Abstract
Diabetes is one of the fastest growing diseases worldwide, projected to affect 693 million adults by 2045. Devastating macrovascular (cardiovascular disease) and microvascular (diabetic kidney disease, diabetic retinopathy, and neuropathy) diabetes complications lead to increased mortality, blindness, kidney failure, and overall decreased quality of life in individuals with diabetes. Clinical risk factors and glycemic control alone cannot predict the development of vascular complications; numerous genetic studies have demonstrated a clear genetic component to both diabetes and its complications. Early research aimed at identifying genetic determinants of diabetes complications relied on familial linkage analysis suited for strong effect loci, candidate gene studies prone to false positives, and under-powered genome-wide association studies (GWAS) limited by sample size. The recent explosion of new genomic datasets, both in terms of biobanks and aggregation of worldwide cohorts, have more than doubled the number of genetic discoveries for both diabetes and diabetes complications. We focus herein on recent genetic discoveries for diabetes and diabetes complications, empowered primarily through GWAS, and emphasize the gaps in research for taking genomic discovery to the next level.
Introduction
Diabetes, a disease of the endocrine system diagnosed by abnormally high blood glucose levels, is one of the most common and fastest growing diseases worldwide, projected to affect 693 million adults by 2045,1 a >50% increase from 2017. Vascular complications of both the macro- and microvascular systems (cardiovascular disease [CVD], diabetic kidney disease [DKD], diabetic retinopathy [DR], and neuropathy) are the leading cause of morbidity and mortality in individuals with diabetes,2 carrying enormous financial burden with unequal healthcare expenditure and access to treatment between developed and developing countries.3–5 While the precise mechanisms of hyperglycemia-induced vascular damage are both complex and not fully understood, it is thought that high levels of intracellular glucose increase the production of reactive oxygen species altering a series of critical downstream pathways, including polyol pathway flux, advanced glycation end product formation and activation, protein kinase C activation, and hexosamine pathway flux.6
Diabetes is not a single disease, but rather a group of conditions broadly categorized by a single diagnostic criterion – hyperglycemia, the final common pathway on which disparate metabolic derangements converge. It is becoming increasingly evident that even type 2 diabetes (T2D), the predominant diabetes subtype making up 90–95% of cases,7 is itself heterogeneous in terms of both the mechanisms of action and the relationships with health outcomes.8 Recent clustering approaches using clinical9 or genetic8 biomarkers have identified subtypes of T2D that are clinically distinct and differentially associated with diabetic complications.8,9 Namely, these studies find an increased risk for decreased renal function among individuals assigned to various insulin resistance clusters, an increased risk for DR among those in the clinical severe insulin deficiency cluster, and an increased risk for coronary artery disease (CAD) among the reduced beta-cell function and lipodystrophy-like fat distribution genetic clusters. Intriguingly, there were no significant differences among the clinical clusters of Ahlqvist et al. for coronary events after adjusting for age and sex.8,9 Furthermore, vascular damage can occur through non-hyperglycemic mechanisms, some of which are also diabetes comorbidities such as hypertension and obesity, further complicating genetic research, diagnosis, and potentially management of hyperglycemia-induced vascular damage.
Current approaches to diabetes complications do not reverse the process, but rely almost exclusively on imperfect attempts at prevention or management of established pathology. On top of landmark studies that show clear decline in both onset and progression of vascular diabetes complications through intensive glucose-lowering treatments,10–12 individuals with diabetes can further reduce their risk for complications by lowering blood pressure13 and taking antihypertensive medications (angiotensin-converting enzyme inhibitors and angiotensin II receptor blockers)14,15 that inhibit the renin-angiotensin-aldosterone system and reduce the risk of complications via blood pressure-dependent and independent mechanisms. More recently, certain classes of glucose-lowering agents (SGLT2 inhibitors and GLP1 receptor agonists) have demonstrated marked reductions in ESKD and CVD-related outcomes in patients with T2D apparently due to both glucose-dependent and independent mechanisms.16–18 Interestingly, loss-of-function mutations in the SLC5A2 gene encoding SGLT2, the major glucose cotransporter in the proximal tubule of the kidney, are known to cause familial renal glycosuria, a disease characterized by decreased renal glucose reabsorption and increased glucose excretion.19 This paradigm illustrates that exploring the genetics of glucose dysfunction diseases as well as the interaction between genetics and treatment may provide insight into the clinical success of novel therapeutics. Beyond medication and lifestyle changes that slow progression of disease, the majority of treatments of late-stage disease involve either dialysis or transplant for kidney disease or other surgical interventions (i.e. laser photocoagulation eye surgery or amputation).
Diabetes and its complications are complex multifactorial conditions with both major environmental and genetic components. When early studies identified differences in diabetic complication susceptibility in patients who seemed otherwise equal in regards to their diabetes glucose control, clinical features, and management,20 family studies were able to demonstrate clear and remarkable differences in incidence of both microvascular and macrovascular complications among individuals with family members with both diabetes and diabetes complications versus those with diabetes but free from complications.21–24 While family studies have demonstrated clear genetic components to diabetes and its complications, early genetic studies suffered serious flaws limiting genetic discovery. As with most complex traits, linkage analysis was unable to identify loci with robust large effects, candidate gene studies were prone to false positives through the adoption of loose statistical thresholds, and early genome-wide association studies (GWAS) lacked large enough sample sizes to detect the modest effect sizes that underlie most complex traits. All of this was further confounded by studying a disease within a disease, namely diabetes-induced micro- and macrovascular complications for which there are multiple intertwined risk factors, indirect disease diagnostics, and unclear disease progression. Nevertheless, in this review we will cover recent advances in genomic analysis, with a focus on GWAS, that have enabled novel genetic discoveries and more than doubled the number of genetic loci associated with T2D and uncovered several novel candidate genes for both micro- and macrovascular complications.
Diabetes
Diabetes is a chronic metabolic disorder characterized by high blood glucose levels that result from absolute or relative insulin deficiency, in the context of beta-cell dysfunction, insulin resistance, or both. Though it’s classically divided into an early-onset autoimmune form (type 1 diabetes or T1D) and a late-onset non-autoimmune form (T2D), additional clinically recognizable subtypes exist, such as monogenic diabetes (e.g. Maturity-onset Diabetes of the Young [MODY] or neonatal diabetes), gestational diabetes, and possibly a late-onset autoimmune form (latent autoimmune diabetes in the adult or LADA). Indeed, the label of T2D is essentially applied to any diabetes that is not autoimmune or monogenic in nature, and it is increasingly recognized that it may represent a conglomerate of varied pathophysiological states. Regardless of this heterogeneity, all of these diabetes forms have a notable genetic component.25–28 Reviewed elsewhere,29,30 genetic exploration of T1D has been heavily focused on the HLA region, though GWAS has identified over 50 loci contributing to T1D risk thus far.31–33 Though no large-scale sequencing efforts in individuals with T1D have been successfully undertaken, targeted sequencing of known loci has additionally identified a cluster of rare variants in PTPN22 that disrupt mRNA splicing.34 T2D, on the other hand, is typically characterized by insulin insensitivity interfering with insulin’s ability to activate glucose transport intracellularly (insulin resistance), while insulin production is unable to compensate for this resistance (relative deficiency). Earlier work on the genetic exploration of T2D has been covered extensively in the literature;35–37 we focus herein on the latest ground-breaking advances in GWAS and sequencing studies of T2D.
The largest T2D GWAS to date was a meta-analysis of 32 European cohorts of ~74K cases and ~824K controls.27 This study identified 243 loci reaching genome-wide significance, including 403 distinct association signals, 152 loci after adjustment for body mass index (BMI; 231 in the unadjusted model), 135 novel loci, and 56 low frequency (frequency <5%) and 24 rare (frequency <0.5%) lead variants across 60 loci, 14 of which have odds ratio (OR) >2. Together these top GWAS signals explain over 17% of the phenotypic variance in T2D, and extensive fine-mapping analysis on these loci found 73 signals implicating a single causal variant. While current polygenic scores, which aggregate the genetic risk for T2D over multiple genetic loci, are not better than clinical markers for predicting T2D, individuals in the top 2.5% of the polygenic score distribution are at a 3.4-fold and 9.4-fold increased risk for having T2D when compared to the median and the bottom 2.5% of the distribution, respectively, highlighting the potential for genetics in T2D precision medicine.
Numerous studies conducted in diverse populations have greatly advanced our knowledge of T2D genetics across populations.38–47 The most recent exome-array association analysis in ~81K T2D cases and ~370K controls from five population groups identified 40 coding variant association signals in 38 loci with P < 2.2 × 10−7, 16 of which were novel.48 Nearly all associations were shared to some degree between both the European-only and the trans-ethnic meta-analyses with 25 reaching study-wide significance in both analyses, 14 with P<0.05 in the complementary meta-analysis (East Asian-specific PAX4 was the exception), and all demonstrating minimal differences in effect size across populations. Interestingly, low-frequency variants were not prominent and did not have strong effects, with only 5/40 with frequency<5% and OR ranging from 1.09–1.29. Genome-wide trans-ethnic meta-analysis has similarly shown minimal differences in effect size across populations,44 with the latest two trans-ethnic GWAS identifying a total of 475 T2D association signals in 250 loci (59 novel) presented at the American Diabetes Association 2019 Meeting49 and 589 T2D associations and a handful of significant SNP × T2D interactions with diabetes complications presented at the American Society of Human Genetics 2019 Meeting.50 Furthermore, studies on the genetics of T2D in isolated populations (reviewed elsewhere)51,52 where population bottlenecks, genetic drift and selection pressure reduce background genetic variability and potentially increase specific allele frequencies have made substantial contributions to genetic discovery. While GWAS in non-European populations have identified relatively few novel loci and have demonstrated mostly homogenous effects across populations, genetic analysis of T2D in diverse and isolated populations may identify variants unique to those populations or achieve genome-wide significance where their low frequency in Europeans leads them to evade detection, and importantly facilitates fine-mapping efforts to identify causal variants to aid downstream functional studies.
Sequencing studies of complex diseases, like T2D, have demonstrated little success in identifying novel genetic loci associated with disease. The largest sequencing study conducted to date interrogated multi-ethnic exome sequencing of 21K cases and 24K non-diabetic controls and identified just four exome-wide significant gene-level associations using a novel method aggregating multiple gene-level analysis approaches.53 Though three of these genes had established links to T2D, the SLC30A8 signal was interestingly driven by 90 missense variants supporting the notion that loss of function reduces T2D risk, a direction of effect recently confirmed in detailed human phenotypic and experimental analyses.54 In addition, 12/16 T2D-related gene sets had a significant enrichment of statistical significance in the gene level analysis, including gene sets for T2D drug targets, mouse models of non-insulin dependent diabetes, impaired glucose tolerance, genes with common T2D variants (after conditioning on the common effect), and MODY genes. The authors also find directional consistency between T2D drug target therapeutic direction and their respective gene ORs, but no such consistency between mouse model knockout effects and their known genes, emphasizing the limits of using mice to model human physiology. Gene-set enrichment and directional consistency analysis emphasizes the need for larger sample sizes to identify more gene-level signals at exome-wide significance. Sequencing studies, which enable a more comprehensive characterization of variants within a locus, are technologically best suited for rare variant analysis, yet they still require equally large sample sizes to their GWAS counterparts;55 with continuously improving imputation and their cost-effectiveness they will most likely remain at the forefront of variant-disease association discovery in years to come.
Microvascular Complications
Diabetic Kidney Disease
Diabetic kidney disease, often referred to as diabetic nephropathy, is a progressive disorder defined by reduced renal function due to hyperglycemia, often co-occurring with albuminuria.56 Individuals with diabetes can also present with non-specific kidney disease, for which their reduced renal function is due to risk factors independent of or indirectly related to their diabetes, such as hypertension, obesity or dyslipidemia. Though DKD is primarily diagnosed by two clinical markers, increased albuminuria and decreased estimated glomerular filtration rate (eGFR), the temporal relationship between diabetes diagnosis and onset of kidney disease can help distinguish between diabetes-specific and non-specific DKD. However, kidney biopsy, which is rarely necessary for kidney disease management and seldom obtained, is the gold standard for making this distinction.57 Differences between diabetes-specific and non-specific DKD contribute greatly to the challenges of studying diabetes complications and were recently reviewed by Anders et al.57 There is no cure to DKD; treatment involves managing blood glucose levels, proteinuria, and progressive kidney damage until late stages of DKD, in which dialysis or kidney transplant are typically necessary for survival. DKD remains the most common cause of end-stage kidney disease (ESKD), which is itself associated with increased mortality.
The progressive nature of DKD coupled with indirect diagnostic measures (albuminuria and eGFR) and heterogeneous risk factors has complicated defining DKD as a phenotype in genetic analyses, and likely contribute to limited and inconsistent findings (Figure 1). Notwithstanding, early heritability studies of DKD found strong familial clustering of both T1D and T2D DKD;21,22,58–60 probands with diabetes of siblings with DKD had approximately 2–4 times the risk of developing DKD than probands with diabetes of diabetic siblings without DKD.21,22,59,60 Recent heritability analysis of DKD estimate that 34–59% of variance in T1D DKD after accounting for sex, diabetes duration, and age at diabetes diagnosis is due to common genetic variants (24–42% unadjusted), depending on the precise definition of DKD using both proteinuria and eGFR levels.61,62 Of note, a similar unadjusted analysis of DKD in individuals with T2D estimates SNP heritability to be only 8%, likely due to the much larger phenotypic heterogeneity of kidney disease in T2D versus T1D.63
Figure 1. Phenotype complexity to diabetic kidney disease.
