ABSTRACT
Clinical microbiology has possessed a marvellous past, an important present and a bright future. Western medicine modernization started with the discovery of bacterial pathogens, and from then, clinical bacteriology became a cornerstone of diagnostics. Today, clinical microbiology uses standard techniques including Gram stain morphology, in vitro culture, antigen and antibody assays, and molecular biology both to establish a diagnosis and monitor the progression of microbial infections. Clinical microbiology has played a critical role in pathogen detection and characterization for emerging infectious diseases as evidenced by the ongoing COVID-19 pandemic. Revolutionary changes are on the way in clinical microbiology with the application of “-omic” techniques, including transcriptomics and metabolomics, and optimization of clinical practice configurations to improve outcomes of patients with infectious diseases.
KEYWORDS: Clinical microbiology, nucleic acid amplification, genomics, transcriptomics, proteomics, metabolomics, point of care, COVID-19
The unprecedented outbreak of the coronavirus disease 2019 (COVID-19) pandemic has highlighted the necessity for readily available, accurate and fast diagnostic testing methods to detect and characterize emerging pathogens. The basic work of the clinical microbiology laboratory is to provide evidence for the diagnosis, treatment, and control of infectious diseases by detecting the presence of specific pathogenic microorganisms in clinical specimens. When a pathogen is detected, it is then subjected to a series of further analyses, including identification, typing, quantification, and antimicrobial susceptibility testing [1,2].
A long and uneven past
The discipline of clinical microbiology has been evolving for more than two hundred years since Dutchman, Anton van Leeuwenhoek invented the microscope (Table 1). The development of the light microscope was the foundational technological advancement in modern medicine for the direct visualization of microorganisms. By the 1830s, Lister introduced the “achromatic” lens to eliminate the blurring and colour distortion known as “chromatic aberration,” which had previously limited resolution at higher magnification (i.e. bacterial level) [3]. These technical advancements in microscopy converged with seminal concepts in bacteriology from the work of Pasteur and Koch. Together, they allowed the microscope to serve as a powerful instrument for physicians and microbiologists to directly visualize pathogens in human specimens, especially with the application of (still universal) techniques such as the Gram and acid-fast staining procedures, also developed in the late nineteenth century [2,4].
Table 1.
Historical figures in clinical microbiology.
| Scientist | Year of birth and death | Major contributions in clinical microbiology | |
|---|---|---|---|
 |
Anton van Leeuwenhoek | 1632–1723 | Invented microscope to observe microorganisms |
 |
Louis Pasteur | 1822–1895 | Found microorganisms in infected patients |
 |
Robert Koch | 1843–1910 | Established three criteria for determining a causative relationship between a microbe and a disease |
 |
Hans Christian Gram | 1853–1938 | Invented the Gram stain |
 |
Julius Richard Petri | 1852–1921 | Invented Petri dishes to start in vitro culture |
 |
John Franklin Enders | 1897–1985 | Applied cell culture techniques for isolation and growth of viruses |
 |
Feifan Tang | 1897–1958 | Discovered and cultured the pathogen causing trachoma |
 |
Kary Banks Mullis | 1944–2019 | Invented polymerase chain reaction for in vitro nucleic acid amplification |
Equally important during this period were newfound abilities to culture microorganisms from human sources. While early bacterial cultures were accomplished with slices of raw potato, the 1887 invention of the petri dish facilitated the direct observation of colonies with gelatine or agar, allowing for the morphological description of species in pure culture [5]. It likewise allowed for their biochemical characterization, creating the phenotypic profiles that have served as the basis for taxonomic identification in clinical laboratories until the twenty-first century. Culture is likewise a pre-requisite for phenotypic antimicrobial susceptibility testing (AST), which remains the gold standard for predicting the response of a patient’s bacterial/fungal infection to treatment [1,4,6].
Shortly after the development of axenic bacterial/fungal culture, methods for propagating human viruses using ex vivo cellular substrates, fertilized chicken embryos, and in vitro cell lines became available. In the late 1940s, John Franklin Enders first applied cell culture techniques to isolate and grow poliovirus, initiating modern clinical virology [7]. In 1956, Feifan Tang, a Chinese microbiologist and virologist, discovered and isolated Chlamydia trachomatis, which clarified the cause of trachoma [8]. For decades, tissue and cell culture-based methods have played critical roles in rapid identification of emerging pathogens and exploration of pathogenesis. This has been demonstrated in three unprecedented outbreaks of emerging human coronavirus (SARS-CoV, MERS-CoV and SARS-CoV-2) infections since the beginning of the twenty-first century [9]. More importantly, cell culture techniques, along with chicken egg embryos, have become the main tools to reproduce large quantities of viruses for vaccine manufacturing for fighting emerging infectious diseases [10].
Alongside these abilities to visualize and cultivate organisms came new techniques that assess the body’s humoral response to infection. This principle was first established in 1892 by Sternberg using the vaccinia virus [11]. It is now recognized that antibodies within serum facilitate identification not just of viruses, but multiple types of pathogens by the clinical laboratory [12,13]. The value of diagnostic serology for infectious diseases is founded on two criteria: (i) antibodies are specific for a particular cellular target and (ii) antibodies are induced by a specific stimulus. Ideally, the definitive evidence for infection by a pathogen would involve recovery of that agent from infected tissue, along with an increase in specific antibodies over time. With some infections, however, the recovery of the causative agent may be difficult, dangerous, impractical, or even impossible [13]. Certainly before the development nucleic acid assays (and in some cases still), the combination of appropriate clinical and serologic findings can serve as sufficient criteria for diagnosis, even if the organism itself is not detected [12]. Seropositivity can likewise function as a marker of functional immunity toward many (but not all) pathogens, and these tests facilitate population monitoring within the fields of epidemiology and public health [12,13]. Moreover, the purification of pathogen-specific antibodies allows for the development of “antigen-based” diagnostics that detect microbe-specific biomolecules other than nucleic acids, typically carbohydrates and proteins.
Kary Banks Mullis, an American biochemist and Nobel Prize winner, was credited with the invention of the polymerase chain reaction (PCR) in the 1980s for nucleic acid amplification, thus applying rapid, sensitive and specific molecular biology techniques to the practice of clinical microbiology [14,15]. A quick application of reverse transcriptase (RT), an enzyme recovered in 1970 simultaneously by Baltimore and Temin [16,17], resulted in RT–PCR which has been widely used in the detection and characterization of RNA targets. Further modification of PCR into a quantitative format (qPCR) by Higuchi et al. in 1993 has enabled accurate determination of pathogen loads in clinical specimens [18]. Additionally, the automation of the Sanger sequencing method by Leroy Hood and Michael Hunkapillar at Applied Biosystems in 1987 rendered the rapid generation of complete genome sequences of Haemophilus influenzae [19] and Mycoplasma genitalium [20] a reality [21].
