Abstract
Streptococcus agalactiae is a leading cause of neonatal sepsis and meningitis. Adherence to extracellular matrix proteins is considered an important factor in the pathogenesis of infection, but the genetic determinants of this process remain largely unknown. We identified and sequenced a gene which codes for a putative lipoprotein that exhibits significant homology to the streptococcal LraI protein family. Mutants of this locus were demonstrated to have substantially reduced adherence to immobilized human laminin. The nucleotide sequence of the gene was subsequently designated lmb (laminin binding) and shown to be present in all of the common serotypes of S. agalactiae. To determine the role of Lmb in the adhesion of S. agalactiae wild-type strains to laminin, a recombinant Lmb protein harboring six consecutive histidine residues at the C terminus was cloned, expressed, and purified from Escherichia coli. Preincubation of immobilized laminin with recombinant Lmb significantly reduced adherence of the wild-type strain O90R to laminin. These results indicate that Lmb mediates the attachment of S. agalactiae to human laminin, which may be essential for the bacterial colonization of damaged epithelium and translocation of bacteria into the bloodstream.
The expression of cell surface receptors determines adhesive properties of streptococci, which include binding to eukaryotic extracellular matrix (ECM) proteins, epithelial cells, and endothelial cells, as well as to other bacteria. The LraI (lipoprotein receptor antigen I) family of surface-associated lipoproteins is involved in the coaggregation of Streptococcus gordonii with Actinomyces naeslundii, the adherence of S. sanguis to the salivary pellicle, the binding of S. parasanguis to a platelet fibrin matrix (14, 37), and the adherence of S. pneumoniae to type II pneumocytes (3). Previously identified members of this family are PsaA from S. pneumoniae, FimA from S. parasanguis, SsaB from S. sanguis, EfaA from Enterococcus faecalis, ScbA from S. crista, and ScaA from S. gordonii. Proteins of this family appear to serve a dual role in adhesion and transport; they are located in ABC transporter-type operons and code for lipoproteins. Similarities between the deduced proteins of lraI genes and MntC, an Mn2+ transporter of Synechocystis, have been described (1), and recently Mn2+ transporter activity was demonstrated for PsaA of S. pneumoniae (5) and ScaA of S. gordonii (17). It has been proposed that the LraI proteins together with other proteins constitute a large family of metal transporters (5). With regard to pathogenicity, PsaA of S. pneumoniae and FimA of S. parasanguis have been shown to be essential for virulence in animal models (3, 37), and immunogenic properties were demonstrated for EfaA (19), FimA (37), and PsaA (35), indicating their potential use as vaccine candidates.
S. agalactiae (group B streptococcus [GBS]) is one of the most important neonatal pathogens, causing 1.8 cases of septicemia or meningitis per 1,000 live births (40). Despite adequate antimicrobial therapy, mortality rates still range between 5 and 30% (38). In addition, recent studies have found an increasing number of serious infections in adults (7, 8). Several virulence factors that contribute to the pathogenesis of the disease have been identified: capsular polysaccharides (39), CAMP factor (9, 28), hemolysin (24), and C proteins (23). The adherence of S. agalactiae to immobilized fibronectin has been implicated in the pathogenesis of disease (34), but genetic determinants for the adherence of S. agalactiae to ECM proteins have not been identified.
Laminin, a 900-kDa glycoprotein, is a major component of the basement membrane. It is composed of three distinct polypeptide chains (A, B1, and B2) which reversibly assemble to form the macromolecular structure. Functions of laminin include the formation of the basement membrane by interaction with other basement membrane components and the development and maintenance of cellular organization. S. agalactiae has been demonstrated to damage the pulmonary epithelium (24), a process that leads to the exposure of underlying basement membrane structures. Thus, the adhesion to basement membrane components may be critical for the bacterial colonization of damaged epithelium and invasion of bacteria into the bloodstream.
In this paper, we describe the identification of a putative lipoprotein with homology to the streptococcal LraI family in GBS. We show that mutants of the genetic locus are deficient in adherence to immobilized laminin and that the recombinant protein inhibits adherence of the wild-type strain to human laminin.
MATERIALS AND METHODS
Bacterial strains.
The Escherichia coli and S. agalactiae strains used in this study are listed in Table 1. E. coli DH5α served as the host for recombinant plasmid pG+host5, E. coli BL21 was used for the expression of recombinant protein from plasmid pET21a, and E. coli XL1-Blue MRF and XLOLR (Stratagene, Heidelberg, Germany) were used as hosts for phages Lambda ZAP Express and ExAssist, respectively.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Description | Source or referencea |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | endA1 hsdR17 supE44 ΔlacU169(φ80lacZΔM15) recA1 gyrA96 thi-1 relA1 | Boehringer |
| XL1-Blue MRF | Δ (mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac [F′proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| BL21(DE3) | E. coli B, F−dcm ompT hsdS gal λ(DE3), T7 polymerase gene under control of the lacUV5 promoter | Novagen |
| S. agalactiae | ||
| R268 | Serotype III, Hly− | Aachen collection |
| O90R (ATCC 12386) | Serotype Ia, Lancefield grouping strain | ATCC |
| ATCC 12400 | Serotype Ia | ATCC |
| ATCC 12401 | Serotype Ib | ATCC |
| ATCC 27591 | Serotype Ia/c | ATCC |
| 18 RS 26 | Serotype II | R. Lancefield collection |
| CNCTC 13/63 | Serotype III | CNCTC |
| CNCTC 1/82 | Serotype IV | CNCTC |
| M 1A-00008 | Serotype V | P. Ferrieri collection |
| 92-085 | Serotype VI | P. Ferrieri collection (original NCTC 2/86) |
| 87-603 | Serotype VII | P. Ferrieri collection (‘7271’ original Perch) |
| JM9-130013 | Serotype VIII | P. Ferrieri collection |
| Lmb-k1 | Strain O90R with pG+host5 integrated into the lmb gene at codon 165 | This study |
| Lmb-k2 | Strain O90R with pG+host5 integrated into the lmb gene at codon 259 | This study |
| Plasmids | ||
| pUC18 | Apr, ColE1, lacI, φ80dlacZ | Boehringer |
| pG+host5 | Eryr, pBR, Ts | Appligene |
| pET21a | Apr, T7lac, pBR, His tag | Novagen |
| pBS1876 | pET21a vector carrying the 918-bp coding region of lmb | This study |
| pBS1817 | pG+host5 derivative carrying an internal 201-bp fragment of lmb | This study |
| pBS1815 | pG+host5 derivative carrying an internal 572-bp fragment of lmb | This study |
ATCC, American Type Culture Collection; CNCTC, Czechoslovak National Collection of Type Cultures.
