BACKGROUND:
Methods that can automate, support, and streamline the preanesthesia evaluation process may improve resource utilization and efficiency. Natural language processing (NLP) involves the extraction of relevant information from unstructured text data. We describe the utilization of a clinical NLP pipeline intended to identify elements relevant to preoperative medical history by analyzing clinical notes. We hypothesize that the NLP pipeline would identify a significant portion of pertinent history captured by a perioperative provider.
METHODS:
For each patient, we collected all pertinent notes from the institution’s electronic medical record that were available no later than 1 day before their preoperative anesthesia clinic appointment. Pertinent notes included free-text notes consisting of history and physical, consultation, outpatient, inpatient progress, and previous preanesthetic evaluation notes. The free-text notes were processed by a Named Entity Recognition pipeline, an NLP machine learning model trained to recognize and label spans of text that corresponded to medical concepts. These medical concepts were then mapped to a list of medical conditions that were of interest for a preanesthesia evaluation. For each condition, we calculated the percentage of time across all patients in which (1) the NLP pipeline and the anesthesiologist both captured the condition; (2) the NLP pipeline captured the condition but the anesthesiologist did not; and (3) the NLP pipeline did not capture the condition but the anesthesiologist did.
RESULTS:
A total of 93 patients were included in the NLP pipeline input. Free-text notes were extracted from the electronic medical record of these patients for a total of 9765 notes. The NLP pipeline and anesthesiologist agreed in 81.24% of instances on the presence or absence of a specific condition. The NLP pipeline identified information that was not noted by the anesthesiologist in 16.57% of instances and did not identify a condition that was noted by the anesthesiologist’s review in 2.19% of instances.
CONCLUSIONS:
In this proof-of-concept study, we demonstrated that utilization of NLP produced an output that identified medical conditions relevant to preanesthetic evaluation from unstructured free-text input. Automation of risk stratification tools may provide clinical decision support or recommend additional preoperative testing or evaluation. Future studies are needed to integrate these tools into clinical workflows and validate its efficacy.
KEY POINTS.
Question: Can natural language processing (NLP) technology be used to identify pertinent preanesthesia history using free text from the electronic medical record?
Findings: The NLP pipeline and anesthesiologist agreed in 81.2% of instances on the presence of medical conditions, but did capture 16.6% of instances in which the anesthesiologist did not find.
Meaning: NLP may be a useful tool to aid preoperative anesthesia providers in screening and evaluation of surgical patients.
See Article, page 1159
Increases in surgical volume and associated costs of care are important challenges faced by the US health care system.1–3 An aging population with higher levels of comorbidity further complicates efforts to improve quality and decrease costs.4,5 Lee6 first introduced the concept of an anesthetic assessment clinic over 70 years ago. Increasing ambulatory surgical volumes and concerted efforts to reduce perioperative complications catalyzed widespread adoption of preoperative clinics.2,3,7 The benefits of a coordinated preoperative patient assessment include a reduction in unnecessary testing, surgical cancelation, and postoperative mortality, as well as increased patient satisfaction and optimized resource utilization.8–12
The American Society of Anesthesiologists practice advisory for preanesthesia evaluation places the responsibility for this process with the anesthesiologist.13 However, implementation of a preoperative evaluation workflow is heterogeneous across institutions. A shortage of anesthesiologists coupled with resource constraints has contributed to development of models where surgical patients may undergo an early triaging process to determine subsequent assessment (eg, preoperative visit, telephone interview, or day-of-surgery evaluation). These evaluations are often performed by nurses, nurse practitioners, or physician assistants working under various degrees of anesthesiologist supervision.1,14 Methods that can automate, support, and streamline this process may improve resource utilization and perioperative efficiency.
The widespread adoption of electronic health records (EHRs) has significantly increased the creation and accessibility of clinical data, which can be further analyzed with new technologies.15 Natural language processing (NLP) is one of many applications within the domain of artificial intelligence.16 NLP involves the extraction of relevant information from the contextual and semantic properties of spoken or written human language. The use of NLP in medical research is increasingly described but clinical applications remain uncommon.17–19 As a significant amount of EHR information relevant to the clinician evaluation is contained within longitudinal free-text (“unstructured”) narratives, NLP may assist in perioperative workflows by improving clinician efficiency and increasing the inclusiveness of the preoperative assessment. We described the development of a clinical NLP pipeline (including a machine learning model and rules-based components) intended to identify elements relevant to preoperative medical history by analyzing clinical notes. In this proof-of-concept study, we implemented an NLP pipeline to extract salient features from unstructured data that are relevant to a preanesthetic evaluation and compared its output to that of an anesthesiologist evaluating the same data. We hypothesized that the NLP pipeline would identify a significant portion of pertinent history captured by a perioperative provider; and if so, it would be a useful tool to support clinicians (but not replace) in the preoperative evaluation process.
METHODS
This study was approved by our institutional review board (Human Research Protections Program) at the University of California, San Diego, and the board waived the requirement for written consent. Data were collected retrospectively from the institutional EHRs of the patients from a single-day census (n = 93) of the Anesthesia Preparedness Clinic, which is our institution’s preoperative care clinic, in January of 2020. Our institution is a quaternary academic medical center with surgical patients presenting both as internal and external referrals, possessing a range of existing clinical documentation from no available health records to extensive EHR data. The clinic is tasked to screen every patient who will undergo elective surgery in advance; therefore, this can range from patients undergoing low-risk outpatient surgery to high-risk major surgery and with patients with low-to-severe comorbidity burden. The clinic does not routinely review inpatients who were added onto the operating room schedule. All patients from this day were included in the analysis, and all were planned to undergo elective surgery. This observational study adheres to the Enhancing the Quality and Transparency of Health Research (EQUATOR) guidelines.
