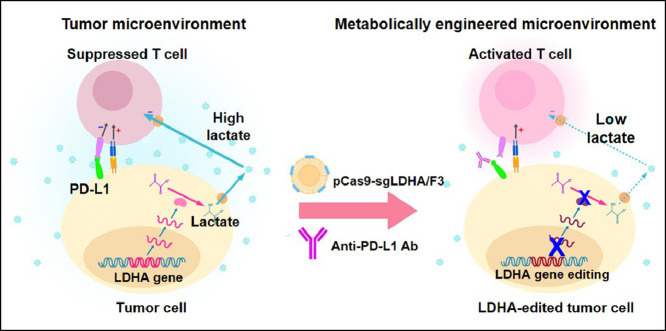
Abstract
Metabolic engineering of the tumor microenvironment has emerged as a new strategy. Lactate dehydrogenase A (LDHA) is a prominent target for metabolic engineering. Here, we designed a cationic lipid nanoparticle formulation for LDHA gene editing. The plasmid DNA delivery efficiency of our lipid nanoparticle formulations was screened by testing the fluorescence of lipid nanoparticles complexed to plasmid DNA encoding green fluorescence protein (GFP). The delivery efficiency was affected by the ratios of three components: a cationic lipid, cholesterol or its derivative, and a fusogenic lipid. The lipid nanoparticle designated formulation F3 was complexed to plasmid DNA co-encoding CRISPR-associated protein 9 and LDHA-specific sgRNA, yielding the lipoplex, pCas9-sgLDHA/F3. The lipoplex including GFP-encoding plasmid DNA provided gene editing in HeLa-GFP cells. Treatment of B16F10 tumor cells with pCas9-sgLDHA/F3 yielded editing of the LDHA gene and increased the pH of the culture medium. pCas9-sgLDHA/F3 treatment activated the interferon-gamma and granzyme production of T cells in culture. In vivo, combining pCas9-sgLDHA/F3 with immune checkpoint-inhibiting anti-PD-L1 antibody provided a synergistic antitumor effect and prolonged the survival of tumor model mice. This study suggests that combining metabolic engineering of the tumor microenvironment with immune checkpoint inhibition could be a valuable antitumor strategy.
Keywords: Gene editing, Lipid nanoparticle, Metabolic engineering, Lactate dehydrogenase A, Tumor microenvironment
Graphical abstract
1. Introduction
Immunotherapy has quickly emerged as a prominent method of cancer treatment and is now considered a standard treatment strategy, alongside surgery, radiation therapy, chemotherapy, and targeted pathway inhibition [1,2]. The immune checkpoint inhibitors (ICIs) that are used in cancer immunotherapy function to block programmed cell death protein 1/programmed cell death ligand 1 (PD-1/PD-L1) or cytotoxic T lymphocyte-associated protein 4 (CTLA-4) to prevent the interactions that lead to T cell inhibition, and thereby promote the antitumor activity of effector T cells.
Although ICIs are currently used to treat various types of cancer, the current response rates and long-term effects of ICIs are far from ideal. Only a small fraction of patients treated with ICIs respond to them: ICI monotherapies are currently estimated to benefit less than 13% of cancer patients [3], only a minority of patients gain life-altering durable survival [4], and a portion of the non-responders end up developing progressive, refractory disease [5]. In part, this lack of efficacy reflects the effect of the tumor microenvironment (TME), which is being increasingly recognized as a promising target in cancer therapy. The TME includes multiple elements, such as immune components, blood vessels, the extracellular matrix, signaling molecules, and metabolites [2]. Factors such as dysfunctional vasculature, hypoxia, and altered metabolism are known to induce immunosuppression and impede the antitumor activity of immune cells. Various strategies thus have been developed to alter the TME and increase the efficacy of ICIs [6,7].
Targeting tumor cell metabolism is a newly emerging strategy for TME modulation, and related research has been gaining traction in the last few years [8]. One of the metabolic pathways receiving the most attention is glycolysis and subsequent lactate production. Tumor cells exhibit the Warburg effect, wherein an abnormally high rate of glycolysis occurs under aerobic conditions, resulting in a high glucose uptake and excessive excretion of lactate into the TME [2,9]. The neighboring immune cells are exposed to high lactate levels in the absence of nutrients; this creates an immunosuppressive niche that favors tumor growth over antitumor immune surveillance [2,8].
A possible method for reducing tumor lactate production is to target the lactate-producing enzyme, lactate dehydrogenase A (LDHA). It has been reported that an inducible mouse model of LDHA deletion (a tamoxifen-regulated cre-recombinase conditional mouse) exhibits inhibition of tumorigenesis and tumor progression [10]. A previous study reported that model animals receiving LDHA-knockdown tumor cells exhibited reduced tumor growth and extended tumor-free survival compared to those receiving wild-type tumor cells [11]. Lactate-induced derangements of T cell activity are considered to contribute to ICI resistance, and high serum lactate dehydrogenase levels were shown to correlate with primary resistance and poor outcomes under blockade of CTLA-4 and PD-1 [5]. Therefore, the long-term inhibition of LDHA expression may be an effective strategy to alter tumor cell metabolism for tumor therapy, especially when used in conjunction with ICI therapy.
The clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR associated protein 9 (CRISPR/Cas9) system is a gene editing tool that enables the easy and efficient editing of desired genes [12,13]. CRISPR/Cas9 is a relatively simple two-component system consisting of a single guide RNA (sgRNA), which is a short RNA sequence that guides Cas9 to the target sequence, and Cas9, which acts as an endonuclease to cleave the target sequence [14]. Since CRISPR/Cas9 can be utilized to freely edit select gene sequences in the genome, is has been proposed that CRISPR/Cas9-mediated editing of the LDHA-encoding sequence could be an effective method to modulate lactate in the TME for cancer therapy. However, despite the high expectations for gene-targeted therapeutics, delivery remains a hurdle in the clinical translation of CRISPR/Cas9 [15]. A delivery system capable of effectively delivering CRISPR/Cas9 in vivo must be developed if we hope to achieve LDHA gene editing for tumor therapy.
Here, we designed a lipid nanoparticle formulation optimized for the transfection of plasmid DNA co-encoding CRISPR/Cas9 and an sgRNA specific for LDHA (sgLDHA). This lipid nanoparticle formulation was used for the in vivo delivery of LDHA-targeting CRISPR/Cas9 plasmid DNA to a tumor mouse model, in combination with anti-PD-L1 antibody (PD-L1 Ab) therapy. We report that the lipid nanoparticle-mediated editing of the LDHA gene in tumor cells activates T cells and provides synergistic antitumor effects in combination with PD-L1 Ab.
2. Materials and methods
2.1. Materials
1,2-dioleoyl-3-trimethylammonium-propane (DOTAP), 1,2-dioleoyl-sn‑glycero-3-ethylphosphocholine (EPC), cholesterol, 3β-[N-(N',N'-dimethylaminoethane)-carbamoyl]cholesterol (DC—Chol), 1,2-dioleoyl-sn‑glycero-3-phosphoethanolamine (DOPE), Cy5-conjugated DOPE and 1,2-diphytanoyl-sn‑glycero-3-phosphoethanolamine (DPhPE) were purchased from Avanti Polar Lipids (Alabaster, AL, USA). 4-(2-hydroxyethyl)−1-piperazineethanesulfonic acid (HEPES), dimethyl sulfoxide, (3-(4,5-dimethylthizol-2-yl)−2,5-diphenyltetrazolium bromide (MTT) and collagenase were purchased from Sigma-Aldrich (St. Louis, MO, USA). Cas9 expression plasmid pSpCas9 (BB) was purchased from Addgene (Watertown, MA, USA). DH5α competent cells ECOS™ 101 was purchased from Yeastern Biotech. Plasmid Miniprep kit was purchased from Qiagen (Hilden, Germany). Murine B16F10 cell line was purchased from Korean Cell Line Bank (Seoul, South Korea). HeLa-GFP cells were purchased from GenTarget Inc., (San Diego, CA, USA). Dulbecco's modified Eagle's medium (DMEM) was purchased from Welgene (Gyeongsan, South Korea). Fetal bovine serum (FBS) was purchased from GenDEPOT (Katy, TX, USA). Penicillin-streptomycin was purchased from Capricorn Scientific (Ebsdorfergrund, Germany). 24-well plate was purchased from SPL Life Sciences (Pocheon, South Korea). Radioimmunoprecipitation assay lysis buffer was purchased from Rockland (Limerick, PA, USA). Roche cOmplete, Mini, EDTA-free Protease Inhibitor Tablet and polyvinylidene fluoride membrane Immobilon -PSQ transfer membrane were purchased from MilliporeSigma (Burlington, MA, USA). Anti-β-actin mouse, anti-GFP rabbit and horseradish peroxidase (HRP)-conjugated secondary antibody were purchased from Cell Signaling Technology (Danvers, MA, USA). The Amersham ECL Western Blotting detection reagent was purchased from GE Healthcare (Chicago, IL, USA). QuickExtract DNA Extraction Solution was purchased from Lucigen (Middleton, WI, USA). T7 endonuclease I was purchased from New England Biolabs (Ipswich, MA, USA). Anti-CD3 antibody, anti-CD28 antibody and anti-PD-L1 antibody were purchased from Bio X Cell (Lebanon, NH, USA). FITC or APC-conjugated anti-CD3 antibody, PerCP-Cy5.5-conjugated anti-CD8 antibody, PE-conjugated anti-IFN-γ antibody and APC-conjugated anti-granzyme B antibody were purchased from BioLegend (San Diego, CA, USA). Five-week-old C57BL/6 mice were purchased from Raon Bio (Yongin, South Korea). 5-(and-6)-Carboxy SNARF™−1 from Thermo-Fisher Scientific (Waltham, MA, USA).
