Abstract
Background:
Human papillomavirus (HPV) infection is known to promote the development of mucosal squamous cell carcinoma (mSCC), including pathologically high-grade lesions, but its role in cutaneous squamous cell carcinoma (cuSCC) remains unclear, particularly in lesions that are considered high risk.
Objective:
We aimed to determine whether enhanced HPV transcriptional activity can be detected in high-risk cuSCC samples compared with low-grade SCC samples or normal skin.
Methods:
We performed RNA sequencing of cuSCC across 23 risk-stratified skin lesions. A subset of samples was tested for the presence of HPV DNA. High-quality, non-human reads from each sample group were used for viral analysis using Microbiome Coverage Profiler.
Results:
None of the samples analysed had detectable expression of HPV RNA, while 64% of samples tested positive for HPV DNA. All samples were found to have expression of human endogenous retrovirus, and multiple samples showed expression of other viruses.
Conclusions:
Viral and prophage gene expression can be monitored in cuSCC or normal skin biopsies, yet no sample in our study showed evidence of active HPV gene expression despite evidence of HPV genome presence. This suggests HPV transcription does not play a role in differentiating high-risk cuSCCs from low-risk cuSCCs or normal skin.
Keywords: SCC, HPV, high-risk SCC, immunosuppression
1 |. INTRODUCTION
Cutaneous squamous cell carcinoma (cuSCC) is the second most common malignancy in the United States.1 While the majority of cuSCCs are cured with local treatment, a subset of lesions, considered “high-risk” cuSCCs, are associated with increased risk of recurrence, metastasis and mortality. Specific histological findings distinguish these lesions;2 however, the molecular factors driving this behaviour are not understood. The development of cuSCCs is strongly linked to UV-associated genomic damage, with subsequent alterations in gene expression.3,4 However, transcriptomic studies and epidemiologic data also indicate a role of immune dysregulation in cuSCC pathogenesis. Immunosuppression is strongly associated with the development of high-risk cuSCCs, with organ transplant recipients in particular showing a 65- to 100-fold increased risk of cuSCC development and dramatically elevated rate of metastases and mortality.5,6 This association suggests an infectious agent or altered local immunity may play a role in the development of high-risk cuSCCs.
The human papillomaviruses (HPVs) have been hypothesized to play a role in high-risk cuSCC development and progression. The HPV family of double-stranded DNA viruses are the most commonly found viruses on human skin, with over 300 types detected.7,8 While many HPV types likely act as commensal organisms, others are associated with benign growths and some types are strongly linked to non-cutaneous SCC. Specific subtypes of HPV have been demonstrated to play an aetiologic role in the development of cervical and anal mucosal SCCs.9,10 In head and neck SCC involving the oropharynx, HPV is associated with a subset of high-grade cancers, which have a distinct mutational profile and therapeutic response.11,12,13 In these settings, viral infection promotes malignancy through transcription of the HPV DNA to RNA and production of the HPV E6 protein, which targets p53 for degradation, mimicking an inactivating p53 mutation to promote tumorigenesis.14,15
In cuSCC, a connection to HPV has also been suggested based on observational work, but the relationship remains unclear. Multiple studies have reported the presence of HPV genomes in cuSCC samples, with a meta-analysis demonstrating increased frequency of HPV DNA detection in cuSCCs as compared to normal skin and a further increase in detection frequency in cuSCCs from immunosuppressed patients compared with immunocompetent patients.16 Support for a causative role of HPV in tumor development and maintenance comes from case reports of individuals who had reduction in SCC frequency or clearance of existing SCCs following administration of the HPV vaccine.17,18 However, a study of viral gene expression called into question this relationship. In that work, RNA sequencing of multiple types of cuSCCs showed no evidence of HPV transcription in cuSCC despite the presence of viral genomes, unlike in mucosal SCC where it is actively transcribed.19 As the oncogenic potential of HPV is thought to be tied to transcription of viral gene products, these data could suggest that the increased presence of HPV genomes in cuSCC samples is artifactual, or that virus activity is involved in the initiation of cuSCC but dispensable as the tumor progresses (“hit and run”).20,21 Alternatively, this study did not stratify results based on histological features, so it is possible that HPV activity is limited to a subset of cuSCCs, as seen in oropharyngeal HNSCC.11,12
Here, we sought to clarify the relationship between HPV presence, HPV transcription and cuSCC type through RNA sequencing of two sets of cuSCCs: tumors histologically classified as well differentiated in patients with low risk of recurrence or metastasis (ie no immunosuppression or haematologic malignancy); and histologically high-grade tumors in patients with high risk of recurrence or metastasis (ie presence of immunosuppression or haematologic malignancy), in order to determine whether HPV activity may play a pathological role in the high-risk cuSCC lesion phenotype. Both sample sets were compared to three normal skin samples, and a subset of all samples were analysed using PCR to determine infection status.
