Abstract
Versatile genome editing can be facilitated by the insertion of DNA sequences into specific locations. Current protocols involving CRISPR and Cas proteins rely on low efficiency homology-directed repair or non-homologous end joining with modified double-stranded DNA oligonucleotides as donors. Our simple protocol eliminates the need for expensive equipment, chemical and enzymatic donor DNA modification, or plasmid construction by using polyethylene glycol-calcium to deliver non-modified single-stranded DNA oligonucleotides and CRISPR-Cas9 ribonucleoprotein into protoplasts. Plants regenerated via edited protoplasts achieved targeted insertion frequencies of up to 50% in Nicotiana benthamiana and 13.6% in rapid cycling Brassica oleracea without antibiotic selection. Using a 60 nt donor containing 27 nt in each homologous arm, 6/22 regenerated N. benthamiana plants showed targeted insertions, and one contained a precise insertion of a 6 bp HindIII site. The inserted sequences were transmitted to the next generation and invite the possibility of future exploration of versatile genome editing by targeted DNA insertion in plants.
Introduction
To insert DNA into a specific location in the plant genome, a DNA double-strand break (DSB) must be induced at the target position to facilitate the donor DNA (DD) insertion into this position via homology-directed repair (HDR)1–12 or non-homologous end joining (NHEJ).13–15 Many tools for creating DSBs are currently available.16–18 Due to its versatility and simplicity, the combination of CRISPR and Cas proteins has become a favorite approach among genetic engineers.4 In addition to CRISPR-Cas reagents, increasing the amount of delivered DD enhances targeted insertion (TI) efficiency. For example, in maize (Zea mays), targeted mutagenesis using Cas9 and guide RNA (gRNA) has been achieved via different methods, although TI plants were obtained only via biolistic methods rather than Agrobacterium-mediated transformation because the number of DD copies delivered by the latter method was low.2
Protoplasts offer an alternative system for genome editing and genetic transformation, since they enable the delivery of high numbers of DD copies19,20 to enhance TI in the genome before plant regeneration.19 Like the transcription activator-like effector nuclease genome editing system,21 CRISPR reagents can be delivered into protoplasts via polyethylene glycol (PEG)-Ca2+-mediated transfection of ribonucleoprotein (RNP) or plasmid DNA, and targeted mutations can be achieved.22–29 The mutated protoplasts can then be regenerated into plants without chimerism, and the mutated alleles are passed onto the progeny.22,24,25 Here, we describe a simple, high-efficiency TI method based on the protoplast strategy for genome editing in plants using Cas9-gRNA and non-modified synthetic single-stranded oligonucleotide DNA (ssODN) DDs that does not require expensive equipment.
Methods
Protoplast isolation, transfection, and regeneration
Nicotiana benthamiana and rapid cycling Brassica oleracea (RCBO) plants were propagated in half-strength Murashige and Skoog (½ MS) medium supplemented with 30 g/L sucrose and 1% agar under a 12-h/12-h light/dark cycle at 25°C. Protoplast isolation was performed according to Lin et al.24 and Hsu et al.29 except for the digestion solution (½ MS medium supplemented with 1 mg/L 1-naphthaleneacetic acid [NAA], 0.3 mg/L kinetin, 30 g/L sucrose, 0.4 M mannitol [1N0.3K], 1% cellulose, 0.5% macerozyme) and digestion time (3 days) in N. benthamiana. The protoplasts were co-transfected with RNP and 50 μg synthetic ssODN DNA (Genomics) according to Woo et al.22 Transfected protoplasts were incubated in a Petri dish 5 cm in diameter containing liquid callus medium (1N0.3K) for 3 weeks. RCBO calli were additionally incubated in 1 mg/L NAA, 1 mg/L 6-benzyladenine (BA), and 0.25 mg/L 2,4-dichlorophenoxyacetic acid for 3 days in the dark. The calli were transferred to liquid shooting medium (containing 2 mg/L BA for N. benthamiana, 0.1 mg/L thidiazuron for RCBO) in a Petri dish 9 cm in diameter and incubated at 25°C for 3–4 weeks in the light (16-h/8-h light/dark, 3,000 lux). Green explants >5 mm were incubated in solid shooting medium and subcultured every 4 weeks. Shoot clusters with leaves were then transferred to solidified rooting medium (HB1: 3 g/L Hyponex No. 1, 2 g/L tryptone, 20 g/L sucrose, 1 g/L activated charcoal, 10 g/L agar, pH 5.2).
Cas9 protein purification, single-guide RNA synthesis, and Cas9 RNP nucleofection
Preparation of Cas9 protein and single-guide RNA (sgRNA) and Cas9 RNP nucleofection were performed according to Huang et al.30 Cas9 recombinant protein was overexpressed in Escherichia coli BL21 harboring the plasmid pMJ915 (Addgene; #69090). Cas9 protein was purified and stored at −80°C in Cas9 RNP buffer (20 mM HEPES at pH 7.5, 150 mM KCl, 10% glycerol, and 1 mM β-mercaptoethanol). The sgRNAs were synthesized by in vitro transcription (IVT) using T7 RNA polymerase (New England Biolabs; M0251L). The DNA oligonucleotides used for IVT template assembly are listed in Supplementary Table S1. The final sgRNA products were dissolved in Cas9 RNP buffer, quantified using a NanoDrop Lite (Thermo Fischer Scientific), and stored as aliquots at −80°C. Cas9 RNP complexes were assembled immediately before nucleofection by mixing equal volumes of 40 μM Cas9 protein and 88.3 μM sgRNA at a molar ratio of 1:2.2 and incubating at 37°C for 10 min.