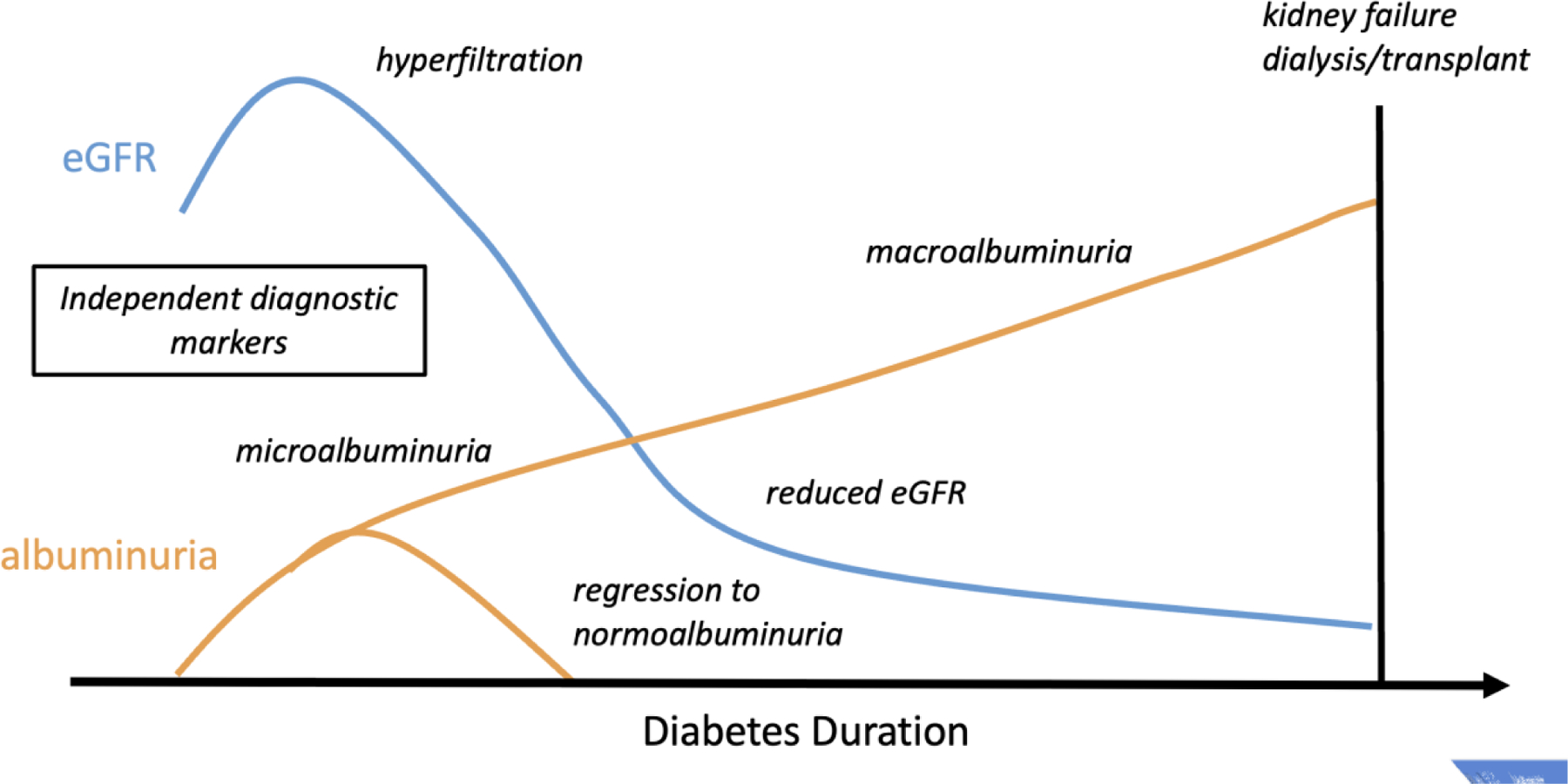
A schematic depicting the complexity to diagnosing diabetic kidney disease based on two primary markers, albuminuria and estimated glomerular filtration rate (eGFR), with increasing diabetes duration. Complications to diagnosis include early increases in eGFR (hyperfiltration), regression of microalbuminuria to normal levels, and independence of the markers such that not all individuals with DKD have both high levels of albuminuria and low eGFR.
The majority of early linkage studies of DKD and related traits identified only several suggestive associations (LOD >3.0),64–66 with the exception of a large linkage analysis in 18 Turkish families with T2D and DKD (chr18q22.3–23, LOD=6.1), later replicated across several ethnicities.65–67 Fine-mapping in this region identified an indel in exon 2 of the carnosine dipeptidase 1 (CNDP1) gene associated with both DKD and serum carnosinase levels. Carnosine possesses anti-glycation activity, blocking glucose-induced increase of extracellular matrix components fibronectin and collagen in podocytes and TGF-beta in mesangial cells.68 Prior to the affordability and feasibility of large-scale GWAS, several candidate gene studies for DKD (summarized elsewhere)69 were also undertaken with mostly limited and inconsistent findings. One promising finding with both robust statistical and functional evidence relates the EPO promoter polymorphism to both proliferative diabetic retinopathy (PDR) and ESKD case status and EPO gene expression.70
Though kidney disease occurs in equal proportions in individuals with T1D and T2D (~30%),71 high rates of co-occurring kidney disease risk factors in individuals with T2D, such as high blood pressure and obesity, increase phenotypic heterogeneity in T2D DKD hindering genetic discovery. Indeed, the majority of genome-wide significant loci from DKD GWAS have been found using T1D cohorts. While no genetic variants achieved genome-wide significance in the first ever GWAS of DKD,72 the strongest association was at the FRMD3 locus (P=5.0×10−7), a gene more recently suggested to affect regulation of the known DKD bone-morphogenetic protein (BMP) signalling pathway.73 The first DKD GWAS to identify robust genome-wide significant loci was conducted by the Genetics of Nephropathy – an International Effort (GENIE) consortium in 2012.74 This study performed a two-stage GWAS meta-analysis examining a total of 12,564 individuals with T1D with or without kidney disease, testing for association with either DKD or ESKD and identified two genome-wide significant loci associated with ESKD (ESKD cases vs. all T1D individuals without ESKD): SNP rs12437854 on chr15q26 in a large gene desert between the RGMA and MCTP2 genes, and intronic SNP rs7583877 on chr2q11 in the AFF3 gene shown to be upregulated in renal endothelial cells when stimulated with pro-fibrotic TGF-β1. Additionally, SNP rs7588550 in intron 1 of ERBB4 was nominally associated (P=2.1×10−7) with DKD, defined as the presence of macroalbuminuria or ESKD in individuals with T1D for at least 10 years vs. controls with T1D for at least 15 years with no clinical evidence for kidney disease. Two neighbouring SNPs in ERBB4 were associated with allele-specific expression of ERBB4 in tubulointerstitial tissue in Pima Indians with T2D DKD. Following the 2012 GENIE DKD GWAS, Sandholm et al. conducted a follow-up sex-specific GWAS of ESKD, and identified a female-specific association with ESKD at a common variant, rs4972593 located on chr2q31.75
The Diabetic Nephropathy Collaborative Research Initiative (DNCRI), an international consortium funded by JDRF, led by the GENIE consortium recently published the largest DKD GWAS conducted to date, tripling their previous sample size to over 19K individuals of European ancestry with T1D and identifying 16 novel genome-wide significant loci associated with various disease definitions (Figure 2–3).76 The strongest association was SNP rs55703767, a common missense variant in exon 17 of the type IV collagen alpha 3 chain (COL4A3) gene, for which the minor allele (T) protects from DKD and several other albuminuria-related phenotypes (Figure 4. Loss of function mutations in COL4A3 cause Alport syndrome.77 The variant was also associated with lower glomerular basement membrane (GBM) thickness in a cohort where ultrastructural information was available for analysis. SNP rs55703767 also demonstrated a significantly stronger effect in women. Notably, as might be predicted for genetic effects expressed in the diabetic context, the protective association was most evident in individuals with higher hemoglobin A1c (HbA1c) levels in an observational study (Figure 5), and in those randomized to a conventional vs. intensive glycemic control in the Diabetes Control and Complications Trial. COL4A3 expression levels were shown to be negatively correlated with GBM surface density in Pima Indians with DKD and glomerulosclerosis in dissected human glomerulus samples.
Figure 2. Expansion of DKD phenotypes for GWAS genetic discovery.
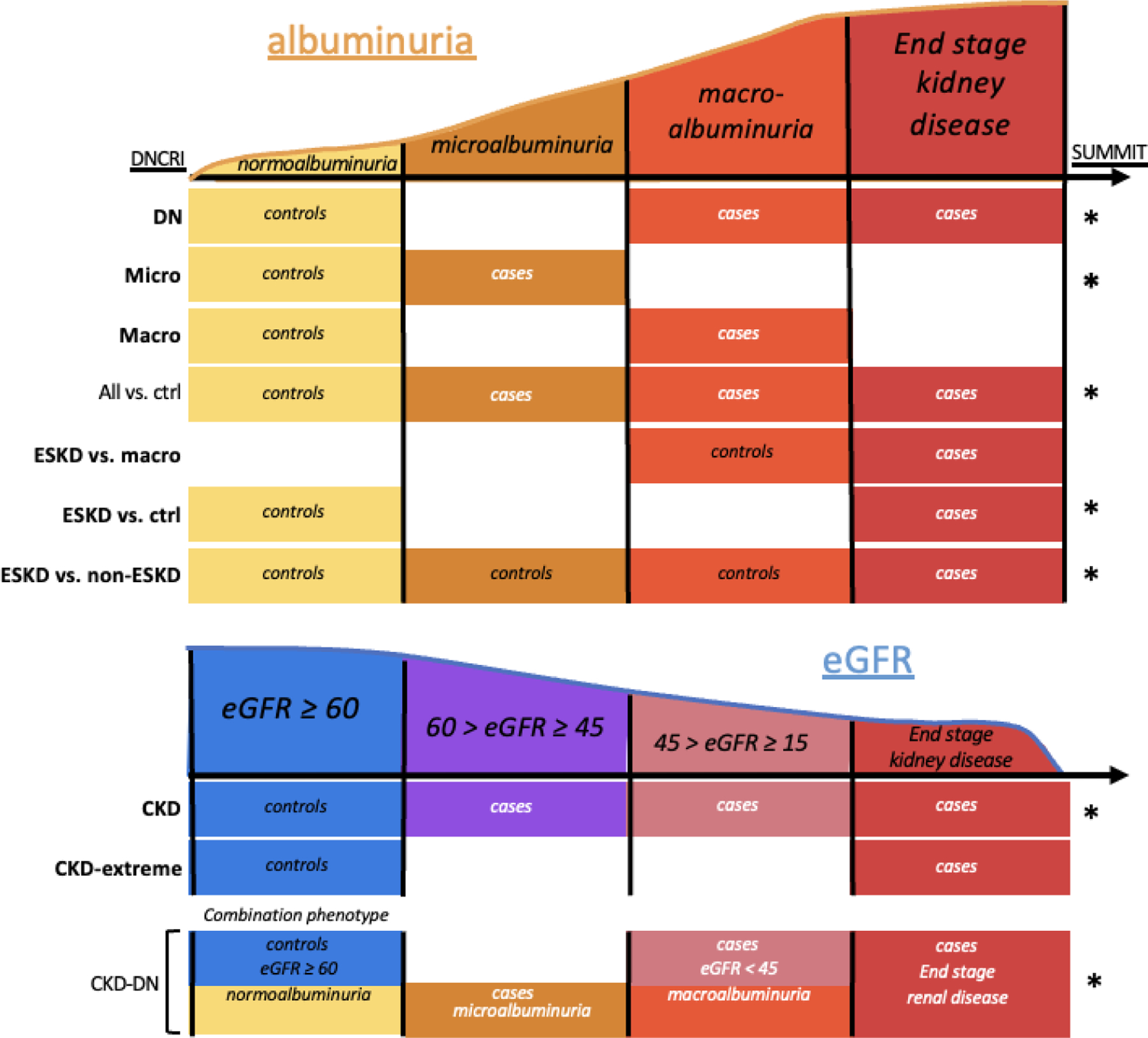
A stylistic representation of the DKD case control phenotype definitions used in both GENIE (all 10 comparisons) and SUMMIT consortiums (those comparisons marked with an asterisk). Phenotype names are taken directly from the DNCRI 2019 GWAS (Salem et al. JASN 2019). All phenotype definitions with a significant genome-wide finding are bolded to highlight the benefit of using multiple definitions.
Figure 3. DNCRI DKD GWAS Manhattan Plot from Salem et al.
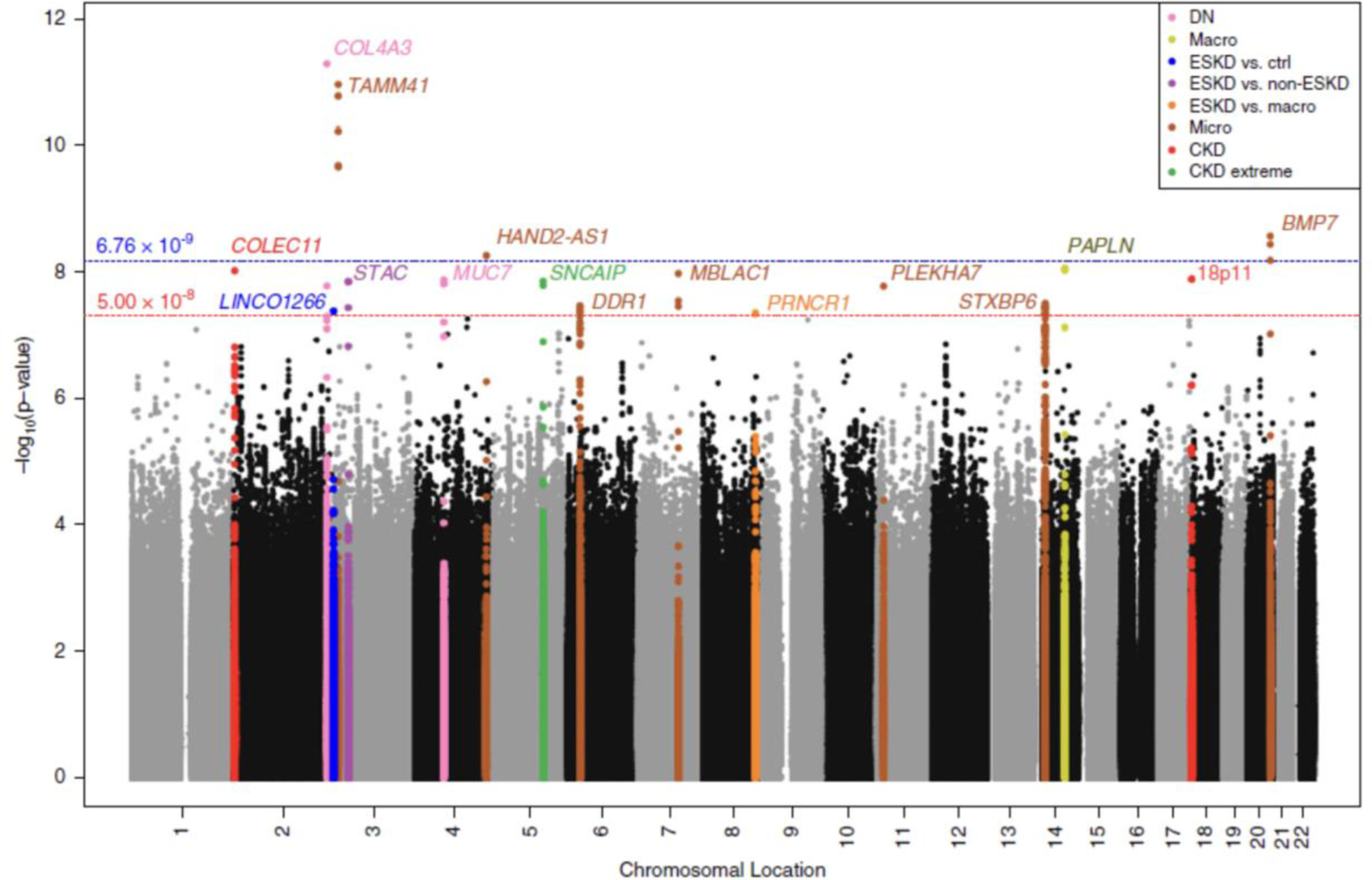
Manhattan plot from Salem et al JASN 2019 publication, highlighting the value in using multiple phenotype definitions of DKD for genetic discovery. Each locus reaching genome-wide significance is colored by its top phenotype. In addition, two distinct significance thresholds used in this study are highlighted.
