Abstract
Glycine-rich proteins (GRPs) are a large family of proteins that play vital roles in cell wall remodeling, metabolism and development, and abiotic stress response. Although the functions of GRPs in cell wall remodeling have been extensively characterized, only a few studies have explored their effects on chlorophyll metabolism and hormone response. Accordingly, we aimed to determine the molecular mechanism of BcGRP23 and its role in chlorophyll metabolism and the BRI1-EMS-SUPPRESSOR 1 (BES1) signaling pathway in flowering Chinese cabbage. The expression levels of BcGRP23 in the leaves and stems gradually decreased with increasing growth and development of flowering Chinese cabbage, while BcGRP23 was barely expressed after flowering. As plant growth continued, the GUS (β-glucuronidase) stain gradually became lighter in hypocotyls and was largely free of growth points. The petioles and stems of BcGRP23-silenced plants lost their green color, and the contents of chlorophyll a (Chl a) and Chl b were significantly reduced. Further research revealed that the expression levels of chlorophyll degradation-related genes were significantly increased in silenced plants compared with the control; however, the opposite was noted for the BcGRP23-overexpressing lines. The BcGRP23 promoter sequence contains numerous hormone-responsive elements. In fact, the expression of BcGRP23 was upregulated in flowering Chinese cabbage following treatment with the hormones indole-3-acetic acid (IAA), gibberellin (GA), 6-benzylaminopurine (6-BA), methyl jasmonate (MeJA), and brassinosteroid (BR). Treatment with BR led to the most significant upregulation. BES1, in response to BRs, directly activated the BcGRP23 promoter. Overall, BcGRP23 regulated the expression of chlorophyll degradation-related genes, thereby affecting the chlorophyll content. Furthermore, the expression of BcGRP23 was significantly regulated by exogenous BR application and was directly activated by BES1. These findings preliminarily suggest the molecular mechanism and regulatory pathway of BcGRP23 in the growth and development of flowering Chinese cabbage plants and their response to environmental stress.
Keywords: glycine-rich protein, chlorophyll, hormone, BES1, transcriptional regulation, flowering Chinese cabbage
Introduction
Glycine-rich proteins (GRPs) are a class of proteins that share a high-glycine-content region and are involved in cellular stress response and signal transduction in plants (Teng et al., 2017; Czolpinska and Rurek, 2018). PtGRP1 was the first glycine-rich cell wall protein to be isolated from petunia (Petunia hybrida) (Condit and Meagher, 1986). More than 150 GRPs have been identified in various plant species, including maize, rice, Arabidopsis thaliana, and tobacco (Cornels et al., 2000; Liu et al., 2003; Zhang et al., 2014; Mangeon et al., 2016). In most plants, GRPs are classified into five groups according to the type and arrangement of their protein motifs (Mangeon et al., 2010). Members of classes I, II, and IV appear to be critically involved in the regulation of plant cells, hormone signaling, stress adaptation, and flower development (Czolpinska and Rurek, 2018). Recently, several studies have sought to determine the functions of GRPs in various plants. It was found that different environmental factors regulate the expression of GRPs, such as temperature, flooding, salinity, ultraviolet light, and hormones (Kang et al., 2013). For example, in Arabidopsis, the expression of AtGRP genes is regulated by biotic and abiotic factors (Sachetto-Martins et al., 2000). In particular, AtGRP2 promotes seed germination and growth under salt stress in Arabidopsis (Fusaro et al., 2007). Both AtGRP2 and AtGRP7 can enhance the cold and freezing tolerance of Arabidopsis and rice, and GRPs that confer stress tolerance may be regulated as RNA chaperones in plants based on the efficiency of messenger RNA (mRNA) translation (Kang et al., 2013).
GRP expression is markedly tissue-specific, and its functions vary with plant growth and development (Lu et al., 2020). Analysis of 11 GRPs in different tissues and organs of Arabidopsis (Ma et al., 2021) revealed that AtGRP1 is weakly expressed in all organs, while other genes had the highest expression in inflorescence cells and protoplasts. In common bean (Phaseolus vulgaris L.), PvGRP1.8 was found to be mainly expressed in the xylem and cambium cells of the stem. In addition, homologous GRP1.8 genes with the same expression pattern were found in tomato, tobacco, petunia, and soybean (Xuan et al., 2010). OsGRP-2 is localized in the cell wall, and its promoter is specifically expressed in vascular bundles in transgenic tobacco and rice (Liu et al., 2003). Notably, some GRP genes are expressed in other tissues. For example, AtGRP5 localizes to the vacuole and is preferentially expressed in developing embryonic cells (Mangeon et al., 2009; Mangeon et al., 2010). Some GRP genes are also specifically expressed in the roots, such as ZmGRP3 and ZmGRP4 in maize and BnGRP22 in rapeseed. The expression of BnGRP22 is induced by brassinolide at high temperatures and may enhance the stress resistance of rapeseed (Dhaubhadel and Krishna, 2008). Overall, GRP genes show different tissue specificities in different plants.
Photosynthesis is a fundamental element of plant growth and development. Many studies have demonstrated the crucial relationship between the photosynthetic pigment content and the growth rate of plants under abiotic stress (Xiang et al., 2016; Suhel et al., 2022). The expression of chlorophyll biosynthesis genes may alter the accumulation of chlorophyll, which causes a significant decline in plant growth and production (Xiong et al., 2018). Chlorophyll biosynthesis is catalyzed by 15 key enzymes encoded by more than 20 genes, which are highly sensitive to environmental influences (Beale, 2005). The hormone signaling pathway plays an important regulatory role in the chlorophyll biosynthesis and degradation pathways (Liu et al., 2022). Brassinosteroids (BRs) regulate plant defense systems by regulating chlorophyll content, which increases the photosynthetic capacity and antioxidant enzyme activities (Yuan et al., 2012). Exogenous application of BRs was found to enhance the expression of the chlorophyll biosynthesis genes—RCBL, RCBS, RCA, SBPase, FBPase, and FBPaldolase—under high root zone temperature in cucumber and pepper under chilly temperatures (Ding et al., 2016). Exogenous jasmonic acid (JA) induces plant leaf yellowing by regulating the expression of multiple senescence-related genes (Buchanan-Wollaston et al., 2005). Chlorophyll degradation and reduced Rubisco levels in barley (Hordeum vulgare) leaves led to barley leaf senescence post-treatment with exogenous JA or methyl jasmonate (MeJA) (He et al., 2002). MYC2, an essential component of the JA signaling pathway, directly interacts with ANAC019 to regulate the expression of NYE1/SGR1 (STAY-GREEN protein), NYE2/SGR2, and NYC1, which are involved in chlorophyll degradation (Zhu et al., 2015). The ethylene signaling pathway transcription factor EIN3 promotes the degradation of chlorophyll by combining with three chlorophyll degradation genes: Non-Yellow Coloring 1 (NYC1), non-yellowing 1 (NYE1), and pheophorbide a oxygenase (PAO) (Cheng and Guan, 2014; Charoenchongsuk et al., 2015). In tomato, the expression levels of the chlorophyll degradation-related genes PPH (Pheophytinase) and PAO were upregulated after abscisic acid (ABA) treatment. Furthermore, the expression of PAO was negatively regulated by nordihydroguaiaretic acid (NDGA) (Mou et al., 2015). In summary, hormones are closely related to the chlorophyll biosynthesis and degradation pathways.
Although several GRPs have been identified and functionally characterized in plants, and their important role in plant development and adaptation to adversity has been revealed, relatively few studies have been conducted on the related functions of GRPs in the chlorophyll pathway in response to hormones. The flowering Chinese cabbage (Brassica campestris L. ssp. chinesis var. utilis Tsen et Lee) is a variant of Chinese cabbage in the Brassica family. It is a key vegetable crop worldwide, especially in South China, and has significant economic value (Zhang et al., 2022). In our previous study on the transcriptome of propiconazole treatment, BcGRP23 was significantly downregulated under exogenous substance stress in the flowering Chinese cabbage ( Supplementary Figure S6 ). GRP23 was hypothesized to be involved in the regulation of the hormone-induced chlorophyll pathway and to play an unknown biological function in the growth regulation and abiotic stress response of flowering Chinese cabbage. Accordingly, BcGRP23 was isolated from flowering Chinese cabbage plants to characterize its function and explore the molecular mechanisms whereby it regulates the growth of flowering Chinese cabbage. The findings of this study will lay the foundation for an in-depth understanding of the molecular mechanisms affecting plant growth and GRP responses to the environment.
