Abstract
Pathogenic enteric microorganisms induce the NF-κB-dependent expression of proinflammatory genes in intestinal epithelial cells. The purpose of the present study was to clarify the contribution of microbial invasion to the degradation of the regulatory protein IκBα and the subsequent activation of NF-κB in cultured intestinal epithelial cells. Caco-2BBe cells were incubated with Salmonella dublin, Salmonella typhimurium, or a weakly invasive strain of E. coli. S. dublin and S. typhimurium (107 organisms/ml) induced equivalent concentration-dependent gel mobility shifts of an NF-κB consensus sequence that was preceded by IκBα degradation. E. coli (107 organisms/ml) did not induce IκBα degradation or NF-κB translocation. Pretreatment with cytochalasin D blocked invasion of all three strains but had no effect on IκBα degradation or NF-κB activation. S. dublin and S. typhimurium adhered to Caco-2BBe cells 3- to 10-fold more than E. coli. NF-κB activation was prevented by physical separation of S. dublin from Caco-2BBe cells by a 0.4-μm-pore-size filter. Our results imply that bacterial adhesion, rather than invasion or release of a secreted factor, is sufficient to induce IκBα degradation and NF-κB activation in intestinal epithelial cells. Our data suggest that strategies to reduce enteric inflammation should be directed to the reduction of bacterial enterocyte adhesion.
The transcription factor NF-κB family plays a significant role in the regulation of immune and inflammatory response genes in intestinal epithelial cells and other tissues (3, 16). The various homo- and heterodimeric combinations of the Rel family, comprising p50, p52, p65 (RelA), RelB, and c-Rel (3), interact with a series of related DNA target sites in the cell nucleus (16). NF-κB dimers exist as inactive complexes in the cytoplasm of unstimulated cells via an interaction with a family of inhibitory proteins collectively designated IκB (2, 8). The prototypic and best-studied member of the IκB family, IκBα, binds to the nuclear translocation sequence of p65 and sequesters NF-κB in the cytoplasm (2). In response to proinflammatory signals, IκBα is phosphorylated, ubiquitinated, and proteolytically degraded (16). In the absence of IκB, NF-κB translocates to the nucleus and subsequently activates the transcription of various proinflammatory genes. NF-κB also upregulates transcription of IκBα, resulting in the replenishment of cytosolic IκBα, which complexes to residual NF-κB and downregulates the immune activation process (15).
Previous studies have demonstrated that NF-κB is induced by a pleiotropic array of agents, including various cytokines, double-stranded RNA, phorbol esters, and several viruses (1, 10, 11). More recently, invasion by pathogenic bacteria, such as Salmonella typhimurium, Shigella flexneri, and enteropathogenic Escherichia coli, has been associated with the activation of NF-κB, leading to the expression of interleukin 8 (IL-8) in a variety of cell types, including cultured enterocytes (4–6, 9, 12, 13). Nonpathogenic microorganisms, which less readily invade enterocytes, do not induce NF-κB activation. Taken together, these studies suggest that an enteroinvasive phenotype is necessary for the microbial activation of NF-κB and subsequent proinflammatory gene expression. Since bacterial adhesion precedes invasion, it is conceivable that microbial adhesion per se, rather than invasion, is a sufficient stimulus to induce the nuclear translocation of NF-κB. The purpose of the present study was to clarify the contribution of microbial invasion to the degradation of IκBα and the subsequent activation of NF-κB in cultured human intestinal epithelial cell monolayers.
MATERIALS AND METHODS
Bacterial culture and LPS preparation.
Salmonella dublin, S. typhimurium (American Type Culture Collection, Rockville, Md.), and E. coli (a gift from Shriner’s Burn Institute, Cincinnati, Ohio) were inoculated into 25 ml of brain heart infusion (BHI) broth (Difco) and incubated overnight at 37°C in a shaking incubator (New Brunswick Scientific Co., Inc., Edison, N.J.). Cultures were centrifuged at 10,000 rpm (model J2-HF; Beckman Instruments, Inc., Palo Alto, Calif.) for 10 min and washed three times with sterile saline. Bacterial pellets were resuspended in sterile saline and adjusted to a final concentration of 107/ml with a Klett densitometer (Klett Manufacturing Co., Long Island, N.Y.). To confirm bacterial concentrations, bacteria were plated on BHI agar at 10-fold serial dilutions and incubated at 37°C overnight, and colonies were counted. Purified lipopolysaccharide (LPS) from S. typhimurium (Sigma Chemical Co., St. Louis, Mo.) was resuspended in phosphate-buffered saline (PBS) and added to Caco-2BBe cells at a final concentration of 1 μg/ml.
