Abstract
Hepatic fibrosis (HF) is a worldwide health problem for which there is no medically effective drug treatment at present, and which is characterized by activation of hepatic stellate cells (HSCs) and excessive extracellular matrix (ECM) deposition. The HF model in cholestatic rats by ligating the common bile duct was induced and the differentially expressed miRNAs in the liver tissues were analyzed by microarray, which showed that miR-22-3p and miR-29a-3p were significantly downregulated in bile-duct ligation (BDL) rat liver compared with the sham control. The synergistic anti-HF activity and molecular mechanism of miR-22-3p and miR-29a-3p by targeting AKT serine/threonine kinase 3 (AKT3) in HSCs were explored. The expression levels of miR-22-3p and miR-29a-3p were downregulated in activated LX-2 and human primary normal hepatic fibroblasts (NFs), whereas AKT3 was found to be upregulated in BDL rat liver and activated LX-2 cells. The proliferation, colony-forming, and migration ability of LX-2 were inhibited synergistically by miR-22-3p and miR-29a-3p. In addition, cellular senescence was induced and the expressions of the LX-2 fibrosis markers COL1A1 and α-SMA were inhibited by miR-22-3p and miR-29a-3p synergistically. Subsequently, these two miRNAs binding to the 3’UTR of AKT3 mRNA was predicted and evidenced by the luciferase reporter assay. Furthermore, the proliferation, migration, colony-forming ability, and the expression levels of COL1A1 and α-SMA were promoted and cellular senescence was inhibited by AKT3 in LX-2 cells. Thus, miR-22-3p/miR-29a-3p/AKT3 regulates the activation of HSCs, providing a new avenue in the study and treatment of HF.
Keywords: Hepatic fibrosis, LX-2, HSC activation, miR-22-3p, miR-29a-3p, AKT3
Impact Statement
Hepatic fibrosis (HF) is a worldwide health problem, and no medically effective drug treatment is available currently. Therefore, understanding the pathogenesis of HF and seeking promising diagnostic and therapeutic targets for HF are important. This study first shows that cell proliferation, migration, and colony-forming ability of LX-2 are synergistically inhibited by miR-22-3p and miR-29a-3p through targeting AKT serine/threonine kinase 3 (AKT3), which is of great significance in the development of HF. Our research offers three potential therapeutic targets for HF and explores the molecular mechanism of the miR-22-3p/miR-29a-3p/AKT3 axis in HF.
Introduction
Hepatic fibrosis (HF) is chronic liver damage caused by viruses, autoimmunity, alcoholism, non-alcoholic fat, and abnormal copper and iron metabolism, and it can develop into cirrhosis in severe cases. Its main features are the activation of hepatic stellate cells (HSCs) and excessive deposition of extracellular matrix (ECM). 1 HSCs that account for approximately 5% of all hepatocytes were first described by Kupffer in 1876, and they exist in the space between parenchymal cells (hepatocytes) and sinusoidal endothelial cells. 2 HSCs not only participate in liver development, differentiation, regeneration, immune regulation, inflammatory response, and liver blood flow control, but also regulate the occurrence and development of various liver diseases, such as in the excessive ECM deposition and scar formation caused by the activation of HSCs. Nevertheless, as the complexity of HSC activation and the pathogenesis of HF are not fully understood, it is therefore critical to clarify the regulatory mechanism in order to improve the treatment of HF.
AKT (protein kinase B, PKB) – comprising AKT1, AKT2, and AKT33 – is a member of the protein kinase family that regulates different cellular functions and is differentially expressed in most human tumors. 4 ErbB2 receptor tyrosine kinase 2 (ErbB2)+ breast cancer cells express low level estrogen receptor alpha (ERα), and its proliferation and endocrine resistance are promoted by AKT3. 5 ErbB2+ERα– mouse breast cancer C4 cell proliferation is significantly reduced by knockout of AKT1 and AKT3, while a slight effect on cell proliferation from knocking out AKT2 is observed. 5 In triple-negative breast cancer, AKT3 is a target of miR-433 and overexpression of AKT3 significantly rescues the inhibitory effect of miR-433 on the proliferative ability of BT-549 cells. 6 The normal expression of AKT3 is essential for brain growth in mice, and AKT3 is often targeted by miRNAs to regulate autophagy to reduce peripheral nerve injury.7,8 Anxiety-like behavior, depression, and memory impairment in mice are found to be related to the decrease or loss of AKT3 expression. 9 Cholesterol ester accumulation and foam cell formation in mouse macrophages are specifically inhibited by AKT3. 10 However, the detailed mechanism of AKT3 upregulation in HF remains to be clarified.
Recently, with improved understanding of the physiology and pathology of HF, the relationship between miRNAs and HF has attracted the attention of many scholars. miRNAs are non-coding single-stranded molecules with lengths of approximately 21–25 nucleotides. They exist stably in body fluids, their sequences are highly conserved in different species, and their expressions are unique to individual tissues or biological states, which makes them valuable biomarkers. 11 An increasing number of miRNA families which are involved in HSC proliferation, activation, apoptosis, and regulation of HF are found. The relationship among miRNA dysregulation, HF, and other human diseases has received increased attention, making miRNAs a focus for the development of new therapeutic strategies. Based on data from miRNA prediction databases TargetScan, DIANA, miRDB, and starBase, AKT3 may be related to two miRNAs (miR-22-3p and miR-29a-3p) involved in HF.
First, the expressions of miR-22-3p and miR-29a-3p were reduced in the livers of bile-duct ligation (BDL) rats, as indicated by microarray analysis. Next, the role of these two miRNAs in HF and their interaction with AKT3 were studied. HSC activation was related to the reduction of these two miRNAs and upregulation of AKT3, which was identified as the common functional target gene of miR-22-3p and miR-29a-3p. In addition, the proliferation, migration, and activation of LX-2 cells were synergistically inhibited by these two miRNAs and were promoted by the common target gene AKT3, thereby affecting liver fibrosis. Thus, the molecular mechanism of miR-22-3p and miR-29a-3p on HF through AKT3 needs to be further clarified.
Materials and methods
BDL rat model
The experimental protocols of all animals involved in this experiment were agreed on by the Laboratory Animal Ethics Committee (LAEC-NCST-2020187). Sprague Dawley male rats (14 in total, aged 2 months, weight 200–240 g) were from Beijing HFK Bioscience Co. Ltd. (License no. SYXK [JI] 2020-007). Rats were kept in a standard clean environment of 23 ± 2°C with sufficient feed and water. Overall, seven rats underwent sham operation, and seven rats underwent BDL, both of which have been described previously. 12 On the 14th day after the operation, the rats were sacrificed by exsanguination through the abdominal aorta after anesthesia, and the liver tissues were taken.
Histology and immunohistochemistry
Paraffin sections were prepared after fixing the liver tissue. Hematoxylin and eosin (H&E; Baso, Zhuhai, China), Sirius red staining kit and Masson staining kit (Servicebio Technology, Wuhan, China) were applied according to the instructions. Paraffin sections were dewaxed in xylene and alcohol and washed with phosphate-buffered saline (PBS) after hydration. Next, the sections were covered with prepared ethylenediaminetetraacetic acid (EDTA) solution (Wanleibio, Shenyang, China), and were repaired under high pressure for 3 min. This was followed by applying peroxidase blocking agent (ReportBio, Hebei, China) for 10 min, and washing with PBS. Next, sections with anti α-SMA antibody (1:200, SQab18108, Arigobio, ShangHai, China) were incubated overnight at 4°C and then incubated with secondary antibody (ZSGB-BIO, Beijing, China) at 37°C for 2 h. Diaminobenzidine kit (SGB-BIO, Beijing, China) was used to visualize antibody binding areas.
miRNA microarray and determination of differentially expressed genes
Differential miRNA expressions between the sham group and BDL group rats were analyzed using a rat gene expression microarray (Agilent Technologies, Palo Alto, CA, USA), using an 8 × 60 K chip in the liver tissue. The TIFF format image data of the Agilent miRNA expressions chip following hybridization scanning were preprocessed and analyzed using feature extraction software.
Cell culture
The use of human primary normal fibroblasts (NFs) was authorized by the Ethics Committee of Tangshan People’s Hospital (RMYY-LLKS-2020-002). 13 Human primary NFs were cultured in Dulbecco’s modified Eagle’s F12 medium (DMEM/F12; Eallbio, Beijing, China) containing 10 ng/mL of epidermal growth factor (EGF; PeproTech, Rocky Hill, NJ, USA), 20% fetal bovine serum (FBS; Biological Industries, Kibbutz Beit Haemek, Israel) and 1% penicillin/streptomycin (PS; Eallbio, Beijing, China). Human LX-2 cells from Peking Union Medical College Hospital were cultured at 37°C and 5% CO2 in DMEM (Eallbio, Beijing, China) containing 10% FBS and 1% PS. LX-2 cells were treated with transforming growth factor-beta 1 (TGF-β1; PeproTech, Rocky Hill, NJ, USA) at a concentration of 5 ng/mL to activate them.
qRT–PCR
Total RNA was extracted from cells and liver tissues by TRIzol reagent, and cDNA was synthesized using the reverse transcription kit (Mei5bio, Beijing, China) and the miRNA First Strand cDNA Synthesis kit (Sangong Biotech, Shanghai, China). Amplification was performed in the ABI 7500 PCR system (Applied Biosystems, Germany) as described. The reaction conditions were as follows: 30 s at 95°C, 5 s at 95°C and 34 s at 60°C for 40 cycles. GAPDH or U6 was used as internal parameter. The relative expression level of genes was calculated by the 2–△△CT method. The primer sequences are listed in Table 1.
Table 1.
