Abstract
One of the least understood aspects of prion diseases is the structure of infectious prion protein aggregates. Here we report a high-resolution cryo-EM structure of amyloid fibrils formed by human prion protein with Y145Stop mutation that is associated with a familial prion disease. This structural insight allows us not only to explain previous biochemical findings, but also provides direct support for the conformational adaptability model of prion transmissibility barriers.
Prion diseases are a group of transmissible neurodegenerative diseases in which the infectious agent is a misfolded aggregate of a prion protein, PrP1–3. Human prion diseases may be sporadic, acquired by infection, or have a hereditary origin due to mutations in the PrP-encoding gene. One of the most intriguing disorders of the latter class is a cerebrovascular amyloidosis associated with the Y145Stop mutation4,5. Amyloid fibrils formed in vitro by mouse Y145Stop PrP fragment (or PrP23–144) have been recently shown to be infectious, causing transmissible disease in mice6. Furthermore, this C-terminally truncated PrP variant has been extensively used as a model for studying molecular aspects of prion propagation, providing important insights into the mechanisms of transmissibility barriers and emergence of new prion strains7–9. However, interpretation of these findings at the structural level has been hampered by the lack of high-resolution information about the architecture of PrP23–144 fibrils. Here, we bridge this critical gap, by determining a cryo-EM structure of human PrP23–144 (huPrP23–144) fibrils at a near-atomic resolution.
huPrP23–144 amyloid fibrils were prepared as described in the Methods section. Inspection of cryo-EM data and subsequent 2D classification revealed only one type of fibrillar aggregates in our sample. The ordered amyloid core of these fibrils has a diameter of ~10 nm, with the remaining, largely disordered parts of huPrP23–144 wrapping around the amyloid core and contributing to a larger apparent diameter of ~20 nm (Extended Data Fig. 1a and b). Atomic force microscopy imaging revealed that fibrils are characterized by left-handed twist (Extended Data Fig. 1c).
Helical reconstruction of the cryo-EM data allowed us to determine a density map for the ordered amyloid core with a nominal resolution of 2.86 Å (Fig. 1a and b, Extended Data Fig. 2 and Supplementary Table 1). This core shows a parallel, in-register architecture, in which each fibril consists of four identical protofilaments with a C2 symmetry (Fig. 1b and c). Four-protofilament fibrils are rather unusual, even though they have been reported for a fragment of TDP-43 low complexity domain10 and recombinant SAA protein11. A near-atomic model was built for the fibril core that maps to residues 108–141 (Fig. 1d and e). Each subunit in this model has an “S” shape, largely extended backbone geometry and encompasses three relatively short β-strands (residues 109–112, 133–135, and 138–140) (Fig. 1c and e). The 113–125 region is rich in rigid turns, with numerous solvent-inaccessible hydrophobic residues participating in intramolecular interactions involving side chains of M112/A116/A117 and A115/A118/V121/L125 (Fig. 1d and f). Another cluster of solvent-inaccessible hydrophobic side chains maps to the C-terminal part of the core and includes M129, A133, I139, and F141 (Fig. 1d and f).
Fig. 1. Cryo-EM structure of huPrP23–144 amyloid fibrils.
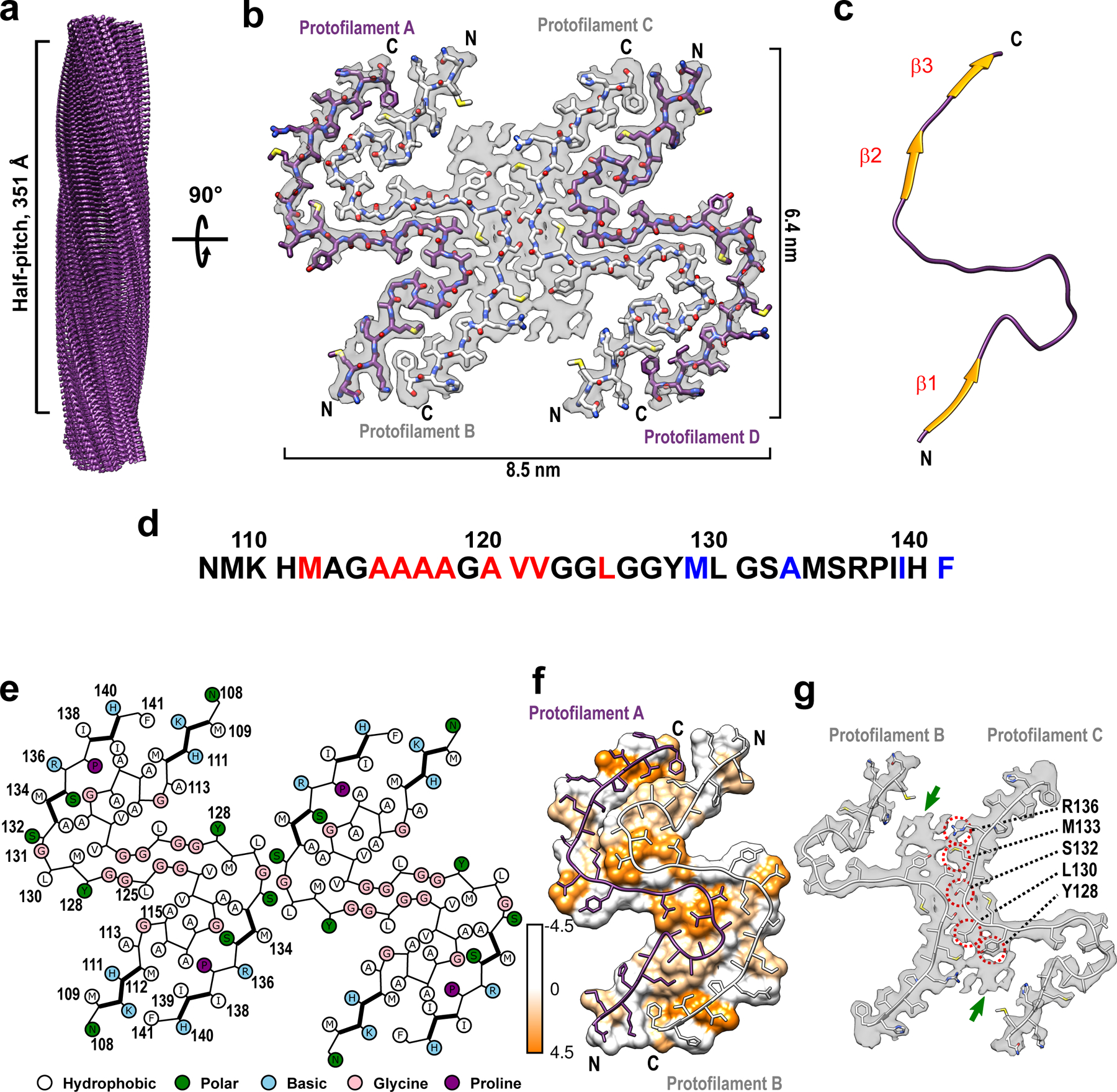
a Cryo-EM density map showing a left-handed twisted helix with a half-pitch of 351 Å and a classical parallel, in-register β-sheet architecture. b The top view of the atomic model superimposed on the cryo-EM density map. The amyloid core contains four identical protofilaments. Two outer protofilaments with greater accessibility to water are depicted in purple; two inner protofilaments are depicted in white. c One representative subunit of the fibril core with β-strands shown in orange. d Amino acid sequence for the highly ordered core of huPrP23–144 fibrils. N- and C-terminal solvent-inaccessible hydrophobic residues are shown in red and blue, respectively. e Schematic representation of one cross-sectional layer of the amyloid core, with β-strands shown as thicker lines. f Hydrophobicity of the cross-section of protofilaments A and B, with hydrophobicity levels colored according to Kyte-Doolittle21 (top view). g One cross-sectional layer of protofilaments B and C shown as atomic model superimposed on the cryo-EM map (top view). Extra unassigned densities (likely representing side chains of residues outside the core region) are indicated by green arrows; residues within the amyloid core involved in the inter-protofilament interactions are indicated by red circles.
