Abstract
This study was initiated to characterize the regulation and secretion of ExoS by Pseudomonas aeruginosa during contact with eukaryotic cells. The production of ExoS was monitored by a sensitive ADP-ribosyltransferase activity assay, and specific activities were calculated for supernatant and cell-associated fractions. Time course analysis indicated that ExoS was produced after a lag period, suggesting that induction of the regulon is necessary for the expression of detectable amounts of enzyme activity. Under tissue culture growth conditions, ExoS was induced when P. aeruginosa was in contact with Chinese hamster ovary (CHO) cells or after growth in tissue culture medium with serum. The serum induction of ExoS appeared to result in generalized type III secretion, while induction by contact with CHO cells appeared to result in polarized type III secretion. Mutants in the type III secretory system that express a null phenotype for ExoS production in bacteriological medium produced but did not secrete the enzyme when P. aeruginosa was grown under inducing conditions in tissue culture medium. These results suggest that both induction and secretion of ExoS may differ when the bacteria are exposed to different growth environments. The putative type III translocation proteins and secretion apparatus of P. aeruginosa were required for translocation of bacterial factors that mediate changes in CHO cell morphology during infection.
Exoenzyme S is an extracellular ADP-ribosyltransferase synthesized by the opportunistic bacterial pathogen Pseudomonas aeruginosa (18). P. aeruginosa produces exoenzyme S as a high-molecular-weight aggregate consisting primarily of two immunologically related polypeptides, ExoS (49 kDa) and ExoT (53 kDa), which are encoded by separate but coordinately regulated genes (27, 38). Although ExoS and ExoT share 75% amino acid identity, ExoT possesses a catalytic defect and expresses only 0.2% of the ADP-ribosyltransferase activity of ExoS (38). The ADP-ribosyltransferase activity of exoenzyme S requires a eukaryotic cofactor termed factor activating exoenzyme S (FAS) (4, 7, 14). FAS is a member of the highly conserved, multifunctional 14-3-3 family of proteins, whose primary function involves the regulation of eukaryotic enzyme activities (1). In the presence of FAS, ExoS preferentially ADP-ribosylates vimentin, several members of the H- and K-Ras family of small (21- to 25-kDa) GTP-binding proteins, apolipoprotein A1, and human immunoglobulin G3 (IgG3) (5, 6, 8, 19). The requirement of a eukaryotic cofactor for activity and the functional importance of in vivo target proteins suggest that ExoS contributes to pathogenesis by disrupting normal cellular processes.
The intoxication of eukaryotic cells by ExoS is postulated to occur by a contact-dependent, type III secretion mechanism (12, 15, 24, 39, 40). The properties of type III-mediated translocation include (i) bacterial and eukaryotic cell contact, (ii) the introduction of translocation proteins into the eukaryotic plasma membrane, and (iii) the formation of a pore or channel through which bacterial effector proteins translocate (23, 25). The Yersinia type III secretion system and effector proteins (Yops), which are encoded on a large plasmid, serve as the prototypical model of type III-mediated intoxication (9). Elegant studies involving immunoprecipitation, immunofluorescence, and localization of enzyme activity have shown that the Yersinia effectors (YopE, YopH, YopM, and YpkA) are delivered directly into host cells from adherent bacteria (3, 17, 29, 34). Evidence supporting the hypothesis that P. aeruginosa delivers ExoS by a similar mechanism includes the observations that (i) secretion of ExoS requires an amino-terminal sequence that is not cleaved (39), (ii) P. aeruginosa encodes a series of homologs to the Yersinia type III secretion and translocation proteins that are coordinately regulated with ExoS (39, 40), (iii) ADP-ribosylation of Ras is observed when HT29 colon carcinoma cells are cocultured with ExoS-producing P. aeruginosa strains but not when purified recombinant ExoS (rExoS) is applied to cells (24), and (iv) the formation of neurite outgrowths by PC12 cells is inhibited when these cells are transfected with plasmid DNA encoding ExoS or when cocultured with P. aeruginosa producing ExoS (15). Functional homology between the type III systems of P. aeruginosa and Yersinia spp. was demonstrated by the translocation of ExoS from Yersinia strains expressing rExoS from multicopy plasmids (12). ExoS translocation was not observed when the yersinia-encoded type III secretion pathway or translocation functions were compromised by a specific mutation (12).
The goal of the present study was to quantitate ExoS production when the native organism, P. aeruginosa is in contact with eukaryotic cells. In this situation, exoS is expressed from a single chromosomal copy and is subject to native regulatory and secretory controls. We show that cell-associated bacteria produce more ExoS per bacterium than supernatant-associated P. aeruginosa. Two tissue culture conditions induce ExoS production: growth in the presence of newborn calf serum and growth in the presence of eukaryotic cells. The regulation of ExoS synthesis differs for bacteria induced by growth in bacteriological medium containing nitrilotriacetic acid (NTA; a chelator of calcium and zinc ions) versus growth in the presence of eukaryotic cells or serum. This difference in induction may reflect changes in the regulation of gene expression that determine whether the bacterium is poised for generalized or polarized type III secretion. Invasion of eukaryotic cells is not required for ExoS expression. Mutations in the P. aeruginosa type III secretion or translocation proteins prevent the translocation of bacterial products involved in cell morphology changes.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
P. aeruginosa strains were maintained as bacterial stocks at −80°C in 10% sterile skim milk. Parental P. aeruginosa 388 (2) and type III secretory mutants 388pscC::Tn1 (39), 388pscN::Tn5Tc (31), 388pscC::Tn5Tc (39), and 388ΔexoS::Tc (21) have been previously described. An interposon insertion within popD that encodes a putative translocator protein, PopD, was constructed in P. aeruginosa 388 (388popD::TcΩ) as described below. ExoS production was measured from bacterial cultures or infections under several growth conditions. For all inoculations P. aeruginosa was initially cultured on Vogel-Bonner minimal medium plates (37) incubated overnight at 37°C. Under these conditions ExoS production is undetectable, indicating that the pathway is off or repressed. To quantitate ExoS production in bacteriological medium, bacteria were transferred from Vogel-Bonner Minimal Medium to a deferrated dialysate of trypticase soy broth supplemented with 1% glycerol, 100 mM monosodium glutamate, and 10 mM NTA and cultivated as previously described (36). Growth in the presence of NTA maximally induces the exoenzyme S regulon. To measure the production of ExoS under tissue culture conditions, bacteria were grown statically in 12-well plates at 37°C under 5% CO2 in tissue culture medium with 5 or 0% serum. To determine whether serum proteins adsorbed to the plastic tissue culture wells induced ExoS expression, 1 ml of complete medium was adsorbed to a well overnight at 37°C in 5% CO2. The well was washed with phosphate-buffered saline (PBS), and a bacterial inoculum (2 × 106) was added in serum-free tissue culture medium. After a 4-h incubation period, viable counts were assessed and the soluble fraction was tested for ADP-ribosyltransferase activity. When P. aeruginosa and CHO cells were cocultivated, ExoS activity was quantitated as described below (see subsequent sections).
