Abstract
HflX is a ubiquitous bacterial GTPase that splits and recycles stressed ribosomes. In addition to HflX, Listeria monocytogenes contains a second HflX homolog, HflXr. Unlike HflX, HflXr confers resistance to macrolide and lincosamide antibiotics by an experimentally unexplored mechanism. Here, we have determined cryo-EM structures of L. monocytogenes HflXr-50S and HflX-50S complexes as well as L. monocytogenes 70S ribosomes in the presence and absence of the lincosamide lincomycin. While the overall geometry of HflXr on the 50S subunit is similar to that of HflX, a loop within the N-terminal domain of HflXr, which is two amino acids longer than in HflX, reaches deeper into the peptidyltransferase center. Moreover, unlike HflX, the binding of HflXr induces conformational changes within adjacent rRNA nucleotides that would be incompatible with drug binding. These findings suggest that HflXr confers resistance using an allosteric ribosome protection mechanism, rather than by simply splitting and recycling antibiotic-stalled ribosomes.
INTRODUCTION
Bacteria have evolved diverse mechanisms by which they can obtain resistance to antibiotics (1,2), which is making our current arsenal of antimicrobial agents obsolete (3). One such mechanism is target protection, where a resistance protein physically interacts with an antibiotic target to rescue it from the antibiotic-mediated inhibition (4). The best characterized target protection mechanisms operate against ribosome-targeting antibiotics and are mediated by so-called ribosome protection proteins (RPPs). Type I RPPs confer resistance by binding to the target at an overlapping site as the drug and thereby inducing its dissociation, as exemplified by the tetracycline RPPs, such as TetO and TetM (4–7). Type II RPPs confer resistance by binding to the target at a distinct site from the drug and promote drug dissociation by inducing allosteric changes within the drug binding site. This latter mechanism is thought to be employed by the large family of antibiotic-resistance (ARE) ATP-binding cassette type F (ABCF) proteins that confer resistance to a range of antibiotics that bind at or near the peptidyl-transferase center (PTC) of the large ribosomal subunit (4,8–13). By contrast, type III RPPs induce conformational changes within the target that restore functionality despite the presence of the bound antibiotic, as illustrated by the fusidic acid resistance proteins such as FusB or FusC (4,14,15).
Recently, a novel resistance protein, HflXr (encoded by the gene lmo0762), has been identified in Listeria monocytogenes and shown to confer resistance to lincosamides, such as lincomycin, as well as macrolides, such as erythromycin—but not to other ribosome-targeting antibiotics, such as chloramphenicol or tetracycline (16). HflXr shares 37% amino acid identity with L. monocytogenes housekeeping HflX protein (encoded by the gene lmo1296) that does not confer antibiotic resistance (16). The effect of HflXr loss on L. monocytogenes antibiotic susceptibility was only observable when the ARE-ABCF VgaL (encoded by the gene lmo0919)—which has a partially-overlapping spectrum of action—was also inactivated (16), implying some redundancy between these two proteins. Unrelated to the ATPase VgaL (11), HflXr is a homolog of the widely-distributed bacterial GTPase HflX, encoded by lmo1296 in L. monocytogenes (16), which is proposed to split and recycle 70S ribosomes that have become stalled under stress conditions (17–20). In Staphylococcus aureus, HflX has also been shown to disassemble hibernating 100S ribosomes (21). Deletion of the gene encoding HflXr also leads to accumulation of 70S ribosomes, suggesting that HflXr may confer resistance by splitting and recycling of antibiotic-stalled ribosomes (16). While structures of Escherichia coli HflX on the 50S subunit have been reported (17,19), as well as more recently for the human mitochondrial HflX homolog GTPBP6 bound to large subunit biogenesis intermediates (22), direct interaction of HflXr with ribosomal particles has not been demonstrated. Since the second HflX homologue in L. monocytogenes—the housekeeping HflX (Lmo1296)—does not appear to confer any notable antibiotic resistance, just like the single E. coli HflX (16), it remains unclear whether splitting and recycling of antibiotic-stalled ribosomes is necessary and sufficient to confer antibiotic resistance, or whether HflXr has an additional target protection role to facilitate drug release from the large 50S subunit. In the structure of the E. coli HflX-GDPNP-50S complex, the loop connecting two helices of subdomain II within the N-terminal domain of HflX is positioned at the PTC in close proximity to the lincomycin binding site (17). Since the L. monocytogenes HflXr loop is two residues longer than E. coli and L. monocytogenes HflX and differs in sequence (Figure 1A), this offers the possibility that a distinct conformation adopted by HflXr could reach deeper into the PTC and overlap with the drug-binding sites (4). Collectively, these observations raise the question as to whether the HflX and HflXr proteins with longer loops confer antibiotic resistance using type I or type II target protection mechanisms (4).
Figure 1.
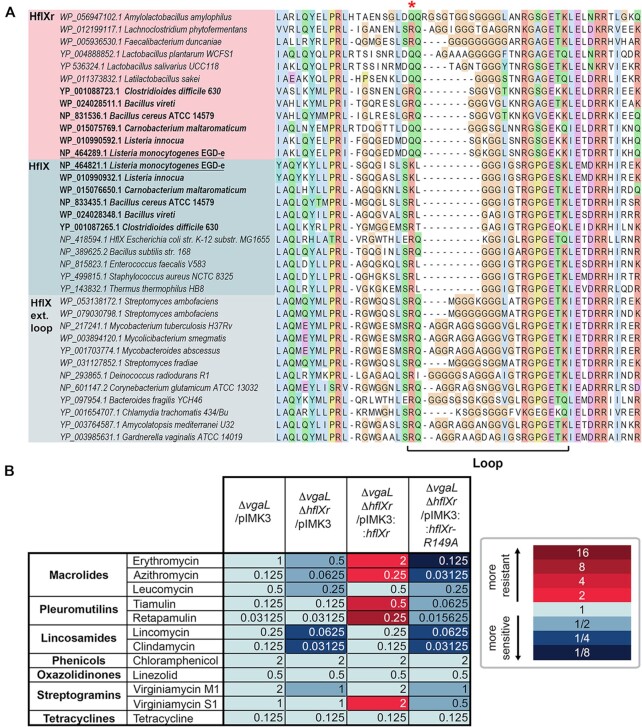
Sequence alignments of HflX and HflXr proteins and MIC data. (A) Sequence alignment of the resistance-associated loop region within the N-terminal domain of selected HflX (blue) and HflXr (pink) representatives, showing independently evolved insertions in HflXr and HflX. Taxa in bold are those with both HflX and HflXr. Conserved R/Q residue (R149 in L. monocytogenes HflXr) is marked with a red asterisk. The full alignment is found in Supplementary Data S1. (B) Minimum inhibitory concentrations (MICs) of ribosome-targeting antibiotics against L. monocytogenes EGDe strains lacking or expressing HflXr or/and VgaL/Lmo0919 ARE-ABCF. The color code is made with respect to the first column that contains the ΔvgaL/pIMK3 MICs.
Here we systematically characterize the resistance spectrum of L. monocytogenes HflXr, demonstrating in agreement with previous reports, that HflXr confers resistance to lincomycin and erythromycin, but not chloramphenicol and tetracycline (16). Further defining the antibiotic classes affected by HflXr, we also observe resistance to the pleuromutilins tiamulin and retapamulin, and the streptogramins virginiamycin M1 and S1, but not the oxazolidinone linezolid. We report a cryo-electron microscopy (cryo-EM) structure of L. monocytogenes HflXr-50S-GDPNP complex ranging in resolution from 2.3 Å for the ribosomal part and 3.0–5.0 Å for HflXr. The structure reveals that while HflXr binds to the 50S subunit analogously to HflX, the N-terminal domain II loop of HflXr penetrates deeper into the peptidyltransferase center (PTC) than that of HflX, and thereby comes into close proximity of the binding sites of the relevant antibiotics. Additionally, we have determined cryo-EM structure of the L. monocytogenes HflX-50S complex, as well as 70S ribosome in the presence and absence of lincomycin at 2.1–3.1 Å, revealing that binding of HflX, or lincomycin, to the 50S subunit or 70S ribosome, respectively, do not induce any conformational changes within the PTC. Comparison of these structures with the L. monocytogenes HflXr-50S-GDPNP complex reveals that in contrast to HflX, HflXr induces conformational changes within the PTC that would be incompatible with drug binding. Our findings lead us to hypothesize that HflXr may confer antibiotic resistance using a type II ribosome protection protein mechanism, analogous to the ARE-ABCF proteins, such as L. monocytogenes VgaL.
MATERIALS AND METHODS
Strains and plasmids
All bacterial strains and plasmids used in the study are listed in Supplementary Table S1.
Gene lmo0762 encoding HflXr was amplified from EGDe genomic DNA using primers VHKT40 (5′-GGGGCCATGGAGAAAAAAGTATTAATCGTTGG) and VHKT41 (5′-GGGGCTGCAGTTACTCAGCTACTTGATATTCTGCT), and cloned into pIMK3 using NcoI and PstI restriction sites, resulting in plasmid VHp838. Variants of lmo0762 were constructed via two step overlap PCR (23). Mutation R149A was introduced to hflXr using primer pairs VHKT236 (5′-GGCGGCCTTAGCAACGCTGGTTCAGGTGAAAA), VHKT41 and VHKT40, VHKT237 (5′-TTTTCACCTGAACCAGCGTTGCTAAGGCCGCC). These overlapping PCR products were then zipped together in a third PCR using primers VHKT41 and VHKT40. The resulting PCR product was cloned into pIMK3 using NcoI and PstI sites resulting in VHp840. The loop deletion ΔS147-E148 were synthesised using primer pairs VHKT238 (5′-GGAAAAGGCGGCCTT_CGTGGTTCAGGTGAA), VHKT41 and VHKT40, VHKT239 (5′-TTCACCTGAACCACG_AAGGCCGCCTTTTCC). These overlapping PCR products were then zipped together in a third PCR using primers VHKT41 and VHKT40. The resulting PCR product was cloned into pIMK3 using NcoI and PstI sites resulting in VHp841.
To construct a vector for the overexpression of HflXr-HTF to enable purification of 50S-HflXr complexes via immunoprecipitation, the lmo0762 locus and HTF tag (HTF stands for His6-TEV-FLAG3) were amplified with primer pairs VHKT40, VHKT165 (5′-ATGATGATGGCCGCCCTCAGCTACTTGATATTCTG) and VHKT166 (5′-TATCAAGTAGCTGAGGGCGGCCATCATCATCATC), VHKT13 (5′-GGGGCTGCAGTTAGCCTTTGTCATCGTC) using EGDe genomic DNA and VHp100 (pCIE::lsa-HTF) plasmid for expression of HTF-tagged LsaA ARE-ABCF (11) as template DNA, respectively, producing fragments with overlapping ends. VHKT40 and VHKT13 were then used to fuse the fragments and the resulting PCR product was cloned into pIMK3 using NcoI and PstI sites resulting in VHp839.
