Abstract
Kirsten rat sarcoma viral oncogene homolog (KRAS) gene encodes a GTPase that acts as a molecular switch for intracellular signal transduction, promoting cell growth and proliferation. Mutations in the KRAS gene represent important biomarkers for NSCLC targeted therapy. However, detection of KRAS mutations in tissues has shown some limitations. During the last years, analyses of circulating free DNA (cfDNA) has emerged as an alternative and minimally invasive, approach to investigate tumor molecular changes. Here, we assessed the diagnostic performance of cfDNA analysis, compared to tissues through a meta-analysis and systematic review of existing literature. From 561 candidate papers, we finally identified 40 studies, including 2,805 NSCLC patients. We extracted values relating to the number of true-positive, false-positive, false-negative, and true-negative. Pooled sensitivity, specificity, positive likelihood ratio, negative likelihood ratio, and diagnostic odds ratio, each with 95% CI, were calculated. A summary receiver operating characteristic curve and the area under curve (AUC) were used to evaluate the overall diagnostic performance. The pooled sensitivity was 0.71 (95% CI 0.68–0.74) and the specificity was 0.93 (95% CI 0.92–0.94). The diagnostic odds ratio was 35.24 (95% CI 24.88–49.91) and the area under the curve was 0.92 (SE = 0.094). These results provide evidence that detection of KRAS mutation using cfDNA testing is of adequate diagnostic accuracy thus offering to the clinicians a new promising screening test for NSCLC patients.
Keywords: cancer risk, diagnostic value, genetic biomarkers, single-nucleotide polymorphisms, liquid biopsy
Introduction
Lung cancer (LC) is the leading cause of cancer-related mortality worldwide representing 18% of the total cancer deaths in 2020 (Sun et al., 2021). Non-small cell lung cancer (NSCLC), the largest group of LC, accounts for approximately 80% of new cases. These patients are often unsuitable for curative surgery duo to the advanced stage at the time of diagnosis (Ganti et al., 2021).
In the last decade, one of the most exciting advances in medical oncology is the application of personalized treatment tailored to the patient’s genetic background, primarily based on mutations in the epidermal growth factor receptor (EGFR) and in the Kirsten rat sarcoma viral oncogene homolog (KRAS) genes (Qiu et al., 2015; Hames et al., 2016). KRAS gene encodes a small GTPase that functions as an intracellular signaling protein promoting cell growth and proliferation (King et al., 2013). Approximately 30% of adenocarcinomas, the most common histological subtype of NSCLC, harbor mutations of the KRAS gene. Mutations in KRAS gene lead to oncogenic conversion ensuing in constitutive activation of downstream signal transduction cascades and thus cancer development and progression as well as specific drug sensitivity. KRAS mutations in NSCLC predominantly occur at codon 12 or codon 13 (Ghimessy et al., 2029) and represent an important biomarker for NSCLC therapy. Moreover, recently, new therapeutic agents (i.e., adagrasib and sotorasib) that target specifically the KRAS G12C variant have been developed and shown promising results in both preclinical and clinical trials (Skoulidis et al., 2021).
The gold standard for detecting cancer mutations has been based on molecular testing of tumor biopsies. However, obtaining a tumor biopsy requires invasive techniques, and is not suitable for real-time monitoring of cancer mutations. Furthermore, it is often difficult to obtain sufficient tissues for molecular testing through biopsy (Overman et al., 2013). During the last years, in order to answer the need of a more accessible and sequentially repeatable approach for tumor genetic analysis, “liquid biopsy” has emerged. The most successful use of liquid biopsy is the analysis of tumor DNA fragments that are released into the bloodstream through apoptosis, necrosis, and/or active secretion processes of cancer cells in the form of circulating free DNA (cfDNA) (Diehl et al., 2008). This approach turned out to be a minimally invasive and efficient method to investigate cancer cells which enables taking multiple blood samples over time (serial sampling) and thus informing about what kind of molecular changes are taking place in the tumor (Castro-Giner et al., 2018).
To date, many meta-analyses have investigated the performance of cfDNA in detecting EGFR mutation status in patients with NSCLC and only two meta-analyses evaluated also KRAS (Fan et al., 2017; Chen et al., 2020). However, the diagnostic accuracy of cfDNA testing for the detection of KRAS mutations remains controversial and not conclusive since the results vary among these two meta-analyses justifying further investigation.
Herein, we conducted a comprehensive systematic review and meta-analysis of available studies that compare the concordance between results on cfDNA within liquid biopsy and genomic DNA within tumor tissue to obtain an all-inclusive evaluation of the diagnostic accuracy of cfDNA testing for detection of KRAS mutations in NSCLC patients.
Materials and methods
Search strategy, inclusion and exclusion criteria and data extraction
We systematically searched the PubMed, Medline, Embase and Web of Science databases up to 7 July 2022 for studies reporting the diagnostic performance of cfDNA comparing sensitivity and specificity between tissue and blood in detecting KRAS mutations in NSCLC patients, using different combinations of the keywords: ‘‘lung neoplasms’’ or ‘‘lung cancer’’ or “NSCLC”, “KRAS,” “re-biopsy” or “repeat biopsy” or “liquid biopsy,” ‘‘serum’’ or ‘‘plasma’’ or ‘‘circulating,’’ and ‘‘mutations,’’ without any restriction. Abbreviations and alternative spellings and were also considered. Eligible publications were evaluated by checking titles and abstracts.
The references of all computer-identified publications were also checked for identifying additional studies that might have been missed in the initial search. Relevant reviews were also manually searched. Publications were checked for overlapping patient populations and, in the case of multiple publications from the same research group on overlapping cohorts, only the largest or most recent study was selected.
All studies evaluating sensitivity and specificity between tissue biopsy and liquid biopsy in KRAS-mutated NSCLC were considered eligible for the meta-analysis. The inclusion criteria were: 1) all NSCLC patients involved should be diagnosed cytologically or histopathologically; 2) tissue and blood biopsies should be paired in the same patient; 3) KRAS mutation status should be detected by circulating cell free DNA and verified in tumor tissues; 4) enough reported data to construct the diagnostic 2 × 2 table. In the process of assessing the eligibility of the studies, only articles written in English were included. We subsequently excluded studies that involved cell lines or artificial samples or where cfDNA was not detected. Finally, articles were excluded if they presented data in a way that did not allow it to be extracted properly (e.g., studies with mixed data from different types of cancers other than NSCLC).
