Abstract
Background
Lack of biomarkers for treatment selection and monitoring in small cell lung cancer (SCLC) patients with the limited therapeutic options, result in poor outcomes. Therefore, new prognostic biomarkers are needed to improve their management. The prognostic value of cell-free DNA (cfDNA) and circulating tumor cells (CTCs) have been less explored in SCLC.
Methods
We quantified cfDNA in 46 SCLC patients at different times during first-line of chemotherapy or chemo-immunotherapy. Moreover, CTCs were analyzed in 21 patients before therapy onset using CellSearch® system. The possible association between both biomarkers and patients’ outcomes was investigated in order to develop a prognostic model.
Results
High cfDNA levels before therapy were associated with shorter progression-free survival (PFS) and overall survival (OS). Furthermore, cfDNA levels at 3 weeks and at progression disease were also associated with patients’ outcomes. Multivariate analyses confirmed the independence of cfDNA levels as a prognostic biomarker. Finally, the three-risk category prognostic model developed included Eastern Cooperative Oncology Group Performance Status (ECOG PS), gender and baseline cfDNA levels was associated with a higher risk of progression and death.
Conclusions
We confirmed the prognostic utility of cfDNA quantitative analysis in SCLC patients before and during therapy. Our novel risk prognostic model in clinical practice will allow to identify patients who could benefit with actual therapies.
Keywords: Small cell lung cancer, liquid biopsy, cell free DNA, circulating tumor cells (CTCs), prognostic biomarkers
Introduction
Small cell lung cancer (SCLC), which accounts for 15% of all lung cancer cases, is characterized by its aggressiveness, its strong association with tobacco and the poor outcome. About 70% of patients present extensive disease SCLC (ED-SCLC) where only 2% survive 5 years after diagnosis (1-3). For many years, chemotherapy was the unique option to treat this tumor type. However, the scenario has changed in the last years (3). New therapies, such as immunotherapy, have been recently incorporated into the management of SCLC patients and, although some survival improvements have been reported in the patients with ED-SCLC (4-8), the majority of them do not benefit from this new treatment (9). The genomic profile of SCLC is characterized by extensive chromosomal rearrangements and a high mutational burden, including in nearly all, inactivation of the tumor suppressor genes TP53 and RB1 (10). However, nowadays the selection of treatment in SCLC patients is not dependent on the characteristics of the tumor (11), and the criteria to stratify patients is not clear, since no predictive biomarkers have been validated for the clinical practice (12). In this context, the use of liquid biopsies as a tool to guide treatment and/or for monitoring the patients’ response represent a valuable alternative (13,14).
Circulating tumor DNA (ctDNA), derived from tissue tumor cells, has demonstrated its clinical utility and represents a promising tool for guiding precision medicine in several cancer types (15,16). In SCLC, different studies have investigated the importance and the clinical value of analyzing ctDNA levels. However, driver mutations known in SCLC are limited to RB1 and TP53 genes (17). In contrast, total cell-free DNA (cfDNA) consists of a heterogeneous and complex DNA fraction released in body fluids by any cell type through cell death mechanisms (18,19). The short half-life of cfDNA enables real-time monitoring for response or relapse, being an easy-to-implement biomarker to monitor cancer evolution and response to therapy (20). In fact, in lung cancer and other solid tumors, cfDNA analysis has been explored as a prognostic marker and surrogate for monitoring treatment response (21,22). High cfDNA level or positive ctDNA detection is clearly correlated tumor burden. Besides pre-treatment levels of ctDNA have value to predict long-term survival in locally advanced non-small cell lung cancer (NSCLC) (23). Early cfDNA/ctDNA changes can be detected at first follow-up to predict radiographic response being their decreasing levels also been associated with improved survival rates as our group previously described in NSCLC (24-26).
Circulating tumor cells (CTCs) are tumor cells originated from the primary or metastatic sites that are able to enter the circulation and disseminate to distant sites, and constitutes another frequent circulating biomarker investigated in cancer (14). As a high metastatic tumor type, SCLC is characterized by a strong release of CTCs, with detection rates of 60.2–94% (17), suggesting that CTCs could be employed as a disease surrogate in SCLC. The analysis of CTCs originated from the primary or metastatic sites (27) as a prognostic biomarker has been reported in different cancer types (28-30) including SCLC. However, the prognostic threshold in SCLC has been not well established (31-35). The low proportion of CTCs in the bloodstream together with the molecular heterogeneity that characterizes these cells is the principal challenge for CTC isolation and detection. For this reason, several platforms have been developed in the last years (14,36). Despite their different nature, the combined analysis of total cfDNA and CTCs in patients with SCLC could provide complementary information for improving SCLC patients’ management.
In this study, we hypothesized that total cfDNA levels can serve as a useful biomarker for prognostic and follow-up of SCLC patients under first line of therapy. For this purpose, we analyzed the total cfDNA levels in a cohort of 46 patients with SCLC prior to the start of therapy, at 3 weeks after the first dose, and at the time of progression of the disease. The additional value of CTCs was investigated in our cohort in order to provide a more complete view of the disease dynamics. Finally, we developed a simple model to segregate patients into three categories based on risk of progression and death (taking into account the cfDNA levels, Eastern Cooperative Oncology Group Performance Status (ECOG PS) and gender of patients). To our knowledge, this study is the first one to examine the possible role of total cfDNA levels as a prognostic and follow-up biomarker in SCLC patients. We present this article in accordance with the REMARK reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-273/rc).
Methods
Patients and blood sample collection
We design a prospective study including newly diagnosed SCLC patients treated with first-line of standard chemotherapy or immunotherapy plus chemotherapy (chemo-immunotherapy). Inclusion criteria were newly diagnosed SCLC patients who received first-line treatment and adequate plasma collection, processing and storage. Forty-six patients treated between June 2017 and June 2021, at the Department of Medical Oncology of Complexo Hospitalario Universitario de Santiago de Compostela were enrolled in the study. In total 111 blood samples were collected at different time points: before therapy onset (baseline) (n=46), 3 weeks after therapy start (n=40) and at the progression of the disease (n=25). A control cohort of 20 healthy individuals was also included in order to select the better assay to quantify total cfDNA.
Data of clinical characteristics such as age, sex, the ECOG PS, smoking status, stage, number of metastasis and liver metastasis were collected, in order to develop a prognostic model together the cfDNA levels and CTCs detected at baseline.
The study was performed in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Santiago de Compostela and Lugo Ethics Committee (Ref: 2017/538). Written informed content was obtained from every participant prior to enrolling in the study and could withdraw their consent at any time.
Clinical endpoints
Progression-free survival (PFS) was defined as the time from the date of initial treatment until the date of progression disease, death or last follow-up, whichever occurred first. Progression date was defined as the date of disease progression based on RECIST (v.1.1), or the date of clinical progression if the patient discontinued the treatment due to clinical deterioration despite not meeting criteria for RECIST progression. Overall survival (OS) was defined as the time from the date of initial treatment to the date of death or the last date of follow-up. For survival analyses about cfDNA levels at progression disease, OS was defined as the time from the date of sample obtained (at progression disease) to the date of death or the last date of follow-up.