For the overexpression of HflX-HTF to enable purification of 50S-HflX complexes via immunoprecipitation, the pIMK3::hflX-HTF plasmid (VHp1068) was constructed by the Protein Expertise Platform (PEP) facility at Umeå University. Using the same cloning strategy as for hflXr, the lmo1296 locus and HTF tag were amplified from EGDe genomic DNA and VHp100 (pCIE::lsa-HTF) plasmid for expression of HTF-tagged LsaA ARE-ABCF (11) as template DNA, respectively. The resulting PCR product was cloned into pIMK3 using NcoI and BamHI sites.
To construct the Δlmo0762 strain VHB680 lacking HflXr, flanking regions of lmo0762 gene were amplified using primers Dlmo0257-Af_BamHI (5′-AATTGGATCCgccaaattgaaatggtggacg) and Dlmo0257-Br_KpnI (5′-AATTGGTACCtgttagcatttctattcctcattttctg), as well as Dlmo0257-Cf_KpnI (5′-AATTGGTACCgcgtgatcaaaaagaagcgg) and Dlmo0257-Dr_NcoI (5′-AATTCCATGGcgtagcccttcatctaagaacc). The PCR products were cloned into BamHI and NcoI sites of pMAD vector and deletion strain (KVA1496) in L. monocytogenes EGDe was obtained as described by Arnaud et al. (23). To construct the double deletion strain VHB685 lacking both VgaL and HflXr (VHB685; EGDe Δlmo0762 Δlmo0919), lmo0919 was deleted from the L. monocytogenes strain VHB680 (EGDe Δlmo0762) lacking HflXr. Vector VHp689 (pMAD::Δlmo0919) (11) was used as per Arnaud et. al. (23) for gene deletion. Resulting colonies were then verified via colony PCR using primers VHKT42 (5′-TTCCCACAATGTAAGCCGTGTATTTCAAAC) and VHKT43 (5′-AGCGAGCCAACAATGACTGC). To construct HflXr complementation strains used for MIC antibiotic susceptibility testing, the pIMK3 integrative plasmid and its derivatives harboring IPTG inducible hflXr or hflXr mutants were transformed into L. monocytogenes via conjugation with E. coli S17.1 as described previously (11). Upon transformation pIMK3 and its derivatives integrate into the tRNAArg locus of the L. monocytogenes genome.
Antibiotic susceptibility testing
Minimum inhibitory concentrations (MIC) of antibiotics were determined by microbroth dilution in 96-well plate format, based on guidelines provided by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (http://www.eucast.org/ast_of_bacteria/mic_determination). L. monocytogenes cells harboring chromosomally integrated pIMK3:hflXr variants, or the control empty vector, were grown in BHI media supplemented with 100 μg/ml kanamycin for plasmid maintenance and 0.5 mM IPTG to induce expression of hflXr and its variants, as well as increasing concentrations of tested antibiotics. The media was inoculated with 5 × 105 CFU/ml (OD600 of approximately 0.0005) of L. monocytogenes strains indicated. After 16–20 h at 37°C without shaking, the presence or absence of bacterial growth was scored by eye.
Preparation of bacterial biomass
For the preparation of immunoprecipitated HflXr-50S and HflX-50S samples: L. monocytogenes strains (i) EGDe ΔhflXr, hflXr-HTF (HTF stands for His6-TEV-FLAG3), harbouring the chromosomal pIMK3 expression construct (either empty vector or expressing wild-type HflXr-HTF) or (ii) EGDe, hflX-HTF harbouring the chromosomal pIMK3 expression construct expressing HflX-HTF, were grown overnight from a single colony in BHI media supplemented with 100 μg/ml kanamycin. Overnight cultures were then used to inoculate 200 ml of BHI media supplemented with 10 μg/ml kanamycin to an initial OD600 of 0.05. Cultures were grown at 37°C shaking at 160 rpm until reaching OD600 0.6 and then induced by 0.5 mM IPTG for an additional 1 h. Cells were harvested by centrifugation (5,000 × g, 15 min, 4°C) and flash frozen in liquid nitrogen.
For the purification of L. monocytogenes 70S ribosomes and 50S subunits: wild-type L. monocytogenes strain EGDe was grown overnight in BHI from a single colony and then used to inoculate 2.5 l of fresh BHI. Cells were grown at 37°C shaking at 160 rpm until OD600 0.5 and harvested by centrifugation (5,000 × g, 15 min, 4°C), yielding a 5 g pellet, which was subsequently flash-frozen in liquid nitrogen.
Preparation of clarified lysates
For the preparation of immunoprecipitated HflXr-50S or HflX-50S samples: Cell pellets were thawed on ice and resuspended in 0.8 ml of 1× HEPES:Polymix buffer (20 mM HEPES:KOH pH 7.5, 5 mM Mg(OAc)2, 95 mM KCl, 15 mM NH4Cl, 0.5 mM CaCl2, 8 mM putrescine, 1 mM spermidine, 0.5 mM EDTA) (24) supplemented with 1 mM DTT, 0.5 mM GTP or GDPNP where indicated as well as 1 tablet of EDTA-free protease inhibitor (cOmplete™ by Roche) per 30 ml of buffer. Cells were lysed by shaking with 200 mg of 0.1 mm zirconium beads using FastPrep cell disrupter (MP Biomedicals) (4 × 20s; 4.5 M/S; cooled on ice for 2 min in between cycles). Cell debris was removed by centrifugation (18,000 × g for 20 min).
For the purification of L. monocytogenes 70S ribosomes: Cell pellets were thawed on ice and resuspended in 50 ml of lysis buffer (20 mM Tris–HCl pH 7.5, 100 mM NH4Cl, 15 mM Mg(OAc)2, 0.5 mM EDTA, 3 mM β-mercaptoethanol) and lysed using Stansted Fluid Power SFPH-10 Stansted Pressure Cell/Homogenizer. Cell debris was removed by centrifugation (18,000 × g for 20 min).
Affinity purification on anti-FLAG M2 affinity gel
50 μl of well-mixed anti-FLAG M2 Affinity Gel aliquots (Sigma) were loaded onto gravity flow columns (Micro Bio-Spin Columns, Bio-Rad) and washed twice with 1 ml of cell opening buffer by gravity flow. All subsequent incubations, wash and elution was performed at 4°C. Cleared lysates were incubated with the pre-washed anti-FLAG resin for 2 h at 4°C with end-over-end mixing. Resin was washed 6 times, with 0.5 ml of 1× HEPES:Polymix cell opening buffer supplemented with the non-ionic detergent Nikkol (0.05%). To elute, resin was incubated with 100 μl of 0.2 mg/ml FLAG peptide solution prepared in cell opening buffer (+0.05% Nikkol) on the column with end over end mixing for 20 min at 4°C. Flow through was collected by centrifugation at 2,000 × g for two min. The resulting pulldown samples were used to prepare cryo-EM grids. 10 μl of both the final wash and elution were retained for resolution on a 12% SDS-PAGE gel, before staining with ‘Blue-Silver’ Coomassie Staining analysis before imaging with LAS4000 (GE Healthcare). L. monocytogenes EGDe 50S ribosomal subunits, purified in small scale, as described previously (25), were used as a loading control and to visually assess sample concentration.
Purification of L. monocytogenes 70S ribosomes
Purification of ribosomes was carried out as previously described (25) with minor modifications. Briefly, cleared lysate was applied to a 37.6% sucrose cushion equilibrated in lysis buffer and ribosomes were pelleted via centrifugation. This was followed by separation on 5–25% sucrose gradients, equilibrated in HEPES:Polymix buffer (24) supplemented with 5 mM Mg(OAc)2 and fractionated on the Gradient Station ip instrument (BioComp Instruments) to isolate 70S. Fractions containing 70S were pooled and applied to a 25% sucrose cushion, and pellets were resuspended in HEPES:Polymix buffer supplemented with 5 mM Mg(OAc)2. The 50S fractions were kept for future use as loading control on SDS-PAGE gels. Resulting samples were flash-frozen and stored at –80°C in single-use aliquots.
Preparation of cryo-EM grids
Elutions from HflX and HflXr pull-downs were kept on ice and applied to grids within 2 h of preparation. 3.5 μl of sample was loaded onto glow discharged cryo-grids (Quantifoil 2/2 Cu300 coated with 2 nm continuous carbon) using the Vitrobot (FEI) under conditions of 100% humidity at 4°C, blotted for 5 s and vitrified by plunge-freezing in liquid ethane. HflXr samples were imaged on a Titan Krios (FEI) operated at 300 kV at a nominal magnification of ×165,000 (0.86 Å/pixel) with a Gatan K2 Summit camera. HflXr-50S samples were imaged at an exposure rate of 5.85 electrons/pixel/s with a 4 s of exposure and 20 frames using the EPU software. HflX-50S samples were imaged at the exposure rate of 6.899 electrons/pixel/s with a 3.2 s of exposure.
Purified 70S L. monocytogenes ribosomes at a concentration of 0.4 μM were incubated with 100 μM lincomycin on ice before 4 μl of sample was loaded onto glow discharged cryo-grids (Quantifoil 2/2 Cu300 coated with 2 nm continuous carbon) using the Vitrobot (FEI) as described above. Data was collected on a Titan Krios (FEI) operated at 300 kV at a nominal magnification of ×270,000 (0.502 Å/pixel) with a Gatan K2 Summit camera at an exposure rate of 3.38 electrons/pixel/s with a 3 s of exposure and 20 frames using the EPU software.
Single-particle reconstruction of LmoHflXr-50S complex
Processing was performed in RELION 3.1 unless otherwise specified (26). RELION’s implementation of MotionCor2 with 5 × 5 patches and CTFFIND4 (using power spectra) were used for motion correction and initial CTF estimation (27,28). 402,944 particles were picked from 4255 micrographs with crYOLO using the general model (29). After 2D classification, all ribosome-like classes were selected, particles extracted with a 3 × reduced pixel size (2.46 Å/pixel), and an initial model at 15 Å created ab initio. After 3D refinement using the ab initio model as a reference (30), 3D classification with six classes and without angular sampling was performed. Two classes with 99% of the particles (204,545 particles), both containing unassigned density in the A-site, with and without E-tRNA density, were combined. Particles were re-extracted at the original pixel size, 3D refined, CTF refined (4th order aberrations, beam-tilt, anisotropic magnification and per-particle defocus value estimation) and Bayesian polished (31). The resulting combined 50S volume was post-processed resulting in a final reconstruction at an average resolution of 2.32 Å. The map was low-pass filtered at 3 Å for modelling. Bsoft was used to estimate local resolution (32) based on the half maps from the final 3D refine (blocres -maxres -box 20 -cutoff 0.143 -verbose 1 -origin 0,0,0 -Mask half_map1 half_map2).