2.2. Preparation of DNA-loaded lipid nanoparticles
Lipid nanoparticles were formed using a thin-film hydration method. Permanently cationic lipids (DOTAP or EPC), cholesterol (or cholesterol derivative DC—Chol), and fusogenic lipids (DOPE or DPhPE) were dissolved in chloroform at the indicated molar ratios to generate eight different formulations (Fig. 2). The samples were vortexed and the chloroform solvent was evaporated using a rotary evaporator (EYELA N-1200A; Tokyo Rikakikai Co. Ltd., Tokyo, Japan), leaving thin lipid films. The films were hydrated with 20 mM HEPES buffer (pH 5.2), then vortexed thoroughly. The suspensions were sonicated for 10 min and centrifuged, and the lipid nanoparticles were collected as the supernatant. Plasmid DNA was loaded onto the lipid nanoparticle surfaces by thorough mixing, which allowed complexation to occur via charge-charge interaction. The plasmid DNA was added to the lipid nanoparticles at an N/P ratio of 1.85 for formulations No. 1 to 4 and 2.44 for formulations No. 5 to 8, and all mixtures were incubated for 10 min at room temperature to allow complexation.
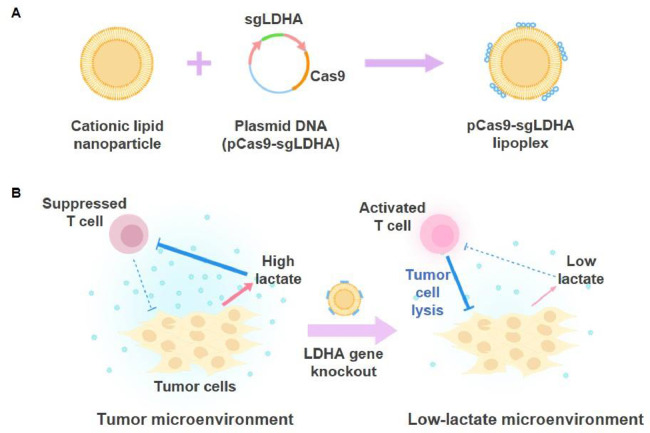
Fig. 1.
Tumor cell gene editing by pCas9-sgLDHA/F3 and activation of T cells. (A) Construction of the pCas9-sgLDHA lipoplex is illustrated. For pCas9-sgLDHA, the DNA sequence encoding sgLDHA was cloned into pCas9 plasmid DNA. pCas9-sgLDHA plasmid DNA was complexed to the cationic lipid nanoparticle to yield pCas9-sgLDHA lipoplexes. (B) Gene editing of tumor cells by pCas9-sgLDHA/F3 may cause LDHA gene knockout in tumor cells, leading to decreased lactate production. Reduced lactate in the tumor microenvironment decreases T cell immune suppression to increase T cell antitumor activity and inhibit tumor growth.
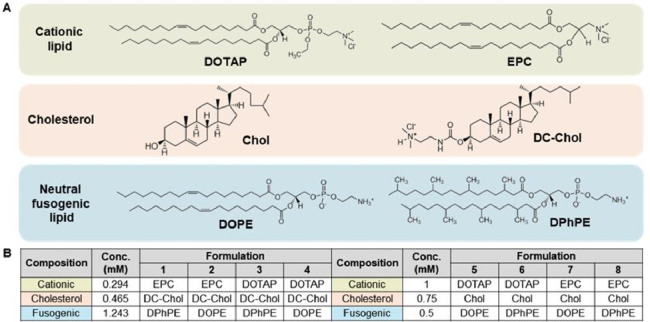
Fig. 2.
Lipid compositions of the lipid nanoparticle formulations. (A) The lipids used to generate the lipid nanoparticles. The following were combined: DOTAP or EPC as a cationic lipid; cholesterol or the cholesterol derivative, DC—Chol; and DOPE or DPhPE as a neutral fusogenic lipid. (B) Lipid nanoparticle formulations. Each lipid nanoparticle formulation was generated by combining a cationic lipid (DOTAP or EPC), cholesterol or DC—Chol, and a neutral fusogenic lipid (DOPE or DPhPE) at a certain molar ratio. A total of eight formulations were designed.
2.3. Characterization and complexation study of lipoplexes
Lipid nanoparticles were characterized by their morphology, size and surface charge. Morphology was observed by transmission electron microscopy (TEM) using a JEM-2100 F (JEOL, Tokyo, Japan). Size was evaluated using an ELSZ-1000 zeta potential and particle size analyzer (Otsuka Electronics Co., Ltd., Osaka, Japan), based on the dynamic light-scattering method. Zeta potential was also evaluated using the ELSZ-1000 analyzer, based on laser Doppler microelectrophoresis. The complexation of lipid nanoparticles and plasmids was analyzed by gel retardation assay. Lipid nanoparticles composed of 0.294 mM DOTAP, 0.465 mM DC—Chol, and 1.243 mM DPhPE were diluted with 20 mM HEPES buffer. Plasmid DNA was complexed to the lipid nanoparticles at N/P ratios ranging from 0.06 to 1.85. Retardation of plasmid DNA in lipoplexes was evaluated by 1% agarose gel electrophoresis followed by visualization using a Gel Doc XR+ Gel Imaging system (Bio-Rad Laboratories Inc., Hercules, CA, USA). Physical stability of F3 or the lipoplexes was evaluated by measuring particle size. F3 nanoparticles or the lipoplexes of pGFP/F3 were dispersed in 20 mM phosphate-buffered saline supplemented with 10% FBS, and stored at 4 °C.
2.4. Cell culture
Murine B16F10 cells and human HeLa-GFP cells were cultured in a humidified atmosphere of 5% CO2/95% air at 37 °C. Both cell lines were cultured in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin.
2.5. Cell viability test
The viability of nanoparticle-treated cells was assessed through an MTT assay [16]. HeLa-GFP cells were seeded to a 24-well plate (5 × 105 cells/well) and incubated for 24 h. The cells were then treated with nanoparticle samples for 4 h. The medium was subsequently switched to fresh DMEM, and the cells were incubated for 24 h. The cells were then treated with MTT (250 µg/ml) for 1 h at 37 °C. The intracellular formazan crystals were dissolved using dimethyl sulfoxide and absorbance at 570 nm was measured using a Sunrise absorbance microplate reader (Tecan, Männedorf, Switzerland). Cell viabilities for each group were calculated by normalizing the absorbance of the untreated group to 100%.
2.6. Construction and amplification of plasmid DNA encoding Cas9 and sgRNA
The plasmid DNA encoding Cas9 and sgRNA was constructed as previously reported [17]. Briefly, sgRNA oligos specific for the sequences of green fluorescence protein (GFP) and LDHA were phosphorylated and annealed in a thermocycler (MyCycler thermal cycler; Bio-Rad), then cloned into pSpCas9 (BB) to generate plasmid DNA co-encoding Cas9 and sgGFP (pCas9-sgGFP), and Cas9 and sgLDHA (pCas9-sgLDHA). In some experiments, plasmid DNA encoding only Cas9 (pCas9) or Cas9 with a scrambled sgRNA sequence (pCas9-sgScr) were used as controls. pCas9-sgGFP and pCas9-sgLDHA were amplified using ECOS™ 101 and extracted using a plasmid Miniprep kit. The proper insertion of sgRNA sequences in the plasmid DNA was verified by Sanger sequencing (Macrogen, Seoul, Republic of Korea). Table S2 shows information on the maps and sizes of plasmid DNA constructs used in the study.
