Abstract
In the medical field, a doctor must have a comprehensive knowledge by reading and writing narrative documents, and he is responsible for every decision he takes for patients. Unfortunately, it is very tiring to read all necessary information about drugs, diseases and patients due to the large amount of documents that are increasing every day. Consequently, so many medical errors can happen and even kill people. Likewise, there is such an important field that can handle this problem, which is the information extraction. There are several important tasks in this field to extract the important and desired information from unstructured text written in natural language. The main principal tasks are named entity recognition and relation extraction since they can structure the text by extracting the relevant information. However, in order to treat the narrative text we should use natural language processing techniques to extract useful information and features. In our paper, we introduce and discuss the several techniques and solutions used in these tasks. Furthermore, we outline the challenges in information extraction from medical documents. In our knowledge, this is the most comprehensive survey in the literature with an experimental analysis and a suggestion for some uncovered directions.
Keywords: Electronic medical records, Information extraction, Medical named entities recognition, Medical relation extraction, Section detection
General introduction
For centuries, physicians play an important role in ensuring good health. Indeed, a physician must be well trained and must be able to manage the disease and patient information in order to find the right treatment and make the right decision. In medicine, different types of information are used for treatment and can be found in narrative documents written by humans. For example, to make a medical prescription, the patient record and medication manufacturer are used [91]. Thanks to the development of information technologies and Hospital Information System (HIS), medical information is digitized into electronic records named Electronic Medical Record (EMR) [116] or Electronic Health Record (EHR). Digital records could be stored, managed, transmitted and reproduced efficiently. The widespread adoption of HIS has contributed to billions of records [90], and they are recognized as valuable resources for large-scale analysis.
Similarly, the number of diseases and medications is gradually increasing. In medical prescription, for example, the doctor must know all the indications and contraindications to prescribe a drug. Also, he has to take a lot of time to read a whole unstructured narrative medical leaflet especially for new doctors [25, 91, 92]. Today, medical records are available on the Internet even for any person. Nevertheless, the physician must manage the large amount of information that is written in natural language and must select the important information from the narrative documents. Many medical researchers are overwhelmed in the huge amount of medical data in their studies [158]. Indeed, the main source of errors in medicine is related to drug prescriptions. In 2006, there were 3900 prescription errors in Germany [91]. For 18 years, the Institute of Medicine in the United States reported that there were 237 million medication errors per year in England that were costly for the country [92]. Also, it reported that there were between 1700 and 22303 deaths per year due to adverse drug reactions. In addition, there are more than 234 million cases and 4 million deaths caused by coronavirus disease (COVID-19) [141]. Likewise, the information of this virus’s symptoms need to be analyzed by efficient tools in order to inform risk assessment, prevention and treatment strategy development and outcome estimation.
All these facts make the use of these available electronic documents more than a necessity. Hence, extracting useful information will be greatly helpful. Unfortunately, this process is not trivial mainly due to the huge number of documents, and consequently requires models able to deal with big data. This is hampered by the unstructured (or semi-structured) nature of such documents. In order to make Information Extraction (IE) feasible and efficient, structuring these documents in a more abstract form that is easily readable by machines/algorithms becomes a fundamental step. The main technologies of IE are the recognition of named entities and the extraction of relations between these entities. Entity recognition involves recognizing references to different types of entities such as medical problems, tests, allergies, risks, adverse events, drugs and treatments. In addition, detecting different sections in a document which can improve IE tasks by providing more context. Generally, medical text mining and IE help in medical decision and disease risk prediction.
In fact, several reviews related to IE in medical field have been published. Meystre et al.[98] have focused on research about IE from clinical narrative from 1995 to 2008. However, they have not discussed the research on IE from biomedical literature. Liu et al. [83] have introduced a novel IE paradigm Open IE (OpenIE) which begins to attract great attention in Biomedical IE (BioIE). Biomedical OpenIE (Bio-OpenIE) aims to extract tuples with any relation types with no, or little, supervision. Their review focuses mainly on recent advances in deep learning-based approaches such as Convolutional Neural Network (CNN) and Recurrent Neural Network (RNN). However, they have focused only on the main deep learning techniques in Bio-OpenIE. Wang et al. [144] have presented researches on clinical IE applications from 2009 to 2016 for a discussion in terms of publication venues, data sources, clinical IE tools, methods, applications in the areas of disease and drug-related studies, and clinical workflow optimizations. Also, they have gained a more concrete understanding of underlying reasons for the gap between clinical studies using EHR and studies using clinical IE. However, their research is made before deep learning was really adopted as mainstream in the informatics community. Sun et al. [129] focuses on the process of medical document processing and analyzes the key techniques involved include Named-Entity Recognition (NER) and Relation Extraction (RE) for IE. They make an in-depth study on the text mining applications, the open challenges and research issues for future work. Pomares-Quimbaya et al. [106] have reported the results of a systematic review concerning section identification in narrative medical documents. It was the first understanding review which focuses on this concept, its existing methods and its growing contribution on the IE tasks. Hahn and Oleynik [46] have discussed the contributions of recent publications from 2017 to 2020 in medical IE and foreshadow future directions of research. They have focused on the methodological paradigm shift from standard machine learning techniques to deep learning. Also, they have selected only two the diseases and drugs semantic classes and the relation between them. Nasar et al. [101] have focused specifically on NER and RE with major focus on advances via deep learning approaches. They have presented recent trends in domain of IE along with open research areas.
In our paper, we have made a comprehensive and up-to-date review about the medical IE domain. We have discussed different techniques for each task in this area such as NER, RE and section identification. Also, we have presented some issues related to the nature of entities with current solutions. Furthermore, we have discussed the data used for IE such as the nature of medical data in general and information about useful resources and datasets used in many published studies. Also, we have made an experimental analysis about the current methods based on their results. We also present the current state and propose some future directions in this field. To the best of our knowledge, this is the most comprehensive survey to be presented in the literature.
Our paper is organized as follow: Sect. 1 is a discussion about the medical data and its problematics; Sect. 2 presents the general classic data processing steps to treat structured data before passing to the unstructured data; Sect. 3 shows the text mining procedure that uses an unstructured text in order to show the position of the IE step in this process; Sect. 4 classify and discusses the methods of IE tasks according to the techniques they use; Sect. 5 presents specifically the section detection task and classifies several methods according to the used techniques; Sect. 6 presents some applications used to discover new knowledge which is useful for some medical tasks; Sect. 7 presents useful data and benchmark datasets in IE; Sect. 8 shows some shared tasks in the field; Sect. 9 is employed for experimental studies; Sect. 10 is a general discussion of the current state and its limits; and in the Conclusion, we suggest some research directions to be further developed.
Problem setting
In fact, there are different sources of medical information, such as daily activities, Internet and clinical staff [25]. The rapid development of hospital information technology has resulted in a rapid accumulation of medical data, and a significant amount of this data is in the form of free text written by the author [158]. Note that it is very likely to have long and maybe useless parts in a narrative content. A clinical narrative is a report-style free-text that is found in the medical document, used for clinical documentation. It is a rich source of information for medical research and analysis, and this source of data is needed to make health care decisions. There are several examples of clinical narratives that vary between full-fledged documents or clinical notes: Discharge Summaries, Radiology Reports, Emergency Reports, Pathology Reports, Urology Reports, Letter of Communication, History or Family History, Physical Exam, Medical Dictation, Admission Notes, Nursing Notes, Progress Notes, Operative or Procedure Notes and Clinic Visit Notes. Thus, there are several sources for these narratives: analytical repositories [81, 87], EMR/EHR systems [75, 135], speech recognition or dictation systems [55, 117], external sources provided by competitions [29, 135], or other open data sources [21]. EMR or EHR are digital representations of medical information. These records are popularized thanks to the development of information technologies and the HIS. They allow medical staff to record digital information, such as texts, symbols, diagrams, graphs, data, etc. Thus, they allow medical institutions to record the patient’s condition, such as diagnostic information, procedures performed and treatment results. These records are therefore sources of clinical information such as demographic data, diagnostic history, medications, laboratory test results and vital signs. Digital records could be stored, managed, transmitted and reproduced efficiently. Widespread adoption of HIS has contributed to billions of records [90], and they are recognized as valuable resources for large-scale analysis.
Many medical researchers are overwhelmed by the huge amount of medical data in their studies, because it is difficult for them to discover the hidden knowledge lying in the massive amount of medical texts [158]. Indeed, there are three types of data [128], structured data contains basic information such as medications taken, allergies and vital signs. This data requires traditional pre-processing technologies that include data cleansing, integration, transformation and reduction. As well, semi-structured or unstructured data contains more health information and requires more complex and difficult processing methods such as text mining. Semi-structured data usually has a flow chart format, similar to Resource Description Files (RDF), and includes names, values and timestamps. Unstructured data includes narrative data and stores a lot of valuable medical information, but it lacks common structural frameworks. The processing complexity of this data is increased by grammatical misuse, misspellings, local dialects, and semantic ambiguities.
Electronic drug prescription helps to control the prescription and monitors the consumption of drugs. To make the appropriate treatment decision, the patient’s EHR and medication leaflets are used [91]. The medication leaflet has information to avoid interactions with other medications, diseases, allergies or the patient’s conditions [25]. For example, SmPC is a legal document approved by the European Medicines Agency, used to represent the package insert, and it is also available in electronic form. According to physicians, the most preferred SmPC section titles are: “4.3 Contraindications”, “4.1 Therapeutic indications”, “4.2 Posology and method of administration”, “4.8 Undesirable effects”, “5.3 Preclinical safety data”, “4.5 Interaction with other medicinal products and other interactions”, “5.2 Pharmacokinetic properties”, “4.4 Special warnings and precautions of use” and “5.1 Pharmacodynamic properties”. Sorted from most to least preferred [47]. Also, the most important sections of the medical record for establishing the appropriate treatment are as follows: Contraindications, Therapeutic indications and Dosage [91].
Indeed, the main source of errors in medicine is related to the prescription of drugs, where the number of diseases and drugs is gradually increasing, and the doctor must know all the indications and contraindications to prescribe a drug. For this reason, there are several problems especially for new doctors, as it needs a lot of time to read all the unstructured instructions [25, 91, 92]. Also, it is necessary to find out about new diseases that require effective treatments. Also, have to find out what new treatments or drugs the doctors need to access. So, doctors may prescribe the wrong treatments or drugs. In 2006, there were 3900 prescribing errors in Germany [91]. For the last 18 years, the U.S. Institute of Medicine has reported that there are 237 million medication errors per year in England, also with costs to the country. Also, it reported that there were between 1700 and 22303 deaths per year due to adverse drug reactions. Current solutions to avoid these problems are the creation of written procedures, improved training for health care professionals, automation of support or research operations, quality control in medicine, improving communication between physicians and encouraging cooperation between medical departments [92]. It is a great challenge to effectively use text in medical documents [158]. Automatic document analysis is affected by ambiguity, format diversity, brevity, sloppy writing, redundancy, complex longitudinal information [147]. Also, obstacles to data mining are diversity, incompleteness, redundancy, and privacy.
General steps of data pre-processing
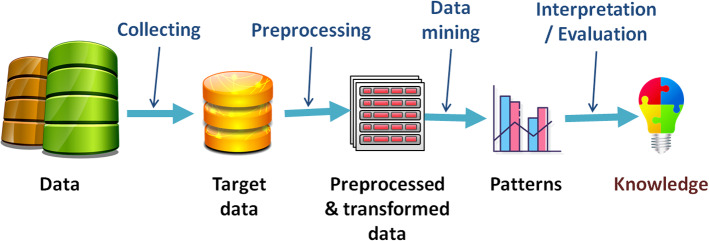
Before we pass to the medical unstructured text processing, which is the object of this survey, we introduce the classic data processing [24, 113, 129] in general which was initially aimed for structured data and is adaptable for unstructured texts. The general procedure for processing medical data consists of five steps as outlined in Fig. 1. The first step is the collection of the data by the government or professional medical institutes [52, 93, 133]. The second step is pre-processing which transforms the unstructured data into a useful format. In the third step, a data mining method is used considering the nature of the dataset, such as classification, clustering, association rules, regression, etc. The fourth step consists of some tests to understand the performance of the model. The last step is the knowledge application which is the goal of the data processing and also its driving force. Knowledge translation is more involved in medical management and arrangement of processing programs. In this step, the extracted patterns and knowledge must be analyzed and improved. A knowledge model is relatively better when it contains interactive iterations and requires continuous corrective feedback.
Fig. 1.
The general steps of the EMR data processing
The classic EMR data pre-processing step ensures that the data is accurate, complete, and consistent, and protects privacy, so as not to affect data mining. In particular for medical data, pre-processing methods should be chosen reasonably. Pre-processing facilitates the analysis of complex EMR data. In addition, high quality data is more likely to yield high quality results. Noting that the workload of the pre-processing step is over 60%. The classical pre-processing of the data consists of five steps: data cleaning, integration, data reduction, data transformation and privacy protection.
Data cleaning
The first step is data cleaning where different operations can be performed. The incomplete data filling operation is used when some data attributes are lost due to manual errors and system failure. Usually, missing data that have a significant influence on the processing will be ignored. Alternatively, if the dataset is relatively small, the defaults can be filled in manually, but this is time-consuming and expensive. In addition, defaults can be filled in by averaging the attribute values when the data distribution is uniform and when the cost budget is not significant. Also, machine learning models such as regression, formal Bayesian methods and decision tree induction can be used to determine the optimal value. But in extreme cases, they can show a relatively large deviation. Another operation for cleaning is noise processing which consists of correcting illegal values in the data source. There are several methods such as binning methods to examine the values around the data in order to smooth these values, regression methods to change the value of noise by adjusting to the value of the attribute and outlier analysis by making groups for similar attributes. The last operation is the correction of data inconsistency which consists in avoiding inconsistency in different sources or homologous data by analyzing the correlation between the data. All these cleaning operations can also be performed by recovering data from other sources.
Integration
The second step is integration, which can improve the speed and accuracy of data mining. It consists of dealing with heterogeneous data, and its redundancy. In fact, there are different forms of redundancy such as attributes that can derived or inferred from others or same medical records that can be come from multiple sources. Thus, redundant attributes should be cleaned up and the duplicate records should be purged out or consolidated and merged if they have complementing information. Most redundant data can be detected by correlation analysis. For nominal data, the chi-square test is used for detection.
Data reduction
The third step is data reduction, where a large amount of data is added every day, and reducing the size of the dataset lead for convenient and efficient data mining. There are different reduction techniques. Dimension reduction is relatively popular and easy to achieve with better effect. It makes the projection to a smaller dataset using Principal Component Analysis and Wavelet Transform methods. Thus, it selects subset of attributes by detecting and removing irrelevant, weakly correlated or redundant attributes or dimensions. There are also the reduction in the quantity and compression of data.
Data transformation
The fourth step is data transformation, which involves converting the dataset into a unified form suitable for data mining. This way, data analysis can be more efficient through directional and targeted data aggregation. This step consists of smoothing out noise, aggregating the data, and normalizing the data to avoid dependency of data attributes on units of measurement. Normalization into a smaller common space [111] can be achieved by different methods such as Min-Max, ZeroMean and Fractional Scale. It is more beneficial for neural networks and also for classification algorithms based on distance measures.
Privacy protection
The last step is privacy protection, where sensitive information about the patient’s private life can cause a serious problem when obtained by the offenders. This step has attracted a lot of attention with the popularity of Software Defined Networking (SDN) technology that facilitates network management [149, 152, 157]. The two main methods of this stage are the use of data protection and access control protocols, where the technical issues are encryption, privacy anonymity processing [82] and access control.
Challenges of processing EMR
In this section, we describe the text mining procedure and its application to the medical data and especially the EMR together with its challenges in the medical field. In fact, text mining is similar to classical data processing where the difference between them is that data mining in classical data processing is aimed for the analysis of structured data, but text mining is used for the analysis of unstructured texts. Text mining plays a major role in the health sector for the prediction of diseases, it allows to examine the data and join them into useful information. Its main objective is the prediction or the description of a medical information. Medical text mining extracts hidden knowledge from unstructured medical text [130], and its general procedure consists of four steps. First, it retrieves information to obtain the desired texts. Then, it extracts pre-defined information from these texts, this step is similar to pre-processing. Then, it explores the knowledge and extracts new knowledge. At the end, it applies the inferred facts to practice. The main strategy used for medical text mining is to convert the texts into structured computer-readable data using IE and Natural Language Processing (NLP) technologies. NLP technology has become a popular application of text classification and clustering, information retrieval and filtering, IE and question/answer (Q & A) system, machine translation, and new information detection. The application of NLP technology to clinical medicine has become a common topic of interest in academia and medicine. NLP for medical research started in 1960, but its research lags behind other fields because of the lack of data, corpus labeling costs too much and mismatches between research and development with the current needs of medical institutions [143]. Also, text structuring helps medical researchers to use EMR text in an easy way to examine the clinical knowledge that resides in the text [158]. The processed text data can furthermore be easily used by computer-aided analysis according to specific needs such as providing effective treatment to patients, creating more structured information, and extracting and structuring important information [25].
Medical text mining technology has several applications in the medical field that can be summarized in a few general applications. This technology can be applied for medical decision support and disease risk prediction. Decision support [11, 84, 160] is especially beneficial for physicians who have less clinical experience and can also advise medical experts on the treatment plan of symptoms. Risk prediction helps physicians judge the possibility of disease deterioration or improvement, and it also reduces costs for patients. In addition, this technology is applied for mobile health [88], networked medical treatment and personalized health care. Mobile health and networked medical treatment facilitate the consultation of doctor’s advice and make the capture of physical quality more accurate. Personalized health care develops treatment plans and nursing methods based on the actual situation of the patient. There are other applications that are prediction of disease evolution and detection of drug reactions. They help to quickly discover the medical trajectory of the disease over time, and detect adverse drug events in a cost-effective manner [59].
