Abstract
Genomes can be edited by homologous recombination stimulated by CRISPR/Cas9 [clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated peptide 9]-induced DNA double-strand breaks. However, this approach is inefficient for inserting or deleting long fragments in mammalian cells. Here, we describe a simple genome-editing method, termed transcription-coupled Cas9-mediated editing (TEd), that can achieve higher efficiencies than canonical Cas9-mediated editing (CEd) in deleting genomic fragments, inserting/replacing large DNA fragments and introducing point mutations into mammalian cell lines. We also found that the transcription on DNA templates is crucial for the promotion of homology-directed repair, and that tethering transcripts from TEd donors to targeted sites further improves editing efficiency. The superior efficiency of TEd for the insertion and deletion of long DNA fragments expands the applications of CRISPR for editing mammalian genomes.
INTRODUCTION
Precision and efficiency are two major considerations when editing genomes for life science research and clinical applications. However, most genome-altering techniques are intrinsically inefficient because they rely on the homologous recombination (HR) pathway, which is much less efficient than the prevalent non-homologous end joining (NHEJ) repair pathway in human cells (1,2). Genome editing usually begins with making a double-stranded break (DSB) near the site in the genome targeted for editing (3). After a DSB is introduced, cells repair the break primarily via the NHEJ repair pathway. Genome editing via the HR pathway, an auxiliary cellular repair pathway (4), usually requires an HR donor harboring left and right homology arms (HAs) (5–7). A critical step in the HR pathway is the processing of DSB ends into single-stranded ends by EXO1 nuclease to ensure annealing of the HR donor fragment to the target site (8,9). CRISPR (clustered regularly interspaced short palindromic repeats) technology easily induces a DSB at the targeted site using a custom single guide RNA (sgRNAs) carried by a nuclease, such as CRISPR-associated peptide 9 (Cas9). The development of the CRISPR/Cas9 system has greatly facilitated targeted genome editing (7,10–13).
To edit genomes more efficiently and accurately, CRISPR-based methods have been developed to bypass HR and NHEJ pathways or to favor the HR pathway (7,10,14,15). For example, the CRISPR/Cas9-based prime-editing method uses an engineered Cas9 nickase and reverse transcriptase fusion (nCas9–RT) to introduce directly short edits encoded by a prime-editing guide RNA (pegRNA) (15,16). Recently, some improved strategies have been reported, but these methods are still inefficient and too imprecise for the insertion of long fragments such as green fluorescent protein (GFP) tags, even when donor templates have > 500 bp long HAs (17–20). HR-independent editing methods, such as microhomology-mediated end joining (MMEJ) and homology-mediated end joining (HMEJ), can integrate long DNA fragments into the genome. However, these methods require extra cleavage of the donor DNA, potentially resulting in off-target effects, and are prone to introducing errors at the boundaries of targeted sites. Also, the MMEJ method integrates long fragments inefficiently in some cultured cells (2,21–25). Thus, considerable research has been expended to develop more efficient and precise methods for the editing of long fragments in the genome. One approach to increase editing efficiency has been to leverage transcription, which is known to profoundly affect HR repair. A previous study demonstrated that active transcription of the donor DNA increases gene correction frequency (26). Transcription around DSBs forms DR-loops comprising DNA–DNA and DNA–RNA hybrids (27). R-loops determine whether a DSB is prone to be repaired by HR or by NHEJ since R-loops preferentially recruit HR proteins (28), such as Rad51 (29), Rad52 (30) and CSB (31), but not NHEJ proteins (27,30,32). A critical step leading to the formation of an R-loop and determining whether the repair proceeds via HR or NHEJ is the processing of DSB ends into single-stranded DNA (ssDNA) ends by EXO1 nuclease (33,34). After EXO1 resects the 5′ ssDNA at the DSB site, a transient R-loop can be formed by the newly transcribed RNA that hybridizes to the 3′ overhang ssDNA (35). This structure ensures the DSB will be repaired by the HR pathway (8,9).
In this study, we describe a transcription-coupled Cas9-mediated editing (TEd) method that can introduce point mutations, short fragments and long fragments more efficiently than the canonical Cas9-mediated editing (CEd) method. The increased editing efficiency was mainly attributed to transcription-coupled HR donor DNAs (TC donors), and the efficiency could be further enhanced by tethering transcribed TC donors to DSBs. Our findings indicate that TEd could substantially improve the efficiency of genome editing for use in basic and clinical sciences.
MATERIALS AND METHODS
Plasmid construction
To construct Cas9/gRNA plasmids, the parental Cas9 plasmid (Addgene plasmid #166033) was digested by restriction enzymes XbaI and PciI to remove the standard sgRNA. The standard sgRNA was replaced with designed sgRNAs by Golden Gate cloning. The sgRNA inserts were flanked by BsaI restriction sites to allow cloning of other additional sgRNAs. All sgRNA sequences used in this study are listed in Supplementary Table S1. To construct Cas9-MCP plasmids, the parent Cas9 plasmid was digested by restriction enzymes SalI and NotI to remove the original backbones, and then purified. Two copies of MCP (codon optimized for expression in human cells) were synthesized (GeneWiz), digested by restriction enzymes and ligated into the vector backbone by using T4 ligase (M0569S, New England BioLabs).
To construct the HR donor plasmids, HAs corresponding to all loci were designed by using the human genomic DNA sequence from NCBI. HR donor sequences are listed in Supplementary Table S2. HAs were PCR amplified from human embryonic kidney (HEK) 293T genomic DNA by using either Q5 High-Fidelity DNA Polymerase (M0494S, New England Biolabs) or KOD DNA Polymerase (KOD-101, Toyobo). HA inserts were cloned into KpnI and BamHI restriction sites of pCDH. To construct the TC donor plasmids, cytomegalovirus (CMV) or H1 promoter sequences were either obtained from the parental Cas9 plasmid or were synthesized (GeneWiz). Promoters were cloned into the EcoRI and NheI restriction sites at the 3′ ends of HAs. Promoters were oriented to transcribe the insert. Self-cleaving ribozymes and MS2 sequences (Addgene; plasmid #86196) were synthesized (GeneWiz) and then cloned into either the 5′ or 3′ end of the TC donor HAs. Primer sequences used for cloning in this study are listed in Supplementary Table S3.
Cell cultures and transfection
HEK293T and U2OS cells were cultured in Dulbecco's-modified Eagle's medium (DMEM; 11995065, Gibco) supplemented with 10% fetal bovine serum (FBS; 10100147, Gibco), 100 U/ml penicillin and 100 μg/ml streptomycin (60162ES76, Yeasen) at 37°C with 5% CO2. Cells were seeded into 24-well plates (∼4–5 × 105 cells/well). Plasmids were extracted by using the Endo-Free Mini Plasmid Kit (DP118-02, Tiangen) before transient transfection by using Lipofactamine 2000 (11668030, Life Technologies) following the manufacturer's protocols. For each well, 350 ng of Cas9 plasmid (or the associated plasmid) and 400 ng of TC donor and/or HR donor were used in TEd1/3 or CEd. For TEd2/4, 350 ng of Cas9 plasmid (or the associated plasmid) and 200 ng of TC donor and 200 ng of HR donor were used (Supplementary Figure S6C–E). The medium was replaced 12–18 h after transfection, and cells were harvested for flow cytometric analysis and microscopy at 72 h post-transfection.
Flow cytometry analysis
For each GFP knock-in (KI) experiment, at least 10 000 cells were analyzed by using either a BD LSR Fortessa flow cytometer (BD Biosciences) or a Beckman Coulter CytoFLEX S (Beckman Coulter Life Sciences). Either a BD FACS Aria III or a BD FACS Aria Fusion (BD Biosciences) were used to sort gated cells. Single cells that harbored the GFP KI were directly sorted into single wells of a 96-well plate for expansion. Batch processing of fluorescence-activated cell sorting (FACS) data was performed by using FlowJo software, version 10.
Genomic PCR
Genomic DNA was extracted from cells by using QuickExtract DNA extraction solution 1.0 (QE0905T, Lucigen) or Genomic DNA Extraction Kit (DP304, Tiangen) following the manufacturers’ protocols. Briefly, cells were harvested 72 h after transfection and washed with phosphate-buffered saline (PBS) three times. Lysis buffer was added to the cells, and samples were incubated at 65°C for 1 h and then at 98°C for 10 min. Genomic polymerase chain reaction (PCR) was usually performed in a mixture containing Ex Taq DNA polymerase (RR006Q, Takara), 0.5 μl of 4 μM duplex DNA substrate (400 nM final), 10 pmol (0.2 μM) primers, 0.5 mM dNTP mix, 20 mM HEPES-K, pH 7.5, 100 mM KCl, 5% glycerol, 0.2 mM EDTA, pH 8.0, 3 mM MgCl2 and 5 mM dithiothreitol (DTT). PCRs were performed as follows: 95°C for 3 min, 28 cycles of (95°C for 30 s, 60°C for 30 s and 72°C for 30 s) followed by a final extension at 72°C for 5 min. PCR primers are listed in Supplementary Table S3. PCR products were analyzed by Sanger sequencing (GeneWiz).
Analysis of KI frequencies by PCR
Common PCR was conducted to amplify the targeted region from extracted genomic DNA by using primers flanking the HAs. Junction PCR was conducted to amplify the junction region by using site-specific primers. Wild-type and truncated genomic fragments were resolved by gel electrophoresis. All PCR primer sequences are listed in Supplementary Table S3.
Immunofluorescence assay and imaging
For immunostaining, cells were washed with PBS three times and then fixed in 4% paraformaldehyde. Cells were frozen in liquid nitrogen and fixed on slides. Slides were blocked in PBSTA [PBS with 0.1% Triton X-100 and 3% bovine albumin (BSA)] for 1 h at room temperature; with primary antibodies, overnight at 4°C; and with secondary antibodies, for 1 h at room temperature. Cell nuclei were stained by using Vectashield mounting medium with 4′,6-diamidino-2-phenylindole (DAPI; H-1800–2, Vector Laboratories). Anti-lamin A/C [rabbit monoclonal antibody (mAb), A19524] was purchased from ABclonal Technology (WuHan, China). Anti-GFP (mouse mAb, ab1218) and anti-fibrillarin (mouse mAb, ab4566) were purchased from Abcam Technology. Immunofluorescence images were collected by using a Zeiss LSM 980 Upright laser scanning confocal microscope with a ×63 objective. Live-cell images were collected by using a Nikon ECLIPSE Ti-E inverted microscope with either ×40 or ×20 objectives.