Figure 4. Visual representation of the COL4A3 gene and missense coding SNP rs55703767 associated with DKD.
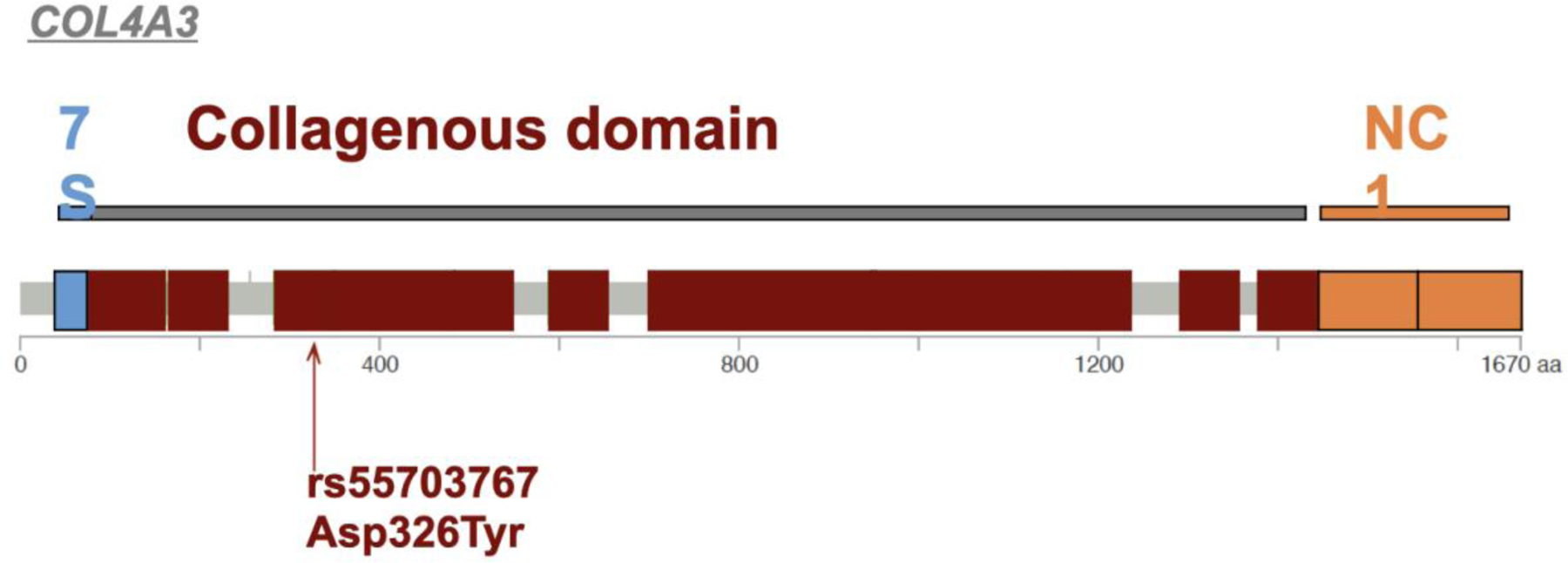
COL4A3 missense coding variant rs55703767 (G → T; Aspartic acid to Tyrosine) in exon 17 in the collagenous domain of COL4A3 (between the triple-helical 7S domain and the non-collagenous NC1 domain).
Figure 5. Association at COL4A3 SNP stratified by hyperglycemia status in the FinnDiane Study published in Salem et al.
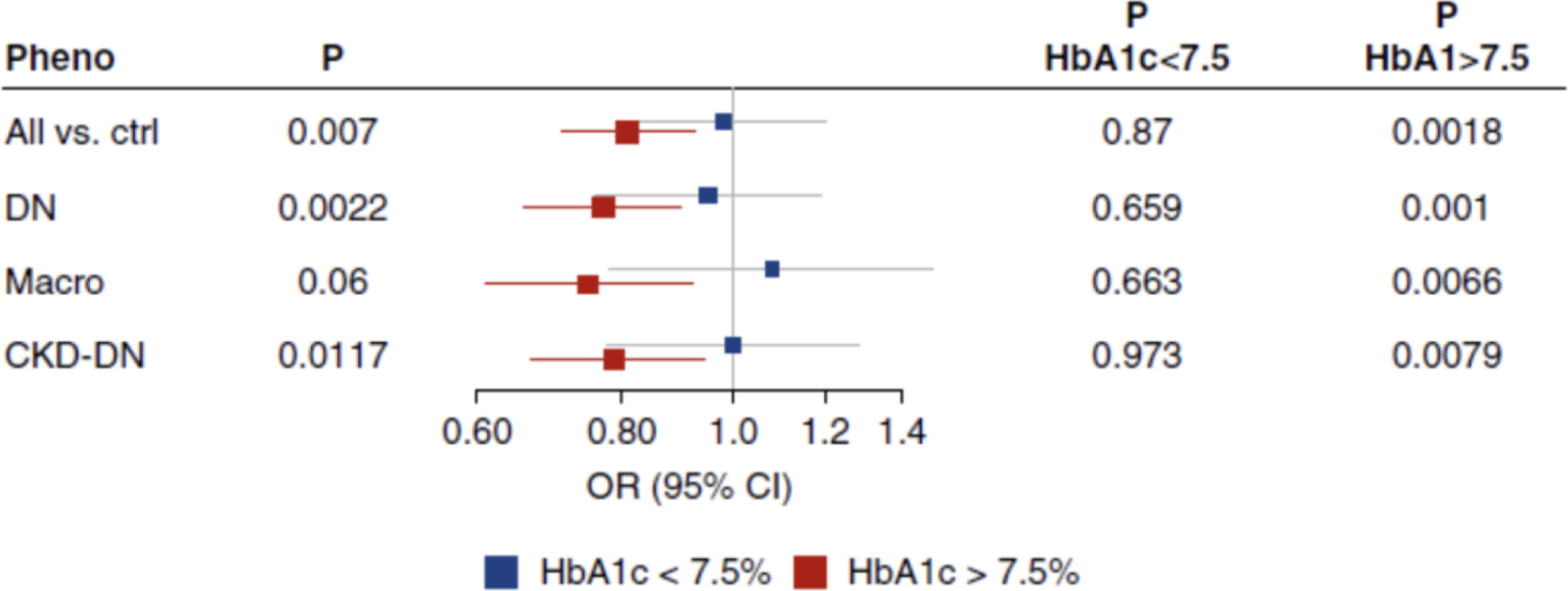
Association of rs55703767 COL4A3 SNP with various DKD disease definitions from the DNCRI DKD GWAS stratified by HbA1c levels in the FinnDiane Study cohort. Though the confidence intervals overlap due to the small sample size, the effect of this SNP on DKD appears to be much stronger in a diabetic context.
Furthermore, this effort identified three additional genetic loci surpassing a strict study-wide significance threshold after correction for multiple testing. SNP rs144434404 in intron 1 of BMP7, a gene involved in renal morphogenesis and almost exclusively expressed in podocytes in mice, was associated with microalbuminuria. SNP rs142823282 near TAMM41 and SNP rs145681168 in intron 3 of HAND2-AS1 were both also associated with microalbuminuria at study-wide significance; the TAMM41 signal was also associated with expression of the nearby gene PPARG (an expression quantitative trait locus or eQTL). Other genome-wide significant signals in this GWAS highlight the involvement of kidney collagen in DKD pathophysiology: DDR1 is a collagen receptor that is highly expressed in the kidneys, particularly upon renal injury, and COLEC11 encodes a collectin protein with both a collagen-like domain and carbohydrate recognition domain. In addition, COL20A1 emerged from a gene-level analysis comparing eGFR extremes. Table 1 reports all genome-wide significant GWAS loci for diabetes complications.
Table 1.
Diabetes Complications GWAS loci reaching genome-wide significance.
| SNP | Reported Gene | P-value | Complication | Reported Phenotype (Total N or Ncases/Ncontrols) | Diabetes Population | Note | Reference |
|---|---|---|---|---|---|---|---|
| rs7583877 | AFF3 | 1.2 × 10−8 | DKD | ESKD (1,399/5,253) | T1D | Sandholm, et al. PLoS Genet. 2012 | |
| rs12437854 | 15q26 intergenic –RGMA, MCTP2 | 2.0 × 10−9 | DKD | ESKD (1,399/5,253) | T1D | Sandholm, et al. PLoS Genet. 2012 | |
| rs4972593 | 2q31 intergenic – SP3, CDCA7 | 3.9 × 10−8 | DKD | ESKD (688/2,009) | T1D | Female-specific | Sandholm, et al. J. Am. Soc. Nephrol. 2013 |
| rs1564939 | GLRA3 | 4.3 × 10−10 | DKD | 24-hour urinary albumin excretion rate (3,612) | T1D | Replicated in Finnish cohort and not other cohorts of European ancestry | Sandholm, et al. Diabetologia 2014; Sandholm, et al. Sci. Rep. 2018 |
| rs12523822 | 6q25 intergenic –SCAF8, CNKSR3 | 1.3 × 10−8 | DKD | DKD (5,226/8,510) | T1D + T2D (not all controls had diabetes) | trans-ethnic meta-analysis | Iyengar, et al. PLoS Genet. 2015 |
| rs13329952 | UMOD | 2.5 × 10−8 | DKD | eGFR (16,477) | T1D + T2D | Also significant in the primary analysis in the general population | Pattaro, et al. Nat. Commun. 2016 |
| rs56094641 | FTO | 7.7 × 10−10 | DKD | DN (4,022/6,890) | T2D | GWAS in Japanese, not replicated in European cohort | Taira, et al. PLoS One. 2018 |
| rs9942471 | GABRR1 | 4.5 × 10−8 | DKD | Microalbuminuria (1,989/2,238) | T2D | Van Zuydam, et al. Diabetes 2018 | |
| rs72858591 | RND3/RBM43 | 4.5 × 10−8 | DKD | ESKD (3,432/6,977) | T2D cases vs. nondiabetic controls | Remained significant after removing loci with P<0.05 with T2D | Guan, et al. Hum. Genomics 2019 |
| rs58627064 | SLITRK3 | 6.8 × 10−10 | DKD | ESKD (3,432/6,977) | T2D cases vs. nondiabetic controls | Remained significant after removing loci with P<0.05 with T2D | Guan, et al. Hum. Genomics 2019 |
| rs142563193 | ENPP7 | 1.2 × 10−8 | DKD | ESKD (3,432/6,977) | T2D cases vs. nondiabetic controls | Remained significant after removing loci with P<0.05 with T2D | Guan, et al. Hum. Genomics 2019 |
| rs142671759 | ENPP7 | 5.5 × 10−9 | DKD | ESKD (3,432/6,977) | T2D cases vs. nondiabetic controls | Remained significant after removing loci with P<0.05 with T2D | Guan, et al. Hum. Genomics 2019 |
| rs4807299 | GNG7 | 3.2 × 10−8 | DKD | ESKD (3,432/6,977) | T2D cases vs. nondiabetic controls | Remained significant after removing loci with P<0.05 with T2D | Guan, et al. Hum. Genomics 2019 |
| rs9622363 | APOL1 | 1.4 × 10−10 | DKD | ESKD (3,432/6,977) | T2D cases vs. nondiabetic controls | Remained significant after removing loci with P<0.05 with T2D | Guan, et al. Hum. Genomics 2019 |
| rs55703767 | COL4A3 | 5.3 × 10−12 | DKD | DN (4,948/12,076), All vs. ctrl (7,247/12,053), CKD+DN (2,897/11,766), macroalbuminuria (2,751/12,124) | T1D | Salem, et al. J. Am. Soc. Nephrol. 2019 | |
| rs12615970 | COLEC11 | 9.4 × 10−9 | DKD | CKD (4,266/14,838) | T1D | Salem, et al. J. Am. Soc. Nephrol. 2019 | |
| rs142823282 | TAMM41 | 1.1 × 10−11 | DKD | Microalbuminuria (2,447/12,113) | T1D | Salem, et al. J. Am. Soc. Nephrol. 2019 | |
| rs145681168 | HAND2-AS1 | 5.4 × 10−9 | DKD | Microalbuminuria (2,447/12,113) | T1D | Salem, et al. J. Am. Soc. Nephrol. 2019 | |
| rs118124843 | DDR1 | 3.4 × 10−8 | DKD | Microalbuminuria (2,447/12,113) | T1D | Salem, et al. J. Am. Soc. Nephrol. 2019 | |
| rs77273076 | MBLAC1 | 1.0 × 10−8 | DKD | Microalbuminuria (2,447/12,113) | T1D | Salem, et al. J. Am. Soc. Nephrol. 2019 | |
| rs551191707 | PRNCR1 | 4.4 × 10−8 | DKD | ESKD vs. macroalbuminuria (2,187/2,725) | T1D | Salem, et al. J. Am. Soc. Nephrol. 2019 | |
| rs144434404 | BMP7 | 4.7 × 10−9 | DKD | Microalbuminuria (2,447/12,113) | T1D | Salem, et al. J. Am. Soc. Nephrol. 2019 | |
| rs115061173 | LINC01266 | 4.1 × 10−8 | DKD | ESKD vs. ctrl (2,187/12,101) | T1D | Salem, et al. J. Am. Soc. Nephrol. 2019 | |
| rs116216059 | STAC | 1.4 × 10−8 | DKD | ESKD vs. non-ESKD (2,187/17,219) | T1D | Salem, et al. J. Am. Soc. Nephrol. 2019 | |
| rs191449639 | MUC7 | 1.3 × 10−8 | DKD | DN (4,948/12,076) | T1D | Salem, et al. J. Am. Soc. Nephrol. 2019 | |
| rs149641852 | SNCAIP | 1.4 × 10−8 | DKD | CKD extreme (2,235/14,993) | T1D | Salem, et al. J. Am. Soc. Nephrol. 2019 | |
| rs183937294 | PLEKHA7 | 1.7 × 10−8 | DKD | Microalbuminuria (2,447/12,113) | T1D | Salem, et al. J. Am. Soc. Nephrol. 2019 | |
| rs61983410 | 14q12 intergenic – STXBP6, NOVA1 | 3.1 × 10−8- | DKD | Microalbuminuria (2,447/12,113) | T1D | Salem, et al. J. Am. Soc. Nephrol. 2019 | |
| rs113554206 | PAPLN | 8.5 × 10−9 | DKD | Macroalbuminuria (2,751/12,124) | T1D | Salem, et al. J. Am. Soc. Nephrol. 2019 | |
| rs185299109 | chr18p11intergenic – LINC00470, METTL4 | 1.3 × 10−8 | DKD | CKD (4,266/14,838) | T1D | Salem, et al. J. Am. Soc. Nephrol. 2019 | |
| rs149131600 | HPN | Pdiabetes = 3.5 × 10−8 | DKD | urinary albumin-to-creatinine ratio (554,659 general population, 46,939 individuals with diabetes) | unspecified | Trans-ethnic meta-analysis; significant in the primary meta-analysis in the general population, though with a larger effect in diabetes subset | Teumer, et al. Nat. Commun. 2019 |
| rs6688849 | 1p33 intergenic – FOXD2, TRABD2B | Pdiabetes =4.1 × 10−9 | DKD | urinary albumin-to-creatinine ratio (564,135 general population, 51,215 individuals with diabetes) | Unspecified | Trans-ethnic meta-analysis; significant in the primary meta-analysis in the general population | Teumer, et al. Nat. Commun. 2019 |
| rs74375025 | CUBN | Pdiabetes =1.1 × 10−24 | DKD | urinary albumin-to-creatinine ratio (558,518 general population, 50,641 individuals with diabetes) | Unspecified | Trans-ethnic meta-analysis; significant in the primary meta-analysis in the general population, though with a larger effect in diabetes subset and replicated in other studies | Teumer, et al. Nat. Commun. 2019 |
| rs790093 | GCKR | Pdiabetes = 1.5 × 10−13 | DKD | urinary albumin-to-creatinine ratio (563,291 general population, 51,515 individuals with diabetes) | Unspecified | Trans-ethnic meta-analysis; significant in the primary meta-analysis in the general population | Teumer, et al. Nat. Commun. 2019 |
| Rs59825600 | KAZN | Pdiabetes =3.6 × 10−8 | DKD | urinary albumin-to-creatinine ratio (549,562 general population, 40,668 individuals with diabetes) | Unspecified | Trans-ethnic meta-analysis; Not significant in the primary meta-analysis in the general population | Teumer, et al. Nat. Commun. 2019 |
| Rs6706313 | MIR4432HG-BCL11A | Pdiabetes =2.8 × 10−8 | DKD | urinary albumin-to-creatinine ratio (564,068 general population, 51,162 individuals with diabetes) | unspecified | Trans-ethnic meta-analysis; Not significant in the primary meta-analysis in the general population | Teumer, et al. Nat. Commun. 2019 |
| Rs17137004 | FOXP2 | Pdiabetes = 2.7 × 10−8 | DKD | urinary albumin-to-creatinine ratio (563,167 general population, 51,294 individuals with diabetes) | unspecified | Trans-ethnic meta-analysis; Not significant in the primary meta-analysis in the general population | Teumer, et al. Nat. Commun. 2019 |
| Rs4258701 | CDH2 | Pdiabetes = 1.1 × 10−8 | DKD | urinary albumin-to-creatinine ratio (564,246 general population, 51,328 individuals with diabetes) | unspecified | Trans-ethnic meta-analysis; Not significant in the primary meta-analysis in the general population | Teumer, et al. Nat. Commun. 2019 |
| rs9896052 | GRB2 | 4.2 × 10−8 | DR | sight-threatening DR (1175/1319) | T1D + T2D | Trans-ethnic meta-analysis | Burdon, et al. Diabetologia 2015 |
| rs80028505 | MAPK14 | 2.5 × 10−8 | neuropathy | foot ulcers in diabetic neuropathy cases vs. no history of foot ulcers in diabetic neuropathy controls (699/2695) | T1D + T2D | Identified in a single study with no replication | Meng, et al. Br. J. Dermatol. 2018. |
| rs13417783 | SCN2A | 7.9 × 10−12 | Neuropathy | Diabetic peripheral neuropathy (5,175/942) | T2D | Tang, et al. Diabetes 2019 | |
| rs10911021 | GLUL | 2.0 × 10−8 | CVD | CHD (1,517/2,671) | T2D | Qi, et al. JAMA 2013 | |
| rs9299870 | MGMT | 9.8 × 10−9 | CVD | cardiovascular mortality (2,667) | T2D under intensive glycemic control | Shah, et al. Diabetes Care 2016 | |