Generally, the challenges of text mining in the medical field include the lack of sufficient annotated public corpus and this is an obstacle especially for Chinese EMR studies, where English corpus studies are more mature and systematic [129]. Another challenge is the lack of personal and knowledge base dictionaries, where there is little useful content of medical dictionaries that is not sufficient, and the quality of dictionaries requires increased evaluation and certification of specialized institutions. In addition, privacy protection is another challenge especially as the transmission of EMR data will become more and more frequent and the market demand needs a more manageable data protection system. There is also the challenge of reasonable selection of processing tools, where a designed method may have poor performance in the biomedical field. Finally, a larger scale and more complex structure of EMR makes text mining difficult to process but more socially and economically beneficial.
Information extraction
Information extraction is a step in text mining that is similar to pre-processing in the classical data processing procedure. This step is mainly based on automatic NLP and machine learning [144]. The main technologies of EMR IE are NER and RE. Indeed, there are three main steps for the clinical IE that start with the extraction of medical problems, tests and treatments from discharge summaries and progress notes. Then, the classification of the assertions made about the medical problems. At the end, the classification of relationships between medical concepts [30]. In addition, the main tasks in the recognition of named entities are the identification of clinical events and temporal expressions. In addition, the main task in the biomedical literature is the recognition of bio-entity names [155].
Named entity recognition
NER was introduced in 1995, its general role is to identify types of names and symbols [43], but its role in the medical domain is to identify medical entities that are important for treatment, such as disease names, symptoms and drug names. It is a task of IE and it consists of recognizing references to different types of entities such as medical problems, tests, allergies, risks, adverse events, drugs and treatments. Indeed, the complexity of natural language increases with negated expressions [114], co-references [162], misspellings [69, 110], different language structures [131], detection of acronyms or abbreviations, expansion and disambiguation [64], anaphoric relations [23], etc. There are two main steps for this task which are the identification of the entity boundary followed by the determination of the entity class. The metrics used for evaluating NER methods are precision, recall, and f1-score, that can be computed based on token level or entity level. For entity level, there are two methods, partial matching and exact matching [12]. These methods are affected by the physician’s writing style, different writing forms of medical terms, ambiguity in term abbreviations, and compound or modified medical terms especially in Chinese. NER methods are classified into three main classes: rule-based; dictionary-based; and machine learning models. Hereafter, we review briefly state-of-the-art approaches within each category.
Rule-based methods
Actually, hand-coded rules give a better result than machine learning methods. However, these methods are valid only in specific datasets [111] and require manual construction of rules by the assistance of a medical expert.
Xu et al. [150] have proposed an unsupervised method to detect boundaries and classify medical entities mentioned in a medical Chinese text, and link mentions to their entities. Initially, the method exploits the part-of-speech and dependency relations, and maps the text to concepts in offline and online lexical resources to detect mentions of medical entities. Then, it classifies these mentions into categories and gives a Word2vec representation for each category. Next, the approach selects candidate entities from a knowledge base that are most similar to medical entity mentions based on the characters. In addition, candidates that are not similar to the category representation will be removed. Finally, the method calculates the similarity between the mention and its candidates according to the common characters, the popularity of the entity and the similarity between the words in the context that have a dependency relationship with the mention and the words in the description of each candidate. In addition, semantic correlation knowledge is added by computing the character similarity between the linking entity descriptions of the context mentions and the description of each candidate. Thus, the target entity is the candidate with best similarity. As an advantage, the method is unsupervised and generalizable. Also, it can recognize nested entities and better cover medical entities. In addition, it outperforms the state-of-the-art methods in terms of performance. Furthermore, the efficiency of online detection to solve the limitation problem of the dictionaries is well proved. However, the linking approach lacks semantic analysis. In addition, the detection may obtain inexact entities in the boundary detection step, and the filtering of non-medical terms may lose medical entities.
Alex et al. [5] have used hand-crafted rules and lexicons made by experts for NER from tokenized and POS tagged medical text. This method can recognize named entities reliably and accurately using brain imaging reports from Edinburgh Stroke Study (ESS) data and perform very well on the new data. In fact, ESS was the first set to be annotated by experts in this domain. Furthermore, this method can outperform machine learning approaches. However, hand-crafted rules are costly and time-consuming, especially when adapted to new and different data.
Zhao et al. [161] propose a weakly supervised method where they manually prepare some seeding rules and automatically extract all possible rules from unlabeled text for each of the six rule types, and connect them in a graph using cosine similarity. Note that the rule is represented by the average contextual embedding of its matched candidate entities. Then, propagate the labeling confidence from seeding rules in the graph to obtain new rules. These new rules are applied on the text to obtain a label matrix in order to estimate noisy labels using Linked Hidden Markov Model (LinkedHMM) generative model. Finally, Bidirectional Long Short-Term Memory-Conditional Random Field (BiLSTM-CRF) discriminative model is trained with noisy labels using Bidirectional Encoder Representations from Transformers (BERT) [34] as token embedding to label tokens by their entity types. As advantage, high-quality new labeling rules can be automatically learned from only a few manually constructed rules and an unlabeled text. In addition, with a limited number of manual rules, a substantially better performance can be achieved. Furthermore, they defined six useful types of rules for entity recognition. Moreover, the performance is improved using a graph neural network with a new class distance-based loss function to maximize the distance between positive and negative rules. However, some rules are overlapped and are not helpful to improve the recognition. Also, some rules learned from training data can’t be applied on testing data due to the mismatch between these datasets.
Dictionary-based methods
These methods are widely used in large-scale annotation and indexing of clinical medical texts. The most common techniques of this approach are Fuzzy Dictionary Matching and Post-processing. However, these methods require manual construction of dictionaries with the assistance of a medical expert and it’s difficult to cover several variants of medical terminology with a single dictionary.
Quimbaya et al. [110] have proposed a combined approach by preprocessing the text of electronic health records to apply exact and fuzzy NER techniques by the help of a knowledge base. In addition, a lemmatized recognition is applied after lemmatizing the target text and the knowledge base. Then, the overlapped entities recognized by these three techniques are combined to decide the final result. As advantage, the use of the Fuzzy Gazetteer match approach can find more instances of the dictionary concepts and even mistyped instances. Furthermore, the combination of these techniques improve the recall. However, this method does not take into consideration the context and surrounding words that appear near a candidate named entity.
The work of Nayel and ShashrekhaH. [102] consists in exploiting a dictionary information with the representation vector of the word and its characters to predict the named entity by a Bidirectional Long Short-Term Memory (BiLSTM) model. For this purpose, the authors used a Skip-gram model to build the word representation with. Also, they gave an initial representation vector for each character to train a BiLSTM to generate orthographic features from the characters of a word. Noting that all numbers are replaced by “NUM” and characters are changed to lowercase. In addition, the method adds dictionary information by using a merged disease vocabulary (MEDIC) as a dictionary. Thus, it represents dictionary information for each word in a binary vector to indicate whether a word is an abbreviation or synonym of a disease or is part of a multi-word disease name. Then, all these 3 types of features were passed to a BiLSTM model to do the learning. Thus, the contextual representation generated by this model is passed to a Conditional Random Field (CRF) layer to classify and select the most appropriate feature to annotate the input word from a sequence of words. As an advantage, Skip-gram learning improves the semantic and syntactic representation of words especially with a huge biomedical and generic text corpus. Also, the character representation significantly increases the performance. Moreover, the addition of dictionary information improves the result according to f1-score.
Sun and Bhatia [127] is based on sequence tagging by fine-tuning model [85] on Medical Information Mart for Intensive Care III (MIMIC-III) dataset [58] as a NER tagger, and train a gazetteer tagger (w/NER tagger) on clinical datasets. They have prepared a dictionary of medical conditions and drugs from the Unified Medical Language System (UMLS) [19] meta-thesaurus. Thus, NER tagger is trained using word and character BERT embedding, while gazetteer tagger is trained separately using gazetteer embedding, where their outputs before their Softmax layers are merged to return the entity type of the word such as medication, treatment, etc. As advantage, the fusion of the separate taggers lead to a better interpretability and flexibility. Also, its worth noting that even without the gazetteer tagger during inference, the NER tagger can preserve the gains. Furthermore, the fusion model is data efficient, interpretable and able to improve NER systems and to easily adapt to new entities mentions in gazetteers. In addition, this model can benefit from different clinical NER datasets. Also, the use of name knowledge from gazetteer leads to an improvement. Furthermore, this method is effective on handling non-stationary gazetteers and limited data. However, this method can be improved by extending to structured knowledge to further improve NER systems and for more interpretability.
Machine learning-based methods
For this approach, different models are used such as Hidden Markov Model (HMM), Support Vector Machine (SVM), CRF [68] and maximum entropy. The CRF model is the most popular and allows the incorporation of various features, which is appropriate for sequence annotation. Although the rule-based methods are widely used and are high performing, based on recent shared tasks, machine learning methods tend to perform best [1]. However, the algorithms and features largely affect the performance of the model, and these methods require standard annotations of the training data.
Lei et al. [76] have tested multiple features such as bag-of-characters, word segmentation, Part of Speech (PoS), and section information by training multiple machine learning models on manually annotated Chinese clinical documents to predict named entities such as clinical problems, procedures, laboratory test, and medications. They conclude that Structural SVM (SSVM) and CRF sequence-labeling models outperforms others, where SSVM is the highest. In addition, most features are gainful for the Chinese NER systems even with limited improvements. Moreover, the fusion of word segmentation and section information lead for the highest performance, and they complement each other. Also, they found that domain knowledge is important for Chinese word segmentation. However, the NER on English clinical text is more difficult than the Chinese because it contains many more entities mentions and the boundaries of its entities are harder to detect. Furthermore, the most errors occurs in long entities where they are not often completely detected. In addition, the training set can’t cover all concepts and some errors caused by unseen samples.
The work of Song et al [121] consists in evaluating learning models for NER in the biomedical domain. For this, the authors use a model based on CRF, and another one based on RNN. Also, different word representations are tested with these models, such as Word2vec, Global Vector (Glove) and Canonical Correlation Analysis (CCA). In addition, these models have been compared with other state-of-the-art methods. In this work, a dataset is annotated by biomedical categories such as protein, DNA, RNA, cell type and cell line. Thus, the models train on the annotations in order to predict the correct categories for the named entities. Indeed, the results show that the CRF model with Word2vec features outperforms all other models. As an advantage, word features are automatically built thanks to unsupervised learning by Word2vec which exploits the juxtaposition of words to extract their context. Thus, feature construction does not require manual annotation, dictionary, domain knowledge or other external resources. In addition, CRF can consider the context and neighbors of the entered word. However, CRF still needs manual annotation for learning. Moreover, the models proposed in this work are not optimized enough.
Li et al. [80] have improved a deep learning model based on the Deep Belief Network (DBN) to predict the named entity of a word using its PoS with its Word2vec feature vector. Indeed, using the PoS of the word leads to the best performance. Thus, this method is beneficial for NER. Moreover, Word2vec vectors capture useful semantic information. Moreover, with the improvement, DBN outperforms state-of-the-art methods. But, this requires manual annotation of the training data. Also, the content of the corpus knowledge is not rich enough.
Ghiasvand and Kate [41] use Decision Forest classifier to find named entities and their boundaries, while seed terms from UMLS are used for an unsupervised annotation. They have used 3 words before and after the target named entity which are presented by PoS, lemmatize form, stemming form and UMLS semantic types as features. Initially, training samples are collected using exact matched unambiguous terms from UMLS. Then, new samples are gathered by applying back the trained method on the corpus for self-training. To select samples for the model, they extract noun phrases that have at least one medical word which is included in any UMLS term. Then, another step is added to learn if a word can expand the boundary of its named entity using the same classifier and features with UMLS. As advantage, this method does not require any manual annotations. In addition, this method can determine the correct boundaries for the detected named entities. Moreover, it performs better than unsupervised methods and competitive to supervised methods. However, using data from different sources may reduce the performance. Furthermore, the automatically obtained noun phrases are not always perfect and can lead to errors.
The method of Chirila et al. [26] consists in extracting the most important information, which are named entities, from the sections of a semi-structured drug package insert. For this purpose, the authors trained the Stanford NER Tagger model on leaflets to predict word entities using distributional similarity based features in addition to other word features. According to the results, the accuracy with drugs of the same type is the highest. But, the method is tested only for the “Therapeutic indications” section and for the Roman language.
Deng et al. [33] combine BiLSTM with CRF to be trained on crawled and manually annotated Chinese TCM patents’ abstract texts using character embedding, in order to recognize entity types such as herb names, disease names, symptoms and therapeutic effects. Firstly, each character embedding vector is an input of a BiLSTM time step. A pre-trained embedding matrix is used to represent each character one-hot vector and is fine-tuned during the back-propagation, in order to extract sentence features. Finally, CRF layer learns the potential relationship between sequences and returns the optimal labeling sequence. As advantages, this method can learn semantic information in the context without feature engineering. Mainly, the use of characters instead of words leads to better performance, while the characters contain a lot of linguistic information and are able to mostly avoid errors caused by poor segmentation. Furthermore, BiLSTM can easily learn about contextual relationships in the text to provide more comprehensive contextual information. Likewise, the CRF layer complements the BiLSTM by optimizing the recognition comprehensively from the sentence level. However, the used data scale affects the learning model and cannot well support its requirements. Also, this method is restricted by the entity labeling granularity where some entities are nested within other entities.
Zhou et al. [163] have pre-trained two deep contextualized language models on clinical corpus from the PubMed Central (PMC): Clinical Embeddings from Language Model (C-ELMo) for word-level features and C-FLAIR clinical contextual string embeddings for character-level features. Then, each of the two embeddings is concatenated with Glove embedding and passed to BiLSTM-CRF model to extract entity types. As advantage, the models gain dramatic improvements compared to domain-generic language models and static word embeddings. In addition, these models can support different applications. Results show that C-Flair can handle entities that do not appear in word-level vocabulary. As well, C-ELMo can better capture the relationship between the word-level contextual features. Also, word-level models may be more robust than purely character-level models. However, it is hard for the two models to correctly recognize complex phrase-level entities.
Relation extraction
In RE technology, the relationship represents the correlation between two named entities appearing in the same sentences. Indeed, understanding the semantic relations between entities is required for many applications in IE such as semantic knowledge bases construction to infer the relationships between entities, and development of question answering systems for text summarization or concepts taxonomy construction. Semantic relations can be classified into two families according to their types, paradigmatic relations and syntagmatic relations [15]. Mainly, paradigmatic relations are operating on concepts of the same class. Usually, concepts are organized as a tree, where these relations are represented hierarchically by vertical links. These relations include the relation of synonymy, antonymy and hypernymy. Likewise, syntagmatic relations rely between two or more medical entities. They represent semantic links in an expression and rely between multiple linguistic units. They can be found by analyzing the syntactic forms in text and by a predicate. According to Uzuner et al. [139], we can also categorize these relations depending on the type of relationships, such as disease relationships, disease-medical examination relationships, and disease-treatment relationships. Generally, the cause-effect or causal RE has received ongoing attention in many medical fields [151] which can assist doctors, by supporting the construction of a knowledge graph, to quickly find causality and customize treatment plans. There are many examples of causal relations such as diseases-cause-symptoms, diseases-bring-complications and treatments-improve-conditions. The most commonly used techniques in this task are rule-based methods and machine learning-based methods [154].
Machine learning-based methods
Usually, this approach uses supplement NLP tools [51, 96, 109] to generate high-level features for a large quantity of examples which are fed to the machine learning model for a classification task. These methods are the most widely used while the biomedical data have an increasing growth. In fact, approaches based on this technique extract useful information from syntactic structures rather than applying manually constructed patterns [44]. Indeed, deep learning methods in particular can deal with the feature sparsity problem by transforming features into low-dimensional dense vectors. Thus, deep learning models have exhibited superior performances compared to the traditional machine learning-based and rule-based models [10, 67]. The most used machine learning models are Long Short-Term Memory (LSTM), CRF, Graph Convolutional Network (GCN) and SVM. However, the quality and the quantity of data have a big impact on this task while data must have sufficient examples. Usually, a manually annotated dataset by medical expert is needed.
Tran and Kavuluru [136] propose a novel distant supervision approach to extract medical treatment predication relations in PubMed abstracts by training a BiLSTM model after leveraging MeSH sub-headings and preparing training sentences with entities using NLM’s MetaMap [9] and UMLS Semantic Network. Indeed, this method uses a variant of BiLSTM with a modified noise-resistant loss function, where the input is word embeddings and learnable position vectors. As advantage, the position vectors can enhance the RE performance. Furthermore, the automatically generated training data is of reasonable quality without the costs of human involvement. In addition, MeSH sub-headings are precisely utilized while it leads for a better filtering of treatment and drug entities. However, this method focuses only on treatment predications. Furthermore, this method may have difficulties dealing with trivially negative cases as it is not trained on them. Also, linguistic phrasing is understandably difficult when there is a weak connecting word. Moreover, false negatives may occur when a unique concept entity is mentioned several times in the same sentence.
Shi et al. [119] train an end-to-end deep learning method to identify people’s pandemic concerns, and extract “co-occurrence” and “cause-effect” relations between them in tweets. The authors consider 8 types of concerns which are finance, government, disease, medicine, person, location, food, and date and time. This method uses BiLSTM-CRF to detect concern entities combined by Bidirectional GCN (BiGCN) model to extract relations, where a hidden state of BiLSTM is shared with BiGCN. Accordingly, each tweet is represented by sequential features using BERT embeddings and regional features using Concern Graph (CG) module. In the CG, each node represents a concern associated by its score and type. The concern score is calculated by sentiment polarity and retweet count of the tweet. To represent the regional features, 3 vectors merge for each concern word to represent PoS and syntactic dependency relation, concern score and type, and relation features. For that, an automated deep learning-based framework [118] was used to detect and construct a concern knowledge graph in order to get the concern types and relations. Likewise, the same framework is used to annotate the training set. As advantage, the state sharing enhance the influences from concerns to improve the performance of the RE. Note that this relation can reveal people’s thoughts behind the expressed concerns or identify the cause of public concerns. Furthermore, the regional features from CG improve the concern identification effectiveness and lead to a high noise-tolerance. In addition to contextual information, this method captures specific features of entities by a designated CG to perform better on tweets. However, this method is not the best for high-quality and manually annotated datasets.