RNA extraction and qRT-PCR
Total RNA was extracted using Direct-zol RNA Miniprep Plus (R2073, Zymo Research). cDNA was synthesized using the PrimeScript II 1st Strand cDNA Synthesis Kit (6210A, Takara) according to the manufacturer's protocol. Quantitative PCR (qPCR) was performed using 2× SYBR Green qPCR Master Mix (HY-K0501A, Bimake) following the manufacturer's protocol. Specifically, each PCR (30 μl) contained 0.2 μM of each forward and reverse primer pair, 10 ng of cDNA and 15 μl of SYBR Green qPCR Master Mix. qPCR was carried out as follows: 95°C for 5 min, 40 cycles of (95°C for 15 s and 60°C for 30 s), followed by a melt curve stage: 95°C for 15 s, 60°C for 30 s and 95°C for 15 s. The qPCR experiments were performed using a real-time PCR system, QuantStudio 7 Flex (Life Technologies). Each experiment included biological triplicates and technical duplicates. To measure GFP expression, quantitative reverse transcription–PCR (RT–qPCR) was performed to measure the GFP (EGFP-RT-F/EGFP-RT-R) transcript level, which was then normalized to that of glyceraldehyde phosphate dehydrogenase (GAPDH; GAPDH-RT-F/ GAPDH-RT-R). The primer sequences used are listed in Supplementary Table S3.
Genome qPCR
Genomic DNA was isolated by using the Genomic DNA Extraction Kit (DP304, Tiangen). Genomic DNA concentration was measured by NanoDrop. Genomic DNA was diluted to 50 ng/μl. qPCR was performed using SYBR Green Master Mix (HY-K0501A, Bimake). Specifically, each PCR (30 μl) contained 0.2 μM of each forward and reverse primer pair, 75 ng of cDNA and 15 μl of SYBR Green qPCR Master Mix. qPCR was carried out as follows: 95°C for 5 min, 40 cycles of (95°C for 15 s and 60°C for 30 s), followed by a melt curve stage: 95°C for 15 s, 60°C for 30 s and 95°C for 15 s. The qPCR experiments were performed using a real-time PCR system, QuantStudio 7 Flex (Life Technologies). Each experiment included biological triplicates and technical duplicates. qPCR was performed to measure the levels of GFP at LMNA (LMNA-F/LMNA-R) and fibrillarin (FBL; FBL-F/FBL-R) loci, which were then normalized to the level of 36B4 (36B4-F/36B4-R). The primer sequences used are listed in Supplementary Table S3.
TA cloning
To measure editing precision, fragments were amplified from genomic DNA by using PrimeSTAR HS DNA Polymerase (R045Q, Takara) and cloned into T-vector by using the pEASY-T1 Sample Cloning Kit (CT101, Transgene). Clones were analyzed by Sanger sequencing (GeneWiz).
Stable cell line construction
To generate stable reporter cell lines, cells were seeded at 4–5 × 105 cells/well into 6-well cell culture plates. After 24 h, reporter constructs (pLenti-CMV-mCherry-EGFP-PURO, a gift from Dr Wensheng Wei Lab, Peking University of China) and two viral packaging plasmids (psPAX2 and pMD2G) were co-transfected using Lipo2000 transfection reagent at a mass ratio of 1:3 (DNA:Lipo2000), together with the respective plasmids, into HEK293T cells. Medium was replaced by fresh medium 24 h after transfection. After another 24 h period, the supernatant was harvested, clarified by passage through a 0.45 μm membrane and stored at −80°C. After 72 h, the supernatant was harvested again, clarified by passage through a 0.45 μm membrane and stored at −80°C. Viable virus titers were determined by flow cytometric analysis of mCherry expression of transduced HEK293T cells. Briefly, the HEK293T cells were seeded at 4–5 × 105 cells/well into 6-well plates. After 24 h, the cell supernatant was replaced with 0.2 ml of the supernatant dilutions in fresh DMEM with 10% (v/v) FBS. At 72 h post-lentiviral transduction, the cells were harvested and analyzed for mCherry fluorescence by flow cytometry and the virus titers were calculated. The mCherry-positive and GFP-negative cells were sorted via FACS and cultured to isolate single cells for expansion into cell lines stably expressing dual fluorescence reporters (with undetectable background EGFP signal).
Western blot analysis
The 293 cells were collected and lysed in 1× loading buffer (9156, Takara). After sonication, lysates were centrifuged at 14 000 × g for 30 min. Total proteins were resolved with 7.5% TGX polyacrylamide gels (1610171, Bio-Rad) and transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore) by reverse electrophoresis. After being blocked, membranes were stained with anti-GFP (1:2000 dilution; ab1218, Abcam), anti-lamin A/C (1:10000 dilution; A19524, ABclonal) or anti-GAPDH (1:2000 dilution; 2118, Cell Signaling Technology) antibodies. Chemiluminescence signals were collected using Amersham Imager 800 (GE).
Next-generation sequencing library preparation
At 72 h after transfection, genomic DNA was extracted using a DNA Extraction Kit (Tiangen). Total DNA (250 ng) was used for next-generation sequencing (NGS) library preparation. LMNA (LMNA-ISCE-F13/ LMNA-ISCE-R13), mCherry-linker–GFP (mCherry-GFP-F11/mCherry-GFP-R11) and PRNP (PRNP-F12/PRNP-R12) were amplified using specific primers in a first-round PCR. For each sample, > 50 ng of purified PCR fragment was used for library preparation. PCRs (50 μl) contained 0.5 μM of each forward and reverse primer, 1 μl of genomic DNA extract and 25 μl of PrimeSTAR® HS Premix (R040Q, Takara). PCRs were carried out as follows: 98°C for 2 min, 28 cycles of (98°C for 10 s, 61°C for 30 s and 72°C for 30 s), followed by a final 72°C extension for 2 min. PCR products were treated in a single reaction with End Prep Enzyme Mix to repair ends, to phosphorylate 5′ ends and to add dA tails to 3′ ends. Then, T-A ligation was performed to add adapters to both ends. Adapter-ligated DNA was purified using DNA Clean Beads (A63882, Beckman Coulter). A second PCR was performed with P5 and P7 primers carrying sequences that anneal with flow cells (for bridge PCR) and indexes (for multiplexing). Specifically, PCRs (25 μl) contained 0.5 μM of each unique forward and reverse Illumina barcoding primer pair (I7/I5), 1 μl of purified adapter-ligated DNA and 12.5 μl of PrimeSTAR® HS Premix. The PCRs were carried out as follows: 98°C for 2 min, 10 cycles of (98°C for 10 s, 61°C for 30 s and 72°C for 30 s), followed by a final 72°C extension for 2 min. The final library product for sequencing was then purified by beads and qualified. The qualified libraries were pair-end sequenced (300 bp) on the Illumina MiSeq System. The primer sequences used are listed in Supplementary Table S3.
High-throughput sequencing data analysis
Alignment of amplicon sequences to a reference sequence was performed using CRISPResso2 (36). The quantification window was increased to 10 bp around the expected cut site to better capture diverse editing outcomes. Only reads containing no mismatches to the expected amplicon were considered for correct editing; reads containing indels that differed from the expected amplicons and reference sequence were included in error editing.
Ligation-mediated PCR (LM-PCR) and analysis
Genome-wide, off-target integration analyses of CEd and TEd were performed by using LM-PCR as detailed below. For each sample, 200 ng of genomic DNA was randomly fragmented to 500–700 bp by sonication (Covaris S220). The fragments were treated with End Prep Enzyme Mix for end repairing, 5′ phosphorylation and dA tailing (in one reaction). Then, T-A ligation was performed to add adapters to both ends. Two successive nested PCRs were performed to increase specificity. Adapter-ligated DNA was then selected on the basis of size by using beads (Beckman Coulter), and ∼400 bp long fragments were recovered. For each PCR product, 50 ng was treated with End Prep Enzyme Mix in a single reaction to repair ends, to phosphorylate 5′ ends and to add dA tails to 3′ ends. Then, T-A ligation was performed to add adapters to both ends. Ligation products were cleaned, and each sample was amplified by PCR using P5 and P7 primers. The PCRs were carried out as follows: 98°C for 2 min, 10 cycles of (98°C for 10 s, 61°C for 30 s and 72°C for 30 s), followed by a final 72°C extension for 2 min. Each primer had flow-cell-annealing sequences (for performing bridge PCR) and indexing sequences (for multiplexing). PCR products were purified by using beads and validated by using a Qsep100 (Bioptic, Taiwan, China). DNA concentrations were measured by using a Qubit 3.0 Fluorometer (Invitrogen, Carlsbad, CA, USA). The final library product for sequencing was then purified by beads, and concentrations were measured. Libraries were pair-end sequenced (250 bp) on the Illumina MiSeq System. Cutadapt (37) was applied to trim reads and generate sequences in fastq format. Fastq files were converted to fasta format using bioawk (https://github.com/lh3/bioawk) for further blastn (38) alignment. The sequences were aligned with hg19 blastdb, and only reads with the lowest e-value were formatted using the -outfmt 6 option. The minimal support of reads was > 3, since the use of a minimal support of one or two reads results in many false positives (39–41). Annotation of gene regions for integration sites was performed using the annotatePeakInBatch function in R package (42).
