Abstract
Background:
Long noncoding RNAs (lncRNAs) have been recognized as the main modulatory molecules in various cancers and perform as competing endogenous RNAs (ceRNAs). The nuclear hormone receptor superfamily of ligand-activated transcription factors (NR3C1) regulates numerous proliferative and metabolic processes such as tumorigenesis and metabolic diseases. Furthermore, X-linked inhibitor of apoptosis protein (XIAP) belongs to a family of the inhibitors of apoptosis proteins, is located downstream of the glucocorticoid receptor (GR or NR3C1) pathway, and cooperates with GR to suppress apoptosis. However, the underlying mechanisms of NR3C1 and XIAP in colorectal cancer (CRC) remain mainly unclear. This research aims to clarify the potential RNA biomarkers and to construct a novel ceRNA network in CRC.
Materials and Methods:
Multistep bioinformatics methods such as Lnc2cancer and miRDB databases were applied to identify candidate lncRNAs and miRNAs. The interaction energy between lncRNAs, NR3C1, and XIAP genes was analyzed by the LncRRIsearch database. Plus, microRNAs and lncRNA were evaluated via the Diana tools database to select microRNAs with the most binding scores. Quantitative reverse transcription–polymerase chain reaction (QRT-PCR) was applied to verify RNA molecules’ expression levels and their association with the clinicopathological factors in 30 CRC tissues compared to 30 adjacent tissues.
Results:
QRT-PCR showed upregulation of KCNQ1OT1, NR3C1, and XIAP and downregulation of miR-421. The ceRNA network was constructed with 17 lncRNAs, 2 mRNAs, and 42 miRNAs. Thus, we explained the potential interactions between KCNQ1OT1 and miR-421 with NR3C1 and XIAP genes.
Conclusion:
Our study represents potential prognostic biomarkers and a new ceRNA network for further study in CRC.
Keywords: Colorectal cancer, competing endogenous RNA, KCNQ1OT1, miR-421, X-linked inhibitor of apoptosis protein, NR3C1
INTRODUCTION
Colorectal cancer (CRC) is the third most frequent kind of cancer and the fourth leading cause of cancer-related death worldwide.[1,2,3] Consequently, it is critical to comprehend the precise molecular processes behind carcinogenesis and to develop novel biomarkers and molecular networks capable of significantly improving therapy efficacy.[4] Long noncoding RNAs (lncRNAs) are described as transcripts having a length of <200 nucleotides. They are involved in a variety of biological processes, including chromosomal remodeling, epigenetics control, the immunological response, and apoptosis, as well as acting as competing endogenous RNAs (ceRNAs) or miRNA sponges, among others. lncRNAs influence gene expression via collaborating with mRNAs to share miRNA.[5] Several studies have shown that lncRNAs are more often expressed in various organs and malignancies, including hepatocellular carcinoma, nonsmall cell lung cancer, human breast cancer, and CRC.[4,6,7,8] A worse survival rate has been indicated by the upregulation of lncRNA FOXD3-AS1 in CRC patients. The downregulation of FOXD3-AS1 has been associated with invasiveness, cell proliferation, and metastasis, according to certain theories.[9] Furthermore, miRNAs are ncRNAs that may regulate the expression of target mRNAs and affect many cell physiological processes including metabolism, cell proliferation, invasion, and death. Aberrant miRNAs also contribute to the development and progression of numerous kinds of malignancies.[10,11] miRNA-149 may be utilized as a method for early diagnosis in CRC.[12] NR3C1 is a nuclear hormone receptor superfamily of ligand-activated transcription factors.[13] NR3C1 has a key function in regulating various proliferative and metabolic processes, including cancer and metabolic disorders, such as diabetes and obesity. NR3C1 serves as a TF that binds to GC response elements in the target genes, both for the mitochondrial and nuclear DNA.[14] In metastatic CRC cells, the glucocorticoid-GR-CDK1 signaling pathway acts as a proliferation mediator.[15] The inhibitors of apoptosis proteins (IAPs) family includes the X-linked Inhibitor of Apoptosis Protein (XIAP). XIAP has been discovered to be overexpressed in a variety of malignancies.[16] For instance, a previous research indicated that XIAP expression was increased in breast cancer, and its level of expression might be regarded as a novel biomarker in breast cancer.[17]
Numerous researches have examined the profiles and roles of lncRNAs in CRC, but few have examined aberrant lncRNAs associated with sex, age, stage, or other clinical variables. In addition, less studies have been conducted to establish a putative ceRNA network in CRC. To address the above problems, we evaluated and constructed a new ceRNA network based on NR3C1 and XIAP mRNAs using several bioinformatics techniques and datasets. To confirm our results’ reproducibility, we assessed the profiles of lncRNA, miRNA, and mRNA from the ceRNA network by quantitative reverse transcription–polymerase chain reaction (qRT-PCR). The link between the expression levels and the clinicopathological characteristics of these RNAs was also identified by qRT-PCR. We also offered a notion that the different RNAs in the newly produced ceRNA in this work may interact with each other, resulting in inhibiting apoptosis. However, additional investigations are required to establish the underlying processes of this idea.