A flourishing present
In the past decades, clinical microbiology laboratories have undergone important changes with the introduction of molecular biology techniques [22] and laboratory automation [23]. Diagnostic methods in clinical microbiology are currently divided into the following five categories (Table 2) [1]. The first one is the morphological observation under the naked eye or microscope. This is a fundamental method, which is currently used mainly for initial screening and for guiding the next step of testing. Specific morphological findings by microscopy can quickly identify the pathogenetic agent in some cases. For example, the finding of Gram-negative diplococci in urethral exudates from males is a reliable indication of Neisseria gonorrhoeae infection. The second is the antigen test for pathogenic microorganisms. It is widely used in clinical practice for outpatient testing because of its rapidity, simplicity, and specificity [24,25]. Examples for this include laboratory testing for the urine pneumococcal and Legionella antigen tests [26]. On the other hand, the disadvantage of antigen tests lies in its often poor sensitivity. Specimens that are antigen-negative in clinical practice usually need to be retested with more sensitive methods, such as culture or/and molecular methods to avoid a missed diagnosis. Examples for this include laboratory testing for Streptococcus pyogenes [24] and influenza virus [25].
Table 2.
Microbiomic technology main contents. MALDI-TOF MS, matrix assisted laser desorption ionization time of flight mass spectrometry.
| Technology | Target molecule | Question addressed | Output | Main methods | Selected references | |
|---|---|---|---|---|---|---|
 |
Genomics | DNA | Infection potential | DNA sequences | DNA sequencing | [69,71,72] |
 |
Transcriptomics | RNA | Infection strategy | Transcription of text | RNA sequencing, quantitative transcription PCR | [77,82,84] |
 |
Proteomics | Protein | Infection process | Protein profiles | MALDI-TOF MS | [83,89,90] |
 |
Metabolomics | Metabolites | Infection outcome | Metabolic text | Gas or liquid phase mass spectrometry | [78–80] |
During the ongoing COVID-19 pandemic, the performance of rapid antigen tests for COVID-19 diagnosis has been widely evaluated and found to be varied in different settings [27]. Rapid antigen tests are cheaper and provide faster results, thus potentially enabling prompt isolation of positive cases and quarantine of close contacts. A recent literature review covering a total of 16 studies reported on the effectiveness of rapid antigen testing for screening of asymptomatic individuals to limit the transmission [9,28]. Eight included studies examining the effectiveness of rapid antigen testing for population-level screening, four for pre-event screening and four for serial testing. Overall, there was no evidence regarding the effectiveness of rapid antigen testing for the screening of asymptomatic individuals to limit the transmission of SARS-CoV-2. This uncertainty is due to the inconsistent results, the relatively low number of studies identified, the predominantly observational and/or uncontrolled nature of the study designs used, and concerns regarding methodological quality. Given this uncertainty, more real-world research evidence in relevant settings, which is of good quality and timely, as well as economic evaluation, is required to inform public policy on the widespread use of rapid antigen tests in asymptomatic individuals [28].
The third category is the culture method which remains the gold standard for culturable pathogenic microorganisms [1]. It mainly includes inanimate media such as agar or broth and animate media such as tissues and cells. The former is mainly used for culturing extracellular pathogens such as bacteria and fungi, while the latter is mainly used for culturing intracellular pathogens such as viruses/chlamydia. Cultivation for bacteriology and mycology has gradually transitioned into a totally automated continuous monitoring mode of blood culture/liquid culture from the manual method. Viral cultures are still an important research and clinical laboratory tool when the potential viral agent is not known. Other application niches include documenting active infection, performing antiviral susceptibility testing and developing a vaccine and therapeutic agents.
Viral culture has been a valuable tool for studying COVID-19 pathogenesis, resulting in developing more effective disease prevention, diagnosis, and control. To combat COVID-19, since SARS-CoV-2 was first isolated from nasopharyngeal and oropharyngeal specimens from a patient with COVID-19 in Vero-CCL81 and Vero E6 cells [29], several in vitro and ex vivo cell culture systems have progressively been used and described [30]. Matsuyama et al engineered a Vero E6 cell line expressing TMPRSS2 for culturing of SARS-CoV-2 showing more than 100 times greater production of viral RNA copies than Vero E6 cells alone [31]. Cell culture remains the definitive assay to determine viral infectivity and transmission. Using cell culture methods, Wölfel et al revealed that infectious viruses were readily isolated from samples derived from the throat or lung, but not from stool samples, in spite of high concentrations of virus RNA. Blood and urine samples never yielded virus [32]. A recent review by Bhat et al summarized that infectious virus was generally not shed beyond 20 days of the onset of symptoms in most COVID-19 patients, including severely ill and immunocompromised, as indicated by failure to isolate replication-competent virus by viral culture [33]. Cell culture-based plaque reduction neutralization assays and derivatives are the most reliable and accurate methods to determine SARS-CoV-2 neutralization antibody activities [34,35].
The fourth category is the serological technique to detect the specific host response to pathogenic microorganisms causing infection. However, due to the lag time to a detectable host response and the cross-reactivity between similar pathogens, serological methods are rarely used for the rapid detection of infections. Successful examples are the detection of pathogen-specific IgM antibodies for hepatitis A virus and perinatal infections, such as Zika virus. In addition to antibodies (humoral immunity), clinical tests are now available for cellular immunity in infectious diseases, including interferon-γ release assays tuberculosis and cytomegalovirus infections [36,37].
Serological detection has been recognized for its sensitivity in convalescent patients with COVID-19 and plays an important role for understanding the epidemiology of SARS-CoV-2 and emerging variants, including the burden and role of asymptomatic infections [9]. A meta-analysis of diagnostic performance of the serological tests for COVID-19 revealed the impact of assay design and post-symptom-onset intervals. Using combined nucleocapsid (N) and spike (S) protein had a better sensitivity compared to either N or S protein only. Serological tests played an important role in the clinical diagnosis for the later-stage COVID-19 patients. Enzyme-linked immunosorbent assays (ELISA) for detecting total antibodies or targeting combined N and S proteins had a higher diagnostic sensitivity compared to other methods [38], which have been used successfully for contact tracing in the early COVID-19 pandemic [39]. Lichtenegger et al developed a faster live virus assay to quantitatively detect neutralizing antibodies through the early measurement of antibody-mediated intracellular virus reduction by SARS-CoV-2 real-time PCR [40]. Cell culture-based plaque reduction neutralization assays and their derivatives are the most reliable and accurate methods to determine SARS-CoV-2 neutralization antibody activities [34,35].