Information collected from all eligible articles included the study characteristics (authors’ names, publication date and journal, country of study, number of patients); the clinical data (histological analysis, clinical stage); the results (method for tissue and blood biopsy; number of true positives (TP), true negatives (TN), false positives (FP), and false negatives (FN) between tissue and blood, and concordance between the two methods).
The original data from the eligible studies reporting partial information on TP, TN, FP, and FN, in comparison to sensitivity and specificity between tissue and cfDNA analysis were obtained by contacting the corresponding authors. Data on individual patients were collected by requesting the completion of a standardized form. The sensitivity equals [TP / (TP + FN)], while specificity equals [TN / (TN + FP)]. If not present, we computed the concordance rate as [(TP + TN)/n]. All records were reviewed and checked by two authors independently (MP and EF) and reached consensus at each eligible study.
Statistical analyses
Meta-analysis was carried out using the Rev-Man v.5.4 (provided by The Cochrane Collaboration, Oxford, England), Meta-DiSc and R packages (version 4.2.1). The combined sensitivity, specificity, positive likelihood ratio (PLR = sensitivity/(1-specificity)), negative likelihood ratio (NLR = (1-sensitivity)/specificity), positive predicted value, negative predicted value, diagnostic odds ratio (DOR = PLR/NLR) and corresponding 95% confidence intervals (95% CI) were calculated by the accuracy data (TP, TN, FP, and FN) collected from each studies. Based on these data, the summary receiver operating characteristic (SROC) was created and the area under the curve (AUC) was calculated.
A random-effects model, fitted via the general linear (mixed-effects) model, was used for all analyses, recognizing that its use can reduce the effect of larger studies and minimize the possible presence of heterogeneity among the studies accounting for the variation both within a study or between the many different studies included in the meta-analysis. Homogeneity of study results in different groupings was assessed using the Q and I 2 statistic. Publication bias was estimated by visual inspection of funnel plots, and a p value <0.05 indicated the occurrence of publication bias. Spearman correlation coefficient and p value were calculated to assess the threshold effect.
Results
Study selection
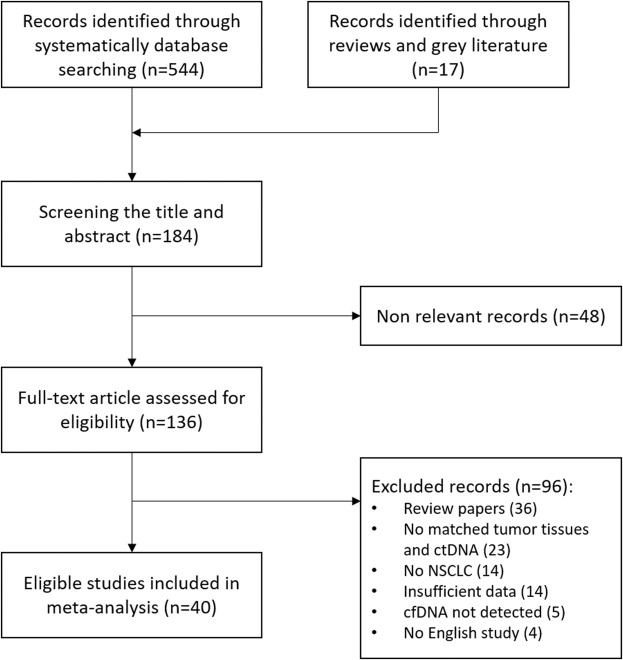
A total of 561 potential studies were initially evaluated for the meta-analysis on the results of the bibliographic search. After primary screening checking titles and abstracts, 136 full-text articles were selected for further evaluation of eligibility and scanned rigorously in full text. The main reasons for exclusion were reviews, not human studies and incorrect or mixed tumor type. After exclusion of studies, a total of 40 eligible studies were identified and finally included in our meta-analysis, comprising 2,805 NSCLC cases. A flowchart of the literature selection is shown in Figure 1.
FIGURE 1.
Flow diagram of literature screening and study selection.
Characteristics of eligible studies
The forty-one eligible studies for meta-analysis were published between 2007 and 2022 and included 2,805 NSCLC patients from 13 countries. The mean number of patients for each study was 70 (range 8–200). Notably, the majority of studies were prospective (28/40) and only 12 out of 40 were retrospective. Various types of methods have been applied for the detection of KRAS mutation in cfDNA, and the targeted NGS sequencing was the most common method (21/40). The median age was 62.8 years (range 20–100), 54.4% of patients were male and 69.7% had a history of smoking (former or current). Most of the patients were at advanced stage (TNM III–IV). The main characteristics of the 40 included studies are shown in Table 1.
TABLE 1.
Characteristics of eligible studies included in the meta- and pooled-analyses.