MATERIALS AND METHODS
Screening of long noncoding RNAs in colorectal cancer
All candidate lncRNAs were selected with the following criteria: (1) high differentially expressed levels in CRC and (2) their roles already admitted by quantitative PCR (qPCR), Western blot, and luciferase assay methods in CRC. LncRNAs were selected from the Lnc2cancer database «http://www.bio-bigdata.net/lnc2cancer2.0».[18] Then, those with the two mentioned criteria were evaluated by the LncRRIsearch database «http://rtools. cbrc. jp/LncRRIsearch/glist. cgi,»[19] to find the lncRNA which had the most interaction energy with NR3C1 and XIAP genes.
Prediction of miRNA-mRNA and miRNA-long noncoding RNA interaction
miRdb database «http://www.mirdb.org» was used to identify the target microRNAs of NR3C1 and XIAP genes with a target score above 50. Common microRNAs between NR3C1 and XIAP genes that had 0.2 < expression levels were adopted by the cBioPortal database. Finally, the correlation between microRNAs and lncRNA was evaluated via the Diana tools database «http://carolina.imis.athenainnovation.gr/diana_tools» to select microRNAs with the most binding scores. Figure 1 illustrates the framework used in this research.
Figure 1.
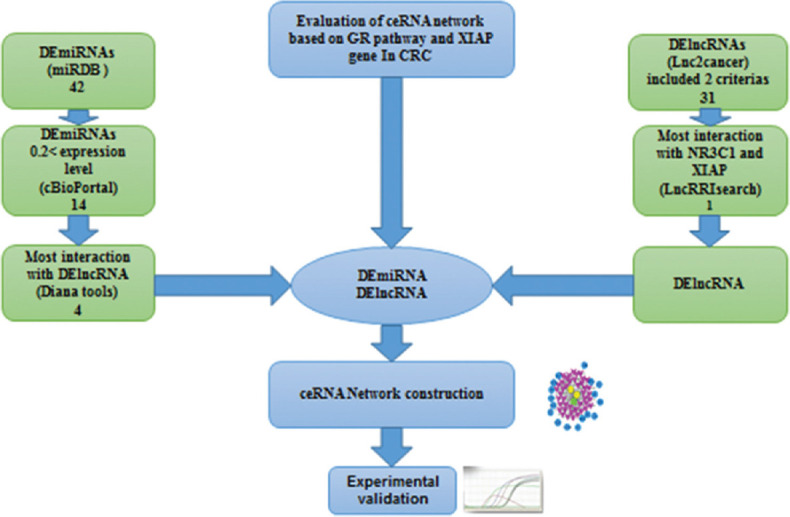
Bioinformatics analysis flowchart of the current research
Competing endogenous RNAs network construction
ceRNA network is a complex posttranscriptional modulatory network. ceRNA network is based on this theory, via using shared microRNA response elements, LncRNAs perform as miRNA sponges to modulate the expression of mRNAs competitively. A ceRNA network in terms of the recognized prognostic lncRNAs, miRNAs, and mRNAs was established. By using miRDB database to identify miRNA-mRNA interactions and Lnc2cancer database to identify LncRNA-mRNA interactions. We constructed and visualized the coexpression network using Cytoscape 3.7.1.
Tissue collection
CRC tissues and their paired adjacent nontumor samples were collected from 30 patients with CRC in different age groups (aged 29–87 years) at the Poursina Hakim Gastrointestinal Disease Research Center (60 samples in total). When people referred to Poursina Hakim Institute with different CRC symptoms, colonoscopy and clinical tests were used to diagnose the CRC patients from healthy people. Hence, the patients were selected quite randomly. All patients have not received radiotherapy or chemotherapy before tissue sampling. Then, tissues were transferred and frozen in liquid nitrogen directly. Tumor samples were admitted by a pathologist to decline heterogeneity. Written informed consent was acquired from patients, and this study was authenticated by the Ethics Boards of the Isfahan University of Medical Sciences.