The work of Yang et al. [154] consists of developing a series of RE models based on 3 transformer architectures, namely BERT [34], RoBERTa [85], and XLNet [156] to identify relation like “Drug-Adverse events” and “Drug-Reason”. This method uses already annotated entities to select candidate entity pairs for classification. Some rules are used as a strategy to generate these candidates, where the 2 entities must be a valid combination according to the annotation guidelines. Thus, the strategy selects only 2 entities in a same sentence or in 2 consecutive sentences as a candidate entity pair. Another strategy is to use the same rules but by using cross-sentence distance to select the number of consecutive sentences. Hence, it applies a separate model for each group of candidates which have different distance value. The results show that clinical pre-trained transformers consistently achieved better performance. In addition, XLNet and RoBERTa achieved the best performance for 2 different datasets. Furthermore, binary classification strategy consistently outperformed the multi-class classification strategy. Moreover, adding positional information to entities as features is critical to learn useful representations. However, there is no significant difference between the 2 generating candidates strategies, but there is still lack of an efficient method to solve both missing samples and sample distribution bias issues. Furthermore, there is a quite small number of training examples for some relation categories which significantly lead to a training difficulty and decent performance. Indeed, this work only focused on RE task, while this task is highly dependent on the NER result.
The work of Tran et al. [137] is aimed to acquire knowledge from COVID-19 scientific papers. For that, an enormous number of relations between entities are extracted by combining several methods. ReVerb [39] is based on verb-based relation phrases. OLLIE [97] extracts relations mediated by verb, nouns, adjectives, and more. ClausIE [31] is a clause-based approach. Relink [120] extracts relations from connected phrases. OpenIE [6] finds the maximally simple relations after breaking a long sentence into short and coherent clauses. Thus, the extracted relations are tagged by biomedical entity types recognized by SciSpacy models [103] trained on different corpus. Finally, the extracted relations are clustered and scored for their informativeness over the corpus to construct the retrieval system. As advantage, higher extraction coverage could be obtained by combining several methods. In addition, various specialized entity information can be obtained by covering different sets of biomedical entities. Also, knowledge can be rapid and efficient across a large number of scientific papers. However, some wrong results are caused by the complexity of the biomedical text where there are long sentences, and conjunctions and nested clauses are commonly used.
Rule-based methods
For rule-based systems, the commonly used strategies include dependency parsing [40], concept co-occurrence detection [57] and pattern matching [77]. Usually, rules are defined manually by domain experts [70] or even automatically generated by using machine learning techniques [122] on annotated data. The co-occurrence based methods are the most straightforward techniques. The key idea is when there are 2 entities mentioned together with higher frequency, there is a stronger chance to be related together. In fact, rule-based methods rely on a set of patterns, procedures or heuristic algorithms in order to directly identify candidate relations. Additionally, this technique can use supplement knowledge resources and apply some matching and mapping techniques. Indeed, sentence structure analysis can be used to explore patterns while it improves the performance and the extraction of implicit causal relations. However, preparing rules and supplement resources is labor-intensive and needs a domain expert for manual effort. Moreover, this approach severely restricts the generalizability and portability of RE for other types of data. Also, some difficult cases can be found in sentences and phrases which prevent rules to be correctly applied.
Ben Abdessalem Karaa et al. [14] extract relations between drug and disease entities such as “cure”, “no cure”, “prevent”, “side effect”, and “other relations”. This method use a combination of features for each sentence by the help of UMLS meta-thesaurus to provide semantic annotations by configuring MetaMap system in order to extract concepts and semantic types. By combination of NLP technique and UMLS knowledge, many features can be extracted such as frequency, lexical, morphological, syntactic and semantic features. Thus, these features are passed to an SVM model to predict the relation associated with the sentence using MEDLINE 2001 [115] as a standard training set. As advantage, this method can extract correct and adequate features while they are relevant to discover interesting relationships between concepts. Furthermore, it outperforms other methods for all types of relations especially in terms of f1-score and for the “cure” relation. Note that this performs better in a multidimensional context and is suitable for semantic relations in natural language texts. However, the lack of training data for “no cure” relation leads to low performance for all comparison methods. In addition, this method requires an annotated training set.
Kim et al. [60] propose a sequence labeling hybrid method to recognize family members and observations entities in EHR text notes, and extract relations between them in addition to living status. A rule based system is used to select family member entities by matching relevant noun terms by the help of PoS. Then, a number of BiLSTM models trained on dependency-based embeddings [62] as static embedding and Embeddings from Language Models (ELMo) [105] as context-dependent embedding. In addition, MetaMap [9] maps semantic types from UMLS and aligns them with entities to choose relevant ones. the family members and observations recognized by these BiLSTM models are ranked and have been voted based on models’ f1-scores. The heuristic rules are used to normalize the family member entities by a simple dictionary-based mapping, and determine family side by looking at cue words considering the degree of relatives. Thus, two Online Gradient Descent (OGD) [20] models are trained on lexical features based on the identified entities to determine living status and observations associated with family members. Hence, alive and healthy scores are assigned for living status phrases using cue words. Likewise, negation attribute is assigned to observations using ConText algorithm [22] with customized trigger terms. As advantage, voting ensemble of BiLSTM models contributes in terms of diversity to achieve better performance, and provides efficient and convenient integration of individual LSTM models which are not deterministic. In addition, this method is substantially benefited from a combination of 2 datasets. Integrating heuristics and advanced IE models lead to high level of performance. The performance is improved especially on RE and benefited by the large training set and the pre-trained embeddings. However, choosing the voting ensemble threshold can achieve best performance for one task but not the highest accuracy for other task. Also, some positive relations which rely on 2 entities in different sentences can be missed by using a carriage return character to filter examples.
Section detection
Generally, section detection can improve IE methods. By definition, a section is a segment of text that groups together consecutive clauses, sentences or phrases. It shares the description of a patient dimension, patient interaction or clinical outcome. In fact, unstructured text has sections defined by the author. There are two types of sections which are explicit sections that have titles, and implicit sections that do not have titles [81]. In addition, the section has a level, where it can be a section or a sub-section. The problem is that the author is free and even the precise defined templates are often ignored, which leads to less uniform titles and finds many sections without titles [134]. In addition, there are different types of documents due to the lack of a standardized typology.
Section detection aims to improve the performance of clinical retrieval tasks that deal with natural language such as entity recognition [76], abbreviation resolution [164], cohort retrieval [37] and temporal RE [66]. Section titles and orders in a prospectus may differ from one source to another. For this, standardizing the sections can help avoid this problem [25]. There are several applications of section detection. Generally, it consists in giving more context to the task by knowing the specific position of a concept. Section detection is used for information retrieval to support cohort selection and identification of patients with risk factors. In addition, some tasks can be improved like co-NER and reference resolution by adding the section as a feature. Another application is that this detection is useful for distinguishing sensitive terms in de-identification. Also, section detection methods can improve tasks that consider the order of events by identifying temporal sections. In addition, this detection can be applied for document quality assessment. Furthermore, these methods can select supporting educational resources by extracting relevant concepts. The main challenges of section detection are the production of a benchmark dataset, the integration of texts from different institutions, the selection of the types of measures to be evaluated, the adaptation of different natural languages and the integration of the most robust methods into high-impact clinical tasks.
According to an analysis of 39 section detection methods [106], there are several methods that adapt pre-built methods for this task. In addition, almost all section detection methods have custom dictionaries created by the authors, and most of them use a controlled terminology named UMLs Meta-thesaurus which is the most common. Also, precision and recall are very high in these methods, but recall is always lower. In addition, almost all methods detect sections that have titles, and half of them can detect sections without titles. Also, more than half of the methods do not detect sub-sections. Concerning the type of narration, most of the methods work on Discharge Summaries in the first class, maybe because of the predefined internal structure or because of the fact that this type of document is frequently transferred. Thus, Radiology Reports are in the second place. There are a few methods that deal with other than the English language. Generally, when the genre of the text, the medical specialty or the institution imposes more restrictions, the methods may have better results.
The methods of section detection can be classified according to the technique used into rule-based methods, machine learning-based methods and hybrid methods.
Rule-based methods
In this approach, there are three types of rules which are exact match, regular expressions and probabilistic rules. This approach requires dictionaries as additional information. The dictionaries can be title flats which are lists containing possible terms used as titles, hierarchical title dictionaries, or synonym and word variant dictionaries containing modifications or abbreviations of standard titles. In addition, the methods in this approach can handle different features such as the most frequent concepts/terms in each section, probabilities assigned to headings or formatting features that represent the shape of the section e.g., number of words, capitalization, a question mark, enumeration pattern, size, etc. But let us keep in mind that feature values may vary depending on the corpus, e.g., the size of the section. Most methods use non-probabilistic rules and require titled sections. With exact matching, recall decreases when the level of standardization of headings is low. Regular expressions can search and match the expected text even with variants. Indeed, the minority of methods is based on probabilistic rules. These rules complement exact matching and regular expressions. They are based on text features to give very high results. These rules are more advanced to detect even unannotated sections. Most methods are based on this approach.
Beel et al. [13] have proved that style information, specifically font size, is very useful for detecting titles in scientific PDF documents in many cases. The authors used a tool to extract formatting style information from a PDF file such as font size and text position. Then, they used a simple heuristic rule to select the three largest font sizes on the first page. Thus, identifying the texts that have these sizes as titles. In fact, this method outperformed an approach based on SVM, which uses only text, in accuracy and even in runtime. Moreover, this technique is independent of the text language because it only considers the font size. However, this method depends on the font size and requires the existence of formatting information.
The approach of Edinger et al. [37] is to identify sections in medical documents and use them in queries for information retrieval. To do this, the authors prepared a list of variations of all section titles for each document type. Variations in terminology, punctuation and spelling were selected to identify the most common section titles using a set of documents for each type. To identify titles in a document, a simple exact search is applied to find them by their variations. Then, the headings are annotated and the document text has been segmented according to these headings. As an advantage, the use of sections in the query instead of searching the whole document increased the accuracy of the search. Thus, this method can avoid retrieval of irrelevant documents. However, it has a smaller recall where the other method can retrieve more relevant documents. In addition, the exact search is accurate enough for section detection.
Lupşe and Stoicu-Tivadar [91] have proposed a method that supports prescribing by extracting and structuring information from medical records. The principle of this approach is to detect sections of the text and unify their titles using regular expressions and a set of section titles. Then, it removes empty words and applies the Stemming algorithm on the sections to root the words without touching medical terms. Thus, this method can suggest drugs that match the patient’s disease, are not contraindicated and do not conflict with other diseases, treatments or allergies of the patient. Indeed, this approach reduces medical errors in drug prescriptions and structures the necessary drug information. However, there are some medical terms that are still modified by Stemming. Also, this method is tested only with the Roman language.
Zhang et al. [158] have tried to effectively use temporal information in the text of electronic medical documents to structure them and help medical researchers to examine clinical knowledge and to facilitate computer-aided analysis. This method is based on rules to perform a few successive steps. These steps consist of correcting pronunciation errors, dividing texts according to grammatical rules, describing medical facts and events and finalizing by processing temporal expressions. However, these texts have little temporal information. In addition, the method gives the same weighting for different speech words.
Machine learning-based methods
This approach uses learning models, where the most popular are CRF, SVM and Viterbi. These methods also require training and testing datasets. These data can be created manually, with rules, with an automated method or with a combination of active learning and remote supervision. The size of the datasets used in these methods analyzed is between 60 and 25842 texts. There are also methods that use the same data for training and evaluation, and can be called cross-validation. This approach also can handle different types of features for this approach such as syntactic features that represent the structure of the sentence, grammatical roles and PoS. Semantic features represent the meaning of words and terms in a sentence. Contextual features are relative or absolute features of a line or section in the text such as the position of the section in the document and layout features. Lexical features are at the word level that can be extracted directly, e.g., the entire word, its prefix, suffix, capitalization, its type (e.g., number or stop word), among others. Indeed, the predictive value of the features had a great positive impact. These methods are not easily adaptable to other contexts. Size variation impacts performance and adaptability to new data. There is a lack of large datasets for learning and evaluation. Also, the construction of training data is time consuming. Non-standard abbreviations in titles and inappropriate phrases in sections result in incorrect classifications. The use of very specific terms for subheadings results in accuracy degradation.
The method of Haug et al. [49] consists in annotating each section in a medical document by its main concept. This approach is based on Tree Augmented Naive Bayesian Networks (TAN BN) to associate topics with sections as their semantic features. For this purpose, this method was trained using features generated by extracting N-grams from the text of section titles, in combination with the document type. In fact, the identification of the section topic improves the accuracy and avoids errors when extracting a specific information. Thus, this task can reduce the natural language processing effort and prepares the document for more targeted IE. However, n-grams have limitations with complex and large documents. In addition, this Bayesian model does not consider the consistent sequencing of section topics.
The method of Deléger and Névéol [32] classifies each line in French clinical documents into its specific high-level sections such as header, content and footer. Thus, a statistical CRF model is trained based on some information about the line taking into account the first token in surrounding lines, first two tokens in the current line, the first token is in uppercase, relative position of the line, number of tokens, presence of preceding empty lines, digits and e-mail addresses. As advantage, the performance is very high especially for content and header lines. It is well noted that the headers and footers are very present in the document and should be identified to focus on the core medical content. However, the granularity level of sections is very high while there are more useful sections within the content that are not identified.
Lohr et al. [86] have trained a logistic regression model on a manually annotated German clinical discharge summaries, short summaries and transfer letters to automatically identify sections using BoW statistics as features for each sentence. As advantage, this method achieves promising results in terms of f1-score. Furthermore, these authors have chosen a set of feasible and relevant categories for annotation. In addition, a sentence was chosen as an annotation unit while it has an appropriate granularity. However, the method does not perform well for categories that barely appear in the corpus.
Lupşe and Stoicu-Tivadar [92] have made a method that consists of homogenizing the sections of drug package inserts by standardizing the section names. At first, the method collects all section names from all drug package inserts, and prepares unique and common reference names that represent different kinds of sections. Then, machine learning is used to find the appropriate reference for each section name. Through this method, access to drug information has been improved for better processing. Moreover, this technique can be used in clinical decision applications to provide the necessary data to physicians. Thus, it helps especially new young doctors or those who start a new specialty. Neural network leads to highest results in the extraction of relevant information and outperforms cosine similarity according to the f1-score metric. Moreover, this model can be generalized to any language or domain. However, this model is appropriate only for records where sections are defined by headings.
The method of Chirila et al. [25] consists in supporting the prescription of drugs by structuring and categorizing the text into sections. For this, a machine learning model was trained to associate each part of the text with its appropriate section. According to the results of this method, the accuracy of the CNN based model is more superior especially with uniform name sections. Moreover, this method was applied on the Roman language where there is no dataset in this language with fully structured information. However, the execution time of CNN increases significantly when compared with a model based on Naive Bayesian classification. Moreover, this method is applied only on the Roman language.
Goenaga et al. [42] have tested rules and machine learning based methods for section identification on Spanish Electronic Discharge Summaries. They have found that machine learning-based method gives the best results. This method is based on transfer learning using FLAIR model [3] and generates character embeddings for sequence of tokens to annotate them by a BiLSTM-CRF model. Indeed, the rules have the lower results especially when an incorrectly marked section affects the surrounding sections even by carefully designing rules which is a time-consuming process. In contrast, the FLAIR method can identify sections even with variations or where headings are absent while it can learn from the headings and the vocabulary inside the sections. Also, training the method on data with more variability is useful to keep obtaining higher efficiency on different types of data. However, the results degrade when testing the trained model on different data and the degradation may be drastic in some cases. Furthermore, errors can be caused by high variability with the lack of training. In addition, implicit and mixed sections are the cause of several errors.
Nair et al. [100] have proposed a method to classify the sentences of i2b2 2010 clinical notes into different major SOAP sections using BiLSTM model with the fusion of Glove, Cui2Vec and ClinicalBERT embeddings. As advantage, the contextual embeddings and the transfer learning provide an efficient solution to this task. Also, the authors have found that 500 sentences per section is a sufficient starting point to achieve a high performance. However, they have considered only 4 sections and ignore other sections and sub-sections. Moreover, they have not considered the context-sensitivity of clinical sentences.
Hybrid methods
These methods use rules often for the construction of training data. These methods are weaker than rule-based methods, more ambitious than rule-based methods in dealing with different types of documents, and better than Machine Learning methods.
Jancsary et al. [55] have trained CRF to recognize (sub)sections in report dictations giving lexical, syntactic categories, BoW, semantic type and relative position features for each word. The training data is constructed by aligning the corrected and formatted medical reports with the text from automatic speech recognition while the annotations are generated by mapping (sub)headings to the (sub)section labels using regular heading grammar. As advantage, this method can detect various structural elements even without explicit dictated clues. Furthermore, it can automatically assign meaningful types for (sub)sections even in the absence of headings. In addition, it is still effective under ideal conditions and can deal with the errors of real-life dictation. However, the manual correction is required to solve the errors of the automatically generated annotations which impact the segmentation results.
Apostolova et al. [7] have constructed a training set by hand-crafted rules to train SVM to classify each medical report sentence into a semantic section using multiples sentence features such as orthography, boundary, cosine vector distance to sections and exact header matching. As advantage, a high-confidence training set is created automatically. Also, the classification of semantically related sections is significantly improved by boundary and formatting features. Furthermore, the segmentation problem could be solved when the NLP techniques are applied. Moreover, using SVM classifier outperforms a rules-based approach. However, it is hard to classify a section when its sentences are often interleaved with other sections.