Sample processing and cell free DNA isolation
Peripheral blood was obtained by direct venepuncture in CellSave tubes (Menarini, Silicon Biosystems, Bologna, Italy) and processed within 96 hours after blood collection, since CellSave tubes allow to maintain blood samples up to 96 hours without affect cfDNA levels (37). Plasma and cellular components were separated by centrifugation at 1,600 g for 10 minutes at room temperature. Plasma was centrifugated a second time at 5,500 g for 10 minutes at room temperature in order to remove any remaining cellular debris and aliquoted for storage at −80 ℃ until the time of cfDNA extraction. cfDNA was isolated from 3 mL of plasma using the QIAamp Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany) using a vacuum pump, according to the manufacturer’s instructions and eluted in LoBind® tubes (Eppendorf AG, Hamburg, Germany).
Total cell free DNA quantification
CfDNA levels were quantified using two different approaches: Qubit 4 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA) and quantitative PCR (qPCR) method by analyzing the telomerase reverse transcriptase (hTERT) single-copy gene (Thermo Fisher Scientific, Waltham, MA, USA).
Two µL of the sample were employed to quantify by the fluorometric instrument Qubit 4 using the Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA).
In the other hand, samples were quantified by a qPCR assay, described previously (26). Briefly, each qPCR reaction was carried out in a final volume of 20 µL: 10 µL of TaqMan Universal Mastermix (Thermo Fisher Scientific, Waltham, MA, USA), 1 µL of hTERT hydrolysis probe and 2 µL of the sample. Each sample was analysed in duplicate. In addition, each plate included a calibration curve and negative controls. The calibration curve calculated based on a dilution series of a commercial standard human genomic DNA (Roche Diagnostics, Mannheim, Germany), was fragmented in 184 bp using Covaris® E220 Focused-ultrasonicator (Covaris, Massachusetts, USA) using the following protocol: 430s duration, peak incident power of 175 Watts, duty factor of 10% and 200 cycles per burst. Fragments size were then determined using a TapeStation 4700 (Agilent, Santa Clara, CA, USA) and the High Sensitivity DNA ScreenTape® (Agilent, Santa Clara, CA, USA). Amplification was performed under the following cycling conditions using a QuantStudioTM 3 real-time PCR system (Thermo Fisher Scientific, Waltham, MA, USA): 50 ℃ for 2 min; 95 ℃ for 10 min; 40 cycles of 95 ℃ 15 s; and 60 ℃ for 1 min. Data were analyzed with QuantStudioTM Design & Analysis software, version 2.5.1 (Thermo Fisher Scientific, Waltham, MA, USA).
The final concentration of each sample was calculated by interpolation of the mean of cycle quantification values (Cq) with the calibration curve. Values with a Cq confidence interval (CI) less than 0.95 were discarded. Moreover, only assays with R2 values greater than 0.98 for the standard curve and with an efficiency ≥88.8% were used. Results obtained from both approaches (Qubit vs. hTERT qPCR) were compared.
CTC detection and enumeration
CTCs analyses were performed using the CellSearch® system (Menarini, Silicon Biosystems, Bologna, Italy). Peripheral whole blood of each patient was collected in CellSave preservative tubes (Menarini, Silicon Biosystems, Bologna, Italy), stored at room temperature and processed within 96 hours after the blood was drawn.
Briefly, 7.5 mL of whole blood were mixed with 6 mL of buffer and centrifugated at 800 g for 10 minutes at room temperature. Next, samples were processed in the CellTracks Autoprep system using the Circulating Tumor Cell Kit (Menarini, Silicon Biosystems, Bologna, Italy). The kit consists of ferrofluids coated with epithelial cell-specific anti-EpCAM antibodies to immunomagnetically enrich epithelial cells; a mixture of antibodies directed to cytokeratins (CKs) 8, 18, and 19 conjugated to phycoerythrin (PE); an antibody to CD45 conjugated to allophycocyanin (APC); nuclear dye 4',6-diamidino-2-phenylindole (DAPI) to fluorescently label the cells as well as buffers to fix, permeabilize, wash and resuspend the cells. Finally, samples were analyzed with the CellTracks Analyzer II according to the manufacturer’s instructions. The CTCs were identified as round or oval cells with an intact nucleus (DAPI+), CK positive and CD45 negative.
Statistical analysis
Continuous data were summarized as mean, median and range whereas frequency and percentage were presented for categorical variables. Categorical variables were compared using the chi-square test or Fisher’s exact test. Swimmer plot was provided to visualize the times of sample collection, every patient’s therapy and clinical outcomes. Pearson test was used to evaluate a pairwise correlation between the different strategies to quantify the cfDNA, by fluorometry and qPCR. The Mann-Whitney-Wilcoxon U-Test was used to compare continuous variables between groups. Receiver operating characteristics (ROC) curves were computed based on cfDNA levels of SCLC patients and healthy controls, representing the area under the curve (AUC) values and computing the CI at 95% confidence levels. ROC curves were also constructed to evaluate the thresholds of cfDNA levels for PFS and OS analyses. In order to determine the CTCs cut-off, we analysed the prognostic value of different levels of CTCs to discriminate progression or death using the Evaluate Cutpoints tool (available in http://wnbikp.umed.lodz.pl/Evaluate-Cutpoints/). Kaplan-Meier method was used to plot the survival curves applying the log-rank test. Univariate and multivariate Cox regression analyses were used to evaluate factors independently associated with PFS and OS. A final prognostic model for PFS and OS was developed. Comparisons of Cox proportional hazard regression models were made using the Akaike information criterion (AIC) technique (38), with a smaller AIC value indicating the better model. A stepwise backward elimination procedure was performed to minimize the AIC. All statistical analyses were performed using R version 4.1.1. The following R packages were used: survival (39), survminer, ggplot2 (40), pROC (41), gtsummary (42), swimplot, stats, rstatix.
Results
Patient characteristics and sample collection
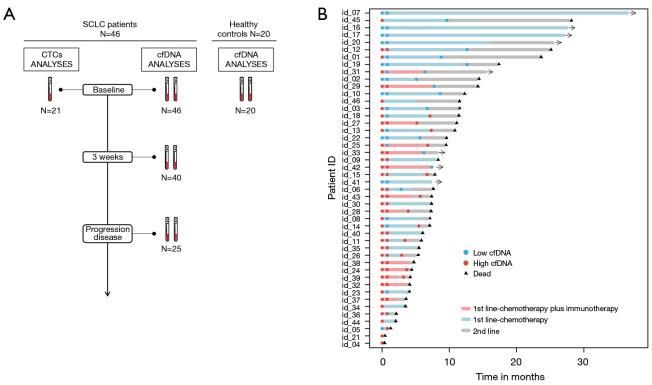
Forty-six SCLC patients were included in the study. Their clinicopathological characteristics are presented in Table 1. The median age was 67 (range, 47–83) years, all the patients were current or former smokers, and most were males (84.78%) and stage IV tumors (89.13%). The ECOG PS <2 accounted for 67.39% of cases and 52.17% of patients had a number of metastases >2. The majority of patients included into the study were treated with chemotherapy (carboplatin and etoposide) (n=33; 71.74%), while 13 (28.26%) patients were treated with chemotherapy in combination with immunotherapy (n=9 carboplatin, etoposide and atezolizumab; n=4 carboplatin, etoposide and durvalumab). The median number of chemotherapy or chemotherapy/immunotherapy treatment cycles was 5 (range, 1–11). At the time of analysis, 41 of the 46 (89.13%) evaluable patients had experienced disease progression and 38 of the 46 (82.6%) evaluable patients had died. Sample collection was performed before therapy onset (n=46), 3 weeks after initiation of therapy (n=40; 2 patients died and in 4 patients sample collection was not possible) and at the time of progression disease (n=25; 4 patients not progressed, 10 patients died and in 7 patients sample collection was not possible) (Figure 1). Median PFS and OS were 174 (range, 4–483) and 229 (range, 4–748) days, respectively.