Quantitative reverse transcription–polymerase chain reaction analyses
TRIzol reagent was used (Invitrogen Life Technologies, Carlsbad, CA) to extract the total RNA from sample tissues. Agarose gel electrophoresis was applied to estimate RNA integrity, and thermo ND-1000 ultramicro nucleic acid protein meter (OD 260 nm; Thermo, Waltham, MA) was used to evaluate RNA quantity. cDNA was constructed using a Transcriptor First Strand cDNA Synthesis Kit (Bio fact, Korea), and 9 μl of total RNA for each sample was applied. Expression analysis was performed by real-time PCR with SYBR Green (Bio fact, Korea) and using the Rotor-Gene 6000 system, with the following conditions: (a) 95°C for 15 min; (b) 40 cycles of 95°C for 20 s and 60°C for 30 s; and (c) 72°C for 30 s. Then, the relative gene expression was calculated using the 2−ΔΔCt method normalized to GAPDH. Plus, each sample was examined in triplicate.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 9 and also to plot the figures. A two-tailed P < 0.05 was regarded as statistically noticeable. The samples parametric and nonparametric t-test, Mann–Whitney test, and Wilcoxon matched-pairs signed rank test were applied to identify the difference in gene expression between the two groups and investigating the correlation between expression of RNA molecules with clinicopathologic features. All the hypothesis testing is two-sided.
RESULTS
KCNQ1OT1 was upregulated in colorectal cancer tissues
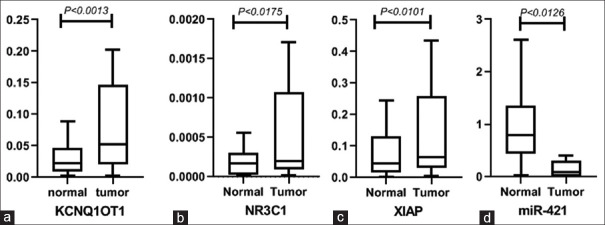
After analyzing all the lncRNAs, 31 lncRNAs matched the two key criteria: (1) they were highly differentially expressed in CRC and (2) their involvement in CRC had previously been shown by qPCR, Western blot, and luciferase assay methodologies [Table 1]. LncRNAs were discovered via the use of the LncRRIsearch database, which was used to identify which lncRNAs had the highest base-pairing interaction energies with the NR3C1 and XIAP Genes, respectively. KCNQ1OT1 exhibited the lowest base-pairing interaction energies with the NR3C1 and XIAP genes (NR3C1 = −671.34 kcal/mol) and the highest base-pairing interaction energies with the NR3C1 and XIAP genes (NR3C1 = −2228.15 kcal/mol). This work focused on KCNQ1OT1 to facilitate future research. With (log2 fold change = 2.5), QRT-PCR analysis revealed that KCNQ1OT1 was overexpressed in tumor tissues as compared to neighboring nontumor tissues [Figure 2a; 95% confidence interval (CI): −1.956 to −0.5262, P = 0.0013] with (log2 fold change = 2.5). Males exhibited (P = 0.038205) and females demonstrated (P = 0.013) a positive connection between KCNQ1OT1 upregulation and gender in CRC, according to the statistical analysis. KCNQ1OT1 expression did not seem to have a statistically significant relationship with any of the other clinical variables [Table 2].
Table 1.
Evaluation of local baseEvaluation of local base-pairing interaction energies with nuclear hormone receptor superfamily of ligandactivated transcription factors and X-linked inhibitor of apoptosis protein genes
| lncRNA | Number of local base-pairing interactions predicted by RIblast (NR3C1) | Total of local base-pairing interaction energies with NR3C1 gene (kcal/mol) | Number of local base-pairing interactions predicted by RIblast (XIAP) | Total of local base-pairing interaction energies with XIAP gene (kcal/mol) |
|---|---|---|---|---|
| ANRIL | No interaction | No interaction | No interaction | No interaction |
| B3GALT5-AS1 | No interaction | No interaction | −21.47 | 1 |
| CCAT1 | −33.12 | 2 | −36.9 | 2 |
| circ_0079662 | No interaction | No interaction | No interaction | No interaction |
| circACAP2 | No interaction | No interaction | No interaction | No interaction |
| CYTOR | No interaction | No interaction | No interaction | No interaction |
| DACOR1 | No interaction | No interaction | No interaction | No interaction |
| FER1L4 | No interaction | No interaction | No interaction | No interaction |
| H19 | −122.8 | 6 | −33.62 | 2 |
| HNF1A-AS1 | −17.07 | 1 | −225.3 | 8 |
| HOTAIR | −16.95 | 1 | No interaction | No interaction |
| HULC | No interaction | No interaction | No interaction | No interaction |
| KCNQ1OT1 | −671.34 | 36 | −2228.15 | 84 |
| LEF1-AS1 | No interaction | No interaction | −540.91 | 15 |
| LINC-ROR | −37.78 | 2 | No interaction | No interaction |
| LINC00152 | No interaction | No interaction | No interaction | No interaction |
| LINC00662 | No interaction | No interaction | −499.31 | 13 |
| LINC00858 | No interaction | No interaction | No interaction | No interaction |
| LINC01234 | −35.3 | 2 | −170.7 | 7 |
| LNAPPCC | No interaction | No interaction | No interaction | No interaction |
| LOC285194 | No interaction | No interaction | No interaction | No interaction |
| MALAT1 | −16.74 | 1 | No interaction | No interaction |
| MBNL1-AS1 | −167.04 | 9 | −115.75 | 5 |
| NEAT1 | −52.12 | 3 | 89.77 | 5 |
| OIP5-AS1 | −18.71 | 1 | 1104.41 | 40 |
| PVT1 | No interaction | No interaction | −482.85 | 16 |
| SNHG16 | −33.66 | 2 | −763.3 | 28 |
| TUG1 | −16.51 | 1 | No interaction | No interaction |
| UCA1 | No interaction | No interaction | No interaction | No interaction |
| XIRP2-AS1 | No interaction | No interaction | No interaction | No interaction |
| XIST | −539.14 | 28 | −284.81 | 15 |
NR3C1=Nuclear hormone receptor superfamily of ligand-activated transcription factors; XIAP=X-linked inhibitor of apoptosis protein; lncRNA=Long noncoding RNAs
Figure 2.