S. agalactiae isolates were cultured on Columbia agar (Oxoid, Basingstoke, England) supplemented with 3% sheep blood, in Todd-Hewitt broth (THB) (Oxoid) or in THB supplemented with 0.5% yeast (THY) at 37°C. Mutant strains harboring chromosomally integrated pG+host5 vectors were maintained in medium containing 5 mg of erythromycin per liter at a temperature of ≥37°C. Growth rates of wild-type and mutant strains were determined by measuring optical density at 600 nm (OD600) in THY or THY supplemented with MnCl2.
General DNA techniques.
Standard recombinant DNA techniques were used for nucleic acid preparation and analysis. PCR was carried out with Taq polymerase as specified by the manufacturer (Boehringer, Mannheim, Germany), with 35 cycles of amplification steps of 1 min at 94°C, 1 min at 50 to 56°C, and 1 to 3 min at 72°C, depending on product size. For PCR with degenerate primers, PCR conditions were 95°C for 5 min, followed by 35 cycles of 95°C for 30 s, 45°C for 30 s, and 72°C for 1 min, with primer 5′-GGG GGG ATC CRT SNN SGA YRA YGG-3′ and 5′-GGG GGG ATC CAR SCC SAV SCC SNN SC-3′ (R = A or G; S = G or C; N = A, G, C, or T; Y = C or T; V = G, A, or C). Genomic streptococcal DNA was isolated as described previously (22). To confirm the presence of lmb in different serotypes, Southern blot analysis was performed after digestion of genomic DNA with EcoRI. Hybridization was performed at 65°C overnight. The hybridization probe was generated by PCR of strain R268 with primers 5′-ACC GTC TGT AAA TGA TGT GG-3′ and 5′-GAT TGA CGT TGT CTT CTG-3′, and the resulting PCR products were labeled by adding Dig (digoxigenin)-dUTP (Boehringer) at a final concentration of 5 μM. Hybridizing fragments were visualized by disodium 3-{4-methoxyspiro[1,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.1.13,7) decan]-4-yl}phenyl phosphate CSPD; Serva, Heidelberg, Germany) as instructed by the manufacturer. Plasmid DNA was isolated and purified by using a Qiaprep Spin Miniprep kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Plasmid and PCR products were sequenced on an ABI 373 automated DNA sequencer, using an ABI PRISM dye terminator cycle sequencing kit (PE Applied Biosystems, Weiterstadt, Germany). GBS strains were transformed according to the protocol of Ricci et al. (30).
Immunofluorescence. For the immunofluorescence test, bacteria were grown overnight in THY. Incubation with anti-Lmb antibody was performed at a dilution of 1:100 in phosphate-buffered saline–2% goat serum for 30 min. The secondary antibody fluorescein isothiocyanate-labeled goat anti-rabbit immunoglobulin G) (Sigma Chemical Co., St. Louis, Mo.) was used at a concentration of 1:100 in phosphate-buffered saline–2% goat serum. Fluorescence was assessed visually under a fluorescence microscope and measured in a FACScan flow cytometer (Becton Dickinson, San Jose Calif.) equipped with a standard argon laser.
Phage techniques.
A Lambda ZAP Express library of strain O90R was created as described by Podbielski et al. (29). Briefly, 200 μg of genomic DNA was digested with 0.2 U of Sau3A (Boehringer) for 30 min at 37°C. The resulting DNA fragments were separated according to size by a salt gradient technique (12). Fractions containing fragments 2 to 9 kb in length were ligated with BamHI-digested λ arms and packaged by using a Gigapack II packaging kit (Stratagene). Further processing and plaque lifting were done according to the manufacturer’s instructions. The library was screened by hybridization with PCR products at 65°C overnight. The PCR products were labeled by adding Dig-dUTP (Boehringer) at a final concentration of 5 μM to the reaction mixture. Detection of positive plaques by CSPD (Serva) was done as instructed by the manufacturer.
Construction of lmb mutants.
Plasmid pG+host5 was used for targeted genetic mutagenesis of lmb. Two mutants of the wild-type strain O90R (Lmb-k1 and Lmb-k2) were created by plasmid insertion at nucleotides 495 and 777, respectively, of the lmb gene. Internal fragments of the lmb gene were amplified by PCR with primers 5′-ACC GTC TGT AAA TGA TGT GG-3′ plus 5′-GAT TGA CGT TGT CTT CTG C-3′ and 5′-GCC GCC ACTAGT ACC GTC TGT AAA TGA TGT GG-3′ plus 5′-GAC GAC GAA TTCGAT TGA CGT TGT CTT CTG C-3′ (the newly introduced SpeI and EcoRI restriction sites are underlined). Resulting PCR products and the vector were digested with BamHI and XbaI and with SpeI and EcoRI, respectively, ligated, and transformed into E. coli. Chromosomal integration into strain O90R was performed as previously described (21). To confirm correct chromosomal insertion of the plasmid, genomic DNA of mutant Lmb-k1 and wild-type strain O90R was digested with the restriction endonuclease EcoRI or XbaI and probed with a nucleotide probe directed to the duplicated fragment of lmb. PCR with primers annealing to vector sequences and genomic nucleotide sequence upstream or downstream of the duplication site followed by DNA sequencing of PCR products was used to confirm chromosomal insertion for both mutants.
Expression of Lmb in E. coli.
