Abstract
This study was designed to synthesize hybridizing molecules from ciprofloxacin and norfloxacin by enhancing their biological activity with tetrazoles. The synthesized compounds were investigated in the interaction with the target enzyme of fluoroquinolones (DNA gyrase) and COVID-19 main protease using molecular similarity, molecular docking, and QSAR studies. A QSAR study was carried out to explore the antibacterial activity of our compounds over Staphylococcus aureus a QSAR study, using descriptors obtained from the docking with DNA gyrase, in combination with steric type descriptors, was done obtaining suitable statistical parameters (, , and ) to support our results. The binding interaction of our compounds with CoV-2-Mpro was done by molecular docking and were compared with different covalent and non-covalent inhibitors of this enzyme. For the docking studies we used several crystallographic structures of the CoV-2-Mpro. The interaction energy values and binding mode with several key residues, by our compounds, support the capability of them to be CoV-2-Mpro inhibitors. The characterization of the compounds was completed using FT-IR, 1H-NMR, 13C-NMR, 19F-NMR and HRMS spectroscopic methods. The results showed that compounds 1, 4, 5, 10 and 12 had the potential to be further studied as new antibacterial and antiviral compounds
Keywords: Hybrid tetrazole-fluoroquinolone, Multicomponent reaction, Molecular docking, QSAR, Covid-19 Main protease, DNA gyrase
1. Introduction
One of the challenges in pharmaceutical research is the development of multi-target drugs, which involves the incorporation of two or more pharmacophores into a single molecule [1, 2]. Computational tools, such as in silico molecular docking, and combinatorial chemistry, can be applied to make the discovery process easier. The SARS-CoV-2 pandemic is significantly affecting human health and severely restricting human activities, and thus, it is urgent to search for drugs to combat SARS-Cov-2, also named COVID-19. No effective drugs are commercially available; however, there are some examples of heterocyclic compounds such as chloroquine, remdesivir, nelfinavir, which are being used for the treatment of COVID-19 with moderate effectiveness [3], [4], [5] and recently, Pfizer's drug paxlovid, has significantly reduced hospitalization and death [6, 7]. Several potential drug targets have been identified, for example, viral proteases such as main and papain-like proteases [4], [5], [6]. This work; explores drug derivatives that can act by inhibiting two conventional targets simultaneously.
Many of the current drugs currently being used to treat COVID-19 are immune system modulators [8]. In the search for anti-SARS medications, the main goal is to keep the response to new species and mutations, where the key target enzymes in coronaviruses reveal some sequence similarities. The genome of SARS-CoV-2 is based on + ssRNA of approximately 32,000 base pairs [8]. During the intracellular life cycle, coronaviruses express and replicate their genomic RNA to produce full-length copies incorporated into newly created viral particles. Among the enzymes involved in replication and transcription, NSP5 is the main protease (Mpro) involved principally in the assembly process of the polyprotein in charge of RNA translation. In addition, NSP5 acts in the post-translational modification of viral proteins via ADP ribose phosphatase [7, 8]. The Mpro and the SARS-CoV counterpart possess 96% sequence homology, and their volumes and electrostatic profiles are highly homogeneous [9]. Thus, a specific inhibitor towards this enzyme would significantly influence the replication cycle of coronaviruses even in future mammalian variants. As indicated, Mpro is an attractive target for anti-COVID drug development [8, 9].
New reports about the 3D structure of SARS-CoV-2 Mpro reveal a complex dimer consisting of two protomers composed of 306 residues in three domains: residues 8-101 are in chymotrypsin-like domain I, residues 102-184 and residues 201-303 are in picornavirus 3C protease-like domain II and globular cluster domain III, respectively. Domains I and II connect with domain III by a loop of residues 185-200 consisting of five helices α. Mpro has a substrate-recognition pocket that is highly conserved among all coronaviruses, located between domains I and II. The substrate-recognition pocket contains a base pair with charged residues, such as the nucleophilic sulfur of Cys145 and the imidazole ring of Hist 41 as a general base. Thus, inhibition of the function of one of these residues will hinder the enzymatic activity of CoV-2Mpro and the infectious process [10, 11].
Several crystallographic structures of Mpro reveal that this pocket could serve as a drug target for the design of broad-spectrum inhibitors. Inhibitors used in MERS-Co-V 3CL protease shown with piperidine remarkable interactions with Cys148, which is equivalent to Cys145 in SARS-Co-V-2, which supports the use of this site for drug development. Computational studies can determine which drugs are most effective in the target site [10], [11], [12]. Several lists of drugs have been published, and many are candidates for in vivo testing, even for clinical trials. Among the suggested drugs are antibiotics with reported efficacy in secondary infections, such as fluoroquinolones, tetracycline, and macrolides [13]. For this reason, using computational studies to analyze FDA-approved drugs is a logical method to examine COVID-19.
Fluoroquinolones have been used for the treatment of several types of infections. They are considered a broad-spectrum antibiotic with activity against infections in the prostate [14], urinary skin [15], intra-abdomen [16], and bone and joints [17]. In addition, fluoroquinolones have exhibited atypical activity such as antitubercular [18], antimalarial [19] and anticancer [20, 21]. Some derivatives have demonstrated antiviral activity against single-stranded RNA viruses such as zika, dengue, hepatitis C and rhinovirus [22, 23], although the mechanism of action is not fully understood. On the other hand, tetrazoles have been used as isosteres for cis-amide bond peptide mimics and exhibit good resistance to biological degradation, reduced secondary drug effects and improved pharmacological properties. Quinolone tetrazole hybrids are a new type of pharmacophores in development for drug-resistant bacteria and are a possible alternative for treating COVID-19.
A large number of fluoroquinolones have been tested in vitro against SARS-CoV-2. The FDA approved enoxacin and levofloxacin as positive antiviral control, which showed to be better SARS-CoV-2 inhibitors compared with arbidol. Furthermore, molecular docking studies identified enoxacin, ciprofloxacin and moxifloxacin as possible SARS-CoV-2 inhibitors [24]. Studies on the pharmacokinetic properties, anti-inflammatory activity and binding towards SARS-CoV-2 protease of levofloxacin and moxifloxacin have allowed its use for the treatment of COVID-19 pneumonia [3]. Given that SARS-CoV-2 and MERS-CoV are both single-stranded, it seems probable that ciprofloxacin, levofloxacin and moxifloxacin may suppress the replication of both beta-coronaviruses [25].
Several fluoroquinolone derivatives have been used in silico studies as inhibitors due to their interaction with COVID-19 Mpro. In silico studies, such as molecular docking and molecular dynamics techniques, have demonstrated that several fluoroquinolone derivatives strongly bind to COVID-19 Mpro. For the analysis, chloroquine and nelfinavir were used as positive controls. The results presented by Marciniec [12] demonstrated that ciprofloxacin and moxifloxacin bind more strongly than the native ligand. Even in comparison with positive controls, the tested fluoroquinolones have a more significant number of protein interactions.
Quinolones are essential synthetic drug classes used for treating community- or hospital-acquired infectious diseases like urinary tract, respiratory, gastrointestinal, chronic osteomyelitis, and sexually transmitted infections [26, 27]. Quinolones successfully inhibit the replication of DNA and functionally exert their effect by inhibiting two types of bacterial topoisomerases II, namely DNA gyrase and topoisomerase IV [28]. Moreover, some derivatives of the fluoroquinolone (FQs) family exhibited antiproliferative activity [29, 30]. For instance, ciprofloxacin (CP) showed antiproliferative and apoptosis-inducing activities on prostate and bladder cancer cells [31, 32]. In addition, fleroxacin, ofloxacin and levofloxacin were also revealed to inhibit the growth of transitional cell bladder carcinoma cell lines [20].
In the present work, we have investigated the synthesis, characterization and molecular docking of 12 new fluoroquinolone derivatives with a tetrazole moiety. The syntheses were performed through fluoroquinolone scaffolds using the Ugi reaction to incorporate the tetrazole moiety and generate a compound library. Docking studies were performed to understand the binding mechanism using the protein receptor for DNA gyrase S aureus and COVID-19 main protease. Our research is based upon the hypothesis that the tetrazole and the fluoroquinolone moieties will act as topoisomerase and main protease Covid-19 inhibitors. Binding affinities and the orientation of the docked hybrid derivatives, ligand efficiency (LE) and hydrogen bonding energies were used to evaluate the binding modes. The potential antibacterial activity −minimal inhibitory concentration (MIC)− of new fluoroquinolone derivatives was evaluated by means of a QSAR approximation, using commercial drugs and compounds previously synthesized by Chauhan [33]. Furthermore, crystal structures of CoV-2-Mpro co-crystallized with covalent and non-covalent inhibitors were used to evaluate the potential antiviral activity of CoV-2-Mpro. The inhibition study was focused on the analysis of intramolecular interactions of our derivatives with selected amino acids (aa) of CoV-2-Mpro: Leu27, His41, Met49, Cys145, His164, Met165, Arg188, Gln189 and Gln 192. The selection was based on the fact that these amino acids have several interaction profiles of CoV-2-Mpro with inhibitors.
2. Results and discussion
2.1. Synthesis
The conventional method of synthesizing tetrazoles is a 1,3-dipolar cycloaddition of an azide (sodium azide, hydrazoic acid or trimethylsilyl azide) to imidoyl chlorides, amides, thioamides, nitriles, isocyanates and ketene imines as starting materials [34, 35]. Brönsted or Lewis acids were used to activate the substrates, or phase-transfer conditions were used [36], [37], [38]. However, many of these reported methods suffer from one or more of the following drawbacks: use of toxic or explosive reagents, a stoichiometric amount of metal catalyst or inorganic salt, long reaction times, harsh reaction conditions, high temperatures or poor selectivity.
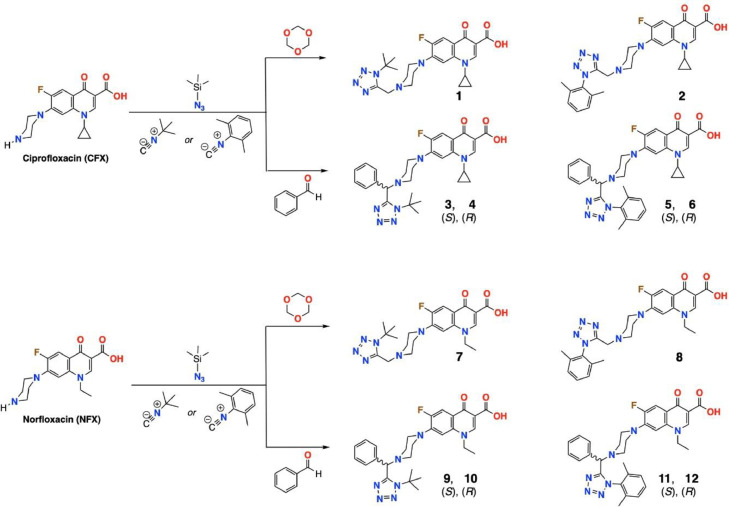
The synthesis of 1,5-disubstituted tetrazoles has been reported by the Ugi-azide reaction using four components simultaneously: an aldehyde or ketone, an amine, trimethylsilyl azide and an isocyanide synthesis [34, 35, 39]. This general, concise, novel strategy can also use Aldo/keto-acids/esters in the Ugi-azide reaction to access many new scaffolds [39]. The synthetic routes for ciprofloxacin or norfloxacin-tetrazole hybrids are depicted in Fig. 1 . The structures of the synthesized compounds were elucidated based on their spectroscopic analysis. The 13C-NMR spectra of derivatives revealed a signal at 156.06-151.53 ppm, typical for the tetrazole ring, which was assigned with the use of 13C-1H HSQC and HMBC experiments. The piperazine methylene or methine bridge in the 13C-NMR spectra exhibited a signal at 64.10-49.54 ppm. The methylene or methine hydrogens were found at 3.96 and 5.63-3.69 ppm, respectively, and the assignments were confirmed by HSQC.
Fig. 1.
Molecular structures of the synthesized quinolones.
2.2. Selection and construction of molecules
All the synthesized molecules exist as zwitterions by their protonation state at physiological pH (7.4) and racemic mixture (where appropriate).
The commercially available fluoroquinolones shown in Fig. 2 were utilized as reference compounds to study the potential bactericidal activity of these quinolone-tetrazole derivatives. The bactericidal activity value against Staphylococcus aureus is the Minimum Inhibitory Concentration (MIC) available for the reference compounds (Table 1 ) [40], [41], [42], [43]. All these molecules were constructed and evaluated as zwitterions for the same reasons as for the synthesized quinolones (vide supra).
Fig. 2.
Commercial bactericidal fluoroquinolone drugs were used as references in this study.
Table 1.
ΔV and values for fluoroquinolone derivatives.
| Molecule |
(kcal/atom) |
(Å3) |
(Å3) |
(µg/mL) |
|
|---|---|---|---|---|---|
| 1 | -255.09 | 187.22 | 35051.33 | 4.535 | – |
| 2 | -182.03 | 234.15 | 54826.22 | 4.679 | – |
| 3 | -202.12 | 269.87 | 72829.82 | 5.11 | – |
| 4 | -253.94 | 269.46 | 72608.69 | 5.11 | – |
| 5 | -255.56 | 316.18 | 99969.79 | 5.273 | – |
| 6 | -245.68 | 282.65 | 79891.02 | 5.273 | – |
| 7 | -244.44 | 171.74 | 29494.63 | 4.56 | – |
| 8 | -229.45 | 224.89 | 50575.51 | 4.685 | – |
| 9 | -212.93 | 258.3 | 66718.89 | 5.124 | – |
| 10 | -251.86 | 253.37 | 64196.36 | 5.124 | 0.39 |
| 11 | -182.36 | 306.76 | 94101.70 | 5.272 | – |
| 12 | -258.04 | 302.32 | 91397.38 | 5.272 | – |
| 13 | -288.46 | 251.53 | 63267.34 | 5.101 | 1.56 |
| 14 | -295.77 | 339.74 | 115423.27 | 6.167 | 1.56 |
| 15 | -268.76 | 236.66 | 56007.96 | 4.865 | 0.78 |
| 16 | -255.32 | 261.96 | 68623.04 | 5.327 | 0.78 |
| 17 | -260.05 | 308.82 | 95369.79 | 5.356 | 1.56 |
| 18 | -256.6 | 283.96 | 80633.28 | 5.324 | 1.56 |
| 19 | -220.73 | 263.3 | 69326.89 | 5.069 | 0.78 |
| TSF | -245.38 | 88.15 | 7770.42 | 3.369 | 0.1 |
| CFX | -151.16 | 59.63 | 3555.74 | 2.822 | 0.5 |
| TFX | -222.39 | 49.28 | 2428.52 | 2.84 | 0.125 |
| GFX | -205.29 | 83.71 | 7007.36 | 3.05 | 0.25 |
| LFX | -189.19 | 71.26 | 5077.99 | 2.747 | 0.125 |
| MFX | -198.77 | 117.15 | 13724.12 | 3.161 | 0.6 |
| LMF | -199.73 | 60.46 | 3655.41 | 3.026 | 0.2 |
| NFX | -192.34 | 55.18 | 3044.83 | 2.843 | 0.39 |
| PFX | -177.99 | 57.77 | 3337.37 | 3.071 | 0.5 |
| SFX | -221.08 | 97.88 | 9580.49 | 3.23 | 0.125 |
| SIT | -228.22 | 94.17 | 8867.99 | 2.787 | 0.5 |
| TFX | -200.56 | 99.02 | 9804.96 | 3.008 | 0.6 |
In addition, we selected seven molecules synthesized by K. Chauhan et al. in 2014, based on the norfloxacin structure with a tetrazole moiety incorporated (Fig. 3 ); in this work, they previously synthesized Compound 10 [33]. These molecules were selected based on their MIC value and the structural similarities that they share with our synthesized tetrazole-fluoroquinolones.