(a-d) aberrant expression levels of KCNQ1OT1, NR3C1, XIAP, and miR-421 in tumor tissues compared to normal samples evaluated by qPCR, respectively. (a) The mean of the tumor and normal groups are −0.3918 and 0.8495. The standard error of the tumor and normal groups are 0.3771 and 0.3493. (b) The mean of the tumor and normal groups are −1.547 and -0.1197. The standard error of the tumor and normal groups are 0.5343 and 0.4275. (c) The mean of the tumor and normal groups are −1.081 and −0.01075. The standard error of the tumor and normal groups are 0.4134 and 0.4036. (d) The mean of the tumor and normal groups are 1.576 and −0.3578. The standard error of the tumor and normal groups are 0.7161 and 0.5280. XIAP: X-linked inhibitor of apoptosis protein, qPCR: Quantitative polymerase chain reaction
Table 2.
Correlation between expression of KCNQ1OT1 in colorectal cancer with clinicopathologic features
| Clinicopathological parameters | Number of cases | KCNQ1OT1 | P | |
|---|---|---|---|---|
|
| ||||
| Low | High | |||
| Total | 30 | 23 | 7 | 0.0013 |
| Age (years) | ||||
| ≤60 | 9 | 6 | 3 | 0.887 |
| >60 | 21 | 17 | 4 | |
| Gender | ||||
| Female | 15 | 9 | 6 | 0.013 |
| Male | 15 | 14 | 1 | 0.05 |
| Location | ||||
| Colon | 20 | 14 | 6 | 0.4457 |
| Rectum | 10 | 9 | 1 | |
| Differentiation | ||||
| Well/moderately | 24 | 18 | 6 | 0.4361 |
| Poorly | 6 | 5 | 1 | |
| TNM stage | ||||
| I–II | 15 | 10 | 5 | 0.583 |
| III–IV | 15 | 13 | 2 | |
TNM=Tumor, nodes, and metastases
NR3C1 and X-linked inhibitor of apoptosis protein were upregulated in colorectal cancer tissues, and XIAP predicted a positive association with gender
According to the findings, NR3C1 expression was significantly higher in CRC tissues when compared to neighboring nontumor pairs [Figure 2b; 95% CI: −2.586 to −0.2689; P = 0.0175] and (log2 fold change: 7.7%). The relationship between clinical characteristics such as gender, stage, location, and differentiation and overexpression of NR3C1 was investigated to discover a beneficial relationship. There were, however, no correlations found between these characteristics and the expression of NR3C1.
QRT PCR analysis revealed that XIAP expression was significantly higher in CRC tissues as compared to nontumor samples (95% CI: −1.866 −0.2748; P < 0.0101) and (log2 fold change = 2.85) in CRC tissues [Figure 2c]. There was no association between these clinical indicators and the expression of XIAP, as shown by the multivariate analysis of the data.