Primer sequence.
| Primer | Sequences (5′→3′) |
|---|---|
| α-SMA (human) | F: CATGAAGTGTGACATCGACATC R: TGATCTTGATCTTCATGGTGCT |
| COL1A1 (human) | F: AAAGATGGACTCAACGGTCTC R: CATCGTGAGCCTTCTCTTGAG |
| AKT3 (human) | F: AGATGCAGCCACCATGAAGACATTC R: GTCTACTGCTCGGCCATAGTCATTATC |
| GAPDH (human) | F: CCGCATCTTCTTGGGCAGTG R: TCCCGTTGATGACCAGCTTC |
| α-SMA (rat) | F: GCGTGGCTATTCCTTCGTGACTAC R: CATCAGGCAGTTCGTAGCTCTTCTC |
| COL1A1 (rat) | F: TGTTGGTCCTGCTGGCAAGAATG R: GTCACCTTGTTCGCCTGTCTCAC |
| AKT3 (rat) | F: AGATGCAGCCACCATGAAGACATTC R: GTCTACTGCTCGGCCATAGTCATTATC |
| GAPDH (rat) | F: GACATGCCGCCTGGAGAAAC R: AGCCCAGGATGCCCTTTAGT |
| miR-22-3p (human/rat) | F: CAAGCTGCCAGTTGAAGAACTGT |
| miR-29a-3p (human/rat) | F: CCGTAGCACCATCTGAAATCGGTTA |
| U6 (human/rat) | F: CTCGCTTCGGCAGCACATA |
Western blotting
The proteins in the cells and liver tissue were extracted with phenylmethylsulfonyl fluoride (PMSF)-containing radioimmunoprecipitation assay (RIPA) lysate (Applygen, Beijing, China), and the bicinchoninic acid (BCA) analysis kit (Beyotime Biotechnology, Shanghai, China) was used to detect the concentration. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis was used to isolate proteins which were then transferred to polyvinylidene fluoride membrane (PVDF; Millipore, Bedford, MA, USA). The membrane was sealed with 5% skimmed milk for 2 h, and the first antibody was applied overnight at 4°C. The next day, the secondary antibody was incubated at room temperature for 2 h. Finally, the protein strength was detected by ECL reagent (Applygen, Beijing, China). The antibodies used were as follows: GAPDH (ab 9485; Abcam; 1:5000), AKT3 (ab 2839860; Affinity; 1:3000), COL1A1 (ab 64883; Abcam; 1:1500) and α-SMA (ab 244177; Abcam; 1:1000).
Mature miRNAs and siRNA transfection
miR-22-3p mimics, miR-29a-3p mimics, mimics negative controls (NC), miR-22-3p inhibitor, miR-29a-3p inhibitor, inhibitor NC (Zhongshi Tongchuang, Tianjin, China), si-AKT3, and si-NC (AnHui General Biotechnology, Anhui, China) were transfected into LX-2 at a final concentration of 50 nM. Transfection was performed according to the instructions of the transfection reagent Lipofectamine® 2000 (Invitrogen, Carlsbad, CA, USA). The pcDNA3.1-AKT3 and pcDNA3.1 plasmids were synthesized by AnHui General Biotechnology Company (Anhui, China), and the transfection dose was 2 µg/well (6-well plate).
Cell counting kit-8 assay
The transfected cells were inoculated into 96 well plates at a density of 2000 cells/well and at 37°C and 5% CO2. In addition, 100 µL of medium and 10 μL of cell counting kit-8 (CCK-8) reagent (Zoman Biotechnology, Beijing, China) were added at 24, 48, 72, and 96 h. The absorbance value at 450 nm in each group was detected after incubation for 2 h.
Transwell assay
Transfected LX-2 cells were inoculated in a Transwell (Corning, NY, USA) upper chamber, and serum-free DMEM medium was used. The culture medium containing 20% FBS was added in the lower chamber and incubated for 24 h. The cells were fixed and stained with crystal violet for 5 min. After cleaning and drying, the cells were observed under a microscope (Olympus, Tokyo, Japan).
Wound healing assay
After the transfected LX-2 were inoculated on the six-well plates, scratches were made on the plate with a pipette suction. After cleaning, serum-free medium was added to continue the culture. Photos were taken at 0, 24, and 48 h. ImageJ software (National Institutes of health, Bethesda, Maryland, USA) was used to analyze cell migration.
Colony formation
The transfected cells were inoculated on a six-well plate and cultured for 1 week with 3000 cells/well. After the cells were fixed, they were stained with crystal violet (Solarbio, Beijing, China), then washed, dried, and photographed for counting.
Senescence-associated β-galactosidase (SA-β-gal) staining
The transfected cells were treated following the instructions of the SA-β-gal staining kit (Beyotime, Shanghai, China). The randomly selected areas were analyzed by light microscope to calculate the percentage of positive cells.
miRNA target gene prediction
Four databases were used to predict the common target genes of miRNAs, as shown in Table 2. The miRNAs and genes were analyzed by Gene Ontology (GO) and Kyoto Encyclopedia Genes and Genomes (KEGG) databases. RNAhybrid was used to analyze the binding possibility of miRNA and common target gene. 14 The ECR browser was used to analyze the species conservation of miRNAs. String was used to analyze the protein–protein interaction (PPI).
Table 2.
Database website.
| Name | Database URL |
|---|---|
| StarBase | https://starbase.sysu.edu.cn/ |
| miRDB | http://www.mirdb.org/ |
| DIANA | http://diana.imis.athena-innovation.gr/DianaTools/index.php |
| Targetscan | http://www.targetscan.org/vert_72/ |
| RNAhybrid | http://bibiserv.techfak.uni-bielefeld.de/rnahybrid |
Plasmid construction and luciferase reporter assay
The sequences of the common target gene and the two miRNA binding sites were inserted into the psiCHECK2.0 vector (AnHui General Biotechnology, Anhui, China), located between the XhoI-NotI sites. Three mutant plasmids and one wild-type plasmid were constructed, which lacked or contained miRNA targets in the 3’UTR of AKT3, as shown in Table 3. Overall, 500 ng plasmids, 20 pmol miRNAs, and 1 μL Lip2000 were added into a 24-well culture plate to transfect LX-2, and the blank medium was supplemented to 500 μL. After 48 h, the activities of firefly luciferase and renilla luciferase in cells were detected by use of the dual luciferase assay kit (Beyotime, Shanghai, China), and their ratio was calculated.
Table 3.
Plasmids used in the luciferase reporter assay.
| Plasmid | miRNAs target sites | Location |
|---|---|---|
| 1 | miR-22-3p (contained) miR-29a-3p (contained) |
position 2004–2011 (+)/position 1667–1674 (+) |
| 2 | miR-22-3p (mutated) miR-29a-3p (contained) |
position 2004–2011 (–)/position 1667–1674 (+) |
| 3 | miR-22-3p (contained) miR-29a-3p (mutated) |
position 2004–2011 (+)/position 1667–1674 (–) |
| 4 | miR-22-3p (mutated) miR-29a-3p (mutated) |
position 2004–2011 (–)/position 1667–1674 (–) |
miRNA: micro ribonucleic acid.
Immunofluorescence assay
After high-pressure repair or cell fixation, the samples were infiltrated with 0.1% Triton X-100 (Solarbio, Beijing, China) for 10 min. After sealing treatment, the primary antibody was treated at 4°C overnight. The next day, the experimental samples were kept away from the light and incubated with Alexa fluor 555 labeled donkey anti rabbit IgG or Alexa fluor 488 labeled goat anti mouse IgG (Beyotime Biotechnology, Shanghai, China, 1:500) for 2 h. The samples were then stained with DAPI (Beyotime Biotechnology, Shanghai, China) for 5 min, washed and dried, and then sealed. Images were taken under a fluorescence microscope (Olympus, Tokyo, Japan). α-SMA, AKT3, p16, and p21 (Affinity, Cincinnati, OH, USA; 1:300) antibodies were used in the study.
Statistical analysis
One-way analysis of variance (ANOVA) was used to evaluate the synergistic effect of miRNAs, and Student’s t-test was used to analyze the differences between two groups. The above experiments were conducted 3 times. GraphPad Prism software (GraphPad software, Inc.) was used for analysis and graph plotting. The data are represented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, and NS.