These two clusters of hydrophobic resides play a key role in stabilizing the four-protofilament structure of the fibrils. In this structure, protofilaments A and B (as well as C and D) are arranged in a head-to-tail manner, with a large dry interface between them that is stabilized largely by intermolecular interaction between the N-terminal hydrophobic residues (M112, A116, A117, A120, and V122) and C-terminal hydrophobic residues (M129, A133, I139, and F141) (Fig. 1e and f). Interestingly, no classical steric zipper motif is observed within this dry interface. The above-described intermolecular interactions, together with intramolecular interactions involving hydrophobic residues within the 112–125 region, result in a structure in which most (13 out of 18) hydrophobic side chains are buried within the amyloid core interior. The interface between inner protofilaments B and C is much shorter, with intermolecular hydrogen bonds involving only L130 and S132. The latter interface is additionally stabilized by side chain-side chain interactions involving nearby residues Y128, L130, M133, and R136 (Fig. 1e and g).
Protofilaments A and B (and C and D) assembled through large hydrophobic interfaces are arranged in such a manner that N- and C-terminal hydrophobic residues within each monomeric subunit interact with two different subunits in the adjacent protofilament, as illustrated in Extended Data Fig. 3. Such a non-planar assembly of subunits results in rugged surfaces of fibril ends, with solvent exposure of several hydrophobic side chains. Specifically, at the top end of the fibril, two clusters of N-terminal hydrophobic residues (M112/A116/A117 and A120/V122) are exposed to the solvent. Depending on the stage of fibril elongation, these residues are either from protofilaments B and C (Fig. 2a) or protofilaments A and D (Fig. 2b). Similarly, two clusters of hydrophobic C-terminal residues (M129/A133 and I139/F141) from protofilaments A and D (Fig. 2c) or B and C (Fig. 2d) are exposed at the bottom end of the fibril. These exposed hydrophobic residues likely play a key role in the recruitment and conformational conversion of the incoming monomers.
Fig. 2. Solvent-exposed amino acid residues at the top and bottom ends of huPrP23–144 fibrils.
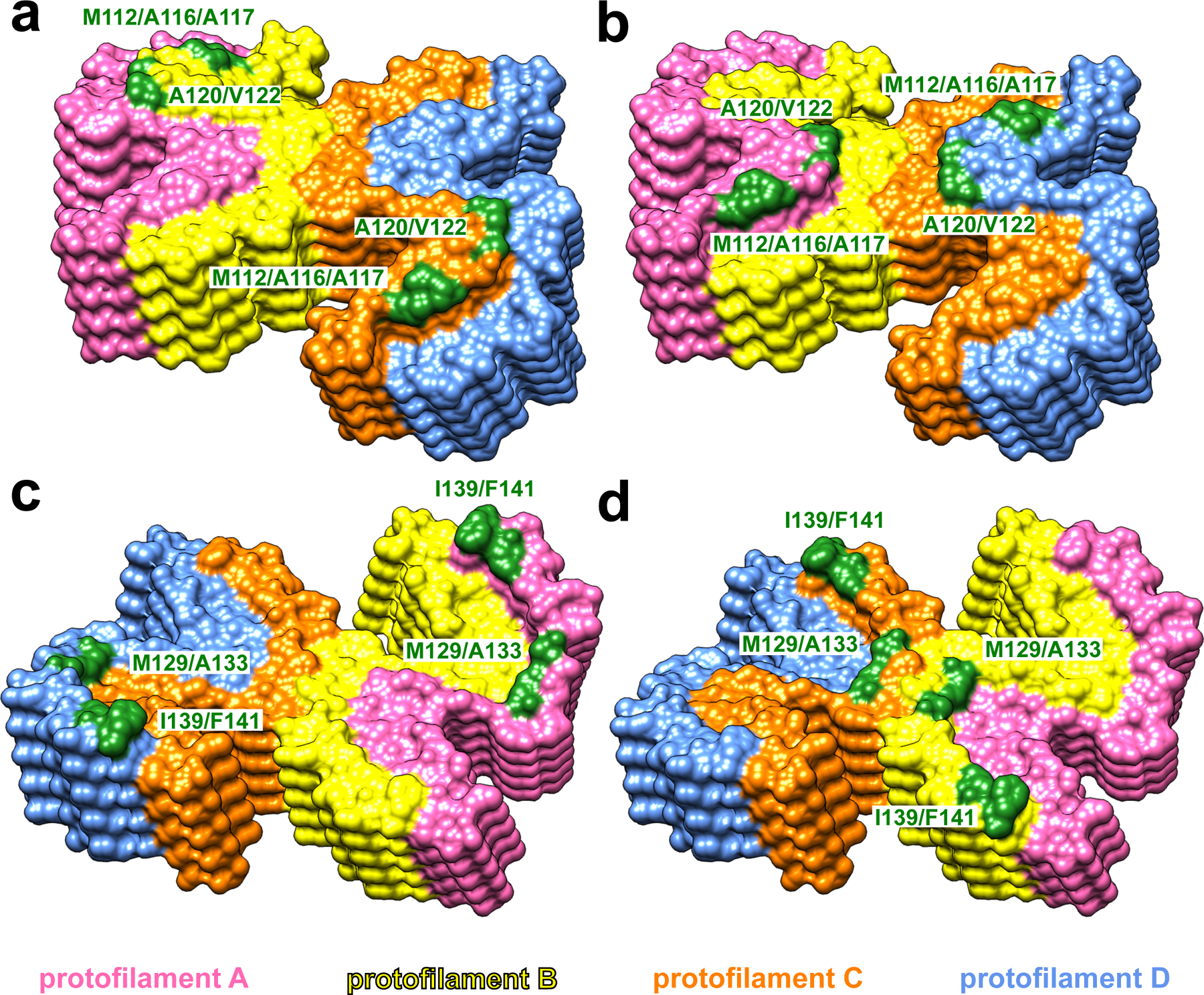
At the top end of the fibril, two clusters of hydrophobic residues (M112/A116/A117 and A120/V122) in protofilaments B and C (yellow and orange, a) or protofilaments A and D (pink and blue, b) are exposed to the solvent. At the bottom end of the fibril, two clusters of hydrophobic residues (M129/A133 and I139/F141) in protofilaments A and D (pink and blue, c) or protofilaments B and C (yellow and orange, d) are exposed to the solvent.
The identity of huPrP23–144 fibril core region in the present cryo-EM-based model is very similar to that suggested by previously reported solid-state NMR studies, even though the fibril samples in those studies appear to be composed of two rather than four protofilaments9,12. The reason for this difference is not fully clear. Remarkably, however, the same sets of hydrophobic residues that are involved in the above-described interactions in the cryo-EM structure were detected in the two-protofilament fibril sample by solid-state NMR9,12 (Extended Data Fig. 4), although the intermolecular interactions in the cryo-EM structure were interpreted as intramolecular in solid-state NMR experiments9,12. Thus, if the preliminary ssNMR-based model is confirmed, this would suggest that there might be two closely related polymorphic forms of the huPrP23–144 fibril core that are stabilized by key interactions between the same residues.
While the disease associated with the Y145Stop PrP mutation is a human disorder, infectivity of amyloid fibrils generated from this PrP variant was established only for the mouse counterpart6. Therefore, we extended our structural studies to moPrP23–144 fibrils, finding that they exist in two distinct polymorphic forms. Importantly, the structure of one of these polymorphs was identical to that described herein for huPrP23–144, even though the resolution of cryo-EM structural data for moPrP-23–144 was somewhat lower (Extended Data Fig. 5 and Supplementary Table 1). The structure of the second polymorph appeared quite different, but the quality of data obtained for this polymorph in many different sample preparations was insufficient for structural modeling.
The present structural data have a number of important implications for understanding the propagation and seeding specificity of huPrP23–144 amyloid fibrils. First, earlier studies revealed that neither huPrP23–137 nor huPrP23–139 fragments are able to form amyloid fibrils (even in the presence of PrP23–144 amyloid seeds), and that the shortest C-truncated variant of huPrP23–144 showing fibril forming capacity is huPrP23–14113. This may be rationalized by the present high-resolution structural model, as this model reveals that the C-terminal hydrophobic residues I139 and F141 are directly involved in the dry interface between the protofilaments, playing an important role in fibril-stabilizing interactions.