Data Collection
For each patient, we collected all pertinent notes from the institution’s electronic medical record system (Epic Hyperspace, Epic Systems Corporation) that were available no later than 1 day before their preoperative anesthesia clinic appointment (the actual preoperative note created by the anesthesiology provider on day of appointment was not included as input into the NLP pipeline). The earliest note would date back as far as 2007, as this was when our institution adopted the current electronic medical record system. Pertinent notes included free-text notes consisting of history and physical, consultation, outpatient, inpatient progress, and previous preanesthetic evaluation notes. These notes were then processed in the NLP pipeline described in more detail below.
Overview
In summary, (1) clinical notes were inputted into the pipeline, and a Named Entity Recognition model (described below) extracted pertinent “entities” based on its machine learning model trained to label spans of text (KAID Health); (2) these entities were then mapped to medical “concepts”; (3) we created a list of pertinent medical comorbidities—or “conditions”—that were of interest to a preanesthesia evaluation (Table 1). Each of the medical concepts extracted from the NLP pipeline was then mapped to one of these pertinent conditions; and (4) the final output of the NLP pipeline was a list of conditions that were associated with a given patient’s medical history. We summarize a few terms and their definition that were used throughout the manuscript.
Table 1.
Description How Each Concept Is Flagged Into (1) History, (2) Negation, (3) Uncertainty‚ and (4) Hypotheticals
| Flagged criteria | Types | Examples |
|---|---|---|
| History | PMH | “PMH: Myocardial infarction (MI), stroke, splenectomy at age 53…” “Patient has a history of post-operative nausea and vomiting (PONV) and difficult intubation…” |
| Family history | “Father died at age 73 of cerebrovascular accident (CVA)...” “Mother had liver disease from alcoholic (EtOH) cirrhosis…” |
|
| Negations | Pertinent negatives | “Patient denies hematuria, fevers, chills night sweats, chest pain, shortness of breath (SOB), nausea…” “Negative for: palpitations, syncope, chest pain, orthopnea…” “Patient has no weight loss, fevers, or chills…” |
| Rule-outs/exclusions | “Negative sputum culture renders an infectious etiology unlikely…” “Chest x-ray (CXR) rules out the possibility of pneumonia…” |
|
| Uncertainties | Differential diagnosis/abstractions | “CXR suggestive of lobar pneumonia of the right upper lobe (RUL)” “Patient presents with symptoms likely due to ____” “If symptoms persist, consider oral glucocorticoid therapy…” “Iron deficiency anemia unlikely given labs” |
| Recommendations and referrals | “Patient recommended to start AEDs for seizure prophylaxis” | |
| Qualifiers | “Patient’s lab indicate borderline anemia…” | |
| Hypotheticals | List of potential adverse outcomes following a procedure | “Patient was informed about the possible anesthetic complications including DVT, heart attack, stroke, and death.” |
| Risk factors | “Patient has uncontrolled HTN and diabetes, which are risk factors for stroke…” | |
| Patient education | “We discussed the risks of testosterone replacement therapy including polycythemia with stroke, MI, and recurrent PE…” |
Those that are considered family history, negation, or hypothetical were removed as pertinent results.
Highlighted cells correspond to the flags that were excluded from the model output for the study.
Abbreviations: AED, anti-epileptic drug; DVT, deep vein thrombosis; EtOH, alcohol; HTN, hypertension; PE, pulmonary embolism; PMH, past medical history; RUL, right upper lobe.
Category—a broader categorization scheme under which multiple conditions may be collated (ie, cardiovascular system and hematology).
Concept—any medical term or idea that is labeled based on the entities that the NLP pipeline identifies from free text (ie, body mass index, depression, blood pressure, and temperature).
Concept unique identifier (CUI)—the CUI for a Metathesaurus (the Metathesaurus is a large biomedical thesaurus organized by concept or meaning, and it links similar names for the same concept from nearly 200 different vocabularies) concept to which strings with the same meaning are linked. The CUI is an identifier that uniquely represents a meaning, and the meaning of a CUI does not change over time.
Condition—distinct clinical diagnosis of pathologic state (ie, coronary artery disease, asthma). A condition is more granular descriptions derived from concepts. They are what comprises the pertinent medical history in the preanesthesia elements’ list.
Entity—the output from the Named Entity Recognition model. These entities, which are captured from the free text, are subsequently labeled as medical concepts.
Unified Medical Language System (UMLS)—large biomedical thesaurus that is organized by concepts, which links similar names for the same concept from nearly 200 different vocabulary systems.
NLP Pipeline
The free-text notes were processed by a Named Entity Recognition model, an NLP machine learning model trained to recognize and label spans of text and extract entities that subsequently correspond to medical concepts (KAID Health). This component captured misspelled entities. The misspelled entities were then coded to a UMLS CUI; depending on the degree of corruption, misspelled entities can be appropriately coded, not coded, or, in much fewer cases, the wrong code may be applied (if the corruption somehow made the misspelling look more similar to another concept). This approach allowed for a significant number of misspellings to be correctly coded and associated with the patient. The NLP pipeline assigned these concepts into 1 of 3 categories: problems, which include diagnoses, syndromes, or chief complaints; tests such as labs or imaging studies; and treatments including medications, procedures, and/or supportive therapy.