2 |. METHODS
2.1 |. Sample Collection
All subjects provided informed consent according to procedures approved by the University of California, Los Angeles, IRB #12–001195. Specimens were collected from patients during Mohs micrographic surgery or biopsy. Tissues were snap-frozen in liquid nitrogen for immediate RNA extraction or placed in RNAlater (Thermo Fisher).
2.2 |. Histological Scoring
Surgical samples acquired during biopsy or Mohs surgery were stratified according to histological grading of differentiation. Differentiation was defined by epithelial appearance of cells and degree of keratinization. Tumors with an epithelial appearance and forming keratin were considered well differentiated. Tumors composed of keratinocytes that appeared epithelioid or spindled/mesenchymal with a lack of keratinization were considered poorly differentiated.
2.3 |. RNA Sequencing
Total RNA was extracted using TRIzol (Thermo) according to manufacturer’s protocol, and total RNA was purified using QIAshredder (Qiagen), and RNeasy (Qiagen) purification columns were used according to manufacturer’s protocols. RNA-Seq library construction was performed using the TruSeq V2 Kit (Illumina). Samples were sequenced paired-end 2 × 100bp on a HiSeq 2000.
2.4 |. Viral gene expression analysis
For viral gene expression analysis, the ROP protocol, v1.0.4, was used to profile viral RNA among the unmapped reads.22 Briefly, ROP filters low-quality, low-complexity, rRNA reads and reads likely to be human. ROP then uses Megablast (BLAST+2.2.30) to align the reads onto viral genomes downloaded from NCBI ftp://ftp.ncbi.nih.gov/ on 1 February 2015. The alignment profiles of the viral reads were examined with Microbiome Coverage Profiler (MCP) v1.0. MCP reports coverage of viral genomes passing stringent criteria. MCP filters out reads mapped due to sequencing or PCR artifacts. For reads mapped to the multiple genomes, MCP averages coverage of each genome from uniquely mapped reads to assign the read to one genome. Each viral genome identified was then analysed by its coverage. Genomes with less than 20 reads were graded as failed. Partially covered genomes were run through BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to determine whether the covered regions are unique to the virus. If not unique, virus was considered failed.
2.5 |. Data access
The accession number for the genome-wide sequencing data set reported in this paper is GEO: GSE111354.
2.6 |. Detection of HPV gDNA
The presence of HPV DNA was evaluated using degenerate primers to HPV with single-round PCR, forward primer CCWGATCCHAATMRRTTTGC and reverse primer ACATTTGIAITTGTTTDGGRTCAA.23 Cycling conditions were as follows: 94C for 2 minutes, followed by 25 cycles of 94C for 30 seconds, 50C for 1 minute and 72C for 1 minute.
3 |. RESULTS
3.1 |. Patient characteristics
A total of 23 skin biopsies were collected, sectioned and graded histologically, summarized in Table 1. Ten samples were classified as high risk, ten as low risk and three as normal skin based on grade of differentiation (Figure 1). Patient sex was evenly distributed between samples classified as low risk and high risk. None of the samples classified as low risk were from immunosuppressed patients, while all as high-risk samples were obtained from immunosuppressed patients. The majority of patients (7/10) were iatrogenically immunosuppressed due to organ transplant, while 3 of 10 were immunosuppressed due to chronic lymphocytic leukaemia.
TABLE 1.