The last category of infectious disease diagnostics is molecular biology techniques for the detection of pathogen-specific nucleic acids. PCR, an enzyme-mediated in vitro nucleic acid amplification method, has become the dominant method in clinical microbiology services, especially for viral infections [15,22]. A group of simple, rapid and integrated molecular devices is gradually replacing antigen testing for immediate diagnosis at the point of care [41,42]. The “syndromic panel” kits incorporating multiplex-PCR have been widely used to identify a range of pathogens with similar symptoms [42,43]. Quantitative pathogenic testing plays a key role in the monitoring of infection treatment. Next-generation nucleic acid sequencing and gene editing technologies are also finding their way into the detection and identification of specific nucleic acids of pathogenic microorganisms [44,45].
Beyond nucleic acids, matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS), which targets microbial proteomic profiles, has been widely used in the clinical microbiology laboratory for rapid and accurate identification [46,47]. In addition, MALDI-TOF MS has been explored for determining epidemic relatedness and antibiotic resistance of microbial isolates. The value of MALDI-TOF MS for microbial typing was investigated in several studies involving Staphylococcus aureus [48,49]. Garrigos and colleagues built a database using MALDI-TOF MS allowing rapid and accurate species identification and determination of the multi-resistant epidemic clones of Achromobacter species in French cystic fibrosis centers [50]. These data indicated that this technology is a potential rapid screening tool for nosocomial infection investigations. Recently, the MALDI-TOF MS technique has been extended to functional identification including determination of antibiotic resistance [51,52]. Youn et al used the MALDI-TOF MS for the rapid detection of potentially blaKPC-containing carbapenem-resistant isolates, providing early and clinically actionable results [53]. MALDI-TOF MS profiling in combination with growth media containing isotopically labelled amino acids was reported for the detection of resistant microorganisms after hours of incubation [54]. Scientists in Bruker Daltonik and clinical collaborators reported the rapid detection of antibiotic resistance by MALDI-TOF MS using a direct-on-target microdroplet growth assay [55–57] Detection of colistin resistance became rapid and reliable by use of the MALDI Biotyper Sirius system in both E. coli and P. aeruginosa [58,59]. These methods being developed based MALDI-TOF MS technology provide an alternative approach to timely monitoring microbial infections [52].
The COVID-19 pandemic ignited the development of numerous nucleic acid amplification methods for the diagnosis and monitoring of SARS-CoV-2 infections. Molecular tests such as real-time PCR are highly sensitive and specific at detecting viral RNA and are now recommended by World Health Organization (WHO) for confirming the diagnosis in individuals who are symptomatic as well as for informing public health decisions. Several newer molecular methods including digital droplet PCR [60] and CRISPR-based assays [61] have been used in COVID-19 diagnostics. Culture accompanied with mNGS increased pathogen diagnostic rate of secondary infections in severe and critical ill COVID-19 patients [62]. Integrated, random-access, point-of-care molecular devices have been developed for fast and accurate diagnosis of SARS-CoV-2 infections in local hospitals and clinics bearing the burden of identifying and treating patients [63,64]. Molecular methods have played significant roles in the discovery and characterization of emerging pathogens such as new world hantavirus [65], influenza A (H7N9) virus [66] and most recently Langya henipavirus [67].
A bright and evolving future
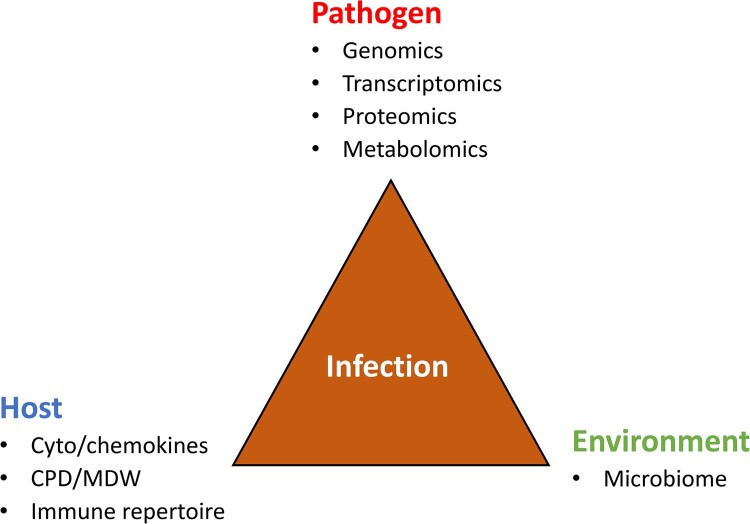
The development and progression of an infectious disease can be thought of as the result of a tripartite interaction between the pathogen, the host, and the environment (Figure 1). On the pathogen side, techniques will continue to evolve to expand and enhance the capacity for detection and characterization of emerging microbial pathogens. Since the genomes of an increasing number of microbial species/strains have been sequenced, the door has been opened wide for “omic” technologies to be used more broadly [68]. Metagenomic next-generation sequencing (mNGS)-enabled surveillance methods offer the opportunity to improve the detection of both known and yet-to-emerge pathogens such as SARS-CoV-2 and the new Langya henipavirus reported recently [67,69,70]. Wilson et al in 2014 reported the use of NGS to provide an actionable diagnosis of neuroleptospirosis [71]. NGS-based system has been commercially available to determine for HIV-1 genotypic resistance directly in clinical samples [72]. The mNGS testing is a powerful tool that can aid in aetiology diagnosis especially in complicated cases as a rule-out. With limitations understood, mNGS data can be useful with host response information incorporated. Currently, turnaround times remain a major hurdle, although same-day result can be produced using faster system such as Nanopore sequencing [73]. Finally, the clinical relevance of the presence of pathogen-specific nucleic acids in a clinical specimen, which may or may not indicate infection, can be adjudicated by a panel of clinical microbiologists and physicians, much like in done in other areas of pathology [74] or a sequencing stewardship panel [75].
Figure 1.
Infection is the result of the interaction between the pathogen, the host and the environment. CPD, cell population data; MDW, monocyte distribution width.
Besides genomics, the laboratory may use transcriptomics, proteomics, and metabolomics [76], each of which may carry potential diagnostic utility. For instance, in Mycobacterium tuberculosis infections, because the half-life of mRNA is extremely short as compared with rRNA or genomic DNA, assays that target mycobacterial mRNA better reflect mycobacterial viability, which may be used to monitor the efficacy of anti-TB therapy [77]. Regarding metabolomics, Koo et al reported the use of breath fungal secondary metabolite signatures to diagnose invasive aspergillosis infections [78]. On 14 April 2022, the US FDA granted an emergency use authorization for the InspectIR COVID-19 Breathalyzer (InspectIR Systems, Frisco, TX, USA), the first COVID-19 diagnostic test using gas chromatography/gas mass-spectrometry to identify chemical mixtures of five volatile organic compounds (VOCs) associated with SARS-CoV-2 infection in exhaled breath, which provides a SARS-CoV-2 identification result in less than three minutes (https://www.fda.gov/media/157720/download). A series of metabolomic profiles were reported to detect and characterize culturable and unculturable bacterial pathogens in clinical settings [79–81]. In these and numerous other research studies, multi-omic techniques have been leveraged to generate molecular profiles for the surveillance and management of emerging infections [82,83].
