Abstract
Background
The delayed diagnosis of acute kidney injury (AKI) episodes and the lack of specificity of current single AKI biomarkers hamper its management. Urinary peptidome analysis may help to identify early molecular changes in AKI and grasp its complexity to identify potential targetable molecular pathways.
Methods
In derivation and validation cohorts totalizing 1170 major cardiac bypass surgery patients and in an external cohort of 1569 intensive care unit (ICU) patients, a peptide-based score predictive of AKI (7-day KDIGO classification) was developed, validated, and compared to the reference biomarker urinary NGAL and NephroCheck and clinical scores.
Results
A set of 204 urinary peptides derived from 48 proteins related to hemolysis, inflammation, immune cells trafficking, innate immunity, and cell growth and survival was identified and validated for the early discrimination (< 4 h) of patients according to their risk to develop AKI (OR 6.13 [3.96–9.59], p < 0.001) outperforming reference biomarkers (urinary NGAL and [IGFBP7].[TIMP2] product) and clinical scores. In an external cohort of 1569 ICU patients, performances of the signature were similar (OR 5.92 [4.73–7.45], p < 0.001), and it was also associated with the in-hospital mortality (OR 2.62 [2.05–3.38], p < 0.001).
Conclusions
An overarching AKI physiopathology-driven urinary peptide signature shows significant promise for identifying, at an early stage, patients who will progress to AKI and thus to develop tailored treatments for this frequent and life-threatening condition. Performance of the urine peptide signature is as high as or higher than that of single biomarkers but adds mechanistic information that may help to discriminate sub-phenotypes of AKI offering new therapeutic avenues.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13054-022-04193-9.
Keywords: Acute kidney injury, Cardiac surgery, Intensive care unit, Urinary peptidomics, Prediction
Background
Acute kidney injury (AKI) is a life-threatening disease with an incidence of 13.5 million patients and an estimated 1.7 million deaths per year worldwide [1]. In survivors, the risk of chronic kidney disease (CKD) increases ninefold [2]. Worldwide, more than 850 million people suffer from AKI and CKD or require renal replacement therapy (RRT) [1]. There is a clear need for early detection (< 12 h after the injury) to reduce the severity of the AKI. However, despite recent and intensive efforts, AKI is still detected at a late stage [1 to 3 days after the injury]. In addition, although treatments exist to reduce the impact of full-blown AKI, they are not specific and do not focus on the molecular mechanisms of AKI. The International Society of Nephrology has formally recognized this alarming situation of AKI as a major challenge and has launched the “0by25” objective, to eliminate preventable deaths from AKI by 2025 in low- and high-income countries [1], echoed by the European Renal Association [3].
The late detection of AKI is largely related to the assessment of kidney dysfunction by serum creatinine, which is inherently downstream of advanced AKI, rather than based on earlier signs of kidney damage [4–8]. Furthermore, even the recently discovered individual molecular markers of kidney injury (e.g., FGF-23, NGAL, IL-18, KIM-1, [TIMP-2].[IGFBP7]) detect AKI optimally at 12–24 h post-injury, when often irreversible damage is already present [4–7, 9]. These individual biomarkers are mainly dependent on the injury to a specific tubular segment, appear non-specific, and do not provide information on the timing and mechanisms of kidney injury [4–8]. To grasp the molecular complexity of AKI, high-throughput strategies using multi-dimensional molecular markers should therefore be proposed, as already employed in other acute conditions such as acute respiratory distress syndrome, acute heart failure, or sepsis [10–16].
Urinary peptidomics has emerged as a powerful method to noninvasively assess characteristics of the kidney parenchyma [17, 18] and to stratify patients according to their risk of progressing to kidney fibrosis [19–21], but also to assess specific risks of systemic diseases, including acute life-threatening conditions such as COVID-19 [22]. In the context of AKI, early preliminary studies using a small number of patients (n = 80–120) have shown the feasibility of using urinary peptidome analysis to predict the development of AKI, outperforming NGAL (neutrophil gelatinase-associated lipocalin) and KIM-1 (kidney injury molecule-1) [23, 24]. However, the samples were collected at a late stage after kidney injury.
In the current study, we determined whether urinary peptide signatures can identify, at an early stage, patients developing AKI in different at-risk clinical settings including cardiac bypass (CBP) surgery and after admission to the intensive care unit (ICU) and grasp the molecular complexity of AKI to identify potential targetable pathways.
Methods
Study design
A multi-step strategy was developed to identify a potential urinary peptide signature of AKI after CBP surgery. First, clinical characteristics were assessed in a derivation cohort of patients referred for cardiac surgery with cardiac bypass and used to identify clinically derived predictive factors of AKI. Urinary peptidomes were then characterized using mass spectrometry, and differentially abundant peptides between AKI and non-AKI patients with a significant Benjamini–Hochberg adjusted Wilcoxon signed-rank testing were identified. The link of one of the peptides to the AKI pathophysiology was confirmed by studying the full-length protein (calprotectin) in urine, in an animal AKI model and in vitro. A set of sequenced peptides was used to build a support vector machine-based predictor. Results were extracted as a calibrated score based on the derivation and then tested in two validation cohorts of patients referred for CBP surgery or admitted to the general ICU.
Patients
CBP surgery patients were prospectively recruited at the University Hospital of Toulouse, France, during two distinct time periods (March 2016–January 2017 for the derivation cohort (n = 509) and January 2019–March 2020 for the validation cohort n = 661). All patients with CBP surgery were eligible. Patients under 18, that underwent unscheduled CBP surgery or who required chronic dialysis before surgery were excluded.
A third cohort of patients admitted to the general ICU were also studied to obtain external validation of the peptide signature. Detailed clinical characteristics of this European multicenter cohort that included 1569 patients admitted to the ICU for sepsis, heart failure, cardiac arrest, or urgent surgery were already reported in previous studies (FROG-ICU) [25–27]. Urine samples were collected during the first 24 h following admission. AKI KDIGO classification [7 days period] was used to define AKI severity.
Characteristics, definitions, and endpoints
Pre-, per-, and postoperative clinical data were gathered retrospectively for all patients based on hospital records. Baseline estimated glomerular filtration rate (eGFR) was estimated using the CKD-EPI formula based on standardized creatinine measurement (IDMS) before cardiac surgery [28]. EuroSCORE-II was calculated as recommended [29].
Surgery was divided into coronary artery bypass (CAB), valvular surgery (valvuloplasty or replacement), combined CAB and valvular surgery, surgery with replacement of the ascending aorta with or without CAB (aortic surgery); surgery that directly affects the cardiac myocardial wall, such as interatrial communication, interventricular communication, ventricular aneurism, or cardiac transplantation (myocardium).
The main endpoint, AKI, was defined according to the AKI kidney disease/improving global outcome (KDIGO) 2012 criteria [30] evaluated during the first 7 days after surgery. Briefly, AKI was defined as a significant increase in serum creatinine (> 1.5 times baseline or > 26.5 μmol/L increase) or a reduced urine output (< 0.5 mL/kg/h for at least 6 h) or RRT requirement. In the external ICU cohort (FROG-ICU cohort), AKI definition only relied on the serum creatinine criteria [25, 26].
CSA-AKI, Ng, Cleveland, AKICS, and SRI scores were calculated as described [31–35]. Some parameters were approximated as follows: History of congestive heart failure was approximated as left ventricular ejection fraction (LVEF) < 60%; preoperative capillary glucose was assimilated with diabetic status regardless of the treatment received; central venous pressure was considered the maximum pressure during the first 24 h after surgery; and low cardiac output was defined as the need for vasopressive or inotropic drugs.
Urinary peptidome analysis
In the CBP surgery cohorts, urine samples were collected 2.5 to 4 h after surgery and immediately frozen (− 20 ℃) before subsampling and re-frozen for long-term conservation (− 80 ℃). In the external ICU validation cohort, urine was collected in the first 24 h of admission to the ICU, immediately frozen (− 20 ℃) before subsampling and re-frozen for long-term conservation (− 80 °C).
Peptide extraction and CEMS processing were performed as previously described (the extended methodology is given in Additional file 1: S1) [36].
Development of peptide-based score
For peptidome analysis, among the 5862 peptides, only peptides with less than 70% of missing data in at least one group were considered for analysis, resulting in a set of 1255 peptides. For those peptides, missing values were replaced by 0 before further analysis. Univariate testing between AKI and non-AKI patients was performed using the Wilcoxon signed-rank test, followed by Benjamini–Hochberg false discovery rate adjustment. Correlations were performed according to the Pearson method.
For score derivation, all patients from the CBP surgery discovery cohort with available peptidome data were considered (n = 446). A set of 328 differentially abundant peptides with a significant Benjamini–Hochberg adjusted Wilcoxon signed-rank testing was identified in this cohort. The amino acid sequence could be obtained for 204 peptides of the 328 peptides. These 204 peptides were used to build a support vector machine-based predictor (MosaCluster software [37]). Results were extracted as a calibrated score based on derivation. Score performances in the CBP surgery derivation cohort were estimated using the leave-one-out procedure. Score validation was then obtained in the CBP surgery validation cohort in patients with available peptidome data; n = 480. External validation in the ICU context was obtained on all patients from the FROG-ICU cohort with available AKI status and peptidome data (n = 1569).
Additional statistical analyses, including scores comparisons and the use of genetic algorithms, are described in Additional file 1: text S1.