Ni et al. [104] have classified medical document sections into pre-defined section types. These authors applied two advanced machine learning techniques: One is based on supervised learning and the other on unsupervised learning. For the supervised technique, a heuristic model pre-trained on old annotated documents is used to select a number of new candidate documents that will be annotated by people and will be used for learning. For the unsupervised technique, a mapping method was used to find and annotate sequences of words, which represent section titles, by their corresponding section types using a knowledge base. A maximum entropy Markov model was used for section classification. The chosen model is faster in learning and allows richer features. In fact, the techniques used can reduce the cost of annotation and allow a quick adaptation on new documents for section classification. In addition, both techniques can achieve high accuracy. However, the supervised technique requires more annotation cost than the other technique. In addition, the performance of the unsupervised technique is highly dependent on the quality of the knowledge base.
Dai et al. [29] have proposed a token-based sequential labeling method with the CRF model for section heading recognition using a set of word features such as affix, orthographic, lexicon, semantic and especially the layout features. To construct training data, they have employed section heading strings from terminology to make candidate annotations. Then, three experts are used to manually correct the annotations of top most section headings. As advantage, this was the first work which treats section detection as a token-based sequential labeling task and outperforms sentence-based formulation and dictionary-based approaches. This method has an integrated solution which avoids the development of heuristics rules to isolate heading from content. Also, layout features improve the results and can recognize section headings that are not appearing in the training set. However, it is difficult to recognize rare or nonstandard topmost section headings. In addition, subsections are not taken into consideration. Furthermore, some section headings are not the topmost in some records. Also, the absence of layout information can decrease the recall.
Sadoughi et al. [117] have applied section detection on clinical dictations in real time. They used automatic speech recognition to transform the speech into plain text. Also, a unidirectional LSTM model, which tracks short and long term dependencies, is run on the text to annotate its section boundaries using Word2vec vectors to represent the input words. To do this, regular expressions were applied on a set of reports to annotate the headings. Each time, a post-processing task is applied on each section to transform the text into a written report. As advantage, the post-processing task can become faster with the processing of a complete section each time, instead of re-executing after each dictated word. Thus, the post-processing of the previous section happens in parallel with the dictation of the current section without disturbing the user. Moreover, the post-processor can benefit from the full context of the section during the transformation. Thus, real-time section detection ensures that the medical report is directly usable for other processes after dictation. However, the detection of a section depends only on the words dictated so far without seeing the whole document. This prevents it from exploiting all the information in the document to provide a better quality result.
Methods dealt with issues related to the nature of entities
In this section, we highlight some solutions proposed by the state-of-the-art methods to solve problems related by the nature of the medical NER. Thus, we have cited and classified some methods into four categories based on the main problems: Ambiguity, Boundary detection, Name variation and Composed entities. Table 1 shows some of the methods used to deal with these various problems.
Table 1.
Some methods that addresses the issues related to the nature of named entities. The abbreviation "NI" in this table means Not Included
| Publication | Ambiguity | Boundary | Name variation | Composed entities |
|---|---|---|---|---|
| Lei et al. [76] | + Word segment and section information. | + Medical dictionary to segment words. | NI | NI |
| - Most of errors are in long entities. | ||||
| Quimbaya et al. [110] | - Ignore the context and surrounding words. | NI | + Edit distance, exact and lemmatized matching by a knowledge base. | NI |
| Xu et al. [150] | + Category Word2vec, PoS and dependence relations, and semantic correlation knowledge. - Filtering may miss some medical entities. | + Medical native noun phrases. + Based on knowledge base. - May obtain some inexact entities. | NI | + All medical native noun phrases. |
| Ghiasvand and Kate [41] | + Exact matching of unambiguous words from UMLS. | + Boundary expansion model trained on UMLS words. + Classify all possible noun phrases. - Noun phrase extraction not always perfect. - There are some nonnoun phrase entities. | + Lemma and stem forms as features. | + Complete parsing to extract all noun phrases. - Automatic noun phrase extraction is not always perfect. - Some entities not belong to noun phrases. |
| Zhou et al. [163] | + Word and character embeddings. - Capture the contextual relation on word-level. | - Can’t treat complex entities in phrase-level. | + Character representation can capture out-of-vocabulary words. | NI |
| Deng et al. [33] | + Learn contextual semantic information without feature engineering. + BiLSTM can learn the contextual dependences. + CRF can improve the annotation in phrase-level. | + Ensures the integrity and the accuracy of the entity by bidirectional storage of textual information. + IOB annotation format. + Avoid segmentation errors by character embeddings. - Nested entities results in unclear boundaries. | + Character embedding. | - Limited by the entity annotation granularity. |
| Zhao et al. [161] | + Extract lexical, contextual and syntactic clues. + Fine-tune BERT with BiLSTM-CRF. + Rules contextual embedding using ELMO model. | + Extract noun phrases in sentence by PoS patterns. | + Clues-based rules. - Rules not appropriate for other domains. | NI |
| Li et al. [78] | + Word2vec is improved by BiLSTM to capture contextual information. + BERT is better and can capture the context without BiLSTM. | + Relation classification between pair of spans is able to recognize discontinuous entities. | + ELMo character-level embedding. - Word-level embedding is needed to capture the whole meaning of words. | + Enumerates and represents all text spans and apply a relation classification. |
| Sui et al. [126] | + Interactions between the words, entity triggers and the whole sentence semantics. | NI | + Entity triggers to recognize entity by cue words. - Manual effort is required to prepare entity triggers. | + Cast the problem into a graph node classification task. |
Ambiguity
The ambiguity is when a medical named entity can belong to more than one class depending on the context. Thus, for some entities, we should explore the preceding words and the position of the entity in the text in order to know its meaning. Generally, that is the most important problem in entity recognition while most studies focus on how to provide more context to recognize the entity. As a special case, the abbreviations are likely to be ambiguous. Moreover, it is hard to determine the expansion of an abbreviation with a very small number of characters. Thus, an abbreviation mostly may have different meanings depending on the context. As common solutions, works try to enrich context information by word or character embedding, knowledge base and word position in the text such as section, surrounding words, PoS, etc. Also, they try to capture the contextual dependence and relation. Lei et al. [76] have reached a higher performance by merging the word segmentation and the section information. Ghiasvand and Kate [41] have benefited from UMLS which provide a lot of entity terms which are declared as unambiguous. Thus, they do an exact matching for these terms to annotate a maximum number of unambiguous entities to partially solve the ambiguity. Xu et al. [150] have benefited from more context to solve the ambiguity problem by using categories representation by Word2vec, PoS, dependency relation and semantic correlation knowledge. However, the method may miss some medical entities in the non-medical terms filtering step. The method of Zhou et al. [163] can solve the ambiguity problem by making two types of embeddings for more context, which are C-ELMo for word-level features and C-Flair for character-level. Likewise, the relationship between word-level contextual features can be captured by the C-ELMo model. However, this method fails to detect the boundary of complex phrase-level entities. Deng et al. [33] have used character embedding with BiLSTM-CRF to avoid feature engineering by learning the semantic information in the context. Thus, BiLSTM can provide more comprehensive contextual information and easily learn about contextual dependencies. Moreover, the CRF optimizes the result from the sentence level. However, this method is restricted by the entity labeling granularity where we can find some nested entities. The method of Zhao et al. [161] avoids the ambiguity as it automatically propagates some seed rules based on lexical or contextual clues which are strong indicators of entity recognition. In addition, the authors have fine-tuned a pre-trained contextual embedding model BERT in the biomedical domain. Also, they used a pre-trained contextual embedding model ELMO to give an average embedding for each rule to estimate the semantic relatedness between rules. Li et al. [78] have tested Word2vec by the help of BiLSTM to improve the results by capturing the contextual information. Indeed, BERT embedding alone is more effective than Word2vec and ELMo and it does not need BiLSTM since it has already captured the contextual information. The method of Sui et al. [126] is based on the interactions among the words, entity triggers and the whole sentence semantics to recognize the entity from its context.
Boundary detection
A method can recognize a part of a named entity and fail to determine the exact words which construct that entity. Thus, the method can miss some words from the full named entity, or may add surrounding words that do not belong to this entity. Indeed, searching for the presence of named entities and their approximate positions is easier than finding entities with their exact beginning and ending positions. Thus, these positions are named by the named entity boundary, where the boundary detection is known as an important challenge for medical named recognition methods. The most popular solutions to this problem is the extraction of noun phrases while the most of named entities are noun phrases or overlapped with noun phrases [159]. However, the noun phrase extraction is not always perfect and there are still some rare entities that do not belong to noun phrases. Also, the sequence tagging with the IOB annotation format, especially by BiLSTM-CRF model, can learn well how to determine the entity limit. However, this annotation is sensitive for nested entities. Lei et al. [76] use a Chinese medical dictionary as a knowledge source for word segmentation. Indeed, most errors appear in long entities. Xu et al. [150] have extracted medical native noun phrases in a boundary detection step. In addition, they have exploited a knowledge-driven method to detect boundary, by mapping text to concepts in offline and online lexical resources. Thus, the recognition performance is significantly improved. However, this method may still obtain inexact entities which show some decline in precision and recall. Ghiasvand and Kate [41] have trained a classifier by the medical terms found in UMLS to learn how to expand the boundary of words. Thus, the classifier is applied to all noun phrases in which the detected entity occurs in order to select the entity with the highest score. However, automatically obtaining noun phrases can make mistakes. Also, sometimes we may find named entities that are not noun phrases. In order to avoid incorrect identification for the entity boundary, Deng et al. [33] have ensured the integrity and accuracy of the named entity by the bidirectional storage of text information. In addition, the IOB labeling method is introduced. Also, they have used character-level embedding which can avoid poor segmentation. However, the phenomenon of nesting entities leads to unclear definition of boundary and results in poor accuracy. Zhao et al. [161] have extracted all noun phrases from each sentence as candidate entity mentions based on a set of PoS patterns. The method of Li et al. [78] apply a relation classification on each pair of candidate entity fragments to determine if it is a discontinuous entity or not.
Name variation
The same named entity can be written in different forms by adding and deleting some characters, changing the order of its component words or changing some words by synonyms. Also, we may have typos on a narrative text written by humans. Thus, an exact searching for named entities cannot cover all the forms of named entities. As common solutions, the preprocessing step is often used to transform words on unified form. Thus, we need some techniques that are able to recognize the named entity in any form in the text. Also, considering the surrounding words or characters may be useful to determinate the named entity. Another solution is to use the character embedding. The method of Quimbaya et al. [110] can solve the variation of named entities using exact, fuzzy and lemmatized matching by a knowledge base. However, it cannot take into consideration the context and the words surrounding the named entity. In the work of Ghiasvand and Kate [41], the lemmatize and stemming form of the words surrounding the entity in addition to other features are fed to a decision tree based classifier. Thus, that can tackle the variability problem. The method of Deng et al. [33] is based on the character embedding which can avoid the variability problem while it is not restricted by a vocabulary of words. Zhou et al. [163] can solve the variability by using character embedding to handle out-of-vocabulary words. Zhao et al. [161] can avoid the name variation problem where they define different types of rules which considers the lexical, contextual and syntax information based on the clues to find entities. However, some rules may not be applicable due to the mismatch between the training set and a different dataset. Li et al. [78] have tested the ELMo character-level embedding which is able to represent out-of-vocabulary words. However, the characters can’t capture the whole meaning of words and should be merged with word-level embedding. Sui et al. [126] have added entity triggers to help the model recognizing the entity by the surrounding cue words. However, this method requires manual effort by annotators to annotate a large group of words to prepare entity triggers.
Composed entity
The named entity can be composed by multiple words and may be a long phrase. Hence, the different named entities are able to overlap with each other. Consequently, we may find a nested named entity which is included in another one and belong to a different class. Thus, the granularity level should be considered to recognize all the named entities that can be found in one longer named entity. To solve this problem, some works extracts all possible noun phrases including the nested ones. But automatically extracting noun phrase is still not perfect. Also, few named entities are not overlapping with noun phrases. Xu et al. [150] can handle nested entities by identifying entity candidates based on the dependency relationships between words. Thus, medical native noun phrases, such as single nouns and maximum noun phrases, are extracted. The method of Ghiasvand and Kate [41] is able to detect nested entities by obtaining all noun phrases, with nested ones, using a full parsing. The method of Li et al. [78] enumerates and represents all possible text spans [89] to recognize the overlapped entities. Thus, a relation classification is applied to judge whether a pair of entity fragments to be overlapping or succession. Sui et al. [126] have proposed a cost-effective and efficient trigger-based graph neural network to cast the problem into a graph node classification task.
Knowledge discovery methods
In this section, we cite examples of studies that applicate text mining methods to discover new knowledge in medical field. Thus, this knowledge can be used to make medical decisions or to gather useful information and conclusions for some medical tasks. Indeed, knowledge discovery is the final and the most important step in medical data processing. This step consists in discovering a needed knowledge from the extracted information which directly supports the medical staff.
The method of Sudeshna et al. [125] is able to predict the probability of having heart disease by using the supervised SVM algorithm to classify the data. This method takes particular symptoms, given by the patient, and the patient’s health record. Based on this data, this method can suggest diseases, treatments, medications and dietary habits for the doctor. The latter can confirm and send the information to the patient. As advantage, this system is reliable, easy to understand and adaptive. Also, the disease is automatically analyzed more efficiently and is easily identified. In addition, this method can identify the best medication for the disease.
Aich et al. [2] have proposed a method to analyze abstracts of articles related to Parkinson’s disease to automatically find the relationship between walking and Parkinson’s disease. This method is based on text mining. It does a simple pre-processing on the text. Then, it provides a graphical visualization to indicate the word frequencies through a Word Cloud. Also, it categorizes the similar terms using a hierarchical classification and K-means based on a distance calculation between words. Indeed, this approach has a great potential to classify a text into different groups. However, this method can be improved by providing more articles to be analyzed or by exploring other classification approaches.
The review of Al-Dafas et al. [4] have shown that applying algorithms of data mining techniques on patients’ health data is able to help doctors to make the right decision at the right time. Thus, these techniques can detect the cancer disease early without surgical intervention. As advantage, they can reduce treatment costs and medical errors in diagnosing the disease. The authors suggest to adapt machine learning-based data mining in the medical diagnosis process.
Benchmark datasets and supplement resources
In this section, we cite the most important benchmark datasets used in the IE task which are manually annotated and considered as gold standard. Most of these datasets are used in shared tasks and used to train and evaluate IE state-of-the-art methods. Table 2 shows some details about these datasets. Generally, the annotation focus on information related to diseases, medicament and chemical entities. Many datasets are constructed from PubMed articles and especially the abstracts because they are easier to collect. Other datasets are obtained from discharge summaries and clinical reports which are de-identified to hide personal information. However, the available datasets are in textual form while the medical documents are originally available on PDF form. Thus, useful information is not available such as formatting style, which is important to define the document structure. Indeed, there is some recent datasets Cohen et al. [28], Islamaj et al. [53] obtained from full-text article which can provide richer information rather than just an abstract.
Table 2.
Benchmark datasets used in IE.
| Dataset | Size | Data type | Information | Statistics | Data source | Annotation method |
|---|---|---|---|---|---|---|
| i2b2 2009 [138] (Shared task) | 170 train, 251 test, 679 un-annotated | Discharge summary | Medication-related information: medications, dosages, modes, frequencies, durations, reason for administration. | 27589 mentions: - 12773 medications - 4791 dosages - 3552 modes - 4342 frequencies - 597 duration - 1534 reasons | Partners Healthcare | Annotation by physician and revision by researcher |
| i2b2 2010 [139] (Shared task) | 394 train, 477 test, 877 un-annotated | Discharge summaries | - Entity types: Medical problem, Treatment and Test. - Assertions: Present, Absent, Possible, Conditional, Hypothetical and Not associated with the patient. - Relations (11): Treatment improves medical problem, Treatment causes medical problem, Test reveals medical problem, Medical problem indicates medical problem, etc. | - 30518 tests - 20268 problems - 22060 treatments - 14333 relations | Partners Healthcare, Beth Israel Deaconess Medical Center and University of Pittsburgh Medical Center. | Partnered with VA Salt Lake City Health Care System. |
| i2b2 2014 [123, 124] (Shared task) | 296 patients: 790 train, 514 test records | longitudinal records for patients | Protected health information (18): name, geographic, date, phone, e-mail, etc. | 28872 mentions | Partners Healthcare | 6 annotators: double annotation per patient followed by adjudication. |
| n2c2 2018 [50] (Shared task) | 303 train, 202 test | Discharge summaries | - Entity types: Concepts related to medications, their signature information, and Adverse drug events (allergic reactions, drug interactions, overdoses, and medication errors). - Relation types: Linking concepts to their medication. | - 83869 concepts. - 59810 relations | MIMIC-III clinical care database. | 2 independent annotators while a third annotator resolved conflicts. |
| n2c2 2019 [145] (Shared task) | 113000 notes: 1642 train, 412 test sentence pairs | Clinical notes | Clinical semantic textual similarity | 2054 sentence pairs | n2c2 2018 + 2 electronic health record systems, GE and Epic | 2 clinical experts for independent annotation |
| SemEval 2014 task 7 [108] (Shared task) | 199 train, 99 valid, 133 test | Clinical reports: Discharge summaries, echo-cardiogram reports, electrocardiograph reports and radiology reports. | Entity type: Disease/Disorder | 19165 mentions | ShARe (clinical notes from MIMIC II) | 21 participants. |
| SemEval 2015 task 14 [38] (Shared task) | 298 train, 133 valid, 100 test | Discharge summaries and radiology reports | Disorder attributes: mentions, normalization, negation, subject, uncertainty, course, severity, conditional, generic, body location | 19111 mentions | ShARe | Double annotation and adjudication by professional trained coders. |
| SemEval 2016 task 12 [16] (Shared task) | 293 train, 147 valid, 151 test | Clinical notes and pathology reports | - Time expression identification - Event expression identification - Temporal relation identification | - 7863 time expressions - 78854 event expressions - 23243 temporal relations | Mayo Clinic cancer patients | Manually annotated and revised by the THYME project |
| SemEval 2017 Task 12 [17] (Shared task) | 621 train, 296 valid, 299 test | Clinical notes and pathology reports | - Time expression identification - Event expression identification - Temporal relation identification | - 18623 time expressions - 130293 event expressions - 31169 temporal relations | Mayo Clinic cancer patients | Manually annotated and revised by the THYME project |
| SemEval 2018 Task 6 [72] (Shared task) | 232 train, 35 valid, 141 test | Clinical notes | Parsing time normalization | 27362 time entities | THYME | Linguistic students |
| Semeval 2021 task 10A [73] (Shared task) | 10259 train, 5545 valid, 9580 test, 622703 un-annotated instances | Clinical notes | Negation detection | 22409 asserted, 2975 negated | SHARP Seed, i2b2 2010, MIMIC II | Manual annotation |
| Semeval 2021 task 10B [73] (Shared task) | 278 train, 99 valid, 17 test, 47 un-annotated | Clinical notes | Time expression recognition | 22151 time entities | THYME, TimeBank | Two independent annotators and an adjudicator |
| NCBI-disease 2014 [35] | 593 train, 100 valid, 100 test | PubMed abstracts | Entity type: Disease | 6892 mentions | MeSH, OMIM | 14 annotators, 2 per document: annotation in 3 phases with checking for corpus-wide consistency of annotations. |
| BC5CDR 2015 [79] (Shared task) | 500 train, 500 valid, 500 test | PubMed abstracts | - Entity types: Disease and Chemical. - Relation types: chemical-induced disease. | - 4409 disease - 5818 chemical - 3116 chemical-induced disease relations | Most were selected from an existing CTD-Pfizer collaboration-related dataset. | MeSH annotators were recruited for manual annotation. |
| CRAFT 2017 [27] | 97 | PubMed full-text articles | - Entity types: Gene, Chemical, Protein, Organism, Cell and Taxonomy. - Structural annotation : syntax and document structure. - Co-reference annotation. | 140000 concept mentions | PMC Open Access subset | Manual annotation using an annotation model and guidelines by the help of a realist ontologies. |
| NLM Chem 2021 [53] (Shared task) | 80 train, 20 valid, 50 test | PubMed full-text journal articles | - Entity type: Chemical. - Segments: sections, paragraphs, figure caption, titles, etc. | 38342 mentions | PMC Open Access dataset | Doubly annotated by ten expert NLM indexers. |
Also, many methods use supplement resources such as dictionaries and ontologies. The most popular resources that used in IE are UMLS [19] meta-thesaurus and Systematized Nomenclature of Medicine-Clinical Terms (SNOMED-CT) [36] ontology. The UMLS meta-thesaurus in its 2021 update can cover 16543671 terms in different languages (11755677 in English) which are associated with 4536653 medical concepts. This large biomedical thesaurus is collected from different sources such as SNOMED-CT, MEDLINE, MeSH, NCBI, etc. Also, it provides 127 semantic types of the concepts such as Disease/Syndrome, Clinical drug, Therapeutic or preventive procedure, laboratory or test result, etc. Furthermore, it provides 54 semantic relations between these semantic types, such as Process of, Result of, Property of, Part of, Associated with, Complicates, Affects, Causes, etc.