Results
Downregulated expression of miR-22-3p and miR-29a-3p in BDL rat liver and activated HSCs
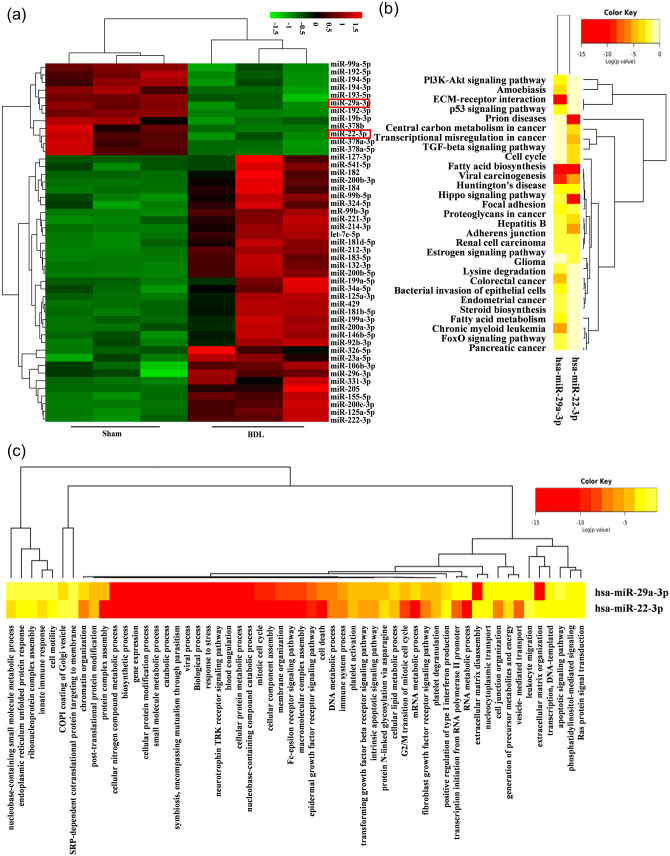
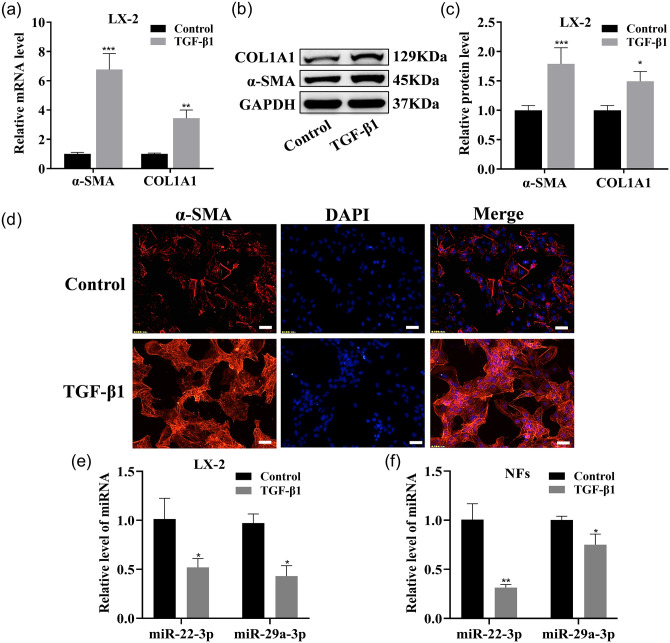
To systematically identify the miRNAs involved in the pathophysiology of liver fibrosis (HF), the miRNA expression profiles in the sham and BDL rat livers were obtained using microarray analysis. Two major clusters, one set closely associated with the sham group and the other with the BDL group, were shown by unsupervised hierarchical cluster analysis. Compared with the sham group, in the BDL group there were 179 upregulated miRNAs and 137 downregulated miRNAs, which included miR-22-3p and miR-29a-3p (Figure 1(a)). These two miRNAs are related to the phosphatidylinositol 3-kinase-AKT (PI3K-AKT) signaling pathway, p53 signaling pathway, TGF-β signaling pathway, and HF occurrence and development, as analyzed by KEGG and GO (Figure 1(b) and (c)). COL1A1 and α-SMA expression levels were significantly upregulated in activated LX-2 with TGF-β1 (Figure 2(a) to (c)). Accordingly, increased α-SMA expression in activated LX-2 was shown by immunofluorescence (IF) analysis (Figure 2(d)). The expression levels of these two miRNAs in activated LX-2 and NFs were significantly downregulated (Figure 2(e) to (f)).
Figure 1.
Differential expression of genes in BDL rat liver. (a) Microarray analysis for miRNAs was used in sham operated or BDL rat liver tissue, n = 3. Hierarchical cluster analysis was performed for differential expression miRNAs. Black, no difference in expression; bright green, low expression; bright red, high expression. (b and c) KEGG and GO analyses of miR-22-3p and miR-29a-3p. (A color version of this figure is available in the online journal.)
Figure 2.
miR-22-3p and miR-29a-3p expressions in activated LX-2, activated NFs. (a to c) qRT–PCR and Western blotting were used to detect COL1A1 and α-SMA mRNA and protein expression in LX-2. (d) IF images of activated LX-2 stained for α-SMA (red, ×200 magnification). Scale bars = 50 μm. (e and f) These two miRNAs were tested by qRT–PCR in LX-2 and NFs. (A color version of this figure is available in the online journal.)
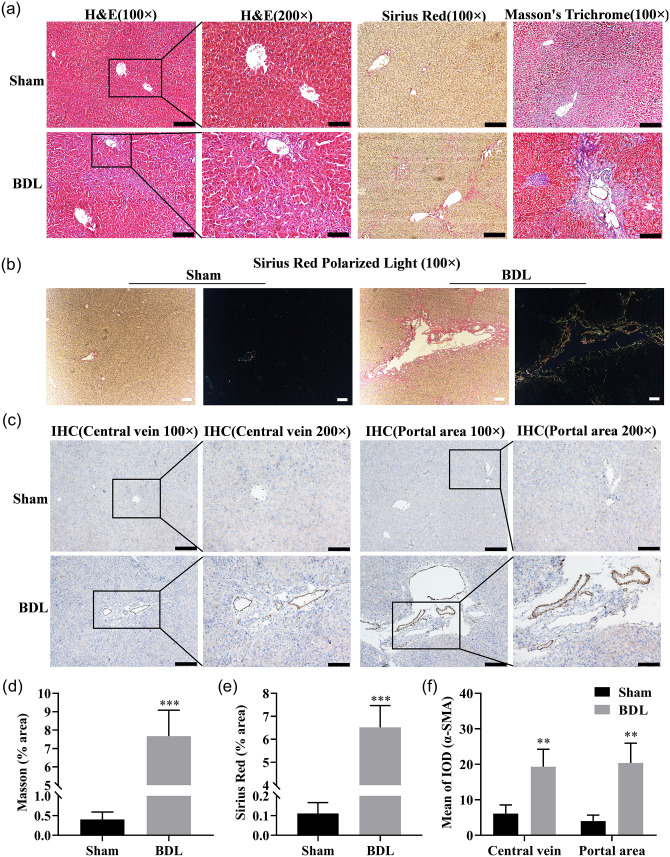
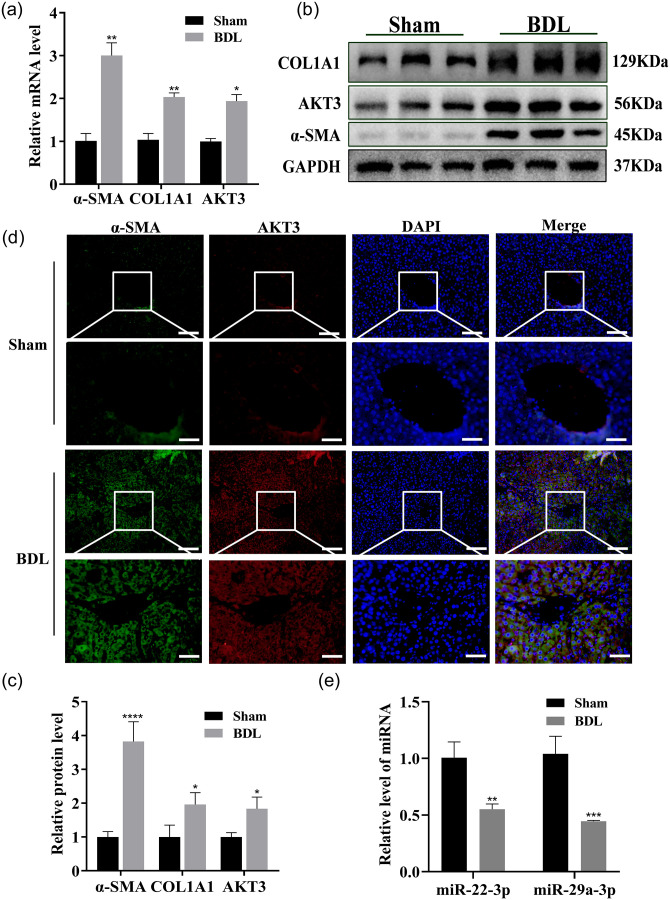
The levels of deposition of collagen, the number of collagen fibers, and Type I and III collagen were increased in BDL rat liver tissue sections, as detected by Sirius Red staining, Masson’s trichrome staining, and Sirius Red polarization, respectively, compared with those in the sham rat liver tissue sections (Figure 3(a), (b), (d) and (e)). In addition, upregulation of α-SMA in the central venous and portal regions in liver tissue were observed, based on immunohistochemical analysis (Figure 3(c) and (f)). The mRNA and protein expression levels of COL1A1 and α-SMA were significantly upregulated, as measured by qRT-PCR and western blot, and the α-SMA expression level was enhanced in the liver tissue of BDL rats, as shown by IF (Figure 4(a) to (d)). The expression levels of these two miRNAs in the liver tissue of BDL rats were significantly downregulated (Figure 4(e)). Thus, these two miRNAs likely play critical roles in HF.
Figure 3.
Histological analysis of rat liver in sham operation group and BDL group (n = 7 per group). (a) The rat liver tissue sections were subjected to H&E (hematoxylin-eosin staining), Sirius Red staining, and Masson’s trichrome (100 ×, scale bars = 200 µm; 200 ×, scale bars = 100 µm). (b) Representative pictures of Sirius Red polarized light on rat liver tissue sections (100 ×, scale bars = 100 µm; Type I collagen: red or yellow, Type III collagen: green). (c) Immunohistochemical staining for α-SMA in the central venous and portal regions of rat liver tissue sections (100 ×, scale bars = 200 µm; 200 ×, scale bars = 100 µm). (d and e) Quantification of the Masson and Sirius Red positive area. (f) Quantitation of mean integrated optical density (IOD) of α-SMA positive stained tissues. (A color version of this figure is available in the online journal.)