Table 1. Association between patients’ demographics/clinical characteristics and baseline cfDNA levels.
| Characteristics | N (%) | Baseline cfDNA, median (GE/mL) | P value |
|---|---|---|---|
| Age (years), median [range]; mean ± SD | 67 [47–83]; 66.83±8.1 | ||
| Below median | 24 (52.17) | 6,488.23 | 0.79 |
| Above median | 22 (47.82) | 8,403.79 | |
| Gender | |||
| Male | 39 (84.78) | 6,130.59 | 0.61 |
| Female | 7 (15.22) | 15,699.93 | |
| Stage | |||
| III | 5 (10.87) | 1,409.11 | 0.031 |
| IV | 41 (89.13) | 7,658.99 | |
| ECOG PS | |||
| <2 | 31 (67.39) | 4,652.13 | 0.025 |
| ≥2 | 15 (32.61) | 22,459.61 | |
| Smoking | |||
| Smoker | 26 (56.52) | 6,488.23 | 0.94 |
| Former smoker | 20 (43.48) | 6,155.56 | |
| Number of metastasis | |||
| ≤2 | 22 (47.82) | 4,563.83 | 0.05 |
| >2 | 24 (52.17) | 12,973.33 | |
| Liver metastases | |||
| No | 23 (50.0) | 3,355.91 | 0.0002 |
| Yes | 23 (50.0) | 24,759.83 | |
| Bone metastases | |||
| No | 18 (39.13) | 3,265.055 | 0.17 |
| Yes | 28 (60.87) | 3,355.91 | |
| Lymph node metastases | |||
| No | 10 (21.74) | 4,563.83 | 0.12 |
| Yes | 36 (78.26) | 7,658.99 | |
| Treatment | |||
| Chemotherapy | 33 (71.74) | 6,130.59 | 0.36 |
| Chemotherapy plus immunotherapy | 13 (28.26) | 10,676.99 |
cfDNA, cell-free DNA; GE, genomic equivalents; SD, standard deviation; ECOG PS, Eastern Cooperative Oncology Group Performance Status.
Figure 1.
Study schema and clinical course of all patients included in the study. (A) Study sampling points and cohorts. (B) Swimmers’ plot showing each patient therapy and the different times of sample collection. The total length of each bar indicates the duration of survival from the diagnoses. Alive patients are represented with the black arrow head. SCLC, small cell lung cancer; cfDNA, cell free DNA; CTCs, circulating tumor cells.
CfDNA levels are specifically increased in SCLC patients
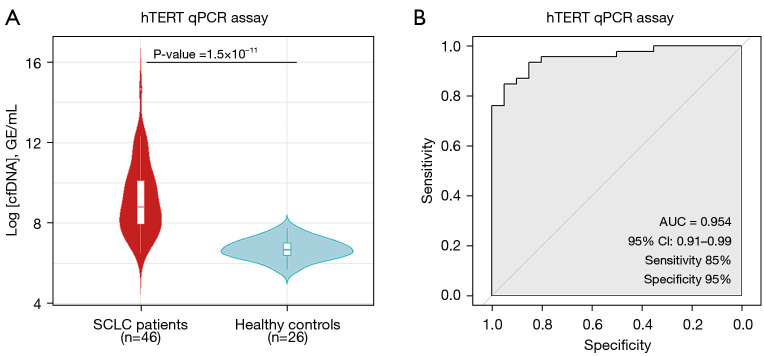
First, we chose the qPCR assay by analyzing the hTERT single-copy gene (Thermo Fisher Scientific, Waltham, MA, USA) as the standard method to quantify total cfDNA levels, obtained similar results in comparison with Qubit 4 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA) (Figure S1). In addition, 20 healthy controls were included in order to compare their cfDNA levels with our patient cohort. We found that total cfDNA levels in healthy controls were statistically lower than in those found in the SCLC cohort (Wilcoxon test P=1.5×10−11) (Figure 2A). Our qPCR assay showed an excellent AUC =0.954 (specificity 95% and sensibility 85%) (Figure 2B). These results evidenced that cfDNA levels were increased as a result of the malignant disease in SCLC patients showing the clear association between cfDNA levels and the high disease burden that characterized SCLC disease and reinforced their interest as a potential biomarker to follow-up the disease evolution.
Figure 2.
cfDNA levels are higher in SCLC patients than healthy controls. (A) Total cfDNA levels in healthy controls and patients with SCLC. Statistical analysis between both groups was performed by the Mann-Whitney-Wilcoxon U-Test. (B) ROC curve for qPCR assay shows high sensitivity and specificity to discriminate healthy controls and SCLC patients. qPCR, quantitative PCR; cfDNA, cell free DNA; CI, confidence interval; GE, genomic equivalents; SCLC, small cell lung cancer; AUC, area under the curve; ROC, receiver operating characteristic.
Clinical interest of cfDNA analysis at baseline
Total cfDNA was quantified by qPCR assay at different time points in our patient cohort with the goal to evaluate its potential as a monitoring tool (Table S1). CfDNA levels at baseline were significantly higher in patients with stage IV cancer, poor performance status, an elevated number of sites of metastasis and presence of liver metastases (P value ≤0.05) (Table 1). There was no statistically significant association with respect to age, gender, smoking status, presence of bone metastasis, presence of lymph node metastases and the treatment used.