| rs57922 | 5q13 intergenic – ARHGEF28, LINC01335 | 2.0 × 10−8 | CVD | cardiovascular mortality (2,667) | T2D under intensive glycemic control | Shah, et al. Diabetes Care 2016 |
Though met with less success, genetic studies of DKD in T2D have begun to overcome the noise created by phenotypic heterogeneity by greatly increasing sample size. The most recent T2D DKD GWAS was conducted in ~27K individuals with T2D (~13K with DKD) by the SUrrogate markers for Micro- and Macrovascular hard endpoints for Innovative diabetes Tools (SUMMIT) consortium in 2018, using phenotype definitions similar to the DNCRI 2019 GWAS(Figure 3).63 This study identified one genome-wide significant locus associated with microalbuminuria in individuals with T2D of European ancestry. The lead SNP rs9942471 is near GABRR1, and the major allele is associated with decreased GABRR1 expression. Additionally, two loci (UMOD and PRKAG2) previously associated with eGFR in the general population were also associated with eGFR in the SUMMIT combined T1D + T2D meta-analysis of 31K subjects of European and Asian ancestries. An additional recent GWAS in Japanese individuals with T2D identified genome-wide significant markers in the FTO gene locus associated with DKD (lead SNP rs56094641), interestingly unaffected by adjustment for BMI (though BMI was collected after diabetes diagnosis).78 Notably, this association was not seen in the SUMMIT consortium dataset of European T2D individuals.
After exhausting nearly all available T1D cohorts of European ancestry in the DNCRI, future analyses aimed at genetic discovery must rely on combining genetic data across diabetes subtypes and populations, such that massive increases in sample size can outweigh the introduction of phenotypic heterogeneity. Unpublished preliminary work in the SUMMIT consortium, combining both individuals with T1D and T2D (including those from DNCRI) demonstrates the potential for large-scale upcoming efforts.
Several additional GWAS using different populations conducted over the years are of interest and have been reviewed elsewhere.37,79,80 Briefly, a population-specific signal associated with urinary albumin excretion rate was detected in the GLRA3 gene in Finnish individuals with T1D.62 While the association did not replicate in non-Finnish populations, meta-analysis conducted years later with an additional independent collection of Finnish individuals with T1D yielded a genome-wide significant P-value, though with a significant difference in effect size which could be due to differences in urinary albumin excretion rate collection methods.81 Interestingly, this signal appears to be specific to high HbA1c levels above 7%. The Family Investigation of Nephropathy and Diabetes (FIND) consortium multi-ethnic GWAS in individuals with T1D and T2D identified SNPs located on chr6q25 between genes SCAF8 and CNKSR3 associated with DKD in the meta-analysis of individuals of European American, American Indian, and Mexican Indian ancestry.82 Guan et al. conducted an African American GWAS comparing individuals with T2D and ESKD to non-diabetic non-DKD controls, followed by a discrimination analysis to remove all SNPs nominally associated with T2D; they identified six independent genome-wide significant associations with ESKD (rs58627064 at chr3q26, independent SNPs rs142563193 and rs142671759 near ENPP7, rs4807299 in GNG7, rs72858591 at chr2q23, and rs9622363 in APOL1 [though this SNP is in moderate linkage disequilibrium with a non-diabetic ESKD signal]).83
Finally, several genetic studies of kidney-related traits in the general population have identified numerous kidney-related genetic loci,84–94 and interrogation by diabetes status has and will continue to help distinguish between DKD and NDKD specific and shared mechanisms. The most recent GWAS of urinary albumin-to-creatinine ratio (UACR) identified eight genome-wide significant signals in the subset of 51K individuals with diabetes, four of which were specific to the diabetes-only subset (KAZN, MIR4432HG-BCL11A, FOXP2, and CDH2).94 Furthermore, SNPs in CUBN associated with UACR have 3–4 times stronger effects in individuals with diabetes versus individuals without diabetes,93,94 and while two loci did not quite achieve genome-wide significance in individuals with diabetes (rs649529 between RAB38 and CTSC and rs13427836 in HS6ST1), they demonstrated significant gene × diabetes interactions.88 GWAS of eGFR in the general population have identified 19 loci that were nominally significant when limited to individuals with diabetes (P<0.05), with UMOD retaining genome-wide significance89 and independently replicating in the DNCRI and SUMMIT consortia.63,76
While at present GWAS arrays and current imputation methods are optimized to capture common genetic variation (frequency ≥1%), sequencing approaches (either whole-exome or whole-genome) can be used to ascertain rarer genetic variation contributing to DKD, though these efforts are currently constrained by cost. In addition, the potential gain in power attained with the presumably larger effect sizes of rare variants is typically offset by their much lower frequency, such that samples sizes comparable to those employed in GWAS are typically necessary.55 Increasing the number of observations for rare variants might be achieved by leveraging family designs. One whole-exome sequencing study has been published to date in individuals with T1D, identifying one non-coding intronic variant with a 0.2% minor allele frequency reaching exome-wide significance (rs188427269 in the NVL gene P=3.3×10−7).61 Only until we collect large enough sample sizes and integrate more sophisticated gene aggregation analyses will we be able to identify novel rare sequence variants associated with DKD.
Diabetic Retinopathy
Hyperglycemia can induce progressive damage to the blood vessels in the retina, which can lead to hemorrhage, retinal detachment and blindness. Diabetic retinopathy (DR) can be classified into an early, more common non-proliferative (NPDR) form, characterized by weakened blood vessels, and the more severe, late-stage PDR form characterized by the growth of new fragile and leaky blood vessels throughout the retina and into the vitreous in the eye. A distinct form of DR involves direct damage to the macula, defined as clinically significant macular edema. DR is the most common diabetes complication, with the overall prevalence in individuals with diabetes of ~35%,95 with wide variation among ethnic groups and populations around the world.96 Furthermore, DR is the leading cause of blindness in adults in the US97 and England;98 the severity of DR is associated with diabetes duration, age of diagnosis, HbA1c levels, blood pressure, insulin use, and presence of proteinuria.99
Early family clustering studies which suggested a genetic component to DR have generally found significant concordance in the presence of DR among DR-positive family members compared with DR-negative family members, depending on diabetes subtype and additional subgroup characteristics.100,101 Specific family study heritability estimates range from 18 to 52%,102–104 with a more recent estimation of SNP heritability due to common genetic variants alone calculated from distantly related individuals of 7%. Overall, however, there seems to be a clearer genetic contribution to the severity of DR, rather than the presence/absence of DR in general.101,105
Reviewed elsewhere,106,107 early genetic studies of DR, including linkage analyses, candidate gene studies, and underpowered GWAS, have uncovered few if any robust genetic signals. The only study conducted to date to report a genome-wide significant result at the discovery + replication meta-analysis stage was conducted by Burdon et al. in 2015 by combining two T2D cohorts and one T1D cohort of European ancestry with one T2D Indian cohort. They identified a significant association between sight-threatening DR (severe NPDR, PDR, or macular edema) and genotypes at SNP rs9896052 (P=4.15×10−8), a variant 17 kb upstream of the GRB2 gene encoding an epidermal growth factor receptor binding protein (Table 1), which they then demonstrated was both expressed in normal human retina and upregulated in the retina of a transgenic retinal stress mouse model.108 Two additional large-scale GWAS of DR worthy of mention have been conducted since, yet were still unable to provide robust evidence for genetic association in meta-analysis. Meng et al. identified a genome-wide significant intronic variant, SNP rs3913535, in NOX4 in the Scottish GoDARTS discovery cohort associated with severe DR (severe background DR or PDR), but were unable to replicate it across multiple cohorts (total N=14,031).109 Interestingly, NOX4 encodes a gene that functions as the catalytic subunit to NADPH oxidase complex reducing oxygen to various reactive oxygen species, and has been previously shown to play a functional role in oxygen-induced retinopathy mouse and rat models.110,111 The most recent GWAS conducted to date expanded both the DR phenotype definitions and the sample to include nearly 6K individuals with T2D of either African American or European ancestry in the discovery cohort and 20 additional replication cohorts totalling 43,565 individuals of diverse ancestries.112 Though not retaining genome-wide significance in the meta-analysis, intronic SNP rs142293996 in NVL (a gene reported above in WES of DKD) was associated with a phenotypic extremes analysis in the European discovery cohort (PDR vs. no DR; N effective = 523). As phenotypic extremes analyses tend to have the smallest samples sizes, this finding highlights the importance of using both endophenotypes and meaningful phenotypic comparisons for genetic discovery. While the current state of DR GWAS has started gaining traction, these studies are still greatly limited by sample size and T2D phenotype heterogeneity, emphasizing the need still for larger cohorts with deeper phenotyping.
