, 2020; Greaney et al, 2021). Briefly, 2‐cell embryos were quickly washed in PHEM buffer, pre‐permeabilised in 0.25% Triton‐X in PHEM before being fixed in 3.7% paraformaldehyde solution in PHEM for 20 min. Embryos were then permeabilised by 0.25% Triton‐X in PBS for 10 min at room temperature and blocked in 3% BSA in PBS containing 0.05% Tween‐20 at 4°C overnight. Primary antibodies and concentrations used are as follows: rabbit anti‐lamin (ab108922, Abcam, 1:200) and mouse anti‐β‐Tubulin (T4026, Sigma‐Aldrich, 1:400). Incubation was carried out in ambient temperature for 2 h. Following three 5‐min washes in PBS containing 0.5% BSA and 0.05% Tween‐20, embryos were incubated with Alexa Fluor 488‐conjugated or 546‐conjugated secondary antibodies (ThermoFisher, 1:200) for 2 h at ambient temperature. Alexa Fluor 488 phalloidin (A12379) was incubated with secondary antibodies according to the manufacturer's instructions. After three washes, embryos were incubated in Hoechst 33342 (Thermo Fisher Scientific, H3570, 10 μg/ml) for 10 min to label the DNA. Embryos were then placed in glass‐bottom dishes within 2 μl droplets of PBS with 0.5% BSA under mineral oil for imaging. Images of immunostained embryos were acquired with a 63×/1.20 NA water‐immersion objective. All images were acquired at a pixel dimension of 1,024 × 1,024, covering a 50 μm‐Z stack (every 5 μm).
To estimate microtubule density in the cortex‐ or centre‐facing portion of forming 2‐cell nuclei, we defined a region of interest extending to each side of the nucleus, as shown in Fig 7. Confocal images of fluorescent β‐tubulin staining were used to generate a maximum projection image for each embryo, from which mean grey values were calculated in Fiji and exported to Excel. Background subtraction was performed by subtracting the fluorescence intensity of an equal‐sized region outside the embryo from the corresponding cytoplasmic measurement.
To quantify the relative F‐actin density in cytoplasmic and perinuclear cortex regions (Appendix Fig S4F), the intensity of Alexa Fluor 488 phalloidin was measured from a single z‐slice and in a same‐sized region outside the embryo for background subtraction. The background‐subtracted intensity was then normalised to the mean intensity measured for DMSO‐control cells.
Image analysis and quantifications on nuclear movement
Post‐acquisition image processing was performed in the Leica LAS X software. Movies and images were exported as the merge of maximum projection in the fluorescence channels with a mid‐Z stack plane of the brightfield channel. Only embryos that underwent post‐anaphase nuclear displacement in the horizontal plane were used for analyses.
To generate kymographs of spindles and chromosomes, a time‐lapse movie containing maximally projected SiR‐Tubulin and H2B‐RFP images was exported to Fiji. A line was drawn across the spindle long axis at metaphase, and the fluorescence across this line at each movie frame was plotted against time using the Multi‐kymograph function.
To calculate the distance between nucleus and cell centre (d), the nucleus and cell on a mid‐Z stack plane are delineated using a freehand selection tool in Fiji (Schindelin et al, 2012). Centroid coordinates were determined and exported to Excel. The Euclidean distance between the two coordinates was calculated as follows:
whereby x1 and y1 were the x, y coordinates of the cell and x2 and y2 were the x, y coordinates of the nucleus. Spindle distance to the cell centre was calculated in a similar manner, whereby x2 and y2 were the x, y coordinates of the spindle centroid instead.