Cell culture and infection.
Caco-2BBe cells (ATCC) between passages 5 and 15 were cultured in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum, 2 mM glutamine, 1% nonessential amino acids, and antibiotics. For analysis of cellular proteins, 6 × 105 cells were seeded into a six-well or a 10-cm tissue culture plate and cultured to confluency. Growth medium was replaced by Dulbecco modified Eagle medium without fetal bovine serum or antibiotics immediately prior to studies. Microbial adhesion and invasion were assayed by coculture of Caco-2BBe cells with 107 S. dublin, S. typhimurium, or E. coli cells for 20, 30, or 60 min, followed by washing in PBS and incubation for 6 h in growth medium containing 50 μg of gentamicin per ml to kill extracellular microorganisms. Intracellular bacteria were counted by lysing Caco-2BBe cells in 0.1% sodium dodecyl sulfate (SDS) and culturing extracts on BHI agar.
For studies of bacterial adhesion, Caco-2BBe cells were pretreated with cytochalasin D for 1 h to prevent microbial invasion and then washed in PBS prior to the addition of bacteria. To examine the effect of soluble bacterial factors on epithelial signal transduction, Caco-2BBe cells were grown in the lower compartment of a six-well Transwell bicameral chamber separated by a 0.4-μm-pore-size polycarbonate filter (Corning Costar Corporation, Cambridge, Mass.) from 107 S. dublin cells, which were added to the upper chamber for 20, 30, 60, or 120 min. To establish the role of bacterium-derived proteins in adhesion-dependent gene expression, S. dublin and S. typhimurium were cultured overnight, washed with sterile saline, and resuspended in sterile saline with 100 μM chloramphenicol (Sigma Chemical Co.) for 3 h. Cultures of S. dublin and S. typhimurium were then pelleted by centrifugation, washed four times in sterile saline, and added at 107 cells/ml at 37°C for 30 and 60 min to six-well culture plates of confluent Caco-2BBe cells that had been pretreated for 1 h with cytochalasin D.
Protein analysis.
Caco-2BBe cells grown in six-well culture plates were stimulated by S. dublin, S. typhimurium, or E. coli for 20, 30, or 60 min or LPS for 10, 20, 30, or 60 min. Caco-2BBe cells were then washed once in PBS and lysed in cold buffer containing 50 mM Tris (pH 8.0), 110 mM NaCl, 5 mM EDTA, 1% Triton X-100, and 0.1 mM phenylmethylsulfonyl fluoride (PMSF). The Bradford assay (Bio-Rad, Hercules, Calif.) was used to determine protein concentrations of each sample. Cell lysates were boiled in an equal volume of loading buffer (4% SDS, 20% glycerol, 125 mM Tris-HCl [pH 6.8], and 10% 2-mercaptoethanol), and 50 μg of each protein sample was loaded per lane on an 8-to-16% Tris-glycine gradient gel (Novex, San Diego, Calif.). Electrophoresed proteins were transferred to a nitrocellulose membrane (Novex) with the Novex Xcell Mini-Gel system. Membranes were blocked with 10% nonfat dried milk in Tris-buffered saline (TBS) for 1 h prior to exposure to rabbit polyclonal anti-IκBα antiserum (Santa Cruz Biotechnology Inc., Santa Cruz, Calif.) at a dilution of 1:200 for 45 min. Blots were washed twice in TBS containing 0.1% Tween 20 (TTBS), followed by the addition of peroxidase-conjugated anti-rabbit immunoglobulin G (Sigma) at a dilution of 1:10,000 for 30 min. Blots were washed three times for 5 min per wash in TTBS, incubated in commercial enhanced chemiluminescence reagents (ECL kit; Amersham, Little Chalfont, Buckinghamshire, England), and exposed to photographic film.
Nuclear protein extraction.
Nuclear protein extracts were prepared after cells were incubated with S. dublin, S. typhimurium, or E. coli for 1 h. Cells were washed twice with ice-cold PBS and harvested by scraping into 1 ml of PBS. After pelleting at 6,000 rpm (model J2-HF; Beckman Instruments, Inc.) for 5 min, cells were washed twice with cold PBS, resuspended in one cell pellet volume of lysis buffer (1.5 mM MgCl2, 0.2% [vol/vol] Nonidet P-40, 10 mM HEPES [pH 7.9], 10 mM KCl, 0.1 mM EDTA, 1 mM dithiothreitol [DTT], and 0.1 mM PMSF), and incubated on ice for 5 min with intermittent vortexing. Nuclei were collected from the cell lysates by centrifugation (6,000 rpm for 5 min) and then resuspended in one cell pellet volume of extraction buffer (1.5 mM MgCl2, 25% [vol/vol] glycerol, 20 mM HEPES [pH 7.9], 420 mM NaCl, 0.1 M EDTA, 1 mM DTT, and 0.5 mM PMSF). After incubation on ice for 15 min with intermittent vortexing, nuclear proteins were collected by centrifugation (14,000 rpm for 15 min). Supernatants also were collected to ensure the elimination of nuclear debris.