The SNOMED-CT ontology covers more than 1314668 clinical terms with their description and they are associated with different concepts. There are 19 top-level concepts which are body structure, finding, event, environment / location, linkage concept, observable entity, organism, product, physical force, physical object, procedure, qualifier value, record artifact, situation, social context, special concept, specimen, staging scale and substance. In addition, it provides more than 3092749 relation mentions of 122 types of relations such as “is a”, “finding site”, “associated morphology”, “method”, “causative agent”, etc.
Experimental analysis
In this section, we have collected results of some methods for each IE task such as NER, RE and section detection. Thus, we have organized these methods according to the used technique, dataset and features. Hence, we can make an experimental analysis according to their available results. As a look at all the results in general, we find that most methods use the machine learning technique. For the NER, the best result is achieved by a machine learning-based method, while a rule-based method gets the best result for RE and a hybrid method is the best for section detection.
Named Entity Recognition: Tables 3, 4 and 5 show the results of rule-based, dictionary-based and machine learning-based named entity recognition methods, respectively. The A, P, R and F symbols refer to Precision, Recall and f1-score, respectively. The best f1-score (96.05%) was for the method of Popovski et al. [107] which uses rules based on computational linguistics and semantic information to describe food concepts in order to annotate entities in 1000 food recipes. This method is aimed to extract food entities which can be useful in medical field such as food safety. For disease and symptoms and some other named entity types, the method of Deng et al. [33] , based on machine learning and tested on 20% characters from 1600 TCM patents’ abstracts, gets the best result in terms of f1-score (94.48%). This method is based on fine-tuning character embeddings with the use of BiLSTM-CRF model. For the i2b2-2010 benchmark dataset, a model based on machine learning gets the best result in terms of f1-score (87.45%), which uses LSTM-CRF model with contextualized and GloVe embeddings. Thus, machine learning-based model using context-specific embedding with the model based on LSTM and CRF can get better results. In fact, the unsupervised models [41, 159], predictably, get the worst results but it was acceptable (51.06% and 59.43% f1-score), and it is worth avoiding manual effort.
Table 3.
Rule-based NER methods
| Publication | Dataset | Dataset size | Method | Features | P | R | F |
|---|---|---|---|---|---|---|---|
| Popovski et al. [107] | FoodBase corpus annotated by food concepts | 1000 recipes | Pre-processing + morphological analysis + rule engine | Computational linguistics + semantic information | 97.80% | 94.37% | 96.05% |
| Zhao et al. [161] | NCBI-disease 2014 | 593 train, 100 valid, 100 test | Propagate seeding rules in graph + LinkedHMM + BiLSTM-CRF | Contextual embedding + BERT | 89.9% | 73.2% | 80.2% |
| Zhao et al. [161] | BC5CDR 2015 | 500 train, 500 valid, 500 test | Propagate seeding rules in graph + LinkedHMM + BiLSTM-CRF | Contextual embedding + BERT | 88.2% | 84.6% | 86.3% |
| Zhao et al. [161] | LaptopReview 2016 laptop aspect terms | 3048 train, 800 test sentences | Propagate seeding rules in graph + LinkedHMM + BiLSTM-CRF | Contextual embedding + BERT | 82.4% | 64.2% | 72.2% |
Table 4.
Dictionary-based NER methods
| Publication | Dataset | Dataset size | Method | Features | P | R | F |
|---|---|---|---|---|---|---|---|
| Sun and Bhatia [127] | i2b2 2009 | 170 train, 256 test | BiLSTM-CRF and gazetteer model combined on a shared tagger | Word/character embedding RoBERTa-mimic + Gazetteer embedding | - | - | 92.35% |
| Zhang and Elhadad [159] | i2b2 2010 | 170 train, 477 test | Unsupervised annotation and boundary detection: noun phrase chunker + IDF filter + Cosine similarity | UMLS + TF-IDF signature vector | 49.73% | 54% | 51.06% |
| Ghiasvand and Kate [41] | i2b2 2010 | 349 train, 477 test | Unsupervised Decision Forest classifier for annotation and boundary detection | UMLS + PoS + lemma + stem | 52.66% | 68.86% | 59.43% |
| Sun and Bhatia [127] | i2b2 2010 | 170 train, 256 test | BiLSTM-CRF and gazetteer model combined on a shared tagger | word/character embedding RoBERTa-mimic + Gazetteer embedding | - | - | 87.41% |
| Kim et al. [60] | n2c2 2019 for family history extraction | 99 train, 117 test | Voting ensemble of BiLSTM models + heuristic rules + OGD + ConText | UMLS + PoS + dependency-based embeddings + static embedding + context-dependent embedding + lexical | 84.83% | 87.24% | 86.02% |
| Ghiasvand and Kate [41] | SemEval 2014 Task 7 | 199 train, 133 test | Unsupervised Decision Forest classifier for annotation and boundary detection | UMLS + PoS + lemma + stem | 88.1% | 69% | 77.3% |
| Sun and Bhatia [127] | DCN [18] de-identified clinical notes with medications and medical conditions annotations | 1500 | BiLSTM-CRF and gazetteer model combined on a shared tagger | Word/character embedding RoBERTa-mimic + Gazetteer embedding | - | - | 84.59% |
Table 5.
Machine learning-based NER methods
| Publication | Dataset | Dataset size | Method | Features | P | R | F |
|---|---|---|---|---|---|---|---|
| Tang et al. [132] | i2b2 2010 | 349 train, 477 test | SSVM | Word + context + sentence + section + cTAKES + MetaMap + ConText + Brown clustering | 87.38% | 84.31% | 85.82% |
| Wu et al. [148] | i2b2 2010 | 349 train, 477 test | LSTM | Word embedding | 85.33% | 86.56% | 85.94% |
| Lee et al. [74] | i2b2 2010 | 170 train, 256 test | BERT model | Pre-trained and fine-tuned BioBERT | - | - | 86.46% |
| Zhou et al. [163] | i2b2 2010 | 349 train, 477 test | LSTM-CRF | Pre-trained contextualized embeddings + Glove embedding | - | - | 87.45% |
| Zhou et al. [163] | NCBI-disease 2014 | 593 train, 100 valid, 100 test | LSTM-CRF | Pre-trained contextualized embeddings + Glove embedding | - | - | 87.88% |
| Lee et al. [74] | NCBI-disease 2014 | 593 train, 100 valid, 100 test | BERT model | Pre-trained and fine-tuned BioBERT | - | - | 89.36% |
| Yang et al. [153] | n2c2 2019 for family history extraction | 99 train, 117 test | majority voting of LSTM-CRF models with BERT fine-tuning | Fasttext embedding + pre-trained BERT | 79.69% | 79.20% | 79.44% |
| Uzuner et al. [139] | SemEval 2014 Task 7A | 199 train, 99 valid, 133 test | CRF models | Textual features enhanced with a rule-based system | 91.1% | 85.6% | 88.3% |
| Vunikili et al. [140] | CANTEMIST-NER sub-task with tumor morphology mentions | 501 train, 500 valid, 5232 test | Transfer learning to fine-tune the BERT model | BERT contextual embeddings pre-trained on general domain Spanish text | 72.7% | 74.1% | 73.4% |
| Deng et al. [33] | crawled TCM patents’ abstract texts annotated with herb names, disease names, symptoms, and therapeutic effects. | 1600 copies: 60% train, 20% valid, 20% test characters | BiLSTM-CRF | Pre-trained and fine-tuned character embedding | 94.63% | 94.47% | 94.48% |
| Zhou et al. [163] | MACCROBAT 2018 case reports | 160 train, 20 valid, 20 test | LSTM-CRF | Pre-trained contextualized embeddings + Glove embedding | - | - | 65.75% |
| Lee et al. [74] | MACCROBAT 2018 case reports | 160 train, 20 valid, 20 test | BERT model | Pre-trained and fine-tuned BioBERT | - | - | 64.38% |
Relation Extraction: Tables 6 and 7 show the results of machine learning-based and rule-based RE methods, respectively. The P, R and F symbols refer to Precision, Recall and f1-score, respectively.. The best method [154] gets the best f1-score (96.06%) which have used rules to generate candidates and classify them based on clinical BERT model. It was trained and tested on n2c2 2018 benchmark dataset and thus it is the best method applied on this dataset which is annotated by relations linking concepts to their medication. In fact, this method can be considered as hybrid-based method while it uses the rules with machine learning. The best machine learning-based methods [95, 146] have achieved 94% f1-score and have trained and evaluated on n2c2 2018 benchmark dataset to link concepts to their medication. These methods have used a fine-tuned BERT as embedding. However, BERT embedding is not suitable neither for i2b2 2010 dataset, which is annotated by relations between medical problem, treatment and test entities, nor for n2c2 2019 dataset which is annotated by relations between family members, observations and living status. The methods [146, 153] applied on these datasets with fine-tuned BERT have almost the worst results. Indeed, there is machine learning based method [119] applied on the text of tweets collected from Twitter social media. As expected, it gets the worst result in terms of f1-score (56.7%) while it is a very difficult task to apply a medical IE on the social media text. This type of text is less structured compared to professional documents and articles while the writer of tweets is often not an expert on medicine and does not pay attention to correct writing. Thus, getting 54% of precision and 63% of recall and 56.7% of f1-score in this type of text is an appropriate result.
Table 6.
Machine learning-based RE methods
| Publication | Dataset | Dataset size | Method | Features | P | R | F |
|---|---|---|---|---|---|---|---|
| Wei et al. [146] | n2c2 2018 | 303 train, 202 test | Multi-class classification with BERT model | BERT fine-tuned on MIMIC-III | 98.38% | 90.15% | 94.09% |
| Mahendran and McInnes [95] | n2c2 2018 | 303 train, 202 test | Binary classification with BERT model | Fine-tuned BERT | 93% | 96% | 94% |
| Wei et al. [146] | i2b2 2010 | 170 train, 256 test | multi-class classification with BERT model | BERT fine-tuned on MIMIC-III | 76.24% | 77.34% | 76.79% |
| Hasan et al. [48] | i2b2 2010 | 170 train, 256 test | BiLSTM | Word2vec, relative distances, PoS, Concept embedding, dependency tree | - | - | 88.08% |
| Yang et al. [153] | n2c2 2019 for family history extraction | 99 train, 117 test | Majority voting of LSTM-CRF models with BERT fine-tuning | Fasttext embedding + pre-trained BERT | 69.95% | 61.84% | 65.44% |
| Kim et al. [60] | n2c2 2019 for family history extraction | 99 train, 117 test | Voting ensemble of BiLSTM models + heuristic rules + OGD + ConText | UMLS + PoS + dependency-based embeddings + static embedding + context-dependent embedding + lexical | 73.27% | 71.70% | 72.48% |
| Ben Abdessalem Karaa et al. [14] | MEDLINE 2001 abstracts from biomedical journals annotated by cure and side effect relations | 4600+ abstracts: 75% train, 25% test sentences | SVM model | UMLS + frequency + lexical + morphological + syntactic + semantic | 86.51% | 91.56% | 88.78% |
| Shi et al. [119] | COVID-19 Twitter dataset with concern categories | 1418 train, 355 test | BiLSTM-CRF to detect entities + BiGCN with shared BiLSTM hidden state | BERT embeddings + PoS + syntactic dependency relation + concern score and type + sentiment polarity and retweet count | 54.5% | 63% | 56.7% |
Table 7.
Rule-based RE methods
| Publication | Dataset | Dataset size | Method | Features | P | R | F |
|---|---|---|---|---|---|---|---|
| Yang et al. [154] | n2c2 2018 | 303 train, 202 test | Rules to generate candidates + binary classification | Clinical BERT + RoBERTa + XLNet | 97.01% | 95.12% | 96.06% |
| Yang et al. [154] | MADE1.02018 [54] fully de-identified longitudinal EHR notes | 876 train, 213 test | Rules to generate candidates + binary classification | Clinical BERT + RoBERTa + XLNet | 91.26% | 87.99% | 89.59% |
| Wang et al. [142] | MEDLINE 2001 abstracts from biomedical journals annotated by cure and side effect relations | 4600+ abstracts: 75% train, 25% test sentences | Pattern to extract candidate pairs + generate the degree of correlation | UMLS + lexical + network embedding | 91.75% | 86.55% | 89.025% |
Section Detection: Tables 8, 9 and 10 show the results of rule-based, machine learning-based and hybrid section detection methods, respectively. The A, P, R and F symbols refer to Accuracy, Precision, Recall and F1-score, respectively. In fact, the section identification methods are not well experimented while this task needs a more standardized evaluation method such as a benchmark dataset. Moreover, some methods use different segmentation and granularity level which make comparing state-of-the-art so complicated. However, according to the results, the best method according to the accuracy metric is a rule-based method [91] which has used a simple regular expression to match titles from a set of section titles and is tested on 3 datasets. The maximum accuracy score (90%) was achieved using 1630 prospectuses. This method outperforms a machine learning based method in terms of accuracy, and this is the only available accuracy result we have. However, when a larger dataset is used the result may decrease significantly. Also, it may differ from a dataset to another while the regular expressions may be inappropriate for other types of documents. For example, the method applied on 3814 prospectuses gets 88% of accuracy while the same method applied on 3002 prospectuses from another source get 66% of accuracy. The best method in terms of f1-score (94.19%) is a hybrid method [29] which uses a terminology to make a semi-supervised annotation of the training set and train a CRF model on layout, affix, orthographic, lexicon and semantic features for sentence tagging. This method is trained and evaluated on a dataset which is already considered as a benchmark for another different IE task and contains patient records such as discharge summaries and procedural notes, etc. Also, this method has the best result in terms of f1-score among the methods applied on discharge summaries. In second place, a method based on machine learning has achieved 93.03% f1-score which uses BiLSTM-CRF with FLAIR character embedding fine-tuning on reports of hospital discharges. Thus, the use of CRF model can lead to the best results in section detection task.
Table 8.
Rule-based section detection methods
| Publication | Dataset | Dataset size | Segment | Method | Features | A | P | R | F |
|---|---|---|---|---|---|---|---|---|---|
| Lupşe and Stoicu-Tivadar [91] | Pagina Farmacistilor prospectuses | 1630 | Text | Regular expressions | Set of section titles | 90% | - | - | - |
| Lupşe and Stoicu-Tivadar [91] | HelpNet prospectuses | 3002 | Text | Regular expressions | Set of section titles | 66% | - | - | - |
| Lupşe and Stoicu-Tivadar [91] | CSID prospectuses | 3814 | Text | Regular expressions | Set of section titles | 88% | - | - | - |
| Lee and Choi [75] | Korean discharge summaries of rheumatism patients | 50 train, 15 valid, 30 test | Chunk | Collect patterns + multiple pattern matchings | text | - | 84.1% | 88.2% | 86% |
Table 9.
