Abstract
Inflammation is an essential process of host defense against infections, illness, or tissue damage. Polymorphonuclear neutrophils (PMN) are among the first immune cells involved in acute inflammatory responses and are on the front line in the fight against bacterial infections. In the presence of bacterial fragments, PMN release inflammatory mediators, enzymes, and microvesicles in the extracellular milieu to recruit additional immune cells required to eliminate the pathogens. Recent evidence shows that platelets (PLTs), initially described for their role in coagulation, are involved in inflammatory responses. Furthermore, upon activation, PLT also release functional mitochondria (freeMitos) within their extracellular milieu. Mitochondria share characteristics with bacterial and mitochondrial damage‐associated molecular patterns, which are important contributors in sterile inflammation processes. Deep sequencing transcriptome analysis demonstrates that freeMitos increase the mitochondrial gene expression in PMN. However, freeMitos do not affect the mitochondrial‐dependent increase in oxygen consumption in PMN. Interestingly, freeMitos significantly induce the release of PMN‐derived microvesicles. This study provides new insight into the role of freeMitos in the context of sterile inflammation.
Keywords: extracellular vesicles, freeMitos, sterile inflammation
Subject Categories: Immunology, Organelles, Signal Transduction
Mitochondria are better appreciated for their role in bioenergy production; however, they are active participants in sterile inflammation. This study reports the transcriptomic profile of human neutrophils modulated by extracellular mitochondria (freeMitos).

Introduction
Platelets are small mitochondria‐containing anucleate blood cells that maintain homeostasis by preventing blood loss and promoting wound repair (Davì & Patrono, 2007). When exposed to physiological agonists, platelets also release small cytoplasmic extensions known as platelet‐derived microvesicles into the extracellular milieu. In addition to the production of microvesicles, activated platelets also shed fully functional mitochondria into their extracellular milieu, known as freeMitos(Boudreau et al, 2014; Marcoux et al, 2017). Consequently, freeMitos and mitochondrial function contribute to the inflammatory response (Puhm et al, 2021).
While mitochondria actively participate in the cell's bioenergy production (i.e., adenosine triphosphate (ATP) generation; Saraste, 1999), they are also involved in immune responses. In fact, when mitochondrial components are released in the extracellular milieu, they elicit damage‐associated molecular patterns (DAMPs) and trigger inflammatory responses, which involve immune cells (Zhang et al, 2010; Boudreau et al, 2014). On the other hand, the inflammatory response is also initiated by polymorphonuclear leukocyte (PMN) activation by pathogen‐associated molecular patterns (PAMPs) expressed by foreign and invading microorganisms. Therefore, PMN are widely considered as the first line of defense against pathogens and important players of innate immunity (Nicolás‐Ávila et al, 2017). In addition to their well‐known phagocytosis and netosis functions, PMN also release microvesicles in the presence of physiological agonists or bacterial components, such as N‐formyl‐methionyl‐leucyl‐phenylalanine (fMLP; Hong et al, 2012) and lipopolysaccharide (Pluskota et al, 2008). While underappreciated in comparison to other cell types, microvesicles of PMN origin are found elevated in circulation of patients with cardiovascular diseases (Sionis et al, 2018) and contribute to the disease by delivering bioactive content such as microRNAs to endothelial cells (Gomez et al, 2020). Mechanistically, pattern recognition receptors, such as formyl peptide receptors (FPRs) and toll‐like receptors on immune cells, recognize PAMPs and DAMPs, which also include mitochondrial components (Gray et al, 1999), during cellular trauma in the context of sterile inflammation (McDonald et al, 2010; Zhang et al, 2010; Zindel & Kubes, 2020). Given that mitochondria encompass many biochemical hallmarks reminiscent of bacteria such as unmethylated CpG circular DNA (Zhang et al, 2016), N‐formylated peptides (Pittman & Kubes, 2013) and inner membrane‐associated cardiolipin (Schlame, 2008), we hypothesized that freeMitos represent a new mechanism of PMN activation and inflammatory response in the context of sterile inflammation.
In this study, we investigate the potential of freeMitos in triggering PMN‐initiated inflammatory responses in comparison to a mitochondrial DAMPs preparation. Our results show that freeMitos induced PMN intracellular calcium mobilization, activated the calpain pathway, and subsequently induced the release of PMN‐derived microvesicles (PMN‐MVs).
Results and Discussion
Platelet‐derived freeMitos associate with PMN and modulate gene expression
The first series of experiments showed using flow cytometry that freeMitos interacted with PMN (Fig 1A and B) with confirmation by confocal microscopy where freeMitos were either found at the surface or localized in the cytosol of PMN (Fig 1C and Movie EV1). No differences were observed between resting or activated PMN.
Figure 1. Modulatory effects of freeMitos on human polymorphonuclear leukocytes.
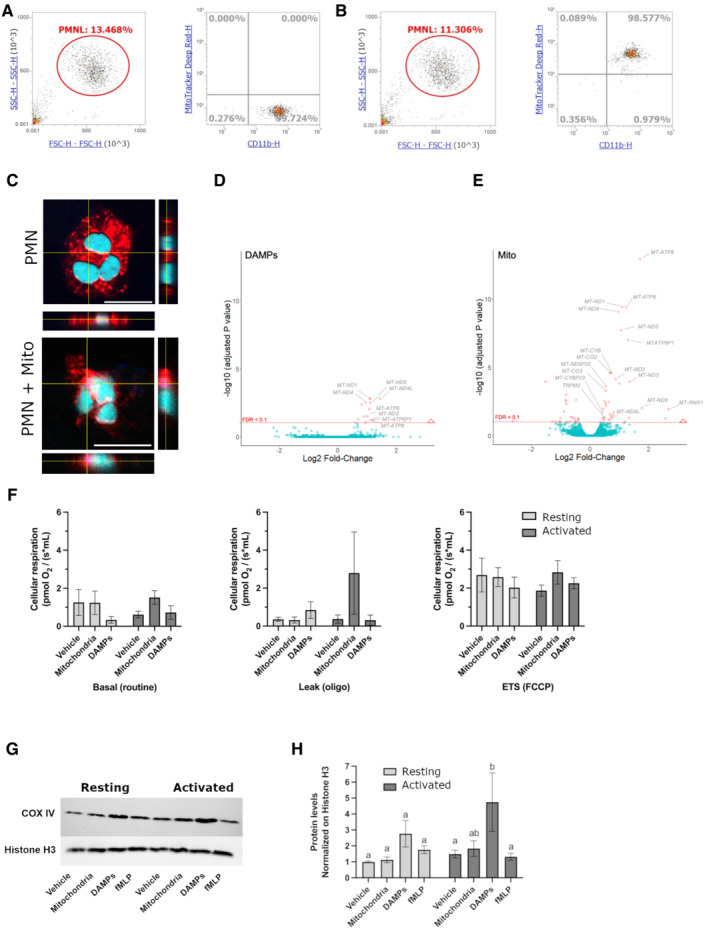
-
A, BFunctional freeMitos association with PMN. Overlay dot plot population, representative of four biological replicates of the flow cytometry gating strategy used to quantify the interaction between PMN (anti‐CD11b‐PE) and freeMitos. Freshly isolated platelets were labeled with MitoTracker™ Deep Red and freeMitos isolation was subsequently performed. CD11b‐labeled PMN were incubated in absence (A) or presence (B) of freeMitos at a 1:5 ratio (PMN:mitochondria).