EMSAs.
An NF-κB oligonucleotide probe (5′-AGT TGA GGG GAC TTT CCC AGG-3′; Santa Cruz Biotechnology, Inc.) was labeled with [γ-32P]ATP by using T4 polynucleotide kinase (Gibco, BRL) and then purified on a Bio-Spin chromatography column (Bio-Rad). Nuclear protein extracts (10 μg) were preincubated with electromobility gel shift assay (EMSA) buffer [1 mM EDTA, 1 mM DTT, 12 mM HEPES (pH 7.9), 4 mM Tris-HCl (pH 7.9), 5 mM MgCl2, 25 mM KCl, 12% glycerol, 50 ng of poly(dI-dC) per ml, and 0.2 mM PMSF] on ice for 10 min, followed by the addition of the radiolabeled probe for 20 min. The specificity of the binding reaction was determined by incubating duplicate nuclear protein samples with a 100-fold molar excess of unlabeled probe. Samples were resolved on a nondenaturing polyacrylamide gel containing 5% acrylamide and run in 0.5× TBE (1 mM EDTA, 45 mM boric acid, and 45 mM Tris-HCl) at a constant current (30 mA) for 1 h. Gels were transferred to Whatman 3M paper, dried under a vacuum at 80°C for 1 h, and exposed to film by using an intensifying screen at −70°C for approximately 3 h.
Statistical analysis.
Results are reported as means ± standard errors of the means of several experiments. Student’s t test was used to compare mean values. Statistical differences were declared significant for P values of <0.05.
RESULTS
Bacterial invasion of Caco-2BBe cells.
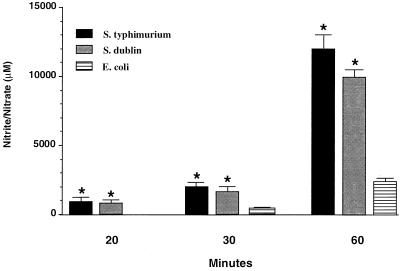
Cellular invasion by pathogenic bacteria has been associated with the NF-κB-dependent induction of various proinflammatory mediators. In order to determine whether microbial invasion of enterocytes is required for NF-κB activation, Caco-2BBe cells were infected with S. dublin, S. typhimurium, or E. coli. At selected times after infection, cells were lysed and intracellular bacteria were quantified. As expected, cellular invasion by both microorganisms increased in a time-dependent manner (Fig. 1). The rate of bacterial invasion by S. dublin or S. typhimurium was greater than that of E. coli (P < 0.05), with relatively no difference between S. dublin and S. typhimurium invasion. After 30 min of infection the ratio of S. dublin and S. typhimurium to Caco-2BBe cells was 1:500. Invasion by E. coli was significantly less (1:25,000 at 30 min). Thus, despite the relatively greater invasion by the Salmonella spp., the microbial invasion of Caco-2BBe cells by E. coli and Salmonella spp. was a rare event.
FIG. 1.
Bacterial invasion of intestinal epithelial cells. Cultures of confluent Caco-2BBe cells were infected with 107 S. dublin, S. typhimurium, or E. coli cells per ml for 20, 30, or 60 min. Bacterial invasion was determined following the killing of all extracellular bacteria with gentamicin. S. dublin and S. typhimurium showed the greatest degree of invasion. Data are presented as means plus standard errors of the means. ∗, P < 0.05 compared with E. coli value.
Effects of bacteria on IκBα degradation and NF-κB activation in Caco-2BBe cells.