miR-421 expression was downregulated in colorectal cancer tissues
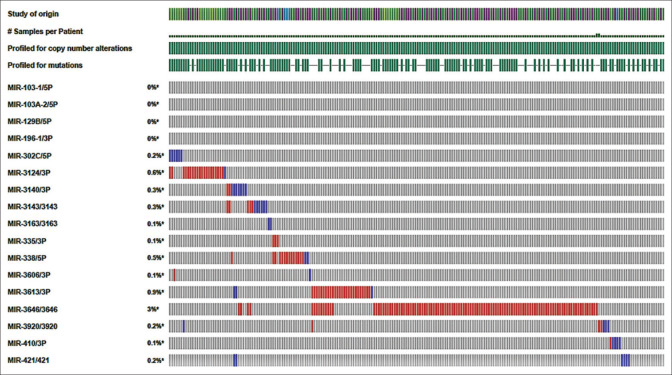
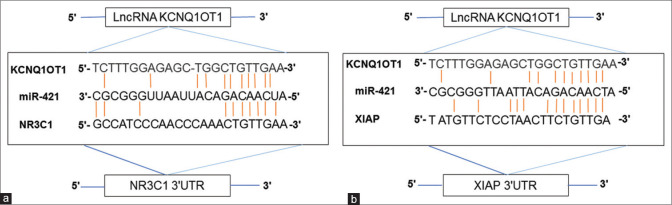
Complementary bioinformatics analysis, such as those performed on the miRDB database, were used to forecast which NR3C1 and XIAP targets will be targeted. The cBioPortal database then identified 14 miRNAs that were shared by both NR3C1 and XIAP mRNAs and were chosen based on their expression levels of more than 0.2 in the cBioPortal database [Figure 3]. In this study, the interactions of miRNAs with KCNQ1OT1 were examined using the Diana tools database (http://carolina. imis.athenainnovation.gr/dianatools/web/index.php? r = lncbasev2 percent 2Findex-predicted) and the lncbasev2 percent 2Findex-predicted [Table 3]. The miR-302c-5p, miR-3920, miR-421, and miR-580-5p obtained the highest binding scores (above 0.013) with KCNQ1OT1, followed by miR-3920 and miR-421. In this respect, we concentrated our efforts on miR-421 for future investigation due to the shown role of miR-421 in various malignancies as compared to other miRNAs. The miRDB database was able to identify the binding sites between miR-421 and the transcription factors KCNQ1OT1/NR3C1/XIAP [Figure 4a and b]. Using quantitative reverse transcription–polymerase chain reaction (QRT-PCR), we discovered that miR-421 was downregulated in tumor tissues as compared to their corresponding normal tissues (P = 0.0126 and log2 fold change = 0.44) [Figure 2d]. A positive correlation between miR-421 expression and the risk of prostate cancer in males was also found (P = 0.031), although this was not statistically significant. There was no association found between other clinical features and low miR-421 expression in CRC.
Figure 3.
The heatmap of 17 differentially expressed miRNAs identified by the cBioPortal database
Table 3.
The binding score of microRNAs to KCNQ1OT1
| MicroRNAs | Expression level in CRC | Binding category (mer) | Binding score with KCNQ1OT1 |
|---|---|---|---|
| Has-mir-302c-5p | 0.2 | 8 | 0.016 |
| Has-mir-3124-3p | 0.6 | 8 | 0.009 |
| Has-mir-3140-3p | 0.3 | 7 | 0.011 |
| Has-mir-3143 | 0.3 | 8 | 0.009 |
| Has-mir-338-5p | 0.5 | 8 | 0.012 |
| Has-mir-3613-3p | 0.9 | 7 | 0.017 |
| Has-mir-3646 | 3 | 7 | 0.011 |
| Has-mir-3920 | 0.2 | 8 | 0.013 |
| Has-mir-421 | 0.2 | 8 | 0.013 |
| Has-mir-4666a-3p | 0.3 | 8 | 0.008 |
| Has-mir-4789-5p | 0.3 | 8 | 0.007 |
| Has-mir-548t-3p | 0.3 | 8 | 0.009 |
| Has-mir-593-5p | 0.2 | 7 | 0.005 |
| Has-mir-580-5p | 0.4 | 8 | 0.015 |
CRC=Colorectal cancer
Figure 4.
(a and b) The binding sites of KCNQ1OT1/miR-421/NR3C1 and KCNQ1OT1/miR-421/XIAP were identified with miRDB. XIAP: X-linked inhibitor of apoptosis protein
Construction of the global -long noncoding RNA-miRNA-mRNA triple network
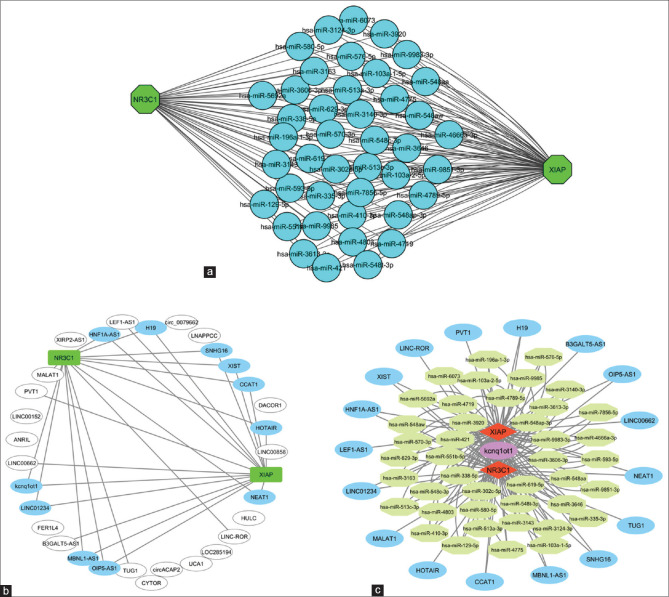
Lnc2cancer and miRDB databases were consulted in order to validate the interaction link between lncRNA, microRNA, and messenger RNA indicators in order to develop the ceRNA network. A total of 42 common miRNAs interact with the NR3C1 and XIAP mRNAs, according to the findings [Figure 5a]. Furthermore, 31 long noncoding RNAs (LncRNAs) were isolated from Lnc2cancer [Figure 5b]. As seen in Figure 5c, miRNA-mRNA and lncRNA-miRNA pairings were merged to form a unique global triple network, which was then visualized.