-
CConfocal microscopy imaging of PMN cell membrane (red) labeled with CellMask Orange in absence (upper panel) or presence (lower panel) of MitoTracker™ Deep Red‐labeled freeMitos (magenta). PMN nuclei were stained with DAPI (cyan). The white scale bars shown in the lower right of the images represent 10 μm in size.
-
D, EFreeMitos associate with PMN and modulate gene expression. Volcano plot, of four biological replicates, representing the gene expression modulation induced by mitochondrial DAMPs preparation (D) and freshly isolated functional freeMitos (E) in PMN.
-
F–HFreeMitos effect on the respiratory state of PMN. O2 consumption rates of resting or activated (incubated with TNF‐a and GM‐CSF). PMN were measured using high‐resolution O2 respirometry (Oroboros Oxygraph) and revealed no significant differences between the basal state (left panel), leak state (middle panel), and the maximum electron transport system capacity (ETS, right panel) of cellular respiration (F). Immunoblot of mitochondrial cytochrome c oxidase (COX IV) and histone H3 as a loading control of PMN after co‐incubations with freeMitos (G) with subsequent densitometry analysis (H).
Data information: For panels (F) and (H), data are shown as means ± SEM, of three biological replicates. Two‐way ANOVA tests followed by Tukey's multiple comparisons tests for each sample was performed for panels (F) and (H). Values without at least one common superscript letter are different (P < 0.05).
Source data are available online for this figure.
Deep RNA sequencing showed that DAMPs only modulated 15 genes (Fig 1D), all of which were upregulated (Appendix Tables S1 and S2, Fig EV1), whereas freeMitos (Fig 1E) modulated a total of 61 genes in PMN (37 upregulated and 24 downregulated). Interestingly, several mitochondrial genes were significantly upregulated by both treatments. Therefore, PMN mitochondrial function was evaluated under these conditions through the analysis of the cell's respiratory state.
Figure EV1. Validation of the transcriptomic analysis by qPCR.
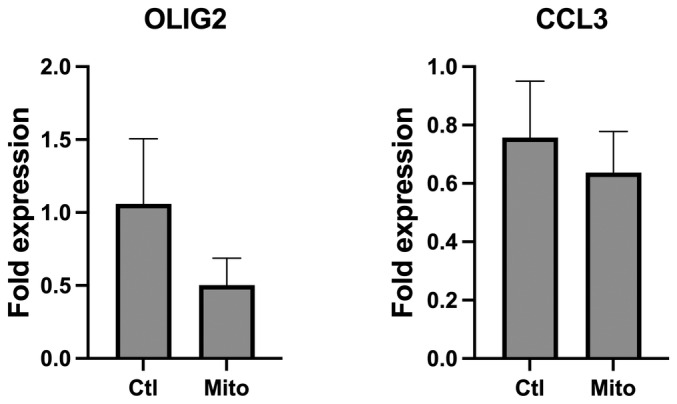
Transcript levels of oligodendrocyte transcription factor 2 (OLIG2) and the C–C motif chemokine ligand 3 (CCL3) in PMNL incubated with platelet‐derived mitochondria (Mito) or absence (Ctl) were assessed by qRT‐PCR. Results are expressed as the mean ± SEM of three biological replicates, each performed in three technical replicates. A paired sample t‐test was performed to determine P‐values of 0.1711 (OLIG2) and 0.4504 (CCL3).
Effect of freeMitos on the respiratory state of PMN
In the next set of experiments, we used both resting PMN, to simulate their circulating state, or activated PMN, to replicate an inflammatory milieu. Since PMN are known to have limited mitochondrial respiratory activity (Chacko et al, 2013), co‐incubation with freeMitos could increase the mitochondrial respiratory capacity of the PMN via an internalization process. Therefore, basal respiration, leak respiration, and maximal electron transport system (ETS) capacity were measured to evaluate mitochondrial oxygen consumption (Fig 1F). While PMN respiration rates were not significantly affected by the co‐incubations with either freeMitos or DAMPs, it is interesting to underline that non‐significant increases in both the basal respiration rate (2.5‐fold, P = 0.0922) and the leak respiration state (fourfold, P = 0.3249) were measured in activated PMN co‐incubated with freeMitos compared to activated PMN incubated with the vehicle. Similarly, the expression of cytochrome c oxidase subunit 4 (COX‐IV), the final enzyme in the respiratory electron transport chain, was not affected in PMN treated with freeMitos, whereas DAMPs treatments induced a 4.8‐fold increase in COX IV levels when compared to vehicle control (Fig 1G and H). These results indicate that despite their uptake by PMN, freeMitos do not modulate the oxygen consumption rate (OCR) of PMN, further supporting evidence that freeMitos may not convey respiratory competent mitochondria to recipient cells (Al Amir Dache et al, 2020; Stier, 2021).
FreeMitos induce intracellular calcium release in PMN
Having shown that internalization of freeMitos and the upregulation of mitochondrial genes did not affect the OCR of PMN, we next examined the ability of freeMitos to modulate other physiological functions of PMN. As shown in Appendix Table S2, the transient receptor potential melastatin 2 (TRPM2) protein, involved in PMN calcium influx signaling(Sumoza‐Toledo & Penner, 2011), was significantly upregulated in PMN treated with freeMitos compared to untreated controls. Therefore, we investigated whether freshly isolated freeMitos induced the release of intracellular calcium in a similar way as DAMPs, a known inducer intracellular calcium influx of PMN via FPR1 and FPR2 (Hauser et al, 2000; Forsman & Dahlgren, 2010; Zhang et al, 2010; Pittman & Kubes, 2013). Mobilization of intracellular calcium in PMN was induced by freeMitos, detectable as early as 5‐s post‐incubation (Fig 2A). Functional FreeMitos and DAMPs showed similar fluorescent signals, with 44.7 and 44.3%, respectively, when compared to the vehicle (HBSS; Fig 2B). Further studies will be required to confirm the mechanism by which freeMitos trigger the intracellular calcium influx and whether cell surface receptors are the same as those activated by DAMPs.
Figure 2. Modulation of the inflammatory state of PMN by freeMitos.