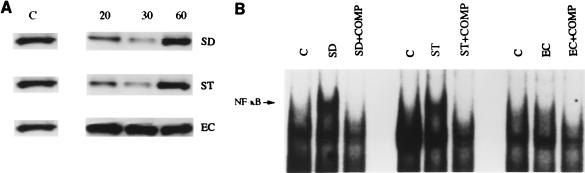
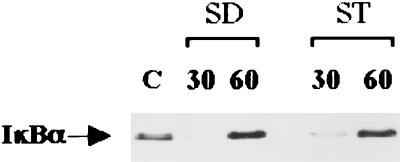
Previous studies of cultured enterocytes have implicated enteroinvasion as a prerequisite for microbial activation of NF-κB (5, 6). The nuclear translocation of NF-κB is regulated by the cytoplasmic inhibitory species IκBα via its binding to the nuclear localization sequence of p65 (2). Degradation of IκBα induced by cytokines and other proinflammatory stimuli triggers the activation of NF-κB (3, 10, 11). In order to relate microbial invasion to the stability of IκBα, Caco-2BBe cells were infected with S. dublin, S. typhimurium, or E. coli which vary in their degrees of invasiveness. Uninfected Caco-2BBe cells had stable levels of IκBα expression (Fig. 2A). Cells infected with invasive pathogens (S. dublin or S. typhimurium) experienced IκBα degradation after 20 min with a virtual loss of immunoreactivity at 30 min. As expected, IκBα immunoreactivity reappeared at 60 min (Fig. 2A), a consequence of NF-κB activation of the IκBα promoter (15). In contrast, no alteration in the level of IκBα expression at any time point was observed in Caco-2BBe cells stimulated with the weakly invasive nonpathogenic strain of E. coli.
FIG. 2.
(A) Representative Western blot analysis demonstrating IκBα degradation in a time-dependent manner in infected Caco-2BBe cells. Cultures of confluent Caco-2BBe cells were infected with 107 S. dublin (SD), S. typhimurium (ST), or E. coli (EC) cells per ml for 20, 30, or 60 min. Unstimulated Caco-2BBe cells were used as controls (C). Total protein was extracted, and 40 μg of protein was electrophoresed on an SDS-polyacrylamide gel followed by immunoblotting of IκBα. Cells infected with S. dublin and S. typhimurium showed IκBα degradation at 20 min, with virtual loss at 30 min and reappearance of IκBα by 60 min. IκBα concentrations were not affected in E. coli-infected cells. (B) Representative EMSA demonstrating NF-κB activation in bacterially stimulated Caco-2BBe cells. EMSAs using nuclear extracts from nonstimulated cells were used as a control (C). Cultures of confluent Caco-2BBe cells were infected with 107 S. dublin, S. typhimurium, or E. coli cells per ml for 60 min, and nuclear extracts were incubated with 32P-labeled NF-κB oligonucleotides to determine NF-κB activation. In order to confirm the specificity of the NF-κB signal, a 100-fold excess of cold oligonucleotide was used as a competitor to inhibit the binding activity of NF-κB (COMP).
In order to correlate the degradation of IκBα with the nuclear translocation of NF-κB, an electrophoretic mobility shift analysis was performed 1 h after infection of Caco-2BBe cells. DNA protein binding to the canonical NF-κB sequence was induced in Caco-2BBe cells stimulated by S. dublin and S. typhimurium (Fig. 2B). To confirm the specificity of the DNA-protein interaction, a 100-fold excess of cold oligonucleotide was used as a competitor to inhibit the binding activity of NF-κB (Fig. 2B). Corresponding to the prediction that NF-κB activation would not be observed in the absence of IκBα degradation, E. coli infection of Caco-2BBe cells did not induce nuclear protein which bound the NF-κB probe.
Invasion of microbes to Caco-2BBe cells is not required to stimulate IκBα degradation and NF-κB activation.
Although S. dublin and S. typhimurium invaded Caco-2BBe cells at a higher rate than E. coli, bacterial invasion was low in absolute terms. Since IκBα degradation was virtually complete at 30 min postinfection, at a time when invasion by S. dublin and S. typhimurium was <1:500, a requirement for enteroinvasion in the signal transduction of NF-κB activation appeared improbable.