Fig. 3.
2D representation of the quinolone-tetrazole derivatives synthesized by K. Chauhan et al. [33].
2.3. Molecular docking over DNA gyrase
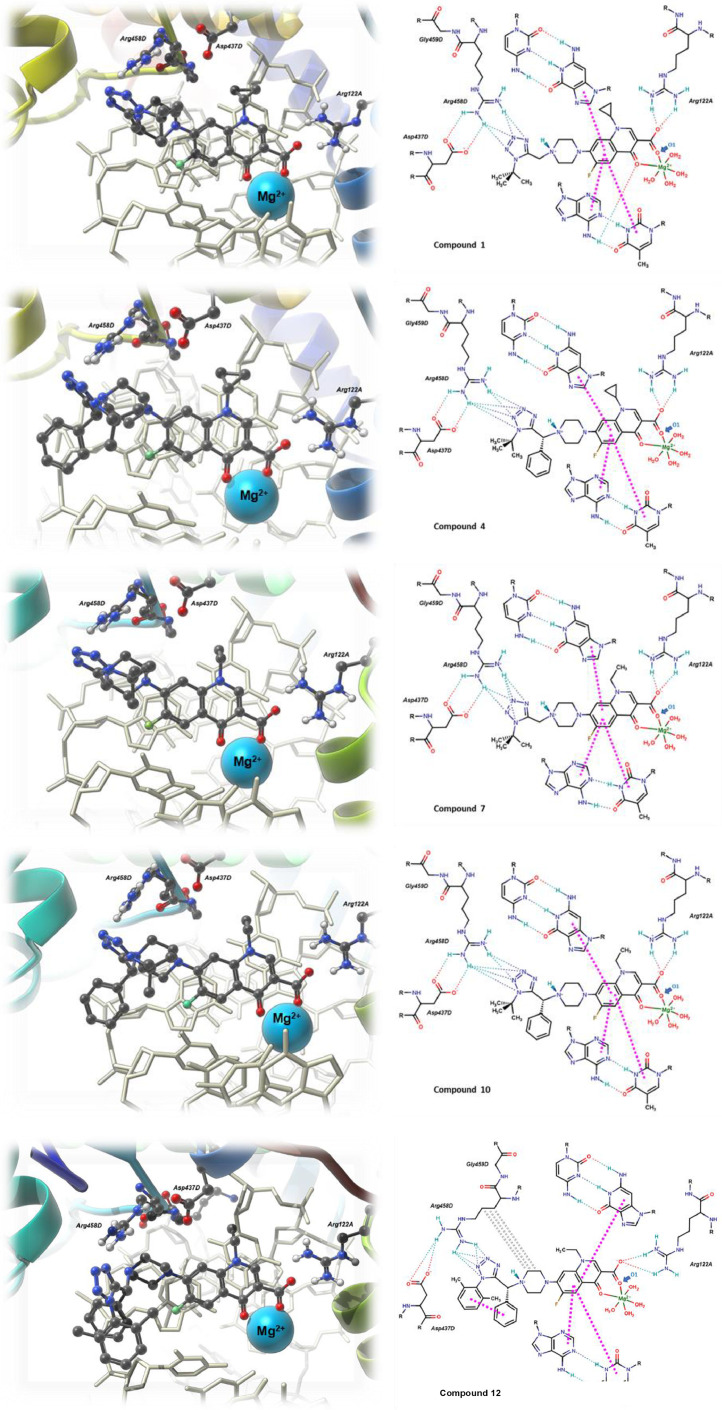
Table S1 gives the interaction energy value (MolDock Score) of each compound with the DNA gyrase. Also, each fluoroquinolone's ligand efficiency (LE) is shown, corresponding to the coefficient of the interaction energy per number of atoms in the molecule (excluding hydrogen atoms). As can be seen from the results, 12 displays the highest MolDock Score of -258.04 kcal/mol, with 1 and 5 being below a slight difference in their values. Compared to the known fluoroquinolones, 1, 4, 5, 10 and 12 show a better interaction profile. Most of the synthesized compounds are predicted to form strong hydrogen bonding interactions between Arg458D and the tetrazole moiety, as well as the Arg122A and the carboxylate group (Fig. 4 ).
Fig. 4.
3D structure representation of the complex formed by the fluoroquinolone-DNA-gyrase. 2D schematic representations of compounds 1, 4, 7, 10 and 12, displaying the most relevant interactions. Dotted lines = hydrogen bonds; dashed pink = π–π interactions; grey dotted bonds = hydrophobic interactions.
Indeed, from Table S1, almost all new compounds exhibit strong H-bonding interactions, with compounds 1, 4, 7 and 10 having the highest total HBond energy values, which correlates well with their MolDock Score values. The hydrogen bonds for 1, 4, 7 and 10 range between calculated values of –16.4 and –15.3 kcal/mol. For 2, 3, 5, 6, and 9, the hydrogen bonds are not as strong when compared to those of the compounds 1, 4, 7 and 10, having calculated total HBond values between -9.1 and -5.4 kcal/mol; however, the existing hydrogen bonds impart necessary stabilization to the complex.
Because of the missing linker and the tetrazole moiety in the known fluoroquinolones (CFX to TFX), no extra hydrogen bonding is possible with the Arg residue. Hence, low energy interactions are observed, which correlates with their MolDock score values. On the other hand, weak hydrogen bonding interactions are predicted for 11 and the rest of the known fluoroquinolones with calculated values below -4.0 kcal/mol. Also, weak hydrogen bonds are seen between the amine protons of the nitrogenous bases with the carbonyl group for some fluoroquinolones (9 and 11). Stabilizing π-π interactions between the aromatic nitrogenous bases with the aromatic moiety of the fluoroquinolones are seen in all compounds. These interactions are well explored for various known compounds that intercalate within the DNA [44], [45], [46]. However, most available fluoroquinolones (CFX to TFX) exhibit higher LE values than the new compounds 1-12, suggesting better occupancy at the DNA site due to less torsional strain in their structures.
Oxygen atoms in the synthesized fluoroquinolones have different energy contributions to the total energy interaction between the ligand and the receptor than Chauhan reported previously. In Table S2, the calculated total (ETotal) and electrostatic energy (EElec) values for the oxygen atoms (O1), which contribute the most to the interaction energy of the complex, are shown. Compounds 4, 6 and 7 display higher ETotal values (-21.20, -21.62 and -21.26 kcal/mol, respectively), while compounds 1, 10 and 12 have predicted values of -20.98 and -20.89 kcal/mol, slightly less when compared to those of 4, 6 and 7. Values above -15 kcal/mol are seen for compounds 8 and 11, while the rest have values below -14.7 kcal/mol. The observed results for 4, 6 and 7 are their preference to coordinate the divalent cation to enhance the energy interactions. For the rest of the molecules, coordination to the Mg2+ is not preferred, perhaps due to steric repulsion of the ligand with adjacent residues.
Compound 5 is predicted to have poor interactions with Mg2+, while compound 9 does not display any interaction as the molecule is docked. Hence, the coordination site of fluoroquinolone is far from the cation. The poor predicted ability to coordinate Mg2+ is compensated with strong to medium hydrogen bonds and electrostatic interactions with positively charged residues, especially with Arg122A, as seen in Fig. 4. Furthermore, no predicted hydrogen bonding is caught between the tetrazole ring and the Arg458D in compound 9, which is low values for this molecule. It is noteworthy to mention that compound 5 has an exceptionally low ETotal value, although MolDock and LE values are among the highest of all. On the other hand, 9 has the lowest ETotal value, which correlates well with its low MolDock and LE values.
Notably, the high predicted MolDock, LE and HBond energy values correspond to the (R) stereoisomers. These results may arise due to steric hindrance between the ligand and the pocket. While the (R) isomers are arranged in such a way as to minimize steric effects, the (S) isomers exhibit significant steric effects with the Arg458D residues that destabilize the formed complex. For example, compound 9 has the (S) stereochemistry. Although hydrogen bond interactions between the tetrazole ring and Arg458D residue are observed in most cases, some other favourable interactions generate a stronger interaction for some compounds. For example, in compounds 6 and 12, intramolecular π-π type interactions between aromatic rings of the ligand are only favoured when the stereochemistry of the chiral carbon is (R). This configuration also allows Van der Waals type interactions between the piperazine ring and the hydrophobic chain fragment of Arg458D. In compounds 2 and 3, the absence of the second aromatic ring leads to significant steric effects that arise due to the lack of π-π interactions, making the molecule adopt a conformation that destabilizes the resulting complex.
For compounds 1, 4, 7 and 10, the repulsions with nearby residues are minimized since the aromatic ring in their structures is pointing toward the solvent. At the same time, the tert-butyl group is assembled to prevent other conformational forms that may cause steric clashing with the residues and the nitrogenous bases. Some of these conformations are only achieved when the stereochemistry is (R) and not (S), as in compounds 4 and 10. No substantial differences between the cyclopropyl and the ethyl groups at the nitrogen are observed.
Two of the reported compounds by Chauhan et al. [33] have the N-tert-butyl-tetrazole moiety incorporating a phenyl and a 4F-phenyl group between the piperazin-1-yl group and the tetrazole, respectively. These compounds exhibit the most potent antibacterial activity with MIC values of 0.78 µ/mL. Replacement of hydrogen with fluorine at the para position does not decrease its antibacterial activity. However, substituting the phenyl ring with another group increases the MIC values, suggesting possible steric hindrance or poor stabilizing interactions. Because of the similarity with the compounds of this study, it is suggested that compounds 1 to 12 should exhibit good to excellent bioactivity.
The compounds reported by Chauhan et al. were investigated as a racemic mixture. No mention of the stereochemistry nor the effects of using the mixture and the possible interactions within the protein are explained. According to our results, the (R) isomers should exhibit more significant interactions within the enzyme than the (S) isomers. To further support this proposal, we performed molecular docking experiments of selected (R)/(S) isomers of compounds 13 to 19. From the results, the (R) isomers (Fig. 5 ) are predicted to form strong hydrogen bonding interactions from the tetrazole moiety of all compounds with the Arg458D residue of the protein. In contrast, for the (S) isomers, these interactions are too far to be formed.
Fig. 5.
3D structure representation of the complex formed by the fluoroquinolone-DNA-gyrase. 2D schematic representation of 5, 6 and 9, displaying the most relevant interactions. Dotted lines = hydrogen bonds; pink dashed = π–π interactions.
Also, the carboxylate moiety of the fluoroquinolones interacts strongly with the Arg122A residue through hydrogen bonds. Furthermore, these fluoroquinolones are predicted to coordinate with the Mg2+ through the lone pairs of the oxygen atoms (Fig. 6 ). All these results agree with the molecular docking of 1 to 12. The supplementary data section gives all the interaction energy values obtained from the molecular docking studies (Table S3).
Fig. 6.
3D structure representation of the complex formed by the fluoroquinolone-DNA-gyrase of compounds 13 to 19. Only the (R) isomers of the fluoroquinolones are displayed, with the nitrogen of tetrazole in light green and the carboxylate oxygens in red.
2.4. QSAR of quinolone-tetrazole derivatives
MIC experiments depend on the compound's pharmacodynamics and pharmacokinetics; the former can be covered by the molecular docking study (interaction energy), and the latter can be approached using molecular descriptors related to the pharmacokinetics of the compounds, such as logP, logS, molecular volume, etc. Therefore, it is unsurprising that a correlation between the docking and the MIC results was not obtained. Consequently, to achieve a correlation between the MIC and the structure of the quinolone-tetrazole derivatives, a QSAR study was done.
In Eq. 1, the best mathematical model that exceeds all statistical parameters and provides a great biological description is displayed.
| (1) |
| (2) |
| (3) |
The significant factors that influence the antibacterial activity of the quinolones are , which corresponds to the MolDock Score, which states for the Kier alpha-modified shape indices and ΔV, which represents the difference in volume compared with a template structure (Fig. 7 ) and is calculated using Eq. 4.
| (4) |
Fig. 7.
Template structure of quinolone derivatives. An equation to obtain . and state for the molecular volume of compound X+ and the molecular volume of the scaffold structure, respectively.
is a topological descriptor defined in terms of the number of graph vertices and the number of paths with length k equal to three in an H-depleted molecular graph [47]. These descriptors evaluate the molecular shape, even considering the different shape contributions of heteroatoms and hybridization states. The S3K index encodes information about the centrality of branching. The α parameter used to calculate the Kier shape indices is derived from the ratio of the covalent radius R i of the ith atom relative to the sp3 carbon atom. This descriptor fits exceptionally well in our study since fluoroquinolones are structurally related, and some differences between them are the number or type of halogen atoms. In this exact point, some of the compounds differ in the molecular cycle bind to the fluoroquinolone template. For these reasons, S3K is an excellent element for our QSAR model. According to our model, fluoroquinolones are better antibacterial agents (minor MIC) if they possess greater values of S3K, like for our new fluoroquinolone derivative.
can be related to the pharmacodynamics of the compounds and, according to the coefficient symbol in the QSAR equation [48], if a quinolone derivative has a greater value (negative), this compound will be more active against S. aureus.
Additionally, is a molecular descriptor that can be related to the pharmacodynamics and pharmacokinetics of the compounds since molecular volume is related to the antibacterial activity of fluoroquinolones in many works, especially by H. Koga and M. Ohta [49, 50]. Based on this work, many others have identified the importance of the molecular volume and size of the quinolone derivatives to their antibacterial activity. Chauhan et al. also observed this relationship through a CoMFA and CoMSIA analysis [33].
In our QSAR results, increasing the quinolone template's molecular volume (Fig. 7), increases the antibacterial activity of the compound. Nevertheless, the quadratic term of the descriptor has a positive and a small coefficient value, indicating that by increasing the molecular volume beyond some point (, considering only in the equation), the antibacterial activity will be negatively affected. QSAR approximations of this type were established and explained by Hansch and Fujita [51], [52], [53].
Because the synthesized molecules in this work (1-12) have a greater volume and number of atoms compared to the commercial fluoroquinolones, our data, combined with the data by Chauhan et al., who studied structurally related compounds, covers an extensive range of values for the volume of the molecules. We now have molecular values that can be interpolated in the QSAR. All values of the molecular descriptors present in the QSAR are shown in Table 1.
It is evident in Table 1 that commercial fluoroquinolones possess lower Eint values (more positive), in some cases lower than -190 kcal/mol (for example, PFX, CFX, and NFX). In addition, some of these compounds have ΔV values lower than the ΔVo value, following their high antibacterial activity. Therefore, an adequate combination of descriptors values may indicate of antibacterial activity in fluoroquinolones. From this perspective, fluoroquinolones presented in this work (1-12) may be potent antibacterial agents. For this reason, we predicted the MIC value of fluoroquinolones (1-12) using the QSAR model. To validate the predictive ability of the model, four different experiments (Fig. 8 ) were performed. In each experiment, the MIC value of an external set of molecules (30%) was predicted and adequate prediction results were obtained ().