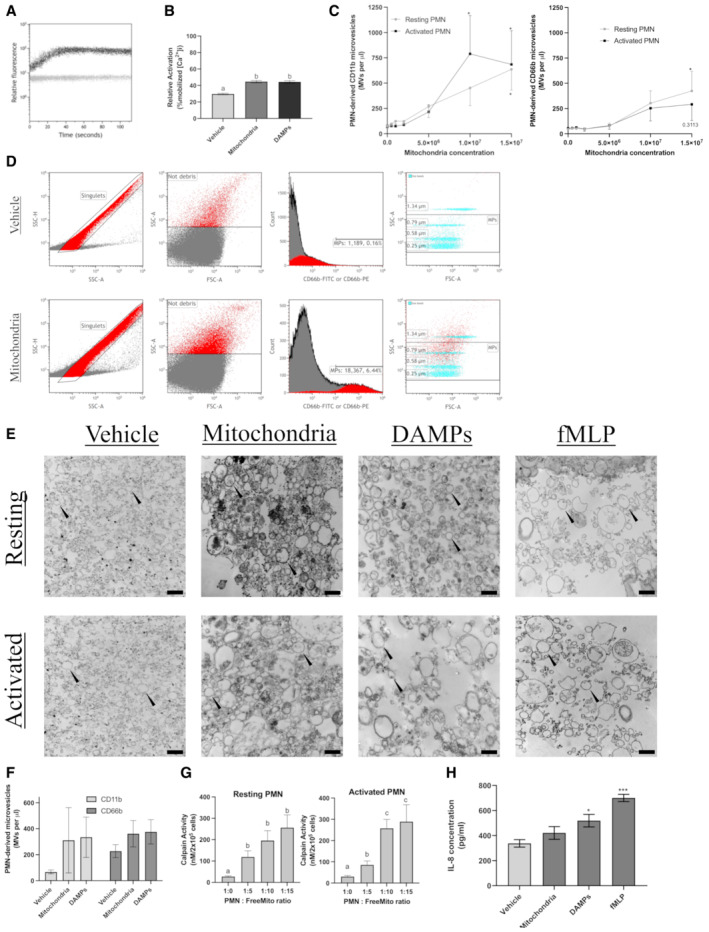
-
A, BFreeMitos induce PMN intracellular calcium release in PMN. Representative visualization of intracellular calcium monitoring by flow cytometry of 1 x 106 PMN co‐incubated with 5 × 106 mitochondria (A). Calcium mobilization was monitored by flow cytometry for 120 s using the fluorescent probe Fluo‐3AM. The gray line represents PMN with the vehicle (HBSS), whereas the dark line represents PMN incubated with mitochondria. PMN intracellular calcium mobilization induced by either the vehicle (HBSS), freeMitos or mitochondrial DAMPs and quantified by flow cytometry (B).
-
C–FFunctional freeMitos induce the release of PMN‐derived microvesicles. A dose–response analysis of various mitochondria to PMN ratios was performed to measure the release of PMN microvesicles. PMN microvesicles labeling was performed with either anti‐CD66b‐FITC labeling (C, left panel) or anti‐CD11b‐PE (C, right panel). PMN‐derived microparticle gating strategy is shown in panel D. Flow cytometry panel established to distinguish the PMNL‐MVs from the background using the leukocyte‐specific integrin alpha‐M beta‐2 (CD11b), the granulocyte‐specific carcinoembryonic antigen‐related cell adhesion molecule 8 (CD66b), and size reference beads. PMN were either incubated with the vehicle (Tyrode's Buffer, upper panels) or freeMitos (lower panels). Following co‐incubation, PMN were washed twice and resuspended in HBSS before processing on flow cytometer. Only single events, identified by the singlets gate (left upper and lower panels) using side scatter area (SSC‐A) versus side scatter heights (SSC‐H) were analyzed. Light scatter (side scatter (SSC) and forward scatter (FSC)) approach for determining the background noise in HBSS. Histogram displaying the fluorescence intensity of samples labeled with either CD11b‐FITC or CD66b‐PE. Overlay dot plot of PMNL‐MVs population and size reference beads, representative of four biological replicates, each performed in two technical replicates.
-
EVisual confirmation of PMN and PMN‐derived microvesicles. Transmission electron microscopy (TEM) imaging of PMN‐derived microvesicles (black arrowhead), representative of three biological replicates, from resting or activated PMN (106 cells) incubated with either mitochondria, mitochondrial DAMPs or fMLP as positive control. The black scale bars are shown in the lower right of the images for the vehicles (400 nm), mitochondria (600 nm), DAMPs (600 nm), and fMLP (500 nm) conditions.
-
FQuantification of resting PMN microvesicles produced by PMN (106 cells) incubated with 107 freeMitos or 25 μg of mitochondrial DAMPs for 2 h.
-
GQuantification of calpain activity, by fluorescence assay, in cell lysate of either resting (left panel) or activated (right panel) PMN (2 × 105 cells) incubated in presence of freeMitos for 2 h.
-
HELISA quantification of IL‐8 concentrations present in the supernatant following the incubation of PMN (106 cells) with freeMitos (107), DAMPs (25 μg) or fMLP (1 μM) for 2 h.
Data information: Data are shown as means ± SEM of three biological replicates (panels B, F, and H) or four biological replicates (panels C and G). One‐way ANOVA tests, followed by Tukey's multiple comparisons tests were performed for panels (B), (F), and (G). Values without one common superscript letter were different (P < 0.05). One‐way (panel H) or two‐way (panel C) ANOVA tests, followed by Dunnett's multiple comparisons test were performed and compared to the vehicle (HBSS); *P < 0.05, **P < 0.01 and ***P < 0.001.
FreeMitos induce the release of PMN‐derived microvesicles
The release of microvesicles from activated cells can be triggered by different stimuli, and in all cases, the cascade requires an increase in the intracellular calcium concentration. Since bacteria induce the release of cell‐derived microvesicles (Timár et al, 2013), we hypothesized that functional freeMitos, given their ancestral bacterial origin (Thrash et al, 2011) and their ability to increase intracellular calcium concentrations, could trigger the release of PMN‐MVs. Using a custom set of flow cytometry panels (Fig 2C and D ), we found that freeMitos induced the release of PMN‐MVs when compared to the vehicle and to the same extent as DAMPs (Fig 2E and F). Of interest, PMN‐MVs size was highly dependent on the condition as the vehicle and freeMitos condition (Fig EV2A and B) had smaller vesicles than the DAMPs and fMLP conditions (Fig EV2C and D). To further support these observations, we found that freeMitos triggered the activation of calpains, a well‐known calcium dependent pathway for the generation of microvesicles, in both resting and activated PMN. Of interest, the calpain's activity in activated PMN (Fox et al, 1991; Zaldivia et al, 2017), but not resting PMN, was detected in the PMN:freeMitos ratio of 1:5 (Fig 2G). This could indicate that activated PMN are more susceptible to release extracellular vesicles under inflammatory conditions, while limiting their ability to release their cargo in the circulation. Since the production of cell‐derived microvesicles is highly dependent on the inducing physiological agonists (Perez‐Pujol et al, 2007; Connor et al, 2010; Boudreau et al, 2014), investigating the bioactive content of PMN‐MVs initiated by freeMitos could provide further insight on the role of PMN‐MVs in the context of sterile inflammation. Finally, we evaluated the capacity of freeMitos to modulate the innate response through PMN production of IL‐8. Interestingly, no changes in IL‐8 levels were detected in PMN treated with freeMitos in contrast to both fMLP and DAMPs, which are known CXCL8 inducers in PMN (Zhang et al, 2010; Fig 2H).
