Abstract
Wastewater surveillance of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) may be useful for monitoring population-wide coronavirus disease 2019 (COVID-19) infections, especially given asymptomatic infections and limitations in diagnostic testing. We aimed to detect SARS-CoV-2 RNA in wastewater and compare viral concentrations to COVID-19 case numbers in the respective counties and sewersheds. Influent 24-hour composite wastewater samples were collected from July to December 2020 from two municipal wastewater treatment plants serving different population sizes in Orange and Chatham Counties in North Carolina. After a concentration step via HA filtration, SARS-CoV-2 RNA was detected and quantified by reverse transcription droplet digital polymerase chain reaction (RT-ddPCR) and quantitative PCR (RT-qPCR), targeting the N1 and N2 nucleocapsid genes. SARS-CoV-2 RNA was detected by RT-ddPCR in 100 % (24/24) and 79 % (19/24) of influent wastewater samples from the larger and smaller plants, respectively. In comparison, viral RNA was detected by RT-qPCR in 41.7 % (10/24) and 8.3 % (2/24) of samples from the larger and smaller plants, respectively. Positivity rates and method agreement further increased for the RT-qPCR assay when samples with positive signals below the limit of detection were counted as positive. The wastewater data from the larger plant generally correlated (⍴ ~0.5, p < 0.05) with, and even anticipated, the trends in reported COVID-19 cases, with a notable spike in measured viral RNA preceding a spike in cases when students returned to a college campus in the Orange County sewershed. Correlations were generally higher when using estimates of sewershed-level case data rather than county-level data. This work supports use of wastewater surveillance for tracking COVID-19 disease trends, especially in identifying spikes in cases. Wastewater-based epidemiology can be a valuable resource for tracking disease trends, allocating resources, and evaluating policy in the fight against current and future pandemics.
Keywords: Wastewater-based epidemiology, Sewage surveillance, Coronavirus, COVID, ddPCR
Graphical abstract
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of coronavirus disease 2019 (COVID-19) and is responsible for a global pandemic exceeding 430 million cases and almost 6 million deaths worldwide (JHU COVID-19 Dashboard, 2/27/22). The United States leads the world in both COVID-19 cases and deaths, with 79 million cases and 950,000 deaths (JHU COVID-19 Dashboard, 2/27/22). While SARS-CoV-2 is primarily transmitted through large respiratory droplets and aerosols, it is also shed in the feces of both symptomatic and asymptomatic infected individuals (Y. Chen et al., 2020b; Gao et al., 2020; Holshue et al., 2020; Tang et al., 2020). This provides an opportunity for surveilling populations for COVID-19 infection by monitoring municipal wastewaters - a form of environmental surveillance known as wastewater-based epidemiology (WBE; Bivins et al., 2020; Xagoraraki and O’Brien, 2020). WBE has been proposed as an efficient method for monitoring population-wide infections given limitations in diagnostic testing and identification of asymptomatic infections (Bivins et al., 2020).
There is precedence for using WBE for population-level surveillance of viruses. For example, WBE has become instrumental in the endgame strategy for eradication of poliomyelitis (Asghar et al., 2014; Hovi et al., 2012; WHO, 2019). Similar to SARS-CoV-2, many poliovirus infections are subclinical, and viral shedding into feces may still occur in these cases (Long et al., 2020; Martinez-Bakker et al., 2015; Nathanson and Kew, 2010). Detection of viral RNA in sewage has helped to identify outbreaks and tailor strategies for polio eradication, including targeted mobilization of vaccine campaigns (Brouwer et al., 2018; P. Chen et al., 2020a; Ivanova et al., 2019).
Like poliovirus, SARS-CoV-2 is a +ssRNA virus which can be shed by infected individuals prior to onset of symptoms or while remaining asymptomatic. Some individuals continue to shed virus in feces up to 26 days post-infection, even while remaining negative for SARS-CoV-2 in respiratory swabs (Falman et al., 2019; O’Reilly et al., 2020; Zhang et al., 2020). Estimates of the total asymptomatic proportion of COVID-19 infections prior to vaccination were 16–18 % (He et al., 2020; Mizumoto et al., 2020). Clinical surveillance may fail to capture these infections as asymptomatic individuals or those with mild cases may not seek diagnostic testing. Even among symptomatic individuals, the median incubation period of approximately five days among positive individuals implies a delay in testing and diagnosis (Lauer et al., 2020). More reliable methods are needed for real-time viral infection estimation and tracking of population-level COVID-19 disease burden (Peccia et al., 2020).
Early COVID-19 WBE work from around the world proved the potential of RT-qPCR for detecting SARS-CoV-2 in wastewater, albeit using small numbers of samples over short time periods (Ahmed et al., 2020a; Medema et al., 2020; Wu et al., 2020b). More robust studies followed, with Gonzalez et al. sampling through the first 5 months of lockdown in the U.S. and capturing the beginning of the surge in cases in Summer 2020 (Gonzalez et al., 2020). Though these studies faced issues with method sensitivity and data scatter, they often found concentrations of SARS-CoV-2 in wastewater that suggested a higher incidence of COVID-19 infections than were reported by diagnosed cases, highlighting gaps in clinical monitoring. As research in this field progressed, scientists employed these methods to detect SARS-CoV-2 in cruise ship and passenger aircraft wastewater and to contain outbreaks in college dormitories (Ahmed et al., 2020b; Betancourt et al., 2021; Gibas et al., 2021). In all studies, the authors have acknowledged a need to refine the approach for wastewater surveillance of COVID-19, particularly for improving assay sensitivity, optimizing controls, and relating wastewater measurements to case numbers (Ahmed et al., 2022).
In this study, detection of SARS-CoV-2 by reverse transcription polymerase chain reaction (RT-PCR) was performed for 24-hour composite waste samples from two publicly owned wastewater treatment plants (WWTPs) in Orange and Chatham Counties, North Carolina, USA. These two testing sites allow for the comparison of SARS-CoV-2 detection in two different sewersheds. The served towns are only 16 miles apart but differ in population size, treatment plant size, and unique inputs to the sewershed such as the proximity of University of North Carolina (UNC) Hospitals and the return of students to the UNC campus in the case of the Orange County treatment plant. In addition to comparing results between these two plants, the paper includes a comparison of PCR methods and a comparison of gene targets for detecting SARS-CoV-2 RNA in wastewater. The paper also includes analysis of data across geographic and temporal scales, and it introduces an approach for estimating sewershed-level case counts useful in wastewater-based epidemiology. Together, this work helps to advance methods for wastewater surveillance of COVID-19 with recommendations that can be useful as wastewater surveillance systems are moved to practice across the globe.
2. Materials and methods
2.1. Description of municipal wastewater treatment plants
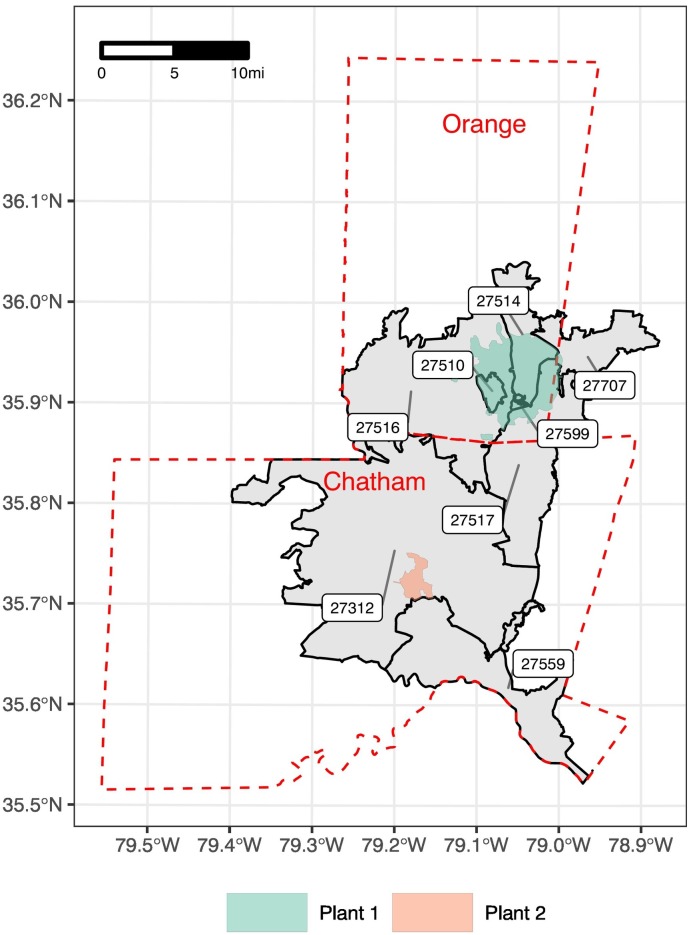
Two wastewater treatment plants of differing profiles were sampled (Fig. 1 ). Plant 1 is an urban treatment plant located in Orange County, NC adjacent to the University of North Carolina-Chapel Hill (UNC-CH) serving the towns of Chapel Hill, Carrboro, and the University, an area of 76.2 km2 (29.4 mi2) with a population of ~83,000 (~55 % of the Orange County population). Plant 2 is a rural treatment plant in neighboring Chatham County, NC serving ~4000 people (~5 % of the Chatham County population) in an area of 10.3 km2 (3.98 mi2). While Plant 2 receives primarily residential and retail inputs to the sewershed, Plant 1 receives residential, industrial, hospital, and retail inputs.