Validation of TIs in protoplasts and regenerated plants
Genomic DNA was extracted from pooled protoplasts and regenerated plants using a Geno Plus Mini Genomic DNA Extraction Kit (GG2002; Viogene). To amplify the genomic region targeted by the sgRNA, the corresponding pairs of primers were designed. Primer sequences are shown in Supplementary Table S1. The polymerase chain reaction (PCR) conditions were 94°C for 5 min, 35 cycles of 94°C for 30 s, annealing at 55–63°C for 30 s, polymerization at 72°C for 30 s, followed by 72°C for 3 min. The PCR products were digested using the appropriate restriction enzyme or RNP and subjected to electrophoresis. The PCR products that could not be digested by restriction enzymes at target sites near the protospacer adjacent motif (PAM) sequence (BstNI in NbPDS1 E target site) or RNP were defined as edited. The PCR products that could be digested by the restriction enzyme in the DD and for which sequencing was confirmed are defined as TI. The PCR products were cloned into the T&A vector (FYC002-20P; Yeastern Biotech). Putative colonies containing the edited DNA were confirmed by Sanger sequencing.
Whole-genome sequencing for off-target DD insertion analysis
Leaves of N. benthamiana protoplast regenerated plants were collected for genomic DNA purification. Genomic DNA for genome sequencing was extracted using a Plant DNA Purification Kit (DP320; Tiangen). Paired-end libraries of DNA were constructed by the NEBNext Ultra II DNA Library Prep for Illumina Kit (New England Biolabs; E7645L) with 2 × 150 bp with an average insert size of ∼900 bp and sequenced on a NovaSeq 6000 platform (Illumina; 20028312). Three technical replicates were performed for each sample. Total reads were 120 Gbp per regenerated plant, and the sequencing depth was more than 30 × . To ensure the read quality, the first 10 bases of Illumina reads were removed, and the last 141 bases were retained for further analysis. High-quality Illumina reads were aligned with the N. benthamiana genome (genome assembly v.1.0.1) by BWA (v.0.7.17) with default setting. Single nucleotide polymorphisms (SNPs) and insertions and deletions (indels) were identified by DeepVariant (v.1.1.0-GPU, WGS model) and subsequently processed by GLnexus (v.1.2.7, DeepVariantWGS model) and bcftools (v.1.10.2, FMT/GQ< = 20 and GT = “RA”) for a joint variant calling. Off-target sites were predicted by Cas-OFFinder (v.2.4.1) with default settings. Sample specific SNPs and indels were compared to predicted off-target sites from Cas-OFFinder to identify coincident sites. To identify the potential off-target insertion sites, target sequence (Exp. 1 DD sequence, TTTGCGATGCCTAACAAGCTTCAGGGGGAGTTCAGCCGCTT) was used as the query in a high sensitivity BLASTN search strategy (-dust no -soft_masking false -word_size 4 -gapopen 1 -gapextend 2 -penalty -1 -reward 1 -evalue 5000 -perc_identity 80 -num_alignments 50000) against the Illumina reads. The high-scoring segment pairs of Illumina reads were filtered according to the known edited sequence lengths in different samples. Candidate Illumina reads were then retrieved to examine the exact location in the N. benthamiana genome further by BLASTN (-dust no -soft masking false -task blastn-short -evalue 0.1 -perc_identity 90 -num_descriptions 1 -num_alignments 1). If the Illumina read was identical to the published genome sequence, it was concluded that these sequences were the same as the DD already existed and were not caused by TI. If there was a difference from the published wild-type (WT) genome sequence, and the difference was the same as the DD, it was regarded as an off-targeted insertion. The raw reads were deposited in the NCBI SRA database (BioProject: PRJNA667297; https://www.ncbi.nlm.nih.gov/bioproject/PRJNA667297).
Results and Discussion
The protoplast regeneration protocol (Fig. 1) used in the present study was modified from previously published protocols.24,25,29 A key step to our approach was the observation that the phase of the cell cycle largely governs the choice of pathway used for DNA repair: NHEJ is the major DNA repair pathway during the G1, S, and G2 phases, whereas HDR occurs only during the late S and G2 phases.31,32 Cell-cycle synchronization is an effective strategy for enhancing TI efficiency in human embryonic kidney 293T cells.33 Here, 5-ethynyl-2′-deoxyuridine (EdU) staining was used for detection of S-phase cell-cycle progression. To increase the number of cells in the late S and G2 phases, N. benthamiana leaves were incubated in 1N0.3K solid medium for 3 days before protoplast isolation (Supplementary Fig. S1A). In comparison with 1N0.3K treatments, no EdU signal was identified in protoplasts incubated in ½ MS, 0.4 M mannitol solid medium (Supplementary Fig. S1B).34 Based on single cell analysis,24 the TI efficiency increased after incubation in 1N0.3K (Supplementary Fig. S1C). For N. benthamiana, to simplify the procedure, we used 1N0.3K for digestion solution preparation and incubated the cut leaf material for 3 days.