The lmb gene was cloned into the pET21a expression vector (Novagen, Madison, Wis.) in E. coli BL21(DE3) (Novagen) for high-level expression and purification over a Ni2+ column. To construct the pET21a::lmb vector, nucleotides coding for amino acids 19 to 306 were amplified by PCR using primers 5′-GCC GCG CAT ATG TGT GAT AAG TCA GCA AAC CCC A-3′ and 5′-GCC GCG CTC GAG CTT CAA CTG TTG ATA GAG CAC TTC C-3′ (the newly introduced restriction sites NdeI and XhoI are underlined). The resulting PCR product was purified by using a QiaQuick PCR purification kit (Qiagen) according to the manufacturer’s instructions. The purified product and pET21a plasmid were digested with NdeI and XhoI, ligated, and introduced into E. coli by using standard molecular biology techniques. For the expression of recombinant protein, E. coli BL21(DE3) (Novagen) harboring the pET21a::lmb construct was grown to an OD600 of 0.6 in Luria-Bertani medium, and protein expression was induced by 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 2 h; cells were pelleted, resuspended in 1 ml of lysis buffer (50 mM sodium phosphate [pH 8.0], 300 mM NaCl, 20 mM imidazole, 1 mg of lysozyme per ml, 1 mM phenylmethylsulfonyl fluoride), placed on ice for 30 min, and subjected to sonication. Recombinant Lmb was purified from lysed E. coli cells by passage over a commercial nickel affinity matrix (Ni-NTA [nitrilotriacetic acid] Spin kit; Qiagen) and eluted under native conditions according to the manufacturer’s instructions. The eluate was subjected to 8 to 25% gradient sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using the Phast system (Pharmacia LKB, Uppsala, Sweden) and visualized by silver staining.
Subcellular localization.
Polyclonal antibodies for Lmb were obtained from Eurogentec (Brussels, Belgium). Antibodies were raised in New Zealand White rabbits by intradermic injection of 100 μg of recombinant Lmb at days 0, 14, 28, and 56. Subcellular fractions were prepared from GBS strain O90R. Bacteria were grown to late logarithmic phase in THB supplemented with 3% sheep blood. Cells were disrupted at a pressure of 16,000 to 20,000 PSI lb/in2 in a high-pressure homogenizer (Avestin Inc., Ottawa, Ontario, Canada); cytoplasmic and membrane fractions were separated by centrifugation at 100,000 × g for 60 min. Cytoplasmic proteins were precipitated by trichloroacetic acid, whereas the pellets containing the membranes were used directly. Samples of both fractions containing 6 μg of protein each were solubilized in SDS-gel electrophoresis buffer, separated by denaturing SDS-PAGE on a 8 to 25% gradient gel, and transferred to an Immobilon P polyvinylidene difluoride membrane (Millipore, Eschborn, Germany) with the Phast system (Pharmacia LKB) according to the manufacturer’s instructions. The blots were probed with a polyclonal anti-Lmb antibody at a dilution of 1:1,000. Controls were probed with preimmune rabbit serum at a dilution of 1:1,000. Primary antibodies were detected by an alkaline phosphatase-labeled anti-rabbit immunoglobulin G secondary antibody (Pierce, St. Augustin, Germany) at a dilution of 1:5,000. Bound secondary antibodies were visualized by chemiluminescent CSPD (Serva) according to the manufacturer’s instructions.
RNA preparation and analysis.
Total RNA was prepared from GBS strains R268 grown to an OD of 0.8 in THB. Cells were lysed mechanically by glass beads in a cell disrupter (Dianova, Hamburg, Germany) in the presence of 1 ml of Trizol (Gibco BRL, Eggenstein, Germany). Purification of the RNA was done according to the manufacturer’s instructions. Reverse transcription RT was carried out with 1 μg of RNA as template, 2 pmol of primer 5′-GCAGCAGCAGCAGGACAGCACTGATTTGATCC-3′, 0.1 M dithiothreitol, 10 mM deoxynucleoside triphosphate mix, and 200 U of Superscript II reverse transcriptase (Gibco BRL) in 50 mM Tris-HCl (pH 8.3)–75 mM KCl–3 mM MgCl2 at 42°C for 50 min in a 20-μl reaction volume; 5 μl of the reaction mixture was used as the template for a subsequent PCR with primers 5′-ACCGTCTGTAAATGATGTGG-3′ and 5′-CAGCACTGATTTGATCC-3′.
Adherence assay.
To study adherence of S. agalactiae wild-type strain O90R and lmb mutant strains Lmb-k1 and Lmb-k2 to immobilized laminin, 60-well Terasaki plates were coated with human placental laminin (100 μg/ml; Gibco BRL) reconstituted in Dulbecco’s phosphate-buffered saline (DPBS). Plates were incubated with laminin for 18 h at room temperature. Streptococci were grown in THB, harvested in mid-logarithmic phase, and washed twice in DPBS. Bacteria were labeled with fluorescein isothiocyanate and resuspended in DPBS. To obtain single cells, bacterial suspensions were sonicated. Laminin-coated wells were washed with DPBS, and then 10 μl of DPBS containing 5 × 106 bacteria was added to each well. For the Mn2+ supplementation studies, MnCl2 was added to a concentration of 10 μM to DPBS. To investigate the effect of recombinant Lmb on the adherence of the wild-type strain O90R, Terasaki wells were preincubated for 20 min at 37°C with 1 μg of recombinant Lmb before the bacterial suspension was added. After incubation for 60 min at 37°C, nonadherent bacteria were removed by being washed five times with DPBS. Adherent bacteria were quantified in a fluorescence counter (Cytofluor II; Perseptive Biosystems Inc.).
Nucleotide sequence accession number.
The nucleotide sequence of the coding regions for the S. agalactiae lmb gene has been submitted to the EMBL/Genbank/DDBJ nucleotide sequence data libraries and assigned accession no. AF062533.
RESULTS
Identification of Lmb.