Fig. 8.
Linear correlation of Ypredvs Y of four experiments. Blue colored points represent the molecules of the training set and yellow-colored points represent the external set. The R2, , , and values are indicated.
The experimental (Y), calculated (Ycal) and predicted (Ypred) MIC values are displayed in Table 2 . The absolute value of the differences between each Ycal and Ypred with Y, represented by the and terms respectively, are shown. In addition, the standard deviation of error in calculation (SDEC) and standard deviation error in prediction (SDEP) are displayed.
Table 2.
Experimental (Y), calculated (Ycal) and predicted (Ypred) MIC values of fluoroquinolones are shown. Calculated and predicted residual values ( and ) are also given. standard deviation of error in calculation (SDEC) and standard deviation error in prediction (SDEP) are displayed.
| Molecule | |||||||
|---|---|---|---|---|---|---|---|
| 1 | – | – | 0.18 | – | – | – | – |
| 2 | – | – | 0.87 | – | – | – | – |
| 3 | – | – | 1.12 | – | – | – | – |
| 4 | – | – | 0.97 | – | – | – | – |
| 5 | – | – | 1.69 | – | – | – | – |
| 6 | – | – | 1.12 | – | – | – | – |
| 7 | – | – | 0.03 | – | – | – | – |
| 8 | – | – | 0.62 | – | – | – | – |
| 9 | – | – | 0.90 | – | – | – | – |
| 10 | 0.39 | 0.76 | 0.86 | 0.37 | 0.47 | ||
| 11 | – | – | 1.71 | – | – | – | – |
| 12 | – | – | 1.43 | – | – | – | – |
| 13 | 1.56 | – | – | – | – | – | – |
| 14 | 1.56 | 1.62 | 1.72 | 0.06 | 0.16 | 0.43 | 1.25 |
| 15 | 0.78 | 0.61 | 0.55 | -0.17 | -0.23 | -0.9 | -1.22 |
| 16 | 0.78 | 0.77 | 0.77 | -0.01 | -0.01 | -0.06 | -0.08 |
| 17 | 1.56 | 1.55 | 1.55 | -0.01 | -0.01 | -0.04 | -0.07 |
| 18 | 1.56 | 1.13 | 1.06 | -0.43 | -0.5 | -2.1 | -2.47 |
| 19 | 0.78 | 1.02 | 1.14 | 0.24 | 0.36 | 1.35 | 1.99 |
| TSF | 0.1 | 0.05 | 0.02 | -0.05 | -0.08 | -0.29 | -0.46 |
| CLIN | 0.5 | 0.44 | 0.4 | -0.06 | -0.1 | -0.35 | -0.6 |
| CFX | 0.13 | 0.28 | 0.32 | 0.15 | 0.2 | 0.79 | 1.01 |
| GFX | 0.25 | 0.31 | 0.31 | 0.06 | 0.06 | 0.28 | 0.3 |
| LFX | 0.13 | 0.46 | 0.53 | 0.34 | 0.4 | 1.67 | 1.98 |
| MFX | 0.6 | 0.46 | 0.42 | -0.14 | -0.18 | -0.7 | -0.92 |
| LMF | 0.2 | 0.24 | 0.24 | 0.04 | 0.04 | 0.18 | 0.22 |
| NFX | 0.39 | 0.34 | 0.33 | -0.05 | -0.06 | -0.24 | -0.29 |
| PFX | 0.5 | 0.26 | 0.17 | -0.24 | -0.33 | -1.28 | -1.75 |
| SFX | 0.13 | 0.24 | 0.26 | 0.12 | 0.13 | 0.57 | 0.65 |
| SIT | 0.5 | 0.45 | 0.42 | -0.05 | -0.08 | -0.28 | -0.46 |
| TFX | 0.6 | 0.43 | 0.4 | -0.1 | -0.2 | -0.85 | -1.01 |
As indicated in Table 2, LFX, TFX, 18 and 19 have greater . The predictive ability of the QSAR model is shown in Fig. 8. According to the values of R2 and considering the size and nature of this system, the QSAR model provides a good description and predictive ability (based on its and values). Also, molecular descriptors (, , and ) presented in the model can be used to explain the antibacterial activity of fluoroquinolones based on their pharmacodynamics and pharmacokinetics.
We used the 4-quinolone structure as a reference to analyze the effect of the structural modifications for each compound through the electron density map, polar surface area (PSA) value, and the molecular volume variations for each one [51]. The electrostatic potential maps of commercial fluoroquinolones show that an electron-rich region (colored red) is concentrated over the carbonyl and carboxylate ion (ketoacid group), and the electron-deficient zone (colored blue) is located over the ring containing the quaternary nitrogen. Also, a neutral site is displayed (yellow-green) over the quinolone base structure (Fig. S1).
Similar features were noted in the MEP map analysis of the synthesized fluoroquinolones. Negative, positive and zero MEP values zones were located over the ketoacid group, quaternary nitrogen, and the quinolone moieties of the compounds, respectively (Fig. S2). Nevertheless, an increase in the MEP values of the neutral MEP zones is noted, primarily over the hydrophobic substituents of the tetrazole ring (phenyl or methyl). Regarding PSA and molecular volume values, the synthesized compounds showed higher PSA values (87.48 Å2 to 92.32 Å2) compared to the commercial drugs (52.64 Å2 to 80.64 Å2). In addition, the synthesized compounds possess a higher molecular volume (461.16 Å3 to 590.12 Å3) (Table S4).
Analysis of the molecular descriptors and the MIC values of the commercial drugs revealed that the most active compounds (TSF and SFX) have the highest PSA values: 80.39 Å2 and 80.64 Å2, respectively. Drugs with low activity (with higher MIC) had a lower value for the PSA: for example, PFX, TFX and MFX have PSA values of 52.64, 70.86 and 66.72 Å2, respectively. Some commercial drugs showed an average molecular volume (concerning all the analyzed compounds) and had a higher activity with lower MIC values. For example, the most active compound (TSF, MIC of 0.1 µg/mL) has a volume of 362.09 Å3, and the compound with the highest volume (MFX, V=391.09 Å3) has a MIC value of 0.6 µg/mL. NFX has the lowest volume, 329.12 Å3, and its MIC value is 0.39 µg/mL. These drugs are derived from CFX, and analysis of the structural relationship between these two compounds revealed that having groups that increase the molecular volume benefits the biological activity (showing lower MIC values).
However, it was also noted that these groups must include specific atoms, such as nitrogen and halogens (F and Cl), because in compounds where nonpolar groups (-CH3, -CH2) were added, in some cases, their volume increases as does their MIC value (for example see MFX and TFX). Other structural features that negatively affected bactericidal activity include adding rotatable bonds. For instance, GFX (MIC= 0.25 µg/mL) has a cyclopropyl group attached to the nitrogen of its quinolone structure, and when it is changed to an ethyl group, such as in NFX, its MIC value increases (0.39 µg / mL). Molecules with several rotatable bonds can adopt different geometries, some of which can be unfavorable for biological activity [54]. For this reason, one of the most common structural modifications is the rigidification of flexible molecules.
2.5. Molecular docking with COVID-19 main protease
To evaluate the potential antiviral activity of the fluoroquinolone-tetrazole derivatives as inhibitors of CoV-2-MPro, we used four crystal structures of CoV-2-MPro co-crystallized with two types of inhibitors (Fig. 9 ), a covalent inhibitor for 6lu7 [11] and a non-covalent inhibitor for 6m2n [54], 6w63 [55] and 7l0d [56]. Using previous results on noncovalent inhibitors [57, 58] and combining these results with information regarding the catalytic mechanism of CoV-2-MPro [59] and the inhibition mechanism of a covalent inhibitor [60], the analysis of the intermolecular interaction of our compounds (1-12) with the following aa of CoV-2-MPro: Leu27, His41, Met49, Cys145, His164, Met165, Arg188, Gln 189 and Gln 192 was set as a starting point (Fig. 9). These aa are conserved in many of the interaction profiles of CoV-2-MPro with inhibitors or their substrate.
Fig. 9.
Structural alignment of CoV-2-MPro crystals: 6lu7 (yellow), 6m2n (blue), 6w63 (cyan) and 7l0d (grey). Leu27, His41, Met49, Cys145, His164, Met165, Arg188, Gln189 and Gln 192 are shown in stick representation.
In addition to the interaction as mentioned earlier parameters, the interaction profiles of all the ligands co-crystallized with CoV-2-MPro in the crystal complexes were used for the docking calculations (Fig. 10 ). These include 6lu7 (peptide-like inhibitor), 6m2n (baicalein, flavonoid-type compound), 6w63 (designed inhibitor) and 7l0d (designed inhibitor). The selection of these crystal structures of CoV-2-MPro was based on the co-crystallized ligands (different sizes and types) since a better analysis of the inductive effect can be done. Furthermore, more and different initial conformations of CoV-2-MPro can be explored.
Fig. 10.
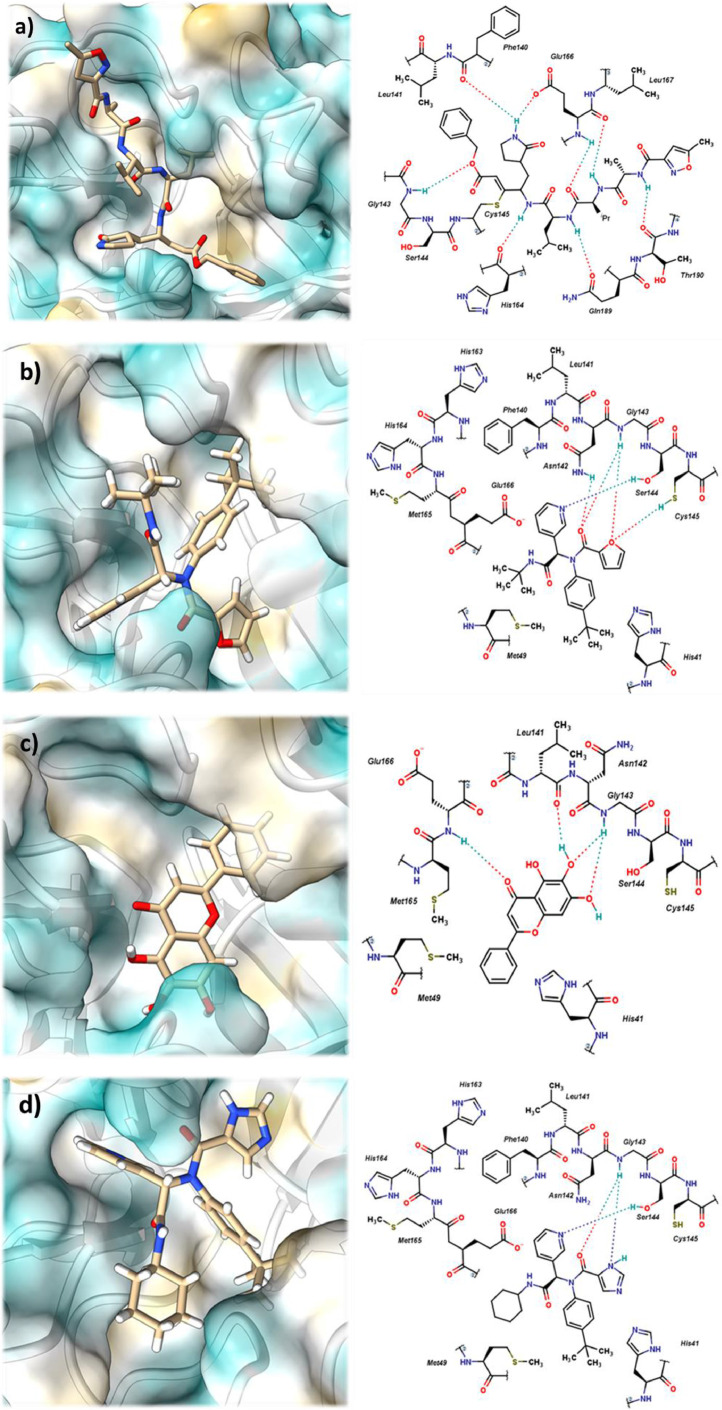
Non-covalent interactions of different inhibitors with CoV-2-Mpro. A) 6lu7, b) 7l0d, c) 6m2n, and d) 6w63. Surface plots of amino acid lipophilicity, the Kyte-Doolittle scale.
From Fig. 10, shows that the noncovalent inhibitors interact strongly by means of hydrogen bonds with the aa located near the catalytic dyad: Phe140, Leu141, Asn142, Gly143 and Ser144. Also, a high energy interaction value occurs with Glu166. These six interactions are present in the four co-crystallized inhibitors, with the except for 6m2n, where the interaction with Phe140 is absent (Table 4). The interactions of the inhibitors with the aa were also established as necessary in previous research [55], [56], [57], [58].
Table 4.
Interaction energy values (kcal/mol) of specific aa of CoV-2-Mpro and fluoroquinolone templates, CFX and NFX.
| aa |
CFX |
NFX |
||||||
|---|---|---|---|---|---|---|---|---|
| 6lu7 | 7l0d | 6m2n | 6w63 | 6lu7 | 7l0d | 6m2n | 6w63 | |
| Leu27 | – | – | – | – | – | – | – | – |
| His41 | -5.48 | -16.44 | -16.17 | -31.73 | -4.55 | -17.09 | -18.33 | -28.32 |
| Met49 | -2.23 | -10.32 | -8.82 | -13.01 | -2.46 | -9.13 | -13.67 | -12.63 |
| Phe140 | -0.32 | – | -3.24 | – | -0.32 | – | -1.86 | -0.41 |
| Leu141 | -3.54 | – | -4.05 | -3.93 | -3.03 | – | -2.97 | -5.01 |
| Asn142 | -8.04 | – | -11.85 | -12.21 | -5.64 | – | -11.74 | -10.20 |
| Gly143 | -6 | – | -1.27 | -4.32 | -4.21 | – | -4.66 | -3.33 |
| Ser144 | -6.65 | – | -1.55 | -4.68 | -5.96 | – | -5.57 | -4.86 |
| Cys145 | -9.11 | – | -4.87 | -3.86 | -7.87 | – | -11.01 | -5.51 |
| His164 | -7.48 | -2.43 | -8.41 | -6.09 | -7.98 | -3.22 | -8.35 | -6.40 |
| Met165 | -23.88 | -12.32 | -19.66 | -5.48 | -24.95 | -11.86 | -21.01 | -11.78 |
| Glu166 | -19.75 | -5.78 | -20.18 | -4.23 | -18.02 | -5.85 | -8.08 | -18.20 |
| Arg188 | -5.55 | -14.55 | -9.27 | -7.10 | -6.48 | -16.54 | -5.77 | -7.03 |
| Gln189 | -17.91 | -16.42 | -21.41 | -10.67 | -19.88 | -15.059 | -9.03 | -12.43 |
| Gln192 | -0.65 | -9.18 | – | – | -3.08 | -11.24 | – | – |
| MDSG | -117.11 | -102.49 | -125.63 | -126.39 | -118.64 | -102.82 | -119.88 | -133.19 |
| HBtot | -13.87 | -5 | -8.05 | -7.72 | -12.89 | -5.52 | -14.44 | -16.15 |
| LE | -4.88 | -4.27 | -5.23 | -5.27 | -5.16 | -4.47 | -5.21 | -5.79 |
The inhibitors, regardless of their structural differences, share common interaction profiles. It is worth mentioning that enzyme inhibition in other studies (Table 2) is involved in the interaction of five aa: Phe140, Leu141, Asn142, Gly143, Ser144 and Glu166. From Table 3 shows that these inhibitors interact strongly with specific aa, such as His41, Met165, Asn142 and Glu166.