Fig. 1.
Map of the study areas. Geographic areas are shown for the sewersheds of Plants 1 (green polygon) and 2 (orange polygon) and their respective counties (red dotted lines) and ZIP codes (black solid lines) to illustrate the difference between populations served by the plants and populations for which epidemiological data is publicly available.
2.2. Sample collection and storage
With the assistance of staff at Plant 1 and Plant 2, ≥250 mL samples of 24-h composite wastewater influent and 30 mL grab samples of primary solids were collected weekly from 7/10/20 to 12/15/20, corresponding to 24 sampling dates. Operators from each treatment plant collected wastewater samples the morning of each sampling day. Samples were stored and transported on ice for processing within 4 h.
2.3. Sample processing
Composite influent samples were heat pasteurized at 75 °C for 30 min followed by addition of a Bovine Coronavirus (BCoV) modified live vaccine (PBS Animal Health 16445, Massillon, OH) as a processing control at a ratio of 1 μL:1 mL sample for 17 sampling dates (07/10/20 through 10/27/20) or 1 μL:10 mL for 7 sampling dates (11/03/20 through 12/15/20). The copy number in the BCoV stock was estimated via reverse transcription droplet digital PCR (RT-ddPCR) to be 29.5 copies/μL (see Supplementary Materials). The samples were then processed via filtration to adsorb virus onto HA filters. Briefly, 200 mL of heat pasteurized influent was acidified to pH 3.5 using 10 M HCl and amended with 2 mL 2.5 M MgCl2 (final concentration 25 mM). Ten replicate samples were then vacuum filtered in 20 mL volumes through mixed cellulose ester 0.45 μm membrane filters fitted on single-use funnels (MilliporeSigma EFHAW100I, Burlington, MA), using a new funnel and filter for each replicate. Filter funnels were rinsed with phosphate buffered saline (PBS) and filters were aseptically transferred using forceps to cryovials containing 600 μL lysis buffer (n = 8) or no buffer (n = 2). Based on availability, NucliSENS lysis buffer (bioMérieux 280,134, Marcy-l'Étoile, France) was used for 5 sampling dates (07/10/20 through 08/06/20), while QIAGEN (Hilden, Germany) RLT Plus lysis buffer was used for the remaining 19 sampling dates (08/11/20 through 12/15/20). At the conclusion of filtering, 50 mL of sterile PBS acidified to pH 3.5 and amended with 500 μL 2.5 M MgCl2 (final concentration 25 mM) and BCoV (1 μL:1 mL or 1 μL:10 mL) was filtered as above and stored in 600 μL lysis buffer to serve as a method control. Primary solids samples were not concentrated and were simply aliquoted into 1:1 by volume mixtures with lysis buffer (NucliSENS or RLT Plus, following the same dates as above) or no buffer. All samples were stored at −80 °C until RNA extraction, performed within 10 days.
2.4. RNA extraction
Frozen samples were extracted using a QIAGEN RNeasy Plus Mini Kit according to manufacturer protocol with the addition of an initial bead-beating step. Briefly, the contents of stored filters (i.e. filter plus 600 μL lysis buffer) were aseptically transferred to garnet bead tubes, with prior removal of endogenous buffer from the QIAGEN DNeasy PowerSoil Kit. Tubes were bead beaten for 120 s at 5000 rpm in a Biospec Products (Bartlesville, OK) Mini-BeadBeater 3110 BX followed by centrifugation at 8000 ×g for 30 s. The resulting supernatant was passed through a gDNA eliminator spin column (Qiagen RNeasy Plus Mini Kit) and the flow through was mixed with an equivalent volume of 70 % ethanol before being passed through the RNeasy spin column at ≥8000 x g. The spin column was washed once with 700 μL RW1 buffer, twice with 500 μL RPE buffer working solution per manufacturer instructions, and then eluted with 75 μL RNase-free water that was collected and passed through the spin column an additional time. Extracts were aliquoted into 25 μL volumes and stored at −80 °C. RNA concentration was quantified using the Qubit RNA HS assay kit and Qubit 4 Fluorometer. Total RNA from the Qubit assay for samples 7/10/20 to 10/13/20 averaged 32.6 ± 16.8 ng/μL for Plant 1 and 40.8 ± 23.6 ng/μL for Plant 2 (data not shown). Later dates were not assayed. All samples were spiked with 5 μL armored Hepatitis G (Asuragen 42,024, Austin, TX) at the beginning of the extraction protocol to serve as an extraction control. The copy number in the HepG stock was estimated via RT-ddPCR to be 459.4 copies/μL (see Supplementary Materials). The corresponding PBS method control prepared during filtration and a negative extraction control containing only lysis buffer were included in each extraction.
2.5. Detection and quantification by RT-ddPCR
“Absolute” quantification of SARS-CoV-2, i.e. quantification using partitioned reactions rather than standard curves, was performed on RNA extracted from influent wastewater samples according to the dMIQE guidelines (minimum information for publication of quantitative digital PCR experiments, see Supplementary Material), using the Bio-Rad QX200 Droplet Digital PCR System (dMIQE Group and Huggett, 2020). The Bio-Rad One-Step RT-ddPCR Advanced Kit for Probes (1,864,021, Hercules, CA) was used per manufacturer protocol but modified to increase the total reaction volume from 20 μL to 22 μL. Each reaction contained 5.5 μL Supermix, 2.2 μL reverse transcription enzyme (RT), 1.1 μL DTT, 3 μL primer/probe mixture (final concentration 913 nM for each primer and 232 nM for probes for all targets), 5.2 μL nuclease-free water, and 5 μL RNA template or nuclease free water (for no template controls). Forward and reverse primers and FAM-labeled probes for SARS-CoV-2 nucleocapsid protein targets N1 and N2 were purchased via the IDT 2019 nCoV Research Use Only Kit (Integrated DNA Technologies 10,006,713, Coralville, IA; Table 1 ). Quantitative Synthetic SARS-CoV-2 RNA: ORF, E, N (American Type Culture Collection VR-3276SD™, Manassas, VA) was used as the positive control, diluted to produce ~15 copies/μL in the RT-ddPCR reaction. The PBS method controls, negative extraction controls, and no template controls were run alongside samples as negative controls. PCR reaction mixtures were briefly vortexed and spun-down in a Fisher Mini-Plate centrifuge spinner for 30 s.
Table 1.
Sequence and reference information for the oligonucleotides used to detect SARS-CoV-2 in municipal wastewaters.
| Target Gene | Primer/Probe | Sequence 5′-3’ | Nucleotide position | Amplicon length | Reference and accession no. |
|---|---|---|---|---|---|
| N1 Nucleocapsid | nCoV N1 Fwd | GACCCCAAAATCAGCGAAAT | 28,303–28,322 | 73 bp |
Lu et al., 2020 SARS-CoV-2 accession no. MN908947 |
| nCoV N1 Rev | TCTGGTTACTGCCAGTTGAATCTG | 28,374–28,351 | |||
| nCoV N1 Probe | FAM-ACCCCGCAT-/ZEN/-TACGTTTGGTGGACC-3IABkFQ | 28,325–28,348 | |||
| N2 Nucleocapsid | nCoV N2 Fwd | TTACAAACATTGGCCGCAAA | 29,180–29,199 | 67 bp |
Lu et al., 2020 SARS-CoV-2 accession no. MN908947 |
| nCoV N2 Rev | GCGCGACATTCCGAAGAA | 29,246–29,228 | |||
| nCoV N2 Probe | FAM-ACAATTTGC-/ZEN/-CCCCAGCGCTTCAG-3IABkFQ | 29,204–29,226 | |||
| Bovine Coronavirus | BCoV Fwd | CTGGAAGTTGGTGGAGTT | 29,026–29,043 | 85 bp |
Decaro et al., 2008 BCoV strain Mebus accession no. U00735 |
| BCoV Rev | ATTATCGGCCTAACATACATC | 29,090–29,110 | |||
| BCoV Probe | FAM-CCTTCATATCTATACACATCAAGTTGTT-BHQ-1 | 29,058–29,085 | |||
| Hepatitis G | HepG Fwd | CGGCCAAAAGGTGGTGGATG | 100–119 | 185 bp |
Schlueter et al., 1996 HGV accession no. U44402 |
| HepG Rev | CGACGAGCCTGACGTCGGG | 285–267 | |||
| HepG Probe | HEX-AGGTCCCTCTGGCGCTTGTGGCGAG-BHQ-1 | 172–196 | |||
| Pepper Mild Mottle Virus | PMMoV Fwd | GAGTGGTTTGACCTTAACGTTTG | 1878–1901 | 67 bp |
Haramoto et al., 2013; Rački et al., 2014 for primers/probe PMMoV isolate S accession no. M81413 |
| PMMoV Rev | TTGTCGGTTGCAATGCAAGT | 1945–1926 | |||
| PMMoV Probe | VIC-CCTACCGAAGCAAATG-MGB-NFQ | 1906–1921 | |||
| PMMoV Pos Control | GAG TGGTTTGACC TTAACGTTTG AGAGGCCTAC CGAAGCAAAT GTCGCACTTG CATTGCAACC GACAA | NA | |||
| SPUD | SPUD Fwd | AACTTGGCTTTAATGGACCTCCA | 449–471 | 101 bp |
Nolan et al., 2006 Solanum tuberosum phyB accession no. Y14572 |
| SPUD Rev | ACATTCATCCTTACATGGCACCA | 549–527 | |||
| SPUD Probe | FAM-TGCACAAGCTATGGAACACCACGT-BHQ1 | 482–505 | |||
| SPUD Pos Control (SPUD-T) | TCAGACGTTACGTACAAACTTGGCTTTAATGGACCTCCAATTTTGAGTGTGCACAAGCTATGGAACACCACGTAAGACATAAAACGGCCACATATGGTGCCATGTAAGGATGAA TGTATCGATCGATCGACTGAGCTA |
NA |
Using a multichannel pipette, 20 μL of each reaction mixture was loaded into an 8-well cartridge for droplet generation on the Bio-Rad QX200 droplet generator along with 70 μL of droplet generation oil. The cartridge was covered with a gasket and placed in the droplet generator per manufacturer instructions. Generated droplets were carefully transferred to a skirted 96-well plate, which was sealed in a Bio-Rad PX1 PCR Plate Sealer. Reverse transcription and PCR were performed in a deep well Bio-Rad C1000 Touch Thermal Cycler with a 3 min 25 °C hold, 1 h 50 °C reverse transcription, 10 min 95 °C enzyme activation, 30 s 95 °C denaturation, 1 min 55 °C annealing, 10 min 98 °C enzyme deactivation, and using 45 denaturation/annealing cycles instead of 40 and a 2 °C/s ramp rate. The plate was then transferred to the Bio-Rad QX200 droplet reader and read for the appropriate fluorophores.