Fragments of chromosomal DNA from GBS strain R268 were amplified by PCR using degenerate primers directed toward the conserved glycine-rich G1 and G2 blocks of bacterial sensor proteins, which resemble nucleotide binding domains (27). This method has previously been used to amplify the nucleotide sequence of cell surface-associated transport proteins in gram-positive organisms (2). The resulting PCR products were purified and subcloned into plasmid pUC18. Nucleotide sequences of the inserts were determined by automated DNA sequencing. Comparison of the deduced amino acid sequence with the GenBank database entries revealed that 3 of 42 clones harbored overlapping fragments of a gene with significant homology to the streptococcal LraI protein family.
Nucleotide and protein sequence analysis.
Nucleotide sequence upstream and downstream of the initial chromosomal fragments was obtained by screening of a λ phage library. Primers to generate PCR products of strain R268 were designed based on the nucleotide sequence of positive plaques. DNA sequencing of the resulting PCR products revealed an open reading frame of 921 nucleotides with a typical ribosome binding site 5 nucleotides upstream of the ATG start codon. Based on sequence similarity, a putative prokaryotic −35 and −10 promoter region was identified approximately 70 nucleotides upstream of the start codon. The deduced protein consists of 306 amino acid residues with a predicted molecular mass of 34.1 kDa. A second open reading frame of 2.4 kb starts 12 nucleotides downstream of the lmb stop codon. The putative start codon, GTG, is preceded by a typical ribosome binding site (GAAGGA). Putative promoter sequences could not be identified in the short intergenic region between lmb and the downstream open reading frame or within the 3′-terminal region of the lmb gene, suggesting that the two genes may comprise an operon. All of the known LraI proteins are lipoproteins with the typical signal peptidase II recognition sequence LxxC for the signal peptidase II at amino acid residue 16 or 17. The GBS homologue has a similar but slightly different sequence at this position (IAGC), with a leucine-to-isoleucine alteration. LplA of Bacillus subtilis, which has been shown to be a lipoprotein by radiolabeling with palmitate, also contains this atypical recognition sequence (32).
Comparison of the deduced amino acid sequence with sequences of previously identified members of the LraI adhesin family revealed 47% homology and 27% identity with PsaA of S. pneumoniae. Similarities to the other LraI proteins were between 36 and 46% (Fig. 1). In addition, there was 30% identity with Adc, a novel Zn transporter of S. pneumoniae which presumably binds Zn through a histidine-rich region of the protein. Lmb, however, does not possess a similar domain. Jenkinson (14) proposed four common structural domains of the LraI proteins: a 20-residue hydrophobic leader sequence that is cleaved off by signal peptidase II; two transmembrane domains, B1 and B2; and the α region, which is assumed to be exposed to the cell surface and comprises the solute binding region of the protein. These domains appear to be conserved in the lmb gene product (Fig. 2). Interestingly, amino acid residues 152 to 197 of Lmb, corresponding to the α domain of the LraI family, exhibit 50% homology to the human laminin B2 chain (Fig. 2).
FIG. 1.
Amino acid sequence alignment of LraI proteins (S. pneumoniae PsaA, S. parasanguis FimA, S. sanguis SsaB, S. gordonii ScaA, S. crista ScbA, and E. faecalis EfaA) and S. agalactiae Lmb. Alignment and determination of consensus sequence were performed with the MultAlin program (http://www.toulouse.inra.fr). Amino acid residues of Lmb matching the consensus sequence are shown in boxes. Highly conserved residues (consensus level of 90%) are represented as capital letters in the consensus sequence; small letters denote a consensus level of ≥50%. !, I or V; $, L or M; %, F or Y; #, N, D, Q, E, or B. Parameters: gap weight, 12; gap length weight, 2.
FIG. 2.
(A) Comparison of Lmb with the LraI domains. The Lmb protein (A) is designated by the open box showing a putative cleavage site for signal peptidase II. At amino acids (aa) 165 and 229, the plasmid insertion sites of the pG+host5 vector in mutants Lmb-k1 and Lmb-k2 are indicated. A region corresponding to the α region of LraI proteins with similarity to the B2 chain of human laminin is represented by a filled rectangle. (B) Structural features of the LraI family as proposed by Jenkinson (14). Numbers refer to amino acid residues, demarcating four regions: leader peptide, cleaved off by signal peptidase II; B1; B2; and the α region, which is presumed to be the solute binding domain. (C) Amino acid sequence alignment of the α region of Lmb with the laminin B2 chain. The alignment was performed with the BLASTp program at the National Center for Biotechnology Information web site.
Presence and expression of the lmb gene in various GBS serotypes.
To determine the distribution of lmb in various S. agalactiae strains, a fragment of the gene was amplified by PCR and used as a hybridization probe for a Southern blot of GBS serotypes Ia, Ib, Ic, and II to VIII. Hybridization of the probe with chromosomal DNA could be detected as a single band in all of the serotypes tested (Fig. 3); immunofluorescence tests performed on the different serotypes confirmed the expression of the protein in these strains.
FIG. 3.
(A) Analysis of the lmb gene in various GBS serotypes by Southern hybridization. Genomic DNA was digested with EcoRI and transferred to a nylon membrane. Hybridization was performed with a Dig-dUTP-labeled fragment of lmb generated by PCR. Lanes: M, molecular size markers (in nucleotides); 1, GBS strain O90R; 2 to 11, specific serotypes as indicated above the lanes. (B) Southern analysis of GBS strain O90R and the isogenic mutant Lmb-k1. Genomic DNA of the parent (lanes 1 and 2) and (lanes 3 and 4) mutant strains was digested with EcoRI (lanes 1 and 3) or XbaI (lanes 2 and 4). Hybridization was performed with a probe directed to the internal fragment of lmb that was used for insertion duplication mutagenesis.
Transcription analysis of the lmb locus.
Several of the homologous streptococcal lraI genes were reported to be polycistronicly transcribed (3, 10, 13, 18). To analyze transcription of the lmb locus in GBS, RT-PCR was performed with RNA isolated from strain R268. For the RT reaction, we used a reverse primer which anneals to nucleotides 191 to 174 of the second open reading frame. The subsequent PCR amplified a 930-bp product consisting of the last 716 nucleotides of lmb, the intergenic region, and the first 191 nucleotides of the second open reading frame (Fig. 4), which supports the hypothesis that lmb and the second open reading frame are transcribed together. To confirm that the specific PCR product originated from mRNA, controls were performed on a portion of the RNA preparation that had not been subjected to an RT reaction.