Table 3.
Interaction energy values (kcal/mol) of the co-crystallized inhibitors with specific aa of CoV-2-Mpro.
| aa | 6lu7 | 7l0d | 6m2n | 6w63 |
| Leu27 | -1.99 | -4.51 | -0.33 | -3.67 |
| His41 | -13.34 | -21.09 | -24.23 | -19.72 |
| Met49 | -5.85 | -8.18 | -13.18 | -7.21 |
| Cys145 | 16.15 | -8.81 | -7.61 | -7.22 |
| His164 | -11.25 | -3.59 | -6.52 | -4.93 |
| Met165 | -21.02 | -10.38 | -7.94 | -12.34 |
| Arg188 | -5.38 | -4.47 | -3.72 | -4.38 |
| Gln189 | -26.68 | -5.04 | -3.17 | -8.21 |
| Gln192 | -4.64 | |||
| *Phe140 | -8.89 | -4.26 | -5.27 | |
| *Leu141 | -9.58 | -7.61 | -1.41 | -7.79 |
| *Asn142 | -14.02 | -18 | -8.41 | -15.24 |
| *Gly143 | -10.62 | -9.78 | -7.68 | -7.99 |
| *Ser144 | -6.3 | -2.65 | -2.19 | -3.27 |
| *Glu166 | -28.62 | -18.61 | -6.59 | -27.03 |
| MDSG | -190.68 | -145.75 | -80.05 | -142.69 |
| HBtot | -18.43 | -11.05 | -10.08 | -8.42 |
| LE | -3.89 | -4.56 | -4.00 | -4.20 |
* Additional aa considered for these inhibitors.
Molecular docking studies of CFX and NFX with the four crystal structures of CoV-2-MPro were examined to analyze the effect of the structural modifications of CFX and NFX on the interactions with CoV-2-MPro.
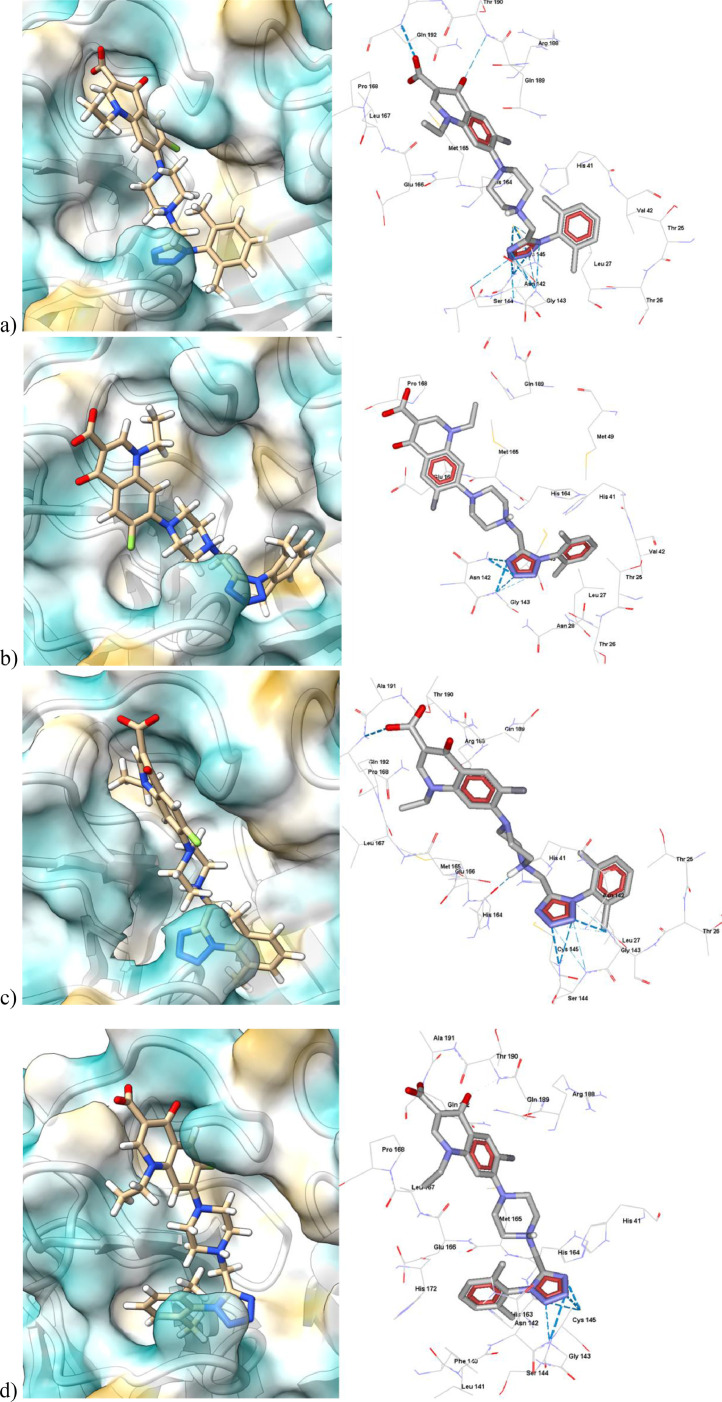
From Fig. 11 , we can observe that CFX and NFX can bind differently to CoV-2-Mpro based on their conformations. The structural rigidity and small size of the quinolone allow for the binding versatility of CFX and NFX. In addition, these compound's electrostatic features (zwitterion) of these compounds facilitate their different binding motifs with other residues. Table 4 lists the interaction energy values of CFX and NFX with CoV-2-Mpro according to its conformation in a given crystal structure.
Fig. 11.
Molecular docking and non-covalent interactions of CFX and NFX with different crystal structures of CoV-2-Mpro. a) 6lu7, b) 7l0d, c) 6m2n, and d) 6w63. Surface plots of amino acid lipophilicity on the Kyte-Doolittle scale.
In addition to the aa that interacts with the co-crystallized inhibitors, the interactions of CFX and NFX with Arg188, Gln189 and Gln192 of CoV-2-Mpro need to be considered (Table 3). These aa are in the loop in the S4 region of the catalytic site. According to the crystal conformation of the enzyme, CFX and NFX will bind in different orientations, especially in the crystals 7l0d, 6m2n, and 6w63, in which the carboxylate moiety is pointing towards the S2 and S4 regions (Fig. 11).
The binding of the tetrazole-fluoroquinolones to the catalytic cavity of CoV-2-Mpro was analyzed. From the calculations, we detected two extra aa must be included in the energy interaction profile: Leu167 and Pro168 (Table 4). The interaction energy parameters for tetrazole-fluoroquinolones (1-12) with each crystal structure of CoV-2-Mpro are given in the Supplementary materials (Table S5-S8). From these results, we selected 5 and 8 as the best candidates to be inhibitors of CoV-2-Mpro. Table 5 shows the most essential residual/molecule energy interactions of the catalytic pocket residues with the fluoroquinolone-tetrazole derivatives 8 and 5.
Table 5.
Interaction energy values (kcal/mol) of specific aa of CoV-2-Mpro and fluoroquinolone templates, 8 and 5.
| aa |
8 |
5 |
||||||
|---|---|---|---|---|---|---|---|---|
| 6lu7 | 7l0d | 6m2n | 6w63 | 6lu7 | 7l0d | 6m2n | 6w63 | |
| Leu27 | -9.76 | -2.40 | -4.91 | – | -1.58 | -1.00 | -1.79 | -0.78 |
| His41 | -8.27 | -15.46 | -5.41 | -3.96 | -13.60 | -15.63 | -6.13 | -20.29 |
| Met49 | -2.97 | -1.62 | -0.58 | – | 8.13 | -2.78 | -4.08 | -12.42 |
| Phe140 | – | – | – | -7.46 | – | -2.35 | -5.72 | -1.64 |
| Leu141 | -1.80 | -1.45 | -2.23 | -9.89 | – | -6.45 | -5.53 | -4.76 |
| Asn142 | -14.31 | -19.35 | -15.98 | -19.45 | -11.16 | -15.12 | -25.52 | -14.93 |
| Gly143 | -10.44 | -12.27 | -12.45 | -10.63 | -5.94 | -10.12 | -6.28 | -5.71 |
| Ser144 | -8.00 | -0.57 | -6.88 | -6.14 | -0.63 | -3.35 | -5.17 | -4.04 |
| Cys145 | -15.99 | -7.07 | -15.00 | -11.45 | -7.18 | -0.73 | -15.54 | -7.02 |
| His164 | -4.66 | -2.05 | -5.18 | -2.27 | -4.49 | -7.21 | 4.79 | -7.04 |
| Met165 | -19.31 | -11.37 | -16.90 | -18.82 | -14.10 | -16.66 | -22.50 | -15.24 |
| Glu166 | -10.55 | -21.25 | -14.84 | -24.23 | -10.12 | -30.24 | -22.53 | -18.77 |
| Leu167 | -10.68 | -9.21 | -7.84 | -2.95 | -6.03 | -9.18 | -4.25 | -6.50 |
| Pro168 | -8.91 | -19.02 | -12.54 | -14.29 | -2.24 | -20.95 | -6.13 | -13.99 |
| Arg188 | -1.10 | -0.51 | -2.33 | -3.21 | 2.76 | -2.26 | -1.13 | -2.32 |
| Gln189 | -27.40 | -4.14 | -16.31 | -23.53 | -26.24 | -14.96 | -20.73 | -22.35 |
| Gln192 | -12.43 | – | -14.08 | -1.17 | -6.70 | -2.11 | -7.63 | -7.51 |
| MDSG | -159.54 | -128.33 | -155.64 | -156.18 | -112.29 | -164.55 | -167.24 | -167.30 |
| HBtot | -25.65 | -10.13 | -15.10 | -13.09 | -3.86 | -11.05 | -16.46 | -10.97 |
| LE | -4.31 | -3.47 | -4.21 | -4.22 | -2.55 | -3.74 | -3.80 | -3.80 |
The interaction analysis reveals that the molecules with higher affinity exhibit three interactions with the binding site: the first is the possibility of a hydrogen bond between the carboxylate and the amino group of Gln192. The second is the possibility of a hydrogen bond between the carbonyl oxygen of the fluoroquinolone and the amino group of Thr190. However, the most important non-covalent interactions are the hydrogen bonds between the tetrazole ring and Phe140, Leu141, Asn142, Gly143, Ser144 and Cys145, as shown in Fig. 12 , an effect that can be inferred as a probable inhibitory activity of CoV-2-MPro.
Fig. 12.
Molecular docking and non-covalent interactions of 8 with different crystal structures of CoV-2-Mpro. A) 6lu7, b) 7l0d, c) 6m2n, and d) 6w63. Hydrogen bond interactions are shown as cyan dashed lines. Surface plots of amino acid lipophilicity, the Kyte-Doolittle scale.
The R enantiomer facilitates the interaction between the tetrazole ring and Cys145; however, in compounds with S chirality, the phenyl substituent is closer to Cys145 but without any strong non-covalent interaction, and the tetrazole ring is further away from interacting with the opposite area of the catalytic site; however, without any strong interactions. The results of the in silico interaction also revealed that the change from cyclopropyl to ethyl on the quinolone nitrogen does not play an important role in the binding affinity (Fig. 11) in the catalytic site (as was observed for the binding with the S. aureus gyrase). Thus, the docking parameters of 1, 2, 3, 4, 5 and 6 are similar to those of 7, 8, 9, 10, 11, and 12, respectively.
From Fig. 12, in all the positions of 8, the tetrazole interacts with the Cys145 and an aa near it, such as Ser144, Gly143, Asn142 and Leu141. The high capacity of the tetrazole ring to form HBs is due to the nitrogen atoms (HBA), which allows this molecule to interact strongly (≈50 kcal/mol) with the catalytic loop of CoV-2-Mpro. In addition, the dimethyl phenyl group binds in the hydrophobic region of the catalytic pocket (over S1 and S1’).
The zwitterionic state of these molecules increases the stability of the interaction by forming strong HBs between the ammonium (HBD) and the carboxylate groups (HBA). The carboxylate group is orientated to the opposite side of the catalytic site. In fact, in most of the positions. This group is exposed to the solvent in most of the positions, except in the 6lu7 crystal, where it binds in a buried conformation interacting by HB with Gln192.
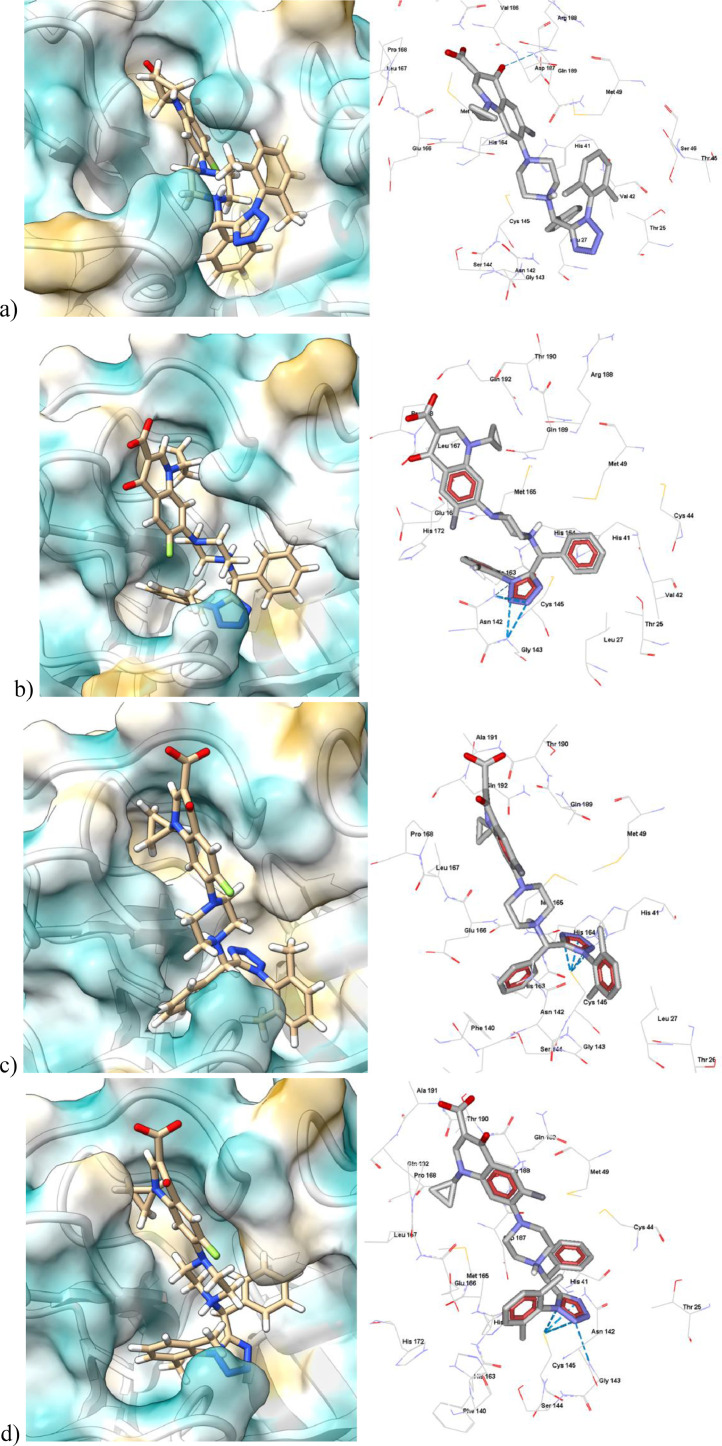
In Fig. 13 , we can see that the molecular structure of 5, especially the fragment where the tetrazole is located, fits into the catalytic cavity of CoV-2-Mpro. Also, tetrazole's hydrophobic character and planarity contribute to stabilizing the binding. The tetrazole interacts with aa located in loop S1’ (Phe140, Leu141, Asn142, Gly143, Ser 144 and Cys145) and forms HBs with Cys145. These interactions explain the high interaction energy value with this aa, with an average value of 10.2 kcal/mol for 8 and –10 kcal/mol for 5.