Materials and methods
Plant materials and growth conditions
The flowering Chinese cabbage cultivar ‘Youlv 501’ was obtained from the Guangzhou Academy of Agricultural Science and cultivated in a greenhouse at the College of Horticulture, South China Agricultural University. Wild-type (WT) and BcGRP23 transgenic plants were grown in the temperature range 23°C–25°C, with 16-h light (100 µmol m−2 s−1) and 8-h dark. Knockdown lines generated by transient virus-induced gene silencing (VIGS) were cultured at 22°C/20°C (day/night). The other environmental conditions were the same as those listed above. Different tissue parts of normal growing flowering Chinese cabbage seedlings were sampled at different growth stages (cotyledon, three-leaf, six-leaf, initial flowering, and full-bloom stages) for quantitative real-time PCR (qRT-PCR). A. thaliana Columbia-0 (Col-0) was used for the genetic transformation of BcGRP23. Transgenic lines and WT Arabidopsis plants were maintained normally in a growth chamber (23°C–25°C, with a light cycle of 16-h light and 8-h dark).
Cloning and bioinformatics analysis of BcGRP23
Total RNA was extracted from the leaves and stems of flowering Chinese cabbage using the Eastep® Super Total RNA Extraction Kit (Promega, Shanghai, China) according to the manufacturer’s instructions. First-strand complementary DNA (cDNA) was synthesized and subjected to reverse transcription using the PrimeScript II 1st Strand cDNA Synthesis Kit (Takara, Dalian, China). The coding sequences (CDS) of BcGRP23 isolated from flowering Chinese cabbage were cloned using gene-homologous primers with reference to the Brassica rapa genome ( Supplementary Table S1 ). The nucleic acid and promoter sequences of Chinese cabbage BrGRP23 (LOC103853913) were obtained from the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov). Amino acid (aa) prediction of BcGRP23 was performed using EditSeq software (DNAStar Inc., Madison, WI, USA). Multiple sequence alignment was performed using MEGA7.0, and a phylogenetic tree was constructed using MEGA7.0 and the neighbor-joining algorithm with 1,000 bootstrap replicates (Kumar et al., 2016). Promoter cis-acting regulatory element searches were conducted on promoters using the PlantCARE software (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Subcellular localization
The full-length BcGRP23 CDS without a stop codon was cloned into the pBI121-GFP vector and fused with the gene sequence of the green fluorescent protein (GFP). The primers used for cloning are listed in Supplementary Table S2 . Onion epidermal cells were first infiltrated with the Agrobacterium tumefaciens strain GV3101 and transformed with the corresponding constructs. The GFP signal was captured using a fluorescence microscope (Axio Imager D2, Zeiss, Oberkochen, Germany). All assays were repeated three times.
Quantitative real-time polymerase chain reaction
Total RNA was isolated from the leaves and petioles of WT and BcGRP23 transgenic plants. Thereafter, qRT-PCR was performed using a SYBR Premix EX Taq kit (Takara, Dalian, China) on a LightCycler 480 Real-Time PCR instrument (Roche Diagnostics, Rotkreutz, Switzerland). Three biological and three technical replicates were used for each sample. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene (XM_033273046.1) was used as the internal control. The primers used for qRT-PCR are listed in Supplementary Tables S2, S3 .
Virus-induced gene silencing assay
The BcGRP23 CDS was cloned into the pTRV2 plasmid to generate the silencing vector, pTRV2:BcGRP23. The specific experimental procedure is described elsewhere (Chen et al., 2011). The pTRV2:BcGRP23 and pTVR1 vectors were co-infiltrated into the cotyledons of young flowering Chinese cabbage seedlings. Two weeks after infection, the second true leaf was collected from each plant, and RNA was extracted for qRT-PCR to determine the efficiency of gene silencing. Plants co-infiltrated with pTRV2 and pTRV1 were used as negative controls (Wang et al., 2022). The primers used for the VIGS assay are listed in Supplementary Table S2 .
Agrobacterium-mediated transformation in flowering Chinese cabbage
Transgenic flowering Chinese cabbage plants overexpressing BcGRP23 were obtained using the PBI121-BcGRP23-GFP vector. Cotyledon transformation of the ‘Youlv 501’ cultivar was performed using our laboratory method (Wang et al., 2022). Positive transgenic plants were identified via qRT-PCR of BcGRP23 in the leaves and petioles.
Arabidopsis thaliana transformation
The BcGRP23 promoter was cloned into the pCAMBIA1391-GUS vector to obtain the pCAMBIA1391-proBcGRP23-GUS recombinant plasmid, which was introduced into the A. tumefaciens strain GV3101. The primers used for cloning are listed in Supplementary Table S2 . Arabidopsis Col-0 transgenic plants expressing pCAMBIA1391-proBcGRP23-GUS were generated using the floral dip method. Positive plants were screened on Murashige–Skoog (MS) medium containing 25 mg/L hygromycin and were identified using PCR.
Histochemical GUS staining analysis
T2 generation transgenic Arabidopsis plant tissues from different parts of the plant at different growth stages were selected, placed in an appropriate amount of β-glucuronidase (GUS) staining solution at 37°C for 12 h, and then decolorized with 50%, 70%, and 95% ethanol in a gradient manner until complete. The samples were stored in 70% ethanol for examination of their expression characteristics.
Exogenous hormone treatments
Seedlings of transgenic Arabidopsis lines in the cotyledon stage were treated with 50 μmol L−1 ABA, indole-3-acetic acid (IAA), gibberellin (GA), 6-benzylaminopurine (6-BA), salicylate (SA), MeJA, and BR and then immersed in water as a control. The concentration of the hormone treatments was the optimal concentration determined during the pre-experiment. After 3 h of treatment, histochemical GUS analysis was performed using the same method described above. Three-week-old seedling leaves of flowering Chinese cabbage (WT) were sprayed with 100 μmol L−1 ABA, IAA, GA, 6-BA, SA, MeJA, or BR (treated with water as a control). After 6 h, the samples were collected and the transcript levels of BcGRP23 determined using qRT-PCR.
Yeast one-hybrid assays
Yeast one-hybrid (Y1H) assays were conducted using the Matchmaker One-Hybrid System (Clontech, Palo Alto, CA, USA) (Wu et al., 2022). The 572-bp region surrounding the BcGRP23 promoter sequence was synthesized with the KnpI and SalI flanking restriction sites and then cloned into a KpnI/SalI-digested pAbAI bait vector. For expression in yeast, the cDNA of BcBES1 was cloned into the pGADT7 prey vector. Saccharomyces Y1H Gold (Clontech, Palo Alto, CA, USA) was used as the host. We first digested the pAbAI-proGRP23 vector using BstBI), and then integrated it into Y1H Gold. The bait strain (Y1H-pAbAI-proGRP23) was screened for self-activated on synthetically defined (SD)/−Ura with aureobasidin A (AbA*). Transformation was confirmed via growth on SD/−Leu medium (Clontech, Palo Alto, CA, USA) containing AbA (Yeasen, Shanghai, China). The primers used are listed in Supplementary Table S2 .
Dual-luciferase reporter assays using N. benthamiana leaves
For analyses of the transcriptional activity of BcBES1 to BcGRP23 in Nicotiana benthamiana leaves, the CDS of BcBES1 was amplified by PCR and inserted into the pGreenII 62-SK vector to generate the effector. To generate the reporter constructs, the promoter fragments of BcGRP23 were inserted into the pGreenII 0800 vector. The empty pGreenII 62-SK vector was used as a control. The constructs were then introduced into tobacco plants via Agrobacterium-mediated transformation. Firefly luciferase (LUC) and Renilla (REN) activities were measured using a dual-luciferase reporter assay kit (Promega, Madison, WI, USA). The relative ratio of LUC to REN was calculated to represent the expression of the reporter genes. The experiment was performed three times, with each sample also tested three times. The primers used for these vectors are listed in Supplementary Table S2 .