FIG. 4.
Transcription analysis of the lmb locus by RT-PCR. RNA was extracted from S. agalactiae R268 and subjected to RT-PCR. Lanes: 1, DNA size marker; 2, PCR with chromosomal DNA as the template; 3, PCR with 1 μg of RNA as the template; 4, control for DNA contamination in which the RNA preparation was subjected to PCR without prior RT-PCR.
Subcellular localization of Lmb.
Based on the homology of the deduced protein with other members of the LraI adhesin family and the presence of a putative signal peptidase II recognition site, we hypothesized that Lmb is localized at the surface of the bacterial cell. The subcellular localization was determined by Western blot analysis with a Lmb-specific antibody after cytoplasmic and membrane fractions of the cells were separated by ultracentrifugation. A single band in the membrane fraction corresponded to the predicted molecular mass of 34 kDa and the size of the recombinant Lmb protein (Fig. 5). Two smaller bands, probably representing degradation products, are present in the lane with the recombinant protein. Preimmune rabbit serum did not react with control blots. The results demonstrate that Lmb is associated with the bacterial membrane fraction. Surface exposure of the protein was investigated with an immunonofluorescence test. Anti-Lmb antibodies were used to detect Lmb on the surface of intact bacterial cells, and a fluorescence-labeled secondary antibody was used to visualize the binding of anti-Lmb to the bacteria. Results demonstrate that the protein is located on the surface and that the most intense fluorescence staining is seen at the margins of the cells (Fig. 6).
FIG. 5.
Subcellular localization of Lmb. Bacterial cells were disrupted by a high-pressure cell homogenizer, separated into membrane and cytoplasmic fractions by centrifugation at 100,000 × g, and subjected to denaturing gel electrophoresis. Western immunoblotting was performed as described in Materials and Methods with a polyclonal anti-Lmb antibody (A) or preimmune serum (B) at a dilution of 1:1,000. Lane 1, cytoplasmic fraction; 2, membrane fraction; 3, recombinant Lmb (3 ng).
FIG. 6.
Analysis of the surface exposure of Lmb by immunofluorescence staining. S. agalactiae cells were labeled with a polyclonal anti-Lmb antibody and viewed by fluorescence microscopy.
Construction of lmb mutants by insertion duplication mutagenesis.
The plasmid pG+host5 (4) was used for targeted genetic mutagenesis of lmb. Two mutants of the wild-type strain O90R (Lmb-k1 and Lmb-k2) were created by insertion duplication mutagenesis at nucleotides 495 and 777, respectively, of the lmb gene (Fig. 2). Correct chromosomal insertion of the plasmid was confirmed by either Southern blot hybridization (Fig. 3) or PCR and subsequent DNA sequencing of both mutants.
Growth properties of S. agalactiae upon Mn2+ substitution.
It was recently reported that the reduced growth rates of psaA mutants which could be improved by the addition of micromolar concentrations of Mn2+ to the culture medium (5). Therefore, growth of the lmb mutants in regular THY and THY medium supplemented with Mn2+ was determined by OD measurements. Growth rates and final growth densities of the wild-type and mutant strains showed no significant differences if cultured in THY or in THY supplemented with Mn2+ to final concentrations of 3 and 10 μM (data not shown). These results indicate that the Mn2+ growth requirements are satisfied in THY, which contains approximately 1 μM Mn2+, and that adhesion deficits of lmb mutants are not attributable to impaired growth rates.
Adherence to ECM proteins.
Since several LraI proteins were reported to be adhesins and Lmb exhibits homologies to the human laminin B2 chain, we tested the adherence of GBS wild-type and mutant strains to immobilized human placental laminin. Adherence of the two independently derived lmb mutants (Lmb-k1 and Lmb-k2) was significantly less than that of the wild-type strain O90R, in both cases reaching only 25% of the wild-type level (Fig. 7). Reduced adherence of lmb mutants was found with a wide range of bacterial concentrations and was consistent across incubation times with human laminin ranging from 30 to 300 min. To investigate the possibility that binding to the immobilized laminin is mediated by a contaminant of the laminin preparation, we tested the binding of wild-type and mutant strains to collagen IV, which is present in trace amounts in the laminin preparation. Neither the wild-type nor the mutant strain exhibited significant binding to collagen IV. Binding of the wild-type strain to collagen IV was less than 2% of the binding observed for laminin (data not shown). A screen for any major contaminants in the laminin preparation performed by protein gel electrophoresis and subsequent silver staining did not reveal the presence of any unexpected proteins bands (data not shown).
FIG. 7.
Adherence of S. agalactiae wild-type (O90R) and lmb mutant (Lmb-k1 and Lmb-k2) strains to immobilized laminin. Adherence was tested in Terasaki wells coated with 100 μg of ECM protein per ml. Assays were carried out as described in Materials and Methods. Bars represent the mean ± standard deviation for six wells. Results are representative for at least three independent experiments performed for each strain. (A) Adherence to laminin with increasing incubation time; (B) adherence to laminin for different bacterial inocula; (C) adherence to laminin after preincubation of immobilized laminin with recombinant Lmb protein; (D) adherence to laminin upon Mn2+ substitution.
Influence of recombinant Lmb protein on the adherence of the wild-type strain.