Figure 5.
The ceRNA network in colorectal cancer. (a) Subnetwork of the interactions between common miRNAs with NR3C1 and XIAP. (b) The subnetwork of lncRNAs and NR3C1 and XIAP mRNAs. (c) The lncRNA–miRNA–mRNA ceRNA network. Octagonal nods indicate lncRNAs, and polygon nodes indicate miRNAs, and Circle nods indicate mRNAs. ceRNA: competing endogenous RNAs, XIAP: X-linked inhibitor of apoptosis protein, lncRNAs: Long noncoding RNAs
DISCUSSION
Several studies have revealed that lncRNAs are involved in miRNA-response elements and may compete for miRNAs with mRNAs. Therefore, lncRNAs may behave as ceRNAs and are related with many biological processes including cancer. lncRNAs have essential benefits, such as diagnostic and prognostic biomarkers than protein-coding genes. Many studies have found that the aberrantly expressed lncRNAs are closely connected with the prognosis and pathophysiology of cancers and may be employed as tumor-related predictors.[20] According to Chen et al., the lncRNA XIST was found to be overexpressed in CRC tumors and was associated with poor overall survival; furthermore, it was discovered that XIST could modulate CRC metastasis and development by competing with the miR200b for the expression of the zinc finger E-box binding homeobox 1.[21] KCNQ1OT1 was shown to be increased in tumor tissues when compared to neighboring normal samples in this investigation, and it has the potential to be used as a prognostic marker in CRC. Another research found that KCNQ1OT1 was overexpressed in CRC cells and tissues, which was consistent with the findings of this one. Knockdown of KCNQ1OT1 lowered CRC invasiveness, migration, and cell proliferation abilities, hindered the cell cycle, and resulted in the development of CRC cell death via inactivation of the PI3K/AKT signal pathway. As a result, KCNQ1OT1 has been proposed as a possible predictive biomarker for CRC.[22] Overexpression of KCNQ1OT1 in CRC was shown to have a positive connection with gender, indicating that girls were more likely than men to have the overexpression of KCNQ1OT1. As a result, upregulation of KCNQ1OT1 might be considered a predictive biomarker for early detection of CRC, particularly in females of both sexes. However, the relevance of KCNQ1OT1 in CRC, as well as the underlying mechanisms that underlie it, are still poorly known.
In this study, miR-421 was shown to be downregulated in tumor tissues when compared to normal samples, suggesting that it may be used as a predictive biomarker and therapeutic target in CRC, especially in males. In a similar vein, Xue et al. discovered that miR-421 was expressed at low levels in CRC. Furthermore, miR-421 has the potential to inhibit CRC invasion and migration through modulating MAT1, which is responsible for the antitumor impact. As a result, the miR-421/MAT1 axis may be considered as a therapeutic target in CRC.[21] In contrast, Zhou et al. found that miR-421 was elevated in cancer tissues when compared to matching paracancerous tissues, and that this resulted in miR-421 having an antiapoptotic effect in CRC.[22] As a result, more investigations should be conducted to determine the precise involvement of miR-421 in CRC.
When comparing tumor tissues to normal samples, the NR3C1 and XIAP genes were found to be overexpressed. In a similar vein, Tian et al. demonstrated that the amount of GR expression in the nucleus was much higher in metastatic T84 cells. Tian demonstrated that GR depletion inhibited the growth of metastatic colon cancer cells.[13] Qiao et al., on the other hand, revealed that the deletion of XIAP significantly increased the sensitivity of CRC cells to PPAR, resulting in a reduction in cell proliferation as well as ligand-induced apoptosis. Accordingly, it is possible that activation of PPAR and inhibition of XIAP at the same time in colon cancer would have a synergistic anticancer impact.[23] Upregulation of the genes XIAP and NR3C1 may be used as prognostic and diagnostic biomarkers in CRC.