RESULTS
HR donor transcription increases CRISPR-mediated GFP knock-in efficiency
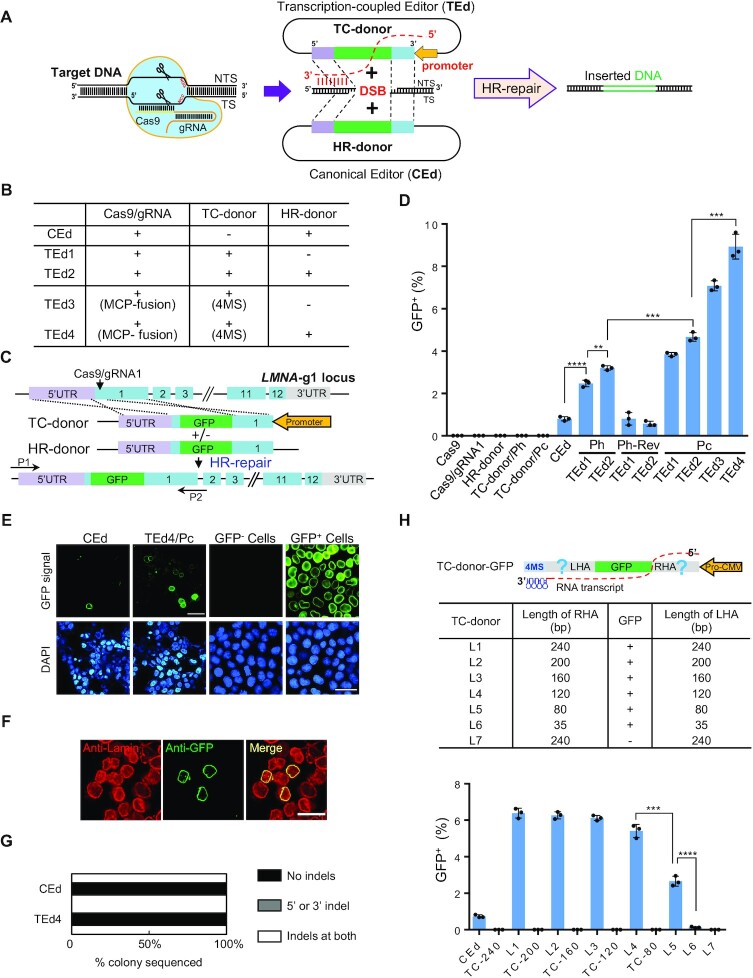
Since earlier studies showed that active transcription of the donor DNA significantly increases the gene correction frequency (26), we decided to test whether coupling of transcription over donor DNA templates to target donor reagent tethering increases the efficiency of CRISPR/Cas9-mediated gene editing. We therefore cloned the H1 promoter (an RNA polymerase III promoter) into either the left or the right end of the HAs in the HR donor plasmid to obtain two TC donors with opposite transcriptional orientations. One TC donor produced a homologous transcription-coupled RNA (TC RNA) complementary to the Cas9/gRNA non-target strand (designated as TEd/Ph) (Figure 1A; Supplementary Figure S1). The other TC RNA is complementary to the target strand (TEd/Ph-Rev). The efficiency and accuracy of gene editing by TEd and CEd were compared by integrating GFP into a sgRNA1 site at lamin A/C (LMNA) (Figure 1B, C). Using FACS analysis, we found that the GFP KI frequencies were significantly higher (3- to 4-fold) with TEd1/Ph (2.7 ± 0.15%) and TEd2/Ph (3.2 ± 0.10%) than with CEd (0.8 ± 0.12%) (Figure 1D). Successfully edited cells expressed GFP–lamin fusion protein localized to nuclear membranes, as expected (Figure 1E). However, the GFP KI efficiency with either TEd1/Ph-Rev or TEd2/Ph-Rev was not higher than that with CEd (Figure 1D), indicating that the direction of TC donor transcription is crucial for increasing GFP KI efficiency. Furthermore, a new LMNA-gRNA2 (adjacent to gRNA1 at the LMNA locus) targeting the opposite strand of the LMNA-gRNA1 locus was designed to test the effects of orientation of TC donor transcription on GFP KI efficiency. We found that the GFP KI frequency of the original TC donor with TEd1/Ph-Rev design (for targeting LMNA-g1) was markedly higher than that of CEd at the LMNA-gRNA2 locus, whereas the GFP KI frequency of the original TEd1/Ph (for targeting LMNA-g1) was not higher than that of CEd at the LMNA-gRNA2 locus (Supplementary Figure S2A, B). These results further confirmed the functionality of the TC RNA complementary to the Cas9/gRNA-non-targeting strand. Thus, we employed TC donors with TC RNA transcripts complementary to Cas9/gRNA non-target strands as the default transcriptional orientation in subsequent experiments. We next tested TEd efficiency of constructs driven by the CMV promoter (designated as TEd/Pc). To prevent TC RNAs from being transported from the nucleus to the cytoplasm for translational processing, we introduced two self-cleaving tRNA systems (hammer head ribozyme and HDV ribozyme) (43) into TEd that cleave the precursor TC RNAs (i.e. the transcripts from the CMV promoter) in the nucleus to release mature TC RNAs and facilitate HR. The KI efficiencies of the constructs with the CMV promoter (TEd1/Pc, 3.6 ± 0.25%; TEd2/Pc, 4.4 ± 0.10%) were higher than those of constructs with the H1 promoter (Figure 1D). Additionally, we further examined the transcripts of TC donors on an agarose gel and evaluated the transcription strengths of the two promoters by qRT-PCR. The results suggested that a higher TC donor transcription level increases KI efficiency (Supplementary Figure S2C). We also found that the TC donor with TC RNA complementary to the Cas9/gRNA- targeting strand [i.e. the CMV promoter in reverse orientation (Pc-Rev)] had a low GFP KI efficiency similar to that of CEd at the LMNA-gRNA1 locus (Supplementary Figure S2D), further confirming that the direction of TC donor transcription is essential for efficient genome editing using the TEd approach. In addition, we found GFP in cells modified by either TEd or CEd localized to nuclear envelopes (Supplementary Figure S2E), consistent with the observations of immunofluorescence using anti-Lamin antibody (Supplementary Figure S2F). Together, these results indicated that strand-specific TC donor transcription increases GFP KI efficiency by > 3-fold at the LMNA locus.
Figure 1.
Schematic overview of the TEd method for gene knock-in in mammalian cells. (A) Comparison of TEd and CEd strategies. A DSB was generated using Cas9/gRNA. TS, Cas9/gRNA target strand; NTS, Cas9/gRNA non-target strand. The TC donor has a promoter inserted into one end of the HAs of the HR donor, which produced homologous TC RNAs complementary to the NTS. (B) Components in the CEd and TEd systems. (C) Schematic overview of TEd-mediated GFP KI at the LMNA locus. The TC donor contains a promoter that directs the transcription of homologous TC RNAs complementary to the NTS of Cas9/gRNA. (D) Efficiencies (percentage of GFP+ cells determined by FACS) of TEd1 to TEd4 for GFP insertion at the LMNA locus in HEK293T cells. TEd/Ph: TEd driven by the TC donor with the H1 promoter directing transcription of the homologous TC RNAs complementary to the NTS of Cas9/gRNA. TEd/Ph-Rev: TEd driven by the TC donor with the opposite H1 promoter transcribing TC RNAs complementary to the TS of Cas9/gRNA. TEd/Pc: TEd driven by the TC donor with the CMV promoter transcribing TC RNAs complementary to NTS of Cas9/gRNA, similar to TEd/Ph. HR donors alone and the corresponding TC donors (without Cas9) were used as controls. (E) Left two columns, living cell images of CEd and TEd4 observed at 3 days post-transfection (unsorted). Right two columns, living cell images of GFP– and GFP+ clones after sorting of TEd4. Scale bar, 50 μm. (F) Representative immunofluorescence images of GFP+ cells in TEd4, showing nuclear membrane localization of GFP signals and co-localization with lamin by anti-lamin immunostaining. GFP, green; nuclei are stained with DAPI, blue. Scale bar, 50 μm. (G) Junction profiles of colonies determined by Sanger sequencing of KI TA clones as in Supplementary Figure S4C. Indels are insertions or deletions detected by sequencing of ∼30 colonies per condition at ± 10 bp around a DSB. (H) KI efficiencies of TEd4 with different HA lengths in HEK293T cells. Upper panel, design of TC donors with various HA lengths. Lower panel, graphs showing the percentage of GFP+ cells determined by FACS for each combination. Corresponding TC donors (without Cas9) were used as control. All GFP KI efficiency data are presented as individual data points with mean ± SD for n = 3 independent biological replicates. P-values were obtained using the two-tailed Student's t-test. **P< 0.01, ***P< 0.001, ****P< 0.0001.
Tethering TC RNAs to DSBs increases GFP KI efficiency
The efficiency of TEd may be limited by the distance between the TC donor and the DSB, and HR repair may be limited if TC RNAs must diffuse randomly from TC donors to DSBs (44). We therefore tested whether we could directly recruit TC RNAs to DSBs by using the MCP–MS2 system, in which the bacteriophage MS2 coat protein (MCP) tightly binds to MS2-binding sites (MBS) in RNA (45). We inserted four MS2s into the 3′ end of TC RNA in TEd1 (to produce TEd3) and TEd2 (to produce TEd4) (Figure 1B; Supplementary Figure S1), and fused MCP to either the N- or C-terminus of Cas9. We then examined the efficiencies of these constructs for inserting GFP into the LMNA locus. We found that the efficiencies of TEd3 and TEd4 (as high as 9.7 ± 0.21%) were ∼10-fold higher than that of the CEd control (0.8 ± 0.01%) and ∼2-fold higher than that of TEd2 (4.6 ± 0.12%) (Figure 1D; Supplementary Figure S3A). The efficiency of C-tagged MCP was slightly higher than that of N-tagged MCP (Supplementary Figure S3A). Moreover, the GFP KI efficiencies of the Cas9-MCP/TC donor (without 4× MS2 aptamers) combination and Cas9/TC donor (with 4× MS2) combination were similar to that of TEd1, but neither was as efficient as that of TEd3 (Supplementary Figure S3B) under the same transfection efficiencies and conditions (Supplementary Figure S3C). We confirmed that the cells were accurately edited by TEd3, TEd4 and CEd, as evidenced by the nuclear membrane localization of GFP (Figure 1E, F; Supplementary Figure S3D, E), and the results of genomic PCR (Supplementary Figure S4A) and junction sequencing (Supplementary Figure S4B). Sequencing TA clones using primers that bind outside the HAs revealed that GFP KI clones obtained by both TEd4 (29/29) and CEd (28/28) were free of indels (Figure 1G; Supplementary Figure S4C). We also observed precise editing (no indels) at both the left and right GFP KI junctions in 21 of 22 single-cell clones derived from sorted GFP-positive cells in TEd4 (Supplementary Figure S4D, E), with heterozygous insertion alleles in most clones. Furthermore, we confirmed that GFP was accurately inserted into the LMNA locus with the expected protein size by immunoblot analysis (Supplementary Figure S4F). Thus, we concluded that tethering the TC RNA to the DSB significantly improves editing efficiency.
To investigate whether the improved editing efficiency of TEd came with more off-target effects, we used the unsorted cells edited by CEd and TEd4 to perform LM-PCR and determine the extent of GFP KI occurring outside the LMNA locus (Supplementary Figure S5A). NGS results revealed that only one off-target site was detected in CEd, with no off-target sites in the case of TEd4 (Supplementary Figure S5B; Table S4), indicating that TEd can effectively improve the KI of long fragments without creating unwanted editing events.
TC donor homology arm lengths affect GFP KI efficiency
We next examined the effect of TC donor HA lengths on HR efficiency. Usually, 0.1–1 kb long HAs are used to insert transgenes (46–50). To target LMNA in HEK293T cells, we co-transfected TEd3-derived TC donors with truncated HAs (35–240 bp; L1–L7) (Figure 1H) and the Cas9/gRNA expression plasmid without the HR donor. TC donor controls with HAs of different lengths (no Cas9/gRNA), which were employed to evaluate the donor background, barely produced GFP signals. The KI efficiency of the donor with 120 bp long HAs (5.4 ± 0.12%) was nearly as high as that of the donor with 240 bp long HAs (6.4 ± 0.11%) (Figure 1H; Supplementary Figure S6A). However, TC donors with HAs < 120 bp exhibited lower editing efficiency (2.7 ± 0.15% for the donor with 80 bp long arms; 0.1 ± 0.02% for the donor with 35 bp long arms) (Figure 1H; Supplementary Figure S6A, B). These results indicated that the longer HAs drive higher integration efficiencies, and that 120 bp is sufficient for efficient TEd.