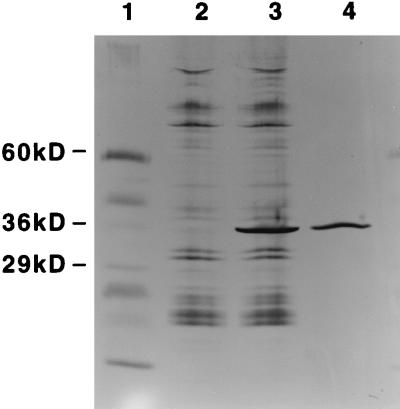
To test the hypothesis that Lmb protein itself interacts with human laminin, a recombinant protein was generated. The recombinant protein could be visualized as a band of approximately 34 kDa upon SDS-PAGE, which is consistent with the predicted molecular mass of Lmb (Fig. 8). To evaluate the influence of Lmb on adherence of the wild-type strain, the immobilized laminin was incubated with recombinant protein prior to adherence assays. Preincubation with recombinant Lmb reduced the adherence of the wild-type strain significantly, to 60% of the initial values (Fig. 7). Interestingly, adherence of the isogenic lmb mutants was slightly increased under the same conditions, possibly because the recombinant protein functions as a bridging molecule between the mutants and laminin.
FIG. 8.
SDS-PAGE analysis of recombinant Lmb. Bacterial lysates and purified recombinant Lmb were separated on an 8 to 25% SDS-polyacrylamide gel along with molecular mass markers and silver stained. Lanes 1, molecular mass marker; 2, crude bacterial lysate of E. coli BL21(DE3)(pET21a::lmb), uninduced; 3, crude bacterial lysate of E. coli BL21(DE3)(pET21a::lmb) after induction of protein expression with IPTG; 4, recombinant Lmb after purification over a Ni2+-NTA affinity matrix.
Influence of Mn2+ supplementation on adherence.
To investigate if supplementation with Mn2+ affected the adherence of bacteria to immobilized laminin, the wild-type strain and mutant strain Lmb-k1 were grown in THB or THB supplemented to a concentration of 10 μM Mn2+ and then subjected to the adherence assay in the presence of 10 μM Mn2+. Under these conditions, adherence of the mutant strain remained significantly less than that of the wild-type strain (Fig. 7); the level for the mutant reached 34% of the wild-type level, demonstrating that the effect on adherence to immobilized human laminin cannot be circumvented by growing cells in the presence of excess Mn2+ or adding Mn2+ to the adherence assay.
DISCUSSION
Pathogenic bacteria frequently express surface proteins that adhere to components of the mammalian ECM. The interaction of microorganisms with ECM proteins can promote bacterial colonization of damaged tissues. Despite considerable understanding of the molecular mechanisms of this process in other streptococcal species, genetic determinants for the adherence of S. agalactiae to ECM proteins have not been identified. In this investigation, we characterized the gene encoding a protein of S. agalactiae with similarities to the LraI adhesin family that mediates the attachment of GBS to laminin and was subsequently designated lmb. Comparison of the deduced amino acid sequence with LraI proteins showed closest homologies to PsaA of S. pneumoniae. However, compared to the conservation between proteins of this family present in other streptococci, the gene of GBS is distinct. Within the group of oral streptococci and the genetically related S. pneumoniae, similarities range between 80 and 93%. The results could reflect the more distant relationship of β-hemolytic streptococci with other streptococcal groups or indicate that the GBS protein has diverged from other LraI proteins.
We found that Lmb is important for the laminin binding properties of S. agalactiae. Isogenic mutants of the lmb locus demonstrated substantially diminished adherence to immobilized laminin, and the recombinant protein inhibited attachment of the wild-type strain. Since the mutants were generated by insertion duplication mutagenesis, the inserted plasmid can lead to polar effects on downstream genes; we cannot rule out that these effects may contribute to some extent to the observed phenomenon. However, the reduction of wild-type adherence by the recombinant protein is a strong indication that the lmb gene product directly mediates the interaction between GBS and laminin.
FimA from S. parasanguis and ScaB from S. gordonii have been shown to be important for the binding of these microorganisms to saliva-coated hydroxylapatite (10, 13). Glycoproteins of the saliva are assumed to be the ligands for these adhesins. Analogous to these findings, our results indicate that Lmb binds to laminin, a high-molecular-weight glycoprotein of the basement membrane. The similarity of the putative α region of Lmb to the human laminin B2 chain could account for an interaction with other polypeptide chains of the laminin molecule. Laminin is a macromolecule that self-assembles in vitro to form large unordered aggregates and can bind a variety of other basement membrane compounds, such as nidogen, collagen IV, and perlecan (6). Interestingly psaA mutants of S. pneumoniae exhibit reduced binding to A549 cells (3), a type II pneumocyte cell line that secrects laminin (16).
The adherence of bacteria to laminin may be a crucial step in the development of invasive GBS infection. Translocation of S. agalactiae into the bloodstream as well as entry of the bacteria into the cerebrospinal fluid, which occurs in the case of meningitis, requires the passage of bacteria through the basement membranes. The interaction of bacterial surface proteins with laminin could be important in this context. In the case of Haemophilus influenzae, adhesion and penetration of the basement membranes, which are open to circulation in the fenestrated endothelium of the choroid plexus, is discussed as the route of entry into the cerebrospinal fluid (36).
The results of the RT-PCR experiments show that lmb is part of an operon and that it is transcribed together with a second open reading frame, similar to the operon structure of other members of this family (10, 13, 31). Several different observations indicate that besides playing a role in adhesion and virulence, proteins of the LraI family function as transporters. Transport systems of gram-positive bacteria usually contain a solute binding component that is lipid modified and associated with the outer region of the cytoplasmic membrane (33). The fimA locus of S. parasanguis encodes an ATP binding membrane transport system (11), and manganese transporter activity has been demonstrated for PsaA of S. pneumoniae (5) and ScaA of S. gordonii (17).
It has been suggested that all members of the LraI family are manganese transporters and that the inhibition of S. sanguis and S. parasanguis adhesion to saliva-coated hydroxylapatite by purified ScaA or FimA is not due to direct binding of these proteins but rather reflects a requirement for Mn2+ (5). To investigate if the effects of an Lmb mutation are influenced by the availability of Mn2+, culture of bacterial cells and adherence assays were performed under supplementation with Mn2+. Mutants grown in THB containing approximately 1 μM Mn2+ and THB supplemented to an Mn2+ concentration of 10 μM both exhibited significantly reduced adherence to immobilized laminin, demonstrating that the reduced binding properties cannot be explained by Mn2+ deficiency alone. Our results indicate a function of Lmb in the adhesion to laminin that is distinct from the putative role as a Mn2+ transporter.