Host response markers also have been explored to facilitate diagnosis of microbial infections. Zhang et al. utilized metatranscriptomics of blood from COVID-19 patients and identified a transcriptional signature of differentially expressed genes significantly associated with immune response to SARS-CoV-2 [84]. Furthermore, Sweeney et al. conducted integrated multi-analyte profiling to yield a three-gene set for testing whole blood specimens that is very robust for diagnosing active tuberculosis (with potential relevance both for diagnosis and treatment monitoring) [82]. Cepheid (Sunnyvale, CA, USA) has recently developed a prototype assay (Xpert MTB® MTB Finger Stick) to detect a three gene host response signature in whole blood specimens using the GeneXpert® system. The gene signature may be of value both for diagnosis and treatment monitoring. Several studies have demonstrated that the assay fulfils the most of the attributes of the WHO target product profile for a point of care triage test for TB. The assay can be performed on fingerstick blood [85–88]. A novel assay called MeMed BV that integrates measurements of blood-borne host-proteins (tumour necrosis factor-related apoptosis-inducing ligand, interferon γ-induced protein-10, and CRP) was developed and manufactured by MeMed Diagnostics (Tirat Carmel, Israel) to assist in differentiation between bacterial and viral disease [83,89]. A recent prospective, multicenter cohort study performed by Papan et al. validated the high diagnostic performance of the MeMed BV assay in a broad paediatric cohort, and supported its potential to reduce antibiotic overuse in children with viral infections [90].
It is now well-established that the gut microbiota plays a critical role in infection pathogenesis [91,92]. The importance of the human microbiome is becoming increasingly evident. While the contributions of individual microbes in the human health are still far from fully understood, the microbiome appears to play a key role in many vital functions, including synthesizing vitamins and amino acids, generating important metabolites, protecting against pathogens, utilizing non-human biochemical pathways and contributing to the immune system. A large body of evidence demonstrated, more than a decade ago, that gut microbial alteration is a key factor in the pathogenesis of many local and systemic disorders, including infections [93]. Characterization of the composition of the gut microbiota as well as a dominant pathogen(s) in patients with microbial infections promises to open new avenues for the development of patient-centered personalized and precision diagnosis [94]. One example was to used robust microbiota-based assay to enable simple diagnostics and disease activity monitoring for inflammatory bowel disease [95]. Laboratory-developed tests are commercially available for microbiome determination in several reference laboratories. For example, the Gut Intelligence Test (Viome Inc., Los Alamos, NM, USA) uses a robust and automated stool metatranscriptomic method offering a rapid and comprehensive taxonomic and functional readout of the gut microbiome [96]. An integrated diagnostic approach combining pathogen, host and microbiome would enhance the speed and accuracy for the laboratory diagnosis and monitoring of microbial infections (Figure 1). Among them, machine learning will gradually apply in clinical microbiology practice especially for unusual emerging pathogens including predicting drug targets or vaccine candidates, diagnosing microorganisms causing epidemics, predicting disease outbreaks and exploring microbial interactions [97–99]. Machine learning has been used in the clinical setting for classifying drug resistance against antimicrobial agents [100,101].
In the future, there will be a need for more rapid diagnoses, increased standardization of testing and greater adaptability to cope with new threats from emerging microbial pathogens. As early as 2004, Didier Raoult, the renowned French medical and clinical microbiologist, jointly with his team, proposed a bipolarization of future clinical microbiology services [102] (Figure 2). On one side, clinical microbiology practice will follow the general trend in the life sciences for large, centralized laboratories with the capacity to analyze large numbers of samples and to carry out a wide range of techniques. Total automation has been gradually achieved for bacterial culture, identification and antimicrobial susceptibility testing [23]. For molecular diagnosis, molecular platforms are increasingly designed with an emphasis on automation and sample-to-result capabilities. These include technologies by Roche (4800/6800/8800 platforms) [103], Abbott Laboratories (m2000 and Alinity m) [104], Hologic (Panther and Panther-Fusion) [105], Becton Dickinson (BDMax and BD COR) [106], and Cepheid (GeneXpert Infinity) [107].
Figure 2.
Polarized clinical microbiology practice in the near future with rapid, random-access tests done at point of care (left) and with batched, large volumes of tests done at central laboratory (right).
On the other side, rapid and on-demand testing is performed at point of care (POC) based with relatively low throughput testing volumes. These include rapid screening for influenza in the emergency room and rapid mixed-sample screening for new coronaviruses in the field [41,108]. The current widely accepted definition of POC testing includes testing that occurs at or near the point of patient care, such that the results drive patient care decisions made during that encounter [109,110]. POC tests can be performed in a variety of settings including physician offices, emergency department, urgent care facilities, school health clinics and pharmacies. Recently, the COVID-19 pandemic has shined a spotlight on Clinical and Laboratory Improvement Amendments (CLIA)-waived diagnostic testing. Some SARS-CoV-2 tests have received Emergency Use Authorization (EUA) from the U.S. Food and Drug Administration (FDA) for use in CLIA-waived testing sites [108]. Further optimization and validation, new technologies, as well as studies to determine clinical and epidemiological impact of SARS-CoV-2 POC tests are needed. Nevertheless, random-access, integrated devices available at the point of care with scalable capacities will increase its weight in the rapid and accurate diagnosis and monitoring of emerging pathogens in the near future.
In summary, the discipline of clinical microbiology, with a long and uneven past, has been thriving at present and is ready to embrace the future. There have been substantial changes in the role of clinical microbiology laboratories over the past decade. The ongoing technological revolution has rapidly transformed research, diagnostic and therapeutic tools. In the near future, clinical microbiology practice will be able to help clinicians implement real-time evidence-based treatments or significantly shorten the process from empirical to evidence-based treatments. While continuously strengthening its scientific attribution, clinical microbiology will be endowed with a more distinct, more profound, more ambitious medical landscape, social value and management significance. Full of challenges and uncertainties, though, the practice of clinical microbiology will keep abreast of the times, promising and never fading, for its demand-oriented, significance-driven, logic and science-based, and humanitarian characteristics.
