Abstract
Standalone ring nucleases are CRISPR ancillary proteins, which downregulate the immune response of Type III CRISPR-Cas systems by cleaving cyclic oligoadenylates (cA) second messengers. Two genes with this function have been found within the Sulfolobus islandicus (Sis) genome. They code for a long polypeptide composed by a CARF domain fused to an HTH domain and a short polypeptide constituted by a CARF domain with a 40 residue C-terminal insertion. Here, we determine the structure of the apo and substrate bound states of the Sis0455 enzyme, revealing an insertion at the C-terminal region of the CARF domain, which plays a key role closing the catalytic site upon substrate binding. Our analysis reveals the key residues of Sis0455 during cleavage and the coupling of the active site closing with their positioning to proceed with cA4 phosphodiester hydrolysis. A time course comparison of cA4 cleavage between the short, Sis0455, and long ring nucleases, Sis0811, shows the slower cleavage kinetics of the former, suggesting that the combination of these two types of enzymes with the same function in a genome could be an evolutionary strategy to regulate the levels of the second messenger in different infection scenarios.
INTRODUCTION
The discovery of an adaptive prokaryotic immune system called Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR), in which the repeats associate with Cas (CRISPR associated) proteins, has constituted a revolution in life sciences. Their discovery (1–3)- and straightforward development into versatile nucleases (4–6) by guide RNA exchange paved the way for modifications à la carte that can be employed in biomedicine (7) and biotechnology (8–10). CRISPR-Cas systems are ribonucleoprotein (RNP) complexes that are highly diverse because of their different evolutionary origins. The CRISPR immune response consists of three stages mediated by a distinct subset of Cas proteins involving adaptation, CRISPR (cr) RNA maturation and interference, concluding with the recognition and cleavage of the target DNA or RNA (10–14).
The CRISPR-Cas systems are divided into two classes according to the Cas proteins composing the nuclease module (15). Class 1 interference modules consist of a multi-subunit protein complex, while Class 2 modules comprise a single multidomain protein. The two classes are further divided into six types and many subtypes depending on which other Cas proteins are present in other functional modules.
The Class 1 Type III constitutes a complex CRISPR immune system of particular interest, as its members deploy an intricated response controlled by a multipronged regulatory pathway to degrade both the mRNA and DNA of the invader (16–18). The large Type III interference complexes are characterized by the presence of the multidomain Cas10 signature protein, which commonly harbours two active sites for ssDNA cleavage (18–21) and a cyclase domain for cyclic oligoadenylate (cA) synthesis (22–24). The recognition of a target RNA triggers the catalytic activities of Cas10, and the cyclase domain polymerises ATP into cA species ranging between 3- and 6-AMP subunits (cAn) (22–25). These molecules act as a second messenger promoting the activation of CRISPR ancillary nucleases (Csx1/Csm6, Can1/Can2 and NucC families), which are the peons of the Type III immune response (22,23,26–28), degrading both host and invading nucleic acids in the cell, resulting in viral clearance, cell dormancy or cell death (29). These enzymes are key components of cyclic oligonucleotide-based antiphage signalling systems (CBASS). They contain CRISPR Associated Rossmann Fold (CARF) domains, which are involved in ligand binding. The CARF superfamily includes numerous domain architectures, in most of which the CARF domain is fused to a nuclease (30).
The control of cAn levels require a regulatory system that can modulate or stop the activity of the indiscriminate nucleases. Although the CARF domains of some Csm6 proteins have been shown to slowly degrade cA4 or cA6, thereby self-limiting their ribonuclease activity (31–34), a major part of Type III systems includes standalone cyclic oligoadenylate degrading enzymes, also termed CRISPR ring nucleases (Crn), to fulfil this function. The first two members of this family were described in Sulpholobus solfataricus (Sso), where the Sso1393 (UniProt ID: Q97YD2) and Sso2081 (UniProt ID: Q7LYJ6) proteins, were shown to bind and degrade cA4 using a metal-independent mechanism, thus returning cells to a basal uninfected state (31). The same function has been assigned to the Sulfolobus islandicus (Sis) Sis0811 (UniProt ID: F0NH89) and Sis0455 (UniProt ID: F0NGX6) enzymes (32,35). These cyclic oligoadenylate degrading enzymes are CARF-domain proteins of the Crn1 family. From the structural point of view, two types of enzymes can be observed in this family. One of them includes a HTH domain joined to a CARF domain that belongs to the CARF7 major clade (36) (Sis0811 and Sso1393, Crn1 Large Standalone Ring Nucleases, LSRN), while the second one displays a small insertion in the C-terminal region of the CARF domain and belongs to the CARF_m13 minor clade (Sis0455 and Sso2081, Crn1 Small Standalone Ring Nucleases, SSRN) (Supplementary Figure S1). We have recently deciphered the catalytic mechanism of Sis0811 (37). However, no structural or mechanistic details are available for Sis0455, thus precluding our understanding of the SSRN group.
Here, we determine the structure of Sis0455 in its apo and substrate-bound form. The structure reveals the dimeric assembly of the enzyme and the unique structural feature of the insertion in the C-terminus of the polypeptide. The substrate-bound conformation unveils the molecular details of cA4 recognition by the standalone ring nuclease. We observe that cA4 binding induces a conformational change from an open to a closed state. The conformational change cages the second messenger and avoids its dissociation by closing the catalytic site with the insertion located at the C-terminal of the CARF domains. Our time course mass spectrometry and binding experiments show that Sis0455 possesses a very high affinity for the substrate and suggest a slower catalytic mechanism than Sis0811. Our study provides the first structural insight into the substrate recognition and processing of this type of standalone ring nucleases, which together with the other ancillary proteins, regulate the Type III CRISPR defence system.
MATERIALS AND METHODS
Construction of expression plasmids
Sis0455 wild type sequence from Sis REY15A was synthesized by the Integrated DNA Technology (IDT, USA). The gene was then cloned by In-Fusion HD Cloning Plus (Tanaka) into pET-21 with a C-terminal extension encoding a TEV (Tobacco Etch Virus) protease target site and a 6× His-tag (histidine tag). The different mutants employed for the biochemical characterization of the enzyme were derived from this plasmid by site-directed mutagenesis carried out by the genomics service company Genewiz.
Purification of ring nucleases
His-tagged Sis0455 and all its variants were expressed and purified from Escherichia coli BL21 pRARE cells. Cells were grown in LB media containing ampicillin (1 mM) and chloramphenicol (34 μg/ml) at 37 °C until an OD at 600nm wavelength of 0.6 was reached. Expression was induced by adding 0.5 mM of isopropyl β-d-1-thiogalactopyranoside (IPTG) at 37°C during 3h. The cells were harvested and re-suspended in lysis buffer (50 mM HEPES pH 7.5, 2 M NaCl, 5 mM MgCl2) in a ratio of about 10 ml buffer/1g cells supplemented with 1 protease inhibitor tablet (Roche Diagnostics GmbH), lysozyme and 1 μl Benzonase. Cells were lysed by sonication for 8 min at 35% amplitude with 15 seconds on and 20 s off cycle and then cell debris and insoluble particles were removed by centrifugation at 11 000 rpm for 45 min at 4 °C (Thermo Fisher Scientific, Multifuge X Pro). The supernatant was separated from the pellet and diluted to 500 mM NaCl (Dilution buffer: 50 mM HEPES pH 7.5, 5 mM MgCl2). This sample was then loaded onto a 5 ml Crude HisTrap FF column (GE Healthcare) equilibrated in buffer A (50 mM HEPES pH 7.5, 5 mM MgCl2, 500 mM NaCl). Elution of the proteins was performed by a stepwise gradient of buffer B (50 mM HEPES pH 7.5, 5 mM MgCl2, 500 mM NaCl, 500 mM Imidazole). Enriched protein fractions were applied onto a 5 ml HiTrap Q HP column (GE Healthcare) equilibrated with buffer A2 (20 mM Tris–HCl pH 8.0, 50 mM NaCl). The protein was eluted with a linear gradient of 0–100% buffer B2 (20 mM Tris–HCl pH 8.0, 1M NaCl). Protein-rich fractions were loaded onto a HiLoad 16/600 75 Superdex column (GE Healthcare) equilibrated in gel filtration buffer GF (25 mM HEPES pH 8.0, 300 mM KCl). The protein fractions were concentrated to ∼12 mg/ml (using 10 kDa MWCO Centriprep Amicon Ultra devices) and aliquots were flash-frozen in liquid nitrogen and subsequently stored at -80°C.
Following the expression and purification protocol described above but growing cells in a Selenomethionine-enriched media, Selenomethionine-labeled Sis0455 protein was also purified for further crystallization experiments. Samples purity were monitored by SDS-PAGE gels.