Second, one of the most puzzling aspects of prion diseases is the phenomenon of species-dependent transmissibility barriers. Animal experiments14 and studies in vitro using the PrP23–144 model7,8 have led to the hypothesis that the key determinant of prion transmission is the ability of monomeric PrP substrate of the host to adapt to the structure of the donor prion aggregate. However, in the absence of high-resolution structural data, this model remains largely conceptual in nature. As part of our studies using the PrP23–144 system, we have established that there is a strong cross-seeding barrier between huPrP23–144 and Syrian hamster PrP23–144 (ShaPrP23–144), and that this barrier is fully controlled by the identity of just one amino acid residue at position 139 (Ile and Met in huPrP and ShaPrP, respectively)8,9. Previous attempts to rationalize this barrier were based on crystal structures for short (6–7 residues) peptide fragments, suggesting the role of structural differences at the backbone level15. The present data for fibrils formed by the entire PrP23–144 protein reveal, however, that this barrier results largely from side-chain packing effects, since the replacement of Ile139 with a longer Met side chain would likely disallow ShaPrP23–144 to adapt to the structure of the huPrP23–144 seed due to steric clashes between side chains of M139 and M112 in the adjacent subunits (Extended Data Fig. 6 and 7). Thus, this structural insight provides direct, molecular-level support for the “conformational adaptability” model of prion transmissibility barriers.
Finally, it should be noted that recent studies reported the structures of (non-infectious) amyloid fibrils formed by the recombinant full-length huPrP16,17 as well as brain-derived infectious prions18,19. While in the latter structure residues 108–141 are part of a much longer core region, the hairpin-like fold of this internal core segment is quite different from that observed for huPrP23–144 fibrils (Extended Data Fig. 8). More intriguingly, the structure of huPrP23–144 fibrils is also very different from that of fibrils formed by a non-physiological huPrP94–178 fragment20, even though the core regions of both fibrils are similar. This strongly suggests that residues outside the core region play an important role in conformational conversion of the prion protein, and thus, caution should be exercised when interpreting structural data for fibrils formed by short, non-physiological fragments of PrP.
Methods
Protein expression, purification, and fibril formation
The plasmids encoding huPrP23–144 or moPrP23–144 with N-terminal linker containing a 6xHis tag and a thrombin cleavage site were transformed into E. coli BL21(DE3) cells (Invitrogen) 13,22. The protein was expressed overnight after induction with 1 mM isopropyl β-D-1-thiogalactopyranoside. Cells were collected by centrifugation, lysed by sonication in Buffer G (10 mM Tris-HCl buffer, pH 8, 6 M guanidinium hydrochloride, 100 mM K2HPO4), and loaded on a Ni-charged nitrilotriacetic acid column. Guanidinium hydrochloride was removed by a slow gradient of 0–100% buffer B (10 mM Tris-HCl buffer, pH 8, 100 mM K2HPO4) in buffer G. The column was further washed by buffer W (10 mM Tris-HCl buffer, pH 8, 50mM imidazole, 100 mM K2HPO4) and the protein was eluted using buffer E (100 mM potassium phosphate buffer, pH 5.8, 500mM imidazole). The N-terminal 6xHis tag was cleaved with biotinylated thrombin (Novagen), followed by removal of thrombin using streptavidin-agarose beads. The protein was dialyzed against distilled water to remove free 6xHis tag, lyophilized and store at 4 ˚C. Lyophilized protein was re-suspended at a concentration of 400 μM in 50 mM potassium phosphate buffer containing 0.01% NaN3, pH 6.5. Initially, we attempted to prepare fibrils in an identical way as for previous solid-state NMR studies9,12, by incubating the solution of monomeric protein at 25˚C under quiescent conditions for 7 days. However, atomic force microscopy images of the end product showed heavily clumped fibrils that were not suitable for cryo-EM studies. Therefore, to prepare a better dispersed sample, these fibrils were sonicated and then used to seed a diluted solution (40 μM) of monomeric proteins, again under quiescent conditions. The final product of the seeded reaction was briefly sonicated and, when examined by AFM, showed relatively well-dispersed fibrils that were immediately used for cryo-EM studies.
Atomic force microscopy
For AFM analysis, 20 μl of fibril suspension was deposited on freshly cleaved mica substrate and incubated for 5 min. The surface was then washed three times with Milli-Q H2O and dried under N2. The images were obtained by NanoScope 9.1 using scan assist mode and a silicon probe (spring constant, 40 newtons/m) on a Bruker multimode atomic force microscope equipped with Nanoscope V controller.
Cryo-EM
Three hundred mesh lacey carbon grids (Ted Pella) were coated with 4 μl of 0.1 mg/ml graphene oxide (Sigma-Aldrich) followed by 4 μl of 0.1% poly-lysine (Sigma-Aldrich) 23,24. Three microliters of freshly sonicated fibril suspension (40 μM) was applied to the coated grid, blotted for 7 s, and plunge-frozen in liquid ethane using a Vitrobot Mark IV (ThermoFisher Scientific). Movies were collected on a Titan Krios G3i microscope (ThermoFisher Scientific) equipped with a BioQuantum K3 camera (Gatan, Inc.), with 0.414 Å/pixel in super resolution mode. A total of 3,604 movies for huPrP23–144 fibrils and 3,280 movies for moPrP23–144 fibrils were automatically collected using SerialEM25 with 7 shots per position. Further details are listed in Supplementary Table 1.
Data processing
Movies were first corrected for drifting and binned by a factor of 2 using MotionCor226. Contrast Transfer Functions (CTF) were estimated by CTFFIND 4.1.327. All further processing was carried out using RELION 3.128–30. Individual fibrils were manually picked, and overlapping segments were extracted using an inter-box distance of 10 Å and a box size of 512 pixels. Segments were first subjected to three rounds of reference-free two-dimensional (2D) classification using T = 8 and K = 200 to remove poorly defined classes, resulting in only segments contributing to clear 2D averages.
For huPrP23–144 fibrils, these segments selected from 2D classifications were then used for subsequent three-dimensional (3D) classification employing an initial model of a featureless cylinder generated by relion_helix_toolbox29. The initial helical rise (4.70 Å) was calculated from the 2D class layer line profile, and the initial helical twist (−2.4°) was calculated from the crossover distance. Two rounds of three-dimensional (3D) classification were performed using K = 8 and T = 4, resulting in 46,803 segments that contributed to a high-resolution reconstruction. The reconstructed map showed a C2 symmetry, which was imposed in the subsequent processing. After 3D classification, segments were re-extracted from micrographs with a box size of 256 pixels and then subjected to iterative high-resolution gold-standard 3D refinement, Bayesian polishing30, and CTF refinement31 to reconstruct the final high-resolution map. The overall resolution was calculated to be 2.86 Å from Fourier shell correlations at 0.143 between two independently refined half-maps. Refined helical symmetry (twist = −2.41°, rise = 4.71 Å) was imposed on the post-processed map for further model building.
For moPrP23–144 fibrils, 76,825 segments contributing to polymorph 1 and 231,951 segments contributing to polymorph 2 were processed separately, following the same approach as that of huPrP23–144 fibrils. A total of 10,628 segments were used to reconstruct the final map for polymorph 1 of moPrP23–144 fibrils. The overall resolution of this map was calculated to be 3.92 Å from Fourier shell correlations at 0.143 between two independently refined half-maps. Refined helical symmetry (twist = −2.36°, rise = 4.74 Å) was imposed on the post-processed map for further model building. For polymorph 2, a total of 28,818 segments were selected from 3D classifications for the final reconstruction. However, the quality of this map was not sufficient for further model building.
Model building
For huPrP23–144 fibrils, an initial model was built in Coot32 using Glycine-rich region of residues 121–130 as a guide. Five cross-sectional layers (20 chains) were built at the central region of the density map. The model was then subjected to iterative real-space refinements in PHENIX33,34. β-strands were identified manually from the density map and such secondary structure restraints were implemented in the real-space refinements. After real-space refinement, side-chain orientations were further adjusted in Coot32 to ensure energy-favored geometry. The final model was validated using the comprehensive validation method in PHENIX35. The amino acid sequence in this huPrP23–144 fibril model was then substituted with moPrP23–144 sequence, providing a starting point for modeling the structure of polymorph 1 of moPrP23–144 fibrils. Further refinements and validation of the model for this polymorph were performed as described for huPrP23–144 fibrils. For both models, maps containing twenty subunits was extracted manually from the reconstructed map using the UCSF Chimera package36 to calculate Fourier shell correlations between the map and the atomic model in PHENIX35.