Due to the high sample size demands for novel gene discovery in sequencing analyses (see above), preliminary whole-exome sequencing studies of DR in approximately 100 individuals with T2D require independent confirmation and functional follow-up.113,114
Diabetic Neuropathy
Diabetes is a leading cause of nerve damage, particularly for the longer peripheral nerves that innervate the lower limbs.115 Generally, diabetic neuropathies can be broken into several subtypes including the most common form, distal symmetric polyneuropathy (a type of peripheral neuropathy), autonomic neuropathies, atypical neuropathies, and also nondiabetic neuropathies common in diabetes.116 On top of excess pain and decreased quality of life associated with diabetic neuropathy, individuals with diabetes have a 15–25% lifetime risk of foot ulcerations and a 15-fold increased risk for lower-extremity amputation vs. individuals without diabetes.117,118 Though diabetic neuropathy has the largest lifetime risk of any diabetes complication, affecting approximately 30% of individuals with diabetes overall and >50% of individuals with diabetes over the age of 50,119–121 it is one of the least studied diabetes complications as it is difficult to measure directly and accurately, and treatment relies solely on prevention with glucose control and management of pain and symptoms.
Similar to other diabetes vascular complications, diabetic neuropathy is a multifactorial condition associated with several risk factors such as HbA1c levels, hypertension, smoking status, and BMI, which also has a genetic component.122 After early familial clustering analysis demonstrated a 2.2-fold increased risk for developing diabetic neuropathy in families for which the proband had neuropathy,123 GWAS estimated the SNP heritability of diabetic neuropathic pain and foot ulcers to be 11–15%124,125 and 6%,126 respectively.
Though several candidate genes have been studied and reviewed elsewhere,127,128 there have only been a handful of GWAS published to date on diabetic neuropathy. Three were conducted in the same Scottish study population (GoDARTS) without replication. The two primary GWAS conducted in GoDARTS found three signals nominally associated with diabetic nerve pain (10−8 < P < 10−7), SNP rs71647933 in ZSCAN20 in females only, SNP rs6986153 at chr8q23 in males only, and SNP rs17428041 in one mRNA transcript of DOK2 in a sex-combined analysis.124,125 A third GWAS in the same GoDARTS dataset investigated the presence of foot ulcers in individuals with diabetic neuropathy vs. diabetic controls with neuropathy and vs. diabetic controls without neuropathy. The authors identified intronic SNP rs80028505 in MAPK14 associated with foot ulcers when comparing both cases and controls with diabetic neuropathy (Table 1).126 Of note, these studies are small and require replication.
One GWAS that did achieve a genome-wide significant result with independent replication in a locus with compelling biological plausibility was conducted in the ACCORD clinical trial, and replicated in the BARI-2D clinical trial.129 The minor allele at lead SNP rs13417783 at chr2q24 demonstrated a strong protective effect for T2D peripheral neuropathy (OR=0.57) in individuals of European ancestry (with consistent direction but non-significant P-value in the African American subset of ACCORD). Endophenotypes within the ACCORD trial that also reached nominal significance (P<0.05) with SNP rs13417783 include self-reported DR, triglycerides, eGFR, and UACR (though some additional micro and macro-vascular complications were not significant). The authors report interesting findings from GTEx that the minor allele is associated with higher expression of the gene SCN2A in tibial nerve tissue, though this gene is over 1MB away.
Furthermore, while no large-scale sequencing effort has been undertaken for diabetic neuropathy likely due to sample size and phenotype limitations, there is some evidence for the presence of rare variants in SCN9A in individuals with painful vs. painless diabetic neuropathy.130
Macrovascular Complications
Cardiovascular Disease
Despite the well-known increased risk for CVD among individuals with diabetes, the pathophysiology linking the two conditions is poorly understood. Depending on the cardiovascular event or disease (i.e. coronary heart disease [CHD], myocardial infarction, heart failure, stroke, etc.) and diabetes subtype, individuals with diabetes have anywhere from a two to ten-fold increased risk of a cardiovascular event when compared to individuals free of diabetes.24,131–133 Additional risk factors for CVD among individuals with diabetes include the presence of other microvascular complications as well as sex, age, BMI, glucose control and HbA1c levels, blood pressure, and smoking status.23,133–138
The evidence for genetic differences attributing to CVD risk among individuals with diabetes is limited. While CHD in the general population has a twin-based heritability of ~40% and a SNP-based heritability of ~30%,139–141 the only evidence for a genetic component for CVD in individuals with diabetes are small family studies of coronary artery calcification, c-reactive protein levels, and carotid intra-medial thickness.142–144
To date, there are over 150 loci associated with CAD in the general population,145 and a handful of these loci have specifically been shown to contribute to CVD risk in individuals with diabetes.146–148 Of note, two SNPs in high linkage disequilibrium at 9p21 in the CDKN2B-AS1 gene display marginal interaction effects with T2D on myocardial infarction (rs10757274) and poor glycemic control on CAD (rs2383206).149,150 One GWAS focused on identifying genetic determinants of CHD in individuals with T2D has been published. Qi et al. found SNP rs10911021 near the glutamate-ammonia ligase (GLUL) gene to be associated with CHD in individuals with T2D, and found no evidence of association with CHD in a population free of diabetes (Table 1); in addition, the same SNP is associated with expression levels of the enzyme.151 Follow-up of this work in several independent cohorts also found an association between SNP rs10911021 and cardiovascular mortality among individuals with T2D.152,153
A more recent study on the genetics of cardiovascular mortality among individuals in the intensive glycemic control treatment arm of the ACCORD trial identified two genome-wide significant loci, SNP rs9299870 in the MGMT gene and SNP rs57922 at 5q13.154 SNP rs9299870 is a significant eQTL for MGMT in the pancreas, spleen, aorta, and subcutaneous adipose tissue. Interestingly, neither SNP was associated with non-cardiovascular mortality nor cardiovascular mortality among participants randomized to the standard glycemic control treatment arm, and genotypes at each SNP or using a combined genetic risk score of both SNPs had a significant interaction with treatment on cardiovascular mortality, highlighting the modulation of these genetic effects by glycemia. Follow-up analyses testing for an association between the above two-SNP genetic risk score and biomarker levels between baseline and 12-months identified a significant association between the score and change in GLP-1 levels in the intensive treatment arm, suggesting a potential cardiovascular protective role for GLP-1 under intensive glucose control,155 in line with recent evidence from CVD outcome trials that reveal the beneficial cardiovascular effects of GLP-1 receptor agonist therapy.156,157 Thus, evidence has begun to emerge supporting a unique genetic component of CVD in individuals with diabetes.
Genetics can also help establish causal inference for the role of glycemia in CVD. The benefits of glucose control for CVD protection is well established for type 1 diabetes,12 but this benefit has been more difficult to demonstrate for T2D, with several randomized clinical trials failing to show a significant CVD benefit of intensive glycemic control.11,158–161 However, subsequent meta-analyses of these trials,162–164 long-term follow-up studies,165–167 and recent CVD outcome trials for specific T2D drug classes156,157,168 all point to a modest but clinically significant effect of glucose control on CVD protection. Whether this is uniquely a class effect or also a global effect of glycemia on CVD remains a matter of debate, but a recent MR instrumental variable analysis has shown that genetically lowered glycemia has a discernible impact on CHD.169
Shared Genetic Architecture
Complications of diabetes, both microvascular and macrovascular, tend to occur together. The co-occurrence of DKD and DR is well studied; In both T1D and T2D, decreased kidney function is associated with higher rates of DR. Specifically, in individuals with T2D, those with microalbuminuria are twice as likely to have DR, and those with macroalbuminuria are six times as likely.170 A similar study in individuals with T1D found that nearly all individuals with eGFR <60 mL/min (>90%) had DR.171 Severity of disease, captured by both disease stage and several quantitative measures of retinal and glomerular structure, is also highly correlated.103,172,173 Retinopathy score is correlated with GBM width,172 even among individuals with T1D who have normal renal function.173 Notably, subgroups of T2D as defined by clinical markers in Ahlqvist et al, found DKD and DR most significantly associated with two distinct clusters, representing insulin resistance and insulin deficiency, respectively.9 Though studied to a much lesser extent, diabetic neuropathy is a prominent microvascular complication which is significantly associated with both DKD and DR.174 Individuals with both diabetes and ESKD have a two-fold increase in foot complications vs. non-nephrotic individuals with diabetes, and a ten-fold higher rate of amputations when compared to the general diabetes population at large.175,176
Microvascular complications, specifically DKD and DR, have also been associated with cardiovascular outcomes. Though DKD diagnosis, lower eGFR, and higher albuminuria are all associated with increased CVD risk,23,177,178 recent work using a genetic instrument for albuminuria in the general population in a MR analysis demonstrates a significant and bidirectional causal effect between albuminuria and hypertension, suggesting a complex feedback loop, while demonstrating only a weak causal effect of albuminuria levels on stroke and heart failure.92 Several studies have also found significant associations between DR and stroke, CHD, myocardial infarction, cardiovascular-related mortality, systolic blood pressure, diastolic blood pressure, and mean blood pressure in individuals with diabetes,172,179–181 though there is also some evidence for a lack of association between DR and blood pressure measurements in individuals with diabetes who have normal renal function.173 Overall, while there is evidence for the co-occurrence of microvascular and macrovascular complications in individuals with diabetes, the biological relationships of how and why they co-occur is less clear.
While there is no overlap between the top genetic loci for diabetes and those for diabetes complications, genetic correlation analysis between T2D and additional health outcomes identified 85/182 significant genetic correlations, including several cardiovascular and kidney-related traits including CHD, adiponectin levels, lipid levels, UACR, CKD, serum creatinine levels, and serum cystatin C.27 Out of 19 of the top coding variants associated with T2D, six were directionally consistent and nominally associated (P<0.05) with CHD, including QSER1, PATJ, HNF1A (2 loci), POC5, and SLC30A8. Thus far, the extent of our knowledge on the shared genetic overlap among microvascular complications includes some evidence for familial clustering123 and early linkage analyses with weakly suggestive signals that may affect more than one diabetes complication.64,182 A lookup of the three suggestive DR SNPs (rs9896052, rs3913535, rs142293996) in the recent DNCRI DKD GWAS yielded a P-value of 0.01 for SNP rs3913535 in NOX4 associated with macroalbuminuria, though this signal would not survive multiple testing across the full GWAS (10 correlated phenotypes and 2 covariate models). Whether shared associations represent a common mechanistic pathway for various microvascular complications or simply denote the co-occurrence of these complications in the same individual, where the associated allele is causal for only one of them, cannot be easily disentangled. Larger sample sizes, deeper phenotyping, more robust findings and functional experiments will be needed to conduct the stringent sensitivity, mediation and mechanistic analyses that will help determine which loci represent overlapping versus complication-specific biology.
Conclusion
Overall there has been a recent surge of novel genetic findings for both diabetes and diabetes complications, largely due to massive sample size increases made possible only by the inclusion of large-scale biobanks, like UK Biobank, and the aggregation and meta-analysis of diabetes cohorts across the world. The most recent T2D GWAS identified twice as many loci (from 113 to 243 loci) as the previous effort in individuals of European ancestry,183 primarily due to two key improvements. The latest GWAS increased the effective sample size by more than three-fold to nearly 900K individuals (9% cases), and also imputed all cohort-level data to a denser imputation panel with a focus on low-frequency variants in Europeans (Haplotype Reference Consortium).184 Even more recently, preliminary trans-ethnic meta-analysis with 1.3M individuals reports hundreds of novel loci associated with T2D.49,50 Whole-exome sequencing analysis of T2D highlights key lessons for studying the genetics of diabetes, complications, and complex traits at large. Together with the latest exome-array association analysis conducted in T2D relatively few novel low-frequency coding variants appear to be detectable at our current sample sizes.48,53 The value of sequencing data lies in its ability to comprehensively characterize genetic variation in genes of interest, serving as an indispensable resource for downstream functional interrogation.
The successes of recent GWAS of diabetes complications have emphasized the need for both larger sample sizes and improved phenotyping. Both the tripling of sample size and the large expansion of phenotype definitions in the latest DNCRI DKD GWAS allowed for novel discovery of the most genome-wide significant loci associated with DKD to date.76 Even larger sample sizes were required to identify a genome-wide significant locus in SUMMIT, where individuals with T2D have a more heterogeneous mixture of kidney disease.63 Expanding analysis to both quantitative normal variation in individuals with diabetes as well as discrimination analysis when using non-diabetic controls has also aided in genetic discovery. Though recent DR GWAS have aggregated numerous cohorts, success came from phenotypic comparisons (e.g. PDR extremes) with relatively small sample sizes, emphasizing both a need for larger samples and thoughtful phenotypic comparisons. The state of GWAS for neuropathy emphasizes these research gaps even more.
Diabetes, which is diagnosed by the presence of hyperglycemia, arises from several distinct biological processes and genetic pathways. Furthermore, complications in patients with diabetes may not be solely related to their hyperglycemia, and attempts to study only individuals with diabetes-mediated complications using diabetes duration criteria subject the study to arbitrary binary cutoffs. Use of “diabetes duration” as a covariate only partially overcomes this, as the relationship with progression of diabetes complications is likely non-linear or not constant, as secular trends continue to escalate glycemic control. Contributing to this challenge is the progressive nature of diabetes complications and indirect diagnostic measures (e.g. pain medication use for neuropathy). Accurate phenotyping is also undermined by the effects of confounders or modifiers, such as co-morbidities (e.g. hypertension and obesity), medication use and adherence, or preventive strategies. Assessing the effects of these confounders via post-hoc sensitivity analyses requires well-curated datasets, which is typically not the case among the largest cohorts; merely increasing sample size may not resolve the issue if the necessary additional variables are not available for analysis. Diseases associated with mortality, like ESKD or CAD, pose an additional difficulty: the competing risk for death may hinder accurate collection of the event of interest. While methods such as competing risk analysis can take this into account when conducting survival comparisons, most genetic studies and large biobanks are biased towards healthier individuals, potentially affecting results and diminishing power. Finally, while studying diabetes complications together may point to shared biology, their high co-occurrence hinders teasing apart complication-specific processes, and only large datasets with deep phenotypic information will allow investigators to distinguish between shared and unique mechanisms.