Single-particle reconstruction of Lmo70S ribosome
For the 70S-only volume, which was processed from a previously described VgaL-immunoprecipitation sample (EMPIAR-10684) (11), processing was performed with RELION 4.0 beta (33). CTFFIND4 was used to estimate CTFs for dose-weighted micrographs which had been motion-corrected with MotionCor2 (27,28). Micrographs with a CtfMaxResolution <5 Å, 1,385 in total, were selected for further processing. Particles were picked with the RELION LoG picker, resulting in 110,725 particles which were extracted with a pixel size of 2.46 Å/pixel and subjected to 2D classification. 54,430 particles were selected and used to create a starting model with the 3D initial model job type. The resulting volume was used as a reference for 3D refinement. 3D classification with four classes and without angular sampling was then performed. Two classes, with 26,143 particles and that contained resolved 70S but no VgaL, were selected for further processing. After another 3D refinement with a generous mask around the E site (generated with the volume eraser tool in UCSF ChimeraX) was used for partial signal subtraction followed again by 3D classification with four classes and without angular sampling. Three classes that did not contain VgaL-like density in the E site, totaling 14,097 particles, were combined and re-extracted with the original pixel size. 3D refinement, followed by refinements for anisotropic magnification, per-particle defocus and per-micrograph astigmatism, fourth order aberrations, and beam-tilt and trefoil (31), and another 3D refinement were performed, prior to the final post-processing.
Single-particle reconstruction of Lmo50S-Lnc and Lmo70S-Lnc complexes
Processing was performed in RELION 3.1 unless otherwise specified (26). RELION’s implementation of MotionCor2 with 5 × 5 patches and CTFFIND4 (with power spectra) were used for motion correction and initial CTF estimation (27,28). 561,654 particles were picked from 14,982 micrographs with crYOLO using the general model (29). After 2D classification, all ribosome-like classes were selected, particles extracted with a 3× reduced pixel size (2.46 Å/pixel). After 3D refinement using the Lmo70S-VgaL map (EMDB-12334) (11) as a reference, 3D classification with six classes and without angular sampling was performed. Two classes were further processed, one with 56.4% of particles (285,330 particles) with E-site tRNA and one with 34% (172,106 particles) with P-tRNA. Particles were re-extracted at the original pixel size, 3D refined, CTF refined (fourth order aberrations, beam-tilt, anisotropic magnification and per-particle defocus value estimation) and Bayesian polished (31). The resulting 70S sub-stoichiometric P-site tRNA volume was post-processed resulting in a masked final reconstruction at an average resolution of 2.25 Å. For the E-site tRNA volume the 50S was masked, to exclude the sub-stoichiometric 30S subunit, resulting in a masked final reconstruction at an average resolution of 2.08 Å. Masks were generated based on the 3D refined volumes with 6 px soft-edge and 15 Å low pass filter applied. Bsoft was used to estimate local resolution (32) based on the half maps from the final 3D refine for both volumes. (blocres -sampling 0.82 -maxres -box 20 -cutoff 0.143 -verbose 1 -origin 0,0,0 -Mask half_map1 half_map2).
Single-particle reconstruction of LmoHflX-50S complex
Processing was performed in RELION 3.1 unless otherwise specified (26). RELION's implementation of MotionCor2 with 5 × 5 patches and CTFFIND4 (using power spectra) were used for motion correction and initial CTF estimation (27,28). 268,147 particles were picked from 4,480 micrographs with crYOLO using the general model (Wagner et al., 2019). After 2D classification, all ribosome like classes were selected, particles extracted with a 3× reduced pixel size (2.46 Å/pixel). After 3D refinement using the Lmo50S-Lnc volume as a reference, 3D classification with six classes and without angular sampling was performed. Three classes with 83.9% of the particles (203,878 particles), all containing density in the A-site that could be assigned to HflX, were combined. Particles were re-extracted at the original pixel size, 3D refined, CTF refined (fourth order aberrations, beam-tilt, anisotropic magnification and per-particle defocus value estimation) and Bayesian polished (31). The resulting combined 50S volume was post processed resulting in a final reconstruction at an average resolution of 2.54 Å. The resulting volume was further subsorted with masked (generated from a molmap of HflXr aligned with 6 px extension) particle subtraction and 31.5% of the particles containing high resolution HflX density were re-extracted at the original pixel size and 3D refined. The resulting 50S volume was post processed resulting in a masked final reconstruction at an average resolution of 2.76 Å, however, the maps exhibited strong orientation bias, presumably due to preferred orientations on the cryo-EM grid. All maps were lowpass filtered at 3 Å for structural analysis. Bsoft was used to estimate local resolution (32) based on the half maps from the final 3D refine (blocres -maxres -box 20 -cutoff 0.143 -verbose 1 -origin 0,0,0 -Mask half_map1 half_map2).
Molecular modelling of the Lmo ribosome complexes
The molecular models for the L. monocytogenes 50S subunit and 70S ribosome were based on the L. monocytogenes VgaL structure (PDB ID 7NHN) (11). An initial molecular model for L. monocytogenes HflXr was prepared using alphafold (34,35). Lincomycin and other molecular restraints were prepared using aceDRG (36). Starting models were rigid body fitted and modelled using Coot 0.9.8.1 (37) from the CCP4 software suite v.8.0 (38). Initial refinement was done in Phenix 1.20–4487 (39) and final refinement in Refmac5 within Servalcat (40) and validated with Phenix comprehensive Cryo-EM validation (41,42).
Sequence analysis
HflX and HflXr sequences were identified via a BlastP v 2.11 (43) search against a diverse set of bacterial NCBI proteomes (E value cut-off 1e-60, L. monocytogenes NCBI RefSeq protein NP_464289.1 as the query). Additional representative HflX/HflXr homologous sequences were identified from Duval et al. (16) supplementary datasets, and clustered with MMseqs2 version 13-45111 using ‘mmseqs cluster’ with a minimum sequence identity value of 0.5 (44). ABCF subfamilies were identified using HMM models made previously (45), scanned with HMMER version 3.1b2 HMMSCAN with an E value cut-off of 1e−70. Sequences were aligned with MAFFT L-INS-i version 7.453 (46). HflXr sequences are classified based on phylogenetic analysis (below, and Supplementary Figure S1). All NCBI accessions are recorded in Supplementary Data S1, along with the full alignment. After removing alignment positions with >50% gaps with TrimAl v1.2 (47), phylogenetic analysis was carried out using IQ-TREE v1.6.12 (http://iqtree.cibiv.univie.ac.at/, (48)) with default parameters and 1000 ultrafast bootstrap replicates (49). Within the analysis, the program determined and used ‘LG + I + G’ as the best-fit substitution model. JalView v.2.11.2.0 (50) and FigTree v.1.4.4 (https://github.com/rambaut/figtree/) were used respectively for alignment and phylogenetic tree visualization.
Figures
UCSF ChimeraX 1.3 (51) was used to isolate density and visualize density images and structural superpositions. Models were aligned using PyMol v2.4 (Schrödinger, LLC). Figures were assembled with Adobe Illustrator (Adobe Inc.) and Inkscape (latest development release, regularly updated).
RESULTS
Phyla that contain ARE-ABCFs often have HflX/HflXr homologs with extended loop regions
HflXr is extended in the loop region relative to E. coli and L. monocytogenes HflX by typically two amino acids (Figure 1A, Supplementary Figure S1, and Data S1). In addition to L. monocytogenes, we observe that for bacteria containing two HflX homologs, usually one homolog is part of the paraphyletic HflX group in the phylogeny and has a shorter loop, and the other is within the HflXr clade and has a longer loop, as seen for example in Listeria innocua, Clostridioides difficile, Carnobacterium maltaromaticum and some Bacillus species, such as B. cereus and B. vireti, but not B. subtilis, the latter having only the HflX homolog (Figure 1A). Streptomyces ambofaciens is unusual in having two relatively closely related HflX homologs with insertions of the same length (Figure 1A). With only 50% bootstrap support for the Faecalibacterium duncaniae homolog branching with the HflXr clade, the classification of this divergent protein—and therefore the boundary of the HflXr clade—is ambiguous. However, this protein has one of the longest insertions in the family, and we tentatively classify it as HflXr. Interestingly, Lactobacillus sakei appears to have HflXr and no corresponding HflX homolog (Figure 1A). In some bacteria containing only a single HflX homolog, we observe independent insertions of distinct sequence and length, for example, Mycobacterium species (M. abscessus, M. smegmatis, M. tuberculosis), Bacteroides fragilis, Amycolatopsis mediterranei and Deinococcus radiodurans have eight amino acid insertions (relative to the E. coli HflX sequence), Cornebacterium glutamacium and Gardnerella vaginalis have seven, Streptomyces fradiae has five and Chlamydia trachomatis has four (Figure 1A and Supplementary Figure S1). Interestingly, the S. fradiae hflX gene resides within the biosynthetic gene cluster for the macrolide spiramycin (52), supporting a possible role for loop-extended HflX homologues as antibiotic resistance determinants. The single HflX homologs present in Mycobacterium abscessus and M. smegmatis have been shown to confer resistance to macrolides and lincosamides, but not to linezolid, chloramphenicol or tetracycline (53). Despite the longer loop with 8aa insertion, M. smegmatis HflX was reported to have no influence on the binding of radiolabelled erythromycin to the ribosome, leading to the suggestion that M. smegmatis HflX does not use target protection mechanism, but that the mechanism involves ribosome recycling (53). The phyla with long loop homologues (regardless of whether they are HflX or HflXr), Bacillota and Actinomycetota, are also those that tend to carry one or more ARE versions of ABCFs (Supplementary Data S1). However, this is not a strict association. For example, Chlamydia trachomatis carries an extended HflX loop while only harboring a single, likely housekeeping ABCF (YbiT), while some isolated Bacillota and Actinomycetota species carry ARE-ABCFs but no extended HflX/HflXr (Supplementary Data S1).