Patient and clinical characteristics.
| Normal skin | Low risk | High risk | |
|---|---|---|---|
| Average age (years) | 72 | 78 | 67.2 |
| Sex | |||
| Male | 2 | 7 | 7 |
| Female | 1 | 3 | 3 |
| Histological category | |||
| Well differentiated | 0 | 8 | 0 |
| Moderately differentiated | 0 | 2 | 0 |
| Poorly differentiated | 0 | 0 | 10 |
| % immunosuppressed | 0 | 0 | 100 |
| Indication for immunosuppression | NA | NA | Organ transplant (6) CLL (3) |
| Renal transplant (1) | |||
| % recurrent | 0 | 0 | 20% |
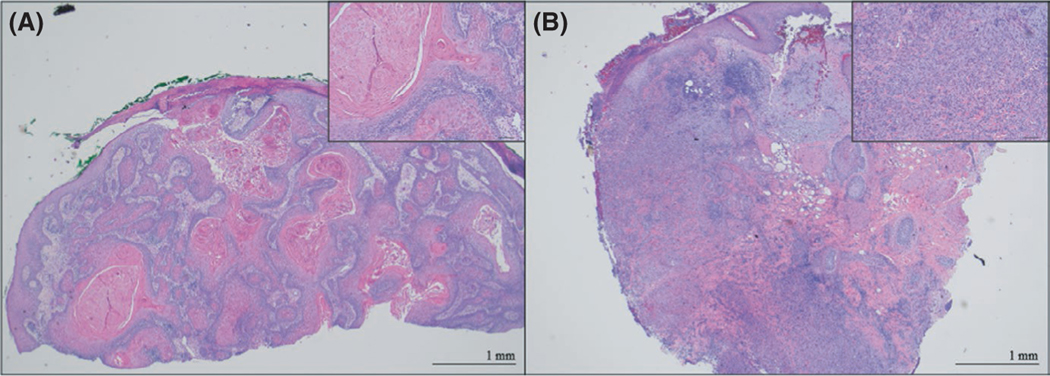
FIGURE 1.
Representative histopathology. Shown are haematoxylin and eosin staining of representative samples graded as histologically low risk (A) and histologically high risk (B). All scale bars denote 1 mm
3.2 |. Detection of HPV infection in normal skin and cutaneous SCC
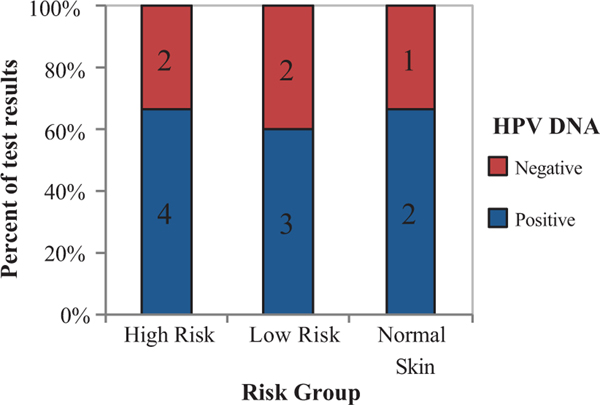
To determine HPV infection status of specimens obtained, DNA was extracted from a subset of 14 samples and the presence of HPV DNA was assessed using an established PCR method.23 Of the 14 samples tested for HPV DNA in human biopsy tissue, nine samples tested positive (64%). HPV infection rates were similar across risk groups as determined by chi-squared test (p-value 0.969) (Figure 2).
FIGURE 2.