2.7. Evaluation of genome editing in GFP-expressing cells
The in vitro genome editing of plasmid DNA in lipoplexes was evaluated in Hela-GFP cells using flow cytometry, fluorescence microscopy, and Western blot analysis. HeLa-GFP cells were seeded to 24-well plates at 1 × 105 cells/well, and incubated at 37 °C for 24 h to reach ∼70% cell confluency. Each well was treated with lipoplexes for 4 h, and the medium was replaced with fresh DMEM containing 10% FBS and 1% penicillin-streptomycin. After 72 h, the cells were harvested and the intensity of GFP fluorescence was analyzed by flow cytometry (FACSCalibur Flow cytometer; BD Biosciences, Franklin Lakes, NJ, USA). For fluorescence microscopy, cells were fixed with 4% formaldehyde in PBS for 10 min and stained with DAPI. Cellular fluorescence was observed using a confocal laser-scanning microscope (Leica, Wetzlar, Germany). For Western blot analysis, cellular proteins were extracted using radioimmunoprecipitation assay lysis buffer containing a dissolved Roche cOmplete, Mini, EDTA-free Protease Inhibitor Tablet. The protein samples were resolved by 10% SDS-polyacrylamide gel electrophoresis, then transferred onto a polyvinylidene fluoride membrane. The membranes were incubated at 4 °C overnight with anti-β-actin mouse and anti-GFP rabbit primary antibody solutions. Then, the membranes were incubated at room temperature for 2 h with horseradish peroxidase (HRP)-conjugated secondary antibody solutions. The targeted proteins were visualized using a ImageQuant LAS 4000 biomolecular imager (GE Healthcare, Chicago, IL, USA) and the Amersham ECL Western Blotting detection reagent.
2.8. Evaluation of in vitro genome editing by T7E1 assay
In vitro genome editing by lipoplexed plasmid DNA was also assessed using a T7E1 assay. Cultured Hela-GFP or B16F10 cells were treated with lipoplexed pCas9-sgGFP or pCas9-sgLDHA. Genomic DNA was extracted from the cells using the QuickExtract DNA Extraction Solution. Sequences of GFP, LDHA or potential off-targets were amplified by PCR using the primers (Table S1). Next, the PCR products were annealed with a thermocycler (MyCycler thermal cycler; Bio-Rad), mixed with 1 µl of T7 endonuclease I, and incubated at 37 °C for 15 min. The cleavage reaction was stopped by adding 1.5 µl of 0.25 M EDTA. Finally, the digested DNA was analyzed by 2% agarose gel electrophoresis.
2.9. Cytokine analysis of T cells cultured in LDHA-edited cancer cell medium
The effect of the LDHA-edited microenvironment on T cell antitumor activity was assessed. T cells were cultured with the conditioned media of LDHA-edited cancer cells, and cytokine levels were measured. Cultured B16F10 cells were transfected with pCas9-sgLDHA in lipoplexes for 4 h, and incubated in DMEM for 48 h, and the medium was collected. Meanwhile, T cells were isolated from the spleens of C57BL/6 mice using nylon wool columns, and then activated for 48 h in a 24-well plate coated with anti-CD3 and CD28 antibodies (anti-CD3 Ab: 10 µg/ml, anti-CD28 Ab: 2 µg/ml). The activated T cells were incubated in the LDHA-edited cancer cell-conditioned medium for 48 h, stained with anti-CD3, anti-interferon gamma (IFN-γ), and anti-granzyme B antibodies, and analyzed by flow cytometry (FACSCalibur Flow cytometer; BD Biosciences). The data from the cytokine-positive T cell populations were further evaluated using the FlowJo software (BD Biosciences).
2.10. In vivo antitumor efficacy
The in vivo antitumor efficacy of pCas9-LDHA in lipoplexes was evaluated by monitoring B16F10 tumor growth and survival rates. Five-week-old C57BL/6 mice were maintained under standard pathogen-free conditions at the Animal Center for Pharmaceutical Research, Seoul National University. B16F10 cells (5 × 105) were subcutaneously inoculated into the right flanks of 5-week-old C57BL/6 mice. For in vivo disruption of LDHA, pCas9-sgLDHA-loaded lipid nanoparticles were injected intratumorally at a dose of 5 µg plasmid DNA/mouse on Day 7, 9 and 11. Anti-PD-L1 antibodies were injected intratumorally at a dose of 100 µg/mouse on days 13, 15, and 17. Tumor sizes were measured in two dimensions using a slide caliper every 2 d Tumor volumes were calculated as a × b × b × 0.5, a and b being the lengths of the largest and smallest dimensions, respectively [18]. Survival rates were monitored for 52 d
2.11. Measurement of in vivo tumor extracellular pH
Tumor extracellular pH was measured using 5-(and-6)-Carboxy SNARF™−1, a cell impermeant dye with pH-responsive fluorescence intensities. Following intratumoral administration of the fluorescent dye in 5% glucose, the fluorescence intensity was measured at 580 and 640 nm emission wavelengths (excitation wavelength: 500 nm) using an IVIS Spectrum In Vivo Imaging System (PerkinElmer, Waltham, MA, USA). Standard curve was generated by the fluorescence intensity ratio at 580 nm and 640 nm emission wavelength at various pH conditions [19].
2.12. In vivo antitumor immune response
The in vivo antitumor immune response was evaluated by cytotoxic T cell infiltration, IFN-γ and granzyme B expression level. One day after the last injection of anti-PD-L1, tumors were extracted and digested with serum-free DMEM supplemented with 1 mg/ml collagenase for single cell isolation. The single cell was collected by centrifugation at 10 000× g for 3 min and stained for flow cytometry. The population of cytotoxic T cell was analyzed by staining with FITC-conjugated anti-CD3 antibody and PerCP-Cy5.5-conjugated anti-CD8 antibody. The population of IFN-γ expressing T cells was evaluated with APC-conjugated anti-CD3 antibody and PE-conjugated anti-IFN-γ antibody. The population of granzyme B expressing T cells was measured with FITC-conjugated anti-CD3 antibody and APC-conjugated anti-granzyme B antibody. Flow cytometry was used to analyze stained cell populations.
2.13. In vivo tumor retention and cellular uptake of pCas9-sgLDHA/F3
Tumor retention of pCas9-LDHA/F3 was evaluated using Cy5-labeled F3. The Cy5-labeled F3 was prepared by adding 0.02 mM of Cy5-conjugated DOPE (18:1 Cy5 PE) during lipid nanoparticle preparation. pCas9-sgLDHA was loaded to the fluorescent lipid nanoparticles and injected intratumorally at a dose of 5 µg plasmid DNA/mouse. The retention of nanoparticle was measured with IVIS Spectrum In Vivo Imaging System for 48 h. Cellular uptake of pCas9-LDHA/F3 was also measured by using the Cy5-labeled F3. For uptake test, T cells were isolated from murine spleen using nylon wool columns [18]. Macrophage and neutrophil were differentiated from murine bone marrow [20, 21]. pCas9-LDHA loaded fluorescent F3 was treated to T cell, neutrophil, macrophage and B16F10 for 4 h and cellular uptake was measured by flow cytometry (FACSCalibur Flow cytometer).
2.14. In vivo safety study
The safety of pCas9-LDHA/F3 was evaluated by histological staining and biochemical parameters. One day after the last injection of pCas9-LDHA/F3, major organs were extracted and stained with hematoxylin and eosin. Blood samples were collected, and biochemical parameters including alanine aminotransferase (ALT), aspartate transaminase (AST), and blood urea nitrogen (BUN) were measured using a clinical chemical analyzer (DRICHEM 2500s; Fujifilm, Tokyo, Japan).
2.15. Statistics
Results are presented as the mean ± standard error of the mean (SEM). Statistical significance was determined by one-way analysis of variance (ANOVA) with the Student−Newman−Keuls post-hoc test. Statistical analyses were conducted with the SigmaStat software (version 12.0; Systat Software, Richmond, CA, USA). For comparison of survival rate differences, log-rank test was used in the Kaplan-Meier plots using GraphPad Prism 8 (GraphPad Software, San Diego, CA, USA). Statistically significant differences are symbolized as ***P < 0.001, **P < 0.01, and *P < 0.05.
3. Results and discussion
3.1. Formulation screening of lipid nanoparticles for plasmid DNA delivery
The lipid nanoparticle formulations comprised complexes of three components: a cationic lipid, cholesterol or a cholesterol derivative, and a neutral fusogenic lipid. As a cationic lipid, either DOTAP or EPC was used. As a neutral fusogenic lipid, DOPE or DPhPE was used. Eight different lipid nanoparticle formulations were prepared representing different combinations of cationic lipid, cholesterol (or its derivative), and fusogenic lipid (Fig. 2).
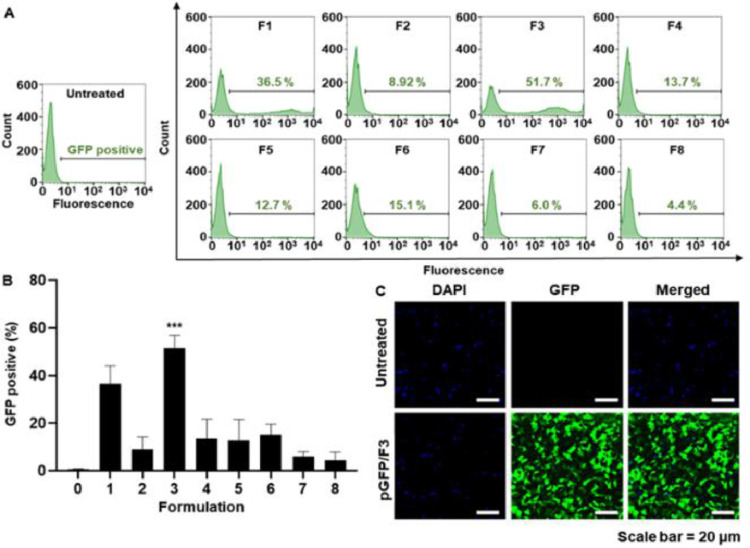
The plasmid DNA delivery efficiencies of the different lipid nanoparticle formulations were affected by the lipid compositions. Among the eight lipid nanoparticles tested, the formulation composed of DOTAP, DC—Chol, and DPhPE provided the greatest delivery efficiency of plasmid DNA encoding GFP and yielded the highest percentage of GFP-positive cells at 51.7% ± 5.2% (Fig. 3A&3B). Consistent with the results of our flow cytometric analysis, confocal microscopy showed that GFP expression was highest for cells treated with lipoplexes composed of DOTAP, DC—Chol, and DPhPE (Fig. 3C). We designated this lipid nanoparticle formulation as F3, and used it for further studies.