Data Availability
Cryo-EM density map and the atomic models of human PrP23–144 fibrils have been deposited to the Electron Microscopy Data Bank and Protein data bank with the accession codes EMD-24514 and 7RL4, respectively. Those for mouse PrP23–144 fibrils have been deposited with the accession codes EMD-27458 and 8DJA. Coordinates used in structure comparison are available at the Protein Data Bank with accession code 6UUR, 7LNA, 6LNI, and 7DWV. All main data supporting the findings of this study are available within the article, Extended Data and Supplementary Information.
Extended Data
Extended Data Fig. 1. Morphology of huPrP23–144 amyloid fibrils.
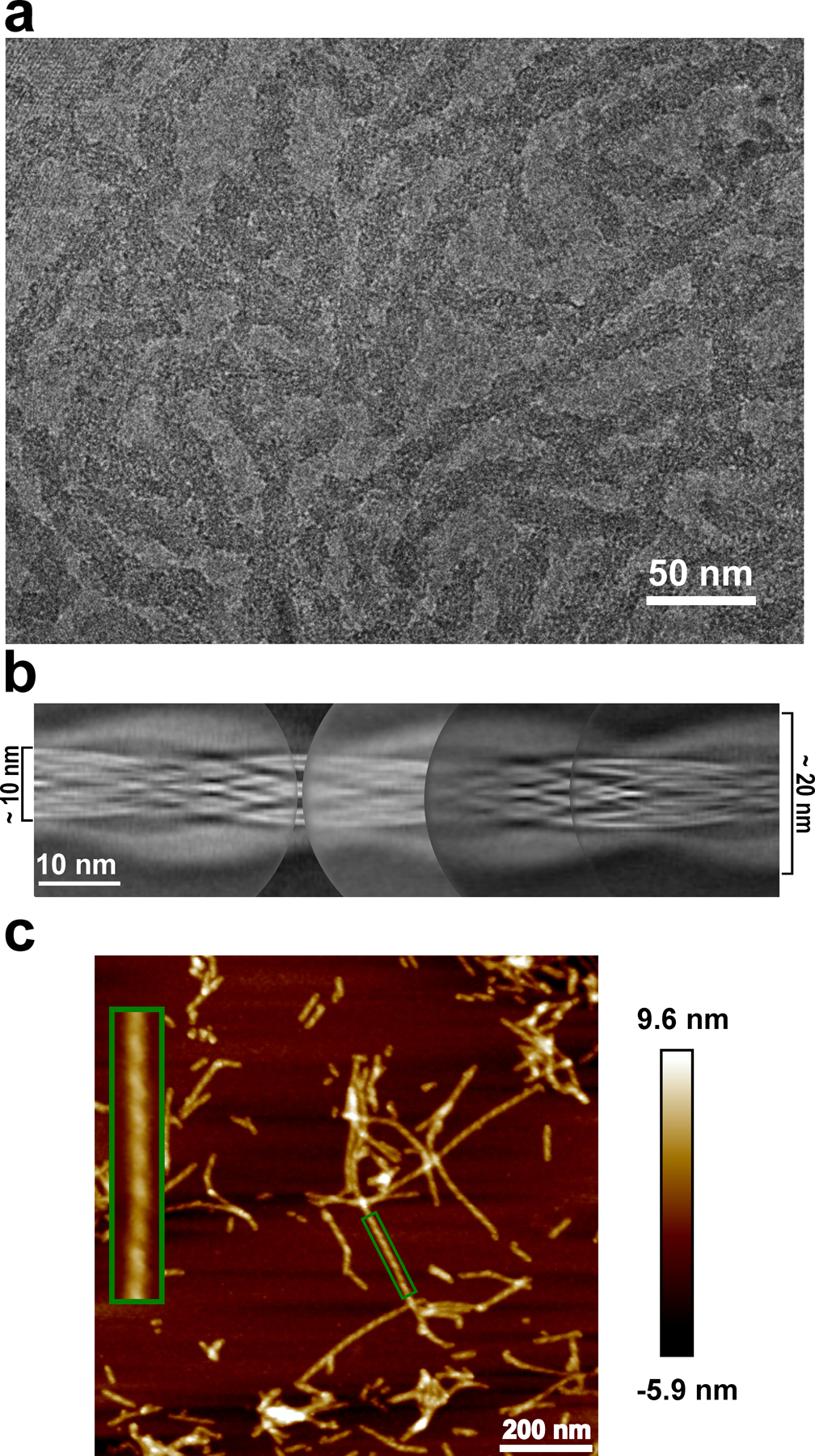
a, Cryo-EM image showing the sole morphology with an apparent twist observed for these fibrils, the same fibril morphology was observed in at least 600 images. b, Manually assembled full pitch of huPrP23–144 fibrils from multiple 2D class averages. The entire fibril has a thickness of ~20 nm and the highly ordered amyloid core has a thickness of ~10 nm. c, AFM image of huPrP23–144 fibrils. A representative fibril in the green box is enlarged to show the left-handed twist, the same fibril morphology was observed in at least 20 images.
Extended Data Fig. 2. Fourier shell correlation curves for huPrP23–144 amyloid fibrils.
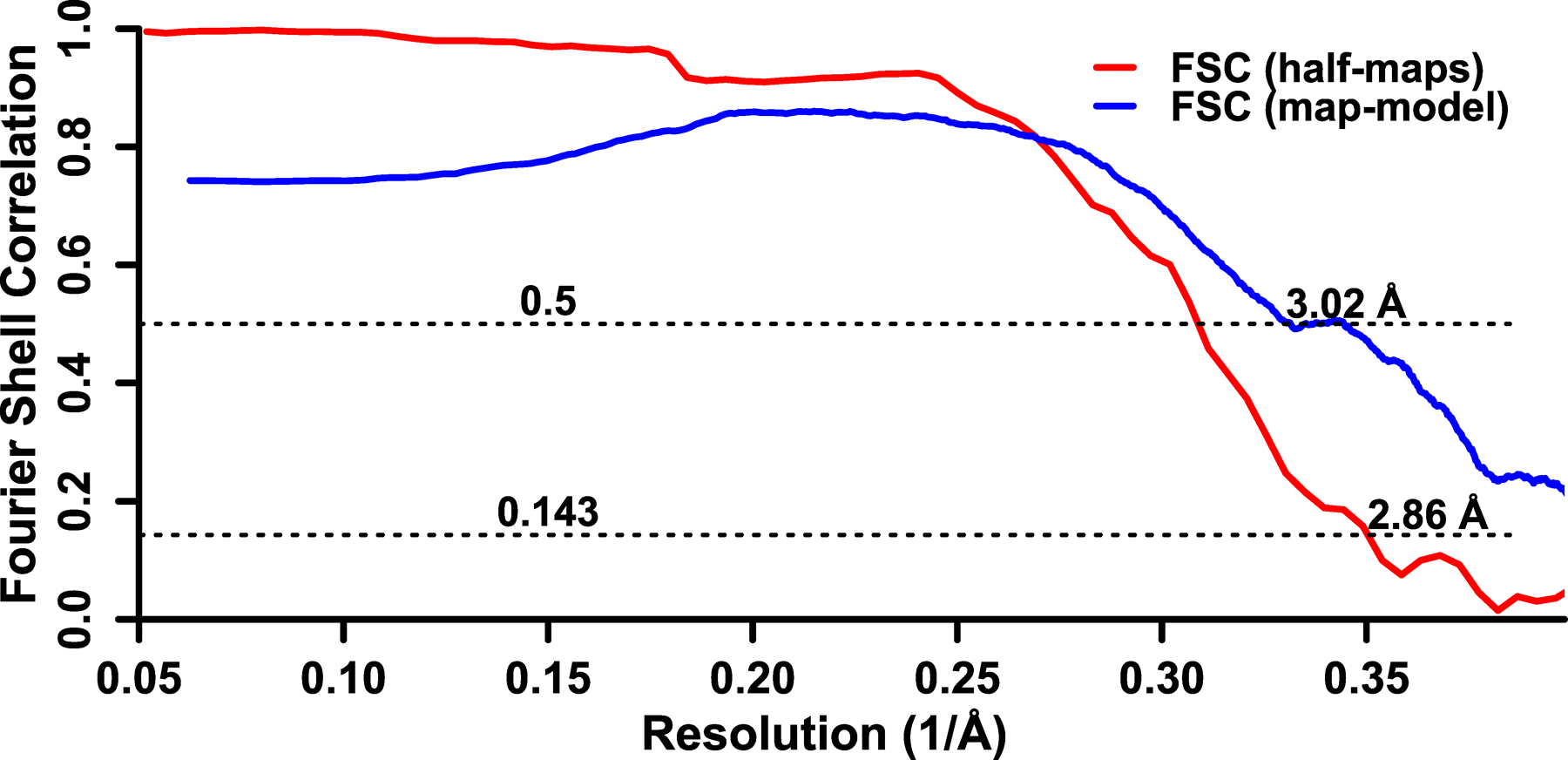
Fourier shell correlation between two independently refined half-maps is shown in red and Fourier shell correlation between the map and the atomic model is shown in blue.