Size-exclusion chromatography–multi-angle light scattering (SEC-MALS)
SEC-MALS experiments were performed using a Dionex (Thermo Scientific) HPLC system connected in-line to a UV detector (Thermo Scientific Dionex Ultimate 3000, MWD-3000), a Wyatt Dawn8 + Heleos 8-angle light-scattering detector and a Wyatt Optilab T-rEX refractive index detector. SEC was performed using a Superdex 200 Increase 10/300 GL column (GE Healthcare) at 20°C in a buffer containing 50 mM HEPES pH 8.0, 300 mM KCl. For the analysis, 50 μl of Sis0455 were injected at 5.0 mg/ml and 0.5 ml/min flow rate. ASTRA (version 8.0.2.5) software was used to collect the data from the UV, refractive index, and light scattering detectors. The weight average molecular masses, Mw, were determined across the elution profile from static LS measurements using ASTRA software and a Zimm model, which relates the amount of scattered light to the weight average molecular weight of the solute, the concentration of the sample, and the square of the refractive index increment (dn/dc) of the sample.
Crystallization
Initial crystallization screenings with Sis0455 sample were performed at 293 K using the sitting-drop vapor-diffusion method and testing a collection of commercially available crystallization screens. The initial drops consisted of 0.15 μl of protein solution (13.54 mg/ml in 25 mM HEPES pH 8.0, and 300 mM KCl) and 0.15 μl well solution, and were equilibrated against 70 μl of well solution. After 60 days, the extensive initial screening only rendered plate-like crystals in 25% PEG 4000 and 0.05 M tricine pH 8.0. These crystals were subsequently scaled up and optimized using a dragonfly (TTP) screen optimizer yielding plate-like crystals grown in 22% PEG 4000 and 0.05 M tricine pH 8.0. Crystals were cryo-protected by adding 20% (v/v) glycerol to the mother liquor before flash-freezing in liquid nitrogen.
Since selenomethionine-modified crystals were not reproducible under the native conditions mentioned above, initial crystallization screenings were performed from the scratch using the selenomethionine sample previously incubated with cA4 substrate before setting up the crystallization trials. After optimizing initial hits found in reservoir conditions based on 40% PEG 400, best diffracting crystals were grown in 41% PEG 400, 0.1M sodium acetate pH 4.5, 0.1M Li2SO4. As crystals were grown in cryo-conditions, they were directly flash-freeze in liquid nitrogen.
X-ray data collection
All data were collected from frozen crystals at 100 K with EIGER and PILATUS detectors at beamlines PXI and PXIII (SLS, Villigen, Switzerland) and at BioMax (MAX-IV, Lund, Sweden). Data processing and scaling were accomplished using XDS (38), POINTLESS and AIMLESS (39) as implemented in autoPROC (40). Statistics for the crystallographic data and structure solution are summarized in Table 1.
Table 1.
X-ray crystallographic data collection and refinement statistics
| Sis0455 | Sis0455: cA4 | |
|---|---|---|
| Apo | Substrate bound form | |
| Data collection | ||
| Space group | P21 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 42.42, 51.00, 80.35 | 42.98, 79.53, 97.74 |
| α, β, γ (°) | 90, 98.70, 90 | 90, 90, 90 |
| Wavelength | 1.00 | 0.98 |
| Resolution (Å) | 79.43–2.27 (2.31–2.27)* | 61.24–1.66 (1.69–1.66)* |
| R pim | 0.04 (0.31) | 0.02 (0.32) |
| CC(1/2) | 0.99 (0.70) | 0.99 (0.79) |
| Mean I / σI | 11.7 (2.5) | 20.7 (2.2) |
| Completeness (%) | 98.8 (93.5) | 99.8 (89.9) |
| Redundancy | 6.8 (6.0) | 27.4 (24.3) |
| Refinement | ||
| Resolution (Å) | 79.43–2.27 | 47.92–1.66 |
| No. reflections | 14957 | 36894 |
| R work/Rfree | 0.18/0.25 | 0.21/0.25 |
| Molecules by a.u. | 2 | 2 |
| No. atoms | ||
| Protein | 2853 | 3009 |
| cA4 | 0 | 88 |
| Water | 30 | 36 |
| Ramachandran | ||
| Favored/allowed (%) | 100 | 99.73 |
| Disallowed (%) | 0 | 0.27 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.013 | 0.014 |
| Bond angles (°) | 1.642 | 1.649 |
| PDB code | 7Z56 | 7Z55 |
*Values in parentheses are for highest-resolution shell. One crystal was used to solve the structure.
Crystal structure solution, model building and refinement
Sis0455:cA4 complex structure was solved from selenomethionine-modified crystals by experimental phasing using single-wavelength anomalous diffraction (SAD) method, as implemented in the program CRANK2 (41). Then, using a monomer from the Sis0455:cA4 complex as a searching model, Sis0455 apo structure was solved by molecular replacement method, as implemented in the program PHASER (42). The quality of the electron density maps provided from this molecular replacement solution allowed us to manually retrace the full model of Sis0455 apo structure. Both models were subjected to iterative cycles of model building and refinement with COOT (43), PHENIX (44) and REFMAC (45) yielding the refinement and data collection statistics summarized in the Table 1. The apo and complex final models have a Rwork/Rfree of 18/25 and 21/25 with 0.00 and 0.27% of the residues in disallowed regions of the Ramachandran plot, respectively. Figures were generated using PyMOL (The PyMOL Molecular Graphics System, Version 2.0 Schrodinger, LLC) and ChimeraX (46,47).
Isothermal titration calorimetry (ITC)
Cyclic tetraadenylate (cA4; catalog number, C355) was acquired from BIOLOG Life Science Institute (Bremen, Germany). Prior to ITC experiments both the proteins (Sis0455 and its variants) and the cA4 were extensively dialyzed against ITC buffer (50 mM HEPES pH 8.0, 300 mM KCl). Protein concentrations were determined using a spectrophotometer by measuring the absorbance at 280 nm and applying values for the extinction coefficients computed from the corresponding sequences by the ProtParam program (http://web.expasy.org/protparam/). The cA4 concentration was determined as well by measuring the absorbance at 260 nm and using a extinction coefficient of 54000 M-1 cm-1. cA4 at approximately 50 or 100 μM concentration was loaded into the syringe and titrated into the calorimetric cell containing the Sis0455 proteins at ∼5 or 10 μM, respectively. The reference cell was filled with distilled water. All ITC experiments were performed on an Auto-iTC200 instrument (Microcal, Malvern Instruments Ltd) at 25°C. The titration sequence consisted of a single 0.4 μl injection followed by 19 injections, 2 μl each, with 150 s spacing between injections to ensure that the thermal power returns to the baseline before the next injection. The stirring speed was 750 rpm. Control experiments with the cyclic tetra-adenylate injected in the sample cell filled with buffer were carried out under the same experimental conditions. These control experiments showed heats of dilution negligible in all cases. The heats per injection normalized per mole of injectant versus the molar ratio [cA4]/[Sis0455 variants] were fitted to a single-site model. Data were analysed with MicroCal PEAQ-ITC (version 1.1.0.1262) analysis software (Malvern Instruments Ltd).
Activity assay and liquid chromatography (LC) ESI-MS
Reactions of 50 μl containing 40 μM cA4, 2 μM Sis0455 in reaction buffer (20 mM Tris pH 8.0, 100 mM NaCl, 1 mM EDTA) were set and incubated at 70°C for one hour in a thermocycler. As a control, a reaction without any enzyme was also set. After this time, the reactions were stopped by lowering the temperature up to 4°C. Then the protein from each of the reactions was removed by performing a phenol extraction. This step was achieved by adding 50 μl of phenol to each of the reactions and followed by vortexing until the solution turns cloudy. Then the organic and aqueous phases were separated by centrifugation (60 s at 7000 rpm) and the aqueous phase dried down in a SpeedVac. The pellet containing the cyclic oligoadenylate compounds was resuspended in 8.6 mM TEA pH 8.3 (LC-MS buffer A) for further analysis.
For the time course experiments, an initial larger volume of reaction was used and the reaction stopped at different time points (0, 2, 5, 10, 20, 30, 40, 50 and 60 min) and processed as above.
All experiments were done, at least, in duplicates.
The solutions containing the cyclic oligoadenylate compounds were injected into a UPLC system (UltiMate 3000, Dionex) using a Kinetex® EVO C18 reverse phase column (Phenomenex, 2.1 mm × 100 mm, 5.0 m particle size) at 30°C. The elution of the column was then analysed with a microOTOF-Q II mass spectrometer (Bruker Daltonik GmhH) equipped with an electrospray ionization (ESI) source (capillary voltage 4500 V, end plate offset –500 V, nebulizer gas (nitrogen) pressure 2.0 bar, flow 9 l/min and drying gas temperature 200°C). The mobile phase of the chromatography was applied at a flow rate of 0.2 mL/min and consisted of a gradient of Buffer A (8.6 mM TEA pH 8.3) and B (8.6 mM TEA pH 8.3, 5% acetonitrile) as follows: 0–4 min 0% B, 4–6 min 100% B, 6–8 min 0% B. Mass data acquisition was performed in negative-ion mode with a mass resolving power of 10 000 and a scan range of 300–1500. Data acquisition was done under the control of the module Hystar 3.2-SR 2 from Bruker Compass 1.3 software that integrates both the LC chromatographic separation and MS methods. Data analysis was done with DataAnalysis Version 4.0 SP5 (Bruker Daltonik GmhH). For the time courses series, the areas under the respective peaks were integrated using QuantAnalysis Version 2.0 SP5 (Bruker Daltonik GmbH).
