Abstract
Enhancer of Zeste Homolog 2 (EZH2) and androgen receptor (AR) are crucial chromatin/gene regulators involved in the development and/or progression of prostate cancer, including advanced castration-resistant prostate cancer (CRPC). To sustain prostate tumorigenicity, EZH2 establishes non-canonical biochemical interaction with AR for mediating oncogene activation, in addition to its canonical role as a transcriptional repressor and enzymatic subunit of Polycomb Repressive Complex 2 (PRC2). However, the molecular basis underlying non-canonical activities of EZH2 in prostate cancer remains elusive, and a therapeutic strategy for targeting EZH2:AR-mediated oncogene activation is also lacking. Here, we report that a cryptic transactivation domain of EZH2 (EZH2TAD) binds both AR and AR spliced variant 7 (AR-V7), a constitutively active AR variant enriched in CRPC, mediating assembly and/or recruitment of transactivation-related machineries at genomic sites that lack PRC2 binding. Such non-canonical targets of EZH2:AR/AR-V7:(co-)activators are enriched for the clinically relevant oncogenes. We also show that EZH2TAD is required for the chromatin recruitment of EZH2 to oncogenes, for EZH2-mediated oncogene activation and for CRPC growth in vitro and in vivo. To completely block EZH2’s multifaceted oncogenic activities in prostate cancer, we employed MS177, a recently developed proteolysis-targeting chimera (PROTAC) of EZH2. Strikingly, MS177 achieved on-target depletion of both EZH2’s canonical (EZH2:PRC2) and non-canonical (EZH2TAD:AR/AR-V7:co-activators) complexes in prostate cancer cells, eliciting far more potent antitumor effects than the catalytic inhibitors of EZH2. Overall, this study reports a previously unappreciated requirement for EZH2TAD for mediating EZH2’s non-canonical (co-)activator recruitment and gene activation functions in prostate cancer and suggests EZH2-targeting PROTACs as a potentially attractive therapeutic for the treatment of aggressive prostate cancer that rely on the circuits wired by EZH2 and AR.
INTRODUCTION
Prostate cancer is the most frequently diagnosed, non-cutaneous malignancy in men, causing ∼30 000 deaths annually in the USA (1). Androgen deprivation therapy (ADT) is the standard treatment for prostate cancer (2), yet it is almost inevitable that patients receiving ADT become refractory and eventually develop castration-resistant prostate cancer (CRPC) (3,4). Both genetic and epigenetic deregulations play critical roles during the development of prostate cancer and its stepwise progression into more advanced and aggressive forms, notably CRPC. Androgen receptor (AR) and Enhancer of Zeste Homolog 2 (EZH2) are among the most relevant prostate cancer-promoting oncoproteins. Amplification of the AR gene, mutation of AR’s ligand-binding domain (LBD) and/or AR cofactor-directed mechanisms can lead to a hyperactive AR signaling during prostate tumorigenesis (5,6); in addition, the AR gene undergoes alternative splicing with inclusion of cryptic exons in CRPC, giving rise to the truncated, hormone-independent, constitutively active AR variants such as AR splice variant 7 (AR-V7) (7,8).
EZH2 is widely known as a catalytic subunit of Polycomb Repressive Complex 2 (PRC2), which induces trimethylation of histone H3 lysine 27 (H3K27me3) to maintain repression of a battery of tumor-suppressive, differentiation-promoting and immunity-related transcripts (9–14). Overexpression of EZH2 due to genomic loss of EZH2-targeting microRNA is associated with prostate cancer progression, correlating to poor clinical outcomes (15). Increasing evidence demonstrated that, in prostate cancers including CRPC, EZH2 also has non-conventional functions in binding to non-PRC2 partners (16), such as AR and fibrillarin for potentiating transcriptional activation (17–20) and translation (21), respectively. Studies of other types of cancer such as breast cancer also showed EZH2 overexpression not correlated with the increase of H3K27me3 (22), consistent with a PRC2-independent role for EZH2. Indeed, a recent study of MLL-rearranged acute myeloid leukemia (AML) (23) demonstrated that EZH2 directly binds cMyc and p300 through a cryptic transactivation domain (TAD), independently of its PRC2 function, to promote oncogenesis (23,24). However, the role of the TAD of EZH2 (EZH2TAD) in prostate cancer remains unexplored to date.
Previously, it has been shown that knockdown (KD) or knockout (KO) of EZH2 in various prostate tumor models suppressed cancer cell proliferation (15,17–19,21), laying a strong foundation for therapeutically targeting EZH2 as an attractive strategy for the treatment of prostate cancer. Small-molecule inhibitors that selectively target the methyltransferase activity harbored within EZH2’s Su(var)3–9, Enhancer-of-zeste and Trithorax (SET) domain have been developed, some of which are currently under clinical development (15,25). However, it is most likely that these EZH2 enzymatic inhibitors cannot block EZH2’s methyltransferase-independent functions such as those related to gene activation and/or scaffolding (e.g. recruiting/binding non-PRC2 factors), some of which have been suggested to be equally critical for oncogenesis (18,21,23). Strategies for completely blocking the multilevel activities of EZH2 in prostate cancer need to be developed.
MATERIALS AND METHODS
Cell lines
Cell lines used in the study included 293T [American Tissue Culture Collection (ATCC), CRL-3216], 22Rv1 (ATCC, CRL-2505), LNCaP (ATCC, CRL-1740), C4-2 (ATCC, CRL-3314), PC3 (ATCC, CRL-1435), DU145 (ATCC, HTB-81) and RWPE-1 (ATCC, CRL-11609), which were cultured and maintained according to the vendor-provided protocols. Cell lines of LNCaP-abl and LNCaP_RB–/–/P53–/– were kindly provided by Dr Zoran Culig (Innsbruck Medical University, Austria) and Dr Charles Sawyers (Memorial Sloan Kettering Cancer Center), respectively. Identity of cell lines was ensured by UNC’s Tissue Culture Facility with genetic signature analyses and examination of mycoplasma contamination performed routinely by using commercial detection kits (Lonza, #LT27-286).
Chemicals
MS177, C24, MS177N1 and MS177N2 were synthesized and used as described previously (23). Pomalidomide (#36471) was purchased from AstaTech. GSK126 (#S7061), EPZ6438 (#S7128), MLN4924 (#S7109) and A-485 (#S8740) were purchased from Selleck Chemical. CPI-1205 and UNC1999 were synthesized as previously reported (26,27). UNC6852 (an EED PROTAC) was used as before (23).
Antibodies
Antibodies used in the work included mouse anti-AR-V7 (Precision, Cat # AG10008), mouse anti-EZH2 (BD, Cat # 612666), rabbit anti-H3K27me3 (Millipore, Cat # 07-449), sheep anti-EED (R&D, Cat # AF5827), mouse anti-M2 Flag tag (Sigma, Cat #F1804), glutathione S-transferase–horseradish peroxidase (GST–HRP; GeneTex, Cat # GTX114099), anti-full-length (FL) AR (against the AR-FL C-terminus, C19) (Santa Cruz, Cat # sc-815), rabbit anti-AR (against the AR N-terminus, N20) (Santa Cruz, Cat # sc-816), mouse anti-ubiquitin (Santa Cruz, Cat # SC-8017), rabbit anti-H3K27ac (Abcam, Cat # ab4729), rabbit anti-H3 (Abcam, Cat # ab1791), rabbit anti-EZH2 (Cell Signaling Technology, Cat # 5246), rabbit anti-H3K27me3 (Cell Signaling Technology, Cat # 9733), rabbit anti-hemagglutinin (HA) tag (Cell Signaling Technology, Cat # 3724), rabbit anti-glyeraldehyde phosphate dehydogenase (GAPDH; Cell Signaling Technology, Cat # 5174), rabbit anti-SUZ12 (Abcam, Cat # ab12073), rabbit anti-AR (Cell Signaling Technology, Cat # 5153), rabbit anti-beta-Actin (Cell Signaling Technology, Cat # 4970), rabbit anti-CRBN (against the AR N-terminus, Cell Signaling Technology, Cat # 71810), rabbit anti-poly (ADP-ribose) polymerase (PARP; Cell Signaling Technology, Cat # 9532), rabbit anti-Cleaved Caspase-3 (Cell Signaling Technology, Cat # 9661), rabbit anti-Cleaved Caspase-7 (Cell Signaling Technology, Cat # 8438) and normal rabbit IgG (Cell Signaling Technology, Cat # 2729). HRP-linked secondary antibodies, either anti-mouse IgG (Cat # 7076) or anti-rabbit IgG (Cat # 7074), were obtained from Cell Signaling Technology.
Cleavage under targets & release using nuclease (CUT&RUN)
CUT&RUN was performed as previously described (23). Briefly, 0.5 × 106 cells were first collected, washed in the CUT&RUN wash buffer and then bound to activated concanavilin A (ConA) beads (Bangs Laboratories, #BP531). Next, the cell:bead sample was incubated with antibodies against the protein target (1:100 dilution) and then permeabilized in digitonin-containing buffer, which was followed by washing in digitonin buffer, incubation with pAG-micrococcal nuclease (MNase) and another wash in digitonin buffer to remove unbound pAG-MNase. After the final wash, cells were subjected to digestion following the pAG-MNase activation by addition of pAG-MNase digestion buffer, followed by incubation on a rotator for 2 h at 4°C. Solubilized chromatin was then released using the CUT&RUN stop buffer, in which an equal amount ofDrosophila spike-in chromatin (0.5 ng/sample) was added across all samples for spike-in normalization, and DNA purification was carried out with a polymerase chain reaction (PCR) cleanup kit (Qiagen). About 5 ng of the purified CUT&RUN DNA was used for preparation of multiplexed libraries with the NEB Ultra II DNA Library Prep Kit as per the manufacturer's instruction. Deep sequencing was conducted using an Illumina NextSeq 500 Sequencing System (available at the core facility of the Department of Pharmacology, University of North Carolina).
Chromatin immunoprecipitation (ChIP) followed by quantitative PCR (ChIP-qPCR)
ChIP-qPCR was performed as previously described (23,28,29). Information of primers used for ChIP-qPCR is given in Supplementary Table S4.
ChIP sequencing (ChIP-seq) and CUT&RUN data analysis
ChIP-seq datasets downloaded from the NCBI Gene Expression Omnibus (GEO) were re-analyzed as previously described (23). AR CUT&RUN was conducted with a pan-AR antibody, which recognizes an N-terminal region of AR-FL and AR-V7 (Cell Signaling Technology, Cat # 5153). CUT&RUN data analysis including spike-in normalization was conducted as before (23). In brief, raw reads were mapped to the reference genome (hg19) by using bowtie v2.3.5. The non-primary alignment, PCR duplicates and blacklist regions were removed from aligned data by using Samtools (v1.9), Picard ‘MarkDuplicates’ function (v2.20.4) and bedtools (v2.28.0), respectively. Peak calling was performed by using MACS2 (macs2 callpeak -f BAMPE -g hs/mm –keep-dup 1). Deeptools (v3.3.0) was used to generate bigwig files. Genomic binding profiles were generated by using the deepTools ‘bam-Compare’ functions. Profiles of CUT&RUN read densities were displayed in Integrative Genomics Viewer (IGV, Broad Institute). Heatmaps of ChIP-seq signals were generated by using the deepTools ‘computeMatrix’ and ‘plotHeatmap’ functions. Distribution of peaks was analyzed by the ‘annotatePeaks.pl’ function of HOMER (Hypergeometric Optimization of Motif Enrichment) (30). Motif analysis was performed by the SeqPos tool in Cistrome to uncover the motifs that are enriched close to peak centers by taking peak locations as the input (31).
RNA sequencing (RNA-seq) and data analysis
RNA-seq was performed as described (23,28). Total RNA was first purified by using the RNeasy Plus Mini Kit (Qiagen, #74136) and then treated with the Turbo DNA-free kit (Thermo, #AM1907) to remove genomic DNA. Multiplexed RNA-seq libraries were generated and subjected to deep sequencing. Reads were mapped to the reference genome followed by analysis of differentially expressed genes (DEGs) as before (28,32). Fastq files were aligned to the GRCh38 human genome (GRCh38.d1.vd1.fa) using STAR v2.4.2 (33) with parameters: –outSAMtype BAM Unsorted –quantMode TranscriptomeSAM. Transcript abundance was estimated with salmon v0.1.19 (34) to quantify the transcriptome defined by Gencode v22. Gene level counts were summed across isoforms, and genes with low counts (maximum expression <10) were filtered for downstream analyses. Raw read counts were used for DEG analysis by DESeq2 v1.38.2 (35) where the size normalization factor was estimated based on median-of-ratios. GSEA (36) was performed as described (23,32). Expression heatmaps were generated using mean-centered log2-converted TPM (transcripts per million) sorted in descending order based on expression values in R’s package ‘gplots’ v3.0.3 with either no clustering or column hierarchical clustering by average linkage. Volcano plots visualizing DEGs were produced by using R’s package ‘EnhancedVolcano’ v3.11. Annotation of DEGs was conducted by using Metascape (37). DisGeNET (38) was used to annotate genes associated with human disease.