Extended Data Fig. 3. Non-planar architecture of two representative huPrP23–144 protofilaments assembled through large hydrophobic interfaces.
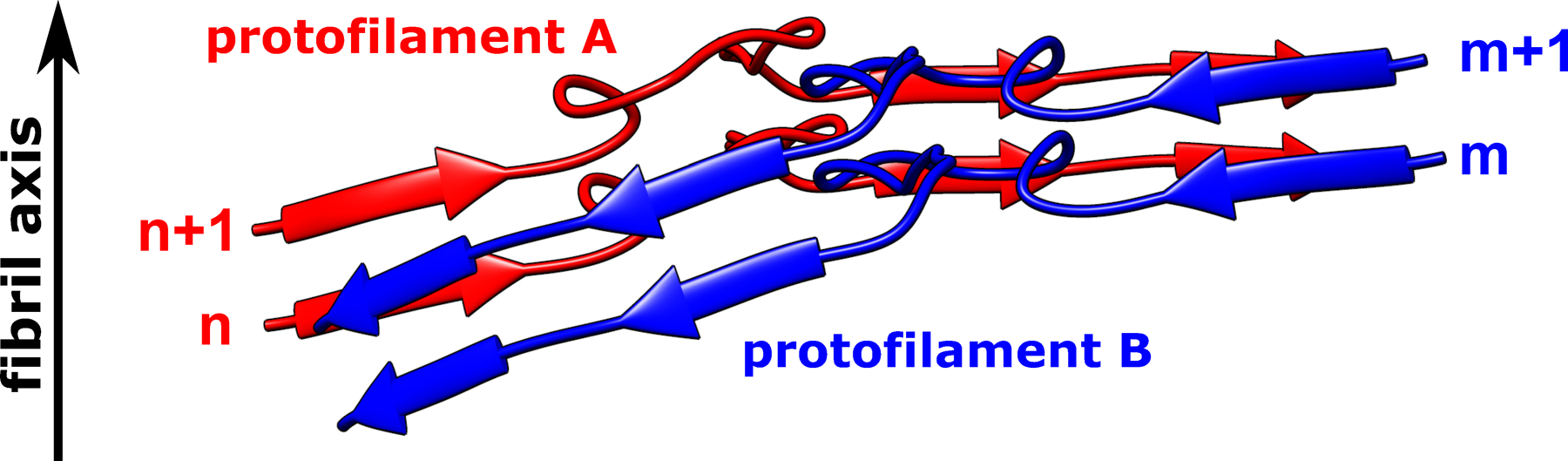
Subunit n in protofilament A interacts with two subunits in protofilament B (subunits m and m+1); subunit m+1 in protofilament B interacts with two subunits in protofilament A (subunits n and n+1). Such a non-planar conformation results in rugged surfaces at fibril ends, with N-terminal residues at the top end and C-terminal residues at the bottom end exposed to water. A similar non-planar assembly is observed for subunits in protofilaments C and D.
Extended Data Fig. 4. Comparison of the structural model for huPrP23–144 fibrils determined herein by cryo-EM (a) with the low-resolution model previously suggested based on ssNMR data9,12 (b).
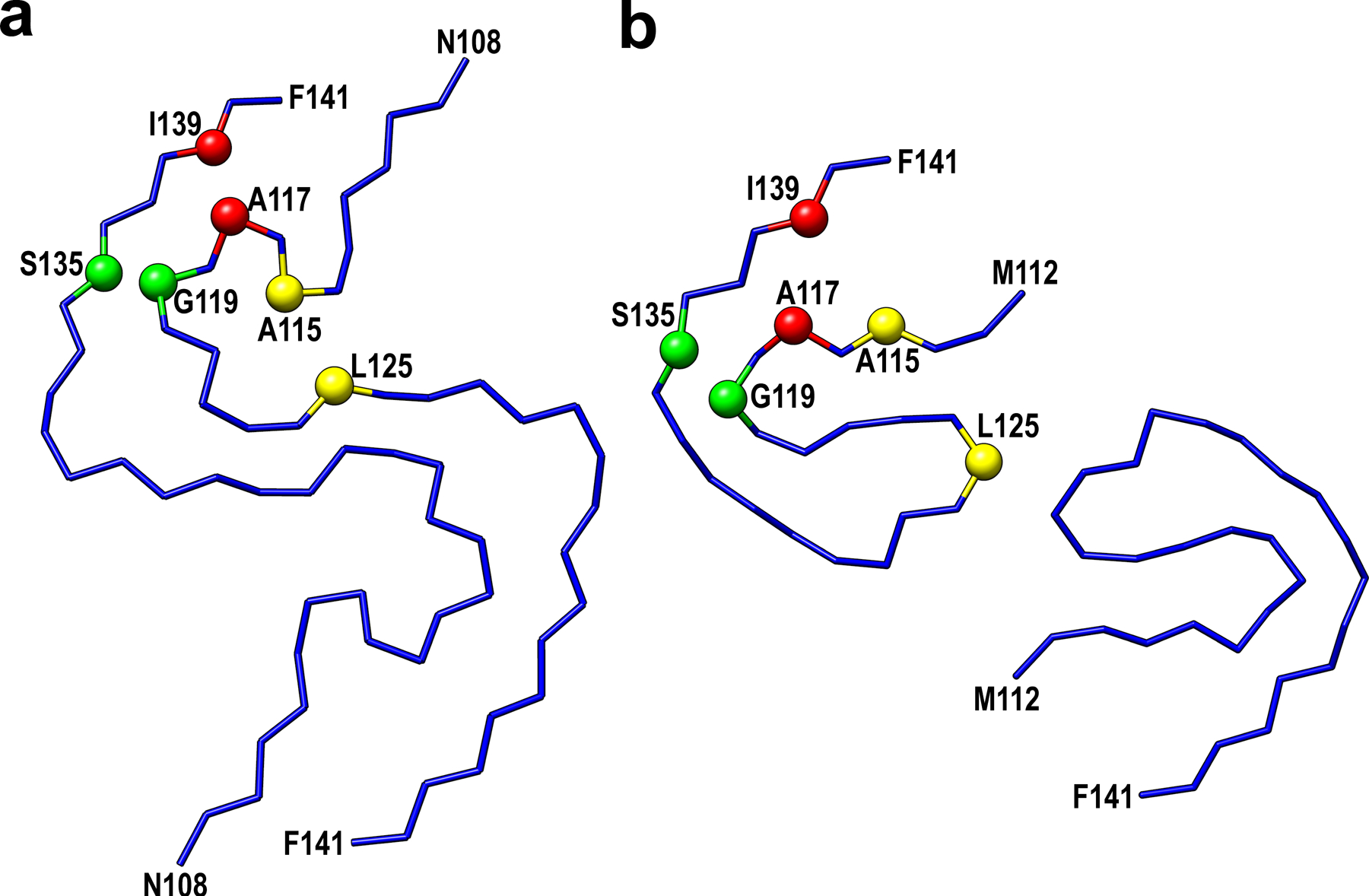
Selected residues involved in close interactions between side-chains in these two models are labeled in red (Ala117-Ile139), yellow (Ala115-Leu125), and green (Gly119-Ser135).
Extended Data Fig. 5. Cryo-EM structure of amyloid fibrils generated from moPrP23–144.