RESULTS
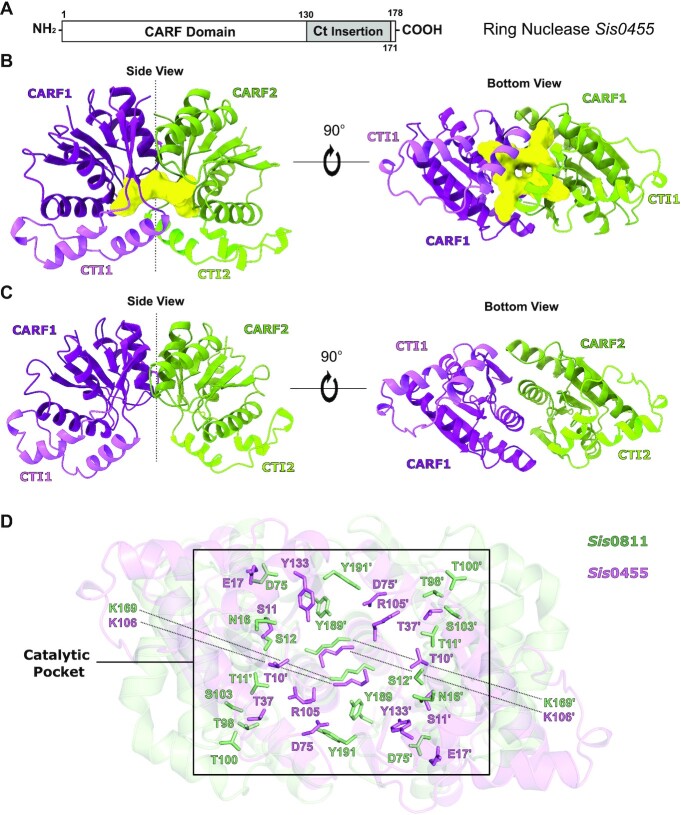
Crystal Structure of Sis0455
Sis0455 is composed of a CRISPR Associated Rossmann Fold (CARF) domain (residues 1–178), which contains a unique insertion fragment at the C-terminal (Ct) region (residues 130–170) (Figure 1A, Supplementary Figure S1). After optimising initial crystallization hits from high throughput commercial screenings, the isolated Sis0455 protein (Supplementary Figure S2) yielded high-quality crystals that diffracted up to 2.27 Å resolution (Materials and Methods, Table 1). Molecular replacement trials using Sis0811 or other CARF domains as models failed in solving the phase problem. Therefore, we prepared a selenomethionine-modified sample to solve the phase problem using the SAD method. Selenium derivative crystals were obtained only in the presence of cA4. These crystals diffracted to 1.67 Å resolution (Materials & Methods, Table 1) and provided a high anomalous signal allowing the determination of the structure. The resulting electron density map permitted the unambiguous building of the Sis0455:cA4 structure (PDB code: 7Z55) (Figure 1B, Supplementary Figure S3a, b). Finally, the Sis0455 apo structure was solved by molecular replacement using this model as a searching model with the native protein data set (PDB code: 7Z56) (Figure 1C, Supplementary Figure 3Sc, d).
Figure 1.
Structure of Sis0455 in its apo form and in complex with its substrate. (A) Domain architecture of Sis0455. (B) Cartoon model of the dimeric Sis0455 crystal structure in complex with cA4. (C) Cartoon model of the dimeric apo Sis0455 crystal structure. The regions of each monomer are coloured with a different intensity. (D) Structural comparison of Sis0455 (violet) and Sis0811 (pale green) active sites depicting their corresponding key residues in binding/catalysis as sticks.
Two monomers of Sis0455 form a dimer within the asymmetric unit (Figure 1B, C, Supplementary Figure S3c, d), in agreement with the molar mass, observed in solution in SEC-MALS experiments (Supplementary Figure S2b, Materials and Methods). The CARF domains shape the catalytic pocket at the interface of the dimer assembly, displaying the Ct insertion fragments oppositely oriented along the 2-fold dimer axis (Figure 1B, C). Hence, the 2-fold axis bisects the cA4 catalytic pocket and the Ct insertion fragments. The dimeric assembly is stabilized by the interaction between the α4 helix of each monomer (Supplementary Figure S1), and interactions between the α3 and β6 segments. The core of the CARF domain consists of a 6-stranded Rossmann-like fold, as in other related proteins (15), but unlike in other CARFs the core β5 and β6 strands do not form a ß-hairpin, as the Ct insertion fragment is located between them, adding the α5, η1, α 6 and η2 helices (Supplementary Figure S1). In addition, two disulphide bridges (C55–C155 and C56–C164) interconnect the CARF domain with the Ct insertion stabilizing the folding of each monomer (Supplementary Figure S3e).
Sequence and structural alignments with other standalone ring nucleases, especially with its ‘cousin’ Sis0811 (PDB code: 7PQ2), reveal the conservation of K106 and S11 in Sis0455 (Figure 1D, Supplementary Figure S1). These residues have been proposed to play key roles in the enzymatic activity of these nucleases (32,37). The equivalent K169 and S12 in Sis0811 display a similar configuration in the apo structure (Figure 1D, Supplementary Figure S3c, d). As previously observed in Sis0811 and Sso1393, the conserved lysine residues are present at the very bottom of the catalytic pocket, while the conserved serine residues are located on each side of the active site (Supplementary Figure S3c, d). However, in Sis0455 the cavity is more positively charged by the presence of R105, which is exclusive of this type of ring nucleases (Supplementary Figures S1 and S3d).
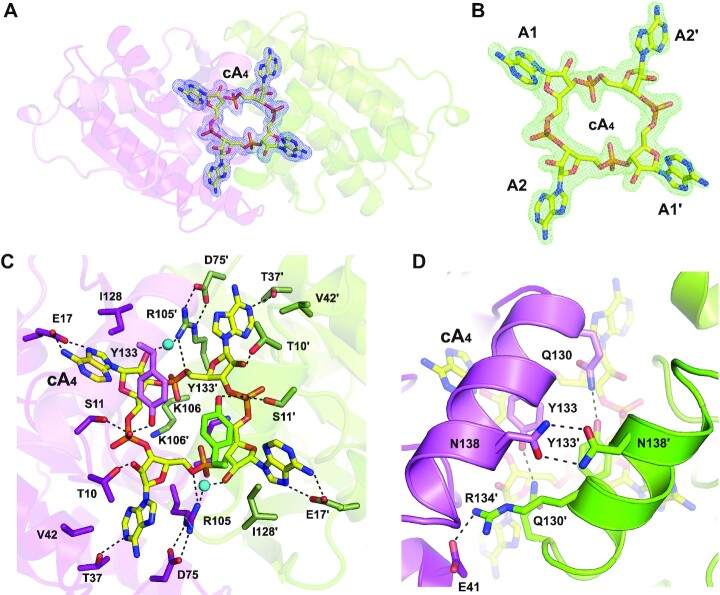
Structure of the Sis0455: cA4 complex
To understand cA4 binding and degradation mechanism, we determined and analysed the structure of Sis0455 in complex with its substrate cA4 (Figure 1C, Supplementary Figure S3a, b, Table 1). The quality of the 2FoFc and omit electron density maps unambiguously revealed the presence of the cyclic oligoadenylate inside the catalytic pocket in a non-processed state (Figure 2A–C, Supplementary Figure S3a, b). The structure revealed that the substrate is trapped in the catalytic pocket isolating the cA4 molecule from the solvent (Figures 1B and 2, Supplementary Figure S3a, b). The structure revealed a closed state conformation of the enzyme, which is stabilized by interactions between the α5 helix in the Ct region of each protomer through polar interactions between N138-N138’, Q130-Y133’/Q130’-Y133 and E41-R134’ (Figure 2D, Supplementary Figure S3a, b).
Figure 2.
Substrate location and pattern interaction with Sis0455. (A) 2Fo – Fc map at the substrate binding pocket superimposed onto its corresponding refined structure. Map displayed at 2.0 σ contour value. (B) Fo – Fc omit map at the substrate binding pocket superimposed onto its corresponding cA4 non-processed structure. FoFc omit map is displayed at 4.0 σ contour value. (C) Zoom view at the substrate binding pocket within the dimer Sis0455 in complex with its substrate, depicting their key interacting residues. (D) Zoom view at the key residues involved in stabilizing the closed dimer state, caging the substrate.