| References | No. of cases | Country | Study design | Males/Females | Median age (range) | % Smoker (former or current) | NSCLC stage (I-IV) | Specimen type for ctDNA | Detection assay in ctDNA |
|---|---|---|---|---|---|---|---|---|---|
| Gautschi et al. (2007) | 15 | Switzerland | Pro | 125/55 | 61 (36–81) | 69 | I-IV | Plasma | RFLP-PCR and Sequencing |
| Wang et al. (2010) | 120 | China | Pro | 158/115 | NA | 41.4 | IIIB, IV | Plasma | RFLP-PCR |
| Narayan A et al. (2012) | 21 | United States | Retro | NA | NA | NA | I-IV | Plasma | Targeted NGS |
| Punnoose EA et al. (2012) | 25 | United States, Australia | Retro | 21/16 | NA | 83.8 | IV | Plasma | In house TaqMan Assay |
| Nygaard et al. (2013) | 10 | Denmark | Pro | 151/95 | 66 (40–80) | NA | II-IV | Plasma | ARMS-qPCR |
| Oxnard GR et al. (2014) | 31 | United States | Retro | NA | NA | NA | NA | Plasma | ddPCR |
| Couraud S et al. (2014) | 68 | France | Pro | 13/93 | NA | 0 | I-IV | Plasma | Targeted NGS |
| Freidin MB et al. (2015) | 82 | United Kingdom | Pro | 52/41 | NA | NA | I-IV | Plasma | Cold PCR-HMR |
| Guibert N et al. (2016) | 32 | France | Pro | 21/11 | NA | NA | IV | Plasma | ddPCR |
| Guo N et al. (2016) | 41 | China | Pro | 22/19 | 52 (38–73) | NA | I-IV | Plasma | Targeted NGS |
| Paweletz C et al. (2016) | 48 | United States | Pro | 19/29 | 57 | NA | IV | Plasma | Targeted NGS |
| Thompson JC et al. (2016) | 50 | United States | Pro | 33/69 | 64 (34–85) | 50 | II-IV | Plasma | Paired-end sequencing |
| Chen KZ et al. (2016) | 58 | China | Retro | 33/25 | 64 (40–84) | 37.9 | I-II | Plasma | Targeted NGS |
| Sacher AG et al. (2016) | 87 | United States | Pro | 68/112 | 62 | NA | IIIB, IV | Plasma | ddPCR |
| Pécuchet N et al. (2016) | 109 | France | Pro | 49/60 | NA | 67 | III-IV | Plasma | Ultradeep-targeted NGS |
| Han JY et al. (2016) | 135 | South Korea | Pro | 136/72 | 58 (29–82) | 63 | III-IV | Plasma | PNA champ-assisted melting curve |
| Del Re M et al. (2017) | 8 | Italy | Retro | 13/20 | 62 (41–75) | 33.3 | III-IV | Plasma | ddPCR |
| Yao T et al. (2017) | 39 | China | Retro | 19/20 | 62 (28–78) | 25.6 | IIIa-IV | Plasma | Targeted NGS |
| Mellert et a. (2017) | 42 | United States | Pro/Retro | NA | NA | NA | NA | Plasma | ddPCR |
| Wang Z et al. (2017) | 103 | China | Pro | 48/55 | 64 (21–87) | 32 | III-IV | Plasma | cSMART |
| Wang X et al. (2017) | 200 | China | Pro | 138/62 | 57 (NA) | NA | NA | Urine | ddPCR |
| Garcia J et al. (2018) | 20 | France | Retro | NA | NA | NA | IV | Plasma | Targeted NGS |
| Liu L et al. (2018) | 72 | China | Pro | 44/28 | 59 (40–83) | 40.3 | IIIa-IV | Plasma | Targeted NGS |
| Papadopoulou E et al. (2019) | 36 | Greece | Pro | 82/39 | NA | NA | NA | Plasma | Targeted NGS |
| Tran LS et al. (2019) | 40 | Vietnam | Pro | 33/25 | 62 (37–90) | 43 | IIIB, IV | Plasma | ddPCR and Ultradeep-targeted NGS |
| Leighl NB et al. (2019) | 89 | United States | Pro | 129/153 | 69 (26–100) | 76.2 | IIIB, IV | Plasma | Targeted NGS |
| Remon J et al. (2019) | 104 | France | Pro | 126/88 | NA | 83.3 | IIIB, IV | Plasma | Targeted NGS |
| Li BT et al. (2019) | 110 | United States | Pro | 47/80 | 66 (23–85) | NA | IIIB, IV | Plasma | Targeted NGS |
| Pritchett MA et al. (2019) | 147 | United States | Pro | 84/94 | NA | 86.6 | IIIB, IV | Plasma | Targeted NGS |
| Cho MS et al. (2020) | 36 | South Korea | Pro | 25/11 | 66 (33–81) | NA | I-IV | Plasma | PANAmutyper™ |
| Jiang J et al. (2020) | 47 | Germany | Retro | 45/26 | 62 (NA) | 85 | IIIa-IV | Plasma | Targeted NGS |
| Zulato E et al. (2020) | 58 | Italy | Pro | 31/27 | 68 (61–73) | 86.2 | NA | Plasma | ddPCR |
| Michaelidou K et al. (2020) | 96 | Greece | Pro | 99/22 | NA | 88.6 | III-IV | Plasma | ddPCR |
| Mehta A et al. (2021) | 21 | India | Retro | 14/7 | 54 (28–79) | 47.62 | IIIB, IV | Plasma | Targeted NGS |
| Qvick A et al. (2021) | 24 | Sweden | Pro | 32/28 | 72 (39–85) | 80 | I-IV | Plasma | Targeted NGS |
| Wahl SGF et al. (2021) | 60 | Norway | Retro | 23/37 | 69 (47–83) | 100 | I-IV | Plasma | ddPCR |
| Lam VK et al. (2021) | 76 | United States | Retro | 66/78 | 64 (28–96) | NA | I-IV | Plasma | Targeted NGS |
| Jiao XD et al. (2021) | 185 | China | Pro | 110/75 | 64 (24–84) | NA | IIIB, IV | Plasma | Targeted NGS |
| Xie X et al. (2022) | 71 | China | Pro | 54/16 | 60 (20–79) | 46.5 | I-IV | Sputum | Targeted NGS |
| Bauml JM et al. (2022) | 189 | United States | Pro | 63/63 | 63.5 (56–70) | 92.9 | I-IV | Plasma | Targeted NGS |
| Total of cases | 2,805 |
NA, not available; Pro, prospective study; Retro, retrospective study; RFLP, Restriction Fragment Length Polymorphism; PCR, Polymerase Chain Reaction; NGS, Next Generation Sequencing; ddPCR, Droplet Digital PCR; ARMS-PCR, Amplification Refractory Mutation System PCR; PCR-HMR, High-resolution Melting PCR; PNA, Peptide Nucleic Acid; cSMART, circulating single-molecule amplification and re-sequencing technology; PANAmutyperTM, PNA Clamping-assisted Fluorescence Melting Curve Analysis.