Immunogenic properties have been demonstrated for three members of the LraI adhesin family, PsaA, EfaA, and FimA (3, 19, 37). A recent publication demonstrates induction of protective antibodies against S. parasanguis endocarditis in an animal model after vaccination with recombinant FimA (37). Current vaccine approaches for GBS favor the use of proteins linked to polysaccharide structures of the capsule (15, 20, 25, 26). None of the presently identified surface proteins of GBS are found in every known serotype; given the presence of Lmb in all of these serotypes, the protein may be a good candidate for GBS vaccine.
ACKNOWLEDGMENTS
We thank P. Ferrieri for generously providing S. agalactiae strains and W. Dott for the determination of Mn2+ in THY and THB. We are grateful to B. Leonard and C. Brandt for helpful discussions and critical review of the manuscript.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (Sp 511/2-1 and Po 391/6-1) to B.S. and A.P.
REFERENCES
- 1.Bartsevich V V, Paktrasi H B. Molecular identification of an ABC transporter complex for manganese: analysis of a cyanobacterial mutant strain impaired in the photosynthetic evolution process. EMBO J. 1995;14:1845–1853. doi: 10.1002/j.1460-2075.1995.tb07176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bayles K W. The use of degenerate, sensor gene specific, oligodeoxyribonucleotide primers to amplify DNA fragments from Staphylococcus aureus. Gene. 1993;123:99–103. doi: 10.1016/0378-1119(93)90546-f. [DOI] [PubMed] [Google Scholar]
- 3.Berry A M, Paton J C. Sequence heterogeneity of PsaA, a 37-kilodalton putative adhesin essential for virulence of Streptococcus pneumoniae. Infect Immun. 1996;64:5255–5262. doi: 10.1128/iai.64.12.5255-5262.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biswas I, Gruss A, Ehrlich D, Maguin E. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J Bacteriol. 1993;175:3628–3635. doi: 10.1128/jb.175.11.3628-3635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dintilhac A, Alloing G, Granadel C, Claverys J P. Competence and virulence of Streptococcus pneumoniae: Adc and PsaA mutants exhibit a requirement for Zn and Mn resulting from inactivation of putative ABC metal permeases. Mol Microbiol. 1997;25:727–739. doi: 10.1046/j.1365-2958.1997.5111879.x. [DOI] [PubMed] [Google Scholar]
- 6.Engel J. Laminins and other strange proteins. Biochemistry. 1992;31:10643–10651. doi: 10.1021/bi00159a001. [DOI] [PubMed] [Google Scholar]
- 7.Farley M. Group B streptococcal infection in older patients. Spectrum of disease and management strategies. Drugs Aging. 1995;6:293–300. doi: 10.2165/00002512-199506040-00004. [DOI] [PubMed] [Google Scholar]
- 8.Farley M M, Harvey R C, Stull T, Smith J D, Schuchat A, Wenger J D, Stephens D S. A population based assessment of invasive disease due to group B Streptococcus in nonpregnant adults. N Engl J Med. 1993;328:1807–1811. doi: 10.1056/NEJM199306243282503. [DOI] [PubMed] [Google Scholar]
- 9.Fehrenbach F, Jürgens D, Rühlmann J, Sterzik B, Özel M. Role of CAMP-factor (protein B) for virulence. Zentbl Bakteriol Hyg Suppl. 1988;17:351–357. [Google Scholar]
- 10.Fenno J C, LeBlanc D J, Fives-Taylor P M. Nucleotide sequence analysis of a type 1 fimbrial gene of Streptococcus sanguis FW213. Infect Immun. 1989;57:3527–3533. doi: 10.1128/iai.57.11.3527-3533.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fenno J C, Shaikh A, Spatafora G, Fives-Taylor P. The fimA locus of Streptococcus parasanguis encodes an ATP-binding membrane transport system. Mol Microbiol. 1995;15:849–863. doi: 10.1111/j.1365-2958.1995.tb02355.x. [DOI] [PubMed] [Google Scholar]
- 12.Fink P. Using sodium chloride step gradients to fractionate DNA fragments. BioTechniques. 1991;10:447–449. [PubMed] [Google Scholar]
- 13.Ganeshkumar N, Hannam P M, Kolenbrander P E, McBride B C. Nucleotide sequence of a gene coding for a saliva-binding protein (SsaB) from Streptococcus sanguis 12 and possible role of the protein in coaggregation with Actinomyces. Infect Immun. 1991;59:1093–1099. doi: 10.1128/iai.59.3.1093-1099.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenkinson H F. Cell surface protein receptors in oral streptococci. FEMS Microbiol Lett. 1994;121:133–140. doi: 10.1111/j.1574-6968.1994.tb07089.x. [DOI] [PubMed] [Google Scholar]
- 15.Kasper D L, Paoletti L C, Wessels M R, Guttormsen H K, Carey V J, Jennings H J, Baker C J. Immune response to type III group B streptococcal polysaccharide-tetanus toxoid conjugate vaccine. J Clin Investig. 1996;98:2308–2314. doi: 10.1172/JCI119042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kikkawa Y, Sanzen N, Sekiguchi K. Isolation and characterization of laminin-10/11 secreted by human lung carcinoma cells. Laminin-10/11 mediates cell adhesion through integrin alpha3beta1. J Biol Chem. 1998;273:15854–15859. doi: 10.1074/jbc.273.25.15854. [DOI] [PubMed] [Google Scholar]