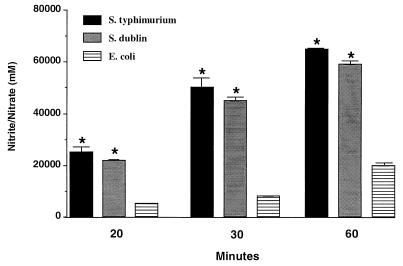
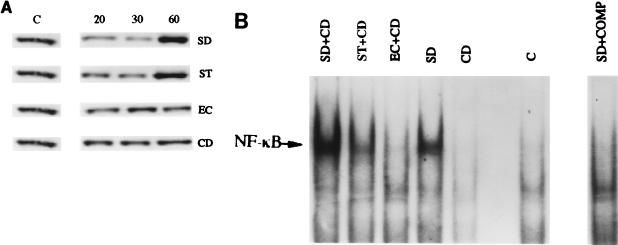
These data imply that adhesion, rather than invasion, might be the critical determinant of NF-κB activation. In order to test this possibility, Caco2-BBe cells were pretreated with the microfilament inhibitor cytochalasin D (25 μM) prior to infection, in order to block microbial invasion. All bacterial strains showed a time-dependent association with Caco-2BBe cells (Fig. 3). The extent of adhesion was 7- to 100-fold greater than the extent of invasion (compare Fig. 1). In parallel with the large difference in the rate of invasion between E. coli and Salmonella spp., the rate of adhesion of S. dublin and S. typhimurium was 3- to 10-fold higher than that of E. coli (Fig. 3) (P < 0.05). Treatment with cytochalasin D had no effect on the level of adhesion of S. dublin or S. typhimurium to Caco-2BBe cells. To ensure that bacterial invasion was completely eliminated by cytochalasin treatment, Caco-2BBe cells were washed with gentamicin prior to cell lysis and bacterial culture. Treatment with cytochalasin D blocked enteroinvasion by all bacterial strains. Cytochalasin D did not interfere with IκBα degradation and NF-κB activation in Caco-2BBe cells exposed to S. dublin or S. typhimurium (Fig. 4), implying that bacterial adhesion per se was sufficient to fully activate the NF-κB pathway.
FIG. 3.
Bacterial adhesion to Caco-2BBe cells. Cultures of confluent Caco-2BBe cells were pretreated for 1 h with 25 μM cytochalasin D. Cytochalasin D was then removed, followed by the addition of 107 S. dublin, S. typhimurium, or E. coli cells for 20, 30, or 60 min. Caco-2BBe cells were lysed and the number of extracellular organisms was calculated. S. dublin and S. typhimurium showed significant adhesion compared to E. coli. Data are presented as means plus standard errors of the means (P < 0.05). ∗, P < 0.05 compared with E. coli value.
FIG. 4.
(A) Representative Western blot analysis demonstrating IκBα degradation in a time-dependent manner in infected Caco-2BBe cells pretreated with 25 μM cytochalasin D (CD), followed by the addition of 107 S. dublin (SD), S. typhimurium (ST), or E. coli (EC) cells per ml for 20, 30, or 60 min. Unstimulated Caco-2BBe cells were used as controls (C). Total protein was extracted, and 40 μg of protein was subjected to SDS-polyacrylamide gel electrophoresis followed by immunoblotting of IκBα. Pretreatment with cytochalasin D did not affect IκBα degradation in S. dublin- and S. typhimurium-infected cells. (B) Representative EMSA demonstrating NF-κB activation in bacterially stimulated Caco-2BBe cells. EMSAs using nuclear extracts from nonstimulated cells were used as a control (C). Cultures of confluent Caco-2BBe cells were pretreated with 25 μM cytochalasin D and then infected with 107 S. dublin, S. typhimurium, or E. coli cells per ml for 60 min, and their nuclear extracts were incubated with 32P-labeled NF-κB oligonucleotides to determine NF-κB activation. To confirm the specificity of the NF-κB signal, a 100-fold excess of cold oligonucleotide was used as a competitor to inhibit the binding activity of NF-κB (COMP). Pretreatment with cytochalasin D did not affect NF-κB activation in S. dublin- and S. typhimurium-infected cells.
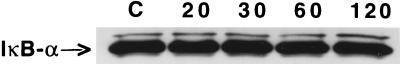
In order to exclude a role for Salmonella LPS in the induction of IκBα degradation, Caco-2BBe cells were incubated with purified LPS (1 μg/ml) for various times comparable to the bacterial adhesion experiment in this study and cell lysates were analyzed for IκBα degradation by Western blotting. LPS did not induce IκBα degradation at any of the tested time points when compared to untreated cells (Fig. 5). Further, to eliminate the possibility that S. dublin or S. typhimurium secreted a soluble protein(s), other than LPS, that could induce IκBα degradation, S. dublin and S. typhimurium were pretreated with chloramphenicol, a protein synthesis inhibitor, and then added to cytochalasin D-treated Caco-2BBe cells for 30 or 60 min. Western blot analysis showed that S. dublin- and S. typhimurium-stimulated cells maintained the capability of inducing IκBα degradation at 30 min (Fig. 6), indicating that a secreted protein was not involved in the induction of IκBα degradation. To lend further support to this evidence, S. dublin cells were cocultured with Caco-2BBe cells in a bicameral chamber, in which a filter impermeable to bacteria was interposed between the cell monolayer, located in the bottom chamber, and the microorganisms, added to the upper chamber. Western blot analysis showed that physical separation of Caco-2 cells from S. dublin blocked IκBα degradation (Fig. 7).