Eukaryotic cell culture.
CHO cells (ATCC 61-CCL) were cultivated at 37°C in 5% CO2 in Ham’s F-12 nutrient mixture supplemented with 10% newborn calf serum, 50 U of penicillin and streptomycin per ml, 2 mM l-glutamine, 0.12% sodium bicarbonate, and 2.5 mM HEPES (complete medium; Life Technologies, Grand Island, N.Y.). Cells were routinely maintained in T-25 culture flasks (Corning, Cambridge, Mass.) and passaged at confluence. To measure ExoS production from cells cultivated under reduced serum conditions, CHO cells were adapted by sequential passage in CHO III PFM (protein-free medium; Life Technologies) with progressive reduction in the amount of complete medium added. The final concentration of serum components in passaged cultures was 0.15%.
CHO cell infection and coculture fractionation.
CHO cells were seeded in 12-well tissue culture plates (Falcon, Franklin Lakes, N.J.) and allowed to grow for 16 to 18 h to obtain 90 to 95% monolayer confluency (4.0 × 105 cells/well). Culture supernatants were removed, the monolayer was washed once with PBS (Life Technologies), and the cells were treated for 30 min with 10 μg of cytochalasin D per ml to prevent bacterial uptake. The medium was removed, and 1 ml of the bacterial inoculum (approximately 2.0 × 106 bacteria) in tissue culture medium with cytochalasin D was placed into the well. The multiplicity of infection from experiment to experiment ranged from 5:1 to 12:1 as determined by measuring the CFU. In initial experiments P. aeruginosa and CHO cells were cocultured in medium with (5%) or without (0%) serum for a 4-h time course at 37°C in 5% CO2. In subsequent experiments, the amount of ExoS activity was compared at single 4-h time points.
After appropriate incubation periods, the medium from infected cell cultures was mixed gently in the well to remove nonadherent or loosely adherent bacteria and transferred to a microfuge tube. An aliquot was removed for viable counts, and the remainder was subjected to centrifugation at 12,000 × g at 4°C for 15 min. A portion of the soluble fraction was stored at −80°C to quantitate ADP-ribosyltransferase activity (supernatant-associated activity). The CHO cell monolayer was washed twice with PBS, and 1 ml of tissue culture medium containing 200 μg of gentamycin (Sigma Chemical Corporation, St. Louis, Mo.), 100 μg of ciprofloxacin (Bayer Corporation, West Haven, Conn.), and 10 μg of cytochalasin D per ml was placed into the well to reduce the contribution of bacterium-associated ExoS activity. Incubation was continued for 2 h at 37°C in 5% CO2, a period which consistently resulted in complete killing of cell-associated P. aeruginosa. The medium was removed, and the cells were washed twice with PBS and then lysed with 150 μl of distilled water. The contents of the well was removed with a cell scraper and subjected to centrifugation at 12,000 × g at 4°C for 15 min. A portion of the soluble fraction was retained at −80°C to quantitate the ADP-ribosyltransferase activity (cell-associated activity). Duplicate wells not treated with antibiotics were used to determine the number of cell-associated bacteria existing at each time point. All of the experiments were performed in triplicate.
ExoS enzymatic activity assays.
Supernatant and lysate fractions from P. aeruginosa strains grown in bacteriological medium were assayed for ADP-ribosyltransferase activity as described previously (16). Infected CHO cell cultures were fractionated into supernatant and cell-associated samples. ExoS activity was measured in reaction mixtures (20 μl) containing 10 μl of the coculture sample, 0.25 M sodium acetate (pH 6.0), 45 μM soybean trypsin inhibitor (SBTI; target for ADP-ribosylation), 33 μM [32P-adenylate phosphate]NAD (specific activity, 2 × 105 cpm/pmol; NEN), and 40 nM recombinant FAS. Reaction mixtures were incubated for 1 h at room temperature and stopped with 6.6 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer containing β-mercaptoethanol. The reaction mixtures were boiled for 5 min, and 5 μl was loaded onto SDS-polyacrylamide gels (13.5% acrylamide separating gels and 4% stacking gels) (22). After being stained with Coomassie blue, the gels were dried and subjected to autoradiography. Labeled SBTI bands were removed from dried gels and analyzed by scintillation counting. Data were normalized to CFU or optical density units at 540 nm (OD540) and reported as 10−18 or 10−19 moles of ADPRT (ADP-ribose transferred to SBTI) as appropriate.
Construction of 388popD::TcΩ.
To test the contribution of the putative translocation protein, PopD, on ExoS cell-associated activity we constructed a chromosomal interposon insertion in popD. Portions of the P. aeruginosa pcrGVHpopBD locus were subcloned into M13mp19. The NotI sites within the locus were removed by site-specific mutagenesis (Sculptor Kit; Amersham, Buckinghamshire, United Kingdom) and returned to the allelic replacement vector pNOT19 (33). A partial SspI digest was performed to insert the interposon cartridge encoding tetracycline resistance (TcΩ) within the first five amino acids of the open reading frame encoding popD. The mobilization cartridge from pMOB3 (33) was subcloned into the interposon-containing constructs by using the unique NotI site of pNOT19. The mobilization cartridge encodes chloramphenicol resistance, mobilization functions for conjugative transfer, and the counter-selectable marker sacBR. Selection for allelic replacement derivatives of P. aeruginosa 388 and Southern blot analyses were performed as previously described (11). As assessed by SDS-PAGE and Coomassie staining of extracellular protein profiles, 388popD::TcΩ secretes parental levels of ExoT, PopN, PcrV, and ExoY. PopD is not detectable by Western blot analysis, and the expression of PopB, a second putative translocator protein encoded 5′ of popD, is reduced in 388popD::TcΩ.