Bio-Rad QuantaSoft Analysis Pro version 1.0.596 software was used to calculate droplet count and gene-copy concentrations. Thresholds for positive droplets were set between the positive and negative droplet bands, visually inspecting for an area of least density when there was no clear distinction (see Fig. S1 for examples of positive and negative samples). Duplicate wells were merged in the software and only reactions containing ≧20,000 droplets were included in the analyses. Concentrations were then adjusted for RNA elution volume and membrane filter volume and reported as gene-copies/μL or gene-copies/L (Eq. (1)). A RT-ddPCR minimum threshold of 3 positive droplets was set for positive detection of targets (Ciesielski et al., 2021). For samples with fewer than 3 positive droplets in merged duplicate wells, the limit of detection (LOD) was determined by calculating the concentration (gene-copies/μL) in the 10 μL of RNA eluate for a 3 positive droplet threshold in the combined droplet count. Non-detects are reported as less than the calculated LOD. Additional droplet details are reported in Supplemental Materials. Positive and negative controls produced expected results for all targets. Methods for BCoV, HepG, PMMoV, and the SPUD inhibition assay are described in Supplemental Materials.
| (1) |
2.6. Detection and quantification by RT-qPCR
SARS-CoV-2 targets N1 and N2 were additionally quantified by RT-qPCR, using the same primers and probes as for RT-ddPCR and following MIQE guidelines (see Supplementary Materials; Bustin et al., 2009). The same synthetic SARS-CoV-2 was also used as a positive control and for producing standard curves. The iTaq universal probes One-Step kit for RT-qPCR (Bio-Rad #1725141) was used per manufacturer instructions. Amplifications were performed in 20 μL reaction mixtures containing 10 μL of Super Mix, 0.5 μL of reverse transcriptase enzyme (RT), 1.5 μL of primer/probe mixture (503 nM of forward primer, 503 nM of reverse primer, 128 nM of probe), 3 μL of nuclease free water, and 5 μL of RNA template. Reactions were run under the following conditions: 50 °C for 10 min for reverse transcription and 95 °C for 1 min for denaturation. Then, 40 cycles were run of 95 °C for 10 s and 60 °C for 30 s on a Bio-Rad CFX96 Touch thermal cycler. All reactions were run in duplicate, with each RT-qPCR plate containing a five-point standard curve in triplicate and at least three non-template controls. Positive and negative controls were as expected for all targets.
To determine the limit of detection (LOD) and limit of quantification (LOQ) for RT-qPCR, 3 RT-qPCR runs containing 8 standard concentrations in replicates of 4 were run for each target. This provided 12 replicates for each of the 8 standard concentrations across all plates. The 8 concentrations ranged from 6645 copies/μL (Std. 1) to 0.30 copies/μL (Std. 8). A detection rate of 75 % was needed across the 12 replicates to include the standard concentration in the LOD calculations. Standard 8 failed to qualify, so Standard 7 represented the lower limit of the linear range (0.70 copies/μL- 6645 copies/μL; see Table S1 for linearity). The Cq for Standard 7 varied from 35.3 to 37.2 for N1 and 35.15 to 36.92 for N2 (Table S2). The LOD was determined by averaging the Ct values for the lowest qualifying concentration for each target, and then calculating the related concentration of SARS-CoV-2 per reaction. The LOQ was determined by subtracting 2 standard deviations of the lowest standard's Ct values from the LOD Ct value, and again calculating the related concentration per reaction. Signals detected below the LOD and LOQ were kept in the data set to facilitate methods comparison but are reported separately in tables and figures.
SARS-CoV-2 concentrations present in influent wastewater samples were calculated from measured RT-qPCR Ct values. A standard curve equation was derived for each target on each plate from the Ct values of the known standards. A calibrator Ct was calculated by averaging the Ct values for the first standard concentration. The average target amplification factor (AF) was calculated via Eq. (2). Concentrations of SARS-CoV-2 per reaction were adjusted for reaction efficiency by multiplying the ratio of the calibrator to unknown samples (derived in Eq. (3)) by the concentration of the calibrator (e.g. Std. 1 concentration). This value was then multiplied by an elution correction factor and divided by the volume filtered to yield concentrations in gene-copies/mL or gene-copies/μL. Methods for BCoV and the SPUD inhibition assay are described in Supplemental Materials. HepG and PMMoV were not run on RT-qPCR due to a lack of original RNA extract remaining.
| (2) |
| (3) |
2.7. Epidemiological data
We obtained daily ZIP code-level COVID-19 case counts from the WRAL online repository (https://github.com/wraldata/nc-covid-data/tree/master/zip_level_data/time_series_data/csv) and county-level case counts from the Johns Hopkins University Center for Systems Science and Engineering repository (https://github.com/CSSEGISandData/COVID-19/tree/master/csse_covid_19_data/csse_covid_19_time_series). Both data sources reported cumulative cases by geography on each date; where dates were missing in the WRAL archive, we substituted data that we had manually downloaded from the NC DHHS COVID-19 dashboard (https://covid19.ncdhhs.gov/dashboard/data-behind-dashboards) on a daily basis for a portion of the study period. Daily incident cases by county and ZIP code were calculated by subtracting the cumulative cases reported the previous day (or the most recent date with case reports) from the cumulative cases reported on each date during the study period.
The populations of each ZIP code and sewershed were estimated from the 2019 American Community Survey (ACS) 5-year block group population estimates, which were distributed among the constituent blocks according to the 2010 population distribution. Blocks were associated with the ZIP code and sewershed that contained their centroids and block populations were summed to obtain ZIP code and sewershed population estimates. Anticipating substantial spatial misalignment between ZIP code and sewershed boundaries (Fig. 1), we weighted the number of cases in each intersecting ZIP code in two ways: by the proportion of the ZIP code area overlapping the sewershed and by the proportion of the ZIP code population living within the sewershed. For both weighting approaches, we summed the weighted daily cases across all intersecting ZIP codes to estimate sewershed case counts. Three-day and seven-day rolling averages were calculated for the incident sewershed cases obtained by each of the three (county-level, area-weighted, population-weighted) approaches.
Effluent flow data was reported by each WWTP and compiled by NC DEQ. Detected SARS-CoV-2 N1/N2 gene copies were normalized by plant-reported effluent flow in millions of gallons per day (MGD), by estimated population within the sewershed, and by prevalence ratio as compared to PMMoV gene copies/L. Influent data is not reported to the state and can vary by plant due to in-plant recycling of flow streams. Results are reported as SARS-CoV-2 N1/N2 gene-copies per person per day (PPPD).
2.8. Statistical analyses
Spearman's rho (ρ) correlations were estimated between N1 concentration (gene-copies/L) and daily, 3-day, and 7-day rolling average COVID-19 case counts at the three levels of geographic case aggregation for Plant 1 and Plant 2. Correlations were examined for wastewater samples collected up to 7 days prior to and after the date the cases were reported to assess timeliness of wastewater measurements. Correlations with a p-value <0.05 were considered statistically significant.
3. Results
3.1. Detection of N1 and N2 using RT-ddPCR
Over a 24-week period from July to December 2020, SARS-CoV-2 was regularly detected in wastewaters from both Plant 1 and Plant 2 via RT-ddPCR. At least one of the SARS-CoV-2 targets (N1 or N2) was detected in 90 % (N = 48) of influent samples (Table 2 ), including 100 % of Plant 1 samples and 79 % of Plant 2 samples. N1 was detected in 85 % of samples (96 % Plant 1, 75 % Plant 2), and N2 was detected in 73 % of samples (83 % Plant 1, 63 % Plant 2). Detection status of N1 and N2 agreed in 79 % of samples in both Plant 1 and Plant 2, with 69 % of samples being positive for both targets. A sample was considered positive if at least one of its replicate extractions was positive.