Fig. 3.
Transfection efficiency of the various lipid nanoparticle formulations. (A,B) Flow cytometry results showing GFP fluorescence intensities of B16F10 cells transfected with pGFP-loaded lipid nanoparticle formulations 1–8. (A) Histograms showing the percentage of GFP-positive cells. (B) Mean percentage of GFP-positive cells for each group (n = 3 per group). (C) Fluorescence microscopy images showing B16F10 cells that were either untreated or transfected with pGFP-loaded lipid nanoparticles (F3). ***P < 0.001.
3.2. Characterization of lipid nanoparticles and lipoplexes
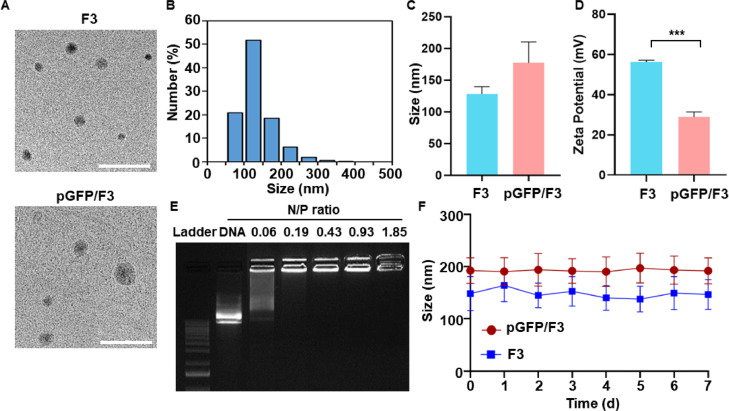
The physical characteristics of F3 were analyzed in the following categories: morphology, particle size, zeta potential, DNA complexation and stability. TEM revealed spherical shapes for both F3 and pGFP/F3 (Fig. 4A). Size analysis revealed that F3 lipid nanoparticles showed a homogeneous size distribution (Fig. 4B). The mean sizes of plain F3 were 128.0 ± 11.7 nm, and this parameter increased to 177.1 ± 33.2 nm after complexation with pGFP plasmid DNA (Fig. 4C). The zeta potentials significantly decreased after complexation with pGFP plasmid DNA (Fig. 4D). Gel retardation assay revealed that the plasmid DNA showed complete complexation to F3 lipid nanoparticles at N/P ratios of 0.19 and above (Fig. 4E). The sizes of either F3 nanoparticles or pGFP/F3 lipoplexes did not significantly change upon storage at 4 °C (Fig. 4F)
Fig. 4.
Characterization of lipid nanoparticles and lipoplexes. (A) Morphology of F3 lipid nanoparticles and pGFP/F3 lipoplex was observed by TEM. Scale bar: 500 nm. (B) The particle size distribution of F3 lipid nanoparticles were measured by dynamic light scattering. (C) Mean sizes of plain F3 lipid nanoparticles and those complexed with pGFP plasmid DNA (n = 3 per group). (D) Zeta potentials were measured by laser Doppler microelectrophoresis (n = 3 per group). (E) F3 lipid nanoparticles were complexed with pGFP plasmid DNA at various N/P ratios and electrophoresed on a 1% agarose gel. (F) The sizes of F3 or pGFP/F3 were monitored by dynamic light scattering over 7 d (n = 3 per group). ***P < 0.001.
3.3. Cas9-mediated GFP gene editing
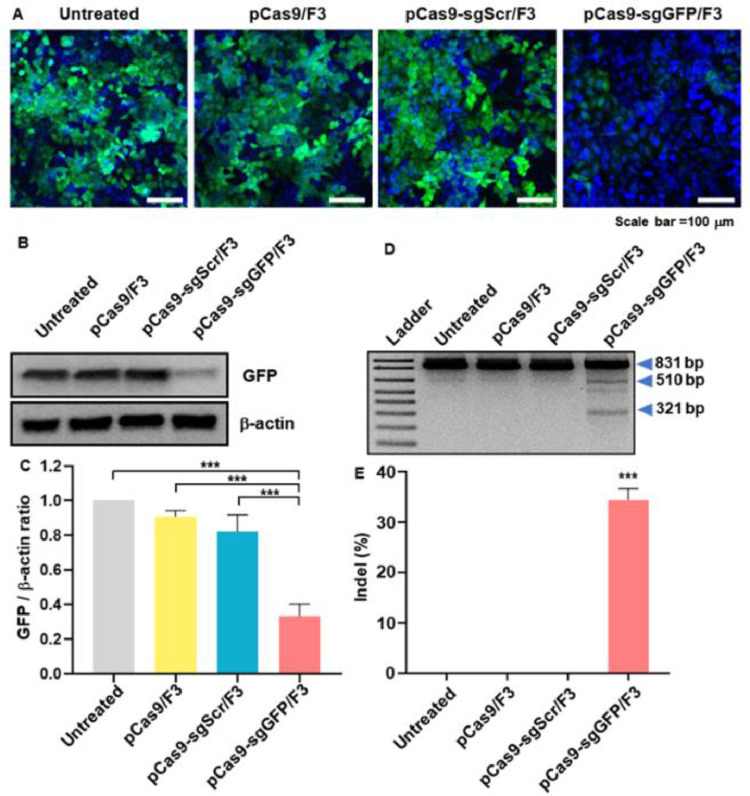
The ability of F3 lipid nanoparticles to efficiently deliver the pCas9-sgGFP plasmid DNA was evaluated in GFP-expressing HeLa-GFP cells. Treatment of HeLa-GFP cells with pCas9/F3 lipoplex or pCas9-sgScr did not significantly decrease the fluorescence of GFP, whereas treatment with pCas9-sgGFP/F3 reduced the expression of GFP (Fig. 5A). Consistent with the fluorescence microscopy images, Western blot analysis showed that the GFP protein level was decreased in the group treated with pCas9-sgGFP/F3 compared to the other groups (Fig. 5B). Density analysis of Western blot images revealed that there was a 3.0-fold lower GFP/β-actin ratio in the pCas9-sgGFP/F3-treated group compared with the untreated group (Fig. 5C). Gene editing of GFP was observed in genomic level. In T7E1 assay, pCas9-sgGFP/F3-treated group showed distinct cleavage of GFP gene due to the GFP -specific gene editing (Fig. 5D). The indel frequency of the group treated with pCas9-sgGFP/F3 was 31.3% ± 2.2%, significantly higher than those of other groups (Fig. 5E).
Fig. 5.
In vitro gene disruption of GFP by pCas9-sgGFP/F3. HeLa-GFP cells were treated with lipid nanoparticles loaded with various plasmids (pCas9, pCas9-sgScr, pCas9-sgRNA), and evaluated for decreased GFP expression. (A) Fluorescence microscopy images showing GFP (green) and DAPI (blue) fluorescence. Scale bar: 100 µm. (B) Western blot image showing GFP expression levels. (C) Intensity ratios of GFP signals and β-actin signals in the Western blot image (n = 3 per group). (D) Gene editing of GFP was tested by T7E1 assay. (E) Indel frequency of GFP gene editing was calculated by the T7E1 assay. *** P < 0.001.
3.4. Cytotoxicity
F3 lipoplexes did not induce significant cytotoxicity. The treated HeLa-GFP cells retained similar levels of viability regardless of the plasmid DNA applied, with the pCas9/F3-, pCas9-sgScr/F3-, and pCas9-sgGFP/F3-treated groups exhibiting viabilities of 85.3% ± 5.8%, 84.0% ± 2.6% and 85.7% ± 9.9% respectively, relative to the untreated group (Fig. 6).
Fig. 6.
Cytotoxicity of various F3 lipoplexes. HeLa-GFP cells were treated with various lipoplexes for 4 h and cell viability was tested by MTT assay (n = 3 per group). n.s.: not significant.