Figure 4.
miR-22-3p and miR-29a-3p expressions in BDL rat liver. (a to c) The COL1A1, α-SMA, and AKT3 mRNA and protein expression in the liver tissues of sham and BDL rats were examined by qRT–PCR (a) and Western blotting (b and c) n = 7 per group. (d) The α-SMA and AKT3 expression were examined by co-IF (α-SMA: green, AKT3: red, nuclear: blue; 200 ×, scale bars = 50 μm). (e) These two miRNAs were examined by qRT–PCR in liver tissues from sham and BDL rats. (A color version of this figure is available in the online journal.)
miR-22-3p/miR-29a-3p synergistically suppress LX-2 proliferation and activation
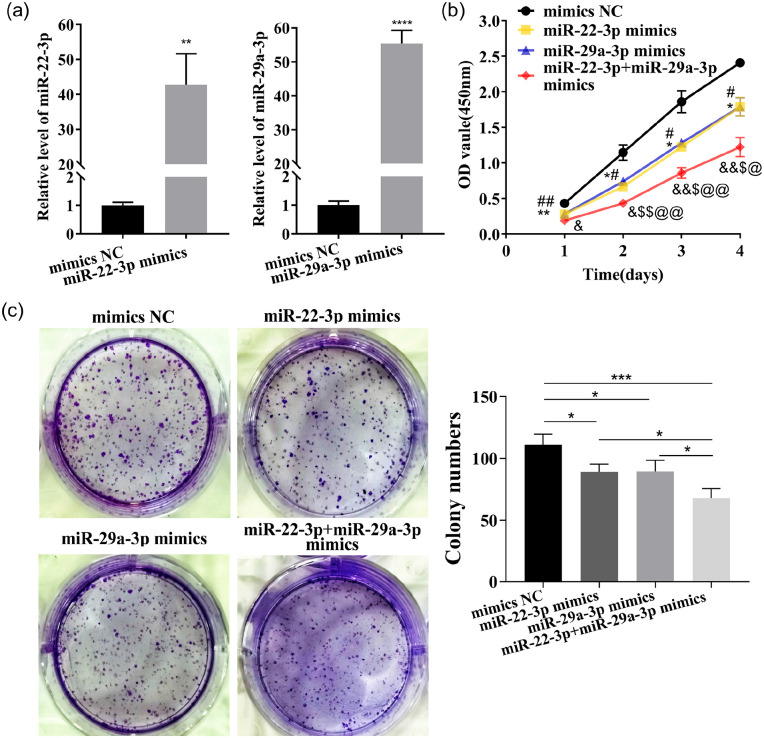
The transfected LX-2 cells were divided into mimics NC, miR-22-3p mimics, miR-29a-3p mimics, miR-22-3p+miR-29a-3p mimics (the combination of both at half dose), inhibitor NC, miR-22-3p inhibitor, miR-29a-3p inhibitor, miR-22-3p+miR-29a-3p inhibitor (the combination of both at half dose). miR-22-3p and miR-29a-3p expressions in LX-2 were dramatically enhanced or decreased by the transfection of their mimics or inhibitors (Figure 5(a) and Supplementary Figure 1(a)).
Figure 5.
Overexpression of miR-22-3p and miR-29a-3p synergistically restrain LX-2 proliferation. (a) These two miRNAs expressions were detected by qRT–PCR in LX-2 transfected with their mimics. (b)The proliferation of LX-2 ability was detected by CCK-8 assay. (c) Colony-forming ability was tested. (A color version of this figure is available in the online journal.)
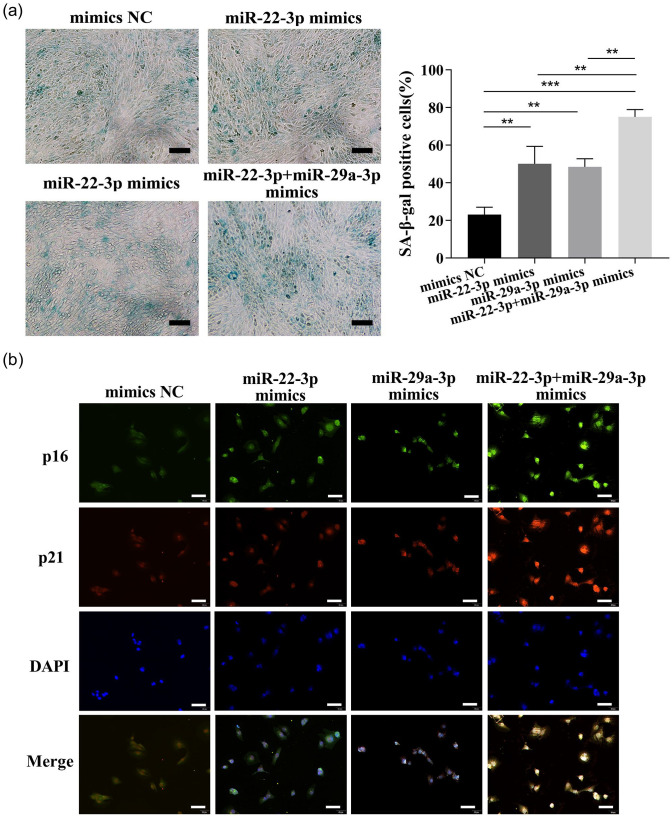
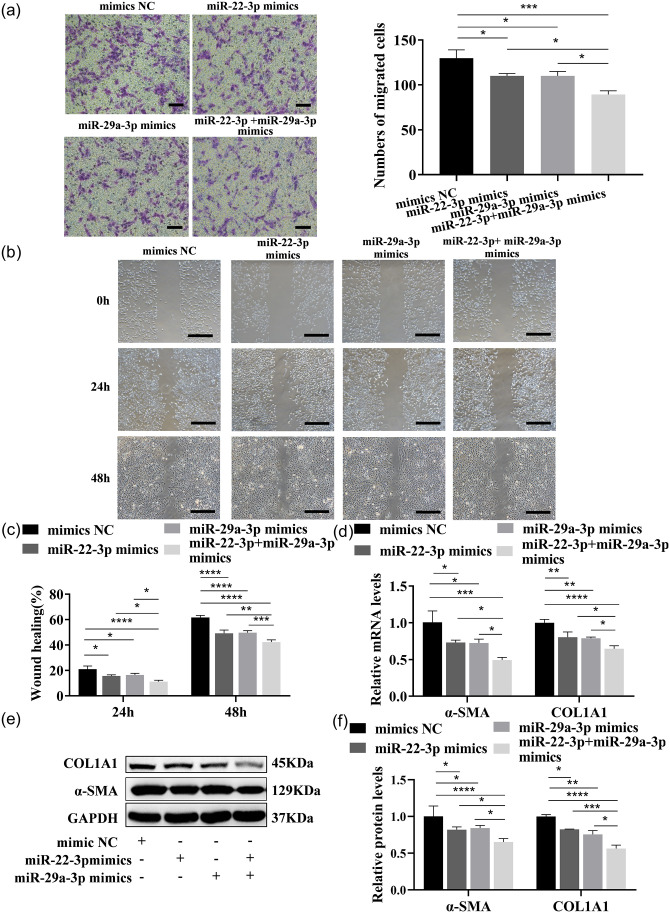
The proliferation and the colony-forming ability of LX-2 were strikingly inhibited by both these two miRNAs, as detected by CCK-8 analysis or the colony formation assay (Figure 5(b) and (c) and Supplementary Figure 1(b) to (c)). Cellular senescence was induced, and the expressions of senescence markers p16 and p21 were enhanced by these two miRNAs, as detected by β-galactosidase staining and cell IF (Figure 6(a) and (b) and Supplementary Figure 2(a) to (b)). Cell migration was inhibited by these two miRNAs, as tested by Transwell and wound healing assays (Figure 7(a) to (c) and Supplementary Figure 3(a) to (c)). In addition, COL1A1 and α-SMA expressions were significantly reduced or induced by these two miRNAs mimics or inhibitors (Figure 7(d) to (f) and Supplementary Figure 3(d) to (f)). Interestingly, a better inhibitory effect on the cell proliferation, migration, and activation was shown by the combination of both at half-dose transfection, compared with full-dose single miRNAs treatments. Thus, these two miRNAs likely function separately or synergistically to inhibit LX-2 activation.
Figure 6.
Overexpression of miR-22-3p and miR-29a-3p synergistically induce cellular senescence. (a) The function of these two miRNAs mimics on cellular senescence was detected by SA-β-gal staining analysis. Scale bars = 200 μm. (b) The p16 and p21 expressions were examined by co-IF (p16: green, p21: red, nuclear: blue; 200 ×, scale bars = 50 μm). (A color version of this figure is available in the online journal.)
Figure 7.
miR-22-3p and miR-29a-3p overexpression synergistically inhibit LX-2 migration and activation. (a) The migration ability of each group of cells transfected with these two miRNAs mimics was tested by Transwell assays. Scale bars = 200 μm. (b and c) The migration ability was tested in each group of cells transfected with these two miRNAs mimics. Scale bars = 500 μm. (d to f) Expressions of COL1A1 and α-SMA mRNA and protein were measured by qRT–PCR (d) and Western blot (e and f) in LX-2 transfected with these two miRNAs mimics. (A color version of this figure is available in the online journal.)
Synergistic mechanism of miR-22-3p and miR-29a-3p
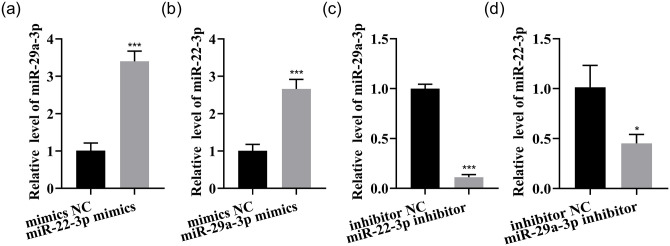
Given that both of these miRNAs inhibit LX-2 proliferation and activation and that the half-dose combination of these two miRNAs has a better inhibitory effect than a single miRNA, the synergistic function of these miRNAs was investigated. An approximately 2.4-fold increase of miR-29a-3p was caused by miR-22-3p overexpression, and an approximately 1.7-fold increase of miR-22-3p was caused by miR-29a-3p overexpression (Figure 8(a) and (b)). In addition, a reduction of mature miR-29a-3p by 89% was caused by miR-22-3p inhibition, and a reduction of mature miR-22-3p by 55% was caused by inhibition of miR-29a-3p (Figure 8(c) and (d)). Thus, these two miRNAs may influence each other through specific genes.