Microscopy.
CHO cells grown in 12-well tissue culture plates were infected with parental (388), type III secretion (388pscC::Tn1, 388pscC::Tn5Tc, and 388pscN::Tn5Tc), and translocation (388popD::TcΩ) mutants of P. aeruginosa. After 4 h of infection, the CHO cells were fixed in 2% paraformaldehyde in PBS (EM Sciences, Fort Washington, Pa.). Phase-contrast photographs were obtained with a Diaphot 200 inverted microscope (×300; Nikon).
RESULTS
Growth and production of ExoS by P. aeruginosa during coculture.
ExoS is an ADP-ribosyltransferase that is secreted from P. aeruginosa by a type III mechanism (39). We wanted to develop a quantitative system to measure the translocation of ExoS into eukaryotic cells to begin to dissect the mechanistic aspects of type III secretion and translocation in P. aeruginosa. ExoS was chosen as an indicator of translocation because it is a highly active enzyme providing a sensitive assay for the assessment of activity when P. aeruginosa is or is not in contact with eukaryotic cells. During the development of the assay system, we noted that substantial amounts of ExoS activity was measured from bacteria that remain associated with cells during the lysis procedure. These results indicated that the difference between translocated ExoS activity versus bacterial associated activity could not be accurately assessed. To measure only the amount of translocated ExoS, we prevented the uptake of bacteria by using cytochalasin D during the infection process. In addition, we used antibiotics to kill residual bacteria after the coculture period and a lysis procedure (distilled water) that failed to release ExoS from P. aeruginosa (data not shown).
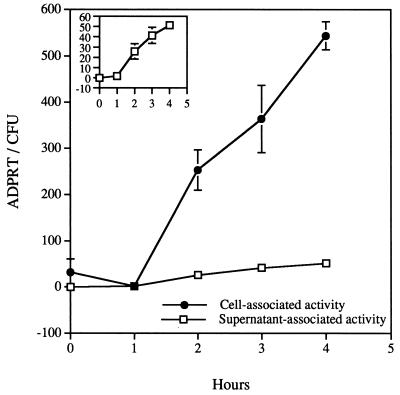
Initial experiments were designed to measure ExoS activity when P. aeruginosa was cocultivated with CHO cells in tissue culture medium with 5% serum. Activity was normalized to the number of viable bacteria and measured during a 4-h time course in the supernatant-associated and CHO cell-associated fractions. After an initial lag time of approximately 1 h, ExoS ADP-ribosyltransferase activity accumulated in both cell-associated and supernatant fractions (Fig. 1). Statistically higher levels of activity were measured from the cellular fractions after 2 h of incubation, and by the end of the incubation period approximately 10-fold more activity per bacterium was associated with the cellular fraction than with the supernatant fractions. At this time point approximately 90% of the bacteria are found in the supernatant fraction yet approximately 90% of the total ExoS activity is cell associated. These data suggest that most of the ExoS activity is translocated into eukaryotic cells but under these conditions, some activity is released into the culture medium.
FIG. 1.
Cell- and supernatant-associated ExoS ADP-ribosyltransferase activity from infections performed in the presence of serum. Cytochalasin D-treated CHO cells were infected with P. aeruginosa 388. Fractions consisting of soluble supernatant material (supernatant-associated activity) and CHO cell lysates (cell-associated activity) were collected and processed as detailed in Materials and Methods. ExoS activity is reported as 10−19 moles of ADP-ribosyltransferase activity that is normalized to CFUs and is presented as the mean and standard deviation of experiments performed in triplicate. The inset of Fig. 1 changes the scale of the y axis to demonstrate the accumulation of ExoS activity in supernatant fractions.
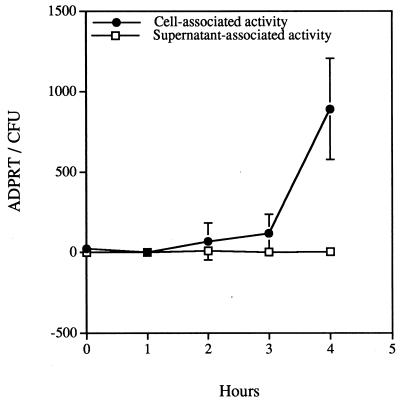
To determine the effect of serum on the expression of ExoS activity in the coculture system, the infection was performed in medium in the absence of serum. Under these conditions the accumulation of ExoS activity was not detected in the supernatant samples during the entire 4-h incubation period (Fig. 2). The accumulation of cell-associated activity was slower, but the total accumulation at 4 h was similar to values obtained with infections in the presence of serum. These results suggest that ExoS production can be induced by cellular contact in the absence of serum components. In addition, when infections were done in the absence of serum, it appeared that type III secretion was limited to polarized translocation; ExoS activity was not detected in the extracellular medium.
FIG. 2.
Cell- and supernatant-associated ExoS ADP-ribosyltransferase activity from infections performed in the absence of serum. Cytochalasin D-treated CHO cells were infected with P. aeruginosa 388. Fractions consisting of soluble supernatant material (supernatant-associated activity) and CHO cell lysates (cell-associated activity) were collected and processed as detailed in Materials and Methods. ExoS activity is reported as 10−19 moles of ADP-ribosyltransferase activity that is normalized to CFUs and is presented as the mean and standard deviation of experiments performed in triplicate. ExoS activity in supernatant-associated fractions was not detected.
Adaptation of CHO cells to reduced serum growth and ExoS production during cocultivation.