Machine learning-based section detection methods
| Publication | Dataset | Dataset size | Segment | Method | Features | A | P | R | F |
|---|---|---|---|---|---|---|---|---|---|
| Lohr et al. [87] | German discharge summaries | 1106 | Sentence | Machine learning baseline classifier | BoW statistics | - | 82% | 84% | 82% |
| Chirila et al. [25] | Pagina Farmacistilor + HelpNet + CSID prospectuses | 8147 prospectuses: 70% train, 30% test sentences | Sentence | Uniform section names + CNN | Embedding information | 86.55% | - | - | - |
| Goenaga et al. [42] | Clinical reports of long-term hospital discharges | 100 train, 100 dev, 100 test | Token | BiLSTM-CRF + FLAIR model fine-tuning | FLAIR character embeddings | - | 93.40% | 92.55% | 93.03% |
| Nair et al. [100] | i2b2 2010 | 427 notes: 80% train, 10% valid, 10% test sentences | Sentence | BiLSTM to classify sentences | Glove + Cui2Vec + ClinicalBERT | - | - | - | 88.68% |
Table 10.
Hybrid section detection methods
| Publication | Dataset | Dataset size | Segment | Method | Features | P | R | F |
|---|---|---|---|---|---|---|---|---|
| Dai et al. [29] | i2b2 2014 shared task (track 2) discharge summaries, procedural notes, emails between the primary physician and the consultant | 521 train, 269 valid, 514 test | Sentence | Employ heading strings from terminology to make train set with little manual correction + CRF | Terminology + layout + affix + orthographic + lexicon + semantic | 96.04% | 92.4% | 94.19% |
| Sadoughi et al. [117] | Medical reports with their parallel ASR hypotheses | 9073 train, 575 valid, 597 test | Token | Regular expressions to annotate headings + unidirectional LSTM | Word2vec | 84.4% | 70.3% | 76.7% |
Literature review
In the last years, we can see that the machine learning methods are well studied in the IE, where many models are tested with many types of features. Generally, CRF is the most popular model which is a sequence of tagging model while the problems are mostly defined as tokens labeling especially in the named recognition task. Recent works have showed that the fusion between BiLSTM and CRF give better result [33, 119, 161], because BiLSTM can consider the order from double directions which makes it able to well understand a sequence of features. Hence, CRF can perform an accurate labeling using the features provided by BiLSTM.
However, the manual effort problem is one of the most important challenges in this field. Many methods are destined to search an automatized or semi-automatized techniques to obtain a high quality annotated dataset due to the lack of training data [41, 65, 161]. In addition, there are different fields and different types of data, and the medical field is evolving day after day, which makes one annotated training set not enough to be used permanently. A manually annotating dataset need much effort and is time consuming, and that is why we need to automatize this step to enable the model to easily adapt new types of data. Comparing to rules, a machine learning model can learn implicit and deep rules using automatically generated features which make possible to explore and predict the best output in many new cases, whereas the rules usually only deal with pre-defined cases although they can be smooth to exploit the context and knowledge. In fact, rules are more precise when they can be exactly matched on the data, but cannot discover new knowledge beyond the defined rules. Also, rules fail in many cases, especially for variable data.
Indeed, some methods try to automatize the construction of rules. An important recent study [161] uses graph propagation method to find new rules giving few seeding manual rules that are easily constructed by experts. Thus, even rules can be automatically generated. In addition, the graph based techniques can be exploited, while they are able to treat the relationships inside data. The synonyms and antonyms relations, contextual similarity and many other relation types between words and phrases are very important and can be well exploited using a graph technique. In addition, even the social media can be very critical area for this task, where the data is more noised and contains many grammatical mistakes, and especially false information by which people can be influenced. Some recent works are aimed for social media [61, 119], for example, Shi et al. [119] have used machine learning models by the help of Concern Graph to apply entity recognition on pandemic concern entities in Twitter. Recently, the social media have played an important role during the COVID-19 pandemic period and it is crucial to automatically understand and supervise a lot of people’s interactions. Indeed, social media is a rich source of information which mostly contains unstructured and confused textual and other multimedia data. Thus, some studies are applied to extract information from that data in order to perform some tasks such as associating tags to posts [71], identifying relevant information [94], etc. It is worth noting that the graph of users’ relations such as following relations can be exploited too to enhance the medical IE. Thus, some important tasks such as community detection and influence identification [45, 56, 63, 99] can be combined with medical IE tasks in social media.
Talking about supplement resources, is good to construct a big resource which tries to cover all things, but it cannot be. Some studies try to automatically update these resources by an online phase, for example, by using a search engine [150]. Indeed, the most used additional resources are UMLS [19] and SNOMED-CT [36] that can cover a lot of concepts, languages, semantic types and much information about terms, which make it possible to even annotate a raw text or extract more information using some matching techniques. Some studies tried to find a smooth technique to use the information inside dictionary in order to overcome the limited coverage. For example, new relationships can be learned using the synonym relationships provided by the SNOMED-CT ontology using CNN model [8]. Also, we can make a gazetteer tagger using the power of machine learning which learns features from a dictionary to provide an output which improves another machine learning model [127]. Indeed, the relation between machine learning and dictionary based methods is well studied, while using machine learning can make the use of a dictionary more robust to override the limits of the resource. Thus, these studies try to find a better way to exploit the knowledge of ontologies. Furthermore, the knowledge inside the dictionary can be used as additional feature for machine learning models [102] and it gives a useful information about the context.
Talking about features, most recent works focus on distributed representation for words as well for characters such as BERT and Word2vec, which can well extract context information for machine learning methods. Furthermore, the models which generate these embeddings can be used in different manners, where they can be pre-trained from other sources, trained during the whole model training or fine-tuned. In particular pre-trained and fine-tuned language models such as BERT have achieved state-of-the-art performance on many natural language processing tasks. For example, the work of Yang et al. [154] have showed that clinical pre-trained transformers achieve better performance for RE. In addition, other types of features can be beneficial such as knowledge and syntactic features. Indeed, there is some information that is not well exploited although that has showed a good potential to improve the IE task. Generally, this task can be improved by giving more context. Indeed, the study of Lei et al. [76] have proved that section information is able to improve the entity recognition task, but it is not well exploited in this field. In addition, Tran and Kavuluru[136] have used sub-headings to improve the RE. Furthermore, the formatting style of the document can be very important, while it have proved its ability to detect section titles in the study of Beel et al. [13] using only font size information. A medical document is generally created on PDF format which provides more useful information than a raw text. However, all available benchmark datasets are provided only as an annotated raw text. Furthermore, most datasets which contain medical articles are available only with annotated titles and abstracts. Recently, some researches [53, 65] are trying to construct new annotated datasets especially for NER with full-text articles which frequently contain more detailed information. These articles are usually collected from PMC [112].
Concerning the named entities, most works focus on the disease entity type, even most datasets [35, 79, 108] and the widely used meta-thesaurus [19] provide annotations and information about this type of entity. Thus, extract information about disease in medical texts is a very important task in the medical field. Indeed, the named entity boundary problem is another challenge [33, 41, 150], while solving this problem lead to obtain a better result especially according to the exact matching metric. However, this metric always give a very low score comparing to partial matching metric. Generally, this problem is related to the noun phrase chunking which is adopted by most works.
A lot of studies in the medical IE are destined for the Chinese language. Although English language is more suitable for this task, the Chinese researchers are trying to improve this task for them while they are very interested by the evolution in medical field generally. However, the Chinese language in medical field is more difficult compared to English especially on the segmentation while it has complicated syntax rules, and on the lack of Chinese data. Hence, many studies are destined to deal with this language problems [33, 76, 150]. The segmentation is generally used to provide samples and extract features from them. It can be performed on word, phrase, sentence and section level. Indeed, Deng et al. [33] found that making features for a sequence of characters is more suitable especially for Chinese language.
Conclusion and Future Research Directions
The IE in the medical field is very interesting especially to find information about diseases. Generally, this task provides more knowledge, supports persons to find relevant information and helps doctors to release the best decision, for example, to choose the right treatment, make the appropriate drug prescription or discover causes and effects of some diseases. A large amount of unstructured medical textual information is terrible to be manually analyzed by doctors while is considered as a heavy treasure of information.
In our survey, we conclude that the rule-based and hybrid methods are generally the promising techniques for IE where they have shown the best results. However, the rules depend highly on a specific domain. Thus, it is difficult to adapt these methods on new type of data since a manual effort by domain experts is needed. Thus, generating rules that are constructed dynamically to adapt the data type is a promising direction. Most methods focus on the combination of CRF and BiLSTM models which is very beneficial for sequence-tagging tasks. However, the CRF model is usually used for flat NER, which is not appropriate for nested and discontinuous named entities. Domain-specific embeddings, especially which are provided by BERT model, are used by many methods and give better results. There is a lack of the medical data where these data contain more privacy compared to other fields. The manual annotation is also a big problem for machine learning and many recent methods are going to automate it. Thus, it is a challenging issue to provide a high quality data for training and other supplement data which can cover the new medical terms, several types of data, all possible cases, multiple languages, different types of labels, etc. Also, all datasets are available in textual format while the formatting style is very important and should be more exploited. Indeed, it adds a very useful information on the text which is especially used to understand the structure of the document and even the meaning of words.
Indeed, section detection is a challenging issue and showed a positive impact on the performance of several IE tasks. Mainly, the position of a concept in a document can provide more contextual information. However, this task is not well covered by the research work and methods. Thus, benchmark datasets should be constructed for it. Another issue is combining rule-based, dictionary-based and deep-learning approaches to well benefit from them in one hybrid method. Many ideas have been proposed and can be exploited further. Indeed, we can use rules to prepare data for machine learning or we can use machine learning to generate rules. Besides, we can ameliorate the dictionary matching by machine learning or we can annotate a lot of data by a dictionary to train a machine learning model. Also, we can use the rules by constructing regular expressions to perform a dictionary matching or we can use dictionary as a supplement resource to support the rules. In addition, we can make features by using dictionary and rules based techniques.
Some important directions about IE in medical documents are not well covered by our analysis. Thus, the research themes in these directions can be further developed since they have provided promising results. Indeed, the difficulty of handling multi-language documents is an interesting issue which makes it hard to adapt the model for data with different languages. Hence, it is good if we can benefit from data of multiple languages. For example, we can use the transfer learning in order to benefit from the knowledge learned from a pre-trained model. Consequently, the model will be able to adapt the knowledge on another language rather than training the model from scratch on one language. Thus, we can combine the learned knowledge from more than one dataset of different languages. Generally, adapting a method on different languages can solve the lack of data and benefit from the knowledge gathered from different data sources. In addition, while most of researches are aimed for English and some for Chinese language, the other languages have a poor chance to be well treated. Therefore, unifying multiple languages in one method would be a really important achievement.
In fact, document summarization is another major and very challenging issue. This task can depend on the NER while it consists in making a text summarization which contains only the most important information. Hence, we can easily recognize a relevant and a very reduced readable part of text in a document instead of reading a whole text. Thus, the useless part of text can be eliminated even for other tasks of IE. Due to the diversity and the growing quantity of medical information, persons need to quickly assimilate and determine the content of a medical document. Thus, document summarization helps persons to quickly determine the main points of a document. However, this research field has not yet reached maturity, while variety of challenges still needs to be overcome such as handling large scale data, providing sufficient annotated data, etc.
Another important issue and possible research direction, which has been discovered especially during the COVID-19 pandemic, is about analyzing the propagated medical information in social media. The content on social media is very different from documents especially when the information is written by normal users and not medical experts. Thus, the natural language processing will be much more difficult while we can find unstructured texts and many typos. As well, we can easily find a big number of users influenced by false medical information which represents a critical problem. Likewise, the social media environment is rich of useful information more than just a document. Therefore, we can easily benefit from the reactions on the post, the owner profile, contacts, etc. Thus, IE in social media is very challenging more than in medical documents and is needed to detect the spread of false information and understand the people’s interactions with medical information.
Acknowledgements
Not applicable.
Biographies
Mohamed Yassine Landolsi
(medyassine.landolsi@isitc.u-sousse.tn; corresponding author) is currently a PhD student at the Higher Institute of Computer Science and Telecom of Hammam Sousse (ISIT’COM), University of Sousse, Tunisia. He received his Bachelor’s degree in computer science and his Master of Research degree in distributed computing from ISIT’COM, University of Sousse, Tunisia, in 2016 and 2019, respectively. His current research interests include text analysis and structuring in the medical field. He is a member of the research Laboratory MARS (Modeling of Automated Reasoning Systems).
Lobna Hlaoua
(lobna.hlaoua@essths.u-sousse.tn) obtained her PhD in computer science from the University Toulouse-III Paul-Sabatier in France. Since September 2008, she has been an associate professor of computer science at the University of Sousse in Tunisia, and since 2011, she has been a member of MARS (Modeling of Automated Reasoning Systems) Research Laboratory. Her research group focuses on information retrieval, social network analysis, and data mining. She co-supervised five PhD and 10 master theses. She is reviewer of the Journal of the Association for Information Science and Technology (JASIST).
Lotfi Ben Romdhane
(lotfi.benromdhane@isitc.u-sousse.tn) holds a PhD degree from the University of Sherbrooke, QC/Canada, and an engineering degree from ENSI/Tunisia; both in computer science. He is currently a professor in computer science at ISIT’COM, University of Sousse, Tunisia and heads MARS (Modeling of Automated Reasoning Systems) Research Lab. His areas of expertise span the general area of data science and includes reasoning, distributed computing, knowledge discovery, and data mining. He has published more than 70 papers on these topics in international conferences and journals.
Author Contributions
M.Y.L. analyzed the literature and wrote the manuscript. All authors read, revised and approved the final manuscript.
Funding
Not applicable.
Availability of data and materials
Not applicable.
Declarations
Abbreviations
Not applicable.
Ethics approval and consent to participate
Not applicable.
Conflict of interest
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Authors’ information
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mohamed Yassine Landolsi, Email: medyassine.landolsi@isitc.u-sousse.tn.
Lobna Hlaoua, Email: lobna.hlaoua@essths.u-sousse.tn.
Lotfi Ben Romdhane, Email: lotfi.BenRomdhane@isitc.u-sousse.tn.