The cA4 is strongly stabilized in the catalytic pocket via a large interaction network (Figure 2C, Supplementary Figure S4), which involves polar interactions of the phosphate groups with S11-S11’ and Y133-Y133’, and the 2’OH group of the riboses with the T10–T10’ in each protomer. In addition, the cyclic molecule is also associated with R105–R105’ via water-mediated interactions, and polar interactions of the bases with T37–T37’ and E17–E17’. Furthermore, the bases are also stabilized through hydrophobic interactions with V42–V42’ and I128–I128’. As observed in other ring nucleases (32,37), the strictly conserved lysine residue (K106–K106’) at the bottom of the catalytic pocket generates the electropositive environment to accommodate the phosphate groups of cA4. Noteworthy is the conformation of the D75–D75’ residues in the active site, which seem to position the R105–R105’ to favour their interaction with the phosphate groups of the cyclic molecule (Figure 2C, Supplementary Figures S3a, b and S4).
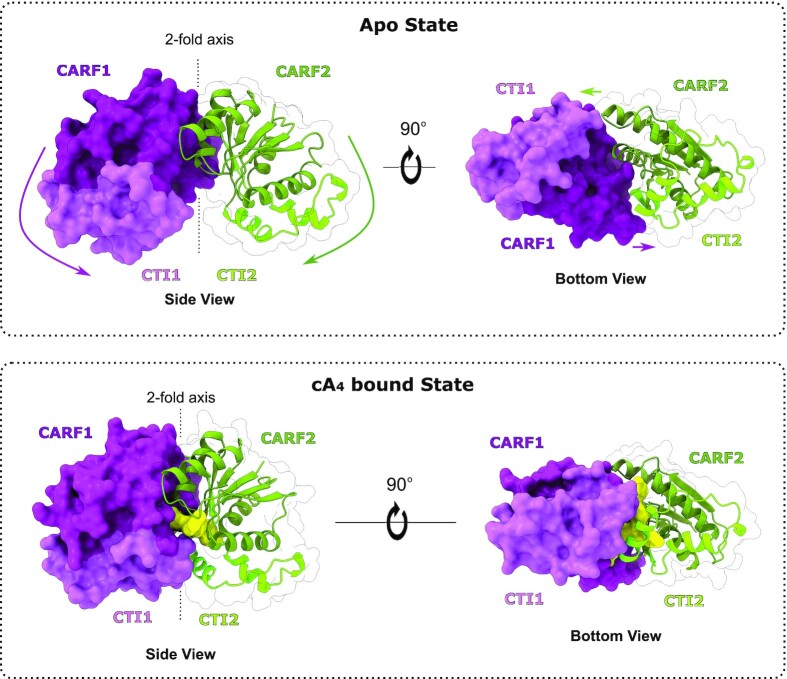
cA4 induces a conformational change in Sis0455
The binding of cA4 in the cataytic pocket induces a large conformational change in the dimeric assembly (Figure 3, Supplementary Video S1). The two monomers undergo a jaw-like movement confining the substrate in the catalytic pocket. The Ct region of each protomer forms the closure of the shared catalytic pocket, while the CARF domains fold inwards, trapping and isolating the substrate into the positively charged active site. The extent of this conformational movement is reflected in a RMSD of 4.42 Å for the 325 Cα between the apo and ligand bound structures, which buries a surface of 1450 Å2.
Figure 3.
Conformational change of the ring nuclease Sis0455 from its apo form to its substrate-bound state. Cartoon/surface models comparing the cA4 non-bound (upper panels) and its cA4 bound state forms (bottom panels). Coloured arrows represent the conformational change path of each corresponding domain from apo structure towards the product reaction bound form.
The key residues in the catalytic pocket undergo a conformational change between the apo and cA4 bound-state (Supplementary Figure S3a–d). The K106 and K106’ in Sis0455 change their arrangement to accommodate the cyclic substrate molecule, while S11 and S11’ residues move their Cα ∼5 Å to interact with cA4. These strictly conserved residues are supposed to be key during catalysis by homology with Sis0811 and Sso1393 nucleases (Supplementary Figure S1). Based in this comparison, the lysines would play an important role in the reaction intermediate stabilization, while the serines would be involved in the stabilization of the reaction product (37).
However, the Sis0455:cA4 complex structure revealed a different cA4 recognition pattern, suggesting aditional residues playing important roles in the cleavage reaction. For instance, R105 and R105’ establish strong water-mediated interactions with the 2′OH of the riboses, thus positioning the substrate for the catalytic reaction (Figure 2C, Supplementary Figure S3a, b). Interestingly, the rotamer of these arginine residues in that substrate-interacting conformation is arranged by a salt-bridge interaction with D75, suggesting an indirect role in catalysis of the aspartic acid, as in the apo structure no interaction is observed between D75 and R105. The presence of the substrate induces a conformational change where both aspartic residues drive the arginines to configurate the pre-catalytic complex (Figure 2C, Supplementary Figure S3a, b).
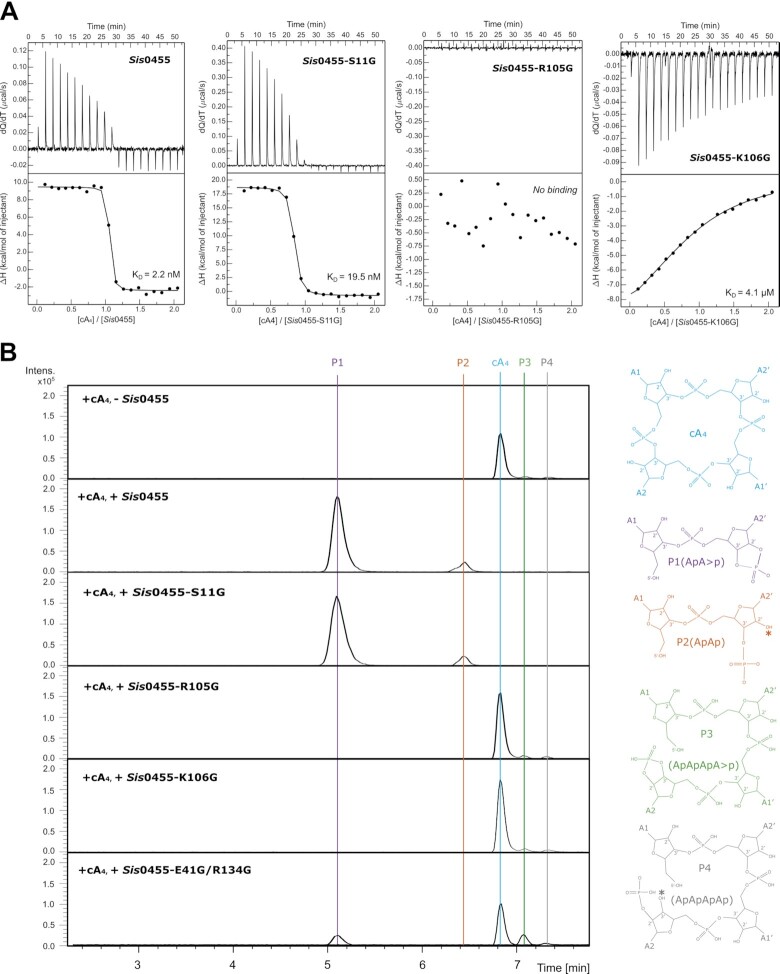
Sis0455 displays a high affinity binding of cA4
To understand the role of the amino acids involved in cA4 binding, we performed isothermal titration calorimetry (ITC) binding assays with the wild type and mutants. The Sis0455 ring nuclease binds cA4 with a very high affinity (KD of 2.2 nM), which is almost five-fold higher than Sis0811 (KD 9.1 nM) (37) (Figure 4A). Interestingly, unlike Sis0811, which displayed an exothermic binding of cA4, the association of the cyclic molecule by Sis0455 is endothermic, suggesting that the conformational jaw movement observed upon ligand binding contributes to a solvent entropy change (Figure 3). The extensive protein surface burial to trap the substrate should result in solvent release in the polar active site upon cA4 binding, thus contributing favourably to the association.
Figure 4.
Biophysical and cleavage activity characterization of Sis0455. (A) Substrate binding assays of Sis0455, Sis0455-S11G, Sis0455-R105G and Sis0455-K106G analysed by ITC. (B) Substrate cleavage assays of Sis0455, Sis0455-S11G, Sis0455-R105G, Sis0455-K106G and Sis0455-E41G/R134G analysed by Mass Spectrometry.
Based in the structural data and the sequence conservation, we selected key residues to perform glycine substitutions to examine the effect of the absence of the side chains in cA4 binding. The S11G and K106G substitutions diminished cA4 binding. The S11G variant displayed a decrease in the binding affinity of ∼10 fold (KD of 19.5 nM), while the binding of the K106G mutant severely affected the association with the ligand (KD of 4.1 μM) (Figure 4a), thus highlighting the importance of the lysine residues in cA4 binding. Unlike Sis0455, the K106G mutant displayed an exothermic binding profile, in agreement with the reduced polarity in the active site due to the lack of the lysine amino groups. The R105G as well as the D75G mutations abolished cA4 binding (Figure 4A, Supplementary Figure S5a), thus supporting the proposed role of D75 positioning R105, and confirming the key role of the arginine in ligand binding (Supplementary Figure S4a). The T10G mutant, which affects the 2’OH stabilization of the ribose, displays a substantial reduction of the cA4 binding affinity (KD of 35.3 nM), but still associates with the ligand. Mutations in the residues interacting with the bases, such as E17G/T37G, do not affect binding substantially (KD of 3.5 nM) (Supplementary Figure S5a).