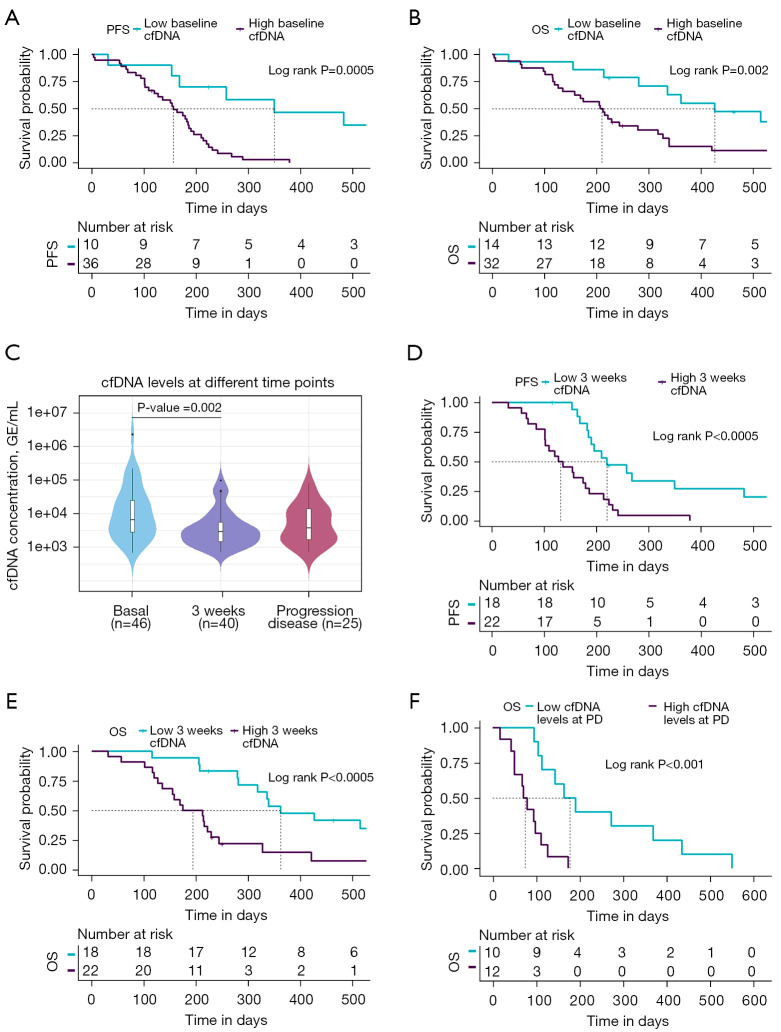
In addition, the possible role of cfDNA as a prognostic biomarker before therapy in SCLC was investigated. Thus, the cfDNA levels were log-transformed and the patients were dichotomized into high and low cfDNA level groups based on ROC analysis (Table S2). The thresholds of baseline log cfDNA levels were chosen at 7.650 [~2,100.65 genomic equivalents (GE)/mL plasma] and 8.077 (~3,219.56 GE/mL plasma) for PFS and OS analyses, respectively. We found that patients with high levels of total cfDNA at baseline presented shorter PFS [log-rank P=0.0005, hazard ratio (HR), 5.06; 95% CI: 1.89–13.6] and OS (log-rank P=0.002, HR, 3.32; 95% CI: 1.50–7.37) than those with low levels of total cfDNA (Figure 3A,3B). The median PFS of patients in the low baseline cfDNA group was 350 days versus 156 days in the high baseline cfDNA group, whereas the median OS was 426 and 210 days in the two respective groups (low vs. high baseline total cfDNA levels) (Table 2).
Figure 3.
cfDNA levels as a prognostic biomarker at different time points of therapy. (A,B) Kaplan-Meier survival analysis of cfDNA levels at baseline for PFS (A) and OS (B). (C) CfDNA levels in SCLC patients at different time-points (baseline, 3 weeks after treatment onset and at progression disease). Total cfDNA levels at baseline were significantly higher than at 3 weeks after the therapy onset (Wilcoxon test P=0.002). (D,E) Kaplan-Meier survival analysis of cfDNA levels at 3 weeks for PFS (D) and OS (E). (F) Kaplan-Meier survival analysis of cfDNA levels at progression disease for OS. cfDNA, cell free DNA; PFS, progression-free survival; OS, overall survival; GE, genomic equivalents; PD, progression disease; SCLC, small cell lung cancer.
Table 2. Univariate and multivariate Cox regression analyses of cfDNA levels, CTC counts and clinical parameters.
| Variable | Univariate | Multivariate | |||
|---|---|---|---|---|---|
| P value | HR (95% CI) | P value | HR (95% CI) | ||
| PFS | |||||
| Baseline log cfDNA (high vs. low cfDNA) | 0.001 | 5.06 (1.89–13.6) | 0.005 | 46.0 (3.16–672) | |
| Baseline CTC count, CellSearch (≥150 vs. <150) | 0.028 | 3.47 (1.14–10.6) | |||
| ECOG PS (≥2 vs. <2) | <0.001 | 3.57 (1.72–7.40) | 0.04 | 17.9 (1.11–289) | |
| Sex (male vs. female) | 0.2 | 1.76 (0.74–4.22) | |||
| Age (continue) | 0.5 | 1.01 (0.97–1.05) | |||
| Stage (IV vs. III) | 0.07 | 3.00 (0.91–9.91) | |||
| Number of metastasis (>2 vs. ≤2) | 0.1 | 1.70 (0.90–3.19) | |||
| Liver metastasis (yes vs. no) | 0.08 | 1.74 (0.93–3.23) | |||
| Smoking (smoker vs. former smoker) | 0.8 | 0.92 (0.49–1.70) | |||
| 3 weeks log cfDNA (high vs. low cfDNA)* | <0.0001 | 3.50 (1.69–7.23) | 0.004 | 3.49 (1.50–8.12) | |
| OS | |||||
| Baseline log cfDNA (high vs. low cfDNA) | 0.003 | 3.32 (1.50–7.37) | 0.004 | 32.4 (3.05–344) | |
| Baseline CTC count, CellSearch (≥150 vs. <150) | 0.07 | 2.71 (0.93–7.88) | |||
| ECOG PS (≥2 vs. <2) | <0.001 | 4.54 (2.13–9.68) | |||
| Sex (male vs. female) | 0.2 | 1.80 (0.75–4.36) | |||
| Age (continue) | 0.5 | 1.01 (0.97–1.06) | |||
| Stage (IV vs. III) | 0.08 | 2.86 (0.86–9.47) | |||
| Number of metastasis (>2 vs. ≤2) | 0.5 | 1.23 (0.65–2.33) | |||
| Liver metastasis (yes vs. no) | 0.3 | 1.45 (0.77–2.76) | |||
| Smoking (smoker vs. former smoker) | 0.5 | 0.82 (0.43–1.55) | |||
| 3 weeks log cfDNA (high vs. low cfDNA)* | <0.001 | 3.67 (1.72–7.82) | 0.002 | 4.35 (1.68–11.3) | |
| PD log cfDNA (high vs. low cfDNA)* | <0.001 | 5.73 (1.93–17.0) | 0.006 | 9.24 (1.87–45.6) | |
*, multivariate Cox regression model including sex, age, ECOG PS, stage, number of metastases, presence of liver metastasis and smoking status. The levels of cfDNA were determined as low (< cut-off) or high (≥ cut-off) based on the cut-off obtained from the ROC curve analyses. cfDNA, cell free DNA; CI, confidence interval; CTC, circulating tumor cell; ECOG PS, Eastern Cooperative Oncology Group Performance Score; PD, progression disease; HR, hazard ratio; OS, overall survival; PFS, progression-free survival; ROC, receiver operating characteristics.
Longitudinal analysis of total cfDNA levels during therapy
To determine whether cfDNA levels can be employed to monitor patients’ evolution during therapy, we quantified longitudinal cfDNA levels at 3 weeks after initiation of treatment (n=40) and at progression disease (n=25). We found that levels of total cfDNA were significantly higher before therapy than at 3 weeks after therapy onset (Wilcoxon test P=0.002; Figure 3C), suggesting clearance of ctDNA after therapy start which impact on cfDNA levels. However, no significant differences between cfDNA levels at baseline and at progression disease were found (Figure 3C).
To analyze the possible prognostic role of cfDNA monitoring during therapy, cfDNA levels were also log-transformed and the patients were dichotomized into high and low cfDNA level groups based on ROC analysis (Table S2) in each sample time. The threshold of three weeks post-treatment log cfDNA levels was chosen at 7.879 (~2,641.23 GE/mL plasma) both for PFS and OS analyses. Thus, the possible prognostic role of monitoring cfDNA levels at 3 weeks after initiation of treatment was investigated. cfDNA levels were defined as high or low depending the threshold obtained with ROC curves analyses described above. Patients with low cfDNA levels at 3 weeks showed a shorter PFS (log-rank P<0.0005, HR, 3.5; 95% CI: 1.69–7.23) and OS (log-rank P<0.0005, HR, 3.67; 95% CI: 1.72–7.82) (Figure 3D,3E) than the patients with high cfDNA levels.