Acknowledgements
The authors thank Monica Wang, Yongzhong Ning and Jingwen Ai for their assistance and Charles Stratton and Sherry Dunbar for their constructive discussion and critical reviewing the manuscript. The Xpert MTB® MTB Finger Stick Prototype is a product in development, which is not for use in diagnostic procedures and not reviewed by any regulatory body.
Disclosure statement
Y.-W.T. is an employee of Cepheid, the commercial manufacturer of the GeneXpert system and Xpert cartridges. No potential conflict of interest was reported by the rest authors.
References
- 1.Cintron M, Hauser JR, Otto C, et al. Diagnostic microbiology. In: Schmidt TM, editor. Encyclopedia of microbiology, 4th ed. Oxford: Elsevier Press; 2019. p. 1–17. [Google Scholar]
- 2.Isenberg HD. Clinical microbiology: past, present, and future. J Clin Microbiol. 2003;41:917–918. doi: 10.1128/JCM.41.3.917-918.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hodgkin T, Lister JJ.. Notice of some microscopic observations of the blood and animal tissues. Phil Mag. 1827;32:130–138. [Google Scholar]
- 4.Tang YW, Sussman M, Liu D, et al. Molecular medical microbiology, 2 ed. Boston (MA: ): Elsevier; 2014. [Google Scholar]
- 5.Hitchens AP, Leikind MC.. The introduction of agar-agar into bacteriology. J Bacteriol. 1939;37:485–493. doi: 10.1128/jb.37.5.485-493.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lagier JC, Edouard S, Pagnier I, et al. Current and past strategies for bacterial culture in clinical microbiology. Clin Microbiol Rev. 2015;28:208–236. doi: 10.1128/CMR.00110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enders JF, Weller TH, Robbins FC.. Cultivation of the lansing strain of poliomyelitis virus in cultures of various human embryonic tissues. Science. 1949;109:85–87. doi: 10.1126/science.109.2822.85. [DOI] [PubMed] [Google Scholar]
- 8.Tang F, Zhang X, Huang Y, et al. Study on the pathogen of trachoma IV. Attempt to isolate the virus in the embryonated hens eggs. Acta Microbiol Sin. 1956;2:189–210. [Google Scholar]
- 9.Loeffelholz MJ, Tang YW.. Laboratory diagnosis of emerging human coronavirus infections – the state of the art. Emerg Microbes Infect. 2020;9:747–756. doi: 10.1080/22221751.2020.1745095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khalil N, Bernstein DI.. Influenza vaccines: where we are, where we are going. Curr Opin Pediatr. 2022;34:119–125. doi: 10.1097/MOP.0000000000001103. [DOI] [PubMed] [Google Scholar]
- 11.Sternberg GM. Practical results of bacteriological researches. Tr A Am Physicians. 1892;7:68–86. [Google Scholar]
- 12.Ayres JC, Feemster RF.. Serologic tests in the diagnosis of infectious diseases. Concluded. N Engl J Med. 1950;243:1034–1043. doi: 10.1056/NEJM195012282432606. [DOI] [PubMed] [Google Scholar]
- 13.Ayres JC, Feemster RF.. Serologic tests in the diagnosis of infectious diseases. Part 1. N Engl J Med. 1950;243:996–1002. doi: 10.1056/NEJM195012212432505. [DOI] [PubMed] [Google Scholar]
- 14.Mullis KB, Faloona FA.. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- 15.Schmitz JE, Stratton CW, Persing DH, et al. Forty years of molecular diagnostics for infectious diseases. J Clin Microbiol. 2022;19:02446–02421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baltimore D. RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature. 1970;226:1209–1211. doi: 10.1038/2261209a0. [DOI] [PubMed] [Google Scholar]
- 17.Temin HM, Mizutani S.. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature. 1970;226:1211–1213. doi: 10.1038/2261211a0. [DOI] [PubMed] [Google Scholar]
- 18.Higuchi R, Fockler C, Dollinger G, et al. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnol (NY). 1993;11:1026–1030. doi: 10.1038/nbt0993-1026. [DOI] [PubMed] [Google Scholar]
- 19.Fleischmann RD, Adams MD, White O, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 20.Fraser CM, Gocayne JD, White O, et al. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 21.Sanger F, Coulson AR.. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975;94:441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- 22.Tang YW, Procop GW, Persing DH.. Molecular diagnostics of infectious diseases. Clin Chem. 1997;43:2021–2038. [PubMed] [Google Scholar]
- 23.Bourbeau PP, Ledeboer NA.. Automation in clinical microbiology. J Clin Microbiol. 2013;51:1658–1665. doi: 10.1128/JCM.00301-13. Epub 2013 Mar 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerber MA. Diagnosis of group A beta-hemolytic streptococcal pharyngitis. Use of antigen detection tests. Diagn Microbiol Infect Dis. 1986;4:5S–15S. doi: 10.1016/s0732-8893(86)80038-4. [DOI] [PubMed] [Google Scholar]
- 25.Smith TF, Wold AD, Espy MJ, et al. New developments in the diagnosis of viral diseases. Infect Dis Clin North Am. 1993;7:183–201. [PubMed] [Google Scholar]
- 26.Bellew S, Grijalva CG, Williams DJ, et al. Pneumococcal and legionella urinary antigen tests in community-acquired pneumonia: prospective evaluation of indications for testing. Clin Infect Dis. 2019;68:2026–2033. doi: 10.1093/cid/ciy826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khalid MF, Selvam K, Jeffry AJN, et al. Performance of rapid antigen tests for COVID-19 diagnosis: A systematic review and meta-analysis. Diagnostics (Basel). 2022;12:110. doi: 10.3390/diagnostics12010110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walsh KA, Broderick N, Ahern S, et al. Effectiveness of rapid antigen testing for screening of asymptomatic individuals to limit the transmission of SARS-CoV-2: A rapid review. Rev Med Virol. 2022;29:e2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harcourt J, Tamin A, Lu X, et al. Severe acute respiratory syndrome coronavirus 2 from patient with coronavirus disease, United States. Emerg Infect Dis. 2020;26:1266–1273. doi: 10.3201/eid2606.200516. Epub 2020 Jun 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heinen N, Klöhn M, Steinmann E, et al. In vitro lung models and their application to study SARS-CoV-2 pathogenesis and disease. Viruses. 2021;13:792. doi: 10.3390/v13050792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuyama S, Nao N, Shirato K, et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci U S A. 2020;117:7001–7003. doi: 10.1073/pnas.2002589117. Epub 2020 Mar 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. Epub 2020 Apr 1. [DOI] [PubMed] [Google Scholar]