- 17.Kolenbrander P E, Andersen R N, Baker R A, Jenkinson H F. The adhesion-associated sca operon in Streptococcus gordonii encodes an inducible high-affinity ABC transporter for Mn2+ uptake. J Bacteriol. 1998;180:290–295. doi: 10.1128/jb.180.2.290-295.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolenbrander P E, Andersen R N, Ganeshkumar N. Nucleotide sequence of the Streptococcus gordonii PK488 coaggregation adhesin gene, scaA, and ATP-binding cassette. Infect Immun. 1994;62:4469–4480. doi: 10.1128/iai.62.10.4469-4480.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lowe A M, Lambert P A, Smith A W. Cloning of an Enterococcus faecalis endocarditis antigen: homology with adhesins from some oral streptococci. Infect Immun. 1995;63:703–706. doi: 10.1128/iai.63.2.703-706.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madoff L C, Paoletti L C, Tai J Y, Kasper D L. Maternal immunization of mice with group B streptococcal type III polysaccharide-beta C protein conjugate elicits protective antibody to multiple serotypes. J Clin Investig. 1994;94:286–292. doi: 10.1172/JCI117319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maguin E, Prevost H, Ehrlich S, Gruss A. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J Bacteriol. 1996;178:931–935. doi: 10.1128/jb.178.3.931-935.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin N J, Kaplan E L, Gerber M A, Menegus M A, Randolph M, Bell K, Cleary P P. Comparison of epidemic and endemic group G streptococci by restriction enzyme analysis. J Clin Microbiol. 1990;28:1881–1886. doi: 10.1128/jcm.28.9.1881-1886.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michel J L, Madoff L C, Kling D E, Kasper D L, Ausubel F M. C proteins of group B streptococci. In: Dunny G M, Cleary P P, McKay L L, editors. Genetics and molecular biology of streptococci, lactococci, and enterococci. Washington, D.C: American Society for Microbiology; 1991. pp. 214–218. [Google Scholar]
- 24.Nizet V, Gibson R, Chi E, Framson P, Hulse M, Rubens C. Group B streptococcal beta-hemolysin expression is associated with injury of lung epithelial cells. Infect Immun. 1996;64:3818–3826. doi: 10.1128/iai.64.9.3818-3826.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paoletti L C, Kennedy R C, Chanh T C, Kasper D L. Immunogenicity of group B Streptococcus type III polysaccharide-tetanus toxoid vaccine in baboons. Infect Immun. 1996;64:677–679. doi: 10.1128/iai.64.2.677-679.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paoletti L C, Wessels M R, Rodewald A K, Shroff A A, Jennings H J, Kasper D L. Neonatal mouse protection against infection with multiple group B streptococcal (GBS) serotypes by maternal immunization with tetravalent GBS polysaccharide-tetanus toxoid conjugate vaccine. Infect Immun. 1994;62:3236–3243. doi: 10.1128/iai.62.8.3236-3243.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parkinson J, Kofoid E. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 28.Podbielski A, Blankenstein O, Lütticken R. Molecular characterization of the cfb gene encoding group B streptococcal CAMP-factor gene. Med Microbiol Immunol. 1994;183:239–256. doi: 10.1007/BF00198458. [DOI] [PubMed] [Google Scholar]
- 29.Podbielski A, Flosdorff A, Weber-Heynemann J. The group A streptococcal virR49 gene controls expression of four structural vir regulon genes. Infect Immun. 1995;63:9–20. doi: 10.1128/iai.63.1.9-20.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ricci M L, Manganelli R, Berneri C, Orefici G, Pozzi G. Electrotransformation of Streptococcus agalactiae with plasmid DNA. FEMS Microbiol Lett. 1994;119:47–52. doi: 10.1111/j.1574-6968.1994.tb06865.x. [DOI] [PubMed] [Google Scholar]
- 31.Sampson J S, O’Connor S P, Stintson A R, Tharpe J A, Russel H. Cloning and nucleotide sequence analysis of psaA, the Streptococcus pneumoniae gene encoding a 37-kilodalton protein homologous to previously reported Streptococcus sp. adhesins. Infect Immun. 1994;62:319–324. doi: 10.1128/iai.62.1.319-324.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sutcliffe I C, Russel R R B. Lipoproteins of gram-positive bacteria. J Bacteriol. 1995;177:1123–1128. doi: 10.1128/jb.177.5.1123-1128.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tam R, Milton H S J. Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol Rev. 1993;57:320–346. doi: 10.1128/mr.57.2.320-346.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamura G S, Rubens C E. Group B streptococci adhere to a variant of fibronectin attached to a solid phase. Mol Microbiol. 1995;15:581–589. doi: 10.1111/j.1365-2958.1995.tb02271.x. [DOI] [PubMed] [Google Scholar]
- 35.Tharpe J A, Russel H. Purification and seroreactivity of pneumococcal surface adhesin A (PsaA) Clin Diagn Lab Immunol. 1995;3:227–229. doi: 10.1128/cdli.3.2.227-229.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Virkola R, Lähteenmäki K, Eberhard T, Kuusela P, van Alphen L, Ullberg M, Korhonen T. Interaction of Haemophilus influenzae with the mammalian extracellular matrix. J Infect Dis. 1996;173:1137–1147. doi: 10.1093/infdis/173.5.1137. [DOI] [PubMed] [Google Scholar]
- 37.Viscount H B, Munro C L, Burnette-Curley D, Peterson D, Macrina F L. Immunization with FimA protects against Streptococcus parasanguis endocarditis in rats. Infect Immun. 1997;65:994–1002. doi: 10.1128/iai.65.3.994-1002.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weisman L E, Stoll B J, Cruess D F, Hall R T, Merenstein G B, Hemming V G, Fischer G W. Early-onset group B streptococcal sepsis: a current assessment. J Pediatr. 1992;121:428–433. doi: 10.1016/s0022-3476(05)81801-3. [DOI] [PubMed] [Google Scholar]
- 39.Wessels M R, Rubens C E, Benedi V J, Kasper D L. Definition of a bacterial virulence factor: sialylation of the group B streptococcal capsule. Proc Natl Acad Sci USA. 1989;86:8983–8987. doi: 10.1073/pnas.86.22.8983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zangwill K, Schuchat A, Wenger J D. Group B streptococcal disease in the United States, 1990: report from a multistate active surveillance system. Morbid Mortal Weekly Rep. 1992;41(SS-6):25–32. [PubMed] [Google Scholar]