Finally, cfDNA quantification at the time of progression disease was performed in 25 SCLC patients. Our results reported that patients with high cfDNA levels at this time point survive fewer days than patients with low cfDNA levels (log-rank P<0.001, HR, 5.73; 95% CI: 1.93–17.0; 177 days in the high cfDNA levels group versus 74.5 days in the low cfDNA levels group) (Figure 3F).
CTCs analyses and prognostic value
CTCs analyses were performed in 21 patients with SCLC before starting the treatment. 85.71% of patients (18 of 21 patients) presented at least 1 CTC with a median of 26 CTCs (range, 0–4,796) (Figure S2A,S2B). Different cut-offs were analyzed in order to determine the possible prognostic value of CTCs (Table S3). Thus, we found that the presence of ≥150 CTCs/7.5 mL of blood was significantly associated with shorter PFS rates (log-rank P=0.02, HR, 3.47; 95% CI: 1.14–10.6) (Table 2; Figure S2C). We also found CTC clusters (≥2 CTCs or CTC with white blood cells) in all patients with >10 CTCs/7.5 mL of blood, however no additional prognostic value was found.
In another hand, CTCs number at baseline was significantly higher in patients with extensive disease (stage IV) and with poor ECOG PS (Figure S3A-S3J). Also, the presence of CTCs and high cfDNA levels were significantly associated (Figure S3K), indicating that both markers are reflecting the tumor burden.
Multivariate analyses and prognostic model for PFS and OS
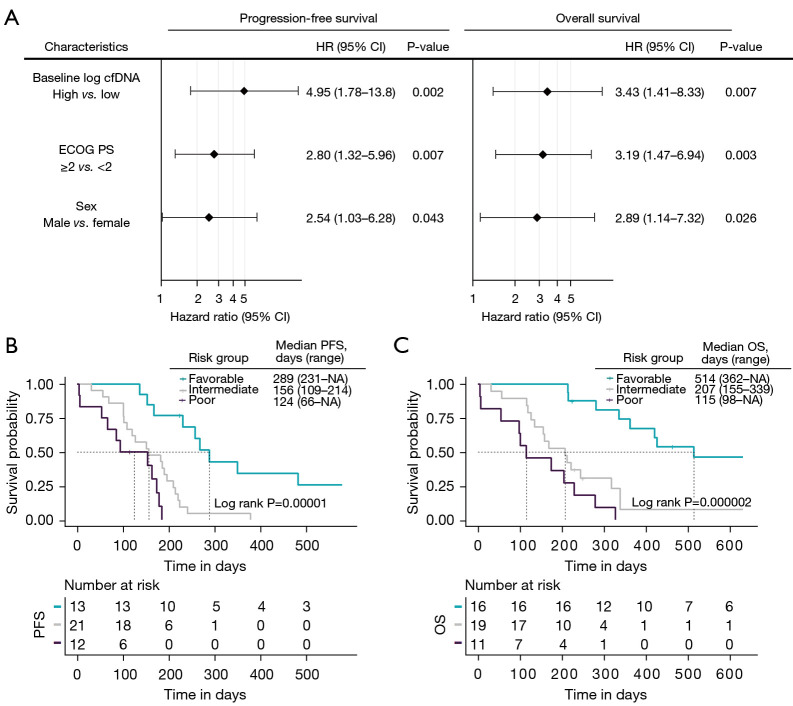
Univariate and multivariate Cox regression analyses of PFS and OS were performed considering various clinical and demographic variables (ECOG PS, sex, age, stage, number of metastases, presence of liver metastasis and smoking status) (Table 2). Multivariate regression analyses confirmed the value of the cfDNA levels at baseline as an early independent predictor biomarker for PFS and OS (HR, 46.0; 95% CI: 3.16–672; P=0.005 and HR, 32.4; 95% CI: 3.05–344; P=0.004, respectively). Regarding the cfDNA at 3 weeks, multivariate regression analyses also confirm its independence as a prognostic biomarker of PFS and OS (HR, 3.49.0; 95% CI: 1.50–8.12; P=0.004 and HR, 4.35; 95% CI: 1.68–11.3; P=0.002, respectively) (Table 2). In contrast, multivariate analysis in CTCs did not show value as an independent predictive biomarker of PFS and OS.
Next, an independent prognostic model for both PFS and OS was developed. Three variables were retained in the final prognostic model: cfDNA levels (high vs. low levels), ECOG PS (<2 vs. ≥2) and sex (male vs. female). The detailed results of the multivariate analyses are shown in Figure 4A. Subsequently, we segregated patients into three risk categories: patients with all adverse prognostic factors were classified in the poor-risk category (high cfDNA levels, ECOG PS ≥2 and male gender), patients with two adverse prognostic factors were classified in the intermediate-risk category, and patients with one or none adverse prognostic factor were classified in the favorable-risk category. The Kaplan-Meier curves representing the three risk categories and median PFS and OS are presented in Figure 4B,4C. Median PFS ranged from 124 to 289 days based on the number of adverse prognostic factors present before therapy. Median OS ranged from 115 to 514 days.
Figure 4.
Final multivariate Cox regression prognostic model for PFS and OS (A) and Kaplan-Meier survival analysis according to risk-group for PFS and OS (B,C). cfDNA, cell free DNA; HR, hazard ratio; CI, confidence interval; ECOG PS, Eastern Cooperative Oncology Group Performance Score; PFS, progression-free survival; OS, overall survival; NA, not available.
Our prognostic model indicates an increased risk of 5 times to present disease recurrence (HR =5.37, 95% CI: 2.32–12,4; P=3×10−5) and 6 times the risk of death (HR =6.02, 95% CI: 2.66–13.6; P=3×10−6) in the unfavorable category (intermediate and poor groups) than in the favorable group (Table S4).
Discussion
Precision medicine has the objective of optimizing the selection of the best therapy for each patient. In this context, liquid biopsy has emerged as a promising and minimally invasive tool for this due to its ability to provide a total image of primary and metastatic tumors at different times across therapy (43). Recently, the management of SCLC has changed and new therapies, such as immunotherapy among others, have been investigated and approved for clinical use (17,44,45). Nevertheless, the necessity to find a prognostic biomarker for helping to select the therapy prescribed and to monitor the evolution of the disease during the treatment, remains a challenge in SCLC patients. In this study, we report for the first time the possibility to employ the cfDNA and its quantification as a prognostic biomarker in SCLC prior to starting therapy and at different time points. Our analyses allow us to identify a group of low-risk patients characterized by low cfDNA levels at baseline who probably will benefit from both: chemotherapy in monotherapy or the combination of immunotherapy and chemotherapy. The study of another common circulating biomarker, CTCs, also provided us information for the prognostic of patients before starting therapy, although the results were less clear. We found a significant association between presence of a high number of CTCs (≥150 CTCs) and worse PFS.