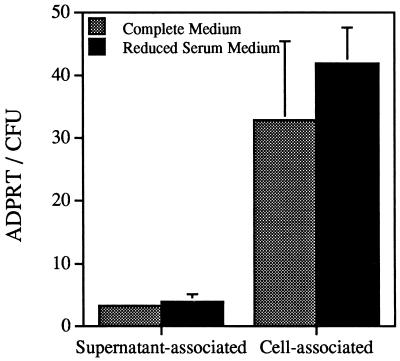
The infection of CHO cells in the absence of serum led to the accumulation of cell-associated enzyme. The induction of ExoS synthesis, however, could not be solely attributed to contact with eukaryotic cells since the CHO cells had been previously propagated in serum containing medium. Thus, even though the cells were washed before incubation, serum proteins either associated with the cells or tissue culture wells could serve to induce ExoS. To address whether contact with cells was an inducing signal, CHO cells were sequentially adapted to medium with reduced serum. After adaptation the cells were passaged for several generations in reduced serum medium (0.15%). Bacteria emulsified in serum-free medium were used to infect CHO cells that had been maintained in 10% serum of CHO cells that were adapted to the reduced serum medium. ExoS activity was measured after a standard 4-h coculture. The levels of supernatant- and cell-associated ExoS activities were not significantly different when the two types of infections were compared (Fig. 3). A 60-fold reduction in the presence of serum components without a significant effect on the amount of cell-associated activity suggested that cellular contact induces ExoS production in P. aeruginosa.
FIG. 3.
Comparison of ExoS activity from CHO cells propagated in complete (10% serum) or reduced (0.15%) serum medium. CHO cells were cultured in complete medium with 10% serum (gray bar) or adapted by sequential passage in CHO III protein-free medium with progressive reduction in the amount of complete medium added. The final concentration of serum components that resulted in consistent CHO cell growth and viability was 0.15%. A bacterial inoculum was prepared in serum-free medium, and the cocultivation was allowed to proceed for 4 h. Fractions were prepared, and ExoS ADP-ribosyltransferase activity was normalized to CFUs as described in Materials and Methods.
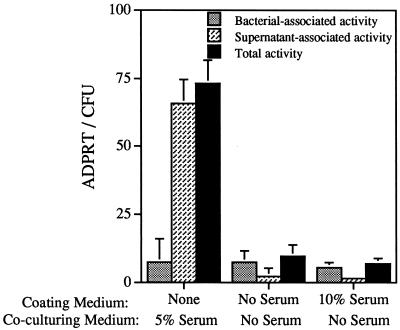
To address whether serum proteins adsorbed to the tissue culture well play a role in inducing ExoS production during coculture, empty wells were filled with medium containing or lacking serum and allowed to incubate overnight. The wells were washed once with PBS and incubated with bacteria emulsified in serum-free medium for 4 h. A well was also incubated with bacteria emulsified in medium with 5% serum. ExoS activity was not detectable in wells precoated with medium containing serum but was present in wells in which bacteria were grown in the presence of serum (Fig. 4). These data indicate that growth in the presence of serum components induces ExoS production. Serum components nonspecifically adsorbed to plastic appear to play no role in the induction of ExoS production.
FIG. 4.
The effect of adsorbed serum components on ExoS production. Tissue culture wells (in the absence of CHO cells) were left untreated or were pretreated overnight at 37°C with medium either containing or not containing newborn calf serum. The wells were washed once with PBS, and a bacterial inoculum with or without serum was added to each well. After a 4-h incubation period, the culture was harvested, and ExoS activity was measured in bacterial lysates (bacterium-associated activity) or in supernatant (supernatant-associated activity) fractions. The total activity measured is the sum of the bacterium- and supernatant-associated activities.
ExoS synthesis and secretion in type III secretory mutants.
In vitro, maximal ExoS synthesis and secretion by P. aeruginosa is induced by growth in bacteriological medium in the presence of NTA. In this study, two additional growth environments were found to induce ExoS expression; growth in the presence of CHO cells and growth in the presence of serum. Based on the high homology exhibited between the type III secretory and translocation components of Yersinia spp. and P. aeruginosa, we predicted that mutations introduced within the P. aeruginosa secretory loci would abrogate the ability to translocate ExoS. Phenotypic analysis of secretory mutants, however, indicated that ExoS synthesis was completely repressed (39). In these experiments ExoS synthesis was induced by growth in the bacteriological medium containing NTA. To determine whether ExoS expression was altered when the bacteria were grown under different inducing conditions, we compared ExoS production after growth in bacteriological medium in the presence of NTA and in tissue culture medium in the presence of serum. ADP-ribosyltransferase activity was measured in both extracellular and bacterial lysate fractions. In bacteriologic medium ExoS expression was detectable in the parental strain and in a popD::Ω insertional mutant (Table 1). The level of ExoS activity from 388popD::Ω was consistently lower than the levels measured in the parental strain, suggesting that PopD may have a minor regulatory effect on ExoS expression. The majority of the activity was associated with the extracellular medium (97%), indicating that most of the ExoS is secreted under these growth conditions (Table 1). ExoS activity was not detectable in either supernatant or lysate fractions when secretory mutants were analyzed after growth in bacteriological medium (Table 1). These data indicate that when the secretory apparatus was rendered nonfunctional, ExoS expression is repressed.
TABLE 1.
In vitro ADP-ribosyltransferase activities of wild-type and mutant strains of P. aeruginosa
| Strain | Activities (ADPRT/OD540)a in:
|
|||
|---|---|---|---|---|
| Bacteriological mediumb
|
Tissue culture mediumc
|
|||
| Supernatantd | Lysatee | Supernatant | Lysate | |
| 388 | 138.6 ± 28.1 | 3.5 ± 1.2 | 56.0 ± 17.7 | 70.7 ± 39.9 |
| 388pscC::Tn1 | 0 | 0.1 ± 0.1 | 3.7 ± 2.1 | 88.2 ± 16.2 |
| 388pscC::Tn5Tc | 0 | 0.1 ± 0.1 | 3.2 ± 2.9 | 73.7 ± 13.5 |
| 388pscN::Tn5Tc | 0 | 0 | 3.9 ± 1.9 | 105.6 ± 7.4 |
| 388popD::TcΩ | 58.0 ± 7.9 | 1.6 ± 0.3 | 32.0 ± 11.6 | 35.6 ± 12.9 |
Values provided are the average and standard deviation for experiments performed in triplicate and are reported as 10−18 moles of ADPRT per OD540.