Results
Sequence analysis and subcellular localization of BcGRP23 in flowering Chinese cabbage
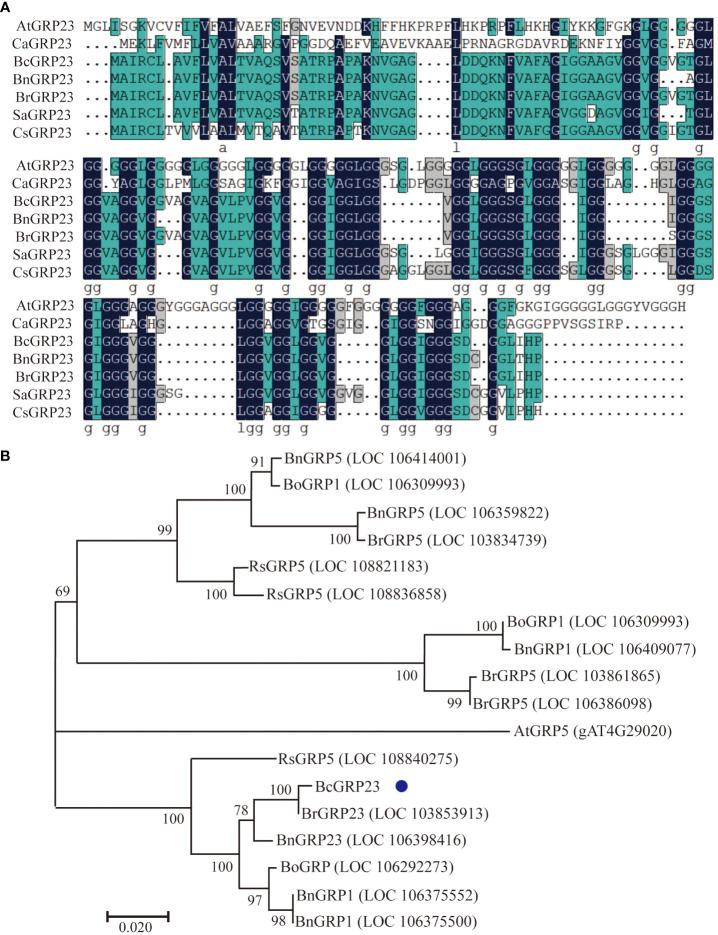
Specific primers were designed for the BcGRP23 gene (CDS) based on the sequence of the BrGRP23 gene from Chinese cabbage. The BcGRP23 gene was then isolated from flowering Chinese cabbage and confirmed using PCR. Based on the sequencing results, a BcGRP23 open reading frame (ORF) with 444 bp was obtained ( Supplementary Figure S1 ). The ORF of BcGRP23 was identified to encode 147 amino acids ( Supplementary Figure S1A ). Subsequently, the aa sequence alignment of GRP23 homologous proteins from Arabidopsis, Chinese cabbage, and flowering Chinese cabbage was performed using MEGA7 software ( Figure 1A ). The results showed that the homology of the BcGRP23 and BrGRP23 (LOC103853913) aa sequences was 99.32%, and both contained multiple GGX or GGGX tandem repeats. In contrast, the homology between BcGRP23 and AtGRP23 (AT2G32690) aa sequences was only 29.85%, with AtGRP23 containing multiple tandem repeats of GGGX and GGGGGX.
Figure 1.
Protein sequence alignments of BcGRP23 in flowering Chinese cabbage and other species. (A) Multiple sequence alignment performed using MEGA7 software. Amino acids with blue background are highly conserved. The sequences were markedly similar between Brassica rapa, Cucumis sativus, Brassica napa, Sinapis alba, Camelina sativa, and Arabidopsis thaliana. (B) Phylogenetic analysis of the glycine-rich protein (GRP) sequences of different plant species performed using the neighbor joining method in MEGA7, with 1,000 bootstrap replicates. Bo, B. oleracea; Bn, B. napus; Bc, B. campestris; Rs, Raphanus sativus.
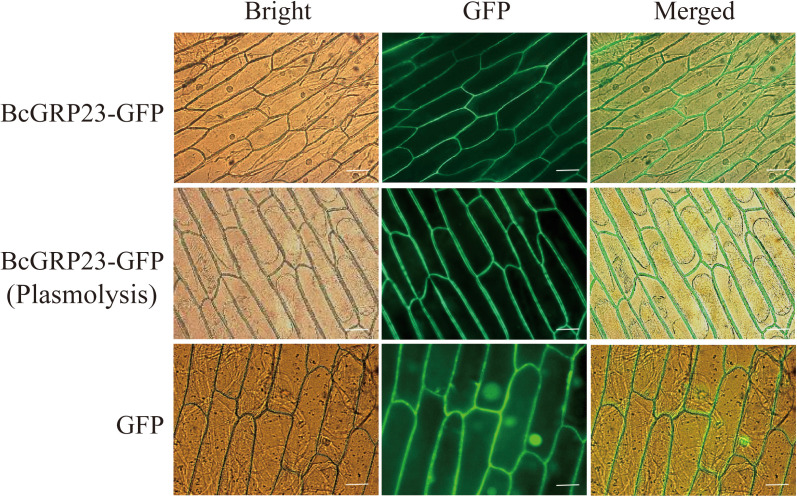
To explore the evolutionary relationships among the BcGRP genes, a maximum likelihood (ML) phylogenetic tree was constructed according to the GRP protein sequences from different species ( Figure 1B ). The evolutionary relationship analysis indicated that BcGRP23 is closely related to GRP23 in other cruciferous species (B. rapa and Brassica napus). In fact, BcGRP23 showed 99.95% homology with BrGRP23 and 94.59% homology with BnGRP23, whereas BcGRP23 showed 92.57% homology with RsGRP5. BcGRP23 had a homology of 90.13% and 84.75% with BnGRP1 and BnGRP5, respectively ( Figures 1A, B ). When the pBI121-BcGRP23-GFP plasmid was transformed into the onion epidermis, an obvious GFP signal was observed in the cell wall of the plasmolysis onion epidermis, but not on the cell membrane ( Figure 2 ). This result indicates that BcGRP23 was localized in the cell wall.
Figure 2.
Subcellular localization of BcGRP23. The vectors 35S::GFP (pBI121-GFP, positive control) and 35S:BcGRP23-GFP (pBI121-BcGRP23-GFP) were injected into onion epidermal cells and visualized with a fluorescence microscope after 24 h of bombardment. Bar, 25 μm.
Expression analysis of BcGRP23 in different tissues at various developmental periods
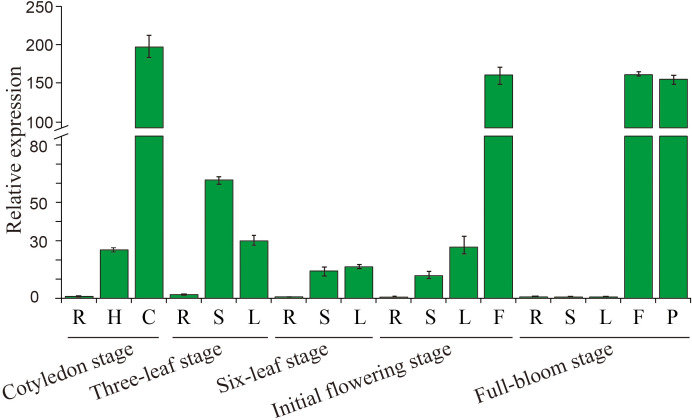
To assess the tissue-specific expression of BcGRP23, qRT-PCR was performed ( Figure 3 ). Noticeable differences were identified when the transcript levels of BcGRP23 in different organs of flowering Chinese cabbage were assayed. The expression level of BcGRP23 in the roots was low during the entire growth period; however, in the cotyledon stage, flowers in the initial flowering stage and the flowers and seeds in the full-bloom stage were the highest, with values 197-, 159.4-, 159.7-, and 153.6-fold higher than those of the roots in the cotyledon stage, respectively. When the stems at different stages were analyzed, the highest expression of BcGRP23 was found at the three-leaf stage, with a value 2.4-fold higher than that at the cotyledon stage; this level was significantly higher than that in the six-leaf stage and the initial flowering stems. Notably, almost no expression was observed in flowering stems. The expression level of BcGRP23 in leaves was the highest in the cotyledon stage, with a value 6.3-fold higher than that of the leaves in the three-leaf stage. No significant difference in the expression of BcGRP23 was found in the three-leaf stage, six-leaf stage, and the initial flowering stage. Furthermore, BcGRP23 was not expressed in the flowering stage.
Figure 3.
Expression analysis of BcGRP23 in different tissues at various developmental stages. R, root; H, hypocotyl; C, cotyledon; S, stem; L, leaf; F, flower; P, pod. Bar on top of the columns represents the standard deviation.