Diagnostic accuracy
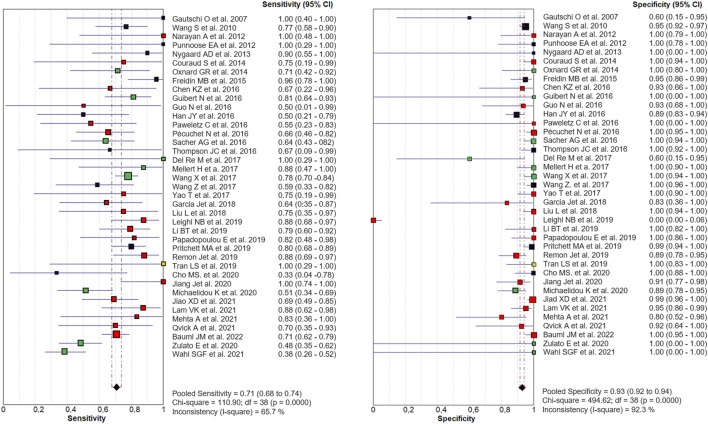
Out of 40 eligible papers, 1 was excluded from our meta-analysis (Xie et al., 2022) since only TP and FN were reported in published data. The diagnostic accuracy of cfDNA analysis for the detection of KRAS mutations in NSCLC, compared with tumor tissue was shown in the Forest Plot for 39 studies, including 2,666 NSCLC patients. The combined sensitivity and specificity in the meta-analysis were 0.71 (95% CI 0.68–0.74) and 0.93 (95% CI 0.92–0.94), respectively (Figure 2).
FIGURE 2.
Paired Forest Plot of sensitivity and specificity of cfDNA testing in detecting KRAS mutation in NSCLC for 39 studies in the meta-analysis. Random-effects (RE) model used. The square and horizontal bars represented study-specific sensitivity and specificity and 95% confidence interval (CI). Squares in different colors represent a different diagnostic method (red = Targeted NSG; blue = ddPCR; yellow = both; black = other method). Diamonds represented the overall results. The pooled sensitivity was 0.71 (95% CI 0.68–0.74) and the specificity was 0.93 (95% CI 0.92–0.94).
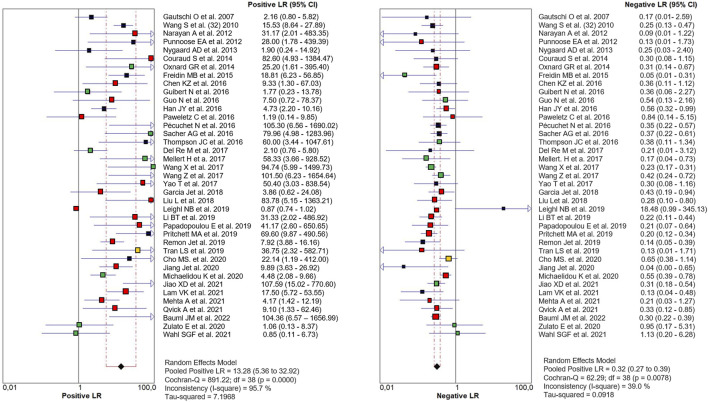
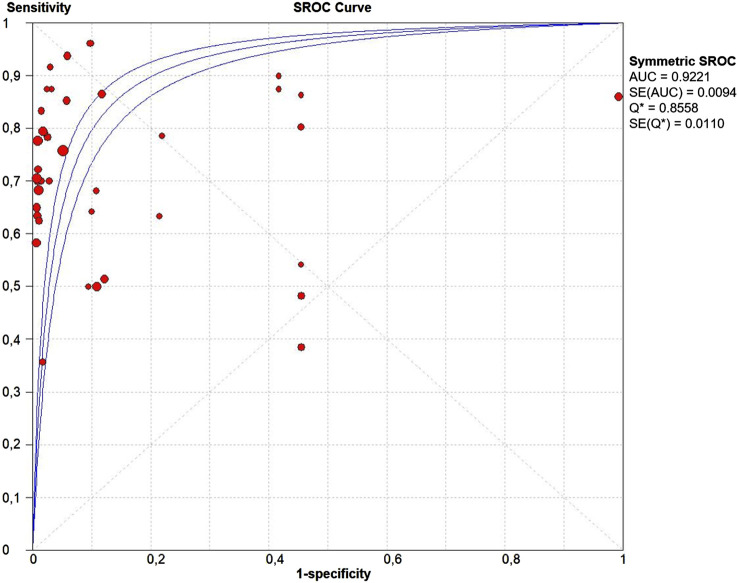
The PLR and NLR of cfDNA were 8.32 (95% CI 6.93–9.99) and 0.29 (95% CI 0.26–0.33), respectively in the meta-analysis (Figure 3). The DOR was 35.24 (95% CI 24.88–49.91). Figure 4 shows the SROC plot with AUC of 0.92 (SE = 0.0094), indicating a high diagnostic accuracy of cfDNA test.
FIGURE 3.
Paired Forest plot of PLR and NLR of cfDNA testing in detecting KRAS mutation in NSCLC for 39 studies in the meta-analysis. Random-effects (RE) model used. The square and horizontal bars represented study-specific PLR and NLR and 95% confidence interval (CI). Squares in different colors represent a different diagnostic method (red = Targeted NSG; blue = ddPCR; yellow = both; black = other method). Diamonds represented the overall results. The pooled PLR and NLR were 8.32 (95% CI 6.93–9.99) and 0.29 (95% CI 0.26–0.33), respectively.
FIGURE 4.
The summary receiver operating characteristic (SROC) curve. Points represent the pair of sensitivity and specificity at a given threshold for each study. The area under the curve was 0.92 (SE = 0.094).
Heterogeneity and publication bias
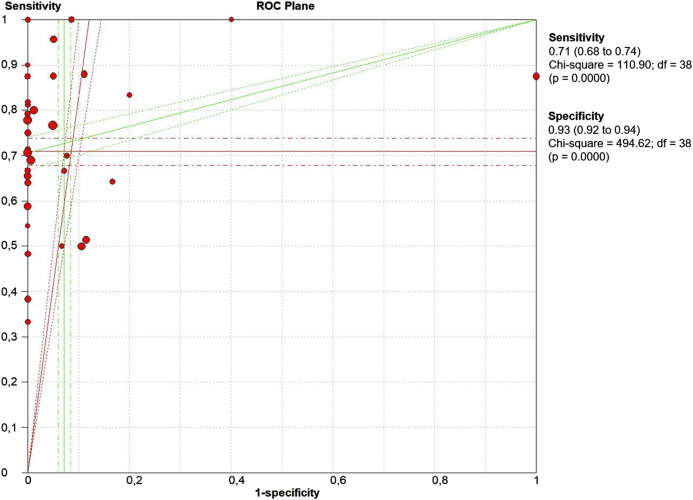
The threshold effect is a major source of heterogeneity among studies. Visual assessment of the ROC plane did not reveal significant threshold effect (Figure 5). Spearman correlation coefficient was 0.332 and the p value was 0.06, confirming no significant evidence of threshold effect.