Plasmid construction
Wild-type (WT) or the serially deleted version of AR was fused in-frame with an N-terminal Flag tag and subsequently cloned into the mammalian expression vector pcDNA3.1 (Invitrogen). GST fusion constructs with full-length EZH2 or only EZH2TAD, either WT or the TAD transactivation-dead mutant (23) [including F145A + F171A (FA) or F145K + F171K (FK)], and the pCDH-EF1a-neo-based lentiviral constructs containing HA-tagged EZH2 (WT or TAD transactivation-dead mutant) were previously described and used as before (23). EZH2 SET domain deletion was generated by using a site-directed mutagenesis kit (Agilent, 200521). All plasmids were verified by direct Sanger sequencing. Information on primers used for plasmid construction is provided in Supplementary Table S4.
Transient transfection
The 293T cells were seeded in a 100 mm dish with fresh medium, and then transfected on the next day following a previously described protocol using polyethylenimine (PEI; Sigma, # 408727) (39). Cells were harvested 48 h after transfection for downstream analysis such as co-IP or immunoblotting.
The siRNA- or shRNA-mediated gene knockdown (KD)
Cells were transfected with small interfering RNA (siRNA) using the Lipofectamine RNAi MAX reagent according to the manufacturer's instruction. The MISSION® esiRNA for human CRBN (Sigma, # EHU047571) was ordered and used as per the vendor's guideline. The pLKO.1-based lentiviral short hairpin RNA (shRNA) plasmids for KD of EZH2 (23), AR or ARV7 (28) or a scrambled control (40) were described and used as before.
Viral production and stable cell line generation
Lentivirus was prepared with the packaging system in 293T cells. In brief, 293T cells were co-transfected with lentiviral vector and the packaging plasmids (psPAX2 and pMD2.5G), and supernatant containing virus was harvested at 48 and 72 h post-transfection. After filtration with a 0.45 μM filter, virus-containing supernatant was used to infect cells in the presence of 8 μg/ml polybrene. At 48 h post-infection, cells were selected with either 1 μg/ml puromycin (Gibco) or 1 mg/ml geneticin (Gibco) for 7 days to establish stable expression cell lines.
Immunoblotting
Cells were collected and lysed in EBC buffer (50 mM Tris pH 8.0, 120 mM NaCl, 0.5% NP-40, 0.1 mM EDTA and 10% glycerol) freshly supplemented with a complete protease inhibitor cocktail (Roche) and phosphatase inhibitor (Roche). Protein concentration of the cell lysate was measured by Bradford assay (BioRad). Equal amounts of protein lysates were separated by sodium dodecylsulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore). Quantification of the protein band intensity was performed by ImageJ software after normalization to that of GAPDH.
Co-immunoprecipitation (co-IP) and GST pull-down
IP was performed as described previously (23,41). Cell pellets were lysed in EBC buffer (freshly supplemented with protease and phosphatase inhibitor cocktail) on ice for 30 min. After sonication, debris was removed by centrifugation at 12 000 g for 15 min at 4°C. For IP of HA-tagged protein, HA-conjugated beads (Roche, # 11815016001) were incubated with lysate overnight at 4°C. GST-tagged protein was purified as previously described (23). GST pull-down was conducted with cell lysate and 1 μg of GST fusion recombinant protein as described (23,41).
Reverse transcription followed by qPCR (RT–qPCR)
RNA was isolated by using the RNeasy PLUS Mini Kit (Qiagen). Reverse transcription was performed with 1 μg of total RNA using the cDNA Reverse Transcription kit according to the manufacturer's protocols (Invitrogen), followed by qPCR using SYBR Green Master Mix (BioRad) on a QuantStudio 6 Flex Real-Time PCR System (Thermo). Relative abundance of gene expression was calculated using the comparative CT method which compares Ct value of the target gene with that of GAPDH. Primers used for RT–qPCR are listed in Supplementary Table S4.
Cell proliferation assays
Cells were seeded at a density of 1000–2500 cells per well in a 96-well plate in triplicate and incubated with medium containing the tested compound. Medium with fresh compound was changed every 2 days. At each time point, MTT reagent (Promega) was added to the cell culture medium and incubated for 1–2 h before measurement of the absorbance at 490 nm by using the CYTATION-5 imaging reader (BioTek). The EC50 (effective control to 50% growth inhibition) values were calculated by using a non-linear regression analysis of the mean ± standard deviation (SD) from at least triplicated datasets for each biological assay.
2D colony formation assay
The 2D colony formation assay was performed as previously described (23). Briefly, 5000 cells were plated in a 6-well plate and incubated with medium containing the compound. After incubation for 2–3 weeks, cells were fixed by 100% methanol and stained with crystal violet.
Soft agar assay
Soft agar-based colony formation assay was performed as described previously (41). Briefly, 22Rv1 cells were plated at a density of 24 000 cells/ml in complete medium supplemented with 0.4% agarose, which was added onto a bottom layer composed of medium with 1% agarose. Every 4 days, 0.5 ml of fresh complete medium containing the tested compound was added onto cells. After incubation for 3–4 weeks, cell culture plates were stained with 100 μg/ml iodonitrotetrazolium chloride solution (Sigma) and, after incubation overnight, the numbers of cell colonies were counted.
Cell fractionation
A total of 1 × 106 cells were harvested, washed with cold phospate-buffered saline (PBS) and resuspended in 200 μl of CSK buffer (10 mM PIPES pH 7.0, 300 mM sucrose, 100 mM NaCl, 3 mM MgCl2 and 0.1% Triton X-100, freshly supplemented with protease/phosphatase inhibitor cocktail), followed by incubation on ice for 30 min, as described (32). Then, the sample was subject to centrifugation at 1300 g for 5 min at 4°C to collect the supernatant (which refers to the soluble fraction) and pellet fraction (which refers to the chromatin fraction). Cell pellets were then dissolved in 1.5× SDS loading buffer. The same amounts of protein sample were used for immunoblotting.
Ubiquitination assay
The 22Rv1 cells treated with the tested compound were harvested and extracted in 100 μl of EBC buffer containing 1% SDS. Cell extract was heat-denatured at 95°C for 5 min and then diluted with 900 μl of EBC buffer. After brief sonication and centrifugation, lysate was subjected to IP with antibody of the target protein, followed by anti-ubiquitin immunoblotting.
Tumor growth in xenografted animal models
All animal experiments were approved and performed in accord with the guidelines of the Institutional Animal Care and Use Committee (IACUC) of the University of North Carolina at Chapel Hill. A total of 1 × 106 22Rv1 cells were suspended in 100 μl of a 1:1 mixture of PBS and Matrigel (BD Biosciences) and subcutaneously (s.c.) injected into dorsal flanks of NOD/SCID/gamma-null (NSG) male mice bilaterally (carried out by the Animal Studies Core, UNC Lineberger Comprehensive Cancer Center). Measurement of xenografted tumors was performed three times per week by using a caliper, and the tumor volume was calculated. Mice were sacrificed when tumors reached the maximum allowed size.
Statistics and reproducibility
Statistical analyses were performed by using GraphPad Prism (version 9). Unpaired two-tailed Student's t-test was used for comparing two sets of data with assumed normal distribution. Data are presented as the mean ± SD from at least three independent experiments. *, ** and *** denote P-values <0.05, 0.01 and 0.005, respectively. P <0.05 was considered to be statistically significant. NS denotes not significant. No statistical methods were used to pre-determine sample size. All data from representative experiments (such as imaging and micrograph) were repeated at least 2–3 times independently, which showed similar results.
RESULTS
EZH2 non-canonically binds genomic sites with the gene-active markers, RNA polymerase II (Pol II) and AR or AR-V7 in prostate cancer cells
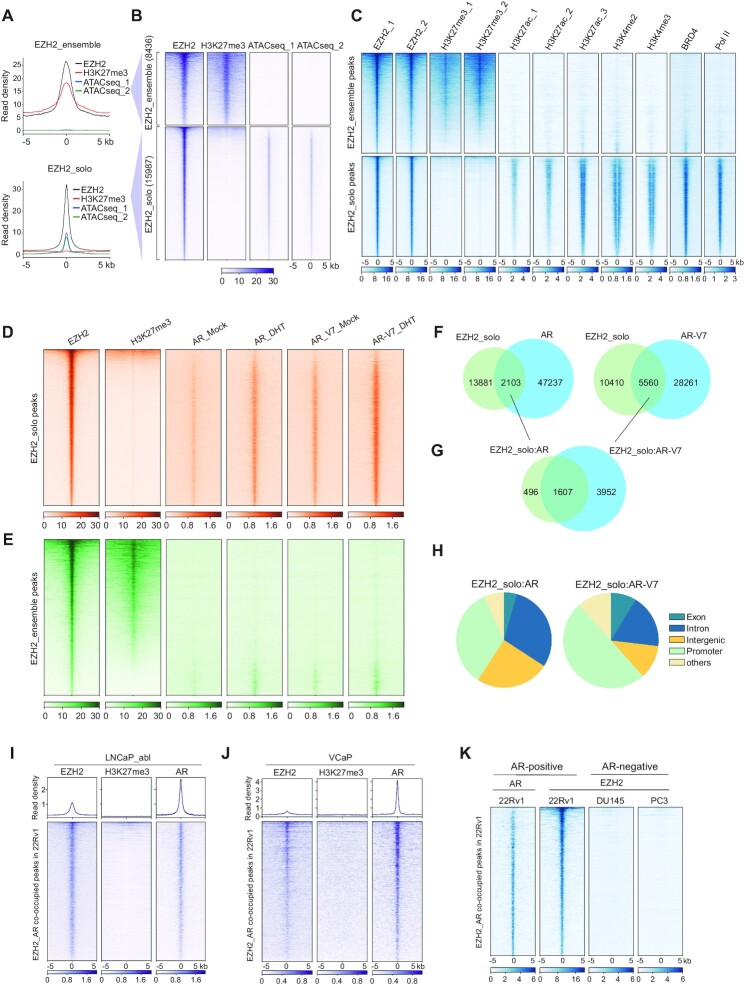
We sought to systematically define genome-wide binding patterns of EZH2 in prostate cancer by performing CUT&RUN (42) for EZH2 and H3K27me3, a gene-repressive histone mark characteristic of the canonical EZH2:PRC2 complex, in 22Rv1 cells, a commonly used CRPC model. Replicated mapping profiles of EZH2 or H3K27me3 were highly correlated (Supplementary Figure S1A), which were additionally correlated to the Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq) dataset of the same cells. A portion of the called EZH2 peaks (8436 or 34.5%) overlapped H3K27me3 and lacked chromatin accessibility (Figure 1A, B; Supplementary Figure S1B); thus, we termed these EZH2+/H3K27me3+/low-accessibility sites as EZH2-ensemble or canonical targets of EZH2:PRC2. Meanwhile, a larger proportion of EZH2-binding sites (15 987 or 65.5%) lacked H3K27me3 and, instead, were enriched for high ATAC-seq signals (Figure 1A, B; Supplementary Figure S1B), indicating a feature of open chromatin and a non-canonical function of EZH2; thus, we defined these EZH2+/H3K27me3−/high-accessibility sites as EZH2-solo targets. Next, we interrogated additional chromatin marks at EZH2-solo sites. Reminiscent of what was observed in MLL-rearranged AML (23), the EZH2-solo peaks identified from 22Rv1 CRPC cells were overwhelmingly enriched for a set of gene activation-related markers including H3K27ac, H3K4me2, H3K4me3, BRD4 (a histone acetylation reader and transcriptional coactivator) and RNA Pol II (Figure 1C, bottom). Such a correlational pattern was in stark contrast to what was seen with the EZH2+/H3K27me3+-co-bound peaks (Figure 1C, top). Furthermore, overall expression of the genes associated with EZH2-solo sites was significantly higher than that with EZH2:PRC2-ensemble peaks, based on RNA-seq data of 22Rv1 cells (28) (Supplementary Figure S1C).
Figure 1.