References
- 1.Abacha AB, Zweigenbaum P (2011) Medical entity recognition: a comparaison of semantic and statistical methods. In: Proceedings of BioNLP 2011 workshop, pp 56–64
- 2.Aich S, Sain M, Park J, Choi KW, Kim HC (2017) A text mining approach to identify the relationship between gait-parkinson’s disease (pd) from pd based research articles. In: 2017 international conference on inventive computing and informatics (ICICI), IEEE, pp 481–485
- 3.Akbik A, Bergmann T, Blythe D, Rasul K, Schweter S, Vollgraf R (2019) Flair: an easy-to-use framework for state-of-the-art nlp. In: Proceedings of the 2019 conference of the north american chapter of the association for computational linguistics (Demonstrations), pp 54–59
- 4.Al-Dafas M, Albujeer A, Hussien SA, Ibrahim RK. On the adaption of data mining technology to categorize cancer diseases. Int J Artif Intell Inform. 2022;3(2):80–91. [Google Scholar]
- 5.Alex B, Grover C, Tobin R, Sudlow C, Mair G, Whiteley W. Text mining brain imaging reports. J Biomed Semant. 2019;10(1):1–11. doi: 10.1186/s13326-019-0211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Angeli G, Premkumar MJJ, Manning CD (2015) Leveraging linguistic structure for open domain information extraction. In: Proceedings of the 53rd annual meeting of the association for computational linguistics and the 7th international joint conference on natural language processing vol 1: Long Papers, pp 344–354
- 7.Apostolova E, Channin DS, Demner-Fushman D, Furst J, Lytinen S, Raicu D (2009) Automatic segmentation of clinical texts. In: 2009 Annual international conference of the IEEE engineering in medicine and biology society, IEEE, pp 5905–5908 [DOI] [PubMed]
- 8.Arbabi A, Adams DR, Fidler S, Brudno M. Identifying clinical terms in medical text using ontology-guided machine learning. JMIR Med Inform. 2019;7(2):e12,596. doi: 10.2196/12596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aronson AR, Lang FM. An overview of metamap: historical perspective and recent advances. J Am Med Inform Assoc. 2010;17(3):229–236. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aydar M, Bozal O, Ozbay F (2020) Neural relation extraction: a survey. arXiv e-prints pp arXiv–2007
- 11.Barnett GO, Cimino JJ, Hupp JA, Hoffer EP. Dxplain: an evolving diagnostic decision-support system. Jama. 1987;258(1):67–74. doi: 10.1001/jama.1987.03400010071030. [DOI] [PubMed] [Google Scholar]
- 12.Batista DS (2018) Named-entity evaluation metrics based on entity-level. http://www.davidsbatista.net/blog/2018/05/09/Named_Entity_Evaluation
- 13.Beel J, Gipp B, Shaker A, Friedrich N. International conference on theory and practice of digital libraries. Cham: Springer; 2010. Sciplore xtract: extracting titles from scientific pdf documents by analyzing style information (font size) pp. 413–416. [Google Scholar]
- 14.Ben Abdessalem Karaa W, Alkhammash EH, Bchir A. Drug disease relation extraction from biomedical literature using nlp and machine learning. Mob Inf Syst. 2021 doi: 10.1155/2021/9958410. [DOI] [Google Scholar]
- 15.Berrazega I. Temporal information processing: a survey. Int J Nat Lang Comput. 2012;1(2):1–14. doi: 10.5121/ijnlc.2012.1201. [DOI] [Google Scholar]
- 16.Bethard S, Savova G, Chen WT, Derczynski L, Pustejovsky J, Verhagen M (2016) Semeval-2016 task 12: clinical tempeval. In: Proceedings of the 10th international workshop on semantic evaluation (SemEval-2016), pp 1052–1062
- 17.Bethard S, Savova G, Palmer M, Pustejovsky J (2017) SemEval-2017 task 12: clinical tempEval. In: Proceedings of the 11th international workshop on semantic evaluation (SemEval-2017), Association for Computational Linguistics, Vancouver, Canada, pp 565–572. 10.18653/v1/S17-2093
- 18.Bhatia P, Celikkaya B, Khalilia M (2019) Joint entity extraction and assertion detection for clinical text. In: Proceedings of the 57th conference of the association for computational linguistics, ACL 2019, Florence, Italy, vol 1: Long Papers, Association for Computational Linguistics, pp 954–959. 10.18653/v1/p19-1091
- 19.Bodenreider O. The unified medical language system (umls): integrating biomedical terminology. Nucleic Acid Res. 2004;32(suppl 1):D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bottou L. On-line learning and stochastic approximations. USA: Cambridge University Press; 1999. pp. 9–42. [Google Scholar]
- 21.Bramsen P, Deshpande P, Lee YK, Barzilay R (2006) Finding temporal order in discharge summaries. In: AMIA annual symposium proceedings, American Medical Informatics Association, vol 2006, p 81 [PMC free article] [PubMed]
- 22.Chapman W, Dowling J, Chu D. Biological, translational, and clinical language processing. Pittsburgh, PA: University of Pittsburgh; 2007. Context: an algorithm for identifying contextual features from clinical text; pp. 81–88. [Google Scholar]
- 23.Chapman WW, Savova GK, Zheng J, Tharp M, Crowley R. Anaphoric reference in clinical reports: characteristics of an annotated corpus. J Biomed Inform. 2012;45(3):507–521. doi: 10.1016/j.jbi.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 24.Chaves L, Marques G. Data mining techniques for early diagnosis of diabetes: a comparative study. Appl Sci. 2021;11(5):2218. doi: 10.3390/app11052218. [DOI] [Google Scholar]
- 25.Chirila OS, Chirila CB, Stoicu-Tivadar L. Improving the prescription process information support with structured medical prospectuses using neural networks. Stud Health Technol Inform. 2019;264:353–357. doi: 10.3233/SHTI190242. [DOI] [PubMed] [Google Scholar]
- 26.Chirila OS, Chirila CB, Stoicu-Tivadar L. Named entity recognition and classification for medical prospectuses. Stud Health Technol Inform. 2019;262:284–287. doi: 10.3233/SHTI190074. [DOI] [PubMed] [Google Scholar]
- 27.Cohen KB, Lanfranchi A, Choi MJY, Bada M, Baumgartner WA, Panteleyeva N, Verspoor K, Palmer M, Hunter LE. Coreference annotation and resolution in the colorado richly annotated full text (craft) corpus of biomedical journal articles. BMC Bioinform. 2017;18(1):1–14. doi: 10.1186/s12859-017-1775-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cohen KB, Verspoor K, Fort K, Funk C, Bada M, Palmer M, Hunter LE. Handbook of linguistic annotation. Cham: Springer; 2017. The colorado richly annotated full text (craft) corpus: multi-model annotation in the biomedical domain; pp. 1379–1394. [Google Scholar]
- 29.Dai HJ, Syed-Abdul S, Chen CW, Wu CC. Recognition and evaluation of clinical section headings in clinical documents using token-based formulation with conditional random fields. BioMed Res Int. 2015 doi: 10.1155/2015/873012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Bruijn B, Cherry C, Kiritchenko S, Martin J, Zhu X. Machine-learned solutions for three stages of clinical information extraction: the state of the art at i2b2 2010. J Am Med Inform Assoc. 2011;18(5):557–562. doi: 10.1136/amiajnl-2011-000150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Del Corro L, Gemulla R (2013) Clausie: clause-based open information extraction. In: Proceedings of the 22nd international conference on world wide web, pp 355–366
- 32.Deléger L, Névéol A (2014) Automatic identification of document sections for designing a french clinical corpus (identification automatique de zones dans des documents pour la constitution d’un corpus médical en français) [in french]. In: TALN
- 33.Deng N, Fu H, Chen X. Named entity recognition of traditional chinese medicine patents based on bilstm-crf. Wirel Commun Mob Comput. 2021 doi: 10.1155/2021/6696205. [DOI] [Google Scholar]
- 34.Devlin J, Chang M, Lee K, Toutanova K (2019) BERT: pre-training of deep bidirectional transformers for language understanding. In: Proceedings of the 2019 conference of the north american chapter of the association for computational linguistics: human language technologies, NAACL-HLT 2019, Minneapolis, MN, USA, Vol 1 (Long and Short Papers), Association for Computational Linguistics, pp 4171–4186. 10.18653/v1/n19-1423
- 35.Doğan RI, Leaman R, Lu Z. Ncbi disease corpus: a resource for disease name recognition and concept normalization. J Biomed Inform. 2014;47:1–10. doi: 10.1016/j.jbi.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Donnelly K. Snomed-ct: the advanced terminology and coding system for ehealth. Stud Health Technol Inform. 2006;121:279. [PubMed] [Google Scholar]
- 37.Edinger T, Demner-Fushman D, Cohen AM, Bedrick S, Hersh W (2017) Evaluation of clinical text segmentation to facilitate cohort retrieval. In: AMIA annual symposium proceedings, American Medical Informatics Association, vol 2017, p 660 [PMC free article] [PubMed]
- 38.Elhadad N, Pradhan S, Gorman S, Manandhar S, Chapman W, Savova G (2015) Semeval-2015 task 14: analysis of clinical text. In: Proceedings of the 9th international workshop on semantic evaluation (SemEval 2015), pp 303–310
- 39.Fader A, Soderland S, Etzioni O (2011) Identifying relations for open information extraction. In: Proceedings of the 2011 conference on empirical methods in natural language processing, pp 1535–1545
- 40.Fundel K. Küffner R, Zimmer R. Relex-relation extraction using dependency parse trees. Bioinformatics. 2007;23(3):365–371. doi: 10.1093/bioinformatics/btl616. [DOI] [PubMed] [Google Scholar]
- 41.Ghiasvand O, Kate RJ. Learning for clinical named entity recognition without manual annotations. Inform Med Unlocked. 2018;13:122–127. doi: 10.1016/j.imu.2018.10.011. [DOI] [Google Scholar]
- 42.Goenaga I, Lahuerta X, Atutxa A, Gojenola K. A section identification tool: towards hl7 cda/ccr standardization in spanish discharge summaries. J Biomed Inform. 2021;121(103):875. doi: 10.1016/j.jbi.2021.103875. [DOI] [PubMed] [Google Scholar]
- 43.Grishman R, Sundheim BM (1996) Message understanding conference-6: A brief history. In: Coling 1996 vol 1: the 16th international conference on computational linguistics
- 44.Guo F, He R, Dang J. Implicit discourse relation recognition via a bilstm-cnn architecture with dynamic chunk-based max pooling. IEEE Access. 2019;7:169,281–169,292. doi: 10.1109/ACCESS.2019.2954988. [DOI] [Google Scholar]
- 45.Hafiene N, Karoui W, Romdhane LB. Influential nodes detection in dynamic social networks: a survey. Expert Syst Appl. 2020;159(113):642. [Google Scholar]
- 46.Hahn U, Oleynik M. Medical information extraction in the age of deep learning. Yearb Med Inform. 2020;29(01):208–220. doi: 10.1055/s-0040-1702001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hallersten A, Fürst W, Mezzasalma R. Physicians prefer greater detail in the biosimilar label (smpc)-results of a survey across seven european countries. Regul Toxicol Pharmacol. 2016;77:275–281. doi: 10.1016/j.yrtph.2016.03.021. [DOI] [PubMed] [Google Scholar]
- 48.Hasan F, Roy A, Pan S (2020) Integrating text embedding with traditional nlp features for clinical relation extraction. In: 2020 IEEE 32nd international conference on tools with artificial intelligence (ICTAI), IEEE, pp 418–425
- 49.Haug PJ, Wu X, Ferraro JP, Savova GK, Huff SM, Chute CG (2014) Developing a section labeler for clinical documents. In: AMIA annual symposium proceedings, American Medical Informatics Association, vol 2014, p 636 [PMC free article] [PubMed]
- 50.Henry S, Buchan K, Filannino M, Stubbs A, Uzuner O. 2018 n2c2 shared task on adverse drug events and medication extraction in electronic health records. J Am Med Inform Assoc. 2020;27(1):3–12. doi: 10.1093/jamia/ocz166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Honnibal M, Montani I. spaCy 2: natural language understanding with Bloom embeddings, convolutional neural networks and incremental parsing. Appear. 2017;7:411–420. [Google Scholar]
- 52.Huang M, Liu A, Wang T, Huang C. Green data gathering under delay differentiated services constraint for internet of things. Wirel Commun Mob Comput. 2018 doi: 10.1155/2018/9715428. [DOI] [Google Scholar]
- 53.Islamaj R, Leaman R, Kim S, Kwon D, Wei CH, Comeau DC, Peng Y, Cissel D, Coss C, Fisher C, Guzman R, Kochar PG, Koppel S, Trinh D, Sekiya K, Ward J, Whitman D, Schmidt S, Lu Z. Nlm-chem, a new resource for chemical entity recognition in pubmed full text literature. Sci Data. 2021;8(1):1–12. doi: 10.1038/s41597-021-00875-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jagannatha A, Liu F, Liu W, Yu H. Overview of the first natural language processing challenge for extracting medication, indication, and adverse drug events from electronic health record notes (made 1.0) Drug Saf. 2019;42(1):99–111. doi: 10.1007/s40264-018-0762-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jancsary J, Matiasek J, Trost H (2008) Revealing the structure of medical dictations with conditional random fields. In: Proceedings of the 2008 conference on empirical methods in natural language processing, pp 1–10
- 56.Jaouadi M, Romdhane LB (2019) Influence maximization problem in social networks: an overview. In: 2019 IEEE/ACS 16th international conference on computer systems and applications (AICCSA), IEEE, pp 1–8
- 57.Jelier R, Jenster G, Dorssers LC, van der Eijk CC, van Mulligen EM, Mons B, Kors JA. Co-occurrence based meta-analysis of scientific texts: retrieving biological relationships between genes. Bioinformatics. 2005;21(9):2049–2058. doi: 10.1093/bioinformatics/bti268. [DOI] [PubMed] [Google Scholar]
- 58.Johnson AE, Pollard TJ, Shen L, Li-Wei HL, Feng M, Ghassemi M, Moody B, Szolovits P, Celi LA, Mark RG. Mimic-iii, a freely accessible critical care database. Sci Data. 2016;3(1):1–9. doi: 10.1038/sdata.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karlsson I, Boström H (2016) Predicting adverse drug events using heterogeneous event sequences. In: 2016 IEEE international conference on healthcare informatics (ICHI), IEEE, pp 356–362
- 60.Kim Y, Heider PM, Lally IR, Meystre SM. A hybrid model for family history information identification and relation extraction: development and evaluation of an end-to-end information extraction system. JMIR Med Inform. 2021;9(4):e22,797. doi: 10.2196/22797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Komariah KS, Shin BK (2021) Medical entity recognition in twitter using conditional random fields. In: 2021 international conference on electronics, information, and communication (ICEIC), IEEE, pp 1–4
- 62.Komninos A, Manandhar S (2016) Dependency based embeddings for sentence classification tasks. In: Proceedings of the 2016 conference of the north american chapter of the association for computational linguistics: human language technologies, pp 1490–1500
- 63.Kouni IBE, Karoui W, Romdhane LB (2021) WLNI-LPA: detecting overlapping communities in attributed networks based on label propagation process. In: Proceedings of the 16th international conference on software technologies, ICSOFT 2021, Online Streaming, July 6 SCITEPRESS, pp 408–416. 10.5220/0010605904080416
- 64.Kreuzthaler M, Schulz S. Detection of sentence boundaries and abbreviations in clinical narratives. BMC Med Inform Decis Mak. 2015;15:S4–S4. doi: 10.1186/1472-6947-15-S2-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kroll H, Pirklbauer J, Ruthmann J, Balke W (2020) A semantically enriched dataset based on biomedical NER for the COVID19 open research dataset challenge. CoRR https://arxiv.org/abs/2005.08823
- 66.Kropf S, Krücken P, Mueller W, Denecke K. Structuring legacy pathology reports by openehr archetypes to enable semantic querying. Method Inform Med. 2017;56(03):230–237. doi: 10.3414/ME16-01-0073. [DOI] [PubMed] [Google Scholar]
- 67.Kumar S (2017) A survey of deep learning methods for relation extraction. arXiv preprint arXiv:1705.03645
- 68.Lafferty JD, McCallum A, Pereira FCN (2001) Conditional random fields: probabilistic models for segmenting and labeling sequence data. In: Proceedings of the eighteenth international conference on machine learning, Morgan Kaufmann Publishers Inc., San Francisco, CA, USA, ICML ’01, pp 282–289
- 69.Lai KH, Topaz M, Goss FR, Zhou L. Automated misspelling detection and correction in clinical free-text records. J Biomed Inform. 2015;55:188–195. doi: 10.1016/j.jbi.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 70.Lan M, Wang J, Wu Y, Niu ZY, Wang H (2017) Multi-task attention-based neural networks for implicit discourse relationship representation and identification. In: Proceedings of the 2017 conference on empirical methods in natural language processing, pp 1299–1308
- 71.Landolsi MY, Mohamed HH, Romdhane LB. Image annotation in social networks using graph and multimodal deep learning features. Multimed Tool Appl. 2021;80(8):12,009–12,034. doi: 10.1007/s11042-020-09730-8. [DOI] [Google Scholar]
- 72.Laparra E, Xu D, Elsayed A, Bethard S, Palmer M (2018) Semeval 2018 task 6: parsing time normalizations. In: SemEval@ NAACL-HLT, pp 88–96
- 73.Laparra E, Su X, Zhao Y, Uzuner O, Miller T, Bethard S (2021) Semeval-2021 task 10: source-free domain adaptation for semantic processing. In: Proceedings of the 15th international workshop on semantic evaluation (SemEval-2021), pp 348–356
- 74.Lee J, Yoon W, Kim S, Kim D, Kim S, So CH, Kang J. Biobert: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 2020;36(4):1234–1240. doi: 10.1093/bioinformatics/btz682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee W, Choi J. Temporal segmentation for capturing snapshots of patient histories in korean clinical narrative. Healthc Inform Res. 2018;24(3):179–186. doi: 10.4258/hir.2018.24.3.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lei J, Tang B, Lu X, Gao K, Jiang M, Xu H. A comprehensive study of named entity recognition in chinese clinical text. J Am Med Inform Assoc. 2014;21(5):808–814. doi: 10.1136/amiajnl-2013-002381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Leroy G, Chen H. Biocomputing 2002. Singapore: World Scientific; 2001. Filling preposition-based templates to capture information from medical abstracts; pp. 350–361. [PubMed] [Google Scholar]
- 78.Li F, Lin Z, Zhang M, Ji D (2021) A span-based model for joint overlapped and discontinuous named entity recognition. CoRR abs/2106.14373, arXiv:2106.14373
- 79.Li J, Sun Y, Johnson RJ, Sciaky D, Wei CH, Leaman R, Davis AP, Mattingly CJ, Wiegers TC, Lu Z. Biocreative v cdr task corpus: a resource for chemical disease relation extraction. Database. 2016 doi: 10.1093/database/baw068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li W, Shi S, Gao Z, Wei W, Zhu Q, Lin X, Jiang D, Gao S (2018) Improved deep belief network model and its application in named entity recognition of chinese electronic medical records. In: 2018 IEEE 3rd international conference on big data analysis (ICBDA), IEEE, pp 356–360
- 81.Li Y, Lipsky Gorman S, Elhadad N (2010) Section classification in clinical notes using supervised hidden markov model. In: Proceedings of the 1st ACM international health informatics symposium, pp 744–750
- 82.Liu F, Li T. A clustering-anonymity privacy-preserving method for wearable iot devices. Secur Commun Netw. 2018 doi: 10.1155/2018/4945152. [DOI] [Google Scholar]
- 83.Liu F, Chen J, Jagannatha A, Yu H (2016a) Learning for biomedical information extraction: methodological review of recent advances. CoRR abs/1606.07993, arXiv:1606.07993