3.5. LDHA gene-editing of cancer cells using pCas9-sgLDHA/F3
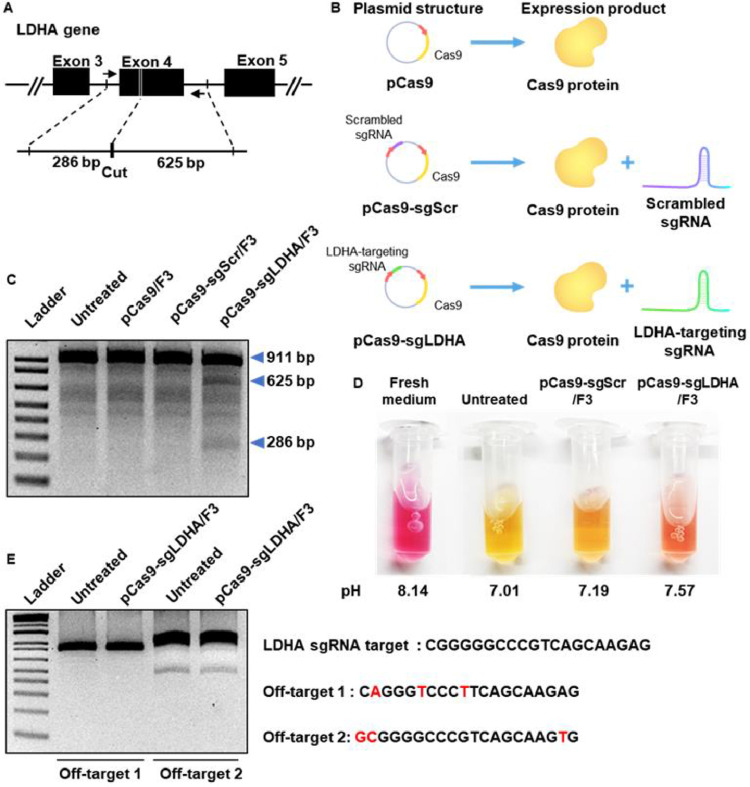
The gene-editing efficiency of F3 lipoplexes was further tested with LDHA as a target gene. As illustrated in Fig. 7A, sgLDHA was designed to cut exon 4 of the LDHA gene, resulting in fragments of 286 and 625 base pairs (bp). The construction and expression products of pCas9, pCas9-sgScr, and pCas9-sgLDHA are illustrated (Fig. 7B). T7E1 assay showed that the LDHA gene was edited to two fragments in the group treated with pCas9-sgLDHA/F3, but not in those treated with pCas9/F3 or pCas9-sgScr/F3 (Fig. 7C).
Fig. 7.
In vitro LDHA gene-editing of cancer cells by pCas9-sgLDHA/F3. (A) sgLDHA was designed to edit exon 4 of the LDHA gene. (B) The construction schemes of various plasmid DNA and expression products are illustrated. (C) B16F10 cells were treated with various lipoplexes, and genomic DNA was extracted for T7E1 assay. (D) B16F10 cells were treated with lipoplexes for 48 h, and the colors and pH values of culture media were assessed. (E) pCas9-sgLDHA/F3 mediated off-target gene editing was confirmed by T7E1 assay. Predicted top 2 potential off target sequence was suggested by Wellcome Sanger Institute Genome Editing (https://wge.stemcell.sanger.ac.uk//).
Since the editing of LDHA was expected to decrease the secretion of lactate from tumor cells, the color of the culture medium was compared. The fresh medium had the reddest color, indicating the highest pH, while the conditioned medium of untreated cells was the yellowest, indicating the lowest pH (Fig. 7D). The conditioned medium of B16F10 cells treated with pCas9-sgLDHA/F3 was reddish orange in color, and direct pH measurement revealed that its pH was higher than that of the cell culture medium conditioned by the untreated group. While pCas9-sgLDHA/F3 treated group showed the LDHA gene editing effect, there was no significant off-target effect compared to untreated group (Fig. 7E).
3.6. Modulation of T cell activity by LDHA gene-edited cancer cells
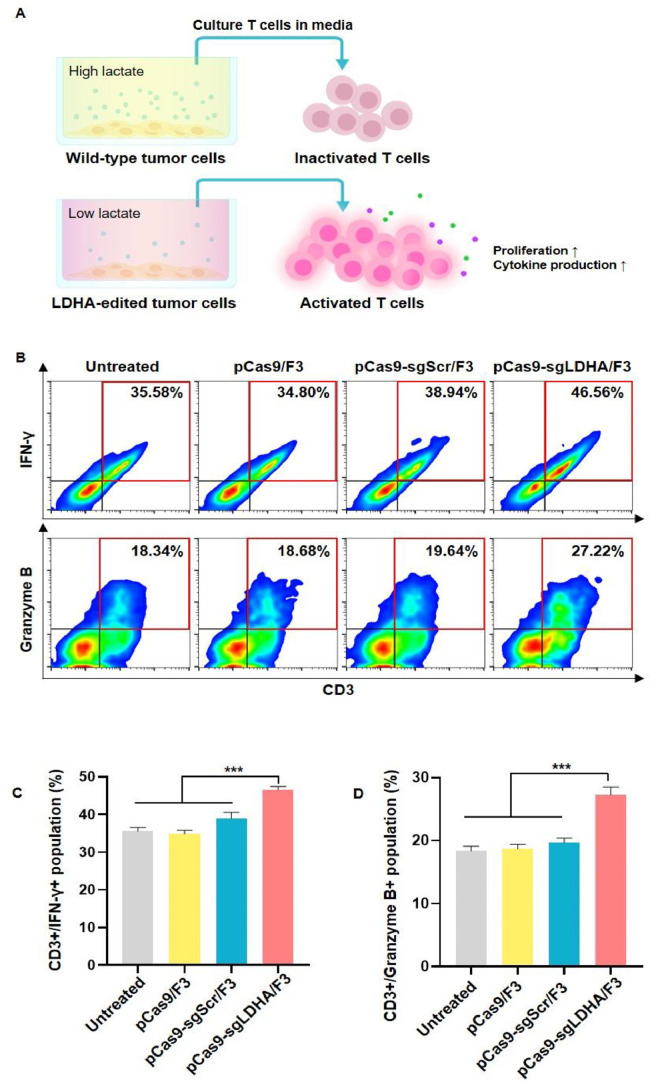
We hypothesized that the activity of T cells can be modulated by the level of lactate in the microenvironment, and that the down-regulation of lactate by LDHA gene editing of cancer cells may activate the function of T cells (Fig. 8A). To test this hypothesis in vitro, we treated T cells with the conditioned media of tumor cells pretreated with various lipoplexes and analyzed IFN-γ and granzyme B, which are major cytokines related to T cell antitumor activity, as T cell activity markers. Flow cytometry revealed that the T cells exposed to medium conditioned by tumor cells treated with pCas9-sgLDHA/F3 showed the highest positivity for IFN-γ and granzyme B (Fig. 8B). In the pCas9-sgLDHA/F3 group, the mean CD3+/IFN-γ+ population was 46.5% ± 0.8%, which was the highest among all of the groups (Fig. 8C). The CD3+/granzyme B+ population was also significantly higher in the pCas9-sgLDHA/F3 group compared to other groups (Fig. 8D).
Fig. 8.
Activity of T cells after culture with LDHA gene-edited culture media. (A) Tumor cells treated with pCas9-sgLDHA/F3 are expected to produce less lactate due to LDHA gene editing, and the lower lactate level is expected to result in greater activation of T cells exposed to conditioned medium of pCas9-sgLDHA/F3-treated cells compared to that of untreated tumor cells. B16F10 cells were treated with various lipoplexes and T cells were incubated in their conditioned media. (B) The percentages of T cells positive for IFN-γ or granzyme B were analyzed by flow cytometry. (C, D) T cell populations positive for IFN-γ (C) and granzyme B (D) were compared (n = 5 per group). ***P < 0.001.
3.7. In vivo antitumor effect of LDHA gene editing
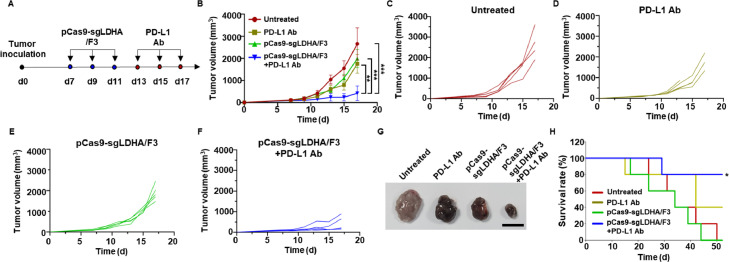
In a B16F10 tumor model in vivo, LDHA gene editing by pCas9-sgLDHA/F3 suppressed tumor growth. Notably, a synergistic antitumor effect was observed when this formulation was combined with an anti-PD-L1 antibody (PD-L1 Ab). B16F10 tumor-bearing mice were treated three times with pCas9-sgLDHA/F3, and then three times with PD-L1 Ab. The treatment scheme is depicted in Fig. 9A.
Fig. 9.