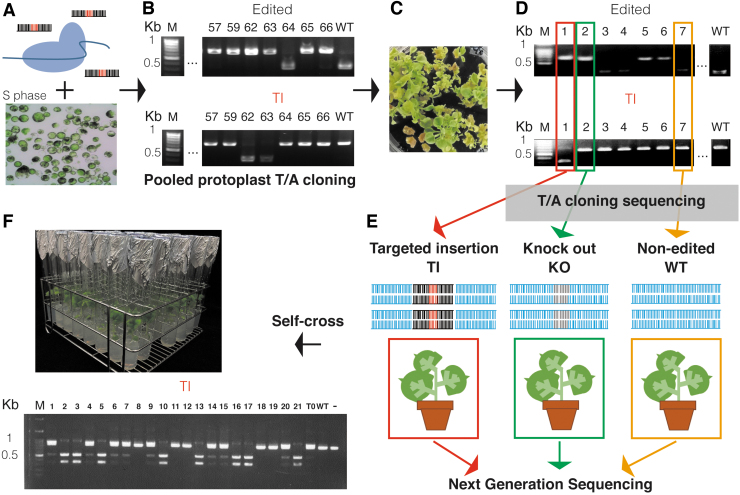
FIG 1.
Strategy for targeted DNA insertion using ribonucleoprotein (RNP) and single-stranded oligonucleotide DNA (ssODN) in Nicotiana benthamiana. (A) RNP and ssODN are delivered to S-phase protoplasts by polyethylene glycol-Ca2+–mediated transfection (Supplementary Fig. S1). (B) Pooled transfected protoplasts DNA was isolated, and the target gene was amplified by polymerase chain reaction (PCR) and cloned into the T/A vector for genotyped using restriction enzymes or RNP. (C) Transfected protoplasts are regenerated. (D) DNA from the regenerated plants is amplified by the PCR and genotyped using restriction enzymes or RNP. (E) DNA is purified from targeted insertion (TI; red), knockout (green), and non-edited (yellow) regenerated plants and sequenced to determine whether ssODN was inserted into other positions. (F) Offspring are genotyped and sequenced to test whether the inserted sequence is heritable. Color images are available online.
To bypass the need for plasmid construction, we used RNP as the Cas9-gRNA reagent. For the DD, we used short non-modified synthetic ssODN, which is relatively inexpensive and easy to obtain. The RNP and ssODN were delivered into protoplasts using PEG-Ca2+-mediated transfection (Fig. 1A).20,22 After 3 days of incubation in 1N0.3K liquid callus medium, genomic DNA was isolated from the protoplasts, and the target gene was amplified by PCR and cloned into the T/A vector for genotyping and Sanger sequencing to assess the TI efficiency (Fig. 1B). These protoplasts were cultured and regenerated (Fig. 1C).24,25,29 The rooted plants were incubated in the growth chamber and genotyped (Fig. 1D). These regenerated plants grew normally and produced seeds. DNA from two types of edited and regenerated plants, carrying TI or knockout (KO), and one non-edited (WT) and regenerated plant were purified for genome-wide sequencing to assess the presence or absence of off-target DD insertion as well as genome stability through the protoplast regeneration processes (Fig. 1E). DNA from the TI T1 progeny was extracted for genotyping to test whether the inserted fragment was heritable (Fig. 1F).
For the regenerated plants with TI, we conducted experiments using PHYTOENE DESATURASE1 (NbPDS1) as a target site in N. benthamiana (Fig. S2).35 Based on single cell analysis,24 the TI only occurred when protoplasts were transfected with DD and RNP (Supplementary Fig. S2A). To evaluate the effect of the length of the homologous arms and the total length of ssODN on TI efficiency at the expected sgRNA target position, we synthesized the DD of 20 nt, 40 nt, or 60 nt ssODN carrying a HindIII site and added 7, 17, or 27 homologous arms, respectively, on the left and right sides (Supplementary Fig. S2B). We genotyped NbPDS1 PCR products from the edited and regenerated plants. No TI were identified with the 20 nt ssODN in regenerated plants, whereas the 40 and 60 nt ssODNs produced TI in regenerated plants with efficiencies of 27.3–31.8% without antibiotic or phenotypic selection (Supplementary Fig. S2C and D). These results suggested that the length of the ssODN donor is an important factor in determining the TI efficiency. In a previous study, a 59 nt ssODN (ssADHE) failed to give rise to successful insertions in 23 T0 rice (Oryza sativa) plants.13 There were no homologous arms in ssADHE, and a lower DD concentration was used than was applied to the protoplasts during RNP transfection in the current study. Based on these results, we selected 40 nt, which had highest TI efficiency, as the length of the ssODN in our subsequent experiments, except for experiment (exp.) 2 (44 nt). Interestingly, one of the regenerated N. benthamiana TI lines mediated by the 60 nt ssODN had a precise insertion of 6 bp HindIII sequence (8.3%) in the predicted target site (+27#6; Supplementary Fig. S2E and F).