BcGRP23 silencing accelerates chlorophyll degradation in flowering Chinese cabbage
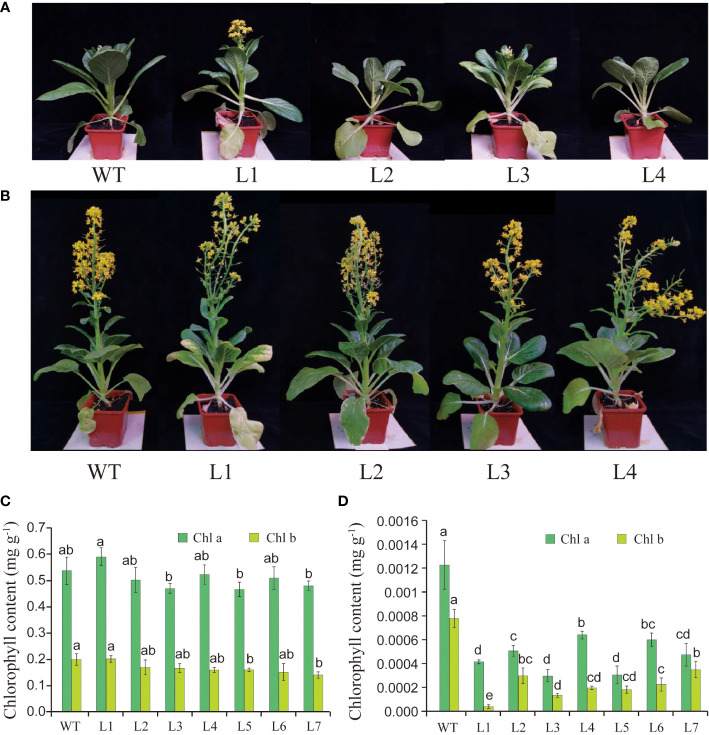
To explore the biological function of BcGRP23, the phenotypes of the pTRV2:BcGRP23 knockdown lines were analyzed. Most interestingly, the pTRV2:BcGRP23 knockdown lines showed whitening of the stems and petioles compared with the WT (pTRV2 empty vector) in the initial flowering stage ( Figure 4A ). In fact, the stems and petioles of the pTRV2:BcGRP23 knockdown lines lost their green color. However, the stems, petioles, and flowers of the upper half of the knockdown lines were not significantly different from those of the WT after entering the flowering and fruiting stages ( Figure 4B ). Overall, the silencing of BcGRP23 could lead to white stems and petioles; however, only the stems and petioles of the lower half turned white during the flowering and fruiting stages. This result may be due to the timeliness of the VIGS knockdown lines. The whitening phenomenon may be related to the decrease in chlorophyll content. Therefore, the contents of chlorophyll a (Chl a) and Chl b in the leaves and petioles were determined in the pTRV2:BcGRP23 knockdown lines and in WT. Chl a and Chl b in the leaves of the pTRV2:BcGRP23 knockdown lines were not significantly different from those of the WT ( Figure 4C ). However, the contents of Chl a and Chl b in the petioles of the pTRV2:BcGRP23 knockdown lines were significantly lower than those in WT plants ( Figure 4D ). Thus, the silencing of BcGRP23 could reduce the contents of photosynthetic pigments such as Chl a and Chl b, which may be the reason for the whitening of the petioles and stems.
Figure 4.
(A, B) Effect of BcGRP23 knockdown on the phenotypes of flowering Chinese cabbage at the initial flowering stage (A) and the full-bloom stage (B). Transgenic lines carrying the empty pTRV2 vector (EV) were used as wild type (WT). L1–L4 represent pTRV2:BcGRP23. (C, D) Chlorophyll contents of the leaves (C) and petioles (D) in the BcGRP23 knockdown lines. L1–L7 represent different BcGRP23 knockdown lines. Error bars represent standard errors. Different lowercase letters indicate significant differences at the p < 0.05 level.
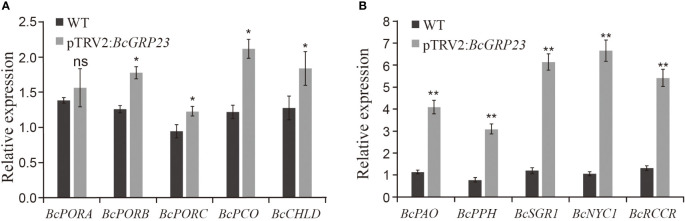
Silencing of BcGRP23 significantly induces the expression of chlorophyll degradation-related genes in flowering Chinese cabbage
To further explore the role of BcGRP23 in chlorophyll biosynthesis and degradation, the transcript levels of five chlorophyll biosynthesis-related genes (BcPORA, BcPORB, BcPORC, BcCAO, and BcCHLD) and four chlorophyll degradation genes (BcPAO, BcPPH, BcNYC1, BcRCCR, and BcSGR1) were measured in pTRV2:BcGRP23 and WT lines. Compared to the WT, the expression levels of the chlorophyll biosynthesis-related genes—BcCAO, BcPORC, BcCHLD, and BcPORB—increased by 1.73-, 1.29-, 1.44-, and 1.41-fold, respectively, in the petioles of the knockdown lines ( Figure 5A ). However, the expression level of BcPORA did not differ significantly.
Figure 5.
Expression of the genes related to chlorophyll biosynthesis (A) and degradation (B) in the petioles of the pTRV2:BcGRP23 knockdown lines. *Significant difference at the p < 0.05 level; **significant difference at the p < 0.01 level; ns, no significant difference (Student’s t-test).
The expression levels of the chlorophyll degradation genes were significantly increased in the pTRV2:BcGRP23 knockdown line compared with the WT. Furthermore, the magnitude of the upregulation of these genes was greater than that of the chlorophyll biosynthesis-related genes. Among them, the expression levels of BcNYC1, BcPAO, BcPPH, BcRCCR, and BcSGR1 were upregulated by 6.26-, 3.59-, 4-, 4-, and 5.13-fold, respectively, in the petioles ( Figure 5B ). Therefore, the silencing of BcGRP23 significantly upregulated the expression of the chlorophyll degradation-related genes in the petioles, accelerated chlorophyll degradation, and whitened the petioles of flowering Chinese cabbage.
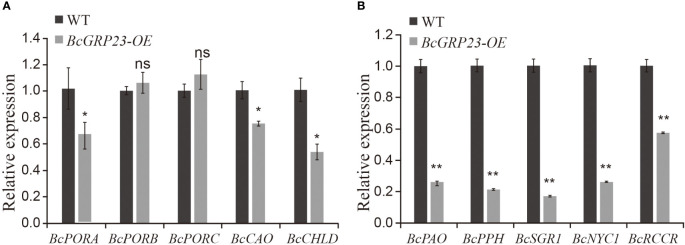
BcGRP23 overexpression might induce an increase in chlorophyll accumulation by downregulating chlorophyll degradation-related genes
To explore the role of BcGRP23 overexpression in flowering Chinese cabbage, the cotyledons of flowering Chinese cabbage plants were infected via Agrobacterium-mediated transformation. Four positive transgenic plants were obtained by screening seeds of the T0 generation. We determined the expression of BcGRP23 in the transgenic lines and found that the expression in L1 was significantly higher (i.e., 14.7-fold higher) than that in WT plants. The expression levels of BcPORB and BcPORC in the petioles were not significantly different between the overexpressing and WT plants. However, the levels of BcPORA, BcCAO, BcCHLD, BcPAO, BcPPH, BcNYC1, and BcRCCR showed a significant downward trend. Among them, the expression levels of BcPORA, BcCAO, and BcCHLD were found to decrease by 35%, 25%, and 46%, respectively, in the overexpression line compared with the WT ( Figure 6A ). In addition, the expression levels of the chlorophyll degradation genes (BcPAO, BcPPH, BcNYC1, BcRCCR, and BcSGR1) decreased by 73%, 78%, 82%, 74%, and 42%, respectively, in the overexpression line compared to the WT ( Figure 6B ). Altogether, these results suggest that BcGRP23 could inhibit the expression of the chlorophyll biosynthesis and degradation-related genes, with a more significant inhibitory effect on the degradation-related genes than the biosynthesis-related genes.
Figure 6.
Expression of the chlorophyll biosynthesis-related (A) and degradation-related (B) genes in the petioles of BcGRP23-overexpressing plants. *Significant difference at the p < 0.05 level; **significant difference at the p < 0.01 level; ns indicates that the difference between the two lines is not statistically significant (Student’s t-test).
Cloning and functional analysis of the BcGRP23 promoter
Analysis of the cis-acting elements contained in the promoter region of BcGRP23 revealed that the BcGRP23 promoter contains different hormone-responsive elements (ethylene-, ABA-, and brassinolide-responsive elements, GA-related elements, MeJA-responsive elements, cytokinin response elements, and salicylic acid-inducible response elements) and some adversity stress response elements ( Supplementary Table S1 ). Such results suggest that BcGRP23 may interact with these hormone signaling pathways when exposed to various hormones and abiotic stresses. The BcGRP23 promoter sequence also had a large number of light regulatory elements, some chlorophyll-related regulatory elements (CPBCSPOR and GATABOX), and different tissue-specific expression regulatory elements [guard cell-specific gene expression elements, mesophyll-specific regulatory elements, one each of multifunctional transcription factors related to flowering gene transcription, pollen late gene promoter, flower organ-specific element, DOF (DNA binding with One Finger) gene binding site, tissue-specific expression-related cis-elements, seed-specific expression elements, and other organ-specific regulatory elements] ( Supplementary Table S1 ). Therefore, the expression of BcGRP23 may be tissue-specific and related to the chlorophyll pathway in flowering Chinese cabbage plants.