Fig. 13.
Molecular docking and non-covalent interactions of 5 with different crystal structures of CoV-2-Mpro. A) 6lu7, b) 7l0d, c) 6m2n, and d) 6w63. Hydrogen bond interactions are shown as cyan dashed lines. Surface plots of amino acid lipophilicity, the Kyte-Doolittle scale.
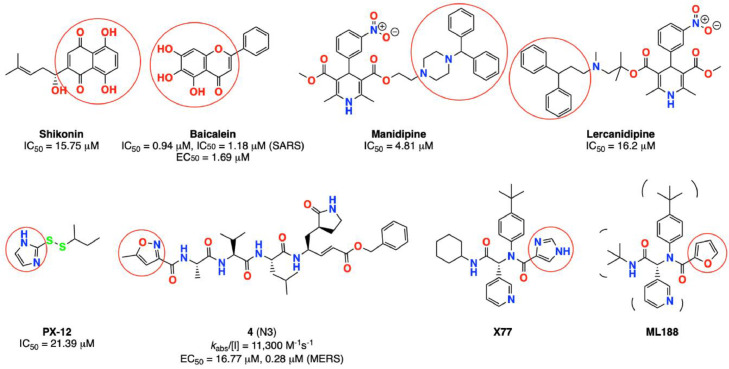
The strong energy interaction obtained by 5 and 8 tetrazole-fluoroquinolone derivatives and CoV-2-Mpro, and their potential inhibitory activity, and therefore, possible antiviral SARS CoV-2 activity, can be supported by the experimental biological evaluation done by other groups [59], [60], [61], [62], [63], [64] using similar compounds. As can be seen in Fig. 14 , some of the experimentally reported inhibitors have a piperazine ring, a bicyclic aromatic ring, or a diphenyl methyl fragment, similar to the tetrazole-fluoroquinolones in this study.
Fig. 14.
Molecules are used as inhibitors of CoV-2-Mpro [59], [60], [61], [62], [63], [64]. Similar molecular fragments are highlighted in red color.
Molecular size is also similar between the tetrazole-fluoroquinolones and some reported inhibitors, especially manidipine and lercanidipine. Furthermore, some reported inhibitors have a five-membered heterocyclic ring, such as imidazole or furan ring, interacting with the aa near Cys145 using HBs. These data follow the interactions between the tetrazole ring of the fluoroquinolones and this region.
Finally, we analyzed the pharmacokinetic properties of our compounds through of the SwissADME server, and the results are displayed in Table S5X. The results indicate that 7 is the most promising candidate considering its pharmacokinetics properties, specifically, the non-toxicity compared to to CYP enzymes. Nevertheless, 10, 11 and 12 show increased bioavailability, even more, significant than the bioavailability of commercial fluoroquinolones. Taking these results and the excellent interaction energy values, we can consider fluoroquinolones with a tetrazole moiety promising candidates for in vitro inhibition experiments.
3. Conclusion
The hybridization of fluoroquinolones with tetrazoles represents a strategy to develop new biologically active compounds with potential therapeutic applications. The results of the molecular docking studies identified the structural features that can influence the binding affinity of the compounds to the enzymes: the volume, the types of interactions, including hydrophilic and hydrophobic interactions, as well as hydrogen bonding and the stereochemistry of the C-7 substituent on the fluoroquinolone scaffold.
The design strategy adopted for the new series of compounds was to introduce different groups at position 7 of the quinolone nucleus since substitution at that location was found to affect potency, bioavailability, and physicochemical properties as well as affinity to DNA gyrase (Gram-negative bacteria) and/or Topoisomerase IV (Gram-positive bacteria) [65, 66]. In addition, introducing an aryl group at the piperazine moiety of the fluoroquinolone shifted the activity from antibacterial to antiviral and anticancer [66, 67].
Some electronic properties, such as the molecular electrostatic potential (MEP), polar surface area (PSA) and molecular volume, were calculated to realize the structure-activity relationship (SAR) of the hybrid compounds. The electrostatic potential maps of the compounds in Figs. S1, S2 and S3 show that an electron-rich region (colored red) is concentrated over the carbonyl and carboxylate ion (ketoacid group). The electron-deficient zone (colored blue) is located over the ring with the quaternary nitrogen, and a neutral site (yellow-green) is situated over the quinolone base structure.
Molecular docking studies were carried out to understand the binding mechanism of the newly synthesized compounds with the protein receptor (DNA gyrase of S aureus and Covid-19 main protease). The score, ligand efficiency (LE) and hydrogen bond energy formed in the active site of the protein receptor are used to predict the binding modes, the binding affinities, and the orientation of the docked hybrid derivatives. All the tested compounds have an affinity for the DNA gyrase, with a MolDock Score between -258.04 to -182.03 kcal/mol. The most potent compound is 12; other compounds with scores in a similar range are 1, 4, 5 and 10. Derivatization of the ciprofloxacin and norfloxacin molecules leads to compounds with better affinities for the DNA gyrase enzyme, considering the potentially more extensive surface contact.
The docking study also showed that the compounds ligated to the magnesium ion via the C-3 carboxylic group and C-4 carbonyl functionalities also have existing interactions with the active site of the gyrase enzyme through hydrogen bonding with amino acid residues (Arg458D, Arg122A), intermolecular π-stacking with nucleotide bases through the quinolone moiety and the tetrazole moiety. π-π interactions are prevalent when the stereochemistry of the chiral carbon is R. Furthermore, compounds 6 and 12 promote additional Van der Waals interactions with gyrase amino acids. The results demonstrated that tetrazole moiety improves biological activity due to the high electron density of the nitrogen atoms in the tetrazole ring.
In our compounds, the structural components added to the fluoroquinolone main structure, increase the interaction energy with the CoV-2-Mpro, specially the tetrazole moiety that makes many hydrogen bonds within the catalytic site, and promotes the orientation of the phenyl and alkyl components to the lipophilic regions. The similarity in the binding mode and the energy interaction with crucial residues of our compounds, compared to approved CoV-2-Mpro inhibitors, support our hypothesis to propose our compounds (particularly5 and 8) as CoV-2-Mpro inhibitors.
4. Methods and materials
4.1. Experimental section
General. Norfloxacin and ciprofloxacin were obtained in pharmaceutical dosage form, and thus, purification was required before the reaction. The remaining reagents were commercially available and were used without further purification. The IR spectra were recorded on a Nicolet model iS10FT-IR spectrometer in ATR mode. 1H-,13C-, and 19F- NMR spectra were recorded on Varian Inova 600 (I600) spectrometer using tetramethylsilane (TMS) as an external standard. The chemical shift is on the δ scale, and coupling constants (J) are in hertz.
Mass spectra analysis was performed using a Thermo Scientific DFS (Double Focusing Sector). All samples were analyzed by Electronic Impact (EI) as suspended solids in a capillary tube filled with acetone. The DFS equipment works in the range of 0-1200 m/z. High-resolution mass spectrometry (HRMS) was also performed to confirm the elemental composition of the synthesized products.
Synthesis of 7-(4-((1-(tert-butyl)-1H-tetrazol-5-yl)methyl)piperazin-1-yl)-1-cyclopropyl-6-fluoro-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (1).
General procedure for the synthesis of 1. As depicted in Fig. 15 , the synthetic pathway leading to the quinolone derivatives is quite similar. 100 mg (0.302 mmol) of ciprofloxacin (CFX) was placed in a flask with 6 mL of MeOH. The mixture was heated to 75-80 °C under constant stirring; once it was completely dissolved, 54.4 mg (0.604 mmol) of paraformaldehyde was added to the flask with one drop of 10% HCl. The mixture was stirred until all the solvent evaporated. The reaction product was re-dissolved in MeOH, then 32.6 mg (0.392 mmol) of tert-butyl isocyanide and 45.2 mg (0.392 mmol) of trimethylsilyl azide were added to the flask at a temperature of 45-50 °C. The resulting mixture was heated (80-85 °C) for 3 h. The final product was washed and rinsed with cold water and then recrystallized with acetone. Ivory solid; yield 30 % mp 210 °C (decomposition). ATR FT-IR (cm−1): 2828 (C-H aliph), 1725 (C=O acid), 1630 (C=O ketone), 1464 (C=C Ar), 1494, 1389, 1337 (C-(CH3)3), 1262 (C-O), 1011 (C-F), 940, 888 (2RC=CR-H), 804 (C-H oop). 1H NMR (600 MHz, DMSO-d6, δ ppm): δ 15.20 (s, 1H), 8.66 (s, 1H), 7.92 (d, J = 12.0 Hz, 1H), 7.56 (d, J = 6.0 Hz, 1H), 4.00 (s, 2H), 3.82 (br s, 1H), 3.36 (br s, 4H), 2.65 (br s, 4H), 1.75 (s, 9H), 1.31 (m, 2H), 1.17 (m, 2H). 13C NMR (151 MHz, DMSO-d6, δ ppm) δ 176.78, 166.33, 151.53, 148.43, 139.59, 111.45, 111.30, 107.17, 106.98, 62.27, 52.42, 51.46, 49.72, 36.31, 29.35, 28.93, 8.00. 19F NMR (376 MHz, DMSO-d6, δ ppm): δ -121.44. ESI-MS C23H28FN7O3 (m/z): 469.25 [M] +; HRMS: calc.: 469.2237 (M+); exp.: 469.2219 (M+). COSY (DMSO-d6, δ ppm) 3 J H,H (3.79, 1.28), (3.29, 2.62). HSQC (DMSO-d6, δ ppm) 1 J C,H (8.63, 148.88), (7.87, 111.31), (7.54, 106.34), (3.96, 51.47), (3.78, 36.34), (3.29, 48.71), (2.62, 52.44), (1.72, 29.36), (1.27, 7.97), (1.27, 7.97), (1.14, 8.02). HMBC (DMSO-d6, δ ppm) J C,H 15.31 (132.52), 8.63 (176.73, 166.33, 139.59, 36.97), 7.89 (154.61, 145.45, 139.55), 7.54 (119.10, 139.61, 145.45, 152.54), 3.97 (151.52, 52.42), 1.72 (52.27, 29.36).
Fig. 15.
General procedure for synthesis of fluoroquinolone-tetrazole hybrids.
Synthesis of 1-cyclopropyl-7-(4-((1-(2,6-dimethylphenyl)-1H-tetrazol-5-yl)methyl)piperazin-1-yl)-6-fluoro-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (2)
For the synthesis of 2, 100 mg (0.302 mmol) of ciprofloxacin (CFX) was placed in a flask along with 6 mL of MeOH. The mixture was heated to 75-80 °C under constant stirring; once it was completely dissolved, 54.4 mg (0.604 mmol) of paraformaldehyde was added to the flask with one drop of HCl (10%). The mixture was stirred until all the solvent was evaporated. The reaction product was re-dissolved in MeOH, then 51.5 mg (0.392 mmol) of 2,6-dimethylphenyl isocyanide and 45.2 mg (0.392 mmol) of trimethylsilyl azide were added to the flask keeping the temperature at 45-50 °C. The resulting mixture was heated (80-85 °C) for 3 h. The final product was washed and rinsed with cold water before recrystallizing with acetone. Ivory solid; yield 29 % mp 160 °C (decomposition). ATR FT-IR (cm−1): 3066 (C-H arom), 2838 (C-H aliph), 1725 (C=O acid), 1625 (C=O ketone), 1541 (C=C), 1493 (C=C Ar), 1467, 1338, 1301, 1258 (C-O), 1130, 1004 (Ar-F), 942, 807 (2RC=CR-H), 781 (C-H oop). 1H NMR (600 MHz, DMSO-d6, δ ppm): 15.16 (s, 1H), 8.62 (s, 1H), 7.86 (d, J = 12.0 Hz, 1H), 7.47 (dd 1H), 7.44 (d, J = 6.0 Hz, 1H),7.33 (d, J = 6.0 Hz, 2H), 3.77 (m, 1H), 3.71 (s, 2H), 3.16 (4H) 1.90 (s, 6H), 1.26 (m, 2H), 1.13 (m, 2H). 13C NMR (151 MHz, DMSO-d6) 194.15, 176.74, 153.57, 148.43, 135.77, 131.39, 129.12, 111.46, 107.15, 106.84, 52.63, 49.61, 49.54, 40.36, 40.22, 40.08, 39.94, 39.80, 39.66, 39.52, 36.29, 31.12, 17.35, 7.98. 19F NMR (564 MHz, DMSO-d6): -121.46. ESI-MS C27H28FN7O3 (m/z): 517.17 [M] +; HRMS: calc.: 517.2237 (M+); exp.: 517.2222 (M+). COSY (DMSO-d6, δ ppm) 3 J H,H (2.57, 3.16), 4 J H,H (7.33, 1.90). HSQC (DMSO-d6, δ ppm) 1 J C,H (3.70, 49.56), (3.16, 49.05), (2.57, 49.70), (2.56, 52.66), (1.89, 17.91), (1.89, 17.35), (1.89, 16.78), (1.89, 16.21). HMBC (DMSO-d6, δ ppm) J C,H 8.61 (176.77, 166.42, 139.57, 36.30), 7.49 (152.47), 7.44 (135.76), 7.32(132.05, 17.29), 7.31(129.11), 3.70 (153.56, 52.63), 3.16(145.17), 1.89 (135.75, 132.09, 129.12).