We also evaluated the effect of mutations in the α5 interacting helices of the Ct insertion in the single N138G, and double E41G/R134G and Q130G/Y133G mutants. Both N138G and E41G/R134G do not affect cA4 binding substantially (KD of 10.5 nM and 1.9 nM); however, the double mutant Q130G/Y133G severely affected the affinity of Sis0455 for the cyclic compound (KD of 2.6 μM). This double mutation destabilizes the closed state conformation by disturbing the interactions observed in the Ct insertion in the ligand bound structure (Figure 2D, Supplementary Figures S4 and S5a).
cA4 degradation by Sis0455 reveals key residues for catalysis
Next, we examined the effect of the mutants in the enzyme cA4 cleavage activity. We performed activity assays incubating the mutants with cA4 as in (37). The reaction mixture was analysed by mass spectrometry revealing the degradation products (Figure 4B, Supplementary Figure S5b, Materials and Methods). Five different species were detected: the intact cA4, P1 (ApA > P), P2 (hydrolysis product of P1, ApAp), P3 (intermediate reaction product, ApApApA > p) and P4 (hydrolysis of P3, ApApApAp).
The assay confirmed that besides its important role in binding the conserved K106is essential for catalysis. The K106G substitution abolished the cleavage activity and did not generate any reaction product (Figure 4b). This result is similar to that observed in Sis0811(36), thus supporting its possible role as the key residue in stabilizing the transition state of the cleavage rection in both types of standalone ring nucleases. In addition, the R105G mutant also abrogated the nuclease activity confirming its important role in the reaction (Figure 4b). Therefore, both K106 and R105 are fundamental for cA4 degradation, however, the latter is only conserved in the SSRN group.
By contrast, the conserved S11, which is present in both groups, does not seem to be critical in binding and catalysis for Sis0455, as it displayed a similar mass spectrometry pattern compared to the wild type (Figure 4B). This behaviour is different from that observed in the conserved S12 and S11 in Sis0811 and Sso1393, the standalone ring nucleases lacking the Ct insertion (32,37). Collectively, these observations suggest that while the role of the lysine residues is conserved between the two types of standalone ring nucleases, the serine does not have the relevance in catalysis which has been observed in the LSRN type. In addition, the important function of the conserved R105 in Sis0455 and Sso2081 is exclusive of the standalone ring nucleases including the Ct insertion.
We also checked the effect on the enzyme activity of those amino acids that interact with cA4, either through the 2’OH of ribose (T10) or the base (E17/E37). In both cases, the substitution by glycine did not affect catalysis, the product profile of the reaction mixture was similar than the wild type (Supplementary Figure S5b). Finally, glycine substitutions in residues involved in the closing of the catalytic pocket in the Ct insertion were tested. The single N138G and double Q130G/Y133G substitutions did not affect catalysis (Supplementary Figure S5b). However, the E41G/R134G double mutant, which disturbs the polar interaction between the Ct insertions (Figure 2D), showed cA4 as major specie, also presenting P1 and the P3 linear products in a minor extent. Collectively, these observations indicate that the E41/R134 interaction is not relevant for binding the ligand, as its affinity is identical to the wild type. However, they seem important for stabilizing the closed state of the enzyme-substrate complex facilitating the configuration of the active site for catalysis.
Comparison of Sis0455 active site with other CARF domain proteins
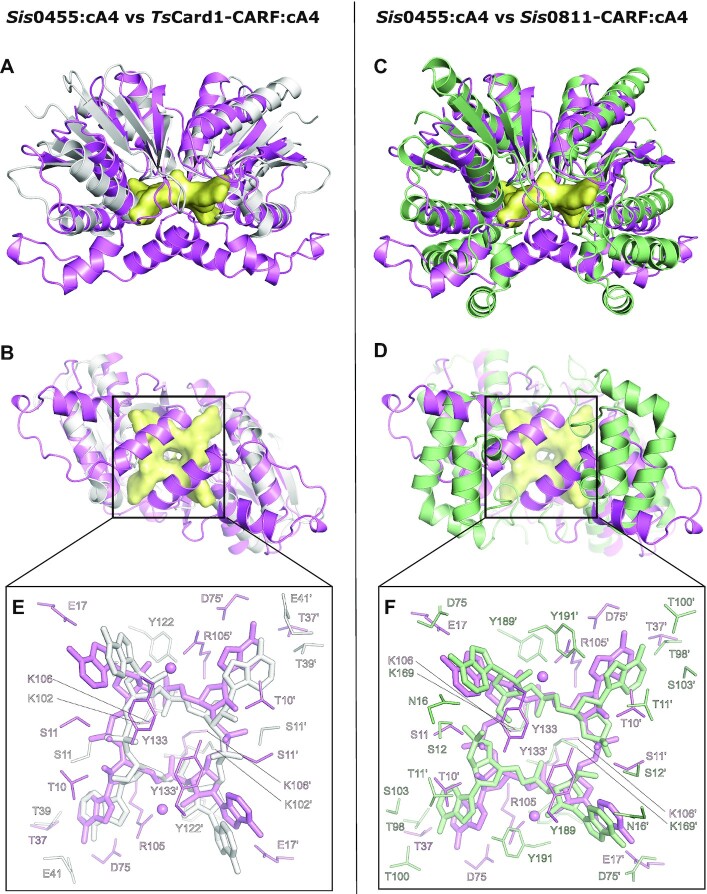
The closest structural homologue of Sis0455 found by the DALI server (38) is the TsCard1:cA4 (48) complex (PDB code: 6WXX) (RMSD 2.7 Å, aligning 124 Cα out of 373 residues of TsCard1). The homology lies within the CARF domains of Card1 (UniProt Entry: F2NWD3) and Sis0455. The main difference arises from the Ct insertion in Sis0455, which includes the two extra helices (α5-α6) and the connecting loops, thus shaping different substrate-binding pockets from its dimeric assembly (Figure 5A, B). This suggests why the TsCard1-CARF dimer requires the action of residues in the Restriction Endonuclease (REase) domains to trap cA4 (48), while in the case of Sis0455 the Ct insertion closes the catalytic pocket. The superimpositon of the dimeric structures of Sis0455 and Sis0811 CARFs in complex with 2A2 (PDB code: 7PQ3) (RMSD: 2.2 Å) also shows that the catalytic pocket is not closed upon binding of the second messenger in Sis0811 (Figure 5C, D). The comparison of the active site of the cA4-bound structures show the conservation of the previuosly reported key residues for cA4 degradation in Sis0811 (S12 and K169) (37) (Figure 5E, F), which are also conserved in TsCard1 (S11 and K102). Although these key residues keep the same structural configuration in Sis0455, Sis0811 and TsCard1-CARF (Figure 5C), Sis0455 and Sis0811 degrade cA4, while TsCard1 does not cleave the cyclic ligand, suggesting differences in the active site. Interestingly, our activity assays (Figure 4B) and previous cleavage assays in Sso2081(32) show that the conserved R105, which is not present in TsCard1, is involved in the reaction (Figures 2C and 5E, F, Supplementary Figure S1). However, while the presence of this conserved arginine could justify the differences with SSRNs and TsCard1, LSRNs do not conserve this residue. On the other hand, TsCard1 conserves key residues involved in the bases recognition such as T39 (T37 in Sis0455 and T98 in Sis0811) and Y122 (Y133 in Sis0455 and Y191 in Sis0811) (Supplementary Figure S1). Hence, the contribution of other residues has to be taken into account to fully understand the molecular basis of cA4 recognition or cleavage.
Figure 5.
Comparison of Sis0455:cA4 with structural homologs. (A) Side view superimposition of the Sis0455:cA4 and TsCard1-CARF:cA4 structures. (B) Bottom view of the superimposition of the Sis0455:cA4 and TsCard1-CARF:cA4 structures. (C) Comparison at the active sites zoom view between Sis0455:cA4 and TsCard1-CARF:cA4 structures. (D) Side view superimposition of the Sis0455:cA4 and Sis0811:2A2 structures. (E) Bottom view of the superimposition of the Sis0455:cA4 and Sis0811:2A2 structures. (F) Comparison at the active sites zoom view between Sis0455:cA4 and Sis0811:2A2 structures.
Collectively, the comparison of the active sites and biochemical properties indicate that a set of catalytic residues are conserved between the two groups of standalone ring nucleases; however, those containing the Ct insertion display extra residues which reshape the properties of the cleavage reaction.