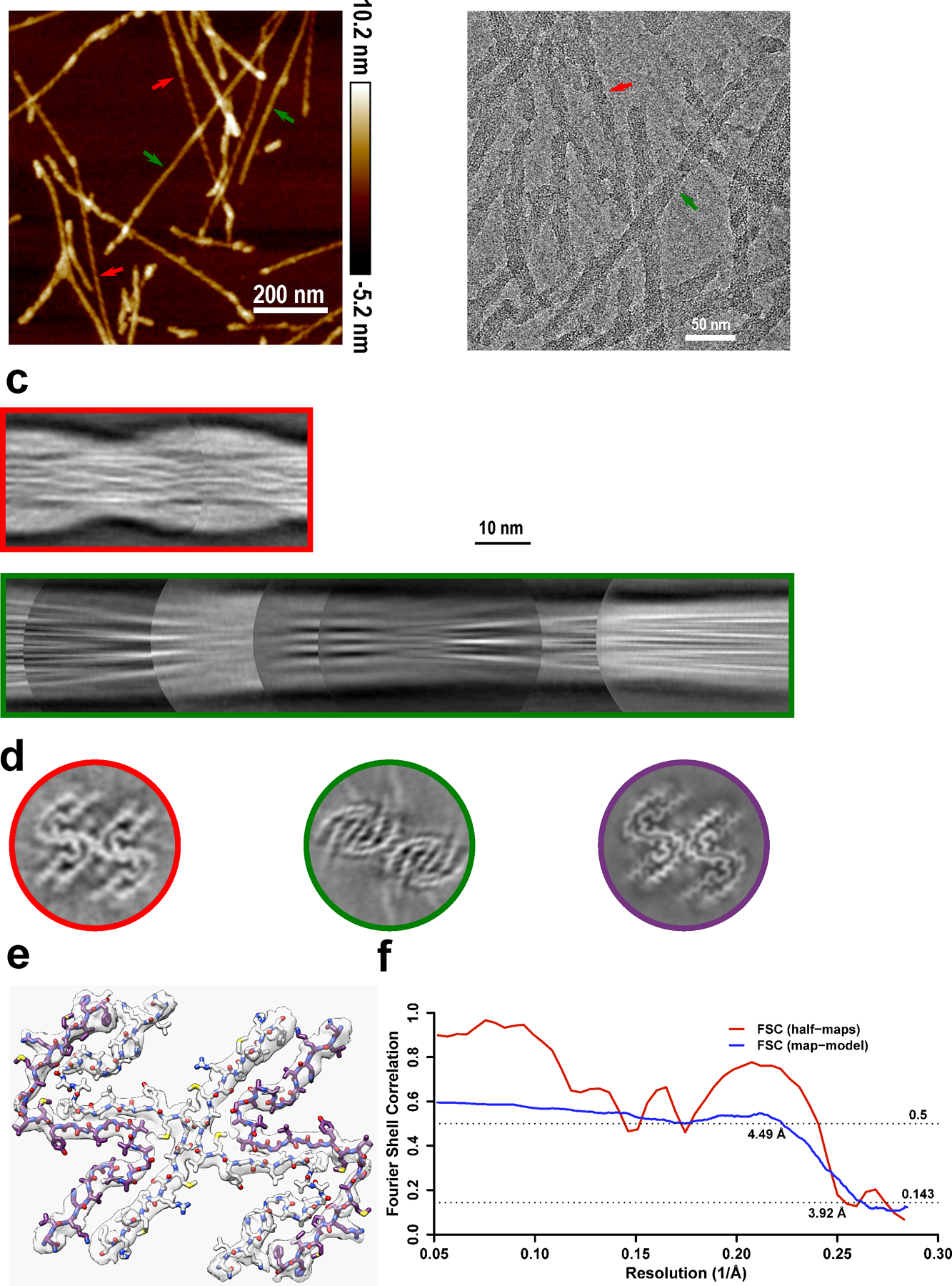
a-b, Two types of polymorphs can be found in AFM images (a) and cryo-EM micrographs (b). Polymorph 1 (red arrows) showed a larger left-handed twist like huPrP23–144 fibrils, and polymorph 2 (green arrows) showed a much smaller right-handed twist. The same fibril morphologies were observed in at least 1,000 cryo-EM images or 20 AFM images for each type of sample. c, Manually assembled half-pitch of both polymorphs from multiple 2D class averages. d, 3D maps of moPrP23–144 and huPrP23–144 fibrils represented by a central slice perpendicular to the fibril axis. Polymorph 1 of moPrP23–144 fibrils (red) showed the same fold as that in huPrP23–144 fibrils (purple). Polymorph 2 of moPrP23–144 fibrils (green) showed a distinctly different fold. e-f, An atomic model built based on the map for polymorph 1 of moPrP23–144 fibrils, with a map resolution of 3.92 Å and model resolution of 4.49 Å. The backbone fold in this model is identical to that in huPrP23–144 fibrils. The structure of polymorph 2 of moPrP23–144 fibrils could not be determined due to poor quality of cryo-EM data for this polymorphic form.
Extended Data Fig. 6. The structural model illustrating seeding reaction at the top end of huPrP23–144 fibrils in the presence of huPrP23–144 and ShaPrP23–144 substrates.
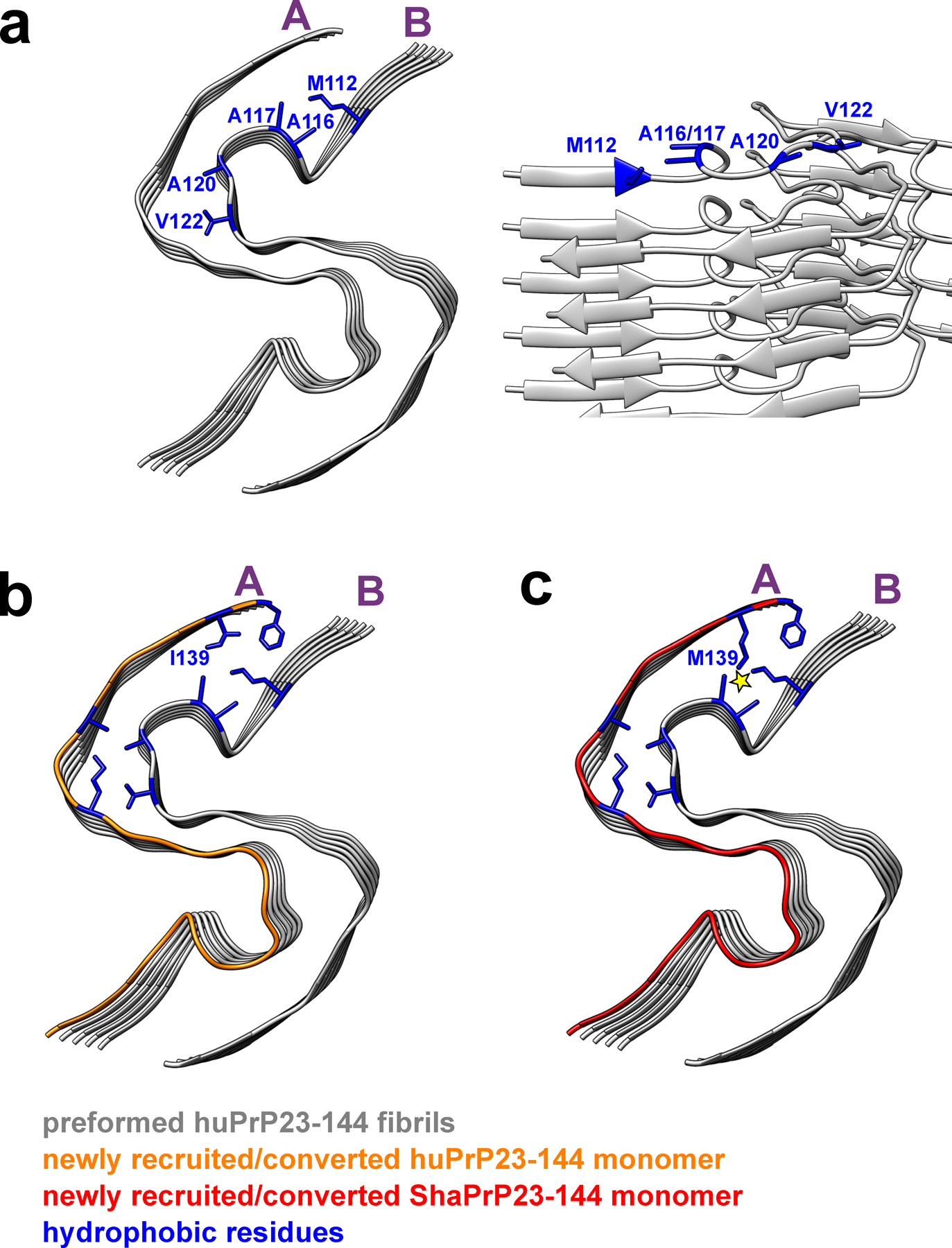
Two representative protofilaments (A and B) are shown only. a, Top and side views of a preformed huPrP23–144 fibril (seed) with solvent-exposed hydrophobic side chains shown in blue. b, Top view of a preformed huPrP23–144 fibril (grey) with a newly recruited and converted subunit of huPrP23–144 (orange). c, Top view of a preformed huPrP23–144 fibril (grey) with a newly recruited subunit of ShaPrP23–144 (red). Adaptation of ShaPrP23–144 to the structure of huPrP23–144 seed would lead to significant intermolecular steric clashes between bulky, elongated side chains of M112 and M139 (as indicated by the yellow star), explaining a cross-seeding barrier.