The concepts extracted by the NLP pipeline from patient charts were further processed by a rules-based system to flag each as (1) pertinent history—concept was associated with patient medical history; (2) negations—included a negative review of systems where a physician notes the absence of various pathologies; (3) uncertainties—suggestions of medical history; or (4) hypotheticals—possible differential diagnoses based on presenting symptoms, laboratories, and/or imaging studies, or a consent form disclaimer that lists possible adverse outcomes of a procedure. For concepts labeled as tests, the rules-based component extracted and assigned the corresponding laboratory values or quantitative data to the test concept (eg, ejection fraction = 36%, Hgb = 14 g/dL).
The remaining concepts (those not removed due to negation or hypotheticals) were then each linked to a UMLS CUI, a meta taxonomy that unifies International Classification of Diseases 10, Systemized Nomenclature of Medicine, Current Procedural Terminology codes, and other clinical ontologies.
We created a dictionary that maps each condition outlined in our institutional preoperative anesthetic evaluation checklist to a set of concepts that would indicate the presence of the condition (Table 2). Each concept in the dictionary was then coded to UMLS CUIs, which were manually vetted and pruned by an anesthesiologist (J.T.) to ensure the CUIs corresponded to conditions relevant to the preoperative evaluation.
Table 2.
List of Conditions Pertinent to Our Institution’s Preanesthetic Evaluation and Example Concepts That Would Map to Each Condition
| Conditions | Example concepts |
|---|---|
| Cardiac | |
| Valve abnormality | Mitral valve regurgitation and aortic stenosis |
| History of heart transplant | Heart transplant |
| Coronary stents | LAD stent and RCA stent |
| Coronary artery disease | CAD and coronary artery atherosclerotic disease |
| Peripheral vascular disease | Deep venous thrombosis and occlusive thrombosis of peripheral vasculature |
| Pacemaker | Pacemaker |
| Implantable cardioverter-defibrillator | Implantable cardioverter-defibrillator |
| Heart murmur | Systolic/diastolic murmur and holosystolic ejection murmur |
| Left ventricular failure | Heart failure with reduced ejection fraction and reduced ejection fraction |
| Myocardial infarction | Heart attack and ST-elevation myocardial infarction |
| Hypertension | Hypertension |
| Congenital heart disease | Ebstein anomaly and coarctation of the aorta |
| Congestive heart failure | Congestive heart failure and jugular venous distension |
| History of CABG | CABG |
| Cardiac arrhythmia | Heart block, atrial fibrillation, and supraventricular tachycardia |
| History of angina | Ischemic chest pain and angina |
| Anticoagulants | Aspirin and clopidogrel |
| Heart disease | Cardiomegaly and cardiomyopathy |
| Central nervous system | |
| Traumatic brain injury | Traumatic brain injury |
| Seizure history | Seizures and epilepsy |
| History of stroke/TIA | Cerebrovascular accident, cerebral infarct, and transient ischemic attack |
| Spine disease | Degenerative disk disease, scoliosis, and herniated disk |
| Spinal cord injury | Compression fracture, disk injury, and hemiplegia |
| Psychiatric disease | Depression, anxiety disorder, and posttraumatic stress disorder |
| Neuromuscular disease | Multiple sclerosis, myasthenia gravis, and muscular dystrophy |
| Cognitive impairment | Dementia, amnesia, and Huntington’s disease |
| Developmental delay | Autism, delay in motor/cognitive development |
| Gastrointestinal | |
| Pancreatic disease | Pancreatitis and pancreatic ductal dilation |
| Liver disease | Nonalcoholic fatty liver disease and cirrhosis |
| Hepatitis | Hepatitis A, B, and C |
| Gastroesophageal reflux disease | Gastroesophageal reflux disease, Barrett’s esophagus, and Schatzki rings |
| Bowel preparation | Bowel preparation |
| Bowel/intestinal obstruction | Bowel/intestinal obstruction |
| General | |
| Weight loss | Rapid weight loss |
| Postoperative nausea/vomiting | Postoperative nausea or vomiting |
| Obesity | Obesity, BMI >30 |
| METS <4 | Inability to climb stairs or exercise, exertional dyspnea, and decreased functional status |
| Risk of falls | Recent fall and ataxia |
| Inability to dress themselves | Compromised activities of daily living |
| Congenital abnormalities | Cystic fibrosis, down syndrome, and Fragile X |
| Chronic pain | Chronic pain with opioid use and neuropathic pain |
| History of anesthesia complication | Anaphylaxis to anesthetics and aspiration pneumonitis |
| Airway issues | Tracheal abnormalities and laryngeal stenosis |
| Hematology/oncology | |
| Radiation therapy | Radiation therapy and radiation treatment |
| Sickle cell anemia | Sickle cell disease |
| Coagulopathy | Hemophilia and thrombocytopenia |
| Chemotherapy | Chemotherapy |
| Cancer | Leukemia, colorectal cancer, and lymphoma |
| Anemia | Iron deficiency anemia and megaloblastic anemia |
| Infectious disease | |
| Vancomycin-resistant enterococcus | Vancomycin-resistant enterococcus |
| Tuberculosis | Miliary tuberculosis and latent tuberculosis |
| Sepsis | Sepsis, bacteremia, and septic shock |
| Pneumonia | Lobar pneumonia, Streptococcus pneumoniae infection of lungs, and bronchopulmonary pneumonia |
| Open wound | Open wound and compromised healing |
| Methicillin-resistant S. aureus | Methicillin-resistant Staphylococcus aureus |
| Human immunodeficiency virus | HIV |
| Clostridium difficile | Clostridium difficile |
| Pulmonary | |
| Upper respiratory tract infection | Epiglottitis, laryngitis, pharyngitis, and common cold |
| Tracheotomy/tracheostomy | Tracheotomy and tracheostomy |
| Pulmonary hypertension | Idiopathic pulmonary hypertension and pulmonary arterial hypertension |
| Obstructive sleep apnea | Obstructive sleep apnea |
| Chronic obstructive pulmonary disease | Emphysema and chronic bronchitis |
| Chronic lung disease | Interstitial lung disease, bronchiectasis, and pneumoconiosis |
| Asthma | Asthma and status asthmaticus |
| Renal | |
| Renal insufficiency/failure | Renal artery stenosis, polycystic kidney disease, and acute kidney injury |
| Electrolyte disorders | Hyperkalemia, hypernatremia, and hypocalcemia |
| Other | |
| Thyroid/parathyroid disease | Hyperthyroidism, goiter, hyperparathyroidism, and Graves’ disease |
| Substance abuse | Alcohol use disorder and IV drug user |
| Steroid use | Hydrocortisone, prednisone, and dexamethasone |
| Rheumatoid disease | Rheumatoid arthritis and Sjogren’s disease |
| Malignant hyperthermia | Malignant hyperthermia |
| Lupus | Systemic lupus erythematosus |
| Diabetes mellitus | Diabetes mellitus |
| Cushing’s disease | Cushing’s disease and iatrogenic Cushing’s disease |
| Back pain | Lumbago and lower back pain |
| Arthritis | Osteoarthritis, gonococcal arthritis, and gout |
All concepts that are deemed pertinent from the NLP engine are then mapped to one of these conditions and reported as the final output.