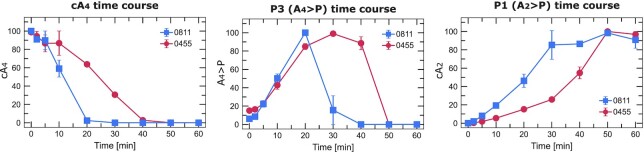
Sis0455 degrades cA4 slower than Sis0811
In order to compare the degradation of cA4 by both ring nucleases from S.islandicus, we performed a time course experiment monitoring the reaction products. The cleavage reaction was stopped at different times, and the comsumption of cA4 and the generation of the linear P3 and P1 dinucleotide products were quantified at different times (Figure 6). Sis0455 and Sis0811 fully degrade cA4 producing 100% P1 product in 50 min. The overall view of the reaction suggest that both enzymes cleave one bond to generate the linear product before the second phosphodiester is hydrolyzed to produce the P1 product. However, the kinetics followed by these enzymes to cleave the two phosphodiester bonds is different. While the hydrolysis of the first phosphodiester is faster for Sis0811, converting almost 100% of the cyclic molecule in the P3 linear product in 20 min, the Sis0455 enzyme takes double time. Then Sis0811 employs 30 min to cleave the second phosphodiester bond converting all the linear product in the P1 dinucleotide. The reaction follows an overall similar scheme in Sis0455; however, the clevage of both phosphodiesters is slower in order to acomplish the complete degradation of the cyclic molecule in the same time. Therefore, Sis0455 linearizes cA4 slowly and follows a more progressive generation of P1 compared to Sis0811. The functional outcome of these different processing kinetics is unknown so far.
Figure 6.
Time course reaction products of Sis0811 and Sis0455. Time dependence of the amount of cA4 and products of reaction for the ring nucleases Sis0455 and Sis0811. The amount of compound was quantitated by integrating the area under the EIC curve and each series normalized (100%) to the largest value within the series.
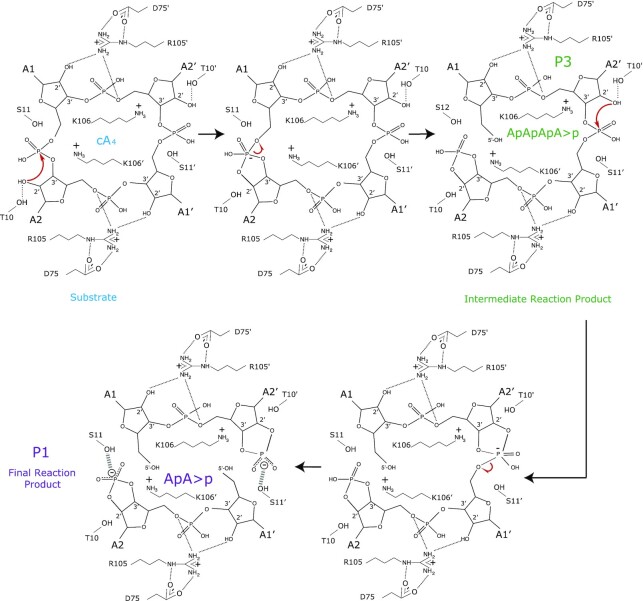
Sis0455 cleavage mechanism
Our structure-function study provides experimental evidence of the catalytic mechanism of a member of the SSRN family (Figure 7). We propose that the cleavage reaction in Sis0455 will be initated by the 2’OH of the ribose, which would initiate the nucleophilic attack on the phosphate. Then, K106, which is conserved in the LSRN and SSRN enzymes, would stabilize the pentacovalent phosphorous formed in the transition state. The 2’–3’cyclic phosphate could be stabilized either by the S11 or T10 hydroxyl groups. These stages of the reaction are similar between the two groups of standalone ring nucleases. The structure of the Sis0455:cA4 complex suggest that T10 will favour the positioning of the 2’OH of the ribose for the nucleophilic attack. The cyclic 2’,3’-cyclic phosphate can be destabilized and disrupted over time (49), as it has been observed in our assays where some P2 product (ApAp) can be detected (Figure 4B and Supplementary Figure S5b). In addition, our analysis of the reaction products suggest that the cleavage reaction of the phosphodiester bonds does not occur in a concerted manner, as in the case of Sis0811 (37). The linear P3 product (ApApApA > p) was detected during our time course experiments (Figure 6), and the presence of the linear intermediate was also observed in a minor extent in the case of R105G and the K106G mutants. In addition, the abundance of P3 was increased for the E41G/R134G mutant, suggesting that disrupting the proper caging the compound in the active site would disturb the processing of the second phosphodiester; therefore, affecting the conversion of the linear P3 product into the P1 dinucleotide.
Figure 7.
Model of Sis0455 catalytic mechanism. The D75 positions the R105, which plays a key role in binding the ligand. The S11 positions the 2’OH of a ribose to initiate the nucleophilic attack on the corresponding phosphate and subsequently K106’ stabilize the pentavalent phosphorous formed in the transition state. Then, the 2’-3’cyclic phosphate is stabilized by the S11 OH group, producing the intermediate reaction product P3 (ApApApA > p). Next, the other cleavable phosphodiester bond is attacked by the 2’-OH of the ribose which is close to S11’ that positions it. Thus, K106 stabilize the pentacovalent phosphorous formed in the transition state and the cyclic 2’,3’-cyclic phosphate is stabilised by the S11’ OH group generating two molecules of P1 (ApA > p) as the final reaction product.
Collectively, the data indicate that overall, the catalytic mechanism in the LSRN and the SSRN enzymes in S. islandicus is very similar. However, the processing of the first phosphodiester hydrolysis is faster in Sis0811 than in Sis0455, thus quickly removing the cyclic compound. These differences between the two enzymes in the cleavage reaction could arise from the requirement of the D75 to position R105 properly for triggering cA4 degradation.
DISCUSSION
A major part of the characterized Type III CRISPR-Cas systems utilizes cA4 as second messenger to trigger their immune response, while some other systems use a cyclic hexa-oligoadenylate (cA6) instead (23,25). The CARF domain architecture of the auxiliary proteins of each system is designed to specifically recognize one of these signals (33,35,48). The most common CARF family proteins are the Csx1/Csm6 ribonucleases, which are activated by their association with cA4 or cA6. These indiscriminate nucleases cleave RNA using their Higher Eukaryotes and Prokaryotes Nucleotide-binding (HEPN) domain (22,23), unleashing an indiscriminate RNase activity, which has been shown to result in cell growth arrest. Cellular growth is reestablished when infection is cleared from cells (29). A possible transcriptional role of cA4 has been suggested, as it was shown to bind to the CARF family transcription regulator Csa3, upregulating CRISPR loci and cas gene expression (50,51).
The regulation of the indiscriminate degradation of cellular DNA and RNA by the cA activated Csx1/Csm6 nucleases is a crucial issue for the cell, as the activity of the indiscriminate nucleases confers Type III CRISPR systems a potential for self-destruction. The strategy of host nucleic acid degradation induces cell dormancy to slow viral propagation, but it can seriously affect cell survival if it is not strictly regulated. To address this problem the cell can switch off the production of the second messenger, however, this strategy will not eliminate the previously synthesized cA, which will remain residing in the cytoplasm. Therefore, the degradation of the signalling molecule is needed to downregulate the levels of the second messenger to switch off the Csx1/Csm6 nucleases. The development of such a regulatory strategy led prokaryotes to develop enzymes, such as the standalone ring nucleases, that cleave the phosphodiester bonds of the cyclic compound. Noteworthy, phages have also developed countermeasures, such as the degradation of these cyclic molecules (52,53), as strategies of immune evasion in phage biology to defeat host defence systems.
The standalone ring nucleases are found in the crenarchaea, and the Sulfolobales usually encode several standalone ring nucleases orthologues alongside Csx1/Csm6 (54). Sso2081 and Sso1393 were the first members of the LSRN and SSRN identified in cellular lysates (32). Protein sequence analyses suggested that these orthologues most likely originated from gene duplication events (55). The genome of S. islandicus REY15A encodes two standalone ring nucleases, Sis0811 and Sis0455, which degrade the cA4 second messenger made by its Type III-B CRISPR system.
Our Sis0455 structures reveals the jaw-like conformational change in the enzyme to accommodate the cyclic tetra-oligoadenylate in the active site, resembling the cA4 trapping mechanism described for the anti-CRISPR viral ring nuclease AcrIII-1 (52). The structural data together with our binding and activity experiments show that Sis0455 follows a mechanism similar to Sis0811 to degrade cA4. The same reaction products were identified in our mass spectrometry assay. Furthermore, the binding assays reveal that Sis0455 displays 5-fold higher afinity for cA4 than Sis0811 (37). While both enzymes degrade the cyclic compound in a non-concerted manner, generating a linear intermediate before its conversion in dinucleotides, the reaction in Sis0455 proceeds in a more progressive way than in Sis0811 (Figure 6). Sis0455 follows a slower kinetics to cleave the first phosphodiester bond of the cyclic compound, while Sis0811 linearizes cA4 very fast. These results suggest that the catalysis of the small ring nuclease would allow a longer lifetime of the intact substrate and the intemediate linear product in the cytoplasm before they are converted to dinucleotides. By contrast, Sis0811 would ‘linearise’ the major part of cA4 much faster. Overall, this result suggests that the different kinetics of these enzymes could be an evolutionary response in S. islandicus to regulate the levels of cA4 in different cellular scenarios during infection. This difference in cA4 degradation have been also observed in the enzymes from S. solfataricus, where Sso2081, the homolog of Sis0455, processes cA4 10-fold faster than Sso1393, the homolog of Sis0811 (32). This may reflect the regulatory needs of a given Type III CRISPR system.