Figure EV2. Sizing of the vesicles released from PMN.
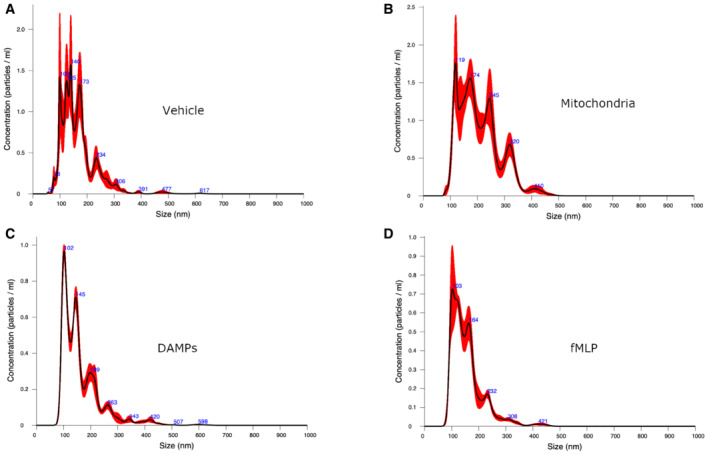
-
A–DPMN‐MVs sizing by nanoparticles tracking analysis (Nanosight) of the supernatant of PMN incubated with either the vehicle (HBSS, panel A), platelet‐derived mitochondria (panel B), platelet‐derived DAMPs (panel C) or fMLP (panel D). Data shown are representative of three biological replicates. The size distribution and concentration of MVs from PMN supernatant were measured using NanosightNS300 (Malvern Panalytical). Sample were prepared for nanoparticle tracking analysis by diluting the stock material in particle‐free water until the concentration was between 1 × 108 and 1 × 109 particles/ml and six videos of 30 s were captured. At least two different dilutions resulting from the stock concentration were analyzed for each sample. The screen gain was set to 10 and the camera level to 16. After capture, the videos were analyzed by the Nanosight Software v3.2 with a detection threshold of 5.
We demonstrated that PMN‐MVs generation was an important process initiated by functional platelet‐derived mitochondria as well as mitochondrial DAMPs. In the current study, we obtained functional freeMitos directly from lysed platelets. While these mitochondria exhibited the same hallmarks of mitochondria released by thrombin activated platelets, there will, nevertheless, be a need to characterize the integrity and bioactive content of mitochondria released from activated platelets in subsequent in vivo studies. While both freeMitos and mitochondrial DAMPs induce similar microparticle release responses in PMN, it is important to consider that both are abundantly present in circulation (Zhang et al, 2010; Al Amir Dache et al, 2020) and their impact on promoting sterile inflammation should be studied individually and/or synergically. The bioactive content of cell‐derived microvesicles is highly dependent on the physiological agonists(Perez‐Pujol et al, 2007; Connor et al, 2010; Brown & McIntyre, 2011; Aatonen et al, 2012), therefore, investigating the specific content found in PMN‐MVs induced by extracellular mitochondria could provide important insight on their role as cargo carriers for intercellular communication under sterile inflammation conditions.
Of interest, a recent study by Al Amir Dache and colleagues further suggests the presence of circulating cell‐free functional mitochondria (Al Amir Dache et al, 2020), thus demonstrating the importance of investigating the interactions and outcomes between respiratory competent mitochondria and immune cells. While cell‐free mitochondria did not transfer their respiratory competent phenotype to PMN in this study, other components within the freeMito organelle may be involved in the initiation of sterile inflammation and release of PMN‐MVs following freeMito/PMN interaction. For instance, it was recently reported that protein markers of PMN origin were significantly enriched in extracellular vesicles present in the synovial fluid of rheumatoid arthritis patients (Foers et al, 2020). The presence of freeMitos in the synovial fluid of rheumatoid patients (Boudreau et al, 2014) could contribute to this increase in PMN‐MVs in vivo, but will require additional investigation to confirm their impact in clinical settings.
Materials and Methods
Isolation of platelet‐derived mitochondria
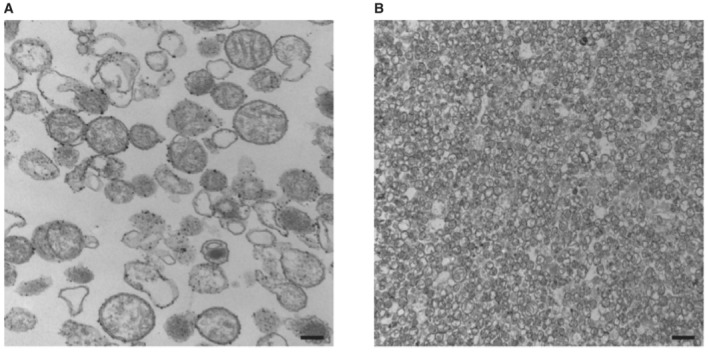
Whole blood was collected from healthy donors in tubes containing the anticoagulant acid citrate dextrose (ACD) and then centrifuged at 275 g for 15 min at room temperature (RT). Platelet‐rich plasma (PRP) was then centrifuged at 400 g for 2 min at RT to remove the contaminating erythrocytes and the supernatant was subsequently centrifuged at 1,300 g for 10 min at RT to pellet the platelets. The cells were then resuspended in Isolation Buffer (IB; sucrose 0.2 M, Tris 11 mM, EDTA 1 mM, pH 7.5) and platelet‐derived mitochondria were isolated as previously described (Fukami & Salganicoff, 1973; Léger et al, 2020). Briefly, platelets, in IB buffer, were homogenized using a Teflon homogenizer with 150 μg/ml of proteinase K (Sigma‐Aldrich). Protease inhibitors (Roche) were added following the procedures of the manufacturer, then the cell lysate was centrifuged at 1,300 g for 10 min at 4°C to pellet remaining platelets and cell debris. The supernatant was collected and centrifuged at 8,000 g for 10 min at 4°C to pellet the crude mitochondrial extract. Platelet membrane debris was removed using a discontinuous sucrose gradient with a density shift using Percoll (GE Healthcare) as previously described (Léger et al, 2020). Briefly, the crude mitochondrial extract in IB was carefully layered on a 15% Percoll layer (15% Percoll, 10% sucrose 2.5 M, 75% IB) in a 1.5 ml conical centrifuge tube. The Percoll gradient containing tube was then centrifuged at 21,000 g at 4°C for 8 min. The pellet was then retrieved with a pipette while being careful not to disrupt the debris layer. The pellet was then resuspended in IB and centrifuged at 13,000 g at 4°C for 10 min. Purification of platelet‐derived mitochondria is shown in Fig EV3A and B, as previously developed by our group (Léger et al, 2020, 2021).