The bioinformatic research revealed that KCNQ1OT1 may interact with the transcription factors XIAP and NR3C1, as well as direct interactions between KCNQ1OT1 and the microRNA miR-421. Furthermore, miR-421 was shown to interact with the transcription factors XIAP and NR3C1. We propose the notion that KCNQ1OT1 may have an effect on the expression of XIAP and NR3C1 via the miR-421 gene. QRT-PCR study revealed that KCNQ1OT1, XIAP, and NR3C1 were overexpressed in CRC tissues, whereas miR-421 was found to be underexpressed. These findings support our hypothesis that KCNQ1OT1 directly interacted with miR-421, resulting in the downregulation of miR-421 in a dose-dependent manner. On the other hand, the transcription of XIAP and NR3C1 was increased as a result of the activity of KCNQ1OT1. However, further research is needed to fully understand the processes and mechanisms that are involved in a uniquely built ceRNA network in CRC.
Overall, this study has developed a comprehensive bioinformatics analysis of CRC. It also looked at several biomarkers for CRC. Additional genetic research, a larger cohort, and clinical procedure validation would all be beneficial to these findings; nonetheless, we are hopeful that our findings will lead to the identification of new prognostic and therapeutic targets for the treatment of CRC.
CONCLUSION
Our results suggest a new approach to identifying lncRNAs, microRNAs, and mRNAs as biomarkers for the progression, modulation, and prognosis of CRC. The current work used QRT-PCR to establish that the candidate genes’ expression was dysregulated. RNA molecules were also linked to clinicopathological traits, which we looked at to see whether that data may be used in future studies. With NR3C1 and XIAP mRNAs as the foundation, an entirely new ceRNA network has been built to study the possible roles of suspected CRC genes. It also aids in understanding the genetic basis of CRC. In order to confirm the link between CRC and the given RNA molecules and to identify novel therapeutic targets, more functional studies of the indicated lncRNA, miRNA, and mRNAs are necessary.
Financial support and sponsorship
Simin Hemati (M.D) Associate Professor of Radiooncology.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Dai GP, Wang LP, Wen YQ, Ren XQ, Zuo SG. Identification of key genes for predicting colorectal cancer prognosis by integrated bioinformatics analysis. Oncol Lett. 2020;19:388–98. doi: 10.3892/ol.2019.11068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Safari M, Mahjub H, Esmaeili H, Abbasi M, Roshanaei G. Specific causes of recurrence after surgery and mortality in patients with colorectal cancer: A competing risks survival analysis. J Res Med Sci. 2021;26:13. doi: 10.4103/jrms.JRMS_430_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao J, Xu J, Shang AQ, Zhang R. A six-LncRNA expression signature associated with prognosis of colorectal cancer patients. Cell Physiol Biochem. 2018;50:1882–90. doi: 10.1159/000494868. [DOI] [PubMed] [Google Scholar]
- 4.Bian Z, Zhang J, Li M, Feng Y, Wang X, Zhang J, et al. LncRNA-FEZF1-AS1 promotes tumor proliferation and metastasis in colorectal cancer by regulating PKM2 signaling. Clin Cancer Res. 2018;24:4808–19. doi: 10.1158/1078-0432.CCR-17-2967. [DOI] [PubMed] [Google Scholar]
- 5.Jafari D, Noorbakhsh F, Delavari A, Tavakkoli-Bazzaz J, Farashi-Bonab S, Abdollahzadeh R, et al. Expression level of long noncoding RNA NKILAmiR103-miR107 inflammatory axis and its clinical significance as potential biomarker in patients with colorectal cancer. J Res Med Sci. 2020;25:41. doi: 10.4103/jrms.JRMS_943_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He M, Lin Y, Xu Y. Identification of prognostic biomarkers in colorectal cancer using a long non-coding RNA-mediated competitive endogenous RNA network. Oncol Lett. 2019;17:2687–94. doi: 10.3892/ol.2019.9936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hui Z, Zhanwei W, Xi Y, Jin L, Jing Z, Shuwen H. Construction of ceRNA coexpression network and screening of molecular targets in colorectal cancer. Dis Mark. 2020 doi: 10.1155/2020/2860582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X, Li B, Ran P, Wang L. Identification of ceRNA network based on a RNA-seq shows prognostic lncRNA biomarkers in human lung adenocarcinoma. Oncol Lett. 2018;16:5697–708. doi: 10.3892/ol.2018.9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Q, Shi M, Meng W, Wang Y, Hui P, Ma J. Long noncoding RNA FOXD3-AS1 promotes colon adenocarcinoma progression and functions as a competing endogenous RNA to regulate SIRT1 by sponging miR-135a-5p. J Cell Physiol. 2019;234:21889–902. doi: 10.1002/jcp.28752. [DOI] [PubMed] [Google Scholar]