FIGURE 5.
The receiver operative curve (ROC) plane plot. Points represent the pair of sensitivity and specificity at a given threshold for each study.
As revealed by the Forest Plots of accuracy data, significant heterogeneity was detected according to the I2 values. Therefore, we performed a meta-regression to detect the source of heterogeneity analyzing the impact of country, study design, sample size, clinical stage, and detection methods. However, none of the above covariates contributed to heterogeneity (Table 2).
TABLE 2.
Resuts of meta-regression analysis.
| Variable | Coefficient | Standard error | p value a | RDOR | 95% CI |
|---|---|---|---|---|---|
| Country | −0.077 | 0.0857 | 0.3742 | 0.93 | 0.78–1.10 |
| Sample Size | 0.000 | 0.0060 | 0.9404 | 1.00 | 0.99–1.01 |
| Clinical Stage | −0.349 | 0.1732 | 0.0537 | 0.71 | 0.49–1.01 |
| Detection Method | 0.097 | 0.1444 | 0.5074 | 1.10 | 0.82–1.48 |
Tau-squared estimate.
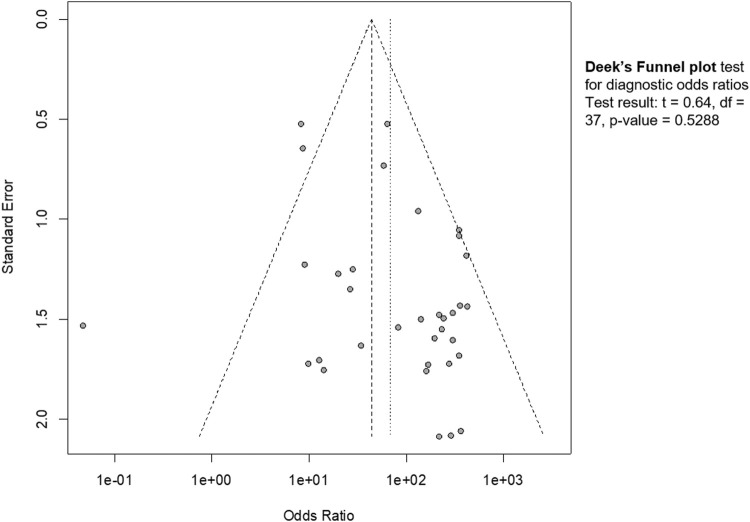
Publication bias was estimated by funnel plot (Figure 6). The visual inspection revealed a partial symmetry with p value = 0.5288, showing no evidence of publication bias.
FIGURE 6.
Funnel plot. The graphical representation showed no significant publication bias (p = 0.53).
Discussion
The need to identify a minimally invasive, efficient and repeatable over time approach to detect tumor mutations in NSCLC patients has always been the greatest challenge in the medical clinic. Serial sampling of traditional biopsies is in fact not feasible. Serial sampling may provide information on 1) relapse or progression of disease, 2) treatment response, 3) survival and 4) temporal and spatial tumor heterogeneity. These elements are of pivotal importance for the application of personalized therapies tailored to the patient’s genetic background. Recently, cfDNA analysis, namely “liquid biopsy”, has emerged as a non-invasive, feasible and reliable approach to investigate cancer DNA. However, its diagnostic accuracy for the detection of KRAS mutations remained controversial as different studies report varying degrees of sensitivity and specificity. As over one-third of lung adenocarcinomas harbour KRAS mutations, KRAS is therefore an important biomarker that might be used in monitoring treatment, as a biomarker for relapse/progression (disease monitoring), and as a prognostic biomarker.
We performed a large-scale meta-analysis of 40 papers and 2,805 NSCLC patients, reviewing controversial evidence for the diagnostic role of cfDNA testing for the detection of KRAS mutations in NSCLC. In our study, the pooled sensitivity for cfDNA was 0.71 (95% CI 0.68–0.74) and the specificity was 0.93 (95% CI 0.92–0.94) (Figure 2). None of the properties analyzed in the patient cohort appear to be specifically related to sensitivity. We are aware that the resulting sensitivity is not high enough as a diagnostic method. However, as a cancer screening test, if the test guarantees non-invasiveness, a high specificity is required instead of sensitivity. In this case, our meta-analysis identified high specificity, and AUC more than 0.9, indicating an overall high diagnostic efficiency in detection of KRAS mutations by cfDNA analysis (Figure 4). Also the values of the Likelihood ratios (PLR, and NLR) confirmed the informativeness of the test on cfDNA (Figure 3).
Conducting our meta-analysis, however, we ran into some noteworthy issues. First of all, there is high variability among clinical stages and consequently in the treatment among the 40 studies (Table 1). Second, different methods with different diagnostic performances were used to assess KRAS mutations in cfDNA in different studies highlighting the importance of method standardization. However, different studies using the same method have obtained diametrically opposite results from the point of view of diagnostic performance (see for example Leighl et al., 2019 and Jiao et al., 2021 both using Targeted NGS). This is due to the fact that numerous variables beyond the method affect the accuracy of mutation detection. Another issue is represented by the small size of some studies that might lead to bias. Last but not least, in most of the eligible studies, the analyzed tissues were formalin-fixed, paraffin-embedded (FFPE) which determine DNA degradation. All these factors represent important limitations that potentially increase the detection bias. Furthermore, it is not possible to overlook the difficulty in defining true negative (TN) or false positive (FP) when comparing detection rates of cfDNA analyses across different studies. A result can turn out to be “negative” in cfDNA if there really is no tumor DNA present or if tumor DNA is present but not detectable due to low amounts and the method is not sensitive enough to detect it. Similarly, mismatch between mutations found in cfDNA and mutations not found in tissue DNA does not necessarily imply FP in cfDNA. A false positive result in cfDNA could be due to DNA degradation of the tumor tissue; to a sampling problem in tissue; or to setting a limit of detection (LOD) threshold too low. Data on FP rates for each method and how the limit of detection (LOD) was determined should be available for each study so that they can be taken into account for cross-study comparison.