Transcription of the donor DNA primarily determines GFP KI efficiency
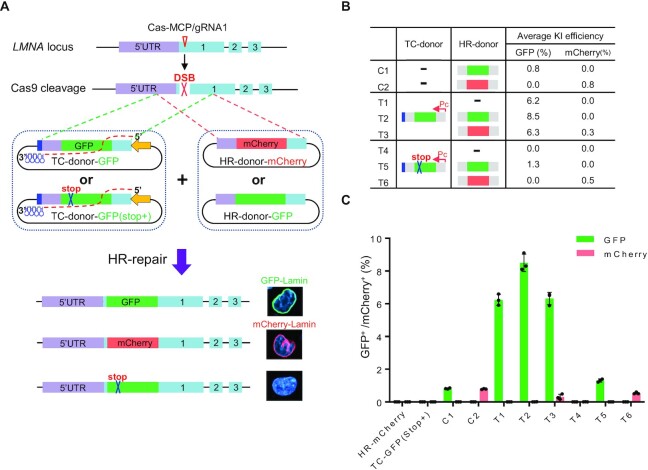
We then asked whether transcription of the TC donor or the transcribed TC RNA itself increased the insertion efficiency, i.e. by acting in cis (only the insertion efficiency of the transcribed donor is increased) or by acting in both cis and trans (the insertion efficiency of the non-transcribed donor could also be increased). We introduced a point mutation into the GFP gene to create a premature stop codon and cloned it into TEd3 and TEd4 constructs [TC donor–GFP (Stop+)] (Figure 2A). We hypothesized that this single point mutation would affect neither transcription of the TC donor nor the hybridization of the TC RNA to the targeted DNA strand at LMNA. Compared with the low background of HR donor or TC donor controls (no Cas9/gRNA), we found that the GFP KI efficiency obtained by co-transfecting Cas9 plasmids with TC donor–GFP and HR donor–GFP (8.5 ± 0.05%; combination T2 in Figure 2B) was higher than that obtained by co-transfection with TC donor–GFP (Stop+) and HR donor–GFP (1.3 ± 0.05%; combination T5 in Figure 2B, C). This suggested that TC donors were responsible for the majority of the GFP KI events, and that TEd operates in cis. To further determine whether the indel with the GFP stop mutation was integrated into the genome, we performed junction PCR and sequenced the targeted insertion. Analysis of the sequencing results showed no detectable GFP (Stop+) signal in TEd4-T2, whereas in TEd4-T5, ∼79% of GFP KI (Stop+) arose from the TC donor and 17% of GFP KI without the stop mutation arose from the HR donor (Supplementary Figure S7A), indicating that the TC donor with the stop mutation was still integrated into the genome and further implying that the preferred DNA template in the TEd method was the TC donor, not the HR donor. To further confirm this conjecture, we compared the efficiency of GFP and mCherry KI with two combinations of HR donor and TC donor (Figure 2A; Supplementary Figure S7B). We first co-transfected cells with TC donor–GFP and HR donor–mCherry (combination T3 in Figure 2B). The efficiency of GFP KI (6.3 ± 0.14%) in T3 was higher than that of mCherry KI (0.3 ± 0.02%) (Figure 2B, C). This result suggests that HR repair is highly prone to the TC donor DNA template. We observed that GFP KI efficiency from co-transfection with HR donor–GFP and TC donor–GFP (Stop+) (1.3 ± 0.04%; T5 combination) was ∼60% greater than that from transfection with HR donor–GFP alone (0.8 ± 0.01%; C1 combination) (Figure 2B, C). This suggested that free TC RNA transcripts might also increase the KI efficiency of the HR donor template in trans. The result was also consistent with our observation that the GFP KI efficiency of the TC donor–GFP (6.2 ± 0.21%) increased by 36% upon co-transfection with both TC donor–GFP and HR donor–GFP (8.5 ± 0.30%; T2 efficiency:T1 efficiency) (Figure 2B). We obtained similar results by using TC donor–mCherry and HR donor–GFP (Supplementary Figure S7C–G). Additionally, the mCherry KI efficiencies obtained by co-transfection with either HR donor–mCherry and TC donor–GFP (0.3 ± 0.02%, T3 combination) or HR donor–mCherry and TC donor–GFP (Stop+) (0.5 ± 0.06%; T6 combination) were significantly lower than that obtained by HR donor–mCherry alone (0.8 ± 0.07%; C2 combination), indicating that transcripts with sequences that do not match the HR donor sequence interfere with HR repair. Altogether, these results suggested that transcription of the donor template is critical for efficient HR-induced gene editing.
Figure 2.
HR repair occurs preferentially on TC donor templates. (A) Schematics showing CEd- and TEd-mediated KI at the LMNA locus with different GFP/mCherry donors. HR donor–GFP contains GFP (green) and HR donor–mCherry contains mCherry (red). TC donor–GFP (Stop+) contains GFP with a stop codon (blue cross). (B) Summary of the GFP/mCherry KI efficiencies by CEd and TEd with different donors. (C) GFP+/mCherry+ KI efficiencies of samples listed in (B). Graphs showing the percentage of GFP+/mCherry+ cells (y-axis, as determined by FACS) for each combination (x-axis). HR donor alone and the TC donor alone were used as controls. All data are presented with individual data points and mean ± SD for n = 3 independent biological replicates.
Other factors that may affect TEd-mediated GFP KI efficiency
To further explore the broad-spectrum applications of TEd, we next tested the effects of the TC donor with premature termination of transcription, plasmid backbone transcription of the TC donor and the transcriptional activity of the insert itself on the editing efficiency at the LMNA-gRNA1 locus. For the H1 promoter (pol III), a poly(T) sequence (9T) was inserted at the junction between the HA and the GFP-coding sequence in the TC donor (Supplementary Figure S8A) to terminate transcription prematurely without introducing a frameshift. Interestingly, TC donors with premature termination of transcription did not have a lower GFP KI efficiency (3.85 ± 0.07%) than the full-length transcribed TC donors (3.92 ± 0.02%), suggesting that complete transcription of TC donors may not be required for TEd-mediated increases in KI levels (Supplementary Figure S8B). To investigate the influence of backbone transcription on gene editing, we constructed a TC donor with transcription oriented toward the plasmid backbone (i.e. transcription into the plasmid backbone instead of into the donor template) (Supplementary Figure S8C). We found that transcription from the plasmid backbone (TEd3/Pc-BT) resulted in a low GFP KI efficiency (0.82 ± 0.01%) comparable with that using the CEd method, suggesting that transcription from the backbone did not increase GFP KI efficiency, and that the TEd strategy requires at least partial transcription of the template DNA (Supplementary Figure S8D). Finally, to investigate the effect of a TC donor template containing a regulatory element on the efficiency of GFP KI, we modified TC donors by inserting a functional H1 promoter into the 3′ end of the GFP-coding sequence (without generating a frameshift) in two different transcription orientations (i.e. TEd3-HG1 and TEd3-HG2) (Supplementary Figure S8E). Compared with the transcriptionally inactive insert (GFP KI in TEd3: 6.3 ± 0.02%), both modified TC donors resulted in reduced GFP KI efficiencies (TEd3-HG1, KI: 3.7 ± 0.19%; TEd3-HG1, KI: 2.9 ± 0.22%) (Supplementary Figure S8F), indicating that insertion of a transcriptionally active element may adversely affect TEd efficiency. However, the GFP KI efficiencies of these modified TC donors with the H1 promoter insertion were still significantly higher than that of CEd. This prompts the need for careful evaluation of the transcriptional properties of inserts in future applications of the TEd method.
TEd accurately and efficiently edits diverse loci in both HEK293T and U2OS cell lines
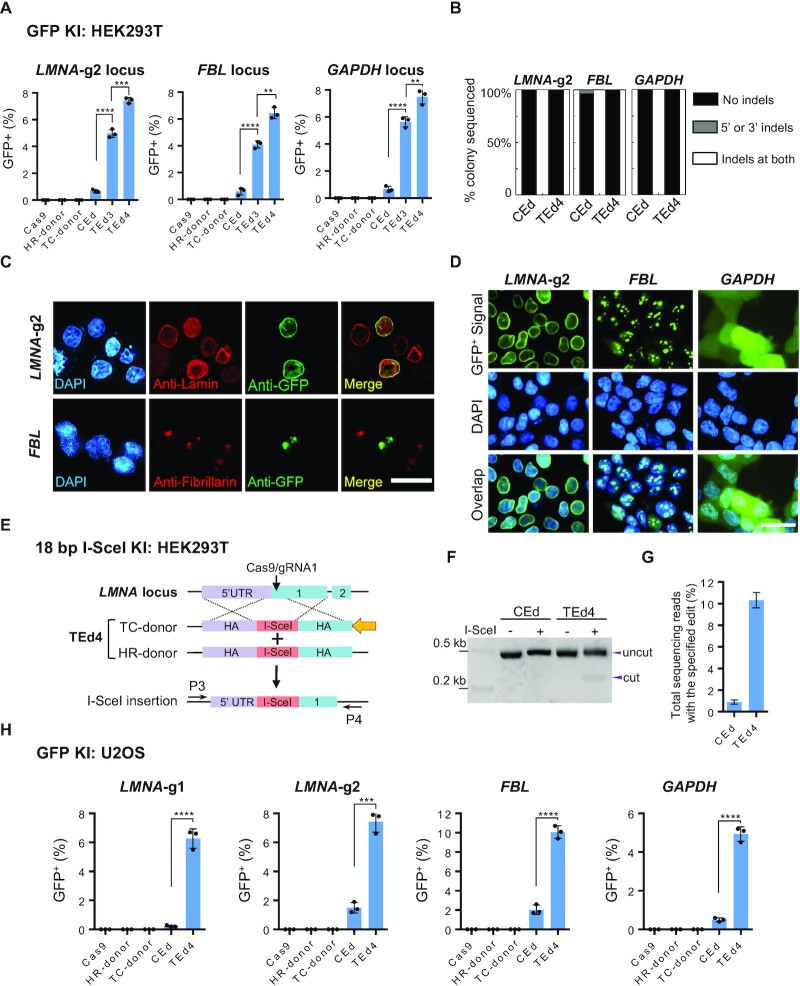
We tested whether TEd was more efficient than CEd for editing diverse sites in the genomes of both HEK293T and U2OS cells. In HEK293T cells, TEd was more efficient than CEd for inserting GFP into LMNA-g2 (TEd, 7.5 ± 0.18%; CEd, 0.6 ± 0.01%); GAPDH (TEd, 7.2 ± 0.17%; CEd, 0.9 ± 0.05%); and FBL (TEd, 6.9 ± 0.13%; CEd, 0.7 ± 0.09%) loci (Figure 3A; Supplementary Figure S9A). qPCR analysis revealed that the TEd efficiency of LMNA-g2 and FBL loci was ∼6-fold higher than the CEd efficiency (Supplementary Figure S9B). To assess whether the increased editing efficiencies of TEd at three loci compromise the precision of KIs (defined by indel-free KI events), genomic DNA of unsorted cells was amplified with PCR primers flanking HAs, followed by TA cloning and colony Sanger sequencing to check the junction sequences. Junction analysis (∼30 colonies per gene per condition) showed that the TEd method had very low indels at 5′ and/or 3′ KI junctions, similar to CEd (Figure 3B). We further confirmed the KI accuracy by verifying the expression and localization of GFP in cells edited at different loci (Figure 3C, D), as well as by Sanger sequencing of KI site amplicons from the sorted single-cell clones (Supplementary Figure S9C, D). For GFP KI at the FBL locus, LM-PCR and NGS-based analysis revealed the expected on-target KI site and the undetected off-target site in both CEd and TEd4 (Supplementary Figure S9E; Table S4). To further compare TEd with CEd for KIs, we validated KI of the I-SceI recognition site (18 bp) into the LMNA locus (Figure 3E). We demonstrated successful KI of the I-SceI fragment by I-SceI digestion of PCR products (amplified with primers flanking the HAs) (Figure 3F). NGS revealed that KI efficiency of TEd4 was up to 10-fold higher than that of CEd (Figure 3G), indicating that TEd significantly increased the KI efficiency of short fragments. Additionally, we obtained similar results for GFP KI into these four loci in U2OS cells; KI efficiencies using TEd were at least 8- to 10-fold higher than those using HR (Figure 3H; Supplementary Figure S9F, G). Collectively, the results show that TEd is an efficient and accurate method for inserting genes into diverse genomic sites in two cell lines.