EZH2 non-canonically binds genomic sites with gene-active markers, Pol II and AR or AR-V7, in prostate cancer cells. (A, B) Averaged intensities (A) and heatmaps (B) of EZH2 or H3K27me3 CUT&RUN and ATAC-seq signals ± 5 kb from the centers of canonical EZH2+/H3K27me3+/low-accessibility peaks (i.e. EZH2-ensemble, 8436 peaks; top panels) or non-canonical EZH2+/H3K27me3−/high-accessibility peaks (i.e. EZH2-solo, 15 987 peaks; bottom panels) in 22Rv1 cells. (C) Heatmap showing the signals of EZH2 (in duplicate), H3K27me3 (in duplicate), H3K27ac (in triplicate), H3K4me2, H3K4me3, BRD4 and RNA Pol II ± 5 kb from the centers of EZH2-solo (top) or EZH2:PRC2-ensemble peaks (bottom). Except H3K4me2, H3K4me3 and Pol II data which were from LNCaP-abl cells, all the others were generated in 22Rv1 cells. EZH2 and H3K27me3 datasets were from CUT&RUN experiments and all the others from ChIP-seq. (D, E) Heatmap showing the EZH2, H3K27me3, AR and AR-V7 signals ± 5 kb from the centers of either EZH2-solo (D) or EZH2:PRC2-ensemble (E) peaks identified in 22Rv1 cells. Mock, ligand-stripped condition; DHT, ligand-treated condition. (F) Venn diagram showing the overlapping of EZH2-solo peaks with either AR (left) or AR-V7 peaks (right) in 22Rv1 cells. (G) Venn diagram showing a significant overlap between EZH2-solo:AR- and EZH2-solo:AR-V7-co-occupied sites in 22Rv1 cells. (H) Pie chart showing genomic annotation of EZH2-solo:AR (left) or EZH2-solo:AR-V7 (right) peaks in 22Rv1 cells. (I, J) Averaged intensities (top) and heatmaps (bottom) of EZH2, H3K27me3 and AR ChIP-seq signals ± 5 kb from the centers of those EZH2-solo:AR-co-occupied sites identified from 22Rv1 cells, in LNCaP-abl (I) or VCaP (J) cells. (K) Heatmap showing the signals of AR (only 22Rv1 cells shown; left) or EZH2 binding, ± 5 kb from the centers of those EZH2-solo:AR-co-bound peaks identified in 22Rv1 cells (second column), in the AR-negative DU145 and PC3 cells (two right columns).
The above observations highlighted the existence of both classic EZH2:PRC2 sites and non-canonical EZH2-solo targets in prostate cancer, in agreement with previous reports that EZH2 forms interactions with AR, mediating target gene activation instead of repression (17,18,20,43). Consistently, a significant portion of EZH2-solo sites overlapped binding of AR and/or AR-V7 (Figure 1D), both expressed in 22Rv1 cells (28), whereas AR/AR-V7 binding at those canonical PRC2 sites was found to be generally minimal (Figure 1E). Notably, there was an elevation of AR/AR-V7 enrichment at EZH2-solo sites in the presence of dihydrotestosterone (DHT), compared with the ligand-deprived condition (Figure 1D), presumably due to formation of AR:AR-V7 heterodimers that enable more robust binding of AR/AR-V7 to the genome (44). Approximately 2100 AR sites and 5500 AR-V7 sites were co-bound with EZH2 and lacking H3K27me3, thus termed as EZH2-solo:AR/AR-V7 sites (Figure 1F). In agreement with the notion that AR-V7 acts as a constitutively active form of AR, the majority of EZH2-solo:AR peaks overlapped EZH2-solo:AR-V7 peaks (Figure 1G). Overall signals of AR/AR-V7 binding at EZH2_solo:AR- or EZH2_solo:AR-V7-co-bound peaks were found on the lower side among signals of all called peaks (Supplementary Figure S1D). Androgen-responsive element (ARE) and the binding motif of FOXA1, a known cofactor of AR, were most enriched at both EZH2-solo:AR and EZH2-solo:AR-V7 sites (Supplementary Figure S1E-F), indicating an AR/cofactor (FOXA1)-driven recruitment. Gene Ontology (GO) and DisGeNET analyses of the sites commonly bound by EZH2-solo, AR and AR-V7 uncovered the enrichment for genes involved in cell proliferation, tissue development, stress response and prostate cancer metastasis (Supplementary Figure S1G, H). Approximately 50% of EZH2-solo:AR-V7 and 33% of EZH2-solo:AR sites were localized at gene promoters (Figure 1H). To assess whether or not the aforementioned EZH2-solo:AR co-binding is conserved across different prostate cancer cell models, we next related those EZH2-solo:AR sites identified from 22Rv1 cells with the publicly available datasets of various traditional prostate cancer cell lines (18,45). While EZH2-solo:AR-co-bound peaks showed similar co-occupancy by both oncoproteins in the two additional AR-positive prostate cancer lines, namely LNCaP-abl (Figure 1I) and VCaP cells (Figure 1J), such binding of EZH2 to the same genomic sites was lacking in the two AR-negative cells, PC3 and DU145 (Figure 1K). These observations suggested potential recruitment of EZH2 by AR to a subset of AR-binding sites in a PRC2-independent fashion.
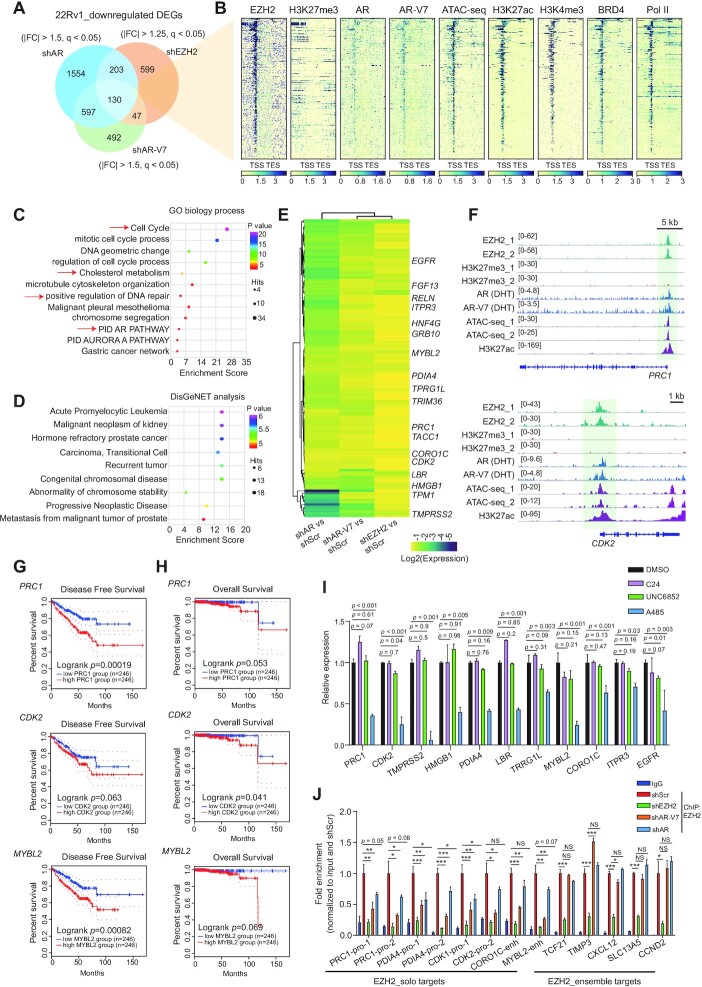
EZH2, AR and AR-V7 cooperate to activate transcription of a set of the clinically relevant oncogenes in prostate cancer
The above genome-wide profiling showed that EZH2-solo sites in prostate cancer exhibit co-localization with AR and/or AR-V7, RNA Pol II and prominent gene-active histone marks, which pointed to a gene activation-related role for EZH2, differing from its canonical EZH2:PRC2 function seen at sites exhibiting high H3K27me3 and low chromatin accessibility. To further determine EZH2-mediated gene regulation in prostate cancer, we performed RNA-seq after EZH2 KD in 22Rv1 cells (Supplementary Table S1; Figure S2A). As expected, depletion of EZH2 greatly inhibited 22Rv1 cell growth (Supplementary Figure S2B). Analysis of DEGs showed more transcripts down-regulated than up-regulated upon EZH2 KD relative to control (Supplementary Figure S2C), consistent with more EZH2-binding sites showing high chromatin accessibility in 22Rv1 cells (Figure 1A–C). GO analysis of DEGs down-regulated following EZH2 depletion showed the enrichment of pathways related to cell cycle, DNA replication and cell metabolism (Supplementary Figure S2D). Furthermore, we compared the transcriptomic profiles of 22Rv1 cells after EZH2 KD with those following either full-length AR- or AR-V7-specific KD (28). Here, we identified 130 genes to be co-activated by AR, AR-V7 and EZH2 (Figure 2A; Supplementary Figure S2E; Supplementary Table S2). Notably, 329 genes were co-activated by AR, AR-V7 and EZH2 with a relaxed cut-off (adjusted P or q value <0.05; Supplementary Figure S2F); here, about half of EZH2-activated genes (46.8%) overlapped those activated by AR, while around one-third of EZH2-activated genes (32.3%) overlapped those activated by AR-V7, suggesting substantial overlapping among EZH2-, AR- and AR-V7-up-regulated transcripts. Conversely, only one gene was found co-repressed by AR, AR-V7 and EZH2 (Supplementary Figure S2G), indicating that the EZH2:AR/AR-V7 complex mainly functions to activate target gene expression. In addition, those EZH2:AR:AR-V7-co-up-regulated genes were generally bound directly by EZH2, AR, AR-V7, coactivators (BRD4), RNA Pol II and gene-active histone marks, but devoid of H3K27me3 (Figure 2B), which again pointed to a gene activation effect by EZH2 and AR/AR-V7 at these targets. GO (Figure 2C) and DisGeNET (Figure 2D) analyses also showed the EZH2-solo:AR:AR-V7-co-activated genes enriched for the pathways associated with cell cycle, DNA repair, metabolism, and prostate cancer progression and metastasis. Through integration of genomic binding and RNA-seq profiles, we determined genes that were co-up-regulated by EZH2, AR and AR-V7 and also exhibited direct co-binding of the three (Figure 2E, labeled on the right), as exemplified by PRC1, CDK2, MYBL2, HMGB1 and PDIA4 (Figure 2F; Supplementary Figure S2H). High expression of EZH2-solo:AR:AR-V7-co-activated transcripts was found to be correlated with poor prognosis of prostate cancer: the patients with higher expression of PRC1, CDK2 or MYBL2 in tumor cells exhibited significantly worse clinical outcomes (Figure 2G, H); additionally, the expression of these genes was found to be positively associated with the EZH2 and AR levels in the TCGA prostate cancer patients (Supplementary Figure S2I). RT–qPCR confirmed that the high expression of select EZH2-solo:AR:AR-V7 target genes indeed relied on the presence of AR, AR-V7 and EZH2 (Supplementary Figure S2J–L), but was generally unaffected (or even slightly increased) upon treatment with C24, a potent EZH2 enzymatic inhibitor (which completely suppressed H3K27me3), or UNC6852, a PROTAC degrader of EED (46) (which depleted the EED-associated PRC2 and thus H3K27me3) (Figure 2I, purple and green; Supplementary Figure S2M, N). In stark contrast, expression of the same genes was significantly inhibited by treatment with the p300/CBP catalytic inhibitor A-485, which suppressed histone acetylation (47) (Figure 2I; Supplementary Figure S2O). Compared with mock treatment, KD of AR or AR-V7 significantly reduced the direct binding of EZH2 onto the EZH2-solo:AR:AR-V7-co-targeted sites, but not canonical EZH2:PRC2 targets (Figure 2J). As a control, AR or AR-V7 loss did not affect EZH2 protein levels in these cells (Supplementary Figure S2P). Thus, establishment of EZH2-solo binding relies on the presence of AR/AR-V7, in agreement with a lack of EZH2-solo:AR:AR-V7 sites in the AR-negative prostate cancer cells (Figure 1K). Together, our results substantiated a PRC2-independent function of EZH2 at its solo targets in prostate cancer, which acts in concert with AR and/or AR-V7 for target gene activation, with p300/CBP and BRD4 potentially serving as co-activators.
Figure 2.