Synthesis of 7-(4-((1-(tert-butyl)-1H-tetrazol-5-yl)(phenyl)methyl)piperazin-1-yl)-1-cyclopropyl-6-fluoro-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (3, 4)
A methodology similar to that used for the synthezing1 was employed to obtain 3, 4; however, instead of paraformaldehyde, 0.332 mmol (35.2 mg) of benzaldehyde was added to the mixture under the same conditions. Ivory solid; yield 44 % mp 220°C (decomposition). ATR FT-IR (cm−1): 3044 (C-H arom), 2815 (C-H aliph), 1729 (C=O acid), 1615 (C=O ketone), 1588 (C=C), 1474 (C=C Ar), 1450, 1376 (C-(CH3)3), 1282, 1258 (C-O), 1141, 1008 (Ar-F), 933, 832 (2RC=CR-H), 804, 722 (C-H oop). 1H NMR (600 MHz, DMSO-d6, δ ppm): 8.62 (s, 1H), 7.86 (d, J = 12.0 Hz, 1H), 7.83 (d, J = 6.0 Hz, 1H), 7.50 (d, J = 12.0 Hz, 2H), 7.37 (dd, J = 6.0 Hz y J = 12.0 Hz, 2H), 7.32 (dd, J = 6.0 Hz y J = 12.0 Hz, 1H), 5.63 (s, 1H), 3.80 (m, 1H), 3.27 (m, 2H), 3.21 (m, 2H), 2.87 (m, 2H), 1.64 (s, 9H), 1.27 (m, 2H), 1.14 (m, 2H). 13C NMR (151 MHz, DMSO-d6, δ ppm) 176.76, 166.37, 154.43, 154.26, 152.61, 148.36, 146.11, 145.48, 139.68, 135.80, 130.12, 128.82, 118.82, 118.76, 111.40, 107.12, 106.51, 63.18, 62.03, 51.12, 51.09, 50.18, 49.52, 45.76, 39.97, 36.27, 30.01, 7.98. 19F NMR (564 MHz, DMSO-d6, δ ppm): -121.64. ESI-MS C29H32FN7O3 (m/z): 545.24 [M] +; HRMS: calc.: 545.2550 (M+); exp.: 545.2552 (M+). COSY (DMSO-d6, δ ppm) 2 J H,H (1.27, 1.15), 3 J H,H δ (7.50, 7.37), (7.39, 7.32), (7.39, 7.50), (7.36, 7.32), (7.36, 7.50), (3.80, 1.28), (3.78, 1.15), (3.21, 2.87), (2.56, 2.85), (2.57, 3.25), (1.28, 3.8), (1.26, 3.74), (1.14, 3.78), (1.30, 1.15) 4 J H,H (8.63, 3.80), (5.62, 7.50), (2.86, 2.56). HMBC (DMSO-d6, δ ppm) J C,H 8.62 (176.75, 166.36, 139.61, 107.04, 36.28), 7.86 (152.62), 7.85 (176.74, 152.62, 145.80, 139.63), 7.84 (154.29), 7.50 (154.28, 146.08, 139.62, 130.09, 128.72, 118.91, 63.18), 7.37 (135.80, 130.11), 5.63 (154.43, 135.81, 130.12, 49.53), 3.26 (50.01), 3.22 (45.73), 3.20 (51.10), 2.86 (45.77, 49.61, 51.11), 2.56 (49.62), 1.64 (30.01, 62.04), 1.28 (8.04, 36.26), 1.14 (36.26).
Synthesis of 1-cyclopropyl-7-(4-((1-(2,6-dimethylphenyl)-1H-tetrazol-5-yl)(phenyl)methyl)piperazin-1-yl)-6-fluoro-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (5, 6)
A methodology similar to that used for the synthesizing 2 was employed to obtain 5, 6; however, instead of paraformaldehyde, 0.332 mmol (35.2 mg) of benzaldehyde was added to the mixture under the same conditions. Ivory solid; yield 25 % mp 190°C (decomposition). ATR FT-IR (cm−1): 3060 (C-H arom) 2841 (C-H aliph), 1727 (C=O acid), 1611 (C=O ketone), 1588 (C=C), 1494 (C=C Ar), 1450, 1383, 1332, 1285, 1252 (C-O), 1142, 1007 (Ar-F), 937, 891, 832 (2RC=CR-H), 728 (C-H oop). 1H NMR (600 MHz, DMSO-d6, δ ppm): 8.62 (s, 1H), 7.85 (d, J = 12.0 Hz, 1H), 7.50 (d, J = 6.0 Hz, 1H), 7.46 (dd, J = 6.0 Hz y J = 12.0 Hz, 1H), 7.38 (d, J = 6.0 Hz, 1H), 7.30-7.31 (m, 3H), 7.17-7.15 (m, 3H), 4.57 (s, 1H), 3.80 (m, 1H), 3.21 (t, J = 6.0 Hz, 4H), 2.64 (d, J = 6.0 Hz, 2H), 2.60 (d, J = 6.0 Hz, 2H), 1.98 (s, 3H), 1.27 (m, 2H), 1.14 (m, 2H), 1.04 (s, 3H). 13C NMR (151 MHz, DMSO-d6, δ ppm) 176.75, 166.38, 156.05, 155.62, 152.65, 151.45, 148.36, 146.17, 139.68, 139.56, 136.16, 135.33, 134.72, 131.65, 131.43, 129.66, 129.44, 129.19, 129.09, 119.16, 118.77, 111.40, 111.25, 107.16, 107.12, 106.53, 64.10, 51.10, 50.42, 49.81, 45.76, 39.96, 36.27, 17.50, 16.35, 7.99. 19F NMR (564 MHz, DMSO-d6, δ ppm): -121.61. ESI-MS C33H32FN7O3 (m/z): 593.25 [M] +; HRMS: calc.: 593.2550 (M+); exp.: 593.2562 (M+). COSY (DMSO-d6, δ ppm) 3 J H,H (3.80, 1.28), (3.17, 2.57), (1.27, 3.79), (1.14, 3.78), 4 J H,H (8.62, 3.79), (7.38, 1.98), (7.38, 1.04), (7.14, 1.94), (7.14, 1.04), (4.57, 7.15), 6 J H,H (1.04, 1.98). HMBC (DMSO-d6, δ ppm) J C,H 8.62 (176.75, 166.36, 139.63, 107.13, 36.31), 7.86 (152.62, 146.18), 7.85 (176.76, 146.15, 139.61), 7.50 (154.28, 176.72, 146.15, 139.65, 118.87), 7.51 (152.61), 7.45 (135.31), 7.38 (131.39), 7.30 (134.70, 129.69), 7.15 (131.41, 64.19), 4.57 (134.70, 129.66, 56.06, 50.44), 3.22 (51.11, 45.77), 2.88 (45.82, 45.75), 2.64 (50.44), 2.60 (50.17), 1.98 (135.33, 131.43, 129.42), 1.28 (8.01), 1.27 (36.23), 1.15 (36.23), 1.14 (7.82), 1.04 (136.16, 131.40, 129.22).
General procedure for the synthesis of 7-(4-((1-(tert-butyl)-1H-tetrazol-5-yl)methyl)piperazin-1-yl)-1-ethyl-6-fluoro-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (7)
As depicted in Fig. 15, the synthetic pathway leading to the remaining quinolone derivatives is quite similar. To obtain compound 7, 100 mg (0.313 mmol) of norfloxacin (NFX) was placed in a flask along with 6 mL of MeOH. The mixture was heated to 75-80 °C under constant stirring; once it was completely dissolved, 56.4 mg (0.626 mmol) of paraformaldehyde was added to the flask with one drop of HCl 10%. The mixture was stirred until the solvent was evaporated. The reaction product was re-dissolved with MeOH, then 33.8 mg (0.407 mmol) of tert-butyl isocyanide and 46.9 mg (0.407 mmol) of trimethylsilyl azide were added to the flask at a temperature of 45-50 °C. The resulting mixture was heated (80-85 °C) for 3 h. The final product was washed and rinsed with cold water before being recrystallizing with acetone. Ivory solid; yield 29 % mp 240 °C (decomposition). ATR FT-IR (cm−1): 2820 (C-H aliph), 1721 (C=O acid), 1624 (C=O ketone), 1463 (C=C Ar), 1442, 1269 (C-O), 1019 (C-F), 874 (2RC=CR-H), 787, 758, 700 (C-H oop). 1H NMR (600 MHz, DMSO-d6, δ ppm): 15.30, (s, 1H), 8.91 (s, 1H), 7.89 (d, J = 12.0 Hz, 1H), 7.56 (d, J = 6.0 Hz, 1H), 4.55 (q, J = 6.0 Hz, 2H), 3.96 (s, 2H), 3.28 (br s, 4H), 2.60 (t, J = 6.0 Hz, 4H), 1.72 (s, 9H), 1.37 (t, J = 6.0 Hz, 3H). 13C NMR (151 MHz, DMSO-d6, δ ppm): 176.60, 166.47, 154.17, 152.51, 151.53, 148.91, 145.70, 137.64, 119.82, 111.67, 111.52, 107.57, 106.49, 62.25, 52.46, 51.50, 49.82, 49.78, 49.46, 40.05, 29.36, 14.80. 19F NMR (376 MHz, DMSO-d6, δ ppm): -121.33. ESI-MS C22H28FN7O3 (m/z): 457.26 [M] +; HRMS: calc.: 457.2237 (M+); exp.: 457.2240 (M+). COSY (DMSO-d6, δ ppm) 3 J H,H (4.55, 1.37), (3.30, 2.70), (3.28, 2.60), 5 J H,H (7.90, 7.15).). HSQC (DMSO-d6, δ ppm) 1 J C,H (8.90, 148.94), (7.87, 11.55), (7.15, 106.51), (4.54, 49.48), (3.95, 51.49), (3.28, 49.81), (2.60, 52.46), (1.72, 29.34), (1.37, 14.83). HMBC (DMSO-d6, δ ppm) J C,H 8.91 (176.59, 166.51, 139.61, 49.48), 7.89 (176.56, 152.50, 145.63, 137.63) 7.15 (176.52, 154.16, 152.48, 145.70, 137.62, 119.81), 4.55 (148.96, 137.60, 14.84), 3.96 (151.53, 52.45) 3.32 (52.07), 2.60 (51.32, 52.47, 49.79), 1.37 (49.48).
Synthesis of 7-(4-((1-(2,6-dimethylphenyl)-1H-tetrazol-5-yl)methyl)piperazin-1-yl)-1-ethyl-6-fluoro-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (8)
For the synthesis of 8, 100 mg (0.313 mmol) of norfloxacin (NFX) were placed in a flask along with 6 mL of MeOH. The mixture was heated to 75-80 °C under constant stirring; once it was completely dissolved, 56.4 mg (0.626 mmol) of paraformaldehyde was added to the flask with one drop of HCl (10%). The mixture was stirred until the solvent was evaporated. The reaction product was re-dissolved in MeOH, then 53.4 mg (0.407 mmol) of 2,6-dimethyl phenyl isocyanide with 46.9 mg (0.407 mmol) of trimethylsilyl azide were added to the flask keeping the temperature at 45-50 °C. The resulting mixture was heated (80-85 °C) for 3 h. The final product was washed and rinsed with cold water before being recrystallizing with acetone. Ivory solid; yield 46 %; mp 240 °C (decomposition). ATR FT-IR (cm−1): 3058 (C-H arom), 2837 (C-H aliph), 1726 (C=O acid), 1625 (C=O ketone), 1540 (C=C), 1497 (C=C Ar), 1466, 1383, 1340, 1300, 1248 (C-O), 1197, 1131, 1093, 1042, 1004 (Ar-F), 955, 933, 816, 806 (2RC=CR-H), 784, 626 (C-H oop). 1H NMR (600 MHz, DMSO-d6, δ ppm): 15.30 (s, 1H), 8.91 (s, 1H), 7.87 (d, J = 12.0 Hz, 1H), 7.45 (dd, J = 6.0 Hz y J = 12.0 Hz,1H), 7.32 (d, J = 6.0 Hz, 2H), 7.10 (d, J = 6.0 Hz, 1H), 4.54 (q, J = 6.0 Hz, 2H), 3.69 (s, 2H), 3.17 (br s, 4H), 2.65 (br s, 4H), 1.90 (s, 6H), 1.36 (t, J = 6.0 Hz, 3H). 13C NMR (151 MHz, DMSO-d6, δ ppm) 176.56, 166.52, 154.05, 153.56, 152.40, 148.95, 145.57, 137.60, 135.78, 132.08, 131.39, 129.12, 119.76, 119.71, 111.68, 111.53, 107.51, 106.35, 52.69, 49.72, 49.68, 49.54, 49.47, 17.33, 14.80. 19F NMR (564 MHz, DMSO-d6, δ ppm): -121.31. ESI-MS C26H28FN7O3 (m/z): 505.19 [M] +; HRMS: calc.: 505.2237 (M+); exp.: 505.2252 (M+). COSY (DMSO-d6, δ ppm) 3 J H,H (7.43, 7.32), (4.54, 1.36), (2.55, 3.16), 4 J H,H (7.33, 1.90), (1.89, 7.32). HSQC (DMSO-d6, δ ppm) 1 J C,H (8.90, 148.95), (7.85, 115.54), (7.44, 131.38), (7.31, 129.13), (7.10, 106.36), (4.53, 49.47), (3.69, 49.54), (3.16, 49.70), (2.55, 52.68), (1.89, 17.34), (1.36, 14.80). HMBC (DMSO-d6, δ ppm) J C,H 8.90 (176.54, 166.48, 139.57, 49.38), 7.45 (135.75), 7.33 (132.00, 17.21), 7.32 (131.95, 17.21), 7.10 (152.36), 4.53 (148.88, 137.55, 14.64), 3.69 (153.52, 52.61), 1.89 (135.73, 132.03, 129.08), 1.35 (49.41).
Synthesis of 7-(4-((1-(tert-butyl)-1H-tetrazol-5-yl)(phenyl)methyl)piperazin-1-yl)-1-ethyl-6-fluoro-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (9, 10)
A methodology similar to that used for the synthesis of 7 was employed to obtain 9, 10; however, instead of paraformaldehyde, 0.344 mmol (36.6 mg) of benzaldehyde was added to the mixture under the same conditions. Ivory solid; yield 34 % mp 200 °C (decomposition). ATR FT-IR (cm−1): 3044 (C-H arom) 2836 (C-H aliph), 1723 (C=O acid), 1628 (C=O ketone), 1520 (C=C), 1478 (C=C Ar), 1453, 1377 (C-(CH3)3), 1263 (C-O), 1110, 1009 (Ar-F), 939, 831 (R2C=CR-H), 804, 753, 744, 698 (C-H oop). 1H NMR (600 MHz, DMSO-d6, δ ppm): 15.29 (s, 1H), 8.90 (s, 1H), 7.86 (d, J = 12.0 Hz, 1H), 7.50 (d, J=12 Hz, 2H), 7.37 (dd, J = 6.0 Hz, 2H), 7.32 (dd, J = 6.0 Hz y J = 12.0 Hz, 1H), 7.12 (d, J = 6.0 Hz, 1H), 5.62 (s, 1H), 4.53 (q, J = 6.0 Hz, 2H), 3.25 (t, J = 6.0 Hz, 4H), 2.83 (q, J = 6.0 Hz, J = 6.0 Hz, 2H), 2.55 (q, 2H), 1.64 (s, 9H), 1.34 (t, J = 6.0 Hz, 3H). 13C NMR (151 MHz, DMSO-d6, δ ppm) 176.60, 166.52, 154.43, 148.97, 145.89, 137.57, 135.79, 130.12, 128.80, 128.71, 119.80, 111.46, 107.50, 106.58, 63.16, 62.03, 50.28, 50.25, 49.53, 49.49, 39.96, 30.00, 14.75. 19F NMR (564 MHz, DMSO-d6, δ ppm): -121.51. ESI-MS C28H32FN7O3 (m/z): 533.22 [M] +; HRMS: calc.: 533.2550 (M+); exp.: 533.2563 (M+). COSY (DMSO-d6, δ ppm) 3 J H,H (4.52, 1.34), (3.25, 2.56), (3.25, 2.82). HSQC (DMSO-d6, δ ppm) 1 J C,H (8.90, 148.98), (7.87, 109.78), (7.49, 130.12), (7.36, 128.78), (7.32, 128.74), (7.12, 106.54), (5.62, 62.71), (5.62, 63.61), (4.52, 49.94), (3.25, 50.26), (3.25, 49.79), (2.83, 49.55), (2.55, 49.95), (1.64, 30.47), (1.64, 30.01), (1.64, 25.54), (1.34, 14.76). HMBC (DMSO-d6, δ ppm) J C,H 8.90 (176.57, 166.52, 137.58, 49.50), 7.50 (63.17), 7.87 (152.54, 145.82), 7.86 (137.57), 7.32 (130.16), 7.13 (152.51, 137.56), 7.12 (154.12, 145.81, 119.77), 5.62 (154.43, 135.79, 130.12, 49.51), 4.53 (148.97, 137.57), 4.52 (14.76), 3.25 (145.97, 50.25), 2.83 (49.47), 2.55 (50.38), 1.64 (62.04, 30.00), 1.34 (49.49).