Tracking of nuclear movement was done using the Fiji plugin, TrackMate (Tinevez et al, 2017), selecting a LoG detector for H2B‐positive structures of 15 μm diameter, and a threshold value of 1.0 to eliminate spurious spots. After completing automatic spot detection at each time frame of the time‐lapse movie, labelled spots were manually reviewed to ensure only chromosomes/nuclei were detected. The HyperStack displayer was then used to overlay spots and tracks generated using a Linear Assignment Problem (LAP) tracker. The parameters used for the LAP tracker were as follows: 15.0 μm linking max distance, 15.0 μm gap‐closing max distance, 2 gap‐closing max frame gap, track splitting allowed. The tracks were colour‐coded with relative velocity information and exported as an image. Quantitative tracking data were exported in the .xml format for MSD analysis. In MATLAB, the .xml files were analysed using the @msdanalyzer class (https://tinevez.github.io/msdanalyzer/) following the Bioimage data analysis workflows (Tarantino et al, 2014; Tinevez et al, 2017).
Particle‐tracking microrheology of living cells and viscosity calculation
The principles and applications of particle‐tracking microrheology of living cells followed here are detailed in an extensive review and tutorial (Wirtz, 2009; McGlynn et al, 2020).
Following the manufacturer's instructions, 1 μm diameter fluorescent carboxylate modified nanospheres (ThermoFisher, F8823) were prepared by three repeated low‐speed centrifugation and washes to remove traces of sodium azide and diluted 50 times in phosphate buffered saline (ThermoFisher, 10010023). Before microinjection, the bead solution was vortexed for 3 min to avoid aggregates and the microinjection needles were pulled to have larger diameters that allow bead passage.
Using the Leica SP8 resonant scanner (8,000 Hz) and 63×/1.20 NA water‐immersion objective, single‐plane regions of 256 × 256 pixel containing in‐focus fluorescent beads were recorded at 50‐ms intervals for a total duration of 20 s (400 frames total). The embryo was illuminated with the 488 nm laser at 0.1% intensity.
Tracking of bead movements was performed using TrackMate with the following parameters: 1 μm diameter for spot detection, threshold value of 1.0 to eliminate spurious spots, 3 μm max distance and 3 μm gap‐closing max distance. Beads that were out‐of‐focus, aggregated or did not complete an entire recording duration of 20 s were excluded for analysis due to violations to assumptions involved in inferring viscosity from particle movement (McGlynn et al, 2020). To obtain statistical averaging, > 200 beads were tracked to calculate the ensemble‐averaged MSD as a function of lag time:
D is the diffusion coefficient of beads and was estimated from a linear fit on the first 25% (100 data points) of the MSD curve to avoid undersampling at larger values (Michalet, 2010). Subsequently, viscosity of the suspending cytoplasm η is calculated using the Stokes–Einstein equation:
where kb is the Boltzmann's constant, T is the temperature, and a is the radius of the particle.
Statistical analysis
All experiments were repeated ≥ 3 times, using samples from ≥ 3 mice. The number of individual data points is specified in figure legends.
Prism 9 (GraphPad Software Inc) was used to calculate mean, standard error of the mean (SEM) and other descriptive data. The normal distribution of data was first assessed using a D'Agostino and Pearson test. Details of subsequent statistical tests used and sample size are provided in individual figure legends. Investigators were not blinded to treatment groups during experiments. Sample size estimates were not used.
Author contributions
Yunan Ye: Conceptualization; resources; data curation; software; formal analysis; validation; investigation; visualization; methodology; writing – original draft; project administration; writing – review and editing. Hayden A Homer: Conceptualization; resources; software; supervision; funding acquisition; validation; visualization; methodology; writing – original draft; project administration; writing – review and editing.
Declaration of competing interests statement
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Movie EV1
Movie EV2
Movie EV3
Movie EV4
Movie EV5
Movie EV6
Movie EV7
Source Data for Figure 5
Acknowledgements
We are grateful to Jean‐Yves Tinevez (Institut Pasteur, Paris) for helpful discussions and assistance on the implementation of TrackMate and MSD analysis. We thank the staff at UQCCR animal facility for assisting with animal welfare. This work was funded by the Professor Christopher Chen Endowment Fund, Start‐up funding from the Faculty of Medicine, University of Queensland and National Health and Medical Research Council Project Grants APP1078134, APP1103689 and APP1122484 to HH. YY was supported by the University of Queensland Research Training Stipend. Open access publishing facilitated by The University of Queensland, as part of the Wiley ‐ The University of Queensland agreement via the Council of Australian University Librarians.