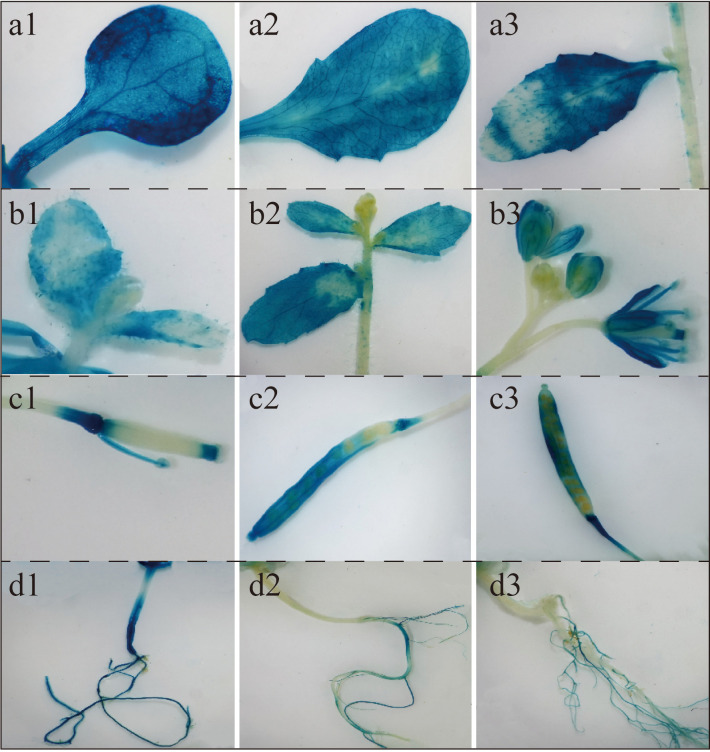
A chemical staining analysis was performed using proBcGRP23-GUS transformed Arabidopsis plants at various growth stages and organs. GUS staining was observed in the roots and leaves of the proBcGRP23::GUS transgenic lines throughout the growth period ( Figure 7, a1–a3, b1–b3, d1–d3 ), with the darkest staining observed in the cotyledons ( Figure 7, a1 ). During the bolting, bud, and flowering stages, a weak GUS stain was observed in the stems and growing points, whereas an intense GUS stain was observed in the petals and stamens of the 35S::GUS transgenic lines ( Figure 7, b1–b3 ). The GUS gene was mainly expressed at the top of the silique during the podding stage, in the direct junction between the stalk and silique in the early stage, and then mainly in the pod ( Figure 7, c1 ). The GUS stain was also observed in the stalk after the seeds were almost mature ( Figure 7, c2, c3 ). An intense GUS stain was observed in the hypocotyl; however, the intensity became weaker with time. After anthesis, the GUS stain was undetectable ( Figure 7, d1–d3 ).
Figure 7.
Localization of BcGRP23 expression at different stages using pBcGRP23::GUS for β-glucuronidase (GUS) staining. a1–a3, cotyledons, rosette leaves, and cauline leaves; b1–b3, flowers, leaves, and stems in the bolting, bud, and flowering stages; c1–c3, pods in the pod setting, seed expansion, and seed maturity stages; d1–d3, roots in the bolting, flowering, and fruiting stages.
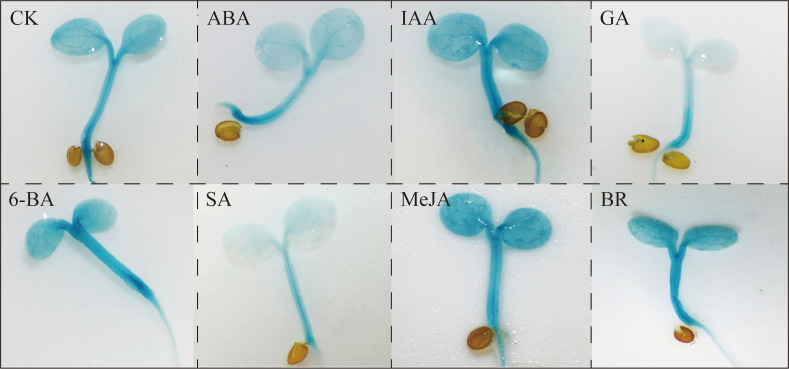
Effects of phytohormones on the expression of GUS driven by the BcGRP23 promoter in transgenic Arabidopsis
To determine whether exogenous phytohormones regulate the transcription level of BcGRP23, histochemical analysis of GUS was performed on ProBcGRP23::GUS transgenic Arabidopsis seedlings in the cotyledon stage treated with 50 μM phytohormones (ABA, IAA, GA, 6-BA, SA, MeJA, and BR). Compared to the control plants (treated with water), the GUS stain became stronger in plants treated with IAA, 6-BA, MeJA, and BR, but it became lighter in plants treated with ABA, GA, and SA ( Figure 8 ). There was no significant difference in the GUS staining intensity after 3 and 6 h of treatment. These results indicate that BcGRP23 is involved in the regulation of various phytohormones.
Figure 8.
Patterns of β-glucuronidase (GUS) staining in Arabidopsis carrying pBcProGRP23::GUS and reporter constructs and treated with phytohormones.
BES1 acts as a transcriptional activator of BcGRP23
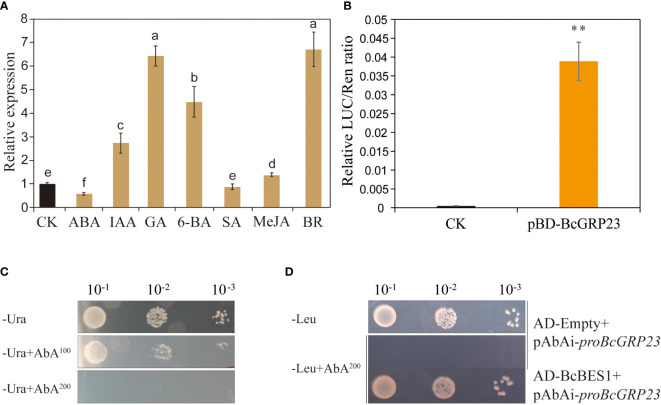
To determine the effect of hormones on the transcription level of BcGRP23 in flowering Chinese cabbage, the expression of BcGRP23 was examined in 3-week-old seedlings treated with exogenous hormones. Compared with the control (treated with water), the transcription level of BcGRP23 significantly increased after hormone (IAA, GA, 6-BA, MeJA, and BR) treatments in flowering Chinese cabbage, but significantly decreased after ABA treatment. Notably, the difference between the control and the SA groups was not significant, as shown in Figure 9A . The expression of BcGRP23 has been reported to be maximal under BR treatment. Thus, the transcription level of BcGRP23 is regulated by various hormones, particularly BR ( Figure 9A ). To assess the regulation of BcGRP23 by the transcription factor BES1, using the BcGRP23 promoter was verified in a yeast expression system. A 572-bp BcGRP23 promoter containing three BES1-binding elements (CANNTG) was selected for the Y1H assay. The results showed that BES1 can bind to the BcGRP23 promoter in a yeast expression system ( Figures 9C, D ). When the interaction of BcBES1 with the BcGRP23 promoter was analyzed, the expression of the positive control (62-BES1-SK+pGreenII 0800-proBcGRP23-LUC), BES1, was found to result in a higher LUC/REN ratio than that of the negative control ( Figure 9B ). These results indicate that BES1 is a transcriptional activator of BcGRP23 and enhances the activity of the BcGRP23 promoter.
Figure 9.
Analysis of the BES1 transcription factor that regulates BcGRP23 promoter activity. (A) Effects of different hormones on the expression of BcGRP23 in flowering Chinese cabbage. Different lowercase letters indicate significant differences at the p < 0.05 level. (B) Luciferase (LUC)/Renilla (REN) ratio indicating the relative luciferase activity in the dual-LUC assay. **Significant difference at the p < 0.01 level. (C, D) Yeast one-hybrid analysis. (C) Autoactivation of the BcGRP23 promoters was tested on synthetically defined (SD) medium without Ura in the presence of aureobasidin A (AbA). AbA100 , 100 ng/ml AbA; AbA200 , 200 ng/ml AbA. (D) Interactions were determined on SD medium without Leu in the presence of 200 ng/ml AbA (−Leu+AbA200). (a) Negative control: AD-Empty+BcGRP23 promoter.
Discussion
GRPs are part of a large and complex family sharing the common feature of a (Gly)nX repeat-rich glycine domain that may be involved in protein–protein interactions, RNA binding, and nuclear targeting (Giarola et al., 2016). GRPs are involved in multiple independent physiological processes, such as cell wall building, growth and development, and stress resistance in plants (Czolpinska and Rurek, 2018). In this study, we investigated the role of BcGRP23 in the flowering Chinese cabbage. Previously, transient expression in the onion epidermis verified the localization of BcGRP23 in the cell wall; it contained (GGX) n repeats that are specific to class I GRPs. These results are consistent with a previous study showing thDat AtGRP23 was located in the cell wall and contained more than 60% of the total amino acids with glycine (Park et al., 2008). BcGRP23 was preliminarily determined to be a class I GRP.