Extended Data Fig. 7. The structural model illustrating seeding reaction at the bottom end of huPrP23–144 fibrils in the presence of huPrP23–144 and ShaPrP23–144 substrates.
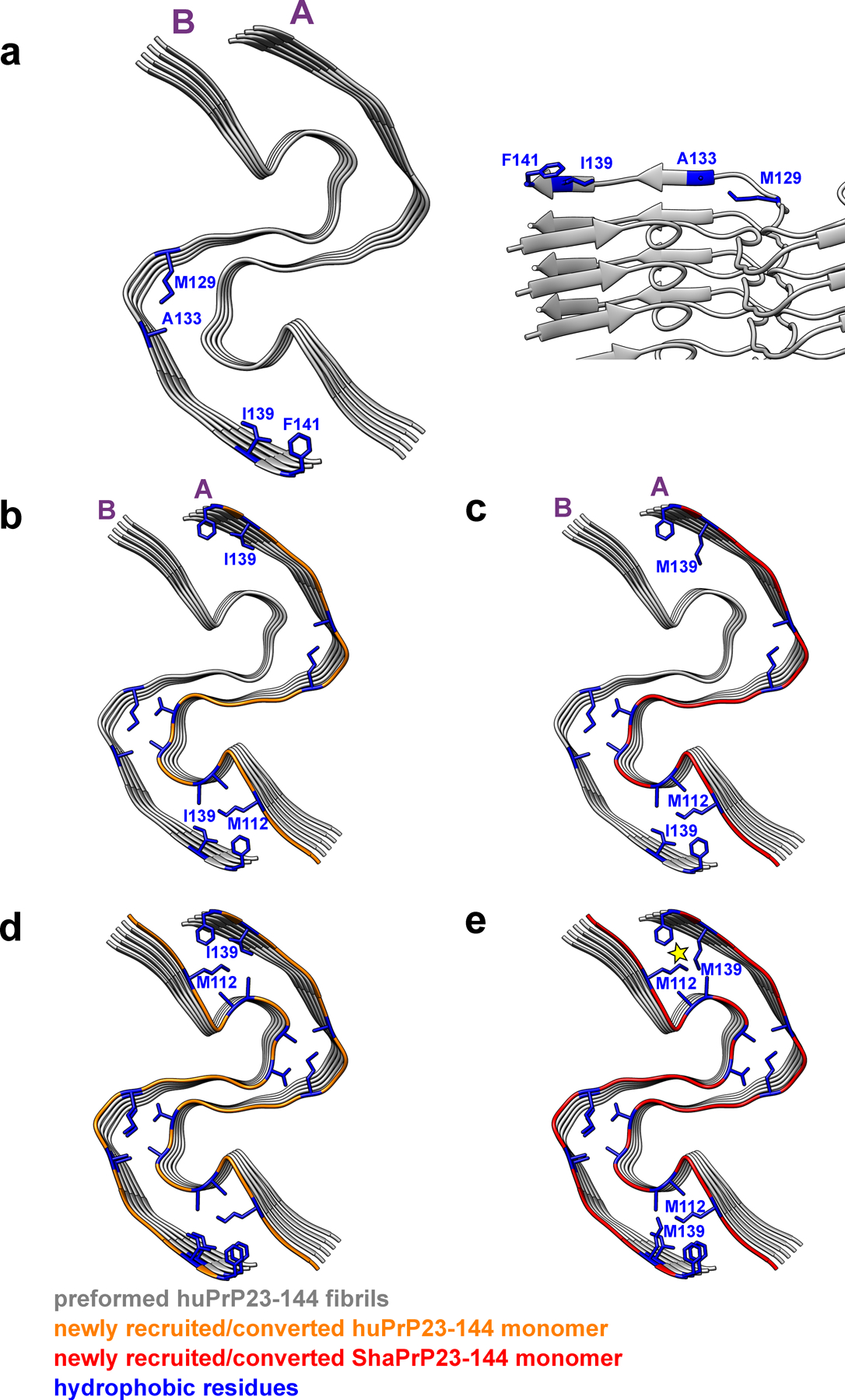
Two representative protofilaments (A and B) are shown for illustrative purposes. a, Bottom and side views of a preformed huPrP23–144 fibril (seed) with solvent-exposed hydrophobic side chains shown in blue. b, Bottom view of a preformed huPrP23–144 fibril (grey) with a newly recruited (to protofilament A) and converted subunit of huPrP23–144 (orange). c, Bottom view of a preformed huPrP23–144 fibril (grey) with a newly recruited (to protofilament A) and converted first subunit of ShaPrP23–144 (red). Due to non-planar structure, C-terminal hydrophobic residues at this end of protofilament B are protruding to water. Thus, recruitment of the first ShaPrP subunit would not result in any steric clashes. d, Bottom view of a preformed huPrP23–144 fibril (grey) with a second newly recruited (to protofilament B) and converted subunit of huPrP23–144 (orange). e, Bottom view of a preformed huPrP23–144 fibril (grey) with a second recruited (to protofilament B) subunit of ShaPrP23–144 (red). Adaptation of this subunit to the structure of the huPrP23–144 seed would lead to intermolecular steric clashes between side chains of M139 and M112 (as indicated by the yellow star), explaining a cross-seeding barrier.
Extended Data Fig. 8. The backbone cryo-EM structures of PrP amyloid fibrils including recombinant huPrP23–144 fibrils determined herein (a, PDB 7RL4), recombinant huPrP94–178 fibrils20 (b, PDB 6UUR), brain-derived 263K prions18 (c, PDB 7LNA), recombinant huPrP23–231 fibrils16 (d, PDB 6LNI), and recombinant E196K huPrP23–231 fibrils17 (e, PDB 7DWV).