Table 2.
Detection of N1 and N2 gene targets of SARS-CoV-2 by RT-ddPCR in raw wastewater samples from two central NC treatment plants (n = 48).
| N1 |
||||
|---|---|---|---|---|
| Positive | Negative | Total | ||
| N2 | Positive | 33 | 2 | 35 |
| Negative | 8 | 5 | 13 | |
| Total | 41 | 7 | 48 | |
Consistent detection of SARS-CoV-2 was successful in wastewater influents but not in primary solids (data not shown) with the RNeasy Plus Mini kit and RT-ddPCR. Other labs (including some in the NC Wastewater Pathogen Research Network) have been able to detect the SARS-CoV-2 N1/N2 targets in primary solids with other RNA extraction kits (Kim et al., 2021; Peccia et al., 2020). For our study, the 20 mL filtered influent samples provided more reliable surveillance above detection limits for RT-ddPCR as compared to direct extraction methods of primary solids. Therefore, we focused our analysis and reporting on the influent wastewater samples.
Concentrations of SARS-CoV-2 from Plant 1 positive samples ranged from 1.7 × 103 to 1.4 × 105 N1 gene-copies/L and 2.4 × 103 to 5.9 × 104 N2 gene-copies/L (Table S3). Plant 2 samples displayed a similar range of 1.9 × 103 to 2.1 × 105 N1 gene-copies/L and 1.9 × 103 to 2.8 × 105 N2 gene-copies/L (Table S4). Virus concentrations normalized by flow, population, and PMMoV helped compare the disparate population density and sewersheds serviced by the two plants. The average 24-h composite flow was 3.65 (SD: 2.21; n = 24) MGD for Plant 1 and 0.34 (SD: 0.19; n = 24) MGD for Plant 2. Flow- and population-normalized SARS-CoV-2 gene copies were calculated using daily matched flow data and sewershed populations of 82,772 for Plant 1 and 3814 for Plant 2. As seen in Tables S3 and S4, Plant 1 normalized values ranged from 2.9 × 105 to 4.1 × 107 N1 gene-copies per person per day and Plant 2 normalized values ranged from 8.0 × 105 to 4.5 × 107 N1 gene-copies per person per day. Normalizing virus concentrations tended not to change virus estimates by more than an order of magnitude (Tables S3 and S4). However, changes greater than an order of magnitude were occasionally observed for the smaller treatment plant.
The total process recovery determined via BCoV concentrations was 39 %, with a range of 3 %–107 %. The extraction recovery determined via HepG concentrations was 42 %, with a range of 0 %–184 %. These recoveries were not used to correct wastewater data.
3.2. Comparison of wastewater and case data
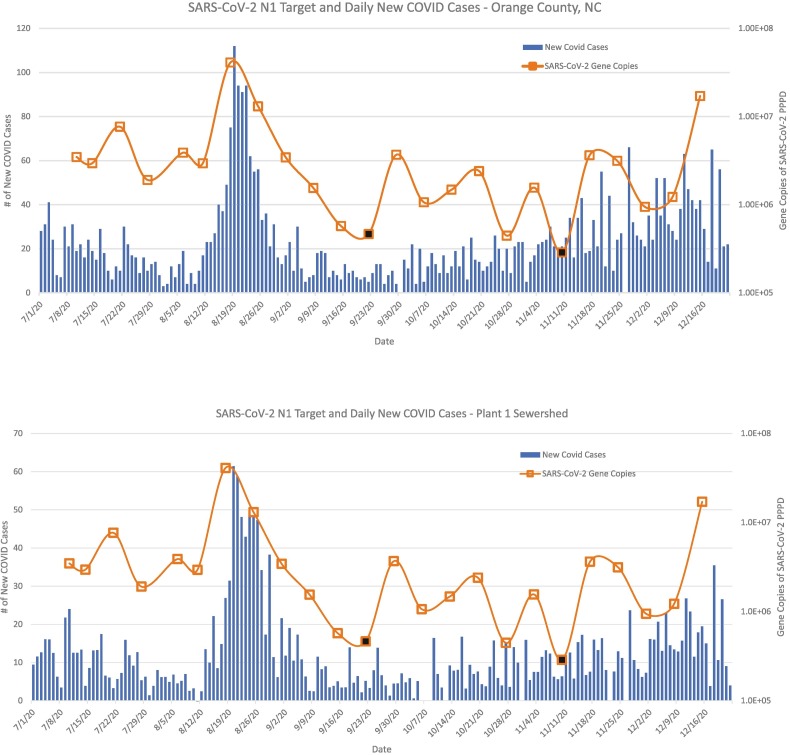
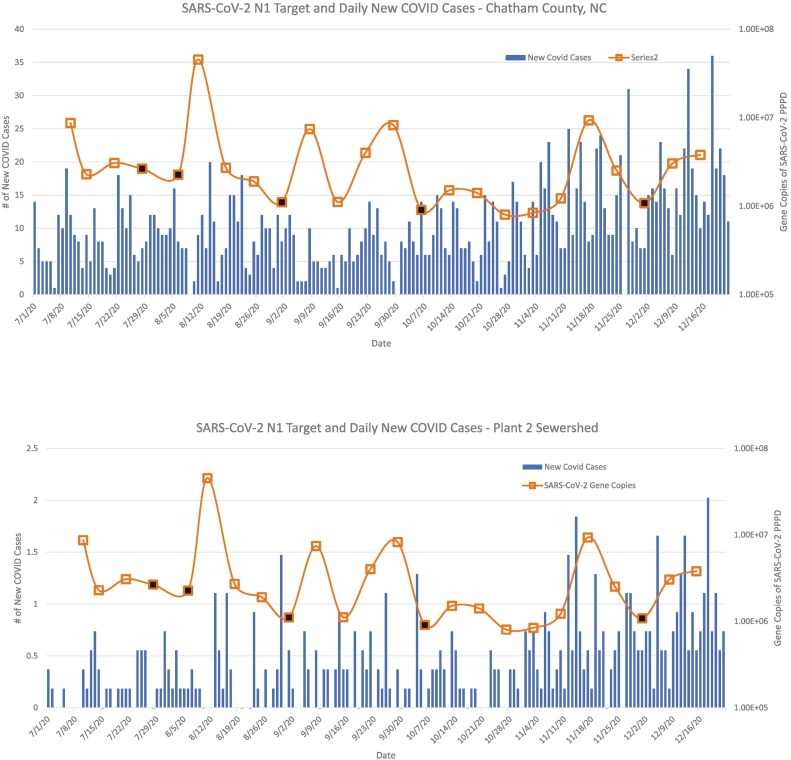
Data from RT-ddPCR analyses were matched by date and location to daily new reported cases for each county and sewershed (Fig. 2, Fig. 3 ). Reported daily new COVID cases in Chatham County and the Plant 2 sewershed were generally lower than cases in Orange County and the Plant 1 sewershed. For the Plant 1 sewershed, the peak in daily COVID cases on 8/20/20 corresponded with the greatest concentration of gene copies for SARS-CoV-2 N1 on 8/18/20. Peak reported daily new COVID case counts for Orange County, as reported by NC DHHS, was 112 and occurred on 8/19/20, with a 3-day running average of 94 cases. These wastewater and case spikes coincided with the return of UNC students to campus. Trends for N2 were similar to those of N1 (Figs. S2-S3).
Fig. 2.
Detection of SARS-CoV-2 N1 in wastewater and new COVID-19 cases for county and sewershed serviced by Plant 1. Bars represent daily new COVID-19 cases for Orange County (top) or the Plant 1 sewershed (bottom; estimated in Section 2.7). Boxes connected by the line graph represent SARS-CoV-2 gene copies per person per day (PPPD) in wastewater. With the assistance of staff at Plant 1, 24-h composite wastewater samples were collected weekly from 7/10/2020 to 12/15/2020, corresponding to 24 sampling dates. SARS-CoV-2 detection data reflects merged duplicate RT-ddPCR reactions for the nucleocapsid gene (N1). Black boxes represent a SARS-CoV-2 detection event below the limit of quantification.
Fig. 3.
Detection of SARS-CoV-2 N1 in wastewater and new COVID-19 cases for the county and sewershed serviced by Plant 2. Bars represent daily new COVID-19 cases for Chatham County (top) or the Plant 2 sewershed (bottom; estimated in Section 2.7). Boxes connected by the line graph represent SARS-CoV-2 gene copies per person per day (PPPD) in wastewater. With the assistance of staff at the Plant 2, 24-h composite wastewater samples were collected weekly from 7/10/2020 to 12/15/2020, corresponding to 24 sampling dates. SARS-CoV-2 detection data reflects merged duplicate RT-ddPCR reactions for the nucleocapsid gene (N1). Black boxes represent a SARS-CoV-2 detection event below the limit of quantification.