FIG. 5.
Representative Western blot analysis demonstrating IκBα immunoreactivity in confluent Caco-2BBe cells stimulated with 1 μg of purified S. typhimurium LPS per ml for 10, 20, 30, or 60 min. Unstimulated cells were used as a control (C). Total protein was extracted, and 40 μg of protein was subjected to SDS-polyacrylamide gel electrophoresis, followed by immunoblotting of IκBα. LPS had no effect on IκBα degradation as determined by comparison of stimulated and unstimulated cells.
FIG. 6.
Representative Western blot analysis demonstrating IκBα degradation in confluent Caco-2BBe cells pretreated for 1 h with 25 μM cytochalasin D. Cells were stimulated with 107 S. dublin (SD) or S. typhimurium (ST) cells that were pretreated with 100 μM chloramphenicol. Unstimulated cells (C) showed no IκBα degradation. Chloramphenicol-treated S. dublin and S. typhimurium stimulated IκBα degradation at 30 min with a reappearance at 60 min.
FIG. 7.
Representative Western blot analysis demonstrating IκBα degradation in confluent Caco-2BBe cells grown on the bottom of a six-well Transwell 0.4-μm-pore-size filter plate followed by the addition of 107 S. dublin cells to the top chamber and incubation for 20, 30, 60, or 120 min. Unstimulated cells were used as a control (C). S. dublin-derived soluble species which permeated the interposed filter did not induce IκBα degradation in Caco-2BBe cells.
DISCUSSION
NF-κB activation is stimulated in intestinal epithelial cells by exposure to pathogenic microbes, including Shigella (5), S. typhimurium (6), and enteropathogenic E. coli (14). Because NF-κB is involved in the expression of multiple proinflammatory genes, its induction by microorganisms suggests an innate mucosal immune response. The nature of the stimulus by which pathogenic bacteria induce NF-κB activation has not been determined. In this study, we demonstrate that invasion is not required for stimulation of IκBα degradation and NF-κB activation in human intestinal epithelial cells. Additionally, we excluded the contribution of a secreted soluble bacterial protein as a mediator of NF-κB activation. These data imply that bacterial adhesion per se may be a sufficient stimulus of IκBα degradation and NF-κB activation in intestinal epithelial cells. NF-κB is an important transcription factor for many proinflammatory genes, and its activation by bacterial adhesion may have profound implications for the pathophysiologic activities of intestinal epithelial cells. Indeed, bacterial enteroadhesion resulting in NF-κB activation is expected to lead to the expression of a pleiotropic group of genes that play a role in the initial host immune response produced by gut epithelial cells or in the pathogenesis of mucosal disease.
In the present study, we observed that highly invasive bacteria, S. dublin and S. typhimurium, stimulated IκBα degradation and NF-κB activation in cultured intestinal epithelial cells. In contrast, a nonpathogenic strain of E. coli did not induce IκBα degradation or NF-κB activation. The correlation between the level of invasiveness, IκBα degradation, and the nuclear translocation of NF-κB suggested a causal relationship, but the level of bacterial invasion was extremely low in absolute terms. Indeed, cultures of Caco-2BBe cells infected for 30 min showed a bacterium/Caco-2BBe cell ratio of only 1:1,000 with S. dublin and S. typhimurium. E. coli showed a significantly lower rate of invasion, only 1:25,000 at 30 min postinfection. It was therefore surprising that S. dublin and S. typhimurium were able to strongly induce IκBα degradation and NF-κB activation when less than 1 of 500 cells was invaded.
Other studies have reported that NF-κB activation by pathogenic microbial species is associated with a much higher rate of invasion (5, 6). In order to address the relative prevalence of enteroadhesion and enteroinvasion, bacterial invasion was blocked by pretreating cells with the microfilament inhibitor cytochalasin D before bacterial infection. We observed that adhesion to the cell surface by S. dublin and S. typhimurium was several orders of magnitude greater than invasion and was fully able to induce IκBα degradation and NF-κB activation. Thirty minutes after infection, the measured ratio of bacteria to Caco-2BBe cells was 1:33 for S. dublin and S. typhimurium. In contrast, the adhesion of E. coli was significantly lower. These values reflect only the measured association of bacteria and enterocytes at the moment of cell extraction. It is likely that over time a single bacterium may interact with and thereby stimulate multiple enterocytes. Thus, the actual ratio of bacterial enterocyte adhesion events during the period of incubation may be greater, perhaps approaching unity.