HPV gDNA is detected in cuSCC skin biopsies across risk groups. HPV genotyping was performed using degenerate primer PCR. % and number of samples testing positive (blue) or negative (red) are indicated on the y-axis, sorted by sample group on the x-axis
3.3 |. HPV RNA is not detected in normal skin or cutaneous SCC
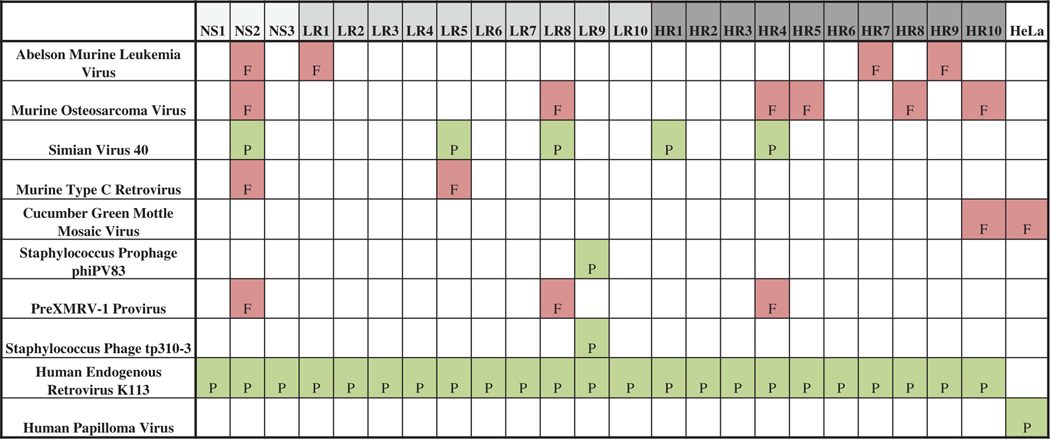
HPV activity was assessed in all samples by measuring viral gene expression using RNA sequencing. An average of 47 million reads per sample were obtained, of which an average of 79.2% of reads were identified as homo sapiens (Table S1). Viral genes were identified and classified using the recently described Read Origin Protocol (ROP) alongside a Microbiome Coverage Profiler (MCP).22,24 This analytical pipeline identifies high-quality RNA transcripts that do not map to the human genomes but do map to a catalogue of published viral and prophage sequencing data. Through this method, we detected viral gene expression from a human endogenous retrovirus K113 in all samples analysed, as well as SV40 in four samples and Staphylococcus prophage phiPV83 in one sample (Figure 3). Other viral gene transcripts were detected but were determined to be not valid due to either poor coverage of the viral transcript or high identity with murine viral transcripts (Table S2). No samples in this study contained detectable HPV viral gene expression including those samples that had evidence of HPV DNA presence. By contrast, when applied to data from a HeLa cell line known to have active HPV transcription,25 our data analysis pipeline detected HPV viral transcripts. An additional computational pipeline, sequence-based ultra-rapid pathogen identification (SURPI),26 also failed to detect the presence of HPV RNA, but was able to detect HPV RNA from a positive control sample obtained from a HeLa cell line.
FIGURE 3.
Detection of non-human transcripts. Viral genomes detected in RNA sequencing by Read Origin Protocol (ROP) and Microbiome Coverage Profiler (MCP) analytical pipeline. Shown are the detected viral transcripts by sample. Samples are categorized as normal skin (NS1–3), low risk (LR1–10), high risk (HR1–10) or HeLa cell line. Viral gene expression was graded pass (green, P) or fail (red, F) based on the viral expression coverage over the length of the transcript
4 |. DISCUSSION
Despite multiple reports of HPV detection within cuSCC samples, the role of HPV in cuSCC development and progression remains unclear, particularly in the context of high-risk cuSCCs. Here, we tested the hypothesis that HPV activity may differentiate high-risk cuSCCs from low-risk cuSCCs by using RNA sequencing to analyse the expression of viral genes across risk-stratified SCCs and normal skin. We focused on SCC with high-grade histopathologic features from immunocompromised patients as viral oncogenes often result in higher-grade neoplasms and diminished immunity would result in a higher likelihood of skin/SCC harbouring HPV. While we detect HPV viral genomes by PCR in two thirds of all sample groups and demonstrate that our detection algorithm can identify viral gene expression, we find no evidence of HPV gene expression in cuSCC, regardless of risk stratification or in normal skin samples.