EMBO reports (2022) 23: e55251
Data availability
This study includes no data deposited in external repositories.
References
- Ahmed WW, Fodor E, Almonacid M, Bussonnier M, Verlhac MH, Gov N, Visco P, van Wijland F, Betz T (2018) Active mechanics reveal molecular‐scale force kinetics in living oocytes. Biophys J 114: 1667–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almonacid M, Ahmed WW, Bussonnier M, Mailly P, Betz T, Voituriez R, Gov NS, Verlhac MH (2015) Active diffusion positions the nucleus in mouse oocytes. Nat Cell Biol 17: 470–479 [DOI] [PubMed] [Google Scholar]
- Almonacid M, Al Jord A, El‐Hayek S, Othmani A, Coulpier F, Lemoine S, Miyamoto K, Grosse R, Klein C, Piolot T et al (2019) Active fluctuations of the nuclear envelope shape the transcriptional dynamics in oocytes. Dev Cell 51: 145–157.e10 [DOI] [PubMed] [Google Scholar]
- Almonacid M, Terret ME, Verlhac MH (2014) Actin‐based spindle positioning: new insights from female gametes. J Cell Sci 127: 477–483 [DOI] [PubMed] [Google Scholar]
- Bovellan M, Romeo Y, Biro M, Boden A, Chugh P, Yonis A, Vaghela M, Fritzsche M, Moulding D, Thorogate R et al (2014) Cellular control of cortical actin nucleation. Curr Biol 24: 1628–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet S, Verlhac MH (2011) Positioning to get out of meiosis: the asymmetry of division. Hum Reprod Update 17: 68–75 [DOI] [PubMed] [Google Scholar]
- Cartagena‐Rivera AX, Logue JS, Waterman CM, Chadwick RS (2016) Actomyosin cortical mechanical properties in nonadherent cells determined by atomic force microscopy. Biophys J 110: 2528–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaigne A, Campillo C, Voituriez R, Gov NS, Sykes C, Verlhac MH, Terret ME (2016) F‐actin mechanics control spindle centring in the mouse zygote. Nat Commun 7: 10253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugh P, Clark AG, Smith MB, Cassani DAD, Dierkes K, Ragab A, Roux PP, Charras G, Salbreux G, Paluch EK (2017) Actin cortex architecture regulates cell surface tension. Nat Cell Biol 19: 689–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clift D, Schuh M (2013) Restarting life: fertilization and the transition from meiosis to mitosis. Nat Rev Mol Cell Biol 14: 549–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin A, Letort G, Razin N, Almonacid M, Ahmed W, Betz T, Terret ME, Gov NS, Voituriez R, Gueroui Z et al (2020) Active diffusion in oocytes nonspecifically centers large objects during prophase I and meiosis I. J Cell Biol 219: e201908195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois A, Schuh M, Ellenberg J, Hiiragi T (2012) The transition from meiotic to mitotic spindle assembly is gradual during early mammalian development. J Cell Biol 198: 357–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels BR, Hale CM, Khatau SB, Kusuma S, Dobrowsky TM, Gerecht S, Wirtz D (2010) Differences in the microrheology of human embryonic stem cells and human induced pluripotent stem cells. Biophys J 99: 3563–3570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels BR, Masi BC, Wirtz D (2006) Probing single‐cell micromechanics in vivo: the microrheology of C. elegans developing embryos. Biophys J 90: 4712–4719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai N, Goldberg JM, Austin C, Falcone T (2018) Are cleavage anomalies, multinucleation, or specific cell cycle kinetics observed with time‐lapse imaging predictive of embryo developmental capacity or ploidy? Fertil Steril 109: 665–674 [DOI] [PubMed] [Google Scholar]
- Deshpande O, Telley IA (2021) Nuclear positioning during development: pushing, pulling and flowing. Semin Cell Dev Biol 120: 10–21 [DOI] [PubMed] [Google Scholar]