Figure 8.
Synergistic mechanism of miR-22-3p and miR-29a-3p. (a to d) The expressions of these two miRNAs were measured by qRT–PCR.
Bioinformatics analysis predicts miR-22-3p and miR-29a-3p target genes
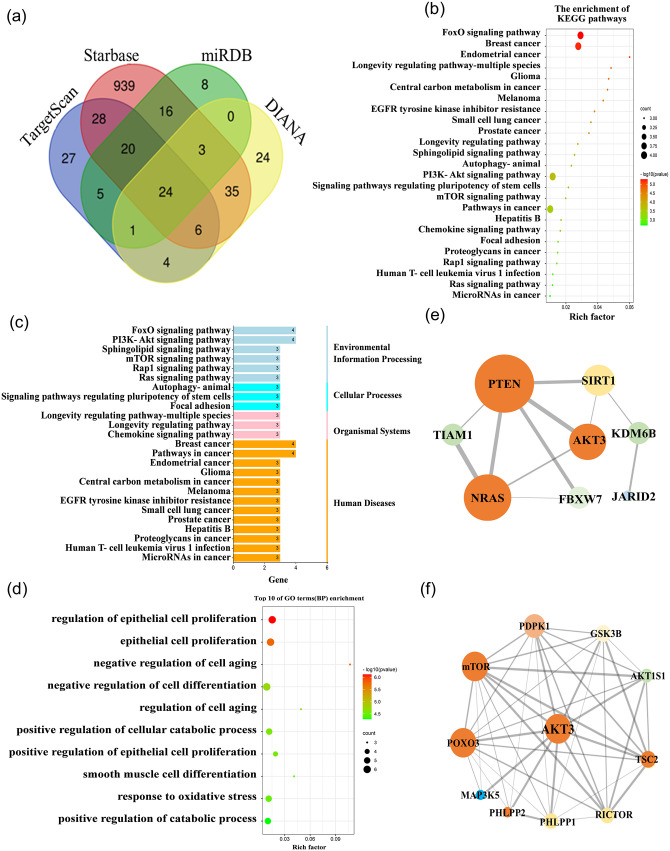
In total, 24 possible candidate common target genes for these two miRNAs were predicted by TargetScan, DIANA, miRDB, and starBase (Figure 9(a)). KEGG pathway analysis and GO analysis were performed on 24 candidate common target genes, which were associated with the PI3K-AKT signaling pathway, and AKT3 was a critical mediator (Figure 9(b) to (d)). Next, the research status, site prediction scores, species conservation, and other comprehensive conditions were combined to initially determine AKT3 as a possible common target gene for these two miRNAs. Proteins of possible target genes of these two miRNAs associated with AKT3 were further analyzed by STRING, which generated a PPI diagram (Figure 9(e)), and the connections between AKT3 and other proteins were predicted (Figure 9(f)).
Figure 9.
Bioinformatics screening for the common bona fide target of miR-22-3p and miR-29a-3p. (a) Common target of these two miRNAs in four databases. (b to e) KEGG (b), pathway enrichment (c), Gene Ontology (d), and STRING (e) analysis of the 24 candidate target genes. (f) PPI network of AKT3. (A color version of this figure is available in the online journal.)
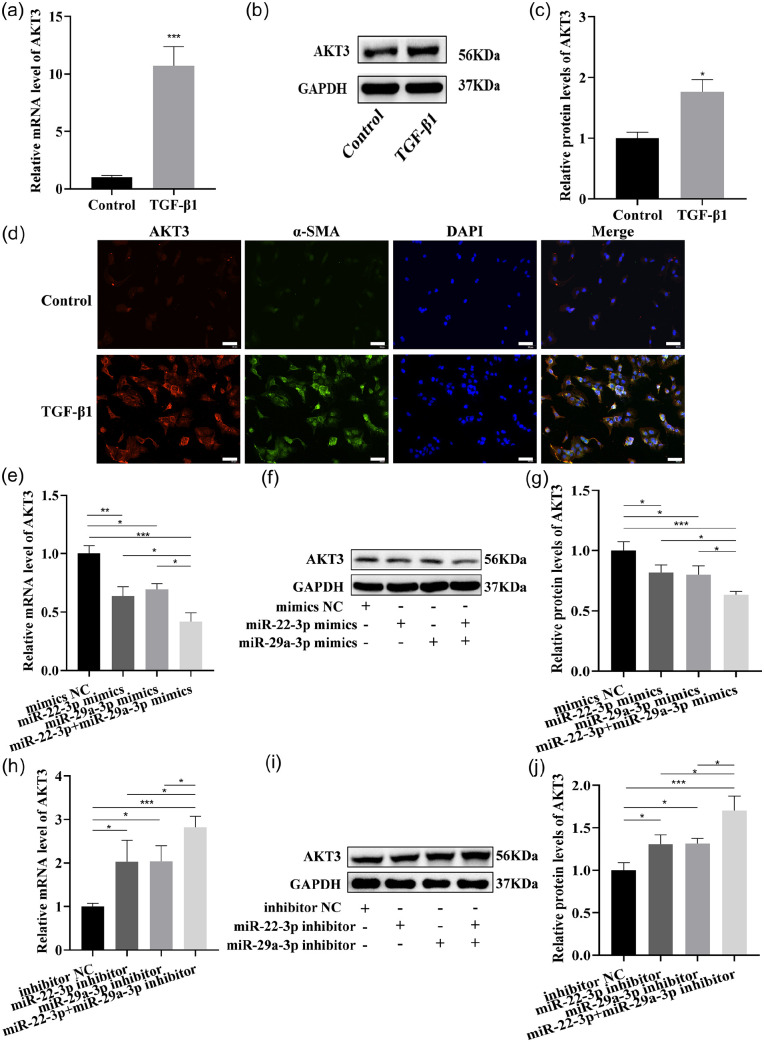
miR-22-3p and miR-29a-3p both target AKT3 and regulate its expression
The upregulation of AKT3 expression was verified in the liver tissue of BDL rats and activated LX-2 cells by qRT-PCR (Figures 4(a) and 10(a)), Western blot (Figures 4(b) and (c) and 10(b) and (c)), as well as co-IF analysis of α-SMA and AKT3 (Figures 4(d) and 10(d)). Next, LX-2 was treated with these two miRNAs mimics, revealing that the expression of AKT3 mRNA and protein were significantly inhibited by these two miRNAs mimics (Figure 10(e) to (g)). Correspondingly, the AKT3 expression was significantly enhanced by these two miRNAs inhibitor treatments (Figure 10(h) to (j)).
Figure 10.
miR-22-3p and miR-29a-3p regulate AKT3 expression in LX-2. (a to c) AKT3 expression was examined in activated LX-2 by qRT–PCR (a) and Western blotting (b and c). (d) co-IF analysis of α-SMA and AKT3 (AKT3: red, α-SMA: green; nuclear: blue; 200 ×, scale bars = 50 μm). (e to g) AKT3 expression was examined in LX-2 transfected with these two miRNAs mimics by qRT–PCR (e), Western blotting (f and g). (h to j) AKT3 expression was examined in LX-2 transfected with these two miRNAs inhibitors by qRT–PCR (h), Western blotting (i and j). (A color version of this figure is available in the online journal.)
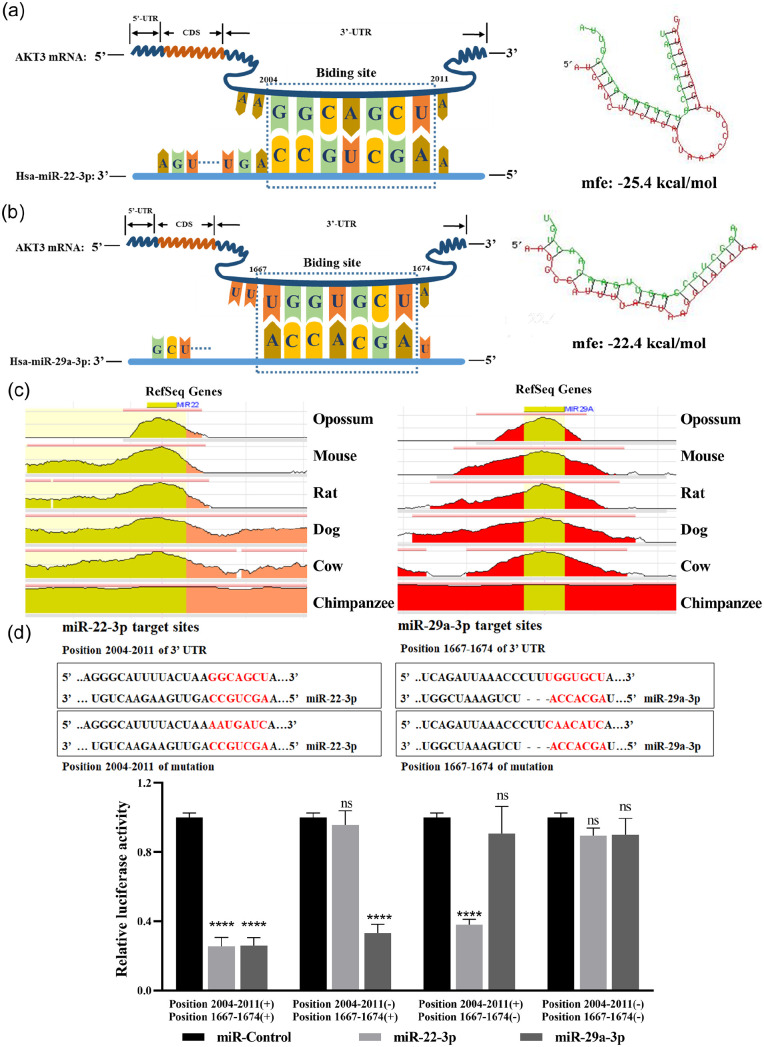
The TargetScan database and RNAhybrid database were used to find the potential binding sites and minimum free energy between these two miRNAs and AKT3, respectively (Figure 11(a) and (b)). These two miRNAs are highly conserved in primates and mammals based on the ECR browser (Figure 11(c)). The Renilla luciferase activity was dramatically decreased in the presence of miR-22-3p or miR-29a-3p and Plasmid 1, and this plasmid contained the 3’UTR of wild-type AKT3 (position 2004–2011 [+] position 1667–1674 [+]). The Renilla luciferase activity was decreased when miR-29a-3p and Plasmid 2 co-existed, but no significant difference was noted in the presence of miR-22-3p and Plasmid 2, which contained the mutated binding sites of miR-22-3p and the wild-type binding sites of miR-29a-3p (position 2004–2011 [–] position 1667–1674 [+]). The Renilla luciferase activity was decreased when miR-22-3p and Plasmid 3 co-existed, but no significant difference was noted in miR-29a-3p and Plasmid 3 co-transfection, which contained the wild-type binding sites of miR-22-3p and the mutated binding sites of miR-29a-3p (position 2004–2011 [+] position 1667–1674 [–]). In addition, Plasmid 4 contained both the mutated binding sites of miR-22-3p and miR-29a-3p (position 2004–2011 [–] position 1667–1674 [–]), and when co-transfected with miR-22-3p/miR-29a-3p mimics, the Renilla luciferase activity was not statistically significant (Figure 11(d)). These results reveal that AKT3 is the common bona fide target gene of these two miRNAs.