HflXr confers resistance to macrolides and lincosamides, but also pleuromutilins and streptogramins
HflXr-mediated resistance is naturally masked in L. monocytogenes by resistance mediated by VgaL (16), an ARE-ABCF that confers resistance to lincosamides, pleuromutilins and streptogramin A antibiotics (11,16,54–56). Thus, in order to specifically characterize the antibiotic resistance profile of HflXr we used a set of hflXr+ and hflXr– ΔvgaL L. monocytogenes strains for our antibiotic sensitivity assays. We have constructed a markerless double-deletion L. monocytogenes ΔvgaL ΔhflXr strain that lacks both VgaL and HflXr, and transformed it either with the empty pIMK3 plasmid or the pIMK3 plasmid expressing HflXr. As a reference strain we used our previously reported L. monocytogenes ΔvgaL strain (11) transformed with an empty pIMK3 plasmid. For the three strains described above we determined minimum inhibitory concentration (MIC) for the macrolides erythromycin and azithromycin, the lincosamides lincomycin and clindamycin, the pleuromutilins tiamulin and retapamulin, the streptogramins virginiamycin M1 and S1, as well as chloramphenicol, linezolid and tetracycline (Figure 1B). In comparison to the ΔvgaL L. monocytogenes, we observed that the ΔvgaL ΔhflXr strain exhibited a slight increase in sensitivity to erythromycin (2-fold) and lincomycin (4-fold), but remained unchanged for chloramphenicol and tetracycline, as reported previously (16). We also observed similar increases in sensitivity to azithromycin, clindamycin as well as virginiamycin M1 (streptogramin A), suggesting that in addition to macrolides and lincosamides, HflXr may also confer some resistance to streptogramin A compounds. By contrast, the MIC remained unchanged for tiamulin, retapamulin, linezolid and virginiamycin S (streptogramin B). As expected we were able to rescue the antibiotic sensitivities by overexpression of HflXr; for example, compared to the pIMK3 control, overexpression of HflXr in the ΔvgaL ΔhflXr strain led to a 4-fold increase in MIC for the macrolides erythromycin and azithromycin and the lincosamides lincomycin and clindamycin, as well as a 2-fold increase in resistance to virginiamycin M (Figure 1B). Additionally, we observed a 2-fold increase in MIC for virginiamycin S and more strikingly, 4- and 8-fold MIC increases for the pleuromutilins tiamulin and retapamulin, respectively. No change in MIC was observed for chloramphenicol, linezolid or tetracycline, suggesting that the observed MIC increases are specific for distinct classes of ribosome-targeting antibiotics. In summary, our findings expand the spectrum of antibiotic classes that HflXr can confer resistance to from macrolides and lincosamides to include pleuromutilins and streptogramins.
Cryo-EM structure of a L. monocytogenes HflXr-50S complex
Our attempts to reconstitute HflXr-50S complexes in vitro were unsuccessful due to difficulties obtaining active soluble L. monocytogenes HflXr protein. Therefore, to generate homogenous L. monocytogenes HflXr-50S complexes for structural analysis, we employed an in vivo pull-down approach using the L. monocytogenes ΔhflXr strain overexpressing a C-terminally FLAG3-tagged HflXr protein, as used recently to generate ARE-ABCF–ribosome complexes (11,12). As a specificity control, we also performed the in vivo pull-down experiment using the L. monocytogenes ΔhflXr strain transformed with the empty pIMK3 plasmid. Affinity purification via the FLAG3-tag was performed in the presence of 0.5 mM GTP, or the non-hydrolysable analog GDPNP, from clarified lysates and the analysis of the elution fractions from the purifications indicated that HflXr was bound stably to the 50S subunit in the presence of both GTP and GDPNP (Supplementary Figure S2). Since the yield was slightly higher in the presence of GDPNP, the HflXr-GDPNP-50S complex was subjected to structural analysis using single-particle cryo-EM.
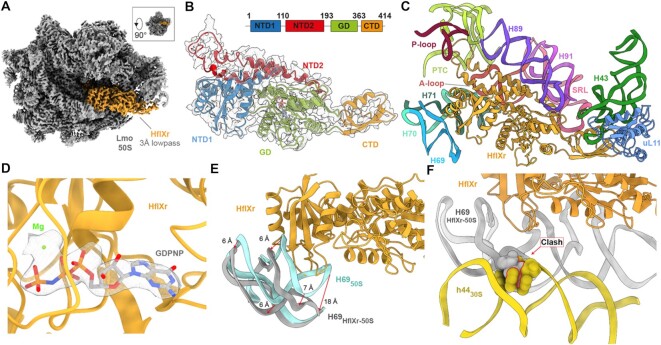
Using a Titan Krios transmission electron microscope with a K2 direct electron detector, we collected 4,255 micrographs which, after 2D classification, yielded 206,159 ribosomal 50S particles (Supplementary Figure S3). In silico sorting revealed that the vast majority (99%) of these particles contained an additional density for HflXr, with or without the presence of E-site tRNA. All HflXr-containing classes were combined and after 3D refinement resulted in a cryo-EM map of L. monocytogenes HflXr-50S complex with an average resolution of 2.3 Å (Supplementary Figure S4 and Table S2). While this number reflected well the resolution at the core of the 50S subunit, the HflXr factor was significantly less well-resolved and visualized better in low-pass filtered maps at 3.0 Å. With the exception of the loop connecting the two α-helices of the N-terminal subdomain II, which exhibited flexibility, the density for HflXr was sufficiently resolved to build a molecular model for the two NTD subdomains, the central GTPase domain (GD) and CTD (Figure 2A, B). As expected based on high sequence conservation, the overall conformation of HflXr is very similar to that reported previously for E. coli HflX in complex with the 50S subunit (17), as well as more recently for the human mitochondrial HflX homolog GTPBP6 bound to a large subunit biogenesis intermediate (22) (Supplementary Figure S5). Briefly, the NTD subdomain I interacts with H69-H71, the α-helices of subdomain II run parallel to H89 and H91 and extend into the PTC, and the CTD interacts with uL11 located at the stalk base (Figure 2C). Like HflX, the GTPase domain of HflXr is positioned differently than other translational GTPases, such that its nucleotide-binding pocket is oriented away from the sarcin-ricin loop (Figure 2C). Careful inspection of the G-domain reveals density within the nucleotide-binding pocket consistent with the presence of a nucleotide triphosphate (Figure 2D), which we presume is the GDPNP that was added to the lysate during sample preparation. Binding of HflXr causes a shift in H69 that would be incompatible with binding of the 30S subunit to form a 70S ribosome, analogous to that observed with HflX (17) (Figure 2E, F). Thus, the similarity between the binding position of HflXr and that observed for HflX (17), suggests that HflXr is likely to induce splitting of the L. monocytogenes 70S ribosome with an analogous mechanism as that reported for splitting of E. coli 70S ribosomes by HflX (17).
Figure 2.
Cryo-EM structure of the L. monocytogenes HflXr-50S complex. (A) Cryo-EM density (3 Å low-pass filtered) of the 50S ribosomal subunit (grey) with HflXr (orange). Inset shows relative orientation of (A) to crown view. (B) Isolated cryo-EM map density (grey mesh; 3 Å lowpass filtered) with a molecular model for HflXr colored according to domains: N-terminal subdomain I (NTD1, blue), N-terminal subdomain II (NTD2, red), G-domain (GD, green) and C-terminal domain (CTD, orange). (C) Interactions of HflXr (orange) NTD1 with H69 (blue), H70 (cyan), H71 (dark green), the α-helices of NTD2 with H89 (purple) and H91 (light purple), NTD2-loop with the PTC (lime) and the CTD with uL11 (light blue). (D) GDPNP (grey) in extracted density (grey mesh) within the binding pocket of the GD (orange) with a putative coordinated magnesium ion (green). (E) Binding of HflXr (orange) causes a shift in H69 (grey) compared to the vacant L. monocytogenes ribosome (light blue, aligned on 23S rRNA). (F) H69 (grey) movement caused by HflXr would sterically clash with h44 of L. monocytogenes 30S subunit (yellow, PDBID: 7NHN; (11)) on the 70S ribosome, when aligned on the basis of 23S rRNA.
Interaction of the NTD subdomain II of HflXr at the PTC
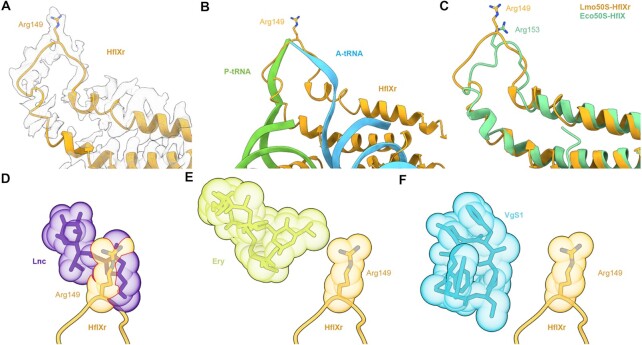
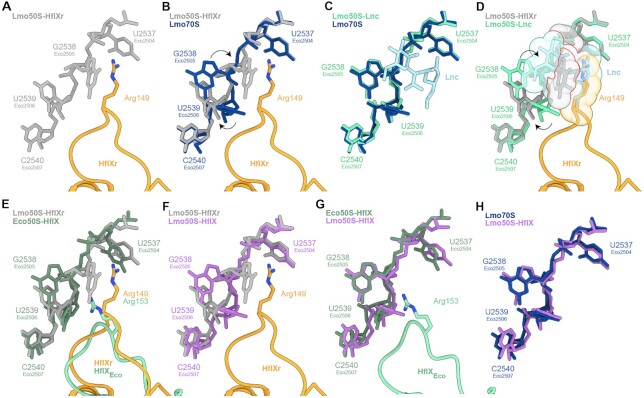
Because the loop connecting the two α-helices of the N-terminal subdomain II (NTD2-Loop) was poorly resolved, we were only able to trace a backbone model for this region (Figure 3A). Nevertheless, the overall position of the NTD2-loop at the PTC overlaps significantly with that of the CCA-ends of both A- and P-site tRNAs (Figure 3B), thus illustrating that binding of HflXr is incompatible with actively translating ribosomes, analogous to HflX (17). However, in the case of HflXr, the NTD2-loop reaches deeper into the PTC than observed previously for HflX (17) (Figure 3C), which is likely due to the NTD2-loop in HflXr being longer by two residues than that of E. coli HflX (4) (Figure 1A). As mentioned, the NTD2-loop of L. monocytogenes HflX is also shorter by two residues compared to L. monocytogenes HflXr (4). Although we cannot be certain of the register of specific amino acids within the NTD2-loop, there is strong density located at the tip of the NTD2-loop of HflXr that would be consistent with the sidechain of Arg149 (Figure 3A). We note that structural alignment of the E. coli HflX from the E. coli HflX-50S structure (17) placed the equivalent Arg153 in a similar location (Figure 3C), although the Arg149 of HflXr is placed somewhat deeper in the PTC (Figure 3A,B), presumably due to the additional two preceding amino acids that are absent in E. coli HflX (Figure 1). To understand the spatial relationship between HflXr and PTC binding antibiotics, we first aligned the Staphylococcus aureus 50S-lincomycin structure (57) to our HflXr-50S complex, revealing significant overlap between the drug and the tip of the NTD2-loop where we predict Arg149 to be located (Figure 3D). To ascertain whether Arg149 of HflXr contributes to antibiotic resistance, we generated an Arg149 to Ala mutation (R149A) in HflXr and monitored antibiotic sensitivities when overexpressed in the ΔvgaL ΔhflXr strain (Figure 1B). Compared with the overexpression of the wild-type HflXr that led to increased MICs against macrolides, pleuromutilins, lincosamides and streptogramins, overexpression of the HflXr-R149A mutant did not increase the MIC for any of the antibiotics tested (Figure 1B), indicating that Arg149 of HflXr plays a critical role for conferring antibiotic resistance. Despite the overlap between HflXr and PTC antibiotics, such as lincomycin (Figure 3D), structural alignments suggest that HflXr does not overlap with macrolides, such as erythromycin (Figure 3E), nor streptogramin B antibiotics, such as virginiamycin S1 (Figure 3F). This raises the question as to whether HflXr confers resistance to macrolides and streptogramin B antibiotics using an allosteric, rather than steric overlap mechanism, as proposed for many ARE-ABCF proteins (11–13,58).