Figure EV3. Visualization of platelet‐derived mitochondria.
Polymorphonuclear leukocyte isolation
Polymorphonuclear leukocytes (PMN) were isolated as previously described (Böyum, 1968). Briefly, blood collected from healthy donors in tubes containing ACD and then centrifuged at 275 g for 15 min at RT. The hematocrits were processed to remove erythrocytes by dextran sedimentation and subsequent centrifugation step at 800 g for 10 min at RT on a lymphocyte separation medium cushion (density of 1.077 g/ml, Corning). Remaining erythrocytes were lysed by hypotonic lysis and PMN purity (≥ 95%) was confirmed by flow cytometry. PMN were resuspended at 107 cells/ml in HBSS supplemented with 1.6 mM CaCl2 and 0.3 U/ml adenosine deaminase (Sigma‐Aldrich).
Polymorphonuclear neutrophils were either left unstimulated (native) or primed for 30 min at 37°C with 100 U/ml tumor necrosis factor‐alpha (TNF‐α), (Cedarlane) and 10 ng/ml granulocyte–macrophage colony‐stimulating factor (GM‐CSF), (Cedarlane) to simulate PMN in tissue inflammatory tissues (Duchez et al, 2015). Cells were then washed and subsequently activated for 2 h in presence of either functional platelet‐derived mitochondria, 25 μg/ml MTD (Zhang et al, 2010) or 1 μM N‐formyl‐methionyl‐leucyl‐phenylalanine (fMLP; Sigma‐Aldrich; Surette et al, 1996) at indicated concentrations in HBSS supplemented with 1.6 mM CaCl2.
Co‐Localization
Platelets were isolated as indicated above and resuspended in Tyrode's Buffer pH 7.4 (134 mM NaCl, 2.9 mM KCl, 0.34 mM Na2HPO4, 12 mM NaHCO3, 20 mM HEPES, 1 mM MgCl2, 5 mM glucose, and 0.5 mg/ml bovine serum albumin) at 108 cells/ml in the presence of 5 mM of CaCl2. Cells were then stained with 100 nM MitoTracker™ Deep Red for 15 min in the dark at 37°C. Platelet‐derived mitochondria were then isolated as described above. Activated or resting PMN were resuspended at 107 cells/ml in HBSS supplemented with 1.6 mM CaCl2, and membranes were stained with CD11b‐FITC (BioLegend) for 15 min in the dark at 37°C. Stained platelet‐derived mitochondria and stained neutrophils were washed, then co‐incubated for 2 h in HBSS supplemented with 1.6 mM CaCl2 in the dark at 37°C. Cells were then washed twice with HBSS and analyzed by flow cytometry. Fluorescent labeling of platelet‐derived mitochondria was only performed for mitochondrial count and for co‐localization assays, including flow cytometry and confocal microscopy as described below. Due to the effects of MitoTracker™ Deep Red on the integrity of the metabolic pathway of platelet mitochondria (Sargiacomo et al, 2021), all PMN functional assays were performed with co‐incubated unlabeled mitochondria.
Detection of PMN‐derived microvesicles
Resting or activated PMN (107 cells/ml) were incubated with freeMitos for 2 h at 37°C. Supernatants were obtained following centrifugation (1,500 g; 10 min) at room temperature (RT) and PMN‐MVs events between 0.25 and 1 μm in size and positive for cell surface markers CD11b or CD66b were measured by flow cytometry.
Transcriptomic analysis
Polymorphonuclear neutrophils were lysed in RiboZol (Amresco) and stored at −80°C until processing. Total RNA was extracted and purified using the RNeasy Mini Kit (Qiagen), following the procedures described by the manufacturer. Tran Quality of the RNA was assessed using the high‐sensitivity RNA (15 nt) kit on the 5200 Fragment Analyzer (Agilent, DNF‐472‐500). Concentration, integrity, and size distribution were determined using version 3.0 of the Agilent Prosize analysis software. Samples with a RIN ≥ 2 were used for library preparation.
Library was prepared using 25 ng RNA input and the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (NEB). First, ribosomal RNA was removed using the NEBNext rRNA Depletion Kit v2 (Human/Mouse/Rat; NEB) as indicated by the manufacturer's instructions. Briefly, samples with a RIN between 2 and 6 were fragmented for 8 min at 94°C, whereas samples with a RIN ≥ 7 were fragmented for 15 min at 94°C for a target insert size of 200 bp. Next, the first and the second cDNA strand were synthesized, followed by an Ampure XP bead (Beckman Coulter) clean up. Then, the samples were end‐prepped, and the adaptor was ligated. The adaptor was diluted following the manufacturer's instructions. The adaptor‐ligated samples were purified with Ampure XP beads before being amplified. For the PCR enrichment, a different Unique Dual Index Primer Pair (NEB) was added to each sample and 14 cycles were used. A last purification using Ampure XP beads was done to obtain the final library. If needed, libraries were pooled at the same concentration and the last bead purification was done a second time to remove the adapter‐dimer. The quality of the library was assessed as follows; size distribution was determined with the D1000 assay on the Tapestation (Agilent). The dsDNA HS assay from Qubit (ThermoFisher Scientific) was used to evaluate the concentration. Samples were loaded on an SP flowcell and a paired‐end sequencing of 2X80 was used. A total of 1.04 billion reads were obtained before alignment.