These findings are consistent with the previous report of minimal HPV viral gene expression in cuSCC samples, in contrast to mucosal SCCs.19 Our study both validates the prior work and clarifies that HPV transcriptional activity is not present even in the least differentiated, most aggressive subset of cuSCCs, unlike what has been reported in HNSCCs.13 Together, our work and the previous study strongly suggest that while HPV genomes are frequently found in cuSCC, viral activity is not required for maintenance and progression, even in the most aggressive lesions. Multiple potential theories for the role of HPV in cuSCC have been purported, which could be consistent with this observation, in particular the idea that HPV is merely a bystander to cuSCC development 19 or the theory that HPV is involved in only the initiation of cancer and then becomes dispensable (“Hit and run”).20,21 While our study does not test these theories, it provides intriguing data points. First, the prevalence of HPV detection is the same in all sample groups. While our sample size is small, the absence of any enrichment in cuSCCs, particularly in the high-risk lesions, argues against a strong pathogenic link. Second, the absence of viral activity even in the subset of high-risk cuSCCs confirms that viral activity does not play a central role in the progression of even the most clinically high-risk cuSCCs, but other factors, including UV exposure compounded with immunodeficiency, as well as genetic polymorphisms, may contribute to high-risk cuSCC development.27,28
These results are puzzling in the light of the recent observational reports of cuSCC clearance following HPV vaccination,18 as inducing a viral immune response should not have curative benefit if viral activity is not promoting cancer progression. One possible explanation comes from mouse studies in which infection with a murine papillomavirus was shown to be protective against skin cancers through induction of protective T cell–mediated immunity.29 While this system is distinct from human cuSCC carcinogenesis in many ways, a similar mechanism could underlie the response of cuSCCs to HPV vaccination in the absence of viral oncogenesis.
5 |. CONCLUSIONS
We demonstrate that HPV viral gene expression is not detected in a set of twenty high-risk and low-risk cuSCC samples or in three normal skin samples despite the presence of the viral genome. These data support previous reports that HPV expression is not necessary for cuSCC progression and argues against a strong association between HPV infection and high-risk cuSCCs.
Supplementary Material
ACKNOWLEDGEMENTS
This research was supported by funding from the Skin Cancer Foundation Paul Silverberg Memorial Award, a generous gift from Michael and Linda Keston, and NIH (K08-AR066545) to PS. AV is supported by T32AR071307 from the National Institute of Arthritis and Musculoskeletal And Skin Diseases.
Funding information
National Institute of Arthritis and Musculoskeletal and Skin Diseases, Grant/Award Number: K08-AR066545 and T32AR071307; Skin Cancer Foundation, Grant/Award Number: Paul Silverberg Memorial Award
CONFLICT OF INTERESTS
Dr. Scumpia has served on an advisory board and received a research grant (grant paid to the institution) from Castle Biosciences, Inc.
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section.
DATA AVAILABILITY STATEMENT
The data that supports this study are openly available in the GEO data base, accession number: GSE111354.
REFERENCES
- 1.Miller DL, Weinstock MA. Nonmelanoma skin cancer in the United States: incidence. J Am Acad Dermatol. 1994;30(5 Pt 1):774–778. [DOI] [PubMed] [Google Scholar]
- 2.Rowe DE, Carroll RJ, Day CL Jr. Prognostic factors for local recurrence, metastasis, and survival rates in squamous cell carcinoma of the skin, ear, and lip. Implications for treatment modality selection. J Am Acad Dermatol. 1992;26(6):976–990. [DOI] [PubMed] [Google Scholar]
- 3.Zheng Q, Capell BC, Parekh V, et al. Whole-exome and transcriptome analysis of UV-exposed epidermis and carcinoma in situ reveals early drivers of carcinogenesis. J Invest Dermatol. 2021;141(2):295–307 e213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Das Mahapatra K, Pasquali L, Sondergaard JN, et al. A comprehensive analysis of coding and non-coding transcriptomic changes in cutaneous squamous cell carcinoma. Sci Rep. 2020;10(1):3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindelof B, Sigurgeirsson B, Gabel H, Stern RS. Incidence of skin cancer in 5356 patients following organ transplantation. Br J Dermatol. 2000;143(3):513–519. [PubMed] [Google Scholar]
- 6.Carucci JA, Martinez JC, Zeitouni NC, et al. In-transit metastasis from primary cutaneous squamous cell carcinoma in organ transplant recipients and nonimmunosuppressed patients: clinical characteristics, management, and outcome in a series of 21 patients. Dermatol Surg. 2004;30(4 Pt 2):651–655. [DOI] [PubMed] [Google Scholar]