- Fakhri N, Wessel AD, Willms C, Pasquali M, Klopfenstein DR, MacKintosh FC, Schmidt CF (2014) High‐resolution mapping of intracellular fluctuations using carbon nanotubes. Science 344: 1031–1035 [DOI] [PubMed] [Google Scholar]
- Greaney J, Subramanian GN, Ye Y, Homer H (2021) Isolation and in vitro culture of mouse oocytes. Bio Protoc 11: e4104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen GG, Worman HJ (2013) Nuclear positioning. Cell 152: 1376–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardarson T, Hanson C, Sjogren A, Lundin K (2001) Human embryos with unevenly sized blastomeres have lower pregnancy and implantation rates: Indications for aneuploidy and multinucleation. Hum Reprod 16: 313–318 [DOI] [PubMed] [Google Scholar]
- Hasley A, Chavez S, Danilchik M, Wuhr M, Pelegri F (2017) Vertebrate embryonic cleavage pattern determination. Adv Exp Med Biol 953: 117–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetrick B, Han MS, Helgeson LA, Nolen BJ (2013) Small molecules CK‐666 and CK‐869 inhibit actin‐related protein 2/3 complex by blocking an activating conformational change. Chem Biol 20: 701–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoing S, Yeh TY, Baumann M, Martinez NE, Habenberger P, Kremer L, Drexler HCA, Kuchler P, Reinhardt P, Choidas A et al (2018) Dynarrestin, a novel inhibitor of cytoplasmic dynein. Cell Chem Biol 25: 357–369.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holubcova Z, Howard G, Schuh M (2013) Vesicles modulate an actin network for asymmetric spindle positioning. Nat Cell Biol 15: 937–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde KJ, Schust DJ (2015) Genetic considerations in recurrent pregnancy loss. Cold Spring Harb Perspect Med 5: a023119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ierushalmi N, Malik‐Garbi M, Manhart A, Abu Shah E, Goode BL, Mogilner A, Keren K (2020) Centering and symmetry breaking in confined contracting actomyosin networks. Elife 9: e55368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iljas JD, Wei Z, Homer HA (2020) Sirt1 sustains female fertility by slowing age‐related decline in oocyte quality required for post‐fertilization embryo development. Aging Cell 19: e13204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illukkumbura R, Bland T, Goehring NW (2020) Patterning and polarization of cells by intracellular flows. Curr Opin Cell Biol 62: 123–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam K, Chin HF, Olivares AO, Saunders LP, De La Cruz EM, Kapoor TM (2010) A myosin V inhibitor based on privileged chemical scaffolds. Angew Chem Int Ed Engl 49: 8484–8488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri D, Brugiere T, Athale CA, Delattre M (2022) Evolutionary divergence of anaphase spindle mechanics in nematode embryos constrained by antagonistic pulling and viscous forces. Mol Biol Cell 33: ar61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kole TP, Tseng Y, Huang L, Katz JL, Wirtz D (2004) Rho kinase regulates the intracellular micromechanical response of adherent cells to rho activation. Mol Biol Cell 15: 3475–3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolega J (2004) Phototoxicity and photoinactivation of blebbistatin in UV and visible light. Biochem Biophys Res Commun 320: 1020–1025 [DOI] [PubMed] [Google Scholar]