Urine NGAL, IGFBP7, TIMP2, calprotectin, and creatinine measurement
Urine samples were centrifuged for 10 min at 2500 rpms. NGAL was measured using ELISA (Human Lipocalin-2/NGAL DuoSet ELISA, R&D, DY1757) in diluted supernatant (1/10 or 1/100) according to the manufacturer’s protocol. Calprotectin was measured using ELISA (Human S100A8/S100A9 Heterodimer DuoSet ELISA, DY8226-05) according to the manufacturer protocol. Creatinine was measured using the QuantiChrom Creatinine Assay Kit (BioAssay Systems, DICT 500) according to the manufacturer protocol. Creatinine-normalized NGAL and creatinine-normalized calprotectin concentrations (microg/g) were used for performance evaluations. TIMP-2/IGFBP7 was measured in urine supernatants using the VITROS NephroCheck immunoassay on a VITROS 5600 Integrated System (Ortho Clinical Diagnostics) according to the manufacturer’s instructions. The Vitros NephroCheck Test result is a single numerical, which is a product of the measured concentrations of the two analytes in the sample divided by 1000.
Certified laboratory technicians blinded to clinical data performed the analyses.
Mouse model of ischemic AKI
Ischemic AKI was induced in C57Bl6 male mice using warm renal ischemia/reperfusion (bilateral clamping of renal arteries for 20 min) (see Additional file 1: text S1). Kidney samples were analyzed and blood urea nitrogen was measured at 6, 24, and 48 h.
Animal experiments were approved by the local and national ethical committees (CREFRE Inserm/UPS, agreement C31 55,507; Protocol APAFIS#122–2015-23).
Cell culture
MCT cells were grown under standard conditions (21% O2, 5% CO2, 37 °C) and submitted to hypoxia for 48 h or TNFα or IL-1β exposure for 8 h. mRNA of KIM1 and calprotectin was quantified in each condition using quantitative polymerase chain reaction (see Additional file 1: Text S1).
Statistics
Statistical analyses were performed with the R (v4.0.3) software using RStudio interface with additional packages caret [38], pROC [39], ROCR [40], mgcv [41], and GA [42]. Detailed statistical procedures can be found in the Additional file 1: text S1.
Study approval: All the patients were orally informed of the inclusion during the anesthetic consultation performed during the weeks before surgery and the non-opposition of the patients to being included in the clinical and biological collection of the University Hospital of Toulouse was obtained before inclusion (agreement number French national ethical committee DC-2008–463). The study was performed according to the Declaration of Helsinki, as revised in 2004. Patients in the external ICU validation cohort were included in the Frog-ICU cohort [25–27].
Results
Cardiac surgery cohorts and AKI incidence
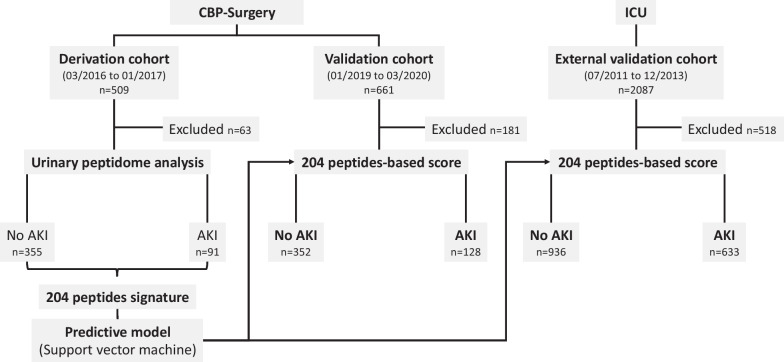
We prospectively included a total of 1170 patients (mean age 66.7 ± 12.0 years) referred for CBP surgery in a single center (Toulouse, France). Inclusion was performed during two different time periods resulting in two independent cohorts: a derivation cohort (n = 509) with patients included between March 2016 and January 2017 and a validation cohort (n = 661) with patients included between January 2019 and March 2020 (Fig. 1).
Fig. 1.
Patient flowchart for the identification and validation of a predictive AKI urinary peptide signature. Three independent cohorts were used: a derivation CBP surgery cohort (n = 509), a validation CBP surgery cohort (n = 661)—both recruited in the University Hospital of Toulouse (France), but during different time periods—and an external ICU cohort (external ICU validation multicenter cohort [25], n = 2087). Sixty-three patients from the derivation and 181 from the validation CBP surgery cohorts were excluded because of missing urine samples or failure of the urinary peptidome analysis pipeline. Five hundred eighteen patients from the external ICU validation cohort were excluded because of missing urine samples, failure of the urinary peptidome analysis pipeline, or missing information with respect to the development or presence of AKI. CBP surgery, cardiac bypass surgery; ICU, intensive care unit
Patient characteristics are summarized in Table 1. Patients from the validation cohort had better baseline kidney function (eGFR 77.6 ± 20.6 vs. 70.3 ± 20.2 mL/min/1.73m2, p < 0.001), were more frequently affected by hypertension (p = 0.029) and chronic obstruction pulmonary disease (COPD) (p < 0.001) and had more frequently undergone previous cardiac surgery (p = 0.022). Due to anesthetic procedure changes between the two inclusion periods, the number of red blood cell (RBC) transfusions during surgery was lower (p = 0.001) and the use of vasoactive agents was more frequent (p = 0.003) in the validation cohort. During the postoperative period, patients included in the validation cohort received iodine contrast agents less frequently (p = 0.001) and had a shorter length of stay within the ICU (p < 0.0001). However, this did not lead to significant differences in AKI incidence between the derivation and validation cohort (~ 23%, KDIGO stage 1–3). Severe AKI (KDIGO stage 2 or 3) was identified in 42 (8.3%) and 93 (14.0%) patients, respectively. Eighteen (3.5%) and 14 (2.1%) patients required RRT in the derivation and validation cohorts, respectively.
Table 1.
Characteristics of patients included in the derivation and validation CBP cohorts
| Parameters CBP cohort | Derivation (n = 509) | Validation (n = 661) | Adjusted univariate p value |
|---|---|---|---|
| Preoperative features | |||
| Male, n (%) | 381 (74.9) | 501 (75.8) | 0.836 |
| Age (years), mean ± SD | 67.6 ± 11.4 | 66.1 ± 12.4 | 0.071 |
| BMI (kg/m2), mean ± SD | 26.9 ± 4.4 | 26.8 ± 4.7 | 0.811 |
| Diabetes, n (%) | 127 (25) | 179 (27.1) | 0.571 |
| Hypertension, n (%) | 269 (52.8) | 399 (60.4) | 0.029 |
| PAOD, n (%) | 49 (9.6) | 59 (8.9) | 0.836 |
| Stroke, n (%) | 33 (6.5) | 57 (8.6) | 0.312 |
| COPD, n (%) | 29 (5.7) | 97 (14.7) | < 0.001 |
| EuroSCORE-II, mean ± SD | 2.4 ± 2.6 | 2.7 ± 4.3 | 0.150 |
| LVEF (%), mean ± SD | 55.9 ± 11.3 | 56.1 ± 11 | 0.844 |
| Serum Creatinine (μmol/L), mean ± SD | 98.3 ± 41.2 | 88.3 ± 30.1 | < 0.001 |
| eGFR (mL/min.1.73m2), mean ± SD | 70.3 ± 20.2 | 77.6 ± 20.6 | < 0.001 |
| Kidney graft recipients, n (%) | 7 (1.4) | 6 (0.9) | 0.771 |
| Per-operative features | |||
| Surgery | |||
| CAB, n (%) | 196 (38.5) | 245 (37.1) | |
| Valvular, n (%) | 175 (34.4) | 199 (30.1) | 0.220 |
| Combined, n (%) | 71 (13.9) | 105 (15.9) | |
| Thoracic aorta, n (%) | 58 (11.4) | 87 (13.2) | |
| Myocardium, n (%) | 9 (1.8) | 25 (3.8) | |
| Previous cardiac surgery, n (%) | 23 (4.5) | 57 (8.6) | 0.022 |
| CBP time (min), mean ± SD | 85.3 ± 36.2 | 88.5 ± 36.5 | 0.246 |
| RBC transfusion, n (%) | 91 (17.9) | 59 (8.9) | < 0.001 |
| Number, mean ± SD | 0.4 ± 0.9 | 0.2 ± 0.8 | 0.001 |
| Vasoactive agents, n (%) | 456 (89.6) | 627 (94.9) | 0.003 |
| Postoperative features | |||
| RBC transfusion, n (%) | 151 (29.7) | 155 (23.4) | 0.045 |
| Number, mean ± SD | 0.8 ± 1.8 | 0.7 ± 1.9 | 0.368 |
| Vasoactive agents, n (%) | 277 (54.4) | 445 (67.3) | < 0.001 |
| Duration (day), mean ± SD | 1.2 ± 2.1 | 1.6 ± 2.6 | 0.004 |
| Infection, n (%) | 83 (16.3) | 112 (16.9) | 0.858 |
| Iodinated contrast agents, n (%) | 25 (4.9) | 7 (1.1) | 0.001 |
| Mechanical ventilation duration (d), mean ± SD | 19.5 ± 77.2 | 12.4 ± 42.6 | 0.122 |
| ICU stay duration (day), mean ± SD | 5.6 ± 6.7 | 4.2 ± 5.2 | < 0.001 |
| Outcomes | |||
| AKI KDIGO | |||
| 0, n (%) | 389 (76.4) | 486 (73.5) | |
| 1, n (%) | 78 (15.3) | 82 (12.4) | 0.005 |
| 2, n (%) | 23 (4.5) | 69 (10.4) | |
| 3, n (%) | 19 (3.7) | 24 (3.6) | |
| In-hospital mortality, n (%) | 15 (2.9) | 26 (3.9) | 0.571 |
This table presents the clinical characteristics and outcome of all CBP patients irrespective of the availability of a urine sample.