- 10.Yuan C. miR-616 promotes breast cancer migration and invasion by targeting TIMP2 and regulating MMP signaling. Oncol Lett. 2019;18:2348–55. doi: 10.3892/ol.2019.10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang G, Pian C, Chen Z, Zhang J, Xu M, Zhang L, et al. Identification of cancer-related miRNA-lncRNA biomarkers using a basic miRNA-lncRNA network. PLoS One. 2018;13:e0196681. doi: 10.1371/journal.pone.0196681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perchepied S, Eskenazi N, Giangrande C, Camperi J, Fournier T, Vinh J, et al. Erratum to “Development of immobilized enzyme reactors for the characterization of the glycosylation heterogeneity of a protein” [Talanta 206 (2020) 120171] Talanta. 2020;209:120568. doi: 10.1016/j.talanta.2019.120568. [DOI] [PubMed] [Google Scholar]
- 13.Gu Y, Deng B, Kong J, Yan C, Huang T, Yang J, et al. Functional polymorphisms in NR3C1 are associated with gastric cancer risk in Chinese population. Oncotarget. 2017;8:105312–9. doi: 10.18632/oncotarget.22172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riabovol OO, Tsymbal DO, Minchenko DO, Lebid-Biletska KM, Sliusar MY, Rudnytska OV, et al. Effect of glucose deprivation on the expression of genes encoding glucocorticoid receptor and some related factors in ERN1-knockdown U87 glioma cells. Endocr Regul. 2019;53:237–49. doi: 10.2478/enr-2019-0024. [DOI] [PubMed] [Google Scholar]
- 15.Tian D, Tian M, Han G, Li JL. Increased glucocorticoid receptor activity and proliferation in metastatic colon cancer. Sci Rep. 2019;9:1–7. doi: 10.1038/s41598-019-47696-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitre-Aguilar IB, Barrios-Garcia T, Ruiz-Lopez VM, Cabrera-Quintero AJ, Mejia-Dominguez NR, Ventura-Gallegos JL, et al. Glucocorticoid-dependent expression of IAP participates in the protection against TNF-mediated cytotoxicity in MCF7 cells. BMC Cancer. 2019;19:356. doi: 10.1186/s12885-019-5563-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitre-Aguilar IB, Barrios-Garcia T, Ruiz-Lopez VM, Cabrera-Quintero AJ, Mejia-Dominguez NR, Ventura-Gallegos JL, et al. Glucocorticoid-dependent expression of IAP participates in the protection against TNF-mediated cytotoxicity in MCF7 cells. BMC Cancer. 2019;19:1–17. doi: 10.1186/s12885-019-5563-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao Y, Wang P, Wang Y, Ma X, Zhi H, Zhou D, et al. Lnc2Cancer v2.0: Updated database of experimentally supported long non-coding RNAs in human cancers. Nucleic Acids Res. 2019;47:D1028–33. doi: 10.1093/nar/gky1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukunaga T, Iwakiri J, Ono Y, Hamada M. LncRRIsearch: A web server for lncRNA-RNA interaction prediction integrated with tissue-specific expression and subcellular localization data. Front Genet. 2019;10:462. doi: 10.3389/fgene.2019.00462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H, Wang Z, Wu J, Ma R, Feng J. Long noncoding RNAs predict the survival of patients with colorectal cancer as revealed by constructing an endogenous RNA network using bioinformation analysis. Cancer Med. 2019;8:863–73. doi: 10.1002/cam4.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen DL, Chen LZ, Lu YX, Zhang DS, Zeng ZL, Pan ZZ, et al. Long noncoding RNA XIST expedites metastasis and modulates epithelial-mesenchymal transition in colorectal cancer. Cell Death Dis. 2017;8:e3011. doi: 10.1038/cddis.2017.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duan Q, Cai L, Zheng K, Cui C, Huang R, Zheng Z, et al. lncRNA KCNQ1OT1 knockdown inhibits colorectal cancer cell proliferation, migration and invasiveness via the PI3K/AKT pathway. sOncol Lett. 2020;20:601–10. doi: 10.3892/ol.2020.11619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiao L, Dai Y, Gu Q, Chan KW, Zou B, Ma J, et al. Down-regulation of X-linked inhibitor of apoptosis synergistically enhanced peroxisome proliferator-activated receptor γ ligand-induced growth inhibition in colon cancer. Molecular cancer therapeutics. 2008;7:2203–11. doi: 10.1158/1535-7163.MCT-08-0326. [DOI] [PubMed] [Google Scholar]