Figure 11.
The common bona fide target gene of miR-22-3p and miR-29a-3p is AKT3. The minimum free energy (MFE) of AKT3 and miR-22-3p (a) or miR-29a-3p (b) was predicted by RNAhybrid and the respective binding sites. (c) Evolutionary conservation of these two miRNAs assessed using the ECR browser. (d) These two miRNAs binding sites on AKT3. Four plasmids were used in the luciferase reporter assay. Including the putative target sites of these two miRNAs: Position 2004–2011 (+) Position 1667–1674 (+); deletion of putative miR-22-3p target site: Position 2004–2011 (–) Position 1667–1674 (+); deletion of putative miR-29a-3p target site: Position 2004–2011 (+) Position 1667–1674 (–); deletion of putative target sites of both these two miRNAs: Position 2004–2011 (–) Position 1667–1674 (–). (A color version of this figure is available in the online journal.)
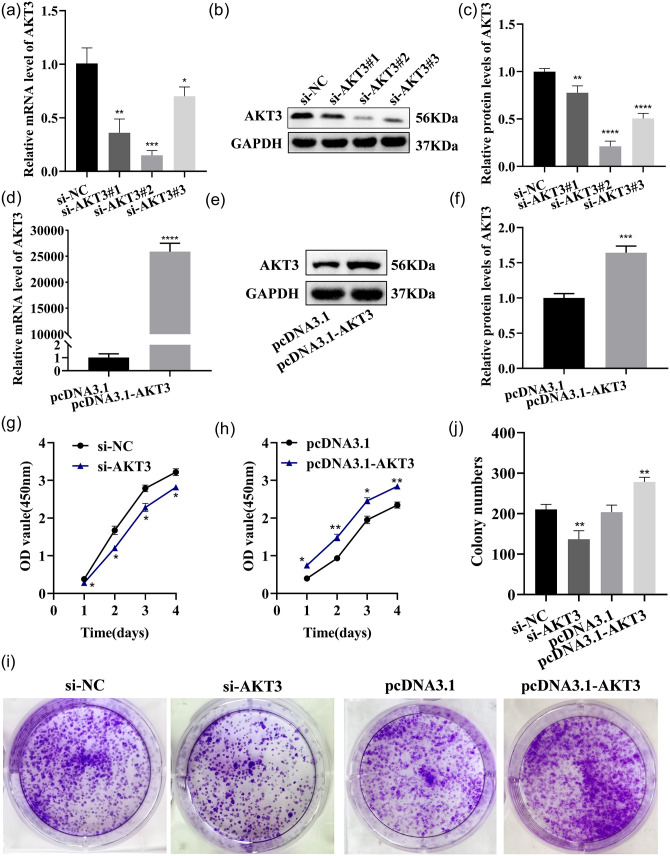
AKT3 promotes LX-2 cell proliferation and activation
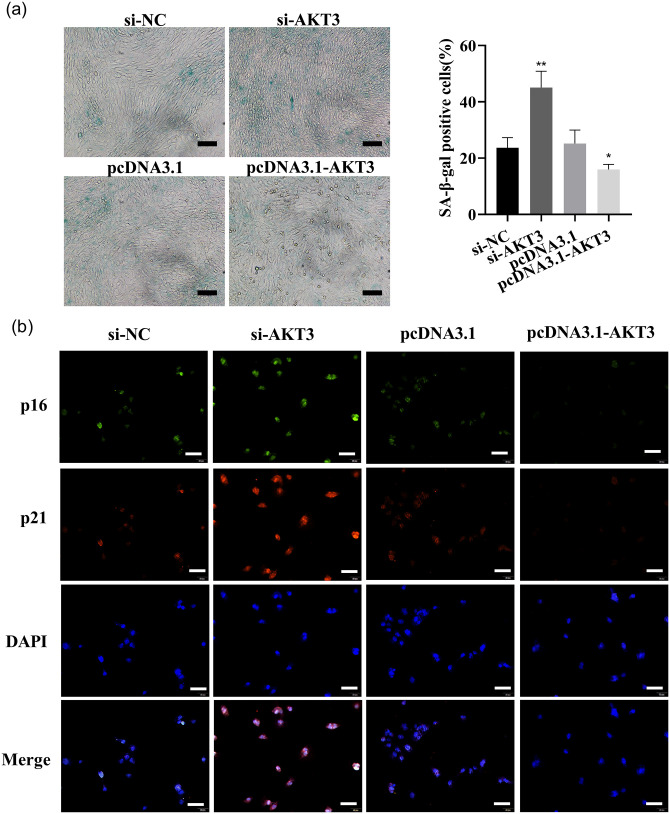
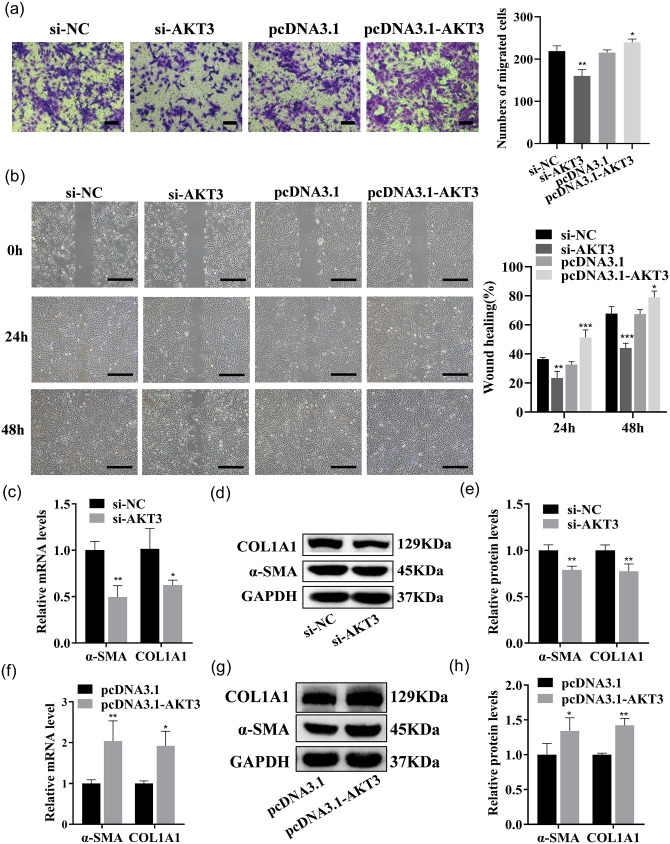
LX-2 cells were transfected with si-AKT3, si-NC, pcDNA3.1-AKT3, and pcDNA3.1, respectively. AKT3 expression in LX-2 was strikingly reduced by si-AKT3, and the knockdown effect of si-AKT3#2 was the most significant, so it was selected as the transfection group for subsequent experiments (Figure 12(a) to (c)). AKT3 mRNA and protein expression in the cells were dramatically increased by pcDNA3.1-AKT3 plasmids (Figure 12(d) to (f)). The above results suggested that AKT3 mRNA and protein expression were effectively reduced or increased by the transfection of si-AKT3 or pcDNA3.1-AKT3 plasmids. The proliferation and the colony-forming ability of LX-2 were significantly accelerated by AKT3, as detected by CCK-8 analysis or the colony formation assay (Figure 12(g) to (j)). Cellular senescence was inhibited, and the fluorescence intensity of senescence markers p16 and p21 was reduced by AKT3, as detected by β-galactosidase staining and cell IF (Figure 13(a) and (b)). Cell migration was promoted by AKT3, which was confirmed by Transwell and wound healing experiments (Figure 14(a) and (b)). In addition, α-SMA and COL1A1 expressions were dramatically decreased or raised in LX-2 transfected with si-AKT3 or pcDNA3.1-AKT3 (Figure 14(c) to (h)). The above results indicated that AKT3 promoted LX-2 cell proliferation and activation.
Figure 12.