Abbreviations: BMI, body mass index; CABG, coronary artery bypass graft; CAD, coronary artery disease; HIV, human immunodeficiency virus; IV, intravenous; LAD, left anterior descending artery; METs, metabolic equivalents; NLP‚ natural language processing; RCA, right coronary artery; TIA, transient ischemic attack.
From the output of the NLP pipeline, concepts flagged as negated, part of family history, or hypothetical were removed from the master list (examples provided in Table 3). For 3 tests—metabolic equivalent of task (objective measure of the ratio of the rate at which a person expends energy while performing a specific task compared to a reference), body mass index, and left ventricular ejection fraction—we filtered for values falling above or below specified thresholds, assigning parent conditions to patients meeting the testing criteria (eg, obesity for body mass index >30, systolic heart failure for left ventricular ejection fraction <40%). We then filtered the NLP output containing all the entities extracted from the notes for only those CUIs associated with conditions included in the preoperative checklist. We created pivot tables for the remaining concepts so that for each condition on the preoperative evaluation checklist, each patient was represented as a binary result of either having or not having it. The final output of the NLP pipeline was a table of patient conditions determined to be of interest in our anesthesia preoperative care clinic, with information on the note where the reference occurred, and location within the note.
Table 3.
Sample Medical Concepts Extracted by the Model and Subsequently Excluded According to the Flagged Criteria
| Components | Sample concept 1 | Sample concept 2 | Sample concept 3 | Sample concept 4 | Sample concept 5 |
|---|---|---|---|---|---|
| Text | Low back pain | Diabetes mellitus | Cancer | BMI | Dialysis |
| Start character | 406 | 63 | 119 | 295 | 657 |
| End character | 419 | 80 | 132 | 298 | 669 |
| Label | PROBLEM | PROBLEM | PROBLEM | TEST | TREATMENT |
| Negate | TRUE | ||||
| Family | TRUE | ||||
| Allergy | |||||
| History | TRUE | ||||
| Uncertain | TRUE | TRUE | |||
| Hypothetical | |||||
| Laboratory value | 32.73 | ||||
| Snippet | Patient denies lower back pain | Past medical history: has a past medical history of borderline diabetes mellitus, high blood pressure… | Family history: diabetes father, heart attack father, and cancer mother | Spo2 100% BMI 32.73 kg/m | If no improvement tomorrow will need to discuss whether this can be managed as nondialysis CKD V or whether dialysis will need to be considered |
| Note ID | X | X | X | X | X |
| Patient ID | X | X | X | X | X |
| CUI | C0024031 | C0011849 | C0006826 | C0005893 | C0011946 |
| Entity | Back pain | Diabetes mellitus | Hx of cancer | Obesity | Renal disease |
| Category | Endocrine/other | Endocrine/other | Heme/onc | General | Renal |
Highlighted cells correspond to medical concepts extracted by model that are excluded from the output based on flagged criteria (eg, negate, family, and hypothetical).
Abbreviations: BMI, body mass index; CKD‚ chronic kidney disease; CUI, concept unique identifier.
Statistical Analysis
All analyses were performed using R Statistical Programming Language (v4.1.2) and Python (v3.9.7). Our primary evaluation was to compare the output of the NLP pipeline to that of an anesthesiologist. An anesthesiologist (M.N.M.), who frequently staffed the anesthesia preoperative care clinic, was given the same list of 93 patients and asked to perform a preanesthetic evaluation utilizing all the available EHR data before and including the date of their preoperative care clinic appointment. Of note, the anesthesiologist was also able to review the preoperative anesthesia note that was created during their anesthesia evaluation (unlike the NLP pipeline). The chart review process ranged from 5 to 20 minutes, on average taking 15 minutes per patient. Once chart review was completed, the anesthesiologist indicated whether they did or did not identify each of the dictionary terms in the patient’s EHR.