The major research needs in studying the genetics of diabetes complications can be summarized into a few key areas of future research (Box 1). Larger sample sizes of diabetes complications datasets will be needed to overcome phenotypic heterogeneity and moderate heritability to make novel genetic discoveries. Improved phenotyping will assuredly also increase signal to noise. Future approaches could focus on more objective diagnostic measures to remove complications not due to hyperglycemia, more quantitative and morphometric measurements of the microvascular system, new phenotypic comparisons and groupings, and better use of longitudinal data. Furthermore, as both GWAS of T2D and CVD have been undertaken in hundreds of thousands of individuals, complementary approaches aimed at dissecting the genetic signals identified among the general population using previously defined genetic clusters may help characterize the biological differences between individuals. Finally, additional future progress should rely on complementary datasets. While most multi-ethnic datasets for diabetes have demonstrated limited heterogeneity across populations, population differences regarding diabetes complications is less clear. Furthermore, genetic studies using diverse populations will also enhance discovery of population-specific signals, aid in fine-mapping of known loci, and improve accuracy of risk prediction in the future of precision medicine. Sequencing datasets will also be of use when needed to explore the full spectrum of genetic variation contributing to disease, a key step for improving drug development. In light of a growing diabetes epidemic and a recent boom of novel genetic discoveries with diabetes complications, future work filling these research gaps is both highly promising and urgently needed.
Box 1. Genetics of diabetes complications research needs.
Larger sample sizes for detection of common genetic variants in GWAS
Development of more sequencing datasets for interrogating rare variation
Genetic studies in diverse populations for fine-mapping and population-specific associations
Improved phenotyping for studying heterogeneous diabetes complications
New research efforts aimed at the shared genetic component of diabetes complications
Key Points.
There is a moderate genetic component and significant genetic overlap to diabetes and diabetes micro- and macrovascular complications.
Large biobanks and aggregation of diabetes cohorts have more than doubled the number of GWAS associations with diabetes and diabetes complications.
Sequencing studies remain limited by sample size, though recent work in T2D highlights their use in gene variant characterization
Future genetic discovery of diabetes and complications will rely on larger sample sizes, interrogation of sequencing datasets, diverse populations, and better phenotyping and sub-phenotyping.
References
- 1.Cho NH et al. IDF Diabetes Atlas: Global estimates of diabetes prevalence for 2017 and projections for 2045. Diabetes Res. Clin. Pract 138, 271–281, doi: 10.1016/j.diabres.2018.02.023 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Morrish NJ et al. Mortality and causes of death in the WHO multinational study of vascular disease in diabetes. Diabetologia 44, S14, doi: 10.1007/PL00002934 (2001). [DOI] [PubMed] [Google Scholar]
- 3.da Rocha Fernandes J et al. IDF Diabetes Atlas estimates of 2014 global health expenditures on diabetes. Diabetes Res. Clin. Pract 117, 48–54, doi: 10.1016/j.diabres.2016.04.016 (2016). [DOI] [PubMed] [Google Scholar]
- 4.American Diabetes Association. Economic costs of diabetes in the US in 2017. Diabetes Care, dci180007 (2018). [Google Scholar]
- 5.Liyanage T et al. Worldwide access to treatment for end-stage kidney disease: A systematic review. Lancet 385, 1975–1982, doi: 10.1016/S0140-6736(14)61601-9 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Brownlee M Biochemistry and molecular cell biology of diabetic complications. Nature 414, 813–820, doi: 10.1038/414813a (2001). [DOI] [PubMed] [Google Scholar]
- 7.American Diabetes Association. Classification and diagnosis of diabetes: Standards of medical care in diabetes. Diabetes Care 41, S13–S27, doi: 10.2337/dc18-S002 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Udler MS et al. Type 2 diabetes genetic loci informed by multi-trait associations point to disease mechanisms and subtypes: A soft clustering analysis. PLoS Med 15, e1002654, doi: 10.1371/journal.pmed.1002654 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahlqvist E et al. Novel subgroups of adult-onset diabetes and their association with outcomes: a data-driven cluster analysis of six variables. Lancet Diabetes Endocrinol 6, 361–369, doi: 10.1016/S2213-8587(18)30051-2 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Nathan DM et al. The effect of intensive treatment of diabetes on the development and progression of long-term complications in insulin-dependent diabetes mellitus. N. Engl. J. Med 329, 977–986, doi: 10.1056/nejm199309303291401 (1993). [DOI] [PubMed] [Google Scholar]
- 11.UK Prospective Diabetes Study (UKPDS) Group. Intensive blood-glucose control with sulphonylureas or insulin compared with conventional treatment and risk of complications in patients with type 2 diabetes (UKPDS 33). UK Prospective Diabetes Study (UKPDS) Group. Lancet 352, 837–853 (1998). [PubMed] [Google Scholar]
- 12.Nathan DM et al. Intensive diabetes treatment and cardiovascular disease in patients with type 1 diabetes. N. Engl. J. Med 353, 2643–2653, doi: 10.1056/NEJMoa052187 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prospective UK Diabetes Study Group. Tight blood pressure control and risk of macrovascular and microvascular complications in type 2 diabetes: UKPDS 38. UK Prospective Diabetes Study Group. BMJ (Clinical research ed.) 317, 703–713 (1998). [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis EJ, Hunsicker LG, Bain RP & Rohde RD The effect of angiotensin-converting-enzyme inhibition on diabetic nephropathy. N. Engl. J. Med 329, 1456–1462, doi: 10.1056/nejm199311113292004 (1993). [DOI] [PubMed] [Google Scholar]
- 15.Brenner BM et al. Effects of Losartan on renal and cardiovascular outcomes in patients with type 2 diabetes and nephropathy. N. Engl. J. Med 345, 861–869, doi: 10.1056/NEJMoa011161 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Zelniker TA et al. Comparison of the Effects of Glucagon-Like Peptide Receptor Agonists and Sodium-Glucose Cotransporter 2 Inhibitors for Prevention of Major Adverse Cardiovascular and Renal Outcomes in Type 2 Diabetes Mellitus. Circulation 139, 2022–2031, doi:doi: 10.1161/CIRCULATIONAHA.118.038868 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Mahaffey Kenneth W et al. Canagliflozin and Cardiovascular and Renal Outcomes in Type 2 Diabetes Mellitus and Chronic Kidney Disease in Primary and Secondary Cardiovascular Prevention Groups. Circulation 140, 739–750, doi: 10.1161/CIRCULATIONAHA.119.042007 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perkovic V et al. Canagliflozin and renal outcomes in type 2 diabetes and nephropathy. N. Engl. J. Med 380, 2295–2306 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Santer R et al. Molecular Analysis of the SGLT2 Gene in Patients with Renal Glucosuria. J. Am. Soc. Nephrol 14, 2873–2882, doi: 10.1097/01.Asn.0000092790.89332.D2 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Deckert T & Poulsen J Diabetic nephropathy: fault or destiny? Diabetologia 21, 178–183 (1981). [DOI] [PubMed] [Google Scholar]
- 21.Quinn M, Angelico MC, Warram JH & Krolewski AS Familial factors determine the development of diabetic nephropathy in patients with IDDM. Diabetologia 39, 940–945, doi: 10.1007/BF00403913 (1996). [DOI] [PubMed] [Google Scholar]
- 22.Seaquist ER, Goetz FC, Rich S & Barbosa J Familial clustering of diabetic kidney disease. N. Engl. J. Med 320, 1161–1165 (1989). [DOI] [PubMed] [Google Scholar]
- 23.Earle K, Walker J, Hill C & Viberti G Familial clustering of cardiovascular disease in patients with insulin-dependent diabetes and nephropathy. N. Engl. J. Med 326, 673–677 (1992). [DOI] [PubMed] [Google Scholar]
- 24.Tuomilehto J et al. Incidence of cardiovascular disease in type 1 (insulin-dependent) diabetic subjects with and without diabetic nephropathy in Finland. Diabetologia 41, 784–790, doi: 10.1007/s001250050988 (1998). [DOI] [PubMed] [Google Scholar]
- 25.Redondo MJ, Jeffrey J, Fain PR, Eisenbarth GS & Orban T Concordance for islet autoimmunity among monozygotic twins. N. Engl. J. Med 359, 2849–2850 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Fajans SS & Bell GI MODY: history, genetics, pathophysiology, and clinical decision making. Diabetes Care 34, 1878–1884 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahajan A et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat. Genet 50, 1505–1513, doi: 10.1038/s41588-018-0241-6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Udler MS Type 2 diabetes: multiple genes, multiple diseases. Curr. Diab. Rep 19, 55, doi: 10.1007/s11892-019-1169-7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robertson CC & Rich SS Genetics of type 1 diabetes. Curr. Opin. Genet. Dev 50, 7–16, doi: 10.1016/j.gde.2018.01.006 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Noble JA & Erlich HA Genetics of type 1 diabetes. Cold Spring Harb. Perspect. Med 2, doi: 10.1101/cshperspect.a007732 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barrett JC et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet 41, 703 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cooper NJ et al. Type 1 diabetes genome-wide association analysis with imputation identifies five new risk regions. bioRxiv, 120022, doi: 10.1101/120022 (2017). [DOI] [Google Scholar]
- 33.Onengut-Gumuscu S et al. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat. Genet 47, 381, doi: 10.1038/ng.3245 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ge Y et al. Targeted deep sequencing in multiple-affected sibships of European ancestry identifies rare deleterious variants in PTPN22 that confer risk for type 1 diabetes. Diabetes 65, 794–802, doi: 10.2337/db15-0322 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ingelsson E & McCarthy Mark I Human genetics of obesity and type 2 diabetes mellitus. Circ. Genom. Precis. Med 11, e002090, doi: 10.1161/CIRCGEN.118.002090 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prasad RB & Groop L Genetics of type 2 diabetes—Pitfalls and possibilities. Genes 6, 87–123 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Florez JC The genetics of type 2 diabetes and related traits Switzerland: Springer International Publishing; (2016). [Google Scholar]
- 38.Chen J et al. Genome-wide association study of type 2 diabetes in Africa. Diabetologia 62, 1204–1211, doi: 10.1007/s00125-019-4880-7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ng MCY et al. Meta-analysis of genome-wide association studies in African Americans provides insights into the genetic architecture of type 2 diabetes. PLoS Genet 10, e1004517, doi: 10.1371/journal.pgen.1004517 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Imamura M et al. Genome-wide association studies in the Japanese population identify seven novel loci for type 2 diabetes. Nat. Commun 7, 10531, doi:10.1038/ncomms10531 https://www.nature.com/articles/ncomms10531#supplementary-information (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qi Q et al. Genetics of Type 2 Diabetes in U.S. Hispanic/Latino Individuals: Results From the Hispanic Community Health Study/Study of Latinos (HCHS/SOL). Diabetes 66, 1419–1425, doi: 10.2337/db16-1150 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cho YS et al. Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in East Asians. Nat. Genet 44, 67 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kooner JS et al. Genome-wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nat. Genet 43, 984, doi:10.1038/ng.921 https://www.nature.com/articles/ng.921#supplementary-information (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mahajan A et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat. Genet 46, 234–244, doi: 10.1038/ng.2897 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams AL et al. Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico. Nature 506, 97–101, doi: 10.1038/nature12828 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Estrada K et al. Association of a low-frequency variant in HNF1A with type 2 diabetes in a Latino population. JAMA 311, 2305–2314, doi: 10.1001/jama.2014.6511 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spracklen CN et al. Identification of type 2 diabetes loci in 433,540 East Asian individuals. bioRxiv, 685172, doi: 10.1101/685172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mahajan A et al. Refining the accuracy of validated target identification through coding variant fine-mapping in type 2 diabetes. Nat. Genet 50, 559–571, doi: 10.1038/s41588-018-0084-1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mahajan A et al. 303-OR: ADA Presidents’ Select Abstract: Transethnic Association Study of Type 2 Diabetes in More than a Million Individuals. Diabetes 68, 303-OR, doi: 10.2337/db19-303-OR (2019). [DOI] [Google Scholar]
- 50.Vujkovic M et al. Discovery of 310 novel loci for type-2 diabetes and related complications involving 1.4 million participants in a multi-ethnic meta-analysis. PgmNr 228 American Soceity of Human Genetics. (2019). [Google Scholar]
- 51.Andersen MK et al. Genetics of Type 2 Diabetes: the Power of Isolated Populations. Curr. Diab. Rep 16, 65, doi: 10.1007/s11892-016-0757-z (2016). [DOI] [PubMed] [Google Scholar]
- 52.Langenberg C & Lotta LA Genomic insights into the causes of type 2 diabetes. Lancet 391, 2463–2474, doi: 10.1016/S0140-6736(18)31132-2 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Flannick J et al. Exome sequencing of 20,791 cases of type 2 diabetes and 24,440 controls. Nature 570, 71–76, doi: 10.1038/s41586-019-1231-2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dwivedi OP et al. Loss of ZnT8 function protects against diabetes by enhanced insulin secretion. Nat. Genet 51, 1596–1606, doi: 10.1038/s41588-019-0513-9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zuk O et al. Searching for missing heritability: Designing rare variant association studies. PNAS, 201322563, doi: 10.1073/pnas.1322563111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martínez-Castelao A, Navarro-González J, Górriz J & de Alvaro F The concept and the epidemiology of diabetic nephropathy have changed in recent years. J. Clin. Med 4, 1207–1216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anders HJ, Huber TB, Isermann B & Schiffer M CKD in diabetes: diabetic kidney disease versus nondiabetic kidney disease. Nat. Rev. Nephrol, 1 (2018). [DOI] [PubMed] [Google Scholar]