BMI Body mass index, PAOD Peripheral artery obliterans disease, COPD Chronic obstructive pulmonary disease, LVEF Left ventricular ejection fraction, eGFR Estimated glomerular filtration rate, CBP Cardiac bypass, RBC Red blood cells, SD Standard deviation, AKI Acute kidney injury, KDIGO Kidney disease/improving global outcome classification
Pre- and per-operative clinical parameters are moderate predictors of AKI after CBP surgery
In the 509 patients of the derivation cohort, the univariate analysis identified several clinical features that were associated with the development of AKI (Table 2). These included preoperative (age, hypertension, kidney transplantation, baseline eGFR) and per-operative (CBP surgery length, RBC transfusion) parameters. The majority remained significantly associated with the development of AKI after multivariate logistic regression adjustment.
Table 2.
Predictive factors of AKI after CBP surgery (derivation cohort, n = 509)
| Parameters (derivation CBP cohort) | Overall (n = 509) | Acute kidney injury | |||
|---|---|---|---|---|---|
| No (n = 389) | Yes (n = 120) | Adjusted univariate p value | Multivariate odds ratio (95% CI) | ||
| Preoperative features | |||||
| Male, n (%) | 381 (74.9) | 295 (75.8) | 86 (71.7) | 0.504 | – |
| Age (years), mean ± SD | 67.6 ± 11.4 | 66.7 ± 12 | 70.3 ± 9.1 | 0.002 | 1.03 [1.00–1.05] |
| BMI (kg/m2), mean ± SD | 26.9 ± 4.4 | 26.8 ± 4.2 | 27.5 ± 5.1 | 0.237 | – |
| Diabetes, n (%) | 127 (25) | 93 (23.9) | 34 (28.3) | 0.492 | – |
| Hypertension, n (%) | 269 (52.8) | 194 (49.9) | 75 (62.5) | 0.033 | 1.55 [0.98–2.50] |
| PAOD, n (%) | 49 (9.6) | 32 (8.2) | 17 (14.2) | 0.116 | 1.34 [0.63–2.72] |
| Stroke, n (%) | 33 (6.5) | 25 (6.4) | 8 (6.7) | 1.000 | – |
| COPD, n (%) | 29 (5.7) | 23 (5.9) | 6 (5) | 0.911 | – |
| EuroSCORE-II, mean ± SD | 2.4 ± 2.6 | 2.1 ± 2.2 | 3.4 ± 3.3 | < 0.001 | – |
| LVEF (%), mean ± SD | 55.9 ± 11.3 | 56 ± 11.4 | 55.7 ± 11.2 | 0.902 | – |
| Serum Creatinine (μmol/L), mean ± SD | 98.3 ± 41.2 | 92.5 ± 25.1 | 117.4 ± 68.7 | 0.001 | – |
| eGFR (mL/min.1.73m2), mean ± SD | 70.3 ± 20.2 | 72.9 ± 18.3 | 61.8 ± 23.7 | < 0.001 | 0.80 [0.71–0.91] |
| Kidney graft recipients, n (%) | 7 (1.4) | 1 (0.3) | 6 (5) | 0.002 | 18.75 [2.64–385.1] |
| Per-operative features | |||||
| Surgery | 0.504 | – | |||
| CAB, n (%) | 196 (38.5) | 156 (40.1) | 40 (33.3) | ||
| Valvular, n (%) | 175 (34.4) | 135 (34.7) | 40 (33.3) | ||
| Combined, n (%) | 71 (13.9) | 50 (12.9) | 21 (17.5) | ||
| Thoracic aorta, n (%) | 58 (11.4) | 41 (10.5) | 17 (14.2) | ||
| Myocardium, n (%) | 9 (1.8) | 7 (1.8) | 2 (1.7) | ||
| Previous cardiac surgery, n (%) | 23 (4.5) | 13 (3.3) | 10 (8.3) | 0.062 | 1.23 [0.39–3.65] |
| CBP time (min), mean ± SD | 85.3 ± 36.2 | 79.3 ± 30.8 | 104.8 ± 44.8 | < 0.001 | 1.02 [1.01–1.03] |
| RBC transfusion, n (%) | 91 (17.9) | 56 (14.4) | 35 (29.2) | 0.001 | 0.50 [0.003–3.33] |
| Number, mean ± SD | 0.4 ± 0.9 | 0.3 ± 0.8 | 0.7 ± 1.2 | 0.003 | 1.34 [0.61–2.93] |
| Vasoactive agents, n (%) | 456 (89.6) | 345 (88.7) | 111 (92.5) | 0.403 | – |
| Postoperative features | |||||
| RBC transfusion, n (%) | 151 (29.7) | 90 (23.1) | 61 (50.8) | < 0.001 | – |
| Number, mean ± SD | 0.8 ± 1.8 | 0.5 ± 1.1 | 1.8 ± 2.9 | < 0.001 | – |
| Vasoactive agents, n (%) | 277 (54.4) | 194 (49.9) | 83 (69.2) | 0.001 | – |
| Duration (day), mean ± SD | 1.2 ± 2.1 | 0.9 ± 1.3 | 2.2 ± 3.6 | 0.001 | – |
| Infection, n (%) | 83 (16.3) | 44 (11.3) | 39 (32.5) | < 0.001 | – |
| Iodinated contrast agents, n (%) | 25 (4.9) | 12 (3.1) | 13 (10.8) | 0.003 | – |
| Mechanical ventilation | |||||
| Duration (d), mean ± SD | 19.5 ± 77.2 | 11.1 ± 35.1 | 46.9 ± 143.4 | 0.014 | – |
| ICU stay duration (day), mean ± SD | 5.6 ± 6.7 | 4.6 ± 3.8 | 8.8 ± 11.4 | 0.001 | – |
| In-hospital mortality, n (%) | 15 (2.9) | 1 (0.3) | 14 (11.7) | < 0.001 | – |
BMI Body mass index, PAOD Peripheral artery obliterans disease, COPD Chronic obstructive pulmonary disease, LVEF Left ventricular ejection fraction, eGFR Estimated glomerular filtration rate, CBP Cardiac bypass, RBC Red blood cells, SD Standard deviation, AKI Acute kidney injury, KDIGO Kidney disease/improving global outcome classification
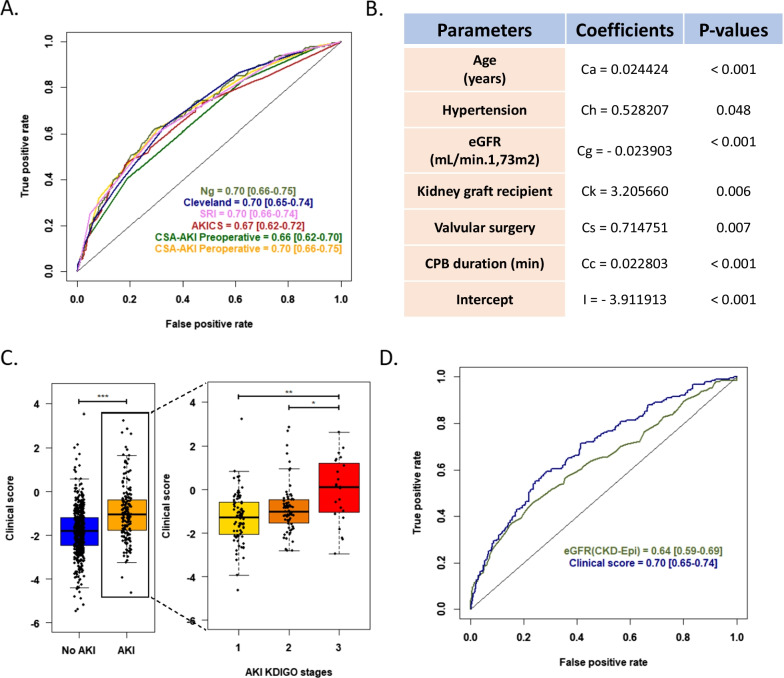
These parameters, both used as single markers (e.g., eGFR) or included in more complex, previously published scores (i.e., CSA-AKI [31], Ng [32], Cleveland [33], AKICS [34] or SRI [35] scores) had poor discriminative power (area under the received operating curves (AUC) of 0.64–0.70 in the validation cohort) (Fig. 2). We therefore built a local clinical score using six pre- and per-operative variables identified by stepwise selection in the derivation cohort (age, hypertension, eGFR, kidney transplantation, valvular surgery, and CBP surgery length) (Fig. 2). However, although there was a clear difference in the local score of patients developing AKI (p < 0.001), this score was not significantly better than the previously established scores (p > 0.1 for all). It is therefore evident that complementary strategies are needed to improve the prediction of patients developing AKI following CBP surgery at an early stage.
Fig. 2.