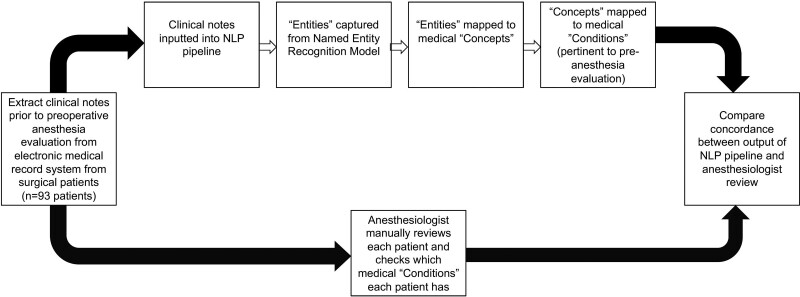
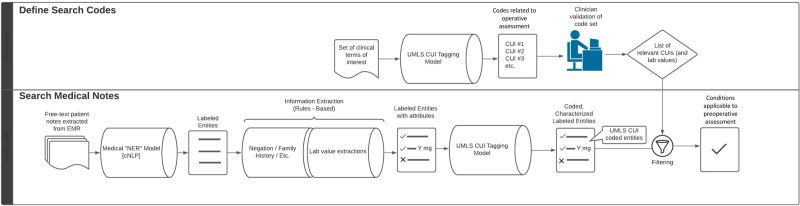
We then compared the concordance rates for each condition on the preoperative checklist by comparing the output for each patient of both the NLP pipeline and the anesthesiologist review (Figure 1). For each condition, we calculated the percentage of time across all patients in which: (1) the NLP pipeline and the anesthesiologist both captured the condition; (2) the NLP pipeline captured the condition but the anesthesiologist did not; and (3) the NLP pipeline did not capture the condition but the anesthesiologist did. Patients identified as having a concept by the anesthesiologist but not the NLP pipeline were investigated further to manually differentiate whether these were either “true” or “false positives” for the clinician, or “true” or “false negatives” for the NLP pipeline. We performed a subsequent review of each patient specifically assessing the condition of interest. The medical entities (eg, diabetes and heart failure) where the NLP pipeline marked >10% of patients having the condition but the anesthesiologist did not were manually reviewed to parse either “true” or “false positives” for the NLP pipeline, or “true” or “false negatives” for the clinician. We looked through the notes that the NLP pipeline noted as containing the diagnosis for the patient to verify the validity of the output. Figure 2 illustrates the overall workflow of the NLP pipeline and clinician review of the same set of patients.
Figure 1.
Workflow of the study, in which free-text clinical notes from 93 patients were extracted from the electronic medical record system. These notes were processed by an NLP pipeline, and its output was compared to that captured by an anesthesiologist. NLP indicates natural language processing.
Figure 2.
Illustration of the algorithm followed by the NLP pipeline (KAID Health). cNLP indicates clinical natural language processing; CUI, concept unique identifier; EMR, electronic medical record; NER, named entity recognition; NLP‚ natural language processing; UMLS, Unified Medical Language System.
RESULTS
A total of 93 patients were included in the NLP pipeline input. Free-text notes were extracted from the EHRs of these patients for a total of 9765 history and physical, consultation, outpatient, inpatient progress, and previous preanesthetic evaluation notes before the actual date of their preoperative anesthesia evaluation. The median (25%–75% quartiles) number of notes per patient was 45 (14.5–151.5) notes. Across these notes, the NLP pipeline captured 221,764 medical concepts. Of these, 17,560 medical concepts were pertinent to our preanesthesia elements and were then mapped to 76 separate conditions in the preoperative evaluation criteria. The dictionary that was used to map these concepts to the preoperative criteria contained 1880 terms that each corresponded to 1 of the 76 concepts (Table 1).
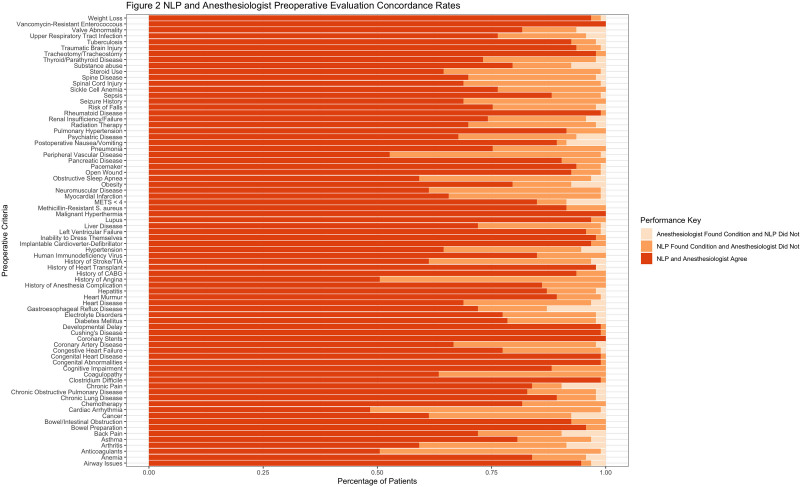
The NLP pipeline and anesthesiologist agreed in 81.24% of instances on the presence or absence of a specific condition. The NLP pipeline identified information that was not noted by the anesthesiologist in 16.57% of instances and did not identify a condition that was noted by the anesthesiologist’s review in 2.19% of instances (Figure 3). The most common conditions that the NLP pipeline captured that the anesthesiologist did not included: cardiac arrhythmias (50.5% of cases with this condition were captured by NLP and not the anesthesiologist), angina (49.5%), anticoagulation (48.4%), peripheral vascular disease (46.2%), obstructive sleep apnea (37.6%), and neuromuscular disease (37.6%). The most common conditions that the NLP pipeline did not capture but the anesthesiologist did included: chronic pain (9.7% of cases with this condition were not captured by NLP but was by the anesthesiologist), back pain (9.7%), arthritis (8.6%), postoperative nausea/vomiting (8.6%), and metabolic equivalents (METs) <4 (8.6%). The most common conditions at which both the NLP pipeline and anesthesiologist captured included: (1) cardiac stents (100% of cases with this condition were captured by both the anesthesiologist and NLP), (2) rheumatoid disease (98.9%), (3) Cushing’s disease (98.9%), (4) developmental delay (98.9%), and (5) congenital heart disease (98.9%).