- 58.Pettitt DJ, Saad MF, Bennett PH, Nelson RG & Knowler WC Familial predisposition to renal disease in two generations of Pima Indians with Type 2 (non-insulin-dependent) diabetes mellitus. Diabetologia 33, 438–443, doi: 10.1007/BF00404096 (1990). [DOI] [PubMed] [Google Scholar]
- 59.Harjutsalo V, Katoh S, Sarti C, Tajima N & Tuomilehto J Population-Based Assessment of Familial Clustering of Diabetic Nephropathy in Type 1 Diabetes. Diabetes 53, 2449 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Borch-Johnsen K et al. Is diabetic nephropathy an inherited complication? Kidney Int 41, 719–722 (1992). [DOI] [PubMed] [Google Scholar]
- 61.Sandholm N et al. The Genetic Landscape of Renal Complications in Type 1 Diabetes. J. Am. Soc. Nephrol, doi: 10.1681/asn.2016020231 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sandholm N et al. Genome-wide association study of urinary albumin excretion rate in patients with type 1 diabetes. Diabetologia 57, 1143–1153, doi: 10.1007/s00125-014-3202-3 (2014). [DOI] [PubMed] [Google Scholar]
- 63.van Zuydam NR et al. A genome-wide association study of diabetic kidney disease in subjects with type 2 diabetes. Diabetes 67, 1414–1427 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Imperatore G et al. Sib-pair linkage analysis for susceptibility genes for microvascular complications among Pima Indians with type 2 diabetes. Pima Diabetes Genes Group. Diabetes 47, 821–830, doi: 10.2337/diabetes.47.5.821 (1998). [DOI] [PubMed] [Google Scholar]
- 65.Iyengar SK et al. Genome-wide scans for diabetic nephropathy and albuminuria in multiethnic populations. Diabetes 56, 1577–1585 (2007). [DOI] [PubMed] [Google Scholar]
- 66.Schelling JR et al. Genome-Wide Scan for Estimated Glomerular Filtration Rate in Multi-Ethnic Diabetic Populations. Diabetes 57, 235–243, doi: 10.2337/db07-0313 (2008). [DOI] [PubMed] [Google Scholar]
- 67.Vardarli I et al. Gene for susceptibility to diabetic nephropathy in type 2 diabetes maps to 18q22.3–23. Kidney Int 62, 2176–2183, doi: 10.1046/j.1523-1755.2002.00663.x (2002). [DOI] [PubMed] [Google Scholar]
- 68.Janssen B et al. Carnosine as a protective factor in diabetic nephropathy: association with a leucine repeat of the carnosinase gene CNDP1. Diabetes 54, 2320–2327 (2005). [DOI] [PubMed] [Google Scholar]
- 69.Mooyaart AL et al. Genetic associations in diabetic nephropathy: a meta-analysis. Diabtologia 54, 544–553, doi: 10.1007/s00125-010-1996-1 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tong Z et al. Promoter polymorphism of the erythropoietin gene in severe diabetic eye and kidney complications. PNAS 105, 6998–7003 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reutens AT Epidemiology of diabetic kidney disease. Medical Clinics 97, 1–18 (2013). [DOI] [PubMed] [Google Scholar]
- 72.Pezzolesi MG et al. Genome-Wide Association Scan for Diabetic Nephropathy Susceptibility Genes in Type 1 Diabetes. Diabetes 58, 1403 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martini S et al. From Single Nucleotide Polymorphism to Transcriptional Mechanism: A Model for FRMD3 in Diabetic Nephropathy. Diabetes 62, 2605–2612, doi: 10.2337/db12-1416 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sandholm N et al. New susceptibility loci associated with kidney disease in type 1 diabetes. PLoS Genet 8, e1002921 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sandholm N et al. Chromosome 2q31. 1 associates with ESRD in women with type 1 diabetes. J. Am. Soc. Nephrol 24, 1537–1543 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Salem RM et al. Genome-Wide Association Study of Diabetic Kidney Disease Highlights Biology Involved in Glomerular Basement Membrane Collagen. J. Am. Soc. Nephrol., ASN 2019030218, doi: 10.1681/asn.2019030218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Longo I et al. COL4A3/COL4A4 mutations: From familial hematuria to autosomal-dominant or recessive Alport syndrome. Kidney Int 61, 1947–1956, doi: 10.1046/j.1523-1755.2002.00379.x (2002). [DOI] [PubMed] [Google Scholar]
- 78.Taira M et al. A variant within the FTO confers susceptibility to diabetic nephropathy in Japanese patients with type 2 diabetes. PLoS One 13, e0208654, doi: 10.1371/journal.pone.0208654 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sandholm N & Groop PH Genetic basis of diabetic kidney disease and other diabetic complications. Curr. Opin. Genet. Dev 50, 17–24, doi: 10.1016/j.gde.2018.01.002 (2018). [DOI] [PubMed] [Google Scholar]
- 80.Ahlqvist E, Van Zuydam NR, Groop LC & McCarthy MI The genetics of diabetic complications. Nat. Rev. Nephrol 11, 277 (2015). [DOI] [PubMed] [Google Scholar]
- 81.Sandholm N et al. Confirmation of GLRA3 as a susceptibility locus for albuminuria in Finnish patients with type 1 diabetes. Sci. Rep 8, 12408, doi: 10.1038/s41598-018-29211-1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Iyengar SK et al. Genome-Wide Association and Trans-ethnic Meta-Analysis for Advanced Diabetic Kidney Disease: Family Investigation of Nephropathy and Diabetes (FIND). PLoS Genet 11, e1005352, doi: 10.1371/journal.pgen.1005352 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Guan M et al. Genome-wide association study identifies novel loci for type 2 diabetes-attributed end-stage kidney disease in African Americans. Hum. Genomics 13, 21, doi: 10.1186/s40246-019-0205-7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Köttgen A et al. Multiple loci associated with indices of renal function and chronic kidney disease. Nat. Genet 41, 712, doi:10.1038/ng.377 https://www.nature.com/articles/ng.377#supplementary-information (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Böger CA et al. CUBN is a gene locus for albuminuria. J. Am. Soc. Nephrol 22, 555–570, doi: 10.1681/asn.2010060598 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pattaro C et al. Genome-Wide Association and Functional Follow-Up Reveals New Loci for Kidney Function. PLoS Genet 8, e1002584, doi: 10.1371/journal.pgen.1002584 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gorski M et al. Genome-wide association study of kidney function decline in individuals of European descent. Kidney Int 87, 1017–1029, doi: 10.1038/ki.2014.361 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Teumer A et al. Genome-wide Association Studies Identify Genetic Loci Associated With Albuminuria in Diabetes. Diabetes 65, 803–817, doi: 10.2337/db15-1313 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pattaro C et al. Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nat. Commun 7, 10023, doi:10.1038/ncomms10023 http://www.nature.com/articles/ncomms10023#supplementary-information (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li M et al. SOS2 and ACP1 Loci Identified through Large-Scale Exome Chip Analysis Regulate Kidney Development and Function. J. Am. Soc. Nephrol 28, 981–994, doi: 10.1681/asn.2016020131 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gorski M et al. 1000 Genomes-based meta-analysis identifies 10 novel loci for kidney function. Sci. Rep 7, 45040, doi:10.1038/srep45040 https://www.nature.com/articles/srep45040#supplementary-information (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Haas ME et al. Genetic Association of Albuminuria with Cardiometabolic Disease and Blood Pressure. Am. J. Hum. Genet 103, 461–473, doi: 10.1016/j.ajhg.2018.08.004 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ahluwalia TS et al. A novel rare CUBN variant and three additional genes identified in Europeans with and without diabetes: results from an exome-wide association study of albuminuria. Diabetologia 62, 292–305, doi: 10.1007/s00125-018-4783-z (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Teumer A et al. Genome-wide association meta-analyses and fine-mapping elucidate pathways influencing albuminuria. Nat. Commun 10, 4130, doi: 10.1038/s41467-019-11576-0 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yau JWY et al. Global Prevalence and Major Risk Factors of Diabetic Retinopathy. Diabetes Care 35, 556–564, doi: 10.2337/dc11-1909 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sivaprasad S, Gupta B, Crosby-Nwaobi R & Evans J Prevalence of Diabetic Retinopathy in Various Ethnic Groups: A Worldwide Perspective. Surv. Ophthalmol 57, 347–370, doi: 10.1016/j.survophthal.2012.01.004 (2012). [DOI] [PubMed] [Google Scholar]
- 97.Centers for Disease Control Prevention. National diabetes fact sheet: national estimates and general information on diabetes and prediabetes in the United States, 2011. Atlanta, GA: US department of health and human services, centers for disease control and prevention 201, 2568–2569 (2011). [Google Scholar]
- 98.Bunce C & Wormald R Causes of blind certifications in England and Wales: April 1999–March 2000. Eye 22, 905 (2008). [DOI] [PubMed] [Google Scholar]
- 99.Klein R, Klein BE & Moss SE Visual impairment in diabetes. Ophthalmology 91, 1–9 (1984). [PubMed] [Google Scholar]
- 100.Leslie R & Pyke D Diabetic retinopathy in identical twins. Diabetes 31, 19–21 (1982). [DOI] [PubMed] [Google Scholar]
- 101.The Diabetes Control and Complications Trial Research Group. Clustering of Long-Term Complications in Families With Diabetes in the Diabetes Control and Complications Trial. Diabetes 46, 1829–1839, doi: 10.2337/diab.46.11.1829 (1997). [DOI] [PubMed] [Google Scholar]
- 102.Looker HC et al. Genome-wide linkage analyses to identify loci for diabetic retinopathy. Diabetes 56, 1160–1166 (2007). [DOI] [PubMed] [Google Scholar]
- 103.Arar NH et al. Heritability of the severity of diabetic retinopathy: The FIND-Eye Study. Invest. Ophthalmol. Vis. Sci 49, 3839–3845, doi: 10.1167/iovs.07-1633 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hietala K, Forsblom C, Summanen P & Groop PH Heritability of Proliferative Diabetic Retinopathy. Diabetes 57, 2176–2180, doi: 10.2337/db07-1495 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hallman DM et al. Familial aggregation of severity of diabetic retinopathy in Mexican Americans from Starr County, Texas. Diabetes Care 28, 1163–1168 (2005). [DOI] [PubMed] [Google Scholar]
- 106.Hampton BM, Schwartz SG, Brantley MA Jr. & Flynn HW Jr. Update on genetics and diabetic retinopathy. Clin. Ophthalmol 9, 2175–2193, doi: 10.2147/OPTH.S94508 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cho H & Sobrin L Genetics of diabetic retinopathy. Curr. Diab. Rep 14, 515, doi: 10.1007/s11892-014-0515-z (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Burdon KP et al. Genome-wide association study for sight-threatening diabetic retinopathy reveals association with genetic variation near the GRB2 gene. Diabetologia 58, 2288–2297, doi: 10.1007/s00125-015-3697-2 (2015). [DOI] [PubMed] [Google Scholar]
- 109.Meng W et al. A genome-wide association study suggests new evidence for an association of the NADPH Oxidase 4 (NOX4) gene with severe diabetic retinopathy in type 2 diabetes. Acta Ophthalmol 96, e811–e819, doi: 10.1111/aos.13769 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li J, Wang JJ & Zhang SX NADPH Oxidase4-derived H2O2 Promotes Aberrant Retinal Neovascularization via Activation of VEGF Receptor 2 Pathway in Oxygen-induced Retinopathy. Invest. Ophthalmol. Vis. Sci 55, 2955–2955 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang H, Yang Z, Jiang Y & Hartnett ME Endothelial NADPH oxidase 4 mediates vascular endothelial growth factor receptor 2-induced intravitreal neovascularization in a rat model of retinopathy of prematurity. Mol. Vis 20, 231–241 (2014). [PMC free article] [PubMed] [Google Scholar]
- 112.Pollack S et al. Multiethnic Genome-Wide Association Study of Diabetic Retinopathy Using Liability Threshold Modeling of Duration of Diabetes and Glycemic Control. Diabetes 68, 441–456, doi: 10.2337/db18-0567 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shtir C et al. Exome-based case–control association study using extreme phenotype design reveals novel candidates with protective effect in diabetic retinopathy. Hum. Genet 135, 193–200, doi: 10.1007/s00439-015-1624-8 (2016). [DOI] [PubMed] [Google Scholar]
- 114.Ung C et al. Whole exome sequencing identification of novel candidate genes in patients with proliferative diabetic retinopathy. Vision Res 139, 168–176, doi: 10.1016/j.visres.2017.03.007 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Johannsen L et al. Evaluation of Patients With Symptoms Suggestive of Chronic Polyneuropathy. J. Clin. Neuromuscul. Dis 3, 47–52 (2001). [DOI] [PubMed] [Google Scholar]
- 116.Pop-Busui R et al. Diabetic Neuropathy: A Position Statement by the American Diabetes Association. Diabetes Care 40, 136–154, doi: 10.2337/dc16-2042 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Singh N, Armstrong DG & Lipsky BA Preventing Foot Ulcers in Patients With Diabetes. JAMA 293, 217–228, doi: 10.1001/jama.293.2.217 (2005). [DOI] [PubMed] [Google Scholar]
- 118.Most RS & Sinnock P The Epidemiology of Lower Extremity Amputations in Diabetic Individuals. Diabetes Care 6, 87–91, doi: 10.2337/diacare.6.1.87 (1983). [DOI] [PubMed] [Google Scholar]
- 119.Young MJ, Boulton AJM, Macleod AF, Williams DRR & Sonksen PH A multicentre study of the prevalence of diabetic peripheral neuropathy in the United Kingdom hospital clinic population. Diabetologia 36, 150–154, doi: 10.1007/BF00400697 (1993). [DOI] [PubMed] [Google Scholar]
- 120.Maser RE et al. Epidemiological Correlates of Diabetic Neuropathy: Report From Pittsburgh Epidemiology of Diabetes Complications Study. Diabetes 38, 1456–1461, doi: 10.2337/diab.38.11.1456 (1989). [DOI] [PubMed] [Google Scholar]
- 121.Tesfaye S et al. Prevalence of diabetic peripheral neuropathy and its relation to glycaemic control and potential risk factors: the EURODIAB IDDM Complications Study. Diabetologia 39, 1377–1384 (1996). [DOI] [PubMed] [Google Scholar]
- 122.Tesfaye S et al. Vascular Risk Factors and Diabetic Neuropathy. N. Engl. J. Med 352, 341–350, doi: 10.1056/NEJMoa032782 (2005). [DOI] [PubMed] [Google Scholar]