In order to establish whether bacterial adhesion per se was sufficient to stimulate IκBα degradation and NF-κB translocation, an inhibitor of enteroinvasion, cytochalasin D, was utilized. This agent had no effect on adhesion but totally inhibited bacterial uptake by Caco-2BBe cells. Cytochalasin D had no effect on the ability of S. dublin or S. typhimurium to induce IκBα degradation and NF-κB translocation. Thus, invasion of Caco-2BBe cells is not a required stimulus for NF-κB activation by Salmonella spp. These data suggested that bacterial stimulation of the cultured enterocytes was mediated by a physical association, i.e., adhesion, or by the release of a soluble activating factor. Utilizing a bicameral chamber, in which bacteria and enterocytes were cocultured yet physically separated by a barrier which could be permeated by macromolecules but not intact organisms, we determined that S. dublin or S. typhimurium alone, and not a secreted factor, induced IκBα degradation. Taken together, our data imply that pathogenic bacterial adhesion to the cell surface, rather than invasion or release of a secreted factor, induces IκBα degradation and NF-κB activation in intestinal epithelial cells.
It is well established that a number of factors induce the nuclear translocation of NF-κB, including various cytokines, viruses, and phorbol esters (1). More recently, it has been appreciated that pathogenic bacteria induce NF-κB activation followed by the upregulation of proinflammatory mediators, such as IL-8, in a variety of cultured epithelial cells (4–6). These studies have reported that bacterial invasion is necessary to stimulate the nuclear translocation of NF-κB in nonprofessional phagocytic cells, such as epithelial cells, by comparing highly invasive and invasion-deficient organisms. Utilizing a noninvasive mutant, Dyer et al. (5) observed that Shigella induction of NF-κB activation in HeLa cells required an invasive phenotype. These studies, however, did not examine whether the invasive phenotype may have been associated with altered adhesion properties. Hauf et al. (9), for example, demonstrated that pretreatment of macrophages with the actin-depolymerizing drug cytochalasin B abolished phagocytic uptake of Listeria but had no effect on bacterial adherence to the cell surface. Inhibition of the internalization of Listeria did not affect NF-κB activation, suggesting that bacterial adhesion to the surface of macrophages induces the nuclear translocation of NF-κB.
We observed a parallel mechanism of NF-κB activation in cultured enterocytes, a nonprofessional phagocytic cell type. Adhesion of S. dublin alone was sufficient to stimulate NF-κB activation, without the direct uptake of the organism into the cell. It is conceivable that these pathogenic organisms are stimulating a receptor(s) on the surface of cells, leading to NF-κB nuclear translocation with subsequent activation of inflammatory responses in the intestinal epithelia. Therefore, intracellular invasion of pathogens does not appear to be necessary to induce an inflammatory response in enterocytes.
In a variety of in vitro reductionist models of the intestinal epithelium, nonpathogenic microorganisms and bacterial LPS fail to stimulate cytokine production. In Caco-2BBe cells, for example, S. dublin invA does not induce IL-8 expression (5, 6, 12). Savkovic et al. observed that infection of intestinal epithelial cells by enteropathogenic E. coli, but not nonpathogenic bacteria, induces the nuclear translocation of NF-κB, resulting in the generation of IL-8 (14). In support of this finding, we found that nonpathogenic E. coli did not adhere to or invade Caco-2BBe cells sufficiently to induce NF-κB activation, in contrast to S. dublin or S. typhimurium. Therefore, gut epithelial cells appear to be able to respond to pathogenic and nonpathogenic bacteria according to the extent of bacterial enterocyte adhesion. Mechanisms which govern microbial enterocyte adhesion are active at the levels of both the microbe and the enterocyte (7, 13). The nature of this adhesion, rather than the act of invasion, appears to determine whether the interaction is commensal or pathologic. Presumably, in most instances bacterial enterocyte adhesion is weak and highly restricted; otherwise, the bowel mucosa would be in a persistent state of NF-κB activation, since it is continuously exposed, particularly in the colon, to an enormous reservoir of gram-negative organisms.
In summary, our data suggest that pathogenic bacterial adhesion, rather than invasion, is sufficient to stimulate NF-κB activation in intestinal epithelial cells. Although previous studies have shown that invasion by pathogenic organisms elicits inflammatory responses in intestinal epithelial cells, we have clarified this signal transduction pathway by showing that bacterial adhesion per se is a sufficient stimulus for NF-κB activation. Our data suggest that strategies to reduce enteric inflammation should be directed to the reduction of bacterial enterocyte adhesion.