Figure 3.
Stacked bar plot illustrating the concordance rates between the NLP pipeline output and anesthesiologist review. CABG indicates coronary artery bypass graft; METs‚ metabolic equivalents; NLP, natural language processing; TIA‚ transient ischemic attack.
DISCUSSION
In this proof-of-concept study, we utilized an NLP pipeline to extract pertinent preanesthesia conditions from unstructured free-text notes from the EHR. The extracted conditions were then compared to what was captured from an anesthesiologist. We demonstrated that among 93 patients and 9765 clinical notes, the NLP pipeline and anesthesiologist agreed in 81.24% of instances on the presence or absence of a specific condition. The NLP pipeline identified information that was not noted by the anesthesiologist in 16.57% of instances and did not identify a condition that was noted by the anesthesiologist’s review in 2.19% of instances. We demonstrated that utilization of NLP produced an output that identified the presence or absence of conditions relevant to preanesthetic evaluation from unstructured free-text input derived from EHR notes, and did so in a manner often in concordance with an anesthesiologist reviewing the same information. While the literature has previously described the use of NLP to extract data from clinical notes,20 to our knowledge, this is the first application to focus on the preanesthetic evaluation.
The ideal preanesthetic evaluation is a longitudinal process, which begins with the surgeon’s decision to operate and ends with the day-of-surgery assessment by the anesthesiologist who will be caring for the patient in the operating room. In between these events lie a continuum of risk stratification tools, institution-specific protocols, and workflows, and optimization of factors including nutrition and cardiopulmonary status. Gaps in this process can result in the omission of critical history information, failure to obtain recommended studies and testing, or inadequate communication between providers, and may contribute to costly surgical delays or cancelations.21,22 Development of tools to assist in the preanesthetic evaluation may reduce the likelihood of such adverse outcomes and may offset the impact of limited personnel or resources available for this process.23–25
It is important to note that while the preanesthetic evaluation is more than just a “chart review,” a significant amount of workflow at our preoperative clinic involved interrogating a patient’s EHR to ensure that information contained is consistent with the history obtained by our clinicians. Patients may be referred by surgeons and other providers who have not manually added history elements or problems to the formal EHR profile.
NLP may thus be used as a tool to aid clinicians in a preoperative care clinic to more efficiently identify high-risk patients, triage resources (eg, screen for healthy patients that may not need a separate preoperative evaluation), ensure EHR information is up-to-date, and reduce workloads/burnout especially in an institution with a high-volume preoperative care clinic. However, further studies are needed to determine its efficacy in producing said benefits.
Construction of the dictionary used by the NLP pipeline to map terms to conditions was designed to be broadly encompassing so as to prioritize “too much” over “too little” data. This approach accepted the higher risk of the inclusion of extraneous or nonclinically relevant information over the potential to miss a crucial history element. For example, the NLP pipeline was taught to recognize numerous kinds of tumors—from basal cell carcinomas to small cell lung cancer—as being terms indicating the presence of the condition “cancer.” Whereas the potential comorbidities of treatments for the latter can be critical to the anesthetic evaluation, the former may have less clinical relevance and, therefore, not included as a positive element in a clinician’s review. The advantages of such an approach can be apparent with other significant conditions. By constructing a definition for “cardiac angina” that included less specific terms such as “chest pain” or “chest tightness,” the patient with musculoskeletal pain may be identified but the likelihood of missing a patient at true risk for intraoperative cardiac ischemia may also decrease.
Certain situations we encountered in analyzing the NLP output provided insights into the limitations of the NLP pipeline’s ability to extrapolate and contextualize from free text. Homonyms (interpreting “falling” in the phrase “trouble falling asleep” as a potential fall risk) and syntax (typographic or formatting errors) were examples of 2 areas of challenge. Medical abbreviation or shorthand could be similarly confusing to the NLP pipeline. The phrase “start AEDs for seizure prophy” (sic) resulted in a positive identification of a seizure disorder when the NLP pipeline was unable to distinguish the truncation of the word prophylaxis. A formatting issue caused the abbreviation “pHTN” for pulmonary hypertension to incorrectly map “HTN” to “pHTN,” which subsequently led to the NLP pipeline missing a number of cases of arterial hypertension marked with the plain abbreviation. However, it should be noted that machine learning models can become more robust and learn to differentiate variations in notation styles as they are trained with additional data.26 Therefore, these and the previously mentioned grammatical and contextual issues are challenges that may resolve as the NLP pipeline is exposed to a wider variety of clinical notes over time.
Furthermore, the NLP pipeline was agnostic to chronology, which may have resulted in the identification of entities deemed to be not clinically relevant if the pathology had since resolved or was particularly remote, such as pneumonia 10 years ago or childhood cancer. Additionally, input to the NLP pipeline did not include numeric laboratory studies or vital signs present in EHR flowsheets outside of free-text clinical notes, nor did it include reports from radiologic studies, all of which were available to the anesthesiologist in their review. The choice was made for this project to emphasize free-text NLP from clinical notes and to avoid the influence of potential variations on reference ranges and diagnostic criteria. However, the underlying characteristics of the NLP pipeline make it competent at integrating these data in a way that would boost performance if numeric data were incorporated in future iterations according to institutional or societal guidelines. Radiology reports and other nonclinical note unstructured free text could also be included in future pipeline input, as could notes obtained from outside institutions via data-sharing agreements such as health information exchanges.