- 33.Bhat V, Chavan P, Khattry N, et al. Dynamics of viral RNA load, virus culture, seroconversion & infectivity in COVID-19 patients: implications on isolation policy. Indian J Med Res. 2021;153:585–590. doi: 10.4103/ijmr.IJMR_3564_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Banga Ndzouboukou JL, Zhang YD, Fan XL.. Recent developments in SARS-CoV-2 neutralizing antibody detection methods. Curr Med Sci. 2021;41:1052–1064. doi: 10.1007/s11596-021-2470-7. Epub 2021 Dec 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muruato AE, Fontes-Garfias CR, Ren P, et al. A high-throughput neutralizing antibody assay for COVID-19 diagnosis and vaccine evaluation. Nat Commun. 2020;11:4059. doi: 10.1038/s41467-020-17892-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manuel O, Husain S, Kumar D, et al. Assessment of cytomegalovirus-specific cell-mediated immunity for the prediction of cytomegalovirus disease in high-risk solid-organ transplant recipients: a multicenter cohort study. Clin Infect Dis. 2013;56:817–824. doi: 10.1093/cid/cis993. Epub 2012 Nov 29. [DOI] [PubMed] [Google Scholar]
- 37.Pai M, Denkinger CM, Kik SV, et al. Gamma interferon release assays for detection of Mycobacterium tuberculosis infection. Clin Microbiol Rev. 2014;27:3–20. doi: 10.1128/CMR.00034-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H, Ai J, Loeffelholz MJ, et al. Meta-analysis of diagnostic performance of serology tests for COVID-19: impact of assay design and post-symptom-onset intervals. Emerg Microbes Infect. 2020;9:2200–2211. doi: 10.1080/22221751.2020.1826362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peeling RW, Wedderburn CJ, Garcia PJ, et al. Serology testing in the COVID-19 pandemic response. Lancet Infect Dis. 2020;20:e245–e249. doi: 10.1016/S1473-3099(20)30517-X. Epub 2020 Jul 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lichtenegger S, Saiger S, Hardt M, Kulnik S, Wagner GE, Kleinhappl B, Assig K, Zauner A, Ober M, Kimpel J, von Laer D, Zatloukal K, Steinmetz I.. Development of a Rapid Live SARS-CoV-2 Neutralization Assay Based on a qPCR Readout. J Clin Microbiol. 2022;60:e0037622. doi: 10.1128/jcm.00376-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kozel TR, Burnham-Marusich AR.. Point-of-Care testing for infectious diseases: past, present, and future. J Clin Microbiol. 2017;55:2313–2320. doi: 10.1128/JCM.00476-17. Epub 2017 May 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ling L, Kaplan SE, Lopez JC, et al. Parallel validation of three molecular devices for simultaneous detection and identification of influenza A and B and respiratory syncytial viruses. J Clin Microbiol. 2018;56:e01691–17. doi: 10.1128/JCM.01691-17. Print 2018 Mar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abbott AN, Fang FC.. Clinical impact of multiplex syndromic panels in the diagnosis of bloodstream, gastrointestinal, respiratory, and central nervous system infections. Clin Microbiol Newsl. 2017;39:133–142. [Google Scholar]
- 44.Gootenberg JS, Abudayyeh OO, Lee JW, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. Epub 2017 Apr 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiu CY, Miller SA.. Clinical metagenomics. Nat Rev Genet. 2019;20:341–355. doi: 10.1038/s41576-019-0113-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bizzini A, Greub G.. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry, a revolution in clinical microbial identification. Clin Microbiol Infect. 2010;16:1614–1619. doi: 10.1111/j.1469-0691.2010.03311.x. [DOI] [PubMed] [Google Scholar]
- 47.Torres-Sangiao E, Leal Rodriguez C, García-Riestra C.. Application and perspectives of MALDI-TOF mass spectrometry in clinical microbiology laboratories. Microorganisms. 2021;9:1539. doi: 10.3390/microorganisms9071539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schlebusch S, Price GR, Hinds S, et al. First outbreak of PVL-positive nonmultiresistant MRSA in a neonatal ICU in Australia: comparison of MALDI-TOF and SNP-plus-binary gene typing. Eur J Clin Microbiol Infect Dis. 2010;29:1311–1314. [DOI] [PubMed] [Google Scholar]
- 49.Wolters M, Rohde H, Maier T, et al. MALDI-TOF MS fingerprinting allows for discrimination of major methicillin-resistant staphylococcus aureus lineages. Int J Med Microbiol. 2011;301:64–68. [DOI] [PubMed] [Google Scholar]
- 50.Garrigos T, Dollat M, Magallon A, et al. Distribution of Achromobacter species in 12 French cystic fibrosis centers in 2020 by a retrospective MALDI-TOF MS spectrum analysis. J Clin Microbiol. 2022;60:e0242221. doi: 10.1128/jcm.02422-21. Epub 2022 May 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hrabak J, Walkova R, Studentova V, et al. Carbapenemase activity detection by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2011;49:3222–3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoon EJ, Jeong SH.. MALDI-TOF mass spectrometry technology as a tool for the rapid diagnosis of antimicrobial resistance in bacteria. Antibiotics (Basel). 2021;10:982. doi: 10.3390/antibiotics10080982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Youn JH, Drake SK, Weingarten RA, et al. Clinical performance of a matrix-assisted laser desorption ionization-time of flight mass spectrometry method for detection of certain blaKPC-containing plasmids. J Clin Microbiol. 2016;54:35–42. doi: 10.1128/JCM.01643-15. Epub 2015 Sep 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sparbier K, Lange C, Jung J, et al. MALDI biotyper-based rapid resistance detection by stable-isotope labeling. J Clin Microbiol. 2013;51:3741–3748. doi: 10.1128/JCM.01536-13. Epub 2013 Sep 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Correa-Martínez CL, Idelevich EA, Sparbier K, et al. Rapid detection of extended-spectrum β-lactamases (ESBL) and AmpC β-lactamases in enterobacterales: development of a screening panel using the MALDI-TOF MS-based direct-on-target microdroplet growth assay. Front Microbiol. 2019;10:13. doi: 10.3389/fmicb.2019.00013. eCollection 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Correa-Martínez CL, Idelevich EA, Sparbier K, et al. Development of a MALDI-TOF MS-based screening panel for accelerated differential detection of carbapenemases in enterobacterales using the direct-on-target microdroplet growth assay. Sci Rep. 2020;10:4988. doi: 10.1038/s41598-020-61890-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Idelevich EA, Sparbier K, Kostrzewa M, et al. Rapid detection of antibiotic resistance by MALDI-TOF mass spectrometry using a novel direct-on-target microdroplet growth assay. Clin Microbiol Infect. 2018;24:738–743. doi: 10.1016/j.cmi.2017.10.016. Epub 2017 Oct 24. [DOI] [PubMed] [Google Scholar]