- Levasseur MD, Thomas C, Davies OR, Higgins JMG, Madgwick S (2019) Aneuploidy in oocytes is prevented by sustained CDK1 activity through degron masking in cyclin B1. Dev Cell 48: 672–684.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukinavicius G, Mitronova GY, Schnorrenberg S, Butkevich AN, Barthel H, Belov VN, Hell SW (2018) Fluorescent dyes and probes for super‐resolution microscopy of microtubules and tracheoles in living cells and tissues. Chem Sci 9: 3324–3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton GW, Gomes ER, Folker ES, Vintinner E, Gundersen GG (2010) Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science 329: 956–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macklon NS, Geraedts JP, Fauser BC (2002) Conception to ongoing pregnancy: the ‘black box’ of early pregnancy loss. Hum Reprod Update 8: 333–343 [DOI] [PubMed] [Google Scholar]
- McGlynn JA, Wu N, Schultz KM (2020) Multiple particle tracking microrheological characterization: fundamentals, emerging techniques and applications. J Appl Phys 127: 201101 [Google Scholar]
- Michalet X (2010) Mean square displacement analysis of single‐particle trajectories with localization error: brownian motion in an isotropic medium. Phys Rev E Stat Nonlin Soft Matter Phys 82: 041914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogessie B, Schuh M (2017) Actin protects mammalian eggs against chromosome segregation errors. Science 357: eaal1647 [DOI] [PubMed] [Google Scholar]
- Munro E, Nance J, Priess JR (2004) Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior‐posterior polarity in the early C. elegans embryo. Dev Cell 7: 413–424 [DOI] [PubMed] [Google Scholar]
- O'Connell CB, Wang YL (2000) Mammalian spindle orientation and position respond to changes in cell shape in a dynein‐dependent fashion. Mol Biol Cell 11: 1765–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paim LMG, FitzHarris G (2019) Tetraploidy causes chromosomal instability in acentriolar mouse embryos. Nat Commun 10: 4834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauerova T, Radonova L, Kovacovicova K, Novakova L, Skultety M, Anger M (2020) Aneuploidy during the onset of mouse embryo development. Reproduction 160: 773–782 [DOI] [PubMed] [Google Scholar]
- Racowsky C, Stern JE, Gibbons WE, Behr B, Pomeroy KO, Biggers JD (2011) National collection of embryo morphology data into Society for Assisted Reproductive Technology Clinic Outcomes Reporting System: Associations among day 3 cell number, fragmentation and blastomere asymmetry, and live birth rate. Fertil Steril 95: 1985–1989 [DOI] [PubMed] [Google Scholar]
- Reichmann J, Nijmeijer B, Hossain MJ, Eguren M, Schneider I, Politi AZ, Roberti MJ, Hufnagel L, Hiiragi T, Ellenberg J (2018) Dual‐spindle formation in zygotes keeps parental genomes apart in early mammalian embryos. Science 361: 189–193 [DOI] [PubMed] [Google Scholar]
- Rizvi SA, Neidt EM, Cui J, Feiger Z, Skau CT, Gardel ML, Kozmin SA, Kovar DR (2009) Identification and characterization of a small molecule inhibitor of formin‐mediated actin assembly. Chem Biol 16: 1158–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto R, Tanabe M, Hiraiwa T, Suzuki K, Ishiwata S, Maeda YT, Miyazaki M (2020) Tug‐of‐war between actomyosin‐driven antagonistic forces determines the positioning symmetry in cell‐sized confinement. Nat Commun 11: 3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Limouze J, Combs CA, Straight AF, Sellers JR (2005) Blebbistatin, a myosin II inhibitor, is photoinactivated by blue light. Biochemistry 44: 584–588 [DOI] [PubMed] [Google Scholar]