A significant limitation of this study was the absence of information from the anesthesiologist performing the initial review of the patient charts as to why and how they identified the presence and absence of each condition. In the situation of disagreement between model and anesthesiologist, it can be challenging to ascertain what criteria were used by the anesthesiologist to identify a condition if it is one that relies on loose associations or abstract reasoning. Furthermore, there may be disagreement between the anesthesiologist and the concepts included in the dictionary with respect to how a particular pathology may be classified. For example, sodium dyscrasias were classified as an electrolyte abnormality in the dictionary, but may be considered a kidney problem by the anesthesiologist if the primary etiology arises from renal dysfunction. In this proof-of-concept project, the primary goal was to evaluate the concordance rate between the NLP pipeline and an anesthesiologist to assess the feasibility of NLP as a tool to be used in the preanesthesia care workflow as opposed to a full substitute for a thorough chart review. As such, we refrained from complex analysis of the potentially subjective clinical judgment of an anesthesiologist in selecting or not selecting options for clinical relevance that the potentially more binary NLP pipeline would. Future pilot studies will focus more on the applicability and relevance of NLP-derived output by having a design in which any clinician input will include subjective descriptions of relative importance. Furthermore, future analysis should include comparison of the performance of NLP to multiple types and numbers of clinical providers.
Similar NLP techniques may be used in the future to integrate additional data from the EHR into preoperative workflows. Incorporation of information from previous anesthetic records, including past physical examinations, airway histories, and intraoperative hemodynamic events, may be contextualized to provide additional predictive or preparatory benefit.27–29 Integration of laboratory values, imaging results, and other data not included in this analysis can further improve performance of this NLP pipeline. Automation of risk stratification tools may provide clinical decision support or recommend additional preoperative testing or evaluation.30 Future studies are needed to integrate these tools into clinical workflows and validate their use.
DISCLOSURES
Name: Harrison S. Suh, BS.
Contribution: This author is responsible for the study design, analysis and interpretation of data, and drafting/finalizing the article.
Conflicts of Interest: H. S. Suh was a paid research intern in summer 2020 for KAID Health (Boston, MA).
Name: Jeffrey L. Tully, MD.
Contribution: This author is responsible for the study design, analysis and interpretation of data, and drafting the article.
Conflicts of Interest: None.
Name: Minhthy N. Meineke, MD.
Contribution: This author is responsible for the study design, analysis and interpretation of data, and drafting the article.
Conflicts of Interest: None.
Name: Ruth S. Waterman, MD, MS.
Contribution: This author is responsible for analysis and interpretation of data and drafting/finalizing the article.
Conflicts of Interest: None.
Name: Rodney A. Gabriel, MD, MAS.
Contribution: This author is responsible for the study design, analysis and interpretation of data, and drafting/finalizing the article.
Conflicts of Interest: The University of California has received funding and/or product for other research projects from Epimed International (Farmers Branch, TX); Infutronics (Natick, MA); Precision Genetics (Greenville, SC); and SPR Therapeutics (Cleveland, OH) for R. A. Gabriel. The University of California San Diego is a consultant for Avanos (Alpharetta, GA), in which R. A. Gabriel represents.
This manuscript was handled by: Richard C. Prielipp, MD.
GLOSSARY
- AED
- anti-epileptic drug
- BMI
- body mass index
- CABG
- coronary artery bypass graft
- CAD
- coronary artery atherosclerotic disease
- CKD
- chronic kidney disease
- CUI
- concept unique identifier
- DVT
- deep vein thrombosis
- EHR
- electronic health record
- EQUATOR
- Enhancing the Quality and Transparency of Health Research
- EtOH
- alcohol
- HIV
- human immunodeficiency virus
- HTN
- hypertension
- IV
- intravenous
- LAD
- left anterior descending
- MET
- metabolic equivalent
- NLP
- natural language processing
- PE
- pulmonary embolism
- RCA
- right coronary artery
- RUL
- right upper lobe
- TIA
- transient ischemic attack
- UMLS
- Unified Medical Language System
Funding: None.
Conflicts of Interest: See Disclosures at the end of the article.
Reprints will not be available from the authors.