- 58.Furniss RCD, Dortet L, Bolland W, et al. Detection of colistin resistance in Escherichia coli by use of the MALDI biotyper sirius mass spectrometry system. J Clin Microbiol. 2019;57:e01427–19. doi: 10.1128/JCM.01427-19. Print 2019 Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jeannot K, Hagart K, Dortet L, et al. Detection of colistin resistance in Pseudomonas aeruginosa using the MALDIxin test on the routine MALDI biotyper sirius mass spectrometer. Front Microbiol. 2021;12:725383. doi: 10.3389/fmicb.2021.725383. eCollection 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suo T, Liu X, Feng J, et al. ddPCR: a more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg Microbes Infect. 2020;9:1259–1268. doi: 10.1080/22221751.2020.1772678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Broughton JP, Deng X, Yu G, et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. Epub 2020 Apr 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang H, Zhang Y, Wu J, et al. Risks and features of secondary infections in severe and critical ill COVID-19 patients. Emerg Microbes Infect. 2020;9:1958–1964. doi: 10.1080/22221751.2020.1812437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Loeffelholz MJ, Tang YW.. Detection of SARS-CoV-2 at the point of care. Bioanalysis. 2021;13:1213–1223. doi: 10.4155/bio-2021-0078. Epub 2021 Jul 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hou H, Chen J, Wang Y, et al. Multicenter evaluation of the cepheid xpert xpress SARS-CoV-2 assay for the detection of SARS-CoV-2 in oropharyngeal swab specimens. J Clin Microbiol. 2020;58:e01288–20. doi: 10.1128/JCM.01288-20. Print 2020 Jul 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nichol ST, Spiropoulou CF, Morzunov S, et al. Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness. Science. 1993;262:914–917. doi: 10.1126/science.8235615. [DOI] [PubMed] [Google Scholar]
- 66.Gao R, Cao B, Hu Y, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368:1888–1897. doi: 10.1056/NEJMoa1304459. Epub 2013 Apr 11. [DOI] [PubMed] [Google Scholar]
- 67.Zhang XA, Li H, Jiang FC, et al. A zoonotic henipavirus in febrile patients in China. N Engl J Med. 2022;387:470–472. doi: 10.1056/NEJMc2202705. [DOI] [PubMed] [Google Scholar]
- 68.Lederberg J, McCray AT.. Ome sweet ‘omics – a genealogical treasury of words. The Scientist. 2001;15:8. [Google Scholar]
- 69.Ko KKK, Chng KR, Nagarajan N.. Metagenomics-enabled microbial surveillance. Nat Microbiol. 2022;7:486–496. doi: 10.1038/s41564-022-01089-w. Epub 2022 Apr 1. [DOI] [PubMed] [Google Scholar]
- 70.Carbo EC, Sidorov IA, Zevenhoven-Dobbe JC, et al. Coronavirus discovery by metagenomic sequencing: a tool for pandemic preparedness. J Clin Virol. 2020;131:104594. doi: 10.1016/j.jcv.2020.104594. Epub 2020 Aug 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wilson MR, Naccache SN, Samayoa E, et al. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N Engl J Med. 2014;370:2408–2417. doi: 10.1056/NEJMoa1401268. Epub 2014 Jun 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Raymond S, Nicot F, Carcenac R, et al. HIV-1 genotypic resistance testing using the vela automated next-generation sequencing platform. J Antimicrob Chemother. 2018;73:1152–1157. doi: 10.1093/jac/dky003. [DOI] [PubMed] [Google Scholar]
- 73.Stratton CW, Tang YW.. Diagnosing bacteremia in real time using next-generation sequencing-based technology. J Mol Diagn. 2020;22:301–303. doi: 10.1016/j.jmoldx.2020.01.002. Epub 2020 Jan 21. [DOI] [PubMed] [Google Scholar]
- 74.Wilson MR, Sample HA, Zorn KC, et al. Clinical metagenomic sequencing for diagnosis of meningitis and encephalitis. N Engl J Med. 2019;380:2327–2340. doi: 10.1056/NEJMoa1803396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stratton CW, Schutzbank TE, Tang YW.. Use of metagenomic next-generation sequencing in the clinical microbiology laboratory: A step forward, but not an end-all. J Mol Diagn. 2021;23:1415–1421. doi: 10.1016/j.jmoldx.2021.09.003. [DOI] [PubMed] [Google Scholar]
- 76.Kiechle FL, Holland-Staley CA.. Genomics, transcriptomics, proteomics, and numbers. Arch Pathol Lab Med. 2003;127:1089–1097. doi: 10.5858/2003-127-1089-GTPAN. [DOI] [PubMed] [Google Scholar]
- 77.Mdivani N, Li H, Akhalaia M, et al. Monitoring therapeutic efficacy by real-time detection of mycobacterium tuberculosis mRNA in sputum. Clin Chem. 2009;55:1694–1700. doi: 10.1373/clinchem.2009.124396. Epub 2009 Jul 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Koo S, Thomas HR, Daniels SD, et al. A breath fungal secondary metabolite signature to diagnose invasive aspergillosis. Clin Infect Dis. 2014;59:1733–1740. doi: 10.1093/cid/ciu725. Epub 2014 Oct 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.La Rosa R, Johansen HK, Molin S.. Convergent metabolic specialization through distinct evolutionary paths in Pseudomonas aeruginosa. mBio. 2018;9:e00269–18. doi: 10.1128/mBio.00269-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Horrocks V, Hind CK, Wand ME, et al. Nuclear magnetic resonance metabolomics of symbioses between bacterial vaginosis-associated bacteria. mSphere. 2022;7:e0016622. doi: 10.1128/msphere.00166-22. Epub 2022 May 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Slupsky CM, Rankin KN, Fu H, et al. Pneumococcal pneumonia: potential for diagnosis through a urinary metabolic profile. J Proteome Res. 2009;8:5550–5558. doi: 10.1021/pr9006427. [DOI] [PubMed] [Google Scholar]
- 82.Sweeney TE, Braviak L, Tato CM, et al. Genome-wide expression for diagnosis of pulmonary tuberculosis: a multicohort analysis. Lancet Respir Med. 2016;4:213–224. doi: 10.1016/S2213-2600(16)00048-5. Epub 2016 Feb 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.van Houten CB, de Groot JAH, Klein A, et al. A host-protein based assay to differentiate between bacterial and viral infections in preschool children (OPPORTUNITY): a double-blind, multicentre, validation study. Lancet Infect Dis. 2017;17:431–440. doi: 10.1016/S1473-3099(16)30519-9. Epub 2016 Dec 22. [DOI] [PubMed] [Google Scholar]