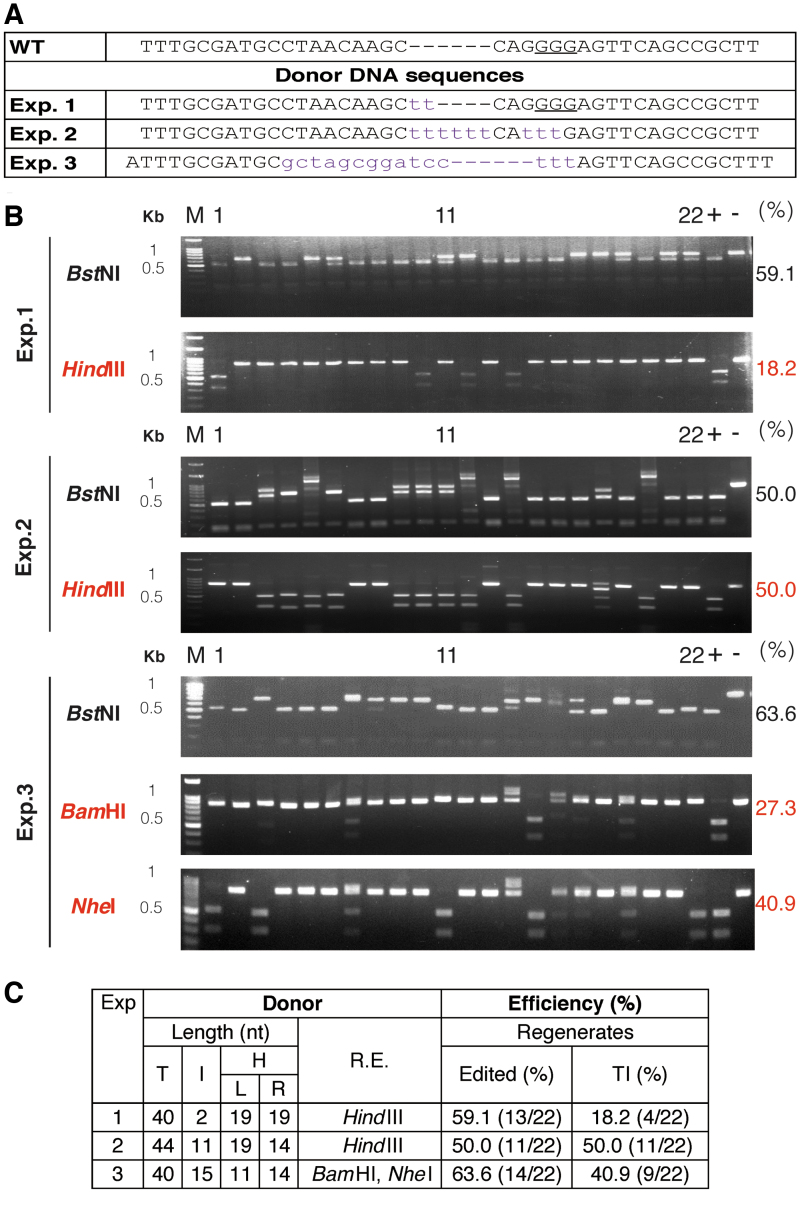
Next, we examined the effect of the insertion length in a 40 nt DD on TI efficiency (Fig. 2). In exp. 1, the HindIII site was generated with additional 2 nt insertion in the DD (Fig. 2A), and TI efficiency in the regenerated plants was 18.2% (Fig. 2B and C). In exp. 2, the PAM sequence in DD was mutated, 6 nt was added, and the TI efficiency increased to 50%. We also increased the insertion length to 15 nt from 6 nt in exp. 3, including sites for NheI and BamHI endonucleases in the insertion, which enabled us to confirm the integrity of TI genotyping by restriction enzyme digestion. As the insertion length increased and the length of the homologous arms decreased (11 and 14 bp, respectively), the TI efficiency decreased slightly (40.9%). Three regenerated plants contained only the NheI site, indicating partial DD insertion (Fig. 2B). Perhaps the ssODN DD was unstable in protoplasts and had been partially degraded before insertion, which would cause the inserted sequence to be incomplete. There was no phosphorothioate-linkage modification in the DD we used in this study, which may have led to the partial degradation of DD before or during the TI process.13
FIG. 2.
TI in NbPDS1 facilitated by short ssODN with different sequences and homology arm length in regenerated Nicotiana benthamiana plants. (A) Donor sequences used in experiments (exp.) 1–3. Lowercase: insertion or replacement nucleotides. The protospacer adjacent motif (PAM) is underlined. (B) PCR of the target gene in the regenerated plants was performed in each experiment, and different restriction enzymes were used for genotyping (black: BstNI, edited; red: HindIII, TI). M, marker; -, wild-type control; +, restriction enzyme control. TI PCR product sequences are shown in Supplementary Tables S2–S4. Because the insertion DNA could be cleaved to ∼20 bp by restriction enzymes, which we designed in DD, all of the TI regenerated plants had the same digested band pattern. In most of cases, the BstNI site in the target site is disrupted by targeted mutagenesis and TI. However, if cytosine is the last nucleobase of TI, for example exp. 1#1, 10, 14, the BstNI site is retained (CCAGG), and PCR products could be cleaved. (C) Edited and TI efficiencies of different ssODN donors. T, total length of DD; I, insertion; H, homologous arm; L, left arm; R, right arm; R.E., restriction enzyme. Color images are available online.