AKI prediction in CBP surgery patients based on clinical pre- and per-operative features. A. ROC curves and corresponding AUROC [95% confidence interval] of published clinical scores for the prediction of AKI (KDIGO 1, 2, or 3) in the CBP surgery validation cohort. B. Parameters and associated coefficients of a local clinical model defined in the derivation cohort. The clinical score is calculated as follows: logit(p(AKI)) = Ca x Age (Years) + Ch x Hypertension (0/1) + Cg x eGFR (mL/min.1.73m2) + Ck x Kidney Graft Recipient (0/1) + Cs x Valvular Surgery (0/1) + Cc x CPB Duration (min) + I. C. Association of the local clinical score with the development of AKI in the validation cohort (all stages, left; according to KDIGO stages, right). * p < 0.05; ** p < 0.01; *** p < 0.001. D. ROC curves and corresponding AUROC [95% confidence interval] of the local clinical score compared to baseline eGFR for the prediction of all stages of AKI in the validation cohort. The AUROCs of the local clinical score and eGFR were significantly different (Delong test; p value = 0.007). ROC, receiver operating characteristic curve; AUROC, area under the receiver operating characteristic curve; CI, confidence interval; AKI, acute kidney injury; CBP, cardiac bypass; eGFR, estimated glomerular filtration rate
AKI leads to early changes in the urinary peptidome
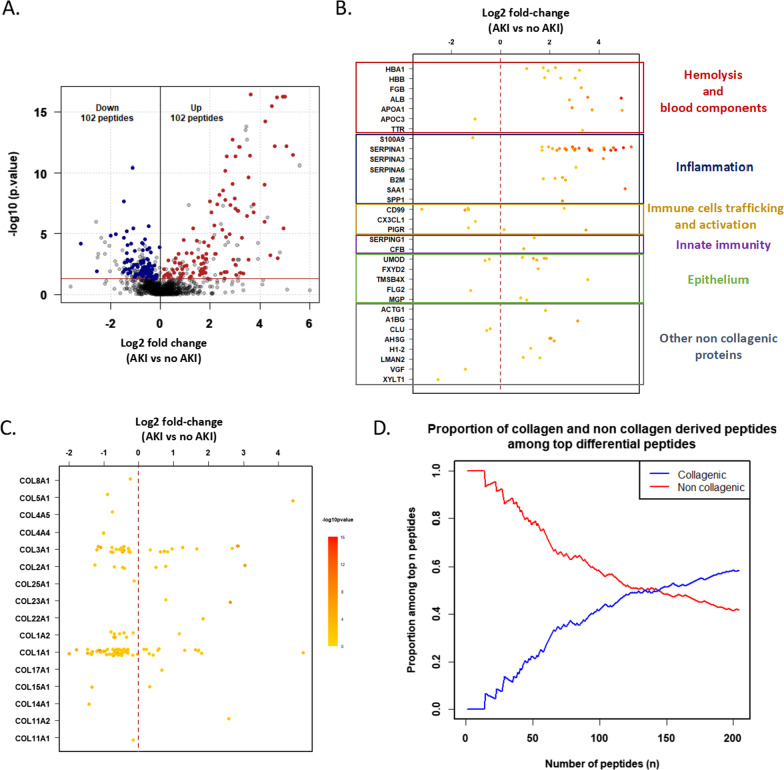
We analyzed for the first time at a very early stage (< 4 h after CBP surgery) the urinary peptide content in this large CBP surgery cohort to identify candidate biomarkers predictive of AKI that could help with clinical decision-making and stratifying patients for further interventional studies. Using the 509 patients in the derivation cohort, 204 displayed a significantly different abundance (Benjamini–Hochberg adjusted Wilcoxon univariate testing (p < 0.05)) in the urine of patients developing AKI (up-regulated n = 102; down-regulated n = 102; Fig. 3). Peptides associated with AKI were derived from 48 proteins, including 16 collagens and 32 non-collagenous proteins. Interestingly, when ranked according to their adjusted univariate p values, top differential peptides were mainly derived from non-collagenous proteins (Fig. 3). These non-collagenous proteins were related to hemolysis (e.g., hemoglobin α and β-subunit), inflammation (e.g., S100A9 (a calprotectin subunit), SERPINA1 (α1-antitrypsin), B2M (β2-microglobulin)), immune cell trafficking and activation (i.e., CD99, CX3CL1, PIGR), innate immunity (e.g., complement factor B), kidney epithelium (e.g., UMOD (uromodulin), FXYD2) and cell growth and survival (e.g., ACTG1).
Fig. 3.
Urinary peptidome changes during CBP-surgery-induced AKI. A. Peptides with significantly different abundances in AKI patients. The volcano plot displays log10-transformed and adjusted univariate p values as a function of log2-transformed fold changes of urinary peptides amplitudes. Sequenced peptides with differential abundances (significant after Benjamini–Hochberg adjusted Wilcoxon univariate testing (p < 0.05)) are represented in color (more abundant: red; less abundant: blue). B. Peptides with significantly different abundances (in respect to distribution frequency and amplitude signal) derived from non-collagenic proteins (y position), log2 fold changes (x position), and –log10 adjusted p values (color scale). The brown dashed line represents log2 fold change = 0. Peptides are ranked according to their functional role during the AKI progression (inflammation, epithelium, blood component, other non-collagenic proteins). C. Peptides with significantly different abundances derived from collagenic proteins (y position), log2 fold changes (x position), and –log10 adjusted p values (color scale). The brown dashed line represents log2 fold change = 0. D. Collagenic and non-collagenic proteins-derived peptides proportions among top differential peptides according to the Benjamini–Hochberg adjusted p value ranking. The red line corresponds to non-collagen and the blue line to collagen-derived peptides
To confirm the relevance of the urinary signature, we measured the expression of S100A9, alone or combined in the form of calprotectin (S100A8/S100A9) in in vitro and in vivo models of epithelial injuries, as well as in the urine of CBP surgery patients. As shown in Additional file 1: Fig. S1, following cardiac surgery, calprotectin was increased in the urine of patients who will develop AKI (Fig. S1A). In C57Bl/6 mice, 20-min bilateral renal ischemia/reperfusion injury induced AKI characterized by increased BUN. In mice, AKI was associated with the upregulation of renal S100A9 mRNA expression as well as S100A9 protein expression within the kidney cortico-medullary junction (Fig. S1B-E). Renal expression of the kidney injury molecule Kim1 was also increased in this model (Fig. S1D). Finally, in mouse proximal tubule cells (MCT cell line), 48 h of hypoxia and/or 8 h exposure to the pro-inflammatory cytokine tumor necrosis factor-α or interleukin-1β induced S100a9 and Kim1 mRNA expression (Fig. S1G-J).
A urinary peptide-based signature predicts CBP-surgery-induced AKI and significantly outperforms clinical parameters.
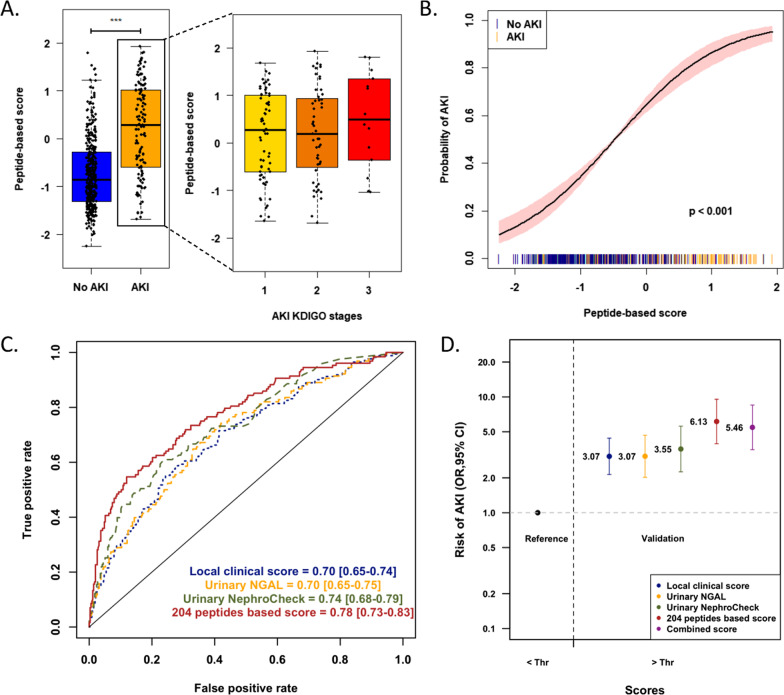
Next, the 204 peptides were included using machine learning in a support vector machine-based mathematical model that was trained on the derivation cohort and applied in the validation cohort. In the validation cohort, the signature clearly identified patients developing AKI (p < 0.001), with an increase in AKI risk at a higher peptide-based score (p < 0.001; Fig. 4).
Fig. 4.
Independent validation of the predictive value of the urinary peptide signature for early AKI diagnosis after CBP surgery. A Association of the urinary peptide-based score with the development of AKI (all stages, left; according to KDIGO stages, right) in the CBP surgery validation cohort (n = 480). *** p < 0.001. B Spline plot of the association between the peptide-based score and the risk of developing AKI. A univariate logistic generalized additive model was used. The black line indicates the estimated risk of AKI with respective 95% confidence intervals. The spikes show the distribution of the peptide-based scores. C ROC curves with corresponding AUROC and 95% confidence intervals for the 204 peptides-based score, the local clinical score (and reference urinary AKI biomarkers NGAL and TIMP2*IGFBP7 for the prediction of AKI (all stages) in the validation cohort. D Odds ratios and corresponding 95% confidence intervals in the validation cohort using a multivariate logistic regression model including the local clinical score, reference urinary AKI biomarkers NGAL and TIMP2*IGFBP7, the 204 peptides-based score or a combination of the local clinical and peptide-based scores as a qualitative value according to the selected threshold (optimal Youden index in the derivation cohort). ROC, receiver operating characteristics curve; AUROC, area under the receiver operating characteristics curve; CI, confidence interval; Thr, threshold
Pre- and per-operative characteristics (diabetes mellitus, COPD, baseline eGFR, surgery indication, redux surgery, CBP surgery length) partly correlated with the peptide-based score (Additional file 1: Table S1). However, the urinary peptide signature contained complementary information with respect to the clinical characteristics, as evidenced by (i) an adjusted R-squared of the multivariate model of 0.246, (ii) the significant correlation between AKI development and the peptide score after adjustment for baseline eGFR (p < 0.001), clinical score (p < 0.001) and individual clinically significant covariates (p < 0.001 for all), (iii) the significantly higher AUC of the peptide score (0.78 [0.73–0.83]) compared to that of the local clinical score (p < 0.001) or the baseline eGFR (p < 0.001), and (iv) the lack of improved performance when combining clinical and peptides-based scores (see below and Fig. S2). A peptide-based score above the threshold was associated with an increased risk of AKI (OR = 6.13 [3.95–9.59], p < 0.001; positive and negative predictive value 0.59 and 0.81, respectively, and positive net reclassification index (0.19) compared to clinical score Fig. 4), even after adjustment for the local clinical score or preoperative clinical features. Performances were also superior to the reference urinary biomarkers NGAL (0.70 [0.65–0.75], p = 0.004) and similar to the [IGFBP7]*[TIMP2] product (0.74 [0.68–0.79], p = 0.14). However, a [IGFBP7]*[TIMP2] product over the recommended cutoff of 0.3 was associated with an increased risk of AKI but with a lower prediction compared to the peptide-based score (OR 3.55 [2.28–5.65], p < 0.001). The urinary peptide score was also significantly associated with severe AKI stage 2 or 3, similarly to the [IGFBP7]*[TIMP2] product (AUC 0.74 [0.68–0.80] vs. 0.73 [0.67–0.79], respectively, p = 0.6), and outperformed NGAL (0.67 [0.61–0.74], p = 0.04).