Validation of the Next Gen Sequence was performed on the same RNA extracts. cDNA was generated using the SensiFAST™ cDNA Synthesis Kit (Bioline), following the procedures described by the manufacturer. Quantitative real‐time PCR was then performed on the CFX Connect Real‐Time PCR Detection System (Bio‐Rad), using SensiFAST™ SYBR No‐ROX Kit (Bioline) and using already designed and tested primers as previously described for the following genes: Primers for the detection of tropomyosin 4 (OLIG2) gene: forward primer sequence 5’‐CTCCTCAAATCGCATCCAGA‐3′ and reverse primer sequence 5’‐GAAAAAGGTCATCGGGCTCT‐3′. Primers for the detection of transient receptor potential cation channel, subfamily M, member 2 (CCL3) gene: forward primer sequence 5′‐TGGTTTCAGACTTCAGAAGGAC‐3′ and reverse primer sequence 5′‐ATGATTCAGAGCAGGTGACG‐3′. Primers for the detection of hypoxanthine‐guanine phosphoribosyl transferase 1 (HPRT) gene: forward primer sequence 5′‐TTGTTGTAGGATATGCCCTTGA‐3′ and reverse primer sequence 5′‐ GCGATGTCAATAGGACTCCAG‐3′. Primers for the detection of beta‐actin (ACTB) gene: forward primer sequence 5′‐ACAGAGCCTCGCCTTTG‐3′ and reverse primer sequence 5′‐CCTTGCACATGCCGGAG‐3′. HPRT and ACTB were used as endogenous controls for normalization for the relative quantification. Cycle conditions were 95°C for 2 min, followed by 40 cycles of 95°C for 5 s, 62°C for 10 s and 72°C for 15 s. The quality of the PCR amplification was confirmed with a melt curve analysis and the efficiency of PCR was confirmed with a standard curve analysis for primer efficiency, as previously described.
Bioinformatics analysis
FASTQ reads from PMNs were assessed using fastqc version 0.11.9 and quality trimmed using Trimmomatic version 0.39 prior to alignment to the hg38 genome using STAR version 2.7.6 with the following optional parameters: ‘—sjdbGTFfile’. Feature quantification was performed using featureCounts version 2.0.1 with the gencode human annotation version 37 using the following parameters: featureCounts ‐p ‐t “exon” ‐g “gene_id” ‐a “gencode.v37.annotation.gff3” ‐‐fracOverlap 0.5 ‐s 2. Differential gene expression analysis was performed using the edgeR package version 3.32.1 for R version 4.0.4 (Robinson et al, 2010; McCarthy et al, 2012). Adjustments for batch effects and other latent sources of variability in sequencing data were done using SVAseq version 3.38.0. The computer code used for analysis of this data is available on github (https://github.com/SupriseCestEric/ErAll_Git/tree/master/Projects/LH_Boudreau). Sequencing data generated by this study are available in the Gene Expression Omnibus (GEO) with the accession number GSE186166.
Oxygen consumption
Resting or activated PMN (107 cells/ml) were incubated in the presence of absence of mitochondria. Mitochondrial oxygen consumption of PMN was assessed by high‐resolution respirometry (Oxygraph‐2k, Oroboros Instruments) as previously reported (Jougleux et al, 2021). The routine respiration state of mitochondrial respiration of PMN was determined as the basal mitochondrial oxygen consumption. Then, 2.5 μM of oligomycin (Sigma‐Aldrich) was injected in the chamber to achieve the leak respiration state by inhibiting complex V. Injections of 0.25 μM of carbonyl cyanide‐p‐trifluoromethoxyphenylgydrazone (FCCP, Sigma‐Aldrich) were performed sequentially to assess the maximal capacity of the electron transport system (ETS). Finally, 0.5 μM of rotenone (Sigma‐Aldrich) and 2.5 μM of antimycin A (Sigma‐Aldrich) were added to inhibit complex I and III, respectively, to measure the residual oxygen consumption (ROX).
Confocal microscopy
Polymorphonuclear neutrophils membranes were stained with 5 μg/ml of Cell Mask Orange (Molecular Probes) for 15 min in the dark at 37°C. Stained neutrophils were washed, resuspended in HBSS supplemented with 1.6 mM CaCl2, then co‐incubated for 2 h, in the dark at 37°C, in presence of MitoTracker™ Deep Red stained platelet‐derived mitochondria. Cells were then fixed with 4% (vol/vol) paraformaldehyde (Alfa Aesar) for 15 min in the dark on ice and 106 cells were centrifuged on microscopy slides at 140 g for 5 min at RT with the StatSpin Cytofuge (Beckman Coulter). Fluoroshield with DAPI (GeneTex) was added, then slides were stored in the dark at 4°C until visualization. Images were captured using FV31S‐SW software on an Olympus Fluoview FV3000 confocal microscope (Olympus Corporation).
Transmission electron microscopy
Samples were fixed in 2.5% glutaraldehyde (Sigma‐Aldrich) in PBS at 4°C for 2 h. The preparation was then additionally post‐fixed in 1% osmium tetroxide (OsO4; Sigma‐Aldrich) at 4°C for 2 h. The preparation was then centrifuged at 350 g for 10 min the cells and at 17,800 g for 90 min for the PMN‐MPs. Pellets were resuspended in PBS and processed by the Dalhousie University's EM Facility Core for dehydration, sectioning, and imaging. Briefly, the samples were dehydrated in a graded series of acetone and were then embedded in epon araldite resin. Ultrathin sections of 100 nm were cut with an ultramicrotome and placed on 300 mesh copper grids, which were stained with 2% uranyl acetate, rinsed, and treated with lead citrate, then rinsed and air‐dried. Samples were examined using a JEOL JEM 1230 transmission electron microscope at 80 kV while images were captured with a Hamatsu ORCA‐HR digital camera attached to the microscope.
Immunoblotting
Polymorphonuclear neutrophils were lysed in 500 μl nonyl‐phenoxypolyethoxyethanol (NP‐40) supplemented with 6× Laemmli sample buffer and boiled for 10 min. Proteins were quantified using bicinchoninic acid assay (BCA; Pierce Chemical) following the procedures described by the manufacturer, then 15 μg of proteins were loaded on a 10% (wt/vol) polyacrylamide gels and transferred to polyvinylidene difluoride (PVDF) membranes. After blocking for 2 h in tris‐buffered saline with Tween 20 (TBS‐T; 20 mM Tris, 150 mM NaCl, 0.1% Tween 20, pH 7.5), containing 5% (wt/vol) skim milk, blots were incubated with rabbit anti‐human mitochondrial cytochrome c oxidase (COX IV; Cell Signaling Technology) or rabbit anti‐human histone H3 (Cell Signaling Technology) in 1:1,000 dilution in TBS‐T. Bound antibodies were detected with enhanced chemiluminescence using a horseradish peroxidase‐conjugated goat anti‐rabbit IgG secondary antibodies (Abcam and Cell Signaling Technology) used in 1:10,000 dilution. Immunolabelings were visualized using a ChemiDoc Imager (Bio‐Rad) and quantified by densitometry analysis using Image Lab (Bio‐Rad).
Calpain activity assay
Polymorphonuclear neutrophils pellets were resuspended in a lysis buffer where protein concentration was determined by Micro BCA Protein Assays. Calpain activity was measured with a calpain activity assay kit (MilliporeSigma) by adding 100 μl of activation buffer to 50 μl of each sample (i.e., standards, samples, and controls), and subsequently mixed with 50 μl of the calpain substrates. Microplates were incubated for 15 min at RT and analyzed by fluorescence spectrometry (Ex370/Em450 nm) using a Synergy H1 spectrometer (Biotek).