- Scheffler K, Uraji J, Jentoft I, Cavazza T, Monnich E, Mogessie B, Schuh M (2021) Two mechanisms drive pronuclear migration in mouse zygotes. Nat Commun 12: 841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda‐Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B et al (2012) Fiji: an open‐source platform for biological‐image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider I, Ellenberg J (2019) Mysteries in embryonic development: how can errors arise so frequently at the beginning of mammalian life? PLoS Biol 17: e3000173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian GN, Greaney J, Wei Z, Becherel O, Lavin M, Homer HA (2020) Oocytes mount a noncanonical DNA damage response involving APC‐Cdh1‐mediated proteolysis. J Cell Biol 219: e201907213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarantino N, Tinevez JY, Crowell EF, Boisson B, Henriques R, Mhlanga M, Agou F, Israel A, Laplantine E (2014) TNF and IL‐1 exhibit distinct ubiquitin requirements for inducing NEMO‐IKK supramolecular structures. J Cell Biol 204: 231–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinevez JY, Perry N, Schindelin J, Hoopes GM, Reynolds GD, Laplantine E, Bednarek SY, Shorte SL, Eliceiri KW (2017) TrackMate: An open and extensible platform for single‐particle tracking. Methods 115: 80–90 [DOI] [PubMed] [Google Scholar]
- van Heesbeen RG, Tanenbaum ME, Medema RH (2014) Balanced activity of three mitotic motors is required for bipolar spindle assembly and chromosome segregation. Cell Rep 8: 948–956 [DOI] [PubMed] [Google Scholar]
- Vera‐Rodriguez M, Chavez SL, Rubio C, Reijo Pera RA, Simon C (2015) Prediction model for aneuploidy in early human embryo development revealed by single‐cell analysis. Nat Commun 6: 7601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang ZB, Zhang X, FitzHarris G, Baltz JM, Sun QY, Liu XJ (2008) Brefeldin a disrupts asymmetric spindle positioning in mouse oocytes. Dev Biol 313: 155–166 [DOI] [PubMed] [Google Scholar]
- Wei Z, Greaney J, Loh WN, Homer HA (2020) Nampt‐mediated spindle sizing secures a post‐anaphase increase in spindle speed required for extreme asymmetry. Nat Commun 11: 3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Greaney J, Zhou C, Homer HA (2018) Cdk1 inactivation induces post‐anaphase‐onset spindle migration and membrane protrusion required for extreme asymmetry in mouse oocytes. Nat Commun 9: 4029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel AD, Gumalla M, Grosshans J, Schmidt CF (2015) The mechanical properties of early Drosophila embryos measured by high‐speed video microrheology. Biophys J 108: 1899–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz D (2009) Particle‐tracking microrheology of living cells: principles and applications. Annu Rev Biophys 38: 301–326 [DOI] [PubMed] [Google Scholar]
- Wuhr M, Tan ES, Parker SK, Detrich HW 3rd, Mitchison TJ (2010) A model for cleavage plane determination in early amphibian and fish embryos. Curr Biol 20: 2040–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata K, FitzHarris G (2013) 4D imaging reveals a shift in chromosome segregation dynamics during mouse pre‐implantation development. Cell Cycle 12: 157–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Skibbens RV, Cheng JW, Salmon ED, Bloom K (1995) Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J Cell Biol 130: 687–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi K, Li R (2012) Actin cytoskeleton in cell polarity and asymmetric division during mouse oocyte maturation. Cytoskeleton 69: 727–737 [DOI] [PubMed] [Google Scholar]
- Yi K, Unruh JR, Deng M, Slaughter BD, Rubinstein B, Li R (2011) Dynamic maintenance of asymmetric meiotic spindle position through Arp2/3‐complex‐driven cytoplasmic streaming in mouse oocytes. Nat Cell Biol 13: 1252–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan Q, Ye Z, Clarke R, Rosenwaks Z, Zaninovic N (2016) Direct unequal cleavages: embryo developmental competence, genetic constitution and clinical outcome. PLoS One 11: e0166398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Hancock JL, Khanna KK, Homer HA (2019) First meiotic anaphase requires Cep55‐dependent inhibitory Cdk1 phosphorylation. J Cell Sci 132: jcs233379 [DOI] [PubMed] [Google Scholar]