Figure 3.
Interaction of the NTD subdomain II of HflXr at the PTC. (A) Arg149 of NTD2-loop of HflXr (orange) in extracted cryo-EM density. (B) HflXr loop (orange) superimposed with A- (blue) and P-site tRNAs (green)(73). (C) The loop of HflXr (orange) extends deeper into the PTC than E. coli HflX (17). Alignment in (B) and (C) are based on 23S rRNA. D-F, Binding position of HflXr NTD2-loop (orange) with Arg149 shown as sphere, superimposed with the binding site of (D) Lincomycin (Lnc, purple, PDB ID 5HKV)(57), (E) erythromycin (Ery, yellow, PDB ID 4V7U) (59) and (F) virginiamycin S1 (VgS1, blue, PDB ID 1YIT)(60). Predicted steric clashes are indicated by red lines at the overlap of spheres.
Cryo-EM structures of L. monocytogenes 70S complexes
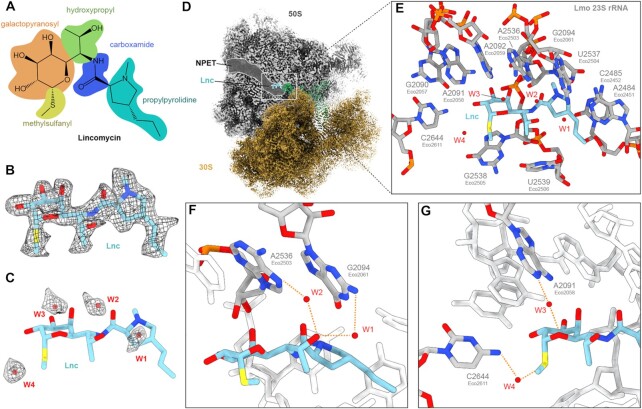
To investigate whether HflXr induces conformational changes within the PTC upon binding, we determined structures of L. monocytogenes 70S ribosomes in the absence of HflXr. First, we determined a structure of the L. monocytogenes 70S ribosome with a P-site tRNA at 3.1 Å (Supplementary Figure S6 and Table S2) by refining a minor subpopulation of L. monocytogenes ribosomal particles from our previous study where the L. monocytogenes ARE-ABCF VgaL was absent (11). In addition, we prepared a new sample where L. monocytogenes 70S ribosomes were incubated with lincomycin and subjected to single particle cryo-EM analysis. Two major populations were identified and refined, enabling structures of a L. monocytogenes 70S-lincomycin complex, with and without P-site tRNA at 2.3 Å and 2.1 Å, respectively, to be determined (Supplementary Figures S7, S8 and Table S2). In both structures, lincomycin was bound within the A-site crevice at the PTC (Figure 4A–D), as observed previously for lincomycin (57) and its closely-related semi-synthetic derivative clindamycin (59,60). While the conformations of PTC nucleotides forming the lincomycin binding site were also identical with that observed previously for lincomycin in complex with the S. aureus 50S subunit (57), small differences were observed for conformation of lincomycin antibiotic itself, namely, a slight rotation of methyl-sulfanyl group on the galactopyranosyl ring and a different orientation of the pyrrolidinyl ring (Supplementary Figure S9A, B). Surprisingly, the differences for lincomycin on the S. aureus 50S subunit at 3.66 Å (57) were not evident when comparing with the available clindamycin-ribosome structures on the E. coli 70S at 3.3 Å (59) and H. marismortui 50S at 3.0 Å (60) (Supplementary Figure S9A–F), however, this may simply arise from limitations in the resolution. An additional exception is the structure of clindamycin bound to the Deinococcus radiodurans 50S at 3.1 Å, where the propyl pyrrolidinyl tail was reported to be rotated by 180° degrees compared to other clindamycin-ribosome structures (61) (Supplementary Figure S9G,H). Another major difference with all available lincomycin and clindamycin structures is that with the higher resolution of the L. monocytogenes 70S-lincomycin complex reported here, we observe densities for four putative water molecules (W1-W4) that can facilitate interactions between the drug and nucleotides of the 23S rRNA (Figure 4E). Specifically, we observe two water-mediated interactions (W1-W2) between the C7-hydroxyl of lincomycin with the N7 A2536 (EcA2503) and N2 of G2094 (EcG2061) of the 23S rRNA (Figure 4F). In addition, two water-mediated (W3-W4) interactions are possible from the galactopyranosyl moiety, namely, between the sulphur atom and the N4 of C2644 (EcC2611) as well as the non-bridging oxygen and the N1 of A2091 (EcA2058) (Figure 4G). While the importance of these water-mediated interactions is unclear, we note that displacement of a single water via dimethylation of 23S rRNA nucleotide A2058 prevents erythromycin binding and leads to macrolide resistance (62).
Figure 4.
Cryo-EM structure of the L. monocytogenes lincomycin-70S complex. (A) Chemical structure of lincomycin, with galactopyranosyl (orange), methylsulfanyl (yellow), hydroxypropyl (green), carboxamide (blue) and propylpyrrolidine (cyan) moieties highlighted. (B) Cryo-EM density (mesh) with molecular model of lincomycin (Lnc, blue). (C) Molecular model of Lnc (blue) with cryo-EM density (mesh) and model for waters W1-W4 (red). (D) Cryo-EM density of L. monocytogenes Lnc-70S complex with Lnc bound at the PTC, adjacent to the ribosomal NPET in the 50S (grey) subunit. (E) Lnc (blue) with surrounding waters W1-W4 (red) and 23S rRNA nucleotides (grey). (F) Water-mediated interaction of the hydroxypropyl-group of Lnc (blue) with N2 of G2094 (EcoG2061, grey) through W1 (red) and N7 of A2536 (EcoA2503, grey) through W2 (red). (G) Water-mediated interaction of Lnc (blue) with N1 of A2091 (EcoA2058, grey) and W3 and N4 of C2644 (EcoC2611, grey) with W4 (red).
HflXr-induced conformation at the PTC is incompatible with drug binding
Comparison of the L. monocytogenes HflXr-50S complex (Figure 5A) with the vacant L. monocytogenes 70S ribosome (Figure 5B) reveals very localized conformational changes within the PTC upon HflXr binding. In particular, the nucleobase of U2539 (EcU2506) appears to shift and rotate, which is necessary to avoid clashing with the NTD2-Loop of HflXr (Figure 5B). The shifted position of U2539 (EcU2506) would in turn clash with the position of G2538 (EcG2505), presumably causing the dramatic shift (up to 10 Å) in G2538 (EcG2505) that is observed upon HflXr binding (Figure 5B). We also observe a change in the position of A2095 (EcA2062), similar to that observed previously in ribosomes bearing peptidyl-tRNA mimics (63) (Supplementary Figure S10). This rotated conformation of A2095 (EcA2062) is likely to be stabilized by direct contact with the NTD2-Loop of HflXr, however, the interactions are not resolved sufficiently to enable a molecular description. Within the limits of resolution, the conformation of the PTC is identical when comparing the structure of L. monocytogenes 70S ribosome in the presence and absence of lincomycin (Figure 5C), indicating that binding of lincomycin to the ribosome does not cause any conformational changes in the rRNA. Superimposition of other antibiotic-ribosome structures suggests that the position of G2538 (EcG2505) observed in HflXr would also clash with the macrolides erythromycin and azithromycin, the pleuromutilins tiamulin and retapamulin, and the streptogramin A antibiotic virginiamycin M1 (Supplementary Figure S11), consistent with our MIC data indicating that HflXr confers some level of resistance to these drugs (Figure 1B). The streptogramin B antibiotic virginiamycin S1 does not directly clash with the HflXr-bound G2538 (EcG2505), although our data indicate that overexpression of HflXr can confer a 2-fold increase in MIC against this drug (Figure 1B). We note here that A2095 (EcA2062) contributes to virginiamycin S1 binding by forming stacking interactions with the aromatic C18 moiety (60,64), which would not be possible with the rotated conformation of A2095 (EcA2062) stabilized by HflXr (Supplementary Figure S10). Conversely, we note that the shifted position of G2538 (EcG2505) that is observed upon HflXr binding significantly overlaps with the binding sites of both linezolid and chloramphenicol (Supplementary Figure S11), yet we observe no effect of HflXr on these classes of antibiotics (Figure 1B). Superimposing the L. monocytogenes HflXr-50S and lincomycin-70S structures reveals that the position of G2538 (EcG2505) observed upon HflXr binding is incompatible with the presence of lincomycin (Figure 5D), suggesting that HflXr may confer resistance to lincomycin by altering the conformation of the PTC to perturb drug binding. Indeed, these conformational changes at the PTC induced by HflXr are not observed when E. coli HflX interacts with the ribosome (Figure 5E) (17), indicating that they are likely to be specific for antibiotic-resistance HflXr proteins in contrast to the non-resistance HflX proteins.
Figure 5.
HflXr-induced conformational changes at the PTC are incompatible with lincomycin binding. (A) NTD2-loop Arg149 of HflXr (orange) with selected L. monocytogenes 23S rRNA nucleotides (grey). (B) as (A), but superimposed with L. monocytogenes 23S rRNA nucleotides (dark blue) from vacant 70S ribosome. Nucleotide rearrangements induced by HflXr are indicated with black arrows. (C) Comparison of L. monocytogenes 23S rRNA nucleotides in presence (green) and absence (dark blue) of lincomycin (Lnc, light blue). (D) as (A), but superimposed with Lnc (light blue) bound to the L. monocytogenes 23S rRNA nucleotides (cyan) with nucleotide rearrangements induced by HflXr indicated with black arrows and shown as spheres with red lines indicating steric clashes. (E, F) as (A), but superimposed with (E) E. coli HflX-50S complex (PDB ID 5ADY) (17), and (F) 23S rRNA nucleotides (purple) from the L. monocytogenes 70S refined into the L. monocytogenes HflX-50S complex. (G, H) Comparison of 23S rRNA nucleotides from L. monocytogenes HflX-50S complex with (G) E. coli HflX-50S complex (PDB ID 5ADY) (17), and (H) L. monocytogenes 70S ribosome.