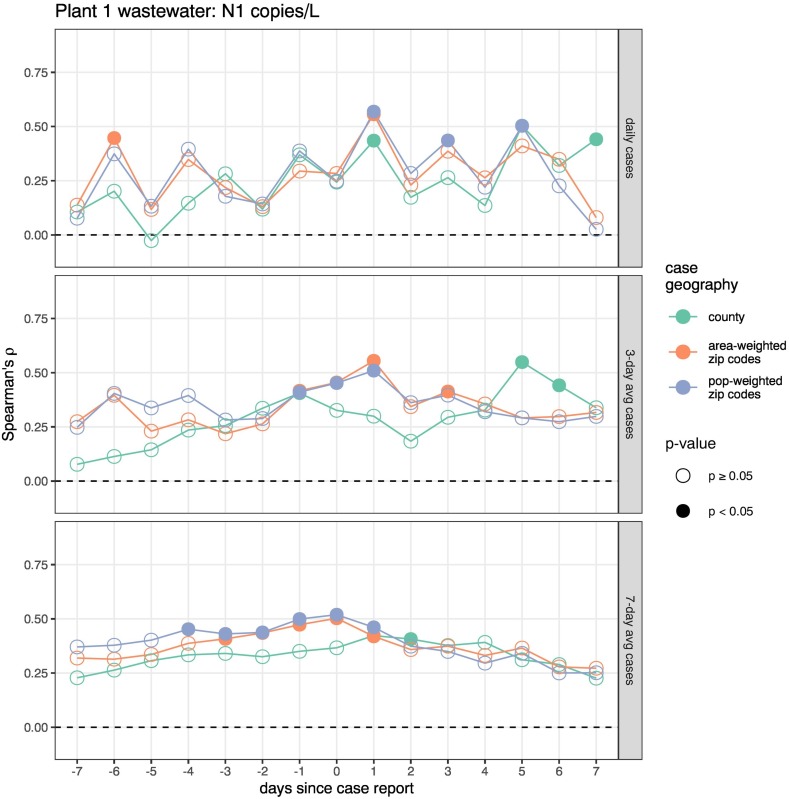
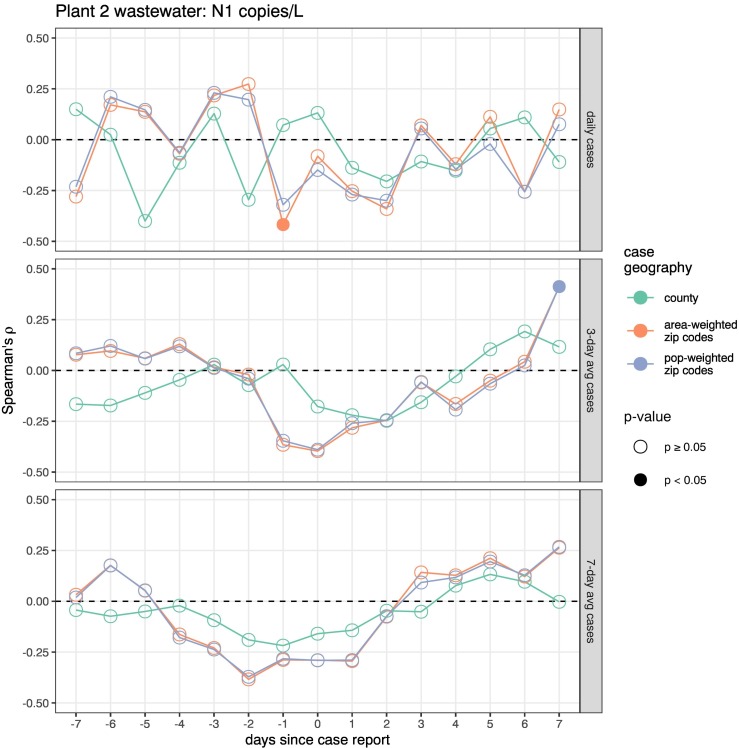
Spearman's correlations between SARS-Cov-2 wastewater concentrations and COVID-19 cases were examined in Figs. 4 (N1) and S4 (N2) for Plant 1 and Figs. 5 (N1) and S5 (N2) for Plant 2. Correlations were stronger at the larger WWTP (Plant 1, ⍴ ~0.5) compared to the smaller WWTP (Plant 2, ⍴ ~ 0.25). At Plant 2, both targets were only sporadically correlated with unsmoothed and 3-day rolling average case counts, and neither was significantly correlated with 7-day rolling average case counts at any geographic scale or temporal offset. Plant 1 correlations were more consistently significant as the smoothing window for cases was increased, with higher variability in associations with daily case counts and more stable relationships with a 7-day rolling average of cases. Correlations were weaker for county-level reported cases than for sewershed-level cases, which were similar whether derived from ZIP code reports by area- or population-weighting. Plant 1 sewershed cases with 7-day smoothing were significantly associated with N1 concentrations measured up to four days before the case report date, and with N2 concentrations measured three to seven days before the case report date. By contrast, smoothed case counts at the county level were only associated with N1 concentrations measured two days after and with N2 concentrations from one day before to two days after being reported.
Fig. 4.
Spearman's correlations between Plant 1 (Orange County) N1 concentrations (copies/L) and publicly reported Covid-19 cases. Each point represents the correlation between case counts and the N1 wastewater concentrations in samples collected x days after the case reporting. For example, −3 represents correlating all wastewater concentrations with the case counts from 3 days after each wastewater sampling. Correlations were calculated separately for daily case counts, 3-day rolling-average, and 7-day rolling average case counts (panel rows) at three levels of geographic case aggregation: all cases reported for the service county (green points), sum of ZIP code cases weighted by area of sewershed overlap (orange points), and sum of ZIP code cases weighted by sewershed population overlap (blue points). Significant correlations (p < 0.05) are indicated with filled circles.
Fig. 5.
Spearman's correlations for Plant 2 (Chatham County) N1 concentrations (copies/L) and publicly reported Covid-19 cases. Each point represents the correlation between case counts and the N1 wastewater concentrations in samples collected x days after the case reporting. For example, −3 represents correlating all wastewater concentrations with the case counts from 3 days after each wastewater sampling. Correlations were calculated separately for daily case counts, 3-day rolling-average, and 7-day rolling average case counts (panel rows) at three levels of geographic case aggregation: all cases reported for the service county (green points), sum of ZIP code cases weighted by area of sewershed overlap (orange points), and sum of ZIP code cases weighted by sewershed population overlap (blue points). Significant correlations (p < 0.05) are indicated with filled circles.
3.3. Comparison of RT-qPCR and RT-ddPCR
For RT-qPCR, the average limit of detection (LOD) for N1 was determined to be 3.3 gene-copies/reaction and the limit of quantification (LOQ) was 8.9 gene-copies/reaction. For N2, the LOD was set at 3.7 gene-copies/reaction and the LOQ set at 9.5 gene-copies/reaction. See Table S2 for the data used to calculate the LOD/LOQ. Plate summary statistics for each RT-qPCR plate can be found in Table S1. For the N1 target, reaction efficiencies ranged from 97 %–102 % and all R2 values exceeded 0.98. For the N2 target, reaction efficiencies ranged from 88 %–110 % and all R2 values exceeded 0.98. Process recovery using BCoV was 2.65 % (range: 0.0 %–58.9 %).
SARS-CoV-2 gene target concentrations were quantified via RT-qPCR in influent wastewater samples from the WWTPs for 24 sampling dates in duplicate, resulting in a total of 48 data points for each target per site (except for Plant 2 N2, which only had 47; Tables S5-S6). Replicate concentrations were classified based on LOD/LOQ levels. See Tables S7-S8 for count data for each category. Table 3 compares detection count data for the two methods. Classifying all detects found below the LOD as non-detects results in a detection rate for Plant 1 of 48 % for N1 and 31 % for N2. For Plant 2 this was 25 % for N1 and 21 % for N2. When considering only detects above the LOD/LOQ, 10 of 24 dates (41.7 %) were positive for at least one SARS-CoV-2 target in Plant 1, and 2 of 24 dates (8.3 %) were positive for Plant 2.
Table 3.
Comparison of replicate counts for sample classifications between RT-qPCR and RT-ddPCR. Count numbers include both targets and compare classifications of replicate samples. N = 188 for both methods.
| RT-ddPCR |
||||
|---|---|---|---|---|
| DETECTS Positive Sample (≥3 positive droplets) |
NON-DETECTS Negative Sample (<3 positive droplets) |
Total | ||
| RT-qPCR | DETECTS Positive Sample |
24 | 1 | 25 |
| NON-DETECTS Negative Sample |
60 | 22 | 82 | |
| DETECTS Signal below LOD |
29 | 17 | 46 | |
| DETECTS Signal below LOQ |
23 | 12 | 35 | |
| Total | 136 | 52 | 188 | |
Almost all (96 %) detects above the LOD/LOQ for RT-qPCR corresponded with detects from RT-ddPCR when comparing the same target and WWTP. 66 % of RT-qPCR samples with signal detected below the LOQ and 63 % of RT-qPCR samples detected below the LOD corresponded with RT-ddPCR detections. Close to half (44 %) of positive samples for RT-ddPCR had no level of detection for RT-qPCR. Only one replicate out of 188 (0.5 %) was positive using RT-qPCR and a non-detect via RT-ddPCR. When considering results below the LOD to be non-detects, McNemar's chi-squared statistic was 55.2 (df = 1, p = 1.11 × 10−13).