- 123.Monti MC et al. Familial Risk Factors for Microvascular Complications and Differential Male-Female Risk in a Large Cohort of American Families with Type 1 Diabetes. J. Clin. Endocrinol. Metab 92, 4650–4655, doi: 10.1210/jc.2007-1185 (2007). [DOI] [PubMed] [Google Scholar]
- 124.Meng W et al. A Genome-wide Association Study Provides Evidence of Sex-specific Involvement of Chr1p35.1 (ZSCAN20-TLR12P) and Chr8p23.1 (HMGB1P46) With Diabetic Neuropathic Pain. EBioMedicine 2, 1386–1393, doi: 10.1016/j.ebiom.2015.08.001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Meng W et al. A genome-wide association study suggests an association of Chr8p21.3 (GFRA2) with diabetic neuropathic pain. Eur. J. Pain 19, 392–399, doi: 10.1002/ejp.560 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Meng W et al. A genome-wide association study suggests that MAPK14 is associated with diabetic foot ulcers. Br. J. Dermatol 177, 1664–1670, doi: 10.1111/bjd.15787 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Politi C et al. Recent advances in exploring the genetic susceptibility to diabetic neuropathy. Diabetes Res. Clin. Pract 120, 198–208, doi: 10.1016/j.diabres.2016.08.006 (2016). [DOI] [PubMed] [Google Scholar]
- 128.Witzel II et al. Identifying Common Genetic Risk Factors of Diabetic Neuropathies. Front. Endocrinol 6, doi: 10.3389/fendo.2015.00088 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tang Y et al. A Genetic Locus on Chromosome 2q24 Predicting Peripheral Neuropathy Risk in Type 2 Diabetes: Results From the ACCORD and BARI 2D Studies. Diabetes 68, 1649–1662, doi: 10.2337/db19-0109 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Blesneac I et al. Rare NaV1.7 variants associated with painful diabetic peripheral neuropathy. Pain 159, 469–480, doi: 10.1097/j.pain.0000000000001116 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Stamler J, Vaccaro O, Neaton JD & Wentworth D Diabetes, Other Risk Factors, and 12-Yr Cardiovascular Mortality for Men Screened in the Multiple Risk Factor Intervention Trial. Diabetes Care 16, 434–444, doi: 10.2337/diacare.16.2.434 (1993). [DOI] [PubMed] [Google Scholar]
- 132.Haffner SM, Lehto S, Rönnemaa T, Pyörälä K & Laakso M Mortality from Coronary Heart Disease in Subjects with Type 2 Diabetes and in Nondiabetic Subjects with and without Prior Myocardial Infarction. N. Engl. J. Med 339, 229–234, doi: 10.1056/nejm199807233390404 (1998). [DOI] [PubMed] [Google Scholar]
- 133.Soedamah-Muthu SS et al. High Risk of Cardiovascular Disease in Patients With Type 1 Diabetes in the UK. Diabetes Care 29, 798–804, doi: 10.2337/diacare.29.04.06.dc05-1433 (2006). [DOI] [PubMed] [Google Scholar]
- 134.Turnbull FM et al. Intensive glucose control and macrovascular outcomes in type 2 diabetes. Diabetologia 52, 2288–2298, doi: 10.1007/s00125-009-1470-0 (2009). [DOI] [PubMed] [Google Scholar]
- 135.Parry HM et al. Both high and low HbA1c predict incident heart failure in type 2 diabetes mellitus. Circ. Heart Fail 8, 236–242, doi: 10.1161/circheartfailure.113.000920 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fall T et al. The role of adiposity in cardiometabolic traits: a Mendelian randomization analysis. PLoS Med 10, e1001474, doi: 10.1371/journal.pmed.1001474 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.De Ferranti SD et al. Type 1 diabetes mellitus and cardiovascular disease: a scientific statement from the American Heart Association and American Diabetes Association. Circulation 130, 1110–1130 (2014). [DOI] [PubMed] [Google Scholar]
- 138.Klein BEK et al. Cardiovascular Disease, Mortality, and Retinal Microvascular Characteristics in Type 1 Diabetes: Wisconsin Epidemiologic Study of Diabetic Retinopathy. Arch. Intern. Med 164, 1917–1924, doi: 10.1001/archinte.164.17.1917 (2004). [DOI] [PubMed] [Google Scholar]
- 139.Zdravkovic S et al. Heritability of death from coronary heart disease: a 36‐year follow‐ up of 20 966 Swedish twins. J. Intern. Med 252, 247–254 (2002). [DOI] [PubMed] [Google Scholar]
- 140.Gusev A et al. Partitioning Heritability of Regulatory and Cell-Type-Specific Variants across 11 Common Diseases. Am. J. Hum. Genet 95, 535–552, doi: 10.1016/j.ajhg.2014.10.004 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Simonson MA, Wills AG, Keller MC & McQueen MB Recent methods for polygenic analysis of genome-wide data implicate an important effect of common variants on cardiovascular disease risk. BMC Med. Genet 12, 146, doi: 10.1186/1471-2350-12-146 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wagenknecht Lynne E et al. Familial Aggregation of Coronary Artery Calcium in Families with Type 2 Diabetes. Circulation 103, 1353–1353, doi: 10.1161/circ.103.suppl_1.9998-11 (2001). [DOI] [PubMed] [Google Scholar]
- 143.Lange LA et al. Heritability and Expression of C-Reactive Protein in Type 2 Diabetes in the Diabetes Heart Study. Ann. Hum. Genet 70, 717–725, doi: 10.1111/j.1469-1809.2006.00280.x (2006). [DOI] [PubMed] [Google Scholar]
- 144.Lange LA et al. Heritability of Carotid Artery Intima-Medial Thickness in Type 2 Diabetes. Stroke 33, 1876–1881, doi: 10.1161/01.STR.0000019909.71547.AA (2002). [DOI] [PubMed] [Google Scholar]
- 145.Erdmann J, Kessler T, Munoz Venegas L & Schunkert H A decade of genome-wide association studies for coronary artery disease: the challenges ahead. Cardiovasc. Res 114, 1241–1257, doi: 10.1093/cvr/cvy084 (2018). [DOI] [PubMed] [Google Scholar]
- 146.Qi L et al. Genetic susceptibility to coronary heart disease in type 2 diabetes. J. Am. Coll. Cardiol 58, 2675–2682, doi: 10.1016/j.jacc.2011.08.054 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Parry HM et al. Genetic variants predicting left ventricular hypertrophy in a diabetic population: a Go-DARTS study including meta-analysis. Cardiovasc. Diabetol 12, 109, doi: 10.1186/1475-2840-12-109 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cox AJ et al. Genetic risk score associations with cardiovascular disease and mortality in the Diabetes Heart Study. Diabetes Care 37, 1157–1164, doi: 10.2337/dc13-1514 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Doria A et al. Interaction Between Poor Glycemic Control and 9p21 Locus on Risk of Coronary Artery Disease in Type 2 Diabetes. JAMA 300, 2389–2397, doi: 10.1001/jama.2008.649 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhang LW et al. Interaction of type 2 diabetes mellitus with Chromosome 9p21 rs10757274 polymorphism on the risk of myocardial infarction: a case–control study in Chinese population. BMC Cardiovasc. Disord 14, 170, doi: 10.1186/1471-2261-14-170 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Qi L et al. Association Between a Genetic Variant Related to Glutamic Acid Metabolism and Coronary Heart Disease in Individuals With Type 2 Diabetes. JAMA 310, 821–828, doi: 10.1001/jama.2013.276305 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Prudente S et al. Genetic Variant at the GLUL Locus Predicts All-Cause Mortality in Patients With Type 2 Diabetes. Diabetes 64, 2658–2663, doi: 10.2337/db14-1653 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.The Look AHEAD Research Group. Prospective Association of GLUL rs10911021 With Cardiovascular Morbidity and Mortality Among Individuals With Type 2 Diabetes: The Look AHEAD Study. Diabetes 65, 297–302, doi: 10.2337/db15-0890 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Shah HS et al. Genetic Predictors of Cardiovascular Mortality During Intensive Glycemic Control in Type 2 Diabetes: Findings From the ACCORD Clinical Trial. Diabetes Care 39, 1915–1924, doi: 10.2337/dc16-0285 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Shah HS et al. Modulation of GLP-1 Levels by a Genetic Variant That Regulates the Cardiovascular Effects of Intensive Glycemic Control in ACCORD. Diabetes Care 41, 348–355, doi: 10.2337/dc17-1638 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Marso SP et al. Semaglutide and Cardiovascular Outcomes in Patients with Type 2 Diabetes. N. Engl. J. Med 375, 1834–1844, doi: 10.1056/NEJMoa1607141 (2016). [DOI] [PubMed] [Google Scholar]
- 157.Marso SP et al. Liraglutide and Cardiovascular Outcomes in Type 2 Diabetes. N. Engl. J. Med 375, 311–322, doi: 10.1056/NEJMoa1603827 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.The ADVANCE Collaborative Group. Intensive Blood Glucose Control and Vascular Outcomes in Patients with Type 2 Diabetes. N. Engl. J. Med 358, 2560–2572, doi: 10.1056/NEJMoa0802987 (2008). [DOI] [PubMed] [Google Scholar]
- 159.Gerstein HC et al. Effects of intensive glucose lowering in type 2 diabetes. N. Engl. J. Med 358, 2545–2559, doi: 10.1056/NEJMoa0802743 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Investigators OT Basal insulin and cardiovascular and other outcomes in dysglycemia. N. Engl. J. Med 367, 319–328 (2012). [DOI] [PubMed] [Google Scholar]
- 161.Gerstein HC et al. Basal insulin and cardiovascular and other outcomes in dysglycemia. N. Engl. J. Med 367, 319–328, doi: 10.1056/NEJMoa1203858 (2012). [DOI] [PubMed] [Google Scholar]
- 162.Ray KK et al. Effect of intensive control of glucose on cardiovascular outcomes and death in patients with diabetes mellitus: a meta-analysis of randomised controlled trials. Lancet 373, 1765–1772, doi: 10.1016/s0140-6736(09)60697-8 (2009). [DOI] [PubMed] [Google Scholar]
- 163.Tkac I Effect of intensive glycemic control on cardiovascular outcomes and all-cause mortality in type 2 diabetes: Overview and metaanalysis of five trials. Diabetes Res. Clin. Pract 86 Suppl 1, S57–62, doi: 10.1016/s0168-8227(09)70011-7 (2009). [DOI] [PubMed] [Google Scholar]
- 164.Boussageon R et al. Effect of intensive glucose lowering treatment on all cause mortality, cardiovascular death, and microvascular events in type 2 diabetes: meta-analysis of randomised controlled trials. BMJ 343, d4169, doi: 10.1136/bmj.d4169 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Holman RR, Paul SK, Bethel MA, Matthews DR & Neil HAW 10-Year Follow-up of Intensive Glucose Control in Type 2 Diabetes. N. Engl. J. Med 359, 1577–1589, doi: 10.1056/NEJMoa0806470 (2008). [DOI] [PubMed] [Google Scholar]
- 166.Gerstein HC et al. Effects of intensive glycaemic control on ischaemic heart disease: analysis of data from the randomised, controlled ACCORD trial. Lancet 384, 1936–1941, doi: 10.1016/s0140-6736(14)60611-5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hayward RA, Reaven PD & Emanuele NV Follow-up of Glycemic Control and Cardiovascular Outcomes in Type 2 Diabetes. N. Engl. J. Med 373, 978, doi: 10.1056/NEJMc1508386 (2015). [DOI] [PubMed] [Google Scholar]
- 168.Zinman B, Lachin JM & Inzucchi SE Empagliflozin, Cardiovascular Outcomes, and Mortality in Type 2 Diabetes. N. Engl. J. Med 374, 1094, doi: 10.1056/NEJMc1600827 (2016). [DOI] [PubMed] [Google Scholar]
- 169.Merino J et al. Genetically Driven Hyperglycemia Increases Risk of Coronary Artery Disease Separately From Type 2 Diabetes. Diabetes Care 40, 687–693, doi: 10.2337/dc16-2625 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Rani PK et al. Albuminuria and diabetic retinopathy in type 2 diabetes mellitus Sankara Nethralaya Diabetic Retinopathy Epidemiology and Molecular Genetic Study (SN-DREAMS, report 12). Diabetol. Metab. Syndr 3, 9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Groop PH et al. The Presence and Severity of Chronic Kidney Disease Predicts All-Cause Mortality in Type 1 Diabetes. Diabetes 58, 1651–1658, doi: 10.2337/db08-1543 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Chavers BM, Mauer SM, Ramsay RC & Steffes MW Relationship between retinal and glomerular lesions in IDDM patients. Diabetes 43, 441–446, doi: 10.2337/diab.43.3.441 (1994). [DOI] [PubMed] [Google Scholar]
- 173.Klein R et al. The Relationship of Diabetic Retinopathy to Preclinical Diabetic Glomerulopathy Lesions in Type 1 Diabetic Patients. Diabetes 54, 527–533, doi: 10.2337/diabetes.54.2.527 (2005). [DOI] [PubMed] [Google Scholar]
- 174.Dyck PJ et al. The prevalence by staged severity of various types of diabetic neuropathy, retinopathy, and nephropathy in a population‐based cohort. Neurology 43, 817-817, doi: 10.1212/wnl.43.4.817 (1993). [DOI] [PubMed] [Google Scholar]
- 175.Hill MN et al. Risk of foot complications in long-term diabetic patients with and without ESRD: a preliminary study. ANNA J 23, 381–389 (1996). [PubMed] [Google Scholar]
- 176.Eggers PW, Gohdes D & Pugh J Nontraumatic lower extremity amputations in the Medicare end-stage renal disease population. Kidney Int 56, 1524–1533, doi: 10.1046/j.1523-1755.1999.00668.x (1999). [DOI] [PubMed] [Google Scholar]
- 177.Drury PL et al. Estimated glomerular filtration rate and albuminuria are independent predictors of cardiovascular events and death in type 2 diabetes mellitus: the Fenofibrate Intervention and Event Lowering in Diabetes (FIELD) study. Diabetologia 54, 32–43, doi: 10.1007/s00125-010-1854-1 (2011). [DOI] [PubMed] [Google Scholar]
- 178.Nichols GA, Déruaz-Luyet A, Hauske SJ & Brodovicz KG The association between estimated glomerular filtration rate, albuminuria, and risk of cardiovascular hospitalizations and all-cause mortality among patients with type 2 diabetes. J. Diabetes Complications 32, 291–297, doi: 10.1016/j.jdiacomp.2017.12.003 (2018). [DOI] [PubMed] [Google Scholar]
- 179.Fuller JH, Stevens LK, Wang SL & and the WHO Multinational Study Group. Risk factors for cardiovascular mortality and morbidity: The WHO multinational study of vascular disease in diabetes. Diabetologia 44, S54, doi: 10.1007/PL00002940 (2001). [DOI] [PubMed] [Google Scholar]
- 180.Cheung N et al. Is diabetic retinopathy an independent risk factor for ischemic stroke? Stroke 38, 398–401 (2007). [DOI] [PubMed] [Google Scholar]
- 181.Cheung N et al. Diabetic retinopathy and the risk of coronary heart disease: the Atherosclerosis Risk in Communities Study. Diabetes Care 30, 1742–1746 (2007). [DOI] [PubMed] [Google Scholar]
- 182.Mukhopadhyay N, Noble JA, Govil M, Marazita ML & Greenberg DA Identifying genetic risk loci for diabetic complications and showing evidence for heterogeneity of type 1 diabetes based on complications risk. PLoS One 13, e0192696, doi: 10.1371/journal.pone.0192696 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Scott RA et al. An Expanded Genome-Wide Association Study of Type 2 Diabetes in Europeans. Diabetes 66, 2888–2902, doi: 10.2337/db16-1253 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.McCarthy S et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet 48, 1279 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]