REFERENCES
- 1.Schubert A, Eckhout GV, Ngo AL, Tremper KK, Peterson MD. Status of the anesthesia workforce in 2011: evolution during the last decade and future outlook. Anesth Analg. 2012;115:407–427. [DOI] [PubMed] [Google Scholar]
- 2.Cullen KA, Hall MJ, Golosinskiy A. Ambulatory surgery in the United States, 2006. Natl Health Stat Report. 2009; 11:1–25. [PubMed] [Google Scholar]
- 3.White PF, Smith I. Ambulatory anesthesia: past, present, and future. Int Anesthesiol Clin. 1994;32:1–16. [PubMed] [Google Scholar]
- 4.Dall TM, Gallo PD, Chakrabarti R, West T, Semilla AP, Storm MV. An aging population and growing disease burden will require a large and specialized health care workforce by 2025. Health Aff. 2013;32:2013–2020. [DOI] [PubMed] [Google Scholar]
- 5.Caley M, Sidhu K. Estimating the future healthcare costs of an aging population in the UK: expansion of morbidity and the need for preventative care. J Public Health (Oxf). 2011;33:117–122. [DOI] [PubMed] [Google Scholar]
- 6.Lee JA. The anaesthetic out-patient clinic. Anaesthesia. 1949;4:169–174. [DOI] [PubMed] [Google Scholar]
- 7.Yen C, Tsai M, Macario A. Preoperative evaluation clinics. Curr Opin Anaesthesiol. 2010;23:167–172. [DOI] [PubMed] [Google Scholar]
- 8.Parker BM, Tetzlaff JE, Litaker DL, Maurer WG. Redefining the preoperative evaluation process and the role of the anesthesiologist. J Clin Anesth. 2000;12:350–356. [DOI] [PubMed] [Google Scholar]
- 9.Rai MR, Pandit JJ. Day of surgery cancellations after nurse-led pre-assessment in an elective surgical centre: the first 2 years. Anaesthesia. 2003;58:692–699. [DOI] [PubMed] [Google Scholar]
- 10.Knox M, Myers E, Hurley M. The impact of pre-operative assessment clinics on elective surgical case cancellations. Surgeon. 2009;7:76–78. [DOI] [PubMed] [Google Scholar]
- 11.Harnett MJ, Correll DJ, Hurwitz S, Bader AM, Hepner DL. Improving efficiency and patient satisfaction in a tertiary teaching hospital preoperative clinic. Anesthesiology. 2010;112:66–72. [DOI] [PubMed] [Google Scholar]
- 12.Trinh LN, Fortier MA, Kain ZN. Primer on adult patient satisfaction in perioperative settings. Perioper Med (Lond). 2019;8:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Apfelbaum JL, Connis RT, Nickinovich DG, et al. ; Committee on Standards and Practice Parameters. Practice advisory for preanesthesia evaluation: an updated report by the American Society of Anesthesiologists task force on preanesthesia evaluation. Anesthesiology. 2012;116:522–538. [DOI] [PubMed] [Google Scholar]
- 14.Varughese AM, Byczkowski TL, Wittkugel EP, Kotagal U, Dean Kurth C. Impact of a nurse practitioner-assisted preoperative assessment program on quality. Paediatr Anaesth. 2006;16:723–733. [DOI] [PubMed] [Google Scholar]
- 15.Adler-Milstein J, Jha AK. HITECH act drove large gains in hospital electronic health record adoption. Health Aff (Millwood). 2017;36:1416–1422. [DOI] [PubMed] [Google Scholar]
- 16.Lluís Marquez JGS. Machine Learning and Natural Language Processing. 2000. Accessed February 25, 2022. https://citeseerx.ist.psu.edu/viewdoc/summary?doi=10.1.1.31.3498.
- 17.Costea EA. Machine learning-based natural language processing algorithms and electronic health records data. Linguistic Philos Investig. 2020;19: 93–99. [Google Scholar]
- 18.Hasan SA, Farri O. Clinical natural language processing with deep learning. Consoli S, Reforgiato Recupero D, Petković M, eds. In: Data Science for Healthcare: Methodologies and Applications. Springer International Publishing, 2019:147–171. [Google Scholar]
- 19.Juhn Y, Liu H. Artificial intelligence approaches using natural language processing to advance EHR-based clinical research. J Allergy Clin Immunol. 2020;145:463–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spasic I, Nenadic G. Clinical text data in machine learning: systematic review. JMIR Med Inform. 2020;8:e17984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nelson O, Quinn TD, Arriaga AF, et al. A model for better leveraging the point of preoperative assessment: patients and providers look beyond operative indications when making decisions. A A Case Rep. 2016;6:241–248. [DOI] [PubMed] [Google Scholar]
- 22.Liu S, Lu X, Jiang M, et al. Preoperative assessment clinics and case cancellations: a prospective study from a large medical center in China. Ann Transl Med. 2021;9:1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chow VW, Hepner DL, Bader AM. Electronic care coordination from the preoperative clinic. Anesth Analg. 2016;123:1458–1462. [DOI] [PubMed] [Google Scholar]
- 24.Vetter TR, Boudreaux AM, Ponce BA, Barman J, Crump SJ. Development of a preoperative patient clearance and consultation screening questionnaire. Anesth Analg. 2016;123:1453–1457. [DOI] [PubMed] [Google Scholar]
- 25.Alvis BD, King AB, Pandharipande PP, et al. Creation and execution of a novel anesthesia perioperative care service at a veterans affairs hospital. Anesth Analg. 2017;125:1526–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sung SF, Hsieh CY, Hu YH. Early prediction of functional outcomes after acute ischemic stroke using unstructured clinical text: retrospective cohort study. JMIR Med Inform. 2022;10:e29806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang AR, Lee J, Jung W, et al. Development of a prediction model for hypotension after induction of anesthesia using machine learning. PLoS One. 2020;15:e0231172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Solomon SC, Saxena RC, Neradilek MB, et al. Forecasting a crisis: machine-learning models predict occurrence of intraoperative bradycardia associated with hypotension. Anesth Analg. 2020;130:1201–1210. [DOI] [PubMed] [Google Scholar]
- 29.Miyaguchi N, Takeuchi K, Kashima H, Morita M, Morimatsu H. Predicting anesthetic infusion events using machine learning. Sci Rep. 2021;11:23648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borab ZM, Lanni MA, Tecce MG, Pannucci CJ, Fischer JP. Use of computerized clinical decision support systems to prevent venous thromboembolism in surgical patients: a systematic review and meta-analysis. JAMA Surg. 2017;152:638–645. [DOI] [PMC free article] [PubMed] [Google Scholar]