The combination of the local clinical and the peptide-based scores did not significantly improve overall performances compared to the peptide-based score alone (Fig. 4). Last, the initial signature of 204 peptides could be reduced to 17 peptides using advanced feature selection methods such as genetic algorithms, with similar performances compared to the full 204 peptide signature in the validation cohort (AUC = 0.77 [0.72–0.82], p = 0.676 for comparison between the two AUC; OR = 5.67 [3.68–8.82], p < 0.001) (Additional file 1: Fig. S2 and Additional file 1: Table S2).
Validation in an external cohort of patients admitted to the ICU
CBP surgery AKI is considered a standardized at-risk setting of conditions with the potential to induce AKI. To study the potential generalization of the use of peptide signatures in AKI, we also evaluated the performance of the urinary peptide signature in 1569 patients admitted to an intensive care unit (ICU), with various causes of AKI, including sepsis, unplanned surgery, and trauma (FROG-ICU cohort) [25–27].
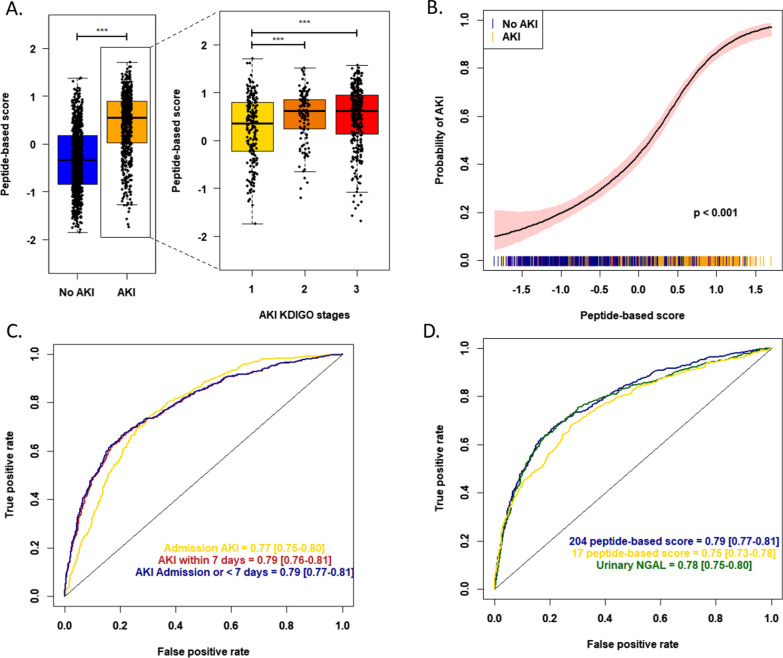
Interestingly, the urinary peptide-based signature assessed in patients at their admission in the ICU was significantly associated with the development of AKI (Fig. 5), and its performance characteristics were equal to those obtained in the case of CBP surgery AKI (AUC 0.79 [0.77–0.81] versus 0.78 [0.73–0.83], p = 0.634; OR = 5.92 [4.73–7.45], p < 0.001). These findings were confirmed when patients were stratified according to the time until AKI diagnosis: diagnosis of AKI at admission AUC 0.77 [0.75–0.80]; AKI within 7 days of admission AUC 0.79 [0.76–0.81]. Subgroup analysis, according to the underlying disease that justified ICU admission, showed quite good performances among groups, despite some heterogeneity. The best AUCs were obtained in patients admitted after surgery (n = 146; AUC 0.86 [0.80–0.92]), for sepsis (n = 378; 0.78 [0.74–0.83]) or for impaired hemodynamic status (n = 193; AUC 0.81 [0.75–0.88]) (Additional file 1: Table S3).
Fig. 5.
External validation of the predictive value of the urinary peptide signature for AKI diagnosis in an intensive care unit (ICU) cohort of 1,569 patients. A Peptide-based score according to AKI status (all stages (left part); according to KDIGO stages, right part) in the external ICU validation cohort. *** p < 0.001. B Spline plot of the association between the peptide-based score and the risk of developing AKI in the external ICU cohort. A univariate logistic generalized additive model was used. The black line indicates the estimated risk of AKI with respective 95% confidence intervals. The spikes show the distribution of the peptidome-based scores. C ROC curves and corresponding AUROC 95% confidence intervals of the 204 peptides-based score in the external ICU validation cohort, according to the time until AKI diagnosis. D ROC curves and corresponding AUROC 95% confidence intervals of the 204 peptides-based score and reference urinary biomarker NGAL in the external ICU validation cohort
When compared with reference urinary AKI biomarkers, such as NGAL, those performances were broadly equivalent (AUC = 0.78 [0.75–0.80], p = 0.426, Fig. 5) for both AKI at admission and AKI within seven days of admission (Additional file 1: Fig. S3). Evaluation of the [IGFBP7]*[TIMP2] product was not available in this cohort. It is worth noting that the reduced 17 peptide signature exhibited a significantly decreased performance compared to the full 204 peptide signature in this ICU setting of mixed AKI etiologies (AUC = 0.75 [0.73–0.78], p < 0.001).
The urinary peptide-based AKI signature provides insights on early mortality
Among the 1170 patients of the CBP surgery cohort, 41 (3.5%) died during hospitalization, including 30 (2.5%) during the first month. The development of AKI was strongly associated with in-hospital mortality (OR 16.2 [7.5–40.1]; p < 0.001), even after adjustment of the preoperative mortality score (EuroSCORE-2: OR 13.8 [6.3–34.5]; p < 0.001) or after multivariate adjustment using a propensity score (OR 5.6 [2.4–14.8]; p < 0.001) (Additional file 1: Fig S4). In both the derivation and validation cohorts, the peptide-based score obtained in samples collected < 4 h after the surgery was associated with higher in-hospital mortality (AUC 0.77 [0.66–0.88]; p < 0.001; OR when peptidome score was above threshold: 6.2 [2.6–16.2]; p < 0.001). Similarly, in the external ICU validation cohort, the peptide-based score assessed at admission was also associated with the in-hospital mortality (OR 2.62 [2.05–3.38], p < 0.001).
Discussion
In this study that included > 2400 patients, we identified a urinary peptide signature that predicts the development of AKI as early as 3–4 h after the initial insult in a variety of high-risk clinical situations, including CBP surgery and ICU admission. Cardiac surgery with cardiac bypass is the prototypical cause of AKI, with a pathophysiology based on various interrelated mechanisms (inflammation, ischemia, hemolysis, oxidative stress, tubular cell injury) [43]. The nature of the peptides identified can be clearly linked to these different mechanisms [8, 43–47] and potentially explains the good performance of the urinary peptide signature in predicting future AKI at this early stage. Furthermore, these mechanisms are shared by various other etiologies of AKI [8, 44–47], which may account for the excellent transferability of the urinary peptide signature to the ICU setting. Hence, our findings highly suggest that such an omic signature may help to refine, a posteriori, the molecular pathways that may predict the response to a dedicated therapeutic.
In the CBP surgery setting, the urine peptide signature outperformed all clinical parameters and urinary NGAL, while the predictive value of the peptide signature was similar to NGAL in an ICU setting. This is most likely due to differences in the time window of the initial event leading to the development of AKI. While in the CBP surgery cohort all samples were obtained at the latest 4 h after the initial injury, in the ICU validation cohort, urine samples were collected within 24 h after ICU admission. In addition, a number of ICU patients had already full-blown AKI on admission to the ICU [25, 27]. This more advanced stage of AKI in the ICU cohort may explain the comparable performance of the urine peptide signature and NGAL in this setting.
In the ICU cohort, the performance of the urine peptide signature was associated with a diagnosis warranting admission to ICU. As expected, because the peptides were identified in the CBP surgery setting, excellent performances were observed in the postoperative subgroup of ICU patients, but surprisingly also in the groups with hemodynamic failure or sepsis. In contrast, performance was poorer after cardiac arrest and respiratory failure, probably due to the involvement of other pathophysiological mechanisms.
A number of peptides in the signature can readily be linked to the pathophysiology of AKI (e.g., alpha-1-antitrypsin (SERPINA1), calprotectin (S100A9), serum amyloid A (SAA) for inflammation, uromodulin (UMOD), albumin (ALB)). However, some other peptides, including thymosin β4 (TMSB4X), CD99, and Na–K-ATPase γ-subunit (FXYD2), are not currently known to be involved in AKI pathophysiology and may represent good candidates for further exploration of their role in AKI. Finally, the identified peptides also suggest candidate targets for specific treatment of AKI. For example, the increased urinary abundance of the complement factor B (CFB)-derived peptide in AKI argues for targeting the alternative complement pathway with recently developed complement inhibitors [48, 49]. We also observed here that calprotectin is dramatically induced in kidney after epithelial injury in a preclinical model. Targeting calprotectin signaling with paquinimod demonstrated beneficial effects to prevent the development of ischemic AKI [50]. This exemplifies the fact that such a urinary peptide-based strategy may, in addition to detecting at an early stage patients who will develop AKI, also furnish druggable targets.