Figure 3.
Targeted insertions with TEd at diverse loci in HEK293T and U2OS cell lines. (A) Comparison of GFP KI efficiencies of TEd and CEd at three genomic loci (LMNA-gRNA2, FBL and GAPDH loci) in HEK293T cells. Graphs showing percentages of GFP+ cells (y-axis, as determined by FACS) for each combination (x-axis). (B) Percentage of colonies with indicated junction profiles determined by Sanger sequencing of KI clones in (A). Indels represent insertions or deletions detected from sequencing of ∼30 colonies per condition per locus at ± 10 bp around the DSB. (C) Immunofluorescence analysis to determine the accuracy of TEd4-mediated GFP KI at LMNA-gRNA2 and FBL loci in HEK293T cells. Upper panel, GFP KI at the LMNA-gRNA2 locus identified by anti-Lamin immunostaining (unsorted); lower panel, GFP KI signal at the FBL locus observed by anti-Fibrillarin immunostaining (unsorted). GFP, green; nuclei are stained with DAPI (blue). Scale bar, 50 μm. (D) Living cell imaging verification of GFP KI at three different loci in sorted GFP-positive cells. GFP, green; DNA, blue. Scale bar, 50 μm. (E) Schematic showing TEd-mediated KI of the I-SceI recognition sequence at the LMNA-gRNA1 locus in HEK293T cells. (F) PCR amplicons incubated with or without I-SceI enzyme were analyzed on an agarose gel. PCR primers were designed outside the HAs [P3 and P4 in (E)]. Digested products are marked as ‘cut’. The original amplicon is marked as ‘uncut’. (G) I-SceI insertion efficiencies of (E) determined by NGS. This test was performed for two independent biological replicates. (H) Comparison of GFP KI efficiencies at four genomic loci in U2OS cells. All data are presented with individual data points and mean ± SD for n = 3 independent biological replicates. P-values are obtained using the two-tailed Student's t-test. **P< 0.01, ***P< 0.001, ****P< 0.0001.
TEd efficiently and accurately inserts longer fragments (up to 10 kb)
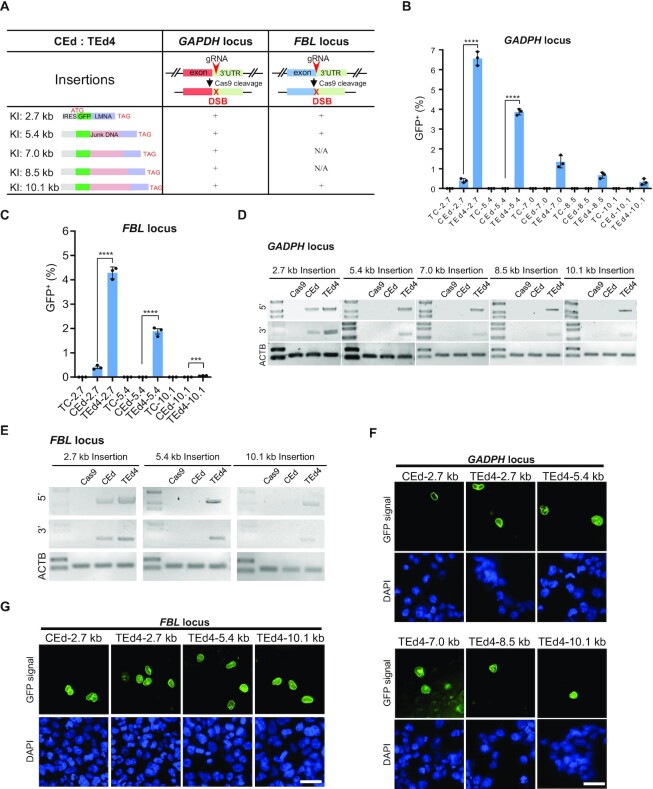
Inserting large DNA fragments efficiently and precisely is challenging for current genome editing methods. To measure the KI ability of TEd for the insertion of differently sized DNA fragments (2.7–10.1 kb) into the 3′ untranslated regions (UTRs) of GAPDH and into the FBL loci of both HEK293T and U2OS cell lines (Figure 4A), we designed DNA fragments of different sizes by inserting different lengths of spacer DNA (in frame) between GFP and LMNA (Figure 4A). To track KI events easily by fluorescence microscopy, we also added a nuclear localization signal to the N-terminus of GFP to visualize localization of the GFP–LMNA fusion protein to the nuclear envelope. For HEK293T cells, the KI efficiency of TEd was higher than that of CEd for the 2.7 kb (TEd efficiency, 6.3 ± 0.15%; CEd efficiency, 0.3 ± 0.02%), 5.4 kb (TEd, 3.9 ± 0.31%; CEd, 0.01 ± 0.00%), 7.0 kb (TEd, 1.2 ± 0.02%; CEd, undetected in > 105 cells), 8.5 kb (TEd, 0.6 ± 0.02%; CEd, undetected in > 105 cells) and 10.1 kb fragment (TEd, 0.1 ± 0.00%; CEd, undetected in > 105 cells) at the GAPDH locus (Figure 4B; Supplementary Figure S10A, B). For the FBL locus, we measured the editing efficiencies of three differently sized fragments (2.7, 5.4 and 10.1 kb). Efficiency of KI by TEd was significantly higher than that by CEd for the 2.7 kb (TEd efficiency, 4.2 ± 0.23%; CEd efficiency, 0.4 ± 0.01%), 5.4 kb (TEd, 1.88 ± 0.21%; CEd, undetected in > 105 cells) and 10.1 kb fragment (TEd, 0.05 ± 0.00%; CEd, undetected in > 105 cells) (Figure 4C; Supplementary Figure S10A, B). We confirmed the accuracy of KI by verifying the nuclear membrane expression of GFP in edited cells and by sequencing the junctions of the KIs (Figure 4D–G; Supplementary Figure S10C-E). Moreover, TEd was more efficient at inserting long fragments into U2OS loci (Supplementary Figure S10F). These results suggested that TEd is more efficient than CEd for the insertion of long DNA fragments.
Figure 4.
Targeted insertions of long fragments by TEd in HEK293T cells. (A) Design of long fragment KI at GAPDH and FBL loci by CEd and TEd4. gRNA was targeted to the 3′UTRs of genes. Insertions included the internal ribosome entry site (IRES, light gray), GFP (green), different lengths of junk DNA (brown), LMNA gene cDNA sequence (purple) and a stop codon (TAG) at the end. The GFP-junk DNA-LMNA sequence was designed with in-frame translational fusion with a fusion protein that is expected to exhibit nuclear membrane localization after successful insertion. (B and C) Comparison of the KI efficiencies of TEd and CEd with targeted long fragments (2.7–10.1 kb) inserted at the GAPDH locus (B) and FBL locus (C). Graphs show percentages of GFP+ cells for each combination. Corresponding TC donor alone (without Cas9) was used for each editing control. All data are presented with individual data points and mean ± SD for n = 3 independent biological replicates. P-values were obtained using the two-tailed Student's t-test. ***P< 0.001, ****P< 0.0001. (D and E) Representative PCR amplicons of 5′ and 3′ junction sites for KI fragments of different lengths at the GAPDH locus (D) and FBL locus (E). For 5′ or 3′ junction PCR amplicons, one primer was designed outside the donor HAs and the other inside the insert (Supplementary Figure S10). The ACTB PCR amplicon was used as an internal control. (F and G) Living cell images of cells with different KI segment lengths at the GAPDH locus (F) and FBL locus (G) before sorting. GFP, green; nuclei, stained by DAPI (blue). Scale bar, 50 μm.