According to many studies, GRPs have tissue and spatiotemporal specificity in plants; however, their expression is strongly suppressed in matured tissues (Lei and Wu, 1991). OsGRP-2 was found to be barely expressed at the seedling stage in rice, but was significantly expressed after the seedling stage. Thereafter, its expression was found to gradually increase, reaching the highest level at the jointing stage and then gradually decreasing at the booting stage (Liu et al., 2003). In this study, the expression of BcGRP23 was found to significantly vary in the different stages and tissues of flowering Chinese cabbage, as shown in Figure 3 . The expression level of BcGRP23 was the highest in the leaves in the cotyledon stage, the flowers in the cotyledon stage, and the flowers and seeds in the flowering stage. Furthermore, its expression was slightly lower in the roots throughout the growth cycle and in the stems and leaves in the flowering stage ( Figure 3 ). These findings are consistent with the predicted results of the BcGRP23 promoter sequence. Numerous tissue-specific cis-elements, such as leaves, flowers, and seeds, were found in the BcGRP23 promoter ( Supplementary Table S1 ). Moreover, BcGRP23 was recognized to have spatiotemporal specificity in the aboveground parts of flowering Chinese cabbage.
The whitening phenomenon in plant tissues is mainly caused by a significant reduction in chlorophyll content. However, this phenomenon may have occurred due to inhibition of the biosynthesis of chlorophyll, or acceleration of metabolism, and the abnormal development of the tissue structure of chloroplast (Thomas et al., 2001). Based on previous studies, SGR1 is a positive regulator of chlorophyll degradation during Arabidopsis senescence (Gao et al., 2016). Under natural senescence and dark-induced conditions, the sgrl-1 mutant Arabidopsis displayed a stay-green phenotype, whereas SGR1-overexpressing Arabidopsis leaves exhibited an early yellowing trait (Sakuraba et al., 2013; Li et al., 2017). In the present study, BcGRP23-silenced plants displayed whitening of the stems and petioles as the levels of Chl a and Chl b decreased ( Figure 4 ). These findings are consistent with those of a previous study that revealed that SGRl controls the leaf color in Arabidopsis. Accordingly, we speculate that BcGRP23 is a previously unknown regulator of chlorophyll degradation and biosynthesis (Li et al., 2017).
PAO is involved in chlorosis and can catalyze the opening of the chlorophyll porphyrin ring to form a quaternary linear pyrrole derivative; the absence of PAO leads to the stay-green trait in plants (Hörtensteiner, 2009). During ripening, PAO interacts with the chlorotic gene SGR, and its activity on demagnesium chelatase gradually increases, with the highest level achieved at the ripening stage, causing rapid degradation of chlorophyll and chlorosis in plants (Costa et al., 2002; Hörtensteiner, 2013). The proteins translated by NYCI and NOL colocalize on the thylakoid membrane of the chloroplast to form a Chl b reductase complex, which catalyzes the degradation of Chl b during the mature stage, resulting in rapid chlorophyll degradation and chlorosis in plants (Sato et al., 2009).
The expression levels of the genes related to the chlorophyll biosynthesis and degradation pathways in BcGRP23-silenced plants were significantly different from those in WT plants. In fact, the expression levels of five chlorophyll degradation genes, namely, BcPAO, BcPPH, BcNYC1, BcRCCR, and BcSGR1, were significantly increased. The expression levels of five chlorophyll biosynthesis-related genes—BcPORA, BcPORB, BcPORC, BcCAO, and BcCHLD—were also upregulated, but to a lesser extent than those of the chlorophyll degradation genes ( Figure 5 ). Notably, changes in these genes led to a reduction in chlorophyll content in the petioles of flowering Chinese cabbage, which may be responsible for the whitening of the petioles and stems. The expression levels of the chlorophyll degradation genes were significantly reduced in plants overexpressing BcGRP23; however, the expression levels of some of the chlorophyll biosynthesis-related genes were not found to differ significantly ( Figure 6 ). Overall, the increase in BcGRP23 had a greater effect on the chlorophyll biosynthesis genes than on the chlorophyll degradation genes, which might be the reason for the increased chlorophyll content in overexpressed plants.
A total of 15 core cis-elements (TATABOX) responsible for initiating transcription were identified, including 13 elements (CAATBOX) that regulate the transcription initiation frequency and two initiation factors (INRNTPSADB) in the BcGRP23 promoter region. These elements play a role in the promotion of transcription, which suggests that the BcGRP23 promoter has strong activity (Butler and Kadonaga, 2001). Notably, the BcGRP23 promoter sequence also contained hormone response elements (ethylene, cytokinin, abscisic acid, brassinolide, GA, and MeJA), indicating that BcGRP23 is associated with the hormone signaling pathways and might interact with these hormones during plant growth and development.
In Arabidopsis, drought and salt stress were found to upregulate the transcription level of AtGRP1, but to downregulate the transcription levels of AtGRP4 and AtGR7. AtGRP7 was also found to be inhibited by endogenous ABA and osmotic stress (Cao et al., 2006). The expression of NtGRP1 and NtGRP3 in tobacco can be disrupted by high salinity, low temperature, drought, high temperature, and mechanical damage; however, their expression levels are low (Lee et al., 2009; Xuan et al., 2010). HvGRP2 and HvGRP3 in barley are induced by fungal pathogens and are regulated by MeJA and ethylene (Molina et al., 1997). Here, several ionic stressors (copper, phosphorus, and sulfur), drought, salicylic acid, injury-specific stress, salt stress, low-temperature stress, and other stress-related regulatory elements were found, indicating that BcGRP23 may be involved in the regulation of various abiotic stresses. A total of 64 light regulatory elements, including I-BOX, REALPHALGLHCB21, GT1 motif, -10PEHVPSBD, and chlorophyll gene-related regulatory elements (3 CPBCSPOR and 26 GATABOX), were also found in the BcGRP23 promoter region, which suggests that the expression of BcGRP23 was affected by light and is related to the chlorophyll cycle ( Supplementary Figure S1 ). The BcGRP23 promoter was transformed into Arabidop1sis, and GUS histochemical staining was carried out using transgenic Arabidopsis at different time points in different plant parts. GUS staining was observed in the roots and leaves throughout the growth period. Notably, only a small amount of GUS stain was observed at the growth points and stems in the bolting, bud, and flowering stages ( Figure 7 ). The GUS activity of BcGRP23 was inconsistent between the tissue expression in flowering Chinese cabbage and the heterologous expression of the BcGRP23 promoter in Arabidopsis; species differences may have influenced the gene expression.
Plant hormones play an important role as signaling molecules in plant development and in response to stressful environments (Ton et al., 2009). During response to biotic stress, the SA and JA hormone signaling pathways play a vital role in regulating the expression of disease resistance genes (He et al., 2016). IAA and ABA are vital hormones involved in plant growth and development (Emenecker and Strader, 2020), while BR and ABA act as stress hormones and play key roles in plant stress resistance (Bari and Jones, 2009). In the present study, the GUS stain became more intense after treatments with IAA, 6-BA, MeJA, and BR, but became lighter after treatments with ABA, GA, and SA. Analogously, normal-growing flowering Chinese cabbage was treated with the hormones IAA, GA, 6-BA, MeJA, and BR. Compared to the control, the transcription level of BcGRP23 increased significantly after 6 h, with the most significant increase observed with BR treatment, and decreased following treatment with ABA. In conclusion, BcGRP23 is regulated by various hormones, especially BR ( Figure 8 ). Thus, BcGRP23 is strongly correlated with plant hormones, resulting in a significant increase in its transcript levels ( Figure 9A ).
It is known that the application of BR regulates plant growth, the net photosynthetic rate, and the antioxidant system capacity. Epi-brassinolide (EBL) treatment significantly increased the Chl a and Chl b contents, photosynthetic rate (P n), and water use efficiency in tomato under Cd stress, thereby alleviating the inhibitory effects of Cd (Guo et al., 2018). BR-insensitive mutants of Arabidopsis and Brassica exhibited dwarfing, dark green leaves, late flowering, and male sterility (Guo et al., 2018). Brassinazole-resistant 1 (BZR1) and BES1 are considered important transcription factors in the BR signal transduction pathway that can bind to BR-responsive genes and play a regulatory role (Lee et al., 2015). The BR-responsive transcriptome data revealed that the promoter regions of the BR-inducible genes are rich in E-box (CANNTG) binding sites, while those of the BR-repressor genes are rich in BRRE cis-acting elements (Guo et al., 2018). In this study, eight E-boxes were found in the BcGRP23 promoter. In addition, the BR transcription factor, BES1, could bind to the BcGRP23 promoter in the yeast expression system ( Figures 9C, D ). The dual-luciferase assay revealed positive regulation of the BcGRP23 promoter by the BR transcription factor, BES1 ( Figure 9B ). Overall, our findings indicate that BR may respond to various stresses by regulating the transcription of BcGRP23 and activating different physiological and molecular mechanisms to promote plant growth and development and improve plant stress tolerance. The response of GRPs to environmental stress in plants has been widely explored over the past few decades. The findings of this study provide new insights into the involvement of GRPs in the response to environmental stress, growth, and the developmental mechanisms of flowering Chinese cabbage.