Synthesis of 7-(4-((1-(2,6-dimethylphenyl)-1H-tetrazol-5-yl)(phenyl)methyl)piperazin-1-yl)-1-ethyl-6-fluoro-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (11, 12)
A methodology similar to that used for the synthesis of 8 was employed to obtain 11, 12; however, instead of paraformaldehyde, 0.344 mmol (36.6 mg) of benzaldehyde was added to the mixture under the same conditions. Ivory solid; yield 21 % mp 210°C (decomposition). ATR FT-IR (cm−1): 3060 (C-H arom), 2828 (C-H aliph), 1720 (C=O acid), 1625 (C=O ketone), 1609 (C=C) 1539 and 1505 (C=C Ar), 1476 (C=C Ar), 1444, 1362, 1298, 1268, 1250 (C-O), 1198, 1139, 1130, 1007 (Ar-F), 933, 896, 780 (2RC=CR-H), 747, 698 (C-H oop). 1H NMR (600 MHz, DMSO-d6, δ ppm): 15.29 (s, 1H), 8.90 (s, 1H), 7.86 (d, J = 18.0 Hz, 1H), 7.45 (dd, J = 6.0 Hz, J = 12.0 Hz, 1H), 7.38 (d, J = 6.0 Hz, 1H), 7.31-7.28 (m, 3H), 7.16-7.14 (m, 3H), 7.12 (d, J = 6.0 Hz, 1H), 4.56 (s, 1H) 4.53 (q, J = 6.0 Hz, 2H), 3.27 (br s, 4H), 2.61 y 2.58 (m, 4H), 1.98 (s, 3H), 1.35 (t, J = 6.0 Hz, 3H), 1.04 (s, 3H). 13C NMR (151 MHz, DMSO-d6, δ ppm) 176.57, 166.53, 156.06, 154.13, 152.47, 148.96, 145.73, 145.67, 137.56, 3800 136.15, 135.34, 134.71, 131.65, 131.41, 129.65, 129.44, 129.19, 129.07, 119.82, 119.77, 111.47, 107.49, 106.52, 64.06, 50.41, 49.92, 49.89, 49.52, 39.93, 17.48, 16.36, 14.76. 19F NMR (564 MHz, DMSO-d6, δ ppm): -121.48. ESI-MS C32H32FN7O3 (m/z): 581.28 [M] +; HRMS: calc.: 581.2550 (M+); exp.: 581.2554 (M+). COSY (DMSO-d6, δ ppm) 3 J H,H (4.52, 1.35), (2.59, 3.27), 4 J H,H (8.89, 4.53), (7.16, 4.55), (7.38, 1.04), (7.38, 1.04), (7.16, 1.98), (7.14, 1.04). HSQC (DMSO-d6, δ ppm) 1 J C,H (8.89, 148.94), (7.85, 111.56), (7.36, 129.43), (7.44, 131.65), (7.29, 129.12), (7.15, 129.65), (7.14, 129.15), (7.11, 106.52), (4.54, 64.07), (4.51, 49.52), (3.26, 49.89), (2.59, 50.42), (1.97, 17.47), (1.34, 14.75), (1.03, 16.34). HMBC (DMSO-d6, δ ppm) J C,H 8.90 (176.58, 166.53, 137.56, 108.33, 49.52), 7.89 (152.48), 7.85 (176.54, 137.57), 7.84 (154.12), 7.45 (136.14), 7.36 (129.21, 131.39, 17.47), 7.30 (131.39, 134.69, 129.68), 7.16 (131.40, 129.25), 7.15 (64.11, 16.36), 7.13 (152.45, 119.79), 7.12 (176.64, 154.11, 145,69, 137.57), 4.56 (156.06, 134.70, 129.65, 50.42), 4.52 (148.97, 137.54, 14.75), 3.27 (50.13), 2.61 (50.31), 2.57 (50.20), 1.98 (135.34, 131.42, 129.41), 1.35 (49.52), 1.04 (136.15, 131.39, 129.22).
5. Computational details
5.1. Conformational analysis and geometry optimization
A systematic conformational analysis using the MMFFaq force field was performed [68] to obtain the most stable conformer of each zwitterionic fluoroquinolone. Then, an equilibrium geometry optimization, without symmetric restrictions, using the semi-empirical quantum-mechanic level of theory with the Parametric Method 6 (PM6) approximation was done [69]. A vibrational frequency analysis was performed to validate the molecular geometry as a minimum on the potential energy surface. All the calculations were carried out using Spartan’18 [70].
5.2. MEP and SPR analysis
Density Functional Theory (DFT) single-point energy calculations were performed using the M06 hybrid functional [71] and the 6-311 + G** basis set [72] for all the fluoroquinolones. From these calculations, the molecular electrostatic potential (MEP) was mapped onto an iso-density surface (0.002 e−/Å3) for each fluoroquinolone. MEP maps are used to study the molecular size and location of electron-rich and electron-deficient zones in a compound series [73]. All the calculations and molecular graphics were done in SPARTAN'18.
5.2. Molecular docking with DNA gyrase
We evaluated the interaction of the synthesized compounds with the DNA-gyrase of S. aureus. We compared the results with the commercial fluoroquinolones to study the potential ability of the hybrids as topoisomerase poisons. For this purpose, all new tetrazole derivatives and the known fluoroquinolones, were docked into the DNA gyrase/DNA structure from S. aureus (PDB:5CDQ) [74]. Electrostatic partial charge types were used for all the ligands in this study. The search algorithm used was MolDock SE with a number of 15 runs, 3500 iterations and a population of 150. MolDock Score GRID was used as the scoring function with a radius of 13Å for the search sphere.
Flexible docking was performed using parameters mentioned above. All the residues with a distance of 6Å from the binding site of the fluoroquinolones were set as flexible; for this purpose, only residues with two or more torsions were considered (Torsion type (TT) = 5, with 0.5 of strength; TT = 4, with 0.6 of strength; TT = 3 with 0.7 of strength and TT = 2 with 0.8 of strength; for all TT, tolerance of 1). The methodology for the docking experiments was validated by reproducing the Moxifloxacin co-crystallized ligand conformation in 5CDQ with an RMSD value of 0.414 (Supplementary material Fig. S4). Docking studies were carried out in Molegro Virtual Docker 6.0 [75].
5.3. QSAR construction
A QSAR study was performed to define the chemical characteristics of fluoroquinolones related to their antibacterial activity on S. aureus. For this purpose, we used the interaction energy results obtained from our molecular docking as descriptors: , and LE. These descriptors have been used to understand and predict the biological activity of compounds [74, 75]. Also, the physicochemical properties of the compounds (1-12), such as diffusion in aqueous solvents, 1-octanol-water partition coefficient () and aqueous solubility calculation (LogS) [76], were also calculated. Molecular volume was employed as a molecular descriptor (V). Using these values, we obtained additional molecular descriptors, including ΔV, corresponding to the molecular volume difference compared to an established template (Fig. 7). Finally, some topological and constitutional descriptors were calculated, those previously used in other works with similar structures [77].
QSAR model construction was done using a multiple linear regression techniques, employing the Excel Office program; molecular descriptors were used as independent variables (). For the dependent variable (), we used the experimental MIC. QSAR model validation was based on the value of its squared multiple correlation coefficient (), explained variance in prediction obtained by the leave many out technique (), standard deviation (), and Fisher function () [78]. Additionally, overfitting () and, to guarantee its future predictive ability, the asymptotic Q2 rule and the external validation technique (Q2 ext) were applied [76, 78].
5.4. Molecular docking with COVID-19 main protease
The details of the selection of the crystal structures used in this work are described in the results section. To validate our docking method, we reproduced the crystal conformation of the co-crystallized compound in the four crystals enzymes (Supplementary materials Figs. S6-S9) using the search algorithm MolDock Optimizer: 50 runs, 4000 iterations and a population of 200, and MolDock Score GRID function with a search sphere of 15Å radius over the catalytic site. After the rigid docking, a flexible docking approach was made considering all the residues of the catalytic site (using the same consideration for DNA/gyrase docking)
Credit author statement
Jaime Cardoso-Ortiz. Conceptualization, synthesis investigation and spectroscopy characterization.
Socorro Leyva-Ramos. Conceptualization, funding adquisition and writing.
Kim M. Baines. Spectroscopy characterization and writing review.
César Fernando Azael Gómez-Durán. Conceptualization, writing original draft, review and editing..
Hiram Hernández-López. Experimental synthesis and characterization.
Francisco José Palacios-Can. Formal analysis, data curation, writing original draft, review and editing.
José Antonio Valcarcel-Gamiño. Data curation, and formal analysis
Mario Alberto Leyva-Peralta. Software, formal analysis, data Curation, writing original draft.
Rodrigo Said Razo Hernández. Conceptualization, methodology, formal analysis, data curation, supervision, validation, writing original draft, review & editing.
Supporting Information
Full experimental details, spectral data of the products, and 1H, 13C and 19F NMR and mass spectra of all the new compounds can be found via the supplementary materials section of this article's Web page.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We would like to thank CONACYT (SEP-82585) for its financial support. Rodrigo Said Razo Hernández thanks CONACyT for the project grant No 320243. In addition, RSRH thanks Dr. Zeferino Gómez-Sandoval of the University of Colima for the software facilities. Dr. Kim M. Baines wishes to thank the Natural Sciences and Engineering Research Council (NSERC) of Canada for funding (RGPIN-2017-06023).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.molstruc.2022.134507.
Appendix. Supplementary materials
Data Availability
Data will be made available on request.
References
- 1.Fedorowicz J., Sączewski J. Modifications of quinolones and fluoroquinolones: hybrid compounds and dual-action molecules. Monatsh. Chem. 2018;149(7):1199–1245. doi: 10.1007/s00706-018-2215-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castro W., Navarro M., Biot C. Medicinal potential of ciprofloxacin and its derivatives. Future Med. Chem. 2013;5(1):81–96. doi: 10.4155/fmc.12.181. [DOI] [PubMed] [Google Scholar]
- 3.Karampela I., Dalamaga M. Could respiratory fluoroquinolones, levofloxacin and moxifloxacin, prove to be beneficial as an adjunct treatment in COVID-19? Arch. Med. Res. 2020;51(7):741–742. doi: 10.1016/j.arcmed.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luan B., Huynh T., Cheng X., Lan G., Wang H.R. Targeting proteases for treating COVID-19. J. Proteome Res. 2020;19(11):4316–4326. doi: 10.1021/acs.jproteome.0c00430. [DOI] [PubMed] [Google Scholar]
- 5.Gil C., et al. COVID-19: Drug targets and potential treatments. J. Med. Chem. 2020;63(21):12359–12386. doi: 10.1021/acs.jmedchem.0c00606. [DOI] [PubMed] [Google Scholar]
- 6.Wen W., et al. Efficacy and safety of three new oral antiviral treatment (molnupiravir, fluvoxamine and paxlovid) for COVID-19: a meta-analysis. Ann. Med. 2022;54(1):516–523. doi: 10.1080/07853890.2022.2034936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mahase E. Covid-19: Pfizer's paxlovid is 89% effective in patients at risk of serious illness, company reports. BMJ. 2021;375:n2713. doi: 10.1136/bmj.n2713. [DOI] [PubMed] [Google Scholar]
- 8.Lim Y.X., Ng Y.L., Tam J.P., Liu D.X. Human coronaviruses: a review of virus–host interactions. Diseases. 2016;4(3) doi: 10.3390/diseases4030026. 26,(1-28) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mukherjee P., Shah F., Desai P., Avery M. Inhibitors of SARS-3CLpro: virtual screening, biological evaluation, and molecular dynamics simulation studies. J. Chem. Inf. Model. 2011;51(6):1376–1392. doi: 10.1021/ci1004916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Canal B., et al. Identifying SARS-CoV-2 antiviral compounds by screening for small molecule inhibitors of nsp15 endoribonuclease. Biochem. J. 2021;478(13):2465–2479. doi: 10.1042/BCJ20210199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin Z., et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 12.Marciniec K., Beberok A., Pęcak P., Boryczka S., Wrześniok D. Ciprofloxacin and moxifloxacin could interact with SARS-CoV-2 protease: preliminary in silico analysis. Pharmacol. Rep. 2020;72(6):1553–1561. doi: 10.1007/s43440-020-00169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Damanhouri Z.A., Alkreathy H.M., Ali A.S., Karim S. The potential role of Fluoroquinolones in the management of Covid-19 a rapid review. J. Adv. Pharm. Educ. Res. 2021;11(1):128–134. [Google Scholar]
- 14.Aranha O., Grignon R., Fernandes N., McDonnell T.J., Wood D.P., Sarkar F.H. Suppression of human prostate cancer cell growth by ciprofloxacin is associated with cell cycle arrest and apoptosis. Int. J. Oncol. 2003;22(4):787–794. [PubMed] [Google Scholar]
- 15.Langtry H.D., Lamb H.M. Errata: Levofloxacin: Its use in infections of the respiratory tract, skin, soft tissue and urinary tract (Drugs (1999) 56 (487-515)) Drugs. 1999;58(2):296. doi: 10.2165/00003495-199856030-00013. [DOI] [PubMed] [Google Scholar]
- 16.Falagas M.E., Matthaiou D.K., Bliziotis I.A. Systematic review: fluoroquinolones for the treatment of intra-abdominal surgical infections. Aliment. Pharmacol. Ther. 2007;25(2):123–131. doi: 10.1111/j.1365-2036.2006.03154.x. [DOI] [PubMed] [Google Scholar]
- 17.Desplaces N., Acar J.F. New quinolones in the treatment of joint and bone infections. Rev. Infect. Dis. 1988;10(suppl. 1):S179–S183. doi: 10.1093/clinids/10.supplement_1.s179. [DOI] [PubMed] [Google Scholar]
- 18.Berning S.E. The role of fluoroquinolones in tuberculosis today. Drugs. 2001;61(1):9–18. doi: 10.2165/00003495-200161010-00002. [DOI] [PubMed] [Google Scholar]
- 19.Gaillard T., Madamet M., Tsombeng F.F., Dormoi J., Pradines B. Antibiotics in malaria therapy: which antibiotics except tetracyclines and macrolides may be used against malaria? Malar. J. 2016;15(1) doi: 10.1186/s12936-016-1613-y. 556 (1–10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yadav V., Talwar P. Repositioning of fluoroquinolones from antibiotic to anti-cancer agents: an underestimated truth. Biomed. Pharmacother. 2019;111:934–946. doi: 10.1016/j.biopha.2018.12.119. December 2018. [DOI] [PubMed] [Google Scholar]