SPUD assay results suggested that RT-ddPCR experienced higher rates of inhibition from the sample matrix than RT-qPCR (Fig. S6). Negative inhibition percentages may have resulted from excess minute volumes of the spike solution being introduced to the samples due to adhesion onto the outside of the pipette tip. However, these differences with the positive control were not statistically significant. There was also no significant deviance in Ct values for spiked samples from those of the positive control for RT-qPCR.
4. Discussion
Results from this study show consistent detection of SARS-CoV-2 from wastewater and support use of wastewater-based epidemiology to track COVID-19 disease trends. As a pooled sampling strategy, WBE requires fewer resources than individual testing, and may be a valuable tool to complement clinical diagnostic testing in assessing population disease burden. In the present study, SARS-CoV-2 was detected by RT-ddPCR in 100 % (24/24) and 79 % (19/24) of influent wastewater samples from Plants 1 and 2, respectively, over a period of 24 weeks between July and December 2020.
Detected concentrations of SARS-CoV-2 in the studied wastewaters tended to track the trends seen in case counts. The wastewater data were fairly noisy, but a peak in the Plant 1 data concentrations coinciding with and, importantly, preceding by two days, a peak in sewershed case counts shortly after the return of university students to the area provides additional evidence toward the benefits of WBE for COVID-19 control (Fig. 2). Between 8/10/20 and 8/16/20, campus COVID-19 testing positivity rates increased from 2.8 % to 13.6 %, triggering university officials to substantially reduce residence hall capacity for undergraduates and move courses to remote learning (Guskiewicz and Blouin, 2020).
Significant (p < 0.05) positive correlations were observed between wastewater and cases in Plant 1, though such correlations were more elusive in Plant 2 (Fig. 4, Fig. 5). These correlations and visual inspection of the overlaid datasets suggest that wastewater tracked case counts quite well for Plant 1. Plant 2 serves a smaller township with a greater rural proportion and thus a smaller percentage of residents with municipal wastewater service. Potential relationships between detected gene copies in Plant 2 wastewater and case counts are complicated by periods of zero reported cases followed by spikes in new cases, which are partially artifacts of irregular access to testing and test results reporting. Additionally, in this smaller served population, a slight change in cases results in a larger change in incidence rate than for plants serving a larger population, an issue referred to as the small number problem. Noise in the data may have also been due to differences across time such as wastewater residence time in piped infrastructure or variance between the plants such as storage conditions or other plant-level factors. For both plants, associations might have been stronger with more frequent sampling. Feng et al. noted that sampling at least twice weekly was needed to maintain accuracy in COVID-19 trend analysis (Feng et al., 2021).
Our finding that smoothed COVID-19 case counts correlated with SARS-CoV-2 concentrations in Plant 1 wastewater four to seven days before the cases were reported is largely consistent with other reports in the literature. A study of 12 WWTPs in Wisconsin found that correlations with 7-day smoothed case counts were typically higher when wastewater was sampled 0–6 days prior to case specimen collection, although the temporal offset with the strongest correlation differed by plant (Feng et al., 2021). Plant size was not found to influence correlation patterns, but their smallest plant served an estimated 11,000 individuals, nearly three times the service population of Plant 2 in our study, for which we did not observe significant correlation between cases and wastewater. Multiple studies from Massachusetts using varied approaches found wastewater viral loads correlated with reported case counts as much as 10–11 days later (Omori et al., 2021; Wu et al., 2020a). In New Haven, Connecticut, SARS-CoV-2 titers in primary sludge predicted new cases up to two days before specimen collection date and 6–8 days before case report date under a distributed lag modeling approach (Peccia et al., 2020).
This work provides multiple insights toward improving WBE for SARS-CoV-2. First, sensitivity is a concern in detection of environmental viruses - information is lost below the lower limits of detection of our tools. This is particularly challenging in wastewater, which often contains substantial concentrations of PCR inhibitors. Our findings suggest an improved sensitivity of RT-ddPCR over RT-qPCR – particularly when considering the increased inhibition seen in RT-ddPCR – which is consistent with recent studies on SARS-CoV-2 WBE (Ciesielski et al., 2021; D’Aoust et al., 2021). Potential limits to widespread adoption of RT-ddPCR include higher costs and longer run times compared to RT-qPCR. When minimal lag times are seen between infection and case reporting, a rapid turnaround time would be needed to make WBE an attractive alternative to clinical testing. This speed may be more achievable with RT-qPCR. However, this study demonstrates the value of increased sensitivity offered by the RT-ddPCR assay in detecting viruses in wastewaters.
SARS-CoV-2 nucleocapsid targets N1 and N2 have arisen as commonly used genetic indicators of the virus in wastewater. Relying on only one target would be best for minimizing resource use, so this work and others have sought to compare the two. Because the two targets each appear once in the virus’s genome, they theoretically should be detected equally. However, we found that they agreed in only 79 % of samples, suggesting differences between the two assays. Between the two, N1 was detected more often (85 % vs 73 %). Working near the lower limit of detection, this suggests greater sensitivity in our N1 assay. As such, we have opted to largely limit N2 results to the Supplemental Information, though we recognize the potential value in the increased anticipation of case data by N2 data if this trend were substantiated. Previous studies have also found discrepancies between N1 and N2, but they conflict on which is preferred for use in wastewater (Gonzalez et al., 2020; Medema et al., 2020; Peccia et al., 2020). Results of this study suggest that N1 may be preferred for programs seeking the efficiencies of a single-target assay.
This study found process control Bovine Coronavirus and extraction control Hepatitis G to both show substantial variability, at times being detected at concentrations substantially above what was calculated to have been spiked into the samples, and sometimes not being detected at all. As such, we did not correct our quantified SARS-CoV-2 concentrations by these controls. Our low and variable HepG extraction recovery is consistent with those of others in the literature (Barua et al., 2022; Gonzalez et al., 2020). Recent WBE research has similarly found BCoV and other coronaviruses to have low and highly variable recovery (Betancourt et al., 2021; Ciesielski et al., 2021). Gonzalez et al. found a 59 % BCoV recovery with no concentration step, and this dropped to 5.5 % with centrifugation or 4.8 % with filtration, and Graham et al. found low but less variable recovery when using BCoV as only an extraction control (Gonzalez et al., 2020; Graham et al., 2021). This suggests that the filtration step may be the weak point in our workflow for BCoV recovery. The heat pasteurization step required by lab safety protocols at the time may have also affected virus recovery. If BCoV is an appropriate analog for SARS-CoV-2 behavior, the body of literature may support higher concentrations of SARS-CoV-2 in wastewater than have been reported. It is unclear if this is the case, however, so research directly comparing BCoV and other common controls to SARS-CoV-2 would be valuable. Producing viral concentrations accurately will be difficult for wastewater samples until processing and extraction controls are improved.
Our approach for estimating sewershed-level case counts will be valuable for evaluating the accuracy of WBE for future COVID-19 surveillance. In the absence of sewershed data, using block group data to estimate ZIP code and sewershed populations, and then weighting ZIP code case data by intersected area or population allowed us to produce more accurate estimates of the number of COVID-19 cases served by the WWTPs. We recommend that others assessing WBE follow this approach, because case data may not be available for rapid WBE at a more precise level than county and ZIP code due to privacy concerns. As a sewershed may represent only a fraction of the ZIP code or county population, its wastewater will provide inadequate geographic resolution to reliably anticipate higher geographic-level incidence.
Our study allowed us to witness sewage viral concentrations clearly responding to an unexpected large-scale outbreak in a college community. Provided a rapid turnaround from sampling to reporting, WBE may allow extra days for decision makers to assess trends and reduce the health impacts of outbreaks like this. Even without a rapid turnaround, WBE could be valuable in assessing policy impacts (e.g., lifting mask mandates) or economic consequences. For example, these measurements could also help evaluate effects of infection rates on school attendance, travel rates, or consumer trends.
Despite these important developments, further research will be needed to realize the potential of WBE. Improving sensitivity of detection methods may improve assessment of infection trends, particularly in less populated sewersheds. This study enabled comparison of results from plants serving different population sizes, but only included two plants. Results should be pooled from a larger number of plants to test more rigorously the effect of plant size on estimating viral concentrations and disease trends. It will also be critical to identify and account for the most important factors impacting the accuracy of data being compared in WBE. For wastewater, this could be viral degradation, rainfall, and flow-related factors and for case data, the willingness and ability of populations to be tested. These improvements may help position wastewater as a cost-effective proxy for case data whenever such data are unavailable or unreliable. Results of this and other studies show that WBE is proving to be helpful in documenting COVID-19 trends and could be a valuable tool for fighting current and future pandemics.
5. Conclusions
-
•
SARS-CoV-2 wastewater concentrations anticipated case counts in the larger treatment plant and sewershed, especially preceding a spike in cases.
-
•
The 7-day average of both N1 and N2 concentrations in wastewater anticipated increases in sewershed cases several days before they were reported. For this study, N1 was detected more frequently than N2 and may be a preferred target if seeking to measure only one target.
-
•
RT-ddPCR was more sensitive than RT-qPCR in the detection of SARS-CoV-2 in wastewater. Method agreement increased when including positive signals noted below the limit of detection for the RT-qPCR assay. Programs may want to consider reporting this type of RT-qPCR data in categories separate from positive or negative.