ACKNOWLEDGMENT
Funding for this study was provided in part by the NIH (1RO1 GM57407-01) to A. L. Salzman.
REFERENCES
- 1.Baeuerle P A. The inducible transcription activator NF-κB: regulation by distinct protein subunits. Biochim Biophys Acta. 1991;1072:63–80. doi: 10.1016/0304-419x(91)90007-8. [DOI] [PubMed] [Google Scholar]
- 2.Baeuerle P A, Baltimore D. IκB: a specific inhibitor of NF-κB transcription factor. Science. 1988;242:540–544. doi: 10.1126/science.3140380. [DOI] [PubMed] [Google Scholar]
- 3.Baeuerle P A, Henkle T. Function and activation of NF-κB in the immune system. Annu Rev Physiol. 1994;12:141–179. doi: 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- 4.Botteri F M, Ballmer-Hofer K, Rajput B, Nagamine Y. Disruption of cytoskeletal structures results in the induction of the urokinase-type plasminogen activator gene expression. J Biol Chem. 1990;265:13327–13334. [PubMed] [Google Scholar]
- 5.Dyer R B, Collaco C R, Niesel D W, Kerzog N. Shigella flexneri invasion of HeLa cells induces NF-κB DNA-binding activity. Infect Immun. 1993;61:4427–4433. doi: 10.1128/iai.61.10.4427-4433.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eckmann L, Kagnoff M F, Fierer J. Epithelial cells secrete the chemokine interleukin-8 in response to bacterial entry. Infect Immun. 1993;61:4569–4574. doi: 10.1128/iai.61.11.4569-4574.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finlay B B, Heffron F, Falkow S. Epithelial cell surfaces induce Salmonella proteins required for bacterial adherence and invasion. Science. 1989;243:940–943. doi: 10.1126/science.2919285. [DOI] [PubMed] [Google Scholar]
- 8.Gilmore T D, Morin P J. The IκB protein: members of a multi-functional family. Trends Genet. 1993;9:427–433. doi: 10.1016/0168-9525(93)90106-r. [DOI] [PubMed] [Google Scholar]
- 9.Hauf N, Goebel W, Serfling E, Kuhn M. Listeria monocytogenes infection enhances transcription factor NF-κB in P388D1 macrophage-like cells. Infect Immun. 1994;62:2740–2747. doi: 10.1128/iai.62.7.2740-2747.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hohmann H P, Remy R, Poschl B, vanLoon A P G M. Tumor necrosis factor-α and -β bind to the same two types of tumor necrosis factor receptors and maximally activate the transcription factor NF-κB at low receptor occupancy and within minutes after receptor binding. J Biol Chem. 1990;265:15183–15188. [PubMed] [Google Scholar]
- 11.Jobin C, Haskill S, Mayer L, Panja A, Sartor R B. Evidence for altered regulation of IκBα degradation in human colonic epithelial cells. J Immunol. 1997;158:226–234. [PubMed] [Google Scholar]
- 12.Jung H C, Eckmann C L, Yang S, Panja A, Fierer E, Morzycka-Wroblewska F, Kagnoff M F. A distinct array of proinflammatory cytokines is expressed in human colon epithelial cells in response to bacterial invasion. J Clin Invest. 1995;95:55–65. doi: 10.1172/JCI117676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCormick B A, Miller S I, Carnes D, Madara J L. Transepithelial signaling to neutrophils by Salmonellae: a novel virulence mechanism for gastroenteritis. Infect Immun. 1995;63:2302–2309. doi: 10.1128/iai.63.6.2302-2309.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarkovic S D, Koutsouris A, Hecht G. Infect. Immun. 64:4480–4487. 1996. Attachment of a noninvasive enteric pathogen, enteropathogenic Escherichia coli, to cultured human intestinal epithelial monolayers induces transmigration of neutrophils. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun S C, Ganchi D, Ballard W, Greene C. NF-κB controls expression of inhibitor IκBα: evidence for an inducible autoregulatory pathway. Science. 1993;259:1912–1915. doi: 10.1126/science.8096091. [DOI] [PubMed] [Google Scholar]
- 16.Thanos D, Maniatis T. NF-κB: a lesson in family values. Cell. 1995;80:529–532. doi: 10.1016/0092-8674(95)90506-5. [DOI] [PubMed] [Google Scholar]