Significant heterogeneity was detected as revealed by the Forest Plots of accuracy data. However, Spearman Correlation and ROC plane plot (Figure 5) suggested that this heterogeneity was not caused by the threshold effect. The results of meta-regression showed that none of the analyzed variables was the source of heterogeneity (Table 2).
In conclusion, in the present meta-analysis of 40 studies including more than 2,800 NSCLC patients, on the basis of accumulated data, the detection of KRAS mutation in cfDNA proves to be of adequate diagnostic accuracy. This is a novel finding regarding KRAS since the previous published meta-analyses focused on the diagnostic role of EGFR detection. Targeted treatment against KRAS G12C is soon a reality, not only for NSCLC. For this reason, we need to search and find cancer patients with this mutation and we need sensitive and accurate methods for detecting KRAS in cfDNA. The present meta-analysis reveals that cfDNA analysis might be a valid alternative for molecular analysis when tumor biopsy or cytological specimens are not available. Given its non-invasive nature and the resulted high specificity, cfDNA testing represents a promising screening assay for detecting KRAS mutations in cancer patients.
Acknowledgments
The authors wish to thank the University of Siena for the contribution granted to support the open access publication process of this study.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
EF designed the meta-analysis; MP and EF searched databases, collected full-text papers, extracted and analysed data; all the authors contributed to the drafting and revision of the article and gave definitive approval to the version to be published.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Bauml J. M., Li B. T., Velcheti V., Govindan R., Curioni-Fontecedro A., Dooms C., et al. (2022). Clinical validation of Guardant360 CDx as a blood-based companion diagnostic for sotorasib. Lung cancer 166, 270–278. 10.1016/j.lungcan.2021.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Giner F., Gkountela S., Donato C., Alborelli I., Quagliata L., Ng C., et al. (2018). Cancer diagnosis using a liquid biopsy: Challenges and expectations. Diagn. (Basel, Switz. 8 (2), 31. 10.3390/diagnostics8020031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K. Z., Lou F., Yang F., Zhang J. B., Ye H., Chen W., et al. (2016). Circulating tumor DNA detection in early-stage non-small cell lung cancer patients by targeted sequencing. Sci. Rep. 6, 31985. 10.1038/srep31985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Miao H., Zeng Q., Xu S., Chen Z., Liu K. (2020). Circulating cell-free DNA as a diagnostic and prognostic biomarker for non-small-cell lung cancer: A systematic review and meta-analysis. Biomark. Med. 14 (7), 587–597. 10.2217/bmm-2018-0093 [DOI] [PubMed] [Google Scholar]
- Cho M. S., Park C. H., Lee S., Park H. S. (2020). Clinicopathological parameters for circulating tumor DNA shedding in surgically resected non-small cell lung cancer with EGFR or KRAS mutation. PloS one 15 (3), e0230622. 10.1371/journal.pone.0230622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couraud S., Vaca-Paniagua F., Villar S., Oliver J., Schuster T., Blanché H., et al. (2014). Noninvasive diagnosis of actionable mutations by deep sequencing of circulating free DNA in lung cancer from never-smokers: A proof-of-concept study from BioCAST/IFCT-1002. Clin. Cancer Res. 20 (17), 4613–4624. 10.1158/1078-0432.CCR-13-3063 [DOI] [PubMed] [Google Scholar]
- Del Re M., Tiseo M., Bordi P., D'Incecco A., Camerini A., Petrini I., et al. (2017). Contribution of KRAS mutations and c.2369C > T (p.T790M) EGFR to acquired resistance to EGFR-TKIs in EGFR mutant NSCLC: A study on circulating tumor DNA. Oncotarget 8 (8), 13611–13619. 10.18632/oncotarget.6957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl F., Schmidt K., Choti M. A., Romans K., Goodman S., Li M., et al. (2008). Circulating mutant DNA to assess tumor dynamics. Nat. Med. 14 (9), 985–990. 10.1038/nm.1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G., Zhang K., Ding J., Li J. (2017). Prognostic value of EGFR and KRAS in circulating tumor DNA in patients with advanced non-small cell lung cancer: A systematic review and meta-analysis. Oncotarget 8 (20), 33922–33932. 10.18632/oncotarget.15412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freidin M. B., Freydina D. V., Leung M., Montero Fernandez A., Nicholson A. G., Lim E. (2015). Circulating tumor DNA outperforms circulating tumor cells for KRAS mutation detection in thoracic malignancies. Clin. Chem. 61 (10), 1299–1304. 10.1373/clinchem.2015.242453 [DOI] [PubMed] [Google Scholar]
- Ganti A. K., Klein A. B., Cotarla I., Seal B., Chou E. (2021). Update of incidence, prevalence, survival, and initial treatment in patients with non-small cell lung cancer in the US. JAMA Oncol. 7 (12), 1824–1832. 10.1001/jamaoncol.2021.4932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia J., Forestier J., Dusserre E., Wozny A. S., Geiguer F., Merle P., et al. (2018). Cross-platform comparison for the detection of RAS mutations in cfDNA (ddPCR Biorad detection assay, BEAMing assay, and NGS strategy). Oncotarget 9 (30), 21122–21131. 10.18632/oncotarget.24950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautschi O., Huegli B., Ziegler A., Gugger M., Heighway J., Ratschiller D., et al. (2007). Origin and prognostic value of circulating KRAS mutations in lung cancer patients. Cancer Lett. 254 (2), 265–273. 10.1016/j.canlet.2007.03.008 [DOI] [PubMed] [Google Scholar]
- Ghimessy A. K., Gellert A., Schlegl E., Hegedus B., Raso E., Barbai T., et al. (2019). KRAS mutations predict response and outcome in advanced lung adenocarcinoma patients receiving first-line bevacizumab and platinum-based chemotherapy. Cancers 11 (10), 1514. 10.3390/cancers11101514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guibert N., Pradines A., Farella M., Casanova A., Gouin S., Keller L., et al. (2016). Monitoring KRAS mutations in circulating DNA and tumor cells using digital droplet PCR during treatment of KRAS-mutated lung adenocarcinoma. Lung cancer 100, 1–4. 10.1016/j.lungcan.2016.07.021 [DOI] [PubMed] [Google Scholar]