Bacterial cultures were grown at 32°C in a deferrated dialysate of trypticase soy broth as detailed in Materials and Methods.
Bacterial cultures were grown in the presence of serum at 37°C.
Activity is measured from the soluble portion of a bacterial culture subjected to centrifugation.
Activity measured from the soluble portion of a bacterial extract. Bacteria were lysed by sonication.
A different pattern emerged when the same series of strains was examined after growth in tissue culture medium with serum as the induction signal. Under these conditions, both the parental and the popD::Ω strains synthesized and secreted ExoS, which appeared to be equally distributed between the supernatant and lysate fractions (Table 1). Strains defective in genes encoding secretory components synthesized but appeared not to secrete ExoS; approximately 96% of the total activity was measured in lysate fractions (Table 1). These results suggest that the induction signals mediating ExoS expression may differ depending on the growth conditions. On the other hand, ExoS repression may also be affected by environmental stimuli.
The role of the type III secretion and translocation proteins in the delivery of ExoS.
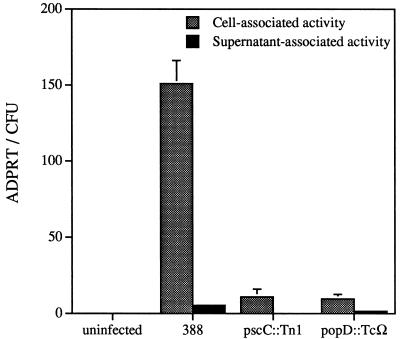
To determine the requirement of the P. aeruginosa type III secretion and translocation proteins in the expression and localization of ExoS activity, specific mutants were examined in the coculture system. ExoS production was monitored from parental and mutant strains after cocultivation with CHO cells for 4 h in serum-free medium in the presence of cytochalasin D. Cell-associated fractions were prepared after antibiotic treatment to eliminate extracellular bacteria. After cultivation with CHO cells, similar levels of viable bacteria were found for each strain (data not shown), indicating that none of the mutations affected P. aeruginosa replication under tissue culture conditions. Approximately 15-fold more ExoS activity was detected in cell-associated fractions from infections performed with strain 388 compared to the cell-associated activity measured from 388pscC::Tn5Tc or 388popD::TcΩ (Fig. 5). The amount of cell-associated activity measured in infections with the secretory or translocation mutants may be associated with the presence of dead bacteria in the sample (containing a stable form of ExoS) and/or the high sensitivity of the ADP-ribosyltransferase assay. These results confirm that the P. aeruginosa type III secretion and translocation components are functionally analogous to those of Yersinia spp. (12, 13). Small amounts of supernatant associated activity were detectable in the medium from infections with strains 388 and 388popD::TcΩ but not from 388pscC::Tn5Tc. These results indicate that PscC is required for secretion and that PopD functions after secretion.
FIG. 5.
Cell- and supernatant-associated ExoS activities of P. aeruginosa strains. Wild-type (388) or mutant P. aeruginosa strains defective in type III secretion (pscC::Tn1) or translocation (popD::TcΩ) were cocultured with cytochalasin D-treated CHO cells for 4 h in serum-free medium. Cellular fractions were prepared, and ExoS activity was measured and normalized to CFUs as described in Materials and Methods.
To control for the passive diffusion of ExoS into cells after membrane damage or the binding of ExoS to cellular membranes, two additional experiments were performed. CHO cells were infected with wild-type, type III secretion, or translocation mutant bacteria for 4 h, washed, and stained with trypan blue to detect membrane damage. After the 4-h incubation period there was no difference in trypan blue permeability between uninfected CHO cells and those infected with wild-type or mutant bacteria suggesting that membrane damage was not occurring at this time point (data not shown). In another experiment, ExoS was produced by growth of P. aeruginosa 388 in tissue culture medium with serum (in the absence of CHO cells). The spent medium was harvested, filter sterilized, and added to CHO cells alone or to CHO cells infected with 388ΔexoS::Tc. Supernatant and cell-associated fractions were prepared after a 4-h incubation with spent medium containing soluble ExoS. Under these conditions, ExoS activity was detected in supernatant-associated fractions of both uninfected and 388ΔexoS::Tc-infected CHO cells. In contrast, activity was not detectable in either cell-associated fraction, indicating that the activity being detected in our assays does not originate from ExoS bound to CHO cell membranes (data not shown).
CHO cell morphology changes are associated with type III-mediated translocation.
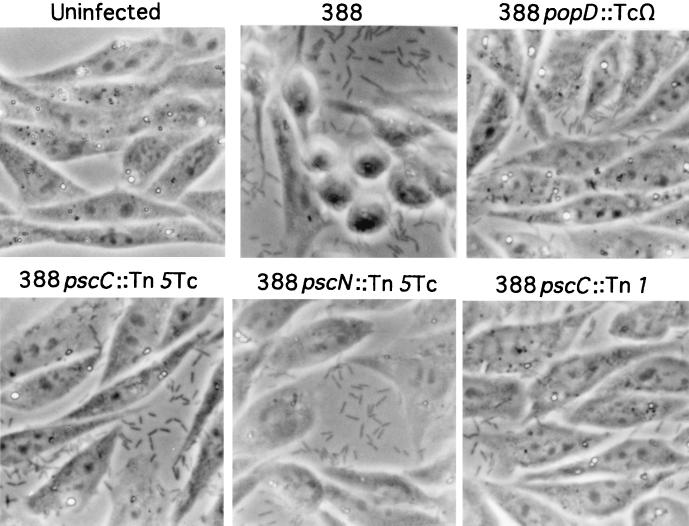
In coculture experiments we observed a rounding of the CHO cells at 3.5 to 4.0 h postinfection with strain 388. Others have reported that the delivery of either YopE or ExoS from Yersinia spp. into HeLa cells results in the collapse of the actin cytoskeleton, which was manifested as a visible rounding of HeLa cells (12, 32). To determine if the P. aeruginosa secretory or translocation mutants were defective in translocation, a change in cellular morphology was used as an independent indicator of effector transfer. CHO cell morphology was monitored after infection with strain 388, 388pscC::Tn5Tc, 388pscC::Tn1, 388pscN::Tn5Tc, or 388popD::TcΩ. CHO cells cocultured with strain 388 appeared by phase-contrast microscopy to be rounded, while cells cocultured with either secretory-defective (388pscC::Tn1 or 388pscN::Tn5Tc) or translocation-defective (388popD::TcΩ) strains were indistinguishable from uninfected control cells (Fig. 6). These results indicate that P. aeruginosa is unable to translocate effectors that cause changes in cell morphology when the type III secretion or translocation proteins are nonfunctional. In addition, this experiment indicates that PopD functions as a translocation protein, since neither type III effector protein production nor secretion was affected by an insertional mutation in popD (Table 1). Because more than one effector may be translocated by P. aeruginosa 388, it is unclear whether the rounding phenomenon is due to ExoS, ExoT, another coordinately regulated and type III secreted protein, or a combination of proteins.