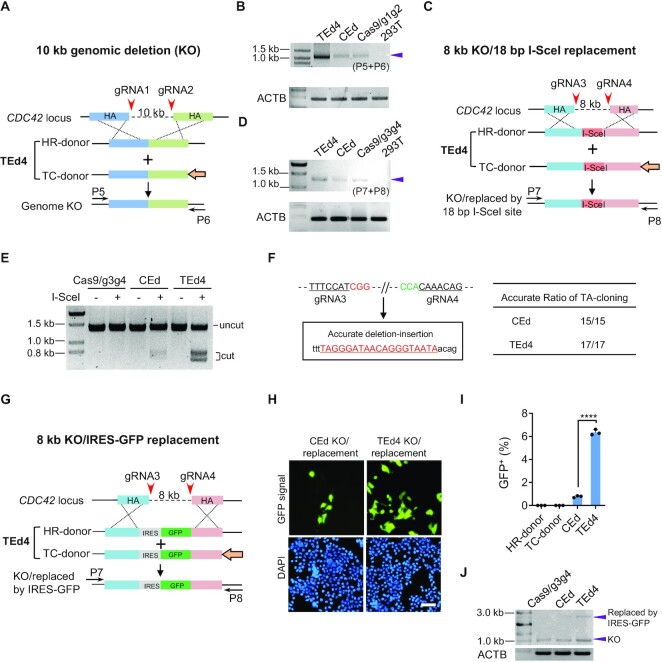
TEd efficiently deletes and replaces long fragments
We next compared TEd4 and CEd efficiencies for deleting a 10 kb long fragment and an 8 kb long fragment of CDC42 in HEK293T cells. We verified deletion of the 10 kb long fragments (Figure 5A) by junction PCR with primers flanking the targeted regions (Figure 5B). We found that the relative efficiency of the TEd4-mediated deletion was higher than that of the traditional knock-out method. We also checked the accuracy of genomic deletion by the TEd4 method by sequencing TA clones from junction amplicons (7 of 10 TA clones with error-free end-to-end ligation sequences) (Supplementary Figure S11A, B). For the replacement of an 8 kb genomic fragment with the 18 bp I-SceI site (Figure 5C), TEd4-mediated deletion was more efficient than the traditional CEd method (Figure 5D) based on the intensities of junction amplicons. Digestion of the PCR products of both TEd4-edited and CEd-edited loci with I-SceI resulted in the expected restriction fragment lengths, indicating that I-SceI replacement was accurate using these methods (Figure 5E). Random sequencing of TA clones revealed that TEd was more efficient for seamless I-SceI replacement than CEd (Supplementary Figure S11C, D). Besides, we found no indel at I-SceI junctions in either CEd-derived clones with I-SceI KI (15 of 15 clones had expected sequences) or TEd4-derived clones with I-SceI KI (17 of 17 clones had expected sequences) (Figure 5F). We also assessed replacement of the 8 kb long genomic fragment with an internal ribosome entry site (IRES)–GFP fragment (Figure 5G). FACS analysis revealed a significantly higher replacement frequency in TEd4-edited cells (6.2 ± 0.24%) than in CEd-edited cells (0.8 ± 0.08%) (Figure 5H, I; Supplementary Figure S11E). In addition, we observed clear pan-cellular GFP expression and expected junction PCR product lengths in TEd4-edited cells (Figure 5J), further confirming accurate replacement of the 8 kb long genomic region with IRES–GFP. These results further demonstrate that the TEd method is more efficient than conventional methods for deleting and replacing long fragments of genomic DNA.
Figure 5.
TEd-mediated long genomic deletion and replacement in HEK293T cells. (A) Schematic presentation of a 10 kb genome knock-out (KO) at the CDC42 locus by TEd4. Paired gRNAs spaced ∼10 kb (gRNA1 + gRNA2) apart were designed to target the CDC42 locus. Primers used to amplify the target genomic deletion are marked as (P5 + P6). (B) Representative PCR amplicons of the junction region using the primers indicated in (A). HEK293T cells served as a negative control. The representative amplicon of the targeted deletion is marked with an arrow. (C) Schematic presentation of an 8 kb genome KO at the CDC42 locus and simultaneous replacement with an I-SceI recognition site (18 bp) at the target site by TEd4. Paired gRNAs spaced ∼8 kb (gRNA3 + gRNA4) apart were designed as indicated to target the CDC42 locus. Primers used to amplify the target genomic regions are marked as (P7 + P8). (D) Representative PCRs in the junction region using the primers indicated in (C). HEK293T cells served as a negative control. The representative amplicon of the targeted deletion is marked with an arrow. (E) Amplicons from CEd and TEd were incubated with or without I-SceI enzyme and analyzed on an agarose gel. Digested products are marked as ‘cut’ while the original amplicon is marked as ‘uncut’. (F) Evaluation of accurate replacement of 18 bp I-SceI recognition sequence in CEd and TEd4 by TA cloning sequencing (CEd, 15/15; TEd, 17/17). Two PAM sequences of gRNAs are shown in red and green. The 18 bp intended insertion is underlined. (G) Schematic presentation of an 8 kb genome KO at the CDC42 locus and simultaneous replacement with IRES–GFP (∼1.2 kb) at the target site by TEd4. (H) Living cell fluorescence images of the GFP pan-cellular signal of CEd and TEd4 in (G) observed at 3 days after transfection (unsorted). Scale bar, 50 μm. (I) Efficiencies of IRES–GFP replacement in (G) were evaluated by FACS. (J) Representative PCR amplicons of the target region using the primers indicated in (G). The deletion amplicons are marked with two bands in CEd and TEd4. The lower bands are the expected KO sizes, the upper bands of CEd and TEd4 are marked with their expected sizes after IRES–GFP insertion (upper arrow). All data are presented with individual data points and mean ± SD for n = 3 independent biological replicates. P-values were obtained using the two-tailed Student's t-test. ***P< 0.001, ****P< 0.0001.
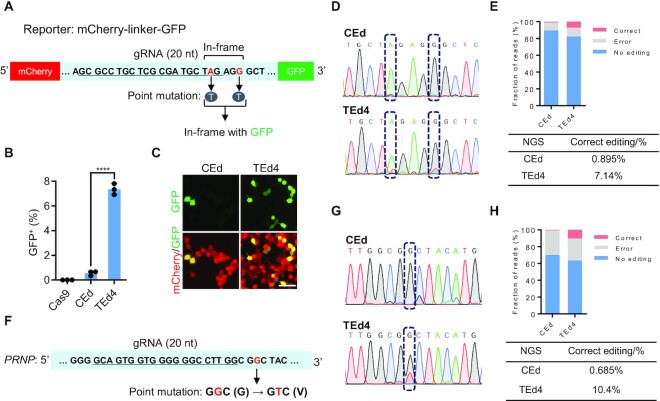
TEd efficiently and accurately introduces point mutations
To compare the efficiency of TEd with CEd for introducing point mutations, we introduced an mCherry–GFP reporter system by fusing GFP to mCherry with an in-frame linker containing a TAG stop codon (51) (Figure 6A). In this system, a CRISPR-introduced point mutation in the linker TAG stop codon allows GFP expression. As shown by FACS analysis, the maximal editing efficiency of TEd4 (7.8 ± 0.15%) was ∼15-fold higher than that of CEd (0.6 ± 0.00%) (Figure 6B; Supplementary Figure S11F). We also confirmed mCherry and GFP fluorescence in living cells (Figure 6C), and verified the editing accuracy by both Sanger sequencing and NGS of the TAG stop codon (Figure 6D, E). We also measured the efficiencies of introducing point mutations with TEd at disease-related sites. PrP misfolding can lead to progressive and fatal neurodegenerative prion diseases, which can occur spontaneously through genetic dominant mutations in PRNP (52). A naturally occurring mutant allele of PRNP (G127V) confers resistance to prion diseases in humans and mice. We used TEd4 and CEd to introduce the G127V allele (which requires a G·C to T·A transversion, GGC to GTC) in HEK293T cells (Figure 6F) and compared the efficiencies of generating the point mutation by Sanger sequencing and NGS. The efficiency of TEd4 was > 10 times greater than that of CEd (Figure 6G, H). Thus, TEd is more efficient than conventional methods for introducing point mutations.
Figure 6.
Targeted point mutations mediated by TEd in HEK293T cells. (A) Schematic presentation of point mutations in the mCherry–GFP reporter generated by fusing GFP to mCherry with an in-frame linker containing a TAG stop codon. The targeted TAG is mutated to TTG, followed by in-frame GFP, and AGG (PAM of gRNA) is mutated to AGT to avoid occasional cleavages in donor plasmids. (B) Graphs showing percentages of GFP+ cells by FACS analysis in (A). All data are shown as individual data points and mean ± SD for n = 3 independent biological replicates. P-values were obtained using the two-tailed Student's t-test. ****P< 0.0001. (C) Living cell fluorescence images of cells with point mutations introduced by CEd and TEd4. Scale bar, 50 μm. (D) Sanger sequencing analysis of point mutations at the linker region of the mCherry–GFP reporter showing the overlapping peaks of targeted single nucleotide editing (dashed boxes). (E) Percentages of accurate editing analyzed by NGS of PCR amplicons using primers outside the HAs. Correct (red) represents edits with the desired point mutations. Error (gray) represents NHEJ-derived indels at the target site. (F) Design of the point mutation at the PRNP locus. The gRNA sequence is underlined. The targeted GGC is expected to be mutated to GTC after G to V transversion. (G) Sanger sequencing analysis of PCR amplicons using primers completely outside the HAs at the PRNP locus by CEd and TEd4 showing overlapping peaks of single nucleotide editing (dashed boxes). (H) Percentages of accurate editing in (F) were analyzed by NGS of PCR amplicons. Correct (red) represents edits with the desired point mutations. Error (gray) represents NHEJ-derived indels at the target site.
DISCUSSION
Efficient KI of exogenous DNA is a highly desirable technology, especially for studies carried out in human cells (17,18,53–55). In this study, we found that transcription of the donor DNA significantly increased the efficiency of CRISPR/Cas9-mediated genome editing. Based on this key finding, we developed a simple gene-editing method, TEd, which is up to 10 times more efficient than the canonical Cas9-mediated HR-editing method.
Our study revealed two key principles about the effect of donor DNA transcription on the efficiency of CRISPR-mediated HR editing. First, transcription of the TC donor appears to play an important mechanistic role in HR-mediated genome editing. Indeed, previous reports have shown that HR preferentially occurs in transcriptionally active regions of the genome (56,57). We observed that transcribed donor DNAs (in cis) are more often used as gene-editing templates than non-transcribed donor DNA templates (in trans) (Figure 2B, C). Interestingly, TEd2/4 (in which the HR donor was added to TEd1/3) exhibited higher genome editing efficiency than TEd1/3, suggesting that the interactions between TC donor, HR donor and targeted genomic region may be complex in cells undergoing TEd2/4 editing. However, the underlying mechanisms for the higher editing efficiency of TEd2/4 remain unclear. Since TC RNAs only partially contributed to the increased editing efficiency of TEd (Figure 2B, C), we hypothesize that transcription unwinds the TC donor to facilitate invasion by the 3′ overhang (derived from the DSB) for the HR pathway. Second, tethering the TC RNA to the DSB significantly increases KI efficiency. Close positioning of TC RNAs and DSBs may help to form DR-loops (between template and target) to stimulate the HR (27). We showed that tethering of TC RNA to the DSB significantly increased the TEd4 editing efficiency when the TC RNA sequence matched the DNA donor sequence (Figures 1H and 2B). One possible explanation is that the Cas9–MCP fusion protein may help recruit TC RNA–donor complexes to the vicinity of DSB sites by facilitating Cas9–MCP-specific binding to TC RNA with the 4× MS2 aptamer, thereby increasing the local concentration of TC donor template around DSBs, which would potentially facilitate HR.