We sequenced all of the NbPDS1 genes with TI in the regenerated plant produced in exp. 1, 2, and 3 (Supplementary Tables S2–S4). In a few regenerated plants with TIs, one end matched the ODN precisely, whereas the other end did not (exp. 1#12 and exp. 2#18). In these regenerated plants, the insertion size was 29–445 bp. These differences were caused by insertion of 1–13 repeats of the ssODN, although some of the repeats were incomplete. In rice, only 20% of TI plants had repeat insertions when using modified double-stranded DD.13 Both orientations were observed for the TI in our regenerated plants. Forward and reverse insertions have also been identified in rice.13
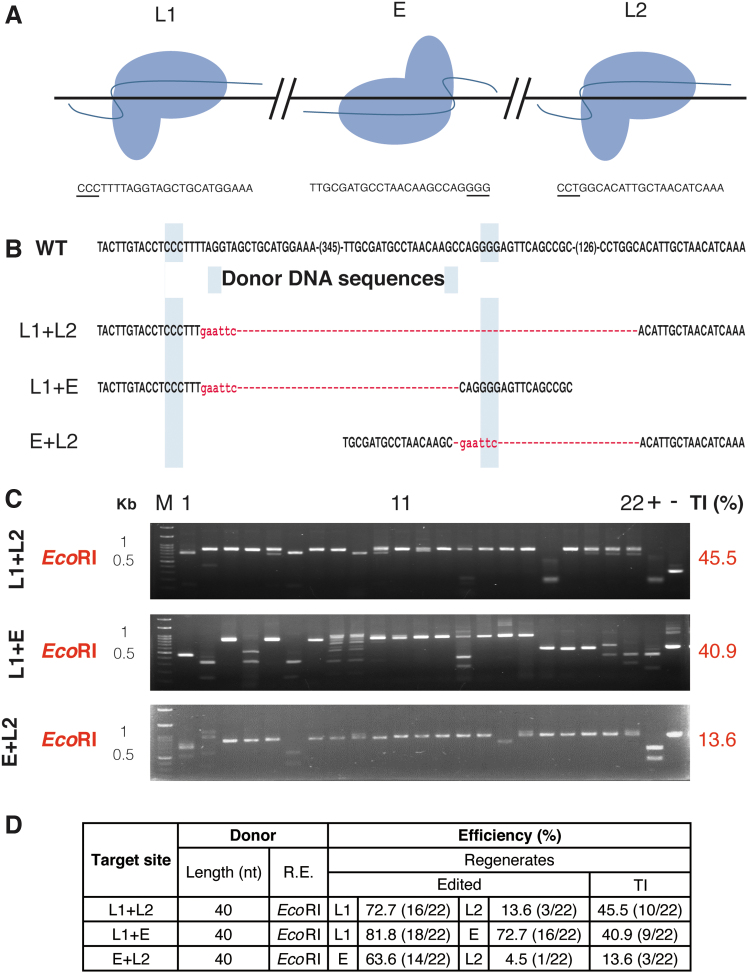
Accurate insertion or replacement of DNA fragments is vital for gene editing. Co-expressing two sgRNAs can lead to fragment deletion in protoplasts.14,24 Therefore, two sgRNAs can be designed at both ends of an exon for exon replacement.14 To aid in TI and DNA replacement, we designed two sgRNAs (L1 and L2) based on both sides of the complementary strands of the original target site (E). These sgRNAs could form a combination of RNPs, including tail-to-head (L1 + L2), tail-to-tail (L1 + E), or head-to-head (E + L2) orientation (Fig. 3A). The ssODN DDs were all located in the same strand of target site E (Fig. 3B). The L1 + E experiment had a higher fragment deletion rate than the others (Fig. 3C and Supplementary Fig. S3). Except for a decrease in E + L2, the overall TI efficiency was similar to that using a single RNP (Fig. 2C, exp. 2 and 3). Compared to the use of E RNP only, there was a decline in the TI/edited ratio when two sgRNA RNPs were co-transfected into protoplasts (Fig. 3D). The insertion sequences are shown in Supplementary Tables S5–S7. From the results, it can be seen that by using two RNPs, a region of DNA can be successfully removed and inserted with DD. Therefore, this method is confirmed as applicable to exon replacement.14
FIG. 3.
TI using two ribonucleoproteins in Nicotiana benthamiana. (A) Relative positions and directions of the three RNPs used. The target site is shown below. The PAM is underlined. (B) Donor DNA (DD) used in each combination. Lowercase: inserted EcoRI site. (C) Restriction enzyme analysis of the target efficiency of regenerated plants derived from different RNP combinations and DD transfected protoplasts. (D) Summary of edited and TI efficiencies of RNP and ssODN. Color images are available online.
To determine whether these insertions were heritable, we analyzed the progeny of five N. benthamiana regenerated plants (Supplementary Fig. S4A). We genotyped TI regenerated plant T1 seedlings and determined that all TI alleles were inherited (Supplementary Fig. S4B and C). Thus, these protoplast regenerated plants were not chimeric at the target gene. By contrast, in rice, most T0 plants appeared to be chimeric.13 In the current study, no Cas9 gene was present in the genomes of the regenerated plants because we used RNP as the Cas9-gRNA reagent, and therefore no new edited alleles were generated.