FIG. 6.
Phase-contrast micrographs of CHO cells infected with parental strain 388 and isogenic mutant strains of P. aeruginosa (popD::TcΩ, pscC::Tn5Tc, pscN::Tn5Tc, and pscC::Tn1). CHO cells were left uninfected or were infected for 4 h with various bacterial strains in the absence of serum. Cells were washed, fixed, and photographed to assess cellular morphology in response to infection.
DISCUSSION
The ability of bacteria to deliver proteins into the cytoplasm of eukaryotic cells by using type III secretion represents a new mechanism of intoxication. ExoS is the first member of the family of bacterial ADP-ribosyltransferases for which type III-mediated translocation has been shown (12, 15, 24, 28). The focus of this study was to establish a quantitative experimental system to study the production of ExoS from P. aeruginosa in contact with eukaryotic cells, where ExoS is expressed from a single-copy chromosomal gene under native regulatory and secretory controls. Cocultivation studies in the presence or absence of serum during infection suggest two patterns of ExoS production. When infections are performed in the presence of serum, bacteria can secrete ExoS into the medium and translocate ExoS into cells, suggesting that serum mediates the induction from both free and cell-associated bacteria. Similar results have been obtained when type III secretion substrates of Salmonella and Shigella strains are monitored under different growth conditions (26, 41). In the absence of serum, ExoS activity was predominantly associated with cellular extracts; little to no activity was detected in supernatant fractions. We interpret these results as suggesting that ExoS is translocated into eukaryotic cells in a polarized fashion during coculture in the absence of serum but can be both translocated and secreted into the medium when P. aeruginosa and CHO cells are grown in the presence of serum. This conclusion is supported by further experiments showing that ExoS activity is secreted into the external medium when P. aeruginosa is grown in serum-containing medium in the absence of cells.
The role of cellular contact in ExoS expression was examined by adapting CHO cells to a reduced serum medium. Similar amounts of ExoS activity were measured in supernatant and cell-associated fractions when infections were performed with cells pregrown in medium containing 10 or 0.15% serum. Although our CHO cells could not be fully adapted to serum-free medium, the reduction of serum by over 60-fold with no change in the distribution of ExoS activity suggests that cellular contact is important in the induction of the exoenzyme S regulon. It should also be noted that the absolute levels of ExoS activity measured in different experiments vary over a wide range, making the comparison of absolute activities between experiments inappropriate. The variation in activity could be due to several factors associated with the cocultivation of cells and bacteria. Although there was variation in the absolute levels of ExoS activity, the general pattern of the translocation of 90% or more of the activity suggested vectoral transfer of the enzyme from the bacterium to the cell’s interior.
It is interesting to note that after growth in the presence of NTA, almost all of the activity is localized extracellularly, with little activity being associated with the bacterial lysates. In contrast, after growth in the presence of serum as the induction factor, ExoS activity appears to be equally distributed between the extracellular and bacterial compartments in secretion-competent strains. Based on the type III secretory Yersinia model, this differential distribution of activities may represent the efficiency of “unplugging” or changing the conformation or association of PopN with the secretion pore. This hypothesis would argue that PopN or YopN cannot stably associate with the secretion pore during cultivation in medium containing NTA or low concentrations of calcium.
ExoS production by P. aeruginosa grown in the presence of NTA also differs from bacteria grown under tissue culture conditions when data from type III secretory mutants are considered. The production of ExoS is undetectable when type III secretory mutants are grown under inducing conditions in bacteriological medium (with NTA) but is detectable after growth with eukaryotic cells or after growth in tissue culture medium with serum. After growth in tissue culture medium, type III secretory mutants were unable to release ExoS into the medium, confirming the requirement of the Psc proteins for the extracellular localization of ExoS. We concluded that induction or repression of the exoenzyme S regulon in P. aeruginosa may differ when the bacteria are cultivated under various growth conditions. In Yersinia spp., Yop production is negatively regulated when the type III apparatus is compromised due to the retention of a secreted repressor or antiactivator, LcrQ/YscM1 or YscM2 (30, 35). The mechanism of repression is unclear but appears to involve transcriptional inhibition. Sequence analysis to date (10; Pseudomonas Genome Project) has not resulted in the identification of a homolog to the LcrQ, YscM1, or YscM2 proteins, indicating that P. aeruginosa may lack a homologous repressor for the exoenzyme S regulon (10, 39). The production of ExoS in tissue culture by secretory mutants also argues against a model involving the secretion of a repressor. Thus, growth in the presence of NTA may represent a different induction mechanism than growth in the presence of serum or CHO cells.
Differences in induction may be reflected as stages in pathogenesis. In the initial stage of pathogenesis, during the colonization of epithelial surfaces, ExoS and/or other coordinately regulated and secreted determinants are probably translocated directly into cells. Effectors translocated into cells are postulated to function by disrupting normal cellular processes to allow the spread of the bacterium past the epithelial barrier (25). In acute pneumonia, P. aeruginosa disseminates from epithelial colonization sites to the pleural fluids, leading to rapid bacterial replication, bacteremia, and fatal septicemia (20). Our data suggest that growth in the presence of serum leads to generalized type III secretion, signaling perhaps a second stage of pathogenesis where ExoS and perhaps other secreted proteins are released into the environment. In support of this notion, Knight et al. have shown that the cofactor required for ExoS ADP-ribosyltransferase activity is present in both serum and pleural fluids and that extracellular targets for ADP-ribosylation exist (human IgG3 and apolipoprotein A1) (19). The ADP-ribosylation of extracellular targets may or may not be of significance in the later stages of infection, as type III-independent toxins (exotoxin A), proteases, and endotoxin surely contribute to the increased mortality associated with septicemia.