EZH2, AR and AR-V7 cooperate to activate transcription of a set of the clinically relevant oncogenes in prostate cancer. (A) Venn diagram using DEGs, identified by RNA-seq to be down-regulated in 22Rv1 cells post-depletion of either EZH2, AR or AR-V7 (n = 2 biologically independent experiments). FC, fold change; q, adjusted P-value. (B) Heatmap showing overall binding of the indicated protein at EZH2:AR:AR-V7-co-upregulated genes (n = 130; defined in A). TSS, transcriptional start site; TES, transcriptional end site. (C, D) GO analysis (C) and enrichment of the DisGeNet category (D) using the EZH2:AR:AR-V7-co-upregulated genes defined in (A). (E) Heatmap using the indicated RNA-seq sample comparison shows log2-converted ratios of the 130 genes co-up-regulated by EZH2, AR and AR-V7 in 22Rv1 cells. Genes with direct co-binding by EZH2, AR and AR-V7 are labeled on the right. Scr, scramble. (F) Integrative Genomics Viewer (IGV) image showing the enrichment for the indicated factor at PRC1 (top) or CDK2 (bottom) in 22Rv1 cells. (G, H) Kaplan–Meier disease-free survival (G) and overall survival (H) analysis based on the PRC1 (top), CDK2 (middle) or MYBL2 (bottom) gene expression in patient samples from the prostate cancer cohort of The Cancer Genomics Atlas (TCGA). Statistical significance was determined by log-rank test and shown. (I) RT–qPCR for the indicated EZH2:AR:AR-V7-co-upregulated genes in 22Rv1 cells, treated with 2.5 μM UNC6852, C24 or A-485, relative to DMSO, for 24 h. The y-axis shows averaged signals after normalization to those of GAPDH and to mock-treated samples (n = 3; mean ± SD; unpaired two-tailed Student's t-test). (J) ChIP-qPCR for EZH2 binding at the promoter (pro) or enhancer (enh) of the indicated EZH2 target genes, either those EZH2-‘solo’ ones co-targeted by EZH2:AR:AR-V7 (left) or the canonical EZH2:PRC2 targets (right), in 22Rv1 cells after depletion of EZH2, AR or AR-V7, relative to mock (red). The y-axis shows averaged signals after normalization to those of input and then to controls (n = 3; mean ± SD; unpaired two-tailed Student's t-test). IgG [dark blue; using shRNA empty vector (shScr)-transduced samples] serves as a non-specific antibody control.
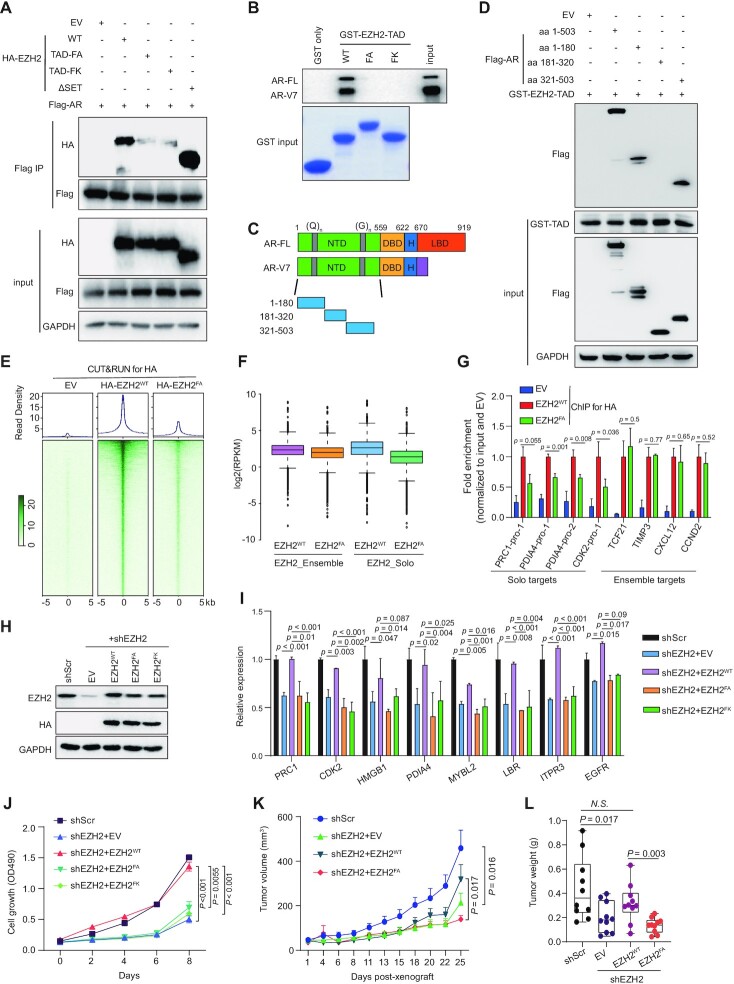
Interaction between EZH2TAD and AR is required for establishment of EZH2-solo binding at AR sites and for malignant growth of prostate cancer
EZH2 associates with AR (17,18,20,48). We validated interaction of EZH2 with AR and an AR variant by using both co-IP (Figure 3A, lane 2 versus 1) and GST pull-down (Supplementary Figure S3A). The biochemical basis underlying EZH2:AR/variant interaction, however, remains murky. Recently, a cryptic transactivation domain of EZH2 (EZH2TAD) was reported to interact directly with cMyc and co-activators (such as p300) for promoting target gene activation (23,24,43). We thus queried whether or not EZH2TAD also interacts with AR and AR variant in prostate cancer. First, GST pull-down using recombinant EZH2TAD readily detected interaction with both AR and AR-V7, expressed either endogenously or exogenously (Figure 3B; Supplementary Figure S3B; lane 2 versus 1). Such interaction with AR or AR-V7 was not observed using the EZH2TAD mutant that carries substitution of the two key hydrophobic residues (Phe145 and Phe171) with either alanine or a charged lysine residue, which dramatically reduced the transactivation effect by EZH2TAD (23,24) (Figure 3B; Supplementary Figure S3B; see lanes of FA or FK versus WT). In agreement, full-length AR interacted with both WT EZH2 and its SET-domain-deleted form, but not the two EZH2TAD transactivation-dead mutants, in co-IP (Figure 3A; lanes 3 and 4 versus 2 and 5), supporting an essential requirement for EZH2TAD for mediating EZH2:AR interaction.
Figure 3.
Interaction between EZH2TAD and AR is required for establishment of EZH2-solo binding at AR sites and for the malignant growth of prostate cancer cells. (A) Anti-Flag immunoprecipitation (IP; top panel) for Flag-tagged AR and the indicated HA-tagged EZH2, either WT or with the indicated EZH2TAD-dead mutation (FA or FK) or the SET deletion, in 293T cells. FA, F145A + F171A; FK, F145K + F171K. (B) GST pull-down using the indicated GST protein, either GST alone or fused with WT or mutant EZH2TAD, and the total lysate of 22Rv1 cells, followed by anti-AR immunoblotting (top). AR-FL, full-length AR. (C) Scheme of the domain organization of AR-FL and AR-V7. NTD, N-terminal domain; DBD, DNA-binding domain; LBD, ligand-binding domain; (Q)n, poly(Q) region; (G)n, poly(G) region. (D) Pull-down using GST-tagged WT EZH2TAD and total lysate of 293T cells transfected with the indicated Flag-tagged AR, followed by anti-Flag immunoblotting (top). (E) Averaged intensity (top) and heatmap (bottom) showing the EZH2 binding [exogenously expressed HA-tagged EZH2, either WT or TAD-mutated (FA), as assayed by anti-HA CUT&RUN], ± 5 kb from the center of called peaks in 22Rv1 cells. Cells with empty vector (EV) served as a CUT&RUN control. (F) Boxplot showing log2-transformed FPKM counts of EZH2, WT or TAD mutated, at EZH2-‘solo’ (right) or EZH2:PRC2-ensemble (left) target sites in 22Rv1 cells. The boundaries of box plots indicate the 25th and 75th percentiles, the center line indicates the median, and the whiskers (dashed) indicate 1.5× the interquartile range. (G) ChIP-qPCR of binding by HA-tagged EZH2, either WT or TAD mutant (FA), at the indicated EZH2 target genes, either those EZH2-‘solo’ ones co-targeted by EZH2:AR:AR-V7 (left) or the canonical EZH2:PRC2 targets (right). The y-axis shows signals after normalization to those of input (n = 3; mean ± SD; unpaired two-tailed Student's t-test). EV-transduced cells served as a negative control. (H–J) Immunoblotting for EZH2 (H), RT–qPCR for EZH2-solo:AR:AR-V7-co-activated genes (I), and measurement for 22Rv1 cell growth (J) after mock treatment [scramble (Scr)] or depletion of endogenous EZH2 (shEZH2) in 22Rv1 cells, pre-rescued with exogenous shEZH2-resistant HA-EZH2, either WT or TAD mutant (FA or FK). For (I) and (J), n = 3, mean ± SD; unpaired two-tailed Student's t-test. (K, L) Averaged size (K) and weight (L) of xenografted tumors by using 22Rv1 cells, which were pre-rescued with exogenous shEZH2-resistant HA-EZH2, either WT or TAD mutant, and then subjected for mock treatment (Scr) or KD of endogenous EZH2. Tumor weight was measured at day 25 post-xenograft in NSG mice (n= 10, mean ± SD; unpaired two-tailed Student's t-test).
We next queried what region within AR mediates EZH2TAD interaction. Here, we found that GST–EZH2TAD bound both full-length AR and its C-terminal-truncated form (Supplementary Figure S3C), showing that AR’s N-terminal domain (amino acids 1–503) is sufficient to mediate EZH2TAD interaction. Moreover, we constructed a set of AR serial deletion constructs for GST pull-down and further narrowed down the EZH2TAD-interacting regions to two segments of AR: its amino acids 1–180, which is enriched for glutamine [poly(Q)]; and amino acids 321–503, which is enriched for glycine [poly(G)] (Figure 3C-D).
To discern the role of EZH2TAD in mediating genomic recruitment of EZH2, we conducted CUT&RUN for HA-tagged WT or TAD-mutated EZH2 following its stable expression into 22Rv1 cells. Compared with the WT, the TAD-mutated EZH2 exhibited significantly reduced genomic binding (Figure 3E), as exemplified by what was seen at PRC1 and MYBL2 (Supplementary Figure S3D). Notably, there was a preferential decrease of binding of EZH2 at its solo target sites, compared with EZH2:PRC2 ensemble sites (Figure 3F, right versus left). ChIP-qPCR corroborated that, relative to the WT, the EZH2TAD mutant exhibited significantly reduced binding at the tested EZH2-solo targets, and not those canonical targets of EZH2:PRC2 (Figure 3G). Lastly, we evaluated the requirement of EZH2TAD for prostate cancer growth. By using 22Rv1 cells with endogenous EZH2 depleted for gene rescue (Figure 3H), we found that, compared with WT EZH2, its TAD transactivation-dead mutants failed to rescue the decreased expression of EZH2-solo target genes (Figure 3I) and failed to rescue the EZH2 loss-related growth inhibition in vitro (Figure 3J; Supplementary Figure S3E). Meanwhile, EZH2’s SET domain was also found to be essential for sustaining 22Rv1 cell proliferation (Supplementary Figure S3F, G), consistent with previous reports supporting an involvement of EZH2:PRC2 during prostate oncogenesis (49,50). Additionally, WT EZH2, but not its TAD transactivation-dead form, rescued the in vivo defects caused by endogenous EZH2 depletion, as assayed by growth of 22Rv1 cell xenografted tumors and the expression of EZH2-solo target genes in xenografted tumors (Figure 3K, L; Supplementary Figure S3H, I).
Overall, our observations demonstrated a previously unappreciated, indispensable requirement for EZH2TAD for mediating EZH2 interaction with AR and AR variant, for EZH2’s chromatin recruitment to EZH2-solo sites and for promoting prostate cancer cell growth in vitro and in vivo.