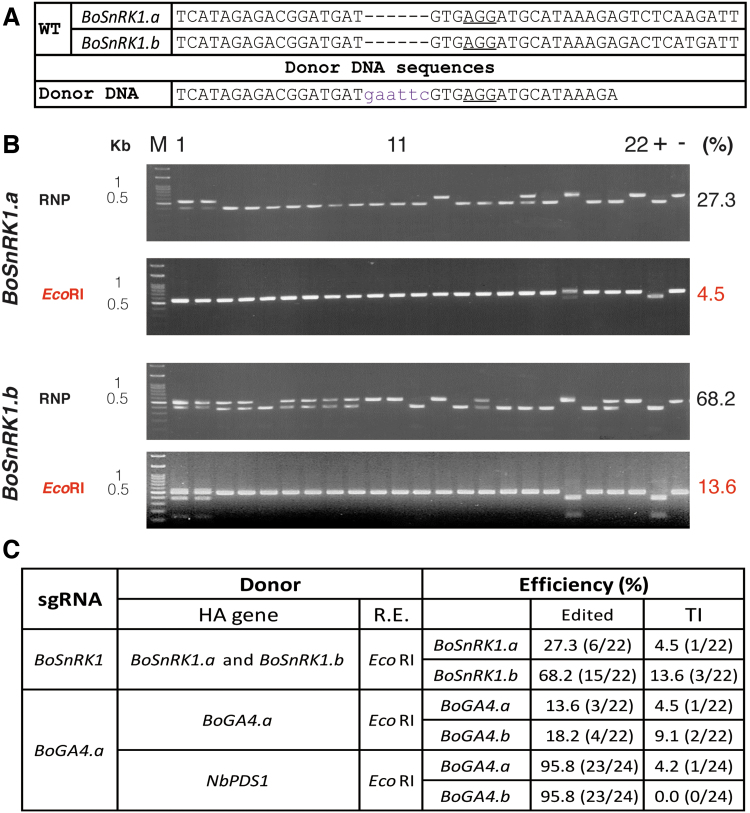
We also examined the TI efficiency in RCBO, targeting BoSnRK1 and BoGA4.a36 (Fig. 4). RCBO contains two BoSnRK1 genes: BoSnRK1.a and BoSnRK1.b (Fig. 4A). DD was inserted into the target sites (Fig. 4B and C and Supplementary Tables S8 and S9). Sequencing indicated that DD was inserted at an efficiency of 4.5–13.6% (Fig. 4C), which is lower than that demonstrated in N. benthamiana.
FIG. 4.
TI using ssODN DD in rapid cycling Brassica oleracea. (A) Target site of BoSnRK1 and the donor sequence. The PAM is underlined. Lowercase: insertion sequence. (B) Genotyping of the regenerated plants derived from RNP and ssODN transfected protoplasts. The edited efficiency by RNP was assessed. TI was assessed using the EcoRI site inserted by the ssODN. (C) Summary of edited and TI efficiencies. HA, homologous arm. Color images are available online.
According to the results, we reasoned that the presence of a homologous arm on the DD designed for TI might not be necessary. To investigate whether ssODN DD with a non-homologous arm could be used for TI, we co-transfected RCBO protoplasts with BoGA4.a sgRNA RNP and NbPDS1 DD. TI was observed in BoGA4.a in only 4.2% of the 24 regenerated plants (1/24), and none was observed in BoGA4.b, even though the editing efficiency was 95.8% (Fig. 4C and Supplementary Table S10). NHEJ is typically guided by short homologous DNA sequences (microhomologies), which affect joining efficiency greatly.37 The lower efficiency when using NbPDS1 DD in RCBO in comparison with N. benthamiana is likely due to variations within genomes, target genes, or the microhomologies between DD and target site. It would require further investigation to understand the exact mechanisms.
N. benthamiana contains two NbPDS genes—Niben101Scf01283g02002.1 (NbPDS1) and Niben101Scf14708g00023.1 (NbPDS2)—and the sgRNA matched NtPDS1 but not NtPDS2 (1 bp mismatch). The NtPDS2 genes in the regenerated plants were analyzed, revealing no off-target mutagenesis or DD insertion. To explore the off-target DD insertion that occurred in the presence of DSBs without homologous sequences with the target site, we performed whole-genome sequencing of three types of plants, in which the regenerated plant NbPDS1 gene was (1) the same as the WT, (2) a heterozygous mutant (KO) but without TI, or (3) a bi-allelic TI (TI; Fig. 1). The ssODN insertion did not occur in the WT or KO genome. In the genome of the TI regenerated plants, the DD insertion occurred not only in the DSB position created by the Cas9-gRNA RNP (Supplementary Fig. S5A), but also in Niben101Scf00150 (Supplementary Fig. S5B) and Niben101Scf06966 (Supplementary Fig. S5C). Using PCR, the Niben101Scf06966 and Niben101Scf00150 DNA fragments were amplified. Only exp. 1#1 regenerated plant contained the extra TI DNA in these regions. It was not found in the other TI (#10) or KO (#2 and #6) regenerated plants (Supplementary Fig. S5D). Using T/A cloning for Niben101Scf00150 PCR products (Supplementary Fig. S5E) and Poly Peak Parser38 for Niben101Scf06966 (Supplementary Fig. S5F), these TI sequences were found to be identical to the TI sequences from whole-genome sequencing results. We genotyped exp. 1#1 regenerated plant T1 seedlings and determined these two off-target TI alleles were inherited. In rice that had been transformed using Cas9-gRNA expressed from plasmid DNA via the biolistic method, quantitative PCR revealed multiple copies (2–10 per plant) of the donor inserts in T1 plants, which suggested that frequent off-target DD insertion occurred in addition to the intended target site insertions.13 The BoGA4 sgRNA matched BoGA4.a but not BoGA4.b (2 bp mismatch). Not only targeted mutagenesis but also off-target DD insertion occurred in BoGA4.b in RCBO (Fig. 4C). These results indicated that the off-target DD insertion was caused by RNP and unexpected DSBs that provided an opportunity for DD insertion.