To ensure that the lack of conformational changes upon HflX binding are not specific for E. coli, we also determined a cryo-EM structure of L. monocytogenes HflX-50S complex using the same approach as used for L. monocytogenes HflXr (Supplementary Figure S2B). As for HflXr, the majority of 50S particles contained additional density for HflX bound within the A- and P-sites as expected. All HflX-containing classes were combined and after 3D refinement resulted in a cryo-EM map of L. monocytogenes HflX-50S complex with an average resolution of 2.6 Å (Supplementary Figure S12 and Table S2). However, unlike the HflXr-50S maps, the HflX-50S map exhibited strong bias resulting from preferred orientation of particles on the cryo-EM grids. Further subsorting identified more stable classes where the HflX was better resolved (Supplementary Figure S12), however, the bias remained. For this reason, we did not generate and refine a molecular model for HflX-50S complex, but rather fitted and adjusted the molecular model of the L. monocytogenes 50S subunit from 70S ribosome structure to ascertain the conformation of the PTC. A comparison of the position of the 23S rRNA nucleotides located at the PTC revealed that there were no conformational changes induced upon HflX binding as observed for HflXr (Figure 5F and with density images shown in Supplementary Figure S13), but rather the conformation of the PTC was similar, if not identical, to that observed in the E. coli HflX-50S determined previously (17) (Figure 5G) and lincomycin-70S complexes (Figure 5H) determined in this study.
DISCUSSION
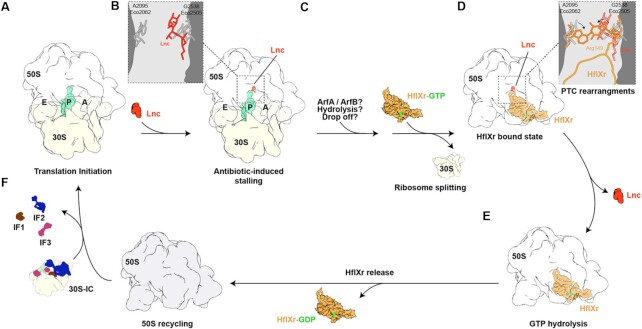
HflXr has been shown to confer resistance to lincomycin and erythromycin by splitting ribosomes and recycling them for subsequent rounds of translation (16), however, direct interaction with ribosomal particles has not been conclusively demonstrated. Here, we demonstrate using affinity tagging and in vivo pull-downs that HflXr can stably interact with the 50S subunit in the presence of GTP or GDPNP (Supplementary Figure S2). A cryo-EM structure of the HflXr-GDPNP-50S complex reveals that HflXr occupies an analogous binding site on the large subunit, as observed previously for E. coli HflX (17) (Figure 2). However, we observe that the NTD2-Loop of HflXr penetrates deeper into the PTC than that of HflX and, unlike HflX, binding of HflXr induces conformational changes within the PTC that would be incompatible with antibiotic binding. Collectively, these observations lead us to expand on the model for the mechanism of action of HflXr (16) by proposing HflXr as a type II ribosome protection protein (Figure 6): Antibiotic-bound 70S ribosomes are recognized and split into their component 30S and 50S subunits by HflXr (Figure 6A), with HflXr remaining stably bound to the 50S subunit (Figure 6B). Interaction of the NTD2-Loop of HflXr with the PTC causes conformational rearrangements of 23S rRNA nucleotides that are incompatible with drug binding, thereby causing dissociation of the drugs (Figure 6C). Hydrolysis of GTP to GDP facilitates subsequent dissociation of HflXr, and enables the free 50S subunit to be recycled for the next round of translation (Figure 6D).
Figure 6.
Proposed mechanism of action of HflXr. (A) An initiating 70S ribosome with initiator tRNA (teal) in the P-site (A) is stalled by an antibiotic (Lnc, red, B) with G2538 (Eco2505) and A2095 (Eco2062) in their canonical position. (C) The P-tRNA dissociates through an unknown mechanism (e.g. ArfA/ArfB, hydrolysis or tRNA drop off) which allows HflXr-GTP to recognize the resulting 70S ribosome, triggering ribosome splitting and subunit dissociation. (D) The HflXr NTD2 loop induces PTC rearrangement of G2538 (Eco2505) and A2095 (Eco2062), leading to antibiotic dissociation. (E) GTP hydrolysis allows HflXr-GDP release and the resulting free 50S subunit (F) is available for reinitiation (A).
Despite these initial insights into the action of HflXr, many questions remain to be answered during each step of the process. What is the initial substrate for HflXr action? It is hard to imagine that HflXr can directly recognize and split antibiotic-stalled ribosomes since the binding site of HflXr overlaps with both A- and P-site tRNAs. Indeed, HflX was shown to have low splitting activity on polysomes or ribosomes bearing peptidyl-tRNAs, but was more efficient at splitting vacant 70S ribosomes, or post-release ribosomes where the deacylated P-site tRNA can adopt a hybrid P/E conformation and thereby free-up the HflX binding (17). Zhang et al. 2015 (17) have also raised the question whether another factor, such as ArfA or ArfB, could be involved prior to HflX splitting – which might also be the case for HflXr too. Notably, the classes of antibiotics to which HflXr confers resistance can trigger peptidyl-tRNA ‘drop-off’, the result of which could be a substrate for HflXr (65,66). Alternatively, HflXr might also act directly on antibiotic-bound 50S subunits. In our model, we indicated that GTP hydrolysis is not required for splitting, but for dissociation of HflXr from the 50S subunit, which was based on the analogous mode of action of E. coli HflX (17) and Mycobacterium HflX (53), however, this hypothesis also needs to be validated for HflXr. Although we have demonstrated here that binding of HflXr to the 50S induces conformational changes at the PTC that would be incompatible with drug binding, it remains to be directly shown whether HflXr binding to antibiotic-50S complexes leads to drug release, and whether drug release requires GTP hydrolysis. While this has been performed previously for other RPPs, such as TetO/TetM (67–69) and the ARE-ABCFs LsaA (70) and MsrE (58), such in vitro assays require soluble active protein, which has so far proven a stumbling block for HflXr. Additionally, it remains unclear as to what prevents rebinding of the drugs once HflXr has dissociated. One possibility is that the conformational changes induced in the PTC by HflXr persist after dissociation of the factor, as proposed for TetM/TetO (69,71,72). However, this would require the conformational changes to remain following initiation, and, for some of the antibiotic classes, throughout multiple rounds of elongation. A more likely (but not mutually exclusive) scenario is that HflXr works in conjunction with efflux pumps. We propose that, unlike short loop HflX (which can also split ribosomes), extended loop HflX/HflXr variants—such as L. monocytogenes HflXr and mycobacterial HflX—confer resistance because in addition to splitting ribosomes they also stimulate the antibiotic disassociation from the 50S. This, in turn, facilitates subsequent drug efflux from the cell.
Generally, the allosteric mechanism proposed here for HflXr is reminiscent of that proposed for some ARE-ABCF proteins, in particular, proteins like L. monocytogenes VgaL that confer resistance to PTC inhibitors, such as pleuromutilins, lincosamides and streptogramin A antibiotics (4,11). Although VgaL has a distinct binding site compared to HflXr, being located predominantly within the ribosomal E-site, VgaL, like HflXr, also has a loop that encroaches into both the A- and P-sites at the PTC (Supplementary Figure S14) (11). However, while VgaL also induces conformational changes within the PTC that are proposed to promote drug release (11), the mechanisms appear to be unrelated i.e. there is no similarity in the types of movements induced by HflXr and ARE-ABCFs. Indeed, the movements induced by VgaL do not affect macrolide or streptogramin B binding, as we observe for HflXr. Instead, resistance to macrolides and streptogramin B is specifically associated with Msr-type ARE-ABCF proteins, such as Pseudomonas aeruginosa MsrE, which has a longer loop that penetrates deeper into the exit tunnel to locally perturb macrolide binding (10,58). Interestingly, VgaL, like HflXr, does not confer resistance to oxazolidinones or phenicols, despite these antibiotics occupying overlapping binding sites with other PTC inhibitors to which they do confer resistance, such as lincomycin and tiamulin (11). In the case of HflXr, one possible explanation is that the chloramphenicol and linezolid-stalled ribosomes are refractory to peptidyl-tRNA release and therefore do not become a substrate for HflXr-mediated splitting, while lincosamides and macrolides can stimulate peptidyl-tRNA drop-off (65,66). However, further investigations will be required to elucidate this.
Lastly, it needs to be re-iterated that the level of resistance conferred by HflXr is relatively modest and no increase in antibiotic sensitivity is observed upon HflXr loss in the presence of VgaL (16). For example, while VgaL confers 4–8-fold increases in MIC for lincomycin and 128-fold increase in MIC for tiamulin (11), HflXr confers MIC increases of 2-fold for macrolides and streptogramins, 4-fold for lincomycin, and increased resistance (4-fold) to pleuromutilins is only observable upon overexpression of HflXr (Figure 1B). One possible explanation for this might be that following antibiotic release and dissociation of VgaL, ribosomes that continue translating could enter into the elongation phase and thereby become immune to the effects of translation initiation inhibitors such as pleuromutilins, lincosamides and streptogramin A antibiotics (4,11). By contrast, following antibiotic release and dissociation of HflXr, the 50S subunit needs to re-enter into translation via the initiation phase, thereby providing more time for rebinding of the antibiotic to the ribosome. The low level of resistance of HflXr compared to ARE-ABCFs does raise the question as to why some bacteria have retained the HflXr protein. One possible explanation is that in addition to its function in antibiotic resistance, HflXr may retain a stress factor function analogous to that of HflX, but, perhaps, acting on another set of stalled ribosomal substrates.
DATA AVAILABILITY
Cryo-EM maps have been deposited in the Electron Microscopy Data Bank (EMDB) with accession codes EMD-15161 [https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-15161] (L. monocytogenes HflXr-50S complex), EMD-15204 [https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-15204] (L. monocytogenes 70S ribosome), EMD-15175 [https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-15175] (L. monocytogenes lincomycin-70S complex), EMD-15864 [https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-15864] (L. monocytogenes lincomycin-50S map) and EMD-15670 [https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-15670] (L. monocytogenes HflX-50S map). Molecular models have been deposited in the Protein Data Bank with accession codes 8A57 [https://doi.org/10.2210/pdb8A57/pdb] (L. monocytogenes HflXr-50S complex), 8A63 [https://doi.org/10.2210/pdb8A63/pdb] (L. monocytogenes 70S ribosome), 8A5I [https://doi.org/10.2210/pdb8A5I/pdb] (L. monocytogenes lincomycin-50S subunit).
ADDITIONAL INFORMATION
Correspondence and requests for materials should be addressed to V.H. or D.N.W.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Michael Hall for help with cryo-EM data collection as well as the Protein Expertise Platform (PEP) at Umeå University and Mikael Lindberg for constructing plasmids VHp1067 and VHp1068.