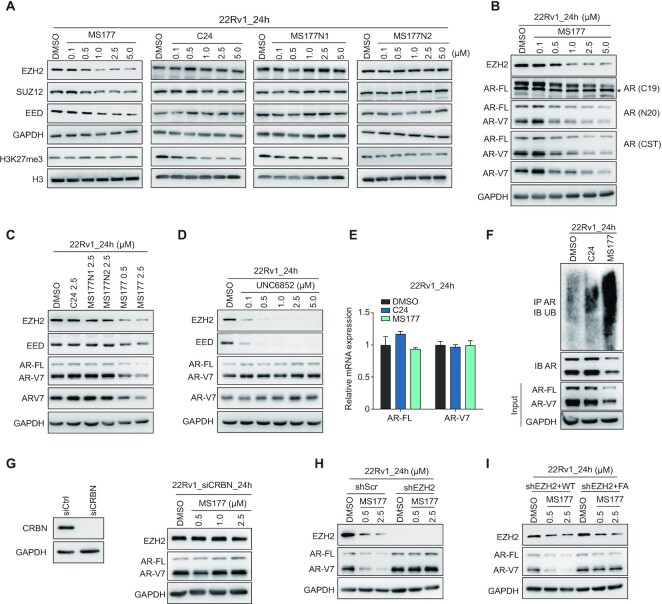
MS177, an EZH2 degrader, efficiently degrades both EZH2:PRC2 and EZH2:AR/AR-V7 complexes in prostate cancer cells
A handful of small molecules have been developed for blocking the catalytic activity harbored within EZH2’s catalytic domain (SET), some of which are under clinical evaluation (49). However, we predicted efficacies of these EZH2 enzymatic inhibitors to be low due to failure in targeting non-canonical, SET(PRC2)-independent functions of EZH2 (23), such as EZH2:AR-mediated prostate oncogene activation, which we showed are TAD dependent and also clinically relevant. This notion was corroborated by a generally mild or a lack of effect by C24, a potent EZH2 SET inhibitor (51), on growth of prostate cancer cells in vitro (Supplementary Figure S4A). Using Proteolysis Targeting Chimera (PROTAC) technology, we recently developed MS177, an EZH2-targeting PROTAC/degrader, that can simultaneously deplete EZH2 and EZH2-associated factors in cells (23). Thus, we examined the EZH2-degrading effect by MS177 in prostate cancer cells. As expected, MS177 treatment in 22Rv1 cells dose-dependently depleted cellular EZH2, regardless of being in the soluble or chromatin-associated fraction (Supplementary Figure S4B), and its PRC2 partners (EED and SUZ12), an effect not observed following comparable treatment with C24 or either of the two inactive analog compounds of MS177, namely MS177N1 (E3 binding-dead) and MS177N2 (C24 binding-dead) (23) (Figure 4A). The half-maximal degradation concentration (DC50) value of MS177 in 22Rv1 cells was measured to be 0.86 ± 0.12 μM and maximum degradation (Dmax) value 77% (Supplementary Figure S4C), supporting that MS177 is indeed a valuable tool for degrading EZH2 in prostate cancer. More excitingly, we observed that, unlike the enzymatic inhibitor C24 or PROTAC-inactive controls (MS177N1 or MS177N2), MS177 also concentration-dependently degraded AR and/or AR-V7 across multiple tested lines of prostate cancer including 22Rv1, C4-2 and LNCaP (Figure 4B, C; Supplementary Figure S4D). The effects by MS177 on degradation of EZH2:PRC2 and AR/AR-V7 were found to be generally comparable (Figure 4B, C; Supplementary Figure S4D). In contrast, the EED degrader UNC6852 efficiently depleted EED and EED-associated EZH2:PRC2 but did not alter the AR and AR-V7 levels in 22Rv1 cells (Figure 4D), consistent with a PRC2-independent association of EZH2 with AR and AR variant as we observed at EZH2-solo sites.
Figure 4.
MS177, an EZH2 degrader, efficiently degrades both EZH2:PRC2 and EZH2:AR/AR-V7 complexes in prostate cancer. (A) Immunoblotting of PRC2 subunits (EZH2, EED and SUZ12) and H3K27me3 in 22Rv1 cells, treated with the indicated concentration of MS177, C24, MS177N1 or MS177N2 versus DMSO for 24 h. (B) Immunoblotting of EZH2, full-length AR (AR-FL) and AR-V7 (probed with different antibodies recognizing either AR-FL only, AR-V7 only or both) in 22Rv1 cells treated with the indicated concentration of MS177 versus DMSO for 24 h. *Non-specific band. (C,D) Immunoblotting of EZH2, EED, AR and AR-V7 in 22Rv1 cells treated with 2.5 μM C24, MS177N1, MS177N2 or different concentrations of MS177 (C), or different indicated concentrations of an EED-targeting PROTAC UNC6852 (D), versus DMSO for 24 h. (E,F) RT–qPCR (E) for AR-FL and AR-V7 mRNA levels and immunoblotting (F) for AR ubiquitination in 22Rv1 cells post-treatment with DMSO or 2.5 μM C24 or MS177 for 24 h. The y-axis in (E) shows averaged signals after normalization to GAPDH and to mock-treated samples (n = 3; mean ± SD). Top panels in (F) used samples from anti-AR pull-down followed by immunoblotting with anti-ubiquitin antibodies, and bottom panels showed input sample immunoblotting. (G) Left: immunoblotting of CRBN in 22Rv1 cells transfected with either control (siCtrl) or CRBN-targeting siRNA (siCRBN). Right: immunoblotting for EZH2 and AR in 22Rv1 cells transfected with CRBN-targeting siRNA, followed by treatment with the indicated concentration of MS177 versus DMSO for 24 h. (H, I) Immunoblotting for EZH2 and AR in 22Rv1 cells expressing either scramble (Scr) or EZH2-targeting (shEZH2) shRNA (H), or in those EZH2-depleted 22Rv1 cells rescued with exogenously expressed WT or TAD-mutated (FA) EZH2 (I), followed by treatment with different doses of MS177 for 24 h compared with DMSO.
We next explored MS177’s mechanism of action (MOA) for degradation of AR and AR variant. First, cellular AR was found to be ubiquitinated and then depleted post-treatment with MS177 but not C24, while the transcriptional levels of AR and AR-V7 remained unchanged (Figure 4E, F), suggesting a proteasome-dependent degradation mechanism underlying AR depletion. Additionally, MS177-induced EZH2 degradation was effectively blocked by pre-treatment of cells with pomalidomide (the E3 ligase ligand module of MS177; Supplementary Figure S4E) or MLN4924 (a NEDD8 neddylation inhibitor that suppresses assembly of Cullin-based E3 ligase; Supplementary Figure S4F). Depletion of CRBN, the E3 ligase that MS177 recruits, also almost completely abrogated MS177-induced depletion of EZH2, AR and AR variant (Figure 4G versus B). Lastly, depletion of EZH2 also completely blocked the MS177-mediated degradation of AR and AR variant (Figure 4H, right versus left), and, in 22Rv1 cells with endogenous EZH2 depleted, MS177 was able to degrade the TAD-mutated form of EZH2 (harboring the intact SET, to which MS177 binds) but not the AR or AR variant anymore (Figure 4I), supporting that the AR and AR variant degradation by MS177 requires the presence of EZH2 and EZH2TAD-directed binding of AR and AR variant.
Together, MS177, an EZH2-targeting PROTAC, effectively degrades EZH2 and EZH2-associated canonical (EZH2:PRC2) and non-canonical (EZH2:AR/AR-V7) partners in prostate cancer, which has not been reported before.
Genomics profiling further substantiates the effect of MS177 on inhibiting both EZH2:PRC2- and AR/AR-V7-related oncogenic nodes
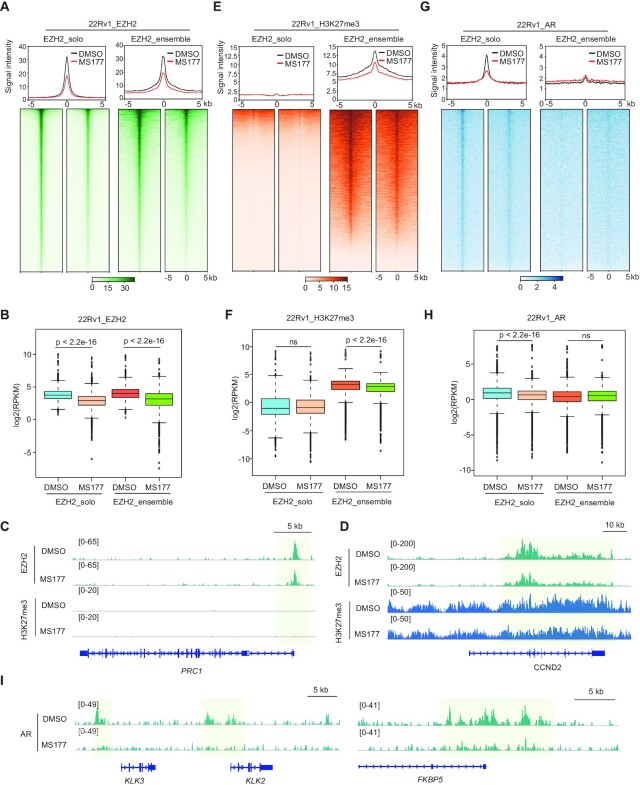
To further delineate the gene regulatory effect of MS177 in prostate cancer, we performed spike-in-controlled CUT&RUN for EZH2, H3K27me3 and AR (by using an antibody that recognizes full-length AR and AR variants) after treatment of 22Rv1 cells with MS177. Compared with mock treatment, MS177 significantly decreased genome-wide binding of EZH2 at both EZH2:PRC2-ensemble sites and EZH2-solo sites (Figure 5A, B; Supplementary Figure S5A), exemplified by what was seen at non-canonical EZH2-solo targets such as PRC1, MYBL2 and CDK2 (Figure 5C; Supplementary Figure S5B) and classic EZH2:PRC2 targets such as CCND2, MYT1 and WNT2B (Figure 5D; Supplementary Figure S5C). As expected, MS177 treatment also decreased the overall H3K27me3 level from EZH2:PRC2 sites (Figure 5E, F; Supplementary Figure S5D; also see bottom panels of Figure 5D and Supplementary Figure S5C). Relative to mock, MS177 treatment caused the expected decrease of AR binding at EZH2-solo sites where AR co-bound (Figure 5G, H); more interestingly, MS177’s inhibitory effect on AR binding was found to be genome-wide and extended to those AR sites where EZH2 does not bind at all (Supplementary Figure S5E), as observed at classic targets of AR and AR variant such as KLK3,KLK2 and FKBP5 (Figure 5I; Supplementary Figure S5F). Such a genome-wide decrease of AR binding is probably due to a ‘drainage’ effect by MS177 on global AR and AR variant proteins in cells (see also the Discussion), highlighting a potential advantage of EZH2-targeting PROTAC degrader (MS177) over conventional enzymatic inhibitors of EZH2.
Figure 5.
CUT&RUN-based profiling demonstrates effect by MS177 on decreasing genomic binding of both EZH2 and AR/AR-V7 in prostate cancer cells. (A, E, G) Average intensity (top) and heatmap (bottom) showing CUT&RUN signals of EZH2 (A), H3K27me3 (E) and AR (G) (normalized against spike-in control and sequencing depth) ± 5 kb around the peak centers of EZH2-solo (left) or EZH2:PRC2-ensemble target sites (right) identified in 22Rv1 cells, treated for 24 h with DMSO (black) or 2.5 μM MS177 (red). AR CUT&RUN (G) was conducted with a pan-AR antibody recognizing the N-terminal region of both AR-FL and AR-V7. (B, F, H) Log2-transformed RPKM counts for CUT&RUN signals of EZH2 (B), H3K27me3 (F) and AR (H) at EZH2-solo (left) or EZH2:PRC2-ensemble target sites (right) identified in 22Rv1 cells, treated for 24 h with DMSO or 2.5 μM MS177. (C, D) IGV views of EZH2 and H3K27me3 binding (spike-in control and depth normalized) at PRC1 (C) or CCND2 (D) post-treatment of 22Rv1 cells with DMSO or 2.5 μM MS177. (I) IGV views of AR binding (spike-in control and depth normalized) at KLK3 and KLK2 (left) and FKBP5 (right) post-treatment of 22Rv1 cells with DMSO or 2.5 μM MS177.