Conclusion
In this study, we used protoplast regeneration, RNP, and ssODN to establish a simple and inexpensive DNA TI method for plant genome editing that can be used in N. benthamiana and RCBO by PEG-Ca2+-mediated protoplast transfection. In stable transformation systems, the expression of Cas protein is important for knock-in,3 but in this study, we used the Cas9-gRNA RNP to enhance expression together with high DD concentration to increase TI efficiency. This insertion method should be applicable to any gene target site in the genome of any plant species that can be regenerated from protoplasts without the need for antibiotic selection or phenotypic screening. Although the efficiency was low, we still obtained precise insertion regenerated plants. In the future, we will use tandem-repeat HDR13 and other methods to improve the precision and efficiency of TI and understand the precise mechanisms using ssODN.
Supplementary Material
Acknowledgments
We thank Yu-Jung Cheng for tissue culture. We thank Yu-Lin Wu for the Illumina whole-genome sequencing library preparation. We thank Miranda Loney and plant editors for editing. We thank Ting-Li Wu for help with the figures. We thank Wen Jiang for sharing RNP-ssODN assembly protocol. We thank the Academia Sinica Advanced Optics Microscope Core Facility for microscope imaging technical support.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
This research was supported by Academia Sinica, Innovative Translational Agricultural Research Administrative Office (AS-KPQ-107-ITAR-10; AS-KPQ-108-ITAR-10; AS-KPQ-109-ITAR-10), Academia Sinica Core Facility and Innovative Instrument Project (AS-CFII-108-116), and the Ministry of Science and Technology (105-2313-B-001-007-MY3; 108-2313-B-001 -011 -; 109-2313-B-001 -011 -), Taiwan.
Supplementary Material
References
- 1. Čermák T, Baltes NJ, Čegan R, et al. High-frequency, precise modification of the tomato genome. Genome Biol 2015;16:232. DOI: 10.1186/s13059-015-0796-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Svitashev S, Young JK, Schwartz C, et al. Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol 2015;169:931–945. DOI: 10.1104/pp.15.00793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Endo M, Mikami M, Toki S. Biallelic gene targeting in rice. Plant Physiol 2016;170:667–677. DOI: 10.1104/pp.15.01663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sauer NJ, Narváez-Vásquez J, Mozoruk J, et al. Oligonucleotide-mediated genome editing provides precision and function to engineered nucleases and antibiotics in plants. Plant Physiol 2016;170:1917–1928. DOI: 10.1104/pp.15.01696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dahan-Meir T, Filler-Hayut S, Melamed-Bessudo C, et al. Efficient in planta gene targeting in tomato using geminiviral replicons and the CRISPR/Cas9 system Plant J 2018;95:5–16. DOI: 10.1111/tpj.13932. [DOI] [PubMed] [Google Scholar]
- 6. Miki D, Zhang W, Zeng W, et al. CRISPR/Cas9-mediated gene targeting in Arabidopsis using sequential transformation. Nat Commun 2018;9:1967. DOI: 10.1038/s41467-018-04416-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wolter F, Klemm J, Puchta H. Efficient in planta gene targeting in Arabidopsis using egg cell-specific expression of the Cas9 nuclease of Staphylococcus aureus. Plant J 2018;94:735–746. DOI: 10.1111/tpj.13893. [DOI] [PubMed] [Google Scholar]
- 8. Wolter F, Puchta H. In planta gene targeting can be enhanced by the use of CRISPR/Cas12a. Plant J 2019;100:1083–1094. DOI: 10.1111/tpj.14488. [DOI] [PubMed] [Google Scholar]
- 9. Li S, Li J, He Y, et al. Precise gene replacement in rice by RNA transcript-templated homologous recombination. Nat Biotechnol 2019;37:445–450. DOI: 10.1038/s41587-019-0065-7. [DOI] [PubMed] [Google Scholar]
- 10. Vu TV, Sivankalyani V, Kim EJ, et al. Highly efficient homology-directed repair using CRISPR/Cpf1-geminiviral replicon in tomato. Plant Biotechnol J 2020;18:2133–2143. DOI: 10.1111/pbi.13373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ali Z, Shami A, Sedeek K, et al. Fusion of the Cas9 endonuclease and the VirD2 relaxase facilitates homology-directed repair for precise genome engineering in rice. Commun Biol 2020;3:44. DOI: 10.1038/s42003-020-0768-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schindele A, Dorn A, Puchta H. CRISPR/Cas brings plant biology and breeding into the fast lane. Curr Opin Biotechnol 2020;61:7–14. DOI: 10.1016/j.copbio.2019.08.006. [DOI] [PubMed] [Google Scholar]
- 13. Lu Y, Tian Y, Shen R, et al. Targeted, efficient sequence insertion and replacement in rice. Nat Biotechnol 2020;38:1402–1407. DOI: 10.1038/s41587-020-0581-5. [DOI] [PubMed] [Google Scholar]
- 14. Li JY, Meng X, Zong Y, et al. Gene replacements and insertions in rice by intron targeting using CRISPR-Cas9. Nat Plants 2016;2:16139. DOI: 10.1038/nplants.2016.139. [DOI] [PubMed] [Google Scholar]