-
•
Wastewater representing only a subset of the county population provides inadequate geographic resolution to reliably anticipate county-level incidence. Sewershed-level estimates were better correlated with wastewater measurements.
-
•
Capacity built from these efforts may be instrumental in responding to current and future pandemics. WBE shows promise for identifying outbreaks and documenting disease trends.
Funding
This project was supported by the North Carolina Policy Collaboratory at the University of North Carolina at Chapel Hill with funding from the North Carolina Coronavirus Relief Fund established and appropriated by the North Carolina General Assembly. Collin Knox Coleman, Connor LaMontagne, and David Holcomb were supported in part by a training grant from the National Institute of Environmental Health Sciences (T32ES007018). Alyssa Grube was supported by the UNC Graduate School Dissertation Completion Fellowship and National Science Foundation Graduate Research Fellowship Program. Nikhil Kothegal was supported in part by a training grant from the National Institute for Occupational Safety and Health (T42-OH008673).
CRediT authorship contribution statement
Alyssa M. Grube: Methodology, Formal analysis, Investigation, Data curation, Writing – original draft. Collin K. Coleman: Methodology, Formal analysis, Investigation, Data curation, Writing – original draft, Writing – review & editing, Visualization. Connor D. LaMontagne: Methodology, Formal analysis, Investigation, Data curation, Writing – original draft, Writing – review & editing. Megan E. Miller: Methodology, Formal analysis, Investigation, Data curation, Writing – original draft, Visualization. Nikhil P. Kothegal: Formal analysis, Investigation, Data curation, Writing – original draft, Writing – review & editing. David A. Holcomb: Methodology, Formal analysis, Investigation, Data curation, Writing – original draft, Writing – review & editing, Visualization. A. Denene Blackwood: Methodology, Validation, Resources, Writing – review & editing, Supervision. Thomas J. Clerkin: Methodology, Validation. Marc L. Serre: Conceptualization, Methodology, Writing – review & editing, Supervision. Lawrence S. Engel: Conceptualization, Methodology, Writing – review & editing, Supervision. Virginia T. Guidry: Conceptualization, Resources, Writing – review & editing, Project administration, Funding acquisition. Rachel T. Noble: Conceptualization, Methodology, Validation, Resources, Writing – review & editing, Supervision, Project administration, Funding acquisition. Jill R. Stewart: Conceptualization, Methodology, Writing – original draft, Writing – review & editing, Project administration, Supervision, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research began during the devastation and unease of the COVID-19 pandemic, and we are grateful to many people who stepped up during such difficult times to support the effort. We would like to acknowledge Brent Wishart, Cathy Brennan, and the larger UNC employee safety and support system for their guidance and assistance to ensure safe access to the laboratories. We would especially like to thank Dr. Jayne Boyer for support ordering supplies despite global shortages and with consideration to not compete with health care needs. We would also like to thank all the health care heroes on the front line of the pandemic and all the mental health professionals who provided essential care during this time.
We would like to thank all collaborating labs and the following individuals for their research contributions: Monica Dodson, Jennifer Hunter, and Ronnie Weed (Plant 1); Jamie McLaurin (Plant 2); and Dr. Eric Johnson (Bio-Rad). A special thank you to the Noble Lab for leading the NC Wastewater Pathogen Research Network (NC WW PATH) and assisting with methods development. We conducted this work as part of NC WW PATH - a joint effort among academic institutions, state agencies (NC Department of Health and Human Services (NC DHHS), NC Department of Environmental Quality (NC DEQ)) and over 20 municipal WWTPs across North Carolina to develop methods for surveillance of SARS-CoV-2 in wastewaters, and to relate the wastewater data to COVID-19 infections. The research effort matured into the NC Wastewater Monitoring Network, led by NC DHHS, which became one of the first programs to join the US CDC National Wastewater Surveillance System (NWSS) and the first to submit data to the CDC. These efforts are building the capacity for wastewater surveillance of SARS-CoV-2, and we’d like to acknowledge the hard work of those working to move wastewater surveillance from research to practice across the globe.
Editor: Warish Ahmed
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2022.159996.
Appendix A. Supplementary data
Supplementary material
Data availability
Data will be made available on request.
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Angel N., Bibby K., Bivins A., Dierens L., Edson J., Ehret J., Gyawali P., Hamilton K.A., Hosegood I., Hugenholtz P., Jiang G., Kitajima M., Sichani H.T., Shi J., Shimko K.M., Simpson S.L., Smith W.J.M., Symonds E.M., Mueller J.F. Detection of SARS-CoV-2 RNA in commercial passenger aircraft and cruise ship wastewater: a surveillance tool for assessing the presence of COVID-19 infected travellers. J. Travel Med. 2020;27 doi: 10.1093/jtm/taaa116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Simpson S.L., Bertsch P.M., Bibby K., Bivins A., Blackall L.L., Bofill-Mas S., Bosch A., Brandão J., Choi P.M., Ciesielski M., Donner E., D’Souza N., Farnleitner A.H., Gerrity D., Gonzalez R., Griffith J.F., Gyawali P., Haas C.N., Hamilton K.A., Shanks O.C. Minimizing errors in RT-PCR detection and quantification of SARS-CoV-2 RNA for wastewater surveillance. Sci. Total Environ. 2022;805 doi: 10.1016/j.scitotenv.2021.149877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asghar H., Diop O.M., Weldegebriel G., Malik F., Shetty S., El Bassioni L., Akande A.O., Al Maamoun E., Zaidi S., Adeniji A.J., Burns C.C., Deshpande J., Oberste M.S., Lowther S.A. Environmental surveillance for polioviruses in the Global Polio Eradication Initiative. J. Infect. Dis. 2014;210(Suppl. 1):S294–S303. doi: 10.1093/infdis/jiu384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barua V.B., Juel M.A.I., Blackwood A.D., Clerkin T., Ciesielski M., Sorinolu A.J., Holcomb D.A., Young I., Kimble G., Sypolt S., Engel L.S., Noble R.T., Munir M. Tracking the temporal variation of COVID-19 surges through wastewater-based epidemiology during the peak of the pandemic: a six-month long study in Charlotte, North Carolina. Sci. Total Environ. 2022;814 doi: 10.1016/j.scitotenv.2021.152503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt W.Q., Schmitz B.W., Innes G.K., Prasek S.M., Pogreba Brown K.M., Stark E.R., Foster A.R., Sprissler R.S., Harris D.T., Sherchan S.P., Gerba C.P., Pepper I.L. COVID-19 containment on a college campus via wastewater-based epidemiology, targeted clinical testing and an intervention. Sci. Total Environ. 2021;779 doi: 10.1016/j.scitotenv.2021.146408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., North D., Ahmad A., Ahmed W., Alm E., Been F., Bhattacharya P., Bijlsma L., Boehm A.B., Brown J., Buttiglieri G., Calabro V., Carducci A., Castiglioni S., Cetecioglu Gurol Z., Chakraborty S., Costa F., Curcio S., de Los Reyes F.L., Delgado Vela J., Bibby K. Wastewater-based epidemiology: global collaborative to maximize contributions in the fight against COVID-19. Environ. Sci. Technol. 2020;54:7754–7757. doi: 10.1021/acs.est.0c02388. [DOI] [PubMed] [Google Scholar]
- Brouwer A.F., Eisenberg J.N.S., Pomeroy C.D., Shulman L.M., Hindiyeh M., Manor Y., Grotto I., Koopman J.S., Eisenberg M.C. Epidemiology of the silent polio outbreak in Rahat, Israel, based on modeling of environmental surveillance data. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E10625–E10633. doi: 10.1073/pnas.1808798115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., Vandesompele J., Wittwer C.T. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Chen P., Liu Y., Wang H., Liu G., Lin X., Zhang W., Ji F., Xu Q., Tao Z., Xu A. Environmental surveillance complements case-based surveillance of acute flaccid paralysis in polio endgame strategy 2019–2023. Appl. Environ. Microbiol. 2020;86 doi: 10.1128/AEM.00702-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Chen L., Deng Q., Zhang G., Wu K., Ni L., Yang Y., Liu B., Wang W., Wei C., Yang J., Ye G., Cheng Z. The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J. Med. Virol. 2020;92:833–840. doi: 10.1002/jmv.25825. [DOI] [PubMed] [Google Scholar]