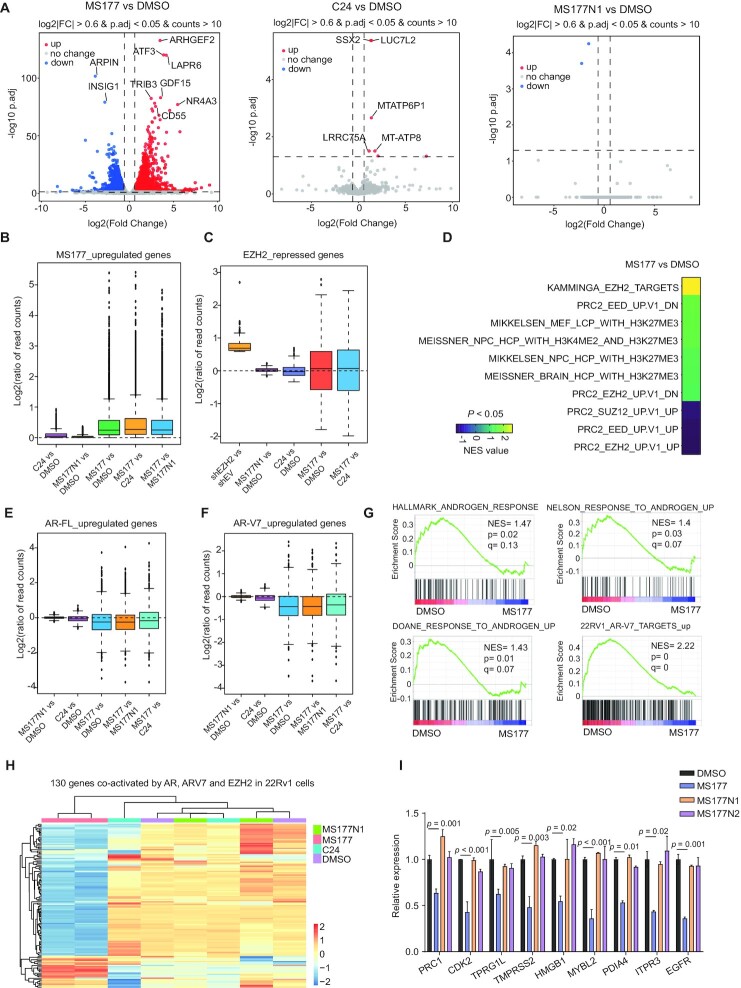
Next, we evaluated the transcriptome-modulatory effect of MS177, compared with its inactive control compounds. Here, we treated 22Rv1 cells with either dimethylsulfoxide (DMSO), MS177, C24 or MS177N1, followed by RNA-seq (Supplementary Table S3). Dramatic transcriptomic changes were observed only after treatment with MS177, but not C24 or MS177N1 (Figure 6A). MS177-activated DEGs only showed mild changes following comparable treatment with C24 or MS177N1 (Figure 6B). Likewise, those EZH2-repressed genes, defined as DEGs re-activated upon EZH2 KD, were found to be de-repressed by treatment with MS177, and not C24 or MS177N1 (Figure 6C), again demonstrating a superior and unique effect of MS177 on altering the transcriptome of prostate cancer cells. Gene Set Enrichment Analysis (GSEA) revealed treatment of MS177 associated with re-activation of known H3K27me3- or PRC2-repressed genes, reminiscent of what was seen with RNA-seq after EZH2 KD (Figure 6D; Supplementary Figure S6A). Meanwhile, the AR/AR-V7 signaling genes were found to be significantly down-regulated upon MS177 treatment relative to mock treatment (Figure 6E–G; Supplementary Figure S6B). Moreover, those genes co-activated by EZH2, AR and AR-V7, as we defined by KD studies (Figure 2A), were significantly inhibited by treatment with MS177 and not C24 or MS177N1 (Figure 6H; Supplementary Figure S6C). By RT–qPCR, we further confirmed the unique effect of MS177 on down-regulating select EZH2-solo:AR/AR-V7 target genes in 22Rv1 cells (Figure 6I).
Figure 6.
RNA-seq further substantiates a unique and superior effect of MS177 on inhibiting both EZH2:PRC2- and AR/AR-V7-related oncogenic programs in prostate cancer cells. (A) Volcano plots showing transcriptomic alteration in 22Rv1 cells following treatment with 2.5 μM MS177 (left), C24 (middle) or MS177N1 (right), compared with DMSO, for 24 h. DEGs with significant expression changes are highlighted. (B, C) Box plots showing log2 ratios for DEGs up-regulated in 22Rv1 cells treated with MS177 versus DMSO (B), or DEGs up-regulated after EZH2 KD in 22Rv1 cells (i.e. EZH2-repressed genes; C). Comparison was indicated on the x-axis, including C24 versus DMSO, MS177N1 versus DMSO, MS177 versus DMSO and MS177 versus C24. (D) Heatmap of GSEA-normalized enrichment score (NES) values revealing that MS177 treatment is highly correlated with de-repression of PRC2- or H3K27me3-repressed genes. (E, F) Box plots showing log2 ratios for DEGs significantly down-regulated in 22Rv1 cells after AR-specific (E) or AR-V7-specific (F) KD relative to control. Comparison was indicated on the x-axis, including C24 versus DMSO, MS177N1 versus DMSO, MS177 versus DMSO, MS177 versus MS177N1 and MS177 versus C24. (G) GSEA revealing that, relative to the control, MS177 treatment in 22Rv1 cells is correlated with down-regulation of the indicated gene set known to be activated by AR or AR-V7. (H) Heatmap showing expression of the 130 genes co-activated by EZH2, AR and AR-V7 in 22Rv1 cells, treated with MS177, C24 or MS177N1 compared with DMSO. Treatment condition was indicated by colored bars on the top of the heatmap, with the key shown on the right. (I) RT–qPCR for the indicated EZH2:AR:AR-V7-co-activated target genes in 22Rv1 cells, treated with DMSO or 2.5 μM C24, MS177N1 or MS177 for 24 h. The y-axis shows RT–qPCR signals after normalization to those of GAPDH and then to DMSO-treated cells (n = 3 independent experiments; presented as the mean ± SD). For (B), (C), (E) and (F), the boundaries of box plots indicate the 25th and 75th percentiles, the center line indicates the median and the whiskers (dashed) indicate 1.5× the interquartile range.
Collectively, our integrated CUT&RUN and RNA-seq profiling following pharmacological treatment or genetic depletion of EZH2 lent strong support for an on-target effect of MS177, resulting in simultaneous suppression of both EZH2:PRC2- and AR/AR-V7-directed oncogenic circuitries in prostate cancer.
Compared with the EZH2 enzymatic inhibitors, MS177 elicits much more potent antitumor effects in prostate cancer cells
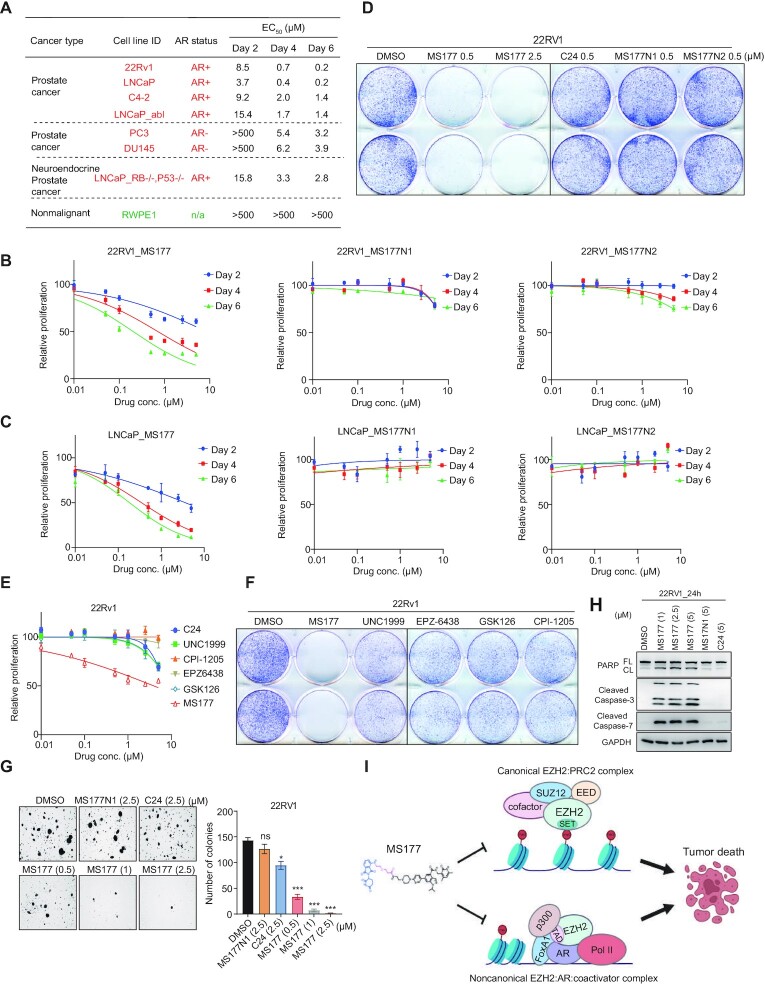
Next, we sought to evaluate the antitumor effect of MS177 in a panel of cell lines representing different stages of prostate cancer, and EC50 values of MS177 were measured (Figure 7A). First, MS177 had little growth-inhibitory effect in a non-malignant prostate epithelial line RWPE1 (Figure 7A bottom; Supplementary Figure S7A), suggesting that MS177 is not generally cytotoxic. In contrast, MS177 demonstrated a consistent and fast-acting anti-proliferation effect in all examined prostate cancer cell line models (Figure 7A–C; Supplementary Figure S7B, C). Notably, MS177 exhibited a magnitude increase of efficacy in inhibiting growth of 22Rv1 and LNCaP cells, compared with its non-PROTAC controls, C24, MS177N1 or MS177N2 (Figure 7B–D; see Supplementary Figure S4A for C24). We also confirmed MS177-induced EZH2 degradation in these cells (Supplementary Figure S7E, F). It appears that MS177 displayed higher anti-growth efficacies in AR-positive prostate cancer cells compared with those in AR-negative cells (Figure 7A). MS177 also showed a superior effect on inhibiting proliferation of late-stage, highly plastic prostate cancer cells known to be resistant to ADT (52,53) (Supplementary Figure S7D), indicating a broader application of MS177 to various prostate cancer stages. Furthermore, MS177 was much more potent in inhibiting 22Rv1 cell growth than a large panel of existing catalytic inhibitors of EZH2, including UNC1999 (27,54), CPI-1205 (26), EPZ-6438 (55), and GSK126 (56) and the EED degrader UNC6852 (Figure 7E, F; Supplementary Figure S7G). In addition, MS177 dose-dependently inhibited colony-forming capability (Figure 7G) and induced apoptosis of 22Rv1 cells, as assayed by immunoblotting for apoptosis markers (Figure 7H), whereas the non-PROTAC controls, MS177N1 or C24, had little effect (Figure 7G, H). Together, the EZH2-targeting PROTAC MS177 robustly induces growth inhibition in a range of human prostate cancer cell lines and, importantly, its cancer-killing effects are superior to those of EZH2 enzymatic inhibitors.
Figure 7.
MS177 exhibits much more potent anti-proliferation effects in prostate cancer cell lines, compared with the enzymatic inhibitors of EZH2. (A) EC50 values of MS177 in the indicated prostate cell lines after a 2, 4 and 6 day treatment. EC50 values are the means of three independent experiments. (B, C) Plots showing the growth-inhibitory effect of various used concentrations (x-axis; in log10 converted value) of either MS177, MS177N1 or MS177N2 in 22Rv1 (B) and LNCaP (C) cells, treated for 2, 4 or 6 days. The y-axis shows relative growth after normalization to DMSO-treated cells (n = 3 independent treatment experiments; presented as the mean ± SD). (D) Colony formation assay using 22Rv1 cells treated with different doses of MS177 or 0.5 μM C24, MS177N1 or MS177N2, compared with DMSO. (E) Plots showing the growth-inhibitory effect of various used concentration (x-axis; in log10 converted value) of MS177 or various EZH2 inhibitors in 22Rv1 cells treated for 2 days. The y-axis shows relative cell growth after normalization to DMSO-treated cells (n = 3 independent treatment experiments; presented as the mean ± SD). (F) Colony formation assay using 22Rv1 cells treated with 2.5 μM MS177 or the indicated EZH2 enzymatic inhibitors, compared with DMSO. (G) Representative view of soft agar-based assay (left) and quantification of colony formation (right; colony numbers counted by ImageJ and presented as the average ± SD of two independent experiments) using 22Rv1 cells, treated with DMSO, 2.5 μM C24 or MS177N1, or three indicated concentrations of MS177 (n = 2 independent treatment experiments; presented as the mean ± SD). (H) Immunoblotting for the indicated apoptotic markers in 22Rv1 cells, treated with either 5 μM MS177N1 or C24 or different indicated concentration of MS177, compared with DMSO, for 24 h. (I) A model showing that EZH2 forms canonical (EZH2:PRC2; top) and non-canonical (EZH2:AR:co-activators; bottom) complexes, both of which mediate tumorigenicity of advanced prostate cancer. EZH2-targeting PROTAC MS177 simultaneously suppresses oncogenic circuitries enforced by the above EZH2-associated onco-complexes, as well as those enforced by AR and AR variant signaling, thus leading to slowed proliferation and apoptosis of prostate cancer cells.