Total cfDNA refers to a heterogeneous and complex DNA fraction free released in body fluids by any cell type (not only tumoral) through several cell death mechanisms such as secretion, apoptosis and necrosis (18,19). Of note, the source of cfDNA is an intriguing question in cancer. According to previous studies, the fraction of ctDNA varies from 0.1–89% of cfDNA but it increases it accordance with the tumor burden. Therefore, although cfDNA content is not tumor specific, it can be assumed that total cfDNA in cancer patients originates from tumor cells, cells in the tumor microenvironment or from cells involved in the antitumor response (20). Therefore, cfDNA can be uses as a surrogate of ctDNA, but always taking into account that they are different entities that can provide us different information. In line with our results, it’s well reported that cancer patients present higher cfDNA levels than healthy controls (46,47) but few studies have investigated the possible prognostic and predictive value of total cfDNA quantification in patients with SCLC. In contrast, the ctDNA, the tumor-derived fraction of this cfDNA, has been reported as a prognostic and predictive biomarker in several works (48-52). Almodovar et al. reported that changes in the mutant allele frequencies on ctDNA were associated with response to treatment and relapse. Twenty-seven patients with SCLC were analysed by next-generation sequencing (NGS) custom panel, however, the lack of driver mutations known in SCLC, limited the number of genes analyzed (48). In another work, Devarakonda et al. analyzed 564 patients using a larger NGS panel, including 73 genes, and, according to previous results into the bibliography, RB1 and TP53 were the most frequent mutant, however, no prognostic or predictive value was reported in this study (53).
In this way, total cfDNA quantification allows to detect the total DNA released from normal and also tumor cells into the blood. Thus, despite the few known driver mutations found in SCLC, cfDNA quantification allows to quantify the total levels before treatment and monitor the changes during therapy. Recently, our group demonstrated the feasibility to quantify cfDNA levels in NSCLC patients and its association with patients’ outcomes (26), suggesting its possible utility in SCLC.
Thus, in the present work, we quantified total cfDNA levels using two different technologies, a fluorometric assay, Qubit and a more specific one, the qPCR assay analyzing the hTERT gene. CfDNA quantification by both technologies showed good concordance. Furthermore, the concordance of cfDNA levels at any time point of therapy using both methods also showed a good concordance. Therefore, both methods could be used to robustly measure the cfDNA content. However, to complete our study we chose the qPCR assay, which is a high sensitive and specific assay for cfDNA quantification in SCLC patients, and was previously employed in studies focused on NSCLC (26,54-56).
Regarding the clinical meaning of cfDNA content, we found that high levels were significantly associated with shorter PFS and OS before therapy onset, being a robust independent prognostic biomarker in newly diagnosed SCLC patients. Also, cfDNA levels at baseline were higher in patients with stage IV that could be a consequence of more aggressive disease. This can be partially explained by an increase of ctDNA levels released from the tumor cells to the bloodstream, increasing the total cfDNA fraction. Moreover, analyses showed that cfDNA levels at 3 weeks are associated with patients’ outcomes, being those patients with high values at 3 weeks, the ones with the worst prognosis. In addition, high cfDNA levels at the time of disease recurrence were associated with a higher risk of death. Of note, multivariate analyses showed the independence of cfDNA levels at 3 weeks and at progression disease as a prognostic biomarker. These results suggest that cfDNA monitoring could provide valuable information for the management of SCLC as our group previously reported in NSCLC (26). Thus, in clinical practice, in those SCLC patients with high levels of cfDNA at the time of disease recurrence, the selection of a more aggressive therapy or the intensification of clinical visits would be considered.
Besides cfDNA levels, we investigated the prognostic value of additional biomarkers such as CTCs and clinical characteristics. CTCs were analyzed in a cohort of 21 SCLC patients using the CellSearch® system, the only Food and Drug Administration (FDA)-approved device for CTC enumeration in prostate, breast and colorectal cancer. According to the literature (17), a detection rate of 85.71% was found in our study. Moreover, the CTC count at baseline determined using the CellSearch® system was significantly associated with PFS and OS (31,34,35,57,58). For example, Naito et al. reported that the presence of ≥8 CTCs/7.5 mL of blood was associated with worse OS (31), however, another study employed 50 CTCs as cut-off for PFS and OS (35). In fact, a consensus regarding the optimal cut-off of CTCs and the prognostic value remains a challenge (17). In this work, we found a discrete association between the presence of ≥150 CTCs and a shorter PFS, however multivariate analyses did not show independent value for the CTC count. Interestingly, high cfDNA levels and the presence of CTCs at baseline were significantly associated, reporting the clear association between both circulating biomarkers and disease burden. CTCs release in the bloodstream is related to the intravasation process of potentially metastatic cancer cells. Nevertheless, cfDNA is released by any cell type including tumoral and normal cells, however, how cfDNA release relates to tumor biology is currently unknown. In another hand, we evaluate several factors that could influence the patients’ outcomes. Thus, we proposed a simple model to segregate patients into three categories based on risk of progression and death (taking into account the cfDNA levels, ECOG PS and gender of patients). We found that patients with one or less adverse prognostic factors were classified in the favorable-risk category and present a longer PFS and OS. Regarding the clinical relevance of these results, some limitations in our design should be considered. First, the sample size of our CTC cohort was relatively small and CTC monitoring during therapy could provide more valuable information. Second, our patient cohort are not homogenous regarding treatment regimen, 71.74% SCLC patients were treated with while 28.26% SCLC patients were treated with chemo-immunotherapy. In addition, a validation study of our prognostic model in a larger independent cohort is needed.
In conclusion, we describe an important potential role of cfDNA levels as a prognostic biomarker in newly diagnosed SCLC patients and also could provide useful information about disease evolution. Finally, a prognostic model based on cfDNA levels and some clinical characteristics (ECOG PS and gender) would allow us to stratify patients and detect those who could particularly benefit from the treatment.
Supplementary
The article’s supplementary files as
Acknowledgments
This project could not be possible without the kindly collaboration of all patients.
Funding: This study was financed by all the donors who participated in the Liquid Biopsy Crowdfunding campaign in 2017. LM-R is supported by the Miguel Servet from ISCIII (CP20/00120) contract, funded by the Instituto de Salud Carlos III (ISCIII) and co-funded by the European Union. RD-P is supported by the Miguel Servet (CP21/00003) contract, funded by the ISCIII and co-funded by the European Union. The funding source did not have any influence on the design, conduction, and report of the results for this study.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was performed in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Santiago de Compostela and Lugo Ethics Committee (Ref: 2017/538). Written informed content was obtained from every participant prior to enrolling in the study and could withdraw their consent at any time.
Footnotes
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-273/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-273/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-273/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-273/coif). JGG reports consultant or advisory role fees from AstraZeneca, Boehringer-Ingelheim, Bristol-Myers Squibb, MSD, Novartis, Roche, Sanofi, and Takeda; honoraria for presentations and speaker bureaus from AstraZeneca, Bristol-Myers Squibb, Pierre-Fabre, Roche, Rovi, and Takeda; non-financial support from AstraZeneca, Bristol-Myers Squibb, Ippsen Pharma, Lilly, MSD, Roche, and Sanofi outside the submitted work. SA reports grants or contracts from Merk, BMS and MSD; consulting fees from Merk, BMS and MSD; payment or honoraria for lectures from BMS and MSD; support for attending meeting from MSD and participation on a data safety monitoring board from Merk and BMS outside the submitted work. LLM receives honoraria for lectures from Pfizer, Boehringer, Novartis, AstraZeneca, Sanofi, Bristol, MSD, Takeda; for advisory board from Sanofi, Lilly, Novartis, Boehringer, Amgen and receives support for attending meetings from MSD and AstraZeneca, outside the submitted work. RLL reports grants from Astellas, Janssen, Sanofi, Bayer, Ipsen, Roche, Novartis, Pfizer; consulting fees from Sanofi, Janssen, Astellas, Pfizer, Bayer, Roche, Ipsen, Novartis, Eisai, EUSA Pharma, BMS; payment or honoraria for lectures from Astellas, Janssen, Sanofi, Bayer, Ipsen, Pfizer, Roche, Novartis; travel, accommodations and expenses from Pharmamar, Roche, BMS and Pfizer; honoraria for participation in advisory boards from Roche, AstraZeneca, Merck, MSD, Bayer, BMS, Novartis, Janssen, Lilly, Pfizer and Leo, outside the submitted work. The other authors have no conflicts of interest to declare.