- Ciesielski M., Blackwood D., Clerkin T., Gonzalez R., Thompson H., Larson A., Noble R. Assessing sensitivity and reproducibility of RT-ddPCR and RT-qPCR for the quantification of SARS-CoV-2 in wastewater. J. Virol. Methods. 2021 doi: 10.1016/j.jviromet.2021.114230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Aoust P.M., Mercier E., Montpetit D., Jia J.-J., Alexandrov I., Neault N., Baig A.T., Mayne J., Zhang X., Alain T., Langlois M.-A., Servos M.R., MacKenzie M., Figeys D., MacKenzie A.E., Graber T.E., Delatolla R. Quantitative analysis of SARS-CoV-2 RNA from wastewater solids in communities with low COVID-19 incidence and prevalence. Water Res. 2021;188 doi: 10.1016/j.watres.2020.116560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Elia G., Campolo M., Desario C., Mari V., Radogna A., Colaianni M.L., Cirone F., Tempesta M., Buonavoglia C. Detection of bovine coronavirus using a TaqMan-based real-time RT-PCR assay. J. Virol. Methods. 2008;151:167–171. doi: 10.1016/j.jviromet.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggett J.F., dMIQE Group The digital MIQE guidelines update: minimum information for publication of quantitative digital PCR experiments for 2020. Clin. Chem. 2020;66:1012–1029. doi: 10.1093/clinchem/hvaa125. [DOI] [PubMed] [Google Scholar]
- Falman J.C., Fagnant-Sperati C.S., Kossik A.L., Boyle D.S., Meschke J.S. Evaluation of secondary concentration methods for poliovirus detection in wastewater. Food Environ. Virol. 2019;11:20–31. doi: 10.1007/s12560-018-09364-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Roguet A., McClary-Gutierrez J.S., Newton R.J., Kloczko N., Meiman J.G., McLellan S.L. 2021. Evaluation of sampling frequency and normalization of SARS-CoV-2 wastewater concentrations for capturing COVID-19 burdens in the community. medRxiv. [DOI] [Google Scholar]
- Gao Q.Y., Chen Y.X., Fang J.Y. 2019 novel coronavirus infection and gastrointestinal tract. J. Dig. Dis. 2020;21:125–126. doi: 10.1111/1751-2980.12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibas C., Lambirth K., Mittal N., Juel M.A.I., Barua V.B., Roppolo Brazell L., Hinton K., Lontai J., Stark N., Young I., Quach C., Russ M., Kauer J., Nicolosi B., Chen D., Akella S., Tang W., Schlueter J., Munir M. Implementing building-level SARS-CoV-2 wastewater surveillance on a university campus. Sci. Total Environ. 2021;782 doi: 10.1016/j.scitotenv.2021.146749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R., Curtis K., Bivins A., Bibby K., Weir M.H., Yetka K., Thompson H., Keeling D., Mitchell J., Gonzalez D. COVID-19 surveillance in Southeastern Virginia using wastewater-based epidemiology. Water Res. 2020;186 doi: 10.1016/j.watres.2020.116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham K.E., Loeb S.K., Wolfe M.K., Catoe D., Sinnott-Armstrong N., Kim S., Yamahara K.M., Sassoubre L.M., Mendoza Grijalva L.M., Roldan-Hernandez L., Langenfeld K., Wigginton K.R., Boehm A.B. SARS-CoV-2 RNA in wastewater settled solids is associated with COVID-19 cases in a large urban sewershed. Environ. Sci. Technol. 2021;55:488–498. doi: 10.1021/acs.est.0c06191. [DOI] [PubMed] [Google Scholar]
- Guskiewicz K.M., Blouin R.A. Carolina Together; 2020. Campus email - UNC Chapel Hill [WWW Document]https://carolinatogether.unc.edu/2020/08/17/campus-email-8-17-20/ (accessed 11.23.21). [Google Scholar]
- Haramoto E., Kitajima M., Kishida N., Konno Y., Katayama H., Asami M., Akiba M. Occurrence of pepper mild mottle virus in drinking water sources in Japan. Appl. Environ. Microbiol. 2013;79:7413–7418. doi: 10.1128/AEM.02354-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Guo Y., Mao R., Zhang J. Proportion of asymptomatic coronavirus disease 2019: a systematic review and meta-analysis. J. Med. Virol. 2020;93:820–830. doi: 10.1002/jmv.26326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox L., Patel A., Gerber S.I., Kim L., Tong S., Lu X., Lindstrom S., Pallansch M.A., Washington State 2019-nCoV Case Investigation Team First case of 2019 novel coronavirus in the united states. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovi T., Shulman L.M., van der Avoort H., Deshpande J., Roivainen M., de Gourville E.M. Role of environmental poliovirus surveillance in global polio eradication and beyond. Epidemiol. Infect. 2012;140:1–13. doi: 10.1017/S095026881000316X. [DOI] [PubMed] [Google Scholar]
- Ivanova O.E., Yarmolskaya M.S., Eremeeva T.P., Babkina G.M., Baykova O.Y., Akhmadishina L.V., Krasota A.Y., Kozlovskaya L.I., Lukashev A.N. Environmental surveillance for poliovirus and other enteroviruses: long-term experience in Moscow, Russian Federation, 2004–2017. Viruses. 2019;11 doi: 10.3390/v11050424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Kennedy L.C., Wolfe M.K., Criddle C.S., Duong D.H., Topol A., White B.J., Kantor R.S., Nelson K.L., Steele J.A., Langlois K., Griffith J.F., Zimmer-Faust A.G., McLellan S.L., Schussman M.K., Ammerman M., Wigginton K.R., Bakker K.M., Boehm A.B. 2021. SARS-CoV-2 RNA is enriched by orders of magnitude in solid relative to liquid wastewater at publicly owned treatment works. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer S.A., Grantz K.H., Bi Q., Jones F.K., Zheng Q., Meredith H.R., Azman A.S., Reich N.G., Lessler J. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann. Intern. Med. 2020;172:577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q.-X., Tang X.-J., Shi Q.-L., Li Q., Deng H.-J., Yuan J., Hu J.-L., Xu W., Zhang Y., Lv F.-J., Su K., Zhang F., Gong J., Wu B., Liu X.-M., Li J.-J., Qiu J.-F., Chen J., Huang A.-L. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- Lu X., Wang L., Sakthivel S.K., Whitaker B., Murray J., Kamili S., Lynch B., Malapati L., Burke S.A., Harcourt J., Tamin A., Thornburg N.J., Villanueva J.M., Lindstrom S. US CDC real-time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerg.Infect. Dis. 2020;26 doi: 10.3201/eid2608.201246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Bakker M., King A.A., Rohani P. Unraveling the transmission ecology of polio. PLoS Biol. 2015;13 doi: 10.1371/journal.pbio.1002172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environ. Sci. Technol. Lett. 2020 doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Mizumoto K., Kagaya K., Zarebski A., Chowell G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.10.2000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathanson N., Kew O.M. From emergence to eradication: the epidemiology of poliomyelitis deconstructed. Am. J. Epidemiol. 2010;172:1213–1229. doi: 10.1093/aje/kwq320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan T., Hands R.E., Ogunkolade W., Bustin S.A. SPUD: a quantitative PCR assay for the detection of inhibitors in nucleic acid preparations. Anal. Biochem. 2006;351:308–310. doi: 10.1016/j.ab.2006.01.051. [DOI] [PubMed] [Google Scholar]
- O’Reilly K.M., Allen D.J., Fine P., Asghar H. The challenges of informative wastewater sampling for SARS-CoV-2 must be met: lessons from polio eradication. Lancet Microbe. 2020;1:e189–e190. doi: 10.1016/S2666-5247(20)30100-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omori R., Miura F., Kitajima M. Age-dependent association between SARS-CoV-2 cases reported by passive surveillance and viral load in wastewater. Sci. Total Environ. 2021;792 doi: 10.1016/j.scitotenv.2021.148442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., Warren J.L., Weinberger D.M., Arnold W., Omer S.B. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38:1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rački N., Dreo T., Gutierrez-Aguirre I., Blejec A., Ravnikar M. Reverse transcriptase droplet digital PCR shows high resilience to PCR inhibitors from plant, soil and water samples. Plant Methods. 2014;10:42. doi: 10.1186/s13007-014-0042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlueter V., Schmolke S., Stark K., Hess G., Ofenloch-Haehnle B., Engel A.M. Reverse transcription-PCR detection of hepatitis G virus. J. Clin. Microbiol. 1996;34:2660–2664. doi: 10.1128/jcm.34.11.2660-2664.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang A., Tong Z.-D., Wang H.-L., Dai Y.-X., Li K.-F., Liu J.-N., Wu W.-J., Yuan C., Yu M.-L., Li P., Yan J.-B. Detection of novel coronavirus by RT-PCR in stool specimen from asymptomatic child, China. Emerg. Infect. Dis. 2020;26:1337–1339. doi: 10.3201/eid2606.200301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . Global Polio Eradication Initiative. World Health Organization; Geneva: 2019. Polio Endgame Strategy 2019-2023: eradication, integration, certification and containment. [Google Scholar]
- Wu F., Xiao A., Zhang J., Moniz K., Endo N., Armas F., Bonneau R., Brown M.A., Bushman M., Chai P.R., Duvallet C., Erickson T.B., Foppe K., Ghaeli N., Gu X., Hanage W.P., Huang K.H., Lee W.L., Matus M., McElroy K.A., Alm E.J. 2020. SARS-CoV-2 titers in wastewater foreshadow dynamics and clinical presentation of new COVID-19 cases. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Poyet M., Moniz K., Washburne A.D., Erickson T.B., Chai P.R., Thompson J., Alm E.J. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 2020;5 doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xagoraraki I., O’Brien E. In: Women in Water Quality: Investigations by Prominent Female Engineers, Women in Engineering And Science. O’Bannon D.J., editor. Springer International Publishing; Cham: 2020. Wastewater-based epidemiology for early detection of viral outbreaks; pp. 75–97. [DOI] [Google Scholar]
- Zhang N., Gong Y., Meng F., Bi Y., Yang P., Wang F. 2020. Virus shedding patterns in nasopharyngeal and fecal specimens of COVID-19 patients. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Data will be made available on request.