Author contributions: G.C.A. and J.A.N. performed sequence conservation analyses. K.J.T. and K.V. cloned the HflXr constructs, performed genetic manipulations of L. monocytogenes, performed polysome fractionations and immunoblotting as well as performed MICs. J.J. assisted in genetic manipulations of L. monocytogenes and provided materials. K.J.T. and M.R. prepared L. monocytogenes 70S ribosomes. K.J.T., T.K. and O.B. performed immunoprecipitations. K.J.T. and C.J. prepared cryo-EM grids and collected cryo-EM datasets. T.O.K. and C.C.M. processed the microscopy data, generated and refined the molecular models and made the structure figures. D.N.W. and V.H. wrote the manuscript with input from all authors. D.N.W and V.H. conceived and supervised the project.
Contributor Information
Timm O Koller, Institute for Biochemistry and Molecular Biology, University of Hamburg, Martin-Luther-King-Platz 6, 20146 Hamburg, Germany.
Kathryn J Turnbull, Department of Molecular Biology and Umeå Centre for Microbial Research (UCMR), Laboratory for Molecular Infection Medicine Sweden (MIMS), Umeå University, 90187 Umeå, Sweden; Department of Clinical Microbiology, Rigshospitalet, 2200 Copenhagen, Denmark.
Karolis Vaitkevicius, Department of Molecular Biology and Umeå Centre for Microbial Research (UCMR), Laboratory for Molecular Infection Medicine Sweden (MIMS), Umeå University, 90187 Umeå, Sweden.
Caillan Crowe-McAuliffe, Institute for Biochemistry and Molecular Biology, University of Hamburg, Martin-Luther-King-Platz 6, 20146 Hamburg, Germany.
Mohammad Roghanian, Department of Molecular Biology and Umeå Centre for Microbial Research (UCMR), Laboratory for Molecular Infection Medicine Sweden (MIMS), Umeå University, 90187 Umeå, Sweden; Department of Clinical Microbiology, Rigshospitalet, 2200 Copenhagen, Denmark; Department of Experimental Medical Science, Lund University, 221 00 Lund, Sweden.
Ondřej Bulvas, Institute of Organic Chemistry and Biochemistry, Academy of Sciences of the Czech Republic, v.v.i., Flemingovo nam. 2, 166 10 Prague 6, Czech Republic; Department of Biochemistry and Microbiology, University of Chemistry and Technology Prague, Technicka 5, 166 28 Prague 6, Czech Republic.
Jose A Nakamoto, Department of Experimental Medical Science, Lund University, 221 00 Lund, Sweden.
Tatsuaki Kurata, Department of Molecular Biology and Umeå Centre for Microbial Research (UCMR), Laboratory for Molecular Infection Medicine Sweden (MIMS), Umeå University, 90187 Umeå, Sweden; Department of Experimental Medical Science, Lund University, 221 00 Lund, Sweden.
Christina Julius, Department of Molecular Biology and Umeå Centre for Microbial Research (UCMR), Laboratory for Molecular Infection Medicine Sweden (MIMS), Umeå University, 90187 Umeå, Sweden.
Gemma C Atkinson, Department of Experimental Medical Science, Lund University, 221 00 Lund, Sweden.
Jörgen Johansson, Department of Molecular Biology and Umeå Centre for Microbial Research (UCMR), Laboratory for Molecular Infection Medicine Sweden (MIMS), Umeå University, 90187 Umeå, Sweden.
Vasili Hauryliuk, Department of Molecular Biology and Umeå Centre for Microbial Research (UCMR), Laboratory for Molecular Infection Medicine Sweden (MIMS), Umeå University, 90187 Umeå, Sweden; Department of Experimental Medical Science, Lund University, 221 00 Lund, Sweden; University of Tartu, Institute of Technology, 50411 Tartu, Estonia.
Daniel N Wilson, Institute for Biochemistry and Molecular Biology, University of Hamburg, Martin-Luther-King-Platz 6, 20146 Hamburg, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft (DFG) [WI3285/8-1 to D.N.W.]; Swedish Research Council (Vetenskapsrådet) [2017-03783, 2021-01146 to V.H., 2019-01085 to G.C.A., 2020-020053 to J.J.]; Stiftelsen Olle Engkvist Byggmästare (to J.J.); Ragnar Söderbergs Stiftelse (to V.H.); European Union from the European Regional Development Fund through the Centre of Excellence in Molecular Cell Engineering [2014-2020.4.01.15-0013 to V.H.]; Estonian Research Council [PRG335 to V.H.]; Interreg ÖKS grant through the ‘Hanseatic League of Science (HALOS)’ project NYPS 20201844 (to V.H.); Crafoord foundation [20220562 to V.H.]; D.N.W. and V.H. groups are also supported by the Deutsche Zentrum fuür Luft- und Raumfahrt [DLR01Kl1820 to D.N.W.]; Swedish Research Council [2018-00956 to V.H.] within the RIBOTARGET consortium under the framework of JPIAMR; G.C.A. and V.H. were also supported by a project grant from the Knut and Alice Wallenberg Foundation [2020-0037 to G.C.A.]; the electron microscopy data was collected at the Umeå Core Facility for Electron Microscopy, a node of the Cryo-EM Swedish National Facility, funded by the Knut and Alice Wallenberg, Family Erling Persson and Kempe Foundations, SciLifeLab, Stockholm University and Umeå University. Funding for open access charge: University of Hamburg.
Conflict of interest statement. None declared.
REFERENCES
- 1. Wilson D.N. Ribosome-targeting antibiotics and bacterial resistance mechanisms. Nat. Rev. Microbiol. 2014; 12:35–48. [DOI] [PubMed] [Google Scholar]
- 2. Munita J.M., Arias C.A.. Mechanisms of antibiotic resistance. Microbiol Spectr. 2016; 4: 10.1128/microbiolspec.VMBF-0016-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Antimicrobial Resistance Collaborators Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022; 399:629–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wilson D.N., Hauryliuk V., Atkinson G.C., O’Neill A.J. Target protection as a key antibiotic resistance mechanism. Nat. Rev. Microbiol. 2020; 18:637–648. [DOI] [PubMed] [Google Scholar]
- 5. Dönhöfer A., Franckenberg S., Wickles S., Berninghausen O., Beckmann R., Wilson D.N.. Structural basis for tetm-mediated tetracycline resistance. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:16900–16905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li W., Atkinson G.C., Thakor N.S., Allas U., Lu C.C., Chan K.Y., Tenson T., Schulten K., Wilson K.S., Hauryliuk V.et al.. Mechanism of tetracycline resistance by ribosomal protection protein Tet(O). Nat. Commun. 2013; 4:1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Arenz S., Nguyen F., Beckmann R., Wilson D.N.. Cryo-EM structure of the tetracycline resistance protein TetM in complex with a translating ribosome at 3.9-Å resolution. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:5401–5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sharkey L.K.R., O’Neill A.J.. Antibiotic resistance ABC-F proteins: bringing target protection into the limelight. ACS Infectious Diseases. 2018; 4:239–246. [DOI] [PubMed] [Google Scholar]
- 9. Murina V., Kasari M., Hauryliuk V., Atkinson G.C.. Antibiotic resistance ABCF proteins reset the peptidyl transferase centre of the ribosome to counter translational arrest. Nucleic Acids Res. 2018; 46:3753–3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ero R., Kumar V., Su W., Gao Y.G.. Ribosome protection by ABC-F proteins-Molecular mechanism and potential drug design. Protein Sci. 2019; 28:684–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Crowe-McAuliffe C., Murina V., Turnbull K.J., Kasari M., Mohamad M., Polte C., Takada H., Vaitkevicius K., Johansson J., Ignatova Z.et al.. Structural basis of ABCF-mediated resistance to pleuromutilin, lincosamide, and streptogramin a antibiotics in Gram-positive pathogens. Nat. Commun. 2021; 12:3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Crowe-McAuliffe C., Murina V., Turnbull K.J., Huch S., Kasari M., Takada H., Nersisyan L., Sundsfjord A., Hegstad K., Atkinson G.C.et al.. Structural basis for poxta-mediated resistance to phenicol and oxazolidinone antibiotics. Nat. Commun. 2022; 13:1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mohamad M., Nicholson D., Saha C.K., Hauryliuk V., Edwards T.A., Atkinson G.C., Ranson N.A., O’Neill A.J.. Sal-type ABC-F proteins: intrinsic and common mediators of pleuromutilin resistance by target protection in staphylococci. Nucleic. Acids. Res. 2022; 50:2128–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cox G., Thompson G.S., Jenkins H.T., Peske F., Savelsbergh A., Rodnina M.V., Wintermeyer W., Homans S.W., Edwards T.A., O’Neill A.J.. Ribosome clearance by fusb-type proteins mediates resistance to the antibiotic fusidic acid. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:2102–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tomlinson J.H., Thompson G.S., Kalverda A.P., Zhuravleva A., O’Neill A.J.. A target-protection mechanism of antibiotic resistance at atomic resolution: insights into fusb-type fusidic acid resistance. Sci. Rep. 2016; 6:19524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Duval M., Dar D., Carvalho F., Rocha E.P.C., Sorek R., Cossart P.. HflXr, a homolog of a ribosome-splitting factor, mediates antibiotic resistance. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:13359–13364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang Y., Mandava C.S., Cao W., Li X., Zhang D., Li N., Zhang Y., Zhang X., Qin Y., Mi K.et al.. HflX is a ribosome-splitting factor rescuing stalled ribosomes under stress conditions. Nat. Struct. Mol. Biol. 2015; 22:906–913. [DOI] [PubMed] [Google Scholar]
- 18. Coatham M.L., Brandon H.E., Fischer J.J., Schummer T., Wieden H.-J.. The conserved GTPase HflX is a ribosome splitting factor that binds to the E-site of the bacterial ribosome. Nucleic Acids Res. 2016; 44:1952–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dey S., Biswas C., Sengupta J.. The universally conserved GTPase HflX is an RNA helicase that restores heat-damaged Escherichia coli ribosomes. J. Cell Biol. 2018; 217:2519–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Srinivasan K., Dey S., Sengupta J.. Structural modules of the stress-induced protein hflx: an outlook on its evolution and biological role. Curr. Genet. 2019; 65:363–370. [DOI] [PubMed] [Google Scholar]