Conclusions
In this study, we preliminarily determined the biological functions and potential regulatory pathways of BcGRP23 during development, interaction with hormones, and adverse stress in flowering Chinese cabbage and Arabidopsis mutants. BcGRP23 localized on the cell wall was found to regulate the expression of chlorophyll degradation-related genes, thus affecting chlorophyll content. BcGRP23 was highly expressed in the leaves, flowers, and seeds at the cotyledon stage; however, its expression levels gradually decreased in the leaves and stems with the growth of flowering Chinese cabbage. BcGRP23 exhibited a significant response to the exogenous hormones IAA, GA, 6-BA, MeJA, and BR and showed enhanced transcript levels. The Y1H assay and relative luciferase activity revealed a potential interaction between BcGRP23 and BES1. Collectively, our findings provide new insights into the role of GRP in the hormonal regulatory network and stress response in flowering Chinese cabbage. Such findings provide a fundamental basis for further studies on the GRP gene family and will be helpful for crop genetic modification.
Data availability statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding authors.
Author contributions
SZ, KC, and WS conceived and designed the experiments. SZ, KC, and SY conducted the experiments and analyzed the data. SZ and KC prepared the manuscript. AA and YW participated in the experiments and revised the manuscript. SS, WS, and RC offered experimental guidance. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (32072656); the Key-Area Research and Development Program of Guangdong Province, China (2020B0202010006); the Guangdong Provincial Special Fund for Modern Agriculture Industry Technology Innovation Teams (2022KJ131); and the China Agriculture Research System of MOF and MARA.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1010470/full#supplementary-material
References
- Bari R., Jones J. D. (2009). Role of plant hormones in plant defence responses. Plant Mol. Biol. 69 (4), 473–488. doi: 10.1007/s11103-008-9435-0 [DOI] [PubMed] [Google Scholar]
- Beale S. I. (2005). Green genes gleaned. Trends Plant Sci. 10 (7), 309–312. doi: 10.1016/j.tplants.2005.05.005 [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V., Page T., Harrison E., Breeze E., Lim P. O., Nam H. G., et al. (2005). Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in arabidopsis. Plant J. 42 (4), 567–585. doi: 10.1111/j.1365-313X.2005.02399.x [DOI] [PubMed] [Google Scholar]
- Butler J. E., Kadonaga J. T. (2001). Enhancer–promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 15 (19), 2515–2519. doi: 10.1101/gad.924301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S., Jiang L., Song S., Jing R., Xu G. (2006). AtGRP7 is involved in the regulation of abscisic acid and stress responses in arabidopsis. Cell. Mol. Biol. Lett. 11 (4), 526–535. doi: 10.2478/s11658-006-0042-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charoenchongsuk N., Ikeda K., Itai A., Oikawa A., Murayama H. (2015). Comparison of the expression of chlorophyll-degradation-related genes during ripening between stay-green and yellow-pear cultivars. Sci. Hortic. 181, 89–94. doi: 10.1016/j.scienta.2014.10.005 [DOI] [Google Scholar]
- Chen C., Chen G., Hao X., Cao B., Chen Q., Liu S., et al. (2011). CaMF2, an anther-specific lipid transfer protein (LTP) gene, affects pollen development in capsicum annuum l. Plant Sci. (Amsterdam Neth) 181 (4), 439–448. doi: 10.1016/j.plantsci.2011.07.003 [DOI] [PubMed] [Google Scholar]
- Cheng Y., Guan J. (2014). Involvement of pheophytinase in ethylene-mediated chlorophyll degradation in the peel of harvested ‘Yali’pear. J. Plant Growth Regul. 33 (2), 364–372. doi: 10.1007/s00344-013-9383-z [DOI] [Google Scholar]
- Condit C. M., Meagher R. B. (1986). A gene encoding a novel glycine-rich structural protein of petunia. Nature 323 (6084), 178–181. doi: 10.1038/323178a0 [DOI] [Google Scholar]
- Cornels H., Ichinose Y., Barz W. (2000). Characterization of cDNAs encoding two glycine-rich proteins in chickpea (Cicer arietinum l.): accumulation in response to fungal infection and other stress factors. Plant Sci. (Amsterdam Neth) 154 (1), 83–88. doi: 10.1016/S0168-9452(00)00193-X [DOI] [PubMed] [Google Scholar]
- Costa M. L., Civello P. M., Chaves A. R., Martínez G. A. (2002). Characterization of mg-dechelatase activity obtained from fragaria× ananassa fruit. Plant Physiol. Biochem. 40 (2), 111–118. doi: 10.1016/S0981-9428(01)01358-4 [DOI] [Google Scholar]
- Czolpinska M., Rurek M. (2018). Plant glycine-rich proteins in stress response: An emerging, still prospective story. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhaubhadel S., Krishna P. (2008). Identification of differentially expressed genes in brassinosteroid-treated brassica napus seedlings. J. Plant Growth Regul. 27 (3), 297–308. doi: 10.1007/s00344-008-9056-5 [DOI] [Google Scholar]
- Ding X., Jiang Y., He L., Zhou Q., Yu J., Hui D., et al. (2016). Exogenous glutathione improves high root-zone temperature tolerance by modulating photosynthesis, antioxidant and osmolytes systems in cucumber seedlings. Sci. Rep. 6 (1), 1–12. doi: 10.1038/srep35424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emenecker R. J., Strader L. C. (2020). Auxin-abscisic acid interactions in plant growth and development. Biomolecules 10 (2), 281. doi: 10.3390/biom10020281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusaro A. F., Bocca S. N., Ramos R. L. B., Barrôco R. M., Magioli C., Jorge V. C., et al. (2007). AtGRP2, a cold-induced nucleo-cytoplasmic RNA-binding protein, has a role in flower and seed development. Planta 225 (6), 1339–1351. doi: 10.1007/s00425-006-0444-4 [DOI] [PubMed] [Google Scholar]
- Gao S., Gao J., Zhu X., Song Y., Li Z., Ren G., et al. (2016). ABF2, ABF3, and ABF4 promote ABA-mediated chlorophyll degradation and leaf senescence by transcriptional activation of chlorophyll catabolic genes and senescence-associated genes in arabidopsis. Mol. Plant 9 (9), 1272–1285. doi: 10.1016/j.molp.2016.06.006 [DOI] [PubMed] [Google Scholar]
- Giarola V., Krey S., von den Driesch B., Bartels D. (2016). The craterostigma plantagineum glycine-rich protein CpGRP1 interacts with a cell wall-associated protein kinase 1 (CpWAK1) and accumulates in leaf cell walls during dehydration. New Phytol. 210 (2), 535–550. doi: 10.1111/nph.13766 [DOI] [PubMed] [Google Scholar]
- Guo J., Zhou R., Ren X., Jia H., Hua L., Xu H., et al. (2018). Effects of salicylic acid, epi-brassinolide and calcium on stress alleviation and cd accumulation in tomato plants. Ecotoxicol. Environ. Saf. 157, 491–496. doi: 10.1016/j.ecoenv.2018.04.010 [DOI] [PubMed] [Google Scholar]
- He Y., Fukushige H., Hildebrand D. F., Gan S. (2002). Evidence supporting a role of jasmonic acid in arabidopsis leaf senescence. Plant Physiol. 128 (3), 876–884. doi: 10.1104/pp.010843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Zhu L., Xu L., Guo W., Zhang X. (2016). GhATAF1, a NAC transcription factor, confers abiotic and biotic stress responses by regulating phytohormonal signaling networks. Plant Cell Rep. 35 (10), 2167–2179. doi: 10.1007/s00299-016-2027-6 [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S. (2009). Stay-green regulates chlorophyll and chlorophyll-binding protein degradation during senescence. Trends Plant Sci. 14 (3), 155–162. doi: 10.1016/j.tplants.2009.01.002 [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S. (2013). Update on the biochemistry of chlorophyll breakdown. Plant Mol. Biol. 82 (6), 505–517. doi: 10.1007/s11103-012-9940-z [DOI] [PubMed] [Google Scholar]
- Kang H., Park S. J., Kwak K. J. (2013). Plant RNA chaperones in stress response. Trends Plant Sci. 18 (2), 100–106. doi: 10.1016/j.tplants.2012.08.004 [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 (7), 1870–1874. doi: 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.-O., Kim K. P., Kim B.-G., Hahn J.-S., Hong C. B. (2009). Flooding stress-induced glycine-rich RNA-binding protein from nicotiana tabacum. Mol. Cells 27 (1), 47–54. doi: 10.1007/s10059-009-0004-4 [DOI] [PubMed] [Google Scholar]