References
- 1.Rudin CM, Brambilla E, Faivre-Finn C, et al. Small-cell lung cancer. Nat Rev Dis Primers 2021;7:3. 10.1038/s41572-020-00235-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thai AA, Solomon BJ, Sequist LV, et al. Lung cancer. Lancet 2021;398:535-54. 10.1016/S0140-6736(21)00312-3 [DOI] [PubMed] [Google Scholar]
- 3.Gazdar AF, Bunn PA, Minna JD. Small-cell lung cancer: what we know, what we need to know and the path forward. Nat Rev Cancer 2017;17:725-37. 10.1038/nrc.2017.87 [DOI] [PubMed] [Google Scholar]
- 4.Esposito G, Palumbo G, Carillio G, et al. Immunotherapy in Small Cell Lung Cancer. Cancers (Basel) 2020;12:2522. 10.3390/cancers12092522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Facchinetti F, Di Maio M, Tiseo M. Adding PD-1/PD-L1 Inhibitors to Chemotherapy for the First-Line Treatment of Extensive Stage Small Cell Lung Cancer (SCLC): A Meta-Analysis of Randomized Trials. Cancers (Basel) 2020;12:2645. 10.3390/cancers12092645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horn L, Mansfield AS, Szczęsna A, et al. First-Line Atezolizumab plus Chemotherapy in Extensive-Stage Small-Cell Lung Cancer. N Engl J Med 2018;379:2220-9. 10.1056/NEJMoa1809064 [DOI] [PubMed] [Google Scholar]
- 7.Paz-Ares L, Dvorkin M, Chen Y, et al. Durvalumab plus platinum-etoposide versus platinum-etoposide in first-line treatment of extensive-stage small-cell lung cancer (CASPIAN): a randomised, controlled, open-label, phase 3 trial. Lancet 2019;394:1929-39. 10.1016/S0140-6736(19)32222-6 [DOI] [PubMed] [Google Scholar]
- 8.Rudin CM, Awad MM, Navarro A, et al. Pembrolizumab or Placebo Plus Etoposide and Platinum as First-Line Therapy for Extensive-Stage Small-Cell Lung Cancer: Randomized, Double-Blind, Phase III KEYNOTE-604 Study. J Clin Oncol 2020;38:2369-79. 10.1200/JCO.20.00793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dumoulin DW, Dingemans AC, Aerts JGJV, et al. Immunotherapy in small cell lung cancer: one step at a time: a narrative review. Transl Lung Cancer Res 2021;10:2970-87. 10.21037/tlcr-20-630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.George J, Lim JS, Jang SJ, et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015;524:47-53. 10.1038/nature14664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalemkerian GP, Loo BW, Akerley W, et al. NCCN guidelines® insights: Small cell lung cancer, version 2.2018 featured updates to the NCCN guidelines. JNCCN J Natl Compr Cancer Netw 2018;16:1171-82. 10.6004/jnccn.2018.0079 [DOI] [PubMed] [Google Scholar]
- 12.Ortega-Franco A, Ackermann C, Paz-Ares L, et al. First-line immune checkpoint inhibitors for extensive stage small-cell lung cancer: clinical developments and future directions. ESMO Open 2021;6:100003. 10.1016/j.esmoop.2020.100003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blackhall F, Frese KK, Simpson K, et al. Will liquid biopsies improve outcomes for patients with small-cell lung cancer? Lancet Oncol 2018;19:e470-81. 10.1016/S1470-2045(18)30455-8 [DOI] [PubMed] [Google Scholar]
- 14.Herath S, Sadeghi Rad H, Radfar P, et al. The Role of Circulating Biomarkers in Lung Cancer. Front Oncol 2022;11:801269. 10.3389/fonc.2021.801269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng ML, Pectasides E, Hanna GJ, et al. Circulating tumor DNA in advanced solid tumors: Clinical relevance and future directions. CA Cancer J Clin 2021;71:176-90. 10.3322/caac.21650 [DOI] [PubMed] [Google Scholar]
- 16.Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24. 10.1126/scitranslmed.3007094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mondelo-Macía P, García-González J, León-Mateos L, et al. Current Status and Future Perspectives of Liquid Biopsy in Small Cell Lung Cancer. Biomedicines 2021;9:48. 10.3390/biomedicines9010048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keller L, Belloum Y, Wikman H, et al. Clinical relevance of blood-based ctDNA analysis: mutation detection and beyond. Br J Cancer 2021;124:345-58. 10.1038/s41416-020-01047-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aucamp J, Bronkhorst AJ, Badenhorst CPS, et al. The diverse origins of circulating cell-free DNA in the human body: a critical re-evaluation of the literature. Biol Rev Camb Philos Soc 2018;93:1649-83. 10.1111/brv.12413 [DOI] [PubMed] [Google Scholar]
- 20.Kustanovich A, Schwartz R, Peretz T, et al. Life and death of circulating cell-free DNA. Cancer Biol Ther 2019;20:1057-67. 10.1080/15384047.2019.1598759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.García-Pardo M, Makarem M, Li JJN, et al. Integrating circulating-free DNA (cfDNA) analysis into clinical practice: opportunities and challenges. Br J Cancer 2022;127:592-602. 10.1038/s41416-022-01776-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathios D, Johansen JS, Cristiano S, et al. Detection and characterization of lung cancer using cell-free DNA fragmentomes. Nat Commun 2021;12:5060. 10.1038/s41467-021-24994-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Romero A, Nadal E, Serna R, et al. OA20.02 Pre-Treatment Levels of ctDNA for Long-Term Survival Prediction in Stage IIIA NSCLC Treated With Neoadjuvant Chemo-Immunotherapy. J Thorac Oncol 2021;16:S883-4. 10.1016/j.jtho.2021.08.102 [DOI] [Google Scholar]
- 24.Moding EJ, Liu Y, Nabet BY, et al. Circulating Tumor DNA Dynamics Predict Benefit from Consolidation Immunotherapy in Locally Advanced Non-Small Cell Lung Cancer. Nat Cancer 2020;1:176-83. 10.1038/s43018-019-0011-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng ML, Lau CJ, Milan MSD, et al. Plasma ctDNA Response Is an Early Marker of Treatment Effect in Advanced NSCLC. JCO Precis Oncol 2021;5:ePO. 10.1200/PO.20.00419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mondelo-Macía P, García-González J, León-Mateos L, et al. Clinical potential of circulating free DNA and circulating tumour cells in patients with metastatic non-small-cell lung cancer treated with pembrolizumab. Mol Oncol 2021;15:2923-40. 10.1002/1878-0261.13094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pantel K, Speicher MR. The biology of circulating tumor cells. Oncogene 2016;35:1216-24. 10.1038/onc.2015.192 [DOI] [PubMed] [Google Scholar]
- 28.Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med 2004;351:781-91. 10.1056/NEJMoa040766 [DOI] [PubMed] [Google Scholar]
- 29.Cohen SJ, Punt CJ, Iannotti N, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol 2008;26:3213-21. 10.1200/JCO.2007.15.8923 [DOI] [PubMed] [Google Scholar]