In vivo antitumor effects of pCas9-sgLDHA/F3. (A) Tumor inoculation and dosing schedules are illustrated. Mice bearing B16F10 tumors were injected intratumorally with pCas9-sgLDHA/F3 on Day 7, 9 and 11, and with PD-L1 Ab on Day 13, 15 and 17. (B) Mean tumor volumes of various groups were measured (n = 5 per group). (C-F) Tumor volume changes of each mouse are presented for the untreated group (C), the PD-L1 Ab-treated group (D), the pCas9-sgLDHA/F3-treated group (E) and the PD-L1 Ab plus pCas9-sgLDHA/F3-treated group (F). (G) Tumor tissues of various groups were dissected at Day 17 after tumor inoculation. Scale bar: 1 cm. (H) Survival of mice was monitored after tumor inoculation at day 0. *P < 0.05; **P < 0.01; ***P < 0.001.
The application of PD-L1 Ab or pCas9-sgLDHA/F3 alone reduced tumor growth, but the combination of PD-L1 Ab plus pCas9-sgLDHA/F3 yielded a significantly higher antitumor effect (Fig. 9B). When assessed on a mouse-by-mouse basis, compared with the tumor growth of untreated mice (Fig. 9C), those treated with PD-L1 Ab (Fig. 9D) and those treated with pCas9-sgLDHA/F3 (Fig. 9E), the combination of PD-L1 Ab plus pCas9-sgLDHA/F3 (Fig. 9F) more effectively inhibited tumor growth.
Representative images of dissected tumor tissues showed the smallest size of tumors in the group treated with pCas9-sgLDHA/F3 plus PD-L1 Ab. The combination of PD-L1 Ab with pCas9-sgLDHA/F3 also prolonged the survival of mice (Fig. 9G). In untreated or pCas9-sgLDHA/F3-treated group, 100% of mice died by 52 d after tumor inoculation. The treatment with PD-L1 Ab alone provided 40% of survival at 52 d after tumor inoculation. In contrast, 80% of mice survived after combination of PD-L1 Ab with pCas9-sgLDHA/F3.
3.8. In vivo ldha gene editing-mediated modulation of tumor extracellular pH
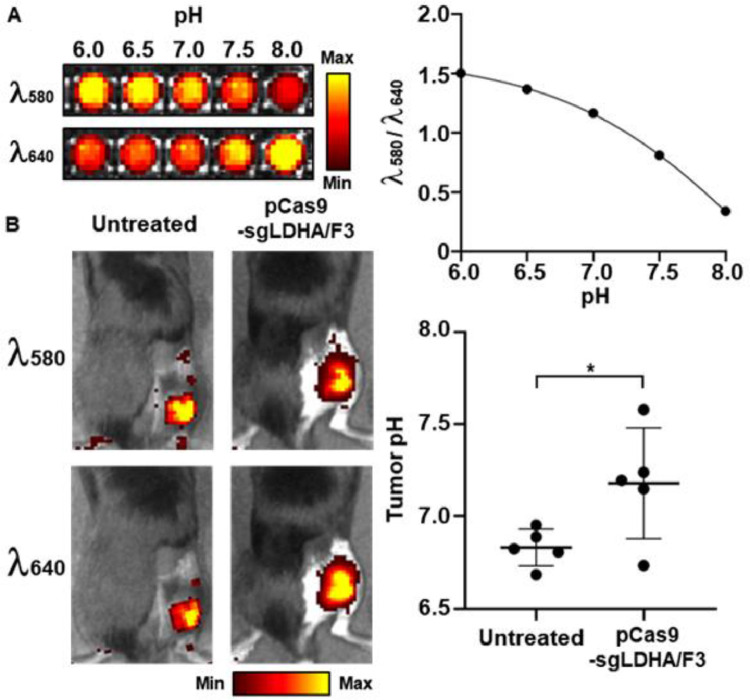
Treatment of pCas9-sgLDHA/F3 could alter the extracellular pH of tumor microenvironment. Intratumorally administered pH-reporting dye exhibited that tumor extracellular pH was significantly increased upon pCas9-sgLDHA/F3. In untreated mice, the extracellular pH values were 6.8 ± 0.1. In contrast, the pH of pCas9-sgLDHA/F3-treated tumors was 7.2 ± 0.3 (Fig. 10A&10B).
Fig. 10.
In vivo tumor pH modulation of pCas9-sgLDHA/F3. (A) Intensities of pH-indicating dye at various pH conditions were measured at 580 and 640 nm. The ratios of fluorescence intensity at 580 nm over 640 nm wavelength were plotted at various pH conditions. (B) Tumor extracellular pH was calculated following intratumoral administration of the pH indicator dye (n = 5 per group). *P < 0.05.
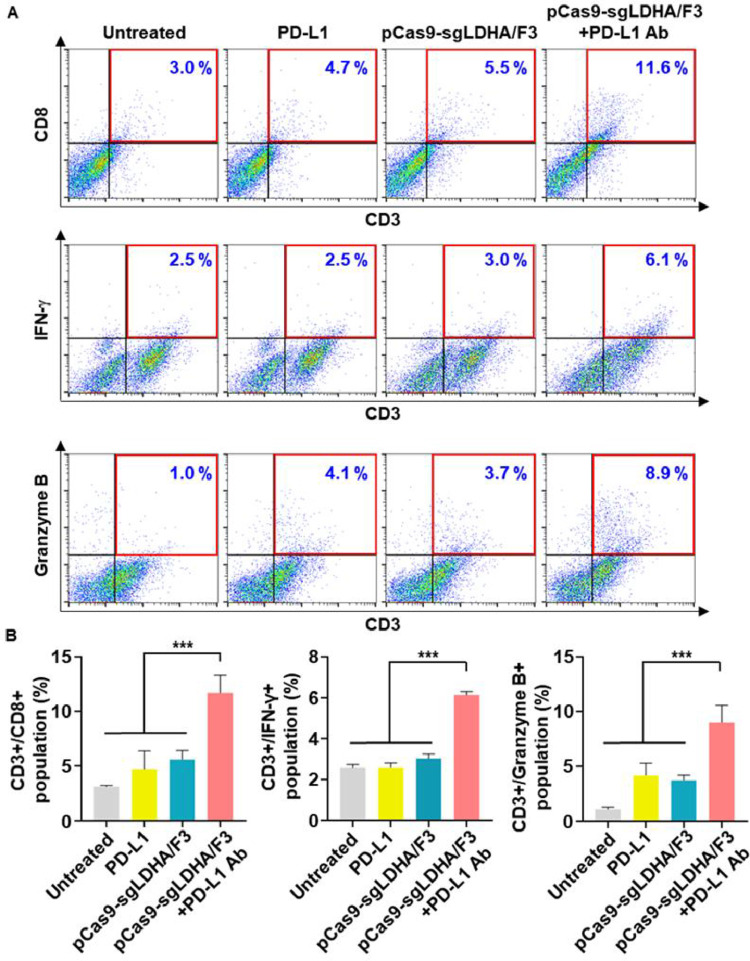
3.9. In vivo LDHA gene editing-mediated anti-tumor immune response
Combination of PD-L1 antibody and pCas9-sgLDHA/F3 showed modulation of immune cell populations in tumor microenvironment. Treatment of mice with PD-L1 antibody plus pCas9-sgLDHA/F3 increased the infiltration of cytotoxic T cells to tumor microenvironment. In tumor tissues, the population of CD3+CD8+ T cells were 3.7-fold higher following the combination of PD-L1 and pCas9-sgLDHA/F3 compared to untreated mice. Moreover, the combination of PD-L1 and pCas9-sgLDHA/F3 enhanced the activity of T cells in tumor microenvironment, showing 2.3-fold higher IFN-γ expression and 8.4-fold higher granzyme B expression compared to T cells in untreated tumor tissues.
3.10. In vivo distribution and cellular uptake of pCas9-sgLDHA/F3
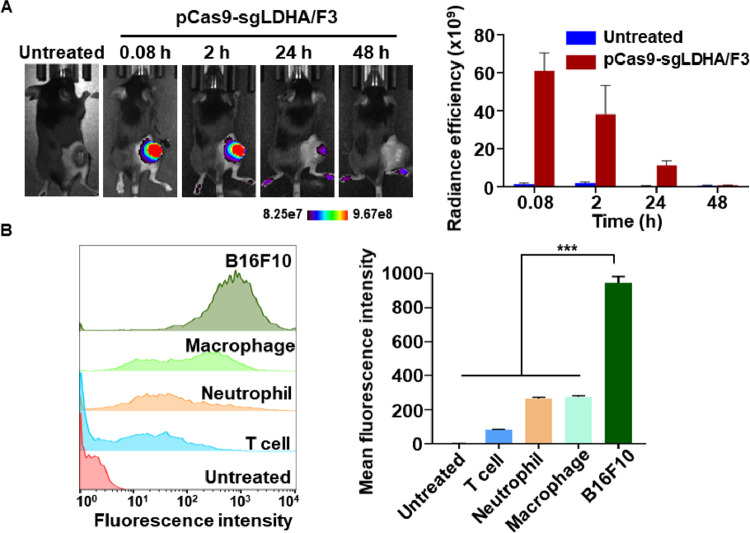
Intratumorally injected pCas9-sgLDHA/F3 showed tumor retention over 24 h. The intensity of fluorescently labeled pCas9-sgLDHA/F3 was the highest at 5 min post-dose, and gradually decreased (Fig. 12A). No signal was observed at 48 h post injection. Cellular uptake of pCas9-sgLDHA/F3 was significantly higher in B16F10 cells compared to other cells including T cells, neutrophils and macrophages. Following treatment with fluorescent Cy5 dye-labelled pCas9-sgLDHA/F3, the fluorescence intensity of B16F10 cells was 3.4-, 3.5-, and 11.3-fold greater than T cells, neutrophils and macrophages, respectively (Fig. 12B).