Collectively, the data suggest that presence of the two orthologues is a regulatory strategy which will overlap temporally or synergistically the activity of both enzymes for the control of cA4 levels. We propose that Sis0455, which displays a remarkable high affinity for the second messenger (KD 2 nM) is performing a continuous surveillance of the cA4 levels in the cytoplasm, as it will bind and slowly degrade low levels of the compound which could be present in the cell. This mechanism could be kept resident in the cell, avoiding the deleterious effect of a very early activation of SisCsx1, whose affinity for cA4 is 10-fold lower (KD 20 nM) (35). However, large quantities of cOA could be synthesized even at low levels of infection (56), and in that situation Sis0455 could not be able to eliminate cA4, either by binding or degrading the ligand. In this scenario Sis0811, whose afinity (KD 9.9 nM) (37) is 2-fold higher than that of SisCsx1, would kick-in eliminating the cyclic compound faster to avoid an early activation of the indiscriminate RNase.
Type III CRISPR-Cas systems have tailored a complex regulatory system to control their immune response, which could have deleterious implications for the cell. Evolution has combined auxiliary proteins with different binding affinities and degradation rates to adapt the intensity of the immune response in different infection scenarios. Further functional studies are needed to fully explore and confirm how the Type III CRISPR-Cas standalone nucleases coordinate their action with the effector complexes and indiscriminate RNases to modulate bacterial defence.
DATA AVAILABILITY
Atomic coordinates and structure factors have been deposited in the Protein Data Bank under the accession codes 7Z55 and 7Z56 accordingly. The rest of the data are available from the corresponding author upon reasonable request.
Supplementary Material
ACKNOWLEDGEMENTS
We thank SLS and MAX-IV synchrotrons for help during X-ray diffraction data collection. Data processing has been performed at the Computerome, the Danish National Computer for Life Sciences. GM is a member of the Integrative Structural Biology Cluster (ISBUC) at the University of Copenhagen.
Author contributions: R.G.M., A.L.G.J., J.M.H. and S.S. perform cloning and protein purifications. B.L.M. and R.G.M. carried out the substrate binding assays and mass spectrometry experiments to analyze Sis0455 reaction products. R.M. obtained initial apo and cA4 bound form crystals, which were later refined by R.M., J.R.C.T. and R.G.M. B.L.M. performed MALLS measurements. R.M. collected and processed diffraction data. R.M. solved and refined the crystal structures. R.M. and G.M. coordinated and supervised the entire project and wrote the manuscript with input from all the authors.
Notes
Present address: Rafael Molina, Department of Crystallography and Structural Biology, Institute of Physical-Chemistry Rocasolano, Spanish National Research Council, Madrid, Spain.
Present address: Stefano Stella, Twelve Bio ApS, Ole Maaløes Vej 3, Copenhagen 2200, Denmark.
Contributor Information
Rafael Molina, Structural Molecular Biology Group, Novo Nordisk Foundation Centre for Protein Research, Faculty of Health and Medical Sciences University of Copenhagen, Blegdamsvej 3-B, 2200 Copenhagen, Denmark.
Ricardo Garcia-Martin, Structural Molecular Biology Group, Novo Nordisk Foundation Centre for Protein Research, Faculty of Health and Medical Sciences University of Copenhagen, Blegdamsvej 3-B, 2200 Copenhagen, Denmark.
Blanca López-Méndez, The Novo Nordisk Foundation Center for Protein Research, Protein Structure & Function Programme, Faculty of Health and Medical Sciences, University of Copenhagen, Blegdamsvej 3B, 2200 Copenhagen, Denmark.
Anne Louise Grøn Jensen, Structural Molecular Biology Group, Novo Nordisk Foundation Centre for Protein Research, Faculty of Health and Medical Sciences University of Copenhagen, Blegdamsvej 3-B, 2200 Copenhagen, Denmark.
J Rafael Ciges-Tomas, Structural Molecular Biology Group, Novo Nordisk Foundation Centre for Protein Research, Faculty of Health and Medical Sciences University of Copenhagen, Blegdamsvej 3-B, 2200 Copenhagen, Denmark.
Javier Marchena-Hurtado, Structural Molecular Biology Group, Novo Nordisk Foundation Centre for Protein Research, Faculty of Health and Medical Sciences University of Copenhagen, Blegdamsvej 3-B, 2200 Copenhagen, Denmark.
Stefano Stella, Structural Molecular Biology Group, Novo Nordisk Foundation Centre for Protein Research, Faculty of Health and Medical Sciences University of Copenhagen, Blegdamsvej 3-B, 2200 Copenhagen, Denmark.
Guillermo Montoya, Structural Molecular Biology Group, Novo Nordisk Foundation Centre for Protein Research, Faculty of Health and Medical Sciences University of Copenhagen, Blegdamsvej 3-B, 2200 Copenhagen, Denmark; The Novo Nordisk Foundation Center for Protein Research, Protein Structure & Function Programme, Faculty of Health and Medical Sciences, University of Copenhagen, Blegdamsvej 3B, 2200 Copenhagen, Denmark.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Novo Nordisk Foundation Center for Protein Research is supported financially by the Novo Nordisk Foundation [NNF14CC0001]; Integrative Structural Biology Cluster (ISBUC) at the University of Copenhagen [NNF0024386, NNF17SA0030214, Distinguished Investigator (NNF18OC0055061) to G.M.]. Funding for open access charge: NNF core grant (to C.P.R.).
Conflict of interest statement. G.M. and S.S. are co-founders and members of the BoD of Twelve BIO. The rest of the authors declare no competing interests.
REFERENCES
- 1. Ishino Y., Shinagawa H., Makino K., Amemura M., Nakata A.. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in escherichia coli, and identification of the gene product. J. Bacteriol. 1987; 169:5429–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mojica F.J.M., Juez G., Rodriguez-Valera F.. Transcription at different salinities of haloferax mediterranei sequences adjacent to partially modified PstI sites. Mol. Microbiol. 1993; 9:613–621. [DOI] [PubMed] [Google Scholar]
- 3. Hermans P.W., van Soolingen D., Bik E.M., de Haas P.E., Dale J.W., van Embden J.D.. Insertion element IS987 from mycobacterium bovis BCG is located in a hot-spot integration region for insertion elements in mycobacterium tuberculosis complex strains. Infect. Immun. 1991; 59:2695–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A.et al.. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gasiunas G., Barrangou R., Horvath P., Siksnys V.. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:2579–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E.. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science (1979). 2012; 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cox D.B.T., Platt R.J., Zhang F.. Therapeutic genome editing: prospects and challenges. Nat. Med. 2015; 21:121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Doudna J.A., Charpentier E.. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014; 346:1258096. [DOI] [PubMed] [Google Scholar]
- 9. Wright A.v., Nuñez J.K., Doudna J.A.. Biology and applications of CRISPR systems: harnessing nature's toolbox for genome engineering. Cell. 2016; 164:29–44. [DOI] [PubMed] [Google Scholar]
- 10. Barrangou R., Horvath P.. A decade of discovery: CRISPR functions and applications. Nat Microbiol. 2017; 2:17092. [DOI] [PubMed] [Google Scholar]
- 11. Amitai G., Sorek R.. CRISPR–Cas adaptation: insights into the mechanism of action. Nat. Rev. Microbiol. 2016; 14:67–76. [DOI] [PubMed] [Google Scholar]
- 12. Jackson S.A., McKenzie R.E., Fagerlund R.D., Kieper S.N., Fineran P.C., Brouns S.J.J.. CRISPR-Cas: adapting to change. Science. 2017; 356:eaal5056. [DOI] [PubMed] [Google Scholar]
- 13. Koonin E.v, Makarova K.S., Zhang F.. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017; 37:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mohanraju P., Makarova K.S., Zetsche B., Zhang F., Koonin E.v, van der Oost J.. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science. 2016; 353:aad5147. [DOI] [PubMed] [Google Scholar]
- 15. Makarova K.S., Wolf Y.I., Iranzo J., Shmakov S.A., Alkhnbashi O.S., Brouns S.J.J., Charpentier E., Cheng D., Haft D.H., Horvath P.et al.. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020; 18:67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Molina R., Sofos N., Montoya G.. Structural basis of CRISPR-Cas type III prokaryotic defence systems. Curr. Opin. Struct. Biol. 2020; 65:119–129. [DOI] [PubMed] [Google Scholar]