ACKNOWLEDGMENTS
This work was supported by grants AI-31665 and AI-01289 to D.W.F. and grants AI-30162 and AI-01087 to J.T.B. from the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
REFERENCES
- 1.Aitken A, Jones D, Soneji Y, Howell S. 14-3-3 proteins: biological function and domain structure. Biochem Soc Trans. 1995;23:605–611. doi: 10.1042/bst0230605. [DOI] [PubMed] [Google Scholar]
- 2.Bjorn M J, Pavlovskis O R, Thompson M R, Iglewski B H. Production of exoenzyme S during Pseudomonas aeruginosa infections of burned mice. Infect Immun. 1979;24:837–842. doi: 10.1128/iai.24.3.837-842.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boland A, Sory M P, Iriarte M, Kerbourch C, Wattiau P, Cornelis G R. Status of YopM and YopN in the Yersinia Yop virulon: YopM of Y. enterocolitica is internalized inside the cytosol of PU5-1.8 macrophages by the YopB,D,N delivery apparatus. EMBO J. 1996;15:5191–5201. [PMC free article] [PubMed] [Google Scholar]
- 4.Coburn J. Pseudomonas aeruginosa exoenzyme S. Curr Top Microbiol Immunol. 1992;175:133–143. doi: 10.1007/978-3-642-76966-5_7. [DOI] [PubMed] [Google Scholar]
- 5.Coburn J, Dillon S T, Iglewski B H, Gill D M. Exoenzyme S of Pseudomonas aeruginosa ADP-ribosylates the intermediate filament protein vimentin. Infect Immun. 1989;57:996–998. doi: 10.1128/iai.57.3.996-998.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coburn J, Gill D M. ADP-ribosylation of p21ras and related proteins by Pseudomonas aeruginosa exoenzyme S. Infect Immun. 1991;59:4259–4262. doi: 10.1128/iai.59.11.4259-4262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coburn J, Kane V, Feig L, Gill D M. Pseudomonas aeruginosa exoenzyme S requires a eukaryotic protein for ADP-ribosyltransferase activity. J Biol Chem. 1991;266:6438–6446. [PubMed] [Google Scholar]
- 8.Coburn J, Wyatt R T, Iglewski B H, Gill D M. Several GTP-binding proteins, including p21c-H-ras, are preferred substrates of Pseudomonas aeruginosa exoenzyme S. J Biol Chem. 1989;264:1–4. [PubMed] [Google Scholar]
- 9.Cornelis G R, Wolf-Watz H. The Yersinia Yop virulon: a bacterial system for subverting eukaryotic cells. Mol Microbiol. 1997;23:861–867. doi: 10.1046/j.1365-2958.1997.2731623.x. [DOI] [PubMed] [Google Scholar]
- 10.Frank D W. The exoenzyme S regulon of Pseudomonas aeruginosa. Mol Microbiol. 1997;26:621–629. doi: 10.1046/j.1365-2958.1997.6251991.x. [DOI] [PubMed] [Google Scholar]
- 11.Frank D W, Nair G, Schweizer H P. Construction and characterization of chromosomal insertional mutations of the Pseudomonas aeruginosa exoenzyme S trans-regulatory locus. Infect Immun. 1994;62:554–563. doi: 10.1128/iai.62.2.554-563.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frithz-Lindsten E, Du Y, Rosqvist R, Forsberg Å. Intracellular targeting of exoenzyme S of Pseudomonas aeruginosa via type III-dependent translocation induces phagocytosis resistance, cytotoxicity and disruption of actin microfilaments. Mol Microbiol. 1997;25:1125–1139. doi: 10.1046/j.1365-2958.1997.5411905.x. [DOI] [PubMed] [Google Scholar]
- 13.Frithz-Lindsten E, Holmström A, Jacobsson L, Soltani M, Olsson J, Rosqvist R, Forsberg Å. Functional conservation of the effector protein translocators PopB/YopB and PopD/YopD of Pseudomonas aeruginosa and Yersinia pseudotuberculosis. Mol Microbiol. 1998;29:1155–1165. doi: 10.1046/j.1365-2958.1998.00994.x. [DOI] [PubMed] [Google Scholar]
- 14.Fu H, Coburn J, Collier R J. The eukaryotic host factor that activates exoenzyme S of Pseudomonas aeruginosa is a member of the 14-3-3 protein family. Proc Natl Acad Sci USA. 1993;90:2320–2324. doi: 10.1073/pnas.90.6.2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ganesan A, Frank D W, Misra R P, Schmidt G, Barbieri J T. Pseudomonas aeruginosa exoenzyme S ADP-ribosylates Ras at multiple sites. J Biol Chem. 1998;273:7332–7337. doi: 10.1074/jbc.273.13.7332. [DOI] [PubMed] [Google Scholar]
- 16.Goranson J, Hovey A K, Frank D W. Functional analysis of exsC and exsB in regulation of exoenzyme S production by Pseudomonas aeruginosa. J Bacteriol. 1997;179:1646–1654. doi: 10.1128/jb.179.5.1646-1654.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Håkansson S, Galyov E, Rosqvist R, Wolf-Watz H. The Yersinia YpkA Ser/Thr kinase is translocated and subsequently targeted to the inner surface of the Hela cell plasma membrane. Mol Microbiol. 1996;20:593–603. doi: 10.1046/j.1365-2958.1996.5251051.x. [DOI] [PubMed] [Google Scholar]
- 18.Iglewski B H, Sadoff J, Bjorn M J, Maxwell E S. Pseudomonas aeruginosa exoenzyme S: an adenosine diphosphate ribosyltransferase distinct from toxin A. Proc Natl Acad Sci USA. 1978;75:3211–3215. doi: 10.1073/pnas.75.7.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knight D A, Barbieri J T. Ecto-ADP-ribosyltransferase activity of Pseudomonas aeruginosa exoenzyme S. Infect Immun. 1997;65:3304–3309. doi: 10.1128/iai.65.8.3304-3309.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kudoh I, Wiener-Kronish J P, Hashimoto S, Pittet J, Frank D W. Exoproduct secretions of Pseudomonas aeruginosa strains influence severity of alveolar epithelial injury. Am J Physiol. 1994;267:L551–L556. doi: 10.1152/ajplung.1994.267.5.L551. [DOI] [PubMed] [Google Scholar]