Fig. 12.
Tumor retention and cellular uptake of pCas9-sgLDHA/F3. (A) After intratumoral injection of Cy5 labeled pCas9-sgLDHA/F3, the nanoparticle retention was measured by molecular imaging. (B) Various cells were treated with Cy5 labeled pCas9-sgLDHA/F3 for 4 h. Cellular uptake of the nanoparticle was analyzed by flow cytometry. (n = 3 per group). ***P < 0.001.
3.11. In vivo safety of pCas9-sgLDHA/F3
Repeated injections of pCas9-sgLDHA/F3 did not induce detectable sign of toxicities. Histochemical staining did not reveal any pathophysiological changes in heart, lung, liver, spleen and kidney following treatment with pCas9-sgLDHA/F3 (Fig. 13A). In addition, the levels of ALT, AST and BUN were in the normal ranges for both untreated and pCas9-sgLDHA/F3-treated mice (Fig. 13B).
Fig. 13.
Histological staining and biochemical parameters. (A) One day after the last pCas9-sgLDHA/F3 injection, major organs were extracted and sectioned for hematoxylin and eosin staining. Scale bar: 50 µm (B) Blood was extracted and biochemical parameters were analyzed (n = 5 per group). The normal ranges were referred from Charles River Laboratories C57BL/6 mice data (https://www.criver.com/products-services/research-models- services).
3.12. Discussion
In this study, we formulated a cationic lipid nanoparticle for the delivery of plasmid DNA co-encoding Cas9 and sgRNA. We provide evidence that the gene editing of LDHA can activate T cells and exert antitumor effects, and that the combination of LDHA gene editing and PD-L1 Ab treatment yields enhanced antitumor effects.
The benefits of lipid-based nanoparticles include the ability of the composition to be flexibly adapted for the desired purpose [22]. For instance, positively charged lipids can allow nucleic acids to be loaded to a lipid nanoparticle Polyethylene glycol-modification has been reported to increase physical stability of nanoparticles during storage by conferring steric hindrance. In biological environments, surface modification with polyethylene glycol can reduce nonspecific adsorption of serum protein, and reduce clearance of nanoparticles by reticuloendothelial system [23,24]. Functional lipids, such as monophosphoryl lipid A or beta-sitosterol, can be formulated into lipid nanoparticles for immune modulation [25,26]. Recently, various ionizable lipids have been intensively developed to aid in the efficient delivery of mRNA vaccines [27]. When various lipids are included in a single formulation, it is critical to fine tune the ratio of each component to achieve the final goal.
Here, we generated lipid nanoparticles using a cationic lipid for plasmid loading, cholesterol or a cholesterol derivative for nanoparticle stabilization, and a fusogenic lipid to induce endosomal escape, and then screened the formulations for their genome editing potential. We observed that the transfection efficiency was substantially influenced by the ratio of each lipid component. We selected F3 as the final formulation, as it offered the highest transfection efficacy among the tested nanoparticles (Fig. 2). F3 formulation did not induce significant cytotoxicity when complexed with various plasmid DNA (Fig. 6).
The exact mechanism by which F3 exerted the highest transfection efficacy needs to be studied further. F3 is composed of DOTAP, DC—Chol, and DPhPE. We speculate that the combination of a fusogenic DPhPE with DC—Chol may provide higher uptake and endosomal escape of plasmid DNA. A previous study reported that DPhPE in the lipid bilayer could form an inverted cone-shape and promote endosomal escape [28], [29], [30].
We used pCas9-sgLDHA/F3 for gene editing of LDHA in tumor cells. High lactate levels in the tumor microenvironment have been found to have multiple immunosuppressive and tumor-promoting effects [2,8]. Lactic acidosis is known to induce immune cell polarization such as tumor associated macrophage or tumor associated neutrophil which have pro-tumorigenic phenotypes [31, 32]. It has been also reported that lactic acidosis can directly impairs the effector function of cytotoxic T cells by inhibiting various signal pathways associated with pro-inflammatory cytokine production [33]. Lactate-mediated acidic environment is known to promote the development of myeloid-derived suppressor cells, tolerogenic dendritic cells and regulatory T cells, which can suppress immune response in tumor tissue [5], [34], [35], [36]. pCas9-sgLDHA/F3 treatment provided significant elevation of pH in tumor by LDHA gene editing (Fig. 10). We observed that LDHA gene editing and elevation of tumor extracellular pH alone were not sufficient to provide anti-tumor immune response (Fig. 11). The T cell activity upon the combination with immune checkpoint blockade might be attributed by that LDHA gene editing-mediated pH elevation could provide favorable immune microenvironment.
Fig. 11.
In vivo antitumor immune response of pCas9-sgLDHA/F3. The percentages of T cells positive for CD8, IFN-γ or granzyme B were analyzed by flow cytometry. (B) CD3+ T cell populations positive for CD8, IFN-γ or granzyme B were analyzed (n = 5 per group). ***P < 0.001.
We observed that cancer cells in tumor microenvironments showed significantly higher uptake of pCas9-sgLDHA/F3 compared to noncancer cells. The exact mechanism why cancer cell showed significantly higher pCas9-sgLDHA/F3 uptake needs to be studied further. However, we cannot exclude the possibility that higher negative charge on cancer cell membrane compared to normal cells membrane may contribute to higher uptake of positively charged nanoparticle, as previously studied [30].
Previous studies involving the inhibition of LDHA in tumor cells involved LDHA sequence editing of tumor cells in vitro prior to inoculation [11], used LDHA knockout mice [10], or involved the administration of siRNA [37] or small-molecule inhibitors of LDHA [35]. The use of small-molecule inhibitors produced short-term effects and tended to yield weaker antitumor effects compared to studies that incorporated gene editing [35]. Using siRNA also had a transient effect and required multiple repeated dosages for longer treatment schedules [30]. Compared to transient silencing, in vivo genomic editing of the LDHA gene sequence in tumors may offer the advantage of providing sustained antitumor effects.
An interesting takeaway from the in vivo results is that the delivery of LDHA-targeting CRISPR/Cas9 exhibited a synergistic antitumor effect in combination with PD-L1 Ab therapy. The synergistic effect has stemmed from the simultaneous action on T cells, where both therapies blocked pathways of T cell immunosuppression and consequentially promoted T cell activation. This implies that combining tumor metabolic engineering with cancer immunotherapy may maximize the effects of cancer immunotherapy.
4. Conclusion
This study demonstrated that customized lipid nanoparticles could be used for the in vivo delivery of CRISPR/Cas9 for the metabolic engineering of tumor cells. Although the current study focused on targeting LDHA, the liposomal CRISPR/Cas9 delivery system is a versatile platform that can be used to target various components of the tumor microenvironment. In conclusion, the lipid nanoparticle-mediated CRISPR/Cas9 delivery platform developed in this study can be utilized for the in vivo genome editing of diverse tumor-related targets, facilitating the metabolic engineering of tumors and modification of the tumor microenvironment.
Ethical approval
All animal experiments were performed under the Guidelines for the Care and Use of Laboratory Animals of the Institute of Laboratory Animal Resources, Seoul National University (approval number, SNU-210106-4(E)).
Conflict of interest
All authors declare that they have no conflicts of interest.
Acknowledgements
This research was funded by grants from the National Research Foundation, Ministry of Science and ICT, Republic of Korea (NRF-2021R1A2B5B03002123; NRF-2018R1A5A2024425; NRF-2021K2A9A2A06044515; 2022M3E5F1017919), Ministry of Education, Republic of Korea (NRF-2021R1A6A3A01086428), and Korean Health Technology R&D Project (No. HI19C0664), Ministry of Health & Welfare, Republic of Korea.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ajps.2022.07.005.
Contributor Information
Dongyoon Kim, Email: mmamic@snu.ac.kr.
Yu-Kyoung Oh, Email: ohyk@snu.ac.kr.