DISCUSSION
Increasing evidence pointed to multifaceted functions of EZH2 in cancer. In particular, EZH2 forms the so-called EZH2-solo sites in a PRC2-independent fashion and binds AR and AR variant in prostate cancer, eliciting a non-canonical gene activation-related effect (18). In this report, we unveiled, for the first time, (i) that EZH2 utilizes a hidden EZH2TAD to form an interaction with AR and its constitutively active variant, AR-V7; here, we further mapped the EZH2TAD-interacting interfaces to at least two unstructured regions of AR and AR-V7: the intrinsically disorganized poly(Q) and poly(G) protein sequences (57–59). These results suggest a multivalent protein–protein interaction involving unstructured protein regions (TADs themselves often being unstructured) and, possibly, liquid–liquid phase separation as seen with other transcription factors such as OCT4 and ER (60), which merits future studies. (ii) Integrated genomic profiling (CUT&RUN, ATAC-seq and RNA-seq), in combination with mutagenesis of EZH2TAD, clearly demonstrated that EZH2TAD-mediated interaction with AR and AR variant is crucial for establishment of the EZH2-solo:AR/AR-V7-co-bound pattern, for transcriptional activation of their common targets, and for the potentiation of advanced CRPC cell growth in vitro and in xenografted animal models. (iii) Furthermore, we show the higher expression of EZH2:AR:AR-V7-co-upregulated transcripts to be correlated with poorer outcomes of prostate cancer patients, thus demonstrating a clinical relevance for the EZH2-mediated non-canonical pathway studied by this work. (iv) By using a battery of biochemical, CUT&RUN and RNA-seq assays, we also observed that MS177, an EZH2-targeting PROTAC/degrader, efficiently causes on-target degradation of EZH2 and EZH2-associated canonical (EZH2:PRC2) and non-canonical (EZH2:AR:AR-V7) onco-complexes in prostate cancer cells (Figure 7I); here, we conducted careful MOA studies to show that such MS177-mediated degradation of AR and AR variant indeed relies on EZH2 binding by MS177 (as demonstrated by EZH2 depletion assay) and relies on EZH2TAD-mediated AR interaction (demonstrated by using an AR interaction-defective mutant of EZH2TAD). (v) Lastly, MS177 is superior to existing catalytic inhibitors of EZH2 in suppressing the in vitro growth of prostate cancer cells. Please note that, in individual gene KD experiments, we only knocked down each one of them (EZH2 alone, AR alone or AR-V7 alone) to generate a list of commonly affected genes, which were also affected by treatment with MS177 but not its PROTAC-inactive analogs (Figure 3I). However, the gene-modulatory effect of MS177 is far more dramatic, which is most probably due to MS177-induced simultaneous degradation of EZH2, AR and AR-V7 (as supported by our RNA-seq results, Supplementary Figure S6A, B; please also see the below Discussion section for a possible mechanism). We have also attempted to assess in vivo efficacy of MS177 in a 22Rv1 cell xenografted mouse model; however, we could not achieve sufficient exposure levels for MS177 in such an animal model (data not shown). Thus, we could not pursue this direction further with MS177. Optimization of MS177 into an improved EZH2 PROTAC/degrader with better in vivo pharmacokinetic properties is needed to demonstratethe in vivo efficacy of this therapeutic approach.
Interplays between EZH2 and AR/AR variant can be complex. Previous studies also showed that EZH2 directly binds the AR gene promoter in a PRC2-independent manner to activate the transcription of AR and thus AR’s downstream targets (such as PSA) and that EZH2 KD led to a significantly down-regulated AR protein level (20,24,48). In this study, we ruled out involvement of such a reported pathway in the MS177-induced degradation of AR and AR variant—indeed, within a short period of time of MS177 treatment (within 24 h), we found the AR and AR variant mRNA levels unaltered, and yet a vast majority of AR and AR variant proteins were already subjected to ubiquitination and proteasomal degradation. Given the existence of various cofactors of AR and AR variants, it is conceivable that only a subset of AR and AR variant proteins in cells forms interactions with EZH2 at a given time, a notion also supported by their common and distinctive genome-binding patterns (Figure 1F). Thus, the observed dramatic and global effect of EZH2-targeting PROTAC (MS177) on degradation of AR and AR variant is most probably due to the fast-acting and recycling nature of MS177, which operates to essentially ‘drain’ the majority of cellular AR and AR variant proteins [which exhibit a half-life time of ∼6–7 h (61)] in repeated cycles of degrading the target (EZH2) and their partners from various AR- and AR variant-containing complexes in prostate cancer cells. A similar ‘drainage’ effect of MS177 was observed with cMyc, another EZH2 non-canonical partner, in AML cells (23). It appears that MS177 is more potent in the treatment of AR-positive prostate cancer cells than of AR-negative ones, pointing to the relevance of the AR-degrading effect of MS177. Please note that EZH2TAD-mediated non-canonical gene activation functions may act in parallel with other EZH2-directed activities in prostate cancer, e.g. EZH2 was recently shown to regulate 2′-O-methylation of rRNA by directly interacting with fibrillarin, thereby promoting global protein translation of prostate cancer cells (21); moreover, EZH2 was also shown to bind to mutant p53 mRNA, increasing its stability and cap-independent protein translation in a methyltransferase-independent manner (62). Further, EZH2 was reported to methylate FOXA1, leading to stabilization of FOXA1 and activation of cell cycle-related genes (63). In theory, EZH2-targeting PROTACs represent a potentially attractive strategy for completely eliminating all of the multifaceted functions of EZH2 in prostate cancer, which awaits further investigation.
DATA AVAILABILITY
Genomic datasets of this study, including those of CUT&RUN and RNA-seq, have been deposited in the NCBI Gene Expression Omnibus (GEO) database under the accession number GSE205107. Human prostate cancer datasets were derived from the TCGA Research Network (http://cancergenome.nih.gov/). Publicly available datasets used in the work included those from NCBI GEO accession numbers GSE99378 (ATAC-seq data of 22Rv1 cells), GSM2827408 and GSE85558 (ChIP-seq for H3K27ac in 22Rv1 cells), GSE94013 [ChIP-seq for BRD4, AR and AR-V7 in 22Rv1 cells, cultured under either ligand-stripped (mock) or ligand-stimulated (DHT) conditions], GSE39459 (ChIP-seq for EZH2, H3K27me3, AR, H3K4me2, H3K4me3 and RNA Pol II in LNCaP-abl cells), GSE28950 (ChIP-seq for EZH2 and AR in VCaP cells), GSE14092 (ChIP-seq for H3K27me3 in VCaP cells), GSE123204 (ChIP-seq for EZH2 in PC3 cells), GSE57498 (ChIP-seq for H3K27me3 in PC3 cells), GSE135623 (ChIP-seq for EZH2 in DU145 cells), GSE82260 (ChIP-seq for H3K27me3 in DU145 cells) and GSE96652 (ChIP-seq for FOXA1 in 22Rv1 cells). Other datasets supporting the findings of this study are available upon request.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all members of the Wang, Jin and Cai laboratories and X. Liu for technical support and helpful discussion. We thank UNC core facilities, including the Tissue Culture Facility, High-throughput Sequencing Facility (HTSF), Bioinformatics Core Facility and Animal Studies Core Facility, for professional assistance and support of this work.
Contributor Information
Jun Wang, Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill School of Medicine, Chapel Hill, NC 27599, USA; Department of Biochemistry and Biophysics, University of North Carolina at Chapel Hill School of Medicine, Chapel Hill, NC 27599, USA.
Kwang-Su Park, Mount Sinai Center for Therapeutics Discovery, Departments of Pharmacological Sciences and Oncological Sciences, Tisch Cancer Institute, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA.
Xufen Yu, Mount Sinai Center for Therapeutics Discovery, Departments of Pharmacological Sciences and Oncological Sciences, Tisch Cancer Institute, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA.
Weida Gong, Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill School of Medicine, Chapel Hill, NC 27599, USA.
H Shelton Earp, Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill School of Medicine, Chapel Hill, NC 27599, USA; Department of Pharmacology, University of North Carolina at Chapel Hill School of Medicine, Chapel Hill, NC 27599, USA; Department of Medicine, University of North Carolina at Chapel Hill School of Medicine, Chapel Hill, NC, 27599, USA.
Gang Greg Wang, Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill School of Medicine, Chapel Hill, NC 27599, USA; Department of Biochemistry and Biophysics, University of North Carolina at Chapel Hill School of Medicine, Chapel Hill, NC 27599, USA; Department of Pharmacology, University of North Carolina at Chapel Hill School of Medicine, Chapel Hill, NC 27599, USA.
Jian Jin, Mount Sinai Center for Therapeutics Discovery, Departments of Pharmacological Sciences and Oncological Sciences, Tisch Cancer Institute, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA.
Ling Cai, Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill School of Medicine, Chapel Hill, NC 27599, USA; Department of Genetics, University of North Carolina at Chapel Hill School of Medicine, Chapel Hill, NC 27599, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Core facilities affiliated to UNC Lineberger Comprehensive Cancer Center (LCCC) are supported partly by UNC LCCC Core Support Grant [P30-CA016086]. This work was also supported in part by grants from the US National Institutes of Health (NIH) [R01CA262903 to L.C.]; [R01CA218600 to J.J. and G.G.W.]; [R01CA268519 to G.G.W. and J.J.]; [R01CA211336 to G.G.W.]; and [R01CA230854 to J.J.], and UNC LCCC’s UCRF Stimulus Initiative Grant (to L.C.). G.G.W. is a Leukemia and Lymphoma Society (LLS) Scholar. This work utilized the NMR Spectrometer Systems at Mount Sinai acquired with funding from NIH SIG grants 1S10OD025132 and 1S10OD028504.
Conflict of interest statement. Icahn School of Medicine at Mount Sinai filed a patent application (WO 2018/081530 A1) covering EZH2 degraders that lists J.J. as an inventor. The Jin laboratory received research funds from Celgene Corporation, Levo Therapeutics, Cullgen, Inc. and Cullinan Oncology. J.J. is a co-founder and equity shareholder in Cullgen Inc. and a consultant for Cullgen Inc., EpiCypher Inc. and Accent Therapeutics Inc. The remaining authors declare no competing interests.