AKT3 promotes LX-2 proliferation. (a to c) AKT3 expression was tested by qRT–PCR (a) and Western blot (b and c) in LX-2 transfected with AKT3-siRNA. (d to f) AKT3 expression was tested by qRT–PCR (d) and Western blot (e and f) in LX-2 transfected with pcDNA3.1-AKT3. (g and h) The CCK-8 assay detected the proliferation ability of each group of cells after the transfection of AKT3-siRNA (g) or pcDNA 3.1-AKT3 (h). (i) The colony-forming ability was detected in each group of cells after the transfection of AKT3-siRNA or pcDNA 3.1-AKT3. (A color version of this figure is available in the online journal.)
Figure 13.
AKT3 inhibits LX-2 cellular senescence. (a) The function of AKT3-siRNA or pcDNA3.1-AKT3 on cellular senescence was detected by SA-β-gal staining analysis. Scale bars = 200 μm. (b) The p16 and p21 expressions were examined by co-IF (p16: green, p21: red, nuclear: blue; 200 ×, scale bars = 50 μm). (A color version of this figure is available in the online journal.)
Figure 14.
AKT3 promotes LX-2 migration and activation. (a and b) The migration ability of each group of cells transfected with AKT3-siRNA or pcDNA3.1-AKT3 was tested by Transwell assays (a; scale bars = 200 μm) and wound healing assays (b; scale bars = 500 μm). (c to e) COL1A1 and α-SMA expressions were examined by qRT–PCR (c) and Western blot (d and e) in LX-2 transfected with AKT3-siRNA. (f to h) COL1A1 and α-SMA expressions were examined by qRT–PCR (f) and Western blot (g and h) in LX-2 transfected with pcDNA 3.1-AKT3. (A color version of this figure is available in the online journal.)
Discussion
Differential expression and pathway analysis of miRNAs provided a comprehensive understanding of HF, and miR-29a-3p and miR-22-3p expressions were detected to be dramatically downregulated in BDL rat liver tissue, the activated human immortalized LX-2 and NFs, which indicates that these two miRNAs are related to HF. Previous studies revealed that miR-22-3p was a tumor inhibitor in several cancers, including breast, 15 ovarian, 16 liver, 17 prostate, 18 and glioma cancers. 19 Exosome-miR-22-3p inhibits the PI3K/AKT pathway by inhibiting the expression of Ras-related protein Rap-2B (RAP2B), inhibiting the proliferation and invasion of colorectal cancer cells. 20 In addition to studies on tumors, Xiaolin Jia et al. 21 found that miR-22-3p strikingly inhibited osteoclast proliferation and induced osteoclast apoptosis. Similarly, our results revealed low expression of miR-22-3p in activated HSCs.
The miR-29 family that comprises the widely expressed miR-29a (-b, -c) has been proven to play various biological functions in the pathogenesis of health conditions and different diseases. Regarding liver diseases, miR-29 is significantly downregulated in liver cancer and functions as a tumor suppressor. 22 Wang et al. 23 confirmed that miR-29b induced HSC apoptosis and inhibited HSC activation to prevent HF, and that this was achieved through the PI3K/AKT pathway. In addition, miR-29a regulates HSC activation through histone deacetylase (HDAC). 24 Pharmacological inhibitors of HDAC inhibit the activation of HSCs and show a strong miR-29 upregulation effect. Similarly, our results revealed a low expression of miR-29a-3p in activated HSCs.
In this study, miRNAs with differential expressions were selected by a miRNA microarray of liver tissue from BDL rats, and the expressions of these two miRNAs were downregulated. KEGG and GO analyses of these two miRNAs revealed that these two miRNAs were associated with the development of HF. Next, these two miRNAs were significantly downregulated in BDL rat liver tissue, activated LX-2, and NFs, and the combination of half doses of these two miRNA mimics showed excellent inhibitory function on the proliferation, migration, colony formation, and fibrosis marker expressions. By contrast, the combination of half doses of miR-22-3p and miR-29a-3p inhibitors demonstrated a promoting function on the proliferation, migration, colony formation, and fibrosis marker expressions.
In the present study, miR-29a-3p was increased or reduced in LX-2 transfected with miR-22-3p mimics or inhibitors. Similar results were obtained in LX-2 transfected with miR-29a-3p mimics or inhibitors, which indicated that there is effective synergy between these two miRNAs. miRNAs may coordinate the expressions of common target genes that are widely expressed in humans, which is more economical and effective for gene regulation. 25 Combined with the prediction of bioinformatics and the key role of AKT3 in miRNA biogenesis, the presence of these two miRNAs-binding sites in the 3‘UTR of AKT3 mRNA, AKT3 was indicated to be a good candidate common target gene.
To further investigate the mechanisms of these two miRNAs, KEGG, GO, and PPI analyses were performed on the 24 selected possible common target genes, combined with minimum free energy (MFE), multiple scores of database target genes, species conservation, and current relevant research progress. The expression of AKT3 was dramatically reduced by these two miRNAs, and AKT3 was the bona fide common target of miR-22-3p and miR-29a-3p in LX-2.
AKT/protein kinase B (PKB) protein includes AKT1, AKT2, and AKT3, which belong to the mammalian serine/threonine-specific protein kinase family. 3 AKT3 is highly expressed in the brain, and AKT2 is highly expressed in the liver, 26 but these two miRNAs did not target AKT2 during our target gene screen. Also, in the microarray data set (GSE11954) obtained from gene expression synthesis (GEO), AKT3 expression is decreased in senescent HSCs, while there is no difference of AKT2 expression between growing and senescent human activated HSCs. 27 Therefore, we speculated that these two miRNAs might regulate HSCs through AKT3 in the liver fibrosis-related signaling PI3K-AKT pathway, rather than AKT2. For example, mice lacking AKT1 are stunted, 28 mice lacking AKT2 have a Type 2 diabetes-like syndrome, 29 and mice lacking AKT3 have impaired brain development. 30 Tingting Zhang et al. 31 found that the long-term spatial memory ability of AKT3-KO mice was impaired, and AKT3-KO mice exhibited synaptic transmission characteristics. In addition, during delayed skin wound healing, tissue remodeling is prolonged, and AKT3 expression is downregulated. Regarding cancer tumors, miR-217 significantly inhibits the growth of thyroid cancer cells by targeting AKT3, and suppresses the migration and invasion of thyroid cancer cells. 32 However, the detailed mechanism of AKT3 upregulation in HF remains to be fully elucidated. Next, AKT3 overexpression plasmid and AKT3-siRNA were transfected into LX-2 to upregulate or knock down the expression level of AKT3 to explore its function. The proliferation, migration, colony formation, and expression of fibrosis markers were promoted, and cellular senescence was restrained by AKT3 in LX-2.
Presently, our research has mainly revealed that these two miRNAs synergistically inhibited HF in vitro by targeting AKT3. However, whether the repression of these two miRNAs on the proliferation and activation of HSCs can be rescued by AKT3 warrants further study. Human HF biopsy samples will be collected, and human primary HSCs will be extracted in future studies to unravel the regulatory function of these two miRNAs on AKT3 in human HF tissues, and human primary HSCs. The synergistic regulatory function of these two miRNAs on AKT3 will also be further studied in multiple HF models in mice or rats. The main cells of collagen in BDL which induced fibrosis come from HSCs and portal vein fibroblasts. 33 Another study has revealed that >70% of myofibroblasts are derived from portal fibroblasts in BDL-induced cholestatic liver injury. 34 This point inspired us to think further about the role of miRNAs and various cell types in HF, and to consider further exploration of which cell type is responsible for the target miRNAs. In future studies, the question of whether miRNAs are involved in relevant biological functions in relevant cells in the liver environment (such as ductal cells in the portal region) and fibroblasts will be addressed.
Conclusions
Our research demonstrates that these two miRNAs act as fibrosis inhibitors and synergistically inhibit HF, and that AKT3 is the common bona fide target gene of these two miRNAs. AKT3 expression was regulated by these two miRNAs, in a coordinated fashion. The activation and proliferation of LX-2 were enhanced, and the process of HF was affected by AKT3. Our findings suggest new insights into AKT3 expression regulation in HF, as well as a novel possibility of miRNA-based treatment protocols of HF.
Supplemental Material
Supplemental material, sj-pdf-1-ebm-10.1177_15353702221108379 for MiR-22-3p and miR-29a-3p synergistically inhibit hepatic stellate cell activation by targeting AKT3 by Yitong Wang, Ronghua Zhang, Jingwu Li, Xiangyang Han, Hongjian Lu, Jinghui Su, Yutan Liu, Xiaoli Tian, Meimei Wang, Yanan Xiong, Tao Lan, Guangling Zhang and Zhiyong Liu in Experimental Biology and Medicine
Acknowledgments
The authors thank Mary Hager (mlhager@umich.edu) and Madaline McPherson (mmcp@umich.edu) in the College of Literature, Sciences, and the Arts of the University of Michigan for advice on English style grammar, syntax, word choice, and sentence structure.
Footnotes
Authors’ Contributions: YW and RZ contributed equally to this paper. YW and RZ performed the cell biology experiments and co-wrote the paper. XH, HL, JS, and YL performed animal experiments. JL, XT, and TL assisted with data analysis. MW and YX assisted with cell biology and animal experiments. GZ and ZL designed the experiments, provided funding, and edited the paper.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Government Funded Clinical Medicine Talents Training Hebei (grant no. [2020] 397); the Natural Science Foundation of Hebei Province (grant no. H2021209026); Returned Overseas Students of Hebei Province (grant no. C20210340); Key Research and Development, Innovation and Improvement Projects of Hebei Province (grant no. 213777115D and 205A7701H); and the Science and Technology Project of Tangshan (grant no. 19130204C).