- Lee H.-S., Kim Y., Pham G., Kim J. W., Song J.-H., Lee Y., et al. (2015). Brassinazole resistant 1 (BZR1)-dependent brassinosteroid signalling pathway leads to ectopic activation of quiescent cell division and suppresses columella stem cell differentiation. J. Exp. Bot. 66 (15), 4835–4849. doi: 10.1093/jxb/erv316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M., Wu R. (1991). A novel glycine-rich cell wall protein gene in rice. Plant Mol. Biol. 16 (2), 187–198. doi: 10.1007/BF00020551 [DOI] [PubMed] [Google Scholar]
- Liu Y., Wang X., Lv H., Cao M., Li Y., Yuan X., et al. (2022). Anabolism and signaling pathways of phytomelatonin. J. Exp. Bot. 73 (17), 5801–5817. doi: 10.1093/jxb/erac158 [DOI] [PubMed] [Google Scholar]
- Liu Z., Wang J., Wang Q., Huang X., Xu W., Zhu L., et al. (2003). Structure, expression pattern and chromosomal localization of the rice Osgrp-2 gene. Sci. China Ser. C.: Life Sci. 46 (6), 584–594. doi: 10.1360/02yc0172 [DOI] [PubMed] [Google Scholar]
- Li Z., Wu S., Chen J., Wang X., Gao J., Ren G., et al. (2017). NYEs/SGRs-mediated chlorophyll degradation is critical for detoxification during seed maturation in Arabidopsis . Plant J. 92 (4), 650–661. doi: 10.1111/tpj.13710 [DOI] [PubMed] [Google Scholar]
- Lu X., Cheng Y., Gao M., Li M., Xu X. (2020). Molecular characterization, expression pattern and function analysis of glycine-rich protein genes under stresses in Chinese cabbage (Brassica rapa l. ssp. pekinensis). Front. Genet. 11. doi: 10.3389/fgene.2020.00774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Cheng K., Li J., Deng Z., Zhang C., Zhu H. (2021). Roles of plant glycine-rich rna-binding proteins in development and stress responses. Int. J. Mol. Sci. 22 (11), 5849. doi: 10.3390/ijms22115849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangeon A., Magioli C., Menezes-Salgueiro A. D., Cardeal V., de Oliveira C., Galvão V. C., et al. (2009). AtGRP5, a vacuole-located glycine-rich protein involved in cell elongation. Planta 230 (2), 253–265. doi: 10.1007/s00425-009-0940-4 [DOI] [PubMed] [Google Scholar]
- Mangeon A., Magioli C., Tarré É., Cardeal V., Araujo C., Falkenbach E., et al. (2010). The tissue expression pattern of the AtGRP5 regulatory region is controlled by a combination of positive and negative elements. Plant Cell Rep. 29 (5), 461–471. doi: 10.1007/s00299-010-0835-7 [DOI] [PubMed] [Google Scholar]
- Mangeon A., Pardal R., Menezes-Salgueiro A. D., Duarte G. L., de Seixas R., Cruz F. P., et al. (2016). AtGRP3 is implicated in root size and aluminum response pathways in arabidopsis. PloS One 11 (3), e0150583. doi: 10.1371/journal.pone.0150583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina A., Mena M., Carbonero P. (1997). Differential expression of pathogen-responsive genes encoding two types of glycine-rich proteins in barley. Plant Mol. Biol. 33 (5), 803–810. doi: 10.1023/A:1005712803130 [DOI] [PubMed] [Google Scholar]
- Mou W., Li D., Luo Z., Mao L., Ying T. (2015). Transcriptomic analysis reveals possible influences of ABA on secondary metabolism of pigments, flavonoids and antioxidants in tomato fruit during ripening. PloS One 10 (6), e0129598. doi: 10.1371/journal.pone.0129598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. H., Suh M. C., Kim T. H., Kim M. C., Cho S. H. (2008). Expression of glycine-rich protein genes, AtGRP5 and AtGRP23, induced by the cutin monomer 16-hydroxypalmitic acid in arabidopsis thaliana. Plant Physiol. Biochem. 46 (11), 1015–1018. doi: 10.1016/j.plaphy.2008.06.008 [DOI] [PubMed] [Google Scholar]
- Sachetto-Martins G., Franco L. O., de Oliveira D. E. (2000). Plant glycine-rich proteins: a family or just proteins with a common motif? Biochim. Biophys. Acta (BBA)-Gene Structure Expression 1492 (1), 1–14. doi: 10.1016/S0167-4781(00)00064-6 [DOI] [PubMed] [Google Scholar]
- Sakuraba Y., Rahman M. L., Cho S. H., Kim Y. S., Koh H. J., Yoo S. C., et al. (2013). The rice faded green leaf locus encodes protochlorophyllide oxidoreductase b and is essential for chlorophyll synthesis under high light conditions. Plant J. 74 (1), 122–133. doi: 10.1111/tpj.12110 [DOI] [PubMed] [Google Scholar]
- Sato Y., Morita R., Katsuma S., Nishimura M., Tanaka A., Kusaba M. (2009). Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. Plant J. 57 (1), 120–131. doi: 10.1111/j.1365-313X.2008.03670.x [DOI] [PubMed] [Google Scholar]
- Suhel M., Husain T., Prasad S. M., Singh V. P. (2022). GABA requires nitric oxide for alleviating arsenate stress in tomato and brinjal seedlings. J. Plant Growth Regul. 1-14. doi: 10.1007/s00344-022-10576-7 [DOI] [Google Scholar]
- Teng K., Tan P. H., Xiao G. Z., Han L. B., Chang Z. H., Chao Y. H. (2017). Heterologous expression of a novel zoysia japonica salt-induced glycine-rich RNA-binding protein gene, ZjGRP, caused salt sensitivity in arabidopsis. Plant Cell Rep. 36 (1), 179–191. doi: 10.1007/s00299-016-2068-x [DOI] [PubMed] [Google Scholar]
- Thomas H., Ougham H., Hörtensteiner S. (2001). Recent advances in the cell biology of chlorophyll catabolism. Adv. Bot. Res. 35, 1–52. doi: 10.1016/S0065-2296(01)35003-6 [DOI] [Google Scholar]
- Ton J., Flors V., Mauch-Mani B. (2009). The multifaceted role of ABA in disease resistance. Trends Plant Sci. 14 (6), 310–317. doi: 10.1016/j.tplants.2009.03.006 [DOI] [PubMed] [Google Scholar]
- Wang Y. D., Huang X., Huang X. M., Su W., Hao Y. W., Liu H. C., et al. (2022). BcSOC1 promotes bolting and stem elongation in flowering Chinese cabbage. Int. J. Mol. Sci. 23 (7), 3459. doi: 10.3390/ijms23073459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Yang Y., Su D., Yu C., Xian Z., Pan Z., et al. (2022). The SlHB8 acts as a negative regulator in tapetum development and pollen wall formation. Horticult. Res. doi: 10.1093/hr/uhac185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang L., Hu L., Xu W., Zhen A., Zhang L., Hu X. (2016). Exogenous γ-aminobutyric acid improves the structure and function of photosystem II in muskmelon seedlings exposed to salinity-alkalinity stress. PloS One 11 (10), e0164847. doi: 10.1371/journal.pone.0164847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J.-L., Wang H.-C., Tan X.-Y., Zhang C.-L., Naeem M. S. (2018). 5-aminolevulinic acid improves salt tolerance mediated by regulation of tetrapyrrole and proline metabolism in brassica napus l. seedlings under NaCl stress. Plant Physiol. Biochem. 124, 88–99. doi: 10.1016/j.plaphy.2018.01.001 [DOI] [PubMed] [Google Scholar]
- Xuan C., Zeng Q., Lu X.-P., Yu D.-Q., Li W.-Z. (2010). Characterization and expression analysis of four glycine-rich RNA-binding proteins involved in osmotic response in tobacco (Nicotiana tabacum cv. xanthi). Agr. Sci. China 9 (11), 1577–1587. doi: 10.1016/S1671-2927(09)60254-6 [DOI] [Google Scholar]
- Yuan L., Shu S., Sun J., Guo S., Tezuka T. (2012). Effects of 24-epibrassinolide on the photosynthetic characteristics, antioxidant system, and chloroplast ultrastructure in cucumis sativus l. under Ca (NO3) 2 stress. Photosynth. Res. 112 (3), 205–214. doi: 10.1007/s11120-012-9774-1 [DOI] [PubMed] [Google Scholar]
- Zhang S., Zhang Y., Wang Y., Hao Y., Su W., Sun G., et al. (2022). Nitrogen absorption pattern detection and expression analysis of nitrate transporters in flowering Chinese cabbage. Horticulturae 8 (3), 188. doi: 10.3390/horticulturae8030188 [DOI] [Google Scholar]
- Zhang J., Zhao Y., Xiao H., Zheng Y., Yue B. (2014). Genome-wide identification, evolution, and expression analysis of RNA-binding glycine-rich protein family in maize. J. Integr. Plant Biol. 56 (10), 1020–1031. doi: 10.1111/jipb.12210 [DOI] [PubMed] [Google Scholar]
- Zhu X., Chen J., Xie Z., Gao J., Ren G., Gao S., et al. (2015). Jasmonic acid promotes degreening via MYC 2/3/4-and ANAC 019/055/072-mediated regulation of major chlorophyll catabolic genes. Plant J. 84 (3), 597–610. doi: 10.1111/tpj.13030 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding authors.