IL‐8 assay
Polymorphonuclear neutrophils (106 cells/ml) were co‐incubated for 8 h with either mitochondria, MTD or fMLP. Interleukin‐8 (IL‐8, CXCL8) was measured by ELISA (hIL‐8 ELISA MAX™, BioLegend) using spectrometry (Synergy H1) at an absorbance of 450 nm according to the manufacturer's instructions.
Ethical approval
Blood was obtained from healthy consenting volunteers. This research project was approved by the Comité d'éthique de la recherche avec les êtres humains at the Université de Moncton.
Statistical analysis
One‐way or two‐way ANOVA followed by Tukey's or Dunnett's multiple comparison analysis were performed as described in the Figure legends. Data were analyzed using GraphPad Prism software (version 9.0, GraphPad Software Inc.) and R version 4.0.4. P‐values of <0.05 were considered significant.
Author contributions
Jacob L Léger: Conceptualization; formal analysis; validation; investigation; methodology; writing—original draft; writing—review and editing. Marie‐France N Soucy: Conceptualization; formal analysis; validation; investigation; methodology; writing—review and editing. Vanessa Veilleux: Validation; investigation; visualization; methodology. Robert D Foulem: Investigation; methodology. Gilles A Robichaud: Conceptualization; formal analysis; investigation; methodology; writing—review and editing. Marc E Surette: Formal analysis; supervision; validation; methodology; writing—review and editing. Eric P Allain: Conceptualization; data curation; formal analysis; validation; methodology. Luc H Boudreau: Conceptualization; data curation; formal analysis; supervision; funding acquisition; validation; investigation; methodology; writing—original draft; writing—review and editing.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Movie EV1
PDF+
Source Data for Figure 1
Acknowledgements
The authors wish to thank Jacynthe Lacroix, Woodson Shaw, and Simi Chacko, of the Atlantic Cancer Research Institute, for their help with whole transcriptome sequencing. The authors also would like to thank Mary Ann Trevors, from the EM Core Facility at Dalhousie University, for the processing and subsequent TEM imaging of our samples. JLL is a recipient of a master's scholarship from the Canadian Institutes of Health Research (CIHR). M‐FNS is a recipient of a summer scholarship from the New Brunswick (NB) Health Research Foundation (NBHRF). VV is a recipient of a PhD scholarship from the NBHRF. LHB is supported by the CIHR (149044), NBHRF, the NB Innovation Foundation (NBIF) and the Natural Sciences and Engineering Research Council of Canada (NSERC, RGPIN‐2019‐05740). MES is the recipient of a research chair from the NBIF and acknowledges support from the CIHR (165965) and NBHRF. GAR is supported by the NBIF; the NBHRF; the DUO research grant program from the Vitalité Health Network NB; The Leukemia and Lymphoma Society of Canada (LLSC); and a NSERC Discovery Development Grants no. DDG‐2020‐00005. EPA is the recipient of a NBHRF New Investigator Award offered in partnership with the Beatrice Hunter Cancer Research Institute. The synopsis image was created with BioRender.com.
EMBO reports (2022) 23: e54910
Data availability
Sequencing data generated by this study are available in the Gene Expression Omnibus (GEO) with the accession number GSE186166 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE186166).
References
- Aatonen M, Grönholm M, Siljander PR‐M (2012) Platelet‐derived microvesicles: multitalented participants in intercellular communication. Semin Thromb Hemost 38: 102–113 [DOI] [PubMed] [Google Scholar]
- Al Amir Dache Z, Otandault A, Tanos R, Pastor B, Meddeb R, Sanchez C, Arena G, Lasorsa L, Bennett A, Grange T et al (2020) Blood contains circulating cell‐free respiratory competent mitochondria. FASEB J 34: 3616–3630 [DOI] [PubMed] [Google Scholar]
- Boudreau LH, Duchez A‐C, Cloutier N, Soulet D, Martin N, Bollinger J, Paré A, Rousseau M, Naika GS, Lévesque T et al (2014) Platelets release mitochondria serving as substrate for bactericidal group IIA‐secreted phospholipase A2 to promote inflammation. Blood 124: 2173–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böyum A (1968) Isolation of leucocytes from human blood. A two‐phase system for removal of red cells with methylcellulose as erythrocyte‐aggregating agent. Scand J Clin Lab Invest Suppl 97: 9–29 [PubMed] [Google Scholar]
- Brown GT, McIntyre TM (2011) Lipopolysaccharide signaling without a nucleus: Kinase cascades stimulate platelet shedding of proinflammatory IL‐1β rich microparticles. J Immunol 186: 5489–5496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacko BK, Kramer PA, Ravi S, Johnson MS, Hardy RW, Ballinger SW, Darley‐Usmar VM (2013) Methods for defining distinct bioenergetic profiles in platelets, lymphocytes, monocytes, and neutrophils, and the oxidative burst from human blood. Lab Invest 93: 690–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor DE, Exner T, Ma DDF, Joseph JE (2010) The majority of circulating platelet‐derived microparticles fail to bind annexin V, lack phospholipid‐dependent procoagulant activity and demonstrate greater expression of glycoprotein Ib. Thromb Haemost 103: 1044–1052 [DOI] [PubMed] [Google Scholar]
- Davì G, Patrono C (2007) Platelet activation and atherothrombosis. N Engl J Med 357: 2482–2494 [DOI] [PubMed] [Google Scholar]
- Duchez A‐C, Boudreau LH, Naika GS, Bollinger J, Belleannée C, Cloutier N, Laffont B, Mendoza‐Villarroel RE, Lévesque T, Rollet‐Labelle E et al (2015) Platelet microparticles are internalized in neutrophils via the concerted activity of 12‐lipoxygenase and secreted phospholipase A2‐IIA. Proc Natl Acad Sci U S A 112: E3564–E3573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foers AD, Dagley LF, Chatfield S, Webb AI, Cheng L, Hill AF, Wicks IP, Pang KC (2020) Proteomic analysis of extracellular vesicles reveals an immunogenic cargo in rheumatoid arthritis synovial fluid. Clin Transl Immunol 9: e1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsman H, Dahlgren C (2010) The FPR2‐induced rise in cytosolic calcium in human neutrophils relies on an emptying of intracellular calcium stores and is inhibited by a gelsolin‐derived PIP2‐binding peptide. BMC Cell Biol 11: 52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JE, Austin CD, Reynolds CC, Steffen PK (1991) Evidence that agonist‐induced activation of calpain causes the shedding of procoagulant‐containing microvesicles from the membrane of aggregating platelets. J Biol Chem 266: 13289–13295 [PubMed] [Google Scholar]