- 7.Foulongne V, Sauvage V, Hebert C, et al. Human skin microbiota: high diversity of DNA viruses identified on the human skin by high throughput sequencing. PLoS One. 2012;7(6):e38499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bzhalava D, Muhr LS, Lagheden C, et al. Deep sequencing extends the diversity of human papillomaviruses in human skin. Sci Rep. 2014;4:5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burd EM. Human papillomavirus and cervical cancer. Clin Microbiol Rev. 2003;16(1):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoff PM, Coudry R, Moniz CM. Pathology of Anal Cancer. Surg Oncol Clin N Am. 2017;26(1):57–71. [DOI] [PubMed] [Google Scholar]
- 11.Agrawal N, Frederick MJ, Pickering CR, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333(6046):1154–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nature. Cancer Genome Atlas N: Comprehensive genomic characterization of head and neck squamous cell carcinomas. 2015;517(7536):576–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chernock RD, El-Mofty SK, Thorstad WL, Parvin CA, Lewis JS Jr. HPV-related nonkeratinizing squamous cell carcinoma of the oropharynx: utility of microscopic features in predicting patient outcome. Head Neck Pathol. 2009;3(3):186–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lechner MS, Laimins LA. Inhibition of p53 DNA binding by human papillomavirus E6 proteins. J Virol. 1994;68(7):4262–4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas M, Pim D, Banks L. The role of the E6–p53 interaction in the molecular pathogenesis of HPV. Oncogene. 1999;18(53):7690–7700. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Aldabagh B, Yu J, Arron ST. Role of human papillomavirus in cutaneous squamous cell carcinoma: a meta-analysis. J Am Acad Dermatol. 2014;70(4):621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nichols AJ, Allen AH, Shareef S, Badiavas EV, Kirsner RS, Ioannides T. Association of Human Papillomavirus Vaccine With the Development of Keratinocyte Carcinomas. JAMA Dermatol. 2017;153(6):571–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nichols AJ, Gonzalez A, Clark ES, et al. Combined Systemic and Intratumoral Administration of Human Papillomavirus Vaccine to Treat Multiple Cutaneous Basaloid Squamous Cell Carcinomas. JAMA Dermatol. 2018;154(8):927–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arron ST, Ruby JG, Dybbro E, Ganem D, Derisi JL. Transcriptome sequencing demonstrates that human papillomavirus is not active in cutaneous squamous cell carcinoma. J Invest Dermatol. 2011;131(8):1745–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rollison DE, Viarisio D, Amorrortu RP, Gheit T, Tommasino M. An emerging issue in oncogenic virology: the role of beta human papillomavirus types in the development of cutaneous squamous cell carcinoma. J Virol. 2019;93(7):e01003–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hasche D, Stephan S, Braspenning-Wesch I, et al. The interplay of UV and cutaneous papillomavirus infection in skin cancer development. PLoS Pathog. 2017;13(11):e1006723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mangul S, Yang HT, Strauli N, et al. ROP: dumpster diving in RNA-sequencing to find the source of 1 trillion reads across diverse adult human tissues. Genome Biol. 2018;19(1):36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forslund O, Ly H, Higgins G. Improved detection of cutaneous human papillomavirus DNA by single tube nested ‘hanging droplet’ PCR. J Virol Methods. 2003;110(2):129–136. [DOI] [PubMed] [Google Scholar]
- 24.Microbiome Coverage Profiler. In., 2007. edn.
- 25.Strong MJ, Baddoo M, Nanbo A, Xu M, Puetter A, Lin Z. Comprehensive high-throughput RNA sequencing analysis reveals contamination of multiple nasopharyngeal carcinoma cell lines with HeLa cell genomes. J Virol. 2014;88(18):10696–10704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bzhalava D, Johansson H, Ekstrom J, et al. Unbiased approach for virus detection in skin lesions. PLoS One. 2013;8(6):e65953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lobl MB, Hass B, Clarey D, Higgins S, Wysong A. Next-generation sequencing identifies novel single nucleotide polymorphisms in high-risk cutaneous squamous cell carcinoma: A pilot study. Exp Dermatol. 2020;29(7):667–671. [DOI] [PubMed] [Google Scholar]
- 28.Lobl MB, Clarey D, Higgins S, Thieman T, Wysong A. The correlation of immune status with ultraviolet radiation-associated mutations in cutaneous squamous cell carcinoma: A case-control study. J Am Acad Dermatol. 2020;82(5):1230–1232. [DOI] [PubMed] [Google Scholar]
- 29.Strickley JD, Messerschmidt JL, Awad ME, et al. Immunity to commensal papillomaviruses protects against skin cancer. Nature. 2019;575(7783):519–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that supports this study are openly available in the GEO data base, accession number: GSE111354.