- 21.Huang C.Y., et al. Fluoroquinolones suppress TGF-β and PMA-induced MMP-9 production in cancer cells: implications in repurposing quinolone antibiotics for cancer treatment. Int. J. Mol. Sci. 2021;22(21) doi: 10.3390/ijms222111602. 11602 (1-22) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scroggs S.L.P., et al. Old drugs with new tricks: efficacy of fluoroquinolones to suppress replication of flaviviruses. Viruses. 2020;12(9) doi: 10.3390/v12091022. 1022 (1-23) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kojima H., Kaita K.D.E., Hawkins K., Uhanova J., Minuk G.Y. Use of fluoroquinolones in patients with chronic hepatitis C virus-induced liver failure. Antimicrob. Agents Chemother. 2002;46(10):3280–3282. doi: 10.1128/AAC.46.10.3280-3282.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alaaeldin R., Mustafa M., Abuo-Rahma G.E.D.A., Fathy M. In vitro inhibition and molecular docking of a new ciprofloxacin- chalcone against SARS- CoV- 2 main protease. Fundam Clin. Pharmacol. 2022;36(1):160–170. doi: 10.1111/fcp.12708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scroggs S.L.P., et al. Fluoroquinolones antibiotics exhibit low antiviral activity against SARS-CoV-2 and MERS-CoV. Viruses. 2021;13(1) doi: 10.3390/v13010008. 8 (1–12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pham T.D.M., Ziora Z.M., Blaskovich M.A.T. Quinolone antibiotics. Med. Chem. Comm. 2019;10(10):1719–1739. doi: 10.1039/c9md00120d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jia Y., Zhao L. The antibacterial activity of fluoroquinolone derivatives: An update (2018–2021) Eur. J. Med. Chem. 2021;224 doi: 10.1016/j.ejmech.2021.113741. 113741, (1-13) [DOI] [PubMed] [Google Scholar]
- 28.Dighe S.N., Collet T.A. Recent advances in DNA gyrase-targeted antimicrobial agents. Eur. J. Med. Chem. 2020;199 doi: 10.1016/j.ejmech.2020.112326. [DOI] [PubMed] [Google Scholar]
- 29.Dileep K., Polepalli S., Jain N., Buddana S.K., Prakasham R.S., Murty M.S.R. Synthesis of novel tetrazole containing hybrid ciprofloxacin and pipemidic acid analogues and preliminary biological evaluation of their antibacterial and antiproliferative activity. Mol. Divers. 2018;22(1):83–93. doi: 10.1007/s11030-017-9795-y. [DOI] [PubMed] [Google Scholar]
- 30.Gao F., Xiao J., Huang G. Current scenario of tetrazole hybrids for antibacterial activity. Eur. J. Med. Chem. 2019;184 doi: 10.1016/j.ejmech.2019.111744. [DOI] [PubMed] [Google Scholar]
- 31.Aranha O., Wood D.P., Sarkar F.H. Ciprofloxacin mediated cell growth inhibition, S/G2-M cell cycle arrest, and apoptosis in a human transitional cell carcinoma of the bladder cell line. Clin. Cancer Res. 2000;6(3):891–900. [PubMed] [Google Scholar]
- 32.El-Rayes B.F., Grignon R., Aslam N., Aranha O., Sarkar F.H. Ciprofloxacin inhibits cell growth and synergises the effect of etoposide in hormone resistant prostate cancer cells. Int. J. Oncol. 2002;21(1):207–211. [PubMed] [Google Scholar]
- 33.Chauhan K., Singh P., Kumar V., Shukla P.K., Siddiqi M.I., Chauhan P.M.S. Investigation of Ugi-4CC derived 1H-tetrazol-5-yl-(aryl) methyl piperazinyl-6-fluoro-4-oxo-1,4-dihydroquinoline-3-carboxylic acid: synthesis, biology and 3d-qsar analysis. eur. j. med. chem. 2014;78:442–454. doi: 10.1016/j.ejmech.2014.03.069. [DOI] [PubMed] [Google Scholar]
- 34.Leyva-Ramos S., Cardoso-Ortiz J. Recent Developments in the Synthesis of Tetrazoles and their Pharmacological Relevance. Curr. Org. Chem. 2020;25(3):388–403. [Google Scholar]
- 35.Sarvary A., Maleki A. A review of syntheses of 1,5-disubstituted tetrazole derivatives. Mol. Divers. 2015;19(1):189–212. doi: 10.1007/s11030-014-9553-3. [DOI] [PubMed] [Google Scholar]
- 36.Safaei-Ghomi J., Paymard-Samani S., Zahedi S., Shahbazi-Alavi H. Sonochemical synthesis of 5-substituted 1H-tetrazoles catalyzed by ZrP2O7 nanoparticles and regioselective conversion into new 2,5-disubstituted tetrazoles. Zeitschrift fur Naturforsch. - Sect. B. 2015;70(11):819–828. [Google Scholar]
- 37.Halder M., Islam M.M., Singh P., Singha Roy A., Islam S.M., Sen K. Sustainable generation of Ni(OH)2 nanoparticles for the green synthesis of 5-substituted 1 H-tetrazoles: a competent turn on fluorescence sensing of H2O2. ACS Omega. 2018;3(7):8169–8180. doi: 10.1021/acsomega.8b01081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kazemnejadi M., Mahmoudi B., Sharafi Z., Nasseri M.A., Allahresani A., Esmaeilpour M. Copper coordinated-poly(α-amino acid) decorated on magnetite graphene oxide as an efficient heterogeneous magnetically recoverable catalyst for the selective synthesis of 5- and 1-substituted tetrazoles from various sources: a comparative study. Appl. Organomet. Chem. 2020;34(2) e5273 (1-15) [Google Scholar]
- 39.Neochoritis C.G., Zhao T., Dömling A. Tetrazoles via multicomponent reactions. Chem. Rev. 2019;119(3):1970–2042. doi: 10.1021/acs.chemrev.8b00564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanaka M., Wang T., Onodera Y., Uchida Y., Sato K. Mechanism of quinolone resistance in Staphylococcus aureus. J. Infect Chemother. 2000;6(3):131–139. doi: 10.1007/s101560070010. [DOI] [PubMed] [Google Scholar]
- 41.Lemaire S., Tulkens P.M., Van Bambeke F. Contrasting effects of acidic pH on the extracellular and intracellular activities of the anti-gram-positive fluoroquinolones moxifloxacin and delafloxacin against Staphylococcus aureus. Antimicrob. Agents Chemother. 2011;55(2):649–658. doi: 10.1128/AAC.01201-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matera M.G. Pharmacologic characteristics of prulifloxacin. Pulm. Pharmacol. Ther. 2006;19(Suppl. 1):20–29. doi: 10.1016/j.pupt.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 43.Schedletzky H., Wiedemann B., Heisig P. The effect of moxifloxacin on its target topoisomerases from Escherichia coli and Staphylococcus aureus. J. Antimicrob. Chemother. 1999;43(Suppl. B):31–37. doi: 10.1093/jac/43.suppl_2.31. [DOI] [PubMed] [Google Scholar]
- 44.Llorente B., Leclerc F., Cedergren R. Using SAR and QSAR analysis to model the activity and structure of the quinolone-DNA complex. Bioorg. Med. Chem. 1996;4(1):61–71. doi: 10.1016/0968-0896(96)83749-7. [DOI] [PubMed] [Google Scholar]
- 45.Noble C.G., Barnard F.M., Maxwell A. Quinolone-DNA interaction: sequence-dependent binding to single-stranded DNA reflects the interaction within the gyrase-DNA complex. Antimicrob. Agents Chemother. 2003;47(3):854–862. doi: 10.1128/AAC.47.3.854-862.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skauge T., Turel I., Sletten E. Interaction between ciprofloxacin and DNA mediated by Mg2+-ions. Inorganica Chim. Acta. 2002;339:239–247. [Google Scholar]
- 47.Pérez D.J., et al. In silico receptor-based drug design of X,Y-benzenesulfonamide derivatives as selective COX-2 inhibitors. Comptes Rendus Chim. 2017;20(2):169–180. [Google Scholar]
- 48.Kier L.B. Distinguishing atom differences in a molecular graph shape index. Quant.Struct.-Act.Relat. 1986;5:7–12. [Google Scholar]
- 49.Ohta M., Koga H. Three-dimensional structure-activity relationships and receptor mapping of N1-substituents of quinolone antibacterials. J. Med. Chem. 1991;34(1):131–139. doi: 10.1021/jm00105a020. [DOI] [PubMed] [Google Scholar]
- 50.Koka H., Ohta M. Three-dimensional structure activity relationship and receptor mapping of quinolone antibacterials. QSAR Drug Des. - New Dev. Appl. 1995:299–320. [Google Scholar]
- 51.Martin Y.C. Exploring QSAR: hydrophobic, electronic, and steric constants C. Hansch, A. Leo, and D. Hoekman. Publisher: American Chemical Society, vol. 2, 1995. Exploring QSAR: Fundamentals and Applications in Chemistry and Biology. C. Hans and A. Leo. J. Med. Chem. 1996;39(5):1189–1190. [Google Scholar]
- 52.Koga H., Fujita T. 1982. Structure-Activity Relationships-Quantitative Approaches; Applications to Drug Design and Mode-of-Action Studies. [Google Scholar]
- 53.Kubinyi H. Chemical similarity and biological activities. J. Braz. Chem. Soc. 2002;13(6):717–726. [Google Scholar]
- 54.Su H.X., et al. Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients. Acta Pharmacol. Sin. 2020;41(9):1167–1177. doi: 10.1038/s41401-020-0483-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mesecar A.D. 2020. 6W63 Structure of COVID-19 Main Protease Bound-Spectrum Non-Covalent Inhibitor X77. [Google Scholar]
- 56.Lockbaum G.J., et al. Crystal structure of SARS-CoV-2 main protease in complex with the non-covalent inhibitor ML188. Viruses. 2021;13(2) doi: 10.3390/v13020174. 174 (1-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang J., et al. Structure-based discovery of novel nonpeptide inhibitors targeting SARS-CoV-2 Mpro. J. Chem. Inf. Model. 2021;61(8):3917–3926. doi: 10.1021/acs.jcim.1c00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang C.H., et al. Potent noncovalent inhibitors of the main protease of SARS-CoV-2 from molecular sculpting of the drug perampanel guided by free energy Perturbation Calculations. ACS Cent. Sci. 2021;7(3):467–475. doi: 10.1021/acscentsci.1c00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Świderek K., Moliner V. Revealing the molecular mechanisms of proteolysis of SARS-CoV-2 Mpro by QM/MM computational methods. Chem. Sci. 2020;11(39):10626–10630. doi: 10.1039/d0sc02823a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arafet K., et al. Mechanism of inhibition of SARS-CoV-2 Mpro by N3 peptidyl Michael acceptor explained by QM/MM simulations and design of new derivatives with tunable chemical reactivity. Chem. Sci. 2021;12(4):1433–1444. doi: 10.1039/d0sc06195f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cortés-García C.J., Chacón-García L., Mejía-Benavides J.E., Díaz-Cervantes E. Tackling the SARS-CoV-2 main protease using hybrid derivatives of 1,5-disubstituted tetrazole-1,2,3-triazoles: an in silico assay. PeerJ Phys. Chem. 2020;2 e10 (1-16) [Google Scholar]
- 62.Ghahremanpour M.M., et al. Identification of 14 known drugs as inhibitors of the main protease of SARS-CoV-2. ACS Med. Chem. Lett. 2020;11(12):2526–2533. doi: 10.1021/acsmedchemlett.0c00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ullrich S., Nitsche C. The SARS-CoV-2 main protease as drug target. Bioorg. Med. Chem. Lett. 2020;30(17) doi: 10.1016/j.bmcl.2020.127377. 127377 (1-25) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ahmad S., et al. Fragment-based in silico design of SARS-CoV-2 main protease inhibitors. Chem. Biol. Drug Des. 2021;98(4):604–619. doi: 10.1111/cbdd.13914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mohammed G. E.-D. A. A. Abuo-Rahma H.H.H., Abbas S.H., Abdelhafez E.-S.M.M. Current trends and future directions of fluoroquinolones. Curr. Med. Chem. 2019;26(17):3132–3149. doi: 10.2174/0929867325666180214122944. [DOI] [PubMed] [Google Scholar]
- 66.Ahadi H., Emami S. Modification of 7-piperazinylquinolone antibacterials to promising anticancer lead compounds: Synthesis and in vitro studies. Eur. J. Med. Chem. 2020;187 doi: 10.1016/j.ejmech.2019.111970. 111970 (1-57) [DOI] [PubMed] [Google Scholar]
- 67.Richter S., Parolin C., Palumbo M., Palù G. Antiviral properties of quinolone-based drugs. Curr. Drug Targets - Infect. Disord. 2004;4(2):111–116. doi: 10.2174/1568005043340920. [DOI] [PubMed] [Google Scholar]
- 68.Halgren T.A. Merck Molecular Force Field. I. Basis, form, scope, parameterization and performance of MMFF94. J. Comput. Chem. 1996;17(5–6):490–519. [Google Scholar]
- 69.Stewart J.J.P. Optimization of parameters for semiempirical methods V: Modification of NDDO approximations and application to 70 elements. J. Mol. Model. 2007;13(12):1173–1213. doi: 10.1007/s00894-007-0233-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wavefunction Inc; United States: 2019. Spartan’18. [Google Scholar]
- 71.Zhao Y., Truhlar D.G. The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: Two new functionals and systematic testing of four M06-class functionals and 12 other function. Theor. Chem. Acc. 2008;120(1):215–241. [Google Scholar]
- 72.Petersson G.A., Tensfeldt T.G., Montgomery J.A. A complete basis set model chemistry. III. The complete basis set-quadratic configuration interaction family of methods. J. Chem. Phys. 1991;94(9):6091–6101. [Google Scholar]
- 73.Sosa-García D., et al. Synthesis, structural investigation, antibacterial and DFT studies of complexes derived from a cholesteryl dithiophosphonate ligand with some thio-metallolane and thio-metallocane heterocycles of As(III) and Sb(III) Inorg. Chim. Acta. 2019;495 118943 (1-12) [Google Scholar]
- 74.Chan P.F., et al. Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin. Nat. Commun. 2015;6(10048):1–13. doi: 10.1038/ncomms10048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thomsen R., Christensen M.H. MolDock: a new technique for high-accuracy molecular docking. J. Med. Chem. 2006;49:3315–3321. doi: 10.1021/jm051197e. [DOI] [PubMed] [Google Scholar]
- 76.Tetko I.V., Tanchuk V.Y. Application of associative neural networks for prediction of lipophilicity in ALOGPS 2.1 program. J. Chem. Inf. Comput. Sci. 2002;42(5):1136–1145. doi: 10.1021/ci025515j. [DOI] [PubMed] [Google Scholar]
- 77.Leyva-Peralta M.A., Robles-Zepeda R.E., Razo-Hernández R.S., Berber L.P.Á., Lara K.O., Ruiz-Bustos E., Gálvez-Ruíz J.C. Berberine as source of antiproliferative hybrid compounds: in vitro antiproliferative activity and quantitative structure-activity relationship. Anticancer Agents Med. Chem. 2019;19:1820–1834. doi: 10.2174/1871520619666190503121820. [DOI] [PubMed] [Google Scholar]
- 78.Hernández-López H., et al. Synthesis of 1,4-biphenyl-triazole derivatives as possible 17β-HSD1 inhibitors: an in silico study. ACS Omega. 2020;5(23):14061–14068. doi: 10.1021/acsomega.0c01519. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.