- Fukami MH, Salganicoff L (1973) Isolation and properties of human platelet mitochondria. Blood 42: 913–918 [PubMed] [Google Scholar]
- Gomez I, Ward B, Souilhol C, Recarti C, Ariaans M, Johnston J, Burnett A, Mahmoud M, Luong LA, West L et al (2020) Neutrophil microvesicles drive atherosclerosis by delivering miR‐155 to atheroprone endothelium. Nat Commun 11: 214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW, Burger G, Lang BF (1999) Mitochondrial evolution. Science 283: 1476–1481 [DOI] [PubMed] [Google Scholar]
- Hauser CJ, Fekete Z, Livingston DH, Adams J, Garced M, Deitch EA (2000) Major trauma enhances store‐operated calcium influx in human neutrophils. J Trauma 48: 592–598 [DOI] [PubMed] [Google Scholar]
- Hong Y, Eleftheriou D, Hussain AAK, Price‐Kuehne FE, Savage CO, Jayne D, Little MA, Salama AD, Klein NJ, Brogan PA (2012) Anti‐neutrophil cytoplasmic antibodies stimulate release of neutrophil microparticles. J Am Soc Nephrol 23: 49–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jougleux J‐L, Léger JL, Djeungoue‐Petga M‐A, Roy P, Soucy M‐FN, Veilleux V, Hébert MPA, Hebert‐Chatelain E, Boudreau LH (2021) Evaluating the mitochondrial activity and inflammatory state of dimethyl sulfoxide differentiated PLB‐985 cells. Mol Immunol 135: 1–11 [DOI] [PubMed] [Google Scholar]
- Léger JL, Jougleux J‐L, Savadogo F, Pichaud N, Boudreau LH (2020) Rapid isolation and purification of functional platelet mitochondria using a discontinuous Percoll gradient. Platelets 31: 258–264 [DOI] [PubMed] [Google Scholar]
- Léger JL, Pichaud N, Boudreau LH (2021) Purification of functional platelet mitochondria using a discontinuous Percoll gradient. Methods Mol Biol 2276: 57–66 [DOI] [PubMed] [Google Scholar]
- Marcoux G, Duchez A‐C, Rousseau M, Lévesque T, Boudreau LH, Thibault L, Boilard E (2017) Microparticle and mitochondrial release during extended storage of different types of platelet concentrates. Platelets 28: 272–280 [DOI] [PubMed] [Google Scholar]
- McCarthy DJ, Chen Y, Smyth GK (2012) Differential expression analysis of multifactor RNA‐seq experiments with respect to biological variation. Nucleic Acids Res 40: 4288–4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald B, Pittman K, Menezes GB, Hirota SA, Slaba I, Waterhouse CCM, Beck PL, Muruve DA, Kubes P (2010) Intravascular danger signals guide neutrophils to sites of sterile inflammation. Science 330: 362–366 [DOI] [PubMed] [Google Scholar]
- Nicolás‐Ávila JÁ, Adrover JM, Hidalgo A (2017) Neutrophils in homeostasis, immunity, and cancer. Immunity 46: 15–28 [DOI] [PubMed] [Google Scholar]
- Perez‐Pujol S, Marker PH, Key NS (2007) Platelet microparticles are heterogeneous and highly dependent on the activation mechanism: Studies using a new digital flow cytometer. Cytometry A 71: 38–45 [DOI] [PubMed] [Google Scholar]
- Pittman K, Kubes P (2013) Damage‐associated molecular patterns control neutrophil recruitment. J Innate Immun 5: 315–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluskota E, Woody NM, Szpak D, Ballantyne CM, Soloviev DA, Simon DI, Plow EF (2008) Expression, activation, and function of integrin alphaMbeta2 (mac‐1) on neutrophil‐derived microparticles. Blood 112: 2327–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhm F, Boilard E, Machlus KR (2021) Platelet extracellular vesicles: beyond the blood. Arterioscler Thromb Vasc Biol 41: 87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste M (1999) Oxidative phosphorylation at the fin de siècle. Science 283: 1488–1493 [DOI] [PubMed] [Google Scholar]
- Sargiacomo C, Stonehouse S, Moftakhar Z, Sotgia F, Lisanti MP (2021) MitoTracker deep red (MTDR) is a metabolic inhibitor for targeting mitochondria and eradicating cancer stem cells (CSCs), with anti‐tumor and anti‐metastatic activity in vivo . Front Oncol 11: 678343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlame M (2008) Cardiolipin synthesis for the assembly of bacterial and mitochondrial membranes. J Lipid Res 49: 1607–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sionis A, Suades R, Sans‐Roselló J, Sánchez‐Martínez M, Crespo J, Padró T, Cubedo J, Ferrero‐Gregori A, Vila‐Perales M, Duran‐Cambra A et al (2018) Circulating microparticles are associated with clinical severity of persistent ST‐segment elevation myocardial infarction complicated with cardiogenic shock. Int J Cardiol 258: 249–256 [DOI] [PubMed] [Google Scholar]
- Stier A (2021) Human blood contains circulating cell‐free mitochondria, but are they really functional? Am J Physiol Endocrinol Metab 320: E859–E863 [DOI] [PubMed] [Google Scholar]
- Sumoza‐Toledo A, Penner R (2011) TRPM2: a multifunctional ion channel for calcium signalling. J Physiol 589: 1515–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surette ME, Nadeau M, Borgeat P, Gosselin J (1996) Priming of human peripheral blood mononuclear cells with lipopolysaccharides for enhanced arachidonic acid release and leukotriene synthesis. J Leukoc Biol 59: 709–715 [DOI] [PubMed] [Google Scholar]
- Thrash JC, Boyd A, Huggett MJ, Grote J, Carini P, Yoder RJ, Robbertse B, Spatafora JW, Rappé MS, Giovannoni SJ (2011) Phylogenomic evidence for a common ancestor of mitochondria and the SAR11 clade. Sci Rep 1: 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timár CI, Lorincz AM, Csépányi‐Kömi R, Vályi‐Nagy A, Nagy G, Buzás EI, Iványi Z, Kittel A, Powell DW, McLeish KR et al (2013) Antibacterial effect of microvesicles released from human neutrophilic granulocytes. Blood 121: 510–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaldivia MTK, McFadyen JD, Lim B, Wang X, Peter K (2017) Platelet‐derived microvesicles in cardiovascular diseases. Front Cardiovasc Med 4: 74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Deng S, Zhao S, Ai Y, Zhang L, Pan P, Su X, Tan H, Wu D (2016) Intra‐peritoneal administration of mitochondrial DNA provokes acute lung injury and systemic inflammation via toll‐like receptor 9. Int J Mol Sci 17: E1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, Junger W, Brohi K, Itagaki K, Hauser CJ (2010) Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature 464: 104–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zindel J, Kubes P (2020) DAMPs, PAMPs, and LAMPs in immunity and sterile inflammation. Annu Rev Pathol 15: 493–518 [DOI] [PubMed] [Google Scholar]