For the 10 kb genomic deletion, regular PCR amplification and sequencing cannot determine the relative contributions of NHEJ and HR to TEd and CEd, since the NHEJ pathway can also result in error-free end-to-end ligation. Therefore, we could only assess the difference in the efficiencies of TEd- and CEd-mediated 10 kb genome deletion based on the relative intensities of the junction amplicons. Similarly, for the 8 kb deletion/I-SceI replacement, the TEd method was more efficient for deletion than CEd based on amplicon intensity. Intriguingly, the proportion of correct I-SceI replacements was higher with TEd than with CEd, implying that the HR pathway accounts for a significant proportion of deletions/replacements in the TEd system, although NHEJ is the most commonly used DNA repair pathway in mammalian cells. However, the underlying mechanism deserves further study.
We propose a model to explain TEd action (Supplementary Figure S12). In this model, high TEd efficiency (compared with CEd efficiency) depends on which strand of the TC donor is transcribed because the CRISPR/Cas9 complex may occupy the DSB site in a particular orientation after generating the DSB (58–60). Transcription of the TC donor may not only open up the double-stranded DNA, but also provide the opportunity for the TC RNA to anneal to the non-target strand (existing as ssDNA), thereby facilitating the formation of the transient RNA–DNA hybrid around the DSB and making it accessible to the repair machinery (Supplementary Figure S12). Thereafter, the 3′ ssDNA of the DSB may invade the displaced HA ssDNA, produced by transcription of the TC donor. Tethering of TC donor and target DSBs by the MCP–MS2 system further increases the efficiency of the HR pathway. However, the molecular mechanisms via which the TC donor is transcribed and via which the TC RNA acts on the HR pathway require further investigation.
Overall, our results show that transcription of donor DNA substantially improves the efficiency of genome editing. Recent studies have shown that recombination-related genes can enhance the efficiency of HR. For example, upon complementation with BRCA1, HR donor integration increases in HCC1937 cells, and RAD52 overexpression significantly improves HR in Pichia pastoris (61,62). Therefore, complementation of the TEd system with these HR-related genes could further increase the efficiency of gene integration. Additional strategies, such as combining TEd with RNP–gRNA complex delivery, long HAs and other Cas enzymes, or ZFN or TALEN tools could further increase the efficiency and accuracy of TEd. We anticipate that the TEd method will have many biotechnological applications (63–68).
DATA AVAILABILITY
All data supporting the findings of this study are available in the article or in the supplementary data files, or are available from the corresponding author upon request. All next-generation sequencing data have been deposited in the NCBI Sequence Read Archive database under the accession code PRJNA798537. Customized scripts for data analysis have been deposited at Github and are available at https://github.com/josephwxy/CEd_TEd_analysis. Complete and annotated plasmid sequences used in the study have been deposited at Github and are available at https://github.com/KaixuanG/Plasmids-used-in-TEd-editing-method.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs Tian Chi, Hanhui Ma, Wensheng Wei, Jia Chen, Xingxu Huang and Yongqiang Wu for kindly providing reagents. We thank Dr Ji-long Liu for critical suggestions during manuscript preparation. We thank the Molecular Imaging Core Facility (MICF) and Molecular and Cell Biology Core Facility (MCBCF) at the School of Life Science and Technology, ShanghaiTech University for providing technical support.
Contributor Information
Kaixuan Gao, Gene Editing Center, School of Life Science and Technology, ShanghaiTech University, Shanghai 201210, China.
Xuedi Zhang, Gene Editing Center, School of Life Science and Technology, ShanghaiTech University, Shanghai 201210, China.
Zhenwu Zhang, Gene Editing Center, School of Life Science and Technology, ShanghaiTech University, Shanghai 201210, China.
Xiangyu Wu, Gene Editing Center, School of Life Science and Technology, ShanghaiTech University, Shanghai 201210, China.
Yan Guo, Gene Editing Center, School of Life Science and Technology, ShanghaiTech University, Shanghai 201210, China.
Pengchong Fu, Gene Editing Center, School of Life Science and Technology, ShanghaiTech University, Shanghai 201210, China.
Angyang Sun, Gene Editing Center, School of Life Science and Technology, ShanghaiTech University, Shanghai 201210, China.
Ju Peng, Gene Editing Center, School of Life Science and Technology, ShanghaiTech University, Shanghai 201210, China.
Jie Zheng, Gene Editing Center, School of Life Science and Technology, ShanghaiTech University, Shanghai 201210, China.
Pengfei Yu, Gene Editing Center, School of Life Science and Technology, ShanghaiTech University, Shanghai 201210, China.
Tengfei Wang, Gene Editing Center, School of Life Science and Technology, ShanghaiTech University, Shanghai 201210, China.
Qinying Ye, Gene Editing Center, School of Life Science and Technology, ShanghaiTech University, Shanghai 201210, China.
Jingwei Jiang, Gene Editing Center, School of Life Science and Technology, ShanghaiTech University, Shanghai 201210, China.
Haopeng Wang, Gene Editing Center, School of Life Science and Technology, ShanghaiTech University, Shanghai 201210, China.
Chao-Po Lin, Gene Editing Center, School of Life Science and Technology, ShanghaiTech University, Shanghai 201210, China.
Guanjun Gao, Gene Editing Center, School of Life Science and Technology, ShanghaiTech University, Shanghai 201210, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by the National Natural Science Foundation of China [32070846 and 91740107 to G.G.]; National Postdoctoral Fellowship [2019M661652 to Z.X.]; and Science, and Technology Commission of Shanghai Municipality [19JC1413600 to G.G.].
Conflict of interest. The authors have submitted a patent application based on the results reported in this paper.
REFERENCES
- 1. Chu V.T., Weber T., Wefers B., Wurst W., Sander S., Rajewsky K., Kuhn R.. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 2015; 33:543–548. [DOI] [PubMed] [Google Scholar]
- 2. Mao Z.Y., Bozzella M., Seluanov A., Gorbunova V.. Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair (Amst.). 2008; 7:1765–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jasin M., Haber J.E.. The democratization of gene editing: insights from site-specific cleavage and double-strand break repair. DNA Repair (Amst.). 2016; 44:6–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ceccaldi R., Rondinelli B., D’Andrea A.D. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016; 26:52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. He X.J., Tan C.L., Wang F., Wang Y.F., Zhou R., Cui D.X., You W.X., Zhao H., Ren J.W., Feng B.. Knock-in of large reporter genes in human cells via CRISPR/Cas9-induced homology-dependent and independent DNA repair. Nucleic Acids Res. 2016; 44:e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Paix A., Folkmann A., Goldman D.H., Kulaga H., Grzelak M.J., Rasoloson D., Paidemarry S., Green R., Reed R.R., Seydoux G.. Precision genome editing using synthesis-dependent repair of Cas9-induced DNA breaks. Proc. Natl Acad. Sci. USA. 2017; 114:E10745–E10754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Doudna J.A., Charpentier E.. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014; 346:1077. [DOI] [PubMed] [Google Scholar]
- 8. Daley J.M., Niu H.Y., Miller A.S., Sung P.. Biochemical mechanism of DSB end resection and its regulation. DNA Repair (Amst.). 2015; 32:66–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Renkawitz J., Lademann C.A., Jentsch S.. DNA damage mechanisms and principles of homology search during recombination. Nat. Rev. Mol. Cell. Bio. 2014; 15:369–383. [DOI] [PubMed] [Google Scholar]
- 10. Hsu P.D., Lander E.S., Zhang F.. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014; 157:1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ramalingam S., Annaluru N., Chandrasegaran S.. A CRISPR way to engineer the human genome. Genome Biol. 2013; 14:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chapman J.R., Taylor M.R.G., Boulton S.J.. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell. 2012; 47:497–510. [DOI] [PubMed] [Google Scholar]
- 13. Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F.. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013; 8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim H., Kim J.S.. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 2014; 15:321–334. [DOI] [PubMed] [Google Scholar]
- 15. Anzalone A.V., Randolph P.B., Davis J.R., Sousa A.A., Koblan L.W., Levy J.M., Chen P.J., Wilson C., Newby G.A., Raguram A.et al.. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019; 576:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lin Q.P., Zong Y., Xue C.X., Wang S.X., Jin S., Zhu Z.X., Wang Y.P., Anzalone A.V., Raguram A., Doman J.L.et al.. Prime genome editing in rice and wheat. Nat. Biotechnol. 2020; 38:582. [DOI] [PubMed] [Google Scholar]
- 17. Pickar-Oliver A., Gersbach C.A.. The next generation of CRISPR-Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 2019; 20:490–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang D., Zhang F., Gao G.. CRISPR-based therapeutic genome editing: strategies and in vivo delivery by AAV vectors. Cell. 2020; 181:136–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Danner E., Bashir S., Yumlu S., Wurst W., Wefers B., Kühn R.. Control of gene editing by manipulation of DNA repair mechanisms. Mamm. Genome. 2017; 28:262–274. [DOI] [PubMed] [Google Scholar]
- 20. Merkle F.T., Neuhausser W.M., Santos D., Valen E., Gagnon J.A., Maas K., Sandoe J., Schier A.F., Eggan K.. Efficient CRISPR-Cas9-mediated generation of knockin human pluripotent stem cells lacking undesired mutations at the targeted locus. Cell Rep. 2015; 11:875–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pawelczak K.S., Gavande N.S., VanderVere-Carozza P.S., Turchi J.J.. Modulating DNA repair pathways to improve precision genome engineering. ACS Chem. Biol. 2018; 13:389–396. [DOI] [PubMed] [Google Scholar]
- 22. Paquet D., Kwart D., Chen A., Sproul A., Jacob S., Teo S., Olsen K.M., Gregg A., Noggle S., Tessier-Lavigne M.. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. 2016; 533:125. [DOI] [PubMed] [Google Scholar]
- 23. Richardson C.D., Ray G.J., DeWitt M.A., Curie G.L., Corn J.E.. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 2016; 34:339. [DOI] [PubMed] [Google Scholar]