- Guo N., Lou F., Ma Y., Li J., Yang B., Chen W., et al. (2016). Circulating tumor DNA detection in lung cancer patients before and after surgery. Sci. Rep. 6, 33519. 10.1038/srep33519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hames M. L., Chen H., Iams W., Aston J., Lovly C. M., Horn L. (2016). Correlation between KRAS mutation status and response to chemotherapy in patients with advanced non-small cell lung cancer☆. Lung cancer (Amsterdam, Neth., 92, 29–34. 10.1016/j.lungcan.2015.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J. Y., Choi J. J., Kim J. Y., Han Y. L., Lee G. K. (2016). PNA clamping-assisted fluorescence melting curve analysis for detecting EGFR and KRAS mutations in the circulating tumor DNA of patients with advanced non-small cell lung cancer. BMC cancer 16, 627. 10.1186/s12885-016-2678-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Adams H. P., Lange M., Siemann S., Feldkamp M., McNamara S., et al. (2020). Plasma-based longitudinal mutation monitoring as a potential predictor of disease progression in subjects with adenocarcinoma in advanced non-small cell lung cancer. BMC cancer 20 (1), 885. 10.1186/s12885-020-07340-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X. D., Ding L. R., Zhang C. T., Qin B. D., Liu K., Jiang L. P., et al. (2021). Serum tumor markers for the prediction of concordance between genomic profiles from liquid and tissue biopsy in patients with advanced lung adenocarcinoma. Transl. Lung Cancer Res. 10 (7), 3236–3250. 10.21037/tlcr-21-543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King P. D., Lubeck B. A., Lapinski P. E. (2013). Nonredundant functions for Ras GTPase-activating proteins in tissue homeostasis. Sci. Signal. 6 (264), re1. 10.1126/scisignal.2003669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam V. K., Zhang J., Wu C. C., Tran H. T., Li L., Diao L., et al. (2021). Genotype-specific differences in circulating tumor DNA levels in advanced NSCLC. J. Thorac. Oncol. 16 (4), 601–609. 10.1016/j.jtho.2020.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leighl N. B., Page R. D., Raymond V. M., Daniel D. B., Divers S. G., Reckamp K. L., et al. (2019). Clinical utility of comprehensive cell-free DNA analysis to identify genomic biomarkers in patients with newly diagnosed metastatic non-small cell lung cancer. Clin. Cancer Res. 25 (15), 4691–4700. 10.1158/1078-0432.CCR-19-0624 [DOI] [PubMed] [Google Scholar]
- Li B. T., Janku F., Jung B., Hou C., Madwani K., Alden R., et al. (2019). Ultra-deep next-generation sequencing of plasma cell-free DNA in patients with advanced lung cancers: Results from the actionable genome consortium. Ann. Oncol. 30 (4), 597–603. 10.1093/annonc/mdz046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Liu H., Shao D., Liu Z., Wang J., Deng Q., et al. (2018). Development and clinical validation of a circulating tumor DNA test for the identification of clinically actionable mutations in nonsmall cell lung cancer. Genes Chromosom. Cancer 57 (4), 211–220. 10.1002/gcc.22522 [DOI] [PubMed] [Google Scholar]
- Mehta A., Kumar Sharma S., Kumar D., Vasudevan S. (2021). Plasma biopsy by tag-sequencing: An acceptable alternative to tumor tissue profiling in non-small-cell lung cancer. Pol. J. Pathol. 72 (2), 117–125. 10.5114/pjp.2021.109514 [DOI] [PubMed] [Google Scholar]
- Mellert H., Foreman T., Jackson L., Maar D., Thurston S., Koch K., et al. (2017). Development and clinical utility of a blood-based test service for the rapid identification of actionable mutations in non-small cell lung carcinoma. J. Mol. Diagn. 19 (3), 404–416. 10.1016/j.jmoldx.2016.11.004 [DOI] [PubMed] [Google Scholar]
- Michaelidou K., Koutoulaki C., Mavridis K., Vorrias E., Papadaki M. A., Koutsopoulos A. V., et al. (2020). Detection of KRAS G12/G13 mutations in cell free-DNA by droplet digital PCR, offers prognostic information for patients with advanced non-small cell lung cancer. Cells 9 (11), 2514. 10.3390/cells9112514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan A., Carriero N. J., Gettinger S. N., Kluytenaar J., Kozak K. R., Yock T. I., et al. (2012). Ultrasensitive measurement of hotspot mutations in tumor DNA in blood using error-suppressed multiplexed deep sequencing. Cancer Res. 72 (14), 3492–3498. 10.1158/0008-5472.CAN-11-4037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygaard A. D., Garm Spindler K. L., Pallisgaard N., Andersen R. F., Jakobsen A. (2013). The prognostic value of KRAS mutated plasma DNA in advanced non-small cell lung cancer. Lung cancer (Amsterdam, Neth. 79 (3), 312–317. 10.1016/j.lungcan.2012.11.016 [DOI] [PubMed] [Google Scholar]
- Overman M. J., Modak J., Kopetz S., Murthy R., Yao J. C., Hicks M. E., et al. (2013). Use of research biopsies in clinical trials: Are risks and benefits adequately discussed? J. Clin. Oncol. 31 (1), 17–22. 10.1200/JCO.2012.43.1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxnard G. R., Paweletz C. P., Kuang Y., Mach S. L., O'Connell A., Messineo M. M., et al. (2014). Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin. Cancer Res. 20 (6), 1698–1705. 10.1158/1078-0432.CCR-13-2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulou E., Tsoulos N., Tsantikidi K., Metaxa-Mariatou V., Stamou P. E., Kladi-Skandali A., et al. (2019). Clinical feasibility of NGS liquid biopsy analysis in NSCLC patients. PloS one 14 (12), e0226853. 10.1371/journal.pone.0226853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paweletz C. P., Sacher A. G., Raymond C. K., Alden R. S., O'Connell A., Mach S. L., et al. (2016). Bias-corrected targeted next-generation sequencing for rapid, multiplexed detection of actionable alterations in cell-free DNA from advanced lung cancer patients. Clin. Cancer Res. 22 (4), 915–922. 