- 84.Liu Y, Wei L, Yao Z, Fei X. The practice and experience of emergency information system construction. China Dig Med. 2016;11(5):53–55. [Google Scholar]
- 85.Liu Y, Ott M, Goyal N, Du J, Joshi M, Chen D, Levy O, Lewis M, Zettlemoyer L, Stoyanov V (2019) Roberta: a robustly optimized BERT pretraining approach. CoRR abs/1907.11692, arXiv:1907.11692
- 86.Lohr C, Luther S, Matthies F, Hahn U (2018a) Cda-compliant section annotation of german-language discharge summaries: guideline development, annotation campaign, section classification. In: AMIA 2018, american medical informatics association annual symposium, San Francisco, CA, AMIA [PMC free article] [PubMed]
- 87.Lohr C, Luther S, Matthies F, Modersohn L, Ammon D, Saleh K, Henkel AG, Kiehntopf M, Hahn U (2018b) Cda-compliant section annotation of german-language discharge summaries: guideline development, annotation campaign, section classification. In: AMIA annual symposium proceedings, American Medical Informatics Association, vol 2018, p 770 [PMC free article] [PubMed]
- 88.Lomotey RK, Deters R (2013) Efficient mobile services consumption in mhealth. In: Proceedings of the 2013 IEEE/ACM international conference on advances in social networks analysis and mining, pp 982–989
- 89.Luan Y, Wadden D, He L, Shah A, Ostendorf M, Hajishirzi H (2019) A general framework for information extraction using dynamic span graphs. In: Proceedings of the 2019 conference of the north american chapter of the association for computational linguistics: human language technologies, Vol 1 (Long and Short Papers), Association for Computational Linguistics, Minneapolis, Minnesota, pp 3036–3046. 10.18653/v1/N19-1308
- 90.Ludwick DA, Doucette J. Adopting electronic medical records in primary care: lessons learned from health information systems implementation experience in seven countries. Int J Med Inform. 2009;78(1):22–31. doi: 10.1016/j.ijmedinf.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 91.Lupşe O, Stoicu-Tivadar L. Extracting and structuring drug information to improve e-prescription and streamline medical treatment. Appl Med Inform. 2018;40(1–2):7–14. [Google Scholar]
- 92.Lupşe O, Stoicu-Tivadar L. Supporting prescriptions with synonym matching of section names in prospectuses. Stud Health Technol Inform. 2018;251:153–156. [PubMed] [Google Scholar]
- 93.Ma F, Liu X, Liu A, Zhao M, Huang C, Wang T. A time and location correlation incentive scheme for deep data gathering in crowdsourcing networks. Wirel Commun Mob Comput. 2018 doi: 10.1155/2018/8052620. [DOI] [Google Scholar]
- 94.Mabrouk O, Hlaoua L, Omri MN. Exploiting ontology information in fuzzy svm social media profile classification. Appl Intell. 2021;51(6):3757–3774. doi: 10.1007/s10489-020-01939-2. [DOI] [Google Scholar]
- 95.Mahendran D, McInnes BT (2021) Extracting adverse drug events from clinical notes. In: AMIA annual symposium proceedings, American Medical Informatics Association, vol 2021, p 420 [PMC free article] [PubMed]
- 96.Manning CD, Surdeanu M, Bauer J, Finkel JR, Bethard S, McClosky D (2014) The stanford corenlp natural language processing toolkit. In: Proceedings of 52nd annual meeting of the association for computational linguistics: system demonstrations, pp 55–60
- 97.Mausam-Schmitz M, Bart R, Soderland S, Etzioni O (2012) Open language learning for information extraction. In: Proceedings of the 2012 joint conference on empirical methods in natural language processing and computational natural language learning, Association for Computational Linguistics, USA, p 523-534
- 98.Meystre SM, Savova GK, Kipper-Schuler KC, Hurdle JF. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform. 2008;17(01):128–144. doi: 10.1055/s-0038-1638592. [DOI] [PubMed] [Google Scholar]
- 99.Mnasri W, Azaouzi M, Romdhane LB. Parallel social behavior-based algorithm for identification of influential users in social network. Appl Intell. 2021 doi: 10.1007/s10489-021-02203-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nair N, Narayanan S, Achan P, Soman K (2022) Clinical note section identification using transfer learning. In: Proceedings of sixth international congress on information and communication technology, Springer, pp 533–542
- 101.Nasar Z, Jaffry SW, Malik MK. Named entity recognition and relation extraction: state-of-the-art. ACM Comput Surv (CSUR) 2021;54(1):1–39. doi: 10.1145/3445965. [DOI] [Google Scholar]
- 102.Nayel HA, ShashrekhaH L (2019) Integrating dictionary feature into a deep learning model for disease named entity recognition. https://arxiv.org/abs/1911.01600
- 103.Neumann M, King D, Beltagy I, Ammar W (2019) Scispacy: fast and robust models for biomedical natural language processing. In: Proceedings of the 18th BioNLP workshop and shared task, BioNLP@ACL 2019, Florence, Italy, Association for Computational Linguistics, pp 319–327. 10.18653/v1/w19-5034
- 104.Ni J, Delaney B, Florian R. Fast model adaptation for automated section classification in electronic medical records. Stud Health Technol Inform. 2015;216:35–39. [PubMed] [Google Scholar]
- 105.Peters ME, Neumann M, Iyyer M, Gardner M, Clark C, Lee K, Zettlemoyer L (2018) Deep contextualized word representations. In: Proceedings of the 2018 conference of the north american chapter of the association for computational linguistics: human language technologies, NAACL-HLT, New Orleans, Louisiana, USA, Vol 1 (Long Papers), Association for Computational Linguistics, pp 2227–2237. 10.18653/v1/n18-1202
- 106.Pomares-Quimbaya A, Kreuzthaler M, Schulz S. Current approaches to identify sections within clinical narratives from electronic health records: a systematic review. BMC Med Res Methodol. 2019;19(1):155. doi: 10.1186/s12874-019-0792-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Popovski G, Seljak BK, Eftimov T. A survey of named-entity recognition methods for food information extraction. IEEE Access. 2020;8:31,586–31,594. doi: 10.1109/ACCESS.2020.2973502. [DOI] [Google Scholar]
- 108.Pradhan S, Elhadad N, Chapman WW, Manandhar S, Savova G (2014) Semeval-2014 task 7: analysis of clinical text. In: SemEval@ COLING, pp 54–62
- 109.Qi P, Zhang Y, Zhang Y, Bolton J, Manning CD (2020) Stanza: a python natural language processing toolkit for many human languages. In: Proceedings of the 58th annual meeting of the association for computational linguistics: system demonstrations, Association for Computational Linguistics, pp 101–108. 10.18653/v1/2020.acl-demos.14
- 110.Quimbaya AP, Múnera AS, Rivera RAG, Rodríguez JCD, Velandia OMM, Peña AAG, Labbé C. Named entity recognition over electronic health records through a combined dictionary-based approach. Procedia Comput Sci. 2016;100:55–61. doi: 10.1016/j.procs.2016.09.123. [DOI] [Google Scholar]
- 111.Rebholz-Schuhman D, Jimeno-Yepes A, Li C, Kafkas S, Lewin I, Kang N, Corbett P, Milward D, Buyko E, Beisswanger E, Hornbostel K, Kouznetsov A, Witte R, Laurila J, Baker C, Kuo CJ, Clematide S, Rinaldi F, Farkas R, Hahn U. Assessment of ner solutions against the first and second calbc silver standard corpus. J Biomed Semant. 2011;2(5):1–12. doi: 10.1186/2041-1480-2-S5-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Roberts RJ. Pubmed central: the genbank of the published literature. Proc Natl Acad Sci. 2001;98:381–382. doi: 10.1073/pnas.98.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Robson B, Boray S, Weisman J. Mining real-world high dimensional structured data in medicine and its use in decision support. Some different perspectives on unknowns, interdependency, and distinguishability. Comput Biol Med. 2022;141:105,118. doi: 10.1016/j.compbiomed.2021.105118. [DOI] [PubMed] [Google Scholar]
- 114.Rokach L, Romano R, Maimon O. Negation recognition in medical narrative reports. Inf Retr. 2008;11(6):499–538. doi: 10.1007/s10791-008-9061-0. [DOI] [Google Scholar]
- 115.Rosario B, Hearst MA (2004) Classifying semantic relations in bioscience texts. In: Proceedings of the 42nd annual meeting of the association for computational linguistics (ACL-04), pp 430–437
- 116.Rundo L, Pirrone R, Vitabile S, Sala E, Gambino O. Recent advances of hci in decision-making tasks for optimized clinical workflows and precision medicine. J Biomed Inform. 2020;108(103):479. doi: 10.1016/j.jbi.2020.103479. [DOI] [PubMed] [Google Scholar]
- 117.Sadoughi N, Finley GP, Edwards E, Robinson A, Korenevsky M, Brenndoerfer M, Axtmann N, Miller M, Suendermann-Oeft D (2018) Detecting section boundaries in medical dictations: toward real-time conversion of medical dictations to clinical reports. In: International conference on speech and computer, Springer, pp 563–573
- 118.Shi J, Li W, Yang Y, Yao N, Bai Q, Yongchareon S, Yu J. Pacific rim knowledge acquisition workshop. Cham: Springer; 2021. Automated concern exploration in pandemic situations-covid-19 as a use case; pp. 178–185. [Google Scholar]
- 119.Shi J, Li W, Yongchareon S, Yang Y, Bai Q. Graph-based joint pandemic concern and relation extraction on twitter. Expert Syst Appl. 2022;195(116):538. doi: 10.1016/j.eswa.2022.116538. [DOI] [Google Scholar]
- 120.Sohrab MG, Duong K, Miwa M, Topić G, Masami I, Hiroya T (2020) Bennerd: a neural named entity linking system for covid-19. In: Proceedings of the 2020 conference on empirical methods in natural language processing: system demonstrations, pp 182–188
- 121.Song HJ, Jo BC, Park CY, Kim JD, Kim YS. Comparison of named entity recognition methodologies in biomedical documents. Biomed Eng Online. 2018;17(2):1–14. doi: 10.1186/s12938-018-0573-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sorgente A, Vettigli G, Mele F. Automatic extraction of cause-effect relations in natural language text. DART @AI *IA. 2013;2013:37–48. [Google Scholar]
- 123.Stubbs A, Kotfila C, Uzuner Ö. Automated systems for the de-identification of longitudinal clinical narratives: overview of 2014 i2b2/uthealth shared task track 1. J Biomed Inform. 2015;58:S11–S19. doi: 10.1016/j.jbi.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Stubbs A, Kotfila C, Xu H, Uzuner Ö. Identifying risk factors for heart disease over time: overview of 2014 i2b2/uthealth shared task track 2. J Biomed Inform. 2015;58:S67–S77. doi: 10.1016/j.jbi.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sudeshna P, Bhanumathi S, Hamlin MA. 2017 international conference on computation of power. IEEE: Energy Information and Commuincation (ICCPEIC); 2017. Identifying symptoms and treatment for heart disease from biomedical literature using text data mining; pp. 170–174. [Google Scholar]
- 126.Sui Y, Bu F, Hu Y, Yan W, Zhang L (2022) Trigger-gnn: a trigger-based graph neural network for nested named entity recognition. arXiv:2204.05518
- 127.Sun Q, Bhatia P. Findings of the association for computational linguistics: ACL/IJCNLP 2021. Stroudsburg, Pennsylvania: Association for Computational Linguistics; 2021. Neural entity recognition with gazetteer based fusion; pp. 3291–3295. [Google Scholar]
- 128.Sun W, Cai Z, Liu F, Fang S, Wang G. 2017 IEEE 19th international conference on e-health networking. IEEE: Applications and Services (Healthcom); 2017. A survey of data mining technology on electronic medical records; pp. 1–6. [Google Scholar]
- 129.Sun W, Cai Z, Li Y, Liu F, Fang S, Wang G. Data processing and text mining technologies on electronic medical records: a review. J Healthc Eng. 2018 doi: 10.1155/2018/4302425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sun W, Cai Z, Li Y, Liu F, Fang S, Wang G. Security and privacy in the medical internet of things: a review. Secur Commun Netw. 2018;2018:1–30. [Google Scholar]
- 131.Suominen HJ, Salakoski TI. Supporting communication and decision making in finnish intensive care with language technology. J Healthc Eng. 2010;1(4):595–614. doi: 10.1260/2040-2295.1.4.595. [DOI] [Google Scholar]
- 132.Tang B, Cao H, Wu Y, Jiang M, Xu H. Recognizing clinical entities in hospital discharge summaries using structural support vector machines with word representation features. BMC Med Inform Decis Mak BioMed Cent. 2013;13:1–10. doi: 10.1186/1472-6947-13-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tang J, Liu A, Zhao M, Wang T. An aggregate signature based trust routing for data gathering in sensor networks. Secur Commun Netw. 2018;2018:1–30. doi: 10.1155/2018/6328504. [DOI] [Google Scholar]
- 134.Tchraktchiev D, Angelova G, Boytcheva S, Angelov Z, Zacharieva S. Patient safety informatics. Amsterdam: IOS Press; 2011. Completion of structured patient descriptions by semantic mining; pp. 260–269. [PubMed] [Google Scholar]
- 135.Tepper M, Capurro D, Xia F, Vanderwende L, Yetisgen-Yildiz M (2012) Statistical section segmentation in free-text clinical records. In: Lrec, pp 2001–2008
- 136.Tran T, Kavuluru R. Distant supervision for treatment relation extraction by leveraging mesh subheadings. Artif Intell Med. 2019;98:18–26. doi: 10.1016/j.artmed.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tran V, Tran VH, Nguyen P, Nguyen C, Satoh K, Matsumoto Y, Nguyen M (2021) Covrelex: a covid-19 retrieval system with relation extraction. In: Proceedings of the 16th conference of the european chapter of the association for computational linguistics: system demonstrations, pp 24–31
- 138.Uzuner Ö, Solti I, Cadag E. Extracting medication information from clinical text. J Am Med Inform Assoc. 2010;17(5):514–518. doi: 10.1136/jamia.2010.003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Uzuner Ö, South BR, Shen S, DuVall SL. 2010 i2b2/va challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc. 2011;18(5):552–556. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Vunikili R, Supriya H, Marica VG, Farri O (2020) Clinical ner using spanish bert embeddings. In: IberLEF@ SEPLN, pp 505–511
- 141.Wang L, Foer D, MacPhaul E, Lo YC, Bates DW, Zhou L. Pasclex: a comprehensive post-acute sequelae of covid-19 (pasc) symptom lexicon derived from electronic health record clinical notes. J Biomed Inform. 2021;125:103951. doi: 10.1016/j.jbi.2021.103951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang P, Hao T, Yan J, Jin L. Large-scale extraction of drug-disease pairs from the medical literature. J Assoc Inf Sci Technol. 2017;68(11):2649–2661. doi: 10.1002/asi.23876. [DOI] [Google Scholar]
- 143.Wang S, Ren F, Lu H (2018) A review of the application of natural language processing in clinical medicine. In: 2018 13th IEEE conference on industrial electronics and applications (ICIEA), pp 2725–2730
- 144.Wang Y, Wang L, Rastegar-Mojarad M, Moon S, Shen F, Afzal N, Liu S, Zeng Y, Mehrabi S, Sohn S, Liu H. Clinical information extraction applications: a literature review. J Biomed Inform. 2018;77:34–49. doi: 10.1016/j.jbi.2017.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wang Y, Fu S, Shen F, Henry S, Uzuner O, Liu H. The 2019 n2c2/ohnlp track on clinical semantic textual similarity: overview. JMIR Med Inform. 2020;8(11):e23375. doi: 10.2196/23375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wei Q, Ji Z, Si Y, Du J, Wang J, Tiryaki F, Wu S, Tao C, Roberts K, Xu H (2019) Relation extraction from clinical narratives using pre-trained language models. In: AMIA annual symposium proceedings, American Medical Informatics Association, vol 2019, p 1236 [PMC free article] [PubMed]
- 147.Weiskopf NG, Hripcsak G, Swaminathan S, Weng C. Defining and measuring completeness of electronic health records for secondary use. J Biomed Inform. 2013;46(5):830–836. doi: 10.1016/j.jbi.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wu Y, Jiang M, Xu J, Zhi D, Xu H (2017) Clinical named entity recognition using deep learning models. In: AMIA annual symposium proceedings, American Medical Informatics Association, vol 2017, p 1812 [PMC free article] [PubMed]
- 149.Xia J, Cai Z, Hu G, Xu M. An active defense solution for arp spoofing in openflow network. Chin J Electron. 2019;28(1):172–178. doi: 10.1049/cje.2017.12.002. [DOI] [Google Scholar]
- 150.Xu J, Gan L, Cheng M, Wu Q. Unsupervised medical entity recognition and linking in chinese online medical text. J Healthc Eng. 2018 doi: 10.1155/2018/2548537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yang J, Han SC, Poon J (2021a) A survey on extraction of causal relations from natural language text. CoRR abs/2101.06426, arXiv:2101.06426
- 152.Yang L, Cai ZP, Xu H. Llmp: exploiting lldp for latency measurement in software-defined data center networks. J Comput Sci Technol. 2018;33(2):277–285. doi: 10.1007/s11390-018-1819-2. [DOI] [Google Scholar]
- 153.Yang X, Zhang H, He X, Bian J, Wu Y. Extracting family history of patients from clinical narratives: exploring an end-to-end solution with deep learning models. JMIR Med Inform. 2020;8(12):e22982. doi: 10.2196/22982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yang X, Yu Z, Guo Y, Bian J, Wu Y (2021b) Clinical relation extraction using transformer-based models. CoRR abs/2107.08957, arXiv:2107.08957
- 155.Yang Z, Lin H, Li Y. Exploiting the performance of dictionary-based bio-entity name recognition in biomedical literature. Comput Biol Chem. 2008;32(4):287–291. doi: 10.1016/j.compbiolchem.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 156.Yang Z, Dai Z, Yang Y, Carbonell J, Salakhutdinov RR, Le QV. Xlnet: generalized autoregressive pretraining for language understanding. Adv Neural Inf Process Syst. 2019;32:1–10. [Google Scholar]
- 157.Zhang H, Cai Z, Liu Q, Xiao Q, Li Y, Cheang CF. A survey on security-aware measurement in sdn. Secur Commun Netw. 2018 doi: 10.1155/2018/2459154. [DOI] [Google Scholar]
- 158.Zhang R, Chu F, Chen D, Shang X. A text structuring method for chinese medical text based on temporal information. Int J Environ Res Public Health. 2018;15(3):402. doi: 10.3390/ijerph15030402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhang S, Elhadad N. Unsupervised biomedical named entity recognition: experiments with clinical and biological texts. J Biomed Inform. 2013;46(6):1088–1098. doi: 10.1016/j.jbi.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhang Y, Yan X, Gao X, Chen Q, Hu HP. Demand analysis of decision support system of grass-roots health. Chin Gen Pract. 2016;19:2636–2639. doi: 10.3969/j.issn.1007-9572.2016.22.005. [DOI] [Google Scholar]
- 161.Zhao X, Ding H, Feng Z (2021) Glara: graph-based labeling rule augmentation for weakly supervised named entity recognition. In: Proceedings of the 16th conference of the european chapter of the association for computational linguistics: main volume, EACL 2021, association for computational linguistics, pp 3636–3649. 10.18653/v1/2021.eacl-main.318
- 162.Zheng J, Chapman WW, Crowley RS, Savova GK. Coreference resolution: a review of general methodologies and applications in the clinical domain. J Biomed Inform. 2011;44(6):1113–1122. doi: 10.1016/j.jbi.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zhou Y, Ju C, Caufield JH, Shih K, Chen CY, Sun Y, Chang K, Ping P, Wang W (2021) Clinical named entity recognition using contextualized token representations. CoRR abs/2106.12608, arXiv:2106.12608
- 164.Zweigenbaum P, Deléger L, Lavergne T, Névéol A, Bodnari A (2013) A supervised abbreviation resolution system for medical text. In: CLEF (Working Notes)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.