- 84.Zhang H, Ai JW, Yang W, et al. Metatranscriptomic characterization of coronavirus disease 2019 identified a host transcriptional classifier associated With immune signaling. Clin Infect Dis. 2021;73:376–385. doi: 10.1093/cid/ciaa663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Moreira FMF, Verma R, Pereira Dos Santos PC, et al. Blood-based host biomarker diagnostics in active case finding for pulmonary tuberculosis: A diagnostic case-control study. EClinicalMedicine. 2021;33:100776. doi: 10.1016/j.eclinm.2021.100776. eCollection 2021 Mar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Södersten E, Ongarello S, Mantsoki A, et al. Diagnostic accuracy study of a novel blood-based assay for identification of tuberculosis in people living with HIV. J Clin Microbiol. 2021;59:e01643–20. doi: 10.1128/JCM.01643-20. Print 2021 Feb 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sutherland JS, van der Spuy G, Gindeh A, et al. Diagnostic accuracy of the cepheid 3-gene host response fingerstick blood test in a prospective, multi-site study: interim results. Clin Infect Dis. 2022;74:2136–2141. doi: 10.1093/cid/ciab839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zimmer AJ, Schumacher SG, Södersten E, et al. A novel blood-based assay for treatment monitoring of tuberculosis. BMC Res Notes. 2021;14:247. doi: 10.1186/s13104-021-05663-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Oved K, Cohen A, Boico O, et al. A novel host-proteome signature for distinguishing between acute bacterial and viral infections. PLoS One. 2015;10:e0120012. doi: 10.1371/journal.pone.0120012. eCollection 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Papan C, Argentiero A, Porwoll M, et al. A host signature based on TRAIL, IP-10, and CRP for reducing antibiotic overuse in children by differentiating bacterial from viral infections: a prospective, multicentre cohort study. Clin Microbiol Infect. 2021;10:00621–00622. [DOI] [PubMed] [Google Scholar]
- 91.Boyd JH, Russell JA, Fjell CD.. The meta-genome of sepsis: host genetics, pathogens and the acute immune response. J Innate Immun. 2014;6:272–283. doi: 10.1159/000358835. Epub 2014 Feb 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Taur Y, Pamer EG.. Microbiome mediation of infections in the cancer setting. Genome Med. 2016;8:40. doi: 10.1186/s13073-016-0306-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Libertucci J, Young VB.. The role of the microbiota in infectious diseases. Nat Microbiol. 2019;4:35–45. doi: 10.1038/s41564-018-0278-4. Epub 2018 Dec 13. [DOI] [PubMed] [Google Scholar]
- 94.Gebrayel P, Nicco C, Al Khodor S, et al. Microbiota medicine: towards clinical revolution. J Transl Med. 2022;20:111. doi: 10.1186/s12967-022-03296-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Eck A, de Groot EFJ, de Meij TGJ, et al. Robust microbiota-based diagnostics for inflammatory bowel disease. J Clin Microbiol. 2017;55:1720–1732. doi: 10.1128/JCM.00162-17. Epub 2017 Mar 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hatch A, Horne J, Toma R, et al. A robust metatranscriptomic technology for population-scale studies of diet, gut microbiome, and human health. Int J Genomics. 2019;2019:1718741. doi: 10.1155/2019/1718741. eCollection 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chiu HR, Hwang CK, Chen SY, et al. Machine learning for emerging infectious disease field responses. Sci Rep. 2022;12:328. doi: 10.1038/s41598-021-03687-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Goodswen SJ, Barratt JLN, Kennedy PJ, et al. Machine learning and applications in microbiology. FEMS Microbiol Rev. 2021;45:fuab015. doi: 10.1093/femsre/fuab015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wong OGW, Ng IFY, Tsun OKL, et al. Machine learning interpretation of extended human papillomavirus genotyping by onclarity in an Asian cervical cancer screening population. J Clin Microbiol. 2019;57:e00997–19. doi: 10.1128/JCM.00997-19. Print 2019 Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Anahtar MN, Yang JH, Kanjilal S.. Applications of machine learning to the problem of antimicrobial resistance: an emerging model for translational research. J Clin Microbiol. 2021;59:e0126020. doi: 10.1128/JCM.01260-20. Epub 2021 Jun 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nguyen M, Long SW, McDermott PF, et al. Using machine learning To predict antimicrobial MICs and associated genomic features for nontyphoidal salmonella. J Clin Microbiol. 2019;57:e01260–18. doi: 10.1128/JCM.01260-18. Print 2019 Feb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Raoult D, Fournier PE, Drancourt M.. What does the future hold for clinical microbiology? Nat Rev Microbiol. 2004;2:151–159. doi: 10.1038/nrmicro820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cobb B, Simon CO, Stramer SL, et al. The cobas® 6800/8800 system: a new era of automation in molecular diagnostics. Expert Rev Mol Diagn. 2017;17:167–180. doi: 10.1080/14737159.2017.1275962. [DOI] [PubMed] [Google Scholar]
- 104.Hirschhorn JW, Kegl A, Dickerson T, et al. Verification and validation of SARS-CoV-2 assay performance on the Abbott m2000 and alinity m systems. J Clin Microbiol. 2021;59:e03119–20. doi: 10.1128/JCM.03119-20. Print 2021 Apr 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Stellrecht KA, Cimino JL, Wilson LI, et al. Panther fusion® respiratory virus assays for the detection of influenza and other respiratory viruses. J Clin Virol. 2019;121:104204. doi: 10.1016/j.jcv.2019.104204. Epub 2019 Nov 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dalpke AH, Hofko M, Zorn M, et al. Evaluation of the fully automated BD MAX Cdiff and Xpert C. difficile assays for direct detection of clostridium difficile in stool specimens. J Clin Microbiol. 2013;51:1906–1908. doi: 10.1128/JCM.00344-13. Epub 2013 Mar 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jang D, Ratnam S, Gilchrist J, et al. Comparison of workflow, maintenance, and consumables in the GeneXpert infinity 80 and panther instruments while testing for Chlamydia trachomatis and Neisseria gonorrhoeae. Sex Transm Dis. 2016;43:377–381. doi: 10.1097/OLQ.0000000000000444. [DOI] [PubMed] [Google Scholar]
- 108.Loeffelholz MJ, Tang YW.. Detection of SARS-CoV-2 infections at the point of care. Bioanalysis. 2021;13:1213–1223. doi: 10.4155/bio-2021-0078. Epub 2021 Jul 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chen H, Liu K, Li Z, et al. Point of care testing for infectious diseases. Clin Chim Acta. 2019;493:138–147. doi: 10.1016/j.cca.2019.03.008. Epub 2019 Mar 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Drancourt M, Michel-Lepage A, Boyer S, et al. The point-of-care laboratory in clinical microbiology. Clin Microbiol Rev. 2016;29:429–447. doi: 10.1128/CMR.00090-15. [DOI] [PMC free article] [PubMed] [Google Scholar]