REFERENCES
- 1. Siegel R., Ma J., Zou Z., Jemal A.. Cancer statistics, 2014. CA Cancer J. Clin. 2014; 64:9–29. [DOI] [PubMed] [Google Scholar]
- 2. Attard G., Cooper C.S., Bono J.S.. Steroid hormone receptors in prostate cancer: a hard habit to break?. Cancer Cell. 2009; 16:458–462. [DOI] [PubMed] [Google Scholar]
- 3. Debes J.D., Tindall D.J.. Mechanisms of androgen-refractory prostate cancer. N. Engl. J. Med. 2004; 351:1488–1490. [DOI] [PubMed] [Google Scholar]
- 4. Feldman B.J., Feldman D.. The development of androgen-independent prostate cancer. Nat. Rev. Cancer. 2001; 1:34–45. [DOI] [PubMed] [Google Scholar]
- 5. Wasmuth E.V., Broeck A.V., LaClair J.R., Hoover E.A., Lawrence K.E., Paknejad N., Pappas K., Matthies D., Wang B., Feng W.et al.. Allosteric interactions prime androgen receptor dimerization and activation. Mol. Cell. 2022; 82:2021–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Watson P.A., Arora V.K., Sawyers C.L.. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat. Rev. Cancer. 2015; 15:701–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hu R., Dunn T.A., Wei S., Isharwal S., Veltri R.W., Humphreys E., Han M., Partin A.W., Vessella R.L., Isaacs W.B.et al.. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. 2009; 69:16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Guo Z., Yang X., Sun F., Jiang R., Linn D.E., Chen H., Chen H., Kong X., Melamed J., Tepper C.G.et al.. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res. 2009; 69:2305–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xu B., Konze K.D., Jin J., Wang G.G.. Targeting EZH2 and PRC2 dependence as novel anticancer therapy. Exp. Hematol. 2015; 43:698–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Morin R.D., Johnson N.A., Severson T.M., Mungall A.J., An J., Goya R., Paul J.E., Boyle M., Woolcock B.W., Kuchenbauer F.et al.. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat. Genet. 2010; 42:181–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sneeringer C.J., Scott M.P., Kuntz K.W., Knutson S.K., Pollock R.M., Richon V.M., Copeland R.A.. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc. Natl Acad. Sci. USA. 2010; 107:20980–20985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Souroullas G.P., Jeck W.R., Parker J.S., Simon J.M., Liu J.Y., Paulk J., Xiong J., Clark K.S., Fedoriw Y., Qi J.et al.. An oncogenic ezh2 mutation induces tumors through global redistribution of histone 3 lysine 27 trimethylation. Nat. Med. 2016; 22:632–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Beguelin W., Popovic R., Teater M., Jiang Y., Bunting K.L., Rosen M., Shen H., Yang S.N., Wang L., Ezponda T.et al.. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell. 2013; 23:677–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Burr M.L., Sparbier C.E., Chan K.L., Chan Y.C., Kersbergen A., Lam E.Y.N., Azidis-Yates E., Vassiliadis D., Bell C.C., Gilan O.et al.. An evolutionarily conserved function of polycomb silences the MHC class I antigen presentation pathway and enables immune evasion in cancer. Cancer Cell. 2019; 36:385–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim K.H., Roberts C.W.. Targeting EZH2 in cancer. Nat. Med. 2016; 22:128–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang J., Wang G.G.. No easy way out for EZH2: its pleiotropic, noncanonical effects on gene regulation and cellular function. Int. J. Mol. Sci. 2020; 21:9501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Davies A., Nouruzi S., Ganguli D., Namekawa T., Thaper D., Linder S., Karaoglanoglu F., Omur M.E., Kim S., Kobelev M.et al.. An androgen receptor switch underlies lineage infidelity in treatment-resistant prostate cancer. Nat. Cell Biol. 2021; 23:1023–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xu K., Wu Z.J., Groner A.C., He H.H., Cai C., Lis R.T., Wu X., Stack E.C., Loda M., Liu T.et al.. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012; 338:1465–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dardenne E., Beltran H., Benelli M., Gayvert K., Berger A., Puca L., Cyrta J., Sboner A., Noorzad Z., MacDonald T.et al.. N-Myc induces an EZH2-mediated transcriptional program driving neuroendocrine prostate cancer. Cancer Cell. 2016; 30:563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim J., Lee Y., Lu X., Song B., Fong K.W., Cao Q., Licht J.D., Zhao J.C., Yu J.. Polycomb- and methylation-independent roles of EZH2 as a transcription activator. Cell Rep. 2018; 25:2808–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yi Y., Li Y., Meng Q., Li Q., Li F., Lu B., Shen J., Fazli L., Zhao D., Li C.et al.. A PRC2-independent function for EZH2 in regulating rRNA 2′-O methylation and IRES-dependent translation. Nat. Cell Biol. 2021; 23:341–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Anwar T., Arellano-Garcia C., Ropa J., Chen Y.C., Kim H.S., Yoon E., Grigsby S., Basrur V., Nesvizhskii A.I., Muntean A.et al.. p38-mediated phosphorylation at T367 induces EZH2 cytoplasmic localization to promote breast cancer metastasis. Nat. Commun. 2018; 9:2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang J., Yu X., Gong W., Liu X., Park K.S., Ma A., Tsai Y.H., Shen Y., Onikubo T., Pi W.C.et al.. EZH2 noncanonically binds cMyc and p300 through a cryptic transactivation domain to mediate gene activation and promote oncogenesis. Nat. Cell Biol. 2022; 24:384–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jiao L., Shubbar M., Yang X., Zhang Q., Chen S., Wu Q., Chen Z., Rizo J., Liu X.. A partially disordered region connects gene repression and activation functions of EZH2. Proc. Natl Acad. Sci. USA. 2020; 117:16992–17002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bhat K.P., Umit Kaniskan H., Jin J., Gozani O.. Epigenetics and beyond: targeting writers of protein lysine methylation to treat disease. Nat. Rev. Drug Discov. 2021; 20:265–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vaswani R.G., Gehling V.S., Dakin L.A., Cook A.S., Nasveschuk C.G., Duplessis M., Iyer P., Balasubramanian S., Zhao F., Good A.C.et al.. Identification of (R)-N-((4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-2-methyl-1-(1-(1-(2,2,2-trifluoroethyl)piperidin-4-yl)ethyl)-1H-indole-3-carboxamide (CPI-1205), a potent and selective inhibitor of histone methyltransferase EZH2, suitable for phase I clinical trials for B-cell lymphomas. J. Med. Chem. 2016; 59:9928–9941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Konze K.D., Ma A., Li F., Barsyte-Lovejoy D., Parton T., Macnevin C.J., Liu F., Gao C., Huang X.P., Kuznetsova E.et al.. An orally bioavailable chemical probe of the lysine methyltransferases EZH2 and EZH1. ACS Chem. Biol. 2013; 8:1324–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cai L., Tsai Y.H., Wang P., Wang J., Li D., Fan H., Zhao Y., Bareja R., Lu R., Wilson E.M.et al.. ZFX mediates non-canonical oncogenic functions of the androgen receptor splice variant 7 in castrate-resistant prostate cancer. Mol. Cell. 2018; 72:341–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li J., Galbo P.M. Jr, Gong W., Storey A.J., Tsai Y.H., Yu X., Ahn J.H., Guo Y., Mackintosh S.G., Edmondson R.D.et al.. ZMYND11-MBTD1 induces leukemogenesis through hijacking nua4/TIP60 acetyltransferase complex and a PWWP-mediated chromatin association mechanism. Nat. Commun. 2021; 12:1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu T., Ortiz J.A., Taing L., Meyer C.A., Lee B., Zhang Y., Shin H., Wong S.S., Ma J., Lei Y.et al.. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 2011; 12:R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yu X., Li D., Kottur J., Shen Y., Kim H.S., Park K.S., Tsai Y.H., Gong W., Wang J., Suzuki K.et al.. A selective WDR5 degrader inhibits acute myeloid leukemia in patient-derived mouse models. Sci. Transl. Med. 2021; 13:eabj1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C.. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017; 14:417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S.et al.. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K.. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019; 10:1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pinero J., Bravo A., Queralt-Rosinach N., Gutierrez-Sacristan A., Deu-Pons J., Centeno E., Garcia-Garcia J., Sanz F., Furlong L.I.. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017; 45:D833–D839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. DeRose Y.S., Gligorich K.M., Wang G., Georgelas A., Bowman P., Courdy S.J., Welm A.L., Welm B.E.. Patient-derived models of human breast cancer: protocols for in vitro and in vivo applications in tumor biology and translational medicine. Curr. Protoc. Pharmacol. 2013; Chapter 14:Unit14.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang J., Wu T., Simon J., Takada M., Saito R., Fan C., Liu X.D., Jonasch E., Xie L., Chen X.et al.. VHL substrate transcription factor ZHX2 as an oncogenic driver in clear cell renal cell carcinoma. Science. 2018; 361:290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu X., Simon J.M., Xie H., Hu L., Wang J., Zurlo G., Fan C., Ptacek T.S., Herring L., Tan X.et al.. Genome-wide screening identifies SFMBT1 as an oncogenic driver in cancer with VHL loss. Mol. Cell. 2020; 77:1294–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Skene P.J., Henikoff J.G., Henikoff S.. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat. Protoc. 2018; 13:1006–1019. [DOI] [PubMed] [Google Scholar]
- 43. Liao Y., Chen C.H., Xiao T., de la Pena Avalos B., Dray E.V., Cai C., Gao S., Shah N., Zhang Z., Feit A.et al.. Inhibition of EZH2 transactivation function sensitizes solid tumors to genotoxic stress. Proc. Natl Acad. Sci. USA. 2022; 119:e2105898119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xu D., Zhan Y., Qi Y., Cao B., Bai S., Xu W., Gambhir S.S., Lee P., Sartor O., Flemington E.K.et al.. Androgen receptor splice variants dimerize to transactivate target genes. Cancer Res. 2015; 75:3663–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chng K.R., Chang C.W., Tan S.K., Yang C., Hong S.Z., Sng N.Y., Cheung E.. A transcriptional repressor co-regulatory network governing androgen response in prostate cancers. EMBO J. 2012; 31:2810–2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Potjewyd F., Turner A.W., Beri J., Rectenwald J.M., Norris-Drouin J.L., Cholensky S.H., Margolis D.M., Pearce K.H., Herring L.E., James L.I.. Degradation of polycomb repressive complex 2 with an EED-targeted bivalent chemical degrader. Cell Chem. Biol. 2020; 27:47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lasko L.M., Jakob C.G., Edalji R.P., Qiu W., Montgomery D., Digiammarino E.L., Hansen T.M., Risi R.M., Frey R., Manaves V.et al.. Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature. 2017; 550:128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu Q., Wang G., Li Q., Jiang W., Kim J.S., Wang R., Zhu S., Wang X., Yan L., Yi Y.et al.. Polycomb group proteins EZH2 and EED directly regulate androgen receptor in advanced prostate cancer. Int. J. Cancer. 2019; 145:415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Park S.H., Fong K.W., Mong E., Martin M.C., Schiltz G.E., Yu J.. Going beyond polycomb: EZH2 functions in prostate cancer. Oncogene. 2021; 40:5788–5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Varambally S., Dhanasekaran S.M., Zhou M., Barrette T.R., Kumar-Sinha C., Sanda M.G., Ghosh D., Pienta K.J., Sewalt R.G., Otte A.P.et al.. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002; 419:624–629. [DOI] [PubMed] [Google Scholar]
- 51. Yang X., Li F., Konze K.D., Meslamani J., Ma A., Brown P.J., Zhou M.M., Arrowsmith C.H., Kaniskan H.U., Vedadi M.et al.. Structure–activity relationship studies for enhancer of zeste homologue 2 (EZH2) and enhancer of zeste homologue 1 (EZH1) inhibitors. J. Med. Chem. 2016; 59:7617–7633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ku S.Y., Rosario S., Wang Y., Mu P., Seshadri M., Goodrich Z.W., Goodrich M.M., Labbe D.P., Gomez E.C., Wang J.et al.. Rb1 and trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science (New York, N.Y. 2017; 355:78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mu P., Zhang Z., Benelli M., Karthaus W.R., Hoover E., Chen C.C., Wongvipat J., Ku S.Y., Gao D., Cao Z.et al.. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science. 2017; 355:84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Xu B., On D.M., Ma A., Parton T., Konze K.D., Pattenden S.G., Allison D.F., Cai L., Rockowitz S., Liu S.et al.. Selective inhibition of EZH2 and EZH1 enzymatic activity by a small molecule suppresses MLL-rearranged leukemia. Blood. 2015; 125:346–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Knutson S.K., Wigle T.J., Warholic N.M., Sneeringer C.J., Allain C.J., Klaus C.R., Sacks J.D., Raimondi A., Majer C.R., Song J.et al.. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat. Chem. Biol. 2012; 8:890–896. [DOI] [PubMed] [Google Scholar]
- 56. McCabe M.T., Ott H.M., Ganji G., Korenchuk S., Thompson C., Van Aller G.S., Liu Y., Graves A.P., Della Pietra A. 3rd, Diaz E.et al.. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012; 492:108–112. [DOI] [PubMed] [Google Scholar]
- 57. Orafidiya F.A., McEwan I.J.. Trinucleotide repeats and protein folding and disease: the perspective from studies with the androgen receptor. Future Sci. OA. 2015; 1:FSO47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tan M.H., Li J., Xu H.E., Melcher K., Yong E.L.. Androgen receptor: structure, role in prostate cancer and drug discovery. Acta Pharmacol. Sin. 2015; 36:3–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. van der Lee R., Buljan M., Lang B., Weatheritt R.J., Daughdrill G.W., Dunker A.K., Fuxreiter M., Gough J., Gsponer J., Jones D.T.et al.. Classification of intrinsically disordered regions and proteins. Chem. Rev. 2014; 114:6589–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Boija A., Klein I.A., Sabari B.R., Dall’Agnese A., Coffey E.L., Zamudio A.V., Li C.H., Shrinivas K., Manteiga J.C., Hannett N.M.et al.. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell. 2018; 175:1842–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Astapova O., Seger C., Hammes S.R.. Ligand binding prolongs androgen receptor protein half-life by reducing its degradation. J. Endocr. Soc. 2021; 5:bvab035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhao Y., Ding L., Wang D., Ye Z., He Y., Ma L., Zhu R., Pan Y., Wu Q., Pang K.et al.. EZH2 cooperates with gain-of-function p53 mutants to promote cancer growth and metastasis. EMBO J. 2019; 38:e99599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Park S.H., Fong K.W., Kim J., Wang F., Lu X., Lee Y., Brea L.T., Wadosky K., Guo C., Abdulkadir S.A.et al.. Posttranslational regulation of FOXA1 by polycomb and BUB3/USP7 deubiquitin complex in prostate cancer. Sci. Adv. 2021; 7:eabe2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genomic datasets of this study, including those of CUT&RUN and RNA-seq, have been deposited in the NCBI Gene Expression Omnibus (GEO) database under the accession number GSE205107. Human prostate cancer datasets were derived from the TCGA Research Network (http://cancergenome.nih.gov/). Publicly available datasets used in the work included those from NCBI GEO accession numbers GSE99378 (ATAC-seq data of 22Rv1 cells), GSM2827408 and GSE85558 (ChIP-seq for H3K27ac in 22Rv1 cells), GSE94013 [ChIP-seq for BRD4, AR and AR-V7 in 22Rv1 cells, cultured under either ligand-stripped (mock) or ligand-stimulated (DHT) conditions], GSE39459 (ChIP-seq for EZH2, H3K27me3, AR, H3K4me2, H3K4me3 and RNA Pol II in LNCaP-abl cells), GSE28950 (ChIP-seq for EZH2 and AR in VCaP cells), GSE14092 (ChIP-seq for H3K27me3 in VCaP cells), GSE123204 (ChIP-seq for EZH2 in PC3 cells), GSE57498 (ChIP-seq for H3K27me3 in PC3 cells), GSE135623 (ChIP-seq for EZH2 in DU145 cells), GSE82260 (ChIP-seq for H3K27me3 in DU145 cells) and GSE96652 (ChIP-seq for FOXA1 in 22Rv1 cells). Other datasets supporting the findings of this study are available upon request.