The limitations of the direct clinical application of the peptide signature in this context of AKI are the specific equipment currently required (capillary electrophoresis coupled with mass spectrometry (CEMS)) and the time needed for the sample preparation and analysis (currently about 12 h). Future improvements on sample preparation, by avoiding the 6-h-long lyophilization step and reducing instrument time, should allow the analysis time window to be reduced to a few hours, compatible with the timely decision-making necessary in patients at risk for AKI. Alternatively, peptides could also be measured using strategies such as multiplex ELISA, if peptide abundance correlates with the parental protein abundance. Targeted mass spectrometry (e.g., multiple reaction monitoring) is another alternative [51]. However, such strategies require a significant reduction in the number of peptides. We observed that reducing the signature from 204 to 17 peptides is possible without significant loss of performance in the CPB cohort, but it led to a significant reduction in the efficiency of AKI prediction in the ICU cohort. This may be due to the fact that the overall signature of 204 peptides incorporates information that is redundant in CBP-surgery-induced AKI, but is essential in other risk situations encountered in the ICU. As some peptides correlate well with clinical features (data not shown), one way to further reduce the size of the signature would be to identify a score complementary to the readily available clinical information. Finally, further improvement in the prediction and reduction in the number of urinary peptides could come from future studies focusing on the discovery of novel molecular markers in, for example, plasma, characterization of immune cell populations, and genetic predisposition for AKI in CBP surgery patients. Strong and complementary molecular markers identified from those sources could help, using machine learning, to further reduce the signature but also further improve the efficacy of the prediction.
Other limitations included the lack of assessment of the urinary [IGFBP7]*[TIMP2] product in all cohorts that would have allowed to robustly compare it to the peptide signature in clinical settings beyond CBP surgery. Also, we did not assess other single biomarkers like KIM-1 or L-FABP. However, the aim of our study was not only to test the AKI predictive value of urinary peptides but also to test whether this approach may identify new, targetable, molecular players of AKI to ultimately develop personalized medicine.
In conclusion, we have identified and validated a urine peptide signature predictive of AKI in a variety of situations at risk. This overarching signature, which contains numerous peptides directly associated with the pathophysiology of AKI, holds great promise for identifying patients developing AKI early after injury and developing tailored treatment.
Supplementary Information
Additional file 1: Supplementary Figure S1: S100A9 expression after epithelial injury. A. Urinary calprotectin (S100A8/A9) abundance 4 hours after cardiac bypass-surgery. B–F. Blood urea nitrogen (B), mRNA S100a9 (C), S100A9 immunostaining (D-E) and mRNA Kim1 (F) in sham mice and after bilateral renal ischemia/reperfusion (hours 6, 24 and 48). G–J. mRNA expression of S100a9 and Kim1 in MCT cells submitted to interleukin-1b (IL1β, 10 ng/mL) or tumor necrosis factor-1a (TNFa, 10 ng/mL) (G-H) or hypoxia (I-J). AKI, acute kidney injury; BUN, blood urea nitrogen; Norm, normoxia; Hyp, hypoxia. Supplementary Figure S2: Performances of the peptide-based signature to identify AKI that developed within the first 2 days following cardiac surgery. ROC curves with corresponding AUROC and 95% confidence intervals of the local clinical score (blue, pointed), the full 204 peptides-based score (red), the urinary NGAL level (yellow, pointed) and the nephrocheck ([IGFBP7].[TIMP2] product) in the validation cohort. Supplementary Figure S3: Reduction and combination of the peptide-based signature. A. ROC curves with corresponding AUROC and 95% confidence intervals of the local clinical score (blue, pointed), the full 204 peptides-based score (red), the reduced 17 peptides-based score (black, dashed) and the combination of local clinical and full peptide-based score in the validation cohort. B. List of peptides included in the reduced 17-peptides signature according to their parental protein. LMAN2, Lectin mannose binding 2 ; MGP, Matrix gla protein. Supplementary Figure S4: Performances of the 204 peptides-based signature and the reference urinary biomarker NGAL for AKI prediction in the external ICU validation cohort. A. ROC curves with corresponding AUROC and 95% confidence intervals of the 204 peptides-based score and the reference urinary biomarker NGAL to predict AKI after ICU admission. B. ROC curves with corresponding AUROC and 95% confidence intervals of the 204 peptides-based score and reference urinary biomarker NGAL to predict the development of AKI within seven days after admission. Supplementary Figure S5: Performances of the 204 peptides-based score for in-hospital mortality prediction. Odds-ratio (OR) of in-hospital mortality were calculated with unadjusted, Euroscore-II-adjusted or propensity score-adjusted logistic regression. Supplementary Table S1: Correlations between clinical characteristics and the 204-peptides-based score. BMI, body mass index; PAOD, peripheral artery obliterans disease; COPD, chronic obstructive pulmonary disease; LVEF, left ventricular ejection fraction; eGFR, estimated glomerular filtration rate; CBP, cardiac bypass; RBC, red blood cells. Supplementary Table S2: Performance of the peptide-based score to predict acute kidney injury in the external ICU validation cohort, according to the cause of admission to the intensive care unit. ICU, intensive care unit; AUROC, area under the receiver operating characteristics curve. Supplementary file S1: Methodology. Urinary peptidomics and statistical analyses.
Acknowledgements
We would like to thank Yatma Thiao for his assistance with the mouse and MCT cell experiments. We acknowledge the Phenotyping Department of the UMS006 CREFRE-Anexplo Platform (Toulouse, France) for plasma analysis in mice and the GeT-Santé facility (UMR1297-Toulouse, France) for technical assistance with qPCR analysis. We greatly acknowledge the members of the Kidney Attack study group (Prof. Stanislas FAGUER, Department of Nephrology and Organ Transplantation, University Hospital of Toulouse, Toulouse, France; Prof. Bertrand MARCHEIX, Department of Cardiac and Vascular Surgery, University Hospital of Toulouse, Toulouse, France; Dr. Etienne GRUNENWALD, Department of Cardiac and Vascular Surgery, University Hospital of Toulouse, Toulouse, France; Prof. Vincent MINVILLE, Department of Anesthesiology and Critical Care Medicine, University Hospital of Toulouse, Toulouse, France; Dr. Julia GROSSAC, Department of Anesthesiology and Critical Care Medicine, University Hospital of Toulouse, Toulouse, France; Dr. François LABASTE, Department of Anesthesiology and Critical Care Medicine, University Hospital of Toulouse, Toulouse, France; Dr. Nicolas MAYEUR, Department of Anesthesiology and Critical Care Medicine, University Hospital of Toulouse, Toulouse, France; Dr. Elsa TARDIF, Department of Anesthesiology and Critical Care Medicine, University Hospital of Toulouse, Toulouse, France; Joost P. SCHANSTRA, Institute for Metabolic and Cardiovascular Disease, National Institute of Health and Medical Research, Toulouse, France; Julie KLEIN, Institute for Metabolic and Cardiovascular Disease, National Institute of Health and Medical Research, Toulouse, France; Benjamin BREUIL, Institute for Metabolic and Cardiovascular Disease, National Institute of Health and Medical Research, Toulouse, France; Melinda ALVES, Institute for Metabolic and Cardiovascular Disease, National Institute of Health and Medical Research, Toulouse, France; Audrey CASEMAYOU, Institute for Metabolic and Cardiovascular Disease, National Institute of Health and Medical Research, Toulouse, France; Guylène FEUILLET, Institute for Metabolic and Cardiovascular Disease, National Institute of Health and Medical Research, Toulouse, France; Marie BULÉON, Institute for Metabolic and Cardiovascular Disease, National Institute of Health and Medical Research, Toulouse, France; Manon BRUNET, Institute for Metabolic and Cardiovascular Disease, National Institute of Health and Medical Research, Toulouse, France; Stéphane GAZUT, Université Paris-Saclay, CEA, List, F-91120, Palaiseau, France; Alberto ORTIZ, Instituto de Investigación Sanitaria-Fundación Jiménez Díaz-Universidad Autónoma de Madrid, Madrid, Spain; Justyna SIWY, Mosaiques Diagnostics, Hannover, Germany; Jochen METZGER, Mosaiques Diagnostics, Hannover, Germany; Harald MISCHAK, Mosaiques Diagnostics, Hannover, Germany
Author’s information
AP, JS and JK are co-first author owing to their relative contributions to conceptualization, data collection, urinary samples analyses, statistical analyses, and manuscript writing.
Abbreviations
- AKI
Acute kidney injury
- CBP
Cardiopulmonary bypass
- CKD
Chronic kidney disease
- COPD
Chronic obstructive pulmonary disease
- eGFR
Estimated glomerular filtration rate
- KDIGO
Kidney diseases improving global outcomes
- ICU
Intensive care unit
- OR
Odds ratio
- RBC
Red blood cells
Author contributions
SF, JS, JPS, AP, JK, AO, HM, JM, and SG contributed to conceptualization. SG, AP, SF, AA, and JS contributed to methodology. BM, FD, SF, AP, VM, JG, LMH, LF, BB, ABS, AAG, MA, and ET investigated the study. SF, HM, SG, JK, JM, and JPS provided funding acquisition. SF, AP, and JPS performed writing—original draft. SF, JPS, JS, JK, SG, and FD performed writing—review and editing. All authors approved the final version of the manuscript.