- 21. Basu A., Yap M.N.. Disassembly of the staphylococcus aureus hibernating 100S ribosome by an evolutionarily conserved GTPase. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E8165–E8173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hillen H.S., Lavdovskaia E., Nadler F., Hanitsch E., Linden A., Bohnsack K.E., Urlaub H., Richter-Dennerlein R.. Structural basis of GTPase-mediated mitochondrial ribosome biogenesis and recycling. Nat. Commun. 2021; 12:3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Arnaud M., Chastanet A., Debarbouille M.. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 2004; 70:6887–6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Takada H., Roghanian M., Murina V., Dzhygyr I., Murayama R., Akanuma G., Atkinson G.C., Garcia-Pino A., Hauryliuk V.. The C-Terminal RRM/ACT domain is crucial for fine-tuning the activation of ‘Long’ RelA-SpoT homolog enzymes by ribosomal complexes. Front. Microbiol. 2020; 11:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jurenas D., Payelleville A., Roghanian M., Turnbull K.J., Givaudan A., Brillard J., Hauryliuk V., Cascales E.. Photorhabdus antibacterial rhs polymorphic toxin inhibits translation through ADP-ribosylation of 23S ribosomal RNA. Nucleic Acids Res. 2021; 49:8384–8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zivanov J., Nakane T., Forsberg B.O., Kimanius D., Hagen W.J., Lindahl E., Scheres S.H.. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife. 2018; 7:e42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., Agard D.A.. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017; 14:331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rohou A., Grigorieff N.. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015; 192:216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wagner T., Merino F., Stabrin M., Moriya T., Antoni C., Apelbaum A., Hagel P., Sitsel O., Raisch T., Prumbaum D.et al.. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2019; 2:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Scheres S.H. RELION: implementation of a bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012; 180:519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zivanov J., Nakane T., Scheres S.H.W.. A bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ. 2019; 6:5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Heymann J.B. Guidelines for using bsoft for high resolution reconstruction and validation of biomolecular structures from electron micrographs. Protein Sci. 2018; 27:159–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kimanius D., Dong L., Sharov G., Nakane T., Scheres S.H.W.. New tools for automated cryo-EM single-particle analysis in RELION-4.0. Biochem. J. 2021; 478:4169–4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Varadi M., Anyango S., Deshpande M., Nair S., Natassia C., Yordanova G., Yuan D., Stroe O., Wood G., Laydon A.et al.. AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022; 50:D439–D444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Zidek A., Potapenko A.et al.. Highly accurate protein structure prediction with alphafold. Nature. 2021; 596:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Long F., Nicholls R.A., Emsley P., Graaeulis S., Merkys A., Vaitkus A., Murshudov G.N.. AceDRG: a stereochemical description generator for ligands. Acta Crystallogr. D Struct. Biol. 2017; 73:112–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of coot. Acta. Crystallogr. D Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A.et al.. Overview of the CCP4 suite and current developments. Acta. Crystallogr. D Biol. Crystallogr. 2011; 67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Moriarty N.W., Grosse-Kunstleve R.W., Adams P.D.. electronic Ligand builder and optimization workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta. Crystallogr. D Biol. Crystallogr. 2009; 65:1074–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yamashita K., Palmer C.M., Burnley T., Murshudov G.N.. Cryo-EM single-particle structure refinement and map calculation using servalcat. Acta Crystallogr. D Struct. Biol. 2021; 77:1282–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen V.B., Arendall W.B. 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C.. MolProbity: all-atom structure validation for macromolecular crystallography. Acta. Crystallogr. D Biol. Crystallogr. 2010; 66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liebschner D., Afonine P.V., Baker M.L., Bunkoczi G., Chen V.B., Croll T.I., Hintze B., Hung L.W., Jain S., McCoy A.J.et al.. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in phenix. Acta Crystallogr. D Struct. Biol. 2019; 75:861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L.. BLAST+: architecture and applications. BMC Bioinf. 2009; 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Steinegger M., Soding J.. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 2017; 35:1026–1028. [DOI] [PubMed] [Google Scholar]
- 45. Murina V., Kasari M., Takada H., Hinnu M., Saha C.K., Grimshaw J.W., Seki T., Reith M., Putrins M., Tenson T.et al.. ABCF ATPases involved in protein synthesis, ribosome assembly and antibiotic resistance: structural and functional diversification across the tree of life. J. Mol. Biol. 2019; 431:3568–3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Katoh K., Standley D.M.. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013; 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Capella-Gutierrez S., Silla-Martinez J.M., Gabaldon T.. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009; 25:1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nguyen L.T., Schmidt H.A., von Haeseler A., Minh B.Q.. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015; 32:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hoang D.T., Chernomor O., von Haeseler A., Minh B.Q., Vinh L.S.. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018; 35:518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Waterhouse A.M., Procter J.B., Martin D.M., Clamp M., Barton G.J.. Jalview version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009; 25:1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., Ferrin T.E.. UCSF chimerax: meeting modern challenges in visualization and analysis. Protein Sci. 2018; 27:14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Karray F., Darbon E., Oestreicher N., Dominguez H., Tuphile K., Gagnat J., Blondelet-Rouault M.H., Gerbaud C., Pernodet J.L.. Organization of the biosynthetic gene cluster for the macrolide antibiotic spiramycin in streptomyces ambofaciens. Microbiology (Reading). 2007; 153:4111–4122. [DOI] [PubMed] [Google Scholar]
- 53. Rudra P., Hurst-Hess K.R., Cotten K.L., Partida-Miranda A., Ghosh P.. Mycobacterial HflX is a ribosome splitting factor that mediates antibiotic resistance. Proc. Natl. Acad. Sci. U.S.A. 2020; 117:629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chesneau O., Ligeret H., Hosan-Aghaie N., Morvan A., Dassa E.. Molecular analysis of resistance to streptogramin a compounds conferred by the vga proteins of staphylococci. Antimicrob. Agents Chemother. 2005; 49:973–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dar D., Shamir M., Mellin J.R., Koutero M., Stern-Ginossar N., Cossart P., Sorek R.. Term-seq reveals abundant ribo-regulation of antibiotics resistance in bacteria. Science. 2016; 352:aad9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Brodiazhenko T., Turnbull K.J., Wu K.J.Y., Takada H., Tresco B.I.C., Tenson T., Myers A.G., Hauryliuk V.. Synthetic oxepanoprolinamide iboxamycin is highly active against human pathogen Listeria monocytogenes. JAC Antimicrob. Resist. 2022; 4:dlac061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Matzov D., Eyal Z., Benhamou R.I., Shalev-Benami M., Halfon Y., Krupkin M., Zimmerman E., Rozenberg H., Bashan A., Fridman M.et al.. Structural insights of lincosamides targeting the ribosome of Staphylococcus aureus. Nucleic. Acids. Res. 2017; 45:10284–10292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Su W., Kumar V., Ding Y., Ero R., Serra A., Lee B.S.T., Wong A.S.W., Shi J., Sze S.K., Yang L.et al.. Ribosome protection by antibiotic resistance ATP-binding cassette protein. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:5157–5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dunkle J.A., Xiong L., Mankin A.S., Cate J.H.. Structures of the escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:17152–17157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tu D., Blaha G., Moore P., Steitz T.. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell. 2005; 121:257–270. [DOI] [PubMed] [Google Scholar]
- 61. Schlünzen F., Zarivach R., Harms J., Bashan A., Tocilj A., Albrecht R., Yonath A., Franceschi F.. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature. 2001; 413:814–821. [DOI] [PubMed] [Google Scholar]
- 62. Svetlov M.S., Syroegin E.A., Aleksandrova E.V., Atkinson G.C., Gregory S.T., Mankin A.S., Polikanov Y.S.. Structure of Erm-modified 70S ribosome reveals the mechanism of macrolide resistance. Nat. Chem. Biol. 2021; 17:412–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Syroegin E.A., Flemmich L., Klepacki D., Vazquez-Laslop N., Micura R., Polikanov Y.S.. Structural basis for the context-specific action of the classic peptidyl transferase inhibitor chloramphenicol. Nat. Struct. Mol. Biol. 2022; 29:152–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Li Q., Pellegrino J., Lee D.J., Tran A.A., Chaires H.A., Wang R., Park J.E., Ji K., Chow D., Zhang N.et al.. Synthetic group a streptogramin antibiotics that overcome vat resistance. Nature. 2020; 586:145–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tenson T., Lovmar M., Ehrenberg M.. The mechanism of action of macrolides, lincosamides and streptogramin b reveals the nascent peptide exit path in the ribosome. J. Mol. Biol. 2003; 330:1005–1014. [DOI] [PubMed] [Google Scholar]
- 66. Menninger J.R., Coleman R.A.. Lincosamide antibiotics stimulate dissociation of peptidyl-transfer RNA from ribosomes. Antimicrob. Agents Chemother. 1993; 37:2027–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Burdett V. Tet(M)-promoted release of tetracycline from ribosomes is GTP dependent. J. Bacteriol. 1996; 178:3246–3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Trieber C.A., Burkhardt N., Nierhaus K.H., Taylor D.E.. Ribosomal Protection from Tetracycline Mediated by Tet(O): Tet(O) Interaction with Ribosomes Is GTP-Dependent. Biol. Chem. 1998; 379:847–855. [DOI] [PubMed] [Google Scholar]
- 69. Connell S.R., Trieber C.A., Stelzl U., Einfeldt E., Taylor D.E., Nierhaus K.H.. The tetracycline resistance protein Tet(O) perturbs the conformation of the ribosomal decoding centre. Mol. Microbiol. 2002; 45:1463–1472. [DOI] [PubMed] [Google Scholar]
- 70. Sharkey L.K., Edwards T.A., O’Neill A.J.. ABC-F proteins mediate antibiotic resistance through ribosomal protection. MBio. 2016; 7:e01975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Connell S.R., Trieber C.A., Dinos G.P., Einfeldt E., Taylor D.E., Nierhaus K.H.. Mechanism of Tet(O)-mediated tetracycline resistance. EMBO J. 2003; 22:945–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Connell S.R., Tracz D.M., Nierhaus K.H., Taylor D.E.. Ribosomal protection proteins and their mechanism of tetracycline resistance. Antimicrob. Agents Chemother. 2003; 47:3675–3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Polikanov Y.S., Steitz T.A., Innis C.A.. A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome. Nat. Struct. Mol. Biol. 2014; 21:787–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM maps have been deposited in the Electron Microscopy Data Bank (EMDB) with accession codes EMD-15161 [https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-15161] (L. monocytogenes HflXr-50S complex), EMD-15204 [https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-15204] (L. monocytogenes 70S ribosome), EMD-15175 [https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-15175] (L. monocytogenes lincomycin-70S complex), EMD-15864 [https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-15864] (L. monocytogenes lincomycin-50S map) and EMD-15670 [https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-15670] (L. monocytogenes HflX-50S map). Molecular models have been deposited in the Protein Data Bank with accession codes 8A57 [https://doi.org/10.2210/pdb8A57/pdb] (L. monocytogenes HflXr-50S complex), 8A63 [https://doi.org/10.2210/pdb8A63/pdb] (L. monocytogenes 70S ribosome), 8A5I [https://doi.org/10.2210/pdb8A5I/pdb] (L. monocytogenes lincomycin-50S subunit).