- 21.Kulich S M, Frank D W, Barbieri J T. Expression of recombinant exoenzyme S of Pseudomonas aeruginosa. Infect Immun. 1995;63:1–8. doi: 10.1128/iai.63.1.1-8.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Lee C A. Type III secretion systems: machines to deliver bacterial proteins into eukaryotic cells? Trends Microbiol. 1997;5:148–156. doi: 10.1016/S0966-842X(97)01029-9. [DOI] [PubMed] [Google Scholar]
- 24.McGuffie E M, Frank D W, Vincent T S, Olson J C. Modification of Ras in eukaryotic cells by Pseudomonas aeruginosa exoenzyme S. Infect Immun. 1998;66:2607–2613. doi: 10.1128/iai.66.6.2607-2613.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mecsas J, Strauss E J. Molecular mechanisms of bacterial virulence: type III secretion and pathogenicity islands. Emerg Infect Dis. 1996;2:271–288. doi: 10.3201/eid0204.960403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ménard R, Sansonsetti P, Parsot C. The secretion of the Shigella flexneri Ipa invasins is activated by epithelial cells and controlled by IpaB and IpaD. EMBO J. 1994;13:5293–5302. doi: 10.1002/j.1460-2075.1994.tb06863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicas T I, Iglewski B H. Isolation and characterization of transposon-induced mutants of Pseudomonas aeruginosa deficient in production of exoenzyme S. Infect Immun. 1984;45:470–474. doi: 10.1128/iai.45.2.470-474.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olson J C, McGuffie E M, Frank D W. Effects of differential expression of the 49-kilodalton exoenzyme S by Pseudomonas aeruginosa on cultured eukaryotic cells. Infect Immun. 1997;65:248–256. doi: 10.1128/iai.65.1.248-256.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Persson C, Nordfelth R, Hölmstrem A, Håkansson S, Rosqvist R, Wolf-Watz H. Cell-surface-bound Yersinia translocate the protein tyrosine phosphatase YopH by a polarized mechanism into the target cell. Mol Microbiol. 1995;18:135–150. doi: 10.1111/j.1365-2958.1995.mmi_18010135.x. [DOI] [PubMed] [Google Scholar]
- 30.Petterson J, Nordfelth R, Dubinina E, Bergman T, Gustafsson M, Magnusson K E, Wolf-Watz H. Modulation of virulence factor expression by pathogen target cell contact. Science. 1996;273:1231–1233. doi: 10.1126/science.273.5279.1231. [DOI] [PubMed] [Google Scholar]
- 31.Rooney P J, Frank D W. Abstracts of the 94th General Meeting of the American Society for Microbiology 1994. Washington, D.C: American Society for Microbiology; 1994. Isolation and characterization of insertional mutations affecting exoenzyme S production in Pseudomonas aeruginosa, abstr. D-188; p. 129. [Google Scholar]
- 32.Rosqvist R, Forsberg Å, Wolf-Watz H. Intracellular targeting of the Yersinia YopE cytotoxin in mammalian cells induces actin microfilament disruption. Infect Immun. 1991;59:4562–4569. doi: 10.1128/iai.59.12.4562-4569.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schweizer H P. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol Microbiol. 1992;6:1195–204. doi: 10.1111/j.1365-2958.1992.tb01558.x. [DOI] [PubMed] [Google Scholar]
- 34.Sory M P, Cornelis G R. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into Hela cells. Mol Microbiol. 1994;14:583–594. doi: 10.1111/j.1365-2958.1994.tb02191.x. [DOI] [PubMed] [Google Scholar]
- 35.Stainer I, Iriarte M, Cornelis G R. YscM1 and YscM2, two Yersinia enterocolitica proteins causing downregulation of yop transcription. Mol Microbiol. 1997;26:833–843. doi: 10.1046/j.1365-2958.1997.6281995.x. [DOI] [PubMed] [Google Scholar]
- 36.Thompson M R, Bjorn M J, Sokol P A, Lile J D, Iglewski B H. Exoenzyme S: an ADP-ribosyltransferase produced by Pseudomonas aeruginosa. In: Smulson M, Sugimura T, editors. Novel ADP-ribosylations of regulatory enzymes and proteins. Amsterdam, The Netherlands: Elsevier; 1980. pp. 425–433. [Google Scholar]
- 37.Vogel H J, Bonner D M. Acetylornithase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 38.Yahr T L, Barbieri J T, Frank D W. Genetic relationship between the 53- and 49-kilodalton forms of exoenzyme S from Pseudomonas aeruginosa. J Bacteriol. 1996;178:1412–1419. doi: 10.1128/jb.178.5.1412-1419.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yahr T L, Goranson J, Frank D W. Exoenzyme S of Pseudomonas aeruginosa is secreted by a type III pathway. Mol Microbiol. 1996;22:991–1003. doi: 10.1046/j.1365-2958.1996.01554.x. [DOI] [PubMed] [Google Scholar]
- 40.Yahr T L, Mende-Mueller L, Friese M B, Frank D W. Identification of type III secreted products of the Pseudomonas aeruginosa exoenzyme S regulon. J Bacteriol. 1997;179:7165–7168. doi: 10.1128/jb.179.22.7165-7168.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zierler M, Galan J. Contact with cultured epithelial cells stimulates secretion of Salmonella invasion protein InvJ. Infect Immun. 1995;63:4024–4028. doi: 10.1128/iai.63.10.4024-4028.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]