- 15. Dong OX, Yu S, Jain R, et al. Marker-free carotenoid-enriched rice generated through targeted gene insertion using CRISPR-Cas9. Nat Commun 2020;11:1178. DOI: 10.1038/s41467-020-14981-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Weinthal D, Tovkach A, Zeevi V, et al. Genome editing in plant cells by zinc finger nucleases. Trends Plant Sci 2010;15:308–321. DOI: 10.1016/j.tplants.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 17. Fauser F, Roth N, Pacher M, et al. In planta gene targeting. Proc Natl Acad Sci U S A 2012;109:7535–7540. DOI: 10.1073/pnas.1202191109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang Y, Zhang F, Li X, et al. Transcription activator-like effector nucleases enable efficient plant genome engineering. Plant Physiol 2013;161:20–27. DOI: 10.1104/pp.112.205179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Krens FA, Molendijk L, Wullems GJ, et al. In vitro transformation of plant protoplasts with Ti-plasmid DNA. Nature 1982;296:72–74. DOI: 10.1038/296072a0. [DOI] [Google Scholar]
- 20. Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2007;2:1565–1572. DOI: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- 21. Li J, Stoddard TJ, Demorest ZL, et al. Multiplexed, targeted gene editing in Nicotiana benthamiana for glyco-engineering and monoclonal antibody production. Plant Biotechnol J 2016;14:533–542. DOI: 10.1111/pbi.12403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Woo JW, Kim J, Kwon SI, et al. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol 2015;33:1162–1164. DOI: 10.1038/nbt.3389. [DOI] [PubMed] [Google Scholar]
- 23. Andersson M, Turesson H, Nicolia A, et al. Efficient targeted multiallelic mutagenesis in tetraploid potato (Solanum tuberosum) by transient CRISPR-Cas9 expression in protoplasts. Plant Cell Rep 2017;36:117–128. DOI: 10.1007/s00299-016-2062-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lin CS, Hsu CT, Yang LH, et al. Application of protoplast technology to CRISPR/Cas9 mutagenesis: from single-cell mutation detection to mutant plant regeneration. Plant Biotechnol J 2018;16:1295–1310. DOI: 10.1111/pbi.12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hsu CT, Cheng YJ, Yuan YH, et al. Application of Cas12a and nCas9-activation-induced cytidine deaminase for genome editing and as a non-sexual strategy to generate homozygous/multiplex edited plants in the allotetraploid genome of tobacco. Plant Mol Biol 2019;101:355–371. DOI: 10.1007/s11103-019-00907-w. [DOI] [PubMed] [Google Scholar]
- 26. Park SC, Park S, Jeong YJ, et al. DNA-free mutagenesis of GIGANTEA in Brassica oleracea var. capitata using CRISPR/Cas9 ribonucleoprotein complexes. Plant Biotechnol Rep 2019;13:483–489. DOI: 10.1007/s11816-019-00585-6. [DOI] [Google Scholar]
- 27. Yu J, Tu L, Subburaj S, et al. Simultaneous targeting of duplicated genes in Petunia protoplasts for flower color modification via CRISPR-Cas9 ribonucleoproteins. Plant Cell Rep 2021;40:1037–1045. DOI: 10.1007/s00299-020-02593-1. [DOI] [PubMed] [Google Scholar]
- 28. De Bruyn C, Ruttink T, Eeckhaut T, et al. Establishment of CRISPR/Cas9 genome editing in Witloof (Cichorium intybus var. foliosum). Front Genome Ed 2021;2:24. DOI: 10.3389/fgeed.2020.604876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hsu CT, Lee WC, Cheng YJ, et al. Genome editing and protoplast regeneration to study plant–pathogen interactions in the model plant Nicotiana benthamiana. Front Genome Ed 2021;2:39. DOI: 10.3389/fgeed.2020.627803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang RS, Shih HA, Lai MC, et al. Enhanced NK-92 Cytotoxicity by CRISPR genome engineering using Cas9 ribonucleoproteins. Front Immunol 2020;11:1008. DOI: 10.3389/fimmu.2020.01008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hiom K. Coping with DNA double strand breaks. DNA Repair 2010;9:1256–1263. DOI: 10.1016/j.dnarep.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 32. Puchta H, Fauser F. Synthetic nucleases for genome engineering in plants: prospects for a bright future. Plant J 2014;78:727–741. DOI: 10.1111/tpj.12338. [DOI] [PubMed] [Google Scholar]
- 33. Lin S, Staahl BT, Alla RK, et al. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife 2014;3:e04766. DOI: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kotogány E, Dudits D, Horváth GV, et al. A rapid and robust assay for detection of S-phase cell cycle progression in plant cells and tissues by using ethynyl deoxyuridine. Plant Methods 2010;6:5. DOI: 10.1186/1746-4811-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kaya H, Mikami M, Endo A, et al. Highly specific targeted mutagenesis in plants using Staphylococcus aureus Cas9. Sci Rep 2016;6:26871. DOI: 10.1038/srep26871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lawrenson T, Shorinola O, Stacey N, et al. Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol 2015;16:258. DOI: 10.1186/s13059-015-0826-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Budman J, Chu G. Processing of DNA for nonhomologous end-joining by cell-free extract. EMBO J 2005;24:849–860. DOI: 10.1038/sj.emboj.7600563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hill JT, Demarest BL, Bisgrove BW, et al. Poly peak parser: method and software for identification of unknown indels using Sanger sequencing of PCR products. Dev Dyn 2014;243:1632–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.