- 24. Liu M.J., Rehman S., Tang X.D., Gu K., Fan Q.L., Chen D.K., Ma W.T.. Methodologies for improving HDR efficiency. Front Genet. 2019; 9:691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nakade S., Tsubota T., Sakane Y., Kume S., Sakamoto N., Obara M., Daimon T., Sezutsu H., Yamamoto T., Sakuma T.et al.. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat. Commun. 2014; 5:5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Humbert O., Maizels N.. Epigenetic modification of the repair donor regulates targeted gene correction. Mol. Ther. Nucleic Acids. 2012; 1:e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ouyang J., Yadav T., Zhang J.M., Yang H.B., Rheinbay E., Guo H.S., Haber D.A., Lan L., Zou L.. RNA transcripts stimulate homologous recombination by forming DR-loops. Nature. 2021; 594:283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Keskin H., Shen Y., Huang F., Patel M., Yang T., Ashley K., Mazin A.V., Storici F.. Transcript-RNA-templated DNA recombination and repair. Nature. 2014; 515:436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cohen S., Puget N., Lin Y.L., Clouaire T., Aguirrebengoa M., Rocher V., Pasero P., Canitrot Y., Legube G.. Senataxin resolves RNA:DNA hybrids forming at DNA double-strand breaks to prevent translocations. Nat. Commun. 2018; 9:533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yasuhara T., Kato R., Hagiwara Y., Shiotani B., Yamauchi M., Nakada S., Shibata A., Miyagawa K.. Human rad52 promotes XPG-mediated R-loop processing to initiate transcription-associated homologous recombination repair. Cell. 2018; 175:558–570. [DOI] [PubMed] [Google Scholar]
- 31. Teng Y., Yadav T., Duan M., Tan J., Xiang Y., Gao B., Xu J., Liang Z., Liu Y., Nakajima S.et al.. ROS-induced r loops trigger a transcription-coupled but BRCA1/2-independent homologous recombination pathway through CSB. Nat. Commun. 2018; 9:4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ouyang J., Lan L., Zou L.. Regulation of DNA break repair by transcription and RNA. Sci. China Life Sci. 2017; 60:1081–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ohle C., Tesorero R., Schermann G., Dobrev N., Sinning I., Fischer T.. Transient RNA–DNA hybrids are required for efficient double-strand break repair. Cell. 2016; 167:1001–1013. [DOI] [PubMed] [Google Scholar]
- 34. Lu W.T., Hawley B.R., Skalka G.L., Baldock R.A., Smith E.M., Bader A.S., Malewicz M., Watts F.Z., Wilczynska A., Bushell M.. Drosha drives the formation of DNA:RNA hybrids around DNA break sites to facilitate DNA repair. Nat. Commun. 2018; 9:532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu S.J., Hua Y., Wang J.N., Li L.Y., Yuan J.J., Zhang B., Wang Z.Y., Ji J.G., Kong D.C.. RNA polymerase III is required for the repair of DNA double-strand breaks by homologous recombination. Cell. 2021; 184:1314. [DOI] [PubMed] [Google Scholar]
- 36. Clement K., Rees H., Canver M.C., Gehrke J.M., Farouni R., Hsu J.Y., Cole M.A., Liu D.R., Joung J.K., Bauer D.E.et al.. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 2019; 37:224–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.Journal. 2011; 17:10–12. [Google Scholar]
- 38. Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J.. Basic local alignment search tool. J. Mol. Biol. 1990; 215:403–410. [DOI] [PubMed] [Google Scholar]
- 39. Cleal K., Baird D.M.. Dysgu: efficient structural variant calling using short or long reads. Nucleic Acids Res. 2022; 50:e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sedlazeck F.J., Rescheneder P., Smolka M., Fang H., Nattestad M., von Haeseler A., Schatz M.C.. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods. 2018; 15:461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nguyen N.P.D., Deshpande V., Luebeck J., Mischel P.S., Bafna V.. ViFi: accurate detection of viral integration and mRNA fusion reveals indiscriminate and unregulated transcription in proximal genomic regions in cervical cancer. Nucleic Acids Res. 2018; 46:3309–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhu L.J., Gazin C., Lawson N.D., Pages H., Lin S.M., Lapointe D.S., Green M.R.. ChIPpeakAnno: a bioconductor package to annotate chip-seq and chip-chip data. BMC Bioinformatics. 2010; 11:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xie K.B., Minkenberg B., Yang Y.N.. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl Acad Sci. USA. 2015; 112:3570–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Welty S., Teng Y.Q., Liang Z.B., Zhao W.X., Sanders L.H., Greenamyre J.T., Rubio M.E., Thathiah A., Kodali R., Wetzel R.et al.. RAD52 is required for RNA-templated recombination repair in post-mitotic neurons. J. Biol. Chem. 2018; 293:1353–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Coller J., Wickens M.. Tethered function assays: an adaptable approach to study RNA regulatory proteins. Methods Enzymol. 2007; 429:299–321. [DOI] [PubMed] [Google Scholar]
- 46. Byrne S.M., Ortiz L., Mali P., Aach J., Church G.M.. Multi-kilobase homozygous targeted gene replacement in human induced pluripotent stem cells. Nucleic Acids Res. 2015; 43:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yang H., Wang H.Y., Shivalila C.S., Cheng A.W., Shi L.Y., Jaenisch R.. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013; 154:1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Moehle E.A., Rock J.M., Lee Y.L., Jouvenot Y., DeKelver R.C., Gregory P.D., Urnov F.D., Holmes M.C.. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc. Natl Acad. Sci. USA. 2007; 104:6090–6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hockemeyer D., Soldner F., Beard C., Gao Q., Mitalipova M., DeKelver R.C., Katibah G.E., Amora R., Boydston E.A., Zeitler B.et al.. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 2009; 27:851–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Urnov F.D., Miller J.C., Lee Y.L., Beausejour C.M., Rock J.M., Augustus S., Jamieson A.C., Porteus M.H., Gregory P.D., Holmes M.C.. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005; 435:646–651. [DOI] [PubMed] [Google Scholar]
- 51. Qu L., Yi Z.Y., Zhu S.Y., Wang C.H., Cao Z.Z., Zhou Z., Yuan P.F., Yu Y., Tian F., Liu Z.H.et al.. Programmable RNA editing by recruiting endogenous ADAR using engineered RNAs. Nat. Biotechnol. 2019; 37:1380–1380. [DOI] [PubMed] [Google Scholar]
- 52. Collinge J. Prion diseases of humans and animals: their causes and molecular basis. Annu. Rev. Neurosci. 2001; 24:519–550. [DOI] [PubMed] [Google Scholar]
- 53. Leonetti M.D., Sekine S., Kamiyama D., Weissman J.S., Huang B.. A scalable strategy for high-throughput GFP tagging of endogenous human proteins. Proc. Natl Acad. Sci. USA. 2016; 113:E3501–E3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Diao Y.R., Fang R.X., Li B., Meng Z.P., Yu J.T., Qiu Y.J., Lin K.C., Huang H., Liu T., Marina R.J.et al.. A tiling-deletion-based genetic screen for cis-regulatory element identification in mammalian cells. Nat. Methods. 2017; 14:629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mollanoori H., Teimourian S.. Therapeutic applications of CRISPR/Cas9 system in gene therapy. Biotechnol. Lett. 2018; 40:907–914. [DOI] [PubMed] [Google Scholar]
- 56. Aymard F., Bugler B., Schmidt C.K., Guillou E., Caron P., Briois S., Iacovoni J.S., Daburon V., Miller K.M., Jackson S.P.et al.. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat. Struct. Mol. Biol. 2014; 21:366–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wei L., Nakajima S., Böhm S., Bernstein K.A., Shen Z., Tsang M., Levine A.S., Lan L.. DNA damage during the G0/G1 phase triggers RNA-templated, Cockayne syndrome B-dependent homologous recombination. Proc. Natl Acad. Sci. USA. 2015; 112:E3495–E3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nishimasu H., Ran F.A., Hsu P.D., Konermann S., Shehata S.I., Dohmae N., Ishitani R., Zhang F., Nureki O.. Crystal structure of cas9 in complex with guide RNA and target DNA. Cell. 2014; 156:935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhu X., Clarke R., Puppala A.K., Chittori S., Merk A., Merrill B.J., Simonovic M., Subramaniam S.. Cryo-EM structures reveal coordinated domain motions that govern DNA cleavage by cas9. Nat. Struct. Mol. Biol. 2019; 26:679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sternberg S.H., Redding S., Jinek M., Greene E.C., Doudna J.A.. DNA interrogation by the CRISPR RNA-guided endonuclease cas9. Nature. 2014; 507:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cai P., Duan X., Wu X., Gao L., Ye M., Zhou Y.J.. Recombination machinery engineering facilitates metabolic engineering of the industrial yeast Pichia pastoris. Nucleic Acids Res. 2021; 49:7791–7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hussain S.S., Majumdar R., Moore G.M., Narang H., Buechelmaier E.S., Bazil M.J., Ravindran P.T., Leeman J.E., Li Y., Jalan M.et al.. Measuring nonhomologous end-joining, homologous recombination and alternative end-joining simultaneously at an endogenous locus in any transfectable human cell. Nucleic Acids Res. 2021; 49:e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Laboulaye M.A., Duan X., Qiao M., Whitney I.E., Sanes J.R.. Mapping transgene insertion sites reveals complex interactions between mouse transgenes and neighboring endogenous genes. Front. Mol. Neurosci. 2018; 11:385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Weis J., Fine S.M., Sanes J.R.. Integration site-dependent transgene expression used to mark subpopulations of cells in vivo: an example from the neuromuscular junction. Brain Pathol. 1992; 2:31–37. [PubMed] [Google Scholar]
- 65. Goodwin L.O., Splinter E., Davis T.L., Urban R., He H., Braun R.E., Chesler E.J., Kumar V., van Min M., Ndukum J.et al.. Large-scale discovery of mouse transgenic integration sites reveals frequent structural variation and insertional mutagenesis. Genome Res. 2019; 29:494–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhang Z., Nam Y.J.. Generation of MLC-2v-tdTomato knock-in reporter mouse line. Genesis. 2018; 56:e23256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Park A., Won S.T., Pentecost M., Bartkowski W., Lee B.. CRISPR/Cas9 allows efficient and complete knock-in of a destabilization domain-tagged essential protein in a human cell line, allowing rapid knockdown of protein function. PLoS One. 2014; 9:e95101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Menalled L.B., Kudwa A.E., Miller S., Fitzpatrick J., Watson-Johnson J., Keating N., Ruiz M., Mushlin R., Alosio W., McConnell K.et al.. Comprehensive behavioral and molecular characterization of a new knock-in mouse model of huntington's disease: zQ175. PLoS One. 2012; 7:e49838. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available in the article or in the supplementary data files, or are available from the corresponding author upon request. All next-generation sequencing data have been deposited in the NCBI Sequence Read Archive database under the accession code PRJNA798537. Customized scripts for data analysis have been deposited at Github and are available at https://github.com/josephwxy/CEd_TEd_analysis. Complete and annotated plasmid sequences used in the study have been deposited at Github and are available at https://github.com/KaixuanG/Plasmids-used-in-TEd-editing-method.