Funding
This work was supported in part through ERA PerMed-JTC 2018 project KIDNEY ATTACK ANR‐18‐PERM‐0003 (France), 01KU1907 (Germany), and AC18/00 064 (Spain), and by a grant from the “Fondation pour la Recherche Médicale” (grant number DEQ20170336759). AP was supported by a research grant from Toulouse University. JK was supported by a grant from the French National Research Agency (ANR), “Calprotect,” grant number ANR-21-CE14-0013-01.
Availability of data and materials
All data, code, and materials used in the analysis are available upon request.
Declarations
Ethical approval and consent to participate
All the patients were orally informed of the inclusion and the non-opposition of the patients to be included in the clinical and biological collection of the University Hospital of Toulouse was obtained before inclusion (agreement number French national ethical committee DC-2008–463). The study was performed according to the Declaration of Helsinki, as revised in 2004.
Consent for publication
Not applicable.
Competing interests
Justyna Siwy and Jochen Metzger are the employee and former employee at Mosaiques diagnostics GmbH (Hannover, Germany), respectively. Harald Mischak is the CEO of Mosaiques diagnostics GmbH. The other authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Alexis Piedrafita, Justyna Siwy, Julie Klein, Joost P. Schanstra and Stanislas Faguer provided equal contribution to this work
Contributor Information
Joost P. Schanstra, Email: joost-peter.schanstra@inserm.fr
Stanislas Faguer, Email: stanislas.faguer@inserm.fr.
Kidney Attack Study Group:
Etienne Grunenwald, Guylène Feuillet, Marie Buléon, and Manon Brunet
References
- 1.Mehta RL, Cerdá J, Burdmann EA, Tonelli M, García-García G, Jha V, et al. International society of nephrology’s 0by25 initiative for acute kidney injury (zero preventable deaths by 2025): a human rights case for nephrology. The Lancet. 2015;385(9987):2616–2643. doi: 10.1016/S0140-6736(15)60126-X. [DOI] [PubMed] [Google Scholar]
- 2.Coca SG, Singanamala S, Parikh CR. Chronic kidney disease after acute kidney injury: a systematic review and meta-analysis. Kidney Int. 2012;81(5):442–448. doi: 10.1038/ki.2011.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vanholder R, Rondeau E, Anders HJ, Carlson N, Fliser D, Kanbay M, et al. EDTAKI: a nephrology and public policy committee platform call for more European involvement in acute kidney injury. Nephrol Dial Transplant. 2022;37(4):740–748. doi: 10.1093/ndt/gfab018. [DOI] [PubMed] [Google Scholar]
- 4.Vaidya VS, Ferguson MA, Bonventre J, v, Biomarkers of acute kidney injury. Annu Rev Pharmacol Toxicol. 2008;48(1):463–493. doi: 10.1146/annurev.pharmtox.48.113006.094615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teo SH, Endre ZH. Biomarkers in acute kidney injury (AKI ) Best Pract Res Clin Anaesthesiol. 2017;31(3):331–344. doi: 10.1016/j.bpa.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Rizvi MS, Kashani KB. Biomarkers for early detection of acute kidney injury. J Appl Lab Med. 2017;2(3):386–399. doi: 10.1373/jalm.2017.023325. [DOI] [PubMed] [Google Scholar]
- 7.Charlton JR, Portilla D, Okusa MD. A basic science view of acute kidney injury biomarkers. Nephrol Dial Transplant. 2014;29(7):1301–1311. doi: 10.1093/ndt/gft510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ronco C, Bellomo R, Kellum JA. Acute kidney injury. The Lancet. 2019;394(10212):1949–1964. doi: 10.1016/S0140-6736(19)32563-2. [DOI] [PubMed] [Google Scholar]
- 9.Krawczeski CD, Goldstein SL, Woo JG, Wang Y, Piyaphanee N, Ma Q, et al. Temporal relationship and predictive value of urinary acute kidney injury biomarkers after pediatric cardiopulmonary bypass. J Am Coll Cardiol. 2011;58(22):2301–2309. doi: 10.1016/j.jacc.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Z, Ravassa S, Nkuipou-Kenfack E, Yang W, Kerr SM, Koeck T, et al. Novel urinary peptidomic classifier predicts incident heart failure. J Am Heart Assoc. 2017;6(8):5432. doi: 10.1161/JAHA.116.005432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bowler RP, Duda B, Chan ED, Enghild JJ, Ware LB, Matthay MA, et al. Proteomic analysis of pulmonary edema fluid and plasma in patients with acute lung injury. Am J Physiol-Lung Cellular Molecul Physiol. 2004;286(6):L1095–L1104. doi: 10.1152/ajplung.00304.2003. [DOI] [PubMed] [Google Scholar]
- 12.Bhargava M, Becker TL, Viken KJ, Jagtap PD, Dey S, Steinbach MS, et al. Proteomic profiles in acute respiratory distress syndrome differentiates survivors from non-survivors. PLoS ONE. 2014;9(10):e109713. doi: 10.1371/journal.pone.0109713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stenemo M, Nowak C, Byberg L, Sundström J, Giedraitis V, Lind L, et al. Circulating proteins as predictors of incident heart failure in the elderly. Eur J Heart Fail. 2018;20(1):55–62. doi: 10.1002/ejhf.980. [DOI] [PubMed] [Google Scholar]
- 14.Bauzá-Martinez J, Aletti F, Pinto BB, Ribas V, Odena MA, Díaz R, et al. Proteolysis in septic shock patients: plasma peptidomic patterns are associated with mortality. Br J Anaesth. 2018;121(5):1065–1074. doi: 10.1016/j.bja.2018.05.072. [DOI] [PubMed] [Google Scholar]
- 15.Hayashi N, Yamaguchi S, Rodenburg F, Ying Wong S, Ujimoto K, Miki T, et al. Multiple biomarkers of sepsis identified by novel time-lapse proteomics of patient serum. PLoS ONE. 2019;14(9):e0222403. doi: 10.1371/journal.pone.0222403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rogers RS, Hibbert K, Sharma R, Calvo S, Brait KL, Pulido NA, et al. (2020) The proteomic signature of septic shock differs from cardiogenic shock or bacteremia without sepsis or shock. In: D33 Sepsis Immunol. Am Thoracic Society A6507–A6507.
- 17.Marx D, Metzger J, Pejchinovski M, Gil RB, Frantzi M, Latosinska A, et al. Proteomics and metabolomics for AKI diagnosis. Semin Nephrol. 2018;38(1):63–87. doi: 10.1016/j.semnephrol.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 18.Klein J, Bascands JL, Mischak H, Schanstra JP. The role of urinary peptidomics in kidney disease research. Kidney Int. 2016;89(3):539–545. doi: 10.1016/j.kint.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 19.Argilés À, Siwy J, Duranton F, Gayrard N, Dakna M, Lundin U, et al. CKD273, a new proteomics classifier assessing CKD and Its prognosis. PLoS ONE. 2013;8(5):e62837. doi: 10.1371/journal.pone.0062837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nkuipou-Kenfack E, Duranton F, Gayrard N, Argilés À, Lundin U, Weinberger KM, et al. Assessment of metabolomic and proteomic biomarkers in detection and prognosis of progression of renal function in chronic kidney disease. PLoS ONE. 2014;9(5):e96955. doi: 10.1371/journal.pone.0096955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodríguez-Ortiz ME, Pontillo C, Rodríguez M, Zürbig P, Mischak H, Ortiz A. Novel urinary biomarkers for improved prediction of progressive eGFR loss in early chronic kidney disease stages and in high risk individuals without chronic kidney disease. Sci Rep. 2018;8(1):15940. doi: 10.1038/s41598-018-34386-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wendt R, Thijs L, Kalbitz S, Mischak H, Siwy J, Raad J, et al. A urinary peptidomic profile predicts outcome in SARS-CoV-2-infected patients. EClinicalMedicine. 2021;36:100883. doi: 10.1016/j.eclinm.2021.100883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Metzger J, Kirsch T, Schiffer E, Ulger P, Mentes E, Brand K, et al. Urinary excretion of twenty peptides forms an early and accurate diagnostic pattern of acute kidney injury. Kidney Int. 2010;78(12):1252–1262. doi: 10.1038/ki.2010.322. [DOI] [PubMed] [Google Scholar]
- 24.Metzger J, Mullen W, Husi H, Stalmach A, Herget-Rosenthal S, Groesdonk H, v,, et al. Acute kidney injury prediction in cardiac surgery patients by a urinary peptide pattern: a case-control validation study. Crit Care. 2016;20(1):157. doi: 10.1186/s13054-016-1344-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gayat E, Cariou A, Deye N, Vieillard-Baron A, Jaber S, Damoisel C, et al. Determinants of long-term outcome in ICU survivors: results from the FROG-ICU study. Crit Care. 2018;22(1):8. doi: 10.1186/s13054-017-1922-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mebazaa A, Casadio MC, Azoulay E, Guidet B, Jaber S, Levy B, et al. Post-ICU discharge and outcome: rationale and methods of the The French and European outcome reGistry in intensive care units (FROG-ICU) observational study. BMC Anesthesiol. 2015;15(1):143. doi: 10.1186/s12871-015-0129-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nkuipou-Kenfack E, Latosinska A, Yang WY, Fournier MC, Blet A, Mujaj B, et al. A novel urinary biomarker predicts 1-year mortality after discharge from intensive care. Crit Care. 2020;24(1):10. doi: 10.1186/s13054-019-2686-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levey AS, Stevens LA, Schmid CH, Zhang Y, (Lucy), Castro AF, Feldman HI,, et al. A new equation to estimate glomerular filtration rate. Ann Intern Med. 2009;150(9):604. doi: 10.7326/0003-4819-150-9-200905050-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nashef SAM, Roques F, Sharples LD, Nilsson J, Smith C, Goldstone AR, et al. EuroSCORE II. Eur J Cardiothorac Surg. 2012;41(4):734–745. doi: 10.1093/ejcts/ezs043. [DOI] [PubMed] [Google Scholar]
- 30.2012 Kidney disease: improving global outcomes (KDIGO) clinical practice guideline for acute kidney injury (AKI). https://kdigo.org/guidelines/acute-kidney-injury/.