- 30.de Bono JS, Scher HI, Montgomery RB, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res 2008;14:6302-9. 10.1158/1078-0432.CCR-08-0872 [DOI] [PubMed] [Google Scholar]
- 31.Naito T, Tanaka F, Ono A, et al. Prognostic impact of circulating tumor cells in patients with small cell lung cancer. J Thorac Oncol 2012;7:512-9. 10.1097/JTO.0b013e31823f125d [DOI] [PubMed] [Google Scholar]
- 32.Igawa S, Gohda K, Fukui T, et al. Circulating tumor cells as a prognostic factor in patients with small cell lung cancer. Oncol Lett 2014;7:1469-73. 10.3892/ol.2014.1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng Y, Liu XQ, Fan Y, et al. Circulating tumor cell counts/change for outcome prediction in patients with extensive-stage small-cell lung cancer. Future Oncol 2016;12:789-99. 10.2217/fon.15.346 [DOI] [PubMed] [Google Scholar]
- 34.Salgia R, Weaver RW, McCleod M, et al. Prognostic and predictive value of circulating tumor cells and CXCR4 expression as biomarkers for a CXCR4 peptide antagonist in combination with carboplatin-etoposide in small cell lung cancer: exploratory analysis of a phase II study. Invest New Drugs 2017;35:334-44. 10.1007/s10637-017-0446-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aggarwal C, Wang X, Ranganathan A, et al. Circulating tumor cells as a predictive biomarker in patients with small cell lung cancer undergoing chemotherapy. Lung Cancer 2017;112:118-25. 10.1016/j.lungcan.2017.08.008 [DOI] [PubMed] [Google Scholar]
- 36.Radfar P, Aboulkheyr Es H, Salomon R, et al. Single-cell analysis of circulating tumour cells: enabling technologies and clinical applications. Trends Biotechnol 2022;40:1041-60. 10.1016/j.tibtech.2022.02.004 [DOI] [PubMed] [Google Scholar]
- 37.van Dessel LF, Beije N, Helmijr JC, et al. Application of circulating tumor DNA in prospective clinical oncology trials - standardization of preanalytical conditions. Mol Oncol 2017;11:295-304. 10.1002/1878-0261.12037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akaike H. A New Look at the Statistical Model Identification. IEEE, Manhattan, New York, U.S.; 1974:215-22. [Google Scholar]
- 39.Therneau TM, Grambsch PM. Modeling Survival Data: Extending the Cox Model. New York: Springer; 2000. [Google Scholar]
- 40.Hadley W. Ggplot2: Elegrant graphics for data analysis. Cham, Switzerland: Springer; 2016. [Google Scholar]
- 41.Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 2011;12:77. 10.1186/1471-2105-12-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sjoberg Daniel D, Whiting K, et al. Reproducible Summary Tables with the gtsummary Package. R J 2021;13:570. 10.32614/RJ-2021-053 [DOI] [Google Scholar]
- 43.Oliveira KCS, Ramos IB, Silva JMC, et al. Current Perspectives on Circulating Tumor DNA, Precision Medicine, and Personalized Clinical Management of Cancer. Mol Cancer Res 2020;18:517-28. 10.1158/1541-7786.MCR-19-0768 [DOI] [PubMed] [Google Scholar]
- 44.Schwendenwein A, Megyesfalvi Z, Barany N, et al. Molecular profiles of small cell lung cancer subtypes: therapeutic implications. Mol Ther Oncolytics 2021;20:470-83. 10.1016/j.omto.2021.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang S, Zhang Z, Wang Q. Emerging therapies for small cell lung cancer. J Hematol Oncol 2019;12:47. 10.1186/s13045-019-0736-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leon SA, Shapiro B, Sklaroff DM, et al. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res 1977;37:646-50. [PubMed] [Google Scholar]
- 47.van der Drift MA, Hol BE, Klaassen CH, et al. Circulating DNA is a non-invasive prognostic factor for survival in non-small cell lung cancer. Lung Cancer 2010;68:283-7. 10.1016/j.lungcan.2009.06.021 [DOI] [PubMed] [Google Scholar]
- 48.Almodovar K, Iams WT, Meador CB, et al. Longitudinal Cell-Free DNA Analysis in Patients with Small Cell Lung Cancer Reveals Dynamic Insights into Treatment Efficacy and Disease Relapse. J Thorac Oncol 2018;13:112-23. 10.1016/j.jtho.2017.09.1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nong J, Gong Y, Guan Y, et al. Circulating tumor DNA analysis depicts subclonal architecture and genomic evolution of small cell lung cancer. Nat Commun 2018;9:3114. 10.1038/s41467-018-05327-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Du M, Thompson J, Fisher H, et al. Genomic alterations of plasma cell-free DNAs in small cell lung cancer and their clinical relevance. Lung Cancer 2018;120:113-21. 10.1016/j.lungcan.2018.04.008 [DOI] [PubMed] [Google Scholar]
- 51.Mohan S, Foy V, Ayub M, et al. Profiling of Circulating Free DNA Using Targeted and Genome-wide Sequencing in Patients with SCLC. J Thorac Oncol 2020;15:216-30. 10.1016/j.jtho.2019.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iams WT, Kopparapu PR, Yan Y, et al. Blood-Based Surveillance Monitoring of Circulating Tumor DNA From Patients With SCLC Detects Disease Relapse and Predicts Death in Patients With Limited-Stage Disease. JTO Clin Res Rep 2020;1:100024. 10.1016/j.jtocrr.2020.100024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Devarakonda S, Sankararaman S, Herzog BH, et al. Circulating Tumor DNA Profiling in Small-Cell Lung Cancer Identifies Potentially Targetable Alterations. Clin Cancer Res 2019;25:6119-26. 10.1158/1078-0432.CCR-19-0879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alama A, Coco S, Genova C, et al. Prognostic Relevance of Circulating Tumor Cells and Circulating Cell-Free DNA Association in Metastatic Non-Small Cell Lung Cancer Treated with Nivolumab. J Clin Med 2019;8:1011. 10.3390/jcm8071011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sirera R, Bremnes RM, Cabrera A, et al. Circulating DNA is a useful prognostic factor in patients with advanced non-small cell lung cancer. J Thorac Oncol 2011;6:286-90. 10.1097/JTO.0b013e31820189a5 [DOI] [PubMed] [Google Scholar]
- 56.Paci M, Maramotti S, Bellesia E, et al. Circulating plasma DNA as diagnostic biomarker in non-small cell lung cancer. Lung Cancer 2009;64:92-7. 10.1016/j.lungcan.2008.07.012 [DOI] [PubMed] [Google Scholar]
- 57.Hou JM, Greystoke A, Lancashire L, et al. Evaluation of circulating tumor cells and serological cell death biomarkers in small cell lung cancer patients undergoing chemotherapy. Am J Pathol 2009;175:808-16. 10.2353/ajpath.2009.090078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hou JM, Krebs MG, Lancashire L, et al. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J Clin Oncol 2012;30:525-32. 10.1200/JCO.2010.33.3716 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as