10.1158/1078-0432.CCR-15-1627-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pécuchet N., Zonta E., Didelot A., Combe P., Thibault C., Gibault L., et al. (2016). Base-position error rate analysis of next-generation sequencing applied to circulating tumor DNA in non-small cell lung cancer: A prospective study. PLoS Med. 13 (12), e1002199. 10.1371/journal.pmed.1002199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchett M. A., Camidge D. R., Patel M., Khatri J., Boniol S., Friedman E. K., et al. (2019). Prospective clinical validation of the InVisionFirst-lung circulating tumor DNA assay for molecular profiling of patients with advanced nonsquamous non-small-cell lung cancer. JCO Precis. Oncol. 3, 1–15. 10.1200/PO.18.00299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punnoose E. A., Atwal S., Liu W., Raja R., Fine B. M., Hughes B. G., et al. (2012). Evaluation of circulating tumor cells and circulating tumor DNA in non-small cell lung cancer: Association with clinical endpoints in a phase II clinical trial of pertuzumab and erlotinib. Clin. Cancer Res. 18 (8), 2391–2401. 10.1158/1078-0432.CCR-11-3148 [DOI] [PubMed] [Google Scholar]
- Qiu M., Wang J., Xu Y., Ding X., Li M., Jiang F., et al. (2015). Circulating tumor DNA is effective for the detection of EGFR mutation in non-small cell lung cancer: A meta-analysis. Cancer Epidemiol. Biomarkers Prev. 24 (1), 206–212. 10.1158/1055-9965.EPI-14-0895 [DOI] [PubMed] [Google Scholar]
- Qvick A., Stenmark B., Carlsson J., Isaksson J., Karlsson C., Helenius G. (2021). Liquid biopsy as an option for predictive testing and prognosis in patients with lung cancer. Mol. Med. 27 (1), 68. 10.1186/s10020-021-00331-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remon J., Lacroix L., Jovelet C., Caramella C., Howarth K., Plagnol V., et al. (2019). Real-world utility of an amplicon-based next-generation sequencing liquid biopsy for broad molecular profiling in patients with advanced non-small-cell lung cancer. JCO Precis. Oncol. 3, 00211–00214. PO.18. 10.1200/PO.18.00211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher A. G., Paweletz C., Dahlberg S. E., Alden R. S., O'Connell A., Feeney N., et al. (2016). Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer. JAMA Oncol. 2 (8), 1014–1022. 10.1001/jamaoncol.2016.0173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoulidis F., Li B. T., Dy G. K., Price T. J., Falchook G. S., Wolf J., et al. (2021). Sotorasib for lung cancers with KRAS p.G12C mutation. N. Engl. J. Med. 384 (25), 2371–2381. 10.1056/NEJMoa2103695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H., Ferlay J., Siegel R. L., Laversanne M., Soerjomataram I., Jemal A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca. Cancer J. Clin. 71 (3), 209–249. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- Thompson J. C., Yee S. S., Troxel A. B., Savitch S. L., Fan R., Balli D., et al. (2016). Detection of therapeutically targetable driver and resistance mutations in lung cancer patients by next-generation sequencing of cell-free circulating tumor DNA. Clin. Cancer Res. 22 (23), 5772–5782. 10.1158/1078-0432.CCR-16-1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran L. S., Pham H. T., Tran V. U., Tran T. T., Dang A. H., Le D. T., et al. (2019). Ultra-deep massively parallel sequencing with unique molecular identifier tagging achieves comparable performance to droplet digital PCR for detection and quantification of circulating tumor DNA from lung cancer patients. PloS one 14 (12), e0226193. 10.1371/journal.pone.0226193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl S., Dai H. Y., Emdal E. F., Ottestad A. L., Dale V. G., Richardsen E., et al. (2021). Prognostic value of absolute quantification of mutated KRAS in circulating tumour DNA in lung adenocarcinoma patients prior to therapy. J. Pathol. Clin. Res. 7 (3), 209–219. 10.1002/cjp2.200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., An T., Wang J., Zhao J., Wang Z., Zhuo M., et al. (2010). Potential clinical significance of a plasma-based KRAS mutation analysis in patients with advanced non-small cell lung cancer. Clin. Cancer Res. 16 (4), 1324–1330. 10.1158/1078-0432.CCR-09-2672 [DOI] [PubMed] [Google Scholar]
- Wang X., Meng Q., Wang C., Li F., Zhu Z., Liu S., et al. (2017). Investigation of transrenal KRAS mutation in late stage NSCLC patients correlates to disease progression. Biomarkers 22 (7), 654–660. 10.1080/1354750X.2016.1269202 [DOI] [PubMed] [Google Scholar]
- Wang Z., Cheng G., Han X., Mu X., Zhang Y., Cui D., et al. (2017). Application of single-molecule amplification and resequencing technology for broad surveillance of plasma mutations in patients with advanced lung adenocarcinoma. J. Mol. Diagn. 19 (1), 169–181. 10.1016/j.jmoldx.2016.09.008 [DOI] [PubMed] [Google Scholar]
- Xie X., Wu J., Guo B., Wang L., Deng H., Lin X., et al. (2022). Comprehensive characterization reveals sputum supernatant as a valuable alternative liquid biopsy for genome profiling in advanced non-small cell lung cancer. Respir. Res. 23 (1), 175. 10.1186/s12931-022-02097-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Liu J., Li L., Yuan Y., Nan K., Wu X., et al. (2017). Detection of circulating tumor DNA in patients with advanced non-small cell lung cancer. Oncotarget 8 (2), 2130–2140. 10.18632/oncotarget.12883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zulato E., Attili I., Pavan A., Nardo G., Del Bianco P., Boscolo Bragadin A., et al. (2020). Early assessment of KRAS mutation in cfDNA correlates with risk of progression and death in advanced non-small-cell lung cancer. Br. J. Cancer 123 (1), 81–91. 10.1038/s41416-020-0833-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.