- 31.Jiang W, Teng J, Xu J, Shen B, Wang Y, Fang Y, et al. Dynamic predictive scores for cardiac surgery–associated acute kidney injury. J Am Heart Assoc. 2016;5(8):14. doi: 10.1161/JAHA.116.003754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ng SY, Sanagou M, Wolfe R, Cochrane A, Smith JA, Reid CM. Prediction of acute kidney injury within 30 days of cardiac surgery. J Thorac Cardiovasc Surg. 2014;147(6):1875–1883.e1. doi: 10.1016/j.jtcvs.2013.06.049. [DOI] [PubMed] [Google Scholar]
- 33.Thakar C v., Arrigain S, Worley S, Yared JP, Paganini EP. A Clinical Score to Predict Acute Renal Failure after Cardiac Surgery. Journal of the American Society of Nephrology. 2005 Jan;16(1):162–8. [DOI] [PubMed]
- 34.Palomba H, de Castro I, Neto ALC, Lage S, Yu L. Acute kidney injury prediction following elective cardiac surgery: AKICS Score. Kidney Int. 2007;72(5):624–631. doi: 10.1038/sj.ki.5002419. [DOI] [PubMed] [Google Scholar]
- 35.Mehta RH, Grab JD, O’Brien SM, Bridges CR, Gammie JS, Haan CK, et al. Bedside tool for predicting the risk of postoperative dialysis in patients undergoing cardiac surgery. Circulation. 2006;114(21):2208–2216. doi: 10.1161/CIRCULATIONAHA.106.635573. [DOI] [PubMed] [Google Scholar]
- 36.Mischak H, Vlahou A, Ioannidis JPA. Technical aspects and inter-laboratory variability in native peptide profiling: the CE–MS experience. Clin Biochem. 2013;46(6):432–443. doi: 10.1016/j.clinbiochem.2012.09.025. [DOI] [PubMed] [Google Scholar]
- 37.Mischak H, Schanstra JP. CE-MS in biomarker discovery, validation, and clinical application. Proteomics Clin Appl. 2011;5(1–2):9–23. doi: 10.1002/prca.201000058. [DOI] [PubMed] [Google Scholar]
- 38.Max Kuhn. Contributions from Jed Wing, Steve Weston, Andre Williams, Chris Keefer, Allan Engelhardt, Tony Cooper, Zachary Mayer, Brenton Kenkel, the R Core Team, Michael Benesty, Reynald Lescarbeau, Andrew Ziem, Luca Scrucca, Yuan Tang, Can Candan and Tyler Hunt (2019) Caret: classification and regression training. R package version 6.0–84. https://CRAN.R-project.org/package=caret.
- 39.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011;12(1):77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21(20):7881. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- 41.Wood SN (2017) Generalized additive models. Chapman Hall/CRC.
- 42.Scrucca L GA (20130) A package for genetic algorithms in R. J Stat Softw 53(4):1.
- 43.Wang Y, Bellomo R. Cardiac surgery-associated acute kidney injury: risk factors, pathophysiology and treatment. Nat Rev Nephrol. 2017;13(11):697–711. doi: 10.1038/nrneph.2017.119. [DOI] [PubMed] [Google Scholar]
- 44.Bonventre J, v, Yang L, Cellular pathophysiology of ischemic acute kidney injury. J Clinic Invest. 2011;121(11):4210–4221. doi: 10.1172/JCI45161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Devarajan P. Update on mechanisms of ischemic acute kidney injury. J Am Soc Nephrol. 2006;17(6):1503–1520. doi: 10.1681/ASN.2006010017. [DOI] [PubMed] [Google Scholar]
- 46.Bellomo R, Kellum JA, Ronco C. Acute kidney injury. The Lancet. 2012;380(9843):756–766. doi: 10.1016/S0140-6736(11)61454-2. [DOI] [PubMed] [Google Scholar]
- 47.Kellum JA, Prowle JR. Paradigms of acute kidney injury in the intensive care setting. Nat Rev Nephrol. 2018;14(4):217–230. doi: 10.1038/nrneph.2017.184. [DOI] [PubMed] [Google Scholar]
- 48.Morgan BP, Harris CL. Complement, a target for therapy in inflammatory and degenerative diseases. Nat Rev Drug Discov. 2015;14(12):857–877. doi: 10.1038/nrd4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Latuszek A, Liu Y, Olsen O, Foster R, Cao M, Lovric I, et al. Inhibition of complement pathway activation with Pozelimab, a fully human antibody to complement component C5. PLoS ONE. 2020;15(5):e0231892. doi: 10.1371/journal.pone.0231892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yao W, Chen Y, Li Z, Ji J, You A, Jin S, et al. Single cell RNA sequencing identifies a unique inflammatory macrophage subset as a druggable target for alleviating acute kidney injury. Adv Sci. 2022;3:2103675. doi: 10.1002/advs.202103675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doerr A. Mass spectrometry–based targeted proteomics. Nat Methods. 2013;10(1):23–23. doi: 10.1038/nmeth.2286. [DOI] [PubMed] [Google Scholar]
- 52.Neuhoff N, v, Kaiser T, Wittke S, Krebs R, Pitt A, Burchard A,, et al. Mass spectrometry for the detection of differentially expressed proteins: a comparison of surface-enhanced laser desorption/ionization and capillary electrophoresis/mass spectrometry. Rapid Commun Mass Spectromet. 2004;18(2):149–156. doi: 10.1002/rcm.1294. [DOI] [PubMed] [Google Scholar]
- 53.Jantos-Siwy J, Schiffer E, Brand K, Schumann G, Rossing K, Delles C, et al. Quantitative urinary proteome analysis for biomarker evaluation in chronic kidney disease. J Proteome Res. 2009;8(1):268–281. doi: 10.1021/pr800401m. [DOI] [PubMed] [Google Scholar]
- 54.Klein J, Papadopoulos T, Mischak H, Mullen W. Comparison of CE-MS/MS and LC-MS/MS sequencing demonstrates significant complementarity in natural peptide identification in human urine. Electrophoresis. 2014;35(7):1060–1064. doi: 10.1002/elps.201300327. [DOI] [PubMed] [Google Scholar]
- 55.Zürbig P, Renfrow MB, Schiffer E, Novak J, Walden M, Wittke S, et al. Biomarker discovery by CE-MS enables sequence analysisvia MS/MS with platform-independent separation. Electrophoresis. 2006;27(11):2111–2125. doi: 10.1002/elps.200500827. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Figure S1: S100A9 expression after epithelial injury. A. Urinary calprotectin (S100A8/A9) abundance 4 hours after cardiac bypass-surgery. B–F. Blood urea nitrogen (B), mRNA S100a9 (C), S100A9 immunostaining (D-E) and mRNA Kim1 (F) in sham mice and after bilateral renal ischemia/reperfusion (hours 6, 24 and 48). G–J. mRNA expression of S100a9 and Kim1 in MCT cells submitted to interleukin-1b (IL1β, 10 ng/mL) or tumor necrosis factor-1a (TNFa, 10 ng/mL) (G-H) or hypoxia (I-J). AKI, acute kidney injury; BUN, blood urea nitrogen; Norm, normoxia; Hyp, hypoxia. Supplementary Figure S2: Performances of the peptide-based signature to identify AKI that developed within the first 2 days following cardiac surgery. ROC curves with corresponding AUROC and 95% confidence intervals of the local clinical score (blue, pointed), the full 204 peptides-based score (red), the urinary NGAL level (yellow, pointed) and the nephrocheck ([IGFBP7].[TIMP2] product) in the validation cohort. Supplementary Figure S3: Reduction and combination of the peptide-based signature. A. ROC curves with corresponding AUROC and 95% confidence intervals of the local clinical score (blue, pointed), the full 204 peptides-based score (red), the reduced 17 peptides-based score (black, dashed) and the combination of local clinical and full peptide-based score in the validation cohort. B. List of peptides included in the reduced 17-peptides signature according to their parental protein. LMAN2, Lectin mannose binding 2 ; MGP, Matrix gla protein. Supplementary Figure S4: Performances of the 204 peptides-based signature and the reference urinary biomarker NGAL for AKI prediction in the external ICU validation cohort. A. ROC curves with corresponding AUROC and 95% confidence intervals of the 204 peptides-based score and the reference urinary biomarker NGAL to predict AKI after ICU admission. B. ROC curves with corresponding AUROC and 95% confidence intervals of the 204 peptides-based score and reference urinary biomarker NGAL to predict the development of AKI within seven days after admission. Supplementary Figure S5: Performances of the 204 peptides-based score for in-hospital mortality prediction. Odds-ratio (OR) of in-hospital mortality were calculated with unadjusted, Euroscore-II-adjusted or propensity score-adjusted logistic regression. Supplementary Table S1: Correlations between clinical characteristics and the 204-peptides-based score. BMI, body mass index; PAOD, peripheral artery obliterans disease; COPD, chronic obstructive pulmonary disease; LVEF, left ventricular ejection fraction; eGFR, estimated glomerular filtration rate; CBP, cardiac bypass; RBC, red blood cells. Supplementary Table S2: Performance of the peptide-based score to predict acute kidney injury in the external ICU validation cohort, according to the cause of admission to the intensive care unit. ICU, intensive care unit; AUROC, area under the receiver operating characteristics curve. Supplementary file S1: Methodology. Urinary peptidomics and statistical analyses.
Data Availability Statement
All data, code, and materials used in the analysis are available upon request.