Appendix. Supplementary materials
References
- 1.Le Q.V., Choi J., Oh Y.K. Nano delivery systems and cancer immunotherapy. J Pharm Investig. 2018;48:527–539. [Google Scholar]
- 2.Murciano-Goroff Y.R., Warner A.B., Wolchok J.D. The future of cancer immunotherapy: microenvironment-targeting combinations. Cell Res. 2020;30:507–519. doi: 10.1038/s41422-020-0337-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin J.D., Cabral H., Stylianopoulos T., Jain R.K. Improving cancer immunotherapy using nanomedicines: progress, opportunities and challenges. Nat Rev Clin Oncol. 2020;17:251–266. doi: 10.1038/s41571-019-0308-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hegde P.S., Chen D.S. Top 10 challenges in cancer immunotherapy. Immunity. 2020;52:17–35. doi: 10.1016/j.immuni.2019.12.011. [DOI] [PubMed] [Google Scholar]
- 5.Syn N.L., Teng M.W.L., Mok T.S.K., Soo R.A. De-novo and acquired resistance to immune checkpoint targeting. Lancet Oncol. 2017;18:e731–e741. doi: 10.1016/S1470-2045(17)30607-1. [DOI] [PubMed] [Google Scholar]
- 6.Le Q.V., Suh J., Oh Y.-.K. Nanomaterial-based modulation of tumor microenvironments for enhancing chemo/immunotherapy. AAPS J. 2019;21:64. doi: 10.1208/s12248-019-0333-y. [DOI] [PubMed] [Google Scholar]
- 7.Wu Y., Li Q., Shim G., Oh Y.K. Melanin-loaded CpG DNA hydrogel for modulation of tumor immune microenvironment. J Control Release. 2021;330:540–553. doi: 10.1016/j.jconrel.2020.12.040. [DOI] [PubMed] [Google Scholar]
- 8.Bader J.E., Voss K., Rathmell J.C. Targeting metabolism to improve the tumor microenvironment for cancer immunotherapy. Mol Cell. 2020;78:1019–1033. doi: 10.1016/j.molcel.2020.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Certo M., Tsai C.H., Pucino V., Ho P.C., Mauro C. Lactate modulation of immune responses in inflammatory versus tumour microenvironments. Nat Rev Immunol. 2020;21:151–161. doi: 10.1038/s41577-020-0406-2. [DOI] [PubMed] [Google Scholar]
- 10.Xie H., Hanai J., Ren J.G., Kats L., Burgess K., Bhargava P., et al. Targeting lactate dehydrogenase-A inhibits tumorigenesis and tumor progression in mouse models of lung cancer and impacts tumor-initiating cells. Cell Metab. 2014;19:795–809. doi: 10.1016/j.cmet.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brand A., Singer K., Koehl G.E., Kolitzus M., Schoenhammer G., Thiel A., et al. LDHA-associated lactic acid production blunts tumor immunosurveillance by T and NK cells. Cell Metab. 2016;24:657–671. doi: 10.1016/j.cmet.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 12.Adli M. The CRISPR tool kit for genome editing and beyond. Nat Commun. 2018;9:1911. doi: 10.1038/s41467-018-04252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen M., Ren Y.X., Xie Y., Lu W.L. Gene regulations and delivery vectors for treatment of cancer. J Pharm Investig. 2020;50:309–326. [Google Scholar]
- 14.Lee J., Le Q.V., Yang G., Oh Y.K. Cas9-edited immune checkpoint blockade PD-1 DNA polyaptamer hydrogel for cancer immunotherapy. Biomaterials. 2019;218 doi: 10.1016/j.biomaterials.2019.119359. [DOI] [PubMed] [Google Scholar]
- 15.Yin H., Kauffman K.J., Anderson D.G. Delivery technologies for genome editing. Nat Rev Drug Discov. 2017;16:387–399. doi: 10.1038/nrd.2016.280. [DOI] [PubMed] [Google Scholar]
- 16.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ko S., Park J.Y., Oh Y.K. A Microbial siderophore-inspired self-gelling hydrogel for noninvasive anticancer phototherapy. Cancer Res. 2019;79:6178–6189. doi: 10.1158/0008-5472.CAN-19-0975. [DOI] [PubMed] [Google Scholar]
- 18.Kim D., Wu Y., Li Q., Oh Y.K. Nanoparticle‑mediated lipid metabolic reprogramming of T cells in tumor microenvironments for immunometabolic therapy. Nano-Micro Lett. 2021;13:31. doi: 10.1007/s40820-020-00555-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim C.S., Ingato D., Wilder-Smith P., Chen Z. Stimuli-disassembling gold nanoclusters for diagnosis of early stage oral cancer by optical coherence tomography. Nano Converg. 2018;5:3. doi: 10.1186/s40580-018-0134-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodell C.B., Arlauckas S.P., Cuccarese M.F., Garris C.S., Li R., Ahmed M.S., et al. TLR7/8-agonist-loaded nanoparticles promote the polarization of tumour-associated macrophages to enhance cancer immunotherapy. Nat Biomed Eng. 2018;2:578–588. doi: 10.1038/s41551-018-0236-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swamydas M., Luo Y., Dorf M.E., Lionakis M.S. Isolation of mouse neutrophils. Curr Protoc Immunol. 2016;110 doi: 10.1002/0471142735.im0320s110. 3.20.1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tran P., Lee S.E., Kim D.H., Pyo Y.C., Park J.S. Recent advances of nanotechnology for the delivery of anticancer drugs for breast cancer treatment. J Pharm Investig. 2020;50:261–270. [Google Scholar]
- 23.Choi Y.H., Han H.-.K. Nanomedicines: current status and future perspectives in aspect of drug delivery and pharmacokinetics. J Pharm Investig. 2018;48:43–60. doi: 10.1007/s40005-017-0370-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beltrán‑Gracia E., López‑Camacho A., Higuera‑Ciapara I., Velázquez‑Fernández J.B., Vallejo‑Cardona A.A. Nanomedicine review: clinical developments in liposomal applications. Cancer Nano. 2019;10:11. [Google Scholar]
- 25.Das A., Asad M., Sabur A., Didwania N., Ali N. Monophosphoryl lipid A based cationic lipid nanoparticle facilitates vaccine induced expansion of polyfunctional T cell immune responses against visceral leishmaniasis. ACS Appl Bio Mater. 2018;1:999–1018. doi: 10.1021/acsabm.8b00184. [DOI] [PubMed] [Google Scholar]
- 26.Cheng Y., Chen Y., Li J., Qu H., Zhao Y., et al. Dietary b-sitosterol regulates serum lipid level and improves immune function, antioxidant status, and intestinal morphology in broilers. Poult. Sci. 2020;99:1400–1408. doi: 10.1016/j.psj.2019.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hou X., Zaks T., Langer R., Dong Y. Lipid nanoparticles for mRNA delivery. Nat Rev Mater. 2021;6:1078–1094. doi: 10.1038/s41578-021-00358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomita N., Mohammad M.M., Niedzeiwcki D.J., Ohta M., Movileanu L. Does the lipid environment impact the open-state conductance of an engineered β-barrel protein nanopore? Biochim Biophys Acta. 2013;1828:1057–1065. doi: 10.1016/j.bbamem.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kara S., Afonin S., Babii O., Tkachenko A.N., Komarov I.V., Ulrich A.S. Diphytanoyl lipids as model systems for studying membrane-active peptides. Biochim Biophys Acta Biomembr. 2017;1859:1828–1837. doi: 10.1016/j.bbamem.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 30.Kim D., Wu Y., Shim G., Oh Y.K. Genome-editing mediated restructuring of tumor immune microenvironment for prevention of metastasis. ACS Nano. 2021;15:17635–17656. doi: 10.1021/acsnano.1c05420. [DOI] [PubMed] [Google Scholar]
- 31.Coleido O.R., Chu N.Q., Szabo A.L., Chu T., Rhebergen A.M., Jairam V., et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature. 2014;513:559–563. doi: 10.1038/nature13490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diaz F.E., Danntas E., Geffner J. Unravelling the interplay between extracellular acidosis and immune cells. Mediators Inflamm. 2018;2018 doi: 10.1155/2018/1218297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z.H., Peng W.B., Zhang P., Yang X.P., Zhou Q. Lactate in the tumour microenvironment: from immune modulation to therapy. EBioMedicine. 2021;73 doi: 10.1016/j.ebiom.2021.103627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quinn W.J., III, Jiao J., TeSlaa T., Stadanlick J., Wang Z., Wang L., et al. Lactate limits T cell proliferation via the NAD(H) redox state. Cell Rep. 2020;33 doi: 10.1016/j.celrep.2020.108500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cascone T., McKenzie J.A., Mbofung R.M., Punt S., Wang Z., Xu C., et al. Increased tumor glycolysis characterizes immune resistance to adoptive T cell therapy. Cell Metab. 2018;27:977–987. doi: 10.1016/j.cmet.2018.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ward C., Meehan J., Gray M.E., Murray A.F., Argyle D.J., Kunkler I.H., et al. The impact of tumour pH on cancer progression: strategies for clinical intervention. Explor Target Antitumor Ther. 2020;1:71–100. doi: 10.37349/etat.2020.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y.X., Zhao Y.Y., Shen J., Sun X., Liu Y., Liu H., et al. Nanoenabled modulation of acidic tumor microenvironment reverses anergy of infiltrating T cells and potentiates anti-PD-1 therapy. Nano Lett. 2019;19:2774–2783. doi: 10.1021/acs.nanolett.8b04296. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.