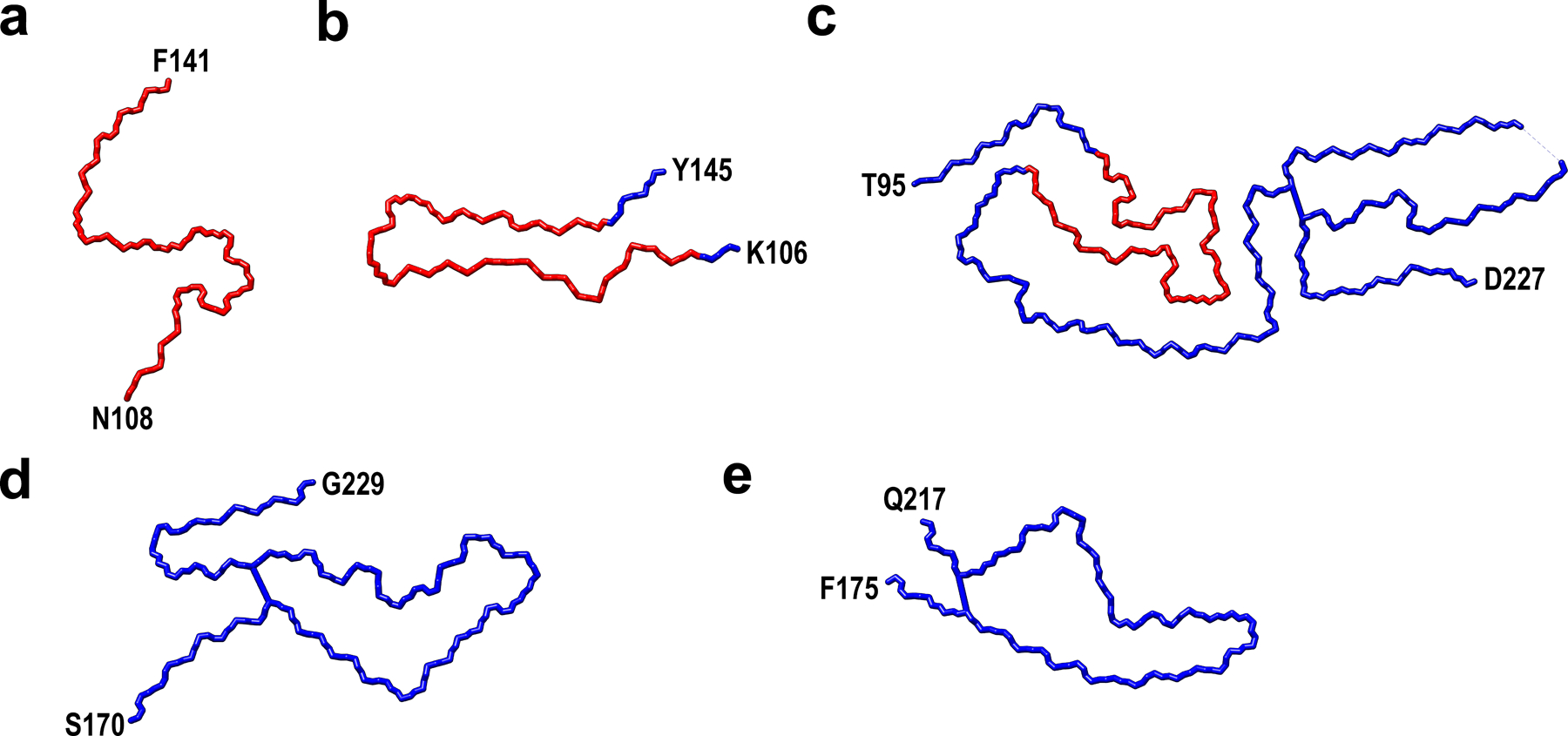
The core of huPrP23–144 fibrils is marked as red.
Supplementary Material
Acknowledgements
This work was supported by NIH grants GM094357 (C.P.J. and W.K.S.) and NS103848 (W.K.S.). We thank Kunpeng Li for help with acquisition of cryo-EM data. We are grateful to the Cryo-EM Core at CWRU School of Medicine (especially Dr. Sudha Chakrapani and Dr. Kunpeng Li) for the access to cryo-EM instrumentation.
Footnotes
Peer Review Information:
Nature Structural and Molecular Biology thanks Cong Liu and Byron Caughey for their contribution to the peer review of this work.
Competing Interests
The authors declare no competing interests.
References
- 1.Prusiner SB Prions. Proceedings of the National Academy of Sciences 95, 13363–13383 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cobb NJ & Surewicz WK Prion diseases and their biochemical mechanisms. Biochemistry 48, 2574–2585 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caughey B & Kraus A Transmissibility versus pathogenicity of self-propagating protein aggregates. Viruses 11, 1044 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kitamoto T, Iizuka R & Tateishi J An amber mutation of prion protein in gerstmann-sträussler syndrome with mutant PrP plaques. Biochemical and Biophysical Research Communications 192, 525–531 (1993). [DOI] [PubMed] [Google Scholar]
- 5.Ghetti B et al. Vascular variant of prion protein cerebral amyloidosis with τ-positive neurofibrillary tangles: The phenotype of the stop codon 145 mutation in PRNP. Proc Natl Acad Sci U S A 93, 744–748 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi JK et al. Amyloid fibrils from the N-terminal prion protein fragment are infectious. Proc Natl Acad Sci U S A 113, 13851–13856 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vanik DL, Surewicz KA & Surewicz WK Molecular basis of barriers for interspecies transmissibility of mammalian prions. Molecular Cell 14, 139–145 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Jones EM & Surewicz WK Fibril conformation as the basis of species- and strain-dependent seeding specificity of mammalian prion amyloids. Cell 121, 63–72 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Theint T et al. Species-dependent structural polymorphism of Y145Stop prion protein amyloid revealed by solid-state NMR spectroscopy. Nature Communications 8, 753 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao Q, Boyer DR, Sawaya MR, Ge P & Eisenberg DS Cryo-EM structures of four polymorphic TDP-43 amyloid cores. Nature Structural and Molecular Biology 26, 619–627 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bansal A et al. AA amyloid fibrils from diseased tissue are structurally different from in vitro formed SAA fibrils. Nature Communications 12, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Theint T et al. Structural Studies of Amyloid Fibrils by Paramagnetic Solid-State Nuclear Magnetic Resonance Spectroscopy. J Am Chem Soc 140, 13161–13166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kundu B et al. Nucleation-dependent conformational conversion of the Y145Stop variant of human prion protein: Structural clues for prion propagation. Proc Natl Acad Sci U S A 100, 12069–12074 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collinge J & Clarke AR A general model of prion strains and their pathogenicity. Science (1979) 318, 930–936 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Apostol MI, Wiltzius JJW, Sawaya MR, Cascio D & Eisenberg D Atomic structures suggest determinants of transmission barriers in mammalian prion disease. Biochemistry 50, 2456–2463 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang LQ et al. Cryo-EM structure of an amyloid fibril formed by full-length human prion protein. Nature Structural and Molecular Biology 27, 598–602 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Wang L, Zhao K, Yuan H-Y, Li X & Dang H-B Familial prion disease-related mutation E196K displays a novel amyloid fibril structure revealed by cryo-EM. bioRxiv 2021.02.18.431846 (2021) doi: 10.1101/2021.02.18.431846. [DOI] [PMC free article] [PubMed]
- 18.Kraus A et al. High-resolution structure and strain comparison of infectious mammalian prions. Molecular Cell 81, 4540–4551.e6 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Artikis E, Kraus A & Caughey B Structural biology of ex vivo mammalian prions. Journal of Biological Chemistry 0, 102181 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glynn C et al. Cryo-EM structure of a human prion fibril with a hydrophobic, protease-resistant core. Nature Structural and Molecular Biology 27, 417–423 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kyte J & Doolittle RF A simple method for displaying the hydropathic character of a protein. Journal of Molecular Biology 157, 105–132 (1982). [DOI] [PubMed] [Google Scholar]
References (to Methods)
- 22.Morillas M, Swietnicki W, Gambetti P & Surewicz WK Membrane environment alters the conformational structure of the recombinant human prion protein. Journal of Biological Chemistry 274, 36859–36865 (1999). [DOI] [PubMed] [Google Scholar]
- 23.Yu G, Li K & Jiang W Antibody-based affinity cryo-EM grid. Methods 100, 16–24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bokori-Brown M et al. Cryo-EM structure of lysenin pore elucidates membrane insertion by an aerolysin family protein. Nature Communications 7, 1–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mastronarde DN Automated electron microscope tomography using robust prediction of specimen movements. Journal of Structural Biology 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Zheng SQ et al. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nature Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rohou A & Grigorieff N CTFFIND4: Fast and accurate defocus estimation from electron micrographs. Journal of Structural Biology 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheres SHW RELION: Implementation of a Bayesian approach to cryo-EM structure determination. Journal of Structural Biology 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He S & Scheres SHW Helical reconstruction in RELION. Journal of Structural Biology 198, 163–176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zivanov J, Nakane T & Scheres SHW A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ 6, 5–17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zivanov J, Nakane T & Scheres SHW Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ 7, 253–267 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallographica Section D: Biological Crystallography 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Afonine PV et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallographica Section D: Structural Biology 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liebschner D et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallographica Section D: Structural Biology 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Afonine PV et al. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallographica Section D: Structural Biology 74, 814–840 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pettersen EF et al. UCSF Chimera - A visualization system for exploratory research and analysis. Journal of Computational Chemistry 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM density map and the atomic models of human PrP23–144 fibrils have been deposited to the Electron Microscopy Data Bank and Protein data bank with the accession codes EMD-24514 and 7RL4, respectively. Those for mouse PrP23–144 fibrils have been deposited with the accession codes EMD-27458 and 8DJA. Coordinates used in structure comparison are available at the Protein Data Bank with accession code 6UUR, 7LNA, 6LNI, and 7DWV. All main data supporting the findings of this study are available within the article, Extended Data and Supplementary Information.