- 17. Peng W., Feng M., Feng X., Liang Y.X., She Q.. An archaeal CRISPR type III-B system exhibiting distinctive RNA targeting features and mediating dual RNA and DNA interference. Nucleic Acids Res. 2015; 43:406–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Samai P., Pyenson N., Jiang W., Goldberg G.W., Hatoum-Aslan A., Marraffini L.A.. Co-transcriptional DNA and RNA cleavage during type III CRISPR-Cas immunity. Cell. 2015; 161:1164–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Elmore J.R., Sheppard N.F., Ramia N., Deighan T., Li H., Terns R.M., Terns M.P.. Bipartite recognition of target RNAs activates DNA cleavage by the type III-B CRISPR–Cas system. Genes Dev. 2016; 30:447–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Estrella M.A., Kuo F.-T., Bailey S.. RNA-activated DNA cleavage by the type III-B CRISPR–Cas effector complex. Genes Dev. 2016; 30:460–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kazlauskiene M., Tamulaitis G., Kostiuk G., Venclovas Č., Siksnys V.. Spatiotemporal control of type III-A CRISPR-Cas immunity: coupling DNA degradation with the target RNA recognition. Mol. Cell. 2016; 62:295–306. [DOI] [PubMed] [Google Scholar]
- 22. Kazlauskiene M., Kostiuk G., Venclovas Č., Tamulaitis G., Siksnys V.. A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science. 2017; 357:605. [DOI] [PubMed] [Google Scholar]
- 23. Niewoehner O., Garcia-Doval C., Rostøl J.T., Berk C., Schwede F., Bigler L., Hall J., Marraffini L.A., Jinek M.. Type III CRISPR–Cas systems produce cyclic oligoadenylate second messengers. Nature. 2017; 548:543–548. [DOI] [PubMed] [Google Scholar]
- 24. Rouillon C., Athukoralage J.S., Graham S., Grüschow S., White M.F.. Control of cyclic oligoadenylate synthesis in a type III CRISPR system. Elife. 2018; 7:e36734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nasef M., Muffly M.C., Beckman A.B., Rowe S.J., Walker F.C., Hatoum-Aslan A., Dunkle J.A.. Regulation of cyclic oligoadenylate synthesis by the staphylococcus epidermidis cas10-csm complex. RNA. 2019; 25:948–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lau R.K., Ye Q., Patel L., Berg K.R., Mathews I.T., Watrous J.D., Whiteley A.T., Lowey B., Mekalanos J.J., Kranzusch P.J.et al.. Structure and mechanism of a cyclic trinucleotide-activated bacterial endonuclease mediating bacteriophage immunity. Mol. Cell. 2020; 77:723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McMahon S.A., Zhu W., Graham S., Rambo R., White M.F., Gloster T.M.. Structure and mechanism of a type III CRISPR defence DNA nuclease activated by cyclic oligoadenylate. Nat. Commun. 2020; 11:500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhu W., Mcquarrie S., Gr S., Mcmahon S.A., Graham S., Gloster T.M., White M.F.. The CRISPR ancillary effector can2 is adual-specificity nuclease potentiating type III CRISPRdefence. Nucleic Acids Res. 2021; 49:2777–2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rostøl J.T., Marraffini L.A.. Non-specific degradation of transcripts promotes plasmid clearance during type III-A CRISPR–Cas immunity. Nat. Microbiol. 2019; 4:656–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Makarova K.S., Anantharaman V., Grishin N.v., Koonin E.v., Aravind L.. CARF and WYL domains: Ligand-binding regulators of prokaryotic defense systems. Front Genet. 2014; 5:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Athukoralage J.S., Graham S., Grüschow S., Rouillon C., White M.F.. A type III CRISPR ancillary ribonuclease degrades its cyclic oligoadenylate activator. J. Mol. Biol. 2019; 431:2894–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Athukoralage J.S., Rouillon C., Graham S., Grueschow S., White M.F.. Ring nucleases deactivate type III CRISPR ribonucleases by degrading cyclic oligoadenylate. Nature. 2018; 562:277–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Garcia-Doval C., Schwede F., Berk C., Rostøl J.T., Niewoehner O., Tejero O., Hall J., Marraffini L.A., Jinek M., Jakob T.et al.. Activation and self-inactivation mechanisms of the cyclic oligoadenylate-dependent CRISPR ribonuclease csm6. Nat. Commun. 2020; 11:1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jia N., Jones R., Yang G., Ouerfelli O., Patel D.J.. CRISPR-Cas III-A csm6 CARF domain is a ring nuclease triggering stepwise cA4 cleavage with apa>p formation terminating RNase activity. Mol. Cell. 2019; 75:944–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Molina R., Stella S., Feng M., Sofos N., Jauniskis V., Pozdnyakova I., López-méndez B., She Q., Montoya G.. Structure of Csx1-coa4 complex reveals the basis of RNA decay in type III-B CRISPR-Cas. Nat. Commun. 2019; 10:4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Makarova K.S., Timinskas A., Wolf Y.I., Gussow A.B., Siksnys V., Venclovas Č., Koonin E.v.. Evolutionary and functional classification of the CARF domain superfamily, key sensors in prokaryotic antivirus defense. Nucleic. Acids. Res. 2020; 48:8828–8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Molina R., Jensen A.L.G., Marchena-Hurtado J., López-Méndez B., Stella S., Montoya G.. Structural basis of cyclic oligoadenylate degradation by ancillary type III CRISPR-Cas ring nucleases. Nucleic. Acids. Res. 2021; 49:12577–12590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kabsch W Xds. Acta. Crystallogr. D Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Evans P.R., Murshudov G.N.. How good are my data and what is the resolution?. Acta. Crystallogr. D Biol. Crystallogr. 2013; 69:1204–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vonrhein C., Flensburg C., Keller P., Sharff A., Smart O., Paciorek W., Womack T., Bricogne G.. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D. 2011; 67:293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Skubák P., Pannu N.S.. Automatic protein structure solution from weak X-ray data. Nat. Commun. 2013; 4:2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 44. Adams P.D., Afonine P.v., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W.et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta. Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Winn M.D., Murshudov G.N., Papiz M.Z.. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods in Enzymology. 2003; 374:Academic Press; 300–321. [DOI] [PubMed] [Google Scholar]
- 46. Schrödinger L. The PyMOL Molecular Graphics System. 2015; LLC; Version 2.0 Schrödinger. [Google Scholar]
- 47. Pettersen E.F., Goddard T.D., Huang C.C., Meng E.C., Couch G.S., Croll T.I., Morris J.H., Ferrin T.E.. UCSF chimerax: structure visualization for researchers, educators, and developers. Protein Sci. 2021; 30:70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rostøl J.T., Xie W., Kuryavyi V., Maguin P., Kao K., Froom R., Patel D.J., Marraffini L.A.. The card1 nuclease provides defence during type III CRISPR immunity. Nature. 2021; 590:624–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yang W. Nucleases: diversity of structure, function and mechanism. Q. Rev. Biophys. 2011; 44:1–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lawrence C.M., Charbonneau A., Gauvin C.. Cyclic tetra-adenylate (cA4) activates CRISPR associated transcription factor csa3, providing feedback activation of protospacer acquisition and crRNA expression. FASEB J. 2020; 34:1. [Google Scholar]
- 51. Ye Q., Zhao X., Liu J., Zeng Z., Zhang Z., Liu T., Li Y., Han W., Peng N.. CRISPR-associated factor csa3b regulates CRISPR adaptation and cmr-mediated RNA interference in sulfolobus islandicus. Front. Microbiol. 2020; 11:2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Athukoralage J.S., McMahon S.A., Zhang C., Grüschow S., Graham S., Krupovic M., Whitaker R.J., Gloster T.M., White M.F.. An anti-CRISPR viral ring nuclease subverts type III CRISPR immunity. Nature. 2020; 577:572–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hobbs S.J., Wein T., Lu A., Morehouse B.R., Schnabel J., Leavitt A., Yirmiya E., Sorek R., Kranzusch P.J.. Phage anti-CBASS and anti-Pycsar nucleases subvert bacterial immunity. Nature. 2022; 605:522–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zink I.A., Wimmer E., Schleper C.. Heavily armed ancestors: CRISPR immunity and applications in archaea with a comparative analysis of CRISPR types in sulfolobales. Biomolecules. 2020; 10:1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rouillon C., Athukoralage J.S., Graham S., Grüschow S., White M.F.. Bailey S. Chapter nine - Investigation of the cyclic oligoadenylate signaling pathway of type III CRISPR systems. CRISPR-Cas Enzymes. 2019; 616:Academic Press; 191–218. [DOI] [PubMed] [Google Scholar]
- 56. Athukoralage J.S., Graham S., Rouillon C., Grüschow S., Czekster C.M., White M.F.. The dynamic interplay of host and viral enzymes in type III CRISPR-mediated cyclic nucleotide signalling. Elife. 2020; 9:e55852. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors have been deposited in the Protein Data Bank under the accession codes 7Z55 and 7Z56 accordingly. The rest of the data are available from the corresponding author upon reasonable request.