ORCID iDs: Yitong Wang  https://orcid.org/0000-0002-9071-2890
https://orcid.org/0000-0002-9071-2890
Guangling Zhang  https://orcid.org/0000-0002-9542-4986
https://orcid.org/0000-0002-9542-4986
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Senoo H, Yoshikawa K, Morii M, Miura M, Imai K, Mezaki Y. Hepatic stellate cell (vitamin A-storing cell) and its relative – past, present and future. Cell Biol Int 2010;34:1247–72 [DOI] [PubMed] [Google Scholar]
- 2. Sufleţel RT, Melincovici CS, Gheban BA, Toader Z, Mihu CM. Hepatic stellate cells – from past till present: morphology, human markers, human cell lines, behavior in normal and liver pathology. Rom J Morphol Embryol 2020;61:615–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Uko NE, Güner OF, Matesic DF, Bowen JP. Akt pathway inhibitors. Curr Top Med Chem 2020;20:883–900 [DOI] [PubMed] [Google Scholar]
- 4. Revathidevi S, Munirajan AK. Akt in cancer: mediator and more. Semin Cancer Biol 2019;59:80–91 [DOI] [PubMed] [Google Scholar]
- 5. Grabinski N, Möllmann K, Milde-Langosch K, Müller V, Schumacher U, Brandt B, Pantel K, Jücker M. AKT3 regulates ErbB2, ErbB3 and estrogen receptor α expression and contributes to endocrine therapy resistance of ErbB2(+) breast tumor cells from Balb-neuT mice. Cell Signal 2014;26:1021–9 [DOI] [PubMed] [Google Scholar]
- 6. Hu X, Wang J, He W, Zhao P, Ye C. MicroRNA-433 targets AKT3 and inhibits cell proliferation and viability in breast cancer. Oncol Lett 2018; 515:3998–4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dobyns WB, Mirzaa GM. Megalencephaly syndromes associated with mutations of core components of the PI3K-AKT-MTOR pathway: PIK3CA, PIK3R2, AKT3, and MTOR. Am J Med Genet C Semin Med Genet 2019;181:582–90 [DOI] [PubMed] [Google Scholar]
- 8. Cai L, Liu X, Guo Q, Huang Q, Zhang Q, Cao Z. MiR-15a attenuates peripheral nerve injury-induced neuropathic pain by targeting AKT3 to regulate autophagy. Genes Genomics 2020;42:77–85 [DOI] [PubMed] [Google Scholar]
- 9. Bergeron Y, Bureau G, Laurier-Laurin M-E, Asselin E, Massicotte G, Cyr M. Genetic deletion of Akt3 induces an endophenotype reminiscent of psychiatric manifestations in mice. Front Mol Neurosci 2017;10:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ding L, Biswas S, Morton R, Smith J, Hay N, Byzova T, Febbraio M, Podrez E. Akt3 deficiency in macrophages promotes foam cell formation and atherosclerosis in mice. Cell Metab 2012;15:861–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Debnath T, Nath NC, Kim EK, Lee KW. Role of phytochemicals in the modulation of miRNA expression in cancer. Food Funct 2017;8:3432–42 [DOI] [PubMed] [Google Scholar]
- 12. Mahmoud MF, Swefy SE, Hasan RA, Ibrahim A. Role of cannabinoid receptors in hepatic fibrosis and apoptosis associated with bile duct ligation in rats. Eur J Pharmacol 2014;742:118–24 [DOI] [PubMed] [Google Scholar]
- 13. Zheng X, Li JW, Liu YK, Ma YF, Gan JH, Han SG, Wang J, Wan ZY, Zhang J, Liu Y, Li YF, Zhang GL. microRNA-10a-5p overexpression suppresses malignancy of colon cancer by regulating human liver cancer fibroblasts. Neoplasma 2021;68:1157–68 [DOI] [PubMed] [Google Scholar]
- 14. Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA 2004;10:1507–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu H, Huang X, Ye T. MiR-22 down-regulates the proto-oncogene ATP citrate lyase to inhibit the growth and metastasis of breast cancer. Am J Transl Res 2018;10:659–69 [PMC free article] [PubMed] [Google Scholar]
- 16. Wu H, Liu J, Zhang Y, Li Q, Wang Q, Gu Z. miR-22 suppresses cell viability and EMT of ovarian cancer cells via NLRP3 and inhibits PI3K/AKT signaling pathway. Clin Transl Oncol 2021;23:257–64 [DOI] [PubMed] [Google Scholar]
- 17. Zeng Z, Dong J, Li Y, Dong Z, Liu Z, Huang J, Wang Y, Zhen Y, Lu Y. The expression level and diagnostic value of microRNA-22 in HCC patients. Artif Cells Nanomed Biotechnol 2020;48:683–6 [DOI] [PubMed] [Google Scholar]
- 18. Dhar S, Kumar A, Gomez CR, Akhtar I, Hancock JC, Lage JM, Pound CR, Levenson AS. MTA1-activated Epi-microRNA-22 regulates E-cadherin and prostate cancer invasiveness. FEBS Lett 2017;591:924–33 [DOI] [PubMed] [Google Scholar]
- 19. Han M, Wang S, Fritah S, Wang X, Zhou W, Yang N, Ni S, Huang B, Chen A, Li G, Miletic H, Thorsen F, Bjerkvig R, Li X, Wang J. Interfering with long non-coding RNA MIR22HG processing inhibits glioblastoma progression through suppression of Wnt/β-catenin signalling. Brain 2020;143:512–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Y, Lin C. Exosomes miR-22-3p derived from mesenchymal stem cells suppress colorectal cancer cell proliferation and invasion by regulating RAP2B and PI3K/AKT pathway. J Oncol 2021;2021: 3874478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jia X, Yang M, Hu W, Cai S. Overexpression of miRNA-22-3p attenuates osteoporosis by targeting MAPK14. Exp Ther Med 2021;22:692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang X, He Y, Mackowiak B, Gao B. MicroRNAs as regulators, biomarkers and therapeutic targets in liver diseases. Gut 2021;70:784–95 [DOI] [PubMed] [Google Scholar]
- 23. Wang J, Chu E, Chen H, Man K, Go M, Huang X, Lan H, Sung J, Yu J. microRNA-29b prevents liver fibrosis by attenuating hepatic stellate cell activation and inducing apoptosis through targeting PI3K/AKT pathway. Oncotarget 2015;6:7325-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mannaerts I, Eysackers N, Onyema OO, Van Beneden K, Valente S, Mai A, Odenthal M, van Grunsven LA. Class II HDAC inhibition hampers hepatic stellate cell activation by induction of microRNA-29. PLoS ONE 2013;8:e55786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Harati R, Mabondzo A, Tlili A, Khoder G, Mahfood M, Hamoudi R. Combinatorial targeting of microRNA-26b and microRNA-101 exerts a synergistic inhibition on cyclooxygenase-2 in brain metastatic triple-negative breast cancer cells. Breast Cancer Res Treat 2021; 187:695–713 [DOI] [PubMed] [Google Scholar]
- 26. Xie X, Shu R, Yu C, Fu Z, Li Z. Mammalian AKT, the emerging roles on mitochondrial function in diseases. Aging Dis 2022;13:157–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krizhanovsky V, Yon M, Dickins RA, Hearn S, Simon J, Miething C, Yee H, Zender L, Lowe SW. Senescence of activated stellate cells limits liver fibrosis. Cell 2008;134:657–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hwang SK, Chang SH, Minai-Tehrani A, Kim YS, Cho MH. Lentivirus-AIMP2-DX2 shRNA suppresses cell proliferation by regulating Akt1 signaling pathway in the lungs of AIMP2+/– mice. J Aerosol Med Pulm Drug Deliv 2013;26:165–73 [DOI] [PubMed] [Google Scholar]
- 29. Cho H, Mu J, Kim J, Thorvaldsen J, Chu Q, Crenshaw E, Kaestner K, Bartolomei M, Shulman G, Birnbaum M. Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKB beta). Science 2001;292:1728–31 [DOI] [PubMed] [Google Scholar]
- 30. DuBois JC, Ray AK, Gruber RC, Zhang Y, Aflakpui R, Macian-Juan F, Shafit-Zagardo B. Akt3-mediated protection against inflammatory demyelinating disease. Front Immunol 2019;10:1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang T, Shi Z, Wang Y, Wang L, Zhang B, Chen G, Wan Q, Chen L. Akt3 deletion in mice impairs spatial cognition and hippocampal CA1 long long-term potentiation through downregulation of mTOR. Acta Physiol 2019;225:e13167 [DOI] [PubMed] [Google Scholar]
- 32. Lin Y, Cheng K, Wang T, Xie Q, Chen M, Chen Q, Wen Q. miR-217 inhibits proliferation, migration, and invasion via targeting AKT3 in thyroid cancer. Biomed Pharmacother 2017;95:1718–24 [DOI] [PubMed] [Google Scholar]
- 33. Pradhan-Sundd T, Kosar K, Saggi H, Zhang R, Vats R, Cornuet P, Green S, Singh S, Zeng G, Sundd P, Nejak-Bowen K. Wnt/β-catenin signaling plays a protective role in the Mdr2 knockout murine model of cholestatic liver disease. Hepatology 2020;71:1732–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sato K, Hall C, Glaser S, Francis H, Meng F, Alpini G. Pathogenesis of Kupffer cells in cholestatic liver injury. Am J Pathol 2016;186:2238–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-ebm-10.1177_15353702221108379 for MiR-22-3p and miR-29a-3p synergistically inhibit hepatic stellate cell activation by targeting AKT3 by Yitong Wang, Ronghua Zhang, Jingwu Li, Xiangyang Han, Hongjian Lu, Jinghui Su, Yutan Liu, Xiaoli Tian, Meimei Wang, Yanan Xiong, Tao Lan, Guangling Zhang and Zhiyong Liu in Experimental Biology and Medicine